
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Cell. Infect. Microbiol., 27 October 2022
Sec. Parasite and Host
Volume 12 - 2022 | https://doi.org/10.3389/fcimb.2022.1037586
This article is part of the Research TopicAdvances in Molecular Biology, Pathogenesis, Diagnosis, Vaccines, and Treatment of Diseases Caused by Apicomplexan ParasitesView all 14 articles
 Yang Zou1,2,3†
Yang Zou1,2,3† Jin-Xin Meng4†
Jin-Xin Meng4† Xin-Yu Wei4,5†
Xin-Yu Wei4,5† Xiao-Yi Gu1
Xiao-Yi Gu1 Chao Chen6
Chao Chen6 Hong-Li Geng4
Hong-Li Geng4 Li-Hua Yang2*
Li-Hua Yang2* Xiao-Xuan Zhang4*
Xiao-Xuan Zhang4* Hong-Wei Cao1*
Hong-Wei Cao1*Toxoplasmosis is an important zoonotic parasitic disease caused by Toxoplasma gondii (T. gondii). However, the functions of circRNAs and miRNAs in response to T. gondii infection in the livers of mice at acute and chronic stages remain unknown. Here, high-throughput RNA sequencing was performed for detecting the expression of circRNAs and miRNAs in livers of mice infected with 20 T. gondii cysts at the acute and chronic stages, in order to understand the potential molecular mechanisms underlying hepatic toxoplasmosis. Overall, 265 and 97 differentially expressed (DE) circRNAs were found in livers at the acute and chronic infection stages in comparison with controls, respectively. In addition, 171 and 77 DEmiRNAs were found in livers at the acute and chronic infection stages, respectively. Functional annotation showed that some immunity-related Gene ontology terms, such as “positive regulation of cytokine production”, “regulation of T cell activation”, and “immune receptor activity”, were enriched at the two infection stages. Moreover, the pathways “Valine, leucine, and isoleucine degradation”, “Fatty acid metabolism”, and “Glycine, serine, and threonine metabolism” were involved in liver disease. Remarkably, DEcircRNA 6:124519352|124575359 was significantly correlated with DEmiRNAs mmu-miR-146a-5p and mmu-miR-150-5p in the network that was associated with liver immunity and pathogenesis of disease. This study revealed that the expression profiling of circRNAs in the livers was changed after T. gondii infection, and improved our understanding of the transcriptomic landscape of hepatic toxoplasmosis in mice.
Toxoplasmosis is a widespread zoonotic disease caused by Toxoplasma gondii (T. gondii) worldwide. T. gondii is an intracellular apicomplexan parasite that can infect almost all warm-blooded animals and humans (Tenter et al., 2000). Gamogony and oocyst can form in the epithelium of small intestine after an ingestion of T. gondii by definitive feline hosts. Then, the unsporulated oocysts can beare released into the intestinal lumen and excrete with feces, leading to contamination of soil and the environment (Tenter et al., 2000). The people who ingested undercooked food or water containing tissue cysts and sporulated oocysts will be infected by T. gondii (Dubey, 2008). The clinical manifestations of toxoplasmosis range from asymptomatic to fatal infection, including abortion, encephalitic illness, and conjunctivitis (Smith et al., 2021). T. gondii can attack the host organs, including livers, lymph nodes, eyes, hearts, and central nervous systems (Montoya and Liesenfeld, 2004; Stauffer et al., 2006; Balasundaram, 2010). In livers, T. gondii infection can cause several pathological changes, e.g. hepatitis, hepatomegaly, granuloma, and necrosis (Karasawa et al., 1981; Ortego et al., 1990; Hassan et al., 1996; Doğan et al., 2007). However, the molecular mechanisms underlying T. gondii infection and liver disease remain poorly understood.
Circular RNAs are one of the novel classes of endogenous noncoding RNAs that are formed by exon-scrambling (Meng et al., 2017). With the development of RNA sequencing (RNA-seq) technology, the abundance, diversity, and dynamic expression patterns of circRNAs in various organisms have been clarified (Conn et al., 2015). CircRNAs can not only antagonize the activity of miRNA through a sponge-like mechanism but also regulate gene expression at the post-transcriptional level (Granados-Riveron and Aquino-Jarquin, 2016). A series of studies showed that circRNAs played roles in the pathological processes of liver disease (Yao et al., 2017; Tang et al., 2020). The circRNAs are used as prognostic biomarkers, owing to remarkably stable characteristics (Lei et al., 2019). In addition, circRNAs are a potential drug target for diseases (Dhamija and Menon, 2018; Zhu et al., 2018). Thus, exploration of circRNA function that connecting with liver disease induced by T. gondii infection will provide a novel perspective for hepatic disease treatment and diagnosis.
In the present study, RNA-seq was performed for identifying the expression of circRNAs and miRNAs in the livers of mice after T. gondii infection, in order to investigate the relationships between circRNAs and miRNAs in the T. gondii infected livers of mice. The simultaneous analyses of the differentially expressed (DE) circRNAs and DE miRNAs were conducted to investigate the relevance of the expression and circRNA-miRNA interactions. Moreover, the potential functional role was predicted. Thus, the correlation networks of circRNAs and miRNAs in the livers of mice after T. gondii infection improved our understanding of the transcriptomic landscape of hepatic toxoplasmosis in mice.
T. gondii cysts were collected from the brains of mice that had been infected with T. gondii for months. In brief, the mice were sacrificed after anesthetization, and the brains were dissected and collected with a mortar for preparing tissue homogenates. The brain homogenates were rinsed with phosphate-buffered saline (PBS), and then were transferred to a 2 mL EP tube. Then, the T. gondii cysts in brain tissues were counted by using a dissection microscope. The 8-10 week-old female BALB/c mice (SPF) were purchased from Spaefer Biotechnology Co., Ltd. (Beijing, China). All mice were housed in cages with an independent ventilation system under a 12-h dark/light cycle, with free food and water ad libitum. The mice (n = 12) were randomly divided into three groups: acute infection group (n = 3); chronic infection group (n = 3); and control group (n=6). In the infection groups, each mouse was infected with 20 T. gondii cysts. In the control group, the mice were treated with PBS. A previous study showed the timing of acute and chronic infection stages in mice infected with T. gondii (Hu et al., 2018). The mice in each group were sacrificed on day 11 (acute infection group) and 33 (chronic infection group) after infection, respectively. The successful establishment of mouse model was examined based on amplification of T. gondii B1 gene as described previously (Hu et al., 2018). At the mentioned time points post infection, the livers of mice in each group were dissected from each mouse. Then, the liver samples were immediately deposited in liquid nitrogen until RNA extraction.
Approximately 50 mg of liver tissue was subjected to RNA extraction using TRIZOL (Life Technologies, Carlsbad, USA). In brief, the samples were firstly homogenized by liquid nitrogen, and the 1 ml TRIZOL reagent was added to the homogenization to lyse sample. Then, the 0.2 ml of chloroform per was added and shake tubes vigorously by hand for 15 s. The sample was incubated at room temperature for 2 to 3 minutes, and then centrifuged at 11,500 g for 15 min at 4°C. The RNA samples remain in the aqueous phase. The aqueous phase was transferred to a new tube, and isopropanol was added to precipitate RNA. The 75% ethanol was used to wash RNA samples; Finally, RNA pellet was redissolved with the water (Chomczynski and Sacchi, 2006). The RNA degradation and contamination were detected with 1% agarose gel test. The RNA Nano 6000 Assay Kit of the Bioanalyzer 2100 system (Agilent Technologies, CA, USA) and Qubit® RNA Assay Kit in Qubit® 2.0 Flurometer (Life Technologies, CA, USA) were used to measure and evaluate the concentration and purity of RNA, respectively. Then, the RNA samples were stored at −80°C for a further analysis.
Approximately 5 μg of RNA sample was used for constructing the circRNA library by using NEBNext® Ultra™ Directional RNALibrary Prep Kit for Illumina® (NEB, USA). In brief, the First-strand cDNA was synthesized using M-MuLV Reverse Transcriptase with random hexamer primer. Then, the cDNA fragments were purified by AMPure XP system (Beckman Coulter, Beverly, USA). The cDNA was used for PCR amplification. Agilent Bioanalyzer 2100 system was employed for assessing library quality (Zhou et al., 2017). The sequencing libraries of circRNAs were performed using Illumina Hiseq 4000 platform, and the 150 bp paired-end reads were generated.
