
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Cell. Infect. Microbiol., 26 October 2022
Sec. Microbes and Innate Immunity
Volume 12 - 2022 | https://doi.org/10.3389/fcimb.2022.1027424
Francisella tularensis, a bacterial causative agent of the zoonosis tularemia, is highly pathogenic to humans. The pathogenicity of this bacterium is characterized by intracellular growth in immune cells, like macrophages, and host immune suppression. However, the detailed mechanism of immune suppression by F. tularensis is still unclear. To identify the key factors causing Francisella-mediated immunosuppression, large-scale screening using a transposon random mutant library containing 3552 mutant strains of F. tularensis subsp. novicida (F. novicida) was performed. Thirteen mutants that caused stronger tumor necrosis factor (TNF)-α production in infected U937 human macrophage cells than the wild-type F. novicida strain were isolated. Sequencing analysis of transposon insertion sites revealed 10 genes, including six novel genes, as immunosuppressive factors of Francisella. Among these, the relationship of the pyrC gene, which encodes dihydroorotase in the pyrimidine biosynthesis pathway, with Francisella-mediated immunosuppression was investigated. The pyrC deletion mutant strain (ΔpyrC) induced higher TNF-α production in U937 host cells than the wild-type F. novicida strain. The ΔpyrC mutant strain was also found to enhance host interleukin-1β and interferon (IFN)-β production. The heat-inactivated ΔpyrC mutant strain could not induce host TNF-α production. Moreover, the production of IFN-β resulting from ΔpyrC infection in U937 cells was repressed upon treatment with the stimulator of interferon genes (STING)-specific inhibitor, H-151. These results suggest that pyrC is related to the immunosuppressive activity and pathogenicity of Francisella via the STING pathway.
Tularemia is a vector-borne zoonosis with severe symptoms, including fever, lymphadenitis, cutaneous lesions, and primary pulmonary involvement in humans (Petersen and Schriefer, 2005). Francisella tularensis, the causative agent of tularemia, is a gram-negative, facultative, and intracellular bacterial pathogen. Tularemia is highly contagious, with subcutaneous infection with as few as 10 bacterial cells and aerosol-mediated inhalation of as few as 25 bacterial cells being able to cause human infection (Sjöstedt, 2007). Accordingly, highly virulent F. tularensis strains have been classified as a Category-A agents of potential bioterrorism by the Centers for Disease Control and Prevention and are required to be handled and contained in BSL-3 laboratories (Shapiro and Schwartz, 2002). F. tularensis is divided into four subspecies, subsp. tularensis, holarctica, mediasiatica, and novicida. Among them, only Francisella tularensis subsp. tularensis (F. tularensis) and Francisellatularensis subsp. holarctica (F. holarctica) cause tularemia in human (Maurin, 2015). F. novicida infection in humans is considerably rare and can be handled in a BSL-2 laboratory. Most cases of human F. novicida infection have involved patients who were immunocompromised or had an underlying disease. However, F. novicida has over 98% identity to F. tularensis at the DNA level and shows many characteristics similar to F. tularensis with regards to its life cycle within macrophages and pathogenicity in mice (Rohmer et al., 2007; Kingry and Petersen, 2014). Therefore, F. novicida has been used as the model bacterium for research on Francisella pathogenicity.
While Francisella research has a long history, the detailed molecular mechanisms of infection by Francisella subspecies remain unknown. Francisella can replicate in immunocompetent cells, such as macrophages, neutrophils, and dendritic cells, which are essentially responsible for the elimination of pathogens from the body (Santic et al., 2006). To survive inside host cells, Francisella species employ various strategies. Francisella enters these cells via phagocytosis, escapes digestion by phagolysosomes and autophagosomes, and finally replicates in the cytoplasm (Chong et al., 2008; Miller and Celli, 2016). Further, Francisella suppresses or evades host pattern recognition receptors (PRRs), which usually initiate the innate immune response to exclude pathogens (Jones et al., 2011; Gillette et al., 2014; Putzova et al., 2017). Although some of the factors responsible for Francisella’s immunosuppressive abilities were identified in previous research (Platz et al., 2010; Nallaparaju et al., 2011; Mahawar et al., 2012), the detailed mechanisms involved in Francisella-mediated immunosuppression remain to be elucidated.
Francisella grows in nutrient-limited host cells, and this nutrient limitation is closely related to its pathogenicity (Best and Abu Kwaik, 2019). To date, a lot of genes responsible for the uptake or biosynthesis of nutrients, such as amino acids, carbon, vitamins, and bases, were reported as crucial factors for the intracellular replication of Francisella (Meibom and Charbit, 2010; Santic and Abu Kwaik, 2013; Feng et al., 2014). Especially, nucleotide biosynthesis is essential for the survival and virulence of bacterial pathogens, including intracellular bacteria, such as Salmonella, Listeria, Brucella, and Francisella (Goncheva et al., 2022). Since pyrimidine nucleotides are essential for all organisms, almost all bacterial species have a de novo pyrimidine biosynthesis pathway, which has highly conserved enzymatic steps. The de novo pyrimidine biosynthesis pathway for synthesizing uridine 5′-monophosphate consists of six steps and employs enzymes encoded by carA/B and pyrB-F (Turnbough Charles and Switzer Robert, 2008). The pyrC gene encodes a putative dihydroorotase that converts carbamoyl N-Carbamoyl-L-aspartate into 4,5-dihydroorotate in the Francisella pyrimidine biosynthesis pathway. Although some reports suggest pyrimidine biosynthesis is important for the intracellular growth of Francisella, it is not well understood how pyrimidine biosynthesis is involved in immunosuppression by Francisella (Qin and Mann, 2006; Horzempa et al., 2010).
In this study, we performed large-scale screening of a F. novicida transposon mutant library to search for the key factors involved in the immunosuppression mechanisms of Francisella. We identified pyrC as a novel F. novicida factor suppressing host innate immune responses and evaluated the immunological characteristics of host cells infected with F. novicida.
F. novicida U112 was obtained from the Pathogenic Microorganism Genetic Resource Stock Center (Gifu University, Gifu, Japan). F. novicida was cultured aerobically at 37°C in a chemically defined medium (CDM) (Nagle et al., 1960) or brain heart infusion broth (Becton, Dickinson and Company, NJ, USA) supplemented with 0.1% cysteine (BHIc) (Mc Gann et al., 2010) or BHIc containing 1.5% agar (Wako Laboratory Chemicals, Osaka, Japan). All experiments were conducted in compliance with the institutional biosecurity guidelines and were approved by Yamaguchi University.
Human monocytic U937 cells were grown in Roswell Park Memorial Institute (RPMI) 1640 medium (Sigma-Aldrich, St. Louis, MO, USA) supplemented with 10% heat-inactivated fetal bovine serum at 37°C in an atmosphere containing 5% CO2.
Supplementary Table 1 lists the primer sets and templates used to construct the plasmids used in this study. Polymerase chain reaction (PCR) was performed using KOD-Plus-Neo polymerase (Toyobo, Osaka, Japan), and ligation was performed using an In-Fusion HD Cloning Kit (Takara Bio, Shiga, Japan). Plasmids were used to transform F. novicida via electroporation. Specifically, bacterial cells were suspended in 0.5 M sucrose with 2 μg of plasmid DNA and were electroporated using a Bio-Rad micropulser (Bio-Rad, Hercules, CA, USA) at 3.0 kV, 10 µF, and 600 Ω with a 0.2 cm cuvette. The transformants were pre-incubated in BHIc medium overnight. To select transformed bacteria, the pre-incubated bacteria were cultured on BHIc agar plates containing 30 μg/ml kanamycin or 2.5 μg/ml chloramphenicol.
The transposon mutant library was constructed using the Ez-Tn5 transposon system (Epicentre Lucigen, Madison, WI, USA), as previously reported (Nakamura et al., 2019). Briefly, the multiple cloning site of pMOD3 was linearized by digestion with Hind III and EcoRI, and the kanamycin resistance cassette of pKEK1140 (Rodriguez et al., 2008) was ligated into the Hind III and EcoRI sites to generate pMOD3-FtKm. The transposon moiety of pMOD3-FtKm was amplified by PCR, purified, mixed with transposase according to the manufacturer’s instructions, and then used to transform F. novicida via electroporation. Transformed bacteria were cultured on BHIc plates containing 30 μg/ml kanamycin.
pMOD3 harbors the E. coli R6Kγ origin of replication. The genomes of F. novicida transposon mutants were purified using a PureLink Genomic DNA Mini Kit (Thermo Fisher Scientific, MA, USA) and digested with a combination of restriction enzymes, including XhoI, BglII, EcoRI, SalI, NotI, and BamHI. The ends of the digested DNAs were then blunted using a DNA Blunting Kit (Takara Bio) and ligated using Ligation High Ver. 2 (Toyobo). The ligated DNA was used to transform λpir Chemically Competent E. coli (Thermo Fisher Scientific). The transformed E. coli were selected for kanamycin resistance, and plasmid DNAs were purified. Sequence analysis was performed using the primer described in the manufacturer’s instructions for the Ez-Tn5 transposon system.
