- 1Clinical Medical College, Yangzhou University, Yangzhou, China
- 2Department of General Surgery, Institute of General Surgery, Clinical Medical College, Yangzhou University, Northern Jiangsu People’s Hospital, Yangzhou, China
The human gut microbiome is important for human health. The development of stable microbial communities in the gastrointestinal tract is closely related to the early growth and development of host immunity. After the birth of a baby, immune cells and the gut microbiome mature in parallel to adapt to the complex gut environment. The gut microbiome is closely linked to the immune system and influences each other. This interaction is associated with various diseases in infants and young children, such as asthma, food allergies, necrotizing colitis, obesity, and inflammatory bowel disease. Thus, the composition of the infant gut microbiome can predict the risk of disease development and progression. At the same time, the composition of the infant gut microbiome can be regulated in many ways and can be used to prevent and treat disease in infants by modulating the composition of the infant gut microbiome. The most important impacts on infant gut microbiota are maternal, including food delivery and feeding. The differences in the gut microbiota of infants reflect the maternal gut microbiota, which in turn reflects the gut microbiota of a given population, which is clinically significant.
Background
Microorganisms have an extremely important impact on human health. In particular, the gut microbiome, consisting of trillions of microorganisms and millions of functional genes, have a mutually beneficial symbiosis with the host (Savage, 1977; Friedrich, 2013). The formation of intestinal micro-ecology in the early life plays a vital role in the healthy growth of life. The initial colonization of the infant gut by microorganisms lays the foundation for a lifelong, relatively stable adult microbiome (Houghteling and Walker, 2015). The host provides essential habitat and nutrients for the gut microbiome that provides the necessary nutrients to support the development of the metabolic system and the maturation of the immune system (Kau et al., 2011). Dietary changes during pregnancy and the early postnatal period can affect the colonization of the gut microbiome in infancy, affecting infant health and perhaps having long-term repercussions later in life (Cerdo et al., 2019). The gut microbiome directs the development and maturation of the immune system, which in turn regulates its composition, distribution and abundance (Garrett et al., 2010; Kayama et al., 2020). When the balance between the gut microbiome and the immune system is disturbed, it can lead to the development of a wide range of diseases (Elson and Alexander, 2015). Dysbiosis of the gut microbiome is closely linked to a number of diseases such as obesity (Turnbaugh et al., 2006), type 2 diabetes (Qin et al., 2012), hypertension (Yang et al., 2015), asthma (Arrieta et al., 2015), allergies (Metsala et al., 2013), necrotizing small bowel colitis (NEC) (Mai et al., 2011) and inflammatory bowel disease (IBD) (Frank et al., 2007; Kau et al., 2011). There is increasing evidence that the gut microbiome and the immune system can interact to have a significant impact on the growth and health of infants.
The intergenerational transmission of the gut microbiome can shape the biology of humans in kinship, leading to differences in their innate and adaptive immune systems and modulating their immunopathological status and risk of disease (Garrett et al., 2010). Systematic changes in the overall dietary consumption patterns of entire populations may lead to changes in the gut microbiota/microbiome, with consequent effects on the nutritional status and immune response of the host (Kau et al., 2011). It is important to understand the factors that influence and modify an individual’s microbiome at different life stages, with a focus on the early years of life (Kumbhare et al., 2019). In this review, we discuss the maturation and interaction of the infant gut microbiota and immune system. Finally, we also summarize infant diseases related to gut dysbiosis and immune system disorders and present our outlook.
Maturation of Immune Cells and Gut Microbiome Parallel Each Other
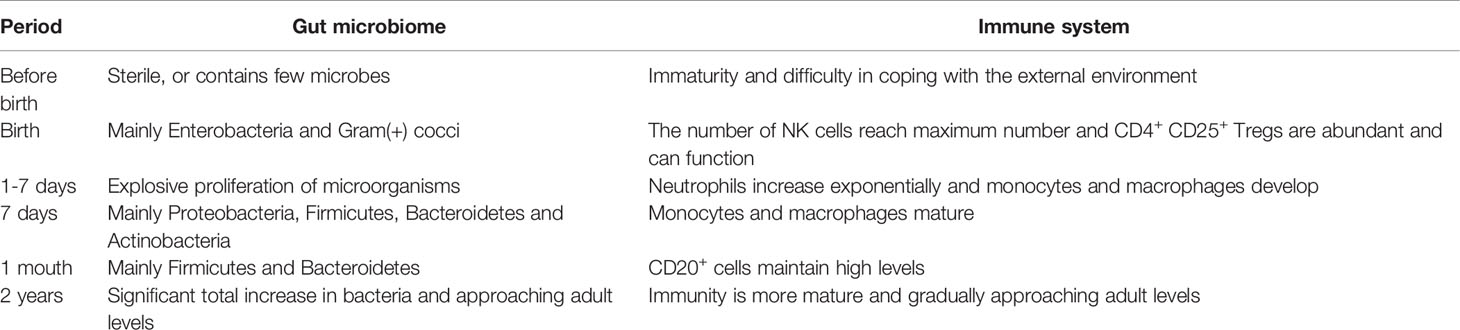
Our gastrointestinal system is sterile or contains few microorganisms when we are born, and it is swiftly colonized by microbes from the outside world (Knoop et al., 2018). From birth to adulthood, gut microbial colonization in healthy individuals is characterized by a dynamic series of events that play critical roles in promoting gut homeostasis and stimulating normal development and responses of the immune system(Table 1) (Del Chierico et al., 2015; Dominguez-Bello et al., 2019). After birth, the fetus is suddenly exposed to an external environment rich in bacteria which immediately colonize the intestinal cavity with a large number of microorganisms, mainly Enterobacteria and Staphylococcus (Koenig et al., 2011; Del Chierico et al., 2015; Matsuki et al., 2016). CD4+ CD25+ Tregs are abundant and can perform their functions well due to microbial colonization. From 1-7 days after birth, the intestinal microbiome proliferates rapidly and in an orderly manner, forming a relatively mature microbial ecosystem dominated by Proteobacteria, Firmicutes, Bacteroidetes and Actinobacteria on the 7th day of birth (Bokulich et al., 2016). At the same time, neutrophils, the immune cells of the gut, increase exponentially, accompanied by the development of monocytes and macrophages, which reach maturity at the same time around the seventh day of birth (Ygberg and Nilsson, 2012). Gut microbial colonization and microbiota diversity lead to the induction of intestinal Tregs and IL-10, which are required to limit the induction of pro-inflammatory Th1 and Th17 cells (Geuking et al., 2011). There is evidence that conventionalization with a standard microbiota or monoclonalization with Bacteroides fragilis in the first 2 weeks of life can prevent abnormal accumulation of colonic iNKT cells and abolishes the increased susceptibility to colitis and asthma (Olszak et al., 2012; Gensollen et al., 2016). Cahenzli et al. demonstrated that microbial colonization early in life, rather than in adulthood, can keep IgE levels low and facilitate a reduction in susceptibility to orally induced allergic reactions (Cahenzli et al., 2013). After the age of 2 years, the abundance of Firmicutes and Bacteroidetes is greater than 90% of the total bacterial load, and the gut microbiome gradually develops to adult levels (Gosalbes et al., 2013; Martin et al., 2016). In summary, postnatal maturation of immune cells and gut microbiota occurs in parallel to adapt to the complex environment of the gut.
Infant Gut Microbiome Promotes Immune System Maturation
The gut microbiome is critical for the immune system development, and its number and variety can influence the development of the intestinal immune system. Commensal microorganisms and their products can interact with immune cells to establish and maintain host tolerance while influencing innate and adaptive immune responses in early life (Li et al., 2014).
