
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
MINI REVIEW article
Front. Cell. Infect. Microbiol. , 09 April 2021
Sec. Virus and Host
Volume 11 - 2021 | https://doi.org/10.3389/fcimb.2021.648055
This article is part of the Research Topic Molecular and Cellular Interactions between the Host and Herpesviruses View all 17 articles
Viruses are known for their ability to alter host gene expression. Kaposi sarcoma-associated herpesvirus has two proteins that obstruct host gene expression. KSHV SOX, encoded by the open reading frame 37 (ORF37), induces a widespread cytoplasmic mRNA degradation and a block on mRNA nuclear export. The other KSHV protein, encoded by the open reading frame 10 (ORF10), was recently identified to inhibit host gene expression through its direct function on the cellular mRNA export pathway. In this review, we summarize the studies on both SOX and ORF10 in efforts to elucidate their mechanisms. We also discuss how the findings based on a closely related rodent virus, murine gammaherpesvirus-68 (MHV-68), complement the KSHV findings to decipher the role of these two proteins in viral pathogenesis.
Kaposi sarcoma-associated herpesvirus (KSHV), or the human herpesvirus-8 (HHV-8), belongs to the gamma subfamily of herpesviruses. KSHV is associated with malignancies and life-threatening diseases (Chang et al., 1994; Soulier et al., 1995; Brambilla et al., 1996; Cesarman et al., 1996; Corbellino et al., 1996a; Corbellino et al., 1996b; Dittmer and Damania, 2013; Goncalves et al., 2017), including Kaposi sarcoma, primary effusion lymphoma, multicentric Castleman’s disease, and KSHV inflammatory cytokine syndrome. KSHV has two distinct phases in its life cycle: latency and lytic replication. Latency is considered to be immunologically silent with few viral genes expressed and no virion production. During lytic replication, over 80 viral genes are expressed in a cascading order, resulting in the assembly and release of infectious particles, generating new infections to replenish the pool of latently infected cells.
Restricted host range limits most KSHV studies to molecular biology experiments. Murine gammaherpesvirus-68 (MHV-68) is closely related to human gammaherpesviruses (Virgin et al., 1997; Barton et al., 2011). Like KSHV, MHV-68 infection in mice leads to acute lytic viral replication followed by the establishment of latency in B-cells. 63 out of the 80 MHV-68 open reading frames (ORFs) share sequence homology with KSHV ORFs with 10-60% shared sequence identity (Virgin et al., 1997). Unlike KSHV, MHV-68 de novo infection readily leads to lytic replication. Therefore, MHV-68 provides a valuable model for in vitro and in vivo functional studies of KSHV viral protein homologs.
Viruses strategically exploit various cellular mechanisms to down-regulate host gene expression for their own benefit. The best studied KSHV protein to inhibit host gene expression is the shutoff and exonuclease (SOX) protein encoded by KSHV ORF37 (Glaunsinger and Ganem, 2004), which accelerates cytoplasmic mRNA degradation. Another common viral strategy, frequently employed by RNA viruses, targets mRNA nuclear export to inhibit host gene expression (Yarbrough et al., 2014). Most RNA viruses synthesize their mRNAs in the cytoplasm and do not depend on cellular mRNA nuclear export mechanism. In contrast, DNA viruses, including herpesviruses, risk hindering the exit of their own mRNAs if they block mRNA nuclear export. Nevertheless, SOX induces mRNA hyperadenylations and their nuclear retention due to global mRNA degradation in the cytoplasm (Kumar and Glaunsinger, 2010). Moreover, we identified KSHV ORF10 as another inhibitor of host gene expression by interacting with a cellular mRNA export factor, Rae1. We review the current knowledge regarding the molecular mechanisms of SOX and ORF10 as well as their roles in the context of viral lytic replication and pathogenesis (Figure 1).
