
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Cell. Infect. Microbiol., 13 December 2021
Sec. Fungal Pathogenesis
Volume 11 - 2021 | https://doi.org/10.3389/fcimb.2021.796929
This article is part of the Research TopicSexual and Parasexual Reproduction of Human Fungal PathogensView all 6 articles
While most fungi have the ability to reproduce sexually, multiple independent lineages have lost meiosis and developed parasexual cycles in its place. Emergence of parasexual cycles is particularly prominent in medically relevant fungi from the CUG paraphyletic group of Candida species. Since the discovery of parasex in C. albicans roughly two decades ago, it has served as the model for Candida species. Importantly, parasex in C. albicans retains hallmarks of meiosis including genetic recombination and chromosome segregation, making it a potential driver of genetic diversity. Furthermore, key meiotic genes play similar roles in C. albicans parasex and highlights parallels between these processes. Yet, the evolutionary role of parasex in Candida adaptation and the extent of resulting genotypic and phenotypic diversity remain as key knowledge gaps in this facultative reproductive program. Here, we present our current understanding of parasex, the mechanisms governing its regulation, and its relevance to Candida biology.
Species survival depends on the generation of reproductive offspring that are capable of competing in their niche or expanding into new niches. While asexual (clonal) and sexual reproduction are the most common forms of reproduction across kingdoms, evidence for the less known parasexual cycle in nature comes exclusively from fungi and unicellular organisms (Alexopoulos et al., 1996). First described in 1953 by the Italian geneticist Guido Pontecorvo during studies on Aspergillus nidulans (Pontecorvo et al., 1953), parasex describes a non-meiotic process of ploidy reduction that produces genetically diverse progeny. Parasex is particularly prevalent in species with no known sexual cycle. While early studies were restricted to filamentous fungi, more recent work has identified parasex in yeast species, with significant relevance to human pathogenic fungi such as Candida albicans (C. albicans) (Bennett and Johnson, 2003; Forche et al., 2008; Seervai et al., 2013).
C. albicans is the most clinically relevant fungal pathogen of humans, owed in large part to being a resident commensal of the oral, dermal, gastrointestinal, and genital niches (Calderone, 2012). Heavy use of steroids or antibiotics, immunosuppression, and autoimmune disorders increase the risk of mucosal and systemic C. albicans infections that most commonly result from overgrowth of the patient’s normal flora (Wisplinghoff et al., 2014; Zhai et al., 2020). Since its identification and first description as a species in 1923 (Barnett, 2004), C. albicans was long believed to strictly reproduce asexually. In fact, prior to its reclassification into the Saccharomycetes, the Candida genus belonged to the Deuteromycetes (imperfect fungi), due to its apparent lack of a sexual cycle (Hibbett et al., 2007).
Early genetic studies of C. albicans used parasex-like processes of protoplast fusion and nuclear hybridization between auxotrophic strains to generate hybrid tetraploid strains. The resulting fusant could often maintain a stable karyotype but would undergo random chromosome loss to become aneuploid in some instances (Sarachek et al., 1981; Poulter et al., 1981). As more genetic tools became available, and with the discovery of the mating-competent opaque cell state (Anderson and Soll, 1987) and the mating-type ‘a’ and ‘α’ loci (Hull and Johnson, 1999), the defining features of a non-meiotic parasexual cycle began to take shape in C. albicans (Bennett and Johnson, 2003). Here, we review the extant knowledge of the parasexual cycle in C. albicans, the underlying molecular mechanisms, parallels to meiosis, and its relevance to natural populations.
The morphological plasticity of C. albicans at both the cellular and colony level has long been a focal point of C. albicans research, stretching back to its first reported isolation from a thrush patient in 1839 (Knoke and Bernhardt, 2006). The ability to switch between distinct phenotypic states is thought to be central to its ability to thrive in multiple host niches with different pH, metabolite, and oxic profiles (reviewed in (Scaduto and Bennett, 2015). Although more focus has been placed on the yeast-to-hyphal transition because of its tight association with disease, the phenotypic switch between the white and opaque state also regulates several key aspects of C. albicans biology.
C. albicans is most commonly isolated from clinical infections in the white state, which is characterized by round cells and white dome-shaped colonies on solid medium (‘Candida’ refers to the white robes worn by Senate candidates in the Roman empire and ‘albicans’ means ‘to whiten’ in Latin) (Bennett and Johnson, 2003). In contrast, opaque cells are elongated and club-shaped, forming flat, dull colonies (Rikkerink et al., 1988; Miller and Johnson, 2002; Lohse and Johnson, 2009). Both states are heritable across many generations, with low-frequency transitions between cell states that are governed by a combination of genetic and epigenetic factors (Lohse and Johnson, 2009). Switching to the opaque state canonically requires cells to be hemi- or homozygous for the a or α idiomorph at the Mating-Type Like (MTL) locus on Chromosome 5 (Chr5), which has strong similarity to the Mating-Type (MAT) locus in S. cerevisiae that determines mating compatibility (Hull and Johnson, 1999; Magee and Magee, 2000). On glucose-containing medium at room temperature and ambient CO2, interconversion between the white and opaque states in MTLa or MTLα homozygous cells is rare, occurring only 1 in 10,000 cell divisions (Bergen et al., 1990; Lockhart et al., 2002). However, certain environmental stimuli induce high rates of unidirectional cell state transitions. Conversion to the opaque state dramatically increases in conditions that mimic certain niches in the gut (acidic pH, ≥ 5% CO2, and the presence of N-acetylglucosamine (GlcNAc) [reviewed in (Lohse and Johnson, 2009; Noble et al., 2017)]. In contrast, growth on glycolytic carbon sources at 37°C induces conversion to the white state en masse (Slutsky et al., 1987; Alkafeef et al., 2018). Interestingly, some heterozygous MTLa/α isolates can undergo low-frequency white-to-opaque switching under in vitro conditions that mimic the gut environment, although the resulting opaque MTLa/α state is less stable than its MTL homozygous counterparts (Xie et al., 2013).
Consistent with in vitro results, in vivo passage through the mouse gut induced opaque cell formation in at least one isolate background, WO-1 (Ramirez-Zavala et al., 2008). WO-1 has an extra copy (trisomy) of Chr1, which carries WOR1, the master regulator of the opaque cell state (Huang et al., 2006). Similarly, artificial overexpression of WOR1 in white cells resulted in a competitive advantage in the mouse gut (Pande et al., 2013). Loss of the third copy of WOR1 decreased recovery of opaque cells from the mouse gut and blocked condition-dependent switching in vitro (Huang et al., 2006). Examination of these GUT (‘gastrointestinally-induced transition’) cells showed that they retained MTL heterozygosity, were phenotypically stable at higher temperatures (37°C) but lacked ‘pimples’ characteristic for true opaque cells in C. albicans. Consequently, this phenotype was considered to be distinct from that of canonical white and opaque cells; its frequency and relevance in natural populations remains unexplored. However, the white state has increased fitness during gut colonization compared to the opaque state in other strain backgrounds and introduced opaque cells will overwhelmingly convert to the white state over time (Pande et al., 2013; Takagi et al., 2019).
White and opaque cell types can be distinguished by distinct physiological, metabolic, transcriptional, and virulence properties. Opaque cells are less metabolically active except when peptides are provided as the sole substrate, which also induce filamentation in opaque cells but not in white cells (Ene et al., 2016). In addition, opaque cells are less susceptible to phagocytosis by macrophages and leukocytes and colonize the heart and spleen more successfully than white cells, potentially resulting in altered pathogenic outcomes (Geiger et al., 2004; Lohse and Johnson, 2008; Sasse et al., 2013; Takagi et al., 2019). The hallmark trait distinguishing white and opaque cells is mating in C. albicans. White cells are ‘sterile’ and incapable of mating, whereas opaque cells are mating competent when they are MTL hemizygous or homozygous. Mating can occur either between opaque cells of the opposite mating type (i.e., a-α; heterothallic) or between cells of the same mating type (i.e., a-a; homothallic) (Alby et al., 2009; Alby and Bennett, 2011; Guan et al., 2019).
Mating in C. albicans requires the molecular sensing of a suitable partner cell followed by a complex series of intracellular events that activate the process of cell-cell fusion. A typical mating cycle in C. albicans requires MTLa and MTLα cells to release and sense sex pheromones encoded by the MFA1 and MFα1 loci, respectively [Figure 1, (Bennett et al., 2003; Lockhart et al., 2003; Panwar et al., 2003)]. MFα1 is constitutively expressed in opaque α cells at higher levels than MFA1 is expressed in opaque a cells. Detection of α-pheromone by opaque a cells induces an increase in MFA1 and MFα1 levels, fully activating the mating response system in both opaque cell populations. Engagement of the pheromone receptors Ste2 and Ste3 on opaque a and α cells, respectively, initiates polarized growth through activation of the mitogen-activated protein kinase (MAPK) cascade. The polarized cells extend towards the source of the detected pheromone to form mating projections or ‘shmoos’ (Bennett et al., 2003; Lockhart et al., 2003; Yi et al., 2009). This process occurs quickly, typically within ~1.5 h, and halts asexual cell division (Bennett et al., 2003; Lockhart et al., 2003; Yi et al., 2009). Physical contact between opaque cells of opposing mating types results in fusion of the cell cytoplasts and nuclei through a process called karyogamy to produce the zygote, which contains the contents of both parental cells. Mating has most often been observed between diploids to produce tetraploid mating products, but successful mating can also occur between haploid cells (Hull et al., 2000; Bennett and Johnson, 2003; Hickman et al., 2013). Mating products are capable of stably maintaining the resultant ploidy by propagating asexually or can be induced to undergo a disordered ploidy reduction called concerted chromosome loss (CCL) to yield recombinant and karyotypically diverse progeny (Forche et al., 2008; Smith and Hickman, 2020).
