
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Cell. Infect. Microbiol. , 30 April 2021
Sec. Molecular Bacterial Pathogenesis
Volume 11 - 2021 | https://doi.org/10.3389/fcimb.2021.647324
 Luciana Hernandez1
Luciana Hernandez1 Enriqueta Bottini2†
Enriqueta Bottini2† Jimena Cadona1†
Jimena Cadona1† Claudio Cacciato2
Claudio Cacciato2 Cristina Monteavaro2
Cristina Monteavaro2 Ana Bustamante1
Ana Bustamante1 Andrea Mariel Sanso1*
Andrea Mariel Sanso1*Streptococcus agalactiae is a pathogen-associated to bovine mastitis, a health disorder responsible for significant economic losses in the dairy industry. Antimicrobial therapy remains the main strategy for the control of this bacterium in dairy herds and human In order to get insight on molecular characteristics of S. agalactiae strains circulating among Argentinean cattle with mastitis, we received 1500 samples from 56 dairy farms between 2016 and 2019. We recovered 56 S. agalactiae isolates and characterized them in relation to serotypes, virulence genes, and antimicrobial susceptibility. Serotypes III and II were the most prevalent ones (46% and 41%, respectively), followed by Ia (7%). In relation to the 13 virulence genes screened in this study, the genes spb1, hylB, cylE, and PI-2b were present in all the isolates, meanwhile, bca, cpsA, and rib were detected in different frequencies, 36%, 96%, and 59%, respectively. On the other hand, bac, hvgA, lmb, PI-1, PI-2a, and scpB genes could not be detected in any of the isolates. Disk diffusion method against a panel of eight antimicrobial agents showed an important number of strains resistant simultaneously to five antibiotics. We also detected several resistance-encoding genes, tet(M), tet(O), ermB, aphA3, and lnu(B) (9%, 50%, 32%, 32%, and 5%, respectively). The results here presented are the first molecular data on S. agalactiae isolates causing bovine mastitis in Argentina and provide a foundation for the development of diagnostic, prophylactic, and therapeutic methods, including the perspective of a vaccine.
Streptococcus agalactiae, or group B Streptococcus (GBS), was first described from bovines (Nocard and Mollereau, 1887) and for seven decades was exclusively associated with mastitis in dairy herds. Later emerged as a leading cause of human neonatal infections (Manning, 2014), and nowadays S. agalactiae is increasingly recognized as an adult invasive pathogen worldwide (Skoff et al., 2009). Also, S. agalactiae infections have been reported in many fish species, particularly is an emerging pathogen in Nile tilapia (Oreochromis niloticus) worldwide (Mian et al., 2009). Recent studies debate if this zoonotic potential remains nowadays and suggest that although S. agalactiae is well adapted to various hosts, interspecies transmission is possible and occurs (Morach et al., 2018). Further, they hypothesize about possible routes through which this bacterium could be transmitted between cattle and humans (Botelho et al., 2018).
According to their epidemiology, mastitis pathogens can be classified as contagious or environmental. Contagious pathogens are those for which the udders of infected cattle act as the main reservoir (Cervinkova et al., 2013). S. agalactiae has been recognized as a highly contagious obligate parasite of the bovine mammary gland, which generally does not survive for long periods outside of the mammary gland (Keefe, 1997). However, at present, it was demonstrated that the bacteria can survive in extramammary sources (Cobo-Ángel et al., 2018). In cattle, intramammary infections are usually chronic and subclinical, with intermittent episodes of clinical mastitis. Bovine mastitis is the dominant health disorder leading to diminished milk quality and production and is responsible for significant economic losses in the dairy industry (Zadoks et al., 2004; Hogeveen et al., 2011).
Despite numerous eradication programs in cattle, S. agalactiae remains a common cause of infections, with high levels of prevalence and contagion in dairy herds in different geographical areas, particularly, in South America countries (Keefe, 2012). In studies conducted in dairy herds of Colombia, the prevalence of S. agalactiae varied from 28 to 35% (Ramírez et al., 2014; Reyes et al., 2015); in Brazil, a research group reported this pathogen as the third most prevalent bacteria causing bovine mastitis (Tomazi et al., 2018).
In Argentina, there is no updated data at the national level, but only for some particular regions, such as Córdoba province (Dieser et al., 2014), or other collected between 1999 and 2007, in a larger region that report an S. agalactiae prevalence of 29% among cows with mastitis from Buenos Aires, Santa Fe and Córdoba (Calvinho, 2017). A bacteriological study of dairy farms located in the Cuenca Mar y Sierras (Buenos Aires province), one of the main dairy regions of the country, showed S. agalactiae to be a frequent etiological pathogen causing subclinical mastitis (Amand De Mendieta et al., 2001). Furthermore, 10% of the sampled dairy farms in this area were positive to this species (Bottini, personal communication).
The pathogenesis of S. agalactiae infection and the severity of the disease have been related to the presence of a series of virulence factors mainly involved in colonization of the host, in the dissemination of the bacteria, the evasion of the immune response and internalization in the mammary gland cells. One of the most important factors involved in virulence is the capsular polysaccharide (Cps) (Slotved et al., 2007). S. agalactiae can be classified into 10 serotypes according to the type of Cps (Ia, Ib, and II to IX).
Antimicrobial therapy remains the main strategy for the control of this bacterium in dairy herds and human infections. Antimicrobial resistance is an area of concern in both human and veterinary medicine (Schwarz et al., 2010). Currently, there is little official information on the use of different antibiotics in veterinary medicine and, therefore, on the resistance models of animal pathogens circulating in the world. Strain characterization and surveillance are important to obtain information that allows evaluating the level and evolution of antimicrobial resistance (OIE - World Organization for Animal Health, 2018). For this reason, and in particular, studies on the antimicrobial activity of mastitis pathogens are necessary for controlling induced resistance and obtaining useful information for therapeutic decisions (Denamiel et al., 2005).
