- 1State Key Laboratory of Oral Diseases & National Clinical Research Center for Oral Diseases & Department of Cariology and Endodontics, West China Hospital of Stomatology, Sichuan University, Chengdu, China
- 2Department of Preventive Dentistry, Academic Center for Dentistry Amsterdam (ACTA), University of Amsterdam and Vrije Universiteit Amsterdam, Amsterdam, Netherlands
The development of periodontitis is associated with an imbalanced subgingival microbial community enriched with species such as the traditionally classified red-complex bacteria (Porphyromonas gingivalis, Tannerella forsythia, and Treponema denticola). Saliva has been suggested as an alternative to subgingival plaque for the microbial analysis due to its easy and non-invasive collection. This systematic review aims to determine whether the levels of red-complex bacteria assessed using saliva reflect those in subgingival plaque from periodontitis patients. The MEDLINE, EMBASE, and Cochrane Library databases were searched up to April 30, 2021. Studies were considered eligible if microbial data of at least one of the red-complex species were reported in both saliva and subgingival plaque from periodontitis patients, based on DNA-based methods. Of the 17 included studies, 4 studies used 16S rRNA gene sequencing techniques, and the rest used PCR-based approaches. The detection frequency of each red-complex species in periodontitis patients was reported to be > 60% in most studies, irrespective of samples types. Meta-analyses revealed that both detection frequencies and relative abundances of red-complex bacteria in saliva were significantly lower than those in subgingival plaque. Moreover, the relative abundances of all 3 bacterial species in saliva showed significantly positive correlation with those in subgingival plaque. In conclusion, current evidence suggests that one-time saliva sampling cannot replace subgingival plaque for microbial analysis of the red-complex bacteria in periodontitis patients. Given the positive microbial associations between saliva and subgingival plaque, a thorough review of longitudinal clinical studies is needed to further assess the role of saliva.
1 Introduction
Periodontitis is one of the most prevalent oral infectious diseases, affecting over 740 million people worldwide (Kassebaum et al., 2014). It is a chronic inflammation, associated with dysbiotic subgingival biofilms, resulting in progressive and irreversible destruction of tooth supporting tissues (Vieira Colombo et al., 2016; Tonetti et al., 2017). Although it is not clear whether dysbiotic biofilms initiate the disease or are a consequence of the disease, it is well established that at diseased status, the subgingival microbiota is enriched with gram-negative, proteolytic bacteria, while in healthy situation, the microbiota is mainly composed of gram-positive bacteria (Curtis et al., 2020; Van Dyke et al., 2020). A recent review collected existing evidence and proposed an “Inflammation-Mediated Polymicrobial-Emergence and Dysbiotic-Exacerbation” (IMPEDE) model, which included the microbial element and complemented the current clinical classification of periodontitis (Van Dyke et al., 2020). According to this model, local inflammation drives an initial shift in microbial composition and the formation of periodontal pocket exacerbates this microbial shift by further enriching disease-associated species.
To determine the compositional shift of periodontitis-related microbiota, subgingival plaque obtained from diseased pockets has been considered as the most representative sample. However, collecting subgingival plaque is invasive and requires specialized training for proper sampling. Moreover, reports have shown that the quality and quantity of collected plaque samples may be greatly influenced by the collection methods (Renvert et al., 1992; van der Horst et al., 2013). Compared to subgingival plaque, saliva is much better accessible and can be collected noninvasively in larger quantity. Since the collection of saliva does not require special sampling tools, sample quality could be less influenced by sampling methods or operators. Saliva has been proposed as an alternative to subgingival plaque for studying the association between oral microbes and periodontal disease since 1998 (Umeda et al., 1998). It was hypothesized that microbes residing in a periodontal pocket could be spread, washed out or spilt over into saliva (Haririan et al., 2014; Li et al., 2015). Multiple clinical studies have compared the levels of important periodontal microbes using samples collected from subgingival pockets and saliva, but the results were inconsistent. For example, Umeda et al. (1998) reported that Porphyromonas gingivalis and Treponema denticola were detected more often in saliva than in subgingival plaque samples, whereas Nickles et al. (2017) reported the opposite. Moreover, the open-ended DNA sequencing techniques developed in the past decades have revealed that different niches in the oral cavity harbor considerably different microbial communities with distinct microbial composition (Huttenhower et al., 2012; Mark Welch et al., 2019). Differential microbial profiles of saliva and subgingival plaque have been demonstrated by several studies (Segata et al., 2012; Simón-Soro et al., 2013). Therefore, a systematic review that includes recent studies using sequencing techniques is needed to objectively assess microbial compositions of saliva and subgingival plaque.
So far, the most frequently examined microbes in clinical samples obtained from periodontitis patients are P. gingivalis, Tannerella forsythia and T. denticola. These 3 species were grouped as red-complex bacteria in 1998 (Socransky et al., 1998) based on the evidence that they were frequently isolated together and were strongly associated with periodontitis. Members of the red-complex group have been the main target in many clinical studies which performed the comparison between saliva and subgingival plaque samples. In the past, techniques employed to examine the levels of these bacteria were mainly targeted approaches, such as bacterial culture, immunological assays, PCR and quantitative real-time PCR (qPCR) (Suchett-Kaye et al., 2001). Among them, PCR and qPCR were used most frequently, since these techniques have better sensitivity and specificity as compared to other methods (Loesche, 1992; Boutaga et al., 2003). In addition, the abovementioned 16S rRNA gene sequencing techniques have been increasingly applied in clinical studies (Li et al., 2015; Belstrøm et al., 2017), since they offer the possibility to profile the entire microbiota besides specific bacterial species of interest.
This systematic review aimed to evaluate clinical evidence on the levels of red-complex bacteria in saliva and subgingival plaque samples collected from patients with periodontitis. We focused on studies which used DNA-based (targeted and open-ended) methods for microbial identification.
2 Materials and Methods
This systematic review was performed following the Preferred Reporting Items for Systematic Reviews and Meta-analysis (PRISMA) statement (Moher et al., 2009), and was registered at the National Institute for Health Research PROSPERO, International Prospective Register of Systematic Reviews (registration number: CRD42020219510).
2.1 Search Strategy
The MEDLINE (via PubMed), EMBASE, and Cochrane Library databases were searched up to April 30, 2021 by two independent researchers (YJ and BS), using the search strategy described in Supplementary Table S1. In addition, a manual search of the reference list of the included studies was conducted.
2.2 Study Selection
Studies that met the following criteria were included: 1) population: humans with periodontitis; 2) exposure: saliva samples; 3) comparison: subgingival plaque samples; 4) outcome: microbial data of at least one of the red-complex bacteria that was obtained using a DNA-based method; 5) study design: clinical studies of any design, except case report and case series.
Exclusion criteria were: 1) studies without periodontal diagnosis of the participants; 2) studies including participants who had an explicit diagnosis of any systemic disease or systemic condition, such as pregnancy; 3) studies including participants who used medication (e.g., antibiotics) or the medication status of participants was not mentioned; 4) studies with no baseline data in case of a prospective or interventional design; 5) not full-text publications (e.g., conference abstracts); 6) studies published in languages other than English. In addition, publications with overlapped data were identified at the phase of full-text screening. These studies were conducted by the same group of authors, and the data (a part or all) reported in different publications were obtained from the same group of subjects. In order to avoid duplicate data extraction, only the publication on a larger number of subjects was included.
The selection of studies was performed in two steps based on the above inclusion and exclusion criteria. In the first step, articles were screened on the basis of title and abstract, using a Web platform (rayyan.qcri.org) (Ouzzani et al., 2016). In the second step, the selected studies underwent full-text evaluation.
2.3 Data Extraction and Methodological Quality Assessment
A customized data extraction form was used to collect the following information from each included study: 1) characteristics of the study (e.g., author, year of publication, and study location); 2) characteristics of the participants (e.g., number, periodontal diagnosis, and clinical parameters); 3) methodological features of the study (e.g., method of saliva and subgingival plaque sample collection, method to evaluate red-complex bacteria); 4) microbial outcomes. For prospective and interventional studies, only data from the baseline measurement were extracted for analysis. When needed, the corresponding author(s) were contacted for the missing data.
The methodological qualities of all included studies were assessed using the 8-item “Critical Appraisal Checklist for Analytical Cross-Sectional Studies” by the Joanna Briggs Institute (JBI) (Moola et al., 2020). The answer to each question was “Yes”, “No”, or “Unclear”, and an overall rating score was given to each study which equals to the total number of “Yes” answers given, ranging from 0 to 8. The scores of 0–3, 4–5, and 6–8 were classified as low, medium, and high quality of studies, respectively (Yazdanian et al., 2020).
The steps mentioned above, including study selection, data extraction and methodological quality assessment were conducted independently by two researchers (YJ and BS), and any disagreement was resolved through discussion. If disagreement persisted, another researcher (DD) was consulted to achieve consensus.
2.4 Summary Outcome Measures and Statistical Analysis
Based on the results of included studies, we summarized 3 types of microbial outcomes: detection frequency, bacterial count and/or relative abundance of each red-complex bacteria.
Detection frequency refers to the percentage of subjects positive for a specific microorganism. It was reported in studies using either targeted PCR-based approaches or 16S rRNA gene sequencing techniques. Bacterial count provides the (semi-) quantity of a specific microorganism in a sample. This outcome parameter was reported in the studies using qPCR techniques. For studies using 16S rRNA gene sequencing techniques, two types of data were extracted: detection frequency and relative abundance.
