- Institute of Microbiology, School of Ecology and Nature Conservation, Beijing Forestry University, Beijing, China
Leptoporus is a rare and remarkable genus, mainly occurring in coniferous forests in the Northern Hemisphere. Recent phylogenetic studies showed that Leptoporus belongs to Irpicaceae in the phlebioid clade. It is worth noting that most species in the phlebioid clade can cause white-rot decay, except for the Leptoporus species, which can cause a brown-rot decay. In this study, we performed phylogenetic and taxonomic studies of Leptoporus and related genera. Molecular phylogenetic analyses were conducted based on sequences from multiple loci including the internal transcribed spacer (ITS) regions, the large subunit of nuclear ribosomal RNA gene (nLSU), the largest subunit of RNA polymerase II gene (RPB1), the second largest subunit of RNA polymerase II gene (RPB2), and the translation elongation factor 1-α gene (TEF1). Combined with morphological characteristics, a new species, Leptoporus submollis sp. nov., is discovered and illustrated from Southwest China.
Introduction
Irpicaceae Spirin & Zmitr. was proposed by Spirin (2003) with Irpex Fr. as type genus. The great majority of the species in Irpicaceae, even in the phlebioid clade, can cause a white rot, except for Leptoporus mollis (Pers.) Quél., which causes a brown rot (Gilbertson and Ryvarden, 1986; Chen et al., 2021). This makes Leptoporus a remarkable genus, which has attracted many mycologists’ attention.
Leptoporus Quél. was established by Quél (1886), with L. mollis as type species, which was described as causing a brown rot on dead conifers and mainly distributed in the Northern Hemisphere (North America, Europe, and Asia) (Gilbertson and Ryvarden, 1986; Ryvarden and Gilbertson, 1993; Núñez and Ryvarden, 2001; Yu et al., 2004; Volobuev, 2019). In North America, L. mollis has been reported in boreal coniferous forests (Gilbertson and Ryvarden, 1986). In Europe, this species was considered as a rare species and needs to be protected (Ryvarden and Gilbertson, 1993; Volobuev, 2019). In Asia, this species has been reported from China and Japan and was also considered as a rare species (Núñez and Ryvarden, 2001; Yu et al., 2004). Previously, Leptoporus was placed in Polyporaceae Fr. ex Corda (Yu et al., 2004; Kirk et al., 2008). Subsequently, some phylogenetic studies showed that Leptoporus was embedded in the phlebioid clade (Binder et al., 2005; Lindner and Banik, 2008; Binder et al., 2013). In recent years, Leptoporus has been proven to belong to Irpicaceae and was closely related to Ceriporia Donk (Justo et al., 2017; Chen et al., 2021). Currently, although the databases Index Fungorum (http://www.indexfungorum.org/) and MycoBank (https://www.mycobank.org/) still record some Leptoporus species, only one species, L. mollis, is accepted in recent studies (Lindner and Banik, 2008; He et al., 2019; Chen et al., 2020; Chen et al., 2021).
During investigations on the diversity of polypores in the Hengduan Mountains of Southwest China, one undescribed species of Leptoporus was discovered. To confirm the affinity of the undescribed species corresponding to Leptoporus, phylogenetic analyses of Irpicaceae were carried out based on the combined sequences datasets of ITS+nLSU and ITS+nLSU+RPB1+RPB2+TEF1.
Materials and methods
Morphological studies
The examined specimens were mostly deposited at the herbarium of the Institute of Microbiology, Beijing Forestry University, China (BJFC), and some specimens were deposited at the Institute of Applied Ecology, Chinese Academy of Sciences, China (IFP). Macromorphological descriptions were based on the field notes and measurements of herbarium specimens. Special color terms followed Petersen (1996). Micromorphological data were obtained from the dried specimens and observed under a light microscope following Ji et al. (2022) and Sun et al. (2022). Sections were studied at a magnification up to ×1,000 using a Nikon Eclipse 80i microscope and phase-contrast illumination (Nikon, Tokyo, Japan). Drawings were made with the aid of a drawing tube. Microscopic features, measurements, and drawings were made from slide preparations stained with Cotton Blue and Melzer’s reagent. Spores were measured from sections cut from the tubes. To present variations in the size of basidiospores, 5% of measurements were excluded from each end of the range and extreme values are given in parentheses.
In the text, the following abbreviations were used: IKI, Melzer’s reagent; IKI–, neither amyloid nor dextrinoid; KOH, 5% potassium hydroxide; CB, Cotton Blue; CB–, acyanophilous; L, mean spore length (arithmetic average of all spores); W, mean spore width (arithmetic average of all spores); Q, variation in the L/W ratios between the specimens studied; n (a/b), number of spores (a) measured from given number (b) of specimens.
Molecular studies and phylogenetic analysis
A cetyl trimethylammonium bromide (CTAB) rapid plant genome extraction kit-DN14 (Aidlab Biotechnologies Co., Ltd., Beijing, China) was used to extract total genomic DNA from dried specimens, and the polymerase chain reaction (PCR) was performed according to the manufacturer’s instructions with some modifications as described by Cui et al. (2019) and Shen et al. (2019). The internal transcribed spacer (ITS) regions were amplified with primer pairs ITS5 and ITS4 (White et al., 1990). The large subunit of nuclear ribosomal RNA gene (nLSU) regions were amplified with primer pairs LR0R and LR7 (http://www.biology.duke.edu/fungi/mycolab/primers.htm). RPB1 was amplified with primer pairs RPB1-Af and RPB1-Cr (Matheny et al., 2002). RPB2 was amplified with primer pairs fRPB2-f5F and bRPB2-7.1R (Matheny, 2005). Part of TEF1 was amplified with primer pairs EF1-983F and EF1-1567R (Rehner, 2001).
The PCR cycling schedule for ITS and TEF1 included an initial denaturation at 95°C for 3 min, followed by 35 cycles at 94°C for 40 s, 54°C for ITS, 55°C for TEF1 for 45 s, 72°C for 1 min, and a final extension at 72°C for 10 min. The PCR cycling schedule for nLSU included an initial denaturation at 94°C for 1 min, followed by 35 cycles at 94°C for 30 s, 51°C for 1 min, 72°C for 1.5 min, and a final extension at 72°C for 10 min. The PCR cycling schedule for RPB1 and RPB2 included an initial denaturation at 94°C for 2 min, followed by 10 cycles at 94°C for 40 s, 60°C for 40 s, and 72°C for 2 min, then followed by 37 cycles at 94°C for 45 s, 55°C–57°C for 1.5 min, 72°C for 2 min, and a final extension of 72°C for 10 min. The PCR products were purified and sequenced at Beijing Genomics Institute (BGI), China, with the same primers. All newly generated sequences were deposited at GenBank (Table 1).