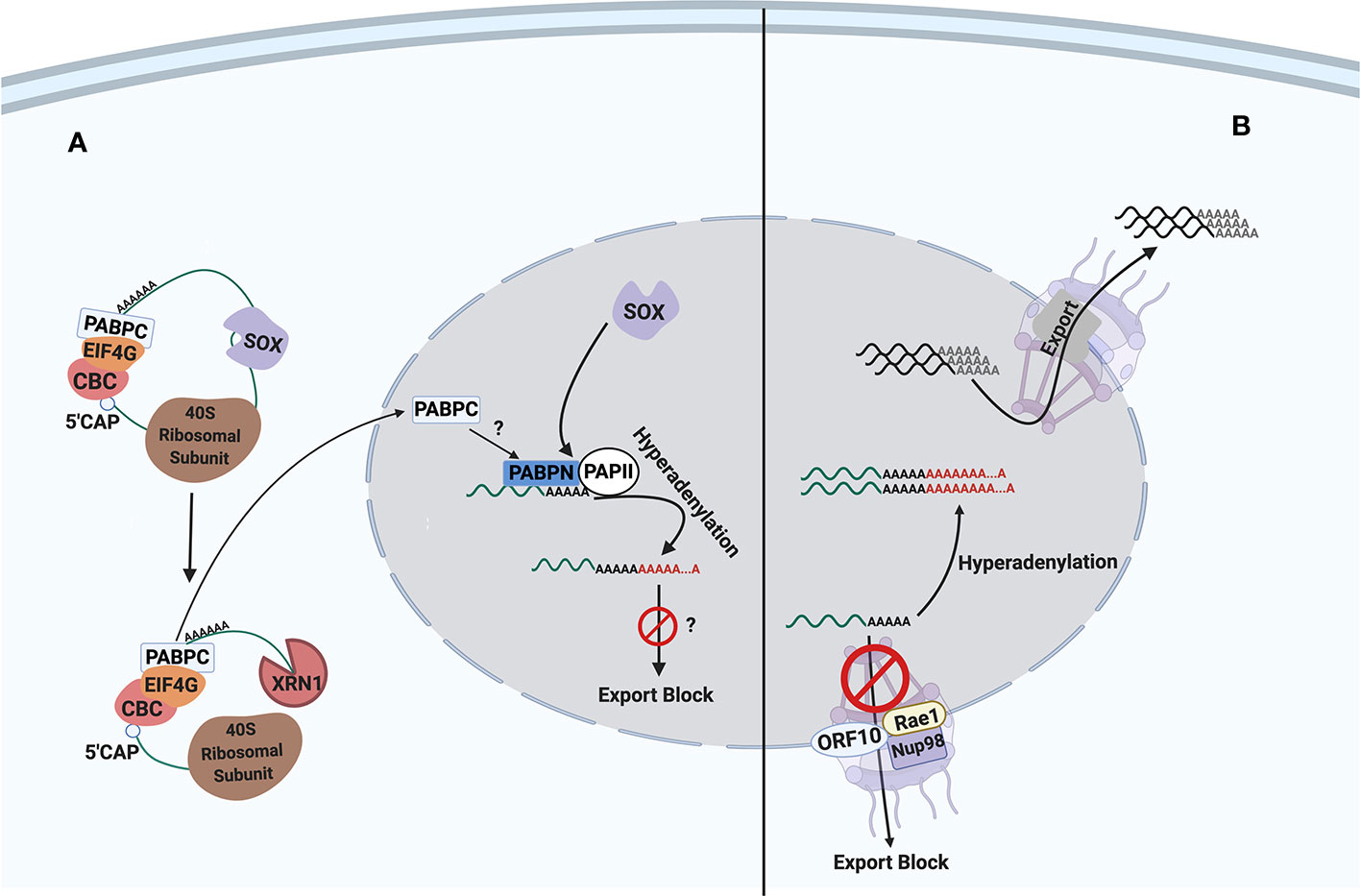
Figure 1 The inhibitory mechanisms of SOX and ORF10 proteins on host gene expression. SOX protein targets transcripts for degradation, affecting both the host and viral transcripts. On the other hand, ORF10 acts solely on cellular transcripts for export inhibition. Interestingly, both SOX and ORF10 cause the hyperadenylation of transcripts within the nucleus. (A) Cytoplasmic SOX protein, as encoded by ORF37, causes host shut off through its endonuclease activity. The viral endonucleolytic action on host transcript then triggers the cellular exonuclease, Xrn1, to complete transcript degradation. (B) At the nuclear envelope, ORF10 inhibits only the export of host transcripts by forming a complex with cellular export factor Rae1 and its partnering nucleoporin, Nup98. The hijacking of Rae1-Nup98 leads to nuclear retention of mRNAs. CBC, cap-binding complex; PABPC, cytoplasmic poly(A)-binding protein; PABPN, nuclear poly(A)-binding protein; PAPII, poly(A) polymerase II. Question marks indicate potential or hypothetical mechanisms and effects within the model.
Herpesviruses undermine mRNA stability through proteins that possess an endoribonuclease activity (Read, 2013). These include the virion host shutoff (vhs) protein encoded by HSV-1 UL41 (Fenwick and McMenamin, 1984; Strom and Frenkel, 1987; Elgadi et al., 1999) and KSHV SOX (Glaunsinger and Ganem, 2004). Unlike vhs, SOX originates from the herpesviral alkaline exonuclease protein family involved in viral DNA genome processing. Only SOX homologs of the gamma subfamily evolved a separate endonuclease activity causing host shutoff (Glaunsinger and Ganem, 2004; Rowe et al., 2007; Covarrubias et al., 2009). The alkaline exonuclease and host shutoff functions of SOX are genetically separable with specific mutations affecting only one of the two functions (Glaunsinger et al., 2005).
SOX causes host shutoff through a sequential action with a cellular 5’-3’ exonuclease, Xrn1 (PACMAN), resulting in cytoplasmic transcript degradation (Covarrubias et al., 2011). The current working model is that SOX internally cleaves the target transcripts in a non-random manner with preferred sites defined by a degenerate sequence motif (Gaglia et al., 2015; Mendez et al., 2018), providing Xrn1 with an entry point to complete degradation. In addition to site preferences, co-sedimentation of SOX with 40S ribosomal subunits indicates that SOX targets transcripts early during translation, further corroborated by SOX’s inability to initiate degradation of translation-incompetent reporters or RNA Pol I and Pol III transcripts (Covarrubias et al., 2011).
Remarkably, SOX targets both cellular and viral mRNAs (Abernathy et al., 2014). The MHV-68 SOX homolog shares the same host shutoff activity as KSHV SOX (Covarrubias et al., 2009). When the shutoff function of SOX is disabled in MHV-68, the majority of MHV-68 genes have higher mRNA levels (Richner et al., 2011). Not all MHV-68 genes with increased transcript levels have higher protein expression. However, this altered viral protein expression unexpectedly impacts the composition of virions produced by the SOX mutant without changing the ratios of particles to plaque forming units. In addition, the resulting changed virion composition affected viral entry. Despite enhanced viral gene expression, the SOX MHV-68 mutant is attenuated in vitro and in vivo. This is a surprising finding since host shutoff is generally thought to help viruses seize cellular resources to maximize their own protein expression for the highest virion production. Instead, SOX controls the viral gene expression to produce virions with a proper composition for optimal viral replication.
