
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Cell. Infect. Microbiol. , 30 November 2022
Sec. Bacteria and Host
Volume 12 - 2022 | https://doi.org/10.3389/fcimb.2022.1082809
This article is part of the Research Topic New Advances in Obligate Intracellular Bacteria: Pathogenesis and Host Interactions View all 9 articles
 HengDuan Zhang†
HengDuan Zhang† Jian Gao†
Jian Gao† Zu Ma†
Zu Ma† Yuan Liu
Yuan Liu Ge Wang
Ge Wang Qing Liu
Qing Liu YuTong Du
YuTong Du Dan Xing
Dan Xing ChunXiao Li
ChunXiao Li Teng Zhao
Teng Zhao YuTing Jiang
YuTing Jiang YanDe Dong
YanDe Dong XiaoXia Guo*
XiaoXia Guo* TongYan Zhao*
TongYan Zhao*Background: Wolbachia is gram-negative and common intracellular bacteria, which is maternally inherited endosymbionts and could expand their propagation in host populations by means of various manipulations. Recent reports reveal the natural infection of Wolbachia in Aedes Aegypti in Malaysia, India, Philippines, Thailand and the United States. At present, none of Wolbachia natural infection in Ae. aegypti has been reported in China.
Methods: A total of 480 Ae. aegypti adult mosquitoes were collected from October and November 2018 based on the results of previous investigations and the distribution of Ae. aegypti in Yunnan. Each individual sample was processed and screened for the presence of Wolbachia by PCR with wsp primers. Phylogenetic trees for the wsp gene was constructed using the neighbour-joining method with 1,000 bootstrap replicates, and the p-distance distribution model of molecular evolution was applied.
Results: 24 individual adult mosquito samples and 10 sample sites were positive for Wolbachia infection. The Wolbachia infection rate (IR) of each population ranged from 0 - 41.7%. The infection rate of group A alone was 0%-10%, the infection rate of group B alone was 0%-7.7%, and the infection rate of co-infection with A and B was 0-33.3%.
Conclusions: Wolbachia infection in wild Ae. aegypti in China is the first report based on PCR amplification of the Wolbachia wsp gene. The Wolbachia infection is 5%, and the wAlbA and wAlbB strains were found to be prevalent in the natural population of Ae. aegypti in Yunnan Province.
Wolbachia are common gram-negative intracellular bacteria that are maternally inherited endosymbionts and can propagate in host populations via various manipulations. Wolbachia was first discovered in the reproductive tissue of Culex pipiens pipiens in 1924 (Hertig, 1924) and later found in field-collected mosquitos (Carvajal et al., 2019). It is estimated to naturally occur in 66% of known insect species, including fruit flies, mosquitos, tsetse flies, bed bugs, ants, kissing bugs, and termites (Hilgenboecker et al., 2008; Werren et al., 2008; Beckmann et al., 2017).
The ecological interactions between Wolbachia and its eukaryotic host cells cover a wide range, including parasitism, symbiosis, and reciprocity (Werren et al., 2008; Hosokawa et al., 2010; Inácio da Silva et al., 2021). Because of the unique ability of Wolbachia to infect and manipulate the reproductive mode of the host, it has deeply influenced not only the ecology and evolution of its host but also the host’s reproductive biology through extensive symbiosis (Landmann, 2019; Ding et al., 2020). The effects of Wolbachia on the reproductive mode of the host mainly include the induction of cytoplasmic incompatibility (CI), parthenogenesis, male feminization, and male-killing increases in male mortality (Werren, 1997). In addition, Wolbachia induces CI during the fusion of male and female gametes (Ross et al., 2019), which not only suppresses mosquito populations but also inhibits the replication of viruses and parasites within mosquitoes, such as dengue virus (DENV), chikungunya virus (CHIKV), yellow fever virus (YFV), Zika virus (ZIKV) and Plasmodium parasites (Moreira et al., 2009; Bian et al., 2010; Walker et al., 2011; van den Hurk et al., 2012; Aliota et al., 2016; Ahmad et al., 2017).
Before 2014, natural Wolbachia infections were mainly concentrated in the Cx. pipiens complex and in Ae. Albopictus (Song She-Wu, 2002a; Song She-Wu, 2002b), and no natural infection was found in Ae. aegypti and Anopheles (Cui Bei-jin, 2015). However, natural Wolbachia infections were recently found in Anopheles gambiae in Burkina Faso, Mali, and areas of West Africa. In addition, Wolbachia infections in Ae. aegypti were found in Malaysia, India, the Philippines, Thailand, and the United States (Baldini et al., 2014; Teo et al., 2017; Hegde et al., 2018; Thongsripong et al., 2018; Balaji et al., 2019; Bennett et al., 2019; Carvajal et al., 2019; Kulkarni et al., 2019). At present, no natural Wolbachia infections in Ae. aegypti have been reported in China. Ae. aegypti is distributed in southern provinces in China, such as Hainan and Guangdong Provinces. Since the first discovery of Ae. aegypti in 2002 at Ruili Port in Yunnan Province, Ae. aegypti larvae and adults have been collected in 9 cities across Yunnan Province, indicating a rapid invasion and spread of this species (Shi et al., 2017). This rapid spread is highly concerning given that Ae. aegypti plays an important role in the transmission of the dengue virus and other mosquito-borne diseases and that Wolbachia infections in Ae. aegypti are linked to multiple invasions. Thus, this study investigated natural Wolbachia infection in this species in the field, especially in the border areas along Yunnan Province, which are the location of the invasion and spread of Ae. aegypti.
The common genes used to detect Wolbachia infection in a host species with polymerase chain reaction (PCR) include wsp (Wolbachia surface protein), ftsZ (filamenting temperature-sensitive mutant Z) and 16S rRNA. Genetic drift in ftsZ and 16S rRNA genes is low; thus, they can be used for stable amplification and classification of partial sequences with large differences at the species level. However, the highly variable marker gene wsp has a very similar genetic relationship, yet evolves faster than the former two and thus cannot be used to distinguish between species. Instead, it is easier to type closely related Wolbachia to determine the phylogenetic relationship of Wolbachia in greater detail (Braig et al., 1998; Zhou et al., 1998a). With the use of PCR and sequencing techniques, Wolbachia has been divided into 17 groups (A-Q) (Augustinos et al., 2011; Wang et al., 2014; Glowska et al., 2015; Wang et al., 2016). Groups A and B are typically capable of reproductive manipulation and are mainly distributed in arthropods (Werren et al., 1995; Ellegaard et al., 2013).
Wolbachia as a new technology to control mosquito and mosquito-borne diseases is more long-lasting and environmentally friendly than traditional insecticide methods. By releasing Wolbachia-infected mosquitoes into target areas, the control of mosquito and mosquito-borne diseases has been applied in the United States, Australia and Mexico (Hoffmann et al., 2011; O’Neill, 2018; Mains et al., 2019; Che-Mendoza et al., 2021; Utarini et al., 2021). Ae. aegypti is the main transmission vector of dengue fever in Yunnan Province which is one of the main provinces for dengue fever outbreaks. With the increase of mosquito resistance, it is important to develop new protection methods. It is important to realize the infection status and types of Wolbachia in major vectors in the region, in order to evaluate the application of Wolbachia in the future. Therefore, this study aimed to evaluate natural Wolbachia infections in Ae. aegypti collected from different sites in Yunnan Province using Wolbachia wsp gene amplification to detect and type the infection.
Yunnan Province is located in southwestern China that comprises 16 prefectures and 129 counties, and extends from 21°8′32″ to 29°15′8″N and 97°31′39″ to 106°11′47″E which shares a 4,060-km border with Laos, Vietnam, and Myanmar. The climate in most regions of this province is fairly mild in winter and rather cool in summer. The temperature information of the sampling site was collected as shown in Table 1.
