
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Cell. Infect. Microbiol. , 13 January 2022
Sec. Molecular Bacterial Pathogenesis
Volume 11 - 2021 | https://doi.org/10.3389/fcimb.2021.803730
Campylobacter jejuni is a major cause of food poisoning worldwide, and remains the main infective agent in gastroenteritis and related intestinal disorders in Europe and the USA. As with all bacterial infections, the stages of adhesion to host tissue, survival in the host and eliciting disease all require the synthesis of proteinaceous virulence factors on the ribosomes of the pathogen. Here, we describe how C. jejuni virulence is attenuated by altering the methylation of its ribosomes to disrupt the composition of its proteome, and how this in turn provides a means of identifying factors that are essential for infection and pathogenesis. Specifically, inactivation of the C. jejuni Cj0588/TlyA methyltransferase prevents methylation of nucleotide C1920 in the 23S rRNA of its ribosomes and reduces the pathogen’s ability to form biofilms, to attach, invade and survive in host cells, and to provoke the innate immune response. Mass spectrometric analyses of C. jejuni TlyA-minus strains revealed an array of subtle changes in the proteome composition. These included reduced amounts of the cytolethal distending toxin (CdtC) and the MlaEFD proteins connected with outer membrane vesicle (OMV) production. Inactivation of the cdtC and mlaEFD genes confirmed the importance of their encoded proteins in establishing infection. Collectively, the data identify a subset of genes required for the onset of human campylobacteriosis, and serve as a proof of principle for use of this approach in detecting proteins involved in bacterial pathogenesis.
Campylobacter jejuni infections are the leading cause of human bacterial gastroenteritis in developed countries, and are characterized by attachment and invasion of the colonic epithelium leading to inflammation of the bowel and diarrheal disease. In severe cases, symptoms can progress to septicemia, post-infectious arthritis, Guillain-Barré syndrome (Yuki et al., 2004), Miller Fisher syndrome (Ang et al., 2002), or chronic bowel disorders such as Crohn’s disease and ulcerative colitis (Johnson et al., 2017). Provoking this range of pathogenic effects undoubtedly requires the coordinated expression of a series of C. jejuni genes that are specifically dedicated to virulence. For instance, this pathogen must first survive the handling and cleaning processes such as those involved in the slaughter of chickens and the preparation of meat products (Hermans et al., 2011; Rasschaert et al., 2020); then, after consumption, the pathogen must remain viable in the human gut to initiate the steps of colonization; and subsequently, when established, the visible symptoms of gastrointestinal disease ensue upon production of virulence factors including toxins (Kreling et al., 2020; Elmi et al., 2021). The molecular mechanisms that direct these steps in C. jejuni pathogenesis are presently largely unknown. We address this problem here using an approach that modulates the expression of subsets of C. jejuni genes, enabling the identification of factors that are necessary for its virulence.
The infectious properties of bacteria pathogens are dependent on the expression of proteinaceous virulence factors on their ribosomes. The process of protein expression is fine-tuned by sets of modified nucleotide residues within the RNA components of the ribosome (Purta et al., 2009; Sergiev et al., 2018). We have previously reported that loss of a single methyl group modification on the ribose of nucleotide C1920 of C. jejuni 23S rRNA severely attenuates the pathogen’s virulence (Sałamaszyńska-Guz et al., 2018). Modification of nucleotide C1920 is catalyzed by the methyltransferase Cj0588, and inactivation of this enzyme results in a wide range of defects including reduced ribosome subunit association, biofilm formation, invasion of epithelial cells, and diminished survival of C. jejuni in macrophages (Sałamaszyńska-Guz et al., 2018; Sałamaszyńska-Guz et al., 2020). The C. jejuni Cj0588 enzyme is homologous to the methyltransferase TlyAI found in certain other bacterial species, where it modifies the same rRNA nucleotide and has also been linked with a range of virulence traits (Monshupanee et al., 2012; Monshupanee, 2013; Keith et al., 2021).
Here, a set of C. jejuni strains defective in nucleotide C1920 methylation are used to identify key genes that are specifically linked with virulence. We have identified most of the potential proteins within the C. jejuni proteome using mass spectrometry, and show that significant downregulation of a limited subset of genes coincides with loss of virulence. Downregulated genes include those encoded the cytolethal distending toxin complex (CDT), and genes of the mlaEFD operon that are necessary for maintaining the asymmetry of the outer membrane (OM) and consequently for outer membrane vesicle (OMV) production. Knockouts of the cdtC and mlaEFD genes were constructed and their effects on biofilm formation (required for C. jejuni survival and transmission in food preparation), epithelial cell attachment and invasion (required for gut colonization), and cytokine stimulation and survival in macrophages (required for disease progression) were followed and compared to a collection of TlyA+ and TlyA- strains. Collectively, the data home in on a limited collection of genes linked with human campylobacteriosis.
The C. jejuni strains used in this study (Table 1) were grown under microaerobic conditions (BD GasPak EZ CO2 sachets, Becton Dickinson) at 37°C on brain-heart infusion (BHI) agar containing 5% (v/v) sheep blood, supplemented when necessary with chloramphenicol at 20 µg ml-1.
Overnight cultures of C. jejuni strains were harvested by centrifugation (Sałamaszyńska-Guz et al., 2018) and resuspended in lysis buffer (7 M urea, 2 M thiourea, 1% DTT, and 0.5% n-octyl-β-D-glucopyranoside) supplemented with protease inhibitors (Roche), and were ultrasonicated for protein extraction. After measuring the protein concentrations (Bradford reagent, Merck), 200 µg samples were alkylated with 45 mM iodoacetamide (IAA) incubated for 30 min in the dark before adding 5 mM DTT and adjusting to pH 8.5 with 30 mM triethylammonium bicarbonate (TEAB) buffer. Proteins were predigested for 3 h at 37°C with 0.005 AU lysyl endopeptidase (Lys-C, FUJIFILM Wako Pure Chemical Corporation). Samples were then further digested at 37°C with methylated trypsin at an enzyme:protein ratio of 1:50 (Heissel et al., 2018) prior to quenching with 10% trifluoroacetic acid (TFA) and centrifuging at 20,000 g for 15 min. Peptides were desalted on R2C8 and R3C18 resin reversed-phase columns (Applied Biosystems, POROS 20 R2; Thermo Fisher Scientific), washed with 0.1% TFA and eluted with 60% acetonitrile/0.1% TFA before drying under vacuum centrifugation and resuspending in 50 mM TEAB buffer.
The peptides were labelled with isobaric tags (iTRAQ 8-plex™, AB SCIEX) according to the supplier’s recommendations; equal amounts of the samples were pooled and acidified prior to desalting on reverse-phase columns. Peptides derived from wild-type cells were tagged with iTRAQ-114 and iTRAQ-117; ΔtlyA peptides with iTRAQ-115 and iTRAQ-118; ΔtlyA::tlyA with iTRAQ-116 and iTRAQ-119; and ΔtlyA::tlyAK188A with iTRAQ-117 and iTRAQ-121.
The iTRAQ-labeled peptides were fractionated by reversed-phase chromatography on a Dionex UltiMate 3000 HPLC+ Focused system (Thermo Fisher Scientific) equipped with an ACQUITY UPLC M-class, CSH C18 130 Å, 1.7 µm, 300 µm ×100 mm analytical column (Waters). Mobile phase A comprised 20 mM ammonium formate in water, pH 9.3; phase B was 80% acetonitrile and 20% solvent A, pH 9.3. Peptides were eluted at a flow rate of 5 µL/min over 71 min in a gradient of solvent B rising from 2% to 40%, followed by 50% solvent B to 122 min, before finally washing with 95% solvent B for 10 min. Twenty fractions were collected for each peptide set.
Fractions were separated on a C18 trap column (100 µm ID × 3 cm, 5 µm, Reprosil-Pur 120 C18-AQ, Dr. Maisch GmbH) and a C18 analytical column (75 µm ID × 18 cm, 3 µm, Reprosil-Pur 120 C18-AQ, Dr. Maisch GmbH) at a flow rate of 250 nL/min in a multi-step gradient of 5% BMS buffer (95% acetonitrile, 0.1% formic acid) raised to 10% BMS over 5 min, to 34% BMS from 5 to 125 min, to 50% BMS from 125 to 135 min, and finally washed with 100% BMS. The EASY-nLC was connected online to a Q Exactive HF mass spectrometer (Thermo Fisher Scientific). MS analyses were performed in data-dependent acquisition mode at a resolution of 120,000 (m/z 200) with the mass range of 350-1600 m/z. The target values were set to 3.00E+06 with a maximum injection time of 100 ms. The fifteen most abundant ions were selected from the MS with an isolation width of 1.2 m/z for fragmentation in the HCD collision cell using a collision energy of 34%. Fragmentation was at a resolution of 30,000 (m/z 200) for a target value of 1.00E+05 with a maximum injection time of 120 ms (Thingholm et al., 2010).
Raw data files were processed by Thermo Proteome Discoverer software v 2.4 (Thermo Fisher Scientific). Protein identification was made from the C. jejuni 81-176 genome sequence (Uniprot databases) using the Sequest search engine. For the search parameters: precursor mass tolerance was set to 10 ppm, with a fragment mass tolerance of 0.02 Da; carbamidomethylation of cysteine, iTRAQ modification of N-termini and of lysine residues were added to dynamic modifications, while acetylation was set to variable modifications; two missed cleavage sites per protein were allowed. For reporter ion quantification, the co-isolation threshold was set to 50, and average reporter ion S/N threshold to 10. Data from Proteome Discoverer was processed using Excel (Microsoft).