Meta-analyses were performed to assess the statistical differences in the detection frequency and relative abundance of each red-complex species between saliva and subgingival plaque samples, using RevMan software version 5.4 (The Cochrane Collaboration; Copenhagen, Denmark). Heterogeneity among studies was assessed by chi-squared test and inconsistency index I2. Values of I2 < 30%, 30%–60%, and > 60% were considered as low, moderate and large heterogeneity, respectively (Higgins et al., 2003). When the heterogeneity was significant (p < 0.1), a random-effects model (DerSimonian-Laird method) was applied to examine the overall effect; otherwise, a fixed-effects model (Mantel-Haenszel method) was used. The odds ratio (OR) for detection frequency and mean difference (MD) for relative abundance were calculated at 95% confidence intervals (CI). Differences were considered statistically significant if p < 0.05.
3 Results
3.1 Results of Search and Study Selection
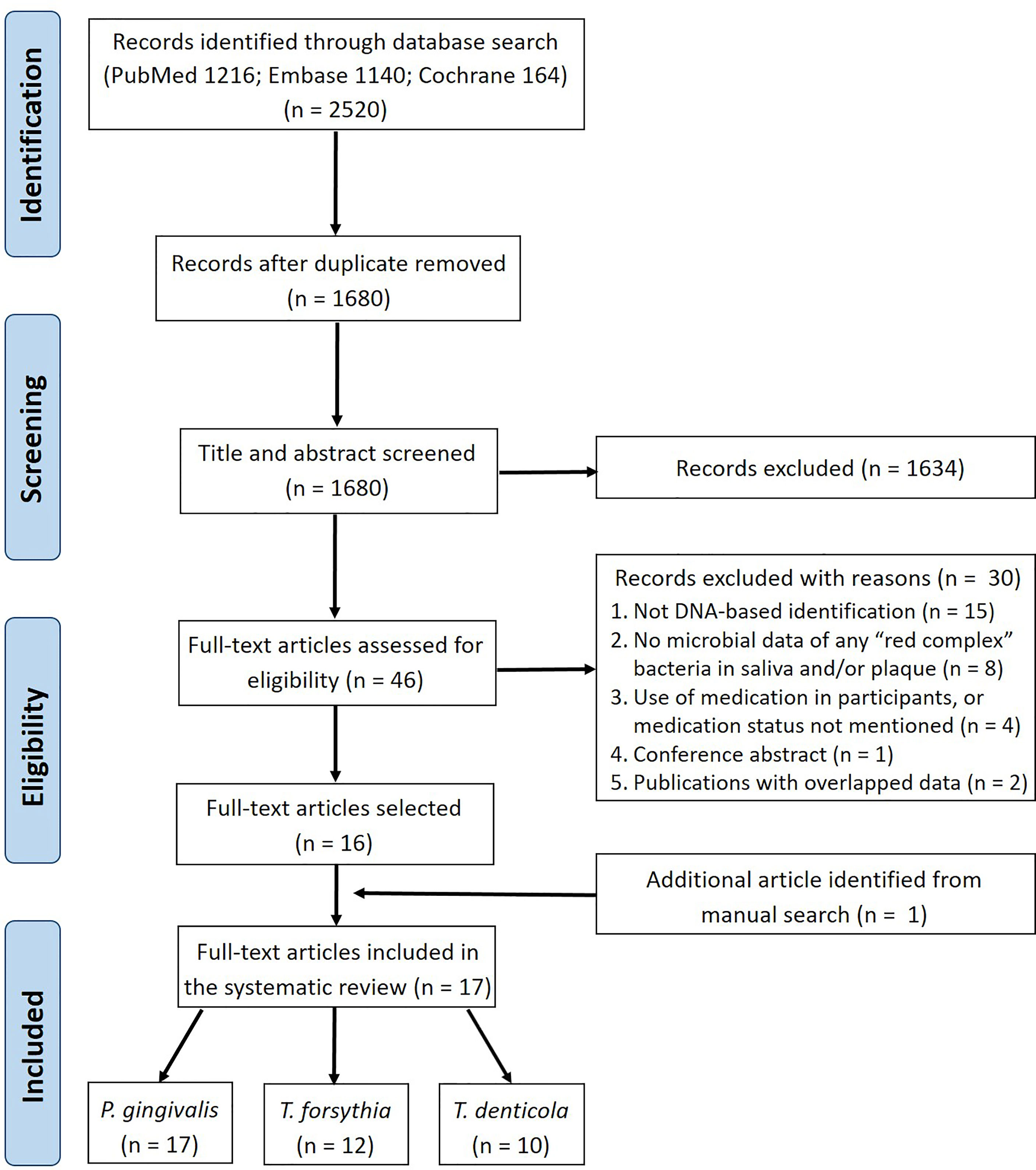
The literature search of three electronic databases identified 2520 records in total (Figure 1). After removing duplicates, 1680 articles were retained for title and abstract screening, from which 1634 articles were excluded and the remaining 46 were assessed further in full-text reading. Of these 46 studies, 30 were excluded based on the eligibility criteria (Supplementary Table S2). One study (Umeda et al., 1998) was identified additionally from the manual search of the reference lists of the selected studies. Therefore, 17 studies were finally included in this systematic review.
3.2 Quality Assessment
Figure 2 shows the methodological quality assessment of the included studies, which presented the answer to each appraisal criteria as well as an overall rating of each study. Nine out of 17 studies had high quality, 8 studies had medium quality and no study had low quality.
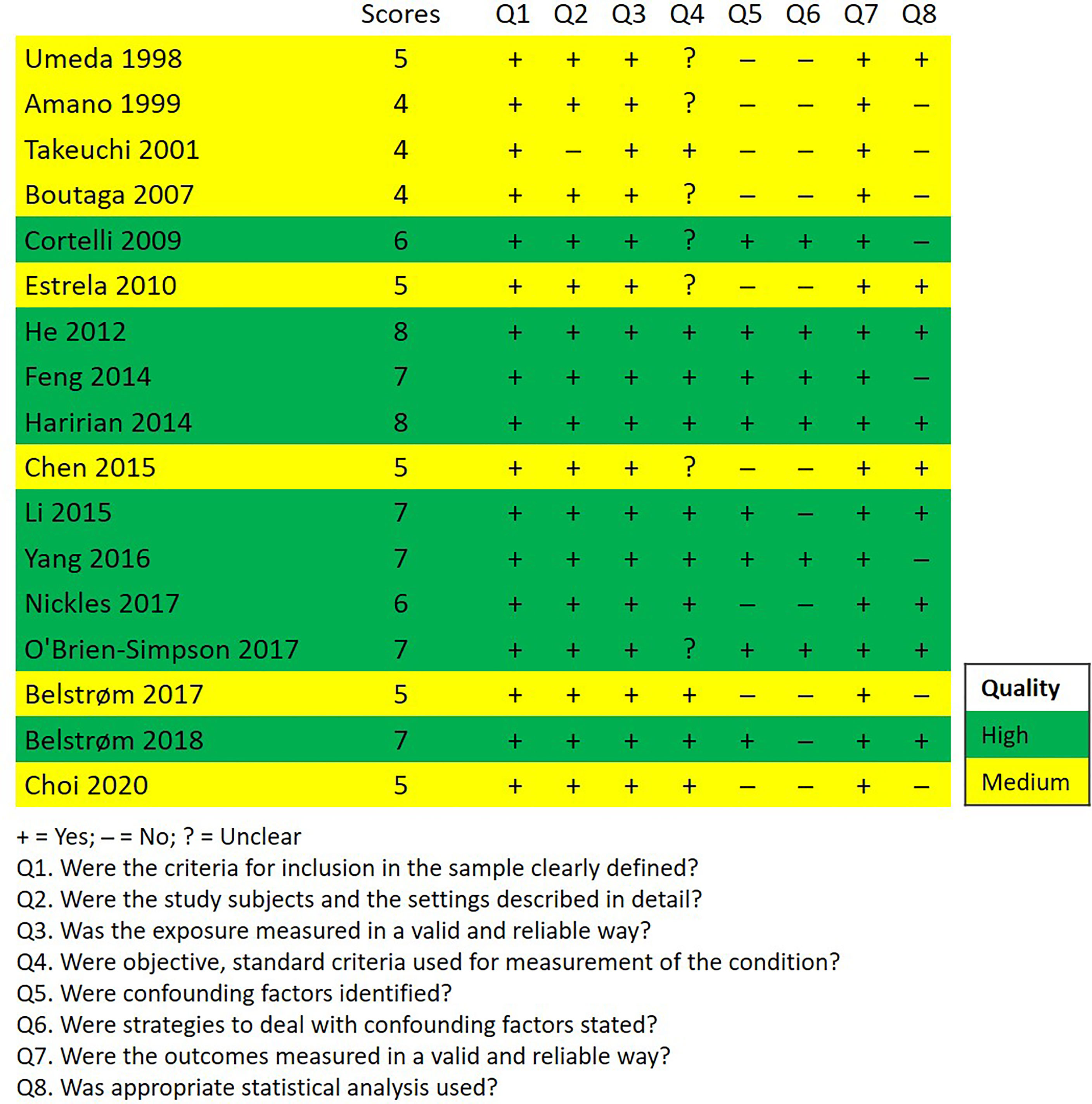
Figure 2 Quality assessment of the included studies according to the JBI Critical Appraisal Checklist for Analytical Cross-Sectional Studies.
