
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Cell. Infect. Microbiol. , 30 June 2022
Sec. Molecular Bacterial Pathogenesis
Volume 12 - 2022 | https://doi.org/10.3389/fcimb.2022.907489
This article is part of the Research Topic Insights into Bee Diseases and Bee Health View all 6 articles
 Giovanni Cilia†
Giovanni Cilia† Simone Flaminio†
Simone Flaminio† Laura Zavatta
Laura Zavatta Rosa Ranalli*
Rosa Ranalli* Marino Quaranta
Marino Quaranta Laura Bortolotti‡
Laura Bortolotti‡ Antonio Nanetti‡
Antonio Nanetti‡Diseases contribute to the decline of pollinator populations, which may be aggravated by the interspecific transmission of honey bee pests and pathogens. Flowers increase the risk of transmission, as they expose the pollinators to infections during the foraging activity. In this study, both the prevalence and abundance of 21 honey bee pathogens (11 viruses, 4 bacteria, 3 fungi, and 3 trypanosomatids) were assessed in the flower-visiting entomofauna sampled from March to September 2021 in seven sites in the two North-Italian regions, Emilia-Romagna and Piedmont. A total of 1,028 specimens were collected, identified, and analysed. Of the twenty-one pathogens that were searched for, only thirteen were detected. Altogether, the prevalence of the positive individuals reached 63.9%, with Nosema ceranae, deformed wing virus (DWV), and chronic bee paralysis virus (CBPV) as the most prevalent pathogens. In general, the pathogen abundance averaged 5.15 * 106 copies, with CBPV, N. ceranae, and black queen cell virus (BQCV) as the most abundant pathogens, with 8.63, 1.58, and 0.48 * 107 copies, respectively. All the detected viruses were found to be replicative. The sequence analysis indicated that the same genetic variant was circulating in a specific site or region, suggesting that interspecific transmission events among honey bees and wild pollinators are possible. Frequently, N. ceranae and DWV were found to co-infect the same individual. The circulation of honey bee pathogens in wild pollinators was never investigated before in Italy. Our study resulted in the unprecedented detection of 72 wild pollinator species as potential hosts of honey bee pathogens. Those results encourage the implementation of monitoring actions aiming to improve our understanding of the environmental implications of such interspecific transmission events, which is pivotal to embracing a One Health approach to pollinators’ welfare.
Pollination is a pivotal ecosystem service to both natural and agricultural environments. Its global economic value is estimated to be on the order of hundreds of billions of dollars per year (Porto et al., 2020). That adds to an invaluable intrinsic contribution to biodiversity (Senapathi et al., 2015). Thus, the decline of pollinator populations is receiving increased attention, with a focus on the role played by pesticides (O’Neal et al., 2018; Siviter et al., 2018; Hrynko et al., 2021), habitat fragmentation (Hung et al., 2021; Librán-Embid et al., 2021), climatic change (Schenk et al., 2018; Duchenne et al., 2020; Cane, 2021), and urbanisation (Fortel, 2014; Choate et al., 2018; Hofmann and Renner, 2020; Wenzel et al., 2020). While the occurrence of interspecific food competition events between wild species and managed honey bee (Apis mellifera) colonies are still debated (Tscharntke and Steffan-Dewenter, 2000; Wojcik et al., 2018; Rasmussen et al., 2021), little is known about the interspecific transmission of pathogens between honey bees and wild pollinators (Nanetti et al., 2021a). This last point is crucial, as the welfare of A. mellifera colonies depends on apicultural management, the associated economic value of apiculture (Ballantyne et al., 2017; Khalifa et al., 2021), and the equilibrium of the ecosystem in which honey bees live.
Interspecific pathogen transmission may occur with arthropods sharing the same environment as the honey bees. The main routes between established and new hosts include direct contact, orofecal exchanges, and the ingestion of pollen contaminated with pathogens (Singh et al., 2010; Cilia et al., 2021; Tehel et al., 2022). The infection may also occur during foraging via contact with pathogen-contaminated pollen, nectar, and floral organs (Chen et al., 2006a; Mazzei et al., 2014; Graystock et al., 2015; Alger et al., 2019b; Schittny et al., 2020). Wasps and hornets predating infected bees (Yañez et al., 2012; Forzan et al., 2017; Mazzei et al., 2018; Mazzei et al., 2019) and ants cannibalising their corpses (Sébastien et al., 2015; Cooling et al., 2017; Gruber et al., 2017) are likely to get infected. Interspecific transmission may affect also organisms that are not expected to come into direct contact with the honey bees, like spiders and beetles (Yue et al., 2007; Erler et al., 2012; Levitt et al., 2013).
Several studies have aimed at elucidating pathogen dynamics beyond interspecific transmission. Bees are considered the most efficient pollinators (Ballantyne et al., 2017). As many pollinators exploit the same floral resources as honey bees, horizontal transmission of pathogens, especially honey bee pathogens (Dalmon et al., 2021), becomes possible with other Hymenoptera species (Santamaria et al., 2018; Purkiss and Lach, 2019; Gusachenko et al., 2020; Ocepek et al., 2021), other pollinators (Bailes et al., 2018), and other arthropods (Nanetti et al., 2021a). Considering the importance of wild pollinators and the adaptive plasticity of pathogens transmitted via regular flower visits (Burnham et al., 2021; Cohen et al., 2022; Manley et al., 2019), our understanding of both ecosystem health and its impact on pollinator decline requires increased research on the interspecific interactions occurring in these ecosystems. The increasing number of studies reporting honey bee pathogens in other host species portrays a scenario consisting of one reservoir species and multiple spillover events.
Population studies might elucidate those aspects (Benjamin-Chung et al., 2018) that, in the case of wild bees, are complicated by those species’ peculiar biological and ecological characteristics (Koh et al., 2016; Drossart and Gérard, 2020; Prendergast et al., 2020). This makes spillover routes generally unknown and undetermined (Yañez et al., 2020).
This study was conducted within a nationwide Italian monitoring project on wild bees (BeeNet project). We aimed to investigate both the occurrence and circulation of the main honey bee pathogens in the wild pollinators of two North-Italian regions, contributing to assessing the risk of possible interspecific transmission and spillover.
Seven sites were chosen (Figure 1) for this study on the occurrence of honey bee pathogens in northern Italy. The geographical and environmental characteristics of each site are reported in Table 1. This study is part of a wider project to monitor wild bees, which aims to compare the communities of these pollinators in different agroecosystems; therefore, the locations were chosen a priori by the project. Two Land Cover Categories were identified based on the management of the agricultural areas: intensive (category 2.1.1—Agricultural areas, non-irrigated arable land) and semi-natural land (category 2.4.3—Heterogeneous agricultural areas, land principally occupied by agriculture, with significant areas of natural vegetation). Finally, a site in the urban area corresponding to the CREA-AA research centre was added and included in category 1.2.1.3 (Industrial, commercial, and transport units).
Sampling sites were located in agricultural agroecosystems with different management practices; both intensively farmed areas and cultivated ones fragmented by natural elements were investigated. All sampling was carried out in field margins, or other landscape elements such as hedges and meadows, but always contiguous to cultivated areas.
Sampling was carried out once a month, from March to September 2021. At each site, an area with a high abundance of entomophilous plant species in anthesis was chosen to ensure that floral resources were sufficient and in high enough abundance to support a pollinator community with high species diversity. The sampling was conducted on sunny and non-windy days, with an average temperature above 15°C.
Sampling focused on wild bees, but honey bees, hoverflies, flies, wasps, and beetles were also collected when caught in the same sweep net action because they were on the same flowers as the targeted bees.
Pollinators were collected by one collector using the sweep net technique during one effective hour of sampling, stopping the timer at each catch. Each specimen was placed, depending on its size, in a sterile single 2-ml microtube or 15-ml tube. The tubes containing captured individuals were then placed in a cooler bag with freezer packs until arrival at the laboratory, where all the specimens were identified.
On each sampling day, captured flower visitors were placed at −80°C for 30 min, after which they were identified to species level whenever possible, otherwise at the genus level. Identification was performed under a stereomicroscope, placing the individual in a Styrofoam container with dry ice to not degrade the RNA. After identification, samples were stored at −80°C until the analysis.
Before extraction, all samples were washed with 95% ethanol to remove external microbial contaminations, and each sample was analysed individually. The sample was placed in a 2-ml microtube with 500 µl of DNA/RNA Shield (Zymo Research, Irvine, CA, USA) and crushed with a TissueLyser II (Qiagen, Hilden, Germany) for 3 min at 30 Hz, as previously reported (Cilia et al., 2019; Nanetti et al., 2021b). The obtained suspensions were split into two aliquots from which DNA and RNA were separately extracted.
The above-described procedures were accomplished by using a Quick DNA Microprep Plus Kit (Zymo Research) and Quick RNA Microprep Plus Kit (Zymo Research), respectively, following the modified manufacturer’s instructions for solid tissue processing (Mazzei et al., 2019; Nanetti et al., 2021c).
The obtained nucleic acids were eluted in 50 µl of DNAase-Rnase-free water, and the extracts were stored at −80°C until the qPCR assays.
The extracted DNA was analysed using Real-Time PCR to quantify the abundance of detected bacteria and trypanosomatids in the samples, using the primers reported in Table 2. For each target, a total reaction volume of 15 µl was prepared as previously described (Cilia et al., 2020) using PowerUp™ SYBR™ Green Master Mix (Thermo Fisher, Waltham, MA, USA) with forward and reverse primers (2 µM) and 3 µl of DNA extract. The Real-Time PCR assay was performed on a QuantStudio™ 3 Real-Time PCR System (Thermo Fisher Scientific), following the protocols for all gene sequences (Dobbelaere et al., 2001; James and Skinner, 2005; Martin-Hernandez et al., 2007; Roetschi et al., 2008; Meeus et al., 2012; Huang et al., 2015; Arismendi et al., 2016; Cilia et al., 2018; Xu et al., 2018). DNA previously extracted from positive honey bee samples was used as positive controls. Sterile water was used as a negative control in all analytical steps. All the analyses were conducted in duplicate.
For each target, a standard curve was generated by amplifying serially diluted recombinant plasmids containing the pathogen-specific DNA fragment from 1 * 101 to 1 * 109 copies in a qPCR assay on a QuantStudio™ 3 Real-Time PCR System (Thermo Fisher Scientific), as previously reported (Cilia et al., 2020; Nanetti et al., 2021c), following the amplification and quantification protocols (Dobbelaere et al., 2001; Martin-Hernandez et al., 2007; Roetschi et al., 2008; Arismendi et al., 2016; Cilia et al., 2018; Xu et al., 2018).
To quantify the virus abundance in the samples, all RNA extracts were analysed through Real-Time PCR using Power SYBR™ Green Cells-to-CT™ Kit (Thermo Fisher Scientific), as previously reported (Cilia et al., 2021). The primers used to amplify the target honey bee viruses considered here are reported in Table 3. The Real-Time PCR assay was performed on a QuantStudio™ 3 Real-Time PCR System (Thermo Fisher Scientific), following the protocols for each gene sequence. RNA previously extracted from positive honey bees was used as the positive control for each investigated virus. All the analyses were conducted in duplicates.
For each target, a standard curve was generated by amplifying the serially diluted recombinant plasmids containing the pathogen-specific RNA fragment from 1 * 101 to 1 * 109 copies in a qPCR assay on a QuantStudio™ 3 Real-Time PCR System (Thermo Fisher Scientific), as previously reported (Mazzei et al., 2019; Cilia et al., 2021), following the amplification and quantification protocols (Chantawannakul et al., 2006; de Miranda et al., 2010; Kajobe et al., 2010; Martin et al., 2012; Hartmann et al., 2015; Garigliany et al., 2017; Mazzei et al., 2018).
The active replication of viruses was evaluated by performing strand-specific RT-PCRs using specific primers, as previously described (Mazzei et al., 2018; Nanetti et al., 2021b). Positive and negative strands previously obtained from positive honey bees were used as positive controls. The obtained cDNAs were amplified by PCR for the viral targets, and the amplicons were visualised on a 2% agarose gel. Subsequently, the amplicons were sequenced (BMR Genomics, Padua, Italy) and analysed using BLAST (Altschul et al., 1990).
A phylogenetical analysis was performed on each viral sequence deposited in GenBank using the maximum likelihood method and Tamura–Nei model (Tamura et al., 2004) (Saitou and Nei, 1987) associating taxa clustered together in the bootstrap test (500 replicates) (Felsenstein, 1985). Evolutionary analyses were conducted in MEGA X (Kumar et al., 2018).
Pathogen prevalence among months, sites, and pollinator taxon were analysed by a chi-square independence test. Since contingency tables contained values <5 and were wider than 2 × 2, Fisher’s exact test could not be applied. A simulated p-value was calculated based on 2,000 replicates.
The pathogen abundance was determined at the individual level by averaging the two technical replicates of each PCR assay. The results are reported in terms of average ± SD. The abundance of 13 pathogens in wild bees and other pollinators was also compared among months, sites, and taxa. Data were tested first for normality using the Shapiro–Wilk test. Since the normality check failed, data were analysed by a Kruskal–Wallis test, followed by a pairwise Wilcoxon test with Bonferroni correction as a post-hoc test. The significance threshold was set at p = 0.05. All the analyses were performed with R version 4.1.2 (R Core Team, 2021).