Samples were collected in July-August 2017, October-November 2018, and September 2019 and 2020. Samples were collected in the months in which Ae. aegypti breed in Yunnan, and the sample collection periods were consecutive. As the samples of Ae. aegypti in 2017, 2019 and 2020 were negative and positive only in the samples of 2018.
In the present study, 19 populations were collected between October and November 2018 based on the results of previous investigations and the distribution of Ae. aegypti in Yunnan Province (Table 2). Larvae were collected and reared to adults. All larvae collected from one breeding container were stored into a bottle, a bottle represents a breeding container. These bottles were brought back to the laboratory for eclosion. According to the standard of the people’s Republic of China: NY/T1964.3-2010, adult mosquitoes and larvae were cultured in 26±1°C and 75±10% humidity in lab. Adult mosquitoes were fed with 8% sugar water, and larval were fed with powder which implement the national standard GB 14924.3-2010 (crude protein≥ 20%, crude fat content≥ 4%, crude fiber content ≤5%). All strains of Aedes aegypti in our insectary have fed with this diet all the time and Wolbachia free. Adult female mosquitoes after 7 days of eclosion were morphologically identified and COI identification and then for Wolbachia detection. According to the sample collection records, the different containers included waste tires, buckets, flowerpot, hydroponic plants and water basins. The distribution of Ae. aegypti in Yunnan is mainly concentrated in Xishuangbanna, Dehong and Lincang, although there are reports of distribution in other counties in Yunnan, but no local Ae. aegypti has been reported for several years. DNA isolation was conducted with the QIAamp® Fast DNA Tissue Kit (Qiagen, Dusseldorf, Germany) according to the manufacturer’s protocol, and all the DNA samples were stored at -80°C (Jiao Jun, 2022).
The DNA samples that were morphologically identified as Ae. aegypti were subjected to molecular identification of the COI gene to ensure that the experimental mosquitoes were Ae. aegypti (Bonacum et al., 2001).
Following the Wolbachia wsp gene classification method established by Zhou et al. (Zhou et al., 1998b), three pairs of diagnostic primers were selected to detect and identify Wolbachia. The downstream primer was 691R (AAAAATTAAACGCTACTCCA), and the upstream primers were 81F (TGGTCCAATAAGTGATGAAGAAAC); 328F (CCAGCAGATACTATTGCG) and 183F (AAGGAACCGAAGTTCATG). 81F is a universal primer that can amplify a fragment of approximately 590-632 bp in all known Wolbachia strains. 328F amplifies a fragment of approximately 380 bp that is specific to Wolbachia Group A, whereas 183F amplifies a fragment of approximately 501 bp that is specific to Wolbachia Group B. The volume of amplified PCR was 50 µL, including 25 µL of Taq DNA polymerase, 16 µL of double-distilled H2O, 2 µL of upstream and downstream primers at a concentration of 10 μmol/L, and 5 µL of the DNA template. The amplification conditions were 95°C predenaturation for 3 min, then [amplification at 94°C for 1 min, 55°C for 1 min, 72°C for 1 min] repeated for 35 cycles with a final extension of 72°C for 7 min. Five microlitres of the above PCR product was used in a 1.2% agarose gel electrophoresis, the results of which were examined under UV light, and the remainder was sent to Tianyi Biotechnology Company, Ltd. for sequencing.
All aligned Wolbachia sequences were compared with other sequences available in the GenBank database to determine the percentage identity using BLAST (http://blast.ncbi.nlm.nih.gov/Blast.cgi). The most similar sequences were downloaded for phylogenetic analysis. The selected sequences of Wolbachia strains (Table S1; Table S2; Table S3) and those obtained in the study then underwent multiple alignments using Clustal W 2.0.10. A phylogenetic tree for the wsp gene was constructed using the neighbour-joining method with 1,000 bootstrap replicates, and the p-distance distribution model of molecular evolution was applied.
Morphological identification and molecular identification of the COI gene determined that all 480 mosquitoes in this study were Ae. aegypti. All sequences have been submitted to the GenBank database with accession numbers ON637917 to ON637937.
Fragments of 500 bp and 380 bp were amplified with Wolbachia A- and Wolbachia B-specific primers, respectively, from each of the 30 samples from each Ae. aegypti population, confirming Wolbachia infection. The sequencing results showed that 24 (5%) adult mosquito samples were positive for Wolbachia infection (Table 3). These individuals were collected from 10 sites: 8 in Xishuangbanna Prefecture and 2 in Dehong Prefecture. The Wolbachia infection rate (IR) of each population ranged from 0% to 41.7%. The infection rate of Group A alone was 0–10%, the infection rate of Group B alone was 0–7.7%, and the rate of coinfection with Groups A and B was 0–33.3% (Figure 1; Table 3).
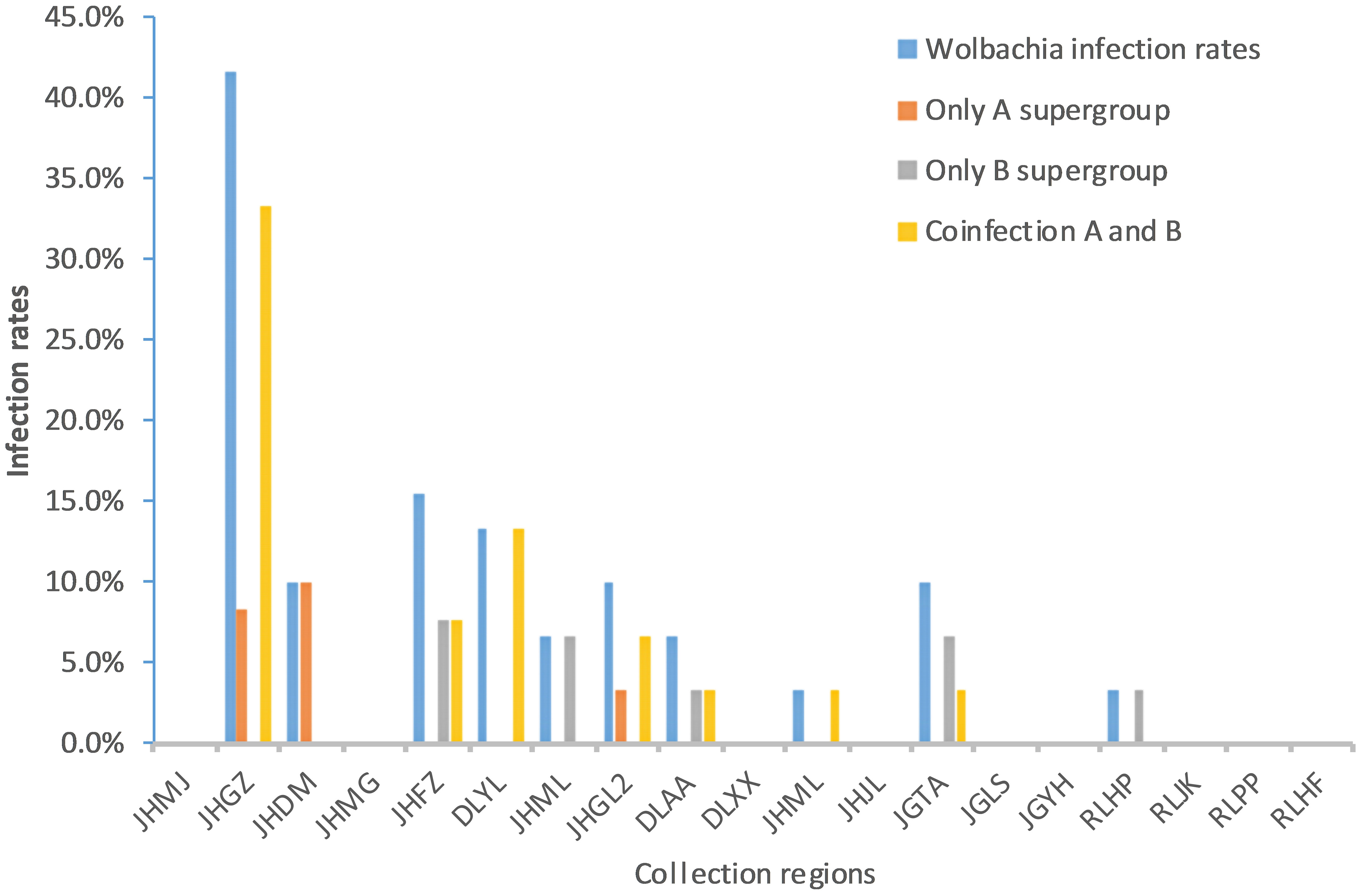
Figure 1 Comparison of Wolbachia infection rates among different collection regions in Yunnan Province. JHMJ、JHGZ、JHDM、JHMG、JHFZ、DLYL、JHML、JHGL2、DLAA、DLXX、JHML and JHJL belong to Xishuangbanna prefecture, Yunnan. JGTA、JGLS、JGYH、RLHP、RLJK、RLPP and RLHF belong to Dehong prefecture, Yunnan.