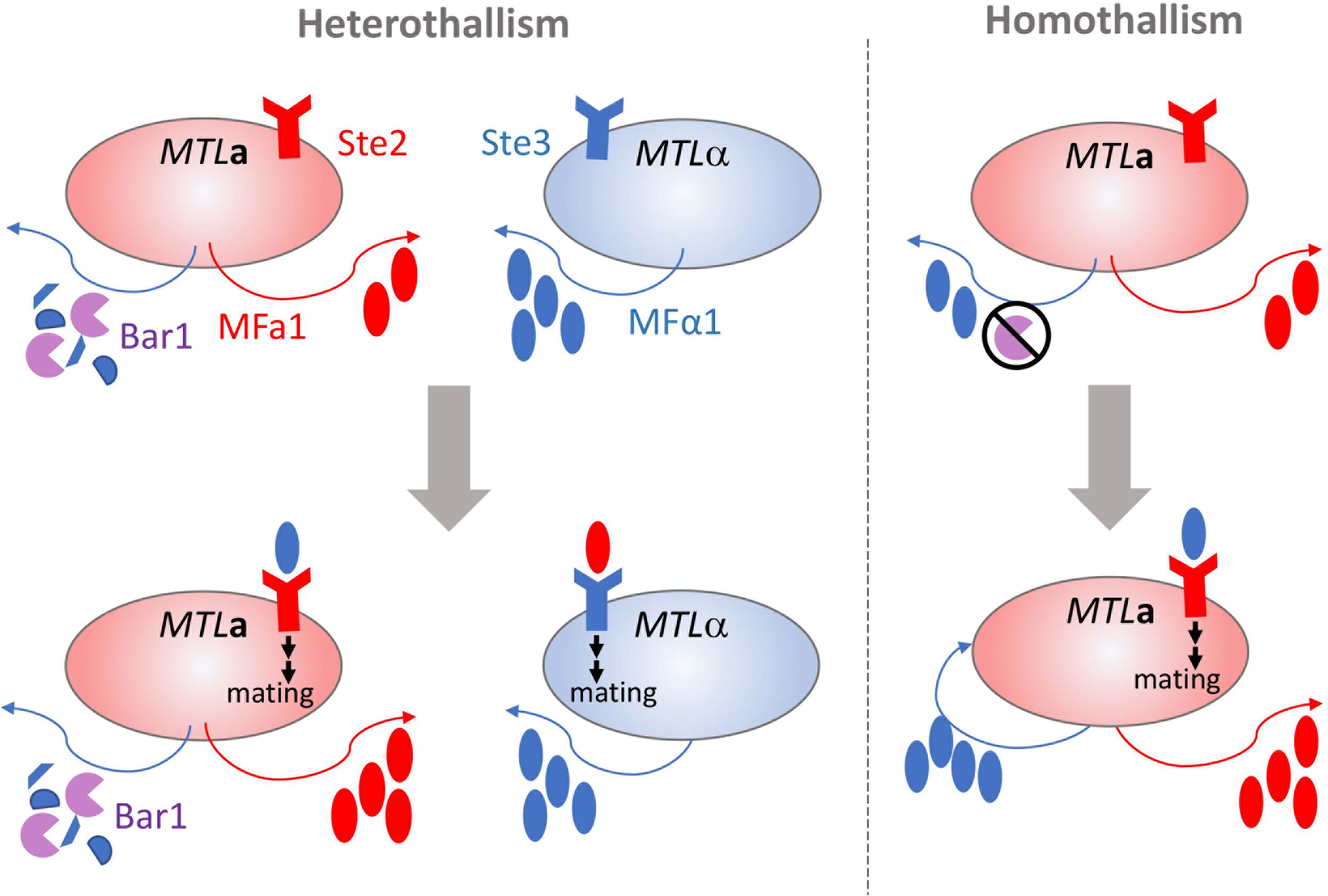
Figure 1 Parasexual mating of opaque cells in C. albicans. Opaque cells of the opposite or same mating types (heterothallism and homothallism, respectively) can undergo mating and cell fusion though production and response to pheromone. In heterothallic mating (shown on left), the pheromone receptor is engaged by mating pheromone produced by cells of the opposite mating type to initiate polarized growth and cell fusion (Ste2 receptor by MFα1 or Ste3 receptor by MFa1). Bar1 protease degrades MFα1 produced by opaque MTLa cells and is overcome by the levels of MFα1 produced by opaque MTLα cells. In homothallic mating (shown on right), the overproduction of MFα1 or inactivation of Bar1 protease in MTLa leads to autocrine pheromone receptor engagement and a-a cell fusion and mating.
Some exceptions exist to the heterothallic mating paradigm for opaque cells. Even though white and opaque cells differentially express ~17% of their transcriptome (Tuch et al., 2010), Scaduto et al. demonstrated that opaque cell fertility was dependent on the activation of only a few genes from the MAPK signaling pathway that are canonically induced by pheromone binding (Scaduto et al., 2017). Ectopic overexpression of the three most transcriptionally silent genes from the MAPK pathway in white cells led to robust pheromone responses and mating at levels comparable to opaque cells. Conversely, disruption of MAPK pathway genes blocks mating in otherwise mating-competent opaque cells even in the presence of pheromones (Magee et al., 2002). This suggests that the MAPK pathway is both necessary and sufficient for parasexual fecundity in C. albicans, challenging the tenet that mating competency is exclusively tied to cell state.
Homothallism offers another exception to the heterothallic mating paradigm. Opaque a cells secrete both a- and α-pheromone, which can stimulate paracrine and autocrine signaling responses by engaging the Ste3 receptor on nearby alpha cells or Ste2 on their own cell surfaces [Figure 1, (Alby et al., 2009)]. Stimulation of a cells by α-pheromone is typically prevented by the Bar1 protease, which degrades the secreted α-pheromone (Schaefer et al., 2007). However, disruption of BAR1 or excessive α-pheromone levels can cause MTLa cells to initiate auto MFα1-activated same-sex mating (Alby et al., 2009). Homothallic mating in opaque cells can also be achieved via ‘ménage à trois’ mechanisms, in which white cells of the opposite mating type produce pheromones that facilitate mating of opaque cells without mating themselves [Figure 2, (Guan et al., 2019)]. Surprisingly, mating pheromone is not necessary for homothallic mating, as opaque MTLa cells can fuse when subjected to environmental stress such as glucose starvation and exposure to oxidative agents (Tao et al., 2014; Guan et al., 2019). Phosphate limitation either through phosphate starvation or mutations in the PHO pathway also increases the mating efficiency of opaque cells, highlighting the myriad environmental conditions that are conducive to initiating the C. albicans parasexual cycle without requiring pheromone production or sensing (Zheng et al., 2020).
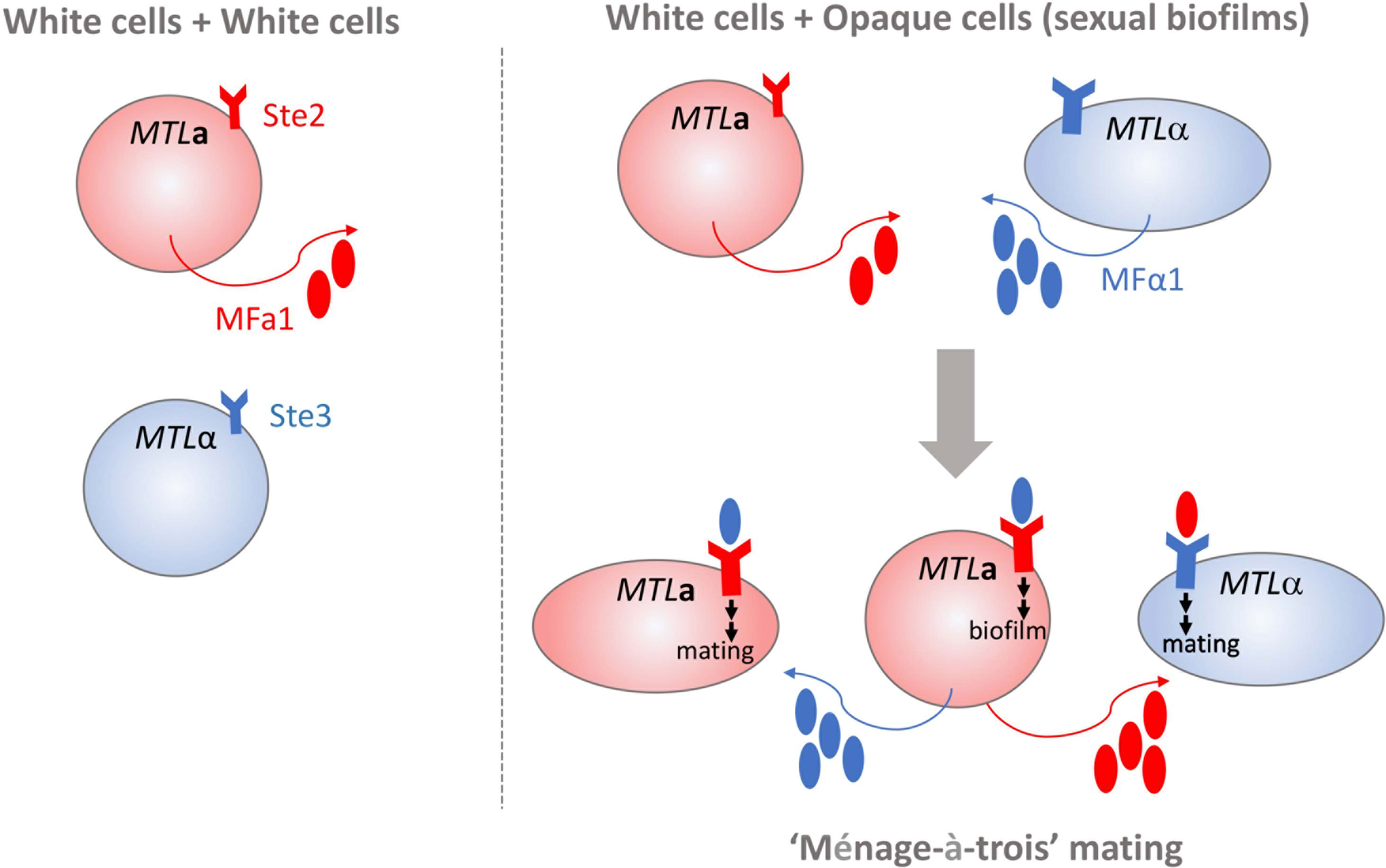
Figure 2 White cells in C. albicans mating. White cells (depicted as spherical cells) of opposite mating types do not produce a mating response because of a lack of pheromone production and receptor engagement (shown on left). MTLa white cells exposed to high levels of MFα1 can initiate formation of complex biofilm structures that support opaque cell mating (shown on right). Pheromone-stimulated MTLa white cells can also engage in “ménage-à-trois” mating by supplying MFα1 and MFa1 pheromones to both opaque MTLa and MTLα opaque cells (depicted as elongated cells) that induces polarized growth and cell fusion.
Pheromone responses also serve to promote the formation of specialized sexual biofilms. Biofilms in C. albicans are ordered structures capable of forming on biotic and abiotic surfaces, such as indwelling catheters and other implanted medical devices. Mature biofilms offer protection from immune cell surveillance and antifungal compounds and can serve as reservoirs for repeated infection by actively dispersing fungal cells throughout the patient’s body (Uppuluri et al., 2010; Gulati and Nobile, 2016; Mitchell et al., 2016; Uppuluri et al., 2018). Dissemination can lead to systemic candidiasis in severely immunocompromised patients that is associated with high mortality rates [reviewed in (Nobile and Johnson, 2015)]. The central role of biofilms in disease has prompted new approaches in antifungal drug design that target initiation of biofilm formation and disruption of mature biofilms (Pierce and Lopez-Ribot, 2013; Atriwal et al., 2021).
Whereas conventional biofilms form through a succession of cell adherence, substrate invasion, hyphal formation, and matrix deposition that is dependent on transcriptional regulators (e.g., BCR1, ACE2) (Fanning et al., 2012; Nobile et al., 2012; Uwamahoro et al., 2012) and cell wall proteins (e.g., Als1, Als3, and Hwp1) (Nobile et al., 2006; Ene and Bennett, 2009; Salgado et al., 2011), sexual biofilm formation initiates in response to the presence of mating pheromone (Lin et al., 2013; Park et al., 2013). Furthermore, conventional and sexual biofilms differ in compactness, antifungal drug resistance, signaling pathways, resilience against host immune responses, and cross-species interactions [reviewed in (Perry et al., 2020)].
