- Institute of Dairy Science, College of Animal Sciences, Zhejiang University, Hangzhou, China
Introduction: Limosilactobacillus reuteri is a gut symbiont with multiple remarkable beneficial effects on host health, and members of L. reuteri are valuable probiotic agents. However, L. reuteri showed obvious host specificity.
Methods: In our study, a novel L. reuteri RGW1 was isolated from feces of healthy calves, and its potential as a probiotic candidate were assessed, by combining in vitro, in vivo experiments and genomic analysis.
Results and discussion: RGW1 was sensitive to all the antibiotics tested, and it did not contain any virulence factor-coding genes. This isolate showed good tolerance to acid (pH 3.0), 0.3% bile salt, and simulated gastric fluid. Moreover, this isolate showed a high hydrophobicity index (73.7 ± 4.6%) and was able to adhere to Caco-2 cells, and antagonize Escherichia coli F5. Treatment of LPS-induced mice with RGW1 elevated TGF-β and IL-10 levels, while RGW1 cell-free supernatant (RCS) decreased TNF-α levels in the sera. Both RGW1 and RCS increased the villus height and villus height/crypt depth ratio of colon. Genomic analysis revealed the mechanism of the probiotic properties described above, and identified the capacity of RGW1 to biosynthesize L-lysine, folate, cobalamin and reuterin de novo. Our study demonstrated the novel bovine origin L. reuteri RGW1 had multiple probiotic characteristics and immunomodulation effects, and provided a deeper understanding of the relationship between these probiotic properties and genetic features.
1. Introduction
Probiotics are defined by the FAO and WHO as “live microorganisms that, when administered in adequate amounts, confer a health benefit on the host” (Hill et al., 2014). Various studies have demonstrated the health-promoting effects of probiotics with mechanisms including anti-pathogen activity and immunomodulation. To achieve these effects, probiotics should exhibit characteristics such as acid, bile and digestive enzyme tolerance, which allows them pass through the stressful environment of the host’s upper gastrointestinal tract, survive in the gut, and adhere to the intestinal wall for immunomodulation. In addition, probiotics must be safe for humans or animals, with the most important concerns being their virulence potential and the possibility to act as a reservoir for mobile antimicrobial resistance genes. Most importantly, probiotics should provide one or more benefits to host health, such as the prevention of enteric infections, diarrhea or inflammatory bowel disease.
Limosilactobacillus reuteri is a well-known probiotic species that naturally colonizes the gastrointestinal tract of a wide range of vertebrates. This species displays multiple beneficial effects on host health, including anti-pathogenic capacity, the production of vitamins and amino acids, modulation of host immune responses and promotion of gut mucosal integrity. However, the effects of probiotics on host health are species-, dose- and disease-specific (Aureli et al., 2011). Furthermore, L. reuteri has developed fundamentally different ecological strategies for different hosts and exhibits obvious host specificity (Walter, 2008; Frese et al., 2011; Duar et al., 2017). Therefore, isolating and characterizing more novel L. reuteri strains especially from relatively unexplored ecological niches, still makes sense to meet the increasing desire to promote human or animal health naturally.
In addition to meeting the prerequisites for being considered as a potential probiotic (described above), additional probiotic properties have been described by Mu et al. (2018), for example, vitamin B9, B12 and exopolysaccharide biosynthesis. Whole genome sequence analysis helps to efficiently identify these potential properties and enhance understanding of the relationships between genotypic and phenotypic profiles. Since previous research have reported the genomes of L. reuteri isolates originated from humans, rodents and poultry (Oh et al., 2010; Frese et al., 2011; Duar et al., 2017), there has been a relative lack of research into bovine L. reuteri strains and their genome. In this study, a new L. reuteri strain originated from bovine, named L. reuteri RGW1, was isolated and identified, and its potential probiotic traits were characterized. Additionally, the draft genome sequence of this strain was present, and probiotic-associated genes were identified.
2. Materials and methods
2.1. Isolation and culture of LAB strains
Fresh fece samples were collected from three one-month-old healthy calves immediately after morning feeding (Zhejiang Yijing Ecological Animal Husbandry Co., Ltd., Shaoxin, China). The feces were fully homogenized, and cultured using anaerobic Man, Rogosa and Sharpe (MRS) broth for 24 h. Then the culture broth was serially diluted 10-fold using sterile anaerobic PBS, spread on MRS agar and incubated at 37 °C anaerobically. After incubation for 48 h, single white colonies were selected and purified by restreaking on MRS agar plates. Their taxonomic status was determined using the 16S rRNA gene sequence with the primers 27F (5′-AGAGTTTGATCCTGGCTCAG-3′) and 1492R (5′-TACGGCTACCTTGTTACGACTT-3′), and compared to those in the National Center Biotechnology Information (NCBI) database using the nucleotide BLAST tool. The phylogenetic tree was constructed using MEGA-X software by the neighbor-joining method, and visualized using iTOL (Letunic and Bork, 2021). The L. reuteri isolate was Gram-stained, and its bacterial morphology was observed by scanning electron microscopy (Hitachi SU-8010, Japan). The isolate was preserved at -80 °C within 20% sterile glycerol dilution for further use.
2.2. In vitro studies on probiotic characteristics
2.2.1. Tolerance to acid, bile salt and simulated gastric fluid
The growth kinetics of L. reuteri RGW1 was determined by measuring the optical density every hour at 600 nm. Acid or bile salt survivability was determined in MRS broth. Briefly, L. reuteri RGW1 was cultured overnight in anaerobic MRS broth at 37 °C, then the bacteria was incubated in MRS broth (2×106 CFU/mL) at different pH values (7.0, 5.0, 4.0 and 3.0) or with different concentrations (0%, 0.1%, 0.3% and 0.5%) of bile salt at 37°C for 3 h, respectively. Tolerance to simulated gastric fluid of L. reuteri RGW1 were examined following the method described by Zárate et al. (2000) with minimal modification. Briefly, overnight-cultured RGW1 cells were incubated in the simulated gastric fluid (2×106 CFU/mL, 3 g/L Pepsin, pH 2.5) at 37 °C for 6 h. The viability was enumerated by colony counts (CFU/mL) on MRS agar plate. Survivability (%) was calculated by the following formula: (final viability/initial viability) × 100. The assay was carried out in three parallel experiments.
2.2.2. Hydrophobicity
Hydrophobicity was determined following the procedure of Nagpal et al. (2010) with minor modification. Briefly, overnight-cultured bacterial cell suspension was mixed with n-hexadecane and vortexed for 2 min. The absorbance of aqueous phase at 600 nm was measured as A0, and the mixture was placed at 37 °C for 1 h. Then the absorbance of aqueous phase at 600 nm was measured as A1. The hydrophobicity (%) was calculated with the following formula: (1-A1/A0) × 100.
2.2.3. Adhesion to Caco-2 cells
Human colorectal adenocarcinoma cell line Caco-2 cells was used to examine the cellular adhesion of L. reuteri RGW1 as described previously (Jacobsen et al., 1999). Briefly, Caco-2 cells were seeded in 12-well plates to grow to approximately 80% confluency. A 100 µL of suspension of RGW1 at concentration of 108 CFU/mL was added to each cell well, and then incubated at 37 °C in a 5% CO2 atmosphere. After 2 h incubation, the medium was removed, and the bacterial cells were washed five times with sterile PBS to remove the nonadherent bacteria. Then the cell monolayers were fixed in a 65 °C incubator, stained with safranin O solution and observed under a microscope.
2.2.4. Antibiotic susceptibility
Antibiotic susceptibility was determined by the disc diffusion method following the procedure of Charteris et al. (1998) with minimal modification. Two hundred microliters of RGW1 cells at a concentration of 108 CFU/mL were coated on MRS agar plates. After the surface was completely dried, the antibiotic discs with ampicillin (10 μg), amoxicillin (30 μg), chloramphenicol (30 μg), cefazolin (30 μg), erythromycin (15 μg), tetracycline (30 μg), or penicillin G (10 μg), respectively were placed on the center of the plate to dispense antibiotic. Then the plates were incubated anaerobically at 37 °C for 24 h. The diameter of inhibition zone was measured, and the results were expressed in terms of resistance, moderate susceptibility, or susceptibility, according to the interpretative standards detailed in Supplementary Table S1.
2.2.5. Virulence genetic amplification
As reported by Casarotti et al. (2017) and Nagpal et al. (2018), virulence genes including the adherent virulence factors esp (Enterococcal surface protein), efaAfs (cell wall adhesins), asa (aggregation substance), and ace (adhesion of collagen), the secretory virulence factors gelE (gelatinase), cylA (protein for activation of cytolysin), hyl (hyaluronidase) and the biogenic amine production factors hdc (histidine decarboxylase), tdc (tyrosine decarboxylase) and odc (ornithine decarboxylase), were detected using PCR. The amplified products were resolved by electrophoresis on a 1% agarose gel. Detailed primer information for these virulence genes is shown in Supplementary Table S2.
