- 1Department of Microbiology, Yongchuan Hospital of Chongqing Medical University, Chongqing, China
- 2Department of Microbiology, Jiaxing Maternity and Child Health Care Hospital, Jiaxing, China
- 3Department of Microbiology, The First Affiliated Hospital of Jiamusi University, Jiamusi, China
- 4Department of Microbiology, Affiliated Hangzhou Xixi Hospital, Zhejiang University School of Medicine, Hangzhou, China
- 5Central Laboratory of Yongchuan Hospital, Chongqing Medical University, Chongqing, China
Background: This study aimed to determine the molecular characteristics of carbapenem-resistant Klebsiella pneumoniae (CRKP) isolates in a hospital in western Chongqing, southwestern China.
Methods: A total of 127 unique CRKP isolates were collected from the Yongchuan Hospital of Chongqing Medical University, identified using a VITEK-2 compact system, and subjected to microbroth dilution to determine the minimal inhibitory concentration. Enterobacteriaceae intergenic repeat consensus polymerase chain reaction and multilocus sequence typing were used to analyze the homology among the isolates. Genetic information, including resistance and virulence genes, was assessed using polymerase chain reaction. The genomic features of the CRKP carrying gene blaKPC-2 were detected using whole-genome sequencing.
Results: ST11 was the dominant sequence type in the homology comparison. The resistance rate to ceftazidime-avibactam in children was much higher than that in adults as was the detection rate of the resistance gene blaNDM (p < 0.0001). Virulence genes such as mrkD (97.6%), uge (96.9%), kpn (96.9%), and fim-H (84.3%) had high detection rates. IncF (57.5%) was the major replicon plasmid detected, and sequencing showed that the CRKP063 genome contained two plasmids. The plasmid carrying blaKPC-2, which mediates carbapenem resistance, was located on the 359,625 base pair plasmid IncFII, together with virulence factors, plasmid replication protein (rep B), stabilizing protein (par A), and type IV secretion system (T4SS) proteins that mediate plasmid conjugation transfer.
Conclusion: Our study aids in understanding the prevalence of CRKP in this hospital and the significant differences between children and adults, thus providing new ideas for clinical empirical use of antibiotics.
Introduction
Carbapenem-resistant Klebsiella pneumoniae (CRKP) has received increasing attention worldwide because of the widespread misuse of carbapenem antibiotics, with CRKP difficult to treat (Gong et al., 2018; Yungyuen et al., 2021). Several factors contribute to the development of CRKP resistance, among which carbapenemase-producing enzymes are the most predominant resistance mechanism (Conte et al., 2016; El-Badawy et al., 2020; Zhang et al., 2021). In 1996, the blaKPC gene was first identified in the United States (Yigit et al., 2001). The CRKP producing KPC enzyme was the most common CRKP isolate in the global outbreak, causing serious endemic epidemics in Europe, America, Asia, and the Middle East (Munoz-Price et al., 2013; Aires-De-Sousa et al., 2019; Rodrigues et al., 2019; Yu et al., 2019). In China, the detection rate of blaKPC-2 is approximately 73% (Zhang et al., 2017). Previous studies have shown that the highest detection rate of blaNDM among K. pneumoniae isolates was in China, where the detection rate was as high as 44.1% (Safavi et al., 2020). The first detection of the OXA-48 enzyme in CRKP was found in a urine sample from a Turkish patient (Poirel et al., 2004), and more than 50 countries reported the OXA-48 outbreak, including Taiwan, Zhejiang, and other areas in China (Lu et al., 2018; Shu et al., 2019).
A case of liver abscess caused by hypervirulent K. pneumoniae (hvKP) infection was first reported in 1986 in Taiwan Province, China (Liu et al., 1986). Since then, hvKP has been reported in many countries, including China, South Korea, Japan, Spain, Madagascar, Cambodia, and Senegal (Cubero et al., 2016; Zhang Y. et al., 2016; Harada et al., 2019; Yoon et al., 2019; Huynh et al., 2020; Parrott et al., 2021). A defining feature of hvKP is its hypermucoid appearance on agar plates. With its unique phenotypic (such as high mucus type and special serotype) and genotypic (such as carrying special virulence genes) characteristics, hvKP has a strong clinical pathogenicity. It can cause infection in patients with low immune function and severe community-acquired infection in young people with normal immune function, and can present migratory spread (Russo and Marr, 2019). HvKP has emerged because of several mechanisms, including pili, lipopolysaccharides, capsular polysaccharides, virulence factors and iron ingestion (Cheng et al., 2010; Lee et al., 2017; Chen et al., 2020). Patients infected with hvKP often show rapid symptom onset and an insidious disease course, making treatment more difficult (Shon and Russo, 2012; Zhang R. et al., 2016; Rahim et al., 2019). Resistance due to CRKP carrying virulence factors increases the failure rate of patient treatment. Therefore, understanding the virulence of CRKP in hospital isolates is vital.
Ceftazidime-avibactam (CAZ-AVI) is currently an effective treatment for carbapenem-resistant Enterobacteriaceae infection and has been approved for treating adults with complicated urinary tract infections (including pyelonephritis), complicated intra-abdominal infections, hospital-acquired pneumonia (including ventilator-associated pneumonia), and other infections caused by aerobic gram-negative bacteria. Avibactam (AVI), an enzyme inhibitor of triethylenediamine, does not contain a beta-lactam ring, and its enzyme inhibition spectrum is broad. AVI acylates the serine residue of beta-lactamase via noncovalent bonding with the beta-lactamase binding region, thus forming an active covalent compound, and the generated product does not undergo hydrolysis. Thus, the intervention has a long-acting enzyme inhibitory effect (Zhanel et al., 2013). In the CAZ-AVI combination, AVI protects the CAZ from degradation via various serine beta-lactamases, thus promoting CAZ-AVI activity. However, many cases of CAZ-AVI resistance have been reported since CAZ-AVI was approved in 2015 (Shields et al., 2016; Wang et al., 2020). The isolates producing metallo-β-lactamases such as blaNDM are not active against CAZ-AVI (Zhang et al., 2020), and the detection rates of blaNDM are different in different populations. Therefore, detecting the in vitro sensitivity of CAZ-AVI to CRKP would aid in understanding the clinical application of CAZ-AVI in CRKP treatment and predicting the application prospects of CAZ-AVI in future clinical treatments.
Sequence type 258 (ST258), an international high-risk epidemic CRKP lineage, is frequent in European and American countries (Wyres et al., 2020), whereas Sequence type 11 is the most common CRKP clone in China, and is part of the clonal complex 258 (CC258) and is a single-locus variant of ST258 (Liao et al., 2020).
We aimed to study a CRKP collection isolated at the Yongchuan Hospital of Chongqing Medical University, revealing their clinical characteristics, determining their resistance and virulence factors, and investigating the possible underlying resistance mechanisms.
Materials and Methods
Sample Collection
A total of 127 non-duplicate clinical CRKP isolates were collected between July 2018 and May 2020 from the Yongchuan Hospital at Chongqing Medical University, a major comprehensive medical center in western Chongqing province with 1480 beds in 47 wards. Age, sex, length of hospital stay, diagnosis, and patient outcomes were collected from the electronic medical records. All isolates were obtained from different clinical departments and were identified using a VITEK-2 automatic microbiological analyzer (bioMérieux, France) and preserved at -80°C for subsequent study. According to the Clinical and Laboratory Standards Institute guidelines (CLSI, 2020), the clinical isolates used in the present study were not susceptible to carbapenems (meropenem, imipenem, or ertapenem). The Ethics Committee determined that patient consent was not required because the present study was retrospective, and the identities of the patients were anonymized.
Antibiotic Susceptibility Testing
Antibiotic susceptibility testing of each isolate was performed using a VITEK-2 compact automatic microbiological analyzer AST-GN card (bioMérieux). The minimum inhibitory concentration refers to the minimum concentration of the compound (µg/mL) required to stop bacterial growth as determined by the microbroth dilution method. Meropenem, imipenem, amikacin, levofloxacin, polymyxin B, tigecycline (TGC), and CAZ-AVI were used to determine the minimum inhibitory concentrations. ATCC 25922, ATCC 700603, and BAA-1705 were used as quality control isolates. The TGC results were interpreted based on the recommendations of the European Committee on Antimicrobial Susceptibility Testing (Testing, 2021), whereas the results for the other antibiotics were interpreted based on the CLSI (2020) criteria.
Enterobacterial Repetitive Intergenic Consensus-Polymerase Chain Reaction (ERIC-PCR)
ERIC-PCR was used for homology analysis with the primer sequences ERIC-1: 5′- ATGTAAGCTCCTGGGGATTCAC-3′ and ERIC-2:5′- AAGTAAGTGACTGGGGTGAGCG-3′. The reaction conditions and system were as previously reported (Ranjbar and Mirsaeed Ghazi, 2013), and the amplified products were electrophoresed on a 1.5% agarose gel and photographed with a UV gel imaging system. The ERIC-PCR electrophoresis cluster analysis was performed using the NTSYS (V2.10e) software. The isolates with a > 90% similarity coefficient of the cluster dendrogram and no significant band differences were determined to have the same genotype.
