- 1Infectious Diseases Translational Research Program, Department of Microbiology and Immunology, National University of Singapore, Singapore, Singapore
- 2Healthy Longevity Translational Research Program, Department of Microbiology and Immunology, National University of Singapore, Singapore, Singapore
The recent COVID-19 pandemic has highlighted the urgency to develop effective antiviral therapies against the disease. Murine hepatitis virus (MHV) is a coronavirus that infects mice and shares some sequence identity to SARS-CoV-2. Both viruses belong to the Betacoronavirus genus, and MHV thus serves as a useful and safe surrogate model for SARS-CoV-2 infections. Clinical trials have indicated that remdesivir is a potentially promising antiviral drug against COVID-19. Using an in vitro model of MHV infection of RAW264.7 macrophages, the safety and efficacy of monotherapy of remdesivir, chloroquine, ivermectin, and doxycycline were investigated. Of the four drugs tested, remdesivir monotherapy exerted the strongest inhibition of live virus and viral RNA replication of about 2-log10 and 1-log10, respectively (at 6 µM). Ivermectin treatment showed the highest selectivity index. Combination drug therapy was also evaluated using remdesivir (6 µM) together with chloroquine (15 µM), ivermectin (2 µM) or doxycycline (15 µM) – above their IC50 values and at high macrophage cell viability of over 95%. The combination of remdesivir and ivermectin exhibited highly potent synergism by achieving significant reductions of about 7-log10 of live virus and 2.5-log10 of viral RNA in infected macrophages. This combination also resulted in the lowest cytokine levels of IL-6, TNF-α, and leukemia inhibitory factor. The next best synergistic combination was remdesivir with doxycycline, which decreased levels of live virus by ~3-log10 and viral RNA by ~1.5-log10. These results warrant further studies to explore the mechanisms of action of the combination therapy, as well as future in vivo experiments and clinical trials for the treatment of SARS-CoV-2 infection.
Introduction
SARS-CoV-2 first emerged in China in December 2019, causing numerous cases of pneumonia, and it was subsequently confirmed to be a novel coronavirus in January 2020. Since then, this highly infectious virus together with its emerging variants have transcended international borders and spread rapidly across the globe, resulting in high infection rates and millions of deaths worldwide. Coronaviruses are positive-sense, single-stranded RNA viruses with a distinct crown-like appearance which are the spike glycoproteins on the viral envelope. SARS-CoV-2 belongs to the Coronavirus family, and Betacoronavirus subgenus. Members of this family often cause respiratory, enteric and neurologic diseases in mammals such as bats and even humans (Sharma et al., 2020a). The SARS-CoV-2 genome is about 29.9 kb, encoding four main structural proteins (spike (S), small envelope (E), membrane (M), and nucleocapsid (N) proteins), as well as other accessory proteins. The S protein is one of the main contributors to SARS-CoV-2 virulence, where it forms homotrimers, promoting the binding of the viral envelope to the ACE2 receptors on the host cell surface for entry (Zheng et al., 2020). ACE2 receptors are mainly expressed on type II alveolar epithelial cells, myocardial cells, kidneys, and are abundantly expressed on intestinal enterocytes. As a result, cell-free and macrophage-associated virus can travel from the lungs to other organs via the circulation, causing systemic infections (Hamming et al., 2004). Patients with critical SARS-CoV-2 infection exhibit multiple organ damage stemming from thromboembolic disease, hyper-coagulation, and the “cytokine storm” arising from overproduction of pro-inflammatory cytokines from infected monocytes and macrophages (Klok et al., 2020; Mondal et al., 2020; Tang et al., 2020).
Remdesivir is proposed to be a nucleoside analogue that inhibits viral RNA-dependent RNA polymerase (RdRp) to prevent viral RNA synthesis, thereby hampering virus replication in host cells. In vitro studies reported up to 6-log10 reduction of MHV with treatment using remdesivir at 11.1 μM (Agostini et al., 2018). A large double-blind, randomized, placebo-controlled trial of remdesivir in 1062 hospitalized adults with COVID-19 revealed shorter recovery time and lower mortality compared with the placebo group (Beigel et al., 2020). Another trial found that remdesivir did not provide significant antiviral effects in seriously ill patients (Wang et al., 2020).
Previous in vitro chloroquine treatment of Vero E6 cells infected with 2003-SARS-CoV demonstrated dose-dependent reduction of viral load by 40-80% (Vincent et al., 2005). Recent clinical trials of SARS-CoV-2-infected patients who received chloroquine phosphate treatment documented clinical improvements such as improved lung imaging, and shorter disease course (Gao et al., 2020). An open-label randomized trial found positive impact of chemoprophylaxis with oral hydroxychloroquine in reducing SARS-CoV-2 infection in young and healthy men (Seet et al., 2021). Chloroquine is proposed to affect the endocytic pathway by increasing pH levels within endosomes and lysosomes, thereby inhibiting proteases to prevent cleavage of the spike protein and subsequent virus entry into host cells (Satarker et al., 2020). SARS-CoV-2-infected patients at high risk of severe disease who were on doxycycline intervention showed improved clinical symptoms and enhanced recovery (Yates et al., 2020).
In vitro experiments observed a 99.8% reduction in viral RNA when SARS-CoV-2-infected Vero/hSLAM cells were treated with 5 μM ivermectin (Caly et al., 2020). One COVID-19 clinical trial reported that intervention with ivermectin achieved earlier virus clearance compared to placebo (9.7 versus 12.7 days), with decreased severity biomarkers (CRP and LDH) by day 7 (Ahmed et al., 2021). Another ivermectin trial involving non-severe COVID-19 patients documented reduction in anosmia/hyposmia, cough, viral loads, and IgG antibody titers (Chaccour et al., 2021). However, one double-blind randomized controlled trial found no significant difference in duration of symptoms (10 versus 12 days) in adults with mild COVID-19 treated with ivermectin versus placebo (Lopez-Medina et al., 2021). More recently, a randomized trial of hospitalized COVID-19 patients revealed that an oral high-dose ivermectin regimen was well-tolerated and led to antiviral activity that was dependent on plasma concentration of the drug (Krolewiecki et al., 2021). A recent meta-analysis of 15 trials indicated moderate-certainty evidence that large reductions in COVID-19 mortality are possible with ivermectin treatment (versus control) – early therapy may also decrease progression to severe disease (Bryant et al., 2021).
Combination therapy can target different pathways and processes during the coronavirus life-cycle to synergistically augment reduction in viral load. Hence, this strongly emphasizes the practical need to develop optimal combination therapies and increase their accessibility to COVID-19 patients worldwide.
This study explored the potential repurposing of four clinically approved drugs (remdesivir, chloroquine, ivermectin, doxycycline) to inhibit murine coronavirus and viral RNA replication, and to dampen excessive cytokine production. Given that it belongs to the same Betacoronavirus genus as SARS-CoV-2, murine hepatitis virus (MHV) was employed as a safe surrogate virus to infect the RAW264.7 mouse macrophage cell line (Pope et al., 1998) to mimic macrophage infection in COVID-19. The specific objectives were: (a) to compare the efficacy of monotherapy versus the efficacy of combined therapy with remdesivir to determine drug synergism; and (b) to investigate the effects of the drugs on cytokine production by infected macrophages.
Materials and Methods
Virus and Cell Cultures
All viral infection experiments were conducted using MHV strain A59 (MHV-A59; GenBank accession number AY910861) (Chiow et al., 2016). The RAW264.7 murine macrophage cell line was maintained at 37°C in RPMI-1640 medium supplemented with 10% fetal bovine serum (FBS). H2.35 murine liver cells were maintained at 35°C in Dulbecco’s modified Eagle medium (DMEM) with 10% FBS. Cells were seeded at a density of 200,000 cells per well on 24-well plates, and at a density of 15,000 cells per well on 96-well plates. Cells subjected to infection and/or drug treatment were monitored daily for any cytopathic effect (CPE).
