
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Cell. Infect. Microbiol. , 16 July 2021
Sec. Clinical Microbiology
Volume 11 - 2021 | https://doi.org/10.3389/fcimb.2021.698807
This article is part of the Research Topic The Application of Phages Against Infectious Diseases View all 15 articles
Enterococcus faecalis is a Gram-positive opportunistic pathogen that could cause pneumonia and bacteremia in stroke patients. The development of antibiotic resistance in hospital-associated E. faecalis is a formidable public health threat. Bacteriophage therapy is a renewed solution to treat antibiotic-resistant bacterial infections. However, bacteria can acquire phage resistance quite quickly, which is a significant barrier to phage therapy. Here, we characterized a lytic E. faecalis bacteriophage Vb_EfaM_LG1 with lytic activity. Its genome did not contain antibiotic resistance or virulence genes. Vb_EfaM_LG1 effectively inhibits E. faecalis growth for a short period, and phage resistance developed within hours. However, the combination of antibiotics and phage has a tremendous synergistic effect against E. faecalis, prevents the development of phage resistance, and disrupts the biofilm efficiently. Our results show that the phage-antibiotic combination has better killing efficiency against E. faecalis.
Enterococci are Gram-positive facultative anaerobes, examples of which include E. faecalis and Enterococcus faecium, which cause bacteremia, pneumonia, endocarditis, and urinary tract infections (Beganovic et al., 2018; Jabbari Shiadeh et al., 2019). In addition, E. faecalis is also one of the major pathogens for pneumonia and bacteremia in stroke patients, and the infection after stroke could lead to the death of the stroke patient (Hannawi et al., 2013; Dyal and Sehgal, 2015; Stanley et al., 2016). Moreover, the intrinsic and acquired antibiotic resistance of Enterococci is a formidable public health threat (Arias et al., 2011; Banla et al., 2018). Enterococci have evolved extensive drug resistance, including that to vancomycin, and could transmit antibiotic resistance among diverse bacteria (Palmer et al., 2010). Therefore, new therapeutic approaches are needed to treat Enterococcal-associated infections (Khalifa et al., 2015; Kortright et al., 2019; Bao et al., 2020).
Phages are viruses that infect and kill bacteria and are used to treat antibiotic-resistant bacteria (Barbu et al., 2016; Kortright et al., 2019; Dion et al., 2020). Phage therapy has several advantages over antibiotics. First, phages have a particular host range and only infect the targeted bacterium, so phage therapy would not affect other bacteria and did not interrupt the commensal microbes (Waters et al., 2017). Second, because of the different phage resistance and antibiotic resistance mechanisms in bacteria, phages could infect multidrug-resistant superbugs. Thus, phage therapy is being proceeded in many countries (Jault et al., 2018; Leitner et al., 2021).
Currently, numerous phages against pathogens had been characterized; however, there are only 63 sequenced E. faecalis bacteriophage deposited in NCBI (Chatterjee et al., 2021), which is relatively understudied compared with phages that infect other pathogens, such as Pseudomonas aeruginosa or Staphylococcus aureus phages (De Smet et al., 2017). More phages need to be characterized to provide more therapeutic options for treating the multidrug-resistant E. faecalis. Moreover, phage resistance is quite common for E. faecalis, which could be quickly selected because of the mutations of cell wall-associated polysaccharide or membrane protein (Duerkop et al., 2016; Banla et al., 2018; Chatterjee et al., 2019). Thus, a better strategy to hinder phage resistance should be investigated. In this study, we identified a phage infecting a broad range of E. faecalis strains and proved that phage-antibiotic synergism effectively inhibits phage resistance and disrupts biofilm.
A phage was isolated from the hospital sewage using E. faecalis strain ef118 as a host. It forms an obvious plaque on the host in the double layer agar plates (Figure 1A). The phage particle was extracted from the bacterial lysate and was observed by transmission electron microscopy. The head of the phage is a regular icosahedral structure with a diameter of approximately 80 nm, and it has a contractable tail with a length of approximately 110 nm (Figure 1B). Thus, the morphology of this phage conforms to the characteristics of the Myoviridae family, and it is named Enterococcus faecalis phage vB_EfaM_LG1 (refer as LG1 hereafter).
