- 1Public Health Research Institute, Rutgers University, New Brunswick, NJ, United States
- 2Department of Microbiology, Biochemistry and Molecular Genetics, Rutgers University, Newark, NJ, United States
- 3Rutgers Center for Lipid Research, Rutgers University, New Brunswick, NJ, United States
Ubiquitin-proteasome mediated protein turnover is an important regulatory mechanism of cellular function in eukaryotes. Extensive studies have linked the ubiquitin-proteasome system (UPS) to human diseases, and an array of proteasome inhibitors have been successfully developed for cancer therapy. Although still an emerging field, research on UPS regulation of fungal development and virulence has been rapidly advancing and has generated considerable excitement in its potential as a target for novel drugs. In this review, we summarize UPS composition and regulatory function in pathogenic fungi, especially in stress responses, host adaption, and fungal pathogenesis. Emphasis will be given to UPS regulation of pathogenic factors that are important for fungal pathogenesis. We also discuss future potential therapeutic strategies for fungal infections based on targeting UPS pathways.
Overview of the Ubiquitin-Proteasome System
Ubiquitin exists in all eukaryotic cells and it modifies proteins for proteasomal degradation and non-proteolytic functions (Finley et al., 2012). The ubiquitin-proteasome system (UPS) is important for a multitude of cellular processes due to its capability to rapidly and selectively turnover intracellular proteins. The UPS involves ubiquitin activation, ubiquitin–substrate conjugation, ubiquitin receptor recognition, and proteasomal degradation (Glickman and Ciechanover, 2002; Finley et al., 2012) (Figure 1). Protein ubiquitination is initiated by the ubiquitin-activating enzyme E1 in an ATP-dependent manner. Activated ubiquitin is transferred to the ubiquitin-conjugating enzyme E2. Thereafter, ubiquitin ligase E3 binds to E2 and catalyzes the covalent attachment of ubiquitin to the lysine residues of target substrates. Polyubiquitin chains are synthesized after several rounds of conjugation. Polyubiquitinated proteins are directly recognized by the 26S proteasome or delivered to the proteasome by ubiquitin receptor proteins (Elsasser and Finley, 2005; Finley et al., 2012), and then are degraded into small peptides and reusable ubiquitin (Glickman and Ciechanover, 2002). Protein ubiquitination is a reversible post-translational modification. Deubiquitinating enzymes (DUB) catalyze protein deubiquitination to prevent proteins from 26S proteasome-mediated degradation and maintain ubiquitin homeostasis in cells (Komander et al., 2009).
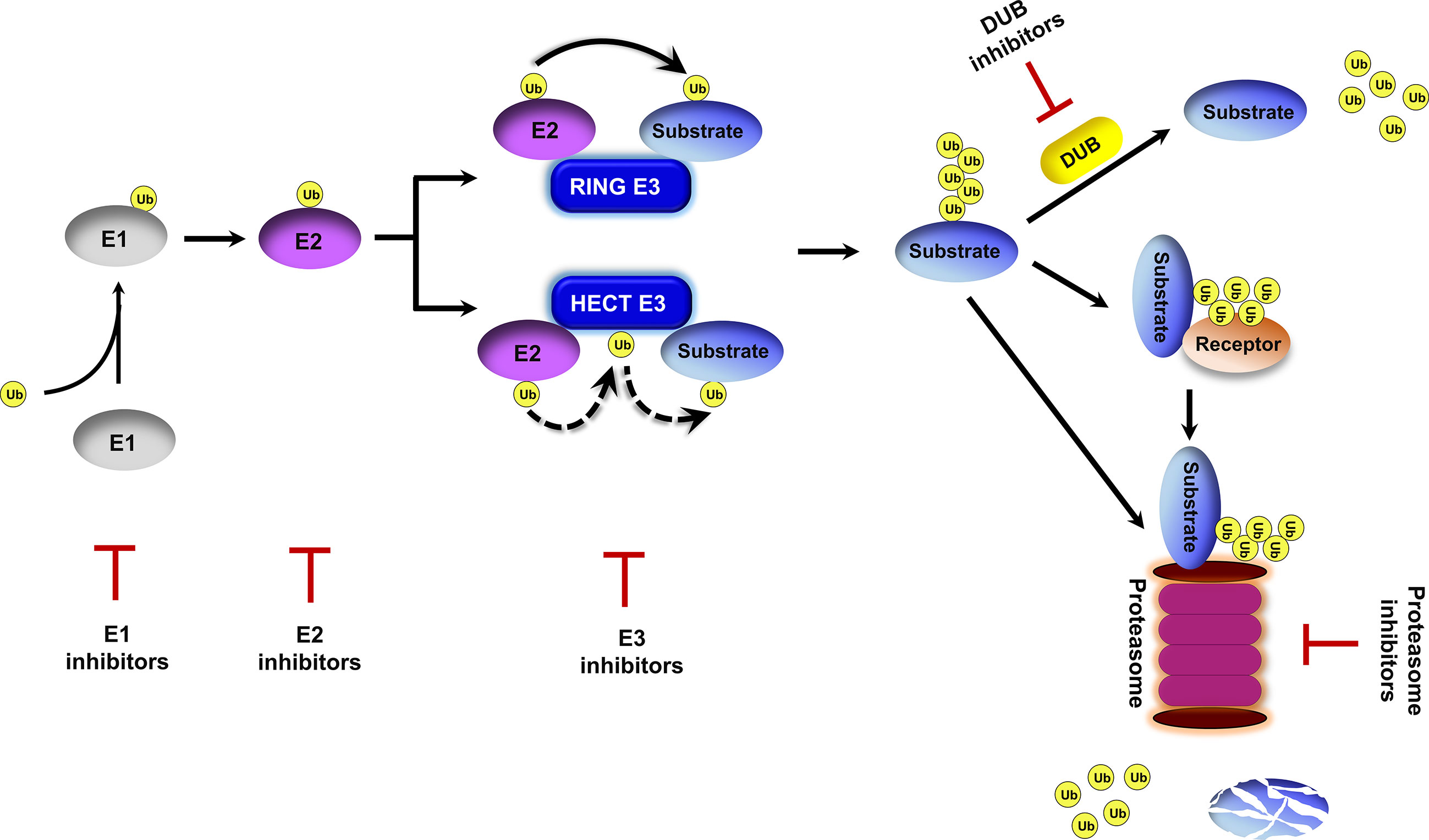
Figure 1 Overview of the ubiquitin proteasome system. Ubiquitin is activated by an E1 enzyme and transferred to an E2 enzyme. The activated ubiquitin is then transferred from the E2 to the substrate in a repeated process mediated by an E3 ubiquitin ligase to form a polyubiquitin chain on the protein substrate. This ubiquitination process can be reversed by deubiquitinating enzymes (DUBs). Polyubiquitinated substrates are either delivered to the proteasome by ubiquitin receptors or directly recognized by the proteasome for cleavage into small peptides and the release of ubiquitin molecules. Inhibitors targeting E1s, E2s, E3s, DUBs and the proteasome have been proposed.
Given the ubiquity of the UPS that targets a wide array of proteins in various processes, it is not surprising that aberrations in the UPS have been implicated in human diseases including cancer, neurodegeneration, metabolic disorders, cardiovascular disorders, and inflammation (Glickman and Ciechanover, 2002; Zheng et al., 2016; Li et al., 2018; Senft et al., 2018; Gupta et al., 2021). Certain cancers arise from stabilization of pro-oncogenic proteins and pathways such as growth-promoting factors (Bahram et al., 2000; Yada et al., 2004; Yumimoto and Nakayama, 2020) or destabilization of tumor suppressors such as p53 and p27 (Slingerland and Pagano, 2000; Devine and Dai, 2013). The accumulation of neurotoxic proteins causes neurodegenerative diseases including Alzheimer’s disease, Parkinson’s disease, and Huntington’s disease, etc. (Popovic et al., 2014; Celebi et al., 2020). Both ubiquitination and the impairment of proteasomal function contribute to ubiquitinated protein accumulation in the cytoplasm (Zheng et al., 2016; Celebi et al., 2020). A number of compounds have been reported to target the UPS as a new class of potential therapeutics for human diseases (Yang et al., 2007; Devine and Dai, 2013; Liu et al., 2015; Hosseini et al., 2019; Wang et al., 2019a). Some proteasome inhibitors have been approved for cancer treatment, e.g., multiple myeloma and mantle cell lymphoma (Sherman and Li, 2020). The roles of various components of the UPS have been widely studied in humans and the model yeast Saccharomyces cerevisiae. However, studies on the role of UPS components in fungal pathogens remain limited.
Fungal diseases are serious threats to both agriculture and human health. They are difficult to treat because fungi are eukaryotic cells that share much of their cellular machinery with hosts. There are currently no vaccines in clinical use to combat fungal infections, and our armamentarium of antifungal drugs is limited compared to antibiotics and even antiviral agents (Rodrigues and Nosanchuk, 2020; Johnson, 2021). Thus, development of new treatment options is critical for controlling mycoses. The UPS tightly regulates degradation of cellular proteins that play important roles in a variety of cellular pathways during the cell life and host adaptation in fungi. Therefore, understanding the regulation of the UPS in fungal pathogenesis may be valuable for the future development of novel therapeutic approaches. Here, we will describe the role of various components of the UPS in fungal morphogenesis, stress response, host adaptation and fungal virulence.
Ubiquitin
Ubiquitin is a small polypeptide of 76 amino acids that is highly conserved in all eukaryotes (Ozkaynak et al., 1987; Hershko and Ciechanover, 1998). Ubiquitin is typically linked to protein lysine residues, as well as lysines on ubiquitin itself, allowing for different types of modification, such as monoubiquitination, multiubiquitination, and polyubiquitination (Peng et al., 2003; Komander and Rape, 2012) (Figure 2). The fates of ubiquitinated proteins are determined by the types of ubiquitin modification: proteins modified by K48-linked polyubiquitin chain can be preferentially degraded by the proteasome, whereas multi- or monoubiquitination often mediates non-proteolytic mechanisms such as DNA repair, protein binding, subcellular localization, and the trafficking of membrane proteins (Hicke, 2001; Kravtsova-Ivantsiv et al., 2009; Ziv et al., 2011).
