
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Cell. Infect. Microbiol. , 03 June 2021
Sec. Molecular Bacterial Pathogenesis
Volume 11 - 2021 | https://doi.org/10.3389/fcimb.2021.682635
 Shoichi Tachiyama1,2,3
Shoichi Tachiyama1,2,3 Ryan Skaar1†
Ryan Skaar1† Yunjie Chang2,3
Yunjie Chang2,3 Brittany L. Carroll2,3
Brittany L. Carroll2,3 Meenakumari Muthuramalingam1
Meenakumari Muthuramalingam1 Sean K. Whittier1
Sean K. Whittier1 Michael L. Barta1†
Michael L. Barta1† Wendy L. Picking1
Wendy L. Picking1 Jun Liu2,3
Jun Liu2,3 William D. Picking1*
William D. Picking1*Shigella flexneri, causative agent of bacillary dysentery (shigellosis), uses a type III secretion system (T3SS) as its primary virulence factor. The T3SS injectisome delivers effector proteins into host cells to promote entry and create an important intracellular niche. The injectisome’s cytoplasmic sorting platform (SP) is a critical assembly that contributes to substrate selection and energizing secretion. The SP consists of oligomeric Spa33 “pods” that associate with the basal body via MxiK and connect to the Spa47 ATPase via MxiN. The pods contain heterotrimers of Spa33 with one full-length copy associated with two copies of a C-terminal domain (Spa33C). The structure of Spa33C is known, but the precise makeup and structure of the pods in situ remains elusive. We show here that recombinant wild-type Spa33 can be prepared as a heterotrimer that forms distinct stable complexes with MxiK and MxiN. In two-hybrid analyses, association of the Spa33 complex with these proteins occurs via the full-length Spa33 component. Furthermore, these complexes each have distinct biophysical properties. Based on these properties, new high-resolution cryo-electron tomography data and architectural similarities between the Spa33 and flagellar FliM-FliN complexes, we provide a preliminary model of the Spa33 heterotrimers within the SP pods. From these findings and evolving models of SP interfaces and dynamics in the Yersinia and Salmonella T3SS, we suggest a model for SP function in which two distinct complexes come together within the context of the SP to contribute to form the complete pod structures during the recruitment of T3SS secretion substrates.
Type III secretion systems (T3SS) are used by many Gram-negative bacterial pathogens to inject host altering proteins into the cytoplasm of target cells (Portaliou et al., 2016; Galan and Waksman, 2018). The Shigella flexneri T3SS apparatus (injectisome) comprises four assemblies that work cooperatively to secrete effectors. The needle tip complex is exposed on the bacterial surface where it can make contact with the host cell membrane and contribute to the formation of a translocon pore (Deane et al., 2006; Espina et al., 2006b). The translocon and tip complex reside at the distal end of a needle that is generated from a polymer of MxiH (Cordes et al., 2003; Kenjale et al., 2005). The needle is then anchored to an envelope-spanning basal body (the injectisome core) (Blocker et al., 1999; Lunelli et al., 2020), which is associated with a cytoplasmic sorting platform (SP) (Hu et al., 2015). The SP contributes to substrate selection and to energizing secretion. The SP was originally proposed as a complex in Salmonella that included SctK, SctQ and SctL (Lara-Tejero et al., 2011) for which the Shigella homologues are MxiK, Spa33 and MxiN, respectively. An increasingly accepted uniform nomenclature for the injectisome components is provided as a guide in Table 1 (Wagner and Diepold, 2020) and will be used here when referring to all non-Shigella T3SS.
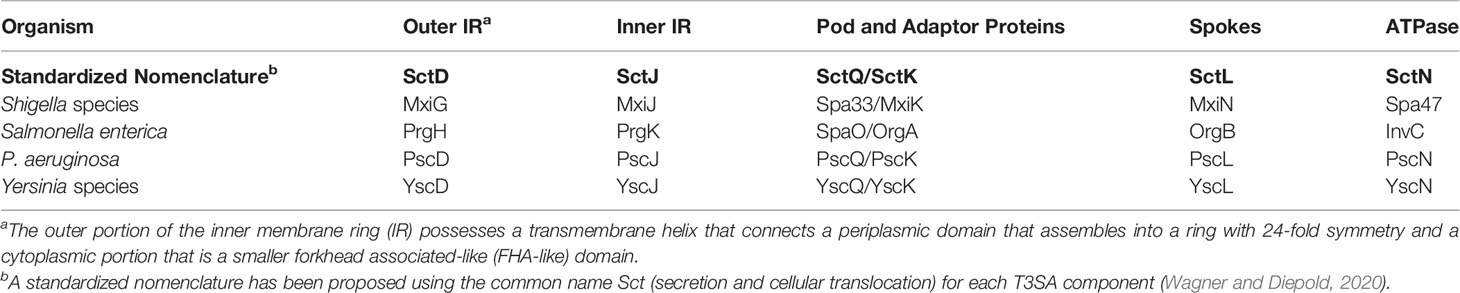
Table 1 The standardized nomenclature for the inner membrane ring (IR) and sorting platform components for injectisome components discussed here.
A key interface between the basal body and the SP in Shigella is formed by interaction of the inner membrane ring (IR) protein MxiG (SctD) with an SP adaptor protein. The cytoplasmic forkhead associated-like (FHA-like) domain of MxiG (MxiGC) and the SP protein MxiK (SctK) mediate this interaction (Tachiyama et al., 2019). A feature of this interface is a switch from the 24-fold symmetry of the IR components to the six-fold symmetry of MxiK, which is in association with the SP “pods”. The pods are the largest densities in the SP and comprise heterooligomeric complexes of Spa33 (SctQ) (Hu et al., 2015; McDowell et al., 2016). Spa33, in turn, interacts with MxiN (SctL) and this links them to the central ATPase Spa47 (SctN) (Supplementary Figure S1) (Hu et al., 2015; Notti et al., 2015). Our understanding of the composition of the SP has evolved with recent cryo-electron tomography (cryo-ET) work using minicells derived from Shigella (Supplementary Figure S1), which are in agreement with structures seen using similar methods for the Salmonella injectisome (Hu et al., 2015; Hu et al., 2017). Interestingly, deletion of mxiK or spa33 results in complete loss of SP assembly, however, mutants in which the radial spoke (and ATPase regulator) protein gene mxiN is deleted continue to allow remnants of the pods to be viewed while Spa47 is absent (Supplementary Figure S2) (Hu et al., 2015).
Recombinant Spa33 (expressed from the wild-type gene) purifies as a heterotrimer comprising one copy of full-length Spa33 and two copies of a C-terminal fragment generated via an alternative internal translation start site (McDowell et al., 2016). The C-terminal fragment (Spa33C) is a “surface presentation of antigens” (SPOA) domain named for alternative forms of SctQ from Salmonella and in Pseudomonas syringae (Fadouloglou et al., 2004; Notti et al., 2015; Notti and Stebbins, 2016). As described in depth for SctQ from Salmonella, full-length Spa33 possesses two SPOA domains at its C-terminus, but an internal translation start site allows for the second of these (SPOA2) to be expressed separately from the full-length protein (Notti et al., 2015). SPOA domains have a significant β-structure component and are structurally equivalent to FliN from Thermotoga maritima, which is a component of the flagellar C-ring (Supplementary Figure S3) (Brown et al., 2005; McDowell et al., 2016). Interestingly, the Spa33 heterotrimer shares significant similarity to the FliM-FliN heterotetramer (FliM-FliN3) where the full-length Spa33 contributes its C-terminal SPOA2 domain and the Spa33C homodimer contributes two additional FliN-equivalent SPOA domains (Fadouloglou et al., 2004; McDowell et al., 2016). This observation prompted the Lea group to logically propose a model in which Spa33 forms a torus structure resembling the flagellar C-ring (McDowell et al., 2016). Subsequent cryo-ET imaging, however, indicated that the bulk of the SP is actually made up of discontinuous pods described above (Hu et al., 2015).
Diepold et al. estimated that there are approximately 22 copies of SctQ in the SP of Yersinia (Diepold et al., 2015). This is consistent with the observation that the crystal structure of Spa33C dimers would only occupy a small portion of the pod densities attributed to Spa33. Based on available data, it now appears that the injectisome pods each consist of multiple copies of Spa33 and Spa33C. Furthermore, Diepold et al. found that during active secretion these SctQ components are in continual exchange between the injectisome SP and a distinct cytoplasmic pool of these proteins (Diepold et al., 2015; Diepold et al., 2017). The dynamics occurring within the SP ultrastructure are considered to be important for type III secretion system activity. Therefore, a better understanding of the structure of the SctQ complexes within the SP should move the field closer to understanding the mechanisms that govern injectisome function.
It was recently suggested that in Salmonella, the SctQ C-terminal domain (SctQC) may not be required for generating a functional injectisome (Lara-Tejero et al., 2019). Instead, it may only stabilize full-length SctQ by occupying a site on the protein that within the functional SP is occupied by SctL. Bernal et al. suggested that a complex consisting of SctQ, SctQC, SctL and SctN can be prepared, perhaps suggesting that SctQC is a required component of the fully functional injectisome (Bernal et al., 2019). Nevertheless, elimination of SctQC genetically still allowed for secretion of the Salmonella invasion proteins (Sips) and only reduced invasiveness by 50%, which is in agreement with the work suggesting SctQC is not essential. On the other hand, Bernal et al. did report that elimination of the internal translation start site still allowed for low-level production of SctQC (Bernal et al., 2019). Work in the Shigella system suggests that Spa33C is indeed required for forming a functional SP because a mutation that eliminates the internal alternative translation start site to limit expression to full-length Spa33 (Spa33FL) fails to restore detectable secretion of translocator and effector proteins (McDowell et al., 2016). The contrasting findings indicate a need to better understand the actual makeup and organization of the SP pods in Shigella. Even more recently, additional Spa33 protein variants were identified, including a 6.5 kDa form called Spa33CC (found to be important for injectisome activity) and two other Spa33 variants (called Spa33N and Spa33X) that have no obvious functions and may be translational artifacts (Kadari et al., 2019). Although we have also observed production of Spa33CC (unpublished observation), it will not be considered further here since it does not associate with the Spa33 heterotrimer.
While the Shigella and Salmonella in situ injectisomes are similar in structure based on cryo-ET imaging (Hu et al., 2015; Hu et al., 2017), there are differences between the two that warrant investigating both. The functional Shigella injectisome requires expression of wild-type spa33 and the production of one Spa33FL and two copies of Spa33C (McDowell et al., 2016). Moreover, we show here that the Spa33 heterotrimer forms stable complexes with MxiK and MxiN that have distinct biophysical behaviors in vitro. Within these complexes, Spa33C acts to stabilize Spa33FL, which is otherwise not soluble, and the Spa33 heterotrimer stabilizes MxiK, which also is not otherwise soluble. Two-hybrid analyses indicate that Spa33FL is able to associate with MxiK, but Spa33C is not, suggesting that Spa33FL mediates formation of the Spa33-MxiK complex. Meanwhile, it was wild-type Spa33 that was found to purify as part of the complex with MxiK, suggesting that Spa33C is required to stabilize this overall complex, as well. Likewise, MxiN specifically interacted with Spa33FL according to two-hybrid analyses but not with Spa33C, and it could associate with the Spa33 heterotrimer to form Spa33-MxiN complexes in vitro. Based on our ability to generate these complexes, their biophysical properties and old and new higher-resolution cryo-ET images, we propose that these two Spa33-containing complexes have critical discrete roles in active type III secretion.
S. flexneri 5a (M90T) was provided from J. Rohde (Dalhousie University, Halifax, NC, Canada). Shigella mxiK and mxiN null strains were from A. Allaoui (Université Libre de Bruxelles, Bruxelles, Belgium) the and spa33 null strain was from C. Lesser (Massachusetts General Hospital, Cambridge, Massachusetts, USA). Mutant strains were grown at 37°C on trypticase soy agar (TSA) plates containing Congo red and then used to inoculate trypticase soy broth (TSB) containing 50 µg/mL kanamycin to select for virulence plasmid-harboring null strains. Ampicillin (100 µg/mL) was added to select wild-type mxiK, mxiN, spa33, or their mutant genes in the pWPsf4 expression plasmid. To generate minicells, wild-type S. flexneri was transformed with pBS58, which is a low-copy number plasmid encoding the spectinomycin-resistance gene. pBS58 continuously expresses the E. coli genes ftsQ, ftsA, and ftsZ, which causes abnormal cell division to produce Shigella minicells (Bi and Lutkenhaus, 1990; Hu et al., 2015; Farley et al., 2016). To produce Shigella minicells, spectinomycin (100 µg/mL) was added into trypticase soy broth (TSB) to grow the strain for overnight. A mL of this overnight culture was added into 100 mL of fresh TSB with spectinomycin (100 µg/mL) and grown at 37°C to an absorbance of 1.0 at 600 nm (A600) (Hu et al., 2015; Tachiyama et al., 2019). To enrich for minicells, the culture was centrifuged at 1000 × g for 5 min to remove the large cells, and the supernatant fraction was further centrifuged at 20,000 × g for 10 min to collect the minicells for subsequent cryo-ET imaging of the SP pods.