2.2.6. Antibacterial activity
The antibacterial activity was determined by the well agar diffusion method and coculture experiment. Escherichia coli F5 (BNCC-125787, also known as K99) was purchased from BeNa Culture Collection (BNCC), and cultured and enriched using Luria-Bertani (LB) broth. For well agar diffusion, briefly, overlaid a petri dish with soft LB agar containing 108 CFU/mL of E. coli F5. After solidification, the wells were punched with Oxford cups and seeded with 150 µL of RGW1 cells (108 CFU/mL). After incubation at 37 °C anaerobically for 16 h, the diameter of inhibition zones was measured. To identify the effective antibacterial compounds, the bacteria-free supernatant of RGW1 (108 CFU/mL) was pretreated by pH adjustment, proteinase K or heating, respectively. Briefly, for pH adjustment, the pH value of supernatant was adjusted to 7.0 using a 1.0 mol/L NaOH solution, then the supernatant was filtered with 0.22 μm filter membrane. For heating pretreatment, the supernatant was heated in a 70 °C water bath for 1 h. When the supernatant was cooled to room temperature, the supernatant was filtered with 0.22 μm filter membrane. For proteinase K pretreatment, the supernatant were treated with proteinase K (0.1mg/mL) in a 37 °C water bath for 30 min. Then the treated supernatant was used to determined antibacterial activity using well agar diffusion method described above. As for the coculture experiment, RGW1 and E. coli F5 were cocultured in a mixture of 5 mL MRS broth and 5 mL LB broth with the initial concentration RGW1 and E. coli F5 cells set at 108 and 107 CFU/mL, respectively. RGW1 monocultured in 10 mL MRS broth at 108 CFU/mL and E. coli F5 monocultured in 10 mL LB broth at 107 CFU/mL were used as controls. All cultures were incubated at 37 °C with shaking at 180 rpm. The viable counts (CFU/mL) of RGW1 and E. coli F5 were enumerated using MRS agar or LB agar at 0, 8, 16, and 24 h of incubation, respectively.
2.3. Oral administration of LAB and sample collection in mouse study
2.3.1. Animals and housing
All procedures were approved by the Animal Care and Use Committee of Zhejiang Provincial Hospital of Chinese Medicine (Hangzhou, China, 60136-38385) and were in accordance with the hospital guidelines for animal research. In the current study, a total of 27 male C57BL/6J mice (5 weeks old, 18.7 ± 0.7 g initial body weight) were purchased from Shanghai SLAC Laboratory Animal Company (Shanghai, China). After three days of adaption, the mice were randomly assigned to three groups, each group contains three cages of three mice in each (n = 9). Following the dose of administration of L. reuteri described in Wu et al. (2020), the three groups of mice were orally administrated (1) 200 µL/d of MRS medium for seven days (control); (2) 200 µL/d of RGW1 (108 CFU) for seven days (RGW1); or (3) 200 µL/d of RGW1 cell-free supernatant for seven days (RCS), respectively. The body weight (BW) and intake of all mice were recorded daily at 9 am before oral administration. Each mouse was challenged with lipopolysaccharide (LPS, 10 mg/kg BW) on day eight. On day nine, the mice were anesthetized and sacrificed by cervical dislocation after collecting blood by eyeball enucleation, and then the intestinal tissues were collected.
2.3.2. Biochemical assays of serum
The concentrations of TNF-α, TGF-β and IL-10 in the serum were determined using ELISA kits (Jianglai Biotechnology Institution, Shanghai, China).
2.3.3. Hematoxylin and eosin staining of the gut
Hematoxylin and eosin (H&E) staining of intestinal tissues was performed as previously described (Zhong et al., 2022). In brief, the cecum and colon samples were fixed in 4% paraformaldehyde, waxed, and cut into 5-μm thick sections before being stained. After deparaffinization, the sections were soaked with alcohols followed by staining with H&E. The villus height and crypt length were measured using Image-Pro Software (Media Cybernetics, Inc., USA) based on photomicrographs obtained by microscopy.
2.4. Complete genome sequencing, assembly and annotation