The 3 μg of RNA sample and NEBNext® Multiplex Small RNA Library Prep Set for Illumina® (NEB, USA) was used for generating miRNA libraries. Briefly, the first strand of cDNA of miRNA was synthesized through M-MuLV Reverse Transcriptase. Then, the LongAmp Taq 2×Master Mix, index (X) primer, and SR primer were used for PCR amplification. Finally, the library preparations were sequenced on an Illumina Hiseq 2500/2000 platform, and 50 bp single-end reads were generated.
The raw reads of fastq format were obtained by the Custom Perl and Python scripts. The ploy-N, with 5′ adapter contaminants, without 3′ adapters, and low reads were removed. The GC content, Q20, Q30, and the error rate were performed to assess quality of the clean data. The HISAT2 v2.0.4 and bowtie2 v2.2.8 were used for building and aligning clean data with the Mus musculus reference genome, respectively (Langmead and Salzberg, 2012; Pertea et al., 2016). The circRNA identification was performed using find_circ (Memczak et al., 2013) and CIRI2 (Gao et al., 2015). circRNA was predicted by the intersection between the two algorithms. Moreover, the small RNA tags were mapped to obtain known miRNAs using MiRBase 20.0 (Griffiths-Jones, 2016). The novel miRNAs were predicted by using miREvo (Wen et al., 2012) and mirdeep2 (Friedlander et al., 2012). The quantification of circRNA and miRNA expression profiles were normalized by TPM (transcript per million) (Zhou et al., 2010). The differential expression analysis was performed using the DESeq R package (1.8.3) (Anders and Huber, 2010). |Log2 fold change (FC)| ≥ 1.0 and P-value < 0.05 were used as thresholds to identify differentially expressed transcripts.
The potential target genes of DE miRNAs were predicted by a combined use of Miranda, PITA, and RNAhybrid softwares. GO enrichment analysis of the potential target genes in the livers of mice infected with T. gondii was conducted using the GOseq R package (Young et al., 2010). The KEGG (Kyoto encyclopedia of genes and genome) pathway functional annotation were performed by using KOBAS 3.0 software (Mao et al., 2005). P-value < 0.05 was considered as significant enrichment.
The DE circRNAs and DE miRNAs were chosen to verify the RNA-Seq results by using qRT-PCR. The qPCR was performed in a LightCycler480 (Roche, Basel, Switzerland) using a ChamQ SYBR qPCR Master Mix kit (Vazyme, Nanjing, China). The reaction was consisted of 40 cycles, circRNA initial degeneration at 95°C for 30 s, and template degeneration in the PCR cycle at 95°C for 10 s, and finally annealing at 60°C for 30 s. The reactive cycle of miRNA was consisted of 95°C for 30 s, then 40 cycles of 95°C for 5 s and 60°C for 34 s. The amplification was ensured by melting curve analysis in each reaction. The primers of miRNAs and circRNAs were listed in Table 1. L13A and U6 were used as the internal controls of circRNA and miRNA, respectively. The relative expression quantity was calculated using the 2-ΔΔCt method (Livak and Schmittgen, 2001).
Compared with the control group, a total of 265 DE circRNAs and 171 DE miRNAs were identified at the acute infection stage, and 97 DE circRNAs and 77 DE miRNAs were detected in the livers at the chronic infection stage (Figure 1 and Supplementary Table S1). A total of 19 circRNAs and 46 miRNAs were commonly dysregulated between the acute and chronic T. gondii-infected groups (Figure 2). Among DE transcripts, the mmu-miR-147-3p was up-regulated 32.94 folds at the acute infection stage, however, it was down-regulated to 3.66 folds at the chronic infection stage. Moreover, mmu-miR-342-3p was up-regulated 8.23 folds at the acute infection stage. Furthermore, mmu-miR-143-3p was down-regulated 4.05 folds and 2.11 folds at the acute and chronic infection stages, respectively (Supplementary Table S1).
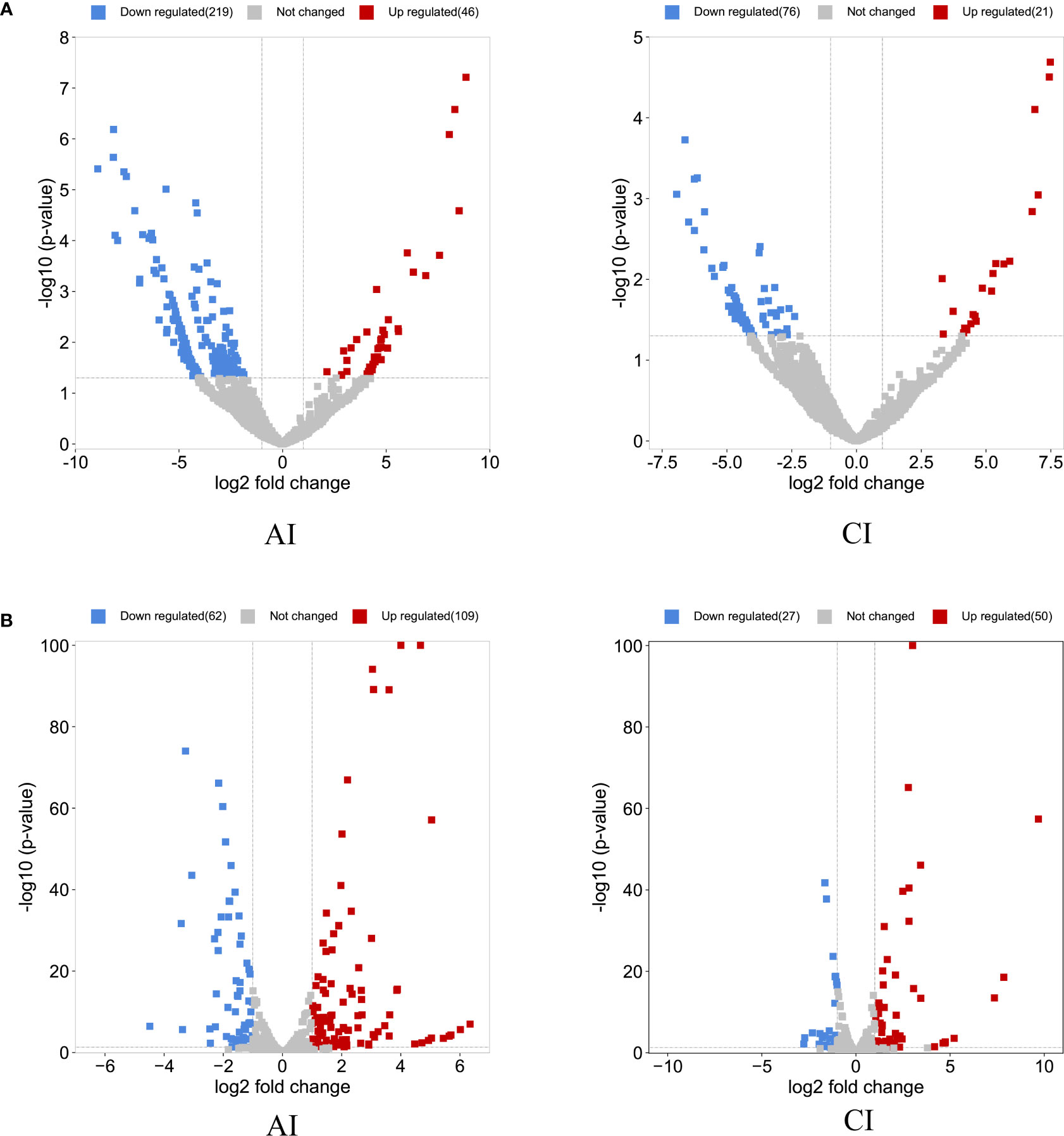
Figure 1 Overview of the differentially expressed (DE) circRNAs and DEmiRNAs. The volcano plots of DEcircRNAs (A) and DEmiRNAs (B) at acute infection (AI) and chronic infection (CI) stages. The horizontal-axis shows the log2 fold change, and the vertical-axis shows the -log10 p-value. The up-regulated are marked in red and the down-regulated RNAs are in blue.