A total of 1,028 flower-visiting insects were captured and analysed altogether in the two considered Italian regions. The samples included the following: Apoidea (N = 835), non-Apoidea Hymenoptera (N = 68), Diptera (N = 107), and pollinators belonging to other taxa (N = 18) (Table S1). The bee specimens were recognised as follows: Halictus spp. (N = 166), Lasioglossum spp. (N = 142), Bombus spp. (N = 134), Andrena spp. (N = 121), Megachile spp. (N = 43), Anthidium spp. (N = 42), Eucera spp. (N = 34), Osmia spp. (N = 27), Chelostoma spp. (N = 20), Ceratina spp. (N = 19), Hylaeus spp. (N = 14), Nomiapis diversipes (N = 13), Heriades spp. (N = 12), A. mellifera (N = 12), Lithurgus cornutus (N = 6), Xylocopa spp. (N = 4), Anthophora spp. (N = 3), Colletes spp. (N = 3), Melitturga clavicornis (N = 3), Stelis breviscula (N = 3), Systropha curvicornis (N = 3), Hoplitis spp. (N = 2), Habropoda tarsata (N = 2), Icteranthidium laterale (N = 2), Epeolus spp. (N = 2), Pseudoanthidium scapulare (N = 1), Sphecodes alternatus (N = 1), and Dasypoda hirtipes (N = 1) (Table S1). The collected wasps and hornets were found to belong to the following: Cerceris spp. (N = 14), Polistes spp. (N = 12), and Vespula spp. (N = 4), plus a number of other specimens belonging to other genera (N = 28). The Diptera were classifies as Syrphus spp. (N = 16), Epistrphus spp. (N = 18), Melanostoma spp. (N = 13), Sphaerophoria spp. (N = 11), Eristalis spp. (N = 9), Bombylius spp. (N = 8), Chloromyia spp. (N = 7), Villa spp. (N = 4), and Volucella spp. (N = 3), plus a number other specimens belonging to other genera (N = 18) (Table S1).
All the samples were negative for Israeli acute bee paralysis virus (IAPV), Apis iridescent virus (AIV), slow paralysis virus (SBPV), Moku virus, Nosema apis, Crithidia mellificae, Paenibacillus larvae, and Melissococcus plutonius. Except for the viruses mentioned above, all the other viruses were found to be present in the analysed samples in their replicative form (i.e., negative strand) (Figure 2).
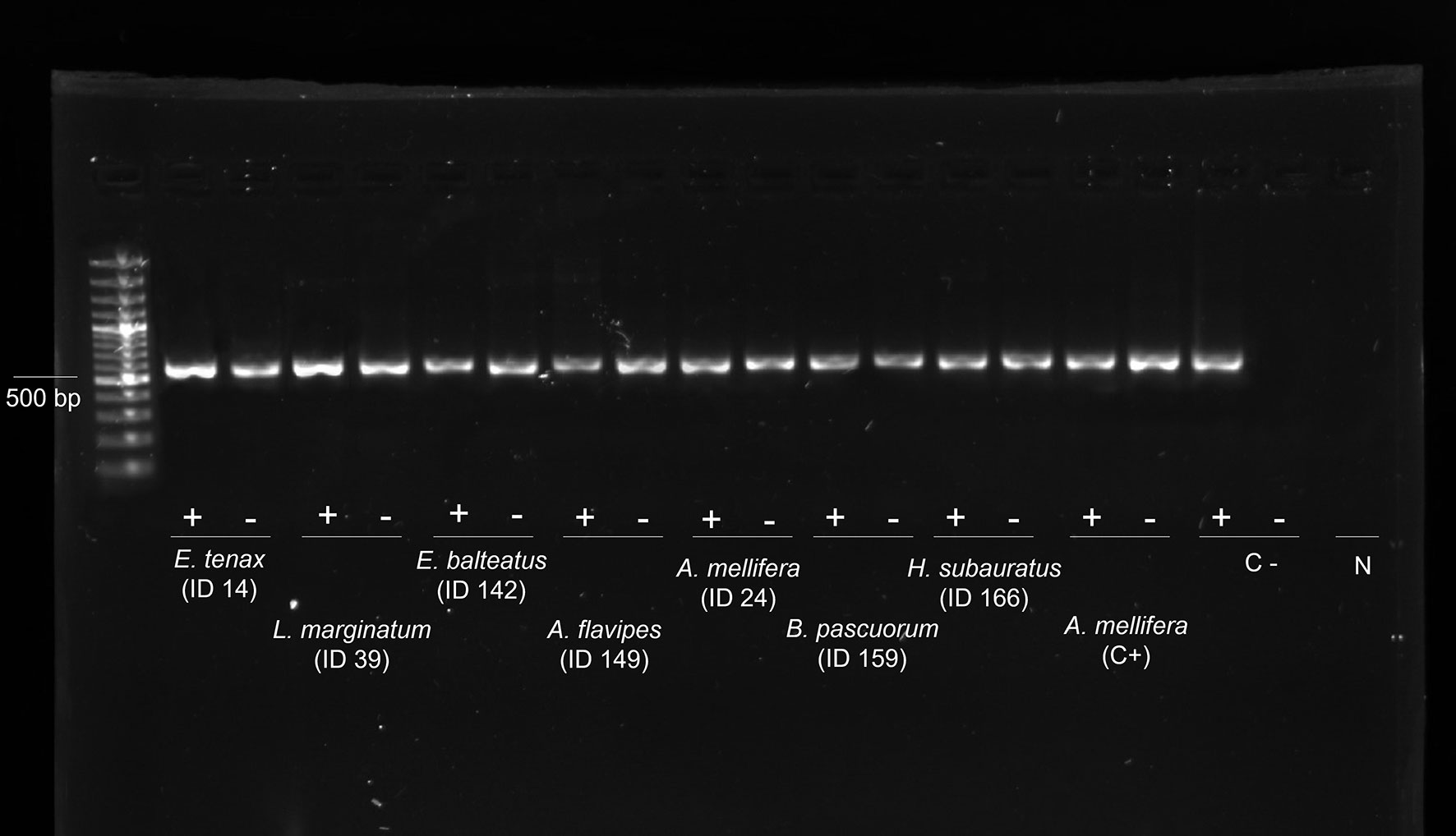
Figure 2 Evidence of genomic and replicative DWV strands. Gel electrophoresis of strand-specific RT-PCR of the cDNA from 8 individuals: genomic strand (+) and replicative strand (−). Positive control (C+): cDNA from replicative DWV of Apis mellifera workers. Negative control (C−): non-replicative DWV of A. mellifera workers. N: DNA- and Rnase-free water. DWV, deformed wing virus.
A total of 712 of the sampled individuals were positive for at least one pathogen (69.3%). Altogether, Nosema ceranae, deformed wing virus (DWV), and chronic bee paralysis virus (CBPV) were the three most prevalent pathogens (16.1%, 15.7%, and 9.5%, respectively). Lower prevalence was observed for black queen cell virus (BQCV), acute bee paralysis virus (ABPV), sac brood virus (SBV), Crithidia bombi, Spiroplasma melliferum, Spiroplasma apis, AmFV, Kashmir bee virus (KBV), Lotmaria passim, and Ascosphaera apis (6.7%, 5.5%, 5.4%, 3.0%, 2.7%, 2.1%, 1.5%, 0.7%, 0.2%, and 0.2%, respectively) (Table 4).
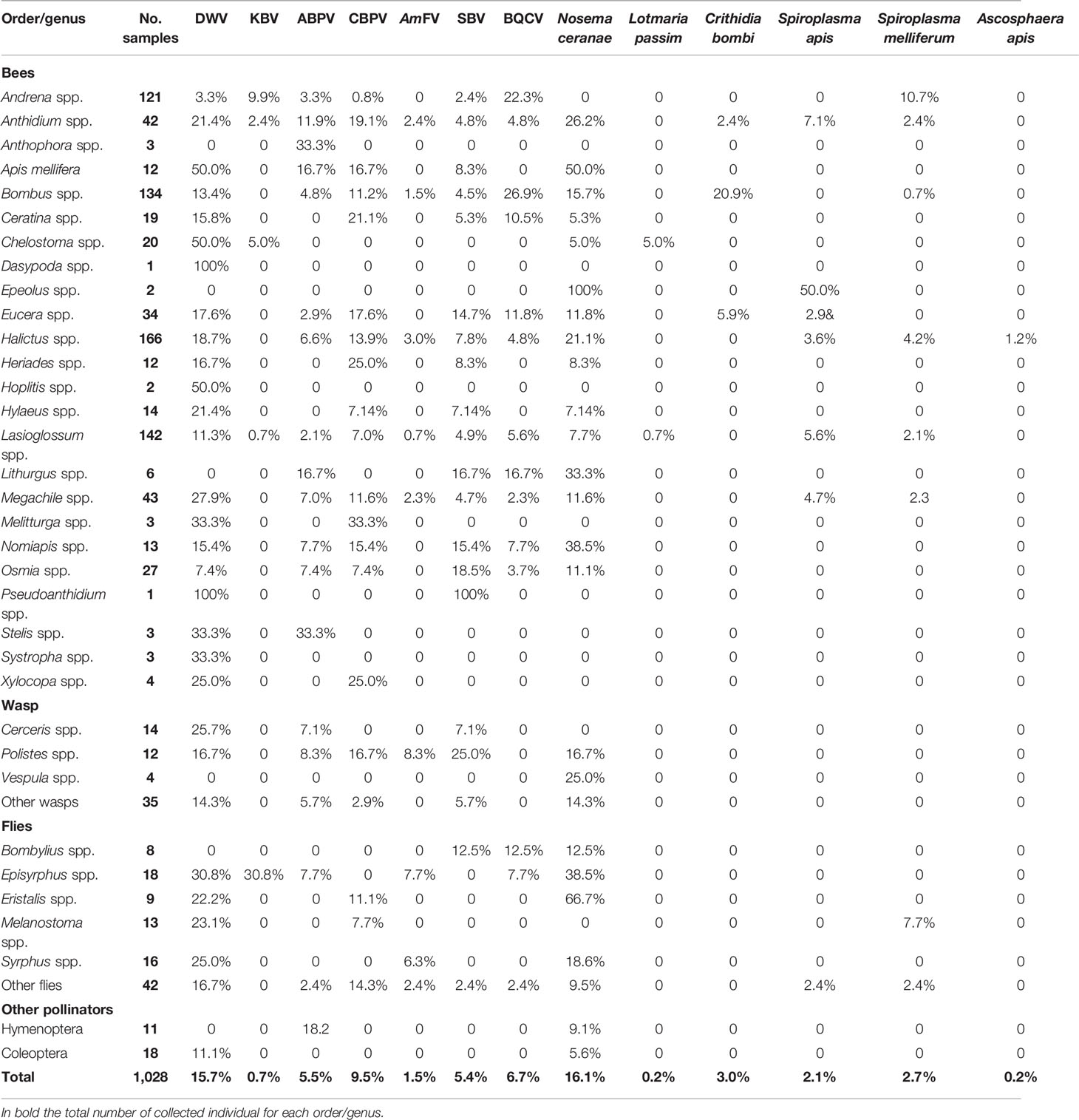
Table 4 Prevalence of the positive samples per order or genus (ranked in alphabetical order) for the investigated pathogens (complete data are provided in Table S1).
Considering all the sampling sites, Chelostoma spp. was found to be frequently infected by DWV (50.0%) and L. passim (5.0%), Episyrphus spp. by KBV (30.8%) and AmFV (7.7%), Anthidium spp. by ABPV (11.9%), Heriades spp. and Ceratina spp. by CBPV (25.0% and 21.1%, respectively), Polistes spp. by AmFV and SBV (8.3% and 25.0%), Osmia spp. by SBV (18.5%), Bombus spp. by BQCV (29.9%) and C. bombi (20.9%), Andrena spp. by BQCV (22.3%) and S. melliferum (10.7%), Eristalis spp. by N. ceranae (66.7%), and Melanostoma spp. by S. melliferum (7.7%) (Table S2).
The pathogen prevalence statistically differed with sampling site (χ2 = 246.35, p < 0.001), genus (χ2 = 615.48, p = 0.026), and month (χ2 = 237.81, p < 0.001).
In general, CBPV was the most abundant pathogen (8.63 * 107 ± 2.72 * 109), followed by N. ceranae (1.58 * 107 ± 3.56 * 108), BQCV (4.84 * 106 ± 1.52 * 108), and DWV (8.40 * 105 ± 1.54 * 107) (Table S3). The same abundance trends were recorded in each region and site (Table S3).
The highest abundance was found in Halictus scabiosae for DWV (10 * 108), in Anthidium loti and Episyrphus balteatus for KBV (10 * 105); in Halictus fulvipes, Andrena hattorfiana, and Osmia bicornis for ABPV (10 * 106); in Eucera eucnemidea for CBPV and SBV (10 * 1010 and 10 * 107, respectively); in Bombus sylvarum for BQCV (10 * 109); in Anthidium florentinum and Lasioglossum villosulum for AmFV (10 * 105); in H. fulvipes and Bombus terrestris for N. ceranae (10 * 109); in Bombus pascuorum and B. terrestris for C. bombi (10 * 107); in A. florentinum and E. eucnemidea for S. apis (10 * 105); and in Halictus simplex and Andrena distinguenda for S. melliferum (10 * 106) (Table S1). Two individuals (Chelostoma rapunculi and Lasioglossum malachurum) sampled in July in ERMO were positive for L. passim (10 * 103) (Table S1), while two individuals sampled in July, H. simplex and Halictus cochlearitarsis, respectively, from PIAI and ERES were found positive for A. apis (10 * 105) (Table S1).
For all pathogens except CBPV, L. passim, and S. apis, the host taxon, sampling site, and month were significant predictors of abundance (Table 5). All three factors mentioned above were significantly correlated with the abundance of DWV, BQCV, N. ceranae, and C. bombi.