As many RNA-binding proteins are capable of nucleo-cytoplasmic shuttling, a SOX-mediated massive decrease in cytoplasmic mRNA can lead to the nuclear relocation of cytoplasmic RNA-binding proteins (Gilbertson et al., 2018). PABPC, a predominantly cytoplasmic poly(A) binding protein, is one such nucleo-cytoplasmic shuttling protein (Afonina et al., 1998). It interacts with the cap-binding complex, eIF4F, through subunit eIF4G, to form a closed loop structure (Tarun and Sachs, 1996; Wells et al., 1998), which promotes the recruitment of ribosome 40S subunit for translation. Upon RNA binding, the PABPC nuclear import signal is masked, and thus when cytoplasmic polyadenylated transcripts drop substantially due to SOX activity, RNA-free PABPC is imported into the nucleus (Kumar et al., 2011). A SOX mutant that cannot accelerate mRNA turnover also fails in PABPC1 nuclear relocalization (Lee and Glaunsinger, 2009). Moreover, overexpression of a cytoplasmic deadenylase to reduce the abundance of cytoplasmic poly(A) RNA also induces nuclear import of PABPC1 (Lee and Glaunsinger, 2009). PABPC nuclear relocalization by SOX is observed during KSHV lytic replication (Lee and Glaunsinger, 2009). HSV-1 vhs can also cause nuclear accumulation of PABPC1 (Kumar and Glaunsinger, 2010), indicating a common impact in cytoplasmic mRNA drop. The major consequences of nuclear relocalization of PABPC1, by SOX, vhs, or the fusion with a nuclear retention signal (NRS) to PABPC1, are hyperadenylation of mRNAs and inhibition on their nuclear export (Kumar and Glaunsinger, 2010). Notably, nuclear PABPC1-NRS does not destabilize GFP reporter mRNA but diminishes GFP protein expression (Kumar and Glaunsinger, 2010), perhaps by retaining the GFP transcript in the nucleus. This indicates that in addition to cytoplasmic mRNA degradation, the impact of ORF37 on nuclear export of mRNA due to PABPC nuclear translocation is a critical part of its host shutoff function.
Nuclear mRNA hyperadenylation has been observed when RNAs are not exported (Hilleren and Parker, 2001; Jensen et al., 2001; Hammell et al., 2002; Qu et al., 2009; Bresson and Conrad, 2013). Since hyperadenylation and nuclear export are intimately linked, it is difficult to determine whether a block on export or hyperadenylation occurs first upon the PABPC nuclear relocation. SOX-induced hyperadenylation depends on PABPC1 and poly(A) polymerase, PAPII (PAPα) (Lee and Glaunsinger, 2009). The involvement of cleavage/polyadenylation CPA machinery is assumed. CPA is carried out by a large complex consisting of multi-subunit of cleavage and polyadenylation specificity factor (CPSF) and PAP that adds adenosines to the cleavage fragment (Proudfoot, 1996). Subsequently, nuclear poly(A)-binding protein (PABPN) associates with the newly-added poly(A) tail and stimulates PAPII to produce a long poly(A) tail of ~250 nucleotides (Bienroth et al., 1993; Kühn et al., 2009; Kühn et al., 2017). It has been proposed that additional adenosines beyond 250 residues cannot support a productive CPSF-PAPII complex for efficient polyadenylation (Wahle, 1995; Kühn et al., 2009). PABPN and CPSF contribute to the recruitment of nuclear export complex to mRNA (Shi et al., 2017). The loss of PABPN can lead to a shortened poly(A) tail and mRNA nuclear retention (Apponi et al., 2010). It is possible that relocating nuclear PABPC1 replaces PABPN on the poly(A) tail of mRNAs and prevents RNA export factor recruitment, interrupting mRNA nuclear export and causing subsequent hyperadenylation.
Despite a predominant nuclear SOX presence, cytoplasmic SOX largely mediates the effects of accelerated cytoplasmic mRNA decay, relocalization of PABC, and nuclear mRNA hyperadenylation (Covarrubias et al., 2009). SOX degrades cytoplasmic RNA, causing the PABPC1 nuclear import, which causes nuclear mRNA hyperadenylation and nuclear retention. Interestingly, the SOX impact on host gene expression in uninfected cells is less substantial compared to the extent of host shut-off during KSHV lytic replication (Clyde and Glaunsinger, 2011). This suggests that KSHV employs additional mechanisms beyond SOX to inhibit host gene expression, as other herpesviruses do (Rivas et al., 2016).
Mature mRNA are associated with a variety of proteins to form export-competent messenger ribonucleoprotein (mRNP) complexes (Carmody and Wente, 2009; Björk and Wieslander, 2017). Export of mRNPs requires the transit of these mRNPs through the nucleopore complex (NPC). An NPC consists of ~30 nucleoporins (Nups), with many containing phenylalanine-glycine (FG) repeats (Strambio-De-Castillia et al., 2010). At the central channel of nuclear pores, the FG-repeats of Nups form a permeability barrier (Terry and Wente, 2009). Majority of mRNA nuclear export through the NPCs involves the TAP-p15 (or NXF1-NXT1) heterodimer (Conti and Izaurralde, 2001; Köhler and Hurt, 2007; Carmody and Wente, 2009). The TAP-p15 heterodimers load mRNPs onto FG-containing Nups (FG-Nups) (Katahira et al., 1999; Bachi et al., 2000). The interaction of TAP with the FG repeats overcomes the permeability barrier of the nuclear pore, enabling mRNP translocation across the central channel (Grünwald et al., 2011; Powers and Forbes, 2012).