The dotU homolog (FTN_1316) deletion mutant (ΔdotU) was previously constructed (Shimizu et al., 2019) through group II intron insertion using a TargeTron Gene Knockout System (Sigma-Aldrich), which was modified for Francisella species (Rodriguez et al., 2008). The pyrC gene (FTN_0024) deletion mutant (ΔpyrC) was generated via homologous recombination using the Francisella suicide vector pFRSU (Shimizu et al., 2019). The upstream and downstream regions of pyrC (1.5 kbp each) were cloned into the BamHI site of pFRSU to generate pFRSU-pyrC. The pFRSU-pyrC vector (2 μg) was used to transform F. novicida; transformants were selected on BHIc plates containing 30 μg/ml kanamycin. Isolated bacteria were cultured in BHIc without antibiotics overnight and then plated on BHIc plates containing 5% sucrose. The deletion of the pyrC gene was confirmed via PCR.
A GFP-expressing plasmid, pOM5-GFP, was constructed according to published procedures (Shimizu et al., 2019). The F. novicida chromosomal pyrC gene was cloned into pOM5 to generate pOM5-pyrC. To construct GFP-expressing strains and pyrC complemented strains, pOM5-pyrC and pOM5-GFP were used to transform the wild-type strain or the ΔpyrC mutant strain of F. novicida via electroporation.
U937 cells (1 × 105 cells/well) were incubated in a 48-well tissue culture plate with 100 nM phorbol myristate acetate (PMA) for 48 h. Then, F. novicida strains were added at a multiplicity of infection (MOI) of 1. Next, the plates were centrifuged for 10 min at 300 × g and incubated for 1 h at 37°C. The cells were then washed three times with RPMI 1640 medium, and extracellular bacteria were killed with gentamicin at 50 μg/ml for 1 h. The cells were then incubated in fresh medium at 37°C for the indicated time durations in figure legends. To measure intracellular growth, the cells were washed with PBS and then lysed with 0.1% Triton X-100 in CDM. The CFUs were determined on BHIc agar plates by plating serial dilutions of cultures.
U937 cells (1 × 105 cells/well) were incubated with 100 nM PMA for 48 h on 12 mm glass coverslips in 24-well tissue culture plates. GFP-expressing F. novicida strains were infected at an MOI of 1. Plates were then centrifuged for 10 min at 300 × g and incubated for 1 h at 37°C. The cells were washed three times with RPMI 1640 medium, and extracellular bacteria were eliminated using gentamicin at 50 μg/ml for 1 h. The cells were then incubated in fresh medium at 37°C for the indicated time durations in figure legends. Cells were fixed with 4% paraformaldehyde at room temperature for 30 min. A FluoView FV1000 confocal laser scanning microscope (Olympus, Tokyo, Japan) was used to obtain images of the cells.
U937 cells (4 × 105 cells/well) were incubated in a 12-well tissue culture plate with 100 nM PMA for 48 h. The medium was exchanged with fresh pre-incubated RPMI 1640 medium one-hour prior to infection. Cells were infected with F. novicida strains at MOI = 1 or stimulated with 100 ng/ml of lipopolysaccharide (LPS) derived from E. coli (O127:B8) (Sigma-Aldrich), or 10 ng/ml 2′3′-cGAMP (InvivoGen, CA, USA). The plates were centrifuged for 10 min at 300 × g and incubated for indicated time the indicated time durations in figure legends. Cells were carefully washed twice with PBS, and total RNA was collected using NucleoSpin RNA kit (Takara Bio). RNA was quantified by determining absorption at 260 nm using NanoDrop 2000 (Thermo Fisher Scientific). Next, qPCR was performed using the RNA-direct Realtime PCR Master Mix (Toyobo) with an RNA concentration of 50 ng per 20 μl reaction. The HPRT1 amplicon was used as an endogenous control to normalize all mRNA expression data. The relative expression levels of genes in various conditions compared with those in the BHIc medium-treated control were calculated using the relative quantification method (∆∆Ct method) (Pfaffl, 2001). Used primer sets are shown in Table S1.
U937 cells (1 × 105 cells/well) were pre-incubated in a 48-well tissue culture plate with 100 nM PMA for 48 h. After exchanging the medium with pre-incubated fresh RPMI medium 1 h prior to infection, cells were infected with bacterial strains (F. novicida transposon mutant strains or deletion mutant strains) at an MOI of 1 or stimulated with 100 ng/ml of LPS derived from E. coli (O127:B8) or 10 ng/ml of 2′3′-cGAMP (InvivoGen). Heat-inactivation of each strain was performed by incubating the bacterial suspension in a heat block at 90°C for 5 min. In the case of STING inhibition, a STING-specific inhibitor, H-151 was used (Haag et al., 2018). U937 cells were treated with H-151 (final concentration 0.5 μM) or the same volume of DMSO 2 h prior to infection. After incubation for the indicated time durations in figure legends, concentrations of tumor necrosis factor (TNF)-α and interleukin (IL)-1β in the supernatants were measured using ELISA MAX Standard Kit (Biolegend, CA, USA) according to the manufacturer’s instructions. Interferon (IFN)-β in the supernatants were measured using VeriKine Human Interferon Beta ELISA Kit (PBL Assay Science, Piscataway, NJ, USA).
Student’s t test or multiple comparisons using the Tukey–Kramer test and Dunnett’s test were used to evaluate the significance of differences compared with the wild-type strain; P < 0.05 indicates a significant difference.
To identify novel immunosuppressive factors of F. novicida in human macrophages, we expanded a previously constructed F. novicida transposon mutant library consist of 750 strains (Nakamura et al., 2019) up to 3552 strains. TNF-α is a cytokine produced through a broad range of innate immune signaling pathways, including Toll-like Receptor (TLR) 4- and TLR2-mediated pathways, and is reported to be suppressed by Francisella infection (Telepnev et al., 2003; Butchar et al., 2008). To identify genes responsible for immunosuppression by F. novicida, U937 cells were infected with a mutant library, and the transposon mutants inducing excessive TNF-α production compared with the wild-type strain were selected through ELISA using the culture supernatant. In the 1st screening, the cut-off was set to a 1.5-fold increase in TNF-α production. In the 2nd screening, U937 cells were infected with mutant strains selected via the first screening, and their TNF-α production was measured three times. Finally, 13 mutants that increased TNF-α production in U937 cells were identified (Figure 1). The Δslt mutant strain, lacking the gene encoding soluble lytic transglycosylase, was used as a positive control (Nakamura et al., 2019). To determine the genes responsible for Francisella-mediated immunosuppression, the transposon insertion sites of the selected mutant strains were evaluated by sequence analysis; 10 unique genes were identified (Table 1). In this study, we focused on pyrC (FTN_0024), the gene putatively encoding dihydroorotase, and the effect of this gene on immunosuppression by Francisella.
