- 1Department of Physiology, Faculty of Medicine, Manipal University College Malaysia, Melaka, Malaysia
- 2Department of Human Physiology, Vidyasagar University, Midnapore, India
- 3Department of Pharmacology, Faculty of Health Sciences, School of Clinical Medicines, University of the Free State, Bloemfontein, South Africa
- 4Department of Human Physiology, Raja NL Khan Women’s College, Midnapore, India
- 5Department of Medical Entomology, Calcutta School of Tropical Medicine, Kolkata, West Bengal, India
Introduction: After being used vigorously for the previous two decades to treat P. falciparum, chloroquine and sulfadoxine-pyrimethamine were replaced in 2009 with an artemisinin-based combination therapy (artesunate-sulfadoxine-pyrimethamine) in an effort to combat multidrug-resistant parasites.
Methods: We set out to assess the genetic variants of sulfadoxine-pyrimethamine resistance and the effectiveness of its treatment in eastern India prior to, during, and 6 to 8 years following the introduction of the new pharmacological regime. In 2008-2009, 318 P. falciparum–positive patients got the recommended doses of sulfadoxine-pyrimethamine. We used 379 additional isolates from 2015 to 2017 in addition to the 106 isolates from 2010. All 803 isolates from two study sites underwent in vitro sulfadoxine-pyrimethamine sensitivity testing and genomic characterisation of sulfadoxine-pyrimethamine resistance (pfdhfr and pfdhps).
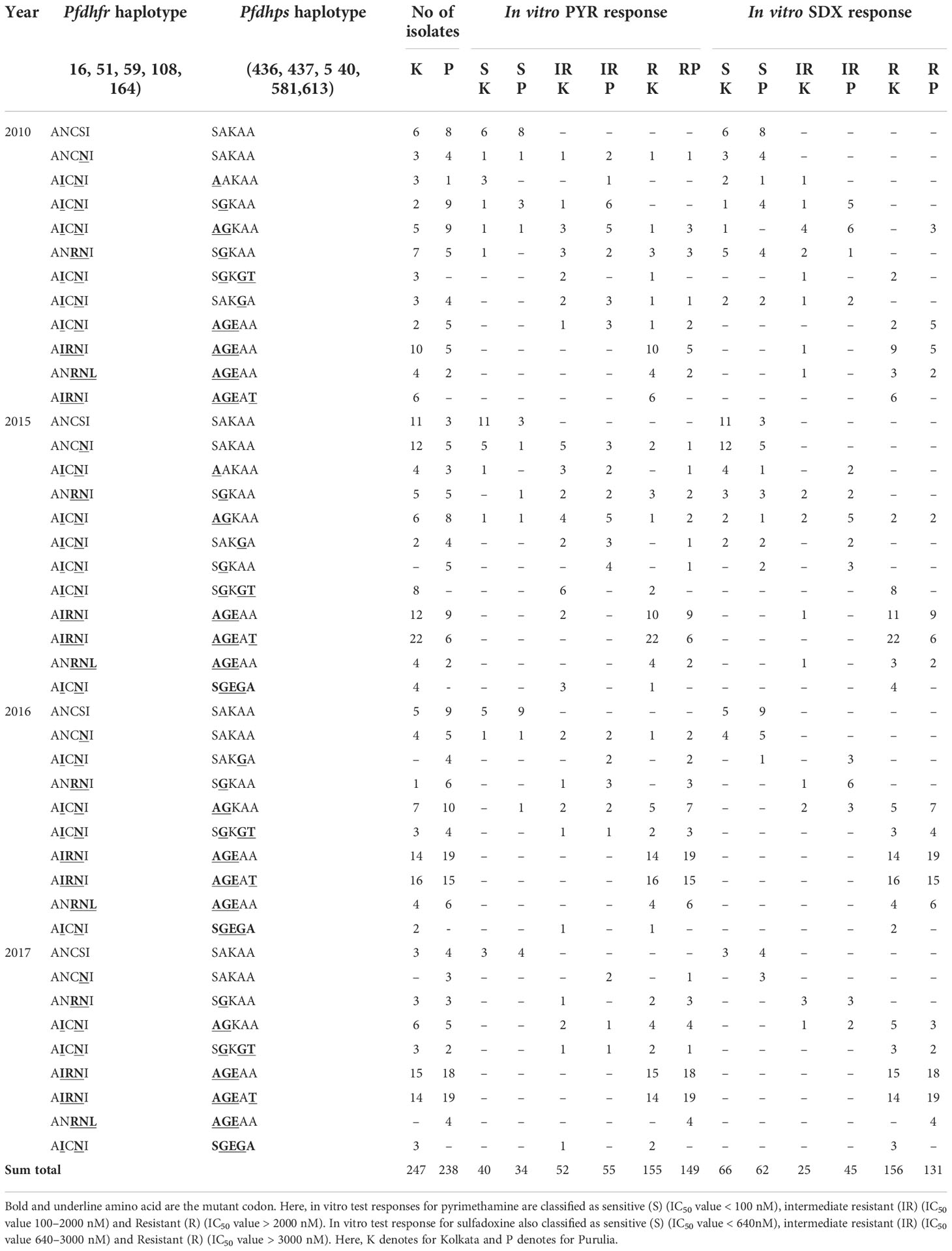
Results: In Kolkata and Purulia, we observed early treatment failure in 30.7 and 14.4% of patients, respectively, whereas recrudescence was found in 8.1 and 13.4% of patients, respectively, in 2008–2009. In 2017, the proportion of in vitro pyrimethamine and sulfadoxine resistance steadily grew in Kolkata and Purulia despite a single use of sulfadoxine-pyrimethamine. Treatment failures with sulfadoxine-pyrimethamine were linked to quintuple or quadruple pfdhfr- pfdhps mutations (AICII-AGKAT, AICII-AGKAA, AICII-SGKGT, AICII-AGKAA, AICNI-AGKAA) in 2008–2009 (p < 0.001). The subsequent spread of mutant-haplotypes with higher in vitro sulfadoxine-pyrimethamine resistance (p < 0.001), such as the sextuple (dhfr-AIRNI+dhps-AGEAA, dhfr-ANRNL+dhps-AGEAA) and septuple (dhfr-AIRNI+dhps-AGEAT), mutations were observed in 2015-2017.
Discussion: This successive spread of mutations with high in vitro sulfadoxine-pyrimethamine resistance confirmed the progressive increase in antifolate resistance even after an 8-year withdrawal of sulfadoxine-pyrimethamine.
Introduction
Globally, 241 million confirmed cases of malaria and an estimated 627,000 deaths were reported in 2020. (WHO, 2021). This represents around 14 million more cases in 2020 compared with 2019 and 69,000 more deaths. Ninety-six percent of malaria-related deaths reported worldwide in 2020 came from Africa (WHO, 2021 prior to 2009, Guidelines for Diagnosis and Treatment of Malaria in India, 2009); sulfadoxine-pyrimethamine combination (SP) was frequently used in India as a second-line therapy against chloroquine-resistant Plasmodium falciparum. However, the spread of SP-resistant parasites has since made SP less effective (Ahmed et al., 2004). Pyrimethamine (PYR), an antifolate drug, targets the parasites’ purine biosynthesis pathway, namely, the dihydrofolate reductase (dhfr) gene, whereas sulfadoxine (SDX) only targets the dihydropteroate synthase (dhps) gene, which prevents parasite DNA replication (Foote and Cowman, 1994; Abdul-Ghani et al., 2014; Heinberg and Kirkman, 2015).
Antifolate resistance is solely caused by point mutations in the genes that produce the proteins DHPS and DHFR (Foote and Cowman, 1994; Juma et al., 2019).
In vivo SP-efficacy and genome wide association studies from south-east Asia to Africa including Middle East countries clearly suggested that polymorphism in the pfdhfr gene especially at codon 108 (Ser-108 to Asn-108) represented the key mutation, whereas additional mutations at codon N51I and C59R conferred high levels of resistance (Juma et al., 2019; Zhao et al., 2020). In addition to the polymorphism in pfdhps-A437G, other alterations (436-Ala, 540-Glu, 581-Gly, and 613-thr) increased the level of resistance to sulphonamides (Sharma, 2012; Abdul-Ghani et al., 2013; Yaqoob et al., 2018; Madkhali et al., 2020). After more than two decades of successful SP use in India, reports indicate the appearance of SP-resistant parasites from various Indian regions (Ahmed et al., 2004; Sharma, 2012; Das et al., 2013a). When at least two pfdhfr mutations and one pfdhps mutation occurred simultaneously, the clinical consequence of SP-resistance was frequently seen (Das et al., 2013a). Except for the north-eastern states of India, artesunate-SP is still the medicine of choice there, despite later reports of SP-resistant parasites in several regions of the country (Das et al., 2013a; Guidelines for Diagnosis and Treatment of Malaria in India, 2014). This study sought to establish a genotype–phenotype correlation before, during, and 6– 8 years after the implementation of new drug policy by evaluating the genetic variations and clinical characteristics of antifolate resistance in a large cohort of eastern Indian population in light of the emergence of SP-resistant parasites.
Material and methods
Selection of patients
In two distinct time periods, we conducted the study in eastern India’s malaria-endemic Kolkata and Purulia. The first phase samples were gathered between April 2008 and November 2010 before and around the time that India’s new drug policy went into effect. After 6–8 years of the new national drug policy, we also collected the samples between March 2015 and December 2017 for the second phase. Patients who experienced a fever with headache, shaking, or vomiting within the previous 24 h and those who had not taken any antimalarial medicine during that time were submitted for a malaria test. We used EDTA-coated vacutainer tubes to collect 3 ml of intravenous blood from 4,652 suspected patients in accordance with the Helsinki procedure.