RGW1 genomic DNA was extracted with the cetyltrimethyl ammonium bromide (CTAB) method (Gagen et al., 2010). The whole genome was sequenced using an Illumina NovaSeq PE150 (Beijing Novogene Bioinformatics Technology Co., Ltd). Illumina PCR adapter reads and low-quality reads (< 40 bp, N > 10 bp) from the paired-end reads were filtered in quality control step using readfq (version 10). All good quality (Q30 = 92.1%, Q20 = 97.26%) paired-end reads were assembled into a number of scaffolds using SOAP denovo (Li et al., 2008), SPAdes (Bankevich et al., 2012) and ABySS (Simpson et al., 2009). Genome component prediction was achieved using the GeneMarkS program (Besemer et al., 2001), and tRNA and rRNA were identified using tRNAscan-SE (Lowe and Eddy, 1997) and rRNAmmer (Lagesen et al., 2007), respectively. The gene functions were predicted using various databases, including NR (Non-Redundant Protein Database, version 201705), GO (Gene Ontology, version 201705), KEGG (Kyoto Encyclopedia of Genes and Genomes, version 201807), COG (Clusters of Orthologous Groups, version 20151214), TCDB (Transporter Classification Database, version 201807), and Swiss-Prot (version 201807). Carbohydrate-active enzymes were predicted by the Carbohydrate-Active EnZymes Database (version 201604). Pathogenicity and drug resistance analyses were performed using PHI (Pathogen Host Interactions, version 20180515) and VFDB (Virulence Factors of Pathogenic Bacteria, version 20180726).
2.5. Statistical analysis
All data are displayed as the mean ± standard error. Student’s t test was applied to compare two groups, and one-way ANOVA was applied to compare more than two groups. Statistical significance was declared at P < 0.05 unless otherwise specified. All analyses were performed using the GraphPad Prism software 9.0.0 (San Diego, CA, USA).
3. Results
3.1. Bacterial isolate and its basic identification
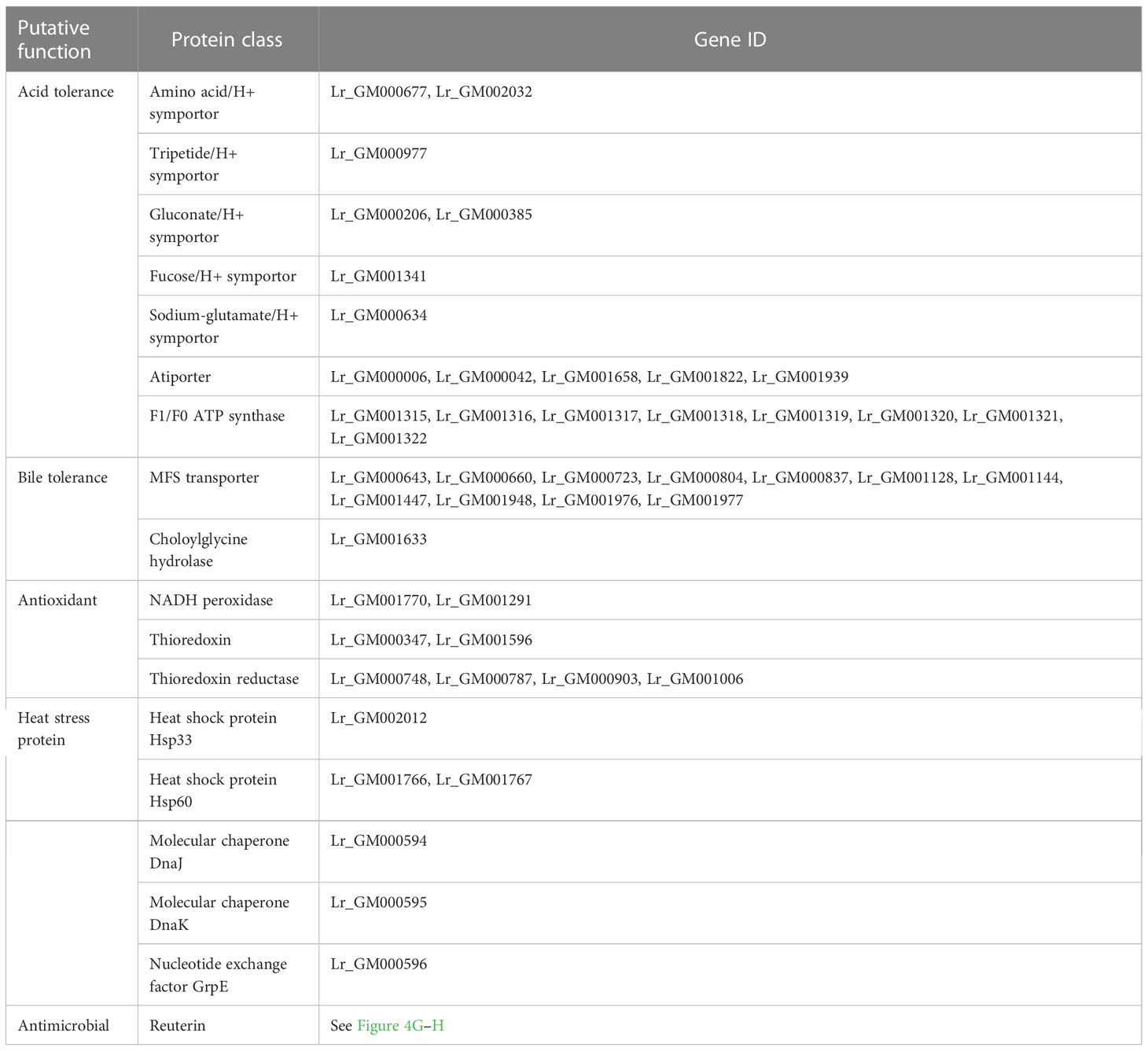
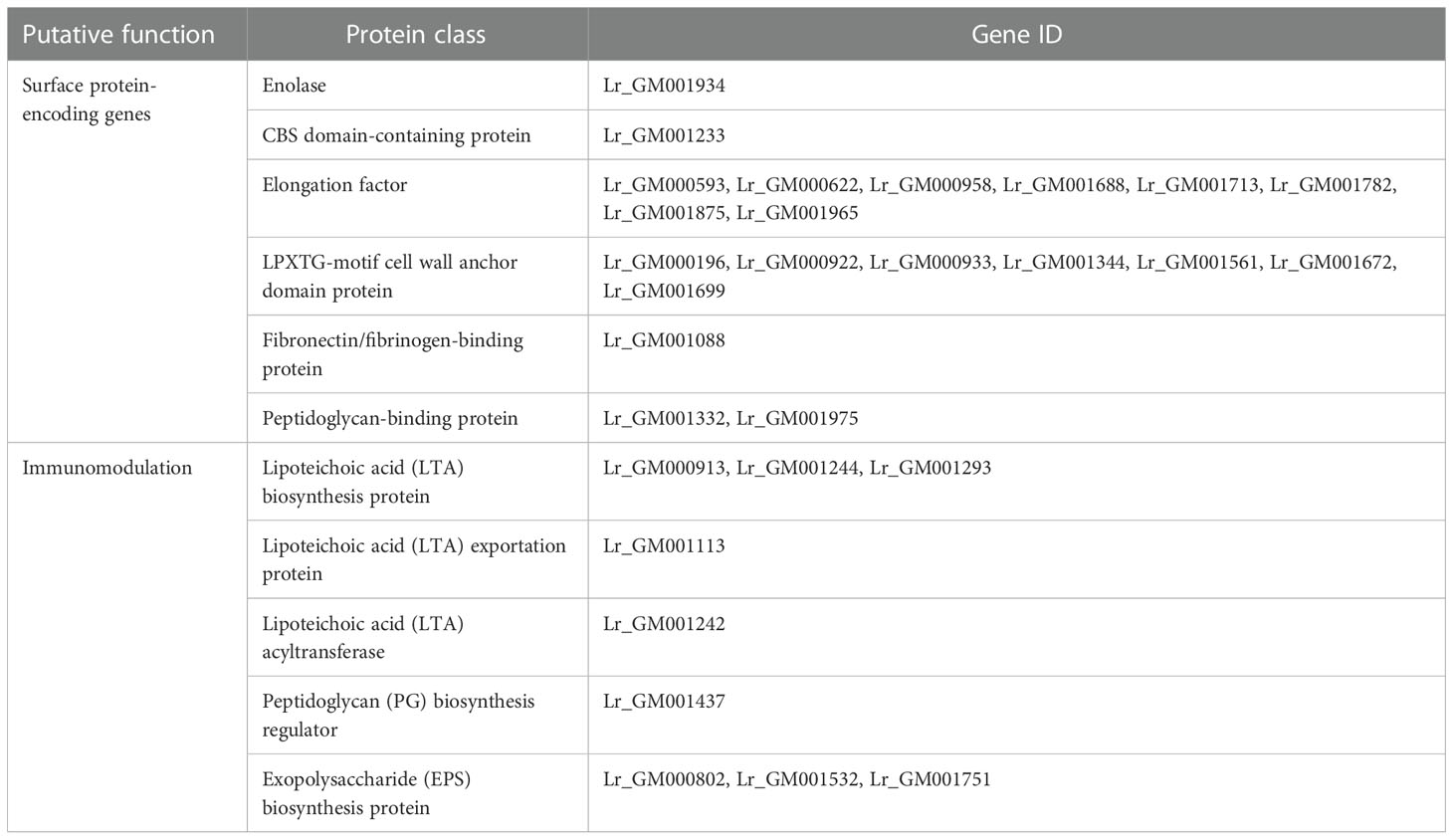
Potential LAB strains were isolated from healthy calf feces, and the 16S rRNA sequence-based phylogenetic tree revealed that the isolate in our study belonged to a subclade of L. reuteri, with more than 99% similarity to other strains within the genus (Figure 1A), thus, the isolate was named L. reuteri RGW1. The strain was identified as gram-positive and produced small white colonies on MRS agar. Scanning electron microscopy images revealed that the strain was rod-shaped (Figure 1B).
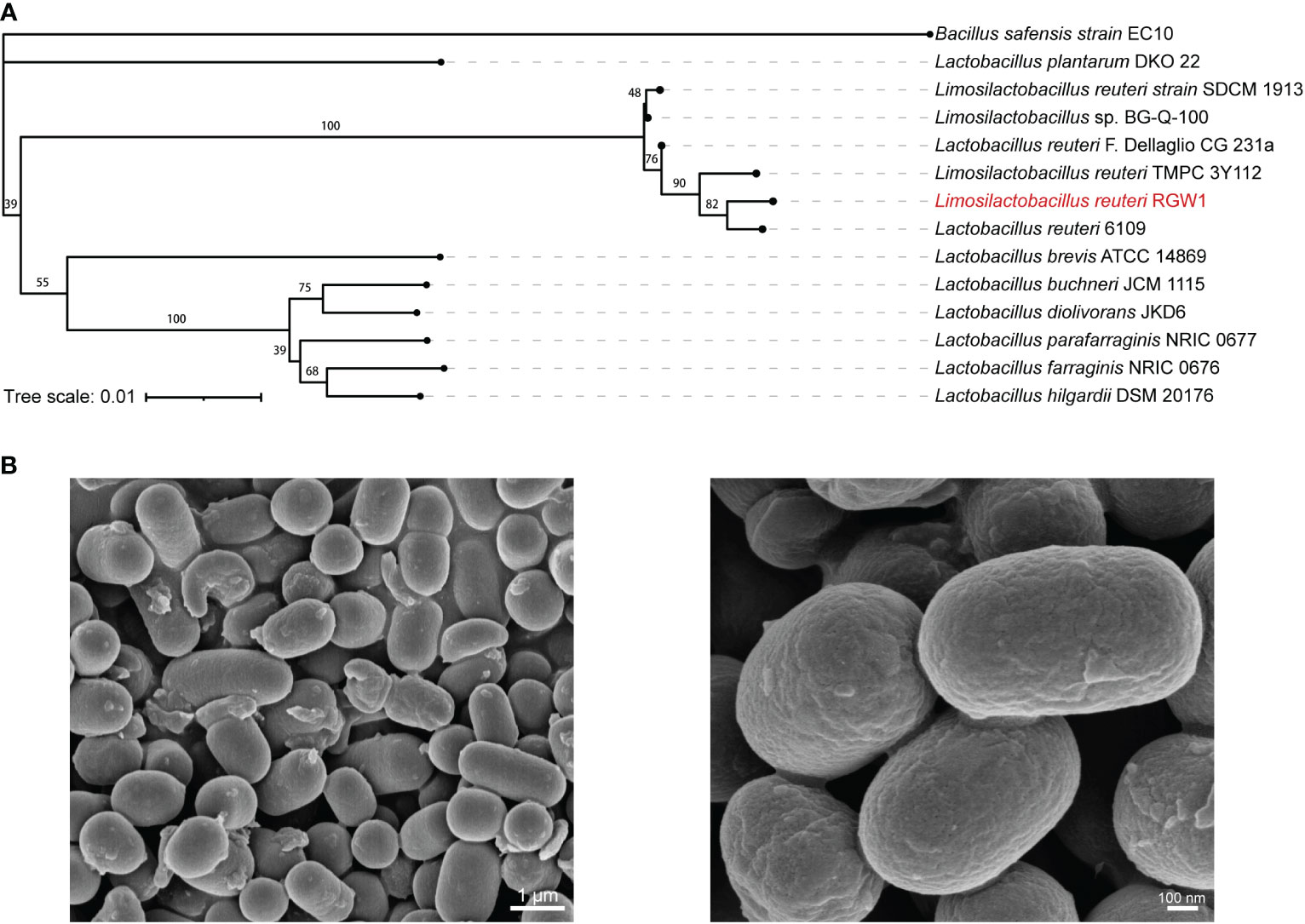
Figure 1 Preparation and characterization of L. reuteri RGW1. (A) Phylogenetic tree of L. reuteri RGW1 (in red color) based on 16S rRNA sequence. The 12 most homologous sequences in the GenBank database were selected for the construction of a phylogenetic tree, Bacillus safensis strain EC10 was selected as outlier bacteria. (B) Representative image of L. reuteri RGW1 using scanning electron microscope.
3.2. In vitro probiotics characteristic of RGW1
3.2.1. Safety of RGW1
The growth kinetics of RGW1 displayed the typical three phases, with lag, log, and stationary phages occurring from 0 to 2 h, 2 to 8 h, and after 8 h, respectively (Figure 2A). Antibiotic susceptibility tests showed that RGW1 was sensitive to ampicillin, amoxicillin, chloramphenicol, cefazolin, and erythromycin, and moderately sensitive to tetracycline and penicillin G (Figure 2B). Virulence genetic analysis showed that RGW1 does not contain virulence factors-coding genes, including gelE, cylA, esp, efaAfs, hyl, asa, hdc, ace, tdc, and odc (Figure 2C).
Figure 2 In vitro probiotic characteristics of L. reuteri RGW1. (A) The growth kinetics of L. reuteri RGW1. (B) Antibiotic susceptibility profile of L. reuteri RGW1 is indicated by the diameters of inhibition zone, the “S” represents sensitivity. (C) Agarose gel electrophoresis results of virulence gene PCR products. (D) Survivability of L. reuteri RGW1 at different pH values (blue), bile salt concentrations (green), and in simulated gastric fluid (orange), a-d represent different superscripted letters (P < 0.05). (E) Representative image of the coculture of L. reuteri RGW1 with Caco-2 cells stained with safranin O solution. (F) Monoculture or coculture of L. reuteri RGW1 with E. coli F5. Each bacterial concentration was counted at 0, 8, 16, 24 h of culture, respectively. (G) The anti E. coli F5 activity of L. reuteri RGW1 indicated by the diameters of inhibition zone, after different pretreatments. Values are expressed as means ± SEM. *P < 0.05; **P < 0.01; ***P < 0.001; ns, not significant.
3.2.2. Tolerance of RGW1 to acid, bile salt and simulated gastric fluid
RGW1 showed a survivability as high as 76.9 ± 5.4% at pH 3.0 for 3 h (Figure 2D). The strain showed good tolerance to 0.3% of bile salt (survivability as high as 151.1 ± 9.5%) after 3 h of exposure (Figure 2D). RGW1 had a high survivability of 124.1 ± 5.7% in simulated gastric fluid (Figure 2D). Accordingly, we conclude that RGW1 has the ability to survive in the host gastrointestinal tract.