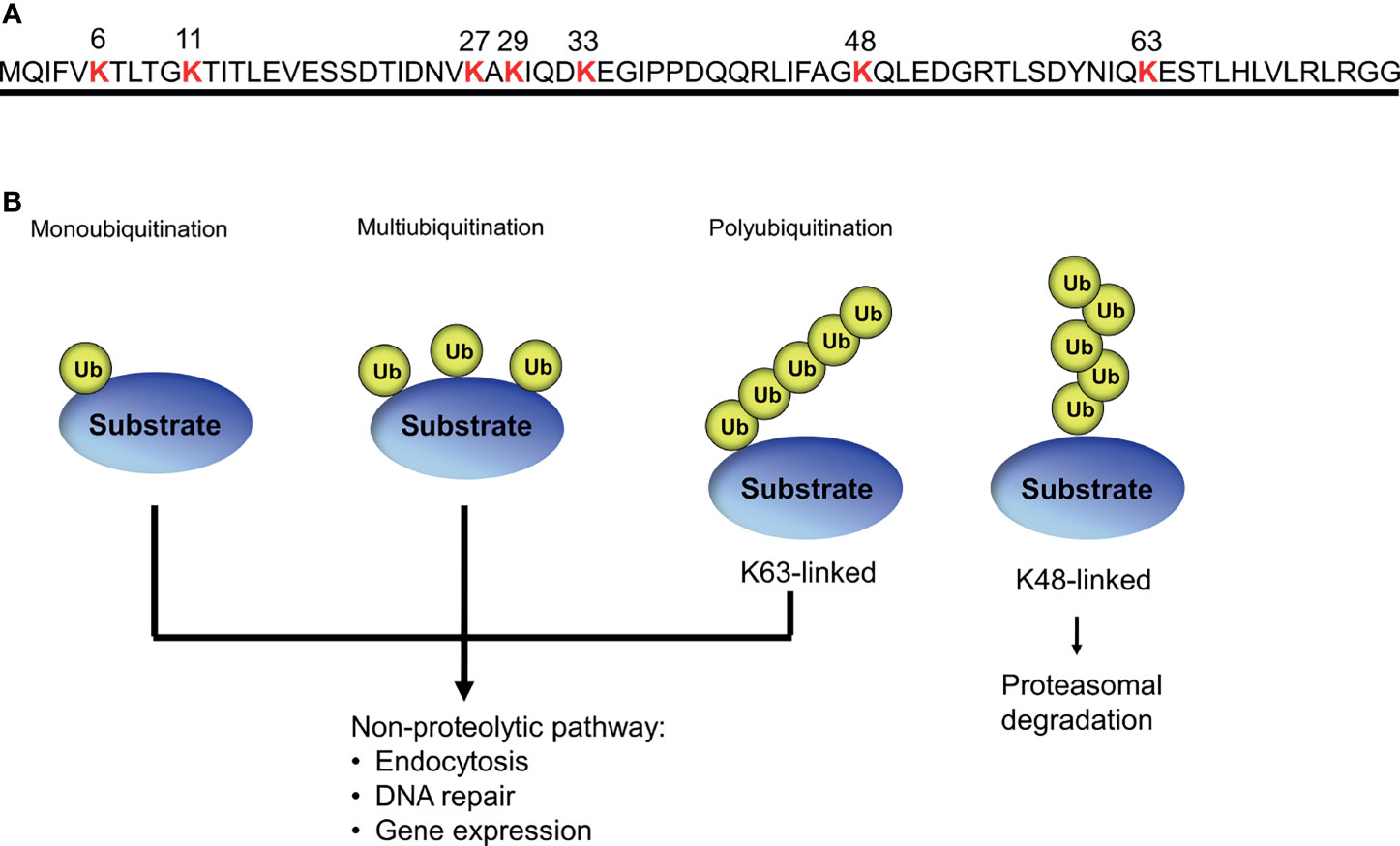
Figure 2 Different forms of ubiquitin modification. (A) Seven specific lysine residues in ubiquitin used for polyubiquitin chain formation. (B) Different types of ubiquitination lead to different biological outcomes.
In S. cerevisiae, ubiquitin is produced by cleavage from precursor proteins that are encoded by a family of natural gene fusions (Ozkaynak et al., 1987). UBI1–3 genes encode hybrid proteins, in which ubiquitin units fuse to unrelated peptide sequences. UBI4 encodes a polyubiquitin that contains five consecutive ubiquitin repeats and is highly induced under stress conditions (Finley et al., 1987; Fraser et al., 1991). In fungal pathogens, ubiquitin genes have been shown to play an important role in fungal development, stress resistance, and fungal virulence (Roig and Gozalbo, 2003; Oh et al., 2012; Chen et al., 2018). Two ubiquitin genes have been identified in Candida albicans (Sepulveda et al., 1996; Roig et al., 2000), an opportunistic fungal pathogen that can switch among different morphologies to adapt to various environmental stimuli (Huang, 2012). The UBI3 gene encodes a hybrid ubiquitin fusion protein which is essential for growth of C. albicans (Roig and Gozalbo, 2002). The polyubiquitin gene UBI4 contains three ubiquitin repeats and is involved in fungal growth and virulence (Sepulveda et al., 1996; Leach et al., 2011). Deletion of UBI4 in C. albicans induces morphological transition from yeast to hyphae, which is important for the pathogenicity of C. albicans (Roig and Gozalbo, 2003; Yang D. et al., 2020). The ubi4∆/∆ mutant displays a cell wall biosynthesis defect when exposed to a number of cell wall stresses including treatment with the anti-fungal drug caspofungin (Leach et al., 2011). The observation that ubi4∆/∆ cells are more sensitive to peroxide is significant because reactive oxygen species contribute to the antimicrobial activity of host immune cells (Miller and Britigan, 1997; Leach et al., 2011). Inactivation of UBI4 significantly attenuates virulence in a murine model of systemic candidiasis (Leach et al., 2011).
There are two ubiquitin encoding genes (UBI1 and UBI4) in Cryptococcus neoformans (Spitzer and Spitzer, 1995), a human pathogen that commonly causes life-threatening meningoencephalitis in immunocompromised individuals (Mitchell and Perfect, 1995). UBI1 encodes a hybrid protein that fuses with ribosomal protein Rpl40a and shares sequence similarity with UBI1 in S. cerevisiae. UBI4 encodes a polyubiquitin precursor containing five ubiquitin repeats (Spitzer and Spitzer, 1995). Deletion of UBI1 results in a vegetative growth defect, morphological changes, a melanin production defect, decreased intracellular survival inside macrophages and virulence attenuation during infection (Zhao et al., 2020). Reconstitution of the full length of UBI1 or the C-terminal Rpl40a could both reverse the phenotypes of ubi1Δ mutation, indicating a role for Ubi1 in cryptococcal ribosome biogenesis (Zhao et al., 2020). Deletion of UBI1 also led to the differential expression of ubiquitin-conjugating enzymes (Zhao et al., 2020), suggesting a regulatory function for Ubi1 in the UPS, but the role of ubiquitin moiety in the pathogenicity of C. neoformans requires further investigation (Zhao et al., 2020).
Deletion of the polyubiquitin gene MGG_01282 resulted in abnormal morphology and virulence defects in the filamentous ascomycete fungus Magnaporthe oryzae, the leading cause of fungal diseases in rice globally (Oh et al., 2012). Likewise, the polyubiquitin gene CpUBI4 is required for fungal development, stress adaptation, and virulence in Cryphonectria parasitica, a phytopathogenic filamentous fungus that causes chestnut blight disease (Chen et al., 2018). The role of polyubiquitin genes in insect fungal pathogens were also explored recently (Wang et al., 2019b; Wang et al., 2020). Polyubiquitin genes were functionally characterized via gene deletion in Metarhizium robertsii and Beauveria bassiana. MrUBI4 is involved in UV stress and the heat-shock response in M. robertsii (Wang et al., 2019b). Deletion of UBI4 leads to a defect in stress tolerance and attenuated virulence in B. bassiana (Wang et al., 2020). In all, ubiquitin genes play important roles in fungal pathogenesis.
Ubiquitin-Activating Enzymes (E1s)
The ubiquitination cascade is initialed by the highly conserved ubiquitin-activating enzyme (E1) that catalyzes covalent bond formation between the active site cysteine of the E1 and the C-terminal glycine residue of ubiquitin (McGrath et al., 1991; Pickart, 2001). Most species have single E1 enzyme, and its activity is necessary for all subsequent steps in ubiquitination (McGrath et al., 1991; Watts et al., 2003; Kulkarni and Smith, 2008). In S. cerevisiae, E1 is encoded by an essential gene UBA1 (McGrath et al., 1991), and inactivation of UBA1 dramatically reduces ubiquitin conjugation (Ghaboosi and Deshaies, 2007). Because of the conservative nature of the E1 in different organisms, their essentiality is likely also shared in pathogenic fungi. Therefore, despite the E1 in S. cerevisiae has been well characterized, there is very limited studies on its function in fungal pathogens. In the yeast pathogen C. neoformans, the cAMP/PKA signaling plays a critical role in capsule and melanin formation, and is important for its pathogenicity (D’Souza et al., 2001). Analysis of the PKA-regulated proteome identified the role of the ubiquitin-proteasome pathway in capsule regulation. Induction of Pka1 leads to decreased protein level of the single E1 in C. neoformans (Geddes et al., 2016).
Ubiquitin-Conjugating Enzymes (E2s)
The ubiquitin-conjugating enzyme E2 is at the center of the E1–E2–E3 enzymatic cascade. E2s interact with E1 and E3 and transfer ubiquitin from E1 to the substrate (Ye and Rape, 2009). E2s belong to the E2 superfamily which contains a highly conserved ubiquitin-conjugating (UBC) domain (Ye and Rape, 2009). A total of 13 E2 genes have been found in S. cerevisiae (Finley et al., 2012), 40 in A. thaliana (Kraft et al., 2005), and 37 in humans (van Wijk and Timmers, 2010). E2 enzymes have the ability to control the topology of ubiquitin conjugates and to determine the fate of labeled proteins to either degradation or nonproteolytic processes (van Wijk and Timmers, 2010). Multiple E2 enzymes can be involved in ubiquitination of a single substrate, such as the Matα2 transcriptional regulator in S. cerevisiae that is controlled by 4 E2s (Chen et al., 1993). A single E2 can combine with different types of E3s that recognize and select distinct substrates (Finley et al., 2012). Interestingly, E2s have also been shown to directly ubiquitinate endogenous substrates independent of an E3 ligase (Berleth and Pickart, 1996; Kraft et al., 2005). Several studies on E2 enzymes in plants have revealed the role of E2 enzymes in host defense against fungal infection (Finley et al., 2012; Liu et al., 2020). Plant E2s are involved in abiotic stress responses, including osmotic stress tolerance (Zhou et al., 2010; Chung et al., 2013), heat shock responses (Feussner et al., 1997), and oxidative stress responses (Zhou et al., 2010). E2 enzymes also play important roles in plant immune responses and DNA repair (Finley et al., 2012; Liu et al., 2020). Inactivation of the ubiquitin-conjugating enzyme 4 (Tau4) in bread wheat Triticum aestivum increases host defense against the phytopathogen Zymoseptoria tritici (Millyard et al., 2016). Ectopic expression of the E2 gene OgUBC1 from rice confers resistance against Botrytis cinerea infection in A. thaliana (Jeon et al., 2012). The ubiquitin-conjugating enzyme Rad6 is conserved amongst eukaryotes (Huang et al., 1997; Worthylake et al., 1998). Rad6 in C. albicans protects the fungus against UV damage and negatively regulates hyphal development (Leng et al., 2000). MoRad6 in M. oryzae is essential for fungal development and pathogenicity (Shi et al., 2016). There is no published work on UBC domain containing proteins in C. neoformans or Aspergillus fumigatus. Sequence homology searching revealed 18 proteins containing the UBC domain in C. neoformans genome (https://fungidb.org).
Ubiquitin Ligases (E3s)
Ubiquitin ligases (E3s) form a large and diverse family that recognize specific substrate proteins. The E3 ligases bind E2s and substrates to facilitate substrate-specific ubiquitination (Hicke and Dunn, 2003; Finley et al., 2012). E3s are classified into two major classes, which catalyze ubiquitin transfer from E2 to substrate via different mechanisms. These two classes are either RING (really interesting new genes) domain-containing E3s, or HECT (homologous to E6-AP C-terminus) domain E3s (Huibregtse et al., 1995; Bernassola et al., 2008). HECT E3s form an intermediate thioester bond between the active site cysteine in the HECT domain and ubiquitin received from E2 and transfer ubiquitin to substrate from the HECT E3-Ub (Huibregtse et al., 1995). RING E3s catalyze the direct transfer of ubiquitin from the E2 to substrates by bridging the interaction between E2 and substrate proteins (Deshaies and Joazeiro, 2009). In addition, RBR proteins (RING-between-RINGs domain E3, a subclass of RING domain E3s) bind E2s with one RING domain and transfer ubiquitin to the other RING domain before its transfer to the substrate, therefore apparently functioning as RING/HECT hybrids (Wenzel et al., 2011; Wenzel and Klevit, 2012). There are only five HECT domain E3s among 60–100 putative E3s in yeast (Finley et al., 2012) that function in diverse processes ranging from protein degradation, metabolic process, endocytosis, and cell cycle progression (Pesin and Orr-Weaver, 2008; Rotin and Kumar, 2009; Finley et al., 2012). Most identified E3s belong to the class of RING domain E3s. Cullin-RING ligases (CRLs) are the largest group of ubiquitin ligases in eukaryotes (Finley et al., 2012). The SCF (Skp1-Cullin-F-box protein) ligase is the typical CRL ligase, in which the cullin protein Cdc53 binds to the small RING domain subunit in Rbx1 with its C-terminal regions and binds to F-box proteins through the linker protein Skp1 with the N- terminal regions (Finley et al., 2012). Substrates of SCF ligases are phosphorylated to create a binding surface for F-box motif recognition (Feldman et al., 1997; Skowyra et al., 1997).