P. falciparum mono-infection determined by microscopic analysis of Giemsa-stained thin and thick blood smears, patients with a parasite density of 1,000–10,000 asexual parasites/l blood, and patients without a history of self-medication with antimalarials were the inclusion criteria for our study (Bloland et al., 2003). Through the use of an allelic family-specific nested PCR (MAD20 and K1 for pfmsp-1 and 3D7 Africa and FC27 for pfmsp-2), monoclonal P. falciparum infections were further evaluated (Das et al., 2014). Pregnant women, newborns, people with non-falciparum malaria, people with severe complicated malaria, and people with mixed infections (P. falciparum + P. vivax) were excluded from the study (Bloland et al., 2003). The study also eliminated any individuals who had used antibiotics or antimalarial medications prior to the diagnosis of P. falciparum infection. We obtained each patient’s or the patient’s legal guardian’s written informed permission. The Vidyasagar University Human Ethical-Committee gave its full approval to the experimental design and protocols (VU/HEC 08-012).
In vivo SP treatment
P. falciparum– positive patients received standard SP doses under the supervision of a medical officer in accordance with the national guideline, with adults receiving a single oral dose of three tablets containing 1000 mg of SDX and 50 mg of PYR, whereas children received a single dose of 20 mg/kg of SDX and 1 mg/kg of PYR (Guidelines for Diagnosis and Treatment of Malaria in India, 2009). Within their expiration dates, we used the quality-assured medications duly provided by the Ministry of Health and Family Welfare, Government of India (IPCA, LTD). Giemsa-stained thin blood smears from each patient were prepared in triplicate at intervals of 12 h until the parasite became undetectable. Two skilled microscopists examined blood smears. If any discrepancies in the results were detected, a senior microscopist was consulted for confirmation. On days 1, 2, 3, 4, 7, 14, 21, and 28, treatment follow-ups were conducted to assess the patient’s clinical status and assure their safety (Bloland et al., 2003). Additionally, if malaria symptoms returned in the intervals between the scheduled visits, unscheduled follow-ups were conducted. By analyzing the msp1, msp2, and glurp genes using family-specific nested PCR, it was possible to distinguish between reinfection and recrudescence (Das et al., 2013b).
According to WHO guidelines, we divided the therapeutic efficacy into three categories: early treatment failure (ETF), late treatment failure (LTF), and adequate clinical parasitological response (ACPR) (WHO, 2013). Patients who did not respond to SP were given a regular dose of artemether-lumefantrine (six tablets, each containing 40 mg AM and 240 mg LF) as a rescue medication and were then monitored for the following 28 days.
In vitro drug sensitivity assay
After adapting clinical isolates to in vitro growth using our standard protocol, the phenotypic parameters of in vitro SP-sensitivity were assessed (Das et al., 2013a). Following our standard laboratory protocol, P. falciparum clinical isolates were grown and maintained in culture (Das et al., 2014). The culture plates were incubated at 37°C with a 95% relative humidity environment and 5% CO2. After 48 h of incubation with SDX and PYR using our routine laboratory procedures, we measured the 50% inhibitory concentrations (IC50) of SP using a hypoxanthine incorporation assay (Das et al., 2014). We used nonlinear probit/logit regression analysis to express the values (Basco and Ringwald, 2000). Stock and dilutions of PYR and SDX were made, respectively, with ethanol and dimethyl sulphoxide (DMSO, 0.1%). The final concentrations ranged from 50 to 25600 nM for both PYR and SDX. We studied each concentration in duplicate. However, we used the PYR-sensitive and SDX-resistant-3D7 strains as control strains and adhered to the established threshold values of in vitro SP sensitivity as previously described (Basco and Ringwald, 2000; Das et al., 2012a).
Genome analyses
We isolated the parasite DNA from parasitized erythrocytes as mentioned earlier (Basco and Ringwald, 2000). Furthermore, we used the P. falciparum wild-sequence (AL844509.2) available in GenBank to create the primers for the pfdhfr and pfdhps genes using the primer design software Primer3. Using nested PCR, followed by restriction fragment length polymorphism and sequence analysis, we evaluated the single nucleotide polymorphisms of the pfdhfr and pfdhps genes (Das et al., 2013a). We used an ABI PRISM big-dye terminator cycle sequencing kit and a 3730-xl genetic analyzer to sequence the pfdhfr and pfdhps amplicons in order to confirm the mutation by reading both the forward and reverse strands. The sequence was translated using the Protein Analysis System proteomic service (http://www.expasy.org), and the translated sequences were aligned using ClustalW2 (http://www.ebi.ac.uk/clustalw).
Molecular modeling of DHFR protein
We have discovered a novel pfdhfr double mutant haplotype. Using the crystal structure of the DHFR protein from Plasmodium falciparum (PDB id: 6A2K) as a template, the double mutant DHFR protein’s 3D structure was modeled in Phyre-2 (Kelley et al., 2015). PyMOL was used to superimpose the wild-type and mutant-type proteins to test the model’s accuracy (The PyMOL Molecular Graphics System, Version 1.8 Schrödinger, LLC, 2016; Badawi et al., 2016).
Statistical analysis
In vitro IC50 values for PYR and SDX coupled with parasitemia were provided as mean SD data. Fisher’s exact test and regression analysis were used to investigate the relationship between SP treatment efficacies and various genotypes. Through the use of the Fisher’s exact test and the Mann–Whitney U test, we evaluated the associations between the IC50s of SP and the dhfr and dhps genotypes. The Kruskal–Wallis test was also used to compare data among more than two groups. The total cure-rate was evaluated using the Kaplan–Meier estimate. The 95% confidence intervals were calculated using the Clopper-Pearson formula. Multivariate logistic regression analysis was used to evaluate the connections between pfdhfr, pfdhps mutations, and in vitro SP resistance. When p < 0.05, a statistical difference was deemed significant. With the help of GraphPad software 3.0. In Stat and Origin 6.1, we conducted the statistical analysis.
Results
Study population
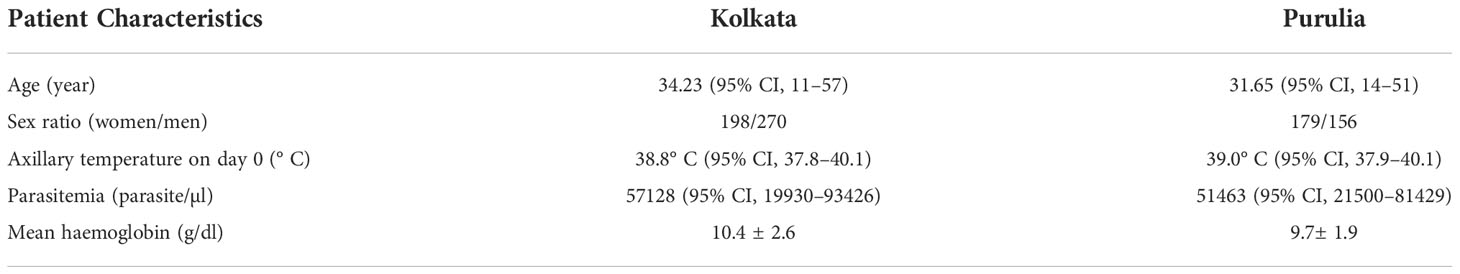
In this investigation, we have paid particular attention to the duration-dependent antifolate susceptibility of P. falciparum isolates. Eight hundred three (17.2%) of the 4,652 suspected cases of malaria had finished the 28-day treatment follow-up. These individuals had uncomplicated P. falciparum monoclonal infections and had a mean age of 27 years (95% CI: 8–57). Of them, 335 patients were from Purulia, whereas the rest 468 patients were from Kolkata (Supplementary Table S1). A total of 761 patients (16.4%) who were P. vivax–positive were eliminated. Due to combined P. falciparum + P. vivax infection, an additional 181 individuals (3.9%) were removed. While men (57.7%) were found to be more affected in Kolkata, women (53.4%) were found to be more impacted in Purulia. Table 1 lists the unique traits of participants upon enrolment in the two study locations. Details of patient selection were presented in Figure 1.