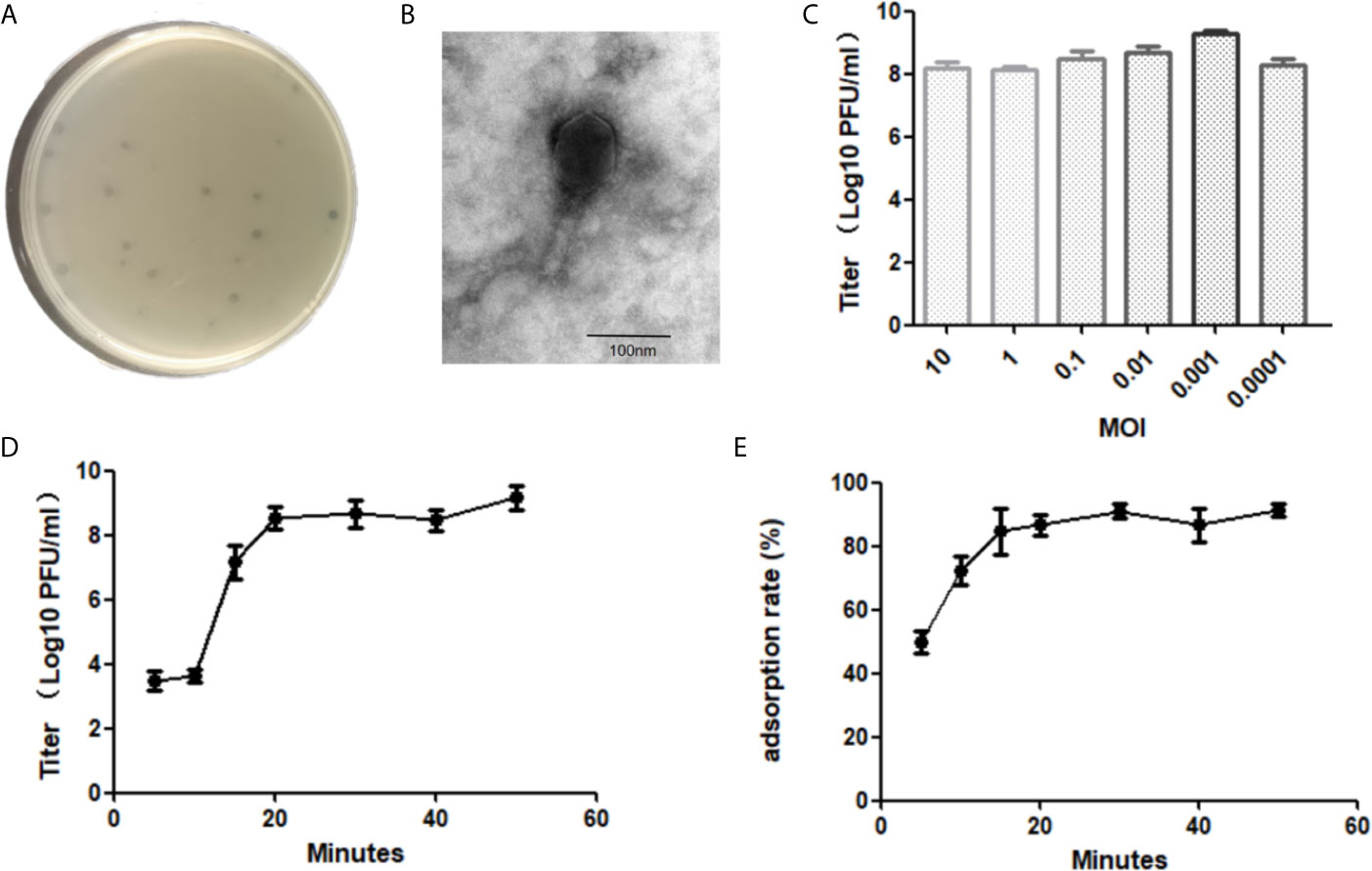
Figure 1 Biological characterization of E. faecalis phage vB_EfaM_LG1. The plaque (A) and transmission electron micrograph (B) of LG1. (C) The optimal MOI test of phage. (D) The one-step growth curve of LG1. (E) The adsorption rate of LG1 against host strain ef118 within 60 min.
The phage titer reached the highest as 4 × 109 PFU/ml when the multiplicity of infection (MOI) was 0.001, the optimal MO of bacteriophage LG1 was 0.001 (Figure 1C). The one-step growth curve of LG1 was shown in Figure 1D. The latent phase was approximately 10 min, and then the titer of phages increased rapidly between 10 and 20 min, indicating a lysis period of approximately 20 min. The burst size was estimated as about 40 pfu per bacterium.
The adsorption rate of LG1 onto the host strain was determined by measuring the remaining phages in the supernatant. LG1 absorbed onto the host ef118 efficiently, and over 50% of the phage particles were adsorbed by the ef118 within 5 min, and approximately 80% of the phage could bind to the host within 20 min (Figure 1E).
Spot agar assays were performed to determine the phage infectivity against 10 E. faecalis clinically isolated strains. The formation of clear plaques indicates that the strain is sensitive to LG1, whereas the formation of blurred plaque or no spots is considered non-sensitive. LG1 infects 50% of the clinical isolated E. faecalis strains, representing a relatively broad host range, but LG1 cannot infect any E. faecium strain (Table 1).
Phage LG1 is a double-stranded (ds) DNA phage with a linear genome of 150,025 base pairs (bp). Its G + C content is 35.88%, and the genome is visualized by CPT Phage Galaxy (Ramsey et al., 2020).
There are 231 putative ORFs predicted by RAST (Overbeek et al., 2014), whereas most of the ORFs are functionally unknown. Also, LG1 encodes five tRNA genes. The annotated ORFs can be categorized into several functional modules, including phage replications, DNA metabolism/modifications, lysis, phage structural protein (Figure 2). LG1 encodes multiple ribonucleotide reductases, implying that LG1 could perform de novo DNA biosynthesis. Moreover, no antibiotic-resistant genes or virulence genes were predicted in the genome of LG1. BlastN searches of the non-redundant database at NCBI reveals that LG1 genome exhibits 90% to 98% nucleotide identity with a group of enterococcal phages, such as Enterococcus phage ECP3 and vB_EfaM_Ef2.1.
