- 1State Key Laboratory of Agricultural Microbiology, College of Veterinary Medicine, Huazhong Agricultural University, Wuhan, China
- 2Key Laboratory of Preventive Veterinary Medicine in Hubei Province, Wuhan, China
The zoonotic protozoa parasites Cryptosporidium spp. and Giardia duodenalis infect a wide range of hosts, including humans. Pigs are reservoir hosts for Cryptosporidium spp. and G. duodenalis, which can transmit cryptosporidiosis and giardiasis to humans and other animals. The current study sought to investigate the infection rates and species/genotypes of Cryptosporidium spp. and G. duodenalis in pigs in Hubei of China. The nested PCR and sequence analyses of the small ribosomal subunit RNA (SSU rRNA) gene for Cryptosporidium spp. and the β-giardin (bg) gene for G. duodenalis was utilized to screen for the infection of those parasites in a total of 826 fresh fecal samples. Both Cryptosporidium spp. and G. duodenalis infection rates were 0.97% (8/826). Of the genotyped positive isolates, 6/8 (75%) were C. scrofarum and 2/8 (25%) were C. suis, while two zoonotic species G. duodenalis assemblage E and assemblage A were also detected in 7/8 (87.5%) isolates and 1/8 (12.5%) isolates, respectively. The findings suggest that both of those parasites in pig in intensive farms of Hubei province, China, pose a potential public health risk.
Introduction
Cryptosporidium spp. and Giardia duodenalis were found worldwide, infecting a wide variety of vertebrate hosts and causing self-limiting diarrhea and other clinical signs in humans and livestock, especially in immunodeficient or immunocompromised individuals (Ryan et al., 2014; Ryan et al., 2019). Each genus comprises a complex of species and genotypes, some of which are zoonotic and some specific to particular hosts (Feng et al., 2018; Naguib et al., 2021). People can be infected by directly or indirectly ingesting infective Cryptosporidium oocysts and Giardia cysts, via contaminated water, food and pasture (Thompson and Ash, 2016). In fact, in the 24 foodborne parasites species listed by Food and Agriculture Organization of the United Nations (FAO), Cryptosporidium spp. and G. duodenalis were ranked fifth and 11th, respectively.
To date, at least 40 Cryptosporidium species and 100 genotypes, as well as nine G. duodenalis assemblages have been identified (Ryan et al., 2021). Based on molecular identification, C. parvum and C. hominis are the most common causes of human cryptosporidiosis. C. meleagridis, C. ubiquitum, C. cuniculus, C. felis, C. canis, C. viatorum, and C. muris are some of the other human-pathogenic Cryptosporidium spp (Feng et al., 2018). Giardia duodenalis is a species complex consisting of nine genetically distinct assemblages (A-H), and the common assemblages A and B usually infect humans and other mammals, whereas assemblages C-H only infect specific host (Caccio and Ryan, 2008). Cryptosporidium and Giardia have been reported in pigs from almost all countries or regions of the world. Six Cryptosporidium species have been isolated in pigs: C. suis, C. parvum, C. muris, C. andersoni, C. scrofarum (formerly Cryptosporidium pig genotype II), and C. tyzzeri (formerly Cryptosporidium mouse genotype I) (Zheng et al., 2019). The main Cryptosporidium species identified in pigs worldwide are C. suis and C. scrofarum (Feng et al., 2018). However, these two pig-adapted Cryptosporidium species have been repeatedly identified in fecal samples from human, which indicated zoonotic potential (Leoni et al., 2006; Chen et al., 2011; Bodager et al., 2015). G. duodenalis assemblage A was commonly reported in humans and has been identified in pig (Langkjaer et al., 2007; Caccio et al., 2008; Armson et al., 2009). Swine dung may pollute the environment through water or other ways, and pasture runoff can introduce enormous quantities of Cryptosporidium oocysts and Giardia spores into streams and rivers. Pigs were infected with zoonotic species and genotypes of Cryptosporidium and G. duodenalis, suggesting that they might be sources of infection for nearby residents.
In China, the pig sector is the most important in animal production, and the Cryptosporidium and G. duodenalis infection in pigs has been recorded in most regions of China (Chen et al., 2011; Zhang et al., 2013; Lin et al., 2015; Wang et al., 2017a; Liu et al., 2019; Zheng et al., 2019). However, no studies have investigated Cryptosporidium spp. and G. duodenalis in pigs in Hubei of China. Therefore, the focus of this study was to determine the prevalence and genotypes of Cryptosporidium and G. duodenalis in domestic pigs of various age groups in Hubei, central region of China, and elucidate the role they play in human and animals health under the One Health concept.