In this study, we present the first molecular data on S. agalactiae isolates causing bovine mastitis in Argentina and provide information in relation to serotypes, virulence, and antimicrobial susceptibility.
A total of 1500 samples recovered from different cows presenting clinical or subclinical mastitis, except one obtained from a milk tank (B3), between December 2016 and August 2019, were received at the lab. Samples came from 56 dairy farms (herds) located in one of the largest milk-producing regions of Argentina, the Cuenca Mar y Sierras, and S. agalactiae isolates could be obtained from seven dairy farms (A, B, C, D, E, G, and H).
Milk samples were collected under aseptic conditions from cows affected by clinical or subclinical mastitis, immediately refrigerated at 4°C and subjected to bacteriological analysis within 24 h of collection. A loopful of milk sample was streaked on trypticase soy agar (TSA) enriched with 5% bovine blood and plates were incubated at 37°C in atmosphere with 5% CO2. Subsequently, the plates were examined for colony morphology, pigmentation and hemolytic characteristics after 24–48 h. Presumptive colonies of Streptococcus species were selected and streaked into a slant agar for 24 h for biochemical tests, and Gram staining. Catalase, NaCl, bile-esculin, Christie-Atkins-Munch-Peterson (CAMP), hippurate hydrolysis, and sorbitol tests were carried out as described by the National Mastitis Council (2017). Sixty-eight isolates were identified as S. agalactiae, being Gram-positive cocci, CAMP reaction-positive, and catalase and esculin activity-negative, and stored at −20°C.
The DNA template was obtained by boiling bacterial colonies suspended in sterile water for 10 min. To confirm the species identification, a region of the monocopy regulatory gene dltR, specific to S. agalactiae, was amplified (Lamy et al., 2006). The capsular type identification, Ia, Ib, II-IX, was determined by PCR according to Imperi et al. (2010). Additionally, a total of ten virulence genes, bac, bca cpsA, cylE, hvgA, hylB, lmb, rib, scpB, spb1, plus three pili genes designated as pilus island 1, PI-1, PI-2a, and PI-2b, were detected according to previous studies. The examined virulence genes have been associated with adhesion and colonization, invasion, tissue damage, and/or immune evasion (Table 1). The PCR products were visualized in 2% agarose gel stained by ethidium bromide.
Taking into account the combinations of the genes detected in the present study, the virulence profiles were defined. A cluster analysis was carried out using the UPGMA clustering method. The dendrogram was generated using the BioNumerics v.6.6 software.
The isolates were tested for susceptibility to eight antibiotics using a disc diffusion method according to the CLSI - Clinical and Laboratory Standards Institute (2019) instructions. The antimicrobial agents were selected taking into account their use for mastitis treatment in cattle (penicillin, oxacillin, kanamycin, pirlimycin, and tetracycline) or/and in human medicine (penicillin, erythromycin, clindamycin, levofloxacin). A bacterial suspension in sterile saline solution from an overnight pure culture, adjusted to a turbidity of 0.5 on the McFarland scale, was inoculated on a Muller-Hinton agar (Britania) plate, supplemented with 5% sheep blood. Antibiotic discs (Britania) were placed on the agar surface and plates were incubated overnight (16–18 h) at 37°C in atmosphere with 5% CO2. The diameters of the zones of inhibition were then measured and data were interpreted by using human breakpoints values for all the antimicrobial agents except for pirlimycin, for which veterinary interpretive criteria for cattle were available (CLSI - Clinical and Laboratory Standards Institute, 2018; CLSI - Clinical and Laboratory Standards Institute, 2019) (Table S1). In relation to the aminoglycoside kanamycin, we used high load antibiotic discs in order to predict lack of synergy when associated with Beta-lactams. Since not kanamycin standards are available, only isolates presenting no zones of inhibition were considered as resistant. The following discs were used: clindamycin (2 μg), pirlimycin (2 μg), erythromycin (15 μg), levofloxacin (5 μg), penicillin (10 units), oxacillin (1 μg), tetracycline (30 μg), and kanamycin (120 μg). Isolates showing resistance against three or more different classes of antibiotics were defined as multi-drug resistant (MDR) (Sweeney et al., 2018).
The macrolide resistance gene ermB was amplified by PCR according to Zhou et al. (2011), mefA, and tetracycline resistance genes tet(M), tet(O), tet(T), and tet(K), according to Lopardo et al. (2003), lincosamide resistance gene lnu(B) (before named linB), according to Bozdogan et al. (1999) and, aminoglycosides resistance genes aphA3 and aad6, according to Poyart et al. (2003). The correlation between phenotype of resistance and resistance genes was analyzed as well as was done by Liu et al. (2017) and Tian et al. (2019).
Out of 1500 samples received during 33 months from 56 dairy farms located in the Cuenca Mar y Sierras, Buenos Aires Province, Argentina, 68 Streptococcus agalactiae strains were identified biochemically. Then, among the original 68 isolates, species-specific PCR (dltR gene amplification) confirmed 56 Streptococcus agalactiae strains arising from seven dairy farms.
The bovine isolates studied here belonged to capsular genotypes Ia, II, III, and three ones were designed as non-typeable (NT), according to the multiplex PCR. Overall, type III and II were the most prevalent accounting for 26 isolates (46%) and 23 isolates (41%), respectively and, type Ia by 4 isolates (7%) (Table 2).
