- 1Department of Clinical Microbiology and Applied Technology, Faculty of Medical Technology, Mahidol University, Nakhon Pathom, Thailand
- 2Department of Clinical Chemistry, Faculty of Medical Technology, Mahidol University, Nakhon Pathom, Thailand
Citrobacter spp. are Gram-negative bacteria commonly found in environments and intestinal tracts of humans and animals. They are generally susceptible to third-generation cephalosporins, carbapenems and colistin. However, several antibiotic resistant genes have been increasingly reported in Citrobacter spp., which leads to the postulation that Citrobacter spp. could potentially be a reservoir for spreading of antimicrobial resistant genes. In this study, we characterized two colistin-resistant Citrobacter spp. isolated from the feces of a healthy individual in Thailand. Based on MALDI-TOF and ribosomal multilocus sequence typing, both strains were identified as Citrobacter sedlakii and Citrobacter amalonaticus. Genomic analysis and S1-nuclease pulsed field gel electrophoresis/DNA hybridization revealed that Citrobacter sedlakii and Citrobacter amalonaticus harbored mcr-3.5 gene on pSY_CS01 and pSY_CA01 plasmids, respectively. Both plasmids belonged to IncFII(pCoo) replicon type, contained the same genetic context (Tn3-IS1-ΔTnAs2-mcr-3.5-dgkA-IS91) and exhibited high transferring frequencies ranging from 1.03×10-4 - 4.6×10-4 CFU/recipient cell Escherichia coli J53. Colistin-MICs of transconjugants increased ≥ 16-fold suggesting that mcr-3.5 on these plasmids can be expressed in other species. However, beside mcr, other major antimicrobial resistant determinants in multidrug resistant Enterobacterales were not found in these two isolates. These findings indicate that mcr gene continued to evolve in the absence of antibiotics selective pressure. Our results also support the hypothesis that Citrobacter could be a reservoir for spreading of antimicrobial resistant genes. To the best of our knowledge, this is the first report that discovered human-derived Citrobacter spp. that harbored mcr but no other major antimicrobial resistant determinants. Also, this is the first report that described the presence of mcr gene in C. sedlakii and mcr-3 in C. amalonaticus.
Introduction
The emergence of antimicrobial resistance (AMR) is one of the most public health concerns. As declared by the World Health Organization (WHO) recently, the most problematic multidrug-resistant (MDR) bacteria is carbapenem-resistant Gram-negative bacilli, in particular Enterobacterales (CRE) (Tacconelli et al., 2018). The presence of such MDR bacteria and the lack of new antimicrobial agents lead to the use of colistin, which has been considered as a last-resort antibiotic (Madec et al., 2017; Zheng et al., 2020; Ouchar Mahamat et al., 2021). Colistin is a cyclic polypeptide antibiotic that targets the lipid A moiety of lipopolysaccharide (LPS), causing destabilization of the bacterial outer membrane, and leading to cell death. Beside clinical usage, colistin was also heavily used as a growth promoter in livestock (Rahal, 2008). As a result of the increased use in clinical practice and inappropriate use in animal production, acquired colistin resistance has emerged (Rahal, 2008; Papp-Wallace et al., 2011). Most of colistin resistant mechanisms are related to chromosomal mutation within two-component systems (TCSs), resulting in modification of LPS by addition of positively charged molecules including phosphoethanolamine (PEtN) and 4-amino-4-deoxy-L-arabinose (Ara4N) to the 1-phosphate or 4-phosphate groups of Lipid A, respectively. Beside chromosomal mutations, plasmid-mediated mobile colistin resistant (mcr) gene has also been reported. The discovery of mcr-1 in 2015 has raised a significant public health concern, since the gene can easily spread by horizontal gene transfer (Liu et al., 2016). Shortly after the discovery of mcr-1, other genetic alleles including mcr-2 to mcr-10 have been identified from various species of Gram-negative bacteria (Xavier et al., 2016; Yin et al., 2017; Borowiak et al., 2017; AbuOun et al., 2018; Wang et al., 2018; Yang et al., 2018; Kieffer et al., 2019; Wang et al., 2020). Currently, mcr genes have been distributed globally. The genes have been identified in at least 70 countries, with mcr-1 being the most prevalent followed by mcr-3 and mcr-4, respectively. They are frequently isolated from E. coli, K. pneumoniae and Salmonella spp. (Mmatli et al., 2022). In Thailand, various mcr alleles including mcr-1, mcr-2, mcr-3, mcr-6, mcr-7, mcr-8, and mcr-9 have been reported. Most of these alleles were found to associate with farmed animals, especially pig and poultry (Mmatli et al., 2022). Beside animals, prevalence of mcr-1 in human patients and co-occurrence of mcr -2 and mcr -3 on chromosome of multidrug-resistant Escherichia coli isolated from a healthy subject were recently reported by our group (Eiamphungporn et al., 2018; Phuadraksa et al., 2022).
Citrobacter spp. are Gram-negative bacteria in the order Enterobacterales. It is commonly found in soil, water, retail meat, and intestines of animals and humans (Liu et al., 2018b). It has been reported to carry several types of antimicrobial resistant genes such as AmpC β-lactamase, extended-spectrum -lactamases, plasmid-mediated quinolone resistant determinants, and carbapenemases (Jacobson et al., 1995; Hanson and Sanders, 1999; Wang et al., 2000; Mohanty et al., 2007; Zhang et al., 2008; Samonis et al., 2009; Shahid, 2010; Kanamori et al., 2011; Lee et al., 2015). Moreover, several variants of mcr genes have been recently reported in many species of Citrobacter, including mcr-1 (Li et al., 2017; Hu et al., 2017; Zhou et al., 2017; Sadek et al., 2021) and mcr-9 (Bitar et al., 2020) in C. freundii, mcr-1 in C. braakii (Sennati et al., 2017; Liu et al., 2018a; Zelendova et al., 2020), and mcr-1.5 in C. amalonaticus (Faccone et al., 2019). Therefore, Citrobacter spp. has been speculated as a potential source for carrying and spreading of antibiotic resistant genes (Jiang et al., 2019).
Herein, colistin-resistant C. sedlakii and C. amalonaticus were isolated from healthy individual under healthcare check-ups program at the Golden Jubilee Medical Center Mahidol University, Nakhon Pathom, Thailand, in 2022. The antimicrobial susceptibility profile, whole genome sequencing, AMR mechanisms, plasmid characteristics and transferring frequencies were investigated.
Materials and methods
Bacterial identification and isolation of colistin-resistant Citrobacter strains
A total of 55 left-over stool samples were obtained from healthcare check-ups program at the Golden Jubilee Medical Center Mahidol University, Nakhon Pathom, Thailand, in 2022. Samples were cultured in MacConkey agar supplemented with 2 mg/L colistin. Citrobacter isolates were identified using traditional biochemical tests (Farmer et al., 1985) and species-level identification was confirmed by Biotyper (matrix-assisted laser desorption/ionization time-of-flight (MALDI-TOF) mass spectrometry) according to the manufacturer’s protocol (Bruker Daltonik, Leipzig, Germany). Colistin-resistant isolates were further confirmed by the gold standard broth-microdilution method defined by the Clinical and Laboratory Standards Institute (CLSI) (Clinical Laboratory and Standards Institute (CLSI), 2020). The presence of mcr-1 to mcr-10 was screened by multiplex PCR using the previously described protocols (Lescat et al., 2018; Wang et al., 2020; Borowiak et al., 2020), and the gene sequence was confirmed by Sanger DNA sequencing.
Antimicrobial susceptibility testing (AST)
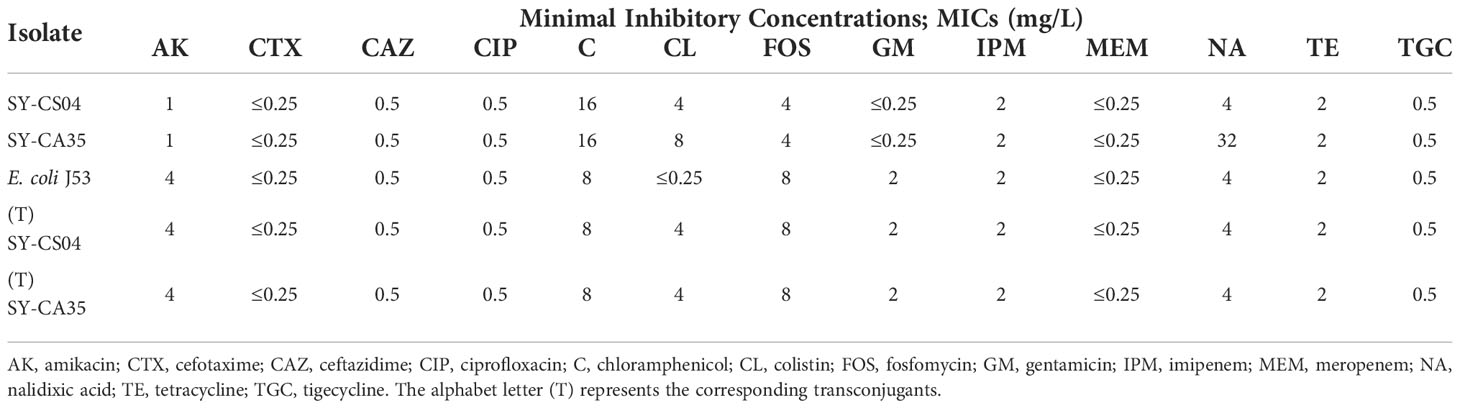
The minimum inhibitory concentrations (MICs) of amikacin, cefotaxime, ceftazidime, ciprofloxacin, chloramphenicol, colistin, gentamicin, imipenem, meropenem, nalidixic acid, tetracycline, and tigecycline were determined by broth microdilution method (BMD). MIC of fosfomycin was investigated by agar dilution method, which is recommended by CLSI. Escherichia coli ATCC 25922 was used as a quality control strain. The results were interpreted according to CLSI guideline.