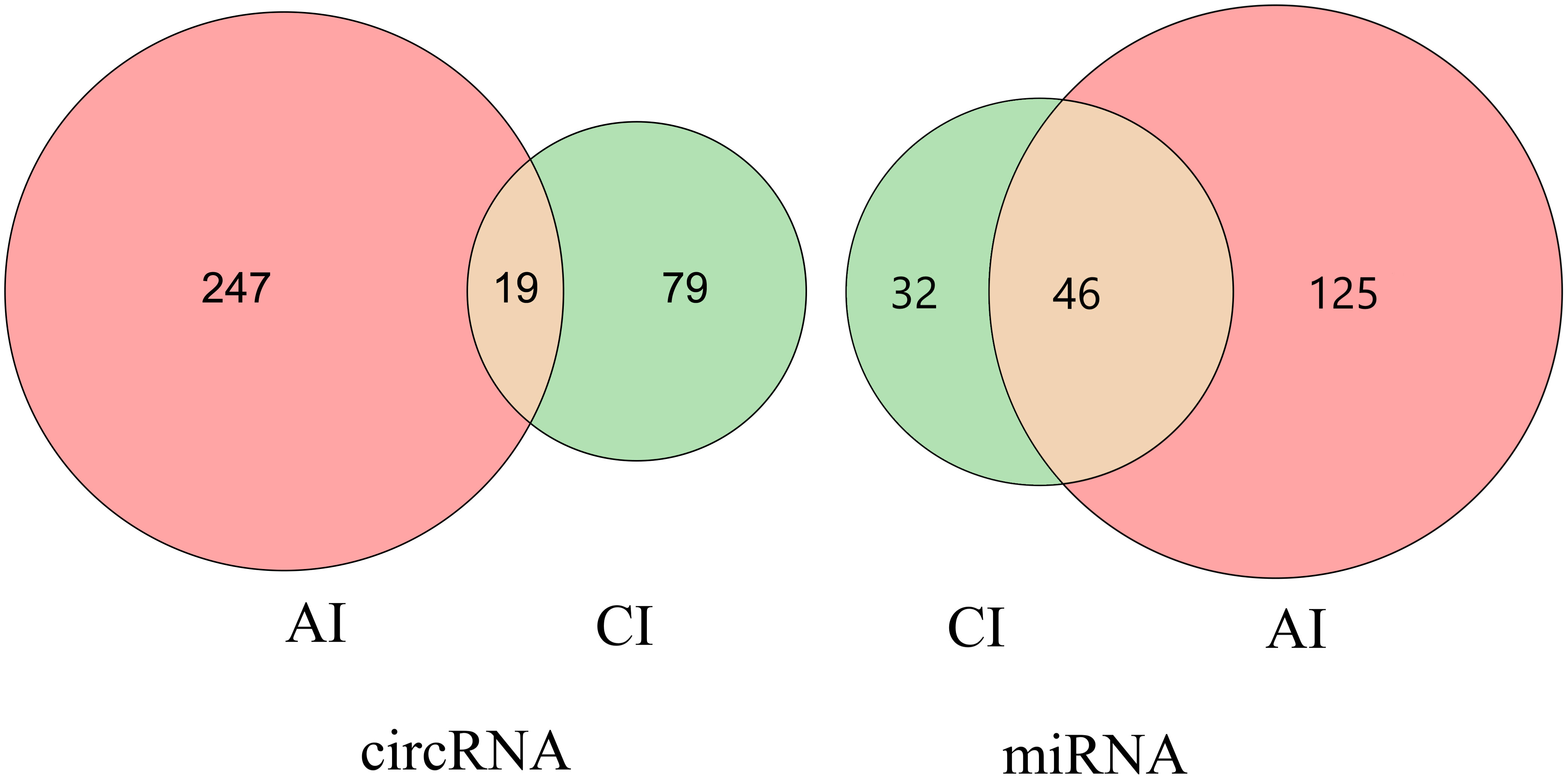
Figure 2 Venn diagram of the differentially expressed (DE) circRNAs and DEmiRNAs. the number of the common or unique DEcircRNAs and DEmiRNAs at two infection stages.
To find the potential biological associations of DE miRNAs, GO and KEGG pathway enrichment analyses for infection-associated transcripts were predicted. The top 30 GO terms were shown in Figure 3. Most of predicted genes were involved in the “fatty acid metabolic process”, “positive regulation of cytokine production”, “positive regulation of response to external stimulus” and “regulation of T cell activation” at the acute infection stage (Figure 3A). Furthermore, the biological process mainly included “leukocyte cell-cell adhesion”, “positive regulation of cytokine production”, and “positive regulation of leukocyte activation”. The cellular component included “membrane raft” and “membrane microdomain”, the molecular function included “immune receptor activity” and “phospholipid binding” at the chronic infection stage (Figure 3B).
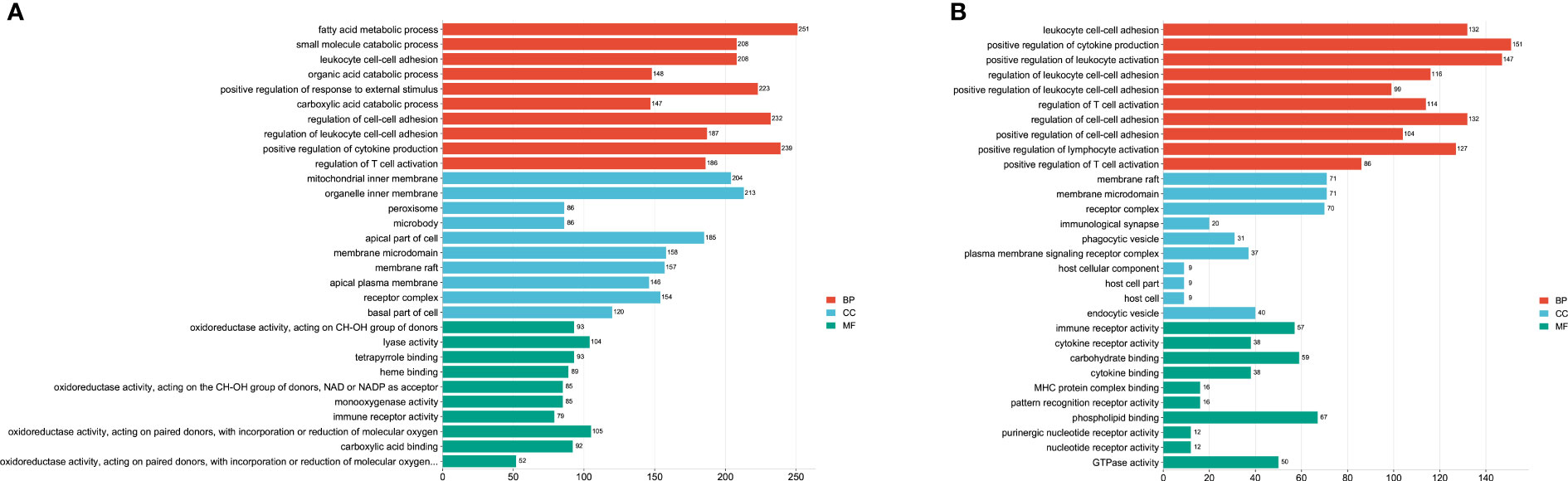
Figure 3 The top 30 GO terms of target genes of the differentially expressed (DE) miRNAs at acute infection (A) and chronic infection (B) stages. The vertical-axis represents the GO terms. The horizontal-axis represents numbers of target genes. The BP represents biological process, CC represents cellular component, and MF represents molecular function.
