- 1Key Laboratory of East China Sea Fishery Resources Exploitation, Ministry of Agriculture and Rural Affair, East China Sea Fisheries Research Institute, Chinese Academy of Fishery Sciences, Shanghai, China
- 2Shanghai Engineering Research Center of Aquaculture, Shanghai Ocean University, Shanghai, China
- 3Shanghai Collaborative Innovation Center for Cultivating Elite Breeds and Green-culture of Aquaculture Animals, Shanghai, China
- 4Fisheries Technology Promotion Station of Fengxian District, Shanghai, China
The microsporidian Enterocytozoon hepatopenaei (EHP) has become a critical threat to the global shrimp aquaculture industry, thus necessitating early detection by screening. Development of a rapid and accurate assay is crucial both for the active surveillance and for the assessment of shrimp with EHP infection. In the present study, a distinct strain of E. hepatopenaei (EHPMr) was found in Macrobrachium rosenbergii. The SWP1 gene analysis revealed it was a new genotype that differed with the common strain isolated from the Litopenaeus vannamei (EHPLv). A nested SWP-PCR method was modified to fix the bug that the original inner primers could not recognize the EHPMr strain. The redesigned inner primers successfully amplified a product of 182 bp for both the EHPMr strain and the EHPLv strain. The new primers also had good specificity and high sensitivity, which may serve as an alternative for EHP genotyping. This study provided a method for detection of EHP in the biosecurity of Macrobrachium rosenbergii farming, and the developed protocol was proposed for the routine investigation and potential carrier screening, especially for molecular epidemiology.
1 Introduction
Enterocytozoon hepatopenaei (EHP) is a microsporidian responsible for hepatopancreatic microsporidiosis (HPM) outbreaks in cultured shrimp (Tourtip et al., 2009; Chaijarasphong et al., 2021). In recent years, EHP has been discovered in several countries, such as Malaysia, Vietnam, India, Indonesia, Thailand, China, and Venezuela, and it has caused huge economic losses (Biju et al., 2016; Tang et al., 2017; Flegel, 2018; Behera et al., 2019; Hou et al., 2021). Its widespread distribution has increased the threat to the global shrimp aquaculture industry. The maintenance of shrimp broodstock and the management of the hatchery, nursery and grow-out are facing enormous challenges in the prevention and control of EHP disease.
The giant freshwater prawn, Macrobrachium rosenbergii, native to the tropical and subtropical areas of Southeast Asia, is one of the important economic prawn species in the world (Azad et al., 2021). It was introduced into China from Japan in 1976 (Dong et al., 2020). Being popular for its large individual size, fast growth, delicious flesh and high nutritional value, it has become one of the important cultured species in China (Wei et al., 2021). Shrimp hosts known to be infected by EHP include P. monodon, L. vannamei, Litopenaeus stylirostris, and a suspected species (Penaeus japonicus) (Chaijarasphong et al., 2021), but there are few reports about M. rosenbergii being infected by EHP.
Specific pathogens screening and detection from the shrimp postlarvae stages have become the important measures taken by farmers to ensure the success of aquaculture. The nested PCR diagnostic technique is widely used because of its high accuracy and low instrument requirements. However, the accuracy and sensitivity of the nested PCR assays established based on different gene sequences are different. Previous research has confirmed that the primers designed based on the EHP SSU rRNA gene can cross-react with other similar microsporidians and generate false positives, but the primers designed based on the spore wall protein (SWP) gene can avoid false positives and are more sensitive than the former (Jaroenlak et al., 2016). Therefore, the nested PCR assay targeting the SWP gene (SWP-PCR) has been adopted by the fishery industry and widely used in the detection of shrimp seedlings. Furthermore, this method has also been selected as the EHP detection standard for the fishery industry in China (SC/T 7232-2020 code of diagnosis for Enterocytozoon hepatopenaei disease).
In March 2020, the above SWP-PCR method was applied by our laboratory to screen for pathogens in M. rosenbergii seedlings. Interestingly, we found that the positive target fragment did amplify by the outer primers, but no band was amplified by the inner primers. In subsequent studies, we confirmed the EHP strain derived from L. vannamei (EHPLv) and the EHP strain derived from M. rosenbergii (EHPMr) were different in SWP gene, although the SSU rDNA sequence of these two EHP strains showed ~99% identity. The above SWP-PCR method was not suitable for the detection of EHPMr. The goal of this study was to develop a sensitive and specific nested PCR method for simultaneous detection of two EHP strains.
2 Material and methods
2.1 Samples collection
The EHP-infected L. vannamei and the EHP-infected M. rosenbergii were collected from the same farm in Fengxian District, Shanghai Province, China (N30°53’18.6”, E121°35’32.3”), in March 2020. The farm suffered severe EHP infection in 2019. The infected L. vannamei were 2.0 ~ 3.0 cm in length. The unnormal M. rosenbergii were 1.0 ~ 1.8 cm in length. Both healthy L. vannamei and healthy M. rosenbergii were collected from the normal ponds on another farm in Fengxian District. Samples were transported to the laboratory with oxygen and then fixed in 95% ethanol for PCR analysis. The handling of shrimps followed the guidelines for the Ethical Committee of Experimental Animal Care at the Shanghai Ocean University of China.