Materials and Methods
Ethical Approval
The Research Ethics Committee of Huazhong Agricultural University reviewed and approved our study. Prior to sample collection, we obtained permission from the head of the animal farms.
Sampling
Between September and December of 2019, 826 fresh fecal specimens of pigs were obtained from nine intensive pig farms located at Xianning, Xiaogan, Wuhan, Huanggang, Ezhou and Xiangyang areas in Hubei (Figure 1). The fecal samples were collected from different age groups, including 153 from pre-weaned piglets (<20 days), 231 from post-weaned piglets (21-70 days), 80 from fattening pigs (71-180 days), 362 from sows and herd boar (>180 days). A veterinarian assisted in the collection of fecal specimens from the rectum or the internal component of a stool specimen found on the ground. All specimens (weighing between 5 and 30 g) were gathered with sterile disposable gloves, labeled with the date, age, and farm, and transported to laboratory in cold containers with ice packs. When fecal specimens were taken froom the pigs, no clear clinical symptoms was observed.
Figure 1 Sampling sites in pigs in Hubei Province, China. Sampling locations are marked in dark colors.
DNA Extraction and PCR Amplification
The TIANamp Stool DNA Kit (TIANGEN BIOTECH(BEIJING) CO., LTD, Beijing, China) was used to extract total genomic DNA from about 200 mg of each fecal material according to the manufacturer’s instructions, each DNA specimen was given 50 μL of elution buffer and stored at −20°C until PCR amplification.
As previously stated, Cryptosporidium and Giardia was screened via using nested PCR amplification of the small ribosomal subunit RNA (SSU rRNA) gene and β-giardin (bg) gene loci, respectively (Xiao et al., 1999; Lalle et al., 2005). For Cryptosporidium and G. duodenalis, rTaq and Extaq (Takara Bio Inc., Dalian, China) were used in a 25 μL PCR amplification system, containing 3.5 μL 10 ×PCR Buffer, 2 μL dNTP Mix (2.5 mmol/L), 0.5 μL of forward and reverse primers (25 μmol/L), 15.3 μL deionized water, 1μL rTaq (1.25 U) or ExTaq (1.25 U), 2 μL genomic DNA for the primary PCR template, and 2 μL primary amplification product used for the secondary PCR template. Positive controls (Cattle-derived C. parvum and G. duodenalis assemblage E DNA) and negative controls (distilled water) were included in each PCR assay. Each sample was analyzed by PCR with two technical replicates for each gene locus. All secondary PCR products were electrophoresed in a 1.5% agarose gel, then visualized via GelRed staining and a UV transilluminator (ProteinSimple Inc., State of California, USA).
Nucleotide Sequencing and Analysis
The target gene’s positive PCR amplicons were direct sequenced bidirectionally using secondary primers of sanger sequencing in TSINGKE Biological Technology (Wuhan, China). To determine the species and subtypes of Cryptosporidium and G. duodenalis, all nucleotide sequences were run through the Basic Local Alignment Search tool (BLAST) and compared to Cryptosporidium and Giardia duodenalis reference sequences downloaded from the National Center for Biotechnology Information (https://www.ncbi.nlm.nih.gov/) using ClustalX 2.1 (2010-11-17) (http://www.clustal.org/).
Statistical Analysis and Nucleotide Sequence Accession Numbers
The χ2 test was utilized to compare the Cryptosporidium or Giardia prevalence from pigs in different sample region or age group using SPSS 22.0. Statistical significance was established at p<0.05. The representative Cryptosporidium and G. duodenalis nucleotide sequences identified in the pigs were submitted to GenBank at the National Center for Biotechnology Information under accession numbers: ON149804-ON149811 for Cryptosporidium and ON168862-ON168869 for Giardia.
Results
Prevalence of Cryptosporidium spp. and Giardia duodenalis
Of the 826 fecal specimens collected from pigs, the infection of Cryptosporidium 0.97% (8/826), and G. duodenalis were also 0.97% (8/826). Of the 6 sampled regions in this study, only 3 regions were positive for Cryptosporidium, and the prevalence of Cryptosporidium in pigs in different regions was between 0 and 1.66% (Table 1). Among them, Wuhan City is the highest (0.66%, 5/302), followed by Huanggang (1.61%, 1/62), Xianning (1.35%, 2/148), and no Cryptosporidium were detected in pig farms from Xiaogan, Ezhou and Xiangyang. In addition, in four pigs age group, the highest infection rate was in nursery pigs (3.03%, 7/231), followed by gilts (0.28%, 1/362), and no Cryptosporidium infection was found in the other age pig herds (Table 2). Significantly difference of Cryptosporidium infection rates was observed among sampled regions (χ2 = 4140, p < 0.001) and pigs aged (χ2 = 4130, p < 0.001) (Figure 2).
