- Centre for Infectious Diseases, Heidelberg University Hospital, Heidelberg, Germany
Regulating the number of progeny generated by replicative cell cycles is critical for any organism to best adapt to its environment. Classically, the decision whether to divide further is made after cell division is completed by cytokinesis and can be triggered by intrinsic or extrinsic factors. Contrarily, cell cycles of some species, such as the malaria-causing parasites, go through multinucleated cell stages. Hence, their number of progeny is determined prior to the completion of cell division. This should fundamentally affect how the process is regulated and raises questions about advantages and challenges of multinucleation in eukaryotes. Throughout their life cycle Plasmodium spp. parasites undergo four phases of extensive proliferation, which differ over three orders of magnitude in the amount of daughter cells that are produced by a single progenitor. Even during the asexual blood stage proliferation parasites can produce very variable numbers of progeny within one replicative cycle. Here, we review the few factors that have been shown to affect those numbers. We further provide a comparative quantification of merozoite numbers in several P. knowlesi and P. falciparum parasite strains, and we discuss the general processes that may regulate progeny number in the context of host-parasite interactions. Finally, we provide a perspective of the critical knowledge gaps hindering our understanding of the molecular mechanisms underlying this exciting and atypical mode of parasite multiplication.
Introduction
Cell types containing multiple genome copies emerged independently on various branches of the eukaryotic tree of life (Comai, 2005), which suggests convergent evolution of this trait in order to accommodate very different adaptive needs (Campbell et al., 2016). Multinucleated cells are prominent in filamentous fungi (Roper et al., 2013; Mela et al., 2020). In some cases those induce polyploidy as an adaptation to stress (Selmecki et al., 2010; Selmecki et al., 2015) or individual nuclei can be degraded as a nutrient source under starvation conditions (Shoji et al., 2010). Having multiple genome copies within one cell can further provide a buffering function, allowing diversification of the genome by structural variation or single nucleotide polymorphisms, without suffering deleterious consequence of those mutations. Hence, filamentous fungi establish long-lived compartments in which genetically divergent nuclei can co-exist, compete and even undergo horizontal gene transfer by fusions of nuclei (Mela et al., 2020). The range of unicellular organisms being multinucleated stretches from yeasts to algae (Coneva and Chitwood, 2015; Mitchison-Field et al., 2019). A striking example is Caulerpa spp., a single-celled alga that can reach a length of meters and contains myriads of nuclei (Jacobs, 1994). Within multicellular organisms the mammalian placenta, bones, and muscles are known to form multinucleated cell types (Mintz and Baker, 1967; Marks, 1983; Mi et al., 2000; Płachno and Światek, 2011). In plants, appearance of multinucleated cells has been associated with the increase of cell size and thereby growth of the organism as a whole (Płachno and Światek, 2011; Coneva and Chitwood, 2015). In those cases multinucleation is necessary to ensure the scaling of genome copies with cytoplasmic volume, which facilitates homeostatic protein expression or gene dosage (Marguerat and Bähler, 2012) (see also Machado et al. in this issue). Plants can sometimes even form giant cells containing up to 100 nuclei, when infected by nematodes (Favery et al., 2016). Interestingly, an organism with a more Plasmodium-related multiplication mode is the free-living unicellular marine Ichthyosporea, a putative predecessor of multicellular animals (Ondracka et al., 2018). It generates up to 128 nuclei by synchronous divisions before completing daughter cell formation and releasing them through bursting. We, however, argue that malaria parasites use multinucleation in very different adaptive context. When transmitting between the mosquito vector and the human host, malaria parasites encounter important population bottlenecks and react to those steep decreases of population size by phases of extensive proliferation. Plasmodium parasites undergo multiple cycles of nuclear division, while cytokinesis only occurs prior to egress from the host cell. Hence, the final number of nuclei determines how many daughter cells will emerge from a single progenitor. The plasticity of Plasmodium spp. division with respect to the total number of daughter cells produced within one replicative cycle is remarkable (Matthews et al., 2018). In this review we i) summarize the importance of multinucleation, ii) discuss how progeny number might be regulated, and iii) define critical cellular parameters that remain to be assessed to better understand this process.
Multinucleation, Polyploidy, Syncytium, and Coenocyte
Since the terminology around multi-genome containing cells is sometimes contradictory, we want to define our usage of those terms. Ploidy refers to the number of chromosomes sets per cell. In a normally haploid cell, any genome copy number higher than two, which is the post-replicative state, would be considered polyploid. Multinucleation refers to the number of physically separated nuclei within a common cytoplasm. Polyploidy and multinucleation are not per se mutually exclusive. In principle a multinucleated cell can arise by two ways. If multinucleation is a result of cell-cell fusion, which has not been reported in malaria parasites, it is referred to as syncytium (Daubenmire, 1936). Multiple rounds of nuclear division that occur within a common cytoplasm cause the formation of a coenocyte.