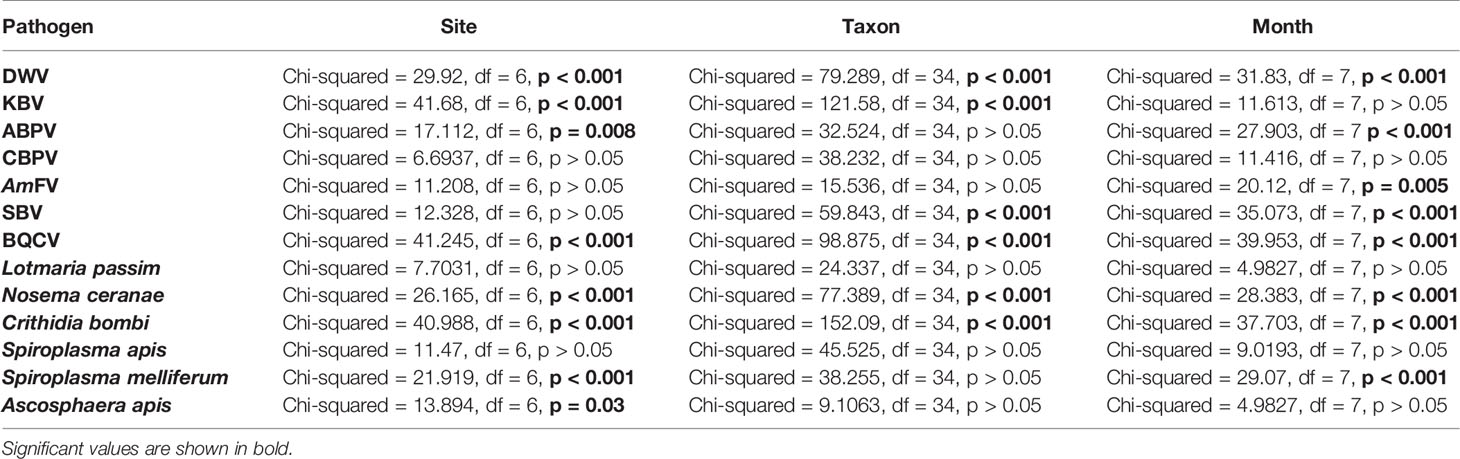
Table 5 Significativity of the sampling site, taxon, and month of the collection as predictors of pathogen abundance (Kruskal–Wallis analysis).
Virus abundance is shown in Figure S1. The results of a post-hoc analysis are reported in Table S4. Briefly, the abundance of DWV was significantly higher in A. mellifera, Megachile spp. Chelostoma spp., Dasypoda spp., and Pseudoanthidium spp. compared to Andrena spp. (p < 0.005). PIES and PIAI reported the highest DWV abundance compared to ERESP (p < 0.005). Episirphus spp. showed higher KBV abundance compared to Halictus spp., Bombus spp., and Andrena spp. (p < 0.000), whereas KBV abundance was higher in PIAI compared to ERES, CREA, and PIES (p < 0.005). ABPV abundance was higher in ERAI related to ERESP (p < 0.001). SBV abundance was significantly lower in Andrena spp. compared to Pseudoanthidium spp., Polistes spp., Osmia spp., Lithurgus spp., and Nomiapis spp. (p < 0.01). BQCV was significantly higher in Bombus spp. than in Andrena spp., Halictus spp., and Lasioglossum spp. (p < 0.005), whereas in ERESP, a lower BQCV abundance was detected compared to ERAI and ERMO (p < 0.05). Host genus, sampling site, and month were not significant predictors of CBPV and AmFV abundance (p > 0.05).
The abundance of DNA pathogens is shown in Figure S2. The results of a post-hoc analysis are reported in Table S4. Briefly, a higher N. ceranae abundance was recorded in Eristalis spp. compared to Lasioglossum spp. and A. mellifera (p < 0.005), while in PIES, its abundance was higher than that in ERESP and ERMO (p < 0.05). The abundance of C. bombi was higher in Bombus spp. than Halictus spp., Lasioglossum spp., and Andrena spp. (p < 0.001), with significantly higher values in ERMO compared to ERES and PIES (p < 0.005). S. melliferum was significantly less abundant in ERMO than in ERESP (p < 0.005). Host genus, sampling site, and month were not significant predictors of L. passim and S. apis abundance (p > 0.05).
In March, a high prevalence of pathogens was detected, mainly due to the frequent occurrence of both N. ceranae and DWV. After a steep decrease, both N. ceranae and DWV resumed increasing, reaching a peak in September. However, the overall pathogen abundance peaked in July, after a steady increase during the previous seasons (Figure 3).
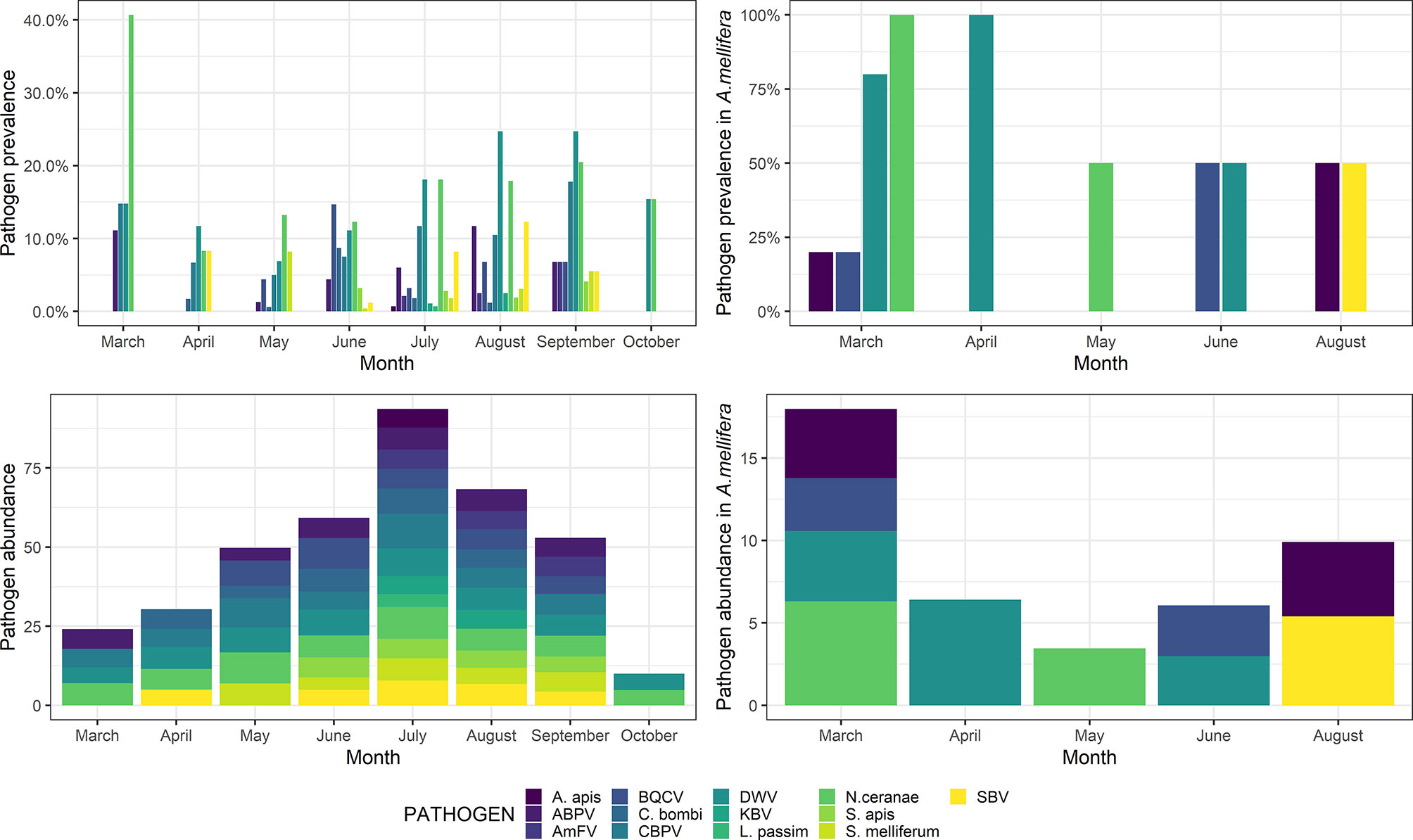
Figure 3 Pathogen prevalence (top) and abundance (bottom) throughout the sampling season (March–October) in wild species (left) and Apis mellifera (right). Abundance is shown as a decimal logarithm.
Detailed post-hoc comparisons are reported in Table S4. Statistical differences in pathogen abundance are shown in Figures S1, S2. Briefly, in March, a significantly higher abundance of ABPV and N. ceranae was detected, whereas the abundance of S. melliferum was significantly higher in May. Also, the abundance of BQCV and C. bombi was significantly higher in June, the abundance of DWV and SBV was significantly higher in August, and the abundance of AmFV was significantly higher in September. No significant differences were observed for KBV, CBPV, L. passim, S. apis, and A. apis (p > 0.05).
The viral sequences were studied to elucidate possible structural spatial and/or temporal similarities. All the sequenced viruses isolated from the different pollinator species belonged to the same strains. Comparing the positive individuals for each viral sequence resulted in perfect homology. Therefore, the subsequent analyses were conducted at the level of the host species rather than individually. All the viral sequences were similar to the European virus sequences available in GenBank.
All the sequences belonging to ABPV (n = 33) (Figure 4) and AmFV (n = 14) (Figure 5) that were recognised in this study clustered in a large single clade. The sequences did not differ among the various host species and sampling sites.
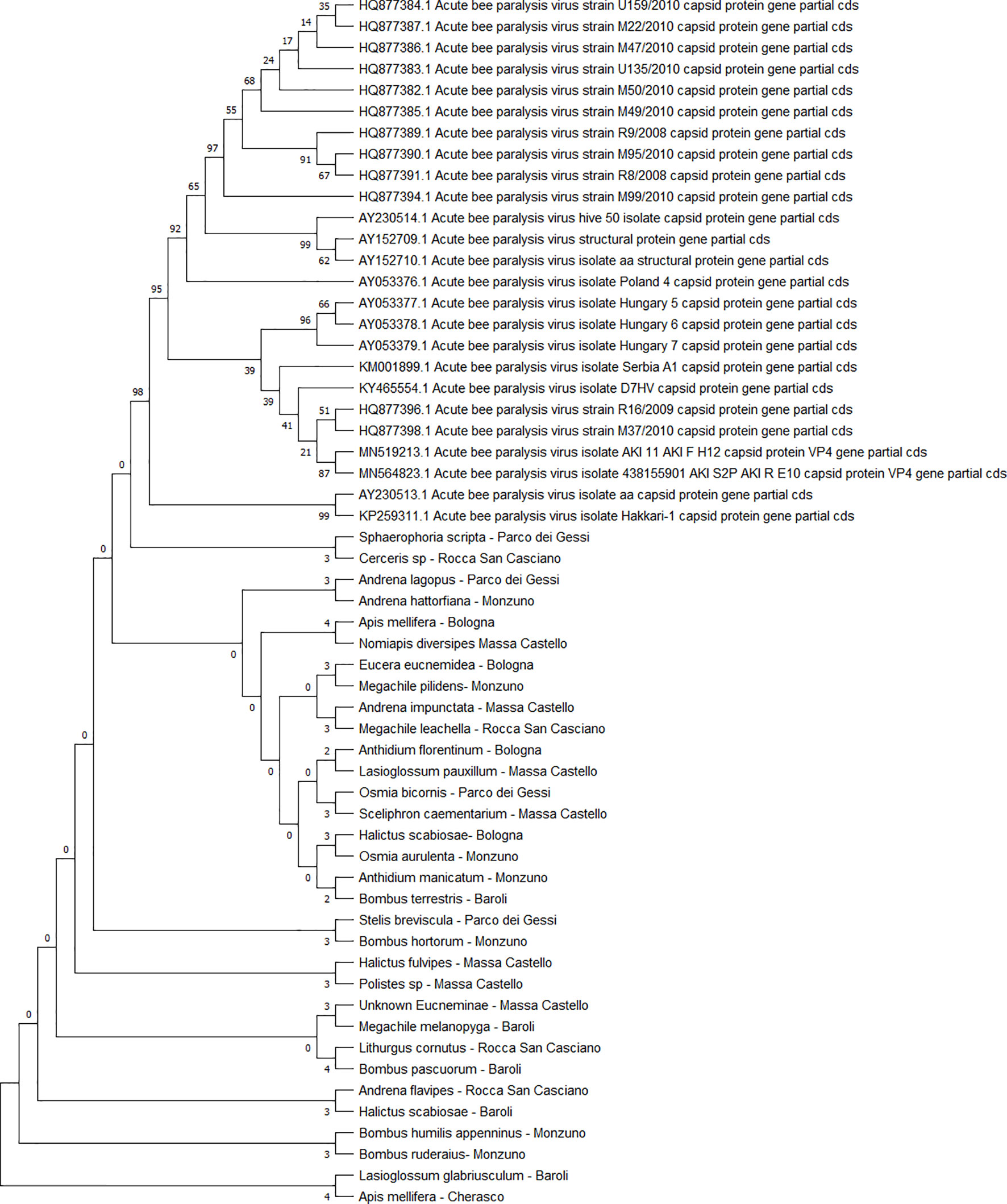
Figure 4 Maximum likelihood nucleotide phylogeny of ABPV capsid protein gene. This analysis involved 57 nucleotide sequences. There were a total of 879 positions in the final dataset. Only those with bootstrap > 50% are reported. Species and sampling sites are reported for the sequences identified in this study. ABPV, acute bee paralysis virus.