3.2.3. Potential probiotic properties
3.2.3.1 Hydrophobicity and adhesion to Caco-2 cells
RGW1 showed a hydrophobicity index as high as 73.7 ± 4.6%, which significantly higher than L. paralimentarius DSM 13238 (P < 0.001, Supplementary Figure 1A). RGW1 was able to adhere to the cell surface of Caco-2 cells after 2 h coculture (Figure 2E).
3.2.3.2 Antagonize E. coli F5 capacity
RGW1 showed antibacterial activity (inhibition zone was 19.2 ± 0.3 mm) against E. coli F5. The antibacterial activities were not significantly different between RGW1 and L. paralimentarius DSM 13238, as well as the antibacterial activities of both RGW1 and L. paralimentarius DSM 13238 were not affected by preheating, but was eliminated by pH adjustment (Figure 2F, Supplementary Figure 1B-D). However, the antibacterial activity of L. paralimentarius DSM 13238 was significantly downregulated (P < 0.05), while of RGW1 was not affected by proteinase K pretreatment (Supplementary Figure 1B-D). With the coculture of RGW1 (108 CFU/mL) and E. coli F5 (107 CFU/mL), the growth of both E. coli F5 and RGW1 was inhibited at 8 h, this inhibition was maintained in E. coli F5 but not in RGW1 after 24 h of coculture (Figure 2G).
3.3. Immunomodulatory effect of RGW1 in inflammatory mice challenged by LPS
Mice were given RGW1 or its cell-free supernatant (RCS group) before undergoing LPS induced inflammation (Figure 3A). The intake and weight were not significantly different from day 1 to 7 among control, RGW1 and RCS groups, but the mouse weights significantly decreased one day after LPS challenge (Figure 3B). We measured the cytokines in the serum (Figure 3C) and found that administration of RCS significantly lowered the level of the pro-inflammatory cytokine TNF-α (P < 0.05), whereas administration of RGW1 significantly elevated the levels of the anti-inflammatory cytokines TGF-β (P < 0.05) and IL-10 (P < 0.05). According to the histological analysis of colon tissues (Figure 3D), administration of RGW1 and RCS reduced the occurrence of intestinal injury, such as a thinner mucosa, deformed crypts and swelled and shed villi. Additionally, administration of RGW1 significantly increased the villus height, crypt depth, and the villus height/crypt depth ratio of colon, while administration of RCS significantly increased the villus height and the villus height/crypt depth ratio of colon (Figure 3E). Both administration of RGW1 and RCS did not change the villus height, crypt depth and the villus height/crypt depth ratio of cecum Figure 3F).
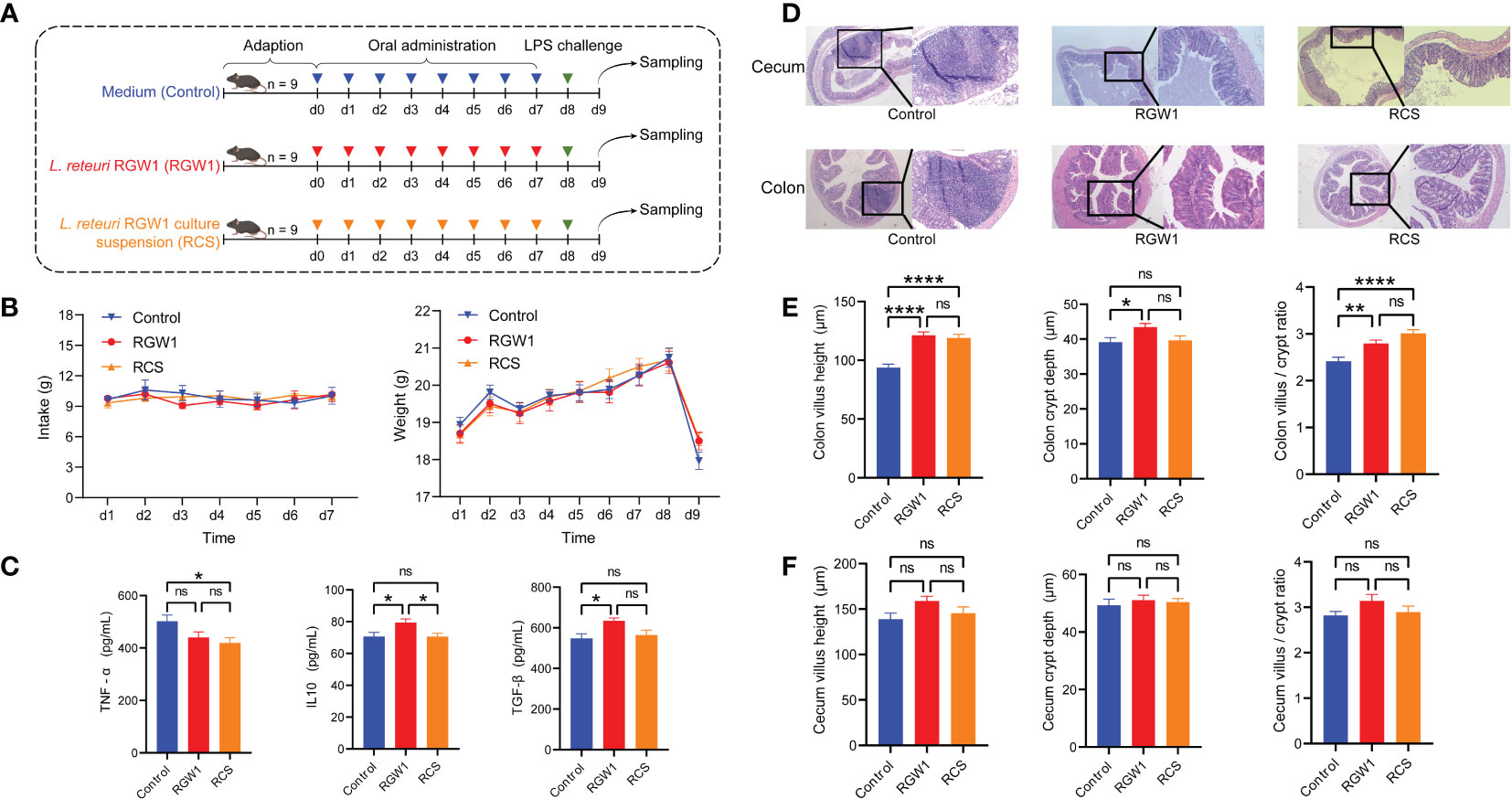
Figure 3 Immunomodulatory effects of L. reuteri RGW1 on inflammation in mice challenged with LPS. (A) Experimental schedule for LPS challenge and administration of L. reuteri RGW1 and RCS. LPS was intraperitoneally injected on day 8, L. reuteri RGW1 or RCS was orally administrated 7 times. (B) The intake and weight of mice. (C) The levels of serum IFN-α, TGF-β and IL-10. (D) Representative images of the colon stained with H&E. (E, F) Intestinal morphology analysis based on measurements of the villus height, crypt depth, and the ratio of villus height to crypt depth (VH/CD) in colon or cecum tissues. Values are expressed as means ± SEM. *P < 0.05; **P < 0.01; ****P < 0.0001; ns, not significant.
3.4. Genomic features and putative probiotic properties of RGW1
3.4.1. Genomic features and functional class of RGW1
This draft genome of RGW1 is 2,090,361 bp in size with 39.43% GC content, and the numbers of predicted coding gene, tRNA, and 5S, 16S and 23S rRNA are 2098, 64, and 5, 1, 1, respectively (Figure 4A). Genome sequencing did not identify any plasmid sequences. Annotation using the NR database found that L. reuteri accounted for 94.9% of all organisms that showed sequence homology with the RGW1 strain. Annotation using KEGG database identified a total of 1970 genes in the genome of RGW1, most of which involved in membrane transport, translation, nucleotide, cofactors and vitamins, carbohydrates, and amino acid metabolism (Figure 4B). Annotation of the genome of RGW1 using COG database predicted 1417 genes, which were assigned to 22 categories of orthologous groups (COG) classes, the “translation, ribosomal structure and biogenesis” group had the highest number of genes, followed by “amino acid transport and metabolism”, “general function prediction only”, and “carbohydrate transport and metabolism” (Figure 4C). A total of 69 predicted carbohydrate-active enzymes (CAZymes) were identified in the RGW1 genome and divided into three subgroups, including 31 genes for glycoside hydrolases, 28 genes for glycosyltransferases, and 13 genes for carbohydrate-binding molecules (Figure 4D). Additionally, PHI and VFDB data annotation of the RGW1 genome did not reveal any virulence genes.