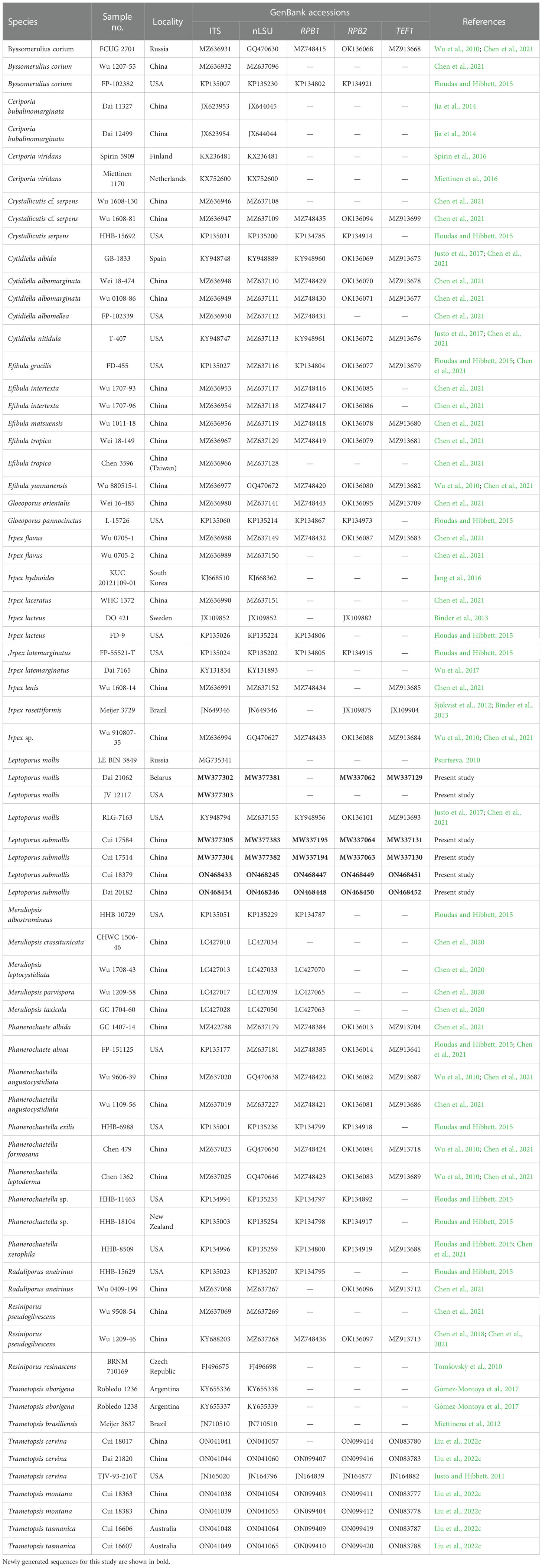
Table 1 A list of species, specimens, and GenBank accession number of sequences used for phylogenetic analyses in this study.
Additional sequences were downloaded from GenBank (Table 1). All sequences of ITS, nLSU, RPB1, RPB2, and TEF1 were respectively aligned in MAFFT 7 (Katoh and Standley, 2013; http://mafft.cbrc.jp/alignment/server/) and manually adjusted in BioEdit (Hall, 1999). Alignments were spliced in Mesquite (Maddison and Maddison, 2017). The missing sequences and ambiguous nucleotides were both coded as “N.”
Most parsimonious phylogenies were inferred from the combined 2-gene dataset (ITS+nLSU) and 5-gene dataset (ITS+nLSU+RPB1+RPB2+TEF1), and their congruences were evaluated with the incongruence length difference (ILD) test (Farris et al., 1994) implemented in PAUP* 4.0b10 (Swofford, 2002) under heuristic search and 1,000 homogeneity replicates. Phylogenetic analyses followed Sun et al. (2020). In phylogenetic reconstruction, the sequences of Phanerochaete albida Sheng H. Wu and P. alnea (Fr.) P. Karst. obtained from GenBank were used as outgroups to root trees following Liu et al. (2022c). Maximum parsimony (MP) analysis was applied to the combined multiple gene datasets, and the tree construction procedure was performed in PAUP* version 4.0b10. All characters were equally weighted, and gaps were treated as missing data. Trees were inferred using the heuristic search option with TBR branch swapping and 1,000 random sequence additions. Max-trees were set to 5,000, branches of zero length were collapsed, and all parsimonious trees were saved. Clade robustness was assessed using a bootstrap (BT) analysis with 1,000 replicates (Felsenstein, 1985). Descriptive tree statistics tree length (TL), consistency index (CI), retention index (RI), rescaled consistency index (RC), and homoplasy index (HI) were calculated for each most parsimonious tree (MPT) generated. RAxmL v.7.2.8 was used to construct a maximum likelihood (ML) tree with a GTR+G+I model of site substitution including estimation of gamma-distributed rate heterogeneity and a proportion of invariant sites (Stamatakis, 2006). The branch support was evaluated with a bootstrapping method of 1,000 replicates (Hillis and Bull, 1993).
MrModeltest 2.3 (Posada and Crandall, 1998; Nylander, 2004) was used to determine the best-fit evolution model for the combined multigene dataset for Bayesian inference (BI). BI was calculated with MrBayes 3.1.2 with a general time-reversible (GTR) model of DNA substitution and a gamma distribution rate variation across sites (Ronquist and Huelsenbeck, 2003). Four Markov chains were run for two runs from random starting trees for 2.5 million generations (ITS+nLSU) and for 4 million generations (ITS+nLSU+RPB1+RPB2+TEF1), and trees were sampled every 100 generations. The first one-fourth generations were discarded as burn-in. A majority rule consensus tree of all remaining trees was calculated. Branches that received BT support for MP, ML, and Bayesian posterior probabilities (BPP) greater than or equal to 75% (MP and ML) and 0.95 (BPP) were considered as significantly supported. Trees were viewed in FigTree v1.4.4 (http://tree.bio.ed.ac.uk/software/figtree/). Sequence alignment was deposited at TreeBase (submission ID: 29921; http://www.treebase.org).
Results
Phylogeny
The combined 2-gene (ITS+nLSU) sequences dataset had an aligned length of 1,556 characters, including gaps (655 characters for ITS, 901 characters for nLSU), of which 998 characters were constant, 78 were variable and parsimony-uninformative, and 480 were parsimony-informative. MP analysis yielded 14 equally parsimonious trees (TL = 2,272, CI = 0.386, RI = 0.760, RC = 0.294, HI = 0.614). The best model for the concatenate sequence dataset estimated and applied in the BI was GTR+I+G with equal frequency of nucleotides. ML analysis resulted in a similar topology as MP and Bayesian analyses, and only the ML topology is shown in Figure 1.
