- 1Department of Disinfection and Infection Control, Chinese People’s Liberation Army (PLA) Center for Disease Control and Prevention, Beijing, China
- 2School of Mathematics and Statistics, Shaanxi Normal University, Xi’an, China
Candida auris (C. auris) is an emerging fungus associated with high morbidity. It has a unique transmission ability and is often resistant to multiple drugs. In this study, we evaluated the ability of different machine learning models to classify the drug resistance and predicted and ranked the drug resistance mutations of C. auris. Two C. auris strains were obtained. Combined with other 356 strains collected from the European Bioinformatics Institute (EBI) databases, the whole genome sequencing (WGS) data were analyzed by bioinformatics. Machine learning classifiers were used to build drug resistance models, which were evaluated and compared by various evaluation methods based on AUC value. Briefly, two strains were assigned to Clade III in the phylogenetic tree, which was consistent with previous studies; nevertheless, the phylogenetic tree was not completely consistent with the conclusion of clustering according to the geographical location discovered earlier. The clustering results of C. auris were related to its drug resistance. The resistance genes of C. auris were not under additional strong selection pressure, and the performance of different models varied greatly for different drugs. For drugs such as azoles and echinocandins, the models performed relatively well. In addition, two machine learning algorithms, based on the balanced test and imbalanced test, were designed and evaluated; for most drugs, the evaluation results on the balanced test set were better than on the imbalanced test set. The mutations strongly be associated with drug resistance of C. auris were predicted and ranked by Recursive Feature Elimination with Cross-Validation (RFECV) combined with a machine learning classifier. In addition to known drug resistance mutations, some new resistance mutations were predicted, such as Y501H and I466M mutation in the ERG11 gene and R278H mutation in the ERG10 gene, which may be associated with fluconazole (FCZ), micafungin (MCF), and amphotericin B (AmB) resistance, respectively; these mutations were in the “hot spot” regions of the ergosterol pathway. To sum up, this study suggested that machine learning classifiers are a useful and cost-effective method to identify fungal drug resistance-related mutations, which is of great significance for the research on the resistance mechanism of C. auris.
Introduction
Candida auris (C. auris) is an emerging fungal pathogen first isolated from the external ear canal of a 70-year-old female inpatient in Tokyo hospital (Satoh et al., 2009). C. auris can persist for weeks in a nosocomial environment, and survive high-end disinfections, thus presenting a serious global health threat (Chaabane et al., 2019; Du et al., 2020). To date, C. auris outbreak has been reported in more than 30 countries worldwide (Rhodes et al., 2018; Tian et al., 2018; Escandon et al., 2019; Rhodes and Fisher, 2019). C. auris, also known as “super fungus”, is a multidrug-resistant species associated with high mortality (Wang et al., 2018).
So far, four specific clades of C. auris have been identified by phylogenetic analysis based on whole-genome sequencing (WGS): South Asia (Clade I), East Asia (Clade II), South Africa (Clade III), and South America (Clade IV). A potential fifth clade of Iranian origin was described by few studies (Chow et al., 2019; Di Pilato et al., 2021). All clades are characterized by distinct single nucleotide polymorphisms (SNPs), highlighting this pathogen’s independent and worldwide emergence (Lockhart et al., 2017). Except for Clade II, the other three clusters have been associated with an outbreak of invasive infection and multiple resistance. Clade II is predominantly an ear canal infection, and presents either single fluconazole resistance or susceptible (Kwon et al., 2019; Welsh et al., 2019).
Clinically, invasive fungal infections are usually treated with three classes of antifungal agents: echinocandins, azoles, and polyenes (ElBaradei, 2020). Fluconazole (FCZ) resistance is the most common. Resistance to other azoles like voriconazole (VCZ), itraconazole (ICZ), and posaconazole (PZ) might vary (Montoya et al., 2019; ElBaradei, 2020).
Ergosterol is a key component of the fungal cell membrane. In Candida, ergosterol is mediated by lanosterol 14-alpha-demethylase (ERG11), which is involved in an important step in the biosynthesis of ergosterol. Antifungal agents effectively inhibit ergosterol biosynthesis by inhibiting the enzyme’s function, thereby compromising membrane integrity (Sanglard et al., 1998). Different mechanisms, including mutations in the ERG11 gene, overexpression of the ATP-binding Cassette (ABC) exogenous pump transporter, which is encoded by the CDR1 gene, and duplication and overexpression of the ERG11 gene, contribute to the reduction of the sensitivity of C. auris to azole drugs (Puri et al., 1999; de Micheli et al., 2002; Coste et al., 2004; Cannon et al., 2009; Noel, 2012; Spampinato and Leonardi, 2013; Medici and Del Poeta, 2015; Nami et al., 2019; Bing et al., 2020). Point mutations in the ERG11 gene, associated with FCZ resistance in Candida albicans, are also one of the mechanisms of FCZ resistance in C. auris. Point mutations in ERG11 can reduce the azole sensitivity of Candida, particularly in the “hot spots” located between 105-165, 266-287, and 405-488 (Lamb et al., 1995; Sanglard et al., 1998; Mellado et al., 2004; Vandeputte et al., 2012). Moreover, Lockhart et al. described three major mutations in ERG11 that influence FCZ resistance, namely, F126T, Y132F, and K143R (Lockhart et al., 2017). Furthermore, Healey et al. found that Y132F mutations significantly reduce the sensitivity of C. auris to azole drugs. Also, it has been reported that these mutations are associated with geographic cues, with mutations leading to Y132F and K143R associated with isolates belonging to South Asian and South American groups (Healey et al., 2018). In addition, Rybak et al. reported new mutations on the zinc-cluster transcription factor-encoding gene (TAC1B) associated with FCZ resistance (Rybak et al., 2020). This study showed that mutations on TAC1B could be produced rapidly in vitro after exposure to FCZ. Most FCZ-resistant isolates have many drug-related TAC1B mutations in a specific global lineage or group of C. auris, and the identification of new resistance determinants has significantly increased the understanding of clinical antifungal resistance in C. auris (Rybak et al., 2020).
C. auris resistance to echinocandins is less common. Caspofungin (CSF), micafungin (MCF), and anidulafungin (AND) are often recommended as first-line treatments for candidemia (ElBaradei, 2020). In vitro studies have demonstrated that CSF and AND have a certain inhibitory effect on the growth of C. auris (Dudiuk et al., 2019). Interestingly, one study reported that among all echinocandins, micafungin has the highest inhibitory effect against C. auris (Kordalewska et al., 2018).
Echinocandins inhibit the 1, 3-beta-D-glucan synthetase required for cell wall synthesis, encoded by the genes FKS1 and FKS2. Several mutations (“hot spots 1 and 2”) in the FKS1 and FKS2 genes in Candida albicans and other non-auris Candida species have been associated with the echinocandins resistance. In the FKS1 gene of C. albicans, these “hot spots” lie between the amino acids 641-649 and 1,345-1,365 (Park et al., 2005). Resistance to the echinocandins involves mutations in the FKS1 gene, with changes in the hot spot 1 region leading to amino acid substitution from serine to proline at 639 (S639P) (Biagi et al., 2019). Moreover, a multicenter study in India reported another mutation in the same position 639 of the FKS1 gene, involving a change from serine to phenylalanine (S639F or S639Y) (Chowdhary et al., 2018). Sharma et al. also found FKS2 in a single copy of the C. auris genome; yet, no mutation associated with echinocandins resistance has been found in this gene (Sharma et al., 2016; Chaabane et al., 2019).
Among polyenes, C. auris and C. lusitaniae have shown high resistance to amphotericin B (AmB). However, the molecular mechanism of polyene drug resistance is not clear (ElBaradei, 2020) and more research may be needed to reveal how non-synonymous mutations promote resistance to AmB in C. auris (Escandon et al., 2019). Kordalewska and Perlin suggested that resistance to AmB is regulated at the transcriptional level rather than mutations (Kordalewska and Perlin, 2019).