KEGG enrichment analysis showed that pathways mainly included “Valine, leucine, and isoleucine degradation”, “Fatty acid metabolism”, “Glycine, serine and threonine metabolism”, and “Tryptophan metabolism” (Figure 4A) at the acute infection stage. These results showed the hepatic metabolism affected the acute infection stage of T. gondii. Moreover, some pathways were related to immunity and inflammation, such as “Cytokine-cytokine receptor interaction”, “Cell adhesion molecules”, “NF-kappa B signaling pathway”, “Primary immunodeficiency”, “Inflammatory bowel disease”, “Th1 and Th2 cell differentiation”, “Th17 cell differentiation”, and “NOD-like receptor signaling pathway”. Interestingly, some pathways were related with intestinal flora, e.g. “Inflammatory bowel disease” and “Intestinal immune network for IgA production” (Figure 4B).
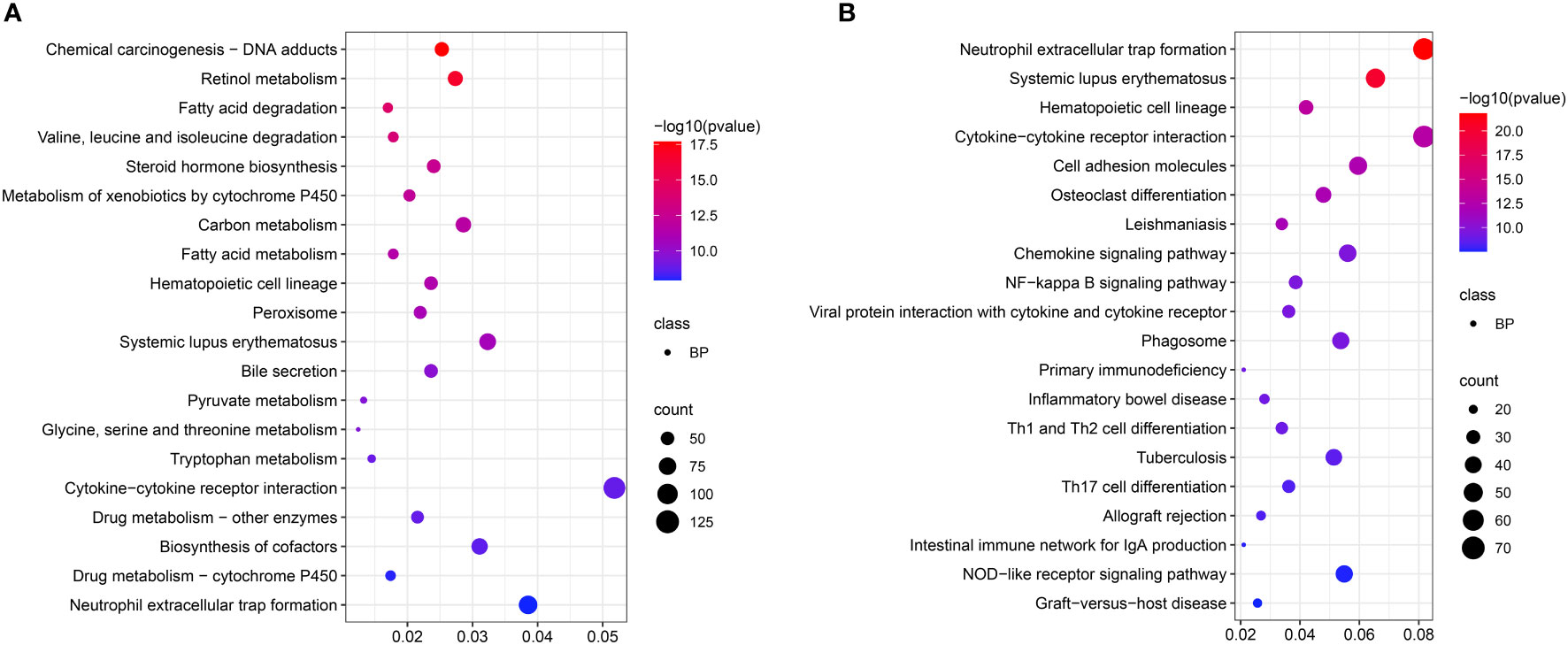
Figure 4 Scatter plots showing KEGG pathway of the target genes. The top 20 enriched pathways at acute infection (A) and chronic infection (B) stages, respectively. The vertical-axis represent pathways, and the horizontal-axis numbers represent the rich factor.
To further reveal the mechanisms underlying the DEcircRNAs and DEmiRNAs in the livers during T. gondii infection, a network was constructed (Figure 5 and Supplementary Table S2). In this network, the DEcircRNA 6:124519352|124575359 was related to DE miRNAs mmu-miR-132-3p, mmu-miR-146a-5p, mmu-miR-150-5p, and other DEmiRNAs (n = 42, Supplementary Table S2). Moreover, DEcircRNA 4:61958498|62052011 was associated with DEmiRNAs (mmu-miR-146a-5p) and 45 other DEmiRNAs. Moreover, DEmiRNA mmu-miR-1247-3p shared 5 DEcircRNAs, including 12:103731961|103897311, 12:103854668|103947209, 5:145708877|145868684, 4:61958498|62052011, and 7:13832577|13909898 (Figure 5). The networks showed that multiple miRNAs were regulated by several circRNAs at the two infection stages, thus suggesting a complex regulatory relationship between DEcircRNAs and DEmiRNAs.
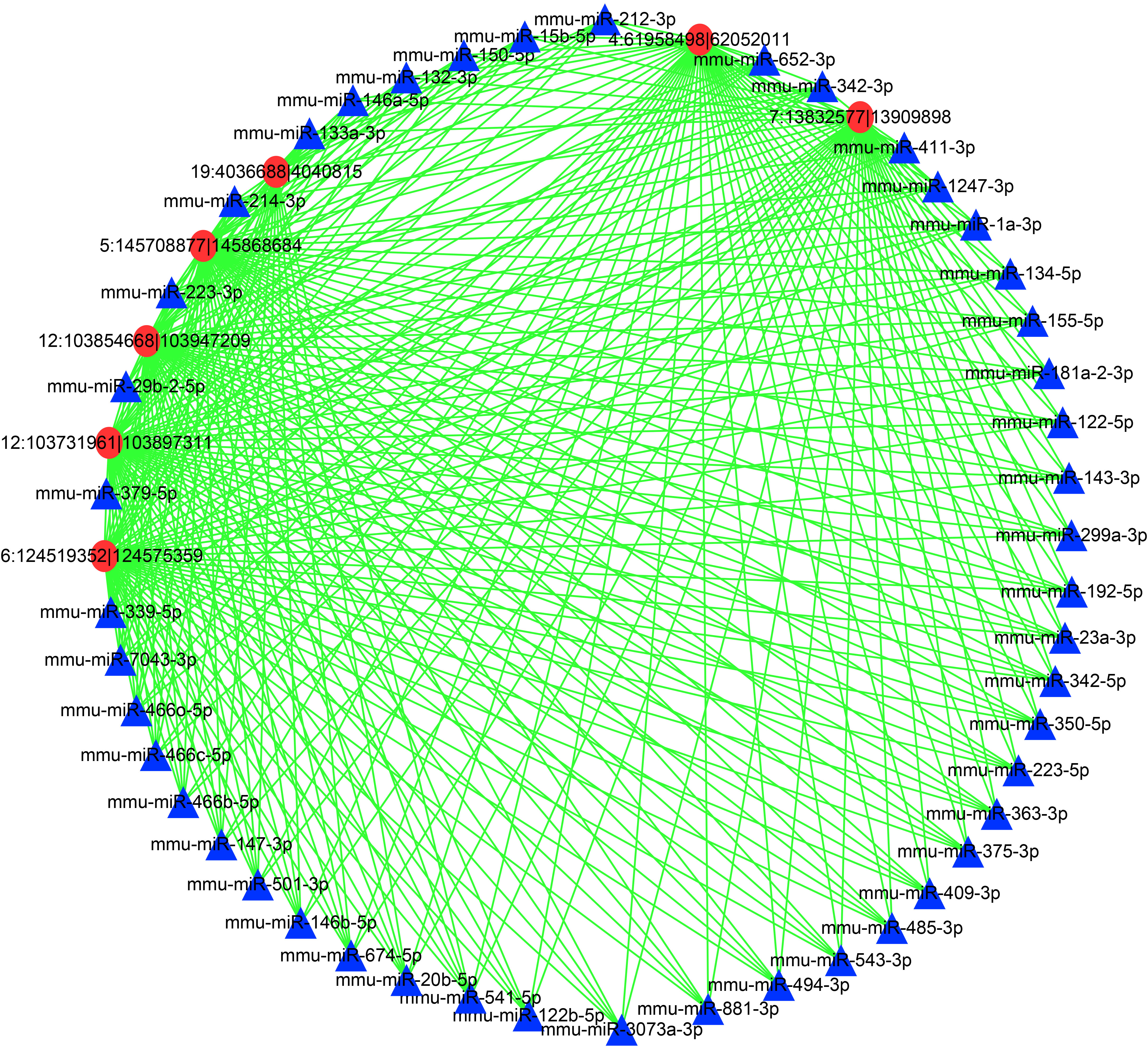
Figure 5 A network of the commonly DEcircRNAs with their associate DEmiRNAs. Different colors show different RNA, with red for DEcircRNAs and blue for DEmiRNAs.