Progeny Numbers in Plasmodium Species Display Remarkable Plasticity
Plasmodium spp. experience polyploid as well as multinucleated stages throughout their life cycle (Francia and Striepen, 2014; Gubbels et al., 2020). The timing of those developmental stages differs significantly and the number of progeny generated by a single progenitor cell varies over three orders of magnitude (Figure 1A). When a mosquito takes up a Plasmodium-infected blood meal it triggers conversion of male gametocytes into gametes. Activated gametocytes undergo three rounds of DNA replication yielding a polyploid nucleus with 8 chromosome sets, which are then packaged into male gamete flagella (Sinden et al., 1978). This process is completed in less than 15 min in P. berghei making it an extreme example of fast DNA replication (Fang et al., 2017; Matthews et al., 2018). The fact that about 60% of gametes are malformed or lack nuclei highlights a potential trade-off between speed and accuracy (Sinden et al., 2010). Once male and female gametes have fused and undergone meiotic division, they embed themselves in the mosquito midgut wall, where oocysts develop. An oocyst takes a few days to mature and can produce up to thousand sporozoites (Beier, 1998; Vaughan, 2007). Polyploidy in this stage is indicated by the presence of multipolar spindles (Schrével et al., 1977; Spreng et al., 2019). When an infected mosquito takes a blood meal it injects sporozoites into the host skin. Those find their way to liver cells inside which tens of thousands of merozoites are produced from a single infected hepatocyte within less than a week (Shortt et al., 1951; Prudêncio et al., 2006; Sturm et al., 2006). Merozoites are then released into the blood where they start the replicative cycles that cause pathogenesis. Once they invade a red blood cell, parasites undergo asynchronous divisions, followed by a final synchronous one. The nuclei alternate between haploid and diploid stages, while polyploidy is usually not observed (Janse et al., 1986; Klaus et al., 2021). The total number of daughter cells resulting from these nuclear division cycles does not only vary within a population but also between different P. falciparum strains. In the Dd2 strain between 8 and 28 merozoites have been counted with an average of 18, while HB3 had an average of 15 (Reilly et al., 2007). In 3D7 average numbers of 22 have been reported (Dorin-Semblat et al., 2008; Garg et al., 2015). In newly isolated Ghanaian parasite strains, values between 16-24 were found, although those were only measured after six months of continuous in vitro culture and once multiplication rates had significantly increased (Stewart et al., 2020). The counted number of merozoites can, however, vary significantly depending on the used visualization method (Garg et al., 2015). Hence, we developed an optimized merozoite counting protocol involving HyVolution-based super-resolution microscopy and applied it to commonly studied P. falciparum strains and laboratory-adapted P. knowlesi (Figure 1B). Contrary to our expectations, this yielded a median of 19 or 20 merozoites for all analyzed P. falciparum strains. Statistical difference was only found between the 3D7, Dd2, and FCR3 strains although their mean values barely deviated by more than 1 merozoite. In P. knowlesi we counted significantly less merozoites, which associates with the shorter cell cycle duration of about 24 hours (while P. falciparum takes about 48 hours). The rodent malaria parasites, which also follow a 24 hour cycle, produce about 12 merozoites in the case of P. berghei (Mancio-Silva et al., 2017), but only around 6 in P. chabaudi (Birget et al., 2019). The convergence of average merozoite numbers for P. falciparum strains potentially hints at the importance of growth conditions rather than of genetic or epigenetic diversity. Nevertheless, the distribution of numbers remains wide reaching from 11 to 30. What causes this variability in progeny number and if and how the parasite counts its nuclei is unknown.
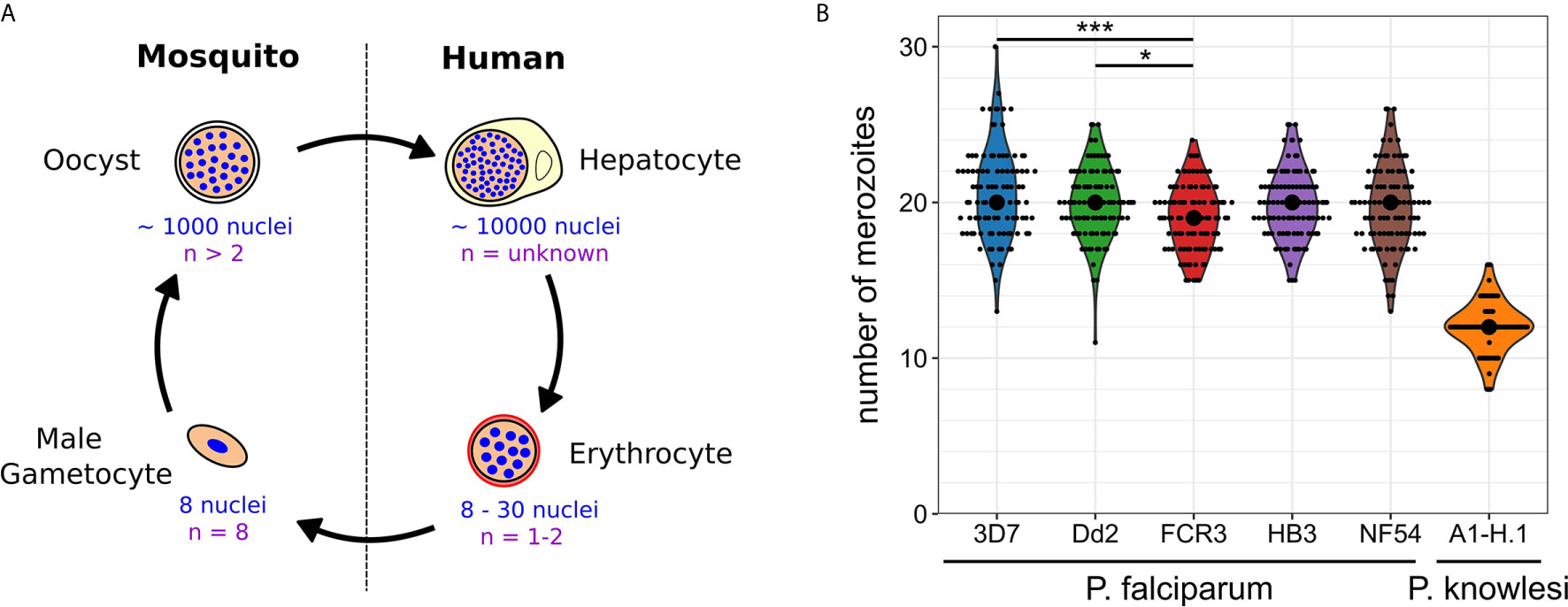
Figure 1 Nuclear and genome copy numbers throughout Plasmodium spp. life cycle stages. (A) Schematic depiction of approximate total number of nuclei produced during the proliferative life cycle stages of P. falciparum as well as the highest observed ploidy (n) or genome copy number within individual nuclei. (B) Number of merozoites produced in P. falciparum and P. knowlesi. To count merozoites, late stage parasites were enriched by magnetic purification or Nycodenz density gradient. Parasites were incubated with the cyclic GMP-dependent protein kinase (PKG) inhibitor ML10 (25 nM in P. falciparum strains; 125 nM in P. knowlesi) for 3 h to prevent egress, increasing the percentage of post-mitotic parasites. After preparation of blood smears parasites were fixed with 4% paraformaldehyde for 20 min at 37°C. Before imaging, cells were stained with Hoechst. Microscopy was performed on a Leica TCS SP8 scanning confocal microscope with Lightning (LNG) module. LNG enables automated adaptive deconvolution after acquisition resulting in super-resolution images. For each Plasmodium strain, 100 cells were imaged. Merozoites were counted in a single-blind mode by three independent researchers using Fiji. The mean merozoite number for each individual cell was taken to create the violin plot. The centered black dot represents the median. High density of nuclei in P. falciparum made it difficult to assess the definitive final number of nuclei. This counting error might contribute to the presence of many uneven numbers in P. falciparum strains, while in P. knowlesi almost exclusively even numbers were counted. For statistical analysis, a Welch-ANOVA test was performed, and individual lines were compared with a post hoc Games-Howell test using R studio. For P. falciparum strains, FCR3 and 3D7 (p = 2.4x10^-4) as well as FCR3 and Dd2 (p = 4.2x10^-2) showed significant differences.
