- 1Institute of Biomedical and Environmental Health Research, School of Health and Life Sciences, University of the West of Scotland, Paisley, United Kingdom
- 2School of Medicine, Dentistry and Nursing, University of Glasgow, Glasgow, United Kingdom
- 3Glasgow Biofilms Research Network, Glasgow, United Kingdom
- 4Department of Biological and Biomedical Sciences, School of Health and Life Sciences, Glasgow Caledonian University, Glasgow, United Kingdom
- 5Centre for Experimental Medicine, Queen’s University Belfast, Belfast, Ireland
Candida albicans is an opportunistic pathogen found throughout multiple body sites and is frequently co-isolated from infections of the respiratory tract and oral cavity with Staphylococcus aureus. Herein we present the first report of the effects that S. aureus elicits on the C. albicans transcriptome. Dual-species biofilms containing S. aureus and C. albicans mutants defective in ALS3 or ECE1 were optimised and characterised, followed by transcriptional profiling of C. albicans by RNA-sequencing (RNA-seq). Altered phenotypes in C. albicans mutants revealed specific interaction profiles between fungus and bacteria. The major adhesion and virulence proteins Als3 and Ece1, respectively, were found to have substantial effects on the Candida transcriptome in early and mature biofilms. Despite this, deletion of ECE1 did not adversely affect biofilm formation or the ability of S. aureus to interact with C. albicans hyphae. Upregulated genes in dual-species biofilms corresponded to multiple gene ontology terms, including those attributed to virulence, biofilm formation and protein binding such as ACE2 and multiple heat-shock protein genes. This shows that S. aureus pushes C. albicans towards a more virulent genotype, helping us to understand the driving forces behind the increased severity of C. albicans-S. aureus infections.
Introduction
Candida albicans is typically found as a commensal organism at mucosal and barrier sites, such as the oral cavity, respiratory and gastrointestinal tracts. Under certain conditions, C. albicans is capable of causing opportunistic infections, ranging from superficial to systemic disease, making it one of the most common fungal infections (Brown et al., 2012). The mechanism behind the opportunistic nature of this pathogen lies in its ability to transition from a budding yeast cell to a highly invasive and filamentous hyphal cell, which are typically associated as being commensal and pathogenic phenotypes, respectively (Brown et al., 2012; Lu et al., 2014). Whether acting as a commensal or pathogen, C. albicans frequently co-exists with various bacterial species at mucosal sites, and the clinical significance of polymicrobial infections is becoming more apparent (Delaney et al., 2018). Interactions that take place within these infections can be synergistic, leading to traits such as increased drug resistance, virulence and biofilm formation (Harriott and Noverr, 2011; Kean et al., 2017).
C. albicans is frequently implicated in polymicrobial infections with bacteria such as Staphylococcus aureus, Staphylococcus epidermidis or Pseudomonas aeruginosa (Peters et al., 2012a; Haiko et al., 2019). A considerable amount of infections involving C. albicans and S. aureus are linked to biofilms in conditions such as angular chelitis, cystic fibrosis and diabetic foot ulcers (Peters et al., 2012a; Tsui et al., 2016).
The relationship between C. albicans and S. aureus appears to be beneficial for the bacterium, which utilises the fungi to augment its own virulence and resistance capabilities. S. aureus has been reported to coat itself in the C. albicans extracellular component, β-1,3-glucan in order to increase tolerance to vancomycin (Kong et al., 2016). Our group has also demonstrated that C. albicans increases the virulence of S. aureus in the Galleria mellonella infection model (Kean et al., 2017). Similar increases in virulence in animal infection models have also been shown (Peters and Noverr, 2013; Todd et al., 2019). Indeed, in a murine infection model, C. albicans was shown to have an ability to augment the agr quorum sensing system of S. aureus, resulting in increased alpha- and delta-toxin production, bacterial burden and mortality rates (Wang et al., 2007; Otto, 2014; Todd et al., 2019). Moreover, because the bacteria adhere directly to the fungal hyphae, S. aureus can more readily invade host cells through its close association with the invasive hyphae, which is akin to a needle-stick injection. In the context of complex multispecies communities, C. albicans has been described as a ‘keystone commensal’, suggesting it plays a critical physical role in promoting and maintaining biofilm stability in complex communities (Janus et al., 2016; Young et al., 2020).
When interacting with C. albicans, S. aureus preferentially adheres to the agglutinin like sequence 3 protein (Als3) (Peters et al., 2012b), which is highly expressed during early stages of C. albicans filamentous growth (Sherry et al., 2014). The Als3 protein plays multiple roles in the C. albicans infection cycle. These include the initial adhesion to host epithelial cells which subsequently induces its own endocytosis (Phan et al., 2007). As well as binding to host tissues, Als3 mediates self-adherence as well as Candida-bacteria binding as deletion of ALS3 results in sparse and disorganised biofilms (Nobile et al., 2006a; Peters et al., 2012b). These Candida-bacteria interactions may be reciprocal, as it has been reported that C. albicans Als3 shares over 80% homology with S. aureus collagen binding factor (Sheppard et al., 2004). Several other genes including, but not limited to, hyphal wall protein 1 (HWP1), enhanced filamentous growth protein 1 (EFG1) and extent of cell elongation 1 (ECE1) have been described as being crucial to C. albicans pathogenesis, and appear to be co-expressed alongside ALS3 (Ramage et al., 2002; Nobile et al., 2006b; Moyes et al., 2016). Of these genes, and arguably one of the most important, is ECE1, which encodes a 29 kDa cytolytic protein (named candidalysin) that is essential for virulence and epithelial cell damage in C. albicans infections. Cells lacking ECE1 show no identifiable morphological differences, do not trigger epithelial cell danger response and are avirulent in animal infection models (Moyes et al., 2016). Recent work has revealed that the global repressor Tup1 and transcription factor Ahr1 are both required for expression of ALS3 and ECE1 (Ruben et al., 2020). Together these data highlight the importance of gene networks controlling pathogenicity, but we are still unclear on how these are controlled in dual-species interactions. Therefore, more in-depth analyses of these interactions are required.
Taken together, it is clear that there is a lack of knowledge surrounding the behaviour of C. albicans in the presence of S. aureus. Therefore, we aimed to address these gaps in the literature regarding the transcriptomic response of C. albicans to S. aureus in a dual-species biofilm using a combination of phenotypic and microscopic analyses in combination with RNA-seq. Secondly, we sought to determine the role that key virulence genes, ALS3 and ECE1 play in the regulation of these interactions.
Materials and Methods
Microbial Storage and Standardisation
Candida albicans SC5314, C. albicans als3Δ/Δ (Silverman et al., 2010), C. albicans ece1Δ/Δ (Moyes et al., 2016) and Staphylococcus aureus NCTC 10833 were used in this study. C. albicans strains and S. aureus were grown on Sabourauds Dextrose (SAB) agar (ThermoFisher, Paisley, UK) and Luria Bertani (LB) agar (ThermoFisher), respectively and stored at 4°C. For long-term storage all organisms were stored in glycerol at -80°C.
To prepare overnight broths of each microorganism, one colony of C. albicans was suspended in Yeast Peptone Dextrose (YPD) media (ThermoFisher) and incubated at 30°C with agitation at 200 rpm. Luria Bertani broth (LB, ThermoFisher) was used to grow overnight broths of S. aureus at 37°C. Overnight cultures were washed by centrifugation and subsequent re-suspension in Phosphate Buffered Saline (PBS, Sigma-Aldrich, Dorset, UK). S. aureus cells were diluted to 0.6 OD600, equating to approximately 1x108 cells/mL as determined by serial dilution and colony counting (data not shown). C. albicans concentrations were determined by cell counting on a Neubauer haemocytometer and diluted to 1x106 cells/mL in growth media.
Media Preparation
Todd Hewitt broth (THB, Sigma-Aldrich) was prepared and supplemented with 10 µM menadione and 4 mg/mL hemin (ThermoFisher) and mixed 1:1 with Roswell Park Memorial Institute media (RPMI). Referred to as sTHB from herein. A similar media has been described elsewhere and has been shown to enable the growth of both fungi and bacteria (Montelongo-Jauregui et al., 2016).
