
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Cell. Infect. Microbiol. , 13 December 2021
Sec. Molecular Bacterial Pathogenesis
Volume 11 - 2021 | https://doi.org/10.3389/fcimb.2021.743390
Faecal E. coli can act as reservoirs for resistance genes. Here, we analyzed prevalence of drug resistance in faecal E. coli isolated from healthy children at a single kindergarten in Beijing, China, then used whole genome sequencing to characterize fluoroquinolone-non-susceptible strains. Our results revealed high resistance to ampicillin (54.0%), trimethoprim/sulphurmethoxazole (47.5%) and tetracycline (58.9%) among 576 faecal E. coli isolates, 49.2% of which exhibited multidrug resistance. A total of 113 E. coli isolates were not susceptible to ciprofloxacin, with four sequence types, namely ST1193 (25.7%), ST773 (13.3%), ST648 (8.8%) and ST131 (7.1%) found to be the most prevalent (54.9%). With regards to resistance to quinolones, we detected chromosomal mutations in gyrA, parC, and parE in 111 (98.2%), 105 (92.9%), and 67 (61.1%) isolates, respectively. blaCTX-M (37.2%) was the major ESBL gene, whereas blaCTX-M-14 (12.4%) and blaCTX-M-27 (11.5%) were the most frequent subtypes. A total of 90 (79.6%) ExPEC and 65 (57.5%) UPEC isolates were classified. Overall, these findings revealed clonal spread of certain prevalent STs, namely ST1193, ST773, ST648 and ST131 E. coli isolates in healthy children within a single kindergarten in Beijing, China, affirming the seriousness of the multidrug resistance problem and potential pathogenicity of E. coli isolates in healthy children. Therefore, there is an urgent need for increased surveillance to enhance control of this problem.
Escherichia coli is an important foodborne opportunistic pathogen, that causes various extraintestinal infections, such as urinary tract infections and septicemia (Zhong et al., 2019; Bonten et al., 2021). Previous studies have reported a high prevalence of fluoroquinolone resistance and extended-spectrum β-lactamase (ESBL) production worldwide (Palma et al., 2017; Boll et al., 2020; Stapleton et al., 2020). In China, more than 50% of E. coli strains isolated from community-acquired infections are resistant to fluoroquinolones (Zhao et al., 2015), while 16% of these strains are reported to be ESBL-producing (Ling et al., 2006). Faecal E. coli can act as reservoirs for resistance genes, and are also considered a useful indicator for the spread of acquired antibiotic resistance genes in the community (Nys et al., 2004; Salyers et al., 2004). While geographical variation had been demonstrated in resistance rates of E. coli isolates in feces of healthy children (Sahoo et al., 2012), resistances to commonly used antibiotics like ampicillin, tetracycline and trimethoprim/sulfamethoxazole were frequently observed, especially in developing countries (Bartoloni et al., 2006; Dyar et al., 2012; Shakya et al., 2013; Ferjani et al., 2018; Mahmoodi et al., 2020). Over 90% of faecal samples from Chinese children tested positive for β-lactam, macrolide, tetracycline, and aminoglycoside resistance genes (Ravensdale et al., 2018), indicating the magnitude of the problem of antimicrobial drug resistance in China. To date, however, only a handful of reports have described antibiotic resistance in faecal E. coli among Chinese healthy children (Lester et al., 1990; Huang et al., 2018). In the present study, we focused on fluoroquinolone resistance in children, since it is restricted in the pediatric population due to concerns about significant adverse effects associated with its use (Jackson and Schutze, 2016). To this end, we determined prevalence of resistance genes in E. coli isolated from rectal swabs from healthy children in China, then applied whole genome sequencing to characterize fluoroquinolone-non-susceptible strains.
A total of 596 nonduplicate E. coli strains were isolated from rectal swab samples collected as part of routine physical examination from 736 children, aged between 3 and 6 years, at a kindergarten in Beijing, China, in October 2018. The strains were identified by matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (MALDI-TOF MS) (Vitek MS; bioMérieux, France). The minimal inhibitory concentrations (MICs) of 10 antimicrobial agents, namely ampicillin, ampicillin/sulbactam, trimethoprim/sulfamethoxazole, ciprofloxacin, chloramphenicol, gentamicin, tetracycline, cefotaxime, ceftazidime, and imipenem, were assessed by the agar dilution method with E. coli strain ATCC 25922 as the control according to CLSI 2018. Multidrug resistance (MDR) was defined as resistance of an isolate to any antibiotic from at least three different antibiotic groups (Magiorakos et al., 2012).
E. coli isolates resistant to ciprofloxacin were selected for whole genome sequencing. Briefly, genomic DNA was extracted from the isolates using a DNeasy Blood and Tissue Kit (Qiagen, Hilden, Germany), confirmed by agarose gel electrophoresis and quantified by a Qubit 4 Fluorometer (Thermo Fisher Scientific, Singapore, Singapore). Whole genome sequencing was performed on the Illumina HiSeq platform (Illumina, San Diego, CA, USA) to generate 2×150 bp pair-end reads. Raw reads were de novo assembled using SOAP (Li et al., 2008), and the genomes annotated using RAST v2.0 (Aziz et al., 2008) and prokka v1.12 (Seemann, 2014).
Resistance genes, chromosomal mutations defining quinolone resistance, serotype, fimH subtype, multilocus sequence typing (MLST), and virulence genes of ciprofloxacin-resistant E. coli strains were analyzed using ResFinder 4.0, PointFinder, SerotypeFinder 2.0, FimTyper 1.0, MLST 2.0, and VirulenceFinder 2.0 tools, from the Center for Genomic Epidemiology (CGE) (https://cge.cbs.dtu.dk/services/). E. coli phylogenetic grouping was performed using the ClermonTyping tool (Beghain et al., 2018). Insertion sequence elements were identified using ISfinder (https://isfinder.biotoul.fr/). All drafts and ST-relevant genomes from Enterobase (Zhou et al., 2020) were submitted to kSNP3.0 (Gardner et al., 2015) for single nucleotide polymorphism (SNP) identification, then used to construct a phylogenetic tree with 100 bootstraps which was later visualized using the iTOL website (https://itol.embl.de/). Sequence comparison and map generation were performed using BLAST and Easyfig v2.1 (Sullivan et al., 2011), respectively. Strains were classified as extraintestinal pathogenic E. coli (ExPEC) if they exhibited positivity ≥2 for the following: papAH and/or papC (P fimbriae), sfa-focDE (S and F1C fimbriae), afa-draBC (Dr-binding adhesins), iutA (aerobactin siderophore system), and kpsM II (group 2 capsules). Strains were classified as uropathogenic E. coli (UPEC) if they showed a positivity ≥2 for chuA (heme uptake), fyuA (yersiniabactin siderophore system), vat (vacuolating toxin), and yfcV (adhesin) (Malberg et al., 2020).