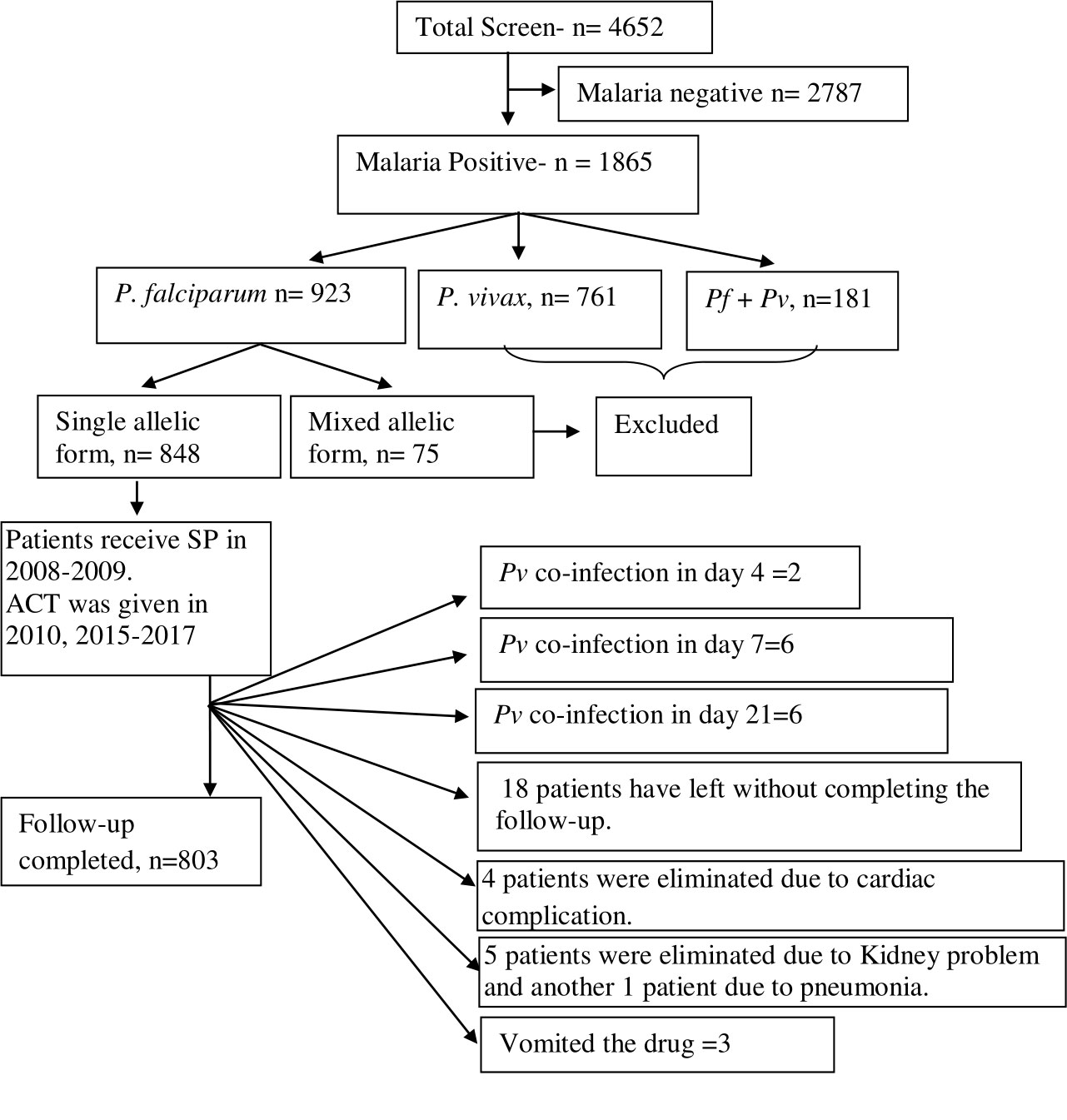
Figure 1 Schematic presentation of patient selection and details inclusion and exclusion criteria for the study. Out of 4,652 screened patients, 2,787 (60.34%) patients were eliminated as they had shown malaria slide negative. A total of 923 patients (19.41%) were found with P. falciparum infection. Of them, 75 isolates (8.64%) were identified as mixed P. falciparum infection and they were eliminated as these isolates contained both mspI and mspII alleles. Forty-five patients had not completed the 28-day treatment follow-up, and they were eliminated from the study.
Clinical efficacy of SP
In 2008–2009, 318 P. falciparum– infected patients had finished the 28-day SP therapy follow-up. A total of 221 of them were from Kolkata, whereas 97 were from Purulia. A summary of the treatment effectiveness following SP therapy is provided in Supplementary Table S2.
In addition, 485 additional P. falciparum infections were treated with the artesunate-SP combination in accordance with the national guidelines between 2010 and 2015–2017; however, we were unable to disclose the clinical outcome data due to a lack of sufficient authorization. The genomic characterization of the antifolate resistant genes in those isolates was presented together with each isolate’s unique phenotypic traits of in vitro SP susceptibility. However, 205 patients (64.5%) out of 318 P. falciparum– infected patients who got SP therapy experienced a successful outcome (ACPR).
More specifically, SP was effective in treating 61.7% of the isolates from Kolkata and 72.1% of the isolates from Purulia. Early treatment failure (ETF) was found in 25.8% of patients based on day 3 positive parasite (parasitemia >1% that of day 0), associated with continuation of fever on day 3, whereas 9.8% of patients were diagnosed as late treatment failure (LTF). Among the study participants, no deaths have been documented. Following the SP treatment, parasite recurrence was seen in three patients on day 4, eight patients on day 7, five patients on day 14, 19 more patients on day 21, and nine more patients on day 28.
All 44 cases of apparent parasite reappearance were subjected to an analysis of merozoite surface proteins 1, 2, and glurp, of which 31 (70.4%) isolates were identified as cases of late SP treatment failure or true recrudescence (Supplementary Table S2). Treatment failure for SP in Purulia (8.9% ETF + 8.9% LTF) was recorded in 17.8% of patients in 2008 and increased to 36.5% (19.2% ETF + 17.3% LTF) in 2009. In Kolkata, treatment failure for SP (6.7% ETF + 8.9% LTF) was only recorded in 15.6% of patients in 2008 and increased sharply to 44.9% (36.9% ETF + 7.9% LTF) in 2009. In contrast to Purulia, Kolkata had a higher prevalence of ETF following SP therapy (Supplementary Table S2). The overall cure rate following SP therapy was reported to be 61.1% in Kolkata (Kaplan–Meier estimate; 95% CI 54.29–67.48) and 72.16% in Purulia (61.98–80.55). All the treatment failure cases were successfully treated after AMLF therapy.
In vitro SP susceptibility
The phenotypic characteristics of in vitro sensitivity for PYR and SDX yielded interpretable results in 795 and 787 isolates, respectively. In Kolkata, the percentage of isolates that were PYR-sensitive in 2008 was 24.4% (mean IC50 = 71.4 nM, 95% CI, 49–91), but it sharply declined to 6.4% by 2017. In Purulia, PYR sensitivity was detected in 46.7% of isolates in 2008 (mean IC50 = 68.8 nM, 95% CI, 51–88 nM), but it dropped to 6.9% in 2017 (mean IC50 = 72 nM, 95% CI, 55–95). In comparison with 2008, the proportion of intermediate PYR resistance significantly decreased at both research locations in 2017. In Purulia, intermediate PYR resistance was just 6.9% in 2017, whereas in Kolkata, intermediate PYR resistance decreased from 28.9% in 2008 to 10.6% in 2017.
In contrast, in Kolkata, despite no usage of SP since 2009, 83% of isolates were found to be extremely resistant to PYR in 2017 (mean IC50 = 9770.65 nM; 95% CI, 7150–12380), up from 46.7% in 2008 (mean IC50 = 5606 nM; 95% CI, 3155–8168). Additionally, the percentage of PYR-resistance in Purulia gradually grew from only 26.7% in 2008 to as high as 86.2% (mean IC50 = 9120.5 nM; 95% CI, 6075–12065) in 2017 (Figure 2A).
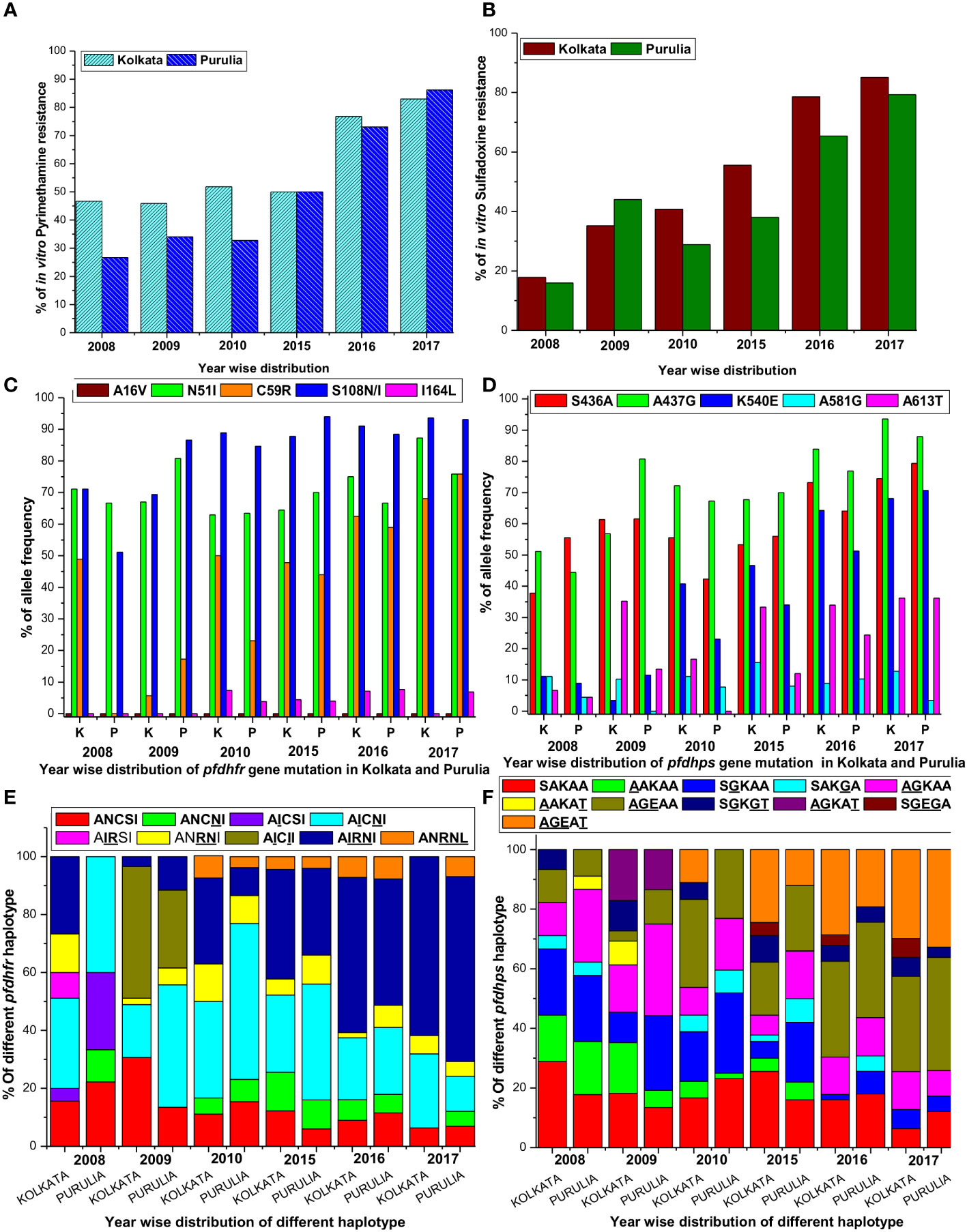
Figure 2 (A) Proportion of in vitro pyrimethamine resistance from 2008 to 2017 in two study sites. (B) Proportion of in vitro sulfadoxine resistance from 2008 to 2017 in two study sites. (C) Single nucleotide polymorphism of different codons of pfdhfr gene in both study sites. (D) Single nucleotide polymorphism of different codons of pfdhps gene in both study sites. (E) Alteration of different pfdhfr haplotype frequencies in different period in both the study sites. (F) Alteration of different pfdhps haplotypes in different time frame at our study sites.
