- 1Department of Medical Microbiology, School of Medicine, University of Pretoria, Pretoria, South Africa
- 2Department of Microbiology and Immunology, Indiana University School of Medicine-Northwest, Gary, IN, United States
- 3Department of Dermatology, School of Medicine, University of Pretoria, Pretoria, South Africa
Background: Mobile colistin resistance (mcr) genes modify Lipid A molecules of the lipopolysaccharide, changing the overall charge of the outer membrane.
Results and discussion: Ten mcr genes have been described to date within eleven Enterobacteriaceae species, with Escherichia coli, Klebsiella pneumoniae, and Salmonella species being the most predominant. They are present worldwide in 72 countries, with animal specimens currently having the highest incidence, due to the use of colistin in poultry for promoting growth and treating intestinal infections. The wide dissemination of mcr from food animals to meat, manure, the environment, and wastewater samples has increased the risk of transmission to humans via foodborne and vector-borne routes. The stability and spread of mcr genes were mediated by mobile genetic elements such as the IncHI2 conjugative plasmid, which is associated with multiple mcr genes and other antibiotic resistance genes. The cost of acquiring mcr is reduced by compensatory adaptation mechanisms. MCR proteins are well conserved structurally and via enzymatic action. Thus, therapeutics found effective against MCR-1 should be tested against the remaining MCR proteins.
Conclusion: The dissemination of mcr genes into the clinical setting, is threatening public health by limiting therapeutics options available. Combination therapies are a promising option for managing and treating colistin-resistant Enterobacteriaceae infections whilst reducing the toxic effects of colistin.
Highlights
● Mcr genes are associated with mobile genetic elements that are facilitating its global dissemination.
● Using colistin as a growth promoter increases the risk of acquiring mcr-positive Enterobacteriaceae in food-producing animals.
● Mcr genes have widely disseminated into clinical settings, threatening public health.
● MCR proteins are phosphoethanolamine (PEtN) transferases that mediate the transfer of PEtN from its primary phosphatidylethanolamine to lipid A.
● Multiple compounds can synergistically restore colistin’s activity, reducing its dosage and toxicity.
● CRISPR-Cas9, endolysins-engineered enzymes, and antimicrobial peptides are promising therapies for colistin resistance
Introduction
Colistin was first introduced into clinical practice in the 1950s (Falagas et al., 2005). It was derived from Bacillus polymyxa and belonged to polymyxins (Nigam et al., 2015), a family of cationic polypeptide antibiotics with a narrow-spectrum antimicrobial activity (Ye et al., 2016). Colistin has a bactericidal effect on Gram-negative bacteria and thus is used for treating Gram-negative bacterial infections (Falagas et al., 2005; Nigam et al., 2015). Cationic polypeptides have a high electrostatic attraction to the anionic lipopolysaccharide (LPS) located on the outer membrane of Gram-negative bacteria. There, it displaces the magnesium and calcium divalent cations (Mg2+ and Ca2+), which stabilize the LPS molecules (Falagas et al., 2005; Nigam et al., 2015). This results in the disruption of the cell’s permeability, leading to cell death. However, due to the adverse side effects such as nephrotoxicity and neurotoxicity seen during colistin therapy (Nigam et al., 2015), in the early 1980s, it was removed from human use and administered to farm-animals as prophylaxis, metaphylaxis, and a therapeutic (Etebu and Arikekpar, 2016). It was further used as a growth promoter in some countries (Dawadi et al., 2021). With the increasing resistance caused by Gram-negative pathogens such as carbapenem-resistant Enterobacteriaceae that threatens global public health (Bulman et al., 2017), colistin was recently reintroduced as a last-line treatment option (Ye et al., 2016; Fyfe et al., 2016; Al Atya et al., 2016).
Colistin resistance was largely associated with chromosomal-encoded mechanisms that involved two-component systems (TCSs) such as PmrAB and PhoPQ, and mutation(s) in the MgrB regulator in Klebsiella pneumoniae (Gunn, 2008; Cannatelli et al., 2013; Paterson and Harris, 2016). These chromosomal mutations mediated colistin resistance by modifying LPS, changing LPS’s overall charge, and reducing the affinity of polymyxins to the outer membrane (Gunn, 2008; Cannatelli et al., 2013; Paterson and Harris, 2016). The types of modifications seen include the addition of phosphoethanolamine (PEtN) and 4-amino-4-deoxy-L-arabinose (Ara4N) to the 1-phosphate or 4-phosphate groups of Lipid A, respectively (Mmatli et al., 2020). The PEtN modification is associated with the PmrAB TCSs and the Ara4N modification with the PhoPQ TCS alongside the MgrB regulator (Mmatli et al., 2020).
Plasmid-mediated colistin resistance (mcr) gene was first identified in both animals and humans by Liu et al. (2016) from Enterobacteriaceae. Mcr-1 gene encodes a PEtN transferase enzyme that adds a PEtN to Lipid A (Liu et al., 2016) at the 4’-phosphate group, thus inducing colistin resistance (Hu et al., 2016; Hu et al., 2016). PEtN transferases have previously been identified in Gram-negative bacteria, these include the lipooligosaccharide PEtN transferase A (LptA) from Neisseria meningitides (Wanty, 2013) and the Campylobacter jejuni PEtN transferase, EptC (Wanty et al., 2013; Fage et al., 2014). These PEtN transferases have similar structural properties to the MCR-1 PEtN transferase (Fage 2014). The lipid A modification is identifiable with a matrix-assisted laser desorption/ionization-time of flight (MALDI-TOF) mass spectrometry (MS) assay with an additional m/z= 1920.5 peak observed in MCR-producing isolates, representing the modified Lipid A molecule (Xu et al., 2018). This activity is seen across the different mcr genes: mcr-2 to mcr-10.
After the discovery of mcr-1, other novel mcr genes i.e., mcr-2 to mcr-10, which are widely distributed within Enterobacteriaceae, have been reported globally. The identification of the mcr genes, mcr-1 to mcr-10, is largely mediated by PCR screening and whole genome sequencing (WGS) tools, which are also used in mcr surveillance programmes (Liu et al., 2016; Xavier et al., 2016; Roer et al., 2017; Yang YQ et al., 2017; Rebelo et al., 2018; Wang Y et al., 2020). Each mcr gene is mostly located on conjugative plasmids, associated with mobile genetic elements (MGEs), and mediates colistin resistance through PEtN transferase activity. mcr are widely distributed within Enterobacteriaceae, including Escherichia coli, K. pneumoniae, Salmonella species, and Enterobacter species (Ye et al., 2016; Liu et al., 2016; Di Pilato et al., 2016; Anjum et al., 2016), and have also been reported within other Gram-negatives such as Pseudomonas aeruginosa and Acinetobacter species (Hsu et al., 2017; Snesrud et al., 2018; Snyman et al., 2021).
Initially, food-producing animals were the reservoir of mcr genes due to the high usage of colistin in livestock (Liu et al., 2016; Kempf et al., 2016; Monte et al., 2017). However, farmers and employees in slaughterhouses are continuously exposed to livestock and their feces and thus are at risk of acquiring MCR-producing isolate (Monte et al., 2017; Runcharoen et al., 2017; Dawadi et al., 2021). Global screening of mcr-1 genes in livestock found increased number of MCR-producing isolates, resulting in a ban of colistin in food-producing animals for both growth promotion and treatment of bacterial infections in some countries (WHO, 2011; Collignon et al., 2016). The World Health Organisation (WHO), thereafter, listed colistin as part of the critically important antimicrobials for human medicine. This was to help preserve the effectiveness of colistin for clinical use and to minimize the transmission of mcr genes from animals, livestock, and the environment to humans (WHO, 2011; Collignon et al., 2016).
Purpose of review
This review provides a map of the dissemination of mcr-1 and other mcr genes. It further evaluates the genomic content of each mcr gene, identifying the possible progenitor and the mobile elements that each is associated with. We largely look at the mcr-1 gene, its structure and function, which enables it to mediate colistin resistance, the fitness cost imposed by mcr-1 expression, and the risk factors enabling the dissemination of mcr genes. The review further summarizes the possible treatment options for MCR-producing colistin-resistant isolates in humans.
Herein, we highlight the evolution of mcr genes by providing insight into the genomic content of each gene, identifying the MGEs that aid in mcr dissemination, the global mcr distribution, mcr crystal structure, and enzymatic activity of MCR proteins. We further discuss promising emerging therapeutics that could manage mcr-positive Enterobacteriaceae infections.
Mcr genes genomic context
The mcr-1 gene is part of a 2,600 base pairs (bp) cassette that is made up of a putative promoter gene responsible for the expression of the mcr-1 gene and the hypothetical protein later identified as pap2 (Poirel et al., 2016; Poirel et al., 2017). Mcr-1 is speculated to have been derived from Moraxella species, which harbors the intrinsic chromosomal encoded mcr-like genes and the pap2 membrane-associated lipid phosphatase (Poirel et al., 2017; Kieffer et al., 2017). The pap2 gene is found in both mcr-1 and mcr-2 cassettes and shares 41% identity with Moraxella oloensis phosphatidic acid phosphatase (Xavier et al., 2016; Poirel et al., 2017). Moraxella mcr-like genes with a significant degree of similarity to mcr-1 and mcr-2 in M. porci and M. osloensis were respectively identified in Genbank. Thus, these genes could be closely related (Kieffer et al., 2017). Poirel et al. identified an mcr-like gene, mcr-2.2 later labelled mcr-6, from an M. pluranimalium strain with an 82% and 99% amino acid identity to mcr-1 and mcr-2, respectively (Poirel et al., 2017).
An analysis of mcr-1 sequences deposited in GenBank in 2017 revealed an mcr-1.10 variant from Moraxella porci MSGI3-CO3 with 97.61% identity to the plasmid-borne mcr-1 gene (AbuOun et al., 2017), additionally an mcr-2.1 variant 99% amino acid identity to plasmid-born mcr-2 was identified in M. pluranimarium (Poirel et al., 2017). This isolate was isolated from the fecal contents of healthy pigs in the United Kingdom in April, 2014 (Snesrud et al., 2018). This data suggests that Moraxella species may have been the likely source of mcr-1 and mcr-2 (Poirel et al., 2017; Poirel et al., 2017). Other evidence that supports this speculation that mcr-1 evolved from the Moraxella species is the identification of an ISApl1 element in M. bovoculi and M. porci (Poirel et al., 2017; Li R et al., 2018). Li et al. suggest ISApl1 integrated into M. bovoculi and thereafter evolved with the mcr-like genes to the point seen today (Li R et al., 2018). This synteny of mcr-pap2 genes across Moraxella species further highlights this genus as a natural reservoir of mcr-like genes and a possible progenitor due to the high amino acid identity (Kieffer et al., 2017; Poirel et al., 2017; AbuOun et al., 2017; Wei W et al., 2018).
To evaluate the phylogenetic relationship between mcr-1 and Moraxella species’ PEA transferase, Wei et al. conducted domain swapping with an intrinsic colistin resistance gene (icr) from M. osloensis (Icr-Mo) (Wei W et al., 2018). PEA transferases are made up of two domains, Transmembrane (TM) and the catalytic domain, discussed later. Thus, Wei et al. created hybrids versions of Icr-Mo gene and found that only TM(MCR-1)-ICR-Mo hybrid had partial activity conferring colistin resistance up to 4 µg/mL and able to modify the Lipid A moiety (Wei W et al., 2018). Therefore, MCR-1 and ICR-Mo are not fully interchangeable, which indicates a fundamental difference between the two proteins (Wei W et al., 2018).
The mobilization of the mcr-pap2 unit was thereafter accomplished through IS elements, but Kieffer et al. identified a replicase gene associated with mcr-1 on IncX4 plasmids (Kieffer et al., 2017). The gene had a 99% identity to M. lacunata, thus showing this species as a possible reservoir of IncX4 plasmids and further speculating that the Moraxella family may encode genetic tools likely involved in the initial mobilization of mcr genes (Kieffer et al., 2017).
Stoesser et al. and Sun et al. suggested that the initial mobilization of mcr-1 genes into Enterobacteriaceae was IS1294-mediated, using a one-ended rolling circle transposition mechanism shown to be capable of mobilizing adjacent sequences (Stoesser et al., 2016; Sun et al., 2018). The IS1294 may have mobilized the mcr-1 cassette into an ISApl1 composite transposon, creating an ISApl1-mcr-1-pap2-IS1294-pap2-ISApl1 cassette, which has been identified on E. coli chromosome (Stoesser et al., 2016; Sun et al., 2018). It was found that the cassette was still flexible enough to jump from the chromosome to a plasmid (Sun et al., 2018) by generating a putative circular intermediate product.
There is increasing evidence, however, that the mcr-1 gene is mobilized primarily as a composite transposon, Tn6330, that is made up of two copies of ISApl1 that bracket cassettes (Figure 1) (Snesrud et al., 2016; Zurfluh et al., 2016). ISApl1 is an IS that was first described in Acinetobacillus pleuropneumoniae and is part of the IS30 family (Tegetmeyer et al., 2008). The IS elements of this family are flanked by 20-30-bp inverted repeats (left IR (IRL), right IR (IRR)), which are essential for transposition (Szabó et al., 2008). The IR contains a 924 bp open-reading frame that encodes a 44.3 kDa transposase protein containing a DDE domain, which encodes three conserved amino acid residues viz., D228, D295, and E648 (DDE), as well as carboxylase residues that help coordinate metal ions for catalysis (Szabó et al., 2008; Snesrud et al., 2016). Analysis of each ISApl1 element flanking the mcr-1-pap2 unit found conserved dinucleotides between the ISApl1 inverted repeats and the mcr-1-pap2 unit, an AT dinucleotide on the IRR of the upstream element and CG on the IRL of the downstream element (Snesrud et al., 2016) (Figure 1). An interesting observation in the mcr-1.10 variant identified in Moraxella sp. MSGI3-CO3 was the presence of these dinucleotides, AT upstream and CG downstream, flanking the mcr-1 structure (Snesrud et al., 2018). These dinucleotides were suggested to represent the ancestral target-site duplications (TSDs) formed during the initial mobilization of mcr-1 during ISApl1 insertion (Snesrud et al., 2018). Therefore, Snesrud et al. suggested that the formation of the composite transposon, Tn6330, was through two independent insertion events of ISApl1 into the TA-rich region of the mcr-1-pap2 unit, generating the conserved interior 2bp TSDs, AT and CG (Snesrud et al., 2018). Subsequent transposition of the Tn6330 would therefore generate new TSDs at the new target site but would retain both internal conserved 2bp dinucleotides (Snesrud et al., 2018).
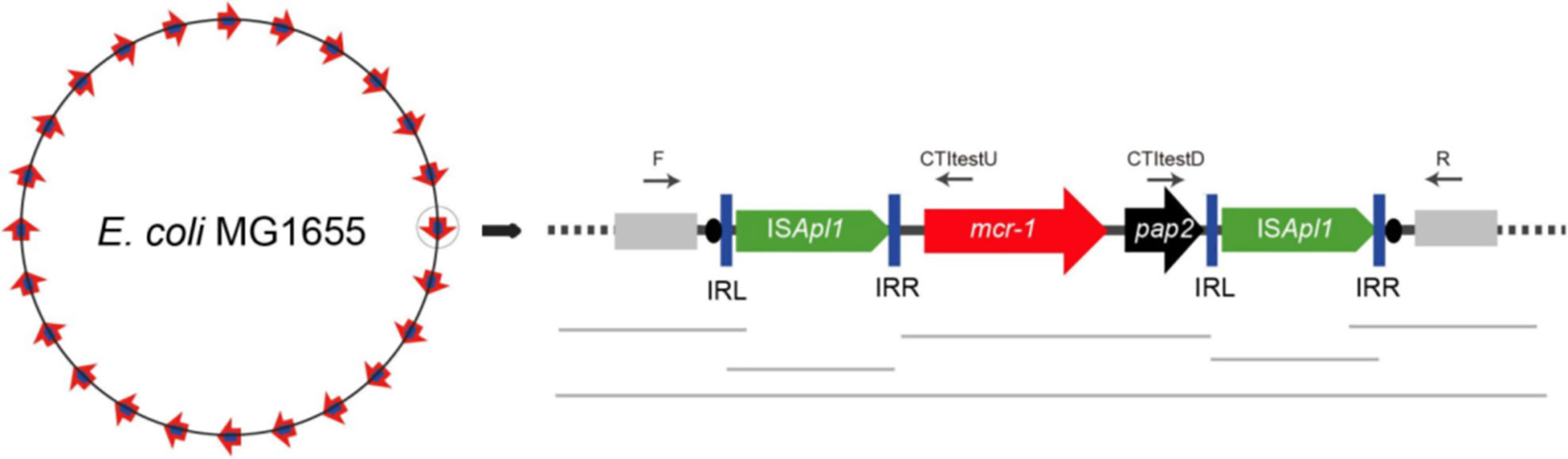
Figure 1 A schematic diagram of the ISApl1 composite transposon identified on the Escherichia coli chromosome retrieved from He et al. (2019). The diagram is made up of mcr-1 gene in red, the hypothetical pap2 protein in black, the inverted repeats (IRL and IRR) in blue vertical bars and the direct repeats as black ovals.
As stated above, the ISApl1 is part of the IS30 family. The family has been shown to mobilize through a copy (out) and paste mechanism, forming circular intermediates of a single IS during transposition. The family is further known to have a high affinity for certain target sites resembling their IR sequence (Tegetmeyer et al., 2008; Szabó et al., 2008; Wang Q et al., 2018). Multiple studies have investigated the mechanisms of ISApl1 in mobilizing the mcr-1 gene and found that during each transposition event, the transposon was a circular intermediate covalently closed doubled stranded DNA, 5 699 bp in size and generated a 2 bp direct repeats at the insertion site which was an AT-rich region (Szabó et al., 2008; Snesrud et al., 2016; Poirel et al., 2017; Snesrud et al., 2017), a similar mechanism seen in the IS30 family. ISApl1 is most likely an important factor responsible for the insertion and fixation of the mcr-1 gene into various classes of self-transmissible plasmids and host chromosomes (Poirel et al., 2017; Zhao et al., 2017; Li R et al., 2018). The formation of an intermediate structure consisting of ISApl1 and mcr-1 during mobilization indicates that the resistance genes have become highly mobilizable in both plasmids and the chromosomes (Li R et al., 2018). After the first identification of mcr-1 in pHNSHP45 (Liu et al., 2016), an Incl2 plasmid, mcr-1 was thereafter detected in a wide range of conjugative plasmids, IncI2, IncHI2, IncX4, IncF, and IncP with the potential to mediate the dissemination of mcr-1 genes into other Gram-negative bacteria (Zhao et al., 2017). Petrillo et al. suggest that the insertion of the complete transposon triggers the rapid mobilization of conjugative plasmids, encouraging their dissemination across the Enterobacteriaceae family (Petrillo et al., 2016). The copy out and paste in mechanisms allow for the transposition of the mcr-1 cassette, and the decay properties of ISApl1 further transfix the resistance gene into a plasmid or chromosome (Snesrud et al., 2017; Sia et al., 2020).
Snesrud et al. and Li et al. discovered that ISApl1 is highly active and that a single copy of ISApl1 can mobilize independently of mcr-1 across the host genome in AT regions with a slight central GC bias (Snesrud et al., 2017; Li et al., 2019). A sequencing analysis of four mcr-1 containing isolates performed by Snesrud et al. identified two to six copies of ISApl1 element throughout the isolates’ genome. The highly active nature of the ISApl1 elements thereafter triggers the deletion of the flanking ISApl1 copies to prevent further plasmid rearrangements (Snesrud et al., 2017; Snesrud et al., 2018). Snesrud et al. analysis of the mcr-1 sequence environment showed that Tn6330 has a strong tendency to decay through deletion, removing parts of, or both copies of ISApl1, thus transfixing mcr-1 into a vector plasmid (Figure 2). This has led to the observation of many sequences lacking one or both of ISApl1 (Snesrud et al., 2016; Poirel et al., 2017). Composite transposons in the IS30 family have been shown to contribute to replicon stabilization through transposition and illegitimate recombination (Szabó et al., 2008; Snesrud et al., 2016). The loss of ISApl1 elements results in the loss of transposability, stabilizing the mcr-1 cassette in plasmids, which facilitates the widespread dissemination of the colistin resistance gene in self-transmissible plasmids (Snesrud et al., 2016; Tarabai et al., 2019; Ji et al., 2019). As discussed, ISApl1 has a significant bias for insertion in AT-regions and generates TSD of two or three bases. The analysis of the mcr-1 cassette in the absence of ISApl1 elements has shown that the cassette is found in similar locations as per plasmid type and flanked by conserved trinucleotides (5’-ATA-3’) that are found immediately downstream of the ISApl1 IRR (Snesrud et al., 2016; Tijet et al., 2017). This is because the deletion event involves one to four flanking nucleotides that remain at the deletion junction (Snesrud et al., 2018).
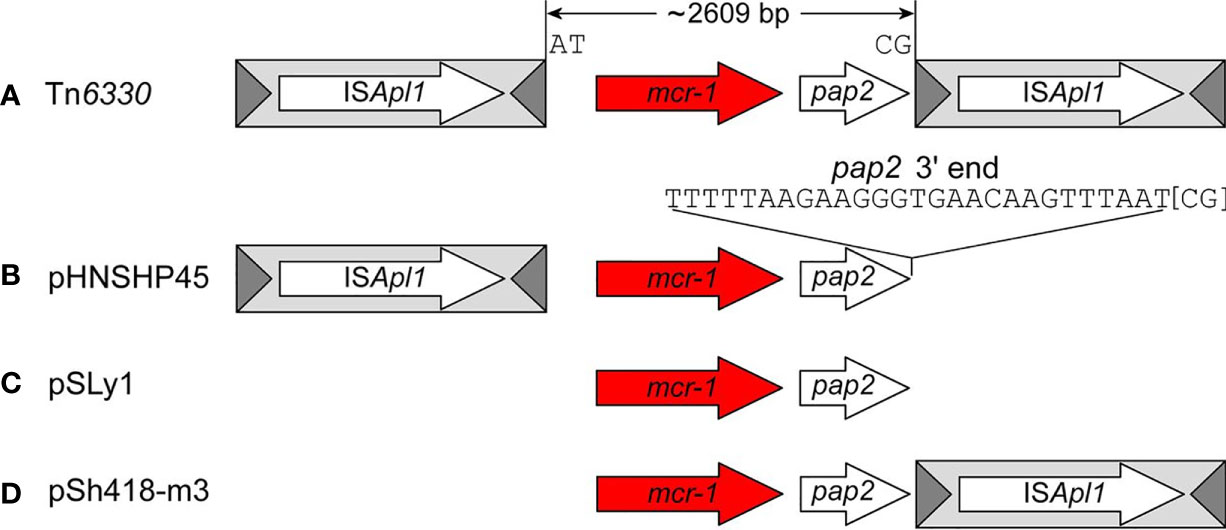
Figure 2 The four general structures of mcr-1 sequences created by the deletion events retrieved from Snesrud et al. (2018). The structures include (A) the complete composite transposon with both copies of ISApl1 elements (Tn6330), (B) Structures with a single copy of ISApl1 located upstream of mcr-1 (pHNSHP45); (C) structures that lost both copies (pSLy1) and (D) a fourth structure with a single copy of ISApl1 located downstream (pSh418-m3).
An analysis of mcr-1 sequences deposited in the public database has shown four general structures of mcr-1 sequences: the complete composite transposon with both copies of ISApl1 elements; structures with a single copy of ISApl1 located downstream of mcr-1; structures that lost both copies and a rare fourth structure with a single copy of ISApl1 located upstream (Snesrud et al., 2016; Snesrud et al., 2018) (Figure 2). In structures with a single copy of ISApl1 found upstream and the IRR sequence of the downstream of deleted ISApl1, Snesrud et al. suggest that the transposase encoded by the upstream ISApl1 can recognize the downstream IRR and thereafter still be able to mobilize the bracketed region without a complete composite transposon (Snesrud et al., 2016). The partially or complete removal of ISApl1 was through an illegitimate recombination that generated mismatches and deletions. Sun et al. found that the 3’ end of the mcr cassette unit was flexible in all IncX4 plasmids and the sequence could match with the perfect IRR of ISApl1, though all IncX4 currently lack ISApl1 elements (Sun et al., 2018). The evidence from this study suggests that the TSD generated, and the six mismatches acquired through illegitimate recombination could be identified as a “relic” to track an insertion event that resulted in the subsequent loss of ISApl1 (Sun et al., 2018). The loss of ISApl1 elements in IncX4 was conducive to maintaining the mcr-1 cassette on the plasmid (Sun et al., 2018), increasing its stability, and thus allowing for the dissemination of resistance genes via the plasmids (Snesrud et al., 2016; Tarabai et al., 2019; Ji et al., 2019). The identification of these IRR in IncX4 plasmids allow for the conclusion that ISApl1 was associated with the transposition of the mcr-1 cassette into IncX4 plasmids (Sun et al., 2017a).
