
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Cell. Infect. Microbiol., 28 July 2022
Sec. Microbiome in Health and Disease
Volume 12 - 2022 | https://doi.org/10.3389/fcimb.2022.896504
This article is part of the Research TopicThe Gut Microbiome and COVID-19View all 9 articles
 Yao Ma1†
Yao Ma1† Mei Luo1†
Mei Luo1† Yusheng Deng2†
Yusheng Deng2† Xiaoman Yang1
Xiaoman Yang1 Xionglue Wang1
Xionglue Wang1 Guozhong Chen1
Guozhong Chen1 Zixin Qin1
Zixin Qin1 Yun Deng1
Yun Deng1 Meiling Nan3
Meiling Nan3 Yang Chen4
Yang Chen4 Peihui Wang3
Peihui Wang3 Hong Wei5
Hong Wei5 Lijuan Han2,4*
Lijuan Han2,4* Xiaodong Fang4*
Xiaodong Fang4* Zhi Liu1*
Zhi Liu1*The gut microbiome profile of COVID-19 patients was found to correlate with a viral load of SARS-CoV-2, COVID-19 severity, and dysfunctional immune responses, suggesting that gut microbiota may be involved in anti-infection. In order to investigate the role of gut microbiota in anti-infection against SARS-CoV-2, we established a high-throughput in vitro screening system for COVID-19 therapeutics by targeting the endoribonuclease (Nsp15). We also evaluated the activity inhibition of the target by substances of intestinal origin, using a mouse model in an attempt to explore the interactions between gut microbiota and SARS-CoV-2. The results unexpectedly revealed that antibiotic treatment induced the appearance of substances with Nsp15 activity inhibition in the intestine of mice. Comprehensive analysis based on functional profiling of the fecal metagenomes and endoribonuclease assay of antibiotic-enriched bacteria and metabolites demonstrated that the Nsp15 inhibitors were the primary bile acids that accumulated in the gut as a result of antibiotic-induced deficiency of bile acid metabolizing microbes. This study provides a new perspective on the development of COVID-19 therapeutics using primary bile acids.
To date, the coronavirus disease (COVID-19) pandemic has infected over 500 million people and resulted in more than six million deaths worldwide (WHO, 2022b). Coronavirus SARS-CoV-2 is the causative agent of COVID-19 with limited specific medicine or treatment. A large number of antiviral agents have been evaluated for SARS-CoV-2 since the breakout of COVID-19 (Izda et al., 2021). However, remdesivir is the only antiviral recommended for COVID-19 treatment in Europe, with efficacy and safety recently questioned (Scavone et al., 2022). The World Health Organization (WHO) reported that only 57 countries have vaccinated over 70% of their population by May 22, 2022, almost all of which are high-income countries. The role of antiviral agents in COVID-19 treatment is still being investigated.
Apart from the common respiratory symptoms, a number of COVID-19 patients showed gastrointestinal symptoms, such as diarrhea (2.0%–55.0%), nausea (1.0%–27.5%), vomiting (1.0%–12.5%), lack of appetite (10.1%–39.7), and abdominal pain (0.98%–5.8%) (Zhang et al., 2021). A study exploring the association between fecal microbiome composition and SARS-CoV-2 transcriptional activity has showed that the patients with active SARS-CoV-2 gastrointestinal tract (GI) infection harbor more opportunistic pathogens and decreased abundance of short-chain fatty acid (SCFA) producing bacteria (Zuo et al., 2021a). Moreover, the gut microbiome profile of COVID-19 patients has been found to be consistent with COVID-19 severity and dysfunctional immune responses, featuring the depletion of Faecalibacterium prausnitzii, Eubacterium rectale, and Bifidobacteria which have been identified with immunomodulatory potential (Yeoh et al., 2021). Taken together, gut microbes may serve as indicators of SARS-CoV-2 infection and may be involved in anti-infection. Therefore, it is important to examine and screen intestinal resources for substances that may be applied in COVID-19 therapy.
The reported targets of anti-coronavirus therapies focus on viral mRNA synthesis and replication, among which, a common target functioning in mRNA synthesis is the coronavirus endoribonuclease (EndoU). EndoU is encoded by nonstructural protein 15 (nsp15), a specific marker for vertebrate nidoviruses, which is involved in viral mRNA production and transcription (Deng and Baker, 2018). The conserved aspartate residue in the coronavirus Nsp15 protein (Asp-298 of Coronavirus humano 229E (HCoV-229E) and its homologous site) is essential for its correct folding and RNA synthesis capacity (Ivanov et al., 2004). In addition to its involvement in regulating viral replication, the Nsp15 protein is also critical in evading host dsRNA sensors in macrophages (Deng et al., 2017). The Nsp15 protein of SARS-CoV exists mainly as a hexamer that cleaves at 3’ of uridylates of RNAs, and preferentially cleaves at the U30 site of the highly conserved RNA element, stem-loop II motif (s2m), and in the 3’ non-translated region (UTR) of the virus genome (Bhardwaj et al., 2006). Since the s2m RNA element is highly conserved in nidoviruses and is not present in the human genome, Nsp15 is considered an important target for antiviral drug development (Robertson et al., 2005; Canal et al., 2021). As for Nsp15, crystal structure determination and endoribonuclease activity revealed that the active site residues of SARS-CoV-2 are conserved to that of SARS-CoV, both in terms of sequence and conformation (Kim et al., 2020). Accordingly, in this study, we chose Nsp15 as a screening target for COVID-19 therapeutics. And we successfully established high-throughput in vitro screening system of Nsp15.
Traditional Chinese patent medicines (CPMs) have been widely used in the clinical treatment of COVID-19 in China, and some of these CPMs have been shown to have anti-viral effects (Zhuang et al., 2020; Huang et al., 2021). In China, three traditional Chinese medicine (TCM) decoctions and three formulated Chinese medicines (as six TCM recipes) have been proven to be the most effective TCMs in treating COVID-19 patients (Luo et al., 2020). Among the six TCM recipes, Jinhua Qinggan granules (JHQGG) have been shown to significantly increase the 7-day viral clearance rate and shorten recovery time from pneumonia (Liu et al., 2020), while Lianhua Qingwen capsules (LHQWC), one of six TCM recipes combined with anti-influenza drugs, more effectively alleviated clinical symptoms and improved treatment efficacy in patients with mild COVID-19 (Huang et al., 2021). A variety of TCM herbal components have been reported to influence the abundance and diversity of gut microbiota, which in turn, affect the efficacy of TCM (Lin et al., 2020). Particularly, TCM ingredients can be transformed by gut microbiota into small chemical molecules to achieve the pharmacological functions. For example, gut microbiota can convert ginsenosides in orally administrated ginseng to compound K, which shows higher anti-tumor, anti-inflammatory, and anti-allergic activity than ginsenoside Rb1 (Wang et al., 2011; Kim et al., 2013). Thus, we evaluated the activity inhibition on the target by substances of intestinal origin using a mouse model and attempted to explore the interaction between gut microbiota and clinical traditional CPMs. The results unexpectedly revealed that it is antibiotic treatment, not CPMs, which induced the appearance of substances with Nsp15 activity inhibition in the intestine of mice. Further analysis revealed that antibiotic treatment led to the depletion of flora functioning in bile acid metabolism, which resulted in the accumulation of primary bile acids and displayed an inhibitory effect on Nsp15 in the gut.
The recombinant Nsp15, with expected molecular weight of 41 KDa, was successfully expressed in E. coli BL21 (DE3) (Figure 1A). Then, we developed the enzymatic activity detection system for Nsp15 based on the fluorescence resonance energy transfer (FRET) mechanism (Boute et al., 2002). Benzopurpurin B was chosen as a Nsp15 inhibitor control according to a study of SARS-CoV (Ortiz-Alcantara et al. 2010). Four fluorogenic RNA substrate candidates were assessed in the construction of the Nsp15 endoribonuclease screening system, based on the characteristic that Nsp15 preferentially cleaves at uridylate bases that are not involved in base-pairing (Bhardwaj et al., 2006). The substrates were 5’6-FAM-GAAGCGAAACCC/rU/AAG-3’BHQ1 (substrate N1), 5’6-FAM-GCGGAGCACGA/rU/CGAG-3’BHQ1 (substrate N2), 5’6-FAM-UACGAUCG-3’BHQ1 (substrate N3), and 5’6-FAM-A/rU/AA-3’BHQ1 (substrate N4). Among them, substrate N4 exhibited the highest enzymatic activity, the minimal non-specific degradation (Figure 1B), thus was selected for subsequent high-throughput screening. The Nsp15 purified with Ni-NTA Agarose exhibited high activity on cleaving fluorogenic RNA substrate N4 (Figures 1C, D). We further utilized the crude enzyme Nsp15 for the high-throughput screening due to the low consumption.
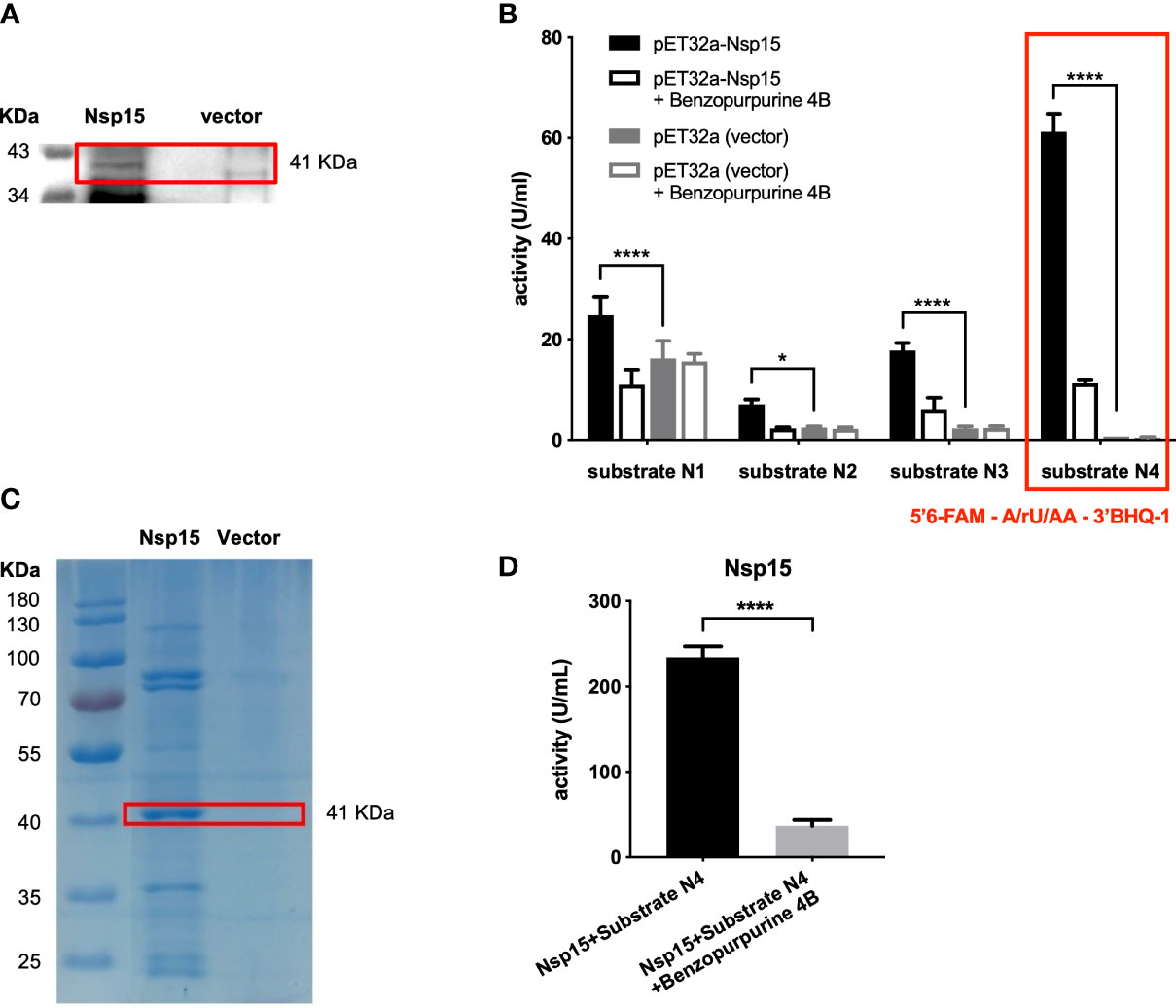
Figure 1 Establishment of in vitro screening system for Nsp15 inhibitors. (A) Nsp15 expression was detected by Western blotting. E. coli BL21(DE3) cells containing pET32a plasmids with or without (vector) recombinant SARS-CoV-2 Nsp15 were grown to OD600 of 0.9 - 1.0 and then induced with 1 mM IPTG. After additional incubation at 18°C for 16 h, cells in 1 ml of bacterial culture were collected for Western blotting with anti-His-6 antiserum. (B) Analysis and comparison of Nsp15 substrates using endoribonuclease assay. Reactions were performed in the reaction buffer containing 50 mM Tris-HCI pH 7.5, 100 mM KCI, 5 mM MnCl2, and 1 mM DTT, at 30°C. Benzopurpurine 4B is an inhibitor of SARS-CoV Nsp15. Enhanced fluorescence due to the cleavage of the fluorogence substrate RNA was monitored at 518 nm with excitation at 492 nm by a fluorescence plate reader. Values expressed are means ± S.D. from three independent experiments. Significance was determined separately for each substrate by One-way ANOVA; p-value: *, < 0.05, ****, < 0.0001. (C) Purified Nsp15 were detected by Coomassie staining. His-tagged Nsp15 proteins were purified by Ni-NTA Agarose and separated by SDS-PAGE followed by Coomassie staining; we spliced the ladder next to the destination protein band. (D) Endoribonuclease assay of purified Nsp15. The endoribonuclease activity of purified Nsp15 was determined by its ability in cleaving fluorogenic RNA substrate N4. Values expressed are means ± S.D. from three independent experiments. Significance was determined by t-test; p-value: ****, < 0.0001.
At this point, the Nsp15 endoribonuclease screening system in vitro was successfully established.
Traditional Chinese patent medicines (CPMs) have been widely used in the clinical treatment of COVID-19 in China, and have a significant role in the treatment, according to a number of clinical reports (Zhuang et al., 2020). Conversely, clinical data and experiments on the macaque model of COVID-19 have suggested that COVID-19 infection reduces the diversity of intestinal microbiome, further affecting the function of the gut microbiome (decreased SCFA synthesis, impaired conversion of primary to secondary bile acids, upregulation of tryptophan metabolism) and promoting intestinal inflammation, which lead to the disruption of gut immune homeostasis (Zuo et al., 2021b). Therefore, we investigated whether CPMs function through intestinal bacteria, using LHQWC (Tao et al., 2013; Ding et al., 2017) and JHQGG (Liu et al., 2020) as examples, since their pharmacological effects are anti-viral (Zhuang et al., 2020) and are recommended for clinical treatment of COVID-19 (Wei, 2020).
By using antibiotic treatment, followed with gavage of CPMs in the mice, we obtained mouse fecal samples with CPM treatment and microbiome depletion (Figure 2A). Similarly, the samples of mice treated with or without CPMs and with or without intestinal microorganisms were obtained. These samples were then subjected to in vitro assay for the inhibitory effect on Nsp15. As a result, fecal samples from the mice treated with the CPMs alone could not inhibit Nsp15 (Figure 2B). Surprisingly, the fecal samples of antibiotic-treated mice, irrespective of CPM treatment, inhibited Nsp15 enzyme activity (Figure 2B). These data lead to the conclusion that CPMs do not inhibit the tested targets via gut microbes, while antibiotic treatment induces the production of Nsp15 inhibitors in the mouse intestine.
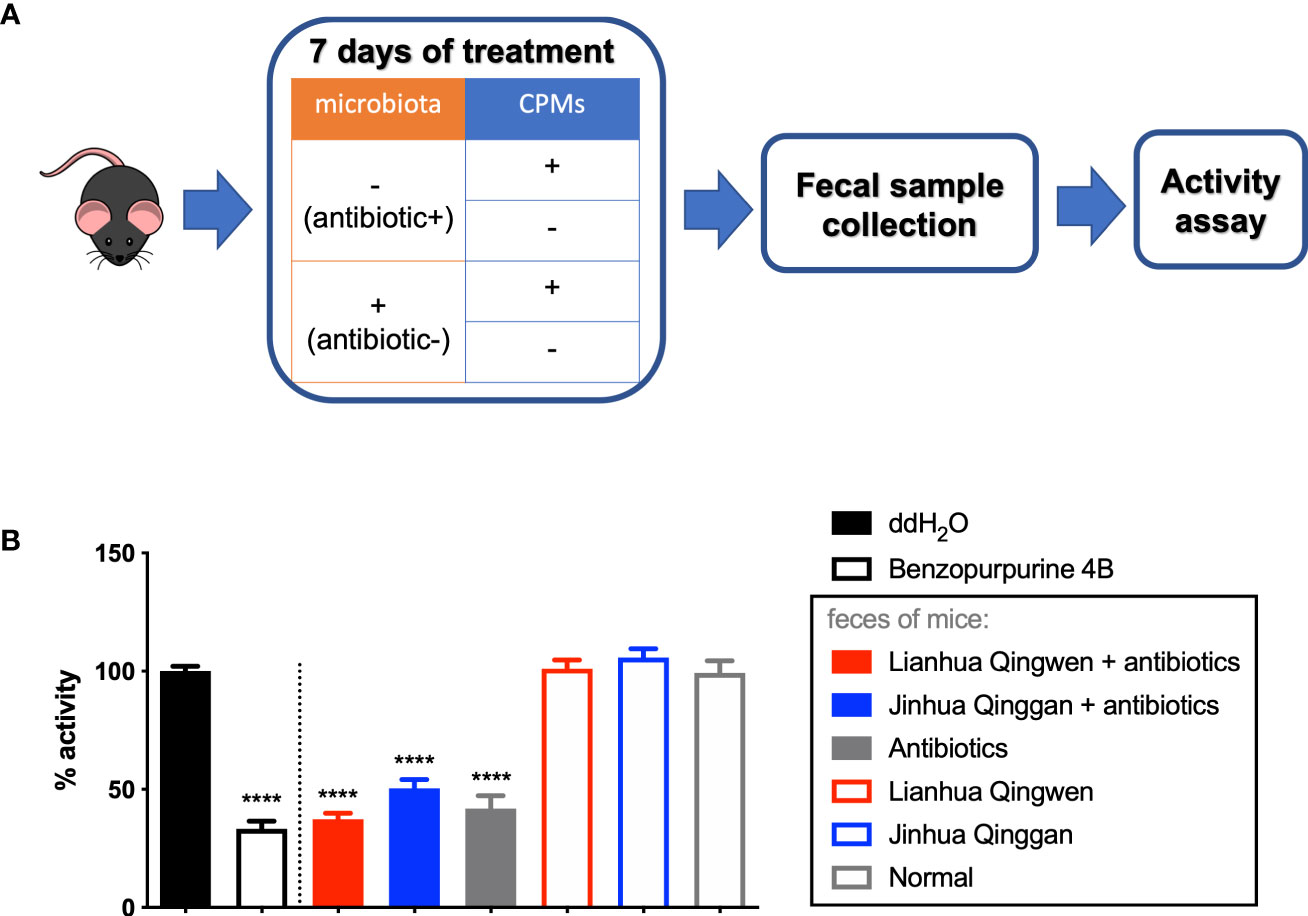
Figure 2 Antibiotic-induced alteration of gut microbiome led to inhibitory effect on Nsp15 activity. (A) Illustration of animal assay. C57BL/6J males were supplied with or without antibiotic cocktail for 7 days to illustrate mouse model with or without gut microbiota. These mice were administrated with or without CPMs at the same time in order to examine the interaction between CPMs and microbiota, by assessing the impact of fecal samples on SARS-CoV-2 Nsp15 activity. Fecal pellets of each mouse were collected at the end of experiment, and supernatant of fecal homogenate in ddH2O was subjected to activity assay. (B) Analysis of the effect of fecal samples on Nsp15 activity. The reaction systems with ddH2O and Benzopurpurine 4B instead of the sample to be tested were negative and positive controls, respectively. Relative activity (% of activity) was calculated for each sample by comparing with the negative control. Values expressed are means ± S.D. from three independent experiments for no less than three biological replicates. Significance was determined by One-way ANOVA; p-value: ****, < 0.0001.
Antibiotic treatment, along with possible residual antibiotics, has been reported to alter gut microbiome and induce changes in metabolic homeostasis in mice (Zhang et al., 2014; Rodrigues et al., 2017; Zarrinpar et al., 2018). Accordingly, we explored the source of Nsp15 inhibitors in the mouse intestine from the following three aspects: antibiotics themselves, intestinal microbiota, and microbiota-involved host metabolism.
To analyze the effect of antibiotics on Nsp15 activity, we conducted in vitro assays on an antibiotic cocktail at 1×, 10×, and 100× doses used in animal experiments. The results showed that the antibiotic cocktail had no effect on the activity of Nsp15, regardless of the dosage (Supplementary Figure 1). Therefore, we turned to examining the association of gut microbiota with Nsp15 inhibitors. Since antibiotic treatment is known to cause microbiome depletion (Xiao et al., 2018; Zarrinpar et al., 2018), we evaluated the perturbation of gut microbiota in mice by the antibiotics used in this study.
We assessed the impact of antibiotic treatment on the composition of gut microbiome by comparing the metagenome of fecal samples from antibiotic-treated mice and vehicle-treated mice. Clean reads generated from the stool-extracted DNA were subjected to quantitative profiling of the taxonomic composition of all samples. The assessment of α-diversity revealed significantly lower within-group diversity in antibiotic-treated mice compared to vehicle-treated mice (p=0.0254) (Figure 3A), while β-diversity measure detected a significant difference in taxonomic composition between antibiotic and vehicle groups (p=0.0193) (Figure 3B). The analyses of taxonomic composition at the species level suggested that the microbiome of the antibiotic-treated mice was dominated by Citrobacter spp. (91.19%), Lactobacillus murinus (7.93%), and Escherichia coli (0.87%), while the bacterial species that were abundant in vehicle-treated mice were Muribaculum spp., Lactobacillus spp., Clostridium spp., and Lachnospiraceae spp. (Figure 3C). However, only Citrobacter spp. and E. coli were enriched in antibiotic-treated mice, as revealed by analyses with LEfSe (Figure 3D).
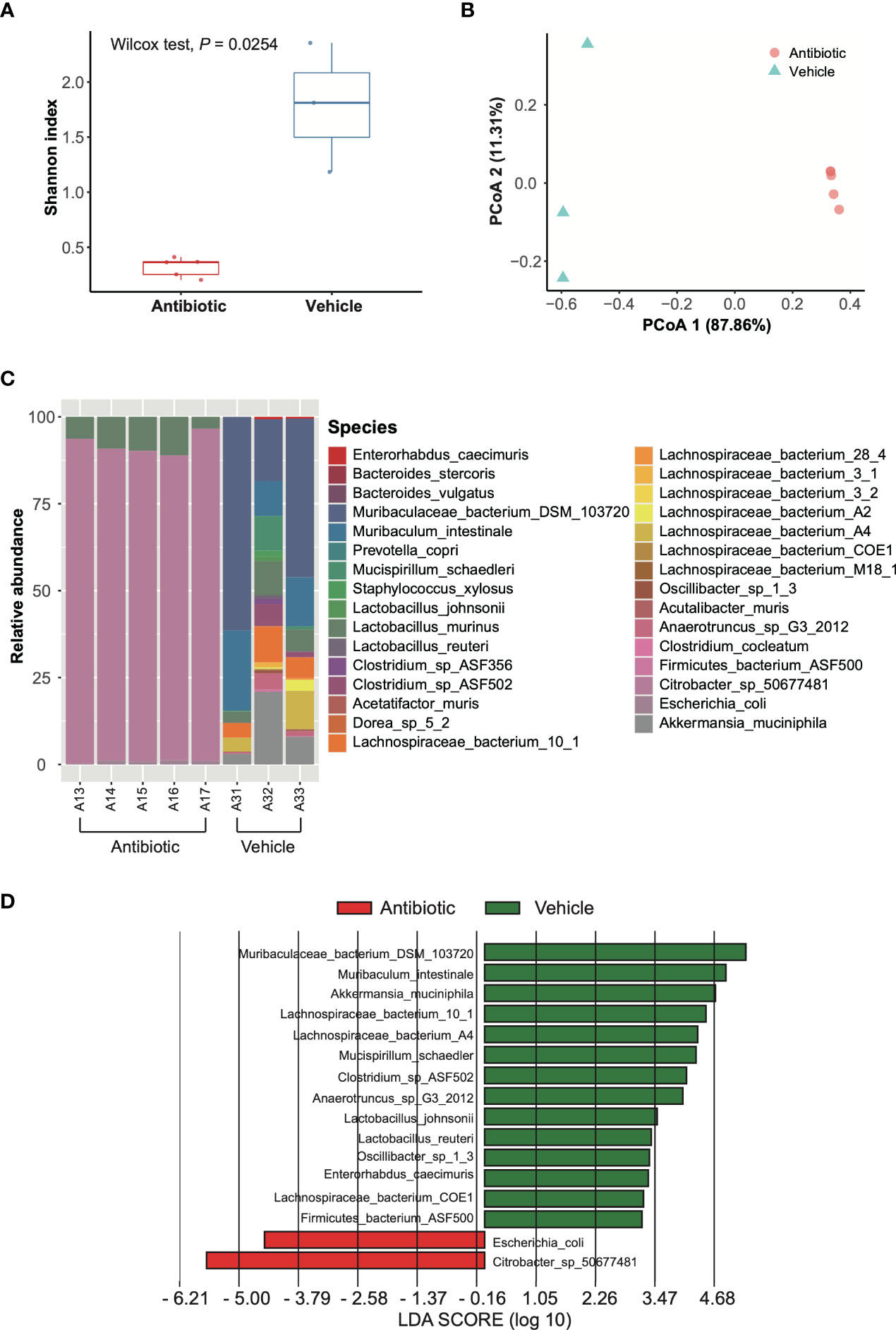
Figure 3 Analysis of gut microbial composition based on metagenomic data. (A) Shannon index. Red and blue boxes indicate antibiotic group and vehicle group, respectively. Wilcoxon test was applied to identify the difference. (B) PCoA based on Bray-Curtis dissimilarity index. The red circle and blue triangles indicate antibiotic individuals and vehicle individuals, respectively. The differences were compared using PERMANOVA with 9,999 permutations. (C) Taxonomic composition at species level. A13-A17 are antibiotic-treated individuals, while A31-A33 are vehicle individuals. (D) LEfSe results for species comparing the antibiotic group and vehicle group. The LDA scores > 2 are listed.
We hypothesized that bacteria enriched by antibiotic treatment have an inhibitory effect on Nsp15 activity, and isolated these bacterial species from mouse feces to determine the impact of their bacterial cultures on Nsp15 activity in vitro. However, results on the most abundant species, Citrobacter spp., rejected the above hypothesis as its pure cultures obtained under aerobic and anaerobic conditions or co-culture with mouse feed, did not inhibit Nsp15 activity (Supplementary Figure 2). These results suggest that the bacteria in relatively high abundance after antibiotic treatment is not the source of Nsp15 inhibition in the mouse gut and, conversely, the Nsp15 inhibitor may be associated with the altered host metabolism due to the depleted bacteria. To verify the association between Nsp15 inhibition and microbiome depletion, we examined the inhibition of Nsp15 activity in the feces of germ-free mice. As expected, fecal supernatant from germ-free mice reduced the activity of Nsp15 by 67%, while antibiotic treatment resulted in a stable level of Nsp15 inhibition in mouse feces after two days of treatment (Figure 4). This result suggested that the missing microbiome play a key role in the inhibition of Nsp15 activity and that they likely act by affecting host metabolism.
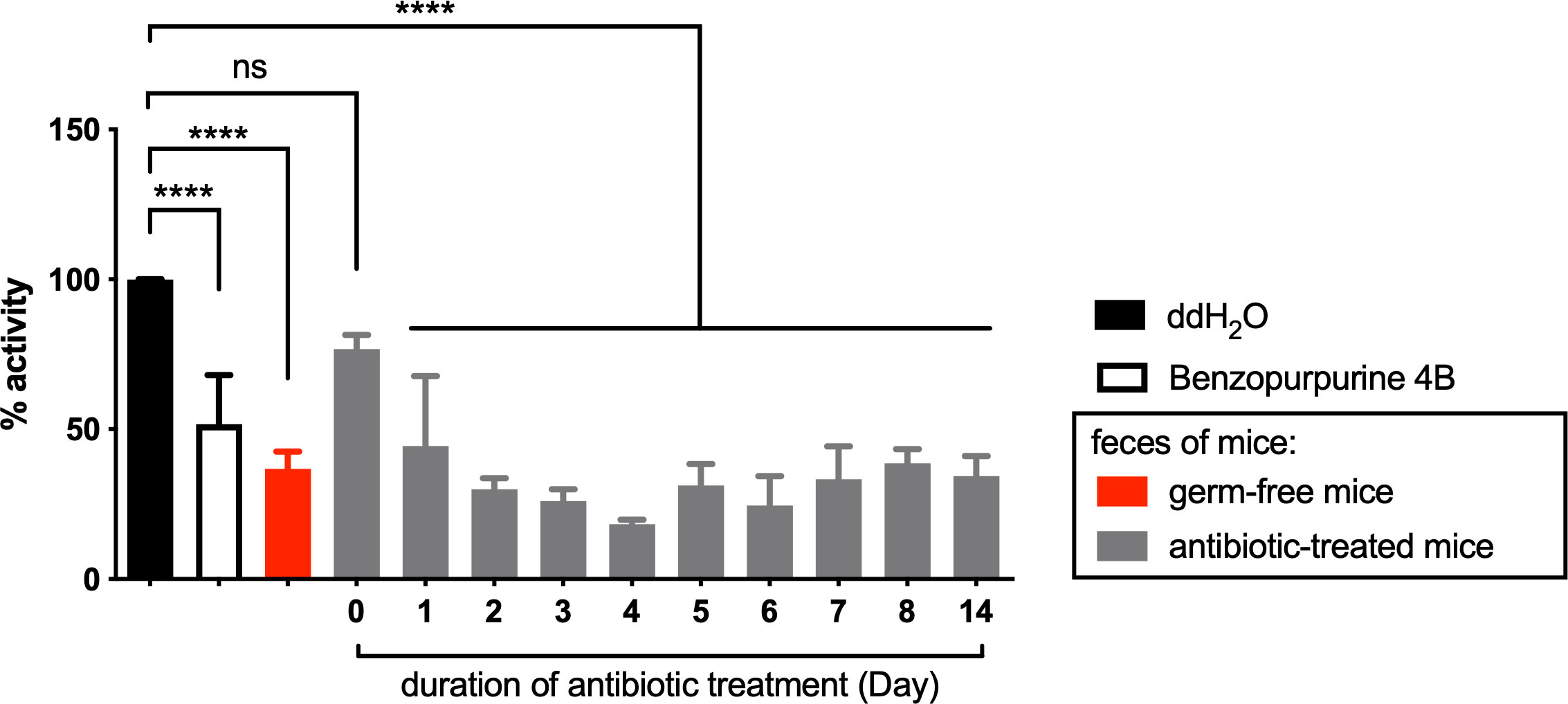
Figure 4 Nsp15 inhibitory activity in the intestine was associated with the impaired microbiota under antibiotic treatment. Supernatant of fecal homogenate of germ-free and antibiotic-treated mice were subjected to in vitro assays for the inhibitory effect on Nsp15. The reaction systems with ddH2O and Benzopurpurine 4B instead of the sample to be tested were negative and positive controls, respectively. Relative activity (% activity) was calculated for each sample by comparing with negative control. Values expressed are means ± S.D. from three independent experiments for no less than three biological replicates. Significance was determined by One-way ANOVA; p-value: ns, not significant, ****, < 0.0001.
To test the above hypothesis, we further identified the pathways that were significantly reduced in the gut of antibiotic-treated mice but were enriched in that of the vehicle mice using LEfSe, with a particular focus on pathways related to microbiota-derived metabolites and microbiota-host co-metabolites. Although metagenomic data revealed global changes in host metabolism with microbiome depletion (Supplementary Figure 3), based on literature, we believed that these changes were associated with impaired metabolism of food-derived plant natural products and bile acids (Quinn et al., 2020). It has been reported that after antibiotic treatment, significant changes in metabolism occur in the intestine of mice: carbohydrates and sugar alcohols other than glucose and mannose is increased, fatty acid metabolism is inhibited, and the availability of short-chain fatty acids (SCFA) is decreased, a shift in amino acid metabolism is observed, the content of amino acids, vitamins, and carboxylic acids is decreased, and the ratio of primary to secondary bile acids is extremely high (Theriot et al., 2014; Rodrigues et al., 2017; Zarrinpar et al., 2018; Gutierrez et al., 2019). Our enrichment analysis agreed with these ideas, showing that the antibiotic treatment affected several metabolism-related functions of the gut microbiota, including the metabolism of bile acids, carbohydrates, fatty acids, amino acids, and vitamins (Table S2, Supplementary Figure 3).
Since carbohydrates and primary bile acids were metabolites enriched in the mouse gut after antibiotic treatment (Rodrigues et al., 2017; Zarrinpar et al., 2018), we examined the inhibition of Nsp15 activity in vitro by commercially available carbohydrates and by taurocholic acid (TCA), a primary bile acid with increased levels in antibiotic-treated mice. The results showed that carbohydrates such as fructose, cellobiose, and maltose had no impact on Nsp15 activity (Supplementary Figure 4A), while low doses of TCA (10 nM~1000 nM) could inhibit Nsp15 activity (Figure 5A). In contrast, taurodeoxycholic acid (TDCA), which is the secondary bile acid derived from TCA, had no inhibitory effect by Nsp15 (Figure 5B). To exclude the reduction of Nsp15 activity as a result of non-targeted aggregation of the protein by TCA (Cremers et al., 2014), we tested the inhibitory effect by TCA or TDCA on RNase A instead of Nsp15. The results showed that neither TCA (10 nM~1000 nM) nor TDCA (10 nM~1000 nM) disrupted substrate from cleavage by RNase A (Figures 5C, D). Further, another primary bile acid, chenodesoxycholic acid (CDCA), was examined on Nsp15 activity inhibition and we found that certain doses of CDCA (5 µM~50 µM) significantly inhibited Nsp15 activity (Supplementary Figure 4B). These results supported the idea that primary bile acids, inhibit Nsp15 activity.
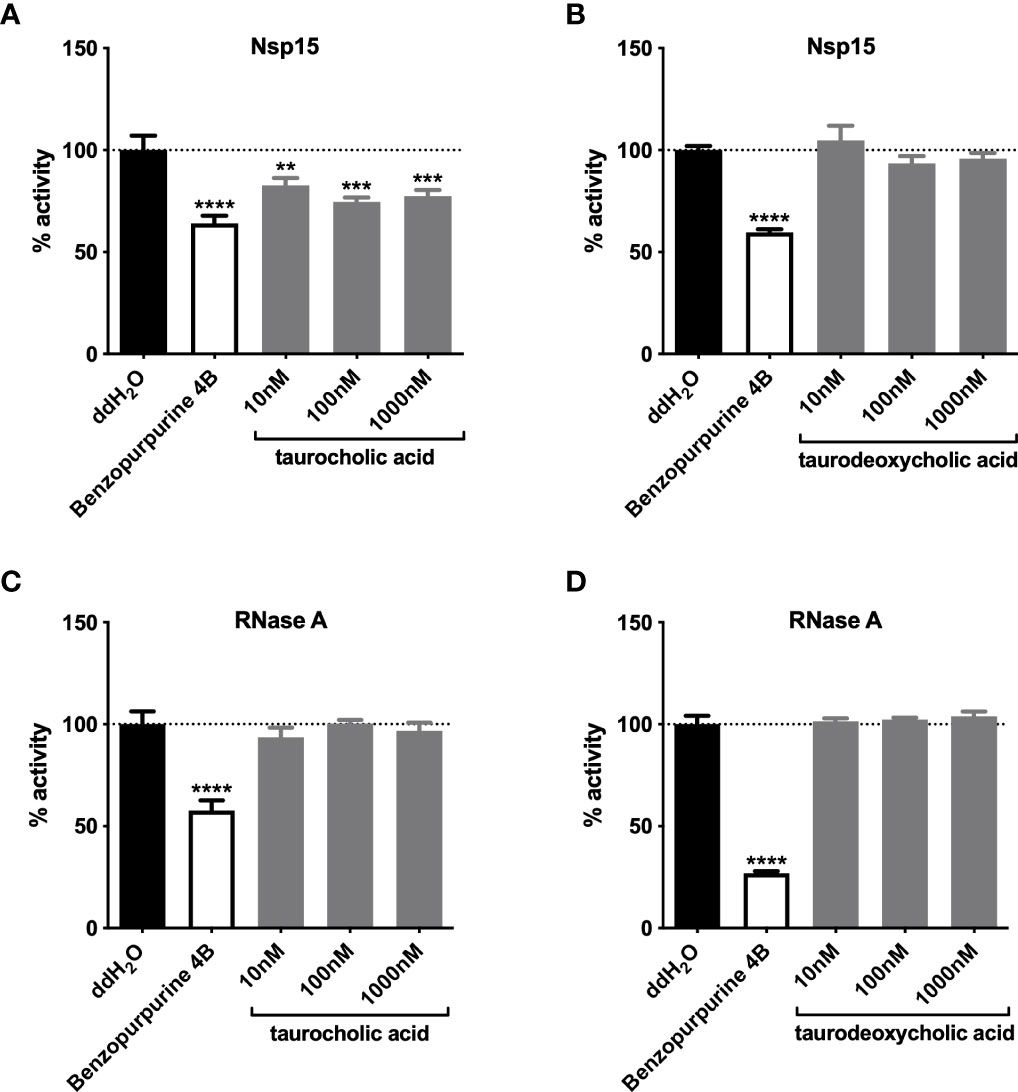
Figure 5 Primary bile acids inhibit Nsp15 activity. The effects of primary (A) and secondary (B) bile acids on the enzymatic activity of Nsp15 were examined. To verify the action specificity of bile acids on Nsp15, further attempts were made to test the inhibition on RNase A activity by primary (C) and secondary (D) bile acids, with each reaction system comprising 35 nM of RNase A. Reaction systems containing ddH2O or Benzopurpurine 4B rather than the sample to be tested were used as negative and positive controls, respectively. Relative activity (% activity) was calculated for each sample by comparison with the negative control. Values expressed are means ± S.D. from three independent experiments. Significance was determined by One-way ANOVA; p-value: **, < 0.01, ***, < 0.001, ****, < 0.0001.
The trend of primary bile acid accumulation in bile acid metabolism was supported by the enrichment analysis of functional genes annotated from fecal metagenomes. Based on gene family annotation against the KEGG Orthology (KO) database, K01442 (choloylglycine hydrolase, also known as bile salt hydrolases (BSHs); EC 3.5.1.24), K22605 (3alpha-hydroxycholanate dehydrogenase (NADP+)), and K15873 (7beta-hydroxy-3-oxochol-24-oyl-CoA 4-desaturase) were identified in the vehicle group, while K00076 (7-alpha-hydroxysteroid dehydrogenase) was found in both antibiotic and vehicle-treated mice (Table S3). K01442, contributed by Lactobacillus johnsonii and Muribaculum intestinale that were enriched in vehicle-treated group in this study, is the only KO that could function in both primary and secondary bile acid biosynthesis and was significantly lower in the antibiotic group compared to the vehicle group (p=0.0168) (Supplementary Figure 5). The other three KOs that only play a role in the secondary bile acid biosynthesis pathway did not differ significantly between the antibiotic and vehicle group. Due to the low abundance of K01442 (BSHs), the abundance of both primary bile acid biosynthesis and secondary bile acid biosynthesis pathways were significantly lower in antibiotic group (p= 0.0168 and 0.0358, respectively) (Table S3). However, considering microbial K01442 (BSHs) present in the gut can catalyze the hydrolysis of conjugated bile salts into deconjugated bile acids (Coleman and Hudson, 1995), our results suggested that antibiotic treatment would induce a shift in the gut microbiota composition and impair the BSH activity of microbiota thus inhibiting the secondary bile acid biosynthesis and accumulating primary bile acids, leading to an inhibitory effect on Nsp15 in the mouse GI tract.
The COVID-19 pandemic caused by SARS-CoV-2 is rapidly spreading and evolving around the world. Though conventional antiviral drugs (Holshue et al., 2020; McKee et al., 2020; NIH, 2022), traditional Chinese medicine (Yang et al., 2020; Wang and Yang, 2021; Shi et al., 2022), antibiotics (Mirzaei et al., 2020; Popp et al., 2021), and vaccines (Poland et al., 2020; Eyre et al., 2022; WHO, 2022a) have been applied in COVID-19 clinical therapy, the screening for drugs targeting SARS-CoV-2 critical proteins remains a long and arduous process
A considerable portion of COVID-19 patients showed liver damage, which might result from direct cholangiocyte injury and consequent bile acid accumulation induced by viral infection (Zhao B. et al., 2020). Bile acids are important substances involved in the regulation of the hepatic-intestinal axis. The liver converts cholesterol into primary bile acids, which are subsequently transported into bile and stored in the gallbladder until released into the intestine. The primary bile acids can be further metabolized by the gut microbiota in the jejunum or terminal ileum or can be circulated through the portal vein reabsorption to the liver (Ridlon et al., 2016).
The human intestinal microbiome can be disrupted by antibiotics (Adamsson et al., 1999; Jernberg et al., 2007; Jakobsson et al., 2010; Subramanian et al., 2014; de Gunzburg et al., 2018) that lead to the imbalance of microbiome-bile acid pool (Theriot et al., 2014; Kuno et al., 2018; Behr et al., 2019). After antibiotic treatment, the fecal microbial communities were dominated by Firmicutes and Proteobacteria (Jakobsson et al., 2010), which is consistent with our result that the microbiota of the antibiotic-treated mice was dominated by Citrobacter spp., Lactobacillus murinus, and Escherichia coli (Figure 3C). Antibiotic-induced microbiome depletion alters the production of bile acids (Ridlon et al., 2014). In particular, cholic acid (CA) and β-muricholic acid (bMCA) are significantly reduced, with an almost two-fold increase in TCA, a three-fold increase in tauro-β-muricholic acid (TbMCA), and a three-fold increase in taurochenodeoxycholic acid (TCDCA) (Zarrinpar et al., 2018). In this study, we found that antibiotic-induced alteration of gut microbiota led to an inhibitory effect on the activity of SARS-CoV-2 Nsp15 via accumulating primary bile acids, suggesting that Nsp15 is a potential drug target for therapies of coronavirus. However, the potential of TCA in the therapy of COVID-19 infection should be investigated in further studies. Follow-up studies could initially focus on assessing the antiviral effects of TCA using cellular models (Canal et al., 2021; Kim et al., 2021).
It is well known that a subset of patients with severe SARS-CoV-2 infection commonly develop a clinically severe hyperinflammatory state or cytokine storm, with respiratory distress syndrome (Ruan et al., 2020; Zhou et al., 2020). Over 70% of the patients with COVID-19 received antibiotics due to symptoms similar to bacterial pneumonia or secondary bacterial infections (Knight et al., 2021; Langford et al., 2021). However, numerous studies suggest that antibiotic prescriptions are excessively applied in the treatment on COVID-19 infection (Lai et al., 2020; Langford et al., 2020; Youngs et al., 2020; RECOVERY Collaborative Group, 2021; PRINCIPLE Trial Collaborative Group, 2021). The chronic overuse of antibiotics not only disrupts the composition of microbiota (Subramanian et al., 2014; Theriot et al., 2016; Kang et al., 2022), but also expands the spectrum of bacterial resistance to antibiotics, posing a threat to human health (Llor and Bjerrum, 2014; Huang et al., 2020). Moreover, antibiotic treatment leads to the imbalance of the microbiome-bile acid pool. In the present study, we found that the composition of gut microbiota changed under antibiotic treatment, as evidenced by a relative increase in Citrobacter and Lactobacillus bacteria, an absence of anti-inflammatory bacteria Akkermansia muciniphila, and the bile acid metabolism-related bacterias Lactobacillus and Clostridium. Although antibiotic treatment is likely to affect the anti-inflammation and bile acid metabolism in the gut, the resulting accumulation of primary bile acids contributes to the inhibitory effect on SARS-Cov-2 Nsp15, according to this study. The above results suggest that antibiotics may not only make the gut microbiome alterative, but also alter the bile acid metabolism of patients, and thus, protect the patients from SARS-CoV-2 infection.
Based on its cytoprotective, anti-inflammatory, and immunomodulatory properties, in particular, its beneficial effects of ursodeoxycholic acid (UDCA) in respiratory diseases such as effective modulation of Th2-derived cytokines and inhibition of apoptosis of airway epithelial cells in mouse models of chronic asthma, along with the stimulation of alveolar fluid clearance in a rat model of acute respiratory distress syndrome via the ALX/cAMP/PI3K pathway, the FDA-approved drug UDCA has been suggested for COVID-19 treatment (Abdulrab et al., 2020; Hirayama et al., 2021). Moreover, bile acids play an important regulatory role in viral replication (Schupp and Graf, 2014). For example, the replication of influenza A virus and hepatitis B virus (HBV) is inhibited by bile acids (Hassan et al., 2012; Mouzannar et al., 2019). Furthermore, bile acids have been found to enhance genotype 1 hepatitis C virus replication through FXR (Scholtes et al., 2008). In a recent study, primary and secondary bile acids and their amino derivatives, such as glycoursodeoxycholic acid, and semisynthetic derivatives, such as obeticholic acid, are found to reduce the binding between the RBD region of SARS-CoV-2 S protein and human ACE2 protein (Carino et al., 2020). Gut microbiota have been suggested as possible indicators of SARS-CoV-2 infection and to be involved in anti-infection (Yeoh et al., 2021; Zuo et al., 2021b). Therefore, it is important and valuable to explore the intestinal resources for applications in COVID-19 therapy. This study revealed that antibiotic treatment can induce a shift in the gut microbiota and lead to impaired microbial BSH activity. This alteration in microbiota-host co-metabolites was characterized by the blockage of secondary bile acid biosynthesis and the accumulation of primary bile acids, which was identified as Nsp15 inhibitors in this study. These findings will benefit the development of choline and its derivatives as SARS-CoV-2 inhibitors.
Bacterial strains and plasmids used in this study are listed in Table S1. All Escherichia coli strains used in this study were propagated in LB media containing appropriate antibiotics (100 μg/ml for ampicillin) at 37°C unless otherwise noted. The Endoribonuclease (nsp15) gene was optimized with E. coli codons and cloned into pET32a (+) plasmid. The pET32a (+) plasmid was transformed into E. coli BL21(DE3) cells to obtain a high yield of recombinant Nsp15 protein.
Recombinant SARS-CoV-2 Nsp15 protein was expressed, as previously described, with slight modification (Bhardwaj et al., 2004; Fan et al., 2004). Briefly, the overnight culture of E. coli BL21(DE3) cells containing the pET32a (+)-nsp15-His plasmids was sub-cultured 1:100 in 250 ml LB containing 100 μg/ml ampicillin. Cells were grown to the optical density at 600nm of 0.9-1.0 and then induced with 1 mM Isopropyl-β-D-1 thiogalactopyranoside (IPTG). After additional incubation at 18°C for 16 h, cells were harvested by centrifugation at 4°C, 8,000×g for 2 min, resuspended in 10 ml of cold lysis buffer, digested with 1 mg/ml lysozyme for 30 min, and finally lysed with sonication. Cell debris was removed by centrifugation at 4°C, 10,000×g for 30 min. The supernatant containing Nsp15 protein (Nsp15 crude enzyme) was added with 50% glycerol and 50 mM Tris-HCl (pH 8.0), and stored at -80°C. The Nsp15 crude enzyme was purified using Ni-NTA Agarose (QIAGEN, Germany), and dialyzed overnight against 20 mM PBS (pH 7.0) buffer containing 25% glycerol by 15 KDa dialysis membrane. Protein purity was determined by 12% polyacrylamide gel electrophoresis.
Fluorogenic RNA substrates 5’6-FAM-GAAGCGAAACCC/rU/AAG-3’BHQ1 (substrate N1), 5’6-FAM- GCGGAGCACG A/rU/CGAG - 3’BHQ1 (substrate N2), 5’6-FAM-UACGAUCG-3’BHQ1 (substrate N3) were synthesized by Tsingke Biotechnology (Beijing, China), 5’6-FAM - A/rU/AA - 3’BHQ1 (substrate N4) was prepared by Sangon Biotech (Shanghai, China). The RNA cleavage assay was performed at 30°C in the reaction buffer (50 mM Tris-HCl pH 7.5, 100 mM KCl, 5 mM MnCl2, 1 mM DTT) containing 1 µM fluorescent RNA substrate and 317 nM of Nsp15. Enhanced fluorescence due to cleavage of the fluorogenic substrate RNA was monitored at 518 nm with excitation at 492 nm (Bhardwaj et al., 2004), using a fluorescence plate reader (Cytation5, Bio Tek, US). Doubling of fluorescence intensity per minute in a reaction system containing the fluorescent RNA substrate N4 and SARS-CoV-2 Nsp15 compared to a reaction system containing only substrate was defined as one activity unit (U). The reaction system containing Benzopurpurine 4B (final concentration of 1 mM) was used as positive control in examination as previously described (Ortiz-Alcantara, 2010).
The antibiotic-treated adult mouse model was used to assess the impact of the intestinal environment on SARS-CoV-2 protein activity. We used male mice in this study in order to minimize potential experimental error. Briefly, six-week-old C57BL/6J males were randomly assigned to each experimental group (6 mice/group) and then supplied with drinking water with or without antibiotic cocktail (0.1 g/L vancomycin, 0.2 g/L neomycin, 0.2 g/L ampicillin, 0.2 g/L metronidazole) and 0.2 g/L aspartame for two weeks. To investigate the interaction between gut microbiota and CPMs used in the clinical treatment of COVID-19, the drug was administered or not by gavage once daily for the last 7 days using either LHQWC and JHQGG. Fecal pellets were collected from each mouse daily throughout the experiment. Samples for metagenomic shotgun sequencing were collected at day 14 and directly frozen at -80°C, while samples for endoribonuclease assay and bacteria isolation were collected at the indicated time-point and homogenized with ddH2O. Germ-free C57BL/6J mice fecal samples were obtained from the College of Animal Sciences and Technology at Huazhong Agricultural University in Wuhan, China.
We used the phenol/chloroform/isoamyl alcohol method to extract microbial DNA from fecal pellets of antibiotic-treated and vehicle-treated mice (Zoetendal et al., 2006). DNA degradation and potential contamination was monitored on 1% agarose gels and the DNA concentration was measured using Qubit® dsDNA BR Assay Kit in Qubit® 2.0 Fluorometer (Life Technologies, CA, USA). The DNA that passed quality control was then subjected to library construction using the TruSeq DNA HT Sample Prep Kit. Paired-end 150 metagenomic shotgun sequencing (mean of 39,350,387 reads per sample) was performed on the Illumina Hi-Seq platform. The sequencing reads subjected to quality control using Kneaddata (https://github.com/biobakery/biobakery/wiki/kneaddata) and host sequences were removed based on the mouse_C57BL_6NJ database. High quality sequences were used for quantitative profiling of the taxonomic composition of the microbial communities of all samples using MetaPhlAn 3.0 (Segata et al., 2012) (https://github.com/biobakery/MetaPhlAn/wiki/MetaPhlAn-3.0), whereas HUMAnN 3.0 (Franzosa et al., 2018) (https://huttenhower.sph.harvard.edu/humann) was used to profile gene-family abundances. Gene families determined by UniRef were mapped to the KEGG Orthology (KO) database and were grouped into the KEGG pathway. The alpha diversity index (Shannon index) and Bray-Curtis dissimilarities were calculated using Vegan package in R. Statistical analysis was performed on Shannon index and KO between groups using a non-parametric Wilcoxon test with a p-value < 0.05. Principal coordinate analysis (PCoA) was performed on beta diversity based on Bray-Curtis distances and the differences were compared using permutational multivariate ANOVA (PERMANOVA) with 9,999 permutations. LEfSe (Segata et al., 2011) was used to quantitate differential taxonomic abundance and pathway abundance using a Kruskal-Wallis test with α value of 0.05 and a log linear discriminant analysis (LDA) score cut-off of 2.
The metagenomic data have been deposited at Sequence Read Archive database of NCBI with the accession number PRJNA734104.
The animal study was reviewed and approved by The Ethical Committee of Huazhong University of Science and Technology. (Permit Number: SYXK (E) 2016-0057).
ZL, YM, and XF conceived the study and designed the experiment. ML, YD, XY, XW, and GC performed the experiment. YSD, LH, YC, PW, HW, and MN analyzed the data. YM, ML, YSD, XY, GC, and ZQ wrote the manuscript. All authors critically revised the manuscript. All authors read and approved the final manuscript.
This work was supported by the National Key R&D Program of China (2019YFA0905600), and the HUST COVID-19 Rapid Response Call (2020kfyXGYJ047), the Specific Fund of State Key Laboratory of Dampness Syndrome of Chinese Medicine (SZ2021ZZ28), and the Science and Technology Planning Project of Guangdong Province (2020B1111100005).
YSD and LH are employed by KMHD, Shenzhen, China.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
We thank Dr. Shengce Tao (Shanghai Jiaotong University) for providing the plasmid expressing recombinant SARS-CoV-2 protein. We thank Dr. Bin Zhu’s lab (Huazhong University of Science and Technology) for providing technical support in purifying Nsp15 and conducting biochemical assays, the Research Core Facilities for Life Science (RCFLS) in Huazhong University of Science and Technology for assistance with fluorescence plate reader analysis.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcimb.2022.896504/full#supplementary-material
Supplementary Figure 1 | Antibiotics had no impact on Nsp15 activity. The effect of antibiotics on Nsp15 activity was evaluated in vitro at 1-100 times the dose used in animal experiments (gray bars). The reaction systems with ddH2O (black bar) and Benzopurpurine 4B (white bar), instead of the sample to be tested, were negative and positive controls, respectively. Relative activity (% activity) was calculated for each sample by comparing with the negative control. Values expressed are means ± S.D. from three independent experiments. Significance was determined by One-way ANOVA; p-value: ****, < 0.0001.
Supplementary Figure 2 | Antibiotic-induced enrichment of Citrobacter spp. did not relate to the Nsp15 inhibition in feces. Supernatant of Citrobacter spp. cultures was prepared aerobically (gray bars) or anaerobically (white bars filled with grey slashes) in LB with or without feed, respectively, to assay the effect on Nsp15 activity in vitro. The reaction systems with ddH2O and Benzopurpurine 4B, instead of the sample to be tested, were used as negative and positive controls, respectively. Relative activity (% activity) was calculated for each sample by comparing with the negative control. Values expressed are means ± S.D. from three experiments. Significance of difference was determined by One-way ANOVA; p-value: ****, < 0.0001.
Supplementary Figure 3 | Heatmap of pathways related to metabolism enriched in the vehicle group. Pathways enriched in the vehicle group were identified using LEfSe with LDA score >2 and Kruskal-Wallis test p-value < 0.05. The color of cells from blue to red corresponds to the scaling for the relative abundance of pathways from low to high. Pathways that were regrouped to carbohydrate and glycan metabolism, vitamin metabolism, bile acid metabolism, amino acid metabolism, fatty acids, and other lipid metabolism, etc. were labeled with different colors. Both rows and columns are clustered using correlation distance and complete linkage.
Supplementary Figure 4 | The effect of antibiotic-enriched metabolites on Nsp15 enzyme activity. (A) Antibiotic-enriched oligosaccharide metabolites did not inhibit Nsp15 enzyme activity. The highly abundant oligosaccharide metabolites such as fructose, cellobiose, and maltose were subjected to in vitro assay for the inhibitory effect on Nsp15. The reaction systems with ddH2O and Benzopurpurine 4B, instead of the sample to be tested, were negative and positive controls, respectively. Relative activity (% activity) was calculated for each sample by comparing with negative control. Values expressed are means ± S.D. from three experiments. Significance was determined by One-way ANOVA; p-value: ****, < 0.0001. (B) The effect of primary bile acids CDCA at different concentrations effect of Nsp15 activity. The reaction systems with ddH2O and Benzopurpurine 4B, instead of the sample to be tested, were negative and positive controls, respectively. Relative activity (% activity) was calculated for each sample by comparing with negative control. Values expressed are means ± S.D. from three independent experiments. Significance was determined by One-way ANOVA; p-value: ****, < 0.0001.
Supplementary Figure 5 | Enrichment analysis of functional genes annotated from metagenomes of mouse fecal samples. Gene families were mapped to the KO database and screened out KOs in the bile acid metabolism pathway (primary bile acid biosynthesis, KEGG map00120; secondary bile acid biosynthesis, KEGG map00121). K01442 (choloylglycine hydrolase; EC 3.5.1.24), K22605 (3alpha-hydroxycholanate dehydrogenase (NADP+)), K00076 (7-alpha-hydroxysteroid dehydrogenase), and K15873 (7beta-hydroxy-3-oxochol-24-oyl-CoA 4-desaturase) were annotated as a result. Red and blue boxes indicated the antibiotic group and vehicle group, respectively. The Wilcoxon test was applied to identify the differentially features. p-value: *, < 0.05.
Table S2 | Annotation of gene families related to metabolism.
Table S3 | Annotation of gene families related to bile acid metabolism.
Abdulrab, S., Al-Maweri, S., Halboub, E. (2020). Ursodeoxycholic Acid as a Candidate Therapeutic to Alleviate and/or Prevent COVID-19-Associated Cytokine Storm. Med. Hypotheses 143, 109897. doi: 10.1016/j.mehy.2020.109897
Adamsson, I., Nord, C. E., Lundquist, P., Sjostedt, S., Edlund, C. (1999). Comparative Effects of Omeprazole, Amoxycillin Plus Metronidazole Versus Omeprazole, Clarithromycin Plus Metronidazole on the Oral, Gastric and Intestinal Microflora in Helicobacter Pylori-Infected Patients. J. Antimicrob. Chemother. 44 (5), 629–640. doi: 10.1093/jac/44.5.629
Behr, C., Slopianka, M., Haake, V., Strauss, V., Sperber, S., Kamp, H., et al. (2019). Analysis of Metabolome Changes in the Bile Acid Pool in Feces and Plasma of Antibiotic-Treated Rats. Toxicol. Appl. Pharmacol. 363, 79–87. doi: 10.1016/j.taap.2018.11.012
Bhardwaj, K., Guarino, L., Kao, C. C. (2004). The Severe Acute Respiratory Syndrome Coronavirus Nsp15 Protein is an Endoribonuclease That Prefers Manganese as a Cofactor. J. Virol. 78 (22), 12218–12224. doi: 10.1128/JVI.78.22.12218-12224.2004
Bhardwaj, K., Sun, J., Holzenburg, A., Guarino, L. A., Kao, C. C. (2006). RNA Recognition and Cleavage by the SARS Coronavirus Endoribonuclease. J. Mol. Biol. 361 (2), 243–256. doi: 10.1016/j.jmb.2006.06.021
Boute, N., Jockers, R., Issad, T. (2002). The Use of Resonance Energy Transfer in High-Throughput Screening: BRET Versus FRET. Trends Pharmacol. Sci. 23 (8), 351–354. doi: 10.1016/s0165-6147(02)02062-x
Canal, B., McClure, A. W., Curran, J. F., Wu, M., Ulferts, R., Weissmann, F., et al. (2021). Identifying SARS-CoV-2 Antiviral Compounds by Screening for Small Molecule Inhibitors of Nsp14/Nsp10 Exoribonuclease. Biochem. J. 478 (13), 2445–2464. doi: 10.1042/BCJ20210198
Carino, A., Moraca, F., Fiorillo, B., Marchianò, S., Sepe, V., Biagioli, M., et al. (2020). Hijacking SARS-CoV-2/ACE2 Receptor Interaction by Natural and Semi-Synthetic Steroidal Agents Acting on Functional Pockets on the Receptor Binding Domain. Front. Chem. 8 (846). doi: 10.3389/fchem.2020.572885
Coleman, J. P., Hudson, L. L. (1995). Cloning and Characterization of a Conjugated Bile Acid Hydrolase Gene From Clostridium Perfringens. Appl. Environ. Microbiol. 61 (7), 2514–2520. doi: 10.1128/aem.61.7.2514-2520.1995
Cremers, C. M., Knoefler, D., Vitvitsky, V., Banerjee, R., Jakob, U. (2014). Bile Salts Act as Effective Protein-Unfolding Agents and Instigators of Disulfide Stress In Vivo. Proc. Natl. Acad. Sci. 111 (16), E1610–E1619. doi: 10.1073/pnas.1401941111
de Gunzburg, J., Ghozlane, A., Ducher, A., Le Chatelier, E., Duval, X., Ruppe, E., et al. (2018). Protection of the Human Gut Microbiome From Antibiotics. J. Infect. Dis. 217 (4), 628–636. doi: 10.1093/infdis/jix604
Deng, X., Baker, S. C. (2018). An "Old" Protein With a New Story: Coronavirus Endoribonuclease is Important for Evading Host Antiviral Defenses. Virology 517, 157–163. doi: 10.1016/j.virol.2017.12.024
Deng, X., Hackbart, M., Mettelman, R. C., O’Brien, A., Mielech, A. M., Yi, G., et al. (2017). Coronavirus Nonstructural Protein 15 Mediates Evasion of dsRNA Sensors and Limits Apoptosis in Macrophages. Proc. Natl. Acad. Sci. 114 (21), E4251–E4260. doi: 10.1073/pnas.1618310114
Ding, Y., Zeng, L., Li, R., Chen, Q., Zhou, B., Chen, Q., et al. (2017). The Chinese Prescription Lianhuaqingwen Capsule Exerts Anti-Influenza Activity Through the Inhibition of Viral Propagation and Impacts Immune Function. BMC Complement Altern. Med. 17 (1), 130. doi: 10.1186/s12906-017-1585-7
Eyre, D. W., Taylor, D., Purver, M., Chapman, D., Fowler, T., Pouwels, K. B., et al. (2022). Effect of Covid-19 Vaccination on Transmission of Alpha and Delta Variants. N Engl. J. Med. 386 (8), 744–756. doi: 10.1056/NEJMoa2116597
Fan, K., Wei, P., Feng, Q., Chen, S., Huang, C., Ma, L., et al. (2004). Biosynthesis, Purification, and Substrate Specificity of Severe Acute Respiratory Syndrome Coronavirus 3C-Like Proteinase. J. Biol. Chem. 279 (3), 1637–1642. doi: 10.1074/jbc.M310875200
Franzosa, E. A., McIver, L. J., Rahnavard, G., Thompson, L. R., Schirmer, M., Weingart, G., et al. (2018). Species-Level Functional Profiling of Metagenomes and Metatranscriptomes. Nat. Methods 15 (11), 962–968. doi: 10.1038/s41592-018-0176-y
PRINCIPLE Trial Collaborative Group. (2021). Azithromycin for Community Treatment of Suspected COVID-19 in People at Increased Risk of an Adverse Clinical Course in UK (PRINCIPLE): A Randomised, Controlled, Open-Label, Adaptive Platform Trial. Lancet 397 (10279), 1063–1074. doi: 10.1016/S0140-6736(21)00461-X
RECOVERY Collaborative Group. (2021). Azithromycin in Patients Admitted to Hospital With COVID-19 (RECOVERY): A Randomised, Controlled, Open-Label, Platform Trial. Lancet 397 (10274), 605–612. doi: 10.1016/S0140-6736(21)00149-5
Gutierrez, D., Weinstock, A., Antharam, V. C., Gu, H., Jasbi, P., Shi, X., et al. (2019). Antibiotic-Induced Gut Metabolome and Microbiome Alterations Increase the Susceptibility to Candida Albicans Colonization in the Gastrointestinal Tract. FEMS Microbiol. Ecol. 96 (1), fiz187 doi: 10.1093/femsec/fiz187
Hassan, I. H., Zhang, M. S., Powers, L. S., Shao, J. Q., Baltrusaitis, J., Rutkowski, D. T., et al. (2012). Influenza A Viral Replication is Blocked by Inhibition of the Inositol-Requiring Enzyme 1 (IRE1) Stress Pathway. J. Biol. Chem. 287 (7), 4679–4689. doi: 10.1074/jbc.M111.284695
Hirayama, M., Nishiwaki, H., Hamaguchi, T., Ito, M., Ueyama, J., Maeda, T., et al. (2021). Intestinal Collinsella may Mitigate Infection and Exacerbation of COVID-19 by Producing Ursodeoxycholate. PloS One 16 (11), e0260451–e0260451. doi: 10.1371/journal.pone.0260451
Holshue, M. L., DeBolt, C., Lindquist, S., Lofy, K. H., Wiesman, J., Bruce, H., et al. (2020). First Case of 2019 Novel Coronavirus in the United States. New Engl. J. Med. 382 (10), 929–936. doi: 10.1056/NEJMoa2001191
Huang, C., Wang, Y., Li, X., Ren, L., Zhao, J., Hu, Y., et al. (2020). Clinical Features of Patients Infected With 2019 Novel Coronavirus in Wuhan, China. Lancet 395 (10223), 497–506. doi: 10.1016/S0140-6736(20)30183-5
Huang, K., Zhang, P., Zhang, Z., Youn, J. Y., Wang, C., Zhang, H., et al. (2021). Traditional Chinese Medicine (TCM) in the Treatment of COVID-19 and Other Viral Infections: Efficacies and Mechanisms. Pharmacol. Ther. 225, 107843. doi: 10.1016/j.pharmthera.2021.107843
Ivanov, K. A., Hertzig, T., Rozanov, M., Bayer, S., Thiel, V., Gorbalenya, A. E., et al. (2004). Major Genetic Marker of Nidoviruses Encodes a Replicative Endoribonuclease. Proc. Natl. Acad. Sci. United States America 101 (34), 12694–12699. doi: 10.1073/pnas.0403127101
Izda, V., Jeffries, M. A., Sawalha, A. H. (2021). COVID-19: A Review of Therapeutic Strategies and Vaccine Candidates. Clin. Immunol. 222, 108634. doi: 10.1016/j.clim.2020.108634
Jakobsson, H. E., Jernberg, C., Andersson, A. F., Sjolund-Karlsson, M., Jansson, J. K., Engstrand, L. (2010). Short-Term Antibiotic Treatment has Differing Long-Term Impacts on the Human Throat and Gut Microbiome. PloS One 5 (3), e9836. doi: 10.1371/journal.pone.0009836
Jernberg, C., Lofmark, S., Edlund, C., Jansson, J. K. (2007). Long-Term Ecological Impacts of Antibiotic Administration on the Human Intestinal Microbiota. ISME J. 1 (1), 56–66. doi: 10.1038/ismej.2007.3
Kang, Y., Chen, S., Chen, Y., Tian, L., Wu, Q., Zheng, M., et al. (2022). Alterations of Fecal Antibiotic Resistome in COVID-19 Patients After Empirical Antibiotic Exposure. Int. J. Hyg Environ. Health 240, 113882. doi: 10.1016/j.ijheh.2021.113882
Kim, Y., Jedrzejczak, R., Maltseva, N. I., Wilamowski, M., Endres, M., Godzik, A., et al. (2020). Crystal Structure of Nsp15 Endoribonuclease NendoU From SARS-CoV-2. Protein Sci. 29 (7), 1596–1605. doi: 10.1002/pro.3873
Kim, K. A., Jung, I. H., Park, S. H., Ahn, Y. T., Huh, C. S., Kim, D. H. (2013). Comparative Analysis of the Gut Microbiota in People With Different Levels of Ginsenoside Rb1 Degradation to Compound K. PloS One 8 (4), e62409. doi: 10.1371/journal.pone.0062409
Kim, Y., Wower, J., Maltseva, N., Chang, C., Jedrzejczak, R., Wilamowski, M., et al. (2021). Tipiracil Binds to Uridine Site and Inhibits Nsp15 Endoribonuclease NendoU From SARS-CoV-2. Commun. Biol. 4 (1), 193. doi: 10.1038/s42003-021-01735-9
Knight, G. M., Glover, R. E., McQuaid, C. F., Olaru, I. D., Gallandat, K., Leclerc, Q. J., et al. (2021). Antimicrobial Resistance and COVID-19: Intersections and Implications. Elife 10. e64139 doi: 10.7554/eLife.64139
Kuno, T., Hirayama-Kurogi, M., Ito, S., Ohtsuki, S. (2018). Reduction in Hepatic Secondary Bile Acids Caused by Short-Term Antibiotic-Induced Dysbiosis Decreases Mouse Serum Glucose and Triglyceride Levels. Sci. Rep. 8 (1), 1253. doi: 10.1038/s41598-018-19545-1
Lai, C.-C., Shih, T.-P., Ko, W.-C., Tang, H.-J., Hsueh, P.-R. (2020). Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2) and Coronavirus Disease-2019 (COVID-19): The Epidemic and the Challenges. Int. J. antimicrobial Agents 55 (3), 105924. doi: 10.1016/j.ijantimicag.2020.105924
Langford, B. J., So, M., Raybardhan, S., Leung, V., Soucy, J. R., Westwood, D., et al. (2021). Antibiotic Prescribing in Patients With COVID-19: Rapid Review and Meta-Analysis. Clin. Microbiol. Infect. 27 (4), 520–531. doi: 10.1016/j.cmi.2020.12.018
Langford, B. J., So, M., Raybardhan, S., Leung, V., Westwood, D., MacFadden, D. R., et al. (2020). Bacterial Co-Infection and Secondary Infection in Patients With COVID-19: A Living Rapid Review and Meta-Analysis. Clin. Microbiol. Infect. 26 (12), 1622–1629. doi: 10.1016/j.cmi.2020.07.016
Lin, T.-L., Lu, C.-C., Lai, W.-F., Wu, T.-S., Lu, J.-J., Chen, Y.-M., et al. (2021). Role of Gut Microbiota in Identification of Novel TCM-Derived Active Metabolites. Protein Cell. 12 (5), 394–410. doi: 10.1007/s13238-020-00784-w
Liu, Z., Li, X., Gou, C., Li, L., Luo, X., Zhang, C., et al. (2020). Effect of Jinhua Qinggan Granules on Novel Coronavirus Pneumonia in Patients. J. Tradit Chin. Med. 40 (3), 467–472. doi: 10.19852/j.cnki.jtcm.2020.03.016
Llor, C., Bjerrum, L. (2014). Antimicrobial Resistance: Risk Associated With Antibiotic Overuse and Initiatives to Reduce the Problem. Ther. Adv. Drug Saf. 5 (6), 229–241. doi: 10.1177/2042098614554919
Luo, H., Gao, Y., Zou, J., Zhang, S., Chen, H., Liu, Q., et al. (2020). Reflections on Treatment of COVID-19 With Traditional Chinese Medicine. Chin. Med. 15, 94. doi: 10.1186/s13020-020-00375-1
McKee, D. L., Sternberg, A., Stange, U., Laufer, S., Naujokat, C. (2020). Candidate Drugs Against SARS-CoV-2 and COVID-19. Pharmacol. Res. 157, 104859. doi: 10.1016/j.phrs.2020.104859
Mirzaei, R., Goodarzi, P., Asadi, M., Soltani, A., Aljanabi, H. A. A., Jeda, A. S., et al. (2020). Bacterial Co-Infections With SARS-CoV-2. IUBMB Life 72 (10), 2097–2111. doi: 10.1002/iub.2356
Mouzannar, K., Fusil, F., Lacombe, B., Ollivier, A., Ménard, C., Lotteau, V., et al. (2019). Farnesoid X Receptor-α is a Proviral Host Factor for Hepatitis B Virus That is Inhibited by Ligands In Vitro and In Vivo. FASEB J. Off. Publ. Fed. Am. Societies Exp. Biol. 33 (2), 2472–2483. doi: 10.1096/fj.201801181R
NIH (2022) Coronavirus Disease 2019 (COVID-19) Treatment Guidelines [Online] (National Institutes of Health). Available at: https://www.covid19treatmentguidelines.nih.gov/ (Accessed May 13 2022).
Ortiz-Alcantara J, B. K., Palaninathan, S., Frieman, M., Baric, R., Kao, C. (2010). Small Molecule Inhibitors of the SARS-CoV Nsp15 Endoribonuclease. Virus Adaptation Treat 2010 (2), 125—133. doi: 10.2147/VAAT.S12733
Poland, G. A., Ovsyannikova, I. G., Crooke, S. N., Kennedy, R. B. (2020). SARS-CoV-2 Vaccine Development: Current Status. Mayo Clinic Proc. 95 (10), 2172–2188. doi: 10.1016/j.mayocp.2020.07.021
Popp, M., Stegemann, M., Riemer, M., Metzendorf, M. I., Romero, C. S., Mikolajewska, A., et al. (2021). Antibiotics for the Treatment of COVID-19. Cochrane Database Systematic Rev. 10 (10), CD015025. doi: 10.1002/14651858.Cd015025
Quinn, R. A., Melnik, A. V., Vrbanac, A., Fu, T., Patras, K. A., Christy, M. P., et al. (2020). Global Chemical Effects of the Microbiome Include New Bile-Acid Conjugations. Nature 579 (7797), 123–129. doi: 10.1038/s41586-020-2047-9
Ridlon, J. M., Harris, S. C., Bhowmik, S., Kang, D.-J., Hylemon, P. B. (2016). Consequences of Bile Salt Biotransformations by Intestinal Bacteria. Gut Microbes 7 (1), 22–39. doi: 10.1080/19490976.2015.1127483
Ridlon, J. M., Kang, D. J., Hylemon, P. B., Bajaj, J. S. (2014). Bile Acids and the Gut Microbiome. Curr. Opin. Gastroenterol. 30 (3), 332–338. doi: 10.1097/MOG.0000000000000057
Robertson, M. P., Igel, H., Baertsch, R., Haussler, D., Ares, M., Jr., Scott, W. G. (2005). The Structure of a Rigorously Conserved RNA Element Within the SARS Virus Genome. PloS Biol. 3 (1), e5–e5. doi: 10.1371/journal.pbio.0030005
Rodrigues, R. R., Greer, R. L., Dong, X., DSouza, K. N., Gurung, M., Wu, J. Y., et al. (2017). Antibiotic-Induced Alterations in Gut Microbiota Are Associated With Changes in Glucose Metabolism in Healthy Mice. Front. Microbiol. 8 (2306). doi: 10.3389/fmicb.2017.02306
Ruan, Q., Yang, K., Wang, W., Jiang, L., Song, J. (2020). Clinical Predictors of Mortality Due to COVID-19 Based on an Analysis of Data of 150 Patients From Wuhan, China. Intensive Care Med. 46 (5), 846–848. doi: 10.1007/s00134-020-05991-x
Scavone, C., Mascolo, A., Rafaniello, C., Sportiello, L., Trama, U., Zoccoli, A., et al. (2022). Therapeutic Strategies to Fight COVID-19: Which is the Status Artis? Br. J. Pharmacol. 179 (10), 2128–2148. doi: 10.1111/bph.15452
Scholtes, C., Diaz, O., Icard, V., Kaul, A., Bartenschlager, R., Lotteau, V., et al. (2008). Enhancement of Genotype 1 Hepatitis C Virus Replication by Bile Acids Through FXR. J. Hepatol. 48 (2), 192–199. doi: 10.1016/j.jhep.2007.09.015
Schupp, A.-K., Graf, D. (2014). Bile Acid-Induced Modulation of Virus Replication. Eur. J. Med. Res. 19 (Suppl 1), S27–S27. doi: 10.1186/2047-783X-19-S1-S27
Segata, N., Izard, J., Waldron, L., Gevers, D., Miropolsky, L., Garrett, W. S., et al. (2011). Metagenomic Biomarker Discovery and Explanation. Genome Biol. 12 (6), R60. doi: 10.1186/gb-2011-12-6-r60
Segata, N., Waldron, L., Ballarini, A., Narasimhan, V., Jousson, O., Huttenhower, C. (2012). Metagenomic Microbial Community Profiling Using Unique Clade-Specific Marker Genes. Nat. Methods 9 (8), 811–814. doi: 10.1038/nmeth.2066
Shi, S. H., Wang, F., Yao, H., Kou, S., Li, W. H., Chen, B. A., et al. (2022). Oral Chinese Herbal Medicine on Immune Responses During Coronavirus Disease 2019: A Systematic Review and Meta-Analysis. Front. Med. 8. doi: 10.3389/fmed.2021.685734
Subramanian, S., Huq, S., Yatsunenko, T., Haque, R., Mahfuz, M., Alam, M. A., et al. (2014). Persistent Gut Microbiota Immaturity in Malnourished Bangladeshi Children. Nature 510 (7505), 417–421. doi: 10.1038/nature13421
Tao, Z., Yang, Y., Shi, W., Xue, M., Yang, W., Song, Z., et al. (2013). Complementary and Alternative Medicine is Expected to Make Greater Contribution in Controlling the Prevalence of Influenza. Biosci. Trends 7 (5), 253–256. doi: 10.5582/bst.2013.v7.5.253
Theriot, C. M., Bowman, A. A., Young, V. B. (2016). Antibiotic-Induced Alterations of the Gut Microbiota Alter Secondary Bile Acid Production and Allow for Clostridium Difficile Spore Germination and Outgrowth in the Large Intestine. mSphere 1 (1), e00045-15. doi: 10.1128/mSphere.00045-15
Theriot, C. M., Koenigsknecht, M. J., Carlson, P. E., Jr., Hatton, G. E., Nelson, A. M., Li, B., et al. (2014). Antibiotic-Induced Shifts in the Mouse Gut Microbiome and Metabolome Increase Susceptibility to Clostridium Difficile Infection. Nat. Commun. 5, 3114–3114. doi: 10.1038/ncomms4114
Wang, L., Liu, Q. M., Sung, B. H., An, D. S., Lee, H. G., Kim, S. G., et al. (2011). Bioconversion of Ginsenosides Rb(1), Rb(2), Rc and Rd by Novel β-Glucosidase Hydrolyzing Outer 3-O Glycoside From Sphingomonas Sp. 2F2: Cloning, Expression, and Enzyme Characterization. J. Biotechnol. 156 (2), 125–133. doi: 10.1016/j.jbiotec.2011.07.024
Wang, Z. L., Yang, L. Y.. (2021). hinese Herbal Medicine: Fighting SARSCoV-2 Infection on All Fronts. J. Ethnopharmacology 270: 113869. doi: 10.1016/j.jep.2021.113869
WHO (2022a) COVID-19 Vaccine Tracker and Landscape. Available at: https://www.who.int/publications/m/item/draft-landscape-of-covid-19-candidate-vaccines (Accessed 24 May 2022).
WHO (2022b) WHO Coronavirus (COVID-19) Dashboard. Available at: https://covid19.who.int (Accessed 27 May 2022).
Xiao, Y., Zhou, K., Lu, Y., Yan, W., Cai, W., Wang, Y. (2018). Administration of Antibiotics Contributes to Cholestasis in Pediatric Patients With Intestinal Failure via the Alteration of FXR Signaling. Exp. Mol. Med. 50 (12), 1–14. doi: 10.1038/s12276-018-0181-3
Yang, Y., Islam, M. S., Wang, J., Li, Y., Chen, X. (2020). Traditional Chinese Medicine in the Treatment of Patients Infected With 2019-New Coronavirus (SARS-CoV-2): A Review and Perspective. Int. J. Biol. Sci. 16 (10), 1708–1717. doi: 10.7150/ijbs.45538
Yeoh, Y. K., Zuo, T., Lui, G. C., Zhang, F., Liu, Q., Li, A. Y., et al. (2021). Gut Microbiota Composition Reflects Disease Severity and Dysfunctional Immune Responses in Patients With COVID-19. Gut 70(4): 698–706. doi: 10.1136/gutjnl-2020-323020
Youngs, J., Wyncoll, D., Hopkins, P., Arnold, A., Ball, J., Bicanic, T. (2020). Improving Antibiotic Stewardship in COVID-19: Bacterial Co-Infection is Less Common Than With Influenza. J. Infect. 81 (3), e55–e57. doi: 10.1016/j.jinf.2020.06.056
Zarrinpar, A., Chaix, A., Xu, Z. Z., Chang, M. W., Marotz, C. A., Saghatelian, A., et al. (2018). Antibiotic-Induced Microbiome Depletion Alters Metabolic Homeostasis by Affecting Gut Signaling and Colonic Metabolism. Nat. Commun. 9 (1), 2872. doi: 10.1038/s41467-018-05336-9
Zhang, J., Garrett, S., Sun, J. (2021). Gastrointestinal Symptoms, Pathophysiology, and Treatment in COVID-19. Genes Dis. 8 (4), 385–400. doi: 10.1016/j.gendis.2020.08.013
Zhang, Y., Limaye, P. B., Renaud, H. J., Klaassen, C. D. (2014). Effect of Various Antibiotics on Modulation of Intestinal Microbiota and Bile Acid Profile in Mice. Toxicol. Appl. Pharmacol. 277 (2), 138–145. doi: 10.1016/j.taap.2014.03.009
Zhao, B., Ni, C., Gao, R., Wang, Y., Yang, L., Wei, J., et al. (2020). Recapitulation of SARS-CoV-2 Infection and Cholangiocyte Damage With Human Liver Ductal Organoids. Protein Cell 11 (10), 771–775. doi: 10.1007/s13238-020-00718-6
Wei, P.-F. (edt.) (2020). "Diagnosis and Treatment Protocol for Novel Coronavirus Pneumonia (Trial Version 7)."Chin. Med. J. 133 (9), 1087–1095. doi: 10.1097/CM9.0000000000000819
Zhou, F., Yu, T., Du, R., Fan, G., Liu, Y., Liu, Z., et al. (2020). Clinical Course and Risk Factors for Mortality of Adult Inpatients With COVID-19 in Wuhan, China: A Retrospective Cohort Study. Lancet (London England) 395 (10229), 1054–1062. doi: 10.1016/S0140-6736(20)30566-3
Zhuang, W., Fan, Z., Chu, Y., Wang, H., Yang, Y., Wu, L., et al. (2020). Chinese Patent Medicines in the Treatment of Coronavirus Disease 2019 (COVID-19) in China. Front. Pharmacol. 11. doi: 10.3389/fphar.2020.01066
Zoetendal, E. G., Booijink, C. C., Klaassens, E. S., Heilig, H. G., Kleerebezem, M., Smidt, H., et al. (2006). Isolation of RNA From Bacterial Samples of the Human Gastrointestinal Tract. Nat. Protoc. 1 (2), 954–959. doi: 10.1038/nprot.2006.143
Zuo, T., Liu, Q., Zhang, F., Lui, G. C., Tso, E. Y., Yeoh, Y. K., et al. (2021a). Depicting SARS-CoV-2 Faecal Viral Activity in Association With Gut Microbiota Composition in Patients With COVID-19. Gut 70 (2), 276–284. doi: 10.1136/gutjnl-2020-322294
Keywords: SARS-CoV-2, Nsp15, antibiotics, gut microbiota, primary bile acids
Citation: Ma Y, Luo M, Deng Y, Yang X, Wang X, Chen G, Qin Z, Deng Y, Nan M, Chen Y, Wang P, Wei H, Han L, Fang X and Liu Z (2022) Antibiotic-Induced Primary Biles Inhibit SARS-CoV-2 Endoribonuclease Nsp15 Activity in Mouse Gut. Front. Cell. Infect. Microbiol. 12:896504. doi: 10.3389/fcimb.2022.896504
Received: 15 March 2022; Accepted: 14 June 2022;
Published: 28 July 2022.
Edited by:
Tao Lin, Baylor College of Medicine, United StatesReviewed by:
Amy Sims, Pacific Northwest National Laboratory (DOE), United StatesCopyright © 2022 Ma, Luo, Deng, Yang, Wang, Chen, Qin, Deng, Nan, Chen, Wang, Wei, Han, Fang and Liu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Zhi Liu, emhpbGl1QGh1c3QuZWR1LmNu; Xiaodong Fang, ZmFuZ3hkQGJnaS5jb20=; Lijuan Han, aGxqQGttaGRnZW5lLmNvbQ==
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.