- Parasite Host Biology, Indian Council of Medical Research-National Institute of Malaria Research, New Delhi, India
The neglected but highly prevalent Plasmodium vivax in South-east Asia and South America poses a great challenge, with regards to long-term in-vitro culturing and heavily limited functional assays. Such visible challenges as well as narrowed progress in development of experimental research tools hinders development of new drugs and vaccines. The leading vaccine candidate antigen Plasmodium vivax Duffy Binding Protein (PvDBP), is essential for reticulocyte invasion by binding to its cognate receptor, the Duffy Antigen Receptor for Chemokines (DARC), on the host’s reticulocyte surface. Despite its highly polymorphic nature, the amino-terminal cysteine-rich region II of PvDBP (PvDBPII) has been considered as an attractive target for vaccine-mediated immunity and has successfully completed the clinical trial Phase 1. Although this molecule is an attractive vaccine candidate against vivax malaria, there is still a question on its viability due to recent findings, suggesting that there are still some aspects which needs to be looked into further. The highly polymorphic nature of PvDBPII and strain-specific immunity due to PvDBPII allelic variation in Bc epitopes may complicate vaccine efficacy. Emergence of various blood-stage antigens, such as PvRBP, PvEBP and supposedly many more might stand in the way of attaining full protection from PvDBPII. As a result, there is an urgent need to assess and re-assess various caveats connected to PvDBP, which might help in designing a long-term promising vaccine for P. vivax malaria. This review mainly deals with a bunch of rising concerns for validation of DBPII as a vaccine candidate antigen for P. vivax malaria.
Preface
Infectious diseases have played an important role in modeling human demography and genetics. Malaria is considered to be one of the most devastating infectious diseases affecting mankind and is believed to be one of the strongest selective pressures in recent human history (Haldane, 2004; Kwiatkowski, 2005). At least nine species of the unicellular eukaryotic parasite of genus Plasmodium are reported to cause infection in humans including P. falciparum, P. vivax, P. malariae, P. ovale curtisi, P. ovale wallikeri (Sutherland et al., 2010), P. knowlesi, P. cynomolgi (Ta et al., 2014), P. simium (Deane, 1992; Brasil et al., 2017), and P. brasilianum (Lalremruata et al., 2015). Out of these nine species, only Plasmodium falciparum and Plasmodium vivax emerge to be the major threats escalating the malaria load globally. Though great progress has been made in the fight against malaria since 2000, an increase in the number of cases in the last few years has placed doubt on the objective of eliminating the illness. In 2020, an estimated 241 million malaria cases were reported in 85 malaria-endemic countries (World Health Organization, 2021). The WHO African Region accounted for around 95% of cases in 2020, with an anticipated 228 million cases (World Health Organization, 2021). There has been an increase in the proportion of malaria caused by P. vivax in co-endemic regions where intensive malaria-control measures have lowered the burden of P. falciparum. In co-endemic areas, there is an increased risk of P. vivax after P. falciparum therapy, suggesting that universal radical cure for both parasites might be beneficial in some situations.
P. vivax is by far the most predominant source of human malaria across Asia and the Asia-Pacific regions, which account for approximately 80% of the worldwide P. vivax burden due to large populations and a diminishing prevalence of P. falciparum infections (Howes et al., 2016). Its existence has also been reported in the horn of Africa, Madagascar, and parts of Central and South America (World Health Organization, 2020). The sensitivity of the present-generation RDTs employed for P. vivax diagnosis is comparable to that of microscopy (Chu and White, 2021). In malaria-endemic areas, it has been found that ultrasensitive PCR technologies detects parasite densities as low as 28/ml (Imwong et al., 2014). This indicates a substantially greater prevalence of asymptomatic P. vivax infection than previously thought. In areas where P. falciparum and P. vivax malaria coexist, the P. vivax burden has overtaken the P. falciparum burden (Battle et al., 2019). P. falciparum, which is well known to cause complicated and fatal malaria, has overshadowed the clinical and public health importance of P. vivax malaria (Baird, 2007; Conway, 2007). Contrary to the belief that P. vivax causes a relatively benign and self-limiting infection, evidence documenting severe and complicated P. vivax malaria are escalating gradually (Price et al., 2007; Herrera et al., 2007; Baird, 2013a; Baird, 2013b). Concomitant or chronic illness could result into a severe P. vivax infection.
It is increasingly becoming visible that efforts towards understanding P. vivax have been inadequate in comparison to those for P. falciparum (Price et al., 2009) and one of the main reasons behind the lag of P. vivax research is the inability of achieving a stable and long-term in-vitro culture for P. vivax leading to significantly restricted laboratory-based experimental studies. Advances in in vitro culture of P. knowlesi in human RBCs, have given critical support for more sophisticated laboratory investigations (Moon et al., 2013; Mohring et al., 2019), allowing some practical functional studies of P. vivax to be conducted. Plasmodium spp. other than P. vivax target almost all stages of RBCs, whereas P. vivax preferentially invades immature RBCs or reticulocytes (Kitchen, 1938), which normally account for 1-2% of the red blood cells in the peripheral blood circulation. Although advancement has been made in understanding the molecular basis underlying P. vivax reticulocyte preference for invasion (Gruszczyk et al., 2018a), still a powerful tool lacks (Krotoski et al., 1982; Mueller et al., 2009) which will help us in overcoming the difficulties in maintaining P. vivax in long-term cultures as it is relatively more difficult to repeatedly obtain and supplement reticulocyte-rich human blood to P. vivax cultures.
Another unique challenge with P. vivax is its ability to produce a dormant liver-stage forms or hypnozoites (Krotoski et al., 1982; Markus, 2011) which are responsible for multiple clinical relapses after a primary infection (Imwong et al., 2007). Prevention of P. vivax relapses is a must for P. vivax malaria to be eliminated. The distinction between relapse, recrudescence, and reinfection, and thus identifying early resistance, is a fundamental difficulty in therapeutic assessment. Chloroquine and the ACT companion medications are very slowly removed, so suppressive blood concentrations can last for weeks post medication (White, 2021).
Clinical intervention of P. vivax malaria requires clinical suspicion, an accurate blood test, and access to an efficient schizonticidal and hypnozoiticidal medication regimens. Although P. vivax is known to be still sensitive to chloroquine combined with primaquine, cases of chloroquine and sulfadoxine-pyrimethamine drug resistant P. vivax have also been reported from many areas of the globe including Australia, Ethiopia, Pakistan, Indonesia, Papua New Guinea, S. Korea and India (Rieckmann et al., 1989; Schunk et al., 2006; Price et al., 2009; Khatoon, 2010; Price, 2014). In addition, the main problem in managing a P. vivax infection is the management of frequent relapses for which both primaquine and its new counterpart, tafenoquine, have problems related to treatment adherence and safety with respect to G6PD deficiency. Thus, the unique clinical biology of P. vivax and restricted progress in the advancement of research tools (Su, 2019), create an obstacle in the way of growth of efficacious drugs and vaccines for vivax malaria. Adding to the above reasons, lack of financing, a paucity of resources and a high cost to create new vaccines contributes to the slow progress in case of development of a successful P. vivax vaccine.
An Ideal Plasmodium vivax Malaria Vaccine
Regardless of decades of continuous efforts, only one vaccine (pre-erythrocytic vaccine RTS, S/ASO1 also known as Mosquirix) for P. falciparum, has been licensed for human use (RTS,S Clinical Trials Partnership, 2014; Laurens, 2020), but no vaccine for P. vivax is available yet. Plasmodium spp. exhibits a unique set of antigens at each stage of its life which makes it difficult for a researcher to identify the best vaccine candidate. The complex biology of P. vivax, its extensive antigenic diversity and its pathway of immune evasion make vaccine development against P. vivax malaria challenging. P. vivax is reported to exhibit greater genetic diversity in comparison to P. falciparum (Neafsey et al., 2012; Winter et al., 2015). While selecting a vaccine candidate for P. vivax, it is highly crucial to focus on those playing a role in invasion and those with a conserved epitope, which can be targeted by neutralizing the strain transcending antibodies. The discovery of broadly conserved inhibitory epitopes provides important new themes for the next generation of P. vivax malaria vaccines, as well as a foundation for rational structure-based vaccine design that will impart global strain-transcending protection (Chen et al., 2016). Multiple clinical isolates of P. vivax were used to investigate a panel of human monoclonal antibodies for their ability to inhibit PvDBP from binding to the DARC, as well as their ability to impede red blood cell invasion and reticulocyte invasion. This led to the discovery of a widely neutralizing human monoclonal antibody that prevented P. vivax invasion in all tested strains (Rawlinson et al., 2019).
Current Status of Candidate P. vivax Malaria Vaccines
The designing and distribution of a successful P. vivax vaccine tends to be a prime concern for speeding up malaria elimination in the Asia-Pacific and the Americas (Tanner et al., 2015). Only a few P. vivax vaccine candidates are close to or have reached different stages of clinical trials (Mueller et al., 2015; Draper et al., 2018). The delay in the development of a CSP-based vaccine for P. falciparum, RTS,S/AS01 (RTS,S) clearly indicates that much more work awaits for a comparable P. vivax vaccine. Although there is potential current research into P. vivax vaccine targets and immunisation tactics, the odds of a P. vivax vaccine becoming available in the near future are low. To date, human clinical trials have only been carried out for three P. vivax antigens namely, the PvCSP-based pre-erythrocytic vaccine, the PvDBP-based blood stage vaccine and the transmission-blocking candidate Pvs25 (Rainbow Tables, WHO). Several novel vaccine candidates are now being studied in a pre-clinical setting and there are excellent reviews discussing them (Galinski and Barnwell, 2008; Valencia et al., 2011).
The VMP001/AS01B vaccine, which encompasses the N- and C- terminal regions of the CSP and a short repeat region comprising of repeat sequences from both the VK210 (type 1) and the VK247 (type 2) genotypes of P. vivax has been shown to clear the Phase I/IIa trial, increasing antibody and cell-mediated immune responses and subsequently resulting in a delay in the pre-patency period in 30 Duffy-positive vaccines (Bennett et al., 2016), but no sterile protection was achieved. However, a combination of PvCSP and PvTRAP provided sterile protection in mice using doses that individually conferred low or no protection (Atcheson et al., 2018). Phase II trials with another candidate, PvRAS (Plasmodium vivax Radiation-Attenuated Sporozoites), showed immunogenic and sterile immunity in only 42% of the Duffy +ve (Fy+) subjects (Arevalo-Herrera et al., 2016). Transmission Blocking Vaccines targeting either a) pre-fertilization antigens expressed by gametocytes (Pvs48/45 and Pvs47) and gametes (Pvs230) (Sauerwein and Bousema, 2015; Tachibana et al., 2015) and b) post-fertilization antigens expressed by zygotes/ookinetes/oocysts (Pvs25 and Pvs28) (Hisaeda et al., 2000; Sauerwein and Bousema, 2015). To date, the Pvs25 protein present on the surface of ookinetes and oocysts (Tsuboi et al., 1998), is one of the best characterized Transmission Blocking Vaccine candidate Blagborough et al. (2016). Phase 1 trial using Pvs25 formulated with Montanide ISA 51 as an adjuvant has demonstrated significant antibody responses in volunteers, but trial was stopped due to frequent local reactogenicity such as erythema, induration, swelling, and tenderness at the site of injection (Wu et al., 2008). However, pre-clinical and clinical studies with P25 proteins shows that inclusion of a carrier protein could potentially boost its immunogenicity (Qian et al., 2007; Parzych et al., 2017; Radtke et al., 2017).