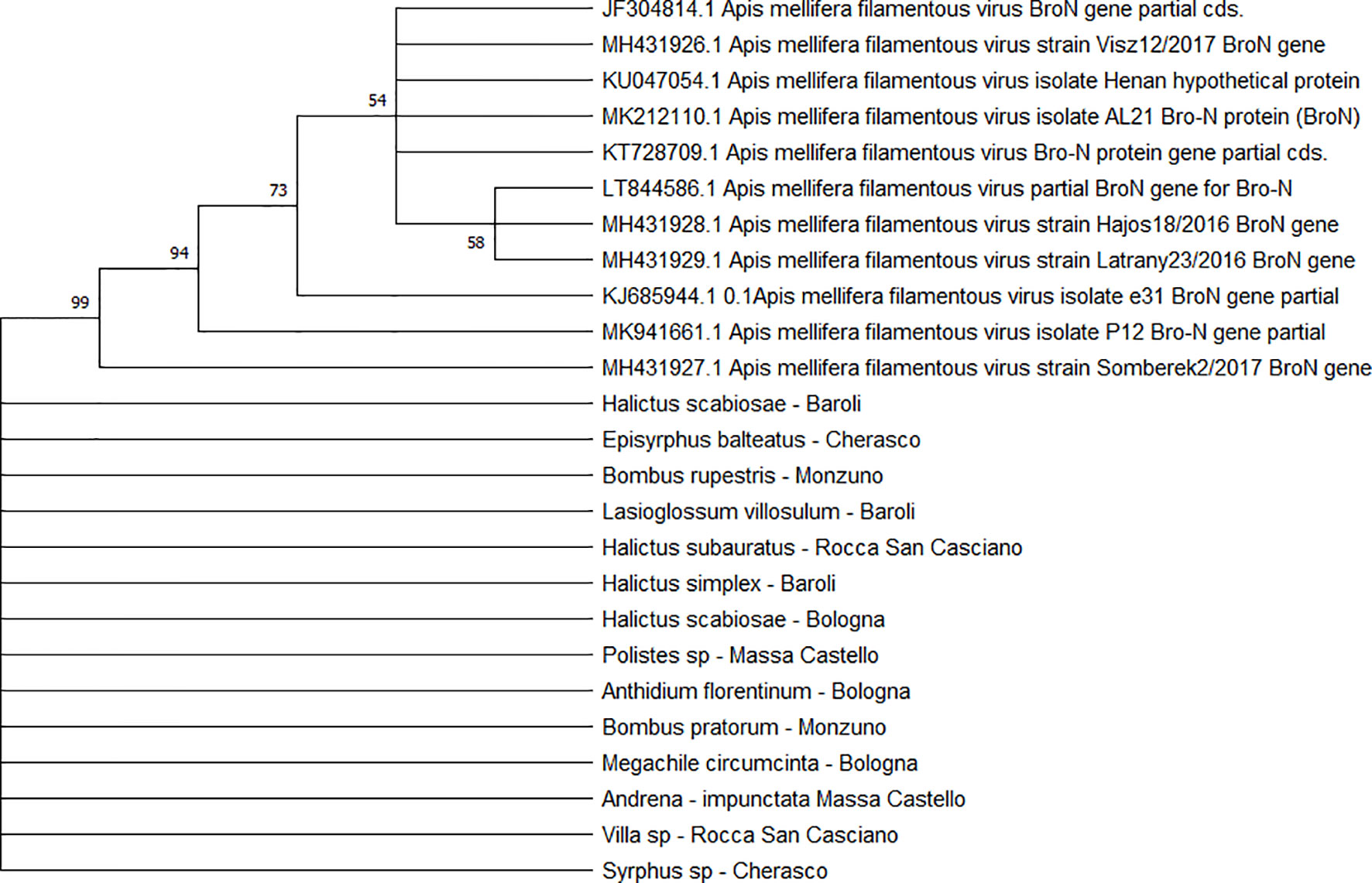
Figure 5 Maximum likelihood nucleotide phylogeny of AmFV BroN gene. This analysis involved 25 nucleotide sequences. There were a total of 896 positions in the final dataset. Only those with bootstrap > 50% are reported. Species and sampling sites are reported for the sequences identified in this study.
Two separate clades were identified for the KBV sequences (n = 4). One of them was associated with three isolates from ERMO sites and the remaining with one PIAI site (Figure 6).

Figure 6 Maximum likelihood nucleotide phylogeny of KBV RNA polymerase gene. This analysis involved 11 nucleotide sequences. There were a total of 421 positions in the final dataset. Only those with bootstrap > 50% are reported. Species and sampling sites are reported for the sequences identified in this study. KBV, Kashmir bee virus.
The detected CBPV sequences (n = 65) (Figure 7) and SBV sequences (n = 37) (Figure 8) clustered in 6 clades each, which were associated with different sampling sites. Both CBPV and SBV sequences had complete homology for the CREA and ERESP isolates.
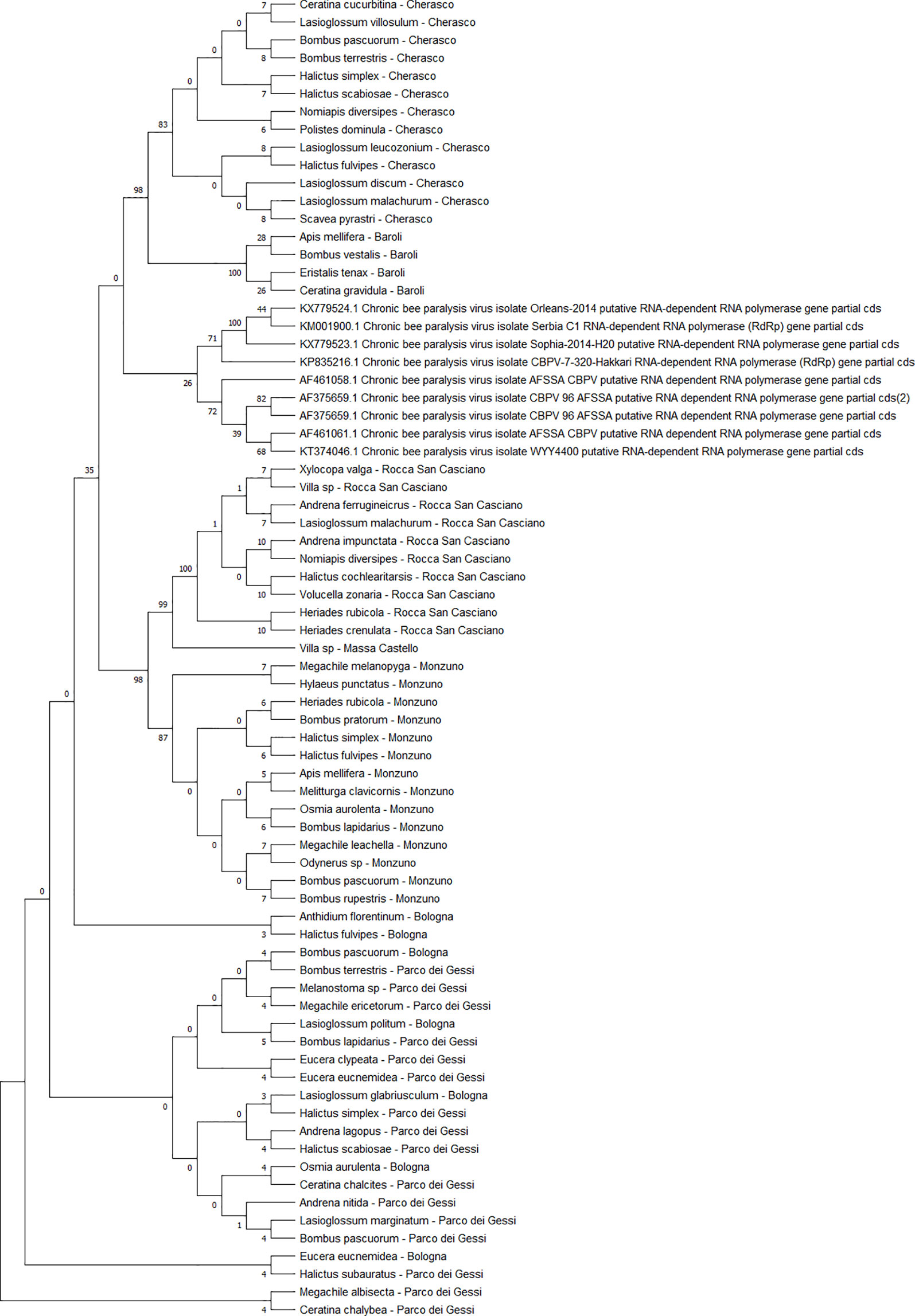
Figure 7 Maximum likelihood nucleotide phylogeny of CBPV RNA-dependent RNA-polymerase gene. This analysis involved 74 nucleotide sequences. There were a total of 583 positions in the final dataset. Only those with bootstrap > 50% are reported. Species and sampling sites are reported for the sequences identified in this study. CBPV, chronic bee paralysis virus.
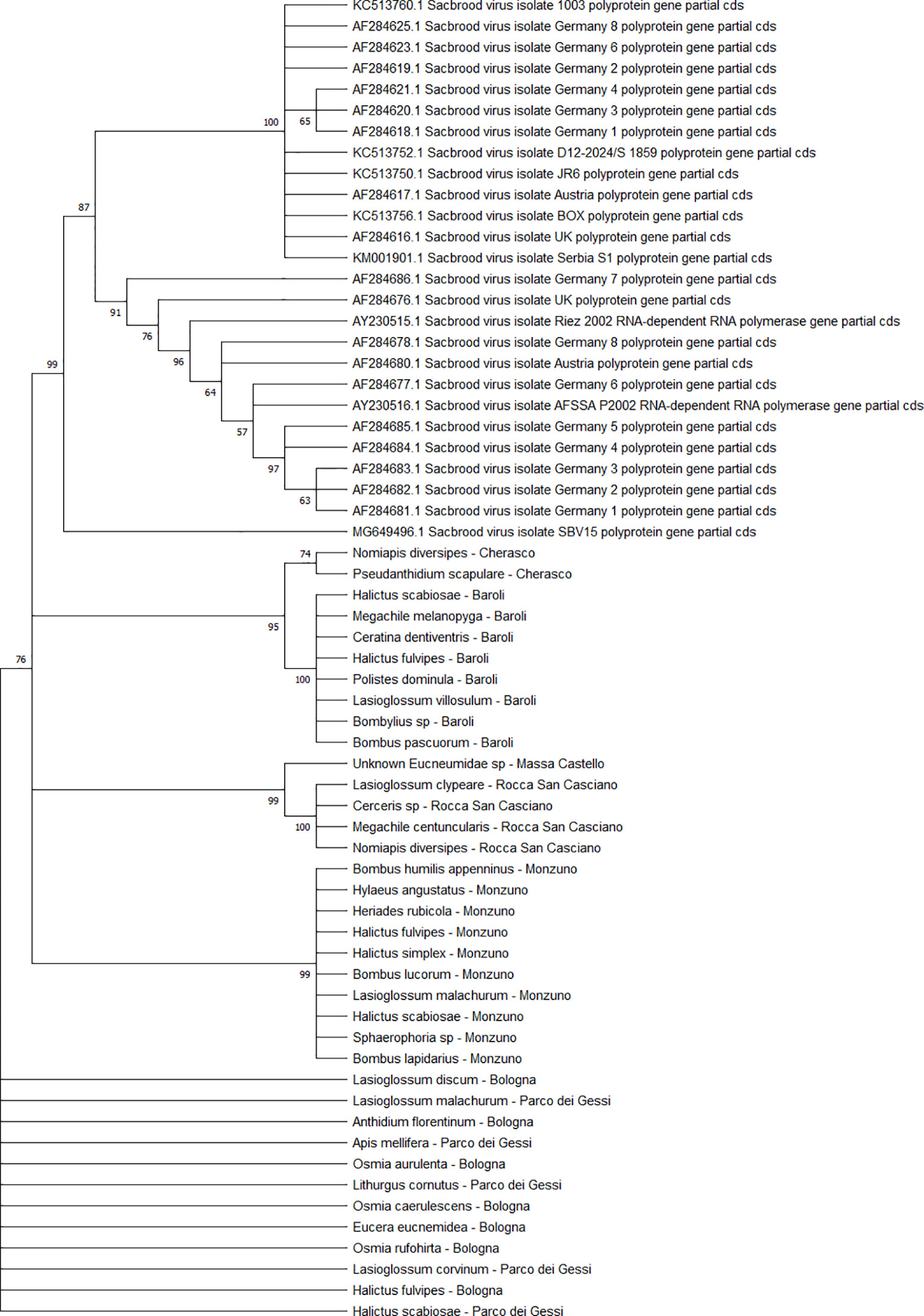
Figure 8 Maximum likelihood nucleotide phylogeny of SBV polyprotein gene. This analysis involved 63 nucleotide sequences. There were a total of 581 positions in the final dataset. Only those with bootstrap > 50% are reported. Species and sampling sites are reported for the sequences identified in this study. SBV, sac brood virus.
All DWV sequences (n = 86) belonged to the DWV-A strain, which is the most frequent and the least virulent variant strain in honey bees, and split into two regional clades (Emilia-Romagna and Piedmont) (Figure 9). Exceptions to this observation were two sequences from PIES (Melanostoma mellinum and L. villosulum; bootstrap = 93%), H. simplex from CREA, and Lasioglossum discum from ERESP (bootstrap = 97%).