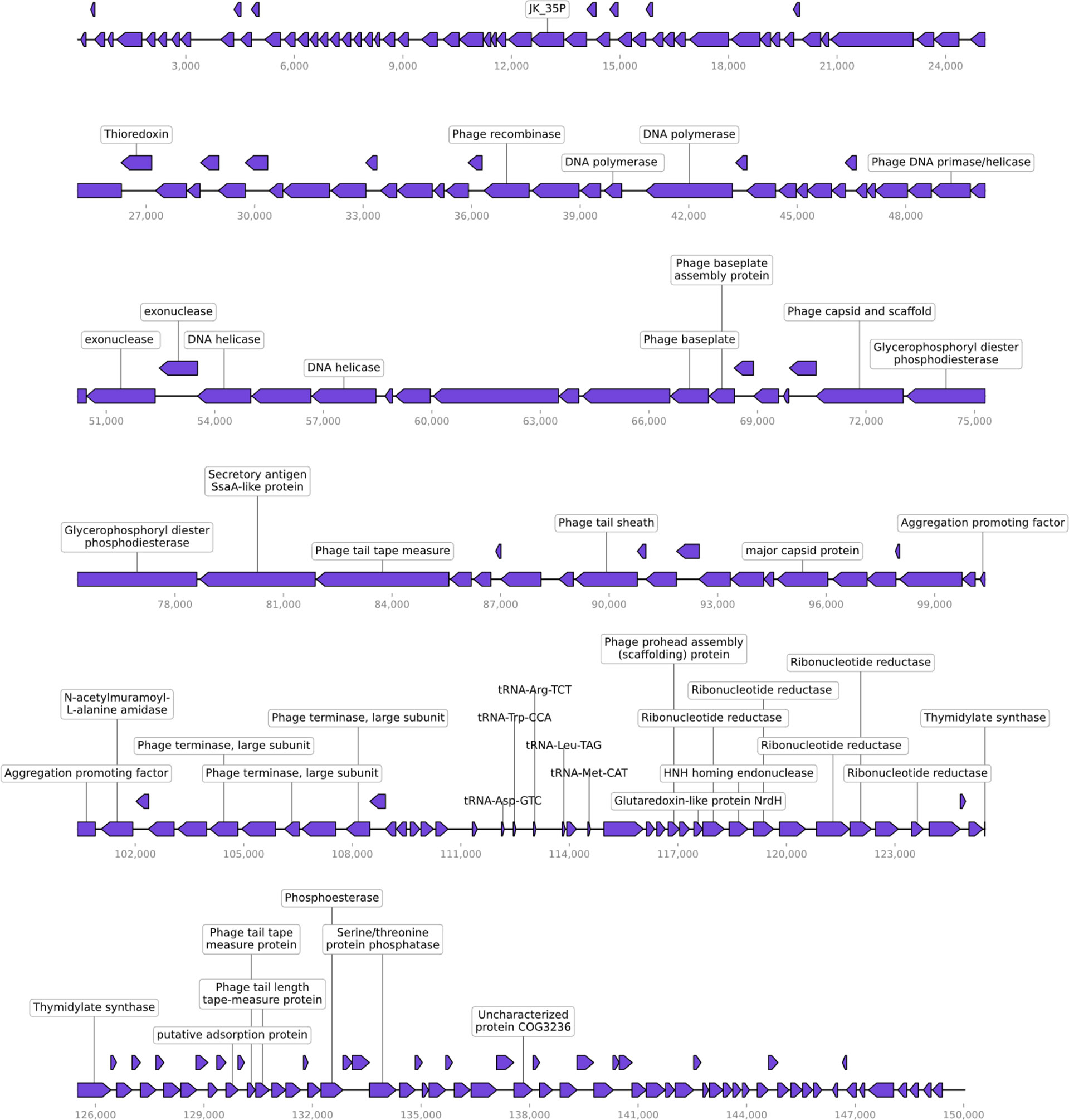
Figure 2 Genomic characterization of vB_EfaM_LG1. LG1 is a dsDNA phage with 231 proteins predicted based on sequence homology and five tRNA genes.
The optimal pH for storing LG1 was 7, and its viability was lost entirely when the pH was lower than 4 or higher than 11 (Figure 3A). The phage titers were further monitored when LG1 was incubated at different temperatures. It was found to be stable at different temperatures, maintained a titer of 105 after 60 min incubation at 70°C (Figure 3B), and was wholly inactivated over 80°C. Besides, chloroform treatment did not affect the phage titer, precluding the presence of lipid components on the phage surface (Figure 3C). Finally, the chloroform-treated phage was stored at 4°C, and its titer was monitored for 3 months (Figure 3D). And the titer of LG1 did not significantly decrease during this period, indicating LG1 was relatively stable at 4°C, and this feature is vital to produce phage agents.
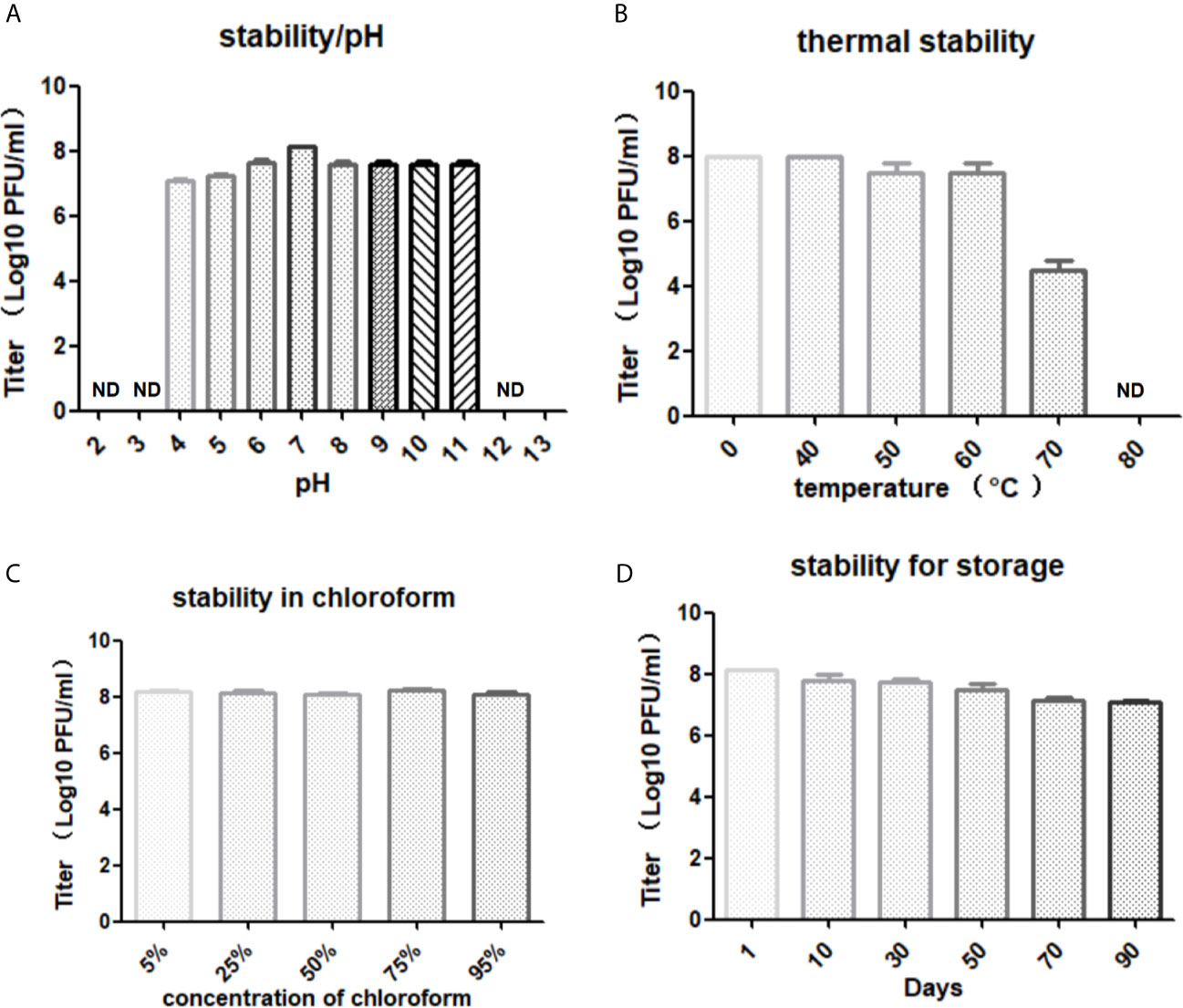
Figure 3 Stability of LG1. (A) Phage LG1 is stable under pH4~11 but significantly inactivated under pH4 or above pH11. (B) LG1 is inactivated by 80°C treatment. (C) LG1 is non-sensitive to chloroform treatment. (D) LG1 is stable for 3 months without a significant decrease of titer when stored at 4°C. ND, not detected.