To prepare expression vectors with wild-type spa33, spa33C, spa33FL (lacking the internal translation start site), mxiK or mxiN, each gene from S. flexneri was amplified using PCR. PCR products were purified and inserted into the pT7HMT expression vector (Geisbrecht et al., 2006; Tachiyama et al., 2019), which encodes His6-tag with TEV protease cleavage site at N-terminal side of the cloning site and a kanamycin resistance gene. To generate E. coli strains for protein expression, these recombinant plasmids were introduced into Tuner (DE3) cells (Novagen). For co-expression of mxiK and spa33, the latter was inserted into multiple cloning site two in pACYCDuet™ (Novagen). E. coli Tuner(DE3) cells were then transformed with both mxiK in pT7HMT and spa33 in pACYCDuet™. To induce protein expression, E. coli strains with recombinant pT7HMT vector were inoculated into LB with kanamycin (50 µg/mL) and grown overnight. The overnight bacterial culture was used to inoculate a larger volume of LB with the antibiotic. When the A600 reached 0.6, the cultures were induced with 0.5 mM isopropyl β-D-1-thiogalactopyranoside (IPTG). The culture was then incubated at 16°C overnight. MxiK and Spa33 production was carried out the same way except that Terrific Broth (TB) with kanamycin and chloramphenicol (34 µg/mL) was used for the large volume culture with induction started at an A600 of 1.0.
The cells expressing the protein of interest were harvested by centrifugation at 4000 rpm for 10 min at 4°C. The cell pellet was suspended in immobilized metal affinity chromatography (IMAC) binding buffer (5 mM imidazole, 0.5 M NaCl, 20 mM Tris) pH 7.5. The lysate was prepared using a microfluidizer at 15,000 psi for three cycles and clarified by centrifugation at 10,000 rpm for 30 min at 4°C. The clarified supernatant was applied to a Ni2+-charged IMAC column with chromatography carried out on an AKTA™ purification system (GE Healthcare, Chicago, IL). The loaded column was washed with additional IMAC binding buffer and then elution carried out using an elution buffer (0.5 M imidazole, 0.5 M NaCl, and 20mM Tris, pH 7.5). The His6 tag was removed using TEV protease after overnight dialysis in IMAC binding buffer. The protein was then applied to a second Ni-IMAC column and the flow-through collected, concentrated and buffer exchanged into 20 mM Tris, 200 mM NaCl, pH 7.5 prior to loading onto an equilibrated Superdex 200 pg (GE) for size exclusion chromatography (SEC). All fractions were analyzed by SDS-PAGE. SEC was also used to purify Spa33-MxiN complexes after mixing the proteins after each had been purified separately. TheMxiN, Spa33-MxiK and Spa33-MxiN samples were made to 10% (v/v) glycerol.
To test for secretion and translocon formation, contact-mediated hemolysis was performed for each Shigella mutant strain. Each strain was grown on a trypticase soy agar (TSA) plate containing Congo-red (Picking et al., 2005; Tachiyama et al., 2019). Isolated colonies were inoculated into TSB for growth at 37°C until mid-log phase with the appropriate antibiotics added for selection. A 10 mL aliquot of each culture was centrifuged at 4,000 rpm for 10 min at 30°C and the pellet was resuspended in 10 mM phosphate, pH 7.4 containing 150 mM NaCl (PBS). The PBS was adjusted to give equal concentrations of bacteria in each case based on the A600. To prepare red blood cells, defibrinated sheep red blood cells (Colorado Serum, Co.) were washed with PBS and resuspended into 3 mL PBS. Then 50 µL of the resuspended bacteria were added into 50 µL of washed red blood cells in a 96-well plate. The plate was centrifuged at 3,500 rpm for 15 min at 30°C to force Shigella contact with red blood cells. The plate was then incubated at 37°C for an hour. The cell mixtures were then vigorously resuspended with 100 µL cold PBS and centrifuged at 3,500 rpm for 15 min at 10°C. The supernatants were then transferred into a new 96-well plate and the amount of released hemoglobin was measured based on the A545. The absorbance was used to calculate relative hemolysis for each strain after comparison to a control for negative hemolysis (incubation in PBS) and a control for positive (complete) hemolysis (incubation in water).
The secondary structure of recombinant proteins and their complexes were analyzed by CD spectroscopy (Espina et al., 2006a; Birket et al., 2007) using a JASCO model J-1500 CD spectropolarimeter (Jasco Inc., Easton, MD). Samples were prepared in 20 mM Tris, pH 7.0, 200 mM NaCl, 5.0% glycerol and the same buffer was used for blank subtraction. For MxiN, tris(2-carboxyethyl)phosphine (TCEP) was added to the buffer to a final concentration of 1 mM. For Spa33 and Spa33C, PBS was used as the buffer. To obtain CD spectra, ellipticities between 190 and 260 nm from each protein sample in 0.1 cm path length quartz cuvettes were obtained at 10°C. Molar ellipticities were then calculated based on protein concentration, molecular weight, and residue numbers. To monitor thermal stability, the molar ellipticity at 222 nm (a minimum seen for proteins having an α-helical component) was monitored every 2.5°C with increasing temperature from 10 to 90°C at a ramp rate of 15°C/h rate. The molar ellipticity was then plotted as a function of temperature to provide a thermal unfolding curve. Thermal transition points or unfolding midpoints (Tm) was determined using Prism software from GraphPad.
A Polara G2 electron microscope (FEI Company) equipped with a field emission gun and a Direct Detection Camera (K2 summit, Gatan) was used to image frozen-hydrate minicells from the wild-type Shigella strain at -170°C. The microscope was operated at 300 kV, with a pixel size of 2.6 Å at the specimen level. The single-axis tilt series was collected at about 5 µm defocus using SerialEM software (Mastronarde, 2005) and with a cumulative dose of ~50 e/Å2 distributed over 35 stacks covering an angle range of -51° to 51° with 3° angle step.
Motioncorr2 (Zheng et al., 2017) was used to do drift correction for each image stack. Then, the tilt series were aligned by IMOD (Kremer et al., 1996), and weighted back-projection (WBP) or Simultaneous Iterative Reconstruction Technique (SIRT) was used to reconstruct the tomograms by Tomo3D software (Agulleiro and Fernandez, 2015). A total of 922 tomograms were reconstructed for the wild-type Shigella minicell sample.
A total of 4,488 injectisome particles from wild-type Shigella were manually picked from the SIRT reconstructed tomograms as previously described (Hu et al., 2015). The particle positions were then used to extract the sub-tomograms from the 1×1×1 binned WBP reconstructed tomograms. I3 software (Winkler, 2007; Winkler et al., 2009) was used for the sub-tomogram averaging. 4×4×4 binned sub-tomograms were used for an initial alignment and 3D classification, and then 2×2×2 (or 1×1×1) binned sub-tomograms were used for refinement. To improve the resolution of the pod structure, a single pod region in the subtomogram average (the green arrow region in Figure 7A) was extracted for additional focused refinement processes.
The SP of the overall T3SA (see Figure 7A) and the refined single pod structures were used to create 3D surface rendering images of the sub-tomogram averaging using UCSF Chimera (Pettersen et al., 2004) and ChimeraX (Goddard et al., 2018) software. For the molecular model, the crystal structure of SctK from P. aeruginosa (Muthuramalingam et al., 2020) was fitted into the MxiK position in the refined pod rendering image using “fit in map” in UCSF Chimera (Pettersen et al., 2004; Hu et al., 2015; Tachiyama et al., 2019; Muthuramalingam et al., 2020). Three FliN (PDB: 1YAB) (Supplementary Figure S3) and a FliM (PDB: 2HP7) (see Supplementary Figure S14) crystal structures from Thermotoga maritima were used to generate a Spa33 heterotrimer model based on a FliN and FliM complex model from the flagellar C-ring (Carroll et al., 2020). Two copies of this model could fit into the bottom part of Spa33 region using the “fit in map’ in UCSF Chimera software (Pettersen et al., 2004). The single pod surface rendering image was then fit back into six pod regions in the 3D surface rendering image of the SP (see Figure 7).
The pod cage structure and the refined images have been deposited in the EM Data Bank (EMD-23440).
A BACTH assay was used to assess possible interactions between the different SP proteins (MxiK, MxiN and Spa33 and their mutants) in an in vivo-like environment (Tachiyama et al., 2019). CyaA subdomain (T25 and T18) fusion proteins were prepared by cloning the proteins of interest from wild-type bacteria. The subdomains were fused to either the N- or C-terminus of the potential interacting protein. The pKT25 expression vector fuses the T25 subdomain to the protein’s N-terminus while the pKNT25 vector fuses the T25 subdomain to the C-terminal end of the protein. The same is achieved for fusion of the T18 subunit using the pUT18 or pUT18C vectors, which fuses the T18 unit to the N- and C-terminus of the protein.
Two compatible plasmids containing the gene fusions of interest were used to co-transform the cyaA- E. coli strain, BTH101. BTH101 was co-transformed with empty plasmids was used for the negative control and the strain was co-transformed with plasmids encoding leucine zipper binding partners for the positive control. Individual colonies were selected on LB plates containing ampicillin and kanamycin and these were inoculated into LB broth containing these antibiotics and 0.5 mM IPTG. The bacteria were grown at 30°C overnight. The overnight bacterial cultures were washed three times with M63 minimum medium and 2 µL was spotted onto indicator plates. Indicator plates could be: a) LB agar plates with ampicillin, kanamycin, 40 µg/mL X-Gal and 0.5 mM IPTG; b) MacConkey agar plates with ampicillin, kanamycin and 1.0% (w/v) maltose; or c) M63 plates with ampicillin, kanamycin, 40 µg/mL X-Gal and 0.5 mM IPTG. These plates were incubated at 30°C for four to five days and color changes over time were compared with that seen for the controls.
Purified MxiN and Spa33 were both dialyzed into 20 mM Tris, pH 7.5, 150 mM NaCl with 10% glycerol (v/v) for SEC. The MxiN and Spa33 were combined with an excess of MxiN and the mixture was concentrated to a final volume of 2 mL for injection onto a Hi Load 26/600 Superdex 200 pg column (GE Healthcare Life Sciences). The protein absorbance at 280 nm was monitored to detect protein elution on an AKTA system (GE Healthcare Life Sciences). The eluted fractions were collected for SDS-PAGE analysis.
The binding of MxiN to the wild-type Spa33 was assessed by biolayer interferometry (BLI) as described elsewhere (Barta et al., 2017). Purified His6-tagged MxiN was dialyzed into PBS with 10% glycerol, 0.01% BSA and 0.002% Tween 20 (BLI buffer). The MxiN was then immobilized onto a Ni-NTA biosensor tip and washed with BLI buffer for 5 min to remove non-immobilized protein. Once a baseline signal was established, the sensors with immobilized protein were immersed into a solution containing untagged Spa33, which was prepared and adjusted to 400, 200, 100, 50, 25 and 12.5 µg/mL. The Octet RED96 system (FortéBIO, Inc., Freemont, CA) was then used to monitor the binding of Spa33 to MxiN that had been immobilized onto a Ni-NTA biosensor tip. The binding of Spa33 at each concentration to the immobilized MxiN was measured in real time to obtain the association rate. The biosensor tips were then placed into BLI buffer lacking Spa33 to measure the dissociation rate. All steps were performed at 25°C. Real-time data were analyzed using Octet Software version 8.2 (ForteBio). Binding kinetics (association and dissociation) as well as steady-state equilibrium concentrations were fitted using a 1:1 Langmuir binding model. Background correction was done by subtracting the data from ligand-loaded biosensor that had been incubated without analyte (Spa33). All steps were carried out with constant shaking at 1000 rpm and each measurement was performed in triplicate with an association time of 300 s followed by 300 s dissociation. The sensorgrams from the experiment were fitted with 1:1 binding equations available for interaction from a global curve fit using Data Analysis software 7.1.0.36 (ForteBio) (Barta et al., 2017).