In Kolkata, the majority of SDX-sensitive parasites were found to be predominant (53.3%) in 2008 but significantly declined to 6.4% in 2013. In 2008, only 17% of isolates were reported to be SDX resistant (mean IC50 = 6015.35 nM; 95% CI, 3988–8050); however, this number rose dramatically to 85.1% in 2017 (mean IC50 = 10220.5 nM; 95% CI, 7800–12500). In Purulia, 55.6% of isolates were found to be SDX-sensitive in 2008, but this figure had dropped to 12.1% in 2017 (mean IC50 = 351.5 nM, 95% CI, 155–560). SDX resistance was gradually increased to 79.3% in 2017 (mean IC50 = 9670 nM; 95% CI, 6980–12390 nM) from only 15.6% in 2008 (Figure 2B). Pyrimethamine and sulfadoxine in vitro responses were highly correlated (r2 = 0.9965, p < 0.019). On the other hand, intermediate SDX resistance in Kolkata fell to 8.5% in 2017 from 29.7% in 2008, whereas intermediate PYR resistance in Purulia was around 8.6% in 2017.
Genetic architecture of pfdhfr and pfdhps before new national drug policy
In 2008, 71.1% of Kolkata isolates had the pfdhfr-S108N mutation, which was slightly reduced to 69.3% in 2009. In 2009, 45.8% of Kolkata isolates and 26.9% of Purulia isolates encoded novel mutant isoleucine at codon108 (108I) instead of wild serine or mutant asparagine or threonine (KP256234, KP256235, KP256236, and KP256237) (Figures 3A–C). This novel dhfr-108I allele was always observed with dhfr-51I mutation and was only discovered after super-cyclone Aila for 6–8 months. After that, we never saw this haplotype again. The superposition of a novel double mutant (108I + 51I) allele with the wild allele revealed structural uniformity except at the 51 and 108 codons (Figure 3D); this resulted in potential conformational changes in the PfDHFR protein, which reduced the binding energy during interaction with pyrimethamine (Figures 3E, F), resulting in resistance to pyrimethamine. In 2009, the proportion of Purulia isolates with the most vulnerable S108N or novel S108I mutation increased from 51.1% to 86.5%. In 2008, the N51I mutation predominated in both Kolkata (71.1%) and Purulia (66.7%). In 2009, the N51I mutation was reduced to 67.1% in Kolkata but increased to 80.7 percent in Purulia. In 2008–2009, no mutations were found in the pfdhfr gene at codon-A16V and I164L (Figure 2C).
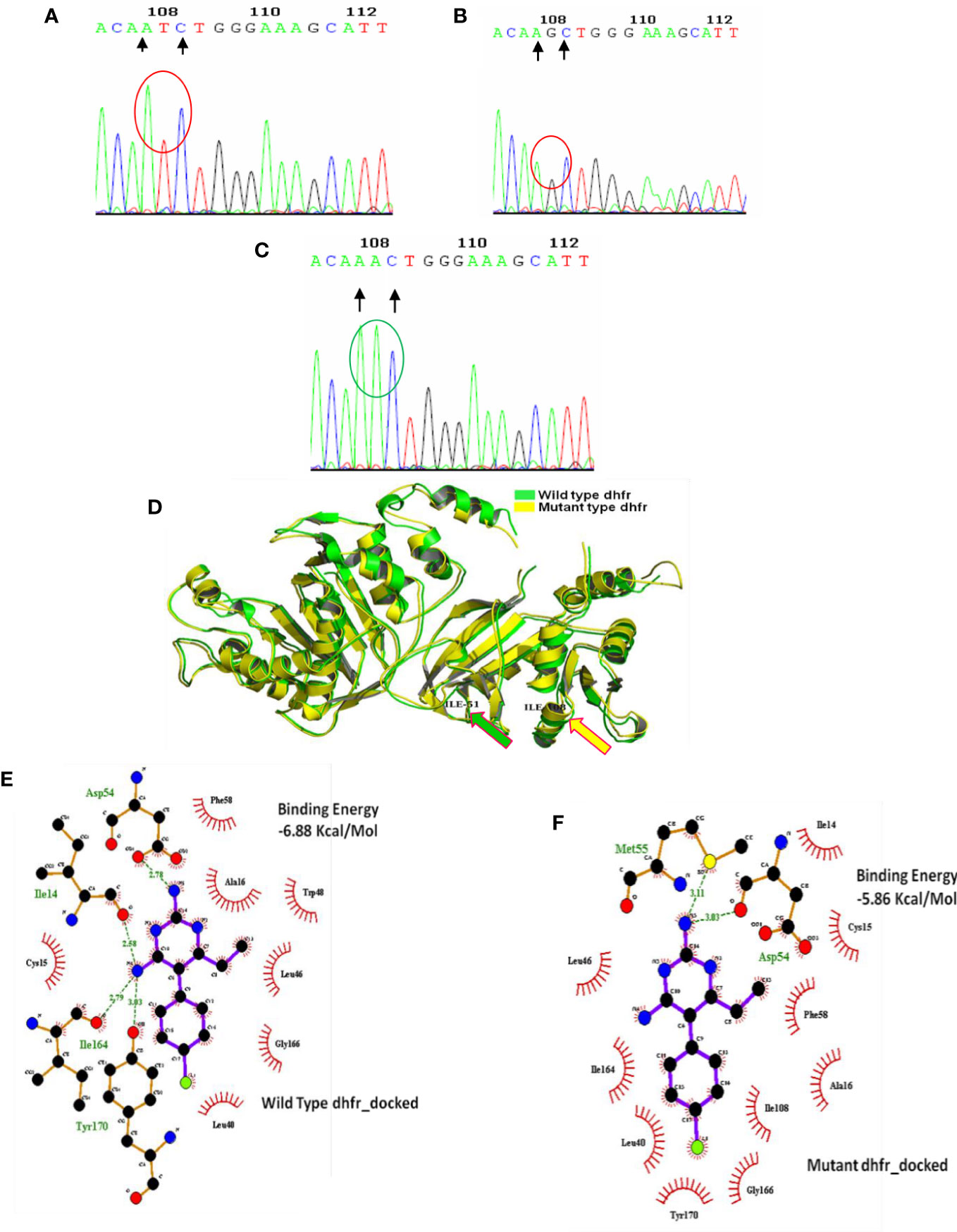
Figure 3 Genomic and proteomic analysis of mutant dhfr 108 codon. Through DNA sequence analysis, an alteration of AGC (B) to ATC (A) was observed at codon 108, which encoded mutant isoleucine (108I) (A). The Expert Protein Analysis System confirmed the presence of isoleucine (ATC). In the wild-type allele, AGC expresses serine at 108 positions of dhfr gene, but after mutation, it was replaced with AAC (encoding asparagine) (C). (D) Superimposition of wild and mutant allele: Super-imposition of wild and novel double dhfr mutation (108I + 51I) proved the uniformity of mutant allele with the wild allele except at 51, and 108 codon and producing a notch having a RMSD of 0·438 Å. (E) Molecular docking of pyrimethamine with wild- type dhfr gene. (F) Molecular docking of pyrimethamine with novel double mutant (dhfr-108I+51I) allele. Docking with wild- type dhfr protein with pyrimethamine produced high binding energy of 6.88 Kcal/Mole, whereas the docking of double mutant dhfr protein with pyrimethamine produced much less binding energy of 5.86 Kcal/Mole. It proves the less affinity of novel mutant dhfr protein toward pyrimethamine.
In Kolkata, the core pfdhps-A437G mutation was found in 51.1% of isolates in 2008 and 56.8% of isolates in 2009, respectively. From 2008 to 2009, Purulia saw a substantial rise in the percentage of isolates with the pfdhps-A437G mutation, going from 55.6 to 80.8%. In Kolkata, the prevalence of the S436A mutation significantly rose from 2008 (37.8%) to 2009 (61.4%). In Purulia, the percentage of the mutant S436A-allele was similar in 2008 (55.6%) and 2009 (61.5%). Only 13.5% of the isolates in Purulia had the A613T mutation, compared with a dramatic increase of 35.2% in Kolkata in 2009 (Figure 2D).