Quantification was based on three biological replicates of each strain, and taking into consideration only the proteins that were identified from at least two unique peptides and with a false discovery rate of less than 1% (Sun et al., 2020; Orsburn, 2021). The abundances of individual proteins that changed ≥ 1.5-fold with p-values below 0.05 (Cottrell, 2011) were considered as being expressed to a significantly different degree in the comparisons of the wild-type with the ΔtlyA strain, the wild-type with the ΔtlyA::tlyAK188A strain, the ΔtlyA with the ΔtlyA::tlyA strain, and the ΔtlyA::tlyA with the ΔtlyA::tlyAK188A strain.
A single-gene knockout of the C. jejuni 81-176 strain was created by inserting a cat chloramphenicol resistance cassette (Cmr) into the cdtC gene. Briefly, the gene was amplified by PCR using the cdtCR and cdtCF primers (all primers are listed in Table S1) and cloned into the pJET1.2 vector (ThermoFisher Scientific), prior to insertion of the cat cassette using the primers cdtC_mutR and cdtC_mutF, replacing 12 bp of the cdtC gene and disrupting its coding sequence. The plasmids lack a C. jejuni replicon and, after electrotransformation into C. jejuni cells, retention of Cmr thus requires allelic exchange with the homologous chromosomal sequence. The null-mutant C. jejuni 81-176 ΔcdtC (Table 1) was used in subsequent experiments. The C. jejuni 81-176 ΔmlaEFD mutant lacking all three of these genes was constructed in a similar manner using the primers 1637F with 1639R, and then 1637_mutR with 1639_mutF. In each of the knockout strains, cat was inserted into the chromosome the same transcriptional direction as the inactivated gene (tlyA, cdtC or mlaEFD). All constructs were verified by PCR sequencing at each stage of the process.
Assays to assess biofilm formation ability of the C. jejuni cdt and mla strains, as well as their adhesion and invasion of Caco-2 epithelial cells, and the immune responses elicited in T84 epithelial cells were performed as previously described for the tlyA strains (Sałamaszyńska-Guz et al., 2020).
OMV were prepared from by filtering overnight cultures through a 0.2 µm-pore filter and passing the supernatants through 10 kDa centrifugal concentrators (Thermo Fisher Scientific) followed by the ExoBacteria™ OMV Isolation Kit (System Biosciences). The purity of OMV samples was ascertained by transmission electron microscopy (TEM), and their protein content was measured spectrophotometrically based on the Bradford method (Zingl et al., 2020). In the adhesion and invasion assays, Caco-2 cells and the wild-type C. jejuni strain (MOI = 100) were co-incubated with OMVs (10 μg). The level of IL-8 secretion in T84 cells after co-incubation with 100 μg OMVs was assessed using ELISA.
Previously, we reported that loss of TlyA methylation at nucleotide C1920 in 23S rRNA attenuates several virulence traits of C. jejuni (Table S2); in the present study, the proteomes of tlyA-defective C. jejuni strains have been analyzed to identify proteins that are linked with virulence. The genome of C. jejuni 81-176 together with the strain’s two resident plasmids encode 1813 open reading frames (ORFs), and using a mass spectrometry-based quantitative proteomic approach we detected 1482 of the protein products (1424 encoded by the chromosome, 35 by plasmid pVir, and 23 by plasmid pTet), representing 82% of the potential proteome. Applying the criteria described in the methods section, the levels of 50 proteins were found to differ significantly in the tlyA null-mutant (lacking C1920 methyltransferase) compared to the wild-type strain, with 27 proteins upregulated and 23 downregulated (Tables 2 and S3). Amongst the upregulated proteins, eleven chromosomally encoded sequences are involved in the assembly and modification of flagella, six are engaged in amino acid biosynthesis, one (CstA) in carbon starvation, one (DnaJ) as a molecular chaperone, and five are not yet characterized. The three remaining proteins are encoded by pVir. The downregulated proteins included five enzymes associated with lipid turnover, four ribosomal proteins, three proteins involved in oxidative phosphorylation, two members of the acetyltransferase GNAT family, proteins associated with ATP production, protein secretion, transport of lipopolysaccharides and capsular polysaccharides, transferase of ions, iron uptake, one toxin-like protein, and one protein that remains uncharacterized (Tables 2 and S3).
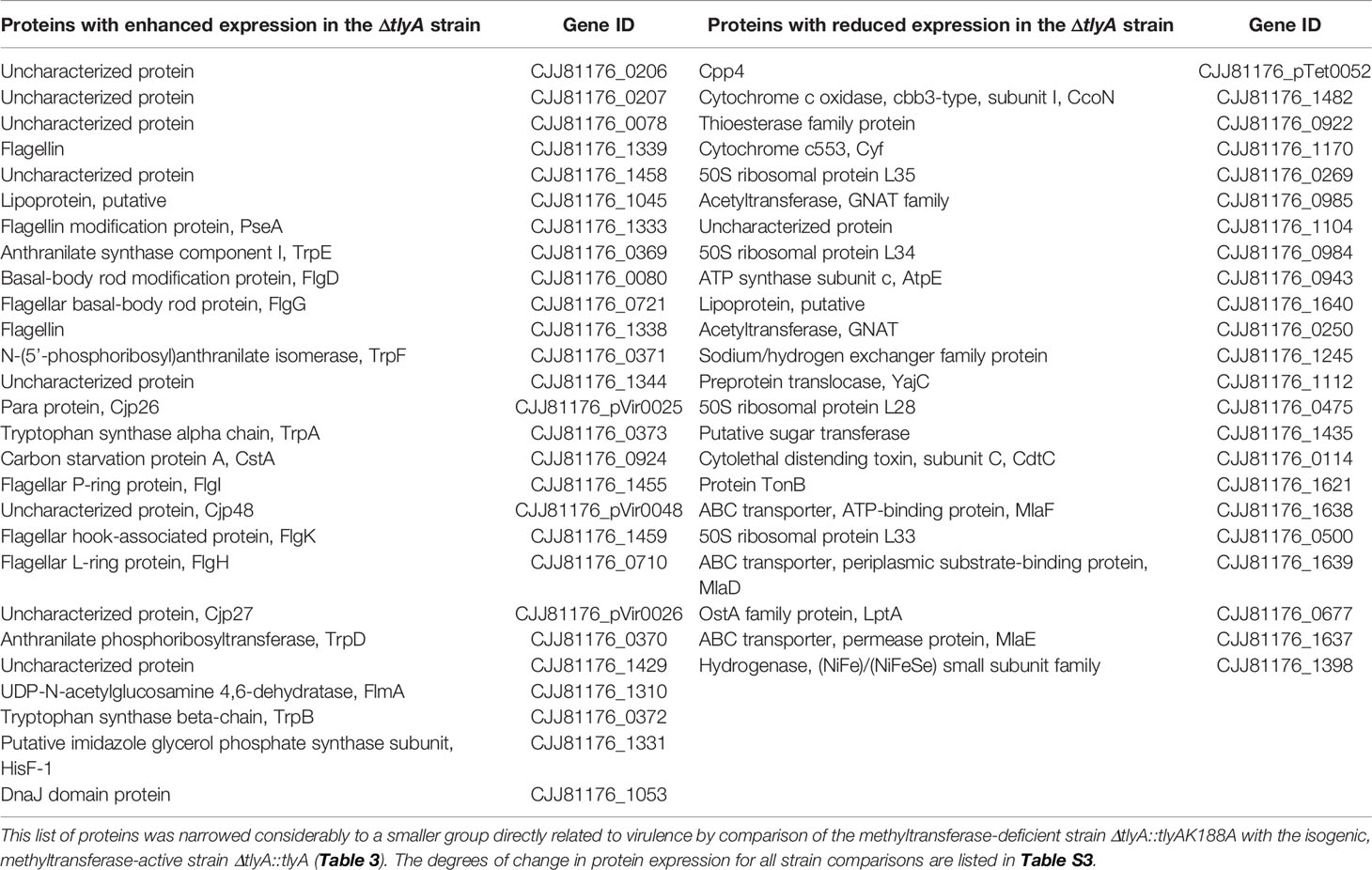
Table 2 Significant changes in the proteome of the C. jejuni ΔtlyA strain compared with the wild-type strain.
Presumably, several of these changes in protein abundance are not be directly related to bacterial virulence, but instead result from other causes such as the introduction of the chloramphenicol resistance cassette into the tlyA gene. The proteome analyses were therefore repeated to include two isogenic strains, the first of which was the tlyA null-mutant complemented with an active copy of tlyA (which restores virulence), and the second was the null-mutant complemented with a methyltransferase-deficient copy of tlyA [the K188A variant, which does not restore virulence (Sałamaszyńska-Guz et al., 2018)]. Comparison of the proteomes of these two strains, which are genetically identical except for the change at the tlyA-188 allele, showed the number of significant changes was limited to eight proteins (Table S3). For the purpose of further analysis, this list was narrowed down to the four proteins that were consistently downregulated in all the different combinations of TlyA+ versus TlyA- strain comparisons (Table 3).