Putative Roles of Multinucleated and Polyploid Cell Stages in Plasmodium Species
Contrary to fungi or plants, the size of the malaria-causing parasite is restricted by its host cell and the duration of a specific multinucleated or polyploid stage is limited by the cyclical nature of the parasite’s lifestyle. Nevertheless, genome diversification via mitotic recombination is frequent in Plasmodium blood stages (Claessens et al., 2014). This effect is, however, concentrated within the clonally variant gene families mostly located at the telomeres, while the core genome is surprisingly stable (Bopp et al., 2013). Recombination events could be augmented in the polyploid mosquito stages where more sister chromatid arms can come in contact with each other. It is, nonetheless, conceivable that generating many nuclei could provide a buffering function for mutations as described in fungi, facilitating a diversified progeny that can yield better adapted offspring. Unlike in most organisms listed above, the number of generated nuclei in Plasmodium directly predicts the number of progeny emerging from an infected cell. Hence, a more manifest purpose of multinucleation is reaching optimal multiplication rates to generate a sufficiently high parasite concentration in the blood, which ultimately increases the probability of transmission upon mosquito bite. Blood-stage cycles of Plasmodium spp. always are multiples of 24 hours even though parasites cultured in vitro can slightly deviate from that (Reilly et al., 2007). This suggests a strong incentive for the parasite to synchronize its growth with the host circadian cycle (Rijo-Ferreira et al., 2020; Smith et al., 2020). Whether there might also be a “safety in numbers”-effect due to the synchronous burst of infected cells and release of huge numbers of merozoites remains to be investigated. We speculate that the parasite uses multinucleation to convert the maximal amount of red blood cell material into merozoites within the given cycle time. This tendency to favor speed over accuracy is underlined by the apparent loss of canonical cell cycle checkpoints involved in error correction such as the spindle assembly and DNA damage checkpoints. Coherently, this lack of cell cycle checkpoints has also been documented in particularly fast growing fungal species, such as Hanseniaspora (Steenwyk et al., 2019) and during the first rapid divisions of zebrafish embryos (Zhang et al., 2015). What regulates the final number of nuclei in malaria parasites is unknown, but general principles, supported by a few studies done in blood stages, can be discussed.
Final Number of Merozoites Could be Predefined by a Counting Mechanism
Multinucleated parasites have to regulate the transition from nuclear division to daughter cell formation. Mechanisms that measure system size before concluding cell division have already been proposed for model organisms (Facchetti et al., 2017; Brownlee and Heald, 2019; Jones et al., 2019). Analogously, nuclear divisions could be limited by a counting mechanism that initiates cytokinesis, or the last synchronous round of division, once a target number of nuclei is reached (Figure 2A). The variability of progeny number within a single parasite strain argues against precise counting. A broader distribution of merozoite numbers could, however, present a competitive advantage. In a bet-hedging strategy the parasite would thereby generate sub-populations, which are better adapted to changing conditions e.g. low merozoite numbers during dropping nutrient levels in the blood (Mancio-Silva et al., 2017). If this is the case merozoite number might be an inherited trait, which predicts that parasites with a high nuclear number produce offspring that also generate high numbers. How the parasite would know how many nuclei there are and how such a mechanism would function is completely speculative. It could comprise an essential division factor available only in limiting amounts and potentially linked to extrinsic resources as discussed below. A factor that more overtly limits nuclear number is the centrosome (Arnot et al., 2011). Each round of division precludes centrosome duplication to form the poles between which chromosomes will be segregated (Simon et al., 2021). Transient localization of the aurora kinase PfArk1 to the centrosome just prior to its duplication has been described (Reininger et al., 2011). Treatment with a canonical aurora kinase inhibitor, hesperadin, causes a reduction in merozoite number, but also the appearance of resistance mutation in the PfNek1 kinase suggesting an epistatic interaction with PfArk1 (Morahan et al., 2020). The centrosome further represents the site where the transition between the nuclear division and daughter cell formation is organized in T. gondii (Chen and Gubbels, 2015; Suvorova et al., 2015). This is achieved by a layered centrosome structure into an inner and outer core, which can independently regulate division and budding cycles. Hence, the depletion of the transcription factor TgAP2IX-5, which acts downstream of centrosome duplication, but is required for the control of daughter cell formation can lead to T. gondii parasites harboring multiple nuclei (Khelifa et al., 2021).
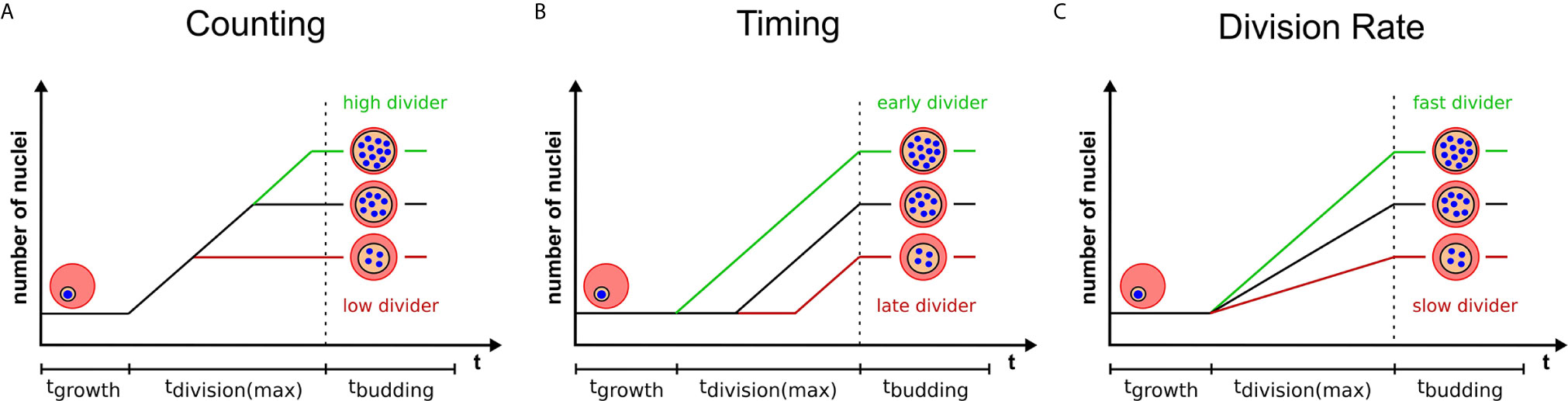
Figure 2 Three models highlighting variables that can contribute to progeny number. Variation in progeny number can be explained by changes in several cellular parameters. Relative number of nuclei are plotted against the total duration of blood stage development, which is divided in growth, division, and budding phase. The longest division time is marked, which starts with the first and ends with the last nuclear division. Time axes are neither to scale nor proportional. Three models provide a visual representation of changing a single determinant which limits the final progeny number and explain the variability within a single parasite population. Extrinsic or intrinsic factors can contribute to either an increase (green) or a decrease (red) of progeny number by (A) influencing the pre-set target number (counting), (B) initiating division at different times (timing) or (C) altering the rate of division of individual parasites.