Multilocus Sequence Typing (MLST)
MLST was performed using seven housekeeping genes of K. pneumoniae that were amplified using primers from online databases (http://bigsdb.pasteur.fr/klebsiella/primers_used.html). PCR products were sequenced, and sequence types (STs) were determined using online database tools https://bigsdb.pasteur.fr/klebsiella/klebsiella.html.
Resistance and Virulence Genes Molecular Detection
All resistance and virulence genes were detected using PCR, and the DNA was extracted using the boiling method (Srisrattakarn et al., 2017; Gong et al., 2018). We detected resistance genes, including carbapenemase genes (blaKPC, blaNDM, blaIMP-4, blaIMP-8, blaVIM-1, blaVIM-2, and blaOXA-48), AmpC beta-lactamase enzymes (blaDHA and blaACC), ESBL genes (blaSHV, blaTEM, blaCTX-M-1, and blaCTX-M-9), and quinolone resistance genes (qnrA, qnrB, qnrS, qepA, and aac(6’)Ib-cr). The virulence genes detected included fim-H, magA, aero, alls, iroNB, kpn, mrkD, rmpA, uge, and wcaG. All primers were obtained from previous studies (Gay et al., 2006; Park et al., 2006; Ma et al., 2009; El Fertas-Aissani et al., 2013; Compain et al., 2014; Wasfi et al., 2016; Jian-Li et al., 2017; Fu et al., 2018; Gong et al., 2018). Positive amplification products were sequenced, and the sequencing results were compared using the Basic Local Alignment Search Tool (BLAST) at https://blast.ncbi.nlm.nih.gov/Blast.cgi.
Identification of Plasmids by PCR Based Replicon Typing (PBRT)
PBRT was performed to classify the plasmids into various incompatibility groups according to a previously described method (Carattoli et al., 2005). Replicons from 18 major plasmid families in Enterobacteriaceae were searched using PBRT: IncK, IncX, IncY, IncF, IncT, IncP, IncI1, IncFIIA, IncFIB, IncL/M, IncN, IncHI2, IncFIA, IncA/C, IncW, IncHI1, IncFIC, and IncB/O. Positive amplification products were sequenced, and the sequencing results were compared using BLAST.
Whole-Genome Sequencing and Analysis
Bacterial genomic DNA was extracted using a MagAttract HWM DNA Mini Kit (cat. no. 67563) from Qiagen. The samples were subjected to third-generation (Nanopore) and second-generation (Illumina Hiseq) whole-genome sequencing by Shanghai Yuanxin Biological Medicine Technology Co., Ltd. Gene prediction for the bacteria was performed using Glimmer3.02. The resistance and virulence genes were identified after the assembled genome was uploaded to ResFinder and the Virulence Factor Database. The PlasmidFinder Database and BLASTn were used to identify the incompatibility groups. Finally, chromosomal and plasmid maps were drawn using the CGView online tool and BRIG (v0.95) respectively. The genetic structure surrounding blaKPC-2 was drawn using Easyfig (v2.2.5). The CRKP063 genomic sequence has been deposited in GenBank under accession no. MZ156798.
Statistical Analysis
Statistical differences among groups were determined using Chi-square and Fisher’s exact tests in SPSS 23.0, and statistical significance was set at p < 0.05.
Results
Analysis of Clinical Information
A total of 127 unique isolates were obtained from 21 hospital departments; 13 isolates were from children and the remainder were from adults. Among these isolates, 56 (44.1%) were from the intensive care unit (ICU), 13 (10.2%) were from the respiratory medicine department, 10 (7.9%) were from the respiratory and critical care medicine department, nine (7.1%) were from the neonatal unit, and the remainder were from the rehabilitation medicine, hepatobiliary surgery, geriatrics, rheumatology, kidney disease, and endocrine departments. Two (1.6%) isolates came from the pediatric ICU, neurological ICU, hematology, infectious disease department, pediatric, orthopedic, and urology departments, and one isolate was obtained from the neurology, neurosurgery, cardiovascular, gastrointestinal surgery, and medical oncology departments. Regarding sources, 72 (56.7%) isolates were obtained from sputum, 23 (18.1%) were obtained from urine, and 19 (15.0%) were obtained from bronchoalveolar lavage fluid; the remainder were obtained from blood, secretions, puncture fluid, and superficial surgery. Most patients had pulmonary disease and were treated with invasive procedures, such as endotracheal intubation or invasive ventilation. The mortality and improvement rates of patients after infection were 8.7% (11/127) and 33.9% (43/127), respectively. Based on the clinical features summarized. most patients with CRKP (61.4%) had used carbapenems.
Results of Antibiotic Susceptibility Testing
Based on the susceptibility testing results, all isolates were identified as multidrug-resistant, defined as resistant to three or more antibiotic classes (Magiorakos et al., 2012), In addition to the high resistance rate of carbapenem antibiotics, CRKPs had high resistance rate to quinolones. The antibiotic resistance rates of polymyxin B, amikacin, and TGC were 42.5%, 38.6%, and 7.9%, respectively. A total of 16 isolates (12.6%) showed resistance to CAZ-AVI, 10 from children (76.9%) and six from adults (5.6%) (p < 0.0001). The susceptibility of CRKP to different antibiotics between children and adults indicated that the resistance rate of CRKP to levofloxacin in adults was significantly higher than that in children (p < 0.05) (Figure 1).
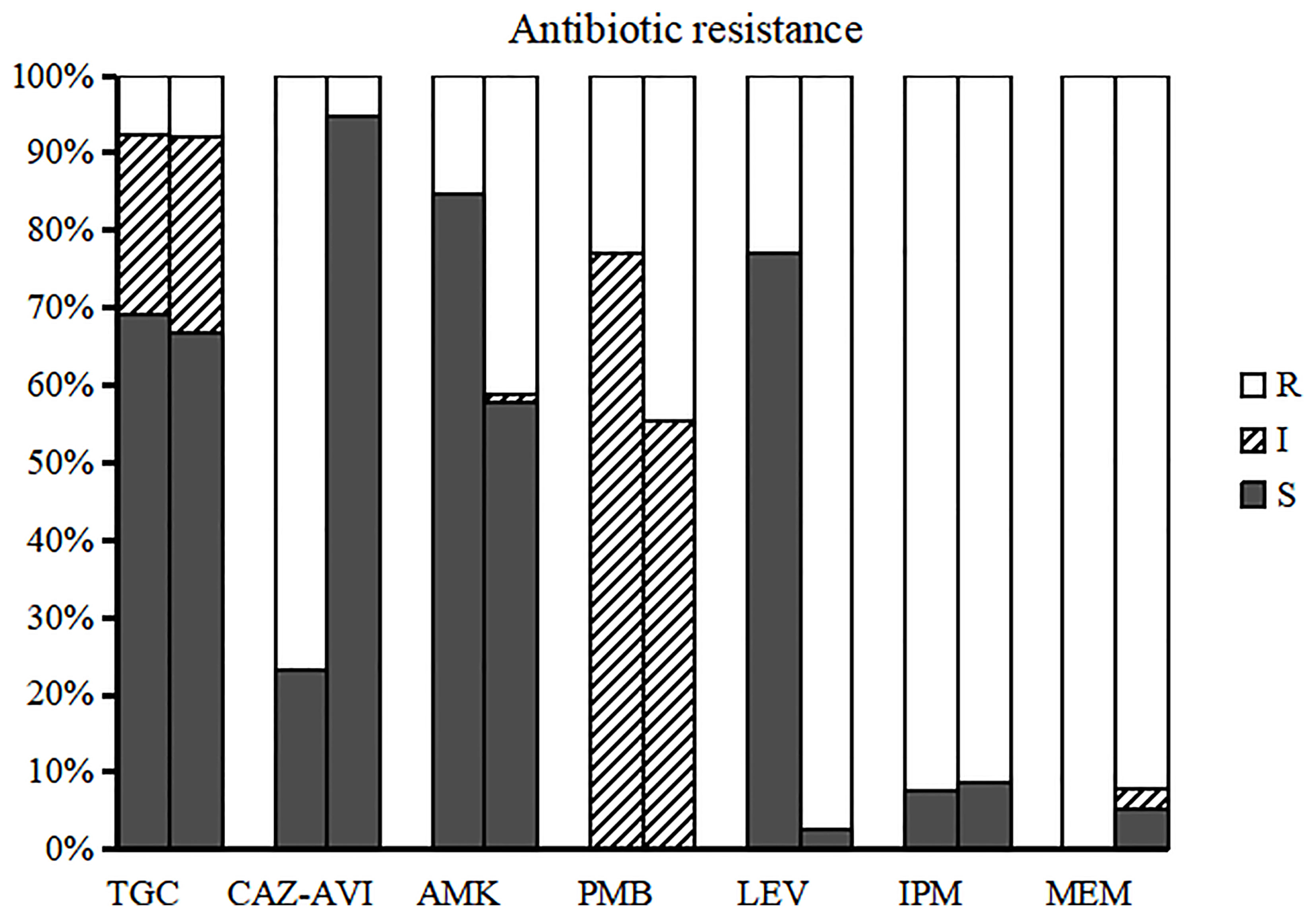
Figure 1 Susceptibility of CRKP to different antimicrobial agents in children and adults. The left bar for each group represents the antibiotic resistance of children and the right bar represents adults. TGC, tigecycline; CAZ-AVI, ceftazidime-avibactam; AMK, amikacin; PMB, polymyxin B; LEV, levofloxacin; IPM, imipenem; MEM, meropenem.