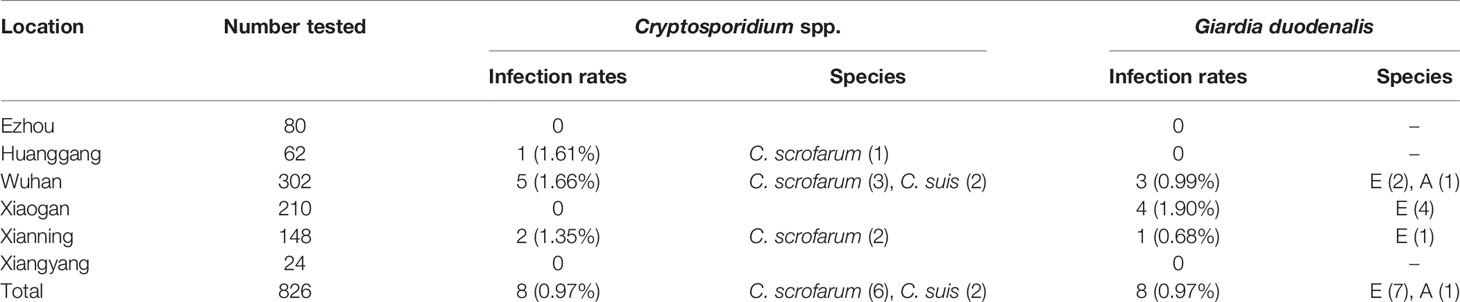
Table 1 Prevalence and genetic characterizations of Cryptosporidium spp. and Giardia duodenalis arranged by sampled location in pigs in Hubei of China.
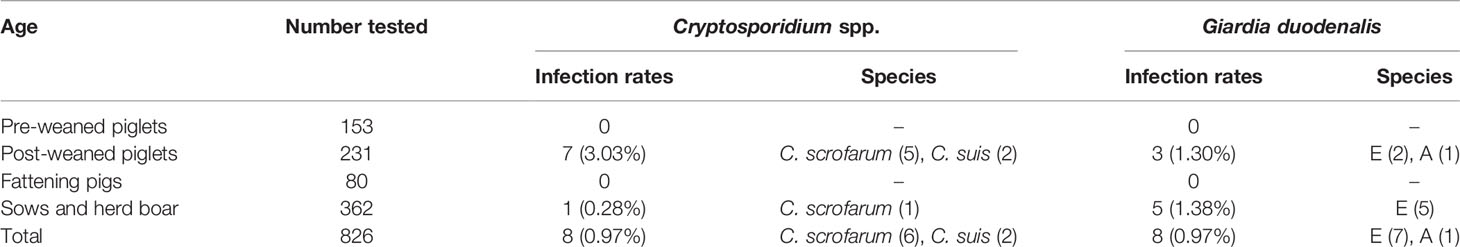
Table 2 Prevalence and genetic characterizations of Cryptosporidium spp. and Giardia duodenalis arranged by age group in pigs in Hubei of China.
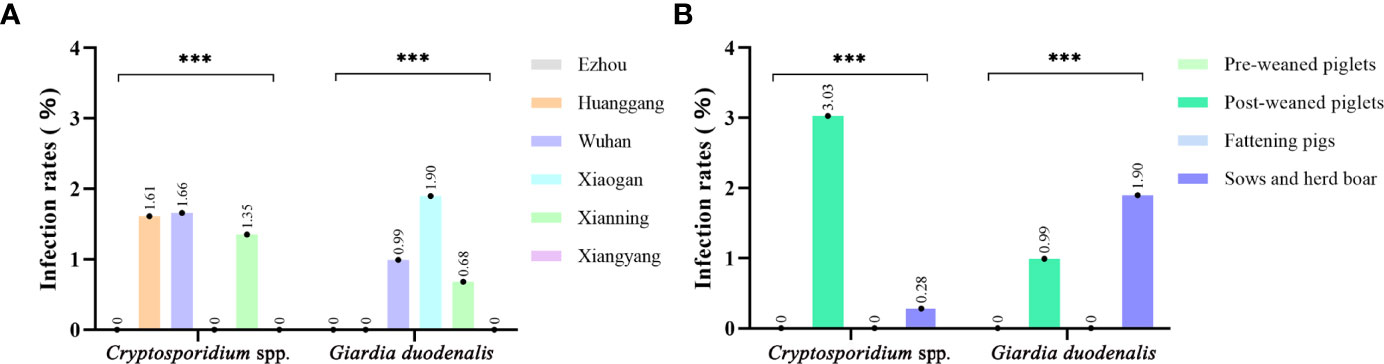
Figure 2 Occurrence of Cryptosporidium spp. and Giardia duodenalis in different sample site (A) and age group (B) of pigs from Hubei Province, China. ***p<0.001.