3.3 Characteristics of the Included Studies
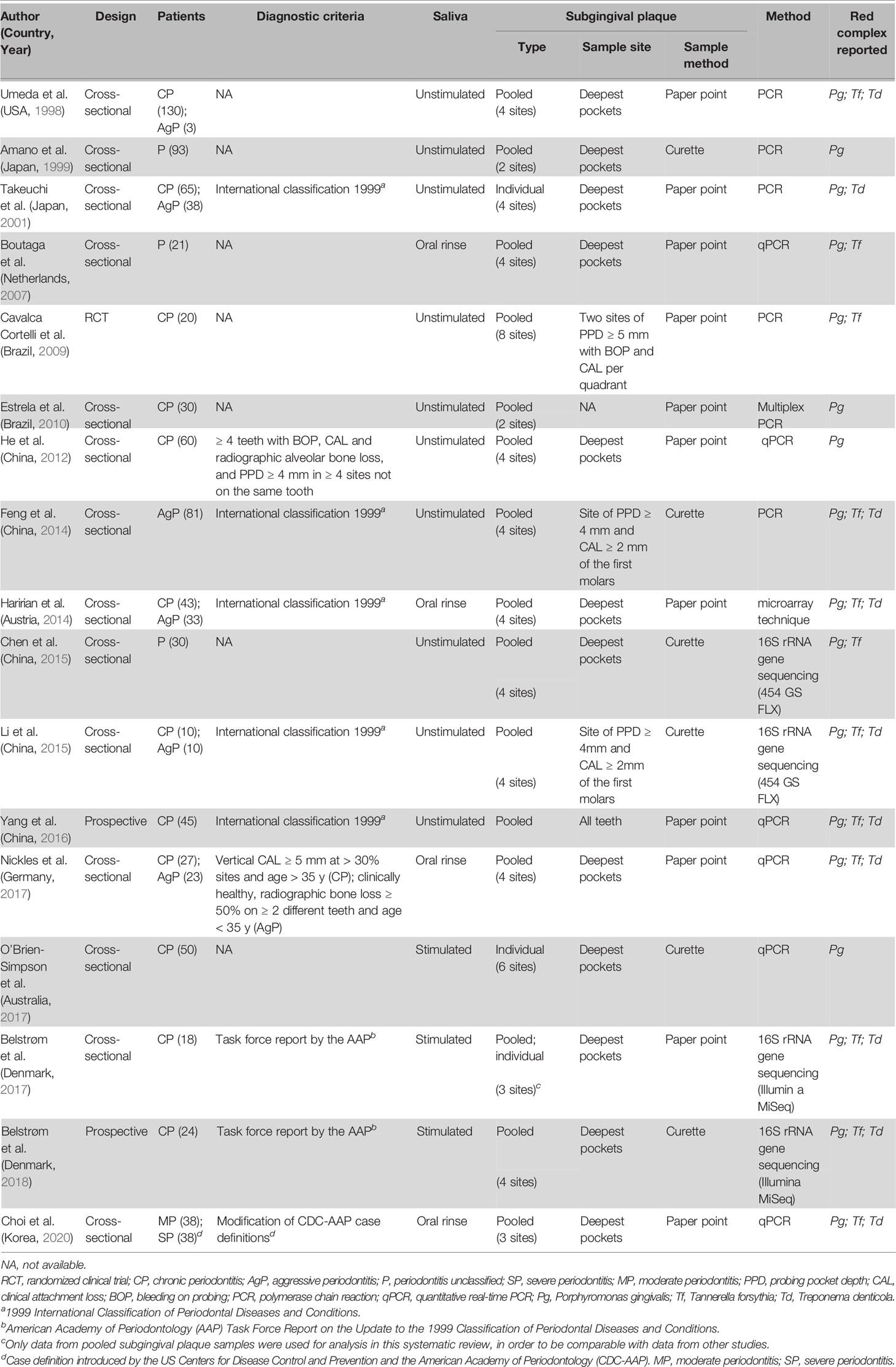
The main characteristics of the included studies are presented in Table 1. Most studies had a cross-sectional design (n = 14); the rest had a prospective (n = 2) or interventional (n = 1) design.
Out of the 17 studies, 14 studies reported the classification of periodontitis: 7 studies included patients with chronic periodontitis (CP) only, 1 study with aggressive periodontitis (AgP) only, and 5 studies with both CP and AgP. Different from the other 13 studies, Choi et al. (2020) classified the disease as moderate and severe periodontitis. Among these 14 studies, only 10 studies specified their diagnostic criteria: 5 studies (Takeuchi et al., 2001; Feng et al., 2014; Haririan et al., 2014; Li et al., 2015; Yang et al., 2016) followed the criteria of the 1999 International Classification of Periodontal Diseases and Conditions (Armitage, 1999), 2 studies (Belstrøm et al., 2017; Belstrøm et al., 2018) used the task force report by the American Academy of Periodontology (Geurs, 2015), 1 study (Choi et al., 2020) used a criteria modified from the case definition by the US Centers for Disease Control and Prevention and the American Academy of Periodontology (Page and Eke, 2007), and 2 studies (He et al., 2012; Nickles et al., 2017) used self-defined criteria. Three studies (Amano et al., 1999; Boutaga et al., 2007; Chen et al., 2015) did not specify the type of periodontitis and the diagnostic criteria.
For sample collection, saliva was collected as unstimulated (10 studies), stimulated (3 studies) or oral rinse sample (4 studies); subgingival plaque was collected by paper point in 11 studies and by curette in 6 studies. Most studies (n = 14) analyzed subgingival plaque samples pooled from multiple periodontal pockets (2 to full mouth). Only 2 studies analyzed plaque samples from individual pocket (Takeuchi et al., 2001; O’Brien-Simpson et al., 2017) and 1 study analyzed both pooled and individual plaque samples (Belstrøm et al., 2017).
With regard to the methods used for microbial identification, 4 studies used open-ended 16S rRNA gene sequencing techniques (Chen et al., 2015; Li et al., 2015; Belstrøm et al., 2017; Belstrøm et al., 2018), and the remaining 13 studies used targeted PCR-based approaches, including PCR, qPCR, and microarray techniques.
3.4 Clinical Data
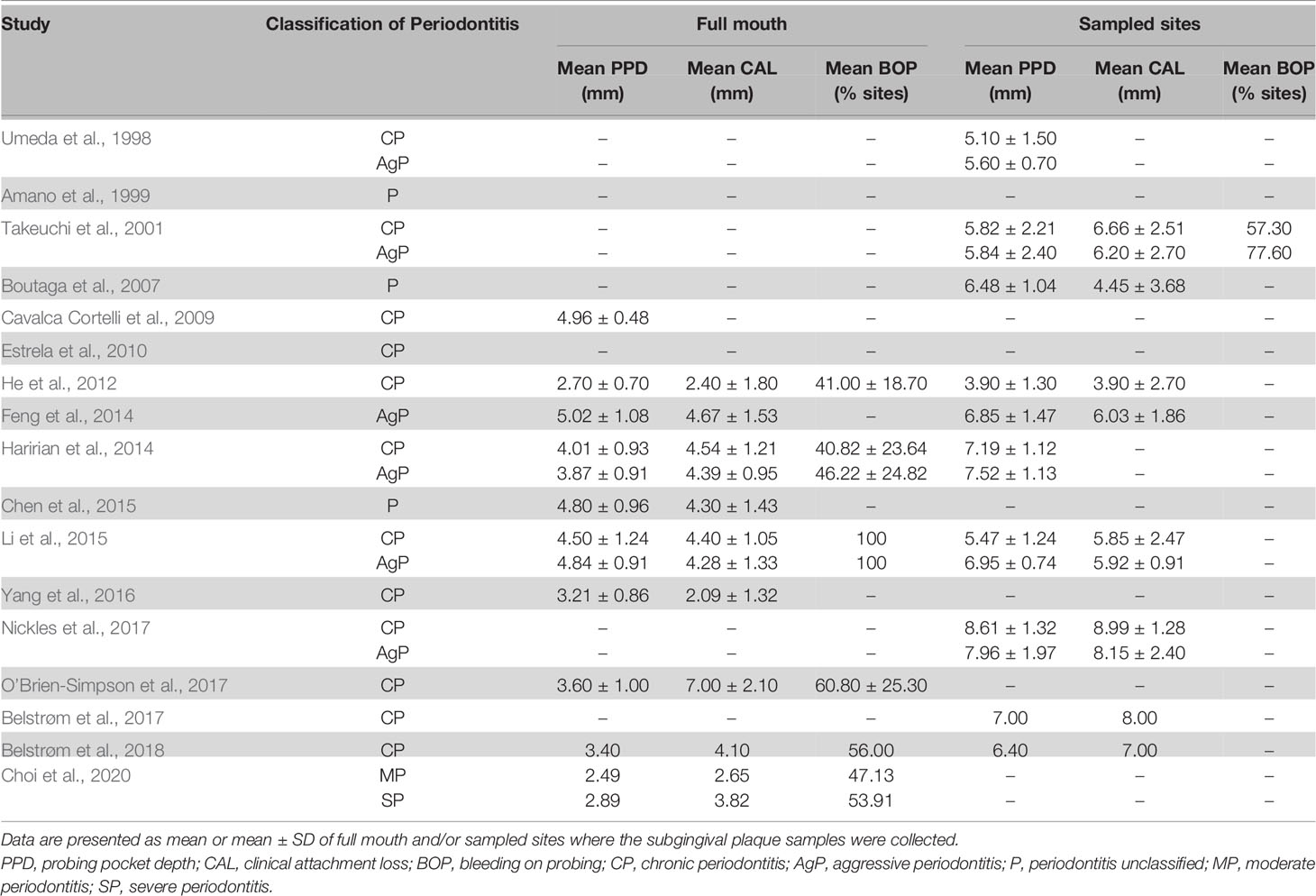
The most frequently reported clinical data in the included studies, mean probing pocket depth (PPD), clinical attachment loss (CAL) and bleeding on probing (BOP) of full mouth and/or the sampled sites, are summarized in Table 2. Two studies did not report any clinical data (Amano et al., 1999; Estrela et al., 2010). Overall, there were no big variations among the reported mean PPD and/or CAL, except that 2 studies reported a mean PPD less than 3 mm (He et al., 2012; Choi et al., 2020). The mean PPD of the sampled sites generally ranged from 4 to 8 mm.
