- 1Department of Pediatric Dentistry, Nanjing Stomatological Hospital, Medical School of Nanjing University, Nanjing, China
- 2Nanjing Stomatological Hospital, Medical School of Nanjing University, Nanjing, China
- 3School/Hospital of Stomatology, Zhejiang Chinese Medical University, Hangzhou, China
- 4Department of Periodontology, Nanjing Stomatological Hospital, Medical School of Nanjing University, Nanjing, China
Different small molecules have been developed to target cariogenic bacteria Streptococcus mutans. Based on target-based designing and in silico screening, a novel diaryl urea derivative, 1,3-bis[3,5-bis(trifluoromethyl)phenyl]urea (BPU), has previously been found effective in inhibiting the growth of S. mutans. However, the exact mechanism remains unclear. This current study aimed to explore the antimicrobial and antibiofilm effects of BPU on S. mutans and locate key enzymes and biological processes affected by the molecule via in silico molecular docking analysis and transcriptomic profile. Our in vitro results confirmed that BPU was capable of inhibiting planktonic growth as well as biofilm formation of S. mutans. The virtual binding analysis predicted that the molecule had strong binding potentials with vital enzymes (3AIC and 2ZID) involved in extracellular exopolysaccharide (EPS) synthesis. The predicted inhibitive binding was further confirmed by in vitro quantification of EPS, which found a decreased amount of EPS in the biofilms. The transcriptomic profile also found differential expression of genes involved in EPS synthesis. Moreover, the transcriptomic profile implied alterations in stress response and nitrogen metabolism in S. mutans treated with BPU. Examination of differentially expressed genes involved in these biological processes revealed that altered gene expression could contribute to impaired growth, biofilm formation, and competitiveness of S. mutans. In conclusion, the novel diaryl urea derivative BPU can inhibit the virulence of S. mutans by affecting different biological processes and serves as a potent anti-caries agent.
Introduction
The prevention and treatment of dental caries have long been associated with the interference of cariogenic bacteria in the oral cavity. Traditional anti-caries agents, including fluoride and antibiotics, have been reported to contribute to local and systematic toxicity, as well as drug resistance (Liao et al., 2017; Qiu et al., 2020). Novel anti-caries chemicals are required to inhibit the growth and metabolism of cariogenic microorganisms while exerting limited side effects.
Various small molecules have been developed to target cariogenic microorganisms including the notorious species, Streptococcus mutans (Cui et al., 2019; Yang et al., 2021). These small molecules usually have unique structures to target specific metabolic pathways and vital enzymes in bacterial cells (Zhang et al., 2015). Different approaches, including drug repositioning, library screening, natural products screening, and target-based designing, have been applied to identify small molecules that have anti-caries potentials (Yang et al., 2021). Among them, target-based designing and in silico screening from the small-molecule library have been of special interest, as they provide hits with good specificity and high throughput (Younson and Kelly, 2004; Nijampatnam et al., 2016; Nijampatnam et al., 2021).
Based on the abovementioned methods, a previous study identified a series of small molecules that have the potential to enhance fluoride toxicity in bacterial cells (Nelson et al., 2015). One of the small molecules, 1,3-bis[3,5-bis(trifluoromethyl)phenyl]urea (BPU; Figure 1A), was found to be especially effective in inhibiting the growth of Escherichia coli and S. mutans when used in combination with fluoride (Nelson et al., 2015). The enhanced antimicrobial ability was suggested to be associated with the trifluoromethyl substituents on the aryl rings, which facilitate fluoride/chloride uptake and/or retention (Busschaert et al., 2012; Nelson et al., 2015). Further investigation of the molecule structure reveals that the trifluoromethyl substituents are not the only structure related to antimicrobial effects. The diaryl rings in the small molecule contribute as hydrogen-bond donors and have been proved to be able to bind and inhibit specific bacterial proteins including DNA gyrase B and penicillin-binding protein 1a (Limban et al., 2020; Sroor et al., 2021). Moreover, similar urea derivatives have been used to target pathogenic bacteria such as Staphylococcus aureus, Pseudomonas aeruginosa, and Enterococcus faecalis (Wu, 1965; Gunduz et al., 2020). The preliminary structure investigation indicated that BPU has the potential to exert an inhibitive effect against S. mutans alone and act as a novel anti-caries agent. However, no in vitro result has been reported to support the antimicrobial effect of BPU alone on S. mutans. Also, the exact inhibition site and affected biological processes remain unknown.
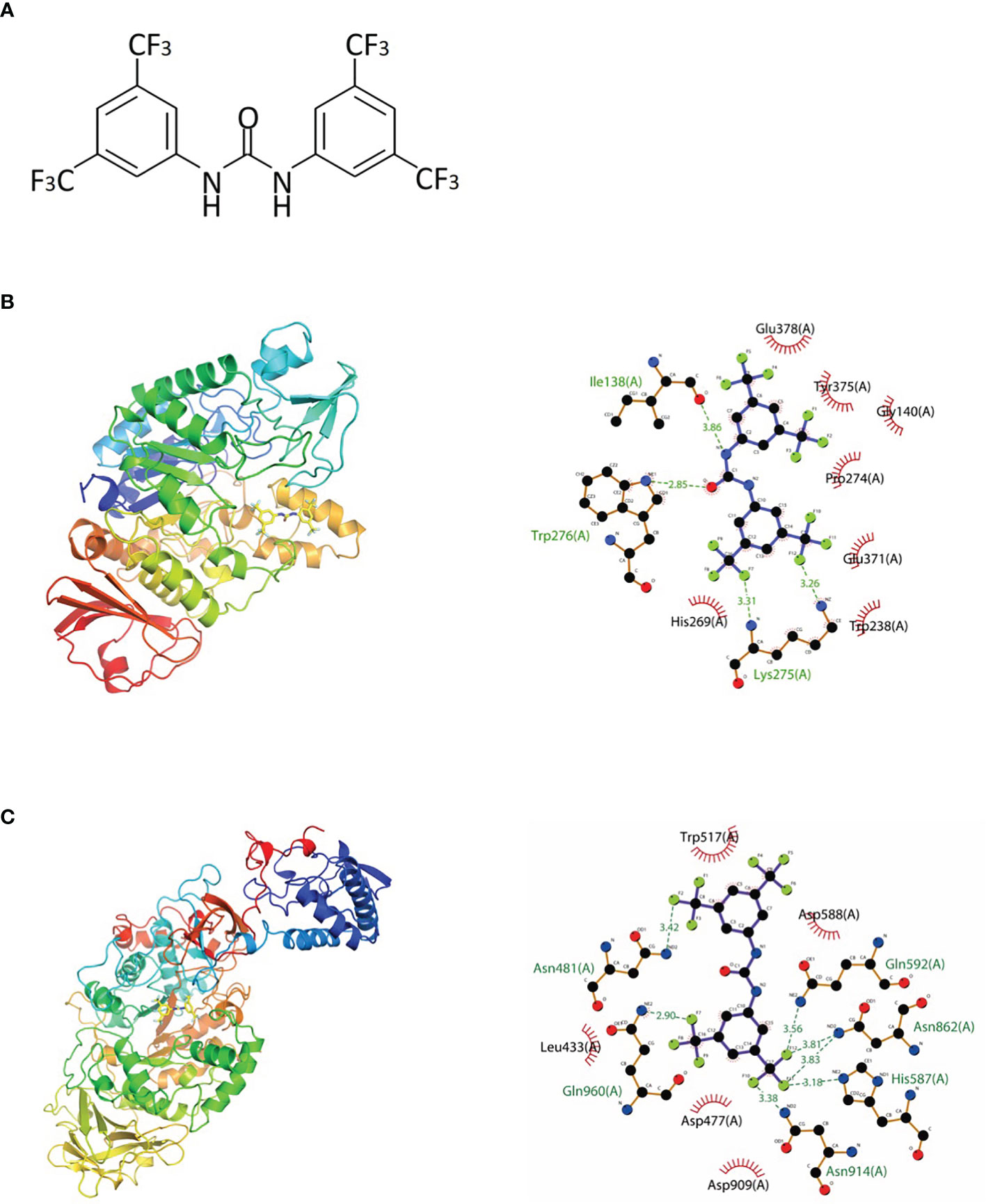
Figure 1 Chemical structure and molecular docking analysis. (A) Chemical structure of 1,3-bis[3,5-bis(trifluoromethyl)phenyl]urea (BPU) molecule. (B, C) Predicted binding mode (left, 3D; right, 2D) of BPU molecule with 2ZID (B) and 3AIC (C). Hydrogen bond is shown with green dashed lines (right). Hydrophobic interaction is shown with red “eyelashes”.
In this study, we investigated the antimicrobial and antibiofilm effects of the novel diaryl urea derivative, namely, BPU, against S. mutans UA159. In silico molecular docking analysis was applied to predict the binding potential and binding site of the small molecule with different proteins in S. mutans cells. Transcriptomic analysis was performed to validate the predictions and locate major biological processes that were affected by the small molecule.
Materials and Methods
In Silico Molecular Docking
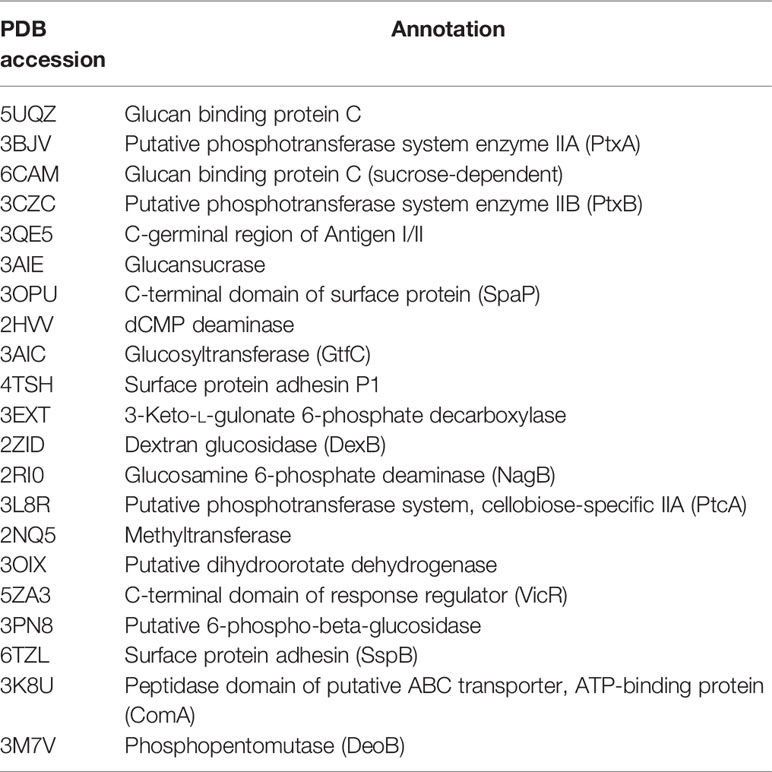
The three-dimensional structure of BPU was downloaded from PubChem (Cas no. 3824-74-6). The structure was geometrically optimized and energy minimized using ChemBio3D Ultra 14.0 software. A total of 20 proteins originating from S. mutans were selected for in silico molecular docking (Table 1). These proteins were selected because they have been reported to be associated with the virulence of S. mutans and their crystal structures were available online. Crystal structures of targeted proteins were obtained from Protein Data Bank (PDB) with accession codes. The protein structures were processed step by step including the removal of original ligands and water molecules, the addition of polar hydrogen atoms, and charge calculation and distribution using PyMOL v2.3.0 and AutoDocktools v1.5.6. Docking analysis was performed using AotoDock Vina v1.1.2 with default parameters. Results with the strongest protein–ligand interactions were visualized and analyzed with PyMOL v2.3.0 and LigPlot v2.2.4.
Experimental Chemicals, Bacterial Strain, and Growth Conditions
Different amount of BPU (Sigma-Aldrich, CA, USA) was dissolved in acetonitrile (Aladdin, Nanjing, China) for in vitro studies. S. mutans UA159 was obtained from the State Key Laboratory of Oral Disease (Sichuan University, Chengdu, China). Bacteria were routinely grown in brain heart infusion (BHI) broth (BD, NJ, USA) or on BHI agar plates at 37°C anaerobically (10% H2, 5% CO2, and 85% N2). For biofilm formation, 1% sucrose (Sigma-Aldrich, CA, USA) was added to the BHI broth.
Planktonic Growth Assay
The antimicrobial effect of BPU on planktonic S. mutans UA159 was tested using a growth curve. S. mutans UA159 was incubated in BHI broth until the early log phase (OD600 = 0.2). Bacterial cells were pelleted and resuspended with fresh BHI broth containing 1, 2, 5, 10, 20, and 50 µg/ml of BPU. For blank control, fresh BHI broth containing the same volume of acetonitrile was used to resuspend bacterial cells. Resuspensions measuring 200 µl were then added into wells of a sterile 96-well microplate. The microplate was sealed with a transparent sealer and incubated at 37°C for 12 h. Growth was monitored by recording the optical density at 600 nm (OD600) every 30 min with Spectra MaxM3 (Molecular Devices, CA, USA).
Crystal Violet Assay
The biomass of S. mutans biofilms with or without BPU treatment was quantified with a crystal violet staining assay. S. mutans UA159 was incubated until mid-log phase (OD600 = 0.5), pelleted, and resuspended with fresh BHI broth supplemented with 1% (w/w) sucrose (BHIS). Resuspensions were transferred to a sterile 96-well microplate, and BPU was added to reach different final concentrations (1, 2, 5, 10, 20, and 50 µg/ml). The same volume of acetonitrile was added to the control group. After anaerobic incubation at 37°C for 24 h, all wells were washed with sterile phosphate-buffered saline (PBS) to remove loose cells. Biofilms attached to the bottom of the wells were fixed with 4% paraformaldehyde (Sigma-Aldrich, CA, USA) for 15 min followed by staining with 0.01% crystal violet solution (Adamas, Shanghai, China) for 8 min. Solutions were then removed, and biofilms were again washed with PBS twice. Acetic acid (33% v/v, Sigma-Aldrich, CA, USA) was added to each well to destain for 10 min. The destaining solution was then transferred to a new 96-well plate, and OD575 was recorded with Spectra MaxM3.
Biofilm Formation on Saliva-Coated Hydroxyapatite Discs
Biofilms of S. mutans UA159 were formed on saliva-coated hydroxyapatite (sHA) discs using a previously described method with a few modifications (Wang et al., 2020). Briefly, HA discs (8 mm in diameter and 2 mm in thickness; Baiameng Bioactive Materials, Chengdu, China) were sonicated in deionized water for 10 min and sterilized at 121°C for 15 min before use. Clarified saliva was prepared by centrifuging whole unstimulated saliva of a healthy human donor at 4,000 rpm for 20 min at 4°C, followed by filtration through 0.22-µm polyethersulfone membrane (Koo et al., 2010). The sterile HA discs were placed into 24-well plates. Each well contained clarified saliva mixed with the same volume of desorption buffer (containing 500 mM of KCl, 10 mM of CaCl2, 1 mM of MgCl2, 6.2 mM of K2HPO4, and 14 mM of KH2PO4). After incubation in clarified saliva and desorption buffer at 37°C for 30 min, the sHA discs were ready for use.
S. mutans UA159 was incubated in BHI broth until the mid-log phase (OD600 = 0.5). Cells were then pelleted and resuspended in BHIS. Resuspended bacterial cells were transferred to sterile 24-well plates with different concentrations of BPU or the same volume of acetonitrile. The abovementioned sHA discs were placed in each well. Plates were incubated anaerobically at 37°C for 24 h. Biofilms formed on the surface of sHA discs were used for further examinations.
Scanning Electron Microscopy
Twenty-four-hour biofilms attached to the surface of sHA discs were harvested and washed twice with cysteine peptone water (CPW; containing yeast extract, peptone, sodium chloride, and cysteine HCl, pH 7.2). Biofilms were fixed with 2.5% glutaraldehyde (Adamas, Shanghai, China) at 4°C overnight, followed by serial dehydration with ethanol (30%, 40%, 50%, 60%, 70%, 80%, 90%, and 100%). Samples were dried and coated with gold before observation with SEM (Quanta 400 FEG, FEI, Hillsboro, OR, USA).
pH Drop Examination
Acid production of S. mutans biofilms was examined with a pH drop of the supernatant. Culture media of the abovementioned biofilms measuring 2 ml formed on sHA discs were taken at 0, 4, 8, 12, and 24 h and centrifuged at 4°C at 4,000 rpm for 10 min. The pH of the supernatant was measured with an electronic pH meter (FiveEasy Plus, FE28-standard, Mettler-Toledo, Schwerzenbach, Switzerland).
Confocal Laser Scanning Microscopy Analysis
A commercial LIVE/DEAD BacLight Viability Kit (Life Technologies, NY, USA) was used to stain viable and dead cells in 24-h biofilms. S. mutans biofilms formed on sHA discs were harvested after 24-h incubation. The staining of bacterial cells was processed according to the manufacturer’s instructions. Viable cells and dead cells were respectively stained with SYTO 9 (excitation, 480 nm; emission, 500 nm) and propidium iodide (PI; excitation, 490 nm; emission, 635 nm). Samples were observed with confocal laser scanning microscopy (CLSM; NikonA1; Nikon Inc., Tokyo, Japan). Images were taken at an interval of 10 µm. Integrated fluorescence density was quantified with ImageJ software (v1.48, National Institutes of Health, USA).
Water-Insoluble Exopolysaccharide Determination
Production of water-insoluble exopolysaccharide (EPS) by S. mutans biofilms was examined qualitatively by fluorescence staining and quantitatively by the anthrone–sulfuric method (Tang et al., 2019). For fluorescence staining, Alexa Fluor 647 (Life Technologies, NY, USA) was used to label EPS, and SYTO 9 was used to label bacterial cells. Alexa Fluor 647 was added to the BHIS medium at the beginning of 24-h biofilm formation. Biofilms were stained with SYTO 9 at the end of the 24-h biofilm formation experiment. Samples were then observed using CLSM with the same procedure as live/dead staining.
To quantify EPS synthesis with the anthrone–sulfuric method, 24-h biofilms were harvested and scraped from sHA discs and washed with PBS buffer. The planktonic cells and suspension were removed by centrifuging the mixture at 4,000 rpm for 10 min at 4°C. The precipitate was mixed with 0.4 mol/L of NaOH and incubated for 2 h at 37°C. The mixture was centrifuged again, and the suspension was collected and transferred to a new EP tube. The suspension was mixed with three volumes of the anthrone–sulfuric acid reagent (Macklin, Shanghai, China) and heated on a heat block at 95°C for 5 min until the reaction was complete. The solution was cooled to room temperature before being transferred to a new 96-well plate. Absorbance at 625 nm was recorded, and the amount of polysaccharide was calculated according to the standard curve.
RNA Sequencing and Data Analysis
For transcriptomic analysis, S. mutans UA159 was grown in BHI until the log phase (OD600 = 0.5). Cells were harvested and resuspended with fresh BHI. Cells were challenged with either a final concentration of 2 µg/ml of BPU or the same volume of acetonitrile for 1 h. This concentration was chosen mainly based on results from crystal violet assay to make sure that there was enough BPU to induce significant alterations in gene expression, while potential toxicity was kept as low as possible. Samples were then harvested with centrifugation and kept at −80°C until used. Total RNA was extracted using TRIzol reagent (Invitrogen, OR, USA). The concentration and quality of RNA were determined with Nanodrop 2000 spectrophotometer (Thermo Scientific, MA, USA). rRNA was removed from total RNA using Zymo-Seq RiboFree Total RNA Library Kit. cDNA library was constructed by Shanghai Personal Biotechnology Co. Ltd (Shanghai, China). RNA sequencing was performed on Novaseq 6000 platform (Illumina, San Diego, CA, USA).
Data were filtered and controlled for quality before analysis. Filtered reads were mapped to the S. mutans UA159 reference genome (RefSeq NC_004350.2) using Bowtie 2 (version 2.2.6). Differentially expressed genes (DEGs) were recognized as transcripts with |log2FoldCHange| > 1 and p < 0.05. Differential expression analysis was performed using DESeq (version 1.30.0). All DEGs were annotated by searching the Gene Ontology (GO) databases. All genes were mapped to terms in the GO database, and numbers of differentially enriched genes in each term were calculated. GO enrichment analysis was performed using the topGO R package on the differential genes. GO terms with significantly enriched differential genes (p < 0.05 according to the hypergeometric distribution method) were identified to determine the main biological functions of DEGs. Three individual samples were sequenced for each group.
Statistical Analyses
Data were analyzed using Prism (version 9.0.0). All experiments were performed at least in triplicate. Student’s t-test or one-way ANOVA was employed to compare data from two or more groups. A p-value of <0.05 was considered statistically significant.
Results
In Silico Screening for Potential Binding Site
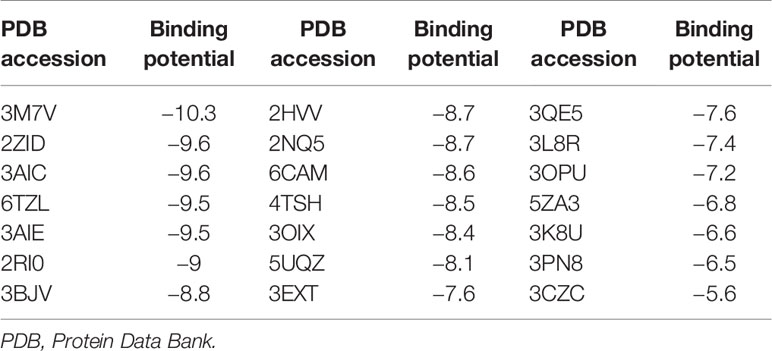
The binding potentials of BPU with 20 selected proteins originating from S. mutans are shown in Table 2. A higher absolute value indicates a stronger binding ability. Proteins with the highest scores were 3M7V (phosphopentomutase), 2ZID (dextran glucosidase), and 3AIC (glucosyltransferase). These three proteins were further visualized to predict the binding mode. Hydrogen bond and hydrophobic interaction acted as the main interaction forces. We further examined the potential binding sites occupied by the BPU molecule. While there was not enough information on 3M7V, we did find interesting results for 2ZID and 3AIC (Figure 1). The molecule can bind several vital amino acid residues of 2ZID, which were involved in the active binding pocket (Lys 275, His269, and Pro274), recognition of substrate (Glu371), and high level of enzyme activity toward long-chain substrates (Trp238) (Hondoh et al., 2008). Also, the molecule may competitively bind several conserved amino acid residues of 3AIC (His587, Asp588, Asp477, Asn481, and Asp909), which were responsible for the recognition of glucosyl moiety of the primary sucrose (Ito et al., 2011). One of the amino acid residues (Gln592) bound by the molecule was found to be involved in the catalytic domains of 3AIC (Ito et al., 2011).
Inhibition of Planktonic Growth
Compared to the control groups (treated with deionized water or acetonitrile), incubation with different levels of BPU significantly inhibited the growth of planktonic S. mutans UA159 in BHI broth (Figure 2). With a trace amount of drug in the culture (1 µg/ml), hardly any growth could be noticed in the bacterial culture.
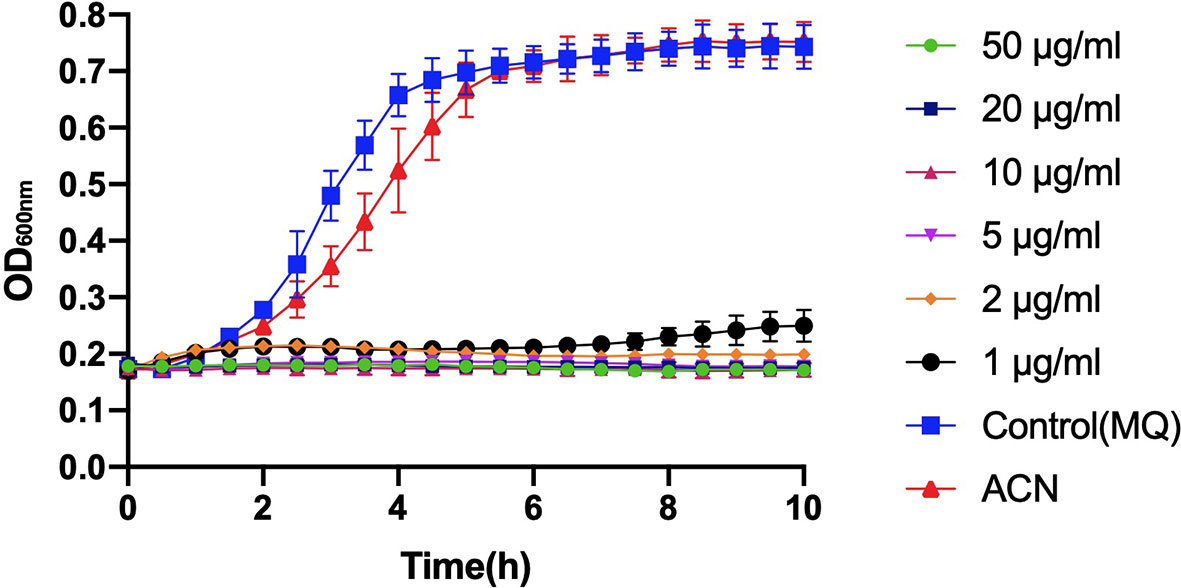
Figure 2 Growth curve of planktonic Streptococcus mutans UA159 incubated with 1, 2, 5, 10, 20, and 50 µg/ml of 1,3-bis[3,5-bis(trifluoromethyl)phenyl]urea (BPU). The same volumes of deionized water (MQ) and acetonitrile (ACN) were added to control groups. Values indicate means ± SDs from three independent experiments.
Inhibition of Biofilm Formation and Acid Production
Biomass of S. mutans UA159 biofilms treated with different levels of BPU was determined with crystal violet assay. As shown in Figure 3A, no significant difference was noticed between groups treated with deionized water and acetonitrile. Treatment of 1 µg/ml of BPU resulted in the loss of half of the biomass as compared to the control groups. Higher concentrations of BPU almost completely inhibited biofilm formation. Similar results were visualized with SEM examination (Figure 3B). Biofilms on sHA discs treated with 1 and 2 µg/ml of BPU were much thinner as compared to the control group. While biofilms of the control group appeared dense and highly stereoscopic, biofilms treated with BPU seemed plainer with more porous structures and less extracellular matrix in the system (Figure 3B).
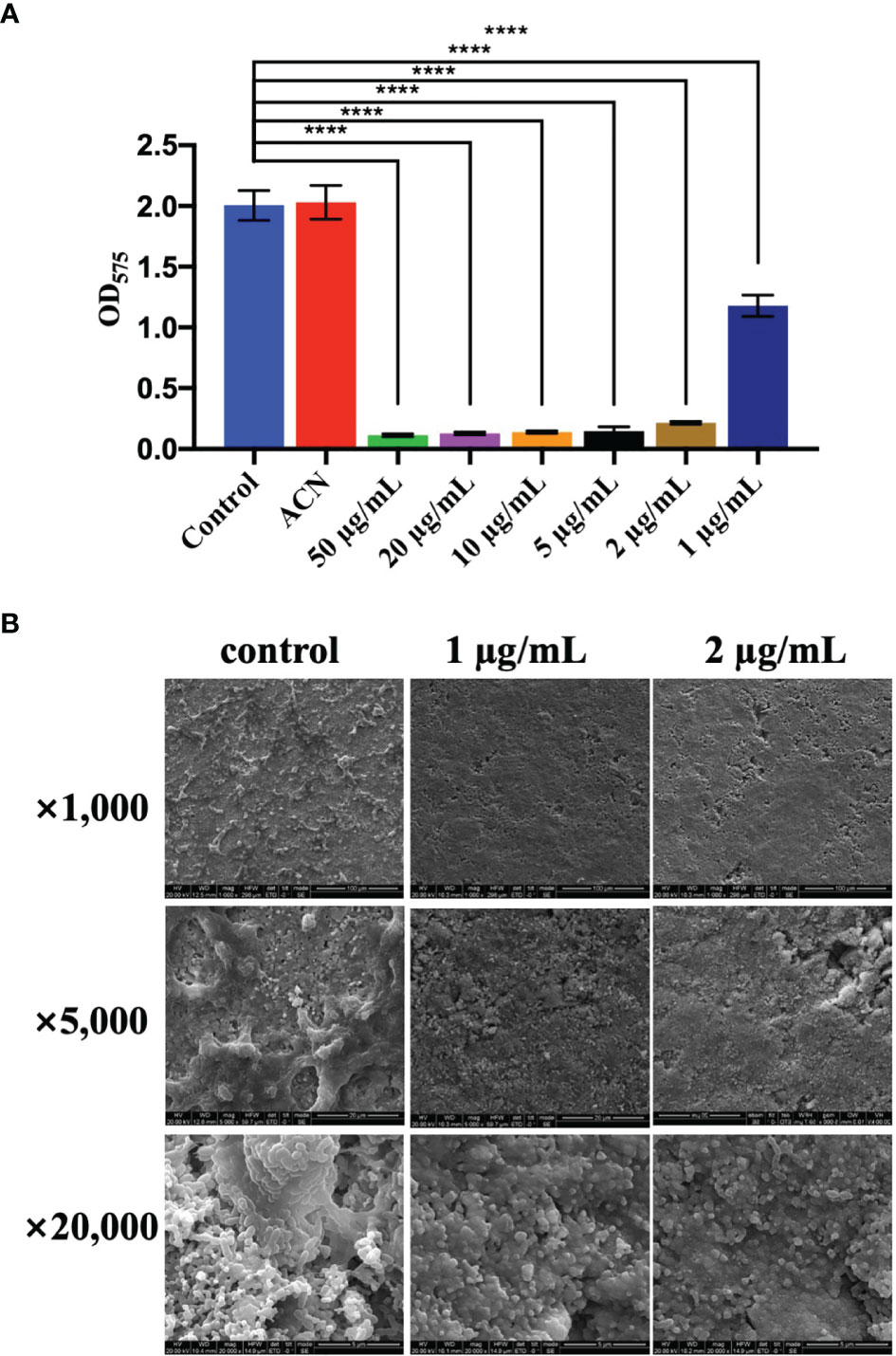
Figure 3 Effect of different levels of 1,3-bis[3,5-bis(trifluoromethyl)phenyl]urea (BPU) on biofilm formation of Streptococcus mutans UA159. (A) Biomass of biofilms quantified with crystal violet staining assay. (B) Representative SEM images of biofilms treated with 1 and 2 µg/ml of BPU or acetonitrile (control). Means ± SDs from three independent experiments are shown. **** indicates p < 0.0001.
CLSM also found severe inhibition of biofilm formation and maturation by BPU (Figure 4). Strong green fluorescence in the control group (treated with acetonitrile) indicated a large number of viable cells in the biofilms. Biofilms treated with BPU exhibited significantly less green fluorescence and enhanced red fluorescence as compared to the control group (Figure 4A). Quantified mean fluorescence intensity confirmed that the signal of green fluorescence was remarkably lower in BPU-treated groups when compared with the control group (Figure 4B).
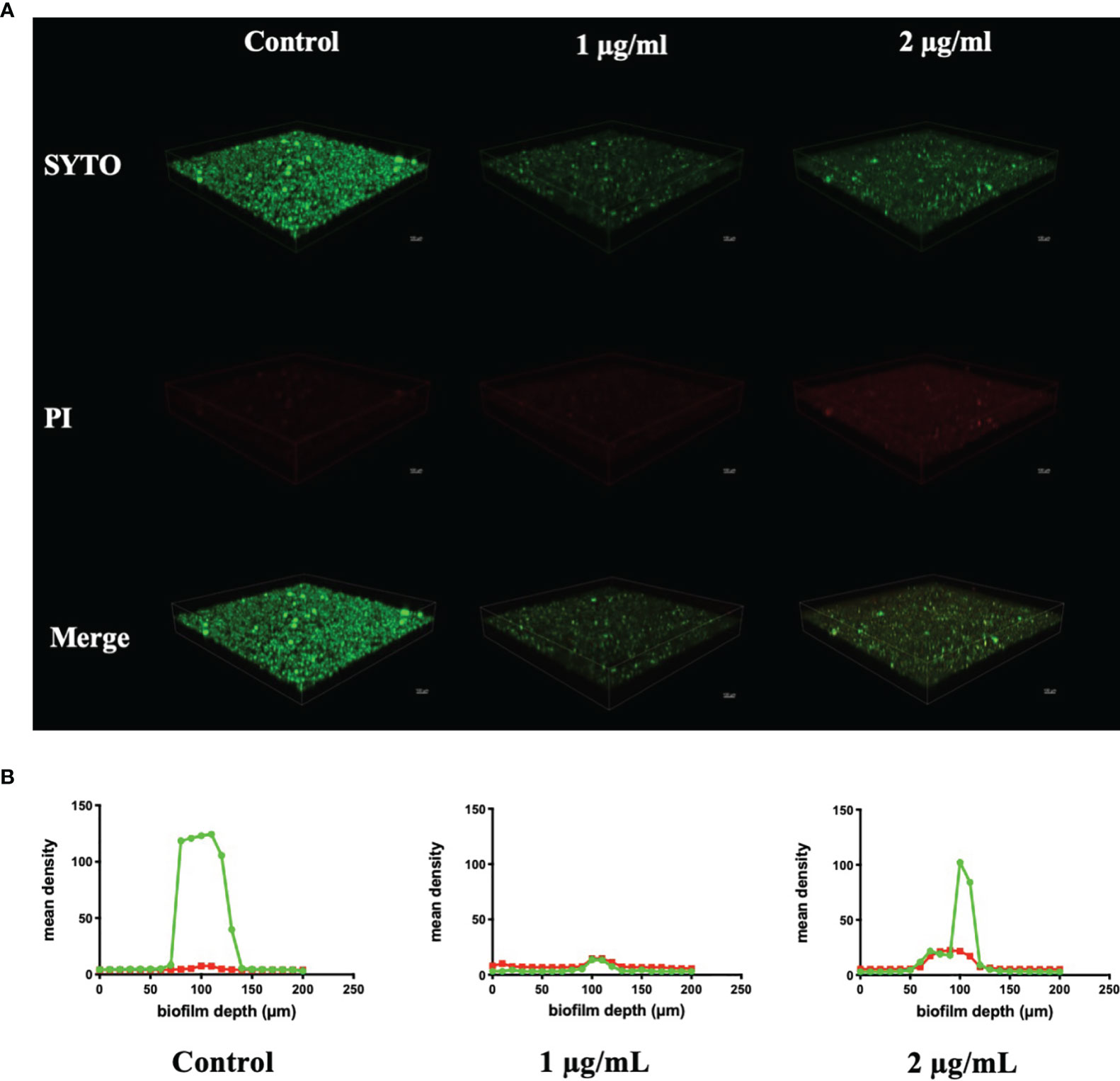
Figure 4 Live/dead staining of Streptococcus mutans UA159 biofilms treated with 1 and 2 µg/ml of 1,3-bis[3,5-bis(trifluoromethyl)phenyl]urea (BPU). (A) Representative three-dimensional visualization of live (green) and dead (red) cells in S. mutans biofilms formed on saliva-coated hydroxyapatite (sHA) discs. (B) Mean density of green and red fluorescence throughout S. mutans biofilms quantified using ImageJ software.
Figure 5 shows the pH change of the supernatant of S. mutans biofilms treated with or without BPU within 24 h. As expected, biofilms treated with acetonitrile (control) were able to decrease environmental pH from 7.40 to 3.58 ± 0.03 within 24 h. Once treated with BPU, very little pH drop was noticed. At 24 h, pH of supernatant was similar for the 1 µg/ml and 2 µg/ml groups (pH 6.84 ± 0.08 and 6.87 ± 0.02, respectively).
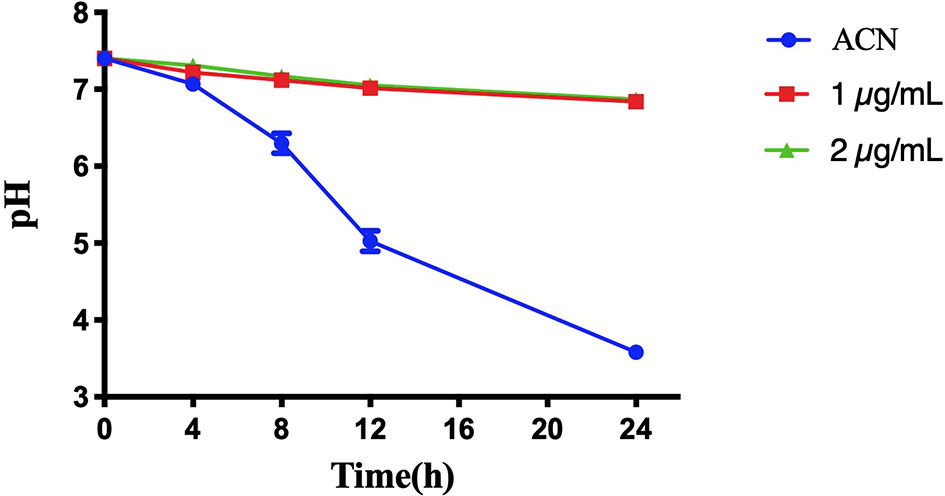
Figure 5 pH change of the supernatant of Streptococcus mutans UA159 biofilms treated with or without 1,3-bis[3,5-bis(trifluoromethyl)phenyl]urea (BPU). Values are presented as means ± SDs from three independent experiments. ACN, acetonitrile.
Inhibition of Exopolysaccharide Synthesis by Streptococcus mutans Biofilms
EPS synthesis by S. mutans biofilms was examined using CLSM analysis and the anthrone–sulfuric method. CLSM images clearly showed that treatment of 1 and 2 µg/ml of BPU disrupted the ability of biofilms to synthesize EPS. Along with the decrease in EPS, the number of bacterial cells in biofilms also reduced significantly (Figure 6). While the control group exhibited thick, dense bacterial aggregations, groups treated with BPU appeared to be more dispersed with much thinner structures and fewer extracellular matrix connections (Figure 6).
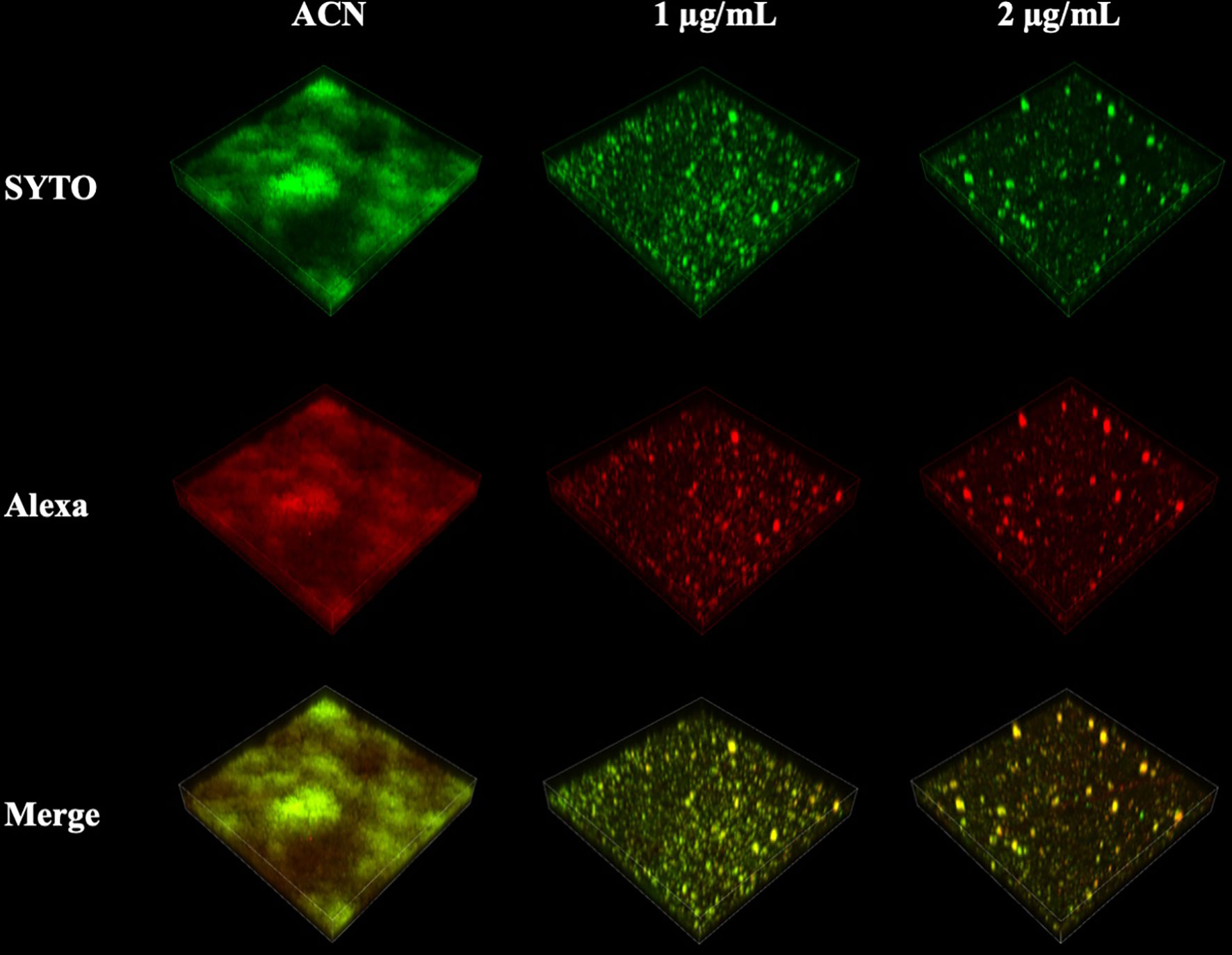
Figure 6 Confocal laser scanning microscopy (CLSM) images of Streptococcus mutans biofilm exopolysaccharide (EPS) synthesis affected by 1,3-bis[3,5-bis(trifluoromethyl) phenyl]urea (BPU). Green, bacterial cells (stained with SYTO 9); red, EPS (stained with Alexa 647). Representative three-dimensional images are exhibited.
The amount of EPS synthesized by biofilms treated with or without BPU was further examined using the anthrone–sulfuric method. Groups treated with deionized water and acetonitrile synthesized 0.53 ± 0.06 and 0.51 ± 0.06 mg/ml of EPS, respectively. The amount of EPS from two experimental groups (treated with 1 and 2 µg/ml of BPU) was both below the detection limit.
Transcriptomic Analysis of Streptococcus mutans Treated With Diaryl Urea Derivative
Transcriptomic analysis revealed that a total of 701 DEGs were identified in the experimental group, including 270 upregulated genes and 431 downregulated genes. GO enrichment analysis indicated that a total of 68 GO terms involved in molecular function and biological processes were affected. A rich factor was calculated for 20 GO terms with the smallest false discovery rate (FDR) values (Figure 7). The results showed that GO term unfolded protein binding, protein folding, and exonuclease activity had the largest rich factors. A directed acyclic graph (DAG) was used to further display the relationship of the enriched GO terms. In cellular component, while not statistically significant, most differences were found in the cytoplasm (GO: 0005737), extracellular region (GO: 0005578), cell wall (GO: 0005618), and primosome complex (GO: 1990077). In molecular function, the difference was mostly concentrated in exonuclease activity (GO: 0004527), unfolded protein binding (GO: 0051082), and nucleic acid binding (GO: 0003676). In biological process, genes associated with protein folding (GO: 0006437), regulation of primary metabolic process (GO: 0080090), regulation of nitrogen compound metabolic process (GO: 0051171), and cellular nitrogen compound metabolic process (0031323) were most significantly differentially expressed (Figure 7).
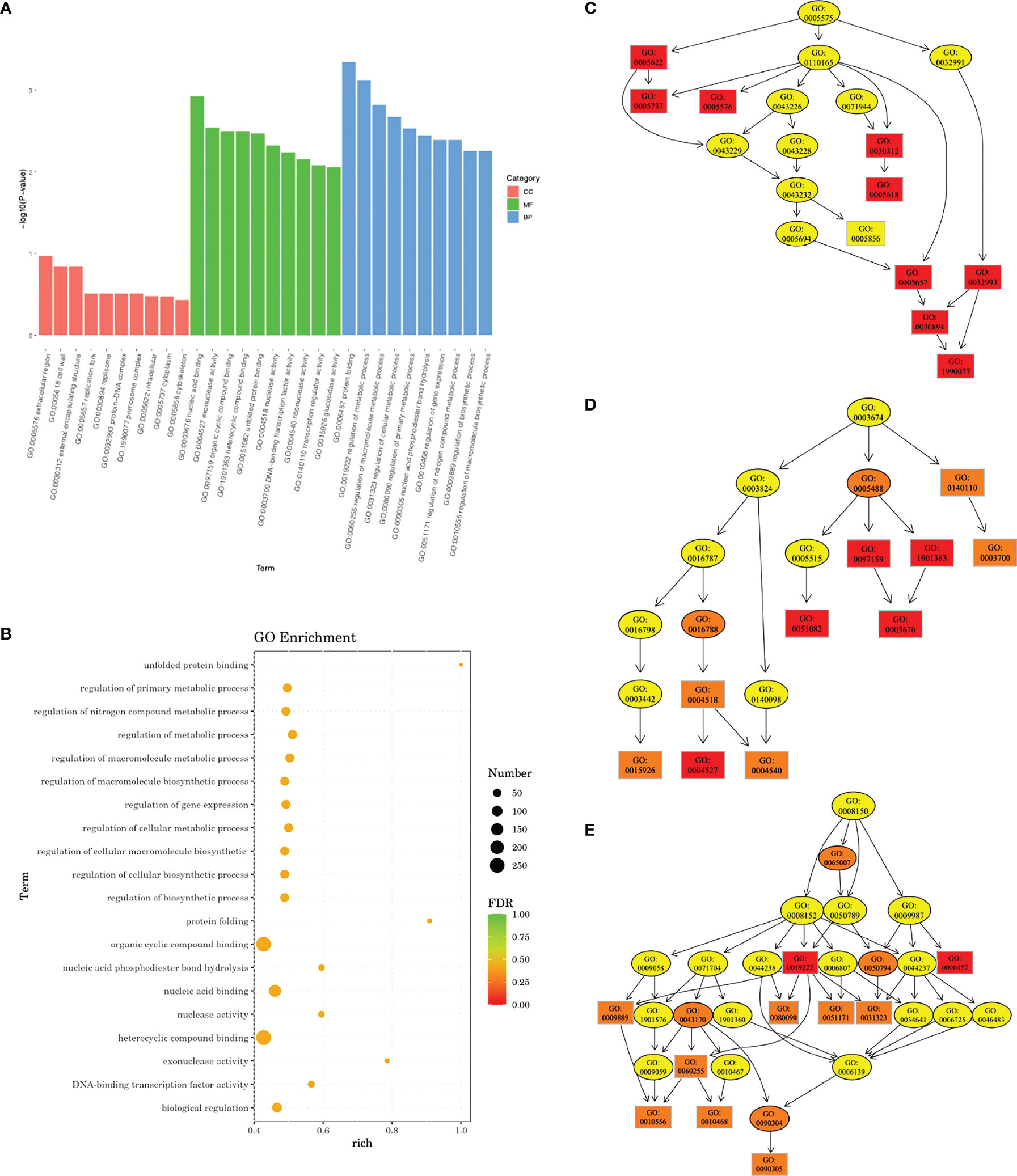
Figure 7 Gene Ontology (GO) enrichment analysis of transcriptomic profile. (A) Bar plot showing the most significantly enriched GO terms. CC, cellular component; MF, molecular function; BP, biological process. (B) Rich factors of 20 GO terms with the smallest false discovery rate (FDR) values. Directed acyclic graph (DAG) in cellular component (C), molecular function (D), and biological process (E). Deeper color indicates a higher level of enrichment. GO accession number is shown.
We further looked into DEGs related to the regulation of metabolic processes and DEGs related to the regulation of macromolecule metabolic processes (Supplementary Figures 1, 2; Supplementary Tables 1, 2). Eleven genes involved in the regulation of metabolic process were upregulated, and 59 genes were downregulated. Most of them were associated with transcriptional regulation, sucrose metabolism, and nitrogen metabolism. For regulation of macromolecule metabolic process, 10 genes were upregulated and 58 downregulated. Among them were genes involved in EPS synthesis (sacR, scrR, and msmR), stress tolerance (rex, perR, spxA, clpE, and ciaR), and nitrogen metabolism (glnB, glnR, ciaR, and clpE).
Differential expression of genes associated with protein functions of 3AIC, 2ZID, and 3M7V was also examined (Figure 8). As 3AIC and 2ZID are both proteins involved in EPS synthesis, genes associated with EPS synthesis (gtfB, gtfC, ftf, dexA, dexB, and scrB) were selected. For 3M7V, its encoding gene (deoB) and two related genes involved in the pentose phosphate pathway (gapN and rpiA) were selected. The heatmap shows that most genes involved in EPS synthesis, including encoding genes of 3AIC and 2ZID (gtfC and dexB), were upregulated. The encoding gene of 3M7V (deoB) was not found differentially expressed. Its related genes showed different regulatory directions (gapN was upregulated and rpiA was downregulated).
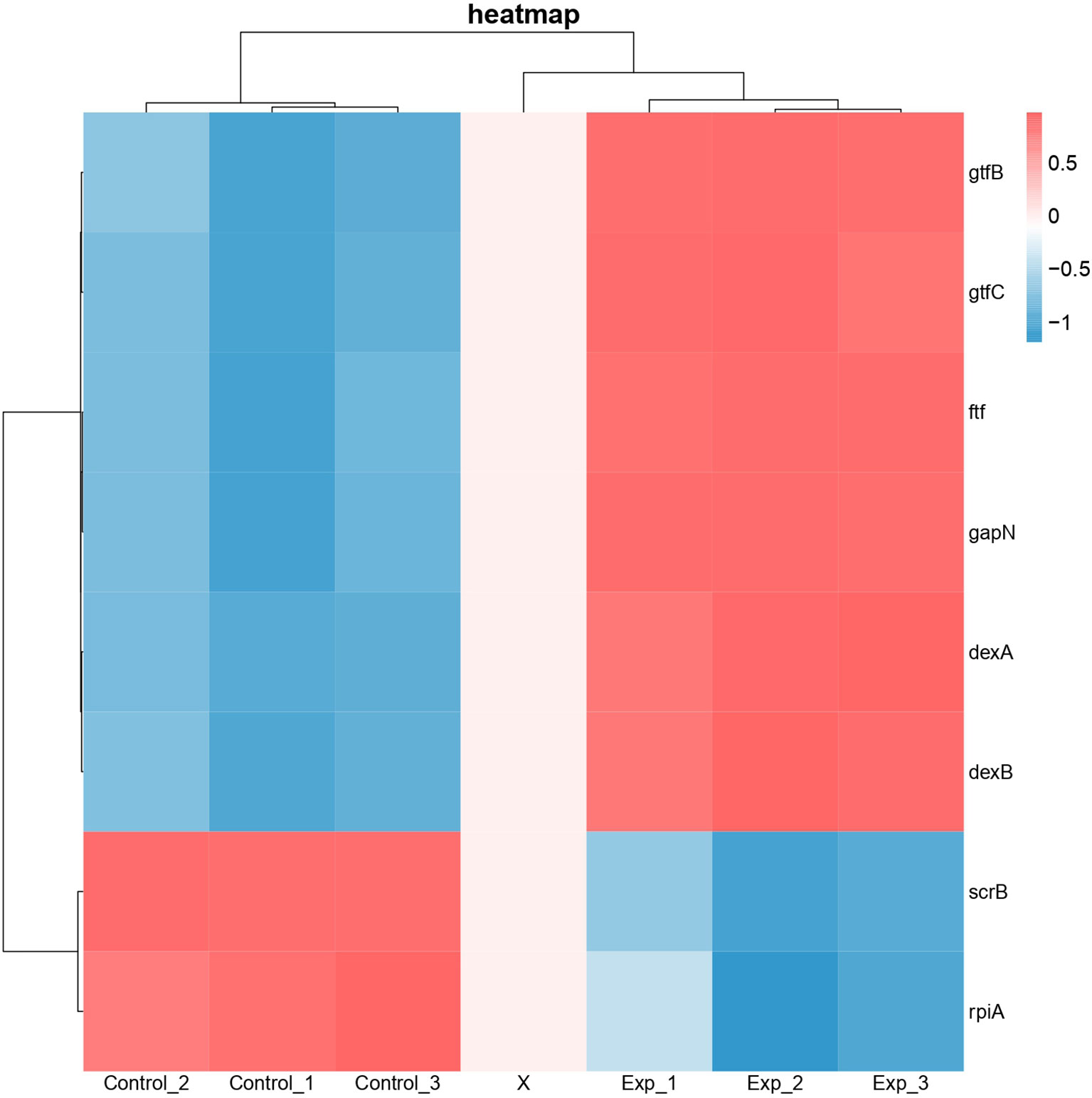
Figure 8 Heatmap of differentially expressed genes associated with protein functions of 3AIC, 2ZID, and 3M7V. Control groups were treated with acetonitrile. Experimental groups were treated with 2 µg/ml of 1,3-bis[3,5-bis(trifluoromethyl)phenyl]urea (BPU). Genes gtfB, gtfC, ftf, dexA, dexB, and scrB are involved in exopolysaccharide biosynthesis. Genes gapN and rplA are involved in pentose phosphate pathway. Blue indicates lower expression, and red indicates higher expression.
Discussion
Small molecule compounds have been extensively explored as potent anti-caries agents targeting caries-associated bacteria. In this study, we described the antimicrobial and antibiofilm effects of a novel diaryl urea derivative, BPU. In silico simulation indicated that the small molecule compound had the potential to bind the key enzymes involved in EPS biosynthesis and the pentose phosphate pathway. The transcriptomic study suggested that except for EPS synthesis, nitrogen metabolism and stress-responsive pathway were also affected by the small molecule.
Diaryl rings have previously been suggested to be able to bind specific bacterial proteins (Limban et al., 2020; Sroor et al., 2021) and act as enzyme inhibitors. Our in silico molecular docking also found strong binding potential between BPU and several enzymes involved in the biological processes of S. mutans. Interestingly, 2 of the top 3 hits (2ZID and 3AIC) are involved in the biosynthesis of EPS. EPS is the main component of the extracellular matrix of bacterial biofilms, and the ability to synthesize EPS is a vital virulence factor of S. mutans (Koo et al., 2013; Krzysciak et al., 2014; Klein et al., 2015). Protein 2ZID is a dextran glucosidase hydrolyzing the α-1,6-glucosidic linkage of α-1,6-D-glucans and derived oligosaccharides (Hondoh et al., 2008). Protein 3AIC is a glucosyltransferase responsible for water-insoluble and water-soluble glucan syntheses in S. mutans (Ito et al., 2011). The binding mode prediction shows that the diaryl urea derivative molecule binds to either substrate binding pocket or catalytic domains of the two proteins. This indicates that the binding of the molecule can competitively inhibit the activity of 3AIC and 2ZID. A review of the literature shows that a number of 3AIC inhibitors have been proved effective in inhibiting biofilm formation of S. mutans and thus caries development (Ren et al., 2016; Zhang et al., 2017; Lin et al., 2021). In this study, we noticed significantly less biofilm formation after BPU treatment with crystal violet assay and SEM observation. CLSM examination and EPS quantification further confirmed that production of EPS was severely impaired by the treatment of BPU. Further transcriptomic analysis revealed that both genes encoding 3AIC and 2ZID (gtfC and dexB) were upregulated in S. mutans treated with BPU. The expression of several genes with associated functions, including gtfB, ftf, and dexA (Li and Burne, 2001; Koo et al., 2013; Yang et al., 2019), was also upregulated. This could be a result of negative feedback of the inhibited activity of corresponding proteins. Accordingly, genes encoding related transcription repressors (sacR and scrR) were downregulated (Supplementary Table 1) (Siegers and Entian, 1995; Hiratsuka et al., 1998), further confirming that bacterial cells were combating the inhibitive effect of BPU on EPS synthesis.
Our results suggest that the treatment of BPU not only inhibits biofilm formation but also affects planktonic growth. An examination of the transcriptomic profile revealed that, except for the EPS synthesis pathway, multiple genes involved in stress response were differentially expressed. Previous studies have found that the function of stress response proteins is important for bacterial physiology. Deficiency of oxidative stress response regulator Rex has been found to lead to an extended lag phase of planktonic growth of S. mutans (Bitoun et al., 2011). Moreover, expression of gtfC was also found significantly differentially expressed due to deficiency of Rex, leading to decreased biofilm formation and altered biofilm morphology (Bitoun et al., 2011). In our study, the downregulation of rex could be associated with decreased planktonic growth and biofilm formation. The regulatory effect of SpxA has been implicated in oxidative stress response, antibiotic response, and biofilm formation (Pamp et al., 2006; Kajfasz et al., 2015; Nilsson et al., 2019). Similarly, two genes involved in oxidative and thermal stress responses, clpE and ciaR, have the ability to affect the biofilm formation of S. mutans (Zhu et al., 2017; Biswas et al., 2021). All three abovementioned genes involved in stress response were found to be downregulated in the current study after BPU treatment (Supplementary Table 2). The involvement of stress response indicates new targets for novel anti-caries agents.
Additionally, relatively abundant changes in nitrogen metabolism in the BPU-treated group were observed in the transcriptomic profile. By metabolizing nitrogen-containing compounds, mainly amino sugars such as glucosamine and N-acetylglucosamine, bacteria are provided with essential materials for macromolecule synthesis (Ardin et al., 2014). The ability to utilize amino sugars is also closely associated with the competitiveness of bacteria to thrive in the oral cavity (Chen et al., 2020). The current study reports the downregulation of genes encoding a membrane ammonium transporter (glnB) (Ardin et al., 2014) and a glutamine synthetase regulator (glnR) (Chen et al., 2010). Results of in silico molecular docking suggest that a key factor affecting cellular entrance of amino acid, NagB (2RI0), has a relatively high binding potential with BPU molecule (−9 kcal/mol, Table 2) (Kawada-Matsuo et al., 2012). The potential inhibition of NagB together with the downregulation of associated genes supports that nitrogen metabolism may be inhibited by the diaryl urea derivative. The resulting decreased synthesis of virulence-related factors, including cell surface protein antigen and glucosyltransferase, can lead to less bacterial aggregation and thus less biofilm formation (Kawada-Matsuo et al., 2012).
The molecular docking analysis suggests that the BPU molecule has the potential to bind to an enzyme involved in the pentose phosphate pathway (3M7V, DeoB). According to previous studies, DeoB acts as a vital role in the pentose phosphate pathway, providing a precursor for phosphoribosyl pyrophosphate, which is involved in histidine and purine biosynthesis (Tozzi et al., 2006; Xue et al., 2012). However, we did not notice significant changes in expression of the encoding gene deoB in groups treated with BPU in the transcriptional profile. Nor did we find an obvious change in the pentose phosphate pathway according to GO enrichment analysis. One explanation is that while a strong binding potential exists between the molecule and the protein, the binding site may not be close to the essential substrate or metal ion binding site. Therefore, the activity of 3M7V and the pentose phosphate pathway may not be significantly affected. Till now, very little information on the functional groups of 3M7V is available, and the effect of the diaryl urea derivative on the protein and its associated pathway requires further validation.
Based on the structure of BPU, previous literature predicted that it may affect cell wall biosynthesis and membrane permeability (Nelson et al., 2015). We were expecting to find changes in GO terms involved in cellular component using GO enrichment analysis. However, no significant alteration was found. We looked specifically into genes involved in the GO term “cell wall” and found 3 DEGs (Supplementary Table 3). What may be of interest is that 2 upregulated genes, spaP and dexA, encode cell surface proteins, which are important for the adherence of S. mutans to the tooth surface (SpaP) and dextran-dependent aggregation (DexA) (Goldschmidt et al., 1990; Yang et al., 2019). This is again in accordance with the upregulation of other EPS synthesis-related genes. The results suggest that multiple steps and factors involved in or associated with EPS synthesis are influenced by the diaryl urea derivative.
In the present study, we noticed a strong inhibitive effect of BPU on growth, biofilm formation, and acidogenesis of S. mutans. Instead of being dosage-dependent, the inhibition seemed to be similar among groups treated with different concentrations of BPU. In other words, the inhibitory effect seemed to be an “ON/OFF” mode. In fact, this is not the first time that we found this kind of inhibition mode in S. mutans. One possible explanation is that the small molecule affects essential factor(s) involved in bacterial growth and/or metabolism. For example, our previous study confirmed that different concentrations of fluoride led to similar levels of inhibition of enolase activity, a key enzyme in glycolysis, in an S. mutans strain (Liao et al., 2018). As we found alterations in several biological signs of progress including sugar metabolism, nitrogen metabolism, and stress response, it is difficult to identify the major switch for the “ON/OFF” inhibition. Further knockout studies could help disclose the inhibitory “switch” of BPU.
In conclusion, our current study has confirmed the strong antimicrobial and antibiofilm effects of a novel urea derivative molecule, BPU, against caries-associated S. mutans. Structure-based in silico prediction together with the transcriptomic study has suggested the involvement of several biological processes affected by the small molecule. It is predicted that the treatment of BPU could alter the expression of genes involved in EPS synthesis, stress response, and nitrogen metabolism, leading to immediate and strong inhibition of bacterial growth, acid production, and biofilm formation. Our work illustrates that BPU is a promising agent to be used in caries prevention.
Data Availability Statement
The transcriptomic data presented in the study are deposited in NCBI SRA database with accession number PRJNA819867.
Author Contributions
YL and FY contributed to the conception and design of the study. YL and MZ conducted the experiments. YL performed the transcriptomic analysis. All authors contributed to the interpretation of the data. YL wrote the first draft of the manuscript. All authors contributed to the manuscript revision and approved the submitted version.
Funding
This work was supported by the National Natural Science Funds (grant number 82001035), the Jiangsu Natural Science Funds (grant number SBK2020041847), the Nanjing Clinical Research Center for Oral Diseases (grant number 2019060009), and the "3456" Cultivation Program for Junior Talents of Nanjing Stomatological School, Medical School of Nanjing University (grant number).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcimb.2022.904488/full#supplementary-material
References
Ardin, A. C., Fujita, K., Nagayama, K., Takashima, Y., Nomura, R., Nakano, K., et al. (2014). Identification and Functional Analysis of an Ammonium Transporter in Streptococcus Mutans. PloS One 9 (9), e107569. doi: 10.1371/journal.pone.0107569
Biswas, S., Dhaked, H. P. S., Keightley, A., Biswas, I. (2021). Involvement of ClpE ATPase in Physiology of Streptococcus Mutans. Microbiol. Spectr. 9 (3), e0163021. doi: 10.1128/Spectrum.01630-21
Bitoun, J. P., Nguyen, A. H., Fan, Y., Burne, R. A., Wen, Z. T. (2011). Transcriptional Repressor Rex Is Involved in Regulation of Oxidative Stress Response and Biofilm Formation by Streptococcus Mutans. FEMS Microbiol. Lett. 320 (2), 110–117. doi: 10.1111/j.1574-6968.2011.02293.x
Busschaert, N., Kirby, I. L., Young, S., Coles, S. J., Horton, P. N., Light, M. E., et al. (2012). Squaramides as Potent Transmembrane Anion Transporters. Angew Chem. Int. Ed Engl. 51 (18), 4426–4430. doi: 10.1002/anie.201200729
Chen, P. M., Chen, Y. Y., Yu, S. L., Sher, S., Lai, C. H., Chia, J. S. (2010). Role of GlnR in Acid-Mediated Repression of Genes Encoding Proteins Involved in Glutamine and Glutamate Metabolism in Streptococcus Mutans. Appl. Environ. Microbiol. 76 (8), 2478–2486. doi: 10.1128/AEM.02622-09
Chen, L., Walker, A. R., Burne, R. A., Zeng, L. (2020). Amino Sugars Reshape Interactions Between Streptococcus Mutans and Streptococcus Gordonii. Appl. Environ. Microbiol. 87 (1), e01459–20. doi: 10.1128/AEM.01459-20
Cui, T., Luo, W., Xu, L., Yang, B., Zhao, W., Cang, H. (2019). Progress of Antimicrobial Discovery Against the Major Cariogenic Pathogen Streptococcus Mutans. Curr. Issues Mol. Biol. 32, 601–644. doi: 10.21775/cimb.032.601
Goldschmidt, R. M., Thoren-Gordon, M., Curtiss, R. (1990). Regions of the Streptococcus Sobrinus spaA Gene Encoding Major Determinants of Antigen I. J. Bacteriol 172 (7), 3988–4001. doi: 10.1128/jb.172.7.3988-4001.1990
Gunduz, M. G., Ugur, S. B., Guney, F., Ozkul, C., Krishna, V. S., Kaya, S., et al. (2020). 1,3-Disubstituted Urea Derivatives: Synthesis, Antimicrobial Activity Evaluation and In Silico Studies. Bioorg Chem. 102, 104104. doi: 10.1016/j.bioorg.2020.104104
Hiratsuka, K., Wang, B., Sato, Y., Kuramitsu, H. (1998). Regulation of Sucrose-6-Phosphate Hydrolase Activity in Streptococcus Mutans: Characterization of the scrR Gene. Infect. Immun. 66 (8), 3736–3743. doi: 10.1128/IAI.66.8.3736-3743.1998