The prevalence of G. duodenalis in pigs in different regions ranged from 0 to 1.90%. Among them, Xiaogan has the highest infection rate (1.90%, 4/210), followed by Wuhan City (0.99%, 3/102), Xianning (0.68%, 1/148), while no G. duodenalis were detected in the pig farms from Huanggang, Ezhou and Xiangyang. Among the pigs of all ages, the highest infection rate was in sows and herd boar (1.38%, 5/362), followed by post-weaned pigs (1.30%, 3/231), and no G. duodenalis infection was found in the other age group. Significantly difference of G. duodenalis infection rates was observed among sampled regions (χ2 = 2481, p < 0.001) and pigs age group (χ2 = 2478, p < 0.001).
Molecular Characterization of Cryptosporidium spp. and Giardia duodenalis
Based on sequence analysis, six and two of eight Cryptosporidium positive samples were identified as C. scrofarum and C. suis, respectively. Among the three positive city, C. scrofarum was identified in Wuhan (n=3), Xianning (n=2) and Huanggang (n=1), while C. suis was only identified in Wuhan (n=2). C. scrofarum were both identified in the two positive age groups, with 5 in post-weaned pigs and 2 in sows and herd boar, while only one C. suis positive samples were identified in post-weaned pigs. Of the eight G. duodenalis positive samples, sequence analysis revealed seven assemblage E and one assemblage A based on BG. Assemblage E was identified in three positive regions, including Wuhan (n=2), Xiaogan (n=4) and Xianning (n=1), while assemblage A was identified in Wuhan (n=1). In addition, assemblage E (n=2) and assemblage A (n=1) were found in post-weaned piglets, and only assemblage E (n=4) was found in Snows and herd boar.
Discussion
The previously reported prevalence of Cryptosporidium and G. duodenalis in pigs from most area of China, and have been reported in most country of the world. The infection rates of Cryptosporidium in pigs were much lower than those reported in Asian countries such as Japan (32.6%) (Yui et al., 2014), Thailand (20.8%) (Thathaisong et al., 2020), Vietnam (14.5%) (Nguyen et al., 2013), and far lower than the total infection rate in China (12.2%), and also lower than those reported in infection rates reported in other provincial areas such as Yunnan (23.0%, 46/200), Zhejiang (14.5%, 18/124), Fujian (11.9%, 16/135), Guangdong (8.3%, 34/217), Jiangxi (4.0%, 41/1036), Shaanxi (3.3%, 44/1337), Henan (3.1%, 28/897), and Heilongjiang (1.6%, 9/568) (Zhang et al., 2013; Lin et al., 2015; Zou et al., 2017; Wang et al., 2018; Wang et al., 2021; Wang et al., 2022). Nested PCR amplification based on the BG gene of G. duodenalis showed that a total of 8 positive samples were amplified with an overall infection rate of 0.97% (8/826), which is much lower than the infection rates previously reported in Denmark (14.0%, 120/857) (Petersen et al., 2015), Korea (14.8%, 110/745) (Lee et al., 2020), and also lower than the previous infection rates reported in other regions of China such as Shanghai (26.9%, 25/93), Zhejiang (10.5%, 13/124), Shaanxi (8.0%, 45/560), Yunnan (5.3%, 21/396; 2.5%, 5/200), Taiwan (4.3%, 6/141), Guangdong (4.2%, 3/72), Xinjiang (2.6%, 21/801), and Henan (1.7%, 15/897) (Wang et al., 2017a; Wang et al., 2018; Jing et al., 2019; Liu et al., 2019; Lam et al., 2021; Zou et al., 2021). There are many reasons for such a large variation in infection rates, which may be related to the number, time and location of sampling, the size and environment of pig farms, animal population density, detection methods and other geographical factors such as temperature and humidity, precipitation and climate (Wang et al., 2021). The main reason for the generally lower infection rates in this study than those previously reported in other regions of China may be that the sampling time of this study was right after the outbreak of African swine fever in China, and most of the pig farms were in the resumption phase as well as in the phase of strict prevention and control of African swine fever, and all large-scale pig farms around the country had greatly improved their biosecurity level, and the frequency and degree of disinfection of the environment in pig farms had been enhanced compared to the previous ones.