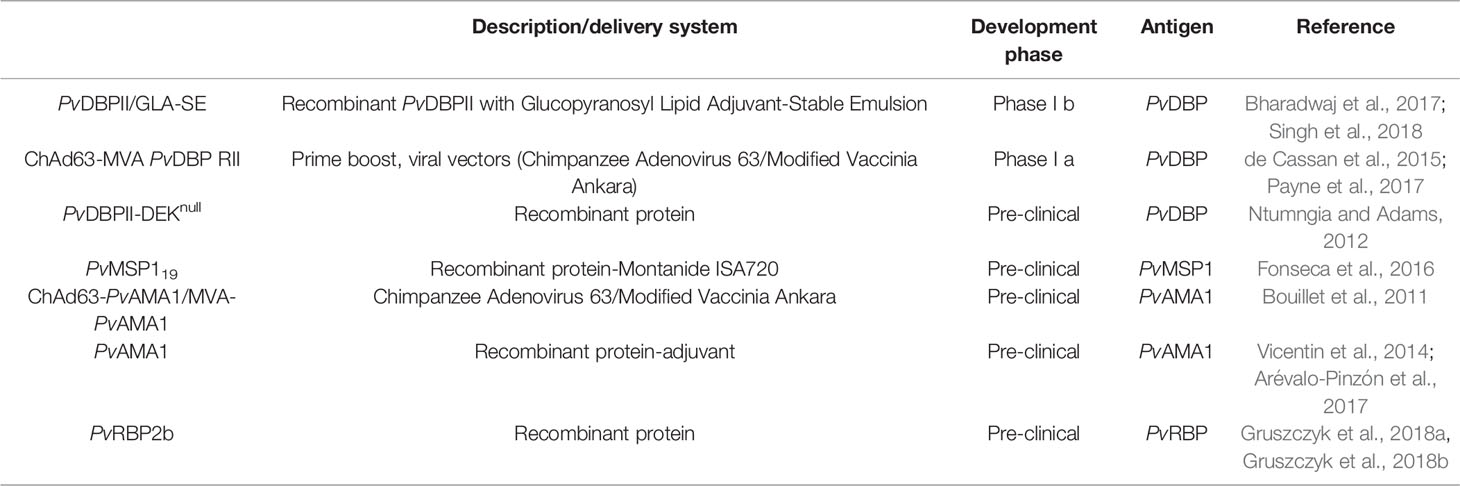
While evaluating a novel vaccine candidate antigen’s eligibility, it should be checked whether the gene that encodes it is required for parasite growth, as targeting a non-essential gene would appear to favour parasites that do not rely on the gene product and hence are immune to the vaccine. Although progress has been observed in identification and antigenic characterization of different P. vivax antigens, this review mainly focuses on the blood stage vaccine candidates and that too on PvDBP, the only to-date blood stage vaccine candidate that has reached Phase 1 clinical trial (de Cassan et al., 2015; Bhardwaj et al., 2017; Payne et al., 2017; Singh et al., 2018). Antigens expressed on the merozoite surface are considered as blood stage vaccine targets. An effective vaccination against P. vivax blood stages would decrease symptoms and pathology associated with such repeated infections, and so potentially play a crucial role in controlling the species. In addition to provision of safety and efficacy, an ideal blood-stage vaccine candidate antigen should be capable of eliciting a strong immune response that inhibits Plasmodium from invading the target host cell. In comparison to 15 P. falciparum’s blood stage vaccine candidates that have been described in literature so far (Illingworth et al., 2019), only a few candidates have been studied in case of P. vivax (Table 1), including Duffy Binding Protein (PvDBP), Merozoite Surface Protein 1 (MSP1), Apical Membrane Antigen 1 (PvAMA1) and Reticulocyte Binding Protein (PvRBP2b), a distant homologue of Reticulocyte Binding Protein Homologue 5 (PfRh5). Utilizing P. knowlesi as a screening model, research on a panel of P. vivax proteins (PvMSP7.1, PvMSP3.10, Pv12, Pv41, PvGAMA, PvCyRPA and PvARP) hypothesized to act in erythrocyte invasion, found an additional erythrocytic stage vaccine candidates (Ndegwa et al., 2021). Taking into account all of the benefits and drawbacks of any model system, it can be concluded that P. knowlesi might serve as an accessible and efficient model to screen for new candidates until a robust and long-term P. vivax culture is produced.
To date, human trials in the erythrocytic stage have only been carried out for PvDBP-based vaccine. P. vivax invasion of human RBCs is restricted to interaction of PvDBP with human reticulocytes (via the Duffy Antigen Receptor for Chemokines, DARC) expressing the Iron Importer, Transferrin Receptor 1 (TfR1) or Cluster of Differentiation 71 (CD71) (Malleret et al., 2015). Till date, only two vaccines targeting the conserved cysteine-rich region II of PvDBP have reached clinical trials, ChAd63/MVA PvDBP RII (Payne et al., 2017) and PvDBPII/GLA-SE (Bhardwaj et al., 2017; Singh et al., 2018).
PvDBP, an Essential Parasite Ligand for Human Reticulocyte Invasion
A number of distinct invasion pathways have been identified by Plasmodium spp. that exploit unique sets of human red blood cell (RBC) receptors for invasion. Two major protein families of Plasmodium, the Erythrocyte-Binding-Like (EBL) family, expressed from the erythrocyte-binding-like (ebl) genes (Fang et al., 1991) and the Reticulocyte-Binding-Like (RBL) protein homologs (RBL or Rh), expressed from reticulocyte binding protein genes (Galinski et al., 1992) are responsible for parasite’s tight interactions with different stages of host’s RBCs. There exists a species-specific variation in the count of EBL proteins, P. falciparum having five members while P. vivax has only a single member (Adams et al., 1992; Adams et al., 2001). The EBL family further consists of the Duffy-Binding-Like (DBL-EBL) and Erythrocyte-Binding Protein sub-families (EBP) (Adams et al., 2001). The DBL-EBL proteins are characterized by presence of two cysteine-rich regions and a Duffy-binding domain in the N-terminal cysteine-rich region (Adams et al., 1992). On the other hand, the members of the RBL family solely target the reticulocytes as well as normocytes, producing parasite proteins which facilitate reticulocyte binding and/or invasion (Ntumngia et al., 2018). Reticulocyte-binding proteins (RBPs) were originally discovered in P. vivax (Galinski et al., 1992) and are the classic instances of reticulocyte binding-like/reticulocyte-binding homolog (RBL/RH) proteins, which have also been discovered in P. cynomolgi (Okenu et al., 2005) and P. yoelii (Ogun et al., 2011). In P. vivax, the RBL determine the reticulocyte restriction of this species. Out of the five PvRBPs, only one (PvRBP2b) is found to bind exclusively to reticulocytes (França et al., 2016).
Duffy-Binding-Like Sub-family of Erythrocyte-Binding-Like Family
A huge macromolecular cascade of proteins is likely involved in host cell selection and invasion activities. However, just a few will be crucial participants in allowing the parasite to retain a significant red blood cell invasion capacity in the face of physiological and immunological changes in the host. This adds to the latency of the infection and, as a result, enhance the possibilities of transmission. These proteins are most likely parasite ligands involved in the erythrocyte surface binding events that contribute to effective invasion. Sequestered in the micronemes of merozoites, the DBL–EBPs are type-I membrane proteins which are supposed to be released during the invasion process (Adams et al., 1990). The first DBL-EBL was identified in P. knowlesi and was called Duffy-Binding Protein (PkDBP) as it was shown to bind the Duffy Antigen Receptor for Chemokines (DARC) on RBCs (Chitnis and Miller, 1994). Subsequently, its orthologues in P. vivax and P. falciparum have also been identified (Haynes et al., 1988). Members of DBL-EBL family are characterized to have six extracellular regions (RI-RVI), subsequently followed by a type I trans-membrane domain, and a short cytoplasmic tail (Adams et al., 1992). Out of the six extracellular regions, the two hydrophobic cysteine-rich regions (N-terminal RII and C-terminal RVI) are functionally conserved in all erythrocyte binding proteins (EBLs) and separated by three low-homology regions (RIII-RV). The N-terminal cysteine-rich region (RII) carries the binding residues responsible for binding to the DARC (Chitnis and Miller, 1994), whereas the C-terminal cysteine-rich region (RVI) has no clear known function, although a high degree of amino acid conservation among the three Plasmodium species (P. falciparum, P. vivax and P. knowlesi) is observed which suggests that this domain might have some importance (Adams et al., 1992). P. falciparum and P. knowlesi exhibit a variety of proteins (PfEBL-1, PfEBA-140, PfEBA-175, PfEBA-181, PfEBA-165, PkDBPα and multiple DBP-like ligands) belonging to DBL-EBL family, creating alternative pathways of RBC invasion, whereas, P. vivax comprises of a single protein, PvDBP (Adams et al., 1992) of the DBL-EBL family. PvEBP, in addition to PvDBP, is a new member to this family (Roesch et al., 2018).