Biofilm Growth and Analysis
Following counting of yeast cells and standardisation of bacteria, all cells were added to sTHB media to give a final concentration of 1x106 CFU/mL. Biofilms were grown by adding inoculated growth media to the desired wells of a flat-bottomed, 96-well microtitre plate and Candida was incubated with or without the presence of bacteria for 4 or 24 h. Each type of biofilm was grown with 8 internal replicates and media only controls were included to test for contamination. Following the incubation step, biofilms were washed with PBS and then incubated with 0.05% crystal violet (CV) as described previously (Sherry et al., 2014). CV absorbance was measured at 570 nm using a multi-mode plate reader (FLUOStar Omega, BMG Labtech, Aylesbury, UK).
DNA Extraction and Biofilm Composition Analysis
Early and mature biofilms of C. albicans mutants with and without S. aureus were grown on polymer coverslips before removing biomass via sonication at 35kHz in an ultrasonic water bath in 1 mL PBS for 10 minutes. DNA was extracted from biofilm cells using the Qiagen DNA mini-kit (Qiagen, Hilden, Germany) following the manufacturer’s instructions. Quantitative PCR (qPCR) was then used to determine the total number of cells within each biofilm as described by Kean et al. (2017). qPCR was carried out using the Step-One plus real time PCR machine (Life Technologies, Paisley, UK). The following profile was used: 50°C for 2 min, 95°C for 2 min, followed by 40 cycles of 95°C for 3 s and 60°C for 30 s. Colony forming equivalents (CFE) were calculated compared to a standard curve of serially diluted DNA of each species as previously described (O’Donnell et al., 2016). Species-specific primer sequences are provided in Table 1.
Visualisation of Inter-Kingdom Biofilm Interactions
C. albicans was standardised to 1x106 cells/mL, as described above, and biofilms grown in chamber slides (ThermoFisher) for 2 h at 37°C to induce hyphal formation. Bacteria were standardised to approximately 1x108 cells/mL, stained with 1.5 mM hexidium iodide (ThermoFisher) and incubated at 37˚C for 1 h. Bacterial cells were pelleted by centrifugation and washed twice with PBS. Candida biofilms were washed with PBS following initial incubation. Bacterial cells at 5x107 cells/mL and 1.5 mM calcofluor white (Sigma-Aldrich) were added to the chamber slide for a further hour at 37°C. After a total growth time of 3h, biofilms were then washed with PBS and imaged using an EVOS cell imaging system (ThermoFisher). Calcofluor white and hexidium iodide fluorescence was detected at excitation/emission wavelengths of 357/447 and 531/593 nm, respectively, before overlaying the images.
RNA Extraction and Sequencing
Dual-species biofilms of C. albicans and S. aureus were grown for 4 and 24 h in 1:1 broth in T-75 cell culture flasks (Corning, UK). At each time point, media was removed and PBS was used to remove any non-adherent cells. Biofilm biomass was harvested using a cell scraper and stored in 1mL RNA later (ThermoFisher). Extractions were performed using RiboPure RNA Extraction Kits (ThermoFisher) following the manufacturer’s instructions. RNA quality and quantity was assessed using a Bioanalyser (Agilent, USA), where a minimum RNA integrity number (RIN) of 7.0 and a minimum quantity of 2.5 µg was achieved for each sample. RNA was sequenced by Edinburgh Genomics (genomics.ed.ac.uk) using a NovaSeq 6000 platform to provide 100bp paired end reads.
FastQC was used to assign quality scores to the produced reads and Illumina adaptors and poor-quality reads were trimmed using Trimmomatic. HISTAT2 was then used to align the resulting reads to a reference C. albicans genome (candidagenomedatabase.org) Assembly22 before the number of sequences that were aligned to each gene were counted using HTSeq. The counted genes were subsequently imported to RStudio (version 3.6.3) in which, the DESeq2 package was used to analyse the differentially expressed genes.
Transcriptome Validation
Firstly, single and dual-species biofilms were grown on coverslips for 24h as described above. Following the growth phase, Biomass was removed by sonication and suspended in 1mL PBS. As per the manufacturers instructions, RNA was extracted using the RNeasy mini kit (Qiagen) and then converted into cDNA using High-Capacity cDNA Reverse Transcription Kit (ThermoFisher). qPCR was carried out using the Step-One plus real time PCR machine (Life Technologies). The following profile was used: 50°C for 2 min, 95°C for 2 min, followed by 40 cycles of 95°C for 3 s and 60°C for 30 s. Expression of each gene-of-interest was measured in relation to expression of ACT1 before comparisons between single and dual-species biofilms. Gene primer sequences are provided in Supplementary Table 1.
Statistical Analysis
Figures depicting differential gene expression between C. albicans and S. aureus biofilms were created using the DESeq2 package in RStudio. Data was also visualised as heatmap as and volcano plots utilising R packages pheatmap and EnhancedVolcano. Principle component analysis (PCA) was performed within R to visualise the dimension within the gene expression data which correspond to the most variance.
Gene interaction networks were created using the ClueGO application in Cytoscape (cytoscape.com). All other graphs and analyses were performed in GraphPad Prism (version 7, La Jolla, California, USA). Non-parametric Kruskal-Wallis tests were used to compare means of corrected raw data following biofilm assays followed by Dunn’s test for multiple comparisons. Differences between means were deemed significant where P < 0.05 and a minimum Log2 fold-change of ±1.5 was applied when analysing gene expression data.
Results
In addition to S. aureus, other staphylococcal species such as Staphylococcus epidermidis has displayed an ability to interact synergistically with C. albicans (El-Azizi et al., 2004; Pammi et al., 2013). The strain of S. aureus used herein was selected purposefully and it lacks the full repertoire of biofilm-forming genes and binds preferentially to C. albicans hyphae, increasing confidence that observed changes in C. albicans is a result of direct fungal-bacterial interactions. ALS3 has been shown to significantly influence staphylococcal interactions and expression of this gene has been linked to the expression of the key virulence gene, ECE1 (Peters et al., 2012b; Ruben et al., 2020). Therefore, to begin identifying the role these genes play in the interkingdom interactions we investigated the impact that losing one of these key biofilm genes has on dual-species biofilm formation with S. aureus. Biofilms were grown for 4 and 24 h as mono- or dual species biofilms with S. aureus and crystal violet (CV) was used to assess total biomass (Figure 1). The presence of S. aureus resulted in significant increases (P < 0.0001) in dual-species biofilm biomass when grown with both WT and ece1Δ/Δ C. albicans at both 4 and 24 h (Figures 1A, B). No significant change in biomass was observed in als3Δ/Δ or S. aureus only (Figures 1A–C) biofilms regardless, of biofilm maturity. No considerable differences in fungal cell morphology were observed, regardless of genotype (data not shown). As expected, S. aureus produced a poor biofilm following 4 and 24 h growth (Figure 1C).
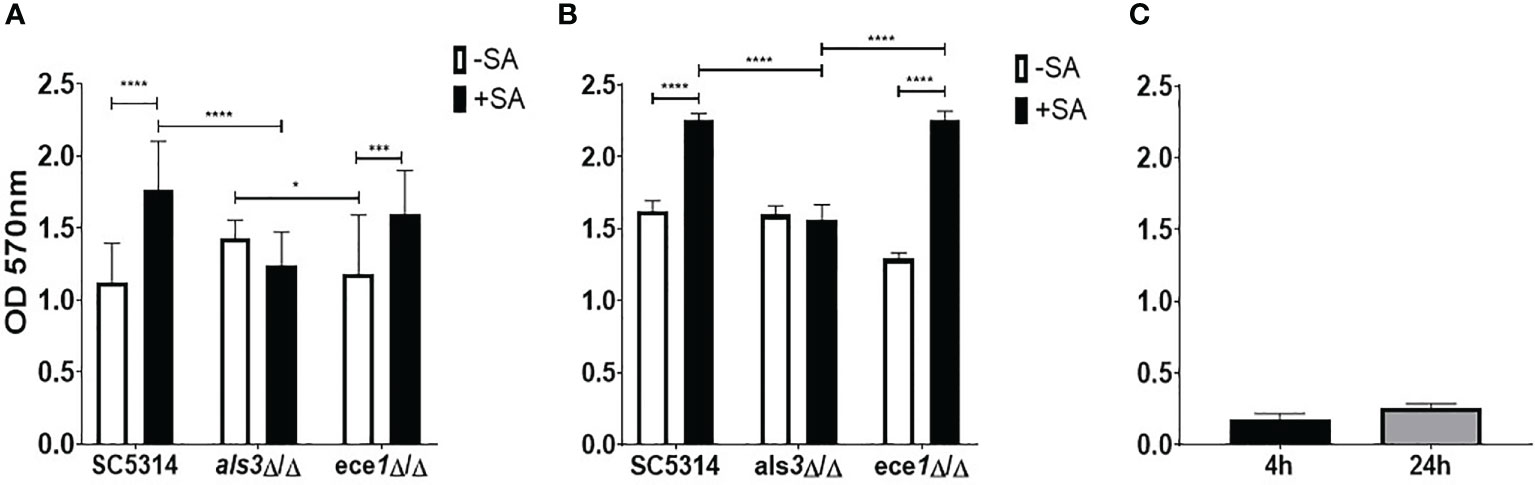
Figure 1 Als3 is responsible for Candida-Staphylococcus interactions. Single and multi-species biofilms containing C. albicans and S. aureus were grown for (A) 4 h and (B) 24 h; (C) single species S. aureus biofilm only. Biomass was quantified by staining using 0.05% crystal violet. Experiments were performed on three separate occasions and error bars represent standard deviation of the mean (*; P < 0.05, ***; P < 0.001, ****; P < 0.0001).
