- 1Laboratory Department of Clinical Laboratory, The Second Hospital of Jilin University, Changchun, China
- 2Department of Dermatology, The Second Hospital of Jilin University, Changchun, China
- 3Department of General Surgery, The Second Hospital of Jilin University, Changchun, China
- 4Changchun Customs Technology Center, Changchun, China
Gram-positive anaerobic cocci (GPAC) are a commensal part of human flora but are also opportunistic pathogens. This is possibly the first study to report a case of Peptoniphilus harei bacteremia in an abdominal aortic aneurysm (AAA) patient. Matrix-assisted laser desorption ionization time-of-flight mass spectrometry (MALDI-TOF MS) failed to identify the isolate and molecular analysis confirmed it as P. harei. A comprehensive literature review revealed that P. harei is an emergent pathogen. This study serves as a reminder for practicing clinicians to include anaerobic blood cultures as part of their blood culture procedures; this is particularly important situations with a high level of suspicion of infection factors in some noninfectious diseases, as mentioned in this publication. Clinical microbiologists should be aware that the pathogenic potential of GPAC can be greatly underestimated leading to incorrect diagnosis on using only one method for pathogen identification. Upgradation and correction of the MALDI-TOF MS databases is recommended to provide reliable and rapid identification of GPAC at species level in medical diagnostic microbiology laboratories.
Introduction
Gram-positive anaerobic cocci (GPAC), a large group of anaerobic bacteria, comprise several bacterial genera. Although they are members of the normal microbiota, they are also known to be important pathogens causing human diseases, accounting for approximately 25%-30% of all isolated anaerobic bacteria from clinical specimens (Murphy and Frick, 2013). Despite the advances in medical practice in recent years, it is well known that the presence of anaerobes in the bloodstream continues to be associated with high mortality and necessitates appropriate treatment (Cobo et al., 2020; Gajdács and Urbán, 2020). The early identification of anaerobes is important for the initiation of appropriate treatment in patients with bacteremia, and the correct identification up to the species level of GPAC often helps to distinguish between clinically relevant microorganisms and culture contaminants. However, the culture and identification of many GPAC strains in diagnostic laboratories remain hampered due to the requirement of prolonged incubation times and time-consuming phenotypic identification procedures (Simmon et al., 2008; Murphy and Frick, 2013; De Keukeleire et al., 2016; Badri et al., 2019). Following the introduction of 16S rRNA for determining the phylogenetic relationships among prokaryotes, this method has been regarded as the “gold standard” for both microbial phylogeny and ecological studies (Prakash et al., 2013) and is also considered a valuable tool for assigning species to newly isolated bacterial strains. Recently, a new method has been positioned at the forefront for bacterial identification in the future–matrix-assisted laser desorption ionization time-of-flight mass spectrometry (MALDI-TOF MS) (La Scola et al., 2011; Veloo et al., 2011a). At present, two MALDI-TOF MS systems are commercially available for routine use: Bruker Biotyper and VITEK MS. The methodology of the two systems is similar, but they contain differences in the composition of databases and the application of software packages for data analysis, as well as Bruker Biotyper provides results in a log(score), while VITEK MS provides results in percentage of similarity (Lee et al., 2017).
Peptoniphilus genus are GPAC that were formerly classified in the genus Peptostreptococcus, currently consisting of 17 valid published species (http://www.bacterio.net/peptoniphilus.html) (Ezaki et al., 2001), which are involved in various opportunistic human infections mainly as part of polymicrobial infections. The diagnosis of Peptoniphilus infection is mainly based on culture, and the identification of strains is usually performed using phenotypic tests, MALDI-TOF MS, and/or molecular methods (Murphy and Frick, 2013). However, many laboratories do not yet have the capacities to use more current (i.e. MALDI-TOF MS and 16S rRNA sequence) technologies in their diagnostic workflow, therefore they heavily rely on biochemical tests, thus P. harei is often misidentified as P. asaccharolyticus, as these two species have the same biochemical characteristics and cannot be differentiated from each other phenotypically (Veloo et al., 2011b). In this study, we report a patient who presented with AAA and was found to have anaerobic bacteremia that was initially missed because anaerobic blood cultures had been eliminated from the standard blood culture protocol of the institution where he was first assessed. Unfortunately, when he was hospitalized again, Bruker Biotyper and VITEK MS failed to identify GPAC at the species level. The isolate was successfully identified using 16S rRNA sequence analysis and a phylogenetic tree was drawn. We supplemented the identification of this isolate with a literature search into previous clinical cases of P. harei and P. asaccharolyticus infections.