Sexual biofilms typically form in an MTL homozygous population of C. albicans cells. Stochastic transitions to the opaque state initiate secretion of MFa and MFα pheromones that influence cell responses of both opaque and white cells. Daniels et al. (2006) discovered that white cells within pheromone-induced populations respond to pheromones by tightly adhering to a substratum that organizes into a biofilm-like community. The resulting three-dimensional cell matrix facilitates opaque cell mating via enhanced chemotropism (Daniels et al., 2006). Mating in sexual biofilms was most efficient with a low percentage of opaque cells (≤ 10%), suggesting that a chemotropism optimum exists to balance formation of a structured environment by white cells with effective migration and mating of opaque cells (Daniels et al., 2006; Park et al., 2013). The immediate presence of white cells may provide additional benefits for mating by promoting homothallism via pheromone secretion and protecting opaque cells from the immune response of the host (Perry et al., 2020). Although a fundamental appreciation for the uniqueness of sexual biofilms exists, key questions addressing their in vivo relevance and potential to enhance fungal survival and/or pathogenesis remain to be answered.
Early population genetic studies of C. albicans suggested that a meiosis-like mechanism may exist in this species based on the presence of non-clonal genotypes (Graeser et al., 1996; Forche et al., 1999). Yet, the capacity of parasexual mating to generate genotypic diversity through the processes of independent segregation and assortment as seen in meiosis remains poorly characterized. Early drafts of the C. albicans genome sequence in the 2000s (Jones et al., 2004; van Het Hoog et al., 2007), genome-wide screens (Magee et al., 2002), and a comparative genomic study (Tzung et al., 2001) provided the groundwork to explore the function of 500 meiosis-related genes from S. cerevisiae in C. albicans. The C. albicans genome retained many mating-related homologs, but some key meiosis regulators were conspicuously absent, such as IME1 (master regulator of meiosis initiation in S. cerevisiae) and SPO13 (regulator of segregation during meiosis I in S. cerevisiae). Subsequent studies identified the presence of several previously overlooked ‘meiosis-specific’ genes in C. albicans although the involvement of most homologs in the parasexual cycle remains unstudied [reviewed in (Sherwood and Bennett, 2009; Usher, 2019)].
As parasex does not undergo the traditional organized halving of DNA content through the meiotic cycle, ploidy reduction (depolyploidization) of tetraploid (or diploid) mating products occurs via concerted chromosome loss albeit without any clear order for which chromosomes are lost. Ploidy reduction via parasex yields highly aneuploid cells with few euploid or near-euploid progeny (Forche et al., 2008; Anderson et al., 2019). Aneuploid progeny preferentially retain extra copies of smaller chromosomes (Chr4-7) that may be less detrimental to the aneuploid cell due to a reduced number of imbalanced genes compared to the larger chromosomes (Forche et al., 2008; Hickman et al., 2015). However, one caveat in most parasexual experiments is that extra copies of Chr1 are selected against because of selection for loss of GAL1 (located on chromosome 1) with 2-deoxygalactose as a proxy for having undergone ploidy reduction (Gorman et al., 1992; Bennett and Johnson, 2003; Forche et al., 2003; Forche et al., 2008; Thomson et al., 2019). Surprisingly, the imbalanced karyotypes of aneuploid progeny are relatively stable under standard laboratory conditions (Magee and Magee, 1997; Hickman et al., 2015; Anderson et al., 2017). However, serial passaging leads to many aneuploids resolving as diploids, reinforcing this ploidy state as the most stable for C. albicans (Hickman et al., 2015). Convergence to diploidy may result from growth advantages of euploid derivatives compared to their aneuploid progenitor in mixed cultures. Indeed, Yang et al. (2021) demonstrated growth defects in rich medium for a panel of strains each trisomic for one of the eight chromosomes. Interestingly, some growth phenotypes were associated with specific chromosome homologs suggesting that inheritance of specific alleles during ploidy reduction can have a larger impact on fitness.
Ploidy reduction via CCL can be induced by growth on specific media including glucose-rich ‘pre-sporulation’ (pre-spo) medium, which induces meiosis in S. cerevisiae (Bennett and Johnson, 2003). Prior work had shown that induction of CCL on pre-spo medium resulted in the death of most tetraploid, but not diploid cells (Bennett and Johnson, 2003). Recent work by Thomson et al. (2019) elegantly showed that the high glucose content of pre-spo medium selectively pushed the tetraploid cells into a metabolically hyperactive state, leading to higher rates of aerobic and anerobic respiration compared to diploid cells. This in turn led to elevated levels of reactive oxygen species (ROS) as a metabolic byproduct, thereby activating the ROS-responsive transcription factor, CAP1 (Wang et al., 2006), and causing DNA double-strand breaks (DSBs). These DSBs then served a dual function of destabilizing aneuploid chromosomes and promoting parasexual recombination. This resulted in rapid and random chromosome loss during cell division, giving rise to parasexual progeny with reduced DNA content and potentially recombinant genomes (Thomson et al., 2019).
Two meiosis-specific genes have defined roles in C. albicans ploidy reduction: SPO11, a topoisomerase-that creates double strand breaks, and REC8, the kleisin subunit of the cohesin complex (Figure 3). Loss of SPO11 activity increases ploidy reduction whereas disruption of REC8 decreases chromosome loss (Anderson et al., 2019). These two genes represent the very limited repertoire of ‘meiosis-specific’ genes studied in the context of parasexual ploidy reduction to date.
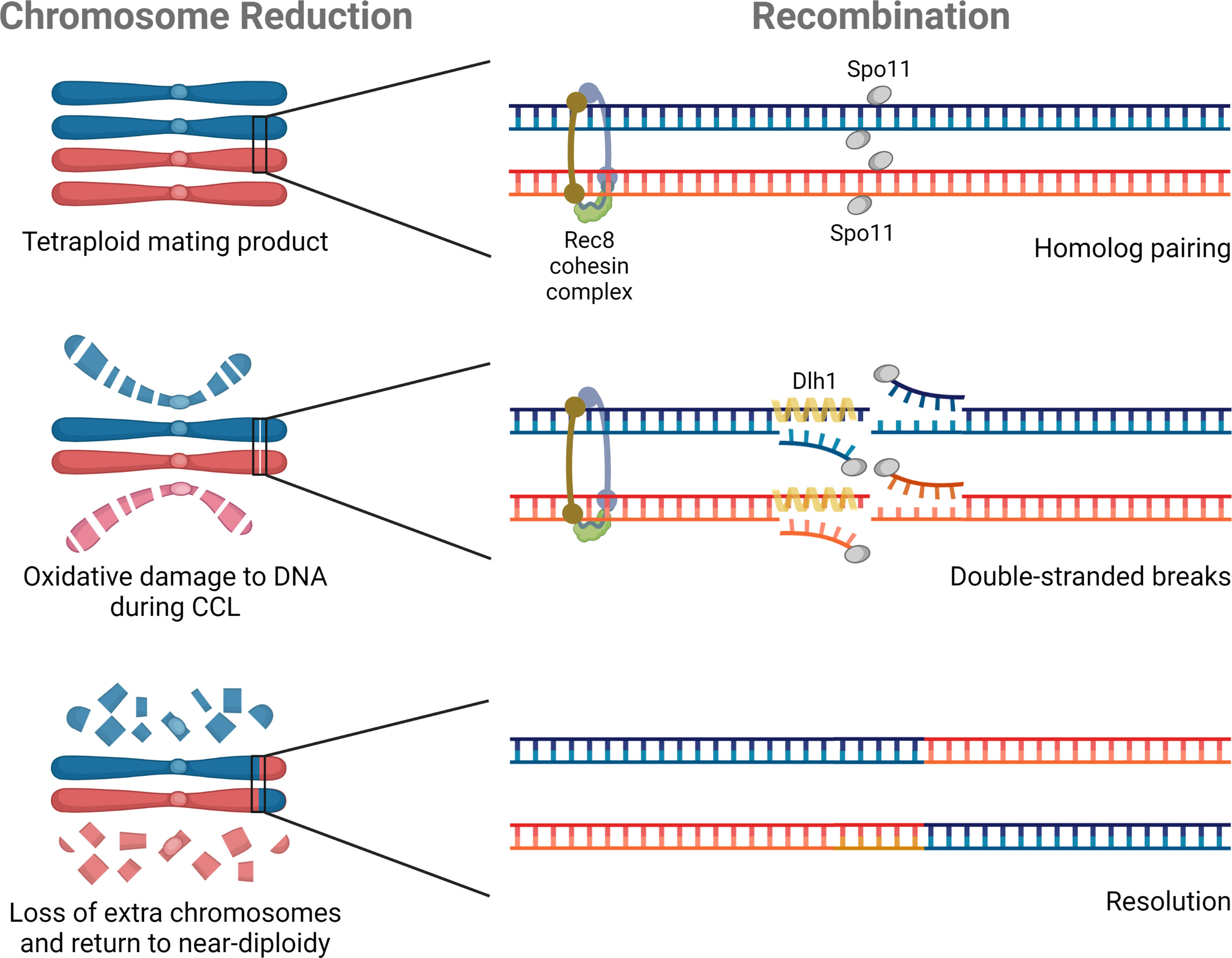
Figure 3 Molecular components of parasexual reproduction. Three genes have experimentally defined roles in C. albicans parasexual processes. SPO11 and REC8 contribute to ploidy reduction (depicted on the left), and all three genes (SPO11, REC8, and DLH1) are involved in recombination during parasex (shown on the right).
Recombination is a hallmark of meiotic ploidy reduction that is also characteristic of parasex during CCL. The first study of genotypes derived from CCL showed clear evidence of mitotic recombination during parasexual reproduction in C. albicans (Forche et al., 2008), with some strains containing multiple recombination events on a single chromosome. Subsequent attempts to quantify the frequency of recombination during parasex demonstrated a greater than 1,000-fold increase in inter-chromosomal recombination than under standard laboratory growth conditions (Anderson et al., 2019). Deletion of SPO11 abolished all evidence of recombination by comparative genome hybridization and served as the first demonstration that components of the meiotic machinery had been co-opted for parasex (Forche et al., 2008). Surprisingly, SPO11 was later shown to promote chromosomal stability and mitotic recombination outside of CCL, although the effect was more prominent under conditions that promoted genome instability (Anderson et al., 2019). In this same study, loss of REC8 similarly decreased parasexual recombination (Anderson et al., 2019). Finally, Ciudad et al. (2020) reported that DLH1, a C. albicans homolog of the meiosis-specific S. cerevisiae factor DMC1, could promote strand invasion thereby promoting long-tract loss of heterozygosity (LOH) during recombination (Figure 3). However, DLH1 function was tested outside of parasex through ectopic expression in mitotic cells so the specific role in CCL remains unclear. Described functional roles among diverse proteins in parasex may point to a common central process that is influenced by all these factors or may suggest nonredundant unique roles during recombination that require the individual contributions of Spo11, Rec8, and Dmc1. It remains to be tested if, like mitotic recombination, parasexual recombination might be further facilitated by the presence of defined repetitive elements and cryptic repeat sequences throughout the C. albicans genome, namely major repeat sequences (MRS), ribosomal DNA (rDNA) repeats, and telomeric elements (Gusa and Jinks-Robertson, 2019; Todd et al., 2019).