A phylogenetic tree was constructed using 44 sequences: 24 sequences collected in this study and 20 GenBank sequences (Figure 2). The phylogenetic analysis indicated that the outgroup (Rickettsia japonica) was independent of one branch. The target taxon was divided into two major branches with bootstrap values of 100%, which indicates that two major branches are plausible. The larger branches were further divided into four subbranches (bootstrap values of 100%, 62%, 99%, and 100%; thus, all four subbranches were plausible). Sixteen sequences (highlighted in red) were from the same node as the Wolbachia strains isolated in Ae. albopictus hosts and had a recent common ancestor. Samples YA10-4, YA17-13, YA8-28, YA5-3, YA14-21, and YA8-30 were from the same node and had high homology (bootstrap value of 100% indicating high support). The Wolbachia strains in these samples were relatively diverse in ancestry and may have shared a recent common ancestor with the Wolbachia strains found in Cx. quinquefasciatus, Ae. aegypti, Drosophila pseudoananassae, and Hofmannophila pseudospretella. Samples YA3-4 and YA9-25 formed a separate branch.
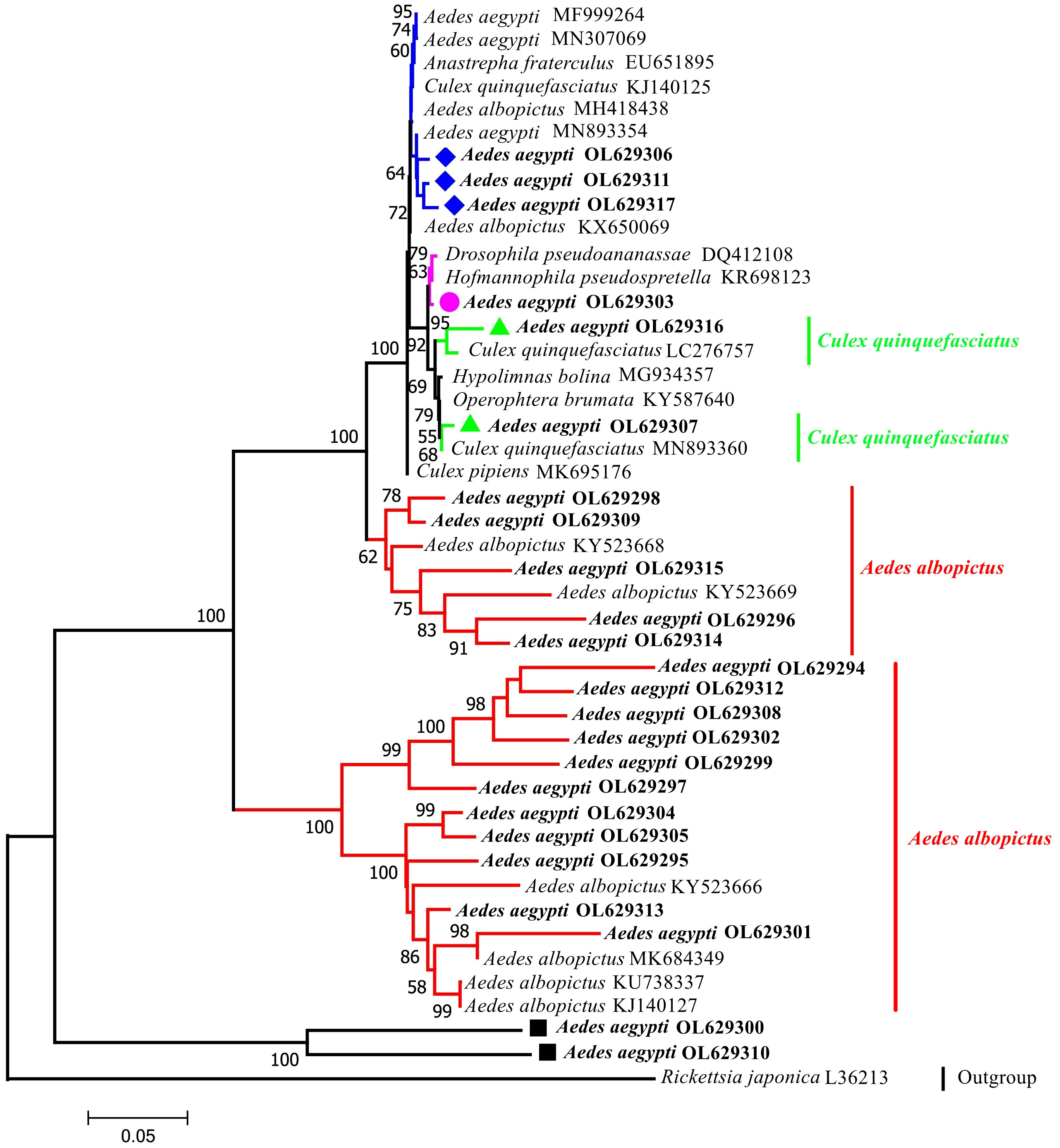
Figure 2 Phylogenetic tree analysis by the neighbor-joining method using the wsp gene (universal primers: 81F,691R). The tree contains 44 nucleotide sequences. 24 from this study; 19 from GenBank searches; 1 outgroup reference sequence (Rickettsia japonica) with Bootstrap values (1000 replicates) marked next to the branches. Taxa were tagged to obtain the host name of the Wolbachia strain and the ID in GenBank. the wsp sequences in this study all start with Aedes aegypti OL. OL629294 to OL629298 belong to JHGZ, Xishuangbanna prefecture. OL629299 to OL629301 belong to JHDM, Xishuangbanna prefecture. OL629302 and OL629303 belong to JHFZ, Xishuangbanna prefecture. OL629304 and OL629305 belong to JHML, Xishuangbanna prefecture. OL629306 and OL629307 belong to JHML, Xishuangbanna prefecture. OL629308 to OL629310 belong to DLAA, Xishuangbanna prefecture. OL629311 and OL629312 belong to DLXX, Xishuangbanna prefecture. OL629313 belongs to JHJL, Xishuangbanna prefecture. OL629314 to OL629316 belong to JGLS, Dehong prefecture. OL629317 belongs to RLJK, Dehong prefecture. The reference sequences used are shown in Table S1.
