- 1South Africa Medical Research Center (SAMRC) Microbial Water Quality Monitoring Centre, University of Fort Hare, Alice, South Africa
- 2Applied and Environmental Microbiology Research Group (AEMREG), Department of Biochemistry and Microbiology, University of Fort Hare, Alice, South Africa
- 3Biology Department, Albion College, Albion, MI, United States
The study investigated the occurrence of antimicrobial resistance genes and virulence determinants in Vibrio species recovered from different freshwater sheds in rustic milieu. A total of 118 Vibrio isolates comprising Vibrio fluvialis (n=41), Vibrio mimicus (n=40) and V. vulnificus (n=37) was identified by amplification of ToxR, vmh and hsp60 genes. The amplification of virulence genes indicated that V. mimicus (toxR, zot, ctx, VPI, and ompU) genes were detected in 12.5%, 32.5%, 45%, 37.5% and 10% respectively. V. fluvialis genes (stn, hupO and vfh) were harboured in 48.8%, 14.6% and 19.5% isolates congruently. The other virulence genes that include vcgC and vcgE were observed in 63.1% and 29% of isolates belonging to V. vulnificus. With the exceptions of imipenem, meropenem and ciprofloxacin, most isolates exhibited more than 50% resistance to antibiotics. The antimicrobial resistance was more prevalent for polymyxin B (100%), azithromycin (100%) and least in ciprofloxacin (16.1%). Multiple antibiotic resistance index range was 0.3 and 0.8 with most isolates showing MARI of 0.8. The blaTEM, AmpC, blaGES, blaIMP, blaOXA-48 and blaKPC genes were detected in 53.3%, 42%, 29.6%, 16.6%, 15%, 11.3% and 5.6% of the isolates. Non-beta lactamases such as streptomycin resistance (aadA and strA), gentamicin resistance (aphA1) and quinolone resistance gene (qnrVC) were found in 5.2%, 44.3%, 26% and 2.8%. Chloramphenicol resistance genes (cmlA1 and catII) were found in 5.2% and 44.3% among the isolates. Our findings reveal the presence of antimicrobial resistance genes and virulent Vibrio species in aquatic environment which can have potential risk to human and animal’s health.
Introduction
Water containing infectious pathogens has been associated with ailments globally amongst human populations (Ramírez-Castillo et al., 2015). The importance and need for better water quality cannot be over-emphasized. Water scarcity is becoming more serious due to urbanization and population growth (Tzanakakis et al., 2020) hence there is an increase in demand for water. However, in most countries a strategy to minimize water shortage has been put in place which involves treating and recycling wastewater (Voulvoulis, 2018). Many treatment plants for wastewater in South Africa discharge their final effluents openly into adjoining streams or rivers used by the nearby villages/suburb for their several domestic needs (Wall, 2018). However, reports suggest that some wastewater are not properly treated and therefore discharge effluent of poor quality (Edokpayi et al., 2015). Wastewater and sewage from several sources contribute to spreading antimicrobial resistance (AMR) to the environment and adversely impacting on the habitation of aquatic animals and their surrounding water (Fouz et al., 2020). Several studies have emphasized on the effect of sewage as a huge environmental pool of AMR, as it signifies a suitable milieu for AMR organisms and antimicrobial resistance genes to thrive (Rizzo et al., 2013: Singer et al., 2016). Numerous resistant pathogens have been identified in various environmental sewage, hospital effluents, surface water and wastewater in treatment plants (Lundborg and Tamhankar, 2017). The aquatic environment has become reservoir of many microbes including Vibrios that are potentially infectious to humans. Vibrio genus is grouped in the family Vibrionaceae, which comprises of opportunistic pathogenic microbes infecting both humans and animals (Takemura et al., 2014). Vibrio species are a group of Gram-negative, curved-rod shaped bacteria being regular constituent of estuarine, freshwater and marine environs (Baker-Austin et al., 2018). Their genomes are separated inside two chromosomes having been shaped by recombination and the acquiring of genes by horizontal gene transfer from other bacteria (Oliveira et al., 2017). Infections caused by pathogenic Vibrio remain severe public health significance. Cholera causing Vibrio (Vibrio cholerae) is implicated in serious diarrhoeal infection which can be rapidly fatal when not treated and is typically disseminated through person-to-person contact as well as infected water (Das et al., 2020). Non-cholera causing Vibrio species (including V. parahaemolyticus, V. anguillarum, V. metchnikovii, Vibrio alginolyticus, V. harveyi, V. mimicus, V. fluvialis, and V. vulnificus) are known to cause vibriosis that is usually contracted via sea water or through ingesting raw or parboiled infected seafood causing necrotizing fasciitis, self-limiting gastroenteritis, severe septicaemia (Gennari et al., 2012). Vibrio species pathogenic to humans produce a variety of virulence genes which contribute to some ailments (Álvarez-Contreras et al., 2021). Some of the virulence trait includes the secretion of extracellular products (ECP), thermostable direct haemolysin (tdh), and tdh-related haemolysin expressed in V. parahaemolyticus, expression of capsular polysaccharides, cytotoxicity, biofilm formation, cholera toxin and the toxin-coregulated pilus (tcp) in Vibrio cholerae (Binesse et al., 2008). The V. fluvialis protease gene (vfp), heme utilization protein gene (hupO) and extracellular haemolysin gene (vfh), are also among the Vibrio virulence genes (Ramamurthy et al., 2014). The emergence of Vibrio strains in aquatic environments have created more apprehension leading to upsurge in resistance among Vibrio isolates against most clinically prescribed antibiotics and this has initiated apprehension in the healthcare sector thwarting current therapeutic outcomes (Banerjee and Farber, 2018). Vibrio species are documented to be very sensitive to many antimicrobials that are clinically recommended for therapeutic purpose and comprises of tetracycline, cephalosporins quinolones, aminoglycosides, and folate pathway inhibitors (Elmahdi et al., 2016). The incessant abuse of antibiotics has over time contributed immensely to the rapid emergence of drug resistance microbial strains including several Vibrio species (Manyi-Loh et al., 2018). Antibiotics are broadly used to manage or treat bacterial ailments in humans as well as in veterinary medicine. The beginning of this millennium signifies the end of the golden era of antibiotics and the commencement of more alertness of the public health alertness on the distribution of antimicrobial resistance. Documented reports have emphasized the significant importance of various aquatic environments in the spread of AMR (Baquero et al., 2008) especially predominantly in wastewater effluents which are regarded as hot spots for horizontal genes transfer (Bouki et al., 2013). South Africa is situated in a semi-dry area and typically experience limited annual rainfalls giving rise to scarcity of portable water. Consequently, most inhabitants in the rural areas resort to the use of untreated water from vulnerable sources, like rivers, and dams for their daily supply (Fobosi, 2013). Over the years, many researchers have focused on the severity of diseases caused by Vibrio cholerae opting out relatively other Vibrio species of medical interest, some of which are emerging pathogens that are able to cause mild to severe human diseases (Osunla and Okoh, 2017). Therefore, the aim of this study was to assess the antibiogram, virulence and resistance genes of non-cholera causing Vibrio species recovered from aquatic sources.
Materials and Methods
Study Site
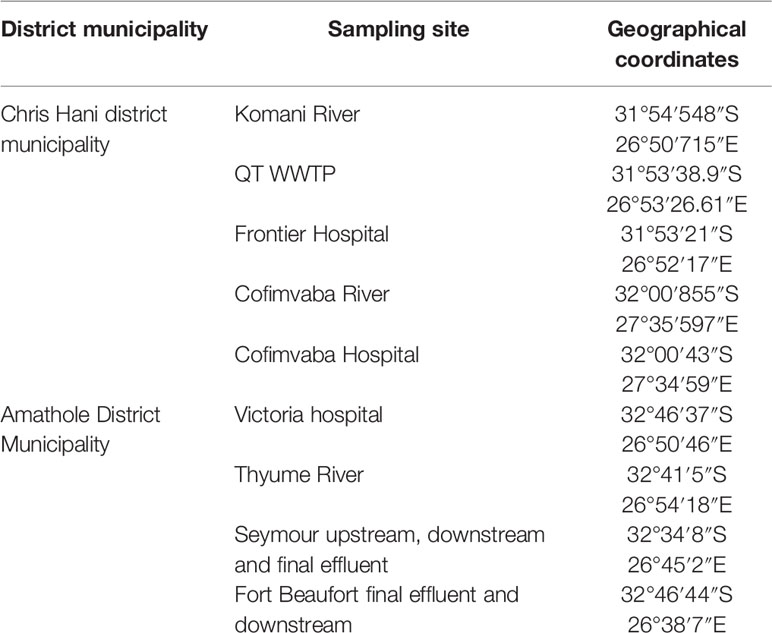
The Eastern Cape is one of the nine provinces within South Africa; it is separated into two Buffalo City and Nelson Mandela Bay and six district municipalities for local government purposes. The district municipalities are in turn split into thirty-one local municipalities. The Eastern Cape mostly consists of rural settlements with minimal basic services. This study was carried out in two of the six district municipalities of this region, specifically at the Amathole District and Chris Hani District Municipalities. The Chris Hani District Municipality is located in the central of the Eastern Cape hinterland, between the Eastern Cape coastline and the Drakensberg mountains and spans an estimate of 37 294 km2. The district municipality is located on the watershed of four river viz. Orange River, Great Fish River, Mbashe River and Great Kei River. Surface water sources supply water for most of the towns in the area while only a few depend on groundwater supplies. The Amathole District Municipality is located in the central part of the Eastern Cape and spans along the Sunshine Coast from the Fish River Mouth and along the Eastern Seaboard to the south of Hole in the Wall alongside the Wild Coast. It is also bordered to the north by the Amathole Mountain Range. The rivers examined in this study were selected due to the fact that they serve as final receiving bodies of water for the release of treated domestic sewage operated in activated sludge systems. The targeted hospitals were also chosen because they serve as referral hospitals for the indigenous inhabitants with various medical conditions, they do not have independent wastewater treatment mechanism but are joined with that of the municipal waste treatment system.