Homology Comparison
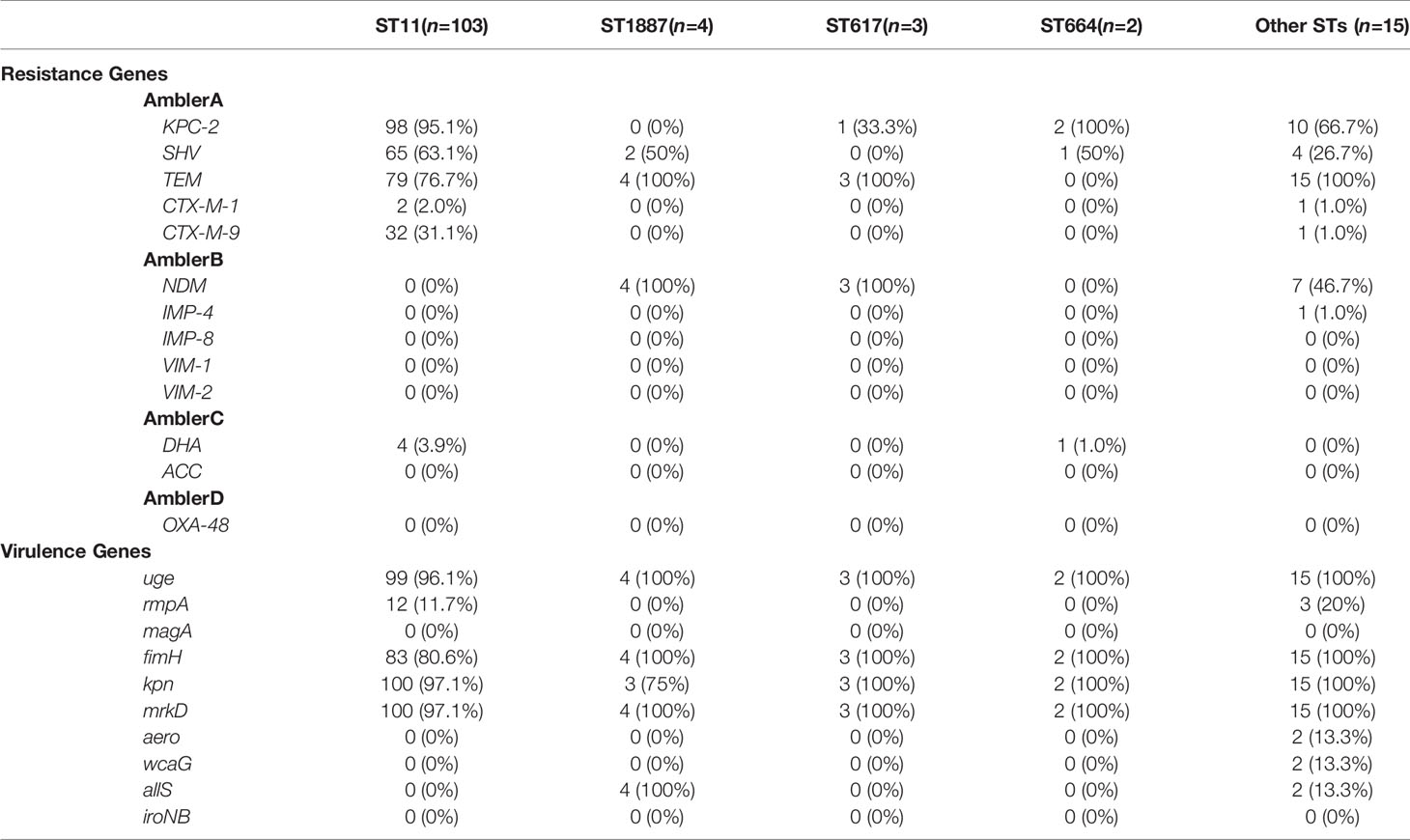
A total of 19 STs were identified among the existing STs in all isolates: 103 (81.1%) belonged to ST11, four (3.1%) belonged to ST1887, three (2.4%) belonged to ST617, two (1.6%) belonged to ST664, and one belonged to each of the other STs. Based on the ERIC-PCR results (Figure 2, Supplementary Figures), six different genotypes (A–F) were observed: 105 (82.7%) were type A, 10 (7.9%) were type B, six (4.7%) were type C, and three pairs were type D, E, and F. The MLST and ERIC-PCR results were not completely consistent; however, both detection methods showed high homology of CRKP in the study hospital.
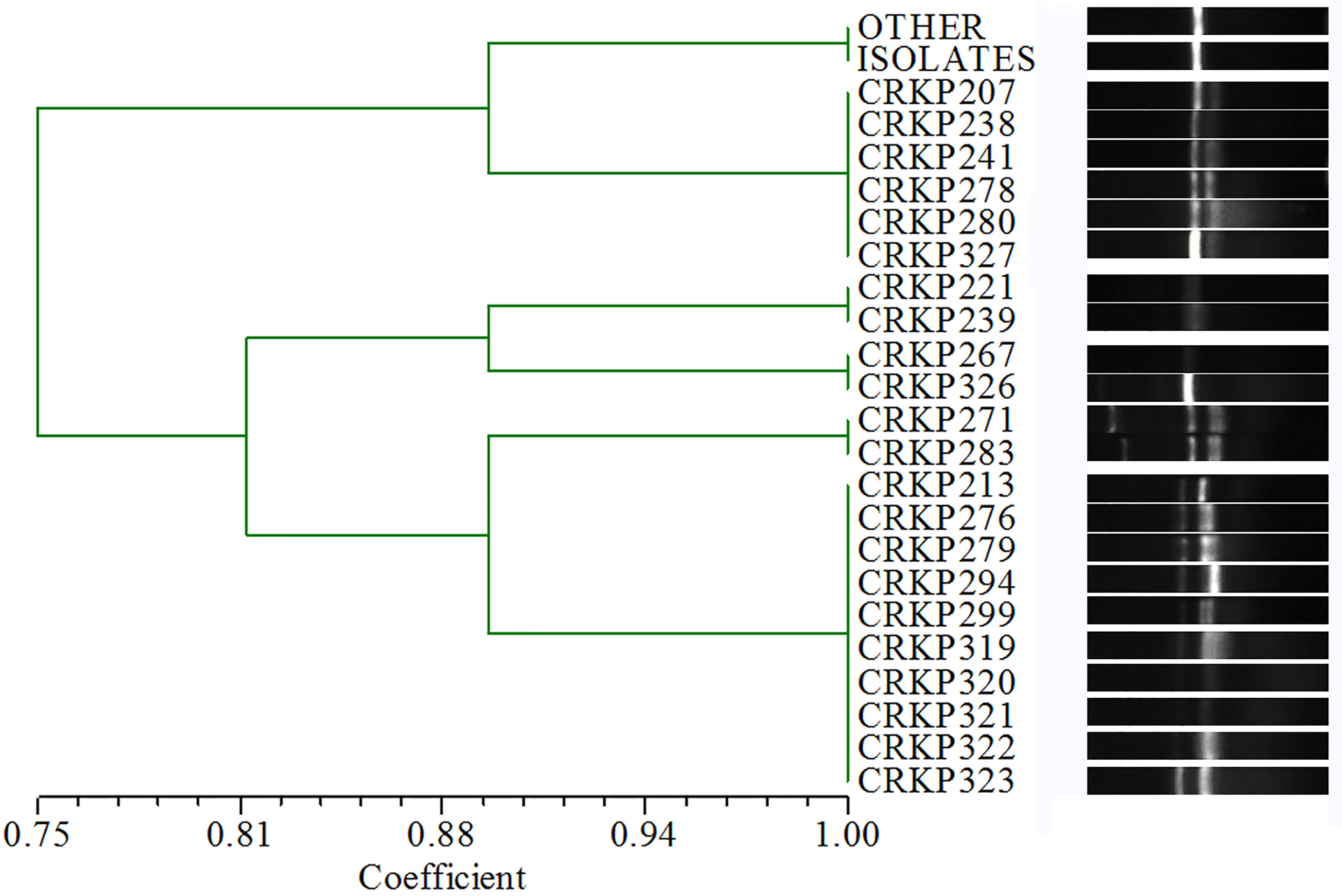
Figure 2 ERIC-PCR results. The isolates with > 90%similarity coefficient for the cluster dendrogram and no significant band difference were determined to have the same genotype. “OTHER ISOLATES” refers to 105 isolates in addition to the 22 isolates listed in the figure.
Detection of Resistance and Virulence Genes
Carbapenem resistance genes such as blaKPC-2 (87.4%), blaNDM (11.0%), and blaIMP-4 were detected. blaTEM (79.5%) and blaSHV (56.7%) also had high detection rates. Other ESBLs, such as blaCTX-M-1 (2.4%), blaCTX-M-9 (25.2%) and blaCTX-M-65 (24.4%), which is a common allelic variant of blaCTX-M-9, were detected. blaDHA (3.9%), an AmpC beta-lactamase, was also detected. qnrB (2.4%), qnrS (13.4%), and aac(6’)-Ib-cr (7.1%) were detected among the plasmid-mediated quinolone resistance genes. Most differences in the detection rates of metalloenzymes or serinases, including blaKPC-2, were not statistically significant between children and adults, except for blaNDM, whose detection rate in children was significantly higher than that in adults (p < 0.0001) (Figure 3). Every isolate in our study had at least two resistance genes detected, except for three isolates with only blaTEM.