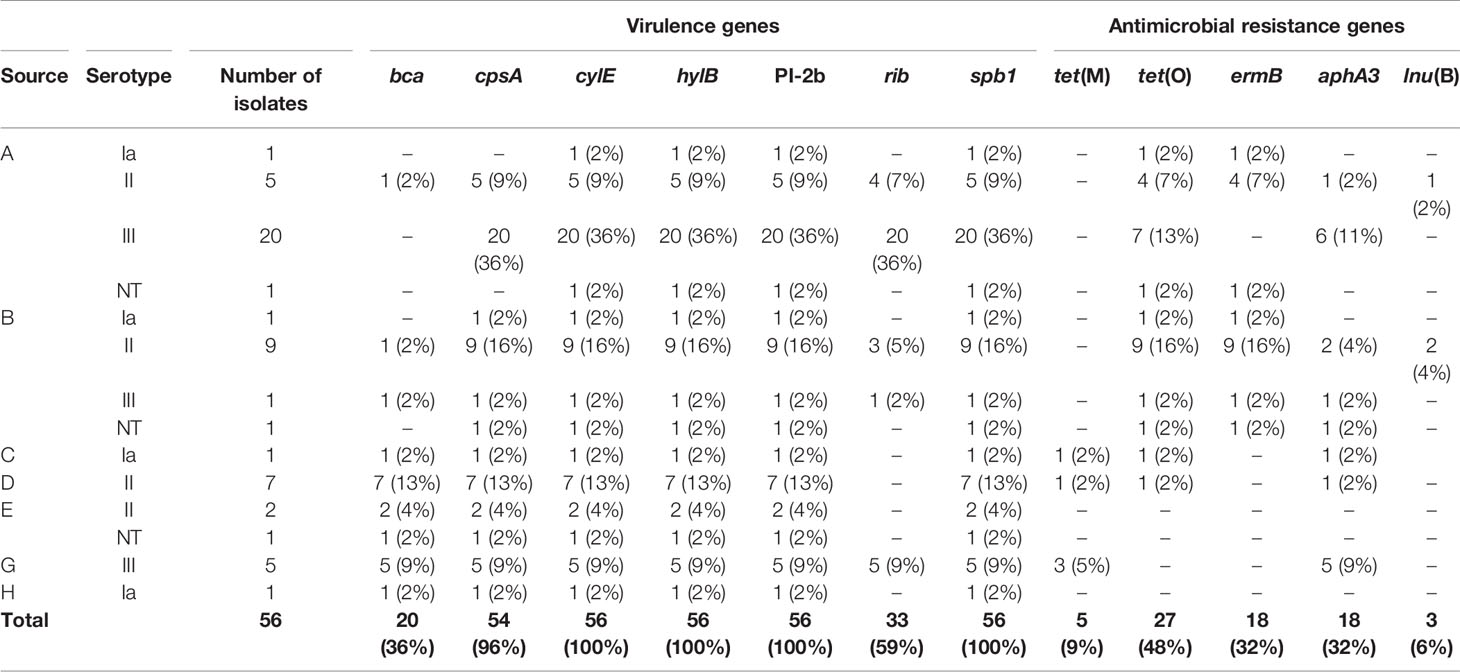
Table 2 Distribution of virulence and antimicrobial resistance genes detected among Streptococcus agalactiae isolates recovered from dairy cattle with mastitis in Argentina.
The virulence genes bac, scpB and lmb could not be detected in any of the isolates. Gene hvgA, marker of ST-17, was also absent in all isolates belonging to serotype III and the remaining ones. Isolates harbored from four to seven of the assayed virulence genes. The genes spb1, hylB and cylE were present in all the isolates meanwhile, bca, cpsA, and rib were detected in different frequencies, 36% (20), 96% (54), and 59% (33), respectively (Table 2). Pilus typing using three PCR assays, showed that PI-1 and PI-2a genes were absent in all the investigated bovine strains. On the other hand, all isolates harbored the PI-2b gene (Table 2).
The cluster analysis taking into account the combinations of the genes detected in the present study showed five virulence profiles shared by isolates from different dairy farm, except one (spB1-hylB-cylE-PI-2b) which was presented only by two isolates from dairy farm A. No one of the profiles could be associated with a particular serotype (Figure 1).
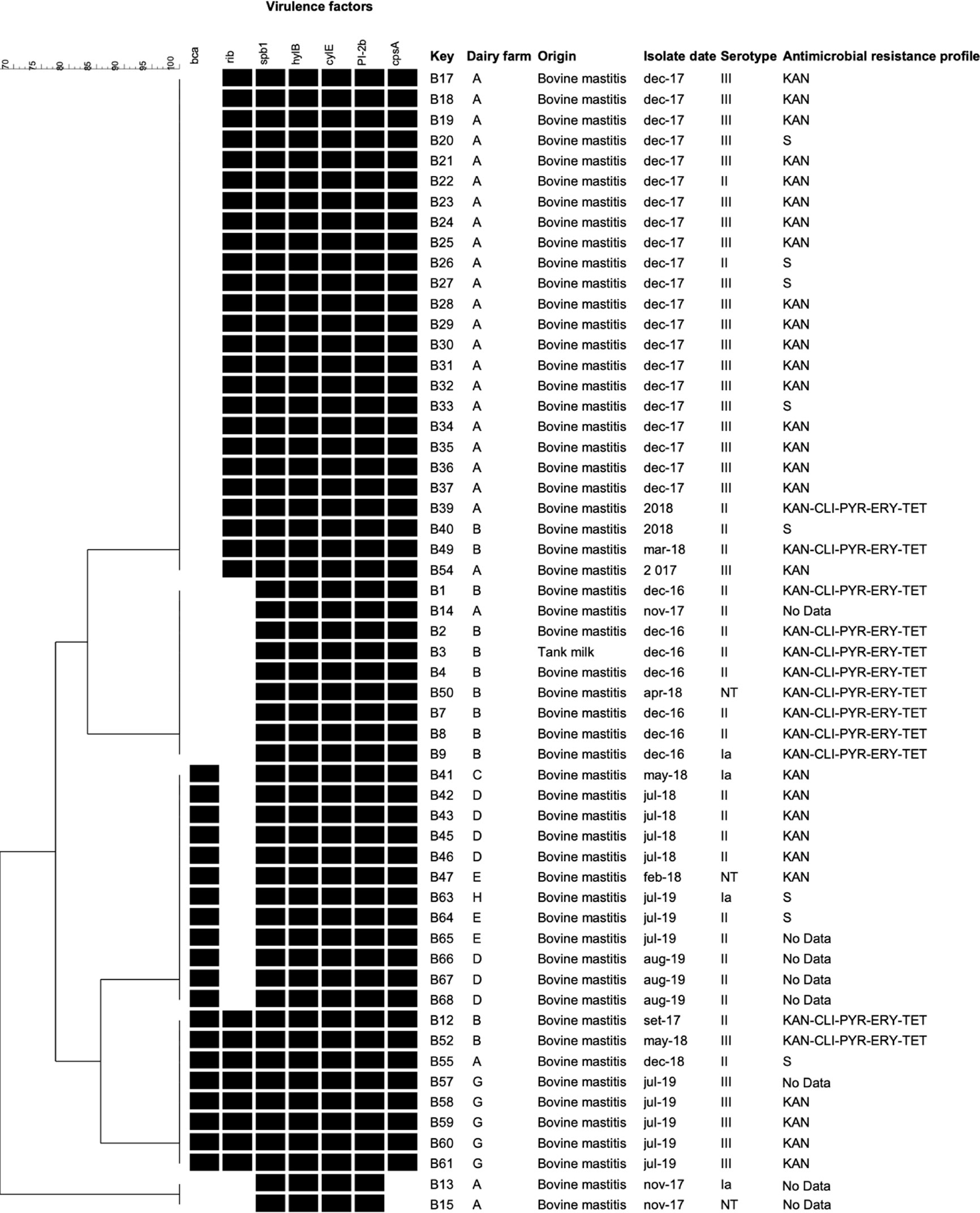
Figure 1 Cluster analysis of Streptococcus agalactiae isolated from dairy cattle with mastitis in Argentina based on virulence-associated genes profiles. The presence (black) or absence (white) of genes, the isolate name, dairy farm, origin, isolation date, and serotype of the isolates are shown. The antimicrobial resistance profiles are indicated on the right. NT, non-typeable. Genes not found in any of the studied isolates: bac, lmb, hvgA, PI, PI-2a, and scpB. S: susceptible to all tested antimicrobial agents.
Regarding antimicrobial resistance, 48 S. agalactiae isolates (86%) could be tested using a disc diffusion method. We did not have data on the remaining eight isolates due to problems with contamination during susceptibility testing. In relation to the aminoglycoside kanamycin, since not reference breakpoints values were available, only isolates presenting no inhibition area were considered as resistant. The isolates without inhibition area were 40, representing 83% of the tested isolates. Twelve isolates, eleven of them obtained from the dairy farm B and one from the farm A, were resistance to five antimicrobials, kanamycin, clindamycin, pirlimycin, erythromycin and, tetracycline (Figure 1).