Phage resistance is quickly developed and is selected in vitro and in vivo (Labrie et al., 2010). And phage resistance in E. faecalis can be achieved through mutations of the receptors on the cell surface (Duerkop et al., 2016). As expected, in the liquid culture, phage LG1 was added to the log phase ef118 (OD600 = 0.5) to a final titer of 5 × 108 pfu/ml, and LG1 could only inhibit E. faecalis ef118 for several hours, and phage-resistant mutants regrow to a high density within 24 h (Figure 4A). The sensitive antibiotic cefotaxime (32 µg/ml) could inhibit the ef118, but the phage-antibiotic combination shows the best killing efficiency (Figure 4A). And in the in vitro biofilm model, cefotaxime (32 µg/ml) is less effective in disrupting the established biofilm than phage (5*108 pfu/ml) alone, and the phage-antibiotic combination has a more significant effect in disrupting the biofilm than single treatment (Figure 4B).
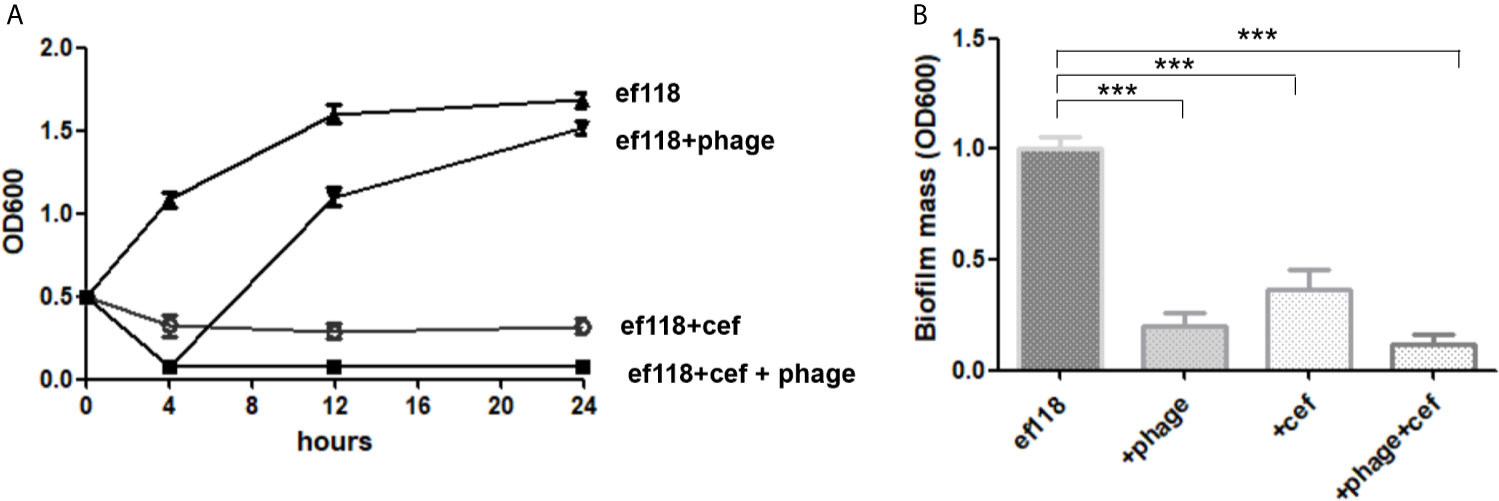
Figure 4 Phage-antibiotic synergism. (A) Phage could only inhibit the growth of E. faecalis for several hours, and then the phage resistance mutant grows to a high density. (B) The phage-antibiotic combination has better efficacy in destroying the biofilm than phage or antibiotic alone (***P < 0.05).
