
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Cell. Infect. Microbiol. , 27 September 2022
Sec. Fungal Pathogenesis
Volume 12 - 2022 | https://doi.org/10.3389/fcimb.2022.944611
This article is part of the Research Topic Alternative Approaches to Antifungal Drugs Against Drug-resistant Fungi View all 5 articles
 Yean Sheng Lee1
Yean Sheng Lee1 Xinyue Chen1
Xinyue Chen1 Tria Widiasih Widiyanto1
Tria Widiasih Widiyanto1 Kanami Orihara1
Kanami Orihara1 Hiroyuki Shibata2
Hiroyuki Shibata2 Susumu Kajiwara1*
Susumu Kajiwara1*Candida albicans is a pathogenic yeast that causes candidiasis in immunocompromised patients. The overuse of antifungal drugs has led to the development of resistance to such drugs by this fungus, which is a major challenge in antifungal chemotherapy. One approach to this problem involves the utilization of new natural products as an alternative source of antifungals. Curcumin, one such natural product, has been widely studied as a drug candidate and is reported to exhibit antifungal activity against C. albicans. Although studies of the mechanism of curcumin against human cancer cells have shown that it inhibits heat shock protein 90 (Hsp90), little is known about its function against C. albicans. In this paper, using a doxycycline-mediated HSP90 strain and an HSP90-overexpressing strain of C. albicans, we demonstrated that the curcumin triggered a decrease in Hsp90 by affecting it at the post-transcriptional level. This also led to the downregulation of HOG1 and CDR1, resulting in a reduction of the stress response and efflux pump activity of C. albicans. However, the inhibition of HSP90 by curcumin was not due to the inhibition of transcription factors HSF1 or AHR1. We also found that curcumin can not only decrease the transcriptional expression of CDR1, but also inhibit the efflux pump activity of Cdr1. Hence, we conclude that disruption of HSP90 by curcumin could impair cell growth, stress responses and efflux pump activity of C. albicans.
Candida albicans is an opportunistic pathogenic fungus, one of many microorganisms including bacteria and other fungi that normally coexist without causing harm within the human body. An infection caused by this fungus is called candidiasis, and healthy people are protected from candidiasis by both innate and acquired immunities. However, this microorganism can cause both superficial and systemic infections in humans, mainly in immunocompromised individuals such as AIDS patients, neonates, people with debilitating disease, or those who have undergone extensive surgery and are hospitalized for an extended period (Eggimann et al., 2003; Hajjeh et al., 2004; Patterson, 2005; Pfaller and Diekema, 2007; Cisterna et al., 2010; Brown et al., 2012). C. albicans has become the most common pathogenic fungi in humans, with the incidence of fungal infections having increased greatly in recent decades. This is due to the growing proportion of the worldwide human population that is immunocompromised and aged. Significant progress has been made in antifungal chemotherapy (Cowen et al., 2002; Carrillo-Munoz et al., 2006; Ortega et al., 2011), but the appearance of antifungal resistant strains and the limits of effective and selective antifungals are major problems in the treatment of clinical fungal infections (Almirante et al., 2005). It is therefore urgent to develop novel antifungal agents that are both safe and effective. One strategy is to combine the search for new potential targets with the screening of promising new antifungal agents (Carrillo-Munoz et al., 2006).
To develop novel antifungals, there have been numerous efforts to identify natural compounds that exhibit effective antifungal activity, and one such compound, curcumin, has been of great interest to the scientific community due to its chemotherapeutic properties (Hatcher et al., 2008). Curcumin is a yellow-pigmented polyphenolic compound that is derived from the roots of the Curcuma longa, a plant native to India and other parts of southeast Asia (Limtrakul et al., 2004; Jurenka, 2009; Rowe et al., 2009). In India and China, this compound has been widely used as a cosmetic and sometimes as medicine for the treatment of wounds and inflammation (Govindarajan and Stahl, 1980; Ammon and Wahl, 1991; Strimpakos and Sharma, 2008). Previous studies have found that curcumin has anti-carcinogenic, antioxidant, anti-inflammatory, and antimicrobial properties, as well as exhibiting hypoglycemic effects in humans (Jurenka, 2009). In addition, the safety of curcumin has been studied in animal models and human clinical trials, which have shown that its toxicity is low even when administered at high doses (Bhavani Shankar et al., 1980; Qureshi et al., 1992). Previous studies using both biochemical, genetic or both approaches have shown that the curcumin exhibits antifungal activity against C. albicans via oxidative stress, inhibiting hyphal development, disrupting cell wall integrity and plasma membrane, modulates proteolytic enzyme activity, altering the membrane-associated properties of ATPase activity, modulating efflux pumps, synergizing with antifungal azoles, and inhibiting biofilm formation (Sharma et al., 2009; Sharma et al., 2010a; Sharma et al., 2010b; Neelofar et al., 2011; Garcia-Gomes et al., 2012; Kumar et al., 2014; Lee and Lee, 2014; Shahzad et al., 2014; Thakre et al., 2016; Alalwan et al., 2017; Chen et al., 2018; Andrade et al., 2019; Hamzah et al., 2020; Cheraghipour et al., 2021; Dong et al., 2021; Lasrado et al., 2022).
However, little is known about the molecular mechanisms of curcumin’s effect on yeast cells. In human cancer therapeutic, a series of recent studies show that curcumin inhibits heat-shock protein 90 (Hsp90) in human cancer cells (Zhang et al., 2007; Giommarelli et al., 2010; Anand et al., 2012; Khan et al., 2012; Li et al., 2012; Bhullar et al., 2015; Lv et al., 2015; Zheng et al., 2016; Ye et al., 2017; Fan et al., 2018; Forouzanfar et al., 2019; Liu et al., 2020; Surma et al., 2022). Due to the high conservation of Hsp90 across species and the high degree of homology between C. albicans and human Hsp90, curcumin is expected to have disruptive effects on Hsp90 in fungal cells as well (Whitesell et al., 2019). But to our knowledge, no study has examined the relationship between fungal HSP90 and curcumin. To illuminate this uncharted area, we examined the effects of curcumin on the HSP90 of C. albicans by utilizing a defective conditional HSP90 mutant and an HSP90-overexpressing strain. Since the HSP90 gene is essential for C. albicans, a doxycycline (Dox)-regulated expression system was adopted in C. albicans by replacing the promoter of HSP90 with the tetO system. This allowed us to control the gene expression of HSP90 by supplementing Dox, which binds to tetR to prevent the transcription of HSP90. To better understand the mechanism of curcumin, we also overexpressed HSP90 by replacing its promoter with the constitutive ADH1 promoter.
In addition, the low bioavailability and poor stability of curcumin have been highlighted as major problems for therapeutic application (Blasius et al., 2004; Anand et al., 2007; Fang et al., 2013). Therefore, many studies have been carried out attempting to improve the bioavailability and stability of curcumin by modification of the molecular structure. GO-Y030, a curcumin derivative designed by Shibata et al. (Shibata et al., 2009), has been reported to possess greater anti-cancer properties than curcumin itself and is significantly less toxic (Gritsko et al., 2006; Cen et al., 2009; Hutzen et al., 2009; Kudo et al., 2011; Mohan Yallapu et al., 2012). However, the antifungal activity of curcumin GO-Y030 has not yet been studied. This study aimed to study the effect of curcumin and its derivative GO-Y030 on Hsp90 of C. albicans.
Curcumin (Wako, Japan) stock solutions were prepared in sterile dimethyl sulfoxide (DMSO). The curcumin analogue GO-Y030 was provided by Akita University, Japan. Its stock solution was prepared in sterile DMSO and stored at 4°C. Nile red (Wako, Japan) stock solution was prepared in ethanol and stored at 4°C. Doxycycline (Wako, Japan) was prepared in distilled water and stored at 4°C. Curcumin compounds and all dye solutions were kept in the dark to prevent light exposure.
C. albicans strains used in this study are listed in the Table 1. All strains were routinely grown in Yeast Extract Peptone Dextrose (YPD; 1% yeast extract, 2% peptone, 2% glucose) or synthetic defined medium (SD; 0.67% yeast nitrogen base without amino acids, 0.079% complete supplement mixture without uracil, 2% glucose) on plates with 2% agar at 37°C. All strains were maintained and stored at 4°C. All strains were stored as frozen stocks with 15% glycerol at −80°C. Construction of the Dox-mediated HSP90 gene mutant and HSP90 overexpression mutant were performed as previously described (Reuß and Morschhäuser, 2006; Lai et al., 2016). The plasmids and primers used in this study are listed in the Tables 2, 3.
The HSP90/hsp90Δ strain was constructed as follows: one of the chromosomal HSP90 alleles in the C. albicans strain THE1 was deleted using the SAT-flipper method, as described previously (Reuß et al., 2004). SAT1 was amplified with primers SAT1.FOR and SAT1.REV. SAT2 was amplified with primers SAT2.FOR and SAT2.REV. From plasmid pSFS2, pSFS2-SAT1/2 was generated by cloning PCR fragments SAT1 and SAT2 into the respective sites. pSFS2-SAT1/2 was digested with KpnI and SacI to release the disruption cassette and transformed into the TR transactivator gene-containing strain THE1 by electroporation (Thompson et al., 1998). Nourseothricin-resistant transformants were selected on YPD agar plates containing 200 µg/mL of nourseothricin. After the induction of FLP recombinase by growing in YPD medium, nourseothricin-sensitive colonies were selected on the YPD plates containing 25 µg/mL of nourseothricin according to their colony size.
The tet-HSP90/hsp90Δ strain was constructed as follows: two regions spanning positions −711 to −138 (HSP1) and positions −7 to +528 (HSP2) relative to the ATG start codon of the HSP90 open reading frame were PCR amplified using primer pairs HSP1.FOR with HSP1.REV and HSP2.FOR with HSP2.REV. These amplification products were then cloned into the respective sites of plasmid p97CAU1 (Nakayama et al., 2000) to form p97CAU1-HSP1/2. This plasmid was then digested with ApaI and SacII to liberate the entire 3 kb promoter-replacing construct and transformed into the HSP90/hsp90Δ strain using electroporation (Thompson et al., 1998). Ura+ transformants were selected on SD agar plates without uracil.
The HSP90-overexpressing mutant (PADH1-HSP90) was constructed as follows: the TR promoter of plasmid p97CAU1-HSP1/2 was replaced by the ADH1 promoter to form plasmid p97CAU1A-HSP1/2. Briefly, the ADH1 promoter was amplified by ADH1.FOR and ADH1.REV using C. albicans SC5314. Then, the ADH1 promoter fragment replaced the TR promoter of plasmid p97CAU1-HSP1/2 at restriction sites SpeI and SmaI to form plasmid p97CAU1A-HSP1/2. Similar to the construction of the tet-HSP90/hsp90Δ strain, this plasmid was then digested with ApaI and SacII to liberate the entire 2.505 kb promoter-replacing construct and transformed into C. albicans THE1 using electroporation (Thompson et al., 1998). Ura+ transformants were selected on SD agar plates without uracil.
Minimum inhibitory concentration (MIC) assays were carried out in flat-bottom, 96-well microtiter plates (Iwaki, Japan) using serial broth microdilution with minor modifications (Rodríguez-Tudela et al., 2003). The MIC80 was defined as the concentration of the antifungal compound that inhibits 80% of the growth of cells as compared with the control. MIC tests were set up in a final volume of 200 µL per well with 2-fold serial dilutions of curcumin compounds in YPD. Gradients of curcumin compounds were diluted from 250 µg/mL down to 0 µg/mL. C. albicans strains SC5314 or PADH1-HSP90 were grown in YPD overnight at 37°C. Then the cells were collected and washed by phosphate-buffered saline (PBS) three times. The number of cells was adjusted to 1 × 103 cells/mL in YPD. 100 µL of each strain was inoculated into each well. The plates were then incubated at 37°C for 24 h. The endpoint of the MIC assay was determined by Varioskan Lux (Thermo Scientific, Japan) at an absorbance of 530 nm and corrected for background from the corresponding medium. Each strain was tested in triplicate for each curcumin compound. The optical densities were averaged for triplicate measurements.
To elucidate the potential mechanism by which curcumin inhibits the growth of C. albicans, HSP90, HSF1, AHR1, HOG1, CDR1, CDR2, and MDR1 gene expression analysis was performed using qRT-PCR. Briefly, the wildtype, tet-HSP90/hsp90Δ, or PADH1-HSP90 strains were cultured at 37°C in YPD until they reached the exponential phase. The cells were then collected, washed, and resuspended in YPD containing curcumin compounds or Dox. A drug-free control was included for each experiment. The samples were then pelleted and washed with Diethyl pyrocarbonate (DEPC)-treated water. The pellet was used to extract the RNA using hot acidic phenol and purified using ethanol (Collart and Oliviero, 1993). The purity and concentration of the extracted RNA were verified using GeneQuant 100 (Biochrom, Japan). RNA was then converted to cDNA using ReverTra Ace™ qRT RT Master Mix with gDNA Remover (Toyobo, Japan) following the manufacturer’s recommended protocol. Gene expression was analyzed by qRT-PCR using the StepOne™ Real-Time PCR System (Thermo Fisher, Japan). The ACT1 housekeeping gene was used as a reference, and the relative gene expression (fold change) was determined by the 2-ΔΔCT method (Livak and Schmittgen, 2001). The primers used for qRT-PCR are listed in Table 4.