Most parasexual progeny will be inhibited from re-entering the parasexual cycle following CCL by containing 2+ copies of Chr5 and being mating incompatible (MTLa/α). A fraction of the progeny, however, could become MTLa or MTLα homozygotes, either during CCL or later through LOH (Popp et al., 2019) and re-enter the mating cycle. Alternatively, haploid progeny could undergo the less commonly observed haploid mating cycle that yields a diploid mating product (Hickman et al., 2013) and/or autodiploidize (i.e., double their DNA content by DNA replication without segregation) to remain mating competent as diploid MTL homozygotes. This progeny could then re-enter the parasexual cycle to generate additional mating products. Thus, genetic diversity is produced via three distinct mechanisms operating simultaneously during CCL: ploidy reduction, segregation of whole chromosomes, and parasexual recombination.
The genetic diversity generated by C. albicans parasex translates to phenotypic variation in mating products and recombinant progeny. The process of polyploidization during mating (via nuclear fusion) serves as a significant driver for phenotypic adaptation by increasing the number of potential karyotypes and genotypes of parasexual progeny as well as changing cell physiology on its own (Harrison et al., 2014; Selmecki et al., 2015). Tetraploid C. albicans cells exhibit unique traits compared to genotypically identical diploids such as reduced growth rates and attenuated virulence in a mouse model of systemic disease (Ibrahim et al., 2005; Hickman et al., 2013).
Among parasexual progeny, phenotypic diversity was immediately apparent in the initial studies of parasex. Colonies with altered morphologies and increased or decreased filamentation in Forche et al. (2008) highlighted the implications of parasex towards pathogenesis. A more recent study assessed the fitness of 32 parasexual progeny for multiple traits such as filamentation, virulence, and biofilm formation in comparison to the parental strains (Hirakawa et al., 2017). Parasexual progeny were phenotypically diverse but showed overall reduced fitness compared to their parents in most environments except for increased fluconazole resistance, which was linked to trisomies of Chr3 and Chr6 (Hirakawa et al., 2017).
Chromosome imbalances are known to confer new characteristics to Candida (Janbon et al., 1998; Forche et al., 2019), serve as a transient solution to stress in multiple fungal species (Yona et al., 2012; Ford et al., 2015), and are key drivers of phenotypic diversity. Moreover, the tolerance and/or fitness advantages associated with aneuploid chromosomes could increase the probability of beneficial mutations to arise while under selection, negating the need to carry additional chromosomes and their associated fitness costs (Pavelka et al., 2010). In this way, parasex provides a potential means for generating a large reservoir of ploidy-diverse strains for selection to act upon. The potential phenotypic diversity of this progeny pool can be expanded through parasexual mating of genotypically distinct parental strains thereby promoting the exchange of beneficial mutations such as those conferring antifungal drug resistance (Popp et al., 2019).
A natural question arises from the in vitro studies: Has parasex played a role in shaping the Candida species phylogeny (Schmid et al., 2016)? In the pre-genomic era, several groups used methods such as enzyme electrophoresis (Pujol et al., 1993), co-dominant molecular markers (Graeser et al., 1996; Forche et al., 1999), and multi-locus enzyme electrophoresis (Arnavielhe et al., 2000) to study the population structure of C. albicans. More recent studies used multi-locus sequence typing (MLST) to expand the number of markers and isolates analyzed (Tavanti et al., 2005; Odds et al., 2007). While most studies did not find widespread evidence of recombination, infrequent instances of intra- and inter-locus variation among C. albicans isolates in a few studies suggested that genetic exchange through a meiosis-like sexual cycle may be possible (Graeser et al., 1996; Forche et al., 1999; Odds et al., 2007). Ambiguity in the C. albicans population structure likely resulted from the study of different populations, different methods, and difficulty in distinguishing parasexual and mitotic recombination in a malleable diploid genome. Difficulty in distinguishing between LOH and parasexual recombination was reinforced in a follow-up a study with 203 isolates (Bougnoux et al., 2008), which concluded that mitotic recombination (leading to LOH events) was widespread among C. albicans isolates but there was negligible evidence of parasexual mating.
Analysis of haploid mitochondrial genomes in C. albicans offered a simplified context to understand the C. albicans population structure without LOH obfuscating inheritance patterns. Contrary to the uniparental transmission of mitochondrial DNA (mtDNA) in most eukaryotes, an early analysis demonstrated low levels of ancient genetic admixture and recombination in the mitochondrial genomes of clinical strains of C. albicans and more recent clonal expansions, suggesting that mitochondrial rearrangements may predate C. albicans speciation or, alternatively, loss of meiosis (Anderson et al., 2001). Indeed, bi-parental inheritance and recombination in the mitochondrial genomes of C. albicans conforms to patterns in other eukaryotes [reviewed in (Barr et al., 2005; White et al., 2008)], which can have some putative evolutionary benefits such as mutational clearance (Neiman and Taylor, 2009). Using the same mitochondrial haplotyping approach, Jacobsen et al. (2008b) reported strong evidence of more recent recombination in a different set of clinical strains. The discordance and inconsistency between the nuclear and mitochondrial genome analyses led to the generally accepted conclusion that, while parasex could be performed in the laboratory, little conclusive evidence existed for it in ‘nature’.
In the post-genomics era, two studies attempted to conclusively address the question of recombination in the C. albicans phylogeny. While both studies reported a predominantly clonal population structure, they also revealed robust and recent genetic admixture between different strain clusters (Ropars et al., 2018) and the existence of mosaic genomes shaped by genetic exchange in both nuclear and mitochondrial genomes based on the presence of identical single nucleotide variant (SNV) patterns found in genetically distant strains (Wang et al., 2018). Therefore, the emergent consensus from these phylogenetic analyses suggests that clonal reproduction remains dominant in most C. albicans clades with strains having intermittently undergone intra-clade or inter-clade exchange of genetic material, possibly through parasexual recombination.
Most C. albicans strains are isolated as mating incompetent MTLa/α heterozygotes and generally restricted to the white state. This bias towards isolation of the white state, even among MTL homozygous strains, may be due to instability of the opaque cell state at the internal human body temperature of 37°C (Alkafeef et al., 2018). Lachke et al. (2003) established that mating can occur on mammalian skin where, at a surface temperature of ~32°C, opaque cells are more stable. Yet, a mixture of MTLa and MTLα opaque cells were stable enough to mate upon simultaneous intravenous inoculation in mice (Hull et al., 2000). Furthermore, specific anaerobic growth conditions that mimic niches in the GI tract promote stability of the opaque cell state at internal body temperatures (Dumitru et al., 2007). Indeed, parasexual mating in the GI tract of a murine host was demonstrated by recovering strains that contained both selectable markers from each singly marked parental strain and were uni-nucleate tetraploids by flow cytometry (Dumitru et al., 2007). Despite the evidence for mating in vivo using murine models, parasexual progeny resulting from CCL have not been observed in these systems. We are hopeful that future studies will reveal host niches and/or specific environmental conditions that facilitate efficient in vivo mating and parasex in C. albicans.
When did parasex emerge in the Candida paraphyletic group? To answer this, most studies have focused on the two species most closely related to C. albicans, C. dubliniensis and C. tropicalis. These three Candida species diverged approximately 20 million years ago (MYA) from the C. orthopsilosis complex (Moran et al., 2012; McManus and Coleman, 2014). All three species are diploid and share similar traits, including white-to-opaque switching and the ability to mate (Pujol et al., 2004; Porman et al., 2011; Seervai et al., 2013). This suggests that key events in the C. albicans parasexual program evolved in the common ancestor before the divergence of these three species.
Consistent with all three Candida species having inherited parasex from a common ancestor, they share many characteristics required for mating and CCL. The C. tropicalis and C. dubliniensis genomes both encode a syntenic MTL locus with a similar organization to that of C. albicans, where the degree of amino acid identity reflects their phylogenetic relationship (i.e., C. dubliniensis being more closely related to C. albicans than C. tropicalis) (Xie et al., 2012). The MTLa and MTLα idiomorphs are present on each homologous chromosome (including all four mating-related genes: a1, a2, α1, and α2) and most isolates are recovered from clinical and environmental settings as MTL heterozygotes (Pujol et al., 2004; Xie et al., 2012). Both these species also share features of the parasexual cycle with C. albicans: mating between diploid cells, a stable tetraploid zygote, CCL, and the ability for diploid progeny to re-enter the parasexual cycle (Seervai et al., 2013). Similar to C. albicans, the population structure of C. tropicalis is predominantly clonal with evidence of low levels of recombination among strains that suggests infrequent but detectable levels of sexual exchange among natural isolates (Jacobsen et al., 2008a).
Each species possesses unique attributes of mating and parasex that highlight the divergence in regulatory features of parasex since speciation. For example, the white-to-opaque switching frequency is about an order of magnitude higher in C. dubliniensis than in C. albicans (Pujol et al., 2004). However, the absence of mating-dependent clumping in C. dubliniensis makes intra-species mating less efficient than inter-species mating with C. albicans both in vitro and in vivo (Pujol et al., 2004; Singh-Babak et al., 2021). Inter-species C. albicans-C. dubliniensis mating products are sterile and unable to either reduce ploidy effectively or re-enter the parasexual cycle. Finally, the grey cell phenotype, found in all three Candida species, represents a mating-competent intermediate state along the white-to-opaque transition only in C. dubliniensis and C. tropicalis (Porman et al., 2011; Anderson et al., 2016; Yue et al., 2016). In C. albicans, grey cell formation requires inactivation of the EFG1 transcription factor, and these cells cannot return to the white state (Liang and Bennett, 2019).
Deeper investigations into C. tropicalis parasex has uncovered some unique traits not found in C. albicans. Pheromone-assisted a-a homothallic mating occurs independent of the white-to-opaque switch, which is required in C. albicans prior to mating (Du et al., 2018). In addition, C. tropicalis homothallic and heterothallic tetraploid mating products can undergo mating with diploid cells to generate hexaploid progeny, something never observed in C. albicans potentially due to the high instability of >4N cells. Taken together, similarities in parasex between these three species likely reflect their common origin from which each Candida species has evolved their own independent parasexual regulatory mechanisms. Retention of parasex over evolutionary time and evidence of recombination in species phylogenies suggests that these mechanisms may be active in vivo to provide fitness advantages in the host albeit under yet unidentified conditions.