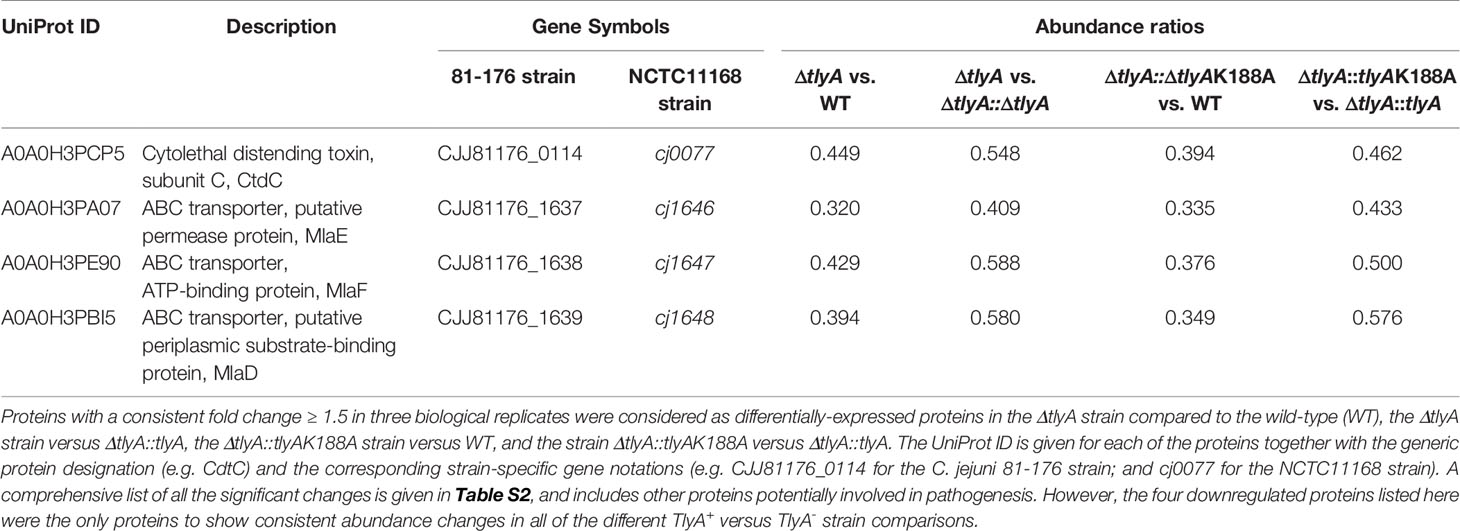
Table 3 Significant differences in the proteome of C. jejuni in the various TlyA+ versus TlyA- strain comparisons.
One of these proteins, CdtC, interacts with the subunits CdtA and CdtB to form the cytolethal distending toxin (CDT). In C. jejuni strains lacking the TlyA modification of the rRNA, the abundance of CdtC protein was reduced to 39% and CdtB to 71% of the wild-type levels (outside the cut-off applied here), while no change was seen in the level of CdtA (Figure 1). Complementation with the active copy of tlyA restored abundance of CdtC and CdtB proteins to 77% and 89% of wild-type levels, respectively. No rescue occurred with the methyltransferase-defective K188A version of TlyA.
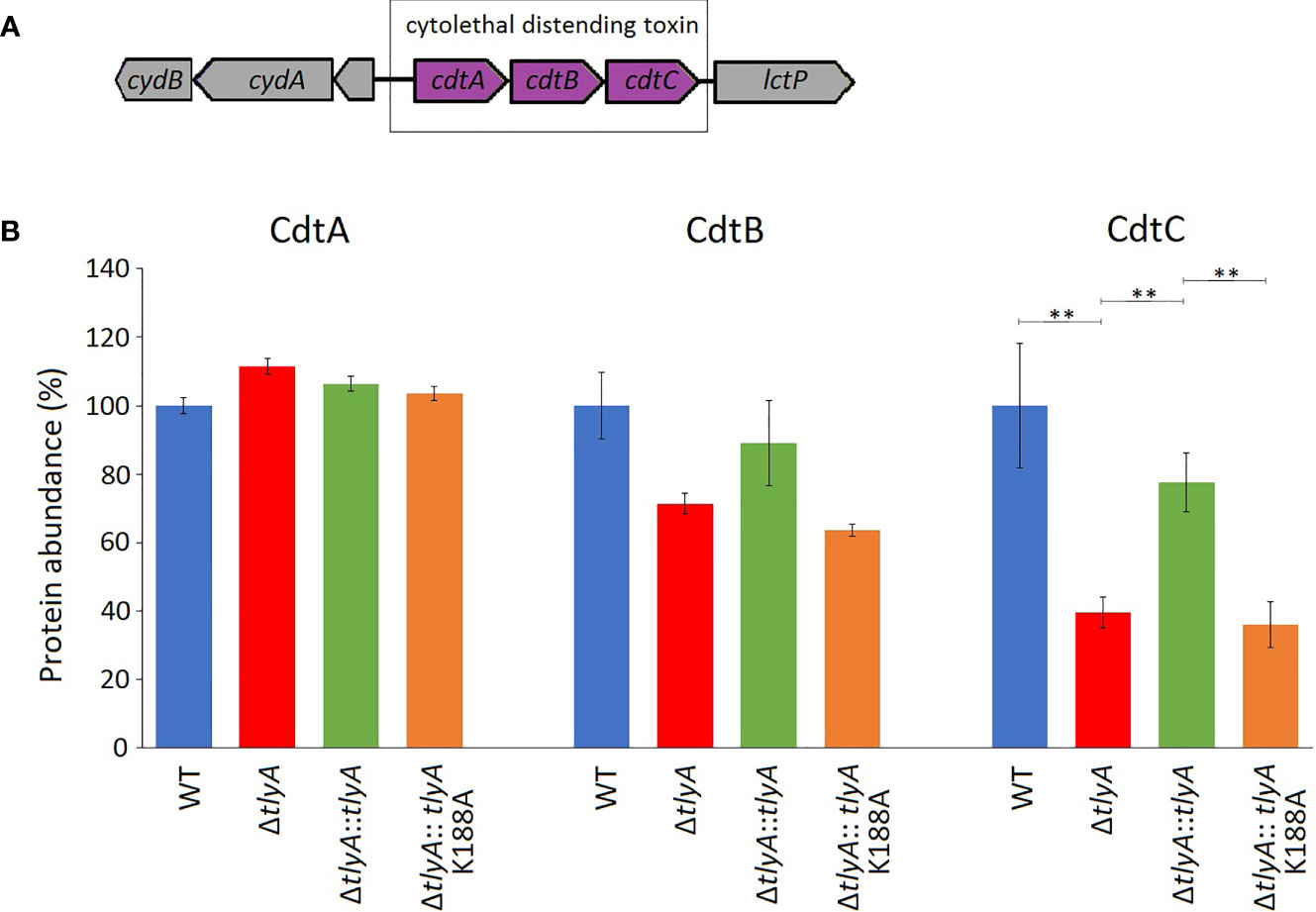
Figure 1 Changes in the abundance of CdtABC proteins upon inactivation of the tlyA gene. (A) Organization of the cdtABC gene cluster in C. jejuni. The strain-specific notations for these genes are given in . (B) Abundances of the individual Cdt proteins in the wild-type (WT) C. jejuni strain, the tlyA knockout strain (ΔtlyA), and the knockout strain complemented with active (ΔtlyA::tlyA) and inactive (ΔtlyA::tlyA K188A) copies of the tlyA gene. The protein amounts in the wild-type strain have been normalized here to 100%. The abundance of CdtC is dependent on a functional tlyA gene; CdtB is also reduced (but outside the significance cut-off applied here); and the upstream encoded protein CdtA remained unchanged. The error bars represent the standard deviations of three biological replicates; **P < 0.001 for these three strain comparisons.
The other three downregulated proteins are encoded by the mla gene cluster (Figure 2), and are homologs of MlaE, MlaF and MlaD that comprise an E. coli ABC-transporter with which they show 28, 32, and 19% respective similarity in their amino acid sequences. In the C. jejuni ΔtlyA strain, the abundances of MlaE, MlaF and MlaD were respectively reduced to 29, 36, and 41% of the wild-type levels. Expression of CJJ81176_1640 (Table 2, and hereafter referred to as cj1640), the next gene downstream, was also lowered by tlyA inactivation, albeit to a lesser degree than mlaEFD; however, expression of tkt and cj1641 immediately upstream and downstream of these genes remained unaffected (Figure 2). The abundances of MlaE, MlaF, MlaD and CJ1640 were partially restored to around two-thirds of the wild-type levels upon complementation with an active copy of tlyA (Figure 2); again, no rescue occurred with the methyltransferase-defective K188A version of this gene.