Materials and Methods
Case Report
A 75-year-old man presented to the emergency department (ED) of the Second Hospital of Jilin University (Changchun, China) with pain in his right lower extremity. His past medical history was notable for gastric cancer 10 years prior to the present admission. Four months before the current presentation, the patient was admitted to our institution because of hematemesis and melena, although CT angiography (CTA) showed the presence of an unruptured AAA (Figure 1), which was treated with endovascular stent–graft implantation. Subsequently, the patient improved clinically, but exhibited unexplained persistent bloody stools and was discharged on day 12 of admission. On the present admission, the patient presented with right lower extremity edema and a bruised with no obvious cause. The patient had no diabetes or coronary heart disease. CTA showed severe stenosis or occlusion of the right lower-extremity artery and his blood chemistries were remarkable; the initial laboratory studies were significant for elevated white blood cells at 17.6 × 109/L (reference range, 3.5−9.5 × 109/L), 93.4% neutrophils (reference range, 40%−75%), a serum C-reactive protein (CRP) level of 194 mg/L (reference range, 0.2−7.44 mg/L), and procalcitonin at 9.39 ng/ml (reference range, <0.05 ng/ml). He was empirically treated with cefminox (1000 mg/i.v/12 h) and admitted for further management of suspected lower-limb ischemia. On day 2 of admission, the patient complained of severe pain in the right lower extremity. On physical examination, the skin defect detected on the proximal part of the great right toe was about 2.0 × 2.5 cm in size with liquefied necrotic tissue. A wound swab was collected from a chronic ulcer on his right foot and 40 mL of blood was collected using a syringe. The blood was cultured using a BacT/Alert three-dimensional (bioMérieux Inc.; Durham, NC) automated blood culture system. Each blood culture consisted of a set of two (FA Plus aerobic, FN Plus anaerobic) bottles. Two sets of blood samples were collected from the patient. The only positive microbiological test was Streptococcus anginosus from the wound secretion, and drug susceptibility testing by disk diffusion (Oxoid, Hampshire, United Kingdom) according to the Clinical and Laboratory Standards Institute (CLSI) recommendations revealed that the isolate was resistant to erythromycin, clindamycin, and tetracycline, and susceptible to cefepime, levofloxacin, linezolid, and vancomycin. These two anaerobic blood cultures were positive after 3 days of incubation, and the broths were subcultured onto Columbia agar plates (Oxoid, Thermo Scientific, Hampshire, United Kingdom) that were incubated anaerobically. The aerobic medium bottles remained negative. Within 1 day, growth on Columbia agar plates revealed pinpoint, smooth glistening gray colonies and Gram staining showed gram-positive cocci (Figure 2). Subsequently, the patient’s antimicrobial therapy was switched to moxifloxacin (400 mg/i.v/24 h) and penicillin G (320 million U/iv/6 h). The bacteremia was ultimately cleared after an additional 5 days, and the patient was discharged home on penicillin G for 2 weeks. Surveillance blood cultures performed 2 weeks after the completion of therapy were negative and the patient had improved clinically.
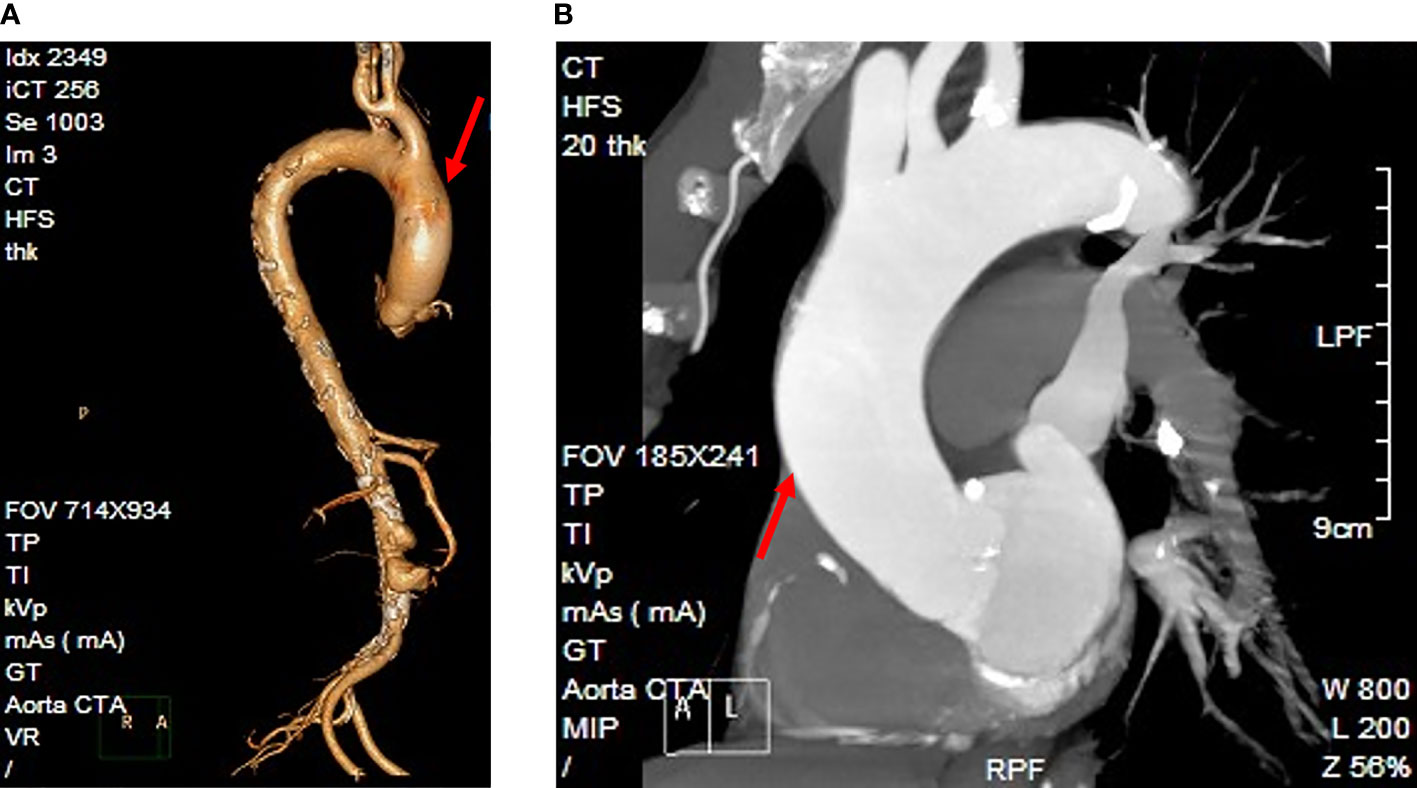
Figure 1 The patient’s diagnostic imaging. (A) Reconstruction of the CT angiography image. (B) CT angiography indicated an unruptured abdominal aortic aneurysm (arrowhead).