Drugs
Remdesivir (HY-104077; MedChemExpress, Monmouth Junction, NJ, USA) was prepared in 100 mM and 10 mM stock solutions in sterile water. Chloroquine (C6628; Sigma-Aldrich, St. Louis, MO, USA) and ivermectin (CAS 70288-86-7; Merck, Burlington, MA, USA) were prepared in 10 mM stock solutions in dimethyl sulfoxide (DMSO). Doxycycline (CAS 24390-14-5; Merck, Burlington, MA, USA) was prepared in 10 mM stock solution in sterile water. For treatment experiments, all drug dilutions were prepared in 0.5% DMSO with the respective cell culture medium.
Cell Viability Assay
Cell viability was assessed using the CellTiter 96 AQueous One Solution Cell Proliferation (MTS) assay (Promega, Madison, WI, USA), according to the manufacturer’s instructions. RAW264.7 cells were incubated in the presence of increasing drug concentrations (0.16 μM, 0.8 μM, 4 μM, 20 μM, 100 μM) for 48 hours. Cell viability was determined using a microplate reader (Tecan, Mannedorf, Switzerland) with values normalized to those of untreated cells.
Evaluation of Antiviral Inhibitory Activities of Monotherapy and Combination Therapy
Sub-confluent monolayers of RAW264.7 cells in 24-well plates were infected with MHV at multiplicity of infection (MOI) of 0.1 for 2 hours. The inoculum was then removed, and the cells were treated with the indicated concentrations of drugs for monotherapy or combination therapy for 48 hours. The cell supernatant was harvested at 48 hours post-infection, and subjected to virus plaque assay using H2.35 mouse liver cells to determine live coronavirus titers, and to real-time PCR to quantify the viral RNA loads. Each experiment was carried out in triplicates.
Live Coronavirus Quantification by Plaque Assay
Live virus was quantified using plaque assay. Sub-confluent H2.35 cells were infected with the diluted supernatant samples for 1 hour. 1.2% Avicel BioPolymer (FMC, Philadelphia, PA, USA) in phosphate-buffered saline (PBS) was added to each well and incubated for 72 hours. The cells were fixed with 4% paraformaldehyde in PBS for 2 hours, and then stained with 1% crystal violet for 15 minutes.
RNA Extraction, Viral RNA Quantification by Real-Time Reverse Transcription- Quantitative PCR
Cell supernatant was harvested from the experiments, and total viral RNA was purified using QIAamp Viral RNA Mini Kit (Qiagen, Hilden, Germany). RNA concentration was determined using the NanoDrop ND-1000 spectrophotometer. Reverse transcription was carried out at 35°C for 1 hour. Quantification of viral RNA was performed using MHV-NF forward and MHV-NR reverse primers for the specific detection of the N gene of MHV (5′-ACGCTTACATTATCWACTTC-3′ and 5′-GATCTAAATTAGAATTGGTC-3′, respectively). The qPCR was performed with FastStart Essential DNA Green Master using the LightCycler 96 instrument (Roche Diagnostics, Basel, Switzerland). To plot the standard curve, a range of positive controls with known plaque-forming units (PFU) of MHV was included. The thermal cycling conditions were as follows: 95°C for 600 s, followed by 55 cycles of 95°C for 10 sec, 40°C for 5 sec, 72°C for 8 sec, and a final 95°C for 10 sec, 65°C for 60 sec and 97°C for 1 sec.
Determining Cytokine Protein Expression Profiles Using Luminex Multiplex Assay
To determine cytokine expression levels, multiplex assays using the Myokine 5-Plex Mouse ProcartaPlex Panel kit (Thermo Fisher Scientific, Waltham, MA, USA) were conducted according to the manufacturer’s instructions. This 5-plex kit quantified the following cytokines: IL-6, IL-10, IL-15, leukemia inhibitory factor (LIF), and TNF-α. Standard curves and values were measured on Luminex MAGPIX, and calculated using xPONENT 4.2 software for MAGPIX (with 30 beads being set as the detection limit).
Statistical Analyses
Non-linear regression and curve-fitting parameter analyses were performed to calculate inhibitory concentration values using GraphPad Prism 7 (GraphPad Software, San Diego, CA, USA). Statistical significance and P-values were calculated by one-way ANOVA using Dunnett’s multiple comparison test. Each error bar of dose-response curves represents the standard error of mean (SEM) of three technical replicates.
Results
Monotherapy of Drugs Inhibits MHV Infection of RAW 264.7 Macrophages With Minimal Toxicity
To evaluate the safety profile of the drugs, cytotoxicity assays were carried out (Figure 1A), and the results expressed as cell viability curves. To evaluate the effectiveness of monotherapy of the individual drugs, increasing concentrations of the drugs were tested against MHV infection (Figure 1B). Table 1 summarizes the 50% cytotoxic concentration (CC50), 50% inhibitory concentration (IC50), and selectivity index (SI) from the monotherapy experiments. In treatment of RAW264.7 cells, ivermectin exhibited the highest SI index, indicating its high safety and efficacy ratio as a treatment option. Based on these determined IC50 and CC50 ranges (Table 1), downstream monotherapy and combination therapy experiments were subsequently performed using the following drug concentrations above their respective IC50 values with high cell viability and low cytotoxicity: remdesivir at 6 µM, chloroquine at 15 µM, ivermectin at 2 µM, and doxycycline at 15 µM.
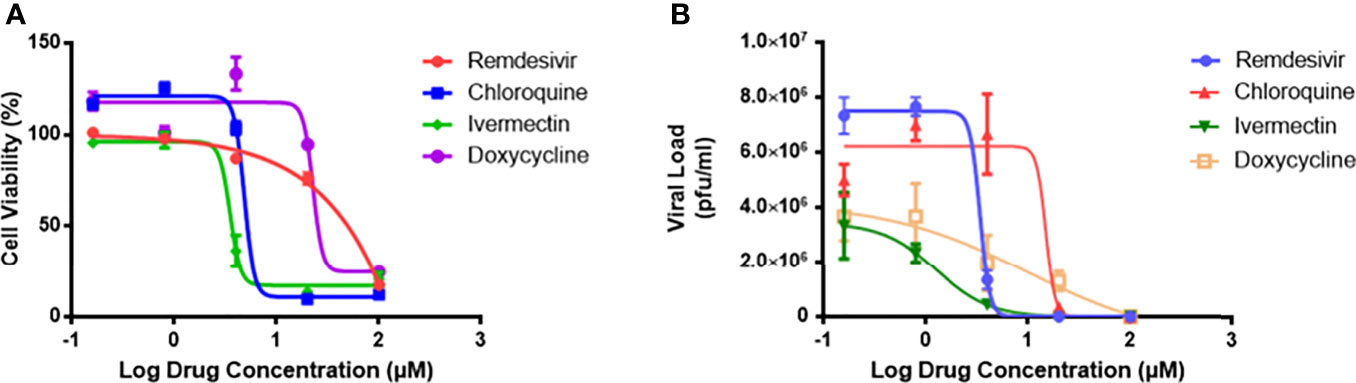
Figure 1 The drug cytotoxicity profiles of RAW264.7 macrophages, and dose-response curves of MHV-infected macrophages. (A) Cytotoxicity profiles of the four individual drugs on RAW264.7 cells as measured by MTS assay after 48 hours exposure. Percentage of cell viability was normalized to untreated cells and blank control. Experiments were performed in quadruplicates. (B) Viral inhibitory activities of the four drugs against MHV infection of RAW264.7 macrophages. Live coronavirus titers (pfu/ml) were quantified by plaque assays performed in triplicates. The dose-response curves were fitted using the non-linear regression method, and IC50 values were calculated using the Prism 7 software. Error bars represent standard error of the mean (SEM).