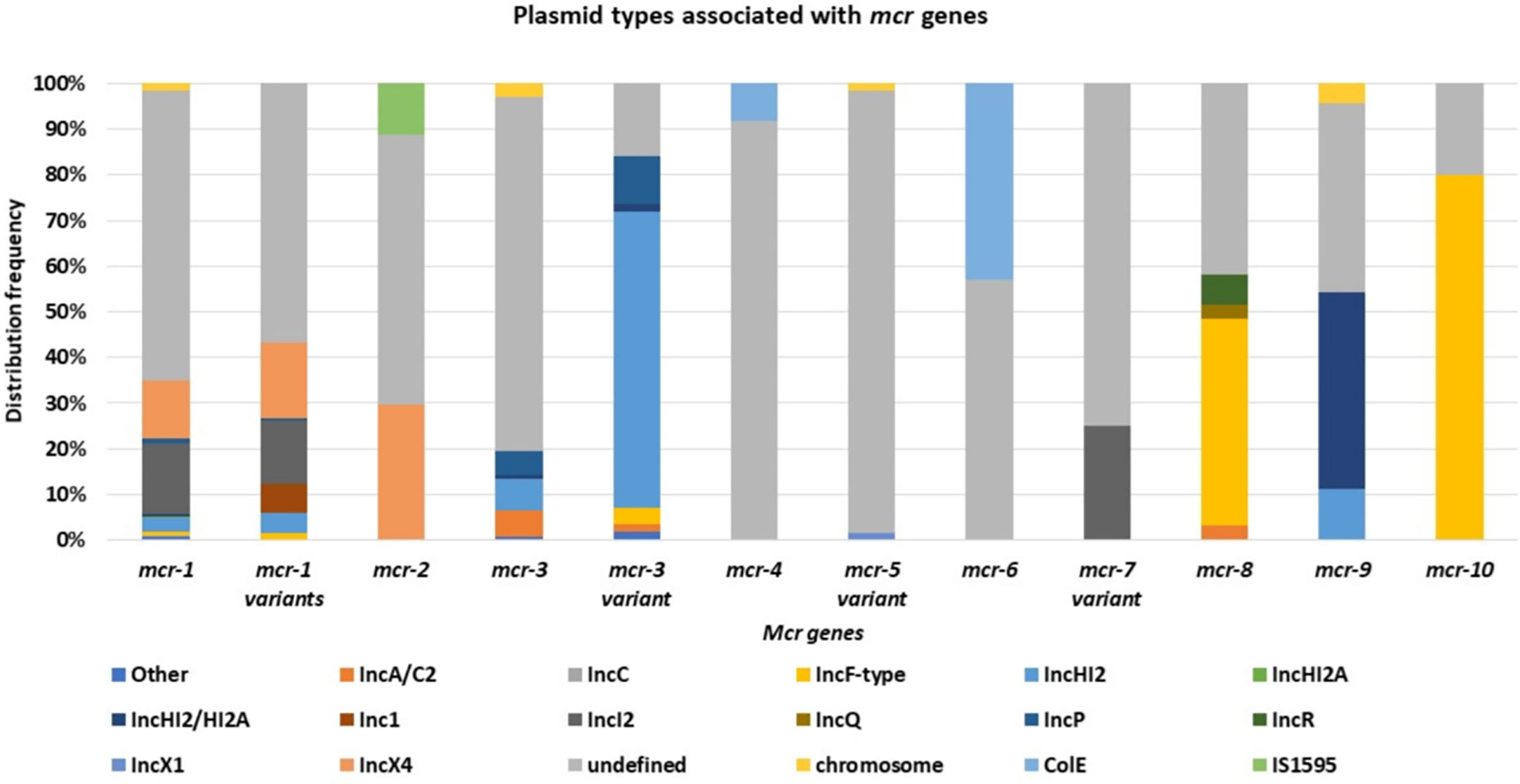
Mcr-1 and its variants (mcr-1 variants) are the most common and well disseminated of all mcr genes (Figure 3). This was later shown to be because of multiple factors such as fitness cost and MGEs. They have spread globally and have been identified in 69 countries, with mcr-1 being the most prevalent mcr gene in most countries (Figure 3). Although the other mcr-genes have spread globally, they have been identified in low numbers compared to mcr-1 (Figure 3A). Mcr-1 and its variants have been identified to be associated with 37 different plasmid incompatibility groups, with IncI2, IncX4, IncHI2, and IncP being the most prevalent (Figure 4, Table S6). These incompatibility groups have also been shown to be broad-spectrum plasmids which further harbor other resistance genes.
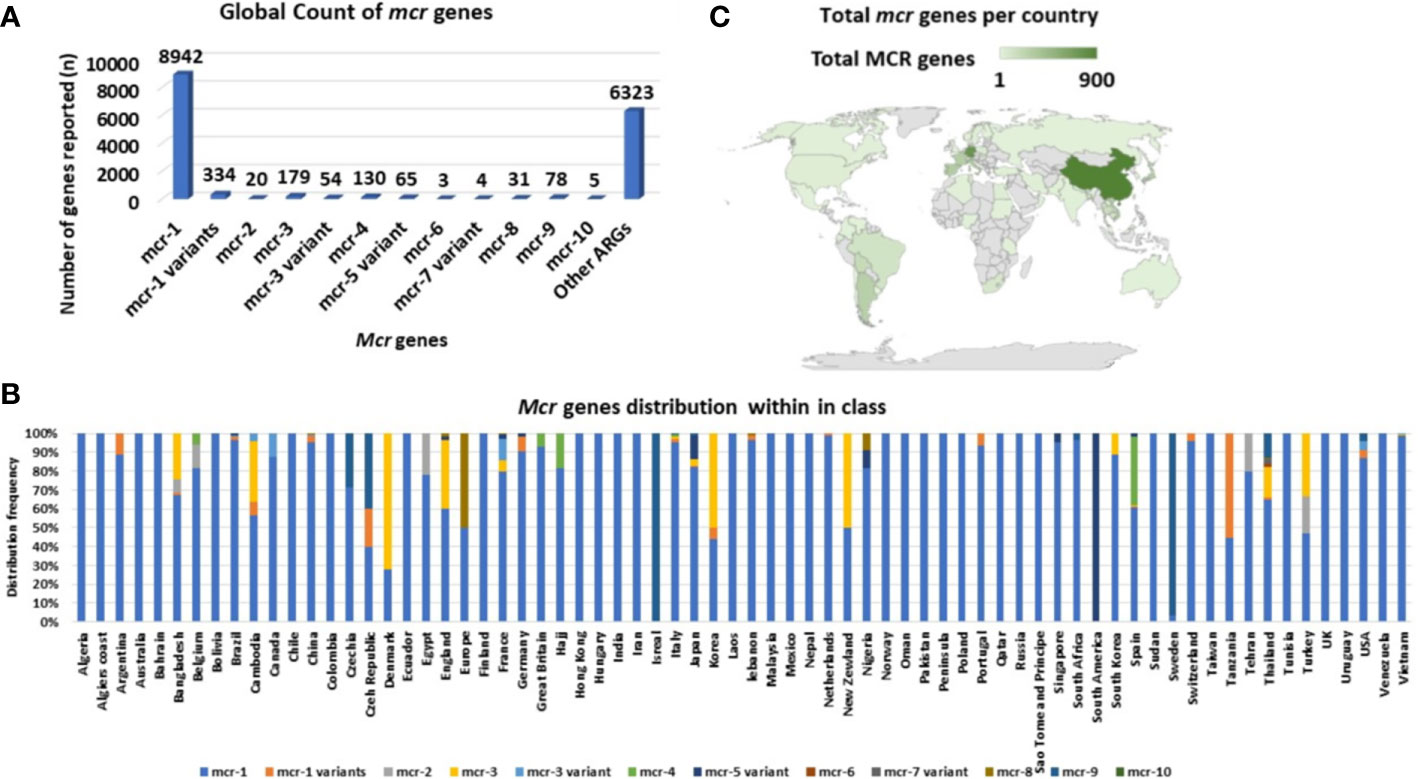
Figure 3 Global distribution and total count of mcr genes. (A) Total number of mcr genes reported globally. (B) The distribution of mcr genes per country. (C) Global map showing the geographical distribution of mcr genes.
The mcr-2 was first identified in ten E. coli isolates during a passive surveillance screening of diarrhea in calves and piglets, and like mcr-1, mcr-2 was located on a conjugative plasmid (IncX4) and was able to confer clinical colistin resistance (4-8 mg/L) (Xavier et al., 2016). The gene was identified due to the presence of a putative membrane protein identified as a PEtN transferase in mcr-1 negative isolates (Xavier et al., 2016). Mcr-2 has a 76.75% and 80.65% nucleotide sequence and amino acid identity, respectively, to mcr-1 (Xavier et al., 2016).
A genetic analysis of the IncX4 plasmid found that mcr-2 was located within two IS1595-like ISs and a 279 bp open reading frame (ORF), located downstream the mcr-2 gene (Xavier et al., 2016). The ORF encodes a PAP2 membrane-associated lipid phosphatase which is related to the PAP2 protein encoded by the ORF of mcr-1 (Xavier et al., 2016; Partridge, 2017). The ORF of mcr-2 has a 41% identity to the phosphatidic acid phosphatase encoded by M. osloensis. When Xavier et al. removed the mcr-2-pap2 from the IS1595 backbone, a BLASTn search produced a single hit to M. bovoculi strain with a 75% identity across a 100% query coverage (Xavier et al., 2016).
The IS1595 element is composed of a transposase gene flanked by two inverted repeats (18 bp each). Mcr-2 is thus present within a complete transposon which allows for horizontal gene transfer (Sun et al., 2017b). Sun et al. found that the IS1595 composite transposon was able to form circularized intermediates, which were identified during an inverse PCR assay (Sun et al., 2017b). Hence, the IS1595 elements are involved in the mobilization of the mcr-2 cassette via homologous recombination events, which like mcr-1, allows for the dissemination of the resistance gene across diversified bacterial hosts (Sun et al., 2017b).
After the first identification of mcr-2 in Belgium on an IncX4 plasmid, an effective vehicle for dissemination of resistance and virulence genes with a high transfer frequency (Xavier et al., 2016; Sun et al., 2017b), it was thereafter identified in Salmonella sp. on IncX4 plasmids in Belgium in 2018 from retail meat collected in 2012 (Garcia-Graells et al., 2018). It has subsequently been found in pig and poultry samples in 19 provinces in China, where the prevalence of mcr-2 in the study was 56.3% in poultry (Zhang et al., 2018). Mcr-2 was then found in clinical settings in Iran, in stool samples collected between 2011-2016 (Ghasemian et al., 2019). In Egypt, mcr-2 was widespread and identified in humans, wild birds, and the environment (water sources) (Ahmed et al., 2019). Compared to mcr-1, mcr-2 has not spread across the world (Figure 3A). and imposed a big threat to public health though found on a low fitness burden plasmid, IncX4 (Figure 4). The gene has only been identified in Bangladesh, Belgium, and Egypt in low numbers (Figure 3B). The gene has however spread well within the Enterobacteriaceae family i.e., E. coli, K. oxytoca, K. pneumoniae and Salmonella sp.
AbuOun et al. (2017) identified an mcr-2.2 variant in Moraxella pluranimalium strain isolated from a pig in Great Britain. The MCR-2.2 protein was found to have an 87.9% amino acid identity to MCR-2, encoding 65 amino acid substitutions (AbuOun et al., 2017). Partridge et al. (2018) suggested that since mcr-2.2 encodes a protein that is 87.9% identical to the original MCR-2, it should be labelled as mcr-6. The mcr-6 gene like mcr-1 and mcr-2 was associated with the PAP2 gene located downstream of mcr-6 (AbuOun et al., 2017). The mcr-6 has only been identified in the M. pluranimalium MSG47-C17 strain isolated in 2012 and has not disseminated into the Enterobacteriaceae family, identified only in E. coli. AbuOun et al. (2017) however highlights that Moraxella species could have been the source of mcr-pap2 genes.
The first identification of mcr-3 was in E. coli isolated from pigs in China (Yin et al., 2017). Thereafter, multiple mcr-3 variants have been identified with one or more amino acid substitutions. Mcr-3 genetic context is made up of a diacylglycerol kinase (dgkA) gene located downstream the mcr-3 gene and is thereafter flanked by a truncated (Δ) ISKpn40 upstream and an intact ISKpn40 downstream (Sia et al., 2020). In some cases, a ΔTnAs2 element is located upstream of the ΔISKpn40 element (Yin et al., 2017; Liu et al., 2017; Sia et al., 2020), and the genetic context (ΔTnAs2-ΔISKpn40-mcr-3-dgkA-ISKpn40) is flanked by an IS26 element upstream and an intact IS15DI element downstream (Creighton et al., 2019; Gomi et al., 2019; Sia et al., 2020). These elements, ISKpn40, TnAs2, IS26 and IS15DI were identified and hypothesized to play a role in the transposition of the mcr-3 cassette among different species or bacterial genera (Wang et al., 2018; Xiang et al., 2018; Sia et al., 2020). Sia et al. identified two circular intermediates of 3535 bp and 5990 bp made up of mcr-3-dgkA-ISKpn40 and ISΔ26-TnAs2-ΔISKpn40-mcr-3-dgkA-ISKpn40, respectively, during conjugation experiments (Sia et al., 2020). Wang et al. evaluated the transferability of the 5990 bp circular intermediate and concluded that the ΔIS26 and intact IS15DI mobilises the mcr-3.1 via homologous recombination through the formation of circular intermediates (Wang et al., 2018). The IS-mediated transposition enables the mobilization of the mcr-3 resistance gene between the chromosome and plasmids, which further contributes to the dissemination of resistance genes (Wang et al., 2018; Gomi et al., 2019). Similar conclusion was reached with the 3535 bp circular intermediates in a study performed by Xiang et al., which showed that mobilization of the mcr-3 core segment (mcr-3-dgkA) through ISKpn40 elements facilitated its spread into various plasmids (Xiang et al., 2018).
The mcr-3.12 variant has a 99% nucleotide sequence identity with an Aeromonas veronii sequence (Kieffer et al., 2018) and significant (77%) amino acid sequence identity to three PEtN transferases found within the genera (Yin et al., 2017; Cabello et al., 2017; Kieffer et al., 2018). It is suggested that mcr-3 genes originated from the Aeromonas genus, or the mcr-3 gene is widely disseminated as an acquired resistance trait and thus also found in Aeromonads (Kieffer et al., 2018). It is however thought that mcr-3 was initially transposed from the Aeromonas genera to the Enterobacteriaceae family through ISKpn40-mediated transposition. The ISKpn40 elements encode two ORF and the second ORF has a 99% identity to both a transposase from Aeromonas sp. and an integrase from Aeromonas caviae (Wang Z et al., 2019). This IS element is found flanking the mcr-3 segment and was also shown to mobilize through homologous recombination (Kieffer et al., 2018; Sia et al., 2020). The last evidence of mcr-3 originating from Aeromonas includes the presence of TnAs2 located upstream of the mcr-3 gene (Yin et al., 2017). The transposon has only been identified in A. salmonicida and because its sequence in pWJ1 (original mcr-3 plasmid) is a partial sequence, it is unlikely to have mobilized the mcr-3 gene (Yin et al., 2017; Liu et al., 2017). This evidence supports the possibility of the Aeromonas genus being a possible progenitor of mcr-3 genes (Yin et al., 2017).
Mcr-3 and its variants are disseminated within Enterobacteriaceae, having been identified in E. coli, Salmonella sp. and K. pneumoniae in healthcare centers, aquaculture, wastewater, and in animals (Roer et al., 2017; Yin et al., 2017; Liu et al., 2017; Cabello et al., 2017; Litrup et al., 2017; Wang et al., 2018a; Gomi et al., 2019). They have further been identified in eighteen countries, with Colombia and Thailand having the highest counts (Table S3, Figure 3C). The resistance genes have been identified in both transmissible and non-transmissible plasmids such as IncA/C2, IncHI2, IncHI2A, IncFII/FIB, IncY, IncR, IncF, and IncP and in many variant mutations with IncHI2 being the most prevalent (Table S6, Figure 4).
Mcr-4 was first identified in S. Typhimurium isolated from the cecal content of a pig at slaughter in Italy in 2013 (Carattoli et al., 2017; Carattoli et al., 2018). The S. Typhimurium was colistin-resistant and negative for known mcr genes; the mcr-4 gene was identified through whole genome sequencing (Carattoli et al., 2017). MCR-4 respectively has an amino acid sequence identity of 34%, 35% and 49% to MCR-1, MCR-2, and MCR-3, and like other PEtN transferases, can mediate colistin resistance through Lipid A modification (Carattoli et al., 2017). Mcr-3 and mcr-4 are suggested to have originated from aquatic environments, as Aeromonas and Shewanella sp. are aquaculture fish pathogens that are intrinsically resistant to colistin (Cabello et al., 2017). MCR-4 has an 82%-99% amino acid sequence identity to one large PEtN transferase found in Shewanella sp., and the mcr-4.3 variant has a 100% nucleotide sequence identity to a chromosomal PEtN transferase encoded by S. frigidimarina, Sfri_3478 (Cabello et al., 2017; Carattoli et al., 2017; Chavda et al., 2018). Zhang et al. suggests a horizontal transfer of mcr-4.3 from S. frigidimarina into a diversified species of Enterobacteriaceae (Zhang et al., 2019).
The Shewanella genus encodes eight variants of PEA transferases called non-mobile colistin resistance genes (nmcr) that confer colistin resistance of up to 16 µg/mL (Zhang et al., 2019; Zhang et al., 2019a). Zhang et al. suggests the intrinsic nmcr-1 enzyme, Sfri_3478, as a progenitor of mcr-4 rather than mcr-1/2 enzymes. Zhang et al. further evaluated the interdomain communication between mcr-4 and nmcr-1 using the domain swapping method (Sun et al., 2017b). The expression of TM(NMCR-1)-MCR-4 and TM(MCR-4)-NMCR-1 in the recipient strain of E. coli MG1655 conferred resistance to colistin up to 8 µg/mL and resulted in LPA modification by an addition of PEA. Thus, the chimeric versions of NMCR-1/MCR-4 are functional in their catalytic activities (Zhang et al., 2019a).With this evidence, Shewanella genus may be a progenitor of MCR-4 and there may be diverse evolutional paths across the entire family of MCR-1 enzymes (Zhang et al., 2019; Zhang et al., 2019a).
Though mcr-4.3 encodes two-point mutations of C179G and V236F which inactivate the PEA transferase activity, Zhang et al. shows that progressive evolution can partially restore its action (Zhang et al., 2019a). This was performed by creating two revertants of mcr-4.3 (G179V and F236V, respectively), each revertant conferred colistin resistance of up to 8 µg/mL and resulted in Lipid A modification (Zhang et al., 2019a).
Mcr-4 was initially identified on an 8749 bp ColE10 plasmid, which encoded a RepB replicase, mobA/L mobilization proteins, a RelE toxin and excl1 gene (Carattoli et al., 2017; Carattoli et al., 2018). In other cases, the ColE plasmid was found to encode a RepA replicase instead of RepB. However, both plasmids showed a 99% nucleotide sequence identity to plasmids from Pantoea species, with a 65% coverage (Carattoli et al., 2017; Chavda et al., 2018; García et al., 2018). The mcr-4 gene was flanked by IS5 elements on some plasmids and on other plasmids, it was flanked by an ΔIS10 element upstream and an IS1294 element downstream (García et al., 2018). Genetic analysis found that the ColE plasmid encodes a conserved 59-bp region upstream the mcr-4 gene, and this region is predicted to encode the -35 (TTATTT) and -10 (AGCTAGTAT) promoter regions (Chavda et al., 2018). This allows for ColE plasmids to replicate independently in different bacterial species and genera (Carattoli et al., 2017). ColE plasmids are broad-range non-conjugative plasmids whose mobilization, however, require a helper plasmid; nevertheless, the mobilization of the mcr-4 gene is hypothesized to be achieved through a transposition event mediated by the IS5 element (Carattoli et al., 2017).
A genomic analysis of the initial mcr-4-producing S. Typhimurium genome found the ColE10 replicon located within the host chromosome integrated within the Type 1 methylation gene. Carattoli et al. suggest a transposition event mediated the chromosomal integration of the ColE10 plasmid (Carattoli et al., 2017). Mcr-4 has been isolated from both humans and animals in Spain, Belgium, Great Britain, Hajj Italy, and China (Figure 3C and Table S3) in E. cloacae, E. coli, L. adecarboxylata and, S. enterica (Table S3) (Carattoli et al., 2017; García et al., 2018; Teo et al., 2018; Sun et al., 2019).
Mcr-5 was first identified in a d-tartrate-fermenting S. Paratyphi isolated from food-producing animals across Germany between 2011 and 2013. Following this discovery of mcr-5, further screening resulted in the identification of fourteen additional mcr-5-producing isolates. These were isolated between 2011 to 2016 in the same study (Borowiak et al., 2017).
Genetic analysis of mcr-5 found that the resistance gene is located within an operon encoding a Chromate B (ChrB) protein domain responsible for regulating the expression of ChrA for chromate resistance and two ORF that encode a major facilitator superfamily (MFS) type transporter (Borowiak et al., 2017; Fleres et al., 2019). The complete operon is located within a 7337bp Tn-3 type transposon named Tn6452 (Borowiak et al., 2017; Sia et al., 2020), that is flanked by inverted repeats (IRs). A genetic analysis of the transposon identified tnpA encoding a Tn3 transposon, tnpR encoding a Tn3 resolvase and a bla gene encoding a β-lactamase (Wang et al., 2018b; Guo et al., 2019; Zhang et al., 2019b). Tn6452 encodes mechanisms for transposition of the mcr-5 cassette, TnpA, and TnpR (Guo et al., 2019), which allows the mcr-5 cassette to be transferred between plasmids and chromosomes (Borowiak et al., 2019).
Mcr-5 has also been identified on Tn3-like structures that encode the ChrB gene but lacked the transposase gene and encoded an imperfect Tn3-like inverted repeats left (IRL) (Hammerl et al., 2018; Borowiak et al., 2019; Kieffer et al., 2019). This structure resembles features of a miniature inverted repeat transposable element, which possess a left or right IRs but lacks a transposase gene (Kieffer et al., 2019). Kieffer et al. suggests the acquisition of the mcr-5 cassette was through a transposition event (Kieffer et al., 2019) and although the Tn3-like structure lacks a transposase gene, it can still mobilize the cassette through a nonautonomous transposition mechanism (Kieffer et al., 2019). The mcr-5 cassette has been identified on the host chromosome (Borowiak et al., 2017), a multicopy ColE-type plasmid (Borowiak et al., 2017; Hammerl et al., 2018; Ma et al., 2018; Borowiak et al., 2019) and on an IncX plasmid (Borowiak et al., 2019). The presence of mcr-5 cassette on a multicopy plasmid (ColE-type) thus means multiple copies of mcr-5 transposon will exist within the cell. Borowiak et al. found that there is a higher degree of colistin resistance (8 mg/L) in isolates harboring ColE-type plasmids encoding mcr-5 than in isolates with a single copy of the mcr-5 integrated within the chromosome (4 mg/L) (Borowiak et al., 2017). The ColE type plasmids are, however, non-conjugative and thus the presence of the mcr-5 cassette on an IncX plasmid allows for horizontal transfer and dissemination of resistance gene between different bacterial genera and species (Guo et al., 2019; Kieffer et al., 2019). Mcr-5 genes are not well disseminated within the Enterobacteriaceae family, identified only in E. coli and S. Typhimurium (Table S4) however these genes have been reported in 10 countries, with Japan, Germany and France being the most prevalent (Table S3)
Mcr-7.1 was first identified in chicken in China (Yang et al., 2018). It has thereafter been identified in environmental samples (Furlan et al., 2020a; Furlan et al., 2020b) and fecal samples in Brazil (Dos Santos et al., 2020). The putative PEtN transferase has a 78% nucleotide sequence identity to part of the mcr-3 gene (Yang et al., 2018). Genetic analysis of mcr-7.1 identified a 381 bp ORF encoding the diacylglycerol kinase, dgkA, downstream of mcr-7.1. The dgkA has an 82% nucleotide sequence identity to Aeromonas (Yang et al., 2018). Yang et al. suggests, like mcr-3, mcr-7.1 originates from Aeromonas (Yang et al., 2018). This was derived based on the close genetic distance between MCR-3 and MCR-7.1 found during a phylogenetic analysis and the presence of dgkA gene downstream both mcr-3 and mcr-7.1 (Yang et al., 2018). The ORF sequence found upstream the mcr-7.1 gene has an 81% nucleotide sequence identity to a putative phosphodiesterase gene found in Aeromonas (Yang et al., 2018). This evidence suggests that the Aeromonas genus is a possible progenitor of mcr-7.1.
Mcr-7.1 was identified on a self-transmissible IncI2 type plasmid with no ISs in its environment; hence, the dissemination of mcr-7.1 may be achieved through plasmid mobilization (Yang et al., 2018). Mcr-7 and its variants, however, are not well disseminated, reported only in three countries, Brazil, China and Thailand (Table S3), identified only in two Enterobacteriaceae species, E. coli and K. pneumoniae (Table S4).
Mcr-8 was initially identified in K. pneumoniae from a swine fecal sample in China (Wang et al., 2018b). Conjugation assays found that it is a functional PEtN transferase, and its expression resulted in a four- and eight-fold increase in colistin MIC (Wang et al., 2018b; Wang X et al., 2019; Bonnin et al., 2020). MCR-8 mediated PEtN modification, which was identified using a MALDI-TOF-based technique that discriminates between colistin resistance mechanisms, i.e mcr-expression or chromosomal-mutations (Bonnin et al., 2020). BLASTn analysis showed that mcr-8 has a 50.23% nucleotide and 39.96% amino acid sequence identity to part of mcr-3 but has 30% to 40% amino acid sequence identity to other MCR proteins.
Ullah et al. performed an in-silico search using an MCR-8 probe to detect putative chromosomally encoded PEA transferases (Ullah et al., 2021). The search identified a nmcr-2 PEA transferases in the plant pathogen Kosakonia pseudosacchari with a 67.3% identity to MCR-8. Ullah et al. further showed that NMCR-2 is a possible progenitor of MCR-8 by evaluating their protein engineering, physiological impact, and biochemical analysis (Ullah et al., 2021). The biochemical analysis found that the functional expression of both mcr-8 and nmcr-2 in E. coli conferred the same level of resistance and physiological impact i.e., retarded growth was observed for expression of both PEA transferase (Ullah et al., 2021). The two PEA transferases were further shown to be domain compatible using the domain-swapping method (Sun et al., 2017b), which showed that the two domains of nmcr-2 were compatible and functionally exchangeable with mcr-8 counterparts (Ullah et al., 2021). This validates the phylogenetic position of nmcr-2 and mcr-8.
Analysis of mcr-8’s genetic environment found that it is usually flanked by two complete IS903 elements, with both 50 bp IRL and IRR located upstream, and downstream, respectively (Wang X et al., 2019; Wu et al., 2020). It is also associated with ISEcl1 and ISKpn26, located upstream and downstream, respectively (Yang et al., 2019). Yang et al. found that the insertion of ISEcl7 was an independent event and may not be involved in mobilizing mcr-8 (Yang et al., 2019). The formation of circular intermediates via IS903B elements to mobilize mcr-8 remains unknown; however, the association of mcr-8 to ISs facilitates both the transmission of mcr-8 and its close association with other resistance genes (Wu et al., 2020); specifically, it has been found associated with aminoglycoside resistance genes (Wu et al., 2020), β-lactamase genes (Wang X et al., 2019) and tmexCD1-toprJ1 genes that encode a novel plasmid-mediated efflux pump that confers resistance to tigecycline (Sun et al., 2020).