Given the changes associated with biofilm biomass, the quantity of each organism in each biofilm was assessed using qPCR (Figure 2). The number of C. albicans cells in 4 h biofilms remained approximately 5x105 cells/mL regardless of presence or absence of S. aureus (Figure 2A). The average number of WT and ece1Δ/Δ C. albicans in 24 h biofilms increased from 4.8x105 to 3.4x106 and 6.5x105 to 2.8x106 cells/mL (equivalent to a 6.9 and 4.2 fold increase), respectively. As expected, there were significantly fewer C. albicans als3Δ/Δ cells compared to the other C. albicans strains (P < 0.05; Figure 2B). Although there were differences in the total number of C. albicans cells when comparing one strain to another, the presence of S. aureus did not affect the number of fungal cells in any biofilm. The significant increases in biomass thus suggested that there must be an increase in S. aureus colonising the biofilm, which was confirmed by the quantification of total bacterial cells in the dual-species biofilms (Supplementary Figure S1). In early dual-species biofilms, the number of S. aureus cells in biofilms with C. albicans als3Δ/Δ was 100 and 345-fold lower than the WT and ece1Δ/Δ strains, respectively (P < 0.05; Supplementary Figure S1A). A similar trend was observed at 24 h, where the concentration of S. aureus recovered from biofilms grown with WT and the ece1Δ/Δ strains was significantly higher than that of als3Δ/Δ (P < 0.05).
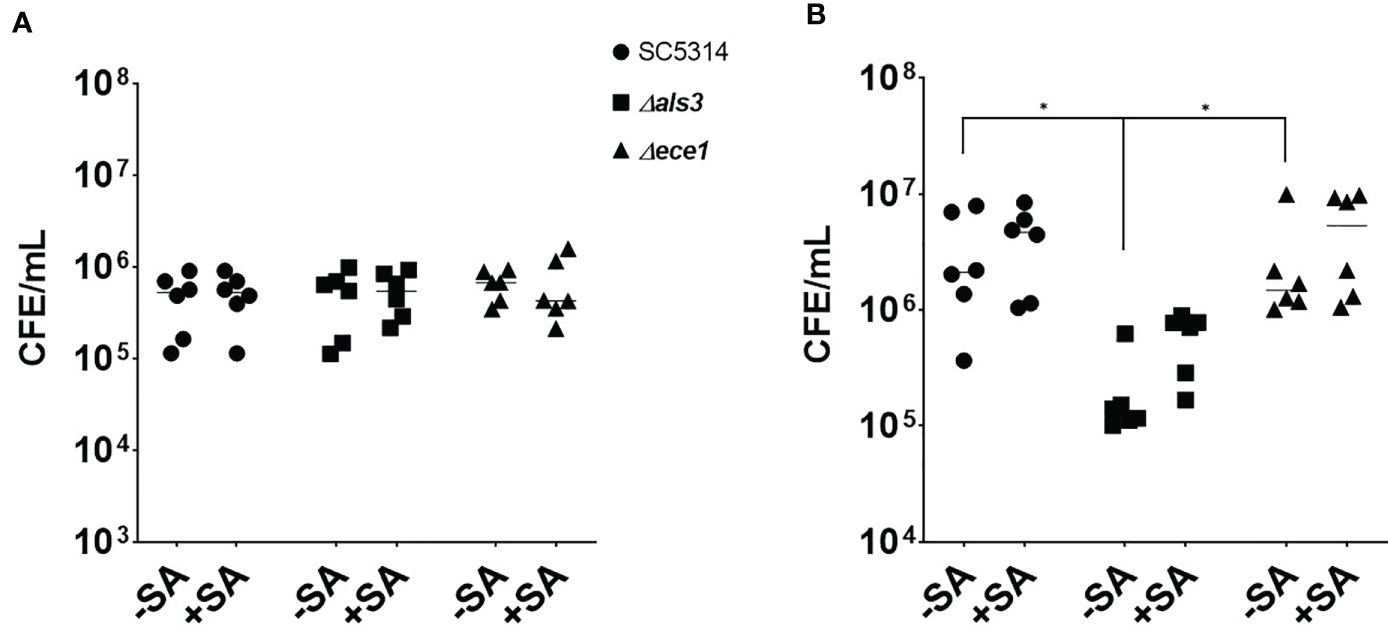
Figure 2 Staphylococcus aureus does not influence the total number of fungal cells in a dual-species biofilm. Biofilm biomass was removed via sonication, DNA was extracted and the total number of fungal cells in each biofilm was quantified using qPCR. The total number of C. albicans cells of (A) 4 h and (B) 24 h biofilms are presented as colony forming equivalents per mL (CFE/mL). Experiments were repeated three times on three separate occasions. Data points represent individual biofilms. CFEs of C. albicans SC5314 and ece1Δ/Δ dual-species biofilms were compared to that of als3Δ/Δ (*, P < 0.05).
The relationship between S. aureus and each C. albicans strain was visualised using fluorescent microscopy. It was observed that S. aureus was able to avidly bind to C. albicans hyphae of WT and ece1Δ/Δ strains (Figure 3) (additional images are provided as part of supplementary data; Supplementary Figure S2). As expected, deletion of ALS3 considerably influenced the relationship between C. albicans and S. aureus, resulting in a decrease in the ability of S. aureus to integrate into the C. albicans biofilm. S. aureus integration was comparable between WT and ece1Δ/Δ strains. Taken together, Figures 1–3 show that when forming dual-species biofilms, S. aureus is closely associated with C. albicans hyphae which is mediated by Als3.
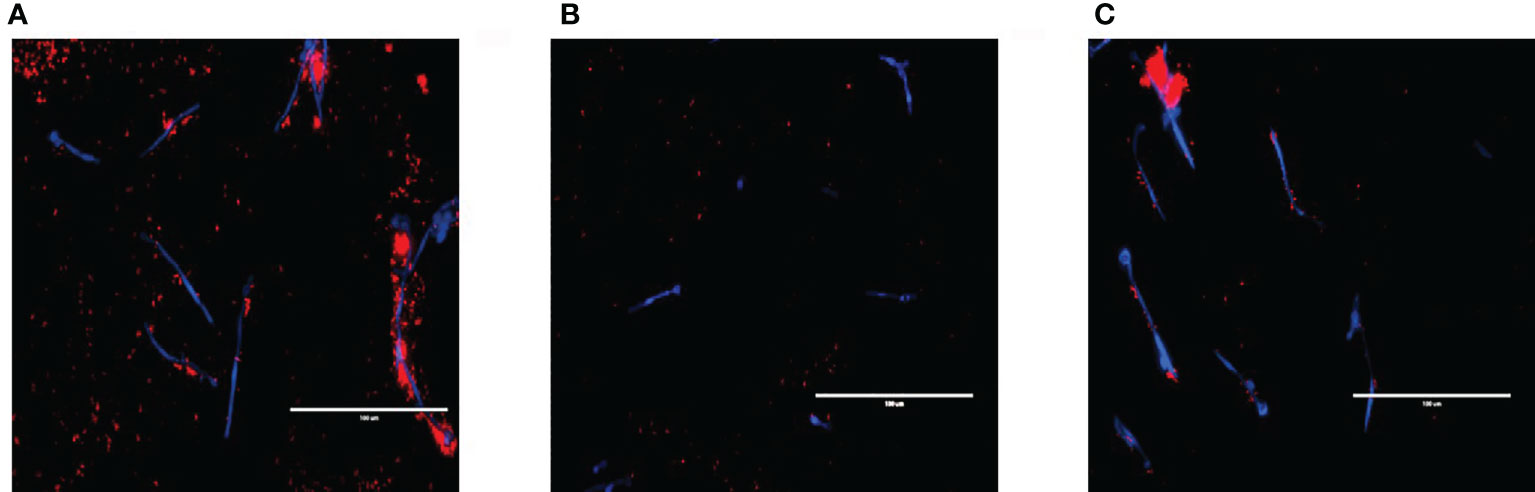
Figure 3 Visualising cell-cell interactions between Candida mutants and S. aureus. C albicans biofilms of (A) wild type SC5314 and (B) ALS3 and (C) ECE1 deletion mutants were grown for 2 h before adding 5x107 cells/mL of S. aureus (which had been pre-stained with hexidium iodide for 1 h prior) for a further hour. Calcofluor white was added to the bacterial inoculum to stain the fungal biofilm before washing the biofilm to remove any non-adherent cells before imaging. Scale bars represent 100μm.