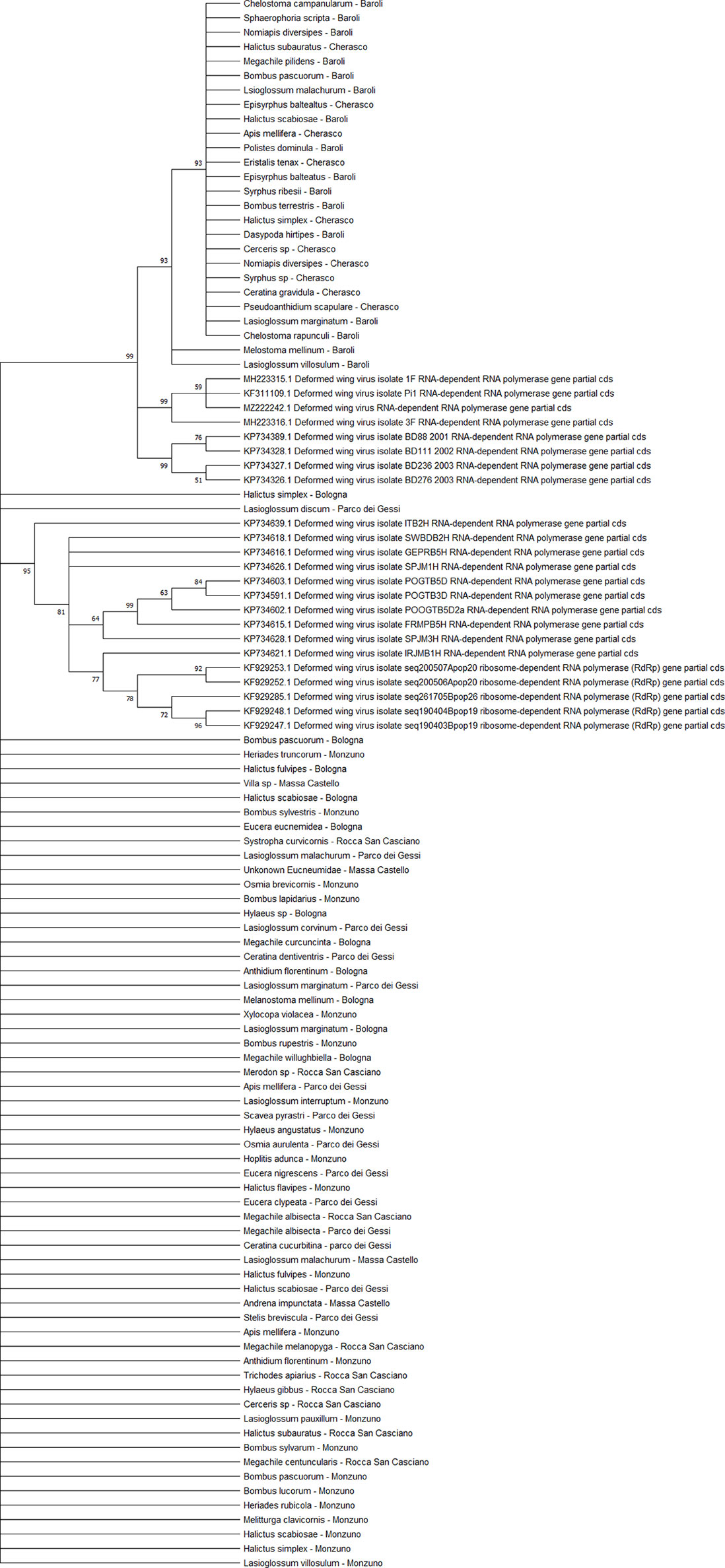
Figure 9 Maximum likelihood nucleotide phylogeny of DWV-A RNA-dependent RNA-polymerase gene. This involved 109 nucleotide sequences. There were a total of 562 positions in the final dataset. Only those with bootstrap > 50% are reported. Species and sampling sites are reported for the sequences identified in this study. DWV, deformed wing virus.
The BQCV sequences (n = 34) clustered in 4 different clades (Figure 10), including the isolates from ERES and ERAI, the sites in the Piedmont region, CREA and ERESP, and ERMO.
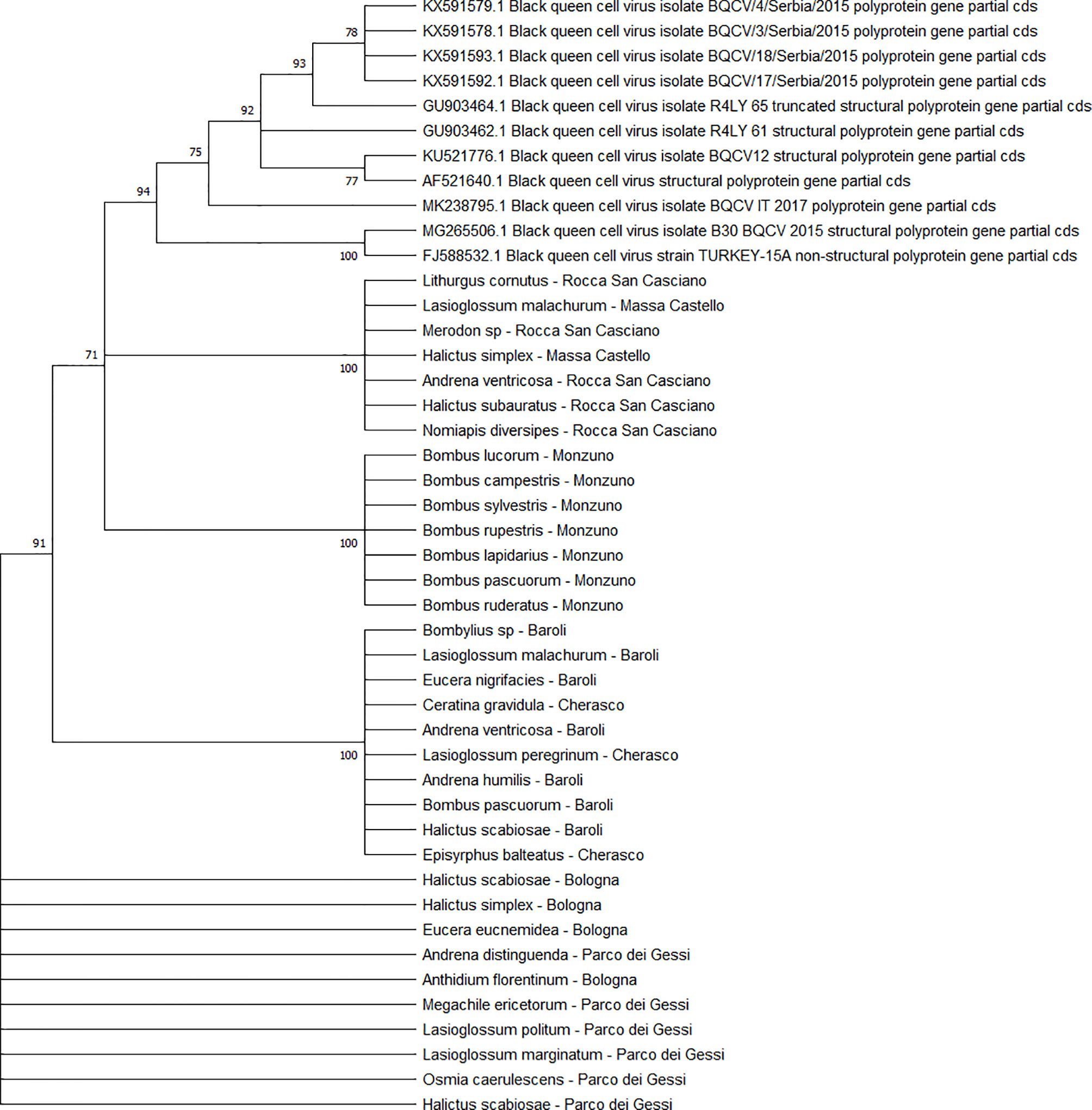
Figure 10 Maximum likelihood nucleotide phylogeny of BQCV polyprotein gene. This analysis involved 45 nucleotide sequences. There were a total of 701 positions in the final dataset. Only those with bootstrap > 50% are reported. Species and sampling sites are reported for the sequences identified in this study. BQCV, black queen cell virus.
Forty-five sampled specimens were found to be co-infected with two or more pathogens. The most complex co-infections were detected in two B. pascuorum individuals that were sampled in PIES. One of them was positive for DWV, SBV, N. ceranae, and C. bombi, and the other was positive for CBPV, SBV, BQCV, and N. ceranae (Table S1).
As reported in Figure 11, the most frequent co-infections included N. ceranae, DWV, CBPV, SBV, and BQCV. Bombus spp. and Halictus spp. were the two genera in which the highest number of multiple infections was found.
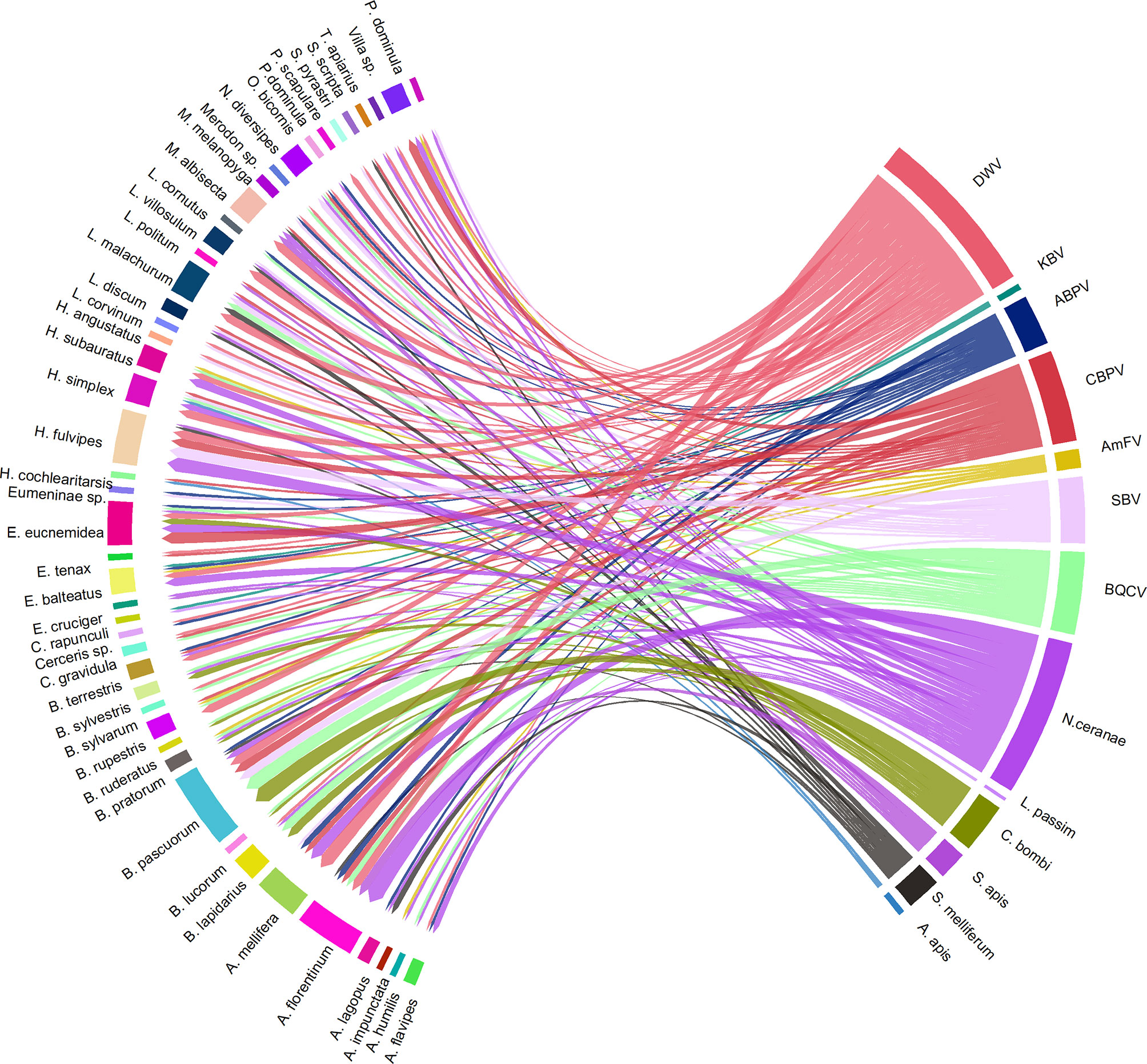
Figure 11 A visual schematisation of the subsample of pathogens that were involved in multiple infections related to their hosts. The arrow thickness denotes the number of co-infections observed within the same host species.
This study was conducted in Italy and confirms that interspecific transmission of pathogens between honey bees and wild pollinators may occur, as reported previously for other countries (Graystock et al., 2013b; Ravoet et al., 2014; Ngor et al., 2020; Pritchard et al., 2021). To the best of our knowledge, this topic has never before been approached in Italian surveys.
Table S6 shows the unprecedented detection of honey bee pathogens in 72 wild bee species, which have shown the potential to act as alternative hosts (Nanetti et al., 2021a). These findings confirm that the considered pathogens show sufficient plasticity to adapt to multiple hosts in nature and the potential to impact the ecosystems involved. The viruses considered were found to be replicative in positive individuals, confirming that their adaptability to alternative host species may result in active infections (Celle et al., 2008; Bailes et al., 2018).
However, our understanding of the pathogenicity of those microorganisms in the alternative hosts is far from complete. Unlike the wing deformations associated with DWV infections in bumblebees, Vespa crabro and Vespa velutina (Genersch et al., 2006; Forzan et al., 2017; Dalmon et al., 2019a; Cilia et al., 2021), the examination of these insects sampled in this study did not show known clinical symptoms that could be ascribed to the pathogens under consideration. This observation is not astounding, as previous attempts to artificially infect bumblebee individuals and other wild bees have resulted in asymptomatic infections (Gusachenko et al., 2020). Nevertheless, artificial inoculations with viruses of the AKI complex group (ABPV, KBV, and IAPV) have reduced the reproductive success of bumblebees and increased the mortality rate in the populations studied (Meeus et al., 2014; Piot et al., 2015). For the time being, the available picture of symptomatic, asymptomatic, and subclinical infections makes it challenging to assess the real impact of honey bee pathogens on the individual wild pollinator species and their communities.
Overall, 69.3% of the individuals sampled in our study were positive for at least one pathogen. That proportion is in line with the prevalence observed in studies conducted in the United States (66% and 80.4%) (Levitt et al., 2013; Dolezal et al., 2016) and in France (79%) where overestimation may have occurred due to pooled sample analysis (Dalmon et al., 2021).
The pathogens that were considered in our study have been reported as transmissible by flowers and pollen (Alger et al., 2019a; Yañez et al., 2020; Nanetti et al., 2021a). Pathogens born by infected foragers (namely, A. mellifera) may persist on flowers, where susceptible pollinators of other species may be infected, with risk dependent on the number of floral visits (Graystock et al., 2013a; Mazzei et al., 2014; Alger et al., 2019b; Burnham et al., 2021).
In this study, DWV, N. ceranae, and CBPV were the pathogens detected with the highest frequency. Likewise, both DWV and microsporidia have been found in a broad range of pollinators (Nanetti et al., 2021a), which implies adaptability to the alternative hosts. As reported in other non-Apis hosts (Forzan et al., 2017; Mazzei et al., 2018; Cilia et al., 2021; Nanetti et al., 2021b), all the detected DWV sequences were type A, which is known as a common and weakly virulent genetic variant in honey bees (McMahon et al., 2016; Mordecai et al., 2016). The frequent occurrence of CBPV could be related to an increased prevalence of this virus in honey bees, which justifies concerns about its agricultural and environmental consequences (Vargas et al., 2017; Budge et al., 2020).