To investigate genetic antimicrobial resistance, PCR assays for genes accounting for resistance to several antibiotics were carried out. In relation to tetracycline resistance, the efflux genes tet(K) and tet(L) and the ribosomal protection genes tet(M) and tet(O) were amplified. The gene tet(O) was recovered in all tetracycline-resistant strains, and, also, in 16 strains which did not present phenotypic susceptibility for tetracycline; the gene tet(M), in 5 isolates (B41, B57, B58, B59 and B57).
In relation to erythromycin resistance, all resistant isolates exhibited resistance associated to ermB gene indicating the presence of a target-site modification by a ribosomal methylase. Six susceptible strains, also carried that gene. Genes aphA3 and aad6, related to aminoglycosides resistance, were also amplified. The gene aad6 was not detected meanwhile aphA3, related to kanamycin resistance, was detected in 18 isolates. The detection of lnuB in clindamycin and pirlimycin-resistant isolates would explain the resistance observed to lincosamides. The positive-isolates were B4, B39, and B49. The average correlation rate between resistant phenotypes and genotypes was 65.62% (Table 3). The calculations were made taking into account only those resistant isolates that were studied by both analyses (phenotypic and genetic resistance). Given each of the antimicrobial classes individually, tetracyclines and macrolides showed the highest correlation (100%), while aminoglycosides and lincosamides presented lower correlations (37.5% and 25% respectively). On the other hand, the 50% of strains that presented phenotypic susceptibility for all antimicrobial classes carried some resistance genes.
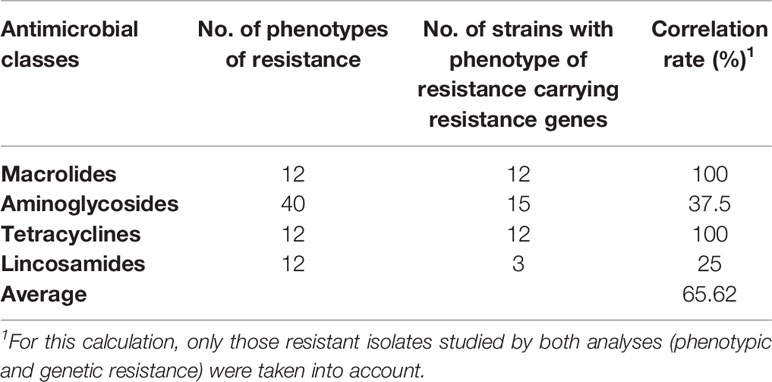
Table 3 The correlations between resistance phenotypes and resistance genes of S. agalactiae isolates.
Streptococcus agalactiae is considered one of the major mastitis pathogens and, for our knowledge, this is the first molecular study that characterizes S. agalactiae isolates circulating among cattle with mastitis in Argentina. Of the 68 original strains identified as S. agalactiae by biochemical tests, only 56 ones were confirmed genetically.