The effect that Spa33 inclusion into complexes with MxiK and MxiN has on particle size and surface charge was determined using dynamic light scattering (DLS). DLS measurements were made on a Malvern ZetaSizer Ultra using a quartz cuvette with a 1-cm path length. Backscattering was detected at an angle of 174.7 degrees, with each measurement performed in triplicate. Size distributions were calculated using the ZS Xplorer software (Malvern). The same instrument was used to determine the zeta-potential for each protein and protein complex.
When Spa33 is expressed in E. coli, it readily purifies as a complex containing one full-length Spa33 (Spa33FL) with two copies of Spa33C, which are expressed from an internal translation initiation site (Supplementary Figure S4). When the alternative start site is mutated to prevent initiation of the internal translation event, giving only Spa33FL, the protein could not be stably expressed and purified from E. coli, due to the fact that it is insoluble and sequestered to inclusion bodies. This is in contrast to expressing a gene that allowed for expression of only Spa33C, which is then easily purified as an apparent homodimer (Supplementary Figure S4). Spa33C was previously used to obtain a crystal structure for this SPOA domain (McDowell et al., 2016). The fact that Spa33 forms an oligomer is further supported by data from a bacterial adenylate cyclase two-hybrid (BACTH) assay where both the bait and prey plasmids contained the wild-type spa33 gene (Table 2 and Supplementary Figure S5, top). The results with wild-type Spa33 are somewhat complicated since when the two adenylate cyclase domains are placed at the Sp33 N-terminus an interaction was observed. This could suggest that multiple Spa33 complexes were forming a higher order complex. We thus felt that it was imperative to look at interactions involving only Spa33FL, only Spa33C, and Spa33FL plus Spa33C.
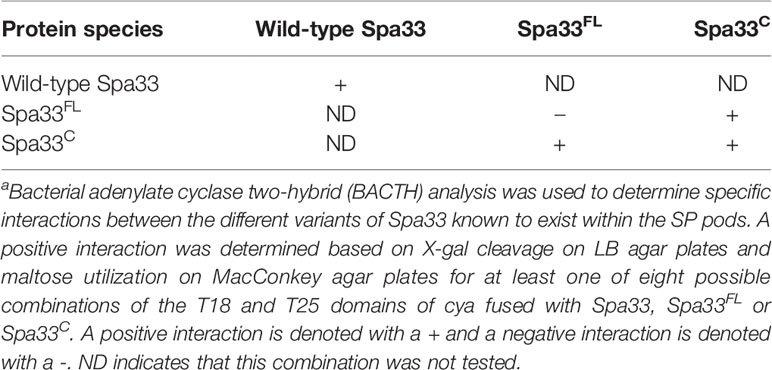
Table 2 Specific Spa33 interactions can be identified by two-hybrid a analysis.
When Spa33FL was used in the same two-hybrid assay, no interaction was detected for any of the eight fusion permutations with the T18 and T25 fragments of the adenylate cyclase (Table 2 and Supplementary Figure S5, bottom). In contrast, when Spa33C was used in the same assay, a clear interaction was again observed (Table 2 and Supplementary Figure S6). It is worth noting that not every fusion permutation between the Spa33C binding pairs gave a positive reaction, but this is not surprising given that the interaction between the binding partners must orient the T18 and T25 fragments of the CyaA adenylate cyclase domains properly for activity to be restored (Supplementary Figure S7). Perhaps more surprising was the observation that all combinations of wild-type Spa33 were positive for interactions in the BACTH system (Supplementary Figure S5). Finally, when Spa33FL was used in the same two-hybrid analysis with Spa33C, an interaction was observed, though not for every fusion permutation, which is consistent for what was seen for the Spa33C-Spa33C interaction (Table 2 and Supplementary Figure S8). These data confirm what is currently believed to occur with respect to the interactions within the Spa33 heterotrimer. However, they also show that interactions within the Spa33-containing pods of the SP do not arise from Spa33FL-Spa33FL interactions, at least not in the absence of Spa33C.
It was previously reported that neither Spa33FL nor Spa33C alone could restore the secretion of Ipa proteins when expressed in a Shigella spa33 null strain (McDowell et al., 2016). In agreement with this prior observation, neither of these forms of Spa33 alone could support contact-mediated hemolysis, although a small amount of residual hemolytic activity was observed for Spa33FL (Figure 1). The reason for the low level of hemolysis for Spa33FL was not entirely clear. It is possible that it could be related to the reported observation by the Lara-Tejero et al. that the Spa33C homologue from Salmonella (SctQC) is not required for secretion of translocator proteins or for cellular invasion (Lara-Tejero et al., 2019) and that it merely contributes to stabilization of SctQFL (the full-length form). They also suggested that SctQC contributed to the stabilization of other SP components (SctK and SctL). Indeed, Spa33C is required for stabilization of Spa33FL since in its absence Spa33FL is insoluble, however, the very minor degree of restoration of contact-mediated hemolysis for Shigella expressing Spa33FL is not at a level considered here to indicate it gives rise to an active injectisome. This is consistent with our observation that when the contact-hemolysis assay is allowed to incubate for long periods of time, the level of hemoglobin release gradually increases (unpublished observation). Thus, there may be degradation of components (i.e. Spa33FL) that allow for undetectable amounts of Spa33C to accumulate and allow residual T3SS activity to be seen as a function of time. This would fit with observations by Bernal et al. who found that SctQFL gave rise to a minor population of SctQC (Bernal et al., 2019), however, even in that report there was a 50% restoration of injectisome function by the SctQFL and we see nothing near that level of contact-mediated hemolysis for Spa33FL (Figure 1).
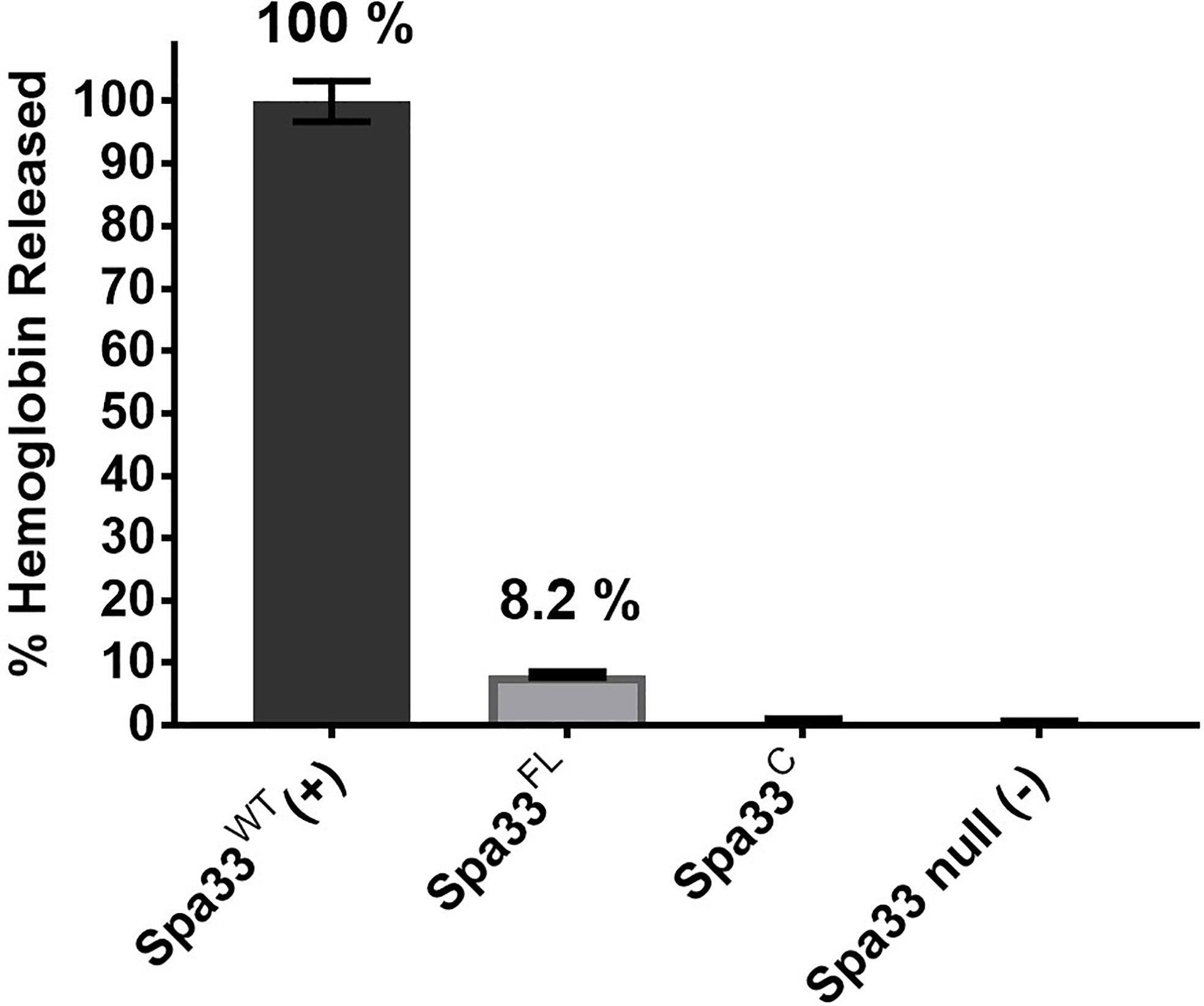
Figure 1 Contact-mediated hemolysis shows that Spa33FL and Spa33C are not active when expressed in a Shigella spa33 null mutant. To test the type III secretion activities of the Spa33 variants in vivo, contact-mediated hemolysis was used to measure of translocon formation. Spa33FL did restore a minor amount of hemolysis activity (about 8%), however, Spa33C was completely inactive. The level of injectisome activity seen for Spa33FL was far below that seen for SctQFL in Salmonella. Statistical differences between wild-type Spa33 (Spa33WT) and other samples (in triplicate) were assessed using one-way ANOVA with Spa33FL, Spa33C and the spa33 null all with p <0.005) and error bars depict mean ± SD.
Circular dichroism (CD) spectroscopy was used to compare the secondary structure properties of the purified Spa33 heterotrimer and Spa33C homodimer. Both complexes possess a dominant β-structure component with only a minor amount of α-helical structure in each (Figure 2). By monitoring the CD signal 222 nm as a function of temperature, we found that Spa33C was highly stable with an unfolding transition midpoint (Tm) of 65°C (Figure 2B and Table 3). Meanwhile, the wild-type Spa33 had a Tm of 62°C. Because Spa33FL could not be purified as a recombinant protein, Spa33C clearly had a significant solubilizing effect on it. In this respect, Spa33C has a similar effect on Spa33FL stability as does SctQC on SctQFL in Salmonella (Lara-Tejero et al., 2019).