Whole−genome sequencing (WGS) and bioinformatics analysis
Genomic DNA (gDNA) of C. amalonaticus and C. sedlakii were extracted using PureLink® Genomic DNA Kits (Invitrogen) according to the manufacturer’s instructions. The DNA samples were subsequently sequenced through NovaSeq 6000-PE150 platform (Illumina, San Diego, CA, USA) to generate paired-end 150-bp reads. The raw reads were then checked for quality and trimming using FastQC and TrimGalore, respectively (Andrews, 2022). De novo assembly was performed by SPAdes genome assembler version 3.15.3 (Prjibelski et al., 2020) to obtain contigs. The assembled contigs were then annotated through PROKKA and RAST server (Aziz et al., 2008; Seemann, 2014). Acquired antimicrobial resistant genes and plasmid replicons were determined using Resfinder (Bortolaia et al., 2020) and PlasmidFinder (Carattoli and Hasman, 2020), respectively. Additionally, the assembled contigs were also used for species identification using ribosomal multilocus sequence typing (rMLST). Based on the seven house-keeping genes (aspC, clpX, fadD, mdh, arcA, dnaG and lysP), the sequence type (ST) was identified using PubMLST server (Jolley and Maiden, 2010). Furthermore, the phylogenetic tree was performed and visualized through Roary (Page et al., 2015) and iTOL (Letunic and Bork, 2021), respectively.
Plasmid characterization
Plasmid profiles of isolates containing mcr genes were characterized by pulsed-field gel electrophoresis with S1 nuclease (S1-PFGE) (Barton et al., 1995). Briefly, bacterial genomic DNA was embedded in plugs and digested with S1 nuclease (Fermentas, USA). Then, the linearized plasmid DNA was separated using a CHEF-DRIII system (Bio-Rad, Hercules, USA). Salmonella braenderup H9812 digested with XbaI was used as a reference DNA size marker. The location of the mcr gene in the plasmids was investigated by Southern blot analysis with a specific probe. The probe was labeled and hybridized using DIG-High Prime DNA Labeling and Detection Starter Kit II (Roche Diagnostics, Indianapolis, IN, USA) according to the manufacturer’s protocol. Transferability of plasmids harboring mcr gene was determined by plasmid conjugation experiment using the filter-mating technique as previously described (Khajanchi et al., 2019). Briefly, Citrobacter isolates harboring mcr gene and Escherichia coli J53, which is resistant to sodium azide were used as donors and recipients, respectively. The donor and recipient were mixed at a ratio of 1:2 on a filter and incubated on LB plate at 37°C for 4 hr. Transconjugants were selected on MacConkey agar containing 2 mg/L of colistin and 150 mg/L of sodium azide. Then, MALDI-TOF MS was used for identification of transconjugants and the presence of mcr gene was investigated by PCR to ensure that the plasmid was successfully transferred to the recipient strain.
Nucleotide sequence accession numbers
The nucleotide sequences of pSY_CA01 and pSY_CS01 have been deposited in in the NCBI database with GenBank accession numbers JALNMK010000021 and JALNML010000026, respectively. The draft genomes of C. amalonaticus SY-CA35 and C. sedlakii SY-CS04 are also available in the NCBI database with accession numbers PRJNA827636 and PRJNA827638, respectively.
Results
Bacterial isolation and identification
Based on MALDI-TOF MS experiment, two bacterial isolates, SY-CS04 and SY-CA35, were identified as C. sedlakii and C. amalonaticus, respectively (Figure 1A). This result is in an agreement with ribosomal multilocus sequence typing (rMLST), which showed that SY-CS04 and SY-CA35 were C. sedlakii and C. amalonaticus, respectively. Sequence alignment with reference strains showed that SY-CS04 and SY-CA35 had high sequence similarity to C. sedlakii (accession no. CP071070) and C. amalonaticus (accession no. CP014070), respectively (Figures 1B, C). Taken together, SY-CS04 and SY-CA35 have been identified as C. sedlakii and C. amalonaticus, respectively.
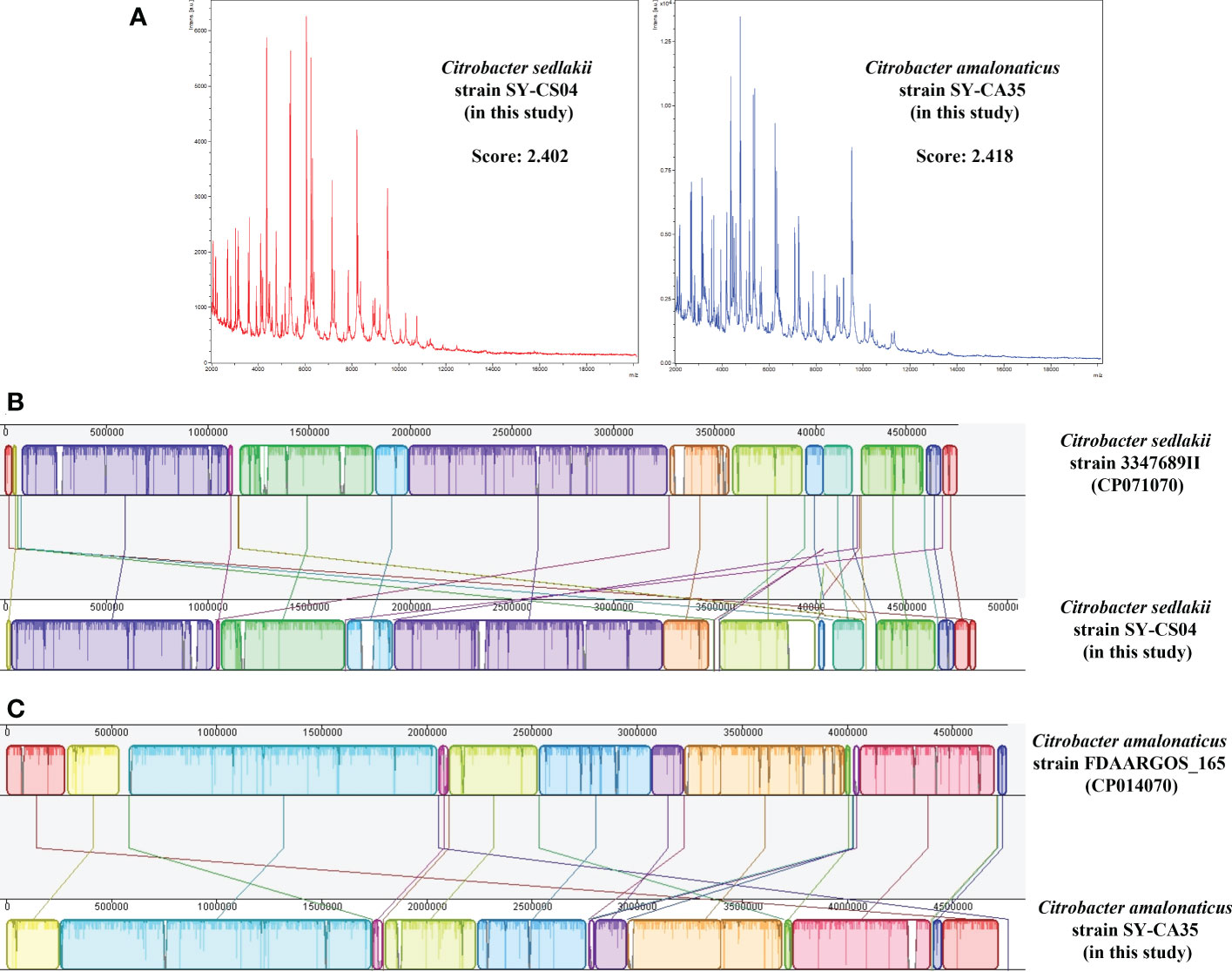
Figure 1 Identification of Citrobacter spp. (A) Mass fingerprinting of C. sedlakii and C. amalonaticus from Matrix-Assisted Laser Desorption/Ionization-Time Of Flight Mass Spectrometry (MALDI-TOF-MS). (B) Sequence alignment of C. sedlakii strain SY-CS04 with C. sedlakii strain 3347689II (accession no. CP071070). (C) Sequence alignment of C. amalonaticus strain SY-CA35 with C. amalonaticus strain FDAARGOS_165 (accession no. CP014070).
Antimicrobial susceptibility testing and screening of mcr genes
Both SY-CS04 and SY-CA35 were susceptible to most of the antibiotics tested except for colistin (Table 1). SY-CA35 also exhibited resistance to nalidixic acid. The presence of mcr genes was sought by multiplex-PCR and the results showed that both isolates were positive for mcr-3. Then, the sequence of the gene was confirmed by Sanger DNA sequencing, which revealed that both isolates harbor mcr-3.5 gene, with 100% identity to the reference sequence (accession number NG_055782.1).
Genomic analysis of Citrobacter isolates
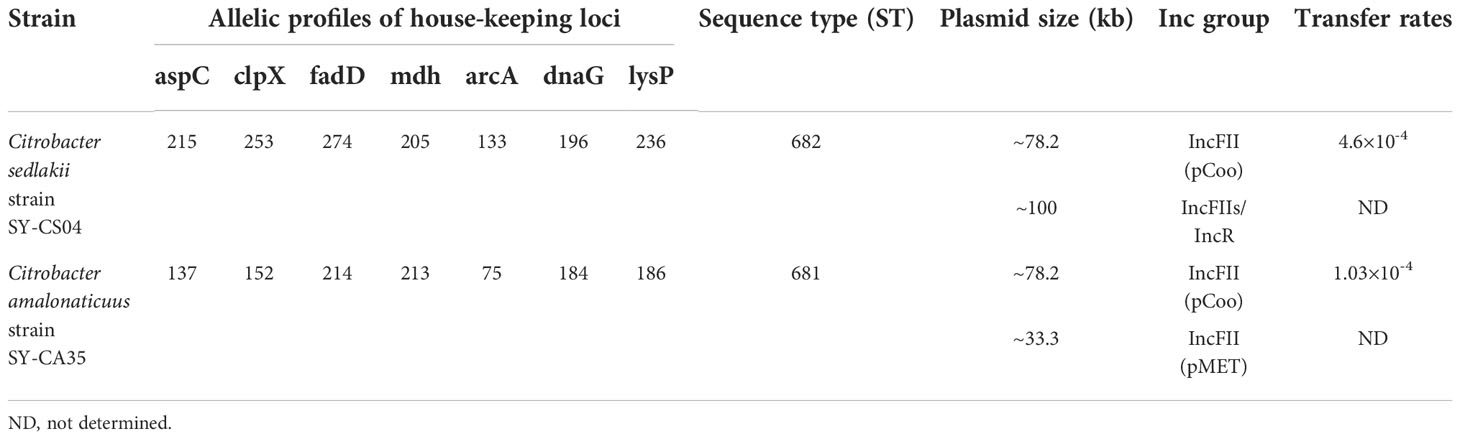
As revealed by whole-genome sequencing, the genomic sizes of SY-CS04 and SY-CA35 were 5,047,858 and 4,851,785-bp, respectively. The GC content of SY-CS04 was 54.46% while that of SY-CA35 was 53.38% (Figure 2). Based on Resfinder analysis, the acquired resistant genes in both isolates were discovered (Figure 3). Both SY-CS04 and SY-CA35 possessed genes conferring resistance to macrolides (erm(B), mph(A)), β-lactams (blaSED-1), and colistin (mcr-3.5). It is worth noting that these two isolates did not contain any other major antimicrobial resistant determinants found in multidrug resistant Enterobacterales. Additionally, SY-CA35 also carried quinolone-resistant gene (qnrS1) and fluoroquinolone-resistant genes (oqxA, oqxB). Virulence factors of SY-CS04 and SY-CA35 were predicted by VirulenceFinder. The presence of genes encoding extracellular nucleation factors (csgA, csgB, csgD), enterobactin (entB, entE), siderophores transportation (fepC, fepD, fepG) were found in both isolates. SY-CA35 also carried enterobactin (entA) while SY-CS04 carried enterobactin (entC). Furthermore, SY-CS04 also contained genes encoding extracellular nucleation factors (csgE, csgG, csgF), yersiniabactin receptor (fyuA), siderophore yersiniabactin (ybtA, ybtE, ybtO, ybtP, ybtS, ybtT, ybtU, ybtX), iron regulatory proteins (irp1, irp2), and outer membrane protein A (ompA). MLST analysis performed by PubMLST revealed that the sequence of SY-CS04 and SY-CA35 did not match with the existed sequences in the database. Therefore, SY-CS04 and SY-CA35 were newly assigned as ST682 and ST681, respectively (Table 2). Then phylogenetic tree was generated through roary bacterial genome analysis. All available genome data of C. sedlakii and C. amalonaticus were retrieved from NCBI genome database. Roary matrix-based gene sequence analysis generated a pangenome consisting of 37,961 gene clusters of 86 whole genomes (Figure 4). The tree revealed that SY-CS04 and SY-CA35 were closely related to a clinical isolate C. sedlakii stain CB00020 (accession no. SAMN10435564) and a clinical isolate C. amalonaticus stain LFYP1 (accession no. SAMEA6160257) from the USA, respectively.