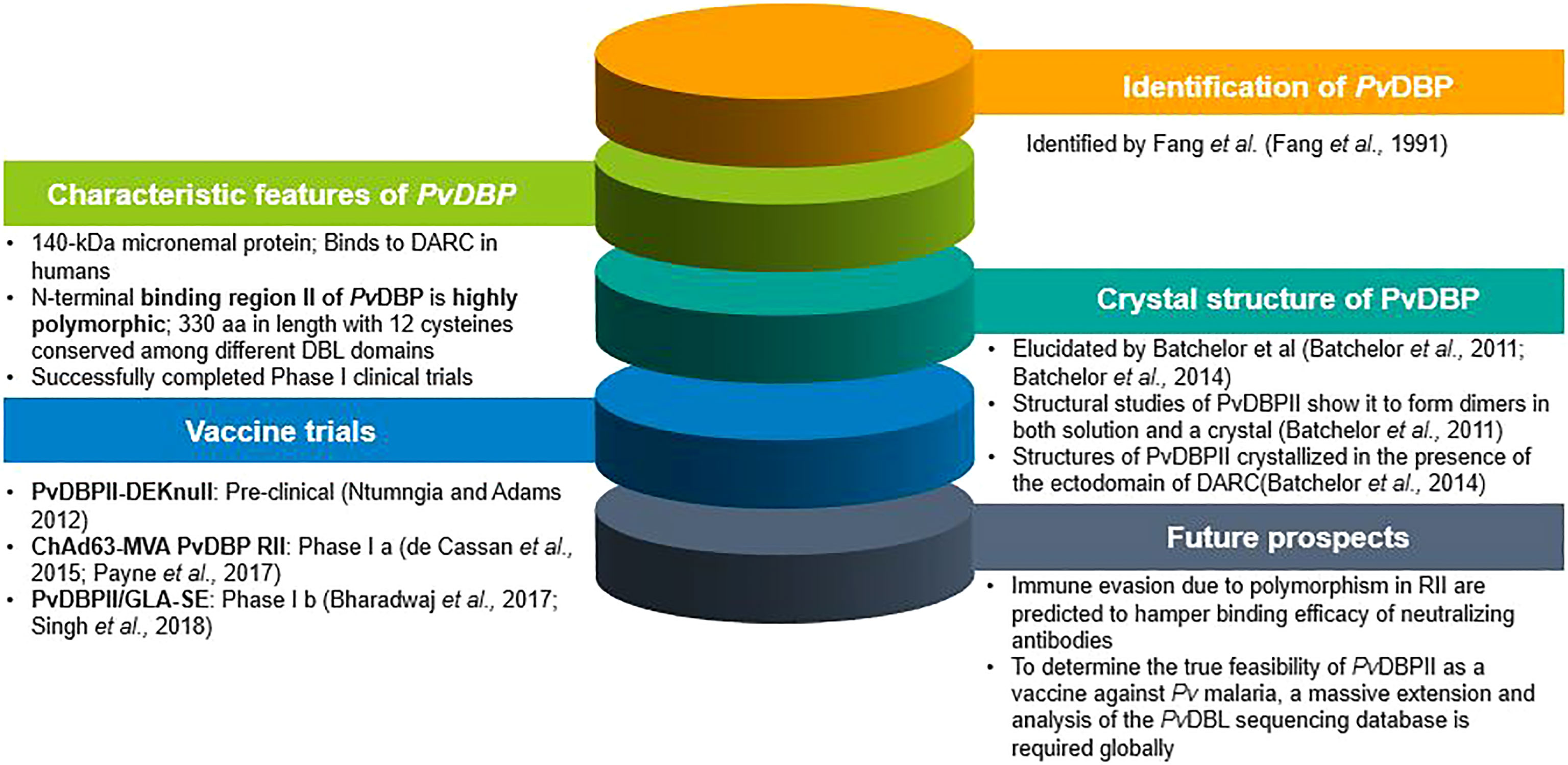
Plasmodium vivax Duffy Binding Protein (PvDBP) is a 140-kDa trans-membrane protein responsible for reticulocyte invasion of P. vivax and is dependent on the host’s Duffy Antigen Receptor for Chemokines (DARC) (Horuk et al., 1993). The Pvdbp gene (PlasmoDB Gene ID = PVX_110810) is present in chromosome 6 of P. vivax spanning a length of 3,762 nucleotides Carlton et al. (2008) (Figure 1A) and comprising of five exons and four introns (Fang et al., 1991). Exon 1 (57 nucleotides) of Pvdbp encodes a signal sequence, exon 2 (2,959 nucleotides) encodes 986 amino acids and covers the six extracellular regions, RI-RVI of the translated protein, exon 3 spans 79 nucleotides and comprises a trans-membrane domain (18 amino acids), exons 4 and 5 spanning 74 and 44 nucleotides, respectively. Exons 4 and 5 and a portion of exon 3 translates into a cytoplasmic tail (45 amino acids) (Adams et al., 1990; Adams et al., 1992) (Figure 1B). The N-terminal cysteine rich region (RII) comprises of DBL domains (Chitnis and Miller, 1994)which contain binding residues responsible for formation of tight junction between PvDBP and DARC. The C-terminal cysteine-rich region (RVI), is separated from RII by three hydrophilic regions III, IV and V and is followed by the trans-membrane domain.
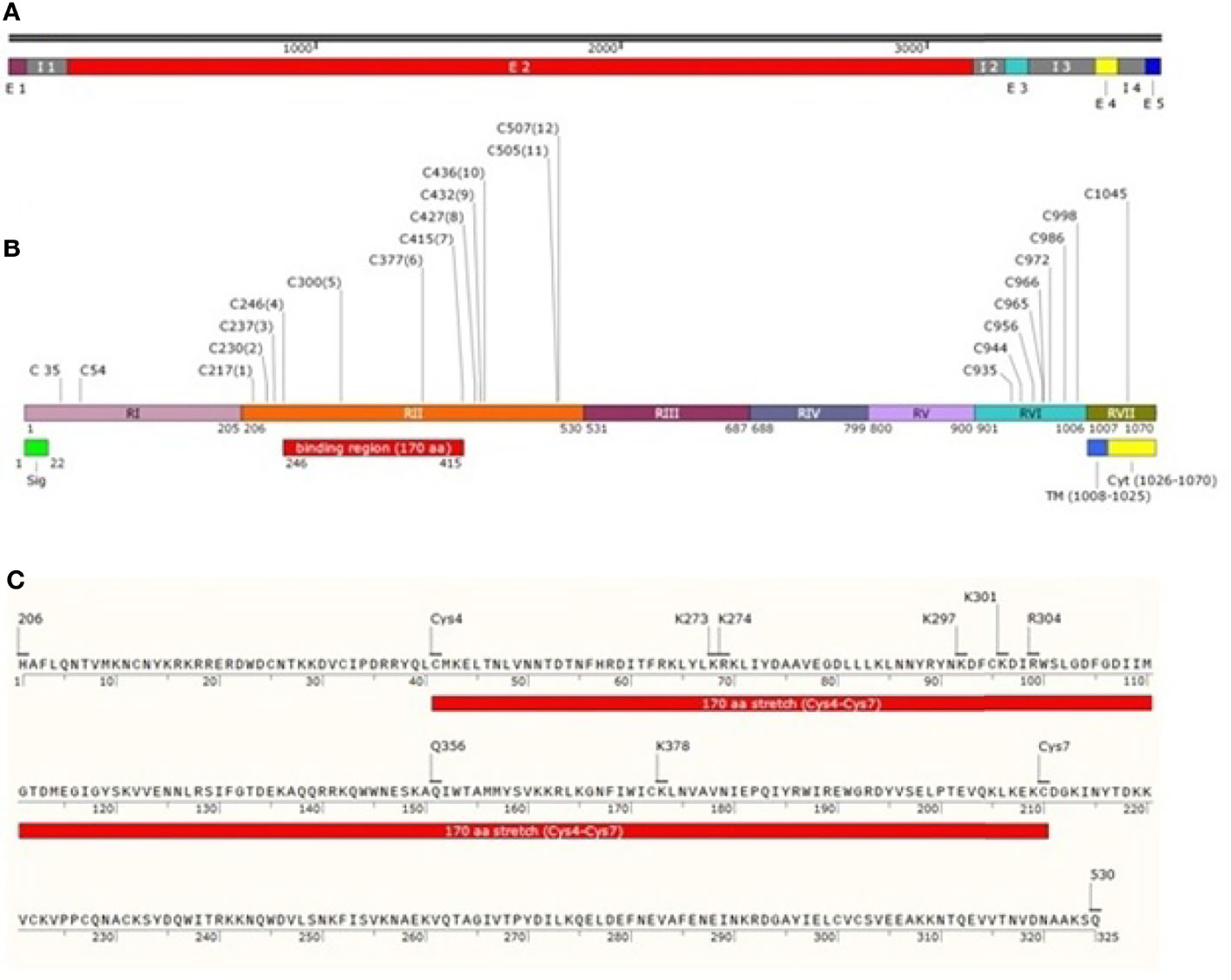
Figure 1 Schematic drawing of PvDBP. (A) Pvdbp gene (3762 nucleotides) with 5 exons (1-57, 193-3151, 3257-3335, 3554-3627 and 3719-3762, respectively) and 4 introns (58-192, 3152-3256, 3336-3553 and 3628-3718, respectively). (B) PvDBP consisting of 1070 amino acid residues. The shared boundaries for regions I-VII (205, 530, 687, 799, 900, 1006 and 1070 amino acids, respectively) (Adams et al., 1992; Okenu et al., 2005). Labelled in it are the positions corresponding to seven regions (RI-RVII) of PvDBP, the cysteine-rich regions (RII and RVI), signal peptide shaded in green (1-22aa), transmembrane domain shaded in blue (1008-1025aa) and cytoplasmic domain shaded in yellow (1026-1070aa). PvDBP consists of 23 cysteines (two in RI, twelve in RII, eight in RVI and one in RVII). The cysteines are labelled along with their amino acid position. The binding residues map to a 170 aa stretch which starts from Cys4 and ends at Cys7 (C246-C415) (C) PvDBPII spans a length of 325 aa from 206-530aa (Adams et al., 1990). The PvDBPII binding residues at sub domain 2: Site 1 (K297, K301, R304 and K378 (VanBuskirk et al., 2004b)) and Site 2 (K273, K274 and Q356 (Hans et al., 2005)) are responsible for reticulocyte binding with respect to 1070 aa residues of PvDBP (NCBI protein id: XP_001608387.1).
The N-terminal cysteine rich region (RII) of PvDBP starts and ends at H206 and Q530, respectively (VanBuskirk et al., 2004b) (Figure 1C). It has been found that RII spans 325 aa residues, and not 330 aa, as it was thought previously. This 325 aa region (RII) comprises of 12 conserved cysteine residues (C217, C230, C237, C246, C300, C377, C415, C427, C432, C436, C505, and C507) (Fang et al., 1991; Adams et al., 1992). The cysteines are reported to contribute to DBL’s structural integrity (Singh et al., 2003; Singh et al., 2006), and so the parasite may not afford changes in these residues. The region deepest within the DBL domain, i.e. between cysteines 4 and 8, have been marked as the portion bearing the prime components for receptor recognition (Tsuboi et al., 1994; Ranjan and Chitnis, 1999; Xainli et al., 2000). The minimal binding region of PvDBPII to the human DARC is localized between cysteines 4 and 7 (Ranjan and Chitnis, 1999; Batchelor et al., 2011). Residues between cysteines 7 and 8 are supposed to be surface-exposed and are not significantly involved in receptor binding (Ranjan and Chitnis, 1999; VanBuskirk et al., 2004b).
Pkα/Pv-DBL is a compact helical, monomeric module spread over three distinct subdomains (SD1, SD2 and SD3). Pkα/Pv-DBL consists of twelve cysteine residues which are stabilized by intra-domain disulfide bonds mostly conserved amongst the DBL family of EBPs (Singh et al., 2006). The indispensable and invariant residues required for DARC recognition (Figure 1C) were mapped within a region on SD2 (Singh et al., 2003; Hans et al., 2005; Singh et al., 2006; Yogavel et al., 2018), which lies between Cys4 (C246) - Cys7 (C415). Cys1 (C217) - Cys3 (C237) and Cys8 (C427) - Cys12 (C507) which flank SD2 (Cys4-Cys7) might play a structural role in the intact DBL domain (Ranjan and Chitnis, 1999; Singh et al., 2003; Singh et al., 2006) (Table 2). SD1 is not required for DBL-DARC interaction (Singh et al., 2003) whereas the functional significance of SD3 is still in question.
Although region II plays a significant role in receptor recognition, this region with respect to the rest of Pvdbp gene is hyper-variable with a high ratio of non-synonymous to synonymous mutations (Tsuboi et al., 1994; Xainli et al., 2000; Cole-Tobian and King, 2003), which might be one of the factors which help the parasite to escape host immunity (Tsuboi et al., 1994; Xainli et al., 2000). Exploration of PvdbpII genetic variation among P. vivax endemic regions showed that PvDBPII is highly polymorphic, however, no changes in the cysteine residues have been reported so far (Tsuboi et al., 1994; Ampudia et al., 1996; Xainli et al., 2000; Kho et al., 2001; Cole-Tobian and King, 2003; Sousa et al., 2006; Gosi et al., 2008; Babaeekhou et al., 2009; Batchelor et al., 2011; Premaratne et al., 2011; Chenet et al., 2012; Ju et al., 2012; Ju et al., 2013.