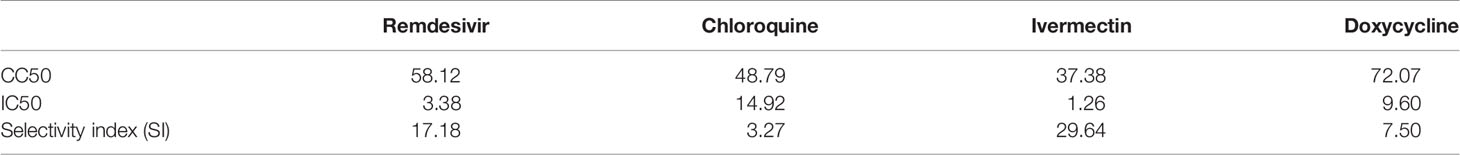
Table 1 Mean values of CC50, IC50 and selectivity index (SI = CC50/EC50) of remdesivir, chloroquine, ivermectin and doxycycline treatment of RAW264.7 macrophages.
To further verify the drug cytotoxicity and antiviral efficacy in another cell line, we selected and assessed remdesivir and chloroquine treatment in H2.35 murine liver cells without and with MHV infection. As shown in Supplementary Figure 1, remdesivir also exhibited a significantly higher selectivity index (204.2) versus that of chloroquine (10.6) in mouse liver cells.
Combination Therapy of Remdesivir With Ivermectin or With Doxycycline Is Synergistic in Reducing Coronavirus Load and Restoring Healthy Cell Morphology
To determine the efficacies of the drug combinations (together with remdesivir), monotherapy experiments at the same concentrations were compared against those of combination treatments. As shown in Figures 2A–C, monotherapy at 6 µM remdesivir yielded a significant 2-log10 reduction in live virus (P < 0.05). Chloroquine monotherapy demonstrated a significant 1-log10 viral reduction (P < 0.05), whereas monotherapy with ivermectin or doxycycline showed no statistically significant difference from untreated infected cells (P > 0.05).
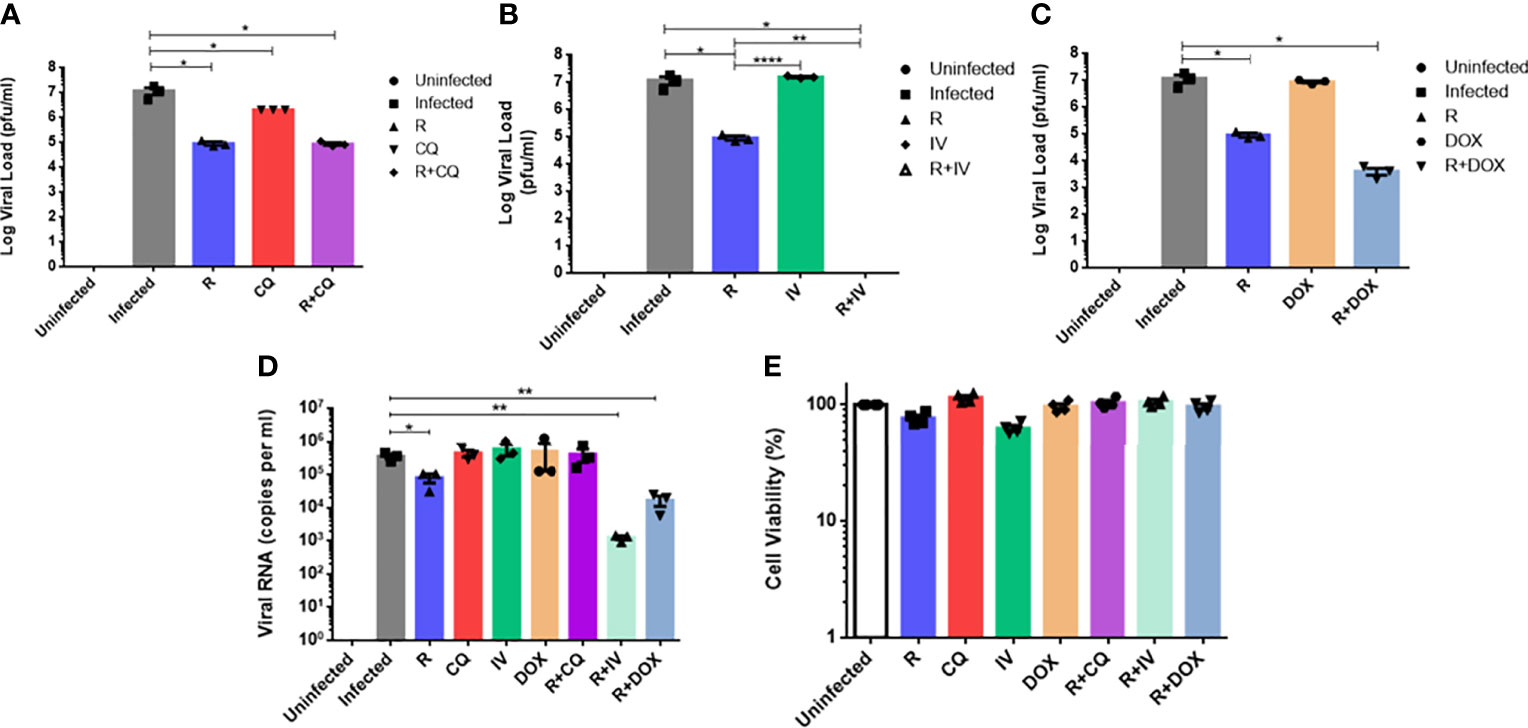
Figure 2 Comparison of antiviral activities of monotherapy versus combination therapy using remdesivir with chloroquine, ivermectin and doxycycline against MHV infection of RAW264.7 macrophages. Remdesivir (R) was tested at a concentration of 6 µM alone or in combination with the other three drugs. Uninfected cells, and infected but untreated cells served as controls. Live coronavirus titers were quantified using plaque assays. (A) Infected cells were treated with 15 µM of chloroquine (CQ) alone or combined with R. (B) Infected cells were treated with 2 µM of ivermectin (IV) alone or combined with R. (C) Infected cells were treated with 15 µM of doxycycline (DOX) alone or combined with R. (D) Quantification of MHV RNA loads (copies per ml) in extracts of supernatants from the above experiments using real-time RT-qPCR. Error bars represent SEM. Statistical significance was determined by one-way ANOVA with Dunnett’s multiple comparison, and P-values denoted by asterisks: *P < 0.05; **P < 0.01; ****P < 0.0001. (E) Cell viability profiles of uninfected and drug-treated cells at tested concentrations. Mean percentages of cell viability were normalized to untreated cells and blank controls. Experiments were carried out in quadruplicates.
There are various primary methods for antimicrobial synergy testing, including the multiple-combination antimicrobial test or MCAT (Doern, 2014). MCAT is an accepted and recognized method for simultaneously assessing combinations of two or more drugs at fixed concentrations against a test organism at a specified inoculum (Aaron et al., 2000). This technique can demonstrate whether the combined effect of two drugs in vitro can exhibit synergism (i.e. an effect larger than the sum of the effect of each independent drug); or additive effect (i.e. an effect equal to the sum of the individual drug effects); or antagonism (i.e. an effect smaller than the sum of the independent drug effects) (Greco et al., 1995).
Combination therapy with remdesivir and chloroquine (Figure 2A) exhibited a significant 2-log10 reduction in live viral load (P < 0.05), which was similar to remdesivir monotherapy, indicating that the antiviral effect of this combination was largely due to remdesivir, without drug synergism. In contrast, combination of remdesivir with ivermectin (Figure 2B) was the most effective and completely inhibited viral replication, with a significantly potent reduction of 7-log10 of live virus, i.e. an additional reduction of 5-log10 of live virus compared to remdesivir monotherapy (P < 0.01), thus revealing evidence of drug synergism. Some synergism was also observed in the combination of remdesivir with doxycycline (Figure 2C), where a further reduction of 1-log10 of live virus titer was achieved compared to remdesivir alone.