Our work identified a novel KSHV post-transcriptional regulator, encoded by ORF10, as an inhibitor of mRNA nuclear export (Gong et al., 2016). ORF10 expression leads to the reduction of cytoplasmic RNA levels for 24% of cellular genes, indicating a role of ORF10 in suppressing host gene expression during KSHV replication. While both ORF37 and ORF10 cause mRNA nuclear retention, ORF10 directly targets the nuclear export pathway by interacting with Rae1. We and others have independently identified, through mass spectrometry, that KSHV and MHV-68 ORF10 interacts with Rae1 (Davis et al., 2015; Gong et al., 2016), which is a highly conserved eukaryotic cellular export factor (Bharathi et al., 1997; Kraemer and Blobel, 1997; Sabri and Visa, 2000). Rae1 is involved in mRNA export by interacting with Nup98 at NPC (Pritchard et al., 1999; Blevins et al., 2003). We engineered a recombinant MHV-68 expressing FLAG-tagged ORF10 and identified Rae1 and Nup98 as ORF10-interacting proteins in the context of infection (Gong et al., 2016). Unlike TAP, Rae1 does not interact with the FG repeats of Nup98. Instead, the interaction is mediated through an evolutionarily conserved sequence within the Nup116/Nup98 family, referred to as Gle2-binding sequence (GLEBS) (Bailer et al., 1998; Pritchard et al., 1999). Rae1 also interacts with TAP (Bachi et al., 2000; Blevins et al., 2003) and thus, rather than functioning in the transit of mRNPs through NPCs, Rae1 potentially facilitates the docking of export-competent mRNPs onto NPCs. KSHV and MHV-68 ORF10s share 19% amino acid identity, highlighting the functional importance of Rae1 interaction during gammaherpesvirus infection. A structural study determined the MHV-68 ORF10 residues involved in the interactions with the Rae1-Nup98 complex (Feng et al., 2020). Some residues are highly conserved across the gammaherpesvirus ORF10 homologs with their mutations impairing interaction with the Rae1-Nup98 complex, causing the loss of mRNA nuclear export inhibition.
We have shown that KSHV and MHV-68 ORF10 induce nuclear accumulation of poly(A) RNA, which is abolished by mutations that disrupt ORF10-Rae1 interaction or Rae1 knockdown. Additionally, ORF10 is enriched at the nuclear rim. This localization depends on Rae1 and Nup98. Our current working model is that ORF10 interacts with Rae1, which in turn interacts with Nup98, to interfere with the Rae1-Nup98 complex function in mRNA export. While global mRNA export is not impacted by the absence of Rae1 (Babu et al., 2003), expression of the GLEBS domain of Nup98 induces nuclear accumulation of poly(A) RNA (Pritchard et al., 1999). The GLEBS domain sequesters Rae1 from binding to the wild type Nup98 at NPCs. ORF10 does not disrupt the interaction between Rae1 and Nup98; instead, it undermines the function of the complex. Both studies support a role of Rae1 in mRNA export, but the precise mechanism remains unknown. Cell fractionation combined with RNA sequencing indicates that the ORF10 impact on mRNA export is not global. While a strong correlation was found between RNA abundance and SOX-mediated degradation (Gilbertson et al., 2018), such correlation is not seen for ORF10-mediated export inhibition. It is possible that the Rae1-Nup98 complex is utilized by a subset of mRNAs for rapid exit of the nucleus. Without Rae1, this subset of mRNAs can still exit the nucleus albeit less efficiently, which can only be observed with careful kinetics studies.
Unlike SOX, we did not find viral transcript inhibition by ORF10. Herpesviral lytic genes are expressed in a cascade order and classified as immediate early, early and late genes. RNA sequencing showed that in the absence of ORF10, the transcription of late genes during KSHV lytic replication was most impacted, resulting in reduced virion production (Gong et al., 2016). This phenotype was recapitulated by the null function ORF10 mutant lacking Rae1 interaction or by Rae1 expression knockdown during KSHV lytic replication. These results indicate that ORF10 facilitates efficient viral late gene expression through its Rae1 interaction. Nevertheless, how the inhibition on nuclear mRNA export by ORF10 affects viral gene transcription requires further investigations.
Gammaherpesviral ORF10 is not the only viral protein targeting Rae1-Nup98. Vesicular stomatitis virus (VSV) and influenza A virus (IAV) encode proteins that interact with Rae1 and inhibit mRNA export (Faria et al., 2005; Satterly et al., 2007), underscoring the importance of Rae1-Nup98 in viral replication. Recently, ORF6 encoded by severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2) was found to interact with Rae1 (Gordon et al., 2020; Li et al., 2020), although the functional consequence is unknown (Miorin et al., 2020). A structural study suggests that the matrix (M) protein of VSV prevents Rae1 from binding to the RNA phosphate backbone, blocking Rae1 function in mRNA export (Quan et al., 2014). MHV-68 ORF10 also binds to a similar interface between Rae1-Nup98 as VSV M (Feng et al., 2020), highlighting a vulnerable structural aspect for viral exploitation. The C-terminal tail of MHV-68 ORF10 interacts with the RNA-binding groove of Rae1-Nup98 but on a different side from the M protein target. The binding of VSV M or gammaherpesviral ORF10 is expected to disrupt the RNA-binding ability of Rae1-Nup98. This prediction is true for the M-Rae1-Nup98 complex but not the ORF10-Rae1-Nup98 complex (Feng et al., 2020). It appears that ORF10 provides an alternative RNA-binding surface for the ORF10-Rae1-Nup98 complex. The IAV NS1 protein forms a complex with Rae1 and Tap-p15 heterodimer (Satterly et al., 2007) but currently no structural information is available on its interaction with Rae1. Embryonic fibroblasts from heterozygous knockout mice of Rae1 (Rae-/+) or Nup98 (Nup98-/+) or both (Rae-/+Nup98-/+) are more susceptible to IAV-induced cell death but produced more virions than the wild type cells (Satterly et al., 2007). This differs from our Rae1 knockdown results that show Rae1 requirement for efficient KSHV lytic replication (Gong et al., 2016). The likely explanation is that nuclear export of cellular genes is impacted differently by the Rae1-Nup98 complex with a subset of genes more dependent than others. mRNAs of several immune-related genes have higher nuclear to cytoplasmic ratios in Nup98-/+ and Rae-/+Nup98-/+ cells. Moreover, Nup98 is upregulated by interferons, and treatment by interferons can overcome VSV M-mediated inhibition on mRNA export (Enninga et al., 2002). Certain cellular genes may require the Rae1-Nup98 complex for exporting their mRNAs into the cytoplasm. Some of these Rae1-dependent genes may encode antiviral proteins, which accounts for the increased replication of IAV in the absence of fully functional Rae1-Nup98. However, there could also be cellular mRNAs that do not need Rae1 for export but recruit it for rapid export under special conditions, such as stress (Izawa et al., 2004). Through Rae1 interaction, ORF10 gains access to Rae1-regulated mRNAs and inhibits their export to promote viral replication.