Collection of Samples
A total of seventy-two (72) wastewater samples were collected aseptically fortnightly from the 9 sampling sites (shown in Table 1) over a period of 4 months (July 2018-October 2018) with sterile 500 mL Borosilicate Glass bottles. The samples were conveyed on ice in a cooler box from the sampling site to the Applied and Environmental Microbiology Research Group (AEMREG) laboratory at the University of Fort Hare for analysis within few hours of collection.
Isolation and Identification of Presumptive Vibrio Species
Standard membrane procedure was employed in processing the wastewater samples (APHA, A. 1998). Each wastewater sample was serially diluted and a 100 ml volume was filtered through 0.45 μm mixed cellulose ester membrane filters (Millipore, Ireland) using a vacuum pump. The membrane filter paper was transferred onto alkaline peptone water as means of enrichment and incubated aerobically at 37°C for 18-24 h. A loopful from the alkaline peptone water was sub-cultured onto thiosulphate citrate bile salts sucrose (TCBS, Darmstadt, Germany) and incubated for another 24 h at 37°C. Four to six colonies were randomly chosen from each plate and consequently purified in fresh TCBS agar plates. Purified isolates were subcultured on Glycerol stocks (50%) and stored at -80 °C sequel to further analysis.
Genomic DNA Extraction
Nucleic acids (DNA) from presumptive Vibrio species were extracted following the method described by Mapipa et al. (2021) with minor modification. The colonies were inoculated in Luria Bertani (LB) broth and incubated at 37°C for 18-24 h. The overnight broths were centrifuged (11,000 rpm for 1 min) to pellet the cells, washed with normal saline (0.85%) and re-suspended in 0.5 ml distilled water. The emulsified pellet suspension was heated at 100°C for 15 min in a heating block to release the DNA. The lysate was centrifuged at 13000 rpm for 5 min to eliminate cell debris and the supernatant was stored (−20°C) until further use.
Presumptive Vibrio Isolates Confirmation
The presumptive confirmation of the Vibrio isolates was achieved with PCR assay using specific primers targeting a variable portion with the 16SrRNA gene. The confirmed isolates of the Vibrio isolates were also delineated into V. mimicus, V. fluvialis and V. Vulnificus. The PCR cocktail comprised of 12 μl of Master-mix (inqaba), 1μl of both forward and reverse oligonucleotide primers (Inqaba, SA), 6 μl of nuclease free water and 5μl of DNA template which summed up to 25 μl reaction mixture. The amplicons were differentiated by electrophoresis in 0.8% agarose gels containing 0.5 μg/mL ethidium bromide (Sigma-Aldrich, St. Louis, USA) run at 90 V for 50 min and visualised under a UV transilluminator (Alliance 4.7 Uvitec, Cambridge). The targeted genes, primer sequences, band sizes and cycling condition of the Vibrio species are shown on Appendix 1 (Supplementary Material).
Antimicrobial Sensitivity Test Performed on the Vibrio Isolates
Antibiotic sensitivity test was carried out using Mueller-Hinton agar (Basingshike, Hampshire, England) by the standard disk diffusion technique (Kirby-Bauer test) as endorsed by the Clinical and Laboratory Standards Institute (Clinical and Laboratory Standards Institute, 2018). Few colonies of confirmed isolates of the Vibrio species were reconstituted on sterile normal saline to give an approximately 0.5 McFarland standard. A lawn of the bacterial suspension was made on Mueller-Hinton agar and allowed to stand for 15 min. A panel of 12 antibiotics which are usually recommended for treating Vibrio infections were selected and impregnated unto the agar plates. The antibiotic disks include: ampicillin (10 μg), ampicillin-sulbactam (10/10 μg), imipenem (10 μg), meropenem (10 μg), kanamycin (30 μg), ciprofloxacin (5 μg), ofloxacin (5 μg), cefuroxime (30 μg), polymyxin B (300 μg), azithromycin (15 μg), nitrofurantoin (300 μg) and Chloramphenicol (30 μg). The plates were incubated for 24 h at 37°C and results documented as sensitive, intermediate or resistant based on the elucidation of the diameter of the zone of inhibition.
Multiple Antibiotic Resistance Index
The Multiple antibiotic resistance index was estimated using the formula: MARI=a/b, where a = is the number of antibiotics to which the isolates exhibited resistance and b = is the total number of antibiotics which the resistant isolates were subjected to. MARI value of more than 0.2 recommends that the isolates are from environ where there are excessive or indiscriminate use of antibiotics (Blanco et al., 2008).
Molecular Detection of Virulence Genes
The identification of virulence genes in the Vibrio species was done through polymerase chain reaction (PCR) analyses. The targeted virulence genes include toxin coregulated pilus (tcp), toxin regulon (toxR), outer membrane protein (ompU), zonula occludens toxin (zot), cholera toxin gene (ctx) and Vibrio pathogenicity island (VPI) in V. mimicus. Others include V. vulnificus virulence genes (vcgC and vcgE) in V. vulnificus, extracellular haemolysin gene (vfh), heme utilization protein gene (hupO), V. fluvialis protease gene (vfpA) and heat stable enterotoxin (stn) in V. fluvialis. Each reaction comprises a preliminary denaturation at 94°C for 4 min, accompanied by 35 cycles each consisting of an initial denaturation at 94°C for 40 sec followed by annealing (52 °C and 62 °C) and extension steps for 90 sec while final elongation was 72°C for 6 min. Primers and their annealing temperatures are presented in Appendix 2 (Supplementary Material).
Detection of Antibiotic Resistance Genes Among the Vibrio Species
Also, PCR analysis was employed to identify various antibiotic resistant genes in the Vibrio species using pairs of specific primers and cycling conditions as follows: initial denaturation of 94°C for 5 min, accompanied by 35 cycles of 94°C for 1 min, annealing temperatures (60-62°C) for 45 s and 72°C for 91 s, final elongation step at 72°C for 5 mins. The selected targets genes were: aminoglycoside resistance genes (strA, aadA, aacC2, apHAI and apHAII), phenicols resistance genes (catI, catII and cmlA1), β-lactamases-encoding genes (ampC, blaTEM and blaZ) and Carbapenem resistance genes (blaGES, blaOXA-48, blaIMP, blaVIM, blaKPC, and qnrVC). The oligonucleotide sequences, amplicon sizes and annealing temperatures of the primers used in this study are presented in Appendix 3 (Supplementary Material).
Results
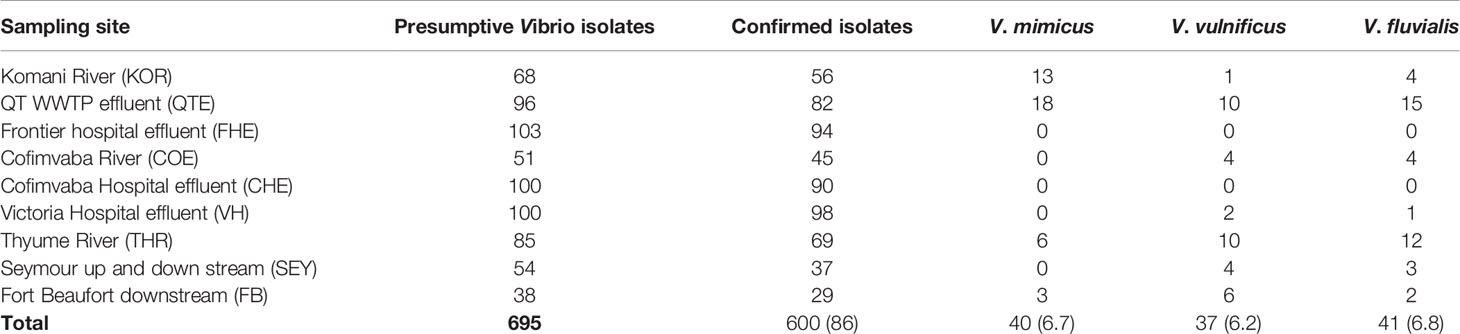
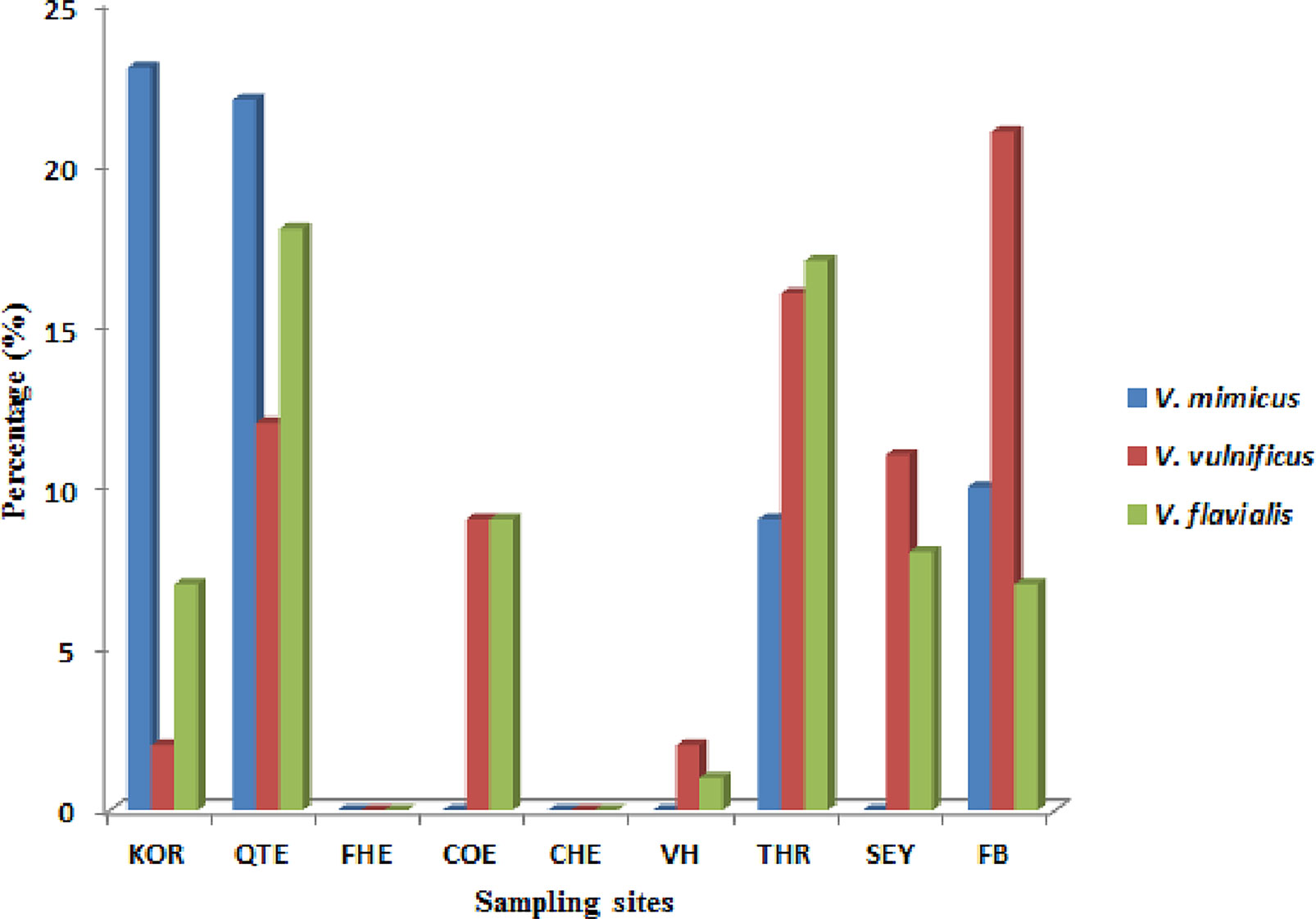
During the study period, a total of 695 presumptive Vibrio isolates were recovered from all the sampling sites. PCR analysis was used to confirm 690 isolates as Vibrio isolates. Of the confirmed Vibrio species: V. mimicus, V. fluvialis and V. vulnificus accounted for 40(6.7%), 41(6.8%) and 37(6.2%) respectively from seven of the nine sampling sites. Other isolates not confirmed could belong to Vibrio species not identified in this study. Confirmation and delineation of Vibrio species recovered from the different sampling sites is shown in Table 2. The frequency of Vibrio species recovered from the different site Figure 1, while the molecular gel electrophoresis delineating Vibrio into species is shown in Figure 2.