In Kolkata pfdhfr-AICNI haplotype was prevalent (31.1%) followed by AIRNI (26.7%) and wild ANCSI (15.6%) allele in 2008, whereas in Purulia pfdhfr-AICNI (40%) was most frequently found haplotype after AICSI (26.7%) and ANCSI (22.2%) allele. In 2009, novel pfdhfr-AICII haplotype (45.5%) was predominant followed by ANCSI (30.7%) and AICNI allele (18.2%) in Kolkata, while in Purulia, AICNI (42.3%) haplotype was commonly found after AICII (26.9%) and ANCSI (13.5%) allele (Figure 2E). In Kolkata, wild pfdhps-SAKAA haplotype (28.9%) was prevalent after SGKAA (22.2%) and AAKAA (15.6%) allele whereas, in Purulia, AGKAA (24.4%) allele was frequently found after SGKAA (22.2%) and AAKAA (17.8%) in 2008. Wild-SAKAA allele (18.2%) was prevalent in Kolkata followed by AGKAT (17.1%), and AAKAA allele (17.1) in 2009 whereas, in Purulia, AGKAA (30.8%) was most common haplotype after SGKAA (25%) (Figure 2F).
Genomic variations of pfdhfr and pfdhps after new drug policy
Since 2009, no single use of SP has been recommended in India. Instead, in the years that followed, the polymorphism at the dhfr-S108N codon grew and peaked in 2017 in Kolkata and Purulia at 93.6% and 931%, respectively. Similar to this, N51I polymorphism grew over the years and peaked in 2017 at 87.2% in Kolkata and 75.9% in Purulia. Following this, the percentage of pfdhfr-C59R mutation rose to 68.1% in Kolkata and 75.9% in Purulia. Codon A16V showed no mutations (Figure 2C).
In addition, pfdhps-S436A and A437G polymorphisms significantly increased from 2010 to 2017 and reached as high as 74.5 and 93.6%, respectively, in Kolkata, whereas 79.3% and 87.9% of isolates, respectively, exhibited mutant-S436A and A437G allele in Purulia. In the following years, the prevalence of the pfdhps-K540E mutation quickly rose, reaching as high as 68.1% in Kolkata and 70.7% in Purulia in 2017. The frequency of the pfdhps-A581G and A613T mutations did not significantly increase (Figure 2D).
Since 2010, wild pfdhfr-ANCSI haplotype rapidly reduced in number, whereas most vulnerable triple mutant AIRNI allele was predominantly observed in Kolkata (61.7%) and in Purulia (63.8%) followed by double mutant AICNI allele (25.5% in Kolkata and 12.1% in Purulia) in 2017. Proportion of single mutant ANCNI allele was gradually decreased in subsequent years. Triple mutant ANRNL allele was observed for first time in 2010 at both study sites and since then it has maintained a low frequency (< 5%) (Figure 2E).
Like pfdhfr, frequencies of wild-pfdhps-SAKAA haplotype also reduced drastically since 2010. Triple mutant AGEAA allele was prevalent in Kolkata (31.9%) and in Purulia (37.9%) in 2017 followed by quadruple mutant AGEAT allele (29.8% in Kolkata and 32.8% Purulia). Frequency of single mutant SGKAA and double mutant AGKAA allele were gradually decreased in consequent years. Triple-mutant SGEGA allele was observed initially in 2015 at Kolkata and thereafter maintained a very low frequency (<5%) (Figure 2F).
Genotypic association with phenotypes of SP efficacy in vivo and in vitro during 2008–2009
In 2008, the number of SP-treatment failure (ETF) cases was very less in both the study sites, but SP-resistant pattern in Purulia was observed differently from Kolkata. In Purulia, ETFs were associated with double dhfr-AICNI+ triple dhps-AGEAA mutation (p < 0.05) whereas, in Kolkata, isolates representing triple dhfr-AIRNI +triple dhps-AGEAA mutation were associated with ETF (p < 0.05) in 2008 (Table 2). Both haplotype-combinations showed very high IC50 for pyrimethamine and sulfadoxine and proved as true in vitro resistant isolates (Figures 4A, B). In Kolkata, two LTF cases were observed with AICNI-AGKAA allele. One each LTF case was reported with AICNI- SGKGT and AIRNI- AGEAA haplotype whereas, in Purulia, one each LTF was found with AICNI-AGEAA, AICNI-AAKAT, AICNI-SAKGA, and ANCNI-AGKAT allele in 2008. Novel dhfr-108I mutation was never observed alone; it was always found with 51I mutation (AICII) and possessed very high IC50s for pyrimethamine (p < 0.001). In 2009, 64 out of 65 (98%) ETF in Kolkata and seven of 10 (70%) ETF in Purulia represented novel AICII haplotype. ETFs in Kolkata were strongly correlated with AICII-AGKAT, AICII-AGKAA, and AICII-SGKGT combination (pfdhfr-pfdhps) mutations (p < 0.001) whereas, in Purulia, ETFs were frequently found with AICII-SGKGT and AICII-AGKAA mutations (p < 0.05). Another two ETFs were reported with AICNI-AGKAA allele in Purulia. On the contrary, in Kolkata, LTFs were frequently found with AICII-AGKAT and AIRNI- AGEAA mutations whereas, in Purulia, LTFs were commonly observed with AIRNI- AGEAA, AICNI-AGKAA, and AICII-SGKGT haplotype (Table 2). Above mentioned haplotypes represented higher IC50s for pyrimethamine and sulfadoxine (p < 0.01) (Figures 4A, B). In multivariate logistic-regression after adjusting parasitemia, age, and dose of SP, an infection linked with pfdhfr AICII-pfdhps SGKGT, pfdhfr AICII-pfdhps AGKAA, and pfdhfr AICII- pfdhps AGKAT mutation was substantially more likely to have ETF (odds ratio, 94.4 P < 0.0001).
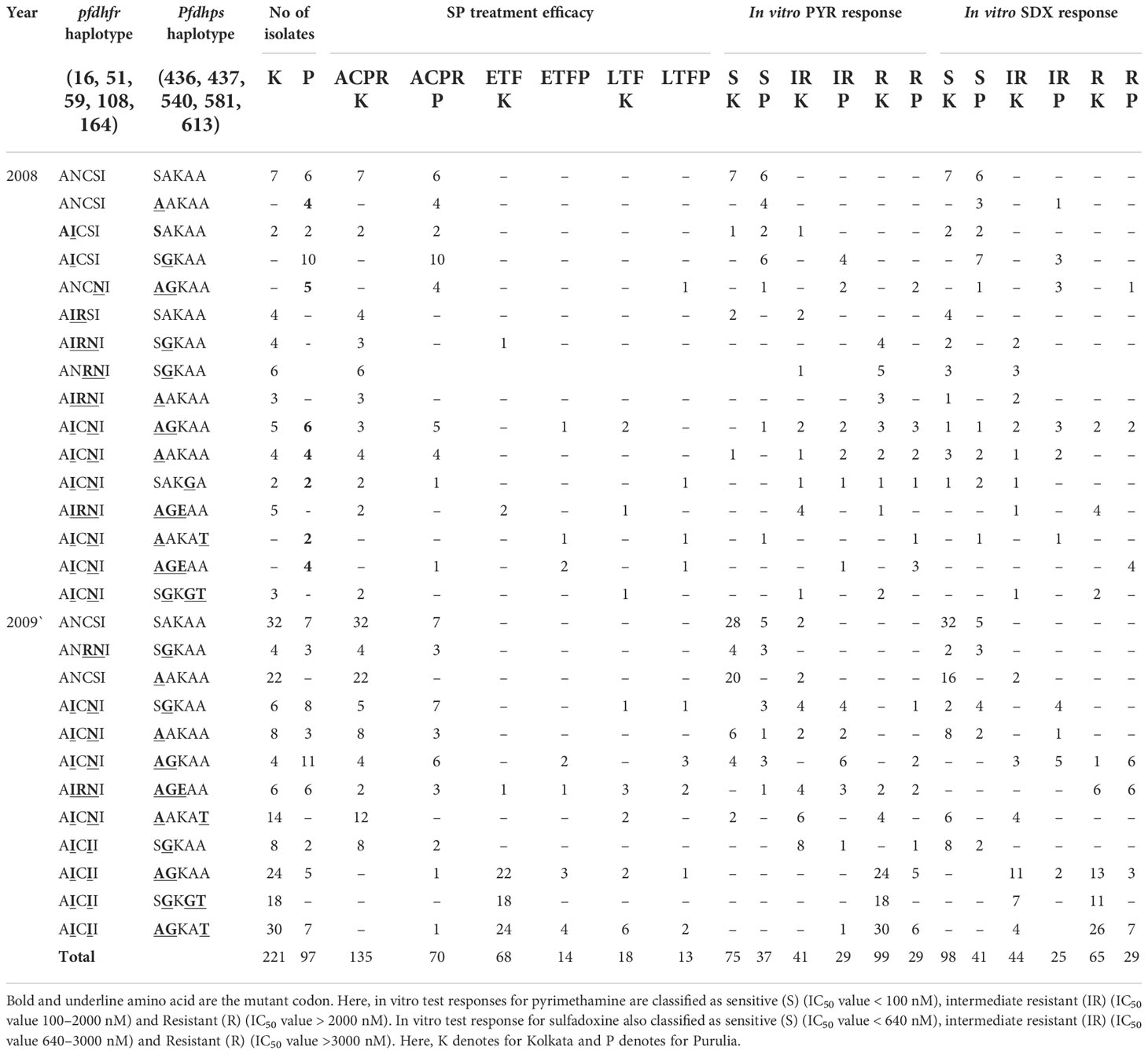
Table 2 Distribution of different pfdhfr and pfdhps haplotype in relation to in vivo SP efficacy and in vitro SP susceptibility in Kolkata and Purulia before new national drug policy.