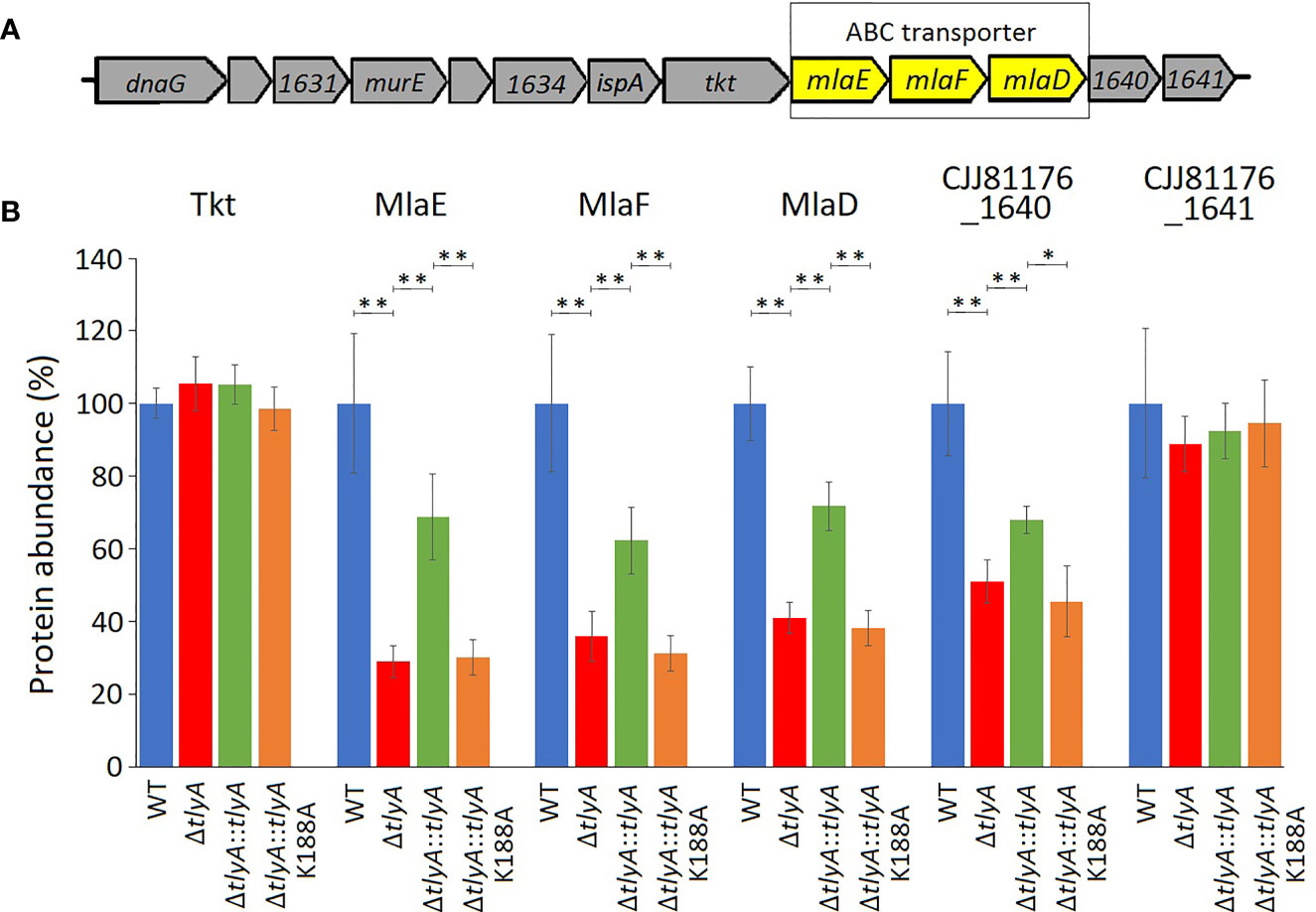
Figure 2 Changes in abundances in the proteins encoded within and adjacent to the C. jejuni mlaEFD operon upon inactivation of tlyA (color-coded as in Figure 1). (A) Organization of the C. jejuni mlaEFD gene cluster. The strain-specific notations for these genes are given in Table 3. (B) The abundance of each of the MlaE, MlaF and MlaD proteins is dependent on a functional C. jejuni tlyA gene **P < 0.001 (strain notation as in Figure 1). The abundances of the upstream-encoded protein Tkt remained unchanged. However, expression of the downstream-encoded protein CJJ81176_1640 (Table 2, and hereafter referred to as CJ1640) was consistently reduced, *P < 0.05, (but outside our cut-off criteria), while its CJ1641 neighbor was unchanged. These three additional proteins are shown here for comparison. The error bars represent standard deviations from three biological replicates.
The lower amounts of the CdtC and MlaEFD proteins correlate with reduced ability of the C. jejuni TlyA-minus strains to adhere, invade and induce IL-8 secretion in host cells (Table S2). These observations were tested further by inactivating cdtC and the mlaEFD gene cluster and reassessing pathogenicity traits. Loss of either CdtC or the MlaEFD proteins decreased bacterial adherence to Caco-2 host cells to the same degree (around 78% of the wild-type strain, Figure 3A). A different picture emerged regarding the invasive properties of the strains, where loss of cdtC elicited a more marked effect that inactivation of the mlaEFD gene cluster (Figure 3B). It should be noted that neither of these deletion mutants adversely affected adherence and invasion to the same extent as the tlyA deletion (Figures 3A, B).
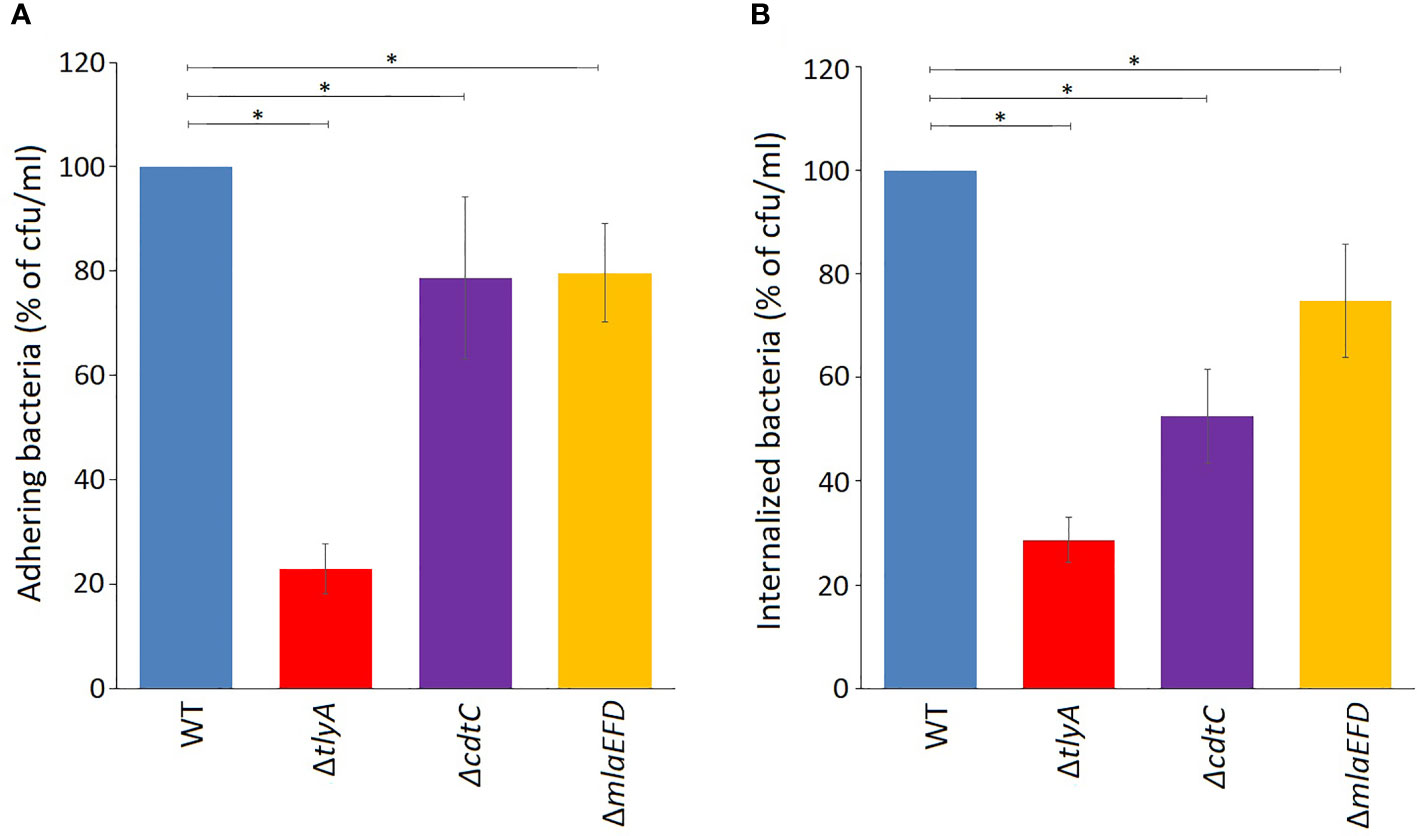
Figure 3 Attenuation of virulence properties in the C. jejuni mutant strains. (A) Adhesion onto Caco-2 epithelial cells by C. jejuni was significantly reduced by the mutations; *P < 0.05 for ΔtlyA, ΔcdtC and ΔmlaEFD versus the wild-type (WT). (B) Invasion of C. jejuni into Caco-2 epithelial cells was also significantly reduced by each of these mutations; *P < 0.05 for ΔtlyA, ΔcdtC and ΔmlaEFD versus the wild-type (WT). Values represent means ± S.E.M. of three independent experiments.