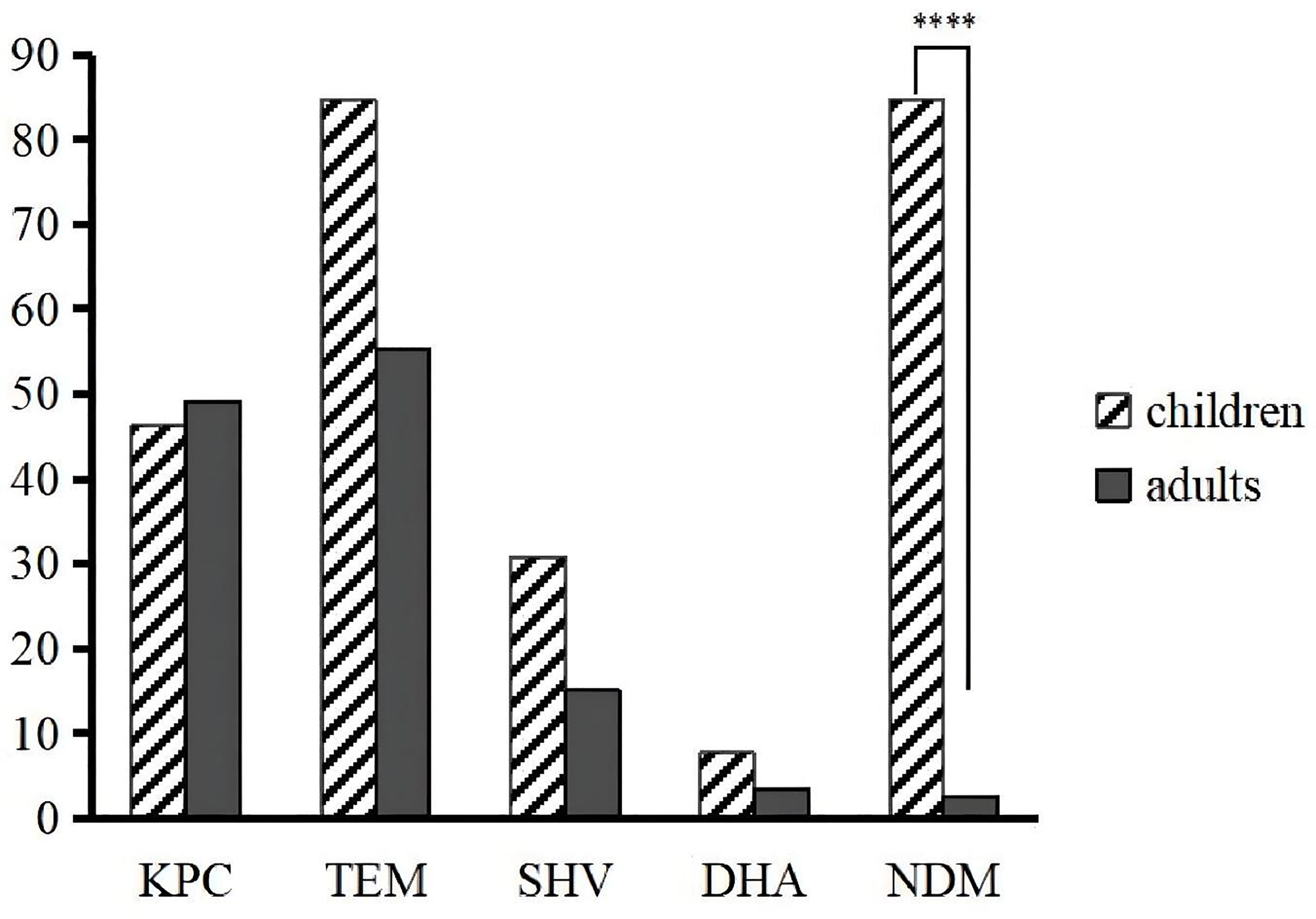
Figure 3 Differences in the prevalence of resistance genes between children and adults. ****p < 0.0001.
Regarding virulence genes, mrkD, uge, kpn, and fim-H had high detection rates in each ST classification. rmpA and allS were detected, including 100% ST1887 isolates with allS. Both aero (1.6%) and wcaG (1.6%) were detected twice. iroNB and magA were not detected in the present study. All isolates were detected to have at least three virulence genes, and the co-detection rate of fim-H, kpn, mrkD, and uge was as high as 63.0%. rmpA and aero were detected simultaneously in two isolates (Table 1).
Identification of Plasmid Incompatibility Groups Using PBRT
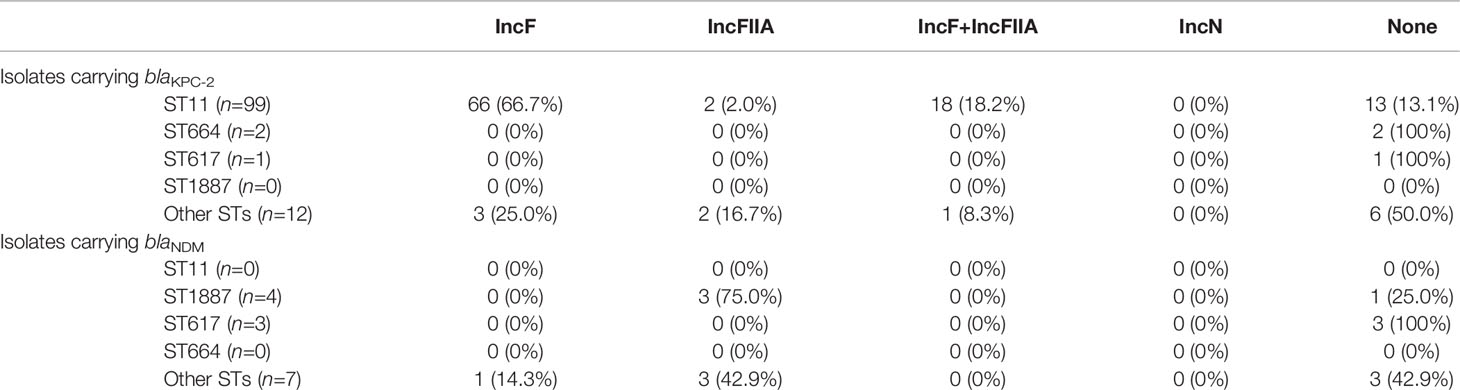
PBRT method revealed that 84 (66.1%) plasmid DNAs belonged to single replicon plasmids, including IncF in 73 (57.5%) isolates, IncFIIA in 10 (7.9%) isolates, and IncN in one isolate. A total of 19 (15.0%) isolates carried multiple Inc groups of plasmids, and IncF + IncFIIA was the only combination detected. No Inc groups could be detected in 24 (18.9%) of isolates. Among the 99 ST11 isolates carrying blaKPC-2, 66 (66.7%) carried IncF plasmids (Table 2).
Genome Sequencing and Analysis
CRKP063, an isolate carrying blaKPC-2, was subjected to genome sequencing and analysis. After nanopore sequencing and filtering of the raw data, the bacterium had 25,901,842 reads with 9,208,426 base pairs (bp), a G + C content of 60.4%, and 7701 annotated protein-coding sequences. The CRKP063 genome consisted of a circular chromosome of 8,074,931 bp and two plasmids. Resistance genes to aminoglycosides (aph(3´)-IIb), fosfomycin (fosA), phenicols (catB7), and beta-lactams (blaKPC-2, blaOXA-396 and blaSHV-182) were identified using whole-genome sequencing. According to our analysis, 37 rRNA genes, 110 tRNA genes, and 42,108 putative open reading frame genes were present on the circular chromosome (Figure 4A). CRKP063 contained a 359,625 bp IncFII plasmid with the siderophore aerobactin genes (iutA and iucABCD); plasmid replication protein (rep B); stabilizing protein (par A); and type IV secretion system (T4SS) proteins traA, traB, traC, traD, and traM, which mediate plasmid conjugation transfer. BLAST comparison revealed similarity of 71% between the plasmid and p205880-2FIIK (accession no. MN824002.1) (Figure 4B). Comparison of the regions surrounding blaKPC-2 revealed that the plasmid contained multiple mobile genetic elements, including ISkpn27, ISkpn6, Tn3-like transposons, and two copies of IS26 (Figure 5). Outside this plasmid, CRKP063 contained other virulence-associated genes, including adhesin gene type 1 fimbriae (fim-H) and type 3 fimbriae (mrkACDF).