Differential expression (DE) analysis was performed to identify transcriptional changes in C. albicans biofilms when interacting with S. aureus. Multivariate analysis by principal component analysis (PCA) showed variance between samples by biofilm maturity and presence of S. aureus (Figure 4A). The greatest amount of variance is observed between 4 h and 24 h with a large number of differentially expressed genes being identified (Supplementary Figure S3). Limited numbers of genes with increased expression were observed in single species biofilms with only 12 upregulated genes in total being specific to C. albicans only biofilms (Figure 4B). Each dual-species biofilm presented its own distinct patterns of up-regulation with 50 and 101 upregulated genes in WT and als3Δ/Δ dual species biofilms (Figure 4C). A depleted response of the ece1Δ/Δ strain was observed with only 27 upregulated genes identified and only 5 of these were specific to the strain. From these data, it can be concluded that any significant, strain specific changes in C. albicans transcription occurred within mature biofilms, and therefore, future analyses were limited to the 24 h time point. Full lists of upregulated genes in single and dual-species biofilms are provided in Supplementary Tables 2–5.
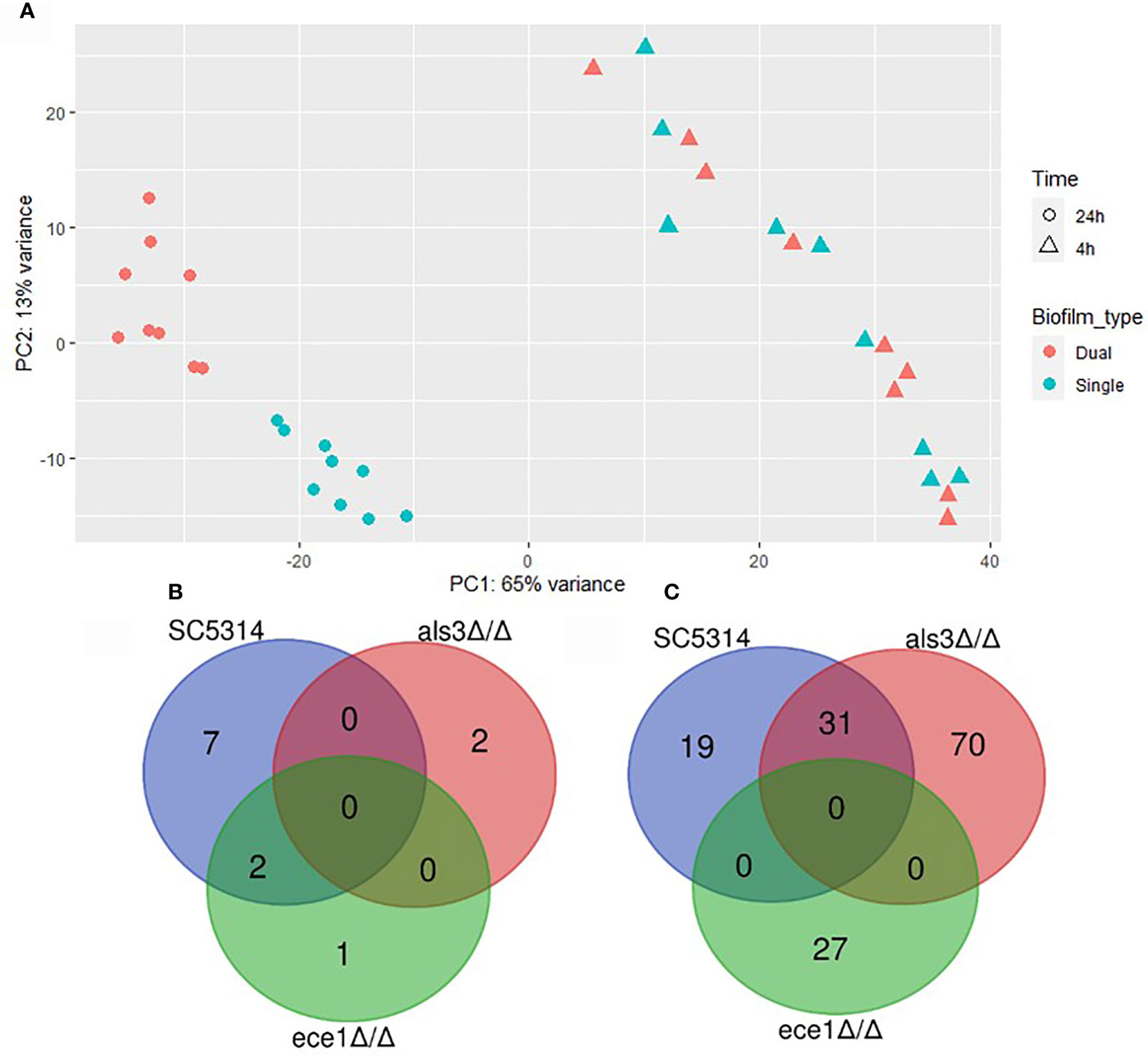
Figure 4 Presence of Staphylococcus aureus determines mature biofilm transcriptome. (A) Principal component analysis plot shows distinct grouping of 24 h samples with the variable of largest variance along PC1 (65%) and second largest along PC2 (13%). Venn diagrams show the number of genes upregulated in 24 h (B) single (C. albicans only) and (C) dual-species (C. albicans and S. aureus) biofilms.
Gene interaction networks show that within a 24h dual-species biofilm there is an upregulation of genes of related function (Figure 5). Genes are grouped by function, taking into account their gene ontology (GO), which forms the nodes (circles) and nodes with similar or related functions are joined by edges (lines). When binding to WT C. albicans (Figure 5A), S. aureus stimulates expression of several genes divided into two distinct groups. Genes in the larger network are classified by functions such as fungal cell wall, biofilm matrix and peptide binding (P < 0.05). As described above, deletion of ALS3 had noticeable effects on biofilm formation, resulting in a vastly different gene expression profile and differentially expressed genes in these biofilms were divided into 4 unrelated groups (Figure 5B). Despite there being a high number of upregulated genes specific to the dual-species als3Δ/Δ biofilm, only 5 gene nodes reached statistical significance (P < 0.05). Genes composing these significant nodes are implicated in processes such as cellular metabolism, protein folding and plasma membrane components. A highly significant yet limited response in genes related to stress responses and protein binding and folding was observed in C. albicans ece1Δ/Δ cells (Figure 5C).