Mcr-8 is commonly found in K. pneumoniae (Wang et al., 2018b; Bonnin et al., 2020; Wu et al., 2020; Sun et al., 2020; Salloum et al., 2020; Ngbede et al., 2020; Nabti et al., 2020), Wang et al. performed a phylogenetic analysis of the mcr-positive K. pneumoniae isolates identified in their study and found that the mcr-positive K. pneumoniae were genetically diverse (Wang et al., 2018b). K. pneumoniae express colistin resistance usually through chromosomal mutations (Mmatli et al., 2020). Wu et al. suggest the expression of mcr-8 may have synergistic effects with the chromosomal mutations within the TCS, resulting in heterogenous colistin resistance mechanisms (Wu et al., 2020). Mcr-8 has also been identified in Klebsiella quasipneumoniae (Yang et al., 2019) and R. ornithinolytica as well as on a diverse range of Inc groups: IncFII, IncQ, IncR, IncFIIK, IncFIA and IncA/C (Table S5); these will expectedly accelerate the dissemination of mcr-8 and other ARGs within and outside Enterobacteriaceae (Wang X et al., 2019; Wu et al., 2020). Mcr -8 genes have reported in seven countries with China being the most prevalent (Table S3) and have been identified in four Enterobacteriaceae species, K. pneumoniae, K. quasipneumoniae, R. ornithinolytica and E. coli.
Mcr-9 gene was identified by Carroll et al. (2019) in a colistin susceptible S. Typhimurium isolated from a human patient in Washington State in 2010. Further, the mcr-9 gene was found associated with wbuC, encoding a cupin fold metalloprotein, located downstream the gene (Yuan et al., 2019; Kieffer et al., 2019; Osei Sekyere et al., 2020). The presence of mcr-9 on a plasmid replicon facilitates its dissemination globally and across Enterobacteriaceae (Carroll et al., 2019). It has thereafter been identified in more clinical settings (Chavda et al., 2019; Yuan et al., 2019; Kananizadeh et al., 2020; Lin et al., 2020; Ngbede et al., 2020; Osei Sekyere et al., 2020; Ding et al., 2021; Ribeiro et al., 2021) and in food-producing animals (Cha et al., 2020; Khanawapee et al., 2021). Among the 355 genomes identified during a BLAST analysis, 65 were associated with a plasmid replicon, which may facilitate their dissemination globally (Carroll et al., 2019). Mcr -9 cassette is usually located either on a IncHI2 and/or IncHI2A replicon or chromosomally encoded (Table S5) (Carroll et al., 2019; Li et al., 2020; Lin et al., 2020; Tyson et al., 2020). It is widely distributed within Enterobacteriaceae, particularly within Enterobacter sp (Chavda et al., 2019; Yuan et al., 2019; Kananizadeh et al., 2020; Lin et al., 2020; Ding et al., 2021)., Citrobacter telavivum (Ribeiro et al., 2021), Salmonella sp (Carroll et al., 2019; Cha et al., 2020; Tyson et al., 2020)., and E. coli (Kieffer et al., 2019; Khanawapee et al., 2021) (Table S4). It has further been reported worldwide in nine countries with Sweden and Thailand having the most reports (Table S3).
A database search using mcr-9 as a template identified a chromosomal-encoded mcr-9-like gene within a Buttiauxella species isolate, a member of the Enterobacteriaceae (Kieffer et al., 2019). The identified mcr-like protein, MCR-BG, was isolated from B. gaviniae, and was found to have an 84% amino acid identity to MCR-9 enzyme.CR-BG, similar to mcr-9, has the wbuC gene located downstream (Kieffer et al., 2019).
Similar results were reported by Yuan et al. with a PEtN transferase isolated from B. brennerae (Yuan et al., 2019). It had 86.83% protein sequence identity to MCR-9 at a 100% coverage (Yuan et al., 2019). Further, the wbuC located adjacent the mcr-9 gene was found to be homologous to that of Buttiauxella with an 86% amino acid identity at a 98% coverage (Yuan et al., 2019). This evidence suggests that mcr-9 may have originated from Buttiauxella species and disseminated to the rest of Enterobacteriaceae family.
The dissemination of mcr-9 may have been aided by its association with IS elements flanking the cassette (Lin et al., 2020). The mcr-9 cassette has been found flanked with IS903 and IS903-like elements upstream as well as IS1R, IS26-like, and IS15DII located downstream in multiple reports (Kieffer et al., 2019; Yuan et al., 2019; Lin et al., 2020; Ding et al., 2021). The first report of the mcr-9 cassette encoded the wbuC gene, a TCS encoding qseC and qseB genes, ΔIS1R and a remnant of ΔsilR, encoding a transcriptional regulatory protein, all of mcr ch are locmcr d downstream of the mcr-9 gene. The mcr-9 cassette is thereafter flanked by intact IS903 elements (Yuan et al., 2019). Though two copies of an intact IS element should be able to form a composite transposon or a circular intermediate, an inverse-PCR assay failed to detect any intermediates (Yuan et al., 2019). However, the identification of IS903-like elements associated with mcr-9 genes suggest the acquisition of the cassette through an IS903 dependent mechanism. In some reports, the 3’region of mcr-9 is adjacent to copS (three copper resistance membrane-spanning proteins that includes rcnA, rcnR and a domain-containing DUF4942) (Li et al., 2020; Ding et al., 2021).
The mcr-10 gene was first identified in an Enterobacter roggenkampii clinical strain in China (Wang C et al., 2020) and has only been reported there since. It has a nucleotide sequence identity and amino acid sequence identity of 79.69% and 82.93% respectively to mcr-9 (Tyson et al., 2020). Therefore, like mcr-9, the mcr-10 gene shares a significant amino acid identity to the chromosomally encoded mcr-like PEtN transferase of Buttiauxella species (Wang C et al., 2020).
The Mcr-10 gene has been identified on IncFIA and IncFIB plasmids (Wang C et al., 2020; Sato et al., 2021; Xu et al., 2021; Yang et al., 2021) and adjacent to a xerC gene, which encodes a XerC type tyrosine recombinase found to mobilize adjacent genetic components such as antimicrobial resistance genes (Wang C et al., 2020). Thus, the mcr-10 gene would be presumed to have disseminated across Enterobacteriaceae family but has only been identified commonly in Enterobacter roggenkampii (Lei et al., 2020; Wang C et al., 2020; Xu et al., 2021) and once in Cronobacter sakazakii (Table S4) (Yang et al., 2021). Wang et al. and Xu et al. suggest that E. roggenkampii is an important reservoir of the mcr-10 gene (Wang C et al., 2020; Xu et al., 2021).
In the initial identification of mcr-10, the gene was adjacent XerC and flanked by two intact IS903 elements which could form a composite transposon with the potential to mediate mobilization of the mcr-10 gene (Wang C et al., 2020). The analysis of the immediate genetic environment of the IS903 element found the absence of direct repeats that are generated during an insertion event; thus, the acquisition of the region bracketed by the IS903 elements was not due to an insertion event (Wang C et al., 2020). The presence of an a ΔISEc36 downstream the mcr-10 gene was reported to make it impossible to identify the recombination sites that the XerC-type tyrosine recombinase recognizes (Wang C et al., 2020). Thus, the acquisition events of mcr-10 into pMCR-10_09006 remains unknown (Wang C et al., 2020). Mcr-10 has thereafter been (Lei et al., 2020) identified with various insertion sequences, including complete and truncated remnants of IS26 and ISKpn26, complete sequences of ISEc36 and IS1, a ΔISEcl1-like elements and recently, a new IS element designated ISCrsa1 (Lei et al., 2020; Yang et al., 2021). The identification of various IS elements downstream mcr-10-xerC suggests that this region might be a hotspot for insertions of MGEs (Wang C et al., 2020; Xu et al., 2021). The acquisition of mcr-10 is presumed to be through site-specific recombination (Yang et al., 2021), however, evidence of mobilization of mcr-10 is scarce.
Dissemination of mcr genes
Mcr genes are well disseminated within Enterobacteriaceae, being frequently identified within E. coli, K. pneumoniae and Salmonella species, in that order. Other Enterobacteriaceae species that have been identified with mcr genes include Cronobacter species, Citrobacter species, Enterobacter spp., Kluyvera species, Leclercia species, Raoultella ornithinolytica, and Shigella species (Figure 5).
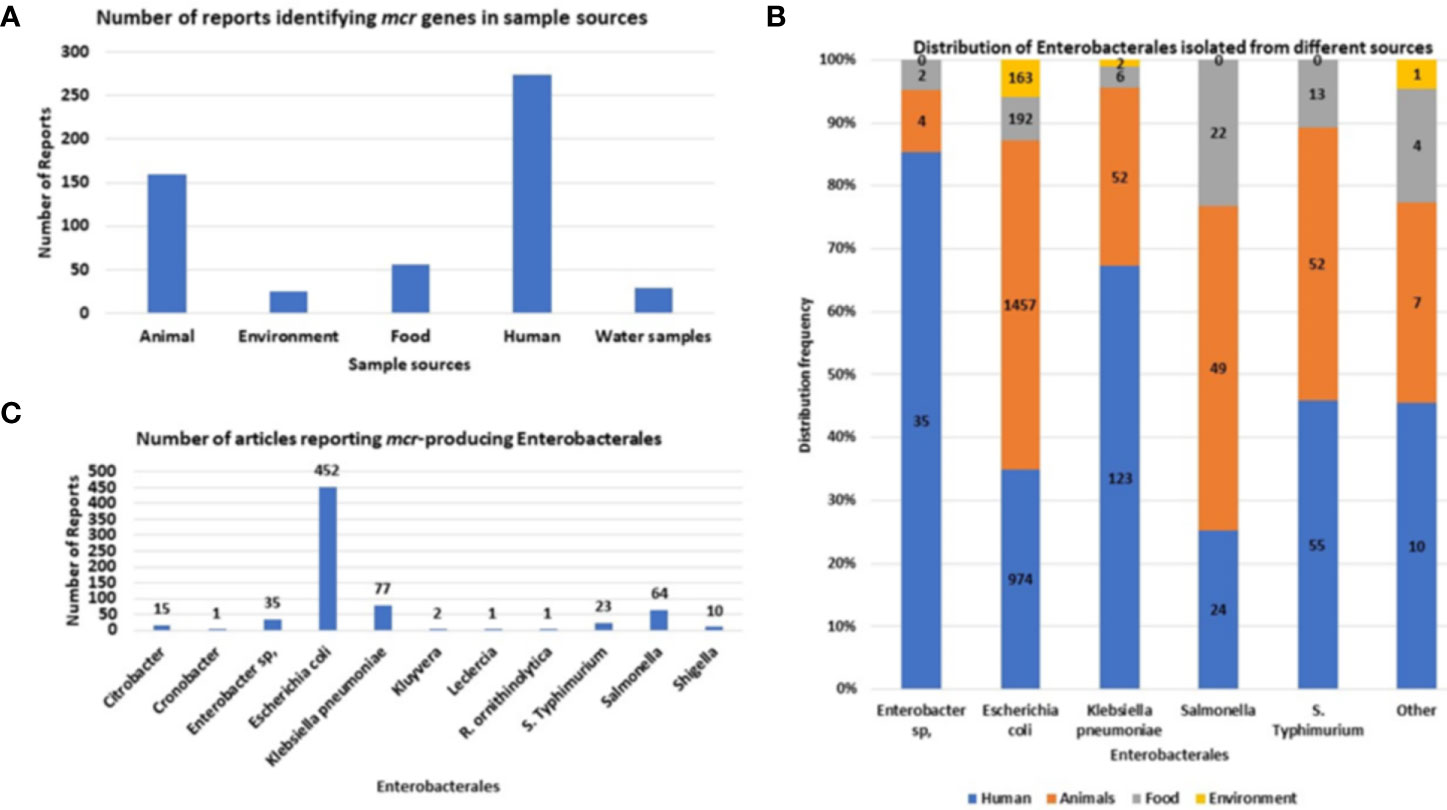
Figure 5 The sources and types of Enterobacteriaceae species identified to be harboring mcr genes. (A) Number of articles reporting samples harboring mcr-positive Enterobacteriaceae isolates. (B) Distribution of MCR-producing Enterobacteriaceae species per sample source. (C) The number of articles reporting each mcr-positive Enterobacteriaceae species.
Initially, mcr genes were commonly identified in animals. However, after evaluating the number of reports collected from 2016 to 2021, the most common report of mcr-positive Enterobacteriaceae isolates is from human specimens (Figure 5A). This is due to the increased use of colistin in the clinical setting and the elimination of colistin use in poultry, shifting the focus away from poultry samples to the dissemination of mcr genes in the clinical setting. Colistin was banned for poultry use in India, China, South Africa, European Union, Thailand, Brazil and many more (Apostolakos and Piccirillo, 2018; Shen et al., 2020), however countries such as Nigeria and Bangladesh, the use of colistin is not regulated (Ahmed et al., 2020; Anyanwu et al., 2021; Anyanwu et al., 2022). E. coli is the most common mcr-positive isolate (Figure 5C) and is usually isolated from animal and human feces samples.
In human specimens, mcr genes are frequently isolated from E. coli, K. pneumoniae, and S. Typhimurium from mostly stool and urine samples (Table S2. In animals, the most common samples that were screened for mcr genes, include feces and meat, which commonly harbored E. coli, K. pneumoniae and Salmonella isolates (Table S2). Other sources identified includes the environment, which are mostly made up of wastewater samples which includes water samples from sewer sheds and wastewater plants treatments facilities. Other environmental samples includes soils and aquaculture such as duck and fish. In the environment, mcr-positive isolates comprised only three species: E. coli, K. pneumoniae, and Kluyvera sp (Table S2). Lastly, mcr-positive species have been also isolated from livestock meat and vegetable samples, being isolated from mostly packaged meat and fresh vegetables in retail stores. These species include: Cronobacter sp., Enterobacter sp., E. coli, K. pneumoniae, Salmonella, and S. Typhimurium (Figure 5B).
Amongst the 70 countries identified with mcr genes, countries such as Thailand harbored six mcr-genes (mcr-1, mcr-3.mcr-6, mcr-7, mcr-8 and mcr-9) and China harbored eight genes (mcr-1, mcr-3, mcr-4, mcr-5, mcr-7, mcr-8, mcr-9 and mcr-10). Other countries such as the USA, Turkey, Spain, Nigeria, Korea, Japan, Italy, France, England, Czech Republic, Cambodia, Brazil, Belgium, and Bangladesh harbored three to four mcr-genes, each inclusive of the mcr-1 variants (Figure 3B, Table S3). Most mcr genes have been reported in studies from China (in Figure 3C), which is due to the large volumes of articles being published on mcr epidemiology from China.
It has thereafter been seen in China, that clones within mcr-positive E. coli, which includes ST744, ST410, ST10, ST43, ST101 and ST206, have been identified in all four sources: food, environment, animals, and humans (Table S4). These clones have, however, also disseminated globally, where ST744 has been identified in humans in eleven countries, in food in five countries, in animals in five countries and in the environment only in China. The mcr-positive E. coli ST10 strain is the most widely distributed within E. coli. It has been identified in animals in seventeen countries, humans in sixteen countries, the environment in five countries, and in food in five countries. Mcr-positive E. coli clones within each country are usually found in both animals and humans, seen with ST744, ST69, ST117, ST131 and ST354 in Italy.
S. Typhimurium is also well distributed globally, with the S. Typhimurium ST34 strain identified in China (Yi et al., 2017), Colombia (Saavedra et al., 2017), Denmark (Litrup et al., 2017), Germany (Borowiak et al., 2019), and the United Kingdom (Doumith et al., 2016), in both animals and humans. In China, the S. Typhimurium ST34 strain has been identified in both humans and animals, and in food samples and human specimens in Germany. Similar results are seen in the monophasic variants of S. Typhimurium serovars (S.) 1,4,[5],12:i:-, and S. 4,[5],12:i:-, where ST34 was the only clone identified with mcr genes (Litrup et al., 2017; Migura-Garcia et al., 2020). The S. 1,4,[5],12:i:- ST34 strain has only been identified in Portugal in animals, food and humans, and the S. 4,[5],12:i:- has been identified in Belgium, Canada, Italy, Switzerland and the United States in animals and humans (Table S4).
The direct transmission of mcr-positive Enterobacteriaceae (MCRPE) isolates from animals and humans is discussed in this review (See Risk factors) and the data mentioned above, highlights colistin-treated livestock as the reservoir of mcr-genes and the route of transmission to humans.
Mcr-positive isolates are commonly identified harboring other resistance genes such as bla genes, mdfA, aadA12, sul123, etc. Thus, similar to mcr genes (Figure 3C), other antimicrobial resistance genes (ARGs) are predominantly located in China (Figure 6), followed by Germany, Denmark, and England. The data however is biased towards China, because the large volumes of mcr epidemiology papers published from China. Notably, several important ARGs are co-hosted by MCRPE isolates (Figure 3A). A Table showing the distribution of mcr-positive isolates hosting other ARGs is shown in Table S5.
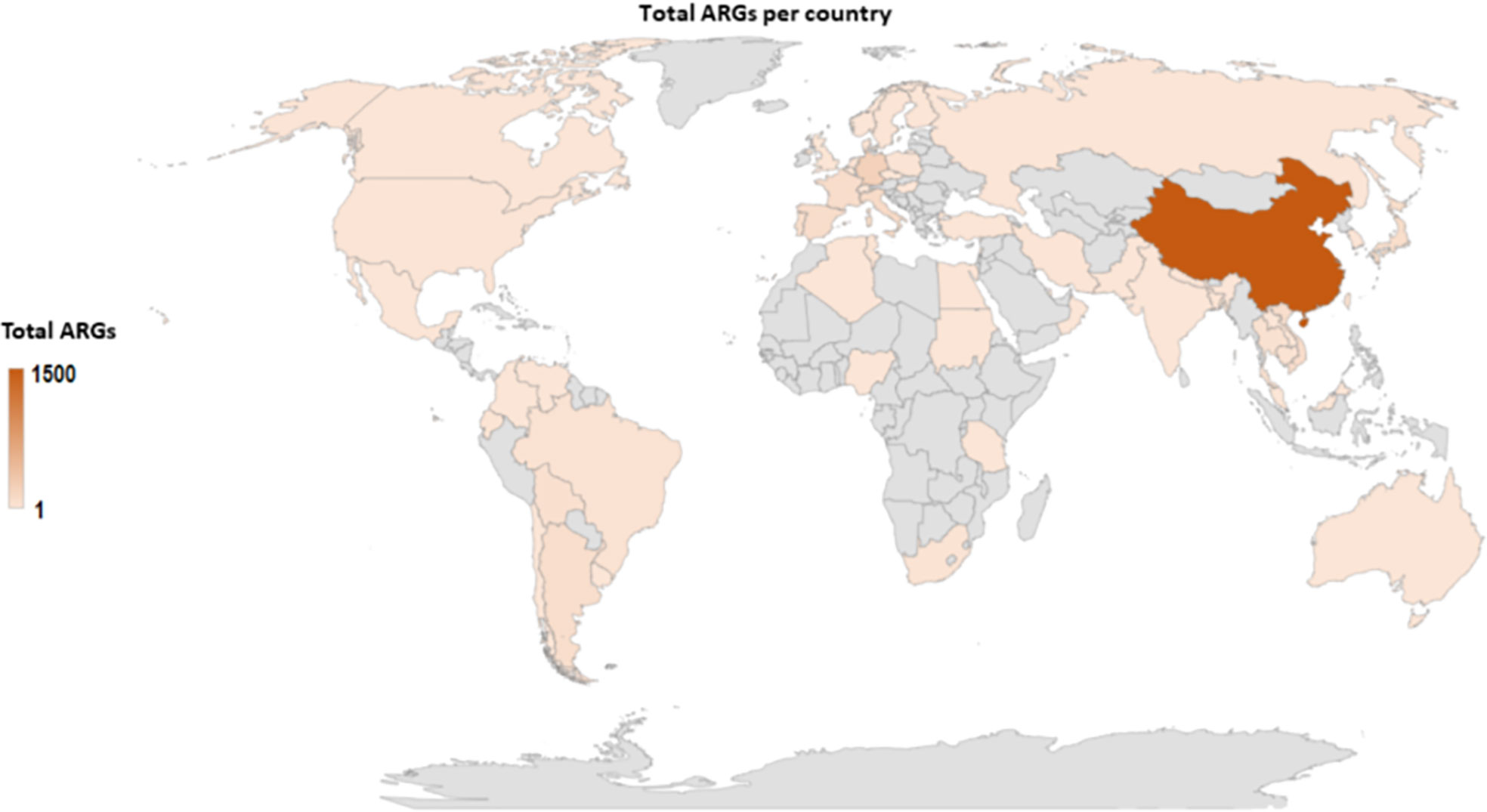
Figure 6 Global map showing the geographical distribution of other antibiotic resistance genes found in identified MCRPE isolates.
MCR -protein structures and mechanisms of action
MCR-1 is a 541 amino acid, integral membrane protein made up of two domains: a C-terminal periplasmic catalytic domain and an N-terminal 5’-helix transmembrane domain (Liu et al., 2018; Wei P et al., 2018). The transmembrane (TM) domain is made up of 5-membrane spanning α-helixes with an overall positive charge because of positively charged residues, which interact with the negatively charged phospholipid head groups of the membrane bilayer (Wei P et al., 2018). The TM domain anchors the protein into the inner membrane and is connected to the catalytic domain through a bridging helix (Wei P et al., 2018)’. Sun et al. found that the acquisition of the transmembrane domain allows for the correct localization of PEtN transferases within the periplasm and that the deletion of this domain, impairs MCR-2’s ability to confer colistin resistance (Sun et al., 2017b). The catalytic domain is made up of both positively and negatively charged residues, where the negatively charged residues create the binding pockets and allow them to be buried within the domain (Wei P et al., 2018).
The overall shape of MCR-1 is a hemispherical shape composed of several β-α-β-α motifs made up of β-strands sandwiched between α-helical structures (Hu et al., 2016; Stojanoski et al., 2016). The catalytic domain has an alkaline phosphatase family α/β/α fold, also with a hemispheric shape equipped with a zinc binding pocket containing a conserved phosphothreonine-285 residue (Figure 7) structure (Hu et al., 2016; Stojanoski et al., 2016). The pocket is common to PEtN transferases and alkaline phosphatase; each enzyme, however, differs in the orientation and number of zinc molecules (Stojanoski et al., 2016). The pocket is proposed to be critical for the nucleophilic attack of the phosphate of the donor PE substrate by stabilizing the alkoxide of the Thr285 (Stojanoski et al., 2016). The Thr285 residue, is the catalytic nucleophile that acts as both a nucleophile and a PEtN acceptor during the catalytic mechanisms. The residue is critical to MCR-1’s function as mutations to this residue significantly decrease MCR-1’s activity (Stojanoski et al., 2016; Hinchliffe et al., 2017). The catalytic domain of MCR-1 is further made up of six cysteine residues that form three disulphide bonds, Cys281/Cys291, Cys356/Cys364 and Cys414/Cys422 (Stojanoski et al., 2016) that stabilize and anchor the β-α-β-α motifs (Hu et al., 2016). These disulphide bonds are conserved, and equivalents are present in both LptA and EptC transferases (Stojanoski et al., 2016), although LptA has four more cysteine residues.
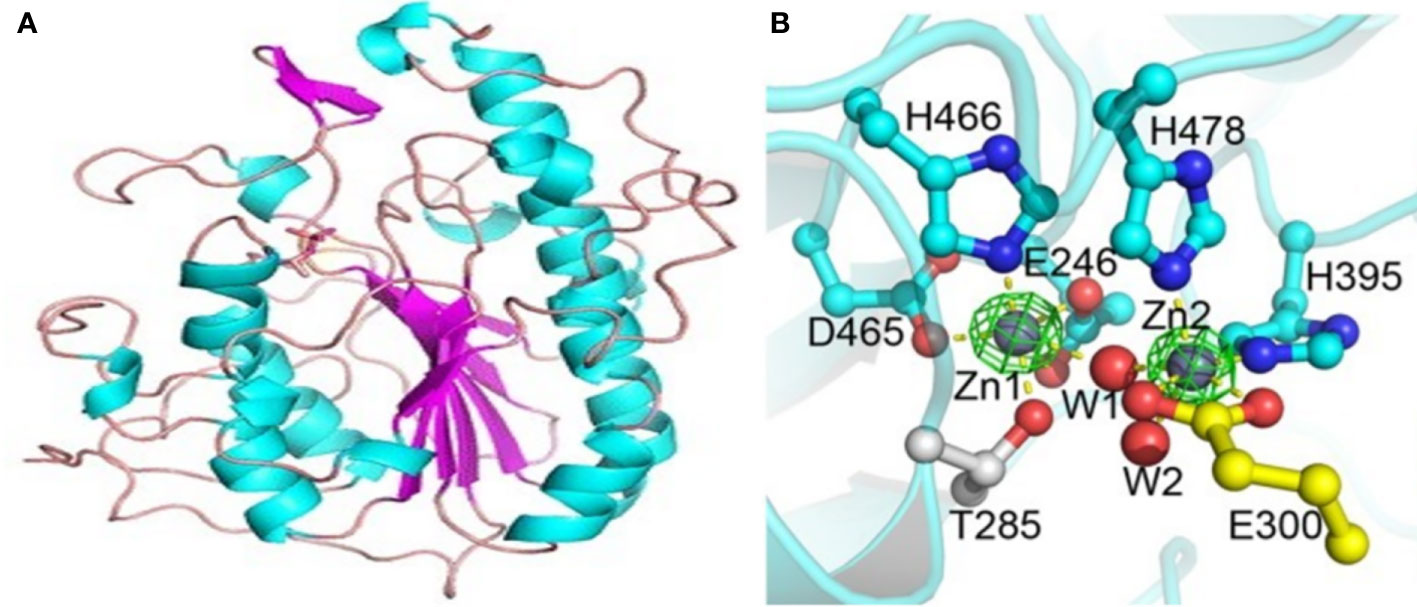
Figure 7 Structure of the catalytic domain of the MCR-1 protein and the five important residues required for its catalytic activity. (A) The MCR-1’s hemispherical shape composed of the β-α-β fold made up of helices (cyan), strands (purple) and loops (salmon). Image was obtained from Stojanoski et al. (2016). (B) A ball and stick model showing the conserved active site residues of MCR-1 consisting of Asp (D)465, Glu (E)246, His (H)466, His(H)395 and Thr (T)285, coordinated to the zinc ions (Zn1 and Zn2) and water molecules (W1 and W2). Image was obtained from Ma et al. (2016).