Predictive models based on machine learning can explore multiple associations between genetic variations. Machine learning is the scientific discipline that focuses on how computers learn from data (Deo, 2015). As an essential component in artificial intelligence (AI), it has been integrated into many fields, such as data generation, analytics and knowledge mining (Handelman et al., 2018; Patel et al., 2020). Several previous studies have used machine learning algorithms to predict microbial resistance. For example, Zhang et al. collected 161 strains of Mycobacterium tuberculosis (MTB) from China and used logistic regression and random forest to find and predict new genes associated with drug resistance of seven drugs (Zhang et al., 2013). Furthermore, using a more geographically diverse data set, Farhat et al. studied the performance of the random forest algorithm based 1,397 isolates (Farhat et al., 2016). Her et al. proposed a pan-genome-based method to characterize antibiotic-resistant microbial strains; the method was tested on Escherichia coli. The drug resistance gene was predicted by identifying the core and accessory gene clusters on Escherichia coli pan-genomic (Her and Wu, 2018). In addition, Yang et al. considered 1,839 bacterial isolates from the UK and compared the performance of more machine learning classifiers, including Logistic Regression, Support Vector Classifier (based on linear and Gaussian kernel functions), product-of-marginals model (PM), Random Forest, gradient tree boosting (GBT), and Adaboost. Finally, mutations associated with drug resistance of MTB ranked and were predicted (Yang et al., 2018; Kouchaki et al., 2019). However, most of the microbes studied were bacteria, while only a few studies applied this method to study fungi. Moreover, currently, there are no studies on the classification of fungi drug resistance and the evaluation of drug resistance mutations by mathematical models.
In this study, we collected C. auris isolates from different countries or regions, analyzed their whole genome sequencing data, constructed the phylogenetic relationship, evaluated the ability of different machine learning models to classify the drug resistance, and predicted and ranked the drug resistance mutations of C. auris.
Materials and Methods
WGS and Pre-processing
As of April 2020, the whole genome sequencing (WGS) data of C. auris published by the European Bioinformatics Institute (EBI, https://www.ebi.ac.uk/) has 796 isolates in total. Among them, 356 strains have undergone antifungal susceptibility testing. According to these results, resistant or susceptible strains were determined according to the Clinical and Laboratory Standards Institute (CLSI) guidelines.
In this study, WGS data of 356 strains containing drug resistance information on the EBI website were collected, and two strains named C1921 and C1922, which showed FCZ resistance from the Chinese PLA Center for Disease Control & Prevention were combined (Chen et al., 2018). This study involved WGS data of 358 C. auris strains (see Supplementary Materials File), all of which were sequenced using Illumina sequencing technology platform; the sequencing data obtained were double-ended WGS data in FASTQ data format. The drug resistance of 358 strains above was collected, including fluconazole, itraconazole, voriconazole, posaconazole, amphotericin B, micafungin, anifenqine and caspofunqine. The statistics of drug resistance of the strains are shown in Table 1.
WGS data of 358 C. auris strains were collected and analyzed using the following steps: FastQC (http://www.bioinformatics.babraham.ac.uk/projects/fastqc/) checked the data quality of each strain’s sequence and divided the data according to different types of sequencing adapters for quality control [Trimmomatic (Bolger et al., 2014)]. All data were aligned and sorted with the reference strain B8441 using Bwa-0.7.17 (Munoz et al., 2018). Duplicates in the file were marked using MarkDuplicates module in GATK (DePristo et al., 2011) v4.1.4.1, and were ignored during the mutation detection. In BaseRecalibrator, 246,258 sites were jointly detected by GATK HaplotypeCaller and Bcftools (Li et al., 2009) mpileup, which were finally used as SNP reference sets.
The recalibration of base mass values mainly involved two steps: GATK BaseRecalibrator and GATK ApplyBQSR. Then, the mutation detection was performed by GATK HaplotypeCaller. Finally, VCFtools (Danecek et al., 2011) software was used to filter the samples and detection sites, respectively. Two samples with high deletion rates (max-missing ≥ 50%) (SRR10461133 and SRR10461145) were removed from the filtering of the samples. The sites with minQ ≤ 30, max-missing ≥ 0.5, mac ≤ 3, and minDP ≤ 3 were deleted, respectively, using VCFtools, and the number of sites after filtering was 229,262. The filtered files were annotated using SNPEff (Cingolani et al., 2012), and the annotated files were used for phylogenetic analysis and machine learning resistance analysis. Three antifungals (FCZ, MCF and AmB) and point mutations (Y132F, K143R and F126L in ERG11, S639Y/S639F and S639P in FKS1) was also depicted in the phylogenetic NJ tree. This process is shown in Figure S1 and Table S1.
Selection and Extraction of Gene Sets
A total of 229,262 SNP mutation sites were found in 358 C. auris isolates. Candidate genes that may have a strong correlation with drug resistance of C. auris in previous studies were selected; this was performed in order to reduce its dimension, facilitate machine learning classification, eliminate redundant sites, and improve the accuracy of the analysis for the complex dimension. In addition, only missense mutations were extracted for further analysis since they accounted for only a small part of the original mutations, but affected the type of amino acids, i.e., the function of proteins.
Three candidate gene sets were selected in this study (Lockhart et al., 2017; Munoz et al., 2018; Chaabane et al., 2019; Rybak et al., 2020) (Table S2). F3 set included genes that were previously reported to be associated with drug resistance and may contain determinants of drug resistance information in C. auris; F2 set was a list of seven genes specific in C. auris, which have been associated with drug resistance in C. albicans, but are highly conserved in C. auris (Munoz et al., 2018). F1 set combined the F2 and F3 genes. All the missense mutations were extracted in the three gene sets and filtered. The samples and sites with too many missing values for each set were deleted, and the dimension of the data set after processing the missing values (samples × mutations) was respectively: F1: 350 x 579; F2: 353 x 202; F3: 352 x 377.
Machine Learning Algorithms
Two algorithms were designed by using Python 3.8.4 (https://www.python.org/downloads/): the classifier on the balanced test set and on the imbalanced test set (Figures S2, S3). The F1, F2, and F3 sets were used as the classification feature sets, and the drug resistance of C. auris was taken as the classification target. Ten machine learning classifiers (Table S3), Logistic Regression (LR), Support Vector Classifier (SVC, including SVC RBF and SVC linear), K-Nearest Neighbors (KNN), Decision Tree (DT), Ensemble Learning (including RandomForest, AdaBoost and GradientBoosting), and Naive Bayes (including BernoulliNB and GaussianNB) were used to build the model (Breiman, 1996; Breiman, 2001) by using Python 3.8.4. For AdaBoost, the Decision Tree Classifier was the base estimator whose number was 200 and the max depth was 1. There were 100 trees set in the random forest classifier. The neighbor was 5 (the value of K) for the KNN classifier. In both algorithms, principal component analysis (PCA) was used to reduce dimensionality based on retaining 99% of the original information. The number of principal components after dimension reduction with PCA method when 99% of the variance is retained is in supplementary material Table S4. Upsampling and downsampling were mainly adopted to balance the data set and repeated sampling 100 times. Downsampling means, for a dataset from the majority classification, creating a new subset with the same sample number as the minority classification from the original set by random sampling. Upsampling means, for a dataset from the minority classification, creating a new dataset with the same sample number as the majority classification from the original set by random sampling. The data were divided into test set and training set according to 5-fold cross-validation (5-CV), which accounted for 20% and 80%, respectively. The model parameters were adjusted on a training set, and the model was retrained using 5-CV. Finally, the model was evaluated on the test set. The area under the ROC (the Receiver operating characteristic curve) curve (AUC), was used as evaluation standard of a model’s performance. A classifier with a larger AUC (closer to 1.0) performed better.
Recursive Feature Elimination With Cross-Validation
The Recursive Feature Elimination with Cross-validation (RFECV) functions in Python’s Scikit-Learn established in mutation sequencing were based on the F1 data set, which contained all candidate genes selected before machine learning modeling. All features were standardized before ranking, and the training model was the classifier above. The standardized method used was StandardScaler() function in Python. The number of features discarded in each iteration was 1, indicating elimination one by one, and the model was built repeatedly through 5-CV.