The drug combinations were studied by means of a two-dimensional broth microdilution checkerboard procedure using two-antifungal agents as described in the Clinical Microbiology Procedures Handbook (Isenberg, 1992). The checkerboard assays were carried out in flat-bottom, 96-well microtiter plates (Iwaki, Japan) using a serial broth microdilution protocol with minor modifications (Chaturvedi et al., 2008). Tests were set up in a final volume of 200 µL per well with 2-fold serial dilutions of curcumin in YPD medium. Gradients of curcumin and Dox were diluted from 250 µg/mL down to 0.24 µg/mL and from 0.1 µg/mL to 0.00156 µg/mL, respectively. C. albicans SC5314 and tet-HSP90/hsp90Δ strains were grown in YPD overnight at 37°C. Then the cells were collected and washed by PBS three times, and the number of cells was adjusted to 1 × 103 cells/mL in YPD. 100 µL of each strain was inoculated into each well, and the plates were incubated at 37°C for 24 h. The endpoint of the MIC was determined by Varioskan Lux (Thermo Scientific, Japan) at an absorbance of 530 nm and corrected for background from the corresponding medium. The data were quantitatively displayed with color using the program Java TreeView 1.2.0.
The heat shock and osmotic stress experiments were performed according to the protocol described previously (Enjalbert et al., 2003). Briefly, cells from an overnight culture were transferred to fresh YPD medium containing curcumin compounds and allowed to grow at 37°C for 2 h. A drug-free control was included for each experiment. For heat shock experiments, the cells were then transferred to 42°C. 100 µL aliquots of the control and stress samples were removed at 0 and 20 min. The cells were then diluted and spread on YPD agar plates. For osmotic stress response, the cells were diluted and spread on YPD plates with or without 1 M NaCl. The plates were then incubated at 37°C for 24 h to observe cell growth. Each experiment was repeated three times.
The Nile red efflux assay was performed according to a previous protocol (Ivnitski-Steele et al., 2009; Keniya et al., 2015; Eldesouky et al., 2018). To determine the relationship between efflux pump activity and curcumin without depleting the HSP90 gene, curcumin and Nile red were supplemented simultaneously. Briefly, exponential phase wildtype strain was harvested, washed twice with PBS, and resuspended in PBS containing 2% glucose. The cells were then incubated with 125 µg/mL of curcumin and 7 µM of Nile red for 10 min. To determine the relationship between efflux pump activity and depletion of the HSP90 gene by curcumin, exponential phase wildtype or PADH1-HSP90 strain were incubated with 125 µg/mL of curcumin for 2 h. Then the cells were washed with PBS, resuspended in PBS containing 2% glucose, and incubated with 7 µM Nile red for 10 min at 37°C. For the tet-HSP90/hsp90Δ mutant, cells were incubated with 0.1 µg/mL of Dox for 2 h before treatment with Nile red. For flow cytometry analysis, the accumulation of Nile red was measured using an EC800 (Sony, Japan) flow cytometer with an excitation wavelength of 488 nm and emission filter of 585/42 nm. At least 10,000 events were analyzed in each experiment.
To determine the in vitro susceptibility of C. albicans SC5314 to curcumin and GO-Y030, broth microdilution assays were performed in YPD medium. Cell growth was determined by absorbance at 530 nm after a 24-h drug treatment. As the concentration of curcumin compounds increased, the cell growth decreased (Figure 1). We found that both curcumin compounds inhibited the growth of C. albicans dose-dependently. The data also revealed that C. albicans was more susceptible to curcumin than to GO-Y030. At a concentration of 250 µg/mL, curcumin inhibited about 80% of cell growth compared to the control (MIC80 = 250 µg/mL). In contrast, GO-Y030 inhibited only about 60% of cell growth.
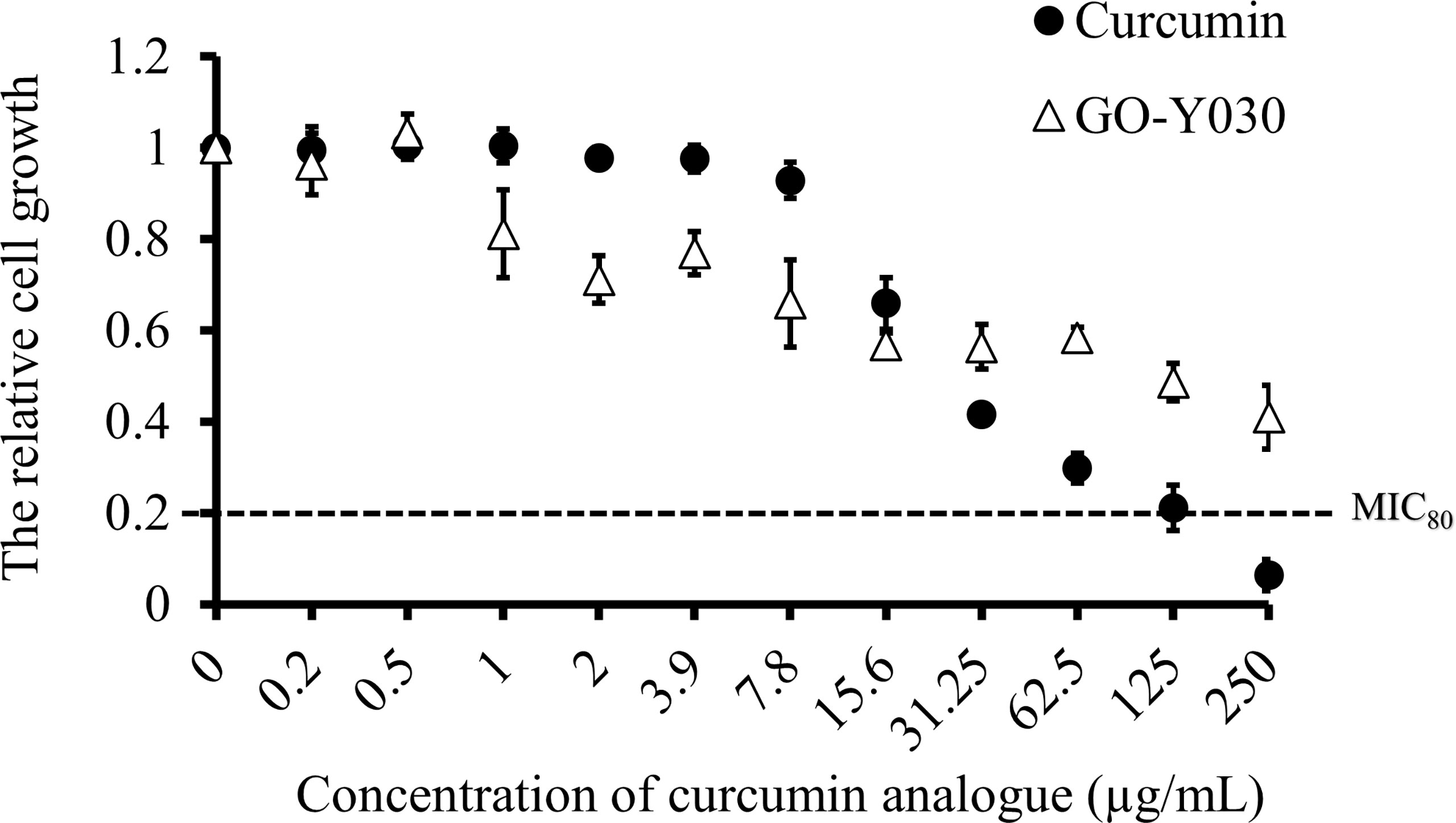
Figure 1 Curcumin compounds exhibit antifungal activity on C. albicans. Susceptibility assays of each curcumin analogue were conducted in YPD medium. Growth was measured by absorbance at 530 nm after 24-h incubation at 37°C. Optical densities were averaged for triplicate measurements and normalized relative to no antifungal compound control. Error bars represent standard deviation from the mean of triplicate measurements.
To investigate the effect of curcumin compounds on the gene expression of HSP90, we determined the transcript levels of HSP90 after mid-log phase C. albicans was treated with these compounds for 2 h in YPD at a sub-MIC80 concentration (125 µg/mL). Figure 2 shows the relative transcript levels of HSP90 in C. albicans SC5314 after treatment with the two curcumin compounds. HSP90 levels in cells with either compound was significantly lower than those in untreated cells. This suggests that both curcumin compounds downregulated HSP90 expression. Our results also showed that curcumin strongly reduced the transcript levels of HSP90 compared to GO-Y030. This might explain why curcumin exhibited greater antifungal activity than GO-Y030. These results suggest that the effect of curcumin might be attributable to the inhibition of HSP90 expression.
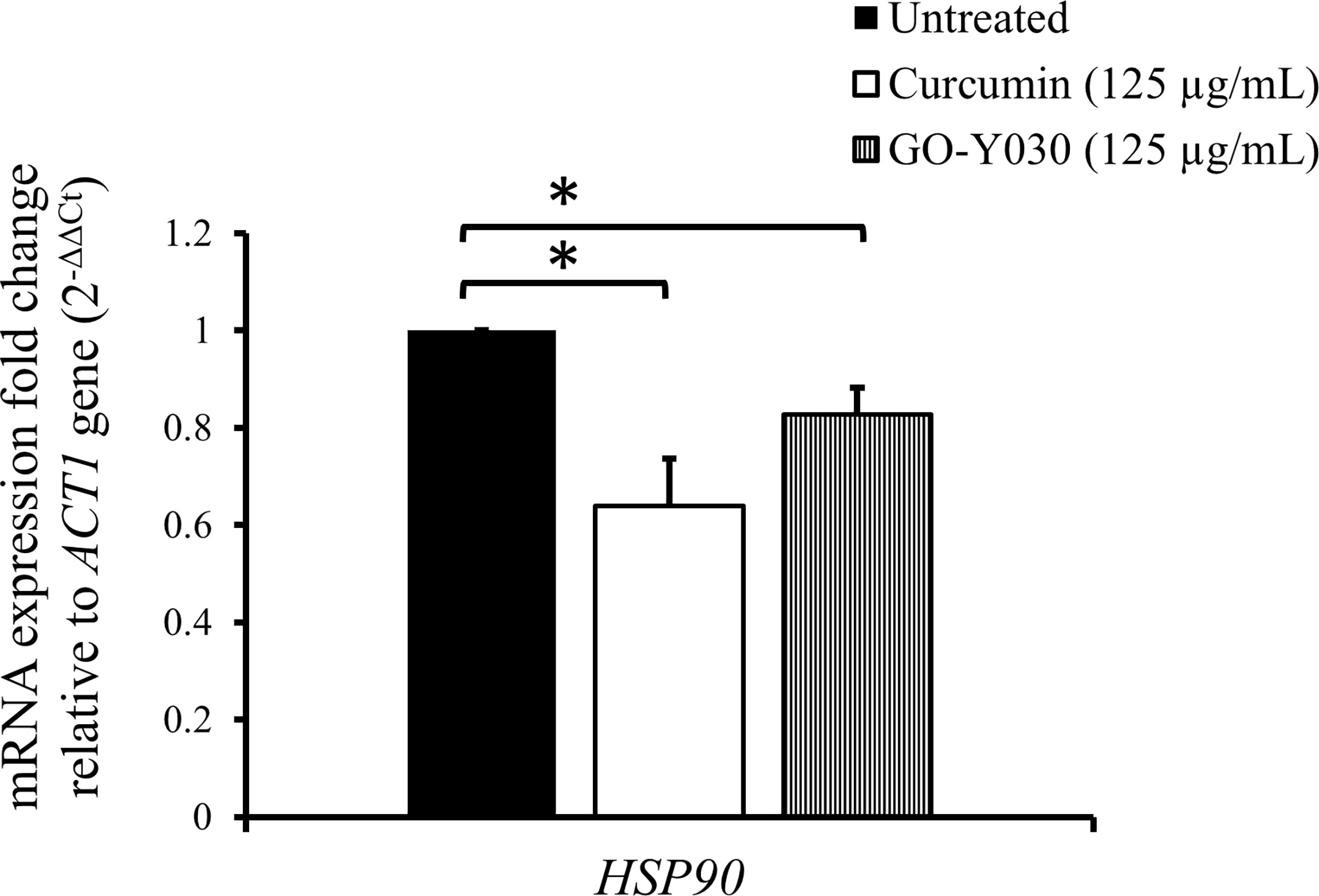
Figure 2 Effect of curcumin compounds on HSP90 gene expression in C. albicans SC5314. Error bars represent standard deviation from the mean of duplicate measurements. * indicates a statistically significant difference (p<0.05) compared to the respective control group.
To determine whether the HSP90 gene is involved in the function of curcumin, we constructed a conditional HSP90 mutant and an HSP90-overexpressing strain of C. albicans. To construct a Dox-repressible allele of HSP90, by using the HSP90/hsp90Δ strain derived from SC5314, we replaced the C. albicans HSP90 promoter on the other allele with a tetO element to obtain the tetO-HSP90/hsp90Δ strain. This strain reduces HSP90 expression in a Dox-dependent manner (Figure 3A). In addition, an HSP90-overexpressing strain, PADH1-HSP90, was produced by replacing the promoter of HSP90 with the ADH1 promoter as shown in Figure 3B.