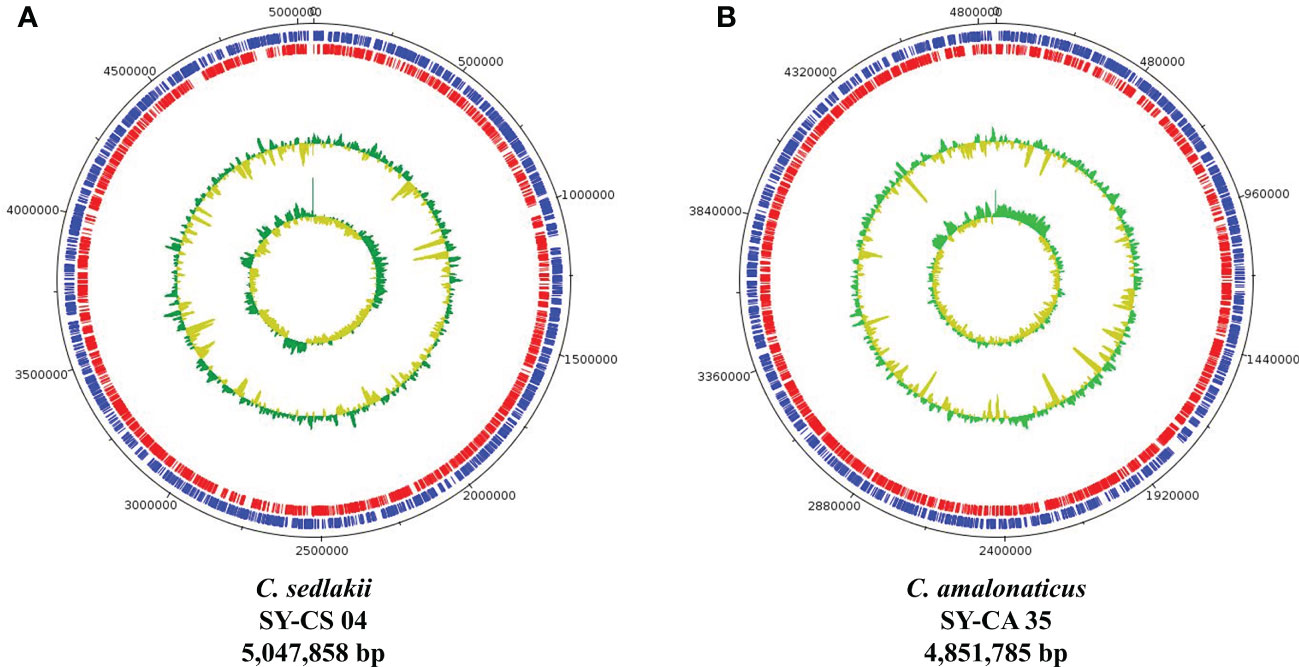
Figure 2 Overview of genomic structure of Citrobacter isolates. (A) Citrobacter sedlakii SY-CS04. (B) Citrobacter amalonaticus strain SY-CA35. The inner circle and outer circle represent GC skew and GC content, respectively. The protein-coding gene on forward strand and reverse strand represent in blue and red, respectively.
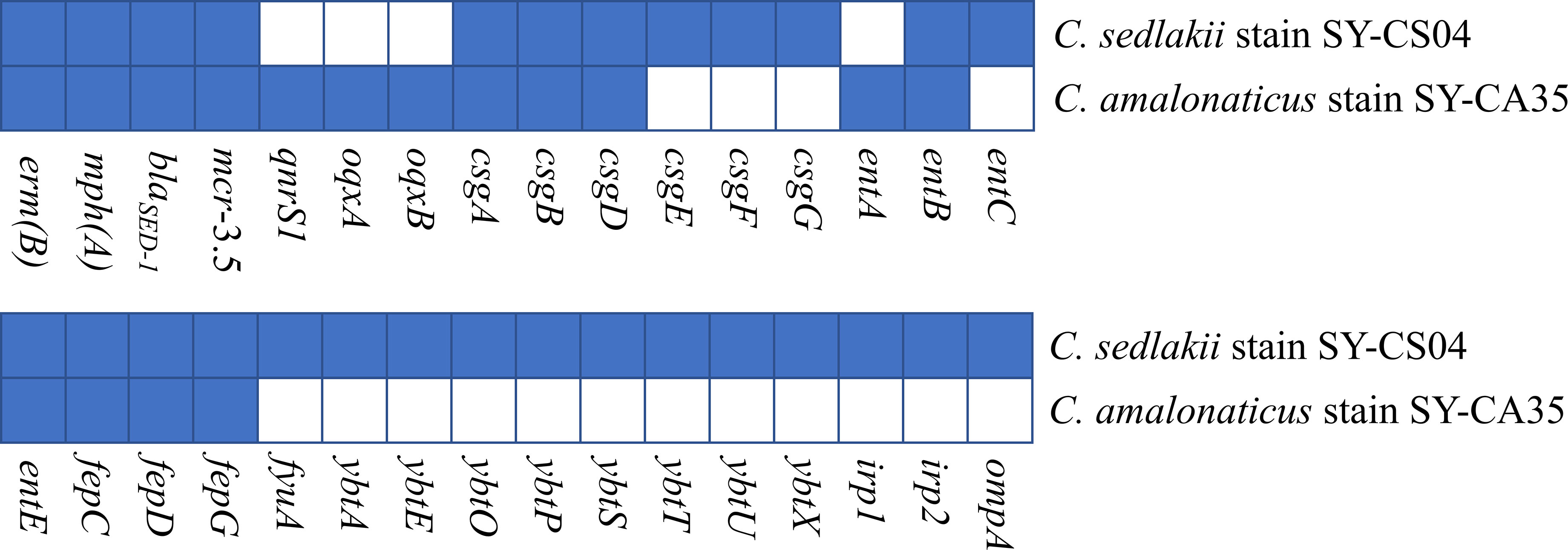
Figure 3 Antimicrobial resistance and virulence-associated profiles of the 2 Citrobacter isolates. Blue squares indicate the presence of genes while white squares represent the absence of genes.
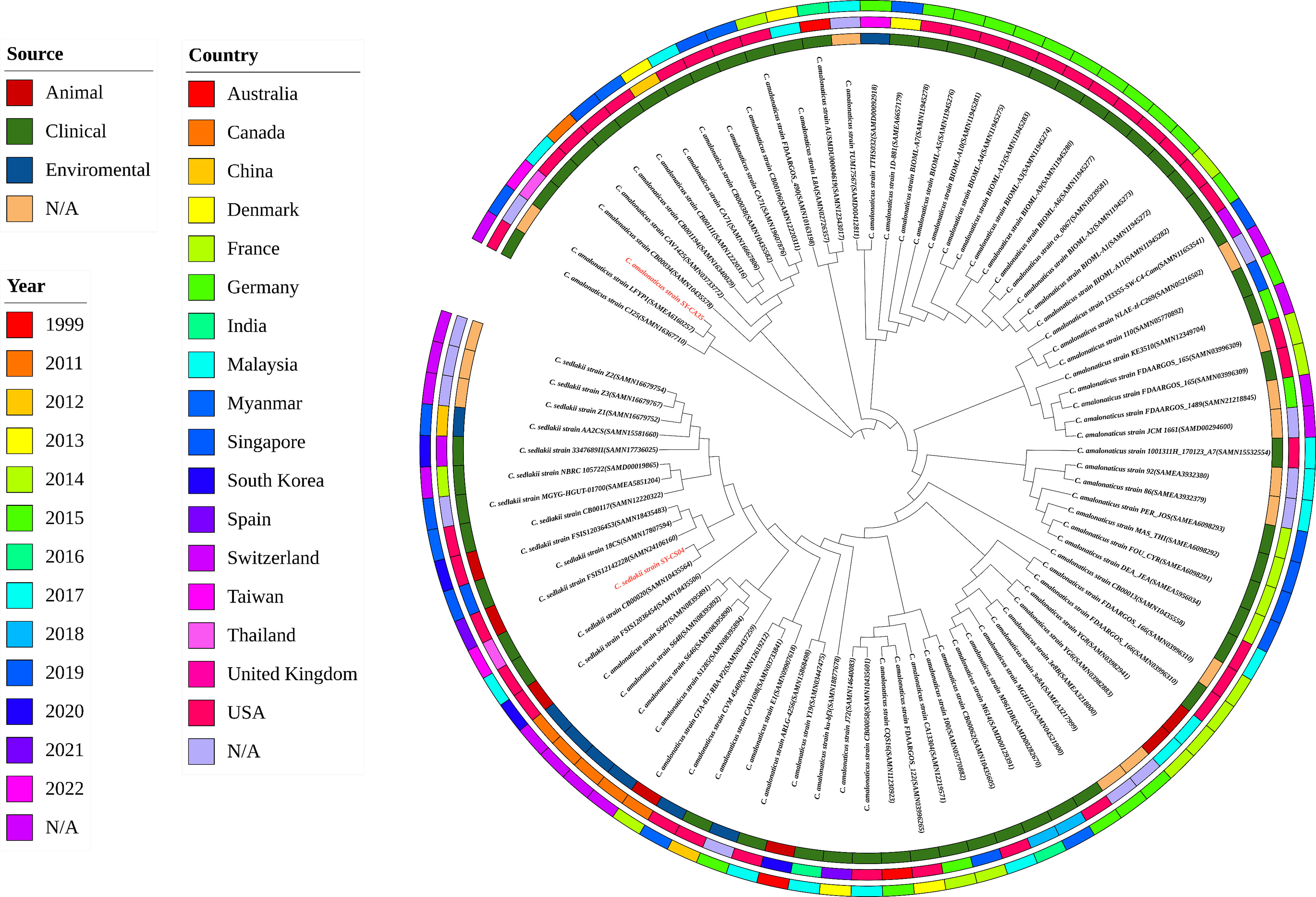
Figure 4 Roary matrix-based gene sequence analysis of 86 Citrobacter isolates. The source of the isolates is shown in the inner ring. The location of the isolates is depicted in the middle ring and the year of the isolates is indicated by the outer ring. Isolates in this study including SY-CS04 and SY-CA35 were colored in red.