Real-time RT-qPCR quantification of MHV RNA displayed a similar trend to the virus plaque assays (Figure 2D). Remdesivir monotherapy attained a significant reduction of about 1-log10 of viral RNA (P < 0.05), whereas monotherapy with the other drugs did not lead to significant reduction in viral RNA. Combination of remdesivir with ivermectin or with doxycycline culminated in the most significant reductions in viral RNA loads of 2.5-log10 and 1.5-log10 respectively (P < 0.01), thus reiterating drug synergism. These results are congruent with the live virus titers which highlighted that remdesivir in combination with ivermectin or doxycycline could significantly decrease both live virus and viral RNA loads. Therefore, the most effective drug combination that exhibited highly potent synergism was remdesivir with ivermectin, followed by remdesivir with doxycycline.
Figure 2E shows high cell viability of RAW264.7 macrophages for monotherapy and combination therapy with the drugs at the tested concentrations. It is notable that the mean cell viability was above 95% for each of the drug combinations evaluated.
As shown in the Supplementary Figure 2, the four monotherapy treatments (Supplementary Figures 2C–F) exhibited cell morphologies that were more similar to the infected untreated control cells (Supplementary Figure 2B) with prominent CPE. In contrast, the combination therapies (Supplementary Figures 2G–J) displayed cell morphologies that resembled the uninfected control (Supplementary Figure 2A), with rounder and smoother cell surface layers. This suggests that the combination therapy (particularly remdesivir and ivermectin) was able to restore cell appearance to a healthy state, in contrast to monotherapy which showed extensive CPE.
Combination Therapy With Remdesivir and Ivermectin Decreases the Levels of Key Cytokines Associated With the Cytokine Storm
Given that certain drugs (individually or in combination) could inhibit live virus and viral RNA, we next analyzed the drug effects on cytokine levels in infected macrophages. The protein expression levels of five key cytokines were quantified, namely IL-6, IL-10, IL-15, LIF and TNF-α (Figure 3). The IL-10 readings were below the detection limit and out of range – thus, the IL-10 results were omitted.
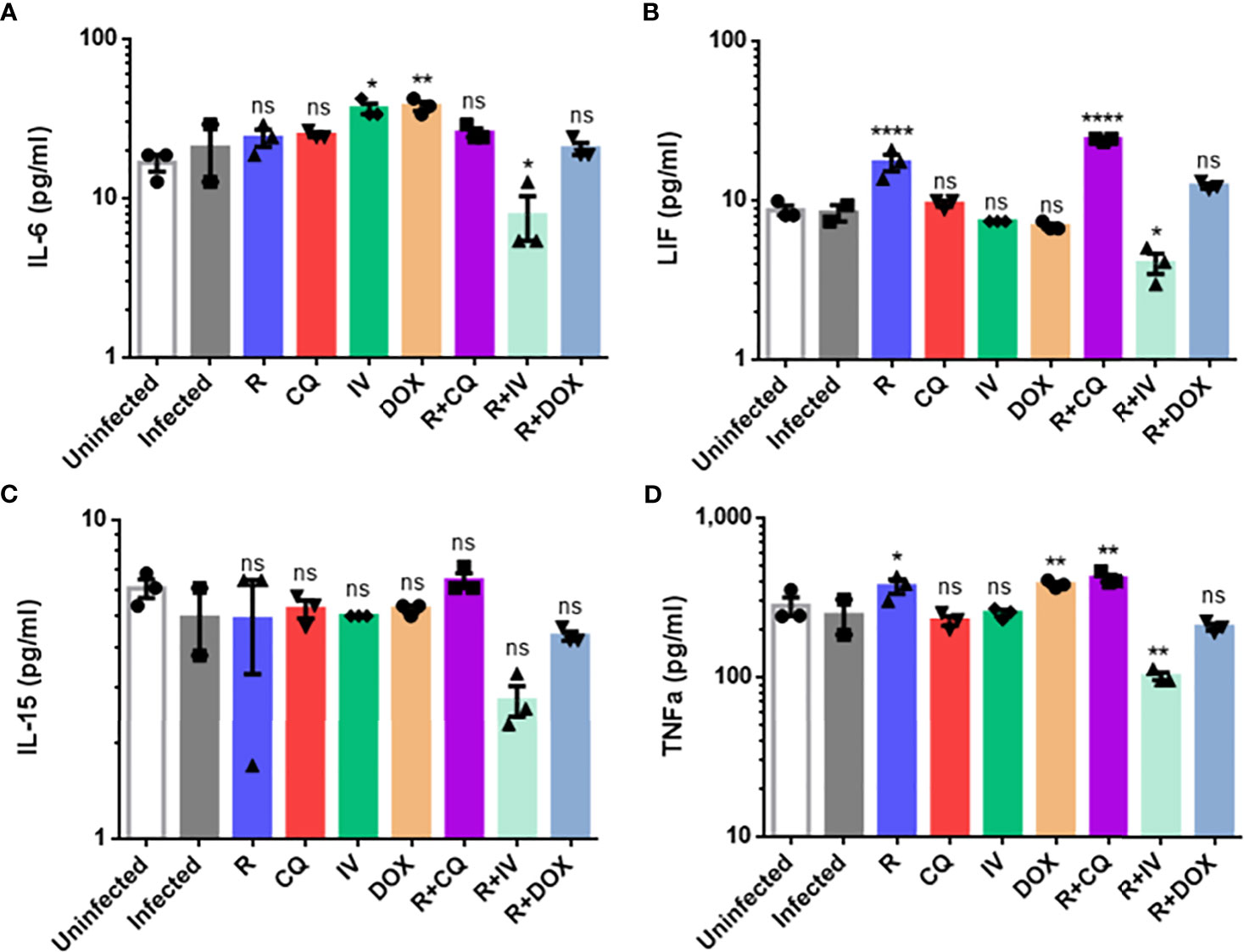
Figure 3 Cytokine protein expression profiles of monotherapy versus combination therapy against MHV infection of RAW264.7 macrophages. Remdesivir (R), chloroquine (CQ), ivermectin (IV) and doxycycline (DOX) treatments were evaluated. Cytokine protein levels (pg/ml) of (A) IL-6, (B) leukemia inhibitory factor or LIF, (C) IL-15, (D) TNF-α were quantified using the Myokine 5-Plex Mouse ProcartaPlex Panel in drug-treated infected samples compared to controls (uninfected cells, and infected but untreated cells). IL-10 cytokine levels were below the detection limit, and were thus omitted. The data represent means and SEM from triplicate experiments. Statistical significance was determined by one-way ANOVA with Dunnett’s multiple comparison against the infected untreated control, and denoted by asterisks: *P < 0.05; **P < 0.01; ****P < 0.0001. ns, not statistically significant.
As shown in Figure 3, combination treatment with remdesivir and ivermectin was found to significantly decrease IL-6 (P < 0.05), LIF (P < 0.05), and TNF-α (P < 0.01). This drug combination also resulted in lower IL-15 level, although it was not statistically significant. To verify these findings, a biological replicate experiment yielded a reproducible trend in the decreased cytokines (data not shown). This suggests that the additional 5-log10 reduction in live virus titer (Figure 2B) was complemented by amelioration of cytokine production to exert anti-inflammatory effects. It is noteworthy that IL-6 and TNF-α are significantly elevated in patients with severe COVID-19, and that LIF is an IL-6 cytokine that can activate the JAK/STAT and MAP kinase pathways (Traber et al., 2017).
Discussion
Bronchoalveolar lavage fluid from severe COVID-19 patients contain abundant mononuclear phagocytes and inflammatory monocyte-derived macrophages. Autopsy analyses of COVID-19 patients reveal the presence of SARS-CoV-2 viral particles and antigen within lymph node and splenic macrophages. Aberrant mononuclear phagocyte activation or macrophage activation syndrome may lead to the cytokine release syndrome or “cytokine storm” which culminates in excessive inflammation and coagulopathy associated with COVID-19 severity and mortality (Merad and Martin, 2020; Sharma et al., 2021b). SARS-CoV-2-infected macrophages thus contribute to viral spread, exaggerated inflammation, and activation-induced lymphocytic cell death (Park, 2020).
