- 1Key Laboratory of Special Animal Epidemic Disease, Ministry of Agriculture, Institute of Special Animal and Plant Sciences, Chinese Academy of Agricultural Sciences, Changchun, China
- 2Shanghai Veterinary Research Institute, Chinese Academy of Agricultural Sciences, Shanghai, China
The molecular epidemiology and biological characteristics of Escherichia coli associated with hemorrhagic pneumonia (HP) mink from five Chinese Provinces were determined. From 2017 to 2019, 85 E. coli strains were identified from 115 lung samples of mink suffering from HP. These samples were subjected to serotyping, antimicrobial susceptibility, detection of virulence genes, phylogenetic grouping, whole-genome sequencing, drug resistant gene, multilocus sequence typing (MLST) and biofilm-forming assays. E. coli strains were divided into 18 serotypes. Thirty-nine E. coli strains belonged to the O11 serotype. Eighty-five E. coli strains were classified into seven phylogenetic groups: E (45.9%, 39/85), A (27.1%, 23/85), B1 (14.1%, 12/85), B2 (3.7%, 3/85), D (3.7%, 3/85), F (2.4%, 2/85) and clade I (1.2%, 1/85). MLST showed that the main sequence types (STs) were ST457 (27/66), All E. coli strains had ≥4 virulence genes. The prevalence of virulence was 98.8% for yijp and fimC, 96.5% for iucD, 95.3% for ompA, 91.8% for cnf-Ⅰ, 89.4% for mat, 82.3% for hlyF, and 81.2% for ibeB. The prevalence of virulence genes iss, cva/cvi, aatA, ibeA, vat, hlyF, and STa was 3.5–57.6%. All E. coli strains were sensitive to sulfamethoxazole, but high resistance was shown to tetracycline (76.5%), chloramphenicol (71.8%), ciprofloxacin (63.5%) and florfenicol (52.9%), resistance to other antibiotics was 35.3–16.5%. The types and ratios of drug-resistance genes were tet(A), strA, strB, sul2, oqxA, blaTEM-1B, floR, and catA1 had the highest frequency from 34%-65%, which were consistent with our drug resistance phenotype tetracycline, florfenicol, quinolones, chloramphenicol, the bla-NDM-I and mcr-I were presented in ST457 strains. Out of 85 E. coli strains, six (7.1%) possessed a strong ability, 12 (14.1%) possessed a moderate ability, and 64 (75.3%) showed a weak ability to form biofilm. Our data will aid understanding of the epidemiological background and provide a clinical basis for HP treatment in mink caused by E. coli.
Introduction
Escherichia coli is a Gram-negative opportunistic pathogen. It is a normal flora of the gastrointestinal tract of humans and warm-blooded animals (Abdollah et al., 2018). However, if the immune system is compromised, E. coli can cause various intestinal and extraintestinal diseases.
Pathogenic E. coli can be divided into enteropathogenic and extraintestinal types. Extraintestinal pathogenic E. coli can cause colisepticemia, urinary-tract infections, and meningitis (Devine et al., 1989; Tibbetts et al., 2003; Gaschignard et al., 2011; Derakhshandeh et al., 2018). Tibbetts and colleagues isolated 40 E. coli strains from the livers, spleens, and lungs of mink (Neogale species) suffering from colisepticemia (Tibbetts et al., 2003). In that study, E. coli was shown, for the first time, to be associated with hemorrhagic pneumonia (HP) in mink. In China, Zhang and colleagues isolated E. coli strains from the lungs of mink suffering from respiratory symptoms (Zhang et al., 2019). That study provided additional evidence of the correlation between E. coli and HP in mink.
HP in mink is almost exclusively seasonal (from September to early December) and is characterized by sudden death (Qi et al., 2014). HP caused by Pseudomonas aeruginosa was described in Denmark in 1953 (Knox, 1953). In 1985, Han and colleagues were the first to report HP outbreaks in mink each year in China (Han et al., 2014). Klebsiella pneumoniae is another pathogen causing HP in mink. Fifteen K. pneumoniae strains were isolated from mink experiencing respiratory distress in China, and most of isolates were identified as serotype K2 (Wang et al., 2017). In that study, the mink showed lung hemorrhage, liver hemorrhage/swelling, slight bleeding in the brain, and liver abscess. The histology lesions of mink with acute HP were found to be associated with infection with P. aeruginosa and E. coli in a study by Hammer et al. (2013). They discovered that P. aeruginosa was most often found surrounding blood vessels and in the alveolar lining, whereas E. coli showed a more diffuse distribution in lung tissue. Although the pathogenesis is different, HP in mink caused by E. coli merits attention, but little information is available.
We investigated the disease characteristics, serotyping, phylogenetic groups, multilocus sequence typing (MLST), whole-genome sequencing, drug-susceptibility, and detection of virulence factors on E. coli strains. Our study will aid understanding of the epidemiological background of HP and provide a clinical basis for its treatment in mink caused by E. coli.
Materials and Methods
Collection of Samples
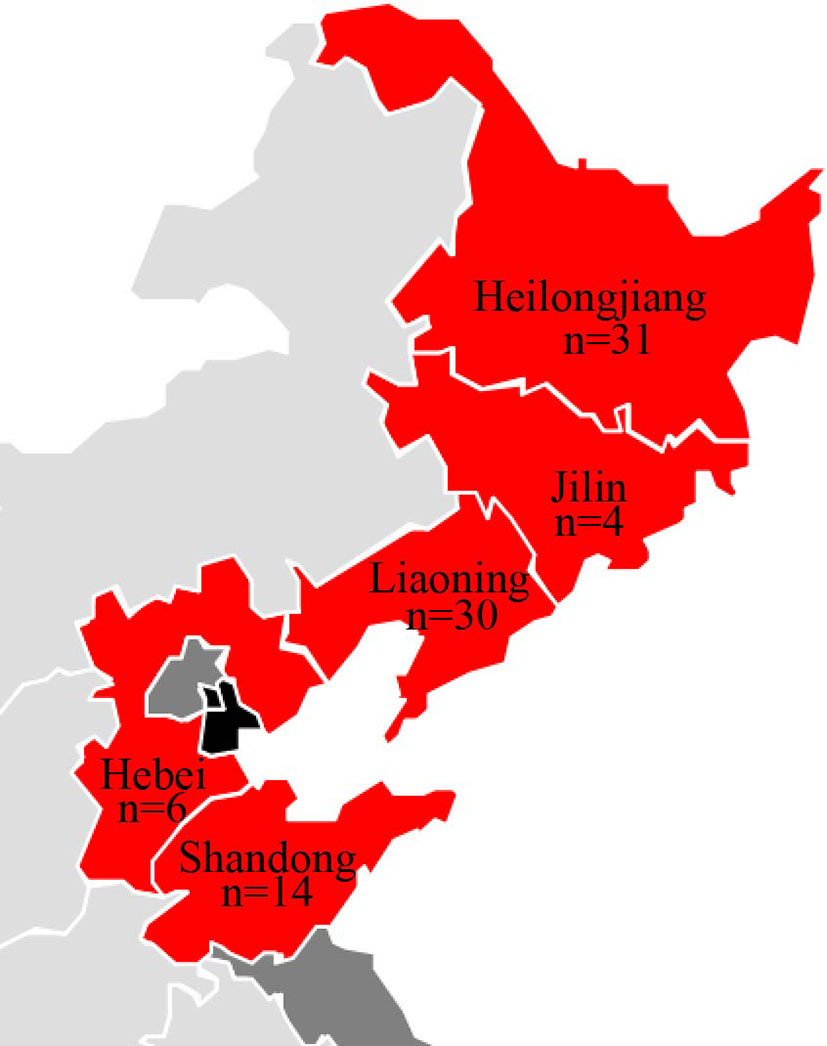
Between March 2017 and February 2019, 115 minks who died of HP were collected from 21 farms in northern China (Jilin, Liaoning, Heilongjiang, Hebei, and Shandong Provinces) (Figure 1). The minks showed clinical pneumonia signs, including: high fever (>40°C), severe depression and anorexia, dyspnea, coughing, as well as hematemesis of the mouth and nose.The dead minks underwent dissection, and lung samples were used for bacterial isolation.
Histopathology
After necropsy, samples were fixed in 10% neutral-buffered formalin. Then, they were embedded in paraffin wax and stained with hematoxylin and eosin for histopathology following standard procedures (Bancroft and Stevens, 1990).
Isolation and Identification of E. coli
Isolation and identification of E. coli strains were undertaken according to standard procedures with some modifications (Qiu et al., 2019). Lung samples were inoculated onto MacConkey agar. Bacterial colonies were selected and cultured in Luria–Bertani (LB) broth at 37°C. These bacterial isolates were identified by classical biochemical methods and confirmed by 16S rRNA-sequencing. All E. coli strains were grown in LB broth at 37°C with aeration and stored at −80°C in 20% glycerol for further use.
DNA Extraction
The genomic DNA of E. coli isolates was extracted using the TIANamp Bacteria DNA Kit according to manufacturer's (Tiangen, Beijing, China) instructions.
Serotyping
Traditional agglutination test and O-genotyping PCR were used to identify the O serotypes of the E. coli isolates. Briefly, serotyping of E. coli isolates was done by agglutination test with specific serum against E. coli O antigens (Statens Serum Institut, Copenhagen, Denmark) according to the manufacturer’s guidelines. The O-genotyping PCR were carried out through the implication of O antigen biosynthesis genes as described previously (Iguchi et al., 2015).
Antimicrobial Susceptibility Testing
The minimal inhibitory concentration of 85 E. coli strains was determined according to the Performance Standards for Antimicrobial Susceptibility Testing of the Clinical Laboratory Standards Institute (CLSI; M100-S23). Each strain was evaluated for susceptibility to the cefotaxime, tetracycline, chloramphenicol, florfenicol, levofloxacin, ciprofloxacin, sulfamethoxazole, colistin, gentamicin, and spectinomycin. The susceptibility of E. coli isolates was interpreted referring to CLSI M100-S23. E. coli ATCC25922 were used as a quality-control strain in antimicrobial susceptibility testing.