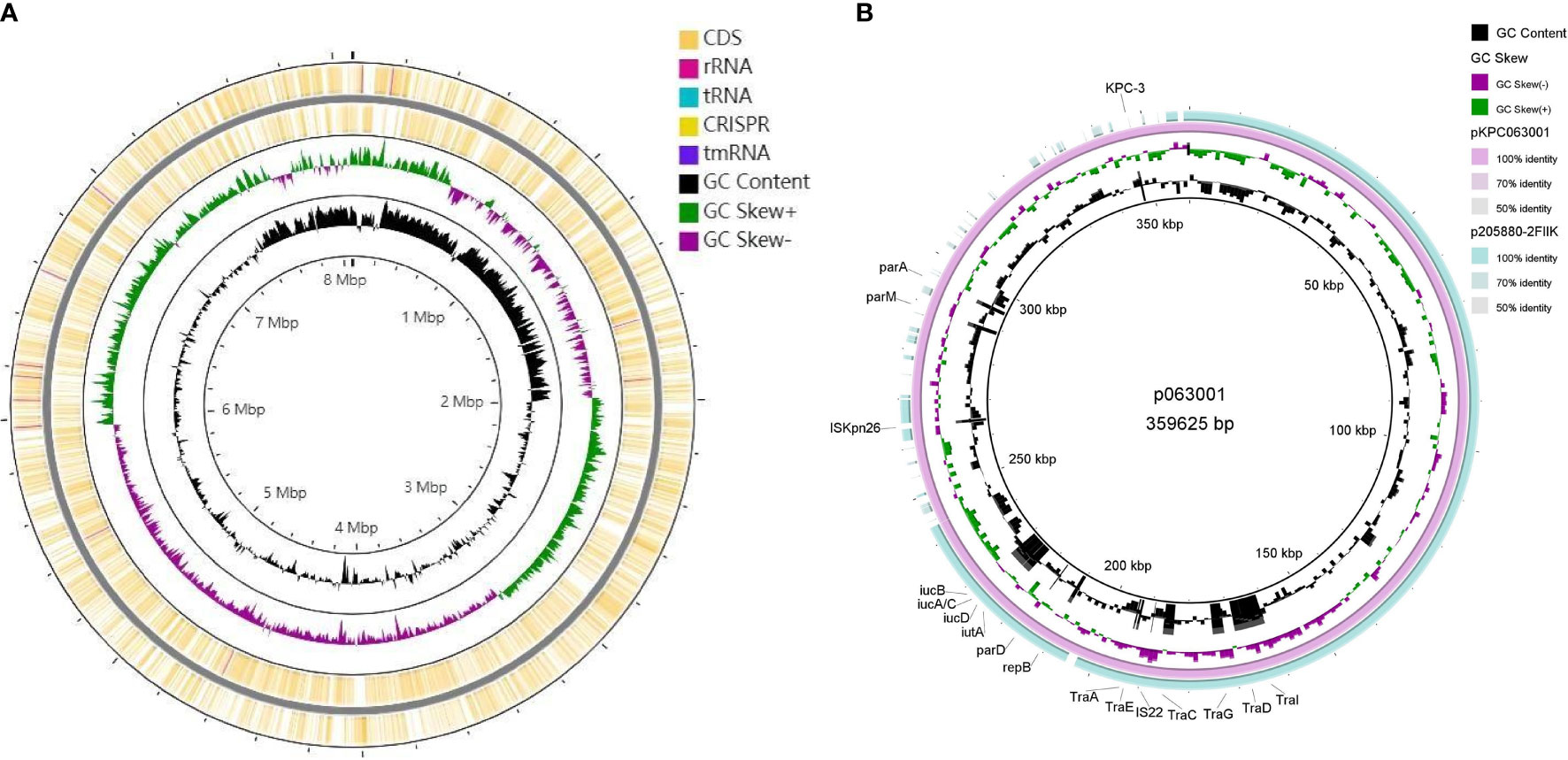
Figure 4 Schematic circular genome of CRKP063. (A) Chromosome circle map. From the outside to the inside, the first two circles represent the coding sequences on the positive and negative chains. The third circle represents the GC skew value. When the value was positive, the positive chain was more likely to transcribe the CDS, and when it was negative, the negative chain was more likely to transcribe the CDS. The fourth circle represents the GC content, and the outer part indicates that the GC content in this region was higher than the average GC content of the whole-genome. (B) Circle map of the IncFII plasmid carrying the blaKPC-2 gene. The outermost circle represents the plasmid p205880-2FIIK (accession no. MN824002.1), possessing high similarity.
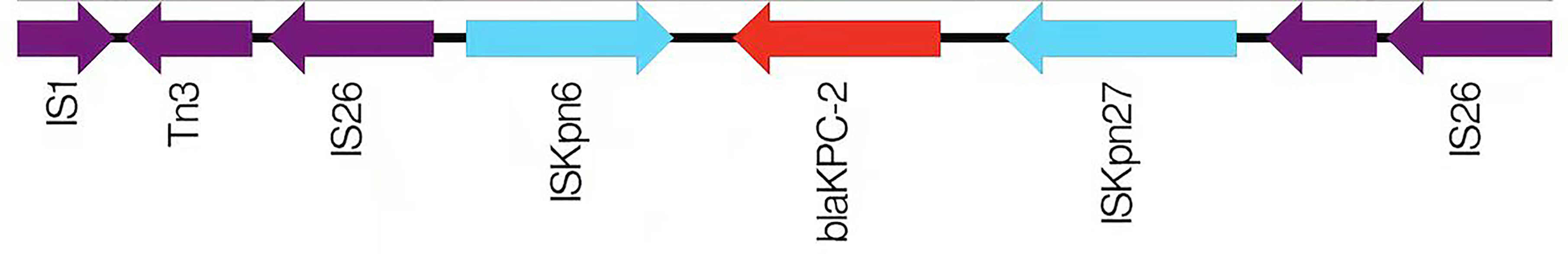
Figure 5 A schematic diagram of the genetic structure surrounding blaKPC-2. The direction of the arrow represents the direction of transcription.
Discussion
As expected, the department with the highest prevalence of CRKP was the ICU, and the major source was sputum (Zhang et al., 2021). The duration of hospital stay in the same department might indicate that the isolates were disseminated within the departments. All mortality occurred in the ICU (10/11) or respiratory and critical care medicine departments; however, the patients rarely had underlying diseases. We hypothesized that mortality was closely associated with complications.
Among the 13 children and 114 adults, the resistance rate to CAZ-AVI was significantly higher in children than in adults. Therefore, in the clinical use of CAZ-AVI, attention must be paid to this result. In addition, the detection rate of blaNDM in children was significantly higher than that in adults. Our results are similar to those of a previous study indicating that acquiring a blaNDM-5-harboring plasmid led to resistance to CAZ-AVI in KPC-2-producing K. pneumoniae during treatment (Huang et al., 2021). With the CAZ-AVI combination, AVI protects CAZ from degradation by various serine beta-lactamases, thus promoting CAZ-AVI activity. Therefore, the two isolates that contained only blaKPC and blaTEM but were resistant to CAZ-AVI require further investigation.
Polymyxins (including colistin and polymyxin B) are the last resort for treating carbapenem-resistant Enterobacteriaceae infections (Macesic et al., 2020); therefore, the resistance in 42.5% of isolates in our study was concerning. A plasmid-mediated polymyxin resistance gene, mcr-1, was first reported in 2015 (Liu et al., 2016), and China had the highest prevalence of mcr-positive isolates (Nang et al., 2019). However, mcr-1 was not detected in any of the isolates. The reasons for this resistance require further study.
Regarding resistance genes, most (98.4%) isolates had more than two genes, indicating that the multiple resistance of CRKP may be because of multiple gene interactions. In the present study, blaKPC-2 was the main carbapenem resistance gene, which agrees with the current prevalence (Hu et al., 2020). Furthermore, the environment surrounding blaKPC-2 as IS26-tnpR-ISkpn27-KPC-2-ISkpn6-IS26-Tn3 was different from the transposon Tn4401 popular abroad (Bowers et al., 2015) and the transposon Tn1721 popular in china (Wang et al., 2015; Yang et al., 2020). It is a typical plasmid-mediated antibiotic resistance gene that is widely distributed in plasmids of different sizes and types (Jain et al., 2013; Pitout et al., 2015), including IncF, IncI, IncA/C, IncN, IncX, IncR, IncP, IncU, IncW, IncL/M, and ColE (Chen et al., 2014). Based on the differences in plasmid replication, blaKPC exists mainly in the IncF plasmid in eastern China (Hu et al., 2020). Our results showed that when blaKPC-2 and plasmids were detected simultaneously, the plasmid was IncF; therefore, the results were consistent with those from previous studies. IncF + IncFIIA appeared in all isolates carrying blaKPC-2, indicating that plasmids have strong mobility and can be transferred between isolates, causing that spread and diffusion of antibiotic resistance.
KPC resistance plasmids simultaneously contain various virulence and retention factors, thus significantly improving the adaptability of KPC-producing strains to the external environment and facilitating the spread of KPC (Dong et al., 2018). uge, mrkD, kpn, and fim-H were present in almost all CRKP isolates in the present study. A previous study showed that fim-H and mrkD encode distinct types of fimbriae that often cause respiratory and urinary tract infections (Yan et al., 2016). Our clinical data showed that 79.0% of the patients had lung diseases, and 71.7% of isolates were from the respiratory tract. Several studies have indicated that isolates coexpressing the mucus phenotype coding gene (rmpA) and aerobin gene (aero) can be considered hvKP (Siu et al., 2012). Two isolates in our study, belonging to ST1373 and ST23 can be defined as hvKP based on this condition. The detection rate of hvKP in the present study was only 1.6%, indicating that the CRKP isolates in this hospital were not yet highly virulent. This result is consistent with those of our previous study in the ICU (Zeng et al., 2021). Several studies have observed that hvKP over time has been susceptible to antibiotics and related mainly to community-acquired infections (Klaper et al., 2021). However, there are reports where this has been reversed and hvKP with antibiotic resistance and involved in infections associated with hospital care are gradually beginning to appear (Gu et al., 2018).