The development of the innate immune system, as well as the generation of immune cells, cytokines, and immunological factors, is influenced by the gut microbiome. The gut microbiome can exert pathogenic and protective functions through specific pattern recognition receptors (PRRs) and microbial pattern signaling pathways (MAMPs) and the subsequent immune responses (Figure 1) (Barko et al., 2018). Because different gram-negative bacteria have different lipopolysaccharide (LPS) alterations, their ability to trigger Toll-like receptors (TLRs) varies, not all bacteria will elicit the same immune response (Barko et al., 2018). Díazropero et al. applied Lactobacillus salivarius CECT5713 and Lactobacillus fermentum CECT5716, both obtained from human milk, to bone marrow-derived macrophages, resulting in the induction of pro-inflammatory cytokine production by Lactobacillus fermentum CECT5716 and, conversely, the promotion of interleukin-10 production by Lactobacillus salivarius CECT5713 (Diaz-Ropero et al., 2007). Gram-negative bacterial peptidoglycan is recognized by nucleotide-binding oligomerization domain (NOD) 1 in intestinal epithelial cells, produces CCL20 and β-defensin 3, and directs B cell uptake into LTi in a cryptocurrency-expressing cluster -DC, thereby reducing sIgA (Maynard et al., 2012). Olzaked et al. demonstrated that natural killer T cells (iNKTs) accumulate in the colonic lamina propria and lungs of microbe-free mice, leading to increased incidence in models of inflammatory bowel disease (IBD) and allergic asthma (Olszak et al., 2012). The regulation of the expansion of iNKT cells was ascribed to reduced expression of the chemokine CXCL16 in the presence of microbiota. The diversity of the gut microbiome determines the degree of immunoglobulin E (IgE) in their adult bodies (Cahenzli et al., 2013). In atopic allergy illness and parasite immunity, IgE antibodies play a critical role (Gould and Sutton, 2008; Allen and Maizels, 2011). Early in infancy, a reduced IgA binding to gut bacteria was linked to an increased susceptibility to the development of asthma and allergies (Dzidic et al., 2017). The gut microbiome can facilitate the process of lymphocyte maturation and secretion of immunoglobulin A (IgA) (Bouskra et al., 2008). Bifidobacterium infantis supplementation inhibited T helper type 2 and T helper type 17 cell responses while stimulating interferon β production in breastfed babies, indicating the establishment of a positive regulatory immunological response (Henrick et al., 2021; Rachid et al., 2021). Beneficial bacteria induce apoptosis stimulation, reactive oxygen species synthesis and toll-like receptor signaling to control the inflammatory response and help establish innate immune defences and improve pathogen recognition (Kaplan et al., 2011; Jakaitis and Denning, 2014).
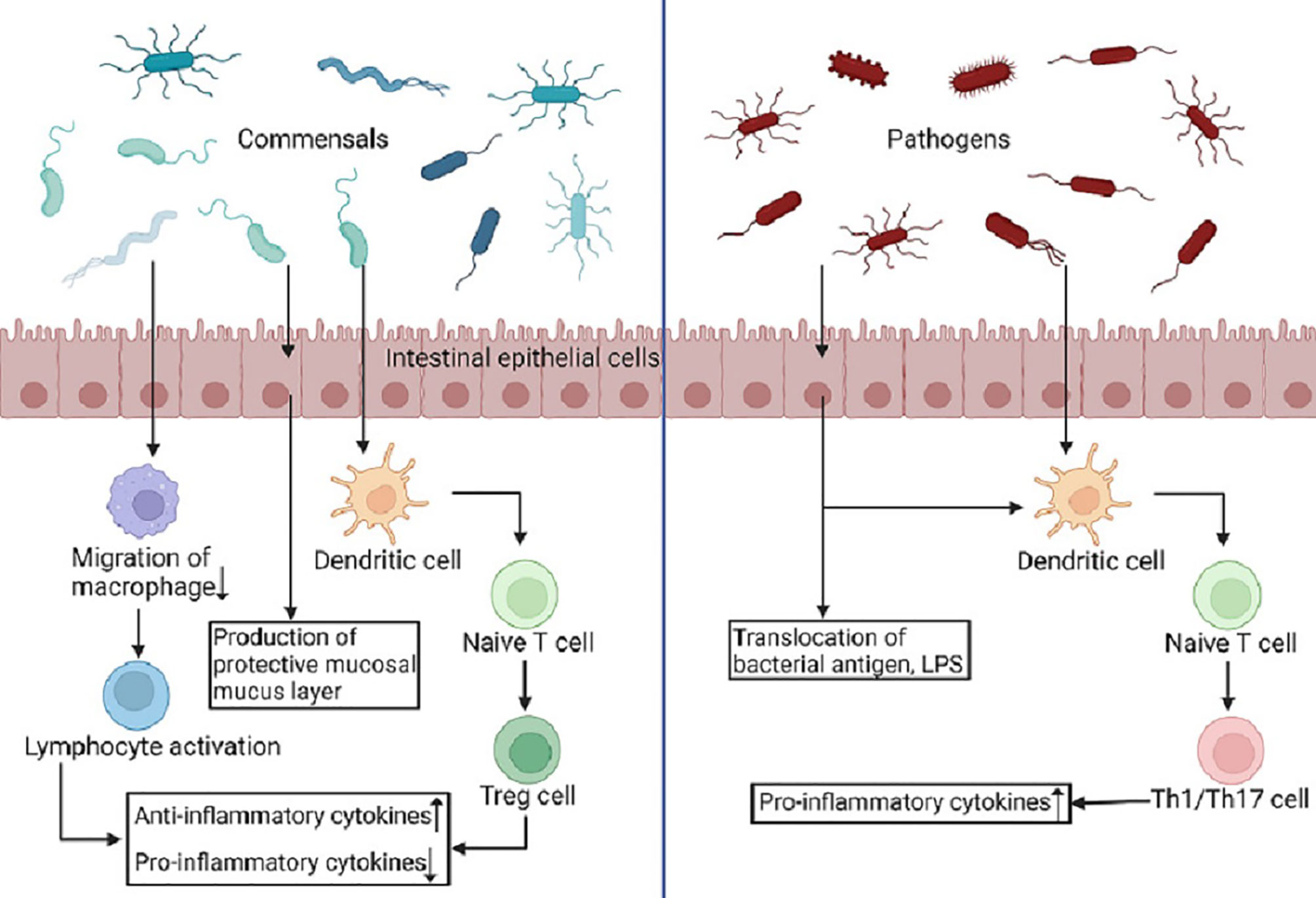
Figure 1 Commensals and pathogens produce different immune responses. PRRs identify MAMPs on pathogens and symbionts and are extensively expressed in intestinal epithelial cells, macrophages, and dendritic cells. Dendritic cells offer antigens to naive T cells as a result of commensals, and these cells differentiate into Treg cells, which emit anti-inflammatory cytokines and spread systemic and local tolerance. Simultaneously, phagocyte migration is inhibited, allowing microbial antigens to reach lymphoid tissue and increasing B- and T-cell activation. Conversely, pathogenic germs cause dendritic cells to secrete pro-inflammatory cytokines, which cause naive T cells to differentiate into Th1 and Th17 cells, resulting in a pro-inflammatory immune response. (Created in BioRender.com).
Therefore, the establishment of gut microbiome in early infancy has a crucial impact on the development of the immune system, its ability to fight off infections and also on its health in adulthood. Zeng et al. demonstrated the ability of gut microbes to induce immunoglobulin G (IgG) production in the organism, thereby inhibiting infection by pathogenic bacteria (Zeng et al., 2016). Schuijt et al. used Streptococcus pneumoniae to infect normal mice and mice with gut microbiome removed and found that the alveolar macrophages of the mice with gut microbiome removed had a reduced ability to phagocytose Streptococcus pneumoniae and a higher mortality rate, suggesting that gut microbes can enhance the function of major alveolar macrophages (Schuijt et al., 2016). Intestinal commensal bacteria can induce the secretion of inhibitory peptides, limit the inflammatory response and promote intestinal mucosal repair by blocking the recognition of TLRs to the intestine or by down regulating the NF-κβ pathway by blocking the degradation of NF-κβ inhibitors (Sampath et al., 2011). However, in immunocompromised hosts, the gut microbiome is not harmless, but may instead trigger an inflammatory response (Hooper et al., 2012). For example, the gut microbiome can contribute to the risk and pathogenesis of malnutrition by affecting nutrient metabolism and immune function (Kau et al., 2011). Cukrowska et al. showed that non-pathogenic Escherichia coli stimulates an immune response during intestinal colonization, but the intestinal mucosal immune response soon diminishes and tolerance forms due to the suppressive mechanisms (oral tolerance) (Cukrowska et al., 2001). Some symbiotic microbiomes are unable to express adhesion and invasion factors, or exhibit only low endotoxicity, and therefore cannot invade the intestinal mucus layer to elicit an immune response (Golenbock et al., 1991). Meanwhile, because the immune system of infants is relatively weakly regulated, exposure to antigens such as the gut microbiome is highly susceptible to lifelong or long-term tolerance, which means a specific non-responsive response to the gut microbiome.