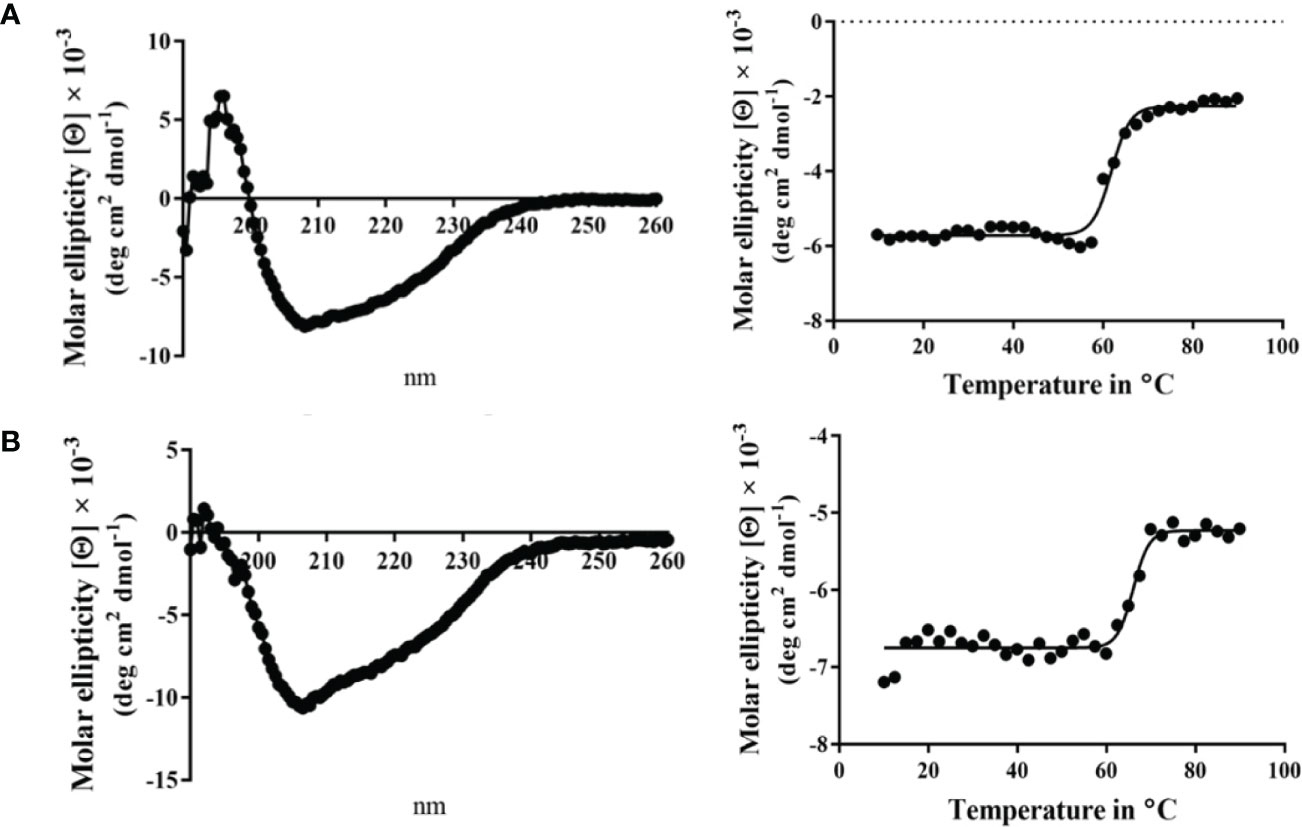
Figure 2 Circular dichroism (CD) spectra and thermal unfolding for wild-type Spa33 and Spa33C. CD spectroscopy was used to analyze the secondary structure content and thermal stabilities of wild-type Spa33 and Spa33C. (A) The CD spectrum of wild-type Spa33 shows a negative peak at 208 nm with a modest shoulder at 222 nm (left). This spectrum is indicative of a significant amount of β-structure. To determine the Tm for wild-type Spa33, the molar ellipticity at 222 nm was monitored as a function of temperature. Based on the large transition observed (right) the Tm was found to be 61.88°C. (B) The CD spectrum of Spa33C also shows more dominant 208 nm minimum suggesting a dominant β-structure component (left). Based on the thermal unfolding curve for Spa33C (right) the Tm was determined to be 65.12°C.
We have shown that MxiK interacts with wild-type Spa33 in the in situ injectisome and this was confirmed by two-hybrid analysis (Tachiyama et al., 2019). As a follow up, we carried out BACTH analyses using MxiK with wild-type Spa33, Spa33FL and Spa33C. While we know that MxiK binds to wild-type Spa33, it has not been shown whether this interaction occurs through Spa33FL or Spa33C (summarized in Table 4). Based on two-hybrid analysis, we found that MxiK was able to interact with Spa33FL, however, we could not detect any interactions with Spa33C (Supplementary Figure S9). Whether this interaction involves the Spa33 N-terminal domain or the SPOA1-SPOA2 domains of Spa33FL is not yet clear. In parallel experiments, we co-expressed wild-type spa33 and mxiK in E. coli and found that the Spa33 heterotrimer could be co-purified as a complex with MxiK possessing an N-terminal His6-tag using IMAC followed by size-exclusion chromatography (Figure 3 and Supplementary Figure S10). The fact that MxiK is part of this complex while it cannot be stably purified as a soluble protein on its own suggests a stabilizing, or minimally a solubilizing, effect for the Spa33 heterotrimer on MxiK.
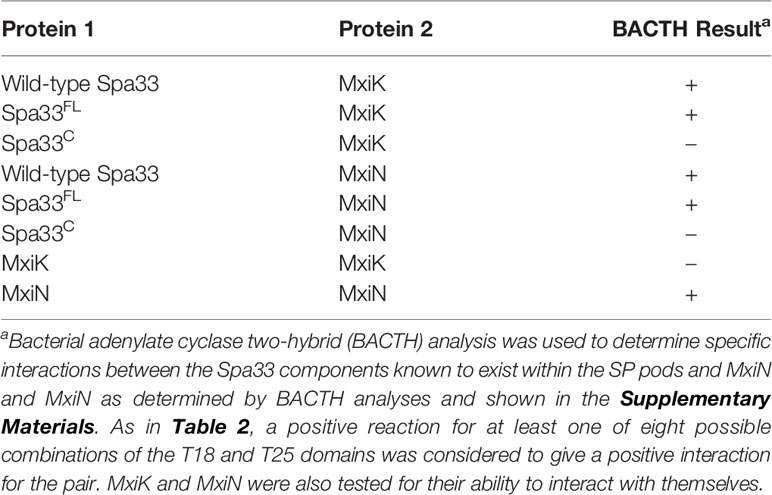
Table 4 Interactions of MxiK and MxiN with Spa33 and the individual components of the Spa33 complex.
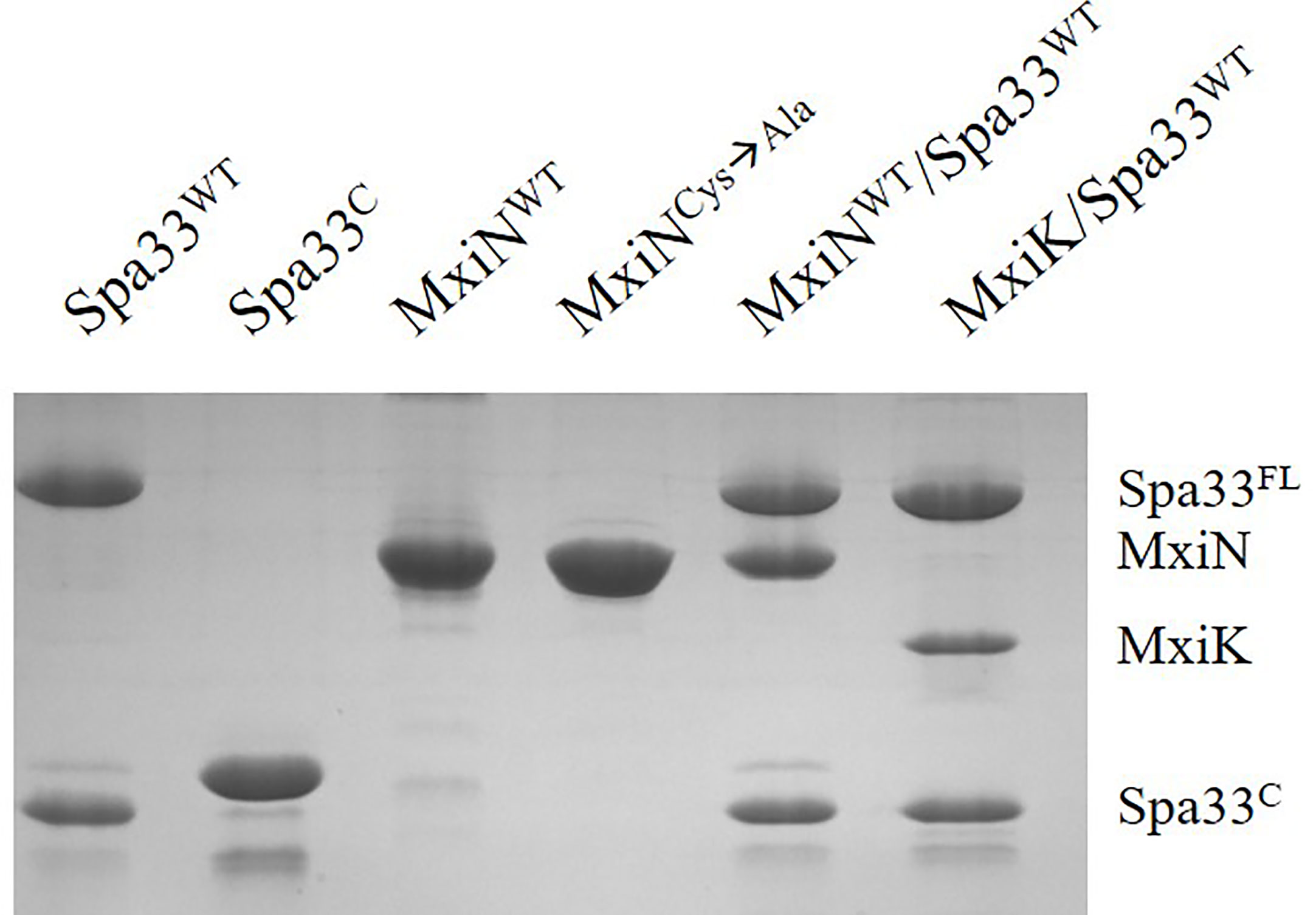
Figure 3 SDS-PAGE analysis for all purified complexes containing Spa33. From the left, wild-type Spa33, Spa33C, MxiN, a MxiN mutant with all Cys residues removed to allow future crystallography screens (fully active in vivo), the MxiN-Spa33 complex formed in vitro and the MxiK-Spa33 complex purified after co-expression in E. coli. The Spa33C (second lane) is slightly larger than the same component in the other lanes because it still possesses an N-terminal His6-tag. Recombinant Spa33FL and MxiK alone are not soluble and could not be purified.
When the CD spectrum of the Spa33-MxiK complex was obtained, it indicated the presence of both α-helical and β-strand components. This is consistent with contributions made to secondary structure by both components of the complex (Figure 4). We know that Spa33 is rich in β-structure and the crystal structure and CD spectrum of the MxiK homologue SctK from P. aeruginosa show that it is rich in α-helical structure (Muthuramalingam et al., 2020). When the signal at 222 nm was followed as a function of temperature, the complex showed a single unfolding transition at 52°C though there may be a second minor transition occurring at about 60°C (Figure 4 and Table 3). The main transition point or Tm is below that observed for the Spa33 heterotrimer, presumably because of structural rearrangements in Spa33 that occur upon complex formation that causes the complex to behave as a single entity with regard to thermal stability. This said, it can’t be ruled out that the minor second transition could be due to some of the Spa33 heterotrimer or Spa33C homodimer escaping in a folded state during the first transition to later unfold at its characteristic Tm. The relative size of this second transition, however, is consistent with this only represented a small subpopulation of the Spa33 present in the original Spa33-MxiK complex. Therefore, because a single dominant Tm is observed and MxiK cannot be prepared as a recombinant protein by itself, the data support the proposal that the Spa33-MxiK complex mostly behaves as a single unit consisting of Spa33FL, Spa33C and MxiK. Although not directly applicable here, MxiK can be purified in small amounts with a C-terminal T4L genetically fused to it (MxiK-T4L-C) and this fusion retains activity when expressed in a Shigella mxiK null mutant (Tachiyama et al., 2019). This purified chimera has a Tm of about 39°C, which compares well with the Tm of SctK P. aeruginosa, which is 40°C (Muthuramalingam et al., 2020). Thus, although the complex has a thermal unfolding transition below that of the Spa33 heterotrimer, it does have a significant stabilizing effect on MxiK. Likewise, the fact that a second transition is not observed supports the idea that it behaves as a single stably folded unit.