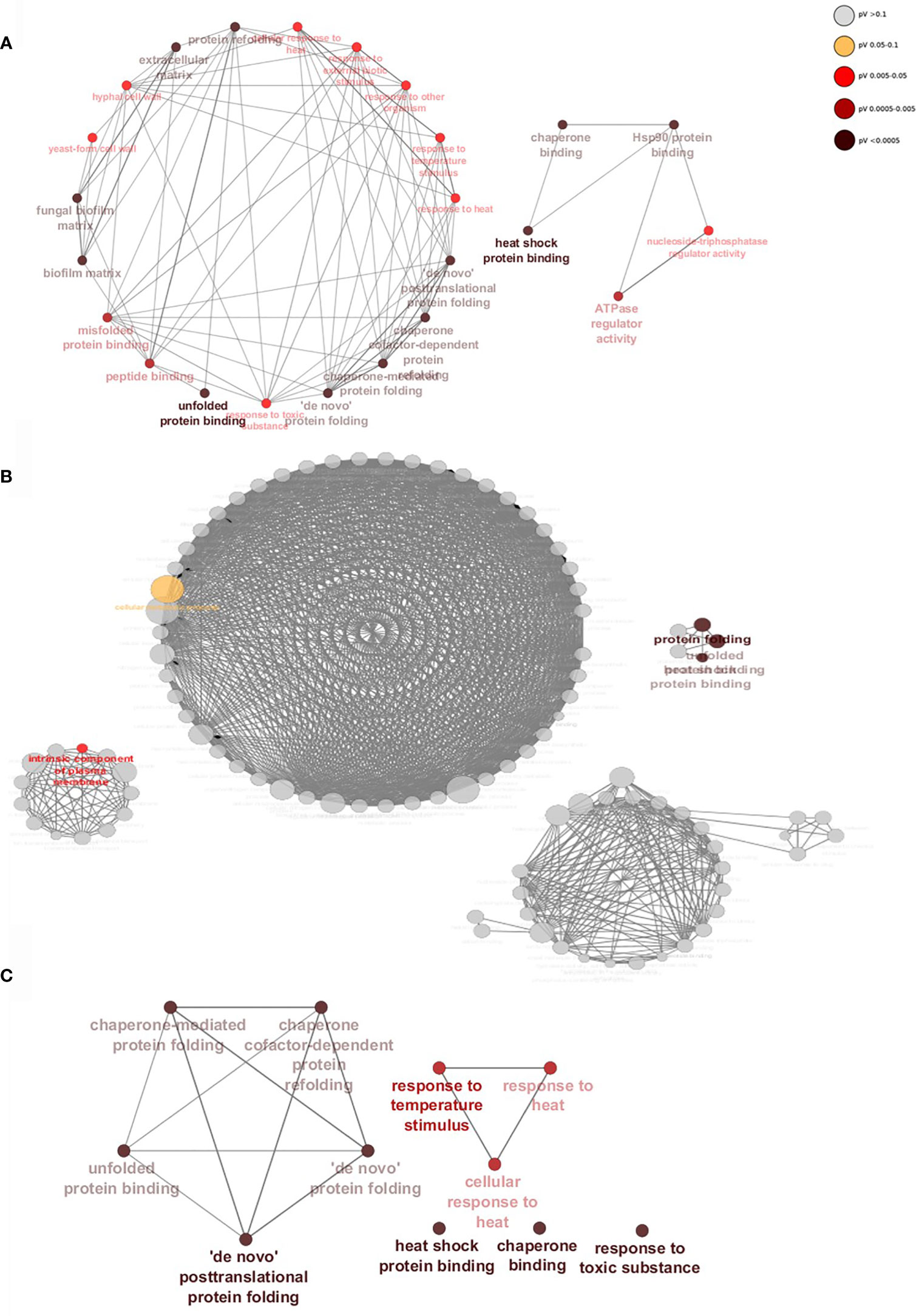
Figure 5 Staphylococcus aureus induces significant upregulation of biofilm-associated genes when binding to Als3. Gene networks show interactions between upregulated genes specific to (A) WT, (B) ALS3 and (C) ECE1 null mutant dual-species, mature biofilms. Genes of similar function are grouped together to form nodes (circles) and nodes with similar functions are linked by edges (lines). Nodes are coloured by levels of significance and node size increases with the number of genes involved in each function. Networks were created using ClueGO.
More in-depth analyses of transcriptional changes at individual gene levels revealed a number of genes involved in biofilm formation and virulence (HSP90, HSP104, FGR41 and ACE2; Figure 6A) to be highly upregulated in WT C. albicans (all over a Log2 fold change of 2). The increased expression of genes related to the als3Δ/Δ strain were found to be typically upregulated in response to external stressors. Some of the genes with the highest increased expression were MDR1, IFD6, HAK1 and CDR4 with a Log2 fold increase of 3.6, 4.6, 5.5 and 4.4, respectively (Figure 6). Genes involved in virulence and biofilm formation (HSP21, HSP104 and ACE2) were also upregulated by more than a Log2 fold increase of 3 in ece1Δ/Δ dual-species biofilms, much like in the WT biofilm (Figure 6C). These findings were confirmed by qPCR analysis (Supplementary Figure S4). Figures 5, 6 show that the response of C. albicans to S. aureus is considerably different when the bacterium is unable to bind to Als3, resulting in the upregulation of stress response genes. Figures 5, 6 suggest that loss of Ece1 does not alter the surface interactions between these two nosocomial pathogens and although a limited transcriptional response is observed in the ece1Δ/Δ strains, 63% of upregulated genes are shared with the WT. Therefore, when bound to the preferred receptor of Als3, S. aureus augments C. albicans virulence through upregulation of virulence and biofilm associated genes.
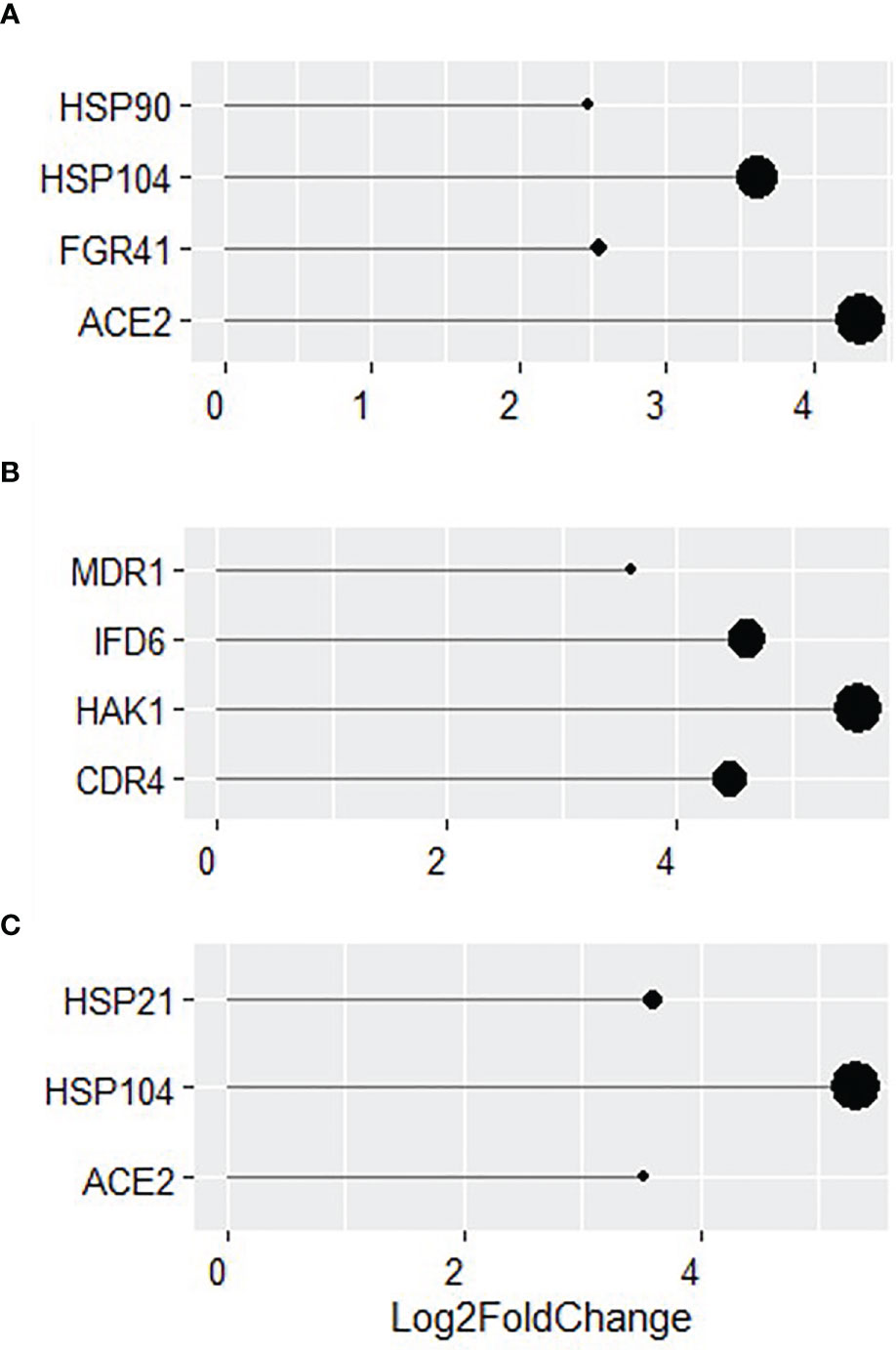
Figure 6 Identification of key genes with increased expression in dual-species biofilms. Log2 fold change of key genes in 24 h dual-species biofilms of (A) WT, (B) als3Δ/Δ and (C) ece1Δ/Δ C albicans with S. aureus.