In addition, the sequences obtained with the Group A primers (wsp136F, wsp691R) were used to conduct a phylogenetic analysis. The phylogenetic analysis was performed by downloading multiple substrain reference sequences (wAlbA, wAegA, wPap, wMel, wRiv, and wMors) from Group A on the NCBI website. The results indicated that the phylogenetic tree had two branches (bootstrap value of 100%); the large branch yielded three subbranches. The Ae. aegypti sample from Yunnan Province (YA series), and the wAlbA and wAegA strains shared a recent common ancestor on one subbranch (Figure 3).

Figure 3 Phylogenetic tree analysis by the neighbor-joining method using the wsp gene (A supergroup primers: 136F,691R). The tree contains 30 nucleotide sequences. 17 from this study and 13 from GenBank searches with Bootstrap values (1000 replicates) marked next to the branches. Taxa were tagged to obtain the host name of the Wolbachia strain and the ID in GenBank. the wsp sequences in this study all start with Aedes aegypti OL. OL629258 to OL629262 belong to JHGZ, Xishuangbanna prefecture. OL629263 to OL629265 belong to JHDM, Xishuangbanna prefecture. OL629266 belongs to JHFZ, Xishuangbanna prefecture. OL629267 and OL629268 belong to JHML, Xishuangbanna prefecture. OL629269 to OL629271 belong to DLAA, Xishuangbanna prefecture. OL629272 belongs to DLXX, Xishuangbanna prefecture. OL629273 belongs to JHJL, Xishuangbanna prefecture. OL629274 belongs to JGLS, Dehong prefecture. The reference sequences used are shown in Table S2.
In addition, the sequences obtained with the Group B primers (wsp183F, wsp691R) were used to conduct a phylogenetic analysis. The phylogenetic analysis of multiple substrain reference sequences of Group B (wAlbB, wtauFJ1, wPip, wPana, wBeph, wBeva_B, Wma, wBani, wDei, WcauB) downloaded from the NCBI website indicated that among the Ae. aegypti samples from Yunnan Province (YA series), 15 sequences (78.9%) were from the same node as the wAlbB strain and shared a recent common ancestor with the Wolbachia strain found in Ae. albopictus. In Clade 2, samples YA8-30-B, YA14-21-B and YA5-3-B were related to the wtauFJ1, wPip, and wPana strains, respectively, and clustered into a single strain. Sample YA2-2-B represented a separate strain (Figure 4).

Figure 4 Phylogenetic tree analysis by the neighbor-joining method using the wsp gene (B supergroup primers: 183F,691R). The tree contains 34 nucleotide sequences. 19 from this study and 15from GenBank searches with Bootstrap values (1000 replicates) marked next to the branches. Taxa were tagged to obtain the host name of the Wolbachia strain and the ID in GenBank. the wsp sequences in this study all start with Aedes aegypti OL. OL629275 to OL629278 belong to JHGZ, Xishuangbanna prefecture. OL629279 and OL629280 belong to JHFZ, Xishuangbanna prefecture. OL629281 and OL629282 belongs to JHML, Xishuangbanna prefecture. OL629283 and OL629284 belong to JHGL2, Xishuangbanna prefecture. OL629285 and OL629286 belong to DLAA, Xishuangbanna prefecture. OL629287 and OL629288 belong to DLXX, Xishuangbanna prefecture. OL629289 belongs to JHJL, Xishuangbanna prefecture. OL629290 to OL629292 belong to JGLS, Dehong prefecture.OL629293 belongs to RLJK, Dehong prefecture. The reference sequences used are shown in TableS3.
This study also recorded the temperature in the sampled areas in 2018 to facilitate subsequent analysis. The diurnal temperature difference in Dehong Prefecture and Xishuangbanna Prefecture was in the range of 15–20°C. Except for Menghai County (MH), the average high temperature was above 26°C and the highest temperatures were above 28°C. Data were provided by the Global Weather Network (www.tianqi.com) (Table 1).
In this study, a heatmap of sequence similarity was generated based on the wsp sequences obtained from the universal primers (Figure 5). The heatmap included Wolbachia sequences from four mosquito species and was divided into the Clade I:B supergroup and Clade II:A supergroup. In the Clade I:B supergroup, the sequences identified in this study were highly homologous to those of Wolbachia found in Ae. aegypti in Malaysia (ID: MN893354) (Wong et al., 2020) and India (ID: MN307069 and MF999264). These sequences were not comparable to the Wolbachia found in Ae. aegypti from the United States (Hegde et al., 2018; Kulkarni et al., 2019); thus, this comparison is not presented on the heatmap. The sequences were also highly homologous with those found in Cx. pipiens quinquefasciatus, Cx. pipiens and Ae. albopictus, but the homology with those found in Ae. albopictus was more easily detected. In Clade II:A supergroup, all sequences had high homology with the Wolbachia found in Ae. albopictus.
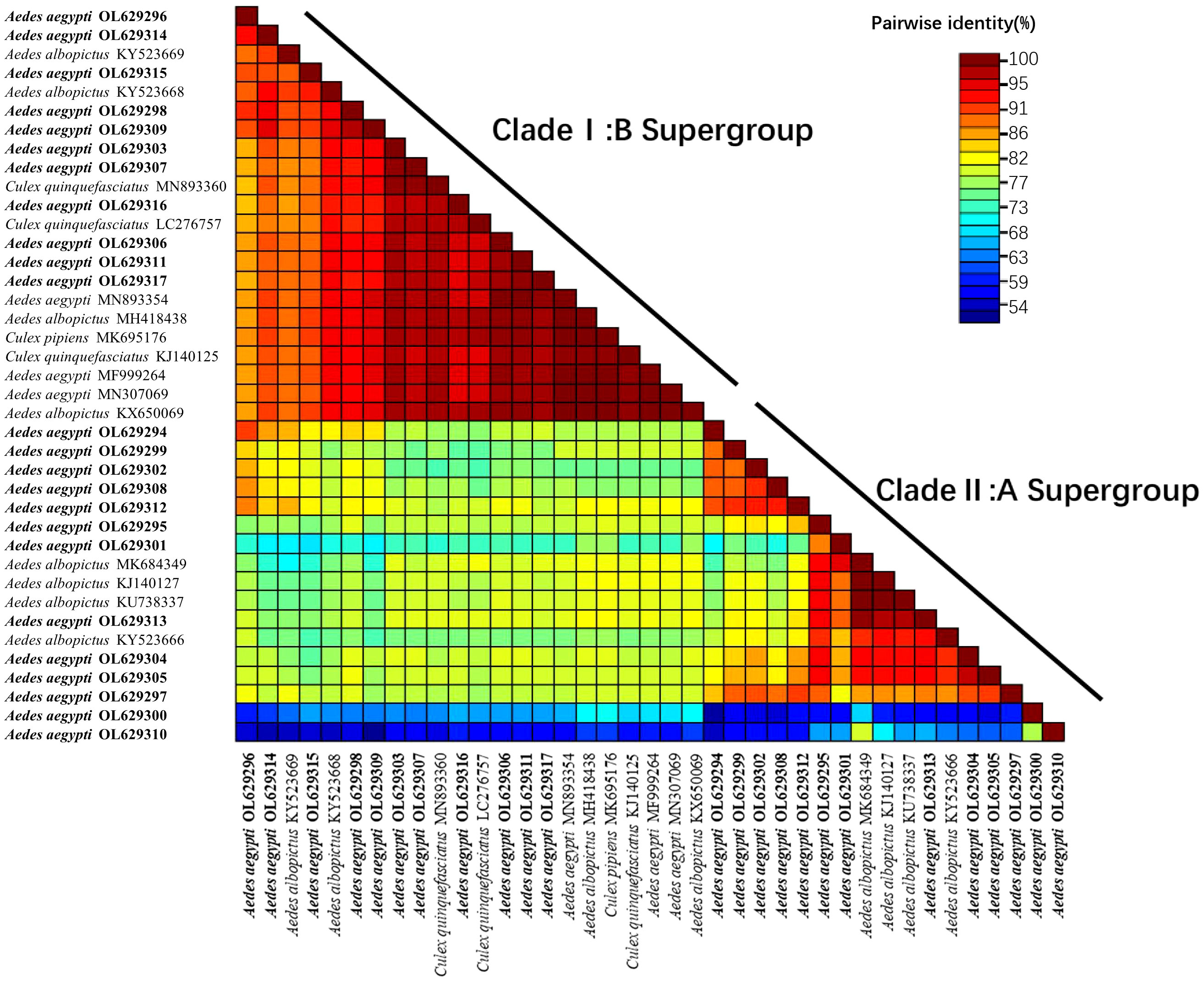
Figure 5 Heat map of sequence similarity between sequences obtained using the wsp gene (universal primers: w81F, w691R) and Wolbachia sequences of different mosquito species. The heat map contains 39 nucleotide sequences (24 from this study; 15 from GenBank search), the 15 sequences from GenBank include: 3 from Ae. aegypti, 8 from Aedes albopictus, 3 from Cx.pipiens quinquefasciatus and 1 from Cx pipiens. The number of reference sequences for these four mosquitoes was chosen based on random selection of sequences with higher homology during homology matching on NCBI. Clustal W alignment were performed for each unique pair of sequences, pairwise similarity scores were calculated, and a color-coded matrix of these scores is displayed. The wsp sequences in this study all start with Aedes aegypti OL. OL629294 to OL629298 belong to JHGZ, Xishuangbanna prefecture. OL629299 to OL629301 belong to JHDM, Xishuangbanna prefecture. OL629302 and OL629303 belong to JHFZ, Xishuangbanna prefecture. OL629304 and OL629305 belong to JHML, Xishuangbanna prefecture. OL629306 and OL629307 belong to JHML, Xishuangbanna prefecture. OL629308 to OL629310 belong to DLAA, Xishuangbanna prefecture. OL629311 and OL629312 belong to DLXX, Xishuangbanna prefecture. OL629313 belongs to JHJL, Xishuangbanna prefecture. OL629314 to OL629316 belong to JGLS, Dehong prefecture. OL629317 belongs to RLJK, Dehong prefecture.