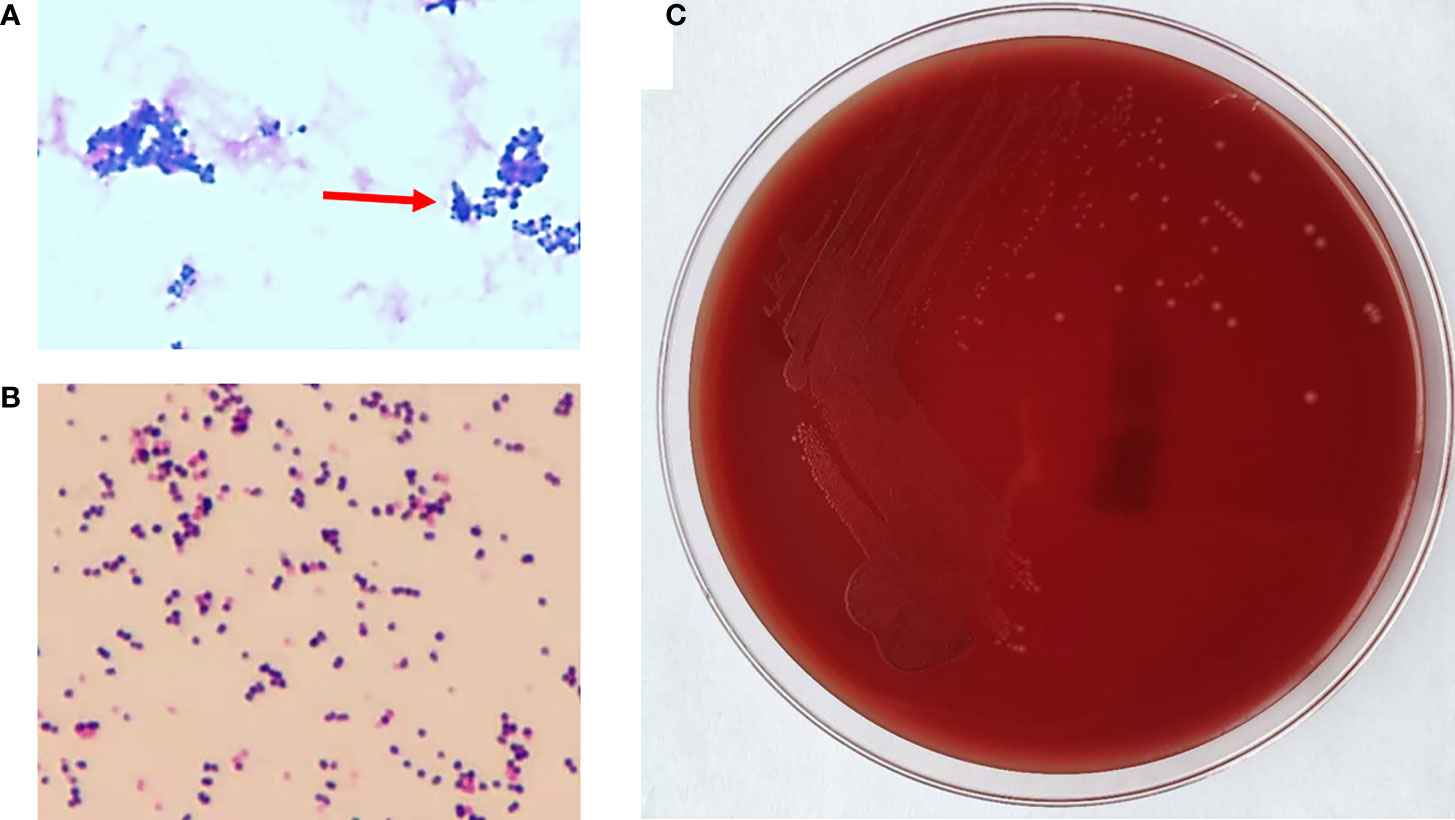
Figure 2 The phenotype of Peptoniphilus harei. (A) Gram staining of P. harei from a positive aerobic blood culture showed gram-positive cocci (1,000× magnification) (arrowhead). (B) Gram-stained colonies showing gram-positive cocci. (C) P. harei colonies on blood agar after 48 h of incubation under anaerobic conditions.
Identification by MALDI- TOF
Pure colonies of the causative bacteria for bacteremia were selected for identification by two MALDI–TOF MS systems, Microflex LT instrument and MALDI Biotyper 3.1 software (Bruker Daltonics, Bremen, Germany) and Vitek MS RUO IVD library (v3.2) (bioMérieux, Marcy I’Étoile, France) in accordance with the manufacturers’ recommendations. Bruker Biotyper reports the identification of each organism with a numerical score, and scores of ≥2.0 and ≥1.7 represent acceptable probable species-and genus-level identifications, respectively. VITEK MS analysis produces a confidence value, reported as a percentage up to 99.9%. The return of an identification of a single taxon, regardless of the confidence value, is considered an acceptable level of identification. If no identification is provided, the isolate is considered unidentified.