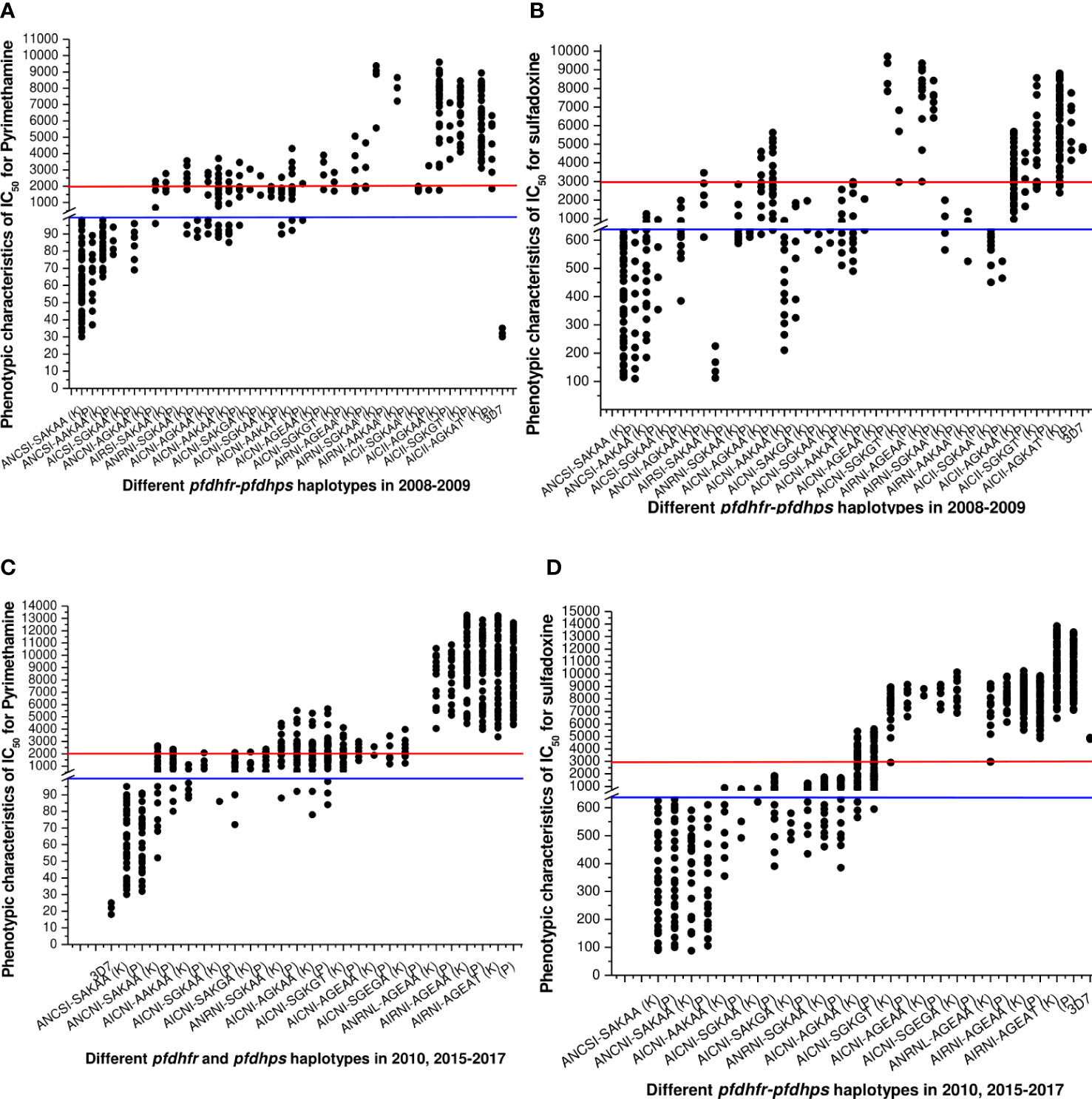
Figure 4 (A) Association of in vitro pyrimethamine sensitivity (expressed as the IC50 of pyrimethamine) with different pfdhfr and pfdhps haplotype combination before new national drug policy. Here, blue line (corresponding to 100 nM of PYR) was hypothetical showed the sensitive levels whereas red line (corresponding to 2000 nM of PYR) was hypothetical showed the resistance levels for in vitro pyrimethamine susceptibility. The isolates presenting pfdhfr-ANCSI, ANCNI, and ANRNI presented low to moderate IC50s for PYR and were not associated with in vitro PYR resistance (P = 0.81, Kruskal–Wallis test) in both study site. (B) Association of in vitro sulfadoxine sensitivity (expressed as the IC50 of sulfadoxine) with different pfdhfr and pfdhps haplotype combination before new national drug policy. Here, blue line (corresponding to 640 nM of SDX) was hypothetical showed the sensitive levels whereas red line (corresponding to 3000 nM of PYR) was hypothetical showed the resistance levels for in vitro pyrimethamine susceptibility. Low IC50s for SDX were mostly associated with wild dhps-SAKAA allele or single mutant AAKAA and SGKAA haplotype. ANCNI-AGKAA, AICSI-SGKAA, and ANCSI-AAKAA allele presented low to moderate in vitro IC50 values for SDX and thereby not associated with in vitro SDX resistance (P = 0.69, Kruskal–Wallis test). (C) Haplotype diversity in relation to in vitro pyrimethamine susceptibility in 2010, 2015–2017. The phenotype of in vitro pyrimethamine-resistance in Kolkata and Purulia was highly correlated with pfdhfr-S108N, N51I, and C5159 mutation (p < 0.01), but the pfdhfr haplotypes were quite different in both study sites. (D) Haplotype diversity in relation to in vitro sulfadoxine sensitivity in 2010, 2015–2017. In Kolkata in vitro sulfadoxine resistance was highly correlated with pfdhps-S436A, A437G, K540E, and A613T mutation (p < 0.01) but not with 581 mutations whereas in Purulia in vitro sulfadoxine resistance was associated with dhps-S436A, A437G, and K540E mutations but not with A613T mutation (p < 0.05).
Haplotype diversity in association with in vitro SP sensitivity after new drug policy
The frequency of wild ANCSI-SAKAA allele subsequently decreased after 2010. The molecular markers associated with in vitro SP-resistance were highly varied in two study sites. In Kolkata, in vitro PYR resistance was highly correlated to pfdhfr-AIRNI and ANRNL allele (p < 0.001), whereas intermediate PYR resistance was associated with AICNI haplotype (p < 0.001). In Purulia, in vitro PYR resistance was strongly associated with pfdhfr-ANRNI and AIRNI haplotype (p < 0.01), whereas intermediate PYR-resistance was correlated with ANCNI and AICNI allele (p < 0.001) (Figure 4C).
In Kolkata, isolates presenting pfdhps-AGEAT, SGKGT, and AGEAA haplotype possessed very high IC50s for sulfadoxine, proving resistant to sulfadoxine (p < 0.01). Furthermore, in vitro SDX resistance in Purulia was highly correlated with AGEAT and AGEAA haplotype (p < 0.01). In both study sites, single mutant pfdhps-AAKAA, SAKGA, SGKAA, and double mutant pfdhps-AGKAA alleles represented low to moderate IC50s for SDX and were associated with intermediate SDX resistant (p < 0.05) (Figure 4D).
Isolates representing AIRNI-AGEAA, AIRNI-AGEAT, and ANRNL-AGEAA combination mutations possessed very high IC50s for PYR and SDX and thereby proving highly resistant to PYR and SDX in vitro (p < 0.001). The isolates containing ANCSI-SAKAA, ANCNI-SAKAA, ANRNI-SGKAA, and AICNI-AGKAA haplotype represented moderate to high IC50s pyrimethamine and sulfadoxine (Table 3). In multivariate logistic-regression after adjusting parasitemia, age, and dose of SP, an infection linked with pfdhfr AIRNI-pfdhps AGEAA and pfdhfr AIRNI-pfdhps AGEAT mutation was substantially more likely to have an in vitro SP resistance (odds ratio, 89.5, P < 0.001; 93.8, P < 0.001, respectively).
Discussion
This study sheds light on the genetic variations and diversity of pfdhfr and pfdhps haplotypes as well as their relationship to phenotypic characteristics SP-treatment efficacy and in vitro SP sensitivity before, at-a-time, and 6–8 years after the implementation of new national drug policy (ACT), allowing us to obtain a thorough understanding of antifolate resistance in eastern India. Although Plasmodium’s genetic diversity was well known, it was still unknown exactly how this diversity affected the clinical manifestation. While there were significantly more LTFs in Purulia than in Kolkata, there were significantly more ETFs in Kolkata. One of the possible explanations for this could be variations in the genetic architecture of pfdhfr and pfdhps.
In Purulia, triple (double-dhfr + single-dhps) and quadruple (double-dhfr + double-dhps) mutations predominated in 2008–2009, but the parasite population in Kolkata exhibited a prevalence of quintuple (double-dhfr + triple-dhps) and quadruple (double-dhfr + double-dhps) mutant haplotypes. Changes in SP drug pressure over the population, which were previously seen with pfcrt mutations in relation to CQ resistance across the same population, may be the cause of the variation of mutation in two separate areas (Das et al., 2013b). For about two decades, SP was the most commonly given medication in the public sector. This usage may be to blame for the unique selective pressures on the parasite population in Kolkata and Purulia brought on by SP medication. Other factors might be attributed to the uncommon drug resistance situation in West Bengal due to its geographical position and the admixture of inhabitants with multiple tribal ethnic origins. Previous reports from North- East India and Andaman Nicobar Island described quintuple and quadruple mutant pfdhfr-pfdhps haplotypes with decreased SP-sensitivity (Das et al., 2010; Das et al., 2012b; Mohapatra et al., 2014). In order to stop the parasite from multiplying, substantially more medicine was needed due to increased mutations that reduced the drug’s ability to connect to the target protein (Yaqoob et al., 2018). We found that isolates with the pfdhfr-AIRNI, pfdhps-AGEAA, and pfdhps-SGKGT haplotypes were linked to pyrimethamine and sulfadoxine resistance in vitro whereas isolates with the pfdhfr-AICNI, pfdhps-AGKAA haplotypes had moderate to high IC50s for PYR and SDX.