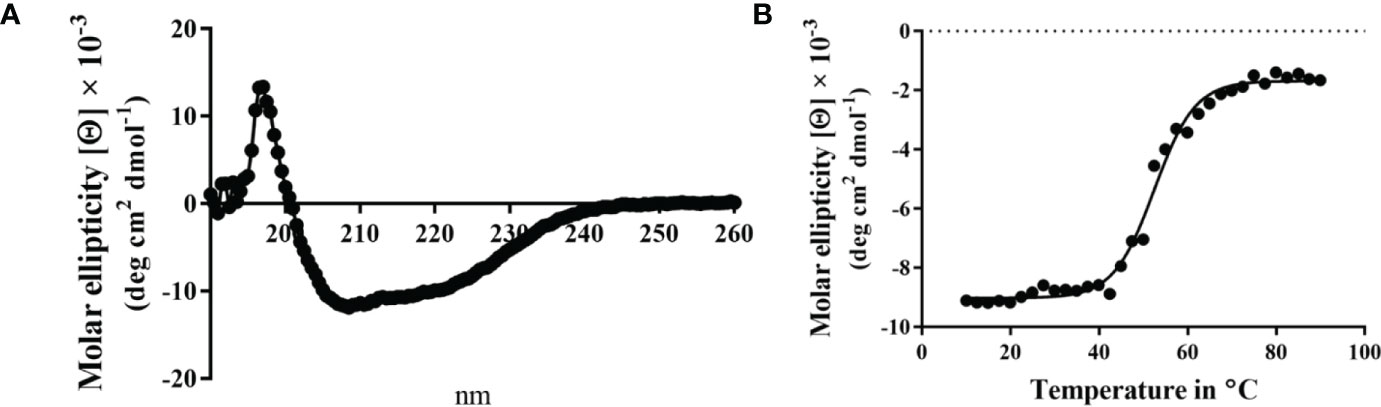
Figure 4 CD spectrum of the wild-type Spa33-MxiK complex. (A) The secondary structure of the Spa33-MxiK complex shows a greater α-helical character than the Spa33 heterotrimer alone. (B) To determine thermal stability, molar ellipticity at 222 nm was monitored as a function of temperature. Based on the large thermal transition observed, the complex had a Tm of approximately 52.46°C. This Tm is lower than that of wild-type Spa33 (62°C) and higher than that of the chimera MxiK-T4L-C (39°C), suggesting that the Spa33 interaction enhances MxiK solubility and thermal stability. R2 = 0.99.
MxiN forms the radial spokes that connect the Spa33 pods of the SP with the hub which is a hexamer of the Spa47 ATPase. MxiN forms a dimer and interacts with wild-type Spa33 as observed in two-hybrid analyses (Tachiyama et al., 2019). When the same two-hybrid system was used to explore MxiN interactions with the two components of the Spa33 heterotrimer (summarized in Table 4), MxiN was found to interact with Spa33FL, but not with Spa33C, which is the same result that was observed for MxiK (Supplementary Figure S9). This is an interesting observation in that it suggests that two proteins found at distant sites within the SP are able to interact with the same component, Spa33FL, within the Spa33 heterotrimer. Nevertheless, this does not rule out the possibility that one or both proteins interact with the two SPOA domains of the Spa33 heterotrimer since Spa33FL contains both a SPOA1 and a SPOA2 domain, much like what was described for Salmonella SctQ (Notti et al., 2015; McDowell et al., 2016). In line with this, Notti et al. were able to solve the structure of a complex consisting of the two SPOA domains from Salmonella SctQ with a fragment of SctL (MxiN homologue) (Notti et al., 2015). Thus, it is interesting that our two-hybrid results did not reveal an interaction with the Spa33C dimer. Perhaps this is because this dimer is generated from two copies of SPOA2 rather than a hybrid SPOA1/SPOA2 structure.
Unlike MxiK, MxiN can be purified as a recombinant protein and when its CD spectrum is obtained, it is clearly rich in α-helical structure with a thermal unfolding transition at 36°C, which agrees with the value reported by others (Figure 5A and Table 3) (Case and Dickenson, 2018). When purified MxiN and Spa33 were mixed (with MxiN being in excess) and the resulting complexes separated by size-exclusion chromatography, MxiN was found to purify in two populations. In addition to MxiN alone (which eluted as the smaller of two eluting peaks of protein by SEC), MxiN also co-purified with the Spa33 complex (Figure 3 and Supplementary Figure S11). As with MxiK-Spa33, the CD spectrum of the Spa33-MxiN complex was a combination of α-helical structure (contributed by MxiN) and β-structure (contributed by Spa33). When the CD signal at 222 nm was monitored as a function of temperature, the Tm of the major transition was found to be 39°C (Figure 5B and Table 3), indicating that Spa33 has at least a minor stabilizing effect on MxiN. However, a second transition at 62°C suggests that a minor population of the MxiK-Spa33 complex, this complex may dissociate at the lower transition temperature with the freed MxiN then unfolding with what appears to be a slightly higher Tm than the MxiN alone. The second transition fairly accurately reflects the thermal unfolding reported above for the stable Spa33 heterotrimer (~60°C). This prompted us to assess the strength of the MxiN-Spa33 interaction using biolayer interferometry (BLI). MxiN possessing an N-terminal His6-tag was immobilized to BLI sensors via bound Ni2+ to allow for monitoring the association and dissociation of the wild-type Spa33 heterotrimer using BLI (Figure 6 and Supplementary Figure S12). From these data, an equilibrium dissociation constant (KD) of 0.55 µM was obtained, suggesting a specific and moderately strong interaction was occurring between them.
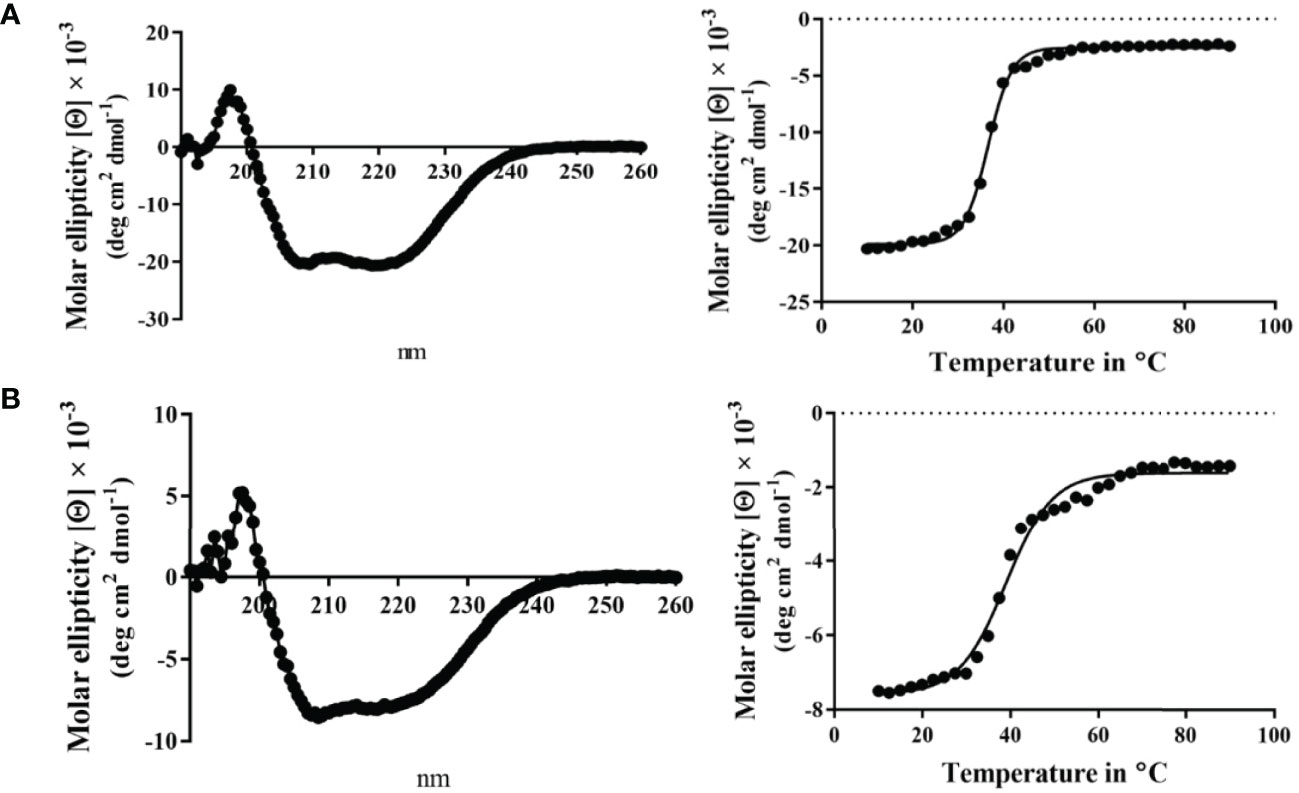
Figure 5 CD spectra from MxiN and the Spa33-MxiN complex. The secondary structures and thermal stabilities of MxiN and its complex with the wild-type Spa33 heterotrimer were analyzed by CD spectroscopy. (A) The CD spectrum of MxiN alone shows two minima that indicate a rich α-helical structure (left). The thermal stability for MxiN was determined by plotting the molar ellipticity at 222 nm as a function of temperature. A single transition was observed giving a Tm value of 36.63°C (right). (B) The CD spectrum for the MxiN-Spa33 complex also had two minima with the stronger minimum at 208 nm, indicating a greater β-structure content than for MxiN alone (left). The thermal stability of the complex was tested by monitoring the molar ellipticity at 222 with increasing temperature. The complex had a major transition with a Tm value of 38.99°C, however, a minor second transition was seen at about 60°C (right). R2 values for these unfolding curves was >0.985.
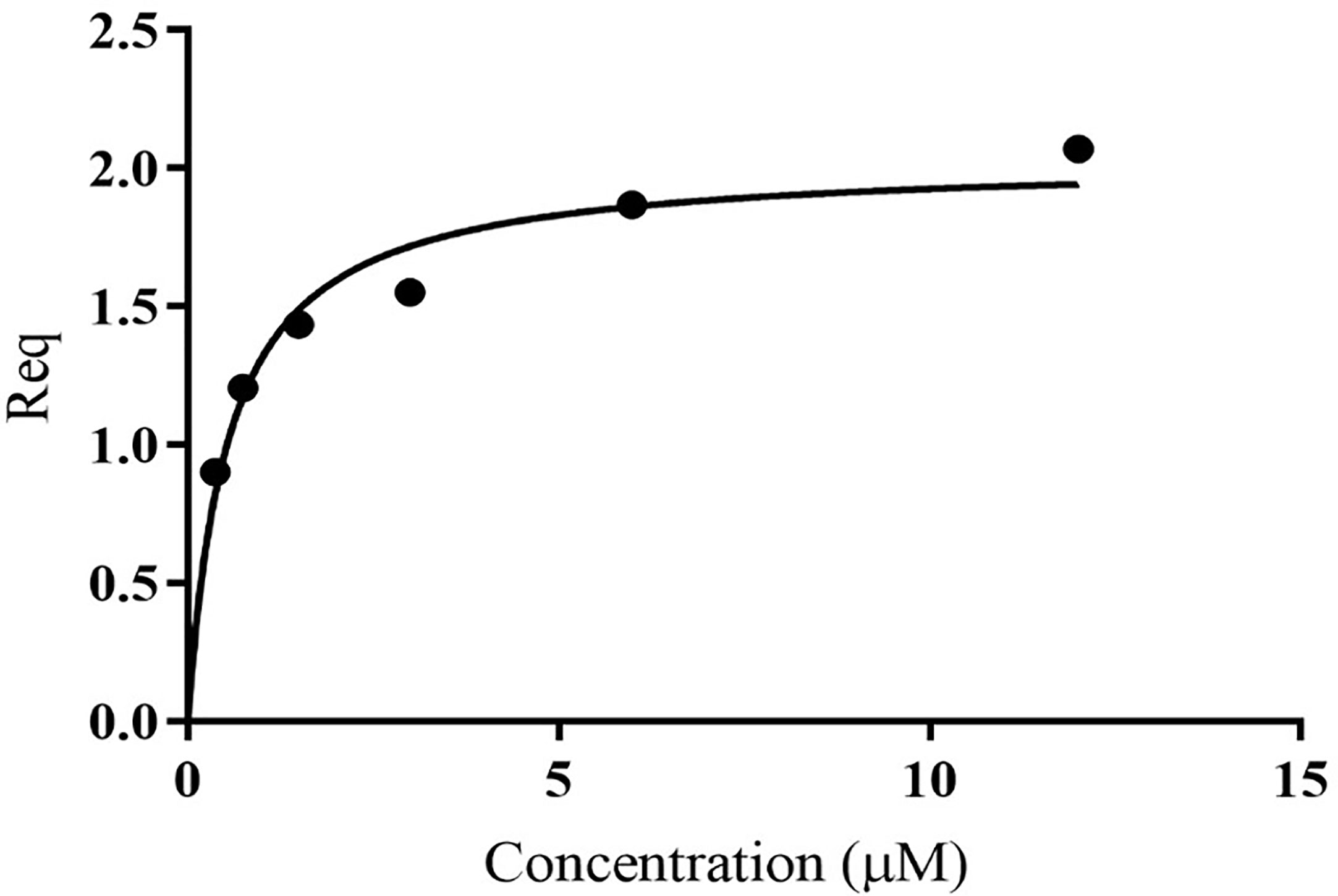
Figure 6 Quantification of the in vitro binding of Spa33 to MxiN using biolayer interferometry (BLI). MxiN with an N-terminal His6-tag was immobilized to sensor tips to measure its ability to bind to wild-type Spa33 heterotrimers. The tips were immersed in different concentrations of Spa33 (lacking a His6-tag) to monitor association and dissociation rates (see Supplementary Materials). The KD was determined based on the equilibrium reaction (Req) at different Spa33 concentrations. The KD value was determined to be 0.55 ± 0.091 µM, which is in agreement with the measured association and dissociation rates. The plotted data were generated using GraphPad prism.