Capsular serotyping is a classical method used for epidemiological studies in S. agalactiae and ten serotypes are identified based on Cps types. Serotypes III and II were the predominant ones among S. agalactiae Argentinean bovine strains (87%), followed by a low frequency of strains classified as Ia and non-typeable. Serotype III, and especially lineage ST-17, is particularly important in human infections because it causes the majority of infections in neonates (Manning, 2014). In this study, we did not detect hvgA, a gene encoding an ST-17–specific surface-anchored protein, critical virulence trait of neonatal disease associated-S agalactiae (Tazi et al., 2010). The distribution of S. agalactiae serotypes involved in bovine mastitis is variable worldwide and certain types appear to predominate within geographical regions. Serotype II was the most prevalent in Canada (Zhao et al., 2006), Eastern-China, USA (together with Ia) (Dogan et al., 2005; Yang et al., 2013) and Germany (together with non-typeable) (Merl et al., 2003). On the other hand, serotypes V and IV were the most prevalent in Norway (Radtke et al., 2012). The dominant serotypes in Poland were Ia and II although also serotypes Ib, III, IV, and V were detected (Kaczorek et al., 2017) while in strains originating from dairy herds in France, non-typeable strains were detected at the highest frequency, followed by serotypes III and IV (Brochet et al., 2006). In Brazil, five capsular types were identified (Ia, Ib, II, III, and IV) (Duarte et al., 2005; Pinto et al., 2014; Carvalho-Castro et al., 2017), being III and II the most prevalent ones, just like our results and, in Iran, only were detected these last two serotypes (Emaneini et al., 2016).
In relation to the virulence genes screened in this study, five virulence profiles were detected, which included spb1, hylB, cylE, and PI-2b. The product of the spb1 gene has been proposed as a factor implicated in adhesion to epithelial cells (Adderson et al., 2003). The gene hylB, encodes an important marker of virulence that degrades hyaluronic acid, facilitating bacterial dissemination (Baker and Pritchard, 2000). Other authors also detected hylB in all the studied samples (Carvalho-Castro et al., 2017; Pang et al., 2017). CylE is a pore-forming toxin, involved in tissue injury and the systemic spread of bacteria (Doran et al., 2002; Doran et al., 2003; Reiß et al., 2011). The expression of this β-hemolysin has proapoptotic, pro-inflammatory, and cytotoxic effects (Randis et al., 2014). Previous studies reported the presence of this gene in 78% of Poland strains (Kaczorek et al., 2017) and in 100% of Chinese ones (Pang et al., 2017).
Pilus structures in S. agalactiae facilitate colonization and invasion of host tissues and participate in biofilm formation. These structures are encoded in islands (PI) and three of them, PI-1, PI-2a, and PI-2b, have been identified in highly virulent strains (Margarit et al., 2009). S. agalactiae strains carry at least one of the three PI and, some studies highlight that strains of different origins usually harbor different pilus variants; while type 2a (PI-2a) is more common in human strains, type 2b (PI-2b) is more frequent in bovine isolates (Martins et al., 2013; Otaguiri et al., 2013; Carvalho-Castro et al., 2017; Pang et al., 2017). Our results agree with previous studies in which all the bovine S. agalactiae strains from China carried only PI-2b (Yang et al., 2013; Pang et al., 2017).
The studied isolates differed mainly by the presence of bca and rib. Someone harbored one of the two genes (bca: 20 isolates, 36%; rib: 33 isolates, 59%) others both (8 isolates, 14%), and some, neither of them (9 isolates, 16%). These genes belong to the alpha-like surface protein family and are important in the pathogenicity. It was not possible associate the presence/absence of these genes with serotype. The protein ą-C, encoded by the bca gene mediates the internalization of the bacteria to host cells (Baron et al., 2004) and Rib is only present in invasive strains (Lindahl et al., 2005). Studies carried out in Poland reported the presence of bca in 37% (Kaczorek et al., 2017) and in Brazil it varied between 3% and 79%, depending on the geographical location (Duarte et al., 2004; Carvalho-Castro et al., 2017). For rib, previous studies reported the presence in bovine strains of 33% in Poland (Kaczorek et al., 2017) and 89% in Iran (Emaneini et al., 2016).
The isolates were negative for the virulence genes bac, lmb, and scpB. Earlier molecular reports showed that most bovine isolates lack surface proteins-encoding genes scpB and lmb, in contrast to human isolates (Franken et al., 2001), but that however they can be detected in some S. agalactiae bovine strains (Rato et al., 2013).
In order to have a global view of antimicrobial susceptibility occurrence, isolates were tested against eight antimicrobial agents selected taking into account their use for mastitis treatment in cattle (penicillin, oxacillin, kanamycin, pirlimycin, and tetracycline) or/and in human medicine (penicillin, erythromycin, clindamycin, levofloxacin). We evaluated their antimicrobial resistance profiles using also antibiotics of human use considering that this information can assist in critical decision making as part of the concept “One health”. The most commonly used antimicrobial classes for the treatment of streptococcal mastitis are β-lactams and macrolides (Denamiel et al., 2005). On the one hand, we did detect resistance to macrolides (erythromycin and clindamycin), and on the other, besides, resistance to the kanamycin. The practice shows the intensive use in Argentinean dairy farms of a product containing this aminoglycoside in association with a β-lactam drug. The absence of resistance to penicillin and oxacillin observed in this study, and in other ones worldwide (Haenni et al., 2018), indicates that β-lactam antibiotics should remain the drugs of choice in the treatment of streptococcal mastitis. However, the high level of resistance to kanamycin detected predicts lack of synergy when associated with β-lactams leading to therapy failure (Chow, 2000).
According to the World Organization for Animal Health (OIE, 2016), between 2010 and 2015, tetracyclines and macrolides were the two classes of antibiotics most commonly used in animals worldwide. We detected resistance to tetracycline and erythromycin, agreeing with previous reports on bovine strains recovered in Brazil, Poland and, Portugal (Rato et al., 2013; Kaczorek et al., 2017; Tomazi et al., 2018). On the other hand, a previous study carried on milk from infected udders reported erythromycin and clindamycin-resistant Argentinean Streptococcus isolates (Denamiel et al., 2005). In addition to resistance to tetracycline, erythromycin, clindamycin, and kanamycin, the multi-drug resistant (MDR) isolates were resistant to pirlimycin, a lincosamide approved only for veterinary use. This antimicrobial agent is available for intramammary administration to treat mastitis caused by gram-positive cocci and pirlimycin resistance among streptococci has been reported previously (Pol and Ruegg, 2007; Tomazi et al., 2018).