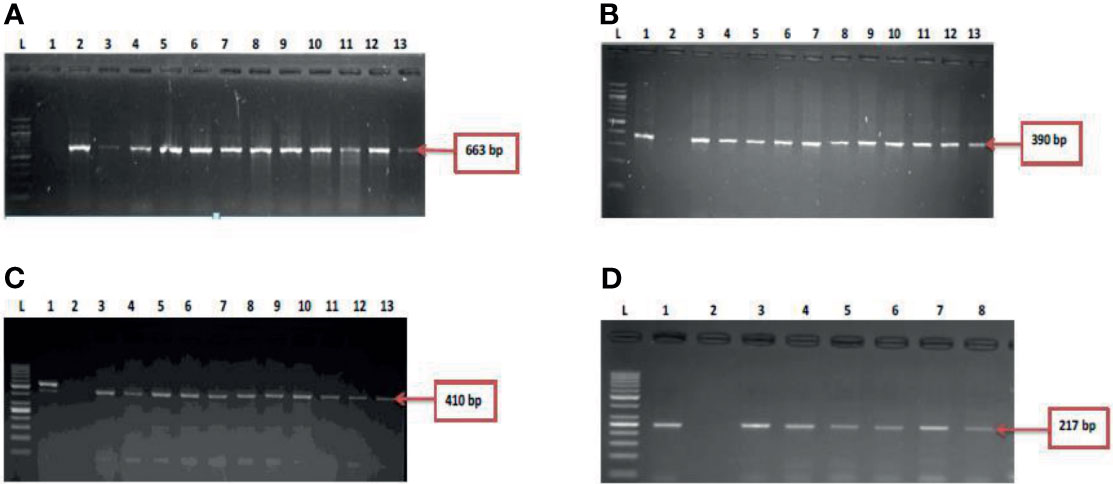
Figure 2 (A) = gel electrophoresis showing the 16s rRNA gene among isolates belonging to the Vibrio genus. Lane L: Molecular DNA-marker (100 bp); Lane 1: Negative control (-ve); Lane 2-13: positive isolates of 16s rRNA gene (663 bp): (B) = Gel electrophoresis showing the vmh gene among isolates belonging to the V. mimicus. Lane L: Molecular DNA-Marker (100 bp), lane 1: positive control (DSM 19130), Lane 2: Negative control (-ve); Lane 3-13: positive isolates of vmh gene (663 bp); (C) = Gel electrophoresis showing the hsp60 gene among isolates belonging to the V. vulnificus. Lane L: Molecular DNA-marker (50 bp), lane 1: positive control (DSM 10143), Lane 2: Negative control (-ve); Lane 3-13: positive isolates of hsp60 gene (410 bp) and (D) = Figure 2D: Gel electrophoresis showing the ToxR gene among isolates belonging to the V. fluvialis. Lane L: Molecular DNA-marker (50 bp), lane 1: positive control (DSM 19283), Lane 2: Negative control (-ve); Lane 3-8: positive isolates of ToxR gene (217 bp).
Distribution of Antimicrobial Susceptibility Profile of the Vibrio Species
All 118 confirmed Vibrio species were subjected to seven antimicrobial classes comprising of twelve antibiotics. The antibiogram of the Vibrio isolates from all sampling sites showed varying degrees of resistance to most of the antibiotics used. A 100% resistance pattern was observed among all 118 isolates to azithromycin and polymyxin B. More than 70% of the isolates also showed resistance to ampicillin, ampicillin-sulbactam, cefuroxime and chloramphenicol. The highest susceptibility among the isolates was observed with imipenem and meropenem antibiotics. Meropenem susceptibility among V. mimicus and V. fluvialis was greater than 60%. A resistance pattern of more than 50% was observed among all the isolates to kanamycin, while fluoroquinolones had variations in the susceptibility pattern. High resistance level was observed with ofloxacin (62%) and ciprofloxacin (16%). The distribution of the antibiotic susceptibility pattern is shown in Table 3. The multiple antibiotic resistance patterns among the Vibrio species showed a range of 0.3 and 0.8. The multi-drug resistance profile among the Vibrio species is shown in Table 4.
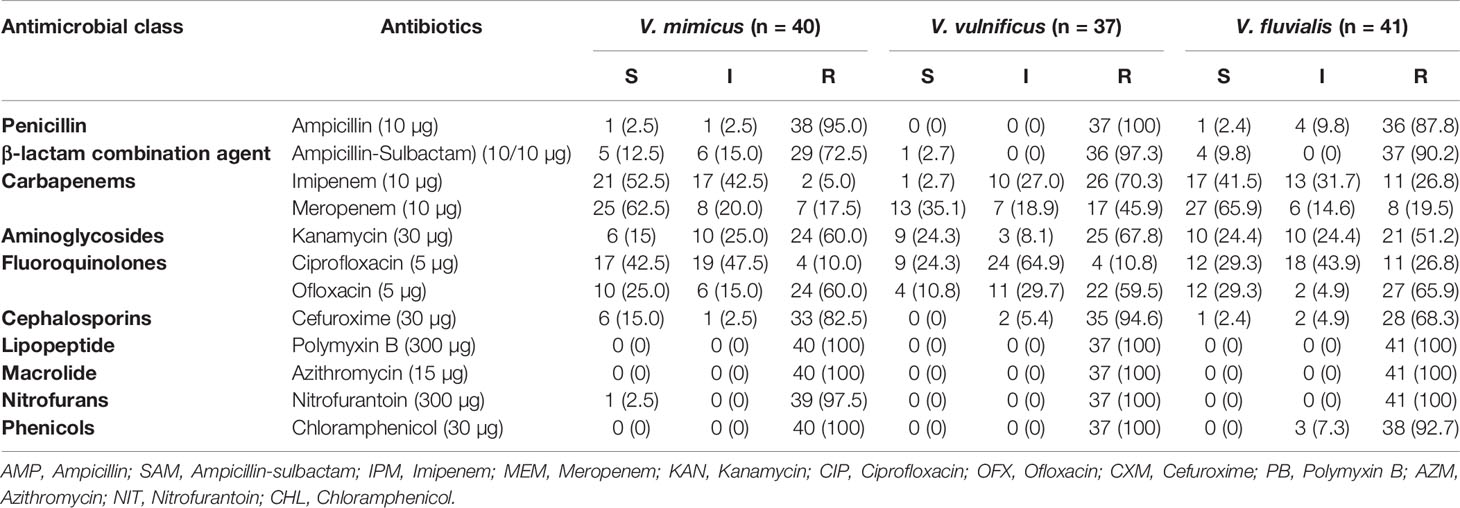
Table 3 Distribution of antimicrobial susceptibility of Vibrio species recovered in all sampling sites.
Frequency of Virulence Genes Among the Vibrio Species
A total of ten of the twelve virulence genes were detected among the three Vibrio species recovered from all the sites. Vibrio mimicus harboured five virulence genes in the following proportion: toxR (12.5%), zot (32.5%), ctx (45%), VPI (37.5%), tcp (0), and ompU (10%). Two virulence genes detected among the V. vulnificus isolates were vcgC (63.1%) and vcgE (29%). Three of the four virulence genes were likewise detected among the V. fluvialis isolates recovered from all the sampling sites in the following proportion: stn (48.8%), hupO (14.6%), vfpA (0) and vfh (19.5%). Figures 3A–C represent the frequency of virulence genes among the three Vibrio species (V. mimicus, V. vulnificus and V. fluvialis) recovered from all sampling sites. The gel representative pictures for the amplification of the virulence genes for the three Vibrio species are shown in Figure 4.