This study is the first to report that populations of Ae. aegypti in Yunnan Province, China were naturally infected with Wolbachia. The average infection rate of Wolbachia in Ae. aegypti populations was 5%, higher than that of Florida (reported at 4.3%) and lower than that of the Philippines (11%) (Carvajal et al., 2019; Kulkarni et al., 2019). Compared with the infection rate in Ae. albopictus and Cx. pipiens, the infection rate of Wolbachia in Ae. aegypti was lower, which may be related to the environmental temperatures and lower density of Wolbachia in Ae. aegypti in the wild. Densities of Wolbachia in Ae. albopictus tended to decrease with increasing temperature, and Wolbachia endosymbionts could be removed from the host by exposure to heat or antibiotics (Hermans et al., 2001; Lau et al., 2020). In China, Ae. aegypti is more concentrated in tropical and subtropical regions than Ae. albopictus and Cx. spp., and the temperature of their habitat was higher than that of Ae. albopictus and Cx. spp. Rearing Wolbachia-infected larvae at 26-37°C reduced the rates of cytoplasmic incompatibility and dramatically decreased the density of Wolbachia in adult mosquitoes. Experiments on the response of Ae. aegypti infected with Wolbachia to cyclical heat stress have suggested that the likelihood of Wolbachia invasion and persistence in populations depends on interactions with environmental conditions, particularly the exposure of larvae to frequent temperature fluctuations and extremes (Ross et al., 2017). In 2018, the average highs and highest temperatures in Dehong Prefecture and Xishuangbanna Prefecture were in the range of 26–37°C (Table 1); thus, the low infection rate of Wolbachia in Ae. aegypti may be caused by the environmental temperatures.
Competition among co-infected microorganisms may result in a decrease in wolbachia titer. Wolbachia usually co-exists with other endosymbiotic bacteria or members of the gut microbiota (Gómez-Valero et al., 2004; Dittmer et al., 2016). Co-infection of multiple bacterial lineages might translate into competition for space and nutrients (Caragata et al., 2014; Geoghegan et al., 2017; Jiménez et al., 2019). The difference in intestinal microbial composition between Ae. aegypti and other mosquitoes resulted in high competition and low titer of Wolbachia. Ae. aegypti ‘s own immune response plays an important role (Masson et al., 2016). Wolbachia is known to activate the basal immune response of Ae. aegypti via the immune deficiency (IMD) and Toll-pathway. Silencing of these immune pathways leads to the reduction of Wolbachia titers (Pan et al., 2018).
Ae. aegypti did not occur in Yunnan Province before 2000, as this species was first found in Ruili Port in 2002 (Dong Xue-shu, 2004). Since then, Ae. aegypti has been found in Mangshi, Mengla, Menghai, Jinghong, and other places in Yunnan Province (Shi et al., 2017). This suggests that Ae. aegypti is an important invasive alien mosquito species in Yunnan Province. Dehong Prefecture borders Myanmar and contains the largest China-Myanmar port, Ruili. Xishuangbanna Prefecture borders Laos and contains the largest China-Laos port, Mohan. Ae. aegypti populations in Southeast Asia may thus invade Yunnan Province from border ports through logistics and the movement of people. The results of this study thus provide valuable evidence for analysing the invasion of Ae. Aegypti in Yunnan Province. The time sequence of Ae. aegypti monitoring reports from different areas of Yunnan Province indicates that Ae. aegypti mosquitoes in Dehong Prefecture and Xishuangbanna Prefecture originated from separate invasion events. This implies a continuous invasion in different locations based on the types and rates of Wolbachia infection of Ae. aegypti populations in the area.
According to the phylogenetic tree results, all Wolbachia Group A (17/17) and 78.9% of Wolbachia Group B (15/19) infections in Ae. aegypti in this study clustered with wAlbA and wAlbB strains isolated from Ae. albopictus. The Wolbachia strains in Ae. aegypti were mainly classified as wAlbA and wAlbB (Coon et al., 2016; Carvajal et al., 2019; Wong et al., 2020), but the detection rates of wAlbB and wAlbA strains in Ae. aegypti differed. In India and the United States, no Group A infections have been found and only Group B was found, which is closely related to the wAlbB strain isolated from Ae. Albopictus (Balaji et al., 2019; Kulkarni et al., 2019). Both Group A and Group B were found in Ae. aegypti in China and the Philippines, but the infection rates of these strains differed. Of the Philippines samples, 60.7% (51/84) were clustered with wAlbB and all (29/29) samples clustered with wAlbA (Carvajal et al., 2019).
The sequence similarity heatmap of the wsp sequences obtained from the universal primers (Figure 5) was split into the Clade I:B and Clade II:A supergroups. In the Clade I:B supergroup, the sequences collected in this study were highly homologous to those of Wolbachia found in Ae. aegypti in Malaysia (ID: MN893354) (Wong et al., 2020) and India (ID: MN307069 and MF999264) but cannot be compared to the Wolbachia sequences in Ae. aegypti in the United States as Group B is not found in this region (Hegde et al., 2018; Kulkarni et al., 2019); thus, this comparison is not presented in the heatmap. The sequences collected in this study were also highly homologous to those of Wolbachia in Cx. pipiens quinquefasciatus, Cx. pipiens and Ae. albopictus, but this strain of Wolbachia occurs more frequently in Ae. albopictus. In Clade II:A supergroup, all sequences were highly homologous to those of Wolbachia found in Ae. albopictus. Comparing the strains found within the same species (Ae. aegypti), the Wolbachia sequence found in this study had high homology with the Wolbachia sequence in Ae. aegypti distributed in countries that are geographically close to China and low homology with geographically distinct populations, such as the Wolbachia sequence in Ae. aegypti distributed in the United States. Comparing the strains found in different mosquitoes, the Wolbachia sequences in Ae. aegypti were highly homologous to those in Ae. Albopictus.