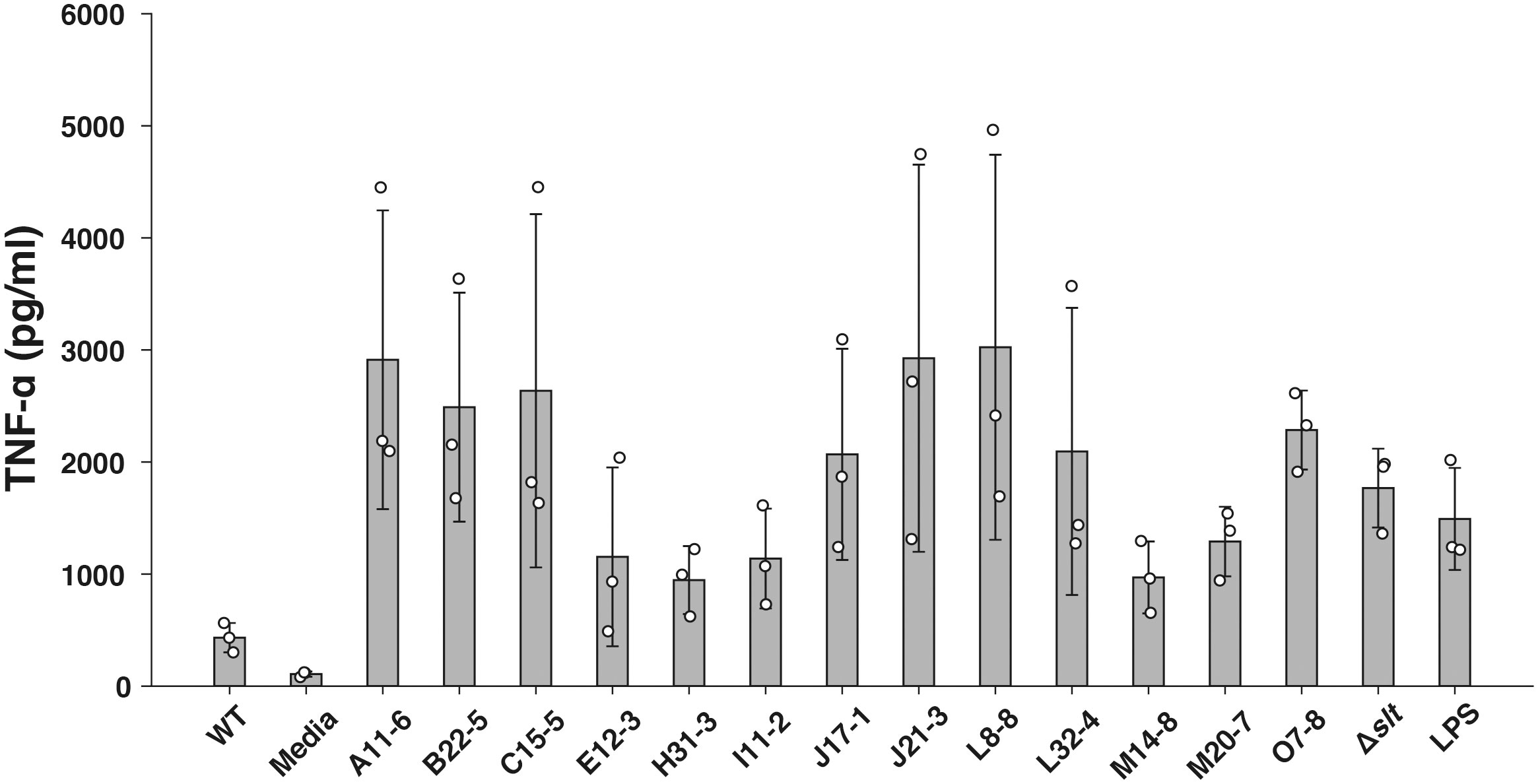
Figure 1 Screening of the F. novicida transposon mutant library using a TNF-α ELISA assay. U937 cells were infected with F. novicida transposon mutants at an MOI of 1 or stimulated with 100 ng/ml of E. coli LPS- or same volume BHIc-containing medium. After 5 h of infection, TNF-α released into the cell supernatant was measured by ELISA. Data shows averages and standard deviations from three independent experiments. Transposon mutant strains that exhibited a 1.5-fold increase in TNF-α production compared with the wild-type strain are shown.
To estimate the effect of pyrC in immunosuppression by F. novicida, we constructed a ΔpyrC mutant strain of F. novicida via homologous recombination. ELISA showed that the ΔpyrC mutant strain induced significantly higher levels of TNF-α production in the cell culture supernatant of U937 cells than the wild-type strain (Figure 2A). Although not significantly different, the pyrC complemented strain tended to show decreased levels of TNF-α induction compared with the ΔpyrC mutant strain. The pyrC complemented strain showed equivalent levels of TNF-α induction to the wild-type strain (Figure 2A). Next, we measured the induction of IL-1β and IFN-β, which are important cytokines for Francisella infection, in the ΔpyrC mutant strain (Gavrilin and Wewers, 2011). As in the case of TNF-α, the ΔpyrC mutant strain-infected U937 cells showed significantly higher levels of IL-1β and IFN-β production than the wild-type strain, and pyrC complementation decreased these levels to that observed upon infection with the wild-type strain (Figures 2B, C). The mRNA expression levels of TNF, IL1B, and IFNB1 in Francisella-infected U937 cells were also examined using real-time PCR. Similar results to those obtained via ELISA were observed for the mRNAs of all these genes (Figures 2D–F). These results indicate that the ΔpyrC mutant strain strongly evokes host innate immune responses compared to the wild-type strain, suggesting that pyrC is a critical factor for the immunosuppression of F. novicida.
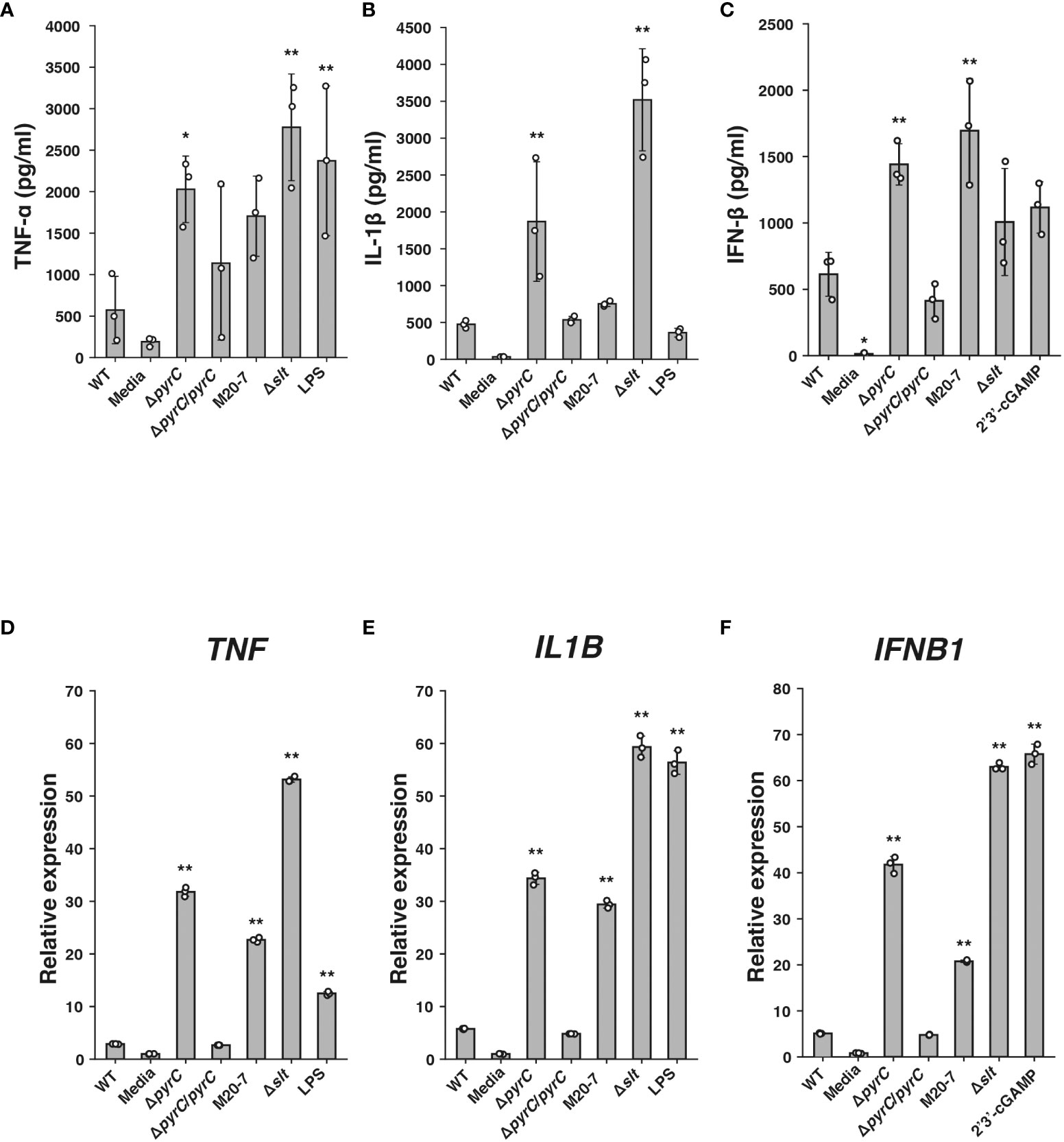
Figure 2 Cytokine induction by the F novicida pyrC deletion mutant. U937 cells were stimulated with 100 ng/ml of E coli LPS-containing medium or 10 ng/ml 2′3′-cGAMP- or same volume BHIc-containing medium or infected with F novicida strains (MOI = 1). Cell supernatants were collected and concentrations of TNF-α at 5 h post infection (p.i.) (A), IL-1β at 12 h p.i. (B), and IFN-β at 24 h p.i. (C) were measured by ELISA. Total RNA was collected, and mRNA expression of TNF at 5 h p.i. (D), IL1B at 5 h p.i. (E), and IFNB1 at 12 h p.i. (F) was measured by qPCR; relative expression normalized to that in the same volume BHIc-containing medium treatment control are shown. Data shows averages and standard deviations from three independent experiments. Differences compared with the wild-type strain were determined by Dunnett’s multiple comparison and are indicated by asterisks **P < 0.01, *P < 0.05.