2.2 DNA extraction
For EHP detection, 10 individual samples of each shrimp and prawn were dissected. The hepatopancreas DNA was extracted using an animal organization DNA Extraction Kit (Tiangen Biotechnology, China) according to the manufacturer’s instructions, and stored at −20°C for PCR assays.
2.3 SWP1 gene amplification and cloning
The target genes were amplified using the nested SWP-PCR method. Briefly: the PCR mixtures (25 μL) for both steps contained 0.625 units of Ex Taq DNA polymerase (Takara Bio) and 0.2 μM of each primer. For the first PCR reaction, outer primers SWP1F and SWP1R (Table 1) were used to amplify a 514 bp fragment. The PCR cycling conditions include an initial denaturation at 95°C for 5 min, followed by 30 cycles of 95°C for 30 s, 58°C for 30 s, and 68°C for 45 s, and a final extension at 68°C for 5 min. The PCR products were checked using 1% agarose gel with DNA ladder.
The DNA fragments of the SWP1 gene from both EHP isolates were purified and cloned into the pMD18-T vector. The resulting plasmids were transferred into competent cells DH5α and cultured in Luria-Bertani (LB) medium. Recombinant colonies were selected by the Blue-White screening. The transformants were identified by colony PCR and then cultured in a shaking incubator for 4 hours. Finally, the positive colonies were sequenced using M13 sequencing primers by the ABI 3730xl DNA Analyzer.
2.4 Sequence and phylogenetic analysis of SWP1 gene
For sequence homology analysis, the full-length SWP1 gene (EhSWP1, GenBank accession nos. MG015710, GenPept accession nos. AVQ09707) previously published by Jaroenlak et al. (2018) was used as the reference sequence, which was amplified from EHP strain isolated from P. vannamei. After cloning and sequencing, the nucleotide sequences obtained were edited and alignments performed using Clustal W and compared with other nucleotide sequences in the GenBank using the BLAST program at National Center for Biotechnology Information (NCBI).
For the SWP1 gene phylogeny, multiple nucleotide sequence alignment was carried out using Clustal W. The “find best DNA/Protein models” program was run to determine the best-fit model with the lowest Bayesian information criterion (BIC). Phylogenetic tree was constructed using Neighbor-Joining method based on the Tamura 3-parameter model in MEGA-X. Bootstrap with 1000 replications was set to assess branch support.
2.5 New inner primers designed for simultaneous detection
To solve the problem that the primers SWP2F and SWP2R could not amplify the SWP1 gene of EHPMr, the new inner primer pairs were designed with the aid of Primer Premier 6.0 software. Based on multiple sequence alignments results, primer positions were derived from the conserved regions of SWP1 gene from all EHP isolates. Specificity of the primers was initially checked using primer blast (www.ncbi.nlm.nih.gov/tools/primer-blast/). The new inner primer pairs, forward primer SWP2F′ (5’-GCAGAGTGTTGTTAAGGGTTTAAG-3’) and reverse primer SWP2R′ (5’-GCTGTTTGTCWCCAACTGTATT-3’), were designed to target 182 bp internally to the first PCR product (Table 1).
2.6 Comparison of original method and modified method for detection of two EHP isolates
To compare the validity of newly designed inner primers and the original inner primers, the external PCR products of three SWP-PCR positive L. vannamei (EHPLv) and three SWP-PCR positive M. rosenbergii (EHPMr) were selected as DNA template respectively.
For the original SWP-PCR method, the inner primers SWP2F and SWP2R (Table 1) were used to generate a 147 bp fragment. The thermal cycling conditions include an initial denaturation at 95°C for 5 min, followed by 20 cycles of 95°C for 20 s, 64°C for 30 s, and 68°C for 20 s, and a final extension at 68°C for 5 min (Jaroenlak et al., 2016).
For the modified SWP-PCR method, 2nd-step (nested) PCR was carried out with the inner primers SWP2F′ and SWP2R′ (Table 1) to amplify a 182 bp product. PCR cycling conditions were initiation denaturation at 95°C for 5 min, followed by 20 cycles of 95°C for 30 s, 55°C for 30 s, and 68°C for 20 s, and a final extension at 68°C for 5 min. All secondary PCR products were analyzed by electrophoresis on a 1% agarose gel.
2.7 Sensitivity of the modified nested PCR assay
The plasmid containing the SWP1 gene of the EHPLv (named pGEM-SWP1) was extracted from the positive colonies as described above. The series of 10-fold dilutions of pGEM-SWP1 were used as positive templates. The single PCR with the two primers sets SWP2F/2R and SWP2F′/2R′ were carried out respectively, for testing the comparative sensitivity of the modified nested PCR and original nested PCR.
2.8 Specificity of the modified nested PCR assay
The genomic DNAs of five different aquatic microsporidians were selected to evaluate the specificity of the designed inner primers. Enterospora epinepheli isolated from Epinephelus spp.; Nucleaspora hippocampi isolated from Hippocampus erectus (Wang et al., 2022); Enterocytospora artemiae isolated from Palaemonetes sinensis; Ameson portunus isolated from Portunus trituberculatus; Potaspora sp. (unidentified) isolated from Exopalaemon carinicauda. Enterospora and Nucleaspora were the closely related genus in the Enterocytozoon group Microsporidia (EGM) that mainly infect gastrointestinal tracts of their hosts (Stentiford et al., 2019). E. artemiae infected the hepatopancreas and gut of crustacean hosts (Rode et al., 2013). Ameson and Potaspora were selected as the representative species of microsporidia infecting the skeletal muscles of hosts (Ding et al., 2016; Wang et al., 2017). The DNA template extracted from EHPMr-infected M. rosenbergii was used as a positive control. The nested PCR conditions referred to the above.