In conclusion, commensal bacteria and the immune system can promote and favor each other and maintain the microecological balance of the gut. Instead, pathogens can disrupt the gut microbiome and stimulate the immune system to mount an inflammatory response. Imbalances in infant gut microbiome can lead to an incompletely developed the immune system in infants. Hence, abnormalities in the microbial composition of the gut of premature newborns may predispose them to postnatal failure in the evolution of key innate defences, leading to disease (Jakaitis and Denning, 2014).
Infant Immune System Regulates the Composition of Gut Microbiome
The immune system influences the composition and abundance of the gut microbiome and can strengthen the epithelial barrier function in infants. The immune system of the intestinal mucosa consists of lymph nodes, lamina propria, and epithelial cells that form a physiological barrier that protects the integrity of the gut (Shi et al., 2017). Innate immune cells use a diversity of PRRs, including Toll-like receptors, NOD-like receptors and leptin-like receptors, to recognize conserved molecular patterns of microbes and signaling of these receptors may then act as developmental triggers for the gut microbiome (Fulde et al., 2018; Price et al., 2018; Dogra et al., 2021). IL-22 produced by innate lymphoid cells is in a position to induce hRegIIIα production in epithelial cells, which binds to bacterial peptidoglycan carbohydrates and kills Gram-positive target bacteria (Mukherjee et al., 2014; Shi et al., 2017). Kubinak et al. showed that the innate adaptor MyD88 in gut T cells is dependent on a signaling pathway and works with IgA to regulate the intestinal microbiome (Kubinak et al., 2015). IgA directly targets intestinal microbes and the absence of MyD88 signaling leads to an abnormal response of IgA antibodies and an imbalance in the intestinal microbiome, leading to inflammatory disease (Kubinak et al., 2015). However, interestingly, Wen et al. showed that non-obese diabetic mice lacking the specific pathogen-free MyD88 protein do not develop type 1 diabetes (Wen et al., 2008). sIgA prevents the adhesion of pathogenic microorganisms by forming complexes with their antigens and stimulating the secretion of mucus by cup cells in the mucosa of the respiratory and digestive tracts (Pulimood et al., 1998). Intestinal epithelial cells influence the composition of the gut microbiome by secreting antimicrobial proteins and alpha-defensins (Maynard et al., 2012). Mice with restricted NLRP6 expression in colonic epithelial cells showed an increased abundance of the bacterial phyla Bacteroidetes in the gut microbiome, as well as an increased susceptibility of intestinal inflammatory cells, thus inducing the development of colitis (Elinav et al., 2011).
During vaginal birth, the newborn is colonized with bacteria from the mother’s intestines and vagina, laying the foundation for a strong immune system (Milani et al., 2017). Infants born by cesarean section have a less complex gut microbiome and are less likely to be colonized than infants born by vaginal delivery (Milani et al., 2017). However, the lack of contact with the mother’s birth canal and inadequate exchange of intestinal microbiome may increase the risk of obesity, diabetes, and asthma in infants born by cesarean section (Algert et al., 2009; Li et al., 2013; Milani et al., 2017). Infants born by cesarean section require breastfeeding or formula feeding for normal and beneficial gut microbiome. In the early years of life, when the immune system is immature, breast milk provides infants with the necessary immune cells and immune related factors to fight infection (Goldman, 2007; Hanson, 2007; Riskin et al., 2012). Macrophages and leukocytes from breast milk can kill enteropathogenic Escherichia coli, Giardia lamblia, Staphylococcus aureus, and Candida albicans, thus contributing to the gradual recovery of infants who have been breastfed (Riskin et al., 2012). Breast milk contains compounds that may affect the immunity of newborns, including a group of oligosaccharides synthesised in the lactose gland, which is known as breast milk oligosaccharides (HMOs) (Bode, 2012; Plaza-Diaz et al., 2018; Singh et al., 2022). HMOs directly modulate the intestinal immune response, ensure enrichment of the gut microbiome and regulate microbial adhesion, thus protecting breastfed infants from microbial infections (Bode, 2012; Chichlowski et al., 2012; Singh et al., 2022). HMOs can stimulate the growth of beneficial microorganisms, mainly Bifidobacterium genus and some strains of Bacteroides and Lactobacillus (Sanchez et al., 2021).
Under normal conditions, the interaction between the immune system and the gut microbiome promotes the development of the infant gut and the immune system, enabling the infant gut to rapidly tolerate food antigens and preventing pathogenic bacteria from invading the mucosa and submucosa. Understanding the tight interplay between the gut microbiome, epithelial cells, and immune cells critical for maintaining gut homeostasis could facilitate advances in diagnostic and therapeutic approaches to various diseases.
Gut Microbiome and Immune System Diseases of Infants
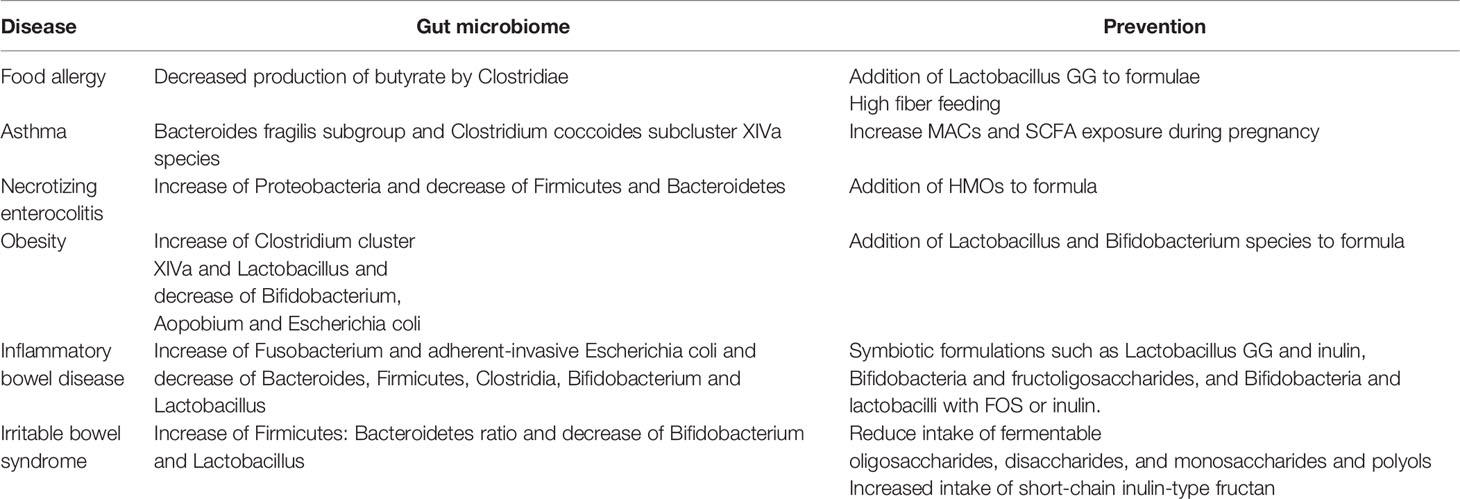
An abnormal microbiome can colonize an infant, impair immune system development after birth, and have long-term effects on health problems such as the development of allergies. Prenatal, inter-pregnant, or neonatal use of antibiotics can adversely affect the development of the neonatal gut microbiome and immune system, contributing to food allergies, asthma, necrotizing enterocolitis, obesity, and more in children (Table 2) (Milliken et al., 2019; Gao et al., 2021). All of these diseases have an overall delay in microbial maturation. Interventions for these disorders may include breastfeeding, changes to the infant’s diet and the use of probiotics (Bokulich et al., 2016; Mendez et al., 2019; Milliken et al., 2019).