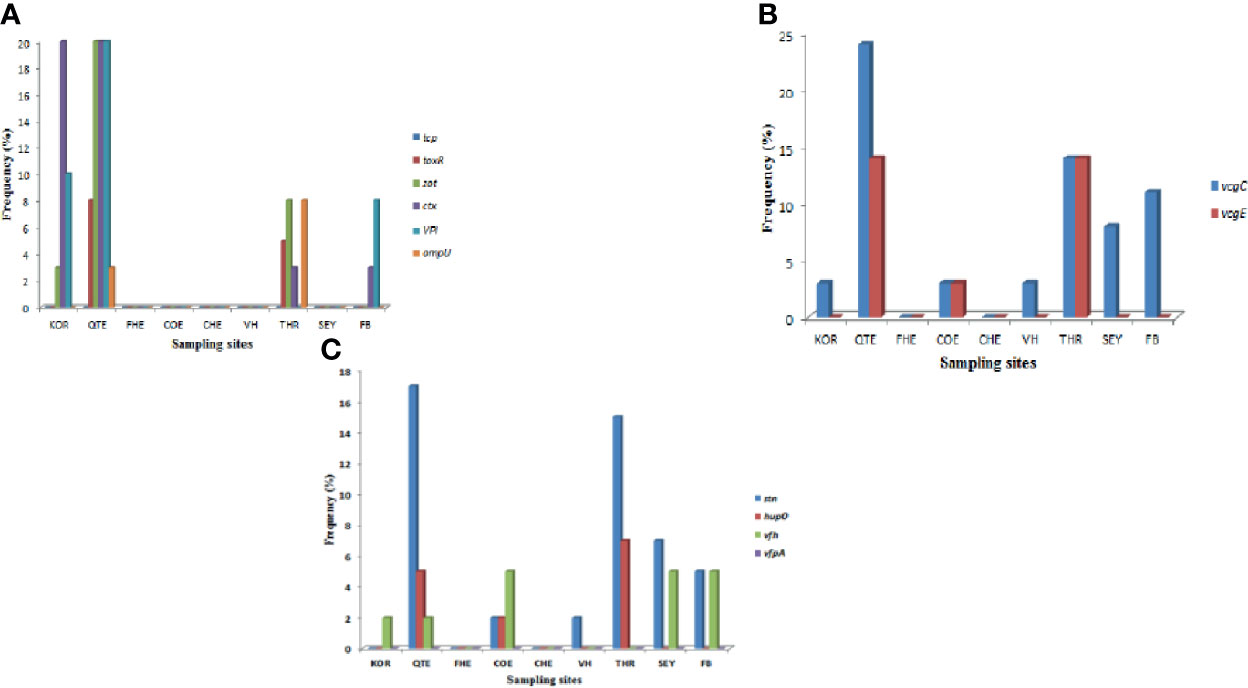
Figure 3 (A) = Frequency of V. mimicus virulence gene from all sampling sites, (B) = Frequency of V. vulnificus virulence genes from all sampling sites and (C) is Frequency of V. fluvialis virulence genes from all sampling sites.
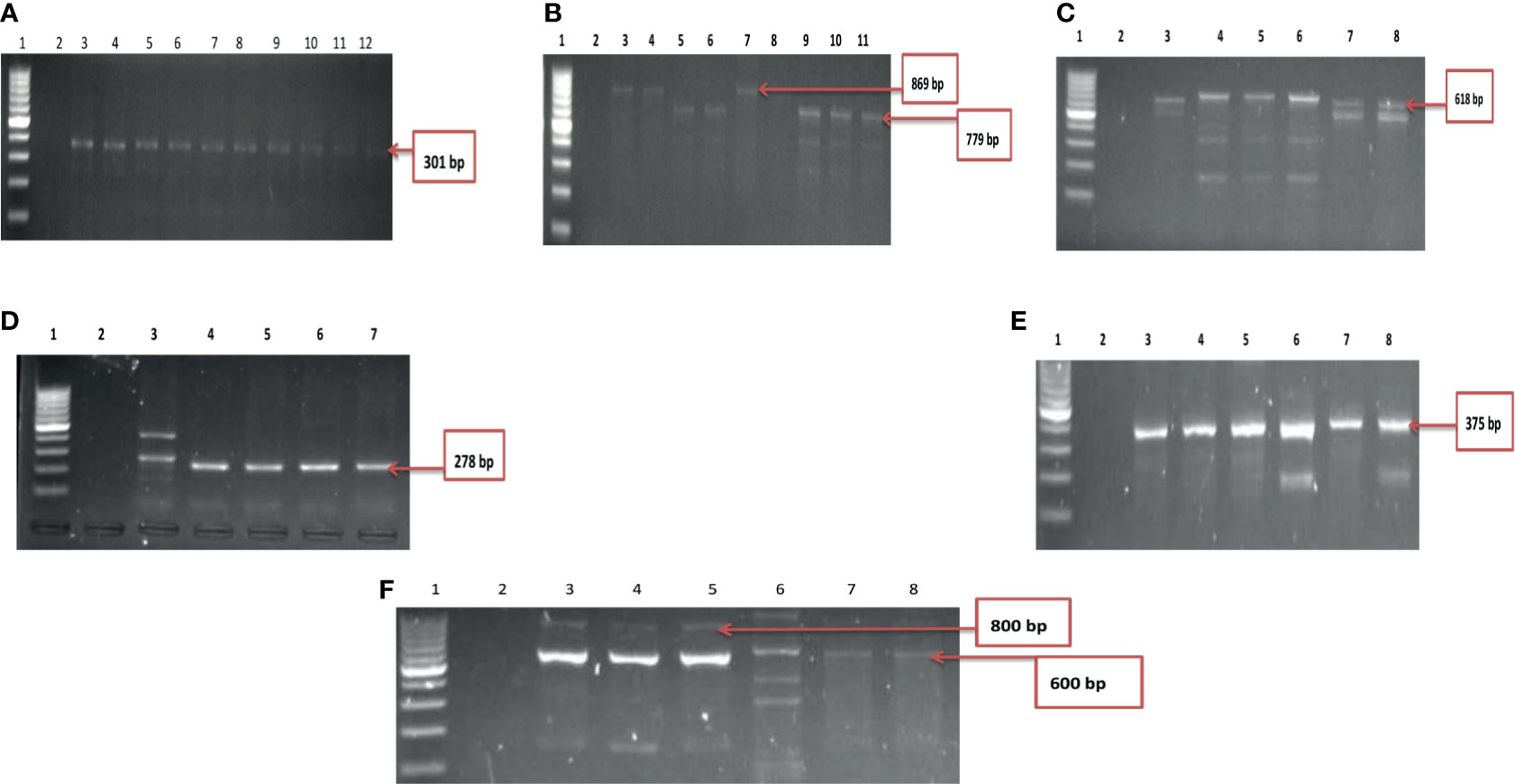
Figure 4 (A) Gel electrophoresis showing the ctx gene among isolates belonging to V. mimicus. Lane 1: Molecular Marker (100 bp), lane 2: Negative control; Lane 3-12: positive isolates of ctx gene (301 bp). (B) Gel multiplex electrophoresis showing the ompU and toxR genes among isolates belonging to V. mimicus. Lane 1: Molecular Marker (100 bp), lane 2: Negative control; Lane 3, 4, 7 and 8: positive isolates of ompU gene (869 bp). Lane 5, 6, 9-11: positive isolates of toxR gene (779 bp). (C) Gel electrophoresis showing the VPI gene among isolates belonging to V. mimicus. Lane 1: Molecular Marker (100 bp), lane 2: Negative control; Lane 3-8: positive isolates of VPI gene (618 bp). (D) Gel electrophoresis showing the vcgC and vcgE gene among isolates belonging to V. vulnificus. Lane 1: Molecular Marker (100 bp), lane 2: Negative control; Lane 3-7: positive isolates of vcgC and vcgE gene (278 bp). (E) Gel electrophoresis showing the stn gene among isolates belonging to V. fluvialis. Lane 1: Molecular Marker (100 bp), lane 2: Negative control; Lane 3-8: positive isolates of stn gene (375 bp). (F) Gel multiplex electrophoresis showing the vfh and hupO gene among isolates belonging to V. fluvialis. Lane 1: Molecular Marker (100 bp), lane 2: Negative control; Lane 3-7: positive isolates of vfh gene (800 bp). Lane 8: positive isolates of hupO gene (600 bp).