3 Results
3.1 Comparison of SWP genes between two strains of EHP
3.1.1 Nucleic acid sequence analysis
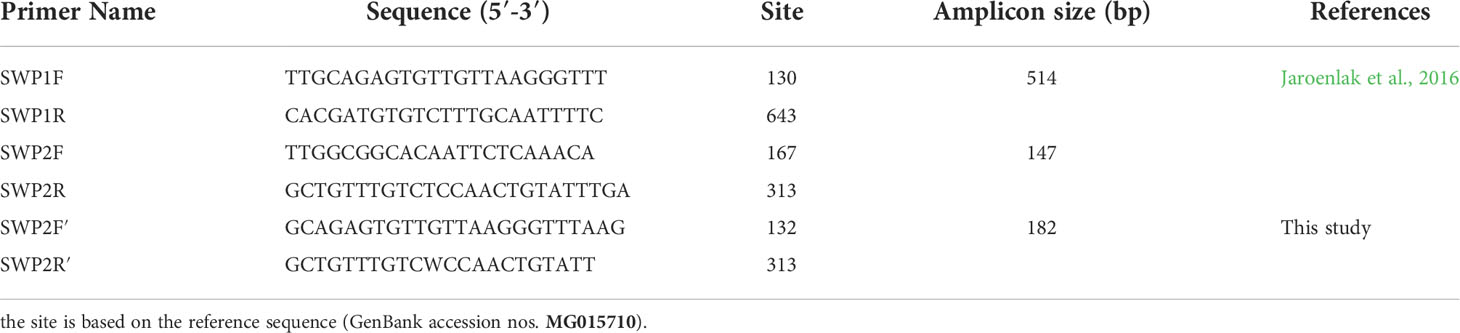
The obtained partial SWP1 gene of EHPLv (EhLvSWP1) and the partial SWP1 gene of EHPMr (EhMrSWP1) were both 514 bp in size (Figure 1). Sequence analysis revealed that the EhLvSWP1 shared a 100% nucleotide sequence identity with the reference EhSWP1. Whereas the EhMrSWP1 shared a 93% nucleotide sequence identity with the EhSWP1. This indicated that the EhMrSWP1 represented the presence of a novel genotype. In comparison with EhLvSWP1 and EhSWP1, the EhMrSWP1 showed 36 single base mutations, including eight transitions and 28 transversions, and no insertion and deletion (Figure 1). The obtained nucleotide sequence of EhMrSWP1 was deposited in GenBank database under accession number MW269619.
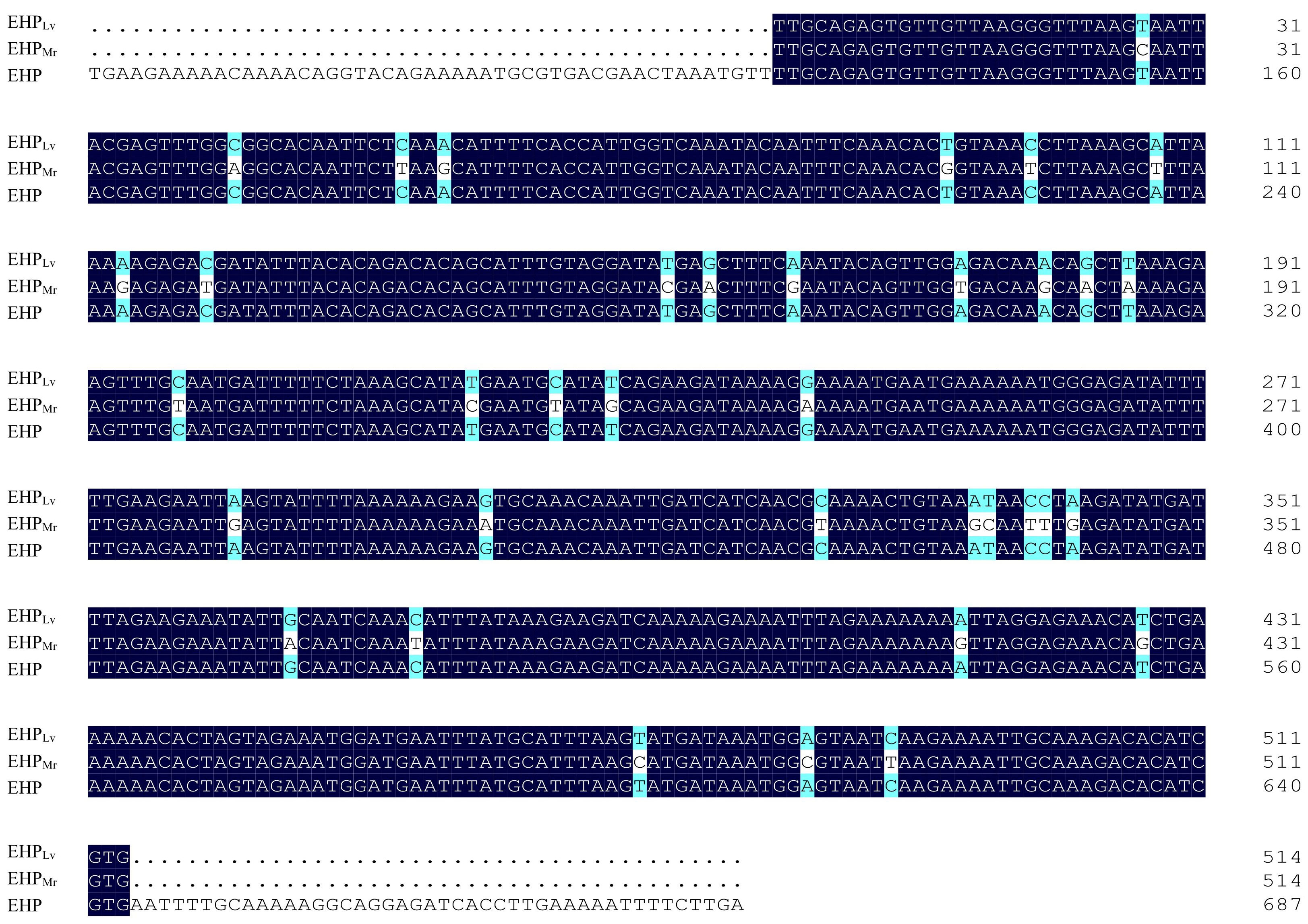
Figure 1 Alignment of partial nucleotide sequences of the SWP1 gene of EHPLv, EHPMr, and reference EHP (MG015710). The nucleobases with white differ from the consensus. The numbers on the right indicate the nucleotide position.
3.1.2 Amino acid sequence comparison