Results
Phylogenetic Analysis of Candida auris
Phylogenetic NJ-tree was constructed using MEGA-X (Kumar et al., 2018), and boostrap test was repeated 500 times. Then, the phylogenetic tree was annotated using the iTOL online tool (https://itol.embl.de/). The phylogenetic NJ tree was divided into four clades starting from the root (Figure 1): Clade I (orange), Clade II (blue), Clade III (purple), and Clade IV (green), which was consistent with the conclusions reported in previous literatures (Lockhart et al., 2017). The clustering results are shown in Table S5. However, strain B16401 (SRR10852068, Kenya) was assigned to Clade I in this study; in a previous study, strain B16401 was assigned to Clade III (Chow et al., 2020). In the NJ tree, C1921 and C1922 from our laboratory were in Clade III, which was consistent with the phylogenetic tree constructed using Internal Transcribed Spacer (ITS) and D1/D2 Large Ribosomal Subunit Region previously (Chen et al., 2018). In addition, the mutations associated with azoles and echinocandins resistance detected were consistent with the previous conclusions (Chow et al., 2020). According to these results, F126L mutation in lanosterol 14-alpha-demethylase ERG11 occurred in C1921 and C1922 strains, which is closely related to their FCZ resistance observed in clinical practice. It was also shown that the phylogenetic tree constructed by the drug-resistant gene set F3 was very similar to the phylogenetic tree constructed by the WGS of C. auris, and there was no difference in the clustering results of the strains (Figure S4), indicating that the evolution of C. auris resistance genes was consistent with the overall evolution of the strains (at the level of the whole genome). It was speculated that the resistance genes of C. auris were not under additional strong selection pressure, which may be related to the clinical use of drugs.
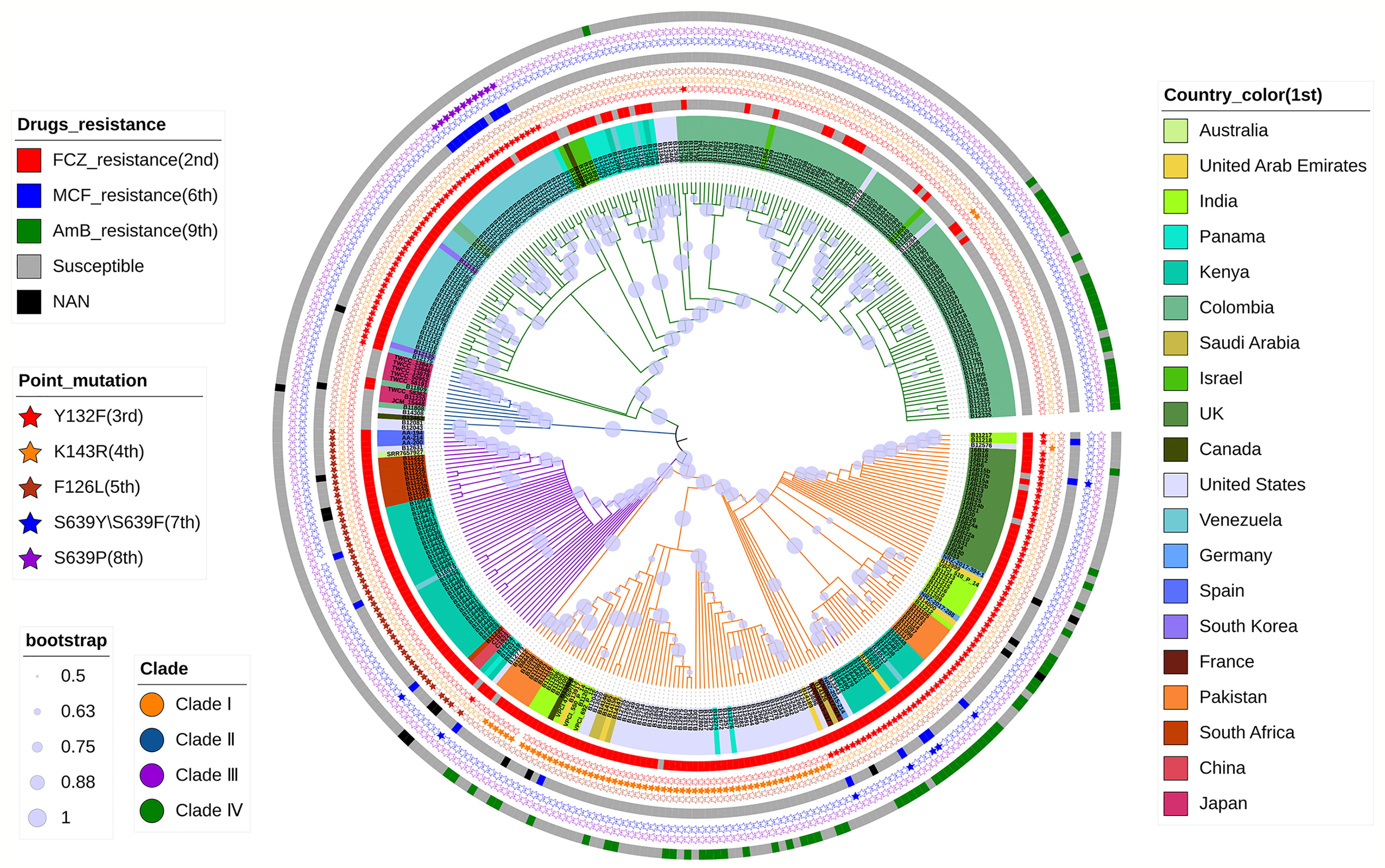
Figure 1 Phylogenetic NJ tree based on WGS of C. auris. The tree describes the phylogeny of 356 strains of C. auris from different regions, divided into four clades. The 1st to 9th indicates a concentric circle from the inner-most to the outer-most, respectively. It also shows the correlation between the resistance of these strains to FCZ, AmB, and MCF and the reported point mutations with Y132F, K143R, and F126L in ERG11, S639Y/S639F, and S639P in FKS1.
Evaluation of Classification Models
The performances of machine learning classifiers, constructed by the two algorithms described above on F1, F2, and F3, were evaluated and compared by several evaluation methods. The best model for each set and drug was listed in Table 2. For most drugs, the evaluation results on the balanced test set were better than on the imbalanced test set. The classifiers established using two algorithms achieved better results for azoles, like FCZ, ICZ and VCZ, since their AUC values were above 0.9. However, compared with other drug models, the evaluation results of AmB needed to be improved; we speculated that this might be closely related to the selection of candidate genes. For well-studied drugs (azoles and echinocandins), the selected three gene sets contained more information about determinants associated with drug resistance, but there were few determinants of polyenes resistance.