Phylogenetic Group Analysis
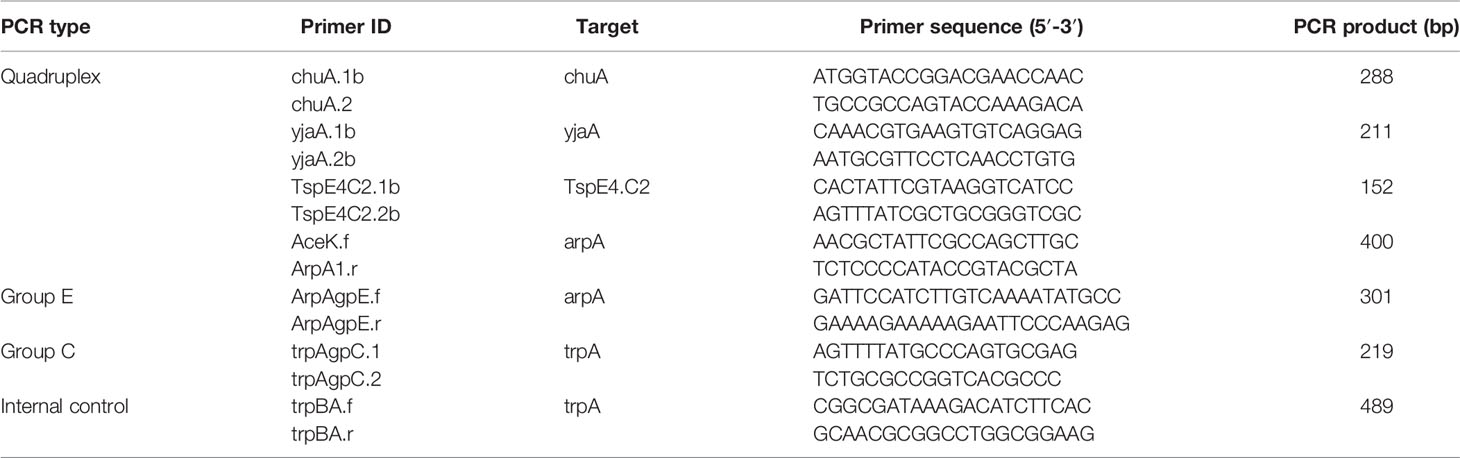
E. coli strains were classified as eight phylogenetic groups using the rapid phylogenetic grouping PCR as described previously. Thus, the phylogenetic groups of E. coli isolates were determined via PCR detection of genes chuA, yjaA, TspE4.C2, arpA, and trpA according to Clermont protocol (Clermont et al., 2013) (Table 1).
Distribution of Virulent Genes
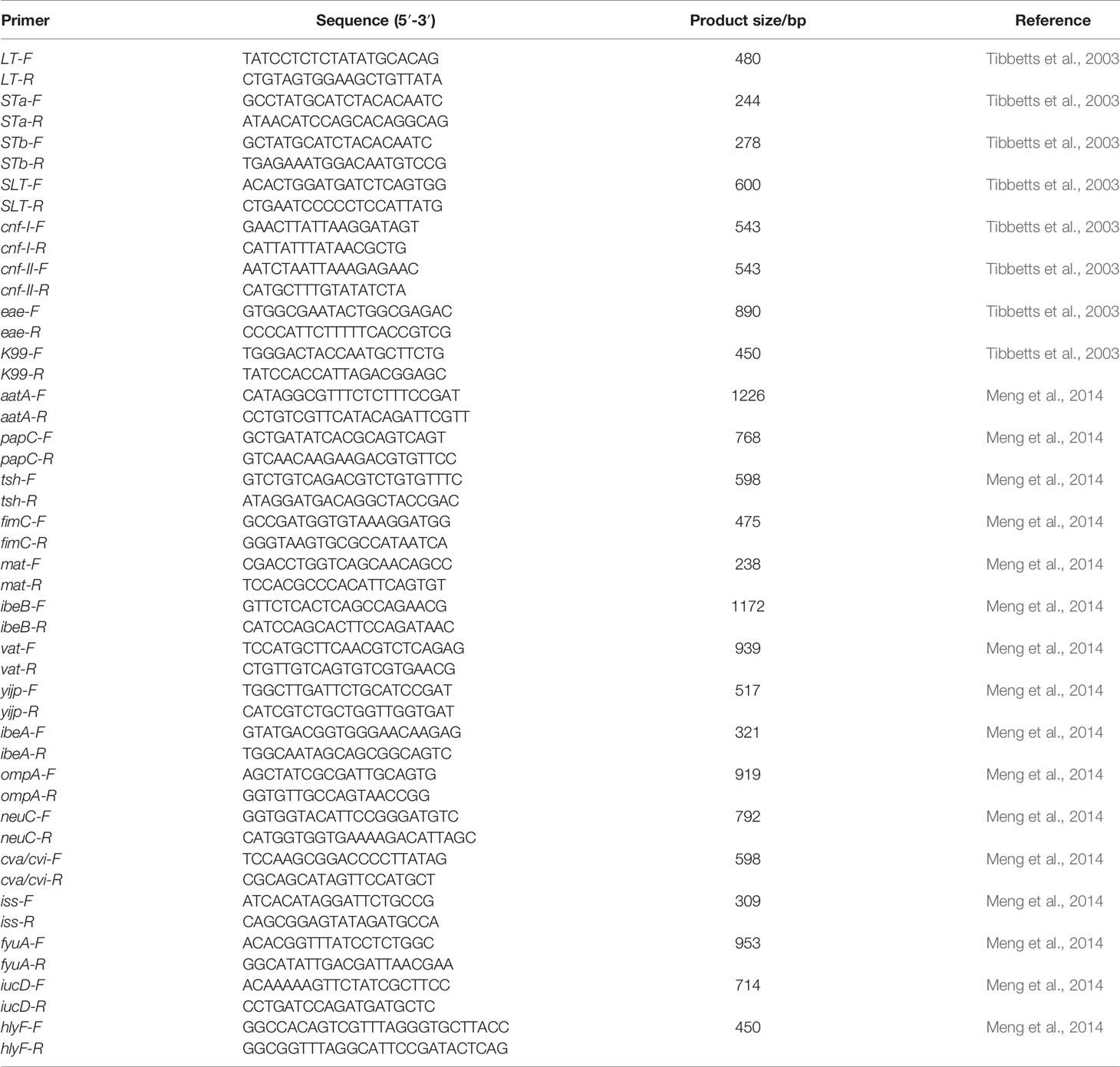
All strains were tested for the presence of 24 virulence-associated genes by simplex PCR and multiplex PCR (Tibbetts et al., 2003; Meng et al., 2014): LT, STa, STb, SLT-Ⅰ, cnf-Ⅰ, cnf-Ⅱ, eae, K99, aatA, papC, tsh, fimC, mat, ibeB, vat, yijp, ibeA, ompA, neuC, cva/cvi, iss, fyuA, iucD and hlyF. The primers used for detection are listed in Table 2. PCRs were conducted under identical reaction conditions according to those described previously (Tibbetts et al., 2003; Meng et al., 2014).The amplification products were visualized by agarose gel (1%) electrophoresis.
Whole-Genome Sequencing
Whole-genome sequencing (WGS) was used to characterize the resistome, MLST and evaluate genetic evolution in 66 representative strains. The whole genome was sequenced by Beijing Novogene Bioinformatics Technology (Beijing, China). The sequence of genomic DNA in cells was identified (PacBio Single Molecule, Real-Time Sequencing; Pacific Biosciences, Menlo Park, CA, USA). The neighbor-joining method (>5 samples) was employed to construct an evolutionary tree using TreeBeST Vision 1.9.2. Resistance genes were predicted by comparing the sequenced strains with a database on antiviral resistance genes (http://katholt.github.io/srst2/).
MLST
Sixty-six E. coli strains were underwent MLST by comparing of seven housekeeping genes (adk, fumC, gyrB, icd, mdh, purA, and recA) according to WGS results and aligned against the allele templates of E. coli retrieved from an online database (http://www.mlst.net/). A minimum spanning tree and clustering tree were generated using BioNumerics 7.6 (www.applied-maths.com/download/software/).
Quantification of Biofilm Formation
Biofilm-formation assays were undertaken, as described previously (Vogeleer et al., 2015) with some modifications. Briefly, E. coli strains were cultured in LB overnight at 37°C and diluted into LB at 1:100. A 200-μL aliquot of each dilution was dispensed into individual wells of a sterile 96-well microtiter plate and incubated for 36 h at 37°C. Wells with sterile LB medium served as negative controls. Then, the culture medium was discarded, and the wells were washed thrice with phosphate-buffered saline. After fixation with methanol for 15 min, biofilms were stained with 0.1% crystal violet for 5 min. The plates were washed five times to remove unbound dye and air-dried. Finally, the biofilms were quantified by measuring the optical density (OD) at 595 nm with a microplate reader(eppendorf, BioPhotomete) after dissolving in 33% glacial acetic acid for 30 min. The capability to form a biofilm was scored in accordance with criteria described previously (Stepanovic et al., 2000): OD< ODc, non-producer; ODc < OD < 2×ODc, weak producer; 2× ODc < OD < 4 × ODc, moderate producer; OD >4ODc, strong producer. “OD” denoted the OD at 595 nm of E. coli, whereas “ODc” denoted three standard deviations above the mean OD of the negative control. Tests were carried out thrice, and the results were averaged.
Results
Organ Symptoms and Hispathological Changes
The onsets of minks were breathing difficulties, blood-like exudates around the nostrils/mouths, and, in some cases, foaming around the mouth, The most common observation were edematous, extensive hemorrhage and pleural effusion. hyperemia or hemorrhagic spots and ecchymosis were observed in lung. Other symptoms were liver hemorrhage, renal hemorrhage, spleen enlargement, and black bleeding spots. The intestinal surface and inner wall were dark-red or pink, which indicated a small amount of bleeding. Swelling and bleeding of inguinal, submandibular, and mesenteric lymph nodes were also documented (Figure 2).