The comparison of predicted protein sequences revealed that EHPLv shared a 100% identity with the reference EHP from L. vannamei (GenPept accession nos. AVQ09707), whereas EHPMr shared a 98.25% amino acid sequence identity to the reference EHP (Figure 2). Remarkably, among the obtained 171 amino acids, three amino acid exchanges: serine mutated to alanine at sites 77 and 143, and asparagine mutated to serine at site 112.

Figure 2 Alignment of partial protein sequences of the SWP1 from EHPLv, EHPMr, and reference EHP (GenPept accession nos. AVQ09707). The amino acids with white differ from the consensus. The numbers on the right indicate the amino acid position in the published sequence.
3.1.3 Phylogenetic analyses
On the phylogenetic tree (Figure 3), all EHP isolated were grouped together in a large branch with a high support (99%). Group 1 is largest group containing most EHP that infecting the L. vannamei. Within Group 2, EHPMrclustered together with an EHP strain obtained from L. vannamei (KY593129).
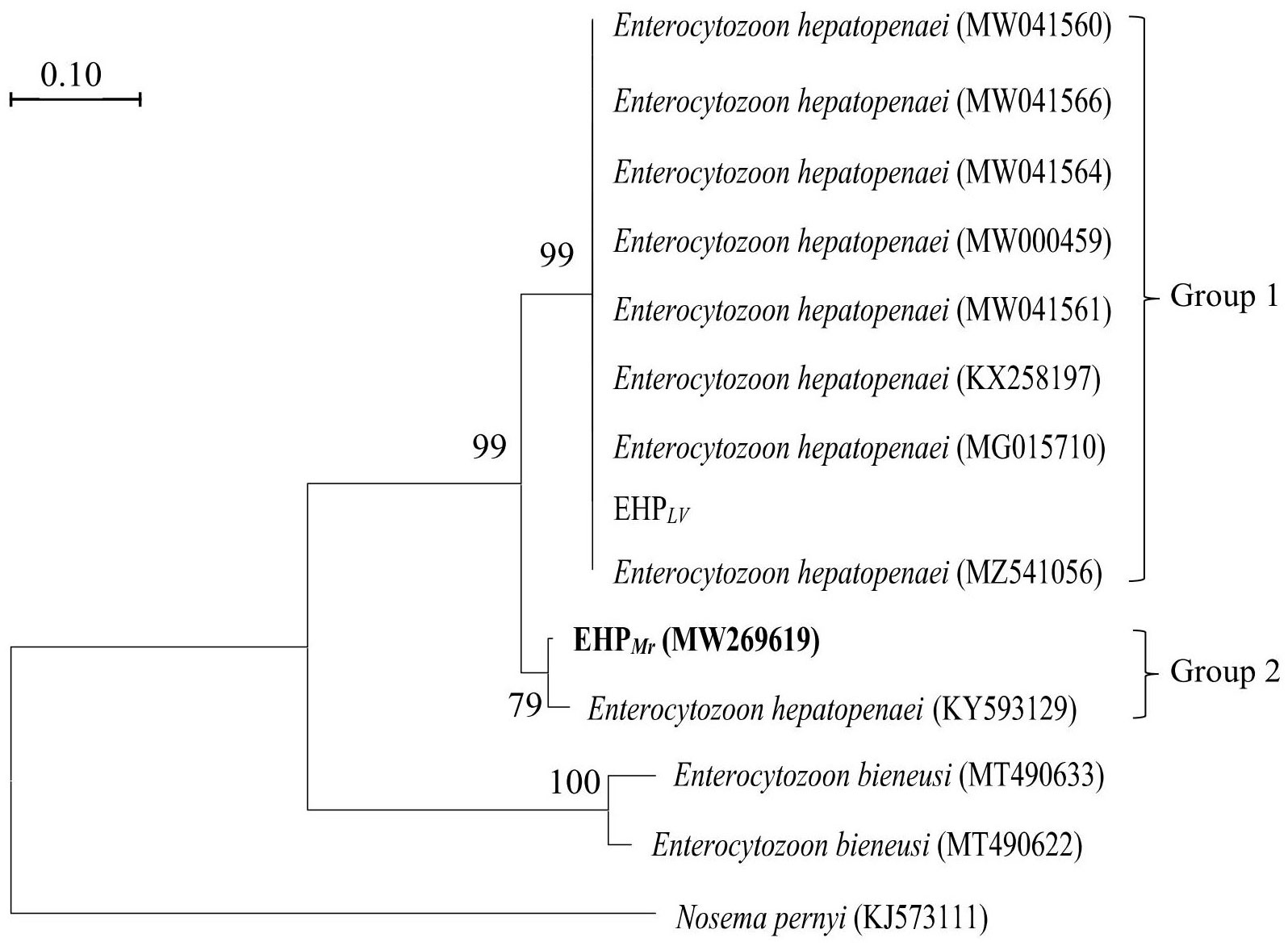
Figure 3 Phylogenetic tree of SWP1 gene of EHP isolates with other microsporidia. Nosema pernyi is used as the outgroup. Bootstrap values are indicated on the branches.