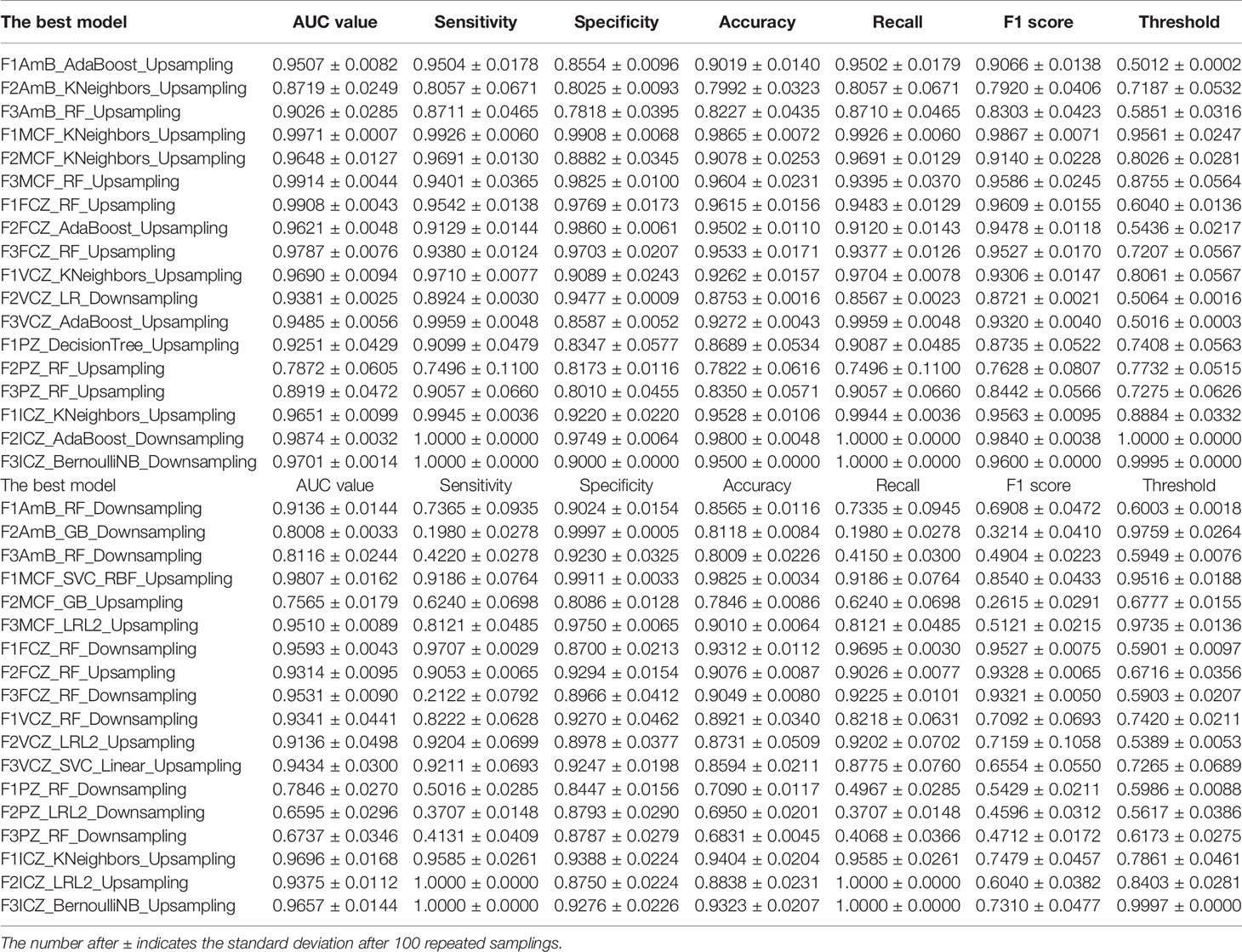
Table 2 Model evaluation results under two different algorithms: on the balanced test set (the upper table) and on the imbalanced test set (the lower table).
The model with the highest AUC value was extracted and compared (Figure 2 and Table S6). Random forest, logistic regression, and K-nearest neighbors ranked in the top and for several times. Under two algorithms, the classifier models performed well on F1 for all drugs, of which the AUC values were above 0.85. While on F2 and F3, classifiers performed well only on some drugs; for example, models performed well on F2 for azoles like FCZ, ICZ, and VCZ, and they performed well on F3 for MCF, but they all had poor classification effect on AmB and PZ. It may be that the correlation between the three sets and classification targets was not very strong, and the information collected for these two drugs was insufficient.
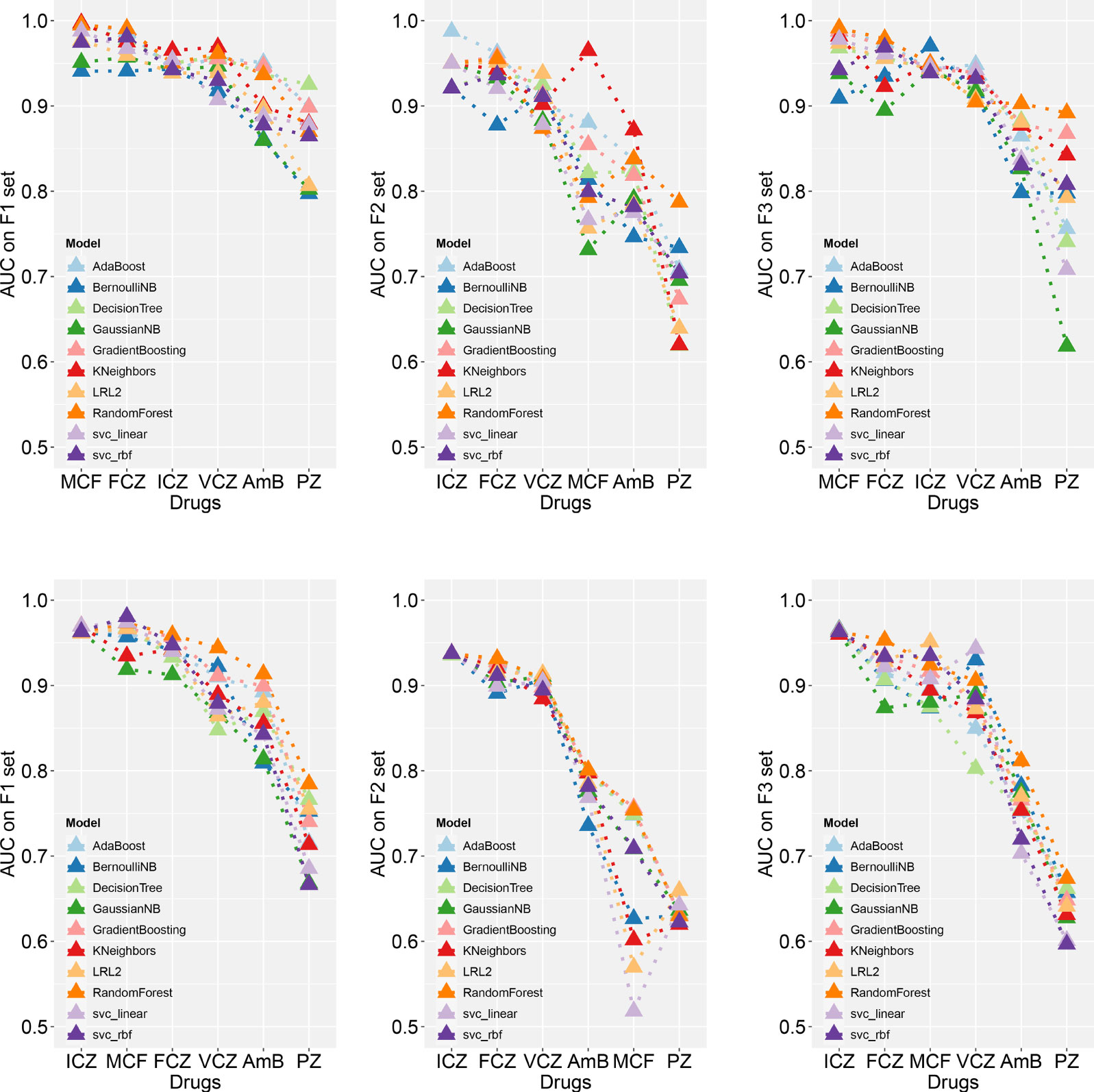
Figure 2 Comparison of the best AUC values using different machine learning classifiers.The best models on the balanced (the upper three) and imbalanced (the lower three) test set are shown, respectively. Please see supplementary materials Table S6 for detailed evaluation results.