Natural infection of Wolbachia is found rarely in Ae. aegypti. Currently, studies on natural infection of Ae. aegypti with Wolbachia have only been reported in Malaysia, India, Thailand, the Philippines, and the U.S. states of Mexico and Florida. Studies of natural infection of Ae. aegypti with Wolbachia on reproduction and physiology and its efficacy on vectors have not been reported and further studies are needed (Teo et al., 2017; Thongsripong et al., 2018; Balaji et al., 2019; Carvajal et al., 2019; Kulkarni et al., 2019).Although not naturally found in Ae. aegypti, wMel strain were stably introduced into this mosquito in 2011 and were shown to reduce the transmission potential of dengue, Zika and chikungunya (Moreira et al., 2009; Walker et al., 2011; Aliota et al., 2016). Ae. aegypti carrying the wMel or wAlbB strains of Wolbachia have the potential to reduce dengue transmission through decreased mosquito vector competence, and there is already good evidence that both strains are having such impacts in Wolbachia-invaded release areas. In 2021, researchers found that long-term storage under warm environment greatly reduces the fertility of hatched females, especially for wAlbB-infected, in which a high proportion of females became infertile (Lau et al., 2021). Prevalence of Ae. aegypti infection with Wolbachia is found in an area, the impact of this situation on arbovirus transmission as well as vector control needs to be considered. Natural infection of Wolbachia by Ae. albopictus and a study showing that naturally occurring strains of Wolbachia can also restrict salivary gland infection of Ae. albopictus with DENV and limit transmission (Mousson et al., 2012; Ciota, 2019). Wolbachia strain wMel transfected into Ae. albopictus can induce cytoplasmic incompatibility and block dengue transmission in Ae. Albopictus (Blagrove et al., 2012). Ae. albopictus is currently facing such a situation, and it seems that the artificial release of Ae. albopictus infected with Wolbachia is also effective in reducing the density of Ae. albopictus already naturally infected with Wolbachia (Zheng et al., 2019).
This was the first study to report Wolbachia infection in wild Ae. aegypti caught in China. Wolbachia was detected in wild populations of this species in Dehong and Xishuangbanna Prefectures of Yunnan Province. A total of 24 mosquitoes (5%) infected with Wolbachia were detected using wsp markers. The strain had high homology with wAlbA and wAlbB, and was prevalent in the wild population of Ae. aegypti in Yunnan Province. This study provides a basis for studying natural Wolbachia infections in wild populations of Ae. aegypti.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: The data presented in the study are deposited in the GenBank database, accession numbers ON637917 to ON637937, OL629258 to OL629317.
HZ, JG, ZM, XG, and TYZ jointly designed and coordinated the study, with contributions from CL, YTD, DX and YDD. HZ and ZM drafted the article with contributions from JG. HZ, JG, YL, GW, QL, YJ, and TZ collected samples from Yunnan Province of China. HZ, JG, and ZM carried out the laboratory work and performed the statistical analysis. All authors contributed to the article and approved the submitted version.
This work was funded by grants from the Infective Diseases Prevention and Cure Project of China (No.2017ZX10303404).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcimb.2022.1082809/full#supplementary-material
Ahmad, N. A., Vythilingam, I., Lim, Y. A. L., Zabari, N., Lee, H. L. (2017). Detection of Wolbachia in Aedes albopictus and their effects on chikungunya virus. Am. J. Trop. Med. Hyg 96 (1), 148–156. doi: 10.4269/ajtmh.16-0516
Aliota, M. T., Peinado, S. A., Velez, I. D., Osorio, J. E. (2016). The wMel strain of Wolbachia reduces transmission of zika virus by Aedes aegypti. Sci. Rep. 6, 28792. doi: 10.1038/srep28792
Augustinos, A. A., Santos-Garcia, D., Dionyssopoulou, E., Moreira, M., Papapanagiotou, A., Scarvelakis, M., et al. (2011). Detection and characterization of Wolbachia infections in natural populations of aphids: is the hidden diversity fully unraveled? PloS One 6 (12), e28695. doi: 10.1371/journal.pone.0028695
Balaji, S., Jayachandran, S., Prabagaran, S. R. (2019). Evidence for the natural occurrence of Wolbachia in Aedes aegypti mosquitoes. FEMS Microbiol. Lett. 366 (6), 9. doi: 10.1093/femsle/fnz055
Baldini, F., Segata, N., Pompon, J., Marcenac, P., Shaw, W. R., Dabiré, R. K., et al. (2014). Evidence of natural Wolbachia infections in field populations of Anopheles gambiae. Nat. Commun. 5, 3985. doi: 10.1038/ncomms4985
Beckmann, J. F., Ronau, J. A., Hochstrasser, M. (2017). A Wolbachia deubiquitylating enzyme induces cytoplasmic incompatibility. Nat. Microbiol. 2, 17007. doi: 10.1038/nmicrobiol.2017.7
Bennett, K. L., Gómez-Martínez, C., Chin, Y., Saltonstall, K., McMillan, W. O., Rovira, J. R., et al. (2019). Dynamics and diversity of bacteria associated with the disease vectors Aedes aegypti and Aedes albopictus. Sci. Rep. 9 (1), 12160. doi: 10.1038/s41598-019-48414-8
Bian, G., Xu, Y., Lu, P., Xie, Y., Xi, Z. (2010). The endosymbiotic bacterium Wolbachia induces resistance to dengue virus in Aedes aegypti. PloS Pathog. 6 (4), e1000833. doi: 10.1371/journal.ppat.1000833
Blagrove, M. S., Arias-Goeta, C., Failloux, A. B., Sinkins, S. P. (2012). Wolbachia strain wMel induces cytoplasmic incompatibility and blocks dengue transmission in Aedes albopictus. Proc. Natl. Acad. Sci. U.S.A. 109 (1), 255–260. doi: 10.1073/pnas.1112021108
Bonacum, J., Desalle, R., O’Grady, P., Oliveira, D., Wintermute, J., Zilversmit, M. (2001). New nuclear and mitochondrial primers for systematics and comparative genomics in drosophilidae. Drosophila Inf. Service 84, 201–204.