3.2 Detection of EHPLv and EHPMr by the existing SWP-PCR and the modified SWP-PCR
To validate the two protocols, the hepatopancreas DNAs, isolated from naturally EHPLv-infected L. vannamei samples and naturally EHPMr-infected M. rosenbergii samples, were subjected to the first round of amplification. The outer primers SWP1F/1R successfully amplified a 514 bp DNA fragment from both infected L. vannamei and infected M. rosenbergii (Figure 4, top).
Figure 4 Nested PCR for the detection of the two EHP stains in Litopenaeus vannamei and Macrobrachium rosenbergii by using the SWP2F/2R primers and SWP2F′/2R′ primers, respectively. Lanes V1~V3: the hepatopancreatic DNA of EHPLv-infected Litopenaeus vannamei; Lanes R1~R3: the hepatopancreatic DNA of EHPMr-infected Macrobrachium rosenbergii; M: molecular weight marker; +: positive control, SWP1 gene plasmid DNA; v-: negative control, the hepatopancreatic DNA of healthy Litopenaeus vannamei, R-: negative control, the hepatopancreatic DNA of healthy Macrobrachium rosenbergii.
In the second round, the inner primers SWP2F/2R amplified the expected 147 bp fragment from all EHPLv-infected L. vannamei but were negative for any of the EHPMr-infected M. rosenbergii (Figure 4, middle), it means the existing SWP-PCR method can only detect the EHPLv. Whereas the novel inner primers SWP2F′/2R′ produced the predicted 182 bp fragment for both infected shrimp and infected prawn (Figure 4, bottom), indicating that the modified SWP-PCR method can detect not only EHPLv but also EHPMr.
In addition, comparing the second-round PCR products by the two methods, the typical products of the existing SWP-PCR method contained an unexpected DNA fragment which migrated very closely together with the target band of 147 bp (Figure 4, middle), similar to previous reports (Jaroenlak et al., 2016; Munkongwongsiri et al., 2022). While only one prominent band of 182 bp was formed by the novel primers, indicating that the modified method improved the specificity of PCR amplification (Figure 4, bottom).
3.3 Sensitivity of the nested PCR
The sensitivity of the two nested PCR was tested using the 10-fold dilution series of pGEM-SWP1 plasmid DNA. The result was shown in Figure 5. In the single PCR, the modified method displayed a high sensitivity identical to the original SWP-PCR method, which could detect as low as 103 copies of pGEM-SWP1 per reaction mix.