Food Allergy
Changes in the microbial composition of the gut are one of the risk factors for food allergies. Hygiene hypothesis suggests that in the extended family, infection at an early age in children due to unhygienic contact with family members can prevent the development of atopic diseases (Strachan, 1989). Reduced family size and reduced opportunities for infection can lead to an increase in the development of atopic diseases (Strachan, 1989). Some bacterial strains as well as microbial diversity supports the maturation of regulatory T lymphocytes and promotes tolerance to food antigens, such as butyrate, a short-chain fatty acid (SCFA) produced from dietary fiber predominantly by Clostridia (Iweala and Nagler, 2019). Conversely, the misuse of antibiotics can disrupt microbiota diversity and thus promote allergy attacks (De Martinis et al., 2020). Tan et al. found that a high fiber diet changed the gut microbial ecology and enhanced SCFA release, especially acetate and butyrate, enhancing oral tolerance and avoiding food allergies in mice (Tan et al., 2016). Intestinal commensals can interact with DCs, macrophages and innate lymphocytes to promote IL-22 production, strengthen the barrier function of intestinal epithelial cells and prevent food allergy (Iweala and Burks, 2016). In a prospective clinical trial, Bernini Canini et al. showed that the addition of Lactobacillus GG to extensively hydrolyzed casein formula increased the tolerance rate to cow’s milk protein in infants with cow’s milk allergy (Berni Canani et al., 2012). Notably, prematurity and low birth weight were not linked with changes in the risk of developing food allergies during childhood (Liem et al., 2007).
Asthma
Asthma is a chronic inflammatory airway disease characterized by reversible airway obstruction and hypersensitivity, generally thought to be caused by a combination of genetic and environmental factors (Nyce and Metzger, 1997; Abdel-Aziz et al., 2019). Penders et al. found that early colonization of Clostridium difficile was associated with all atopic outcomes, including asthma (Penders et al., 2007). The development of the gut microbiome in the first year of life may influence the development of childhood asthma, and adequate microbial maturation during this period may protect children at risk for asthma (Stokholm et al., 2018; Depner et al., 2020). Germ-free mice have been demonstrated to develop an immune defence system against allergic asthma if colonized as a newborn, but not if colonized as an adult (Olszak et al., 2012). The early microbial composition of preterm infants is disturbed compared to full-term infants, which increases their chances of having a disturbed immune system and makes them more susceptible to infections due to a lack of immune maturity (Bokulich et al., 2016; Kumbhare et al., 2019). Vael et al. showed that fecal colonization of Bacteroides fragilis subgroup or Clostridium coccoides subcluster XIVa subgroup within age 3 weeks is an early indicator of possible future asthma (Vael et al., 2011; Shi et al., 2017). Oral administration of Bifidobacterium shortum M-16V has been shown to upregulate transforming growth factor-β1 signaling and promotes intestinal Bifidobacterial colonization in preterm infants, reducing inflammatory and allergic reactions in infants (Fujii et al., 2006; Patole et al., 2014). The World Allergy Organization (WAO) guidelines for the prevention of allergic diseases recommend the use of probiotics to prevent eczema in non-exclusively breastfed infants at high risk of allergy (Fiocchi et al., 2015). Breastfeeding can facilitate the colonization of the infant’s gut with beneficial bacteria, such as Bifidobacteria, and prevents asthma in later childhood (Kull et al., 2010). However, in the case of mothers with asthma, breast milk does not have this effect, probably due to the low level of Bifidobacteria in the mother’s milk (Li et al., 2014). Furthermore, the mother’s diet and exposure to gut bacteria during pregnancy can also influence the development of the fetal immune system and subsequent asthma in the offspring (Daley, 2014; Gray et al., 2017; Ober et al., 2017). Induction mediated by fetal lung regulatory T cells, increased microbiota-accessible carbohydrates (MACs) and SCFA exposure during pregnancy can reduce the risk of asthma in offspring (Gray et al., 2017). Therefore, modulation of the infant gut microbiome through breastfeeding, prebiotics and others during pregnancy or early life may prevent the incidence of asthma in later life.
Necrotizing Enterocolitis
Premature infants are susceptible to sepsis and necrotizing enterocolitis (NEC) due to a compromised intestinal barrier, immature immunity and a dysbiosis of the intestinal microbiota (Daley, 2014; Cuna et al., 2021). Increased relative abundance of Proteobacteria and decreased relative abundance of Firmicutes and Bacteroidetes characterize microbial dysbiosis in preterm infants prior to NEC (Pammi et al., 2017). Changes in the gut microbiome can lead to changes in host immunity and barrier function, which can contribute to NEC clinical signs and symptoms (Li et al., 2014). Human milk oligosaccharides (HMOs) are a specific type of non-digestible carbohydrates found in mother’s milk that aid in the formation of the infant gut microbiome, epithelial barrier, and immune system (Akkerman et al., 2019; Zhang et al., 2022). HMOs are of great importance in the prevention and treatment of NEC. In a single-center randomized control trial, the incidence of NEC was found to be significantly lower in preterm infants fed with formula containing HMOs compared to those fed with formula without HMOs, and the length of hospital stay was significantly shorter (Armanian et al., 2014). Austin et al. found that this may be explained by the disialyllacto-N-tetraose influence on TLR- and selectin-mediated inflammatory pathways in HMOs (Austin et al., 2019). Therefore, breastfeeding has a preventive and therapeutic effect on NEC to some extent. Because of the lack of microbial diversity in infant gut and its ease of control, the study of the infant gut microbiome has significant implications for the survival and prognosis of preterm infants at high risk of NEC.
Antibiotics
Antibiotic abuse can lead to disruption of the infant gut microbiome, which can lead to an imbalanced immune response and eventually the development of many diseases in infants and young children. Epidemiological studies have established an association between the use of antibiotics in early infancy and the development of diseases such as obesity, diabetes and asthma in later life (Marra et al., 2006; Gasparrini et al., 2016). Nogacka et al. demonstrated that on day 7 after perinatal antibiotic administration, the proportion of Actinobacteria and Bacteroides in infant intestine decreased, while the proportion of Aspergillus and Firmicutes increased, resulting in a dysbiosis of the gut microbiome (Nogacka et al., 2017). Bokulich et al. found that antibiotic exposure disrupts microbial succession and can slow down the maturation of the microbiome (Bokulich et al., 2016). In addition, antibiotic exposure may cause an incomplete, yet stable and permanent, recovery of the microbiome, possibly leaving the recipient vulnerable to infectious diseases later in life (Dethlefsen et al., 2008; Gasparrini et al., 2016). Therefore, physicians must pay close attention to the serious consequences of antibiotic abuse, strictly control the indications for antibiotic use, and use antibiotics scientifically and rationally.
Conclusions
There is growing evidence that the infant immune system and the gut microbiome are intimately connected, interact and mature together. Imbalances in the infant microbiome and disturbances in the immune system can also be causally linked to the development and progression of diseases. Theoretically, differences in the gut microbiome of infants at different stages and between individuals could reflect changes in the infant immune system, help define the human genetic evolution, reflect our changes in our eating habits, and enable disease prevention or new treatment strategies (Kau et al., 2011). However, techniques for the quantitative analysis of the innate and adaptive immune system are not yet fully developed, and biomarkers relevant to the infant gut microbiome, including those for mucosal-associated barrier function, are lacking. Furthermore, the current studies are still in the preclinical stage and the results are highly variable and uncontrollable, which still poses a challenge for clinical application. Therefore, there is a need to better understand the composition of the gut microbiome during early life stages, as well as the process of microbial community succession and evolution.