Final Number of Merozoites Could be Regulated by Cell Cycle Timing
Alternatively to counting, nuclear number could be defined by a timing mechanism (Facchetti et al., 2017; Jones et al., 2019) i.e. the duration of the division phase (Figure 2B). Indeed, several conditions have been described that delay schizogony onset (Babbitt et al., 2012; van Biljon et al., 2018; Prior et al., 2020). Time lapse imaging shows that the length of schizogony can vary from 12 to 16 hours (Klaus et al., 2021). If time limits the number of progeny, then a linear correlation between these two factors should be observable. Although Plasmodium spp. have no conserved “clock” genes like mammalians, they seem to possess pathways that regulate cell cycle length (Reilly Ayala et al., 2010; Rijo-Ferreira et al., 2020; Smith et al., 2020). Low nutrient levels or anemic host conditions also increase duration of the entire intraerythrocytic development cycle (Babbitt et al., 2012; Birget et al., 2019) but if these factors are influencing schizogony length is still unclear. Two kinases that have been reported to be involved in regulation of cell cycle length are PfPK7 and Pfcrk-5 (Dorin-Semblat et al., 2008; Dorin-Semblat et al., 2013). Knock-out lines for both proteins, however, exhibited longer asexual cycles and fewer nuclei per schizont suggesting merozoite release to be independent of a fixed progeny number and supporting the notion of a timer. The PfPK7 phosphoproteome further suggests implication of multiple cell-cycle associated kinases that could influence timing (Pease et al., 2018). Several other kinases associated with blood stage replication, such as Crk4, a master regulator for S-phase initiation (Ganter et al., 2017), have been described (Carvalho et al., 2016; Adderley et al., 2021). For male gametogenesis, which is activated from a G0-like phase upon mosquito uptake, actual signaling cascades are investigated (Invergo et al., 2017). Calcium is a secondary messenger activating CDPK4, which acts upstream of many division processes by e.g. phosphorylating the CDK-related kinase Crk5 (Fang et al., 2017). Interfering with Crk5 function by either knock down or depletion of its associated cyclin-like protein SOC2 reduces ploidy and prevents reorganization of mitotic microtubules (Balestra et al., 2020). How additional kinase pathways influence progeny number throughout the life cycle is still under investigation (White and Suvorova, 2018).
Effect of Nuclear Division Rates
The two proposed models are very simplified in that they assume a constant nuclear division rate. But different progeny number could just as well be the result of variable division rates, while the duration of schizogony would remain equal (Figure 2C). Any combination of division rate and schizogony duration is also conceivable. If, however, counting is the dominant source of variability, parasites with increased division rates would just reach the predefined final number of nuclei earlier and no clear correlation between division rates and merozoite numbers should be observed.
Influence of Extrinsic Factors on Progeny Number
Regulation of progeny number could also be influenced by extrinsic host factors. In this case the parasite would adapt its multiplication depending on the presence of certain nutrients or stress factors. Inducing anemia in mice by phenylhydrazine treatment, prior to P. chabaudi infection led to increase of merozoite number from 6-7 to 7-8, depending on the strain (Birget et al., 2019). Sirtuins, like Sir2, are considered metabolic sensors depending on NAD+ levels to activate their histone deacetylation activity. Sir2a in P. falciparum is not only involved in antigenic variation (Merrick et al., 2010; Petter et al., 2011), but its knock-out also causes increase of ribosomal RNA levels. Surprisingly, this correlates with accelerated growth, which is likely caused by the increased number of merozoites per schizont (Mancio-Silva et al., 2013). A more direct link between nutrition and progeny number was, however, demonstrated in a study infecting calorie-restricted mice with P. berghei parasites (Mancio-Silva et al., 2017). This causes them to produce less merozoites, a phenotype that can be rescued by supplementation with Glucose. Importantly, Plasmodium requires KIN kinase to regulate merozoite number in response to host starvation demonstrating the presence of a non-canonical nutrient sensing pathway. The complete depletion of Glucose from the medium, however, induces rapid death of P. falciparum parasites (Babbitt et al., 2012). A more subtle effect is caused by starvation from the essential amino acid isoleucine. If removed prior to schizogony a dormancy-like state is induced, which can be rescued up to 72h later (McLean and Jacobs-Lorena, 2020). Coherently, isoleucine has been postulated as one of the key factors sensed by the parasite for synchronization (Prior et al., 2020). A similar reversible arrest can be induced by addition of DL-α-difluoromethylornithine, which prevents synthesis of polyamines (van Biljon et al., 2018). Another potentially limiting resource that has been discussed, especially in the context of liver stage development, are lipids and the fatty acid synthesis pathway (Tarun et al., 2009). Although the effect on progeny number has not been quantified in most of those latter studies, they, nevertheless, highlight that the parasite can adapt its cell cycle to host cues by inducing something that could be characterized as schizogony-entry checkpoint.
Correlation of Cell Size and Nuclear Number
An indirect way through which an extrinsic factor could affect nuclear number is through regulation of cell growth. Eukaryotic cells have a constant nucleo-to-cytoplasmic volume ratio (N/C-ratio) and an exception to this rule has not been found to date (Cantwell and Nurse, 2019). Our unpublished data suggest that this remains valid for P. falciparum. A simplistic model would posit that as the cytoplasmic volume increases the parasite forms more nuclei keeping the N/C-ratio constant. It is noteworthy, that blood stage parasite volume only increases by a factor of about seven (Waldecker et al., 2017), while nuclear numbers get clearly higher (Figure 1). Nevertheless, nuclear volume can be altered by chromatin compaction. Indeed, when comparing maximal intensities of a DNA stain like Hoechst, it is clearly higher in rings versus trophozoites, which have a greater cell and nuclear volume. Similarly, nuclear surface in late schizonts seems to decrease, while number of nuclei increases. More detailed analysis of the correlation between those parameters would clearly benefit from the discovery of a nuclear envelope marker for Plasmodium spp.