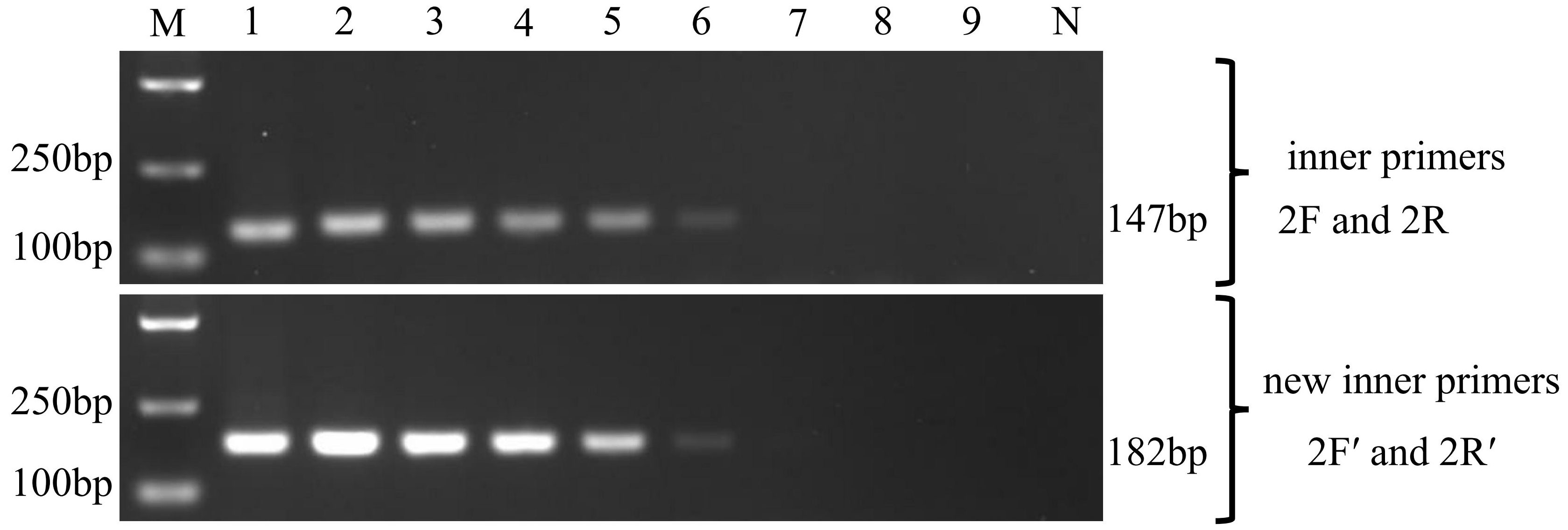
Figure 5 Comparison of sensitivity of the inner primers (SWP2F/2R) and new inner primers (SWP2F′/2R′) to amplify the SWP1 gene. M: molecular weight marker; 1-9:1×108 -1×100 copies of 10-fold dilutions of pGEM-SWP1; N: negative control, DNA samples of healthy P. vannamei.
3.4 Specificity of the nested PCR
In cross-amplification assays, none of the other microsporidian showed any amplification product in the nested PCR (Figure 6). This confirmed the specificity of the designed primers for EHP detection.

Figure 6 Validation of the specificity of improved nested PCR detection. M: molecular weight marker; lane 1, Enterospora epinepheli; lane 2, Nucleaspora hippocampi; lane 3, Enterocytospora artemiae; lane 4, Ameson portunus; lane 5, Potaspora sp.; +: positive control.
4 Discussion
4.1 The application of new nested PCR for discovering the EHP mutants
Microsporidia are known as the obligate intracellular parasite. To adapt to the host cell life, its genome has been extremely compressed (Corradi and Slamovits, 2011). The natural genetic variation of pathogenic microorganisms can determine the success of infecting the host and favorable mutations may help to expand its host range (Bonneaud and Longdon, 2020). Variants have been found in a variety of microsporidia, such as Encephalitozoon cuniculi, Encephalitozoon hellem, Encephalitozoon intestinalis and Enterocytozoon bieneusi (Duzlu et al., 2019; Li et al., 2019). According to the results of this study, a mutant strain of EHP has been detected in M. rosenbergii for the first time, and the difference of its SWP1 gene suggested that EHP is also quietly changing itself to infect other crustacean hosts.
The nested PCR method has become the common method for disease surveillance because of its high specificity and sensitivity. However, due to the strain differentiation of EHP, the single specific primers cannot recognize different strains of EHP. To solve this problem, in this study, we designed a pair of degenerate primers to meet the demand for the EHP mutants’ detection in disease control and prevention.
Furthermore, the combined use of SWP2F′/2R′ primer pairs and SWP2F/2R primer pairs will help us to identify EHPLv and EHPMr strains. In a batch of shrimp infected with EHP, if SWP2F′/2R′ is positive and SWP2F/2R test is negative, it suggests that this batch of samples is infected with EHPMr strain; if SWP2F′/2R′ is positive and SWP2F/2R test is also positive, it shows that this batch of samples is infected with EHPLv strain.
4.2 SWP1 gene can be a recognizing site for EHP genotyping
Most researches on genotyping of microsporidia mutant strains use ITS site as diagnostic target and seldom use SWP gene. However, in recent years, more and more studies demonstrated that SWP gene was a promising target for genotyping. Xiao et al. (2001) found that the SWP1 gene of E. cuniculi had genetic diversity; Ou et al. (2021) confirmed the canine-adapted genotypes (Group 11) of E. bieneusi are one unique group of genotypes, and genetically divergent from other genotype groups by the sequence difference in SWP1 gene. Polonais et al. (2010) found that the EhSWP1 C-terminal of four strains of human microsporidia E. hellem showed significant interspecific and intraspecific polymorphisms, suggesting that SWP gene is more suitable for genotyping than internal transcribed spacer (ITS) or small subunit ribosomal DNA (SSU-rDNA) sequences.
To investigate the spread of EHP in global shrimp aquaculture, genotyping will become an important problem that needs to be solved in epidemiology. The polymorphism of SWP1 gene in different isolates will make it a good marker for studying EHP genotyping. In addition, the mutation of SWP1 gene may help to increase our understanding of the adhesion of EHP spore wall proteins to different host cell surface receptors.
5 Conclusion
In summary, it is the first report on characterization of the SWP1 gene from a new EHP genotype. The mutation of the SWP1 gene will be useful to understand the molecular mechanism that EHP adapts to different hosts. Furthermore, this study provides a modified nested PCR assay for EHP detection in both L. vannamei and M. rosenbergii. The modified method possesses excellent specificity and comparable sensitivity with the previous nested PCR method, which is proposed for the EHP mutants’ investigation in epidemiological studies.