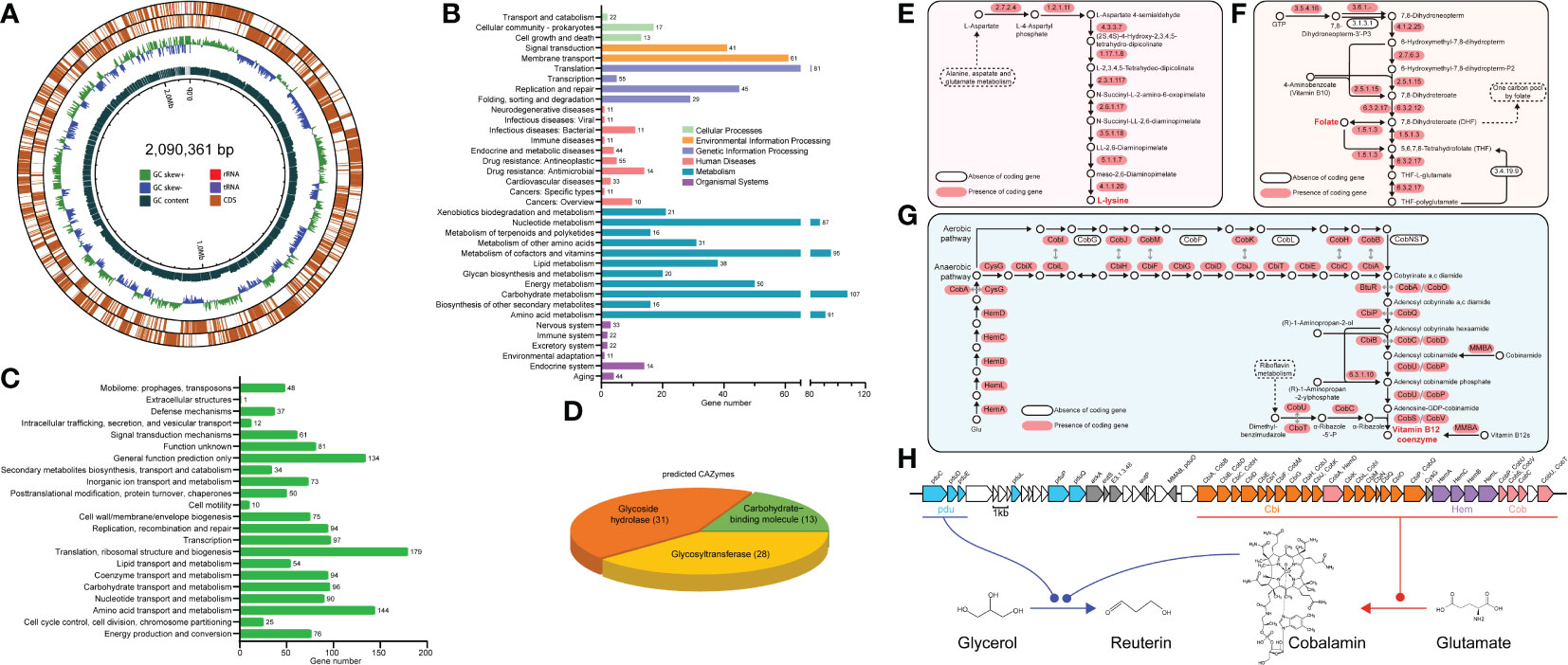
Figure 4 General genome features of L. reuteri RGW1. (A) Circular draft genome maps of the L. reuteri RGW1 chromosome. From outside to the inside: forward CDSs, reverse CDSs, GC skew, GC content. (B) Histogram representing the number of genes included in each KEGG pathway. (C) COG classes of predicted protein sequences of L. reuteri RGW1. (D) Predicted CAZymes of L. reuteri RGW1. (E) L-lysine biosynthesis pathway. (F) Folate biosynthesis pathway. (G) Cobalamin biosynthesis pathway, the sequence homology and functional equivalency between aerobic and anaerobic pathway enzymes are indicated by gray arrows. (E-G) were adapted from KEGG pathway. (H) pdu-cbi-hem-cob gene cluster and the proposed reuterin biosynthesis pathway in L. reuteri RGW1.
3.4.2. Specific metabolism of RGW1
Genomic analysis revealed the genetic potential of RGW1 for de novo biosynthesis of L-lysine (Figure 4E), one of the essential amino acids required in the diets of animals, as well as folate (vitamin B9) (Figure 4F), and cobalamin (vitamin B12) (Figure 4G). Gene locus information is in Supplementary Table S3-5. The genes (Lr_GM000502 - Lr_GM000531) encoding for cobalamin biosynthesis located in cbi-hem-cob cluster (Figure 4H). Moreover, three propanediol dehydratase subunit (pduC, pduD and pduE) encoding genes (Lr_GM000480, Lr_GM000481, Lr_GM000482) were identified (Figure 4H), which have been reported to be the glycerol utilization gene candidates in the pdu operon and are involved in reuterin (a mixture of different forms of 3-hydroxypropionaldehyde, 3-HPA) biosynthesis, when cobalamin is present as a cofactor. Moreover, the pdu operon was located adjacent to the cbi-hem-cob gene cluster (Figure 4H).
3.4.3. Putative probiotic feature
The coding genes involved in acid and bile tolerance, adherence, antimicrobial activity, and intrinsic defense were identified (Table 1) by combining the functional predictions using distinct databases as described in methods. In the genome of RGW1, amino acid/H+ (Lr_GM000677, Lr_GM002032), tripetide/H+ (Lr_GM000977), gluconate/H+ (Lr_GM000206, Lr_GM000385), fucose/H+ (Lr_GM001341), and sodium-glutamate/H+ (Lr_GM000634) symporters that cotransport proton and other compounds across the cellular membrane were identified. The genes coding for the Na+/H+ antiporter (Lr_GM000006, Lr_GM000042, Lr_GM001658, Lr_GM001822, Lr_GM001939) and CPA2 family monovalent cation/H+ antiporter (Lr_GM000790), which exchange sodium ions and protons across the membrane were also documented. F1/F0 ATP synthase is essential to maintain intracellular pH homeostasis, and the genes coding for F1/F0 ATP synthase subunits (Lr_GM001315, Lr_GM001316, Lr_GM001317, Lr_GM001318, Lr_GM001319, Lr_GM001320, Lr_GM001321, Lr_GM001322) were identified in the genome of RGW1. RGW1 harbored the genes that encode MFS transporter (Lr_GM000643, Lr_GM000660, Lr_GM000723, Lr_GM000804, Lr_GM000837, Lr_GM001128, Lr_GM001144, Lr_GM001447, Lr_GM001948, Lr_GM001976, Lr_GM001977) and choloylglycine hydrolase (Lr_GM001633), which are linked to bile tolerance. RGW1 also carries the genes encoding NADH peroxidase (Lr_GM001770, Lr_GM001291), thioredoxin (Lr_GM000347, Lr_GM001596), and thioredoxin reductase (Lr_GM000748, Lr_GM000787, Lr_GM000903, Lr_GM001006), which are associated with antioxidant capacity. RGW1 also carries various heat stress protein-coding genes, including heat shock protein Hsp33 (Lr_GM002012), heat shock protein Hsp60 (Lr_GM001766, Lr_GM001767), molecular chaperone DnaJ (Lr_GM000594), molecular chaperone DnaK (Lr_GM000595), and nucleotide exchange factor GrpE (Lr_GM000596).
3.4.4. Molecular determinants of host-bacteria interactions
A total of 19 surface protein-coding genes were identified from the genome of RGW1 (Table 2). RGW1 contains the genes encoding cell surface proteins involved in adhesion, including CBS domain-containing protein (Lr_GM001233), elongation factors (Lr_GM000593, Lr_GM000622, Lr_GM000958, Lr_GM001688, Lr_GM001713, Lr_GM001782, Lr_GM001875, Lr_GM001965), LPxTG-motif cell wall anchor domain proteins (Lr_GM000196, Lr_GM000922, Lr_GM000933, Lr_GM001344, Lr_GM001561, Lr_GM001672, Lr_GM001699), fibronectin/fibrinogen-binding protein (Lr_GM001088), and peptidoglycan-binding proteins (Lr_GM001332, Lr_GM001975). These surface proteins are involved in colonization by binding to epithelia, epithelial cells, or mucus. RGW1 encodes genes involved in lipoteichoic acid (LTA), peptidoglycan (PG), and exopolysaccharide (EPS) biosynthesis, including LTA biosynthesis proteins (Lr_GM000913, Lr_GM001244, Lr_GM001293), LTA exportation protein (Lr_GM001113), LTA acyltransferase (Lr_GM001242), PG biosynthesis regulator (Lr_GM001437), and EPS biosynthesis proteins (Lr_GM000802, Lr_GM001532, Lr_GM001751), which can activate different pattern recognition receptors present in the epithelial cells and immune cells.
4. Discussion
In this study, one new L. reteuri strain named RGW1 was isolated from calf feces and identified based on 16S rRNA gene sequencing. Combining genomic analysis with experimental studies, we revealed a variety of probiotic effects of L. reteuri RGW1, including resistance to harsh environments, anti-pathogenic activity, adhesion to host cells and immunomodulation. We also identified numerous probiotic-associated genes, which could at least partially explain its probiotic properties. Our study extends the probiotic library with the bovine origins and provides genotypic clues for the probiotic characteristics of RGW1.
4.1. Safety assessment
Antibiotic susceptibility and virulence factors are important indicators of safety. Probiotics must not participate in undesirable antibiotic resistance gene transfer cascades in vivo. Thus, we surveyed the susceptibility of RGW1 to seven antibiotics: ampicillin, amoxicillin, cefazolin, and penicillin G are inhibitors of cell wall synthesis, and chloramphenicol, erythromycin and tetracycline are inhibitors of protein synthesis (Charteris et al., 1998). The virulence genes tested in our study encode (1) adherent virulence factors (esp, efaAfs, asa and ace) which aid the bacterium in adhesion and evasion of the host cell; (2) secretory virulence factors (gelE, cylA, hyl) which help the bacterium wade through the innate and adaptive immune response mounted within the host; and (3) decarboxylase enzymes (hdc, tdc and odc), which act as biogenic amine production factors involved in biogenic amine production. The overproduction of biogenic amines has been proved to be toxic, causing symptoms such as diarrhea, food poisoning, vomiting, sweating or tachycardia (Özogul and Hamed, 2018; Wójcik et al., 2021). RGW1 is not resistant to any antibiotics tested in our study and contains none of the virulence genes tested, these results demonstrate that RGW1 is safe for application.