Categorical variables were assessed using two-tailed chi-square or Fisher exact tests, where appropriate. Data followed by P ≤ 0.05 were considered statistically significant.
The results of resistant rates of individual antibiotics after antimicrobial susceptibility testing among the studied isolates are shown in Table 1. Summarily, a majority of the isolates were highly resistant to tetracycline (58.9%), followed by ampicillin (54.0%) and trimethoprim/sulphurmethoxazole (47.5%). On the other hand, low resistance was observed in the isolates to ceftazidime (0.3%), whereas no isolate was resistant to imipenem (0.0%). Almost half of the isolates (n= 293, 49.2%) were resistant to three or more different antibiotics, typical of multidrug resistance.
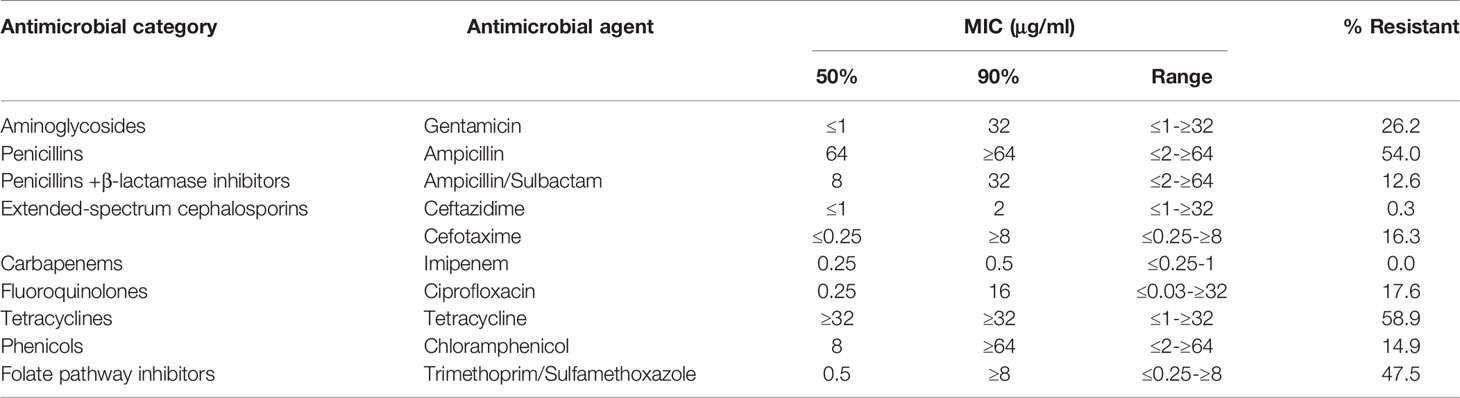
Table 1 MIC profiles of 10 antibiotics against 596 Escherichia coli strains isolated from rectal swab samples collected from 736 children.
A total of 113 ciprofloxacin-non-susceptible E. coli isolates were selected for whole genome sequencing. Phylogenetically, these strains were broadly distributed into the following groups: B2 (38.1%), A (31.0%), F (8.8%), B1 (8.0%), D (8.0%), G (2.7%), C (1.8%), and E (1.8%). The results from in silico MLST analysis revealed 35 different STs, including 3 novel ones (ST11494-11496) which have been submitted to Enterobase. ST1193 (25.7%), ST773 (13.3%), ST648 (8.8%) and ST131 (7.1%) were the most prevalent, accounting for 54.9% of the 113 E. coli isolates. Moreover, ST131 and ST1193 belonged to phylogenetic group B2, while ST648 and ST773 belonged to groups F and group A, respectively (Figure 1).