Other parameters used to evaluate the health of C. jejuni strains, such as growth rates and the ability to form biofilms, indicated that these criteria were not adversely affected by deletion of the cdtC gene (not shown). However, we saw indications that loss of the mlaEFD operon has a potential health cost. While the growth of static cultures and the formation of biofilms by the mlaEFD strain were not adversely affected, agitation of cultures during growth lowered the viable cell count by approximately 30% compared to that of the wild-type. In addition, loss of mlaEFD increased the susceptibility of C. jejuni to polymyxin antibiotics, whereas no discernible effect was seen upon tlyA inactivation (Table S4). Neither of the mutations changed vancomycin susceptibility.
The outer membrane vesicles (OMVs) of C. jejuni play an important role in its ability to adhere to and invade epithelial cells (Elmi et al., 2012). These pathogenic properties of C. jejuni are enhanced by co-incubation with an extra portion of isolated OMVs (Figures 4A, B). Addition of OMVs from the tlyA, cdtC and mlaEFD mutants also improved the adhesion of the C. jejuni wild-type strain to Caco-2 epithelial cells (Figure 4A). Bacterial internalization into epithelial cells was also improved by addition of OMVs from the wild-type and mlaEFD strains (Figure 4B). However, coincubation with extra OMVs derived from the tlyA and the cdtC mutants did not improve internalization of C. jejuni.
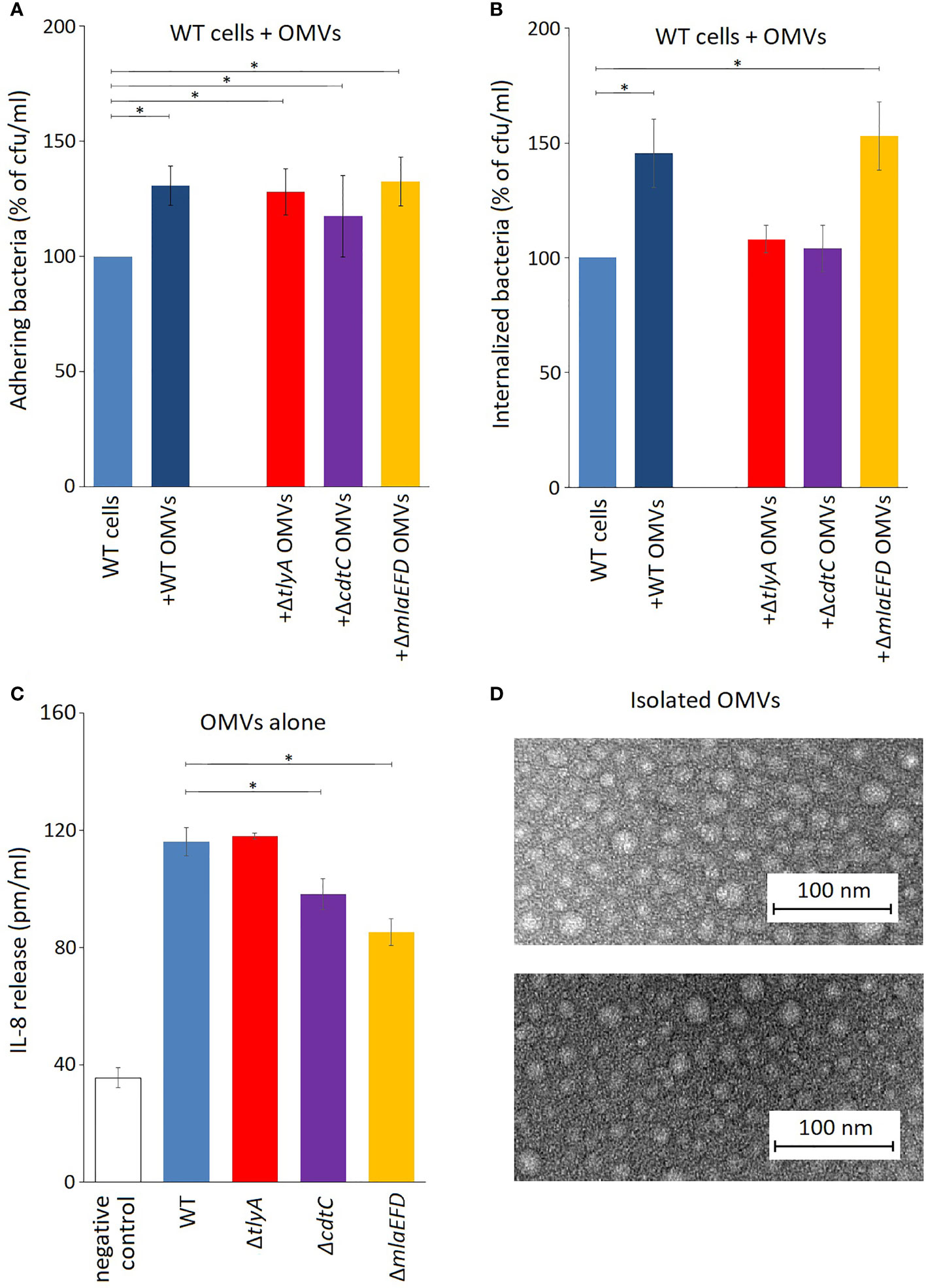
Figure 4 OMVs produced by C. jejuni strains. (A) Influence of C. jejuni OMVs on adhesion to epithelial cells. Purified OMVs (10 μg) from the various strains were added to viable wild-type C. jejuni and incubated for 2 h with Caco-2 cells. Addition of OMVs isolated from wild-type and mutant strains significantly increased adhesion of C. jejuni to Caco-2 cells (*P < 0.05). (B) OMVs from the wild-type and ΔmlaEFD strains (added and incubated as in panel A) significantly improved internalization of wild-type C. jejuni into Caco-2 epithelial cells (*P < 0.05). No significant change in C. jejuni internalization was observed with OMVs from the ΔtlyA or ΔcdtC strains. (C) Secretion of interleukin IL-8 by T84 cells during incubation with 100 μg OMVs from the various strains, in the absence of viable C. jejuni cells. Significantly less IL-8 was produced upon addition of OMVs from the ΔcdtC and ΔmlaEFD strains compared to wild-type OMVs (*P < 0.05); however, no significant difference between wild-type and ΔtlyA OMVs was observed. (D) Representative images of purified OMVs from the C. jejuni wild-type strain (upper panel) and ΔmlaEFD (lower panel) visualized by transmission electron microscopy. No visible differences were observed between OMVs that were isolated from wild-type strain and mutants. The OMV populations, purified by the resin-binding procedure used here, had a size distribution of between 10 to 30 nm in diameter.
Production of interleukin-8 (IL-8) by T84 epithelial cells was assayed using ELISA to measure the induction of an innate immune response. Addition of OMVs from all the C. jejuni strains elicited release of IL-8, although the stimulatory effect was significantly lower with OMVs from the cdtC the mlaEFD mutant strains (Figure 4C).
Loss of some rRNA modifications have been reported to cause a series of pleiotropic effects, presumably due to changes in protein expression, and can result in reduced virulence in pathogenic bacteria (Sergiev et al., 2018). One example of such effects is brought about by inactivation of the methyltransferase TlyAI, which 2´-O-methylates nucleotide C1920 in 23S rRNA (Monshupanee et al., 2012). Loss of this modification in C. jejuni rRNA through inactivation of its TlyAI homolog Cj0588 results in wide-ranging defects in ribosome subunit association, cell motility, biofilm formation, adherence to and invasion of human epithelial cells, and survival within macrophages (Sałamaszyńska-Guz et al., 2018; Sałamaszyńska-Guz et al., 2020). The defective ribosomal subunit association is readily explained by loss of rRNA methylation at the site of subunit interaction (Johansen et al., 2006), although the mechanisms behind the extensive reduction in virulence traits (Table S2) are more complex. We previously proposed that the loss of the ribosome modification interfered with in protein expression (in manner that remained to be defined), and this in turn resulted in attenuation of bacterial virulence (Sałamaszyńska-Guz et al., 2018).
The present study addresses this idea using a mass spectrometric approach to determine what changes occur in the C. jejuni proteome after loss of the Cj0588-catalyzed ribosome modification. Using the stringency criteria described above, the abundances of fifty proteins were significantly different in the tlyA mutant compared with the wild-type strain (Tables 2 and S3). The roles of many of these proteins (including, for example, those involved in the assembly of flagella, the formation lipids, lipopolysaccharides and membranes, and the secretion of toxins) could readily be imagined as being linked to pathogenesis. On the other hand, some proteins (such as enzymes for amino acid biosynthesis and oxidative phosphorylation) are probably only indirectly linked with pathogenesis. Some of the remaining proteins undoubtedly have no connection with the loss of C. jejuni virulence, but rather result from the process used to disrupt the tlyA gene or (in the case of the ribosomal proteins L28, L33, L34 and L35) as a consequence of effects on 50S assembly (Röhl and Nierhaus, 1982) or its association with the 30S ribosomal subunit (Sałamaszyńska-Guz et al., 2018).