We consider of particular interest our results on L. passim and KBV. Presently, scant information about the spread of trypanosomatid infections in bees is available in Italy. However, in northern Italy, L. passim has been recently found both in apiary conditions of the Veneto region (Bordin et al., 2022) and as e-DNA in honey samples from the Emilia-Romagna region (Ribani et al., 2021). The epidemiological picture is still incomplete; nonetheless, the detection of L. passim in two of our wild bee samples collected in ERMO corroborates the effective circulation of this trypanosomatid in the environment. Similarly, the limited Italian reports about KBV refer to central regions (one case in the Tuscany region and two in the Latium region) (Cersini et al., 2013). Active infections in our wild bee samples from ERMO and PIAI suggest a broader spread of this honey bee virus. This finding recommends the use of non-Apis species as potential “sentinels” providing early epidemiological information about the environmental KBV presence.
The average pathogen abundance measured in the wild species considered in this study was lower than the threshold (>1 * 106 copies) generally considered necessary for symptomatic infection in honey bees (Chen et al., 2006b; Mazzei et al., 2014; Martín-Hernández et al., 2018; Cilia et al., 2020), though various individuals exceeded this limit, and some of them reached 1 * 1010 genic copies. However, abundance does not provide definitive epidemiological information, as the symptomatic threshold is unknown yet for most pathogens and host species.
In general, the prevalence of the pathogens in this study showed a seasonal peak in March, followed by a quick decrease and a subsequent gradual increase until late summer. The cumulative abundance had a different trend, as it gradually increased monthly to peak in July when a decrease proceeded to reach a minimum in October. These results are in line with the seasonal pattern the same pathogens show in the A. mellifera colonies, where they often peak in spring/summer with a possible return in the late season (Tentcheva et al., 2004; D’Alvise et al., 2019; Dalmon et al., 2019b; Loeza-Concha et al., 2020; Chen et al., 2021).
In March, the comparatively high prevalence of positive samples was influenced by the frequent cases of N. ceranae. This finding agrees with the infection dynamics often occurring in the honey bees, where that pathogen may have an acute development in early spring depending on the colony development (Ptaszyńska et al., 2021; Ugolini et al., 2021; Formato et al., 2022). The subsequent decrease in nosemosis suggests a dilution due to the increased species richness, as previously reported for some viruses (Fearon and Tibbetts, 2021); however, a similar dilution effect was not common to all pathogens. The highest richness of pathogens corresponded to the peak in abundance recorded in July. That increase may be associated with both the development of the honey bee colonies and the availability of floral resources. Indeed, after the winter, the colony starts the growth of its population, which is intended to peak in summer, with an increased probability both of intra-colonial (Smith et al., 2013; Steinhauer et al., 2018; D’Alvise et al., 2019; Chen et al., 2021) and interspecific environmental transmission (Bartlett et al., 2019; Wilfert, 2021). Furthermore, the reduced flower availability in summer prompts pollinators to concentrate on the limited resources available, increasing the probability of pathogen exchanges among the potential host species.
Finally, our study showed repeated cases of co-infection in the wild bee species, as already reported for A. mellifera and Aethina tumida (Hung et al., 1995; Evans and Schwarz, 2011; Meeus et al., 2014; Nanetti et al., 2021b). Multiple infections are common in natural environments; nonetheless, we have a limited understanding of the interactions occurring among the involved pathogens (Armitage et al., 2022). Often, multiple infections reduce pathogen virulence due to antagonism (Garbutt et al., 2011), but in other cases, synergistic interactions may increase virulence and reduce the host lifespan (Clay and Rudolf, 2019; Armitage et al., 2022).
This study demonstrates the environmental circulation of honey bee pathogens in the wild pollinating entomofauna present in two different North-Italian regions. The presence of the replicative forms of viruses affecting the honey bees suggests effective interspecific transmission between A. mellifera and alternate host species. To elucidate the effects of those infections on wild pollinators, studies are needed on their fitness, behaviour, mating and reproductive success, nesting, pollen stores, and larval development. Spillover of honey bee pathogens may have undetected yet important drawbacks to the health and functioning of an ecosystem. Health management of honey bee colonies is of high importance in this context, and the beekeepers should therefore undertake the consequent responsibility of being an essential component of the One Health concept (Wilfert et al., 2021).
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material. Sequences were submitted to Genbank under the accession numbers ON304218 to ON304221 (KBV); ON304222 to ON304252 (ABPV); ON448627 to ON448640 (AmFV); ON448642 to ON448727 (DWV); ON448728 to ON448764 (SBV); ON448765 to ON448798 (BQCV): ON448799 to ON448863 (CBPV).
The study was designed by GC and LB. Samples were collected by GC, SF, LZ, and RR. The insect specimens were identified by SF, and the laboratory analyses were performed by GC. Data were analysed by GC, SF, LZ, RR, AN, and LB. GC, SF, LZ, and RR wrote the first version of the manuscript, which was revised by all the authors (MQ, AN, and LB). LB managed the project and the funding acquisition. All authors listed have made a substantial, direct, and intellectual contribution to the work and approved it for publication.
This study was supported by the project BeeNet (Italian National Fund under FEASR 2014–2020) from the Italian Ministry of Agricultural, Food and Forestry Policies (MIPAAF).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcimb.2022.907489/full#supplementary-material
Supplementary Figure 1 | Virus abundance related to the site, month, and genus. Data are presented log10 transformation for better visualization. Means are visualized are red dots. Different number of asterisks indicate statistical differences from base average: p-value<0.05 (*); p-value<0.01 (**); p-value<0.001 (***).
Supplementary Figure 2 | DNA pathogen abundance related to the site, month, and genus. Data are presented log10 transformation for better visualization. Means are visualized are red dots. Different number of asterisks indicate statistical differences from base average: p-value<0.05 (*); p-value<0.01 (**); p-value<0.001 (***).
Supplementary Table 1 | Individual pathogen abundance of the collected samples, grouped by genus, species, region, site, and month.
Supplementary Table 4 | Wilcoxon test results. Tables show the significativity level of each paired test for each pathogen in different sites, months and genera. Only p-values<0.05 are shown. All the other comparisons (not shown) have p-values>0.05.
Supplementary Table 5 | Accession number of viral sequences, deposited in GenBank.
Supplementary Table 6 | Associations between wild bee species and honey bee pathogens that were never described before.
Alger, S. A., Burnham, P. A., Boncristiani, H. F., Brody, A. K. (2019a). RNA Virus Spillover From Managed Honeybees (Apis Mellifera) to Wild Bumblebees (Bombus Spp.). PLos One 14, e0217822. doi: 10.1371/journal.pone.0217822
Alger, S. A., Burnham, P. A., Brody, A. K. (2019b). Flowers as Viral Hot Spots: Honey Bees (Apis Mellifera) Unevenly Deposit Viruses Across Plant Species. PLos One 14, e0221800. doi: 10.1371/journal.pone.0221800
Altschul, S. F., Gish, W., Miller, W., Myers, E. W., Lipman, ,. D. J. (1990). Basic Local Alignment Search Tool. J. Mol. Biol. 215, 403–410. doi: 10.1016/S0022-2836(05)80360-2
Arismendi, N., Bruna, A., Zapata, N., Vargas, M. (2016). PCR-Specific Detection of Recently Described Lotmaria Passim (Trypanosomatidae) in Chilean Apiaries. J. Invertebr. Pathol. 134, 1–5. doi: 10.1016/j.jip.2015.12.008
Armitage, S. A., Genersch, E., McMahon, D. P., Rafaluk-Mohr, C., Rolff, J. (2022). Tripartite Interactions: How Immunity, Microbiota and Pathogens Interact and Affect Pathogen Virulence Evolution. Curr. Opin. Insect Sci. 50, 100871. doi: 10.1016/J.COIS.2021.12.011
Bailes, E. J., Deutsch, K. R., Bagi, J., Rondissone, L., Brown, M. J. F., Lewis, O.T. (2018). First Detection of Bee Viruses in Hoverfly (Syrphid) Pollinators. Biol. Lett. 14, 20180001. doi: 10.1098/rsbl.2018.0001
Ballantyne, G., Baldock, K. C. R., Rendell, L., Willmer, P. G. (2017). Pollinator Importance Networks Illustrate the Crucial Value of Bees in a Highly Speciose Plant Community. Sci. Rep. 7, 1–13. doi: 10.1038/s41598-017-08798-x
Bartlett, L. J., Rozins, C., Brosi, B. J., Delaplane, K. S., de Roode, J. C., White, A., et al. (2019). Industrial Bees: The Impact of Apicultural Intensification on Local Disease Prevalence. J. Appl. Ecol. 56, 2195–2205. doi: 10.1111/1365-2664.13461
Benjamin-Chung, J., Arnold, B. F., Berger, D., Luby, S. P., Miguel, E., Colford, J. M., et al. (2018). Spillover Effects in Epidemiology: Parameters, Study Designs and Methodological Considerations. Int. J. Epidemiol. 47, 332–347. doi: 10.1093/ije/dyx201
Bordin, F., Zulian, L., Granato, A., Caldon, M., Colamonico, R., Toson, M., et al. (2022). Presence of Known and Emerging Honey Bee Pathogens in Apiaries of Veneto Region (Northeast of Italy) During Spring 2020 and 2021. Appl. Sci. 2022 12, 2134 12, 2134. doi: 10.3390/APP12042134
Budge, G. E., Simcock, N. K., Holder, P. J., Shirley, M. D. F., Brown, M. A., Van Weymers, P. S. M., et al. (2020). Chronic Bee Paralysis as a Serious Emerging Threat to Honey Bees. Nat. Commun. 2020 111 (11), 1–9. doi: 10.1038/s41467-020-15919-0
Burnham, P. A., Alger, S. A., Case, B., Boncristiani, H., Hébert-Dufresne, L., Brody, A. K. (2021). Flowers as Dirty Doorknobs: Deformed Wing Virus Transmitted Between Apis Mellifera and Bombus Impatiens Through Shared Flowers. J. Appl. Ecol. 58 (10), 2065-2074. doi: 10.1111/1365-2664.13962
Cane, J. (2021). Global Warming, Advancing Bloom and Evidence for Pollinator Plasticity From Long-Term Bee Emergence Monitoring. Insects 12, 1–11. doi: 10.3390/insects12050457
Celle, O., Blanchard, P., Olivier, V., Schurr, F., Cougoule, N., Faucon, J. P., et al. (2008). Detection of Chronic Bee Paralysis Virus (CBPV) Genome and its Replicative RNA Form in Various Hosts and Possible Ways of Spread. Virus Res. 133, 280–284. doi: 10.1016/j.virusres.2007.12.011
Cersini, A., Bellucci, V., Lucci, S., Mutinelli, F., Granato, A., Porrini, C., et al. (2013). First Isolation of Kashmir Bee Virus (KBV) in Italy. J. Apic. Res. 52, 54–55. doi: 10.3896/IBRA.1.52.2.08
Chantawannakul, P., Ward, L., Boonham, N., Brown, M. (2006). A Scientific Note on the Detection of Honeybee Viruses Using Real-Time PCR (TaqMan) in Varroa Mites Collected From a Thai Honeybee (Apis Mellifera) Apiary. J. Invertebr. Pathol. 91, 69–73. doi: 10.1016/j.jip.2005.11.001
Chen, Y., Evans, J., Feldlaufer, M. (2006a). Horizontal and Vertical Transmission of Viruses in the Honey Bee, Apis Mellifera. J. Invertebr. Pathol. 92, 152–159. doi: 10.1016/J.JIP.2006.03.010
Chen, Y. P., Pettis, J. S., Collins, A., Feldlaufer, M. F. (2006b). Prevalence and Transmission of Honeybee Viruses. Appl. Environ. Microbiol. 72, 606–611. doi: 10.1128/AEM.72.1.606-611.2006
Chen, G., Wu, Y., Deng, J., Wen, Z., Wang, S., Chen, Y., et al. (2021). Seasonal Variation of Viral Infections Between the Eastern Honey Bee (Apis Cerana) and the Western Honey Bee (Apis Mellifera). Microbiologyopen 10, e1162. doi: 10.1002/MBO3.1162