The role of E3 ligases has been widely studied in human health and disease, including cancer, neurodegenerative disease, and neurological syndromes, with a view to develop new clinical therapies (Huang et al., 2016; Mao et al., 2018; Pan, 2020; Wang Y. et al., 2020; Goto et al., 2021). Studies on E3s in pathogenic fungi have revealed that many E3s are required for fungal virulence, as reviewed by (Liu and Xue, 2011). These studies focus on the homologs of Grr1 and Cdc4, which are the two best studied SCF ligases in S. cerevisiae. SCF(Grr1) induces degradation of the Mth1 and G1 cyclins Cln1 and Cln2 to control glucose sensing and the cell cycle in S. cerevisiae (Seol et al., 1999; Skowyra et al., 1999; Flick et al., 2003). SCF(Cdc4) controls cell cycle transitions and nutrient responses by recruiting downstream phosphorylated substrates, such as the cyclin-dependent kinase inhibitor Sic1 and the transcription factor Gcn4 (Feldman et al., 1997; Skowyra et al., 1997; Meimoun et al., 2000; Chi et al., 2001). The role of other major subunit of SCF complex, such as cullin Cdc53, has also been reported in pathogenic fungi (Trunk et al., 2009; Sela et al., 2012). Cdc53 is an essential gene in C. albicans and negatively regulates filamentation when under the control of a tetracycline-dependent promoter (Trunk et al., 2009). Modification of Cdc53 by the ubiquitin-related protein NEDD8/Rub1 (neddylation, ubiquitin-related protein) is also involved in morphogenetic phenotype (Sela et al., 2012). Either deletion of CaRUB1 or mutation of the neddylation target site in CaCdc53 showed filamentous growth, indicating that CaCdc53 neddylation regulates Candida dimorphic switch (Sela et al., 2012).
Grr1 contains an F-box domain and a leucine rich repeat (LRR) at its C-terminus. Homologs of Grr1 have been reported in several important fungal pathogens (Liu and Xue, 2011). Fbp1 is essential for Cryptococcus virulence. Even though the fbp1∆ mutant produces normal classical virulence factors, including capsule and melanin production, mice infected with fbp1∆ showed a low persistence of fungal burden throughout infection, (Liu et al., 2011; Liu and Xue, 2014). Further studies found that fbp1Δ-infected mice developed a robust Th1 and Th17 host protective immune response that helps contain yeast cells in the lung and prevent them from dissemination to maintain the long-term survival of the host (Masso-Silva et al., 2018). Mice challenged with heat-killed fbp1Δ cells (HK-fbp1∆) also can develop a robust Th1 response that confers protection against infection with the virulent wild type strain (Masso-Silva et al., 2018). Interestingly, vaccination of HK-fbp1Δ cells induces cross-protection against challenge with other invasive fungal pathogens, including C. gattii and A. fumigatus. The vaccine protection is effective even in mice depleted of CD4+ T cells, a condition mimics HIV/AIDS-induced immune deficiency (Wang Y. et al., 2019). Several potential substrates of Fbp1 have been identified in C. neoformans (Liu et al., 2011; Liu and Xue, 2014; Han et al., 2020; Fan and Liu, 2021). One of them, a Cdk-related kinase Crk1, has been found to be involved in meiosis regulation (Liu et al., 2011; Liu and Shen, 2011). Another substrate Isc1 (the inositol phosphosphingolipid-phospholipase C1) is required for fungal survival inside macrophage cells (Liu and Xue, 2014). Several additional Fbp1 interacting proteins, including mannoprotein Cmp1 and vacuolar morphogenesis protein Vlp1, has also been considered as potential substrates of Fbp1 based on protein pull-down assays, although their functional verification remains missing. Both proteins are required for pathogenicity of C. neoformans, as the vlp1Δ mutant is avirulent and the cmp1Δ mutant or the CMP1 overexpression strain shows virulence attenuation in a murine infection model of systemic cryptococcosis (Han et al., 2020; Fan and Liu, 2021). Besides Fbp1, there are 19 additional F-box proteins in C. neoformans that remain to be studied.
In C. albicans, Grr1 negatively regulates pseudohyphal development (Butler et al., 2006), an important fungal virulence factor implicated in the cell cycle (Bachewich and Whiteway, 2005; Bensen et al., 2005; Chapa y Lazo et al., 2005; Li et al., 2006). A. fumigatus GrrA shares structural similarity to Fbp1 of C. neoformans. Deletion of GRRA in A. fumigatus has no impact on its virulence, indicating the role of Grr1 homologs in virulence might vary in different pathogenic fungi (Johnk et al., 2016). In Gibberella zeae, an important plant pathogen and fungal mycotoxins producer, the Grr1 homolog Fbp1 is a versatile F-box protein involved in the mycelial growth, sexual reproduction, and virulence. Deletion of FBP1 impairs the ability of G. zeae to colonize plant cells, which leads to attenuated virulence with mild disease (Han et al., 2007). Two homologs of Grr1 have been identified in M. oryzae. Both of them are required for fungal pathogenicity based on mutagenesis studies (Sweigard et al., 1998; Guo et al., 2015).
Cdc4 homologs have also been well characterized in a number of fungal pathogens. Cdc4 contains an F-box domain and a WD40 domain that is important for interaction with its downstream substrates. SCF(Cdc4) plays critical roles in C. albicans morphogenesis and biofilm formation (Atir-Lande et al., 2005; Shieh et al., 2005; Chin et al., 2013; Tseng et al., 2015). Deletion of CDC4 promotes hyphae growth, especially of true hyphae rather than pseudohyphal development, which is different from Grr1 in this fungus. Hyphae growth in the cdc4∆/∆ mutant is independent of the transcription factors Efg1 and Cph1 or G1 cyclins (Atir-Lande et al., 2005). Multiple Cdc4 substrates important for Cdc4-mediated morphogenesis have been identified. Sol1, a homologue of Sic1 (a substrate of Cdc4 in S. cerevisiae), has been identified as a substrate of Cdc4 in C. albicans and it involved in C. albicans morphogenesis. Another SCF(Cdc4) substrate, Ume6, has been found to be critical target of SCF(Cdc4) and responsible for the hyphae formation of cdc4∆/∆ mutant in C. albicans. The cdc4∆/∆ume6∆/∆ double mutant has reduced filamentous growth, but more pseudohyphal development than the cdc4∆/∆ single mutant (Mendelsohn et al., 2017). Consistent with this observation, deletion of UME6 blocks the increased expression of hypha-specific genes in the cdc4∆/∆ mutant background. The cdc4∆/∆ume6∆/∆sol1∆/∆ triple mutant lost the ability for filamentous growth, while can be restored by introducing of UME6, indicating the important role of Ume6 in Cdc4-mediated hyphal formation (Mendelsohn et al., 2017). Thr1 has also been identified as a Cdc4 substrate by in vitro affinity purification in C. albicans (Tseng et al., 2010). Thr1 is a homoserine kinase and its stability is dependent on Cdc4. The thr1∆/∆ mutant accumulates toxic homoserine and shows virulence attenuation (Kingsbury and McCusker, 2010a; Kingsbury and McCusker, 2010b). Similar to Sol1, Thr1 positively modulates hyphal formation (Lee et al., 2018). In addition, Thr1 negatively regulates biofilm formation in C. albicans (Tseng et al., 2015).
There are over 20 proteins containing the F-box domain in the genomes of Aspergillus spp. e.g., human pathogen A. fumigatus and mycotoxin producing plant pathogen A. flavus (Frawley and Bayram, 2020). A total of 26 F-box proteins in A. fumigatus and 30 in A. flavus have been detected by immunoprecipitation of the HA-tagged SkpA adaptor protein during vegetative growth. Analysis of fungal F-box proteins has revealed an interaction network in the presence of various exogenous stress conditions, including osmotic and oxidative stresses, the cell wall stressor Congo red, and the antifungal drug Amphotericin B or Miconazole (Frawley and Bayram, 2020). While some F-box proteins were identified to interact with SkpA across all these different stress conditions, other F-box proteins were only detected under one condition, suggesting they may be specific to one particular stress. Two F-box proteins (Fbx20 and Fbx45) in A. fumigatus and three F-box proteins (Fbx1, Fbx11 and Fbx45) in A. flavus interact with SkpA only in response to Amphotericin B. Fbx45 was detected in both species in response to Amphotericin B. Orthologs of Fbx45 are conserved among Aspergillus species and do not exist in humans, suggesting that Fbx45 could be a potential anti-fungal target (Frawley and Bayram, 2020). Fbx15 is specific to filamentous fungi. Deletion of the F-box domain in Fbx15 impairs stress resistance, similar to the fbx15Δ mutant, suggesting that Fbx15 functions as a subunit of the SCF(Fbx15) E3 ligase and is critical for stress tolerance in A. fumigatus. The fbx15Δ mutant cells are cleared at an early stage of infection and the infected animals show no disease symptom in a mouse model of aspergillosis, indicating that Fbx15 is required for Aspergillus virulence (Johnk et al., 2016).
Other F-box proteins, such as Frp1 in the root-infecting fungus Fusarium oxysporum and several F-box proteins in M. oryzae, including MoFwd1, MoCdc4 and MoFbx15, and HECT type E3 Upl3 have also been reported to be required for fungal virulence (Duyvesteijn et al., 2005; Jonkers et al., 2011; Shi et al., 2019; Li et al., 2020).
Overall, because the F-box protein family has large members with diverse functions and the fact that the F-box protein determines the substrate specificity, much of attention on the E3 ligase studies in fungal pathogens have been focusing on function of F-box proteins and their substrates. Understanding the function of F-box proteins and their substrates will lead to a better understanding of the UPS mediated functional regulation, including fungal development and pathogenesis. Identification of F-box proteins that are important for fungal virulence may also lead to development of novel drug targets, hence is highly significant.
Deubiquitinating Enzymes (DUBs)
Deubiquitinating enzymes (DUBs) reverse ubiquitination by hydrolyzing polyubiquitin chains (Amerik and Hochstrasser, 2004; Reyes-Turcu et al., 2009). DUBs are an important part of the UPS in multiple fundamental cellular processes, including DNA damage repair, protein quality control and cell cycle regulation (Komander, 2009; He et al., 2016; Kee and Huang, 2016). DUBs have been associated with human pathologies including cancer, neurodegenerative diseases and infectious diseases (Nanduri et al., 2013; Heideker and Wertz, 2015; He et al., 2016; Das et al., 2020). DUBs are involved in the regulation of various stress responses in S. cerevisiae (Auesukaree et al., 2009; Mat Nanyan et al., 2020) and C. neoformans (Liu et al., 2008; Fang et al., 2012). There are 19 putative DUB proteins in C. neoformans and most of them are required for its pathogenesis (Liu et al., 2008; Fang et al., 2012). Deletion of UBP5 elevates sensitivity to external stress such as high temperature, reactive oxygen species, osmotic changes, or antifungal agents. Ubp5 and Doa4 are important for Cryptococcus virulence factor production (capsule and melanin) and in vivo fungal virulence (Fang et al., 2012). Several studies have identified the role of DUBs in the pathogenic fungus M. oryzae. Deletion of MoUBP14 resulted in virulence attenuation and phenotypic defects, including stress sensitivity and reduced conidiation (Wang et al., 2018). MoUbp4 and MoUbp8 are required for infection-related morphogenesis and pathogenicity. The moubp4Δ and moubp8Δ mutants show reduced mycelial growth, which blocks penetration and invasive growth and reduces pathogenicity (Que et al., 2020; Yang J. et al., 2020). Overall, the deubiquitination process provides another layer of post-translation regulation that has been shown to be important for fungal pathogenesis.