Mutation Ranking
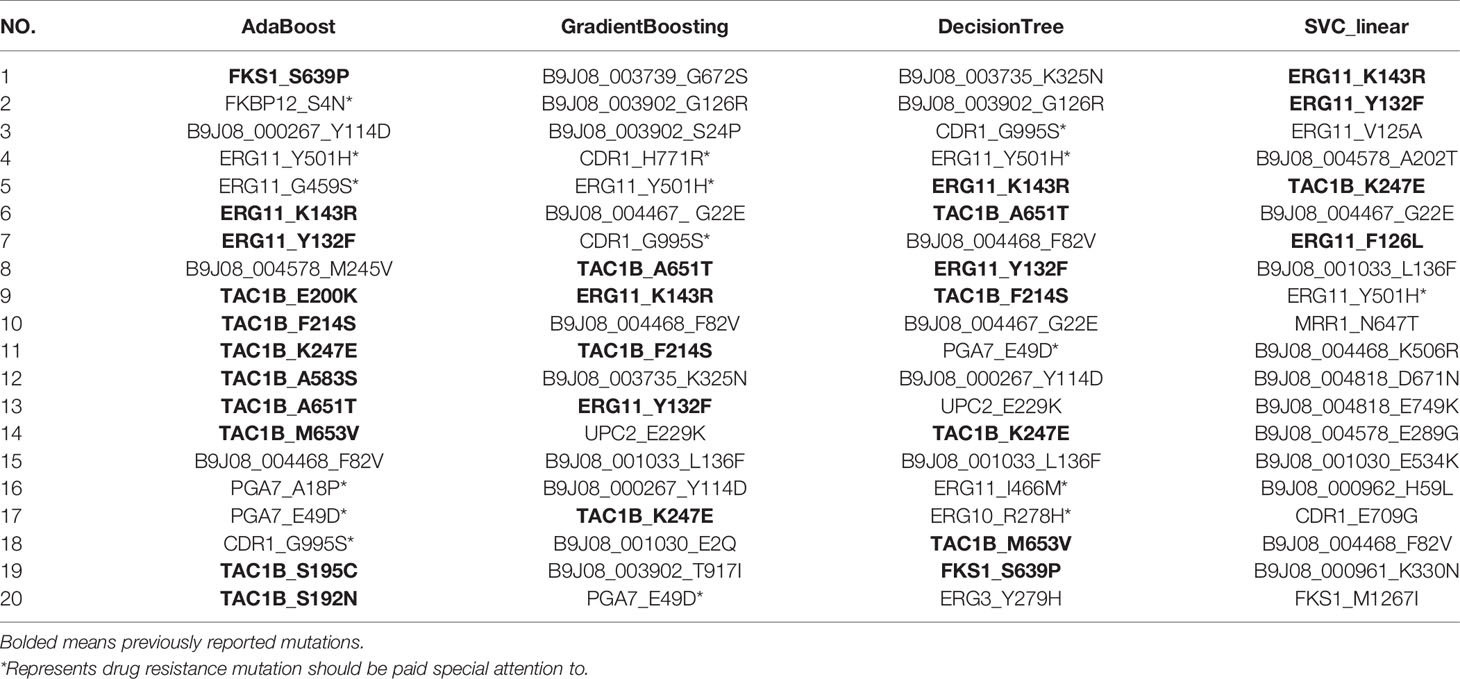
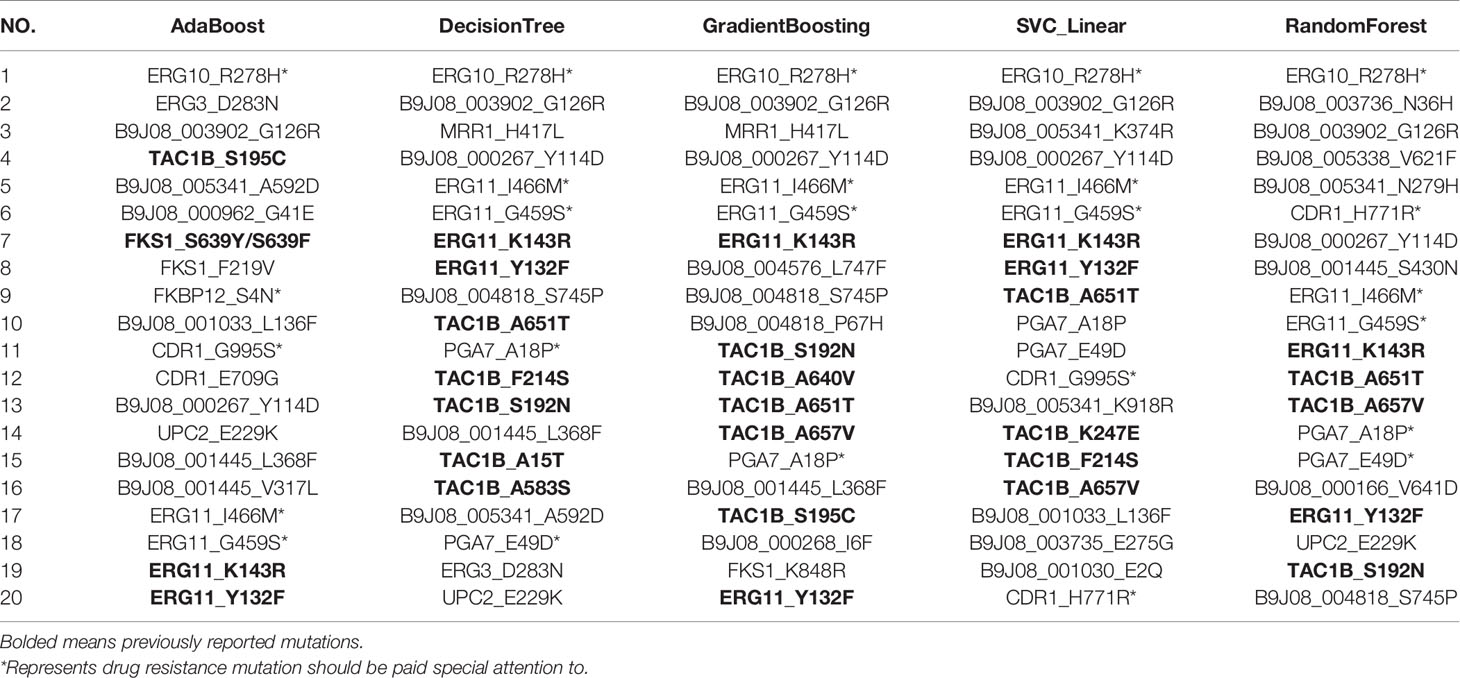
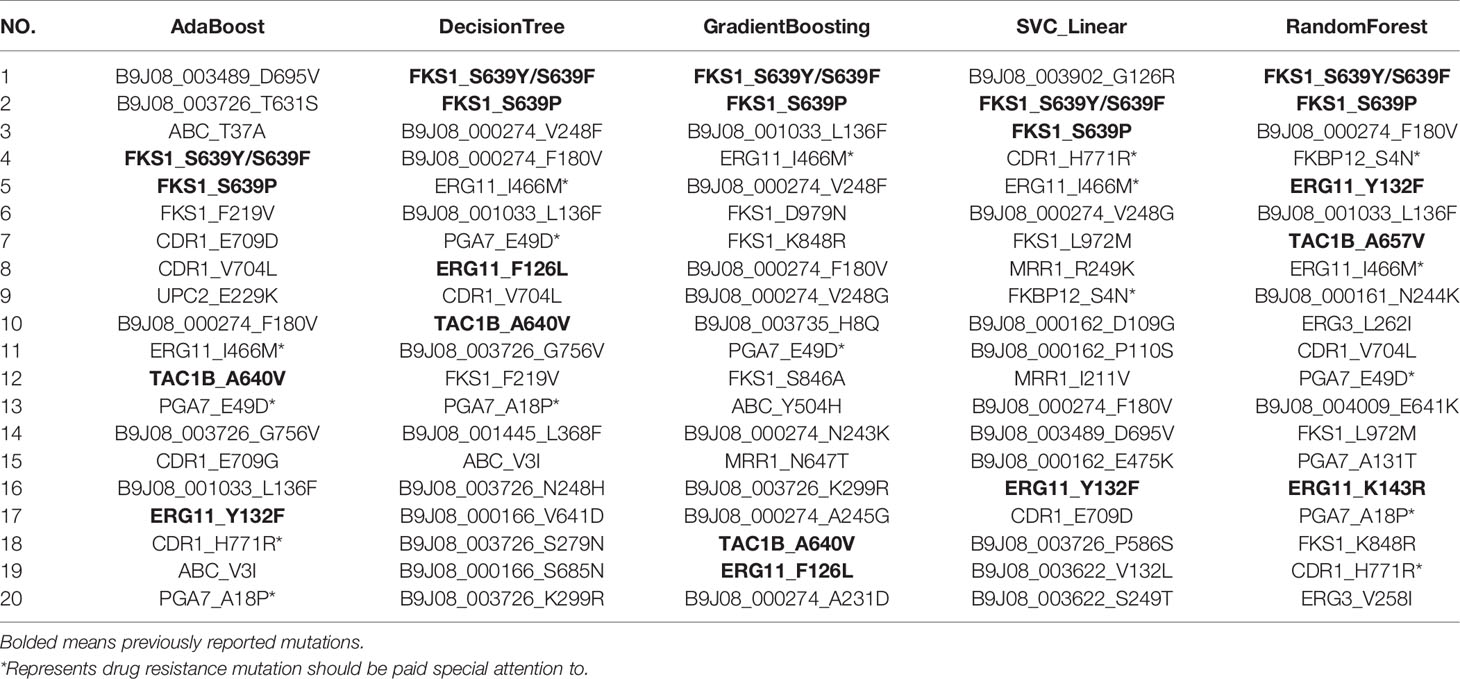
Using RFECV, three antifungal drugs, including FCZ, MCF, and AmB, were ranked and predicted, respectively. The mutation ranking results are shown in Tables 3–5. Previously reported mutations (bolded in the table), such as Y132F, K143R, and F126L on the ERG11, mutations on the TAC1B (Rybak et al., 2020), and S639Y/S639F and S639P on the FKS1 gene, were detected and listed as important mutations. In addition, several novel mutations were detected (marked by an asterisk). Particularly, mutations in the “hot spot” regions of the ergosterol pathway, such as I466M, G459S, and Y501H in ERG11, and R278H in ERG10, were detected. These mutations were frequently and highly ranked mutations. FKBP12 has been reported to be associated with multiple resistance in Candida spp., and the S4N mutation was detected in this gene. Two frequently occurring mutations, H771R and G995S, were identified in CDR1, the gene encoding the ATP-Binding Cassette efflux pump transporter. Two high-frequency mutations, E49D and A18P, were also found in a specific gene (PGA7, C. albicans homolog) of C. Auris. These mutations should be paid special attention to in the following research.