With an ever-increasing amount of antibiotic-resistant strains of E. faecalis found in clinical and the difficulties in the treatment of those caused by the biofilm formation (Arias et al., 2011; Palmer et al., 2011). A better strategy to constrain E. faecalis infection is needed more than ever, and lytic bacteriophage is a promising alternative treatment to fight multidrug-resistant E. faecalis (Al-Zubidi et al., 2019). In this study, we isolated a dsDNA phage LG1, which effectively infects E. faecalis strains with a relatively broad host range. Transmission electron microscopy showed that the phage belongs to the Myoviridae family, and its genome sequence exhibited similarity to other E. faecalis phages in the Myoviridae family.
Phage stability is an essential parameter for manufacturing phage agents (Pires et al., 2020). In Phagoburn project, researchers found that the phage cocktail is significantly inactivated because of long-term storage, and the phage titer is as low as 102 pfu/ml per daily dose, which is one of the reasons for the failure of this phage therapy clinical trial (Jault et al., 2018). LG1 is stable under different conditions, including heat and pH, and it can be stored at 4°C without significant loss of the titer for 3 months, which is an important parameter when LG1 is included in a phage cocktail agent.
Phage resistance is also an issue in phage therapy. Because bacteria are able to obtain phage resistance because of various mechanisms, including mutations of the receptor, restriction and modification systems, CRISPR-cas systems (Labrie et al., 2010; Goldfarb et al., 2015; Shen et al., 2018b; Azam and Tanji, 2019), and phage resistance have been reported in phage therapy cases (El Haddad et al., 2019; Bao et al., 2020), which is a severe issue in phage therapy. E. faecalis phage resistance has been investigated previously, mainly through the mutation of phage receptor, including membrane protein PIP for phage phiVPE25 (Duerkop et al., 2016) and enterococcal polysaccharide antigen for phage (Chatterjee et al., 2019). Various approaches had been suggested to inhibit the development of phage resistance. Phage-antibiotic is the well-acknowledged method in treating other pathogens, such as P. aeruginosa (Oechslin et al., 2017). This study also suggests that phage-antibiotic combination is a better strategy to treat E. faecalis infection.
Formation of biofilm is a severe issue in infections because the established biofilm is extremely difficult to disrupt, and the biofilm increases antibiotic resistance (Pires et al., 2017). Phage effectively disrupts biofilm because phage could penetrate the biofilm, and some phage encodes depolymerase to degrade the biofilm matrix to destroy further the biofilm. Depolymerases can be associated with the phage particle or be released during lysis of the host bacteria. Depolymerases are enzymes to degrade the extracellular polysaccharide. Therefore, it is particularly interesting in the removal of biofilms. (Wu et al., 2019; Ferriol-Gonzalez and Domingo-Calap, 2020).
Moreover, this experiment shows phage-antibiotic combination has better effects in treating biofilms, which would be a better approach to treat chronic E. faecalis when biofilm might have already formed. The phage-antibiotic synergism is mainly because of the different antibacterial targets. And under certain conditions, phages provide an adjuvating effect by lowering the minimum inhibitory concentration for drug-resistant strains to enhancing the effect of antibiotics (Liu et al., 2020). Overall, these data indicate phage-antibiotic synergism has better treating efficiency than single phage therapy.
The bacterial strains in this work were listed in Table 1. Enterococcus strains were collected from the Department of Clinical Laboratory Medicine and grown aerobically on Luria-Bertani (LB) broth at 37°C.
Bacteriophage LG1 was isolated from hospital sewage as previously described (Duerkop et al., 2016). Briefly, the sewage was pelleted, and the supernatant was filtered through a 0.45-µm pore-size filter to remove particles. Then, 50-µl sample was immediately mixed with 200 µl bacterial culture, and 4 ml of molten LB soft agar (0.7%) was added and poured onto LB agar plates, followed by overnight culture. Any formed plaque was picked using a pipette, deposited in 1 ml of LB, followed by 10-fold dilution, and double-layer agar assay to purify the phage. The phage was purified by three consecutive rounds. Then, one plaque from the third round was picked for this study.
Phage particles were dropped on carbon-coated copper grids for 10 min. Then phosphotungstic acid (pH 7.0) was used to stain the sample for 15 s and examined under a Philips EM 300 electron microscope. The sizes of the phage were measured based on five randomly selected images using AxioVision LE.
Phage titer was calculated by standard double-layer agar plate assay. Briefly, 10-fold dilutions of phage suspension were mixed with 200 µl host bacteria, then mixed with 5 ml molten 0.7% LB agar broth. Then poured on a 1.5% agar plate. After overnight incubation at 37°C, one plaque is calculated as a plaque-forming unit (pfu). MOI experiments were performed by mixing log-phase bacteria (OD600 = 0.5) with a different number of phages, and the coculture was incubated at 37°C with shaking for 5 h. Then the titer in the supernatant was calculated using a double-layer agar plate assay.
The one-step growth curve of LG1 was determined as described (Zhong et al., 2020). Briefly, 1 ml of log-phase bacteria and 1 ml of LG1 were mixed at an MOI of 1 and incubated at 37°C for 3 min. Then, the mixture was centrifuged at 4°C for 2min at a speed of 12,000g, and the pellet was resuspended in 10 ml LB medium. And samples were taken at the given time points, which are immediately pelleted, and phage titer in the supernatant was measured by directly using double-layer plate assay.