Ubiquitin Receptors
How do proteasomes specifically acquire ubiquitinated proteins? Studies in S. cerevisiae have revealed that several ubiquitin receptors, including Rad23, Ddi1, and Dsk2, play important roles in shuttling substrates to the proteasome (Verma et al., 2004; Dantuma et al., 2009). These three ubiquitin receptors contain both ubiquitin-like (UBL) domains and ubiquitin-associated (UBA) domains. Ubiquitin receptors specifically bind to polyubiquitin chains of target proteins through UBA domains and interact with the proteasome through their UBL domain (Dantuma et al., 2009). In yeast, Rad23 and Ddi1 are two DNA damage-inducible proteins (Liu et al., 1997; Jelinsky et al., 2000) and Dsk2 is required for the duplication of microtubule-organizing centers (Biggins et al., 1996). Rad23 contributes to both nucleotide excision repair (NER) and protein turnover by directing proteins to the proteasome from yeast to human (Dantuma et al., 2009; Farmer et al., 2010; Lahari et al., 2017). Rad23 has a UBL domain and two UBA domains (UBA1 and UBA2). Its UBA2 domain binds to ubiquitin molecules and the UBL domain binds to proteasome (Dantuma et al., 2009). In addition, it also contains a XPCB (Rad4-binding) domain that is important for NER. In C. albicans, either deletion or overexpression of RAD23 leads to increased UV sensitivity. The rad23∆/∆ mutant showed hypervirulence in a murine infection model (Feng et al., 2020). Rad4 is critical in the Rad23 mediated response to UV and has similar functions regulating cell morphogenesis and biofilm formation, yet it does not play an important role in fungal virulence. Since Rad4 is a key component of the NER pathway and is disposable for fungal virulence, Rad23 mediated virulence suppression is likely due to its role in protein degradation, rather than the NER pathway in C. albicans (Feng et al., 2020). Deletion of different domains to uncouple the two functions of Rad23 has also been reported in C. neoformans. In the wax moth (Galleria mellonella) larvae infection model, the UBA2 domain is required for virulence and the XPCB domain is not involved in fungal virulence in C. neoformans, indicating that the role of Rad23 in virulence is also due to its function in protein turnover rather than NER. How the ubiquitin receptor Rad23 regulates virulence remains to be understood (Verma et al., 2019).
The Proteasome
The proteasome is found in all eukaryotes and has been extensively characterized in S. cerevisiae (Finley et al., 2012). The proteasome is a highly conserved protein complex composed of a catalytic 20S core particle and associated 19S regulatory particles. The regulatory particles recognize ubiquitin–protein conjugates or shuttle receptors with their cargo of ubiquitinated substrates, deubiquitinate substrates, and translocate proteins into the core particle for hydrolysis. The core particle contains interior proteolytic active sites that control proteolysis (Finley, 2009; Finley et al., 2012; Enenkel, 2014). Although the proteasome has not been extensively characterized in pathogenic fungi, it has been implicated in fungal virulence (Geddes et al., 2016; Hossain et al., 2020). Proteasome inhibitors targeting the 20S subunit of the proteasome were used to identify the role of the proteasome in fungal virulence. Commonly used proteasome inhibitors are bortezomib, a boronic acid-based inhibitor widely used for the treatment of multiple myeloma and hematologic malignancy (Nunes and Annunziata, 2017), and MG132, a synthesized peptide aldehyde commonly used for in vitro studies (Lee and Goldberg, 1998).
Among the 33 predicted proteasome subunit genes in C. albicans, 28 are essential subunits. Genetic depletion of most proteasome subunits induces filamentation, demonstrating that proteasome function is essential for C. albicans growth and morphogenesis (Hossain et al., 2020). Proteasome inhibitor treatment also produces filaments that share structural similarities with the hsp90∆/∆ mutant in C. albicans. Inhibition of the proteasome relieves Hsp90-mediated repression of cAMP-PKA signaling to induce morphogenesis (Hossain et al., 2020). To assess the role of the proteasome in the filamentation of other Candida species, including C. dubliniensis, C. tropicalis, C. krusei, C. parapsilosis, and C. auris, Bortezomib has been used to treat these fungi. Bortezomib treatment results indicate that the proteasome has conserved roles in regulating fungal morphogenesis across diverse fungal species (Hossain et al., 2020). Bortezomib treatment impairs the growth of C. neoformans strains and reduces capsule size and cell size, suggesting that the proteasome function is involved in virulence factor production in C. neoformans (Geddes et al., 2016). Because of the conservative nature of the proteasome function in eukaryotes, there is a legitimate concern about the toxicity of utilizing proteasome inhibitors as potential antifungal drugs. Once identified a potent inhibitor, employing medicinal chemistry to further modify the compound to improve its specificity may increase the feasibility for its further development.
UPS-Based Drug Discovery
With improved understanding of the UPS in regulating protein function and its role in human diseases, it has been realized that inhibitors targeting specific components of the UPS may have significant therapeutic potential (Veggiani et al., 2019). The clinical importance of such components has been demonstrated by the success of the proteasome inhibitors Bortezomib (PS341/Velcade, approved by the US Food and Drug Administration (FDA) in 2003) and Carfilzomib (PR-171/Kyprolis, approved by the FDA in 2012) in anti-cancer therapy (Richardson et al., 2005; Rentsch et al., 2013). Given the essential role of E1s in globally controlling protein ubiquitination in cells, inhibition of E1s is predicted to be non-specific (Zhang and Sidhu, 2014). It could be feasible to develop E2 inhibitors that selectively target E2-E3 interaction interfaces, with better specificity than targeting the interfaces of E1–E2 and E2–Ub (Zhang and Sidhu, 2014). E3s determine specific substrates for ubiquitination and thus enable selective targeting of a limited number of proteins. Inhibition of E3s may increase the effectiveness of treatment with fewer side effects compared to general UPS inhibitors. Several small molecule inhibitors of E3 ligases have been reported (Zhang and Sidhu, 2014). MDM2 is a RING E3 ligase that regulates the abundance of the tumor suppressor p53 (Reifenberger et al., 1993). Small molecules such as Nutlins and RITA, disrupt the interaction between MDM2 and p53 to mediate p53 ubiquitination and have significant anti‐tumor effects (Issaeva et al., 2004). Inhibitors of several SCF family members have also been identified, e.g., CpdA prevents the incorporation of Skp2 into the SCFskp2 complex (Chen et al., 2008), while SCF‐I2 and SMER3 inhibit Cdc4 and Met30 (Aghajan et al., 2010), respectively. Significant progress has been made to identify inhibitors targeting DUBs because of the substrate specificity and well-defined catalytic pockets in DUBs (Zhang and Sidhu, 2014). Therefore, it is likely feasible to identify small molecules that selectively target specific UPS components.
Several emerging technologies have been developed to manipulate the UPS, aiding drug discovery in this domain (Veggiani et al., 2019). PROTAC (proteolysis-targeting chimeras) technology represents a promising strategy for drug discovery in cancer research (Sakamoto et al., 2001). PROTACs are hetero-bifunctional molecules that recruit an E3 for precise degradation of drug targets (Toure and Crews, 2016), therefore controlling target specificity and reducing systemic toxicity in vivo (Bondeson et al., 2015). Some PROTACs, such as ARV-110 (an oral protein degradation agent) and ARV-471 (an estrogen receptor (ER) alpha PROTAC molecule), have shown encouraging results in clinical trials (Qi et al., 2021). In yeast, a ligase-trapping system has been developed to aid the identification of E3 ligase targets (Mark et al., 2014; Mark et al., 2016). Although it is not directly utilized for drug discovery, this system will aid the identification of novel E3 substrates, which could be valuable for future drug target discovery. In this system, the E3 is fused with the UBA domain from the ubiquitin receptor proteins Rad23 or Dsk2. The polyubiquitin-binding domain in the E3 ligase increases the binding affinity to its ubiquitinated substrates, allowing identification of weak and transit substrate proteins. In addition, the commercialization of antibodies that recognize the Lys-ϵ-Gly-Gly (K-ϵ-GG) remnant, which is produced by trypsin digestion of proteins containing ubiquitinated lysine residues, improved the ability to analyze the ubiquitinated substrates and identify ubiquitination sites (Udeshi et al., 2013). Although still understudied and no proven inhibitor against fungal UPS in clinical trial, technologies developed in other systems should provide powerful tools for UPS research in fungal pathogens, and may aid the future drug discovery targeting UPS in fungi.
Concluding Remarks
Invasive fungal infections are serious threats to human health with estimated 1.5 million deaths each year (Brown et al., 2012). They are difficult to treat because fungi are eukaryotic cells that share much of their cellular machinery with hosts. The close evolutionary relationship between human and fungi hampers the development of antifungal drugs (Robbins et al., 2016). There is a clear medical need to develop new antifungal drugs. Given that the UPS plays pivotal cellular roles in diverse fungal pathogens and has a track record as a proven drug target in anti-cancer drug development, selective targeting of UPS components provides a promising therapeutic strategy to mitigate antifungal resistance and combat fungal infections.
Studies on UPS components in fungal pathogens remains limited and further studies are warranted to better understand the regulation of UPS in fungal development and virulence. Drug development against targets in the fungal UPS has not been explored extensively. Distinguishing UPS proteins in pathogenic fungi from their hosts and selective targeting of the fungal UPS and its fungal function-specific substrates will be crucial steps to develop a UPS inhibitor into a potential antifungal drug. Improved understanding of the specificity of E3s and their downstream substrates in the fungal UPS will increase the feasibility of targeting the UPS as a promising antifungal strategy. UPS substrates that are involved in some unique processes in fungal pathogens, such as cell wall regeneration, morphologic changes, spore germination and certain virulence factors development, are potential antifungal targets. Therefore, research on the fungal UPS system will not only improve our understanding of the molecular basis of UPS regulation of fungal development and pathogenesis, but also may lead to a valuable new avenue of antifungal drug discovery.