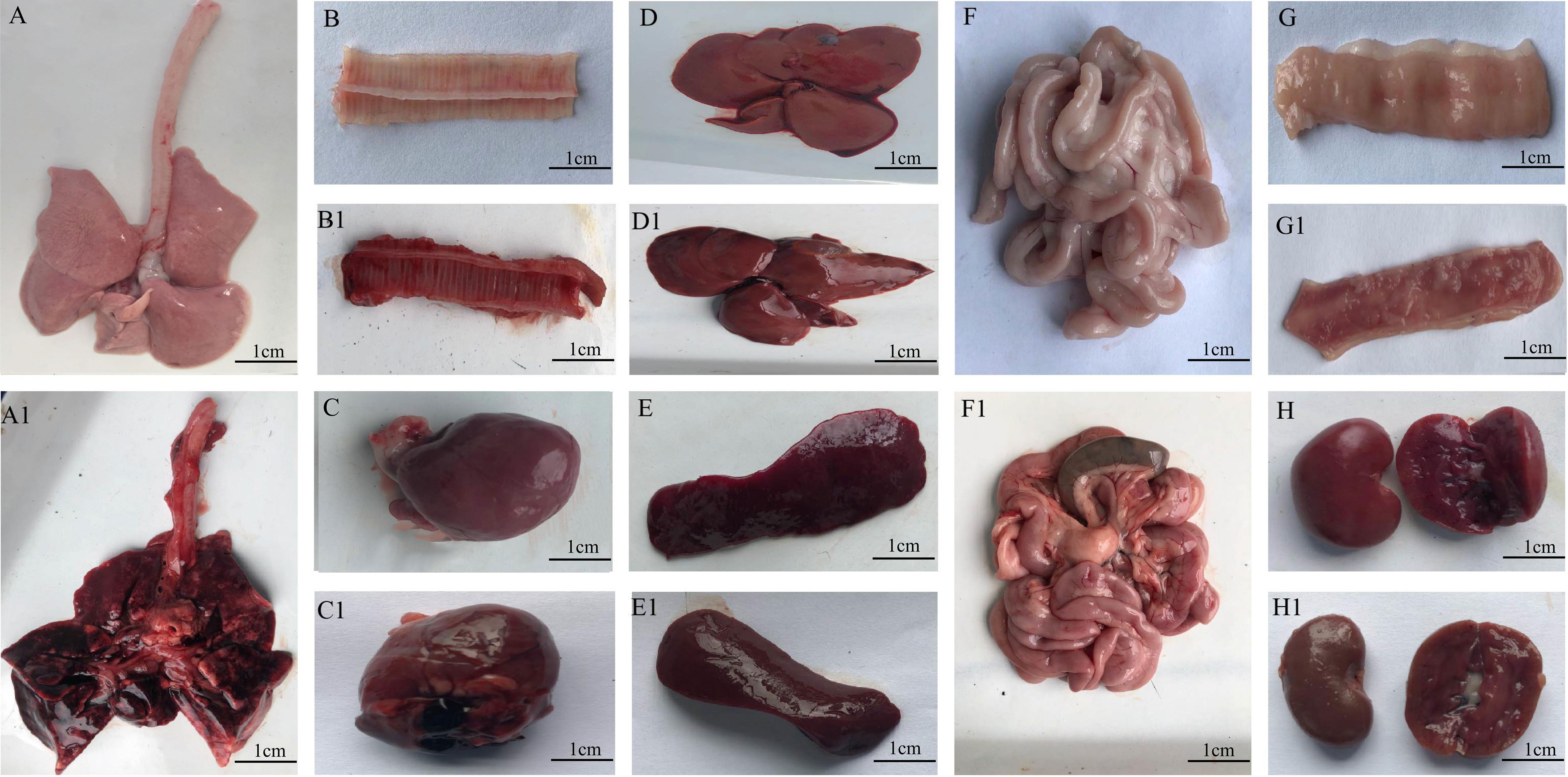
Figure 2 Organs from minks after E. coli infection. (A1–G1) Organs of infected minks. (A–G) Organs of heathy minks. (A, A1) Lung. (B, B1) trachea. (C, C1) Heart. (D, D1) Liver. (E, E1). Spleen. (F, F1) Inner wall of the intestinal tract. (G, G1) Kidney. (H, H1) Renal hemorrhage.
The hispathological changes of organs were as follows, lung, severe pulmonary edema, a large number of eosinophilic serous exudation in alveolar cavity; tracheae, cell necrosis, nuclear fragmentation or dissolution in mucosal layer; heart and liver: lymphocytic infiltration around blood vessels; spleen, massive extramedullary hematopoietic foci and multinucleated giant cells; Intestine, no obvious inflammation; kidney, necrotic shedding of renal tubular epithelial cells (Figure 3).
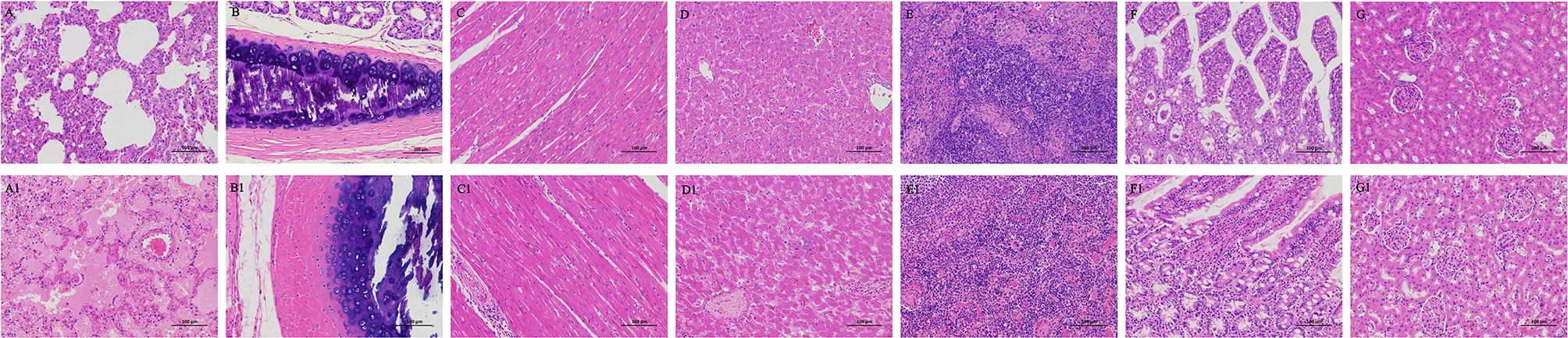
Figure 3 Hispathological changes of Organs from minks after E. coli infection. (A–G) Organs of infected minks. (A1–G1) Organs of heathy minks. (A, A1) Lung. (B, B1) trachea. (C, C1) Heart. (D, D1) Liver. (E, E1). Spleen. (F, F1) the intestinal tract. (G, G1) Kidney.
Isolation and Identification of E. coli Strains
A total of 115 lung samples from 21 mink farms were cultured. Eighty-five E. coli isolates were identified by conventional biochemical methods. 16S rRNA-sequencing indicated that these strains had features consistent with E. coli.
Serotyping
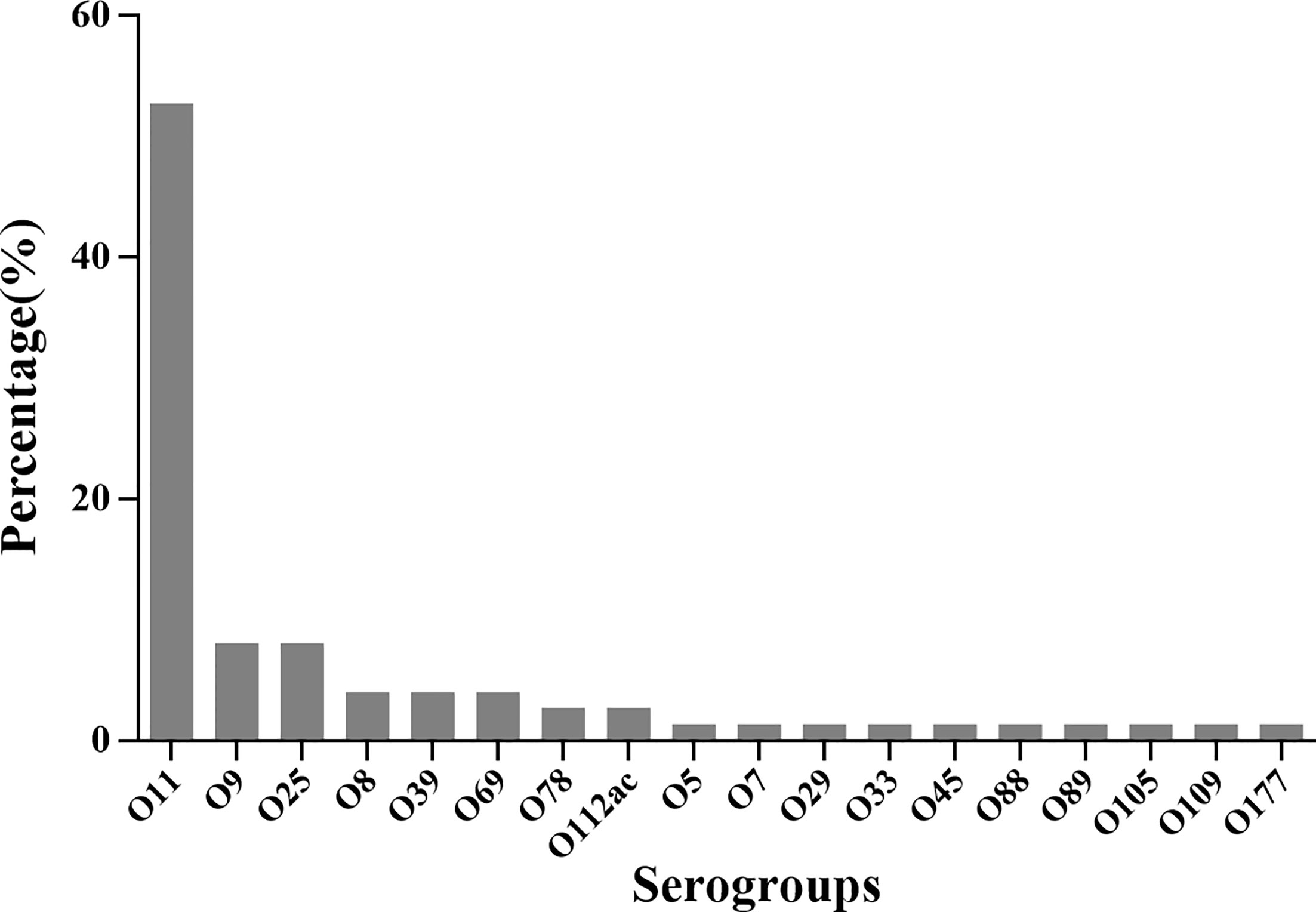
Serotyping showed that 87.1% (74/85) of E. coli isolates could be classified into the single-O type, whereas 11 isolates could not be classified into any serotype. Of these serotyped isolates, 18 O serotypes were identified (Figure 4). The most prevalent serotype were O11 (39/85), followed by O9 (6/85), O25 (6/85), O8 (3/85), O39(3/85), O69(3/85), O78(2/85), and O112ac (2/85). Only one strain belonged to O5, O7, O29, O33, O45, O88, O89, O105, O109, and O177 serotypes, respectively. These results indicated that O11, O9, and O25 were the predominant serotypes of extraintestinal pathogenic E. coli from mink in a particular population in northern China.