Bacteriophage adsorption assay with various time points was performed as previously described (Al-Zubidi et al., 2019). Briefly, the log phase bacterial cultures were pelleted and resuspended in LB medium to a final concentration of 5 × 108 CFU/ml. Then, phage was added to a final titer of 5 × 105 pfu/ml. Then, the samples were cultured at 37°C for 60 min, and a 1-ml sample was collected at the set time point and centrifuged at 16,000g for 1 min. The phages in the supernatant were titered using the double-agar plating assays. At a given time point, the adsorption rate was calculated as (the original phage titer − the remaining phage titer)/the original phage titer.
Ten E. faecalis and five E. faecium strains were selected as test strains. The host range of phage LG1 was determined using spot testing by dropping 1 µl of phage onto the double-layer soft agar premixed with the test strain and cultured at 37°C for 18 h. The formation of a clear plaque is considered as the sensitive host for phage LG1.
The phage DNA extraction is performed as previously described (Khan et al., 2021). Briefly, DNase I and RNase A were added to a final concentration of 5 and 1 μg/ml, respectively, and the purified phage particle was treated for 1 h at 37°C. Proteinase K (final concentration of 50 μg/ml), EDTA (pH 8.0), and 0.5% SDS were added and treated at 56°C for 1 h. Then, phage genome DNA was extracted with saturated phenol (pH 8.0). After centrifugation, the aqueous phase was extracted with chloroform and mixed with the same volume of isopropyl alcohol and stored at −20°C for 1 h. Then, phage DNA was precipitated by centrifugation and was washed with 70% ethanol and absolute ethanol, respectively. After drying, the precipitate was dissolved in TE solution, and the phage DNA was stored at −80°C.
Phage genomic DNA was sequenced using an Illumina Hiseq 2500 platform (~1 Gbp/sample). Fastp (Chen et al., 2018) was used for adapter trimming and quality filtering after demultiplexing the raw reads. The read data were assembled using the de novo assembly algorithm Newbler Version2.9 with default parameters, and the assembled genome was annotated by RAST. The DNA and protein sequences were checked for homologs with BLAST manually. The genome map was drawn by a phage genome visualization online software CPT Phage Galaxy (Ramsey et al., 2020). The sequence data are available in the NCBI under accession number MZ420150.
To test the phage stability under various conditions, 108 pfu of LG1 was treated with different pH, temperature, or chloroform for 60 min, then the titer of the phage was calculated by double-layer agar assay. The LG1 was stored at 4°C, and its titer was determined at the given time points for 3 months.
Biofilms were examined by the crystal violet staining method as previously described (Shen et al., 2018a). Briefly, 0.2 ml of log-phase bacterial culture were added to 96-well polystyrene microplates and incubated for 24 h at 37°C to establish biofilm. Then, the untreated control wells were washed with phosphate-buffered saline (PBS) and stained with crystal violet for 15 min, which was solubilized in 0.2 ml of 95% ethanol, and the biofilm biomass was estimated by measuring the OD 600, which was determined using a SpectraMax M3 multimode microplate reader. For the treatment groups, the wells were washed and PBS, then 0.2 ml of phage or antibiotic was added and incubated at 37°C for 4 h, the biofilm biomass was determined by crystal violet staining method.
All the experiments were performed three times, and statistical analysis was performed using one-way ANOVA or t-test, and statistical significance was assumed if the P value was <0.05.
The data sets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/, MZ420150, PRJNA723057.
All the authors listed have made a substantial, direct and intellectual contribution to this work, and approved the submitted version for publication.
This work was supported by Chongqing Natural Science Foundation(cstc2019jcyj-msxmX0173).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Al-Zubidi, M., Widziolek, M., Court, E. K., Gains, A. F., Smith, R. E., Ansbro, K., et al. (2019). Identification of Novel Bacteriophages With Therapeutic Potential That Target Enterococcus Faecalis. Infect. Immun. 87 (11), e00512–19. doi: 10.1128/IAI.00512-19
Arias, C. A., Panesso, D., McGrath, D. M., Qin, X., Mojica, M. F., Miller, C., et al. (2011). Genetic Basis for In Vivo Daptomycin Resistance in Enterococci. N Engl. J. Med. 365, 892–900. doi: 10.1056/NEJMoa1011138