To evaluate the reliability of RNA-sequence results, the expression profiles of the randomly selected DEcircRNAs and DEmiRNAs were successfully confirmed by qRT-PCR. The results obtained by RNA-Seq and qRT-PCR were consistent in the trend and magnitude of the expression (Supplementary Figure S1).
Previous omics studies have provided a wealth of resources that improved understanding of the pathogenesis of T. gondii (Garfoot et al., 2019; Antil et al., 2021; Menard et al., 2021; Antil et al., 2022), and many studies mainly focused on mRNAs (He et al., 2016; Zhou et al., 2016; Lutshumba et al., 2020). Recently, Zhou and colleagues revealed putative functions of miRNAs and circRNAs in brains of mice after an infection with T. gondii (Zhou et al., 2020). However, the expression levels of circRNAs and miRNAs specific to T. gondii infection in livers of mice were unclear. Thus, the present study explored the expression profiles of circRNAs and miRNAs in the livers of mice at acute and chronic stages after T. gondii infection by using RNA-seq technique.
MiR-147-3p (mmu-miR-147-3p) has been reported to dampen Toll-like receptor (TLR)-signaling in murine macrophages (Liu et al., 2009). It can limit excessive inflammation in the hosts response to influenza A virus infection (Preusse et al., 2017). The miR-147 plays an important role in the negative regulation of TLR/NF-κB-mediated proinflammatory cytokines, and inhibits the expression of proinflammatory cytokines (Zuo et al., 2020). In this study, the miR-147-3p was up-regulated 32.94 folds at the acute infection stage. T. gondii infection triggered liver inflammation at an early infection stage, and the up-regulation of miR-147-3p could limit excessive inflammation for protecting the hosts. Further protection was attenuated with increasing duration of infection, as the expression of miR-147-3p decreased to 3.66-fold during the chronic infection stage. Interestingly, miR-342-3p (up-regulated 8.23 fold) seemed to have the same effect as miR-147-3p at the acute infection stage. Overexpression of miR-342-3p can suppress inflammation response in THP-1 cells (Wang et al., 2019). Moreover, miR-342-3p was considered to be significantly relative with regulating metabolic profiles of Treg cells (Kim et al., 2020), which may explain for its role in inflammation inhibition. Moreover, miR-143-3p could participate in inflammatory pain responses in fibromyalgia patients (Jiang et al., 2015). A previous study showed that miR-143-3p might inhibit inflammatory factors’ levels through regulating the MyD88/NF-κB signaling pathway (Wang et al., 2020). In this study, the miR-143-3p (mmu-miR-143-3p) was down-regulated 4.05 folds and 2.11 folds at the acute and chronic infection stages, respectively. Thus, these findings suggest that miR-143-3p may participate in the inflammation reaction of livers during T. gondii infection, and the down-regulation of miR-143-3p can increase the inflammation of livers for resisting infection. In addition, these findings reveal mediation of pro-inflammatory and anti-inflammatory mechanisms in the livers in T. gondii pathogenesisis.
In this study, some immunity-related GO terms were enriched at the two infection stages, such as “positive regulation of cytokine production”, “regulation of T cell activation”, “leukocyte cell-cell adhesion”, “positive regulation of leukocyte activation”, and “immune receptor activity” (Figure 3), indicating that T. gondii induced the liver immunity reaction of hosts. KEGG analysis showed that a series of pathways (e.g. “Valine, leucine and isoleucine degradation”, “Fatty acid metabolism”, and “Glycine, serine and threonine metabolism”) were involved in liver disease. The reductions of valine, leucine, and isoleucine are considered to be related to hepatic encephalopathy pathogenesis, and impair liver regeneration (Fischer et al., 1975; Marchesini et al., 2003; Nakaya et al., 2007; Urata et al., 2007). Moreover, TNF-α and IL-6 activate branched-chain keto acid dehydrogenase increased valine, leucine, and isoleucine catabolism (Nawabi et al., 1990; Holecek, 1996). IL-6 and TNF-α are proinflammatory cytokines, and metabolic disturbances are strongly related to increased levels of these two cytokines (Popko et al., 2010), thus suggesting T. gondii causes the metabolic disturbance of the livers at the acute infection stage. The other metabolism pathways enriched in the livers at the acute infection stage indicated disorders of metabolic function of the host livers induced by T. gondii infection. The cell adhesion molecules are considered as targets for the bacterial pathogens in establishing intimate contact with the cells and tissues of hosts (Hauck et al., 2006). A previous study reported that targeted disruption of SAG3 gene in T. gondii results in a partial decrease in host cell adhesion, and drastic reduction of virulence in mice (Dzierszinski et al., 2000). The “Cell adhesion molecules” pathway enriched at the chronic infection stage indicated potential relationships between cell adhesion molecules and T. gondii infection. The NF-κB family of transcription factors was closely related to the mediation of innate and adaptive immunities against infection (Caamaño et al., 1999). A previous study showed that the NF-κB2−/− mice could resist T. gondii infection during the acute phase of toxoplasmosis, but displayed a protracted pattern of mortality during the chronic stage of infection (Franzoso et al., 1998). These findings suggest NF-κB signaling pathway is essential for T. gondii infection in livers. Th17 produces AMPs that prevent the dysbiosis and bacterial translocation related to the pathogenic infection through secreting IL-17, and Th17 is crucial for host survival after T. gondii (type II strain) infection (Cervantes-Barragan et al., 2019). Moreover, a few studies reported that IL-17 signaling played a protective role during T. gondii infection (Kelly et al., 2005; Moroda et al., 2017). Thus, the “Th17 cell differentiation” pathway was observed in the livers at the chronic infection stage. An early T. gondii infection can disrupt the resident microbial communities and induce acute inflammation in the ileum (Heimesaat et al., 2006; Raetz et al., 2013). T. gondii infection also induces the increment of abundance of proinflammatory proteobacteria, and decrement of beneficially bacterial communities, causing disruption of the microbial community composition that persists at the chronic infection stage (French et al., 2022). These findings support that T. gondii infection causes microbial imbalance. Notably, “Inflammatory bowel disease” and “Intestinal immune network for IgA production” pathways were also enriched in the livers at the chronic infection stage. These two pathways are significantly associated with microbiota (Federici et al., 2022; Tan et al., 2022; Tchitchek et al., 2022). Thus, we suspect that T. gondii infection could mediate hepatic metabolism, and further affecting microbial balance in mice. How this mechanism works remains to be investigated.