Antimicrobial Susceptibility Testing
The results of the antimicrobial susceptibility test are shown in Table S1 of Supplementary Materials. Eighty-five E. coli strains were sensitive to sulfamethoxazole, but they showed high resistance to tetracycline (76.5%), chloramphenicol (71.8%), ciprofloxacin (63.5%), and florfenicol (52.9%). Resistance to colistin was noted in only 5.9% of E. coli strains. Resistance to the other antibiotics tested was 16.5%–35.3% (Figure 5). We analyzed the susceptibility of E. coli strains to 10 antibiotics, and found that 55 of 85 (64.7%) strains were resistant to ≥3 antibiotics.
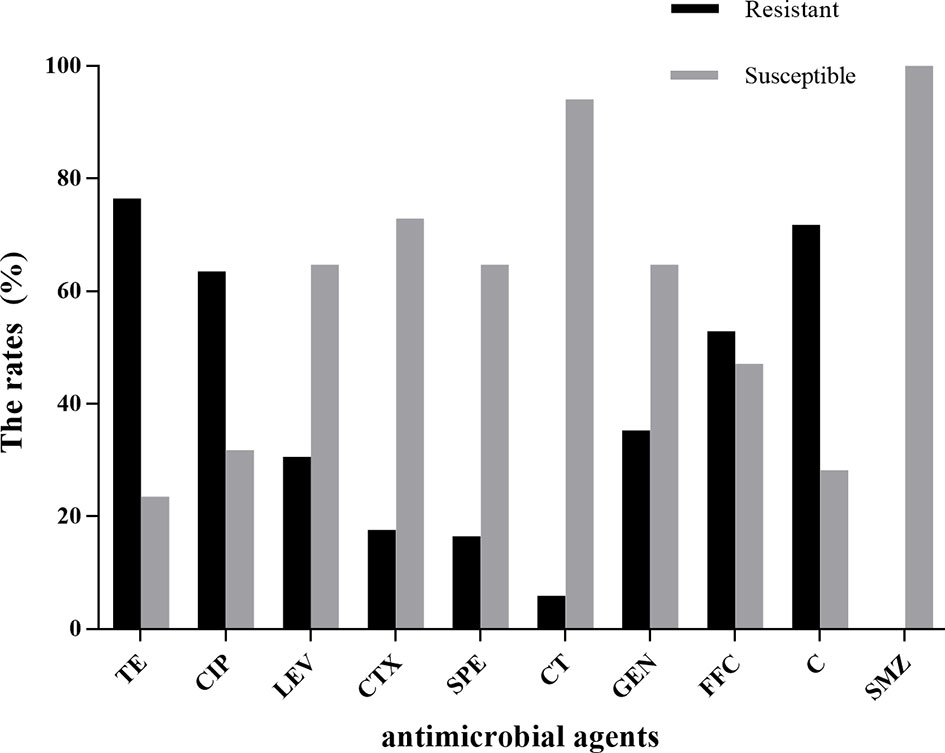
Figure 5 Results of antimicrobial susceptibility testing of 85 E. coli strains. TE, tetracycline; CIP, ciprofloxacin; LEV, levofloxacin; C, chloramphenicol; GEN, gentamicin; FFC, florfenicol; CTX, cefotaxime; SPE, spectinomycin; CT, colistin; SMZ, sulfamethoxazole.
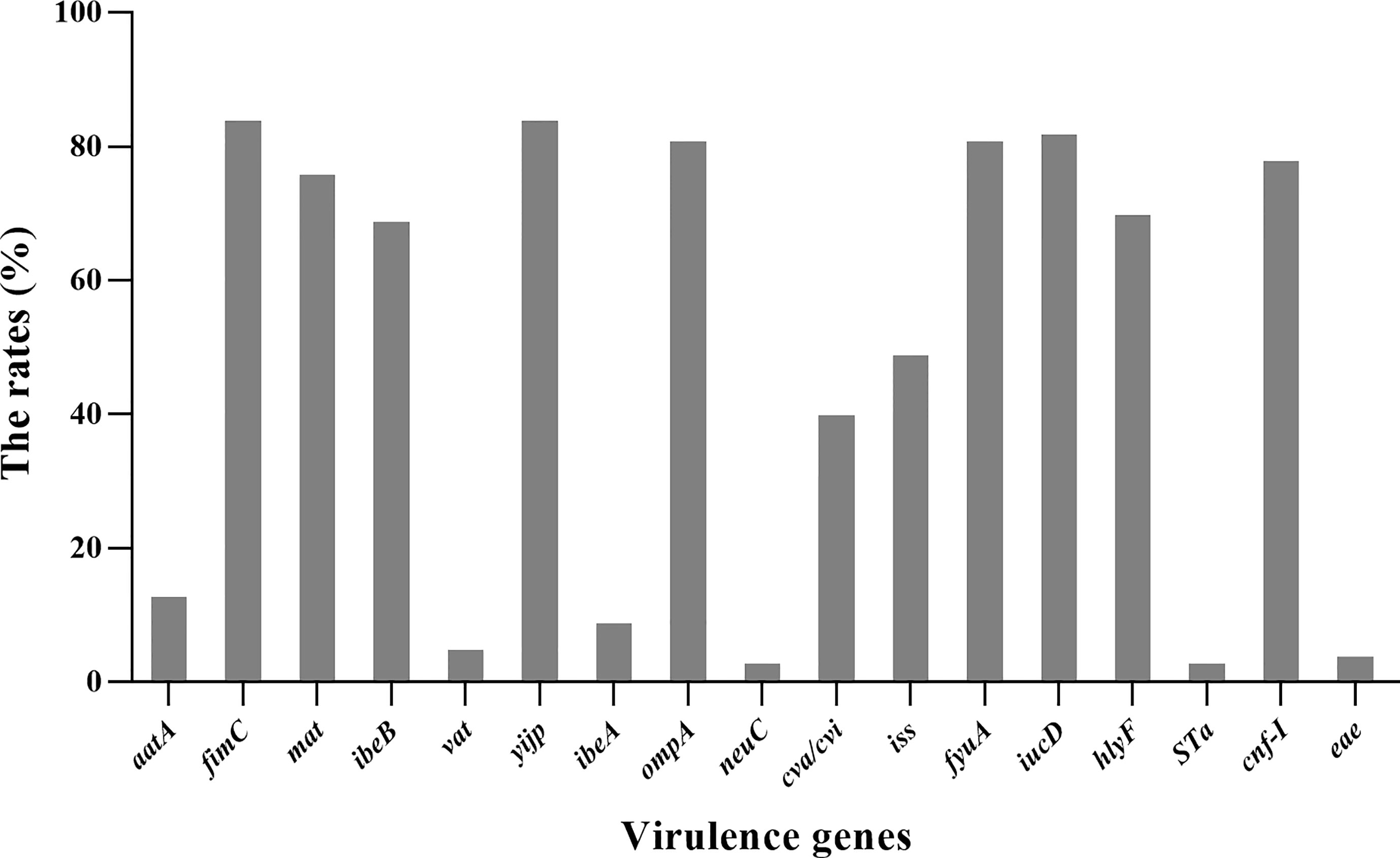
Distribution of Virulence Genes
All 85 E. coli strains were screened for the presence of 24 virulence genes. yijp and fimiC were detected in 84 (98.8%) strains. iucD, ompA, cnf-Ⅰ, and fyuA were detected in 82 (96.5%), 81 (95.3%), 81 (95.3%), and 78 (91.8%) strains, respectively. mat, hlyF, and ibeB were present in 76 (89.4%), 70 (82.3%), and 69 (81.2%) of strains, respectively. iss and cav/cvi were detected in 49 (57.6%) and 40 (47.1%) of strains, respectively. aatA, ibeA, vat, and eae were detected in 13 (15.3%), 9 (10.6%), 5 (5.9%), and 4 (4.7%) strains, respectively. Moreover, three (3.7%) strains harbored STa and nueC. In general, all E. coli strains contained ≥4 virulence genes. However, LT, STb, SLT-Ⅰ, cnf-Ⅱ, K99, papC, and tsh were not detected (Figure 6).
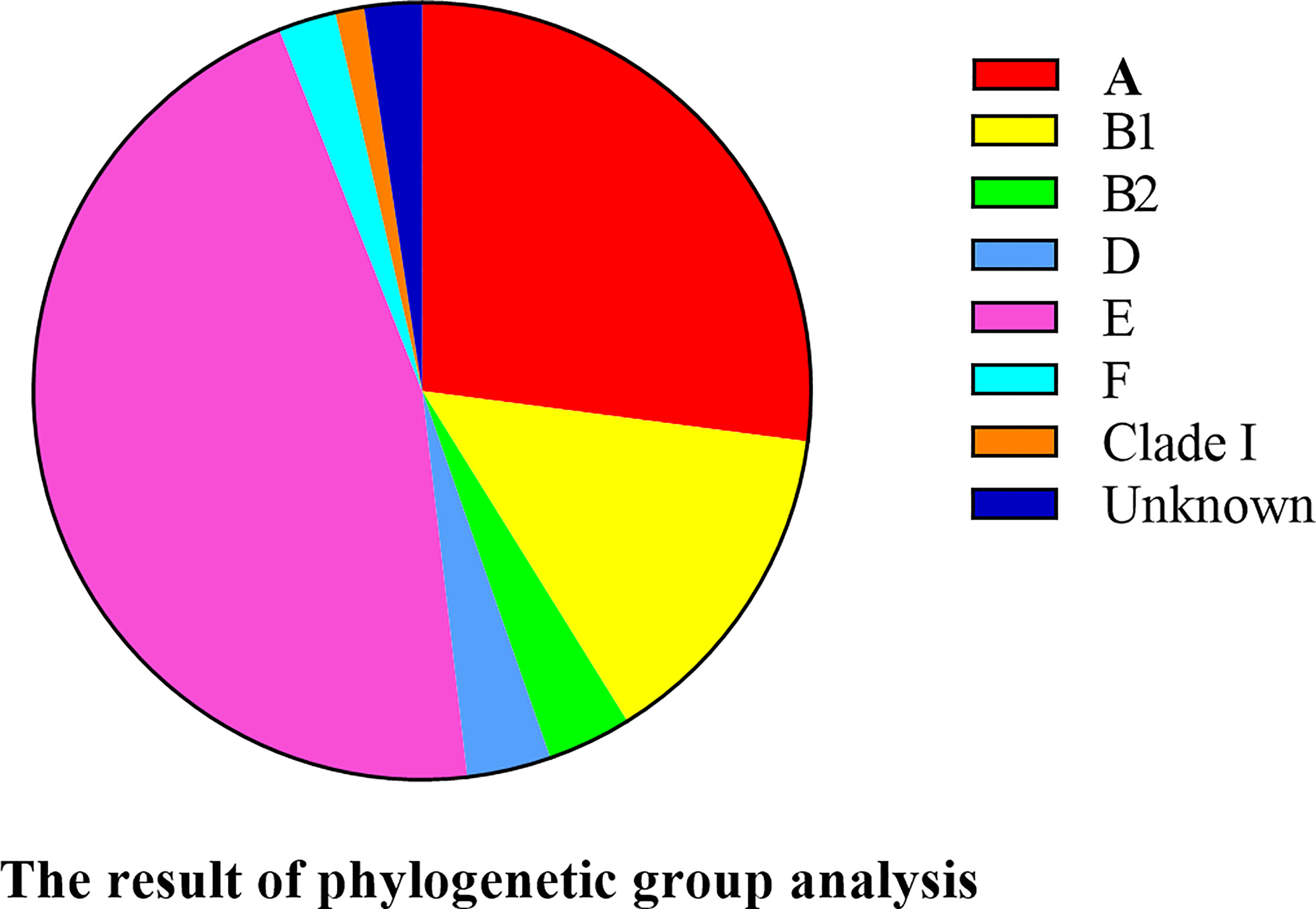
Phylogenetic Grouping
Phylogenetic typing showed that most of E. coli strains belonged to group E (45.9%, 39/85) and A (27.1%, 23/85). Only 3.5% of E. coli strains belonged to groups B2 and D, 2.4% to F, and 1.2% to clade I. However, two E. coli strains were not classified into any phylogenetic group (Figure 7). Thus, the E. coli strains that caused HP in mink were strongly associated with phylogenetic groups A and E.