As with all forms of sexual reproduction, parasex is an energetically expensive process but, like meiosis, can promote removal of deleterious mutations in a population and bring together beneficial mutations from genetically distinct lineages. Before evidence of parasexual mating emerged, it was speculated that the C. albicans genome contained many detrimental recessive alleles and that the irreversible accumulation of deleterious mutations in an exclusively asexual, diploid organism would lead to progressively declining fitness and eventual extinction (Muller, 1964; Schmid et al., 2016). Even after mating was described in C. albicans, the lack of evidence for meiosis-like levels of recombination fueled the hypothesis that rare and cryptic parasex is not enough to confer the benefits of a traditional sexual cycle (Schmid et al., 2004). However, the subsequent discovery of frequent recombination during parasex somewhat disputes this view (Forche et al., 2009; Schmid et al., 2016; Anderson et al., 2019). Furthermore, the genetic and phenotypic diversity generated via parasex may improve fitness in colonized or novel host niches. Parasex has the potential to generate genetic diversity at much higher levels than conventional meiosis because of the added karyotypic diversity (Berman and Hadany, 2012), which makes it a useful mechanism for stress adaptation (Yona et al., 2012; Yang et al., 2021). This argument is bolstered by the strong association between exposure to oxidative damage and CCL (Thomson et al., 2019) as well as the phenomenon of stress-induced mating competence in C. albicans (Popp et al., 2019). Evidence for niche-specific selection of aneuploidies in vivo (Forche et al., 2018; Forche et al., 2019) suggests that parasex may indeed occur in the host but at levels too low to be detected in genetically similar populations without proper tools. In addition, parasex also avoids spore formation that might prove advantageous, especially in vivo, because it could limit activation of the host immune system (Heitman, 2010).
One counterargument for the advantages of parasex in vivo is the dominance of a clonal population structure in parasexual Candida species (Pujol et al., 1993; Boerlin et al., 1996; Forche et al., 1999; Arnavielhe et al., 2000). However, rare sex in otherwise asexually reproducing organisms still has the potential to produce novel genetic variation even from mating between genetically homogenous populations via homozygosis of recessive alleles (Masel and Lyttle, 2011).
Key ancient hybridization events are thought to have played a major role in the evolution of many fungal species and facilitate adaptation to new niches [reviewed in (Steensels et al., 2021)]. Indeed, a hybrid origin was recently proposed for C. albicans (Mixão and Gabaldón, 2020) and hybridization events have been detected for non-albicans Candida species as well (Pryszcz et al., 2015; Schröder et al., 2016; Mixão et al., 2019; Hovhannisyan et al., 2020; O’Brien et al., 2021). Hybridization often restricts ploidy reduction in sexual and parasexual mating products as has been observed in C. dubliniensis-C. albicans hybrids, suggesting that species involved in hybridization events were genetically closely related to allow for chromosome loss and continued mating competency. Ploidy reduction of hybridization and parasexual mating products may be aided by the highly plastic genomes of opportunistic fungal pathogens like C. albicans that may have evolved to not only allow for the generation of high genetic diversity within populations but also support mixing of otherwise genetically distinct lineages. Both, hybridization events and parasexual mating mark periodic breaks from asexual reproduction at microevolutionary and/or macroevolutionary scales, with potential fitness effects for the population via heterosis (hybrid vigor) (Dagilis et al., 2019). The challenge in the future will be to tease apart how and to what extent a variety of evolutionary processes (e.g., hybridization, parasex, aneuploidy, loss of heterozygosity, mitotic recombination, and mutation) have operated independently or in concert to promote the evolution, adaptation, and survival of this species within its mammalian host in general and within specific host niches in particular.
Writing - original draft: AM. Writing - review & editing: AM, AF, and MA. All authors contributed to the article and approved the submitted version.
This work was also supported by the President’s Postdoctoral Scholars Program fellowship from The Ohio State University to AM. This work was supported by National Institutes of Health grant R01AI148788 and NSF CAREER Award 2046863 to MA. AF was supported by National Institutes of Health grant R15 AI090633.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
We would like to thank the Anderson lab for their helpful feedback on aspects of this review and Audra Crouch in particular for assistance in figure design. Figure 3 was created with BioRender.com.
Alby, K., Bennett, R. J. (2011). Interspecies Pheromone Signaling Promotes Biofilm Formation and Same-Sex Mating in Candida Albicans. Proc. Natl. Acad. Sci. 108, 2510–2515. doi: 10.1073/pnas.1017234108
Alby, K., Schaefer, D., Bennett, R. J. (2009). Homothallic and Heterothallic Mating in the Opportunistic Pathogen Candida Albicans. Nature 460, 890–893. doi: 10.1038/nature08252
Alexopoulos, C. J., Mims, C. W., Blackwell, M. (1996). Introductory Mycology (New York: John Wiley and Sons).
Alkafeef, S. S., Yu, C., Huang, L., Liu, H. (2018). Wor1 Establishes Opaque Cell Fate Through Inhibition of the General Co-Repressor Tup1 in Candida Albicans. PloS Genet. 14, e1007176. doi: 10.1371/journal.pgen.1007176
Anderson, M. Z., Porman, A. M., Wang, N., Mancera, E., Huang, D., Cuomo, C. A., et al. (2016). A Multistate Toggle Switch Defines Fungal Cell Fates and Is Regulated by Synergistic Genetic Cues. PloS Genet. 12, e1006353–e1006353. doi: 10.1371/journal.pgen.1006353
Anderson, M. Z., Saha, A., Haseeb, A., Bennett, R. J. (2017). A Chromosome 4 Trisomy Contributes to Increased Fluconazole Resistance in a Clinical Isolate of Candida Albicans. Microbiology 163, 856–865. doi: 10.1099/mic.0.000478
Anderson, J. M., Soll, D. R. (1987). Unique Phenotype of Opaque Cells in the White-Opaque Transition of Candida Albicans. J. Bacteriology. 169, 5579–5588. doi: 10.1128/jb.169.12.5579-5588.1987
Anderson, M. Z., Thomson, G. J., Hirakawa, M. P., Bennett, R. J. (2019). A ‘Parameiosis’ Drives Depolyploidization and Homologous Recombination in Candida Albicans. Nat. Commun. 10, 4388–4388. doi: 10.1038/s41467-019-12376-2
Anderson, J. B., Wickens, C., Khan, M., Cowen, L. E., Federspiel, N., Jones, T., et al. (2001). Infrequent Genetic Exchange and Recombination in the Mitochondrial Genome of Candida Albicans. J. Bacteriol. 183, 865–872. doi: 10.1128/JB.183.3.865-872.2001
Arnavielhe, S., De Meeüs, T., Blancard, A., Mallié, M., Renaud, F., Bastide, J. M. (2000). Multicentric Genetic Study of Candida Albicans Isolates From Non-Neutropenic Patients Using Mlee Typing: Population Structure and Mode of Reproduction. Mycoses 43, 109–117. doi: 10.1046/j.1439-0507.2000.00552.x
Atriwal, T., Azeem, K., Husain, F. M., Hussain, A., Khan, M. N., Alajmi, M. F., et al. (2021). Mechanistic Understanding of Candida Albicans Biofilm Formation and Approaches for Its Inhibition. Front. Microbiol. 12, 638609. doi: 10.3389/fmicb.2021.638609
Barnett, J. A. (2004). A History of Research on Yeasts 7: Enzymic Adaptation and Regulation. Yeast (Chichester England) 21, 703–746. doi: 10.1002/yea.1113
Barr, C. M., Neiman, M., Taylor, D. R. (2005). Inheritance and Recombination of Mitochondrial Genomes in Plants, Fungi and Animals. N. Phytologist. 168, 39–50. doi: 10.1111/j.1469-8137.2005.01492.x
Bennett, R. J., Johnson, A. D. (2003). Completion of a Parasexual Cycle in Candida Albicans by Induced Chromosome Loss in Tetraploid Strains. EMBO J. 22, 2505–2515. doi: 10.1093/emboj/cdg235
Bennett, R. J., Uhl, M. A., Miller, M. G., Johnson, A. D. (2003). Identification and Characterization of a Candida Albicans Mating Pheromone. Mol. Cell Biol. 23, 8189–8201. doi: 10.1128/MCB.23.22.8189-8201.2003
Bergen, M. S., Voss, E., Soll, D. R. (1990). Switching at the Cellular Level in the White-Opaque Transition of Candida Albicans. J. Gen. Microbiol. 136, 1925–1936. doi: 10.1099/00221287-136-10-1925