CircRNAs act as miRNA decoys or sponges in regulating gene expression (Panda, 2018). Thus, a correlation analysis of the expression profiles from the DEcircRNAs and DEmiRNAs predictive interactions was performed. In the network, DE miRNAs mmu-miR-146a-5p and mmu-miR-150-5p were regulated by DEcircRNA 6:124519352|124575359. miR-146a-5p plays a role in different disease contexts and acts as a negative regulator of inflammatory and immune responses (Xu et al., 2012). A previous study reported that HCV-induced increment of miR-146a-5p promoted viral infection and metabolic pathways related to pathogenesis of hepatic disease (Bandiera et al., 2016). Thus, miR-146a-5p is associated with liver immunity and pathogenesis of liver disease. The upregulation of miR-146a-5p at both two infection stages (Supplementary Table S1) revealed a potential role in the T. gondii infection induced liver disease. However, the DEcircRNA 6:124519352|124575359 mediates this process. miR-150-5p regulates target genes IL-10 and PIM1, having an anti-inflammatory effect (Neamah et al., 2019). It was also increased at both acute and chronic infection stages, and was regulated by DEcircRNA 6:124519352|124575359. This illustrates both circRNA 6:124519352|124575359 and miR-150-5p have protective effects on T. gondii-induced liver excessive inflammation. Moreover, mmu-miR-1247-3p was regulated by multiple circRNAs. The treatment of HCC with miR-1247-3p would increase the expression levels of IL-1β, IL-6, and IL-8, thus suggesting the proinflammatory role of miR-1247-3p (Fang et al., 2018). The DEmiRNA mmu-miR-1247-3p was regulated by DEcircRNAs 12:103731961|103897311, 12:103854668|103947209, 5:145708877|145868684, 4:61958498|62052011, and 7:13832577|13909898 in the network, indicating that these DEcircRNAs participated in liver inflammation caused by T. gondii infection. In this study, RNA-seq was performed to detect the differential expression profiles of circRNAs and miRNAs in the livers of mice infected with T. gondii. However, some limits should be addressed, including other organizations of expression profiles should be added, and the potential interaction between circRNA-miRNA and mRNA is needed to validate in the future work.
This study explored the differential expression levels of circRNAs and miRNAs in the livers of mice infected with T. gondii at the acute and chronic stages. The functional enrichment analysis showed that many DEcirciRNAs and DE miRNAs were associated with the inflammation responses of the livers after T. gondii infection. Our results provided valuable data for the understanding of the circRNAs and miRNAs involved in the molecular basis of the hepatic responses to T. gondii infection. The functions of DEcircRNAs and DEmiRNAs in the pathogenesis of T. gondii infection in livers will be further verified.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
The animal study was reviewed and approved by the Animal Ethics Committee of Qingdao Agricultural University.
X-XZ and H-WC conceived and designed the experiments. J-XM and X-YM performed the experiments. X-YG, X-YW, J-XM, CC and H-LC contributed reagents/materials/analysis tools. YZ analyzed the data and wrote the paper. X-YW, J-XM, L-HY, X-XZ and HW-C critically revised the manuscript. All authors read and approved the final version of the manuscript.
This work was supported by the National Natural Science Foundation of China (Grant No. 31902238), Research Foundation for Distinguished Scholars of Qingdao Agricultural University (665-1120044).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcimb.2022.1037586/full#supplementary-material
Supplementary Figure 1 | Validation for the expression of the DEcircRNAs and DEmiRNAs by qRT-PCR at acute infection (AI) and chronic infection (CI) stages.
Supplementary Table 1 | The expression profile of differentially expressed circRNAs and miRNAs in the liver at acute infection and chronic infection stages.
Supplementary Table 2 | The data of DEcircRNA-DEmiRNA network analysis.
Anders, S., Huber, W. (2010). Differential expression analysis for sequence count data. Genome. Biol. 11, R106. doi: 10.1186/gb-2010-11-10-r106
Antil, N., Arefian, M., Kandiyil, M. K., Awasthi, K., Keshava Prasad, T. S., Raju, R. (2022). The core human microRNAs regulated by Toxoplasma gondii. Microrna doi: 10.2174/2211536611666220428130250
Antil, N., Kumar, M., Behera, S. K., Arefian, M., Kotimoole, C. N., Rex, D. A. B., et al. (2021). Unraveling GT1 strain virulence and new protein-coding genes with proteogenomic analyses. OMICS 25, 591–604. doi: 10.1089/omi.2021.0082
Balasundaram, M. B. (2010). Outbreak of acquired ocular toxoplasmosis involving 248 patients. Arch. Ophthalmol. 128, 28. doi: 10.1001/archophthalmol.2009.354
Bandiera, S., Pernot, S., El Saghire, H., Durand, S. C., Thumann, C., Crouchet, E., et al. (2016). Hepatitis c virus-induced upregulation of microRNA miR-146a-5p in hepatocytes promotes viral infection and deregulates metabolic pathways associated with liver disease pathogenesis. J. Virol. 90, 6387–6400. doi: 10.1128/JVI.00619-16
Caamaño, J., Alexander, J., Craig, L., Bravo, R., Hunter, C. A. (1999). The NF-kappa b family member RelB is required for innate and adaptive immunity to Toxoplasma gondii. J. Immunol. 163, 4453–4461.
Cervantes-Barragan, L., Cortez, V. S., Wang, Q., McDonald, K. G., Chai, J. N., Di Luccia, B., et al. (2019). CRTAM protects against intestinal dysbiosis during pathogenic parasitic infection by enabling Th17 maturation. Front. Immunol. 10. doi: 10.3389/fimmu.2019.01423
Chomczynski, P., Sacchi, N. (2006). The single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction: Twenty-something years on. Nat. Protoc. 1, 581–585. doi: 10.1038/nprot.2006.83
Conn, S. J., Pillman, K. A., Toubia, J., Conn, V. M., Salmanidis, M., Phillips, C. A., et al. (2015). The RNA binding protein quaking regulates formation of circRNAs. Cell. 160, 1125–1134. doi: 10.1016/j.cell.2015.02.014
Dhamija, S., Menon, M. B. (2018). Non-coding transcript variants of protein-coding genes - what are they good for? RNA Biol. 15, 1025–1031. doi: 10.1080/15476286.2018
Doğan, N., Kabukçuoğlu, S., Vardareli, E. (2007). Toxoplasmic hepatitis in an immunocompetent patient. Turkiye Parazitol. Derg. 31, 260–263.
Dubey, J. P. (2008). The history of toxoplasma gondii–the first 100 years. J. Eukaryot. Microbiol. 55, 467–475. doi: 10.1111/j.1550-7408.2008.00345.x
Dzierszinski, F., Mortuaire, M., Cesbron-Delauw, M. F., Tomavo, S. (2000). Targeted disruption of the glycosylphosphatidylinositol-anchored surface antigen SAG3 gene in Toxoplasma gondii decreases host cell adhesion and drastically reduces virulence in mice. Mol. Microbiol. 37, 574–582. doi: 10.1046/j.1365-2958.2000.02014.x
Fang, T., Lv, H., Lv, G., Li, T., Wang, C., Han, Q., et al. (2018). Tumor-derived exosomal miR-1247-3p induces cancer-associated fibroblast activation to foster lung metastasis of liver cancer. Nat. Commun. 9, 191. doi: 10.1038/s41467-017-02583-0
Federici, S., Kredo-Russo, S., Valdés-Mas, R., Kviatcovsky, D., Weinstock, E., Matiuhin, Y., et al. (2022). Targeted suppression of human IBD-associated gut microbiota commensals by phage consortia for treatment of intestinal inflammation. Cell 185, 2879–2898.e24. doi: 10.1016/j.cell.2022.07.003
Fischer, J. E., Funovics, J. M., Aguirre, A., James, J. H., Keane, J. M., Wesdorp, R. I., et al. (1975). The role of plasma amino acids in hepatic encephalopathy. Surgery 78, 276–290.