The polymorphic residues adjacent to the binding site are reported to escape the binding inhibitory antibodies thus keeping the binding site of the protein undisturbed. Site-directed mutagenesis of PvDBPII identified several residues which are vital for receptor recognition (VanBuskirk et al., 2004b). The conserved residues present in the binding region of PvDBP were found to be responsible for ligand receptor interaction. The variant residues are reported to flank the functionally important residues. So, changes occurring in the conserved amino acid residues (which are not exposed on the surface, as a result are not detected by hosts immunity) might be accountable for loss of binding activity. The reported polymorphic residues were not found to affect reticulocyte binding as they are found to be mapped in the face opposite to the residues critical for binding to DARC (Chitnis and Sharma, 2008).
Batchelor et al. elucidated the crystal structure of PvDBPII (PDB: 3RRC), which indicates a model for receptor recognition through PvDBP dimerization, facilitating the development of a complex composed of two PvDBP and two DARC molecules, which might pave way towards invasion (Batchelor et al., 2011). The critical binding residues required for reticulocyte binding were found to be structurally and functionally conserved, and are also targets of immune response (Batchelor et al., 2011). Protective antibodies targeting the critical binding regions in PvDBPII were found to disturb dimerization and/or inhibit receptor binding. A step-wise binding model has also been proposed which involves receptor-induced PvDBPII dimerization facilitating the formation of a heterotrimer that eventually employs a second DARC molecule to form a heterotetramer (PDB: 4NUU and 4NUV) Batchelor et al. (2014). Although these structural and biophysical studies provide deep insight into PvDBPII-DARC engagement, further studies are required to assess these models as this region is prone to polymorphisms (Mittal et al., 2020) and as a result, the inherent variability in PvDBL might render the PvDBP-based vaccines inefficacious. Further, Yogavel et al. reported the existence of two binding sites in PvDBPII, a) Site 1 which includes residues K266, K270, R273 and K347 and, b) Site 2 including residues K242, R243 and H325 (from PDB: 3RRC, 4NUU and 4NUV) (Yogavel et al., 2018). The DARC peptide, by means of its sulfated Tyr41 and phosphated Tyr30, engages at sites 1 and 2, respectively on Pv/Pk-DBLs. This is a testable model depicting DARC’s engagement with Pv/Pk-DBP and needs to be experimentally assessed for further confirmation.
Keeping allelic variation in mind, a successful and efficacious DBPII-based vaccine should aim at conserved epitopes which are supposed to be the prospective targets of strain-transcending neutralizing immunity. Naturally acquired binding-inhibitory antibodies to PvDBPII are associated with clinical immunity of the subject to P. vivax malaria and thus potently neutralize the P. vivax invasion mechanism (Grimberg et al., 2007; Chootong et al., 2010; Nicolete et al., 2016)
For the first time, studies using ELISA and flow cytometry confirmed that both rabbit and human antibodies inhibited recombinant PvDBPII-DARC interactions and were found to reduce invasion efficiency of wild P. vivax by up to 64%, while a reduced P. vivax invasion by up to 54% was observed in a combined PvDBPII antisera from people exposed to P. vivax (Grimberg et al., 2007). Polymorphisms in PvDBPII and the presence of multiple strains in endemic regions present unique challenges in the path of vaccine design (VanBuskirk et al., 2004a; Cole-Tobian et al., 2009; Ntumngia et al., 2012). In spite of the variations that exists in PvDBPII, broadly conserved epitopes of three inhibitory murine monoclonal antibodies have been recognized in PvDBPII (subdomain 3) (Chen et al., 2016) which were not found to lie in close vicinity to the dimer interface as well as the DARC-binding site (Chen et al., 2016). Clinical trials in humans using PvDBPII produced antibodies that block in vitro binding of different allelic variants of PvDBPII to the DARC for more than 100 days following three immunization doses (Payne et al., 2017; Singh et al., 2018). Both the vaccine candidates of PvDBP which are in clinical trial (PvDBPII/GLA-SE, ChAd63-MVA PvDBP RII) were found to give rise to strain-transcending antibodies. By means of human mAbs produced in the course of vaccination or through natural P. vivax exposure, a broadly neutralizing human mAb have been identified which inhibited the invasion of all tested strains of P. vivax, thus indicating the molecular basis for inhibition. that will thus aid in the design of successful and efficient DBP-based vaccine for P. vivax malaria (Rawlinson et al., 2019; Urusova et al., 2019). The various landmarks achieved in PvDBP research are shown in Figure 2.
Duffy Negativity and Other Challenges
The Duffy antigens act as receptors for a wide range of chemokines and are therefore called as Duffy Antigen Receptor for Chemokines (DARC). The same Duffy antigens also serve as receptors for P. knowlesi, P. vivax and P. cynomolgi (Kosaisavee et al., 2017). Absence of DARC on the reticulocyte surface is thought to confer protection against blood stage infections caused by P. vivax in Africa (Sanger et al., 1955; Miller et al., 1976). However, Duffy-negative individuals infected with P. vivax have been reported in sub-Saharan Africa (Ryan et al., 2006; Gosi et al., 2008; Ménard et al., 2010; Woldearegai et al., 2013; Djeunang Dongho et al., 2021), which points to the fact that there might be an alternative route of invading human reticulocytes lacking DARC. A recent study conducted in 952 individuals observed the absence of P. vivax infections in Ghana where a high frequency of the Duffy-negative genotype was reported (Brown et al., 2021). Human P. vivax strains have been reported in Madagascar and parts of Africa, which might be due to the re-establishment of this parasite (Culleton and Carter, 2012). However, it is still to be confirmed whether these cases have emerged due to the introduction of a new P. vivax strain which may use an alternative pathway which is independent of DARC. Of late, a novel P. vivax Erythrocyte Binding Protein (PvEBP, also known as DBP2) was reported which may enable interactions with other membrane proteins on erythrocytes (Hester et al., 2013). Pvebp gene belongs to the DBL-EBP family, which harbours all the key features of EBPs, suggesting its ability to bind to human erythrocytes and facilitate RBC invasion (106). This gene was found to be expressed in the blood-stage of P. vivax and was found in all P. vivax strains examined, comprising an extensive geographical span (Hester et al., 2013). Both PvDBP and PvEBP were found to be antigenically distinct. The discovery of PvEBP promptly highlights an alternative invasion pathway and could perhaps illuminate the initial step towards decoding the principle underlying P. vivax infection of the Duffy-negatives. Further investigations on PvEBP revealed its preferential binding to young (CD71high) Duffy positive reticulocytes and minimal binding capacity for Duffy-negative reticulocytes (Ntumngia et al., 2016). This study proposes that PvEBP might not serve as a ligand for Duffy-negative reticulocytes, but may act as an alternative pathway for invading Duffy-positive population. Whole genome sequencing studies show that Pvdbp gene is duplicated (at a higher rate) in Madagascar population, where both Duffy-negative and Duffy-positive individuals co-exist (Menard et al., 2013), and was supposed to be associated with infection in Duffy negatives possibly in response to constraints imposed by Duffy negativity in some human populations. In addition to this another study confirmed the presence of multiple copies of Pvdbp gene (3 and 8 copies) in two Duffy-negative Ethiopian isolates (Gunalan et al., 2016; Roesch et al., 2018). But this observation was contradicted by emergence of widespread Pvdbp gene duplication in malaria endemic areas of South-east Asia comprising Duffy-positive population (Hostetler et al. (2016). Although an excess of nonsynonymous mutations and no synonymous mutations was observed in Pvebp in comparison to Pvdbp, but in terms of allelic diversity, Pvebp was found to be less diverse than Pvdbp in Madagascar (both Duffy-negative and Duffy-positive) and Cambodian population (Duffy-positive). The absence of synonymous mutation in this case clearly marks that the Pvebp gene is under strong positive selection and validates the importance of this protein in reticulocyte invasion as well as Duffy-independent invasion pathways used by Plasmodium vivax (Roesch et al., 2018).
Conclusion
P. vivax has evolved with a variety of mechanisms to overcome immune defense at every step of communication with its host species. Developing an effective vaccine that provides protection and prevents transmission is highly essential in eliminating P. vivax malaria. The Region II of PvDBP, the only blood-stage vaccine candidate, spans 325 aa residues, and not 330 aa, as was previously reported. PvDBPII is the lone vaccine candidate that has entered clinical trial Phase 1b and is crucially involved in P. vivax merozoite invasion of human reticulocytes.
In spite of the fact that PvDBP is crucial for blood-stage infection, its exercise for vaccine development constitutes of major obstacles as cited below.
(i) Polymorphisms in PvDBP seem to be critical for the evasion of host immune response. A brief exposure of PvDBP to the host’s immune system, due to its micronemal location and the rapid kinetics of parasite invasion, allows a short-term exposure of PvDBP to the host immune system. As a result of this phenomenon, the parasite might gain an advantage of escaping the host’s immunity. Immune selection being a major driving force for allelic variation, as even a single amino acid substitution can change the antigenic nature of a pathogen. Also the evolutionary arms race between the parasite and human indicates that the parasite genome is evolving at a higher rate than the latter. The amount of polymorphic data available globally is insufficient for declaring PvDBP as a long-lasting and effective vaccine candidate. Therefore, a global (covering the P. vivax endemic regions) rigorous survey in terms of allelic diversity of PvDBL domain is required. All these points lead to the fact that we are in requirement of a candidate antigen that is less polymorphic.
(ii) The surfacing of P. vivax infection in Duffy negative population is an alarming condition.
(iii) Emergence of newly reported P. vivax ligands targeting RBCs questions the viability of PvDBPII as the lone vaccine candidate and (iv) presence of more than single copy (1-4 reported) of Pvdbp might create an obstacle in the way of attaining an efficacious vaccine.
(iv) A strain transcending PvDBPII-based vaccine demands a globally conserved epitope. Mittal et al., in their global Single Amino Acid Polymorphism (SAAP) data analysis reported that from the four PvDBL–mAb complex structures, 2 out of the 4 purported neutralizing mAbs do not bind near the supposed dimer interface (Mittal et al., 2020). Such discoveries points towards the fact that a vaccine against P. vivax could have more impact if above challenges of PvDBPII (as the lone candidate) were considered.
(v) Considering PvDBPII as a potential vaccine target, its immunodominant variant epitopes deflect immune responses, compromising the vaccine efficacy in triggering high titer neutralising antibodies against conserved strain-transcending functional epitopes (Chootong et al., 2010; Ntumngia and Adams, 2012).
Contributing to the formulation of preventative and/or therapeutic approaches which will assist in minimising the effects of malaria, there is a requirement of deciphering and combining functional and structural investigations (Patarroyo et al., 2020).While PvDBP may still be required for the invasion of Duffy negative erythrocytes (Gunalan et al., 2018; Lo et al., 2019), the only focus on PvDBP as a vaccine candidate certainly needs to be reconsidered, and alternative targets explored as potential substitutes for PvDBP or in conjunction with it. A vaccine targeting only a single-stage parasite antigen faces challenges in retaining similar antibody responses due to the genomic changes in parasite ligands which in turn might improve the fitness of P. vivax isolates. Most of the P. vivax vaccines in pipeline target individual stages and are based on single antigens. Combination allele vaccines in case of PvdbpII achieve greater specificity by targeting a majority of antibody to common epitopes among the constituent alleles that form the vaccine (De et al., 2021; Ntumngia et al., 2013). This suggested that a vaccine with multiple DBPII variant alleles is necessary for broader coverage. For finding new interaction hotspots to which malaria elimination approaches can be directed, a profound analysis is needed to correlate structural, functional (adhesion, invasion, and inhibition), and polymorphism data (Patarroyo et al., 2020). Moreover, a blood stage vaccine has to face a huge number of merozoites in comparison to a few as in case of pre-erythrocytic and transmission blocking stage. So, it is of high importance to combine antigens including multi-stages of parasite life cycle to attain the purpose of developing an effective vaccine for P. vivax malaria.