Molecular Investigations
16S rRNA sequencing is a valuable tool for definitive molecular identification of important clinical isolates that cannot be readily identified by phenotypic methods or MALDI-TOF. The identity of the clinical isolate was further confirmed by amplifying the 16S rRNA gene with the universal primers 27F and 1492R (Lane, 1991). Next, independent phylogenetic analyses of the partial 16S rDNA sequences were performed using the Neighbor-Joining method (Saitou and Nei, 1987) with the MEGA 6 software (Molecular Evolutionary Genetics Analysis Version 6.0) (Tamura et al., 2013), and the robustness of the trees was evaluated by 1,000 bootstrap replicates to infer the evolutionary history.
Antimicrobial Susceptibility Testing
The minimum inhibitory concentrations (MICs) of various antibiotics against the isolated organisms were determined using the E-test method and according the 2021 CLSI criteria.
Literature Search
After the species identity of the causal organism was confirmed, an electronic search was performed using the key words “Peptoniphilus harei”, “Peptoniphilus asaccharolyticus”, and “human infection” in the PubMed database.
Results
Initially, small colonies were observed in pure culture and were subsequently analyzed via the Bruker Biotyper system. The provided identification of P. harei yielded a log score of 1.288, according to Bruker recommendations; however, no reliable identification at genus level (log score ≥1.70) or species level (log score ≥2) was observed with the most recent MALDI Biotyper database (i.e., the MALDI Biotyper database was not able to correctly identify the isolate). The results of mass spectrometry are dependent on the quantity and accuracy of the database; therefore, we reidentified the isolate by Vitek MS as P. asaccharolyticus based on a confidence level of 99.9%. To our surprise, when additional molecular identification via PCR amplification of the 16s rRNA gene, which facilitates the precise identification of the anaerobic bacteria involved, was performed, the clinical isolate exhibited a 16S rRNA similarity of 99% with P. harei (GenBank NR_026358.1). The sequence has been deposited in GenBank under accession number MZ008068. A multiple alignment was created from the consensus sequences of the novel strains as well as the type sequences of other GPACs with nomenclature obtained from NCBI GenBank. A phylogenetic tree (Figure 3) of 19 nucleotide sequences was constructed. Consistent with the sequencing results, the clinical isolate strain clustered most closely with Peptoniphilus harei MK404230. Therefore, the clinical isolate strain was confidently identified as P. harei. The strain was susceptible to all antimicrobials tested and the MICs obtained for this strain were as follows: penicillin G (0.032 μg/mL), ampicillin (0.032 μg/mL), clindamycin (0.064 μg/mL), meropenem (0.008 μg/mL), ceftriaxone (0.125 μg/mL), chloramphenicol (1.5μg/mL) and metronidazole (0.064μg/mL).