Conclusion
To build predictive models for malaria parasite multiplication we need to understand the dynamic relation between cycle time, cell volume and nuclear numbers. Therefore, we need to quantify those very basic cellular parameters in a time-resolved manner. The description of blood-stage development has been dominated by the useful, but limited, separation in ring, trophozoite and schizont stages. The more gradual change of key biophysical parameters, however, is best assessed by dynamic single cell analysis i.e. 4D live cell imaging, a technology that still has potential for a broader application in the malaria field (Gruring et al., 2011; De Niz et al., 2016). By verifying the dependency of merozoite number on schizogony duration we can establish whether timing plays a determinant role. This assumes that we also assess the variability in nuclear division rates. A strict dependency on the concentration of specific extrinsic factors would support a counting mechanism. In this context it will be important to verify whether progeny number generated by a parasite is an inheritable, possibly epigenetic, trait. Investigating this requires tracking progeny number from one to the next generation. Whether this number correlates with final cell size is another relevant parameter. Finally, all these parameters should be linked to parasite multiplication rates to predict how they might correlate with disease severity. Knowing that multiplication rates can increase throughout culture adaptation, we further need to assess the validity of those cellular parameters in samples that have more recently been isolated from patients (Stewart et al., 2020). Ultimately, we advocate for the systematic assessment of merozoite number in Plasmodium mutants that develop a growth phenotype, so that we can expand our list of candidate proteins implicated in defining progeny numbers and advance our understanding of malaria parasites proliferation mechanisms.
Data Availability Statement
The original contributions presented in the study are included in the article/supplementary material. Further inquiries can be directed to the corresponding author.
Author Contributions
Manuscript was written by JG with the help of VS and CS. Experimental data and Figure 1 were generated by CS and VS. Data analysis was assisted by VS and JG. Figure 2 was generated by VS with the help of JG. All authors contributed to the article and approved the submitted version.
Funding
Funded by the Human Frontiers Science Program (CDA00013/2018-C), and the German Research Foundation (DFG) (349355339) to JG.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
Severina Klaus and Markus Ganter for sharing unpublished data, and Silvia Portugal and Markus Ganter for critical reading of the manuscript. The Infectious Diseases Imaging Platform for imaging support (www.idip-heidelberg.org). Michael Lanzer for sharing P. falciparum strains and Robert Moon for P. knowlesi.
References
Adderley, J., Williamson, T., Doerig, C. (2021). Parasite and Host Erythrocyte Kinomics of Plasmodium Infection. Trends Parasitol. doi: 10.1016/j.pt.2021.01.002
Arnot, D. E., Ronander, E., Bengtsson, D. C. (2011). The Progression of the Intra-Erythrocytic Cell Cycle of Plasmodium Falciparum and the Role of the Centriolar Plaques in Asynchronous Mitotic Division During Schizogony. Int. J. Parasitol. 41, 71–80. doi: 10.1016/j.ijpara.2010.07.012
Babbitt, S. E., Altenhofen, L., Cobbold, S. a., Istvan, E. S., Fennell, C., Doerig, C., et al. (2012). Plasmodium Falciparum Responds to Amino Acid Starvation by Entering Into a Hibernatory State. Proc. Natl. Acad. Sci. U. S. A. 109, e3278–e3287. doi: 10.1073/pnas.1209823109
Balestra, A. C., Zeeshan, M., Rea, E., Pasquarello, C., Brusini, L., Mourier, T., et al. (2020). A Divergent Cyclin/Cyclin-Dependent Kinase Complex Controls the Atypical Replication of a Malaria Parasite During Gametogony and Transmission. Elife 9, 1–25. doi: 10.7554/eLife.56474
Beier, J. C. (1998). Malaria Parasite Development in Mosquitoes. Annu. Rev. Entomol. 43, 519–543. doi: 10.1146/annurev.ento.43.1.519
Birget, P. L. G., Prior, K. F., Savill, N. J., Steer, L., Reece, S. E. (2019). Plasticity and Genetic Variation in Traits Underpinning Asexual Replication of the Rodent Malaria Parasite, Plasmodium Chabaudi. Malar. J. 1, 222. doi: 10.1186/s12936-019-2857-0
Bopp, S. E., Manary, M. J., Bright, A. T., Johnston, G. L., Dharia, N. V., Luna, F. L., et al. (2013). Mitotic Evolution of Plasmodium Falciparum Shows a Stable Core Genome But Recombination in Antigen Families. PloS Genet. 9, e1003293. doi: 10.1371/journal.pgen.1003293
Brownlee, C., Heald, R. (2019). Importin α Partitioning to the Plasma Membrane Regulates Intracellular Scaling. Cell 176, 805–815.e8. doi: 10.1016/j.cell.2018.12.001
Campbell, M. A., Ganley, A. R. D., Gabaldón, T., Cox, M. P. (2016). The Case of the Missing Ancient Fungal Polyploids. Am. Nat. 188, 602–614. doi: 10.1086/688763