3.5 Microbial Data
3.5.1 Detection Frequency of the Red-Complex Bacteria
The detection frequency of at least one of the red-complex bacteria could be extracted from 16 out of the 17 included studies. One study (Yang et al., 2016) did not report detection frequency, but bacterial counts only. Figure 3 presents an overview of bacterial detection frequencies in saliva and subgingival plaque samples. Most studies reported the detection frequencies of the red-complex bacteria were more than 60% in both saliva and subgingival samples of periodontitis patients. The only study which reported detection frequencies of less than 60% for all 3 species combined the data of healthy subjects and periodontitis patients (Umeda et al., 1998). Four studies also included healthy subjects, which showed varied detection frequencies of the red-complex bacteria, ranging from 2% to 45%. However, in each study, the detection frequencies of the red-complex bacteria in healthy group were much lower than those in the corresponding periodontitis group, irrespective of the sample types (Takeuchi et al., 2001; He et al., 2012; Feng et al., 2014; O’Brien-Simpson et al., 2017). Generally, most studies reported that the detection frequency of a specific red-complex species in periodontitis patients was lower in saliva than in subgingival plaque. But a few studies reported opposite trends. These studies are Boutaga et al. (2007) and Choi et al. (2020) for P. gingivalis, Boutaga et al. (2007), Choi et al. (2020) and Belstrøm et al. (2017) for T. forsythia, and Choi et al. (2020) for T. denticola.
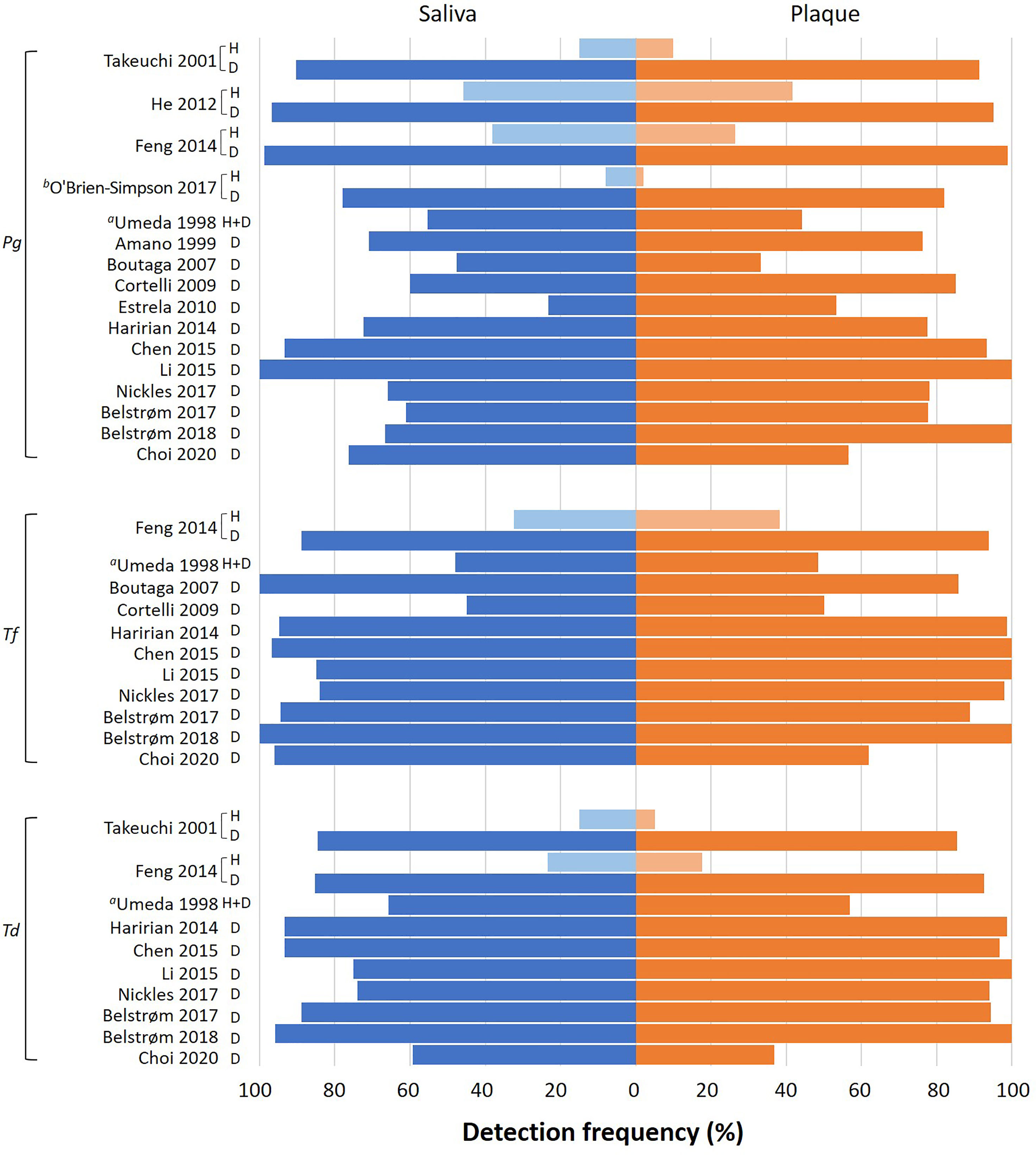
Figure 3 Overview of the detection frequency of P. gingivalis (Pg), T. forsythia (Tf) and T. denticola (Td) in saliva and subgingival plaque samples reported in each study. H: healthy subjects; D: periodontitis patients. Pooled subgingival plaque samples were used for analysis unless specified otherwise. aThis study mentioned 3 groups of subjects (health, gingivitis and periodontitis), but the data of different subject groups were reported together. bThis study analyzed the subgingival plaque samples from 6 periodontal pockets individually.
Next, meta-analyses were performed by summarizing the results from different studies, in order to assess the differences between saliva and subgingival plaque samples statistically. In total, 10 out of 16 studies were included in the meta-analyses. Six studies were excluded due to the following reasons: 1. Umeda et al. (1998) did not report data per healthy, gingivitis and periodontitis group. Only the average data of 3 groups were given; 2. O’Brien-Simpson et al. (2017) analyzed subgingival plaque samples from 6 periodontal pockets individually while other studies used pooled subgingival plaque samples; 3. Since the pocket depth was believed to affect the profile of subgingival microbiota (Van Dyke et al., 2020), studies which did not report PPD (Amano et al., 1999; Estrela et al., 2010) and reported PPD < 3 mm (He et al., 2012; Choi et al., 2020) were excluded. As shown in Figure 4, meta-analyses revealed that the heterogeneity among studies was low for all 3 red-complex bacteria, with I2 ranging from 6% to 17%. The detection frequencies of P. gingivalis [Figure 4A; OR = 0.64, 95% CI: (0.43, 0.93), p = 0.02], T. forsythia [Figure 4B; OR = 0.53, 95% CI: (0.30, 0.95), p = 0.03], and T. denticola [Figure 4C; OR = 0.45, 95% CI: (0.28, 0.72), p = 0.001] were all significantly higher in subgingival plaque samples than that in saliva samples.

Figure 4 Forest plots of meta-analyses comparing the detection frequency of: (A) P. gingivalis; (B) T. forsythia; (C) T. denticola between saliva and subgingival plaque samples from patients with periodontitis.
3.5.2 Bacterial Counts
Six out of the 17 studies reported bacterial counts of at least one of the red-complex species. Meta-analyses could not be performed on the data of bacteria counts due to the varied data format reported among studies (e.g., bacterial cell numbers and bacterial DNA copy numbers). Therefore, only a descriptive summary of the data is presented. As shown in Table 3, 4 studies (Boutaga et al., 2007; Yang et al., 2016; Nickles et al., 2017; O’Brien-Simpson et al., 2017) reported higher red-complex counts in subgingival plaque than in saliva, while the other 2 studies reported the opposite results (He et al., 2012; Choi et al., 2020). Only 1 study (Nickles et al., 2017) performed statistical analysis to confirm the reported higher counts in subgingival plaque.
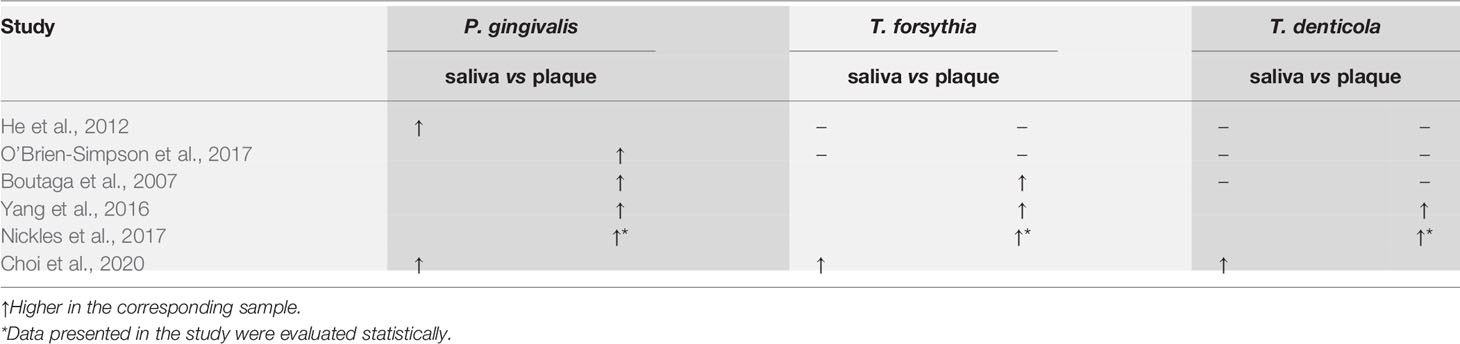
Table 3 Comparisons of bacterial counts of the red complex species between saliva and subgingival plaque samples from periodontitis patients.