Next, we examined whether ΔpyrC is involved in the intracellular growth of F. novicida. The ΔpyrC mutant strain entered the stationary phase slightly earlier than wild-type strain in BHIc medium and failed to grow in CDM (Supplementary Figure1). The GFP-expressing wild-type strain of F. novicida grew intracellularly in U937 cells from 2 to 48 h post infection, while the GFP-expressing ΔpyrC mutant strain did not show remarkable intracellular growth during the same period through fluorescence microscopy (Figure 3A). To support this finding, the number of ΔpyrC and transposon mutant strain cells were significantly decreased to approximately 1/10th that of wild-type strain cells within U937 cells. On the contrary, the ΔpyrC mutant strain showed higher intracellular growth compared to the strain deficient in the type VI secretion system (ΔdotU), which was used as a negative control. The complemented strain restored the ability of the ΔpyrC mutant strain to grow intracellularly (Figure 3B). These results suggest that pyrC is not essential but is important for the intracellular growth of F. novicida in host cells.
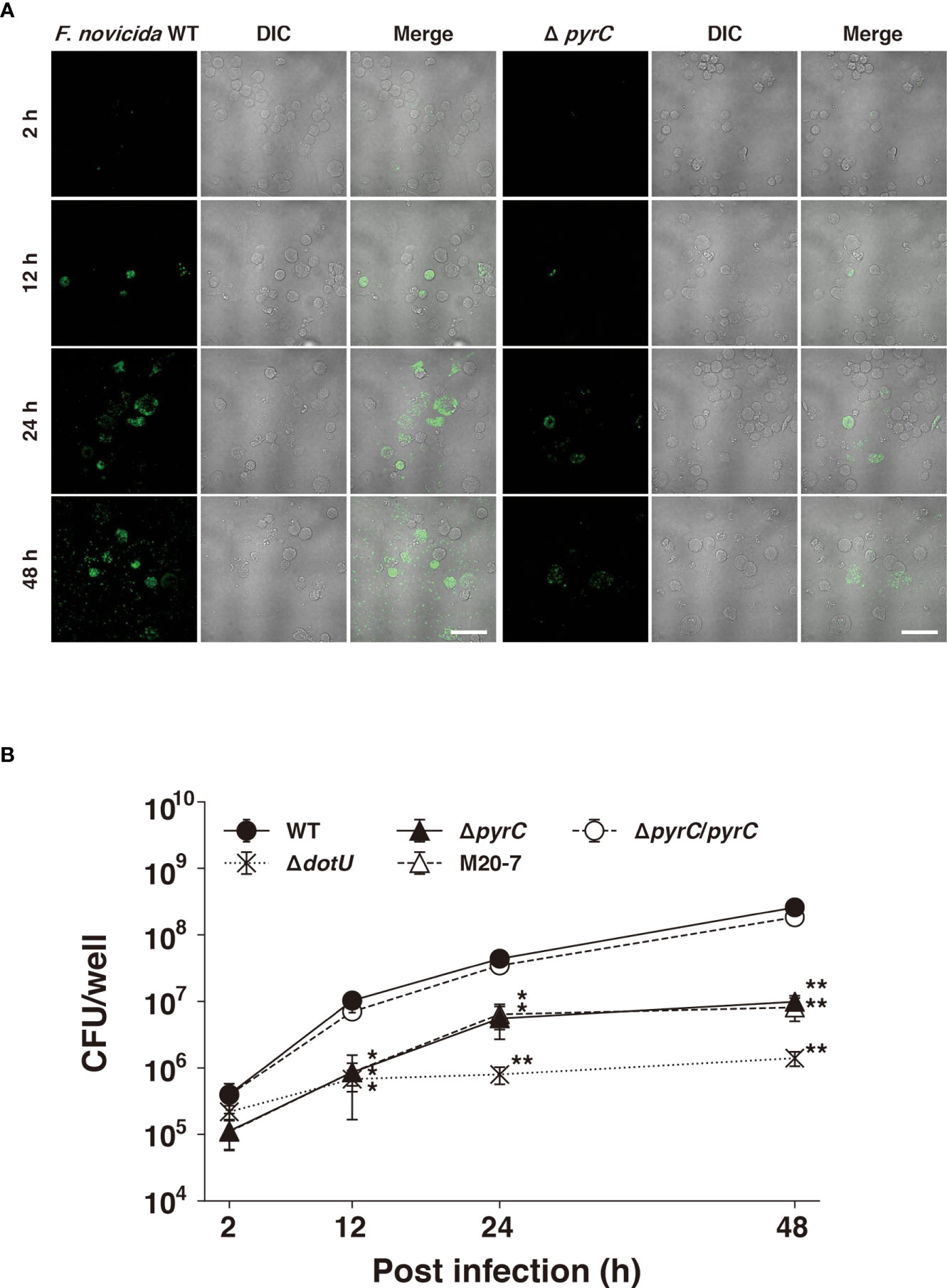
Figure 3 Intracellular growth of the pyrC deletion mutant. (A) U937 cells were infected with GFP-expressing F novicida strains at an MOI of 1 and treated with 50 μg/ml of gentamicin for 1 (h) At 2–48 h post infection, the cells were fixed and observed by FV1000 confocal laser scanning microscopy. Scale bar: 20 μm. (B) U937 cells were infected with F novicida strains at an MOI of 1 and treated with 50 μg/ml of gentamicin for 1 (h) At indicated time points post infection, cells were disrupted with 0.1% Triton X-100 and plated on BHIc agar; colony forming units were then counted. Data shows averages and standard deviations from three independent experiments. Differences compared with the wild-type strain were determined by Dunnett’s multiple comparison and are indicated by asterisks **P < 0.01, *P < 0.05.
Host cells infected with bacteria produce TNF-α due to the recognition of various bacterial ligands, such as LPS, peptidoglycan, and nucleotides, by PRRs (Kawai and Akira, 2006; Li and Wu, 2021). To identify ΔpyrC mutant strain ligands responsible for the induction of TNF-α in the ΔpyrC mutant strain infected U937 cells, U937 cells were treated with heat-inactivated F. novicida mutant strains. TNF-α induction by all heat-inactivated F. novicida strains, including the ΔpyrC mutant strain, was decreased to the same level as that by the negative control; however, LPS retained the ability to induce TNF-α with or without heat inactivation (Figure 4). This result suggests that TNF-α production by F. novicida strains, including the ΔpyrC mutant strain, is induced through biological activities of F. novicida, such as internalization by or proliferation in host cells, and not through heat-stable ligands, such as LPS, peptidoglycan, and nucleotides.
Figure 4 TNF-α induction by heat-inactivated F. novicida pyrC deletion mutant. U937 cells were stimulated with 100 ng/ml of E. coli LPS- or same volume BHIc-containing medium or infected with F. novicida strains (MOI = 1). Heat inactivation of the bacterial suspension was performed at 90℃ for 5 min. At 5 h post infection, cell supernatants were collected, and TNF-α concentrations were measured by ELISA. Data shows averages and standard deviations from three independent experiments. The differences among various conditions were analyzed by Tukey–Kramer multiple comparison and are indicated by asterisks **P < 0.01.