The proteome analyses were repeated to include two isogenic strains, the first of which was the tlyA null-mutant complemented with an active copy of tlyA (which restores the virulence phenotype), and the second was the null-mutant complemented with a methyltransferase-deficient copy of tlyA (the K188A variant, which does not restore virulence). Comparison of the proteomes of these two strains, which are genetically identical except for the change at the tlyA-188 allele, limited the changes to only a few proteins. Each of these proteins was downregulated in the K188A variant, and differed to approximately the same degree as in the comparison of ΔtlyA versus wild-type (Table S3). An array of proteins involved in flagellum construction (including FlgD, FlgG, FlgH, FlgI and FlgK) and the structural flagellins were significantly upregulated after inactivation of tlyA (Table S3). All these proteins were, however, expressed at this same higher level in the tlyA strains after complementation with active and inactive copies of this gene, indicating that flagellar protein overexpression was not linked to rRNA methylation but rather to other factors, such as the intrusion of the resistance cassette at the tlyA locus. Other candidates that were downregulated to a slightly lesser degree in the K188A variant, including ribosomal proteins such as L28, and were dropped in the final analyses for reasons discussed above.
A limited number of candidates were chosen for further genetic analyses to test whether our approach of altering rRNA methylation is in fact a reliable means of identifying bacterial virulence factors. Here, we included only proteins that were consistently significantly different in each of the TlyA+ versus TlyA- strain comparisons (Table S3). Thus, four proteins made the final cut: CdtC, MlaE, MlaF and MlaD, all of which were downregulated (Table 3). The CdtC protein together with CdtA and CdtB constitute the cytolethal distending toxin (CDT), the components of which are encoded by three contiguous genes cdtA, cdtB, cdtC (Pickett et al., 1996) within a single operon (Figure 1A). The CdtB component, whose abundance is also reduced in the tlyA mutants (albeit to a lesser degree), appears to be the cytotoxic component of CDT acting through type I deoxyribonuclease (DNase I) activity to cause DNA breakage at the G2/M phase in epithelial host cells, while the CdtA and CdtC proteins serve as carriers for delivering CdtB subunit across the host cell membrane (Lai et al., 2016). The lower amounts of CdtC and CdtB in the tlyA mutant (Figure 1B) correlate with reduced ability of C. jejuni to attach and invade host cells (Figure 3), and is consistent with previous reports of CDT-production by Campylobacter strains stimulating adherence to and invasion of epithelial cells (Biswas et al., 2006; Ghorbanalizadgan et al., 2018).
After inactivation of the cdtC gene, we observed reduced adherence (Figure 3A) and host cell invasion by C. jejuni (Figure 3B), but to a lesser extent than for the tlyA mutant. While confirming the involvement of CDT in these processes, this at the same time emphasizes that other factors are involved. CdtC and the other CDT components have been detected in the C. jejuni cytoplasmic and the periplasmic space, although they are mainly associated with its OMVs (Lindmark et al., 2009; Elmi et al., 2012), suggesting that CDT localization within the bacterium is of importance for toxin delivery. OMVs play key roles in bacterial physiology and pathogenesis, ranging from secretion and delivery of biomolecules (including toxins, DNA, and quorum sensing molecules), stress response, biofilm formation, adherence to host cells, and immunomodulation (Roier et al., 2016). Consistent with this, the adhesion and invasion properties of C. jejuni can be enhanced by adding extra purified OMVs to the epithelial cell incubation mix (Elmi et al., 2012; Taheri et al., 2018). The addition of purified OMVs to our C. jejuni/Caco-2 cell incubation mixtures showed that while CdtC is important for host attachment (Figure 4A), it appears to play a greater role in host invasion (Figure 4B).
Prior to epithelial cell invasion, C. jejuni elicits an immune response and, in mice, the pathogen is quickly eliminated if its CDT activity is lacking (Fox et al., 2004). Here, we tested our C. jejuni mutants and their OMVs in an in vitro system using T84 epithelial cells assaying for induction of IL-8 release as a proxy for an innate immune response (Hickey et al., 1999; Hickey et al., 2000). The absence of CdtC in purified OMVs reduced their ability to induce IL-8 production (Figure 4C), from which we would infer that without CdtC the entry of the main actor, the CdtB toxin, is not facilitated. OMVs from the tlyA mutant (in which CdtC is present, albeit in reduced amounts) were as effective as wild-type OMVs at stimulating IL-8 release, suggesting that the smaller amount of CdtC facilitates entry of sufficient CDT to provoke the immune response. Interestingly, the lowest IL-8 immune response was seen in the case of the OMVs from cells in which the mla operon had been inactivated.
The mla operon encodes three proteins: a permease, MlaE; an ATP-binding protein, MlaF; and periplasmic substrate-binding, MlaD (Roier et al., 2016). The Mla (maintenance of lipid asymmetry) proteins were initially proposed to traffic phospholipids from the inner membrane (IM) to the outer membrane (OM) and are involved in formation of OMVs for the transport of lipids, cholesterol and other steroids (Ekiert et al., 2017). However, more recent work on Mla in Acinetobacter baumannii supports an alternative model for the directionality of the Mla system being exclusively retrograde (i.e. from the OM to IM) (Powers and Trent, 2019; Powers et al., 2020). It has been proposed that downregulation or mutation of the mla genes can lead to accumulation of phospholipids in the outer leaflet of the OM, which could promote OMV formation (Roier et al., 2016). This fits with the increase in OMV production observed here in the ΔmlaEFD strain (Table S5), which has also been reported upon mlaA mutation (Davies et al., 2019). Despite these effects, Mla proteins are apparently absent within C. jejuni OMVs (Elmi et al., 2012).
Inactivation of tlyA leads to a decrease in the abundance of all three MlaE, MlaF and MlaD proteins (Figure 2), indicating that their expression was being coordinately downregulated. Downstream of the mla operon, expression of cj1640 was also reduced, suggesting its expression is linked to that of mlaEFD. Although the role of protein CJ1640 has not been determined, it shows similarity to proteins linked with cholesterol transport, and bears a vague resemblance to MlaB, a protein that has been reported to be absent in Helicobacter pylori (Roier et al., 2016), which has recently been reassigned with C. jejuni to the class of Campylobacteria (Waite et al., 2017; Parks et al., 2018). Reduced expression in this chromosome region is limited to the genes extending from mlaE to cj1640: expression of tkt (situated immediately upstream) and cj1641 (immediately downstream) remained unaffected (Figure 2). Upon inactivation of the entire mla operon we observed a minor decrease in C. jejuni adherence and invasion of host cells (Figure 3) although, unexpectedly, OMVs from the Δmla strain promoted these abilities in wild-type C. jejuni cells (Figure 4). Sensitivity to polymyxins was increased in the Δmla strain (Table S4), suggesting a weakening of the OM structure that was not apparent in the ΔtlyA strains where the Mla proteins were still present, albeit in reduced amounts (Table 3). Tolerance to vancomycin, an inhibitor of cross-linking of the peptidoglycan matrix, remained unaffected by loss of mlaEFD (Table S4). This would point to the role of the Mla proteins being confined to the OM, and fits with the reduced ability of Δmla OMVs to stimulate IL-8 production (Figure 4C). Counterparts of the MlaEFD proteins are present in Haemophilus influenzae (Davies et al., 2019), and have been shown to possess similar properties, with their loss causing attenuation of virulence (Fernández-Calvet et al., 2018).
The ability of C. jejuni cells to form biofilms and to adhere to abiotic surfaces in food-processing systems, and later to biotic surfaces, is essential for their transmission and ability to cause disease (Walan and Kihlström, 1988; Nguyen et al., 2011; Burnham and Hendrixson, 2018). Biofilm formation was extensively disrupted in the ΔtlyA strain (Sałamaszyńska-Guz et al., 2018), and the lack of such an effect with the ΔmlaEFD or ΔcdtC strains indicates that other cellular components are playing the prominent role in this process. One such candidate would be LptA, found in the periplasm of many Gram-negative bacteria where it transports lipopolysaccharides (LPS) and lower molecular weight lipooligosaccharides (LOS) for OM biogenesis (Ruiz et al., 2009). Depletion of LptA after tlyA inactivation (Table S2) would thus reduce the strain’s ability to maintain the asymmetry of the outer leaflet of the OM, altering cell surface hydrophobicity and attachment to host cells (Walan and Kihlström, 1988; Nguyen et al., 2011). Interrupting the delivery of LOS to the OM reduces Campylobacter invasion of intestinal epithelial cells in vitro (Kanipes et al., 2008) and in vivo in mice (Naito et al., 2010) and chickens (Iwata et al., 2013). The observations fit with studies on Acinetobacter baumannii where a deficiency of LOS was shown to reduce both growth and virulence (May and Grabowicz, 2018).
In conclusion, comparison of the tlyA knockout with the wild-type strain showed about fifty significant changes in protein abundance, over half of which were upregulated (Table S2). More stringent criteria for judging the effects of loss of TlyA methylation were applied by comparing the isogenic strains ΔtlyA::tlyAK188A and ΔtlyA::tlyA, and indicated that key changes causing loss of virulence are linked to fewer proteins, all of which are downregulated. The genes encoding four of these proteins (CdtC, MlaE, MlaF and MlaD), which were consistently downregulated in all TlyA+ and TlyA- strain comparisons (Table 3), were themselves inactivated to test the principle of using an rRNA methyltransferase deficient strain as a means of identifying factors specifically required for virulence. We infer from the cdtC strain that CdtC plays a role in the host cell invasion process (Figure 3B). Inactivation of the mlaEFD operon confirmed the involvement of the Mla proteins in maintaining the integrity of the OM and reducing cell fragility under agitated culturing conditions, and these proteins also play a role in prompting the IL-8 innate immune response (Figure 4C).