KSHV encodes SOX and ORF10, known to inhibit host gene expression through distinct molecular mechanisms. SOX targets viral transcript for degradation, resulting in reduced viral protein expression, maintaining virion production with balanced composition (Abernathy et al., 2014). In contrast, ORF10 does not seem to impact nuclear export of viral mRNAs but is required for efficient expression of viral proteins and virion production (Gong et al., 2016). While SOX and ORF10 are capable of inhibiting host gene expression, their functions during gammaherpesvirus infection do not overlap. Moreover, SOX and ORF10 have different timings of expression during KSHV lytic replication; the former is an early gene and the latter is a late gene (Arias et al., 2014). Does ORF37 coordinate with ORF10 to down-regulate host gene expression for optimal viral replication or does their inhibition on mRNA export serve different purposes? Investigating the roles of ORF10 and ORF37 in the context of infection combined with cell fractionation and RNA sequencing will provide insight into their impact on host gene expression. Additionally, the impact of these two viral genes on the host proteome remained to be determined. Due to the functional conservation of ORF10 and ORF37 in KSHV and MHV-68, a combination of in vitro molecular biology and in vivo infection model with MHV-68 will certainly provide a comprehensive overview of their functions in viral pathogenesis. ORF10 interacts with Rae1 to achieve export inhibition, yet Rae1 functions in RNA export is still largely unclear. Therefore, ORF10 also serves as a valuable tool to understand this cellular pathway that is targeted by multiple viruses.
CP and T-TW wrote the manuscript and prepared the figure. All authors contributed to the article and approved the submitted version.
This work was funded by NIH National Institute of Dental and Craniofacial Research, grant number R01DE028774.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The authors express their gratitude to Drs. Guillaume Chanfreau, Samson Chow, and Christel Uittenbogaart for their critical inputs during the preparation of the manuscript.
Abernathy E., Clyde K., Yeasmin R., Krug L. T., Burlingame A., Coscoy L., et al. (2014). Gammaherpesviral gene expression and virion composition are broadly controlled by accelerated mRNA degradation. PloS Pathog. 10, e1003882. doi: 10.1371/journal.ppat.1003882
Afonina E., Stauber R., Pavlakis G. N. (1998). The human poly(A)-binding protein 1 shuttles between the nucleus and the cytoplasm. J. Biol. Chem. 273, 13015–13021. doi: 10.1074/jbc.273.21.13015
Apponi L. H., Leung S. W., Williams K. R., Valentini S. R., Corbett A. H., Pavlath G. K. (2010). Loss of nuclear poly(A)-binding protein 1 causes defects in myogenesis and mRNA biogenesis. Hum. Mol. Genet. 19, 1058–1065. doi: 10.1093/hmg/ddp569
Arias C., Weisburd B., Stern-Ginossar N., Mercier A., Madrid A. S., Bellare P., et al. (2014). KSHV 2.0: a comprehensive annotation of the Kaposi’s sarcoma-associated herpesvirus genome using next-generation sequencing reveals novel genomic and functional features. PloS Pathog. 10, e1003847. doi: 10.1371/journal.ppat.1003847
Babu J. R., Jeganathan K. B., Baker D. J., Wu X., Kang-Decker N., van Deursen J. M. (2003). Rae1 is an essential mitotic checkpoint regulator that cooperates with Bub3 to prevent chromosome missegregation. J. Cell Biol. 160, 341–353. doi: 10.1083/jcb.200211048
Bachi A., Braun I. C., Rodrigues J. P., Panté N., Ribbeck K., von Kobbe C., et al. (2000). The C-terminal domain of TAP interacts with the nuclear pore complex and promotes export of specific CTE-bearing RNA substrates. RNA 6, 136–158. doi: 10.1017/s1355838200991994
Bailer S. M., Siniossoglou S., Podtelejnikov A., Hellwig A., Mann M., Hurt E. (1998). Nup116p and nup100p are interchangeable through a conserved motif which constitutes a docking site for the mRNA transport factor gle2p. EMBO J. 17, 1107–1119. doi: 10.1093/emboj/17.4.1107
Barton E., Mandal P., Speck S. H. (2011). Pathogenesis and host control of gammaherpesviruses: lessons from the mouse. Annu. Rev. Immunol. 29, 351–397. doi: 10.1146/annurev-immunol-072710-081639
Bharathi A., Ghosh A., Whalen W. A., Yoon J. H., Pu R., Dasso M., et al. (1997). The human RAE1 gene is a functional homologue of Schizosaccharomyces pombe rae1 gene involved in nuclear export of Poly(A)+ RNA. Gene 198, 251–258. doi: 10.1016/s0378-1119(97)00322-3
Bienroth S., Keller W., Wahle E. (1993). Assembly of a processive messenger RNA polyadenylation complex. EMBO J. 12, 585–594. doi: 10.1002/j.1460-2075.1993.tb05690.x
Björk P., Wieslander L. (2017). Integration of mRNP formation and export. Cell. Mol. Life Sci. 74, 2875–2897. doi: 10.1007/s00018-017-2503-3
Blevins M. B., Smith A. M., Phillips E. M., Powers M. A. (2003). Complex formation among the RNA export proteins Nup98, Rae1/Gle2, and TAP. J. Biol. Chem. 278, 20979–20988. doi: 10.1074/jbc.M302061200
Brambilla L., Boneschi V., Berti E., Corbellino M., Parravicini C. (1996) HHV8 cell-associated viraemia and clinical presentation of Mediterranean Kaposi’s sarcoma. Lancet 347, 1338. doi: 10.1016/s0140-6736(96)90989-7
Bresson S. M., Conrad N. K. (2013). The human nuclear poly(a)-binding protein promotes RNA hyperadenylation and decay. PloS Genet. 9, e1003893. doi: 10.1371/journal.pgen.1003893
Carmody S. R., Wente S. R. (2009). mRNA nuclear export at a glance. J. Cell Sci. 122, 1933–1937. doi: 10.1242/jcs.041236
Cesarman E., Nador R. G., Aozasa K., Delsol G., Said J. W., Knowles D. M. (1996). Kaposi’s sarcoma-associated herpesvirus in non-AIDS related lymphomas occurring in body cavities. Am. J. Pathol. 149, 53–57.