Plasmid characterization
The plasmid profiles of SY-CS04 and SY-CA35 were characterized by S1-PFGE (Figure 5A), which revealed the presence of two plasmids in each of the two strains. In SY-CS04, the plasmid sizes were ~78.2 and ~100 kb, while in SY-CA35, the plasmid sizes were ~33.3 and ~78.2 kb. The location of mcr-3.5 gene was then identified using DNA hybridization with a specific probe (Figure 5B), which revealed that the gene was located on the ~78.2 kb plasmid in both SY-CS04 and SY-CA35. The incompatibility group of the plasmids was identified through PlasmidFinder. IncFII(pCoo) plasmid was found in both SY-CS04 and SY-CA35. In addition, an IncFII(S)/IncR plasmid was found in SY-CS04 while an IncFII(pMET) plasmid was found in SY-CA35 (Table 2). In combination with S1-PFGE, these results suggest that the mcr-3.5 gene is located on IncFII(pCoo) plasmid with a size of ~78.2 kb in both strains. IncFII(pMET) is the plasmid with a size of ~33.3 kb in SY-CA35. IncFII(S)/IncR plasmid is a hybrid plasmid with the size of ~100 kb in SY-CS04.
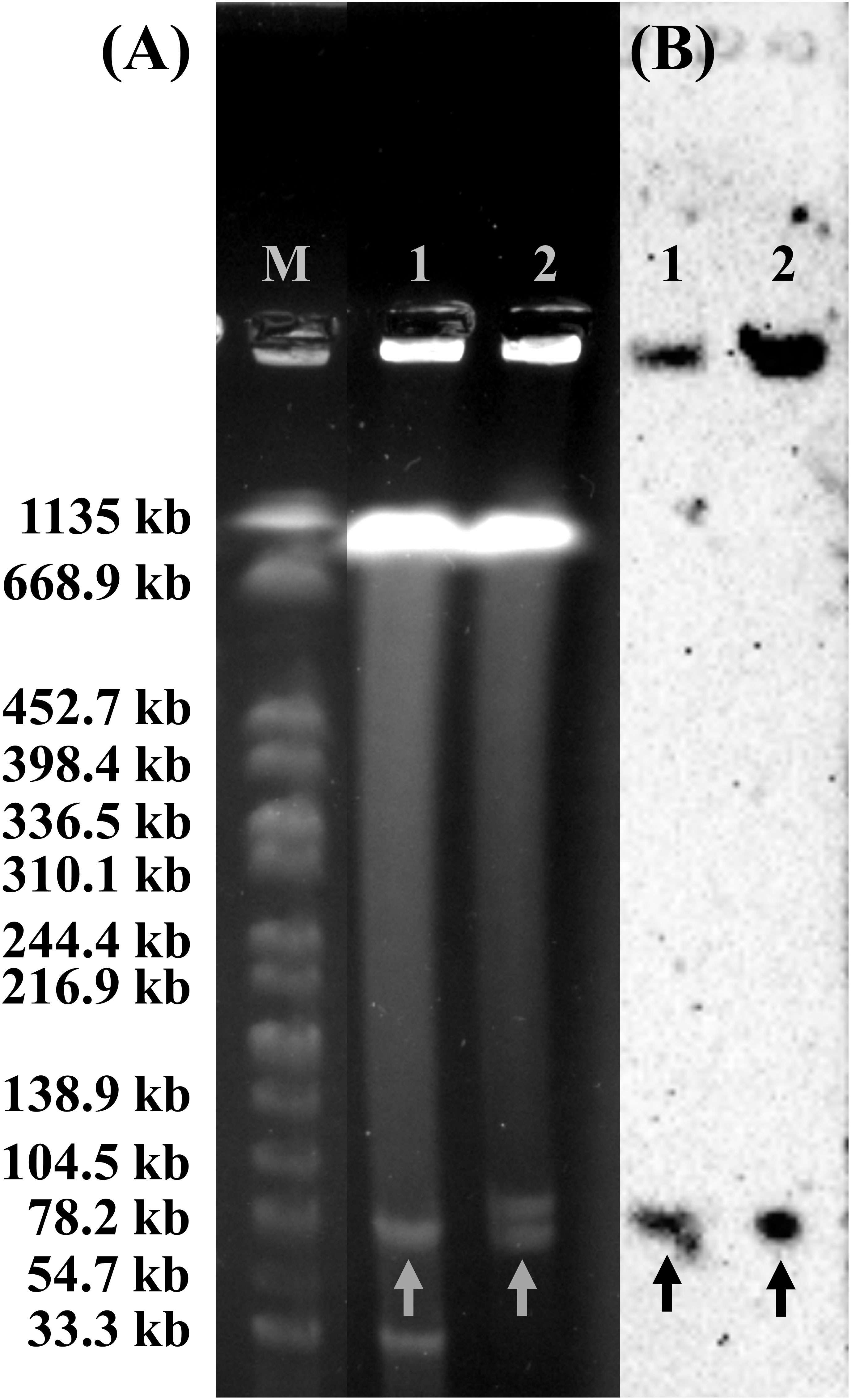
Figure 5 Plasmid profile analysis of Citrobacter isolates harboring mcr-3.5 gene by S1-PFGE and DNA hybridization. (A) The profile of total DNA treated with S1 nuclease and (B) relative hybridization of mcr-3 probe. Lane M, molecular standard, which is Salmonella braenderup H9812 digested with XbaI. Lane 1, Citrobacter amalonaticus strain SY-CA35. Lane 2, Citrobacter sedlakii strain SY-CS04. Arrows indicate the locations of plasmid harboring mcr-3 gene.
Bioinformatic analysis revealed that mcr-3.5 was located on a plasmid of SY-CS04 and SY-CA35, which were then designated as pSY_CS01 and pSY_CA01, respectively. The size of pSY_CS01 and pSY_CA01 were 80,003-bp with 52.59% GC content and 80,445-bp with 52.67% GC content, respectively. Both plasmids belonged to IncFII(pCoo) plasmid replicon type and contained 279 predicted ORFs encoding proteins with over 50 amino acids long (Figure 6). Notably, the genetic environment of mcr-3.5 in pSY_CS01 and pSY_CA01 was the same, which is Tn3-IS1-ΔTnAs2-mcr3.5-dgkA-IS91. Plasmids pSY_CS01 and pSY_CA01 were then blasted through BLASTN and 8 best matches with query cover >75% and identity >99% were identified, these include pVNCEc57 (LC549806.1), pRHBSTW-00122 (CP056847.1), p92944-mph (MG838205.1), p702_18_4 (CP074705.1), pNCYU-26-73-6 (CP042621.1), pECQ4552 (CP077064.1), unnamed3 (CP041102.1), and pVE769 (AP018353.1). In this regard, mcr-3 was identified in only 3 plasmids, which were found in E. coli including pVNCEc57 from Vietnam, pECQ4552 from France, and pVE769 from Vietnam (Figure 7). Then, the sequences of plasmid containing mcr-3 were compared with sequence from our study. As shown in Figures 7, 8, all plasmids shared the same backbone region. However, the surrounding region of mcr-3 from our study was different from the sequences in the database suggesting that insertion of genetic elements had occurred. Moreover, mobile genetic element also contained toxin/antitoxin system indicating the stabilization of mobile genetic element within plasmid (Song and Wood, 2020). In addition, the surrounding region of mcr-3.5 in this study were compared with 13 plasmids harboring mcr-3.5 (Figure 9), which were retrieved from NCBI database. The result showed that ΔTnAs2-mcr-3.5-dgkA region were found in all sequences. Various insertion sequences (IS) such as IS91, Tn3, IS26 were also identified at the upstream or downstream of that region.