Discussion
The presence of polymicrobial biofilms during infection affects patient outcome due to decreased susceptibility to antimicrobial treatments and increased duration of hospital stay (Sancho et al., 2012). With increased understanding of the importance of interactions between fungi and bacteria, polymicrobial interkingdom biofilms have been identified in increasing numbers during infection. Of these interkingdom biofilms, C. albicans and S. aureus are frequently co-isolated from conditions such as cystic fibrosis and periodontitis (Valenza et al., 2008; Carolus et al., 2019). We now have a better understanding of the mechanisms by which these interactions benefit S. aureus (Harriott and Noverr, 2009; Kong et al., 2016; Todd et al., 2019) but what remains unclear is how this relationship affects C. albicans. The work carried out herein has therefore aimed to define the C. albicans transcriptome whilst interacting with S. aureus.
Our findings from analysis of dual-species biofilms describe a synergistic relationship between both organisms. Similar findings have been shown by our group in a previous study (Kean et al., 2017), whereby we showed that C. albicans augments bacterial growth through acting as a biofilm substrate to enhance bacterial colonisation, a process we defined as a mycofilm. These findings were confirmed here through biofilm compositional analysis, which revealed a significant increase in bacterial CFEs whilst concentrations of fungal cells remained unchanged. Close interactions between C. albicans and S. aureus were observed in Figure 3. Loss of Ece1 did not appear to affect this relationship which is in line with that reported by Peters et al. (2012b) who identified Als3 as the main binding receptor for S. aureus and reported a significant decrease in the ability of S. aureus to bind to C. albicans hyphae following ALS3 deletion. These findings were more recently confirmed by (Van Dyck et al., 2021).
Although lacking the ability to elicit a proper immune response in the host, ece1Δ/Δ is still capable of forming robust biofilms (as observed in Figure 1) due to the significant up regulation of protein folding and binding genes as shown by the gene interaction networks in Figure 5. The same strain was also found to not exhibit any significant morphological differences to WT C. albicans (Moyes et al., 2016). It could be hypothesised that these cells may also prove to be more tolerant to external stimuli such as heat, which can be inferred from the significant upregulation of multiple heat-shock genes (HSP104, HSP21 and HSP70).
Data presented in Figure 4 showed that a limited number of transcriptional changes occurred in ece1Δ/Δ cells in comparison to the WT. Based on previous data describing Ece1 as a key virulence protein, it was hypothesised that transcriptional changes would mirror that of WT C. albicans. However, there were considerably fewer differentially regulated genes in the ECE1 null mutant than other strains. Transcriptional analysis revealed that PGA4 was downregulated in single-species ece1Δ/Δ biofilms when compared to WT (Figure S5). Expression of PGA4 is required for proper cell wall biosynthesis whilst playing a minor role in regulating responses to antibiotics (Ene et al., 2012). As shown in Figure S6, expression of PGA4 is linked to expression of several other cell wall related genes such as CHT2, ECM331, BGL2 and MP65 (Heilmann et al., 2011; Gil-Bona et al., 2018). This differential cell wall makeup may provide insight as to why the transcriptome of ECE1 null strains responds differently to S. aureus compared to WT C. albicans.
Binding of S. aureus to WT and ece1Δ/Δ C. albicans induces upregulation of several genes whose functions are closely related to biofilm and hyphal formation. Genes involved in these processes include virulence, adhesion and filamentation genes such as HSP90. HSP90 is the most commonly studied heat shock protein in C. albicans, due to its involvement as a transcription factor in several virulence associated pathways such as biofilm formation and hyphal morphogenesis (O’Meara et al., 2017). Other key upregulated genes include HSP104, FGR41 and ACE2, which also play key roles in effective formation of hyphae and biofilms (Kelly et al., 2004; Fiori et al., 2012; Lan et al., 2017).
GO term analysis of WT strains revealed multiple groups of genes whose functions are related to biofilm formation such as peptide binding and extra-cellular matrix (ECM) formation. ECM is a key factor behind the recalcitrant nature of biofilms towards antimicrobial therapy (Singh et al., 2017). As described by Kong et al. (2016), C. albicans ECM components protect S. aureus. It can therefore, be deduced from this that S. aureus induces an upregulation of genes involved in C. albicans biofilm and ECM production such as GLX3 (Cabello et al., 2018) to protect itself and the fungal cells from antimicrobials. Recent work has also shown that not only does S. aureus promote ECM production in C. albicans but the fungi also decreases expression of S. aureus Nuc. Nuc cleaves extracellular DNA and promotes biofilm dispersal (Vila et al., 2021). From this it can be deduced that C. albicans promotes S. aureus ECM production in S. aureus and vice versa. This supports data presented in Figure 1 and previous findings by Kean et al. (2017) who reported an increase in biofilm formation and virulence in a dual-species inoculum.
As mentioned above, despite deletion of ALS3 significantly altering the binding pattern of S. aureus it was not abolished. This resulted in increased expression of over 100 highly interconnected genes involved in biological processes, such as response to drugs and cellular response to stress as demonstrated in Figure 5. Although several genes were upregulated in both WT and als3Δ/Δ dual-species biofilms, there was minimal crossover of enriched GO terms in gene networks. This suggests that even though similar genes are upregulated in each strain, deletion of ALS3 significantly alters how C. albicans interacts with and responds to S. aureus. Among the genes up regulated were CDR4, MDR1 and CAP1. Although there is no confirmed role for CDR4, it is upregulated in the C. albicans core stress response (Enjalbert et al., 2006). Similar to CDR1, CDR4 belongs to the ABC superfamily of efflux pumps, which can lead to speculation that the protein encoded by CDR4 is also involved in drug resistance. MDR1 and CAP1 are also induced by external stressors such as antifungal drugs and oxidative stress, respectively (Feng et al., 2018). Therefore, it can be deduced that when S. aureus binds to other adhesion proteins other than Als3, it triggers C. albicans stress response pathways. Although this stress does not appear to hinder hyphal formation, virulence may be attenuated through loss of Als3.
Despite this study accurately describing the effects of S. aureus on the transcriptome of C. albicans, it is not without its limitations. For example, this study used a biofilm-deficient strain of S. aureus to focus more on direct fungal-bacterial interactions. The use of a biofilm positive bacterial strain is likely to interact with the C. albicans strains used herein differently, therefore resulting in a different fungal response. This would then help create a more detailed picture of how S. aureus influences the C. albicans transcriptome. Additionally, transcriptomics is not without its drawbacks such as this study only captured the C. albicans transcriptome at 4 and 24 h, which may be significantly different to earlier or late time-points. However, the inclusion of additional permutations in transcriptomics experiments can significantly increase costs. Previous work by Cue and colleagues identified that the S. aureus strain herein presented a biofilm deficient phenotype via secretion of a heat-stable peptide (Cue et al., 2015). This peptide was shown to inhibit biofilm formation in other S. aureus strains and could therefore be a strain specific and contributing factor towards the transcriptomic response observed above.
To conclude, this study is the first to report on the changes in the C. albicans transcriptome caused by the closely associated bacterial pathogen, S. aureus. We describe, under normal circumstances in WT C. albicans, an upregulation of biofilm and virulence associated genes when S. aureus adheres to Als3 that likely leads to a more virulent phenotype. We also show that ECE1 is not required for staphylococci to interact with C. albicans and that the relationship between these two organisms is beneficial for the bacterium as well as the yeast through upregulation of adhesion and biofilm formation genes. S. aureus induces the upregulation of virulence-associated genes in ece1Δ/Δ dual-species that may compensate for the loss of virulence that comes with the loss of candidalysin. However, further experiments are required to discern if a virulent phenotype is indeed restored. Exploring these interactions in more depth will continue to unveil additional mechanisms of interaction, which may help to explain their frequent co-isolation in biofilm-related infections and identify potential, novel antimicrobial therapies to combat these complex biofilm communities in a diverse range of clinically important contexts.
Data Availability Statement
The datasets presented in this study can be found in NCBI-SRA under BioProject number PRJNA731052.
Author Contributions
BS, CD, and EM participated in study design, experimental procedures and data analysis and were also responsible for preparation of the manuscript. JB, RK, GL, CW, and WM participated in study design and preparation of the manuscript. LM participated in preparation and critical appraisal of the manuscript. GR conceived the study, participated in study design and data analysis and was responsible for producing the final manuscript. All authors contributed to the article and approved the submitted version.