Data availability statement
The data presented in the study are deposited in the NCBI GenBank repository, accession number MW269619.
Author contributions
WF and HT designed the experiments. JZ and YW performed the experiments and prepared the manuscript. MY, NY, and YX participated in molecular analyses. WL and XL constructed the figures. JY assisted in sample collection. All authors contributed to the article and approved the submitted version.
Funding
The research was supported by Fundamental Research Funds for Public Research Institutes at central level (East China Sea Fisheries Research Institute) (No.2019M03) and the Central Public-interest Scientific Institution Basal Research Fund, CAFS (No. 2020TD41).
Acknowledgments
The authors acknowledge Liwen Xu for providing Enterospora epinepheli, and Hongbo Jiang for providing Enterocytospora artemiae used in this research.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Azad, M. A. K., Islam, S. S., Amin, M. N., Ghosh, A. K., Hasan, K. R., Bir, J., et al. (2021). Production and economics of probiotics treated Macrobrachium rosenbergii at different stocking densities. Anim. Feed Sci. Technol. 282, 115125. doi: 10.1016/j.anifeedsci.2021.115125
Behera, B. K., Das, A., Paria, P., Sahoo, A. K., Parida, P. K., Abdulla, T., et al. (2019). Prevalence of microsporidian parasite, Enterocytozoon hepatopenaei in cultured pacific white shrimp, Litopenaeus vannamei (Boone 1931) in West Bengal, East coast of India. Aquacult. Int. 27 (2), 609–620. doi: 10.1007/s10499-019-00350-0
Biju, N., Sathiyaraj, G., Raj, M., Shanmugam, V., Baskaran, B., Govindan, U., et al. (2016). High prevalence of Enterocytozoon hepatopenaei in shrimps Penaeus monodon and Litopenaeus vannamei sampled from slow growth ponds in India. Dis. Aquat. Org. 120 (3), 225–230. doi: 10.3354/dao03036
Bonneaud, C., Longdon, B. (2020). Emerging pathogen evolution using evolutionary theory to understand the fate of novel infectious pathogens. EMBO Rep. 21 (9), e51374. doi: 10.15252/embr.202051374
Chaijarasphong, T., Munkongwongsiri, N., Stentiford, G. D., Aldama-Cano, D. J., Thansa, K., Flegel, T. W., et al. (2021). The shrimp microsporidian Enterocytozoon hepatopenaei (EHP): Biology, pathology, diagnostics and control. J. Invertebr. Pathol. 186, 107458. doi: 10.1016/j.jip.2020.107458
Corradi, N., Slamovits, C. H. (2011). The intriguing nature of microsporidian genomes. Brief Funct. Genomics 10 (3), 115–124. doi: 10.1093/bfgp/elq032
Ding, Z., Sun, M., Liu, H., Zhao, Y., Pan, J., Xue, H. (2016). A new microsporidium, Potaspora macrobrachium n.sp. infecting the musculature of pond-reared oriental river prawn Macrobrachium nipponense (Decapoda: Palaemonidae). J. Invertebr. Pathol. 136, 57–64. doi: 10.1016/j.jip.2016.02.006
Dong, X., Liu, Q., Kan, D., Zhao, W., Guo, H., Lv, L. (2020). Effects of ammonia-n exposure on the growth, metabolizing enzymes, and metabolome of Macrobrachium rosenbergii. Ecotoxicol. Environ. Saf. 189, 110046. doi: 10.1016/j.ecoenv.2019.110046
Duzlu, O., Yildirim, A., Onder, Z., Ciloglu, A., Yetismis, G., Inci, A. (2019). Prevalence and genotyping of microsporidian parasites in dogs in Turkey: Zoonotic concerns. J. Eukaryot. Microbiol. 66 (5), 771–777. doi: 10.1111/jeu.12725
Flegel, T. W. (2018). Recent research on acute hepatopancreatic necrosis disease (AHPND) and Enterocytozoon hepatopenaei in Thailand. Asian Fish. Sci. 31, 257–269. doi: 10.33997/j.afs.2018.31.S1.018
Hou, Z., Yu, J., Wang, J., Li, T., Chang, L., Fang, Y., et al. (2021). Development of a PCR assay for the effective detection of Enterocytozoon hepatopenaei (EHP) and investigation of EHP prevalence in Shandong province, China. J. Invertebr. Pathol. 184, 107653. doi: 10.1016/j.jip.2021.107653
Jaroenlak, P., Boakye, D. W., Vanichviriyakit, R., Williams, B. A. P., Sritunyalucksana, K., Itsathitphaisarn, O. (2018). Identification, characterization and heparin binding capacity of a spore-wall, virulence protein from the shrimp microsporidian, Enterocytozoon hepatopenaei (EHP). Parasites Vectors 11, 177. doi: 10.1186/s13071-018-2758-z
Jaroenlak, P., Sanguanrut, P., Williams, B. A. P., Stentiford, G. D., Flegel, T. W., Sritunyalucksana, K., et al. (2016). A nested PCR assay to avoid false positive detection of the microsporidian Enterocytozoon hepatopenaei (EHP) in environmental samples in shrimp farms. PloS One 11 (11), 15. doi: 10.1371/journal.pone.0166320