Two Cryptosporidium species, C. scrofarum and C. suis, were identified in this study, and the results are consistent with those reported in other provinces of China, and these two Cryptosporidium species were frequently identified in pigs worldwide, and both are human-animal species. In addition, C. parvum, which mainly infects humans, has been reported several times in foreign swine surveys, but is rarely reported in China (Ryan et al., 2014). C. suis mainly infects pigs, have also been isolated from water source and tap water samples in Shanghai, China (Feng et al., 2011). A total of two G. duodenalis assemblages were identified, with seven samples being assemblage E and one being assemblage A. This result was similar to that reported in Shaanxi Province (Wang et al., 2017a), assemblage E was the dominant genotype, and like the results reported for G. duodenalis of porcine origin in other regions of China (Liu et al., 2019). In contrast, assemblage A is capable of infecting animals such as humans and domestic animals, and was identified in this study and in swine herds in Shaanxi, Shanghai, and in children with diarrhea in Wuhan, indicating that assemblage A is currently prevalent among both humans and swine (Wang et al., 2017a; Wang et al., 2017b; Liu et al., 2019).
Cases of C. scrofarum and C. suis infections have been reported in immunocompetent diarrhea patients and HIV-positive patients in recent years, suggesting that these two pig-adapted Cryptosporidium species may be zoonotic (Xiao et al., 2006; Bodager et al., 2015). C. suis was first identified in a 24-year-old HIV patient in Peru and China (Xiao et al., 2002; Cama et al., 2007; Wang et al., 2013). C. suis has also been identified in patients with digestive diseases in the UK and Madagascar (Leoni et al., 2006; Bodager et al., 2015). Only one case of human infection with C. scrofarum has been reported, which occurred in the Czech Republic (Kvac et al., 2009). Cryptosporidiosis in pigs should receive more attention because it is not only a veterinary problem but may be important for public health. More important, C. suis has been detected in drinking water source water (Feng et al., 2011; Xiao et al., 2012; Hu et al., 2014). G. duodenalis assemblage A and E were frequently found in pigs around the world (Jing et al., 2019; Zou et al., 2021). In addition, assemblage A is one of the main assemblages of infected people, and assemblage E was identified in humans in Australia (Zahedi et al., 2017; Cai et al., 2021). All these reports indicate that pig populations play an important role in the transmission of Cryptosporidium and G. duodenalis are of great public health importance. Therefore, there is a need to develop better farm management systems to reduce environmental contamination by zoonotic diseases and prevent the occurrence of zoonotic transmission caused by Cryptosporidium and G. duodenalis. The prevention and control of those parasites in large-scale pig farms requires strengthening the nutritional management of the herd, improving the immunity of the herd, selecting drugs with anti-oocyst activity to enhance the disinfection of environmental hygiene as well as taking effective measures to control insects such as flies, enhancing the cleaning of manure and keeping the pen dry.
Conclusion
This study reported the prevalence of Cryptosporidium and G. duodenalis infection in healthy pigs. Due to the zoonotic nature of Cryptosporidium spp. and G. duodenalis, adequate cleaning and sanitary activities in pig farms should be used to prevent probale transfer of infective parasites to humans and other animals.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/supplementary material.
Author Contributions
JZ and LH conceived and designed this study. DL, HD, YZ and HZ conducted the laboratory work. DL drafted the manuscript. JZ, LH and SW critically revised the manuscript. All authors have read and approved the final manuscript.