We emphasize that these proteins (Table 3) by no means constitute a comprehensive list of factors involved in C. jejuni pathogenesis. Other cell components, including for example CdtB and LptA (Tables 2 and S3) that were downregulated to a slightly lesser degree in TlyA- strains, also would play important roles. However, we believe that we can conclude from these initial proteomics analyses that attenuating virulence by modulating ribosome function is a useful tool for defining key virulence factors in C. jejuni, and is an approach which might be applied to other bacterial pathogens.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: http://www.proteomexchange.org/, PXD030125.
AS-G designed the experiments. AS-G, PKR, and MM performed the experiments. AS-G data analysis. SD and AS-G data interpretation. SD and AS-G: writing, review and editing. All authors contributed to manuscript revision, read, and approved the submitted version.
Support from the National Science Centre [2018/30/M/NZ6/00429] to AS-G, and from the Danish Research Agency (FNU-rammebevilling 10-084554) to SD is gratefully acknowledged.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
We thank Martin Røssel Larsen, Arkadiusz Nawrocki and Lene Jakobsen, University of Southern Denmark for their essential advice on the proteomics experiments. The mass spectrometry proteomics data have been deposited at the ProteomeXchange Consortium via the PRIDE (Perez-Riverol et al., 2019) partner repository with the dataset identifier PXD030125.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcimb.2021.803730/full#supplementary-material
Ang, C. W., Laman, J. D., Willison, H. J., Wagner, E. R., Endtz, H. P., De Klerk, M. A., et al. (2002). Structure of Campylobacter jejuni Lipopolysaccharides Determines Antiganglioside Specificity and Clinical Features of Guillain-Barre and Miller Fisher Patients. Infect. Immun. 70, 1202–1208. doi: 10.1128/IAI.70.3.1202-1208.2002
Biswas, D., Fernando, U., Reiman, C., Willson, P., Potter, A., Allan, B. (2006). Effect of Cytolethal Distending Toxin of Campylobacter jejuni on Adhesion and Internalization in Cultured Cells and in Colonization of the Chicken Gut. Avian Dis. 50, 586–593. doi: 10.1637/7514-020706R1.1
Burnham, P. M., Hendrixson, D. R. (2018). Campylobacter jejuni: Collective Components Promoting a Successful Enteric Lifestyle. Nat. Rev. Microbiol. 16, 551–565. doi: 10.1038/s41579-018-0037-9
Cottrell, J. S. (2011). Protein Identification Using MS/MS Data. J. Proteomics 74, 1842–1851. doi: 10.1016/j.jprot.2011.05.014
Davies, C., Taylor, A. J., Elmi, A., Winter, J., Liaw, J., Grabowska, A. D., et al. (2019). Sodium Taurocholate Stimulates Campylobacter jejuni Outer Membrane Vesicle Production via Downregulation of the Maintenance of Lipid Asymmetry Pathway. Front. Cell. Infect. Microbiol. 9, 177. doi: 10.3389/fcimb.2019.00177
Ekiert, D. C., Bhabha, G., Isom, G. L., Greenan, G., Ovchinnikov, S., Henderson, I. R., et al. (2017). Architectures of Lipid Transport Systems for the Bacterial Outer Membrane. Cell 169, 273–285.e217. doi: 10.1016/j.cell.2017.03.019
Elmi, A., Nasher, F., Dorrell, N., Wren, B., Gundogdu, O. (2021). Revisiting Campylobacter jejuni Virulence and Fitness Factors: Role in Sensing, Adapting, and Competing. Front. Cell. Infect. Microbiol. 10, 607704. doi: 10.3389/fcimb.2020.607704
Elmi, A., Watson, E., Sandu, P., Gundogdu, O., Mills, D. C., Inglis, N. F., et al. (2012). Campylobacter jejuni Outer Membrane Vesicles Play an Important Role in Bacterial Interactions With Human Intestinal Epithelial Cells. Infect. Immun. 80, 4089–4098. doi: 10.1128/IAI.00161-12
Fernández-Calvet, A., Rodriguez-Arce, I., Almagro, G., Moleres, J., Euba, B., Caballero, L., et al. (2018). Modulation of Haemophilus influenzae Interaction With Hydrophobic Molecules by the VacJ/MlaA Lipoprotein Impacts Strongly on Its Interplay With the Airways. Sci. Rep. 8, 6872. doi: 10.1038/s41598-018-25232-y
Fox, J. G., Rogers, A. B., Whary, M. T., Ge, Z., Taylor, N. S., Xu, S., et al. (2004). Gastroenteritis in NF-kappaB-Deficient Mice Is Produced With Wild-Type Camplyobacter jejuni But Not With C. jejuni Lacking Cytolethal Distending Toxin Despite Persistent Colonization With Both Strains. Infect. Immun. 72, 1116–1125. doi: 10.1128/IAI.72.2.1116-1125.2004
Ghorbanalizadgan, M., Bakhshi, B., Najar-Peerayeh, S. (2018). Heterogeneity of Cytolethal Distending Toxin Sequence Types of Campylobacter jejuni and Correlation to Invasion/Cytotoxicity Potential: The First Molecular Survey From Iran. Microb. Pathog. 114, 213–218. doi: 10.1016/j.micpath.2017.11.035
Heissel, S., Bunkenborg, J., Kristiansen, M. P., Holmbjerg, A. F., Grimstrup, M., Mortz, E., et al. (2018). Evaluation of Spectral Libraries and Sample Preparation for DIA-LC-MS Analysis of Host Cell Proteins: A Case Study of a Bacterially Expressed Recombinant Biopharmaceutical Protein. Protein Expr. Purif. 147, 69–77. doi: 10.1016/j.pep.2018.03.002
Hermans, D., Van Deun, K., Martel, A., Van Immerseel, F., Messens, W., Heyndrickx, M., et al. (2011). Colonization Factors of Campylobacter jejuni in the Chicken Gut. Vet. Res. 42, 82. doi: 10.1186/1297-9716-42-82
Hickey, T. E., Baqar, S., Bourgeois, A. L., Ewing, C. P., Guerry, P. (1999). Campylobacter jejuni-Stimulated Secretion of Interleukin-8 by INT407 Cells. Infect. Immun. 67, 88–93. doi: 10.1128/IAI.67.1.88-93.1999
Hickey, T. E., Mcveigh, A. L., Scott, D. A., Michielutti, R. E., Bixby, A., Carroll, S. A., et al. (2000). Campylobacter jejuni Cytolethal Distending Toxin Mediates Release of Interleukin-8 From Intestinal Epithelial Cells. Infect. Immun. 68, 6535–6541. doi: 10.1128/IAI.68.12.6535-6541.2000
Iwata, T., Chiku, K., Amano, K., Kusumoto, M., Ohnishi-Kameyama, M., Ono, H., et al. (2013). Effects of Lipooligosaccharide Inner Core Truncation on Bile Resistance and Chick Colonization by Campylobacter jejuni. PloS One 8, e56900. doi: 10.1371/journal.pone.0056900
Johansen, S. K., Maus, C. E., Plikaytis, B. B., Douthwaite, S. (2006). Capreomycin Binds Across the Ribosomal Subunit Interface Using tlyA-Encoded 2′-O-Methylations in 16S and 23S rRNAs. Mol. Cell 23, 173–182. doi: 10.1016/j.molcel.2006.05.044
Johnson, T. J., Shank, J. M., Johnson, J. G. (2017). Current and Potential Treatments for Reducing Campylobacter Colonization in Animal Hosts and Disease in Humans. Front. Microbiol. 8, 487. doi: 10.3389/fmicb.2017.00487
Kanipes, M. I., Tan, X., Akelaitis, A., Li, J., Rockabrand, D., Guerry, P., et al. (2008). Genetic Analysis of Lipooligosaccharide Core Biosynthesis in Campylobacter jejuni 81-176. J. Bacteriol. 190, 1568–1574. doi: 10.1128/JB.01696-07
Keith, B. A., Harding, J. C. S., Loewen, M. E. (2022). Mutational Analysis of TlyA From Brachyspira Hampsonii Reveals Two Key Residues Conserved in Pathogenic Bacteria Responsible for Oligomerization and Hemolytic Activity. Biochim. Biophys. Acta Gen. Subj. 1866, 130045. doi: 10.1016/j.bbagen.2021.130045
Korlath, J. A., Osterholm, M. T., Judy, L. A., Forfang, J. C., Robinson, R. A. (1985). A Point-Source Outbreak of Campylobacteriosis Associated With Consumption of Raw Milk. J. Infect. Dis. 152, 592–596.