In RAW264.7 macrophages, SARS-CoV-2 spike S1 induces pro-inflammatory responses (TNF-α expression) which can be markedly suppressed by RNA interference of TLR4 (Shirato and Kizaki, 2021). The related SARS-CoV spike protein can induce TNF-α and IL-6 expression by activating the NF-κB signaling pathway in RAW264.7 cells (Wang et al., 2007).
Besides MHV, the RAW264.7 line also supports replication of other viral pathogens, including Japanese encephalitis virus, mengovirus, and pneumonia virus of mice (PVM) (Murali-Krishna et al., 1995; Martin et al., 2000; Dyer et al., 2007). On the other hand, the murine lung alveolar macrophage MH-S cell line does not support replication of PVM (Brenner et al., 2016). In one study, SARS‐CoV‐2 challenge of human alveolar macrophages (AMs) does not result in the release of free SARS‐CoV‐2 RNAs and proteins into the cytoplasm, suggesting that AMs are not productively infected (Dalskov et al., 2020).
M1 phenotype (pro-inflammatory) macrophages arise as a result of classical activation via stimulation of toll-like receptors through ligands, including microbial stimuli (e.g. lipopolysaccharide or LPS) and certain cytokines (such as IFN-γ and TNF-α). M1 macrophages produce inflammatory mediators (e.g. IL-6, TNF-α, nitric oxide), and significantly contribute to the immune response (Sica et al., 2015; Smith et al., 2016). For example, one regulatory mechanism of M1 polarization and activation of RAW264.7 macrophages is triggered by activating the TLR4 and NF-κB signaling pathway (Shi et al., 2020). Interestingly, in contrast to M2 AMs, M1 AMs have a lower endosomal pH that favors membrane fusion, permits entry of viral RNA from the endosome into the cytoplasm where SARS-CoV-2 replicates – the virus is then packaged and released to facilitate infection of the lungs (Lv et al., 2021).
RAW264.7 and J774.1 macrophage cell lines are well-established model systems in cell biology and immunology research. Bacterial LPS induces dose-dependent secretion of pro-inflammatory cytokines (IL-6, TNF-α, MCP-1) in both RAW264.7 and J774.1 cells (Nativel et al., 2017). Moreover, in both macrophage cell lines infected with herpes simplex virus 1 and treated with inhibitory CpG, similar viral induction of cytokine expression involving TLR9 is observed (Malmgaard et al., 2004). Sun et al. (2020) also showed that estradiol can promote trained immunity in both RAW264.7 and J774.1 by using TNF-α and IL-1β as the marker of inflammation. Considering the above reasons, we therefore focused on RAW264.7 as our working macrophage cell line model in our experiments.
Efficacious broad-spectrum pan-coronavirus antivirals are needed to treat the highly infectious and virulent SARS-CoV-2, as well as emerging zoonotic coronaviruses to prepare for future epidemics and even pandemics. Although there are many studies and clinical trials testing the efficacy of the four tested drugs, very few explore the combination with remdesivir, which is the most studied antiviral for SARS-CoV-2 treatment.
In this study, remdesivir alone at 6 µM was able to achieve a 2-log10 live viral reduction, but was exceptionally synergistic when combined with ivermectin at 2 µM, culminating in an extremely potent 7-log10 reduction in viral load, with no visible virus plaques even at the highest virus concentration tested. A similar trend was corroborated by the synergistic reduction of 2.5-log10 of viral RNA with the combination of remdesivir and ivermectin, compared to the corresponding monotherapy. Hence, the additional 5-log10 of live virus reduction arising from synergism may be attributed to mechanisms other than inhibition of viral RNA production. It is notable that in hospitalized COVID-19 patients, infectious virus was readily isolated from samples of the throat or lung during the first week of symptoms. However, when symptoms generally subsided after day 8, live virus could not be cultured despite ongoing high viral RNA loads detectable in throat swabs well into the second week (Wölfel et al., 2020). Another study supported the in vitro synergistic interaction between remdesivir and ivermectin resulting in enhanced antiviral activity against SARS-CoV-2 (Jeffreys et al., 2020).
Strikingly, the remdesivir and ivermectin combination culminated in the most significant and consistent reduction of IL-6, LIF and TNF-α, thus highlighting its important impact on inhibiting the production of pro-inflammatory cytokines from infected macrophages. This notable anti-inflammatory characteristic of this drug combination is relevant in view of the need to ameliorate the “cytokine storm” especially in patients with severe coronavirus infections, to improve associated complications, and to delay disease progression (Narasaraju et al., 2020). In addition, the drug combinations also helped to improve morphology of the infected cells (Supplementary Figure 2) compared to the monotherapy and infected controls, which may promote and restore proper cell growth following coronavirus infection. While SARS-CoV-2 can infect human macrophages, it is well-appreciated that these cells are not a primary target of SARS-CoV-2 infection. One mechanism to explain the “cytokine storm” associated with COVID-19 is that SARS-CoV-2 pathogen-associated molecular patterns trigger IL-6 production followed by hyper-activation of the NF-κB pathway in both immune and non-immune cells, culminating in the establishment of the IL-6 amplifier inflammatory circuit (Hojyo et al., 2020).
The mode of action of remdesivir in coronavirus infection is primarily targeting RdRp-mediated RNA synthesis, thus preventing virus replication and diminishing viral load (Agostini et al., 2018). Notably, remdesivir also possesses broad-spectrum antiviral inhibition of human and zoonotic coronaviruses with divergent RdRp (Brown et al., 2019). A proposed antiviral mechanism of ivermectin is the inhibition of nuclear translocation of viral proteins which is mediated by host importin α/β1 heterodimerization (Wagstaff et al., 2012). The preformed heterodimer can also be destabilized by ivermectin. In addition, ivermectin can hinder heterodimer formation via binding to the importin-α armadillo repeat domain to affect thermal stability and α-helicity of importin-α, thereby preventing its binding to importin β1 (Yang et al., 2020). One possible mode of action of ivermectin is through blocking the nuclear localization signal of importin-α (Kinobe and Owens, 2021). Viral protein entry into the nucleus is thus impeded, which impairs viral inhibition of the host anti-viral response, culminating in more effective viral clearance (Caly et al., 2020). Computational modelling of doxycycline and SARS-CoV-2 proteins identified this drug to be a potential inhibitor of the 3C-like main protease that mediates the maturation of non-structural proteins, potentially restricting viral replication (Wu et al., 2020). In mice with influenza virus pneumonia, doxycycline treatment ameliorates acute lung injury with significantly diminished inflammation, protein leakage, and matrix metalloproteinase activity in the lungs (Ng et al., 2012).
Going forward, more studies and trials are warranted to better understand the underlying mechanisms by which ivermectin and doxycycline synergize with remdesivir to exert antiviral activities against coronaviruses in multiple cell lines. Importantly, by targeting different viral and/or host processes, effective drug combinations can lower the therapeutic concentrations of drugs compared to monotherapy, thus reducing treatment toxicity and costs. Moreover, such drug combinations can minimize drug-resistant virus strains which have higher propensity to emerge with monotherapy (Sun et al., 2016). Indeed, combination treatment with remdesivir plus baricitinib (a Janus kinase inhibitor) was superior to remdesivir alone in improving clinical status and recovery among COVID-19 patients (Kalil et al., 2021). A multi-center study reported that four approved drugs against hepatitis C that inhibit the papain-like protease of SARS-CoV-2 can enhance the antiviral effects of remdesivir by ten-fold in vitro (Bafna et al., 2021). Synergy between remdesivir and emetine has also been documented (Choy et al., 2020). Given that remdesivir is relatively more expensive, its potential combination at lower doses together with more affordable drugs can lower the overall treatment cost, thus offering cheaper but effective options especially to lower- and middle-income countries which are already suffering from existing epidemics such as tuberculosis (Bong et al., 2020).