In streptococci, resistance to tetracycline is encoded by ribosome protection genes including tet(M) and tet(O) or by efflux pump genes, tet(K), and tet(L) (Rubio-López et al., 2012). Resistance to macrolide are due to two common mechanisms, a ribosome methylase, encoded by the erm gene and an active efflux pump by a membrane-bound protein, encoded by the mef gene. The former concurrently confers high-level resistance to macrolides, as well as to lincosamide and streptogramin B antibiotics (Ko et al., 2004). All studied macrolide-resistant isolates were also resistant to clindamycin and all of them harbored the ermB gene.
Resistance to tetracycline was attributed to the presence of tet(O), erythromycin resistance to target site modification encoded by the erythromycin ribosome methylase gene ermB, pirlimycin/clindamycin resistance to lnu(B), a gene encoding a lincosamide inactivating nucleotidyl transferase and, kanamycin resistance to aphA3, a gene encoding an aminoglycoside phosphotransferase. The average correlation between resistance phenotypes and resistance genes of S. agalactiae was 65.62%. Particularly, the correlation for tetracycline was 100%. Similar findings were reported by da Silva et al. (2017), in S. agalactiae strains isolated from Brazilian mastitic cows and by Liu et al. (2017), in Staphylococcus aureus strains isolated from raw milk. Although mastitis therapy is commonly initiated before the results of antimicrobial susceptibility tests of the pathogen are known (Guérin-Faublée et al., 2002), the emergence of resistant pathogens (such as the detected in this study) makes essential performing susceptibility tests for the selection of the appropriate chemotherapeutic agents (Minst et al., 2012).
Farm animal disease control often share active substances with human medicines and, excessive use of antibiotics is associated with the risk of the creation of MDR foodborne pathogens (van Duin and Paterson, 2016). Another point to keep in mind when considering antimicrobial resistance of mastitis pathogens is that the interpretive criteria used for categorizing isolates are based on human data for the majority of compounds tested. They may not accurately reflect the efficacy of the drugs in the treatment of bovine mastitis (Rajala-Schultz et al., 2004) and therefore, veterinary-specific breakpoints are necessary (Thomas et al., 2015).
On the other hand, vaccination is one of the strategies most likely to be implemented to prevent GBS infections. Several GBS vaccine candidates are in development, especially for humans (Bianchi-Jassir et al., 2020). Cps and pilus proteins are some of the main targets proposed for the vaccine. In order to guide vaccine development or before of regulating the use of it, it is essential to answer the question about which serotypes are the most implicated in cases of disease in the country and, if possible, identify alternative targets. In this study, we reported that serotypes III and II were the most prevalent ones (87%) and the presence of spb1, hylB, cylE, and PI-2b in all the isolates. These data could be of interest in the perspective of a future vaccine.
The results of this study are the first molecular data on S. agalactiae isolates causing bovine mastitis in Argentina. We detected several virulence and antimicrobial susceptibility profiles associated with S. agalactiae intramammary infections. On the one hand, we found the all the isolates harbored the genes spb1, hylB, cylE, and PI-2b, and the predominance of serotype III and II. On the other hand, we detected strains MDR to clinical and veterinary relevant antimicrobials, and, several resistance-encoding genes. These data present us with the future challenge of closely monitoring the spread of MDR strains, to explore the molecular mechanisms responsible for the antimicrobial resistance, and provide a foundation for the development of diagnostic, prophylactic, and therapeutic methods, including the perspective of a vaccine.
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
Conceptualization and design of the study, AS and AB. Methodology, LH, EB, JC, CC, CM, and AB. Data analysis and interpretation, LH, JC, AB, and AS. Writing original draft preparation, AS and LH. Critical revisions and writing of the revised manuscript, AS. Supervision and administration project, AS. All authors contributed to the article and approved the submitted version.
This research was funded by Fondo para la Investigación Científica y Tecnológica (PICT 1139-17) and Consejo Nacional de Investigaciones Científicas y Técnicas (PIP 365/15, PUE CIVETAN).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcimb.2021.647324/full#supplementary-material
Adderson E. E., Takahashi S., Wang Y., Armstrong J., Miller D. V., Bohnsack J. F. (2003). Subtractive Hybridization Identifies a Novel Predicted Protein Mediating Epithelial Cell Invasion by Virulent Serotype III Group B Streptococcus Agalactiae. Infect. Immun. 71, 6857–6863. doi: 10.1128/IAI.71.12.6857-6863.2003
Amand De Mendieta V., Micheo C., Soriano C., Tabera A., Stefano A., Casasnovas G., et al. (2001). Aislamiento E Identificación De Patógenos Mamarios De Animales Bovinos Lecheros De La Cuenca Mar Y Sierras. Vet. Argent. 18, 499–504.