Choate, B. A., Hickman, P. L., Moretti, E. A. (2018). Wild Bee Species Abundance and Richness Across an Urban–Rural Gradient. J. Insect Conserv. 22, 391–403. doi: 10.1007/s10841-018-0068-6
Cilia, G., Cabbri, R., Maiorana, G., Cardaio, I., Dall’Olio, R., Nanetti, A. (2018). A Novel TaqMan ® Assay for Nosema Ceranae Quantification in Honey Bee, Based on the Protein Coding Gene Hsp70. Eur. J. Protistol. 63, 44–50. doi: 10.1016/j.ejop.2018.01.007
Cilia, G., Garrido, C., Bonetto, M., Tesoriero, D., Nanetti, A. (2020). Effect of Api-Bioxal ® and ApiHerb ® Treatments Against Nosema Ceranae Infection in Apis Mellifera Investigated by Two qPCR Methods. Vet. Sci. 2020 7, 125 7, 125. doi: 10.3390/vetsci7030125
Cilia, G., Sagona, S., Giusti, M., Jarmela dos Santos, P. E., Nanetti, A., Felicioli, A. (2019). Nosema Ceranae Infection in Honeybee Samples From Tuscanian Archipelago (Central Italy) Investigated by Two qPCR Methods. Saudi J. Biol. Sci. 26, 1553–1556. doi: 10.1016/j.sjbs.2018.11.017
Cilia, G., Zavatta, L., Ranalli, R., Nanetti, A., Bortolotti, L. (2021). Replicative Deformed Wing Virus Found in the Head of Adults From Symptomatic Commercial Bumblebee (Bombus Terrestris) Colonies. Vet. Sci. 8, 117. doi: 10.3390/vetsci8070117
Clay, P. A., Rudolf, V. H. W. (2019). How Parasite Interaction Strategies Alter Virulence Evolution in Multi-Parasite Communities. Evol. (N. Y). 73, 2189–2203. doi: 10.1111/EVO.13843
Cohen, H., Ponisio, L. C., Russell, K. A., Philpott, S. M., McFrederick, Q. S. (2022). Floral Resources Shape Parasite and Pathogen Dynamics in Bees Facing Urbanization. Mol. Ecol. 31 (7): 2157-2171. doi: 10.1111/MEC.16374
Cooling, M., Gruber, M. A. M., Hoffmann, B. D., Sébastien, A., Lester, P. J. (2017). A Metatranscriptomic Survey of the Invasive Yellow Crazy Ant, Anoplolepis Gracilipes, Identifies Several Potential Viral and Bacterial Pathogens and Mutualists. Insectes Soc 64, 197–207. doi: 10.1007/s00040-016-0531-x
Dalmon, A., Diévart, V., Thomasson, M., Fouque, R., Vaissière, B. E., Guilbaud, L., et al. (2021). Possible Spillover of Pathogens Between Bee Communities Foraging on the Same Floral Resource. Insects 12, 122. doi: 10.3390/insects12020122
Dalmon, A., Gayral, P., Decante, D., Klopp, C., Bigot, D., Thomasson, M., et al. (2019a). Viruses in the Invasive Hornet Vespa Velutina. Viruses 2019 11, 1041 11, 1041. doi: 10.3390/V11111041
Dalmon, A., Peruzzi, M., Le Conte, Y., Alaux, C., Pioz, M. (2019b). Temperature-Driven Changes in Viral Loads in the Honey Bee Apis Mellifera. J. Invertebr. Pathol. 160, 87–94. doi: 10.1016/j.jip.2018.12.005
D’Alvise, P., Seeburger, V., Gihring, K., Kieboom, M., Hasselmann, M. (2019). Seasonal Dynamics and Co-Occurrence Patterns of Honey Bee Pathogens Revealed by High-Throughput RT-qPCR Analysis. Ecol. Evol. 9, 10241–10252. doi: 10.1002/ECE3.5544
de Miranda, J. R., Dainat, B., Locke, B., Cordoni, G., Berthoud, H., Gauthier, L., et al. (2010). Genetic Characterization of Slow Bee Paralysis Virus of the Honeybee (Apis Mellifera L.). J. Gen. Virol. 91, 2524–2530. doi: 10.1099/vir.0.022434-0
Dobbelaere, W., de Graaf, D. C., Peeters, J. E. (2001). Development of a Fast and Reliable Diagnostic Method for American Foulbrood Disease ( Paenibacillus Larvae Subsp. Larvae ) Using a 16S rRNA Gene Based PCR. Apidologie 32, 363–370. doi: 10.1051/apido:2001136
Dolezal, A. G., Hendrix, S. D., Scavo, N. A., Carrillo-Tripp, J., Harris, M. A., Wheelock, M. J., et al. (2016). Honey Bee Viruses in Wild Bees: Viral Prevalence, Loads, and Experimental Inoculation. PLos One 11, e0166190. doi: 10.1371/journal.pone.0166190
Drossart, M., Gérard, M. (2020). Beyond the Decline of Wild Bees: Optimizing Conservation Measures and Bringing Together the Actors. Insects 11, 1–23. doi: 10.3390/insects11090649
Duchenne, F., Thébault, E., Michez, D., Gérard, M., Devaux, C., Rasmont, P., et al. (2020). Long-Term Effects of Global Change on Occupancy and Flight Period of Wild Bees in Belgium. Glob. Change Biol. 26, 6753–6766. doi: 10.1111/gcb.15379
Erler, S., Popp, M., Wolf, S., Lattorff, H. M. G. (2012). Sex, Horizontal Transmission, and Multiple Hosts Prevent Local Adaptation of Crithidia Bombi, a Parasite of Bumblebees (Bombus Spp.). Ecol. Evol. 2, 930. doi: 10.1002/ECE3.250
Evans, J. D., Schwarz, R. S. (2011). Bees Brought to Their Knees: Microbes Affecting Honey Bee Health. Trends Microbiol. 19, 614–620. doi: 10.1016/j.tim.2011.09.003
Fearon, M. L., Tibbetts, E. A. (2021). Pollinator Community Species Richness Dilutes Prevalence of Multiple Viruses Within Multiple Host Species. Ecology 102, e03305. doi: 10.1002/ECY.3305
Felsenstein, J. (1985). Confidence Limits on Phylogenies: An Approach Using the Bootstrap. Evol. (N. Y). 39, 783. doi: 10.2307/2408678
Formato, G., Rivera-Gomis, J., Bubnic, J., Martín-Hernández, R., Milito, M., Croppi, S., et al. (2022). Nosemosis Prevention and Control. Appl. Sci. 2022 12, 783 12, 783. doi: 10.3390/APP12020783
Fortel, L. (2014). Ecologie Et Conservation Des Abeilles Sauvages Le Long D’un Gradient D’urbanisation. University of Avignon
Forzan, M., Sagona, S., Mazzei, M., Felicioli, A. (2017). Detection of Deformed Wing Virus in Vespa Crabro. Bull. Insectol. 70, 261–265.
Garbutt, J., Bonsall, M. B., Wright, D. J., Raymond, B. (2011). Antagonistic Competition Moderates Virulence in Bacillus Thuringiensis. Ecol. Lett. 14, 765–772. doi: 10.1111/J.1461-0248.2011.01638.X
Garigliany, M., Taminiau, B., El Agrebi, N., Cadar, D., Gilliaux, G., Hue, M., et al. (2017). Moku Virus in Invasive Asian Hornets, Belgium 2016. Emerg. Infect. Dis. 23, 2109–2112. doi: 10.3201/eid2312.171080
Genersch, E., Yue, C., Fries, I., de Miranda, J. R. (2006). Detection of Deformed Wing Virus, a Honey Bee Viral Pathogen, in Bumble Bees (Bombus Terrestris and Bombus Pascuorum) With Wing Deformities. J. Invertebr. Pathol. 91, 61–63. doi: 10.1016/J.JIP.2005.10.002
Graystock, P., Goulson, D., Hughes, W. O. H. (2015). Parasites in Bloom: Flowers Aid Dispersal and Transmission of Pollinator Parasites Within and Between Bee Species. Proc. R. Soc B Biol. Sci. 282. doi: 10.1098/rspb.2015.1371
Graystock, P., Yates, K., Darvill, B., Goulson, D., Hughes, W. O. H. (2013a). Emerging Dangers: Deadly Effects of an Emergent Parasite in a New Pollinator Host. J. Invertebr. Pathol. 114, 114–119. doi: 10.1016/j.jip.2013.06.005
Graystock, P., Yates, K., Evison, S. E. F., Darvill, B., Goulson, D., Hughes, W. O. H. (2013b). The Trojan Hives: Pollinator Pathogens, Imported and Distributed in Bumblebee Colonies. J. Appl. Ecol. 50, 20151371. doi: 10.1111/1365-2664.12134
Gruber, M. A. M., Cooling, M., Baty, J. W., Buckley, K., Friedlander, A., Quinn, O., et al. (2017). Single-Stranded RNA Viruses Infecting the Invasive Argentine Ant, Linepithema Humile. Sci. Rep. 7, 1–10. doi: 10.1038/s41598-017-03508-z
Gusachenko, O. N., Woodford, L., Balbirnie-Cumming, K., Ryabov, E. V., Evans, D. J. (2020). Evidence for and Against Deformed Wing Virus Spillover From Honey Bees to Bumble Bees: A Reverse Genetic Analysis. Sci. Rep. 10, 16847. doi: 10.1038/s41598-020-73809-3
Hartmann, U., Forsgren, E., Charrière, J. D., Neumann, P., Gauthier, L. (2015). Dynamics of Apis Mellifera Filamentous Virus (AmFV) Infections in Honey Bees and Relationships With Other Parasites. Viruses 7, 2654–2667. doi: 10.3390/v7052654
Hofmann, M. M., Renner, S. S. (2020). Bee Species Decrease and Increase Between the 1990s and 2018 in Large Urban Protected Sites. J. Insect Conserv. 24, 637–642. doi: 10.1007/s10841-020-00238-y
Hrynko, I., Kaczyński, P., Łozowicka, B. (2021). A Global Study of Pesticides in Bees: QuEChERS as a Sample Preparation Methodology for Their Analysis – Critical Review and Perspective. Sci. Total Environ. 792, 148385. doi: 10.1016/j.scitotenv.2021.148385
Huang, W. F., Skyrm, K., Ruiter, R., Solter, L. (2015). Disease Management in Commercial Bumble Bee Mass Rearing, Using Production Methods, Multiplex PCR Detection Techniques, and Regulatory Assessment. J. Apic. Res. 54, 516–524, 702-704. doi: 10.1080/00218839.2016.1173352
Hung, A. C., Adams, J. R., Shimanuki, H. (1995). Bee Parasitic Mite Syndrome. (II). The Role of Varroa Mite and Viruses. Am. Bee J. 324, 702-704.
Hung, K. L. J., Sandoval, S. S., Ascher, J. S., Holway, D. A. (2021). Article Joint Impacts of Drought and Habitat Fragmentation on Native Bee Assemblages in a California Biodiversity Hotspot. Insects 12, 1–17. doi: 10.3390/insects12020135
James, R. R., Skinner, J. S. (2005). PCR Diagnostic Methods for Ascosphaera Infections in Bees. J. Invertebr. Pathol. 90, 98–103. doi: 10.1016/J.JIP.2005.08.004
Kajobe, R., Marris, G., Budge, G., Laurenson, L., Cordoni, G., Jones, B., et al. (2010). First Molecular Detection of a Viral Pathogen in Ugandan Honey Bees. J. Invertebr. Pathol. 104, 153–156. doi: 10.1016/J.JIP.2010.02.007
Khalifa, S. A. M., Elshafiey, E. H., Shetaia, A. A., El-Wahed, A. A. A., Algethami, A. F., Musharraf, S. G., et al. (2021). Overview of Bee Pollination and its Economic Value for Crop Production. Insects 12, 1–23. doi: 10.3390/insects12080688
Koh, I., Lonsdorf, E. V., Williams, N. M., Brittain, C., Isaacs, R., Gibbs, J., et al. (2016). Modeling the Status, Trends, and Impacts of Wild Bee Abundance in the United States. Proc. Natl. Acad. Sci. U. S. A. 113, 140–145. doi: 10.1073/pnas.1517685113
Kumar, S., Stecher, G., Li, M., Knyaz, C., Tamura, K. (2018). MEGA X: Molecular Evolutionary Genetics Analysis Across Computing Platforms. Mol. Biol. Evol. 35, 1547–1549. doi: 10.1093/molbev/msy096
Levitt, A. L., Singh, R., Cox-Foster, D. L., Rajotte, E., Hoover, K., Ostiguy, N., et al. (2013). Cross-Species Transmission of Honey Bee Viruses in Associated Arthropods. Virus Res. 176, 232–240. doi: 10.1016/j.virusres.2013.06.013
Librán-Embid, F., Grass, I., Emer, C., Ganuza, C., Tscharntke, T. (2021). A Plant–Pollinator Metanetwork Along a Habitat Fragmentation Gradient. Ecol. Lett. 24, 2700–2712. doi: 10.1111/ele.13892
Loeza-Concha, H., Salgado-Moreno, S., Avila-Ramos, F., Escalera-Valente, F., Lemus-Flores, C., Domínguez-Rebolledo, Á., et al. (2020). Seasonal Variation in the Prevalence of Varroa, Nosema and Acarapis in Hives From Which Queen Bee Mating Nuclei are Produced. J. Apic. Res. 59, 558–563. doi: 10.1080/00218839.2020.1717060
Manley, R., Temperton, B., Doyle, T., Gates, D., Hedges, S., Boots, M., et al. (2019). Knock-On Community Impacts of a Novel Vector: Spillover of Emerging DWV-B From Varroa -Infested Honeybees to Wild Bumblebees. Ecol. Lett. 22, ele.13323. doi: 10.1111/ele.13323
Martín-Hernández, R., Bartolomé, C., Chejanovsky, N., Le Conte, Y., Dalmon, A., Dussaubat, C., et al. (2018). Nosema Ceranae in Apis Mellifera : A 12 Years Postdetection Perspective. Environ. Microbiol. 20, 1302–1329. doi: 10.1111/1462-2920.14103
Martin-Hernandez, R., Meana, A., Prieto, L., Salvador, A. M., Garrido-Bailon, E., Higes, M. (2007). Outcome of Colonization of Apis Mellifera by Nosema Ceranae. Appl. Environ. Microbiol. 73, 6331–6338. doi: 10.1128/AEM.00270-07
Martin, S. J., Highfield, A. C., Brettell, L., Villalobos, E. M., Budge, G. E., Powell, M., et al. (2012). Global Honey Bee Viral Landscape Altered by a Parasitic Mite. Sci. (80-. ) 336, 1304–1306. doi: 10.1126/science.1220941
Mazzei, M., Carrozza, M. L., Luisi, E., Forzan, M., Giusti, M., Sagona, S., et al. (2014). Infectivity of DWV Associated to Flower Pollen: Experimental Evidence of a Horizontal Transmission Route. PLos One 9, e113448. doi: 10.1371/journal.pone.0113448
Mazzei, M., Cilia, G., Forzan, M., Lavazza, A., Mutinelli, F., Felicioli, A. (2019). Detection of Replicative Kashmir Bee Virus and Black Queen Cell Virus in Asian Hornet Vespa Velutina (Lepelieter 1836) in Italy. Sci. Rep. 9, 1–9. doi: 10.1038/s41598-019-46565-2
Mazzei, M., Forzan, M., Cilia, G., Sagona, S., Bortolotti, L., Felicioli, A. (2018). First Detection of Replicative Deformed Wing Virus (DWV) in Vespa Velutina Nigrithorax. Bull. Insectol. 71, 211–216.