Franzoso, G., Carlson, L., Poljak, L., Shores, E. W., Epstein, S., Leonardi, A., et al. (1998). Mice deficient in nuclear factor (NF)-kappa B/p52 present with defects in humoral responses, germinal center reactions, and splenic microarchitecture. J. Exp. Med. 187, 147–159. doi: 10.1084/jem.187.2.147
French, T., Steffen, J., Glas, A., Osbelt, L., Strowig, T., Schott, B. H., et al. (2022). Persisting microbiota and neuronal imbalance following infection reliant on the infection route. Front. Immunol. 13. doi: 10.3389/fimmu.2022.920658
Friedländer, M. R., Mackowiak, S. D., Li, N., Chen, W., Rajewsky, N. (2012). miRDeep2 accurately identifies known and hundreds of novel microRNA genes in seven animal clades. Nucleic. Acids Res. 40, 37–52. doi: 10.1093/nar/gkr688
Gao, Y., Wang, J., Zhao, F. (2015). CIRI: an efficient and unbiased algorithm for de novo circular RNA identification. Genome Biol. 16, 4. doi: 10.1186/s13059-014-0571-3
Garfoot, A. L., Wilson, G. M., Coon, J. J., Knoll, L. J. (2019). Proteomic and transcriptomic analyses of early and late-chronic Toxoplasma gondii infection shows novel and stage specific transcripts. BMC Genomics 20, 859. doi: 10.1186/s12864-019-6213-0
Granados-Riveron, J. T., Aquino-Jarquin, G. (2016). The complexity of the translation ability of circRNAs. Biochim. Biophys. Acta 1859, 1245–1251. doi: 10.1016/j.bbagrm.2016.07.009
Griffiths-Jones, S. (2016). miRBase: the microRNA sequence database. Methods. Mol. Biol. 342, 129–38. doi: 10.1385/1-59745-123-1:129
Hassan, M. M., Farghaly, A. M., Gaber, N. S., Nageeb, H. F., Hegab, M. H., Galal, N. (1996). Parasitic causes of hepatomegaly in children. J. Egypt. Soc Parasitol. 26, 177–189.
Hauck, C. R., Agerer, F., Muenzner, P., Schmitter, T. (2006). Cellular adhesion molecules as targets for bacterial infection. Eur. J. Cell Biol. 85, 235–242. doi: 10.1016/j.ejcb.2005.08.002
Heimesaat, M. M., Bereswill, S., Fischer, A., Fuchs, D., Struck, D., Niebergall, J., et al. (2006). Gram-negative bacteria aggravate murine small intestinal Th1-type immunopathology following oral infection with Toxoplasma gondii. J. Immunol. 177, 8785–8795. doi: 10.4049/jimmunol.177.12.8785
He, J. J., Ma, J., Li, F. C., Song, H. Q., Xu, M. J., Zhu, X. Q. (2016). Transcriptional changes of mouse splenocyte organelle components following acute infection with Toxoplasma gondii. Exp. Parasitol. 167, 7–16. doi: 10.1016/j.exppara.2016.04.019
Holecek, M. (1996). Leucine metabolism in fasted and tumor necrosis factor-treated rats. Clin. Nutr. 15, 91–93. doi: 10.1016/s0261-5614(96)80028-8
Hu, R. S., He, J. J., Elsheikha, H. M., Zhang, F. K., Zou, Y., Zhao, G. H., et al. (2018). Differential brain microRNA expression profiles after acute and chronic infection of mice with oocysts. Front. Microbiol. 9. doi: 10.3389/fmicb.2018.02316
Jiang, Q., Yi, M., Guo, Q., Wang, C., Wang, H., Meng, S., et al. (2015). Protective effects of polydatin on lipopolysaccharide-induced acute lung injury through TLR4-MyD88-NF-κB pathway. Int. Immunopharmacol. 29, 370–376. doi: 10.1016/j.intimp.2015.10.027
Karasawa, T., Shikata, T., Takizawa, I., Morita, K., Komukai, M. (1981). Localized hepatic necrosis related to cytomegalovirus and Toxoplasma gondii. Acta Pathol. Jpn. 31, 527–534.
Kelly, M. N., Kolls, J. K., Happel, K., Schwartzman, J. D., Schwarzenberger, P., Combe, C., et al. (2005). Interleukin-17/interleukin-17 receptor-mediated signaling is important for generation of an optimal polymorphonuclear response against Toxoplasma gondii infection. Infect. Immun. 73, 617–621. doi: 10.1128/IAI.73.1.617-621.2005
Kim, D., Nguyen, Q. T., Lee, J., Lee, S. H., Janocha, A., Kim, S., et al. (2020). Anti-inflammatory roles of glucocorticoids are mediated by Foxp3 regulatory T cells via a miR-342-dependent mechanism. Immunity. 53, 581–596.e5. doi: 10.1016/j.immuni.2020.07.002
Langmead, B., Salzberg, S. L. (2012). Fast gapped-read alignment with bowtie 2. Nat. Methods 9, 357–359. doi: 10.1038/nmeth.1923
Lei, B., Tian, Z., Fan, W., Ni, B. (2019). Circular RNA: a novel biomarker and therapeutic target for human cancers. Int. J. Med. Sci. 16, 292–301. doi: 10.7150/ijms.28047
Liu, G., Friggeri, A., Yang, Y., Park, Y. J., Tsuruta, Y., Abraham, E. (2009). miR-147, a microRNA that is induced upon toll-like receptor stimulation, regulates murine macrophage inflammatory responses. Proc. Natl. Acad. Sci. U.S.A. 106, 15819–15824. doi: 10.1073/pnas.0901216106
Livak, K. J., Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2(-delta delta C(T)) method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262
Lutshumba, J., Ochiai, E., Sa, Q., Anand, N., Suzuki, Y. (2020). Selective upregulation of transcripts for six molecules related to T cell costimulation and phagocyte recruitment and activation among 734 immunity-related genes in the brain during perforin-dependent, CD8 T cell-mediated elimination of Toxoplasma gondii cysts. mSystems 5, e00189-20. doi: 10.1128/mSystems.00189-20
Mao, X., Cai, T., Olyarchuk, J. G., Wei, L. (2005). Automated genome annotation and pathway identification using the KEGG orthology (KO) as a controlled vocabulary. Bioinformatics 21, 3787–3793. doi: 10.1093/bioinformatics/bti430
Marchesini, G., Bianchi, G., Merli, M., Amodio, P., Panella, C., Loguercio, C., et al. (2003). Nutritional supplementation with branched-chain amino acids in advanced cirrhosis: a double-blind, randomized trial. Gastroenterology 124, 1792–1801. doi: 10.1016/s0016-5085(03)00323-8
Memczak, S., Jens, M., Elefsinioti, A., Torti, F., Krueger, J., Rybak, A., et al. (2013). Circular RNAs are a large class of animal RNAs with regulatory potency. Nature 495, 333–338. doi: 10.1038/nature11928
Menard, K. L., Bu, L., Denkers, E. Y. (2021). Transcriptomics analysis of Toxoplasma gondii-infected mouse macrophages reveals coding and noncoding signatures in the presence and absence of MyD88. BMC Genomics 22, 130. doi: 10.1186/s12864-021-07437-0
Meng, S., Zhou, H., Feng, Z., Xu, Z., Tang, Y., Li, P., et al. (2017). CircRNA: functions and properties of a novel potential biomarker for cancer. Mol. Cancer. 16, 94. doi: 10.1186/s12943-017-0663-2
Montoya, J. G., Liesenfeld, O. (2004). Toxoplasmosis. Lancet 363, 1965–1976. doi: 10.1016/S0140-6736(04)16412-X
Moroda, M., Takamoto, M., Iwakura, Y., Nakayama, J., Aosai, F. (2017). Interleukin-17A-deficient mice are highly susceptible to Toxoplasma gondii infection due to excessively induced T. gondii HSP70 and interferon gamma production. Infect. Immun. 85, e00399-17. doi: 10.1128/IAI.00399-17
Nakaya, Y., Okita, K., Suzuki, K., Moriwaki, H., Kato, A., Miwa, Y., et al. (2007). BCAA-enriched snack improves nutritional state of cirrhosis. Nutrition 23, 113–120. doi: 10.1016/j.nut.2006.10.008
Nawabi, M. D., Block, K. P., Chakrabarti, M. C., Buse, M. G. (1990). Administration of endotoxin, tumor necrosis factor, or interleukin 1 to rats activates skeletal muscle branched-chain alpha-keto acid dehydrogenase. J. Clin. Invest. 85, 256–263. doi: 10.1172/JCI114421
Neamah, W. H., Singh, N. P., Alghetaa, H., Abdulla, O. A., Chatterjee, S., Busbee, P. B., et al. (2019). AhR activation leads to massive mobilization of myeloid-derived suppressor cells with immunosuppressive activity through regulation of CXCR2 and microRNA miR-150-5p and miR-543-3p that target anti-inflammatory genes. J. Immunol. 203, 1830–1844. doi: 10.4049/jimmunol.1900291
Ortego, T. J., Robey, B., Morrison, D., Chan, C. (1990). Toxoplasmic chorioretinitis and hepatic granulomas. am. J. Gastroenterology 85, 1418–1420.