Baker J. R., Pritchard D. G. (2000). Action Pattern and Substrate Specificity of the Hyaluronan Lyase From Group B Streptococci. Biochem. J. 348, 465–471. doi: 10.1042/0264-6021:3480465
Baron M. J., Bolduc G. R., Goldberg M. B., Aupérin T. C., Madoff L. C. (2004). Alpha C Protein of Group B Streptococcus Binds Host Cell Surface Glycosaminoglycan and Enters Cells by an Actin-Dependent Mechanism. J. Biol. Chem. 279, 24714–24723. doi: 10.1074/jbc.M402164200
Bianchi-Jassir F., Paul P., To K. N., Carreras-Abad C., Seale A. C., Jauneikaite E., et al. (2020). Systematic Review of Group B Streptococcal Capsular Types, Sequence Types and Surface Proteins as Potential Vaccine Candidates. Vaccine 38, 6682–6694. doi: 10.1016/j.vaccine.2020.08.052
Bidet P., Brahimi N., Chalas C., Aujard Y., Bingen E. (2003). Molecular Characterization of Serotype Iii Group B-Streptococcus Isolates Causing Neonatal Meningitis. J. Infect. Dis. 188, 1132–1137. doi: 10.1086/378517
Botelho A. C. N., Ferreira A. F. M., Fracalanzza S. E. L., Teixeira L. M., Pinto T. C. A. (2018). A Perspective on the Potential Zoonotic Role of Streptococcus Agalactiae: Searching for a Missing Link in Alternative Transmission Routes. Front. Microbiol. 9:608. doi: 10.3389/fmicb.2018.00608
Bozdogan B., Berrezouga L., Kou M. S., Yurek D. A., Farley K. A., Stockman B. J., et al. (1999). A New Resistance Gene, linB, Conferring Resistance to Lincosamides by Nucleotidylation in Enterococcus Faecium HM1025. Antimicrob. Agents Chemother. 43, 925–929. doi: 10.1128/aac.43.4.925
Brochet M., Couvé E., Zouine M., Vallaeys T., Rusniok C., Lamy M. C., et al. (2006). Genomic Diversity and Evolution Within the Species Streptococcus Agalactiae. Microbes Infect. 8, 1227–1243. doi: 10.1016/j.micinf.2005.11.010
Calvinho L. F. (2017). Mastitis Bovina: Evolución Del Control En Argentina Y Nuevos Horizontes De Investigación. An. La Acad. Nac. Agron. y Vet. 70, 148–165.
Carvalho-Castro G. A., Silva J. R., Paiva L. V., Custódio D. A. C., Moreira R. O., Mian G. F., et al. (2017). Molecular Epidemiology of Streptococcus Agalactiae Isolated From Mastitis in Brazilian Dairy Herds. Braz. J. Microbiol. 48, 551–559. doi: 10.1016/j.bjm.2017.02.004
Cervinkova D., Vlkova H., Borodacova I., Makovcova J., Babak V., Lorencova A., et al. (2013). Prevalence of Mastitis Pathogens in Milk From Clinically Healthy Cows. Vet. Med. (Praha) 58, 567–575. doi: 10.17221/7138-VETMED
Chow J. W. (2000). Aminoglycoside Resistance in Enterococci. Clin. Infect. Dis. 31, 586–589. doi: 10.1086/313949
CLSI - Clinical and Laboratory Standards Institute (2018). Performance Standards for Antimicrobial Disk and Dilution Susceptibility Tests for Bacteria Isolated From Animals. 4th ed (Wayne, PA, USA: Clinical and Laboratory Standards Institute).
CLSI - Clinical and Laboratory Standards Institute (2019). Performance Standards for Antimicrobial Susceptibility Testing. 29th ed (Wayne, PA, USA: Clinical and Laboratory Standards Institute).
Cobo-Ángel C., Jaramillo-Jaramillo A. S., Lasso-Rojas L. M., Aguilar-Marin S. B., Sanchez J., Rodriguez-Lecompte J. C., et al. (2018). Streptococcus Agalactiae is Not Always an Obligate Intramammary Pathogen: Molecular Epidemiology of GBS From Milk, Feces and Environment in Colombian Dairy Herds. PloS One 13, 1–14. doi: 10.1371/journal.pone.0208990
da Silva J. R., Castro G. D. A. D. C., Gonçalves M. S., Custódio D. A. D. C., Mian G. F., Da Costa G. M. (2017). In Vitro Antimicrobial Susceptibility and Genetic Resistance Determinants of Streptococcus Agalactiae Isolated From Mastitic Cows in Brazilian Dairy Herds. Semin. Agrar. 38, 2581–2594. doi: 10.5433/1679-0359.2017v38n4Supl1p2581
Denamiel G., Llorente P., Carabella M., Rebuelto M., Gentilini E. (2005). Anti-Microbial Susceptibility of Streptococcus Spp. Isolated From Bovine Mastitis in Argentina. J. Vet. Med. Ser. B. Infect. Dis. Vet. Public Heal. 52, 125–128. doi: 10.1111/j.1439-0450.2005.00830.x
Dieser S. A., Vissio C., Lasagno M. C., Bogni C. I., Larriestra A. J., Odierno L. M. (2014). Prevalence of Pathogens Causing Subclinical Mastitis in Argentinean Dairy Herds. Pak. Vet. J. 34, 124–126.