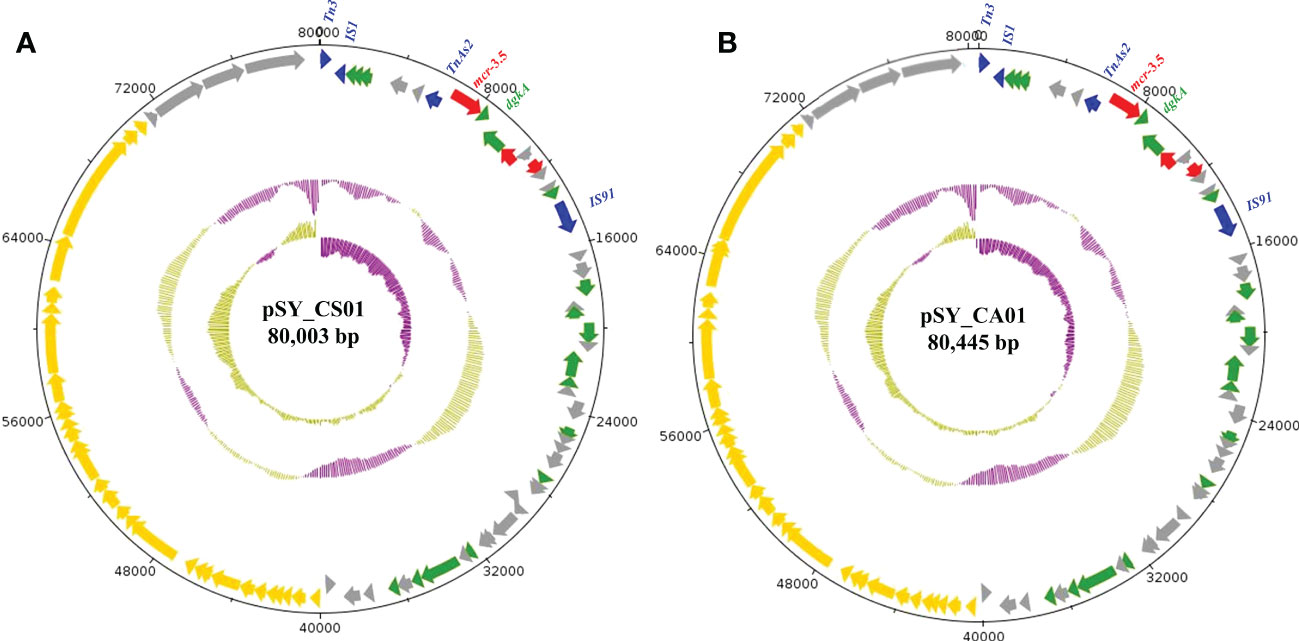
Figure 6 Structure of the IncFII(pCoo) harboring mcr-3.5, including pSY_CS01 (A) and pSY_CA01 (B). The inner circle and outer circle represent GC skew and GC content, respectively. The arrows indicate the directions of gene transcription. The red arrows represent antimicrobial resistance genes, the green arrows show other functional genes, the blue arrows show mobile element-encoding genes, the yellow arrows show IncF plasmid conjugative element and grey arrows for hypothetical protein-encoding genes.
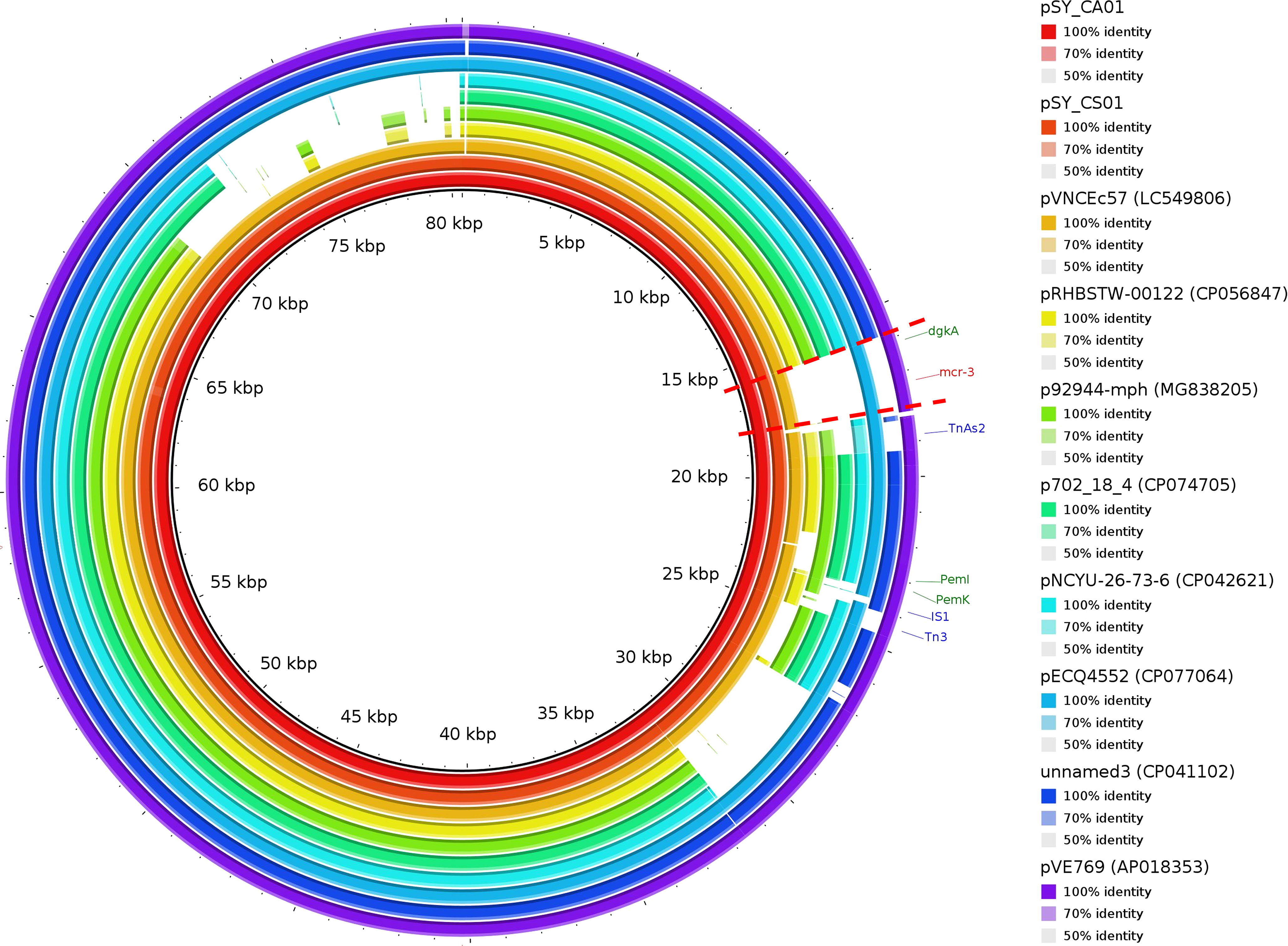
Figure 7 Circular comparison of IncFII(pCoo) harboring mcr-3.5, pSY_CS01 and pSY_CA01 with eight homologous plasmids with considerable query coverage.
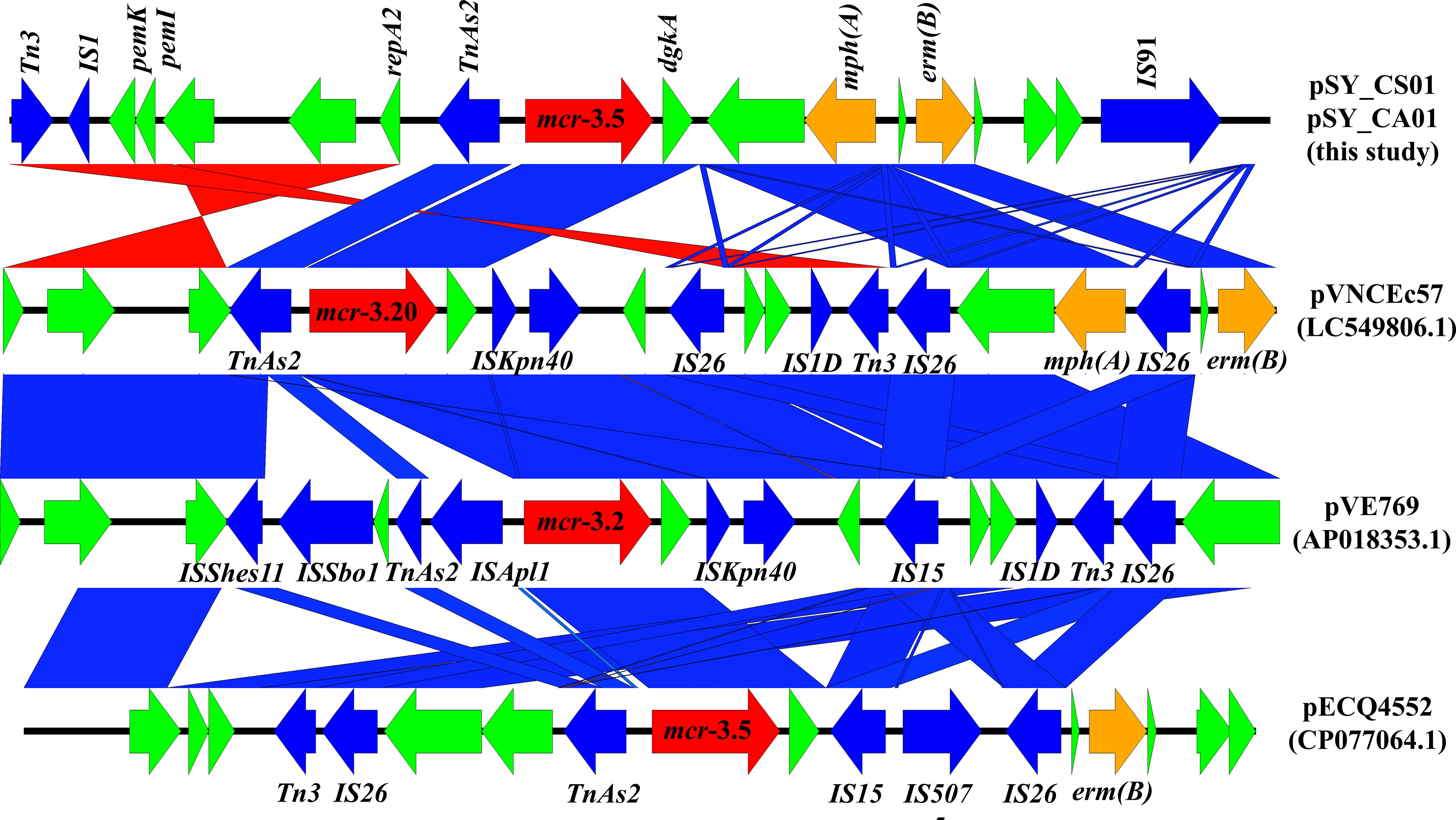
Figure 8 Linear comparison of surrounding regions of mcr-3. The arrows indicate directions of gene transcription. mcr-3 is labeled in red arrow, while other antimicrobial resistance genes are labeled in orange. Mobile genetic elements are indicated in blue and other functional gene are in green.