Author Contributions
HZ and ZZ drafted the manuscript and counted and plotted the diagram and tables. DT critically revised the article for important intellectual content. All authors read and approved the final manuscript. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by grants from the Training Project of Key Talents of Youth Medicine in Jiangsu province, China [No. QNRC2016330], the Key disease standardization diagnosis and treatment project in Jiangsu province [NO. BE2015664], the Academic Science and Technology Innovation Fund for College Students [No. ×20180714], the Social Development-Health Care Project of Yangzhou, Jiangsu Province [No. YZ2018087], the Social Development Project of Yangzhou, Jiangsu Province [No. YZ2021075], the Graduate Research and Practice innovation Plan of Graduate Education Innovation Project in Jiangsu Province [NO. SJCX211644], and High-level talent”six one projects” top talent scientific research project of Jiangsu Province [No. LGY2019034]. The funding bodies had no role in writing the manuscript.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Abbreviations
IBD, Inflammatory bowel disease; iNKT, Invariant natural killer T; IgA, Immunoglobulin A; IgE, Immunoglobulin E; IgG, Immunoglobulin G; NEC, Necrotizing small bowel colitis; NOD, Nucleotide-binding oligomerization domain; PRRs, Pattern recognition receptors; MAMPs, Microbe‐associated molecular patterns; MACs, Microbiota-accessible carbohydrates; SCFA, Short-chain fatty acid; HMOs, Human milk oligosaccharides; TLRs, Toll‐like receptors; LPS, Lipopolysaccharide; WAO, World Allergy Organization.
References
Abdel-Aziz, M. I., Vijverberg, S. J. H., Neerincx, A. H., Kraneveld, A. D., Maitland-van der Zee, A. H. (2019). The Crosstalk Between Microbiome and Asthma: Exploring Associations and Challenges. Clin. Exp. Allergy 49 (8), 1067–1086. doi: 10.1111/cea.13444
Akkerman, R., Faas, M. M., de Vos, P. (2019). Non-Digestible Carbohydrates in Infant Formula as Substitution for Human Milk Oligosaccharide Functions: Effects on Microbiota and Gut Maturation. Crit. Rev. Food Sci. Nutr. 59 (9), 1486–1497. doi: 10.1080/10408398.2017.1414030
Algert, C. S., McElduff, A., Morris, J. M., Roberts, C. L. (2009). Perinatal Risk Factors for Early Onset of Type 1 Diabetes in a 2000-2005 Birth Cohort. Diabetes Med. 26 (12), 1193–1197. doi: 10.1111/j.1464-5491.2009.02878.x
Allen, J. E., Maizels, R. M. (2011). Diversity and Dialogue in Immunity to Helminths. Nat. Rev. Immunol. 11 (6), 375–388. doi: 10.1038/nri2992
Armanian, A. M., Sadeghnia, A., Hoseinzadeh, M., Mirlohi, M., Feizi, A., Salehimehr, N., et al. (2014). The Effect of Neutral Oligosaccharides on Reducing the Incidence of Necrotizing Enterocolitis in Preterm Infants: A Randomized Clinical Trial. Int. J. Prev. Med. 5 (11), 1387–1395. doi: 10.4103/2279-042X.176558
Arrieta, M. C., Stiemsma, L. T., Dimitriu, P. A., Thorson, L., Russell, S., Yurist-Doutsch, S., et al. (2015). Early Infancy Microbial and Metabolic Alterations Affect Risk of Childhood Asthma. Sci. Transl. Med. 7 (307), 307ra152. doi: 10.1126/scitranslmed.aab2271
Austin, S., De Castro, C. A., Sprenger, N., Binia, A., Affolter, M., Garcia-Rodenas, C. L., et al. (2019). Human Milk Oligosaccharides in the Milk of Mothers Delivering Term Versus Preterm Infants. Nutrients 11 (6),1282. doi: 10.3390/nu11061282
Barko, P. C., McMichael, M. A., Swanson, K. S., Williams, D. A. (2018). The Gastrointestinal Microbiome: A Review. J. Vet. Intern. Med. 32 (1), 9–25. doi: 10.1111/jvim.14875
Berni Canani, R., Nocerino, R., Terrin, G., Coruzzo, A., Cosenza, L., Leone, L., et al. (2012). Effect of Lactobacillus GG on Tolerance Acquisition in Infants With Cow's Milk Allergy: A Randomized Trial. J. Allergy Clin. Immunol. 129 (2), 580–582, 582 e581-585. doi: 10.1016/j.jaci.2011.10.004
Bode, L. (2012). Human Milk Oligosaccharides: Every Baby Needs a Sugar Mama. Glycobiology 22 (9), 1147–1162. doi: 10.1093/glycob/cws074
Bokulich, N. A., Chung, J., Battaglia, T., Henderson, N., Jay, M., Li, H., et al. (2016). Antibiotics, Birth Mode, and Diet Shape Microbiome Maturation During Early Life. Sci. Transl. Med. 8 (343), 343ra382. doi: 10.1126/scitranslmed.aad7121
Bouskra, D., Brezillon, C., Berard, M., Werts, C., Varona, R., Boneca, I. G., et al. (2008). Lymphoid Tissue Genesis Induced by Commensals Through NOD1 Regulates Intestinal Homeostasis. Nature 456 (7221), 507–510. doi: 10.1038/nature07450
Cahenzli, J., Koller, Y., Wyss, M., Geuking, M. B., McCoy, K. D. (2013). Intestinal Microbial Diversity During Early-Life Colonization Shapes Long-Term IgE Levels. Cell Host Microbe 14 (5), 559–570. doi: 10.1016/j.chom.2013.10.004
Cerdo, T., Dieguez, E., Campoy, C. (2019). Early Nutrition and Gut Microbiome: Interrelationship Between Bacterial Metabolism, Immune System, Brain Structure, and Neurodevelopment. Am. J. Physiol. Endocrinol. Metab. 317 (4), E617–E630. doi: 10.1152/ajpendo.00188.2019
Chichlowski, M., De Lartigue, G., German, J. B., Raybould, H. E., Mills, D. A. (2012). Bifidobacteria Isolated From Infants and Cultured on Human Milk Oligosaccharides Affect Intestinal Epithelial Function. J. Pediatr. Gastroenterol. Nutr. 55 (3), 321–327. doi: 10.1097/MPG.0b013e31824fb899
Cukrowska, B., Kozakova, H., Rehakova, Z., Sinkora, J., Tlaskalova-Hogenova, H. (2001). Specific Antibody and Immunoglobulin Responses After Intestinal Colonization of Germ-Free Piglets With Non-Pathogenic Escherichia Coli O86. Immunobiology 204 (4), 425–433. doi: 10.1078/0171-2985-00052
Cuna, A., Morowitz, M. J., Ahmed, I., Umar, S., Sampath, V. (2021). Dynamics of the Preterm Gut Microbiome in Health and Disease. Am. J. Physiol. Gastrointest. Liver Physiol. 320 (4), G411–G419. doi: 10.1152/ajpgi.00399.2020
Daley, D. (2014). The Evolution of the Hygiene Hypothesis: The Role of Early-Life Exposures to Viruses and Microbes and Their Relationship to Asthma and Allergic Diseases. Curr. Opin. Allergy Clin. Immunol. 14 (5), 390–396. doi: 10.1097/ACI.0000000000000101
Del Chierico, F., Vernocchi, P., Petrucca, A., Paci, P., Fuentes, S., Pratico, G., et al. (2015). Phylogenetic and Metabolic Tracking of Gut Microbiota During Perinatal Development. PloS One 10 (9), e0137347. doi: 10.1371/journal.pone.0137347