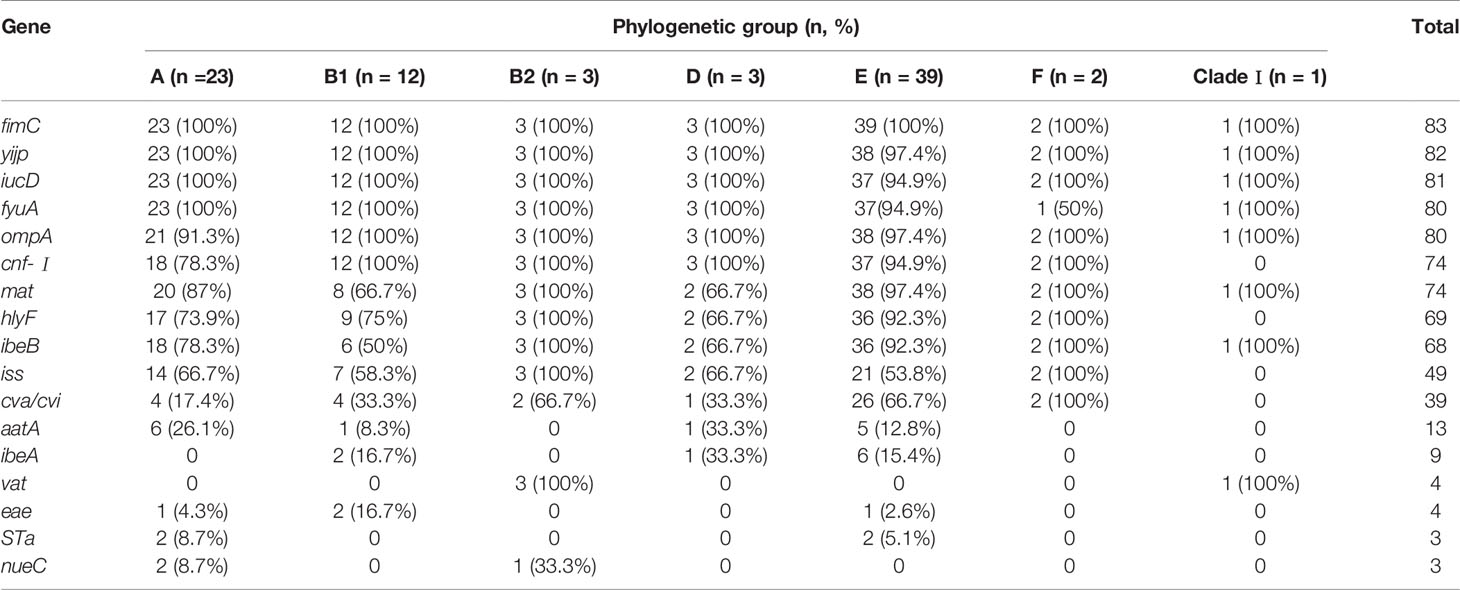
Most virulence genes were associated with phylogenetic groups E and A. yijp, fimC, iucD, ompA, and fyuA showed a wide distribution among all groups (A, B1, B2, D, E, F, and clade I). cnf-Ⅰ, hlyF, iss, and cva/cvi were less prevalent in clade I. vat was associated with group B2 and clade I. STa was found only in groups A and E. nueC was associated only with groups A and group B2 (Table 3).
Whole-Genome Sequencing
Whole-genome sequencing (WGS) was done on the genome data of the new strains in the present study. The complete genome MLST clustering tree showed that the E. coli strains from mink were far from the reference strains. The E. coli strains in mink showed regional aggregation and transmission. The homology of the E. coli strains from Weihai, Dalian, Shangzhi, Harbin, and Weihe reached 100%, respectively. However, the other strains were far from each other (Figure 8).
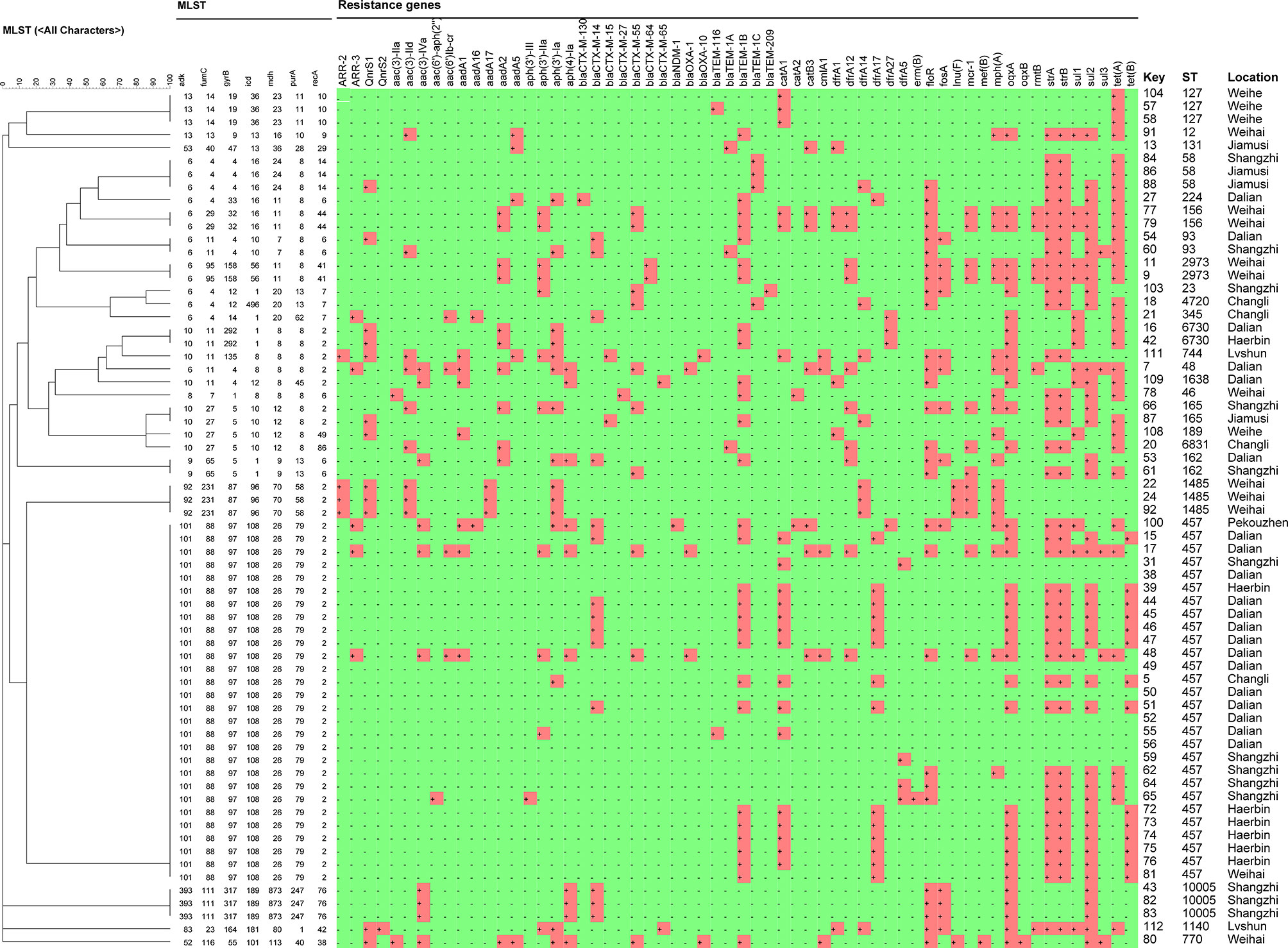
Figure 8 Clustering tree of 66 E. coli strains using multilocus sequence typing by Whole-genome sequencing analysis. The red cube region represents the presence of a drug-resistance gene.
We predicted the types and ratios of drug-resistance genes in 66 strains of E. coli by whole-gene sequencing. tet(A), strA, strB, sul2, oqxA, blaTEM-1B, floR, and catA1 had the highest frequency at 50%, 65.8%, 64.5%, 64.5%, 53.9%, 48.7%, 40.8%, and 34.2%, respectively. The other drug-resistance genes were rangely from 1.3%∼29%, (Figure 9). the highest frequency antibiotic genes (ARGs) were resistant to tetracycline, florfenicol, quinolones, chloramphenicol, which were consistent with our drug resistance phenotype.