Author Contributions
CC and CX designed the review, wrote and edited the manuscript. All authors reviewed and approved the final manuscript. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by the NIH grants R01AI141368, R01AI123315, and R21AI154318 to CX.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Aghajan, M., Jonai, N., Flick, K., Fu, F., Luo, M., Cai, X., et al. (2010). Chemical Genetics Screen for Enhancers of Rapamycin Identifies a Specific Inhibitor of an Scf Family E3 Ubiquitin Ligase. Nat. Biotechnol. 28, 738–742. doi: 10.1038/nbt.1645
Amerik, A. Y., Hochstrasser, M. (2004). Mechanism and Function of Deubiquitinating Enzymes. Biochim. Biophys. Acta 1695, 189–207. doi: 10.1016/j.bbamcr.2004.10.003
Atir-Lande, A., Gildor, T., Kornitzer, D. (2005). Role for the Scfcdc4 Ubiquitin Ligase in Candida Albicans Morphogenesis. Mol. Biol. Cell 16, 2772–2785. doi: 10.1091/mbc.e05-01-0079
Auesukaree, C., Damnernsawad, A., Kruatrachue, M., Pokethitiyook, P., Boonchird, C., Kaneko, Y., et al. (2009). Genome-Wide Identification of Genes Involved in Tolerance to Various Environmental Stresses in Saccharomyces Cerevisiae. J. Appl. Genet. 50, 301–310. doi: 10.1007/BF03195688
Bachewich, C., Whiteway, M. (2005). Cyclin Cln3p Links G1 Progression to Hyphal and Pseudohyphal Development in Candida Albicans. Eukaryot. Cell 4, 95–102. doi: 10.1128/EC.4.1.95-102.2005
Bahram, F., von der Lehr, N., Cetinkaya, C., Larsson, L. G. (2000). C-Myc Hot Spot Mutations in Lymphomas Result in Inefficient Ubiquitination and Decreased Proteasome-Mediated Turnover. Blood 95, 2104–2110. doi: 10.1182/blood.V95.6.2104
Bensen, E. S., Clemente-Blanco, A., Finley, K. R., Correa-Bordes, J., Berman, J. (2005). The Mitotic Cyclins Clb2p and Clb4p Affect Morphogenesis in Candida Albicans. Mol. Biol. Cell 16, 3387–3400. doi: 10.1091/mbc.e04-12-1081
Berleth, E. S., Pickart, C. M. (1996). Mechanism of Ubiquitin Conjugating Enzyme E2-230k: Catalysis Involving a Thiol Relay? Biochemistry-Us 35, 1664–1671. doi: 10.1021/bi952105y
Bernassola, F., Karin, M., Ciechanover, A., Melino, G. (2008). The Hect Family of E3 Ubiquitin Ligases: Multiple Players in Cancer Development. Cancer Cell 14, 10–21. doi: 10.1016/j.ccr.2008.06.001
Biggins, S., Ivanovska, I., Rose, M. D. (1996). Yeast Ubiquitin-Like Genes Are Involved in Duplication of the Microtubule Organizing Center. J. Cell Biol. 133, 1331–1346. doi: 10.1083/jcb.133.6.1331
Bondeson, D. P., Mares, A., Smith, I. E., Ko, E., Campos, S., Miah, A. H., et al. (2015). Catalytic In Vivo Protein Knockdown by Small-Molecule Protacs. Nat. Chem. Biol. 11, 611–617. doi: 10.1038/nchembio.1858
Brown, G. D., Denning, D. W., Gow, N. A., Levitz, S. M., Netea, M. G., White, T. C. (2012). Hidden Killers: Human Fungal Infections. Sci. Transl. Med. 4, 165rv113. doi: 10.1126/scitranslmed.3004404
Butler, D. K., All, O., Goffena, J., Loveless, T., Wilson, T., Toenjes, K. A. (2006). The Grr1 Gene of Candida Albicans Is Involved in the Negative Control of Pseudohyphal Morphogenesis. Fungal Genet. Biol. 43, 573–582. doi: 10.1016/j.fgb.2006.03.004
Celebi, G., Kesim, H., Ozer, E., Kutlu, O. (2020). The Effect of Dysfunctional Ubiquitin Enzymes in the Pathogenesis of Most Common Diseases. Int. J. Mol. Sci. 21, 6335. doi: 10.3390/ijms21176335
Chapa y Lazo, B., Bates, S., Sudbery, P. (2005). The G1 Cyclin Cln3 Regulates Morphogenesis in Candida Albicans. Eukaryot. Cell 4, 90–94. doi: 10.1128/EC.4.1.90-94.2005
Chen, P., Johnson, P., Sommer, T., Jentsch, S., Hochstrasser, M. (1993). Multiple Ubiquitin-Conjugating Enzymes Participate in the In Vivo Degradation of the Yeast Mat Alpha 2 Repressor. Cell 74, 357–369. doi: 10.1016/0092-8674(93)90426-q
Chen, Q., Li, Y., Wang, J., Li, R., Chen, B. (2018). Cpubi4 Is Essential for Development and Virulence in Chestnut Blight Fungus. Front. Microbiol. 9, 1286. doi: 10.3389/fmicb.2018.01286
Chen, Q., Xie, W., Kuhn, D. J., Voorhees, P. M., Lopez-Girona, A., Mendy, D., et al. (2008). Targeting the P27 E3 Ligase Scf(Skp2) Results in P27- and Skp2-Mediated Cell-Cycle Arrest and Activation of Autophagy. Blood 111, 4690–4699. doi: 10.1182/blood-2007-09-112904
Chi, Y., Huddleston, M. J., Zhang, X., Young, R. A., Annan, R. S., Carr, S. A., et al. (2001). Negative Regulation of Gcn4 and Msn2 Transcription Factors by Srb10 Cyclin-Dependent Kinase. Genes Dev. 15, 1078–1092. doi: 10.1101/gad.867501
Chin, C., Lai, W. C., Lee, T. L., Tseng, T. L., Shieh, J. C. (2013). Dissection of the Candida Albicans Cdc4 Protein Reveals the Involvement of Domains in Morphogenesis and Cell Flocculation. J. BioMed. Sci. 20, 97. doi: 10.1186/1423-0127-20-97
Chung, E., Cho, C. W., So, H. A., Kang, J. S., Chung, Y. S., Lee, J. H. (2013). Overexpression of Vrubc1, a Mung Bean E2 Ubiquitin-Conjugating Enzyme, Enhances Osmotic Stress Tolerance in Arabidopsis. PloS One 8, e66056. doi: 10.1371/journal.pone.0066056
Dantuma, N. P., Heinen, C., Hoogstraten, D. (2009). The Ubiquitin Receptor Rad23: At the Crossroads of Nucleotide Excision Repair and Proteasomal Degradation. DNA Repair (Amst.) 8, 449–460. doi: 10.1016/j.dnarep.2009.01.005
Das, S., Ramakrishna, S., Kim, K. S. (2020). Critical Roles of Deubiquitinating Enzymes in the Nervous System and Neurodegenerative Disorders. Mol. Cells 43, 203–214. doi: 10.14348/molcells.2020.2289
Deshaies, R. J., Joazeiro, C. A. (2009). Ring Domain E3 Ubiquitin Ligases. Annu. Rev. Biochem. 78, 399–434. doi: 10.1146/annurev.biochem.78.101807.093809
Devine, T., Dai, M. S. (2013). Targeting the Ubiquitin-Mediated Proteasome Degradation of P53 for Cancer Therapy. Curr. Pharm. Des. 19, 3248–3262. doi: 10.2174/1381612811319180009
D’Souza, C. A., Alspaugh, J. A., Yue, C., Harashima, T., Cox, G. M., Perfect, J. R., et al. (2001). Cyclic Amp-Dependent Protein Kinase Controls Virulence of the Fungal Pathogen Cryptococcus Neoformans. Mol. Cell Biol. 21, 3179–3191. doi: 10.1128/MCB.21.9.3179-3191.2001
Duyvesteijn, R. G., van Wijk, R., Boer, Y., Rep, M., Cornelissen, B. J., Haring, M. A. (2005). Frp1 Is a Fusarium Oxysporum F-Box Protein Required for Pathogenicity on Tomato. Mol. Microbiol. 57, 1051–1063. doi: 10.1111/j.1365-2958.2005.04751.x
Elsasser, S., Finley, D. (2005). Delivery of Ubiquitinated Substrates to Protein-Unfolding Machines. Nat. Cell Biol. 7, 742–749. doi: 10.1038/ncb0805-742
Enenkel, C. (2014). Proteasome Dynamics. Biochim. Biophys. Acta 1843, 39–46. doi: 10.1016/j.bbamcr.2013.03.023
Fang, W., Price, M. S., Toffaletti, D. L., Tenor, J., Betancourt-Quiroz, M., Price, J. L., et al. (2012). Pleiotropic Effects of Deubiquitinating Enzyme Ubp5 on Growth and Pathogenesis of Cryptococcus Neoformans. PloS One 7, e38326. doi: 10.1371/journal.pone.0038326
Fan, C. L., Liu, T. B. (2021). The Vacuolar Morphogenesis Protein Vam6-Like Protein Vlp1 Is Required for Pathogenicity of Cryptococcus Neoformans. J. Fungi (Basel) 7, 418. doi: 10.3390/jof7060418
Farmer, L. M., Book, A. J., Lee, K. H., Lin, Y. L., Fu, H., Vierstra, R. D. (2010). The Rad23 Family Provides an Essential Connection Between the 26s Proteasome and Ubiquitylated Proteins in Arabidopsis. Plant Cell 22, 124–142. doi: 10.1105/tpc.109.072660
Feldman, R. M., Correll, C. C., Kaplan, K. B., Deshaies, R. J. (1997). A Complex of Cdc4p, Skp1p, and Cdc53p/Cullin Catalyzes Ubiquitination of the Phosphorylated Cdk Inhibitor Sic1p. Cell 91, 221–230. doi: 10.1016/s0092-8674(00)80404-3
Feng, J., Yao, S., Dong, Y., Hu, J., Whiteway, M., Feng, J. (2020). ). Nucleotide Excision Repair Protein Rad23 Regulates Cell Virulence Independent of Rad4 in Candida albicans. mSphere 5, e00062–20. doi: 10.1128/mSphere.00062-20
Feussner, K., Feussner, I., Leopold, I., Wasternack, C. (1997). Isolation of a Cdna Coding for an Ubiquitin-Conjugating Enzyme Ubc1 of Tomato–the First Stress-Induced Ubc of Higher Plants. FEBS Lett. 409, 211–215. doi: 10.1016/s0014-5793(97)00509-7
Finley, D. (2009). Recognition and Processing of Ubiquitin-Protein Conjugates by the Proteasome. Annu. Rev. Biochem. 78, 477–513. doi: 10.1146/annurev.biochem.78.081507.101607
Finley, D., Ozkaynak, E., Varshavsky, A. (1987). The Yeast Polyubiquitin Gene Is Essential for Resistance to High Temperatures, Starvation, and Other Stresses. Cell 48, 1035–1046. doi: 10.1016/0092-8674(87)90711-2
Finley, D., Ulrich, H. D., Sommer, T., Kaiser, P. (2012). The Ubiquitin-Proteasome System of Saccharomyces Cerevisiae. Genetics 192, 319–360. doi: 10.1534/genetics.112.140467
Flick, K. M., Spielewoy, N., Kalashnikova, T. I., Guaderrama, M., Zhu, Q., Chang, H. C., et al. (2003). Grr1-Dependent Inactivation of Mth1 Mediates Glucose-Induced Dissociation of Rgt1 From Hxt Gene Promoters. Mol. Biol. Cell 14, 3230–3241. doi: 10.1091/mbc.e03-03-0135
Fraser, J., Luu, H. A., Neculcea, J., Thomas, D. Y., Storms, R. K. (1991). Ubiquitin Gene Expression: Response to Environmental Changes. Curr. Genet. 20, 17–23. doi: 10.1007/BF00312760
Frawley, D., Bayram, O. (2020). Identification of Skpa-Cula-F-Box E3 Ligase Complexes in Pathogenic Aspergilli. Fungal Genet. Biol. 140:103396. doi: 10.1016/j.fgb.2020.103396
Geddes, J. M., Caza, M., Croll, D., Stoynov, N., Foster, L. J., Kronstad, J. W. (2016). Analysis of the Protein Kinase a-Regulated Proteome of Cryptococcus Neoformans Identifies a Role for the Ubiquitin-Proteasome Pathway in Capsule Formation. Mbio 7, e01862–e01815. doi: 10.1128/mBio.01862-15