De Martinis, M., Sirufo, M. M., Suppa, M., Ginaldi, L. (2020). New Perspectives in Food Allergy. Int. J. Mol. Sci. 21 (4), 1474. doi: 10.3390/ijms21041474
Depner, M., Taft, D. H., Kirjavainen, P. V., Kalanetra, K. M., Karvonen, A. M., Peschel, S., et al. (2020). Maturation of the Gut Microbiome During the First Year of Life Contributes to the Protective Farm Effect on Childhood Asthma. Nat. Med. 26 (11), 1766–1775. doi: 10.1038/s41591-020-1095-x
Dethlefsen, L., Huse, S., Sogin, M. L., Relman, D. A. (2008). The Pervasive Effects of an Antibiotic on the Human Gut Microbiota, as Revealed by Deep 16S rRNA Sequencing. PloS Biol. 6 (11), e280. doi: 10.1371/journal.pbio.0060280
Diaz-Ropero, M. P., Martin, R., Sierra, S., Lara-Villoslada, F., Rodriguez, J. M., Xaus, J., et al. (2007). Two Lactobacillus Strains, Isolated From Breast Milk, Differently Modulate the Immune Response. J. Appl. Microbiol. 102 (2), 337–343. doi: 10.1111/j.1365-2672.2006.03102.x
Dogra, S. K., Kwong Chung, C., Wang, D., Sakwinska, O., Colombo Mottaz, S., Sprenger, N. (2021). Nurturing the Early Life Gut Microbiome and Immune Maturation for Long Term Health. Microorganisms 9 (10), 2110. doi: 10.3390/microorganisms9102110
Dominguez-Bello, M. G., Godoy-Vitorino, F., Knight, R., Blaser, M. J. (2019). Role of the Microbiome in Human Development. Gut 68 (6), 1108–1114. doi: 10.1136/gutjnl-2018-317503
Dzidic, M., Abrahamsson, T. R., Artacho, A., Bjorksten, B., Collado, M. C., Mira, A., et al. (2017). Aberrant IgA Responses to the Gut Microbiota During Infancy Precede Asthma and Allergy Development. J. Allergy Clin. Immunol. 139 (3), 1017–1025 e1014. doi: 10.1016/j.jaci.2016.06.047
Elinav, E., Strowig, T., Kau, A. L., Henao-Mejia, J., Thaiss, C. A., Booth, C. J., et al. (2011). NLRP6 Inflammasome Regulates Colonic Microbial Ecology and Risk for Colitis. Cell 145 (5), 745–757. doi: 10.1016/j.cell.2011.04.022
Elson, C. O., Alexander, K. L. (2015). Host-Microbiota Interactions in the Intestine. Dig Dis. 33 (2), 131–136. doi: 10.1159/000369534
Fiocchi, A., Pawankar, R., Cuello-Garcia, C., Ahn, K., Al-Hammadi, S., Agarwal, A., et al. (2015). World Allergy Organization-McMaster University Guidelines for Allergic Disease Prevention (GLAD-P): Probiotics. World Allergy Organ J. 8 (1), 4. doi: 10.1186/s40413-015-0055-2
Frank, D. N., St Amand, A. L., Feldman, R. A., Boedeker, E. C., Harpaz, N., Pace, N. R. (2007). Molecular-Phylogenetic Characterization of Microbial Community Imbalances in Human Inflammatory Bowel Diseases. Proc. Natl. Acad. Sci. U. S. A. 104 (34), 13780–13785. doi: 10.1073/pnas.0706625104
Friedrich, M. J. (2013). Genomes of Microbes Inhabiting the Body Offer Clues to Human Health and Disease. JAMA 309 (14), 1447–1449. doi: 10.1001/jama.2013.2824
Fujii, T., Ohtsuka, Y., Lee, T., Kudo, T., Shoji, H., Sato, H., et al. (2006). Bifidobacterium Breve Enhances Transforming Growth Factor Beta1 Signaling by Regulating Smad7 Expression in Preterm Infants. J. Pediatr. Gastroenterol. Nutr. 43 (1), 83–88. doi: 10.1097/01.mpg.0000228100.04702.f8
Fulde, M., Sommer, F., Chassaing, B., van Vorst, K., Dupont, A., Hensel, M., et al. (2018). Neonatal Selection by Toll-Like Receptor 5 Influences Long-Term Gut Microbiota Composition. Nature 560 (7719), 489–493. doi: 10.1038/s41586-018-0395-5
Gao, Y., Nanan, R., Macia, L., Tan, J., Sominsky, L., Quinn, T. P., et al. (2021). The Maternal Gut Microbiome During Pregnancy and Offspring Allergy and Asthma. J. Allergy Clin. Immunol. 148 (3), 669–678. doi: 10.1016/j.jaci.2021.07.011
Garrett, W. S., Gordon, J. I., Glimcher, L. H. (2010). Homeostasis and Inflammation in the Intestine. Cell 140 (6), 859–870. doi: 10.1016/j.cell.2010.01.023
Gasparrini, A. J., Crofts, T. S., Gibson, M. K., Tarr, P. I., Warner, B. B., Dantas, G. (2016). Antibiotic Perturbation of the Preterm Infant Gut Microbiome and Resistome. Gut Microbes 7 (5), 443–449. doi: 10.1080/19490976.2016.1218584
Gensollen, T., Iyer, S. S., Kasper, D. L., Blumberg, R. S. (2016). How Colonization by Microbiota in Early Life Shapes the Immune System. Science 352 (6285), 539–544. doi: 10.1126/science.aad9378
Geuking, M. B., Cahenzli, J., Lawson, M. A., Ng, D. C., Slack, E., Hapfelmeier, S., et al. (2011). Intestinal Bacterial Colonization Induces Mutualistic Regulatory T Cell Responses. Immunity 34 (5), 794–806. doi: 10.1016/j.immuni.2011.03.021
Goldman, A. S. (2007). The Immune System in Human Milk and the Developing Infant. Breastfeed Med. 2 (4), 195–204. doi: 10.1089/bfm.2007.0024
Golenbock, D. T., Hampton, R. Y., Qureshi, N., Takayama, K., Raetz, C. R. (1991). Lipid A-Like Molecules That Antagonize the Effects of Endotoxins on Human Monocytes. J. Biol. Chem. 266 (29), 19490–19498. doi: 10.1016/S0021-9258(18)55023-7
Gosalbes, M. J., Llop, S., Valles, Y., Moya, A., Ballester, F., Francino, M. P. (2013). Meconium Microbiota Types Dominated by Lactic Acid or Enteric Bacteria are Differentially Associated With Maternal Eczema and Respiratory Problems in Infants. Clin. Exp. Allergy 43 (2), 198–211. doi: 10.1111/cea.12063
Gould, H. J., Sutton, B. J. (2008). IgE in Allergy and Asthma Today. Nat. Rev. Immunol. 8 (3), 205–217. doi: 10.1038/nri2273
Gray, L. E., O'Hely, M., Ranganathan, S., Sly, P. D., Vuillermin, P. (2017). The Maternal Diet, Gut Bacteria, and Bacterial Metabolites During Pregnancy Influence Offspring Asthma. Front. Immunol. 8. doi: 10.3389/fimmu.2017.00365
Hanson, L. A. (2007). Session 1: Feeding and Infant Development Breast-Feeding and Immune Function. Proc. Nutr. Soc. 66 (3), 384–396. doi: 10.1017/S0029665107005654
Henrick, B. M., Rodriguez, L., Lakshmikanth, T., Pou, C., Henckel, E., Arzoomand, A., et al. (2021). Bifidobacteria-Mediated Immune System Imprinting Early in Life. Cell 184 (15), 3884–3898 e3811. doi: 10.1016/j.cell.2021.05.030
Hooper, L. V., Littman, D. R., Macpherson, A. J. (2012). Interactions Between the Microbiota and the Immune System. Science 336 (6086), 1268–1273. doi: 10.1126/science.1223490
Houghteling, P. D., Walker, W. A. (2015). Why Is Initial Bacterial Colonization of the Intestine Important to Infants' and Children's Health? J. Pediatr. Gastroenterol. Nutr. 60 (3), 294–307. doi: 10.1097/MPG.0000000000000597