The zinc binding pocket is in the active site of MCR-1 and is made up of conserved residues viz., Asp465, Glu246, His466, His395, and Thr285, and is made up of zinc molecules (Figure 7) (Gao et al., 2016; Stojanoski et al., 2016). The conserved residues are critical for the substrate binding of MCR-1 (Gao et al., 2016) and mutations in these amino acids abolish the MCR-1 activity, lowering the colistin MIC value down to control levels (Hu et al., 2016). The five conserved residues are well conserved in MCR-1, LptA, EptC and other alkaline phosphatase family members, although the nucleophile residue may differ between enzymes (Hinchliffe et al., 2017). MCR-1 has three zinc molecules clustered around the active site: Zn1 is buried within the active site and coordinated by the phosphate oxygen of Thr285; it is conserved in LptA and EptC (Stojanoski et al., 2016). Zn2 is coordinated by His395, bound to a phosphate oxygen of Thr285 and three water molecules, forming a trigonal bipyramidal configuration, (Figure 7) (Stojanoski et al., 2016). Hu et al. found that Zn2 is not critical for maintaining MCR-1 activity (Hu et al., 2016).
This may be because it is located on the surface of the active site. A structural comparison between LptA, EptC and MCR-1 found that the Zn2 site is less conserved. Hence, Hinchliffe et al. concluded that an intact Zn2 site was not a prerequisite for MCR-1’s catalytic activity (Hinchliffe et al., 2017). However, His395 residue is important for the structure and activity of MCR-1. The Zn3 molecule, unlike the other zinc molecules, is not coordinated by the conserved residues but is tetrahedrally coordinated by four water molecules. The seven water molecules found in the active site, coordinating Zn2 and Zn3, are embedded in the protein through hydrogen bonding (Stojanoski et al., 2016). The catalytic domain is thus a zinc-rich area (Figure 7B), suggesting that MCR-1 may be able to attract zinc ions at different levels into the domain (Hu et al., 2016). The role of zinc in mediating lipid A modification, the catalytic mechanism, was evaluated by measuring the colistin MIC in the presence and absence of EDTA. Hinchliffe et al. found clear reductions in colistin MIC values after EDTA treatment and concluded that the presence of zinc in the active site was important for MCR-1 function (Hinchliffe et al., 2017).
MCR-1 mediates the transfer of PEtN from the primary phosphatidylethanolamine (PE) to lipid A moiety at the 4’-phosphate group through a ‘ping-pong’ mechanism seen across MCR family regardless of their evolutionary placement (Hu et al., 2016; Gao et al., 2016; Xu et al., 2018; Zhang et al., 2019; Zhang et al., 2019a). Wei et al. and Liu et al. investigated the molecular mechanism involved in this process (Wei P et al., 2018; Liu et al., 2018). Wei et al. identified that the active site within the catalytic domain is made up of two binding pockets: the PEtN binding pocket and the Lipid A binding pocket (Wei P et al., 2018). These binding pockets are in close proximity and are integrated within each other (Wei P et al., 2018). Wei et al. observed that ethanolamine was a good analogue of PEtN and D-glucose for lipid A (Wei P et al., 2018). Ethanolamine (ETA) is a good analogue as it mimics the covalent PEtN-enzyme intermediate. Lipid A is made up of two glucosamine units that are attached to acyl chains (Wei P et al., 2018). Thus, monosaccharides and disaccharides can mimic a Lipid A molecule as they are both hexacyclic compounds (Wei P et al., 2018; Liu et al., 2018).
The PEtN binding pocket that accommodates ETA is made up of Glu246, Thr285, Asn329, Lys333, His395, Asp465, His466 and His478 residues (Wei P et al., 2018). This pocket is responsible for the binding of phosphatidyl-ethanolamine (PE), which is stabilized through hydrogen bonding with Asn329, a water molecule, and phosphorylated Thr285 (Wei P et al., 2018). Analysis of the entry of ETA into the pocket showed that the pocket undergoes conformational changes to adjust to the substrate. These included a 50° rotation of the His395, accomplished by breaking a hydrogen bond with a water molecule (wat7) and creating a new hydrogen bond with a new water molecule (wat8) (Wei P et al., 2018). This releases the wat7 molecule from the PEtN binding pocket and thus, collectively, the conformational changes create room to accommodate the substrate, ETA. The entry of ETA further deprotonates the phosphorylated Thr285 residue, forming a PEtN molecule and a nucleophilic state at Thr285 (Wei P et al., 2018). The Zn1 molecule subsequently facilitates the stabilization of the nucleophilic state of Thr285 that allows that residue to thereafter attack the PEtN, creating a PEtN enzyme intermediate. This constitutes the first step of the MCR-1-mediated catalytic process (Wei P et al., 2018).
Wei et al. and Liu et al. investigated the mechanisms behind PEtN-transfer reaction with a lipid A analogue, D-glucose, identified within the Lipid A binding pocket (Wei P et al., 2018; Liu et al., 2018). This pocket is made up of the following residues: Thr283, Ser284, Tyr287, Pro481 and Asn482, that is located near the Thr285 residue. The analysis of D-glucose found that D-glucose was held in the pocket by Thr285, Ser284 and Asn482 and was flanked by Tyr281 and Pro481 forming a sandwich structure of a π-π conjugative interaction (Wei P et al., 2018; Liu et al., 2018). The importance of the residues in both PEtN and Lipid A binding pockets in MCR-1 activity were evaluated through mutation construction. Mutations within Thr285, Asn329, Lys333, Glu246, His398, Asp465, His466 and His478 of the PEtN binding pocket and Tyr281 and Pro481 of the Lipid A binding pocket, resulted in a decrease in colistin MIC values (Wei P et al., 2018). However, these mutations did not disrupt the protein expression and membrane localization of the MCR-1 protein. The mutations of Tyr281 and Pro481 in the lipid A binding pocket highlights the importance of this pocket in the MCR-1 activity. Wei et al. suggests that it could bind to the glucosamine group of Lipid A and allows for the transfer of the reactive phosphate group from PEtN enzyme intermediate to lipid A (Wei P et al., 2018). Thus, the second step of the catalytic process therefore involves the transfer of PEtN to Lipid A that is situated in a nearby pocket (Wei P et al., 2018).
The MCR-1 catalytic domain is therefore made up of a PEtN binding pocket and a Lipid A binding pocket that mediate the transfer of PEtN from the PEtN binding pocket to lipid A moiety at the 4’-phosphate group (Hu et al., 2016; Zhang et al., 2017). This activity is validated through MALDI-TOF mass spectrometry (MS), provides in-vivo evidence of the MCR-1 activity (Gao et al., 2016; Anandan et al., 2017). This is accomplished by evaluating the LPS lipid A MS profile in the presence and absence of mcr-1 expression as adding PEtN increases Lipid A mass units (+123) (Gao et al., 2016; Xu et al., 2018; Dortet et al., 2018). In the absence of mcr-1 expression, there is a single Lipid A peak (m/z = 1797.4) and in the presence of mcr-1 expression, there are two unique peaks. The first peak is the wildtype bis-phosphorylated, hexa-acylated lipid A (m/z = 1797.4) and the second peak is the PPEtN-4’ (m/z = 1920.5) (Xu et al., 2018).
MCR-1 has thus proven to have PEtN transferase activity and has a sequence and structural similarity to EptC and LptA PEtN transferases in Campylobacter jejuni (Fage et al., 2014) and Neisseria meningitidis (Wanty et al., 2013), respectively (Stojanoski et al., 2016; Gao et al., 2016; Hinchliffe et al., 2017). The active sites of the three PEtN transferases are highly conserved, i.e., the phosphorylated catalytic nucleophile Thr285, disulphide bonds, zinc binding pockets and the conserved active site residues (Hinchliffe et al., 2017). Therefore, these three sites are important for MCR-1 function; these conserved mechanistic activities and binding pockets will allow for the use of MCR-1 inhibitors to also inhibit chromosomally encoded PEtN transferases, restoring colistin susceptibility in acquired and intrinsically resistant bacteria (Stojanoski et al., 2016).
All ten MCR proteins (MCR-1 to MCR-10) encode a putative PEtN transferase made up of two domains- transmembrane helix and a periplasmic domain with the catalytic center (Yin et al., 2017; Borowiak et al., 2017; Wang et al., 2018b; Yang et al., 2018; Zhang et al., 2019a; Zhang et al., 2019b; Wang X et al., 2019; Carroll et al., 2019; Xu et al., 2021). These proteins further have the same conversed properties required for PEtN activity, i.e., the active residues (E246, T285, K333, H395, D465, H466, E468 and H478) and the six cysteine residues required for the three-sulphide bonds (Xavier et al., 2016; Sun et al., 2017b; Teo et al., 2018; Wang et al., 2018b; Wang et al., 2018c). A phylogenetic analysis of all MCR proteins shows that MCR-1, MCR-2, MCR-6 and Neisseria LptA were part of a parallel evolutionary event for functional acquisition of colistin resistance during some environmental selection pressure, i.e., the intensive use of colistin in animals feed (Sun et al., 2017b; Wang et al., 2017; Wang Y et al., 2020; Islam et al., 2020). Thereafter, MCR-3, MCR-4, MCR-7, and MCR-9 have similar amino acid sequences (Kieffer et al., 2019; He et al., 2021), and further, are closely related at a structural level (Carroll et al., 2019). However, MCR-5, MCR-8 and MCR-10 have a low identity to the other MCR proteins, though a sequence alignment shows that all ten MCR proteins each encode the conserved active residues, six cysteine residues and each is capable of LPS modifications (Borowiak et al., 2017; Wang et al., 2018b; Wang X et al., 2019; Carroll et al., 2019). Zhang et al. further showed that MCR-5 and MCR-1/2 have different interdomain relationships, and their domains (TM1-MCR-5, TM5-MCR-1, TM2-MCR-5 and TM5-MCR-2) are mostly incompatible (Zhang et al., 2019b). MCR-5, however, has 100% amino acid sequence identities to PEtN transferases identified in Pigmentiphaga genus and Cupriavidus gilardii. The first 404 amino acids of the MCR-5 protein also have a high identity to a hypothetical protein from Pseudomonas aeruginosa (Borowiak et al., 2017).
All MCR proteins therefore, like MCR-1 mediate the transfer of PEtN from the PEtN binding pocket to the Lipid A moiety at the 4’-phosphate group. This reduces the MCR-producing microorganism’s susceptibility to colistin (Kieffer et al., 2018; Long et al., 2019). Thus, the difference in nucleotide and amino acid sequences between the MCR proteins have no impact on their PEtN activity. However, Yang et al., found that mcr genes, i.e., the mcr-variants, may encode compensatory mutations that mitigate the expression of mcr genes or might affect PEtN’s activity (Yang et al., 2020). This was found with mcr-4.3 identified by Teo et al. which encodes two missense mutations, V179G and V236F, that inactivated the PEtN activity (Chavda et al., 2018; Teo et al., 2018). A comparative alignment of the MCR-4.3 to MCR-1 and MCR-2 found that the active residues were conserved in MCR-4.3 (Teo et al., 2018). However, an MS spectrum of lipid A in mcr-4.3-expressing isolates showed mcr-4.3 failed to modify lipid A (Chavda et al., 2018). The transformation of E. coli with an mcr-4.3 expressing vector resulted in no difference in colistin MIC values, suggesting that mcr-4.3 does not confer colistin resistance (Chavda et al., 2018; Teo et al., 2018). Thus, some compensatory mutations in mcr seen in mcr-4.3 significantly altered the PEtN transferase activity of mcr-4.3 (Chavda et al., 2018).
Fitness cost and compensatory evolution
The acquisition of mcr-1 bearing plasmids has been shown to have a beneficial effect on the host, improving bacterial survival in the presence of colistin treatment (Nang et al., 2018). The acquisition and expression of the mcr-1 gene results in the incorporation of MCR-1 into the bacterial membrane and the phosphoethanolamine (PEtN) modification of the lipopolysaccharides (LPS) (Li et al., 2017). However, multiple studies have shown that the expression of mcr-1 imposes a fitness cost by placing an energy burden on the host (He et al., 2017), impairing cell growth, and diminishing bacterial fitness (Li et al., 2017). Andersson et al. explained that a significant fitness cost is seen when the susceptible strain outcompetes the resistant strain in an antibiotic free environment (Andersson, 2006). The imposed fitness cost of a conjugative plasmid harboring a resistance gene has been shown to be because of various factors such as the resistance mechanisms, the bacterial species, and the antibiotic (Nang et al., 2018).
Evaluating the cost of colistin resistance on resistant strains, it is shown that certain chromosomal mutations within genes such as mgrB have no significant fitness cost on K. pneumoniae (Cannatelli et al., 2015) whilst genes such as pmrB, do (Giordano et al., 2019). Giordano et al. found that the expression of the mcr-1 gene imposes less of a burden on K. pneumoniae than the mutated pmrB gene (Giordano et al., 2019). The chromosomal mutations within pmrAB, acrab, phoPQ, and mgrB genes have been shown to mediate colistin resistance through the phosphorylation of lipid A (Cheng et al., 2016; Phan et al., 2017; Cannatelli et al., 2017). The acrab genes encode a multidrug efflux system that confers resistance to a wide variety of agents (Mmatli et al., 2020). Tietgen et al. discovered that the fitness costs imposed by mcr-1 plasmid carriage, such as growth rates and cytotoxicity, could be species-specific (Tietgen et al., 2018). Particularly, the acquisition of an mcr-1-harboring plasmid in K. pneumoniae has been shown to impose a significant fitness cost on the host (Nang et al., 2018), but multiple studies have shown that the acquisition of an IncI2 plasmid, in various sizes, carrying mcr-1 had no significant cost on the host (Zheng et al., 2017; Wang R et al., 2018). This may be plasmid and/or species specific or may be due to acquired compensatory mutations within the IncI2 plasmid. However, the overexpression of mcr-1 was seen to result in profound changes in the architecture of the outer membrane, resulting in the loss of membrane structural integrity and causing leakage of cellular cytoplasm, resulting in cell death (Li et al., 2017; Yang Q et al., 2017).
Yang et al. found that the overexpression of mcr-1 imposes a significant cost by decreasing the growth rate, causing significant membrane degradation and moderate fitness loss’ (Yang Q et al., 2017). They came to this conclusion through evaluating the effects of mcr-1 expression on the relative fitness of E. coli TOP10 and found that with increasing levels of mcr-1 expression resulted in a significant fitness burden on the host (Li et al., 2017; Yang Q et al., 2017). An analysis of the cellular morphology in the mcr-1 overexpressed strains using a transmission electron microscopy showed cell architecture alterations and a complete loss of cellular morphology. The overexpression of mcr-1 altered the structural integrity of the outer membrane and further impaired the cell membrane (Li et al., 2017). They finally found that the embedding of MCR-1 into the outer membrane and the PEtN modifications of Lipid A were the leading factors contributing to fitness cost and membrane degradation. Therefore, the expression of mcr-1 in host strains is tightly controlled to regulate the mcr-1 fitness cost (Yang Q et al., 2017).
This was further seen in mcr-3 expression, where expression was shown to significantly impair the cell wall integrity and decrease the electron density of mcr-3 producing E. coli 94. Yang et al. evaluated the impact of mcr-3 expression on bacterial fitness using two mcr-3 variants, mcr-3.1 and mcr-3.5 (T488I, M23V, A456E) (Yang et al., 2020). The study showed that although the expression of both variants imposed a fitness cost and impaired the cell wall integrity, the expression of mcr-3.5 was less costly (Yang et al., 2020). Yang et al. showed that the amino acid substitutions A457V and T448I, seen in mcr-3.5, had strong compensatory effects when introduced in mcr-3.1 (Yang et al., 2020). The introduction of these substitutions, individually, resulted in an increased fitness of up to 45%. However, double substitutions demonstrated a negative epistasis. Yang et al. further evaluated the fitness cost imposed by mcr-3 expression and compared it to that of mcr-1 and found that the expression of both mcr genes had an impact on the bacterial fitness; however, the mcr-3.1 and mcr-3.5 variants imposed less of a fitness cost (Yang et al., 2020).Zhang et al. compared the physiological effect of mcr-1, mcr-3, and mcr-4 expression in recombinant strain of E. coli MG1655 and observed inhibited bacterial growth with each mcr gene, with mcr-4 having less of a physiological impact (Zhang et al., 2019a).
Expression of mcr gene is therefore quite toxic for the bacterial host, and the acquisition of an mcr-bearing plasmid thereafter results in compensatory adaptation that allows for the maintenance of high-cost conjugative plasmids (Dahlberg and Chao, 2003; Yang YQ et al., 2017; Ma et al., 2018). Dahlberg and Chao found that the cost of plasmid carriage is reduced over long-term culture because of compensatory mutations (Dahlberg and Chao, 2003). The host chromosome or plasmid evolves compensatory mutations that, in the case of plasmids, enhance the fitness of the host, and in the case of chromosomal mutations, aid the host in evolving towards new growth conditions by decreasing the plasmid carriage cost (Dahlberg and Chao, 2003). This has been seen in multiple studies where mcr-1 bearing plasmids initially imposed a biological cost on the transformant. However, overtime, the cost of plasmid carriage in long-term cultures was largely compensated for and plasmids were stably maintained through passages (Zhang et al., 2017; Li et al., 2017; Tietgen et al., 2018; Ma et al., 2018; Giordano et al., 2019).
Ma et al. investigated the potential mechanisms involved in compensatory adaptation through comparative genomics and identified single nucleotide polymorphisms in several genes (Ma et al., 2018). Amongst the genes identified in this study are dnaK, which encodes a molecular chaperon involved in chromosomal DNA replication, and cpoB, which encodes an RHS repeat protein involved in maintaining the cell envelope integrity during cellular division (Ma et al., 2018). These two genes are located on the chromosome and have non-synonymous single nucleotide polymorphisms (SNPs). The role of these genes in reducing plasmid carriage cost is unknown and may represent novel mechanisms (Ma et al., 2018). The study further shows that bacteria may use different strategies to reduce the fitness cost of plasmid carriage under different environmental conditions (Ma et al., 2018).
In the absence of antibiotics (colistin) selective pressure, mcr-1 bearing plasmids have been shown to be less maintained and that the complete elimination of the mcr-1 gene within a population is possible (Nang et al., 2018). Nang et al. performed a plasmid stability assay and found that in the absence of colistin, there was a gradual loss of the mcr-1 plasmid, there was a decrease in its maintenance within the population. This may be due to the instability of mcr-1 harboring plasmids (Nang et al., 2018). Arcilla et al. have shown the complete elimination of mcr-1 bearing bacteria in travelers returning to their home country after a month (Arcilla et al., 2016).
Risk factors for acquiring mcr genes
Use of colistin in livestock
Antimicrobials are commonly administered by farmers to food animals such as pigs, calves, and poultry for prophylaxis and therapeutics purposes, and as a growth promoter (Carrique-Mas et al., 2015; Nguyen et al., 2016; Monte et al., 2017). Studies around the world have shown that the use of colistin as a growth promoter results in a high frequency of mcr-1 positive, colistin-resistant Enterobacteriaceae because of the excessive use of colistin in the farming environment. The selection pressure led to the acquisition and spread of mcr-1 genes (Liu et al., 2016; Nguyen et al., 2016; Monte et al., 2017; Trung et al., 2017; Shen et al., 2018; Hille et al., 2018; Clemente et al., 2019). The presence of mcr-1 in food animals had a serious potential to spread MCRPE to humans via foodborne and direct contact transmission routes (Liu et al., 2016; Trung et al., 2017; Shen et al., 2018).
The pharmaceutical form of colistin is indicated for oral administration and is present in a powder or solution form (Hille et al., 2018). Studies have shown that the drug is poorly absorbed in the digestive tract after oral administration and can be excreted in high levels through the feces (Rhouma et al., 2015; Peng et al., 2021). This results in the presence of colistin or its active metabolites in the environment alongside the MCRPE isolates from the animal feces (Shen et al., 2018). Rhouma et al. found that the movement of colistin through the gastric passage of pigs, produced degradation products of colistin made of several metabolites that have lost some colistin side chains. These metabolites however had increased antimicrobial activity as compared to the original form of colistin (Rhouma et al., 2015). Xia et al. (2019) measured the concentration of colistin in animal feeds and fresh manure in five swine farms and found that the average concentration of colistin in animal feed on farm 4 was 60 mg/kg, which was scientifically higher than other farms. The fresh manure samples collected on farm 4 had the highest concentration of 17,383 µg/kg. Other farms had colistin concentrations in their fresh manures ranging from 140 µg/kg in farms 1, 2 and 5, to 7,529 µg/kg in farm 3 (Xia et al., 2019). The researchers also discovered a strong relationship between colistin concentration and the relative abundance of mcr-1 genes in fresh manure (Xia et al., 2019). The use of this manure as a fertilizer in agriculture or feeding farmed fish can result in the contamination of agriculture and, in some cases, the aquatic environment, polluting rivers and lakes (Luo et al., 2017; Shen et al., 2018).
This therefore led to the ban on colistin use for growth promotion and disease prevention in the food industry to preserve their effectiveness in human medicine as their use contributes to the rising threat of antibiotic resistance (World Health Organization, 2017). This colistin ban however, was only adopted in some countries. Randall et al. (2018) also demonstrated that discontinuing colistin in farming can result in the elimination of mcr-1. This longitudinal study evaluated the presence of mcr-1 in pig feces and slurry at two time points (Randall et al., 2018). At the beginning of the study, the majority of the pigs were colonized by mcr-1 producing E. coli. Twenty months after cessation of colistin use and implementation of hygiene and other measures, mcr-1 producing E. coli was not detected in samples. These results thus highlight that the use of colistin is the driving force behind the spread of colistin resistance genes and its stability in the population (Randall et al., 2018). Mcr-1 has however, been identified on conjugative plasmids encoding other resistance genes such as floR and sul123. This allows for the use of other antibiotics such as chloramphenicol and sulfonamide to simultaneously co-select the mcr-1 gene, allowing for its stability and dissemination within the conjugative plasmid (Nguyen et al., 2016; Haenni et al., 2016; Skov and Monnet, 2016). This, thus, creates severe challenges for controlling the selection and subsequent transmission of mcr-1 genes (Yang et al., 2016).
Dissemination of mcr-contaminated feces into the environment
Luo et al. cultured a mcr-1 producing Raoultella ornithinolytica, a member of the Enterobacteriaceae family commonly found in soil, aquatic, and botanical environments, in retail vegetables in China. They emphasized that using animal excrements contaminated with colistin residuals and/or MCRPE can cause contamination and spread of MCRPE to fresh vegetables (Luo et al., 2017).. Further, in fish-livestock integrated farms, fish are fed animal manure, which acts as a fertilizer for aquaculture ponds (Dang et al., 2011). The deposit of manure/animal excrements with colistin residuals into fish ponds or the environment favors the selection and growth of colistin-resistant bacteria (Shen et al., 2019). In Lebanon, there is evidence of the aquatic environment being affected by anthropogenic waste including sewage and agricultural waste (Hmede and Kassem, 2018; Sulaiman and Kassem, 2019; Hassan et al., 2020; Dandachi et al., 2020; Kassem, 2020). There is, however, little global epidemiological data on the contamination of rivers and lakes by MCRPE-contaminated animal excrements and thus, no evidence of the spread of MCRPE to the aquaculture (Cabello et al., 2017; Shen et al., 2018). However, the presence of mcr-1-producing E. coli has been reported in duck and fish samples in China. Shen et al. (2019) found a potential spread of mcr-1 producing E. coli from aquaculture supply chain to humans through seafood (Shen et al., 2018; Shen et al., 2019; Hassan et al., 2020). Other examples of the spread of MCRPE into the environment includes the identification and isolation of MCRPE in blowflies (Chrysomya sp.) (Yang QE et al., 2019) and black kites (Tarabai et al., 2019). Both cases are of public concern because of the potential transmission into human communities.
Yang et al. (Yang QE et al., 2019) showed that blowflies may serve as an environmental reservoir and vector of MCRPE between animals, humans, the environment, and waste (landfills and sewage water) (Yang QE et al., 2019). The presence of MCRPE in Black kites in Russia highlights the spread of mcr-1 genes into both wildlife and the environment. Tarabai et al. (2019) hypothesized that the acquisition of MCRPE was either through contact with the Biysk Municipal landfill or through their food, which was commonly found along the Biya River near their nest. Black kites can thereafter spread MCRPE along their long migratory pathways (Tarabai et al., 2019). The mcr-1 gene is commonly found on transferable plasmids, and thus the presence of mcr-1 producing isolates in the microbiota of insects and wildlife increases the possibility of horizontal transfer of mcr-1 genes alongside other resistance genes within the microbiota. This increases the environmental gene pool and the spread of multi-drug resistant (MDR) pathogens within insects and wildlife (Yang QE et al., 2019).