Kreling, V., Falcone, F. H., Kehrenberg, C., Hensel, A. (2020). Campylobacter Sp.: Pathogenicity Factors and Prevention Methods-New Molecular Targets for Innovative Antivirulence Drugs? Appl. Microbiol. Biotechnol. 104, 10409–10436. doi: 10.1007/s00253-020-10974-5
Lai, C. K., Chen, Y. A., Lin, C. J., Lin, H. J., Kao, M. C., Huang, M. Z., et al. (2016). Molecular Mechanisms and Potential Clinical Applications of Campylobacter jejuni Cytolethal Distending Toxin. Front. Cell. Infect. Microbiol. 6, 9. doi: 10.3389/fcimb.2016.00009
Lindmark, B., Rompikuntal, P. K., Vaitkevicius, K., Song, T., Mizunoe, Y., Uhlin, B. E., et al. (2009). Outer Membrane Vesicle-Mediated Release of Cytolethal Distending Toxin (CDT) From Campylobacter jejuni. BMC Microbiol. 9, 220. doi: 10.1186/1471-2180-9-220
May, K. L., Grabowicz, M. (2018). The Bacterial Outer Membrane Is an Evolving Antibiotic Barrier. Proc. Natl. Acad. Sci. U. S. A. 115, 8852–8854. doi: 10.1073/pnas.1812779115
Monshupanee, T. (2013). Increased Bacterial Hemolytic Activity Is Conferred by Expression of TlyA Methyltransferase But Not by Its 2′-O-Methylation of the Ribosome. Curr. Microbiol. 67, 61–68. doi: 10.1007/s00284-013-0332-7
Monshupanee, T., Johansen, S. K., Dahlberg, A. E., Douthwaite, S. (2012). Capreomycin Susceptibility Is Increased by TlyA-Directed 2′-O-Methylation on Both Ribosomal Subunits. Mol. Microbiol. 85, 1194–1203. doi: 10.1111/j.1365-2958.2012.08168.x
Naito, M., Frirdich, E., Fields, J. A., Pryjma, M., Li, J., Cameron, A., et al. (2010). Effects of Sequential Campylobacter jejuni 81-176 Lipooligosaccharide Core Truncations on Biofilm Formation, Stress Survival, and Pathogenesis. J. Bacteriol. 192, 2182–2192. doi: 10.1128/JB.01222-09
Nguyen, V. T., Turner, M. S., Dykes, G. A. (2011). Influence of Cell Surface Hydrophobicity on Attachment of Campylobacter to Abiotic Surfaces. Food Microbiol. 28, 942–950. doi: 10.1016/j.fm.2011.01.004
Orsburn, B. C. (2021). Proteome Discoverer-A Community Enhanced Data Processing Suite for Protein Informatics. Proteomes 9, 15. doi: 10.3390/proteomes9010015
Parks, D. H., Chuvochina, M., Waite, D. W., Rinke, C., Skarshewski, A., Chaumeil, P. A., et al. (2018). A Standardized Bacterial Taxonomy Based on Genome Phylogeny Substantially Revises the Tree of Life. Nat. Biotechnol. 36, 996–1004. doi: 10.1038/nbt.4229
Perez-Riverol, Y., Csordas, A., Bai, J., Bernal-Llinares, M., Hewapathirana, S., Kundu, D. J., et al. (2019). The PRIDE Database and Related Tools and Resources in 2019: Improving Support for Quantification Data. Nucleic Acids Res. 47, D442–D450. doi: 10.1093/nar/gky1106
Pickett, C. L., Pesci, E. C., Cottle, D. L., Russell, G., Erdem, A. N., Zeytin, H. (1996). Prevalence of Cytolethal Distending Toxin Production in Campylobacter jejuni and Relatedness of Campylobacter Sp. cdtB Gene. Infect. Immun., 64 2070–2078. doi: 10.1128/iai.64.6.2070-2078.1996
Powers, M. J., Simpson, B. W., Trent, M. S. (2020). The Mla Pathway in Acinetobacter baumannii has No Demonstrable Role in Anterograde Lipid Transport. Elife 9, e56571. doi: 10.7554/eLife.56571
Powers, M. J., Trent, M. S. (2019). Intermembrane Transport: Glycerophospholipid Homeostasis of the Gram-Negative Cell Envelope. Proc. Natl. Acad. Sci. U. S. A. 116, 17147–17155. doi: 10.1073/pnas.1902026116
Purta, E., O’Connor, M., Bujnicki, J. M., Douthwaite, S. (2009). YgdE Is the 2′-O-Ribose Methyltransferase RlmM Specific for Nucleotide C2498 in Bacterial 23S rRNA. Mol. Microbiol. 72, 1147–1158. doi: 10.1111/j.1365-2958.2009.06709.x
Rasschaert, G., De Zutter, L., Herman, L., Heyndrickx, M. (2020). Campylobacter Contamination of Broilers: The Role of Transport and Slaughterhouse. Int. J. Food Microbiol. 322, 108564. doi: 10.1016/j.ijfoodmicro.2020.108564
Röhl, R., Nierhaus, K. H. (1982). Assembly Map of the Large Subunit (50S) of Escherichia coli Ribosomes. Proc. Natl. Acad. Sci. U. S. A. 79, 729–733. doi: 10.1073/pnas.79.3.729
Roier, S., Zingl, F. G., Cakar, F., Durakovic, S., Kohl, P., Eichmann, T. O., et al. (2016). A Novel Mechanism for the Biogenesis of Outer Membrane Vesicles in Gram-Negative Bacteria. Nat. Commun. 7, 10515. doi: 10.1038/ncomms10515
Ruiz, N., Kahne, D., Silhavy, T. J. (2009). Transport of Lipopolysaccharide Across the Cell Envelope: The Long Road of Discovery. Nat. Rev. Microbiol. 7, 677–683. doi: 10.1038/nrmicro2184
Sałamaszyńska-Guz, A., Rose, S., Lykkebo, C. A., Taciak, B., Bącal, P., Uśpieński, T., et al. (2018). Biofilm Formation and Motility Are Promoted by Cj0588-Directed Methylation of rRNA in Campylobacter jejuni. Front. Cell. Infect. Microbiol. 7, 533. doi: 10.3389/fcimb.2017.00533
Sałamaszyńska-Guz, A., Serafińska, I., Bącal, P., Douthwaite, S. (2020). Virulence Properties of Campylobacter jejuni Are Enhanced by Displaying a Mycobacterial TlyA Methylation Pattern in Its rRNA. Cell Microbiol. 22, e13199. doi: 10.1111/cmi.13199
Sergiev, P. V., Aleksashin, N. A., Chugunova, A. A., Polikanov, Y. S., Dontsova, O. A. (2018). Structural and Evolutionary Insights Into Ribosomal RNA Methylation. Nat. Chem. Biol. 14, 226–235. doi: 10.1038/nchembio.2569
Sun, L., Rasmussen, P. K., Bai, Y., Chen, X., Cai, T., Wang, J., et al. (2020). Proteomic Changes of Klebsiella pneumoniae in Response to Colistin Treatment and crrB Mutation-Mediated Colistin Resistance. Antimicrob. Agents Chemother. 64 (6), e02200-19. doi: 10.1128/AAC.02200-19
Taheri, N., Mahmud, A., Sandblad, L., Fallman, M., Wai, S. N., Fahlgren, A. (2018). Campylobacter jejuni Bile Exposure Influences Outer Membrane Vesicles Protein Content and Bacterial Interaction With Epithelial Cells. Sci. Rep. 8, 16996. doi: 10.1038/s41598-018-35409-0
Thingholm, T. E., Palmisano, G., Kjeldsen, F., Larsen, M. R. (2010). Undesirable Charge-Enhancement of Isobaric Tagged Phosphopeptides Leads to Reduced Identification Efficiency. J. Proteome Res. 9, 4045–4052. doi: 10.1021/pr100230q
Waite, D. W., Vanwonterghem, I., Rinke, C., Parks, D. H., Zhang, Y., Takai, K., et al. (2017). Comparative Genomic Analysis of the Class Epsilonproteobacteria and Proposed Reclassification to Epsilonbacteraeota (Phyl. Nov.). Front. Microbiol. 8, 682. doi: 10.3389/fmicb.2017.00682
Walan, A., Kihlström, E. (1988). Surface Charge and Hydrophobicity of Campylobacter jejuni Strains in Relation to Adhesion to Epithelial HT-29 Cells. APMIS 96, 1089–1096. doi: 10.1111/j.1699-0463.1988.tb00986.x
Yuki, N., Susuki, K., Koga, M., Nishimoto, Y., Odaka, M., Hirata, K., et al. (2004). Carbohydrate Mimicry Between Human Ganglioside GM1 and Campylobacter jejuni Lipooligosaccharide Causes Guillain-Barre Syndrome. Proc. Natl. Acad. Sci. U. S. A. 101, 11404–11409. doi: 10.1073/pnas.0402391101
Keywords: Campylobacter jejuni, rRNA methylation, TlyA, OMV, MlaEFD, CDT
Citation: Sałamaszyńska-Guz A, Rasmussen PK, Murawska M and Douthwaite S (2022) Campylobacter jejuni Virulence Factors Identified by Modulating Their Synthesis on Ribosomes With Altered rRNA Methylation. Front. Cell. Infect. Microbiol. 11:803730. doi: 10.3389/fcimb.2021.803730
Received: 28 October 2021; Accepted: 17 December 2021;
Published: 13 January 2022.
Edited by:
Vincenzo Scarlato, University of Bologna, ItalyReviewed by:
Timothy Randall Hoover, University of Georgia, United StatesCopyright © 2022 Sałamaszyńska-Guz, Rasmussen, Murawska and Douthwaite. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Agnieszka Sałamaszyńska-Guz, YWduaWVzemthX3NhbGFtYXN6eW5za2FfZ3V6QHNnZ3cuZWR1LnBs; Stephen Douthwaite, c3JkQGJtYi5zZHUuZGs=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.