Panda, A. C. (2018). Circular RNAs act as miRNA sponges. Adv. Exp. Med. Biol. 1087, 67–79. doi: 10.1007/978-981-13-1426-1_6
Pertea, M., Kim, D., Pertea, G. M., Leek, J. T., Salzberg, S. L. (2016). Transcript-level expression analysis of RNA-seq experiments with HISAT, StringTie and ballgown. Nat. Protoc. 11, 1650–1667. doi: 10.1038/nprot.2016.095
Popko, K., Gorska, E., Stelmaszczyk-Emmel, A., Plywaczewski, R., Stoklosa, A., Gorecka, D., et al. (2010). Proinflammatory cytokines il-6 and TNF-α and the development of inflammation in obese subjects. Eur. J. Med. Res. 15 Suppl 2, 120–122. doi: 10.1186/2047-783x-15-s2-120
Preusse, M., Schughart, K., Pessler, F. (2017). Host genetic background strongly affects pulmonary microRNA expression before and during influenza a virus infection. Front. Immunol. 8. doi: 10.3389/fimmu.2017.00246
Raetz, M., Hwang, S.-H., Wilhelm, C. L., Kirkland, D., Benson, A., Sturge, C. R., et al. (2013). Parasite-induced TH1 cells and intestinal dysbiosis cooperate in IFN-γ-dependent elimination of paneth cells. Nat. Immunol. 14, 136–142. doi: 10.1038/ni.2508
Smith, N. C., Goulart, C., Hayward, J. A., Kupz, A., Miller, C. M., van dooren, G. G. (2021). Control of human toxoplasmosis. Int. J. Parasitol. 51, 95–121. doi: 10.1016/j.ijpara.2020.11.001
Stauffer, W. M., Magill, A., Kain, K. C. (2006). Parasitic central nervous system infections in immunocompromised hosts: clarification of malaria diagnosis. Clin. Infect. Dis. 43, 114–115. doi: 10.1086/504951
Tang, Y., Jiang, M., Jiang, H. M., Ye, Z. J., Huang, Y. S., Li, X. S., et al. (2020). The roles of circRNAs in liver cancer immunity. Front. Oncol. 10. doi: 10.3389/fonc.2020.598464
Tan, J., Ni, D., Taitz, J., Pinget, G. V., Read, M., Senior, A., et al. (2022). Dietary protein increases T-cell-independent sIgA production through changes in gut microbiota-derived extracellular vesicles. Nat. Commun. 13, 4336. doi: 10.1038/s41467-022-31761-y
Tchitchek, N., Nguekap Tchoumba, O., Pires, G., Dandou, S., Campagne, J., Churlaud, G., et al. (2022). Low-dose interleukin-2 shapes a tolerogenic gut microbiota that improves autoimmunity and gut inflammation. JCI. Insight. 7, e159406. doi: 10.1172/jci.insight.159406
Tenter, A. M., Heckeroth, A. R., Weiss, L. M. (2000). Toxoplasma gondii: from animals to humans. Int. J. Parasitol. 30, 1217–1258. doi: 10.1016/s0020-7519(00)00124-7
Urata, Y., Okita, K., Korenaga, K., Uchida, K., Yamasaki, T., Sakaida, I. (2007). The effect of supplementation with branched-chain amino acids in patients with liver cirrhosis. Hepatol. Res. 37, 510–516. doi: 10.1111/j.1872-034X.2007.00081.x
Wang, Y., Li, H., Shi, Y., Wang, S., Xu, Y., Li, H., et al. (2020). miR-143-3p impacts on pulmonary inflammatory factors and cell apoptosis in mice with mycoplasmal pneumonia by regulating TLR4/MyD88/NF-κB pathway. Biosci. Rep. 40, BSR20193419. doi: 10.1042/BSR20193419
Wang, L., Xia, J. W., Ke, Z. P., Zhang, B. H. (2019). Blockade of NEAT1 represses inflammation response and lipid uptake via modulating miR-342-3p in human macrophages THP-1 cells. J. Cell. Physiol. 234, 5319–5326. doi: 10.1002/jcp.27340
Wen, M., Shen, Y., Shi, S., Tang, T. (2012). miREvo: an integrative microRNA evolutionary analysis platform for next-generation sequencing experiments. BMC Bioinf. 13, 140. doi: 10.1186/1471-2105-13-140
Xu, W. D., Lu, M. M., Pan, H. F., Ye, D. Q. (2012). Association of microRNA-146a with autoimmune diseases. Inflammation 35, 1525–1529. doi: 10.1007/s10753-012-9467-0
Yao, T., Chen, Q., Fu, L., Guo, J. (2017). Circular RNAs: Biogenesis, properties, roles, and their relationships with liver diseases. Hepatol. Res. 47, 497–504. doi: 10.1111/hepr.12871
Young, M. D., Wakefield, M. J., Smyth, G. K., Oshlack, A. (2010). Gene ontology analysis for RNA-seq: accounting for selection bias. Genome Biol. 11, R14. doi: 10.1186/gb-2010-11-2-r14
Zhou, C. X., Ai, K., Huang, C. Q., Guo, J. J., Cong, H., He, S. Y., et al. (2020). miRNA and circRNA expression patterns in mouse brain during toxoplasmosis development. BMC Genomics 21, 46. doi: 10.1186/s12864-020-6464-9
Zhou, L., Chen, J., Li, Z., Li, X., Hu, X., Huang, Y., et al. (2010). Integrated profiling of microRNAs and mRNAs: microRNAs located on Xq27.3 associate with clear cell renal cell carcinoma. PLoS One 5, e15224. doi: 10.1371/journal.pone.0015224
Zhou, C. X., Elsheikha, H. M., Zhou, D. H., Liu, Q., Zhu, X. Q., Suo, X. (2016). Dual identification and analysis of differentially expressed transcripts of porcine PK-15 cells and Toxoplasma gondii during in vitro infection. Front. Microbiol. 7. doi: 10.3389/fmicb.2016.00721
Zhou, J., Xiong, Q., Chen, H., Yang, C., Fan, Y. (2017). Identification of the spinal expression profile of non-coding RNAs involved in neuropathic pain following spared nerve injury by sequence analysis. Front. Mol. Neurosci. 10. doi: 10.3389/fnmol.2017.00091
Zhu, S., Wang, J., He, Y., Meng, N., Yan, G. R. (2018). Peptides/proteins encoded by non-coding RNA: A novel resource bank for drug targets and biomarkers. Front. Pharmacol. 9. doi: 10.3389/fphar.2018.01295
Keywords: toxoplasma gondii, non-coding RNAs, high-throughput RNA sequencing, liver, networks
Citation: Zou Y, Meng J-X, Wei X-Y, Gu X-Y, Chen C, Geng H-L, Yang L-H, Zhang X-X and Cao H-W (2022) CircRNA and miRNA expression analysis in livers of mice with Toxoplasma gondii infection. Front. Cell. Infect. Microbiol. 12:1037586. doi: 10.3389/fcimb.2022.1037586
Received: 06 September 2022; Accepted: 07 October 2022;
Published: 27 October 2022.
Edited by:
Mingming Liu, Hubei University of Arts and Science, ChinaReviewed by:
Hongchao Sun, Zhejiang Academy of Agricultural Sciences, ChinaCopyright © 2022 Zou, Meng, Wei, Gu, Chen, Geng, Yang, Zhang and Cao. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Li-Hua Yang, amx5bGgxMjNAMTYzLmNvbQ==; Xiao-Xuan Zhang, emhhbmd4aWFveHVhbjE5ODhAMTI2LmNvbQ==; Hong-Wei Cao, Y2FvaHdAeWN0dS5lZHUuY24=
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.