Conclusion
In summary, this in vitro study has provided evidence that the combination of remdesivir and ivermectin was highly synergistic and potent, culminating in striking reduction in live murine coronavirus replication and viral RNA synthesis. Furthermore, the efficacy and beneficial effects of this combination therapy also included immunomodulatory effects via inhibition of cytokine production, and improving cell morphology of infected macrophages.
The limitation of our study is that antiviral activities of drugs were investigated only against MHV in the RAW264.7 macrophage cell culture model. For direct comparison versus MHV, future detailed studies are thus warranted to evaluate the potential utility of this drug combination against SARS-CoV-2 and other coronaviruses (e.g. SARS-CoV, MERS-CoV, HCoV-229E, HCoV-OC43), such as multi-dose checkerboard experiments to ascertain additive effects of drugs (Bakowski et al., 2021). This caveat is also critically important given the observations of cell line-dependent compound efficacy against SARS-CoV-2 infection reported in many previous SARS-CoV-2 antiviral studies. For example, Vero cells, human Huh-7.5 liver cancer cells and human Calu-3 lung epithelial cells exhibit major differences in sensitivity to drugs with antiviral activity due to the distinct entry pathways utilized by SARS-CoV-2 in these cell lines (Dittmar et al., 2021). Also necessary are in vivo experiments using suitable animal model(s) of COVID-19 before progressing onto clinical trials in SARS-CoV-2 infected patients.
Data Availability Statement
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
Author Contributions
VC and YT conceptualized and designed the research project, and analyzed the data. All experiments were carried out by YT. All authors contributed to the article and approved the submitted version.
Funding
This study was supported by a research grant of the National University of Singapore.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We thank S. H. Lau, Joe Ong, J. H. Ch’ng, S. Lee, and other staff members of the National University Health System for their technical, statistical, and other assistance.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcimb.2021.700502/full#supplementary-material
Supplementary Figure 1 | The drug cytotoxicity profiles of H2.35 murine liver cells, and dose-response curves of MHV-infected H2.35 cells. (A) Cytotoxicity profiles of remdesivir and chloroquine on H2.35 cells as measured by MTS assay after 48 hours exposure. Percentage of cell viability was normalized to untreated cells and blank control. Experiments were performed in quadruplicates. (B) Viral inhibitory activities of remdesivir and chloroquine against MHV infection of H2.35 cells. Live coronavirus titers (pfu/ml) were quantified by plaque assays performed in triplicates. The dose-response curves were fitted using the non-linear regression method, and IC50 values were calculated using the Prism 7 software. Error bars represent standard error of the mean (SEM). (C) Mean values of CC50, IC50 and selectivity index (SI = CC50/EC50) of remdesivir and chloroquine treatment of H2.35 cells.
Supplementary Figure 2 | Representative morphological features of RAW264.7 macrophages under different infection and/or treatment conditions. Microscopic images were captured by the EVOS XL microscope at 10× magnification. The scale bar represents 200 μm. (A) Uninfected control cells. (B) MHV-infected control cells. Monotherapy of MHV-infected macrophages using: (C) remdesivir alone, (D) chloroquine alone, (E) ivermectin alone, (F) doxycycline alone. Combination therapy of MHV-infected macrophages using: (G) remdesivir and chloroquine, (H) remdesivir and ivermectin, (J) remdesivir and doxycycline.
References
Aaron, S. D., Ferris, W., Henry, D. A., Speert, D. P., Macdonald, N. E. (2000). Multiple Combination Bactericidal Antibiotic Testing for Patients With Cystic Fibrosis Infected With Burkholderia Cepacia. Am. J. Respir. Crit. Care Med. 161, 1206–1212. doi: 10.1164/ajrccm.161.4.9907147
Agostini, M. L., Andres, E. L., Sims, A. C., Graham, R. L., Sheahan, T. P., Lu, X., et al. (2018). Coronavirus Susceptibility to the Antiviral Remdesivir (GS-5734) is Mediated by the Viral Polymerase and the Proofreading Exoribonuclease. mBio 9, e00221–18. doi: 10.1128/mBio.00221-18
Ahmed, S., Karim, M. M., Ross, A. G., Hossain, M. S., Clemens, J. D., Sumiya, M. K., et al. (2021). A Five-Day Course of Ivermectin for the Treatment of COVID-19 may Reduce the Duration of Illness. Int. J. Infect. Dis. 103, 214–216. doi: 10.1016/j.ijid.2020.11.191
Bafna, K., White, K., Harish, B., Rosales, R., Ramelot, T. A., Acton, T. B., et al. (2021). Hepatitis C Virus Drugs That Inhibit SARS-CoV-2 Papain-Like Protease Synergize With Remdesivir to Suppress Viral Replication in Cell Culture. Cell Rep. 35, 109133. doi: 10.1016/j.celrep.2021.109133
Bakowski, M. A., Beutler, N., Wolff, K. C., Kirkpatrick, M. G., Chen, E., Nguyen, T. H., et al. (2021). Drug Repurposing Screens Identify Chemical Entities for the Development of COVID-19 Interventions. Nat. Commun. 12, 3309. doi: 10.1038/s41467-021-23328-0
Beigel, J. H., Tomashek, K. M., Dodd, L. E., Mehta, A. K., Zingman, B. S., Kalil, A. C., et al. (2020). Remdesivir for the Treatment of Covid-19 - Final Report. N. Engl. J. Med. 383, 1813–1826. doi: 10.1056/NEJMoa2007764
Bong, C. L., Brasher, C., Chikumba, E., McDougall, R., Mellin-Olsen, J., Enright, A. (2020). The COVID-19 Pandemic: Effects on Low- and Middle-Income Countries. Anesth. Analg. 131, 86–92. doi: 10.1213/ANE.0000000000004846
Brenner, T. A., Rice, T. A., Anderson, E. D., Percopo, C. M., Rosenberg, H. F. (2016). Immortalized MH-S Cells Lack Defining Features of Primary Alveolar Macrophages and do Not Support Mouse Pneumovirus Replication. Immunol. Lett. 172, 106–112. doi: 10.1016/j.imlet.2016.02.012
Brown, A. J., Won, J. J., Graham, R. L., Dinnon, K. H., Sims, A. C., Feng, J. Y., et al. (2019). Broad Spectrum Antiviral Remdesivir Inhibits Human Endemic and Zoonotic Deltacoronaviruses With a Highly Divergent RNA Dependent RNA Polymerase. Antiviral Res. 169, 104541. doi: 10.1016/j.antiviral.2019.104541
Bryant, A., Lawrie, T. A., Dowswell, T., Fordham, E. J., Mitchell, S., Hill, S. R., et al. (2021). Ivermectin for Prevention and Treatment of COVID-19 Infection: A Systematic Review, Meta-Analysis, and Trial Sequential Analysis to Inform Clinical Guidelines. Am. J. Ther. 28, e434–e460. doi: 10.1097/MJT.0000000000001402
Caly, L., Druce, J. D., Catton, M. G., Jans, D. A., Wagstaff, K. M. (2020). The FDA-Approved Drug Ivermectin Inhibits the Replication of SARS-CoV-2 In Vitro. Antiviral Res. 178, 104787. doi: 10.1016/j.antiviral.2020.104787
Chaccour, C., Casellas, A., Blanco-Di Matteo, A., Pineda, I., Fernandez-Montero, A., Ruiz-Castillo, P., et al. (2021). The Effect of Early Treatment With Ivermectin on Viral Load, Symptoms and Humoral Response in Patients With non-Severe COVID-19: A Pilot, Double-Blind, Placebo-Controlled, Randomized Clinical Trial. EClinicalMedicine 32, 100720. doi: 10.1016/j.eclinm.2020.100720
Chiow, K. H., Phoon, M. C., Putti, T., Tan, B. K., Chow, V. T. (2016). Evaluation of Antiviral Activities of Houttuynia Cordata Thunb. Extract, Quercetin, Quercetrin and Cinanserin on Murine Coronavirus and Dengue Virus Infection. Asian Pac J. Trop. Med. 9, 1–7. doi: 10.1016/j.apjtm.2015.12.002
Choy, K. T., Wong, A. Y., Kaewpreedee, P., Sia, S. F., Chen, D., Hui, K. P. Y., et al. (2020). Remdesivir, Lopinavir, Emetine, and Homoharringtonine Inhibit SARS-CoV-2 Replication In Vitro. Antiviral Res. 178, 104786. doi: 10.1016/j.antiviral.2020.104786
Dalskov, L., Møhlenberg, M., Thyrsted, J., Blay-Cadanet, J., Poulsen, E. T., Folkersen, B. H., et al. (2020). SARS-CoV-2 Evades Immune Detection in Alveolar Macrophages. EMBO Rep. 21, e51252. doi: 10.15252/embr.202051252
Dittmar, M., Lee, J. S., Whig, K., Segrist, E., Li, M., Kamalia, B., et al. (2021). Drug Repurposing Screens Reveal Cell-Type-Specific Entry Pathways and FDA-Approved Drugs Active Against SARS-Cov-2. Cell Rep. 35, 108959. doi: 10.1016/j.celrep.2021.108959
Doern, C. D. (2014). When Does 2 Plus 2 Equal 5? A Review of Antimicrobial Synergy Testing. J. Clin. Microbiol. 52, 4124–4128. doi: 10.1128/JCM.01121-14
Dyer, K. D., Schellens, I. M., Bonville, C. A., Martin, B. V., Domachowske, J. B., Rosenberg, H. F. (2007). Efficient Replication of Pneumonia Virus of Mice (PVM) in a Mouse Macrophage Cell Line. Virol. J. 4, 48. doi: 10.1186/1743-422X-4-48
Gao, J., Tian, Z., Yang, X. (2020). Breakthrough: Chloroquine Phosphate has Shown Apparent Efficacy in Treatment of COVID-19 Associated Pneumonia in Clinical Studies. Biosci. Trends 14, 72–73. doi: 10.5582/bst.2020.01047
Greco, W. R., Bravo, G., Parsons, J. C. (1995). The Search for Synergy: A Critical Review From a Response Surface Perspective. Pharmacol. Rev. 47, 331–385.