Braig, H. R., Zhou, W., Dobson, S. L., O’Neill, S. L. (1998). Cloning and characterization of a gene encoding the major surface protein of the bacterial endosymbiont Wolbachia pipientis. J. Bacteriol 180 (9), 2373–2378. doi: 10.1128/jb.180.9.2373-2378.1998
Caragata, E. P., Rancès, E., O’Neill, S. L., McGraw, E. A. (2014). Competition for amino acids between Wolbachia and the mosquito host, Aedes aegypti. Microb. Ecol. 67 (1), 205–218. doi: 10.1007/s00248-013-0339-4
Carvajal, T. M., Hashimoto, K., Harnandika, R. K., Amalin, D. M., Watanabe, K. (2019). Detection of Wolbachia in field-collected Aedes aegypti mosquitoes in metropolitan Manila, Philippines. Parasit Vectors 12 (1), 361. doi: 10.1186/s13071-019-3629-y
Che-Mendoza, A., Martin-Park, A., Chávez-Trava, J. M., Contreras-Perera, Y., Delfín-González, H., González-Olvera, G., et al. (2021). Abundance and seasonality of Aedes aegypti (Diptera: Culicidae) in two suburban localities of south Mexico, with implications for Wolbachia (Rickettsiales: Rickettsiaceae)-carrying Male releases for population suppression. J. Med. Entomol 58 (4), 1817–1825. doi: 10.1093/jme/tjab052
Ciota, A. T. (2019). The role of co-infection and swarm dynamics in arbovirus transmission. Virus Res. 265, 88–93. doi: 10.1016/j.virusres.2019.03.010
Coon, K. L., Brown, M. R., Strand, M. R. (2016). Mosquitoes host communities of bacteria that are essential for development but vary greatly between local habitats. Mol. Ecol. 25 (22), 5806–5826. doi: 10.1111/mec.13877
Cui Bei-jin, H. J., Bing-yao, Wu, Sun, L.-x. (2015). Research progress on mosquitoes symbiotic bacteria of the genus Wolbachia. Chin. J. Frontier Health Quarantine 38 (6), 449–452. doi: 10.16408/j.1004-9770.2015.06.019
Ding, H., Yeo, H., Puniamoorthy, N. (2020). Wolbachia infection in wild mosquitoes (Diptera: Culicidae): implications for transmission modes and host-endosymbiont associations in Singapore. Parasit Vectors 13 (1), 612. doi: 10.1186/s13071-020-04466-8
Dittmer, J., Lesobre, J., Moumen, B., Bouchon, D. (2016). Host origin and tissue microhabitat shaping the microbiota of the terrestrial isopod armadillidium vulgare. FEMS Microbiol. Ecol. 92 (5), fiw063. doi: 10.1093/femsec/fiw063
Dong Xue-shu, C.-c., Hong-yu, Z., Xiang, W., Li-min, D., Chao, Wu, Bu-yu, W. (2004). Investigation on mosquitoes at border ports in yunnan province. Chin. J. Vector Biol. Control 15 (2), 4. doi: 10.3969/j.issn.1003-4692.2004.02.022
Ellegaard, K. M., Klasson, L., Näslund, K., Bourtzis, K., Andersson, S. G. (2013). Comparative genomics of Wolbachia and the bacterial species concept. PloS Genet. 9 (4), e1003381. doi: 10.1371/journal.pgen.1003381
Geoghegan, V., Stainton, K., Rainey, S. M., Ant, T. H., Dowle, A. A., Larson, T., et al. (2017). Perturbed cholesterol and vesicular trafficking associated with dengue blocking in Wolbachia-infected Aedes aegypti cells. Nat. Commun. 8 (1), 526. doi: 10.1038/s41467-017-00610-8
Glowska, E., Dragun-Damian, A., Dabert, M., Gerth, M. (2015). New Wolbachia supergroups detected in quill mites (Acari: Syringophilidae). Infect. Genet. Evol. 30, 140–146. doi: 10.1016/j.meegid.2014.12.019
Gómez-Valero, L., Soriano-Navarro, M., Pérez-Brocal, V., Heddi, A., Moya, A., García-Verdugo, J. M., et al. (2004). Coexistence of Wolbachia with buchnera aphidicola and a secondary symbiont in the aphid cinara cedri. J. Bacteriol 186 (19), 6626–6633. doi: 10.1128/jb.186.19.6626-6633.2004
Hegde, S., Khanipov, K., Albayrak, L., Golovko, G., Pimenova, M., Saldaña, M. A., et al. (2018). Microbiome interaction networks and community structure from laboratory-reared and field-collected Aedes aegypti, aedes albopictus, and Culex quinquefasciatus mosquito vectors. Front. Microbiol. 9 2160. doi: 10.3389/fmicb.2018.02160
Hermans, P. G., Hart, C. A., Trees, A. J. (2001). In vitro activity of antimicrobial agents against the endosymbiont Wolbachia pipientis. J. Antimicrob. Chemother. 47 (5), 659–663. doi: 10.1093/jac/47.5.659
Hertig, M. (1924). S Studies on rickettsia-like micro-organisms in insects. BJ. Med. Res. 44 (3), 329–374.327.
Hilgenboecker, K., Hammerstein, P., Schlattmann, P., Telschow, A., Werren, J. H. (2008). How many species are infected with Wolbachia?–a statistical analysis of current data. FEMS Microbiol. Lett. 281 (2), 215–220. doi: 10.1111/j.1574-6968.2008.01110.x
Hoffmann, A. A., Montgomery, B. L., Popovici, J., Iturbe-Ormaetxe, I., Johnson, P. H., Muzzi, F., et al. (2011). Successful establishment of Wolbachia in Aedes populations to suppress dengue transmission. Nature 476 (7361), 454–457. doi: 10.1038/nature10356
Hosokawa, T., Koga, R., Kikuchi, Y., Meng, X. Y., Fukatsu, T. (2010). Wolbachia as a bacteriocyte-associated nutritional mutualist. Proc. Natl. Acad. Sci. U.S.A. 107 (2), 769–774. doi: 10.1073/pnas.0911476107
Inácio da Silva, L. M., Dezordi, F. Z., Paiva, M. H. S., Wallau, G. L. (2021). Systematic review of Wolbachia symbiont detection in mosquitoes: An entangled topic about methodological power and true symbiosis. Pathogens 10 (1), 20. doi: 10.3390/pathogens10010039
Jiao Jun, Y. Y., Peisheng, He, Weiqing, W., Xuan, O., Bohai, W., Yi, S., et al. (2022). First detection of rickettsia aeschlimannii in Hyalomma marginatum in Tibet, China. Zoonoses 2 (1), 8. doi: 10.15212/ZOONOSES-2022-0026
Jiménez, N. E., Gerdtzen, Z. P., Olivera-Nappa, Á., Salgado, J. C., Conca, C. (2019). A systems biology approach for studying Wolbachia metabolism reveals points of interaction with its host in the context of arboviral infection. PloS Negl. Trop. Dis. 13 (8), e0007678. doi: 10.1371/journal.pntd.0007678
Kulkarni, A., Yu, W., Jiang, J., Sanchez, C., Karna, A. K., Martinez, K. J. L., et al. (2019). Wolbachia pipientis occurs in Aedes aegypti populations in new Mexico and Florida, USA. Ecol. Evol. 9 (10), 6148–6156. doi: 10.1002/ece3.5198
Landmann, F. (2019). The Wolbachia endosymbionts. Microbiol. Spectr. 7 (2), 15. doi: 10.1128/microbiolspec.BAI-0018-2019
Lau, M. J., Ross, P. A., Endersby-Harshman, N. M., Hoffmann, A. A. (2020). Impacts of low temperatures on Wolbachia (Rickettsiales: Rickettsiaceae)-infected Aedes aegypti (Diptera: Culicidae). J. Med. Entomol 57 (5), 1567–1574. doi: 10.1093/jme/tjaa074
Lau, M. J., Ross, P. A., Hoffmann, A. A. (2021). Infertility and fecundity loss of Wolbachia-infected Aedes aegypti hatched from quiescent eggs is expected to alter invasion dynamics. PloS Negl. Trop. Dis. 15 (2), e0009179. doi: 10.1371/journal.pntd.0009179