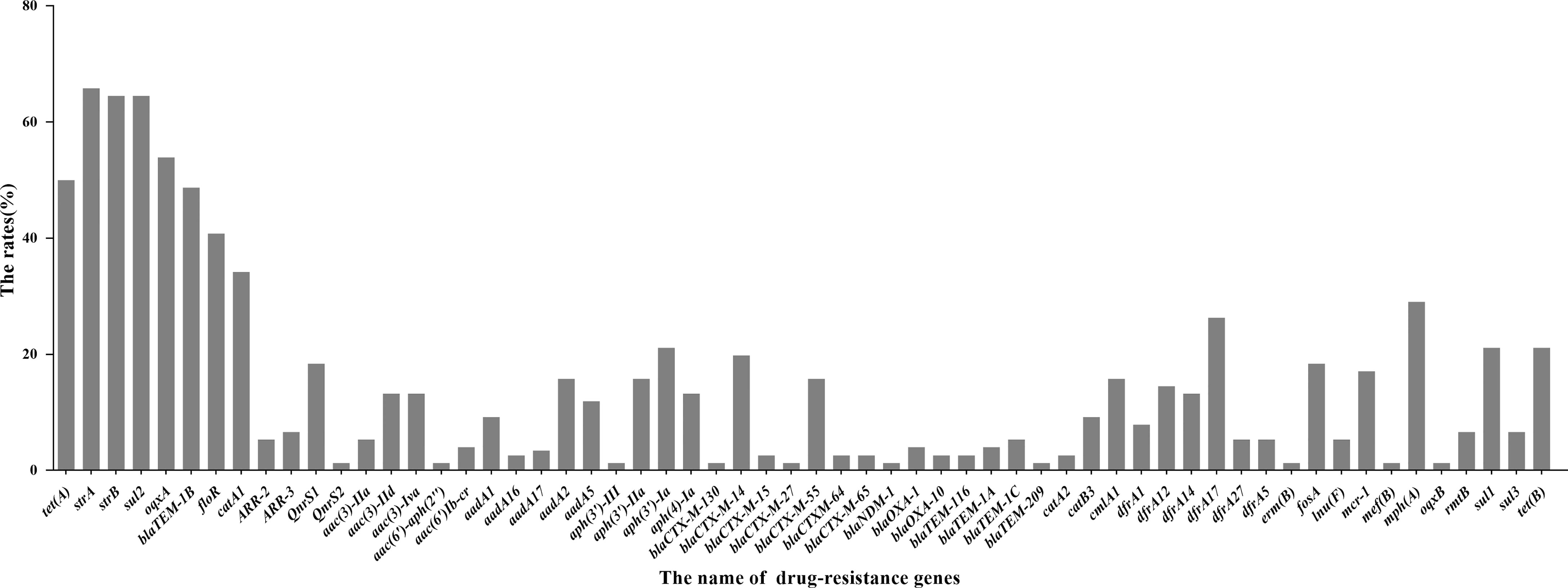
Figure 9 Distribution of drug-resistant genes of 66 E. coli strains by Whole-genome sequencing analysis.
MLST
Sixty-six E. coli strains were classified into 28 sequence types (STs). The dominant types were ST457 (twenty-seven strains from six outbreaks), followed by ST127 (three strains from one outbreak), ST1485 (three strains from one outbreak), ST10005 (three strains from one outbreak), ST58 (two strains from two outbreaks), ST6730 (two strains from two outbreaks), ST93 (two strains from two outbreaks), ST2973 (two strains from two outbreaks), ST162 (two strains from two outbreaks), and ST156 (two strains from one outbreak). The only one strain of ST was ST23, ST58, ST189, ST1638, ST744, ST1140, ST131, ST165, ST4720, ST6831, ST345, ST224, ST48, ST46, ST770, ST7, ST5, and ST12 (Figure 10).
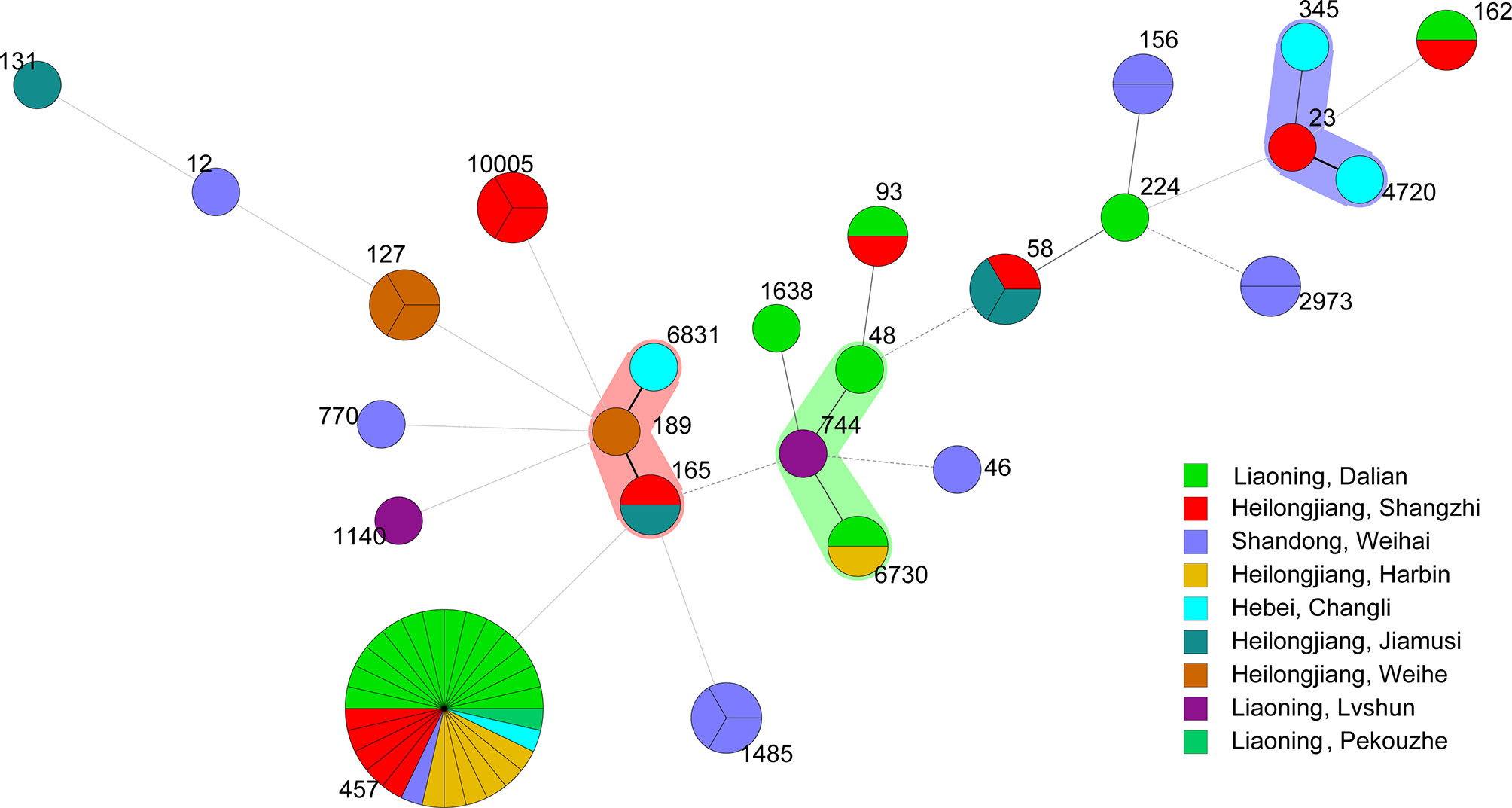
Figure 10 Minimum spanning tree of 66 E. coli strains by Whole-genome sequencing analysis. Each circle represents one ST. The area of the circle corresponds to the number of strains. The color of the circle indicates the area to which the strain belongs.
Quantification of Biofilm Formation
Out of 85 E. coli strains, six (7.1%) possessed strong biofilm-forming ability, 12 (14.1%) possessed moderate biofilm-forming ability, and 64 (75.3%) showed weak biofilm-forming ability. Three E. coli strains could not form biofilms (Table S1 in Supplementary Materials).
Discussion
HP in mink was first discovered in China in 1983 (Han et al., 2014). P. aeruginosa and K. pneumoniae are the important agents of HP; they are the major causes of death and induce economic loss in the mink industry (Wang et al., 2017, Zhao et al., 2018).
We isolated 85 E. coli strains from 115 minks with HP. We assessed the serotype, phylogenetic group, and virulence genes of these E. coli strains. We undertook, whole-genome sequencing, MLST, antimicrobial susceptibility testing and quantification of biofilms to provide a theoretical basis for the prevention and treatment of HP in mink.
O11 was the predominant (45.9%) serotype of the 85 E. coli strains in the lung samples of mink with HP. Serotype O11 has been reported to be the most prevalent among isolates from pigs with postweaning diarrhea and birds with colibacillosis (Chen et al., 2004; Hussein et al., 2013). Our data are consistent with findings from Xu and colleagues showing that two out of five E. coli strains belonged to serotype O11 (Xu et al., 2018). The other serotypes we detected, including O9, O25, and O8, have been described to be most frequently related to urinary-tract infections, bacteremia, and diarrhea (Devine et al., 1989; Tibbetts et al., 2003; Wani et al., 2004). The serotypes of E. coli are not found commonly in other types of disease in mink (Tibbetts et al., 2003). This disparity may be the result of genetic variations and regional differences.
The distribution and frequencies of serotypes varied considerably from region to region (Table S1 in Supplementary Material). Serotype O11 was distributed in four Provinces (Liaoning, Heilongjiang, Shandong, Jilin and Hebei). The other dominant serotypes (O8, O9 and O25) were in Shandong Province. Serotypes O39, O69, O78, O112ac, O5, O7, O29, O33, O45, O88, O89, O105, O109, and O177 were less common.
Phylogenetic analyses have shown that 95% of E. coli strains fall into eight phylogenetic groups: A, B1, B2, C, D, E, F, and clade I (Clermont et al., 2013; Silva et al., 2017). Primary infection with E. coli leading to persistence or relapse of infection is associated with phylogenetic group B2, whereas primary infection with E. coli followed by cure or reinfection is associated with phylogenetic group D (Ejrns, 2011), respectively. We also found that E. coli strains isolated from infected mink belonged mainly to phylogenetic groups A and E, and other E. coli strains belonged to phylogenetic groups B1, B2, D, F, and clade I. This distribution of E. coli strains is different to that reported in bovine mastitis and broiler chickens (Hussein et al., 2013; Zhang et al., 2017), but is consistent with the distribution of human enteroinvasive E. coli (Da Silva et al., 2017). Our results provide helpful references of the ecological distribution and genetic evolution of E. coli in mink in northern China.