Because IFN-β production is induced by recognition of cytosolic DNA by the cyclic-di-nucleotide sensor STING (Jones et al., 2010; Storek et al., 2015) pathway, we next examined whether F. novicida pyrC affects the STING pathway. U937 cells treated with a STING inhibitor H-151 were infected with the ΔpyrC mutant strain, and the IFN-β levels induced in them were measured using ELISA. H-151 showed no significant effect on the growth of both the wild-type and the ΔpyrC mutant strain in BHIc medium (Supplementary Figure 2). IFN-β levels in the supernatant of H-151-treated U937 cells infected with the ΔpyrC mutant or transposon mutants and stimulated with the STING agonist 2′3′-cGAMP were significantly decreased compared to those in the supernatant of the DMSO control cells (Figure 5). Contrarily, H-151-treated U937 cells showed no significant difference in IFN-β induction upon infection with ΔpyrC mutant or transposon mutants compared to the wild-type and pyrC complemented strains. These results indicate that pyrC is involved in the suppression of IFN-β through the STING pathway.
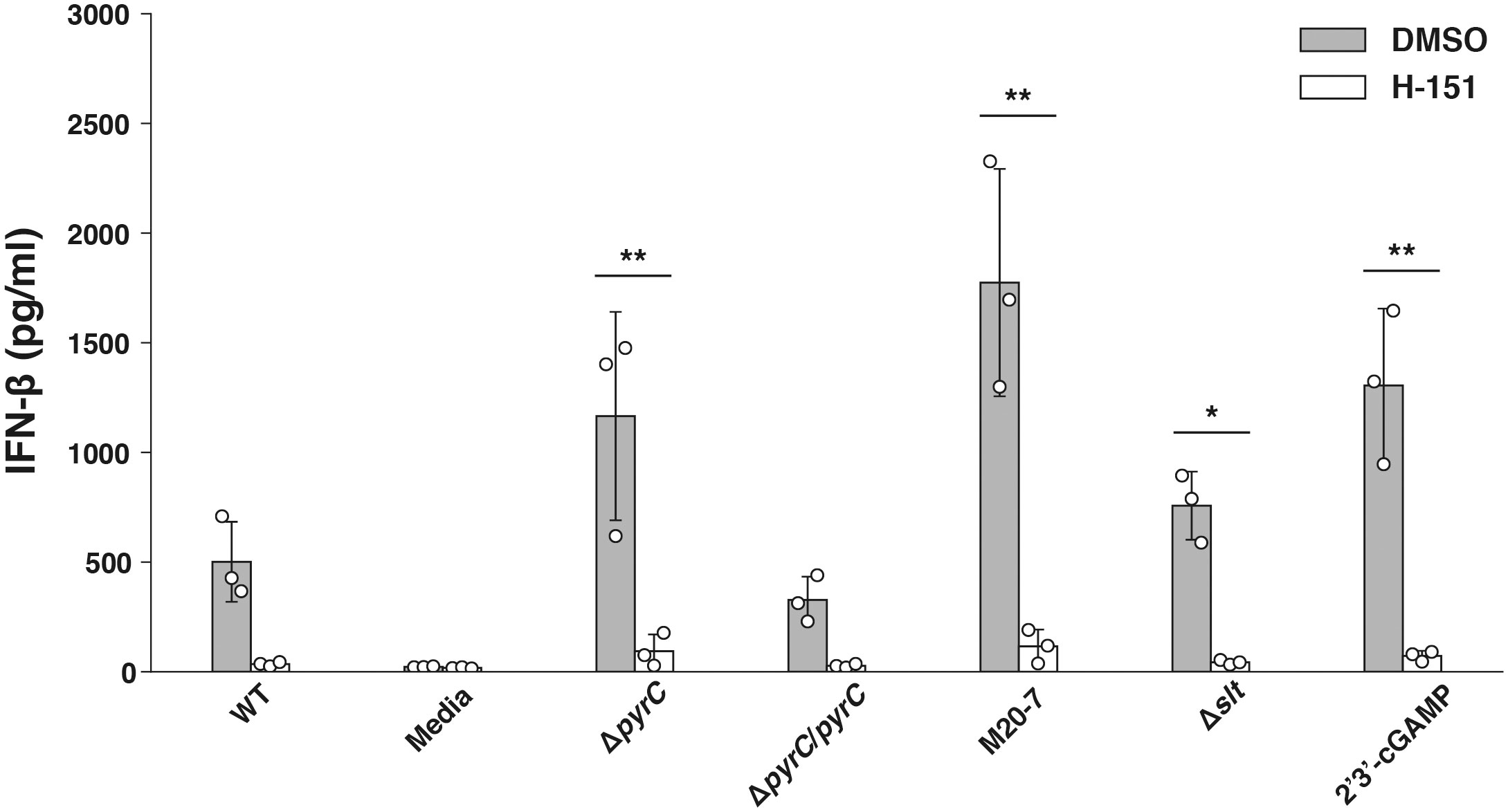
Figure 5 IFN-β induction by the F. novicida pyrC deletion mutant under STING inhibition. U937 cells were treated the H-151 STING inhibitor 2 h prior to infection and then stimulated with 10 ng/ml 2′3′-cGAMP, BHIc medium or infected with F. novicida strains (MOI = 1). At 24 h post infection, cell supernatants were collected, and concentrations of IFN-β were measured by ELISA. Data shows averages and standard deviations from three independent experiments. The differences among various conditions were determined by Tukey–Kramer multiple comparison and are indicated by asterisks **P < 0.01, *P < 0.05.
Intracellular bacteria, including Francisella, have refined their strategy to escape the host immune system and survive in host cells. To date, the importance of immunosuppression and immune evasion in Francisella infection has been well recognized, but their detailed mechanisms are poorly understood. To our knowledge, a large-scale gene screening of Francisella mutants focusing on Francisella immunosuppressive properties has not been performed yet. Therefore, here, we developed a transposon mutant library of F. novicida consisting of 3552 mutants. This library seems to cover the 1731 protein coding genes of the F. novicida U112 strain (Rohmer et al., 2007). Among the 3552 mutant strains in our library, strains that induced higher levels of host immune response than the wild-type F. novicida strain were isolated. Ten genes were determined to be immunosuppression-related. Among these 10 genes, four genes, FTN_0756 (fopA), FTN_1286 (mltA), FTN_0757 (cas9), and FTN_0496 (slt), were previously reported as immunosuppressive factors of Francisella (Peng et al., 2011; Sampson et al., 2013; Nakamura et al., 2019; Nakamura et al., 2021), ensuring the reliability of the screening method employed in this study. The remaining 6 genes, FTN_1641 (ampG), FTN_1199 (capA/B) FTN_1548 (yfgL), FTN_0917 (dacB), FTN_0611 (kdsA), and FTN_0024 (pyrC), were newly identified as Francisella immunosuppressive factors in this study. Because ampG, capA/B, yfgL, dacB, and kdsA are involved in creating the structure of bacterial cells, disruption of these genes can result in the leakage of ligands for intracellular receptor inflammasomes, such as LPS, peptidoglycan, and nucleotides (Peng et al., 2011; Man and Kanneganti, 2015; Tsuchiya, 2020). In contrast, the gene pyrC putatively encodes dihydroorotase, which converts carbamoyl N-Carbamoyl-L-aspartate into 4,5-dihydroorotate through the Francisella pyrimidine biosynthesis pathway (Turnbough Charles and Switzer Robert, 2008). It was previously reported that another pyrimidine biosynthesis gene pyrF is involved in immune suppression by the F. tularensis LVS (Horzempa et al., 2010). Although these results indicate that pyrimidine biosynthesis pathway is strongly related to immunosuppression by Francisella, the detailed mechanisms through which the pyrimidine biosynthesis pathway affects immunosuppression by Francisella remain unclear. Therefore, we focused on pyrC in this study. In this screening method, pyrF was not identified as immune suppressive factor. This may indicate that 3552 mutants are not enough to cover the 1731 protein coding genes of F. novicida or there is a bias in the insertion site of transposon.
To confirm the effects of pyrC on immunosuppression, a pyrC deletion mutant was generated. The ΔpyrC mutant strain induced a high level of TNF-α production in infected U937 macrophage cells compared to that in the wild-type strain, and the complemented strain restored the immunosuppressive property of F. novicida. TNF-α is produced through a broad range of innate immune signaling pathways, including TLR signaling pathways activated by various bacterial ligands, such as LPS, peptidoglycan, and nucleotides (Kawai and Akira, 2006; Li and Wu, 2021). In host cells infected by Francisella, the production of inflammatory cytokines, such as TNF-α and IL-6, is induced by the recognition of Francisella by TLR2, followed by the recognition of Francisella DNA by TLR 9 (Jones et al., 2012). Because pyrC is related to pyrimidine biosynthesis, it may suppress immune responses by modifying nucleotides recognized by TLRs, such as TLR9. However, as discussed below, heat-inactivated F. novicida strains, including the ΔpyrC mutant strain, failed to induce TNF-α production, indicating that biological activities of F. novicida, such as its phagosomal escape, are necessary for it to induce TNF-α and other cytokines.