Author Contributions
AS conceptualized the study, coordinated, drafted and critically revised the manuscript. SK aided in conception, literature search and drafting the whole manuscript. All authors gave final approval for publication and agree to be held accountable for the work performed therein.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
The authors thank and acknowledge ICMR-NIMR for its support and assistance towards providing a working environment.
References
Adams, J. H., Hudson, D. E., Torii, M., Ward, G. E., Wellems, T. E., Aikawa, M., et al. (1990). The Duffy Receptor Family of Plasmodium Knowlesi is Located Within the Micronemes of Invasive Malaria Merozoites. Cell 63 (1), 141–153. doi: 10.1016/0092-8674(90)90295-p
Adams, J. H., Sim, B. K., Dolan, S. A., Fang, X., Kaslow, D. C., Miller, L. H., et al. (1992). A Family of Erythrocyte Binding Proteins of Malaria Parasites. Proc. Natl. Acad. Sci. U.S.A. 89 (15), 7085–7089. doi: 10.1073/pnas.89.15.7085
Adams, J. H., Blair, P. L., Kaneko, O., Peterson, D. S.. (2001). An Expanding Ebl Family of Plasmodium Falciparum. Trends Parasitol. 17 (6), 297–299. doi: 10.1016/s1471-4922(01)01948-1
Ampudia, E., Patarroyo, M. A., Patarroyo, M. E., Murillo, L. A. (1996). Genetic Polymorphism of the Duffy Receptor Binding Domain of Plasmodium Vivax in Colombian Wild Isolates. Mol. Biochem. Parasitol. 78 (1-2), 269–272. doi: 10.1016/s0166-6851(96)02611-4
Arévalo-Herrera, M., Vásquez-Jiménez, J. M., Lopez-Perez, M., Vallejo, A. F., Amado-Garavito, A. B., Céspedes, N., et al. (2016). Protective Efficacy of Plasmodium Vivax Radiation-Attenuated Sporozoites in Colombian Volunteers: A Randomized Controlled Trial. PLoS Negl. Trop. Dis. 10 (10), e0005070. doi: 10.1371/journal.pntd.0005070
Arévalo-Pinzón, G., Bermúdez, M., Hernández, D., Curtidor, H., Patarroyo, M. A. (2017). Plasmodium Vivax Ligand-Receptor Interaction: PvAMA-1 Domain I Contains the Minimal Regions for Specific Interaction With CD71+ Reticulocytes. Sci. Rep. 7 (1), 9616. doi: 10.1038/s41598-017-10025-6
Atcheson, E., Bauza, K., Salman, A. M., Alves, E., Blight, J., Viveros-Sandoval, M. E., et al. (2018). Tailoring a Plasmodium Vivax Vaccine To Enhance Efficacy Through a Combination of a CSP Virus-Like Particle and TRAP Viral Vectors. Infect. Immun. 86 (9), e00114–e00118. doi: 10.1128/IAI.00114-18
Babaeekhou, L., Zakeri, S., Djadid, N. D.. (2009). Genetic Mapping of the Duffy Binding Protein (DBP) Ligand Domain of Plasmodium Vivax From Unstable Malaria Region in the Middle East. Am. J. Trop. Med. Hyg. 80 (1), 112–118. doi: 10.4269/ajtmh.08-0241err
Baird, J. K.. (2007). Neglect of Plasmodium Vivax Malaria. Trends Parasitol. 23 (11), 533–539. doi: 10.1016/j.pt.2007.08.011
Baird, J. K.. (2013a). Malaria Caused by Plasmodium Vivax: Recurrent, Difficult to Treat, Disabling, and Threatening to Life–the Infectious Bite Preempts These Hazards. Pathog. Glob. Health 107 (8), 475–479. doi: 10.1179/2047772413Z.000000000179
Baird, J. K.. (2013b). Evidence and Implications of Mortality Associated With Acute Plasmodium Vivax Malaria. Clin. Microbiol. Rev. 26 (1), 36–57. doi: 10.1128/CMR.00074-12
Batchelor, J. D., Zahm, J. A., Tolia, N. H.. (2011). Dimerization of Plasmodium Vivax DBP is Induced Upon Receptor Binding and Drives Recognition of DARC. Nat. Struct. Mol. Biol. 18 (8), 908–914. doi: 10.1038/nsmb.2088
Batchelor, J. D., Malpede, B. M., Omattage, N. S., DeKoster, G. T., Henzler-Wildman, K. A., Tolia, N. H.. (2014). Red Blood Cell Invasion by Plasmodium Vivax: Structural Basis for DBP Engagement of DARC. PLoS Pathog. 10 (1), e1003869. doi: 10.1371/journal.ppat.1003869
Battle, K. E., Lucas, T., Nguyen, M., Howes, R. E., Nandi, A. K., Twohig, K. A., et al. (2019). Mapping the Global Endemicity and Clinical Burden of Plasmodium Vivax 2000-17: A Spatial and Temporal Modelling Study. Lancet (London England) 394 (10195), 332–343. doi: 10.1016/S0140-6736(19)31096-7
Bennett, J. W., Yadava, A., Tosh, D., Sattabongkot, J., Komisar, J., Ware, L. A., et al. (2016). Phase 1/2a Trial of Plasmodium Vivax Malaria Vaccine Candidate VMP001/AS01B in Malaria-Naive Adults: Safety, Immunogenicity, and Efficacy. PLoS Negl. Trop. Dis. 10 (2), e0004423. doi: 10.1371/journal.pntd.0004423
Bhardwaj, R., Shakri, A. R., Hans, D., Gupta, P., Fernandez-Becerra, C., Del Portillo, H. A., et al. (2017). Production of Recombinant PvDBPII, Receptor Binding Domain of Plasmodium Vivax Duffy Binding Protein, and Evaluation of Immunogenicity to Identify an Adjuvant Formulation for Vaccine Development. Protein Expr. Purif. 136, 52–57. doi: 10.1016/j.pep.2015.06.011
Blagborough, A. M., Musiychuk, K., Bi, H., Jones, R. M., Chichester, J. A., Streatfield, S., et al. (2016). Transmission Blocking Potency and Immunogenicity of a Plant-Produced Pvs25-Based Subunit Vaccine Against Plasmodium Vivax. Vaccine 34 (28), 3252–3259. doi: 10.1016/j.vaccine.2016.05.007
Bouillet, L. É., Dias, M. O., Dorigo, N. A., Moura, A. D., Russell, B., Nosten, F., et al. (2011). Long-Term Humoral and Cellular Immune Responses Elicited by a Heterologous Plasmodium Vivax Apical Membrane Antigen 1 Protein Prime/Adenovirus Boost Immunization Protocol. Infect. Immun. 79 (9), 3642–3652. doi: 10.1128/IAI.05048-11
Brasil, P., Zalis, M. G., de Pina-Costa, A., Siqueira, A. M., Júnior, C. B., Silva, S., et al. (2017). Outbreak of Human Malaria Caused by Plasmodium Simium in the Atlantic Forest in Rio De Janeiro: A Molecular Epidemiological Investigation. Lancet Glob. Health Engl. 5 (10), e1038–e1046. doi: 10.1016/S2214-109X(17)30333-9
Brown, C. A., Pappoe-Ashong, P. J., Duah, N., Ghansah, A., Asmah, H., Afari, E., et al. (2021). High Frequency of the Duffy-Negative Genotype and Absence of Plasmodium Vivax Infections in Ghana. Malar. J. 20, 99. doi: 10.1186/s12936-021-03618-0
Carlton, J. M., Adams, J. H., Silva, J. C., Bidwell, S. L., Lorenzi, H., Caler, E., et al. (2008). Comparative Genomics of the Neglected Human Malaria Parasite Plasmodium Vivax. Nature 455 (7214), 757–763. doi: 10.1038/nature07327
Chen, E., Salinas, N. D., Huang, Y., Ntumngia, F., Plasencia, M. D., Gross, M. L., et al. (2016). Broadly Neutralizing Epitopes in the Plasmodium Vivax Vaccine Candidate Duffy Binding Protein. Proc. Natl. Acad. Sci. U.S.A. 113 (22), 6277–6282. doi: 10.1073/pnas.1600488113
Chenet, S. M., Tapia, L. L., Escalante, A. A., Durand, S., Lucas, C., Bacon, D. J. (2012). Genetic Diversity and Population Structure of Genes Encoding Vaccine Candidate Antigens of Plasmodium Vivax. Malaria J. 11, 68. doi: 10.1186/1475-2875-11-68
Chitnis, C. E., Miller, L. H.. (1994). Identification of the Erythrocyte Binding Domains of Plasmodium Vivax and Plasmodium Knowlesi Proteins Involved in Erythrocyte Invasion. J. Exp. Med. 180 (2), 497–506. doi: 10.1084/jem.180.2.497
Chitnis, C. E., Sharma, A.. (2008). Targeting the Plasmodium Vivax Duffy-Binding Protein. Trends Parasitol. 24 (1), 29–34. doi: 10.1016/j.pt.2007.10.004
Chootong, P., Ntumngia, F. B., VanBuskirk, K. M., Xainli, J., Cole-Tobian, J. L., Campbell, C. O., et al. (2010). Mapping Epitopes of the Plasmodium Vivax Duffy Binding Protein With Naturally Acquired Inhibitory Antibodies. Infect. Immun. 78 (3), 1089–1095. doi: 10.1128/IAI.01036-09
Chu, C. S., White, N. J.. (2021). The Prevention and Treatment of Plasmodium Vivax Malaria. PLoS Med. 18 (4), e1003561. doi: 10.1371/journal.pmed.1003561
Cole-Tobian, J. L., Michon, P., Biasor, M., Richards, J. S., Beeson, J. G., Mueller, I., et al. (2009). Strain-Specific Duffy Binding Protein Antibodies Correlate With Protection Against Infection With Homologous Compared to Heterologous Plasmodium Vivax Strains in Papua New Guinean Children. Infect. Immun. 77 (9), 4009–4017. doi: 10.1128/IAI.00158-09
Cole-Tobian, J., King, C. L.. (2003). Diversity and Natural Selection in Plasmodium Vivax Duffy Binding Protein Gene. Mol. Biochem. Parasitol. 127 (2), 121–132. doi: 10.1016/s0166-6851(02)00327-4
Conway, D. J.. (2007). Molecular Epidemiology of Malaria. Clin. Microbiol. Rev. 20 (1), 188–204. doi: 10.1128/CMR.00021-06
Culleton, R., Carter, R.. (2012). African Plasmodium Vivax: Distribution and Origins. Int. J. Parasitol. 42 (12), 1091–1097. doi: 10.1016/j.ijpara.2012.08.005
Deane, L. M.. (1992). Simian Malaria in Brazil. Mem. Inst. Oswaldo Cruz Brazil 87 Suppl 3, 1–20. doi: 10.1590/s0074-02761992000700001
De, S. L., Ntumngia, F. B., Nicholas, J., Adams, J. H.. (2021). Progress Towards the Development of a P. Vivax Vaccine. Expert Rev Vaccines 20 (2), 97–112. doi: 10.1080/14760584.2021.1880898