Dissemination of mcr genes to humans
There are therefore multiple reservoirs highlighted above with the potential of spreading towards humans because of the close association between humans and the environment. A study in Vietnam showed that the farmers that are in contact with animals exposed to colistin have a high risk of being colonized by MCRPE (Trung et al., 2017). Trung et al. (2017) found that chickens associated with colistin administration had a high carriage of mcr-1 producing isolates and the colonization of humans was through exposure to the chickens. The chicken faecal samples had a high prevalence of mcr-1 (59.4%) and zoonotic transmission prevalence was 34.7% in farmers. Another possible route of transmission of MCRPE from animals exposed to colistin, was foodborne transmission (Liu et al., 2016; Malhotra-Kumar et al., 2016; Agnoletti et al., 2018). Monte et al. found that chicken meat was acting as a reservoir of mcr-1 producing E. coli. A study performed by Shen et al. (2018) found a positive correlation between mcr-1 producing E. coli carriage in the human normal flora and the consumption of colistin-exposed farm animals, which included pork and sheep. The correlation analysis also proposed a possible transmission of mcr-1 producing E. coli from aquatic food to humans (Shen et al., 2018; Hassan et al., 2020). This data is a single example that highlights the presence of MCRPE in food-producing animals and the transmission route of MCRPE from colistin-exposed animals to humans, which impacts public health care.
Risk-factors
After the first detection of mcr-1 by Liu et al. (2016), mcr-1 genes have been detected in animals, the environment, and humans. Initially, the detection of mcr-1 genes in humans was associated with the risk factors mentioned above: contact with farm animals, ingestion of colistin-exposed farm animals, or fresh vegetables. Yang et al. identified mcr-1 producing E. coli and K. pneumoniae in market retail fruits, a significant report because fruits are consumed without cooking and processing unlike meat and vegetables. This is potentially a high-risk source of mcr-1 (Yang F et al., 2019). Epidemiology studies that identified mcr-1 genes in the clinical settings were accompanied with a questionnaire to identify the MCRPE source. In countries such as Finland, where the use of colistin is restricted for rare clinical indications, the first detection of the mcr-1 gene was identified in a healthy male with previous history of travelling abroad, 6 months before fecal sampling (Gröndahl-Yli-Hannuksela et al., 2018). Traveling to other countries has been shown to increase the risk of acquiring mcr-1 positive isolates, particularly in patients who consumed meat while traveling abroad (Gröndahl-Yli-Hannuksela et al., 2018; Henig et al., 2019). The use of antibiotics prior to hospitalization or infection has been seen as an important risk factor for a multidrug resistant organism infection (Henig et al., 2019). An epidemiological and clinical study performed by Wang et al. (2017) reported that mcr-1 producing E. coli can acquire other resistance genes, and thus previous use of antibiotics such as carbapenems and fluroquinolones was associated with an increased risk of infection by mcr-1 producing E. coli (Wang et al., 2017). Thus, the administration of antibiotics in clinics simultaneously promotes the co-selection and the preservation of colistin resistance genes (Lu et al., 2019).
An interesting possible transmission of mcr-1 producing E. coli is between companion animals and humans. A worker at a pet shop with no prior antibiotic use or travelling abroad was identified carrying an mcr-1 producing E. coli (Zhang et al., 2016). A fecal screening of the pets residing where the man worked identified 4 dogs and 1 cat with an mcr-1 positive isolate. Zhang et al. (2016) suggested companion animals as a possible reservoir of colistin-resistant E. coli, facilitating the spread of colistin resistance genes within the community.
Current and future perspectives on treating mcr-mediated colistin resistant infections
Clustered regularly interspaced short palindromic repeats (CRISPR)
Clustered regularly interspaced short palindromic repeats (CRISPR) has been exploited and developed within molecular biology as a site-specific tool for genetic engineering (Hryhorowicz et al., 2017), and in regards to resistance, as a tool to deliver a programmable DNA nuclease (CRISPR-Cas9) into an MDR-pathogen to eliminate the resistance gene or plasmid (Dong et al., 2019). Dong et al. (2019) developed a conjugative CRISPR-Cas9 system that aimed to remove plasmids harboring the mcr-1 gene from bacteria. The system targeted the mcr-1 gene and the authors found that the formation of DSBs during the elimination of the resistance gene resulted in the elimination of the whole plasmid in recipient cells (Dong et al., 2019). This thereafter sensitized the recipient cells to colistin. Dong et al. (2019) further found that the cells are thereafter immune against the mcr-1 gene. Wang et al. (Wang P et al., 2019) used the same concept but developed the tool to remove both mcr-1-harboring plasmids and MDR plasmids present in recipient cells using partial sequences of the targeted plasmids. He et al. (2021) used the tool to eliminate both chromosomal and plasmid-borne mcr-1 genes in E. coli. The CRISPR-Cas9 system was designed to target ISApl1, which eliminated mcr-1 harboring plasmids, and in the case of chromosomally encoded mcr-1, cell death was seen. Similar results were achieved (Dong et al., 2019), where recipient cells further acquired immunity against the acquisition of the exogenous mcr-1 containing plasmid (He et al., 2021). These studies show that CRISPR-Cas9 system is an efficient tool for plasmid curing and to sensitize clinical isolates to antibiotics in vitro (Dong et al., 2019; Wang P et al., 2019). This tool can be optimized for therapeutic application for the elimination of antibiotic genes from resistance reservoirs such as animal guts with prior exposure to antibiotics, the human microbial flora or the bacteria in natural settings such as wastewater, farms or hospital settings (Dong et al., 2019).
The CRISPR-Cas9 system is however made up of highly anionic nucleic acids and proteins that cannot penetrate the cell membrane into cells and thus requires a delivery vehicle. Sun et al. (Sun L et al., 2017) explored Cathelicidins, which are short cationic antimicrobial peptides, as a plasmid delivery system. The study evaluated the use of BMAP-27 to increase the efficiency of plasmid transfer of the CRISPR-Cas9 system and found that pCas::mcr exhibited better, efficient, and specific antimicrobial effects with the help of BMAP-27 (Sun L et al., 2017). The CRISPR-Cas9 system is a powerful tool for genome editing, its development into targeting multiple genes with one use (Wang P et al., 2019), could aid in the removal of mcr genes in resistance reservoirs and in clinical settings to restore colistin activity.
In the meantime, there are multiple studies evaluating existing antibiotics for activity against MCR-producing isolates and synergy combinations, summarized in Table 1.
Antimicrobials agents effective against mcr-1 producing isolates
There are multiple novel antibiotics that have been found to have activity against mcr-1-producing isolates, including eravacycline (Fyfe et al., 2016; Zhao et al., 2019) and plazomicin (Denervaud-Tendon et al., 2017). Eravacycline is a broad-spectrum synthetic tetracycline that has been proven to be effective against clinically important MDR pathogens, including both Gram-negative and Gram-positive bacteria (Zhao et al., 2019). Fyfe et al. (2016) found that eravacycline had a bactericidal effect against MCR-producing isolates from Enterobacteriaceae. The study further showed that although the overexpression of mcr-1 increased colistin MIC values by 64-folds, there was no effect on eravacycline susceptibility (Fyfe et al., 2016). Eravacycline activity was thereafter evaluated against 336 isolates collected from hospitals across China and was found to exhibit a good efficacy against all strains (Zhao et al., 2019). These isolates included Extended spectrum β-lactamase (ESβL) and carbapenemase-producing Enterobacteriaceae, MCR-producing Enterobacteriaceae and A. baumannii, vancomycin-resistant Enterobacteriaceae, β-lactamase producing Haemophilus influenzae, penicillin-resistant Streptococcus pneumoniae and lastly, methicillin-resistant Staphylococcus aureus (MRSA), thus showing a positive potential to treat current drug-resistant bacterial infections (Zhao et al., 2019). This, however, also puts eravacycline at risk of being overused.
Plazomicin is a novel broad-spectrum semi-synthetic aminoglycoside that was derived from Sisomicin. It has been shown to have activity against a broad spectrum of MDR pathogens, including ESβL- and carbapenemase-producing isolates and fluroquinolone-resistant isolates (Denervaud-Tendon et al., 2017). Denervaud-Tendon et al. (2017) evaluated the bactericidal activity of plazomicin against colistin-resistant Enterobacterial strains, which harbored different colistin resistance mechanisms i.e., chromosomal-encoded, mcr-1-expressing, and intrinsically resistant strains. Plazomicin activity was further compared with other aminoglycoside antibiotics viz., amikacin, gentamicin, and tobramycin. Plazomicin displayed potent activity against the clinical colistin-resistant Enterobacterial strains irrespective of their resistance mechanisms. However, those that were intrinsically resistant (Serratia, Proteus, Morganella and Hafnia) had a higher MIC value (Denervaud-Tendon et al., 2017).
Denervaud-Tendon et al. also found that amongst all aminoglycoside antibiotics tested, plazomicin was the most potent (Denervaud-Tendon et al., 2017). They lastly also found that plazomicin’s activity is restricted by aminoglycoside resistance mechanisms such as the 16S rRNA methylase-encoding gene, which resulted in an elevated plazomicin MIC value (>128 mg/L) (Denervaud-Tendon et al., 2017). Plazomicin, however, like other aminoglycosides, has a rapid bactericidal activity with favorable chemical and pharmacokinetics properties and thus could be incorporated into therapeutic treatments against MCR-producing bacterial infections (Denervaud-Tendon et al., 2017).
Artilysin ® Art-175 is a novel engineered enzyme-based experimental therapeutic derived from endolysins produced by lytic bacteriophages. These endolysins degrade the bacterial cell wall resulting in cell lysis, which occurs at the end of the bacteriophage infection cycle (Gerstmans et al., 2016). Artilysin ® Art-175 is made up of the outer membrane destabilizing peptide, which is fused to an endolysin, it targets the anionic lipopolysaccharide molecules of the outer membrane of Gram-negatives. Schirmeier et al. (2018) evaluated the potential of Art-175 against pan-drug resistant isolates, including colistin-resistant mcr-1 producing E. coli and further determined the potential of cross-resistance between colistin and Art-175. Art-175 was highly bactericidal against the tested isolates, which were more susceptible to Art-175 than the colistin-susceptible isolates (Schirmeier et al., 2018). Hence, there is no cross-resistance between the two peptides, highlighting Art-175 as a potent solution against MCR-producing isolates (Schirmeier et al., 2018).
Antimicrobial peptides
Bacteriocins are ribosome-synthesized antimicrobial peptides (AMPs) produced by both Gram-positive and Gram-negative bacteria. Studies have shown that bacteriocins produced by lactic acid bacteria (LAB) are cationic peptides with bactericidal activity that act on the cytoplasmic membrane of susceptible microorganisms (Drider et al., 2016). The LAB bacteriocins create pores in the membrane, resulting in intracellular damage. Al Atya et al. (2016) evaluated the combinations of nisin and enterocin DD14 LAB bacteriocins with colistin to eradicate colistin-resistant E. coli strains in either planktonic states or in biofilm. Whilst colistin and the bacteriocins were ineffective as monotherapy against both planktons and biofilms, colistin-nisin and colistin-enterocin DD14 combination were able to reduce the number of colony-forming units (CFU) of strains in both planktonic and biofilm states. The triple combination of all three molecules, colistin-nisin-enterocin DD14, completely eradicated all E. coli strains, including colistin-resistant phenotype (Al Atya et al., 2016). LAB bacteriocins alone cannot penetrate the outer membrane. However, with the disruption of the LPS via colistin activity, the LAB bacteriocins may thereafter act upon the cytoplasmic membrane (Al Atya et al., 2016). Al Atya et al. (2016) therefore highlights LAB bacteriocins as a potential novel adjunctive treatment for colistin-resistant mcr-1 producing E. coli infections.
There have been multiple reports of AMPs that have been developed for treating MDR pathogens, which have a similar mode of action as colistin (van der Weide et al., 2019). Van der Weider et al. evaluated the antimicrobial activity of two novel AMPs, AA139 and SET-MM3. AA139 originates from a marine lugworm, Arenicola marina, AMP arenicin-3. SET-MM3 is a synthetic tetra-branched peptide linked by a lysine core (van der Weide et al., 2019). The antimicrobial activity of these AMPs was evaluated against a collection of clinically and genotypically diverse K. pneumoniae with different antimicrobial susceptibility profiles. Both AMPs, AA139 and SET-M33, further showed a concentration-dependent bactericidal effect across all isolates, meaning that the susceptibility profile of both AMPs was unaffected by the resistance profile and colistin susceptibility of the isolates (van der Weide et al., 2019). The AMPs were effective against the colistin-resistant strains, and no cross-resistance was observed between the AMPs and colistin. Thus the antimicrobial activity of AA139 and SET-M33 should further be investigated across other Gram-negatives to evaluate its spectrum of bactericidal activity (van der Weide et al., 2019).
Cross-resistance between several human cationic AMPs (CAMPs) and polymyxins has been previously reported in PEtN transferase-producing Haemophilus ducreyi and Campylobacter jeyuni but rarely in Enterobacteriaceae. These CAMPs are part of the intrinsic human immune system and act against Gram-negative bacteria by disrupting the outer and inner membrane through electrostatic interaction, similar to colistin’s mechanism of action (Dobias et al., 2017). Dobias et al. (2017) selected a few widely distributed CAMPs in humans, LL-37, α-defensin 5 (HD5) and β-defensin 3 (HDB3) and evaluated whether cross-resistance between CAMPs and polymyxin would be observed in mcr-1 producing E. coli. Susceptibility of E. coli to the different CAMPs selected varied depending on the CAMP molecule rather than the entire CAMP family. It further showed that the production of MCR proteins did not confer cross-resistance with the human CAMPs selected (Dobias et al., 2017).
However, the CAMPs activity studied by Dobias et al. (2017), compared to other studies (Napier et al., 2013), shows that factors such as bacterial species, level of resistance, and colistin resistance mechanisms may affect the bactericidal levels of CAMPs (Napier et al., 2013; Dobias et al., 2017). Therefore, although mcr production did not affect the bactericidal activity of CAMPs against E. coli, it needs to be evaluated across Enterobacteriaceae and other Gram-negative bacteria to see how different colistin resistance mechanisms affect the CAMPs’ bactericidal effect. Guachalla et al. (2018) further evaluated the effectiveness of other derivates of the human immune system against a clinical mcr-1 producing E. coli strain. This includes humanized monoclonal antibodies that have been previously shown to target the LPS O25b antigen that is associated with E. coli ST131-H30 (Guachalla et al., 2018). The expression of mcr-1 does not affect the binding of ASn-4 to the LPS’s O-antigen and the integration of the complement membrane-attack complex (MAC) into the LPS-modified outer membrane (Guachalla et al., 2018). This means the expression of mcr-1 in E. coli ST131-H30 does not affect the functionality of the human immune system, nor does it not aid the strain via immune evasion. The authors thereafter suggested the use of antibodies targeting the LPS’s O-antigen as an alternative strategy to combatting MDR infections against mcr-1 positive isolates (Guachalla et al., 2018).
Other novel compounds produced to manage MCR-producing isolates are antisense agents with antimicrobial properties; this include phosphorodiamidate morpholino oligomers (PPMOs) and peptide nucleic acids (PNAs) (Nezhadi et al., 2019). These antisense molecules are designed to target mRNA, prevent translation, and restore antimicrobial sensitivity. Daly et al. (2017) designed peptide-conjugated PPMOs targeted to mcr-1 mRNA (MCR-1 PPMO) of clinical mcr-1-positive E. coli strains and showed that these molecules were able to make the strains re-sensitive to polymyxins by MCR-1 inhibition (Daly et al., 2017). Similar results were achieved by Nezhadi et al. (2019) with PNAs, designed for inhibition of mcr-1 translation. They (Nezhadi et al., 2019) showed that the introduction of PNA resulted in a 95% reduction of mcr-1 expression, measured by RT-PCR, highlighting the efficacy of PNAs (Nezhadi et al., 2019). Daly et al. (2017) evaluated the antisense approach in a sepsis model using mice and found that a combined therapy of MCR-1 PPMO with colistin reduced morbidity and bacterial burden in the spleen at 24hr. Thus, antisense agents may be effective therapeutics, either alone by targeting an essential gene resulting in cell death (Daly et al., 2017), or in combination with colistin (Daly et al., 2017; Nezhadi et al., 2019) to treat mcr-1-producing isolates. However, these molecules have varying toxicity effects and more in-vivo research is required prior to clinical application (Nezhadi et al., 2019).
Natural compounds
Other novel compounds that have been reported to have synergistic effects with colistin against mcr-1 producing isolates includes the novel MCR-1 inhibitor, Osthole (7-methoxy-8-(3-methyl-2-buteryl) coumarin. Osthole (OST) is a natural compound derived from the dried root and rhizome of Cnidium monnieri (Zhou et al., 2019). Zhou et al. (2019) evaluated the effects of OST on the inhibition of MCR-1 enzyme and the mechanisms behind the inhibition. The colistin-OST combination had a synergistic effect as a mouse-thigh-infection model showed that the combination exhibits a bactericidal activity against mcr-1-producing Enterobacteriaceae, significantly reducing the bacterial load in the thighs following subcutaneous administration (Zhou et al., 2019). Using a molecular dynamic stimulation, Zhao et al. (Zhou et al., 2019) found that OST could localize in the binding pocket (residue 330-350) of MCR-1, blocking the substrate and reducing MCR-1’s biological activity. OST therefore inhibits MCR-1 in Enterobacteriaceae, making colistin potent (Zhou et al., 2019). Due to the conservation of MCR proteins, OST may have the same inhibition efficiency across the other MCR genes.
Natural/organic compounds identified with a synergistic effect on colistin include Honokiol (Guo et al., 2020), Isoalantolactone (IAL) (Lu et al., 2020), Calycosin (Liu et al., 2020) and Eugenol (Wang et al., 2018). Honokiol was derived from the dry skin and bark of Magnolia officinalis (Guo et al., 2020), IAL is the main sesquiterpene lactone from Radix inulae and other plants (Lu et al., 2020). Calycosin is a flavonoid from a direct root extract of the traditional Chinese medicinal herb, Radix astragali (Liu et al., 2020). Lastly, Eugenol is a phenylpropanoid, an essential oil isolated from plants (Dos Santos et al., 2021). Each of these compounds, when combined with colistin, decreased colistin MICs and increased colistin’s bactericidal activity against colistin-resistant isolates. Eugenol decreased the expression of mcr-1 (Wang et al., 2018) and honokiol bound directly to the active site of MCR-1, inhibiting its activity (Guo et al., 2020). Guo et al. (2020) and Liu et al. (2020) showed that the natural compounds, honokiol and calycosin, in combinations with colistin, respectively reduced the load of bacteria and improved viability in animal models.
Efflux pump inhibitors
Efflux pumps play a role in reducing colistin susceptibility in Enterobacteriaceae (Mmatli et al., 2020). Baron et al. (Baron and Rolain, 2018) found that an efflux pump inhibitor, carbonyl cyanide 3-chlorophenylhydrazone (CCCP), was a good alternative to reverse colistin resistance in colistin-resistant isolates irrespective of their molecular resistance mechanisms. The study shows that CCCP was able to restore colistin activity across a diverse set of Enterobacteriaceae isolates carrying different colistin resistance mechanisms, intrinsic and acquired (mcr-1, pmrAB, mgrB, etc) (Osei Sekyere and Amoako, 2017; Baron and Rolain, 2018). In mcr-1 producing isolates, it was seen that colistin-CCCP combination inhibited the transcription of the mcr-1 gene. The mechanism behind this observation, however, is unknown (Baron and Rolain, 2018).
FDA-approved drugs
The food and drug administration (FDA) has a library of FDA-approved drugs that can be screened for bactericidal activity on MCR-producing Enterobacteriaceae isolates (Prestwick Chemical, Illkirch-Graffenstuden, France). Multiple compounds such as pentamidine, zidovudine (Okdah et al., 2018), sulphonamide compounds (Okdah et al., 2018), a polymyxin derivative, NAB739 (Zhang Y et al., 2019), and compound PFK-185 (Zhang Y et al., 2019), were identified.
Zidovudine is a nucleoside reverse transcriptase inhibitor. In the 1980s, it was used as an anticancer drug (Chow et al., 2009). In 1986, its antibacterial effect was revealed (Elwell et al., 1987) and in 1987, it was used as the first antiretroviral for treatment against HIV infections (Chow et al., 2009; Peyclit et al., 2018). Peyclit et al. (2018) investigated the antibacterial activity of Zidovudine against a large number of characterized MDR Enterobacteriaceae strains isolated from different geographical areas, including carbapenem- and colistin-resistant isolates; Zidovudine was effective against Enterobacteriaceae. Despite their antimicrobial susceptibility profile, the MIC values ranged between 0.05-1.67 µg/mL (Peyclit et al., 2018).
Another antiretroviral drug that is used to treat HIV/AIDS but also has bactericidal activity against Gram-negative bacteria is Azidothymidine (AZT). Hu et al. (2019) shows that AZT, in combination with colistin, was able to eradicate MDR Enterobacteriaceae strains with different antimicrobial susceptibility profiles and enhance the activity of colistin. The combined therapy was also effective therapeutically in a murine peritoneal infection against NDM-1-producing K. pneumoniae and mcr-1 producing E. coli (Hu et al., 2019).
Further, Pentamidine is an antiprotozoal drug that Stokes et al. (2017) identified as an effective antibiotic adjuvant producing synergistic combination activity against a wide range of Gram-negative bacteria. The adjuvant was identified during a library screening of non-lethal, outer membrane-active compounds (Stokes et al., 2017). The study shows that Pentamidine directly associates with the outer membrane by inhibiting the oligosaccharide (OS) core biosynthesis, releasing LPS from the outer membrane (Stokes et al., 2017). Stokes et al. (2017) further showed that Pentamidine has synergistic activity with hydrophobic antibiotics such as rifampicin, novobiocin, and erythromycin but not with hydrophilic and low-molecular weight antibiotics (Stokes et al., 2017). Although the study does not evaluate synergistic combinations with Pentamidine, Stokes et al. (2017) concluded that pentamidine can be used as an antibiotic adjuvant for treating colistin-resistant infections.
Barker et al. (2017) screened a diverse cohort of adjuvants to identify a compound that could both sensitize colistin-resistant and hypersensitize colistin-susceptible bacteria to colistin, to potentially lower the effective dosage of colistin, reducing its toxicity. The study identified three compounds that were potent modulators of colistin resistance and were able to significantly reduce MIC values in both plasmid- and chromosomal-mediated resistance mechanisms, and further hypersensitize colistin-susceptible isolates (Barker et al., 2017)
Another compound identified in the FDA-approved library with synergistic activity with colistin is sulfadiazine (SDI), a sulphonamide compound. Okdah et al. (2018) evaluated the potential of different sulphonamide compounds with potential synergistic and bactericidal activity in combination with colistin; SDI had the highest synergistic effect. The combination of SDI-colistin was effective, independent of colistin resistance mechanism, across the broad range of MDR bacterium strains tested (Okdah et al., 2018).
Zhang et al. (Zhang Y et al., 2019) identified an antitumor drug, PFK-185 and its analogs, PFK-015 and 3PO, during a screening of the clinical compound library. These compounds can exert synergize with colistin against colistin-resistant Enterobacteriaceae despite mcr expression and the antimicrobial susceptibility profile (Zhang Y et al., 2019). Zhang et al. (Zhang Y et al., 2019) found that PFK-185 had no effect on cellular morphology when used alone. However, when in combination with colistin, it enhanced the bacterium-killing effect of colistin, increasing the survival rate to 60%. The combined colistin-PFK-158 therapy had the most significant bactericidal activity, with the most significant reductions in the bacterial burdens post-in vivo experiments and during time-kills studies (Zhang Y et al., 2019).
The last compound reported from the FDA-approved library is a novel polymyxin derivative, NAB739, which carries only three of the five amino groups of colistin, each placed in a strategic position (Tyrrell et al., 2019). Tyrrell et al. (2019) showed that NAB739 sensitizes polymyxin-resistant strains to rifampicin and a combination of the two was synergistic against ten of the eleven colistin-resistant strains. Tyrrell et al. (2019) further showed that NAB739 was also synergistic with meropenem and retapamulin. As well, NAB739 in combination with other compounds has significant bactericidal activity against polymyxin-resistant isolates and compared to polymyxins, it has a better tolerability and efficacy (Tyrrell et al., 2019). Thus, NAB739 combination therapies may replace polymyxin for treating MDR pathogens (Tyrrell et al., 2019).
These studies (Stokes et al., 2017; Peyclit et al., 2018) however, showed that old antimicrobials listed within the FDA-approved library may be re-introduced against bacteria that are resistant to multiple antibacterial in current use (Peyclit et al., 2018).
Combination therapy
There are multiple compounds, either antibiotics or adjuvants, which are synergistic with colistin, thus reducing the dosage of colistin required for treatment, minimizing its toxicity, overcoming colistin resistance, and ensuring maximal therapeutic efficacy (Barker et al., 2017; Hu et al., 2019). Colistin activity has been restored when it is combined with antibiotics such as rifampicin (Li Y et al., 2018; Yu et al., 2019), rifabutin, minocycline (Yu et al., 2019), amikacin (Zhou et al., 2017), tigecycline (Zhou et al., 2020) and clarithromycin (MacNair et al., 2018). In all these reports, the antibiotics were ineffective as monotherapy against MCRPE isolates. However, in combination with colistin, they resulted in significant bactericidal activity. Yu et al. (2019) found that colistin in combination with rifampicin, rifabutin or minocycline was able to eradicate XDR, NDM- and mcr– co-producing E. coli in-vitro and in mouse models. Other combinations that were confirmed effective against mcr-1-producing colistin-resistant E. coli using an animal model include tigecycline-colistin (Zhou et al., 2020), clarithromycin-colistin (MacNair et al., 2018) and a triple combination of colistin-rifampin-azithromycin discovered by Li et al. (Li Y et al., 2018). Another triple combination therapy that was discovered was colistin-aztreonam-amikacin, which was effective against both carbapenemase-producing and mcr-1-producing Enterobacteriaceae isolates (Bulman ZP et al., 2017).
Lastly, a combination therapy that was effective against both carbapenem- and colistin-resistant Enterobacteriaceae isolates was the β-lactam-β-lactamase inhibitor, imipenem-relebactam, combination (Carpenter et al., 2019). This combined therapy was potent against colistin-resistant carbapenemase-producing isolates, making it a potential agent against carbapenemase-producing Enterobacteriaceae; particularly, those that are colistin resistant (Carpenter et al., 2019).