Virulence genes have important roles in pathogenic E. coli. Hence, we detected the distribution of extraintestinal virulence gene markers in E. coli strains from mink (Tenglin et al., 2018). The virulence genes mainly encoded genes related to invasion, toxins, adhesion, antiserum survival, and iron transport. yijp, fimC, iucD, ompA, cnf-Ⅰ, mat, hlyF, and ibeB had a high presence (98.8%, 98.8%, 96.5%, 95.3%, 91.8%, 89.4%, 82.3% and 81.2%, respectively) but some had prevalence <60% (iss, cva/cvi, aatA, ibeA, vat, and eae). The resluts proofed that E.coli strains isolated in this study were extra-intestinal strains. the profiles of virulent genes were the same as other extra-intestinal strains, Tibbetts found that 47.5% of the test strains had cnf-I gene,and 7.5% of the strains had eae gene in mink with colisepticaemia (Tibbetts et al., 2003). Paixão found that the frequency of avian E.coli virulence gene fimC, iucD, cva/cvi, iss was 92.91%, 65.35%, 55.12% and 33.07% (Paixão et al., 2016).
MLST is a bacterial-typing method based on determination of the sequence of nucleic acids. MLST has been used for epidemiological monitoring and evolutionary studies. In our study, 28 STs were found. The results are different from E. coli strains isolated from mink feces in Zhucheng (Qiu et al., 2019), These differences may suggest that, compared with E. coli isolated from mink feces, the genetic information and pathogenicity of E. coli isolated from mink lungs are different. In our study, the dominant type was ST457(27/66, 40.9%). which has the potential cause antimicrobial-resistant extraintestinal infection, almost certainly in dogs, wild animals and possibly in humans (Nicolas-Chanoine et al., 2014). ST457 in E. coli isolated from dog feces contains bla-CMY and bla-CTX-M-15, which are resistant to fluoroquinolone (Guo et al., 2015), ST457 in E. coli isolated from blood contains blaCTX-M-1 and mcr-1 (Kuo et al., 2016), ST457 in E. coli isolated from bovine mammary glands is associated with resistance to aminoglycoside and trimethoprim, respectively (Suzuki et al., 2016). The clustering tree showed that the resistance genes of ST457 in E. coli were similar (Figure 8). ST127 can cause intestinal infections in humans, dogs, and cats (Johnson et al., 2008), and it is a biological model of E. coli infection outside the intestine (Hochhut et al., 2006). ST1485 and ST131 in E. coli are related to human infection. ST131 in E. coli is an extraintestinal pathogen. Pitout and colleagues showed that invasion by ST131 is closely related to colonization, iron-absorption capacity, and the ability to form biofilms (Pitoust and Devinney, 2017).
According to the cgMLST clustering tree, HP outbreak in mink had the characteristics of regional aggregation and transmission. We suggest that farmers should pay attention to detection and isolation during the start of an HP outbreak in mink. We found that the main drug-resistance genes were tet(a), sul2, oqxa, flor, blatem-1b, cata1, stra, and strB, Profiles for antibiotic resistance and resistance genes for ST genotypes with different genetic relationships were documented. The distribution of drug-resistance genes was similar in the same ST type, as shown in Figure 5.
Use of antimicrobial drugs can lead to resistance to their effects (Mackenzie et al., 2014; Wang et al., 2017). A vaccine to prevent extraintestinal pathogenic E. coli in mink is not available. We analyzed the susceptibility of E. coli strains to 10 antibiotics. All E. coli strains were susceptible to sulfamethoxazole but showed high resistance to tetracycline (76.5%), chloramphenicol (71.8%), ciprofloxacin (63.5%), and florfenicol (52.9%). Resistance to multiple drugs was observed, with 55 of 85 (65.7%) isolates resistant to ≥3 antibiotics. Pedersen and colleagues also showed E. coli to be susceptible to sulfamethoxazole and resistant to tetracycline (54.7%) and spectinomycin (18.5%) (Pedersen et al., 2009). However, those findings were not in accordance with those of Tibbetts and coworkers, who reported that the E. coli strains from mink with colisepticemia were sensitive to ciprofloxacin but resistant to sulfamethoxazole (63%), gentamycin (20%), and chloramphenicol (8%) (Tibbetts et al., 2003). The reasons for these inconsistencies may be the different sources and regional differences of E. coli strains.
A biofilm is composed of surface-bound or sessile microbes enclosed in an amorphous extracellular matrix (Donlan and Costerton, 2002). Residence in a biofilm community offers bacteria an enhanced ability to cause disease (Wirth et al., 2006; Wang et al., 2011). We found that 75.3% of E. coli strains had a weak ability to form a biofilm. This weakness is obviously different for strains isolated from other species; toxin-producing strains usually possess a strong ability to form biofilms (Vogeleer et al., 2015). E. coli strains isolated from poultry often possess a moderate ability to form a biofilm (Wang et al., 2016). We found that strains that could form a strong biofilm were resistant to tetracycline, ciprofloxacin, and chloramphenicol.
Conclusions
The E. coli strains isolated from mink were extraintestinal pathogenic and the dominant serotype was O11. A and E were the predominant phylogenetic groups among E. coli strains in the parts of northern China we sampled. E. coli isolated from mink lungs had similar genes, and some strains had the characteristics of regional aggregation and transmission. Although these strains were resistant to multiple drugs, sulfonamides are first-line treatment for HP caused by E. coli in mink in China. Through the study of extraintestinal pathogenic E. coli in mink, we aim to develop a triple vaccine using P. aeruginosa, K. pneumoniae, and extraintestinal pathogenic E. coli against HP in mink.
Data Availability Statement
The data presented in the study are deposited in the NCBI repository, accession number listed in the Supplementary Materials Table S1.
Ethics Statement
The animal study was reviewed and approved by the Ethics Committee of Institute of Special Animal and Plant Sciences, Chinese Academy of Agricultural Sciences. Written informed consent was obtained from the owners for the participation of their animals in this study.
Author Contributions
YY, BH, and HZ conducted the experiments. SW and XB designed the experiments. YY and HF analyzed the data. SZL and HL assisted with the experiments. YY wrote the manuscript. SW and XB revised the manuscript. SW and XB have contributed equally to this work. All authors contributed to the article and approved the submitted version.
Funding
This work was funded by the Science and Technology Development Project of Jilin Province (20200402111NC).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcimb.2021.781068/full#supplementary-material
References
Abdollah, F., Dalela, D., Sood, A., Keeley, J., Alanee, S., Briganti, A., et al. (2018). Impact of Adjuvant Radiotherapy in Node-positive Prostate Cancer Patients: The Importance of Patient Selection. Eur. Urol. 74 (3), 253–256. doi: 10.1016/j.eururo.2018.04.017
Bancroft, J. D., Stevens, A. (1990). Theory and Practice of Histological Techniques, 3rd. Edinburgh: Churchill Livingstone. pp. 740.
Chen, X., Gao, S., Jiao, X., Liu, X. F. (2004). Erratum to “Prevalence of Serogroups and Virulence Factors of Escherichia coli Strains Isolated From Pigs With Postweaning Diarrhoea in Eastern China”. Vet. Microbiol. 103, 13–20. doi: 10.1016/j.vetmic.2004.10.002
Clermont, O., Christenson, J. K., Denamur, E., Gordon, D. M. (2013). The Clermont Escherichia coli Phylotyping Method Revisited: Improvement of Specificity and Detection of New Phylogroups. Environ. Microbiol. Rep. 5, 58–65. doi: 10.1111/1758-2229.12019
Da Silva, L. C., Santos, A. C. D., Silva, R. M. (2017). Uropathogenic Escherichia Coli Pathogenicity Islands and Other ExPEC Virulence Genes May Contribute to the Genome Variability of Enteroinvasive E. coli. BMC Microbiol. 17, 68. doi: 10.1186/s12866-017-0979-5
Derakhshandeh, A., Eraghi, V., Boroojeni, A. M., Niaki, M. A., Zare, S., Naziri, Z. (2018). Virulence Factors, Antibiotic Resistance Genes and Genetic Relatedness of Commensal Escherichia coli Isolates From Dogs and Their Owners. Microbial. Pathog. 116, 241–245. doi: 10.1016/j.micpath.2018.01.041
Devine, D. A., Robinson, L., Roberts, A. P. (1989). Occurrence of K1, K5 and O Antigens in Escherichia coli Isolates From Patients With Urinary Tract Infections or Bacteraemia. J. Med. Microbiol. 30 (4), 295–299. doi: 10.1099/00222615-30-4-295
Donlan, R. M., Costerton, J. W. (2002). Biofilms: Survival Mechanisms of Clinically Relevant Microorganisms. Clin. Microbiol. Rev. 15 (2), 167–193. doi: 10.1111/j.1440-169x.2004.00774.x
Ejrns, K. (2011). Bacterial Characteristics of Importance for Recurrent Urinary Tract Infections Caused by Escherichia coli. Danish Med. Bull. 58 (4), B4187. doi: 10.1016/S0140-6736(10)62229-5
Gaschignard, J., Levy, C., Romain, O., Cohen, R., Bingen, E., Aujard, Y., et al. (2011). Neonatal Bacterial Meningitis: 444 Cases in 7 Years. Pediatr. Infect. Dis. J. 30 (3), 212. doi: 10.1097/INF.0b013e3181fab1e7
Guo, S. Y., Wakeham, D., Brouwers, H., Cobbold, R. N., Abraham, S., Mollinger, J. L., et al. (2015). Human-Associated Fluoroquinolone-Resistant Escherichia coli Clonal Lineages, Including ST354, Isolated From Canine Feces and Extraintestinal Infections in Australia. Microbes Infect. 17 (4), 266–274. doi: 10.1016/j.micinf.2014.12.016
Hammer, A. S., Jensen, T. H., Høiby, N., Boye, M., Salomonsen, C. M. (2013). Comparison of Histological Lesions in Mink With Acute Hemorrhagic Pneumonia Associated With Pseudomonas Aeruginosa or Escherichia coli. Can. J. Vet. Res. 77 (3), 199–204. doi: 10.9775/kvfd.2012.8415
Han, M. M., Mu, L. Z., Liu, X. P., Zhao, J., Liu, H. (2014). ERIC-PCR Genotyping of Pseudomonas Aeruginosa Isolates From Haemorrhagic Pneumonia Cases in Mink. Vet. Rec. Open 1 (1), e000043–e000043. doi: 10.1136/vropen-2014-000043
Hochhut, B., Wilde, C., Balling, G., Middendorf, B., Dobrindt, U., Brzuszkiewicz, E., et al. (2006). Role of Pathogenicity Island-Associated Integrases in the Genome Plasticity of Uropathogenic Escherichia coli Strain 536. Mol. Microbiol. 61 (3), 584–595. doi: 10.1111/j.1365-2958.2006.05255.x
Hussein, A. H., Ghanem, I. A., Eid, A. A., Ali, M. A., Sherwood, J. S., Li, G., et al. (2013). Molecular and Phenotypic Characterization of Escherichia coli Isolated From Broiler Chicken Flocks in Egypt. Avian Dis. 57 (3), 602–611. doi: 10.1637/10503-012513-Reg.1
Iguchi, A., Iyoda, S., Seto, K., Morita-Ishihara, T., Ohnishi, M. (2015). Escherichia coli O-Genotyping PCR: A Comprehensive and Practical Platform for Molecular O Serogrouping. J. Clin. Microbiol. 53 (8), 2427–2432. doi: 10.1128/JCM.00321-15
Johnson, J. R., Johnston, B., Clabots, C. R., Kuskowski, M. A., Roberts, E., Debroy, C. (2008). Virulence Genotypes and Phylogenetic Background of Escherichia coli Serogroup O6 Isolates From Humans, Dogs, and Cats. J. Clin. Microbiol. 46 (2), 417. doi: 10.1128/JCM.00674-07
Knox, B. (1953). Pseudomonas Aeruginosa Som Rsag Til Enzootiske Infektioner Hos Mink. Vet. Med. 4, 731–760.