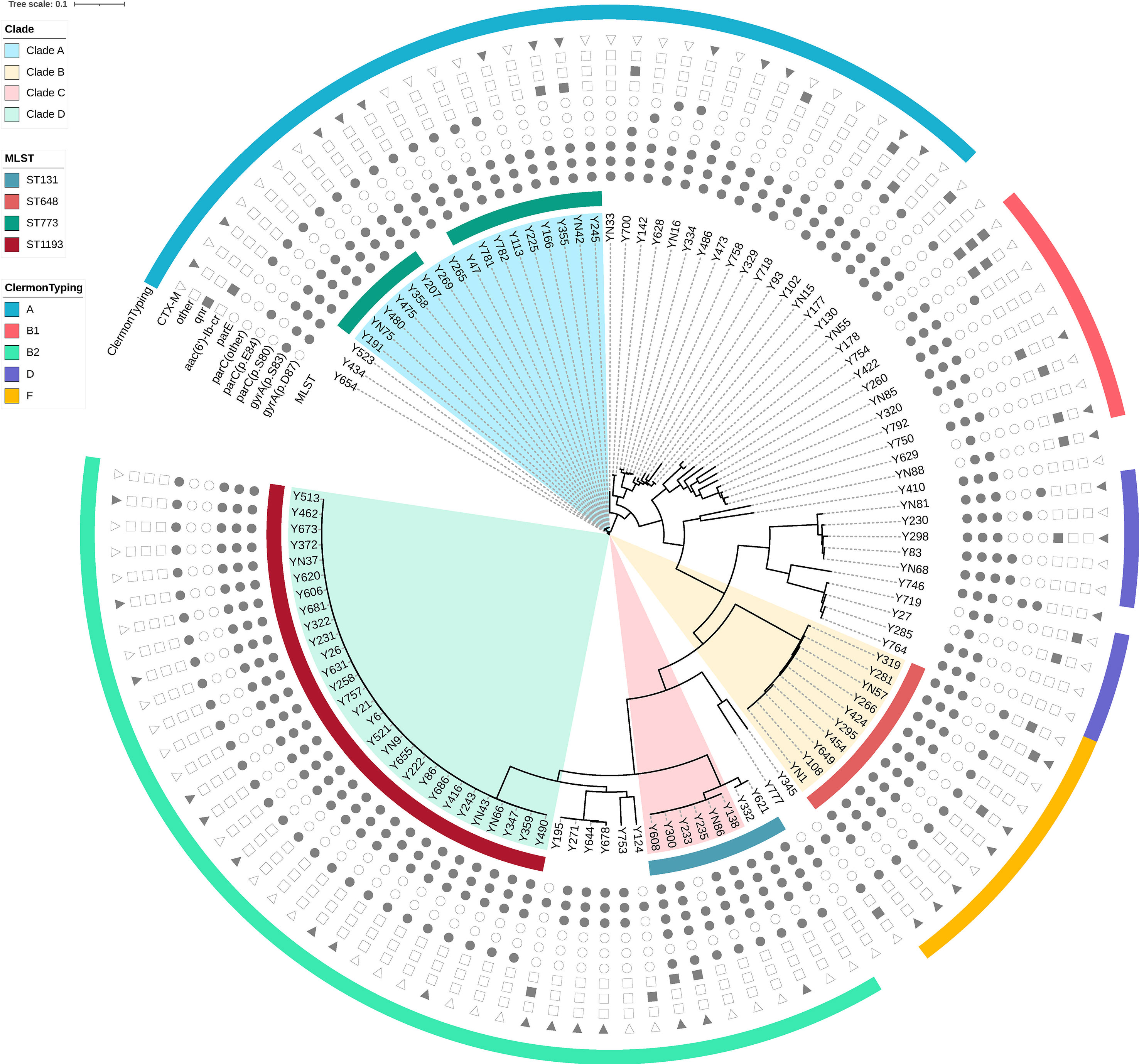
Figure 1 Phylogenetic tree and genomic characteristics of 113 Escherichia coli isolates. Solid and hollow signs indicate presence and absence of acquired resistance genes and chromosomal mutations in quinolone-resistance-determining-regions (QRDR), respectively.
Among 113 E. coli strains, 25 O serogroups and 20 fimH types were identified. The most prevalent O serogroup was O75 (24.8%), followed by O1 (13.3%), O25 (9.7%), and O21 (9.7%). In addition, fimH allele subtyping analysis showed that fimH64 was the most frequent subtype (25.7%), followed by fimH27 (9.7%), fimH54 (6.2%), and fimH30 (5.3%). Notably, almost all of the ST1193 isolates belonged to the O75-fimH64 type, while most of the ST648 and ST773 isolates belonged to O1 and O21 serogroups, respectively. For ST131 isolates, there were two major serogroup-fimH types, namely O25-fimH30 and O16-fimH41.
We adopted ResFinder and PointFinder tools for analysis of antimicrobial resistance genes, and detected resistance to eight groups, namely aminoglycosides, β-lactams, quinolones, macrolides, tetracyclines, phenicols, fosfomycin, and sulphonamides. Notably, all of the 113 ciprofloxacin-resistant E. coli strains carried one or more resistance genes, 103 (91.2%) out of which carried three or more resistances to different antibiotic groups. With regards to resistance to quinolones, we detected chromosomal mutations in gyrA, parC, and parE in 111 (98.2%), 105 (92.9%), and 67 (61.1%) isolates, respectively. The most prevalent chromosomal mutations included gyrA (p.S83L) (98.2%), parC (p.S80I) (90.3%), and gyrA (p.D87N) (81.4%). Moreover, the mutation rate of parE among the four most prevalent STs isolates (ST1193, ST773, ST648 and ST131) was significantly higher than that among other isolates (91.9% vs 23.5%, p=0.000). Notably, parE (p.L416F), parE (p.I529L) and parE (p.S458A) were the unique chromosomal mutation types of parE in ST1193, ST131 and ST648 isolates, respectively. Apart from chromosomal mutations, we also detected aac (6’)-Ib-cr (8.8%), qnrS1 (7.1%), oqxAB (4.4%), qnrS2 (2.7%) and qnrB4 (1.8%). At least one candidate quinolone resistance gene or chromosomal mutation could be found in all of the 113 ciprofloxacin-resistant E. coli strains. With regards to β-lactam resistance, blaCTX-M (37.2%) was the major ESBL gene, of which blaCTX-M-14 (12.4%) and blaCTX-M-27 (11.5%) were the most frequent subtypes. ISEcp1/ISEcp1Δ (100.0%) was found upstream of blaCTX-M in both CTX-M-1 and CTX-M-9 groups. IS903B/IS903BΔ (100.0%) was always found downstream in the CTX-M-9 group, while orf477/orf477Δ (83.3%) was common in the CTX-M-1 group. Besides ISEcp1 and IS903B, IS26 (57.1%) was also found to be adjacent to blaCTX-M genes frequently (Table 2). For aminoglycoside resistance, the main genes carried by these isolates were strA (55.8%), strB (57.5%), aadA5 (53.1%), and aac(3)-IId (40.7%). sul1 (56.6%) and sul2 (57.5%) were the most prevalent genes encoded sulphonamide resistance while fosA3 (4.4%) and fosA7 (1.8%) were the major genes for fosfomycin resistance. The main tetracycline resistance genes detected were tet(A) (61.9%) and tet(B) (17.7%). Neither mcr nor carbapenemase-encoding genes were detected.
The results from VirulenceFinder analysis revealed the presence of 90 (79.6%) ExPEC and 65 (57.5%) UPEC isolates among the 113 ciprofloxacin-resistant E. coli strains. All of the ST131 and ST1193 isolates, which belonged to phylogenetic group B2, could be classified as both ExPEC and UPEC owing to the fact that they harbored iutA, kpsMII/kpsMII_K1/kpsMII_K5, papA, chuA, and fyuA virulence genes. Despite belonging to phylogenetic group A, and different from ST131 and ST1193, all 15 ST773 isolates were identified as ExPEC, and harbored kpsMII_K1, papA, and papC virulence genes (Table 3).