3.5.3 Relative Abundance of the Red-Complex Bacteria
The relative abundance data were extracted from 4 sequencing studies in the following ways: from the published data (Belstrøm et al., 2017), from the data provided by the authors upon request (Li et al., 2015; Belstrøm et al., 2018), and from the raw sequence data that have been uploaded to the Sequence Read Archive (Chen et al., 2015).
As shown in Figure 5, meta-analyses revealed that the relative abundances of P. gingivalis [Figure 5A; MD = -10.27, 95% CI: (-18.15, -2.38), p < 0.00001], T. forsythia [Figure 5B; MD = -1.85, 95% CI: (-2.57, -1.12), p < 0.00001] and T. denticola [Figure 5C; MD = -1.20, 95% CI: (-1.74, -0.66), p < 0.0001] in subgingival plaque were all significantly higher than in saliva. However, considerably high heterogeneities among 4 studies were observed for the data of all 3 bacteria. Taking P. gingivalis as an example, the reported mean relative abundance ranged from 2.5 to 25% in subgingival plaque and from 0.1% to 5% in saliva, with an I2 index of 93%.
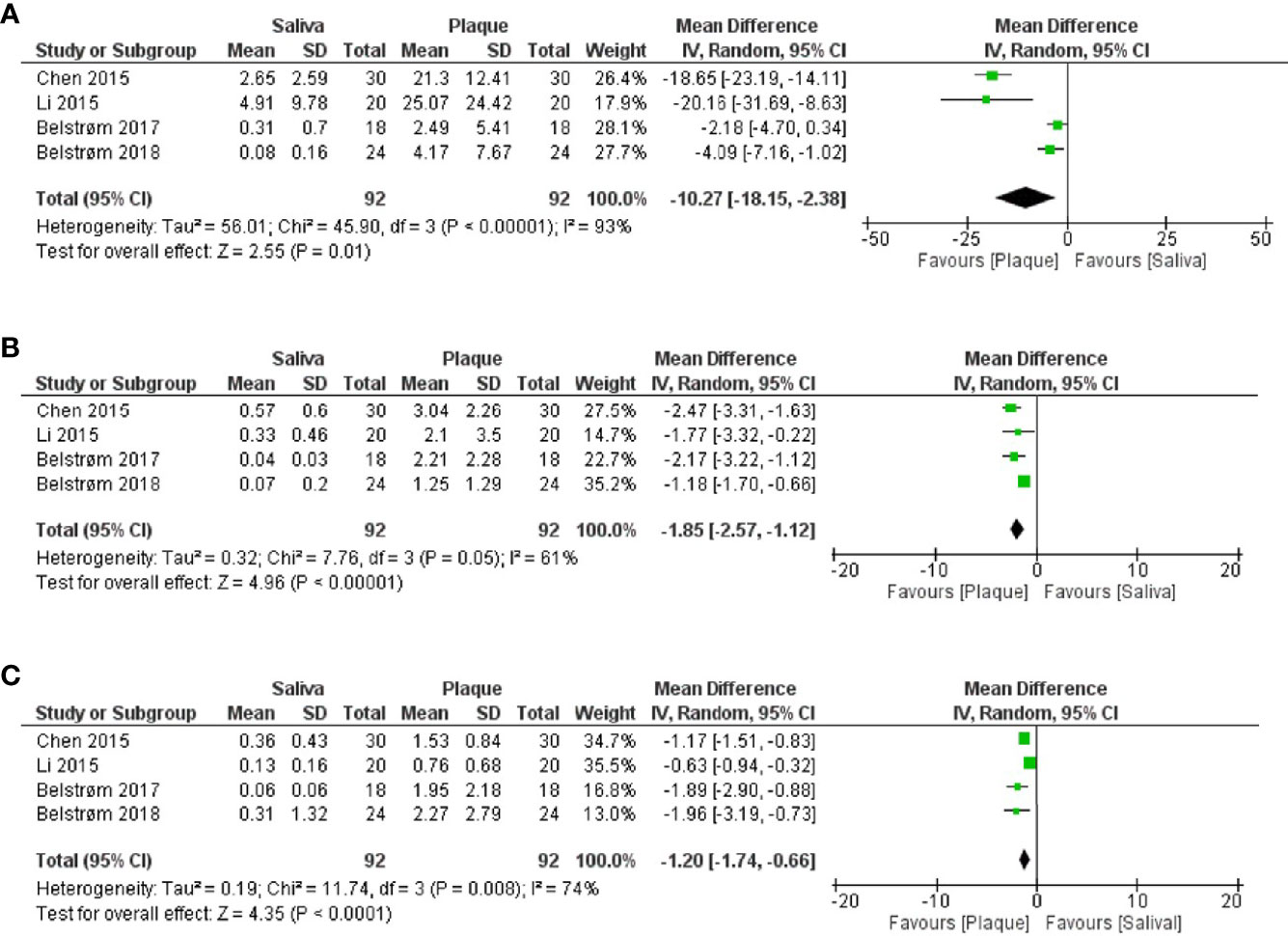
Figure 5 Forest plots of meta-analyses comparing the relative abundances (%) of: (A) P. gingivalis; (B) T. forsythia; (C) T. denticola between saliva and subgingival plaque samples from patients with periodontitis, as determined in studies using 16S rRNA gene sequencing techniques.
Since the relative abundance data of each patient were available from all 4 sequencing studies, we conducted Spearman’s rank correlation analysis in SPSS version 25 (SPSS Inc., Chicago, IL, USA) using the paired data obtained from saliva and subgingival plaque per patient. Figure 6 shows that positive correlations in relative abundance between saliva and subgingival plaque samples were observed for all 3 red-complex species, with a correlation coefficient of 0.73 for P. gingivalis (p < 0.0001), 0.28 for T. forsythia (p = 0.006) and 0.27 for T. denticola (p = 0.01).
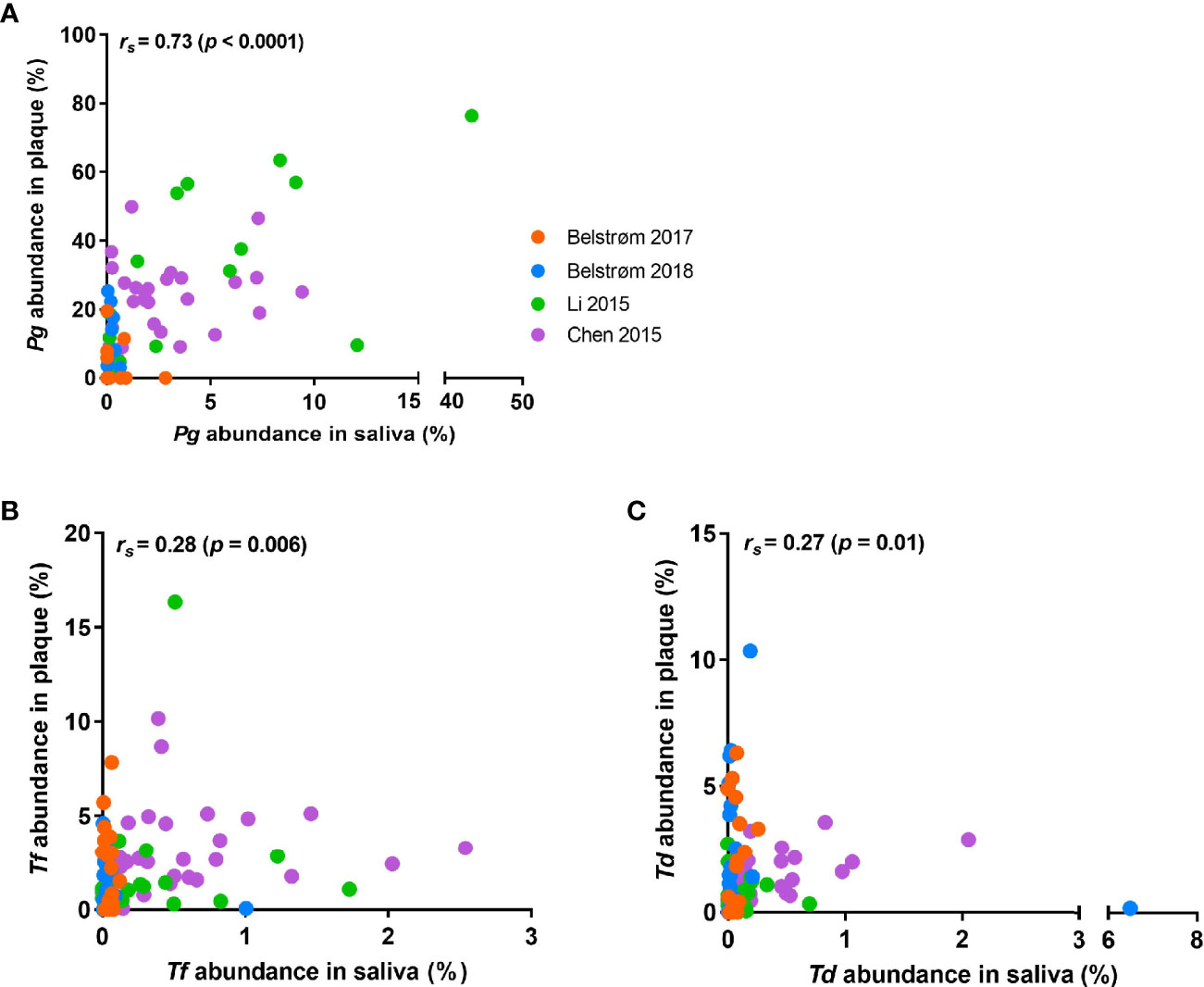
Figure 6 Scatter plots showing the distributions of the relative abundance of: (A) P. gingivalis; (B) T. forsythia; (C) T. denticola in saliva and subgingival plaque samples per patient, as determined in studies using 16S rRNA gene sequencing techniques. Each dot represents one patient, and patients from different studies are indicated by different colors of the dots. Spearman’s rank correlation coefficient (rs) and p value are shown in each plot.