Detection of Resistance Genes Among the Vibrio Species
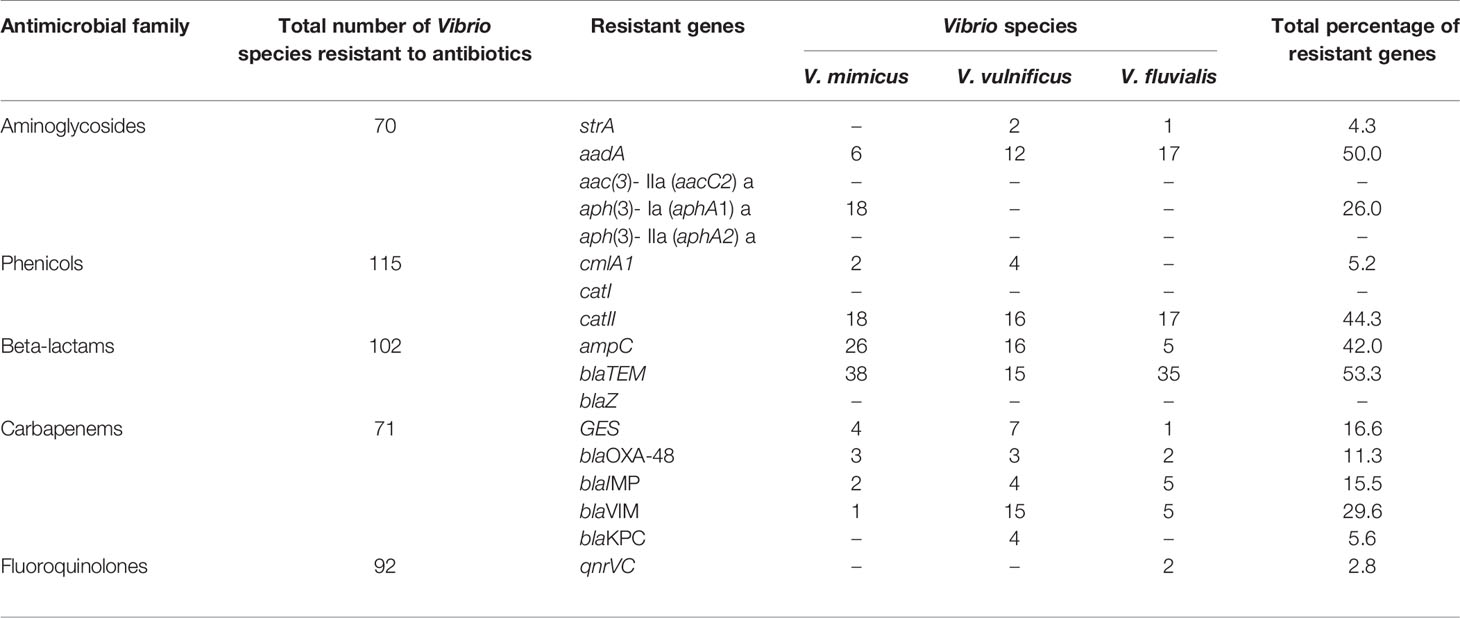
Five resistant genes encoding for aminoglycoside were examined in this study and three were detected of which aadA (50%) was observed as the most prevalent while strA was the least with a prevalence of 4.3%. V. mimicus was devoid of any strA gene. However, 26% of V. mimicus solely harboured aph(3)- Ia (aphA1)a gene among all the Vibrio isolates. For the phenicols, three resistance genes were detected in 5.2% among the confirmed Vibrio species. 44.3% of the Vibrio species harboured the catII gene while catI gene was not detected among the three Vibrio species studied. The ampC gene was identified in 42% of the Vibrio isolates, blaTEM in 53.3% of the Vibrio isolates, and no isolate harboured the blaZ gene. The frequency of resistance genes among the carbapenems class of antibiotics detected in the studied isolates varied from 5.6% for and 29.6%. The frequency of resistance genes with carbapenem was blaGES (16.6%), blaOXA-48 (11.3%), blaIMP (15%), blaVIM (29.6%) and blaKPC (5.6%). Among the resistance genes to fluoroquinolones assayed, only two isolates representing 2.8% harboured the qnrVC gene in all the isolates studied. The distribution of the resistance genes among the Vibrio resistance isolates is shown in Table 5. The gel representative pictures for the amplification of the resistance genes for aminoglycoside, phenicols, beta-lactams, carbapenems and fluoroquinolones are shown in Figure 5 respectively.
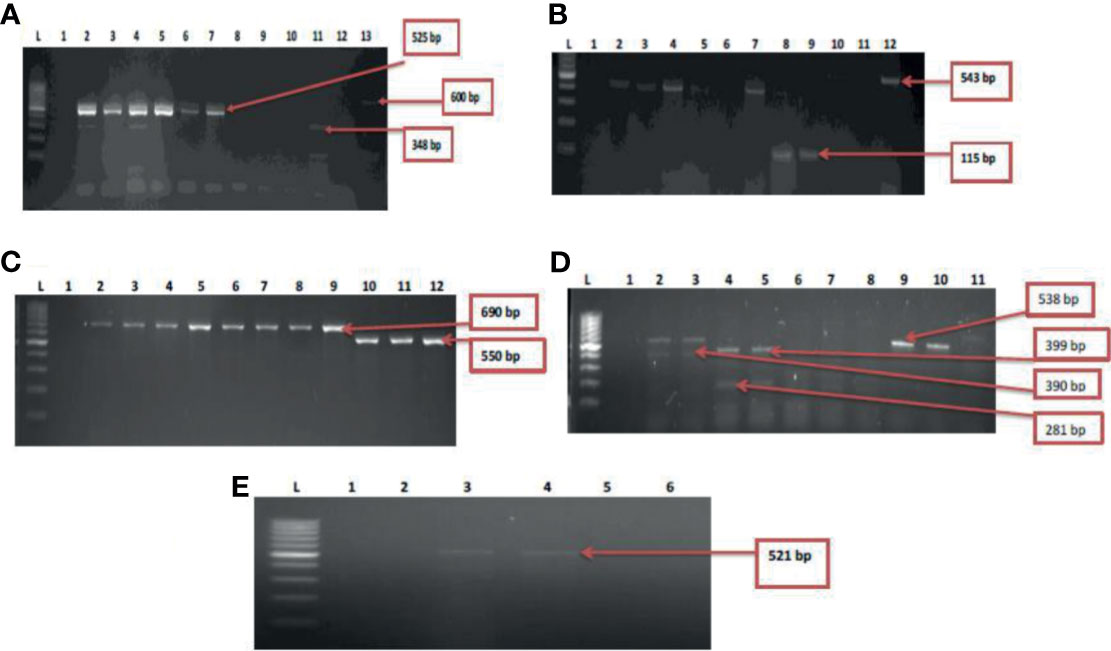
Figure 5 (A) = Gel multiplex electrophoresis showing the aphA1, aadA and strA genes among the Vibrio species. Lane L: Molecular Marker (100 bp), lane 1: Negative control; Lane 2-7: positive isolates of aadA gene (525 bp). Lane 11: positive isolate of strA gene (348 bp) and lane 13: positive isolate of aphA1 gene (600 bp), (B) = Gel multiplex electrophoresis showing the cmlA1 and catII genes among the Vibrio species. Lane L: Molecular Marker (100 bp), lane 1: Negative control; Lane 2-5, 7, 10 and 12: positive isolates of catII gene (543 bp). Lane 8 and 9: positive isolate of cm/A/gene (115 bp), (C) = Gel multiplex electrophoresis showing the blaTEM and ampC genes among the Vibrio species. Lane L: Molecular Marker (100 bp), lane 1: Negative control; Lane 2-9: positive isolates of blaTEM gene (690 bp). Lane 10-12: positive isolate of ampC gene (550 bp), (D) = Gel multiplex electrophoresis showing the blaKPC, blaGES, blaVIM and blaOXA-48 genes among the Vibrio species. Lane L: Molecular DNA-Marker (100 bp), lane 1: Negative control; Lane 2, 3, 9 and 10: positive isolates of blaKPC gene (543 bp). Lane 4 and 5: positive isolate of blaGES gene (399 bp). Lane 2 and 3: positive blaVIM isolates (390 bp). Lane 4 and 5: positive isolates of blaOXA-48 (281 bp), (E) = Gel electrophoresis showing the qnrVC genes among the Vibrio species. Lane L: Molecular DNA-marker (100 bp), lane 1: Negative control; Lane 3 and 4: positive isolates of qnrVC gene (521 bp).
Discussion
Water is considered as one of the basic necessities for the existence of all living organisms. South Africa is a semi-arid region which obtains minimal amount of rain annually. In lieu of this and with the incessant shortages of portable and clean water in the country, most inhabitants, especially in the rural areas have resorted to different alternatives for water sources such as river and dams for their routine domestic activities (Meissner et al., 2018). Additionally, the upsurge in various informal settlements around the country as well as the present poor conditions of some wastewater treatment plants have significantly contributed to the diminished quality of water sources found in most of the rural areas (Govender et al., 2011). Most hospital settings in the rural areas do not treat their effluents and are channelled with municipal wastewater prior (Edokpayi et al., 2017). Often times, compromised sewage pipelines leak their contents into the environment which eventually end up in adjourning streams and rivers.
Our study detected three Vibrio species (V. mimicus, V. fluvialis and V. vulnificus) from all sampling sites through molecular analyses. While culture-based techniques are relied upon for detecting microbial presence in various environments, however current molecular methods are comparatively more specific and sensitive than culture-based methods (Franco-Duarte et al., 2019). Specifically, the confirmation of Vibrio isolates from targeted sites in this study was achieved by molecular amplification of 16SrRNA gene followed by target genes hsp60, toxR and vmh for V. vulnificus, V. fluvialis and V. mimicus isolates. Previous studies have also employed similar molecular methods in the confirmation and delineation of Vibrio isolates into species (Kim et al., 2015; Okoh et al., 2015; Guardioala-Avila et al., 2016). Overall, Vibrio species recovered during this study were absent in two sampling sites (FHE and CHE) and could be Vibrio isolates belonging to other species not identified in this study. The presence of the Vibrio isolates from seven sampling sites is shown in Table 5. The QTE site had the highest recovery of the Vibrio isolates and poor treatment plant performance could have contributed to the recovery of the Vibrio isolates in the final effluent sample. The occurrence of pathogens in waterbodies is often due to the aftereffect from faecal contamination involving humans and animals (Penakalapati et al., 2017). The uncontrolled discharge of sewage as well as nonchalant management of wastewater treatment plants have been incriminated in water pollution of microbial origin especially around the rural settlements in South Africa (Teklehaimanot et al., 2015). The antibiotic sensitivity patterns among the Vibrio isolates recorded showed varying degrees of resistance pattern. All Vibrio species in the study exhibited more than 95% resistance to polymyxin B, chloramphenicol and nitrofurantoin. Results from our findings on the resistance pattern among the Vibrio isolates were slightly higher to those reported by Beshiru et al. (2020). In general, all confirmed Vibrio species from our study showed more than 85% resistance pattern to ampicillin. The carbapenems have been proven to be broad spectrum antibiotic and active against a diverse range of Gram positive and Gram negative bacteria (Papp-Wallace et al., 2011). From our study, a high susceptibility pattern was observed with the carbapenems (imipenem and meropenem) against V. mimicus and V. fluvialis. However, low sensitivity pattern was noticed among V. vulnificus isolates. Kanamycin also exhibited a sensitivity pattern of more than 50% with the test isolates and in strong agreement with previous report by Maje et al. (2020). A third generation antibiotic (cefuroxime) also showed a resistance pattern of more than 75% among the Vibrio isolates and is comparable with the findings by Manjusha and Sarita (2011). Ofloxacin also had a higher sensitivity pattern compared to ciprofloxacin but however, more isolates had an intermediate pattern of sensitivity with ciprofloxacin.