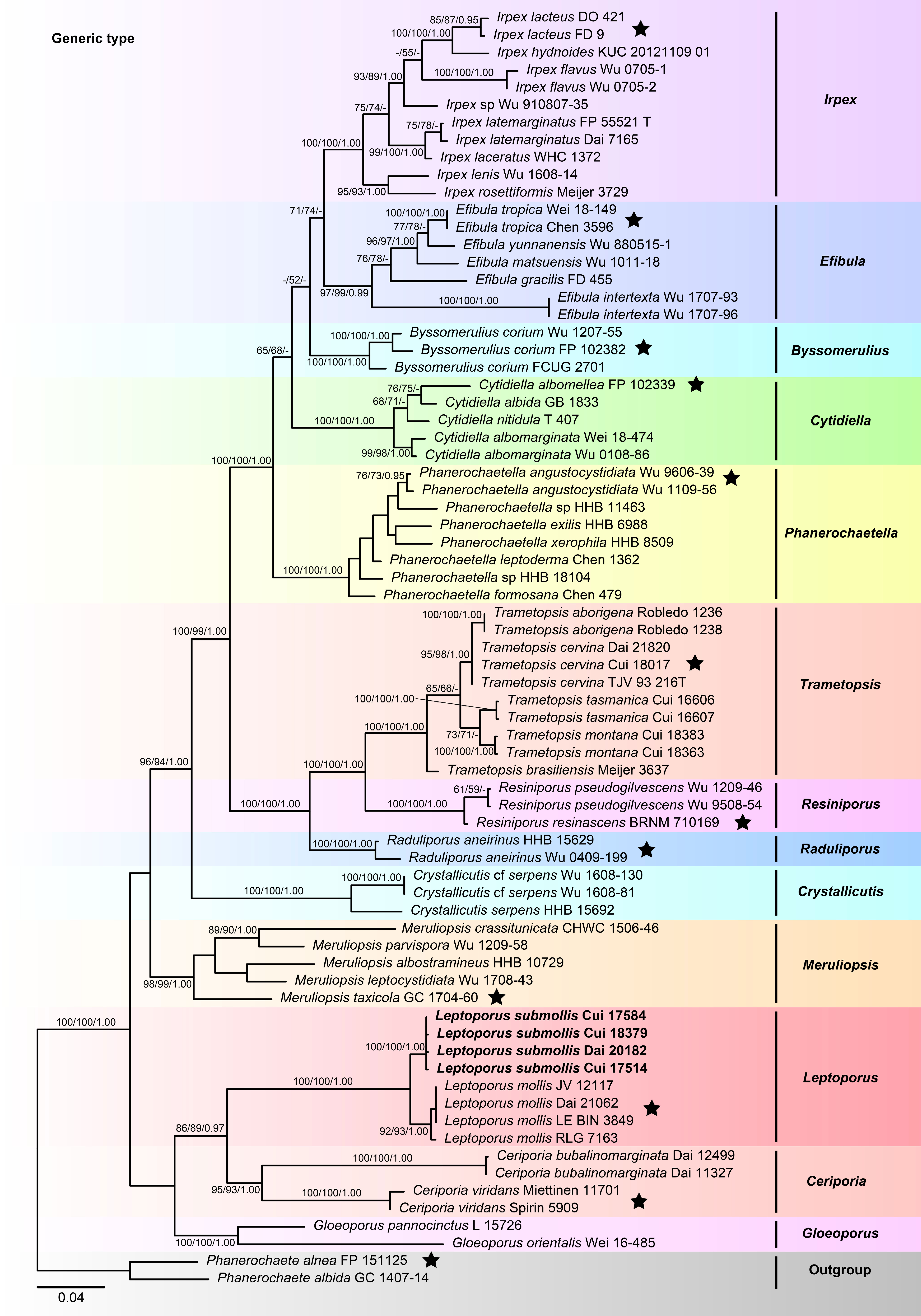
Figure 1 Maximum likelihood tree illustrating the phylogeny of Irpicaceae based on the combined sequence dataset of ITS+nLSU. Branches are labeled with maximum likelihood bootstrap higher than 50%, parsimony bootstrap proportions higher than 50%, and Bayesian posterior probabilities more than 0.90, respectively. Bold names = New species.
The combined 5-gene (ITS+nLSU+RPB1+RPB2+TEF1) sequences dataset had an aligned length of 4,234 characters, including gaps (655 characters for ITS, 901 characters for nLSU, 1,192 characters for RPB1, 1,019 characters for RPB2, 467 characters for TEF1), of which 2,327 characters were constant, 207 were variable and parsimony-uninformative, and 1,700 were parsimony-informative. MP analysis yielded 33 equally parsimonious trees (TL = 10,223, CI = 0.332, RI = 0.665, RC = 0.221, HI = 0.668). The best model for the concatenate sequence dataset estimated and applied in the BI was GTR+I+G with equal frequency of nucleotides. ML analysis resulted in a similar topology as MP and Bayesian analyses, and only the ML topology is shown in Figure 2.
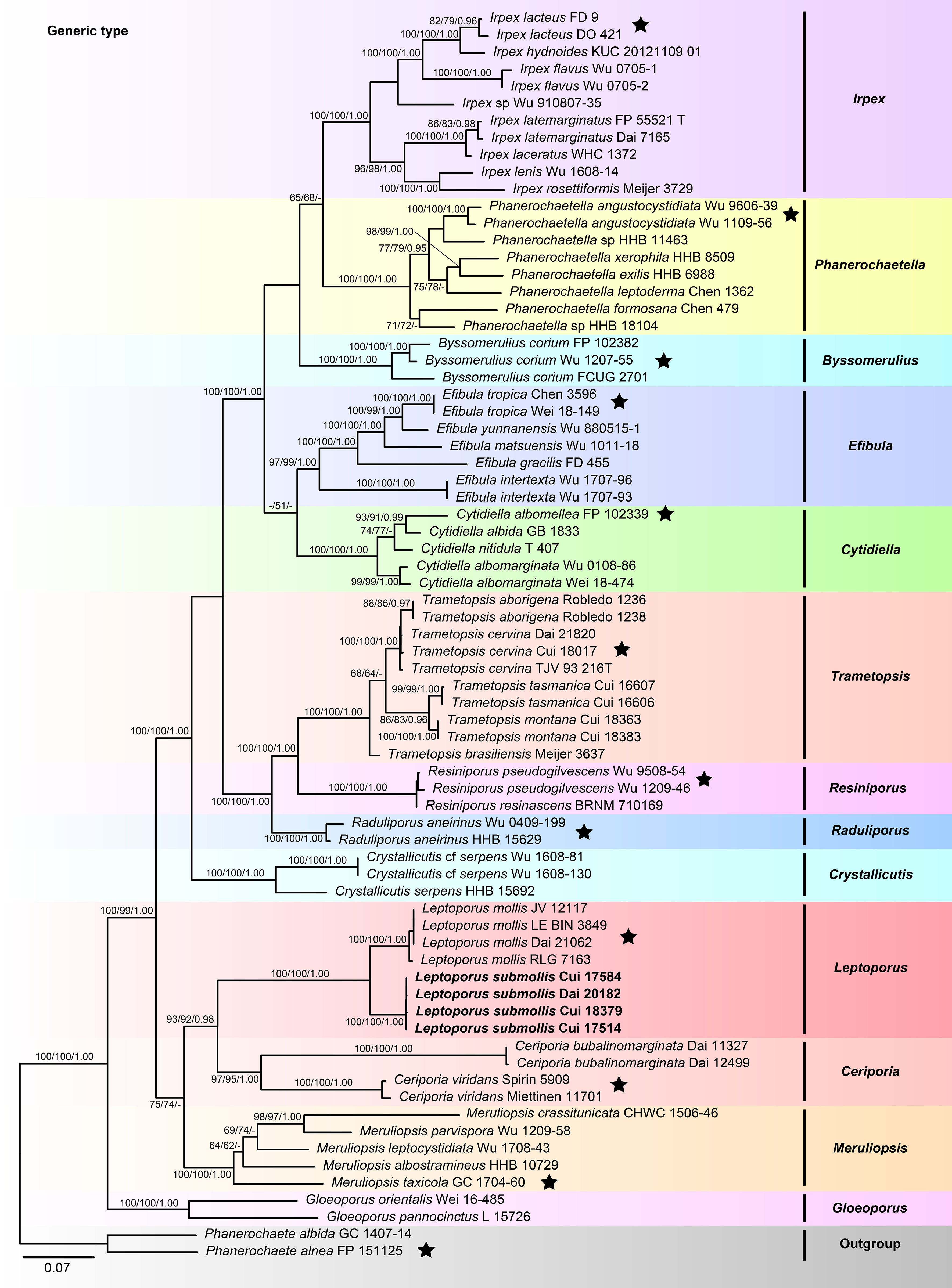
Figure 2 Maximum likelihood tree illustrating the phylogeny of Irpicaceae based on the combined sequence dataset of ITS+nLSU+RPB1+RPB2+TEF1. Branches are labeled with maximum likelihood bootstrap higher than 50%, parsimony bootstrap proportions higher than 50%, and Bayesian posterior probabilities more than 0.90, respectively. Bold names = New species.