3.5.4 Predominant Bacterial Genera in Saliva and Plaque Samples
The open-ended sequencing techniques allow the detection of more bacterial species than the targeted approach. Hence, the microbial compositions were summarized from 4 sequencing studies. The top 5 most abundant bacterial genera of each study are shown in Figure 7. Within each study, the top 5 most abundant bacterial genera in saliva were generally different from those in subgingival plaque, suggesting a major compositional difference between these two sample types. Among the most abundant genera, 4 bacterial genera were shared by all 4 studies: Streptococcus and Prevotella in saliva samples and Porphyromonas and Fusobacterium in subgingival plaque samples.
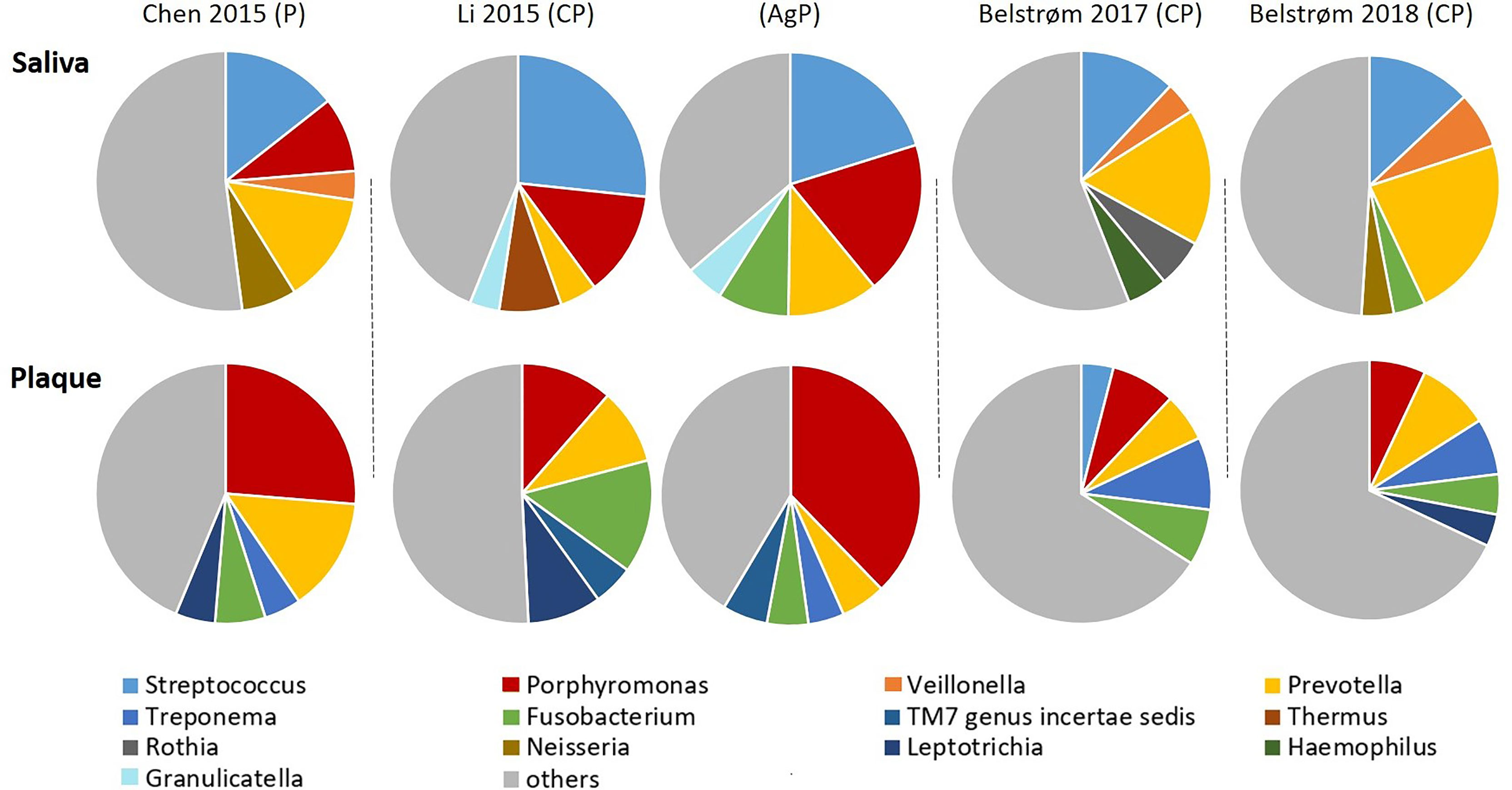
Figure 7 Approximate relative abundance of the top 5 most abundant genera identified in saliva and subgingival plaque from patients with periodontitis, as determined in studies using 16S rRNA gene sequencing techniques. CP, chronic periodontitis; AgP, aggressive periodontitis; P, periodontitis unclassified.
4 Discussion
It has been widely accepted that periodontitis is an inflammatory disease caused by a dysbiotic microbial community (Hajishengallis, 2015). Representative microbial sampling is crucial for disease prevention, diagnosis and treatment. Subgingival plaque in periodontal pockets represents the onset and development of periodontitis the best. However, due to its complicated sampling process, the use of saliva as an alternative has been an interest in many clinical studies. This systematic review identified 17 studies that reported the levels of red-complex bacteria in both saliva and subgingival plaque in periodontitis patients. Three types of outcome parameters, detection frequency, bacterial count and relative abundance, were examined. The meta-analyses on both detection frequency and relative abundance revealed that the levels of the red-complex bacteria in saliva were significantly lower than those in subgingival plaque.
Previously various researchers have claimed, based on their own data, that saliva could be a potential alternative to subgingival plaque for microbiologic analysis in periodontitis patients (Boutaga et al., 2007; Haririan et al., 2014; Belstrøm et al., 2017). Our meta-analysis summarized the results based on the samples obtained from 443 periodontitis patients in 10 studies (Figure 4). We found that saliva samples cannot represent the levels (i.e., detection frequency and relative abundance) of red-complex bacteria in subgingival plaque accurately in these patients. Since all the data analyzed in this review were obtained from samples taken at one time point, our finding indicates that one-time saliva sampling cannot be used to screen patients for the red-complex bacteria. We also examined other factors which might influence the comparison between saliva and subgingival plaque samples. Interestingly, the subgroup analysis (Supplementary Material; Figure S1) based on the collection methods of subgingival plaque, paper point or curette, showed that the results of studies using curette were in line with the finding mentioned above. However, in the studies using paper point, the detection frequencies of P. gingivalis and T. forsythia in saliva and subgingival plaque samples were similar. Possibly, a paper point collects unattached microbes in the periodontal pocket, which are likely spilt over to saliva; whereas a curette collects firmly attached biofilms (Jervøe-Storm et al., 2007). Clinical findings on the collection methods of subgingival plaque are inconsistent: one study (Renvert et al., 1992) stated that paper point sampling presented different microbial information as compared to curette sampling; whereas another study claimed a good agreement for the results of two sampling methods. Moreover, the open-ended sequencing method revealed DNA contamination in paper points, making this collection method unsuitable for sequencing analysis (van der Horst et al., 2013). Taken together, the methods used for collecting subgingival plaque may potentially influence microbial composition comparisons between saliva and subgingival plaque.
It is worth noting that our data analyses demonstrated positive associations between saliva and subgingival plaque in terms of red-complex levels despite limited data. Among the 17 included studies, 4 studies not only examined samples from periodontitis groups, but also from periodontal healthy groups. Although the reported detection frequencies in the healthy subjects varied considerably among studies (2% to 45%), all 4 studies showed that within one study, the detection frequency of each red-complex bacteria was much higher in periodontitis group than that in healthy group, irrespective of the sample type. Moreover, the relative abundances of the red-complex bacteria in saliva were also significantly correlated to that in subgingival plaque (Figure 6). Hence, it is possible that once the red-complex bacteria are enriched in subgingival plaque at diseased state, they could be spilt over or washed out into saliva, which consequently increase their levels in saliva. To this end, sampling saliva at multiple time points might help to trace the compositional shift of subgingival microbiota towards a disease provoking state. The study of Belstrøm et al. (2018) showed the correlation of the levels of salivary red-complex bacteria to that of subgingival plaque before and after treatment, indicating the possibility of such application of saliva sample. However, a thorough review on longitudinal clinical studies is needed to confirm this.