Berman, J., Hadany, L. (2012). Does Stress Induce (Para)Sex? Implications for Candida Albicans Evolution. Trends Genet. 28, 197–203. doi: 10.1016/j.tig.2012.01.004
Boerlin, P., Boerlin-Petzold, F., Goudet, J., Durussel, C., Pagani, J. L., Chave, J. P., et al. (1996). Typing Candida Albicans Oral Isolates From Human Immunodeficiency Virus-Infected Patients by Multilocus Enzyme Electrophoresis and DNA Fingerprinting. J. Clin. Microbiol. 34, 1235–1248. doi: 10.1128/jcm.34.5.1235-1248.1996
Bougnoux, M. E., Pujol, C., Diogo, D., Bouchier, C., Soll, D. R., d’Enfert, C. (2008). Mating Is Rare Within as Well as Between Clades of the Human Pathogen Candida Albicans. Fungal Genet. Biol. 45, 221–231. doi: 10.1016/j.fgb.2007.10.008
Ciudad, T., Bellido, A., Hermosa, B., Andaluz, E., Larriba, G. (2020). Dlh1, the Candida Albicans Homologue of the Meiosis-Specific Dmc1, Is Not Involved in DNA Repair But Catalyses Spontaneous Interhomologue Recombination and Might Promote Non-Crossover Events. Cell. Microbiol. 22, e13137. doi: 10.1111/cmi.13137
Dagilis, A. J., Kirkpatrick, M., Bolnick, D. I. (2019). The Evolution of Hybrid Fitness During Speciation. PloS Genet. 15, e1008125. doi: 10.1371/journal.pgen.1008125
Daniels, K. J., Srikantha, T., Lockhart, S. R., Pujol, C., Soll, D. R. (2006). Opaque Cells Signal White Cells to Form Biofilms in Candida Albicans. EMBO J. 25, 2240–2252. doi: 10.1038/sj.emboj.7601099
Dumitru, R., Navarathna, D. H., Semighini, C. P., Elowsky, C. G., Dumitru, R. V., Dignard, D., et al. (2007). In Vivo and In Vitro Anaerobic Mating in Candida Albicans. Eukaryot Cell. 6, 465–472. doi: 10.1128/ec.00316-06
Du, H., Zheng, Q., Bing, J., Bennett, R. J., Huang, G. (2018). A Coupled Process of Same- and Opposite-Sex Mating Generates Polyploidy and Genetic Diversity in Candida Tropicalis. PloS Genet. 14, e1007377. doi: 10.1371/journal.pgen.1007377
Ene, I. V., Bennett, R. J. (2009). Hwp1 and Related Adhesins Contribute to Both Mating and Biofilm Formation in Candida Albicans. Eukaryot Cell. 8, 1909–1913. doi: 10.1128/ec.00245-09
Ene, I. V., Lohse, M. B., Vladu, A. V., Morschhäuser, J., Johnson, A. D., Bennett, R. J. (2016). Phenotypic Profiling Reveals That Candida Albicans Opaque Cells Represent a Metabolically Specialized Cell State Compared to Default White Cells. mBio 7, e01269–16. doi: 10.1128/mBio.01269-16
Fanning, S., Xu, W., Solis, N., Woolford, C. A., Filler, S. G., Mitchell, A. P. (2012). Divergent Targets of Candida Albicans Biofilm Regulator Bcr1 In Vitro and In Vivo. Eukaryotic Cell. 11, 896–904. doi: 10.1128/ec.00103-12
Forche, A., Alby, K., Schaefer, D., Johnson, A. D., Berman, J., Bennett, R. J. (2008). The Parasexual Cycle in Candida Albicans Provides an Alternative Pathway to Meiosis for the Formation of Recombinant Strains. PloS Biol. 6, e110. doi: 10.1371/journal.pbio.0060110
Forche, A., Cromie, G., Gerstein, A. C., Solis, N. V., Pisithkul, T., Srifa, W., et al. (2018). Rapid Phenotypic and Genotypic Diversification After Exposure to the Oral Host Niche in. Candida Albicans. Genet. 209, 725–741. doi: 10.1534/genetics.118.301019
Forche, A., Magee, P. T., Selmecki, A., Berman, J., May, G. (2009). Evolution in Candida Albicans Populations During a Single Passage Through a Mouse Host. Genetics 182, 799–811. doi: 10.1534/genetics.109.103325
Forche, A., May, G., Beckerman, J., Kauffman, S., Becker, J., Magee, P. T. (2003). A System for Studying Genetic Changes in Candida Albicans During Infection. Fungal Genet. Biol. 39, 38–50. doi: 10.1016/s1087-1845(02)00585-6
Forche, A., Schönian, G., Gräser, Y., Vilgalys, R., Mitchell, T. G. (1999). Genetic Structure of Typical and Atypical Populations of Candida Albicans From Africa. Fungal Genet. Biol. 28, 107–125. doi: 10.1006/fgbi.1999.1164
Forche, A., Solis, N. V., Swidergall, M., Thomas, R., Guyer, A., Beach, A., et al. (2019). Selection of Candida Albicans Trisomy During Oropharyngeal Infection Results in a Commensal-Like Phenotype. PloS Genet. 15, e1008137. doi: 10.1371/journal.pgen.1008137
Ford, C. B., Funt, J. M., Abbey, D., Issi, L., Guiducci, C., Martinez, D. A., et al. (2015). The Evolution of Drug Resistance in Clinical Isolates of Candida Albicans. Elife 4, e00662. doi: 10.7554/eLife.00662
Geiger, J., Wessels, D., Lockhart, S. R., Soll, D. R. (2004). Release of a Potent Polymorphonuclear Leukocyte Chemoattractant Is Regulated by White-Opaque Switching in Candida Albicans. Infect. Immun. 72, 667–677. doi: 10.1128/IAI.72.2.667-677.2004
Gorman, J. A., Gorman, J. W., Koltin, Y. (1992). Direct Selection of Galactokinase-Negative Mutants of Candida Albicans Using 2-Deoxy-Galactose. Curr. Genet. 21, 203–206. doi: 10.1007/BF00336842
Graeser, Y., Volovsek, M., Arrington, J., Schoenian, G., Presber, W., Mitchell, T. G., et al. (1996). Molecular Markers Reveal That Population Structure of the Human Pathogen Candida Albicans Exhibits Both Clonality and Recombination. Proc. Natl. Acad. Sci. U.S.A. 93, 12473–12477. doi: 10.1073/pnas.93.22.12473
Guan, G., Tao, L., Yue, H., Liang, W., Gong, J., Bing, J., et al. (2019). Environment-Induced Same-Sex Mating in the Yeast Candida Albicans Through the Hsf1-Hsp90 Pathway. PloS Biol. 17, e2006966. doi: 10.1371/journal.pbio.2006966
Gulati, M., Nobile, C. J. (2016). Candida Albicans Biofilms: Development, Regulation, and Molecular Mechanisms. Microbes Infect. 18, 310–321. doi: 10.1016/j.micinf.2016.01.002
Gusa, A., Jinks-Robertson, S. (2019). Mitotic Recombination and Adaptive Genomic Changes in Human Pathogenic Fungi. Genes 10, 901. doi: 10.3390/genes10110901
Harrison, B. D., Hashemi, J., Bibi, M., Pulver, R., Bavli, D., Nahmias, Y., et al. (2014). A Tetraploid Intermediate Precedes Aneuploid Formation in Yeasts Exposed to Fluconazole. PloS Biol. 12, e1001815. doi: 10.1371/journal.pbio.1001815
Heitman, J. (2010). Evolution of Eukaryotic Microbial Pathogens via Covert Sexual Reproduction. Cell Host Microbe 8, 86–99. doi: 10.1016/j.chom.2010.06.011
Hibbett, D. S., Binder, M., Bischoff, J. F., Blackwell, M., Cannon, P. F., Eriksson, O. E., et al. (2007). A Higher-Level Phylogenetic Classification of the Fungi. Mycological Res. 111, 509–547. doi: 10.1016/j.mycres.2007.03.004
Hickman, M. A., Paulson, C., Dudley, A., Berman, J. (2015). Parasexual Ploidy Reduction Drives Population Heterogeneity Through Random and Transient Aneuploidy in. Candida Albicans. Genet. 200, 781–794. doi: 10.1534/genetics.115.178020
Hickman, M., Zeng, G., Forche, A., Hirakawa, M. P., Abbey, D., Harrison, B. D., et al. (2013). The ‘Obligate Diploid’ Candida Albicans Forms Mating-Competent Haploids. Nature 494, 55–59. doi: 10.1038/nature11865
Hirakawa, M. P., Chyou, D. E., Huang, D., Slan, A. R., Bennett, R. J. (2017). Parasex Generates Phenotypic Diversity De Novo and Impacts Drug Resistance and Virulence in Candida Albicans. Genetics 207, 1195–1211. doi: 10.1534/genetics.117.300295
Hovhannisyan, H., Saus, E., Ksiezopolska, E., Gabaldón, T. (2020). The Transcriptional Aftermath in Two Independently Formed Hybrids of the Opportunistic Pathogen Candida Orthopsilosis. mSphere 5, e00282–20. doi: 10.1128/mSphere.00282-20
Huang, G., Wang, H., Chou, S., Nie, X., Chen, J., Liu, H. (2006). Bistable Expression of Wor1, a Master Regulator of White-Opaque Switching in Candida Albicans. Proc. Natl. Acad. Sci. USA 103, 12813–12818. doi: 10.1073/pnas.0605270103
Hull, C. M., Johnson, A. D. (1999). Identification of a Mating Type-Like Locus in the Asexual Pathogenic Yeast. Candida Albicans. Science 285, 1271–1275. doi: 10.1126/science.285.5431.1271
Hull, C. M., Raisner, R. M., Johnson, A. D. (2000). Evidence for Mating of the “Asexual” Yeast Candida Albicans in a Mammalian Host. Science 289, 307–310. doi: 10.1126/science.289.5477.307
Ibrahim, A. S., Magee, B. B., Sheppard, D. C., Yang, M., Kauffman, S., Becker, J., et al. (2005). Effects of Ploidy and Mating Type on Virulence of Candida Albicans. Infect. Immun. 73, 7366–7374. doi: 10.1128/iai.73.11.7366-7374.2005
Jacobsen, M. D., Davidson, A. D., Li, S. Y., Shaw, D. J., Gow, N. A., Odds, F. C. (2008a). Molecular Phylogenetic Analysis of Candida Tropicalis Isolates by Multi-Locus Sequence Typing. Fungal Genet. Biol. 45, 1040–1042. doi: 10.1016/j.fgb.2008.03.011
Jacobsen, M. D., Rattray, A. M. J., Gow, N. A. R., Odds, F. C., Shaw, D. J. (2008b). Mitochondrial Haplotypes and Recombination in Candida Albicans. Med. Mycol. 46, 647–54. doi: 10.1080/13693780801986631 pages.