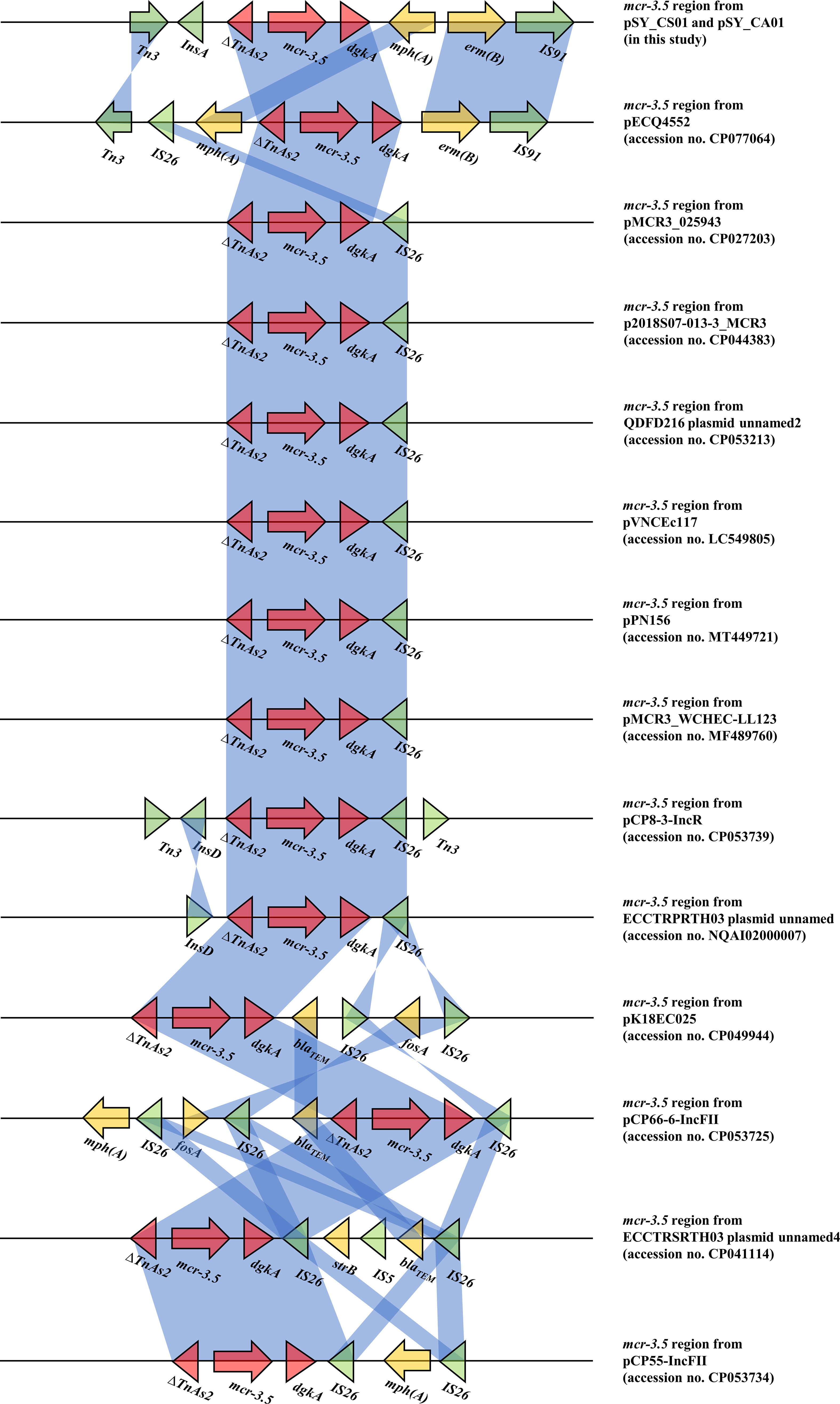
Figure 9 Comparison of 14 mcr-3.5 regions from 16 plasmids. The arrows indicate directions of gene transcription. Shading in light blue denotes regions of homology (nucleotide identity 95%).
Furthermore, the transferability of plasmids harboring mcr-3.5 gene was determined by plasmid conjugation assay. Both plasmids were successfully transferred to E. coli J53 with high transferring efficiency, ranging from 1.03×10-4 - 4.6×10-4 colony forming units (CFU) per recipient cell. Both transconjugants exhibited a 16-fold (4 mg/L) increase in the colistin MICs when compared with that of the recipient cell (E. coli J53) (Table 1). These results suggested that the mcr-3.5 gene on IncFII(pCoo) plasmid can be transferred and expressed in transconjugants.
Discussion
The mcr-3 gene was first reported by Yin W et al. in China (Yin et al., 2017). The gene was located on IncHI2 replicon type plasmid found in E. coli isolated from pig. Currently, more than 40 variants of mcr-3 have been deposited in the NCBI database, indicating that the mcr-3 gene is widespread and genetically diverse. In addition, the gene has been reported to be associated with three replicon types including IncP1, IncFII and IncI1, which can be found in various species of bacteria including Aeromonas spp., E. coli, K. pneumoniae, Salmonella, and Enterobacter spp. Citrobacter spp. are opportunistic bacterial pathogens that can cause both hospital- and community-acquired infections. It has been reported that Citrobacter spp. represent up to 6% of all isolated Enterobacterales from clinical specimens (Oberhettinger et al., 2020). In this study, we identified and characterized two clinical isolates of Citrobacter spp. (SY-CS04 and SY-CA35) harboring mcr-3.5. Identification of these isolates at species level was not possible with biochemical tests. Yet, it has been reported that 16S rRNA sequences displays limited resolution distinguishing only three groups within the genus (Clermont et al., 2015). Therefore, in our study, MALDI-TOF MS has been used for identification and the results yielded a category A identification (score > 2.0), which can be considered a reliable identification. In addition, rMLST, an approach of integrating taxonomy and typing of microbial communities by analyzing variation in 53 genes encoding ribosome protein subunits (rps genes) has been used to confirm the species and the results were in an agreement with MALDI-TOF MS, which identified SY-CS04 and SY-CA35 as C. sedlakii and C. amalonaticus, respectively. For mcr-3.5, it was first identified on IncP1 plasmid found in E. coli in China (Liu et al., 2017). It has also been found in other plasmid replicon types including IncR, IncFII, and IncFII(pCoo). In our study, IncFII(pCoo) harboring mcr-3.5, namely pSY_CS01 and pSY_CA01 were identified in C. sedlakii SY-CS04 and C. amalonaticus SY-CA35, respectively. It was noted that the genetic context of pSY_CS01 and pSY_CA01 were the same. Since both Citrobacter isolates were from the same human subject, the two plasmids might be derived from the same clone. Comparison of 16 mcr-3.5 loci showed that the genetic context of ΔTnAs2-mcr-3.5-dgkA might be the conserved structure of the mcr-3.5 locus. Interestingly, this genetic context has been interrupted by various IS elements at the upstream or downstream, suggesting that the area surrounding this conserved region could be the high-frequency region for insertion of mobile genetic elements. IncFII type is a low-copy number plasmid. It is one of the narrow-host range plasmids that are commonly found in E. coli (Carattoli, 2009). However, IncFII plasmid can disseminate and replicate in a variety of Enterobacterales, which contributes a crucial role for spreading of antimicrobial resistant genes (Chen et al., 2014). As shown in Figures 7, 8, comparison of pSY_CS01 and pSY_CA01 with plasmids containing mcr-3 from E. coli recovered from Vietnam and France showed that these plasmids share a similar backbone. Since these plasmids have been recovered from different species and geographical locations, these results suggest that pSY_CS01 and pSY_CA01 may contribute to the transmission of mcr-3.5 among other Enterobacterales species.
There is an evidence that the presence of mcr genes in food animals significantly increased the risk of direct contact with bacteria harboring mcr genes, in particular transmission of Enterobacterales to humans (Liu et al., 2016; Trung et al., 2017; Shen et al., 2018). In addition, several research groups have proposed the other risk factors with high potential for dissemination of mcr genes to humans, especially environmental contaminations (Liu et al., 2016; Malhotra-Kumar et al., 2016; Trung et al., 2017; Shen et al., 2018; Agnoletti et al., 2018). Based on a meta-analysis of publications in six major databases published between 18 November 2015 and 30 December 2018, environmental samples exhibited the highest cumulative average prevalence of mcr genes, followed by animals, food, and humans. In human, 62% were from clinical patients and 38% were from asymptomatic carriers (Elbediwi et al., 2019). Thus, based on these findings, the presence of Citrobacter spp. carrying mcr gene in healthy individual found in our study may be due to ingestion of contaminated food animals or environmental. Therefore, strategic action plans, such as surveillance programs of human, animal and environmental setting which is the perspective of “One Health” to control and prevent the spread of mcr genes are urgently needed.
In conclusion, in this study, two colistin-resistant Citrobacter spp. were isolated from feces of healthy individuals. The two isolates, C. sedlakii strain SY-CS04 and C. amalonaticus strain SY-CA35 were newly assigned to ST682, and ST681, respectively. Both isolates exhibited resistant phenotype only to colistin, which is mediated by IncFII(pCoo) plasmid harboring mcr-3.5. These plasmids displayed high transferring efficiency and conferred colistin resistance to transconjugant E. coli. These findings suggest the widespread of mcr plasmid-mediated colistin resistance among Enterobacterales species. It is worth noting that both Citrobacter isolates harbored only mcr gene but no any other major antimicrobial resistant determinants found in multidrug resistant Enterobacterales. To the best of our knowledge, this is the first report of mcr alleles in C. sedlakii and mcr-3 in C. amalonaticus. Due to the fact that the two Citrobacter spp. were isolated from the healthy individual and lacked major resistant determinants in multidrug resistant Enterobacterales, our results suggested an ongoing evolution of mcr gene in human under unknown selection. More importantly, since Citrobacter spp. is one of the most abundant intestinal bacteria, our findings supported the theory that Citrobacter may serve as a reservoir of antibiotic resistant genes, which poses a significant public health threat.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/supplementary material.
Ethics statement
Ethical approval in this study was waived by the Mahidol University Central Institutional Review Board (MU-CIRB), Mahidol University (Nakhon Pathom, Thailand) because the sample used is anonymous. All protocols were in accordance with the ethical standards of our institution and with the 1964 Helsinki Declaration and its later amendments or comparable ethical standards.