Li, W., Feng, Y. Y., Santin, M. (2019). Host specificity of Enterocytozoon bieneusi and public health implications. Trends Parasitol. 35 (6), 436–451. doi: 10.1016/j.pt.2019.04.004
Munkongwongsiri, N., Thepmanee, O., Lertsiri, K., Vanichviriyakit, R., Itsathitphaisarn, O., Sritunyalucksana, K. (2022). False mussels (Mytilopsis leucophaeata) can be mechanical carriers of the shrimp microsporidian Enterocytozoon hepatopenaei (EHP). J. Invertebr. Pathol. 187, 107690. doi: 10.1016/j.jip.2021.107690
Ou, Y., Jiang, W., Roellig, D. M., Wan, Z., Li, N., Guo, Y., et al. (2021). Characterizations of Enterocytozoon bieneusi at new genetic loci reveal a lack of strict host specificity among common genotypes and the existence of a canine-adapted Enterocytozoon species. Int. J. Parasitol. 51 (2), 215–223. doi: 10.1016/j.ijpara.2020.09.008
Polonais, V., Mazet, M., Wawrzyniak, I., Texier, C., Blot, N., El Alaoui, H., et al. (2010). The human microsporidian Encephalitozoon hellem synthesizes two spore wall polymorphic proteins useful for epidemiological studies. Infect. Immun. 78 (5), 2221–2230. doi: 10.1128/Iai.01225-09
Rode, N. O., Landes, J., Lievens, E. J. P., Flaven, E., Segard, A., Jabbour-Zahab, R., et al. (2013). Cytological, molecular and life cycle characterization of Anostracospora rigaudi n. g., n. sp. and Enterocytospora artemiae n. g., n. sp., two new microsporidian parasites infecting gut tissues of the brine shrimp artemia. Parasitology 140 (9), 1168–1185. doi: 10.1017/S0031182013000668
Stentiford, G. D., Bass, D., Williams, B. A. P. (2019). Ultimate opportunists-the emergent Enterocytozoon group microsporidia. PloS Path. 15 (5), e1007668. doi: 10.1371/journal.ppat.1007668
Tang, K. F. J., Aranguren, L. F., Piamsomboon, P., Han, J. E., Maskaykina, I. Y., Schmidt, M. M. (2017). Detection of the microsporidian Enterocytozoon hepatopenaei (EHP) and taura syndrome virus in Penaeus vannamei cultured in Venezuela. Aquaculture 480, 17–21. doi: 10.1016/j.aquaculture.2017.07.043
Tourtip, S., Wongtripop, S., Stentiford, G. D., Bateman, K. S., Sriurairatana, S., Chavadej, J., et al. (2009). Enterocytozoon hepatopenaei sp. nov. (Microsporida: Enterocytozoonidae), a parasite of the black tiger shrimp Penaeus monodon (Decapoda: Penaeidae): Fine structure and phylogenetic relationships. J. Invertebr. Pathol. 102 (1), 21–29. doi: 10.1016/j.jip.2009.06.004
Wang, Y., Li, X., Fu, G., Zhao, S., Chen, Y., Wang, H., et al. (2017). Morphology and phylogeny of Ameson portunus n. sp. (Microsporidia) infecting the swimming crab Portunus trituberculatus from China. Eur. J. Protistol. 61, 122–136. doi: 10.1016/j.ejop.2017.09.008
Wang, Y., Ying, N., Huang, Y. Q., Zou, X., Liu, X., Li, L. T., et al. (2022). Nucleospora hippocampi n. sp., an intranuclear microsporidian infecting the seahorse Hippocampus erectus from China. Front. Cell. Infect. Microbiol. 12. doi: 10.3389/fcimb.2022.882843
Wei, J., Tian, L., Wang, Y., Yu, L., Zhu, X. (2021). Effects of salinity, photoperiod, and light spectrum on larval survival, growth, and related enzyme activities in the giant freshwater prawn, Macrobrachium rosenbergii. Aquaculture 530, 735794. doi: 10.1016/j.aquaculture.2020.735794
Keywords: microsporidia, Enterocytozoon hepatopenaei, Macrobrachium rosenbergii, nested PCR, spore wall protein
Citation: Wang Y, Zhou J, Yin M, Ying N, Xiang Y, Liu W, Ye J, Li X, Fang W and Tan H (2022) A modification of nested PCR method for detection of Enterocytozoon hepatopenaei (EHP) in giant freshwater prawn Macrobrachium rosenbergii. Front. Cell. Infect. Microbiol. 12:1013016. doi: 10.3389/fcimb.2022.1013016
Received: 06 August 2022; Accepted: 06 September 2022;
Published: 23 September 2022.
Edited by:
Rongrong Ma, Ningbo University, ChinaReviewed by:
Linxiang Yin, Harvard Medical School, United StatesHongbo Jiang, Shenyang Agricultural University, China
Copyright © 2022 Wang, Zhou, Yin, Ying, Xiang, Liu, Ye, Li, Fang and Tan. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Wenhong Fang, ZndlbmhvbmdAMTYzLmNvbQ==; Hongxin Tan, aHh0YW5Ac2hvdS5lZHUuY24=
†These authors have contributed equally to this work
 Yuan Wang
Yuan Wang Jinyang Zhou
Jinyang Zhou Menghe Yin1
Menghe Yin1 Yang Xiang
Yang Xiang Xincang Li
Xincang Li