Janbon, G., Sherman, F., Rustchenko, E. (1998). Monosomy of a Specific Chromosome Determines L-Sorbose Utilization: A Novel Regulatory Mechanism in Candida Albicans. PNAS 95, 5150–5155. doi: 10.1073/pnas.95.9.5150
Jones, T., Federspiel, N. A., Chibana, H., Dungan, J., Kalman, S., Magee, B. B., et al. (2004). The Diploid Genome Sequence of Candida Albicans. Proc. Natl. Acad. Sci. U.S.A. 101, 7329–7334. doi: 10.1073/pnas.0401648101
Knoke, M., Bernhardt, H. (2006). The First Description of an Oesophageal Candidosis by Bernhard Von Langenbeck in 1839. Mycoses 49, 283–287. doi: 10.1111/j.1439-0507.2006.01237.x
Lachke, S. A., Lockhart, S. R., Daniels, K. J., Soll, D. R. (2003). Skin Facilitates Candida Albicans Mating. Infect. Immun. 71, 4970–4976. doi: 10.1128/IAI.71.9.4970-4976.2003
Liang, S.-H., Bennett, R. J. (2019). The Impact of Gene Dosage and Heterozygosity on the Diploid Pathobiont Candida Albicans. J. Fungi. 6, 10. doi: 10.3390/jof6010010
Lin, C. H., Kabrawala, S., Fox, E. P., Nobile, C. J., Johnson, A. D., Bennett, R. J. (2013). Genetic Control of Conventional and Pheromone-Stimulated Biofilm Formation in Candida Albicans. PloS Pathog. 9, e1003305. doi: 10.1371/journal.ppat.1003305
Lockhart, S. R., Pujol, C., Daniels, K. J., Miller, M. G., Johnson, A. D., Pfaller, M. A., et al. (2002). In Candida Albicans, White-Opaque Switchers Are Homozygous for Mating Type. Genetics 162, 737–745. doi: 10.1093/genetics/162.2.737
Lockhart, S. R., Zhao, R., Daniels, K. J., Soll, D. R. (2003). Alpha-Pheromone-Induced “Shmooing” and Gene Regulation Require White-Opaque Switching During Candida Albicans Mating. Eukaryot Cell. 2, 847–855. doi: 10.1128/EC.2.5.847-855.2003
Lohse, M. B., Johnson, A. D. (2008). Differential Phagocytosis of White Versus Opaque Candida Albicans by Drosophila and Mouse Phagocytes. PloS One 3, e1473. doi: 10.1371/journal.pone.0001473
Lohse, M. B., Johnson, A. D. (2009). White-Opaque Switching in Candida Albicans. Curr. Opin. Microbiol. 12, 650–654. doi: 10.1016/j.mib.2009.09.010
Magee, B. B., Legrand, M., Alarco, A. M., Raymond, M., Magee, P. T. (2002). Many of the Genes Required for Mating in Saccharomyces Cerevisiae Are Also Required for Mating in Candida Albicans. Mol. Microbiol. 46, 1345–1351. doi: 10.1046/j.1365-2958.2002.03263.x
Magee, B. B., Magee, P. T. (1997). Wo-2, a Stable Aneuploid Derivative of Candida Albicans Strain Wo-1, can Switch From White to Opaque and Form Hyphae. Microbiology 143, 289–295. doi: 10.1099/00221287-143-2-289
Magee, B. B., Magee, P. T. (2000). Induction of Mating in Candida Albicans by Construction of Mtl a and Mtl Alpha Strains. Science 289, 310–313. doi: 10.1126/science.289.5477.310
Masel, J., Lyttle, D. N. (2011). The Consequences of Rare Sexual Reproduction by Means of Selfing in an Otherwise Clonally Reproducing Species. Theor. Population Biol. 80, 317–322. doi: 10.1016/j.tpb.2011.08.004
McManus, B. A., Coleman, D. C. (2014). Molecular Epidemiology, Phylogeny and Evolution of Candida Albicans. Infect. Genet. Evol. 21, 166–178. doi: 10.1016/j.meegid.2013.11.008
Miller, M., Johnson, A. (2002). White-Opaque Switching in Candida Albicans Is Controlled by Mating-Type Locus Homeodomain Proteins and Allows Efficient Mating. Cell 110, 293 – 302. doi: 10.1016/S0092-8674(02)00837-1
Mitchell, K. F., Zarnowski, R., Andes, D. R. (2016). The Extracellular Matrix of Fungal Biofilms. Adv. Exp. Med. Biol. 931, 21–35. doi: 10.1007/5584_2016_6
Mixão, V., Gabaldón, T. (2020). Genomic Evidence for a Hybrid Origin of the Yeast Opportunistic Pathogen Candida Albicans. BMC Biol. 18, 48. doi: 10.1186/s12915-020-00776-6
Mixão, V., Hansen, A. P., Saus, E., Boekhout, T., Lass-Florl, C., Gabaldón, T. (2019). Whole-Genome Sequencing of the Opportunistic Yeast Pathogen Candida Inconspicua Uncovers Its Hybrid Origin. Front. Genet. 10, 383. doi: 10.3389/fgene.2019.00383
Moran, G. P., Coleman, D. C., Sullivan, D. J. (2012). Candida Albicans Versus Candida Dubliniensis: Why Is C. Albicans More Pathogenic? Int. J. Microbiol. 2012:205921. doi: 10.1155/2012/205921
Muller, H. J. (1964). The Relation of Recombination to Mutational Advance. Mutat. Res. 106, 2–9. doi: 10.1016/0027-5107(64)90047-8
Neiman, M., Taylor, D. R. (2009). The Causes of Mutation Accumulation in Mitochondrial Genomes. Proc. Biol. Sci. 276, 1201–1209. doi: 10.1098/rspb.2008.1758
Nobile, C. J., Fox, E. P., Nett, J. E., Sorrells, T. R., Mitrovich, Q. M., Hernday, A. D., et al. (2012). A Recently Evolved Transcriptional Network Controls Biofilm Development in Candida Albicans. Cell 148, 126–138. doi: 10.1016/j.cell.2011.10.048
Nobile, C. J., Johnson, A. D. (2015). Candida Albicans Biofilms and Human Disease. Annu. Rev. Microbiol. 69, 71–92. doi: 10.1146/annurev-micro-091014-104330
Nobile, C., Nett, J., Andes, D., Mitchell, A. (2006). Function of Candida Albicans Adhesin Hwp1 in Biofilm Formation. Eukaryotic Cell. 5, 1604 – 1610. doi: 10.1128/EC.00194-06
Noble, S. M., Gianetti, B. A., Witchley, J. N. (2017). Candida Albicans Cell-Type Switching and Functional Plasticity in the Mammalian Host. Nat. Rev. Microbiol. 15, 96–108. doi: 10.1038/nrmicro.2016.157
O’Brien, C. E., Oliveira-Pacheco, J., Haase, M. A. B., Hittinger, C. T., Rogers, T. R., Zaragoza, O., et al. (2021). Population Genomics of the Pathogenic Yeast Candida Tropicalis Identifies Hybrid Isolates in Environmental Samples. PloS Pathog. 17, e1009138. doi: 10.1371/journal.ppat.1009138 Ó.C. E.
Odds, F. C., Bougnoux, M.-E., Shaw, D. J., Bain, J. M., Davidson, A. D., Diogo, D., et al. (2007). Molecular Phylogenetics of Candida Albicans. Eukaryot Cell. 6, 1041–1052. doi: 10.1128/EC.00041-07
Pande, K., Chen, C., Noble, S. M. (2013). Passage Through the Mammalian Gut Triggers a Phenotypic Switch That Promotes Candida Albicans Commensalism. Nat. Genet. 45, 1088–91. doi: 10.1038/ng.2710 advance online publication.
Panwar, S. L., Legrand, M., Dignard, D., Whiteway, M., Magee, P. T. (2003). Mfalpha1, the Gene Encoding the Alpha Mating Pheromone of Candida Albicans. Eukaryot Cell. 2, 1350–1360. doi: 10.1128/EC.2.6.1350-1360.2003
Park, Y. N., Daniels, K. J., Pujol, C., Srikantha, T., Soll, D. R. (2013). Candida Albicans Forms a Specialized “Sexual” as Well as “Pathogenic” Biofilm. Eukaryot Cell. 12, 1120–1131. doi: 10.1128/ec.00112-13
Pavelka, N., Rancati, G., Li, R. (2010). Dr Jekyll and Mr Hyde: Role of Aneuploidy in Cellular Adaptation and Cancer. Curr. Opin. Cell Biol. 22, 1–7. doi: 10.1016/j.ceb.2010.06.003
Perry, A. M., Hernday, A. D., Nobile, C. J. (2020). Unraveling How Candida Albicans Forms Sexual Biofilms. J. Fungi. 6, 14. doi: 10.3390/jof6010014
Pierce, C. G., Lopez-Ribot, J. L. (2013). Candidiasis Drug Discovery and Development: New Approaches Targeting Virulence for Discovering and Identifying New Drugs. Expert Opin. Drug Discov 8, 1117–1126. doi: 10.1517/17460441.2013.807245
Pontecorvo, G., Roper, J. A., Forbes, E. (1953). Genetic Recombination Without Sexual Reproduction in Aspergillus Niger. J. Gen. Microbiol. 8, 198–210. doi: 10.1099/00221287-8-1-198
Popp, C., Ramírez-Zavala, B., Schwanfelder, S., Krüger, I., Morschhäuser, J. (2019). Evolution of Fluconazole-Resistant Candida Albicans Strains by Drug-Induced Mating Competence and Parasexual Recombination. mBio 10, e02740–18. doi: 10.1128/mBio.02740-18
Porman, A. M., Alby, K., Hirakawa, M., Bennett, R. J. (2011). Discovery of a Phenotypic Switch Regulating Sexual Mating in the Opportunistic Fungal Pathogen Candida Tropicalis. Proc. Natl. Acad. Sci. USA 108, 21158–21163. doi: 10.1073/pnas.1112076109
Poulter, R., Jeffery, K., Hubbard, M. J., Shepherd, M. G., Sullivan, P. A. (1981). Parasexual Genetic Analysis of Candida Albicans by Spheroplast Fusion. J. Bacteriology. 146, 833–840. doi: 10.1128/jb.146.3.833-840.1981
Pryszcz, L. P., Németh, T., Saus, E., Ksiezopolska, E., Hegedűsová, E., Nosek, J., et al. (2015). The Genomic Aftermath of Hybridization in the Opportunistic Pathogen Candida Metapsilosis. PloS Genet. 11, e1005626. doi: 10.1371/journal.pgen.1005626
Pujol, C., Daniels, K. J., Lockhart, S. R., Srikantha, T., Radke, J. B., Geiger, J., et al. (2004). The Closely Related Species Candida Albicans and Candida Dubliniensis can Mate. Eukaryot Cell. 3, 1015–1027. doi: 10.1128/EC.3.4.1015-1027.2004
Pujol, C., Reynes, J., Renaud, F., Raymond, M., Tibayrenc, M., Ayala, F. J., et al. (1993). The Yeast Candida Albicans has a Clonal Mode of Reproduction in a Population of Infected Human Immunodeficiency Virus-Positive Patients. Proc. Natl. Acad. Sci. U.S.A. 90, 9456–9459. doi: 10.1073/pnas.90.20.9456
Ramirez-Zavala, B., Reuss, O., Park, Y.-N., Ohlsen, K., Morschhauser, J. (2008). Environmental Induction of White-Opaque Switching in Candida Albicans. PloS Pathog. 4, e1000089. doi: 10.1371/journal.ppat.1000089
Rikkerink, E. H., Magee, B. B., Magee, P. T. (1988). Opaque-White Phenotype Transition: A Programmed Morphological Transition in Candida Albicans. J. Bacteriology. 170, 895–899. doi: 10.1128/jb.170.2.895-899.1988
Ropars, J., Maufrais, C., Diogo, D., Marcet-Houben, M., Perin, A., Sertour, N., et al. (2018). Gene Flow Contributes to Diversification of the Major Fungal Pathogen Candida Albicans. Nat. Commun. 9, 2253. doi: 10.1038/s41467-018-04787-4
Salgado, P. S., Yan, R., Taylor, J. D., Burchell, L., Jones, R., Hoyer, L. L., et al. (2011). Structural Basis for the Broad Specificity to Host-Cell Ligands by the Pathogenic Fungus Candida Albicans. Proc. Natl. Acad. Sci. 108, 15775–15779. doi: 10.1073/pnas.1103496108