Chang Y., Cesarman E., Pessin M. S., Lee F., Culpepper J., Knowles D. M., et al. (1994). Identification of herpesvirus-like DNA sequences in AIDS-associated Kaposi’s sarcoma. Science 266, 1865–1869. doi: 10.1126/science.7997879
Clyde K., Glaunsinger B. A. (2011). Deep sequencing reveals direct targets of gammaherpesvirus-induced mRNA decay and suggests that multiple mechanisms govern cellular transcript escape. PloS One 6, e19655. doi: 10.1371/journal.pone.0019655
Conti E., Izaurralde E. (2001). Nucleocytoplasmic transport enters the atomic age. Curr. Opin. Cell Biol. 13, 310–319. doi: 10.1016/s0955-0674(00)00213-1
Corbellino M., Poirel L., Aubin J. T., Paulli M., Magrini U., Bestetti G., et al. (1996a). The role of human herpesvirus 8 and Epstein-Barr virus in the pathogenesis of giant lymph node hyperplasia (Castleman’s disease). Clin. Infect. Dis. 22, 1120–1121. doi: 10.1093/clinids/22.6.1120
Corbellino M., Poirel L., Bestetti G., Pizzuto M., Aubin J. T., Capra M., et al. (1996b). Restricted tissue distribution of extralesional Kaposi’s sarcoma-associated herpesvirus-like DNA sequences in AIDS patients with Kaposi’s sarcoma. AIDS Res. Hum. Retroviruses 12, 651–657. doi: 10.1089/aid.1996.12.651
Covarrubias S., Richner J. M., Clyde K., Lee Y. J., Glaunsinger B. A. (2009). Host shutoff is a conserved phenotype of gammaherpesvirus infection and is orchestrated exclusively from the cytoplasm. J. Virol. 83, 9554–9566. doi: 10.1128/JVI.01051-09
Covarrubias S., Gaglia M. M., Kumar G. R., Wong W., Jackson A. O., Glaunsinger B. A. (2011). Coordinated destruction of cellular messages in translation complexes by the gammaherpesvirus host shutoff factor and the mammalian exonuclease Xrn1. PloS Pathog. 7, e1002339. doi: 10.1371/journal.ppat.1002339
Davis Z. H., Verschueren E., Jang G. M., Kleffman K., Johnson J. R., Park J., et al. (2015). Global mapping of herpesvirus-host protein complexes reveals a transcription strategy for late genes. Mol. Cell 57, 349–360. doi: 10.1016/j.molcel.2014.11.026
Dittmer D. P., Damania B. (2013). Kaposi sarcoma associated herpesvirus pathogenesis (KSHV)–an update. Curr. Opin. Virol. 3, 238–244. doi: 10.1016/j.coviro.2013.05.012
Elgadi M. M., Hayes C. E., Smiley J. R. (1999). The herpes simplex virus vhs protein induces endoribonucleolytic cleavage of target RNAs in cell extracts. J. Virol. 73, 7153–7164. doi: 10.1128/JVI.73.9.7153-7164.1999
Enninga J., Levy D. E., Blobel G., Fontoura B. M. A. (2002). Role of nucleoporin induction in releasing an mRNA nuclear export block. Science 295, 1523–1525. doi: 10.1126/science.1067861
Faria P. A., Chakraborty P., Levay A., Barber G. N., Ezelle H. J., Enninga J., et al. (2005). VSV disrupts the Rae1/mrnp41 mRNA nuclear export pathway. Mol. Cell 17, 93–102. doi: 10.1016/j.molcel.2004.11.023
Feng H., Tian H., Wang Y., Zhang Q., Lin N., Liu S., et al. (2020). Molecular mechanism underlying selective inhibition of mRNA nuclear export by herpesvirus protein ORF10. Proc. Natl. Acad. Sci. U. S. A. 117, 26719–26727. doi: 10.1073/pnas.2007774117
Fenwick M. L., McMenamin M. M. (1984). Early virion-associated suppression of cellular protein synthesis by herpes simplex virus is accompanied by inactivation of mRNA. J. Gen. Virol. 65 ( Pt 7), 1225–1228. doi: 10.1099/0022-1317-65-7-1225
Gaglia M. M., Rycroft C. H., Glaunsinger B. A. (2015). Transcriptome-Wide Cleavage Site Mapping on Cellular mRNAs Reveals Features Underlying Sequence-Specific Cleavage by the Viral Ribonuclease SOX. PloS Pathog. 11, e1005305. doi: 10.1371/journal.ppat.1005305
Gilbertson S., Federspiel J. D., Hartenian E., Cristea I. M., Glaunsinger B. (2018). Changes in mRNA abundance drive shuttling of RNA binding proteins, linking cytoplasmic RNA degradation to transcription. Elife 7, e37663. doi: 10.7554/eLife.37663
Glaunsinger B., Ganem D. (2004). Lytic KSHV infection inhibits host gene expression by accelerating global mRNA turnover. Mol. Cell 13, 713–723. doi: 10.1016/S1097-2765(04)00091-7
Glaunsinger B., Chavez L., Ganem D. (2005). The exonuclease and host shutoff functions of the SOX protein of Kaposi’s sarcoma-associated herpesvirus are genetically separable. J. Virol. 79, 7396–7401. doi: 10.1128/JVI.79.12.7396-7401.2005
Goncalves P. H., Ziegelbauer J., Uldrick T. S., Yarchoan R. (2017). Kaposi sarcoma herpesvirus-associated cancers and related diseases. Curr. Opin. HIV AIDS 12, 47–56. doi: 10.1097/COH.0000000000000330
Gong D., Kim Y. H., Xiao Y., Du Y., Xie Y., Lee K. K., et al. (2016). A Herpesvirus Protein Selectively Inhibits Cellular mRNA Nuclear Export. Cell Host Microbe 20, 642–653. doi: 10.1016/j.chom.2016.10.004
Gordon D. E., Jang G. M., Bouhaddou M., Xu J., Obernier K., White K. M., et al. (2020). A SARS-CoV-2 protein interaction map reveals targets for drug repurposing. Nature 583, 459–468. doi: 10.1038/s41586-020-2286-9
Grünwald D., Singer R. H., Rout M. (2011). Nuclear export dynamics of RNA-protein complexes. Nature 475, 333–341. doi: 10.1038/nature10318
Hammell C. M., Gross S., Zenklusen D., Heath C. V., Stutz F., Moore C., et al. (2002). Coupling of termination, 3’ processing, and mRNA export. Mol. Cell. Biol. 22, 6441–6457. doi: 10.1128/mcb.22.18.6441-6457.2002
Hilleren P., Parker R. (2001). Defects in the mRNA export factors Rat7p, Gle1p, Mex67p, and Rat8p cause hyperadenylation during 3’-end formation of nascent transcripts. RNA 7, 753–764. doi: 10.1017/s1355838201010147
Izawa S., Takemura R., Inoue Y. (2004). Gle2p is essential to induce adaptation of the export of bulk poly(A)+ mRNA to heat shock in Saccharomyces cerevisiae. J. Biol. Chem. 279, 35469–35478. doi: 10.1074/jbc.M403692200
Jensen T. H., Patricio K., McCarthy T., Rosbash M. (2001). A block to mRNA nuclear export in S. cerevisiae leads to hyperadenylation of transcripts that accumulate at the site of transcription. Mol. Cell 7, 887–898. doi: 10.1016/s1097-2765(01)00232-5
Katahira J., Strässer K., Podtelejnikov A., Mann M., Jung J. U., Hurt E. (1999). The Mex67p-mediated nuclear mRNA export pathway is conserved from yeast to human. EMBO J. 18, 2593–2609. doi: 10.1093/emboj/18.9.2593
Köhler A., Hurt E. (2007). Exporting RNA from the nucleus to the cytoplasm. Nat. Rev. Mol. Cell Biol. 8, 761–773. doi: 10.1038/nrm2255
Kraemer D., Blobel G. (1997). mRNA binding protein mrnp 41 localizes to both nucleus and cytoplasm. Proc. Natl. Acad. Sci. U. S. A. 94, 9119–9124. doi: 10.1073/pnas.94.17.9119
Kühn U., Gündel M., Knoth A., Kerwitz Y., Rüdel S., Wahle E. (2009). Poly(A) tail length is controlled by the nuclear poly(A)-binding protein regulating the interaction between poly(A) polymerase and the cleavage and polyadenylation specificity factor. J. Biol. Chem. 284, 22803–22814. doi: 10.1074/jbc.M109.018226
Kühn U., Buschmann J., Wahle E. (2017). The nuclear poly(A) binding protein of mammals, but not of fission yeast, participates in mRNA polyadenylation. RNA 23, 473–482. doi: 10.1261/rna.057026.116
Kumar G. R., Glaunsinger B. A. (2010). Nuclear import of cytoplasmic poly(A) binding protein restricts gene expression via hyperadenylation and nuclear retention of mRNA. Mol. Cell. Biol. 30, 4996–5008. doi: 10.1128/MCB.00600-10
Kumar G. R., Shum L., Glaunsinger B. A. (2011). Importin alpha-mediated nuclear import of cytoplasmic poly(A) binding protein occurs as a direct consequence of cytoplasmic mRNA depletion. Mol. Cell. Biol. 31, 3113–3125. doi: 10.1128/MCB.05402-11
Lee Y. J., Glaunsinger B. A. (2009). Aberrant herpesvirus-induced polyadenylation correlates with cellular messenger RNA destruction. PloS Biol. 7, e1000107. doi: 10.1371/journal.pbio.1000107
Li J., Guo M., Tian X., Wang X., Yang X., Wu P., et al. (2020). Virus-Host Interactome and Proteomic Survey Reveal Potential Virulence Factors Influencing SARS-CoV-2 Pathogenesis. Med. (N Y) 2 (1), 99–112.e7. doi: 10.1016/j.medj.2020.07.002
Mendez A. S., Vogt C., Bohne J., Glaunsinger B. A. (2018). Site specific target binding controls RNA cleavage efficiency by the Kaposi’s sarcoma-associated herpesvirus endonuclease SOX. Nucleic Acids Res. 46, 11968–11979. doi: 10.1093/nar/gky932
Miorin L., Kehrer T., Sanchez-Aparicio M. T., Zhang K., Cohen P., Patel R. S., et al. (2020). SARS-CoV-2 Orf6 hijacks Nup98 to block STAT nuclear import and antagonize interferon signaling. Proc. Natl. Acad. Sci. U. S. A. 117, 28344–28354. doi: 10.1073/pnas.2016650117
Powers M. A., Forbes D. J. (2012). Nuclear transport: beginning to gel? Curr. Biol. 22, R1006–R1009. doi: 10.1016/j.cub.2012.10.037
Pritchard C. E., Fornerod M., Kasper L. H., van Deursen J. M. (1999). RAE1 is a shuttling mRNA export factor that binds to a GLEBS-like NUP98 motif at the nuclear pore complex through multiple domains. J. Cell Biol. 145, 237–254. doi: 10.1083/jcb.145.2.237