The combined datasets of ITS+nLSU and ITS+nLSU+RPB1+RPB2+TEF1 contained sequences obtained from 74 fungal samples representing 45 taxa within the phlebioid clade (Figures 1, 2). The phylogenetic trees (Figures 1, 2) generated by MP, ML, and Bayesian analyses show that the new species Leptoporus submollis grouped with L. mollis with strong support (100% MP, 100% ML, 1.00 BPP; Figures 1, 2) within Irpicaceae.
Taxonomy
Leptoporus Quél., Enchiridion Fungorum in Europa media et praesertim in Gallia Vigentium: 175, 1886.
Type species: L. mollis (Pers.) Quél.
MycoBank: MB 17951
Basidiomata annual, effused-reflexed to pileate or resupinate, soft corky to corky or fragile. Pileal surface pale vinaceous to milky coffee, azonate, glabrous to tomentose. Pore surface flesh pink to snuff brown; pores circular to angular. Context pinkish buff to buff, corky. Tubes concolorous with pore surface, corky. Hyphal system monomitic; generative hyphae simple-septate, IKI–, CB–. Cystidia absent, cystidioles present. Basidiospores allantoid, cylindrical to oblong-ellipsoid, hyaline, thin-walled, smooth, IKI–, CB–. Causing a brown rot.
Specimen examined: L. mollis. BELARUS. Brestskaya Voblasts, Belavezhskaya Pushcha National Park, on stump of Picea sp., 19 October 2019, Dai 21062 (BJFC 032721). CHINA. Heilongjiang, Yichun, Fenglin Nature Reserve, on fallen trunk of Picea sp., 5 August 2000, Penttilä 13266 (IFP 014914). FINLAND. Koillissmaa, Oulanka National Park, on rotten wood of Picea sp., 17 September 1997, Dai 2674 (IFP 014915).
Leptoporus submollis B.K. Cui & Shun Liu, sp. nov. (Figures 3, 4)
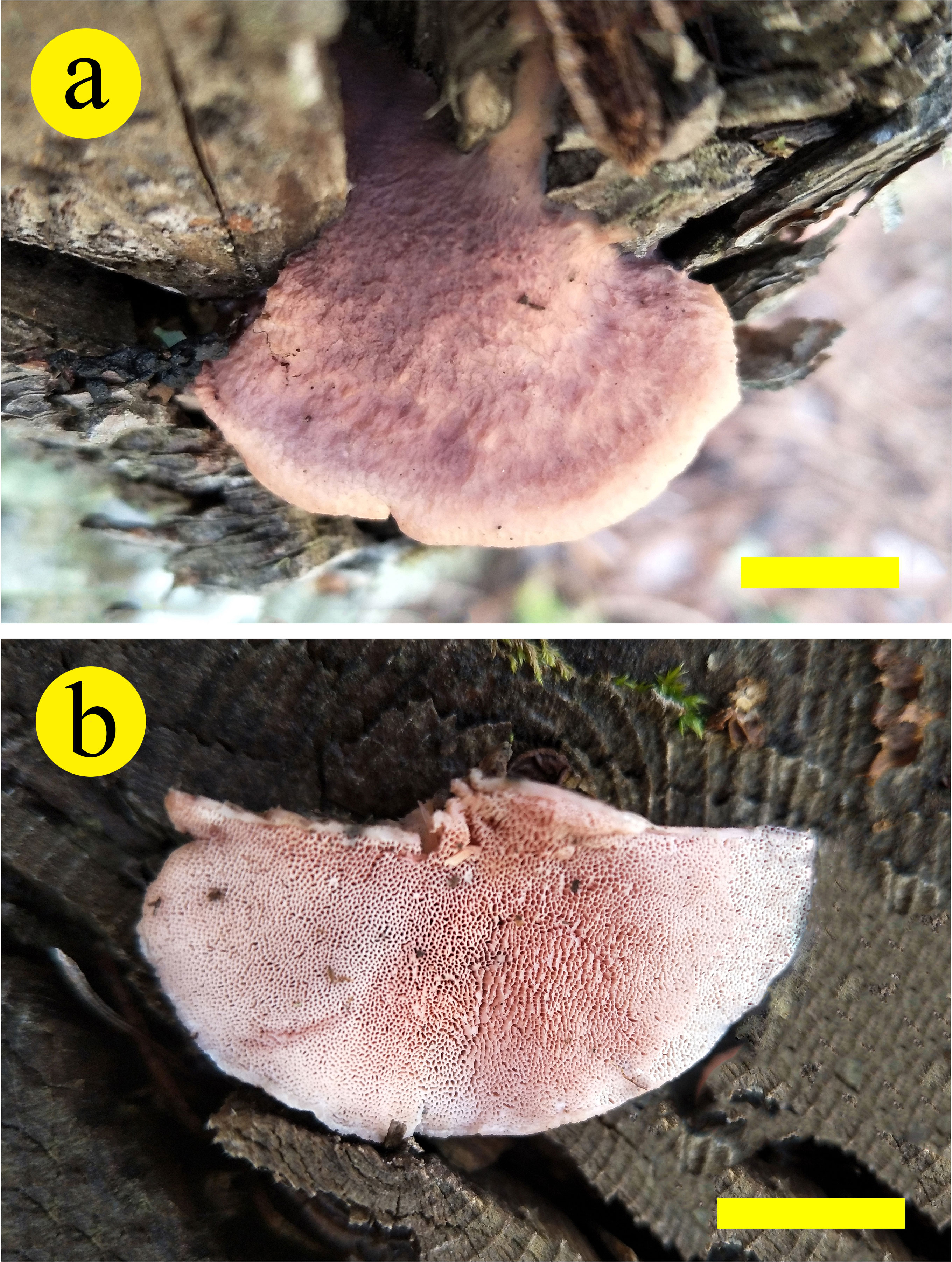
Figure 3 Basidiocarps of Leptoporus submollis (Cui 17514) (scale bar = 1.5 cm). Photo by Bao-Kai Cui.