McMahon, D. P., Natsopoulou, M. E., Doublet, V., Fürst, M., Weging, S., Brown, M. J. F., et al. (2016). Elevated Virulence of an Emerging Viral Genotype as a Driver of Honeybee Loss. Proc. Biol. Sci. 283, 20160811. doi: 10.1098/rspb.2016.0811
Meeus, I., de Miranda, J. R., de Graaf, D. C., Wäckers, F., Smagghe, G. (2014). Effect of Oral Infection With Kashmir Bee Virus and Israeli Acute Paralysis Virus on Bumblebee (Bombus Terrestris) Reproductive Success. J. Invertebr. Pathol. 121, 64–69. doi: 10.1016/J.JIP.2014.06.011
Meeus, I., Vercruysse, V., Smagghe, G. (2012). Molecular Detection of Spiroplasma Apis and Spiroplasma Melliferum in Bees. J. Invertebr. Pathol. 109, 172–174. doi: 10.1016/J.JIP.2011.11.006
Mordecai, G. J., Wilfert, L., Martin, S. J., Jones, I. M., Schroeder, D. C. (2016). Diversity in a Honey Bee Pathogen: First Report of a Third Master Variant of the Deformed Wing Virus Quasispecies. ISME J. 10, 1264–1273. doi: 10.1038/ismej.2015.178
Nanetti, A., Bortolotti, L., Cilia, G. (2021a). Pathogens Spillover From Honey Bees to Other Arthropods. Pathog. 2021 10, 1044 10, 1044. doi: 10.3390/PATHOGENS10081044
Nanetti, A., Ellis, J. D., Cardaio, I., Cilia, G. (2021b). Detection of Lotmaria Passim, Crithidia Mellificae and Replicative Forms of Deformed Wing Virus and Kashmir Bee Virus in the Small Hive Beetle (Aethina Tumida). Pathogens 10, 372. doi: 10.3390/pathogens10030372
Nanetti, A., Ugolini, L., Cilia, G., Pagnotta, E., Malaguti, L., Cardaio, I., et al. (2021c). Seed Meals From Brassica Nigra and Eruca Sativa Control Artificial Nosema Ceranae Infections in Apis Mellifera. Microorganisms 9, 949. doi: 10.3390/microorganisms9050949
Ngor, L., Palmer-Young, E. C., Burciaga Nevarez, R., Russell, K. A., Leger, L., Giacomini, S. J., et al. (2020). Cross-Infectivity of Honey and Bumble Bee-Associated Parasites Across Three Bee Families. Parasitology 147, 1290–1304. doi: 10.1017/S0031182020001018
Ocepek, M. P., Toplak, I., Zajc, U., Bevk, D. (2021). The Pathogens Spillover and Incidence Correlation in Bumblebees and Honeybees in Slovenia. Pathog. 2021 10, 884 10, 884. doi: 10.3390/PATHOGENS10070884
O’Neal, S. T., Anderson, T. D., Wu-Smart, J. Y. (2018). Interactions Between Pesticides and Pathogen Susceptibility in Honey Bees. Curr. Opin. Insect Sci. 26, 57–62. doi: 10.1016/j.cois.2018.01.006
Piot, N., Snoeck, S., Vanlede, M., Smagghe, G., Meeus, I. (2015). The Effect of Oral Administration of dsRNA on Viral Replication and Mortality in Bombus Terrestris. Viruses 7, 3172–3185. doi: 10.3390/v7062765
Porto, R. G., de Almeida, R. F., Cruz-Neto, O., Tabarelli, M., Viana, B. F., Peres, C. A., et al. (2020). Pollination Ecosystem Services: A Comprehensive Review of Economic Values, Research Funding and Policy Actions. Food Secur. 12, 1425–1442. doi: 10.1007/s12571-020-01043-w
Prendergast, K. S., Menz, M. H. M., Dixon, K. W., Bateman, P. W. (2020). The Relative Performance of Sampling Methods for Native Bees: An Empirical Test and Review of the Literature. Ecosphere 11, e03076. doi: 10.1002/ecs2.3076
Pritchard, Z. A., Hendriksma, H. P., St Clair, A. L., Stein, D. S., Dolezal, A. G., O’Neal, M. E., et al. (2021). Do Viruses From Managed Honey Bees (Hymenoptera: Apidae) Endanger Wild Bees in Native Prairies? Environ. Entomol. 50, 455–466. doi: 10.1093/EE/NVAA181
Ptaszyńska, A. A., Latoch, P., Hurd, P. J., Polaszek, A., Michalska-Madej, J., Grochowalski, Ł., et al. (2021). Amplicon Sequencing of Variable 16s rRNA From Bacteria and ITS2 Regions From Fungi and Plants, Reveals Honeybee Susceptibility to Diseases Results From Their Forage Availability Under Anthropogenic Landscapes. Pathog. 2021 10, 381 10, 381. doi: 10.3390/PATHOGENS10030381
Purkiss, T., Lach, L. (2019). Pathogen Spillover From Apis Mellifera to a Stingless Bee. Proc. R. Soc B Biol. Sci. 286, 20191071. doi: 10.1098/rspb.2019.1071
Rasmussen, C., Dupont, Y. L., Madsen, H. B., Bogusch, P., Goulson, D., Herbertsson, L., et al. (2021). Evaluating Competition for Forage Plants Between Honey Bees and Wild Bees in Denmark. PLos One 16, 1–19. doi: 10.1371/journal.pone.0250056
Ravoet, J., De Smet, L., Meeus, I., Smagghe, G., Wenseleers, T., de Graaf, D. C. (2014). Widespread Occurrence of Honey Bee Pathogens in Solitary Bees Journal of Invertebrate Pathology 122, 55–58. doi: 10.1016/j.jip.2014.08.007
R Core Team (2021). R: A Language and Environment for Statistical Computing. R Found Stat. Comput. Vienna, Austria. http://www.r-project.org/index.html
Ribani, A., Utzeri, V. J., Taurisano, V., Galuppi, R., Fontanesi, L. (2021). Analysis of Honey Environmental DNA Indicates That the Honey Bee (Apis Mellifera L.) Trypanosome Parasite Lotmaria Passim is Widespread in the Apiaries of the North of Italy. J. Invertebr. Pathol. 184, 107628. doi: 10.1016/J.JIP.2021.107628
Roetschi, A., Berthoud, H., Kuhn, R., Imdorf, A. (2008). Infection Rate Based on Quantitative Real-Time PCR of Melissococcus Plutonius, the Causal Agent of European Foulbrood, in Honeybee Colonies Before and After Apiary Sanitation. Apidologie 39, 362–371. doi: 10.1051/apido:200819
Saitou, N., Nei, M. (1987). The Neighbor-Joining Method: A New Method for Reconstructing Phylogenetic Trees. Mol. Biol. Evol. 4, 406–425. doi: 10.1093/OXFORDJOURNALS.MOLBEV.A040454
Santamaria, J., Villalobos, E. M., Brettell, L. E., Nikaido, S., Graham, J. R., Martin, S. (2018). Evidence of Varroa-Mmediated Deformed Wing Virus Spillover in Hawaii. J Invertebrate Pathol 151, 126–130. doi: 10.1016/j.jip.2017.11.008
Schenk, M., Krauss, J., Holzschuh, A. (2018). Desynchronizations in Bee–Plant Interactions Cause Severe Fitness Losses in Solitary Bees. J. Anim. Ecol. 87, 139–149. doi: 10.1111/1365-2656.12694
Schittny, D., Yañez, O., Neumann, P. (2020). Honey Bee Virus Transmission via Hive Products. Vet. Sci. 7, 96. doi: 10.3390/vetsci7030096
Sébastien, A., Lester, P. J., Hall, R. J., Wang, J., Moore, N. E., Gruber, M. A. M. (2015). Invasive Ants Carry Novel Viruses in Their New Range and Form Reservoirs for a Honeybee Pathogen. Biol. Lett. 11, 20150610. doi: 10.1098/rsbl.2015.0610
Senapathi, D., Biesmeijer, J. C., Breeze, T. D., Kleijn, D., Potts, S. G., Carvalheiro, L. G. (2015). Pollinator Conservation - The Difference Between Managing for Pollination Services and Preserving Pollinator Diversity. Curr. Opin. Insect Sci. 12, 93–101. doi: 10.1016/j.cois.2015.11.002
Singh, R., Levitt, A. L., Rajotte, E. G., Holmes, E. C., Ostiguy, N., vanEngelsdorp, D., et al. (2010). RNA Viruses in Hymenopteran Pollinators: Evidence of Inter-Taxa Virus Transmission via Pollen and Potential Impact on Non-Apis Hymenopteran Species. PLos One 5, e14357. doi: 10.1371/journal.pone.0014357
Siviter, H., Koricheva, J., Brown, M. J. F., Leadbeater, E. (2018). Quantifying the Impact of Pesticides on Learning and Memory in Bees. J. Appl. Ecol. 55, 2812–2821. doi: 10.1111/1365-2664.13193
Smith, K. M., Loh, E. H., Rostal, M. K., Zambrana-Torrelio, C. M., Mendiola, L., Daszak, P. (2013). Pathogens, Pests, and Economics: Drivers of Honey Bee Colony Declines and Losses. Ecohealth 10, 434–445. doi: 10.1007/S10393-013-0870-2
Steinhauer, N., Kulhanek, K., Antúnez, K., Human, H., Chantawannakul, P., Chauzat, M. P., et al. (2018). Drivers of Colony Losses. Curr. Opin. Insect Sci. 26, 142–148. doi: 10.1016/j.cois.2018.02.004
Tamura, K., Nei, M., Kumar, S. (2004). Prospects for Inferring Very Large Phylogenies by Using the Neighbor-Joining Method. Proc. Natl. Acad. Sci. 101, 11030–11035. doi: 10.1073/PNAS.0404206101
Tehel, A., Streicher, T., Tragust, S., Paxton, R. J. (2022). Experimental Cross Species Transmission of a Major Viral Pathogen in Bees is Predominantly From Honeybees to Bumblebees. Proc. R. Soc B 289, 20212255. doi: 10.1098/RSPB.2021.2255
Tentcheva, D., Gauthier, L., Zappulla, N., Dainat, B., Cousserans, F., Colin, M. E., et al. (2004). Prevalence and Seasonal Variations of Six Bee Viruses in Apis Mellifera L. And Varroa Destructor Mite Populations in France. Appl. Environ. Microbiol. 70, 7185–7191. doi: 10.1128/AEM.70.12.7185-7191.2004
Tscharntke, T., Steffan-Dewenter, I. (2000). Resource Overlap and Possible Competition Between Honey Bees and Wild Bees in Central Europe. Oecologia 122, 288–296. doi: 10.1007/s004420050034
Ugolini, L., Cilia, G., Pagnotta, E., Malaguti, L., Capano, V., Guerra, I., et al. (2021). Glucosinolate Bioactivation by Apis Mellifera Workers and Its Impact on Nosema Ceranae Infection at the Colony Level. Biomolecules 11, 1657. doi: 10.3390/BIOM11111657
Vargas, M., Arismendi, N., Riveros, G., Zapata, N., Bruna, A., Vidal, M., et al. (2017). Viral and Intestinal Diseases Detected in Apis Mellifera in Central and Southern Chile. Chil. J. Agric. Res. 77, 243–249. doi: 10.4067/S0718-58392017000300243
Wenzel, A., Grass, I., Belavadi, V. V., Tscharntke, T. (2020). How Urbanization is Driving Pollinator Diversity and Pollination – A Systematic Review. Biol. Conserv. 241, 108321. doi: 10.1016/j.biocon.2019.108321
Wilfert, L. (2021). Viral Adaptations to Vector-Borne Transmission can Result in Complex Host–Vector–Pathogen Interactions. J. Anim. Ecol. 90, 2230–2233. doi: 10.1111/1365-2656.13570
Wilfert, L., Brown, M. J. F., Doublet, V. (2021). OneHealth Implications of Infectious Diseases of Wild and Managed Bees. J. Invertebr. Pathol. 186, 107506. doi: 10.1016/J.JIP.2020.107506
Wojcik, V. A., Morandin, L. A., Davies Adams, L., Rourke, K. E. (2018). Floral Resource Competition Between Honey Bees and Wild Bees: Is There Clear Evidence and Can We Guide Management and Conservation? Environ. Entomol. 47, 822–833. doi: 10.1093/ee/nvy077
Xu, G., Palmer-Young, E., Skyrm, K., Daly, T., Sylvia, M., Averill, A., et al. (2018). Triplex Real-Time PCR for Detection of Crithidia Mellificae and Lotmaria Passim in Honey Bees. Parasitol. Res. 117, 623–628. doi: 10.1007/s00436-017-5733-2
Yañez, O., Piot, N., Dalmon, A., de Miranda, J. R., Chantawannakul, P., Panziera, D., et al. (2020). Bee Viruses: Routes of Infection in Hymenoptera. Front. Microbiol. 11. doi: 10.3389/fmicb.2020.00943
Yañez, O., Zheng, H.-Q., Hu, F.-L., Neumann, P., Dietemann, V. (2012). A Scientific Note on Israeli Acute Paralysis Virus Infection of Eastern Honeybee Apis Cerana and Vespine Predator Vespa Velutina. Apidologie 43, 587–589. doi: 10.1007/s13592-012-0128-y
Keywords: wild bees, pollinators, pathogen transmission, managed honey bees, honey bee pathogens, Hymenoptera, biodiversity, spillover
Citation: Cilia G, Flaminio S, Zavatta L, Ranalli R, Quaranta M, Bortolotti L and Nanetti A (2022) Occurrence of Honey Bee (Apis mellifera L.) Pathogens in Wild Pollinators in Northern Italy. Front. Cell. Infect. Microbiol. 12:907489. doi: 10.3389/fcimb.2022.907489
Received: 29 March 2022; Accepted: 23 May 2022;
Published: 30 June 2022.
Edited by:
Joseph Horzempa, West Liberty University, United StatesReviewed by:
Hannah Levenson, North Carolina State University, United StatesCopyright © 2022 Cilia, Flaminio, Zavatta, Ranalli, Quaranta, Bortolotti and Nanetti. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Rosa Ranalli, cm9zYS5yYW5hbGxpQGNyZWEuZ292Lml0
†These authors have contributed equally to this work and share first authorship
‡These authors have contributed equally to this work and share last authorship
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.