Kuo, S. C., Huang, W. C., Wang, H. Y., Shiau, Y. R., Cheng, M. F., Lauderale, T. L. (2016). Colistin Resistance Gene MCR-1 in Escherichia coli Isolates From Humans and Retail Meats, Taiwan. J. Antimicrob. Chemother. 71 (8), 2327–2329. doi: 10.1093/jac/dkw122
Mackenzie, M., Rae, N., Nathwani, D. (2014). Outcomes From Global Adult Outpatient Parenteral Antimicrobial Therapy Programmes: A Review of The Last Decade. Int. J. Antimicrob. Agents 43 (1), 7–16. doi: 10.1016/j.ijantimicag.2013.09.006
Meng, Q. M., Wang, S. H., Han, X. G., Han, Y., Ding, C., Dai, J. J., et al. (2014). Multiplex PCR Assay for Detection of Virulence Genes in Avian Pathogenic Escherichia coli. Acta Microbiol. Sin. 54 (6), 696–702. doi: 10.13343/j.cnki.wsxb.2014.06.013
Nicolas-Chanoine, M. H., Bertrand, X., Madec, J. Y. (2014). Escherichia coli ST131, an Intriguing Clonal Group. Clin. Microbiol. Rev. 27, 543–574. doi: 10.1128/CMR.00125-13
Paixão, A. C., Ferreira, A. C., Fontes, M., Themudo, P., Albuquerque, T., Soares, M. C., et al. (2016). Detection of Virulence-Associated Genes in Pathogenic and Commensal Avian Escherichia coli Isolates. Poult. Sci. 95 (7), 1646–1652. doi: 10.3382/ps/pew087
Pedersen, K., Hammer, A. S., Sørensen, C. M., Heuer, O. E. (2009). Usage of Antimicrobials and Occurrence of Antimicrobial Resistance Among Bacteria From Mink. Vet. Microbiol. 133 (1-2), 115–122. doi: 10.1016/j.vetmic.2008.06.005
Pitoust, J. D., Devinney, R. (2017). Escherichia coli ST131: A Multidrug-Resistant Clone Primed for Global Domination. F1000 Reas. 6, F1000 Faculty Rev-195. doi: 10.12688/f1000research.10609.1
Qi, J., Li, L., Du, Y., Wang, S., Wang, J., Luo, Y., et al. (2014). The Identification, Typing, and Antimicrobial Susceptibility of Pseudomonas Aeruginosa Isolated From Mink With Hemorrhagic Pneumonia. Vet. Microbiol. 170, 456–461. doi: 10.1016/j.vetmic.2014.02.025
Qiu, J. H., Jiang, Z. Y., Ju, Z. J., Zhao, X. N., Yang, J., Guo, H. J., et al. (2019). Molecular and Phenotypic Characteristics of Escherichia coli Isolates From Farmed Minks in ZhuCheng, China. BioMed. Res. Int. 1 (40), 1–12. doi: 10.1155/2019/3917841
Silva, L., Carolina, D., Silva, R. M. (2017). Uropathogenic Escherichia coli Pathogenicity Islands and Other Expec Virulence Genes May Contribute to the Genome Variability of Enteroinvasive E. coli. BMC Microbiol. 17 (1), 68. doi: 10.1186/s12866-017-0979-5
Stepanovic, S., Vukovic, D., Dakic, I., Savic, B., Svabic-Vlahovic, M. (2000). A Modified Microtiter-plate Test for Quantification of Staphylococcal Biofilm Formation. J. Microbiol. Meth. 40 (2), 175–179. doi: 10.1016/s0167-7012(00)00122-6
Suzuki, S., Ohnishi, M., Kawanishi, M., Akiba, M., Kuroda, M. (2016). Investigation of A Plasmid Genome Database for Colistin-Resistance Gene Mcr-1. Lancet Infect. Dis. 16 (3), 284–285. doi: 10.1016/S1473-3099(16)00008-6
TengLin, X. U., Xing, G. L., Jiang, C. G., Liu, J. S., Liu, D. F., Jiang, Q., et al. (2018). Phylogenetic Group, Antimicrobial Resistance and Pathogenic Analysis of Extraintestinal Pathogenic Escherichia coli Isolated From Swine, Chicken and Mink. Chin. J. Prev. Vet. Med. 40, 386–391. doi: 10.3969/j.issn.1008-0589.201712017
Tibbetts, R. J., White, D. G., Dyer, N. W., Giddings, C. W., Nolan, L. K. (2003). Characterization of Escherichia coli Isolates Incriminated in Colisepticaemia in Mink. Vet. Res. Commun. 27 (5), 341–357. doi: 10.1023/A:1024741719361
Vogeleer, P., Tremblay, Y. D. N., Jubelin, G., Jacques, M., Harel, J. (2015). Biofilm-Forming Abilities of Shiga Toxin-Producing Escherichia coli Isolates Associated With Human Infections. Appl. Environ. Microbiol. 82, 1448–1458. doi: 10.1128/aem.02983-15
Wang, S., Niu, C., Shi, Z., Xia, Y., Yaqoob, M., Dai, J., et al. (2011). Effects of ibeA Deletion on Virulence and Biofilm Formation of Avian Pathogenic Escherichia coli. Infect. Immun. 79 (1), 279–287. doi: 10.1128/IAI.00821-10
Wang, J. L., Shang, Y. Y., Guo, S. Y., Diao, F. F., Yu, J. Y., Wei, X. H., et al. (2017). Serotype and Virulence Genes of Klebsiella Pneumoniae Isolated From Mink and its Pathogenesis in Mice and Mink. Sci. Rep. 7 (1), 17291. doi: 10.1038/s41598-017-17681-8
Wang, Y., Yi, L., Wang, Y., Wang, Y., Ding, C. (2016). Isolation, Phylogenetic Group, Drug Resistance, Biofilm Formation, and Adherence Genes of Escherichia coli From Poultry in Central China. Poult. Sci. 95 (12), 2895–2901. doi: 10.3382/ps/pew252
Wani, S. A., Samanta, I., Bhat, M. A., Nishikawa, Y. (2004). Investigation of Shiga Toxin-Producing Escherichia coli in Avian Species in India. Lett. Appl. Microbiol. 39, 389–394. doi: 10.1111/j.1472-765X.2004.01586.x
Wirth, D., Falush, R., Lan, R., Colles, F., Mensa, P., Wieler, L. H., et al. (2006). Sex and Virulence in Escherichia coli: An Evolutionary Perspective. Mol. Microbiol. 60 (5), 1136–1151. doi: 10.1111/j.1365-2958.2006.05172.x
Xu, T. L., Xing, G. L., Jiang, C. G., Liu, J. S., Liu, D. F., Jiang, Q., et al. (2018). Phylogenetic Group, Antimicrobial Resistance and Pathogenic Analysis of Extraintestinal Pathogenic Escherichia coli Isolated From Swine, Chicken and Mink. Chin. J. Prev. Vet. Med. 40, 386–391.
Zhang, H., Wang, X., Zhou, Y., Qiao, L., Zhou, M., Yang, Y., et al. (2019). Identification of Mink Hemorrhagic Pneumonia Caused by Mixed Infection of Escherichia coli and Pseudomonas. Heilongjiang Anim. Sci. Vet. Med. 21), 81–86. doi: 10.13881/j.cnki.hljxmsy.2019.03.0098
Zhang, D. X., Zhang, Z. H., Huang, C. C., Gao, X., Wang, Z., Liu, Y. C., et al. (2017). The Phylogenetic Group, Antimicrobial Susceptibility, and Virulence Genes of Escherichia coli From Clinical Bovine Mastitis. J. Dairy Sci. 101 (1), 572–580. doi: 10.3168/jds.2017-13159
Keywords: Escherichia coli, hemorrhagic pneumonia, serotype, virulence, multilocus sequence typing
Citation: Yu Y, Hu B, Fan H, Zhang H, Lian S, Li H, Li S, Yan X, Wang S and Bai X (2021) Molecular Epidemiology of Extraintestinal Pathogenic Escherichia coli Causing Hemorrhagic Pneumonia in Mink in Northern China. Front. Cell. Infect. Microbiol. 11:781068. doi: 10.3389/fcimb.2021.781068
Received: 22 September 2021; Accepted: 05 October 2021;
Published: 28 October 2021.
Edited by:
Yongsheng Liu, Chinese Academy of Agricultural SciencesCopyright © 2021 Yu, Hu, Fan, Zhang, Lian, Li, Li, Yan, Wang and Bai. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xue Bai, eHVlYmFpXzE5ODJAMTI2LmNvbQ==; Shaohui Wang, c2h3YW5nMDgyN0AxMjYuY29t
 Ying Yu
Ying Yu Bo Hu1
Bo Hu1 Huanhuan Fan
Huanhuan Fan Shaohui Wang
Shaohui Wang Xue Bai
Xue Bai