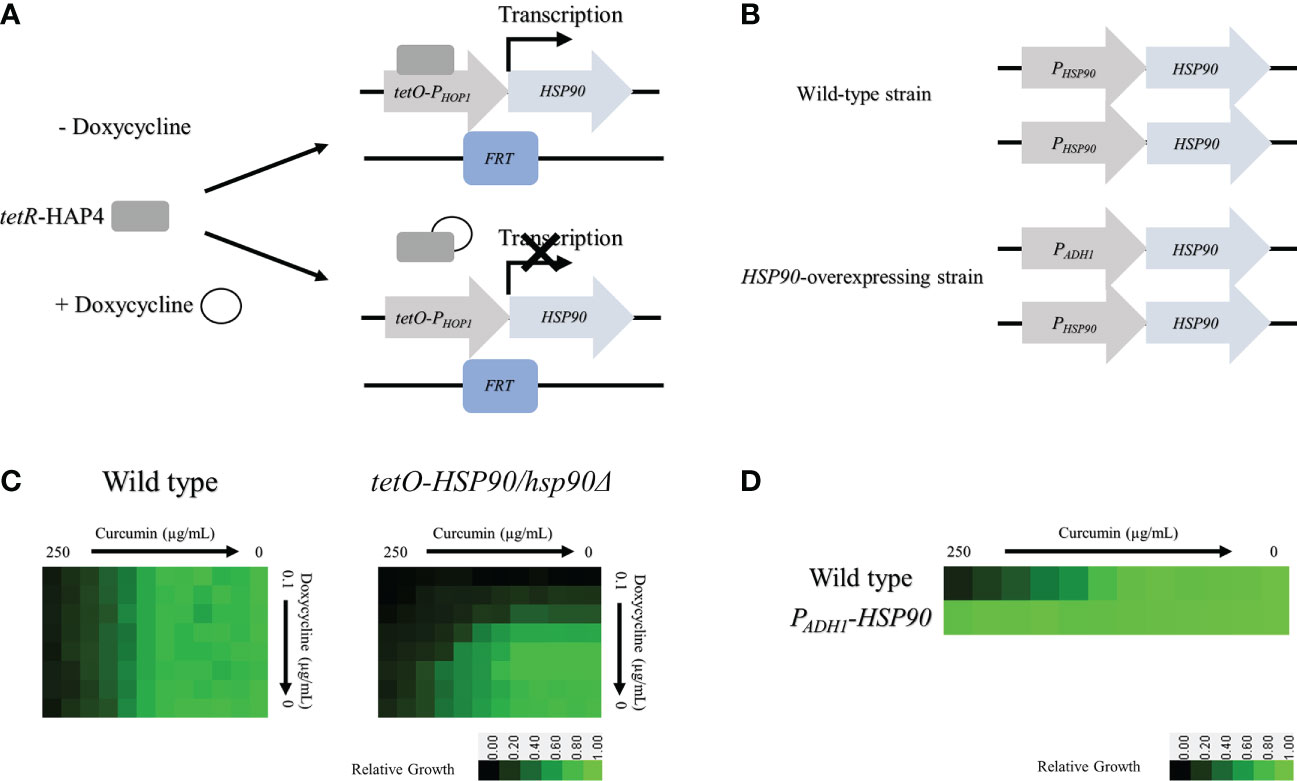
Figure 3 Depletion of HSP90 increases the sensitivity of cells to curcumin. (A) Diagram showing control of the tetR-regulated tetO-HSP90 fusion gene. In the absence of Dox, tetR can bind to and induce HSP90 expression. (B) Checkboard assays for curcumin in combination with Dox tested against C albicans in YPD medium. The optical density of cells at 530 nm (OD530) is colored between dark green (no yeast growth) and light green (maximum growth) relative to the control. Data are quantitatively displayed with color using Java TreeView 1.2.0. (C) Diagram showing the HSP90 promoter replaced with the ADH1 promoter. (D) Diagram of the microbroth dilution assay for curcumin against the overexpression mutant PADH1-HSP90 in YPD medium. The cells were treated with curcumin at 37°C for 24 h.
The levels of curcumin sensitivity of the wildtype, tetO-HSP90/hsp90Δ, and PADH1-HSP90 strains were compared. For the wildtype, 250 µg/mL of curcumin (MIC80) inhibited cell growth completely (Figure 3C). This data also revealed that Dox did not affect the curcumin sensitivity of the wild type. For the tetO-HSP90/hsp90Δ strain, the MIC80 of curcumin was decreased under a high concentration of Dox. At 0.025 µg/mL and 0.05 µg/mL of Dox, the MIC80 of curcumin were 15.6 µg/mL and 7.8 µg/mL, respectively. This result showed that the depletion of HSP90 enhanced sensitivity to curcumin. Hence, the HSP90 gene might play an important role in resistance to curcumin in C. albicans. Moreover, microbroth dilution assays using the HSP90 overexpression strain (PADH1-HSP90) were performed. Figure 3D shows the growth of SC5314 and PADH1-HSP90 after treatment with curcumin. Although growth of the wildtype strain was decreased at a high concentration of curcumin, this compound had no effect on the growth of PADH1-HSP90 cells. This result indicated that overexpression of the HSP90 gene suppressed sensitivity to curcumin. Overall, these results suggest that HSP90 is the key factor in the growth inhibition of C. albicans by curcumin.
We then investigated the effect of curcumin on HOG1 expression as well as that of HSP90. Figure 4A shows that curcumin reduced the transcript levels of both HOG1 and HSP90. This reduction was restored by the overexpression of HSP90 in the presence of curcumin (Figure 4B). To confirm the relationship between the HSP90 and HOG1, the mRNA levels of tetO-HSP90/Δhsp90 strain were also analyzed. When HSP90 expression was suppressed by Dox, the gene expression of HOG1 was decreased (Figure 4C), confirming that the reduction of HSP90 transcripts led to the reduction of HOG1 gene expression in C. albicans. To further validate the stress responses of the wild type, PADH1-HSP90, and tetO-HSP90/hsp90Δ, the growth of these strains under thermal and osmotic stresses were analyzed. Stress response tests were performed with heat shock at 42°C for 30 min and with 1 M NaCl. Curcumin reduced the thermotolerance of the wildtype strain after it was exposed to 42°C for 30 min compared to the control (37°C) (Figure 4D). In contrast to the wild type, HSP90 overexpression in C. albicans resulted in the growth of cells at 42°C (Figure 4E). In addition, the repression of HSP90 led to a reduction in the tolerance of cells at high temperature (Figure 4F). Similarly, curcumin also reduced the osmotic tolerance of C. albicans to exposure to 1 M NaCl (Figure 4D). Contrastingly, the overexpression of HSP90 in C. albicans retained the tolerance when cells were treated with curcumin (Figure 4E). Repression of HSP90 decreased the osmotic tolerance to a high concentration of NaCl (Figure 4F). These results implied that the downregulation of HSP90 by curcumin impaired the thermal and osmotic stress responses of C. albicans.
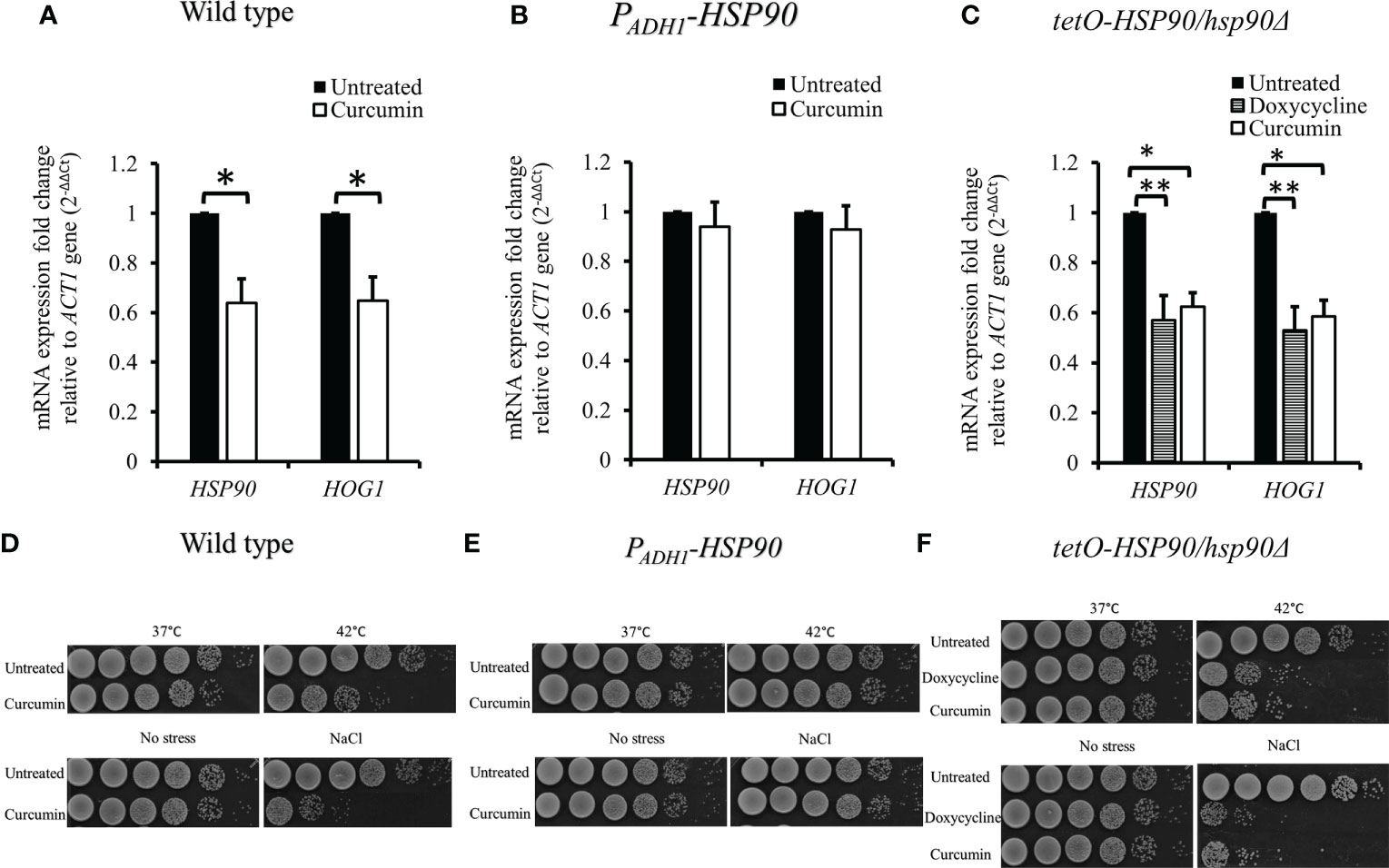
Figure 4 Reduction of HSP90 gene expression impaired the stress response of C. albicans. (A–C) Effect of curcumin on HSP90 and HOG1 gene expression in the wildtype, tetO-HSP90/hsp90Δ, and PADH1-HSP90 strains after treatment with curcumin. Error bars represent standard deviation from the mean of duplicate measurements. * and ** indicate statistically significant differences (p<0.05 and p<0.01) compared to the respective control group. (D–F) For the short-term heat shock stress response, YPD-overnight cultures of the wild type and PADH1-HSP90 mutant, with or without pre-exposure to curcumin, as well as YPD-overnight cultures of the tetO-HSP90/hsp90Δ mutant, with or without pre-exposure to Dox, were serially diluted from 106 to 101 cells (left to right) after exposure to heat shock (42°C for 30 min) or not (control), plated on YPD, and incubated for 24 h at 37°C. For the osmotic stress response, YPD-overnight cultures of the wild type and PADH1-HSP90 mutant, with or without pre-exposure to curcumin, as well as YPD-overnight cultures of the tetO-HSP90/hsp90Δ mutant, with or without pre-exposure to Dox, were serially diluted from 106 to 101 cells (left to right) on YPD plates with or without1 M NaCl. The plates were incubated for 24 h at 37°C.
The mRNA levels of transcription factors HSF1 and AHR1 were analyzed in the presence of curcumin. Unexpectedly, curcumin initiated the transcriptional induction of HSF1, while it did not affect the mRNA of AHR1 (Figure 5A). This suggests that the reduction of the HSP90 mRNA level by curcumin was not due to the inhibition of HSF1 transcriptional expression. We confirmed this using the tetO-HSP90/Δhsp90 mutant. In the presence of the Dox, which suppressed the gene expression of HSP90, the HSF1 transcript level was increased (Figure 5B). Therefore, HSP90 reduction by curcumin was not a result of, but rather led to, the increase of HSF1 mRNA in C. albicans.