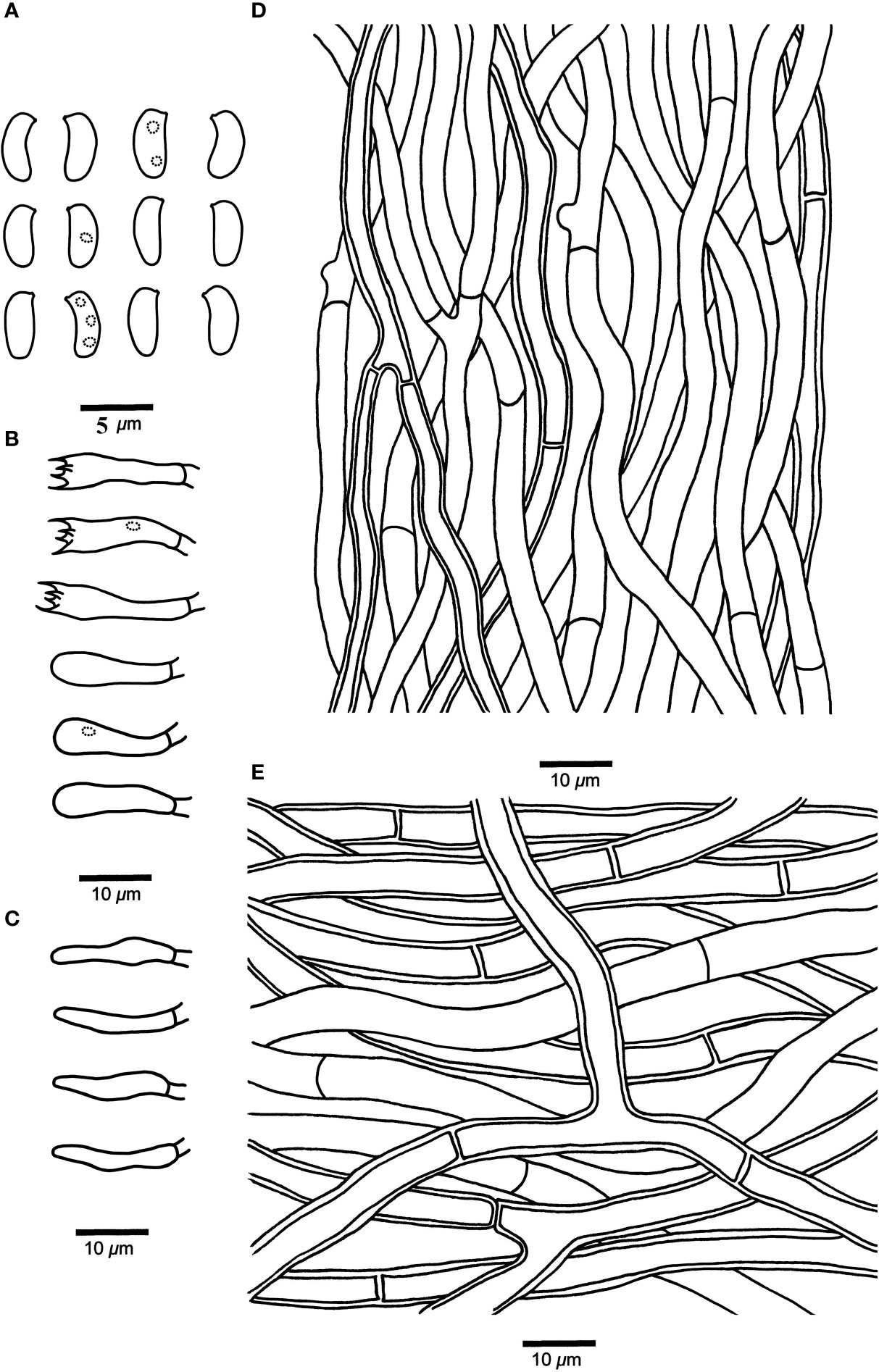
Figure 4 Microscopic structures of Leptoporus submollis (drawn from the holotype). (A) Basidiospores. (B) Basidia and basidioles. (C) Cystidioles. (D) Hyphae from trama. (E) Hyphae from context. Scale bar: A = 5 µm; B–E = 10 µm. Drawings by Shun Liu.
MycoBank: MB 840366
Diagnosis. L. submollis is characterized by its pale vinaceous to pale reddish pileal surface when fresh, becoming grayish brown to milky coffee upon drying, flesh pink to brownish vinaceous pore surface when fresh, becoming isabelline to snuff brown when dry, circular to angular pores (4–6 per mm) and cylindrical to oblong-ellipsoid basidiospores (4–4.8 μm × 1.8–2.3 μm).
Type. CHINA. Sichuan Province, Yanyuan County, on stump of Pinus yunnanensis, elevation 3,100 m, 15 August 2019, Cui 17514 (holotype, BJFC 034373).
Etymology. “submollis” (Lat.) refers to the new species is similar to L. mollis in morphology.
Fruiting body. Basidiomata annual, effused-reflexed to pileate, solitary, soft corky, without odor or taste when fresh, corky and light in weight when dry. Pileus semicircular or irregular, projecting up to 2.5 cm, 5 cm wide, and 2 cm thick at base. Pileal surface pale vinaceous to pale reddish when fresh, becoming grayish brown to milky coffee upon drying, glabrous. Pore surface flesh pink to brownish vinaceous when fresh, becoming isabelline to snuff brown when dry; sterile margin narrow to almost lacking; pores circular to angular, 4–6 per mm; dissepiments slightly thick to thick, entire to lacerate. Context pinkish buff to buff, corky, up to 10 mm thick. Tubes concolorous with pore surface, corky, up to 6 mm long.
Hyphal structure. Hyphal system monomitic; generative hyphae simple-septate, IKI–, CB–; tissues unchanged in KOH.
Context. Generative hyphae hyaline, thin- to slightly thick-walled, occasionally branched, interwoven, 3.5–8.5 μm in diameter.
Tubes. Generative hyphae hyaline, thin- to slightly thick-walled, occasionally branched, 2–5 μm in diameter. Cystidia absent; fusoid cystidioles present, hyaline, thin-walled, 11–17 μm × 2–4 μm. Basidia clavate, bearing four sterigmata and a basal simple-septum, 12–20 μm × 3–5 μm; basidioles dominant, in shape similar to basidia, but smaller.
Spores. Basidiospores cylindrical to oblong-ellipsoid, hyaline, thin-walled, smooth, occasionally with 1–3 small oily inclusions, IKI–, CB–, 4–4.8 μm × 1.8–2.3 μm, L = 4.46 μm, W = 2.06 μm, Q = 2.02–2.13 (n = 90/3).
Type of rot. Brown rot.
Additional specimens examined. CHINA. Sichuan Province, Muli County, on stump of Pinus yunnanensis, elevation 3,050 m, 16 August 2019, Cui 17584 (paratype, BJFC 034443). Xizang Autonomous Region (Tibet), Linzhi, on living gymnosperm tree, elevation 3,100 m, 18 July 2019, Dai 20182 (paratype, BJFC 031853); Mangkang County, on stump of Abies sp., elevation 3,900 m, 8 September 2020, Cui 18379 (paratype, BJFC 035238).
Discussion
Decay mode is one of the most stable characteristics in Polyporales and has been used as the basis for distinguishing genera (Gilbertson and Ryvarden, 1986; Ryvarden, 1991). Among the Polyporales, nearly all of the brown-rot fungi species are clustered in the antrodia clade, which have been widely studied in recent years (Ortiz-Santana et al., 2013; Han et al., 2014; Shen et al., 2014; Song et al., 2014; Han et al., 2015; Han and Cui, 2015; Shen et al., 2015; Chen et al., 2015; Han et al., 2016; Chen and Cui, 2016; Song and Cui, 2017; Song et al., 2018; Shen et al., 2019; Liu et al., 2019; Liu et al., 2021a; Liu et al., 2021b; Liu et al., 2022a; Liu et al., 2022b; Liu et al., 2022d). In the phlebioid clade, most species can produce white-rot decay, with one notable exception, L. mollis, which can produce brown-rot decay (Binder et al., 2013; Chen et al., 2021). This result suggests that brown-rot fungi may have evolved more than once in Polyporales (Floudas and Hibbett, 2015).
In the present study, the phylogenetic analyses of Irpicaceae are inferred from the combined datasets of ITS+nLSU sequences (Figure 1) and ITS+nLSU+RPB1+RPB2+TEF1 sequences (Figure 2). The results show that the genera of Ceriporia and Leptoporus grouped together and formed a highly supported lineage (Figures 1, 2). Morphologically, Ceriporia spp. differs by possessing resupinate basidiomata, absence of cystidioles, and causing a white decay of wood (Chen et al., 2020; Chen et al., 2022). Therefore, Ceriporia and Leptoporus are treated as independent genera in Irpicaceae (Chen et al., 2020; Chen et al., 2021).