Hamming, I., Timens, W., Bulthuis, M. L. C., Lely, A. T., Navis, G. J., van Goor, H. (2004). Tissue Distribution of ACE2 Protein, the Functional Receptor for SARS Coronavirus. A First Step in Understanding SARS Pathogenesis. J. Pathol. 203, 631–637. doi: 10.1002/path.1570
Hojyo, S., Uchida, M., Tanaka, K., Hasebe, R., Tanaka, Y., Murakami, M., et al. (2020). How COVID-19 Induces Cytokine Storm With High Mortality. Inflamm. Regen. 40, 37. doi: 10.1186/s41232-020-00146-3
Jeffreys, L., Pennington, S. H., Duggan, J., Breen, A., Jinks, J., Ardrey, A., et al. (2020). Remdesivir-Ivermectin Combination Displays Synergistic Interaction With Improved In Vitro Antiviral Activity Against SARS-CoV-2. bioRxiv. 12.23.424232. doi: 10.1101/2020.12.23.424232
Kalil, A. C., Patterson, T. F., Mehta, A. K., Tomashek, K. M., Wolfe, C. R., Ghazaryan, V., et al. (2021). Baricitinib Plus Remdesivir for Hospitalized Adults With Covid-19. N. Engl. J. Med. 384, 795–807. doi: 10.1056/NEJMoa2031994
Kinobe, R. T., Owens, L. (2021). A Systematic Review of Experimental Evidence for Antiviral Effects of Ivermectin and an in Silico Analysis of Ivermectin’s Possible Mode of Action Against SARS-CoV-2. Fundam. Clin. Pharmacol. 35, 260–276. doi: 10.1111/fcp.12644
Klok, F. A., Kruip, M. J. H. A., van der Meer, N. J. M., Arbous, M. S., Gommers, D. A. M. P. J., Kant, K. M., et al. (2020). Incidence of Thrombotic Complications in Critically Ill ICU Patients With COVID-19. Thromb. Res. 191, 145–147. doi: 10.1016/j.thromres.2020.04.013
Krolewiecki, A., Lifschitz, A., Moragas, M., Travacio, M., Valentini, R., Alonso, D. F., et al. (2021). Antiviral Effect of High-Dose Ivermectin in Adults With COVID-19: A Proof-of-Concept Randomized Trial. EClinicalMedicine 37, 100959. doi: 10.1016/j.eclinm.2021.100959
López-Medina, E., López, P., Hurtado, I. C., Dávalos, D. M., Ramirez, O., Martínez, E., et al. (2021). Effect of Ivermectin on Time to Resolution of Symptoms Among Adults With Mild COVID-19: A Randomized Clinical Trial. JAMA 325, 1426–1435. doi: 10.1001/jama.2021.3071
Lv, J., Wang, Z., Qu, Y., Zhu, H., Zhu, Q., Tong, W., et al. (2021). Distinct Uptake, Amplification, and Release of SARS-CoV-2 by M1 and M2 Alveolar Macrophages. Cell Discov. 7, 24. doi: 10.1038/s41421-021-00258-1
Malmgaard, L., Melchjorsen, J., Bowie, A. G., Mogensen, S. C., Paludan, S. R. (2004). Viral Activation of Macrophages Through TLR-Dependent and -Independent Pathways. J. Immunol. 173, 6890–6898. doi: 10.4049/jimmunol.173.11.6890
Martin, L. R., Neal, Z. C., McBride, M. S., Palmenberg, A. C. (2000). Mengovirus and Encephalomyocarditis Virus Poly(C) Tract Lengths can Affect Virus Growth in Murine Cell Culture. J. Virol. 74, 3074–3081. doi: 10.1128/jvi.74.7.3074-3081.2000
Merad, M., Martin, J. C. (2020). Pathological Inflammation in Patients With COVID-19: A Key Role for Monocytes and Macrophages. Nat. Rev. Immunol. 20, 355–362. doi: 10.1038/s41577-020-0331-4
Mondal, S., Quintili, A. L., Karamchandani, K., Bose, S. (2020). Thromboembolic Disease in COVID-19 Patients: A Brief Narrative Review. J. Intensive Care 8, 70. doi: 10.1186/s40560-020-00483-y
Murali-Krishna, K., Ravi, V., Manjunath, R. (1995). Japanese Encephalitis Virus Infection of Mouse Cell Lines: Ability to Prime Mice for Generation of Virus Specific Cytotoxic T Lymphocytes and Differences in CTL Recognisable Viral Determinants. Arch. Virol. 140, 127–143. doi: 10.1007/BF01309728
Narasaraju, T., Tang, B. M., Herrmann, M., Muller, S., Chow, V. T. K., Radic, M. (2020). Neutrophilia and NETopathy as Key Pathologic Drivers of Progressive Lung Impairment in Patients With COVID-19. Front. Pharmacol. 11, 870. doi: 10.3389/fphar.2020.00870
Nativel, B., Couret, D., Giraud, P., Meilhac, O., d’Hellencourt, C. L., Viranaïcken, W., et al. (2017). Porphyromonas Gingivalis Lipopolysaccharides Act Exclusively Through TLR4 With a Resilience Between Mouse and Human. Sci. Rep. 7, 15789. doi: 10.1038/s41598-017-16190-y
Ng, H. H., Narasaraju, T., Phoon, M. C., Sim, M. K., Seet, J. E., Chow, V. T. (2012). Doxycycline Treatment Attenuates Acute Lung Injury in Mice Infected With Virulent Influenza H3N2 Virus: Involvement of Matrix Metalloproteinases. Exp. Mol. Pathol. 92, 287–295. doi: 10.1016/j.yexmp.2012.03.003
Park, M. D. (2020). Macrophages: A Trojan Horse in COVID-19? Nat. Rev. Immunol. 20, 351. doi: 10.1038/s41577-020-0317-2
Pope, M., Marsden, P. A., Cole, E., Sloan, S., Fung, L. S., Ning, Q., et al. (1998). Resistance to Murine Hepatitis Virus Strain 3 Is Dependent on Production of Nitric Oxide. J. Virol. 72, 7084–7090. doi: 10.1128/JVI.72.9.7084-7090.1998
Satarker, S., Ahuja, T., Banerjee, M., Balaji, V. E., Dogra, S., Agarwal, T., et al. (2020). Hydroxychloroquine in COVID-19: Potential Mechanism of Action Against SARS-CoV-2. Curr. Pharmacol. Rep. 24, 1–9. doi: 10.1007/s40495-020-00231-8
Seet, R. C. S., Quek, A. M. L., Ooi, D. S. Q., Sengupta, S., Lakshminarasappa, S. R., Koo, C. Y., et al. (2021). Positive Impact of Oral Hydroxychloroquine and Povidone-Iodine Throat Spray for COVID-19 Prophylaxis: An Open-Label Randomized Trial. Int. J. Infect. Dis. 106, 314–322. doi: 10.1016/j.ijid.2021.04.035