Multiple antibiotic resistance (MAR) analysis has been introduced to discern bacteria from sources using antimicrobial agents regularly approved for human therapy. In bacteria, MAR is most frequently associated with the presence of plasmids which harbours single or multiple resistance genes (Ruppé et al., 2015). Multiple antibiotic resistance patterns among the Vibrio species had a range of 0.5 to 0.8 while most of the isolates had a MARI of 0.8. This inferred that the Vibrio isolates originated from sources where antibiotic were greatly utilized.
Microbial virulence factors involve a wide range of molecules produced by pathogenic bacteria with increasing propensity to elude their host defence and cause infection. Also, some secretes microbial products which have the ability to gain entry into the host cell and utilize their mechanism assisting in the attainment of infection (Kitamoto et al., 2016). Virulence genes in Vibrio species have assisted in the initiation of infection ranging from diarrhoea, gastroenteritis, cholera, wound and blood infections (Schirmeister et al., 2014). It is commonly assumed that environmental bacterial strains are devoid of virulence traits that are present in clinical strains. Nonetheless, recent studies have reported that virulence genes, or their homologues could be found in strains from environmental sources and that acquiring such genes may have occurred in the aquatic environment (Persson et al., 2009). From this study, ten virulence genes were detected in varying frequencies among the Vibrio species isolated. The toxR virulence is made up of a set of genes encoding proteins that encourages internal colonization, toxin production and survival in the host cell coordinately expressed and dependent upon transcriptional activator (toxR) (Bina et al., 2003). The toxR gene was detected among V. mimicus isolates from two sampling sites (QTE and THR). This study affirms with previous report (Chen et al., 2011) detecting toxR gene in V.mimicus from sea foods. Zot gene was initially detected in Vibrio cholerae and encoded by ctx prophage while the core part of ctx gene comprises of many structural genes including zot genes which has significance in phage morphogenesis and assembly (Pant et al., 2020). The detection of ctx and zot genes from our study is comparable to the reports by Menezes et al. (2014). The presence of a zot gene in ctx-positive V. mimicus indicates a possible significant role of zot in the toxigenicity of the species. Vibrio pathogenicity island (VPI) gene and ompU (encoding a porin in the outer membrane of the bacteria) were also detected among some V. mimicus isolates. VPI has been attributed to be an essential virulence gene cluster for colonization of the human intestine as well as receptor for infection in most toxigenic Vibrio strains (Labbate et al., 2016). Detection of VPI and ompU genes among some V. mimicus in this study is in agreement with previous study by Guardiola-Avila et al. (2020). The detection of virulence correlated gene in this study among the V. vulnificus isolates can be distinguished apparently as clinical (vcgE) and environmental (vcgC) (Bier et al., 2015). Our finding is in accord with the report by Warner and Oliver (2008) who detected vcgE (47%) and vcgC (53%) from oysters’ isolates and water surrounding oysters harvest in North Carolina. Among the V. fluvialis isolates, three genes detected were stn (48%), hupO (14.6%) and vfh (19.5%). These factors have been reported to assist in haemolysin production which intensifies the pathogenic prowess of V. fluvialis and increase diarrhoea in infected patients (Liang et al., 2013). A similar trend was documented by Fri et al. (2017) that also identified similar virulence genes from a dusky farm and estuary in South Africa.
Antibiotic resistance is one of the imminent challenges to global health and with the current trends in escalating environmental contaminants such as antibiotic resistance genes; natural environments likely to be potential repositories for the spread of antibiotic resistance (Hernando-Amado et al., 2020). Aminoglycosides are potent antibiotics that function via the inhibition of protein synthesis (Wasserman et al., 2015). We found in this study that there was a wide range from relatively moderate to low and eventually no detection of aminoglycoside resistance genes among the Vibrio isolates. The detection of streptomycin resistance genes (aadA, strA) and gentamicin resistance genes (aphA2) from our study varied among the Vibrio species which corresponds with a previous study by Adefisoye and Okoh (2017). Chloramphenicol resistance genes was analysed among all Vibrio species showing phenotypic resistance to the antibiotic. Chloramphenicol resistance is attributed to the enzymatic inhibition of the drug facilitated by chloramphenicol acetyl-transferases (CAT) (Alcala et al., 2020). The detection of cat resistance genes among some Vibrio isolates from our study is akin with other studies (Yoo et al., 2003; Zhang et al., 2019; Håkonsholm et al., 2020). Beta lactams are the most broadly used class of antimicrobial agents, characterized by minimal toxicity and employed in the treatment of various bacterial ailments including those attributed with different Vibrio species (Bush and Bradford, 2016). Apparently, resistance among the beta-lactam drugs continues to be on the surge. The broad spectrum TEM-beta lactamase has been reported to be widely distributed in plasmids (Tooke et al., 2019). AmpC genes are situated chromosomally but nonetheless, plasmid associated variants among some bacteria have become increasingly recognized and disseminated via horizontal transfer (Lan et al., 2017). Our study detected the presence of ampC and blaTEM genes among the Vibrio species. Carbapenems introduction as broader antimicrobial spectrum are usually the last resort for most antimicrobial agents due to the proliferation of resistance among most antimicrobial classes. But resistance has been observed among the carbapenems. Our study detected five beta lactamase resistance genes to carbapenems. Detection of resistance genes from this study correlates with recent report (Elbadawi et al., 2021) identifying carbapenem resistance genes (blaOXA-48, blaIMP, blaVIM and blaKPC) among Gram negative isolates in Sudan. In addition, blaGES was identified among the three Vibrio species in our study. Documented reports have shown limited Vibrio carrying blaGES genes (Dahanayake et al., 2020; Hossain et al., 2020). These beta-lactam resistance genes are frequently identified in Enterobacteriaceae which portends concern on the impact of sewage in aquatic environment, giving rise to drug resistance bacteria and fortified selective pressure increasing the advent and dissemination of antimicrobial resistance (Fresia et al., 2019) The acquisition of these genes in Vibrio isolates in our study may have been through plasmid mediation in the aquatic environment from other bacteria strains. The addition of fluoride to quinolone creates fluoroquinolones with enhanced bactericidal activity. However, the plasmid- mediated quinolone resistance gene (qnrVC) was identified in two V. fluvialis isolates from our study and similar trend have been reported by Vinothkumar et al. (2016).
Conclusion
Our present study is a broad assessment of the antimicrobial susceptibility of various Vibrio species that were successfully recovered from aquatic and hospital environments. Detection of multiple drug resistance Vibrio species, isolates harbouring virulence genes as well as the detection of antimicrobial resistance genes suggests risk for public health concern. The maintenance of dysfunctional wastewater treatment plants as well as burst sewage pipes in rural settlements should be rectified to further prevent the leakages of sewage into the surrounding environments. More importantly, the promotion of proper antibiotic stewardships should be upheld in the society to forestall the continuous propagation and sustenance of antimicrobial resistance among bacteria population in nature.
Data Availability Statement
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
Author Contributions
UN, Conceptualization. OG and BI, methodology. OG and TD, formal analyses. NU, resources. OG and TD, writing—original draft preparation. OG, TD, BI, OO, AO, and UN, writing—review and editing. UN, supervision. All authors contributed to the article and approved the submitted version.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We are grateful to the South Africa Medical Research Council and the University of Fort Hare for financial support.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcimb.2021.732001/full#supplementary-material
References
Adefisoye, M. A., Okoh, A. I. (2017). Ecological and Public Health Implications of the Discharge of Multidrug-Resistant Bacteria and Physicochemical Contaminants From Treated Wastewater Effluents in the Eastern Cape, South Africa. Water 9 (8), 562. doi: 10.3390/w9080562
Alcala, A., Ramirez, G., Solis, A., Kim, Y., Tan, K., Luna, O., et al. (2020). Structural and Functional Characterization of Three Type B and C Chloramphenicol Acetyl-Transferases From Vibrio Species. Prot. Sci. 29 (3), 695–710. doi: 10.1002/pro.3793
Álvarez-Contreras, A. K., Quinones-Ramírez, E. I., Vázquez-Salinas, C. (2021). Prevalence, Detection of Virulence Genes and Antimicrobial Susceptibility of Pathogen Vibrio Species Isolated From Different Types of Seafood Samples at “La Nueva Viga” Market in Mexico City. Antonie van Leeuwenhoek, 1–13. doi: 10.1007/s10482-021-01591-x
APHA, A. (1998). Standard Methods for the Examination of Water and Wastewater Analysis (American Public Health Association).
Baker-Austin, C., Oliver, J. D., Alam, M., Ali, A., Waldor, M. K., Qadri, F., et al. (2018). Vibrio Spp. Infections. Nat. Rev. Dis. Primers 4 (1), 1–19. doi: 10.1038/s41572-018-0005-8
Banerjee, S. K., Farber, J. M. (2018). Trend and Pattern of Antimicrobial Resistance in Molluscan Vibrio Species Sourced to Canadian Estuaries. Antimicrob. Agents Chemother. 62 (10), e00799–e00718. doi: 10.1128/AAC.00799-18
Baquero, F., Martínez, J. L., Cantón, R. (2008). Antibiotics and Antibiotic Resistance in Water Environments. Curr. Opin. Biotechnol. 19 (3), 260–265. doi: 10.1016/j.copbio.2008.05.006
Beshiru, A., Okareh, O. T., Okoh, A. I., Igbinosa, E. O. (2020). Detection of Antibiotic Resistance and Virulence Genes of Vibrio Strains Isolated From Ready-to-Eat Shrimps in Delta and Edo States, Nigeria. J. Appl. Microbiol. 129 (1), 17–36. doi: 10.1111/jam.14590
Bier, N., Diescher, S., Strauch, E. (2015). Multiplex PCR for Detection of Virulence Markers of Vibrio Vulnificus. Lett. Appl. Microbiol. 60 (5), 414–420. doi: 10.1111/lam.12394
Bina, J., Zhu, J., Dziejman, M., Faruque, S., Calderwood, S., Mekalanos, J. (2003). ToxR Regulon of Vibrio Cholerae and its Expression in Vibrios Shed by Cholera Patients. Proc. Natl. Acad. Sci. 100 (5), 2801–2806. doi: 10.1073/pnas.2628026100
Binesse, J., Delsert, C., Saulnier, D., Champomier-Verges, M. C., Zagorec, M., Munier-Lehmann, H., et al. (2008). Metalloprotease Vsm Is the Major Determinant of Toxicity for Extracellular Products of Vibrio Splendidus. Appl. Environ. Microbiol. 74 (23), 7108–7117. doi: 10.1128/AEM.01261-08
Bouki, C., Venieri, D., Diamadopoulos, E. (2013). Detection and Fate of Antibiotic Resistant Bacteria in Wastewater Treatment Plants: A Review. Ecotoxicol. Environ. Saf. 91, 1–9. doi: 10.1016/j.ecoenv.2013.01.016
Bush, K., Bradford, P. A. (2016). β-Lactams and β-Lactamase Inhibitors: An Overview. Cold Spring Harbor Perspect. Med. 6 (8), a025247. doi: 10.1101/cshperspect.a025247
Chen, W., Yu, S., Zhang, C., Zhang, J., Shi, C., Hu, Y., et al. (2011). Development of a Single Base Extension-Tag Microarray for the Detection of Pathogenic Vibrio Species in Seafood. Appl. Microbiol. Biotechnol. 89 (6), 1979–1990. doi: 10.1007/s00253-010-2959-7
Clinical and Laboratory Standards Institute (2018). Performance Standards for Antimicrobial Susceptibility Testing Vol. supplement M100 (CLSI).