4.2. Tolerance
Growth at a pH of 3.0 is usually considered optimal for a successful probiotic to survive, due to the buffering impact of ingested food that raises the pH in the stomach environment to 3.0 (Liu et al., 2013). L. reteuri RGW1 showed a survivability of more than 100% at pH 3.0. Such property coincides with the genome of RGW1, which contains a series of acid resistance genes coding for symporters, Na/H+ antiporters and the F1/F0 ATP synthase subunits. The first two protein familys play important roles in transporting proton, and the latter has a synthase function in the ATP synthesis coupled with proton transport. Previous studies have demonstrated that overproduction of ATP synthase subunit beta is associated with acid resistance in L. pentosus (Pérez Montoro et al., 2018b). In addition to the stomach environment, it is also essential for a probiotic to display tolerance to small intestinal transit, due to the antimicrobial impact of bile salt (Charteris et al., 1998). The MFS (major facilitator superfamily) transporter is a membrane transporter superfamily that can facilitate the movement of small solutes on the cell membrane and expel intracellular antibiotics (Pao et al., 1998; Langton et al., 2005). Three MFS genes have been reported to be bile salt exposure associated genes (Ruiz et al., 2012). Choloylglycine hydrolase (bile salt hydrolase) is essential for bile acid homeostasis in the host and represents a vital contribution of the gut microbiome to host health. Bile salt hydrolase has been reported in Lactobacillus (Long et al., 2017). In our study, RGW1 displayed a degree of tolerance to 0.3% bile salt, and this capacity can be explained by the presence of genes coding for MFS transporters and one bile salt hydrolase in the genome of RGW1. In addition, L. reteuri RGW1 survived well in the simulated gastric fluid. These in vitro results with genomic analysis indicate the potential of L. reteuri RGW1 in nutritional and clinical applications.
4.3. Adhesion
Adhesion of probiotics to the host epithelium is thought to increase host-bacterial interactions, which are linked to both competitive exclusion of pathogens and immunomodulation mechanisms (Hymes et al., 2016; Ku et al., 2016). Previous study has revealed the correlation between hydrophobicity and adhesion of probiotic bacteria (Pan et al., 2006). In our study, the high hydrophobic index of RGW1 was consistent with its strong adhesion to Caco-2 cells. L. reteuri RGW1 encodes most of the known adhesion factors, including elongation factors, an LPxTG-motif protein, fibronectin/fibrinogen-binding proteins, peptidoglycan-binding proteins, and enolase. Muscariello et al. (2020) reviewed the mechanisms by which these adhesion proteins in Lactobacillus are involved in adhesion to the host cell, mucin, or the intestinal epithelia. One possible mechanism for adherence is through binding to the mucus layer. Elongation factors (EFs) function in “translation, ribosomal structure and biogenesis” according to the GO function annotation; however, some EF proteins have been shown to be involved in bacterial adhesion. For example, EF-Tu was demonstrated to be a mucin adhesion factor identified at the surface of some Lactobacillus species (Dhanani and Bagchi, 2013; Nishiyama et al., 2013; Du et al., 2022); furthermore, EF-GreA was reported to be an adhesion biomarker in L. pentosus using immobilized mucin model (Pérez Montoro et al., 2018a). In addition to EF-Tu and EF-GreA, other EF coding genes were identified in the RGW1 genome. LPxTG-motif proteins are a type of precursor protein that carries a conserved LPxTG-motif at the C-terminus, these proteins were reported to promote the adhesion of L. rhamnosus GG to the intestinal mucosa (von Ossowski et al., 2011) and the intestinal survival rate and adhesion characteristics of L. reuteri (Xu et al., 2022). In addition to adhering to the mucus layer, another possible adherent mechanism is binding to a variety of proteins present in the extracellular matrix (ECM) of intestinal epithelial cells, such as fibronectin, collagen, and peptidoglycan. RGW1 carries one gene coding for a fibronectin/fibrinogen-binding protein and two genes coding for peptidoglycan-binding proteins. Besides, the genome of RGW1 contains the gene coding for enolase, which was shown to mediate the adhesion of L. casei BL23 to fibronectin and collagen (Muñoz-Provencio et al., 2011) and L. plantarum to collagen (Salzillo et al., 2015). Enolase is also involved in the immunostimulation of Caco-2 cells and biofilm development (Vastano et al., 2016). The presence of these protein coding genes in the RGW1 genome helps to explain the strong adhesion of this isolate to Caco-2 cells. Because the adhesion can be host-dependent, further testing will be required to assess more RGW1 adhesion characteristics of with different host.
4.4. Inhibition of pathogens
The anti-pathogenic property of probiotics is an important benefit. Here, the anti-pathogenic effects of RGW1 were tested against enterotoxigenic E. coli F5, one of the pathogens that causes diarrhea in calves. L. reteuri RGW1 showed inhibitory activity against E. coli F5. Combining with the properties that RGW1 could adhere to Caco-2 cell, this result suggested the potential probiotic characteristics in vivo of enhancing colonization resistance thus preventing animal severe diarrhea caused by pathogen. Among the treatments to the RGW1 suspension that we tested, only the adjustment of pH reduced its inhibitory activity. These results indicated that the antimicrobial compounds of RGW1 are tolerant to heat therefore are not be peptides or proteins. Reuterin is a well-known antimicrobial compound, which can be produced and excreted by most L. reuteri strains. The synthesis of reuterin is mediated by glycerol dehydratase, which catalyzes the conversion of glycerol to 3-HPA. Previous study revealed that propanediol dehydratase coding genes (pduC, pduD, pduE) act as glycerol utilization candidates and encode the glycerol dehydratase activity in L. reuteri JCM 1112(T) (Morita et al., 2008). Both propanediol and glycerol dehydratase enzymes require cobalamin as cofactor. Cobalamin biosynthesis requires more than 30 enzymatic steps, via aerobic or anaerobic pathways. Herein, we found that RGW1 possesses all of the genes involved in the anaerobic pathways, while cobG, cobF and cobNST involved in the aerobic pathways were absent. These results indicated that RGW1 might synthesize cobalamin in an anaerobic environment rather than aerobically. The gene sets for cobalamin synthesis (cbi, cob, and hem) in RGW1 are located adjacent to the pdu operon, and the structure of this gene cluster probably reflects the requirement for glycerol dehydratase activity. Not surprisingly, the strong anti-pathogenic effects of L. reuteri strains were associated with the presence of the pdu-cbi-cob-hem gene cluster (Lee et al., 2017). Genomic analysis further supports the potential role of RGW1 reuterin in anti-microbiota. While RGW1 showed good anti E. coli F5 activity and the reuterin has a good antibacterial broad-spectrum. The anti-pathogen activities of RGW1 against both gram-positive and negative pathogen, such as Salmonella, Staphylococcus, Streptococcus, should be tested in the future, as well as the in vivo experiments will be conducted.
4.5. Immunomodulation
Feeding mice RGW1 or its cell free supernatant did not affect their weight or intake. We also investigated its immunomodulation properties using mouse module of inflammation induced by the gram-negative bacterial LPS, which is known to induce acute inflammatory responses in the host (Guijarro-Muñoz et al., 2014). Previous research reported that L. reuteri CRL1101 downregulated the levels of cytokines (IL-1β, IL-6 and TNF-α) in the sera of mice stimulated with LPS (Juarez et al., 2013), and L. reuteri ZJ617 and ZJ615 downregulated TNF-α in the sera of mice stimulated with LPS (Gao et al., 2016). Consistent with these findings, the metabolites of L. reuteri RGW1 exerted anti-inflammatory properties by downregulating TNF-α in the sera significantly, while live L. reuteri RGW1 downregulating TNF-α numerically. This phenomenon might be explained by that some compounds in the surface of live L. reuteri RGW1 could stimulate host, thus slightly altering the immune status. As evidenced by Spindler et al. (2022), who revealed that the gut commensals could also stimulate phylogenetically defined innate immune responses, including TNF-α expression.
Exopolysaccharide (EPS) is a self-produced extracellular component for biofilm formation (Salas-Jara et al., 2016) that has anti-inflammatory activity (Ryan et al., 2019). Lipoteichoic acid (LTA) is the major microbe-associated molecular pattern of gram-positive bacteria and the agonist of Toll-like receptors (Claes et al., 2012). LTA of L. reuteri was reported to alleviate inflammatory responses, as evidenced by the altered levels of the inflammatory cytokine TNF-α, IL-6, and IL-10 (Lu et al., 2022). PG are more likely to activate NOD-like receptors (Wolf and Underhill, 2018). Both Toll-like and NOD-like receptors are pattern recognition receptors that carefully regulate downstream cytokine secretion, and are therefore associated with the regulation of intestinal immune conditioning and responses that limit bacterial burden and prevent excessive inflammatory responses (Abraham et al., 2022). The genes coding for EPS, LTA, and PG biosynthesis have been identified in the genome of RGW1, suggesting that RGW1 may alleviate inflammatory responses through these molecules. Folate (vitamin B9) is essential for several metabolic preprocesses involved in the transfer and formation of one-carbon (C1) units, and C1 metabolism is critical for purine and thymidylate synthesis, methylation reactions and the metabolism of several amino acids. Furthermore, folate synthesis and metabolism by some specific L. reuteri strains (such as L. reuteri 6475) were also demonstrated to contribute to the suppression of inflammation (Thomas et al., 2016; Röth et al., 2019). In the current study, all genes for de novo biosynthesis of folate in the presence of vitamin B10 were identified in the genome of RGW1, as a rare exception, vitamin B10 can be synthesized by the genera Lactococcus and Streptococcus (Rossi et al., 2011). These results indicate that RGW1 has the potential to biosynthesis folate in the gastrointestinal tract.
Taken together, the presence of these genes related to EPS, LTA, PG, and folate biosynthesis in the genome of RGW1 provides a clue to the underlying mechanisms of the anti-inflammatory effects of both RGW1 and its metabolites in LPS-reduced mice. Further studies will aim to explore the exact molecules participating in the immunomodulatory effect of RGW1, as well as these molecules associated probiotic properties in vivo, especially the regulation of gut microbiota will be explored.