Conclusion
Herein, mcr genes are commonly identified in E. coli, K. pneumoniae, and Salmonella sp., with clones within these strains being disseminated globally within animals, food, the environment, and humans. Mcr genes were initially identified in animal samples, specifically livestock animals that were treated with colistin as a therapeutic, prophylaxis and metaphylaxis against Gram-negative bacterial infections or used to promote growth. Mcr genes have spread to the environment and humans through contaminated and untreated animal feces used as manure and through contaminated food-producing-animals’ products on markets, respectively. Contact with these colistin-treated, MCRPE-contaminated livestock or their feces by humans was identified as a possible transmission route. This study identified multiple possible routes of transmission of MCRPE to humans, these include routes from livestock, the environment and from livestock-treated meat. The use of colistin as a therapeutic against carbapenem-resistant Enterobacteriaceae infections has also shown to increase the prevalence of MCRPE in humans. The presence of clones such as E. coli ST744 or ST101 in all sources, including animals, the environment, food, and humans, highlights these transmission routes. This makes mcr a concerning antibiotic resistance gene of great interest and priority.
Further, mcr genes are commonly associated with mobile genetic elements. ISApl1 elements have facilitated the horizontal transfer of mcr-1 within cassettes of composite transposons. They have been also associated with IncX4 plasmids, facilitating their horizontal and vertical transfer across species, genera and families.
Although the mcr genes have different possible progenitors viz., Moracella sp., Aeromonas sp., and Shewanella sp. for mcr-1, mcr-3 and mcr-4, respectively, the MCR proteins are very well conserved. Each of the 10 MCR proteins encodes a two-domain integral membrane protein with a C-terminal periplasmic domain and an N-terminal 5’-helix transmembrane domain, each MCR protein further encodes the five conserved residues i.e., E248, T286, H389, D458, and H359, located within the active site. PEtN transferase activity was found for each MCR protein, and each was able to mediate colistin resistance, although mcr-9 expression requires a specific genetic environment to regulate its expression. The conservation seen within MCR proteins, allows for the use of similar therapies for the management of MCRPE isolates, specifically in the cases of identified MCR-1 inhibitors. Research on novel therapeutics is well-summarized in this review (Table 1), with techniques such as CRISPR-Cas9, peptide nucleic acids, and antimicrobial peptides, that can aid manage mcr -positive carbapenem resistant isolates. Other novel therapeutics were identified by reviving old FDA-approved drugs. Some were effective against MCR-producing isolates or were synergistic with colistin. These therapeutics, however, need to be further evaluated for their toxicity in humans. This will aid in alleviating the threat imposed by mcr -positive carbapenem isolates in the public health sector.
Author contributions
MM undertook the systematic search, data collection and manuscript write-up; NM was a co-supervisor to the study and assisted with funding; JO designed and supervised the study, reviewed and edited the manuscript, as well as assisted with analysis of the data. All authors contributed to the article and approved the submitted version.
Funding
This work was funded by the National Health Laboratory Service (NHLS) given to JO under grant number GRANT004 94809 (reference number PR2010486), and the National Research Foundation.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcimb.2022.941358/full#supplementary-material
References
AbuOun, M., Stubberfield, E. J., Duggett, N. A., Kirchner, M., Dormer, L., Nunez-Garcia, J., et al. (2017). Mcr-1 and mcr-2 variant genes identified in moraxella species isolated from pigs in great Britain from 2014 to 2015. J. Antimicrob. Chemother. 72, 2745–2749. doi: 10.1093/jac/dkx286
Agnoletti, F., Brunetta, R., Bano, L., Drigo, I., Mazzolini, E. (2018). Longitudinal study on antimicrobial consumption and resistance in rabbit farming. Int. J. Antimicrob. Agents 51, 197–205. doi: 10.1016/j.ijantimicag.2017.10.007
Ahmed, Z. S., Elshafiee, E. A., Khalefa, H. S., Kadry, M., Hamza, D. A. (2019). Evidence of colistin resistance genes (mcr-1 and mcr-2) in wild birds and its public health implication in Egypt. Antimicrob. Resist. Infect. Control 8, 197. doi: 10.1186/s13756-019-0657-5
Ahmed, S., Das, T., Islam, M. Z., Herrero-Fresno, A., Biswas, P. K., Olsen, J. E.. (2020). High prevalence of mcr-1-encoded colistin resistance in commensal escherichia coli from broiler chicken in Bangladesh. Sci. Rep. 10, 18637. doi: 10.1038/s41598-020-75608-2
Al Atya, A. K., Abriouel, H., Kempf, I., Jouy, E., Auclair, E., Vachée, A., et al. (2016). Effects of colistin and bacteriocins combinations on the in vitro growth of escherichia coli strains from swine origin. Probiotics Antimicrob. Proteins 8, 183–190. doi: 10.1007/s12602-016-9227-9
Anandan, A., Evans, G. L., Condic-Jurkic, K., O’Mara, M. L., John, C. M., Phillips, N. J, et al. (2017). Structure of a lipid a phosphoethanolamine transferase suggests how conformational changes govern substrate binding. Proc. Natl. Acad. Sci. 114, 2218–2223. doi: 10.1073/pnas.1612927114
Andersson, D. I. (2006). The biological cost of mutational antibiotic resistance: any practical conclusions? Curr. Opin. Microbiol. 9, 461–465. doi: 10.1016/j.mib.2006.07.002
Anjum, M. F., Duggett, N. A., AbuOun, M., Randall, L., Nunez-Garcia, J., Ellis, R. J., et al. (2016). Colistin resistance in salmonella and escherichia coli isolates from a pig farm in great Britain. J. Antimicrob. Chemother. 71, 2306–2313. doi: 10.1093/jac/dkw149
Anyanwu, M. U., Anyogu, D. C., Chah, K. F., Shoyinka, V. S. (2022). Mobile colistin resistance (mcr-1) gene-positive escherichia coli from chickens in Nigeria is potentially pathogenic and transfers colistin resistance to other organisms. Comp. Clin. Pathol. 31, 323–332. doi: 10.1007/s00580-022-03336-2
Anyanwu, M. U., Marrollo, R., Paolucci, M., Brovarone, F., Nardini, P., Chah, K. F., et al. (2021). Isolation and characterisation of colistin-resistant enterobacterales from chickens in southeast Nigeria. J. Glob. Antimicrob. Resist. 26, 93–100. doi: 10.1016/j.jgar.2021.04.030
Apostolakos, I., Piccirillo, A. (2018). A review on the current situation and challenges of colistin resistance in poultry production. Avian Pathol. 47, 546–558. doi: 10.1080/03079457.2018.1524573
Arcilla, M. S., van Hattem, J. M., Matamoros, S., Melles, D. C., Penders, J., de Jong, M. D., et al. (2016). Dissemination of the mcr-1 colistin resistance gene. Lancet Infect. Dis. 16, 147–149. doi: 10.1016/S1473-3099(15)00541-1
Barker, W. T., Martin, S. E., Chandler, C. E., Nguyen, T. V., Harris, T. L., Goodell, C., et al. (2017). Small molecule adjuvants that suppress both chromosomal and mcr-1 encoded colistin-resistance and amplify colistin efficacy in polymyxin-susceptible bacteria. Bioorg. Med. Chem. 25, 5749–5753. doi: 10.1016/j.bmc.2017.08.055
Baron, S. A., Rolain, J. M. (2018). Efflux pump inhibitor CCCP to rescue colistin susceptibility in mcr-1 plasmid-mediated colistin-resistant strains and gram-negative bacteria. J. Antimicrob. Chemother. 73, 1862–1871. doi: 10.1093/jac/dky134
Bonnin, R. A., Bernabeu, S., Jaureguy, F., Naas, T., Dortet, L. (2020). MCR-8 mediated colistin resistance in a carbapenem-resistant klebsiella pneumoniae isolated from a repatriated patient from Morocco. Int. J. Antimicrob. Agents 55, 105920. doi: 10.1016/j.ijantimicag.2020.105920
Borowiak, M., Fischer, J., Hammerl, J. A., Hendriksen, R. S., Szabo, I., Malorny, B.. (2017). Identification of a novel transposon-associated phosphoethanolamine transferase gene, mcr-5, conferring colistin resistance in d-tartrate fermenting salmonella enterica subsp. enterica serovar paratyphi b. J. Antimicrob. Chemother. 72, 3317–3324. doi: 10.1093/jac/dkx327
Borowiak, M., Hammerl, J. A., Deneke, C., Fischer, J., Szabo, I., Malorny, B.. (2019). Characterization of mcr-5-Harboring salmonella enterica subsp. enterica serovar typhimurium isolates from animal and food origin in Germany. Antimicrob. Agents Chemother. 63, e00063–00019 . doi: 10.1128/aac.00063-19
Bulman, Z. P., Chen, L., Walsh, T. J., Satlin, M. J., Qian, Y., Bulitta, J. B., et al, et al. (2017) Polymyxin Combinations Combat Escherichia coli Harboring mcr-1 and bla(NDM-5): Preparation for a Postantibiotic Era. mBio 8, e00540–00517. doi: 10.1128/mBio.00540-17
Cabello, F. C., Tomova, A., Ivanova, L., Godfrey, H. P. (2017). Aquaculture and mcr colistin resistance determinants. mBio 8, e01229–e01217. doi: 10.1128/mBio.01229-17
Cannatelli, A., D'Andrea, M. M., Giani, T., Di Pilato, V., Arena, F., Ambretti, S., et al. (2013). In vivo emergence of colistin resistance in klebsiella pneumoniae producing KPC-type carbapenemases mediated by insertional inactivation of the PhoQ/PhoP mgrB regulator. Antimicrob. Agents Chemother. 57, 5521–5526. doi: 10.1128/AAC.01480-13
Cannatelli, A., Giani, T., Aiezza, N., Di Pilato, V., Principe, L., Luzzaro, F., et al. (2017). An allelic variant of the PmrB sensor kinase responsible for colistin resistance in an escherichia coli strain of clinical origin. Sci. Rep. 7, 5071. doi: 10.1038/s41598-017-05167-6
Cannatelli, A., Santos-Lopez, A., Giani, T., Gonzalez-Zorn, B., Rossolini, G. M. (2015). Polymyxin resistance caused by mgrB inactivation is not associated with significant biological cost in klebsiella pneumoniae. Antimicrob. Agents Chemother. 59, 2898–2900. doi: 10.1128/AAC.04998-14
Carattoli, A., Carretto, E., Brovarone, F., Sarti, M., Villa, L. (2018). Comparative analysis of an mcr-4 salmonella enterica subsp. enterica monophasic variant of human and animal origin. J. Antimicrob. Chemother. 73, 3332–3335. doi: 10.1093/jac/dky340
Carattoli, A., Villa, L., Feudi, C., Curcio, L., Orsini, S., Luppi, A., et al. (2017). Novel plasmid-mediated colistin resistance mcr-4 gene in salmonella and escherichia coli, Italy 2013, Spain and Belgium, 2015 to 2016. Euro Surveill. 22. doi: 10.2807/1560-7917.ES.2017.22.31.30589
Carpenter, J., Neidig, N., Campbell, A., Thornsberry, T., Truex, T., Fortney, T., et al. (2019). Activity of imipenem/relebactam against carbapenemase-producing enterobacteriaceae with high colistin resistance. J. Antimicrob. Chemother. 74, 3260–3263. doi: 10.1093/jac/dkz354
Carrique-Mas, J. J., Trung, N. V., Hoa, N. T., Mai, H. H., Thanh, T. H., Campbell, J. I., et al. (2015). Antimicrobial usage in chicken production in the Mekong delta of Vietnam. Zoonoses Public Health 62, 70–78. doi: 10.1111/zph.12165
Carroll, L. M., Gaballa, A., Guldimann, C., Sullivan, G., Henderson, L. O., Wiedmann, M.. (2019). Identification of novel mobilized colistin resistance gene mcr-9 in a multidrug-resistant, colistin-susceptible salmonella enterica serotype typhimurium isolate. mBio 10. doi: 10.1128/mBio.00853-19
Cha, M. H., Woo, G. J., Lee, W., Kim, S. H., Woo, J. H., Kim, J., et al. (2020). Emergence of transferable mcr-9 gene-carrying colistin-resistant salmonella enterica dessau ST14 isolated from retail chicken meat in Korea. Foodborne Pathog. Dis. 17, 720–727. doi: 10.1089/fpd.2020.2810
Chavda, B., Lv, J., Hou, M., Chavda, K. D., Kreiswirth, B. N., Feng, Y., et al. (2018). Coidentification of mcr-4.3 and bla(NDM-1) in a clinical enterobacter cloacae isolate from China. Antimicrob. Agents Chemother. 62: e00649–00618. doi: 10.1128/aac.00649-18
Chavda, K. D., Westblade, L. F., Satlin, M. J., Hemmert, A. C., Castanheira, M., Jenkins, S. G., et al. (2019). First report of bla (VIM-4)- and mcr-9-Coharboring enterobacter species isolated from a pediatric patient. mSphere 4:e00629–00619. doi: 10.1128/mSphere.00629-19
Cheng, Y.-H., Lin, T.-L., Lin, Y.-T., Wang, J.-T. (2016). Amino acid substitutions of CrrB responsible for resistance to colistin through CrrC in klebsiella pneumoniae. Antimicrob. Agents Chemother. 60, 3709–3716. doi: 10.1128/AAC.00009-16
Chow, W. A., Jiang, C., Guan, M. (2009). Anti-HIV drugs for cancer therapeutics: back to the future? Lancet Oncol. 10, 61–71.
Clemente, L., Manageiro, V., Correia, I., Amaro, A., Albuquerque, T., Themudo, P., et al. (2019). Revealing mcr-1-positive ESBL-producing escherichia coli strains among enterobacteriaceae from food-producing animals (bovine, swine and poultry) and meat (bovine and swine), Portugal, 2010-2015. Int. J. Food Microbiol. 296, 37–42. doi: 10.1016/j.ijfoodmicro.2019.02.006
Collignon, P. C., Conly, J. M., Andremont, A., McEwen, S. A., Aidara-Kane, A.. (2016). World health organization ranking of antimicrobials according to their importance in human medicine: a critical step for developing risk management strategies to control antimicrobial resistance from food animal production. Clin. Infect. Dis. 63, 1087–1093. doi: 10.1093/cid/ciw475
Creighton, J., Anderson, T., Howard, J., Dyet, K., Ren, X., Freeman, J.. (2019). Co-Occurrence of mcr-1 and mcr-3 genes in a single escherichia coli in new Zealand. J. Antimicrob. Chemother. 74, 3113–3116. doi: 10.1093/jac/dkz311
Dahlberg, C., Chao, L. (2003). Amelioration of the cost of conjugative plasmid carriage in eschericha coli K12. Genetics 165, 1641–1649. doi: 10.1093/genetics/165.4.1641
Daly, S. M., Sturge, C. R., Felder-Scott, C. F., Geller, B. L., Greenberg, D. E. (2017). MCR-1 inhibition with peptide-conjugated phosphorodiamidate morpholino oligomers restores sensitivity to polymyxin in escherichia coli. mBio 8:e01315–01317. doi: 10.1128/mBio.01315-17
Dandachi, I., Fayad, E., Sleiman, A., Daoud, Z., Rolain, J. M. (2020). Dissemination of multidrug-resistant and mcr-1 gram-negative bacilli in broilers, farm workers, and the surrounding environment in Lebanon. Microb. Drug Resist. 26, 368–377. doi: 10.1089/mdr.2019.0137
Dang, S. T., Petersen, A., Van Truong, D., Chu, H. T., Dalsgaard, A. (2011). Impact of medicated feed on the development of antimicrobial resistance in bacteria at integrated pig-fish farms in Vietnam. Appl. Environ. Microbiol. 77, 4494–4498. doi: 10.1128/AEM.02975-10
Dawadi, P., Bista, S., Bista, S. (2021). Prevalence of colistin-resistant escherichia coli from poultry in south Asian developing countries. Veterinary Med. Int. 2021, 6398838. doi: 10.1155/2021/6398838
Denervaud-Tendon, V., Poirel, L., Connolly, L. E., Krause, K. M., Nordmann, P. (2017). Plazomicin activity against polymyxin-resistant enterobacteriaceae, including MCR-1-producing isolates. J. Antimicrob. Chemother. 72, 2787–2791. doi: 10.1093/jac/dkx239
Ding, M., Shi, J., Ud Din, A., Liu, Y., Zhang, F., Yan, X., et al. (2021). Co-Infections of two carbapenemase-producing enterobacter hormaechei clinical strains isolated from the same diabetes individual in China. J. Med. Microbiol. 70:001316. doi: 10.1099/jmm.0.001316
Di Pilato, V., Arena, F., Tascini, C., Cannatelli, A., Henrici De Angelis, L., Fortunato, S., et al. (2016). Mcr-1.2, a new mcr variant carried on a transferable plasmid from a colistin-resistant KPC carbapenemase-producing klebsiella pneumoniae strain of sequence type 512. Antimicrob. Agents Chemother. 60, 5612–5615. doi: 10.1128/AAC.01075-16
Dobias, J., Poirel, L., Nordmann, P. (2017). Cross-resistance to human cationic antimicrobial peptides and to polymyxins mediated by the plasmid-encoded MCR-1? Clin. Microbiol. Infect. 23, 676.e671–676.e675. doi: 10.1016/j.cmi.2017.03.015
Dong, H., Xiang, H., Mu, D., Wang, D., Wang, T. (2019). Exploiting a conjugative CRISPR/Cas9 system to eliminate plasmid harbouring the mcr-1 gene from escherichia coli. Int. J. Antimicrob. Agents 53, 1–8. doi: 10.1016/j.ijantimicag.2018.09.017
Dortet, L., Bonnin, R. A., Pennisi, I., Gauthier, L., Jousset, A. B., Dabos, L., et al. (2018). Rapid detection and discrimination of chromosome- and MCR-plasmid-mediated resistance to polymyxins by MALDI-TOF MS in escherichia coli: the MALDIxin test. J. Antimicrob. Chemother. 73, 3359–3367. doi: 10.1093/jac/dky330
Dos Santos, A., da Silva, M. F., de Araújo-Júnior, J. X., da Silva-Júnior, E. F. (2021). Revealing insights into natural products against mcr-1-Producing bacteria. Curr. Drug Targets 22, 1964–1985. doi: 10.2174/1389450122666210415102413
Dos Santos, L. D. R., Furlan, J. P. R., Ramos, M. S., Gallo, I. F. L., de Freitas, L. V. P., Stehling, E. G.. (2020). Co-Occurrence of mcr-1, mcr-3, mcr-7 and clinically relevant antimicrobial resistance genes in environmental and fecal samples. Arch. Microbiol. 202, 1795–1800. doi: 10.1007/s00203-020-01890-3
Doumith, M., Godbole, G., Ashton, P., Larkin, L., Dallman, T., Day, M., et al. (2016). Detection of the plasmid-mediated mcr-1 gene conferring colistin resistance in human and food isolates of salmonella enterica and escherichia coli in England and Wales. J. Antimicrob. Chemother. 71, 2300–2305. doi: 10.1093/jac/dkw093
Drider, D., Bendali, F., Naghmouchi, K., Chikindas, M. L. (2016). Bacteriocins: not only antibacterial agents. Probiotics Antimicrob. Proteins 8, 177–182. doi: 10.1007/s12602-016-9223-0
Elwell, L. P., Ferone, R., Freeman, G. A., Fyfe, J. A., Hill, J. A., Ray, P. H, et al. (1987). Antibacterial activity and mechanism of action of 3'-azido-3'-deoxythymidine (BW A509U). Antimicrob. Agents Chemother. 31, 274–280. doi: 10.1128/AAC.31.2.274
Etebu, E., Arikekpar, I. (2016). Antibiotics: Classification and mechanisms of action with emphasis on molecular perspectives. Int. J. Appl. Microbiol. Biotechnol. Res. 4, 90–101.