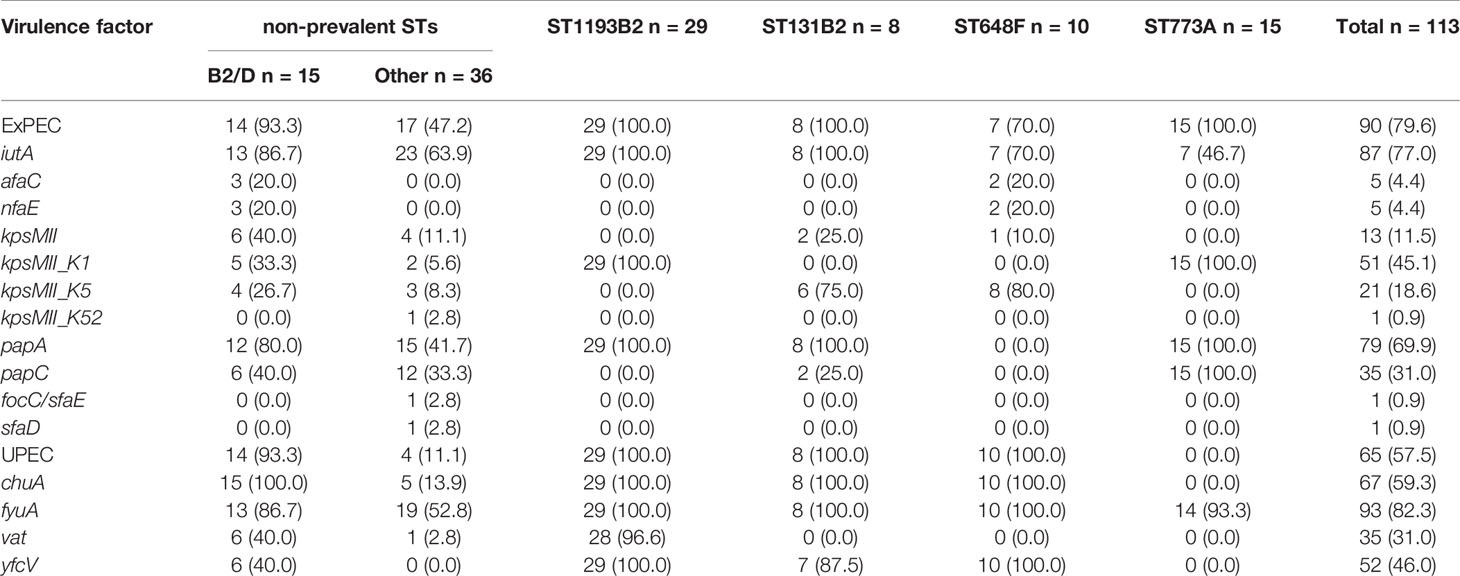
Table 3 Distribution of virulence genes associated with ExPEC/UPEC identification in 113 ciprofloxacin-non-susceptible Escherichia coli isolates (n, %).
The phylogenetic tree obtained by core-genome SNP analysis corroborated the findings from MLST analysis and ClermonTyping phylogenetic group segregation (A, B1, B2, C, D, E, and F). Notably, the four most prevalent STs, ST1193, ST773, ST648 and ST131, were clustered within a monophyletic clade (Figure 1). We also downloaded sequences of draft genomes of 54 ST773 strains isolated across 6 continents (Africa, Asia, Europe, North America, Oceania, and South America) from Enterobase, and incorporated them in our genome analysis to construct a phylogenetic tree comprising 69 ST773 isolates. The results showed that all ST773 isolates belonged to the phylogenetic group A, and revealed 4 serogroups. The most prevalent serogroup was O21:H52 (56.5%), followed by H52 (21.7%), O11:H52 (18.8%), and O11:H4 (2.9%). The phylogenetic tree further clustered the four serogroup isolates into three clades, designated A-C: O11:H52 and H52 serogroups belonged to clade B while O21:H52 serogroup belonged to clade A (Figure 2). Notably, we detected the same chromosomal mutations in all but three of the ST773 isolates, namely gyrA (p.S83L)-gyrA (p.D87N)-parC (p.S80I). In addition, the chromosomal mutation rate of parE (p.S458A) in O11:H52 and H52 isolates (clade B) was significantly higher than that of O21:H52 isolates (clade A) (100.0% vs 38.7%, p=0.000). With regards to resistance to β-lactams, blaCTX-M (37.7%) was the major ESBL gene, of which blaCTX-M-14 (26.1%) and blaCTX-M-15 (7.2%) were the most frequent types.
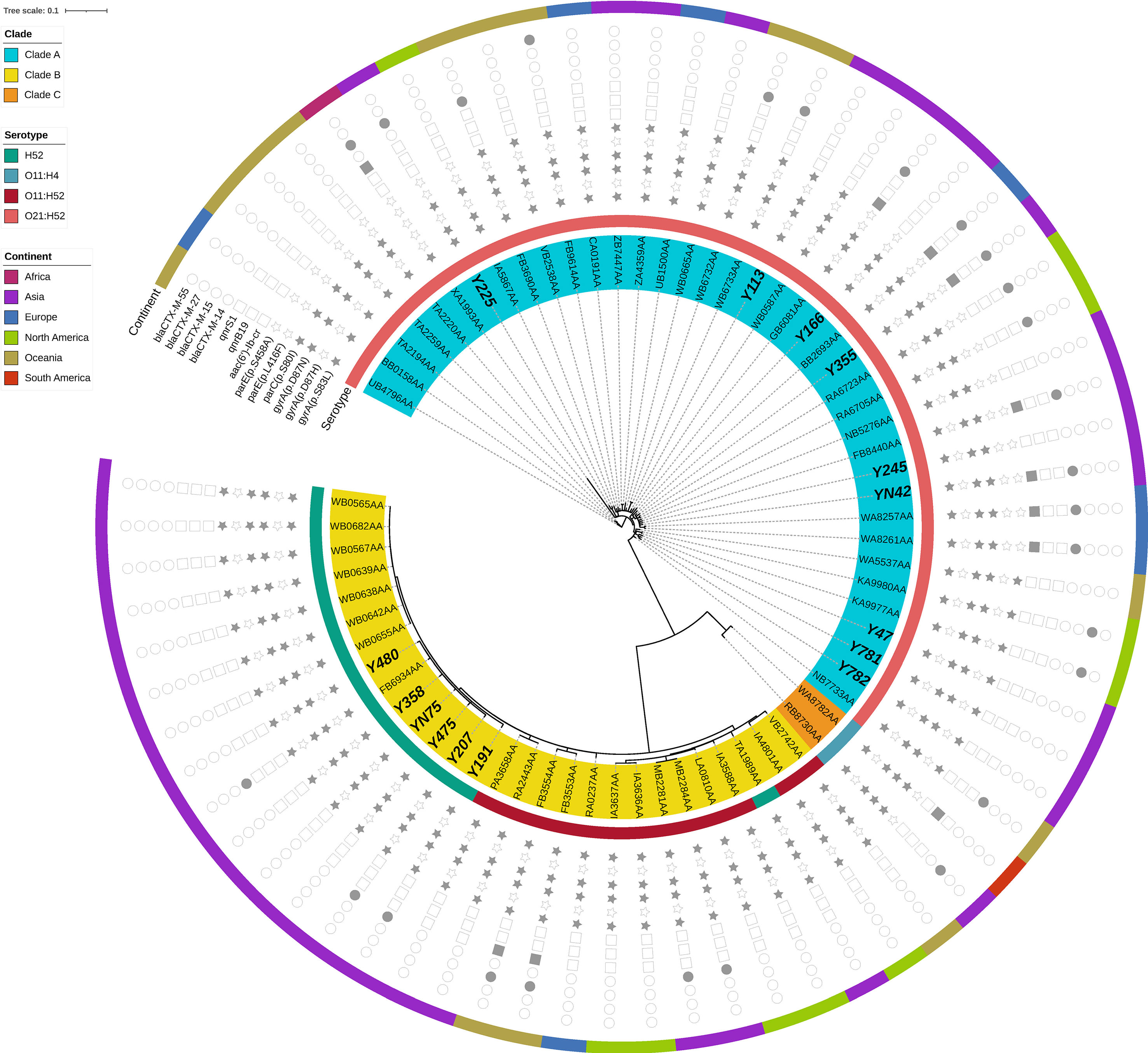
Figure 2 Phylogenetic tree and genomic characteristics of 51 Escherichia coli ST773 isolates. Solid and hollow signs indicate presence and absence of acquired resistance genes and chromosomal mutations in QRDR, respectively.
Antibiotic resistance is a major global public health concern. Faecal E. coli is considered a key indicator for the transmission of acquired antibiotic resistance genes in the community. For instance, a previous systematic review reported that resistance to many primary care prescribed antibiotics was common among E. coli from faecal matter, and this was carried by asymptomatic children, especially in non-Organization for Economic Co-operation and Development countries (Bryce et al., 2016). In addition, multidrug resistance and ESBL production of commensal E. coli isolates were observed in more than 36.2 and 11.7%, respectively, among healthy children under 3 years old in Iran (Mahmoodi et al., 2020). The results from the present study revealed that a majority of the E. coli strains isolated from faecal matter of Chinese healthy children were highly resistant to ampicillin, trimethoprim/sulphurmethoxazole and tetracycline, while almost half of the isolates were multidrug resistant, which was consistent with previous studies (Bartoloni et al., 2006; Shakya et al., 2013; Mahmoodi et al., 2020). Notably, although the use of fluoroquinolones is restricted in the pediatric population because of the associated musculoskeletal adverse effects (Jackson and Schutze, 2016), 17.6% of the E. coli strains exhibited resistance to ciprofloxacin in this study.
Next, we employed whole genome sequencing to determine genetic characteristics of the 113 ciprofloxacin-resistant E. coli strains. Overall, the results indicated that resistance to fluoroquinolones was associated with amino acid substitutions in quinolone-resistance-determining-regions (QRDR), including gyrA, parC, and parE. blaCTX-M, especially blaCTX-M-14 and blaCTX-M-27, contributed to third-generation cephalosporin resistance. In addition, more than 90% of those strains harbored resistance genes for three or more antibiotic groups, while 80% and 60% of them were classified as ExPEC and UPEC strains, respectively. These findings affirmed the seriousness of the problem of multidrug resistance and potential pathogenicity of E. coli isolates in healthy children. Therefore, there is an urgent need for increased surveillance and control of this phenomenon.
The results from our phylogenetic and MLST analyses revealed existence of clonal spread of E. coli isolates, especially for the four most prevalent STs, namely ST1193, ST773, ST648 and ST131, consistent with previous studies that have demonstrated that E. coli strains can be shared within households (Madigan et al., 2015; Johnson et al., 2016). In fact, E. coli ST131 has emerged as a major pathogen of blood-stream and urinary tract infections worldwide (Chen et al., 2019; Birgy et al., 2020; Findlay et al., 2020; Holland et al., 2020). To date, several factors, including resistance to fluoroquinolones, affiliation to phylogroup B2, and high virulence gene contents, have been associated with its successful spread (Valenza et al., 2019). Petty et al. reported that most fluoroquinolone-resistant ST131 strains belonged to a single subclone, designated clade C. Surprisingly, most CTX-M-15-producing ST131 isolates were also derived from a single clade within clade C, named clade C2 (Petty et al., 2014). In the present study, we identified 8 ST131 strains among the 113 ciprofloxacin-resistant E. coli isolates, of which 2 were CTX-M-15 positive. Notably, 6 of the 8 ST131 isolates clustered in clade C (Figure S1), while the 2 CTX-M-15-producing ST131 belonged to clade C2 (Figure S2), which was consistent with the findings of Petty et al. (2014). Since 2012, a new fluoroquinolone-resistant clone namely ST1193, and belonging to phylogenetic group B2, has been reported worldwide (Zhao et al., 2015; Johnson et al., 2019; Tchesnokova et al., 2019). Results of the present study showed that ST1193 was one of the most prevalent STs, accounting for a quarter of all ciprofloxacin-resistant strains. Almost all of the ST1193 isolates belonged to the O75-fimH64 type, and exhibited a set of four conserved mutations in QRDR (gyrA S83L, gyrA D87N, parC S80I and parE L416F), which was consistent with the findings of Valenza et al. (2019). ST648 strains have been reported globally in human patients and more incidentally from animals, because of ESBL phenotype and ExPEC-association (Ewers et al., 2014; Fernandes et al., 2018). Apart from the well-known prevalence of ST131, ST1193 and ST648, ST773 has been considered another key ST, despite having been rarely reported in the past. Despite its affiliation with phylogenetic group A, all 15 ST773 isolates were identified as ExPEC. In fact, almost all of these isolates harbored three conserved mutations that were associated with fluoroquinolone resistance (gyrA D87N, gyrA S83L, parC S80I), while nearly one-third of the isolates carried the blaCTX-M gene. Our phylogenetic tree revealed that 36 ST773 strains isolated across 6 continents exhibited a high degree of homology with our isolates, suggesting the potential of the widespread dissemination of this clone.
In conclusion, we identified four prevalent STs, namely ST1193, ST773, ST648 and ST131, in E. coli isolates. These exhibited clonal transmission across healthy children within a single kindergarten in Beijing, China. Since multidrug resistance and potential pathogenicity of E. coli is a serious problem among healthy children, we envisage that our findings will stimulate relevant discussions to guide urgent development of surveillance approaches to help control this phenomenon.
The datasets generated for this study can be found in GenBank under BioProject no. PRJNA679380.
Neither ethics committee approval, nor informed consent were required as all collected data was fully anonymized, there was no contact with patients and/or their families and no interventions to treatment were made, in accordance with local guidelines.
QZ, YL, SC, and YT conceived and designed the study. YS, GC, and YL acquired the data. QZ drafted the manuscript. SC and YT critically revised the manuscript. All authors contributed to manuscript revision, read, and approved the submitted version.
This work was supported by funds from the National Key Research and Development Program of China (Grant number: 2017YFC1601400).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcimb.2021.743390/full#supplementary-material
Supplementary Figure 1 | Phylogenetic tree and genomic characteristics of 103 Escherichia coli ST131 isolates, including 8 isolates (in bold italics) reported in this study alongside 95 isolates reported by Petty et al. (https://github.com/BeatsonLab-MicrobialGenomics/ST131_99). Solid and hollow signs indicate presence and absence of the acquired resistance genes and chromosomal mutations in QRDR, respectively.
Supplementary Figure 2 | Phylogenetic tree and genomic characteristics of 71 Escherichia coli ST131 isolates that belonged to C clade, including 6 isolates (highlighted by bold italic title) reported in this study alongside 65 isolates reported by Petty et al. (https://github.com/BeatsonLab-MicrobialGenomics/ST131_99).
Supplementary Table 1 | MIC of 10 antibiotics against 596 Escherichia coli strains isolated from rectal swab samples collected from 736 children.
Supplementary Table 2 | Distribution of antimicrobial resistance genes and virulence genes in 113 ciprofloxacin-non-susceptible Escherichia coli isolates.
Supplementary Table 3 | Distribution of antimicrobial resistance genes and virulence genes in 69 Escherichia coli ST773 isolates.
Aziz, R. K., Bartels, D., Best, A. A., DeJongh, M., Disz, T., Edwards, R. A., et al. (2008). The RAST Server: Rapid Annotations Using Subsystems Technology. BMC Genomics 9, 75. doi: 10.1186/1471-2164-9-75
Bartoloni, A., Pallecchi, L., Benedetti, M., Fernandez, C., Vallejos, Y., Guzman, E., et al. (2006). Multidrug-Resistant Commensal Escherichia coli in Children, Peru and Bolivia. Emerg. Infect. Dis. 12 (6), 907–913. doi: 10.3201/eid1206.051258
Beghain, J., Bridier-Nahmias, A., Le, N. H., Denamur, E., Clermont, O. (2018). ClermonTyping: An Easy-to-Use and Accurate in Silico Method for Escherichia Genus Strain Phylotyping. Microb. Genom. 4 (7), e000192. doi: 10.1099/mgen.0.000192
Birgy, A., Madhi, F., Jung, C., Levy, C., Cointe, A., Bidet, P., et al. (2020). Diversity and Trends in Population Structure of ESBL-Producing Enterobacteriaceae in Febrile Urinary Tract Infections in Children in France From 2014 to 2017. J. Antimicrob. Chemother. 75 (1), 96–105. doi: 10.1093/jac/dkz423
Boll, E. J., Overballe-Petersen, S., Hasman, H., Roer, L., Ng, K., Scheutz, F., et al. (2020). Emergence of Enteroaggregative Escherichia coli Within the ST131 Lineage as a Cause of Extraintestinal Infections. mBio 11 (3), e00353-20. doi: 10.1128/mBio.00353-20
Bonten, M., Johnson, J. R., van den Biggelaar, A. H. J., Georgalis, L., Geurtsen, J., de Palacios, P. I., et al. (2021). Epidemiology of Escherichia coli Bacteremia: A Systematic Literature Review. Clin. Infect. Dis. 72 (7), 1211–1219. doi: 10.1093/cid/ciaa210
Bryce, A., Costelloe, C., Hawcroft, C., Wootton, M., Hay, A. D. (2016). Faecal Carriage of Antibiotic Resistant Escherichia coli in Asymptomatic Children and Associations With Primary Care Antibiotic Prescribing: A Systematic Review and Meta-Analysis. BMC Infect. Dis. 16, 359. doi: 10.1186/s12879-016-1697-6
Chen, X., Zou, Q., Zhang, W., Wang, R., Yu, F., Chen, Y. (2019). Clinical Features and Microbiological Characteristics of Hospital- and Community-Onset Escherichia coli Bloodstream Infection. J. Med. Microbiol. 68 (2), 178–187. doi: 10.1099/jmm.0.000904
Dyar, O. J., Hoa, N. Q., Trung, N. V., Phuc, H. D., Larsson, M., Chuc, N. T., et al. (2012). High Prevalence of Antibiotic Resistance in Commensal Escherichia coli Among Children in Rural Vietnam. BMC Infect. Dis. 12, 92. doi: 10.1186/1471-2334-12-92
Ewers, C., Bethe, A., Stamm, I., Grobbel, M., Kopp, P. A., Guerra, B., et al. (2014). CTX-M-15-D-ST648 Escherichia coli From Companion Animals and Horses: Another Pandemic Clone Combining Multiresistance and Extraintestinal Virulence. J. Antimicrob. Chemother. 69 (5), 1224–1230. doi: 10.1093/jac/dkt516
Ferjani, S., Saidani, M., Maamar, E., Harbaoui, S., Hamzaoui, Z., Hosni, H., et al. (2018). Escherichia coli Colonizing Healthy Children in Tunisia: High Prevalence of Extra-Intestinal Pathovar and Occurrence of non-Extended-Spectrum-β-Lactamase-Producing ST131 Clone. Int. J. Antimicrob. Agents 52 (6), 878–885. doi: 10.1016/j.ijantimicag.2018.07.015
Fernandes, M. R., Sellera, F. P., Moura, Q., Gaspar, V. C., Cerdeira, L., Lincopan, N. (2018). International High-Risk Clonal Lineages of CTX-M-Producing Escherichia coli F-ST648 in Free-Roaming Cats, South America. Infect. Genet. Evol. 66, 48–51. doi: 10.1016/j.meegid.2018.09.009
Findlay, J., Gould, V. C., North, P., Bowker, K. E., Williams, M. O., MacGowan, A. P., et al. (2020). Characterization of Cefotaxime-Resistant Urinary Escherichia coli From Primary Care in South-West England 2017-18. J. Antimicrob. Chemother. 75 (1), 65–71. doi: 10.1093/jac/dkz397
Gardner, S. N., Slezak, T., Hall, B. G. (2015). kSNP3.0: SNP Detection and Phylogenetic Analysis of Genomes Without Genome Alignment or Reference Genome. Bioinformatics 31 (17), 2877–2878. doi: 10.1093/bioinformatics/btv271
Holland, M. S., Nobrega, D., Peirano, G., Naugler, C., Church, D. L., JDD, P. (2020). Molecular Epidemiology of Escherichia coli Causing Bloodstream Infections in a Centralized Canadian Region: A Population-Based Surveillance Study. Clin. Microbiol. Infect. 26 (11), 1554.e1–1554.e8. doi: 10.1016/j.cmi.2020.02.019
Huang, I. F., Lee, W. Y., Wang, J. L., Hung, C. H., Hu, H. H., Hung, W. Y., et al. (2018). Faecal Carriage of Multidrug-Resistant Escherichia coli by Community Children in Southern Taiwan. BMC Gastroenterol. 18 (1), 86. doi: 10.1186/s12876-018-0807-x
Jackson, M. A., Schutze, G. E. (2016). The Use of Systemic and Topical Fluoroquinolones. Pediatrics 138 (5), e20162706. doi: 10.1542/peds.2016-2706
Johnson, J. R., Davis, G., Clabots, C., Johnston, B. D., Porter, S., DebRoy, C., et al. (2016). Household Clustering of Escherichia coli Sequence Type 131 Clinical and Faecal Isolates According to Whole Genome Sequence Analysis. Open Forum Infect. Dis. 3 (3), ofw129. doi: 10.1093/ofid/ofw129
Johnson, J. R., Johnston, B. D., Porter, S. B., Clabots, C., Bender, T. L., Thuras, P., et al. (2019). Rapid Emergence, Subsidence, and Molecular Detection of Escherichia coli Sequence Type 1193-Fimh64, a New Disseminated Multidrug-Resistant Commensal and Extraintestinal Pathogen. J. Clin. Microbiol. 57 (5), e01664-18. doi: 10.1128/JCM.01664-18
Lester, S. C., del, P. P. M., Wang, F., Perez, S. I., Jiang, H., O’Brien, T. F. (1990). The Carriage of Escherichia coli Resistant to Antimicrobial Agents by Healthy Children in Boston, in Caracas, Venezuela, and in Qin Pu, China. N. Engl. J. Med. 323 (5), 285–289. doi: 10.1056/NEJM199008023230501
Li, R., Li, Y., Kristiansen, K., Wang, J. (2008). SOAP: Short Oligonucleotide Alignment Program. Bioinformatics 24 (5), 713–714. doi: 10.1093/bioinformatics/btn025
Ling, T. K., Xiong, J., Yu, Y., Lee, C. C., Ye, H., Hawkey, P. M. (2006). Multicenter Antimicrobial Susceptibility Survey of Gram-Negative Bacteria Isolated From Patients With Community-Acquired Infections in the People’s Republic of China. Antimicrob. Agents Chemother. 50 (1), 374–378. doi: 10.1128/AAC.50.1.374-378.2006
Madigan, T., Johnson, J. R., Clabots, C., Johnston, B. D., Porter, S. B., Slater, B. S., et al. (2015). Extensive Household Outbreak of Urinary Tract Infection and Intestinal Colonization Due to Extended-Spectrum β-Lactamase-Producing Escherichia coli Sequence Type 131. Clin. Infect. Dis. 61 (1), e5–12. doi: 10.1093/cid/civ273
Magiorakos, A. P., Srinivasan, A., Carey, R. B., Carmeli, Y., Falagas, M. E., Giske, C. G., et al. (2012). Multidrug-Resistant, Extensively Drug-Resistant and Pandrug-Resistant Bacteria: An International Expert Proposal for Interim Standard Definitions for Acquired Resistance. Clin. Microbiol. Infect. 18 (3), 268–281. doi: 10.1111/j.1469-0691.2011.03570.x
Mahmoodi, F., Rezatofighi, S. E., Akhoond, M. R. (2020). Antimicrobial Resistance and Metallo-Beta-Lactamase Producing Among Commensal Escherichia coli Isolates From Healthy Children of Khuzestan and Fars Provinces; Iran. BMC Microbiol. 20 (1), 366. doi: 10.1186/s12866-020-02051-8
Malberg, T. A. M., Johnson, J. R., Johnston, B. D., Lund, O., Scheutz, F. (2020). In Silico Genotyping of Escherichia coli Isolates for Extraintestinal Virulence Genes by Use of Whole Genome Sequencing Data. J. Clin. Microbiol. 58 (10), e01269-20. doi: 10.1128/JCM.01269-20
Nys, S., Okeke, I. N., Kariuki, S., Dinant, G. J., Driessen, C., Stobberingh, E. E. (2004). Antibiotic Resistance of Faecal Escherichia coli From Healthy Volunteers From Eight Developing Countries. J. Antimicrob. Chemother. 54 (5), 952–955. doi: 10.1093/jac/dkh448
Palma, N., Pons, M. J., Gomes, C., Mateu, J., Riveros, M., García, W., et al. (2017). Resistance to Quinolones, Cephalosporins and Macrolides in Escherichia coli Causing Bacteraemia in Peruvian Children. J. Glob. Antimicrob. Resist. 11, 28–33. doi: 10.1016/j.jgar.2017.06.011
Petty, N. K., Ben, Z. N. L., Stanton-Cook, M., Skippington, E., Totsika, M., Forde, B. M., et al. (2014). Global Dissemination of a Multidrug Resistant Escherichia coli Clone. Proc. Natl. Acad. Sci. U. S. A. 111 (15), 5694–5699. doi: 10.1073/pnas.1322678111
Ravensdale, J. T., Xian, D. T. W., Wei, C. M., Lv, Q., Wen, X., Guo, J., et al. (2018). PCR Screening of Antimicrobial Resistance Genes in Faecal Samples From Australian and Chinese Children. J. Glob. Antimicrob. Resist. 14, 178–181. doi: 10.1016/j.jgar.2018.03.003
Sahoo, K. C., Tamhankar, A. J., Sahoo, S., Sahu, P. S., Klintz, S. R., Lundborg, C. S. (2012). Geographical Variation in Antibiotic-Resistant Escherichia coli Isolates From Stool, Cow-Dung and Drinking Water. Int. J. Environ. Res. Public Health 9 (3), 746–759. doi: 10.3390/ijerph9030746
Salyers, A. A., Gupta, A., Wang, Y. (2004). Human Intestinal Bacteria as Reservoirs for Antibiotic Resistance Genes. Trends Microbiol. 12 (9), 412–416. doi: 10.1016/j.tim.2004.07.004
Seemann, T. (2014). Prokka: Rapid Prokaryotic Genome Annotation. Bioinformatics 30 (14), 2068–2069. doi: 10.1093/bioinformatics/btu153
Shakya, P., Barrett, P., Diwan, V., Marothi, Y., Shah, H., Chhari, N., et al. (2013). Antibiotic Resistance Among Escherichia coli Isolates From Stool Samples of Children Aged 3 to 14 Years From Ujjain, India. BMC Infect. Dis. 13, 477. doi: 10.1186/1471-2334-13-477
Stapleton, A. E., Wagenlehner, F. M. E., Mulgirigama, A., Twynholm, M. (2020). Escherichia coli Resistance to Fluoroquinolones in Community-Acquired Uncomplicated Urinary Tract Infection in Women: A Systematic Review. Antimicrob. Agents Chemother. 64 (10), e00862-20. doi: 10.1128/AAC.00862-20
Sullivan, M. J., Petty, N. K., Beatson, S. A. (2011). Easyfig: A Genome Comparison Visualizer. Bioinformatics 27 (7), 1009–1010. doi: 10.1093/bioinformatics/btr039
Tchesnokova, V. L., Rechkina, E., Larson, L., Ferrier, K., Weaver, J. L., Schroeder, D. W., et al. (2019). Rapid and Extensive Expansion in the United States of a New Multidrug-Resistant Escherichia coli Clonal Group, Sequence Type 1193. Clin. Infect. Dis. 68 (2), 334–337. doi: 10.1093/cid/ciy525
Valenza, G., Werner, M., Eisenberger, D., Nickel, S., Lehner-Reindl, V., Höller, C., et al. (2019). First Report of the New Emerging Global Clone ST1193 Among Clinical Isolates of Extended-Spectrum β-Lactamase (ESBL)-Producing Escherichia coli From Germany. J. Glob. Antimicrob. Resist. 17, 305–308. doi: 10.1016/j.jgar.2019.01.014
Zhao, L., Zhang, J., Zheng, B., Wei, Z., Shen, P., Li, S., et al. (2015). Molecular Epidemiology and Genetic Diversity of Fluoroquinolone-Resistant Escherichia coli Isolates From Patients With Community-Onset Infections in 30 Chinese County Hospitals. J. Clin. Microbiol. 53 (3), 766–770. doi: 10.1128/JCM.02594-14
Zhong, Y. M., Liu, W. E., Meng, Q., Li, Y. (2019). Escherichia coli O25b-ST131 and O16-ST131 Causing Urinary Tract Infection in Women in Changsha, China: Molecular Epidemiology and Clinical Characteristics. Infect. Drug Resist. 12, 2693–2702. doi: 10.2147/IDR.S212658
Keywords: ESBL, Escherichia coli, fluoroquinolone, multidrug resistance, whole genome sequencing
Citation: Zhao Q, Shen Y, Chen G, Luo Y, Cui S and Tian Y (2021) Prevalence and Molecular Characterization of Fluoroquinolone-Resistant Escherichia coli in Healthy Children. Front. Cell. Infect. Microbiol. 11:743390. doi: 10.3389/fcimb.2021.743390
Received: 24 July 2021; Accepted: 25 November 2021;
Published: 13 December 2021.
Edited by:
Guo-bao Tian, Sun Yat-sen University, ChinaReviewed by:
Jian Sun, South China Agricultural University, ChinaCopyright © 2021 Zhao, Shen, Chen, Luo, Cui and Tian. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yaping Tian, dGlhbnlwQDMwMWhvc3BpdGFsLmNvbS5jbg==; Shenghui Cui, Y3Vpc2hlbmdodWlAYWxpeXVuLmNvbQ==
†These authors have contributed equally to this work and share first authorship
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.