Funding
This work was supported by the National Key Research and Development Program Intergovernmental International Cooperation Project of China (Grant No. SQ2021YFE012313), and the Fundamental Research Funds for the Central Universities in China (Project 2662020DKPY016).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Armson, A., Yang, R., Thompson, J., Johnson, J., Reid, S., Ryan, U. M. (2009). Giardia Genotypes in Pigs in Western Australia: Prevalence and Association With Diarrhea. Exp. Parasitol. 121, 381–383. doi: 10.1016/j.exppara.2009.01.008
Bodager, J. R., Parsons, M. B., Wright, P. C., Rasambainarivo, F., Roellig, D., Xiao, L., et al. (2015). Complex Epidemiology and Zoonotic Potential for Cryptosporidium Suis in Rural Madagascar. Veterinary Parasitol. 207, 140–143. doi: 10.1016/j.vetpar.2014.11.013
Caccio, S. M., Beck, R., Lalle, M., Marinculic, A., Pozio, E. (2008). Multilocus Genotyping of Giardia Duodenalis Reveals Striking Differences Between Assemblages A and B. Int. J. Parasitol. 38, 1523–1531. doi: 10.1016/j.ijpara.2008.04.008
Caccio, S. M., Ryan, U. (2008). Molecular Epidemiology of Giardiasis. Mol. Biochem. Parasitol. 160, 75–80. doi: 10.1016/j.molbiopara.2008.04.006
Cai, W. L., Ryan, U., Xiao, L. H., Feng, Y. Y. (2021). Zoonotic Giardiasis: An Update. Parasitol. Res. 120, 4199–4218. doi: 10.1007/s00436-021-07325-2
Cama, V. A., Ross, J. M., Crawford, S., Kawai, V., Chavez-Valdez, R., Vargas, D., et al. (2007). Differences in Clinical Manifestations Among Cryptosporidium Species and Subtypes in HIV-Infected Persons. J. Infect. Dis. 196, 684–691. doi: 10.1086/519842
Chen, Z., Mi, R., Yu, H., Shi, Y., Huang, Y., Chen, Y., et al. (2011). Prevalence of Cryptosporidium Spp. In Pigs in Shanghai, China. Veterinary Parasitol. 181, 113–119. doi: 10.1016/j.vetpar.2011.04.037
Feng, Y., Ryan, U. M., Xiao, L. (2018). Genetic Diversity and Population Structure of Cryptosporidium. Trends Parasitol. 34, 997–1011. doi: 10.1016/j.pt.2018.07.009
Feng, Y., Zhao, X., Chen, J., Jin, W., Zhou, X., Li, N., et al. (2011). Occurrence, Source, and Human Infection Potential of Cryptosporidium and Giardia Spp. In Source and Tap Water in Shanghai, China. Appl. Environ. Microbiol. 77, 3609–3616. doi: 10.1128/AEM.00146-11
Hu, Y., Feng, Y. Y., Huang, C. C., Xiao, L. H. (2014). Occurrence, Source, and Human Infection Potential of Cryptosporidium and Enterocytozoon Bieneusi in Drinking Source Water in Shanghai, China, During a Pig Carcass Disposal Incident. Environ. Sci. Technol. 48, 14219–14227. doi: 10.1021/es504464t
Jing, B., Zhang, Y., Xu, C., Li, D., Xing, J., Tao, D., et al. (2019). Detection and Genetic Characterization of Giardia Duodenalis in Pigs From Large-Scale Farms in Xinjiang, China. Parasite 26, 53. doi: 10.1051/parasite/2019056
Kvac, M., Kvetonova, D., Sak, B., Ditrich, O. (2009). Cryptosporidium Pig Genotype II in Immunocompetent Man. Emerging Infect. Dis. 15, 982–983. doi: 10.3201/eid1506.07621
Lalle, M., Pozio, E., Capelli, G., Bruschi, F., Crotti, D., Caccio, S. M. (2005). Genetic Heterogeneity at the Beta-Giardin Locus Among Human and Animal Isolates of Giardia Duodenalis and Identification of Potentially Zoonotic Subgenotypes. Int. J. Parasitol. 35, 207–213. doi: 10.1016/j.ijpara.2004.10.022
Lam, H. Y. P., Chen, T. T., Tseng, Y. C., Chang, K. C., Yang, T. H., Peng, S. Y. (2021). Detection and Genotyping of Giardia Duodenalis From Cattle and Pigs in Hualien Country, Eastern Taiwan. J. Microbiology Immunology Infection = Wei Mian Yu Gan Ran Za Zhi 54, 718–727. doi: 10.1016/j.jmii.2020.05.009
Langkjaer, R. B., Vigre, H., Enemark, H. L., Maddox-Hyttel, C. (2007). Molecular and Phylogenetic Characterization of Cryptosporidium and Giardia From Pigs and Cattle in Denmark. Parasitology 134, 339–350. doi: 10.1017/S0031182006001533