Based on a comparison of MLST and ERIC-PCR, we concluded that all isolates from the hospital had a high degree of homology, particularly in ST11 (81.1%), in agreement with the prevalence in China (Qi et al., 2011; Liao et al., 2020). ST11 has recently been reported elsewhere in China, causing fatal infections and high mortality rates in other hospitals (Hu et al., 2020; Qiao et al., 2020). We concluded that all isolates had high homology based on the similarity coefficients of different clusters by ERIC-PCR. One novel allele of locus infB in MLST was found in the present study, which was defined as infB-236. In addition, three new STs were identified. ST1887 CRKP is a ST commonly found in our hospital, accounting for 3.1% of cases. All four ST1887 isolates expressed the blaNDM and blaTEM resistance genes, and the virulence genes of uge, fim-H, mrkD, and allS, which were higher than those in ST11 CRKP, particularly blaNDM (p < 0.0001) and allS (p < 0.0001) (Table 1).
In summary, ST11 CRKP isolates with blaKPC-2 remain widespread in western Chongqing, southwestern China, and transmission within the hospital should be monitored, and effectively controlled. Effective measures should be taken to prevent further expansion of multi-drug resistant bacteria.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/, MZ156798.
Ethics Statement
The studies involving human participants were reviewed and approved by The Ethics Committee of Yongchuan Hospital of ChongQing Medical University. Written informed consent to participate in this study was provided by the participants’ legal guardian/next of kin.
Author Contributions
BL and XZ conceived of and designed the study. WH and JZ wrote this paper and contributed equally to this work. LZ, CY, LY, and KH performed the experiments. JW, JL, and XL analyzed the data. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by General projects of Chongqing Natural Science Foundation (cstc2020jcyj-msxm0067), Yongchuan Natural Science Foundation (2021yc-jckx20053) and Talent introduction project of Yongchuan Hospital of Chongqing Medical University (YJYJ202005, YJYJ202004).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We would like to thank the curators of the Institute Pasteur MLST system (Paris, France) for information about novel alleles, profiles, andr isolates available at http://bigsdb.web.pasteur.fr. We would also like to thank Dr. Yu YunSong from Sir Run Run Shaw Hospital affiliated with Zhejiang University for providing the EC600 isolates.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcimb.2021.775740/full#supplementary-material
References
Aires-De-Sousa, M., Ortiz de la Rosa, J. M., Goncalves, M. L., Pereira, A. L., Nordmann, P., Poirel, L. (2019). Epidemiology of Carbapenemase-Producing Klebsiella Pneumoniae in a Hospital, Portugal. Emerg. Infect. Dis. 25, 1632–1638. doi: 10.3201/eid2509.190656
Bowers, J. R., Kitchel, B., Driebe, E. M., Maccannell, D. R., Roe, C., Lemmer, D., et al. (2015). Genomic Analysis of the Emergence and Rapid Global Dissemination of the Clonal Group 258 Klebsiella Pneumoniae Pandemic. PloS One 10, e0133727. doi: 10.1371/journal.pone.0133727
Carattoli, A., Bertini, A., Villa, L., Falbo, V., Hopkins, K. L., Threlfall, E. J. (2005). Identification of Plasmids by PCR-Based Replicon Typing. J. Microbiol. Methods 63, 219–228. doi: 10.1016/j.mimet.2005.03.018
Cheng, H. Y., Chen, Y. S., Wu, C. Y., Chang, H. Y., Lai, Y. C., Peng, H. L. (2010). RmpA Regulation of Capsular Polysaccharide Biosynthesis in Klebsiella Pneumoniae CG43. J. Bacteriol 192, 3144–3158. doi: 10.1128/JB.00031-10
Chen, Y., Marimuthu, K., Teo, J., Venkatachalam, I., Cherng, B. P. Z., De Wang, L., et al. (2020). Acquisition of Plasmid With Carbapenem-Resistance Gene Blakpc2 in Hypervirulent Klebsiella Pneumoniae, Singapore. Emerg. Infect. Dis. 26, 549–559. doi: 10.3201/eid2603.191230
Chen, L., Mathema, B., Chavda, K. D., Deleo, F. R., Bonomo, R. A., Kreiswirth, B. N. (2014). Carbapenemase-Producing Klebsiella Pneumoniae: Molecular and Genetic Decoding. Trends Microbiol. 22, 686–696. doi: 10.1016/j.tim.2014.09.003
CLSI (2020). “Performance Standards for Antimicrobial Suceptibility Testing,” in CLSI Supplement M100, 30th. Ed. Wayne, P. A. (Wayne, PA, USA: Clinical and Laboratory Standards Institute).
Compain, F., Babosan, A., Brisse, S., Genel, N., Audo, J., Ailloud, F., et al. (2014). Multiplex PCR for Detection of Seven Virulence Factors and K1/K2 Capsular Serotypes of Klebsiella Pneumoniae. J. Clin. Microbiol. 52, 4377–4380. doi: 10.1128/JCM.02316-14
Conte, V., Monaco, M., Giani, T., D'ancona, F., Moro, M. L., Arena, F., et al. (2016). Molecular Epidemiology of KPC-Producing Klebsiella Pneumoniae From Invasive Infections in Italy: Increasing Diversity With Predominance of the ST512 Clade II Sublineage. J. Antimicrob. Chemother. 71, 3386–3391. doi: 10.1093/jac/dkw337
Cubero, M., Grau, I., Tubau, F., Pallares, R., Dominguez, M. A., Linares, J., et al. (2016). Hypervirulent Klebsiella Pneumoniae Clones Causing Bacteraemia in Adults in a Teaching Hospital in Barcelona, Spai-2013). Clin. Microbiol. Infect. 22, 154–160. doi: 10.1016/j.cmi.2015.09.025
Dong, N., Lin, D., Zhang, R., Chan, E. W., Chen, S. (2018). Carriage of blaKPC-2 by a Virulence Plasmid in Hypervirulent Klebsiella Pneumoniae. J. Antimicrob. Chemother. 73, 3317–3321. doi: 10.1093/jac/dky358
El-Badawy, M. F., El-Far, S. W., Althobaiti, S. S., Abou-Elazm, F. I., Shohayeb, M. M. (2020). The First Egyptian Report Showing the Co-Existence of Bla (NDM-25), Bla (OXA-23), Bla (OXA-181), and Bla (GES-1) Among Carbapenem-Resistant K. Pneumoniae Clinical Isolates Genotyped by BOX-PCR. Infect. Drug Resist. 13, 1237–1250. doi: 10.2147/IDR.S244064
El Fertas-Aissani, R., Messai, Y., Alouache, S., Bakour, R. (2013). Virulence Profiles and Antibiotic Susceptibility Patterns of Klebsiella Pneumoniae Strains Isolated From Different Clinical Specimens. Pathol. Biol. (Paris) 61, 209–216. doi: 10.1016/j.patbio.2012.10.004
Fu, L., Huang, M., Zhang, X., Yang, X., Liu, Y., Zhang, L., et al. (2018). Frequency of Virulence Factors in High Biofilm Formation blaKPC-2 Producing Klebsiella Pneumoniae Strains From Hospitals. Microb. Pathog. 116, 168–172. doi: 10.1016/j.micpath.2018.01.030
Gay, K., Robicsek, A., Strahilevitz, J., Park, C. H., Jacoby, G., Barrett, T. J., et al. (2006). Plasmid-Mediated Quinolone Resistance in non-Typhi Serotypes of Salmonella Enterica. Clin. Infect. Dis. 43, 297–304. doi: 10.1086/505397
Gong, X., Zhang, J., Su, S., Fu, Y., Bao, M., Wang, Y., et al. (2018). Molecular Characterization and Epidemiology of Carbapenem Non-Susceptible Enterobacteriaceae Isolated From the Eastern Region of Heilongjiang Province, China. BMC Infect. Dis. 18, 417. doi: 10.1186/s12879-018-3294-3