Discussion
C. auris strains C1921 and C1922 sequenced in our laboratory were classified into Clade III from the phylogenetic tree, which was consistent with the tree constructed using Internal Transcribed Spacer and D1/D2 Large Ribosomal Subunit Region in the previous study (Chen et al., 2018). Previous studies classified C. auris into four clades: South Asia (Clade I), East Asia (Clade II), South Africa (Clade III), and South America (Clade IV) (potential fifth clade of Iranian origin), and it was emphasized that each clade has a great relationship with geographical location. The clustering results from the phylogenetic tree in this study illustrated that these strains could be divided into four clades, but the conclusion of clustering according to geographical location was not very prominent.
Machine learning technology has great potential in classifying drug resistance of strains with WGS data and analyzing high-dimensional data sets, which is very important for predicting mutations associated with drug resistance. Our model evaluation results illustrated that the machine learning classifiers performed quite different when testing different drugs. The classifier model showed excellent performance for azoles and echinocandins such as FCZ, ICZ, VCZ, and MCF, but not for others like AmB and PZ. It was speculated that there might be more information about determinants associated with azoles and echinocandins resistance but less for AmB and PZ in the three sets. This was directly indicative of the fact that the correlation between feature sets and classification targets was stronger for azoles and echinocandins, but was weaker for the two drugs. In addition, there were some deficiencies in model optimization so that only several models were optimized in the process of constructing classifier models and adjusting parameters. Therefore, optimizing models through a large number of experiments and tests should be performed in future work in order to achieve better performance.
In this study, RFECV combined with a machine learning classifier was used to predict and rank the mutations of C. auris related to antifungal drug resistance. In the RFECV process, different ranked mutation results were obtained by combining different classifiers. Overall, the results indicated that the RFECV method could not only rank several known mutations as important, especially for well-studied drugs but also predict some new important mutations on the genes closely related to drug resistance. Some of the predicted mutations were known to be important resistance mutations, which to some extent demonstrated the validity of our classification model. The model could obtain more reliable conclusions for well-studied drugs, such as azoles and echinocandins, while for amphotericin B, the model also predicted some resistance-related mutations. Based on these results, further research and verification are needed on the specific mutations and drug resistance mechanisms of C. auris.
Machine learning models can improve the prediction of important genetic mutation sites related to drug resistance in fungi, particularly beneficially for less-studied drugs. The amount of test data, or sample size, is one of the keys to the performance of machine learning methods. We speculate that 500 to 1,000 fungal samples may get satisfactory results according to previous studies. Random forest, logistic regression, and K-nearest neighbors classifier performed relatively better in this study. While in another study, PM (product-of-marginal model) and SVC-RBF ranked as the top two best-performing classifiers on MTB (Yang et al., 2018). The most common issues in machine learning lie around overfitting, underfitting, noisy data and inappropriate validation. Hence, considering all available variants and allowing machine learning methods to reduce the dimension can improve the performance. In the future, it is necessary to conduct systematic verification and related functional studies on these mutations.
This study may help to analyze the drug resistance mechanism of C. auris, and provide a scientific basis for developing prevention and control strategies against drug resistance and the search for possible new drug targets.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Author Contributions
LH and XC conceived the project. JZ and FC collected the samples. DL, YW, and WH conducted the NGS. YW and DL conducted the RNA analysis, analyzed data and wrote manuscript. LH evaluated all results. All authors contributed to the article and approved the submitted version.
Funding
This study was supported by Scientific Research Project of National Natural Science Foundation of China (81971914, 81772163, 82172293), the State Key Program of National Natural Science Foundation of China (12031016), Project of Natural Science Foundation of Liaoning Province (20180550255) and Fundamental Research Funds for the Central Universities (GK201901008).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We thank all the subjects who participated in this study.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcimb.2021.742062/full#supplementary-material
References
Biagi, M. J., Wiederhold, N. P., Gibas, C., Wickes, B. L., Lozano, V., Bleasdale, S. C., et al. (2019). Development of High-Level Echinocandin Resistance in a Patient With Recurrent Candida Auris Candidemia Secondary to Chronic Candiduria. Open Forum Infect. Dis. 6 (7), ofz262. doi: 10.1093/ofid/ofz262
Bing, J., Hu, T., Zheng, Q., Munoz, J. F., Cuomo, C. A., Huang, G. (2020). Experimental Evolution Identifies Adaptive Aneuploidy as a Mechanism of Fluconazole Resistance in Candida Auris. Antimicrob. Agents Chemother. 65 (1). doi: 10.1128/AAC.01466-20
Bolger, A. M., Lohse, M., Usadel, B. (2014). Trimmomatic: A Flexible Trimmer for Illumina Sequence Data. Bioinformatics 30 (15), 2114–2120. doi: 10.1093/bioinformatics/btu170
Cannon, R. D., Lamping, E., Holmes, A. R., Niimi, K., Baret, P. V., Keniya, M. V., et al. (2009). Efflux-Mediated Antifungal Drug Resistance. Clin. Microbiol. Rev. 22 (2), 291–321. doi: 10.1128/CMR.00051-08
Chaabane, F., Graf, A., Jequier, L., Coste, A. T. (2019). Review on Antifungal Resistance Mechanisms in the Emerging Pathogen Candida Auris. Front. Microbiol. 10, 2788. doi: 10.3389/fmicb.2019.02788