Following super cyclone Aila in 2009, unique dhfr-AICII mutations were found in 48.8% of Kolkata isolates and 26.7% of Purulia isolates, and these alterations were strongly linked with both SP treatment failure and in vitro SP resistance. However, the dhfr-AICII haplotype was never discovered after 8 months, suggesting that despite its fitness cost, these changes presumably did not benefit the parasite population. Similar to this, the dhps-K540N mutation was discovered for the first time at Andaman-Nicobar Island in 2004, following the tsunami; however, it only persisted for 6 months due to a lack of fitness (Lumb et al., 2009).
ACT was introduced in late 2009 in place of SP’s single use in an effort to minimize the prevalence of drug-resistant P falciparum malaria in India. Previous research carried out in Malawi demonstrated that the population’s share of mutant haplotypes was lowered and that the emergence of sensitive parasites was elicited by the discontinuation of a resistant treatment for a period of 8 –10 years. As a result, after 12 years of its discontinuation, CQ-sensitive parasites returned in Malawi (Laufer et al., 2006). However, the proportion of in vitro PYR and SDX-resistance was subsequently raised and reached as high as above 85% after 8 years of withdrawal from single use of SP. This was due to steady increases in both study sites in the mutations in the pfdhfr and pfdhps genes. One of the possible causes of this high SP-pressure could be the extensive and careless usage of SP over the previous two decades (Das et al., 2013a). Additionally, with the exception of the north-east states, SP is one of the sole ingredients in ACT (artesunate-SP) in India. These factors may have recently influenced and increased the pressure of SP-drugs on this parasite population.
The vulnerable pfdhfr-164L mutation, which is frequently seen in the surrounding states of Assam and Arunachal Pradesh, has started to develop in both study locations since 2010, which is an alarming sign toward further development of resistant parasite (Sarmah et al., 2017). Despite the single use of SP, emergence and subsequent spread of sextuple (triple-dhfr+ triple-dhps) (dhfr-AIRNI +dhps-AGEAA, dhfr-ANRNL +dhps-AGEAA) and septuple (triple-dhfr +quadruple-dhps) (dhfr-AIRNI +dhps-AGEAT) mutant haplotypes with severe in vitro pyrimethamine and sulfadoxine-resistance confirmed the shift from lower to higher antifolate-resistance in eastern India. The ensuing increase in pfdhfr-pfdhps mutations may reduce the effectiveness of artesunate-SP and thus cause the emergence of partial artemisinin resistance, as has previously been observed in eastern India and southeast Asia (Sarmah et al., 2017; Das et al., 2018; Das et al., 2019). Because of the subsequent elevation of pfdhfr-pfdhps mutant haplotypes, it is believed that these mutations happened gradually and step by step (Miotto et al., 2015). The following increase in dhfr-AIRNI, dhps-AGEAA, and dhps-AGEAT haplotypes between 2010 and 2017 was the best evidence for this hypothesis (Per se dhfr-AICNI and dhfr-ANCNI were much prevalent in 2008–2009). The pfdhfr mutation most likely starts with codon-108, additive mutations at codons N51I, C59R, and I164L provide higher degrees of resistance (McCollum et al., 2006; Sandefur et al., 2007). On the other hand, pfdhps polymorphism likely starts at codon-A437G and progresses through mutations S436A, K540E, A581G, and A613T. However, as evidenced by the large number of isolates with the dhps-SGEGA, dhps-SGKGT, and dhfr-ANRNL haplotypes, the formation of dhfr and dhps haplotypes did not necessarily occur in an orderly, stepwise fashion.
If gene polymorphism always developed in an orderly, stepwise fashion, then the likelihood of receiving the dhps-613 mutation would only be present in a lineage that already had mutations at codons 437, 436, and 540 (Raman et al., 2010). Similarly, the dhfr-164L mutation would only be found in parasites that already had the 108, 51, and 59 mutations. Our results supported the idea that convergent genotypes could arise in an orderly, stepwise manner, through local parasite evolution, through a de novo mutation, or through sequential gene flow as a result of ongoing drug pressure, which eventually contributes to the onset of various pfdhfr-pfdhps haplotype variations (Gregson and Plowe, 2005; Karema et al., 2012; Miotto et al., 2015). We lacked certainty regarding genotypic recombination or the accumulation of subsequent point mutations for haplotypes.
The high parasite transmission intensity in India makes it impossible to rule out the possibility of recombination (Sharma, 2012). The sextuple and septuple pfdhfr-pfdhps haplotypes have become stable and common in eastern, north-eastern, and central India despite single use of SP in recent years (Lumb et al., 2009; Mohapatra et al., 2014; Das et al., 2021), showing that these haplotypes are likely benefiting the parasite population without incurring any fitness costs.
In conclusion, the subsequent elevation of pfdhfr-pfdhps combination mutation even after 8 years of discontinuing the use of single SP and its correlation with a progressive rise in in vitro SP resistance were alarming signs for the country’s efforts to manage and eradicate malaria. In this region of India, antimalarial medications containing SP should not be advised, and the use of alternative antimalarial drug combinations needs to be carefully considered.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/genbank/, KP256234, https://www.ncbi.nlm.nih.gov/genbank/, KP256235, https://www.ncbi.nlm.nih.gov/genbank/, KP256236, https://www.ncbi.nlm.nih.gov/genbank/, KP256237.
Ethics statement
The studies involving human participants were reviewed and approved by Vidyasagar University Human Ethical-Committee. Written informed consent to participate in this study was provided by the participants’ legal guardian/next of kin.
Author contributions
SD, AH, and SR contributed to the designing and conceptualization of the manuscript. AH supervised in vivo ASSP therapy. In vivo data analyses and interpretation were performed by SD, AH, AD, and MS. MS, AD, and AN analyzed the in vitro data. SD, ST, AD, AN, and MS contributed to genome analysis, and interpretation of the data. SD, AH, and SR had written the manuscript and gave input at all stages of the study. All authors have approved the final version of the manuscript.
Funding
There is no external research grant. This first author (SD) has received a personal fellowship of monthly remuneration from the Council of Scientific and Industrial Research, India (sanction no: 09/599(0055)2K1-EMR-I). SR received a personal research grant (PRG_SR2008-PRG_SR2014) from the Vidyasagar University.
Acknowledgments
The authors express gratitude to the Vidyasagar University, West Bengal, India for providing the facilities to execute the research work. We are indeed thankful to the Gautam Laboratories and Imaging, Kolkata, India (NABL accredited laboratory, ISO 15189:2007-M-0423) for their Continuous help. Under their supervision, in vivo tests were done in collaboration with Vidyasagar University, Midnapore. We are undoubtedly grateful to all the patients who participated in this work.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcimb.2022.865814/full#supplementary-material
References
Abdul-Ghani, R., Farag, H. F., Allam, A. F. (2013). Sulfadoxine-pyrimethamine resistance in plasmodium falciparum: A zoomed image at the molecular level within a geographic context. Acta Trop. 125, 163–190. doi: 10.1016/j.actatropica.2012.10.013
Abdul-Ghani, R., Farag, H. F., Allam, A. F. (2014). A better resolution for integrating methods for monitoring Plasmodium falciparum resistance to antimalarial drugs. Acta. Trop. 137, 44–57. doi: 10.1016/j.actatropica.2014.04.031
Ahmed, A., Bararia, D., Vinayak, S., Yameen, M., Biswas, S., Dev, V., et al. (2004). Plasmodium falciparum isolates in India exhibit a progressive increase in mutations associated with sulfadoxine pyrimethamine resistance. Antimicrobial. Agents Chemother. 48, 879–889. doi: 10.1128/AAC.48.3.879-889.2004
Badawi, M. M., Alla, A. A. F., Alla, S. S., Mohamed, W. A., Osman, D. A. N., Ali, S. A. A. A., et al. (2016). Immunoinformatics Predication and in silico Modeling of Epitope-Based Peptide Vaccine Against virulent Newcastle Disease Viruses. Am. J. Infect. Dis. Microbiol. 4 (3), 61–71. doi: 10.12691/ajidm-4-3-3
Basco, K. L., Ringwald, P. (2000). Molecular epidemiology of malaria in Yaounde, Cameroon VI. sequence variations in the plasmodium falciparum dihydrofolate reductase-thymidylate synthase gene and in vitro resistance to pyrimethamine and cycloguanil. Am. J. Trop. Med. Hyg. 62, 271–276. doi: 10.4269/ajtmh.2000.62.271
Bloland, P. B., Ringwald, P., Snow, R. W., Global Partnership to Roll Back Malaria. (2003). Assessment and monitoring of antimalarial drug efficacy for the treatment of uncomplicated falciparium malaria. World Health Organization. Available at: https://apps.who.int/iris/handle/10665/68453.