Dogan B., Schukken Y. H., Santisteban C., Boor K. J. (2005). Distribution of Serotypes and Antimicrobial Resistance Genes Among Streptococcus Agalactiae Isolates From Bovine and Human Hosts. J. Clin. Microbiol. 43, 5899–5906. doi: 10.1128/JCM.43.12.5899-5906.2005
Doran K. S., Chang J. C. W., Benoit V. M., Eckmann L., Nizet V. (2002). Group B Streptococcal β-Hemolysin/Cytolysin Promotes Invasion of Human Lung Epithelial Cells and the Release of Interleukin-8. J. Infect. Dis. 185, 196–203. doi: 10.1086/338475
Doran K. S., Liu G. Y., Nizet V. (2003). Group B Streptococcal β-Hemolysin/Cytolysin Activates Neutrophil Signaling Pathways in Brain Endothelium and Contributes to Development of Meningitis. J. Clin. Invest. 112, 736–744. doi: 10.1172/jci17335
Duarte R. S., Bellei B. C., Miranda O. P., Brito M. A. V. P., Teixeira L. M. (2005). Distribution of Antimicrobial Resistance and Virulence-Related Genes Among Brazilian Group B Streptococci Recovered From Bovine and Human Sources. Antimicrob. Agents Chemother. 49, 97–103. doi: 10.1128/AAC.49.1.97-103.2005
Duarte R. S., Miranda O. P., Bellei B. C., Brito M. A. V. P., Teixeira L. M. (2004). Phenotypic and Molecular Characteristics of Streptococcus Agalactiae Isolates Recovered From Milk of Dairy Cows in Brazil. J. Clin. Microbiol. 42, 4214–4222. doi: 10.1128/JCM.42.9.4214-4222.2004
Emaneini M., Jabalameli F., Abani S., Dabiri H., Beigverdi R. (2016). Comparison of Virulence Factors and Capsular Types of Streptococcus Agalactiae Isolated From Human and Bovine Infections. Microb. Pathog. 91, 1–4. doi: 10.1016/j.micpath.2015.11.016
Franken C., Haase G., Brandt C., Weber-Heynemann J., Martin S., Lämmler C., et al. (2001). Horizontal Gene Transfer and Host Specificity of Beta-Haemolytic Streptococci: The Role of a Putative Composite Transposon Containing Scpb and Lmb. Mol. Microbiol. 41, 925–935. doi: 10.1046/j.1365-2958.2001.02563.x
Guérin-Faublée V., Tardy F., Bouveron C., Carret G. (2002). Antimicrobial Susceptibility of Streptococcus Species Isolated From Clinical Mastitis in Dairy Cows. Int. J. Antimicrob. Agents 19, 219–226. doi: 10.1016/S0924-8579(01)00485-X
Haenni M., Lupo A., Madec J.-Y. (2018). Antimicrobial Resistance in Streptococcus Spp. Antimicrob. Resist. Bact. Livest. Companion Anim., 159–184. doi: 10.1128/microbiolspec.arba-0008-2017
Hogeveen H., Huijps K., Lam T. J. G. M. (2011). Economic Aspects of Mastitis: New Developments. N. Z. Vet. J. 59, 16–23. doi: 10.1080/00480169.2011.547165
Imperi M., Pataracchia M., Alfarone G., Baldassarri L., Orefici G., Creti R. (2010). A Multiplex PCR Assay for the Direct Identification of the Capsular Type (Ia to IX) of Streptococcus Agalactiae. J. Microbiol. Methods 80, 212–214. doi: 10.1016/j.mimet.2009.11.010
Kaczorek E., Małaczewska J., Wójcik R., Siwicki A. K. (2017). Biofilm Production and Other Virulence Factors in Streptococcus Spp. Isolated From Clinical Cases of Bovine Mastitis in Poland. BMC Vet. Res. 13, 1–7. doi: 10.1186/s12917-017-1322-y
Keefe G. (2012). Update on Control of Staphylococcus Aureus and Streptococcus Agalactiae for Management of Mastitis. Vet. Clin. North Am. Food Anim. Pract. 28, 203–216. doi: 10.1016/j.cvfa.2012.03.010
Ko W. C., Yan J. J., Lee N. Y., Wu H. M., Wu J. J. (2004). Polyclonal Spread of Erythromycin-Resistant Streptococcus Agalactiae in Southern Taiwan. Microb. Drug Resist. 10, 306–312. doi: 10.1089/mdr.2004.10.306
Lamy M. C., Dramsi S., Billoët A., Réglier-Poupet H., Tazi A., Raymond J., et al. (2006). Rapid Detection of the “Highly Virulent” Group B Streptococcus ST-17 Clone. Microbes Infect. 8, 1714–1722. doi: 10.1016/j.micinf.2006.02.008
Lindahl G., Stålhammar-Carlemalm M., Areschoug T. (2005). Surface Proteins of Streptococcus Agalactiae and Related Proteins in Other Bacterial Pathogens. Clin. Microbiol. Rev. 18, 102–127. doi: 10.1128/CMR.18.1.102-127.2005
Liu H., Li S., Meng L., Dong L., Zhao S., Lan X., et al. (2017). Prevalence, Antimicrobial Susceptibility, and Molecular Characterization of Staphylococcus Aureus Isolated From Dairy Herds in Northern China. J. Dairy Sci. 100, 8796–8803. doi: 10.3168/jds.2017-13370
Lopardo H. A., Vidal P., Jeric P., Centron D., Paganini H., Facklam R. R., et al. (2003). Six-Month Multicenter Study on Invasive Infections Due to Group B Streptococci in Argentina. J. Clin. Microbiol. 41, 4688–4694. doi: 10.1128/JCM.41.10.4688-4694.2003
Manning S. D. (2014). Emergence of a Hypervirulent Neonatal Pathogen. Lancet Infect. Dis. 14, 1028–1030. doi: 10.1016/S1473-3099(14)70938-7
Margarit I., Rinaudo C. D., Galeotti C. L., Maione D., Ghezzo C., Buttazzoni E., et al. (2009). Preventing Bacterial Infections With Pilus-Based Vaccines: The Group B Streptococcus Paradigm. J. Infect. Dis. 199, 108–115. doi: 10.1086/595564
Martins E. R., Andreu A., Melo-Cristino J., Ramirez M. (2013). Distribution of Pilus Islands in Streptococcus Agalactiae That Cause Human Infections: Insights Into Evolution and Implication for Vaccine Development. Clin. Vaccine Immunol. 20, 313–316. doi: 10.1128/CVI.00529-12
Martins E. R., Melo-Cristino J., Ramirez M. (2010). Evidence for Rare Capsular Switching in Streptococcus Agalactiae. J. Bacteriol. 192, 1361–1369. doi: 10.1128/JB.01130-09
Merl K., Abdulmawjood A., Lämmler C., Zschöck M. (2003). Determination of Epidemiological Relationships of Streptococcus Agalactiae Isolated From Bovine Mastitis. FEMS Microbiol. Lett. 226, 87–92. doi: 10.1016/S0378-1097(03)00564-0
Mian G. F., Godoy D. T., Leal C. A. G., Yuhara T. Y., Costa G. M., Figueiredo H. C. P. (2009). Aspects of the Natural History and Virulence of S. Agalactiae Infection in Nile Tilapia. Vet. Microbiol. 136, 180–183. doi: 10.1016/j.vetmic.2008.10.016
Minst K., Märtlbauer E., Miller T., