
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
MINI REVIEW article
Front. Cell. Infect. Microbiol., 22 April 2022
Sec. Clinical Microbiology
Volume 12 - 2022 | https://doi.org/10.3389/fcimb.2022.866463
This article is part of the Research TopicBeyond Traditional Culture: New Approaches for a Rapid Detection and Identification of Microorganisms and their Antimicrobial ResistanceView all 10 articles
Rapid and accurate identification of pathogens causing infections is one of the biggest challenges in medicine. Timely identification of causative agents and their antimicrobial resistance profile can significantly improve the management of infection, lower costs for healthcare, mitigate ever-growing antimicrobial resistance and in many cases, save lives. Raman spectroscopy was shown to be a useful—quick, non-invasive, and non-destructive —tool for identifying microbes from solid and liquid media. Modifications of Raman spectroscopy and/or pretreatment of samples allow single-cell analyses and identification of microbes from various samples. It was shown that those non-culture-based approaches could also detect antimicrobial resistance. Moreover, recent studies suggest that a combination of Raman spectroscopy with optical tweezers has the potential to identify microbes directly from human body fluids. This review aims to summarize recent advances in non-culture-based approaches of identification of microbes and their virulence factors, including antimicrobial resistance, using methods based on Raman spectroscopy in the context of possible use in the future point-of-care diagnostic process.
Microorganisms play irreplaceable roles in human existence—we can find them everywhere. In fact, human bodies include more microbial cells than their own cells (Sender et al., 2016). At the same time, it is estimated that approximately 1400 pathogens (including bacteria, fungi, viruses, protozoa, and helminths) can cause infections in humans (Balloux and van Dorp, 2017; Franco-Duarte et al., 2019).
Despite enormous progress in medicine during the last decades, accurate and rapid species-level identification of pathogens causing infections and their virulence factors (including antimicrobial resistance and ability to form biofilms) still poses a challenge. It is important to accent that timely identification and characterization of pathogens is essential for choosing a suitable tailored antimicrobial treatment and proper management of patients. This, in turn, leads to the shortening of hospital stays, reducing costs and time to adequate treatment, increasing the wellbeing of patients, reducing the spread of antimicrobial resistance, and, above all, saving the lives of many patients.
Basically, we can divide existing identification methods into two groups: culture-based and direct approaches (without cultivation). Major advantages and disadvantages of established methods are summarized in the Table 1.
Culture-based approaches are widely used in clinical diagnostics; they could be considered “golden standards”. Cultivation provides large amounts of microbial cells for further testing and offers a way to separate different microbes from a mixed culture. This allows the application of various identification and characterization methods separately or in combinations. However, it also makes the culture-based methods relatively time-consuming, expensive, and labor-demanding. Methods commonly used in clinical diagnostics include biochemical testing and mass spectroscopy-based methods. Due to the ever-growing problem of antimicrobial resistance, additional testing of antimicrobial susceptibility (AST) is often required. Conventional AST methods include disk diffusion, gradient diffusion, microdilution, and E-test—all of them require the cultivation step (Khan et al., 2019).
As an alternative to biochemical testing, matrix-assisted laser desorption/ionization-time-of-flight mass spectrometry (MALDI-TOF MS) has become a revolutionary, widely used tool in numerous clinical diagnostic laboratories. The method is based on the ionization of chemical compounds and measurements of their mass to charge (m/z) ratio. These create a specific microbial fingerprint (a peptide mass fingerprint) allowing identification upon comparison with databases. The whole process takes only minutes and can distinguish even between the most closely related microbial species (Bader et al., 2011; Hendrickx et al., 2011; Lee et al., 2017; Rychert, 2019) and detect antimicrobial resistance (Burckhardt and Zimmermann, 2018; Florio et al., 2020). However, the procedure involves multiple-step-sample preparation and relatively costly consumables. The expensive MALDI-TOF MS device makes the method unaffordable for laboratories in developing countries (Yonetani et al., 2016; Peng et al., 2019).
Direct methods, besides characterization of microbes and screening, can be used for the identification of microbes in mixed samples as well as for identification of non-culturable microbes (Ramamurthy et al., 2014; Franco-Duarte et al., 2019). They are predominantly based on microscopy, serology, or molecular analyses and do not require cultivation.
Microscopy techniques (bright-field microscopy, dark-field microscopy) can give some indication of the presence of microbes in a sample (Franco-Duarte et al., 2019). A combination of microscopy techniques with other tools/procedures can be used to increase the identification power. Examples include fluorescent dyes (Amann and Fuchs, 2008; Sabnis, 2015), scanning electron microscopy (SEM) (Relucenti et al., 2021), transmission electron microscopy (TEM) (McCutcheon and Southam, 2018), confocal microscopy (CLSM) (Cardinale et al., 2017), and atomic force microscopy (ATM) (James et al., 2016). These methods are also considered valuable in biofilm studies. A disadvantage of direct microscopy methods is an unspecific result not allowing accurate identification unless combined with additional procedure/tools, which makes the process costly and time-demanding.
Serological methods used in clinical diagnostics include the detection of antigens and antibodies. They are usually highly specific and must be ordered in a goal-directed manner (e.g., confirmation or exclusion of certain infectious agents). The positivity of antibody tests may be delayed due to the dynamics of antibody production in the human body. Besides, the immune response of the hosts usually has polyclonal nature and is influenced by genetic factors as well as environmental factors. Therefore, the reaction of a patient´s serum with an analytical system is not precisely predictable and there might be some variations. During the detection of antigens, antigenic variations (leading to different serotypes) might cause problems (Fierz, 2003).
The advent of the “genomic era” brought an astounding array of techniques incredibly useful for the characterization of biological materials and organisms, including microbes (Spratt, 2004). The development of polymerase chain reaction (PCR) in 1983 was a real breakthrough in (clinical) microbiology. Later on, real-time PCR (RT-PCR) was developed and brought a new wind into diagnostics improving the speed, sensitivity, and specificity of microbial detection (Mackay, 2004). Also, 16S rRNA, 16S – 23S rRNA (bacteria), and 18S rRNA (eukaryotes) PCR-sequencing can be a useful tool for identifying microorganisms combining PCR amplification of 16S (18S) rRNA gene, which is highly specific to each microbial species, and its subsequent sequencing (Reller et al., 2007; Singhal et al., 2015; Peker et al., 2019). When 16S rRNA gene is identical, species identification could rely on other conserved genes, such as gyrA, gyrB, rpoB, tuf, and heat shock proteins (Franco-Duarte et al., 2019). Although PCR-based methods can detect pathogens at early stages of infection and do not require cultivation, clinical samples often contain low numbers of microbial cells, complicating their capturing. They require preprocessing before the PCR reaction (incl. removal of PCR inhibitors, extraction of maximum microbes from the sample without contamination, isolation of nucleic acids). Furthermore, those methods detect the only presence of nucleic acid (or its part), which can be misleading since the human body contains large amounts of microbial genetic material (Franco-Duarte et al., 2019; Kubina and Dziedzic, 2020).
Modern technologies allow miniaturization and automatization of the methods and building more efficient approaches, such as next-generation sequencing (NGS). NGS allows parallel sequencing of enormous numbers of whole genomes or parts of nucleic acids at once, providing reliable identification of microorganisms at the nucleic acid level (Sabat et al., 2017; Boers et al., 2019). To overcome the low numbers of pathogens present in a sample, PCR amplification can be used. Compared with other widely used methods, disadvantages of using NGS in clinical diagnostics of infections include high costs and lower sensitivity, or more specifically, higher recovery of clinically unimportant microorganisms, and often too general identification (e.g., phylum level) (Lee et al., 2020).
We can conclude that all the methods mentioned above have advantages and disadvantages. Thus, different methods might be suitable or combined for different types of infections/expected causative agents. To shorten the time necessary for identification, many approaches have been proposed. Their main goals include shortening/skipping cultivation, shortening the time and reducing consumables needed for the identification, automatizing the process, and/or adding further information about a sample (e.g., virulence factors). These approaches include but are not limited to certain protocols for MALDI-TOF MS (Drancourt, 2010; Meex et al., 2012; Hoyos-Mallecot et al., 2014; Idelevich et al., 2014; Machen et al., 2014; Verroken et al., 2015; Zhou et al., 2017), infrared spectroscopy (FTIR) (Ojeda and Dittrich, 2012; Zarnowiec et al., 2015; Vogt et al., 2019), nuclear magnetic resonance (NMR) spectroscopy (Romaniuk and Cegelski, 2015; Palama et al., 2016), capillary electrophoresis (incl. capillary isoelectric focusing (Ruzicka et al., 2016; Xu and Sun, 2021), electrical field-flow fractionation (Saenton et al., 2000; Reschiglian et al., 2002), microfluidic devices (Kim et al., 2012; Zhou et al., 2019; Pérez-Rodríguez et al., 2022) and Raman spectroscopy (Franco-Duarte et al., 2019). Recently, Raman spectroscopy has been undergoing a boom in microbiology, as many studies suggest its potential for the identification of microbes, their virulence factors, detection of metabolic changes, and last but not least, single-cell analyses of microbial cells.
Raman spectroscopy is an optical method based on inelastic scattering of monochromatic light. In general, when a light beam (laser beam) reaches an object (particles), most of the light is scattered elastically (energy after deexcitation is equal to Rayleigh scattering), part of the beam passes through, part of the beam is absorbed. The last tiny part (approximately 10-5%) is inelastically scattered (Figure 1)—the energy of photons in the beam changes upon interaction with molecular vibrations in a sample and leads to a momentary distortion of electrons in a bond of a molecule. It means that the molecule has an induced dipole and is temporarily polarized. Upon returning to its normal state, the radiation is reemitted (Raman scattering). If a photon passes a part of its energy on a molecule, its frequency gets lower, and the vibrational energy of the molecule participating in a collision gets higher (Stokes Raman scattering). Thanks to thermal energy, it is also possible that a molecule is in an excited state. Then, a photon can gain energy from the molecule: the energy and the photon’s frequency gets higher, the energy of the molecule gets lower (anti-Stokes Raman scattering) (Das and Agrawal, 2011). Therefore, molecular vibrations in a sample play an essential role in Raman scattering: to be Raman active, a molecule must undergo a change in polarizability of an electron cloud around the molecule (a tendency of the electron cloud to be distorted from its original position) during a vibration. The polarizability of molecules decreases with increasing electron density (shorter and stronger bonds). The intensity of the Raman spectrum is dependent on the change of polarizability. Therefore, the most intensive Raman spectra can be acquired from symmetric valence vibrations (Smith and Dent, 2004). To conclude, we can say that the basic principle of Raman spectroscopy is tracking of scattered electrons´ energetic changes against the energy of photons from a source of monochromatic light “mirroring” chemical bonds present in the sample.
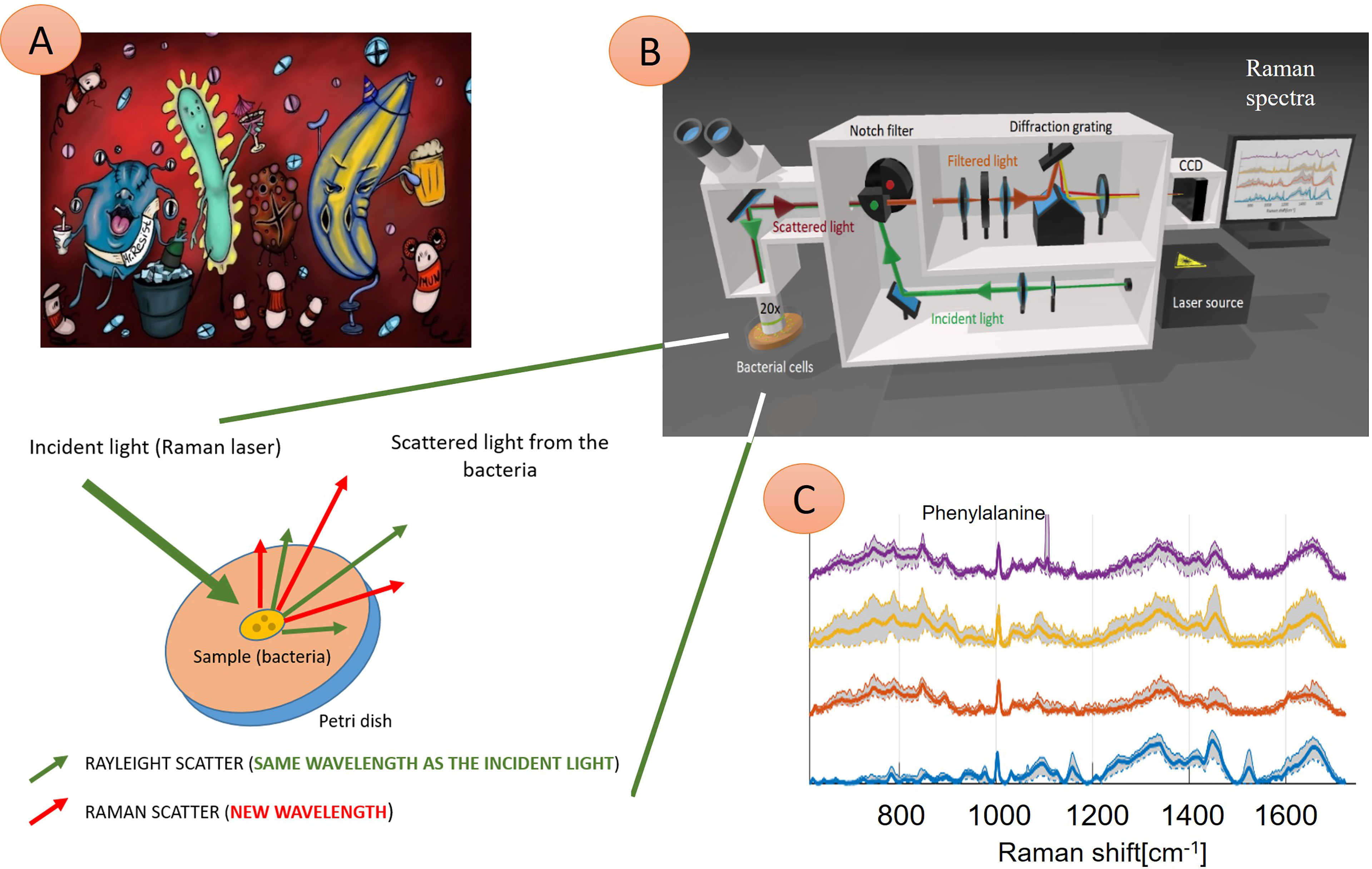
Figure 1 Illustration of four different microbial species interacting with the immune system and their Raman spectra. (A) Microbes are present in the human body, causing an infection. These pathogens can be identified using Raman spectroscopy technique (B)—here: probing laser (green) is focused on the sample (bacteria), and a small amount of light, which transports the chemical structure of analyzed bacteria, is reflected (red) and in the next step further analyzed. Consequently, four Raman spectra (C) show information about molecular bond vibrations of given bacteria, such as phenylalanine at 1005 cm-1. In this example, the naked eye can see differences between the spectra of four samples. Thus, these pathogens can be identified quickly (in minutes) to treat infection with tailored antibiotics. (C) Examples of Raman spectra: Staphylococcus pasteuri (violet curve), Staphylococcus warneri (yellow curve), Streptococcus oralis (red curve), Staphylococcus sciuri (blue trace).
This can be useful in various scientific and industrial fields ranging from archeology arts (Ziemann and Madariaga, 2021) and food industry (Weng et al., 2019), pharmacy (Vankeirsbilck et al., 2002), life sciences (McCreery, 2000; Pimenta et al., 2007; Butler et al., 2016; Kuhar et al., 2018; Li et al., 2020; Wang et al., 2020; Pezzotti, 2021) to medicine. Examples of medical applications include measurements of inflammatory markers including C-reactive protein (Bergholt and Hassing, 2009; Neugebauer et al., 2014), measurements of blood and urine chemicals (Qi and Berger, 2007), measurements of blood coagulation (Poon et al., 2012), determination oxygen saturation in live tissues (Das and Agrawal, 2011), tissue engineering (Ember et al., 2017), in vivo and in vitro diagnostics of various cancers (Chan et al., 2008; Harvey et al., 2008; Dochow et al., 2013; Taleb et al., 2013; Kong et al., 2015; Auner et al., 2018), diagnostics of prenatal diseases (Kim et al., 2018), endometriosis (Parlatan et al., 2019), and osteomyelitis (Khalid et al., 2018). Raman spectroscopy also has a plethora of applications in clinical, experimental, environmental, and technical microbiology.
Raman spectroscopy appears to be a valuable tool for the identification of microorganisms (Maquelin et al., 2003; Samek et al., 2008; Almarashi et al., 2012; Kastanos et al., 2012; Schie and Huser, 2013; Neugebauer et al., 2015; Pahlow et al., 2015; Read and Whiteley, 2015; Tien et al., 2016; Rebrošová et al., 2017; de Siqueira e Oliveira et al., 2021; Rebrošová et al., 2022), even in mixed samples (Yogesha et al., 2019). The identification can be performed from colonies grown on solid agar plates, microcolonies (Choo-Smith et al., 2001; Mathey et al., 2015), or microorganisms in liquid media (Schuster et al., 2000; Samek et al., 2010; Avci et al., 2015; Kotanen et al., 2016; Nakar et al., 2022; Rebrošová et al., 2022) and microbial spectra are highly reproducible within a device (Mlynáriková et al., 2015). Furthermore, Raman spectroscopy can be used for the characterization of microbial virulence factors, including antimicrobial resistance (Wulf et al., 2012; Bernatová et al., 2013; Dekter et al., 2017; Rousseau et al., 2021; Nakar et al., 2022) and the ability to form a biofilm (Samek et al., 2008; Samek et al., 2014; Liu et al., 2014; Hrubanova et al., 2018; Keleştemur et al., 2018; Rebrošová et al., 2019). There are some Raman studies of phenotypic changes caused by exposure to environmental stimuli, including antibiotics (Athamneh et al., 2014), alcohol (Zu et al., 2016), or metabolic stressors (Tanniche et al., 2020). Raman spectroscopy was successfully used to quantify microbes in a sample, too (Escoriza et al., 2006). To gain a stronger signal, the Raman signal can be amplified using surface-enhanced Raman spectroscopy (SERS), which is widely used in microbiological studies (Samek et al., 2021). Recently, there was significant progress in single-cell analyzes employing Raman spectroscopy and other variations of Raman spectroscopy, allowing to skip the cultivation step. The most frequently used approaches are summarized below.
A commonly used method for separating microbes from liquid media/samples is centrifugation. Published works consider centrifugation+Raman spectroscopy to be promising for identification of microbes from human body fluids, namely ascitic fluid (Kloß et al., 2015b), sputum (Kloß et al., 2015a), artificial bronchoalveolar lavage (Wichmann et al., 2021), and urine (Schröder et al., 2015; Rebrošová et al., 2022). Premasiri et al. showed a possibility to combine centrifugation and SERS to identify pathogens and their antimicrobial susceptibility (Premasiri et al., 2017). Together with filtration lysis and SERS, centrifugation was used to identify pathogens from human serum (Kotanen et al., 2016).
Moreover, centrifugation+SERS showed a possibility of identifying Chlamydia trachomatis and Neisseria gonorrhoeae and characterizing their extra-cellular metabolomics (Chen et al., 2018).
Magnetic beads are widely used for separation and isolation during bioprocessing, especially for the isolation of nucleic acids. However, the mechanism itself allows magnetic separation to be applied on various samples: magnetic separation relies upon forces induced in magnetically susceptible materials by magnetic fields (Schwaminger et al., 2019). In biology, primarily magnetic beads coated with synthetical or biological polymers (including antibodies) capable of capturing the target molecules/cells are used. Target molecules/cells bind to a polymer. Afterward, the whole complex (magnetic carrier with polymer + target molecule/cell) is captured by applying magnetic force (Berensmeier, 2006).
Kusić et al. successfully used magnetic beads coated with Legionella spp. specific polyclonal immunoglobulins for isolating single Legionella sp. cells from biofilm and subsequently identifying them with Raman spectroscopy (Kusić et al., 2014). Kearns et al. developed a bionanosensor based on magnetic separation and SERS, which can be used to identify microbes in concentrations as low as 101 CFU/mL in less than one hour (Kearns et al., 2017). Hu et al. showed a possibility of capturing Candida sp. cells from serum and characterizing them using SERS (Hu et al., 2021). Li et al. used polyethyleneimine-modified magnetic microspheres (Fe3O4@PEI) and SERS for bacterial identification and antimicrobial resistance determination from 77 blood samples (Li et al., 2019). A combination of SERS and immunomagnetic beads can also be used to detect Clostridium botulinum toxins A and B (Kim et al., 2019). Since magnetic separation is commonly used for nucleic acid isolation, Hwang et al. applied this method to isolate bacterial genomic DNA and identify it using SERS and fluorescent assay favoring SERS by means of sensitivity (Hwang et al., 2021). Detection of bacterial DNA by SERS using streptavidin-coated magnetic particles was also proved by Qun et al. with a limit of detection of 5 pM (Qun et al., 2015).
Dielectrophoresis (DEP) is defined as a movement of dielectric particles through a medium in response to a non-uniform electric field. A particle becomes polarized, and due to the difference in electric field strength on the two sides of the particle, the particle is moved in the electric field gradient region by net dielectrophoretic force (Tay et al., 2009). This effect can be used for the separation of particles (Fernandez et al., 2017) and is widely used for enrichment and isolation of microbial cells before analysis at a single-cell level (Fernandez et al., 2017; Zhang et al., 2019; Sarno et al., 2021; Weber et al., 2021). Some groups recently suggested its possible combination with Raman spectroscopy for single-cell microbial analyses (Neugebauer et al., 2015). Chen et al. showed the efficiency of DEP-based microfluidic chip for Raman detection and measurements of Shewanella oneidensis cells in HEPES buffer (Chen et al., 2018). From clinically relevant applications, Cheng et al. used DEP with SERS to isolate and identify bacteria in isotonic solution with human blood cells, proposing rapid detection of microbes in human blood from 12h blood cultures (Cheng et al., 2014). To identify pathogens causing urinary tract infections faster, Schröder et al. proposed using a combination of DEP and Raman spectroscopy to identify Escherichia coli and Enterococcus faecalis in human urine, providing results in 35 minutes (Schröder et al., 2013).
Optical tweezers—a Nobel price-winning (2018) groundbreaking invention by Alfred Ashkin—use single-beam gradient force to hold particles/micro-objects (incl. microbial cells) in place and manipulate them (Ashkin, 1997; Wu et al., 2017). With an optical trap, living cells suspended in a liquid cultivation medium can be immobilized in a solution using the forces generated by a tightly focused laser beam. Combined with Raman spectroscopy (usually termed Raman tweezers), it has found numerous applications, especially in analytical and physical chemistry. Raman tweezers is currently starting to be applied in cell and molecular biology. It allows single-cell analyses of microbes, for example, direct identification of microbes from liquid samples, including wastewater (Cui et al., 2021) and human urine (Rebrošová et al., 2022), in less than 10 minutes. This combination can also be used to detect antibiotic resistance (Bernatová et. al 2014; Pilát et al., 2018; Bernatová et al., 2021) and an ability to form a biofilm (Samek et al., 2015). Moreover, it was used to describe metabolic changes in microbes (Singh et al., 2006; Huang et al., 2007) and bacterial lysis (Chen et al., 2009).
Current advantages and disadvantages of the Raman spectroscopy for microbiology are summarized in the Table 1.
Unambiguously, Raman spectroscopy offers a streamlined, fast, non-destructive, and non-invasive approach for identifying microbes and their virulence factors suitable for clinical diagnostics. Compared to conventional and molecular methods, sample preparation is easy and does not necessarily require expensive consumables. Moreover, multiple methods were proposed to isolate or enrich microbes, allowing their single-cell analyses and non-culture-based detection and identification.
However, getting Raman spectroscopy to clinical laboratories might have a long-time coming. At the moment, there are no commercial and/or well-annotated databases of microbial Raman fingerprints, as discussed by Wang et al. (Wang et al., 2021). To make comparisons of acquired Raman spectra across various instruments, one should consider that the quantum efficiency of the given detector and optical elements depends on a wavelength. Therefore, acquired data should be corrected according to an instrument response profile. Ideally, a spectral sensitivity curve should be used (Rebrošová et al., 2017). Thus, Raman spectra presented in different studies are group-specific and custom-tailored, which makes data standardization complicated (Lorenz et al., 2017). Nonetheless, no large-scale studies were performed to compare microbial Raman spectra from different groups.
Microbial studies often employ the SERS technique. Again, there is a problem with the standardization of SERS microbial detection protocols (Witkowska et al., 2020; Samek et al., 2021). Although SERS provides a stronger signal than conventional Raman spectroscopy, several factors influence the signal enhancement and, consequently, the final spectrum. These may include types of culture media, culturing conditions, sample preparation method, and interactions between SERS substrate and individual microbial cells (Witkowska et al., 2020; Samek et al., 2021).
Other problems are (currently) relatively high input costs, the need for complicated instrumentation, and specialized operators. Automatization and miniaturization would be beneficial for potential future clinical use.
If those problems are solved, Raman spectroscopy-based point-of-care device could start a revolution in clinical diagnostic microbiology in the future. Assuming its broad spectrum of applications in medicine, one device equipped with various diagnostic databases might become a complex diagnostic tool. From a microbiological point of view, Raman spectroscopy could be used for rapid pathogen identification and characterization of its virulence factors, which would, in turn, result in tailored antimicrobial treatment, reduced financial burdens associated with healthcare, and above all, improved patient management and reduced mortality from infections.
Raman spectroscopy is an elegant optical method that could significantly contribute to rapid clinical diagnostics of infections. It allows the identification of microbes and detection of certain virulence factors, including antimicrobial resistance and biofilm formation. In combination with techniques for isolation/enrichment of microbes from a liquid sample, it could be used for rapid single-cell analyzes of microbial cells, even directly from human body fluids. Therefore, it could provide an effective solution for identifying microbes in the future.
KR, OS, MK participated in conceptualization and writing of the original draft. SB, VH, and FR participated in review and editing. All authors contributed to the article and approved the submitted version.
This work was supported by the Grant Agency of Masaryk University (MUNI/A/1291/2021) and the Czech Health Research Council (NU21-05-00341).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
We would like to thank the artist Gabriela Samkova for the beautiful illustration of the bacterial infection she prepared for our manuscript.
Almarashi, J. F. M., Kapel, N., Wilkinson, T. S., Telle, H. H. (2012). Raman Spectroscopy of Bacterial Species and Strains Cultivated Under Reproducible Conditions. Spectroscopy: Int. J. 27, 361–365. doi: 10.1155/2012/540490
Amann, R., Fuchs, B. M. (2008). Single-Cell Identification in Microbial Communities by Improved Fluorescence in Situ Hybridization Techniques. Nat. Rev. Microbiol. 6 (5), 339–348. doi: 10.1038/nrmicro1888
Ashkin, A. (1997). Optical Trapping and Manipulation of Neutral Particles Using Lasers. Proc. Natl. Acad. Sci. 94 (10), 4853–4860. doi: 10.1073/pnas.94.10.4853
Athamneh, A. I. M., Alajlouni, R. A., Wallace, R. S., Seleem, M. N., Senger, R. S. (2014). Phenotypic Profiling of Antibiotic Response Signatures in Escherichia Coli Using Raman Spectroscopy. Antimicrob. Agents Chemother. 58 (3), 1302–1314. doi: 10.1128/AAC.02098-13
Auner, G. W., Koya, S. K., Huang, C., Broadbent, B., Trexler, M., Auner, Z., et al. (2018). Applications of Raman Spectroscopy in Cancer Diagnosis. Cancer Metastasis Rev. 37 (4), 691–717. doi: 10.1007/s10555-018-9770-9
Avci, E., Kaya, N. S., Ucankus, G., Culha, M. (2015). Discrimination of Urinary Tract Infection Pathogens by Means of Their Growth Profiles Using Surface Enhanced Raman Scattering. Anal. Bioanal. Chem. 407 (27), 8233–8241. doi: 10.1007/s00216-015-8950-5
Bader, O., Weig, M., Taverne-Ghadwal, L., Lugert, R., Groß, U., Kuhns, M. (2011). Improved Clinical Laboratory Identification of Human Pathogenic Yeasts by Matrix-Assisted Laser Desorption Ionization Time-of-Flight Mass Spectrometry. Clin. Microbiol. Infect. 17 (9), 1359–1365. doi: 10.1111/j.1469-0691.2010.03398.x
Balloux, F., van Dorp, L. (2017). Q&a: What are Pathogens, and What Have They Done to and for Us? BMC Biol. 15 (1), 91. doi: 10.1186/s12915-017-0433-z
Berensmeier, S. (2006). Magnetic Particles for the Separation and Purification of Nucleic Acids. Appl. Microbiol. Biotechnol. 73 (3), 495–504. doi: 10.1007/s00253-006-0675-0
Bergholt, M. S., Hassing, S. (2009). Quantification of C-Reactive Protein in Human Blood Plasma Using Near-Infrared Raman Spectroscopy. Analyst 134 (10), 2123. doi: 10.1039/b903089a
Bernatová, S., Rebrošová, K., Pilát, Z., Šerý, M., Gjevik, A., Samek, O., et al. (2013). Following the Mechanisms of Bacteriostatic Versus Bactericidal Action Using Raman Spectroscopy. Molecules 18 (11), 13188–13199. doi: 10.3390/molecules181113188
Bernatova, S., Samek, O., Pilát, Z., Šerý, M., Ježek, J., Jákl, P., et al. (2014). “Raman Tweezers on Bacteria: Following the Mechanisms of Bacteriostatic Versus Bactericidal Action,” in Spie Photonics Europe. Ed. Popp, J., et al. (Brussels, Belgium: SPIE), 91291Y. doi: 10.1117/12.2052538
Bernatová, S., Samek, O., Pilat, Z., Sery, M., Jezek, J., Jakl, P., et al. (2021). Rapid Detection of Antibiotic Sensitivity of Staphylococcus Aureus by Raman Tweezers. Eur. Phys. J. Plus 136 (2), 233. doi: 10.1140/epjp/s13360-021-01152-1
Boers, S. A., Jansen, R., Hays, J. P. (2019). Understanding and Overcoming the Pitfalls and Biases of Next-Generation Sequencing (NGS) Methods for Use in the Routine Clinical Microbiological Diagnostic Laboratory. Eur. J. Clin. Microbiol. Infect. Dis. 38 (6), 1059–1070. doi: 10.1007/s10096-019-03520-3
Brock, T. D., et al. (2003). ‘Brock Biology of Microorganisms’, in. Upper Saddle River NJ: Prentice hall (10) p, 1019.
Burckhardt, I., Zimmermann, S. (2018). Susceptibility Testing of Bacteria Using Maldi-Tof Mass Spectrometry. Front. Microbiol. 9. doi: 10.3389/fmicb.2018.01744
Butler, H. J., Ashton, L., Bird, B., Cinque, G., Curtis, K., Dorney, J., et al. (2016). Using Raman Spectroscopy to Characterize Biological Materials. Nat. Protoc. 11 (4), 664–687. doi: 10.1038/nprot.2016.036
Cardinale, M., Kaiser, D., Lueders, T., Schnell, S., Egert, M. (2017). Microbiome Analysis and Confocal Microscopy of Used Kitchen Sponges Reveal Massive Colonization by Acinetobacter, Moraxella and Chryseobacterium Species. Sci. Rep. 7 (1), 5791. doi: 10.1038/s41598-017-06055-9
Chan, J. W., Taylor, D. S., Lane, S. M., Zwerdling, T., Tuscano, J., Huser, T. (2008). Nondestructive Identification of Individual Leukemia Cells by Laser Trapping Raman Spectroscopy. Anal. Chem. 80 (6), 2180–2187. doi: 10.1021/ac7022348
Chen, D., Chen, T.-Y., Lu, R.-J., Wu, H.-W. (2009). Laser Tweezers Raman Spectroscopy Potential for Studies of Complex Dynamic Cellular Processes: Single Cell Bacterial Lysis. Anal. Chem. 81 (9), 3227–3238. doi: 10.1021/ac8023476
Chen, X., Liang, Z., Li, D., Xiong, Y., Xiong, P., Guan, Y., et al. (2018). Microfluidic Dielectrophoresis Device for Trapping, Counting and Detecting Shewanella Oneidensis at the Cell Level. Biosensors Bioelectronics 99, 416–423. doi: 10.1016/j.bios.2017.08.017
Cheng, I.-F., Chen, T.-Y., Lu, R.-J., Wu, H.-W. (2014). Rapid Identification of Bacteria Utilizing Amplified Dielectrophoretic Force-Assisted Nanoparticle-Induced Surface-Enhanced Raman Spectroscopy. Nanoscale Res. Lett. 9 (1), 324. doi: 10.1186/1556-276X-9-324
Chen, Y., Premasiri, W. R., Ziegler, L. D. (2018). Surface Enhanced Raman Spectroscopy of Chlamydia Trachomatis and Neisseria Gonorrhoeae for Diagnostics, and Extra-Cellular Metabolomics and Biochemical Monitoring. Sci. Rep. 8 (1), 5163. doi: 10.1038/s41598-018-23562-5
Choo-Smith, L.-P., Maquelin, K., van Vreeswijk, T., Bruining, H. A., Puppels, G. J., Thi, N. A. N., et al. (2001). Investigating Microbial (Micro)Colony Heterogeneity by Vibrational Spectroscopy. Appl. Environ. Microbiol. 67 (4), 1461–1469. doi: 10.1128/AEM.67.4.1461-1469.2001
Cui, L., Li, H.-Z., Yang, K., Zhu, L.-J., Xu, F., Zhu, Y.-G. (2021). Raman Biosensor and Molecular Tools for Integrated Monitoring of Pathogens and Antimicrobial Resistance in Wastewater. TrAC Trends Analy. Chem. 143, 116415. doi: 10.1016/j.trac.2021.116415
Das, R. S., Agrawal, Y. K. (2011). Raman Spectroscopy: Recent Advancements, Techniques and Applications. Vibrational Spectrosc. 57 (2), 163–176. doi: 10.1016/j.vibspec.2011.08.003
Dekter, H. E., Orelio, C. C., Morsink, M. C., Tektas, S., Vis, B., te Witt, R., et al. (2017). Antimicrobial Susceptibility Testing of Gram-Positive and -Negative Bacterial Isolates Directly From Spiked Blood Culture Media With Raman Spectroscopy. Eur. J. Clin. Microbiol. Infect. Dis. 36 (1), 81–89. doi: 10.1007/s10096-016-2773-y
de Siqueira e Oliveira, F. S., da Silva, A. M., Pacheco, M. T. T., Giana, H. E., Silveira, L. (2021). Biochemical Characterization of Pathogenic Bacterial Species Using Raman Spectroscopy and Discrimination Model Based on Selected Spectral Features. Lasers Med. Sci. 36 (2), 289–302. doi: 10.1007/s10103-020-03028-9
Dochow, S., Beleites, C., Henkel, T., Mayer, G., Albert, J., Clement, J., et al. (2013). Quartz Microfluidic Chip for Tumour Cell Identification by Raman Spectroscopy in Combination With Optical Traps. Anal. Bioanal. Chem. 405 (8), 2743–2746. doi: 10.1007/s00216-013-6726-3
Drancourt, M. (2010). Detection of Microorganisms in Blood Specimens Using Matrix-Assisted Laser Desorption Ionization Time-of-Flight Mass Spectrometry: A Review. Clin. Microbiol. Infect. 16 (11), 1620–1625. doi: 10.1111/j.1469-0691.2010.03290.x
Ember, K. J. I., Hoeve, M. A., McAughtrie, S. L., Bergholt, M. S., Dwyer, B. J., Stevens, M. M., et al. (2017). Raman Spectroscopy and Regenerative Medicine: A Review. NPJ Regenerative Med. 2 (1), 12. doi: 10.1038/s41536-017-0014-3
Escoriza, M. F., VanBriesen, J. M., Stewart, S., Maier, J., Treado, P. J. (2006). Raman Spectroscopy and Chemical Imaging for Quantification of Filtered Waterborne Bacteria. J. Microbiol. Methods 66 (1), 63–72. doi: 10.1016/j.mimet.2005.10.013
Fernandez, R. E., Rohani, A., Farmehini, V., Swami, N. S. (2017). Review: Microbial Analysis in Dielectrophoretic Microfluidic Systems. Analytica Chimica Acta 966, 11–33. doi: 10.1016/j.aca.2017.02.024
Fierz, W. (2003). “Basic Problems of Serological Laboratory Diagnosis,” in Molecular Diagnosis of Infectious Diseases. Eds. Decker, J., Reischl, U. (New Jersey: Humana Press), 393–428. doi: 10.1385/1-59259-679-7:393
Florio, W., Baldeschi, L., Rizzato, C., Tavanti, A., Ghelardi, E., Lupetti, A. (2020). Detection of Antibiotic-Resistance by MALDI-TOF Mass Spectrometry: An Expanding Area. Front. Cell. Infect. Microbiol. 10. doi: 10.3389/fcimb.2020.572909
Franco-Duarte, R., Černáková, L., Kadam, S., S. Kaushik, K., Salehi, B., Bevilacqua, A., et al. (2019). Advances in Chemical and Biological Methods to Identify Microorganisms—From Past to Present. Microorganisms 7 (5), 130. doi: 10.3390/microorganisms7050130
Harvey, T. J., Faria, E. C., Henderson, A., Gazi, E., Ward, A. D., Clarke, N. W., et al. (2008). Spectral Discrimination of Live Prostate and Bladder Cancer Cell Lines Using Raman Optical Tweezers. J. Biomed. Optics 13 (6), 064004. doi: 10.1117/1.2999609
Hendrickx, M., Goffinet, J.-S., Swinne, D., Detandt, M. (2011). Screening of Strains of the Candida Parapsilosis Group of the BCCM/IHEM Collection by MALDI-TOF MS. Diagn. Microbiol. Infect. Dis. 70 (4), 544–548. doi: 10.1016/j.diagmicrobio.2011.04.006
Hoyos-Mallecot, Y., Riazzo, C., Miranda-Casas, C., Rojo-Martín, M. D., Gutiérrez-Fernández, J., Navarro-Marí, J. M. (2014). Rapid Detection and Identification of Strains Carrying Carbapenemases Directly From Positive Blood Cultures Using MALDI-TOF MS. J. Microbiol. Methods 105, 98–101. doi: 10.1016/j.mimet.2014.07.016
Hrubanova, K., Krzyzanek, V., Nebesarova, J., Ruzicka, F., Pilat, Z., Samek, O. (2018). Monitoring Candida Parapsilosis and Staphylococcus Epidermidis Biofilms by a Combination of Scanning Electron Microscopy and Raman Spectroscopy. Sensors 18 (12), 4089. doi: 10.3390/s18124089
Huang, S., Chen, D., Pelczar, P. L., Vepachedu, V. R., Setlow, P., Li, Y. (2007). Levels of Ca2+-Dipicolinic Acid in Individual Bacillus Spores Determined Using Microfluidic Raman Tweezers. J. Bacteriol. 189 (13), 4681–4687. doi: 10.1128/JB.00282-07
Hu, S., Kang, H., Gu, F., Wang, C., Cheng, S., Gong, W., et al. (2021). Rapid Detection Method for Pathogenic Candida Captured by Magnetic Nanoparticles and Identified Using SERS via Agnps+’. Int. J. Nanomed. 16, 941–950. doi: 10.2147/IJN.S285339
Hwang, M. J., Jang, A. S., Lim, D.-K. (2021). Comparative Study of Fluorescence and Surface-Enhanced Raman Scattering With Magnetic Microparticle-Based Assay for Target Bacterial DNA Detection. Sensors Actuators B: Chem. 329, 129134. doi: 10.1016/j.snb.2020.129134
Idelevich, E. A., Schüle, I., Grünastel, B., Wüllenweber, J., Peters, G., Becker, K. (2014). Rapid Identification of Microorganisms From Positive Blood Cultures by MALDI-TOF Mass Spectrometry Subsequent to Very Short-Term Incubation on Solid Medium. Clin. Microbiol. Infect. 20 (10), 1001–1006. doi: 10.1111/1469-0691.12640
James, S. A., Powell, L. C., Wright, C. J. (2016). “Atomic Force Microscopy of Biofilms—Imaging, Interactions, and Mechanics,” in Microbial Biofilms - Importance and Applications. Eds. Dhanasekaran, D., Thajuddin, N. (London: InTech). doi: 10.5772/63312
Kastanos, E., Kyriakides, A., Hadjigeorgiou, K., Pitris, C. (2012). A Novel Method for Bacterial UTI Diagnosis Using Raman Spectroscopy. Int. J. Spectrosc. 2012, 1–13. doi: 10.1155/2012/195317
Kearns, H., Goodacre, R., Jamieson, L. E., Graham, D., Faulds, K. (2017). SERS Detection of Multiple Antimicrobial-Resistant Pathogens Using Nanosensors. Analytical Chem. 89 (23), 12666–12673. doi: 10.1021/acs.analchem.7b02653
Keleştemur, S., Avci, E., Çulha, M. (2018). Raman and Surface-Enhanced Raman Scattering for Biofilm Characterization. Chemosensors 6 (1), 5. doi: 10.3390/chemosensors6010005
Khalid, M., Bora, T., Ghaithi, A. A., Thukral, S., Dutta, J. (2018). Raman Spectroscopy Detects Changes in Bone Mineral Quality and Collagen Cross-Linkage in Staphylococcus Infected Human Bone. Sci. Rep. 8 (1), 9417. doi: 10.1038/s41598-018-27752-z
Khan, Z. A., Siddiqui, M. F., Park, S. (2019). Current and Emerging Methods of Antibiotic Susceptibility Testing. Diagnostics 9 (2), 49. doi: 10.3390/diagnostics9020049
Kim, J., Hegde, M., Kim, S. H., Wood, T. K., Jayaraman, A. (2012). A Microfluidic Device for High Throughput Bacterial Biofilm Studies. Lab. Chip 12 (6), 1157. doi: 10.1039/c2lc20800h
Kim, W., Lee, S. H., Kim, J. H., Ahn, Y. J., Kim, Y.-H., Yu, J. S., et al. (2018). Paper-Based Surface-Enhanced Raman Spectroscopy for Diagnosing Prenatal Diseases in Women. ACS Nano 12 (7), 7100–7108. doi: 10.1021/acsnano.8b02917
Kim, K., Choi, N., Jeon, J. H., Rhie, G., Choo, J. (2019). SERS-Based Immunoassays for the Detection of Botulinum Toxins a and B Using Magnetic Beads. Sensors 19 (19), 4081. doi: 10.3390/s19194081
Kloß, S., Rösch, P., Pfister, W., Kiehntopf, M., Popp, J. (2015b). Toward Culture-Free Raman Spectroscopic Identification of Pathogens in Ascitic Fluid. Anal. Chem. 87 (2), 937–943. doi: 10.1021/ac503373r
Kloß, S., Lorenz, B., Dees, S., Labugger, I., Rösch, P., Popp, J. (2015a). Destruction-Free Procedure for the Isolation of Bacteria From Sputum Samples for Raman Spectroscopic Analysis. Anal. Bioanal. Chem. 407 (27), 8333–8341. doi: 10.1007/s00216-015-8743-x
Kong, K., Kendall, C., Stone, N., Notingher, I. (2015). Raman Spectroscopy for Medical Diagnostics — From in-Vitro Biofluid Assays to in-Vivo Cancer Detection. Advanced Drug Delivery Rev. 89, 121–134. doi: 10.1016/j.addr.2015.03.009
Kotanen, C. N., Martinez, L., Alvarez, R., Simecek, J. W. (2016). Surface Enhanced Raman Scattering Spectroscopy for Detection and Identification of Microbial Pathogens Isolated From Human Serum. Sens. Bio-Sensing Res. 8, 20–26. doi: 10.1016/j.sbsr.2016.03.002
Kubina, R., Dziedzic, A. (2020). Molecular and Serological Tests for COVID-19. A Comparative Review of SARS-Cov-2 Coronavirus Laboratory and Point-of-Care Diagnostics. Diagnostics 10(6) p, 434. doi: 10.3390/diagnostics10060434
Kuhar, N., Sil, S., Verma, T., Umapathy, S. (2018). Challenges in Application of Raman Spectroscopy to Biology and Materials. RSC Adv. 8 (46), 25888–25908. doi: 10.1039/C8RA04491K
Kusić, D., Kampe, B., Rösch, P., Popp, J. (2014). Identification of Water Pathogens by Raman Microspectroscopy. Water Res. 48, 179–189. doi: 10.1016/j.watres.2013.09.030
Lee, J., Shin, Y., Kim, S., Rho, K., Park, K. H. (2017). “SVM Classification Model of Similar Bacteria Species Using Negative Marker: Based on Matrix-Assisted Laser Desorption/Ionization Time-of-Flight Mass Spectrometry,” in 2017 IEEE 17th International Conference on Bioinformatics and Bioengineering (BIBE). 2017 IEEE 17th International Conference on Bioinformatics and Bioengineering (BIBE) (Washington, DC: IEEE), 145–150. doi: 10.1109/BIBE.2017.00-64
Lee, R. A., Al Dhaheri, F., Pollock, N. R., Sharma, T. S. (2020). Assessment of the Clinical Utility of Plasma Metagenomic Next-Generation Sequencing in a Pediatric Hospital Population. J. Clin. Microbiol. 58 (7), e00419–20. doi: 10.1128/JCM.00419-20
Li, J., Wang, C., Shi, L., Shao, L., Fu, P., Wang, K., et al. (2019). Rapid Identification and Antibiotic Susceptibility Test of Pathogens in Blood Based on Magnetic Separation and Surface-Enhanced Raman Scattering. Microchimica Acta 186 (7), 475. doi: 10.1007/s00604-019-3571-x
Li, S., Li, Y., Yi, R., Liu, L., Qu, J. (2020). Coherent Anti-Stokes Raman Scattering Microscopy and its Applications. Front. Phys. 8. doi: 10.3389/fphy.2020.598420
Liu, H., Xu, Q., Huo, L., Wei, X., Ling, J. (2014). Chemical Composition of Enterococcus Faecalis in Biofilm Cells Initiated From Different Physiologic States. Folia Microbiologica 59 (5), 447–453. doi: 10.1007/s12223-014-0319-1
Lorenz, B., Wichmann, C., Stöckel, S., Rösch, P., Popp, J. (2017). Cultivation-Free Raman Spectroscopic Investigations of Bacteria. Trends Microbiol. 25 (5), 413–424. doi: 10.1016/j.tim.2017.01.002
Machen, A., Drake, T., Wang, Y. F. (2014). Same Day Identification and Full Panel Antimicrobial Susceptibility Testing of Bacteria From Positive Blood Culture Bottles Made Possible by a Combined Lysis-Filtration Method With MALDI-TOF VITEK Mass Spectrometry and the VITEK2 System. PloS One 9 (2), e87870. doi: 10.1371/journal.pone.0087870. Wayne.
Mackay, I. M. (2004). Real-Time PCR in the Microbiology Laboratory. Clin. Microbiol. Infect. 10 (3), 190–212. doi: 10.1111/j.1198-743X.2004.00722.x
Maquelin, K., Kirschner, C., Choo-Smith, L.-P., Ngo-Thi, N. A., van Vreeswijk, T., Stämmler, M., et al. (2003). Prospective Study of the Performance of Vibrational Spectroscopies for Rapid Identification of Bacterial and Fungal Pathogens Recovered From Blood Cultures. J. Clin. Microbiol. 41 (1), 324–329. doi: 10.1128/JCM.41.1.324-329.2003
Mathey, R., Dupoy, M., Espagnon, I., Leroux, D., Mallard, F., Novelli-Rousseau, A. (2015). Viability of 3h Grown Bacterial Micro-Colonies After Direct Raman Identification. J. Microbiol Methods 109, 67–73. doi: 10.1016/j.mimet.2014.12.002
McCreery, R. L. (2000). Raman Spectroscopy for Chemical Analysis: Mccreery/Raman Spectroscopy. Hoboken (NJ, USA: John Wiley & Sons, Inc). doi: 10.1002/0471721646
McCutcheon, J., Southam, G. (2018). Advanced Biofilm Staining Techniques for TEM and SEM in Geomicrobiology: Implications for Visualizing EPS Architecture, Mineral Nucleation, and Microfossil Generation. Chem. Geol. 498, 115–127. doi: 10.1016/j.chemgeo.2018.09.016
Meex, C., Neuville, F., Descy, J., Huynen, P., Hayette, M.-P., De Mol, P., et al. (2012). Direct Identification of Bacteria From Bact/ALERT Anaerobic Positive Blood Cultures by MALDI-TOF MS: MALDI Sepsityper Kit Versus an in-House Saponin Method for Bacterial Extraction. J. Med. Microbiol. 61 (11), 1511–1516. doi: 10.1099/jmm.0.044750-0
Mlynáriková, K., Samek, O., Bernatová, S., Růžička, F., Ježek, J., Hároniková, A., et al. (2015). Influence of Culture Media on Microbial Fingerprints Using Raman Spectroscopy. Sensors 15 (11), 29635–29647. doi: 10.3390/s151129635
Nakar, A., Pistiki, A., Ryabchykov, O., Bocklitz, T., Rösch, P., Popp, J. (2022). Detection of Multi-Resistant Clinical Strains of E. Coli With Raman Spectroscopy. Anal. Bioanal. Chem. 18, 1481–1492. doi: 10.1007/s00216-021-03800-y
Neugebauer, U., Rösch, P., Popp, J. (2014). Fast Differentiation of SIRS and Sepsis From Blood Plasma of ICU Patients Using Raman Spectroscopy: Spectroscopic Differentiation of SIRS and Sepsis From Blood Plasma. J. Biophotonics 7 (3–4), 232–240. doi: 10.1002/jbio.201400010
Neugebauer, U., Rösch, P., Popp, J. (2015). Raman Spectroscopy Towards Clinical Application: Drug Monitoring and Pathogen Identification. Int. J. Antimicrob. Agents 46, S35–S39. doi: 10.1016/j.ijantimicag.2015.10.014
Ojeda, J. J., Dittrich, M. (2012). “Fourier Transform Infrared Spectroscopy for Molecular Analysis of Microbial Cells,” in Microbial Systems Biology. Ed. Navid, A. (Totowa, NJ: Humana Press (Methods in Molecular Biology), 187–211. doi: 10.1007/978-1-61779-827-6_8
Pahlow, S., Meisel, S., Cialla-May, D., Weber, K., Rösch, P., Popp, J. (2015). Isolation and Identification of Bacteria by Means of Raman Spectroscopy. Advanced Drug Delivery Rev. 89, 105–120. doi: 10.1016/j.addr.2015.04.006
Palama, T. L., Canard, I., Rautureau, G. J. P., Mirande, C., Chatellier, S., Elena-Herrmann, B. (2016). Identification of Bacterial Species by Untargeted NMR Spectroscopy of the Exo-Metabolome. Analyst 141 (15), 4558–4561. doi: 10.1039/C6AN00393A
Parlatan, U., Inanc, M. T., Ozgor, B. Y., Oral, E., Bastu, E., Unlu, M. B., et al. (2019). Raman Spectroscopy as a non-Invasive Diagnostic Technique for Endometriosis. Sci. Rep. 9 (1), 19795. doi: 10.1038/s41598-019-56308-y
Peker, N., Garcia-Croes, S., Dijkhuizen, B., Wiersma, H. H., van Zanten, E., Wisselink, G., et al. (2019). A Comparison of Three Different Bioinformatics Analyses of the 16S–23S Rrna Encoding Region for Bacterial Identification. Front. Microbiol. 10. doi: 10.3389/fmicb.2019.00620
Peng, Y., Zhang, Q., Xu, C., Shi, W. (2019). MALDI−TOF MS for the Rapid Identification and Drug Susceptibility Testing of Filamentous Fungi. Exp. Ther. Med. 414, 4865–4873 doi: 10.3892/etm.2019.8118
Pérez-Rodríguez, S., García-Aznar, J. M., Gonzalo-Asensio, J. (2022). Microfluidic Devices for Studying Bacterial Taxis, Drug Testing and Biofilm Formation. Microbial Biotechnol. 15, 395–414. doi: 10.1111/1751-7915.13775
Pezzotti, G. (2021). Raman Spectroscopy in Cell Biology and Microbiology. J. Raman Spectrosc. 52 (12), 2348–2443. doi: 10.1002/jrs.6204
Pilát, Z., Bernatová, S., Ježek, J., Kirchhoff, J., Tannert, A., Neugebauer, U., et al. (2018). ‘Microfluidic Cultivation and Laser Tweezers Raman Spectroscopy of E. Coli Under Antibiotic Stress’ Sensors 18(5) p, 1623. doi: 10.3390/s18051623
Pimenta, M. A., Dresselhaus, G., Dresselhaus, M. S., Cançado, L. G., Jorio, A., Saito, R. (2007). Studying Disorder in Graphite-Based Systems by Raman Spectroscopy. Phys. Chem. Chem. Phys. 9 (11), 1276–1290. doi: 10.1039/B613962K
Poon, K. W. C., Lyng, F. M., Knief, P., Howe, O., Meade, A. D., Curtin, J. F., et al. (2012). Quantitative Reagent-Free Detection of Fibrinogen Levels in Human Blood Plasma Using Raman Spectroscopy. Analyst 137 (8), 1807. doi: 10.1039/c2an35042d
Premasiri, W. R., Chen, Y., Williamson, P. M., Bandarage, D. C., Pyles, C., Ziegler, L. D. (2017). Rapid Urinary Tract Infection Diagnostics by Surface-Enhanced Raman Spectroscopy (SERS): Identification and Antibiotic Susceptibilities. Anal. Bioanal. Chem. 409 (11), 3043–3054. doi: 10.1007/s00216-017-0244-7
Qi, D., Berger, A. J. (2007). Chemical Concentration Measurement in Blood Serum and Urine Samples Using Liquid-Core Optical Fiber Raman Spectroscopy. Appl. Optics 46 (10), 1726. doi: 10.1364/AO.46.001726
Qun, M., Yan-Le, L., Nian-Chun, G., Xi, J., Shuang-Yan, H. (2015). Surface Enhanced Raman Spectroscopy Sensor Based on Magnetic Beads-Induced Nanoparticles Aggregation for Detection of Bacterial Deoxyribonucleic Acid. Chin. J. Anal. Chem. 43 (11), 1676–1681. doi: 10.1016/S1872-2040(15)60876-3
Ramamurthy, T., Ghosh, A., Pazhani, G. P., Shinoda, S. (2014). Current Perspectives on Viable But non-Culturable (VBNC) Pathogenic Bacteria. Front. Public Health 2. doi: 10.3389/fpubh.2014.00103
Read, D. S., Whiteley, A. S. (2015). Chemical Fixation Methods for Raman Spectroscopy-Based Analysis of Bacteria. J. Microbiol. Methods 109, 79–83. doi: 10.1016/j.mimet.2014.12.008
Rebrošová, K., Šiler, M., Samek, O., Růžička, F., Bernatová, S., Holá, V., et al. (2017). Rapid Identification of Staphylococci by Raman Spectroscopy. Sci. Rep. 7 (1), 14846. doi: 10.1038/s41598-017-13940-w
Rebrošová, K., Šiler, M., Samek, O., Růžička, F., Bernatová, S., Ježek, J., et al. (2019). Identification of Ability to Form Biofilm in Candida Parapsilosis and Staphylococcus Epidermidis by Raman Spectroscopy. Future Microbiol. 14 (6), 509–517. doi: 10.2217/fmb-2018-0297
Rebrošová, K., Bernatová, S., Šiler, M., Uhlirova, M., Samek, O., Ježek, J., et al. (2022). Raman Spectroscopy—a Tool for Rapid Differentiation Among Microbes Causing Urinary Tract Infections. Analytica Chimica Acta 1191, 339292. doi: 10.1016/j.aca.2021.339292
Reller, L. B., Weinstein, M. P., Petti, C. A. (2007). Detection and Identification of Microorganisms by Gene Amplification and Sequencing. Clin. Infect. Dis. 44 (8), 1108–1114. doi: 10.1086/512818
Relucenti, M., Familiari, G., Donfrancesco, O., Taurino, M., Li, X., Chen, R., et al. (2021). Microscopy Methods for Biofilm Imaging: Focus on SEM and VP-SEM Pros and Cons. Biology 10 (1), 51. doi: 10.3390/biology10010051
Reschiglian, P., Zattoni, A., Roda, B., Casolari, S., Moon, M. H., Lee, J., et al. (2002). Bacteria Sorting by Field-Flow Fractionation. Application to Whole-Cell Escherichia Coli Vaccine Strains. Anal. Chem. 74 (19), 4895–4904. doi: 10.1021/ac020199t
Romaniuk, J. A. H., Cegelski, L. (2015). Bacterial Cell Wall Composition and the Influence of Antibiotics by Cell-Wall and Whole-Cell NMR. Philos. Trans. R. Soc. B: Biol. Sci. 370 (1679), 20150024. doi: 10.1098/rstb.2015.0024
Rousseau, A. N., Faure, N., Rol, F., Sedaghat, Z., Le Galudec, J., Mallard, F., et al. (2021). Fast Antibiotic Susceptibility Testing via Raman Microspectrometry on Single Bacteria: An MRSA Case Study. ACS Omega 6 (25), 16273–16279. doi: 10.1021/acsomega.1c00170
Ruzicka, F., Horka, M., Hola, V., Mlynarikova, K., Drab, V. (2016). Capillary Isoelectric Focusing—Useful Tool for Detection and Quantification of Lactic Acid Bacteria in Milk. Food Anal. Methods 9 (12), 3251–3257. doi: 10.1007/s12161-016-0522-6
Rychert, J. (2019). Benefits and Limitations of MALDI-TOF Mass Spectrometry for the Identification of Microorganisms. J. Infectiol. 2 (4), 1–5. doi: 10.29245/2689-9981/2019/4.1142
Sabat, A. J., van Zanten, E., Akkerboom, V., Wisselink, G., van Slochteren, K., de Boer, R. F., et al. (2017). Targeted Next-Generation Sequencing of the 16S-23S Rrna Region for Culture-Independent Bacterial Identification - Increased Discrimination of Closely Related Species. Sci. Rep. 7 (1), 3434. doi: 10.1038/s41598-017-03458-6
Saenton, S., Lee, H., Gao, Y.-S., Ranville, J. F., Williams, S. K. R. (2000). Evaluation of Different Field-Flow Fractionation Techniques for Separating Bacteria. Separation Sci. Technol. 35 (11), 1761–1775. doi: 10.1081/SS-100102492
Samek, O., Bernatová, S., Dohnal, F. (2021). The Potential of SERS as an AST Methodology in Clinical Settings. Nanophotonics 10 (10), 2537–2561. doi: 10.1515/nanoph-2021-0095
Samek, O., Telle, H. H., Harris, L. G., Bloomfield, M., Mack, D. (2008). Raman Spectroscopy for Rapid Discrimination of Staphylococcus Epidermidis Clones Related to Medical Device-Associated Infections. Laser Phys. Lett. 5 (6), 465–470. doi: 10.1002/lapl.200810011
Samek, O., Jonáš, A., Pilát, Z., Zemánek, P., Nedbal, L., Tříska, J., et al. (2010). Raman Microspectroscopy of Individual Algal Cells: Sensing Unsaturation of Storage Lipids In Vivo. Sensors 10 (9), 8635–8651. doi: 10.3390/s100908635
Samek, O., Mlynariková, K., Bernatová, S., Ježek, J., Krzyžánek, V., Šiler, M., et al. (2014). Candida Parapsilosis Biofilm Identification by Raman Spectroscopy. Int. J. Mol. Sci. 15 (12), 23924–23935. doi: 10.3390/ijms151223924
Samek, O., Bernatová, S., Ježek, J., Šiler, M., Šerý, M., Krzyžánek, V., et al. (2015). Identification of Individual Biofilm-Forming Bacterial Cells Using Raman Tweezers. J. Biomed. Optics 20 (5), 51038. doi: 10.1117/1.JBO.20.5.051038
Sarno, B., Heineck, D., Heller, M. J., Ibsen, S. D. (2021). Dielectrophoresis: Developments and Applications From 2010 to 2020. ELECTROPHORESIS 42 (5), 539–564. doi: 10.1002/elps.202000156
Schie, I. W., Huser, T. (2013). Methods and Applications of Raman Microspectroscopy to Single-Cell Analysis. Appl. Spectrosc. 67 (8), 813–828. doi: 10.1366/12-06971
Schröder, U.-C., Ramoji, A., Glaser, U., Sachse, S., Leiterer, C., Csaki, A., et al. (2013). Combined Dielectrophoresis–Raman Setup for the Classification of Pathogens Recovered From the Urinary Tract. Anal. Chem. 85 (22), 10717–10724. doi: 10.1021/ac4021616
Schröder, U.-C., Bokeloh, F., O’Sullivan, M., Glaser, U., Wolf, K., Pfister, W., et al. (2015). Rapid, Culture-Independent, Optical Diagnostics of Centrifugally Captured Bacteria From Urine Samples. Biomicrofluidics 9 (4), 044118. doi: 10.1063/1.4928070
Schuster, K. C., Urlaub, E., Gapes, J. R. (2000). Single-Cell Analysis of Bacteria by Raman Microscopy: Spectral Information on the Chemical Composition of Cells and on the Heterogeneity in a Culture. J. Microbiol. Methods 42 (1), 29–38. doi: 10.1016/S0167-7012(00)00169-X
Schwaminger, S. P., Fraga-García, P., Eigenfeld, M., Becker, T. M., Berensmeier, S. (2019). Magnetic Separation in Bioprocessing Beyond the Analytical Scale: From Biotechnology to the Food Industry. Front. Bioeng. Biotechnol. 7. doi: 10.3389/fbioe.2019.00233
Sender, R., Fuchs, S., Milo, R. (2016). Revised Estimates for the Number of Human and Bacteria Cells in the Body. PloS Biol. 14 (8), e1002533. doi: 10.1371/journal.pbio.1002533
Singhal, N., Kumar, M., Kanaujia, P. K., Virdi, J. S. (2015). MALDI-TOF Mass Spectrometry: An Emerging Technology for Microbial Identification and Diagnosis. Front. Microbiol. 6. doi: 10.3389/fmicb.2015.00791
Singh, G. P., Volpe, G., Creely, C. M., Grötsch, H., Geli, I. M., Petrov, D. (2006). The Lag Phase and G1 Phase of a Single Yeast Cell Monitored by Raman Microspectroscopy. J. Raman Spectrosc. 37 (8), 858–864. doi: 10.1002/jrs.1520
Smith, E., Dent, G. (2004). Modern Raman Spectroscopy - a Practical Approach: Smith/Modern Raman Spectroscopy - a Practical Approach. Chichester (UK: John Wiley & Sons, Ltd). doi: 10.1002/0470011831
Spratt, D. A. (2004). Significance of Bacterial Identification by Molecular Biology Methods. Endodontic Topics 9 (1), 5–14. doi: 10.1111/j.1601-1546.2004.00106.x
Taleb, I., Thiéfin, G., Gobinet, C., Untereiner, V., Bernard-Chabert, B., Heurgué, A., et al. (2013). Diagnosis of Hepatocellular Carcinoma in Cirrhotic Patients: A Proof-of-Concept Study Using Serum Micro-Raman Spectroscopy. Analyst 138 (14), 4006. doi: 10.1039/c3an00245d
Tanniche, I., Collakova, E., Denbow, C., Senger, R. S. (2020). Characterizing Metabolic Stress-Induced Phenotypes of Synechocystis PCC6803 With Raman Spectroscopy. PeerJ 8, e8535. doi: 10.7717/peerj.8535
Tay, F., Yu, L., Iliescu, C. (2009). Particle Manipulation by Miniaturised Dielectrophoretic Devices. Defence Sci. J. 59 (6), 595–604. doi: 10.14429/dsj.59.1564
Tien, N., Chen, H.-C., Gau, S.-L., Lin, T.-H., Lin, H.-S., You, B.-J., et al. (2016). Diagnosis of Bacterial Pathogens in the Dialysate of Peritoneal Dialysis Patients With Peritonitis Using Surface-Enhanced Raman Spectroscopy. Clinica Chimica Acta 461, 69–75. doi: 10.1016/j.cca.2016.07.026
Vankeirsbilck, T., Vercauteren, A., Baeyens, W., van der Weken, G., Verpoort, F., Vergote, G., et al. (2002). Applications of Raman Spectroscopy in Pharmaceutical Analysis. TrAC Trends Anal. Chem. 21 (12), 869–877. doi: 10.1016/S0165-9936(02)01208-6
Verroken, A., Defourny, L., Lechgar, L., Magnette, A., Delmée, M., Glupczynski, Y. (2015). Reducing Time to Identification of Positive Blood Cultures With MALDI-TOF MS Analysis After a 5-H Subculture. Eur. J. Clin. Microbiol. Infect. Dis. 34 (2), 405–413. doi: 10.1007/s10096-014-2242-4
Vogt, S., Löffler, K., Dinkelacker, A. G., Bader, B., Autenrieth, I. B., Peter, S., et al. (2019). Fourier-Transform Infrared (FTIR) Spectroscopy for Typing of Clinical Enterobacter Cloacae Complex Isolates. Front. Microbiol. 10. doi: 10.3389/fmicb.2019.02582
Wang, X., Huang, S.-C., Hu, S., Yan, S., Ren, B. (2020). Fundamental Understanding and Applications of Plasmon-Enhanced Raman Spectroscopy. Nat. Rev. Phys. 2 (5), 253–271. doi: 10.1038/s42254-020-0171-y
Wang, L., Liu, W., Tang, J.-W., Wang, J.-J., Liu, Q.-H., Wen, P.-B., et al. (2021). Applications of Raman Spectroscopy in Bacterial Infections: Principles, Advantages, and Shortcomings. Front. Microbiol. 12. doi: 10.3389/fmicb.2021.683580
Weber, R. E., Petkowski, J. J., Michaels, B., Wisniewski, K., Piela, A., Antoszczyk, S., et al. (2021). Fluid-Screen as a Real Time Dielectrophoretic Method for Universal Microbial Capture. Sci. Rep. 11 (1), 22222. doi: 10.1038/s41598-021-01600-z
Weng, S., Zhu, W., Zhang, X., Yuan, H., Zheng, L., Zhao, J., et al. (2019). Recent Advances in Raman Technology With Applications in Agriculture, Food and Biosystems: A Review. Artif. Intell. Agric. 3, 1–10. doi: 10.1016/j.aiia.2019.11.001
Wichmann, C., Rösch, P., Popp, J. (2021). Isolation of Bacteria From Artificial Bronchoalveolar Lavage Fluid Using Density Gradient Centrifugation and Their Accessibility by Raman Spectroscopy. Anal. Bioanal. Chem. 413 (20), 5193–5200. doi: 10.1007/s00216-021-03488-0
Witkowska, E., Niciński, K., Korsak, D., Dominiak, B., Waluk, J., Kamińska, A. (2020). Nanoplasmonic Sensor for Foodborne Pathogens Detection. Towards Development of ISO-SERS Methodology for Taxonomic Affiliation of Campylobacter Spp. J. Biophotonics 13 (5), e201960227. doi: 10.1002/jbio.201960227
Wu, M., Ling, D., Ling, L., Li, W., Li, Y. (2017). Stable Optical Trapping and Sensitive Characterization of Nanostructures Using Standing-Wave Raman Tweezers. Sci. Rep. 7 (1), 42930. doi: 10.1038/srep42930
Wulf, M. W. H., Willemse-Erix, D., Verduin, C. M., Puppels, G., van Belkum, A., Maquelin, K. (2012). The Use of Raman Spectroscopy in the Epidemiology of Methicillin-Resistant Staphylococcus Aureus of Human- and Animal-Related Clonal Lineages. Clin. Microbiol. Infect. 18 (2), 147–152. doi: 10.1111/j.1469-0691.2011.03517.x
Xu, T., Sun, L. (2021). A Mini Review on Capillary Isoelectric Focusing-Mass Spectrometry for Top-Down Proteomics. Front. Chem. 9. doi: 10.3389/fchem.2021.651757
Yogesha, M., Chawla, K., Bankapur, A., Acharya, M., D’Souza, J. S., Chidangil, S. (2019). A Micro-Raman and Chemometric Study of Urinary Tract Infection-Causing Bacterial Pathogens in Mixed Cultures. Anal. Bioanal. Chem. 411 (14), 3165–3177. doi: 10.1007/s00216-019-01784-4
Yonetani, S., Ohnishi, H., Ohkusu, K., Matsumoto, T., Watanabe, T. (2016). Direct Identification of Microorganisms From Positive Blood Cultures by MALDI-TOF MS Using an in-House Saponin Method. Int. J. Infect. Dis. 52, 37–42. doi: 10.1016/j.ijid.2016.09.014
Zarnowiec, P., Lechowicz, L., Czerwonka, G., Kaca, W. (2015). Fourier Transform Infrared Spectroscopy (FTIR) as a Tool for the Identification and Differentiation of Pathogenic Bacteria. Curr. Med. Chem. 22 (14), 1710–1718. doi: 10.2174/0929867322666150311152800
Zhang, H., Chang, H., Neuzil, P. (2019). DEP-on-a-Chip: Dielectrophoresis Applied to Microfluidic Platforms. Micromachines 10 (6), 423. doi: 10.3390/mi10060423
Zhou, M., Le, J., Chen, Y., Cai, Y., Hong, Z., Chai, Y. (2017). An Improved in-House MALDI-TOF MS Protocol for Direct Cost-Effective Identification of Pathogens From Blood Cultures. Front. Microbiol. 8. doi: 10.3389/fmicb.2017.01824
Zhou, W., Yang, Q., Kudinha, T., Sun, L., Zhang, R., Liu, C., et al. (2019). Recent Advances in Microfluidic Devices for Bacteria and Fungus Research. TrAC Trends Analy. Chem. 112, 175–195. doi: 10.1016/j.trac.2018.12.024
Ziemann, M. A., Madariaga, J. M. (2021). Applications of Raman Spectroscopy in Art and Archaeology. J. Raman Spectrosc. 52 (1), 8–14. doi: 10.1002/jrs.6054
Keywords: Raman spectroscopy, Raman tweezers, identification of microorganisms, antimicrobial resistance, microfluidic devices, magnetic beads, diagnostics
Citation: Rebrosova K, Samek O, Kizovsky M, Bernatova S, Hola V and Ruzicka F (2022) Raman Spectroscopy—A Novel Method for Identification and Characterization of Microbes on a Single-Cell Level in Clinical Settings. Front. Cell. Infect. Microbiol. 12:866463. doi: 10.3389/fcimb.2022.866463
Received: 31 January 2022; Accepted: 07 March 2022;
Published: 22 April 2022.
Edited by:
Nicasio Mancini, Vita-Salute San Raffaele University, ItalyReviewed by:
Anna Chiara De Luca, National Research Council (CNR), ItalyCopyright © 2022 Rebrosova, Samek, Kizovsky, Bernatova, Hola and Ruzicka. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Veronika Hola, dmVyb25pa2EuaG9sYUBmbnVzYS5jeg==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.