Das, S., Chakraborty, S. P., Hati, A., Roy, S. (2013a). Association between prevalence of chloroquine resistance and unusual mutation in pfmdr-i and pfcrt gene in India. Am. J. Trop. Med. Hygiene 88, 828–834. doi: 10.4269/ajtmh.11-0795
Das, S., Chakraborty, S. P., Hati, A., Roy, S. (2013b). Malaria treatment failure with novel mutation in the plasmodium falciparum dihydrofolate reductase (pfdhfr) gene in kolkata, West Bengal, India. Int. J. Antimicrob. Agents 41, 447–451. doi: 10.1016/j.ijantimicag.2013.01.005
Das, S., Chakraborty, S. P., Tripathy, S., Hati, A., Roy, S. (2012a). Novel quadruple mutations in dihydropteroate synthase genes of plasmodium falciparum in West Bengal, India. Trop. Med. Int. Health 17, 1329–1334. doi: 10.1111/j.1365-3156.2012.03071.x
Das, S., Chakraborty, S. P., Tripathy, S., Hati, A., Roy, S. (2012b). Association between prevalence of pyrimethamine resistance and double mutation in pfdhfr gene in West Bengal, India. Asian Pacific J. Trop. Dis. 2, 31–35. doi: 10.1016/S2222-1808(12)60008-5
Das, S., KarMahapatra, S., Tripathy, S., Chattopadhyay, S., Dash, S., Das, B., et al. (2014). Double mutation in the pfmdr1 gene is associated with emergence of chloroquine resistant plasmodium falciparum malaria in eastern India. Antimicrobial. Agents Chemother. 58, 5909–5915. doi: 10.1128/AAC.02762-14
Das, S., Kar, A., Manna, S., Mandal, S., Mandal, S., Das, S., et al. (2021). Artemisinin combination therapy fails even in the absence of plasmodium falciparum kelch13 gene polymorphism in central India. Sci. Rep. 11, 9946. doi: 10.1038/s41598-021-89295-0
Das, M. K., Lumb, V., Mittra, P., Singh, S. S., Dash, A. P., Sharma, Y. D. (2010). High chloroquine treatment failure rates and predominance of mutant genotypes associated with chloroquine and antifolate resistance among falciparum malaria patients from the island of car nicobar India. J. Antimicrob. Chemother. 65, 1258–1261. doi: 10.1093/jac/dkq090
Das, S., Manna, S., Saha, B., Hati, A. K., Roy, S. (2019). Novel pfkelch13 gene polymorphism associates with artemisinin resistance in eastern India. Clin. Infect. Dis. 69, 1144–1152. doi: 10.1093/cid/ciy1038
Das, S., Saha, B., Hati, A. K., Roy, S. (2018). Evidence of artemisinin resistant plasmodium falciparum malaria in Eastern India. N. Engl. J. Med. 379, 1962–1964. doi: 10.1056/NEJMc1713777
Foote, S. J., Cowman, A. F. (1994). The mode of action and the mechanism of resistance to antimalarial drugs. Acta Tropica 56, 157–171. doi: 10.1016/0001-706X(94)90061-2
Gregson, A., Plowe, C. V. (2005). Mechanisms of resistance of malaria parasites to antifolates. Pharmacol. Rev. 57, 117–145. doi: 10.1124/pr.57.1.4
Guidelines for Diagnosis and Treatment of Malaria in India (2009). National institute of malaria research; national vector borne disease control programme (New Delhi: Government of India) Available at: https://nvbdcp.gov.in/Doc/Guidelines_for_Diagnosis:_Treatment.pdf.
Guidelines for Diagnosis and Treatment of Malaria in India. (2014) (New Delhi, India: National Institute of Malaria Research). Available at: http://www.mrcindia.org/Diagnosis%20of%20Malaria%20pdf/Guidelines%202014.pdf.
Heinberg, A., Kirkman, L. (2015). The molecular basis of antifolate resistance in plasmodium falciparum: Looking beyond point mutations. Ann. NY Acad. Sci. 1342 (1), 10–18. doi: 10.1111/nyas.12662
Juma, D. W., Muiruri, P., Yuhas, K., John-Stewart, G., Ottichilo, R., Waitumbi, J., et al. (2019). The prevalence and antifolate drug resistance profiles of Plasmodium falciparum in study participants randomized to discontinue or continue cotrimoxazole prophylaxis. PloS Negl. Trop. Dis. 13 (3), e0007223. doi: 10.1371/journal.pntd.0007223
Karema, C., Imwong, M., Fanello, C. I., Stepniewska, K., Uwimana, A., Dondorp, A., et al. (2012). Molecular correlates of high-level antifolate resistance in Rwandan children with Plasmodium falciparum malaria. Antimicrob. Agent Chemother. 54, 477–483. doi: 10.1128/AAC.00498-09
Kelley, L. A., Mezulis, S., Yates, C. M., Wass, M. N., Sternberg, M. J. (2015). The Phyre2 web portal for protein modeling, prediction and analysis. Nat. Protoc. 10 (6), 845–858. doi: 10.1038/nprot.2015.053
Laufer, M. K., Thesing, P. C., Eddington, N. D., Masonga, R., Dzinjalamala, F. K., Takala, S. L., et al. (2006). Return of chloroquine antimalarial efficacy in Malawi. N. Engl. J. Med. 355, 19959–19960. doi: 10.1056/NEJMoa062032
Lumb, V., Das, M. K., Mittra, P., Ahmed, A., Kumar, M., Kaur, P., et al. (2009). Emergence of an unusual sulfadoxine pyrimethamine resistance pattern and a novel K540N mutation in dihydropteroate-synthetase in plasmodium falciparum isolates obtained from car nicobar island, India, after the 2004 tsunami. J. Infect. Dis. 199, 1064–1073. doi: 10.1086/597206
Madkhali, A. M., Al-Mekhlafi, H. M., Atroosh, W. M., Ghzwani, A. H., Zain, K. A., Abdulhaq, A. A., et al. (2020). Increased prevalence of pfdhfr and pfdhps mutations associated with sulfadoxine-pyrimethamine resistance in plasmodium falciparum isolates from jazan region, southwestern Saudi Arabia: Important implications for malaria treatment policy. Malar. J. 19, 446. doi: 10.1186/s12936-020-03524-x
Malaria Fact Sheet (2013) Pennsylvania Department of health. Available at: https://www.health.pa.gov/topics/Documents/Diseases%20and%20Conditions/Malaria.pdf.
McCollum, A. M., Poe, A. C., Hamel, M., Huber, C., Zhou, Z., Shi, Y. P., et al. (2006). Antifolate resistance in Plasmodium falciparum: Multiple origins and identification of novel dhfr alleles. J. Infect. Dis. 194, 189–197. doi: 10.1086/504687
Miotto, O., Amato, R., Ashley, E. A., MacInnis, B., Almagro-Garcia, J., Amaratunga, C., et al. (2015). Genetic architecture of artemisinin-resistant plasmodium falciparum. Nat. Genet. 47, 226–234. doi: 10.1038/ng.3189
Mohapatra, P. K., Sarma, D. K., Prakash, A., Bora, K., Ahmed Md, A., Sarma, B., et al. (2014). Molecular evidence of increased resistance to anti-folate drugs in plasmodium falciparum in north-East India: A signal for potential failure of artemisinin plus sulfadoxine-pyrimethamine combination therapy. Plose One 9, e105562. doi: 10.1371/journal.pone.0105562
Raman, J., Little, F., Roper, C., Kleinschmidt, I., Cassam, Y., Maharaj, R., et al. (2010). Five years of Large-scale dhfr and dhps mutation surveillance following the phased implementation of artesunate plus sulfadoxine-pyrimethamine in Maputo province, southern Mozambique. Am. J. Trop. Med. Hyg. 82, 788–794. doi: 10.4269/ajtmh.2010.09-0401
Sandefur, C. I., Wooden, J. M., Quaye, I. K., Sirawaraporn, W., Sibley, C. H. (2007). Pyrimethamine-resistant dihydrofolate reductase enzymes of Plasmodium falciparum are not enzymatically compromised in vitro. Mol. Bioche Parasitol. 154, 1–5. doi: 10.1016/j.molbiopara.2007.03.009
Sarmah, N. P., Sarma, K., Bhattacharyya, D. R., Sultan, A. A., Bansal, D., Singh, N., et al. (2017). Antifolate drug resistance: Novel mutations and haplotype distribution in dhps and dhfr from northeast India. J. Biosci. 42, 531–535. doi: 10.1007/s12038-017-9706-5
Sharma, Y. D. (2012). Molecular surveillance of drug-resistant malaria in India. Curr. Sci. 102, 696–703.
WHO (2021) World malaria report. Available at: https://www.who.int/teams/global-malaria-programme/reports/world-malaria-report-2021.
Yaqoob, A., Khattak, A. A., Nadeem, M. F., Fatima, H., Mbambo, G., Ouattara, A., et al. (2018). Prevalence of molecular markers of sulfadoxine-pyrimethamine and artemisinin resistance in plasmodium falciparum from Pakistan. Malar. J. 17, 471. doi: 10.1186/s12936-018-2620-y
Keywords: Plasmodium falciparum, antifolate resistance, pfdhfr mutation, pfdhps mutation, in vitro pyrimethamine resistance, in vitro sulfadoxine resistance
Citation: Das S, Tripathy S, Das A, Sharma MK, Nag A, Hati AK and Roy S (2022) Genomic characterization of Plasmodium falciparum genes associated with anti-folate drug resistance and treatment outcomes in eastern India: A molecular surveillance study from 2008 to 2017. Front. Cell. Infect. Microbiol. 12:865814. doi: 10.3389/fcimb.2022.865814
Received: 30 January 2022; Accepted: 15 November 2022;
Published: 13 December 2022.
Edited by:
Ashley Vaughan, Seattle Children’s Research Institute, United StatesReviewed by:
Siraj Ahmed Khan, Regional Medical Research Centre (ICMR), IndiaBenedicta Ayiedu Mensah, University of Ghana, Ghana
Copyright © 2022 Das, Tripathy, Das, Sharma, Nag, Hati and Roy. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Somenath Roy, cm95c29tZW5hdGhAb3V0bG9vay5jb20=; c3JveV92dUBvdXRsb29rLmNvbQ==
 Sabyasachi Das
Sabyasachi Das Satyajit Tripathy
Satyajit Tripathy Ankita Das
Ankita Das Meenakshi Kumari Sharma2
Meenakshi Kumari Sharma2 Somenath Roy
Somenath Roy