Gu, D., Dong, N., Zheng, Z., Lin, D., Huang, M., Wang, L., et al. (2018). A Fatal Outbreak of ST11 Carbapenem-Resistant Hypervirulent Klebsiella Pneumoniae in a Chinese Hospital: A Molecular Epidemiological Study. Lancet Infect. Dis. 18, 37–46. doi: 10.1016/S1473-3099(17)30489-9
Harada, S., Aoki, K., Yamamoto, S., Ishii, Y., Sekiya, N., Kurai, H., et al. (2019). Clinical and Molecular Characteristics of Klebsiella Pneumoniae Isolates Causing Bloodstream Infections in Japan: Occurrence of Hypervirulent Infections in Health Care. J. Clin. Microbiol. 57 (18), e01206–19. doi: 10.1128/JCM.01206-19
Huang, J., Zhang, S., Zhao, Z., Chen, M., Cao, Y., Li, B. (2021). Acquisition of a Stable and Transferable Bla NDM-5-Positive Plasmid With Low Fitness Cost Leading to Ceftazidime/Avibactam Resistance in KPC-2-Producing Klebsiella Pneumoniae During Treatment. Front. Cell Infect. Microbiol. 11, 658070. doi: 10.3389/fcimb.2021.658070
Hu, Y., Liu, C., Shen, Z., Zhou, H., Cao, J., Chen, S., et al. (2020). Prevalence, Risk Factors and Molecular Epidemiology of Carbapenem-Resistant Klebsiella Pneumoniae in Patients From Zhejiang, Chin-2018. Emerg. Microbes Infect. 9, 1771–1779. doi: 10.1080/22221751.2020.1799721
Huynh, B. T., Passet, V., Rakotondrasoa, A., Diallo, T., Kerleguer, A., Hennart, M., et al. (2020). Klebsiella Pneumoniae Carriage in Low-Income Countries: Antimicrobial Resistance, Genomic Diversity and Risk Factors. Gut Microbes 11, 1287–1299. doi: 10.1080/19490976.2020.1748257
Jain, R., Walk, S. T., Aronoff, D. M., Young, V. B., Newton, D. W., Chenoweth, C. E., et al. (2013). Emergence of Carbapenemaseproducing Klebsiella Pneumoniae of Sequence Type 258 in Michigan, USA. Infect. Dis. Rep. 5, e5. doi: 10.4081/idr.2013.e5
Jian-Li, W., Yuan-Yuan, S., Shou-Yu, G., Fei-Fei, D., Jia-Yu, Y., Xue-Hua, W., et al. (2017). Serotype and Virulence Genes of Klebsiella Pneumoniae Isolated From Mink and its Pathogenesis in Mice and Mink. Sci. Rep. 7, 17291. doi: 10.1038/s41598-017-17681-8
Klaper, K., Wendt, S., Lubbert, C., Lippmann, N., Pfeifer, Y., Werner, G. (2021). Hypervirulent Klebsiella Pneumoniae of Lineage ST66-K2 Caused Tonsillopharyngitis in a German Patient. Microorganisms 9 (18), 133. doi: 10.3390/microorganisms9010133
Lee, C. R., Lee, J. H., Park, K. S., Jeon, J. H., Kim, Y. B., Cha, C. J., et al. (2017). Antimicrobial Resistance of Hypervirulent Klebsiella Pneumoniae: Epidemiology, Hypervirulence-Associated Determinants, and Resistance Mechanisms. Front. Cell Infect. Microbiol. 7, 483. doi: 10.3389/fcimb.2017.00483
Liao, W., Liu, Y., Zhang, W. (2020). Virulence Evolution, Molecular Mechanisms of Resistance and Prevalence of ST11 Carbapenem-Resistant Klebsiella Pneumoniae in China: A Review Over the Last 10 Years. J. Glob Antimicrob. Resist. 23, 174–180. doi: 10.1016/j.jgar.2020.09.004
Liu, Y.-C., Cheng, D.-L., Lin, C.-L. (1986). Klebsiella Pneumoniae Liver Abscess Associated With Septic Endophthalmitis. Arch. Internal Med. 146, 1913–1916. doi: 10.1001/archinte.1986.00360220057011
Liu, Y.-Y., Wang, Y., Walsh, T. R., Yi, L.-X., Zhang, R., Spencer, J., et al. (2016). Emergence of Plasmid-Mediated Colistin Resistance Mechanism MCR-1 in Animals and Human Beings in China: A Microbiological and Molecular Biological Study. Lancet Infect. Dis. 16, 161–168. doi: 10.1016/S1473-3099(15)00424-7
Lu, M. C., Tang, H. L., Chiou, C. S., Wang, Y. C., Chiang, M. K., Lai, Y. C. (2018). Clonal Dissemination of Carbapenemase-Producing Klebsiella Pneumoniae: Two Distinct Sub-Lineages of Sequence Type 11 Carrying blaKPC-2 and blaOXA-48. Int. J. Antimicrob. Agents 52, 658–662. doi: 10.1016/j.ijantimicag.2018.04.023
Macesic, N., Nelson, B., Mcconville, T. H., Giddins, M. J., Green, D. A., Stump, S., et al. (2020). Emergence of Polymyxin Resistance in Clinical Klebsiella Pneumoniae Through Diverse Genetic Adaptations: A Genomic, Retrospective Cohort Study. Clin. Infect. Dis. 70, 2084–2091. doi: 10.1093/cid/ciz623
Magiorakos, A. P., Srinivasan, A., Carey, R. B., Carmeli, Y., Falagas, M. E., Giske, C. G., et al. (2012). Multidrug-Resistant, Extensively Drug-Resistant and Pandrug-Resistant Bacteria: An International Expert Proposal for Interim Standard Definitions for Acquired Resistance. Clin. Microbiol. Infect. 18, 268–281. doi: 10.1111/j.1469-0691.2011.03570.x
Ma, J., Zeng, Z., Chen, Z., Xu, X., Wang, X., Deng, Y., et al. (2009). High Prevalence of Plasmid-Mediated Quinolone Resistance Determinants Qnr, Aac(6')-Ib-Cr, and qepA Among Ceftiofur-Resistant Enterobacteriaceae Isolates From Companion and Food-Producing Animals. Antimicrob. Agents Chemother. 53, 519–524. doi: 10.1128/AAC.00886-08
Munoz-Price, L. S., Poirel, L., Bonomo, R. A., Schwaber, M. J., Daikos, G. L., Cormican, M., et al. (2013). Clinical Epidemiology of the Global Expansion of Klebsiella Pneumoniae Carbapenemases. Lancet Infect. Dis. 13, 785–796. doi: 10.1016/S1473-3099(13)70190-7
Nang, S. C., Li, J., Velkov, T. (2019). The Rise and Spread of Mcr Plasmid-Mediated Polymyxin Resistance. Crit. Rev. Microbiol. 45, 131–161. doi: 10.1080/1040841X.2018.1492902
Park, C. H., Robicsek, A., Jacoby, G. A., Sahm, D., Hooper, D. C. (2006). Prevalence in the United States of Aac(6')-Ib-Cr Encoding a Ciprofloxacin-Modifying Enzyme. Antimicrob. Agents Chemother. 50, 3953–3955. doi: 10.1128/AAC.00915-06
Parrott, A. M., Shi, J., Aaron, J., Green, D. A., Whittier, S., Wu, F. (2021). Detection of Multiple Hypervirulent Klebsiella Pneumoniae Strains in a New York City Hospital Through Screening of Virulence Genes. Clin. Microbiol. Infect. 27, 583–589. doi: 10.1016/j.cmi.2020.05.012
Pitout, J. D., Nordmann, P., Poirel, L. (2015). Carbapenemase-Producing Klebsiella Pneumoniae, a Key Pathogen Set for Global Nosocomial Dominance. Antimicrob. Agents Chemother. 59, 5873–5884. doi: 10.1128/AAC.01019-15
Poirel, L., Heritier, C., Tolun, V., Nordmann, P. (2004). Emergence of Oxacillinase-Mediated Resistance to Imipenem in Klebsiella Pneumoniae. Antimicrob. Agents Chemother. 48, 15–22. doi: 10.1128/AAC.48.1.15-22.2004
Qiao, F., Wei, L., Feng, Y., Ran, S., Zheng, L., Zhang, Y., et al. (2020). Handwashing Sink Contamination and Carbapenem-Resistant Klebsiella Infection in the Intensive Care Unit: A Prospective Multicenter Study. Clin. Infect. Dis. 71, S379–S385. doi: 10.1093/cid/ciaa1515
Qi, Y., Wei, Z., Ji, S., Du, X., Shen, P., Yu, Y. (2011). ST11, the Dominant Clone of KPC-Producing Klebsiella Pneumoniae in China. J. Antimicrob. Chemother. 66, 307–312. doi: 10.1093/jac/dkq431
Rahim, G. R., Gupta, N., Maheshwari, P., Singh, M. P. (2019). Monomicrobial Klebsiella Pneumoniae Necrotizing Fasciitis: An Emerging Life-Threatening Entity. Clin. Microbiol. Infect. 25, 316–323. doi: 10.1016/j.cmi.2018.05.008
Ranjbar, R., Mirsaeed Ghazi, F. (2013). Antibiotic Sensitivity Patterns and Molecular Typing of Shigella Sonnei Strains Using ERIC-PCR. Iran J. Public Health 42, 1151–1157.