de Cassan, S. C., Shakri, A. R., Llewellyn, D., Elias, S. C., Cho, J. S., Goodman, A. L., et al. (2015). Preclinical Assessment of Viral Vectored and Protein Vaccines Targeting the Duffy-Binding Protein Region II of Plasmodium Vivax. Front. Immunol. 6. doi: 10.3389/fimmu.2015.00348
Djeunang Dongho, G. B., Gunalan, K., L'Episcopia, M., Paganotti, G. M., Menegon, M., Efeutmecheh Sangong, R., et al. (2021). Plasmodium Vivax Infections Detected in a Large Number of Febrile Duffy-Negative Africans in Dschang, Cameroon. Am. J. Trop. Med. Hyg. 104 (3), 987–992. doi: 10.4269/ajtmh.20-1255
Draper, S. J., Sack, B. K., King, C. R., Nielsen, C. M., Rayner, J. C., Higgins, M. K., et al. (2018). Malaria Vaccines: Recent Advances and New Horizons. Cell Host Microbe 24 (1), 43–56. doi: 10.1016/j.chom.2018.06.008
Fang, X. D., Kaslow, D. C., Adams, J. H., Miller, L. H. (1991). Cloning of the Plasmodium Vivax Duffy Receptor. Mol. Biochem. Parasitol. 44 (1), 125–132. doi: 10.1016/0166-6851(91)90228-x
Fonseca, J. A., Cabrera-Mora, M., Singh, B., Oliveira-Ferreira, J., da Costa Lima-Junior, J., Calvo-Calle, J. M., et al. (2016). A Chimeric Protein-Based Malaria Vaccine Candidate Induces Robust T Cell Responses Against Plasmodium Vivax MSP119. Sci. Rep. 6, 34527. doi: 10.1038/srep34527
França, C. T., He, W. Q., Gruszczyk, J., Lim, N. T., Lin, E., Kiniboro, B., et al. (2016). Plasmodium Vivax Reticulocyte Binding Proteins Are Key Targets of Naturally Acquired Immunity in Young Papua New Guinean Children. PLoS Negl. Trop. Dis. 10 (9), e0005014. doi: 10.1371/journal.pntd.0005014
Galinski, M. R., Barnwell, J. W. (2008). Plasmodium Vivax: Who Cares? Malar. J. 7, S9. doi: 10.1186/1475-2875-7-S1-S9
Galinski, M. R., Medina, C. C., Ingravallo, P., Barnwell, J. W.. (1992). A Reticulocyte-Binding Protein Complex of Plasmodium Vivax Merozoites. Cell 69 (7), 1213–1226. doi: 10.1016/0092-8674(92)90642-p
Gosi, P., Khusmith, S., Khalambaheti, T., Lanar, D. E., Schaecher, K. E., Fukuda, M. M., et al. (2008). Polymorphism Patterns in Duffy-Binding Protein Among Thai Plasmodium Vivax Isolates. Malaria J. 7, 112. doi: 10.1186/1475-2875-7-112
Grimberg, B. T., Udomsangpetch, R., Xainli, J., McHenry, A., Panichakul, T., Sattabongkot, J., et al. (2007). Plasmodium Vivax Invasion of Human Erythrocytes Inhibited by Antibodies Directed Against the Duffy Binding Protein. PLoS Med. 4 (12), e337. doi: 10.1371/journal.pmed.0040337
Gruszczyk, J., Kanjee, U., Chan, L. J., Menant, S., Malleret, B., Lim, N., et al. (2018a). Transferrin Receptor 1 Is a Reticulocyte-Specific Receptor for Plasmodium Vivax. Science (New York N.Y.) 359 (6371), 48–55. doi: 10.1126/science.aan1078
Gruszczyk, J., Huang, R. K., Chan, L. J., Menant, S., Hong, C., Murphy, J. M., et al. (2018b). Cryo-EM Structure of an Essential Plasmodium Vivax Invasion Complex. Nature 559 (7712), 135–139. doi: 10.1038/s41586-018-0249-1
Gunalan, K., Lo, E., Hostetler, J. B., Yewhalaw, D., Mu, J., Neafsey, D. E., et al. (2016). Role of Plasmodium Vivax Duffy-Binding Protein 1 in Invasion of Duffy-Null Africans. Proc. Natl. Acad. Sci. U.S.A. 113 (22), 6271–6276. doi: 10.1073/pnas.1606113113
Gunalan, K., Niangaly, A., Thera, M. A., Doumbo, O. K., Miller, L.H.. (2018). Plasmodium Vivax Infections of Duffy-Negative Erythrocytes: Historically Undetected or a Recent Adaptation? Trends Parasitol. 34 (5), 420–429. doi: 10.1016/j.pt.2018.02.006
Haldane, J. B.. (2004). The Rate of Spontaneous Mutation of a Human Gene. 1935. J. Genet. 83 (3), 235–244. doi: 10.1007/BF02717892
Hans, D., Pattnaik, P., Bhattacharyya, A., Shakri, A. R., Yazdani, S. S., Sharma, M., et al. (2005). Mapping Binding Residues in the Plasmodium Vivax Domain That Binds Duffy Antigen During Red Cell Invasion. Mol. Microbiol. 55 (5), 1423–1434. doi: 10.1111/j.1365-2958.2005.04484.x
Haynes, J. D., Dalton, J. P., Klotz, F. W., McGinniss, M. H., Hadley, T. J., Hudson, D. E., et al. (1988). Receptor-Like Specificity of a Plasmodium Knowlesi Malarial Protein That Binds to Duffy Antigen Ligands on Erythrocytes. J. Exp. Med. 167 (6), 1873–1881. doi: 10.1084/jem.167.6.1873
Herrera, S., Corradin, G., Arévalo-Herrera, M.. (2007). An Update on the Search for a Plasmodium Vivax Vaccine. Trends Parasitol. 23 (3), 122–128. doi: 10.1016/j.pt.2007.01.008
Hester, J., Chan, E. R., Menard, D., Mercereau-Puijalon, O., Barnwell, J., Zimmerman, P. A., et al. (2013). De Novo Assembly of a Field Isolate Genome Reveals Novel Plasmodium Vivax Erythrocyte Invasion Genes. PLoS Negl. Trop. Dis. 7 (12), e2569. doi: 10.1371/journal.pntd.0002569
Hisaeda, H., Stowers, A. W., Tsuboi, T., Collins, W. E., Sattabongkot, J. S., Suwanabun, N., et al. (2000). Antibodies to Malaria Vaccine Candidates Pvs25 and Pvs28 Completely Block the Ability of Plasmodium Vivax to Infect Mosquitoes. Infect. Immun. 68 (12), 6618–6623. doi: 10.1128/IAI.68.12.6618-6623.2000
Horuk, R., Chitnis, C. E., Darbonne, W. C., Colby, T. J., Rybicki, A., Hadley, T. J., et al. (1993). A Receptor for the Malarial Parasite Plasmodium Vivax: The Erythrocyte Chemokine Receptor. Science (New York N.Y.) 261 (5125), 1182–1184. doi: 10.1126/science.7689250
Hostetler, J. B., Lo, E., Kanjee, U., Amaratunga, C., Suon, S., Sreng, S., et al. (2016). Independent Origin and Global Distribution of Distinct Plasmodium Vivax Duffy Binding Protein Gene Duplications. PloS Negl. Trop. Dis. 10 (10), e0005091. doi: 10.1371/journal.pntd.0005091
Howes, R. E., Battle, K. E., Mendis, K. N., Smith, D. L., Cibulskis, R. E., Baird, J. K., et al. (2016). Global Epidemiology of Plasmodium Vivax. Am. J. Trop. Med. Hyg. 95 (6 Suppl), 15–34. doi: 10.4269/ajtmh.16-0141
Illingworth, J. J., Alanine, D. G., Brown, R., Marshall, J. M., Bartlett, H. E., Silk, S. E., et al. (2019). Functional Comparison of Blood-Stage Plasmodium Falciparum Malaria Vaccine Candidate Antigens. Front. Immunol. 10. doi: 10.3389/fimmu.2019.01254
Imwong, M., Snounou, G., Pukrittayakamee, S., Tanomsing, N., Kim, J. R., Nandy, A., et al. (2007). Relapses of Plasmodium Vivax Infection Usually Result From Activation of Heterologous Hypnozoites. J. Infect. Dis. 195 (7), 927–933. doi: 10.1086/512241
Imwong, M., Hanchana, S., Malleret, B., Rénia, L., Day, N. P., Dondorp, A., et al. (2014). High-Throughput Ultrasensitive Molecular Techniques for Quantifying Low-Density Malaria Parasitemias. J. Clin. Microbiol. 52 (9), 3303–3309. doi: 10.1128/JCM.01057-14
Ju, H. L., Kang, J. M., Moon, S. U., Kim, J. Y., Lee, H. W., Lin, K., et al. (2012). Genetic Polymorphism and Natural Selection of Duffy Binding Protein of Plasmodium Vivax Myanmar Isolates. Malaria J. 11, 60. doi: 10.1186/1475-2875-11-60
Ju, H. L., Kang, J. M., Moon, S. U., Bahk, Y. Y., Cho, P. Y., Sohn, W. M., et al. (2013). Genetic Diversity and Natural Selection of Duffy Binding Protein of Plasmodium Vivax Korean Isolates. Acta Trop. 125 (1), 67–74. doi: 10.1016/j.actatropica.2012.09.016
Khatoon, L.. (2010). Genetic Structure of Plasmodium Vivax and Plasmodium Falciparum in the Bannu District of Pakistan. Malaria J. 9, 112. doi: 10.1186/1475-2875-9-112
Kho, W. G., Chung, J. Y., Sim, E. J., Kim, D. W., Chung, W. C. (2001). Analysis of Polymorphic Regions of Plasmodium Vivax Duffy Binding Protein of Korean Isolates. Korean J. Parasitol. 39 (2), 143–150. doi: 10.3347/kjp.2001.39.2.143
Kitchen, S. F.. (1938). The Infection of Reticulocytes by Plasmodium Vivax. Am. J. Trop. Med. s1-18 (4), 347–359. doi: 10.4269/ajtmh.1938.s1-18.347
Kosaisavee, V., Suwanarusk, R., Chua, A., Kyle, D. E., Malleret, B., Zhang, R., et al. (2017). Strict Tropism for CD71+/CD234+ Human Reticulocytes Limits the Zoonotic Potential of Plasmodium Cynomolgi. Blood 130 (11), 1357–1363. doi: 10.1182/blood-2017-02-764787
Krotoski, W. A., Collins, W. E., Bray, R. S., Garnham, P. C., Cogswell, F. B., Gwadz, R. W., et al. (1982). Demonstration of Hypnozoites in Sporozoite-Transmitted Plasmodium Vivax Infection. Am. J. Trop. Med. Hyg. 31 (6), 1291–1293. doi: 10.4269/ajtmh.1982.31.1291
Kwiatkowski, D. P.. (2005). How Malaria has Affected the Human Genome and What Human Genetics can Teach Us About Malaria. Am. J. Hum. Genet. 77 (2), 171–192. doi: 10.1086/432519
Lalremruata, A., Magris, M., Vivas-Martínez, S., Koehler, M., Esen, M., Kempaiah, P., et al. (2015). Natural Infection of Plasmodium Brasilianum in Humans: Man and Monkey Share Quartan Malaria Parasites in the Venezuelan Amazon. EBioMedicine 2 (9), 1186–1192. doi: 10.1016/j.ebiom.2015.07.033
Laurens, M. B.. (2020). RTS,S/AS01 Vaccine (Mosquirix™): An Overview. Hum. Vaccin. Immunother. 16 (3), 480–489. doi: 10.1080/21645515.2019.1669415