The magnitude and presence of statistical heterogeneity in a meta-analysis are usually explained by heterogeneity of methodological and/or clinical sources (Deeks et al., 2021). In our results, high heterogeneities of the relative abundance data among the 4 sequencing studies were observed, irrespective of the targeted bacterial species (I2: 61%–93%). Since the sequencing method-related heterogeneity in the meta-analysis of microbiome data has been reported before (Lozupone et al., 2013; Duvallet, 2018), the source of high heterogeneity here is likely related to the sequencing techniques. All 4 studies varied in many steps in sample processing as well as analysis, such as the DNA extraction methods, targeted 16S rRNA gene region for sequencing, sequencing platform used and sequencing depth. Lozupone et al. (2013) conducted meta-analysis on human microbiome data extracted from 12 different studies, and revealed that the technical variations in different studies could obscure biologically meaningful compositional differences. However, when the studied parameter had a large effect size (e.g., the body sites), the bias caused by variation in sequencing methodology could be outweighed by the real difference. In our case, despite the high heterogeneity, the relative abundance of red-complex bacteria in saliva was consistently lower than that in subgingival plaque.
During data analysis, we identified a clinical parameter, periodontal pocket depth, which potentially contributed to the high heterogeneity in a meta-analysis. In addition to the results on the basis of 10 studies reported in Figure 4, we also performed a meta-analysis on the detection frequencies reported in 14 studies, where an additional 4 studies with unknown or low periodontal pocket depth were included (Supplementary Material; Figure S2). From these meta-analyses, we observed substantially high heterogeneities (I2: 49%–73%) as compared to those in the 10-study based meta-analyses (I2: 6%–17%). The 14-study based meta-analyses showed no significant difference in the detection frequency of all 3 red-complex bacteria between saliva and subgingival plaque. We suspected that the high heterogeneity of the 14-study based meta-analyses was, at least partly, caused by variations in pocket depths. Van Dyke et al. (2020) stated that the depth of periodontal pocket was one of the crucial elements which determined the dysbiosis of subgingival microbiota, as differentiate microbial profiles were observed in pockets with different depths. For example, the abundance of Bacteriodetes significantly increased as the pocket deepened (Kirst et al., 2015). Our observation showed that the periodontal pocket depth might be an important confounding factor for microbial analysis. Interestingly, in the summary of bacterial count data, the 2 studies that reported a low pocket depth were also the only 2 studies which showed higher salivary red-complex bacterial counts in saliva as compared to subgingival plaque (He et al., 2012; Choi et al., 2020). Likely, the differential levels of red-complex bacteria in saliva and subgingival plaque are associated with the depth of periodontal pockets. Unfortunately, most studies included this review obtained samples from deep pockets, and we could not further illustrate the potential influence of this confounding factor.
In conclusion, this systematic review shows that the levels of red-complex bacteria in saliva were significantly lower than those in subgingival plaque in patients with periodontitis, in terms of the detection frequency and relative abundance. This finding is based on the meta-analyses on the data obtained from 443 patients at one sampling time point. In addition, our analyses reveal positive associations in the levels of red-complex bacteria between saliva and subgingival plaque despite limited data. Therefore, we recommend a thorough review of longitudinal clinical studies to further assess the role of saliva in detecting periodontitis-related microorganisms.
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding authors.
Author Contributions
YJ and DD conceived and designed the study. YJ and BS performed the study and collected the data. YJ, BB, and DD analyzed and interpreted the data. YJ drafted the manuscript. BB, LC, XZ, RE, WC, and DD revised the manuscript critically. All authors contributed to the article and approved the submitted version.
Funding
This study was supported by the ACTA Research Institute and the National Natural Science Foundation of China grant 81870759 (to LC).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We would like to thank dr. Daniel Belstrøm and dr. Xianghui Feng who kindly provided us the requested data in their studies, and dr. Zisheng Tang who kindly responded to our email but was not able to provide the requested data.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcimb.2021.727732/full#supplementary-material
References
Amano, A., Nakagawa, I., Kataoka, K., Morisaki, I., Hamada, S. (1999). Distribution of Porphyromonas Gingivalis Strains With fimA Genotypes in Periodontitis Patients. J. Clin. Microbiol. 37 (5), 1426–1430. doi: 10.1128/JCM.37.5.1426-1430.1999
Armitage, G. C. (1999). Development of a Classification System for Periodontal Diseases and Conditions. Ann. Periodontol. 4 (1), 1–6. doi: 10.1902/annals.1999.4.1.1
Belstrøm, D., Grande, M. A., Sembler-Møller, M. L., Kirkby, N., Cotton, S. L., Paster, B. J., et al. (2018). Influence of Periodontal Treatment on Subgingival and Salivary Microbiotas. J. Periodontol. 89 (5), 531–539. doi: 10.1002/JPER.17-0377
Belstrøm, D., Sembler-Møller, M. L., Grande, M. A., Kirkby, N., Cotton, S. L., Paster, B. J., et al. (2017). Microbial Profile Comparisons of Saliva, Pooled and Site-Specific Subgingival Samples in Periodontitis Patients. PloS One 12 (8), e0182992–e0182992. doi: 10.1371/journal.pone.0182992
Boutaga, K., Savelkoul, P. H. M., Winkel, E. G., van Winkelhoff, A. J. (2007). Comparison of Subgingival Bacterial Sampling With Oral Lavage for Detection and Quantification of Periodontal Pathogens by Real-Time Polymerase Chain Reaction. J. Periodontol. 78 (1), 79–86. doi: 10.1902/jop.2007.060078
Boutaga, K., van Winkelhoff, A. J., Vandenbroucke-Grauls, C. M. J. E., Savelkoul, P. H. M. (2003). Comparison of Real-Time PCR and Culture for Detection of Porphyromonas Gingivalis in Subgingival Plaque Samples. J. Clin. Microbiol. 41 (11), 4950–4954. doi: 10.1128/jcm.41.11.4950-4954.2003
Chen, H., Liu, Y., Zhang, M., Wang, G., Qi, Z., Bridgewater, L., et al. (2015). A Filifactor Alocis-Centered Co-Occurrence Group Associates With Periodontitis Across Different Oral Habitats. Sci. Rep. 5, 9053–9053. doi: 10.1038/srep09053
Cavalca Cortelli, S., Cavallini, F., Regueira Alves, M. F., et al. (2009). Clinical and Microbiological Effects of an Essential-Oil-Containing Mouth Rinse Applied in the “One-Stage Full-Mouth Disinfection” Protocol—A Randomized Doubled-Blinded Preliminary Study. Clin. Oral. Invest. 13 (2), 189–94. doi: 10.1007/s00784-008-0219-3
Choi, J. U., Lee, J. B., Kim, K. H., Kim, S., Seol, Y. J., Lee, Y. M., et al. (2020). Comparison of Periodontopathic Bacterial Profiles of Different Periodontal Disease Severity Using Multiplex Real-Time Polymerase Chain Reaction. Diagn. (Basel Switzerland) 10 (11), 965. doi: 10.3390/diagnostics10110965
Curtis, M. A., Diaz, P. I., Van Dyke, T. E. (2020). The Role of the Microbiota in Periodontal Disease. Periodontol. 2000 83 (1), 14–25. doi: 10.1111/prd.12296
Deeks, J. J., Higgins, J. P. T., Altman, D. G. (2021). “Chapter 10: Analysing Data and Undertaking Meta-Analyses,” in Cochrane Handbook for Systematic Reviews of Interventions Version 6-2 (Updated February 2021). Eds. Higgins, J. P. T., Thomas, J., Chandler, J., Cumpston, M., Li, T., Page, M. J., Welch, V. A. (Cochrane). Available at: www.training.cochrane.org/handbook.