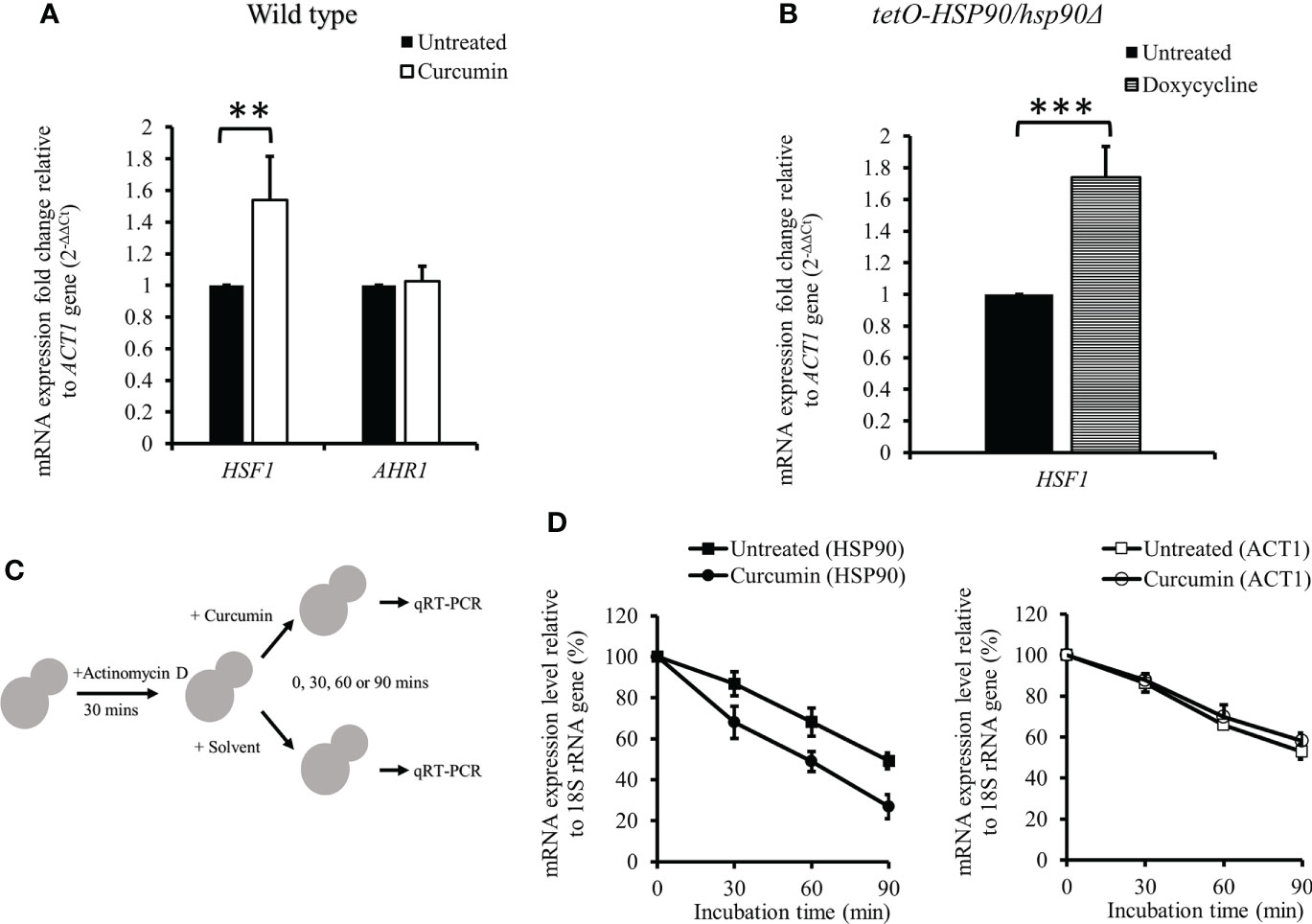
Figure 5 Effect of curcumin on the gene expression of HSF1 and AHR1 and the post-transcriptional level of HSP90. (A, B) Effect of curcumin on HSF1 and AHR1 gene expression in the wildtype and tetO-HSP90/hsp90Δ strains after treatment with curcumin. Error bars represent standard deviation from the mean of duplicate measurements. ** and *** indicate a statistically significant difference (p<0.01 and p<0.001) compared to the respective control group. (C) Diagram showing the post-transcriptional regulation test. Cells were pre-treated with 10 µg/mL of actinomycin D for 30 min, then incubated with curcumin at sub-MIC80 for 0, 30, 60, or 90 min. (D) Effect of curcumin on HSP90 and ACT1 gene expression for the wildtype strain after treatment with actinomycin D and curcumin. Error bars represent standard deviation from the mean of triplicate measurements.
We initially thought that Dox-controlled HSP90 in the tetO-HSP90/Δhsp90 strain would not be affected by curcumin. However, our results showed that curcumin downregulated HSP90 in tetO-HSP90/Δhsp90 as well as in the wild type. We speculated that this might be due to the post-transcriptional regulation of HSP90 by curcumin. To test this, the mRNA level of HSP90 was measured after cells were treated with actinomycin D (ActD), an RNA polymerase inhibitor, as shown in Figure 5C. The cells were then treated with curcumin for 0, 30, 60, or 90 min. After treatment with ActD, HSP90 mRNA of the cells degraded gradually, and in addition, it degraded faster in the presence of curcumin than in the control (Figure 5D). In contrast, the degradation of ACT1 mRNA was not affected by the addition of curcumin (Figure 5D). This showed that curcumin accelerated the degradation of HSP90 mRNA specifically and suggests that curcumin induces the post-transcriptional degradation of HSP90.
Since curcumin downregulated the transcriptional level of HSP90, we expected that curcumin might influence the gene expression of CDR1. Therefore, we investigated CDR1 and CDR2 (another ABC transporter gene) expression in the presence of curcumin or geldanamycin, which is a known Hsp90 inhibitor. Figure 6A shows that CDR1 gene expression was reduced significantly by curcumin and geldanamycin, but that of CDR2 was not. A reduction of the CDRs by curcumin and geldanamycin did not occur in the PADH1-HSP90 strain (Figure 6B). To further confirm that the CDR1 reduction was due to a reduction of HSP90 by curcumin and geldanamycin, the effect of depleting HSP90 on CDR1 was determined using the tetO-HSP90/Δhsp90 strain (Figure 6C). In the presence of Dox, CDR1 transcripts decreased while those of CDR2 remained unchanged. These results suggest that curcumin only downregulated CDR1 expression via a reduction of HSP90 in C. albicans.
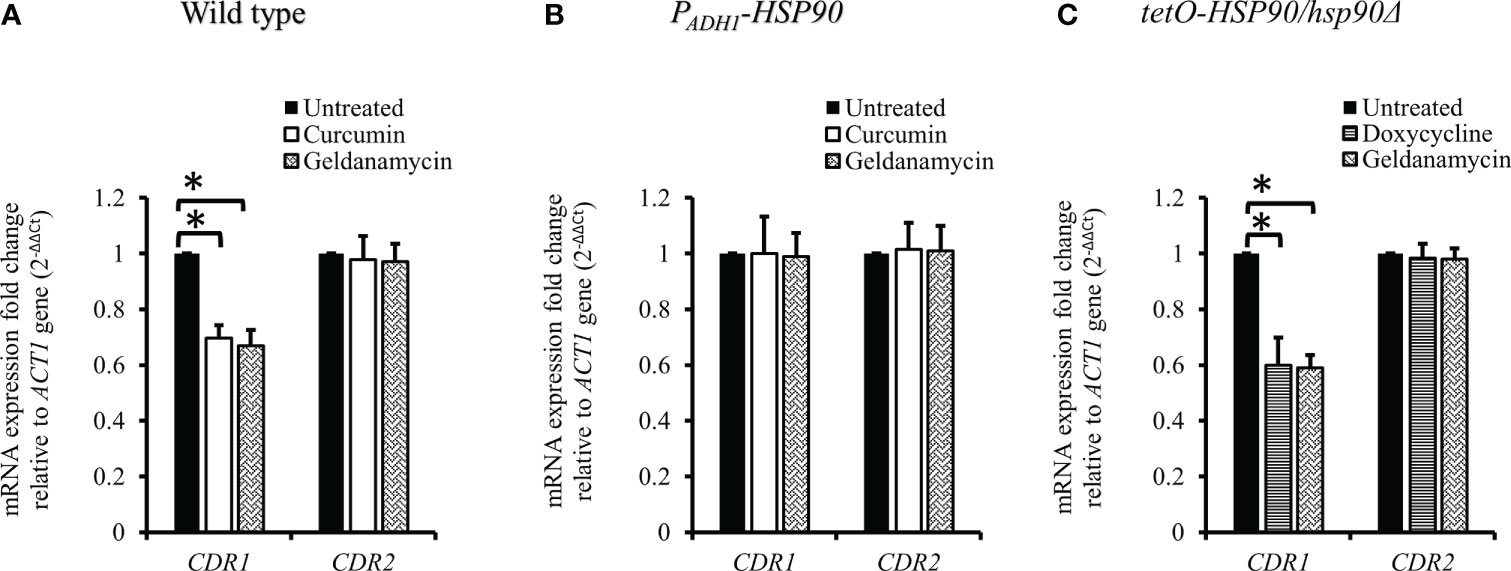
Figure 6 Transcriptional levels of CDR1 and CDR2 in the presence of curcumin and geldanamycin. Effects of curcumin on CDR1 and CDR2 gene expression for wildtype (A), PADH1-HSP90 (B), and tetO-HSP90/hsp90Δ strains (C) after treatment with curcumin or geldanamycin. Error bars represent standard deviation from the mean of duplicate measurements. * indicates a statistically significant difference (p<0.05) compared to the respective control group.
As the gene expression of CDR1 in C. albicans was reduced by curcumin, we assumed that the efflux pump activity of the cells might be affected by curcumin. To test this, the Nile red accumulation assay was performed (Figure 7A). For the wildtype strain, the addition of curcumin led to a high accumulation of Nile red compared to the control (Figures 7B, C). This accumulation was thought to be due to the repression of the CDR1 gene by the reduction of HSP90 (Figure 8). The relationship between the expression of HSP90 and the export of Nile red was confirmed by using the tetO-HSP90/hsp90Δ strain (Figures 7D, E). The overexpression of HSP90 reversed the accumulation of Nile red by curcumin (Figures 7F, G), suggesting that overexpression of the HSP90 gene maintained CDR1 expression and restored Nile red extrusion. These results implied that curcumin repressed HSP90 mRNA resulted in repression of CDR1 which caused the disruption of efflux pump activity.
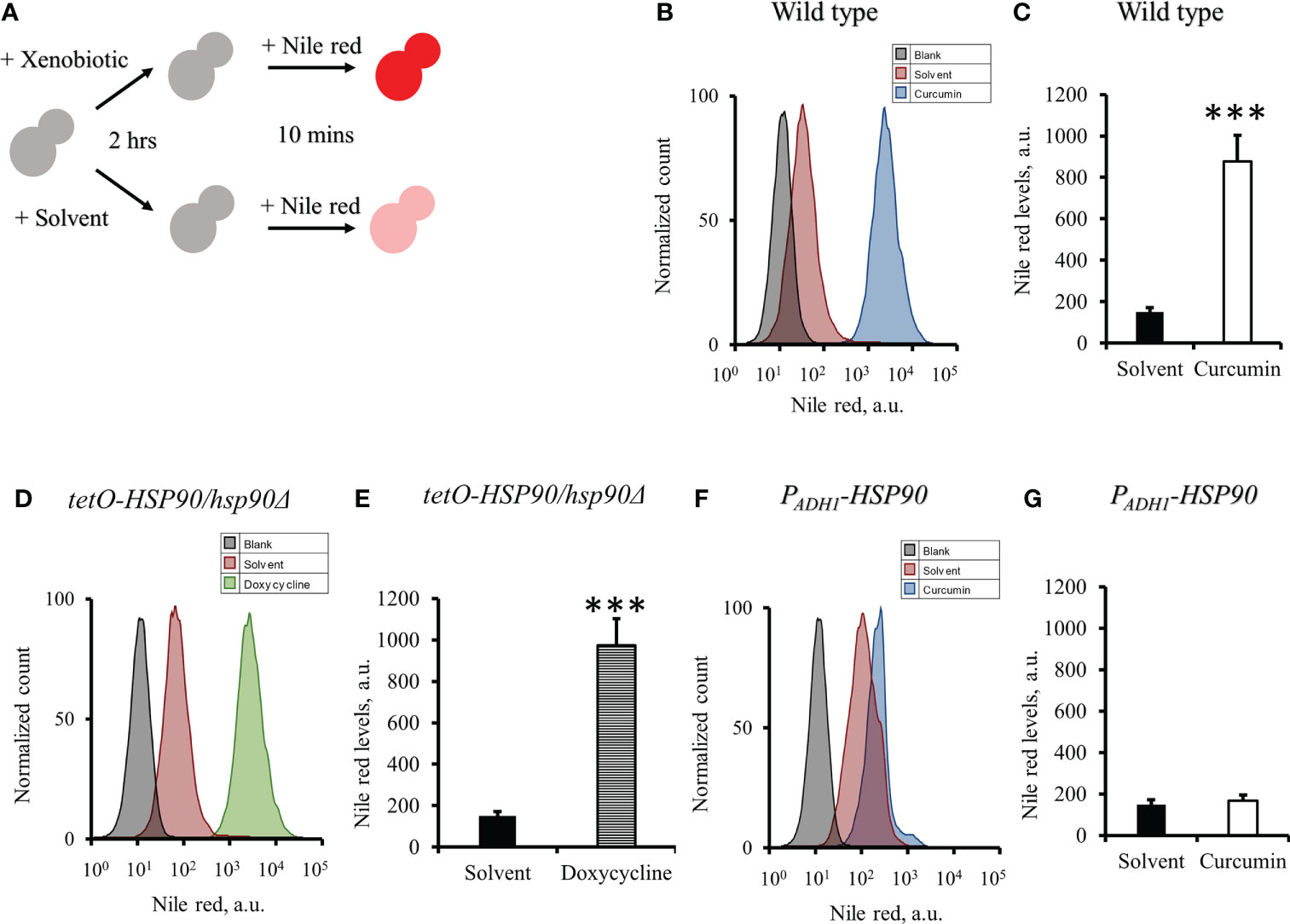
Figure 7 Effect of curcumin on the efflux pump activity of C. albicans. (A) Diagram showing the Nile red accumulation assay to determine the disruption of efflux pump activity by curcumin. The amounts of Nile red accumulated in C. albicans after treatment with curcumin for 2 h were determined for SC5314 (B, C) and the PADH1-HSP90 mutant (F, G), and that in the tetO-HSP90/hsp90Δ mutant was determined after treatment with Dox for 2 h (D, E) Flow cytometry histograms for mean Nile red fluorescence are shown in C, E, G. Error bars represent standard deviation from the mean of duplicate measurements. *** indicates a statistically significant difference (p<0.001) compared to the respective control group.