Our data also revealed that the disruption of the pyrimidine biosynthesis pathway by F. novicida induces higher levels of IL-1β and IFN-β production in U937 macrophage cells. IL-1β is secreted when its precursor is expressed through the activation of TLR or type I IFN signaling followed by cleavage with caspase-1 activated through recognition by inflammasomes (Henry et al., 2007; Man and Kanneganti, 2015). In F. novicida infections, infected host cells exhibit robust inflammasome activation and IL-1β secretion compared with F. holarctica LVS and F. tularensis Schu S4 infections (Jones et al., 2011; Ghonime et al., 2015). In this study, we found that IL-1β production was suppressed in host cells infected by wild-type F. novicida and increased in host cells infected with the ΔpyrC mutant strain. This result is inconsistent with those of previous reports by Horzempa’s and Schulert, which indicate that several genes of F. tularensis Schu S4 and F. holartica LVS strains are related to the suppression of IL-1β production (Schulert et al., 2009; Horzempa et al., 2010). Our results, however, show that the expression of IL-1β mRNA was suppressed upon host cell infection by the wild-type strain of F. novicida and increased upon host cell infection with the ΔpyrC mutant strain. Taken together with the fact that the IL-1β precursor is expressed through TLR or type I IFN signaling, the suppression of IL-1β secretion by pyrC cannot be attributed to the inhibition of inflammasomes but to the inhibition of TLR or type I IFN signaling.
Type I IFNs are secreted through STING-related pathways. The cGAS-STING pathway, known as an intracellular DNA sensor, recognizes bacterial DNA and induces type I IFNs (Ishikawa and Barber, 2008). STING is also activated by direct recognition of cyclic-di-nucleotides released by bacteria (Barber, 2015). Type I IFN secretion resulting from Francisella infection exacerbates the infection. It was reported that type I IFNs suppress host immunity by inhibiting IL-17A expression of γδT cell, and deficiency in type I IFN-related molecules, such as cGAS, STING, IFNAR1, and IRF3, in mice result in resistance to Francisella infection (Henry et al., 2007; Henry et al., 2010; Storek et al., 2015). However, bacterial factors involved in the modulation of host type I IFNs and their mechanisms of action are incompletely understood. In this study as well, INF-β was suppressed in U937 cells infected with F. novicida, and pyrC was related to this suppression. To elucidate the mechanisms involved in IFN-β suppression, the STING inhibitor H-151 was used. H-151 decreased the induction of IFN-β in U937 cells infected by the ΔpyrC mutant strain, indicating that pyrC is involved in the suppression of pathways related to STING. IFN-β production through STING in Francisella infected cells has been observed in human cell line and mouse infection models (Jones et al., 2010; Nandakumar et al., 2019). Several studies revealed that IFN-β secretion by the STING-dependent detection of F. novicida in host cell cytoplasm boosts inflammasome activation, IL-1β release, and pyroptosis by promoting gene expression of gamma guanylate proteins, ZBP1, and pyrin (Henry et al., 2007; Man et al., 2015; Meunier et al., 2015; Lee et al., 2021). In our investigation, IFN-β and IL-1β were induced in U937 cells infected with the ΔpyrC mutant strain, not only at the protein level, but also at the mRNA level. Because STING pathway activation was found to induce IL-1β mRNA expression in a type I IFN-dependent manner (Swanson et al., 2017), our findings suggest that pyrC is crucial for IFN-β suppression followed by the suppression of IL-1β secretion.
To determine the ligands that induce TNF-α in U937 cells infected with the ΔpyrC mutant strain, we treated U937 cells with heat-inactivated Francisella mutant strains, including the ΔpyrC mutant strain. TNF-α production decreased to control levels in all the heat-inactivated strains. This result indicates that TNF-α production is not caused by the recognition of F. novicida by heat-resistant ligands, such as LPS, peptidoglycan, and nucleotides, present outside the cells, but by biological responses of F. novicida, such as intracellular proliferation. A previous report on Pseudomonas aeruginosa indicated that uracil controls biofilm formation via quorum-sensing, and P. aeruginosa mutants lacking genes involved in uracil biosynthesis could not form biofilms (Ueda et al., 2009). A biofilm is a structured community of microbial cells in a matrix formed by extracellular polymeric substances (EPS). The EPS consist of polysaccharides, nucleic acids (extracellular DNA and RNA), proteins, lipids, and other biomolecules (Karygianni et al., 2020). It has also been shown that biofilms formed by Mycobacterium avium and P. aeruginosa and the extracellular DNA of these bacteria have the potential to induce TNF-α production in host cells infected by them (Rose and Bermudez, 2014; Ramirez et al., 2019). In the ΔpyrC mutant strain-infected U937 cells, the production of the TNF-α, IL-1β, and IFN-β cytokines was increased in response to the recognition of bacterial nucleic acids compared with that in wild-type strain-infected U937 cells. These results suggest that pyrC may be involved in the coordination or modification of ligands, such as extracellular DNA in host cytosol, protecting Francisella from TLR or STING recognition and allowing it to grow intracellularly by suppressing immune responses.
Intracellular growth is one of the most important abilities determining Francisella’s pathogenicity, a lot of genes involved in the intracellular growth of Francisella have been identified in previous studies (Qin and Mann, 2006; Su et al., 2007). PyrC is required for de novo pyrimidine biosynthesis (Choi and Zalkin, 1990). Mutants of Francisella genes involved in the pyrimidine pathway (e.g., carA, pyrB, pyrD, and pyrF) become uracil auxotrophs and show deficient growth on complete medium (Maier et al., 2006; Qin and Mann, 2006; Horzempa et al., 2010). Several reports indicate that these pyrimidine pathway-related mutants can grow within epithelial cells but not macrophage cells (Qin and Mann, 2006; Horzempa et al., 2010). In addition, Schulert et al. showed that a pyrimidine biosynthesis pathway transposon mutant of F. novicida was eliminated by monocyte-derived macrophages, in part via phagosomes (Schulert et al., 2009). In our study, the ΔpyrC mutant showed decreased growth in culture medium, and decreased but constant intracellular growth in U937 cells compared with that of the wild-type strain. Francisella mutants that is deficient in the phagosomal escape-related factor such as mglA or type VI secretion system, have no ability to induce IL-1β secretion (Mariathasan et al., 2005; Gavrilin et al., 2006; Jones et al., 2010). These results indicate that the ΔpyrC mutant strain can enter the host cytoplasm and grow intracellularly. Although the intracellular bacterial number of the ΔpyrC mutant strain was relatively low, it increased the production of cytokines compared with that in cells infected with the wild-type strain, suggesting strongly that pyrC contributes to the suppression of host immune responses.
In summary, we performed here, a large-scale screening to search for factors responsible for immunosuppression by F. novicida in human macrophage cells. Ten genes were determined to be responsible for the immunosuppression. Among them, pyrC was identified as a novel F. novicida immunosuppressive factor and was immunologically characterized. Although further studies are needed to elucidate the detailed mechanisms by which pyrC is involved in host immunosuppression by Francisella, research on pyrimidine metabolic pathways involving pyrC may provide new insight into Francisella immunosuppression and pathogenicity and into the mechanisms by which host cells recognize intracellular bacteria.
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
TN contributed to laboratory analysis, investigation, and writing of the original draft. TS contributed to study conceptualization, laboratory analysis, investigation, and writing of the original draft. RI contributed to laboratory analysis. AU contributed to methodology and resources. KW contributed to laboratory analysis, investigation, and validation. MW was involved in study conceptualization, laboratory analysis, supervision, manuscript review, and manuscript editing. All authors contributed to the article and approved the submitted version.
This work was supported by JSPS KAKENHI Grant Number 19K07556 and 22K07054 and Grant-in-Aid for JSPS Fellows Grant Number 22J20065.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcimb.2022.1027424/full#supplementary-material
Barber, G. N. (2015). STING: infection, inflammation and cancer. Nat. Rev. Immunol. 15, 760–770. doi: 10.1038/nri3921
Best, A., Abu Kwaik, Y. (2019). Nutrition and bipartite metabolism of intracellular pathogens. Trends Microbiol. 27, 550–561. doi: 10.1016/j.tim.2018.12.012
Butchar, J. P., Cremer, T. J., Clay, C. D., Gavrilin, M. A., Wewers, M. D., Marsh, C. B., et al. (2008). Microarray analysis of human monocytes infected with francisella tularensis identifies new targets of host response subversion. PloS One 3, e2924. doi: 10.1371/journal.pone.0002924
Choi, K. Y., Zalkin, H. (1990). Regulation of escherichia coli pyrC by the purine regulon repressor protein. J. Bacteriol. 172, 3201–3207. doi: 10.1128/jb.172.6.3201-3207.1990
Chong, A., Wehrly, T. D., Nair, V., Fischer, E. R., Barker, J. R., Klose, K. E., et al. (2008). The early phagosomal stage of francisella tularensis determines optimal phagosomal escape and francisella pathogenicity island protein expression. Infect. Immun. 76, 5488–5499. doi: 10.1128/IAI.00682-08
Feng, Y., Napier, B. A., Manandhar, M., Henke, S. K., Weiss, D. S., Cronan, J. E. (2014). A francisella virulence factor catalyses an essential reaction of biotin synthesis. Mol. Microbiol. 91, 300–314. doi: 10.1111/mmi.12460
Gavrilin, M. A., Bouakl, I. J., Knatz, N. L., Duncan, M. D., Hall, M. W., Gunn, J. S., et al. (2006). Internalization and phagosome escape required for Francisella to induce human monocyte IL-1β processing and release. Proc. Natl. Acad. Sci. 103, 141–146. doi: 10.1073/pnas.0504271103
Gavrilin, M. A., Wewers, M. D. (2011). Francisella recognition by inflammasomes: Differences between mice and men. Front. Microbiol. 2. doi: 10.3389/fmicb.2011.00011
Ghonime, M. G., Mitra, S., Eldomany, R. A., Wewers, M. D., Gavrilin, M. A. (2015). Inflammasome priming is similar for francisella species that differentially induce inflammasome activation. PloS One 10, e0127278. doi: 10.1371/journal.pone.0127278
Gillette, D. D., Curry, H. M., Cremer, T., Ravneberg, D., Fatehchand, K., Shah, P. A., et al. (2014). Virulent type a francisella tularensis actively suppresses cytokine responses in human monocytes. Front. Cell. Infect. Microbiol. 4. doi: 10.3389/fcimb.2014.00045
Goncheva, M. I., Chin, D., Heinrichs, D. E. (2022). Nucleotide biosynthesis: the base of bacterial pathogenesis. Trends Microbiol. 30, 793–804. doi: 10.1016/j.tim.2021.12.007
Haag, S. M., Gulen, M. F., Reymond, L., Gibelin, A., Abrami, L., Decout, A., et al. (2018). Targeting STING with covalent small-molecule inhibitors. Nature 559, 269–273. doi: 10.1038/s41586-018-0287-8
Henry, T., Brotcke, A., Weiss, D. S., Thompson, L. J., Monack, D. M. (2007). Type I interferon signaling is required for activation of the inflammasome during francisella infection. J. Exp. Med. 204, 987–994. doi: 10.1084/jem.20062665
Henry, T., Kirimanjeswara, G. S., Ruby, T., Jones, J. W., Peng, K., Perret, M., et al. (2010). Type I IFN signaling constrains IL-17A/F secretion by γδ T cells during bacterial infections. J. Immunol. 184, 3755–3767. doi: 10.4049/jimmunol.0902065
Horzempa, J., O’Dee, D. M., Shanks, R. M. Q., Nau, G. J. (2010). Francisella tularensis DeltapyrF mutants show that replication in nonmacrophages is sufficient for pathogenesis in vivo. Infect. Immun. 78, 2607–2619. doi: 10.1128/IAI.00134-10
Ishikawa, H., Barber, G. N. (2008). STING is an endoplasmic reticulum adaptor that facilitates innate immune signalling. Nature 455, 674–678. doi: 10.1038/nature07317
Jones, J. W., Broz, P., Monack, D. M. (2011). Innate immune recognition of francisella tularensis: activation of type-I interferons and the inflammasome. Front. Microbiol. 2. doi: 10.3389/fmicb.2011.00016
Jones, J. W., Kayagaki, N., Broz, P., Henry, T., Newton, K., O’Rourke, K., et al. (2010). Absent in melanoma 2 is required for innate immune recognition of francisella tularensis. Proc. Natl. Acad. Sci. U. S. A. 107, 9771–9776. doi: 10.1073/pnas.1003738107
Jones, C. L., Napier, B. A., Sampson, T. R., Llewellyn, A. C., Schroeder, M. R., Weiss, D. S. (2012). Subversion of host recognition and defense systems by francisella spp. Microbiol. Mol. Biol. Rev. 76, 383–404. doi: 10.1128/mmbr.05027-11
Karygianni, L., Ren, Z., Koo, H., Thurnheer, T. (2020). Biofilm matrixome: Extracellular components in structured microbial communities. Trends Microbiol. 28, 668–681. doi: 10.1016/j.tim.2020.03.016
Kawai, T., Akira, S. (2006). TLR signaling. Cell Death Differ. 13, 816–825. doi: 10.1038/sj.cdd.4401850
Kingry, L. C., Petersen, J. M. (2014). Comparative review of francisella tularensis and francisella novicida. Front. Cell. Infect. Microbiol. 4. doi: 10.3389/fcimb.2014.00035
Lee, S., Karki, R., Wang, Y., Nguyen, L. N., Kalathur, R. C., Kanneganti, T.-D. (2021). AIM2 forms a complex with pyrin and ZBP1 to drive PANoptosis and host defence. Nature 597, 425–419. doi: 10.1038/s41586-021-03875-8
Li, D., Wu, M. (2021). Pattern recognition receptors in health and diseases. Signal Transduct Target Ther. 6, 291. doi: 10.1038/s41392-021-00687-0
Mahawar, M., Atianand, M. K., Dotson, R. J., Mora, V., Rabadi, S. M., Metzger, D. W., et al. (2012). Identification of a NovelFrancisella tularensisFactor required for intramacrophage survival and subversion of innate immune response. J. Biol. Chem. 287, 25216–25229. doi: 10.1074/jbc.m112.367672
Maier, T. M., Pechous, R., Casey, M., Zahrt, T. C., Frank, D. W. (2006). In vivo Himar1-based transposon mutagenesis of francisella tularensis. Appl. Environ. Microbiol. 72, 1878–1885. doi: 10.1128/AEM.72.3.1878-1885.2006
Man, S. M., Kanneganti, T.-D. (2015). Regulation of inflammasome activation. Immunol. Rev. 265, 6–21. doi: 10.1111/imr.12296
Man, S. M., Karki, R., Malireddi, R. K. S., Neale, G., Vogel, P., Yamamoto, M., et al. (2015). The transcription factor IRF1 and guanylate-binding proteins target activation of the AIM2 inflammasome by francisella infection. Nat. Immunol. 16, 467–475. doi: 10.1038/ni.3118
Mariathasan, S., Weiss, D. S., Dixit, V. M., Monack, D. M. (2005). Innate immunity against francisella tularensis is dependent on the ASC/caspase-1 axis. J. Exp. Med. 202, 1043–1049. doi: 10.1084/jem.20050977
Maurin, M. (2015). Francisella tularensis as a potential agent of bioterrorism? Expert Rev. Anti Infect. Ther. 13, 141–144. doi: 10.1586/14787210.2015.986463
Mc Gann, P., Rozak, D. A., Nikolich, M. P., Bowden, R. A., Lindler, L. E., Wolcott, M. J., et al. (2010). A novel brain heart infusion broth supports the study of common francisella tularensis serotypes. J. Microbiol. Methods 80, 164–171. doi: 10.1016/j.mimet.2009.12.005
Meibom, K. L., Charbit, A. (2010). Francisella tularensis metabolism and its relation to virulence. Front. Microbiol. 1. doi: 10.3389/fmicb.2010.00140
Meunier, E., Wallet, P., Dreier, R. F., Costanzo, S., Anton, L., Rühl, S., et al. (2015). Guanylate-binding proteins promote activation of the AIM2 inflammasome during infection with francisella novicida. Nat. Immunol. 16, 476–484. doi: 10.1038/ni.3119