Iweala, O. I., Burks, A. W. (2016). Food Allergy: Our Evolving Understanding of Its Pathogenesis, Prevention, and Treatment. Curr. Allergy Asthma Rep. 16 (5), 37. doi: 10.1007/s11882-016-0616-7
Iweala, O. I., Nagler, C. R. (2019). The Microbiome and Food Allergy. Annu. Rev. Immunol. 37, 377–403. doi: 10.1146/annurev-immunol-042718-041621
Jakaitis, B. M., Denning, P. W. (2014). Commensal and Probiotic Bacteria May Prevent NEC by Maturing Intestinal Host Defenses. Pathophysiology 21 (1), 47–54. doi: 10.1016/j.pathophys.2013.11.012
Kaplan, J. L., Shi, H. N., Walker, W. A. (2011). The Role of Microbes in Developmental Immunologic Programming. Pediatr. Res. 69 (6), 465–472. doi: 10.1203/PDR.0b013e318217638a
Kau, A. L., Ahern, P. P., Griffin, N. W., Goodman, A. L., Gordon, J. I. (2011). Human Nutrition, the Gut Microbiome and the Immune System. Nature 474 (7351), 327–336. doi: 10.1038/nature10213
Kayama, H., Okumura, R., Takeda, K. (2020). Interaction Between the Microbiota, Epithelia, and Immune Cells in the Intestine. Annu. Rev. Immunol. 38, 23–48. doi: 10.1146/annurev-immunol-070119-115104
Knoop, K. A., Holtz, L. R., Newberry, R. D. (2018). Inherited Nongenetic Influences on the Gut Microbiome and Immune System. Birth Defects Res. 110 (20), 1494–1503. doi: 10.1002/bdr2.1436
Koenig, J. E., Spor, A., Scalfone, N., Fricker, A. D., Stombaugh, J., Knight, R., et al. (2011). Succession of Microbial Consortia in the Developing Infant Gut Microbiome. Proc. Natl. Acad. Sci. U. S. A. (108 Suppl 1), 4578–4585. doi: 10.1073/pnas.1000081107
Kubinak, J. L., Petersen, C., Stephens, W. Z., Soto, R., Bake, E., O'Connell, R. M., et al. (2015). MyD88 Signaling in T Cells Directs IgA-Mediated Control of the Microbiota to Promote Health. Cell Host Microbe 17 (2), 153–163. doi: 10.1016/j.chom.2014.12.009
Kull, I., Melen, E., Alm, J., Hallberg, J., Svartengren, M., van Hage, M., et al. (2010). Breast-Feeding in Relation to Asthma, Lung Function, and Sensitization in Young Schoolchildren. J. Allergy Clin. Immunol. 125 (5), 1013–1019. doi: 10.1016/j.jaci.2010.01.051
Kumbhare, S. V., Patangia, D. V. V., Patil, R. H., Shouche, Y. S., Patil, N. P. (2019). Factors Influencing the Gut Microbiome in Children: From Infancy to Childhood. J. Biosci. 44 (2), 49. doi: 10.1007/s12038-019-9860-z
Liem, J. J., Kozyrskyj, A. L., Huq, S. I., Becker, A. B. (2007). The Risk of Developing Food Allergy in Premature or Low-Birth-Weight Children. J. Allergy Clin. Immunol. 119 (5), 1203–1209. doi: 10.1016/j.jaci.2006.12.671
Li, M., Wang, M., Donovan, S. M. (2014). Early Development of the Gut Microbiome and Immune-Mediated Childhood Disorders. Semin. Reprod. Med. 32 (1), 74–86. doi: 10.1055/s-0033-1361825
Li, H. T., Zhou, Y. B., Liu, J. M. (2013). The Impact of Cesarean Section on Offspring Overweight and Obesity: A Systematic Review and Meta-Analysis. Int. J. Obes. (Lond) 37 (7), 893–899. doi: 10.1038/ijo.2012.195
Mai, V., Young, C. M., Ukhanova, M., Wang, X., Sun, Y., Casella, G., et al. (2011). Fecal Microbiota in Premature Infants Prior to Necrotizing Enterocolitis. PloS One 6 (6), e20647. doi: 10.1371/journal.pone.0020647
Marra, F., Lynd, L., Coombes, M., Richardson, K., Legal, M., Fitzgerald, J. M., et al. (2006). Does Antibiotic Exposure During Infancy Lead to Development of Asthma?: A Systematic Review and Metaanalysis. Chest 129 (3), 610–618. doi: 10.1378/chest.129.3.610
Martin, R., Makino, H., Cetinyurek Yavuz, A., Ben-Amor, K., Roelofs, M., Ishikawa, E., et al. (2016). Early-Life Events, Including Mode of Delivery and Type of Feeding, Siblings and Gender, Shape the Developing Gut Microbiota. PloS One 11 (6), e0158498. doi: 10.1371/journal.pone.0158498
Matsuki, T., Yahagi, K., Mori, H., Matsumoto, H., Hara, T., Tajima, S., et al. (2016). A Key Genetic Factor for Fucosyllactose Utilization Affects Infant Gut Microbiota Development. Nat. Commun. 7, 11939. doi: 10.1038/ncomms11939
Maynard, C. L., Elson, C. O., Hatton, R. D., Weaver, C. T. (2012). Reciprocal Interactions of the Intestinal Microbiota and Immune System. Nature 489 (7415), 231–241. doi: 10.1038/nature11551
Mendez, R., Banerjee, S., Bhattacharya, S. K., Banerjee, S. (2019). Lung Inflammation and Disease: A Perspective on Microbial Homeostasis and Metabolism. IUBMB Life 71 (2), 152–165. doi: 10.1002/iub.1969
Metsala, J., Lundqvist, A., Virta, L. J., Kaila, M., Gissler, M., Virtanen, S. M. (2013). Mother's and Offspring's Use of Antibiotics and Infant Allergy to Cow's Milk. Epidemiology 24 (2), 303–309. doi: 10.1097/EDE.0b013e31827f520f
Milani, C., Duranti, S., Bottacini, F., Casey, E., Turroni, F., Mahony, J., et al. (2017). The First Microbial Colonizers of the Human Gut: Composition, Activities, and Health Implications of the Infant Gut Microbiota. Microbiol. Mol. Biol. Rev. 81 (4), e00036–17. doi: 10.1128/MMBR.00036-17
Milliken, S., Allen, R. M., Lamont, R. F. (2019). The Role of Antimicrobial Treatment During Pregnancy on the Neonatal Gut Microbiome and the Development of Atopy, Asthma, Allergy and Obesity in Childhood. Expert Opin. Drug Saf. 18 (3), 173–185. doi: 10.1080/14740338.2019.1579795
Mukherjee, S., Zheng, H., Derebe, M. G., Callenberg, K. M., Partch, C. L., Rollins, D., et al. (2014). Antibacterial Membrane Attack by a Pore-Forming Intestinal C-Type Lectin. Nature 505 (7481), 103–107. doi: 10.1038/nature12729
Nogacka, A., Salazar, N., Suarez, M., Milani, C., Arboleya, S., Solis, G., et al. (2017). Impact of Intrapartum Antimicrobial Prophylaxis Upon the Intestinal Microbiota and the Prevalence of Antibiotic Resistance Genes in Vaginally Delivered Full-Term Neonates. Microbiome 5 (1), 93. doi: 10.1186/s40168-017-0313-3
Nyce, J. W., Metzger, W. J. (1997). DNA Antisense Therapy for Asthma in an Animal Model. Nature 385 (6618), 721–725. doi: 10.1038/385721a0
Ober, C., Sperling, A. I., von Mutius, E., Vercelli, D. (2017). Immune Development and Environment: Lessons From Amish and Hutterite Children. Curr. Opin. Immunol. 48, 51–60. doi: 10.1016/j.coi.2017.08.003
Olszak, T., An, D., Zeissig, S., Vera, M. P., Richter, J., Franke, A., et al. (2012). Microbial Exposure During Early Life has Persistent Effects on Natural Killer T Cell Function. Science 336 (6080), 489–493. doi: 10.1126/science.1219328