From the CD data on the Spa33-containing complexes and individual proteins presented above, a great deal of information can be obtained regarding secondary structure content and thermal stability. To begin putting these data into context regarding the solution properties for these proteins and complexes, we carried out dynamic light scattering (DLS) studies. DLS allows us to determine the relative sizes of the proteins and their complexes, along with ascertaining the influence that complex formation has on their relative sizes. It also lets us determine whether the complexes are of discrete sizes or are able to nonspecifically form larger species. Furthermore, we obtained the zeta-potential in each case to determine the effect of complex formation on the electrical potential of the particles being examined. As expected, Spa33C is smaller than the wild-type Spa33 heterotrimer (Table 3) with hydrodynamic diameters (Dh) of 6 and 13 nm, respectively. The size of the Spa33 complex is more than twice the size of the Spa33C dimer, which is consistent with the compared masses of the two. Spa33FL has been reported to be 35.6 kDa and each Spa33C was reported to be 11.6 kDa based on mass spectrometry analysis (McDowell et al., 2016). This means the heterotrimer would have a mass close to 58.8 kDa relative to the Spa33C dimer’s mass of 23.2 kDa. Nevertheless, the Dh of the heterotrimer is smaller than what might be expected for the entire pod structure based on previous reports (Hu et al., 2015). This is consistent with there being multiple copies of Spa33 or the Spa33-containing complexes within each pod as has been suggested by others (Diepold et al., 2015; McDowell et al., 2016).
Interestingly, the Spa33-MxiK complex has a Dh of 10 nm, which is smaller than that of the Spa33 heterotrimer (Table 3), however, the purified Spa33-MxiK complex has all three of the expected components (Spa33FL, Spa33C and MxiK) present (Figure 3). Furthermore, the Spa33-MxiK complex has a dominant transition for thermal unfolding (Figure 4) and this Tm is distinct from that shown for the Spa33 heterotrimer. This suggests that much of the Spa33-MxiK complex acts as a single entity. The absence of multiple complex forms is also evident from the DLS scans when either particle numbers (Supplementary Figure S13) or intensity (not shown) are examined. This also suggests that the Spa33 component undergoes a significant conformational change when in a complex with MxiK and that the overall particle has a more compact globular structure than the elongated Spa33 heterotrimer. The Dh for MxiK could not be determined because this protein is not soluble on its own in solution and could not be purified. Nevertheless, initial reports of the interface between MxiK and the Spa33 pods using cryo-ET imaging suggest an intimate interaction between the two (Tachiyama et al., 2019).
In contrast to MxiK, MxiN can be purified and it was found to have a Dh of almost 10 nm, and this is consistent with its ability to form dimers that become the elongated spokes connecting the pods with Spa47 (Hu et al., 2015). When incubated with Spa33 in vitro, the resulting complex has a Dh of 18.5 nm and this is what might be expected following the association of the two structures (Table 3). This behavior is distinct from that of the Spa33-MxiK complex, but like the Spa33-MxiK complex there is single-sized species that is formed (Supplementary Figure S13). From the two-hybrid, CD and DLS data, we propose that Spa33 forms two distinct complexes that are stable in solution, one incorporating MxiK and one incorporating MxiN. In each of these complexes, it appears that the interaction with Spa33 occurs through the Spa33FL moiety. Whether this means that the interactions occur through the N-terminal portion of Spa33 is not yet clear, however, Spa33C alone has not been found to be directly responsible for the protein-protein interactions giving rise to these complexes though it does contribute to their stabilization.
In addition to Dh, we monitored the surface charge properties (zeta-potential) of these proteins and their complexes. Spa33 and Spa33C both have a negative surface electric potential (Spa33 = -8.87 nV) while MxiN has near neutral surface charge. The fact that the Spa33-MxiN complex has a zeta-potential of -5 mV suggests that this interaction masks some of the surface electric potential of Spa33. In contrast, the complex with MxiK only reduces the negative zeta-potential of Spa33 by about 1.5 mV. While we do not know the zeta-potential of MxiK alone, this suggests that there are significant differences in how it interacts with Spa33 in comparison to MxiN.
Bzymek et al. first described the independent expression of a SctQ C-terminal domain in Yersinia (Bzymek et al., 2012). Notti et al. then identified SPOA domains and described their relationship to flagellar FliN in their work in the Salmonella system (Notti et al., 2015). For the Shigella system, the two SP structures that have been solved thus far include the SPOA domain of Spa33, which forms a dimer (the Spa33C homodimer) (McDowell et al., 2016) and the Spa47 ATPase (Burgess et al., 2016). While MxiK and MxiN have not had their high-resolution structures solved. Our group recently reported the crystal structure of the MxiK homologue from P. aeruginosa, SctK (Muthuramalingam et al., 2020). The latter has a unique fold and is the first structure solved for any protein in the SctK family. No structures are available for the virulence-related T3SS Spa33/SctQ heterotrimer or any MxiN family member. There are, however, structures available for FliN (Supplementary Figure S3) and FliM (Supplementary Figure S14) from T. maritima (Brown et al., 2005). We chose to use these as surrogates for modeling the Spa33 heterotrimer into the pod densities of the SP. FliM-FliN is a heterotetramer (Paul et al., 2006) that has been proposed to be the structural equivalents of the Spa33 heterotrimer. In such a model FliM would be equivalent to the N-terminal portion of Spa33, despite there likely being significant structural differences between them, and the FliN trimer would represent the three copies of Spa33C within the overall Spa33 complex (comprising two copies of Spa33C with a third copy of the SPOA domain contributed by one of the two tandem SPOA domains at the C-terminus of Spa33FL). Using this and the flagellar C-ring of Vibrio alginolyticus (Carroll et al., 2020) as a guide for generating a model of the Spa33 heterotrimer and how such Spa33 complexes might form an interface, we could insert two copies of the modeled Spa33 homotrimer into a newly generated high-resolution image of a single SP pod from the Shigella injectisome (Figure 7). The fit appears to agree with the data on the size and shape of wild-type Spa33 when it is alone and/or incorporated into complexes with MxiN and MxiK (Table 3 and Figures 7C, D).
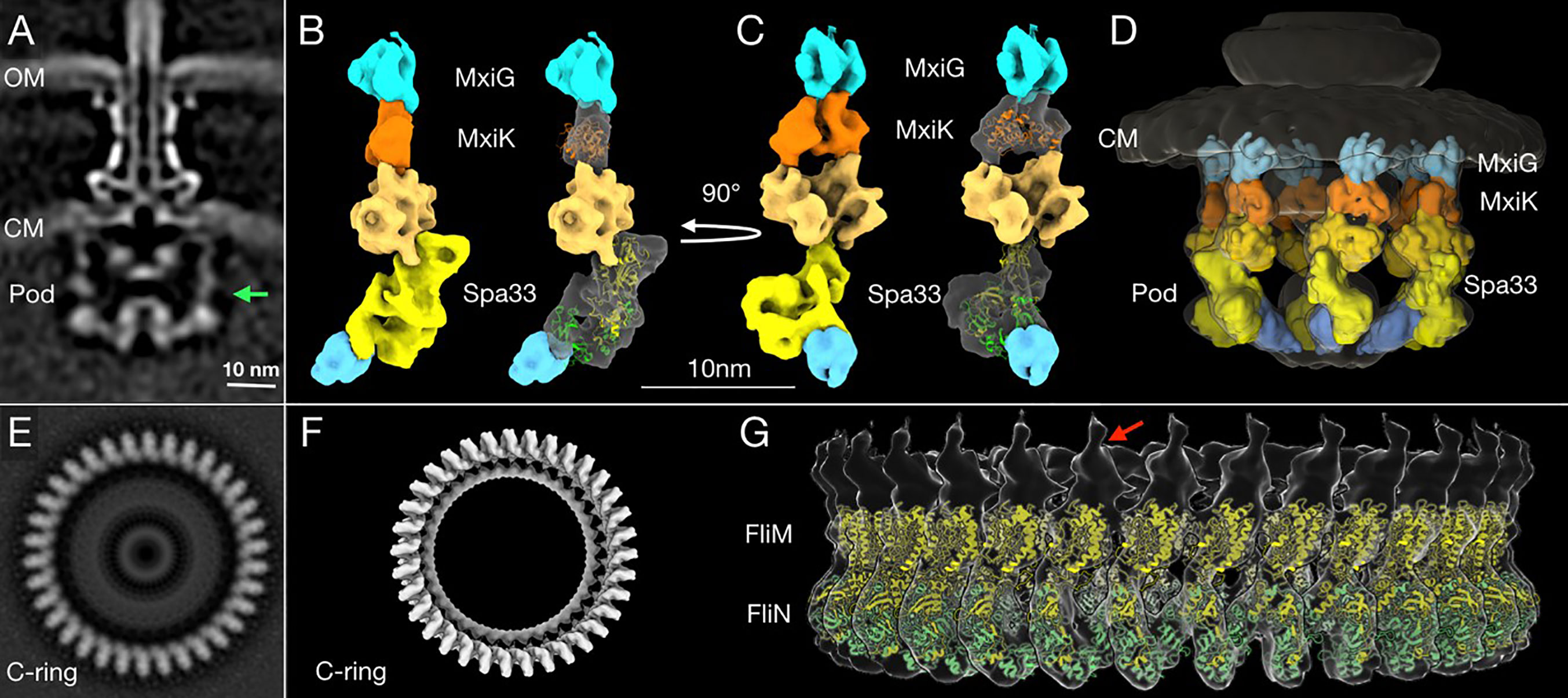
Figure 7 High-resolution structure and proposed model of the in situ SP pods from Shigella. (A) A cross-section image of the subtomogram average structure of the injectisome shows (from top to bottom) the needle, basal body and SP. The two bacterial membranes are indicated by OM (the outer membrane) and CM (the cytoplasmic membrane). The green arrow indicates the right side of the pod region that was used for the additional refinement process to improve its resolution. (B) A 3D surface rendering image of the refined pod structure is shown. From the top down, the refined pod structure associates with the cytoplasmic domain of MxiG (blue) and consists of MxiK (orange) and multiple copies of Spa33 (different shades of yellow), which also associate with MxiN (cyan at bottom) (Hu et al., 2015; Tachiyama et al., 2019). The crystal structure of MxiK’s homologue from P. aeruginosa, SctK (Muthuramalingam et al., 2020) was fit into the density beneath MxiG (right). Two copies of the Spa33 heterotrimer model were fit into the bottom part of Spa33 region in the refined pod structural image (right). Because this is a preliminary model, some regions did not fit precisely into the cryo-ET image (see Supplementary Figure S15). The top of the proposed Spa33 density forms a globular structure that interacts with MxiK. (C) The same images are shown rotated 90° relative to (B). This angle shows the single pod with a view that would be on the inside of the SP cage. (D) The refined pod structure was fit back into a 3D surface rendering of the complete SP. This image shows the six pods forming the cage-like structure of the SP. (E) A bottom view of a subtomogram average structure from the Vibrio alginolyticus flagella C-ring (Carroll et al., 2020). The C-ring is composed FliM and FliN, which are homologous to Spa33 in the pod assembly. (F) A surface rendering of the C-ring with its 34-fold symmetry (EMDB-21837) was generated using the UCSF Chimera (Pettersen et al., 2004) and ChimeraX (Goddard et al., 2018) software. (G) A side view of the 3D surface rendering of the C-ring with a model of a full-length FliM (yellow) and three copies of FliN (green) from I-TASSER (Roy et al., 2010; Yang et al., 2015; Carroll et al., 2020) is shown. The red arrow indicates a single unit of the C-ring structure for comparison with the refined single pod structure in (B, C).