Cantwell, H., Nurse, P. (2019). Unravelling Nuclear Size Control. Curr. Genet. 65, 1281–1285. doi: 10.1007/s00294-019-00999-3
Carvalho, T. G., Morahan, B., John von Freyend, S., Boeuf, P., Grau, G., Garcia-Bustos, J., et al. (2016). The Ins and Outs of Phosphosignalling in Plasmodium: Parasite Regulation and Host Cell Manipulation. Mol. Biochem. Parasitol. 208, 2–15. doi: 10.1016/j.molbiopara.2016.05.006
Chen, C. T., Gubbels, M. J. (2015). Apicomplexan Cell Cycle Flexibility: Centrosome Controls the Clutch. Trends Parasitol. 31, 229–230. doi: 10.1016/j.pt.2015.04.003
Claessens, A., Hamilton, W. L., Kekre, M., Otto, T. D., Faizullabhoy, A., Rayner, J. C., et al. (2014). Generation of Antigenic Diversity in Plasmodium Falciparum by Structured Rearrangement of Var Genes During Mitosis. PloS Genet. 10, e1004812. doi: 10.1371/journal.pgen.1004812
Comai, L. (2005). The Advantages and Disadvantages of Being Polyploid. Nat. Rev. Genet. 6, 836–846. doi: 10.1038/nrg1711
Coneva, V., Chitwood, D. H. (2015). Plant Architecture Without Multicellularity: Quandaries Over Patterning and the Soma-Germline Divide in Siphonous Algae. Front. Plant Sci. 6, 287. doi: 10.3389/fpls.2015.00287
Daubenmire, R. F. (1936). The Use of the Terms Coenocyte and Syncytium in Biology. Science 84, 533. doi: 10.1126/science.84.2189.533
De Niz, M., Burda, P.-C., Kaiser, G., del Portillo, H. A., Spielmann, T., Frischknecht, F., et al. (2016). Progress in Imaging Methods: Insights Gained Into Plasmodium Biology. Nat. Rev. Microbiol. 15, 37–54. doi: 10.1038/nrmicro.2016.158
Dorin-Semblat, D., Carvalho, T. G., Nivez, M.-P., Halbert, J., Poullet, P., Semblat, J.-P., et al. (2013). An Atypical Cyclin-Dependent Kinase Controls Plasmodium Falciparum Proliferation Rate. Kinome 1, 4–16. doi: 10.2478/kinome-2013-0001
Dorin-Semblat, D., Sicard, A., Doerig, C., Ranford-Cartwright, L., Doerig, C. (2008). Disruption of the PfPK7 Gene Impairs Schizogony and Sporogony in the Human Malaria Parasite Plasmodium Falciparum. Eukaryot. Cell 7, 279–285. doi: 10.1128/EC.00245-07
Facchetti, G., Chang, F., Howard, M. (2017). Controlling Cell Size Through Sizer Mechanisms. Curr. Opin. Syst. Biol. 5, 86–92. doi: 10.1016/j.coisb.2017.08.010
Fang, H., Klages, N., Baechler, B., Hillner, E., Yu, L., Pardo, M., et al. (2017). Multiple Short Windows of Calcium-Dependent Protein Kinase 4 Activity Coordinate Distinct Cell Cycle Events During Plasmodium Gametogenesis. Elife 6, 1–23. doi: 10.7554/eLife.26524
Favery, B., Quentin, M., Jaubert-Possamai, S., Abad, P. (2016). Gall-Forming Root-Knot Nematodes Hijack Key Plant Cellular Functions to Induce Multinucleate and Hypertrophied Feeding Cells. J. Insect Physiol. 84, 60–69. doi: 10.1016/j.jinsphys.2015.07.013
Francia, M. E., Striepen, B. (2014). Cell Division in Apicomplexan Parasites. Nat. Rev. Microbiol. 12, 125–136. doi: 10.1038/nrmicro3184
Ganter, M., Goldberg, J. M., Dvorin, J. D., Paulo, J. A., King, J. G., Tripathi, A. K., et al. (2017). Plasmodium Falciparum CRK4 Directs Continuous Rounds of DNA Replication During Schizogony. Nat. Microbiol. 2, 17017. doi: 10.1038/nmicrobiol.2017.17
Garg, S., Agarwal, S., Dabral, S., Kumar, N., Sehrawat, S., Singh, S. (2015). Visualization and Quantification of Plasmodium Falciparum Intraerythrocytic Merozoites. Syst. Synth. Biol. 9, 23–26. doi: 10.1007/s11693-015-9167-9
Gruring, C., Heiber, A., Kruse, F., Ungefehr, J., Gilberger, T. W., Spielmann, T. (2011). Development and Host Cell Modifications of Plasmodium Falciparum Blood Stages in Four Dimensions. Nat. Commun. 2, 165. doi: 10.1038/ncomms1169
Gubbels, M. J., Keroack, C. D., Dangoudoubiyam, S., Worliczek, H. L., Paul, A. S., Bauwens, C., et al. (2020). Fussing About Fission: Defining Variety Among Mainstream and Exotic Apicomplexan Cell Division Modes. Front. Cell. Infect. Microbiol. 5:269. doi: 10.3389/fcimb.2020.00269
Invergo, B. M., Brochet, M., Yu, L., Choudhary, J., Beltrao, P., Billker, O. (2017). Sub-Minute Phosphoregulation of Cell Cycle Systems During Plasmodium Gamete Formation. Cell Rep. 21, 2017–2029. doi: 10.1016/j.celrep.2017.10.071
Janse, C. J., van der Klooster, P. F., van der Kaay, H. J., van der Ploeg, M., Overdulve, J. P. (1986). DNA Synthesis in Plasmodium Berghei During Asexual and Sexual Development. Mol. Biochem. Parasitol. 20, 173–182. doi: 10.1016/0166-6851(86)90029-0
Jones, A. R., Band, L. R., Murray, J. A. H. (2019). Double or Nothing? Cell Division and Cell Size Control. Trends Plant Sci. 24, 1083–1093. doi: 10.1016/j.tplants.2019.09.005
Khelifa, A. S., Guillen Sanchez, C., Lesage, K. M., Huot, L., Mouveaux, T., Pericard, P., et al. (2021). TgAP2IX-5 is a Key Transcriptional Regulator of the Asexual Cell Cycle Division in Toxoplasma Gondii. Nat. Commun. 12, 116. doi: 10.1038/s41467-020-20216-x
Klaus, S., Binder, P., Kim, J., Machado, M., Funaya, C., Schaaf, V., et al. (2021). Asynchronous Nuclear Cycles in Multinucleated Plasmodium Falciparum Enable Rapid Proliferation. bioRxiv 2021. doi: 10.1101/2021.04.15.440016 04.15.440016.
Mancio-Silva, L., Lopez-Rubio, J. J., Claes, A., Scherf, A. (2013). Sir2a Regulates rDNA Transcription and Multiplication Rate in the Human Malaria Parasite Plasmodium Falciparum. Nat. Commun. 4, 1530. doi: 10.1038/ncomms2539
Mancio-Silva, L., Slavic, K., Grilo Ruivo, M. T., Grosso, A. R., Modrzynska, K. K., Vera, I. M., et al. (2017). Nutrient Sensing Modulates Malaria Parasite Virulence. Nature 547, 213–216. doi: 10.1038/nature23009
Marguerat, S., Bähler, J. (2012). Coordinating Genome Expression With Cell Size. Trends Genet. 28, 560–565. doi: 10.1016/j.tig.2012.07.003
Marks, S. C. (1983). The Origin of Osteoclasts : Evidence, Clinial Implications and Investigative Challenges of an Extra-Skeletal Source. J. Oral. Pathol. 12, 226–256. doi: 10.1111/j.1600-0714.1983.tb00337.x
Matthews, H., Duffy, C. W., Merrick, C. J. (2018). Checks and Balances? DNA Replication and the Cell Cycle in Plasmodium. Parasites Vectors 11, 216. doi: 10.1186/s13071-018-2800-1
McLean, K. J., Jacobs-Lorena, M. (2020). The Response of Plasmodium Falciparum to Isoleucine Withdrawal is Dependent on the Stage of Progression Through the Intraerythrocytic Cell Cycle. Malar. J. 19, 147. doi: 10.1186/s12936-020-03220-w
Mela, A. P., Rico-Ramírez, A. M., Glass, N. L. (2020). Syncytia in Fungi. Cells 9, 1–14. doi: 10.3390/cells9102255
Merrick, C. J., Dzikowski, R., Imamura, H., Chuang, J., Deitsch, K., Duraisingh, M. T. (2010). The Effect of Plasmodium Falciparum Sir2a Histone Deacetylase on Clonal and Longitudinal Variation in Expression of the Var Family of Virulence Genes. Int. J. Parasitol. 40, 35–43. doi: 10.1016/j.ijpara.2009.06.012
Mi, S., Lee, X., Li, X., Veldman, G. M., Finnerty, H., Racie, L., et al. (2000). Syncytin is a Captive Retroviral Envelope Protein Involved in Human Placental Morphogenesis. Nature 403, 785–789. doi: 10.1038/35001608
Mintz, B., Baker, W. W. (1967). Normal Mammalian Muscle Differentiation and Gene Control of Isocitrate Dehydrogenase Synthesis. Proc. Natl. Acad. Sci. U. S. A. 58, 592–598. doi: 10.1073/pnas.58.2.592
Mitchison-Field, L. M. Y., Vargas-Muñiz, J. M., Stormo, B. M., Vogt, E. J. D., Van Dierdonck, S., Pelletier, J. F., et al. (2019). Unconventional Cell Division Cycles From Marine-Derived Yeasts. Curr. Biol. 29, 3439–3456.e5. doi: 10.1016/j.cub.2019.08.050
Morahan, B. J., Abrie, C., Al-Hasani, K., Batty, M. B., Corey, V., Cowell, A. N., et al. (2020). Human Aurora Kinase Inhibitor Hesperadin Reveals Epistatic Interaction Between Plasmodium Falciparum PfArk1 and PfNek1 Kinases. Commun. Biol. 3, 1–10. doi: 10.1038/s42003-020-01424-z
Ondracka, A., Dudin, O., Ruiz-Trillo, I. (2018). Decoupling of Nuclear Division Cycles and Cell Size During the Coenocytic Growth of the Ichthyosporean Sphaeroforma Arctica. Curr. Biol. 28, 1964–1969.e2. doi: 10.1016/j.cub.2018.04.074
Pease, B. N., Huttlin, E. L., Jedrychowski, M. P., Dorin-Semblat, D., Sebastiani, D., Segarra, D. T., et al. (2018). Characterization of Plasmodium Falciparum Atypical Kinase PfPK7 - Dependent Phosphoproteome. J. Proteome Res. 17, 2112–2123. doi: 10.1021/acs.jproteome.8b00062
Petter, M., Lee, C. C., Byrne, T. J., Boysen, K. E., Volz, J., Ralph, S. A., et al. (2011). Expression of P. Falciparum Var Genes Involves Exchange of the Histone Variant H2A.Z At the Promoter. PloS Pathog. 7, e1001292. doi: 10.1371/journal.ppat.1001292
Płachno, B. J., Światek, P. (2011). Syncytia in Plants: Cell Fusion in Endosperm-Placental Syncytium Formation in Utricularia (Lentibulariaceae). Protoplasma 248, 425–435. doi: 10.1007/s00709-010-0173-1
Prior, K. F., Middleton, B., Owolabi, A. T. Y., Westwood, M. L., Holland, J., O’Donnell, A. J., et al. (2020). An Essential Amino Acid Synchronises Malaria Parasite Development With Daily Host Rhythms. bioRxiv. doi: 10.1101/2020.08.24.264689
Prudêncio, M., Rodriguez, A., Mota, M. M. (2006). The Silent Path to Thousands of Merozoites: The Plasmodium Liver Stage. Nat. Rev. Microbiol. 4, 849–856. doi: 10.1038/nrmicro1529
Reilly Ayala, H. B., Wacker, M. A., Siwo, G., Ferdig, M. T. (2010). Quantitative Trait Loci Mapping Reveals Candidate Pathways Regulating Cell Cycle Duration in Plasmodium Falciparum. BMC Genomics 11:577. doi: 10.1186/1471-2164-11-577
Reilly, H. B., Wang, H., Steuter, J. A., Marx, A. M., Ferdig, M. T. (2007). Quantitative Dissection of Clone-Specific Growth Rates in Cultured Malaria Parasites. Int. J. Parasitol. 37, 1599–1607. doi: 10.1016/j.ijpara.2007.05.003
Reininger, L., Wilkes, J. M., Bourgade, H., Miranda-Saavedra, D., Doerig, C. (2011). An Essential Aurora-related Kinase Transiently Associates With Spindle Pole Bodies During Plasmodium Falciparum Erythrocytic Schizogony. Mol. Microbiol. 79, 205–221. doi: 10.1111/j.1365-2958.2010.07442.x
Rijo-Ferreira, F., Acosta-Rodriguez, V. A., Abel, J. H., Kornblum, I., Bento, I., Kilaru, G., et al. (2020). The Malaria Parasite has an Intrinsic Clock. Science 368, 746–753 doi: 10.1126/science.aba2658
Roper, M., Simonin, A., Hickey, P. C., Leeder, A., Glass, N. L. (2013). Nuclear Dynamics in a Fungal Chimera. Proc. Natl. Acad. Sci. U. S. A. 110, 12875–12880. doi: 10.1073/pnas.1220842110
Schrével, J., Asfaux-Foucher, G., Bafort, J. M. (1977). Etude Ultrastructurale Des Mitoses Multiples Au Cours De La Sporogonie Du Plasmodium B. Berghei. J. Ultrasructure Res. 59, 332–350. doi: 10.1016/S0022-5320(77)90043-0
Selmecki, A., Forche, A., Berman, J. (2010). Genomic Plasticity of the Human Fungal Pathogen Candida Albicans. Eukaryot. Cell 9, 991–1008. doi: 10.1128/EC.00060-10
Selmecki, A. M., Maruvka, Y. E., Richmond, P. A., Guillet, M., Shoresh, N., Sorenson, A. L., et al. (2015). Polyploidy can Drive Rapid Adaptation in Yeast. Nature 519, 349–352. doi: 10.1038/nature14187
Shoji, J. Y., Kikuma, T., Arioka, M., Kitamoto, K. (2010). Macroautophagy-Mediated Degradation of Whole Nuclei in the Filamentous Fungus Aspergillus Oryzae. PloS One 5, e15650. doi: 10.1371/journal.pone.0015650
Shortt, H. E., Fairley, N. H., Covell, G., Shute, P. G., Garnham, P. C. C. (1951). The Pre-Erythrocytic Stage of Plasmodium Falciparum. Trans. R. Soc Trop. Med. Hyg. 44, 405–419. doi: 10.1016/S0035-9203(51)80019-1
Simon, C. S., Voss, Y., Funaya, C., Machado, M., Penning, A., Klaschka, D., et al. (2021). An Extended DNA-free Intranuclear Compartment Organizes Centrosomal Microtubules in Plasmodium Falciparum. bioRxiv 2021. doi: 10.1101/2021.03.12.435157 03.12.435157.