Ghaboosi, N., Deshaies, R. J. (2007). A Conditional Yeast E1 Mutant Blocks the Ubiquitin-Proteasome Pathway and Reveals a Role for Ubiquitin Conjugates in Targeting Rad23 to the Proteasome. Mol. Biol. Cell 18, 1953–1963. doi: 10.1091/mbc.e06-10-0965
Glickman, M. H., Ciechanover, A. (2002). The Ubiquitin-Proteasome Proteolytic Pathway: Destruction for the Sake of Construction. Physiol. Rev. 82, 373–428. doi: 10.1152/physrev.00027.2001
Goto, J., Otaki, Y., Watanabe, T., Watanabe, M. (2021). The Role of Hect-Type E3 Ligase in the Development of Cardiac Disease. Int. J. Mol. Sci. 22, 6065. doi: 10.3390/ijms22116065
Guo, M., Gao, F., Zhu, X., Nie, X., Pan, Y., Gao, Z. (2015). Mogrr1, a Novel F-Box Protein, Is Involved in Conidiogenesis and Cell Wall Integrity and Is Critical for the Full Virulence of Magnaporthe Oryzae. Appl. Microbiol. Biotechnol. 99, 8075–8088. doi: 10.1007/s00253-015-6820-x
Gupta, A., Behl, T., Aleya, L., Rahman, M. H., Yadav, H. N., Pal, G., et al. (2021). Role of Upp Pathway in Amelioration of Diabetes-Associated Complications. Environ. Sci. Pollut. Res. Int. 28, 19601–19614. doi: 10.1007/s11356-021-12781-5
Han, Y. K., Kim, M. D., Lee, S. H., Yun, S. H., Lee, Y. W. (2007). A Novel F-Box Protein Involved in Sexual Development and Pathogenesis in Gibberella Zeae. Mol. Microbiol. 63, 768–779. doi: 10.1111/j.1365-2958.2006.05557.x
Han, L. T., Wu, L., Liu, T. B. (2020). A Predicted Mannoprotein Cmp1 Regulates Fungal Virulence in Cryptococcus Neoformans. Pathogens 9, 881. doi: 10.3390/pathogens9110881
Heideker, J., Wertz, I. E. (2015). Dubs, the Regulation of Cell Identity and Disease. Biochem. J. 467:191. doi: 10.1042/bj4670191
Hershko, A., Ciechanover, A. (1998). The Ubiquitin System. Annu. Rev. Biochem. 67, 425–479. doi: 10.1146/annurev.biochem.67.1.425
He, M., Zhou, Z., Shah, A. A., Zou, H., Tao, J., Chen, Q., et al. (2016). The Emerging Role of Deubiquitinating Enzymes in Genomic Integrity, Diseases, and Therapeutics. Cell Biosci. 6, 62. doi: 10.1186/s13578-016-0127-1
Hicke, L. (2001). Protein Regulation by Monoubiquitin. Nat. Rev. Mol. Cell Biol. 2, 195–201. doi: 10.1038/35056583
Hicke, L., Dunn, R. (2003). Regulation of Membrane Protein Transport by Ubiquitin and Ubiquitin-Binding Proteins. Annu. Rev. Cell Dev. Biol. 19, 141–172. doi: 10.1146/annurev.cellbio.19.110701.154617
Hossain, S., Veri, A. O., Cowen, L. E. (2020). The Proteasome Governs Fungal Morphogenesis Via Functional Connections With Hsp90 and cAMP-Protein Kinase a Signaling. Mbio 11, e00290–20. doi: 10.1128/mBio.00290-20
Hosseini, S. M., Okoye, I., Chaleshtari, M. G., Hazhirkarzar, B., Mohamadnejad, J., Azizi, G., et al. (2019). E2 Ubiquitin-Conjugating Enzymes in Cancer: Implications for Immunotherapeutic Interventions. Clin. Chim. Acta 498, 126–134. doi: 10.1016/j.cca.2019.08.020
Huang, G. (2012). Regulation of Phenotypic Transitions in the Fungal Pathogen Candida Albicans. Virulence 3, 251–261. doi: 10.4161/viru.20010
Huang, H., Kahana, A., Gottschling, D. E., Prakash, L., Liebman, S. W. (1997). The Ubiquitin-Conjugating Enzyme Rad6 (Ubc2) Is Required for Silencing in Saccharomyces Cerevisiae. Mol. Cell Biol. 17, 6693–6699. doi: 10.1128/MCB.17.11.6693
Huang, X. L., Zhang, L., Duan, Y., Wang, Y. J., Zhao, J. H., Wang, J. (2016). E3 Ubiquitin Ligase: A Potential Regulator in Fibrosis and Systemic Sclerosis. Cell Immunol. 306-307, 1–8. doi: 10.1016/j.cellimm.2016.07.003
Huibregtse, J. M., Scheffner, M., Beaudenon, S., Howley, P. M. (1995). A Family of Proteins Structurally and Functionally Related to the E6-Ap Ubiquitin-Protein Ligase. Proc. Natl. Acad. Sci. U.S.A. 92, 5249. doi: 10.1073/pnas.92.11.5249-a
Issaeva, N., Bozko, P., Enge, M., Protopopova, M., Verhoef, L. G., Masucci, M., et al. (2004). Small Molecule Rita Binds to P53, Blocks P53-Hdm-2 Interaction and Activates P53 Function in Tumors. Nat. Med. 10, 1321–1328. doi: 10.1038/nm1146
Jelinsky, S. A., Estep, P., Church, G. M., Samson, L. D. (2000). Regulatory Networks Revealed by Transcriptional Profiling of Damaged Saccharomyces Cerevisiae Cells: Rpn4 Links Base Excision Repair With Proteasomes. Mol. Cell Biol. 20, 8157–8167. doi: 10.1128/MCB.20.21.8157-8167.2000
Jeon, E. H., Pak, J. H., Kim, M. J., Kim, H. J., Shin, S. H., Lee, J. H., et al. (2012). Ectopic Expression of Ubiquitin-Conjugating Enzyme Gene From Wild Rice, Ogubc1, Confers Resistance Against Uv-B Radiation and Botrytis Infection in Arabidopsis Thaliana. Biochem. Biophys. Res. Commun. 427, 309–314. doi: 10.1016/j.bbrc.2012.09.048
Johnk, B., Bayram, O., Abelmann, A., Heinekamp, T., Mattern, D. J., Brakhage, A. A., et al. (2016). Scf Ubiquitin Ligase F-Box Protein Fbx15 Controls Nuclear Co-Repressor Localization, Stress Response and Virulence of the Human Pathogen Aspergillus Fumigatus. PloS Pathog. 12, e1005899. doi: 10.1371/journal.ppat.1005899
Johnson, M. D. (2021). Antifungals in Clinical Use and the Pipeline. Infect. Dis. Clin. North Am. 35, 341–371. doi: 10.1016/j.idc.2021.03.005
Jonkers, W., JA, V. A. N. K., Tijm, P., Lee, Y. W., Tudzynski, P., Rep, M., et al. (2011). The Frp1 F-Box Gene Has Different Functions in Sexuality, Pathogenicity and Metabolism in Three Fungal Pathogens. Mol. Plant Pathol. 12, 548–563. doi: 10.1111/j.1364-3703.2010.00689.x
Kee, Y., Huang, T. T. (2016). Role of Deubiquitinating Enzymes in DNA Repair. Mol. Cell Biol. 36, 524–544. doi: 10.1128/MCB.00847-15
Kingsbury, J. M., McCusker, J. H. (2010a). Fungal Homoserine Kinase (Thr1delta) Mutants Are Attenuated in Virulence and Die Rapidly Upon Threonine Starvation and Serum Incubation. Eukaryot. Cell 9, 729–737. doi: 10.1128/EC.00045-10
Kingsbury, J. M., McCusker, J. H. (2010b). Homoserine Toxicity in Saccharomyces Cerevisiae and Candida Albicans Homoserine Kinase (Thr1delta) Mutants. Eukaryot. Cell 9, 717–728. doi: 10.1128/EC.00044-10
Komander, D. (2009). The Emerging Complexity of Protein Ubiquitination. Biochem. Soc. Trans. 37, 937–953. doi: 10.1042/BST0370937
Komander, D., Clague, M. J., Urbe, S. (2009). Breaking the Chains: Structure and Function of the Deubiquitinases. Nat. Rev. Mol. Cell Biol. 10, 550–563. doi: 10.1038/nrm2731
Komander, D., Rape, M. (2012). The Ubiquitin Code. Annu. Rev. Biochem. 81, 203–229. doi: 10.1146/annurev-biochem-060310-170328
Kraft, E., Stone, S. L., Ma, L., Su, N., Gao, Y., Lau, O. S., et al. (2005). Genome Analysis and Functional Characterization of the E2 and Ring-Type E3 Ligase Ubiquitination Enzymes of Arabidopsis. Plant Physiol. 139, 1597–1611. doi: 10.1104/pp.105.067983
Kravtsova-Ivantsiv, Y., Cohen, S., Ciechanover, A. (2009). Modification by Single Ubiquitin Moieties Rather Than Polyubiquitination Is Sufficient for Proteasomal Processing of the P105 Nf-Kappab Precursor. Mol. Cell 33, 496–504. doi: 10.1016/j.molcel.2009.01.023
Kulkarni, M., Smith, H. E. (2008). E1 Ubiquitin-Activating Enzyme Uba-1 Plays Multiple Roles Throughout C. Elegans Development. PloS Genet. 4, e1000131. doi: 10.1371/journal.pgen.1000131
Lahari, T., Lazaro, J., Schroeder, D. F. (2017). Rad4 and Rad23/Hmr Contribute to Arabidopsis UV Tolerance. Genes (Basel) 9, 8. doi: 10.3390/genes9010008
Leach, M. D., Stead, D. A., Argo, E., MacCallum, D. M., Brown, A. J. (2011). Molecular and Proteomic Analyses Highlight the Importance of Ubiquitination for the Stress Resistance, Metabolic Adaptation, Morphogenetic Regulation and Virulence of Candida Albicans. Mol. Microbiol. 79, 1574–1593. doi: 10.1111/j.1365-2958.2011.07542.x
Lee, Y. T., Fang, Y. Y., Sun, Y. W., Hsu, H. C., Weng, S. M., Tseng, T. L., et al. (2018). Thr1 Mediates Gcn4 and Cdc4 to Link Morphogenesis With Nutrient Sensing and the Stress Response in Candida Albicans. Int. J. Mol. Med. 42, 3193–3208. doi: 10.3892/ijmm.2018.3930
Lee, D. H., Goldberg, A. L. (1998). Proteasome Inhibitors: Valuable New Tools for Cell Biologists. Trends Cell Biol. 8, 397–403. doi: 10.1016/s0962-8924(98)01346-4
Leng, P., Sudbery, P. E., Brown, A. J. (2000). Rad6p Represses Yeast-Hypha Morphogenesis in the Human Fungal Pathogen Candida Albicans. Mol. Microbiol. 35, 1264–1275. doi: 10.1046/j.1365-2958.2000.01801.x
Li, G., Qi, X., Sun, G., Rocha, R. O., Segal, L. M., Downey, K. S., et al. (2020). Terminating Rice Innate Immunity Induction Requires a Network of Antagonistic and Redox-Responsive E3 Ubiquitin Ligases Targeting a Fungal Sirtuin. New Phytol. 226, 523–540. doi: 10.1111/nph.16365
Liu, O. W., Chun, C. D., Chow, E. D., Chen, C., Madhani, H. D., Noble, S. M. (2008). Systematic Genetic Analysis of Virulence in the Human Fungal Pathogen Cryptococcus Neoformans. Cell 135, 174–188. doi: 10.1016/j.cell.2008.07.046
Liu, Y., Dai, H., Xiao, W. (1997). Uas(Mag1), a Yeast Cis-Acting Element That Regulates the Expression of Mag1, Is Located Within the Protein Coding Region of Ddi1. Mol. Gen. Genet. 255, 533–542. doi: 10.1007/s004380050526
Liu, J., Shaik, S., Dai, X., Wu, Q., Zhou, X., Wang, Z., et al. (2015). Targeting the Ubiquitin Pathway for Cancer Treatment. Biochim. Biophys. Acta 1855, 50–60. doi: 10.1016/j.bbcan.2014.11.005
Liu, K. H., Shen, W. C. (2011). Mating Differentiation in Cryptococcus Neoformans Is Negatively Regulated by the Crk1 Protein Kinase. Fungal Genet. Biol. 48, 225–240. doi: 10.1016/j.fgb.2010.11.005