Miller, C., Celli, J. (2016). Avoidance and subversion of eukaryotic homeostatic autophagy mechanisms by bacterial pathogens. J. Mol. Biol. 428, 3387–3398. doi: 10.1016/j.jmb.2016.07.007
Nagle, S. C., Anderson, R. E., Gary, N. D. (1960). Chemically defined medium for the growth of pasteurella tularensis. J. Bacteriol. 79, 566–571. doi: 10.1128/jb.79.4.566-571.1960
Nakamura, T., Shimizu, T., Inagaki, F., Okazaki, S., Saha, S. S., Uda, A., et al. (2021). Identification of membrane-bound lytic murein transglycosylase a (MltA) as a growth factor for francisella novicida in a silkworm infection model. Front. Cell. Infect. Microbiol. 10. doi: 10.3389/fcimb.2020.581864
Nakamura, T., Shimizu, T., Uda, A., Watanabe, K., Watarai, M. (2019). Soluble lytic transglycosylase SLT of francisella novicida is involved in intracellular growth and immune suppression. PloS One 14, e0226778. doi: 10.1371/journal.pone.0226778
Nallaparaju, K. C., Yu, J.-J., Rodriguez, S. A., Zogaj, X., Manam, S., Guentzel, M. N., et al. (2011). Evasion of IFN-γ signaling by francisella novicida is dependent upon francisella outer membrane protein c. PloS One 6, e18201. doi: 10.1371/journal.pone.0018201
Nandakumar, R., Tschismarov, R., Meissner, F., Prabakaran, T., Krissanaprasit, A., Farahani, E., et al. (2019). Intracellular bacteria engage a STING-TBK1-MVB12b pathway to enable paracrine cGAS-STING signaling. Nat. Microbiol. 4, 701–713. doi: 10.1038/s41564-019-0367-z
Peng, K., Broz, P., Jones, J., Joubert, L.-M., Monack, D. (2011). Elevated AIM2-mediated pyroptosis triggered by hypercytotoxic francisella mutant strains is attributed to increased intracellular bacteriolysis. Cell. Microbiol. 13, 1586–1600. doi: 10.1111/j.1462-5822.2011.01643.x
Petersen, J. M., Schriefer, M. E. (2005). Tularemia: emergence/re-emergence. Vet. Res. 36, 455–467. doi: 10.1051/vetres:2005006
Pfaffl, M. W. (2001). A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 29, e45. doi: 10.1093/nar/29.9.e45
Platz, G. J., Bublitz, D. C., Mena, P., Benach, J. L., Furie, M. B., Thanassi, D. G. (2010). A tolC mutant of francisella tularensis is hypercytotoxic compared to the wild type and elicits increased proinflammatory responses from host cells. Infect. Immun. 78, 1022–1031. doi: 10.1128/IAI.00992-09
Putzova, D., Panda, S., Härtlova, A., Stulík, J., Gekara, N. O. (2017). Subversion of innate immune responses by francisella involves the disruption of TRAF3 and TRAF6 signalling complexes. Cell. Microbiol. 19, 1–11. doi: 10.1111/cmi.12769
Qin, A., Mann, B. J. (2006). Identification of transposon insertion mutants of francisella tularensis tularensis strain schu S4 deficient in intracellular replication in the hepatic cell line HepG2. BMC Microbiol. 6, 69. doi: 10.1186/1471-2180-6-69
Ramirez, T., Shrestha, A., Kishen, A. (2019). Inflammatory potential of monospecies biofilm matrix components. Int. Endod J. 52, 1020–1027. doi: 10.1111/iej.13093
Rodriguez, S. A., Yu, J.-J., Davis, G., Arulanandam, B. P., Klose, K. E. (2008). Targeted inactivation of francisella tularensis genes by group II introns. Appl. Environ. Microbiol. 74, 2619–2626. doi: 10.1128/AEM.02905-07
Rohmer, L., Fong, C., Abmayr, S., Wasnick, M., Larson Freeman, T. J., Radey, M., et al. (2007). Comparison of francisella tularensis genomes reveals evolutionary events associated with the emergence of human pathogenic strains. Genome Biol. 8, R102. doi: 10.1186/gb-2007-8-6-r102
Rose, S. J., Bermudez, L. E. (2014). Mycobacterium avium biofilm attenuates mononuclear phagocyte function by triggering hyperstimulation and apoptosis during early infection. Infect. Immun. 82, 405–412. doi: 10.1128/IAI.00820-13
Sampson, T. R., Saroj, S. D., Llewellyn, A. C., Tzeng, Y.-L., Weiss, D. S. (2013). A CRISPR/Cas system mediates bacterial innate immune evasion and virulence. Nature 497, 254–257. doi: 10.1038/nature12048
Santic, M., Abu Kwaik, Y. (2013). Nutritional virulence of francisella tularensis. Front. Cell. Infect. Microbiol. 3. doi: 10.3389/fcimb.2013.00112
Santic, M., Molmeret, M., Klose, K. E., Abu Kwaik, Y. (2006). Francisella tularensis travels a novel, twisted road within macrophages. Trends Microbiol. 14, 37–44. doi: 10.1016/j.tim.2005.11.008
Schulert, G. S., McCaffrey, R. L., Buchan, B. W., Lindemann, S. R., Hollenback, C., Jones, B. D., et al. (2009). Francisella tularensis genes required for inhibition of the neutrophil respiratory burst and intramacrophage growth identified by random transposon mutagenesis of strain LVS. Infect. Immun. 77, 1324–1336. doi: 10.1128/IAI.01318-08
Shapiro, D. S., Schwartz, D. R. (2002). Exposure of laboratory workers to francisella tularensis despite a bioterrorism procedure. J. Clin. Microbiol. 40, 2278–2281. doi: 10.1128/JCM.40.6.2278-2281.2002
Shimizu, T., Otonari, S., Suzuki, J., Uda, A., Watanabe, K., Watarai, M. (2019). Expression of francisella pathogenicity island protein intracellular growth locus e (IglE) in mammalian cells is involved in intracellular trafficking, possibly through microtubule organizing center. Microbiologyopen 8, e00684. doi: 10.1002/mbo3.684
Sjöstedt, A. (2007). Tularemia: history, epidemiology, pathogen physiology, and clinical manifestations. Ann. N. Y. Acad. Sci. 1105, 1–29. doi: 10.1196/annals.1409.009
Storek, K. M., Gertsvolf, N. A., Ohlson, M. B., Monack, D. M. (2015). cGAS and Ifi204 cooperate to produce type I IFNs in response to francisella infection. J. Immunol. 194, 3236–3245. doi: 10.4049/jimmunol.1402764
Su, J., Yang, J., Zhao, D., Kawula, T. H., Banas, J. A., Zhang, J.-R. (2007). Genome-wide identification of francisella tularensis virulence determinants. Infect. Immun. 75, 3089–3101. doi: 10.1128/IAI.01865-06
Swanson, K. V., Junkins, R. D., Kurkjian, C. J., Holley-Guthrie, E., Pendse, A. A., El Morabiti, R., et al. (2017). A noncanonical function of cGAMP in inflammasome priming and activation. J. Exp. Med. 214, 3611–3626. doi: 10.1084/jem.20171749
Telepnev, M., Golovliov, I., Grundström, T., Tärnvik, A., Sjöstedt, A. (2003). Francisella tularensis inhibits toll-like receptor-mediated activation of intracellular signalling and secretion of TNF-alpha and IL-1 from murine macrophages. Cell. Microbiol. 5, 41–51. doi: 10.1046/j.1462-5822.2003.00251.x
Tsuchiya, K. (2020). Inflammasome-associated cell death: Pyroptosis, apoptosis, and physiological implications. Microbiol. Immunol. 64, 252–269. doi: 10.1111/1348-0421.12771
Turnbough Charles, L., Switzer Robert, L. (2008). Regulation of pyrimidine biosynthetic gene expression in bacteria: Repression without repressors. Microbiol. Mol. Biol. Rev. 72, 266–300. doi: 10.1128/MMBR.00001-08
Keywords: Francisella, tularemia, pyrimidine, immune response, cytokine
Citation: Nakamura T, Shimizu T, Ikegaya R, Uda A, Watanabe K and Watarai M (2022) Identification of pyrC gene as an immunosuppressive factor in Francisella novicida infection. Front. Cell. Infect. Microbiol. 12:1027424. doi: 10.3389/fcimb.2022.1027424
Received: 25 August 2022; Accepted: 13 October 2022;
Published: 26 October 2022.
Edited by:
Santanu Bose, Washington State University, United StatesReviewed by:
Daniel Alford Powell, University of Arizona, United StatesCopyright © 2022 Nakamura, Shimizu, Ikegaya, Uda, Watanabe and Watarai. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Masahisa Watarai, d2F0YXJhaUB5YW1hZ3VjaGktdS5hYy5qcA==
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.