In our current phylogenetic analyses, L. mollis and L. submollis grouped together and formed a well-supported lineage (Figures 1, 2). Morphologically, L. mollis may be confused with L. submollis by possessing annual growth habit, soft to corky basidiomata when fresh, and monomitic hyphal system with simple-septate generative hyphae, while L. mollis differs in having larger pores (2–4 per mm), narrower contextual generative hyphae (3–4 μm), and larger basidiospores (4.7–6 μm × 1.6–2.1 μm; Yu et al., 2004). Geographically, L. mollis has been reported in Asia, Europe, and North America (Gilbertson and Ryvarden, 1986; Ryvarden and Gilbertson, 1993; Núñez and Ryvarden, 2001; Yu et al., 2004). Yu et al. (2004) reported Leptoporus in China for the first time, which is distributed in Heilongjiang Province of China. In their study, the morphological characteristics of the studied specimens fit well with L. mollis. Therefore, there are two species of Leptoporus in China, viz., L. mollis is distributed in Northeast China, while L. submollis is distributed in Southwest China. In terms of ecological habits, Leptoporus species mainly grow on fallen trunk or stump of various coniferous trees (especially on Abies sp., Picea sp., and Pinus sp.) in the alpine plateau and cold temperate zone and cause a brown decay of wood.
Nomenclature
BI, Bayesian inference; BJFC, Herbarium of the Institute of Microbiology, Beijing Forestry University; BGI, Beijing Genomics Institute; BPP, Bayesian posterior probabilities; BT, bootstrap; CB, Cotton Blue; CB–, acyanophilous; GTR+I+G, general time reversible+proportion invariant+gamma; IFP, Herbarium of the Institute of Applied Ecology, Chinese Academy of Sciences; IKI, Melzer’s reagent; IKI–, neither amyloid nor dextrinoid; ILD, incongruence length difference test; ITS, internal transcribed spacer; KOH, 5% potassium hydroxide; L, mean spore length (arithmetic average of all spores); ML, maximum likelihood; MP, maximum parsimony; MPT, most parsimonious tree; n (a/b), number of spores (a) measured from given number (b) of specimens; nLSU, large subunit of nuclear ribosomal RNA; Q, variation in the L/W ratios between the specimens studied; RPB1, DNA-directed RNA polymerase II subunit 1; RPB2, DNA-directed RNA polymerase II subunit 2; TL, tree length; W, mean spore width (arithmetic average of all spores); CI, consistency index; RI, retention index; RC, rescaled consistency index; TBR, tree-bisection-reconnection HI, homoplasy index; TEF1, translation elongation factor 1-α.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Author contributions
B-KC designed the research. B-KC, SL, Y-FS, XJ, C-GS and T-MX prepared the samples. SL, C-GS and T-MX conducted the molecular experiments and analyzed the data. SL, Y-FS and B-KC drafted the manuscript. All authors contributed to the article and approved the submitted version.
Funding
The research is supported by the National Natural Science Foundation of China (Nos. 32270010, U2003211, 31870008), the Scientific and Technological Tackling Plan for the Key Fields of Xinjiang Production and Construction Corps (No. 2021AB004) and Beijing Forestry University Outstanding Young Talent Cultivation Project (No. 2019JQ03016).
Acknowledgments
We express our gratitude to Ms. Yan Wang (China) is grateful for help during field collections and molecular studies. Drs. Yu-Cheng Dai (China), Jun-Zhi Qiu (China), Xiao-Lan He (China), Hai-Xia Ma (China), Yuan-Yuan Chen (China), Shi-Liang Liu (China) and Long-Fei Fan (China) for assistance during field collections.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Binder, M., Hibbett, D. S., Larsson, K. H., Larsson, E., Langer, E., Langer, G. (2005). The phylogenetic distribution of resupinate forms across the major clades of mushroom-forming fungi (Homobasidiomycetes). Syst. Biodivers. 3, 113–157. doi: 10.1017/S1477200005001623
Binder, M., Justo, A., Riley, R., Salamov, A., López-Giráldez, F., Sjökvist, E., et al. (2013). Phylogenetic and phylogenomic overview of the polyporales. Mycologia 105, 1350–1373. doi: 10.3852/13-003
Chen, C. C., Chen, C. Y., Lim, Y. W., Wu, S. H. (2020). Phylogeny and taxonomy of Ceriporia and other related taxa and description of three new species. Mycologia 112, 64–82. doi: 10.1080/00275514.2019.1664097
Chen, C. C., Chen, C. Y., Wu, S. H. (2021). Species diversity, taxonomy and multi−gene phylogeny of phlebioid clade (Phanerochaetaceae, irpicaceae, meruliaceae) of polyporales. Fungal Divers. 6, 337–442. doi: 10.1007/s13225-021-00490-w
Chen, C. C., Wu, S. H., Chen, C.Y. (2018). Four species of polyporoid fungi newly recorded from Taiwan. Mycotaxon 133 (1), 45–54. doi: 10.5248/133.45
Chen, Y. Y., Cui, B. K. (2016). Phylogenetic analysis and taxonomy of the Antrodia heteromorpha complex in China. Mycoscience 57, 1–10. doi: 10.1016/j.myc.2015.07.003
Chen, Y. Y., Li, H. J., Cui, B. K. (2015). Molecular phylogeny and taxonomy of Fibroporia (Basidiomycota) in China. Phytotaxa 203, 47–54. doi: 10.11646/phytotaxa.203.1.4
Chen, J. J., Wang, Y. R., Wang, C. G., Dai, Y. C. (2022). Two new species of Ceriporia (Irpicaceae, basidiomycota) from the Asia pacific area. Mycol. Prog. 21, 39–48. doi: 10.1007/s11557-021-01731-7
Chen, Y. Y., Wu, F., Wang, M., Cui, B. K. (2017). Species diversity and molecular systematics of Fibroporia (Polyporales, basidiomycota) and its related genera. Mycol. Prog. 16, 521–533. doi: 10.1007/s11557-017-1285-1
Cui, B. K., Li, H. J., Ji, X., Zhou, J. L., Song, J., Si, J., et al. (2019). Species diversity, taxonomy and phylogeny of polyporaceae (Basidiomycota) in China. Fungal Divers. 97, 137–392. doi: 10.1007/s13225-019-00427-4
Farris, J. S., Källersjö, M., Kluge, A. G., Kluge, A. G., Bult, C. (1994). Testing significance of incongruence. Cladistics 10, 315–319. doi: 10.1111/j.1096-0031.1994.tb00181.x
Felsenstein, J. (1985). Confidence intervals on phylogenies: An approach using the bootstrap. Evolution 39, 783–791. doi: 10.2307/2408678
Floudas, D., Hibbett, D. S. (2015). Revisiting the taxonomy of Phanerochaete (Polyporales, basidiomycota) using a four gene dataset and extensive ITS sampling. Fungal Biol. 119, 679–719. doi: 10.1016/j.funbio.2015.04.003
Gilbertson, R. L., Ryvarden, L. (1986). North American polypores 1 (Oslo: Fungiflora: Abortiporus - Lindtneria), 1–433.