Dahanayake, P. S., Hossain, S., Wickramanayake, M. V. K. S., Heo, G. J. (2020). Prevalence of Virulence and Extended-Spectrum β-Lactamase (ESBL) Genes Harbouring Vibrio Spp. Isolated From Cockles (Tegillarca Granosa) Marketed in Korea. Lett. Appl. Microbiol. 71 (1), 61–69.
Das, B., Verma, J., Kumar, P., Ghosh, A., Ramamurthy, T. (2020). Antibiotic Resistance in Vibrio Cholerae: Understanding the Ecology of Resistance Genes and Mechanisms. Vaccine 38, A83–A92. doi: 10.1016/j.vaccine.2019.06.031
Edokpayi, J. N., Odiyo, J. O., Durowoju, O. S. (2017). Impact of Wastewater on Surface Water Quality in Developing Countries: A Case Study of South Africa. Water Qual., 401–416. doi: 10.5772/66561
Edokpayi, J. N., Odiyo, J. O., Msagati, T. A., Popoola, E. O. (2015). Removal Efficiency of Faecal Indicator Organisms, Nutrients and Heavy Metals From a Peri-Urban Wastewater Treatment Plant in Thohoyandou, Limpopo Province, South Africa. Int. J. Environ. Res. Pub. Health 12 (7), 7300–7320. doi: 10.3390/ijerph120707300
Elbadawi, H. S., Elhag, K. M., Mahgoub, E., Altayb, H. N., Ntoumi, F., Elton, L., et al. (2021). Detection and Characterization of Carbapenem Resistant Gram-Negative Bacilli Isolates Recovered From Hospitalized Patients at Soba University Hospital, Sudan. BMC Microbiol. 21 (1), 1–9.
Elmahdi, S., DaSilva, L. V., Parveen, S. (2016). Antibiotic Resistance of Vibrio Parahaemolyticus and Vibrio Vulnificus in Various Countries: A Review. Food Microbiol. 57, 128–134. doi: 10.1016/j.fm.2016.02.008
Fobosi, S. (2013). Rural Areas in the Eastern Cape Province, South Africa: The Right to Access Safe Drinking Water and Sanitation Denied. Polity Newspaper January 24.
Fouz, N., Pangesti, K. N., Yasir, M., Al-Malki, A. L., Azhar, E. I., Hill-Cawthorne, G. A., et al. (2020). The Contribution of Wastewater to the Transmission of Antimicrobial Resistance in the Environment: Implications of Mass Gathering Settings. Trop. Med. Infect. Dis. 5 (1), 33. doi: 10.3390/tropicalmed5010033
Franco-Duarte, R., Cernaková, L., Kadam, S., S Kaushik, K., Salehi, B., Bevilacqua, A., et al. (2019). Advances in Chemical and Biological Methods to Identify Microorganisms—From Past to Present. Microorganisms 7 (5), 130. doi: 10.3390/microorganisms7050130
Fresia, P., Antelo, V., Salazar, C., Giménez, M., D’Alessandro, B., Afshinnekoo, E., et al. (2019). Urban Metagenomics Uncover Antibiotic Resistance Reservoirs in Coastal Beach and Sewage Waters. Microbiome 7 (1), 1–9. doi: 10.1186/s40168-019-0648-z
Fri, J., Ndip, R. N., Njom, H. A., Clarke, A. M. (2017). Occurrence of Virulence Genes Associated With Human Pathogenic Vibrios Isolated From Two Commercial Dusky Kob (Argyrosmus Japonicus) Farms and Kareiga Estuary in the Eastern Cape Province, South Africa. Int. J. Environ. Res. Public Health 14 (10), 1111. doi: 10.3390/ijerph14101111
Gennari, M., Ghidini, V., Caburlotto, G., Lleo, M. M. (2012). Virulence Genes and Pathogenicity Islands in Environmental Vibrio Strains non-Pathogenic to Humans. FEMS Microbiol. Ecol. 82 (3), 563–573. doi: 10.1111/j.1574-6941.2012.01427.x
Govender, T., Barnes, J. M., Pieper, C. H. (2011). Contribution of Water Pollution From Inadequate Sanitation and Housing Quality to Diarrheal Disease in Low-Cost Housing Settlements of Cape Town, South Africa. Ameri. J. Pub. Health 101 (7), e4–e9. doi: 10.2105/AJPH.2010.300107
Guardiola-Avila, I., Acedo-Felix, E., Sifuentes-Romero, I., Yepiz-Plascencia, G., Gomez-Gil, B., Noriega-Orozco, L. (2016). Molecular and Genomic Characterization of Vibrio Mimicus Isolated From a Frozen Shrimp Processing Facility in Mexico. PloS One 11 (1), e0144885. doi: 10.1371/journal.pone.0144885
Guardiola-Avila, I., Martínez-Vázquez, V., Juárez-Rendón, K., Alvarez-Ainza, M., Paz-González, A., Rivera, G. (2020). Prevalence and Virulence of Vibrio Species Isolated From Raw Shrimp From Retail Markets in Reynosa, Mexico. Lett. Appl. Microbiol. 71 (3), 280–286. doi: 10.1111/lam.13315
Håkonsholm, F., Lunestad, B. T., Aguirre Sánchez, J. R., Martinez-Urtaza, J., Marathe, N. P., Svanevik, C. S. (2020). Vibrios From the Norwegian Marine Environment: Characterization of Associated Antibiotic Resistance and Virulence Genes. Microbiol. Open 9 (9), e1093. doi: 10.1002/mbo3.1093
Hernando-Amado, S., Coque, T. M., Baquero, F., Martínez, J. L. (2020). Antibiotic Resistance: Moving From Individual Health Norms to Social Norms in One Health and Global Health. Front. Microbiol. 11, 1914. doi: 10.3389/fmicb.2020.01914
Hossain, S., Wickramanayake, M. V. K. S., Dahanayake, P. S., Heo, G. J. (2020). Occurrence of Virulence and Extended-Spectrum β-Lactamase Determinants in Vibrio Spp. Isolated From Marketed Hard-Shelled Mussel (Mytilus Coruscus). Microb. Drug Res. 26 (4), 391–401.
Kim, H. J., Ryu, J. O., Lee, S. Y., Kim, E. S., Kim, H. Y. (2015). Multiplex PCR for Detection of the Vibrio Genus and Five Pathogenic Vibrio Species With Primer Sets Designed Using Comparative Genomics. BMC Microbiol. 15 (1). doi: 10.1186/s12866-015-0577-3
Kitamoto, S., Nagao-Kitamoto, H., Kuffa, P., Kamada, N. (2016). Regulation of Virulence: The Rise and Fall of Gastrointestinal Pathogens. J. Gastroenterol. 51 (3), 195–205. doi: 10.1007/s00535-015-1141-5
Labbate, M., Orata, F. D., Petty, N. K., Jayatilleke, N. D., King, W. L., Kirchberger, P. C., et al. (2016). A Genomic Island in Vibrio Cholerae With VPI-1 Site-Specific Recombination Characteristics Contains CRISPR-Cas and Type VI Secretion Modules. Sci. Rep. 6 (1), 1–7. doi: 10.1038/srep36891
Lan, N. P. H., Hien, N. H., Phuong, T. L. T., Thanh, D. P., Thieu, N. T. V., Ngoc, D. T. T., et al. (2017). Phenotypic and Genotypic Characteristics of ESBL and AmpC Producing Organisms Associated With Bacteraemia in Ho Chi Minh City, Vietnam. Antimicrob. Res. Infect. Cont. 6 (1), 1–9. doi: 10.1186/s13756-017-0265-1
Liang, P., Cui, X., Du, X., Kan, B., Liang, W. (2013). The Virulence Phenotypes and Molecular Epidemiological Characteristics of Vibrio Fluvialis in China. Gut Pathog. 5 (1), 1–11. doi: 10.1186/1757-4749-5-6
Lundborg, C. S., Tamhankar, A. J. (2017). Antibiotic Residues in the Environment of South East Asia. Bmj 358. doi: 10.1136/bmj.j2440
Maje, M. D., Kaptchouang Tchatchouang, C. D., Manganyi, M. C., Fri, J., Ateba, C. N. (2020). Characterization of Vibrio Species From Surface and Drinking Water Sources and Assessment of Biocontrol Potentials of Their Bacteriophages. Int. J. Microbiol. 2020, 8863370. doi: 10.1155/2020/8863370
Manjusha, S., Sarita, G. B. (2011). Plasmid Associated Antibiotic Resistance in Vibrios Isolated From Coastal Waters of Kerala. Int. Food Res. J. 18 (3).