Chen, Y., Zhao, J., Han, L., Qi, L., Fan, W., Liu, J., et al. (2018). Emergency of Fungemia Cases Caused by Fluconazole-Resistant Candida Auris in Beijing, China. J. Infect. 77 (6), 561–571. doi: 10.1016/j.jinf.2018.09.002
Chow, N. A., de Groot, T., Badali, H., Abastabar, M., Chiller, T. M., Meis, J. F. (2019). Potential Fifth Clade of Candida Auris, Ira. Emerg. Infect. Dis. 25 (9), 1780–1781. doi: 10.3201/eid2509.190686
Chow, N. A., Munoz, J. F., Gade, L., Berkow, E. L., Li, X., Welsh, R. M., et al. (2020). Tracing the Evolutionary History and Global Expansion of Candida Auris Using Population Genomic Analyses. mBio 11 (2). doi: 10.1128/mBio.03364-19
Chowdhary, A., Prakash, A., Sharma, C., Kordalewska, M., Kumar, A., Sarma, S., et al. (2018). A Multicentre Study of Antifungal Susceptibility Patterns Among 350 Candida Auris Isolate-17) in India: Role of the ERG11 and FKS1 Genes in Azole and Echinocandin Resistance. J. Antimicrob. Chemother. 73 (4), 891–899. doi: 10.1093/jac/dkx480
Cingolani, P., Platts, A., Wang le, L., Coon, M., Nguyen, T., Wang, L., et al. (2012). A Program for Annotating and Predicting the Effects of Single Nucleotide Polymorphisms, SnpEff: SNPs in the Genome of Drosophila Melanogaster Strain W1118; Iso-2; Iso-3. Fly (Austin) 6 (2), 80–92. doi: 10.4161/fly.19695
Coste, A. T., Karababa, M., Ischer, F., Bille, J., Sanglard, D. (2004). TAC1, Transcriptional Activator of CDR Genes, Is a New Transcription Factor Involved in the Regulation of Candida Albicans ABC Transporters CDR1 and CDR2. Eukaryot Cell 3 (6), 1639–1652. doi: 10.1128/EC.3.6.1639-1652.2004
Danecek, P., Auton, A., Abecasis, G., Albers, C. A., Banks, E., DePristo, M. A., et al. (2011). The Variant Call Format and VCFtools. Bioinformatics 27 (15), 2156–2158. doi: 10.1093/bioinformatics/btr330
de Micheli, M., Bille, J., Schueller, C., Sanglard, D. (2002). A Common Drug-Responsive Element Mediates the Upregulation of the Candida Albicans ABC Transporters CDR1 and CDR2, Two Genes Involved in Antifungal Drug Resistance. Mol. Microbiol. 43 (5), 1197–1214. doi: 10.1046/j.1365-2958.2002.02814.x
Deo, R. C. (2015). Machine Learning in Medicine. Circulation 132 (20), 1920–1930. doi: 10.1161/CIRCULATIONAHA.115.001593
DePristo, M. A., Banks, E., Poplin, R., Garimella, K. V., Maguire, J. R., Hartl, C., et al. (2011). A Framework for Variation Discovery and Genotyping Using Next-Generation DNA Sequencing Data. Nat. Genet. 43 (5), 491–498. doi: 10.1038/ng.806
Di Pilato, V., Codda, G., Ball, L., Giacobbe, D. R., Willison, E., Mikulska, M., et al. (2021). Molecular Epidemiological Investigation of a Nosocomial Cluster of C. Auris: Evidence of Recent Emergence in Italy and Ease of Transmission During the COVID-19 Pandemic. J. Fungi (Basel) 7 (2). doi: 10.3390/jof7020140
Du, H., Bing, J., Hu, T., Ennis, C. L., Nobile, C. J., Huang, G. (2020). Candida Auris: Epidemiology, Biology, Antifungal Resistance, and Virulence. PloS Pathog. 16 (10), e1008921. doi: 10.1371/journal.ppat.1008921
Dudiuk, C., Berrio, I., Leonardelli, F., Morales-Lopez, S., Theill, L., Macedo, D., et al. (2019). Antifungal Activity and Killing Kinetics of Anidulafungin, Caspofungin and Amphotericin B Against Candida Auris. J. Antimicrob. Chemother. 74 (8), 2295–2302. doi: 10.1093/jac/dkz178
ElBaradei, A. (2020). A Decade After the Emergence of Candida Auris: What do We Know? Eur. J. Clin. Microbiol. Infect. Dis. 39 (9), 1617–1627. doi: 10.1007/s10096-020-03886-9
Escandon, P., Chow, N. A., Caceres, D. H., Gade, L., Berkow, E. L., Armstrong, P., et al. (2019). Molecular Epidemiology of Candida Auris in Colombia Reveals a Highly Related, Countrywide Colonization With Regional Patterns in Amphotericin B Resistance. Clin. Infect. Dis. 68 (1), 15–21. doi: 10.1093/cid/ciy411
Farhat, M. R., Sultana, R., Iartchouk, O., Bozeman, S., Galagan, J., Sisk, P., et al. (2016). Genetic Determinants of Drug Resistance in Mycobacterium Tuberculosis and Their Diagnostic Value. Am. J. Respir. Crit. Care Med. 194 (5), 621–630. doi: 10.1164/rccm.201510-2091OC
Handelman, G. S., Kok, H. K., Chandra, R. V., Razavi, A. H., Lee, M. J., Asadi, H. (2018). Edoctor: Machine Learning and the Future of Medicine. J. Intern. Med. 284 (6), 603–619. doi: 10.1111/joim.12822
Healey, K. R., Kordalewska, M., Jimenez Ortigosa, C., Singh, A., Berrio, I., Chowdhary, A., et al. (2018). Limited ERG11 Mutations Identified in Isolates of Candida Auris Directly Contribute to Reduced Azole Susceptibility. Antimicrob. Agents Chemother. 62 (10). doi: 10.1128/AAC.01427-18
Her, H. L., Wu, Y. W. (2018). A Pan-Genome-Based Machine Learning Approach for Predicting Antimicrobial Resistance Activities of the Escherichia Coli Strains. Bioinformatics 34 (13), i89–i95. doi: 10.1093/bioinformatics/bty276
Kordalewska, M., Lee, A., Park, S., Berrio, I., Chowdhary, A., Zhao, Y., et al. (2018). Understanding Echinocandin Resistance in the Emerging Pathogen Candida Auris. Antimicrob. Agents Chemother. 62 (6). doi: 10.1128/AAC.00238-18
Kordalewska, M., Perlin, D. S. (2019). Identification of Drug Resistant Candida Auris. Front. Microbiol. 10, 1918. doi: 10.3389/fmicb.2019.01918
Kouchaki, S., Yang, Y., Walker, T. M., Sarah Walker, A., Wilson, D. J., Peto, T. E. A., et al. (2019). Application of Machine Learning Techniques to Tuberculosis Drug Resistance Analysis. Bioinformatics 35 (13), 2276–2282. doi: 10.1093/bioinformatics/bty949
Kumar, S., Stecher, G., Li, M., Knyaz, C., Tamura, K. (2018). MEGA X: Molecular Evolutionary Genetics Analysis Across Computing Platforms. Mol. Biol. Evol. 35 (6), 1547–1549. doi: 10.1093/molbev/msy096
Kwon, Y. J., Shin, J. H., Byun, S. A., Choi, M. J., Won, E. J., Lee, D., et al. (2019). Candida Auris Clinical Isolates From South Korea: Identification, Antifungal Susceptibility, and Genotyping. J. Clin. Microbiol. 57 (4). doi: 10.1128/JCM.01624-18
Lamb, D. C., Corran, A., Baldwin, B. C., Kwon-Chung, J., Kelly, S. L. (1995). Resistant P45051A1 Activity in Azole Antifungal Tolerant Cryptococcus Neoformans From AIDS Patients. FEBS Lett. 368 (2), 326–330. doi: 10.1016/0014-5793(95)00684-2
Li, H., Handsaker, B., Wysoker, A., Fennell, T., Ruan, J., Homer, N., et al. (2009). The Sequence Alignment/Map Format and SAMtools. Bioinformatics 25 (16), 2078–2079. doi: 10.1093/bioinformatics/btp352