Lo, E., Hostetler, J. B., Yewhalaw, D., Pearson, R. D., Hamid, M., Gunalan, K., et al. (2019). Frequent Expansion of Plasmodium Vivax Duffy Binding Protein in Ethiopia and its Epidemiological Significance. PloS Negl. Trop. Dis. 13 (9), e0007222. doi: 10.1371/journal.pntd.0007222
Malleret, B., Li, A., Zhang, R., Tan, K. S., Suwanarusk, R., Claser, C., et al. (2015). Plasmodium Vivax: Restricted Tropism and Rapid Remodeling of CD71-Positive Reticulocytes. Blood 125 (8), 1314–1324. doi: 10.1182/blood-2014-08-596015
Markus, M. B. (2011). Malaria: Origin of the Term “Hypnozoite”. J. Hist. Biol. 44 (4), 781–786. doi: 10.1007/s10739-010-9239-3
Ménard, D., Barnadas, C., Bouchier, C., Henry-Halldin, C., Gray, L. R., Ratsimbasoa, A., et al. (2010). Plasmodium Vivax Clinical Malaria is Commonly Observed in Duffy-Negative Malagasy People. Proc. Natl. Acad. Sci. U.S.A. 107 (13), 5967–5971. doi: 10.1073/pnas.0912496107
Menard, D., Chan, E. R., Benedet, C., Ratsimbasoa, A., Kim, S., Chim, P., et al. (2013). Whole Genome Sequencing of Field Isolates Reveals a Common Duplication of the Duffy Binding Protein Gene in Malagasy Plasmodium Vivax Strains. PloS Negl. Trop. Dis. 7 (11), e2489. doi: 10.1371/journal.pntd.0002489
Miller, L. H., Mason, S. J., Clyde, D. F., McGinniss, M. H.. (1976). The Resistance Factor to Plasmodium Vivax in Blacks. The Duffy-Blood-Group Genotype, FyFy. N. Engl. J. Med. 295 (6), 302–304. doi: 10.1056/NEJM197608052950602
Mittal, P., Mishra, S., Kar, S., Pande, V., Sinha, A., Sharma, A., et al. (2020). Global Distribution of Single Amino Acid Polymorphisms in Plasmodium Vivax Duffy-Binding-Like Domain and Implications for Vaccine Development Efforts. Open Biol. 10 (9), 200180. doi: 10.1098/rsob.200180
Mohring, F., Hart, M. N., Rawlinson, T. A., Henrici, R., Charleston, J. A., Diez Benavente, E., et al. (2019). Rapid and Iterative Genome Editing in the Malaria Parasite Plasmodium Knowlesi Provides New Tools for P. Vivax Research. eLife 8, e45829. doi: 10.7554/eLife.45829
Moon, R. W., Hall, J., Rangkuti, F., Ho, Y. S., Almond, N., Mitchell, G. H., et al. (2013). Adaptation of the Genetically Tractable Malaria Pathogen Plasmodium Knowlesi to Continuous Culture in Human Erythrocytes. Proc. Natl. Acad. Sci. U.S.A. 110 (2), 531–536. doi: 10.1073/pnas.1216457110
Mueller, I., Galinski, M. R., Baird, J. K., Carlton, J. M., Kochar, D. K., Alonso, P. L., et al. (2009). Key Gaps in the Knowledge of Plasmodium Vivax, a Neglected Human Malaria Parasite. Lancet Infect. Dis. 9 (9), 555–566. doi: 10.1016/S1473-3099(09)70177-X
Mueller, I., Shakri, A. R., Chitnis, C.E.. (2015). Development of Vaccines for Plasmodium Vivax Malaria. Vaccine 33 (52), 7489–7495. doi: 10.1016/j.vaccine.2015.09.060
Ndegwa, D. N., Kundu, P., Hostetler, J. B., Marin-Menendez, A., Sanderson, T., Mwikali, K., et al. (2021). Using Plasmodium Knowlesi as a Model for Screening Plasmodium Vivax Blood-Stage Malaria Vaccine Targets Reveals New Candidates. PLoS Pathog. 17 (7), e1008864. doi: 10.1371/journal.ppat.1008864
Neafsey, D. E., Galinsky, K., Jiang, R. H., Young, L., Sykes, S. M., Saif, S., et al. (2012). The Malaria Parasite Plasmodium Vivax Exhibits Greater Genetic Diversity Than Plasmodium Falciparum. Nat. Genet. 44 (9), 1046–1050. doi: 10.1038/ng.2373
Nicolete, V. C., Frischmann, S., Barbosa, S., King, C. L., Ferreira, M. U.. (2016). Naturally Acquired Binding-Inhibitory Antibodies to Plasmodium Vivax Duffy Binding Protein and Clinical Immunity to Malaria in Rural Amazonians. J. Infect. Dis. 214 (10), 1539–1546. doi: 10.1093/infdis/jiw407
Ntumngia, F. B., Adams, J. H.. (2012). Design and Immunogenicity of a Novel Synthetic Antigen Based on the Ligand Domain of the Plasmodium Vivax Duffy Binding Protein. Clin. Vaccine Immunol. 19 (1), 30–36. doi: 10.1128/CVI.05466-11
Ntumngia, F. B., Schloegel, J., Barnes, S. J., McHenry, A. M., Singh, S., King, C. L., et al. (2012). Conserved and Variant Epitopes of Plasmodium Vivax Duffy Binding Protein as Targets of Inhibitory Monoclonal Antibodies. Infect. Immun. 80 (3), 1203–1208. doi: 10.1128/IAI.05924-11
Ntumngia, F. B., Schloegel, J., McHenry, A. M., Barnes, S. J., George, M. T.. (2013). Immunogenicity of Single Versus Mixed Allele Vaccines of Plasmodium Vivax Duffy Binding Protein Region II. Vaccine 31 (40), 4382–4388. doi: 10.1016/j.vaccine.2013.07.002
Ntumngia, F. B., Thomson-Luque, R., Torres, L., Gunalan, K., Carvalho, L. H., Adams, J. H.. (2016). A Novel Erythrocyte Binding Protein of Plasmodium Vivax Suggests an Alternate Invasion Pathway Into Duffy-Positive Reticulocytes. mBio 7 (4), e01261–e01216. doi: 10.1128/mBio.01261-16
Ntumngia, F. B., Thomson-Luque, R., Galusic, S., Frato, G., Frischmann, S., Peabody, D. S., et al. (2018). Identification and Immunological Characterization of the Ligand Domain of Plasmodium Vivax Reticulocyte Binding Protein 1a. J. Infect. Dis. 218 (7), 1110–1118. doi: 10.1093/infdis/jiy273
Ogun, S. A., Tewari, R., Otto, T. D., Howell, S. A., Knuepfer, E., Cunningham, D. A., et al. (2011). Targeted Disruption of Py235ebp-1: Invasion of Erythrocytes by Plasmodium Yoelii Using an Alternative Py235 Erythrocyte Binding Protein. PLoS Pathog. 7 (2), e1001288. doi: 10.1371/journal.ppat.1001288
Okenu, D. M., Meyer, E. V., Puckett, T. C., Rosas-Acosta, G., Barnwell, J. W., Galinski, M. R., et al. (2005). The Reticulocyte Binding Proteins of Plasmodium Cynomolgi: A Model System for Studies of P. Vivax. Mol. Biochem. Parasitol. 143 (1), 116–120. doi: 10.1016/j.molbiopara.2005.04.010
Parzych, E. M., Miura, K., Ramanathan, A., Long, C. A., Burns, J. M.. (2017). Evaluation of a Plasmodium-Specific Carrier Protein To Enhance Production of Recombinant Pfs25, a Leading Transmission-Blocking Vaccine Candidate. Infect. Immun. 86 (1), e00486–e00417. doi: 10.1128/IAI.00486-17
Patarroyo, M. A., Molina-Franky, J., Gómez, M., Arévalo-Pinzón, G., Patarroyo, M. E., et al. (2020). Hotspots in Plasmodium and RBC Receptor-Ligand Interactions: Key Pieces for Inhibiting Malarial Parasite Invasion. Int. J. Mol. Sci. 21 (13), 4729. doi: 10.3390/ijms21134729
Payne, R. O., Silk, S. E., Elias, S. C., Milne, K. H., Rawlinson, T. A., Llewellyn, D., et al. (2017). Human Vaccination Against Plasmodium Vivax Duffy-Binding Protein Induces Strain-Transcending Antibodies. JCI Insight 2 (12), e93683. doi: 10.1172/jci.insight.93683
Premaratne, P. H., Aravinda, B. R., Escalante, A. A., Udagama, P.V.. (2011). Genetic Diversity of Plasmodium Vivax Duffy Binding Protein II (PvDBPII) Under Unstable Transmission and Low Intensity Malaria in Sri Lanka. Infect. Genet. Evol. 11 (6), 1327–1339. doi: 10.1016/j.meegid.2011.04.023
Price, R. N.. (2014). Global Extent of Chloroquine-Resistant Plasmodium Vivax: A Systematic Review and Meta-Analysis. Lancet Infect. Dis. 14 (10), 982–991. doi: 10.1016/S1473-3099(14)70855-2
Price, R. N., Tjitra, E., Guerra, C. A., Yeung, S., White, N. J., Anstey, N. M.. (2007). Vivax Malaria: Neglected and Not Benign. Am. J. Trop. Med. Hyg. 77 (6 Suppl), 79–87.
Price, R. N., Douglas, N. M., Anstey, N. M.. (2009). New Developments in Plasmodium Vivax Malaria: Severe Disease and the Rise of Chloroquine Resistance. Curr. Opin. Infect. Dis. 22 (5), 430–435. doi: 10.1097/QCO.0b013e32832f14c1
Qian, F., Wu, Y., Muratova, O., Zhou, H., Dobrescu, G., Duggan, P., et al. (2007). Conjugating Recombinant Proteins to Pseudomonas Aeruginosa ExoProtein A: A Strategy for Enhancing Immunogenicity of Malaria Vaccine Candidates. Vaccine 25 (20), 3923–3933. doi: 10.1016/j.vaccine.2007.02.073
Radtke, A. J., Anderson, C. F., Riteau, N., Rausch, K., Scaria, P., Kelnhofer, E. R., et al. (2017). Adjuvant and Carrier Protein-Dependent T-Cell Priming Promotes a Robust Antibody Response Against the Plasmodium Falciparum Pfs25 Vaccine Candidate. Sci. Rep. 7, 40312. doi: 10.1038/srep40312
Ranjan, A., Chitnis, C. E.. (1999). Mapping Regions Containing Binding Residues Within Functional Domains of Plasmodium Vivax and Plasmodium Knowlesi Erythrocyte-Binding Proteins. Proc. Natl. Acad. Sci. U.S.A. 96 (24), 14067–14072. doi: 10.1073/pnas.96.24.14067
Rawlinson, T. A., Barber, N. M., Mohring, F., Cho, J. S., Kosaisavee, V., Gérard, S. F., et al. (2019). Structural Basis for Inhibition of Plasmodium Vivax Invasion by a Broadly Neutralizing Vaccine-Induced Human Antibody. Nat. Microbiol. 4 (9), 1497–1507. doi: 10.1038/s41564-019-0462-1
Registration T. Tables of Malaria Vaccine Projects Globally. Available at: http://www.who.int/immunization/research/development/Rainbow tables/en/.