Azam, A. H., Tanji, Y. (2019). Bacteriophage-Host Arm Race: An Update on the Mechanism of Phage Resistance in Bacteria and Revenge of the Phage With the Perspective for Phage Therapy. Appl. Microbiol. Biotechnol. 103, 2121–2131. doi: 10.1007/s00253-019-09629-x
Banla, I. L., Kommineni, S., Hayward, M., Rodrigues, M., Palmer, K. L., Salzman, N. H., et al. (2018). Modulators of Enterococcus Faecalis Cell Envelope Integrity and Antimicrobial Resistance Influence Stable Colonization of the Mammalian Gastrointestinal Tract. Infect. Immun. 86 (1), e00381-17. doi: 10.1128/IAI.00381-17
Bao, J., Wu, N., Zeng, Y., Chen, L., Li, L., Yang, L., et al. (2020). Non-Active Antibiotic and Bacteriophage Synergism to Successfully Treat Recurrent Urinary Tract Infection Caused by Extensively Drug-Resistant Klebsiella Pneumoniae. Emerg. Microbes Infect. 9, 771–774. doi: 10.1080/22221751.2020.1747950
Barbu, E. M., Cady, K. C., Hubby, B. (2016). Phage Therapy in the Era of Synthetic Biology. Cold Spring Harbor Perspect. Biol. 8 (10), a023879. doi: 10.1101/cshperspect.a023879
Beganovic, M., Luther, M. K., Rice, L. B., Arias, C. A., Rybak, M. J., LaPlante, K. L. (2018). A Review of Combination Antimicrobial Therapy for Enterococcus Faecalis Bloodstream Infections and Infective Endocarditis. Clin. Infect. Dis: Off. Publ. Infect. Dis. Soc. America 67, 303–309. doi: 10.1093/cid/ciy064
Chatterjee, A., Johnson, C. N., Luong, P., Hullahalli, K., McBride, S. W., Schubert, A. M., et al. (2019). Bacteriophage Resistance Alters Antibiotic-Mediated Intestinal Expansion of Enterococci. Infect. Immun. 87 (6), e00085-19. doi: 10.1128/IAI.00085-19
Chatterjee, A., Willett, J. L. E., Dunny, G. M., Duerkop, B. A. (2021). Phage Infection and Sub-Lethal Antibiotic Exposure Mediate Enterococcus Faecalis Type VII Secretion System Dependent Inhibition of Bystander Bacteria. PloS Genet. 17, e1009204. doi: 10.1371/journal.pgen.1009204
Chen, S., Zhou, Y., Chen, Y., Gu, J. (2021). Fastp: An Ultra-Fast All-In-One Fastq Preprocessor. Bioinformatics 34 (17), i884–i890. doi: 10.1093/bioinformatics/bty560
De Smet, J., Hendrix, H., Blasdel, B. G., Danis-Wlodarczyk, K., Lavigne, R. (2017). Pseudomonas Predators: Understanding and Exploiting Phage-Host Interactions. Nat. Rev. Microbiol. 15, 517–530. doi: 10.1038/nrmicro.2017.61
Dion, M. B., Oechslin, F., Moineau, S. (2020). Phage Diversity, Genomics and Phylogeny. Nat. Rev. Microbiol. 18, 125–138. doi: 10.1038/s41579-019-0311-5
Duerkop, B. A., Huo, W., Bhardwaj, P., Palmer, K. L., Hooper, L. V. (2016). Molecular Basis for Lytic Bacteriophage Resistance in Enterococci. mBio 7 (4), e01304-16. doi: 10.1128/mBio.01304-16
Dyal, H. K., Sehgal, R. (2015). The Catastrophic Journey of a Retained Temporary Epicardial Pacemaker Wire Leading to Enterococcus Faecalis Endocarditis and Subsequent Stroke. BMJ Case Rep. 2015, bcr2014206215. doi: 10.1136/bcr-2014-206215
El Haddad, L., Harb, C. P., Gebara, M. A., Stibich, M. A., Chemaly, R. F. (2019). A Systematic and Critical Review of Bacteriophage Therapy Against Multidrug-Resistant ESKAPE Organisms in Humans. Clin. Infect. Dis: Off. Publ. Infect. Dis. Soc. America 69, 167–178. doi: 10.1093/cid/ciy947
Ferriol-Gonzalez, C., Domingo-Calap, P. (2020). Phages for Biofilm Removal. Antibiotics 9 (5), 268. doi: 10.3390/antibiotics9050268
Goldfarb, T., Sberro, H., Weinstock, E., Cohen, O., Doron, S., Charpak-Amikam, Y., et al. (2015). BREX Is a Novel Phage Resistance System Widespread in Microbial Genomes. EMBO J. 34, 169–183. doi: 10.15252/embj.201489455
Hannawi, Y., Hannawi, B., Rao, C. P., Suarez, J. I., Bershad, E. M. (2013). Stroke-Associated Pneumonia: Major Advances and Obstacles. Cerebrovasc Dis. 35, 430–443. doi: 10.1159/000350199
Jabbari Shiadeh, S. M., Pormohammad, A., Hashemi, A., Lak, P. (2019). Global Prevalence of Antibiotic Resistance in Blood-Isolated Enterococcus Faecalis and Enterococcus Faecium: A Systematic Review and Meta-Analysis. Infect. Drug Resist. 12, 2713–2725. doi: 10.2147/IDR.S206084
Jault, P., Leclerc, T., Jennes, S., Pirnay, J. P., Que, Y. A., Resch, G., et al. (2018). Efficacy and Tolerability of a Cocktail of Bacteriophages to Treat Burn Wounds Infected by Pseudomonas Aeruginosa (PhagoBurn): A Randomised, Controlled, Double-Blind Phase 1/2 Trial. Lancet Infect. Dis. 19 (1), 35–45. doi: 10.1016/S1473-3099(18)30482-1
Khalifa, L., Brosh, Y., Gelman, D., Coppenhagen-Glazer, S., Beyth, S., Poradosu-Cohen, R., et al. (2015). Targeting Enterococcus Faecalis Biofilms With Phage Therapy. Appl. Environ. Microbiol. 81, 2696–2705. doi: 10.1128/AEM.00096-15
Khan, F. M., Gondil, V. S., Li, C., Jiang, M., Li, J., Yu, J., et al. (2021). A Novel Acinetobacter Baumannii Bacteriophage Endolysin LysAB54 With High Antibacterial Activity Against Multiple Gram-Negative Microbes. Front. Cell Infect. Microbiol. 11, 637313. doi: 10.3389/fcimb.2021.637313