Fage, C. D., Brown, D. B., Boll, J. M., Keatinge-Clay, A. T., Trent, M. S. (2014). Crystallographic study of the phosphoethanolamine transferase EptC required for polymyxin resistance and motility in campylobacter jejuni. Acta Crystallogr. D Biol. Crystallogr. 70, 2730–2739. doi: 10.1107/S1399004714017623
Falagas, M. E., Kasiakou, S. K., Saravolatz, L. D. (2005). Colistin: the revival of polymyxins for the management of multidrug-resistant gram-negative bacterial infections. Clin. Infect. Dis. 40, 1333–1341. doi: 10.1086/429323
Fleres, G., Couto, N., Schuele, L., Chlebowicz, M. A., Mendes, C. I., van der Sluis, L. W. M., et al. (2019). Detection of a novel mcr-5.4 gene variant in hospital tap water by shotgun metagenomic sequencing. J. Antimicrob. Chemother. 74, 3626–3628. doi: 10.1093/jac/dkz363
Furlan, J. P. R., Dos Santos, L. D.R., Ramos, M. S., Gallo, I. F. L., Moretto, J. A. S., Stehling, E. G.. (2020a). Occurrence of clinically relevant antimicrobial resistance genes, including mcr-3 and mcr-7.1, in soil and water from a recreation club. Int. J. Environ. Health Res. 32, 1–10. doi: 10.1080/09603123.2020.1799953
Furlan, J. P. R., Dos Santos, L. D. R., Moretto, J. A. S., Ramos, M. S., Gallo, I. F. L., Alves, G. A. D., et al. (2020b). Occurrence and abundance of clinically relevant antimicrobial resistance genes in environmental samples after the brumadinho dam disaster, Brazil. Sci. Total Environ. 726, 138100. doi: 10.1016/j.scitotenv.2020.138100
Fyfe, C., LeBlanc, G., Close, B., Nordmann, P., Dumas, J., Grossman, T. H.. (2016). Eravacycline is active against bacterial isolates expressing the polymyxin resistance gene mcr-1. Antimicrob. Agents Chemother. 60, 6989–6990. doi: 10.1128/AAC.01646-16
Gao, R., Hu, Y., Li, Z., Sun, J., Wang, Q., Lin, J., et al. (2016). Dissemination and mechanism for the MCR-1 colistin resistance. PloS Pathog. 12, e1005957. doi: 10.1371/journal.ppat.1005957
García, V., García-Meniño, I., Mora, A., Flament-Simon, S. C., Díaz-Jiménez, D., Blanco, J. E., et al. (2018). Co-Occurrence of mcr-1, mcr-4 and mcr-5 genes in multidrug-resistant ST10 enterotoxigenic and shiga toxin-producing escherichia coli in Spain (2006-2017). Int. J. Antimicrob. Agents 52, 104–108. doi: 10.1016/j.ijantimicag.2018.03.022
Garcia-Graells, C., De Keersmaecker, S. C.J., Vanneste, K., Pochet, B., Vermeersch, K., Roosens, N., et al. (2018). Detection of plasmid-mediated colistin resistance, mcr-1 and mcr-2 genes, in salmonella spp. isolated from food at retail in Belgium from 2012 to 2015. Foodborne Pathog. Dis. 15, 114–117. doi: 10.1089/fpd.2017.2329
Gerstmans, H., Rodríguez-Rubio, L., Lavigne, R., Briers, Y. (2016). From endolysins to artilysin® s: novel enzyme-based approaches to kill drug-resistant bacteria. Biochem. Soc. Trans. 44, 123–128. doi: 10.1042/BST20150192
Ghasemian, A., Mohabati Mobarez, A., Najar Peerayeh, S., Talebi Bezmin Abadi, A., Khodaparast, S., Mahmood, S. S.. (2019). Expression of adhesin genes and biofilm formation among klebsiella oxytoca clinical isolates from patients with antibiotic-associated haemorrhagic colitis. J. Med. Microbiol. 68, 978–985. doi: 10.1099/jmm.0.000965
Giordano, C., Klak, A., Barnini, S., Chlebowicz, M. A., Menconi, M., Rossen, J. W., et al. (2019). Reduced fitness costs of mcr-1.2 compared to mutated pmrB in isogenic colistin-resistant KPC-3-Producing klebsiella pneumoniae. mSphere 4:e00551–00519. doi: 10.1128/mSphere.00551-19
Gomi, R., Matsuda, T., Yamamoto, M., Tanaka, M., Ichiyama, S., Yoneda, M., et al. (2019). Molecular characterization of a multidrug-resistant IncF plasmid carrying mcr-3.1 in an escherichia coli sequence type 393 strain of wastewater origin. Int. J. Antimicrob. Agents 54, 524–526. doi: 10.1016/j.ijantimicag.2019.06.024
Gröndahl-Yli-Hannuksela, K., Lönnqvist, E., Kallonen, T., Lindholm, L., Jalava, J., Rantakokko-Jalava, K., et al. (2018). The first human report of mobile colistin resistance gene, mcr-1, in Finland. APMIS 126, 413–417. doi: 10.1111/apm.12834
Guachalla, L. M., Ramoni, K., Varga, C., Mutti, M., Ghazawi, A., Pál, T., et al. (2018). Retained activity of an O25b-specific monoclonal antibody against an mcr-1-Producing escherichia coli sequence type 131 strain. Antimicrob. Agents Chemother. 62:e00046–00018. doi: 10.1128/AAC.00046-18
Gunn, J. S. (2008). The salmonella PmrAB regulon: lipopolysaccharide modifications, antimicrobial peptide resistance and more. Trends Microbiol. 16, 284–290. doi: 10.1016/j.tim.2008.03.007
Guo, S., Tay, M. Y. F., Thu, A. K., Seow, K. L. G., Zhong, Y., Ng, L. C., et al. (2019). Conjugative IncX1 plasmid harboring colistin resistance gene mcr-5.1 in escherichia coli isolated from chicken rice retailed in Singapore. Antimicrob. Agents Chemother. 63:e01043–01019. doi: 10.1128/aac.01043-19
Guo, Y., Lv, X., Wang, Y., Zhou, Y., Lu, N., Deng, X., et al. (2020). Honokiol restores polymyxin susceptibility to MCR-1-Positive pathogens both In vitro and in vivo. Appl. Environ. Microbiol. 86:e02346–02319. doi: 10.1128/AEM.02346-19
Haenni, M., Métayer, V., Gay, E., Madec, J. Y. (2016). Increasing trends in mcr-1 prevalence among extended-Spectrum-β-Lactamase-Producing escherichia coli isolates from French calves despite decreasing exposure to colistin. Antimicrob. Agents Chemother. 60, 6433–6434. doi: 10.1128/AAC.01147-16
Hammerl, J. A., Borowiak, M., Schmoger, S., Shamoun, D., Grobbel, M., Malorny, B., et al. (2018). Mcr-5 and a novel mcr-5.2 variant in escherichia coli isolates from food and food-producing animals, Germany, 2010 to 2017. J. Antimicrob. Chemother. 73, 1433–1435. doi: 10.1093/jac/dky020
Hassan, J., Eddine, R. Z., Mann, D., Li, S., Deng, X., Saoud, I. P., et al. (2020). The mobile colistin resistance gene, mcr-1.1, is carried on IncX4 plasmids in multidrug resistant e. coli isolated from rainbow trout aquaculture. Microorganisms 8:1636. doi: 10.3390/microorganisms8111636
He, T., Wei, R., Zhang, L., Sun, L., Pang, M., Wang, R., et al. (2017). Characterization of NDM-5-positive extensively resistant escherichia coli isolates from dairy cows. Vet. Microbiol. 207, 153–158. doi: 10.1016/j.vetmic.2017.06.010
He, Y.-Z., Li, X.-P., Miao, Y.-Y., Lin, J., Sun, R.-Y., Wang, X.-P., et al. (2019). The ISApl12 dimer circular intermediate participates in mcr-1 transposition. Front. Microbiol. 10, 15. doi: 10.3389/fmicb.2019.00015
He, Y. Z., Yan, J. R., He, B., Ren, H., Kuang, X., Long, T. F., et al. (2021). A transposon-associated CRISPR/Cas9 system specifically eliminates both chromosomal and plasmid-borne mcr-1 in escherichia coli. Antimicrob. Agents Chemother., Aac0105421. doi: 10.1128/AAC.01054-21
Henig, O., Rojas, L. J., Bachman, M. A., Rudin, S. D., Brennan, B. M., Soehnlen, M. K., et al. (2019). Identification of four patients with colistin-resistant escherichia coli containing the mobile colistin resistance mcr-1 gene from a single health system in Michigan. Infect. Control Hosp. Epidemiol. 40, 1059–1062. doi: 10.1017/ice.2019.177
Hille, K., Roschanski, N., Ruddat, I., Woydt, J., Hartmann, M., Rösler, U., et al. (2018). Investigation of potential risk factors for the occurrence of escherichia coli isolates from German fattening pig farms harbouring the mcr-1 colistin-resistance gene. Int. J. Antimicrob. Agents 51, 177–180. doi: 10.1016/j.ijantimicag.2017.08.007
Hinchliffe, P., Yang, Q. E., Portal, E., Young, T., Li, H., Tooke, C. L, et al. (2017). Insights into the mechanistic basis of plasmid-mediated colistin resistance from crystal structures of the catalytic domain of MCR-1. Sci. Rep. 7, 39392. doi: 10.1038/srep39392
Hmede, Z., Kassem, I. I. (2018). The colistin resistance gene mcr-1 is prevalent in commensal escherichia coli isolated from preharvest poultry in Lebanon. Antimicrob. Agents Chemother. 62:e01304–01318. doi: 10.1128/AAC.01304-18
Hryhorowicz, M., Lipiński, D., Zeyland, J., Słomski, R. (2017). CRISPR/Cas9 immune system as a tool for genome engineering. Archivum. Immunol. Therapiae Experimentalis 65, 233–240. doi: 10.1007/s00005-016-0427-5
Hsu, L. Y., Apisarnthanarak, A., Khan, E., Suwantarat, N., Ghafur, A., Tambyah, P.A. (2017). Carbapenem-resistant acinetobacter baumannii and enterobacteriaceae in south and southeast Asia. Clin. Microbiol. Rev. 30, 1–22. doi: 10.1128/CMR.00042-16
Hu, M., Guo, J., Cheng, Q., Yang, Z., Chan, E. W. C., Chen, S, et al. (2016). Crystal structure of escherichia coli originated MCR-1, a phosphoethanolamine transferase for colistin resistance. Sci. Rep. 6, 38793. doi: 10.1038/srep38793
Hu, Y., Liu, Y., Coates, A. (2019). Azidothymidine produces synergistic activity in combination with colistin against antibiotic-resistant enterobacteriaceae. Antimicrob. Agents Chemother. 63:e01630–01618. doi: 10.1128/AAC.01630-18
Hu, Y., Liu, F., Lin, I. Y., Gao, G. F., Zhu, B. (2016). Dissemination of the mcr-1 colistin resistance gene. Lancet Infect. Dis. 16, 146–147. doi: 10.1016/S1473-3099(15)00533-2
Islam, S., Urmi, U. L., Rana, M., Sultana, F., Jahan, N., Hossain, B., et al. (2020). High abundance of the colistin resistance gene mcr-1 in chicken gut-bacteria in Bangladesh. Sci. Rep. 10, 17292. doi: 10.1038/s41598-020-74402-4
Ji, X., Zheng, B., Berglund, B., Zou, H., Sun, Q., Chi, X., et al. (2019). Dissemination of extended-spectrum β-lactamase-producing escherichia coli carrying mcr-1 among multiple environmental sources in rural China and associated risk to human health. Environ. pollut. 251, 619–627. doi: 10.1016/j.envpol.2019.05.002
Kananizadeh, P., Oshiro, S., Watanabe, S., Iwata, S., Kuwahara-Arai, K., Shimojima, M., et al. (2020). Emergence of carbapenem-resistant and colistin-susceptible enterobacter cloacae complex co-harboring bla(IMP-1) and mcr-9 in Japan. BMC Infect. Dis. 20, 282. doi: 10.1186/s12879-020-05021-7
Kassem, I. I. (2020). First report of the plasmid-borne colistin resistance gene (mcr-1) in Proteus mirabilis isolated from domestic and sewer waters in Syrian refugee camps. Travel Med. Infect. Dis. 33, 101482. doi: 10.1016/j.tmaid.2019.101482
Kempf, I., Jouy, E., Chauvin, C. (2016). Colistin use and colistin resistance in bacteria from animals. Int. J. Antimicrob. Agents 48, 598–606. doi: 10.1016/j.ijantimicag.2016.09.016
Khanawapee, A., Kerdsin, A., Chopjitt, P., Boueroy, P., Hatrongjit, R., Akeda, Y., et al. (2021). Distribution and molecular characterization of escherichia coli harboring mcr genes isolated from slaughtered pigs in Thailand. Microb. Drug Resist. 27, 971–979. doi: 10.1089/mdr.2020.0242
Kieffer, N., Nordmann, P., Moreno, A. M., Zanolli Moreno, L., Chaby, R., Breton, A., et al. (2018). Genetic and functional characterization of an MCR-3-Like enzyme-producing escherichia coli isolate recovered from swine in Brazil. Antimicrob. Agents Chemother. 62:e00278–00218. doi: 10.1128/AAC.00278-18
Kieffer, N., Nordmann, P., Millemann, Y., Poirel, L. (2019). Mcr-9, an inducible gene encoding an acquired phosphoethanolamine transferase in escherichia coli, and its origin. Antimicrob. Agents Chemother. 63:e00559–00519. doi: 10.1128/AAC.00965-19
Kieffer, N., Nordmann, P., Millemann, Y., Poirel, L. (2019). Functional characterization of a miniature inverted transposable element at the origin of mcr-5 gene acquisition in escherichia coli. Antimicrob. Agents Chemother. 63. doi: 10.1128/AAC.00559-19
Kieffer, N., Nordmann, P., Poirel, L. (2017). Moraxella species as potential sources of MCR-like polymyxin resistance determinants. Antimicrob. Agents Chemother. 61:e00129–00117. doi: 10.1128/AAC.00129-17
Lei, C. W., Zhang, Y., Wang, Y. T., Wang, H. N. (2020). Detection of mobile colistin resistance gene mcr-10.1 in a conjugative plasmid from enterobacter roggenkampii of chicken origin in China. Antimicrob. Agents Chemother. 64. doi: 10.1128/aac.01191-20
Li, R., Chen, K., Chan, E. W., Chen, S. (2019). Characterization of the stability and dynamics of Tn6330 in an escherichia coli strain by nanopore long reads. J. Antimicrob. Chemother. 74, 1807–1811. doi: 10.1093/jac/dkz117
Li, R., Xie, M., Zhang, J., Yang, Z., Liu, L., Liu, X., et al. (2017). Genetic characterization of mcr-1-bearing plasmids to depict molecular mechanisms underlying dissemination of the colistin resistance determinant. J. Antimicrob. Chemother. 72, 393–401. doi: 10.1093/jac/dkw411
Li, R., Yu, H., Xie, M., Chen, K., Dong, N., Lin, D., et al. (2018). Genetic basis of chromosomally-encoded mcr-1 gene. Int. J. Antimicrob. Agents 51, 578–585. doi: 10.1016/j.ijantimicag.2017.11.015
Li, Y., Lin, X., Yao, X., Huang, Y., Liu, W., Ma, T., et al. (2018). Synergistic antimicrobial activity of colistin in combination with rifampin and azithromycin against escherichia coli producing MCR-1. Antimicrob. Agents Chemother. 62:e01631–01618. doi: 10.1128/AAC.01631-18
Li, Y., Dai, X., Zeng, J., Gao, Y., Zhang, Z., Zhang, L.. (2020). Characterization of the global distribution and diversified plasmid reservoirs of the colistin resistance gene mcr-9. Sci. Rep. 10, 8113. doi: 10.1038/s41598-020-65106-w
Lin, M., Yang, Y., Yang, Y., Chen, G., He, R., Wu, Y., et al. (2020). Co-Occurrence of mcr-9 and bla (NDM-1) in enterobacter cloacae isolated from a patient with bloodstream infection. Infect. Drug Resist. 13, 1397–1402. doi: 10.2147/IDR.S248342
Litrup, E., Kiil, K., Hammerum, A. M., Roer, L., Nielsen, E. M., Torpdahl, M.. (2017). Plasmid-borne colistin resistance gene mcr-3 in salmonella isolates from human infections, Denmark, 2009-17. Euro Surveill. 22:30587. doi: 10.2807/1560-7917.ES.2017.22.31.30587
Liu, Y. Y., Wang, Y., Walsh, T. R., Yi, L. X., Zhang, R., Spencer, J, et al. (2016). Emergence of plasmid-mediated colistin resistance mechanism MCR-1 in animals and human beings in China: a microbiological and molecular biological study. Lancet Infect. Dis. 16, 161–168. doi: 10.1016/S1473-3099(15)00424-7
Liu, L., Feng, Y., Zhang, X., McNally, A., Zong, Z. (2017). New variant of mcr-3 in an extensively drug-resistant escherichia coli clinical isolate carrying mcr-1 and bla(NDM-5). Antimicrob. Agents Chemother. 61:e01757–01717. doi: 10.1128/AAC.01757-17
Liu, Z.-X., Han, Z., Yu, X.-L., Wen, G., Zeng, C. (2018). Crystal structure of the catalytic domain of MCR-1 (cMCR-1) in complex with d-xylose. Crystals (Basel) 8, 172. doi: 10.3390/cryst8040172
Liu, X., Sun, X., Deng, X., Lv, X., Wang, J. (2020). Calycosin enhances the bactericidal efficacy of polymyxin b by inhibiting MCR-1 in vitro. J. Appl. Microbiol. 129, 532–540. doi: 10.1111/jam.14635
Long, H., Feng, Y., Ma, K., Liu, L., McNally, A., Zong, Z.. (2019). The co-transfer of plasmid-borne colistin-resistant genes mcr-1 and mcr-3.5, the carbapenemase gene bla(NDM-5) and the 16S methylase gene rmtB from escherichia coli. Sci. Rep. 9, 696. doi: 10.1038/s41598-018-37125-1
Lu, X., Zeng, M., Xu, J., Zhou, H., Gu, B., Li, Z., et al. (2019). Epidemiologic and genomic insights on mcr-1-harbouring salmonella from diarrhoeal outpatients in shanghai, China, 2006-2016. EBioMedicine 42, 133–144. doi: 10.1016/j.ebiom.2019.03.006
Lu, N., Lv, Q., Sun, X., Zhou, Y., Guo, Y., Qiu, J., et al. (2020). Isoalantolactone restores the sensitivity of gram-negative enterobacteriaceae carrying MCR-1 to carbapenems. J. Cell Mol. Med. 24, 2475–2483. doi: 10.1111/jcmm.14936
Luo, J., Yao, X., Lv, L., Doi, Y., Huang, X., Huang, S, et al. (2017). Emergence of mcr-1 in raoultella ornithinolytica and escherichia coli isolates from retail vegetables in China. Antimicrob. Agents Chemother. 61:e01139–01117. doi: 10.1128/AAC.01139-17
MacNair, C. R., Stokes, J. M., Carfrae, L. A., Fiebig-Comyn, A. A., Coombes, B. K., Mulvey, M. R., et al. (2018). Overcoming mcr-1 mediated colistin resistance with colistin in combination with other antibiotics. Nat. Commun. 9, 458. doi: 10.1038/s41467-018-02875-z
Ma, S., Sun, C., Hulth, A., Li, J., Nilsson, L. E., Zhou, Y., et al. (2018). Mobile colistin resistance gene mcr-5 in porcine aeromonas hydrophila. J. Antimicrob. Chemother. 73, 1777–1780. doi: 10.1093/jac/dky110
Ma, K., Feng, Y., Zong, Z. (2018). Fitness cost of a mcr-1-carrying IncHI2 plasmid. PloS One 13, e0209706. doi: 10.1371/journal.pone.0209706
Malhotra-Kumar, S., Xavier, B. B., Das, A. J., Lammens, C., Hoang, H. T., Pham, N. T., et al. (2016). Colistin-resistant escherichia coli harbouring mcr-1 isolated from food animals in Hanoi, Vietnam. Lancet Infect. Dis. 16, 286–287. doi: 10.1016/S1473-3099(16)00014-1
Ma, G., Zhu, Y., Yu, Z., Ahmad, A., Zhang, H. (2016). High resolution crystal structure of the catalytic domain of MCR-1. Sci. Rep. 6, 1–7. doi: 10.1038/srep39540
Migura-Garcia, L., González-López, J. J., Martinez-Urtaza, J., Aguirre Sánchez, J. R., Moreno-Mingorance, A., Perez de Rozas, A., et al. (2020). Mcr-colistin resistance genes mobilized by IncX4, IncHI2, and IncI2 plasmids in escherichia coli of pigs and white stork in Spain. Front. Microbiol. 10. doi: 10.3389/fmicb.2019.03072
Mmatli, M., Mbelle, N. M., Maningi, N. E., Osei Sekyere, J. (2020). Emerging transcriptional and genomic mechanisms mediating carbapenem and polymyxin resistance in enterobacteriaceae: a systematic review of current reports. mSystems 5, e00783–e00720. doi: 10.1128/mSystems.00783-20
Monte, D. F., Mem, A., Fernandes, M. R., Cerdeira, L., Esposito, F., Galvão, J. A, et al. (2017). Chicken meat as a reservoir of colistin-resistant escherichia coli strains carrying mcr-1 genes in south America. Antimicrob. Agents Chemother. 61:e02718–02716. doi: 10.1128/AAC.02718-16
Nabti, L. Z., Sahli, F., Ngaiganam, E. P., Radji, N., Mezaghcha, W., Lupande-Mwenebitu, D., et al. (2020). Development of real-time PCR assay allowed describing the first clinical klebsiella pneumoniae isolate harboring plasmid-mediated colistin resistance mcr-8 gene in Algeria. J. Glob. Antimicrob. Resist. 20, 266–271. doi: 10.1016/j.jgar.2019.08.018
Nang, S. C., Morris, F. C., McDonald, M. J., Han, M. L., Wang, J., Strugnell, R. A., et al. (2018). Fitness cost of mcr-1-mediated polymyxin resistance in klebsiella pneumoniae. J. Antimicrob. Chemother. 73, 1604–1610. doi: 10.1093/jac/dky061
Napier, B. A., Burd, E. M., Satola, S. W., Cagle, S. M., Ray, S. M., McGann, P., et al. (2013). Clinical use of colistin induces cross-resistance to host antimicrobials in acinetobacter baumannii. mBio 4, e00021–e00013. doi: 10.1128/mBio.00021-13
Nezhadi, J., Narenji, H., Soroush Barhaghi, M. H., Rezaee, M. A., Ghotaslou, R., Pirzadeh, T., et al. (2019). Peptide nucleic acid-mediated re-sensitization of colistin resistance escherichia coli KP81 harboring mcr-1 plasmid. Microb. Pathog. 135, 103646. doi: 10.1016/j.micpath.2019.103646
Ngbede, E. O., Poudel, A., Kalalah, A., Yang, Y., Adekanmbi, F., Adikwu, A. A., et al. (2020). Identification of mobile colistin resistance genes (mcr-1.1, mcr-5 and mcr-8.1) in enterobacteriaceae and alcaligenes faecalis of human and animal origin, Nigeria. Int. J. Antimicrob. Agents 56, 106108. doi: 10.1016/j.ijantimicag.2020.106108
Nguyen, N. T., Nguyen, H. M., Nguyen, C. V., Nguyen, T. V., Nguyen, M. T., Thai, H. Q, et al. (2016). Use of colistin and other critical antimicrobials on pig and chicken farms in southern Vietnam and its association with resistance in commensal escherichia coli bacteria. Appl. Environ. Microbiol. 82, 3727–3735. doi: 10.1128/AEM.00337-16
Nigam, A., Kumari, A., Jain, R., Batra, S. (2015). Colistin neurotoxicity: revisited. Case Rep. 2015, bcr2015210787. doi: 10.1136/bcr-2015-210787
Okdah, L., Le Page, S., Olaitan, A. O., Dubourg, G., Hadjadj, L., Rolain, J. M.. (2018). New therapy from old drugs: synergistic bactericidal activity of sulfadiazine with colistin against colistin-resistant bacteria, including plasmid-mediated colistin-resistant mcr-1 isolates. Int. J. Antimicrob. Agents 51, 775–783. doi: 10.1016/j.ijantimicag.2018.01.027
Osei Sekyere, J., Amoako, D. G. (2017). Carbonyl cyanide m-chlorophenylhydrazine (CCCP) reverses resistance to colistin, but not to carbapenems and tigecycline in multidrug-resistant enterobacteriaceae. Front. Microbiol. 8, 228. doi: 10.3389/fmicb.2017.00228
Osei Sekyere, J., Maningi, N. E., Modipane, L., Mbelle, N. M. (2020). Emergence of mcr-9.1 in extended-Spectrum-β-Lactamase-Producing clinical enterobacteriaceae in Pretoria, south Africa: Global evolutionary phylogenomics, resistome, and mobilome. mSystems 5:e00148–00120 . doi: 10.1128/mSystems.00148-20
Partridge, S. R. (2017). Mcr-2 in the IncX4 plasmid pKP37-BE is flanked by directly oriented copies of ISEc69. J. Antimicrob. Chemother. 72, 1533–1535. doi: 10.1093/jac/dkw575
Partridge, S. R., et al. (2018). Proposal for assignment of allele numbers for mobile colistin resistance (mcr) genes. J. Antimicrob. Chemother. 73, 2625–2630. doi: 10.1093/jac/dky262
Paterson, D. L., Harris, P. N. (2016). Colistin resistance: a major breach in our last line of defence. Lancet Infect. Dis. 16, 132–133. doi: 10.1016/S1473-3099(15)00463-6
Peng, C., Zuo, S., Qiu, Y., Fu, S., Peng, L. (2021). Determination of colistin in contents derived from gastrointestinal tract of feeding treated piglet and broiler. Antibiotics (Basel) 10:422. doi: 10.3390/antibiotics10040422
Petrillo, M., Angers-Loustau, A., Kreysa, J. (2016). Possible genetic events producing colistin resistance gene mcr-1. Lancet Infect. Dis. 16, 280. doi: 10.1016/S1473-3099(16)00005-0
Peyclit, L., Baron, S. A., Yousfi, H., Rolain, J. M. (2018). Zidovudine: A salvage therapy for mcr-1 plasmid-mediated colistin-resistant bacterial infections? Int. J. Antimicrob. Agents 52, 11–13. doi: 10.1016/j.ijantimicag.2018.03.012
Phan, M.-D., Nhu, N. T.K., Achard, M. E., Forde, B. M., Hong, K. W., Chong, T. M., et al. (2017). Modifications in the pmrB gene are the primary mechanism for the development of chromosomally encoded resistance to polymyxins in uropathogenic escherichia coli. J. Antimicrob. Chemother. 72, 2729–2736. doi: 10.1093/jac/dkx204
Poirel, L., Kieffer, N., Brink, A., Coetze, J., Jayol, A., Nordmann, P.. (2016). Genetic features of MCR-1-Producing colistin-resistant escherichia coli isolates in south Africa. Antimicrob. Agents Chemother. 60, 4394–4397. doi: 10.1128/AAC.00444-16
Poirel, L., Kieffer, N., Fernandez-Garayzabal, J. F., Vela, A. I., Larpin, Y., Nordmann, P.. (2017). MCR-2-mediated plasmid-borne polymyxin resistance most likely originates from moraxella pluranimalium. J. Antimicrob. Chemother. 72, 2947–2949. doi: 10.1093/jac/dkx225
Poirel, L., Kieffer, N., Nordmann, P. (2017). In vitroIn Vitro study of ISApl1-mediated mobilization of the colistin resistance gene mcr-1. Antimicrob. Agents Chemother. 61, e00127–e00117. doi: 10.1128/AAC.00127-17
Randall, L. P., Horton, R. A., Lemma, F., Martelli, F., Duggett, N. A. D., Smith, R. P., et al. (2018). Longitudinal study on the occurrence in pigs of colistin-resistant escherichia coli carrying mcr-1 following the cessation of use of colistin. J. Appl. Microbiol. 125, 596–608. doi: 10.1111/jam.13907
Rebelo, A. R., Bortolaia, V., Kjeldgaard, J. S., Pedersen, S. K., Leekitcharoenphon, P., Hansen, I. M., et al. (2018). Multiplex PCR for detection of plasmid-mediated colistin resistance determinants, mcr-1, mcr-2, mcr-3, mcr-4 and mcr-5 for surveillance purposes. Euro Surveill. 23:17-00672. doi: 10.2807/1560-7917.ES.2018.23.6.17-00672
Rhouma, M., Beaudry, F., Thériault, W., Bergeron, N., Laurent-Lewandowski, S., Fairbrother, J. M., et al. (2015). Gastric stability and oral bioavailability of colistin sulfate in pigs challenged or not with escherichia coli O149: F4 (K88). Res. Vet. Sci. 102, 173–181. doi: 10.1016/j.rvsc.2015.08.005
Ribeiro, T. G., Izdebski, R., Urbanowicz, P., Carmeli, Y., Gniadkowski, M., Peixe, L.. (2021). Citrobacter telavivum sp. nov. with chromosomal mcr-9 from hospitalized patients. Eur. J. Clin. Microbiol. Infect. Dis. 40, 123–131. doi: 10.1007/s10096-020-04003-6