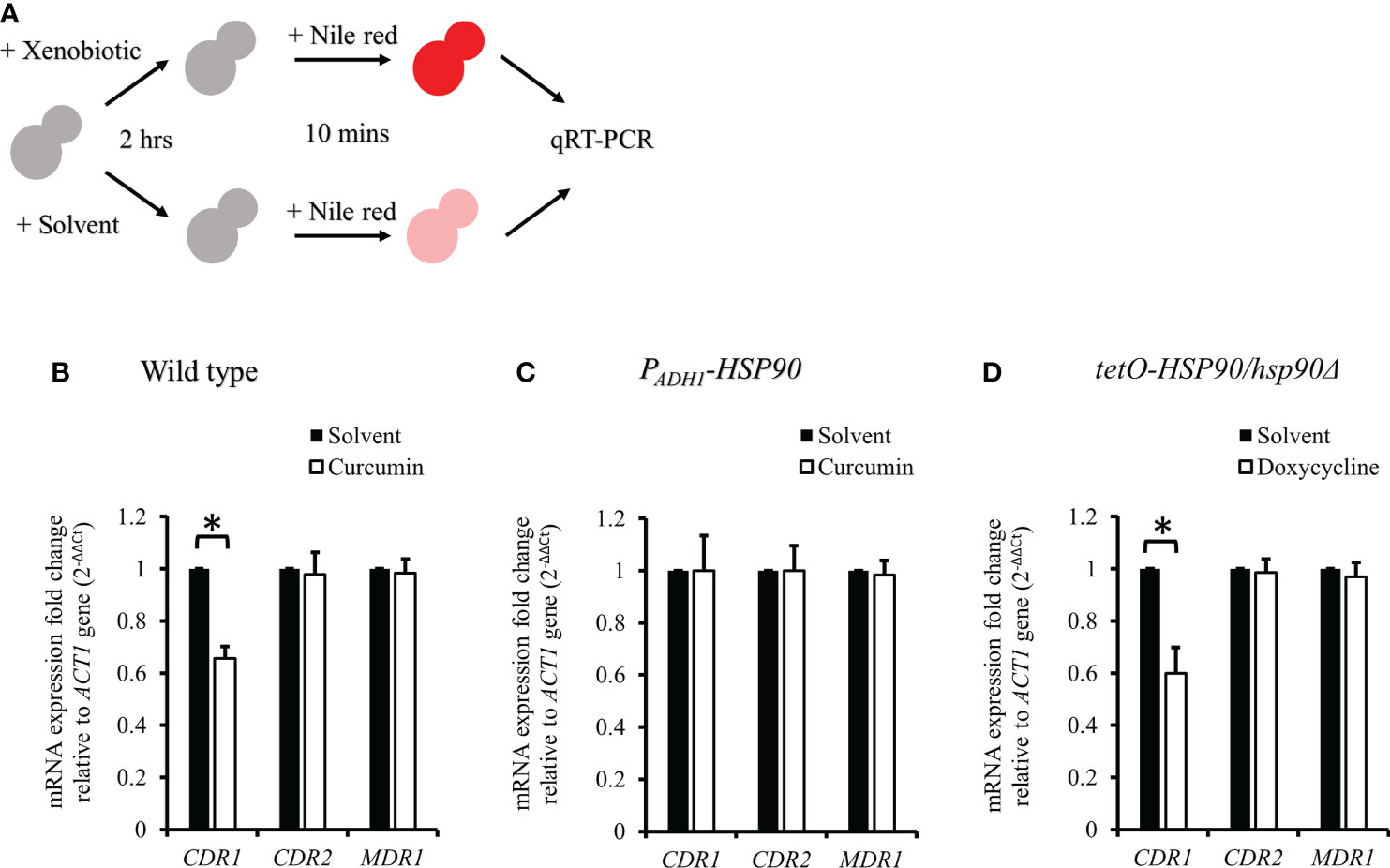
Figure 8 CDR1, CDR2, and MDR1 gene expressions in C. albicans after treatment with xenobiotics for 2 h and Nile red for 10 min. (A) Diagram showing the qRT-PCR experiment. (B–D) mRNA amounts of CDR1, CDR2, and MDR1 were determined by qRT-PCR in wildtype, PADH1-HSP90, and tetO-HSP90/hsp90Δ strains. * indicates a statistically significant difference (p<0.05) compared to the respective control group.
To test the effect of curcumin as an efflux pump inhibitor, cells were exposed to curcumin and Nile red simultaneously for 10 min (Figure 9A). Based on the flow cytometry results, the addition of curcumin led to the accumulation of Nile red compared to the control (Figures 9B, C). However, Nile red did not accumulate in the tetO-HSP90/hsp90Δ strain with Dox (Figures 9D, E). Since the 10-min pretreatment with curcumin would not alter the gene expression of HSP90 and CDR1, Cdr1 was not decreased (Figure 10). These results suggest that, without affecting gene expression of ABC-transporters, curcumin also acts on Cdr1 directly to inhibit its function but Dox does not.
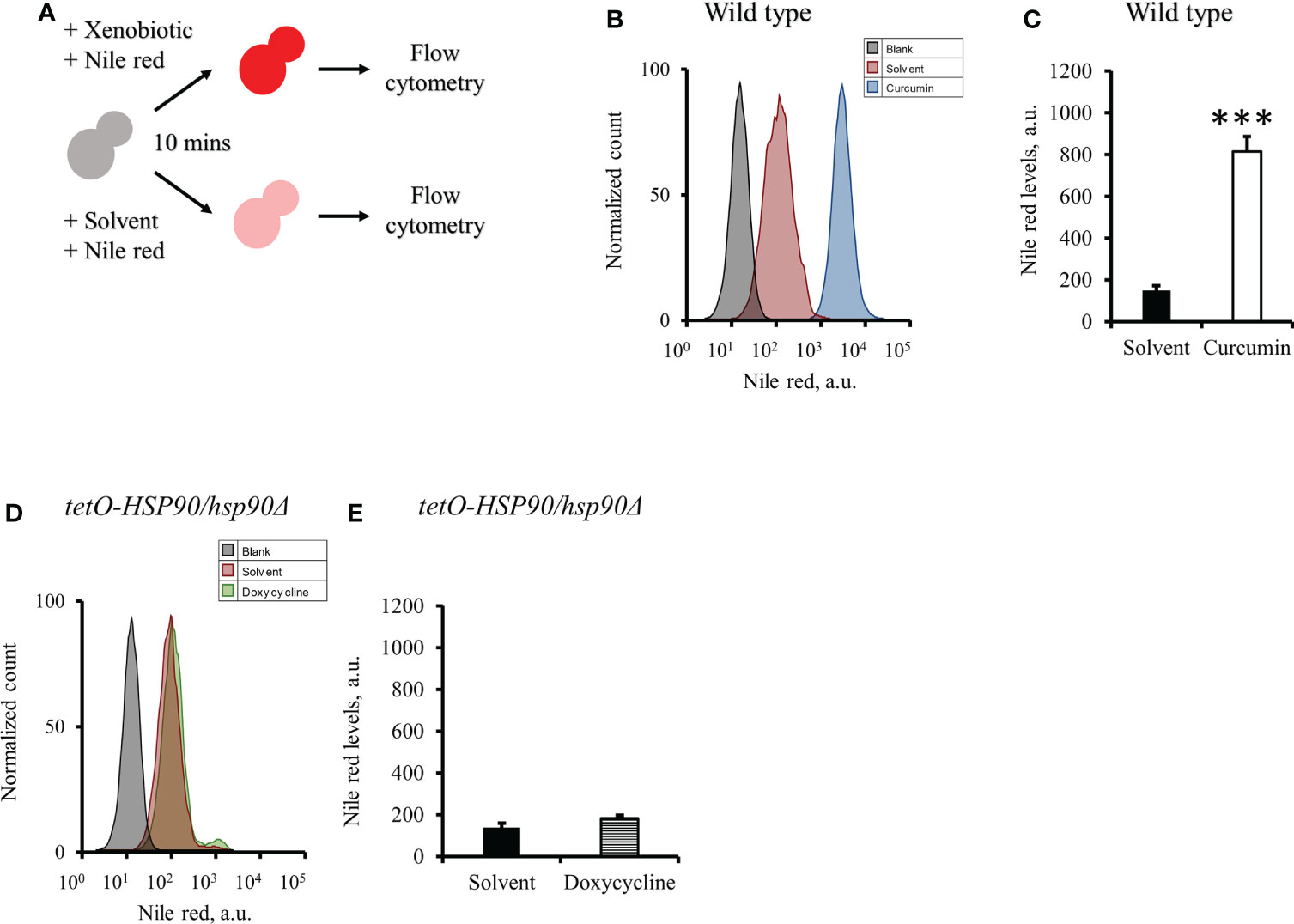
Figure 9 Effect of curcumin on the efflux pump activity of C. albicans. (A) Diagram showing the Nile red accumulation assay for determining the ability of curcumin to inhibit the efflux pump. (B) The amount of the Nile red accumulated in C. albicans SC5314 was determined after it was treated with curcumin for 10 minutes. (C) The flow cytometry histograms for the mean Nile red fluorescence. (D) The amount of the Nile red accumulated in tetO-HSP90/hsp90Δ was determined after it was treated with doxycycline for 10 minutes. (E) The flow cytometry histograms for the mean Nile red fluorescence. Error bars represented standard deviation from the mean of duplicate measurements. *** indicates a statistically significant difference (p<0.001) compared to the respective control group.
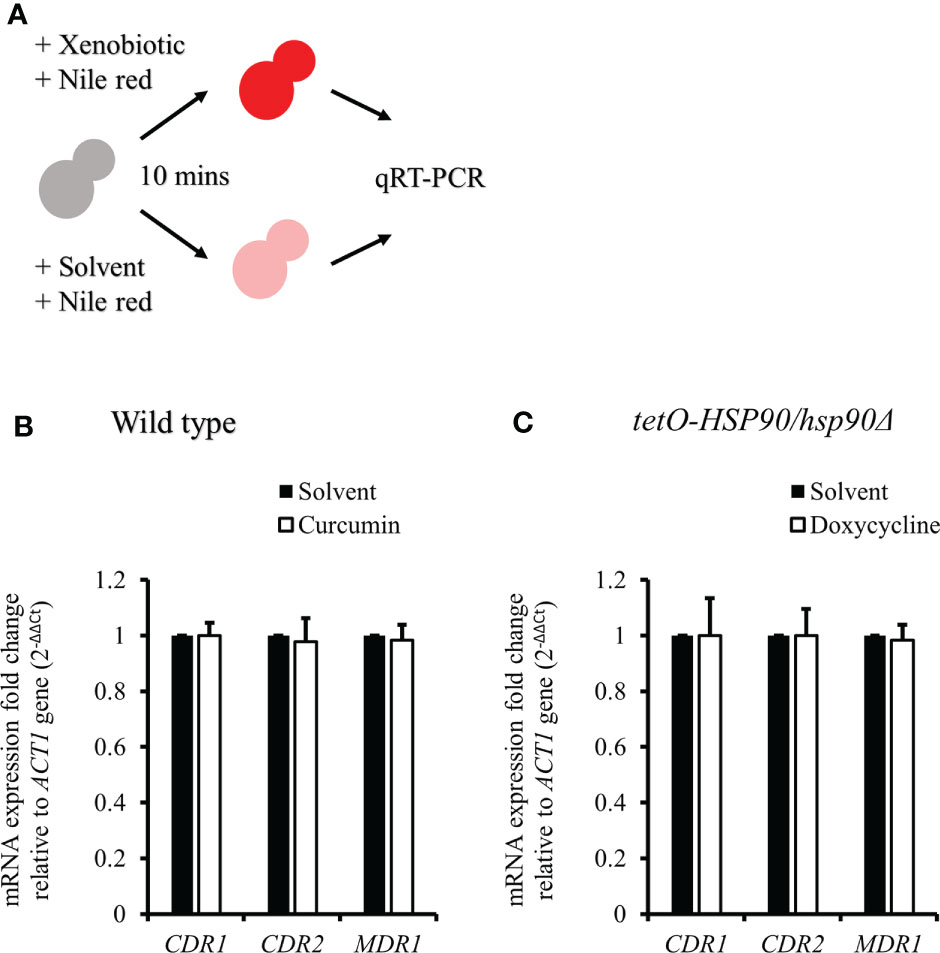
Figure 10 CDR1, CDR2, and MDR1 gene expressions in C. albicans after treatment with xenobiotics and Nile red for 10 min. (A) Diagram showing the qRT-PCR experiment. (B, C) The mRNA amounts of CDR1, CDR2, and MDR1 were determined by qRT-PCR in wildtype and tetO-HSP90/hsp90Δ strains.
Curcumin, a curcuminoid produced from the rhizomes of Curcuma longa (a tiny perennial herb native to India), has been reported to possess anti-inflammatory, anticarcinogenic, and anti-infectious activities (Ravindran et al., 2009; Neelofar et al., 2011). Several shortcomings of curcumin, such as its low bioavailability and poor stability, have been highlighted as major problems in its therapeutic application (Blasius et al., 2004; Anand et al., 2007; Fang et al., 2013). Therefore, many studies have tried to improve the bioavailability and stability of curcumin by structural modification. GO-Y030 reportedly has greater anti-carcinogenic activity and lower toxicity than curcumin (Gritsko et al., 2006; Cen et al., 2009; Hutzen et al., 2009; Kudo et al., 2011; Mohan Yallapu et al., 2012). As the antifungal activity of GO-Y030 has remained unclear, we assessed its inhibitory effect on C. albicans growth. In this study, broth microdilution assays demonstrated that curcumin and GO-Y030 had antifungal inhibitory activity against the growth of C. albicans SC5314 (Figure 1). This result is consistent with previous studies (Andrade et al., 2019; Narayanan et al., 2020) wherein curcumin inhibited the growth of Candida strains in a range between 100 µg/mL and 250 µg/mL. Although GO-Y030 had lower antifungal activity than curcumin in this study, to our knowledge, this is the first report showing that GO-Y030 inhibits the growth of C. albicans.
Despite the numerous cytotoxic effects of curcumin on C. albicans that have been already reported (Sharma et al., 2009; Sharma et al., 2010a; Sharma et al., 2010b; Neelofar et al., 2011; Shahzad et al., 2014; Alalwan et al., 2017; Andrade et al., 2019; Hamzah et al., 2020), the mechanism of this function of curcumin remains unknown. In cancer therapeutics for humans, curcumin has also been reported to be an antitumor compound. This compound influences the HSP90 gene and its gene product in human tumor cells (Zhang et al., 2007; Giommarelli et al., 2010; Anand et al., 2012; Khan et al., 2012; Li et al., 2012; Liu et al., 2014; Bhullar et al., 2015; Lv et al., 2015; Zheng et al., 2016; Ye et al., 2017; Fan et al., 2018; Forouzanfar et al., 2019). Recent studies have shown that curcumin inhibits ATPase activity in Hsp90 of human cancer cells. The expression of HSP90 is higher in tumors compared with normal tissues and is important in the maintenance of the stability, integrity, and function of oncogenic proteins. Curcumin and several curcumin derivatives such as C1206, C0818, and CUR3d, have been shown to inhibit Hsp90 function. This results in the dissociation of complexes with client proteins that are important in cell proliferation, cytotoxic damage survivability, and apoptosis, among other functions (Jung et al., 2007; Giommarelli et al., 2010; Lee and Chung, 2010; Bhullar et al., 2015; Fan et al., 2017; Fan et al., 2018). In addition, curcumin has also been found to downregulate HSP90 gene expression in human cells such as chronic myeloid leukemia cells and human embryonic lung fibroblast cells (Zhang et al., 2007; Lv et al., 2015; Zheng et al., 2016; Ye et al., 2017; Sang et al., 2018). C. albicans Hsp90 has also been studied as a heat shock protein that is essential for maintaining homeostasis by promoting the proper folding of abundant client proteins. According to numerous studies, Hsp90 is involved in thermal stability, morphogenesis, cell cycle control, apoptosis, and drug resistance in C. albicans (Leach et al., 2012b; O’Meara and Cowen, 2014). Hence, interfering with the physiological activity of Hsp90 could be a promising strategy for treating candidiasis. However, there have been no reports about the effects of curcumin on Hsp90 in this pathogenic fungus. Hsp90 is common among many species and has a conserved amino acid sequence between C. albicans and humans (Swoboda et al., 1995), so we expected curcumin to affect Hsp90 in C. albicans as well. This study showed that the exposure of C. albicans to curcumin or GO-Y030 triggered the transcriptional reduction of HSP90 (Figure 2). This is an important finding in understanding the function of curcumin in C. albicans. Unexpectedly, curcumin exhibited greater antifungal activity than GO-Y030 on C. albicans, which is the opposite of findings in human tumor cells.
To alter HSP90 expression in C. albicans, we utilized a Dox-mediated HSP90 strain and an HSP90-overexpressing strain to investigate the effects of curcumin on C. albicans. Our data revealed that the depletion of HSP90 in the tetO-HSP90/hsp90Δ strain increased susceptibility to curcumin dose-dependently with Dox (Figure 3C), and a synergic effect of curcumin and Dox appeared. In contrast, the effect of curcumin disappeared by the overexpression of HSP90 in the PADH1-HSP90 strain (Figure 3D). These findings indicated that curcumin inhibits the growth of C. albicans by repressing HSP90 function.
Previous reports (Diezmann et al., 2012; Leach et al., 2012a; Leach et al., 2012b; O’Meara and Cowen, 2014) have shown that HSP90 plays an important role in the survivability of yeast species at high temperatures and high osmotic pressures. The depletion of HSP90 reduced the thermotolerance of C. albicans and led to the decrease of a mitogen-activated protein kinase, Hog1, which plays an important role in the osmotic stress response. This study showed that the depletion of HSP90 by curcumin resulted in a reduction of HOG1 (Figure 4), suggesting that curcumin also decreased HOG1 expression and impaired the stress response of C. albicans.
In C. albicans, protein kinase CK2 and transcription factor Ahr1 operate upstream of Hsp90 to promote cell growth in many environments. In addition, HSP90 expression is also controlled by the transcription factor Hsf1, whose activation is repressed by Hsp90. The depletion of Hsp90 induces Hsf1 phosphorylation and upregulates Hsf1 targets, and depletion of HSP90 activates HSF1 (Zou et al., 1998; Diezmann et al., 2012; Leach et al., 2012a; Leach et al., 2016; Kijima et al., 2018; Veri et al., 2018). Our results showed that curcumin induced the transcription of HSF1 in the wildtype strain, the same as in the Dox-mediated tetO-HSP90/hsp90Δ strain (Figures 5A,B). These experiments suggested that curcumin reduces HSP90 expression directly, and not dependently on Ahr1 and Hsf1. Interestingly, we thought that HSP90 expression in the tetO-HSP90/hsp90Δ strain would not be influenced by curcumin, because the HSP90 promoter was replaced by the tetO element. However, curcumin reduced HSP90 mRNA in the tetO-HSP90/Δhsp90 strain (Figure 4C). Hence, we assumed that the induction of HSP90 mRNA degradation occurred in the presence of curcumin. After actinomycin D inhibited transcription in the cells, the HSP90 mRNA amount was measured in the presence or absence of curcumin (Figure 5C). Although HSP90 mRNA was degraded gradually after inhibition of mRNA synthesis, curcumin accelerated its degradation (Figure 5D). In contrast, faster degradation of ACT1 mRNA was not observed when adding curcumin (Figure 5D). Curcumin has been reported to change DNA methylation in human cancer cells as an epigenetic modification (Link et al., 2013). This indicates that curcumin might have inhibited HSP90 expression at the post-transcriptional level by DNA methylation changes.
ABC transporters, including Cdr1 and Cdr2, are drug efflux pumps that play an important role in the development of multidrug resistance in C. albicans. Previous studies have shown that curcumin inhibits ABC transporters, including C. albicans Cdr1, Cdr2, and Saccharomyces cerevisiae Pdr5p, competitively (Sharma et al., 2009; Sharma and Prasad, 2011). In addition, recent studies (Diezmann et al., 2012; Leach et al., 2012a) have shown that a reduction of HSP90 reduces the protein level of Cdr1 in C. albicans. In this study, curcumin downregulated CDR1 gene expression, suggesting that the reduction of HSP90 expression by curcumin led to a decrease in CDR1 expression.
Previous study showed that curcumin was able to modulate multidrug resistance (MDR) phenotype of C. albicans (Garcia-Gomes et al., 2012). In this study, the Nile red accumulation assay was used to analyze the efflux pump activity in C. albicans. Nile red is a known substrate of ABC-transporters Cdr1, Cdr2 and Mdr1 in C. albicans (Ivnitski-Steele et al., 2009), the cells can efflux Nile red immediately. Our data showed that curcumin drastically decreased the efflux pump activity of the wildtype strain after a 2-h incubation with curcumin (Figures 7B, C). This was due to the reduction of CDR1 expression by curcumin. The depletion of HSP90 in the tetO-HSP90/Δhsp90 strain also led to a decrease in efflux pump activity, while HSP90 overexpression maintained the efflux pump activity of the PADH1-HSP90 strain in the presence of curcumin (Figures 7D-G). Curcumin is also known to be an inhibitor of Cdr1 activity (Pearson et al., 1999; Falagas et al., 2006; Piddock, 2006; Sharom, 2008; Wu et al., 2011). In this study, without affecting gene expression of ABC-transporters, curcumin also blocked efflux pump activity for Nile red in the wild type (Figures 9, 10). This finding showed that curcumin inhibited the activity of efflux pumps such as Cdr1, while low efflux pump activity remained because Cdr2 and Mdr1 were not inhibited by curcumin.
Taken together, this study sheds new light on the functions of curcumin (Figure 11). Curcumin affects not only drug efflux pumps such as Cdr1 but also HSP90 expression, mainly at the post-transcriptional level. Hence, the natural product curcumin and its derivatives may be used as antifungals to inhibit drug efflux pumps and cell growth of C. albicans. However, the complex mechanism by which curcumin affects C. albicans needs to be further explored.
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding author.
YL and SK conceived the study. YL performed the experiments; YL, TW, and XC collected and analyzed the data. KO and HS gave support YL, XC and SK wrote the manuscript. All authors contributed to the article and approved the submitted version.
We received internal research funds from Tokyo Institute of Technology.
We thank the Open Research Facilities for Life Science and Technology in the Tokyo Institute of Technology for technical support and equipment.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcimb.2022.944611/full#supplementary-material
Alalwan, H., Rajendran, R., Lappin, D. F., Combet, E., Shahzad, M., Robertson, D., et al. (2017). The anti-adhesive effect of curcumin on candida albicans biofilms on denture materials. Front. Microbiol. 8, 659. doi: 10.3389/fmicb.2017.00659
Almirante, B., Rodríguez, D., Park, B. J., Cuenca-Estrella, M., Planes, A. M., Almela, M., et al. (2005). Epidemiology and predictors of mortality in cases of candida bloodstream infection: Results from population-based surveillance, Barcelona, Spain, from 2002 to 2003. J. Clin. Microbiol. 43 (4), 1829–1835. doi: 10.1128/JCM.43.4.1829-1835.2005
Ammon, H. P., Wahl, M. A. (1991). Pharmacology of curcuma longa. Planta Med. 57 (01), 1–7. doi: 10.1055/s-2006-960004
Anand, P., Kunnumakkara, A. B., Newman, R. A., Aggarwal, B. B. (2007). Bioavailability of curcumin: problems and promises. Mol. Pharmaceutics 4 (6), 807–818. doi: 10.1021/mp700113r
Anand, K., Sarkar, A., Kumar, A., Ambasta, R. K., Kumar, P. (2012). Combinatorial antitumor effect of naringenin and curcumin elicit angioinhibitory activities in vivo. Nutr. Cancer 64 (5), 714–724. doi: 10.1080/01635581.2012.686648
Andrade, J. T., de Figueiredo, G. F., Cruz, L. F., de Morais, S. E., Souza, C. D. F., Pinto, F. C. H., et al. (2019). Efficacy of curcumin in the treatment of experimental vulvovaginal candidiasis. Rev. Iberoamericana Micologia 36 (4), 192–199. doi: 10.1016/j.riam.2019.01.003
Bhavani Shankar, T. N., Shantha, N. V., Ramesh, H. P., Murthy, I. A. S., Murthy, V. S. (1980). Toxicity studies of turmeric: Acute toxicity studies in rats, guinea pigs and monkeys [India]. Indian J. Exp. Biol. (India) 18 (1), 73–5.
Bhullar, K. S., Jha, A., Rupasinghe, H. V. (2015). Novel carbocyclic curcumin analog CUR3d modulates genes involved in multiple apoptosis pathways in human hepatocellular carcinoma cells. Chemico-biological Interact. 242, 107–122. doi: 10.1016/j.cbi.2015.09.020
Blasius, R., Duvoix, A., Morceau, F., Schnekenburger, M., Delhalle, S., Henry, E., et al. (2004). Curcumin stability and its effect on glutathione s-transferase P1-1 mRNA expression in K562 cells. Ann. New York Acad. Sci. 1030 (1), 442–448. doi: 10.1196/annals.1329.055
Brown, G. D., Denning, D. W., Gow, N. A., Levitz, S. M., Netea, M. G., White, T. C. (2012). Hidden killers: Human fungal infections. Sci. Trans. Med. 4 (165), 165rv13–165rv13. doi: 10.1126/science.1222236
Carrillo-Munoz, A. J., Giusiano, G., Ezkurra, P. A., Quindós, G. (2006). Antifungal agents: Mode of action in yeast cells. Rev. Esp Quimioter. 19 (2), 130–139.
Cen, L., Hutzen, B., Ball, S., DeAngelis, S., Chen, C. L., Fuchs, J. R., et al. (2009). New structural analogues of curcumin exhibit potent growth suppressive activity in human colorectal carcinoma cells. BMC Cancer 9 (1), 1–8. doi: 10.1186/1471-2407-9-99
Chaturvedi, V., Ramani, R., Ghannoum, M. A., Killian, S. B., Holliday, N., Knapp, C., et al. (2008). Multilaboratory testing of antifungal combinations against a quality control isolate of candida krusei. Antimicrobial. Agents Chemother. 52 (4), 1500–1502. doi: 10.1128/AAC.00574-07
Chen, E., Benso, B., Seleem, D., Ferreira, L. E. N., Pasetto, S., Pardi, V., et al. (2018). Fungal-host interaction: curcumin modulates proteolytic enzyme activity of Candida albicans and inflammatory host response in vitro. Int. J. Dentistry 2018, 1–7. doi: 10.1155/2018/2393146
Cheraghipour, K., Ezatpour, B., Masoori, L., Marzban, A., Sepahvand, A., Rouzbahani, A. K., et al. (2021). Anti-Candida activity of curcumin: A systematic review. Curr. Drug Discovery Technol. 18 (3), 379–390. doi: 10.2174/1570163817666200518074629
Cisterna, R., Ezpeleta, G., Telleria, O., Guinea, J., Regueiro, B., Garcia-Rodriguez, J., et al. (2010). Nationwide sentinel surveillance of bloodstream candida infections in 40 tertiary care hospitals in Spain. J. Clin. Microbiol. 48 (11), 4200–4206. doi: 10.1128/JCM.00920-10
Collart, M. A., Oliviero, S. (1993). Preparation of yeast RNA. Curr. Protoc. Mol. Biol. 23 (1), 13–12. doi: 10.1002/0471142727.mb1312s23
Cowen, L. E., Anderson, J. B., Kohn, L. M. (2002). Evolution of drug resistance in candida albicans. Annu. Rev. Microbiol. 56 (1), 139–165. doi: 10.1146/annurev.micro.56.012302.160907
Diezmann, S., Michaut, M., Shapiro, R. S., Bader, G. D., Cowen, L. E. (2012). Mapping the Hsp90 genetic interaction network in candida albicans reveals environmental contingency and rewired circuitry. PloS Genet. 8 (3), p.e1002562. doi: 10.1371/journal.pgen.1002562
Dong, H. H., Wang, Y. H., Peng, X. M., Zhou, H. Y., Zhao, F., Jiang, Y. Y., et al. (2021). Synergistic antifungal effects of curcumin derivatives as fungal biofilm inhibitors with fluconazole. Chem. Biol. Drug Design 97 (5), 1079–1088. doi: 10.1111/cbdd.13827
Eggimann, P., Garbino, J., Pittet, D. (2003). Epidemiology of candida species infections in critically ill non-immunosuppressed patients. Lancet Infect. Dis. 3 (11), 685–702. doi: 10.1016/S1473-3099(03)00801-6
Eldesouky, H. E., Li, X., Abutaleb, N. S., Mohammad, H., Seleem, M. N. (2018). Synergistic interactions of sulfamethoxazole and azole antifungal drugs against emerging multidrug-resistant candida auris. Int. J. Antimicrobial. Agents 52 (6), 754–761. doi: 10.1016/j.ijantimicag.2018.08.016
Enjalbert, B., Nantel, A., Whiteway, M. (2003). Stress-induced gene expression in candida albicans: Absence of a general stress response. Mol. Biol. Cell 14 (4), 1460–1467. doi: 10.1091/mbc.e02-08-0546
Falagas, M. E., Koletsi, P. K., Bliziotis, I. A. (2006). The diversity of definitions of multidrug-resistant (MDR) and pandrug-resistant (PDR) acinetobacter baumannii and pseudomonas aeruginosa. J. Med. Microbiol. 55 (12), 1619–1629. doi: 10.1099/jmm.0.46747-0
Fang, X., Fang, L., Gou, S., Cheng, L. (2013). Design and synthesis of dimethylaminomethyl-substituted curcumin derivatives/analogues: Potent antitumor and antioxidant activity, improved stability and aqueous solubility compared with curcumin. Bioorganic Medicinal Chem. Lett. 23 (5), 1297–1301. doi: 10.1016/j.bmcl.2012.12.098
Fan, Y., Liu, Y., Zhang, L., Cai, F., Zhu, L., Xu, J. (2017). C0818, a novel curcumin derivative, interacts with Hsp90 and inhibits Hsp90 ATPase activity. Acta Pharm. Sin. B 7 (1), 91–96. doi: 10.1016/j.apsb.2016.05.014
Fan, Y. J., Zhou, Y. X., Zhang, L. R., Lin, Q. F., Gao, P. Z., Cai, F., et al. (2018). C1206, a novel curcumin derivative, potently inhibits Hsp90 and human chronic myeloid leukemia cells in vitro. Acta Pharmacologica Sin. 39 (4), 649–658. doi: 10.1038/aps.2017.160
Forouzanfar, F., Barreto, G., Majeed, M., Sahebkar, A. (2019). Modulatory effects of curcumin on heat shock proteins in cancer: A promising therapeutic approach. BioFactors 45 (5), 631–640. doi: 10.1002/biof.1522
Garcia-Gomes, A. S., Curvelo, J. A. R., Soares, R. A., Ferreira-Pereira, A. (2012). Curcumin acts synergistically with fluconazole to sensitize a clinical isolate of candida albicans showing a MDR phenotype. Med. Mycol. 50 (1), 26–32. doi: 10.3109/13693786.2011.578156
Giommarelli, C., Zuco, V., Favini, E., Pisano, C., Dal Piaz, F., De Tommasi, N., et al. (2010). The enhancement of antiproliferative and proapoptotic activity of HDAC inhibitors by curcumin is mediated by Hsp90 inhibition. Cell. Mol. Life Sci. 67 (6), 995–1004. doi: 10.1007/s00018-009-0233-x
Govindarajan, V. S., Stahl, W. H. (1980). Turmeric–chemistry, technology, and quality. Crit. Rev. Food Sci. Nutr. 12 (3), 199–301. doi: 10.1080/10408398009527278
Gritsko, T., Williams, A., Turkson, J., Kaneko, S., Bowman, T., Huang, M., et al. (2006). Persistent activation of stat3 signaling induces survivin gene expression and confers resistance to apoptosis in human breast cancer cells. Clin. Cancer Res. 12 (1), 11–19. doi: 10.1158/1078-0432.CCR-04-1752
Hajjeh, R. A., Sofair, A. N., Harrison, L. H., Lyon, G. M., Arthington-Skaggs, B. A., Mirza, S. A., et al. (2004). Incidence of bloodstream infections due to candida species and in vitro susceptibilities of isolates collected from 1998 to 2000 in a population-based active surveillance program. J. Clin. Microbiol. 42 (4), 1519–1527. doi: 10.1128/JCM.42.4.1519-1527.2004
Hamzah, H., Hertiani, T., Pratiwi, S. U. T., Nuryastuti, T., Murti, Y. B. (2020). The biofilm inhibition and eradication activity of curcumin againts polymicrobial biofilm. In Bio Web Conferences 28, 04001). EDP Sciences. doi: 10.1051/bioconf/20202804001
Hatcher, H., Planalp, R., Cho, J., Torti, F. M., Torti, S. V. (2008). Curcumin: from ancient medicine to current clinical trials. Cell. Mol. Life Sci. 65 (11), 1631–1652. doi: 10.1007/s00018-008-7452-4
Hutzen, B., Friedman, L., Sobo, M., Lin, L., Cen, L., De Angelis, S., et al. (2009). Curcumin analogue GO-Y030 inhibits STAT3 activity and cell growth in breast and pancreatic carcinomas. Int. J. Oncol. 35 (4), 867–872. doi: 10.3892/ijo_00000401
Ivnitski-Steele, I., Holmes, A. R., Lamping, E., Monk, B. C., Cannon, R. D., Sklar, L. A. (2009). Identification of Nile red as a fluorescent substrate of the candida albicans ATP-binding cassette transporters Cdr1p and Cdr2p and the major facilitator superfamily transporter Mdr1p. Analytical Biochem. 394 (1), 87–91. doi: 10.1016/j.ab.2009.07.001
Jung, Y., Xu, W., Kim, H., Ha, N., Neckers, L. (2007). Curcumin-induced degradation of ErbB2: A role for the E3 ubiquitin ligase CHIP and the Michael reaction acceptor activity of curcumin. Biochim. Biophys. Acta (BBA)-Molecular Cell Res. 1773 (3), 383–390. doi: 10.1016/j.bbamcr.2006.11.004
Jurenka, J. S. (2009). Anti-inflammatory properties of curcumin, a major constituent of curcuma longa: a review of preclinical and clinical research. Altern. Med. Rev. 14 (2), 141–53. Available at: https://altmedrev.com/wp-content/uploads/2019/02/v14-2-141.pdf
Keniya, M. V., Fleischer, E., Klinger, A., Cannon, R. D., Monk, B. C. (2015). Inhibitors of the candida albicans major facilitator superfamily transporter Mdr1p responsible for fluconazole resistance. PloS One 10 (5), p.e0126350. doi: 10.1371/journal.pone.0126350
Khan, M. A., Gahlot, S., Majumdar, S. (2012). Oxidative stress induced by curcumin promotes the death of cutaneous T-cell lymphoma (HuT-78) by disrupting the function of several molecular targets. Mol. Cancer Ther. 11 (9), 1873–1883. doi: 10.1158/1535-7163.MCT-12-0141
Kijima, T., Prince, T. L., Tigue, M. L., Yim, K. H., Schwartz, H., Beebe, K., et al. (2018). HSP90 inhibitors disrupt a transient HSP90-HSF1 interaction and identify a noncanonical model of HSP90-mediated HSF1 regulation. Sci. Rep. 8 (1), 1–13. doi: 10.1038/s41598-018-25404-w
Kudo, C., Yamakoshi, H., Sato, A., Ohori, H., Ishioka, C., Iwabuchi, Y., et al. (2011). Novel curcumin analogs, GO-Y030 and GO-Y078, are multi-targeted agents with enhanced abilities for multiple myeloma. Anticancer Res. 31 (11), 3719–3726.