Sarachek, A., Rhoads, D. D., Schwarzhoff, R. H. (1981). Hybridization of Candida Albicans Through Fusion of Protoplasts. Arch. Microbiol. 129, 1–8. doi: 10.1007/BF00417169
Sasse, C., Hasenberg, M., Weyler, M., Gunzer, M., Morschhaeuser, J. (2013). White-Opaque Switching of Candida Albicans Allows Immune Evasion in an Environment-Dependent Fashion. Eukaryotic Cell. 12, 50–58. doi: 10.1128/ec.00266-12
Scaduto, C. M., Bennett, R. J. (2015). Candida Albicans the Chameleon: Transitions and Interactions Between Multiple Phenotypic States Confer Phenotypic Plasticity. Curr. Opin. Microbiol. 26, 102–108. doi: 10.1016/j.mib.2015.06.016
Scaduto, C. M., Kabrawala, S., Thomson, G. J., Scheving, W., Ly, A., Anderson, M. Z., et al. (2017). Epigenetic Control of Pheromone Mapk Signaling Determines Sexual Fecundity in Candida Albicans. Proc. Natl. Acad. Sci. USA 114, 13780–13785. doi: 10.1073/pnas.1711141115
Schaefer, D., Cote, P., Whiteway, M., Bennett, R. J. (2007). Barrier Activity in Candida Albicans Mediates Pheromone Degradation and Promotes Mating. Eukaryot Cell. 6, 907–918. doi: 10.1128/EC.00090-07
Schmid, J. A. N., Cannon, R. D., Holland, B. (2004). A Futile Act? Thoughts on the Reproductive Biology of Candida Albicans. Mycologist 18, 158–163. doi: 10.1017/S0269-915X(04)00415-X
Schmid, J., Magee, P. T., Holland, B. R., Zhang, N., Cannon, R. D., Magee, B. B. (2016). Last Hope for the Doomed? Thoughts on the Importance of a Parasexual Cycle for the Yeast Candida Albicans. Curr. Genet. 62, 81–85. doi: 10.1007/s00294-015-0516-8
Schröder, M. S., Martinez de San Vicente, K., Prandini, T. H., Hammel, S., Higgins, D. G., Bagagli, E., et al. (2016). Multiple Origins of the Pathogenic Yeast Candida Orthopsilosis by Separate Hybridizations Between Two Parental Species. PloS Genet. 12, e1006404. doi: 10.1371/journal.pgen.1006404
Seervai, R. N. H., Jones, S. K., Hirakawa, M. P., Porman, A. M., Bennett, R. J. (2013). Parasexuality and Ploidy Change in Candida Tropicalis. Eukaryot Cell. 12, 1629–1640. doi: 10.1128/ec.00128-13
Selmecki, A., Maruvka, Y. E., Richmond, P. A., Guillet, M., Shoresh, N., Sorenson, A., et al. (2015). Polyploidy can Drive Rapid Adaptation in Yeast. Nature 519, 349–352. doi: 10.1038/nature14187
Sherwood, R. K., Bennett, R. J. (2009). Fungal Meiosis and Parasexual Reproduction-Lessons From Pathogenic Yeast. Curr. Opin. Microbiol. 12, 599–607. doi: 10.1016/j.mib.2009.09.005
Singh-Babak, S. D., Babak, T., Fraser, H. B., Johnson, A. D. (2021). Lineage-Specific Selection and the Evolution of Virulence in the Candida Clade. Proc. Natl. Acad. Sci. USA 118, e2016818118. doi: 10.1073/pnas.2016818118
Slutsky, B., Staebell, M., Anderson, J., Risen, L., Pfaller, M., Soll, D. R. (1987). “White-Opaque Transition”: A Second High-Frequency Switching System in Candida Albicans. J. Bacteriol. 169, 189–197. doi: 10.1128/jb.169.1.189-197.1987
Smith, A. C., Hickman, M. A. (2020). Host-Induced Genome Instability Rapidly Generates Phenotypic Variation Across Candida Albicans Strains and Ploidy States. mSphere 5, e00433–20. doi: 10.1128/mSphere.00433-20
Steensels, J., Gallone, B., Verstrepen, K. J. (2021). Interspecific Hybridization as a Driver of Fungal Evolution and Adaptation. Nat. Rev. Microbiol. 19, 485–500. doi: 10.1038/s41579-021-00537-4
Takagi, J., Singh-Babak, S. D., Lohse, M. B., Dalal, C. K., Johnson, A. D. (2019). Candida Albicans White and Opaque Cells Exhibit Distinct Spectra of Organ Colonization in Mouse Models of Infection. PloS One 14, e0218037. doi: 10.1371/journal.pone.0218037
Tao, L., Cao, C., Liang, W., Guan, G., Zhang, Q., Nobile, C. J., et al. (2014). White Cells Facilitate Opposite- and Same-Sex Mating of Opaque Cells in Candida Albicans. PloS Genet. 10, e1004737. doi: 10.1371/journal.pgen.1004737
Tavanti, A., Davidson, A. D., Fordyce, M. J., Gow, N. A., Maiden, M. C., Odds, F. C. (2005). Population Structure and Properties of Candida Albicans, as Determined by Multilocus Sequence Typing. J. Clin. Microbiol. 43, 5601–5613. doi: 10.1128/JCM.43.11.5601-5613.2005
Thomson, G. J., Hernon, C., Austriaco, N., Shapiro, R. S., Belenky, P., Bennett, R. J. (2019). Metabolism-Induced Oxidative Stress and DNA Damage Selectively Trigger Genome Instability in Polyploid Fungal Cells. EMBO J. 38, e101597. doi: 10.15252/embj.2019101597
Todd, R. T., Wikoff, T. D., Forche, A., Selmecki, A. (2019). Genome Plasticity in Candida Albicans Is Driven by Long Repeat Sequences. Elife 8, e45954. doi: 10.7554/eLife.45954
Tuch, B. B., Mitrovich, Q. M., Homann, O. R., Hernday, A. D., Monighetti, C. K., de la Vega, F. M., et al. (2010). The Transcriptomes of Two Heritable Cell Types Illuminate the Circuit Governing Their Differentiation. PloS Genet. 6, e1001070. doi: 10.1371/journal.pgen.1001070
Tzung, K. W., Williams, R. M., Scherer, S., Federspiel, N., Jones, T., Hansen, N., et al. (2001). Genomic Evidence for a Complete Sexual Cycle in Candida Albicans. Proc. Natl. Acad. Sci. USA 98, 3249–3253. doi: 10.1073/pnas.061628798
Uppuluri, P., Acosta Zaldívar, M., Anderson, M. Z., Dunn, M. J., Berman, J., Lopez Ribot, J. L., et al. (2018). Candida Albicans Dispersed Cells Are Developmentally Distinct From Biofilm and Planktonic Cells. mBio 9, e01338–18. doi: 10.1128/mBio.01338-18
Uppuluri, P., Chaturvedi, A. K., Srinivasan, A., Banerjee, M., Ramasubramaniam, A. K., Köhler, J. R., et al. (2010). Dispersion as an Important Step in the Candida Albicans Biofilm Developmental Cycle. PloS Pathog. 6, e1000828. doi: 10.1371/journal.ppat.1000828
Usher, J. (2019). The Mechanisms of Mating in Pathogenic Fungi-a Plastic Trait. Genes 10, 831. doi: 10.3390/genes10100831
Uwamahoro, N., Qu, Y., Jelicic, B., Lo, T. L., Beaurepaire, C., Bantun, F., et al. (2012). The Functions of Mediator in Candida Albicans Support a Role in Shaping Species-Specific Gene Expression. PloS Genet. 8, e1002613. doi: 10.1371/journal.pgen.1002613
van Het Hoog, M., Rast, T. J., Martchenko, M., Grindle, S., Dignard, D., Hogues, H., et al. (2007). Assembly of the Candida Albicans Genome Into Sixteen Supercontigs Aligned on the Eight Chromosomes. Genome Biol. 8, R52. doi: 10.1186/gb-2007-8-4-r52
Wang, J. M., Bennett, R. J., Anderson, M. Z. (2018). The Genome of the Human Pathogen Candida Albicans Is Shaped by Mutation and Cryptic Sexual Recombination. mBio 9, e01205–18. doi: 10.1128/mBio.01205-18
Wang, Y., Cao, Y. Y., Jia, X. M., Cao, Y. B., Gao, P. H., Fu, X. P., et al. (2006). Cap1p Is Involved in Multiple Pathways of Oxidative Stress Response in Candida Albicans. FreeRadic Biol. Med. 40, 1201–1209. doi: 10.1016/j.freeradbiomed.2005.11.019
White, D. J., Wolff, J. N., Pierson, M., Gemmell, N. J. (2008). Revealing the Hidden Complexities of Mtdna Inheritance. Mol. Ecology. 17, 4925–4942. doi: 10.1111/j.1365-294X.2008.03982.x
Wisplinghoff, H., Ebbers, J., Geurtz, L., Stefanik, D., Major, Y., Edmond, M. B., et al. (2014). Nosocomial Bloodstream Infections Due to Candida Spp. In the USA: Species Distribution, Clinical Features and Antifungal Susceptibilities. Int. J. Antimicrobial Agents. 43, 78–81. doi: 10.1016/j.ijantimicag.2013.09.005
Xie, J., Du, H., Guan, G., Tong, Y., Kourkoumpetis, T. K., Zhang, L., et al. (2012). N-Acetylglucosamine Induces White-to-Opaque Switching and Mating in Candida Tropicalis, Providing New Insights Into Adaptation and Fungal Sexual Evolution. Eukaryot Cell. 11, 773–782. doi: 10.1128/ec.00047-12
Xie, J., Tao, L., Nobile, C. J., Tong, Y., Guan, G., Sun, Y., et al. (2013). White-Opaque Switching in Natural Mtla/Alpha Isolates of Candida Albicans: Evolutionary Implications for Roles in Host Adaptation, Pathogenesis, and Sex. PloS Biol. 11, e1001525. doi: 10.1371/journal.pbio.1001525
Yang, F., Todd, R. T., Selmecki, A., Jiang, Y.-y., . Cao, Y.-b., Berman, J. (2021). The Fitness Costs and Benefits of Trisomy of Each Candida Albicans Chromosome. Genetics 218, iyab056. doi: 10.1093/genetics/iyab056
Yi, S., Sahni, N., Pujol, C., Daniels, K. J., Srikantha, T., Ma, N., et al. (2009). A Candida Albicans-Specific Region of the α-Pheromone Receptor Plays a Selective Role in the White Cell Pheromone Response. Mol. Microbiol. 71, 925–947. doi: 10.1111/j.1365-2958.2008.06575.x
Yona, A. H., Manor, Y. S., Herbst, R. H., Romano, G. H., Mitchell, A., Kupiec, M., et al. (2012). Chromosomal Duplication Is a Transient Evolutionary Solution to Stress. Proc. Natl. Acad. Sci. USA 109, 21010–21015. doi: 10.1073/pnas.1211150109
Yue, H., Hu, J., Guan, G., Tao, L., Du, H., Li, H., et al. (2016). Discovery of the Gray Phenotype and White-Gray-Opaque Tristable Phenotypic Transitions in Candida Dubliniensis. Virulence 7, 230–242. doi: 10.1080/21505594.2015.1135287
Zhai, B., Ola, M., Rolling, T., Tosini, N. L., Joshowitz, S., Littmann, E. R., et al. (2020). High-Resolution Mycobiota Analysis Reveals Dynamic Intestinal Translocation Preceding Invasive Candidiasis. Nat. Med. 26, 59–64. doi: 10.1038/s41591-019-0709-7
Keywords: Candida, parasex, mating, genetic diversity, recombination
Citation: Mishra A, Forche A and Anderson MZ (2021) Parasexuality of Candida Species. Front. Cell. Infect. Microbiol. 11:796929. doi: 10.3389/fcimb.2021.796929
Received: 18 October 2021; Accepted: 19 November 2021;
Published: 13 December 2021.
Edited by:
Ulrich Kück, Ruhr University Bochum, GermanyReviewed by:
Marie Desnos-Ollivier, Institut Pasteur, FranceCopyright © 2021 Mishra, Forche and Anderson. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Matthew Z. Anderson, YW5kZXJzb24uMzE5NkBvc3UuZWR1
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.