Proudfoot N. (1996). Ending the message is not so simple. Cell 87, 779–781. doi: 10.1016/s0092-8674(00)81982-0
Qu X., Lykke-Andersen S., Nasser T., Saguez C., Bertrand E., Jensen T. H., et al. (2009). Assembly of an export-competent mRNP is needed for efficient release of the 3’-end processing complex after polyadenylation. Mol. Cell. Biol. 29, 5327–5338. doi: 10.1128/MCB.00468-09
Quan B., Seo H.-S., Blobel G., Ren Y. (2014). Vesiculoviral matrix (M) protein occupies nucleic acid binding site at nucleoporin pair (Rae1 • Nup98). Proc. Natl. Acad. Sci. U. S. A. 111, 9127–9132. doi: 10.1073/pnas.1409076111
Read G. S. (2013). Virus-encoded endonucleases: expected and novel functions. Wiley Interdiscip Rev. RNA 4, 693–708. doi: 10.1002/wrna.1188
Richner J. M., Clyde K., Pezda A. C., Cheng B. Y. H., Wang T., Kumar G. R., et al. (2011). Global mRNA degradation during lytic gammaherpesvirus infection contributes to establishment of viral latency. PloS Pathog. 7, e1002150. doi: 10.1371/journal.ppat.1002150
Rivas H. G., Schmaling S. K., Gaglia M. M. (2016). Shutoff of Host Gene Expression in Influenza A Virus and Herpesviruses: Similar Mechanisms and Common Themes. Viruses 8:102. doi: 10.3390/v8040102
Rowe M., Glaunsinger B., van Leeuwen D., Zuo J., Sweetman D., Ganem D., et al. (2007). Host shutoff during productive Epstein-Barr virus infection is mediated by BGLF5 and may contribute to immune evasion. Proc. Natl. Acad. Sci. U. S. A. 104, 3366–3371. doi: 10.1073/pnas.0611128104
Sabri N., Visa N. (2000). The Ct-RAE1 protein interacts with Balbiani ring RNP particles at the nuclear pore. RNA 6, 1597–1609. doi: 10.1017/s1355838200001138
Satterly N., Tsai P.-L., van Deursen J., Nussenzveig D. R., Wang Y., Faria P. A., et al. (2007). Influenza virus targets the mRNA export machinery and the nuclear pore complex. Proc. Natl. Acad. Sci. U. S. A. 104, 1853–1858. doi: 10.1073/pnas.0610977104
Shi M., Zhang H., Wu X., He Z., Wang L., Yin S., et al. (2017). ALYREF mainly binds to the 5’ and the 3’ regions of the mRNA in vivo. Nucleic Acids Res. 45, 9640–9653. doi: 10.1093/nar/gkx597
Soulier J., Grollet L., Oksenhendler E., Cacoub P., Cazals-Hatem D., Babinet P., et al. (1995). Kaposi’s sarcoma-associated herpesvirus-like DNA sequences in multicentric Castleman’s disease. Blood 86, 1276–1280. doi: 10.1182/blood.V86.4.1276.bloodjournal8641276
Strambio-De-Castillia C., Niepel M., Rout M. P. (2010). The nuclear pore complex: bridging nuclear transport and gene regulation. Nat. Rev. Mol. Cell Biol. 11, 490–501. doi: 10.1038/nrm2928
Strom T., Frenkel N. (1987). Effects of herpes simplex virus on mRNA stability. J. Virol. 61, 2198–2207. doi: 10.1128/JVI.61.7.2198-2207.1987
Tarun S. Z., Jr., Sachs A. B. (1996). Association of the yeast poly(A) tail binding protein with translation initiation factor eIF-4G. EMBO J. 15, 7168–7177.
Terry L. J., Wente S. R. (2009). Flexible gates: dynamic topologies and functions for FG nucleoporins in nucleocytoplasmic transport. Eukaryot Cell 8, 1814–1827. doi: 10.1128/EC.00225-09
Virgin H.W. 4., Latreille P., Wamsley P., Hallsworth K., Weck K. E., Dal Canto A. J., et al. (1997). Complete sequence and genomic analysis of murine gammaherpesvirus 68. J. Virol. 71, 5894–5904. doi: 10.1128/JVI.71.8.5894-5904.1997
Wahle E. (1995). Poly(A) tail length control is caused by termination of processive synthesis. J. Biol. Chem. 270, 2800–2808. doi: 10.1074/jbc.270.6.2800
Wells S. E., Hillner P. E., Vale R. D., Sachs A. B. (1998). Circularization of mRNA by eukaryotic translation initiation factors. Mol. Cell. 2, 135–140.
Keywords: gammaherpesvirus, Kaposi sarcoma-associated herpesvirus (KSHV), host mRNA nuclear export, mRNA stability, host gene expression inhibition, virus host interaction
Citation: Pardamean CI and Wu T-T (2021) Inhibition of Host Gene Expression by KSHV: Sabotaging mRNA Stability and Nuclear Export. Front. Cell. Infect. Microbiol. 11:648055. doi: 10.3389/fcimb.2021.648055
Received: 31 December 2020; Accepted: 19 March 2021;
Published: 09 April 2021.
Edited by:
Subhash C. Verma, University of Nevada, Reno, United StatesReviewed by:
Rolf Renne, University of Florida, United StatesCopyright © 2021 Pardamean and Wu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ting-Ting Wu, dHd1QG1lZG5ldC51Y2xhLmVkdQ==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.