Lee, H., Jung, B., Lim, J. S., Seo, M. G., Lee, S. H., Choi, K. H., et al. (2020). Multilocus Genotyping of Giardia Duodenalis From Pigs in Korea. Parasitol. Int. 78, 102154. doi: 10.1016/j.parint.2020.102154
Leoni, F., Amar, C., Nichols, G., Pedraza-Diaz, S., McLauchlin, J. (2006). Genetic Analysis of Cryptosporidium From 2414 Humans With Diarrhoea in England Between 1985 and 2000. J. Med. Microbiol. 55, 703–707. doi: 10.1099/jmm.0.46251-0
Lin, Q., Wang, X. Y., Chen, J. W., Ding, L., Zhao, G. H. (2015). Cryptosporidium Suis Infection in Post-Weaned and Adult Pigs in Shaanxi Province, Northwestern China. Korean J. Parasitol. 53, 113–117. doi: 10.3347/kjp.2015.53.1.113
Liu, H., Xu, N., Yin, J., Yuan, Z., Shen, Y., Cao, J. (2019). Prevalence and Multilocus Genotyping of Potentially Zoonotic Giardia Duodenalis in Pigs in Shanghai, China. Parasitology 146, 1199–1205. doi: 10.1017/S0031182019000349
Naguib, D., Roellig, D. M., Arafat, N., Xiao, L. (2021). Genetic Characterization of Cryptosporidium Cuniculus From Rabbits in Egypt. Pathogens 10(6):775. doi: 10.3390/pathogens10060775
Nguyen, S. T., Fukuda, Y., Tada, C., Sato, R., Huynh, V. V., Nguyen, D. T., et al. (2013). Molecular Characterization of Cryptosporidium in Pigs in Central Vietnam. Parasitol. Res. 112, 187–192. doi: 10.1007/s00436-012-3124-2
Petersen, H. H., Jianmin, W., Katakam, K. K., Mejer, H., Thamsborg, S. M., Dalsgaard, A., et al. (2015). Cryptosporidium and Giardia in Danish Organic Pig Farms: Seasonal and Age-Related Variation in Prevalence, Infection Intensity and Species/Genotypes. Veterinary Parasitol. 214, 29–39. doi: 10.1016/j.vetpar.2015.09.020
Ryan, U., Fayer, R., Xiao, L. (2014). Cryptosporidium Species in Humans and Animals: Current Understanding and Research Needs. Parasitology 141, 1667–1685. doi: 10.1017/S0031182014001085
Ryan, U. M., Feng, Y., Fayer, R., Xiao, L. (2021). Taxonomy and Molecular Epidemiology of Cryptosporidium and Giardia - a 50 Year Perspective (1971-2021). Int. J. Parasitol. 51, 1099–1119. doi: 10.1016/j.ijpara.2021.08.007
Ryan, U., Hijjawi, N., Feng, Y., Xiao, L. (2019). Giardia: An Under-Reported Foodborne Parasite. Int. J. Parasitol. 49, 1–11. doi: 10.1016/j.ijpara.2018.07.003
Thathaisong, U., Siripattanapipong, S., Inpankaew, T., Leelayoova, S., Mungthin, M. (2020). High Prevalence of Cryptosporidium Infection Caused by C. Scrofarum and C. Suis Among Pigs in Thailand. Parasitol. Int. 77, 102122. doi: 10.1016/j.parint.2020.102122
Thompson, R. C. A., Ash, A. (2016). Molecular Epidemiology of Giardia and Cryptosporidium Infections. Infection Genet. evolution: J. Mol. Epidemiol. evolutionary Genet. Infect. Dis. 40, 315–323. doi: 10.1016/j.meegid.2015.09.028
Wang, T., Fan, Y., Koehler, A. V., Ma, G., Li, T., Hu, M., et al. (2017b). First Survey of Cryptosporidium, Giardia and Enterocytozoon in Diarrhoeic Children From Wuhan, China. Infection Genet. evolution: J. Mol. Epidemiol. evolutionary Genet. Infect. Dis. 51, 127–131. doi: 10.1016/j.meegid.2017.03.006
Wang, W., Gong, Q. L., Zeng, A., Li, M. H., Zhao, Q., Ni, H. B. (2021). Prevalence of Cryptosporidium in Pigs in China: A Systematic Review and Meta-Analysis. Transboundary emerging Dis. 68, 1400–1413. doi: 10.1111/tbed.13806
Wang, P., Li, S., Zou, Y., Du, Z. C., Song, D. P., Wang, P., et al. (2022). The Infection and Molecular Characterization of Cryptosporidium Spp. In Diarrheic Pigs in Southern China. Microb. Pathog. 165, 105459. doi: 10.1016/j.micpath.2022.105459
Wang, S. S., Yuan, Y. J., Yin, Y. L., Hu, R. S., Song, J. K., Zhao, G. H. (2017a). Prevalence and Multilocus Genotyping of Giardia Duodenalis in Pigs of Shaanxi Province, Northwestern China. Parasites Vectors 10, 490. doi: 10.1186/s13071-017-2418-8
Wang, H., Zhang, Y., Wu, Y., Li, J., Qi, M., Li, T., et al. (2018). Occurrence, Molecular Characterization, and Assessment of Zoonotic Risk of Cryptosporidium Spp., Giardia Duodenalis, and Enterocytozoon Bieneusi in Pigs in Henan, Central China. J. Eukaryotic. Microbiol. 65, 893–901. doi: 10.1111/jeu.12634
Wang, L., Zhang, H. W., Zhao, X. D., Zhang, L. X., Zhang, G. Q., Guo, M. J., et al. (2013). Zoonotic Cryptosporidium Species and Enterocytozoon Bieneusi Genotypes in HIV-Positive Patients on Antiretroviral Therapy. J. Clin. Microbiol. 51, 557–563. doi: 10.1128/Jcm.02758-12
Xiao, S. M., An, W., Chen, Z. M., Zhang, D. Q., Yu, J. W., Yang, M. (2012). Occurrences and Genotypes of Cryptosporidium Oocysts in River Network of Southern-Eastern China. Parasitol. Res. 110, 1701–1709. doi: 10.1007/s00436-011-2688-6
Xiao, L., Bern, C., Arrowood, M., Sulaiman, I., Zhou, L., Kawai, V., et al. (2002). Identification of the Cryptosporidium Pig Genotype in a Human Patient. J. Infect. Dis. 185, 1846–1848. doi: 10.1086/340841
Xiao, L., Escalante, L., Yang, C., Sulaiman, I., Escalante, A. A., Montali, R. J., et al. (1999). Phylogenetic Analysis of Cryptosporidium Parasites Based on the Small-Subunit rRNA Gene Locus. Appl. Environ. Microbiol. 65, 1578–1583. doi: 10.1128/AEM.65.4.1578-1583.1999
Xiao, L., Moore, J. E., Ukoh, U., Gatei, W., Lowery, C. J., Murphy, T. M., et al. (2006). Prevalence and Identity of Cryptosporidium Spp. In Pig Slurry. Appl. Environ. Microbiol. 72, 4461–4463. doi: 10.1128/AEM.00370-06
Yui, T., Nakajima, T., Yamamoto, N., Kon, M., Abe, N., Matsubayashi, M., et al. (2014). Age-Related Detection and Molecular Characterization of Cryptosporidium Suis and Cryptosporidium Scrofarum in Pre- and Post-Weaned Piglets and Adult Pigs in Japan. Parasitol. Res. 113, 359–365. doi: 10.1007/s00436-013-3662-2
Zahedi, A., Field, D., Ryan, U. (2017). Molecular Typing of Giardia Duodenalis in Humans in Queensland - First Report of Assemblage E. Parasitology 144, 1154–1161. doi: 10.1017/S0031182017000439
Zhang, W., Yang, F., Liu, A., Wang, R., Zhang, L., Shen, Y., et al. (2013). Prevalence and Genetic Characterizations of Cryptosporidium Spp. In Pre-Weaned and Post-Weaned Piglets in Heilongjiang Province, China. PLoS One 8, e67564. doi: 10.1371/journal.pone.0067564
Zheng, S., Li, D., Zhou, C., Zhang, S., Wu, Y., Chang, Y., et al. (2019). Molecular Identification and Epidemiological Comparison of Cryptosporidium Spp. Among Different Pig Breeds in Tibet and Henan, China. BMC veterinary Res. 15, 101. doi: 10.1186/s12917-019-1847-3
Zou, Y., Ma, J. G., Yue, D. M., Zheng, W. B., Zhang, X. X., Zhao, Q., et al. (2017). Prevalence and Risk Factors of Cryptosporidium Infection in Farmed Pigs in Zhejiang, Guangdong, and Yunnan Provinces, China. Trop. Anim. Health production 49, 653–657. doi: 10.1007/s11250-017-1230-y
Keywords: Cryptosporidium, Giardia duodenalis, pigs, infection rates, zoonotic
Citation: Li D, Deng H, Zheng Y, Zhang H, Wang S, He L and Zhao J (2022) First Characterization and Zoonotic Potential of Cryptosporidium spp. and Giardia duodenalis in Pigs in Hubei Province of China. Front. Cell. Infect. Microbiol. 12:949773. doi: 10.3389/fcimb.2022.949773
Received: 21 May 2022; Accepted: 20 June 2022;
Published: 11 July 2022.
Edited by:
Razakandrainibe Romy, Université de Rouen, FranceReviewed by:
Junqiang Li, Henan University of Traditional Chinese Medicine, ChinaNagah Arafat, Mansoura University, Egypt
Copyright © 2022 Li, Deng, Zheng, Zhang, Wang, He and Zhao. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Junlong Zhao, emhhb2p1bmxvbmdAbWFpbC5oemF1LmVkdS5jbg==
 Dongfang Li
Dongfang Li Han Deng1
Han Deng1 Yaxin Zheng
Yaxin Zheng Sen Wang
Sen Wang Lan He
Lan He Junlong Zhao
Junlong Zhao