Hondoh, H., Saburi, W., Mori, H., Okuyama, M., Nakada, T., Matsuura, Y., et al. (2008). Substrate Recognition Mechanism of Alpha-1,6-Glucosidic Linkage Hydrolyzing Enzyme, Dextran Glucosidase From Streptococcus Mutans. J. Mol. Biol. 378 (4), 913–922. doi: 10.1016/j.jmb.2008.03.016
Ito, K., Ito, S., Shimamura, T., Weyand, S., Kawarasaki, Y., Misaka, T., et al. (2011). Crystal Structure of Glucansucrase From the Dental Caries Pathogen Streptococcus Mutans. J. Mol. Biol. 408 (2), 177–186. doi: 10.1016/j.jmb.2011.02.028
Kajfasz, J. K., Rivera-Ramos, I., Scott-Anne, K., Gregoire, S., Abranches, J., Lemos, J. A. (2015). Transcription of Oxidative Stress Genes Is Directly Activated by SpxA1 and, to a Lesser Extent, by SpxA2 in Streptococcus Mutans. J. Bacteriol 197 (13), 2160–2170. doi: 10.1128/JB.00118-15
Kawada-Matsuo, M., Mazda, Y., Oogai, Y., Kajiya, M., Kawai, T., Yamada, S., et al. (2012). GlmS and NagB Regulate Amino Sugar Metabolism in Opposing Directions and Affect Streptococcus Mutans Virulence. PloS One 7 (3), e33382. doi: 10.1371/journal.pone.0033382
Klein, M. I., Hwang, G., Santos, P. H., Campanella, O. H., Koo, H. (2015). Streptococcus Mutans-Derived Extracellular Matrix in Cariogenic Oral Biofilms. Front. Cell Infect. Microbiol. 5. doi: 10.3389/fcimb.2015.00010
Koo, H., Falsetta, M. L., Klein, M. I. (2013). The Exopolysaccharide Matrix: A Virulence Determinant of Cariogenic Biofilm. J. Dent. Res. 92 (12), 1065–1073. doi: 10.1177/0022034513504218
Koo, H., Xiao, J., Klein, M. I., Jeon, J. G. (2010). Exopolysaccharides Produced by Streptococcus Mutans Glucosyltransferases Modulate the Establishment of Microcolonies Within Multispecies Biofilms. J. Bacteriol 192 (12), 3024–3032. doi: 10.1128/JB.01649-09
Krzysciak, W., Jurczak, A., Koscielniak, D., Bystrowska, B., Skalniak, A. (2014). The Virulence of Streptococcus Mutans and the Ability to Form Biofilms. Eur. J. Clin. Microbiol. Infect. Dis. 33 (4), 499–515. doi: 10.1007/s10096-013-1993-7
Liao, Y., Brandt, B. W., Li, J., Crielaard, W., Van Loveren, C., Deng, D. M. (2017). Fluoride Resistance in Streptococcus Mutans: A Mini Review. J. Oral. Microbiol. 9 (1), 1344509. doi: 10.1080/20002297.2017.1344509
Liao, Y., Yang, J., Brandt, B. W., Li, J., Crielaard, W., van Loveren, C., et al. (2018). Genetic Loci Associated With Fluoride Resistance in Streptococcus Mutans. Front. Microbiol. 9. doi: 10.3389/fmicb.2018.03093
Li, Y., Burne, R. A. (2001). Regulation of the gtfBC and Ftf Genes of Streptococcus Mutans in Biofilms in Response to pH and Carbohydrate. Microbiol. (Reading) 147 (Pt 10), 2841–2848. doi: 10.1099/00221287-147-10-2841
Limban, C., Chifiriuc, M. C., Caproiu, M. T., Dumitrascu, F., Ferbinteanu, M., Pintilie, L., et al. (2020). New Substituted Benzoylthiourea Derivatives: From Design to Antimicrobial Applications. Molecules 25 (7), 1478. doi: 10.3390/molecules25071478
Lin, Y., Chen, J., Zhou, X., Li, Y. (2021). Inhibition of Streptococcus Mutans Biofilm Formation by Strategies Targeting the Metabolism of Exopolysaccharides. Crit. Rev. Microbiol. 47 (5), 667–677. doi: 10.1080/1040841X.2021.1915959
Nelson, J. W., Plummer, M. S., Blount, K. F., Ames, T. D., Breaker, R. R. (2015). Small Molecule Fluoride Toxicity Agonists. Chem. Biol. 22 (4), 527–534. doi: 10.1016/j.chembiol.2015.03.016
Nijampatnam, B., Ahirwar, P., Pukkanasut, P., Womack, H., Casals, L., Zhang, H., et al. (2021). Discovery of Potent Inhibitors of Streptococcus Mutans Biofilm With Antivirulence Activity. ACS Med. Chem. Lett. 12 (1), 48–55. doi: 10.1021/acsmedchemlett.0c00373
Nijampatnam, B., Casals, L., Zheng, R., Wu, H., Velu, S. E. (2016). Hydroxychalcone Inhibitors of Streptococcus Mutans Glucosyl Transferases and Biofilms as Potential Anticaries Agents. Bioorg Med. Chem. Lett. 26 (15), 3508–3513. doi: 10.1016/j.bmcl.2016.06.033
Nilsson, M., Jakobsen, T. H., Givskov, M., Twetman, S., Tolker-Nielsen, T. (2019). Oxidative Stress Response Plays a Role in Antibiotic Tolerance of Streptococcus Mutans Biofilms. Microbiol. (Reading) 165 (3), 334–342. doi: 10.1099/mic.0.000773
Pamp, S. J., Frees, D., Engelmann, S., Hecker, M., Ingmer, H. (2006). Spx Is a Global Effector Impacting Stress Tolerance and Biofilm Formation in Staphylococcus Aureus. J. Bacteriol. 188 (13), 4861–4870. doi: 10.1128/JB.00194-06
Qiu, W., Zhou, Y., Li, Z., Huang, T., Xiao, Y., Cheng, L., et al. (2020). Application of Antibiotics/Antimicrobial Agents on Dental Caries. BioMed. Res. Int. 2020, 5658212. doi: 10.1155/2020/5658212
Ren, Z., Cui, T., Zeng, J., Chen, L., Zhang, W., Xu, X., et al. (2016). Molecule Targeting Glucosyltransferase Inhibits Streptococcus Mutans Biofilm Formation and Virulence. Antimicrob. Agents Chemother. 60 (1), 126–135. doi: 10.1128/AAC.00919-15
Siegers, K., Entian, K. D. (1995). Genes Involved in Immunity to the Lantibiotic Nisin Produced by Lactococcus Lactis 6F3. Appl. Environ. Microbiol. 61 (3), 1082–1089. doi: 10.1128/aem.61.3.1082-1089.1995
Sroor, F. M., Othman, A. M., Tantawy, M. A., Mahrous, K. F., El-Naggar, M. E. (2021). Synthesis, Antimicrobial, Anti-Cancer and In Silico Studies of New Urea Derivatives. Bioorg Chem. 112, 104953. doi: 10.1016/j.bioorg.2021.104953
Tang, B., Gong, T., Zhou, X., Lu, M., Zeng, J., Peng, X., et al. (2019). Deletion of Cas3 Gene in Streptococcus Mutans Affects Biofilm Formation and Increases Fluoride Sensitivity. Arch. Oral. Biol. 99, 190–197. doi: 10.1016/j.archoralbio.2019.01.016
Tozzi, M. G., Camici, M., Mascia, L., Sgarrella, F., Ipata, P. L. (2006). Pentose Phosphates in Nucleoside Interconversion and Catabolism. FEBS J. 273 (6), 1089–1101. doi: 10.1111/j.1742-4658.2006.05155.x
Wang, X., Liu, L., Zhou, X., Huo, Y., Gao, J., Gu, H. (2020). Casein Phosphopeptide Combined With Fluoride Enhances the Inhibitory Effect on Initial Adhesion of Streptococcus Mutans to the Saliva-Coated Hydroxyapatite Disc. BMC Oral. Health 20 (1), 169. doi: 10.1186/s12903-020-01158-8
Wu, Y. (1965). [Synthesis of Subtitute Urea Derivative and Its In Vitro Antimicrobial Effect.]. Yao Xue Xue Bao 08), 523–532. doi: 10.16438/j.0513-4870.1965.08.007%WCNKI
Xue, X., Li, J., Wang, W., Sztajer, H., Wagner-Dobler, I. (2012). The Global Impact of the Delta Subunit RpoE of the RNA Polymerase on the Proteome of Streptococcus Mutans. Microbiol. (Reading) 158 (Pt 1), 191–206. doi: 10.1099/mic.0.047936-0
Yang, Y., Mao, M., Lei, L., Li, M., Yin, J., Ma, X., et al. (2019). Regulation of Water-Soluble Glucan Synthesis by the Streptococcus Mutans dexA Gene Effects Biofilm Aggregation and Cariogenic Pathogenicity. Mol. Oral. Microbiol. 34 (2), 51–63. doi: 10.1111/omi.12253
Yang, S., Zhang, J., Yang, R., Xu, X. (2021). Small Molecule Compounds, A Novel Strategy Against Streptococcus Mutans. Pathogens 10 (12), 1540. doi: 10.3390/pathogens10121540
Younson, J., Kelly, C. (2004). The Rational Design of an Anti-Caries Peptide Against Streptococcus Mutans. Mol. Divers. 8 (2), 121–126. doi: 10.1023/b:modi.0000025655.93643.fa
Zhang, Q., Nguyen, T., McMichael, M., Velu, S. E., Zou, J., Zhou, X., et al. (2015). New Small-Molecule Inhibitors of Dihydrofolate Reductase Inhibit Streptococcus Mutans. Int. J. Antimicrob. Agents 46 (2), 174–182. doi: 10.1016/j.ijantimicag.2015.03.015
Zhang, Q., Nijampatnam, B., Hua, Z., Nguyen, T., Zou, J., Cai, X., et al. (2017). Structure-Based Discovery of Small Molecule Inhibitors of Cariogenic Virulence. Sci. Rep. 7 (1), 5974. doi: 10.1038/s41598-017-06168-1
Keywords: dental caries, Streptococcus mutans, transcriptomic study, small molecules, stress response, nitrogen metabolism
Citation: Liao Y, Zhang M, Lin X and Yan F (2022) Diaryl Urea Derivative Molecule Inhibits Cariogenic Streptococcus mutans by Affecting Exopolysaccharide Synthesis, Stress Response, and Nitrogen Metabolism. Front. Cell. Infect. Microbiol. 12:904488. doi: 10.3389/fcimb.2022.904488
Received: 25 March 2022; Accepted: 11 April 2022;
Published: 10 May 2022.
Edited by:
Xin Xu, Sichuan University, ChinaReviewed by:
Zhengwei Huang, Shanghai Jiao Tong University, ChinaYuan Liu, University of Pennsylvania, United States
Copyright © 2022 Liao, Zhang, Lin and Yan. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Fuhua Yan, eWFuZmhAbmp1LmVkdS5jbg==; Xingnan Lin, bGlueGluZ25hbkAxMjYuY29t
†These authors share last authorship
 Ying Liao
Ying Liao Mengyun Zhang
Mengyun Zhang Xingnan Lin
Xingnan Lin Fuhua Yan
Fuhua Yan