Liu, W., Tang, X., Qi, X., Fu, X., Ghimire, S., Ma, R., et al. (2020). The Ubiquitin Conjugating Enzyme: An Important Ubiquitin Transfer Platform in Ubiquitin-Proteasome System. Int. J. Mol. Sci. 21, 2894. doi: 10.3390/ijms21082894
Liu, T. B., Wang, Y., Stukes, S., Chen, Q., Casadevall, A., Xue, C. (2011). The F-Box Protein Fbp1 Regulates Sexual Reproduction and Virulence in Cryptococcus Neoformans. Eukaryot. Cell 10, 791–802. doi: 10.1128/EC.00004-11
Liu, T. B., Xue, C. (2011). The Ubiquitin-Proteasome System and F-Box Proteins in Pathogenic Fungi. Mycobiology 39, 243–248. doi: 10.5941/MYCO.2011.39.4.243
Liu, T. B., Xue, C. (2014). Fbp1-Mediated Ubiquitin-Proteasome Pathway Controls Cryptococcus Neoformans Virulence by Regulating Fungal Intracellular Growth in Macrophages. Infect. Immun. 82, 557–568. doi: 10.1128/IAI.00994-13
Li, W. J., Wang, Y. M., Zheng, X. D., Shi, Q. M., Zhang, T. T., Bai, C., et al. (2006). The F-Box Protein Grr1 Regulates the Stability of Ccn1, Cln3 and Hof1 and Cell Morphogenesis in Candida Albicans. Mol. Microbiol. 62, 212–226. doi: 10.1111/j.1365-2958.2006.05361.x
Li, S., Zhao, J., Shang, D., Kass, D. J., Zhao, Y. (2018). Ubiquitination and Deubiquitination Emerge as Players in Idiopathic Pulmonary Fibrosis Pathogenesis and Treatment. JCI Insight 3, e120362. doi: 10.1172/jci.insight.120362
Mao, X., Sethi, G., Zhang, Z., Wang, Q. (2018). The Emerging Roles of the Herc Ubiquitin Ligases in Cancer. Curr. Pharm. Des. 24, 1676–1681. doi: 10.2174/1381612824666180528081024
Mark, K. G., Loveless, T. B., Toczyski, D. P. (2016). Isolation of Ubiquitinated Substrates by Tandem Affinity Purification of E3 Ligase-Polyubiquitin-Binding Domain Fusions (Ligase Traps). Nat. Protoc. 11, 291–301. doi: 10.1038/nprot.2016.008
Mark, K. G., Simonetta, M., Maiolica, A., Seller, C. A., Toczyski, D. P. (2014). Ubiquitin Ligase Trapping Identifies an Scf(Saf1) Pathway Targeting Unprocessed Vacuolar/Lysosomal Proteins. Mol. Cell 53, 148–161. doi: 10.1016/j.molcel.2013.12.003
Masso-Silva, J., Espinosa, V., Liu, T. B., Wang, Y., Xue, C., Rivera, A. (2018). The F-Box Protein Fbp1 Shapes the Immunogenic Potential of Cryptococcus Neoformans. Mbio 9, e01828–17. doi: 10.1128/mBio.01828-17
Mat Nanyan, N. S. B., Watanabe, D., Sugimoto, Y., Takagi, H. (2020). Effect of the Deubiquitination Enzyme Gene Ubp6 on the Stress-Responsive Transcription Factor Msn2-Mediated Control of the Amino Acid Permease Gnp1 in Yeast. J. Biosci. Bioeng 129, 423–427. doi: 10.1016/j.jbiosc.2019.10.002
McGrath, J. P., Jentsch, S., Varshavsky, A. (1991). Uba1: An Essential Yeast Gene Encoding Ubiquitin-Activating Enzyme. EMBO J. 10, 227–236. doi: 10.1002/j.1460-2075.1991.tb07940.x
Meimoun, A., Holtzman, T., Weissman, Z., McBride, H. J., Stillman, D. J., Fink, G. R., et al. (2000). Degradation of the Transcription Factor Gcn4 Requires the Kinase Pho85 and the Scf(Cdc4) Ubiquitin-Ligase Complex. Mol. Biol. Cell 11, 915–927. doi: 10.1091/mbc.11.3.915
Mendelsohn, S., Pinsky, M., Weissman, Z., Kornitzer, D. (2017). Regulation of the Candida Albicans Hypha-Inducing Transcription Factor Ume6 by the Cdk1 Cyclins Cln3 and Hgc1. mSphere 2, e00248–16. doi: 10.1128/mSphere.00248-16
Miller, R. A., Britigan, B. E. (1997). Role of Oxidants in Microbial Pathophysiology. Clin. Microbiol. Rev. 10, 1–18. doi: 10.1128/CMR.10.1.1
Millyard, L., Lee, J., Zhang, C., Yates, G., Sadanandom, A. (2016). The Ubiquitin Conjugating Enzyme, Tau4 Regulates Wheat Defence Against the Phytopathogen Zymoseptoria Tritici. Sci. Rep. 6:35683. doi: 10.1038/srep35683
Mitchell, T. G., Perfect, J. R. (1995). Cryptococcosis in the Era of Aids–100 Years After the Discovery of Cryptococcus Neoformans. Clin. Microbiol. Rev. 8, 515–548. doi: 10.1128/CMR.8.4.515
Nanduri, B., Suvarnapunya, A. E., Venkatesan, M., Edelmann, M. J. (2013). Deubiquitinating Enzymes as Promising Drug Targets for Infectious Diseases. Curr. Pharm. Des. 19, 3234–3247. doi: 10.2174/1381612811319180008
Nunes, A. T., Annunziata, C. M. (2017). Proteasome Inhibitors: Structure and Function. Semin. Oncol. 44, 377–380. doi: 10.1053/j.seminoncol.2018.01.004
Oh, Y., Franck, W. L., Han, S. O., Shows, A., Gokce, E., Muddiman, D. C., et al. (2012). Polyubiquitin Is Required for Growth, Development and Pathogenicity in the Rice Blast Fungus Magnaporthe Oryzae. PloS One 7, e42868. doi: 10.1371/journal.pone.0042868
Ozkaynak, E., Finley, D., Solomon, M. J., Varshavsky, A. (1987). The Yeast Ubiquitin Genes: A Family of Natural Gene Fusions. EMBO J. 6, 1429–1439. doi: 10.1002/j.1460-2075.1987.tb02384.x
Pan, Z. Q. (2020). Cullin-Ring E3 Ubiquitin Ligase 7 in Growth Control and Cancer. Adv. Exp. Med. Biol. 1217, 285–296. doi: 10.1007/978-981-15-1025-0_17
Peng, J., Schwartz, D., Elias, J. E., Thoreen, C. C., Cheng, D., Marsischky, G., et al. (2003). A Proteomics Approach to Understanding Protein Ubiquitination. Nat. Biotechnol. 21, 921–926. doi: 10.1038/nbt849
Pesin, J. A., Orr-Weaver, T. L. (2008). Regulation of Apc/C Activators in Mitosis and Meiosis. Annu. Rev. Cell Dev. Biol. 24, 475–499. doi: 10.1146/annurev.cellbio.041408.115949
Pickart, C. M. (2001). Mechanisms Underlying Ubiquitination. Annu. Rev. Biochem. 70, 503–533. doi: 10.1146/annurev.biochem.70.1.503
Popovic, D., Vucic, D., Dikic, I. (2014). Ubiquitination in Disease Pathogenesis and Treatment. Nat. Med. 20, 1242–1253. doi: 10.1038/nm.3739
Qi, S. M., Dong, J., Xu, Z. Y., Cheng, X. D., Zhang, W. D., Qin, J. J. (2021). Protac: An Effective Targeted Protein Degradation Strategy for Cancer Therapy. Front. Pharmacol. 12:692574. doi: 10.3389/fphar.2021.692574
Que, Y., Xu, Z., Wang, C., Lv, W., Yue, X., Xu, L., et al. (2020). The Putative Deubiquitinating Enzyme Moubp4 Is Required for Infection-Related Morphogenesis and Pathogenicity in the Rice Blast Fungus Magnaporthe Oryzae. Curr. Genet. 66, 561–576. doi: 10.1007/s00294-019-01049-8
Reifenberger, G., Liu, L., Ichimura, K., Schmidt, E. E., Collins, V. P. (1993). Amplification and Overexpression of the Mdm2 Gene in a Subset of Human Malignant Gliomas Without P53 Mutations. Cancer Res. 53, 2736–2739.
Rentsch, A., Landsberg, D., Brodmann, T., Bulow, L., Girbig, A. K., Kalesse, M. (2013). Synthesis and Pharmacology of Proteasome Inhibitors. Angew. Chem. Int. Ed. Engl. 52, 5450–5488. doi: 10.1002/anie.201207900
Reyes-Turcu, F. E., Ventii, K. H., Wilkinson, K. D. (2009). Regulation and Cellular Roles of Ubiquitin-Specific Deubiquitinating Enzymes. Annu. Rev. Biochem. 78, 363–397. doi: 10.1146/annurev.biochem.78.082307.091526
Richardson, P. G., Sonneveld, P., Schuster, M. W., Irwin, D., Stadtmauer, E. A., Facon, T., et al. (2005). Bortezomib or High-Dose Dexamethasone for Relapsed Multiple Myeloma. N. Engl. J. Med. 352, 2487–2498. doi: 10.1056/NEJMoa043445
Robbins, N., Wright, G. D., Cowen, L. E. (2016). Antifungal Drugs: The Current Armamentarium and Development of New Agents. Microbiol. Spectr. 4 (5), FUNK-0002-2016. doi: 10.1128/microbiolspec.FUNK-0002-2016
Rodrigues, M. L., Nosanchuk, J. D. (2020). Fungal Diseases as Neglected Pathogens: A Wake-Up Call to Public Health Officials. PloS Negl. Trop. Dis. 14, e0007964. doi: 10.1371/journal.pntd.0007964
Roig, P., Gozalbo, D. (2002). The Candida Albicans Ubi3 Gene Encoding a Hybrid Ubiquitin Fusion Protein Involved in Ribosome Biogenesis Is Essential for Growth. FEMS Yeast Res. 2, 25–30. doi: 10.1111/j.1567-1364.2002.tb00065.x
Roig, P., Gozalbo, D. (2003). Depletion of Polyubiquitin Encoded by the Ubi4 Gene Confers Pleiotropic Phenotype to Candida Albicans Cells. Fungal Genet. Biol. 39, 70–81. doi: 10.1016/s1087-1845(03)00004-5
Roig, P., Martinez, J. P., Gil, M. L., Gozalbo, D. (2000). Molecular Cloning and Characterization of the Candida Albicans Ubi3 Gene Coding for a Ubiquitin-Hybrid Protein. Yeast 16, 1413–1419. doi: 10.1002/1097-0061(200011)16:15<1413::AID-YEA632>3.0.CO;2-U
Rotin, D., Kumar, S. (2009). Physiological Functions of the Hect Family of Ubiquitin Ligases. Nat. Rev. Mol. Cell Biol. 10, 398–409. doi: 10.1038/nrm2690
Sakamoto, K. M., Kim, K. B., Kumagai, A., Mercurio, F., Crews, C. M., Deshaies, R. J. (2001). Protacs: Chimeric Molecules That Target Proteins to the Skp1-Cullin-F Box Complex for Ubiquitination and Degradation. Proc. Natl. Acad. Sci. U.S.A. 98, 8554–8559. doi: 10.1073/pnas.141230798
Sela, N., Atir-Lande, A., Kornitzer, D. (2012). Neddylation and Cand1 Independently Stimulate Scf Ubiquitin Ligase Activity in Candida Albicans. Eukaryot. Cell 11, 42–52. doi: 10.1128/EC.05250-11
Senft, D., Qi, J., Ronai, Z. A. (2018). Ubiquitin Ligases in Oncogenic Transformation and Cancer Therapy. Nat. Rev. Cancer 18, 69–88. doi: 10.1038/nrc.2017.105
Seol, J. H., Feldman, R. M., Zachariae, W., Shevchenko, A., Correll, C. C., Lyapina, S., et al. (1999). Cdc53/Cullin and the Essential Hrt1 Ring-H2 Subunit of Scf Define a Ubiquitin Ligase Module That Activates the E2 Enzyme Cdc34. Genes Dev. 13, 1614–1626. doi: 10.1101/gad.13.12.1614
Sepulveda, P., Cervera, A. M., Lopez-Ribot, J. L., Chaffin, W. L., Martinez, J. P., Gozalbo, D. (1996). Cloning and Characterization of a Cdna Coding for Candida Albicans Polyubiquitin. J. Med. Vet. Mycol. 34, 315–322. doi: 10.1080/02681219680000541