Sharma, A., Ahmad Farouk, I., Lal, S. K. (2021a). COVID-19: A Review on the Novel Coronavirus Disease Evolution, Transmission, Detection, Control and Prevention. Viruses 13, 202. doi: 10.3390/v13020202
Sharma, A., Ong, J. W., Loke, M. F., Chua, E. G., Lee, J. J., Choi, H. W., et al. (2021b). Comparative Transcriptomic and Molecular Pathway Analyses of HL-CZ Human Pro-Monocytic Cells Expressing SARS-CoV-2 Spike S1, S2, NP, NSP15 and NSP16 Genes. Microorganisms 9, 1193. doi: 10.3390/microorganisms9061193
Shi, Y., Luo, P., Wang, W., Horst, K., Bläsius, F., Relja, B., et al. (2020). M1 But Not M0 Extracellular Vesicles Induce Polarization of RAW264.7 Macrophages via the TLR4-Nfκb Pathway In Vitro. Inflammation 43, 1611–1619. doi: 10.1007/s10753-020-01236-7
Shirato, K., Kizaki, T. (2021). SARS-CoV-2 Spike Protein S1 Subunit Induces Pro-Inflammatory Responses via Toll-Like Receptor 4 Signaling in Murine and Human Macrophages. Heliyon 7, e06187. doi: 10.1016/j.heliyon.2021.e06187
Sica, A., Erreni, M., Allavena, P., Porta, C. (2015). Macrophage Polarization in Pathology. Cell. Mol. Life Sci. 72, 4111–4126. doi: 10.1007/s00018-015-1995-y
Smith, T. D., Tse, M. J., Read, E. L., Liu, W. F. (2016). Regulation of Macrophage Polarization and Plasticity by Complex Activation Signals. Integr. Biol. 8, 946–955. doi: 10.1039/c6ib00105j
Sun, Z., Pan, Y., Qu, J., Xu, Y., Dou, H., Hou, Y. (2020). 17β-Estradiol Promotes Trained Immunity in Females Against Sepsis Via Regulating Nucleus Translocation of RelB. Front. Immunol. 11, 1591. doi: 10.3389/fimmu.2020.01591
Sun, W., Sanderson, P., Zheng, W. (2016). Drug Combination Therapy Increases Successful Drug Repositioning. Drug Discov. Today 21, 1189–1195. doi: 10.1016/j.drudis.2016.05.015
Tang, Y., Liu, J., Zhang, D., Xu, Z., Ji, J., Wen, C. (2020). Cytokine Storm in COVID-19: The Current Evidence and Treatment Strategies. Front. Immunol. 11, 1708. doi: 10.3389/fimmu.2020.01708
Traber, K. E., Symer, E. M., Allen, E., Kim, Y., Hilliard, K. L., Wasserman, G. A., et al. (2017). Myeloid-Epithelial Cross Talk Coordinates Synthesis of the Tissue-Protective Cytokine Leukemia Inhibitory Factor During Pneumonia. Am. J. Physiol. Lung Cell. Mol. Physiol. 313, L548–L558. doi: 10.1152/ajplung.00482.2016
Vincent, M. J., Bergeron, E., Benjannet, S., Erickson, B. R., Rollin, P. E., Ksiazek, T. G., et al. (2005). Chloroquine is a Potent Inhibitor of SARS Coronavirus Infection and Spread. Virol. J. 2, 69. doi: 10.1186/1743-422X-2-69
Wagstaff, K. M., Sivakumaran, H., Heaton, S. M., Harrich, D., Jans, D. A. (2012). Ivermectin is a Specific Inhibitor of Importin α/β-Mediated Nuclear Import Able to Inhibit Replication of HIV-1 and Dengue Virus. Biochem. J. 443, 851–856. doi: 10.1042/BJ20120150
Wang, W., Ye, L., Ye, L., Li, B., Gao, B., Zeng, Y., et al. (2007). Up-Regulation of IL-6 and TNF-Alpha Induced by SARS-Coronavirus Spike Protein in Murine Macrophages Via NF-KappaB Pathway. Virus Res. 128, 1–8. doi: 10.1016/j.virusres.2007.02.007
Wang, Y., Zhang, D., Du, G., Du, R., Zhao, J., Jin, Y., et al. (2020). Remdesivir in Adults With Severe COVID-19: A Randomised, Double-Blind, Placebo-Controlled, Multicentre Trial. Lancet 395, 1569–1578. doi: 10.1016/S0140-6736(20)31022-9
Wölfel, R., Corman, V. M., Guggemos, W., Seilmaier, M., Zange, S., Muller, M. A., et al. (2020). Virological Assessment of Hospitalized Patients With COVID-2019. Nature 581, 465–469. doi: 10.1038/s41586-020-2196-x
Wu, C., Liu, Y., Yang, Y., Zhang, P., Zhong, W., Wang, Y., et al. (2020). Analysis of Therapeutic Targets for SARS-CoV-2 and Discovery of Potential Drugs by Computational Methods. Acta Pharm. Sin. B 10, 766–788. doi: 10.1016/j.apsb.2020.02.008
Yang, S. N. Y., Atkinson, S. C., Wang, C., Lee, A., Bogoyevitch, M. A., Borg, N. A., et al. (2020). The Broad Spectrum Antiviral Ivermectin Targets the Host Nuclear Transport Importin α/β1 Heterodimer. Antiviral Res. 177, 104760. doi: 10.1016/j.antiviral.2020.104760
Yates, P. A., Newman, S. A., Oshry, L. J., Glassman, R. H., Leone, A. M., Reichel, E. (2020). Doxycycline Treatment of High-Risk COVID-19-Positive Patients With Comorbid Pulmonary Disease. Ther. Adv. Respir. Dis. 14, 1753466620951053. doi: 10.1177/1753466620951053
Keywords: coronavirus, murine hepatitis virus, remdesivir, chloroquine, ivermectin, doxycycline, combination treatment, RAW264.7 macrophage cells
Citation: Tan YL, Tan KSW, Chu JJH and Chow VT (2021) Combination Treatment With Remdesivir and Ivermectin Exerts Highly Synergistic and Potent Antiviral Activity Against Murine Coronavirus Infection. Front. Cell. Infect. Microbiol. 11:700502. doi: 10.3389/fcimb.2021.700502
Received: 26 April 2021; Accepted: 13 July 2021;
Published: 30 July 2021.
Edited by:
Charles T. Spencer, The University of Texas at El Paso, United StatesReviewed by:
Hugues Ouellet, The University of Texas at El Paso, United StatesScott Benjamin Biering, University of California, Berkeley, United States
Copyright © 2021 Tan, Tan, Chu and Chow. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Vincent T. Chow, bWljY3RrQG51cy5lZHUuc2c=
 Yu Ling Tan1
Yu Ling Tan1 Kevin S. W. Tan
Kevin S. W. Tan Justin Jang Hann Chu
Justin Jang Hann Chu Vincent T. Chow
Vincent T. Chow