The proposed orientation of the Spa33 units in our model is different from what is seen in the flagellar C-ring of V. alginolyticus. Within the flagellar C-ring, multiple copies of the FliM-FliN complex align side by side to generate a contiguous ring (Figures 7E–G), however, the Spa33 heterotrimer could be oriented differently so that it is nearly on its side to form a complex with another Spa33 complex that lies above it (Figures 7B, C). This would then give rise to six distinct pod entities evenly placed around the central ATPase to which it is connected via MxiN. Meanwhile, the upper Spa33 heterotrimer may be shaped slightly differently in that it forms a tight complex with MxiK whose density appears more globular than of MxiN in the in situ SP (Figure 7D). The Spa33 component of the Spa33-MxiK complex in this case could undergo a conformational change as it interfaces with MxiK to give rise to a more compact globular structure compared with the Spa33-MxiN complex. This fits with the observation that the Dh of the Spa33-MxiK, which is slightly smaller than that of the Spa33 heterotrimer alone. The structure of SctK from P. aeruginosa is fit into the density occupied by MxiK where it forms an intimate interface with the upper Spa33 heterotrimer (Figures 7C, D and Supplementary Figure S15). Thus, the Spa33 moiety within the Spa33-MxiN and Spa33-MxiK complexes are proposed to undergo conformational changes that give rise to two distinct complexes within the SP – one for interacting with the IR via MxiGC and one for interacting with the Spa47 ATPase via MxiN.
This work provides a detailed biochemical and biophysical exploration of the Shigella T3SS sorting platform (SP). The component protein interactions described here have been proposed before based on different types of analyses, including two-hybrid analysis and pull-down assays (Jouihri et al., 2003; Morita-Ishihara et al., 2006; Johnson and Blocker, 2008), however, the data presented here and related cryo-ET studies are making significant contributions to our detailed understanding of the structures and interfaces that are important for SP assembly (Hu et al., 2015; Hu et al., 2017). The pods are the major component of the Shigella injectisome SP. A difficulty encountered in focusing on these structures in situ is their apparent flexibility and dynamics within the active SP. Nevertheless, based on our findings, we can propose a model for the assembly of the Spa33-containing pods and speculate as to how these individual sub-assemblies contribute to active type III secretion in Shigella. It is accepted that dynamic processes within the SP are critical for recruitment of secretion substrates to the injectisome and that the SctN ATPase has an essential role in presenting these substrates to the export gate (Diepold et al., 2017). As part of this process, SctN has been proposed to act as a rotary motor (Majewski et al., 2019) in guiding substrates to the export gate, perhaps via the central stalk formed by SctO (Spa13 in Shigella). Dielpold et al. reported that Yersinia SctQ, a Spa33 homologue, is present in more than 20 copies within the injectisome SP and that it is in dynamic exchange with a cytoplasmic population during active type III secretion (Diepold et al., 2017). The details of how this dynamic exchange occurs remains to be determined and how this applies precisely to the Shigella system is far from clear.
It was recently reported by Bernal et al. that soluble complexes consisting of the Salmonella SP components SctQFL, SctQC, SctL and SctN could be assembled and the organization of this complex could be determined using small-angle X-ray scattering (SAXS) (Bernal et al., 2019). As with the Spa33 complex, SctQFL was made soluble by the presence of SctQC. The SctQ heterotrimer could associate with a SctL dimer in the same way that Spa33 and MxiN interact. They further showed that the SctQ-SctL complex could associate with the SctN ATPase. This is similar to the work of Notti et al. (2015) who could purify a complex consisting of SctQ, SctL and SctN. We did not attempt to generate a Spa33-MxiN-Spa47 complex, however, this is a relationship that will need to be explored as this work continues. We also found that in addition to MxiN, the Spa33 heterotrimer could associate with MxiK, however, this required that the wild-type spa33 and mxiK be co-expressed prior to purifying the resulting complex. This association has a stabilizing (and solubilizing) effect on MxiK and, based on the poor solubility of MxiK, it seems likely that the MxiK that exists in the Shigella cytoplasm is in fact part of a Spa33-MxiK complex that for the most part behaves as a single folded unit that is highly stable. Moreover, the Spa33-MxiK complex has biophysical properties (thermal stability and hydrodynamic radius) that are distinct from those of the Spa33-MxiN complex, which can be assembled in vitro from purified components.
We have demonstrated that MxiK is an essential SP component that acts as an adaptor between the SP and the cytoplasmic domain of the IR protein MxiG. In the absence of MxiK, the SP is completely absent from the injectisome. Meanwhile, MxiN forms radial spokes that emanate from a central hub that is the Spa47 hexamer (Hu et al., 2015). In the absence of MxiN, remnants of the pods are still visible and these are the result of the interactions between MxiG, MxiK and Spa33 (Supplementary Figure S2) (Hu et al., 2015). This unique observation indicates that a sub-assembly consisting of MxiK and Spa33 can be incorporated into an otherwise absent SP. That cannot be said of MxiN-Spa33 subassemblies and this is why we proposed that the Spa33-MxiK and Spa33-MxiN complexes can form and behave independently in the Shigella cytoplasm. Furthermore, it appears that the Spa33-MxiK units can be associate with the injectisome basal body in the absence of MxiN (Supplementary Figure S2).
From the data presented here and elsewhere, we propose that the Spa33-MxiK and Spa33-MxiN assemblies are physiologically relevant structures with important roles in the mechanisms governing type III secretion in Shigella. These data are also consistent with the large pod structures of the SP being formed from two copies of the Spa33 heterotrimer with an architecture that is altered from the interactions that give rise to a contiguous flagellar C-ring. Thus, in view of the dynamics involving SctQ exchange between the SP and cytoplasm, we propose a model in which Spa33 heterotrimers act at two different locations during the recruitment of secretion cargo. Spa33-MxiK complexes are thus proposed to be required for communication between the injectisome IR and the SP while Spa33-MxiN complexes are active in ushering secretion substrates to the SP where they can then be acted upon by Spa47 for chaperone removal and presentation to the export gate (Akeda and Galan, 2005). It is also possible that the Spa33-MxiN complex is interacting with Spa47 prior to incorporation (with secretion substrate) into the SP based on independent work by Notti et al. and Bernal et al. (Notti et al., 2015; Bernal et al., 2019). The export gate then harnesses the proton-motive forces to actually fuel the secretion process (Shen and Blocker, 2016). It is also likely that the Spa33-MxiK complex is in exchange with a cytoplasmic pool of similar complexes. Interestingly, this complex has the potential to act independent of the Spa33-MxiN complex in interacting with the IR (Hu et al., 2015), perhaps to act in guiding Spa33-MxiN-effector (and possibly Spa47) complexes to the SP where there are up to five other pod complexes comprised of MxiK, Spa33 and MxiN in association with the Spa47 hub. Such a model fits with the cryo-ET based structural fits depicted in Figure 7, which are modeled based on the similarities between the now solved structure of Pseudomonas SctK and a model of the Spa33 heterotrimer based on similarities with the flagellar C-ring components FliM and FliN (Supplementary Figure S15). The model we propose here may be speculative, however, it does appear that the densities beneath MxiK are sufficiently large to accommodate two Spa33 heterotrimers. Ultimately, the ability to precisely fit structures into these densities will require still higher resolution cryo-ET structures and/or a high-resolution structure (i.e crystal structure) of the Spa33 heterotrimer. Clearly, a great deal of work remains to fully understand the events occurring within the SP, however, complementary molecular, biophysical and imaging techniques are beginning to shed light on how the full injectisome functions. Perhaps further revelations will become known when we are able to manipulate the Shigella T3SS so that it adopts specific secretion states.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.emdataresource.org/EMD-23440.
WDP was responsible for conceptualization, funding acquisition, project administration, writing portions of the original draft and supervision of research staff. ST was responsible for conceptualization, investigation, methodology and writing portions of the original draft. RS was responsible for two-hybrid analyses, portions of the biophysical analyses and critical reading of the manuscript. YC was responsible for the cryo-ET analyses and refinements. BC was responsible for data analysis and modeling. MM was responsible for data curation, formal analysis, investigation, methodology and critical review of the manuscript. SW was responsible for conceptualization, formal analysis, investigation, methodology, visualization and writing portions of the original draft. MB was responsible for generating fusion protein mutants and portions of the contact-hemolysis experiments. WLP was responsible for manuscript editing and writing. JL was responsible for conceptualization, overseeing the cryo-ET imaging studies and data analysis and interpretation. All authors contributed to the article and approved the submitted version.
This research was funded by the National Institute of Allergies and Infectious Disease awards NIAID R01 AI123351 and R21 AI146517 to WDP.
Author RS was employed by the company NanoBio Designs, LLC during the writing of this manuscript. The author MB was employed by the company Catalent Pharma Solutions during the writing of this manuscript.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcimb.2021.682635/full#supplementary-material
Agulleiro, J. I., Fernandez, J. J. (2015). Tomo3D 2.0–Exploitation of Advanced Vector Extensions (AVX) for 3D Reconstruction. J. Struct. Biol. 189 (2), 147–152. doi: 10.1016/j.jsb.2014.11.009
Akeda, Y., Galan, J. E. (2005). Chaperone Release and Unfolding of Substrates in Type III Secretion. Nature 437 (7060), 911–915. doi: 10.1038/nature03992
Barta, M. L., Shearer, J. P., Arizmendi, O., Tremblay, J. M., Mehzabeen, N., Zheng, Q., et al. (2017). Single-Domain Antibodies Pinpoint Potential Targets Within Shigella Invasion Plasmid Antigen D of the Needle Tip Complex for Inhibition of Type III Secretion. J. Biol. Chem. 292 (40), 16677–16687. doi: 10.1074/jbc.M117.802231
Bernal, I., Bornicke, J., Heidemann, J., Svergun, D., Horstmann, J. A., Erhardt, M., et al. (2019). Molecular Organization of Soluble Type III Secretion System Sorting Platform Complexes. J. Mol. Biol. 431 (19), 3787–3803. doi: 10.1016/j.jmb.2019.07.004
Bi, E., Lutkenhaus, J. (1990). Interaction Between the Min Locus and ftsZ. J. Bacteriol. 172 (10), 5610–5616. doi: 10.1128/JB.172.10.5610-5616.1990
Birket, S. E., Harrington, A. T., Espina, M., Smith, N. D., Terry, C. M., Darboe, N., et al. (2007). Preparation and Characterization of Translocator/Chaperone Complexes and Their Component Proteins From Shigella Flexneri. Biochemistry 46 (27), 8128–8137. doi: 10.1021/bi700099c