Roer, L., Hansen, F., Stegger, M., Sönksen, U. W., Hasman, H., Hammerum, A. M.. (2017). Novel mcr-3 variant, encoding mobile colistin resistance, in an ST131 escherichia coli isolate from bloodstream infection, Denmark, 2014. Euro Surveill. 22:30584. doi: 10.2807/1560-7917.ES.2017.22.31.30584
Runcharoen, C., Raven, K. E., Reuter, S., Kallonen, T., Paksanont, S., Thammachote, J., et al. (2017). Whole genome sequencing of ESBL-producing escherichia coli isolated from patients, farm waste and canals in Thailand. Genome Med. 9, 81. doi: 10.1186/s13073-017-0471-8
Saavedra, S. Y., Diaz, L., Wiesner, M., Correa, A., Arévalo, S. A., Reyes, J. (2017). Genomic and molecular characterization of clinical isolates of enterobacteriaceae harboring mcr-1 in Colombia, 2002 to 2016. Antimicrob. Agents Chemother. 61. doi: 10.1128/AAC.00841-17
Salloum, T., Panossian, B., Bitar, I., Hrabak, J., Araj, G. F., Tokajian, S.. (2020). First report of plasmid-mediated colistin resistance mcr-8.1 gene from a clinical klebsiella pneumoniae isolate from Lebanon. Antimicrob. Resist. Infect. Control 9, 94. doi: 10.1186/s13756-020-00759-w
Sato, T., Usui, M., Harada, K., Fukushima, Y., Nakajima, C., Suzuki, Y., et al. (2021). Complete genome sequence of an mcr-10-Possessing enterobacter roggenkampii strain isolated from a dog in Japan. Microbiol. Resour. Announc. 10, e00426–e00421. doi: 10.1128/MRA.00426-21
Schirmeier, E., Zimmermann, P., Hofmann, V., Biebl, M., Gerstmans, H., Maervoet, V. E. T, et al. (2018). Inhibitory and bactericidal effect of artilysin(®) art-175 against colistin-resistant mcr-1-positive escherichia coli isolates. Int. J. Antimicrob. Agents 51, 528–529. doi: 10.1016/j.ijantimicag.2017.08.027
Shen, Y., Zhou, H., Xu, J., Wang, Y., Zhang, Q., Walsh, T. R., et al. (2018). Anthropogenic and environmental factors associated with high incidence of mcr-1 carriage in humans across China. Nat. Microbiol. 3, 1054–1062. doi: 10.1038/s41564-018-0205-8
Shen, Y., Lv, Z., Yang, L., Liu, D., Ou, Y., Xu, C., et al. (2019). Integrated aquaculture contributes to the transfer of mcr-1 between animals and humans via the aquaculture supply chain. Environ. Int. 130, 104708. doi: 10.1016/j.envint.2019.03.056
Shen, Y., Zhang, R., Schwarz, S., Wu, C., Shen, J., Walsh, T. R., et al. (2020). Farm animals and aquaculture: significant reservoirs of mobile colistin resistance genes. Environ. Microbiol. 22, 2469–2484. doi: 10.1111/1462-2920.14961
Sia, C. M., Greig, D. R., Day, M., Hartman, H., Painset, A., Doumith, M., et al. (2020). The characterization of mobile colistin resistance (mcr) genes among 33 000 salmonella enterica genomes from routine public health surveillance in England. Microb. Genom. 6:e000331. doi: 10.1099/mgen.0.000331
Skov, R. L., Monnet, D. L. (2016). Plasmid-mediated colistin resistance (mcr-1 gene): three months later, the story unfolds. Euro Surveill. 21, 30155. doi: 10.2807/1560-7917.ES.2016.21.9.30155
Snesrud, E., He, S., Chandler, M., Dekker, J. P., Hickman, A. B., McGann, P, et al. (2016). A model for transposition of the colistin resistance gene mcr-1 by IS Apl1. Antimicrob. Agents Chemother. 60, 6973–6976. doi: 10.1128/AAC.01457-16
Snesrud, E., Ong, A. C., Corey, B., Kwak, Y. I., Clifford, R., Gleeson, T., et al. (2017). Analysis of serial isolates of mcr-1-Positive escherichia coli reveals a highly active ISApl1 transposon. Antimicrob. Agents Chemother. 61:e00056–00017. doi: 10.1128/aac.00056-17
Snesrud, E., Maybank, R., Kwak, Y. I., Jones, A. R., Hinkle, M. K., McGann, P.. (2018). Chromosomally encoded mcr-5 in colistin-nonsusceptible pseudomonas aeruginosa. Antimicrob. Agents Chemother. 62:e00679–00618. doi: 10.1128/AAC.00679-18
Snesrud, E., McGann, P., Chandler, M. (2018). The birth and demise of the IS Apl1-mcr-1-IS Apl1 composite transposon: the vehicle for transferable colistin resistance. mBio 9, e02381–e02317. doi: 10.1128/mBio.02381-17
Snyman, Y., Reuter, S., Whitelaw, A. C., Stein, L., Maloba, M. R. B., Newton-Foot, M.. (2021). Characterisation of mcr-4.3 in a colistin-resistant acinetobacter nosocomialis clinical isolate from cape town, south Africa. J. Glob. Antimicrob. Resist. 25, 102–106. doi: 10.1016/j.jgar.2021.03.002
Stoesser, N., Mathers, A. J., Moore, C. E., Day, N. P., Crook, D. W. (2016). Colistin resistance gene mcr-1 and pHNSHP45 plasmid in human isolates of escherichia coli and klebsiella pneumoniae. Lancet Infect. Dis. 16, 285–286. doi: 10.1016/S1473-3099(16)00010-4
Stojanoski, V., Sankaran, B., Prasad, B. V., Poirel, L., Nordmann, P., Palzkill, T. (2016). Structure of the catalytic domain of the colistin resistance enzyme MCR-1. BMC Biol. 14, 1–10. doi: 10.1186/s12915-016-0303-0
Stokes, J. M., MacNair, C. R., Ilyas, B., French, S., Côté, J.-P., Bouwman, C., et al. (2017). Pentamidine sensitizes gram-negative pathogens to antibiotics and overcomes acquired colistin resistance. Nat. Microbiol. 2, 1–8. doi: 10.1038/nmicrobiol.2017.28
Sulaiman, A. A. A., Kassem, I. I. (2019). First report on the detection of the plasmid-borne colistin resistance gene mcr-1 in multi-drug resistant e. coli isolated from domestic and sewer waters in Syrian refugee camps in Lebanon. Travel Med. Infect. Dis. 30, 117–120. doi: 10.1016/j.tmaid.2019.06.014
Sun, J., Fang, L. X., Wu, Z., Deng, H., Yang, R. S., Li, X. P., et al. (2017a). Genetic analysis of the IncX4 plasmids: Implications for a unique pattern in the mcr-1 acquisition. Sci. Rep. 7, 424. doi: 10.1038/s41598-017-00095-x
Sun, J., Xu, Y., Gao, R., Lin, J., Wei, W., Srinivas, S., et al. (2017b). Deciphering MCR-2 colistin resistance. mBio 8. doi: 10.1128/mBio.00625-17
Sun, L., He, T., Zhang, L., Pang, M., Zhang, Q., Zhou, Y., et al. (2017). Generation of newly discovered resistance gene mcr-1 knockout in escherichia coli using the CRISPR/Cas9 system. J. Microbiol. Biotechnol. 27, 1276–1280. doi: 10.4014/jmb.1611.11021
Sun, J., Li, X. P., Fang, L. X., Sun, R. Y., He, Y. Z., Lin, J., et al. (2018). Co-Occurrence of mcr-1 in the chromosome and on an IncHI2 plasmid: persistence of colistin resistance in escherichia coli. Int. J. Antimicrob. Agents 51, 842–847. doi: 10.1016/j.ijantimicag.2018.01.007
Sun, Q., Wang, H., Shu, L., Dong, N., Yang, F., Zhou, H., et al. (2019). Leclercia adecarboxylata from human gut flora carries mcr-4.3 and bla (IMP-4)-Bearing plasmids. Front. Microbiol. 10, 2805. doi: 10.3389/fmicb.2019.02805
Sun, S., Gao, H., Liu, Y., Jin, L., Wang, R., Wang, X., et al. (2020). Co-Existence of a novel plasmid-mediated efflux pump with colistin resistance gene mcr in one plasmid confers transferable multidrug resistance in klebsiella pneumoniae. Emerg. Microbes Infect. 9, 1102–1113. doi: 10.1080/22221751.2020.1768805
Szabó, M., Kiss, J., Nagy, Z., Chandler, M., Olasz, F. (2008). Sub-Terminal sequences modulating IS30 transposition in vivo and in vitro. J. Mol. Biol. 375, 337–352. doi: 10.1016/j.jmb.2007.10.043
Tarabai, H., Valcek, A., Jamborova, I., Vazhov, S. V., Karyakin, I. V., Raab, R., et al. (2019). Plasmid-mediated mcr-1 colistin resistance in escherichia coli from a black kite in Russia. Antimicrob. Agents Chemother. 63. doi: 10.1128/AAC.01266-19
Tegetmeyer, H. E., Jones, S. C., Langford, P. R., Baltes, N. (2008). ISApl1, a novel insertion element of actinobacillus pleuropneumoniae, prevents ApxIV-based serological detection of serotype 7 strain AP76. Vet. Microbiol. 128, 342–353. doi: 10.1016/j.vetmic.2007.10.025
Teo, J. W. P., Kalisvar, M., Venkatachalam, I., Ng, O. T., Lin, R. T. P., Octavia, S., et al. (2018). Mcr-3 and mcr-4 variants in carbapenemase-producing clinical enterobacteriaceae do not confer phenotypic polymyxin resistance. J. Clin. Microbiol. 56. doi: 10.1128/JCM.01562-17
Tietgen, M., Semmler, T., Riedel-Christ, S., Kempf, V. A. J., Molinaro, A., Ewers, C., et al. (2018). Impact of the colistin resistance gene mcr-1 on bacterial fitness. Int. J. Antimicrob. Agents 51, 554–561. doi: 10.1016/j.ijantimicag.2017.11.011
Tijet, N., Faccone, D., Rapoport, M., Seah, C., Pasterán, F., Ceriana, P., et al. (2017). Molecular characteristics of mcr-1-carrying plasmids and new mcr-1 variant recovered from polyclonal clinical escherichia coli from Argentina and Canada. PloS One 12, e0180347. doi: 10.1371/journal.pone.0180347
Trung, N. V., Matamoros, S., Carrique-Mas, J. J., Nghia, N. H., Nhung, N. T., Chieu, T. T., et al. (2017). Zoonotic transmission of mcr-1 colistin resistance gene from small-scale poultry farms, Vietnam. Emerg. Infect. Dis. 23, 529–532. doi: 10.3201/eid2303.161553
Tyrrell, J. M., Aboklaish, A. F., Walsh, T. R., Vaara, T., Vaara, M. (2019). The polymyxin derivative NAB739 is synergistic with several antibiotics against polymyxin-resistant strains of escherichia coli, klebsiella pneumoniae and acinetobacter baumannii. Peptides 112, 149–153. doi: 10.1016/j.peptides.2018.12.006
Tyson, G. H., Li, C., Hsu, C. H., Ayers, S., Borenstein, S., Mukherjee, S., et al. (2020). The mcr-9 gene of salmonella and escherichia coli is not associated with colistin resistance in the united states. Antimicrob. Agents Chemother. 64:e00573–00520, . doi: 10.1128/AAC.00573-20
Ullah, S., Ji, K., Li, J., Xu, Y., Jiang, C., Zhang, H., et al. (2021). Characterization of NMCR-2, a new non-mobile colistin resistance enzyme: implications for an MCR-8 ancestor. Environ. Microbiol. 23, 844–860. doi: 10.1111/1462-2920.15171
van der Weide, H., Jongh Vermeulen-de, H. D., Meijden der van, M., Boers, A. S., Kreft, A., Marian, D. T., et al. (2019). Antimicrobial activity of two novel antimicrobial peptides AA139 and SET-M33 against clinically and genotypically diverse klebsiella pneumoniae isolates with differing antibiotic resistance profiles. Int. J. Antimicrob. Agents 54, 159–166. doi: 10.1016/j.ijantimicag.2019.05.019
Wang, Y., Tian, G. B., Zhang, R., Shen, Y., Tyrrell, J. M., Huang, X., et al. (2017). Prevalence, risk factors, outcomes, and molecular epidemiology of mcr-1-positive enterobacteriaceae in patients and healthy adults from China: an epidemiological and clinical study. Lancet Infect. Dis. 17, 390–399. doi: 10.1016/S1473-3099(16)30527-8
Wang, Q., Wang, X., Wang, J., Ouyang, P., Jin, C., Wang, R, et al. (2018). Phenotypic and genotypic characterization of carbapenem-resistant enterobacteriaceae: Data from a longitudinal Large-scale CRE study in China (2012-2016). Clin. Infect. Dis. 67, S196–s205.
Wang, X., Zhai, W., Li, J., Liu, D., Zhang, Q., Shen, Z., et al. (2018a). Presence of an mcr-3 variant in aeromonas caviae, Proteus mirabilis, and escherichia coli from one domestic duck. Antimicrob. Agents Chemother. 62. doi: 10.1128/AAC.02106-17
Wang, X., Wang, Y., Zhou, Y., Li, J., Yin, W., Wang, S., et al. (2018b). Emergence of a novel mobile colistin resistance gene, mcr-8, in NDM-producing klebsiella pneumoniae. Emerg. Microbes Infect. 7, 122. doi: 10.1038/s41426-018-0124-z
Wang, X., et al. (2018c). Emergence of the colistin resistance gene mcr-1 and its variant in several uncommon species of enterobacteriaceae from commercial poultry farm surrounding environments. Vet. Microbiol. 219, 161–164. doi: 10.1016/j.vetmic.2018.04.002
Wang, R., Liu, Y., Zhang, Q., Jin, L., Wang, Q., Zhang, Y., et al. (2018). The prevalence of colistin resistance in escherichia coli and klebsiella pneumoniae isolated from food animals in China: coexistence of mcr-1 and bla(NDM) with low fitness cost. Int. J. Antimicrob. Agents 51, 739–744. doi: 10.1016/j.ijantimicag.2018.01.023
Wang, Z., Fu, Y., Schwarz, S., Yin, W., Walsh, T. R., Zhou, Y., et al. (2019). Genetic environment of colistin resistance genes mcr-1 and mcr-3 in escherichia coli from one pig farm in China. Vet. Microbiol. 230, 56–61. doi: 10.1016/j.vetmic.2019.01.011
Wang, X., Wang, Y., Zhou, Y., Wang, Z., Wang, Y., Zhang, S., et al. (2019). Emergence of colistin resistance gene mcr-8 and its variant in raoultella ornithinolytica. Front. Microbiol. 10, 228. doi: 10.3389/fmicb.2019.00228
Wang, P., He, D., Li, B., Guo, Y., Wang, W., Luo, X., et al. (2019). Eliminating mcr-1-harbouring plasmids in clinical isolates using the CRISPR/Cas9 system. J. Antimicrob. Chemother. 74, 2559–2565. doi: 10.1093/jac/dkz246
Wang, Y., Xu, C., Zhang, R., Chen, Y., Shen, Y., Hu, F., et al. (2020). Changes in colistin resistance and mcr-1 abundance in escherichia coli of animal and human origins following the ban of colistin-positive additives in China: an epidemiological comparative study. Lancet Infect. Dis. 20, 1161–1171. doi: 10.1016/S1473-3099(20)30149-3
Wang, C., Feng, Y., Liu, L., Wei, L., Kang, M, Zong, Z., et al. (2020). Identification of novel mobile colistin resistance gene mcr-10. Emerg. Microbes Infect. 9, 508–516. doi: 10.1080/22221751.2020.1732231
Wang, Z., Fu, Y., Du, X. D., Jiang, H., Wang, Y. (2018). Potential transferability of mcr-3 via IS26-mediated homologous recombination in escherichia coli. Emerg. Microbes Infect. 7, 55. doi: 10.1038/s41426-018-0057-6
Wang, Y. M., Kong, L. C., Liu, J., Ma, H. X. (2018). Synergistic effect of eugenol with colistin against clinical isolated colistin-resistant escherichia coli strains. Antimicrob. Resist. Infect. Control 7, 17. doi: 10.1186/s13756-018-0303-7
Wanty, C., Anandan, A., Piek, S., Walshe, J., Ganguly, J., Carlson, R. W, et al. (2013). The structure of the neisserial lipooligosaccharide phosphoethanolamine transferase a (LptA) required for resistance to polymyxin. J. Mol. Biol. 425, 3389–3402. doi: 10.1016/j.jmb.2013.06.029
Wei, W., Srinivas, S., Lin, J., Tang, Z., Wang, S., Ullah, S., et al. (2018). Defining ICR-Mo, an intrinsic colistin resistance determinant from moraxella osloensis. PloS Genet. 14, e1007389. doi: 10.1371/journal.pgen.1007389
Wei, P., Song, G., Shi, M., Zhou, Y., Liu, Y., Lei, J, et al. (2018). Substrate analog interaction with MCR-1 offers insight into the rising threat of the plasmid-mediated transferable colistin resistance. FASEB J. 32, 1085–1098. doi: 10.1096/fj.201700705R
World Health Organization (2017). Stop using antibiotics in healthy animals to prevent the spread of antibiotic resistance (World Health Organization Media Center). Available at: http://www.who.int/mediacentre/news/releases/2017/antibiotics-animalseffectiveness/en.
Wu, B., Wang, Y., Ling, Z., Yu, Z., Shen, Z., Zhang, S, et al. (2020). Heterogeneity and diversity of mcr-8 genetic context in chicken-associated klebsiella pneumoniae. Antimicrob. Agents Chemother. 65. doi: 10.1128/AAC.01872-20
Xavier, B. B., Lammens, C., Ruhal, R., Kumar-Singh, S., Butaye, P., Goossens, H, et al. (2016). Identification of a novel plasmid-mediated colistin-resistance gene, mcr-2, in escherichia coli, Belgium, June 2016. Euro Surveill. 21. doi: 10.2807/1560-7917.ES.2016.21.27.30280
Xia, X., Wang, Z., Fu, Y., Du, X. D., Gao, B., Zhou, Y, et al. (2019). Association of colistin residues and manure treatment with the abundance of mcr-1 gene in swine feedlots. Environ. Int. 127, 361–370. doi: 10.1016/j.envint.2019.03.061
Xiang, R., Liu, B. H., Zhang, A. Y., Lei, C. W., Ye, X. L., Yang, Y. X, et al. (2018). Colocation of the polymyxin resistance gene mcr-1 and a variant of mcr-3 on a plasmid in an escherichia coli isolate from a chicken farm. Antimicrob. Agents Chemother. 62:e00501–00518. doi: 10.1128/AAC.00501-18
Xu, Y., Wei, W., Lei, S., Lin, J., Srinivas, S, Feng, Y, et al. (2018). An evolutionarily conserved mechanism for intrinsic and transferable polymyxin resistance. mBio 9. doi: 10.1128/mBio.02317-17
Xu, T., Zhang, C., Ji, Y., Song, J., Liu, Y., Guo, Y, et al. (2021). Identification of mcr-10 carried by self-transmissible plasmids and chromosome in enterobacter roggenkampii strains isolated from hospital sewage water. Environ. Pollut. 268, 115706. doi: 10.1016/j.envpol.2020.115706
Xu, Y., Lin, J., Cui, T., Srinivas, S., Feng, Y. (2018). Mechanistic insights into transferable polymyxin resistance among gut bacteria. J. Biol. Chem. 293, 4350–4365. doi: 10.1074/jbc.RA117.000924
Yang, Y. Q., Zhang, A. Y., Ma, S. Z., Kong, L. H., Li, Y. X., Liu, J. X, et al. (2016). Co-Occurrence of mcr-1 and ESBL on a single plasmid in salmonella enterica. J. Antimicrob. Chemother. 71, 2336–2338. doi: 10.1093/jac/dkw243
Yang, Y. Q., Li, Y. X., Song, T., Yang, Y. X., Jiang, W., Zhang, A. Y, et al. (2017). Colistin resistance gene mcr-1 and its variant in escherichia coli isolates from chickens in China. Antimicrob. Agents Chemother. 61, e01204–01216,. doi: 10.1128/AAC.01204-16
Yang, Q., Li, M., Spiller, O. B., Andrey, D. O., Hinchliffe, P., Li, H, et al. (2017). Balancing mcr-1 expression and bacterial survival is a delicate equilibrium between essential cellular defence mechanisms. Nat. Commun. 8, 2054. doi: 10.1038/s41467-017-02149-0
Yang, Q. E., Tansawai, U., Andrey, D. O., Wang, S., Wang, Y., Sands, K, et al. (2019). Environmental dissemination of mcr-1 positive enterobacteriaceae by chrysomya spp. (common blowfly): An increasing public health risk. Environ. Int. 122, 281–290. doi: 10.1016/j.envint.2018.11.021
Yang, F., Shen, C., Zheng, X., Liu, Y., El-Sayed Ahmed, M. A.E., Zhao, Z, et al. (2019). Plasmid-mediated colistin resistance gene mcr-1 in escherichia coli and klebsiella pneumoniae isolated from market retail fruits in guangzhou, China. Infect. Drug Resist. 12, 385–389. doi: 10.2147/IDR.S194635
Yang, Q. E., MacLean, C., Papkou, A., Pritchard, M., Powell, L., Thomas, D, et al. (2020). Compensatory mutations modulate the competitiveness and dynamics of plasmid-mediated colistin resistance in escherichia coli clones. ISME J. 14, 861–865. doi: 10.1038/s41396-019-0578-6
Yang, J., Liu, L., Feng, Y., He, D., Wang, C, Zong, Z, et al. (2021). Potential mobilization of mcr-10 by an integrative mobile element via site-specific recombination in cronobacter sakazakii. Antimicrob. Agents Chemother. 65. doi: 10.1128/AAC.01717-20
Yang, Y. Q., Li, Y. X., Lei, C. W., Zhang, A. Y., Wang, H. N. (2018). Novel plasmid-mediated colistin resistance gene mcr-7.1 in klebsiella pneumoniae. J. Antimicrob. Chemother. 73, 1791–1795. doi: 10.1093/jac/dky111
Yang, X., Liu, L., Wang, Z., Bai, L., Li, R. (2019). Emergence of mcr-8.2-bearing klebsiella quasipneumoniae of animal origin. J. Antimicrob. Chemother. 74, 2814–2817. doi: 10.1093/jac/dkz213
Ye, H., Li, Y., Li, Z., Gao, R., Zhang, H., Wen, R, et al. (2016). Diversified mcr-1-harbouring plasmid reservoirs confer resistance to colistin in human gut microbiota. mBio 7, e00177–00116. doi: 10.1128/mBio.00177-16
Yi, L., Wang, J., Gao, Y., Liu, Y., Doi, Y., Wu, R, et al. (2017). Mcr-1-Harboring salmonella enterica serovar typhimurium sequence type 34 in pigs, China. Emerg. Infect. Dis. 23, 291–295. doi: 10.3201/eid2302.161543
Yin, W., Li, H., Shen, Y., Liu, Z., Wang, S., Shen, Z, et al. (2017). Novel plasmid-mediated colistin resistance gene mcr-3 in escherichia coli. mBio 8. doi: 10.1128/mBio.00543-17
Yuan, Y., Li, Y., Wang, G., Li, C., Xiang, L., She, J, et al. (2019). Coproduction of MCR-9 and NDM-1 by colistin-resistant enterobacter hormaechei isolated from bloodstream infection. Infect. Drug Resist. 12, 2979–2985. doi: 10.2147/IDR.S217168
Yu, Y., Walsh, T. R., Yang, R. S., Zheng, M., Wei, M. C., Tyrrell, J. M, et al. (2019). Novel partners with colistin to increase its in vivo therapeutic effectiveness and prevent the occurrence of colistin resistance in NDM- and MCR-co-producing escherichia coli in a murine infection model. J. Antimicrob. Chemother. 74, 87–95.
Zhang, X. F., Doi, Y., Huang, X., Li, H. Y., Zhong, L. L., Zeng, K. J., et al. (2016). Possible transmission of mcr-1-Harboring escherichia coli between companion animals and human. Emerg. Infect. Dis. 22, 1679–1681. doi: 10.3201/eid2209.160464
Zhang, Y., Liao, K., Gao, H., Wang, Q., Wang, X., Li, H., et al. (2017). Decreased fitness and virulence in ST10 escherichia coli harboring bla(NDM-5) and mcr-1 against a ST4981 strain with bla(NDM-5). Front. Cell Infect. Microbiol. 7, 242. doi: 10.3389/fcimb.2017.00242
Zhang, J., Chen, L., Wang, J., Yassin, A. K., Butaye, P., Kelly, P, et al. (2018). Molecular detection of colistin resistance genes (mcr-1, mcr-2 and mcr-3) in nasal/oropharyngeal and anal/cloacal swabs from pigs and poultry. Sci. Rep. 8, 3705. doi: 10.1038/s41598-018-22084-4
Zhang, H., Hou, M., Xu, Y., Srinivas, S., Huang, M., Liu, L, et al. (2019a). Action and mechanism of the colistin resistance enzyme MCR-4. Commun. Biol. 2, 1–14. doi: 10.1038/s42003-018-0278-1
Zhang, H., Zong, Z., Lei, S., Srinivas, S., Sun, J., Feng, Y, et al. (2019b). A genomic, evolutionary, and mechanistic study of MCR-5 action suggests functional unification across the MCR family of colistin resistance. Adv. Sci. 6, 1900034. doi: 10.1002/advs.201900034
Zhang, Y., Wang, X., Li, X., Dong, L., Hu, X., Nie, T, et al. (2019). Synergistic effect of colistin combined with PFK-158 against colistin-resistant enterobacteriaceae. Antimicrob. Agents Chemother. 63. doi: 10.1128/AAC.00271-19
Zhang, H., Wei, W., Huang, M., Umar, Z., Feng, Y. (2019). Definition of a family of nonmobile colistin resistance (NMCR-1) determinants suggests aquatic reservoirs for MCR-4. Adv. Sci. 6, 1900038. doi: 10.1002/advs.201900038
Zhao, C., Wang, X., Zhang, Y., Wang, R., Wang, Q., Li, H, et al. (2019). In vitro activities of eravacycline against 336 isolates collected from 2012 to 2016 from 11 teaching hospitals in China. BMC Infect. Dis. 19, 508. doi: 10.1186/s12879-019-4093-1
Zhao, F., Feng, Y., Lü, X., McNally, A., Zong, Z. (2017). IncP plasmid carrying colistin resistance gene mcr-1 in klebsiella pneumoniae from hospital sewage. Antimicrob. Agents Chemother. 61, e02229–02216. doi: 10.1128/AAC.02229-16
Zheng, B., Yu, X., Xu, H., Guo, L., Zhang, J., Huang, C, et al. (2017). Complete genome sequencing and genomic characterization of two escherichia coli strains co-producing MCR-1 and NDM-1 from bloodstream infection. Sci. Rep. 7, 17885. doi: 10.1038/s41598-017-18273-2
Zhou, Y. F., Tao, M. T., Feng, Y., Yang, R. S., Liao, X. P., Liu, Y. H, et al. (2017). Increased activity of colistin in combination with amikacin against escherichia coli co-producing NDM-5 and MCR-1. J. Antimicrob. Chemother. 72, 1723–1730. doi: 10.1093/jac/dkx038
Zhou, Y., Wang, J., Guo, Y., Liu, X., Liu, S., Niu, X, et al. (2019). Discovery of a potential MCR-1 inhibitor that reverses polymyxin activity against clinical mcr-1-positive enterobacteriaceae. J. Infect. 78, 364–372. doi: 10.1016/j.jinf.2019.03.004
Zhou, Y. F., Liu, P, Dai, S. H., Sun, J., Liu, Y. H, Liao, X. P, et al. (2020). Activity of tigecycline or colistin in combination with zidovudine against escherichia coli harboring tet(X) and mcr-1. Antimicrob. Agents Chemother. 65. doi: 10.1128/AAC.01172-20
Keywords: mcr-1, pandrug resistance, colistin resistance, polymyxins, crystal structure, MCR activity, risk factors, Enterobacteriaceae
Citation: Mmatli M, Mbelle NM and Osei Sekyere J (2022) Global epidemiology, genetic environment, risk factors and therapeutic prospects of mcr genes: A current and emerging update. Front. Cell. Infect. Microbiol. 12:941358. doi: 10.3389/fcimb.2022.941358
Received: 11 May 2022; Accepted: 01 August 2022;
Published: 26 August 2022.
Edited by:
Dongsheng Zhou, Beijing Institute of Microbiology and Epidemiology, ChinaReviewed by:
Hui Li, Beijing Center for Disease Prevention and Control (Beijing CDC), ChinaMohamed Rhouma, Université de Montréal, Canada
Jingjing Quan, Sir Run Run Shaw Hospital, China
Copyright © 2022 Mmatli, Mbelle and Osei Sekyere. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: John Osei Sekyere, ai5vc2Vpc2VreWVyZUB1cC5hYy56YQ==; am9kMTQxMzlAZ21haWwuY29t
 Masego Mmatli
Masego Mmatli Nontombi Marylucy Mbelle1
Nontombi Marylucy Mbelle1 John Osei Sekyere
John Osei Sekyere