5. Conclusion
We isolated a novel L. reuteri strain from healthy calf feces, and systematically characterized the probiotic properties and disclosed the draft genome. The strain RGW1 showed various probiotic properties including tolerance to acid and bile salt, survival in simulated gastric and intestinal fluid, adherence to Caco-2 cells, antipathogenic and immunomodulatory effects, and carried related genes related. In addition, RGW1 could de novo biosynthesize L-lysine, folate, cobalamin, and reuterin. This study provides a considerable insight into the probiotic functions of L. reuteri RGW1 and the corresponding molecular mechanisms, thereby contributing to its application as a probiotic in antipathogen and host health regulation.
Data availability statement
The data presented in the study are deposited in the NCBI repository, accession number PRJNA872372.
Ethics statement
The animal study was reviewed and approved by Animal Care and Use Committee of Zhejiang Provincial Hospital of Chinese Medicine (Hangzhou, China, 60136-38385).
Author contributions
JW designed the experiments. KH and WS performed the experiments. KH, BY, and JW analyzed the data. KH and JW wrote and revised the main manuscript. All authors contributed to the article and approved the submitted version.
Funding
This project was funded by the Project of Zhejiang Science and Technology Plan (2022C04038).
Acknowledgments
We would like to thank the members of the Experimental Teaching Center, College of Animal Sciences, and Bio-ultrastructure Analysis Lab., Agrobiology and Environmental Sciences, Zhejiang University, China for their assistance in assessing the antibacterial activity and imaging with scanning electron microscopy, respectively. We would also like to thank the members of the Animal Care and Use Committee of Zhejiang Provincial Hospital of Chinese Medicine, Hangzhou, China for their assistance in the mice feeding and samples collection.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcimb.2022.1086861/full#supplementary-material
References
Abraham, C., Abreu, M. T., Turner, J. R. (2022). Pattern recognition receptor signaling and cytokine networks in microbial defenses and regulation of intestinal barriers: Implications for inflammatory bowel disease. Gastroenterology 162 (6), 1602-1616. doi: 10.1053/j.gastro.2021.12.288
Aureli, P., Capurso, L., Castellazzi, A. M., Clerici, M., Giovannini, M., Morelli, L., et al. (2011). Probiotics and health: an evidence-based review. Pharmacol. Res. 63 (5), 366–376. doi: 10.1016/j.phrs.2011.02.006
Bankevich, A., Nurk, S., Antipov, D., Gurevich, A. A., Dvorkin, M., Kulikov, A. S., et al. (2012). SPAdes: A new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 19 (5), 455–477. doi: 10.1089/cmb.2012.0021
Besemer, J., Lomsadze, A., Borodovsky, M. (2001). GeneMarkS: A self-training method for prediction of gene starts in microbial genomes. implications for finding sequence motifs in regulatory regions. Nucleic Acids Res. 29 (12), 2607–2618. doi: 10.1093/nar/29.12.2607
Casarotti, S. N., Carneiro, B. M., Todorov, S. D., Nero, L. A., Rahal, P., Pena, A. L. B. (2017). In vitro assessment of safety and probiotic potential characteristics of lactobacillus strains isolated from water buffalo mozzarella cheese. Ann. Microbiol. 67 (4), 289–301. doi: 10.1007/s13213-017-1258-2
Charteris, W. P., Kelly, P. M., Morelli, L., Collins, J. K. (1998). Antibiotic susceptibility of potentially probiotic lactobacillus species. J. Food Protect 61 (12), 1636–1643. doi: 10.4315/0362-028x-61.12.1636
Claes, I. J. J., Segers, M. E., Verhoeven, T. L. A., Dusselier, M., Sels, B. F., De Keersmaecker, S. C. J., et al. (2012). Lipoteichoic acid is an important microbe-associated molecular pattern of lactobacillus rhamnosus GG. Microb. Cell Fact 11, 161. doi: 10.1186/1475-2859-11-161
Dhanani, A. S., Bagchi, T. (2013). The expression of adhesin EF-tu in response to mucin and its role in lactobacillus adhesion and competitive inhibition of enteropathogens to mucin. J. Appl. Microbiol. 115 (2), 546–554. doi: 10.1111/jam.12249
Duar, R. M., Frese, S. A., Lin, X. B., Fernando, S. C., Burkey, T. E., Tasseva, G., et al. (2017). Experimental evaluation of host adaptation of lactobacillus reuteri to different vertebrate species. Appl. Environ. Microb. 83 (12), e00132-17. doi: 10.1128/AEM.00132-17
Du, Y., Li, H., Shao, J., Wu, T., Xu, W., Hu, X., et al. (2022). Adhesion and colonization of the HC-2 in the intestine of are associated with bacterial surface proteins. Front. Microbiol. 13. doi: 10.3389/fmicb.2022.878874
Frese, S. A., Benson, A. K., Tannock, G. W., Loach, D. M., Kim, J., Zhang, M., et al. (2011). The evolution of host specialization in the vertebrate gut symbiont lactobacillus reuteri. PloS Genet. 7 (2), e1001314. doi: 10.1371/journal.pgen.1001314
Gagen, E. J., Denman, S. E., Padmanabha, J., Zadbuke, S., Al Jassim, R., Morrison, M., et al. (2010). Functional gene analysis suggests different acetogen populations in the bovine rumen and tammar wallaby forestomach. Appl. Environ. Microb. 76 (23), 7785–7795. doi: 10.1128/aem.01679-10
Gao, K., Liu, L., Dou, X., Wang, C., Liu, J., Zhang, W., et al. (2016). Doses lactobacillus reuteri depend on adhesive ability to modulate the intestinal immune response and metabolism in mice challenged with lipopolysaccharide. Sci. Rep. 6, 28332. doi: 10.1038/srep28332
Guijarro-Muñoz, I., Compte, M., Álvarez-Cienfuegos, A., Álvarez-Vallina, L., Sanz, L. (2014). Lipopolysaccharide activates toll-like receptor 4 (TLR4)-mediated NF-κB signaling pathway and proinflammatory response in human pericytes. J. Biol. Chem. 289 (4), 2457–2468. doi: 10.1074/jbc.M113.521161
Hill, C., Guarner, F., Reid, G., Gibson, G. R., Merenstein, D. J., Pot, B., et al. (2014). Expert consensus document. the international scientific association for probiotics and prebiotics consensus statement on the scope and appropriate use of the term probiotic. Nat. Rev. Gastroenterol. Hepatol. 11 (8), 506–514. doi: 10.1038/nrgastro.2014.66
Hymes, J. P., Johnson, B. R., Barrangou, R., Klaenhammer, T. R. (2016). Functional analysis of an s-Layer-Associated fibronectin-binding protein in lactobacillus acidophilus NCFM. Appl. Environ. Microb. 82 (9), 2676–2685. doi: 10.1128/AEM.00024-16
Jacobsen, C. N., Rosenfeldt Nielsen, V., Hayford, A. E., Møller, P. L., Michaelsen, K. F., Paerregaard, A., et al. (1999). Screening of probiotic activities of forty-seven strains of lactobacillus spp. by in vitro techniques and evaluation of the colonization ability of five selected strains in humans. Appl. Environ. Microb. 65 (11), 4949–4956. doi: 10.1128/AEM.65.11.4949-4956.1999
Juarez, G. E., Villena, J., Salva, S., de Valdez, G. F., Rodriguez, A. V. (2013). Lactobacillus reuteri CRL1101 beneficially modulate lipopolysaccharide-mediated inflammatory response in a mouse model of endotoxic shock. J. Funct. Foods 5 (4), 1761–1773. doi: 10.1016/j.jff.2013.08.002
Ku, S., Park, M. S., Ji, G. E., You, H. J. (2016). Review on bifidobacterium bifidum BGN4: Functionality and nutraceutical applications as a probiotic microorganism. Int. J. Mol. Sci. 17 (9), 1544. doi: 10.3390/ijms17091544
Lagesen, K., Hallin, P., Rødland, E. A., Staerfeldt, H.-H., Rognes, T., Ussery, D. W. (2007). RNAmmer: Consistent and rapid annotation of ribosomal RNA genes. Nucleic Acids Res. 35 (9), 3100–3108. doi: 10.1093/nar/gkm160
Langton, K. P., Henderson, P. J. F., Herbert, R. B. (2005). Antibiotic resistance: multidrug efflux proteins, a common transport mechanism? Nat. Prod Rep. 22 (4), 439–451. doi: 10.1039/b413734p
Lee, J.-Y., Han, G. G., Choi, J., Jin, G.-D., Kang, S.-K., Chae, B. J., et al. (2017). Pan-genomic approaches in lactobacillus reuteri as a porcine probiotic: Investigation of host adaptation and antipathogenic activity. Microb. Ecol. 74 (3), 709–721. doi: 10.1007/s00248-017-0977-z
Letunic, I., Bork, P. (2021). Interactive tree of life (iTOL) v5: an online tool for phylogenetic tree display and annotation. Nucleic Acids Res. 49 (W1), W293–W296. doi: 10.1093/nar/gkab301
Li, R., Li, Y., Kristiansen, K., Wang, J. (2008). SOAP: short oligonucleotide alignment program. Bioinformatics 24 (5), 713–714. doi: 10.1093/bioinformatics/btn025
Liu, W. J., Chen, Y. F., Kwok, L. Y., Li, M. H., Sun, T., Sun, C. L., et al. (2013). Preliminary selection for potential probiotic bifidobacterium isolated from subjects of different Chinese ethnic groups and evaluation of their fermentation and storage characteristics in bovine milk. J. Dairy Sci. 96 (11), 6807–6817. doi: 10.3168/jds.2013-6582
Long, S. L., Gahan, C. G. M., Joyce, S. A. (2017). Interactions between gut bacteria and bile in health and disease. Mol. Aspects Med. 56, 54–65. doi: 10.1016/j.mam.2017.06.002
Lowe, T. M., Eddy, S. R. (1997). tRNAscan-SE: A program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 25 (5), 955–964. doi: 10.1093/nar/25.5.955
Lu, Q., Guo, Y., Yang, G., Cui, L., Wu, Z., Zeng, X., et al. (2022). Structure and anti-inflammation potential of lipoteichoic acids isolated from strains. Foods 11 (11), 1610. doi: 10.3390/foods11111610
Morita, H., Toh, H., Fukuda, S., Horikawa, H., Oshima, K., Suzuki, T., et al. (2008). Comparative genome analysis of lactobacillus reuteri and lactobacillus fermentum reveal a genomic island for reuterin and cobalamin production. DNA Res. 15 (3), 151–161. doi: 10.1093/dnares/dsn009
Muñoz-Provencio, D., Pérez-Martínez, G., Monedero, V. (2011). Identification of surface proteins from lactobacillus casei BL23 able to bind fibronectin and collagen. Probiotics Antimicro 3 (1), 15–20. doi: 10.1007/s12602-011-9065-8
Muscariello, L., De Siena, B., Marasco, R. (2020). Lactobacillus cell surface proteins involved in interaction with mucus and extracellular matrix components. Curr. Microbiol. 77 (12), 3831–3841. doi: 10.1007/s00284-020-02243-5
Mu, Q., Tavella, V. J., Luo, X. M. (2018). Role of lactobacillus reuteri in human health and diseases. Front. Microbiol. 9. doi: 10.3389/fmicb.2018.00757
Nagpal, R., Kumar, A., Arora, S. (2010). In vitro probiotic potential of lactobacilli isolated from indigenous fermented milk products. Int. J. Probiotics Prebiotics 5 (2), 103–110.