Funding
BS is supported by a matched Ph.D. studentship provided by the University of the West of Scotland to support the Borders and Regions Airways Training Hub project (BREATH; INT-VA/045) which is funded by the European Union (EU), under the INTERREG VA Programme, managed by the Special EU Programmes Body. We would also like to acknowledge the funding support of the BBSRC Industrial CASE PhD studentship for Christopher Delaney (BB/P504567/1).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We would like to thank Dr Angela Nobbs of the University of Bristol and Dr Julian Naglik from Kings College London for gifting the als3Δ/Δ and ece1Δ/Δ C. albicans strains used in this study. Our thanks also go to Dr Bernhard Hube of Friedrich Schiller University for creation of the ece1Δ/Δ strain.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcimb.2021.791523/full#supplementary-material
References
Brown, G. D., Denning, D. W., Gow, N. A. R., Levitz, S. M., Netea, M. G., White, T. C. (2012). Hidden Killers: Human Fungal Infections. Sci. Trans. Med. 4, 165rv13–165rv13. doi: 10.1126/science.1222236
Cabello, L., Gómez-Herreros, E., Fernández-Pereira, J., Maicas, S., Martínez-Esparza, M. C., De Groot, P. W. J., et al. (2018). Deletion of GLX3 in Candida Albicans Affects Temperature Tolerance, Biofilm Formation and Virulence. FEMS Yeast Res. 19. doi: 10.1093/femsyr/foy124
Carolus, H., Van Dyck, K., Van Dijck, P. (2019). Candida Albicans and Staphylococcus Species: A Threatening Twosome. Front. Microbiol. 10. doi: 10.3389/fmicb.2019.02162
Cue, D., Junecko, J. M., Lei, M. G., Blevins, J. S., Smeltzer, M. S., Lee, C. Y. (2015). SaeRS-Dependent Inhibition of Biofilm Formation in Staphylococcus Aureus Newman. PloS One 10, e0123027–e0123027. doi: 10.1371/journal.pone.0123027
Delaney, C., Kean, R., Short, B., Tumelty, M., Mclean, W., Nile, C. J., et al. (2018). Fungi at the Scene of the Crime: Innocent Bystanders or Accomplices in Oral Infections? Curr. Clin. Microbiol. Rep. 5, 190–200. doi: 10.1007/s40588-018-0100-3
El-Azizi, M. A., Starks, S. E., Khardori, N. (2004). Interactions of Candida Albicans With Other Candida Spp. And Bacteria in the Biofilms. J. Appl. Microbiol. 96, 1067–1073. doi: 10.1111/j.1365-2672.2004.02213.x
Ene, I. V., Heilmann, C. J., Sorgo, A. G., Walker, L. A., De Koster, C. G., Munro, C. A., et al. (2012). Carbon Source-Induced Reprogramming of the Cell Wall Proteome and Secretome Modulates the Adherence and Drug Resistance of the Fungal Pathogen Candida Albicans. Proteomics 12, 3164–3179. doi: 10.1002/pmic.201200228
Enjalbert, B., Smith, D. A., Cornell, M. J., Alam, I., Nicholls, S., Brown, A. J. P., et al. (2006). Role of the Hog1 Stress-Activated Protein Kinase in the Global Transcriptional Response to Stress in the Fungal Pathogen Candida Albicans. Mol. Biol. Cell 17, 1018–1032. doi: 10.1091/mbc.e05-06-0501
Feng, W., Yang, J., Yang, L., Li, Q., Zhu, X., Xi, Z., et al. (2018). Research of Mrr1, Cap1 and MDR1 in Candida Albicans Resistant to Azole Medications. Exp. Ther. Med. 15, 1217–1224. doi: 10.3892/etm.2017.5518
Fiori, A., Kucharíková, S., Govaert, G., Cammue, B. P. A., Thevissen, K., Van Dijck, P. (2012). The Heat-Induced Molecular Disaggregase Hsp104 of Candida Albicans Plays a Role in Biofilm Formation and Pathogenicity in a Worm Infection Model. Eukaryotic Cell 11, 1012–1020. doi: 10.1128/EC.00147-12
Gil-Bona, A., Amador-García, A., Gil, C., Monteoliva, L. (2018). The External Face of Candida Albicans: A Proteomic View of the Cell Surface and the Extracellular Environment. J. Proteomics 180, 70–79. doi: 10.1016/j.jprot.2017.12.002
Haiko, J., Saeedi, B., Bagger, G., Karpati, F., Özenci, V. (2019). Coexistence of Candida Species and Bacteria in Patients With Cystic Fibrosis. Eur. J. Clin. Microbiol. Infect. Dis. Off. Publ. Eur. Soc. Clin. Microbiol. 38, 1071–1077. doi: 10.1007/s10096-019-03493-3
Harriott, M. M., Noverr, M. C. (2009). Candida Albicans and Staphylococcus Aureus Form Polymicrobial Biofilms: Effects on Antimicrobial Resistance. Antimicrob Agents chemother 53, 3914–3922. doi: 10.1128/AAC.00657-09
Harriott, M. M., Noverr, M. C. (2011). Importance of Candida-Bacterial Polymicrobial Biofilms in Disease. Trends Microbiol. 19, 557–563. doi: 10.1016/j.tim.2011.07.004
Heilmann, C. J., Sorgo, A. G., Siliakus, A. R., Dekker, H. L., Brul, S., De Koster, C. G., et al. (2011). Hyphal Induction in the Human Fungal Pathogen Candida Albicans Reveals a Characteristic Wall Protein Profile. Microbiol. (Reading) 157, 2297–2307. doi: 10.1099/mic.0.049395-0
Janus, M. M., Willems, H. M., Krom, B. P. (2016). Candida Albicans in Multispecies Oral Communities; A Keystone Commensal? Adv. Exp. Med. Biol. 931, 13–20. doi: 10.1007/5584_2016_5
Kean, R., Rajendran, R., Haggarty, J., Townsend, E. M., Short, B., Burgess, K. E., et al. (2017). Candida Albicans Mycofilms Support Staphylococcus Aureus Colonization and Enhances Miconazole Resistance in Dual-Species Interactions. Front. Microbiol. 8. doi: 10.3389/fmicb.2017.00258
Kelly, M. T., Maccallum, D. M., Clancy, S. D., Odds, F. C., Brown, A. J. P., Butler, G. (2004). The Candida Albicans CaACE2 Gene Affects Morphogenesis, Adherence and Virulence. Mol. Microbiol. 53, 969–983. doi: 10.1111/j.1365-2958.2004.04185.x
Kong, E. F., Tsui, C., Kucharíková, S., Andes, D., Van Dijck, P., Jabra-Rizk, M. A. (2016). Commensal Protection of Staphylococcus Aureus Against Antimicrobials by Candida Albicans Biofilm Matrix. mBio 7, e01365–e01316. doi: 10.1128/mBio.01365-16
Lan, Y.-B., Huang, Y.-Z., Qu, F., Li, J.-Q., Ma, L.-J., Yan, J., et al. (2017). Time Course of Global Gene Expression Alterations in Candida Albicans During Infection of HeLa Cells. Bosnian J. Basic Med. Sci. 17, 120–131. doi: 10.17305/bjbms.2017.1667
Lu, Y., Su, C., Liu, H. (2014). Candida Albicans Hyphal Initiation and Elongation. Trends Microbiol. 22, 707–714. doi: 10.1016/j.tim.2014.09.001
Montelongo-Jauregui, D., Srinivasan, A., Ramasubramanian, A. K., Lopez-Ribot, J. L. (2016). An In Vitro Model for Oral Mixed Biofilms of Candida Albicans and Streptococcus Gordonii in Synthetic Saliva. Front. Microbiol. 7, 686–686. doi: 10.3389/fmicb.2016.00686
Moyes, D. L., Wilson, D., Richardson, J. P., Mogavero, S., Tang, S. X., Wernecke, J., et al. (2016). Candidalysin is a Fungal Peptide Toxin Critical for Mucosal Infection. Nature 532, 64–68. doi: 10.1038/nature17625
Nobile, C. J., Andes, D. R., Nett, J. E., Smith, F. J., Yue, F., Phan, Q. T., et al. (2006a). Critical Role of Bcr1-Dependent Adhesins in C. Albicans Biofilm Formation In Vitro and In Vivo. PloS Pathog. 2, e63. doi: 10.1371/journal.ppat.0020063