Author contributions
SY conceived the project proposal. TP isolated and identified the bacteria and performed antibiotic susceptibility testing. TP and NS performed bioinformatics analysis of WGS. TP and SW performed molecular experiments including PCR and PFGE. SY, ST, and CI-N-A evaluated the data and provided expertise and feedback. TP wrote the preliminary draft of the manuscript. SY edited and finalized the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by the Royal Golden Jubilee PhD Program (grant number PHD/0064/2561) and Research Career Development Grant (grant number RSA6280021) from Thailand Research Fund (TRF) and the National Research Council of Thailand (NRCT). This work was partly supported by the Graduate Scholarship of the Faculty of Medical Technology, Mahidol University.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
AbuOun, M., Stubberfield, E. J., Duggett, N. A., Kirchner, M., Dormer, L., Nunez-Garcia, J., et al. (2018). Mcr-1 and mcr-2 (mcr-6.1) variant genes identified in moraxella species isolated from pigs in great Britain from 2014 to 2015. J. Antimicrob. Chemother. 73, 2904. doi: 10.1093/jac/dky272
Agnoletti, F., Brunetta, R., Bano, L., Drigo, I., Mazzolini, E. (2018). Longitudinal study on antimicrobial consumption and resistance in rabbit farming. Int. J. Antimicrob. Agents 51, 197–205. doi: 10.1016/j.ijantimicag.2017.10.007
Andrews, S. FastQC: A quality control tool for high throughput sequence data. Available at: http://www.bioinformatics.babraham.ac.uk/projects/fastqc/ (Accessed February 2, 2022).
Aziz, R. K., Bartels, D., Best, A. A., DeJongh, M., Disz, T., Edwards, R. A., et al. (2008). The RAST server: rapid annotations using subsystems technology. BMC Genomics 9, 75. doi: 10.1186/1471-2164-9-75
Barton, B. M., Harding, G. P., Zuccarelli, A. J. (1995). A general method for detecting and sizing large plasmids. Anal. Biochem. 226, 235–240. doi: 10.1006/abio.1995.1220
Bitar, I., Papagiannitsis, C. C., Kraftova, L., Chudejova, K., Mattioni Marchetti, V., Hrabak, J. (2020). Detection of five mcr-9-Carrying enterobacterales isolates in four Czech hospitals. mSphere 5, e01008-20. doi: 10.1128/mSphere.01008-20
Borowiak, M., Baumann, B., Fischer, J., Thomas, K., Deneke, C., Hammerl, J. A., et al. (2020). Development of a novel mcr-6 to mcr-9 multiplex PCR and assessment of mcr-1 to mcr-9 occurrence in colistin-resistant salmonella enterica isolates from environment, feed, animals and food, (2011-2018) in Germany. Front. Microbiol. 11. doi: 10.3389/fmicb.2020.00080
Borowiak, M., Fischer, J., Hammerl, J. A., Hendriksen, R. S., Szabo, I., Malorny, B. (2017). Identification of a novel transposon-associated phosphoethanolamine transferase gene, mcr-5, conferring colistin resistance in d-tartrate fermenting salmonella enterica subsp. enterica serovar paratyphi b. J. Antimicrob. Chemother. 72, 3317–3324. doi: 10.1093/jac/dkx327
Bortolaia, V., Kaas, R. S., Ruppe, E., Roberts, M. C., Schwarz, S., Cattoir, V., et al. (2020). ResFinder 4.0 for predictions of phenotypes from genotypes. J. Antimicrob. Chemother. 75, 3491–3500. doi: 10.1093/jac/dkaa345
Carattoli, A. (2009). Resistance plasmid families in enterobacteriaceae. Antimicrob. Agents Chemother. 53, 2227–2238. doi: 10.1128/AAC.01707-08
Carattoli, A., Hasman, H. (2020). PlasmidFinder and in silico pMLST: Identification and typing of plasmid replicons in whole-genome sequencing (WGS). Methods Mol. Biol. 2075, 285–294. doi: 10.1007/978-1-4939-9877-7_20
Chen, L., Mathema, B., Chavda, K. D., DeLeo, F. R., Bonomo, R. A., Kreiswirth, B. N. (2014). Carbapenemase-producing klebsiella pneumoniae: molecular and genetic decoding. Trends Microbiol. 22, 686–696. doi: 10.1016/j.tim.2014.09.003
Clermont, D., Motreff, L., Passet, V., Fernandez, J.-C., Bizet, C., Brisse, S. (2015). Multilocus sequence analysis of the genus citrobacter and description of citrobacter pasteurii sp. nov. Int. J. Syst. Evol. Microbiol. 65, 1486–1490. doi: 10.1099/ijs.0.000122
Clinical Laboratory and Standards Institute (CLSI) (2020). Performance standards for antimicrobial susceptibility testing. 32nd ed (Wayne, PA, USA: Clinical and Laboratory Standards Institute). M100 2022.
Eiamphungporn, W., Yainoy, S., Jumderm, C., Tan-Arsuwongkul, R., Tiengrim, S., Thamlikitkul, V. (2018). Prevalence of the colistin resistance gene mcr-1 in colistin-resistant escherichia coli and klebsiella pneumoniae isolated from humans in Thailand. J. Glob. Antimicrob. Resist. 15, 32–35. doi: 10.1016/j.jgar.2018.06.007
Elbediwi, M., Li, Y., Paudyal, N., Pan, H., Li, X., Xie, S., et al. (2019). Global burden of colistin-resistant bacteria: Mobilized colistin resistance genes study, (1980-2018). Microorganisms 7, 461. doi: 10.3390/microorganisms7100461
Faccone, D., Albornoz, E., Tijet, N., Biondi, E., Gomez, S., Pasterán, F., et al. (2019). Characterization of a multidrug resistant citrobacter amalonaticus clinical isolate harboring blaNDM-1 and mcr-1.5 genes. Infect. Genet. Evol. 67, 51–54. doi: 10.1016/j.meegid.2018.10.020
Farmer, J. J., Davis, B. R., Hickman-Brenner, F. W., McWhorter, A., Huntley-Carter, G. P., Asbury, M. A., et al. (1985). Biochemical identification of new species and biogroups of enterobacteriaceae isolated from clinical specimens. J. Clin. Microbiol. 21, 46–76. doi: 10.1128/jcm.21.1.46-76.1985
Hanson, N. D., Sanders, C. C. (1999). Regulation of inducible AmpC beta-lactamase expression among enterobacteriaceae. Curr. Pharm. Des. 5, 881–894.
Hu, Y.-Y., Wang, Y.-L., Sun, Q.-L., Huang, Z.-X., Wang, H.-Y., Zhang, R., et al. (2017). Colistin resistance gene mcr-1 in gut flora of children. Int. J. Antimicrob. Agents 50, 593–597. doi: 10.1016/j.ijantimicag.2017.06.011
Jacobson, K. L., Cohen, S. H., Inciardi, J. F., King, J. H., Lippert, W. E., Iglesias, T., et al. (1995). The relationship between antecedent antibiotic use and resistance to extended-spectrum cephalosporins in group I beta-lactamase-producing organisms. Clin. Infect. Dis. 21, 1107–1113. doi: 10.1093/clinids/21.5.1107
Jiang, X., Cui, X., Liu, W., Xu, H., Zheng, B. (2019). Genetic characterization of a novel sequence type of multidrug-resistant citrobacter freundii strain recovered from wastewater treatment plant. Infect. Drug Resist. 12, 2775–2779. doi: 10.2147/IDR.S213525
Jolley, K. A., Maiden, M. C. J. (2010). BIGSdb: Scalable analysis of bacterial genome variation at the population level. BMC Bioinf. 11, 595. doi: 10.1186/1471-2105-11-595
Kanamori, H., Yano, H., Hirakata, Y., Endo, S., Arai, K., Ogawa, M., et al. (2011). High prevalence of extended-spectrum β-lactamases and qnr determinants in citrobacter species from Japan: dissemination of CTX-M-2. J. Antimicrob. Chemother. 66, 2255–2262. doi: 10.1093/jac/dkr283
Khajanchi, B. K., Kaldhone, P. R., Foley, S. L. (2019). Protocols of conjugative plasmid transfer in salmonella: plate, broth, and filter mating approaches. Methods Mol. Biol. 2016, 129–139. doi: 10.1007/978-1-4939-9570-7_12
Kieffer, N., Royer, G., Decousser, J.-W., Bourrel, A.-S., Palmieri, M., Ortiz de la Rosa, J.-M., et al. (2019). Mcr-9, an inducible gene encoding an acquired phosphoethanolamine transferase in escherichia coli, and its origin. Antimicrob. Agents Chemother. 63, e00965-19. doi: 10.1128/AAC.00965-19
Lee, C.-H., Lee, Y.-T., Kung, C.-H., Ku, W.-W., Kuo, S.-C., Chen, T.-L., et al. (2015). Risk factors of community-onset urinary tract infections caused by plasmid-mediated AmpC β-lactamase-producing enterobacteriaceae. J. Microbiol. Immunol. Infect. 48, 269–275. doi: 10.1016/j.jmii.2013.08.010
Lescat, M., Poirel, L., Nordmann, P. (2018). Rapid multiplex polymerase chain reaction for detection of mcr-1 to mcr-5 genes. Diagn. Microbiol. Infect. Dis. 92, 267–269. doi: 10.1016/j.diagmicrobio.2018.04.010
Letunic, I., Bork, P. (2021). Interactive tree of life (iTOL) v5: an online tool for phylogenetic tree display and annotation. Nucleic Acids Res. 49, W293–W296. doi: 10.1093/nar/gkab301
Li, X.-P., Fang, L.-X., Jiang, P., Pan, D., Xia, J., Liao, X.-P., et al. (2017). Emergence of the colistin resistance gene mcr-1 in citrobacter freundii. Int. J. Antimicrob. Agents 49, 786–787. doi: 10.1016/j.ijantimicag.2017.04.004
Liu, L., Feng, Y., Zhang, X., McNally, A., Zong, Z. (2017). New variant of mcr-3 in an extensively drug-resistant escherichia coli clinical isolate carrying mcr-1 and blaNDM-5. Antimicrob. Agents Chemother. 61, e01757-17. doi: 10.1128/AAC.01757-17
Liu, Y.-Y., Wang, Y., Walsh, T. R., Yi, L.-X., Zhang, R., Spencer, J., et al. (2016). Emergence of plasmid-mediated colistin resistance mechanism MCR-1 in animals and human beings in China: a microbiological and molecular biological study. Lancet Infect. Dis. 16, 161–168. doi: 10.1016/S1473-3099(15)00424-7