Lockhart, S. R., Etienne, K. A., Vallabhaneni, S., Farooqi, J., Chowdhary, A., Govender, N. P., et al. (2017). Simultaneous Emergence of Multidrug-Resistant Candida Auris on 3 Continents Confirmed by Whole-Genome Sequencing and Epidemiological Analyses. Clin. Infect. Dis. 64 (2), 134–140. doi: 10.1093/cid/ciw691
Medici, N. P., Del Poeta, M. (2015). New Insights on the Development of Fungal Vaccines: From Immunity to Recent Challenges. Mem Inst Oswaldo Cruz 110 (8), 966–973. doi: 10.1590/0074-02760150335
Mellado, E., Garcia-Effron, G., Alcazar-Fuoli, L., Cuenca-Estrella, M., Rodriguez- Tudela, J. L. (2004). Substitutions at Methionine 220 in the 14alpha-Sterol Demethylase (Cyp51A) of Aspergillus Fumigatus Are Responsible for Resistance In Vitro to Azole Antifungal Drugs. Antimicrob. Agents Chemother. 48 (7), 2747–2750. doi: 10.1128/AAC.48.7.2747-2750.2004
Montoya, M. C., Moye-Rowley, W. S., Krysan, D. J. (2019). Candida Auris: The Canary in the Mine of Antifungal Drug Resistance. ACS Infect. Dis. 5 (9), 1487–1492. doi: 10.1021/acsinfecdis.9b00239
Munoz, J. F., Gade, L., Chow, N. A., Loparev, V. N., Juieng, P., Berkow, E. L., et al. (2018). Genomic Insights Into Multidrug-Resistance, Mating and Virulence in Candida Auris and Related Emerging Species. Nat. Commun. 9 (1), 5346. doi: 10.1038/s41467-018-07779-6
Nami, S., Mohammadi, R., Vakili, M., Khezripour, K., Mirzaei, H., Morovati, H. (2019). Fungal Vaccines, Mechanism of Actions and Immunology: A Comprehensive Review. BioMed. Pharmacother. 109, 333–344. doi: 10.1016/j.biopha.2018.10.075
Noel, T. (2012). The Cellular and Molecular Defense Mechanisms of the Candida Yeasts Against Azole Antifungal Drugs. J. Mycol Med. 22 (2), 173–178. doi: 10.1016/j.mycmed.2012.04.004
Park, S., Kelly, R., Kahn, J. N., Robles, J., Hsu, M. J., Register, E., et al. (2005). Specific Substitutions in the Echinocandin Target Fks1p Account for Reduced Susceptibility of Rare Laboratory and Clinical Candida Sp. Isolates. Antimicrob. Agents Chemother. 49 (8), 3264–3273. doi: 10.1128/AAC.49.8.3264-3273.2005
Patel, L., Shukla, T., Huang, X., Ussery, D. W., Wang, S. (2020). Machine Learning Methods in Drug Discovery. Molecules 25 (22). doi: 10.3390/molecules25225277
Puri, N., Krishnamurthy, S., Habib, S., Hasnain, S. E., Goswami, S. K., Prasad, R. (1999). CDR1, A Multidrug Resistance Gene From Candida Albicans, Contains Multiple Regulatory Domains in Its Promoter and the Distal AP-1 Element Mediates Its Induction by Miconazole. FEMS Microbiol. Lett. 180 (2), 213–219. doi: 10.1111/j.1574-6968.1999.tb08798.x
Rhodes, J., Abdolrasouli, A., Farrer, R. A., Cuomo, C. A., Aanensen, D. M., Armstrong-James, D., et al. (2018). Genomic Epidemiology of the UK Outbreak of the Emerging Human Fungal Pathogen Candida Auris. Emerg. Microbes Infect. 7 (1), 43. doi: 10.1038/s41426-018-0045-x
Rhodes, J., Fisher, M. C. (2019). Global Epidemiology of Emerging Candida Auris. Curr. Opin. Microbiol. 52, 84–89. doi: 10.1016/j.mib.2019.05.008
Rybak, J. M., Munoz, J. F., Barker, K. S., Parker, J. E., Esquivel, B. D., Berkow, E. L., et al. (2020). Mutations in TAC1B: A Novel Genetic Determinant of Clinical Fluconazole Resistance in Candida Auris. mBio 11 (3). doi: 10.1128/mBio.00365-20
Sanglard, D., Ischer, F., Koymans, L., Bille, J. (1998). Amino Acid Substitutions in the Cytochrome P-450 Lanosterol 14alpha-Demethylase (CYP51A1) From Azole-Resistant Candida Albicans Clinical Isolates Contribute to Resistance to Azole Antifungal Agents. Antimicrob. Agents Chemother. 42 (2), 241–253. doi: 10.1128/AAC.42.2.241
Satoh, K., Makimura, K., Hasumi, Y., Nishiyama, Y., Uchida, K., Yamaguchi, H. (2009). Candida Auris Sp. Nov., a Novel Ascomycetous Yeast Isolated From the External Ear Canal of an Inpatient in a Japanese Hospital. Microbiol. Immunol. 53 (1), 41–44. doi: 10.1111/j.1348-0421.2008.00083.x
Sharma, C., Kumar, N., Pandey, R., Meis, J. F., Chowdhary, A. (2016). Whole Genome Sequencing of Emerging Multidrug Resistant Candida Auris Isolates in India Demonstrates Low Genetic Variation. New Microbes New Infect. 13, 77–82. doi: 10.1016/j.nmni.2016.07.003
Spampinato, C., Leonardi, D. (2013). Candida Infections, Causes, Targets, and Resistance Mechanisms: Traditional and Alternative Antifungal Agents. BioMed. Res. Int. 2013, 204237. doi: 10.1155/2013/204237
Tian, S., Rong, C., Nian, H., Li, F., Chu, Y., Cheng, S., et al. (2018). First Cases and Risk Factors of Super Yeast Candida Auris Infection or Colonization From Shenyang, China. Emerg. Microbes Infect. 7 (1), 128. doi: 10.1038/s41426-018-0131-0
Vandeputte, P., Ferrari, S., Coste, A. T. (2012). Antifungal Resistance and New Strategies to Control Fungal Infections. Int. J. Microbiol. 2012, 713687. doi: 10.1155/2012/713687
Wang, X., Bing, J., Zheng, Q., Zhang, F., Liu, J., Yue, H., et al. (2018). The First Isolate of Candida Auris in China: Clinical and Biological Aspects. Emerg. Microbes Infect. 7 (1), 93. doi: 10.1038/s41426-018-0095-0
Welsh, R. M., Sexton, D. J., Forsberg, K., Vallabhaneni, S., Litvintseva, A. (2019). Insights Into the Unique Nature of the East Asian Clade of the Emerging Pathogenic Yeast Candida Auris. J. Clin. Microbiol. 57 (4). doi: 10.1128/JCM.00007-19
Yang, Y., Niehaus, K. E., Walker, T. M., Iqbal, Z., Walker, A. S., Wilson, D. J., et al. (2018). Machine Learning for Classifying Tuberculosis Drug-Resistance From DNA Sequencing Data. Bioinformatics 34 (10), 1666–1671. doi: 10.1093/bioinformatics/btx801
Keywords: Candida auris, phylogenetic analysis, drug resistance, whole genome sequencing, machine learning, antifungal drugs, ergosterol pathway
Citation: Li D, Wang Y, Hu W, Chen F, Zhao J, Chen X and Han L (2021) Application of Machine Learning Classifier to Candida auris Drug Resistance Analysis. Front. Cell. Infect. Microbiol. 11:742062. doi: 10.3389/fcimb.2021.742062
Received: 15 July 2021; Accepted: 22 September 2021;
Published: 15 October 2021.
Edited by:
Jianping Xu, McMaster University, CanadaReviewed by:
Yue Wang, McMaster University, CanadaMarie Desnos-Ollivier, Institut Pasteur, France
Kin-Ming (Clement) Tsui, University of British Columbia, Canada
Copyright © 2021 Li, Wang, Hu, Chen, Zhao, Chen and Han. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Li Han, aGFubGljZGNAMTYzLmNvbQ==; Xia Chen, eGNoZW44MEBzbm51LmVkdS5jbg==
†These authors have contributed equally to this work and share first authorship
 Dingchen Li
Dingchen Li Yaru Wang1,2†
Yaru Wang1,2† Wenjuan Hu
Wenjuan Hu Xia Chen
Xia Chen Li Han
Li Han