Nagpal, R., Wang, S., Ahmadi, S., Hayes, J., Gagliano, J., Subashchandrabose, S., et al. (2018). Human-origin probiotic cocktail increases short-chain fatty acid production via modulation of mice and human gut microbiome. Sci. Rep. 8 (1), 12649. doi: 10.1038/s41598-018-30114-4
Nishiyama, K., Ochiai, A., Tsubokawa, D., Ishihara, K., Yamamoto, Y., Mukai, T. (2013). Identification and characterization of sulfated carbohydrate-binding protein from lactobacillus reuteri. PloS One 8 (12), e83703. doi: 10.1371/journal.pone.0083703
Oh, P. L., Benson, A. K., Peterson, D. A., Patil, P. B., Moriyama, E. N., Roos, S., et al. (2010). Diversification of the gut symbiont lactobacillus reuteri as a result of host-driven evolution. Isme J. 4 (3), 377–387. doi: 10.1038/ismej.2009.123
Özogul, F., Hamed, I. (2018). The importance of lactic acid bacteria for the prevention of bacterial growth and their biogenic amines formation: A review. Crit. Rev. Food Sci. Nutr. 58 (10), 1660–1670. doi: 10.1080/10408398.2016.1277972
Pan, W.-H., Li, P.-L., Liu, Z. (2006). The correlation between surface hydrophobicity and adherence of bifidobacterium strains from centenarians' faeces. Anaerobe 12 (3), 148–152. doi: 10.1016/j.anaerobe.2006.03.001
Pao, S. S., Paulsen, I. T., Saier, M. H. (1998). Major facilitator superfamily. Microbiol. Mol. Biol. Rev. 62 (1), 1-34. doi: 10.1128/MMBR.62.1.1-34.1998
Pérez Montoro, B., Benomar, N., Caballero Gómez, N., Ennahar, S., Horvatovich, P., Knapp, C. W., et al. (2018a). Proteomic analysis of lactobacillus pentosus for the identification of potential markers of adhesion and other probiotic features. Food Res. Int. 111, 58–66. doi: 10.1016/j.foodres.2018.04.072
Pérez Montoro, B., Benomar, N., Caballero Gómez, N., Ennahar, S., Horvatovich, P., Knapp, C. W., et al. (2018b). Proteomic analysis of lactobacillus pentosus for the identification of potential markers involved in acid resistance and their influence on other probiotic features. Food Microbiol. 72, 31–38. doi: 10.1016/j.fm.2017.11.006
Rossi, M., Amaretti, A., Raimondi, S. (2011). Folate production by probiotic bacteria. Nutrients 3 (1), 118–134. doi: 10.3390/nu3010118
Röth, D., Chiang, A. J., Hu, W., Gugiu, G. B., Morra, C. N., Versalovic, J., et al. (2019). Two-carbon folate cycle of commensal lactobacillus reuteri 6475 gives rise to immunomodulatory ethionine, a source for histone ethylation. FASEB J. 33 (3), 3536–3548. doi: 10.1096/fj.201801848R
Ruiz, L., Zomer, A., O'Connell-Motherway, M., van Sinderen, D., Margolles, A. (2012). Discovering novel bile protection systems in bifidobacterium breve UCC2003 through functional genomics. Appl. Environ. Microb. 78 (4), 1123–1131. doi: 10.1128/AEM.06060-11
Ryan, P. M., Stolte, E. H., London, L. E. E., Wells, J. M., Long, S. L., Joyce, S. A., et al. (2019). Lactobacillus mucosae DPC 6426 as a bile-modifying and immunomodulatory microbe. BMC Microbiol. 19 (1), 33. doi: 10.1186/s12866-019-1403-0
Salas-Jara, M. J., Ilabaca, A., Vega, M., García, A. (2016). Biofilm forming lactobacillus: New challenges for the development of probiotics. Microorganisms 4 (3), 35. doi: 10.3390/microorganisms4030035
Salzillo, M., Vastano, V., Capri, U., Muscariello, L., Sacco, M., Marasco, R. (2015). Identification and characterization of enolase as a collagen-binding protein in lactobacillus plantarum. J. Basic Microb. 55 (7), 890–897. doi: 10.1002/jobm.201400942
Simpson, J. T., Wong, K., Jackman, S. D., Schein, J. E., Jones, S. J. M., Birol, I. (2009). ABySS: A parallel assembler for short read sequence data. Genome Res. 19 (6), 1117–1123. doi: 10.1101/gr.089532.108
Spindler, M. P., Siu, S., Mogno, I., Li, Z., Yang, C., Mehandru, S., et al. (2022). Human gut microbiota stimulate defined innate immune responses that vary from phylum to strain. Cell Host Microbe 30 (10), 1481–1498.e5. doi: 10.1016/j.chom.2022.08.009
Thomas, C. M., Saulnier, D. M. A., Spinler, J. K., Hemarajata, P., Gao, C., Jones, S. E., et al. (2016). FolC2-mediated folate metabolism contributes to suppression of inflammation by probiotic lactobacillus reuteri. Microbiologyopen 5 (5), 802–818. doi: 10.1002/mbo3.371
Vastano, V., Pagano, A., Fusco, A., Merola, G., Sacco, M., Donnarumma, G. (2016). The lactobacillus plantarum eno A1 enolase is involved in immunostimulation of caco-2 cells and in biofilm development. Adv. Exp. Med. Biol. 897, 33–44. doi: 10.1007/5584_2015_5009
von Ossowski, I., Satokari, R., Reunanen, J., Lebeer, S., De Keersmaecker, S. C. J., Vanderleyden, J., et al. (2011). Functional characterization of a mucus-specific LPXTG surface adhesin from probiotic lactobacillus rhamnosus GG. Appl. Environ. Microb. 77 (13), 4465–4472. doi: 10.1128/AEM.02497-10
Walter, J. (2008). Ecological role of lactobacilli in the gastrointestinal tract: Implications for fundamental and biomedical research. Appl. Environ. Microb. 74 (16), 4985–4996. doi: 10.1128/AEM.00753-08
Wójcik, W., Łukasiewicz, M., Puppel, K. (2021). Biogenic amines: formation, action and toxicity - a review. J. Sci. Food Agr. 101 (7), 2634–2640. doi: 10.1002/jsfa.10928
Wolf, A. J., Underhill, D. M. (2018). Peptidoglycan recognition by the innate immune system. Nat. Rev. Immunol. 18 (4), 243–254. doi: 10.1038/nri.2017.136
Wu, H., Xie, S., Miao, J., Li, Y., Wang, Z., Wang, M., et al. (2020). Lactobacillus reuteri maintains intestinal epithelial regeneration and repairs damaged intestinal mucosa. Gut. Microbes 11 (4), 997–1014. doi: 10.1080/19490976.2020.1734423
Xu, H., Lao, L., Ji, C., Lu, Q., Guo, Y., Pan, D., et al. (2022). Anti-inflammation and adhesion enhancement properties of the multifunctional LPxTG-motif surface protein derived from the lactobacillus reuteri DSM 8533. Mol. Immunol. 146, 38–45. doi: 10.1016/j.molimm.2022.04.004
Zárate, G., Chaia, A. P., González, S., Oliver, G. (2000). Viability and beta-galactosidase activity of dairy propionibacteria subjected to digestion by artificial gastric and intestinal fluids. J. Food Protect 63 (9), 1214–1221. doi: 10.4315/0362-028x-63.9.1214
Keywords: Limosilactobacillus reuteri, probiotic, genome analysis, antibacterial activity, immunomodulation
Citation: Huang K, Shi W, Yang B and Wang J (2023) The probiotic and immunomodulation effects of Limosilactobacillus reuteri RGW1 isolated from calf feces. Front. Cell. Infect. Microbiol. 12:1086861. doi: 10.3389/fcimb.2022.1086861
Received: 04 November 2022; Accepted: 20 December 2022;
Published: 12 January 2023.
Edited by:
Thuy Do, University of Leeds, United KingdomReviewed by:
Zhihong Sun, Inner Mongolia Agricultural University, ChinaPeng Sun, State Key Laboratory of Animal Nutrition (CAAS), China
Copyright © 2023 Huang, Shi, Yang and Wang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jiakun Wang, amlha3Vud2FuZ0B6anUuZWR1LmNu
 Kailang Huang
Kailang Huang Weibing Shi
Weibing Shi Bin Yang
Bin Yang Jiakun Wang
Jiakun Wang