Nobile, C. J., Nett, J. E., Andes, D. R., Mitchell, A. P. (2006b). Function of Candida Albicans Adhesin Hwp1 in Biofilm Formation. Eukaryotic Cell 5, 1604–1610. doi: 10.1128/EC.00194-06
O'donnell, L. E., Smith, K., Williams, C., Nile, C. J., Lappin, D. F., Bradshaw, D., et al. (2016). Dentures are a Reservoir for Respiratory Pathogens. J. Prosthodontics 25, 99–104. doi: 10.1111/jopr.12342
O'meara, T. R., Robbins, N., Cowen, L. E. (2017). The Hsp90 Chaperone Network Modulates Candida Virulence Traits. Trends Microbiol. 25, 809–819. doi: 10.1016/j.tim.2017.05.003
Otto, M. (2014). Staphylococcus Aureus Toxins. Curr. Opin. Microbiol. 17, 32–37. doi: 10.1016/j.mib.2013.11.004
Pammi, M., Liang, R., Hicks, J., Mistretta, T.-A., Versalovic, J. (2013). Biofilm Extracellular DNA Enhances Mixed Species Biofilms of Staphylococcus Epidermidis and Candida Albicans. BMC Microbiol. 13, 1–13. doi: 10.1186/1471-2180-13-257
Peters, B. M., Jabra-Rizk, M. A., O'may, G. A., Costerton, J. W., Shirtliff, M. E. (2012a). Polymicrobial Interactions: Impact on Pathogenesis and Human Disease. Clin. Microbiol. Rev. 25, 193–213. doi: 10.1128/CMR.00013-11
Peters, B. M., Noverr, M. C. (2013). Candida Albicans-Staphylococcus Aureus Polymicrobial Peritonitis Modulates Host Innate Immunity. Infect Immun. 81, 2178–2189. doi: 10.1128/IAI.00265-13
Peters, B. M., Ovchinnikova, E. S., Krom, B. P., Schlecht, L. M., Zhou, H., Hoyer, L. L., et al. (2012b). Staphylococcus Aureus Adherence to Candida Albicans Hyphae is Mediated by the Hyphal Adhesin Als3p. Microbiology 158, 2975. doi: 10.1099/mic.0.062109-0
Phan, Q. T., Myers, C. L., Fu, Y., Sheppard, D. C., Yeaman, M. R., Welch, W. H., et al. (2007). Als3 Is a Candida Albicans Invasin That Binds to Cadherins and Induces Endocytosis by Host Cells. PloS Biol. 5, e64. doi: 10.1371/journal.pbio.0050064
Ramage, G., Vandewalle, K., López-Ribot, J. L., Wickes, B. L. (2002). The Filamentation Pathway Controlled by the Efg1 Regulator Protein is Required for Normal Biofilm Formation and Development in Candida Albicans. FEMS Microbiol. Lett. 214, 95–100. doi: 10.1111/j.1574-6968.2002.tb11330.x
Ruben, S., Garbe, E., Mogavero, S., Albrecht-Eckardt, D., Hellwig, D., Häder, A., et al. (2020). Ahr1 and Tup1 Contribute to the Transcriptional Control of Virulence-Associated Genes in Candida Albicans. mBio 11. doi: 10.1128/mBio.00206-20
Sancho, S., Artero, A., Zaragoza, R., Camarena, J., González, R., Nogueira, J. (2012). Impact of Nosocomial Polymicrobial Bloodstream Infections on the Outcome in Critically Ill Patients. Eur. J. Clin. Microbiol. Infect. Dis. 31, 1791–1796. doi: 10.1007/s10096-011-1503-8
Sheppard, D. C., Yeaman, M. R., Welch, W. H., Phan, Q. T., Fu, Y., Ibrahim, A. S., et al. (2004). Functional and Structural Diversity in the Als Protein Family of Candida Albicans. J. Biol. Chem. 279, 30480–30489. doi: 10.1074/jbc.M401929200
Sherry, L., Rajendran, R., Lappin, D. F., Borghi, E., Perdoni, F., Falleni, M., et al. (2014). Biofilms Formed by Candida Albicans Bloodstream Isolates Display Phenotypic and Transcriptional Heterogeneity That Are Associated With Resistance and Pathogenicity. BMC Microbiol. 14, 182–182. doi: 10.1186/1471-2180-14-182
Silverman, R. J., Nobbs, A. H., Vickerman, M. M., Barbour, M. E., Jenkinson, H. F. (2010). Interaction of Candida Albicans Cell Wall Als3 Protein With Streptococcus Gordonii SspB Adhesin Promotes Development of Mixed-Species Communities. Infect. Immun. 78, 4644–4652. doi: 10.1128/IAI.00685-10
Singh, S., Singh, S. K., Chowdhury, I., Singh, R. (2017). Understanding the Mechanism of Bacterial Biofilms Resistance to Antimicrobial Agents. Open Microbiol. J. 11, 53–62. doi: 10.2174/1874285801711010053
Todd, O. A., Fidel, P. L., Harro, J. M., Hilliard, J. J., Tkaczyk, C., Sellman, B. R., et al. (2019). Candida Albicans Augments Staphyloccus Aureus Virulence by Engaging the Staphylococcal Quorum Sensing System. mBio 10, e00910–e00919. doi: 10.1128/mBio.00910-19
Tsui, C., Kong, E. F., Jabra-Rizk, M. A. (2016). Pathogenesis of Candida Albicans Biofilm. Pathog. Dis. 74. doi: 10.1093/femspd/ftw018
Valenza, G., Tappe, D., Turnwald, D., Frosch, M., König, C., Hebestreit, H., et al. (2008). Prevalence and Antimicrobial Susceptibility of Microorganisms Isolated From Sputa of Patients With Cystic Fibrosis. J. Cystic Fibrosis 7, 123–127. doi: 10.1016/j.jcf.2007.06.006
Van Dyck, K., Viela, F., Mathelié-Guinlet, M., Demuyser, L., Hauben, E., Jabra-Rizk, M. A., et al. (2021). Adhesion of Staphylococcus Aureus to Candida Albicans During Co-Infection Promotes Bacterial Dissemination Through the Host Immune Response. Front. Cell. Infect Microbiol. 10, 624839–624839. doi: 10.3389/fcimb.2020.624839
Vila, T., Kong, E. F., Montelongo-Jauregui, D., Van Dijck, P., Shetty, A. C., Mccracken, C., et al. (2021). Therapeutic Implications of C. Albicans-S. Aureus Mixed Biofilm in a Murine Subcutaneous Catheter Model of Polymicrobial Infection. Virulence 12, 835–851. doi: 10.1080/21505594.2021.1894834
Wang, R., Braughton, K. R., Kretschmer, D., Bach, T.-H. L., Queck, S. Y., Li, M., et al. (2007). Identification of Novel Cytolytic Peptides as Key Virulence Determinants for Community-Associated MRSA. Nat. Med. 13, 1510–1514. doi: 10.1038/nm1656
Keywords: Candida, Staphylococcus, biofilm, transcriptomics, interkingdom interactions
Citation: Short B, Delaney C, McKloud E, Brown JL, Kean R, Litherland GJ, Williams C, Martin SL, MacKay WG and Ramage G (2021) Investigating the Transcriptome of Candida albicans in a Dual-Species Staphylococcus aureus Biofilm Model. Front. Cell. Infect. Microbiol. 11:791523. doi: 10.3389/fcimb.2021.791523
Received: 08 October 2021; Accepted: 27 October 2021;
Published: 23 November 2021.
Edited by:
Jose L Lopez-Ribot, University of Texas at San Antonio, United StatesReviewed by:
Pang Li Mei, National Dental Centre of Singapore, SingaporeIslam Abdelhalim Abdelaziz Ali, The University of Hong Kong, Hong Kong SAR, China
Copyright © 2021 Short, Delaney, McKloud, Brown, Kean, Litherland, Williams, Martin, MacKay and Ramage. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Gordon Ramage, Z29yZG9uLnJhbWFnZUBnbGFzZ293LmFjLnVr
 Bryn Short1,2,3
Bryn Short1,2,3 Jason L. Brown
Jason L. Brown Ryan Kean
Ryan Kean Gary J. Litherland
Gary J. Litherland Craig Williams
Craig Williams S. Lorraine Martin
S. Lorraine Martin William G. MacKay
William G. MacKay Gordon Ramage
Gordon Ramage