Duvallet, C. (2018). Meta-Analysis Generates and Prioritizes Hypotheses for Translational Microbiome Research. Microb. Biotechnol. 11 (2), 273–276. doi: 10.1111/1751-7915.13047
Estrela, C., Pimenta, F. C., Alencar, A. H. G. D., Ruiz, L. F. N., Estrela, C. (2010). Detection of Selected Bacterial Species in Intraoral Sites of Patients With Chronic Periodontitis Using Multiplex Polymerase Chain Reaction. J. Appl. Oral. Sci. 18 (4), 426–431. doi: 10.1590/s1678-77572010000400018
Feng, X., Zhang, L., Xu, L., Meng, H., Lu, R., Chen, Z., et al. (2014). Detection of Eight Periodontal Microorganisms and Distribution of Porphyromonas Gingivalis fimA Genotypes in Chinese Patients With Aggressive Periodontitis. J. Periodontol. 85 (1), 150–159. doi: 10.1902/jop.2013.120677
Geurs, N. (2015). American Academy of Periodontology Task Force Report on the Update to the 1999 Classification of Periodontal Diseases and Conditions. J. Periodontol. 86 (7), 835–838. doi: 10.1902/jop.2015.157001
Hajishengallis, G. (2015). Periodontitis: From Microbial Immune Subversion to Systemic Inflammation. Nat. Rev. Immunol. 15 (1), 30–44. doi: 10.1038/nri3785
Haririan, H., Andrukhov, O., Bertl, K., Lettner, S., Kierstein, S., Moritz, A., et al. (2014). Microbial Analysis of Subgingival Plaque Samples Compared to That of Whole Saliva in Patients With Periodontitis. J. Periodontol. 85 (6), 819–828. doi: 10.1902/jop.2013.130306
He, J., Huang, W., Pan, Z., Cui, H., Qi, G., Zhou, X., et al. (2012). Quantitative Analysis of Microbiota in Saliva, Supragingival, and Subgingival Plaque of Chinese Adults With Chronic Periodontitis. Clin. Oral. Investig. 16 (6), 1579–1588. doi: 10.1007/s00784-011-0654-4
Higgins, J. P. T., Thompson, S. G., Deeks, J. J., Altman, D. G. (2003). Measuring Inconsistency in Meta-Analyses. BMJ (Clin. Res. ed.) 327 (7414), 557–560. doi: 10.1136/bmj.327.7414.557
Huttenhower, C., Gevers, D., Knight, R., Abubucker, S., Badger, J. H., Chinwalla, A. T., et al. (2012). Structure, Function and Diversity of the Healthy Human Microbiome. Nature 486 (7402), 207–214. doi: 10.1038/nature11234
Jervøe-Storm, P.-M., AlAhdab, H., Koltzscher, M., Fimmers, R., Jepsen, S. (2007). Comparison of Curet and Paper Point Sampling of Subgingival Bacteria as Analyzed by Real-Time Polymerase Chain Reaction. J. Periodontol. 78 (5), 909–917. doi: 10.1902/jop.2007.060218
Kassebaum, N. J., Bernabé, E., Dahiya, M., Bhandari, B., Murray, C. J. L., Marcenes, W. (2014). Global Burden of Severe Periodontitis in 1990-2010: A Systematic Review and Meta-Regression. J. Dent. Res. 93 (11), 1045–1053. doi: 10.1177/0022034514552491
Kirst, M. E., Li, E. C., Alfant, B., Chi, Y.-Y., Walker, C., Magnusson, I., et al. (2015). Dysbiosis and Alterations in Predicted Functions of the Subgingival Microbiome in Chronic Periodontitis. Appl. Environ. Microbiol. 81 (2), 783–793. doi: 10.1128/AEM.02712-14
Li, Y., Feng, X., Xu, L., Zhang, L., Lu, R., Shi, D., et al. (2015). Oral Microbiome in Chinese Patients With Aggressive Periodontitis and Their Family Members. J. Clin. Periodontol. 42 (11), 1015–1023. doi: 10.1111/jcpe.12463
Loesche, W. J. (1992). DNA Probe and Enzyme Analysis in Periodontal Diagnostics. J. Periodontol. 63 (12S), 1102–1109. doi: 10.1902/jop.1992.63.12s.1102
Lozupone, C. A., Stombaugh, J., Gonzalez, A., Ackermann, G., Wendel, D., Vázquez-Baeza, Y., et al. (2013). Meta-Analyses of Studies of the Human Microbiota. Genome Res. 23 (10), 1704–1714. doi: 10.1101/gr.151803.112
Mark Welch, J. L., Dewhirst, F. E., Borisy, G. G. (2019). Biogeography of the Oral Microbiome: The Site-Specialist Hypothesis. Annu. Rev. Microbiol. 73, 335–358. doi: 10.1146/annurev-micro-090817-062503
Moher, D., Liberati, A., Tetzlaff, J., Altman, D. G., Group, P (2009). Preferred Reporting Items for Systematic Reviews and Meta-Analyses: The PRISMA Statement. PloS Med. 6 (7), e1000097–e1000097. doi: 10.1371/journal.pmed.1000097
Moola, S., Munn, Z., Tufanaru, C., Aromataris, E., Sears, K., Sfetcu, R., et al. (2020). “Chapter 7: Systematic Reviews of Etiology and Risk,” in JBI Manual for Evidence Synthesis. Eds. Aromataris, E., Munn, Z. (JBI).
Nickles, K., Scharf, S., Röllke, L., Dannewitz, B., Eickholz, P. (2017). Comparison of Two Different Sampling Methods for Subgingival Plaque: Subgingival Paper Points or Mouthrinse Sample? J. Periodontol. 88 (4), 399–406. doi: 10.1902/jop.2016.160249
O’Brien-Simpson, N. M., Burgess, K., Lenzo, J. C., Brammar, G. C., Darby, I. B., Reynolds, E. C. (2017). Rapid Chair-Side Test for Detection of Porphyromonas Gingivalis. J. Dent. Res. 96 (6), 618–625. doi: 10.1177/0022034517691720
Ouzzani, M., Hammady, H., Fedorowicz, Z., Elmagarmid, A. (2016). Rayyan—A Web and Mobile App for Systematic Reviews. Syst. Rev. 5 (1), 210. doi: 10.1186/s13643-016-0384-4
Page, R. C., Eke, P. I. (2007). Case Definitions for Use in Population-Based Surveillance of Periodontitis. J. Periodontol. 78 (7S), 1387–1399. doi: 10.1902/jop.2007.060264
Renvert, S., Wikström, M., Helmersson, M., Dahlén, G., Claffey, N. (1992). Comparative Study of Subgingival Microbiological Sampling Techniques. J. Periodontol. 63 (10), 797–801. doi: 10.1902/jop.1992.63.10.797
Segata, N., Haake, S. K., Mannon, P., Lemon, K. P., Waldron, L., Gevers, D., et al. (2012). Composition of the Adult Digestive Tract Bacterial Microbiome Based on Seven Mouth Surfaces, Tonsils, Throat and Stool Samples. Genome Biol. 13 (6), R42. doi: 10.1186/gb-2012-13-6-r42
Simón-Soro, Á., Tomás, I., Cabrera-Rubio, R., Catalan, M. D., Nyvad, B., Mira, A. (2013). Microbial Geography of the Oral Cavity. J. Dent. Res. 92 (7), 616–621. doi: 10.1177/0022034513488119
Socransky, S. S., Haffajee, A. D., Cugini, M. A., Smith, C., Kent, J. R. L. (1998). Microbial Complexes in Subgingival Plaque. J. Clin. Periodontol. 25 (2), 134–144. doi: 10.1111/j.1600-051X.1998.tb02419.x
Suchett-Kaye, G., Morrier, J.-J., Barsotti, O. (2001). Clinical Usefulness of Microbiological Diagnostic Tools in the Management of Periodontal Disease. Res. Microbiol. 152 (7), 631–639. doi: 10.1016/S0923-2508(01)01242-6
Takeuchi, Y., Umeda, M., Sakamoto, M., Benno, Y., Huang, Y., Ishikawa, I. (2001). Treponema Socranskii, Treponema Denticola, and Porphyromonas Gingivalis Are Associated With Severity of Periodontal Tissue Destruction. J. Periodontol. 72 (10), 1354–1363. doi: 10.1902/jop.2001.72.10.1354
Tonetti, M. S., Jepsen, S., Jin, L., Otomo-Corgel, J. (2017). Impact of the Global Burden of Periodontal Diseases on Health, Nutrition and Wellbeing of Mankind: A Call for Global Action. J. Clin. Periodontol. 44 (5), 456–462. doi: 10.1111/jcpe.12732
Umeda, M., Contreras, A., Chen, C., Bakker, I., Slots, J. (1998). The Utility of Whole Saliva to Detect the Oral Presence of Periodontopathic Bacteria. J. Periodontol. 69 (7), 828–833. doi: 10.1902/jop.1998.69.7.828
van der Horst, J., Buijs, M. J., Laine, M. L., Wismeijer, D., Loos, B. G., Crielaard, W., et al. (2013). Sterile Paper Points as a Bacterial DNA-Contamination Source in Microbiome Profiles of Clinical Samples. J. Dent. 41 (12), 1297–1301. doi: 10.1016/j.jdent.2013.10.008
Van Dyke, T. E., Bartold, P. M., Reynolds, E. C. (2020). The Nexus Between Periodontal Inflammation and Dysbiosis. Front. Immunol. 11, 511. doi: 10.3389/fimmu.2020.00511
Vieira Colombo, A. P., Magalhães, C. B., Hartenbach, F. A. R. R., Martins do Souto, R., Maciel da Silva-Boghossian, C. (2016). Periodontal-Disease-Associated Biofilm: A Reservoir for Pathogens of Medical Importance. Microb. Pathog. 94, 27–34. doi: 10.1016/j.micpath.2015.09.009
Yang, X., Li, C., Pan, Y. (2016). The Influences of Periodontal Status and Periodontal Pathogen Quantity on Salivary 8-Hydroxydeoxyguanosine and Interleukin-17 Levels. J. Periodontol. 87 (5), 591–600. doi: 10.1902/jop.2015.150390
Keywords: periodontitis, Porphyromonas gingivalis, Tannerella forsythia, Treponema denticola, 16S rRNA gene amplicon sequencing, real-time PCR
Citation: Jiang Y, Song B, Brandt BW, Cheng L, Zhou X, Exterkate RAM, Crielaard W and Deng DM (2021) Comparison of Red-Complex Bacteria Between Saliva and Subgingival Plaque of Periodontitis Patients: A Systematic Review and Meta-Analysis. Front. Cell. Infect. Microbiol. 11:727732. doi: 10.3389/fcimb.2021.727732
Received: 19 June 2021; Accepted: 14 September 2021;
Published: 08 October 2021.
Edited by:
Ulvi Kahraman Gürsoy, University of Turku, FinlandCopyright © 2021 Jiang, Song, Brandt, Cheng, Zhou, Exterkate, Crielaard and Deng. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Lei Cheng, Y2hlbmdsZWlAc2N1LmVkdS5jbg==; Dong Mei Deng, ZC5kZW5nQGFjdGEubmw=
 Yaling Jiang
Yaling Jiang Bingqing Song
Bingqing Song Bernd W. Brandt
Bernd W. Brandt Lei Cheng
Lei Cheng Xuedong Zhou
Xuedong Zhou Rob A. M. Exterkate
Rob A. M. Exterkate Wim Crielaard2
Wim Crielaard2 Dong Mei Deng
Dong Mei Deng