Sherman, D. J., Li, J. (2020). Proteasome Inhibitors: Harnessing Proteostasis to Combat Disease. Molecules 25, 671. doi: 10.3390/molecules25030671
Shi, H. B., Chen, G. Q., Chen, Y. P., Dong, B., Lu, J. P., Liu, X. H., et al. (2016). Morad6-Mediated Ubiquitination Pathways Are Essential for Development and Pathogenicity in Magnaporthe Oryzae. Environ. Microbiol. 18, 4170–4187. doi: 10.1111/1462-2920.13515
Shi, H. B., Chen, N., Zhu, X. M., Liang, S., Li, L., Wang, J. Y., et al. (2019). F-Box Proteins Mofwd1, Mocdc4 and Mofbx15 Regulate Development and Pathogenicity in the Rice Blast Fungus Magnaporthe Oryzae. Environ. Microbiol. 21, 3027–3045. doi: 10.1111/1462-2920.14699
Shieh, J. C., White, A., Cheng, Y. C., Rosamond, J. (2005). Identification and Functional Characterization of Candida Albicans Cdc4. J. BioMed. Sci. 12, 913–924. doi: 10.1007/s11373-005-9027-9
Skowyra, D., Craig, K. L., Tyers, M., Elledge, S. J., Harper, J. W. (1997). F-Box Proteins Are Receptors That Recruit Phosphorylated Substrates to the Scf Ubiquitin-Ligase Complex. Cell 91, 209–219. doi: 10.1016/s0092-8674(00)80403-1
Skowyra, D., Koepp, D. M., Kamura, T., Conrad, M. N., Conaway, R. C., Conaway, J. W., et al. (1999). Reconstitution of G1 Cyclin Ubiquitination With Complexes Containing Scfgrr1 and Rbx1. Science 284, 662–665. doi: 10.1126/science.284.5414.662
Slingerland, J., Pagano, M. (2000). Regulation of the Cdk Inhibitor P27 and Its Deregulation in Cancer. J. Cell Physiol. 183, 10–17. doi: 10.1002/(SICI)1097-4652(200004)183:1<10::AID-JCP2>3.0.CO;2-I
Spitzer, E. D., Spitzer, S. G. (1995). Structure of the Ubiquitin-Encoding Genes of Cryptococcus Neoformans. Gene 161, 113–117. doi: 10.1016/0378-1119(95)00231-t
Sweigard, J. A., Carroll, A. M., Farrall, L., Chumley, F. G., Valent, B. (1998). Magnaporthe Grisea Pathogenicity Genes Obtained Through Insertional Mutagenesis. Mol. Plant-Microbe Interact.: MPMI 11, 404–412. doi: 10.1094/MPMI.1998.11.5.404
Toure, M., Crews, C. M. (2016). Small-Molecule Protacs: New Approaches to Protein Degradation. Angew. Chem. Int. Ed. Engl. 55, 1966–1973. doi: 10.1002/anie.201507978
Trunk, K., Gendron, P., Nantel, A., Lemieux, S., Roemer, T., Raymond, M. (2009). Depletion of the Cullin Cdc53p Induces Morphogenetic Changes in Candida Albicans. Eukaryot. Cell 8, 756–767. doi: 10.1128/EC.00332-08
Tseng, T. L., Lai, W. C., Jian, T., Li, C., Sun, H. F., Way, T. D., et al. (2010). Affinity Purification of Candida Albicans Cacdc4-Associated Proteins Reveals the Presence of Novel Proteins Involved in Morphogenesis. Biochem. Biophys. Res. Commun. 395, 152–157. doi: 10.1016/j.bbrc.2010.03.162
Tseng, T. L., Lai, W. C., Lee, T. L., Hsu, W. H., Sun, Y. W., Li, W. C., et al. (2015). A Role of Candida Albicans Cdc4 in the Negative Regulation of Biofilm Formation. Can. J. Microbiol. 61, 247–255. doi: 10.1139/cjm-2014-0526
Udeshi, N. D., Mertins, P., Svinkina, T., Carr, S. A. (2013). Large-Scale Identification of Ubiquitination Sites by Mass Spectrometry. Nat. Protoc. 8, 1950–1960. doi: 10.1038/nprot.2013.120
van Wijk, S. J., Timmers, H. T. (2010). The Family of Ubiquitin-Conjugating Enzymes (E2s): Deciding Between Life and Death of Proteins. FASEB J. 24, 981–993. doi: 10.1096/fj.09-136259
Veggiani, G., Gerpe, M. C. R., Sidhu, S. S., Zhang, W. (2019). Emerging Drug Development Technologies Targeting Ubiquitination for Cancer Therapeutics. Pharmacol. Ther. 199, 139–154. doi: 10.1016/j.pharmthera.2019.03.003
Verma, R., Oania, R., Graumann, J., Deshaies, R. J. (2004). Multiubiquitin Chain Receptors Define a Layer of Substrate Selectivity in the Ubiquitin-Proteasome System. Cell 118, 99–110. doi: 10.1016/j.cell.2004.06.014
Verma, S., Shakya, V. P. S., Idnurm, A. (2019). The Dual Function Gene Rad23 Contributes to Cryptococcus Neoformans Virulence Independently of Its Role in Nucleotide Excision DNA Repair. Gene 717:144043. doi: 10.1016/j.gene.2019.144043
Wang, Y., Argiles-Castillo, D., Kane, E. I., Zhou, A., Spratt, D. E. (2020). Hect E3 Ubiquitin Ligases - Emerging Insights Into Their Biological Roles and Disease Relevance. J. Cell Sci. 133, jcs228072. doi: 10.1242/jcs.228072
Wang, Z., Kang, W., You, Y., Pang, J., Ren, H., Suo, Z., et al. (2019a). Usp7: Novel Drug Target in Cancer Therapy. Front. Pharmacol. 10, 427. doi: 10.3389/fphar.2019.00427
Wang, D. Y., Ren, K., Tong, S. M., Ying, S. H., Feng, M. G. (2020). Pleiotropic Effects of Ubi4, a Polyubiquitin Precursor Required for Ubiquitin Accumulation, Conidiation and Pathogenicity of a Fungal Insect Pathogen. Environ. Microbiol. 22, 2564–2580. doi: 10.1111/1462-2920.14940
Wang, Y., Wang, K., Masso-Silva, J. A., Rivera, A., Xue, C. (2019). A Heat-Killed Cryptococcus Mutant Strain Induces Host Protection Against Multiple Invasive Mycoses in a Murine Vaccine Model. Mbio 10, e02145–19. doi: 10.1128/mBio.02145-19
Wang, Z., Zhang, H., Liu, C., Xing, J., Chen, X. L. (2018). A Deubiquitinating Enzyme Ubp14 Is Required for Development, Stress Response, Nutrient Utilization, and Pathogenesis of Magnaporthe Oryzae. Front. Microbiol. 9:769. doi: 10.3389/fmicb.2018.00769
Wang, Z., Zhu, H., Cheng, Y., Jiang, Y., Li, Y., Huang, B. (2019b). The Polyubiquitin Gene Mrubi4 Is Required for Conidiation, Conidial Germination, and Stress Tolerance in the Filamentous Fungus Metarhizium Robertsii. Genes (Basel) 10, 412. doi: 10.3390/genes10060412
Watts, R. J., Hoopfer, E. D., Luo, L. (2003). Axon Pruning During Drosophila Metamorphosis: Evidence for Local Degeneration and Requirement of the Ubiquitin-Proteasome System. Neuron 38, 871–885. doi: 10.1016/s0896-6273(03)00295-2
Wenzel, D. M., Klevit, R. E. (2012). Following Ariadne’s Thread: A New Perspective on Rbr Ubiquitin Ligases. BMC Biol. 10:24. doi: 10.1186/1741-7007-10-24
Wenzel, D. M., Lissounov, A., Brzovic, P. S., Klevit, R. E. (2011). Ubch7 Reactivity Profile Reveals Parkin and Hhari to Be Ring/Hect Hybrids. Nature 474, 105–108. doi: 10.1038/nature09966
Worthylake, D. K., Prakash, S., Prakash, L., Hill, C. P. (1998). Crystal Structure of the Saccharomyces Cerevisiae Ubiquitin-Conjugating Enzyme Rad6 at 2.6 a Resolution. J. Biol. Chem. 273, 6271–6276. doi: 10.1074/jbc.273.11.6271
Yada, M., Hatakeyama, S., Kamura, T., Nishiyama, M., Tsunematsu, R., Imaki, H., et al. (2004). Phosphorylation-Dependent Degradation of C-Myc Is Mediated by the F-Box Protein Fbw7. EMBO J. 23, 2116–2125. doi: 10.1038/sj.emboj.7600217
Yang, J., Chen, D., Matar, K. A. O., Zheng, T., Zhao, Q., Xie, Y., et al. (2020). The Deubiquitinating Enzyme Moubp8 Is Required for Infection-Related Development, Pathogenicity, and Carbon Catabolite Repression in Magnaporthe Oryzae. Appl. Microbiol. Biotechnol. 104, 5081–5094. doi: 10.1007/s00253-020-10572-5
Yang, D., Hu, Y., Yin, Z., Gao, Q., Zhang, Y., Chan, F. Y., et al. (2020). Candida Albicans Ubiquitin and Heat Shock Factor-Type Transcriptional Factors Are Involved in 2-Dodecenoic Acid-Mediated Inhibition of Hyphal Growth. Microorganisms 8, 75. doi: 10.3390/microorganisms8010075
Yang, Y., Kitagaki, J., Dai, R. M., Tsai, Y. C., Lorick, K. L., Ludwig, R. L., et al. (2007). Inhibitors of Ubiquitin-Activating Enzyme (E1), a New Class of Potential Cancer Therapeutics. Cancer Res. 67, 9472–9481. doi: 10.1158/0008-5472.CAN-07-0568
Ye, Y., Rape, M. (2009). Building Ubiquitin Chains: E2 Enzymes at Work. Nat. Rev. Mol. Cell Biol. 10, 755–764. doi: 10.1038/nrm2780
Yumimoto, K., Nakayama, K. I. (2020). Recent Insight Into the Role of Fbxw7 as a Tumor Suppressor. Semin. Cancer Biol. 67, 1–15. doi: 10.1016/j.semcancer.2020.02.017
Zhang, W., Sidhu, S. S. (2014). Development of Inhibitors in the Ubiquitination Cascade. FEBS Lett. 588, 356–367. doi: 10.1016/j.febslet.2013.11.003
Zhao, J., Yang, Y., Fan, Y., Yi, J., Zhang, C., Gu, Z., et al. (2020). Ribosomal Protein L40e Fused With a Ubiquitin Moiety Is Essential for the Vegetative Growth, Morphological Homeostasis, Cell Cycle Progression, and Pathogenicity of Cryptococcus Neoformans. Front. Microbiol. 11, 570269. doi: 10.3389/fmicb.2020.570269
Zheng, Q., Huang, T., Zhang, L., Zhou, Y., Luo, H., Xu, H., et al. (2016). Dysregulation of Ubiquitin-Proteasome System in Neurodegenerative Diseases. Front. Aging Neurosci. 8:303. doi: 10.3389/fnagi.2016.00303
Zhou, G. A., Chang, R. Z., Qiu, L. J. (2010). Overexpression of Soybean Ubiquitin-Conjugating Enzyme Gene Gmubc2 Confers Enhanced Drought and Salt Tolerance Through Modulating Abiotic Stress-Responsive Gene Expression in Arabidopsis. Plant Mol. Biol. 72, 357–367. doi: 10.1007/s11103-009-9575-x
Keywords: ubiquitin-proteasome system (UPS), ubiquitin, fungal pathogens, E3 ligase, drug discovery
Citation: Cao C and Xue C (2021) More Than Just Cleaning: Ubiquitin-Mediated Proteolysis in Fungal Pathogenesis. Front. Cell. Infect. Microbiol. 11:774613. doi: 10.3389/fcimb.2021.774613
Received: 12 September 2021; Accepted: 25 October 2021;
Published: 10 November 2021.
Edited by:
Tong-Bao Liu, Southwest University, ChinaReviewed by:
James Kronstad, University of British Columbia, CanadaChangbin Chen, Institut Pasteur of Shanghai (CAS), China
Copyright © 2021 Cao and Xue. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Chaoyang Xue, eHVlY2hAbmptcy5ydXRnZXJzLmVkdQ==
 Chengjun Cao
Chengjun Cao Chaoyang Xue
Chaoyang Xue