Gómez-Montoya, N., Drechsler-Santos, E. R., Ferreira-Lopes, V., Tomšovský, M., Urcelay, C., Roble-do, G. L. (2017). New insights on Trametopsis Tomšovský (Polyporales Gäum.) based on phylogenetic evidences and morphological analyses of neotropical species. Phytotaxa 311 (2), 155–166. doi: 10.11646/phytotaxa.311.2.3
Hall, T. A. (1999). Bioedit: a user-friendly biological sequence alignment editor and analysis program for windows 95/98/NT. Nucleic Acids Symp. Ser. 41, 95–98. doi: 10.1021/bk-1999-0734.ch008
Han, M. L., Chen, Y. Y., Shen, L. L., Song, J., Vlasák, J., Dai, Y. C., et al. (2016). Taxonomy and phylogeny of the brown-rot fungi: Fomitopsis and its related genera. Fungal Divers. 80, 343–373. doi: 10.1007/s13225-016-0364-y
Han, M. L., Cui, B. K. (2015). Morphological characters and molecular data reveal a new species of Fomitopsis (Polyporales) from southern China. Mycoscience 56, 168–176. doi: 10.1016/j.myc.2014.05.004
Han, M. L., Song, J., Cui, B. K. (2014). Morphology and molecular phylogeny for two new species of Fomitopsis (Basidiomycota) from south China. Mycol. Prog. 13, 905–914. doi: 10.1007/s11557-014-0976-0
Han, M. L., Vlasák, J., Cui, B. K. (2015). Daedalea americana sp. nov. (Polyporales, basidiomycota) evidenced by morphological characters and phylogenetic analysis. Phytotaxa 204, 277–286. doi: 10.11646/phytotaxa.204.4.4
He, M. Q., Zhao, R. L., Hyde, K. D., Begerow, D., Kemler, M., Yurkov, A., et al. (2019). Notes, outline and divergence times of Basidiomycota. Fungal Divers 99, 105–367. doi: 10.1007/s13225-019-00435-4
Hillis, D. M., Bull, J. J. (1993). An empirical test of bootstrapping as a method for assessing confidence in phylogenetic analysis. Syst. Biodivers. 42, 182–192. doi: 10.1093/sysbio/42.2.182
Ji, X., Zhou, J. L., Song, C. G., Xu, T. M., Wu, D. M., Cui, B. K. (2022). Taxonomy, phylogeny and divergence times of Polyporus (Basidiomycota) and related genera. Mycosphere 13, 1–52. doi: 10.5943/mycosphere/13/1/1
Jang, Y., Jang, S., Lee, J., Lee, H., Lim, Y.W., Kim, C., et al. (2016). Diversity of wood-inhabiting polyporoid and corticioid fungi in Odaesan National Park, Korea. Mycobiology 44 (4), 217–236. doi: 10.5941/MYCO.2016.44.4.217
Jia, B. S., Zhou, L.W., Cui, B. K., Rivoire, B., Dai, Y. C. (2014). Taxonomy and phylogeny of Ceriporia (Polyporales, Basidiomycota) with an emphasis of Chinese collections. Mycol. Prog. 13 (1), 81–93. doi: 10.1007/s11557-013-0895-5
Justo, A., Hibbett, D.S. (2011). Phylogenetic classification of Trametes (Basidiomycota, Polyporales) based on a five-marker dataset. Taxon 60 (6), 1567–1583. doi: 10.1002/tax.606003
Justo, A., Miettinen, O., Floudas, D., Ortiz-Santana, B., Sjökvist, E., Lindner, D., et al. (2017). A revised family-level classification of the polyporales (Basidiomycota). Fungal Biol. 121, 798–824. doi: 10.1016/j.funbio.2017.05.010
Katoh, K., Standley, D. M. (2013). MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 30, 772–780. doi: 10.1093/molbev/mst010
Kirk, P. M., Cannon, P. F., Minter, D. W., Stalpers, J. A. (2008). Dictionary of the fungi. 10th Edn (Oxon: CAB International Wallingford, UK).
Lindner, D. L., Banik, M. T. (2008). Molecular phylogeny of Laetiporus and other brown rot polypore genera in north America. Mycologia 100, 417–430. doi: 10.3852/07-124R2
Liu, S., Chen, Y. Y., Sun, Y. F., He, X. L., Song, C. G., Si, J., et al. (2022a). Systematic classification and phylogenetic relationships of the brown−rot fungi within the polyporales. Fungal Divers. doi: 10.1007/s13225-022-00511-2
Liu, S., Han, M. L., Xu, T. M., Wang, Y., Wu, D. M., Cui, B. K. (2021a). Taxonomy and phylogeny of the Fomitopsis pinicola complex with descriptions of six new species from east Asia. Front. Microbiol. 12, 644979. doi: 10.3389/fmicb.2021.644979
Liu, S., Shen, L. L., Wang, Y., Xu, T. M., Gates, G., Cui, B. K. (2021b). Species diversity and molecular phylogeny of Cyanosporus (Polyporales, basidiomycota). Front. Microbiol. 12, 631166. doi: 10.3389/fmicb.2021.631166
Liu, S., Song, C. G., Cui, B. K. (2019). Morphological characters and molecular data reveal three new species of Fomitopsis (Basidiomycota). Mycol. Prog. 18, 1317–1327. doi: 10.1007/s11557-019-01527-w
Liu, S., Song, C. G., Xu, T. M., Ji, X., Wu, D. M., Cui, B. K. (2022b). Species diversity, molecular phylogeny, and ecological habits of Fomitopsis (Polyporales, basidiomycota). Front. Microbiol. 13, 859411. doi: 10.3389/fmicb.2022.859411
Liu, S., Sun, Y. F., Wang, Y., Xu, T. M., Song, C. G., Chen, Y. Y., et al. (2022c). Taxonomy and molecular phylogeny of Trametopsis (Polyporales, basidiomycota) with descriptions of two new species. MycoKeys 90, 31–51. doi: 10.3897/mycokeys.90.84717
Liu, S., Xu, T. M., Song, C. G., Zhao, C. L., Wu, D. M., Cui, B. K. (2022d). Species diversity, molecular phylogeny and ecological habits of Cyanosporus (Polyporales, basidiomycota) with an emphasis on Chinese collections. MycoKeys 86, 19–46. doi: 10.3897/mycokeys.86.78305
Maddison, W. P., Maddison, D. R. (2017) Mesquite: A modular system for evolutionary analysis, version 3.2. Available at: http://mesquiteproject.org.
Miettinena, O., Larssonb, E., Sjökvist, E., Larssonc, K. H. (2012). Comprehensive taxon sampling reveals unaccounted diversity and morphological plasticity in a group of dimitic polypores (Polyporales, Basidiomycota). Cladistics 28 (3), 251–270. doi: 10.1111/j.1096-0031.2011.00380.x
Miettinen, O., Spirin, V., Vlasák, J., Rivoire, B., Stenroos, S., Hibbett, D. (2016). Polypores and genus concepts in Phanerochaetaceae (Polyporales, Basidiomycota). MycoKeys 17, 1–46. doi: 10.3897/mycokeys.17.10153
Matheny, P. B. (2005). Improving phylogenetic inference of mushrooms with RPB1 and RPB2 nucleotide sequences (Inocybe, agaricales). Mol. Phylogenet Evol. 35, 1–20. doi: 10.1016/j.ympev.2004.11.014
Matheny, P. B., Liu, Y. J., Ammirati, J. F., Hall, B. D. (2002). Using RPB1 sequences to improve phylogenetic inference among mushrooms (Inocybe, agaricales). Am. J. Bot. 89, 688–698. doi: 10.2307/4131413
Nylander, J. A. A. (2004). MrModeltest v2. program. distributed by the author; evolutionary biology center (Uppsala: Uppsala University).
Ortiz-Santana, B., Lindner, D. L., Miettinen, O., Justo, A., Hibbett, D. S. (2013). A phylogenetic overview of the antrodia clade (Basidiomycota, polyporales). Mycologia 105, 1391–1411. doi: 10.3852/13-051
Petersen, J. H. (1996). Farvekort (Greve: The Danish Mycological Society’s colour-chart. Foreningen til Svampekundskabens Fremme).