Kumar, A., Dhamgaye, S., Maurya, I. K., Singh, A., Sharma, M., Prasad, R. (2014). Curcumin targets cell wall integrity via calcineurin-mediated signaling in Candida albicans. Antimicrobial. Agents Chemother. 58 (1), 167–175. doi: 10.1128/AAC.01385-13
Lai, W. C., Sun, H. F. S., Lin, P. H., Lin, H., Shieh, J. C. (2016). A new rapid and efficient system with dominant selection developed to inactivate and conditionally express genes in candida albicans. Curr. Genet. 62 (1), 213–235. doi: 10.1007/s00294-015-0526-6
Lasrado, S., Rao, S., Lal Madathil, P. S., George, R. (2022). Curcumin, the principal compound of turmeric (Curcuma longa Lin) in oral health: A mini. Chem. Inside Spices Herbs: Res. Dev. 2, 2, 85–92. doi: 10.2174/9781681089492122020006
Leach, M. D., Budge, S., Walker, L., Munro, C., Cowen, L. E., Brown, A. J. (2012a). Hsp90 orchestrates transcriptional regulation by Hsf1 and cell wall remodelling by MAPK signalling during thermal adaptation in a pathogenic yeast. PLoS Pathog. 8 (12), p.e1003069. doi: 10.1371/journal.ppat.1003069
Leach, M. D., Farrer, R. A., Tan, K., Miao, Z., Walker, L. A., Cuomo, C. A., et al. (2016). Hsf1 and Hsp90 orchestrate temperature-dependent global transcriptional remodelling and chromatin architecture in candida albicans. Nat. Commun. 7 (1), 1–13. doi: 10.1038/ncomms11704
Leach, M. D., Klipp, E., Cowen, L. E., Brown, A. J. (2012b). Fungal Hsp90: a biological transistor that tunes cellular outputs to thermal inputs. Nat. Rev. Microbiol. 10 (10), 693–704. doi: 10.1038/nrmicro2875
Lee, J. H., Chung, I. K. (2010). Curcumin inhibits nuclear localization of telomerase by dissociating the Hsp90 co-chaperone p23 from hTERT. Cancer Lett. 290 (1), 76–86. doi: 10.1016/j.canlet.2009.08.026
Lee, W., Lee, D. G. (2014). An antifungal mechanism of curcumin lies in membrane-targeted action within. Candida Albicans IUBMB Life 66 (11), 780–785. doi: 10.1002/iub.1326
Limtrakul, P., Anuchapreeda, S., Buddhasukh, D. (2004). Modulation of human multidrug-resistance MDR-1 gene by natural curcuminoids. BMC Cancer 4 (1), 1–6. doi: 10.1186/1471-2407-4-13
Link, A., Balaguer, F., Shen, Y., Lozano, J. J., Leung, H. C. E., Boland, C. R., et al. (2013). Curcumin modulates DNA methylation in colorectal cancer cells. PloS One 8 (2), p.e57709. doi: 10.1371/journal.pone.0057709
Liu, B., Shen, Y., Huang, H., Croce, K. D., Wu, M., Fan, Y., et al. (2020). Curcumin derivative C212 inhibits Hsp90 and eliminates both growing and quiescent leukemia cells in deep dormancy. Cell Communication Signaling 18 (1), 1–15. doi: 10.1186/s12964-020-00652-4
Liu, Y., Ye, M., Wu, Q., Wu, L., Xu, J. (2014). Synthesis and evaluation of 4-arylmethyl curcumin analgues as potent Hsp90 inhibitors. Lett. Drug Design Discovery 11 (8), 993–999. doi: 10.2174/1570180811666140512221037
Livak, K. J., Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2– ΔΔCT method. methods 25 (4), 402–408. doi: 10.1006/meth.2001.1262
Li, Y., Zhang, D., Xu, J., Shi, J., Jiang, L., Yao, N., et al. (2012). Discovery and development of natural heat shock protein 90 inhibitors in cancer treatment. Acta Pharm. Sin. B 2 (3), 238–245. doi: 10.1016/j.apsb.2012.03.009
Lv, Y., Gong, L., Wang, Z., Han, F., Liu, H., Lu, X., et al. (2015). Curcumin inhibits human cytomegalovirus by downregulating heat shock protein 90. Mol. Med. Rep. 12 (3), 4789–4793. doi: 10.3892/mmr.2015.3983
Mohan Yallapu, M., Ray Dobberpuhl, M., Michele Maher, D., Jaggi, M., Chand Chauhan, S. (2012). Design of curcumin loaded cellulose nanoparticles for prostate cancer. Curr. Drug Metab. 13 (1), 120–128. doi: 10.2174/138920012798356952
Nakayama, H., Mio, T., Nagahashi, S., Kokado, M., Arisawa, M., Aoki, Y. (2000). Tetracycline-regulatable system to tightly control gene expression in the pathogenic fungus candida albicans. Infect. Immun. 68 (12), 6712–6719. doi: 10.1128/IAI.68.12.6712-6719.2000
Narayanan, V. S., Muddaiah, S., Shashidara, R., Sudheendra, U. S., Deepthi, N. C., Samaranayake, L. (2020). Variable antifungal activity of curcumin against planktonic and biofilm phase of different candida species. Indian J. Dental Res. 31 (1), p.145. doi: 10.4103/ijdr.IJDR_521_17
Neelofar, K., Shreaz, S., Rimple, B., Muralidhar, S., Nikhat, M., Khan, L. A. (2011). Curcumin as a promising anticandidal of clinical interest. Can. J. Microbiol. 57 (3), 204–210. doi: 10.1139/W10-117
O’Meara, T. R., Cowen, L. E. (2014). Hsp90-dependent regulatory circuitry controlling temperature-dependent fungal development and virulence. Cell. Microbiol. 16 (4), 473–481. doi: 10.1111/cmi.12266
Ortega, M., Marco, F., Soriano, A., Almela, M., Martínez, J. A., López, J., et al. (2011). Candida species bloodstream infection: Epidemiology and outcome in a single institution from 1991 to 2008. J. Hosp. Infect. 77 (2), 157–161. doi: 10.1016/j.jhin.2010.09.026
Patterson, T. F. (2005). Advances and challenges in management of invasive mycoses. Lancet 366 (9490), 1013–1025. doi: 10.1016/S0140-6736(05)67381-3
Pearson, J. P., Van Delden, C., Iglewski, B. H. (1999). Active efflux and diffusion are involved in transport of pseudomonas aeruginosa cell-to-cell signals. J. Bacteriol. 181 (4), 1203–1210. doi: 10.1128/JB.181.4.1203-1210.1999
Pfaller, M. A., Diekema, D. (2007). Epidemiology of invasive candidiasis: A persistent public health problem. Clin. Microbiol. Rev. 20 (1), 133–163. doi: 10.1128/CMR.00029-06
Piddock, L. J. (2006). Multidrug-resistance efflux pumps? not just for resistance. Nat. Rev. Microbiol. 4 (8), 629–636. doi: 10.1038/nrmicro1464
Qureshi, S., Shah, A. H., Ageel, A. M. (1992). Toxicity studies on alpinia galanga and curcuma longa. Planta Med. 58 (02), 124–127. doi: 10.1055/s-2006-961412
Ravindran, J., Prasad, S., Aggarwal, B. B. (2009). Curcumin and cancer cells: How many ways can curry kill tumor cells selectively? AAPS J. 11 (3), 495–510. doi: 10.1208/s12248-009-9128-x
Reuß, O., Morschhäuser, J. (2006). A family of oligopeptide transporters is required for growth of candida albicans on proteins. Mol. Microbiol. 60 (3), 795–812. doi: 10.1111/j.1365-2958.2006.05136.x
Reuß, O., Vik, Å., Kolter, R., Morschhäuser, J. (2004). The SAT1 flipper, an optimized tool for gene disruption in candida albicans. Gene 341, 119–127. doi: 10.1016/j.gene.2004.06.021
Rodriguez-Tudela, J. L., Barchiesi, F., Bille, J., Chryssanthou, E., Cuenca-Estrella, M., Denning, D., et al. (2003). Method for the determination of minimum inhibitory concentration (MIC) by broth dilution of fermentative yeasts. Clin. Microbiol. Infect. 9 (8), i–viii. doi: 10.1046/j.1469-0691.2003.00789.x
Rowe, D. L., Ozbay, T., O’Regan, R. M., Nahta, R. (2009). Modulation of the BRCA1 protein and induction of apoptosis in triple negative breast cancer cell lines by the polyphenolic compound curcumin. Breast Cancer: Basic Clin. Res. 3, BCBCR–S3067. doi: 10.4137/bcbcr.s3067
Sang, Q., Liu, X., Wang, L., Qi, L., Sun, W., Wang, W., et al. (2018). Curcumin protects an SH-SY5Y cell model of parkinson’s disease against toxic injury by regulating HSP90. Cell. Physiol. Biochem. 51 (2), 681–691. doi: 10.1159/000495326
Shahzad, M., Sherry, L., Rajendran, R., Edwards, C. A., Combet, E., Ramage, G. (2014). Utilising polyphenols for the clinical management of candida albicans biofilms. Int. J. Antimicrobial. Agents 44 (3), 269–273. doi: 10.1016/j.ijantimicag.2014.05.017
Sharma, M., Manoharlal, R., Negi, A. S., Prasad, R. (2010a). Synergistic anticandidal activity of pure polyphenol curcumin I in combination with azoles and polyenes generates reactive oxygen species leading to apoptosis. FEMS Yeast Res. 10 (5), 570–578. doi: 10.1111/j.1567-1364.2010.00637.x
Sharma, M., Manoharlal, R., Puri, N., Prasad, R. (2010b). Antifungal curcumin induces reactive oxygen species and triggers an early apoptosis but prevents hyphae development by targeting the global repressor TUP1 in candida albicans. Biosci. Rep. 30 (6), 391–404. doi: 10.1042/BSR20090151
Sharma, M., Manoharlal, R., Shukla, S., Puri, N., Prasad, T., Ambudkar, S. V., et al. (2009). Curcumin modulates efflux mediated by yeast ABC multidrug transporters and is synergistic with antifungals. Antimicrobial. Agents Chemother. 53 (8), 3256–3265. doi: 10.1128/AAC.01497-08
Sharma, M., Prasad, R. (2011). The quorum-sensing molecule farnesol is a modulator of drug efflux mediated by ABC multidrug transporters and synergizes with drugs in candida albicans. Antimicrobial. Agents Chemother. 55 (10), 4834–4843. doi: 10.1128/AAC.00344-11
Sharom, F. J. (2008). ABC Multidrug transporters: structure, function and role in chemoresistance Pharmacogenomics 9(1), 105–27. doi: 10.2217/14622416.9.1.105
Shibata, H., Yamakoshi, H., Sato, A., Ohori, H., Kakudo, Y., Kudo, C., et al. (2009). Newly synthesized curcumin analog has improved potential to prevent colorectal carcinogenesis in vivo. Cancer Sci. 100 (5), 956–960. doi: 10.1111/j.1349-7006.2009.01127.x
Strimpakos, A. S., Sharma, R. A. (2008). Curcumin: Preventive and therapeutic properties in laboratory studies and clinical trials. Antioxidants Redox Signaling 10 (3), 511–546. doi: 10.1089/ars.2007.1769
Surma, S., Sahebkar, A., Urbański, J., Penson, P. E., Banach, M. (2022). Curcumin-the nutraceutical with pleiotropic effects? which cardiometabolic subjects might benefit the most? Front. Nutr. 9. doi: 10.3389/fnut.2022.865497
Swoboda, R. K., Bertram, G., Budge, S., Gooday, G. W., Gow, N. A., Brown, A. J. (1995). Structure and regulation of the HSP90 gene from the pathogenic fungus candida albicans. Infect. Immun. 63 (11), 4506–4514. doi: 10.1128/iai.63.11.4506-4514.1995
Thakre, A. D., Mulange, S. V., Kodgire, S. S., Zore, G. B., Karuppayil, S. M. (2016). Effects of cinnamaldehyde, ocimene, camphene, curcumin and farnesene on candida albicans. Adv. Microbiol. 6 (09), p.627. doi: 10.4236/aim.2016.69062
Thompson, J. R., Register, E., Curotto, J., Kurtz, M., Kelly, R. (1998). An improved protocol for the preparation of yeast cells for transformation by electroporation. Yeast 14 (6), 565–571. doi: 10.1002/(SICI)1097-0061(19980430)14:6<565::AID-YEA251>3.0.CO;2-B
Veri, A. O., Miao, Z., Shapiro, R. S., Tebbji, F., O’Meara, T. R., Kim, S. H., et al. (2018). Tuning Hsf1 levels drives distinct fungal morphogenetic programs with depletion impairing Hsp90 function and overexpression expanding the target space. PloS Genet. 14 (3), p.e1007270. doi: 10.1371/journal.pgen.1007270
Whitesell, L., Robbins, N., Huang, D. S., McLellan, C. A., Shekhar-Guturja, T., LeBlanc, E. V., et al. (2019). Structural basis for species-selective targeting of Hsp90 in a pathogenic fungus. Nat. Commun. 10 (1), 1–17. doi: 10.1038/s41467-018-08248-w
Wu, C. P., Ohnuma, S., V Ambudkar, S. (2011). Discovering natural product modulators to overcome multidrug resistance in cancer chemotherapy. Curr. Pharm. Biotechnol. 12 (4), 609–620. doi: 10.2174/138920111795163887
Ye, M., Huang, W., Wu, W. W., Liu, Y., Ye, S. N., Xu, J. H. (2017). FM807, a curcumin analogue, shows potent antitumor effects in nasopharyngeal carcinoma cells by heat shock protein 90 inhibition. Oncotarget 8 (9), p.15364. doi: 10.18632/oncotarget.14970
Zhang, K. Z., XU, J. H., Huang, X. W., Wu, L. X., Su, Y., Chen, Y. Z. (2007). Curcumin synergistically augments bcr/abl phosphorothioate antisense oligonucleotides to inhibit growth of chronic myelogenous leukemia cells 1. Acta Pharmacologica Sin. 28 (1), 105–110. doi: 10.1111/j.1745-7254.2007.00471.x
Zheng, C., Fan, Y., Wu, S., Cai, X., Shi, Y. (2016). Synergistic effects of curcumin and bortezomib on multiple myeloma cells. Int. J. Clin. Exp. Med. 9 (11), 21787–21793. Available at: https://e-century.us/files/ijcem/9/11/ijcem0017967.pdf
Keywords: antifugal activity, Hsp90, CDR1, post-transcripional control, pathogenic fungus
Citation: Lee YS, Chen X, Widiyanto TW, Orihara K, Shibata H and Kajiwara S (2022) Curcumin affects function of Hsp90 and drug efflux pump of Candida albicans. Front. Cell. Infect. Microbiol. 12:944611. doi: 10.3389/fcimb.2022.944611
Received: 15 May 2022; Accepted: 07 September 2022;
Published: 27 September 2022.
Edited by:
Shahram Mahmoudi, Iran University of Medical Sciences, IranReviewed by:
Sanaz Aghaei Gharehbolagh, Tehran University of Medical Sciences, IranCopyright © 2022 Lee, Chen, Widiyanto, Orihara, Shibata and Kajiwara. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Susumu Kajiwara, a2FqaXdhcmEucy5hYUBtLnRpdGVjaC5hYy5qcA==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.