Kortright, K. E., Chan, B. K., Koff, J. L., Turner, P. E. (2019). Phage Therapy: A Renewed Approach to Combat Antibiotic-Resistant Bacteria. Cell Host Microbe 25, 219–232. doi: 10.1016/j.chom.2019.01.014
Labrie, S. J., Samson, J. E., Moineau, S. (2010). Bacteriophage Resistance Mechanisms. Nat. Rev. Microbiol. 8, 317–327. doi: 10.1038/nrmicro2315
Leitner, L., Ujmajuridze, A., Chanishvili, N., Goderdzishvili, M., Chkonia, I., Rigvava, S., et al. (2021). Intravesical Bacteriophages for Treating Urinary Tract Infections in Patients Undergoing Transurethral Resection of the Prostate: A Randomised, Placebo-Controlled, Double-Blind Clinical Trial. Lancet Infect. Dis. 21, 427–436. doi: 10.1016/S1473-3099(20)30330-3
Liu, C., Green, S., Min, L., Clark, J., Salazar, K., Terwilliger, A., et al. (2020). Phage-Antibiotic Synergy Is Driven by a Unique Combination of Antibacterial Mechanism of Action and Stoichiometry. mbio, 01462–01420. doi: 10.1128/mBio.01462-20
Oechslin, F., Piccardi, P., Mancini, S., Gabard, J., Moreillon, P., Entenza, J. M., et al. (2017). Synergistic Interaction Between Phage Therapy and Antibiotics Clears Pseudomonas Aeruginosa Infection in Endocarditis and Reduces Virulence. J. Infect. Dis. 215, 703–712. doi: 10.1093/infdis/jiw632
Overbeek, R., Olson, R., Pusch, G. D., Olsen, G. J., Davis, J. J., Disz, T., et al. (2014). The SEED and the Rapid Annotation of Microbial Genomes Using Subsystems Technology (RAST). Nucleic Acids Res. 42, D206–D214. doi: 10.1093/nar/gkt1226
Palmer, K. L., Daniel, A., Hardy, C., Silverman, J., Gilmore, M. S. (2011). Genetic Basis for Daptomycin Resistance in Enterococci. Antimicrob. Agents Chemother. 55, 3345–3356. doi: 10.1128/AAC.00207-11
Palmer, K. L., Kos, V. N., Gilmore, M. S. (2010). Horizontal Gene Transfer and the Genomics of Enterococcal Antibiotic Resistance. Curr. Opin. Microbiol. 13, 632–639. doi: 10.1016/j.mib.2010.08.004
Pires, D. P., Costa, A. R., Pinto, G., Meneses, L., Azeredo, J. (2020). Current Challenges and Future Opportunities of Phage Therapy. FEMS Microbiol. Rev. 44, 684–700. doi: 10.1093/femsre/fuaa017
Pires, D. P., Melo, L. D. R., Boas, D. V., Sillankorva, S., Azeredo, J. (2017). Phage Therapy as an Alternative or Complementary Strategy to Prevent and Control Biofilm-Related Infections. Curr. Opin. Microbiol. 39, 48–56. doi: 10.1016/j.mib.2017.09.004
Ramsey, J., Rasche, H., Maughmer, C., Criscione, A., Mijalis, E., Liu, M., et al. (2020). Galaxy and Apollo as a Biologist-Friendly Interface for High-Quality Cooperative Phage Genome Annotation. PloS Comput. Biol. 16, e1008214. doi: 10.1371/journal.pcbi.1008214
Shen, M., Yang, Y., Shen, W., Cen, L., McLean, J. S., Shi, W., et al. (2018a). A Linear Plasmid-Like Prophage of Actinomyces Odontolyticus Promotes Biofilm Assembly. Appl. Environ. Microbiol. 84 (17), e01263-1. doi: 10.1128/AEM.01263-18
Shen, M., Zhang, H., Shen, W., Zou, Z., Lu, S., Li, G., et al. (2018b). Pseudomonas Aeruginosa MutL Promotes Large Chromosomal Deletions Through non-Homologous End Joining to Prevent Bacteriophage Predation. Nucleic Acids Res. 46, 4505–4514. doi: 10.1093/nar/gky160
Stanley, D., Mason, L. J., Mackin, K. E., Srikhanta, Y. N., Lyras, D., Prakash, M. D., et al. (2016). Translocation and Dissemination of Commensal Bacteria in Post-Stroke Infection. Nat. Med. 22, 1277–1284. doi: 10.1038/nm.4194
Waters, E. M., Neill, D. R., Kaman, B., Sahota, J. S., Clokie, M. R. J., Winstanley, C., et al. (2017). Phage Therapy is Highly Effective Against Chronic Lung Infections With Pseudomonas Aeruginosa. Thorax 72, 666–667. doi: 10.1136/thoraxjnl-2016-209265
Wu, Y., Wang, R., Xu, M., Liu, Y., Zhu, X., Qiu, J., et al. (2019). A Novel Polysaccharide Depolymerase Encoded by the Phage SH-KP152226 Confers Specific Activity Against Multidrug-Resistant Klebsiella Pneumoniae via Biofilm Degradation. Front. Microbiol. 10, 2768. doi: 10.3389/fmicb.2019.02768
Keywords: bacteriophage, phage-antibiotic combination, Enterococcus faecalis, antibiotic resistance, phage therapy
Citation: Song M, Wu D, Hu Y, Luo H and Li G (2021) Characterization of an Enterococcus faecalis Bacteriophage vB_EfaM_LG1 and Its Synergistic Effect With Antibiotic. Front. Cell. Infect. Microbiol. 11:698807. doi: 10.3389/fcimb.2021.698807
Received: 22 April 2021; Accepted: 25 June 2021;
Published: 16 July 2021.
Edited by:
Jingmin Gu, Jilin University, ChinaCopyright © 2021 Song, Wu, Hu, Luo and Li. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Gongbo Li, bGlnb25nYm9AaG9zcGl0YWwuY3FtdS5lZHUuY24=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.