Posada, D., Crandall, K. A. (1998). Modeltest: Testing the model of DNA substitution. Bioinformatics 14, 817–818. doi: 10.1093/bioinformatics/14.9.817
Psurtseva, N. V. (2010). Conservation of medicinal mushrooms in the V. L. Komarov Botanical Institute Basidiomycetes Culture Collection (LE-BIN, Russia). Int. J. Med. Mushrooms 12 (12), 193–199. doi: 10.1615/IntJMedMushr.v12.i2.100
Quélet, L. (1886). Enchiridion fungorum in Europa media et praesertim in Gallia vigentium. Octave Dion, Paris, 1–352.
Rehner, S. (2001) Primers for elongation factor 1-a (EF1-a). Available at: http://ocid.nacse.org/research/deephyphae/EF1primer.pdf (Accessed 20 May 2020).
Ronquist, F., Huelsenbeck, J. P. (2003). MRBAYES 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19, 1572–1574. doi: 10.1093/bioinformatics/btg180
Ryvarden, L. (1991). Genera of polypores: Nomenclature and taxonomy. Synop. Fungorum (Oslo, Norway) 5, 363.
Shen, L. L., Cui, B. K., Dai, Y. C. (2014). A new species of Postia (Polyporales, basidiomycota) from China based on morphological and molecular evidence. Phytotaxa 162, 147–156. doi: 10.11646/phytotaxa.162.3.3
Shen, L. L., Liu, H. X., Cui, B. K. (2015). Morphological characters and molecular data reveal two new species of Postia (Basidiomycota) from China. Mycol. Prog. 14, 7. doi: 10.1007/s11557-015-1032-4
Shen, L. L., Wang, M., Zhou, J. L., Xing, J. H., Cui, B. K., Dai, Y. C. (2019). Taxonomy and phylogeny of Postia. multi-gene phylogeny and taxonomy of the brown-rot fungi: Postia (Polyporales, basidiomycota) and related genera. Persoonia 42, 101–126. doi: 10.3767/persoonia.2019.42.05
Sjökvist, E., Larsson, E., Eberhardt, U., Ryvarden, L., Larsson, K. H. (2012). Stipitate stereoid basidiocarps have evolved multiple times. Mycologia 104 (5), 1046–1055. doi: 10.3852/11-174
Song, J., Chen, Y. Y., Cui, B. K., Liu, H. G., Wang, Y. Z. (2014). Morphological and molecular evidence for two new species of Laetiporus (Basidiomycota, polyporales) from southwestern China. Mycologia 106, 1039–1050. doi: 10.3852/13-402
Song, J., Cui, B. K. (2017). Phylogeny, divergence time and historical biogeography of Laetiporus (Basidiomycota, polyporales). BMC Evol. Biol. 17, 102. doi: 10.1186/s12862-017-0948-5
Song, J., Sun, Y. F., Ji, X., Dai, Y. C., Cui, B. K. (2018). Phylogeny and taxonomy of Laetiporus (Basidiomycota, polyporales) with descriptions of two new species from western China. MycoKeys 37, 57–71. doi: 10.3897/mycokeys.37.26016
Spirin, V., Vlasák, J., Rivoire, B., Kout, J., Kotiranta, H., Miettinen, O. (2016). Studies in the Ceriporia purpurea group (Polyporales, Basidiomycota), with notes on similar Ceriporia species. Cryptogamie Mycol 37 (4), 421–435. doi: 10.7872/crym/v37.iss4.2016.421
Stamatakis, A. (2006). RAxML-VI-HPC: Maximum likelihood-based phylogenetic analysis with thousands of taxa and mixed models. Bioinformatics 22, 2688–2690. doi: 10.1093/bioinformatics/btl446
Sun, Y. F., Costa-Rezende, D. H., Xing, J. H., Zhou, J. L., Zhang, B., Gibertoni, T. B., et al. (2020). Multi-gene phylogeny and taxonomy of Amauroderma s. lat. (Ganodermataceae). Persoonia 44, 206–239. doi: 10.3767/persoonia.2020.44.08
Sun, Y. F., Xing, J. H., He, X. L., Wu, D. M., Song, C. G., Liu, S., et al. (2022). Species diversity, systematic revision and molecular phylogeny of ganodermataceae (Polyporales, basidiomycota) with an emphasis on Chinese collections. Stud. Mycol. 101, 287–415. doi: 10.3767/10.3114/sim.2022.101.05
Swofford, D. L. (2002). PAUP∗: Phylogenetic analysis using parsimony (∗and other methods). version 4.0b10 (Sunderland, MA: Sinauer Associates). doi: 10.1111/j.0014-3820.2002.tb00191.x
Tomšovský, M., Menkis, A., Vasaitis, R. (2010). Phylogenetic relationships in European Ceriporiopsis species inferred from nuclear and mitochondrial ribosomal DNA sequences. Fungal Biol. 114 (4), 350–358. doi: 10.1016/j.funbio.2010.02.004
Volobuev, S. (2019). To the study of aphyllophoroid fungi (Agaricomycetes, basidiomycota) in shebekinsky district, belgorod region. Diversity Plant World 3, 21–25. doi: 10.22281/2686-9713-2019-3-21-25
White, T. J., Bruns, T., Lee, S., Taylor, J. (1990). “Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics,” in PCR protocols: A guide to methods and applications. Eds. Innis, M. A., Gelfand, D. H., Sninsky, J. J., White, T. J. (San Diego: Academic Press), 315–322. doi: 10.1016/B978-0-12-372180-8.50042-1
Wu, F., Chen, J. J., Ji, X. H., Vlasák, J., Dai, Y. C. (2017). Phylogeny and diversity of the morphologically similar polypore genera Rigidoporus, Physisporinus, Oxyporus, and Leucophellinus. Mycologia 109, 749–765. doi: 10.1080/00275514.2017.1405215
Wu, S. H., Nilsson, H. R., Chen, C. T., Yu, S. Y., Hallenberg, N. (2010). The white-rotting genus Phanerochaete is polyphyletic and distributed throughout the phleboid clade of the Polyporales (Basidiomycota). Fungal Divers 42 (1), 107–118. doi: 10.1007/s13225-010-0031-7
Keywords: brown-rot fungi, Irpicaceae, macro-fungi, multi-gene phylogeny, taxonomy
Citation: Liu S, Sun Y-F, Ji X, Song C-G, Xu T-M and Cui B-K (2023) Molecular phylogeny and taxonomy of the remarkable genus Leptoporus (Polyporales, Basidiomycota) with description of a new species from Southwest China. Front. Cell. Infect. Microbiol. 12:1116035. doi: 10.3389/fcimb.2022.1116035
Received: 05 December 2022; Accepted: 28 December 2022;
Published: 23 January 2023.
Edited by:
Jia-Jia Chen, Jiangsu Vocational College of Agriculture and Forestry, ChinaReviewed by:
Yu-Guang Fan, Hainan Medical University, ChinaNian-Kai Zeng, Hainan Medical University, China
Yulian Wei, Institute of Applied Ecology (CAS), China
Copyright © 2023 Liu, Sun, Ji, Song, Xu and Cui. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Bao-Kai Cui, Y3VpYmFva2FpQGJqZnUuZWR1LmNu
 Shun Liu
Shun Liu Yi-Fei Sun
Yi-Fei Sun Xing Ji
Xing Ji Chang-Ge Song
Chang-Ge Song Tai-Min Xu
Tai-Min Xu Bao-Kai Cui
Bao-Kai Cui