Sinden, R. E., Canning, E. U., Bray, R. S., Smalley, M. E. (1978). Gametocyte and Gamete Development in Plasmodium Falciparum. Proc. R. Soc. London - Biol. Sci. 201, 375–399. doi: 10.1098/rspb.1978.0051
Sinden, R. E., Talman, A., Marques, S. R., Wass, M. N., Sternberg, M. J. E. (2010). The Flagellum in Malarial Parasites. Curr. Opin. Microbiol. 13, 491–500. doi: 10.1016/j.mib.2010.05.016
Smith, L. M., Motta, F. C., Chopra, G., Moch, J. K., Nerem, R. R., Cummins, B., et al. (2020). An Intrinsic Oscillator Drives the Blood Stage Cycle of the Malaria Parasite Plasmodium Falciparum. Science 368, 754–759. doi: 10.1126/science.aba4357
Spreng, B., Fleckenstein, H., Kübler, P., Di Biagio, C., Benz, M., Patra, P., et al. (2019). Microtubule Number and Length Determine Cellular Shape and Function in Plasmodium. EMBO J. 38, e100984. doi: 10.15252/embj.2018100984
Steenwyk, J. L., Opulente, D. A., Kominek, J., Shen, X. X., Zhou, X., Labella, A. L., et al. (2019). Extensive Loss of Cell-Cycle and DNA Repair Genes in an Ancient Lineage of Bipolar Budding Yeasts. PloS Biol. 17, e3000255. doi: 10.1371/journal.pbio.3000255
Stewart, L. B., Diaz-Ingelmo, O., Claessens, A., Abugri, J., Pearson, R. D., Goncalves, S., et al. (2020). Intrinsic Multiplication Rate Variation and Plasticity of Human Blood Stage Malaria Parasites. Commun. Biol. 3, 624 doi: 10.1038/s42003-020-01349-7
Sturm, A., Amino, R., Van De Sand, C., Regen, T., Retzlaff, S., Rennenberg, A., et al. (2006). Manipulation of Host Hepatocytes by the Malaria Parasite for Delivery Into Liver Sinusoids. Science 313, 1287–1290. doi: 10.1126/science.1129720
Suvorova, E. S., Francia, M., Striepen, B., White, M. W. (2015). A Novel Bipartite Centrosome Coordinates the Apicomplexan Cell Cycle. PloS Biol. 13, e1002093. doi: 10.1371/journal.pbio.1002093
Tarun, A. S., Vaughan, A. M., Kappe, S. H. I. (2009). Redefining the Role of De Novo Fatty Acid Synthesis in Plasmodium Parasites. Trends Parasitol. 25, 545–550 doi: 10.1016/j.pt.2009.09.002
van Biljon, R., Niemand, J., van Wyk, R., Clark, K., Verlinden, B., Abrie, C., et al. (2018). Inducing Controlled Cell Cycle Arrest and Re-Entry During Asexual Proliferation of Plasmodium Falciparum Malaria Parasites. Sci. Rep. 8, 16581 doi: 10.1038/s41598-018-34964-w
Vaughan, J. A. (2007). Population Dynamics of Plasmodium Sporogony. Trends Parasitol. 23, 63–70. doi: 10.1016/j.pt.2006.12.009
Waldecker, M., Dasanna, A. K., Lansche, C., Linke, M., Srismith, S., Cyrklaff, M., et al. (2017). Differential Time-Dependent Volumetric and Surface Area Changes and Delayed Induction of New Permeation Pathways in P. Falciparum-Infected Hemoglobinopathic Erythrocytes. Cell. Microbiol. 19, e1265 doi: 10.1111/cmi.12650
White, M. W., Suvorova, E. S. (2018). Apicomplexa Cell Cycles: Something Old, Borrowed, Lost, and New. Trends Parasitol. 34, 759–771. doi: 10.1016/j.pt.2018.07.006
Keywords: malaria, Plasmodium, fungi, cell division, multinucleated, mitosis, polyploidy
Citation: Simon CS, Stürmer VS and Guizetti J (2021) How Many Is Enough? - Challenges of Multinucleated Cell Division in Malaria Parasites. Front. Cell. Infect. Microbiol. 11:658616. doi: 10.3389/fcimb.2021.658616
Received: 26 January 2021; Accepted: 21 April 2021;
Published: 07 May 2021.
Edited by:
Mathieu Brochet, University of Geneva, SwitzerlandReviewed by:
Lyn-Marie Birkholtz, University of Pretoria, South AfricaMathieu Gissot, Centre National de la Recherche Scientifique (CNRS), France
Copyright © 2021 Simon, Stürmer and Guizetti. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Julien Guizetti, anVsaWVuLmd1aXpldHRpQG1lZC51bmktaGVpZGVsYmVyZy5kZQ==
 Caroline S. Simon
Caroline S. Simon Vanessa S. Stürmer
Vanessa S. Stürmer Julien Guizetti
Julien Guizetti