Liu, L.-H., Wang, N.-Y., Wu, A. Y.-J., Lin, C.-C., Lee, C.-M., Liu, C.-P. (2018b). Citrobacter freundii bacteremia: Risk factors of mortality and prevalence of resistance genes. J. Microbiol. Immunol. Infect. 51, 565–572. doi: 10.1016/j.jmii.2016.08.016
Liu, J., Yang, Y., Li, Y., Liu, D., Tuo, H., Wang, H., et al. (2018a). Isolation of an IncP-1 plasmid harbouring mcr-1 from a chicken isolate of citrobacter braakii in China. Int. J. Antimicrob. Agents 51, 936–940. doi: 10.1016/j.ijantimicag.2017.12.030
Madec, J. Y., Haenni, M., Nordmann, P., Poirel, L. (2017). Extended-spectrum β-lactamase/AmpC- and carbapenemase-producing enterobacteriaceae in animals: a threat for humans? Clin. Microbiol. Infect. 23, 826–833. doi: 10.1016/j.cmi.2017.01.013
Malhotra-Kumar, S., Xavier, B. B., Das, A. J., Lammens, C., Hoang, H. T. T., Pham, N. T., et al. (2016). Colistin-resistant escherichia coli harbouring mcr-1 isolated from food animals in Hanoi, Vietnam. Lancet Infect. Dis. 16, 286–287. doi: 10.1016/S1473-3099(16)00014-1
Mmatli, M., Mbelle, N. M., Osei Sekyere, J. (2022). Global epidemiology, genetic environment, risk factors and therapeutic prospects of mcr genes: A current and emerging update. Front. Cell. Infect. Microbiol. 12. doi: 10.3389/fcimb.2022.941358
Mohanty, S., Singhal, R., Sood, S., Dhawan, B., Kapil, A., Das, B. K. (2007). Citrobacter infections in a tertiary care hospital in northern India. J. Infect. 54, 58–64. doi: 10.1016/j.jinf.2006.01.015
Oberhettinger, P., Schüle, L., Marschal, M., Bezdan, D., Ossowski, S., Dörfel, D., et al. (2020). Description of citrobacter cronae sp. nov., isolated from human rectal swabs and stool samples. Int. J. Syst. Evol. Microbiol. 70, 2998–3003. doi: 10.1099/ijsem.0.004100
Ouchar Mahamat, O., Kempf, M., Lounnas, M., Tidjani, A., Hide, M., Benavides, J. A., et al. (2021). Epidemiology and prevalence of extended-spectrum β-lactamase- and carbapenemase-producing enterobacteriaceae in humans, animals and the environment in West and central Africa. Int. J. Antimicrob. Agents 57, 106203. doi: 10.1016/j.ijantimicag.2020.106203
Page, A. J., Cummins, C. A., Hunt, M., Wong, V. K., Reuter, S., Holden, M. T. G., et al. (2015). Roary: rapid large-scale prokaryote pan genome analysis. Bioinformatics 31, 3691–3693. doi: 10.1093/bioinformatics/btv421
Papp-Wallace, K. M., Endimiani, A., Taracila, M. A., Bonomo, R. A. (2011). Carbapenems: past, present, and future. Antimicrob. Agents Chemother. 55, 4943–4960. doi: 10.1128/AAC.00296-11
Phuadraksa, T., Wichit, S., Arikit, S., Songtawee, N., Yainoy, S. (2022). Co-Occurrence of mcr-2 and mcr-3 genes on chromosome of multidrug-resistant escherichia coli isolated from healthy individuals in Thailand. Int. J. Antimicrob. Agents 60, 106662. doi: 10.1016/j.ijantimicag.2022.106662
Prjibelski, A., Antipov, D., Meleshko, D., Lapidus, A., Korobeynikov, A. (2020). Using SPAdes de novo assembler. Curr. Protoc. Bioinf. 70, e102. doi: 10.1002/cpbi.102
Rahal, J. J. (2008). The role of carbapenems in initial therapy for serious gram-negative infections. Crit. Care 12 Suppl 4, S5. doi: 10.1186/cc6821
Sadek, M., Ortiz de la Rosa, J. M., Abdelfattah Maky, M., Korashe Dandrawy, M., Nordmann, P., Poirel, L. (2021). Genomic features of MCR-1 and extended-spectrum β-Lactamase-Producing enterobacterales from retail raw chicken in Egypt. Microorganisms 9, 195. doi: 10.3390/microorganisms9010195
Samonis, G., Karageorgopoulos, D. E., Kofteridis, D. P., Matthaiou, D. K., Sidiropoulou, V., Maraki, S., et al. (2009). Citrobacter infections in a general hospital: characteristics and outcomes. Eur. J. Clin. Microbiol. Infect. Dis. 28, 61–68. doi: 10.1007/s10096-008-0598-z
Seemann, T. (2014). Prokka: rapid prokaryotic genome annotation. Bioinformatics 30, 2068–2069. doi: 10.1093/bioinformatics/btu153
Sennati, S., Di Pilato, V., Riccobono, E., Di Maggio, T., Villagran, A. L., Pallecchi, L., et al. (2017). Citrobacter braakii carrying plasmid-borne mcr-1 colistin resistance gene from ready-to-eat food from a market in the chaco region of Bolivia. J. Antimicrob. Chemother. 72, 2127–2129. doi: 10.1093/jac/dkx078
Shahid, M. (2010). Citrobacter spp. simultaneously harboring blaCTX-m, blaTEM, blaSHV, blaampC, and insertion sequences IS26 and orf513: an evolutionary phenomenon of recent concern for antibiotic resistance. J. Clin. Microbiol. 48, 1833–1838. doi: 10.1128/JCM.01467-09
Shen, Y., Zhou, H., Xu, J., Wang, Y., Zhang, Q., Walsh, T. R., et al. (2018). Anthropogenic and environmental factors associated with high incidence of mcr-1 carriage in humans across China. Nat. Microbiol. 3, 1054–1062. doi: 10.1038/s41564-018-0205-8
Song, S., Wood, T. K. (2020). A primary physiological role of toxin/antitoxin systems is phage inhibition. Front. Microbiol. 11. doi: 10.3389/fmicb.2020.01895
Tacconelli, E., Carrara, E., Savoldi, A., Harbarth, S., Mendelson, M., Monnet, D. L., et al. (2018). Discovery, research, and development of new antibiotics: the WHO priority list of antibiotic-resistant bacteria and tuberculosis. Lancet Infect. Dis. 18, 318–327. doi: 10.1016/S1473-3099(17)30753-3
Trung, N. V., Matamoros, S., Carrique-Mas, J. J., Nghia, N. H., Nhung, N. T., Chieu, T. T. B., et al. (2017). Zoonotic transmission of mcr-1 colistin resistance gene from small-scale poultry farms, Vietnam. Emerging Infect. Dis. 23, 529–532. doi: 10.3201/eid2303.161553
Wang, J. T., Chang, S. C., Chen, Y. C., Luh, K. T. (2000). Comparison of antimicrobial susceptibility of citrobacter freundii isolates in two different time periods. J. Microbiol. Immunol. Infect. 33, 258–262.
Wang, C., Feng, Y., Liu, L., Wei, L., Kang, M., Zong, Z. (2020). Identification of novel mobile colistin resistance gene mcr-10. Emerg. Microbes Infect. 9, 508–516. doi: 10.1080/22221751.2020.1732231
Wang, X., Wang, Y., Zhou, Y., Li, J., Yin, W., Wang, S., et al. (2018). Emergence of a novel mobile colistin resistance gene, mcr-8, in NDM-producing klebsiella pneumoniae. Emerg. Microbes Infect. 7, 122. doi: 10.1038/s41426-018-0124-z
Xavier, B. B., Lammens, C., Ruhal, R., Kumar-Singh, S., Butaye, P., Goossens, H., et al. (2016). Identification of a novel plasmid-mediated colistin-resistance gene, mcr-2, in escherichia coli, Belgium, June 2016. Euro. Surveill. 21, 27. doi: 10.2807/1560-7917.ES.2016.21.27.30280
Yang, Y.-Q., Li, Y.-X., Lei, C.-W., Zhang, A.-Y., Wang, H.-N. (2018). Novel plasmid-mediated colistin resistance gene mcr-7.1 in klebsiella pneumoniae. J. Antimicrob. Chemother. 73, 1791–1795. doi: 10.1093/jac/dky111
Yin, W., Li, H., Shen, Y., Liu, Z., Wang, S., Shen, Z., et al. (2017). Novel plasmid-mediated colistin resistance gene mcr-3 in escherichia coli. MBio 8, e00543-17. doi: 10.1128/mBio.00543-17
Zelendova, M., Papagiannitsis, C. C., Valcek, A., Medvecky, M., Bitar, I., Hrabak, J., et al. (2020). Characterization of the complete nucleotide sequences of mcr-1-Encoding plasmids from enterobacterales isolates in retailed raw meat products from the Czech republic. Front. Microbiol. 11. doi: 10.3389/fmicb.2020.604067
Zhang, R., Yang, L., Cai, J. C., Zhou, H. W., Chen, G.-X. (2008). High-level carbapenem resistance in a citrobacter freundii clinical isolate is due to a combination of KPC-2 production and decreased porin expression. J. Med. Microbiol. 57, 332–337. doi: 10.1099/jmm.0.47576-0
Zheng, B., Xu, H., Lv, T., Guo, L., Xiao, Y., Huang, C., et al. (2020). Stool samples of acute diarrhea inpatients as a reservoir of ST11 hypervirulent KPC-2-Producing klebsiella pneumoniae. mSystems 5, e00498-20. doi: 10.1128/mSystems.00498-20
Keywords: colistin resistance, citrobacter spp., citrobacter sedlakii, citrobacter amalonaticus, mcr gene, mcr-3
Citation: Phuadraksa T, Wichit S, Songtawee N, Tantimavanich S, Isarankura-Na-Ayudhya C and Yainoy S (2023) Emergence of plasmid-mediated colistin resistance mcr-3.5 gene in Citrobacter amalonaticus and Citrobacter sedlakii isolated from healthy individual in Thailand. Front. Cell. Infect. Microbiol. 12:1067572. doi: 10.3389/fcimb.2022.1067572
Received: 12 October 2022; Accepted: 08 December 2022;
Published: 04 January 2023.
Edited by:
Min Yue, Zhejiang University, ChinaReviewed by:
Jiangang Ma, Zhejiang Academy of Agricultural Sciences, ChinaVittoria Mattioni Marchetti, Charles University, Czechia
Copyright © 2023 Phuadraksa, Wichit, Songtawee, Tantimavanich, Isarankura-Na-Ayudhya and Yainoy. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Sakda Yainoy, c2FrZGEueWFpQG1haGlkb2wuYWMudGg=
 Thanawat Phuadraksa
Thanawat Phuadraksa Sineewanlaya Wichit1
Sineewanlaya Wichit1 Napat Songtawee
Napat Songtawee Sakda Yainoy
Sakda Yainoy