Rieckmann, K. H., Davis, D. R., Hutton, D. C.. (1989). Plasmodium Vivax Resistance to Chloroquine? Lancet (Lond. Engl.) 2 (8673), 1183–1184. doi: 10.1016/s0140-6736(89)91792-3
Roesch, C., Popovici, J., Bin, S., Run, V., Kim, S., Ramboarina, S., et al. (2018). Genetic Diversity in Two Plasmodium Vivax Protein Ligands for Reticulocyte Invasion. PLoS Negl. Trop. Dis. 12 (10), e0006555. doi: 10.1371/journal.pntd.0006555
RTS,S Clinical Trials Partnership. (2014). Efficacy and Safety of the RTS,S/AS01 Malaria Vaccine During 18 Months After Vaccination: A Phase 3 Randomized, Controlled Trial in Children and Young Infants at 11 African Sites. PLoS Med. 11 (7), e1001685. doi: 10.1371/journal.pmed.1001685
Ryan, J. R., Stoute, J. A., Amon, J., Dunton, R. F., Mtalib, R., Koros, J., et al. (2006). Evidence for Transmission of Plasmodium Vivax Among a Duffy Antigen Negative Population in Western Kenya. Am. J. Trop. Med. Hyg. 75 (4), 575–581. doi: 10.4269/ajtmh.2006.75.575
Sanger, R., Race, R. R., Jack, J.. (1955). The Duffy Blood Groups of New York Negroes: The Phenotype Fy (a-B-). Br. J. Haematol. 1 (4), 370–374. doi: 10.1111/j.1365-2141.1955.tb05523.x
Sauerwein, R. W., Bousema, T.. (2015). Transmission Blocking Malaria Vaccines: Assays and Candidates in Clinical Development. Vaccine 33 (52), 7476–7482. doi: 10.1016/j.vaccine.2015.08.073
Schunk, M., Kumma, W. P., Miranda, I. B., Osman, M. E., Roewer, S., Alano, A., et al. (2006). High Prevalence of Drug-Resistance Mutations in Plasmodium Falciparum and Plasmodium Vivax in Southern Ethiopia. Malar. J. 5, 54. doi: 10.1186/1475-2875-5-54
Singh, S. K., Singh, A. P., Pandey, S., Yazdani, S. S., Chitnis, C. E., Sharma, A., et al. (2003). Definition of Structural Elements in Plasmodium Vivax and P. Knowlesi Duffy-Binding Domains Necessary for Erythrocyte Invasion. Biochem. J. 374 (Pt 1), 193–198. doi: 10.1042/BJ20030622
Singh, S. K., Hora, R., Belrhali, H., Chitnis, C. E., Sharma, A.. (2006). Structural Basis for Duffy Recognition by the Malaria Parasite Duffy-Binding-Like Domain. Nature 439 (7077), 741–744. doi: 10.1038/nature04443
Singh, K., Mukherjee, P., Shakri, A. R., Singh, A., Pandey, G., Bakshi, M., et al. (2018). Malaria Vaccine Candidate Based on Duffy-Binding Protein Elicits Strain Transcending Functional Antibodies in a Phase I Trial. NPJ Vaccines 3, 48. doi: 10.1038/s41541-018-0083-3
Sousa, T. N., Cerávolo, I. P., Fernandes Fontes, C. J., Couto, A., Carvalho, L. H., Brito, C. F., et al. (2006). The Pattern of Major Polymorphisms in the Duffy Binding Protein Ligand Domain Among Plasmodium Vivax Isolates From the Brazilian Amazon Area. Mol. Biochem. Parasitol. 146 (2), 251–254. doi: 10.1016/j.molbiopara.2005.11.006
Su, X. Z.. (2019). Plasmodium Genomics and Genetics: New Insights Into Malaria Pathogenesis, Drug Resistance, Epidemiology, and Evolution. Clin. Microbiol. Rev. 32 (4), e00019–e00019. doi: 10.1128/CMR.00019-19
Sutherland, C. J., et al. (2010). Two Nonrecombining Sympatric Forms of the Human Malaria Parasite Plasmodium Ovale Occur Globally. J. Infect. Dis. United States 201 (10), 1544–1550. doi: 10.1086/652240
Tachibana, M., Suwanabun, N., Kaneko, O., Iriko, H., Otsuki, H., Sattabongkot, J., et al. (2015). Plasmodium Vivax Gametocyte Proteins, Pvs48/45 and Pvs47, Induce Transmission-Reducing Antibodies by DNA Immunization. Vaccine 33 (16), 1901–1908. doi: 10.1016/j.vaccine.2015.03.008
Ta, T. H., Hisam, S., Lanza, M., Jiram, A. I., Ismail, N., Rubio, J. M., et al. (2014). First Case of a Naturally Acquired Human Infection With Plasmodium Cynomolgi. Malaria J. 13 (1), 1–7. doi: 10.1186/1475-2875-13-68
Tanner, M., Greenwood, B., Whitty, C. J., Ansah, E. K., Price, R. N., Dondorp, A. M., et al. (2015). Malaria Eradication and Elimination: Views on How to Translate a Vision Into Reality. BMC Med. 13, 167. doi: 10.1186/s12916-015-0384-6
Tsuboi, T., Kappe, S. H., al-Yaman, F., Prickett, M. D., Alpers, M., Adams, J. H., et al. (1994). Natural Variation Within the Principal Adhesion Domain of the Plasmodium Vivax Duffy Binding Protein. Infect. Immun. 62 (12), 5581–5586. doi: 10.1128/iai.62.12.5581-5586.1994
Tsuboi, T., Kaslow, D. C., Gozar, M. M., Tachibana, M., Cao, Y. M., Torii, M., et al. (1998). Sequence Polymorphism in Two Novel Plasmodium Vivax Ookinete Surface Proteins, Pvs25 and Pvs28, That are Malaria Transmission-Blocking Vaccine Candidates. Mol. Med. (Cambridge Mass.) 4 (12), 772–782. doi: 10.1007/BF03401770
Urusova, D., Carias, L., Huang, Y., Nicolete, V. C., Popovici, J., Roesch, C., et al. (2019). Structural Basis for Neutralization of Plasmodium Vivax by Naturally Acquired Human Antibodies That Target DBP. Nat. Microbiol. 4 (9), 1486–1496. doi: 10.1038/s41564-019-0461-2
Valencia, S. H., Rodríguez, D. C., Acero, D. L., Ocampo, V., Arévalo-Herrera, M.. (2011). Platform for Plasmodium Vivax Vaccine Discovery and Development. Mem. Inst. Oswaldo Cruz 106 Suppl 1 (Suppl 1), 179–192. doi: 10.1590/s0074-02762011000900023
VanBuskirk, K. M., Cole-Tobian, J. L., Baisor, M., Sevova, E. S., Bockarie, M., King, C. L., et al. (2004a). Antigenic Drift in the Ligand Domain of Plasmodium Vivax Duffy Binding Protein Confers Resistance to Inhibitory Antibodies. J. Infect. Dis. 190 (9), 1556–1562. doi: 10.1086/424852
VanBuskirk, K. M., Sevova, E., Adams, J. H. (2004b). Conserved Residues in the Plasmodium Vivax Duffy-Binding Protein Ligand Domain are Critical for Erythrocyte Receptor Recognition. Proc. Natl. Acad. Sci. U.S.A. 101 (44), 15754–15759. doi: 10.1073/pnas.0405421101
Vicentin, E. C., Françoso, K. S., Rocha, M. V., Iourtov, D., Dos Santos, F. L., Kubrusly, F. S., et al. (2014). Invasion-Inhibitory Antibodies Elicited by Immunization With Plasmodium Vivax Apical Membrane Antigen-1 Expressed in Pichia Pastoris Yeast. Infect. Immun. 82 (3), 1296–1307. doi: 10.1128/IAI.01169-13
White, N. J.. (2021). Anti-Malarial Drug Effects on Parasite Dynamics in Vivax Malaria. Malaria J. 20 (1), 161. doi: 10.1186/s12936-021-03700-7
Winter, D. J., Pacheco, M. A., Vallejo, A. F., Schwartz, R. S., Arevalo-Herrera, M., Herrera, S., et al. (2015). Whole Genome Sequencing of Field Isolates Reveals Extensive Genetic Diversity in Plasmodium Vivax From Colombia. PLoS Negl. Trop. Dis. 9 (12), e0004252. doi: 10.1371/journal.pntd.0004252
Woldearegai, T. G., Kremsner, P. G., Kun, J. F., Mordmüller, B.. (2013). Plasmodium Vivax Malaria in Duffy-Negative Individuals From Ethiopia. Trans. R. Soc. Trop. Med. Hyg. 107 (5), 328–331. doi: 10.1093/trstmh/trt016
World Health Organisation. (2021). World Malaria Report 2021 (Geneva, Switzerland: World Health Organization).
World Health Organization. (2020). World Malaria Report 2020 (Geneva, Switzerland: World Health Organization).
Wu, Y., Ellis, R. D., Shaffer, D., Fontes, E., Malkin, E. M., Mahanty, S., et al. (2008). Phase 1 Trial of Malaria Transmission Blocking Vaccine Candidates Pfs25 and Pvs25 Formulated With Montanide ISA 51. PLoS One 3 (7), e2636. doi: 10.1371/journal.pone.0002636
Xainli, J., Adams, J. H., King, C. L.. (2000). The Erythrocyte Binding Motif of Plasmodium Vivax Duffy Binding Protein is Highly Polymorphic and Functionally Conserved in Isolates From Papua New Guinea. Mol. Biochem. Parasitol. 111 (2), 253–260. doi: 10.1016/s0166-6851(00)00315-7
Keywords: Malaria, Plasmodium vivax, vaccine, blood stage malaria antigen, PvDBP
Citation: Kar S and Sinha A (2022) Plasmodium vivax Duffy Binding Protein-Based Vaccine: a Distant Dream. Front. Cell. Infect. Microbiol. 12:916702. doi: 10.3389/fcimb.2022.916702
Received: 09 April 2022; Accepted: 21 June 2022;
Published: 13 July 2022.
Edited by:
Tania F. De Koning-Ward, Deakin University, AustraliaReviewed by:
Mohammad Zeeshan, University of London, United KingdomDalma Maria Banic, Oswaldo Cruz Foundation (Fiocruz), Brazil
Copyright © 2022 Kar and Sinha. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Abhinav Sinha, YWJoaW5hdnNpbmhhQGljbXIuZ292Lmlu
 Sonalika Kar
Sonalika Kar Abhinav Sinha
Abhinav Sinha