Rodrigues, A. C. S., Santos, I. C. O., Campos, C. C., Rezende, I. N., Ferreira, Y. M., Chaves, C. E. V., et al. (2019). Non-Clonal Occurrence of pmrB Mutations Associated With Polymyxin Resistance in Carbapenem-Resistant Klebsiella Pneumoniae in Brazil. Mem Inst Oswaldo Cruz 114, e180555. doi: 10.1590/0074-02760180555
Russo, T. A., Marr, C. M. (2019). Hypervirulent Klebsiella Pneumoniae. Clin. Microbiol. Rev. 32 (3), e00001–19. doi: 10.1128/CMR.00001-19
Safavi, M., Bostanshirin, N., Hajikhani, B., Yaslianifard, S., Van Belkum, A., Goudarzi, M., et al. (2020). Global Genotype Distribution of Human Clinical Isolates of New Delhi Metallo-Beta-Lactamase-Producing Klebsiella Pneumoniae; A Systematic Review. J. Glob Antimicrob. Resist. 23, 420–429. doi: 10.1016/j.jgar.2020.10.016
Shields, R. K., Potoski, B. A., Haidar, G., Hao, B., Doi, Y., Chen, L., et al. (2016). Clinical Outcomes, Drug Toxicity, and Emergence of Ceftazidime-Avibactam Resistance Among Patients Treated for Carbapenem-Resistant Enterobacteriaceae Infections. Clin. Infect. Dis. 63, 1615–1618. doi: 10.1093/cid/ciw636
Shon, A. S., Russo, T. A. (2012). Hypervirulent Klebsiella Pneumoniae: The Next Superbug? Future Microbiol. 7, 669–671. doi: 10.2217/fmb.12.43
Shu, L., Dong, N., Lu, J., Zheng, Z., Hu, J., Zeng, W., et al. (2019). Emergence of OXA-232 Carbapenemase-Producing Klebsiella Pneumoniae That Carries a pLVPK-Like Virulence Plasmid Among Elderly Patients in China. Antimicrob. Agents Chemother. 63 (3), e02246–18. doi: 10.1128/AAC.02246-18
Siu, L. K., Yeh, K.-M., Lin, J.-C., Fung, C.-P., Chang, F.-Y. (2012). Klebsiella Pneumoniae Liver Abscess: A New Invasive Syndrome. Lancet Infect. Dis. 12, 881–887. doi: 10.1016/S1473-3099(12)70205-0
Srisrattakarn, A., Lulitanond, A., Wilailuckana, C., Charoensri, N., Wonglakorn, L., Saenjamla, P., et al. (2017). And Chanawong, ARapid and Simple Identification of Carbapenemase Genes, Bla NDM, Bla OXA-48, Bla VIM, Bla IMP-14 and Bla KPC Groups, in Gram-Negative Bacilli by in-House Loop-Mediated Isothermal Amplification With Hydroxynaphthol Blue Dye. World J. Microbiol. Biotechnol. 33, 130. doi: 10.1007/s11274-017-2295-5
The European Committee on Antimicrobial Susceptibility Testing (2021). Breakpoint Tables for Interpretation of MICs and Zone Diameters, Version 11.0. Available at: http://www.eucast.org/clinical_breakpoints/.
Wang, L., Fang, H., Feng, J., Yin, Z., Xie, X., Zhu, X., et al. (2015). Complete Sequences of KPC-2-Encoding Plasmid P628-KPC and CTX-M-55-Encoding P628-CTXM Coexisted in Klebsiella Pneumoniae. Front. Microbiol. 6, 838. doi: 10.3389/fmicb.2015.00838
Wang, Y., Wang, J., Wang, R., Cai, Y. (2020). Resistance to Ceftazidime-Avibactam and Underlying Mechanisms. J. Glob Antimicrob. Resist. 22, 18–27. doi: 10.1016/j.jgar.2019.12.009
Wasfi, R., Elkhatib, W. F., Ashour, H. M. (2016). Molecular Typing and Virulence Analysis of Multidrug Resistant Klebsiella Pneumoniae Clinical Isolates Recovered From Egyptian Hospitals. Sci. Rep. 6, 38929. doi: 10.1038/srep38929
Wyres, K. L., Lam, M. M. C., Holt, K. E. (2020). Population Genomics of Klebsiella Pneumoniae. Nat. Rev. Microbiol. 18, 344–359. doi: 10.1038/s41579-019-0315-1
Yang, Q., Jia, X., Zhou, M., Zhang, H., Yang, W., Kudinha, T., et al. (2020). Emergence of ST11-K47 and ST11-K64 Hypervirulent Carbapenem-Resistant Klebsiella Pneumoniae in Bacterial Liver Abscesses From China: A Molecular, Biological, and Epidemiological Study. Emerg. Microbes Infect. 9, 320–331. doi: 10.1080/22221751.2020.1721334
Yan, Q., Zhou, M., Zou, M., Liu, W. E. (2016). Hypervirulent Klebsiella Pneumoniae Induced Ventilator-Associated Pneumonia in Mechanically Ventilated Patients in China. Eur. J. Clin. Microbiol. Infect. Dis. 35, 387–396. doi: 10.1007/s10096-015-2551-2
Yigit, H., Queenan, A. M., Anderson, G. J., Domenech-Sanchez, A., Biddle, J. W., Steward, C. D., et al. (2001). Novel Carbapenem-Hydrolyzing Beta-Lactamase, KPC-1, From a Carbapenem-Resistant Strain of Klebsiella Pneumoniae. Antimicrob. Agents Chemother. 45, 1151–1161. doi: 10.1128/AAC.45.4.1151-1161.2001
Yoon, J. H., Kim, Y. J., Kim, S. I. (2019). Prognosis of Liver Abscess With No Identified Organism. BMC Infect. Dis. 19, 488. doi: 10.1186/s12879-019-4131-z
Yungyuen, T., Chatsuwan, T., Plongla, R., Kanthawong, S., Yordpratum, U., Voravuthikunchai, S. P., et al. (2021). Nationwide Surveillance and Molecular Characterization of Critically Drug-Resistant Gram-Negative Bacteria: Results of the Research University Network Thailand Study. Antimicrob. Agents Chemother., 65 (9), e0067521. doi: 10.1128/AAC.00675-21
Yu, X., Zhang, W., Zhao, Z., Ye, C., Zhou, S., Wu, S., et al. (2019). Molecular Characterization of Carbapenem-Resistant Klebsiella Pneumoniae Isolates With Focus on Antimicrobial Resistance. BMC Genomics 20, 822. doi: 10.1186/s12864-019-6225-9
Zeng, L., Yang, C., Zhang, J., Hu, K., Zou, J., Li, J., et al. (2021). An Outbreak of Carbapenem-Resistant Klebsiella Pneumoniae in an Intensive Care Unit of a Major Teaching Hospital in Chongqing, China. Front. Cell Infect. Microbiol. 11, 656070. doi: 10.3389/fcimb.2021.656070
Zhanel, G. G., Lawson, C. D., Adam, H., Schweizer, F., Zelenitsky, S., Lagace-Wiens, P. R., et al. (2013). Ceftazidime-Avibactam: A Novel Cephalosporin/Beta-Lactamase Inhibitor Combination. Drugs 73, 159–177. doi: 10.1007/s40265-013-0013-7
Zhang, W. X., Chen, H. Y., Chen, C., Chen, J. H., Wan, F. S., Li, L. X., et al. (2021). Resistance Phenotype and Molecular Epidemiology of Carbapenem-Resistant Klebsiella Pneumoniae Isolates in Shanghai. Microb. Drug Resist. 27 (10), 1312–1318. doi: 10.1089/mdr.2020.0390
Zhang, R., Lin, D., Chan, E. W., Gu, D., Chen, G. X., Chen, S. (2016). Emergence of Carbapenem-Resistant Serotype K1 Hypervirulent Klebsiella Pneumoniae Strains in China. Antimicrob. Agents Chemother. 60, 709–711. doi: 10.1128/AAC.02173-15
Zhang, R., Liu, L., Zhou, H., Chan, E. W., Li, J., Fang, Y., et al. (2017). Nationwide Surveillance of Clinical Carbapenem-Resistant Enterobacteriaceae (CRE) Strains in China. EBioMedicine 19, 98–106. doi: 10.1016/j.ebiom.2017.04.032
Zhang, P., Shi, Q., Hu, H., Hong, B., Wu, X., Du, X., et al. (2020). Emergence of Ceftazidime/Avibactam Resistance in Carbapenem-Resistant Klebsiella Pneumoniae in China. Clin. Microbiol. Infect. 26, 124.e121–124.e124. doi: 10.1016/j.cmi.2019.08.020
Zhang, Y., Zhao, C., Wang, Q., Wang, X., Chen, H., Li, H., et al. (2016). High Prevalence of Hypervirulent Klebsiella Pneumoniae Infection in China: Geographic Distribution, Clinical Characteristics, and Antimicrobial Resistance. Antimicrob. Agents Chemother. 60, 6115–6120. doi: 10.1128/AAC.01127-16
Keywords: carbapenem-resistant Klebsiella pneumoniae, antibiotic susceptibility, molecular epidemiology, whole-genome sequencing, ST1887, ceftazidime-avibactam
Citation: Huang W, Zhang J, Zeng L, Yang C, Yin L, Wang J, Li J, Li X, Hu K, Zhang X and Liu B (2022) Carbapenemase Production and Epidemiological Characteristics of Carbapenem-Resistant Klebsiella pneumoniae in Western Chongqing, China. Front. Cell. Infect. Microbiol. 11:775740. doi: 10.3389/fcimb.2021.775740
Received: 14 September 2021; Accepted: 13 December 2021;
Published: 04 January 2022.
Edited by:
Milena Dropa, University of São Paulo, BrazilReviewed by:
Graciela Castro Escarpulli, Instituto Politécnico Nacional de México (IPN), MexicoZhenbo Xu, South China University of Technology, China
Copyright © 2022 Huang, Zhang, Zeng, Yang, Yin, Wang, Li, Li, Hu, Zhang and Liu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Beizhong Liu, bGl1YmVpemhvbmdAY3FtdS5lZHUuY24=; Xiaoli Zhang, am1zenhsMTIzQDE2My5jb20=
†These authors have contributed equally to this work
 Wan Huang
Wan Huang Jisheng Zhang1†
Jisheng Zhang1† Jianmin Wang
Jianmin Wang Xiaoli Zhang
Xiaoli Zhang