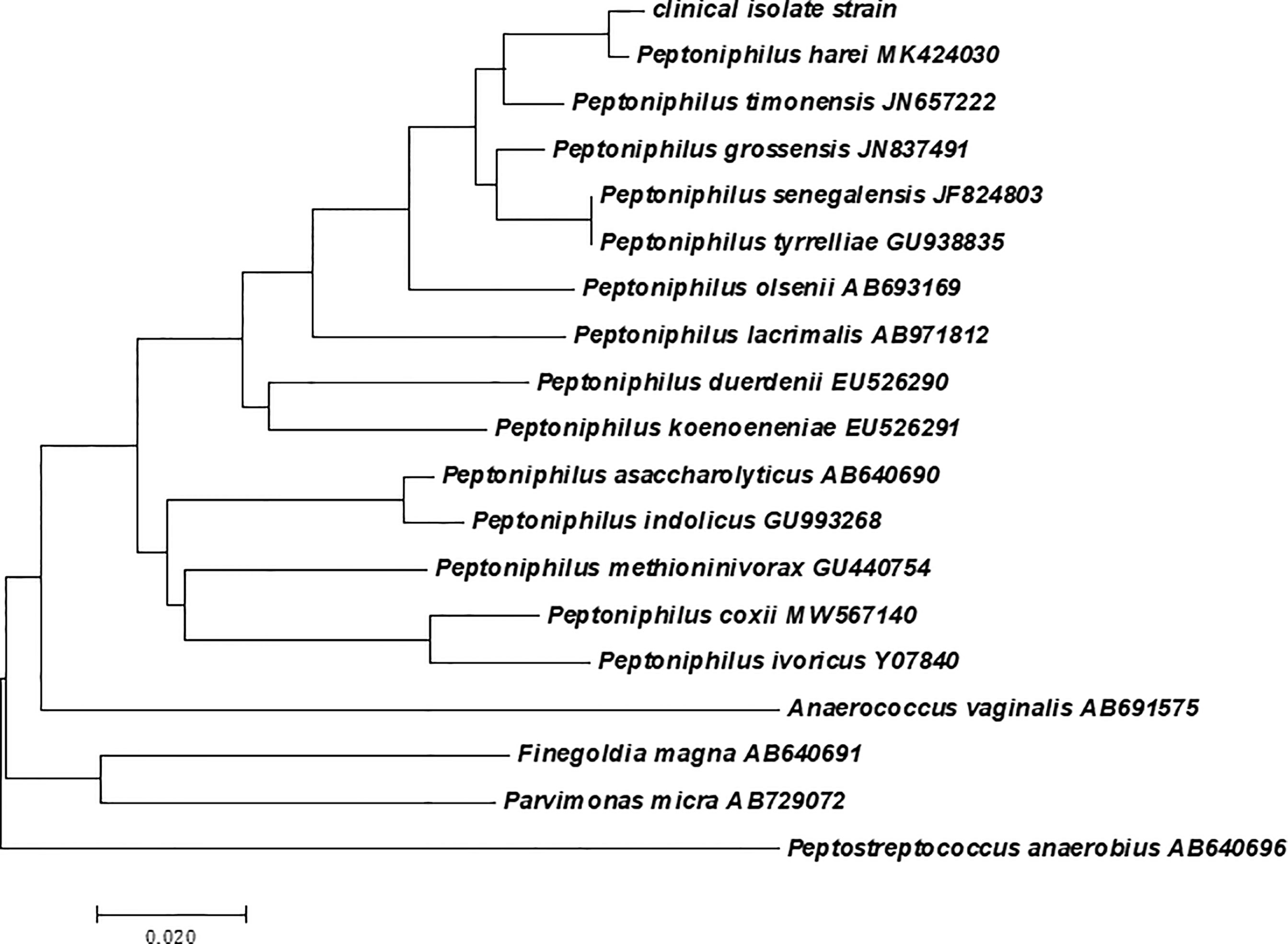
Figure 3 Phylogenetic analysis based on the 16S rRNA gene sequence involving the clinical isolate strain, and the phylogram was constructed using the neighbor-joining method with 1000 bootstrap replicates.
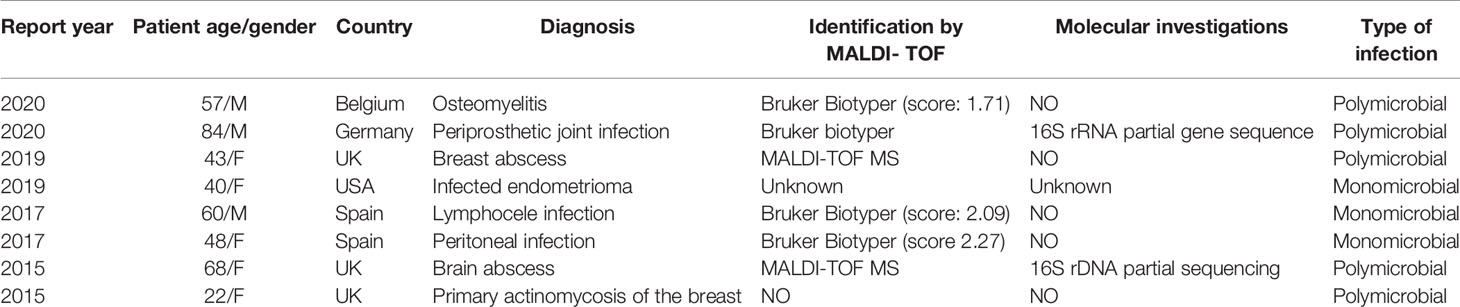
In a comprehensive literature search, we found that the isolation of P. harei in human samples has been mainly observed in the context of polymicrobial infections and that P. harei is an uncommon cause of infection as a single agent. Walter et al. (Walter et al., 2014) identified six strains of P. harei and six strains of P. asaccharolyticus from anaerobic bone and joint infection samples using Biotyper 3.0 software. Cobo et al. employed a Bruker system to identify a case of peritoneal infection (Cobo et al., 2017) and lymphocele infection (Cobo, 2018) due to P. harei, with scores of 2.27 and 2.09, respectively. Another study described a case of Escherichia coli and Peptoniphilus spp. mixed osteomyelitis occurring after trauma; however, the score given by the Bruker system in this case was 1.71, and no additional identification method, such as PCR amplification of the 16S rRNA gene, was used (Costescu Strachinaru et al., 2020). Three strains had been isolated from abscesses in different locations as part of mixed polymicrobial infections (Abdulrahman and Gateley, 2015; Parha et al., 2015; Le Bihan et al., 2019). P. harei is considered a rare pathogen of severe infections, such as infections of the periprosthetic joint (Rieber et al., 2020), breast implants (Seng et al., 2015), or endometrioma (Cornman-Homonoff et al., 2019). Interestingly, in most P. harei cases, species identification was performed using Bruker Biotyper, but infections due to P. asaccharolyticus were diagnosed using Vitek MS (Muller-Schulte et al., 2019). The essential data from some P. harei infections are summarized in Table 1.
Discussion
Although anaerobic bacteria are common pathogens, they are an uncommon cause of bacteremia in humans. A significant minority of bacteremia cases are caused by obligate anaerobic organisms, which are associated with poor clinical outcomes and high rates of mortality. In the first and only case series of Peptoniphilus bloodstream infections presented in 2014, 3 of the 15 cases had a fatal outcome (Brown et al., 2014). P. harei was firstly isolated by Murdoch et al. in 1998 from the pus of various infected sites (Murdoch et al., 1997). It is an uncommon cause of infection as a single agent and is mainly observed in polymicrobial infections in human samples, including pressure ulcers, osteoarticular samples, and skin and soft-tissue infections. Our study was the first report of P. harei bacteremia in a patient with AAA.
The correct identification of pathogens is of paramount importance in clinical practice, epidemiological studies, and the basic research of microorganisms. Previous studies have shown that MALDI-TOF-MS is satisfactory for the genus identification of clinically pathogenic anaerobic bacteria; however, this method still suffers from drawbacks in the precise identification of rare anaerobes at species level. A meta-analysis that included 28 studies covering 6,685 strains of anaerobic bacteria showed that the identification accuracy of MALDI-TOF MS was 84% for species and 92% for genera, meanwhile, the identification accuracy rate was 90% for VITEK MS and 86% for MALDI Biotyper system (Li et al., 2019). A multi-center study that assessed the clinical performance of the VITEK MS system in the identification of anaerobic bacteria found that 651 anaerobic bacterial isolates representing 11 genera and 26 separate species (a total of 91.2%; 594/651) were correctly identified to species level as confirmed by 16S rRNA gene sequencing (Garner et al., 2014). However, Ferrand et al. (2018) reported that the Biotyper system identified more isolates belonging to less commonly encountered anaerobic and facultative anaerobic organisms, including Peptoniphilus, than the Vitek MS system. In this study we observed that the species level was successfully identified by neither of these two systems, although both included P. harei and P. asaccharolyticus in their database. As mentioned earlier, invasive anaerobic infections are life-threatening, and the mortality rate of anaerobic bacteremia can be as high as 40%. Meanwhile, it is difficult to identify rare or newly identified species using conventional phenotyping methods and commercial kits. The causes of the lack of identification by Bruker Biotyper TOF-MS and the erroneous identification by Vitek MS are mainly due to the lack of software or updates in data library databases. It should be mentioned that although the European Network for the Rapid Identification of Anaerobes (ENRIA) project in recent years has successfully increased the number of anaerobic isolates that can be identified with high confidence (Veloo et al., 2018). But the accuracy of both systems should be further increased by expanding, updating, and perfecting the databases.This is essential for improving the identification rate and decreasing misidentifications of Peptoniphilus isolated from clinical specimens and, subsequently, to improve the reporting of uncommon GPAC infections in humans. Hence, we hypothesized that infection due to P. harei may be seriously underestimated depending on the database of the MALDI-TOF MS system, which may allow for additional tests for species identity confirmation.
Our case had several unique clinically interesting characteristics. First, the patient was diagnosed with an AAA 4 months prior to the current presentation. Anaerobic bacteria are an uncommon but important cause of AAA, and various anaerobic bacteria have been found in MAAs (Itzhak and Brook, 2009). MAAs are predominant among males older than 60 years, and especially in those older than 65 years (Wang et al., 1996). The present case of a 75-year-old man who had a history of gastric cancer and surgery exhibited the most significant risk factors. Based on a previous imaging study, the presence of inflammation, and persistent bloody stool of unknown cause, it is possible that the original aneurysm was a mycotic aneurysm that remained undiagnosed because of the lack of an aerobic blood culture. Within the past two decades, the incidence of anaerobic bacteremia among hospitalized patients is thought to have decreased because of the implementation of the recommendation to discontinue routine anaerobic blood cultures in some hospitals. We believe that our case illustrates the need for awareness among infectious disease clinicians and other researchers of the continued importance of anaerobic cultures; we propose that they should be specifically requested in situations of a high level of suspicion of the contribution of anaerobes to the infectious process, and that it is necessary to consider MAAs when diagnosing masses in the context of AAA. In addition, P. harei is generally considered a co-occurring obligate anaerobe in chronic wounds or diabetic ulcers. Our findings were astonishing because the open wound located on the patient’s right great toe was thought to be the source of S. anginosus infection exclusively. Clinicians should consider the independent hematogenous spread of P. harei and P. harei-associated mono-bacteremia in cases of patients with chronic wound infections, as in the present case.
In summary, we report this case to increase the public awareness of P. harei infections, P. harei bacteremia is an uncommon but important human infection, routine use of anaerobic blood cultures can contribute to the diagnosis of a significant number of anaerobic bacteremia cases, resulting in the opportunity to better manage the infections. GPAC have a high pathogenic potential in more severe, invasive infections. To improve reliable identification of GPAC using a combination of multiple technologies will help to better estimate its real prevalence and pathogenicity in a clinical setting.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: MZ008068.
Ethics Statement
This work was approved by the Ethics Committee of the Second Hospital of Jilin University. Written informed consent was obtained from the individual(s) for the publication of any potentially identifiable images or data included in this article.
Author Contributions
XW designed the study, collected the data, and wrote the manuscript. SW and MW were involved in the clinical decision making. JL constructed the tree. YZ reviewed the manuscript and contributed materials. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by research funds from the Project of the Education Department of Jilin Province, (grant no. JJKH20211145KJ), Jilin Province Science and Technology Project of Traditional Chinese Medicine (grant no. 2020031).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
The author apologizes to the authors whose work could not be cited due to space constraints. The author thanks Yufen Jin, Lin Li, and Jing Chang for insightful discussions and critical feedback. We also thank Enago (www.enago.cn/) for the English language review.
References
Abdulrahman, G. O., Jr., Gateley, C. A. (2015). Primary Actinomycosis of the Breast Caused by Actinomyces Trikinis With Associated Peptoniphilus Harei. Breast Dis. 35 (1), 45–47. doi: 10.3233/BD-140381
Badri, M., Nilson, B., Ragnarsson, S., Senneby, E., Rasmussen, M. (2019). Clinical and Microbiological Features of Bacteraemia With Gram-Positive Anaerobic Cocci: A Population-Based Retrospective Study. Clin. Microbiol. Infect. 25 (6), 760.e761–760.e766. doi: 10.1016/j.cmi.2018.09.001
Brown, K., Church, D., Lynch, T., Gregson, D. (2014). Bloodstream Infections Due to Peptoniphilus Spp.: Report of 15 Cases. Clin. Microbiol. Infect. 20 (11), O857–O860. doi: 10.1111/1469-0691.12657
Cobo, F. (2018). Lymphocele Infection Due to Peptoniphilus Harei After Radical Prostatectomy. Med. Mal. Infect. 48 (2), 154–155. doi: 10.1016/j.medmal.2017.10.003
Cobo, F., Aliaga, L., Exposito-Ruiz, M., Navarro-Mari, J. M. (2020). Anaerobic Bacteraemia: A Score Predicting Mortality. Anaerobe 64, 102219. doi: 10.1016/j.anaerobe.2020.102219
Cobo, F., Rodriguez-Granger, J., Sampedro, A., Navarro-Mari, J. M. (2017). Peritoneal Infection Due to Peptoniphilus Harei in a Patient With Intestinal Occlusion. Anaerobe 44, 126–127. doi: 10.1016/j.anaerobe.2017.03.009
Cornman-Homonoff, J., Fenster, T. B., Schiffman, M. H. (2019). Percutaneous Drainage of an Infected Endometrioma. Clin. Imaging 58, 105–107. doi: 10.1016/j.clinimag.2019.06.012
Costescu Strachinaru, D. I., Gallez, J. L., Paridaens, M. S., Djebara, S., Soete, O., Soentjens, P. (2020). A Case of Escherichia Coli and Peptoniphilus Species Mixed Osteomyelitis Successfully Identified by MALDI TOF-MS With a Review of the Literature. Acta Clin. Belg. 18, 1–4. doi: 10.1080/17843286.2020.1783908
De Keukeleire, S., Wybo, I., Naessens, A., Echahidi, F., van der Beken, M., Vandoorslaer, K., et al. (2016). Anaerobic Bacteraemia: A 10-Year Retrospective Epidemiological Survey. Anaerobe 39, 54–59. doi: 10.1016/j.anaerobe.2016.02.009
Ezaki, T., Kawamura, Y., Li, N., Li, Z. Y., Zhao, L., Shu, S. (2001). Proposal of the Genera Anaerococcus Gen. Nov., Peptoniphilus Gen. Nov. And Gallicola Gen. Nov. For Members of the Genus Peptostreptococcus. Int. J. Syst. Evol. Microbiol. 51 (Pt 4), 1521–1528. doi: 10.1099/00207713-51-4-1521
Ferrand, J., Bonnet, I., Alauzet, C., Lozniewski, A. (2018). Evaluation of the Vitek MS and the MALDI Biotyper Systems for the Identification of Less Commonly Isolated But Clinically Relevant Anaerobes and Facultative Anaerobes. Anaerobe 54, 210–216. doi: 10.1016/j.anaerobe.2018.05.014
Gajdács, M., Urbán, E. (2020). Relevance of Anaerobic Bacteremia in Adult Patients: A Never-Ending Story? Eur. J. Microbiol. Immunol. (Bp). 10 (2), 64–75. doi: 10.1556/1886.2020.00009
Garner, O., Mochon, A., Branda, J., Burnham, C. A., Bythrow, M., Ferraro, M., et al. (2014). Multi-Centre Evaluation of Mass Spectrometric Identification of Anaerobic Bacteria Using the VITEK(R) MS System. Clin. Microbiol. Infect. 20 (4), 335–339. doi: 10.1111/1469-0691.12317
Itzhak, Brook (2009). Anaerobic Bacteria as a Cause of Mycotic Aneurysm of the Aorta: Microbiology and Antimicrobial Therapy. Curr. Cardiol. Rev 5 (1), 36–39. doi: 10.2174/157340309787048095
Lane, D. J. (1991). 16s/23s rRNA Sequencing. Nucleic Acid Techniques in Bacterial Systematics. J.Wiley. Sons.
La Scola, B., Fournier, P. E., Raoult, D. (2011). Burden of Emerging Anaerobes in the MALDI-TOF and 16S rRNA Gene Sequencing Era. Anaerobe 17 (3), 106–112. doi: 10.1016/j.anaerobe.2011.05.010
Le Bihan, A., Ahmed, F., O’Driscoll, J. (2019). An Uncommon Cause for a Breast Abscess: Actinomyces Turicensis With Peptoniphilus Harei. BMJ Case Rep. 12 (12), e231194. doi: 10.1136/bcr-2019-231194
Lee, H. S., Shin, J. H., Choi, M. J., Won, E. J., Kee, S. J., Kim, S. H., et al. (2017). Comparison of the Bruker Biotyper and VITEK MS Matrix-Assisted Laser Desorption/Ionization Time-Of-Flight Mass Spectrometry Systems Using a Formic Acid Extraction Method to Identify Common and Uncommon Yeast Isolates. Ann. Lab. Med. 37 (3), 223–230. doi: 10.3343/alm.2017.37.3.223
Li, Y., Shan, M., Zhu, Z., Mao, X., Yan, M., Chen, Y., et al. (2019). Application of MALDI-TOF MS to Rapid Identification of Anaerobic Bacteria. BMC Infect. Dis. 19 (1), 941. doi: 10.1186/s12879-019-4584-0
Muller-Schulte, E., Heimann, K. C., Treder, W. (2019). Peptoniphilus Asaccharolyticus -Commensal, Pathogen or Synergist? Two Case Reports on Invasive Peptoniphilus Asaccharolyticus Infection. Anaerobe 59, 159–162. doi: 10.1016/j.anaerobe.2019.07.001
Murdoch, D. A., Collins, M., Willems, A., Hardie, J. M., Young, K. A., Magee, J. T. (1997). Description of Three New Species of the Genus Peptostreptococcus From Human Clinical Specimens: Peptostreptococcus Harei Sp. Nov., Peptostreptococcus Ivorii Sp. Nov., and Peptostreptococcus Octavius Sp. Nov. Int. J. Syst. Bacteriol. 47 (3), 781–787. doi: 10.1099/00207713-47-3-781
Murphy, E. C., Frick, I. M. (2013). Gram-Positive Anaerobic Cocci–Commensals and Opportunistic Pathogens. FEMS Microbiol. Rev. 37 (4), 520–553. doi: 10.1111/1574-6976.12005
Parha, E., Alalade, A., David, K., Kaddour, H., Degun, P., Namnyak, S. (2015). Brain Abscess Due to Trueperella Bernardiae. Br. J. Neurosurg. 29 (5), 728–729. doi: 10.3109/02688697.2015.1023776
Prakash, O., Jangid, K., Shouche, Y. S. (2013). Carl Woese: From Biophysics to Evolutionary Microbiology. Indian J. Microbiol. 53 (3), 247–252. doi: 10.1007/s12088-013-0401-4
Rieber, H., Frontzek, A., Alefeld, M., Heinrich, S., Barden, B., Jerosch, J., et al. (2020). Sonicate Fluid Inoculated Into Blood Culture Bottles Does Not Improve Diagnosis of Periprosthetic Joint Infection Caused by Anaerobes. A Retrospective Analysis. Anaerobe 62, 102152. doi: 10.1016/j.anaerobe.2020.102152
Saitou, N., Nei, M. (1987). The Neighbor-Joining Method: A New Method for Reconstructing Phylogenetic Trees. Mol. Biol. Evol. 4 (4), 406–425. doi: 10.1093/oxfordjournals.molbev.a040454
Seng, P., Bayle, S., Alliez, A., Romain, F., Casanova, D., Stein, A. (2015). The Microbial Epidemiology of Breast Implant Infections in a Regional Referral Centre for Plastic and Reconstructive Surgery in the South of France. Int. J. Infect. Dis. 35, 62–66. doi: 10.1016/j.ijid.2015.04.010
Simmon, K. E., Mirrett, S., Reller, L. B., Petti, C. A. (2008). Genotypic Diversity of Anaerobic Isolates From Bloodstream Infections. J. Clin. Microbiol. 46 (5), 1596–1601. doi: 10.1128/JCM.02469-07
Tamura, K., Stecher, G., Peterson, D., Filipski, A., Kumar, S. (2013). MEGA6: Molecular Evolutionary Genetics Analysis Version 6.0. Mol. Biol. Evol. 30 (12), 2725–2729. doi: 10.1093/molbev/mst197
Veloo, A. C., Erhard, M., Welker, M., Welling, G. W., Degener, J. E. (2011a). Identification of Gram-Positive Anaerobic Cocci by MALDI-TOF Mass Spectrometry. Syst. Appl. Microbiol. 34 (1), 58–62. doi: 10.1016/j.syapm.2010.11.005
Veloo, A. C., Pierre, H. J., Justesen, Morris, T., Urban, E., Wybo, I., et al. (2018). Validation of MALDI-TOF MS Biotyper Database Optimized for Anaerobic Bacteria: The ENRIA Project. Anaerobe 54, 224–230. doi: 10.1016/j.anaerobe.2018.03.007
Veloo, A. C., Welling, G. W., Degener, J. E. (2011b). Mistaken Identity of Peptoniphilus Asaccharolyticus. J. Clin. Microbiol. 49 (3), 1189. doi: 10.1128/JCM.00043-11
Walter, G., Vernier, M., Pinelli, P. O., Million, M., Coulange, M., Seng, P., et al. (2014). Bone and Joint Infections Due to Anaerobic Bacteria: An Analysis of 61 Cases and Review of the Literature. Eur. J. Clin. Microbiol. Infect. Dis. 33 (8), 1355–1364. doi: 10.1007/s10096-014-2073-3
Keywords: bacteremia, gram-positive anaerobic cocci, Peptoniphilus harei, MALDI–TOF, 16s rDNA sequencing
Citation: Wan X, Wang S, Wang M, Liu J and Zhang Y (2021) Identification of Peptoniphilus harei From Blood Cultures in an Infected Aortic Aneurysm Patient: Case Report and Review Published Literature. Front. Cell. Infect. Microbiol. 11:755225. doi: 10.3389/fcimb.2021.755225
Received: 08 August 2021; Accepted: 22 November 2021;
Published: 22 December 2021.
Edited by:
Max Maurin, Université Grenoble Alpes, FranceReviewed by:
Fernando Cobo, Hospital Universitario Virgen de la Nieve, SpainMárió Gajdács, University of Szeged, Hungary
Copyright © 2021 Wan, Wang, Wang, Liu and Zhang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yu Zhang, zhyky@jlu.edu.cn
 Xue Wan1
Xue Wan1 Yu Zhang
Yu Zhang