Mains, J. W., Kelly, P. H., Dobson, K. L., Petrie, W. D., Dobson, S. L. (2019). Localized control of Aedes aegypti (Diptera: Culicidae) in Miami, FL, via inundative releases of Wolbachia-Infected Male mosquitoes. J. Med. Entomol 56 (5), 1296–1303. doi: 10.1093/jme/tjz051
Masson, F., Zaidman-Rémy, A., Heddi, A. (2016). Antimicrobial peptides and cell processes tracking endosymbiont dynamics. Philos. Trans. R Soc. Lond B Biol. Sci. 371 (1695), 9. doi: 10.1098/rstb.2015.0298
Moreira, L. A., Iturbe-Ormaetxe, I., Jeffery, J. A., Lu, G., Pyke, A. T., Hedges, L. M., et al. (2009). A Wolbachia symbiont in Aedes aegypti limits infection with dengue, chikungunya, and plasmodium. Cell 139 (7), 1268–1278. doi: 10.1016/j.cell.2009.11.042
Mousson, L., Zouache, K., Arias-Goeta, C., Raquin, V., Mavingui, P., Failloux, A. B. (2012). The native Wolbachia symbionts limit transmission of dengue virus in Aedes albopictus. PloS Negl. Trop. Dis. 6 (12), e1989. doi: 10.1371/journal.pntd.0001989
O’Neill, S. L. (2018). The use of Wolbachia by the world mosquito program to interrupt transmission of Aedes aegypti transmitted viruses. Adv. Exp. Med. Biol. 1062, 355–360. doi: 10.1007/978-981-10-8727-1_24
Pan, X., Pike, A., Joshi, D., Bian, G., McFadden, M. J., Lu, P., et al. (2018). The bacterium Wolbachia exploits host innate immunity to establish a symbiotic relationship with the dengue vector mosquito Aedes aegypti. Isme J. 12 (1), 277–288. doi: 10.1038/ismej.2017.174
Ross, P. A., Turelli, M., Hoffmann, A. A. (2019). Evolutionary ecology of Wolbachia releases for disease control. Annu. Rev. Genet. 53, 93–116. doi: 10.1146/annurev-genet-112618-043609
Ross, P. A., Wiwatanaratanabutr, I., Axford, J. K., White, V. L., Endersby-Harshman, N. M., Hoffmann, A. A. (2017). Wolbachia infections in Aedes aegypti differ markedly in their response to cyclical heat stress. PloS Pathog. 13 (1), e1006006. doi: 10.1371/journal.ppat.1006006
Shi, Q. M., Zhang, H. D., Wang, G., Guo, X. X., Xing, D., Dong, Y. D., et al. (2017). The genetic diversity and population structure of domestic Aedes aegypti (Diptera: Culicidae) in yunnan province, southwestern China. Parasit Vectors 10 (1), 292. doi: 10.1186/s13071-017-2213-6
Song, S.-W., T.-Y., Z., Yan-De, D., Shu-Nan, J., Bao-Lin, Lu (2002a). Sequencing and sequence analysis of the wsp gene of Wolbachia in Chinese mosquitoes. Acta Entomologica Sin. 05), 7. doi: 10.16380/j.kcxb.2002.05.003
Song, S.-W., T.-Y, Z., Yan-De, D., Shu-Nan, J., Bao-Lin, Lu (2002b). Study on the infections of wolbachia in mosquitoes in China. Chin. J. Vector Biol. Control 13 (01), 3.
Teo, C. H. J., Lim, P. K. C., Voon, K., Mak, J. W. (2017). Detection of dengue viruses and Wolbachia in Aedes aegypti and Aedes albopictus larvae from four urban localities in Kuala Lumpur, Malaysia. Trop. BioMed. 34 (3), 583–597. Available at: https://www.msptm.org/files/Vol34No3/583-597-Lim-PKC.pdf.
Thongsripong, P., Chandler, J. A., Green, A. B., Kittayapong, P., Wilcox, B. A., Kapan, D. D., et al. (2018). Mosquito vector-associated microbiota: Metabarcoding bacteria and eukaryotic symbionts across habitat types in Thailand endemic for dengue and other arthropod-borne diseases. Ecol. Evol. 8 (2), 1352–1368. doi: 10.1002/ece3.3676
Utarini, A., Indriani, C., Ahmad, R. A., Tantowijoyo, W., Arguni, E., Ansari, M. R., et al. (2021). Efficacy of Wolbachia-infected mosquito deployments for the control of dengue. N Engl. J. Med. 384 (23), 2177–2186. doi: 10.1056/NEJMoa2030243
van den Hurk, A. F., Hall-Mendelin, S., Pyke, A. T., Frentiu, F. D., McElroy, K., Day, A., et al. (2012). Impact of Wolbachia on infection with chikungunya and yellow fever viruses in the mosquito vector Aedes aegypti. PloS Negl. Trop. Dis. 6 (11), e1892. doi: 10.1371/journal.pntd.0001892
Walker, T., Johnson, P. H., Moreira, L. A., Iturbe-Ormaetxe, I., Frentiu, F. D., McMeniman, C. J., et al. (2011). The wMel wolbachia strain blocks dengue and invades caged Aedes aegypti populations. Nature 476 (7361), 450–453. doi: 10.1038/nature10355
Wang, G. H., Jia, L. Y., Xiao, J. H., Huang, D. W. (2016). Discovery of a new Wolbachia supergroup in cave spider species and the lateral transfer of phage WO among distant hosts. Infect. Genet. Evol. 41, 1–7. doi: 10.1016/j.meegid.2016.03.015
Wang, Z., Su, X. M., Wen, J., Jiang, L. Y., Qiao, G. X. (2014). Widespread infection and diverse infection patterns of Wolbachia in Chinese aphids. Insect Sci. 21 (3), 313–325. doi: 10.1111/1744-7917.12102
Werren, J. H. (1997). Biology of Wolbachia. Annu. Rev. Entomol 42, 587–609. doi: 10.1146/annurev.ento.42.1.587
Werren, J. H., Baldo, L., Clark, M. E. (2008). Wolbachia: master manipulators of invertebrate biology. Nat. Rev. Microbiol. 6 (10), 741–751. doi: 10.1038/nrmicro1969
Werren, J. H., Zhang, W., Guo, L. R. (1995). Evolution and phylogeny of Wolbachia: reproductive parasites of arthropods. Proc. Biol. Sci. 261 (1360), 55–63. doi: 10.1098/rspb.1995.0117
Wong, M. L., Liew, J. W. K., Wong, W. K., Pramasivan, S., Mohamed Hassan, N., Wan Sulaiman, W. Y., et al. (2020). Natural wolbachia infection in field-collected anopheles and other mosquito species from Malaysia. Parasit Vectors 13 (1), 414. doi: 10.1186/s13071-020-04277-x
Zheng, X., Zhang, D., Li, Y., Yang, C., Wu, Y., Liang, X., et al. (2019). Incompatible and sterile insect techniques combined eliminate mosquitoes. Nature 572 (7767), 56–61. doi: 10.1038/s41586-019-1407-9
Zhou, W., Rousset, F., O’Neil, S. (1998a). Phylogeny and PCR-based classification of Wolbachia strains using wsp gene sequences. Proc. Biol. Sci. 265 (1395), 509–515. doi: 10.1098/rspb.1998.0324
Keywords: Aedes aegypti, Wolbachia, wsp gene, phylogenetics, China
Citation: Zhang HD, Gao J, Ma Z, Liu Y, Wang G, Liu Q, Du YT, Xing D, Li CX, Zhao T, Jiang YT, Dong YD, Guo XX and Zhao TY (2022) Wolbachia infection in field-collected Aedes aegypti in Yunnan Province, southwestern China. Front. Cell. Infect. Microbiol. 12:1082809. doi: 10.3389/fcimb.2022.1082809
Received: 28 October 2022; Accepted: 10 November 2022;
Published: 30 November 2022.
Edited by:
Jiaqi Fu, Purdue University, United StatesReviewed by:
XiaoLong Zhang, Science and Technology Research Center of China Customs (STRC), ChinaCopyright © 2022 Zhang, Gao, Ma, Liu, Wang, Liu, Du, Xing, Li, Zhao, Jiang, Dong, Guo and Zhao. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: XiaoXia Guo, Z3VveHg5OUAxNjMuY29t; TongYan Zhao, dG9uZ3lhbnpoYW9AMTI2LmNvbQ==
†These authors have contributed equally to this work and share first authorship
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
From Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.