
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
MINI REVIEW article
Front. Cell. Infect. Microbiol. , 12 November 2021
Sec. Microbiome in Health and Disease
Volume 11 - 2021 | https://doi.org/10.3389/fcimb.2021.752304
This article is part of the Research Topic The Host-Microbiome Interplay in Colorectal Cancer View all 8 articles
 Laure Campillo-Gimenez1*
Laure Campillo-Gimenez1* David Rios-Covian1
David Rios-Covian1 Jesus Rivera-Nieves2,3
Jesus Rivera-Nieves2,3 Hiroshi Kiyono2,4,5
Hiroshi Kiyono2,4,5 Hiutung Chu1,4
Hiutung Chu1,4 Peter B. Ernst1,3,4,6*
Peter B. Ernst1,3,4,6*Over the last several years, many advances have been made in understanding the role of bacteria in the pathogenesis of gastrointestinal cancers. Beginning with Helicobacter pylori being recognized as the first bacterial carcinogen and the causative agent of most gastric cancers, more recent studies have examined the role of enteric microbes in colorectal cancer. In the digestive tract, these communities are numerous and have a complex interrelationship with local immune/inflammatory responses that impact the health of the host. As modifying the microbiome in the stomach has decreased the risk of gastric cancer, modifying the distal microbiome may decrease the risk of colorectal cancers. To date, very few studies have considered the notion that mucosal lymphocyte-dependent immune memory may confound attempts to change the microbial components in these communities. The goal of this review is to consider some of the factors impacting host-microbial interactions that affect colorectal cancer and raise questions about how immune memory responses to the local microbial consortium affect any attempt to modify the composition of the intestinal microbiome.
Bacteria – specifically H. pylori - have been implicated in cancers of the gastrointestinal tract (Marshall and Warren, 1983; Sears and Garrett, 2014) and this species has now been described as a Group 1 carcinogen by the WHO (Forman, 1996; Fu and Xie, 2019; Cho et al., 2021). Changes in the gastric microbiome occur in association with H. pylori infection but their impact on colonization, bacterial burden, gene expression and susceptibility to adverse outcomes remain to be determined (Brawner et al., 2014). Removing the infection with anti-microbial protocols decreases the risk of gastric cancer significantly (Crowe, 2019).
As Correa (Correa et al., 1976) predicted, an important factor contributing to cancer caused by H. pylori is the oxidative stress associated with chronic inflammation as it increases oxidative DNA damage and mutations leading to malignant transformation (Ernst et al., 2006; Hoeijmakers, 2009; Bhattacharyya et al., 2014; Chumduri et al., 2016). Other causes of chronic inflammation in the digestive tract are associated with cancer including: duodenal tumors with unmanaged or refractory celiac disease (Sharaiha et al., 2012); and inflammatory bowel diseases (IBD) leading to colorectal cancer (CRC) (Irrazabal et al., 2020; Zouggar et al., 2020). IBD has been associated with dysbiosis (Elson and Cong, 2012; Winter et al., 2013; Schaubeck et al., 2015; Honda and Littman, 2016; Dixit et al., 2021) that favors inflammation and enhances the presence of genotoxic strains of bacteria that impart DNA damage (Arthur et al., 2012). Thus, the host response is inextricably linked to the host’s microbiome with both factors contributing to health or disease, including CRC. The purpose of this review is to propose the hypothesis that immunological memory, conferred by the antigen-specific T helper (Th) and B cell responses in health or disease, “shapes” the composition of the microbiological communities. Furthermore, in trying to alter the microbiome for therapeutic benefit, these memory responses would favor the return of the dysbiosis.
Antigen-presenting cells (especially dendritic cells) recognize infections, process, and present microbial antigens to Th cells leading to a pool of antigen-specific memory T and B lymphocytes with the capacity to rapidly expand an immune response to a previously encountered stimulus. While innate cells contribute to “trained immunity” (Bekkering et al., 2021), adaptive immunity is the focus for this discussion on immunological memory. Evidence for memory responses in the gut include the presence of antigen-specific IgA B cell responses after immunization with cholera toxin in mice (Lange et al., 1980; Lycke and Holmgren, 1989) or humans (Quiding et al., 1991). Memory B cell responses have also been reported for rotavirus (Moser et al., 1998). These studies indicate that secondary responses are accelerated and one can reasonably assume that they occur to most of the persistent microbes throughout the digestive tract.
The immunological challenge in the digestive tract requires a measured response to the enormous antigenic burden such that it will not induce inflammation and damage the epithelial surface. Evidence for IgA’s protective role is found in studies showing that the absence of IgA is associated with inflammatory disease in the gut (Nagaishi et al., 2021). IgA is a preferred effector as it can block adherence or neutralize toxins without activation of complement (Marshall et al., 1989; Cerutti and Rescigno, 2008). Secretory IgA binds microbes and also modulates bacterial gene expression and their colonization (Donaldson et al., 2018; Sterlin et al., 2020; Guo et al., 2021). In this manner, IgA limits acute infections with pathogens and establishes an equilibrium with other organisms that are beneficial, or pose no immediate threat. Thus “antigen-specific” IgA establishes a relatively stable, homeostatic environment shared by the host and local microbes. To be clear, not all host responses that enable a microbial species to colonize will be desirable as illustrated by H. pylori. This organism persists throughout life, but being a pathobiont, it can become pathogenic. Thus, homeostatic memory may enable organisms of varying virulence to persist. This concept is complicated by the fact that a single clone of IgA can bind multiple species of bacteria (Bunker et al., 2017; Pabst and Slack, 2020; Sterlin et al., 2020).
In contrast to the healthy gut, IgG is the predominant mucosal isotype in gastritis associated with H. pylori¸ celiac disease and IBD (Brandtzaeg et al., 1986). IgG responses are coupled with the accumulation of activated complement in the inflamed stomach (Berstad et al., 2001) and colon (Berstad et al., 1997). The persistent change in isotype may reflect an infective attempt to clear the organism driving the response. In humans with Crohn’s disease, intestinal bacteria are coated with IgA and IgG whereas in ulcerative colitis, just IgG (Rengarajan et al., 2020). Inoculation of susceptible mice with antibody-coated microbes induces disease (Palm et al., 2014). Thus, IgA can respond to bacteria that promote health or disease and as such, pre-existing IgA responses may select for the species of bacteria that induced them and favor the return of the previous health condition.
Th cell responses are the major driving force for enabling the preferred B cell response. These include control of isotype switch, for example to IgA, as well as the expansion of antigen-specific B cell clones. Certain species of bacteria shape the B cell repertoire (Li et al., 2020), favor the induction of IgA (Yang et al., 2020), including some T cell-independent responses (Pabst and Slack, 2020), and will impact homeostasis accordingly. Memory T cell responses in the mucosa have been described and include Th cells (Offit et al., 1992) as well as cytotoxic T lymphocytes (CTL) (Offit et al., 1991; Cheroutre and Madakamutil, 2004) recognizing rotavirus. Another example of a memory response would be the acquired “oral tolerance” that develops to antigens persistently administered orally (Tokuhara et al., 2019). While much remains to be learned about how these populations contribute to health, compelling evidence suggests intestinal regulatory T cells (Treg) display a classic memory T cell phenotype (Guo et al., 2008) and favor a homeostatic interaction with persistent colonization by specific microbes.
Another property of tissue-resident memory Th cells is their dependence on adhesion molecules (i.e. α4β7 integrin) (Lawrance, 2012; Ley et al., 2016), chemokines (Dixon et al., 2016; Perez-Jeldres et al., 2019) and chemokine receptors (i.e. CCL25/CCR9) that control the trafficking of memory lymphocytes among mucosal tissues (Wong et al., 2016; Fu et al., 2019). In this manner, gut memory “does not fade” (Szilagyi et al., 2017) and ensures memory lymphocytes encounter reinfections and expedite secondary responses. Evidence that these interactions contribute to host-microbial interactions is found when immunotherapies targeting these interactions in IBD (Lawrance, 2012; Perez-Jeldres et al., 2019) and refractory celiac disease remove the inflammatory cells and decrease inflammation. Thus, in disease, there can be a persistent antigen-host response dialogue and some of these triggers will include microbes.
Since Pasteur, host-microbial relationships have been acknowledged to reflect their respective co-evolution. Under germ-free conditions, there is a paucity of immune and inflammatory cells in the gut (Hapfelmeier et al., 2010). As colonization progresses, inductive sites, such as Peyer’s patches and draining lymph nodes, enlarge and seed lymphoblasts that traffic throughout the gut and other mucosal tissues (McDermott and Bienenstock, 1979; Guy-gr et al., 1984; McDermott et al., 1985; Kurashima and Kiyono, 2017; Zhao and Elson, 2018; Chen et al., 2020). If microbiota are necessary to activate intestinal immune responses, it is reasonable to assume that they will favor immunological memory and homeostasis (Marshall et al., 1989). Further, this relationship may have to be “broken” to enable the colonization of the gut with new microbial species administered for their health benefit (Figure 1A).
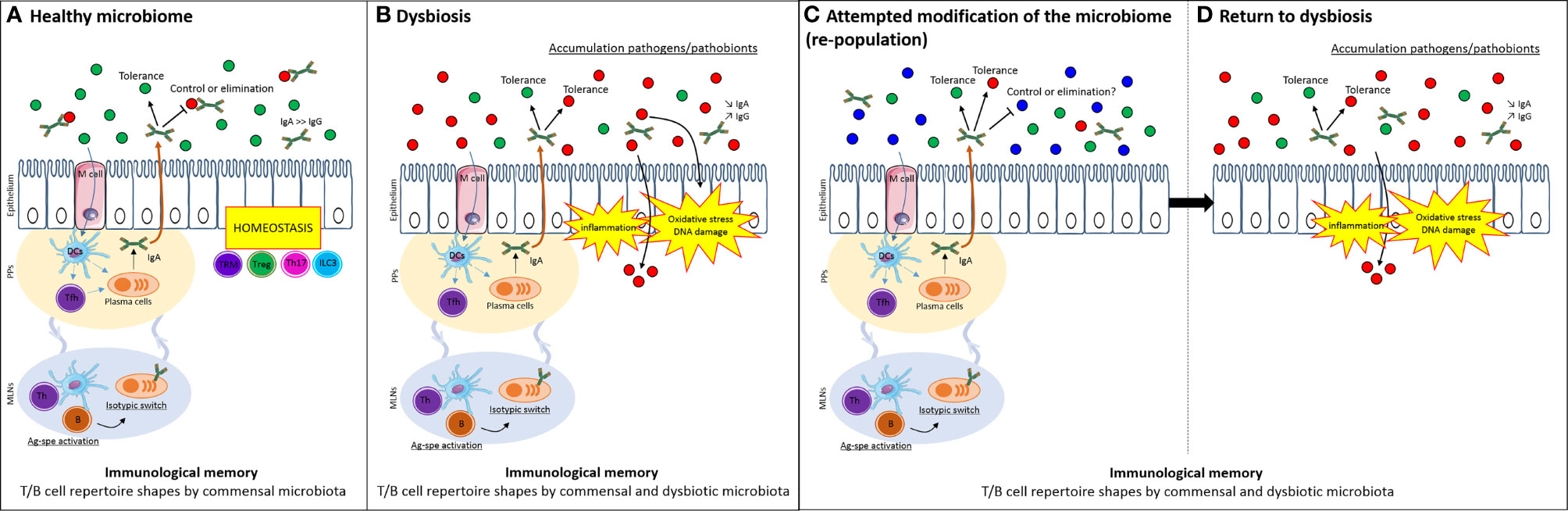
Figure 1 Immunological memory selects for familiar microbiota and limits attempt to modify microbiome. (A) The host can have a balanced homeostatic relationship that inhibits (through any innate or adaptive mechanism) the growth of deleterious bacteria (in particular, those that cause acute disease, red dots) while co-existing with symbiotic species (green dot) or even species that pose no immediate risk but may contribute in the future to chronic disease such as IBD or CRC (e.g. a pathobiont, red dots). Antigen-specific activation of T and B cells imparts the immunological memory to the existing microbiome. (B) In dysbiosis, microbes favoring inflammatory responses and carcinogenesis exist in the presence of host responses that are also antigen-specific. These responses have adapted to favor the persistence of the dysbiotic microbial population. (C) When attempts are made to modulate the microbiota – without altering the immunological memory - existing host responses from the dysbiotic state persist and (D) favor the re-selection of the same community, provided through the therapeutic inoculum (such as a fecal transplant) or additional encounters in the host’s normal environment, e.g. microbes shared from family members, dietary ingredients and so on. DCs, dendritic cells; Tfh, follicular T cells; Th, helper T cells; TRM, tissue-resident memory T cells; Treg, regulatory T cells; B, B cells; IgA, immunoglobulin A; IgG, immunoglobulin G; Ag-spe activation, antigen-specific activation; PPs, Peyer’s patches; MLNs, mesenteric lymph nodes.
T-cell and B-cell receptors (respectively TCR and BCR) recognize antigens that reflect prior exposure to microbes throughout life (Chu et al., 2019; Kreuk et al., 2019; Chen et al., 2020; New et al., 2020). Regionalization of the repertoire is reflected in intestinal intraepithelial lymphocytes as they display a higher oligoclonality compared to peripheral lymphocytes (Regnault et al., 1995). These repertoires comprise some of the evidence for the co-evolution of host responses and local antigenic challenges and demonstrate the specificity of the mucosal memory to specific microbiota.
Microbial colonization of the gut is paired with the development of immunological memory and immune tolerance even before birth, in order to discriminate pathogenic from commensal microbiota and to ensure homeostasis. Mono-colonization of maternal germ-free mice with B. fragilis promotes, in 14-day pups, thymic and splenic T cell development dependent on myeloid cells migrating from the colon (Ennamorati et al., 2020). Similarly, antibiotic treatment impacts the thymic cell distribution and sensitizes the mice to colitis later in life (Ennamorati et al., 2020). Thus, the shaping of memory responses early in life reflects colonization with specific bacterial species as well as disease susceptibility, including those associated with CRC (Figure 1B).
CRC refers to any cancer of the colon or rectum, regardless of their etiology. Clearly, site-specific nuances exist – including in their microbiome - but for this mini-review, the term is used broadly. The gut microbiome evolves and diversifies throughout life depending on the type of birth, diet, lifestyle, geographic factors and medications (Fiorentini et al., 2020; Dixit et al., 2021). The impact of diet and the microbiota on CRC is evidenced by data showing that mice fed a high fat diet leading to changes in their microbial communities have more disease (Yang et al., 2021). Further, the combined effects of the host response and bacteria on intestinal inflammation and CRC were illustrated in a study showing that inoculation of Rag-2-deficient mice with H. hepaticus induced inflammation and colonic tumors (Erdman and Poutahidis, 2017). Moreover, the decrease in the age of onset of human CRC has been associated with changes in the microbial composition in the gut, possibly due to diet or other environmental factors (Couturier-Maillard et al., 2013; Schwabe and Jobin, 2013; Allen-Vercoe and Jobin, 2014; Ray and Kidane, 2016). Based on studies like these, it has been proposed that cancer is “a failure of immune homeostasis” (Erdman and Poutahidis, 2017).
How bacteria contribute to somatic mutations associated with CRC is not well understood (Louis et al., 2014; Thomas and Jobin, 2015; Bullman et al., 2017). However, to establish causation for the microbiota in CRC, it is necessary to envisage a mechanism (Janney et al., 2020). The process of malignant transformation involves many steps including DNA damage and mutations that can be facilitated by genotoxic bacteria (Arthur et al., 2012; Arthur et al., 2014; Vizcaino and Crawford, 2015). Some organisms induce DNA damage directly while others lead to mutations through oxidative stress and changes in DNA methylation (Zouggar et al., 2020). The genetic damage induced by genotoxic bacteria and/or oxidative stress is dealt with by DNA repair pathways. Mutations or changes in the expression and function of DNA repair enzymes that respond to infections – such as AP endonuclease 1 - are associated with cancers (Wang et al., 2003; Fishel et al., 2011; Bhattacharyya et al., 2014; Sevilya et al., 2015), including CRC (Kasahara et al., 2008; Santos et al., 2014) suggesting that when DNA repair becomes compromised, somatic mutations accumulate (Taylor et al., 2003; Nooteboom et al., 2010; Tudek et al., 2010). As key genes are affected, a mutational signature emerges that favors the development of CRC. Other indirect pathways to CRC impacted by microbes and host responses include changes in local metabolites that support cell proliferation and an increase in tumor biomass (Vander Heiden et al., 2009; Datta et al., 2017; Mollace et al., 2021).
Some strains of E. coli produce a genotoxin that is sufficient to increase the risk of CRC (Arthur et al., 2012). Their ability to trigger inflammation may increase their oncogenic potential through effects on bacterial burden or the expression of genotoxic genes (Arthur et al., 2014). In addition, early colonization with genotoxic strains of E. coli decreases the protective oral tolerance response which may explain their ability to increase inflammation (Secher et al., 2015). Other species of bacteria create a dysbiosis that favors inflammation making some people more susceptible to diseases such as IBD (Chu et al., 2016; Dheer et al., 2020; Rengarajan et al., 2020; Torres et al., 2020; Dixit et al., 2021; Galipeau et al., 2021) – and by extension, increases their risk of CRC (Figure 1B). Inflammation in these models may be driven by the loss of bacteria that synthesize anti-inflammatory metabolites, such as short chain fatty acids (SCFAs) (Sommer et al., 2017; Zhao and Elson, 2018); or by the increase in bacteria producing pro-inflammatory molecules. For example, the bacterial production of ATP favors the induction of Th17 cells and enhances colitis (Atarashi et al., 2008; Iwase et al., 2010), while IBD has been associated with fewer SCFAs (Machiels et al., 2014) and their supplementation in animal models favorably regulates the Th17/Treg balance (Ji et al., 2019).
Other bacterial species, such as Fusobacterium nucleatum, have been implicated in CRC without obvious inflammation (Castellarin et al., 2012; Sears and Garrett, 2014; Garrett, 2015; Wang and Jia, 2016; Dejea et al., 2018; Yang and Jobin, 2018). While single species may be able to increase the risk of CRC, complex communities, archaea and biofilms also appear to be important (Tomkovich et al., 2019; Coker et al., 2020; Tomkovich et al., 2020). The combination of genotoxic E. coli and Fusobacterium nucleatum and enterotoxic Bacillus fragilis has been reported to potentiate CRC (Allen-Vercoe and Jobin, 2014; Brennan and Garrett, 2016; Fiorentini et al., 2020). While more direct evidence in humans is being sought, the data to date have led some to suggest that modifying the microbiota can protect against CRC (Dixit et al., 2021; Saeed et al., 2021).
While most of us relate to bacteria within the lumen, microorganisms with either anti- or pro-inflammatory properties have been found in the lamina propria (Sonnenberg and Artis, 2012; Dheer et al., 2020; Hosomi et al., 2020). Relevant to IBD, some microbes exist as symbionts during homeostatic conditions, but under abnormal circumstances, they assume a pathological role (Chow and Mazmanian, 2010; Devkota et al., 2012; Kamada et al., 2013). Such microbes, referred to originally as amphibionts (Blaser and Falkow, 2009) and more recently, pathobionts (Chow and Mazmanian, 2010), depend on innate lymphoid cell function (Goto et al., 2014), or Treg development (Mazmanian et al., 2008; Round and Mazmanian, 2010) linked to the production of anti-inflammatory cytokines IL-22 and IL-10 respectively, to control their pathological potential. Following a disruption in homeostasis, inflammation changes microbial gene expression to alter their colonization and virulence (Arpaia et al., 2011; Barrozo et al., 2013), sensitize host responses (Palm et al., 2014) and leaves a homeostatic “scar” that perpetuates recurrent disease (Fonseca et al., 2015). In other studies, Treg cells have been shown to both inhibit inflammation and enhance IgA (Kawamoto et al., 2014) which may explain Treg modify the microbiota and inhibit colon cancer induced by bacteria (Erdman et al., 2003). Thus, a beneficial role for homeostasis has been established.
These data reveal the direct and indirect impact that local microbes can have on host responses – as evidence of memory responses – and CRC. Should the association of microbial infection and disease extend to direct causal relationships, the argument to modify these communities will strengthen and lead to microbial manipulations for the prevention or management of disease using pre-, pro- or antibiotics (Janney et al., 2020; Dixit et al., 2021). For example, some microbial species may inhibit the growth of certain species of bacteria that express genotoxic machinery that favors DNA damage and mutations that increase the risk of cancer (Arthur et al., 2012; Pope et al., 2017).
Targeting the microbiome alone can have some benefit in modulating diseases that lead to CRC (Sommer et al., 2017) as well as CRC itself (Dixit et al., 2021; Kim and Lim, 2021). Furthermore, it is done to enhance the efficacy of immunotherapy (Ansaldo and Belkaid, 2021). While the evidence may be incomplete, if one assumes that manipulating the microbiota will be beneficial for the prevention of at least some forms of CRC, the implications of the pre-existing immune response should be considered (Figure 1C). Arguments that immunological memory favors the return of the previous bacterial communities include studies showing that the composition of the microbiota rebounds after antibiotic treatment (Sommer et al., 2017). This notion is supported directly by Lindner et al (Lindner et al., 2015) who reported a persistent B cell repertoire over time in humans and mice despite infections or treatment with antibiotics. The interdependence of host-microbial interactions would be predicted by the evolutionary pressures that shape these relationships. The entire homeostatic T and B cell response that leads to the accumulation of IgA has evolved for this to occur. Clearly, memory responses that contribute to this selection could impact the effective colonization by other bacterial species – either protective or deleterious (Figure 1D).
Dr. Belkaid eloquently proposed the concept that a change in microbiota, associated with disease, “scars” immunological homeostasis long after the provocation has been removed (Fonseca et al., 2015). Alternatively, this “scar” may impair any attempt to re-equilibrate host responses by means of modulating the microbiota. Consequently, organisms that favor disease return or persist (Sommer et al., 2017; Lünemann et al., 2020; Galipeau et al., 2021) and trigger disease recurrence (Garrett et al., 2007). This notion is supported by a study in mice showing that anti-TNF-α used to decrease colitis changes the microbiota and attenuates the risk of developing CRC (Yang et al., 2020). Thus, treatment should break immunological barriers and allow colonization by species that favor beneficial homeostasis. This could be achieved by inhibiting “sterilizing” immunity that curtails the survival of beneficial organisms. For example, non-toxicogenic B. fragilis release outer membrane vesicles that induce Treg cells and prevent colitis (Mazmanian et al., 2008; Round and Mazmanian, 2010; Chu et al., 2016) while enterotoxigenic strains can favor the induction of CRC (Geis et al., 2015). As they are immunologically cross-reactive, sparing the memory responses that enable the presence of a protective strain of B. fragilis may be desirable. One would also want to enhance sterilizing immunity to limit colonization by “rogue” strains/species. Limiting infection is not restricted to pathogens as other species can co-exist for decades and may or may not lead to disease including cancer, e.g. H. pylori or genotoxic strains of E. coli. Therefore, altering preexisting immunity to reject unwanted species that inhabited the niche previously, perhaps through vaccination, may be needed to target pathobionts that enhance the risk of CRC and may be re-encountered in the hosts’ environment during attempts to modify the microbiome.
Mucosal antibodies do indeed, shape the microbiota (Kubinak and Round, 2016) and the molecular signature of B cells has been shown to differ in health and disease (Castro-Dopico et al., 2020). These data suggest that B cells, and likely T cells, must be “re-programmed” to allow for the optimal colonization of any transplanted microbiota and a return to homeostasis. While hard evidence is generally lacking, emerging data have led to the notion that the microbiota shape the neonatal immune system as well as following bone marrow reconstitution (Fiorenza and Turtle, 2021; Henig et al., 2021). If the microbiota and local immune response are collaborating in disease, then one would predict that totally reprograming the host response combined with modulating the microbiota would be the preferred strategy to create a new homeostatic equilibrium and a prolonged benefit. Modifying memory responses has been done by using bone marrow reconstitution to reprogram host responses in immune-mediated diseases - not just in mice but also in humans (Sellner and Rommer, 2020; Cencioni et al., 2021).
The vast and diverse microbial community in the intestine makes it challenging to establish a causal role for bacteria in CRC, however there are many reported associations (Castellarin et al., 2012; Sears and Garrett, 2014; Garrett, 2015; Nakatsu et al., 2015; Wang and Jia, 2016; Dejea et al., 2018; Yang and Jobin, 2018; Song and Chan, 2019; Fiorentini et al., 2020; Janney et al., 2020; Nguyen et al., 2020; Kim and Lim, 2021). The first step to establish the importance of a microbial community is to describe it and then determine if the microbe(s) transmit a phenotype. For establishing transmission – an approach that may have implications for the prevention of CRC (Kim and Lim, 2021) – one can manipulate the microbiota and determine how this impacts cancer (Pope et al., 2017). Thus, the use of fecal transplants, probiotics, prebiotics and other strategies, are credible approaches to limit the impact of microbes on CRC (Zhu et al., 2019). However, there are many variables to explore and an effective approach to treat patients by manipulating the microbiota remains to be established (Fong et al., 2020).
If immunological memory is shaping the microbiota, then mapping the antigen-binding repertoires of T and B cells would be informative to prospectively study how changes in repertoire affect the composition of the microbiota. Ideally, these assays would be performed before, during and after CRC development or treatment – with or without any manipulation of the microbiome. Introducing novel microbiota could change a substantial portion of the repertoires (Li et al., 2020) while targeted deletion of antigen-specific responses may favor colonization with new microbial species. One might also predict that a targeted strategy to neutralize a single species or pathogenic element through immunization could limit the ability of CRC-causing microbes to activate disease. As this has been challenging for H. pylori – a species known to cause cancer – it would be premature to propose immunizations for CRC. However, as research exposes the species and mechanisms, it could become an effective approach. As host responses to mucosal antigens are cross reactive with other microbial species, it is worth considering that some vaccines could alter the microbial balance in a deleterious, rather than a beneficial direction.
The situation is expected to be complicated by the unique properties of each microbe, host variation as well as cross-reactive host responses (Fong et al., 2020; Kabbert et al., 2020). Nonetheless, the implications are that the most successful attempt to manipulate the microbiome to prevent CRC may require parallel manipulation of the immune response, either through targeted deletions to favor colonization by beneficial microbes or the induction/maintenance of antigen-specific responses that eliminate pathogenic organisms. Many experimental tools are now in hand to address these interactions and to make targeted changes to the microbiota in CRC as effective as possible.
All authors contributed to the planning of the review. PE wrote the initial draft. LC-G formatted the manuscript and created the figure. LC-G and DR-C participated to the writing of the manuscript. JR-N, HK, and HC provided critical feedback. All authors contributed to the article and approved the submitted version.
This publication was supported by NIH R01-AI079145 (to PE) and the Chiba University-UC San Diego Center for Mucosal Immunology, Allergy and Vaccine Development (PE, HK, HC).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The authors thank Dr. R. Boland for his input related to the pathogenesis of CRC.
Allen-Vercoe, E., Jobin, C. (2014). Fusobacterium and Enterobacteriaceae: Important Players for CRC? Immunol. Lett. 162 (2 Pt A), 54–61. doi: 10.1016/j.imlet.2014.05.014
Ansaldo, E., Belkaid, Y. (2021). How Microbiota Improve Immunotherapy. Science 373 (6558), 966–967. doi: 10.1126/science.abl3656
Arpaia, N., Godec, J., Lau, L., Sivick, K. E., McLaughlin, L. M., Jones, M. B., et al. (2011). TLR Signaling is Required for Salmonella Typhimurium Virulence. Cell 144 (5), 675–688. doi: 10.1016/j.cell.2011.01.031
Arthur, J. C., Gharaibeh, R. Z., Muhlbauer, M., Perez-Chanona, E., Uronis, J. M., McCafferty, J., et al. (2014). Microbial Genomic Analysis Reveals the Essential Role of Inflammation in Bacteria-Induced Colorectal Cancer. Nat. Commun. 5, 4724. doi: 10.1038/ncomms5724
Arthur, J. C., Perez-Chanona, E., Muhlbauer, M., Tomkovich, S., Uronis, J. M., Fan, T. J., et al. (2012). Intestinal Inflammation Targets Cancer-Inducing Activity of the Microbiota. Science 338 (6103), 120–123. doi: 10.1126/science.1224820
Atarashi, K., Nishimura, J., Shima, T., Umesaki, Y., Yamamoto, M., Onoue, M.. (2008). ATP Drives Lamina Propria T(H)17 Cell Differentiation. Nature 455 (7214), 808–812. doi: 10.1038/nature07240
Barrozo, R. M., Cooke, C. L., Hansen, L. M., Lam, A. M., Gaddy, J. A., Johnson, E. M., et al. (2013). Functional Plasticity in the Type IV Secretion System of Helicobacter Pylori. PloS Pathog. 9 (2), e1003189. doi: 10.1371/journal.ppat.1003189
Bekkering, S., Dominguez-Andres, J., Joosten, L. A. B., Riksen, N. P., Netea, M. G. (2021). Trained Immunity: Reprogramming Innate Immunity in Health and Disease. Annu. Rev. Immunol. 39, 667–693. doi: 10.1146/annurev-immunol-102119-073855
Berstad, A. E., Brandtzaeg, P., Stave, R., Halstensen, T. S. (1997). Epithelium Related Deposition of Activated Complement in Helicobacter Pylori Associated Gastritis. Gut 40, 196–203. doi: 10.1136/gut.40.2.196
Berstad, A. E., Hogasen, K., Bukholm, G., Moran, A. P., Brandtzaeg, P. (2001). Complement Activation Directly Induced by Helicobacter Pylori. Gastroenterology 120, 1108–1116. doi: 10.1053/gast.2001.23248
Bhattacharyya, A., Mitra, S., Chattopadhyay, R., Crowe, S. E. (2014). Oxidative Stress: Role in the Pathogenesis of Gastrointestinal Mucosal Diseases. Physiol. Rev. 94, 329–354. doi: 10.1152/physrev.00040.2012
Blaser, M. J., Falkow, S. (2009). What are the Consequences of the Disappearing Human Microbiota? Nat. Rev. Microbiol. 7 (12), 887–894. doi: 10.1038/nrmicro2245
Brandtzaeg, P., Valnes, K., Scott, H., Rognum, T. O., Bjerke, K., Baklien, K. (1986). “The Human Gastrointestinal Secretory Immune System in Health and Disease,” in Basic Science in Gastroenterology: Diseases of the Gut. Eds. Polak, J. M., Bloom, S. R., Wright, N. A., Butler, A. G. (Norwich: Page Bros), 179–200.
Brawner, K. M., Morrow, C. D., Smith, P. D. (2014). Gastric Microbiome and Gastric Cancer. Cancer J. 20 (3), 211–216. doi: 10.1097/PPO.0000000000000043
Brennan, C. A., Garrett, W. S. (2016). Gut Microbiota, Inflammation, and Colorectal Cancer. Annu. Rev. Microbiol. 70, 395–411. doi: 10.1146/annurev-micro-102215-095513
Bullman, S., Pedamallu, C. S., Sicinska, E., Clancy, T. E., Zhang, X., Cai, D. (2017). Analysis of Fusobacterium Persistence and Antibiotic Response in Colorectal Cancer. Science 358 (6369), 1443–1448. doi: 10.1126/science.aal5240
Bunker, J. J., Erickson, S. A., Flynn, T. M., Henry, C., Koval, J. C., Meisel, M. (2017). Natural Polyreactive IgA Antibodies Coat the Intestinal Microbiota. Science 358 (6361), eaan6619. doi: 10.1126/science.aan6619
Castellarin, M., Warren, R. L., Freeman, J. D., Dreolini, L., Krzywinski, M., Strauss, J. (2012). Fusobacterium Nucleatum Infection is Prevalent in Human Colorectal Carcinoma. Genome Res. 22 (2), 299–306. doi: 10.1101/gr.126516.111
Castro-Dopico, T., Colombel, J. F., Mehandru, S. (2020). Targeting B Cells for Inflammatory Bowel Disease Treatment: Back to the Future. Curr. Opin. Pharmacol. 55, 90–98. doi: 10.1016/j.coph.2020.10.002
Cencioni, M. T., Mattoscio, M., Magliozzi, R., Bar-Or, A., Muraro, P. A. (2021). B Cells in Multiple Sclerosis - From Targeted Depletion to Immune Reconstitution Therapies. Nat. Rev. Neurol. 17 (7), 399–414. doi: 10.1038/s41582-021-00498-5
Cerutti, A., Rescigno, M. (2008). The Biology of Intestinal Immunoglobulin A Responses. Immunity 28 (6), 740–750. doi: 10.1016/j.immuni.2008.05.001
Chen, K., Magri, G., Grasset, E. K., Cerutti, A. (2020). Rethinking Mucosal Antibody Responses: IgM, IgG and IgD Join IgA. Nat. Rev. Immunol. 20 (7), 427–441. doi: 10.1038/s41577-019-0261-1
Chen, H., Zhang, Y., Ye, A. Y., Du, Z., Xu, M., Lee, C. S. (2020). BCR Selection and Affinity Maturation in Peyer's Patch Germinal Centres. Nature 582 (7812), 421–425. doi: 10.1038/s41586-020-2262-4
Cheroutre, H., Madakamutil, L. (2004). Acquired and Natural Memory T Cells Join Forces at the Mucosal Front Line. Nat. Rev. Immunol. 4 (4), 290–300. doi: 10.1038/nri1333
Cho, J., Prashar, A., Jones, N. L., Moss, S. F. (2021). Helicobacter Pylori Infection. Gastroenterol. Clin. North Am. 50 (2), 261–282. doi: 10.1016/j.gtc.2021.02.001
Chow, J., Mazmanian, S. K. (2010). A Pathobiont of the Microbiota Balances Host Colonization and Intestinal Inflammation. Cell Host Microbe 7 (4), 265–276. doi: 10.1016/j.chom.2010.03.004
Chu, N. D., Bi, H. S., Emerson, R. O., Sherwood, A. M., Birnbaum, M. E., Robins, H. S., et al. (2019). Longitudinal Immunosequencing in Healthy People Reveals Persistent T Cell Receptors Rich in Highly Public Receptors. BMC Immunol. 20 (1), 19. doi: 10.1186/s12865-019-0300-5
Chu, H., Khosravi, A., Kusumawardhani, I. P., Kwon, A. H. K., Vasconcelos, A. C., Cunha, L. D., et al. (2016). Gene-Microbiota Interactions Contribute to the Pathogenesis of Inflammatory Bowel Disease. Science 352 (6289), 1116–1120. doi: 10.1126/science.aad9948
Chumduri, C., Gurumurthy, R. K., Zietlow, R., Meyer, T. F. (2016). Subversion of Host Genome Integrity by Bacterial Pathogens. Nat. Rev. Mol. Cell Biol. 17 (10), 659–673. doi: 10.1038/nrm.2016.100
Coker, O. O., Wu, W. K. K., Wong, S. H., Sung, J. J. Y., Yu, J. (2020). Altered Gut Archaea Composition and Interaction With Bacteria Are Associated With Colorectal Cancer. Gastroenterology 159 (4), 1459–1470.e1455. doi: 10.1053/j.gastro.2020.06.042
Correa, P., Cuello, C., Duque, E., Burbano, L. C., Garcia, F. T., Bolanos, O., et al. (1976). Gastric Cancer in Colombia. III. Natural History of Precursor Lesions. J. Natl. Cancer Inst. 57 (5), 1027–1035.
Couturier-Maillard, A., Secher, T., Rehman, A., Normand, S., De Arcangelis, A., Haesler, R., et al. (2013). NOD2-Mediated Dysbiosis Predisposes Mice to Transmissible Colitis and Colorectal Cancer. J. Clin. Invest 123 (2), 700–711. doi: 10.1172/JCI62236
Crowe, S. E. (2019). Helicobacter Pylori Infection. N. Engl. J. Med. 380 (12), 1158–1165. doi: 10.1056/NEJMcp1710945
Datta, S., Chowdhury, S., Roy, H. K. (2017). Metabolism, Microbiome and Colorectal Cancer. Aging (Albany NY) 9 (4), 1086–1087. doi: 10.18632/aging.101234
Dejea, C. M., Fathi, P., Craig, J. M., Boleij, A., Taddese, R., Geis, A. L., et al. (2018). Patients With Familial Adenomatous Polyposis Harbor Colonic Biofilms Containing Tumorigenic Bacteria. Science 359, 292–597. doi: 10.1126/science.aah3648
Devkota, S., Wang, Y., Musch, M. W., Leone, V., Fehlner-Peach, H., Nadimpalli, A., et al. (2012). Dietary-Fat-Induced Taurocholic Acid Promotes Pathobiont Expansion and Colitis in Il10-/- Mice. Nature 487 (7405), 104–108. doi: 10.1038/nature11225
Dheer, R., Davies, J. M., Quintero, M. A., Damas, O. M., Deshpande, A. R., Kerman, D. H., et al. (2020). Microbial Signatures and Innate Immune Gene Expression in Lamina Propria Phagocytes of Inflammatory Bowel Disease Patients. Cell Mol. Gastroenterol. Hepatol. 9 (3), 387–402. doi: 10.1016/j.jcmgh.2019.10.013
Dixit, K., Chaudhari, D., Dhotre, D., Shouche, Y., Saroj, S. (2021). Restoration of Dysbiotic Human Gut Microbiome for Homeostasis. Life Sci. 278, 119622. doi: 10.1016/j.lfs.2021.119622
Dixon, B. R., Radin, J. N., Piazuelo, M. B., Contreras, D. C., Algood, H. M. (2016). IL-17a and IL-22 Induce Expression of Antimicrobials in Gastrointestinal Epithelial Cells and May Contribute to Epithelial Cell Defense Against Helicobacter Pylori. PloS One 11 (2), e0148514. doi: 10.1371/journal.pone.0148514
Donaldson, G. P., Ladinsky, M. S., Yu, K. B., Sanders, J. G., Yoo, B. B., Chou, W. C., et al. (2018). Gut Microbiota Utilize Immunoglobulin A for Mucosal Colonization. Science 360 (6390), 795–800. doi: 10.1126/science.aaq0926
Elson, C. O., Cong, Y. (2012). Host-Microbiota Interactions in Inflammatory Bowel Disease. Gut Microbes 3 (4), 332–344. doi: 10.4161/gmic.20228
Ennamorati, M., Vasudevan, C., Clerkin, K., Halvorsen, S., Verma, S., Ibrahim, S., et al. (2020). Intestinal Microbes Influence Development of Thymic Lymphocytes in Early Life. Proc. Natl. Acad. Sci. U. S. A. 117 (5), 2570–2578. doi: 10.1073/pnas.1915047117
Erdman, S. E., Poutahidis, T., Tomczak, M., Rogers, A. B., Cormier, K., Plank, B., et al. (2017). Gut Microbiota Modulate Host Immune Cells in Cancer Development and Growth. Free Radical Biol. Med. 105, 28–34. doi: 10.1016/j.freeradbiomed.2016.11.013
Erdman, S. E., Poutahidis, T., Tomczak, M., et al. (2003). CD4+ CD25+ Regulatory T Lymphocytes Inhibit Microbially Induced Colon Cancer in Rag2-Deficient Mice. Am. J. Pathol. 162 (2), 691–702. doi: 10.1016/S0002-9440(10)63863-1
Ernst, P. B., Peura, D. A., Crowe, S. E. (2006). The Translation of Helicobacter Pylori Basic Research to Patient Care. Gastroenterology 130 (1), 188–206. doi: 10.1053/j.gastro.2005.06.032
Fiorentini, C., Carlini, F., Germinario, E. A. P., Maroccia, Z., Travaglione, S., Fabbri, A. (2020). Gut Microbiota and Colon Cancer: A Role for Bacterial Protein Toxins? Int. J. Mol. Sci. 21 (17), 6201. doi: 10.3390/ijms21176201
Fiorenza, S., Turtle, C. J. (2021). Associations Between the Gut Microbiota, Immune Reconstitution, and Outcomes of Allogeneic Hematopoietic Stem Cell Transplantation. Immunometabolism 3 (1), e210004. doi: 10.20900/immunometab20210004.
Fishel, M. L., Jiang, Y., Rajeshkumar, N. V., Scandura, G., Sinn, A. L., He, Y., et al. (2011). Impact of APE1/Ref-1 Redox Inhibition on Pancreatic Tumor Growth. Mol. Cancer Ther. 10 (9), 1698–1708. doi: 10.1158/1535-7163.MCT-11-0107
Fong, W., Li, Q., Yu, J. (2020). Gut Microbiota Modulation: A Novel Strategy for Prevention and Treatment of Colorectal Cancer. Oncogene 39 (26), 4925–4943. doi: 10.1038/s41388-020-1341-1
Fonseca, D. M., Hand, T. W., Han, S. J., Gerner, M. Y., Zaretsky, A. G., Byrd, A. L., et al. (2015). Microbiota-Dependent Sequelae of Acute Infection Compromise Tissue-Specific Immunity. Cell 163 (2), 354–366. doi: 10.1016/j.cell.2015.08.030
Forman, D. (1996). Helicobacter Pylori and Gastric Cancer. Scand J. Gastroenterol. Suppl 220, 23–26.
Fu, H., Jangani, M., Parmar, A., Wang, G., Coe, D., Spear, S., et al. (2019). A Subset of CCL25-Induced Gut-Homing T Cells Affects Intestinal Immunity to Infection and Cancer. Front. Immunol. 10, 271. doi: 10.3389/fimmu.2019.00271
Fu, L., Xie, C. (2019). A Lucid Review of Helicobacter Pylori-Induced DNA Damage in Gastric Cancer. Helicobacter 24 (5), e12631. doi: 10.1111/hel.12631
Galipeau, H. J., Caminero, A., Turpin, W., Bermudez-Brito, M., Santiago, A., Libertucci, J., et al. (2021). Novel Fecal Biomarkers That Precede Clinical Diagnosis of Ulcerative Colitis. Gastroenterology 160 (5), 1532–1545. doi: 10.1053/j.gastro.2020.12.004
Garrett, W. S. (2015). Cancer and the Microbiota. Science 348 (6230), 80–86. doi: 10.1126/science.aaa4972
Garrett, W. S., Lord, G. M., Punit, S., Lugo-Villarino, G., Mazmanian, S. K., Ito, S., et al. (2007). Communicable Ulcerative Colitis Induced by T-Bet Deficiency in the Innate Immune System. Cell 131 (1), 33–45. doi: 10.1016/j.cell.2007.08.017
Geis, A. L., Fan, H., Wu, X., Wu, S., Huso, D. L., Wolfe, J. L., et al. (2015). Regulatory T-Cell Response to Enterotoxigenic Bacteroides Fragilis Colonization Triggers IL17-Dependent Colon Carcinogenesis. Cancer Discovery 5 (10), 1098–1109. doi: 10.1158/2159-8290.CD-15-0447
Goto, Y., Obata, T., Kunisawa, J., Sato, S., Ivanov, I. I., Lamichhane, A., et al. (2014). Innate Lymphoid Cells Regulate Intestinal Epithelial Cell Glycosylation. Science 345 (6202), 1254009. doi: 10.1126/science.1254009
Guo, Z., Jang, M. H., Otani, K., Bai, Z., Umemoto, E., Matsumoto, M., et al. (2008). CD4+CD25+ Regulatory T Cells in the Small Intestinal Lamina Propria Show an Effector/Memory Phenotype. Int. Immunol. 20 (3), 307–315. doi: 10.1093/intimm/dxm143
Guo, J., Ren, C., Han, X., Huang, W., You, Y., Zhan, J. (2021). Role of IgA in the Early-Life Establishment of the Gut Microbiota and Immunity: Implications for Constructing a Healthy Start. Gut Microbes 13 (1), 1–21. doi: 10.1080/19490976.2021.1908101
Guy-gr, D., Dy, M., Luffau, G., Vassalli, P. (1984). Gut Mucosal Mast Cells. Origin, Traffic, and Differentiattion. J. Exp. Med. 160, 12–28. doi: 10.1084/jem.160.1.12
Hapfelmeier, S., Lawson, M. A. E., Slack, E., Kirundi, J. K., Stoel, M., Heikenwalder, M., et al. (2010). Reversible Microbial Colonization of Germ-Free Mice Reveals the Dynamics of IgA Immune Responses. Science 328 (5986), 1705–1709. doi: 10.1126/science.1188454
Henig, I., Yehudai-Ofir, D., Zuckerman, T. (2021). The Clinical Role of the Gut Microbiome and Fecal Microbiota Transplantation in Allogeneic Stem Cell Transplantation. Haematologica 106 (4), 933–946.
Hoeijmakers, J. H. (2009). DNA Damage, Aging, and Cancer. N Engl. J. Med. 361 (15), 1475–1485. doi: 10.1056/NEJMra0804615
Honda, K., Littman, D. R. (2016). The Microbiota in Adaptive Immune Homeostasis and Disease. Nature 535 (7610), 75–84. doi: 10.1038/nature18848
Hosomi, K., Shibata, N., Shimoyama, A., Uto, T., Nagatake, T., Tojima, Y., et al. (2020). Lymphoid Tissue-Resident Alcaligenes Establish an Intracellular Symbiotic Environment by Creating a Unique Energy Shift in Dendritic Cells. Front. Microbiol. 11, 561005. doi: 10.3389/fmicb.2020.561005
Irrazabal, T., Thakur, B. K., Kang, M., Malaise, Y., Streutker, C., Wong, E. O. Y., et al. (2020). Limiting Oxidative DNA Damage Reduces Microbe-Induced Colitis-Associated Colorectal Cancer. Nat. Commun. 11 (1), 1802. doi: 10.1038/s41467-020-15549-6
Iwase, T., Shinji, H., Tajima, A., Sato, F., Tamura, T., Iwamoto, T., et al. (2010). Isolation and Identification of ATP-Secreting Bacteria From Mice and Humans. J. Clin. Microbiol. 48 (5), 1949–1951. doi: 10.1128/JCM.01941-09
Janney, A., Powrie, F., Mann, E. H. (2020). Host-Microbiota Maladaptation in Colorectal Cancer. Nature 585 (7826), 509–517. doi: 10.1038/s41586-020-2729-3
Ji, J., Ge, X., Chen, Y., Zhu, B., Wu, Q., Zhang, J., et al. (2019). Daphnetin Ameliorates Experimental Colitis by Modulating Microbiota Composition and Treg/Th17 Balance. FASEB J. 33 (8), 9308–9322. doi: 10.1096/fj.201802659RR
Kabbert, J., Benckert, J., Rollenske, T., Hitch, T. C. A., Clavel, T., Cerovic, V., et al. (2020). High Microbiota Reactivity of Adult Human Intestinal IgA Requires Somatic Mutations. J. Exp. Med. 217 (11), e20200275. doi: 10.1084/jem.20200275
Kamada, N., Chen, G. Y., Inohara, N., Nunez, G. (2013). Control of Pathogens and Pathobionts by the Gut Microbiota. Nat. Immunol. 14 (7), 685–690. doi: 10.1038/ni.2608
Kasahara, M., Osawa, K., Yoshida, K., Miyaishi, A., Osawa, Y., Inoue, N., et al. (2008). Association of MUTYH Gln324His and APEX1 Asp148Glu With Colorectal Cancer and Smoking in a Japanese Population. J. Exp. Clin. Cancer Res. 27, 49. doi: 10.1186/1756-9966-27-49
Kawamoto, S., Maruya, M., Kato, L. M., Suda, W., Atarashi, K., Doi, Y., et al. (2014). Foxp3(+) T Cells Regulate Immunoglobulin a Selection and Facilitate Diversification of Bacterial Species Responsible for Immune Homeostasis. Immunity 41 (1), 152–165. doi: 10.1016/j.immuni.2014.05.016
Kim, S. H., Lim, Y. J. (2021). The Role of Microbiome in Colorectal Carcinogenesis and Its Clinical Potential as a Target for Cancer Treatment. Intest Res. doi: 10.5217/ir.2021.00034
Kreuk, L. S., Koch, M. A., Slayden, L. C., Lind, N. A., Chu, S., Savage, H. P., et al. (2019). B Cell Receptor and Toll-Like Receptor Signaling Coordinate to Control Distinct B-1 Responses to Both Self and the Microbiota. eLife 8, e47015. doi: 10.7554/eLife.47015.
Kubinak, J. L., Round, J. L. (2016). Do Antibodies Select a Healthy Microbiota? Nat. Rev. Immunol. 16 (12), 767–774. doi: 10.1038/nri.2016.114
Kurashima, Y., Kiyono, H. (2017). Mucosal Ecological Network of Epithelium and Immune Cells for Gut Homeostasis and Tissue Healing. Annu. Rev. Immunol. 35, 119–147. doi: 10.1146/annurev-immunol-051116-052424
Lange, S., Nygren, H., Svennerholm, A. M., Holmgren, J. (1980). Antitoxic Cholera Immunity in Mice: Influence of Antigen Deposition on Antitoxin-Containing Cells and Protective Immunity in Different Parts of the Intestine. Infect. Immun. 28 (1), 17–23. doi: 10.1128/iai.28.1.17-23.1980
Lawrance, I. C. (2012). Modifying T-Cell Trafficking to the Intestinal as a Potential Management for Inflammatory Bowel Disease. Expert Opin. Investig. Drugs 21 (7), 975–984. doi: 10.1517/13543784.2012.690030
Ley, K., Rivera-Nieves, J., Sandborn, W. J., Shattil, S. (2016). Integrin-Based Therapeutics: Biological Basis, Clinical Use and New Drugs. Nat. Rev. Drug Discovery 15 (3), 173–183. doi: 10.1038/nrd.2015.10
Li, H., Limenitakis, J. P., Greiff, V., Yilmaz, B., Schären, O., Urbaniak, C., et al. (2020). Mucosal or Systemic Microbiota Exposures Shape the B Cell Repertoire. Nature 584 (7820), 274–278. doi: 10.1038/s41586-020-2564-6
Lindner, C., Thomsen, I., Wahl, B., Ugur, M., Sethi, M. K., Friedrichsen, M., et al. (2015). Diversification of Memory B Cells Drives the Continuous Adaptation of Secretory Antibodies to Gut Microbiota. Nat. Immunol. 16 (8), 880–888. doi: 10.1038/ni.3213
Louis, P., Hold, G. L., Flint, H. J. (2014). The Gut Microbiota, Bacterial Metabolites and Colorectal Cancer. Nat. Rev. Microbiol. 12 (10), 661–672. doi: 10.1038/nrmicro3344
Lünemann, J. D., Ruck, T., Muraro, P. A., Bar-Or, A., Wiendl, H. (2020). Immune Reconstitution Therapies: Concepts for Durable Remission in Multiple Sclerosis. Nat. Rev. Neurol. 16 (1), 56–62. doi: 10.1038/s41582-019-0268-z
Lycke, N., Holmgren, J. (1989). Adoptive Transfer of Gut Mucosal Antitoxin Memory by Isolated B Cells 1 Year After Oral Immunization With Cholera Toxin. Infect. Immun. 57 (4), 1137–1141. doi: 10.1128/iai.57.4.1137-1141.1989
Machiels, K., Joossens, M., Sabino, J., De Preter, V., Arijs, I., Eeckhaut, V., et al. (2014). A Decrease of the Butyrate-Producing Species Roseburia Hominis and Faecalibacterium Prausnitzii Defines Dysbiosis in Patients With Ulcerative Colitis. Gut 63 (8), 1275–1283. doi: 10.1136/gutjnl-2013-304833
Marshall, J. S., Bienenstock, J., Perdue, M. H., Stanisz, A. M., Stead, R. H., Ernst, P. B. (1989). Novel Cellular Interactions and Networks Involving the Intestinal Immune System and Its Microenvironment. Acta Pathologica MicrobiolImmunolScand. 97, 383–394. doi: 10.1111/j.1699-0463.1989.tb00805.x
Marshall, B. J., Warren, J. R. (1983). Unidentified Curved Bacilli on Gastric Epithelium in Active Chronic Gastritis. Lancet 8336, 1273–1275.
Mazmanian, S. K., Round, J. L., Kasper, D. L. (2008). A Microbial Symbiosis Factor Prevents Intestinal Inflammatory Disease. Nature 453 (7195), 620–625. doi: 10.1038/nature07008
McDermott, M. R., Bienenstock, J. (1979). Evidence for a Common Mucosal Immunologic System I. Migration of B Immunoblasts Into Intestinal, Respiratory, and Genital Tissues. J. Immunol. 122, 1892–1898.
McDermott, M. R., Horley, B. A., Warner, A. A., Bienenstock, J. (1985). Mesenteric Lymphoblast Localization Throughout the Murine Small Intestine: Temporal Analysis Relating Intestinal Length and Lymphoblast Division. Cell Tissue Kinet 18, 505–519. doi: 10.1111/j.1365-2184.1985.tb00692.x
Mollace, A., Coluccio, M. L., Donato, G., Mollace, V., Malara, N. (2021). Cross-Talks in Colon Cancer Between RAGE/AGEs Axis and Inflammation/Immunotherapy. Oncotarget 12 (13), 1281–1295. doi: 10.18632/oncotarget.27990
Moser, C. A., Cookinham, S., Coffin, S. E., Clark, H. F., Offit, P. A. (1998). Relative Importance of Rotavirus-Specific Effector and Memory B Cells in Protection Against Challenge. J. Virol. 72 (2), 1108–1114. doi: 10.1128/JVI.72.2.1108-1114.1998
Nagaishi, T., Watabe, T., Kotake, K., Kumazawa, T., Aida, T., Tanaka, K., et al. (2021). Immunoglobulin A-Specific Deficiency Induces Spontaneous Inflammation Specifically in the Ileum. Gut. doi: 10.1136/gutjnl-2020-322873
Nakatsu, G., Li, X., Zhou, H., Sheng, J., Wong, S. H., William Ka Kai Wu, W. K. K., et al. (2015). Gut Mucosal Microbiome Across Stages of Colorectal Carcinogenesis. Nat. Commun. 6, 8727. doi: 10.1038/ncomms9727
New, J. S., Dizon, B. L. P., Fucile, C. F., Rosenberg, A. F., Kearney, J. F., King, R. G. (2020). Neonatal Exposure to Commensal-Bacteria-Derived Antigens Directs Polysaccharide-Specific B-1 B Cell Repertoire Development. Immunity 53 (1), 172–186.e176. doi: 10.1016/j.immuni.2020.06.006
Nguyen, L. H., Ma, W., Wang, D. D., Cao, Y., Mallick, H., Gerbaba, T. K., et al. (2020). Association Between Sulfur-Metabolizing Bacterial Communities in Stool and Risk of Distal Colorectal Cancer in Men. Gastroenterology 158 (5), 1313–1325. doi: 10.1053/j.gastro.2019.12.029
Nooteboom, M., Johnson, R., Taylor, R. W., Wright, N. A., Lightowlers, R. N., Kirkwood, T. B. L., et al. (2010). Age-Associated Mitochondrial DNA Mutations Lead to Small But Significant Changes in Cell Proliferation and Apoptosis in Human Colonic Crypts. Aging Cell. 9 (1), 96–99. doi: 10.1111/j.1474-9726.2009.00531.x
Offit, P. A., Cunningham, S. L., Dudzik, K. I. (1991). Memory and Distribution of Virus-Specific Cytotoxic T Lymphocytes (CTLs) and CTL Precursors After Rotavirus Infection. JVirol 65, 1318–1324. doi: 10.1128/jvi.65.3.1318-1324.1991
Offit, P. A., Hoffenberg, E. J., Pia, E. S., Panackal, P. A., Hill, N. L. (1992). Rotavirus-Specific Helper T Cell Responses in Newborns, Infants, Children, and Adults. JInfectDis 165, 1107–1111. doi: 10.1093/infdis/165.6.1107
Pabst, O., Slack, E. (2020). IgA and the Intestinal Microbiota: The Importance of Being Specific. Mucosal Immunol. 13 (1), 12–21. doi: 10.1038/s41385-019-0227-4
Palm, N. W., de Zoete, M. R., Cullen, T. W., Barry, N. A., Stefanowski, J., Hao, L., et al. (2014). Immunoglobulin A Coating Identifies Colitogenic Bacteria in Inflammatory Bowel Disease. Cell 158 (5), 1000–1010. doi: 10.1016/j.cell.2014.08.006
Perez-Jeldres, T., Tyler, C. J., Boyer, J. D., Karuppuchamy, T., Bamias, G., Dulai, P. S., et al. (2019). Cell Trafficking Interference in Inflammatory Bowel Disease: Therapeutic Interventions Based on Basic Pathogenesis Concepts. Inflammation Bowel Dis. 25 (2), 270–282. doi: 10.1093/ibd/izy269
Pope, J. L., Tomkovich, S., Yang, Y., Jobin, C. (2017). Microbiota as a Mediator of Cancer Progression and Therapy. Transl. Res. 179, 139–154. doi: 10.1016/j.trsl.2016.07.021
Quiding, M., Nordstr”m, I., Kilander, A., Andersson, G., Hanson, L. A., Holmgren, J., et al. (1991). Intestinal Immune Responses in Humans. Oral Cholera Vaccination Induces Strong Intestinal Antibody Responses and Interferon- Gamma Production and Evokes Local Immunological Memory. J. Clin. Invest. 88, 143–148. doi: 10.1172/JCI115270
Ray, D., Kidane, D. (2016). Gut Microbiota Imbalance and Base Excision Repair Dynamics in Colon Cancer. J. Cancer 7 (11), 1421–1430. doi: 10.7150/jca.15480
Regnault, A., Kourilsky, P., Cumano, A. (1995). The TCR-Beta Chain Repertoire of Gut-Derived T Lymphocytes. Semin. Immunol. 7 (5), 307–319. doi: 10.1016/1044-5323(95)90012-8
Rengarajan, S., Vivio, E. E., Parkes, M., Peterson, D. A., Elisha D O Roberson, E. D. O., Newberry, R. D., et al. (2020). Dynamic Immunoglobulin Responses to Gut Bacteria During Inflammatory Bowel Disease. Gut Microbes 11 (3), 405–420. doi: 10.1080/19490976.2019.1626683
Round, J. L., Mazmanian, S. K. (2010). Inducible Foxp3+ Regulatory T-Cell Development by a Commensal Bacterium of the Intestinal Microbiota. Proc. Natl. Acad. Sci. U. S. A. 107 (27), 12204–12209. doi: 10.1073/pnas.0909122107
Saeed, M., Shoaib, A., Kandimalla, R., Javed, S., Almatroudi, A., Gupta, R., et al. (2021). Microbe-Based Therapies for Colorectal Cancer: Advantages and Limitations. Semin. Cancer Biol S1044-579X(21)00148-6. doi: 10.1016/j.semcancer.2021.05.018.
Santos, J. C., Funck, A., Silva-Fernandes, I. J., Rabenhorst, S. H., Martinez, C. A., Ribeiro, M. L. (2014). Effect of APE1 T2197G (Asp148Glu) Polymorphism on APE1, XRCC1, PARP1 and OGG1 Expression in Patients With Colorectal Cancer. Int. J. Mol. Sci. 15 (10), 17333–17343. doi: 10.3390/ijms151017333
Schaubeck, M., Clavel, T., Calasan, J., Lagkouvardos, I., Haange, S. B., Jehmlich, N., et al. (2015). Dysbiotic Gut Microbiota Causes Transmissible Crohn's Disease-Like Ileitis Independent of Failure in Antimicrobial Defence. Gut 65 (2), 225–237. doi: 10.1136/gutjnl-2015-309333.
Schwabe, R. F., Jobin, C. (2013). The Microbiome and Cancer. Nat. Rev. Cancer 13 (11), 800–812. doi: 10.1038/nrc3610
Sears, C. L., Garrett, W. S. (2014). Microbes, Microbiota, and Colon Cancer. Cell Host Microbe 15 (3), 317–328. doi: 10.1016/j.chom.2014.02.007
Secher, T., Payros, D., Brehin, C., Boury, M., Watrin, C., Gillet, M., et al. (2015). Oral Tolerance Failure Upon Neonatal Gut Colonization With Escherichia Coli Producing the Genotoxin Colibactin. Infect. Immun. 83 (6), 2420–2429. doi: 10.1128/IAI.00064-15
Sellner, J., Rommer, P. S. (2020). Immunological Consequences of "Immune Reconstitution Therapy" in Multiple Sclerosis: A Systematic Review. Autoimmun Rev. 19 (4), 102492. doi: 10.1016/j.autrev.2020.102492
Sevilya, Z., Leitner-Dagan, Y., Pinchev, M., Kremer, R., Elinger, D., Lejbkowicz, F., et al. (2015). Development of APE1 Enzymatic DNA Repair Assays: Low APE1 Activity is Associated With Increase Lung Cancer Risk. Carcinogenesis 36 (9), 982–991. doi: 10.1093/carcin/bgv082
Sharaiha, R. Z., Lebwohl, B., Reimers, L., Bhagat, G., Green, P. H., Neugut, A. I. (2012). Increasing Incidence of Enteropathy-Associated T-Cell Lymphoma in the United States, 1973-2008. Cancer 118 (15), 3786–3792. doi: 10.1002/cncr.26700
Sommer, F., Anderson, J. M., Bharti, R., Raes, J., Rosenstiel, P. (2017). The Resilience of the Intestinal Microbiota Influences Health and Disease. Nat. Rev. Microbiol. 15 (10), 630–638. doi: 10.1038/nrmicro.2017.58
Song, M., Chan, A. T. (2019). Environmental Factors, Gut Microbiota, and Colorectal Cancer Prevention. Clin. Gastroenterol. Hepatol. 17 (2), 275–289. doi: 10.1016/j.cgh.2018.07.012
Sonnenberg, G. F., Artis, D. (2012). Innate Lymphoid Cell Interactions With Microbiota: Implications for Intestinal Health and Disease. Immunity 37 (4), 601–610. doi: 10.1016/j.immuni.2012.10.003
Sterlin, D., Fadlallah, J., Adams, O., Fieschi, C., Parizot, C., Dorgham, K., et al. (2020). Human IgA Binds a Diverse Array of Commensal Bacteria. J. Exp. Med. 217 (3), e20181635. doi: 10.1084/jem.20181635
Szilagyi, B. A., Triebus, J., Kressler, C., de Almeida, M., Tierling, S., Durek, P., et al. (2017). Gut Memories do Not Fade: Epigenetic Regulation of Lasting Gut Homing Receptor Expression in CD4. Mucosal Immunol. 10 (6), 1443–1454. doi: 10.1038/mi.2017.7
Taylor, R. W., Barron, M. J., Borthwick, G. M., Gospel, A., Chinnery, P. F., Samuels, D. C., et al. (2003). Mitochondrial DNA Mutations in Human Colonic Crypt Stem Cells. J. Clin. Invest 112 (9), 1351–1360. doi: 10.1172/JCI19435
Thomas, R. M., Jobin, C. (2015). The Microbiome and Cancer: Is the 'Oncobiome' Mirage Real? Trends Cancer 1 (1), 24–35. doi: 10.1016/j.trecan.2015.07.005
Tokuhara, D., Kurashima, Y., Kamioka, M., Nakayama, T., Ernst, P., Kiyono, H. (2019). A Comprehensive Understanding of the Gut Mucosal Immune System in Allergic Inflammation. Allergol Int. 68 (1), 17–25. doi: 10.1016/j.alit.2018.09.004
Tomkovich, S., Dejea, C. M., Winglee, K., Drewes, J. L., Chung, L., Housseau, F., et al. (2019). Human Colon Mucosal Biofilms From Healthy or Colon Cancer Hosts are Carcinogenic. J. Clin. Invest 129 (4), 1699–1712. doi: 10.1172/JCI124196
Tomkovich, S., Gharaibeh, R. Z., Dejea, C. M., Pope, J. L., Jiang, J., Winglee, K., et al. (2020). Human Colon Mucosal Biofilms and Murine Host Communicate via Altered mRNA and microRNA Expression During Cancer. mSystems 5 (1), e00451–19. doi: 10.1128/mSystems.00451-19
Torres, J., Hu, J., Seki, A., Eisele, C., Nair, N., Huang, R., et al. (2020). Infants Born to Mothers With IBD Present With Altered Gut Microbiome That Transfers Abnormalities of the Adaptive Immune System to Germ-Free Mice. Gut 69 (1), 42–51. doi: 10.1136/gutjnl-2018-317855
Tudek, B., Winczura, A., Janik, J., Siomek, A., Foksinski, M., Olinski, R. (2010). Involvement of Oxidatively Damaged DNA and Repair in Cancer Development and Aging. Am. J. Transl. Res. 2 (3), 254–284.
Vander Heiden, M. G., Cantley, L. C., Thompson, C. B. (2009). Understanding the Warburg Effect: The Metabolic Requirements of Cell Proliferation. Science 324 (5930), 1029–1033. doi: 10.1126/science.1160809
Vizcaino, M. I., Crawford, J. M. (2015). The Colibactin Warhead Crosslinks DNA. Nat. Chem. 7 (5), 411–417. doi: 10.1038/nchem.2221
Wang, J., Jia, H. (2016). Metagenome-Wide Association Studies: Fine-Mining the Microbiome. Nat. Rev. Microbiol. 14 (8), 508–522. doi: 10.1038/nrmicro.2016.83
Wang, G. S., Wang, M. W., Wu, B. Y., Liu, X. B., You, W. D., Yang, X. Y. (2003). A Gene Encoding an Apurinic/Apyrimidinic Endonuclease-Like Protein is Up-Regulated in Human Gastric Cancer. World JGastroenterol 9 (6), 1196–1201. doi: 10.3748/wjg.v9.i6.1196
Winter, S. E., Lopez, C. A., Baumler, A. J. (2013). The Dynamics of Gut-Associated Microbial Communities During Inflammation. EMBO Rep. 14 (4), 319–327. doi: 10.1038/embor.2013.27
Wong, M. T., Ong, D. E., Lim, F. S., Teng, K. W. W., McGovern, N., Narayanan, S., et al. (2016). A High-Dimensional Atlas of Human T Cell Diversity Reveals Tissue-Specific Trafficking and Cytokine Signatures. Immunity 45 (2), 442–456. doi: 10.1016/j.immuni.2016.07.007
Yang, Y., Gharaibeh, R. Z., Newsome, R. C., Jobin, C. (2020). Amending Microbiota by Targeting Intestinal Inflammation With TNF Blockade Attenuates Development of Colorectal Cancer. Nat. Cancer 1 (7), 723–734. doi: 10.1038/s43018-020-0078-7
Yang, Y., Jobin, C. (2018). Colorectal Cancer: Hand-In-Hand - Colorectal Cancer Metastasizes With Microorganisms. Nat. Rev. Gastroenterol. Hepatol. 15 (3), 133–134. doi: 10.1038/nrgastro.2017.186
Yang, C., Mogno, I., Contijoch, E. J., Borgerding, J. N., Aggarwala, V., Li, Z., et al. (2020). Fecal IgA Levels Are Determined by Strain-Level Differences in Bacteroides Ovatus and Are Modifiable by Gut Microbiota Manipulation. Cell Host Microbe 27 (3), 467–475 e466. doi: 10.1016/j.chom.2020.01.016
Yang, J., Wei, H., Zhou, Y., Szeto, C.-H., Li, C., Lin, Y., et al. (2021). High-Fat Diet Promotes Colorectal Tumorigenesis Through Modulating Gut Microbiota and Metabolites. Gastroenterology. doi: 10.1053/j.gastro.2021.08.041
Zhao, Q., Elson, C. O. (2018). Adaptive Immune Education by Gut Microbiota Antigens. Immunology 154 (1), 28–37. doi: 10.1111/imm.12896
Zhu, W., Miyata, N., Winter, M. G., Arenales, A., Hughes, E. R., Spiga, L., et al. (2019). Editing of the Gut Microbiota Reduces Carcinogenesis in Mouse Models of Colitis-Associated Colorectal Cancer. J. Exp. Med. 216 (10), 2378–2393. doi: 10.1084/jem.20181939
Keywords: microbiome, IBD, CRC, T/B cell repertoire, immune memory, microbiome-editing
Citation: Campillo-Gimenez L, Rios-Covian D, Rivera-Nieves J, Kiyono H, Chu H and Ernst PB (2021) Microbial-Driven Immunological Memory and Its Potential Role in Microbiome Editing for the Prevention of Colorectal Cancer. Front. Cell. Infect. Microbiol. 11:752304. doi: 10.3389/fcimb.2021.752304
Received: 02 August 2021; Accepted: 12 October 2021;
Published: 12 November 2021.
Edited by:
D Scott Merrell, Uniformed Services University, United StatesReviewed by:
Holly Algood, Vanderbilt University, United StatesCopyright © 2021 Campillo-Gimenez, Rios-Covian, Rivera-Nieves, Kiyono, Chu and Ernst. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Peter B. Ernst, cGVybnN0QGhlYWx0aC51Y3NkLmVkdQ==; Laure Campillo-Gimenez, bGNhbXBpbGxvQGhlYWx0aC51Y3NkLmVkdQ==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.