Blocker, A., Gounon, P., Larquet, E., Niebuhr, K., Cabiaux, V., Parsot, C., et al. (1999). The Tripartite Type III Secreton of Shigella Flexneri Inserts IpaB and IpaC Into Host Membranes. J. Cell Biol. 147 (3), 683–693. doi: 10.1083/jcb.147.3.683
Brown, P. N., Mathews, M. A., Joss, L. A., Hill, C. P., Blair, D. F. (2005). Crystal Structure of the Flagellar Rotor Protein FliN From Thermotoga Maritima. J. Bacteriol. 187 (8), 2890–2902. doi: 10.1128/JB.187.8.2890-2902.2005
Burgess, J. L., Burgess, R. A., Morales, Y., Bouvang, J. M., Johnson, S. J., Dickenson, N. E. (2016). Structural and Biochemical Characterization of Spa47 Provides Mechanistic Insight Into Type III Secretion System ATPase Activation and Shigella Virulence Regulation. J. Biol. Chem. 291 (50), 25837–25852. doi: 10.1074/jbc.M116.755256
Bzymek, K. P., Hamaoka, B. Y., Ghosh, P. (2012). Two Translation Products of Yersinia yscQ Assemble to Form a Complex Essential to Type III Secretion. Biochemistry 51 (8), 1669–1677. doi: 10.1021/bi201792p
Carroll, B. L., Nishikino, T., Guo, W., Zhu, S., Kojima, S., Homma, M., et al. (2020). The Flagellar Motor of Vibrio Alginolyticus Undergoes Major Structural Remodeling During Rotational Switching. Elife 9, e61446. doi: 10.7554/eLife.61446
Case, H. B., Dickenson, N. E. (2018). MxiN Differentially Regulates Monomeric and Oligomeric Species of the Shigella Type Three Secretion System Atpase Spa47. Biochemistry 57 (15), 2266–2277. doi: 10.1021/acs.biochem.8b00070
Cordes, F. S., Komoriya, K., Larquet, E., Yang, S., Egelman, E. H., Blocker, A., et al. (2003). Helical Structure of the Needle of the Type III Secretion System of Shigella Flexneri. J. Biol. Chem. 278 (19), 17103–17107. doi: 10.1074/jbc.M300091200
Deane, J. E., Roversi, P., Cordes, F. S., Johnson, S., Kenjale, R., Daniell, S., et al. (2006). Molecular Model of a Type III Secretion System Needle: Implications for Host-Cell Sensing. Proc. Natl. Acad. Sci. U. S. A. 103 (33), 12529–12533. doi: 10.1073/pnas.0602689103
Diepold, A., Kudryashev, M., Delalez, N. J., Berry, R. M., Armitage, J. P. (2015). Composition, Formation, and Regulation of the Cytosolic C-Ring, a Dynamic Component of the Type III Secretion Injectisome. PLoS Biol. 13 (1), e1002039. doi: 10.1371/journal.pbio.1002039
Diepold, A., Sezgin, E., Huseyin, M., Mortimer, T., Eggeling, C., Armitage, J. P. (2017). A Dynamic and Adaptive Network of Cytosolic Interactions Governs Protein Export by the T3SS Injectisome. Nat. Commun. 8, 15940. doi: 10.1038/ncomms15940
Espina, M., Ausar, S. F., Middaugh, C. R., Picking, W. D., Picking, W. L. (2006a). Spectroscopic and Calorimetric Analyses of Invasion Plasmid Antigen D (IpaD) From Shigella Flexneri Reveal the Presence of Two Structural Domains. Biochemistry 45 (30), 9219–9227. doi: 10.1021/bi060625v
Espina, M., Olive, A. J., Kenjale, R., Moore, D. S., Ausar, S. F., Kaminski, R. W., et al. (2006b). IpaD Localizes to the Tip of the Type III Secretion System Needle of Shigella Flexneri. Infect. Immun. 74 (8), 4391–4400. doi: 10.1128/IAI.00440-06
Fadouloglou, V. E., Tampakaki, A. P., Glykos, N. M., Bastaki, M. N., Hadden, J. M., Phillips, S. E., et al. (2004). Structure of HrcQB-C, a Conserved Component of the Bacterial Type III Secretion Systems. Proc. Natl. Acad. Sci. U. S. A. 101 (1), 70–75. doi: 10.1073/pnas.0304579101
Farley, M. M., Hu, B., Margolin, W., Liu, J. (2016). Minicells, Back in Fashion. J. Bacteriol. 198 (8), 1186–1195. doi: 10.1128/JB.00901-15
Galan, J. E., Waksman, G. (2018). Protein-Injection Machines in Bacteria. Cell 172 (6), 1306–1318. doi: 10.1016/j.cell.2018.01.034
Geisbrecht, B. V., Bouyain, S., Pop, M. (2006). An Optimized System for Expression and Purification of Secreted Bacterial Proteins. Protein Expr. Purif. 46 (1), 23–32. doi: 10.1016/j.pep.2005.09.003
Goddard, T. D., Huang, C. C., Meng, E. C., Pettersen, E. F., Couch, G. S., Morris, J. H., et al. (2018). UCSF ChimeraX: Meeting Modern Challenges in Visualization and Analysis. Protein Sci. 27 (1), 14–25. doi: 10.1002/pro.3235
Hu, B., Lara-Tejero, M., Kong, Q., Galan, J. E., Liu, J. (2017). In Situ Molecular Architecture of the Salmonella Type III Secretion Machine. Cell 168 (6), 1065–1074.e1010. doi: 10.1016/j.cell.2017.02.022
Hu, B., Morado, D. R., Margolin, W., Rohde, J. R., Arizmendi, O., Picking, W. L., et al. (2015). Visualization of the Type III Secretion Sorting Platform of Shigella Flexneri. Proc. Natl. Acad. Sci. U. S. A. 112 (4), 1047–1052. doi: 10.1073/pnas.1411610112
Johnson, S., Blocker, A. (2008). Characterization of Soluble Complexes of the Shigella Flexneri Type III Secretion System Atpase. FEMS Microbiol. Lett. 286 (2), 274–278. doi: 10.1111/j.1574-6968.2008.01284.x
Jouihri, N., Sory, M. P., Page, A. L., Gounon, P., Parsot, C., Allaoui, A. (2003). MxiK and MxiN Interact with the Spa47 Atpase and Are Required for Transit of the Needle Components MxiH and MxiI, But Not of Ipa Proteins, Through the Type III Secretion Apparatus of Shigella Flexneri. Mol. Microbiol. 49 (3), 755–767. doi: 10.1046/j.1365-2958.2003.03590.x
Kadari, M., Lakhloufi, D., Delforge, V., Imbault, V., Communi, D., Smeesters, P., et al. (2019). Multiple Proteins Arising From a Single Gene: The Role of the Spa33 Variants in Shigella T3SS Regulation. Microbiologyopen 8 (12), e932. doi: 10.1002/mbo3.932
Kenjale, R., Wilson, J., Zenk, S. F., Saurya, S., Picking, W. L., Picking, W. D., et al. (2005). The Needle Component of the Type III Secreton of Shigella Regulates the Activity of the Secretion Apparatus. J. Biol. Chem. 280 (52), 42929–42937. doi: 10.1074/jbc.M508377200
Kremer, J. R., Mastronarde, D. N., McIntosh, J. R. (1996). Computer Visualization of Three-Dimensional Image Data Using IMOD. J. Struct. Biol. 116 (1), 71–76. doi: 10.1006/jsbi.1996.0013
Lara-Tejero, M., Kato, J., Wagner, S., Liu, X., Galan, J. E. (2011). A Sorting Platform Determines the Order of Protein Secretion in Bacterial Type III Systems. Science 331 (6021), 1188–1191. doi: 10.1126/science.1201476
Lara-Tejero, M., Qin, Z., Hu, B., Butan, C., Liu, J., Galan, J. E. (2019). Role of SpaO in the Assembly of the Sorting Platform of a Salmonella Type III Secretion System. PLoS Pathog. 15 (1), e1007565. doi: 10.1371/journal.ppat.1007565
Lunelli, M., Kamprad, A., Burger, J., Mielke, T., Spahn, C. M. T., Kolbe, M. (2020). Cryo-EM Structure of the Shigella Type III Needle Complex. PLoS Pathog. 16 (2), e1008263. doi: 10.1371/journal.ppat.1008263
Majewski, D. D., Worrall, L. J., Hong, C., Atkinson, C. E., Vuckovic, M., Watanabe, N., et al. (2019). Cryo-EM Structure of the Homohexameric T3SS ATPase-Central Stalk Complex Reveals Rotary ATPase-Like Asymmetry. Nat. Commun. 10 (1), 626. doi: 10.1038/s41467-019-08477-7
Mastronarde, D. N. (2005). Automated Electron Microscope Tomography Using Robust Prediction of Specimen Movements. J. Struct. Biol. 152 (1), 36–51. doi: 10.1016/j.jsb.2005.07.007
McDowell, M. A., Marcoux, J., McVicker, G., Johnson, S., Fong, Y. H., Stevens, R., et al. (2016). Characterisation of Shigella Spa33 and Thermotoga FliM/N Reveals a New Model for C-ring Assembly in T3SS. Mol. Microbiol. 99 (4), 749–766. doi: 10.1111/mmi.13267
Morita-Ishihara, T., Ogawa, M., Sagara, H., Yoshida, M., Katayama, E., Sasakawa, C. (2006). Shigella Spa33 Is an Essential C-Ring Component of Type III Secretion Machinery. J. Biol. Chem. 281 (1), 599–607. doi: 10.1074/jbc.M509644200
Muthuramalingam, M., Whittier, S. K., Lovell, S., Battaile, K. P., Tachiyama, S., Johnson, D. K., et al. (2020). The Structures of SctK and SctD From Pseudomonas Aeruginosa Reveal the Interface of the Type III Secretion System Basal Body and Sorting Platform. J. Mol. Biol. 432 (24), 166693. doi: 10.1016/j.jmb.2020.10.027
Notti, R. Q., Bhattacharya, S., Lilic, M., Stebbins, C. E. (2015). A Common Assembly Module in Injectisome and Flagellar Type III Secretion Sorting Platforms. Nat. Commun. 6, 7125. doi: 10.1038/ncomms8125
Notti, R. Q., Stebbins, C. E. (2016). The Structure and Function of Type III Secretion Systems. Microbiol. Spectr. 4 (1), 10.1128/microbiolspec.VMBF-0004-2015. doi: 10.1128/microbiolspec.VMBF-0004-2015
Paul, K., Harmon, J. G., Blair, D. F. (2006). Mutational Analysis of the Flagellar Rotor Protein FliN: Identification of Surfaces Important for Flagellar Assembly and Switching. J. Bacteriol. 188 (14), 5240–5248. doi: 10.1128/JB.00110-06
Pettersen, E. F., Goddard, T. D., Huang, C. C., Couch, G. S., Greenblatt, D. M., Meng, E. C., et al. (2004). UCSF Chimera–A Visualization System for Exploratory Research and Analysis. J. Comput. Chem. 25 (13), 1605–1612. doi: 10.1002/jcc.20084
Picking, W. L., Nishioka, H., Hearn, P. D., Baxter, M. A., Harrington, A. T., Blocker, A., et al. (2005). IpaD of Shigella Flexneri Is Independently Required for Regulation of Ipa Protein Secretion and Efficient Insertion of IpaB and IpaC Into Host Membranes. Infect. Immun. 73 (3), 1432–1440. doi: 10.1128/IAI.73.3.1432-1440.2005
Portaliou, A. G., Tsolis, K. C., Loos, M. S., Zorzini, V., Economou, A. (2016). Type III Secretion: Building and Operating a Remarkable Nanomachine. Trends Biochem. Sci. 41 (2), 175–189. doi: 10.1016/j.tibs.2015.09.005
Roy, A., Kucukural, A., Zhang, Y. (2010). I-TASSER: A Unified Platform for Automated Protein Structure and Function Prediction. Nat. Protoc. 5 (4), 725–738. doi: 10.1038/nprot.2010.5
Shen, D. K., Blocker, A. J. (2016). MxiA, MxiC and IpaD Regulate Substrate Selection and Secretion Mode in the T3SS of Shigella Flexneri. PLoS One 11 (5), e0155141. doi: 10.1371/journal.pone.0155141
Tachiyama, S., Chang, Y., Muthuramalingam, M., Hu, B., Barta, M. L., Picking, W. L., et al. (2019). The Cytoplasmic Domain of MxiG Interacts With MxiK and Directs Assembly of the Sorting Platform in the Shigella Type III Secretion System. J. Biol. Chem. 294 (50), 19184–19196. doi: 10.1074/jbc.RA119.009125
Wagner, S., Diepold, A. (2020). A Unified Nomenclature for Injectisome-Type Type III Secretion Systems. Curr. Top. Microbiol. Immunol. 427, 1–10. doi: 10.1007/82_2020_210
Winkler, H. (2007). 3D Reconstruction and Processing of Volumetric Data in Cryo-Electron Tomography. J. Struct. Biol. 157 (1), 126–137. doi: 10.1016/j.jsb.2006.07.014
Winkler, H., Zhu, P., Liu, J., Ye, F., Roux, K. H., Taylor, K. A. (2009). Tomographic Subvolume Alignment and Subvolume Classification Applied to Myosin V and SIV Envelope Spikes. J. Struct. Biol. 165 (2), 64–77. doi: 10.1016/j.jsb.2008.10.004
Yang, J., Yan, R., Roy, A., Xu, D., Poisson, J., Zhang, Y. (2015). The I-TASSER Suite: Protein Structure and Function Prediction. Nat. Methods 12 (1), 7–8. doi: 10.1038/nmeth.3213
Keywords: type III secretion system, injectisome, Spa33, MxiN, MxiK, sorting platform, Shigella flexneri
Citation: Tachiyama S, Skaar R, Chang Y, Carroll BL, Muthuramalingam M, Whittier SK, Barta ML, Picking WL, Liu J and Picking WD (2021) Composition and Biophysical Properties of the Sorting Platform Pods in the Shigella Type III Secretion System. Front. Cell. Infect. Microbiol. 11:682635. doi: 10.3389/fcimb.2021.682635
Received: 26 March 2021; Accepted: 17 May 2021;
Published: 03 June 2021.
Edited by:
Matthew S. Francis, Umeå University, SwedenReviewed by:
Tohru Minamino, Osaka University, JapanCopyright © 2021 Tachiyama, Skaar, Chang, Carroll, Muthuramalingam, Whittier, Barta, Picking, Liu and Picking. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: William D. Picking, cGlja2luZ0BrdS5lZHU=
†Present address: Ryan Skaar, NanoBio Designs, LLC., West Des Moines, IA, United States
Michael L. Barta, Catalent Pharma Solutions, Kansas City, MO, United States
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.