Manyi-Loh, C., Mamphweli, S., Meyer, E., Okoh, A. (2018). Antibiotic Use in Agriculture and Its Consequential Resistance in Environmental Sources: Potential Public Health Implications. Molecules 23 (4), 795. doi: 10.3390/molecules23040795
Mapipa, Q., Digban, T. O., Nnolim, N. E., Nwodo, U. U. (2021). Antibiogram Profile and Virulence Signatures of Pseudomonas Aeruginosa Isolates Recovered From Selected Agrestic Hospital Effluents. Sci. Rep. 11 (1), 1–11.
Meissner, R., Steyn, M., Moyo, E., Shadung, J., Masangane, W., Nohayi, N., et al. (2018). South African Local Government Perceptions of the State of Water Security. Environ. Sci. Policy 87, 112–127. doi: 10.1016/j.envsci.2018.05.020
Menezes, F.G.R. de, Soraya da Silva, N., Oscarina Viana de, S., Candida Machado Vieira Maia, V.-N., Rodrigo, M., Grace Nazareth, D. T., et al. (2014). Detection of Virulence Genes in Environmental Strains of Vibrio Cholerae From Estuaries in Northeastern Brazil. Rev. Inst. Med. Trop.de São Paulo 565, 427–432. doi: 10.1590/S0036-46652014000500010
Okoh, A. I., Sibanda, T., Nongogo, V., Adefisoye, M., Olayemi, O. O., Nontongana, N. (2015). Prevalence and Characterization of Non-Cholerae Vibrio Spp. In Final Effluents of Wastewater Treatment Facilities in Two Districts of the Eastern Cape Province of South Africa: Implications for Public Health. Environ. Sci. Pollut. Res. 22 (3), 2008–2017. doi: 10.1007/s11356-014-3461-z
Oliveira, P. H., Touchon, M., Cury, J., Rocha, E. P. (2017). The Chromosomal Organization of Horizontal Gene Transfer in Bacteria. Nat. Communi. 8 (1), 1–11. doi: 10.1038/s41467-017-00808-w
Osunla, C. A., Okoh, A. I. (2017). Vibrio Pathogens: A Public Health Concern in Rural Water Resources in Sub-Saharan Africa. Inter. J. Env. Res. Pub. Health 14 (10), 1188. doi: 10.3390/ijerph14101188
Pant, A., Das, B., Bhadra, R. K. (2020). CTX Phage of Vibrio Cholerae: Genomics and Applications. Vaccine 38, A7–A12. doi: 10.1016/j.vaccine.2019.06.034
Papp-Wallace, K. M., Endimiani, A., Taracila, M. A., Bonomo, R. A. (2011). Carbapenems: Past, Present, and Future. Antimicrob. Agents Chemother. 55 (11), 4943–4960. doi: 10.1128/AAC.00296-11
Penakalapati, G., Swarthout, J., Delahoy, M. J., McAliley, L., Wodnik, B., Levy, K., et al. (2017). Exposure to Animal Faeces and Human Health: A Systematic Review and Proposed Research Priorities. Environ. Sci. Technol. 51 (20), 11537–11552. doi: 10.1021/acs.est.7b02811
Persson, O. P., Pinhassi, J., Riemann, L., Marklund, B. I., Rhen, M., Normark, S., et al. (2009). High Abundance of Virulence Gene Homologues in Marine Bacteria. Environ. Microbiol. 11 (6), 1348–1357. doi: 10.1111/j.1462-2920.2008.01861.x
Ramamurthy, T., Chowdhury, G., Pazhani, G. P., Shinoda, S. (2014). Vibrio Fluvialis: An Emerging Human Pathogen. Front. Microbiol. 5, 91. doi: 10.3389/fmicb.2014.00091
Ramírez-Castillo, F. Y., Loera-Muro, A., Jacques, M., Garneau, P., Avelar-González, F. J., Harel, J., et al. (2015). Waterborne Pathogens: Detection Methods and Challenges. Pathogens 4 (2), 307–334. doi: 10.3390/pathogens4020307
Rizzo, L., Manaia, C., Merlin, C., Schwartz, T., Dagot, C., Ploy, M. C., et al. (2013). Urban Wastewater Treatment Plants as Hotspots for Antibiotic Resistant Bacteria and Genes Spread Into the Environment: A Review. Sci. Total Environ. 447, 345–360. doi: 10.1016/j.scitotenv.2013.01.032
Ruppé, É., Woerther, P. L., Barbier, F. (2015). Mechanisms of Antimicrobial Resistance in Gram-Negative Bacilli. Ann. Intensive Care 5 (1), 1–15. doi: 10.1186/s13613-015-0061-0
Schirmeister, F., Dieckmann, R., Bechlars, S., Bier, N., Faruque, S. M., Strauch, E. (2014). Genetic and Phenotypic Analysis of Vibrio Cholerae Non-O1, Non-O139 Isolated From German and Austrian Patients. Eur. J. Clin. Microbiol. Infect. Dis. 33 (5), 767–778. doi: 10.1007/s10096-013-2011-9
Singer, A. C., Shaw, H., Rhodes, V., Hart, A. (2016). Review of Antimicrobial Resistance in the Environment and its Relevance to Environmental Regulators. Front. Microbiol. 7, 1728. doi: 10.3389/fmicb.2016.01728
Singh, D. V., Sree Renjini, I., Colwell, R. R. (2002). Development of a Hexaplex PCR Assay for Rapid Detection of Virulence and Regulatory Genes in Vibrio Cholerae and Vibrio Mimicus. J. Clin. Microbiol. 40 (11), 4321–4324. doi: 10.1128/JCM.40.11.4321-4324.2002
Takemura, A. F., Chien, D. M., Polz, M. F. (2014). Associations and Dynamics of Vibrionaceae in the Environment, From the Genus to the Population Level. Front. Microbiol. 5, 38. doi: 10.3389/fmicb.2014.00038
Teklehaimanot, G. Z., Genthe, B., Kamika, I., Momba, M. N. B. (2015). Prevalence of Enteropathogenic Bacteria in Treated Effluents and Receiving Water Bodies and Their Potential Health Risks. Sci. Total Environ. 518, 441–449. doi: 10.1016/j.scitotenv.2015.03.019
Tooke, C. L., Hinchliffe, P., Bragginton, E. C., Colenso, C. K., Hirvonen, V. H., Takebayashi, Y., et al. (2019). β-Lactamases and β-Lactamase Inhibitors in the 21st Century. J. Mol. Biol. 431 (18), 3472–3500. doi: 10.1016/j.jmb.2019.04.002
Tzanakakis, V. A., Paranychianakis, N. V., Angelakis, A. N. (2020). Water Supply and Water Scarcity. Water 12 (9), 2347.
Vinothkumar, K., Kumar, G. N., Bhardwaj, A. K. (2016). Characterization of Vibrio Fluvialis Qnrvc5 Gene in Native and Heterologous Hosts: Synergy of Qnrvc5 With Other Determinants in Conferring Quinolone Resistance. Front. Microbiol. 7, 146. doi: 10.3389/fmicb.2016.00146
Voulvoulis, N. (2018). Water Reuse From a Circular Economy Perspective and Potential Risks From an Unregulated Approach. Cur. Opin. Environ. Sci. Health 2, 32–45. doi: 10.1016/j.coesh.2018.01.005
Wall, K. (2018). The Evolution of South African Wastewater Effluent Parameters and Their Regulation: A Brief History of the Drivers, Institutions, Needs and Challenges. TD: J. Transdiscipl. Res. South Afr. 14 (1), 1–11. doi: 10.4102/td.v14i1.581
Warner, E., Oliver, J. D. (2008). Population Structures of Two Genotypes of Vibrio Vulnificus in Oysters (Crassostrea Virginica) and Seawater. Appl. Environ. Microbiol. 74 (1), 80. doi: 10.1128/AEM.01434-07
Wasserman, M. R., Pulk, A., Zhou, Z., Altman, R. B., Zinder, J. C., Green, K. D., et al. (2015). Chemically Related 4, 5-Linked Aminoglycoside Antibiotics Drive Subunit Rotation in Opposite Directions. Nat. Commun. 6 (1), 1–12. doi: 10.1038/ncomms8896
Yoo, M. H., Huh, M. D., Kim, E. H., Lee, H. H., Do Jeong, H. (2003). Characterization of Chloramphenicol Acetyltransferase Gene by Multiplex Polymerase Chain Reaction in Multidrug-Resistant Strains Isolated From Aquatic Environments. Aquaculture 217 (1-4), 11–21. doi: 10.1016/S0044-8486(02)00169-2
Zhang, G., Sun, K., Ai, G., Li, J., Tang, N., Song, Y., et al. (2019). A Novel Family of Intrinsic Chloramphenicol Acetyltransferase CATC in Vibrio Parahaemolyticus: Naturally Occurring Variants Reveal Diverse Resistance Levels Against Chloramphenicol. Int. J. Antimicrob. Agents 54 (1), 75–79. doi: 10.1016/j.ijantimicag.2019.03.012
Keywords: antibiotic resistance, Vibrio isolates, virulence genes, MARI, South Africa
Citation: Gxalo O, Digban TO, Igere BE, Olapade OA, Okoh AI and Nwodo UU (2021) Virulence and Antibiotic Resistance Characteristics of Vibrio Isolates From Rustic Environmental Freshwaters. Front. Cell. Infect. Microbiol. 11:732001. doi: 10.3389/fcimb.2021.732001
Received: 28 June 2021; Accepted: 31 July 2021;
Published: 19 August 2021.
Edited by:
Yang Zhang, University of Pennsylvania, United StatesReviewed by:
Hong Zhang, Johns Hopkins University, United StatesHao Zhang, University of Pennsylvania, United States
Yu Zhou, Institut Pasteur of Shanghai (CAS), China
Copyright © 2021 Gxalo, Digban, Igere, Olapade, Okoh and Nwodo. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Uchechukwu U. Nwodo, VU53b2RvQHVmaC5hYy56YQ==
 Oyama Gxalo1,2
Oyama Gxalo1,2 Tennison O. Digban
Tennison O. Digban Ola A. Olapade
Ola A. Olapade Anthony I. Okoh
Anthony I. Okoh Uchechukwu U. Nwodo
Uchechukwu U. Nwodo