Pammi, M., Cope, J., Tarr, P. I., Warner, B. B., Morrow, A. L., Mai, V., et al. (2017). Intestinal Dysbiosis in Preterm Infants Preceding Necrotizing Enterocolitis: A Systematic Review and Meta-Analysis. Microbiome 5 (1), 31. doi: 10.1186/s40168-017-0248-8
Patole, S., Keil, A. D., Chang, A., Nathan, E., Doherty, D., Simmer, K., et al. (2014). Effect of Bifidobacterium Breve M-16V Supplementation on Fecal Bifidobacteria in Preterm Neonates–a Randomised Double Blind Placebo Controlled Trial. PloS One 9 (3), e89511. doi: 10.1371/journal.pone.0089511
Penders, J., Thijs, C., van den Brandt, P. A., Kummeling, I., Snijders, B., Stelma, F., et al. (2007). Gut Microbiota Composition and Development of Atopic Manifestations in Infancy: The KOALA Birth Cohort Study. Gut 56 (5), 661–667. doi: 10.1136/gut.2006.100164
Plaza-Diaz, J., Fontana, L., Gil, A. (2018). Human Milk Oligosaccharides and Immune System Development. Nutrients 10 (8), 1038. doi: 10.3390/nu10081038
Price, A. E., Shamardani, K., Lugo, K. A., Deguine, J., Roberts, A. W., Lee, B. L., et al. (2018). A Map of Toll-Like Receptor Expression in the Intestinal Epithelium Reveals Distinct Spatial, Cell Type-Specific, and Temporal Patterns. Immunity 49 (3), 560–575 e566. doi: 10.1016/j.immuni.2018.07.016
Pulimood, A. B., Mathan, M. M., Mathan, V. I. (1998). Quantitative and Ultrastructural Analysis of Rectal Mucosal Mast Cells in Acute Infectious Diarrhea. Dig Dis. Sci. 43 (9), 2111–2116. doi: 10.1023/a:1018875718392
Qin, J., Li, Y., Cai, Z., Li, S., Zhu, J., Zhang, F., et al. (2012). A Metagenome-Wide Association Study of Gut Microbiota in Type 2 Diabetes. Nature 490 (7418), 55–60. doi: 10.1038/nature11450
Rachid, R., Stephen-Victor, E., Chatila, T. A. (2021). The Microbial Origins of Food Allergy. J. Allergy Clin. Immunol. 147 (3), 808–813. doi: 10.1016/j.jaci.2020.12.624
Riskin, A., Almog, M., Peri, R., Halasz, K., Srugo, I., Kessel, A. (2012). Changes in Immunomodulatory Constituents of Human Milk in Response to Active Infection in the Nursing Infant. Pediatr. Res. 71 (2), 220–225. doi: 10.1038/pr.2011.34
Sampath, V., Le, M., Lane, L., Patel, A. L., Cohen, J. D., Simpson, P. M., et al. (2011). The NFKB1 (G.-24519delattg) Variant is Associated With Necrotizing Enterocolitis (NEC) in Premature Infants. J. Surg. Res. 169 (1), e51–e57. doi: 10.1016/j.jss.2011.03.017
Sanchez, C., Fente, C., Regal, P., Lamas, A., Lorenzo, M. P. (2021). Human Milk Oligosaccharides (HMOs) and Infant Microbiota: A Scoping Review. Foods 10 (6), 1429. doi: 10.3390/foods10061429
Savage, D. C. (1977). Microbial Ecology of the Gastrointestinal Tract. Annu. Rev. Microbiol. 31, 107–133. doi: 10.1146/annurev.mi.31.100177.000543
Schuijt, T. J., Lankelma, J. M., Scicluna, B. P., de Sousa e Melo, F., Roelofs, J. J., de Boer, J. D., et al. (2016). The Gut Microbiota Plays a Protective Role in the Host Defence Against Pneumococcal Pneumonia. Gut 65 (4), 575–583. doi: 10.1136/gutjnl-2015-309728
Shi, N., Li, N., Duan, X., Niu, H. (2017). Interaction Between the Gut Microbiome and Mucosal Immune System. Mil. Med. Res. 4, 14. doi: 10.1186/s40779-017-0122-9
Singh, R. P., Niharika, J., Kondepudi, K. K., Bishnoi, M., Tingirikari, J. M. R. (2022). Recent Understanding of Human Milk Oligosaccharides in Establishing Infant Gut Microbiome and Roles in Immune System. Food Res. Int. 151, 110884. doi: 10.1016/j.foodres.2021.110884
Stokholm, J., Blaser, M. J., Thorsen, J., Rasmussen, M. A., Waage, J., Vinding, R. K., et al. (2018). Maturation of the Gut Microbiome and Risk of Asthma in Childhood. Nat. Commun. 9 (1), 141. doi: 10.1038/s41467-017-02573-2
Strachan, D. P. (1989). Hay Fever, Hygiene, and Household Size. BMJ 299 (6710), 1259–1260. doi: 10.1136/bmj.299.6710.1259
Tan, J., McKenzie, C., Vuillermin, P. J., Goverse, G., Vinuesa, C. G., Mebius, R. E., et al. (2016). Dietary Fiber and Bacterial SCFA Enhance Oral Tolerance and Protect Against Food Allergy Through Diverse Cellular Pathways. Cell Rep. 15 (12), 2809–2824. doi: 10.1016/j.celrep.2016.05.047
Turnbaugh, P. J., Ley, R. E., Mahowald, M. A., Magrini, V., Mardis, E. R., Gordon, J. I. (2006). An Obesity-Associated Gut Microbiome With Increased Capacity for Energy Harvest. Nature 444 (7122), 1027–1031. doi: 10.1038/nature05414
Vael, C., Vanheirstraeten, L., Desager, K. N., Goossens, H. (2011). Denaturing Gradient Gel Electrophoresis of Neonatal Intestinal Microbiota in Relation to the Development of Asthma. BMC Microbiol. 11, 68. doi: 10.1186/1471-2180-11-68
Wen, L., Ley, R. E., Volchkov, P. Y., Stranges, P. B., Avanesyan, L., Stonebraker, A. C., et al. (2008). Innate Immunity and Intestinal Microbiota in the Development of Type 1 Diabetes. Nature 455 (7216), 1109–1113. doi: 10.1038/nature07336
Yang, T., Santisteban, M. M., Rodriguez, V., Li, E., Ahmari, N., Carvajal, J. M., et al. (2015). Gut Dysbiosis Is Linked to Hypertension. Hypertension 65 (6), 1331–1340. doi: 10.1161/HYPERTENSIONAHA.115.05315
Ygberg, S., Nilsson, A. (2012). The Developing Immune System - From Foetus to Toddler. Acta Paediatr. 101 (2), 120–127. doi: 10.1111/j.1651-2227.2011.02494.x
Zeng, M. Y., Cisalpino, D., Varadarajan, S., Hellman, J., Warren, H. S., Cascalho, M., et al. (2016). Gut Microbiota-Induced Immunoglobulin G Controls Systemic Infection by Symbiotic Bacteria and Pathogens. Immunity 44 (3), 647–658. doi: 10.1016/j.immuni.2016.02.006
Keywords: immune, infant, microbiome, maturation, diseases
Citation: Zhang H, Zhang Z, Liao Y, Zhang W and Tang D (2022) The Complex Link and Disease Between the Gut Microbiome and the Immune System in Infants. Front. Cell. Infect. Microbiol. 12:924119. doi: 10.3389/fcimb.2022.924119
Received: 20 April 2022; Accepted: 23 May 2022;
Published: 15 June 2022.
Edited by:
Theresa Li-Yun Chang, Rutgers, The State University of New Jersey, United StatesReviewed by:
Gonzalo Solís Sánchez, Central University Hospital of Asturias, SpainAnthony Santilli, Lerner Research Institute, United States
Laurel Lagenaur, Osel, United States
Copyright © 2022 Zhang, Zhang, Liao, Zhang and Tang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Dong Tang, ODMzOTI3ODVAcXEuY29t
 Huan Zhang
Huan Zhang Zhilin Zhang
Zhilin Zhang Yiqun Liao2
Yiqun Liao2 Wenjie Zhang
Wenjie Zhang Dong Tang
Dong Tang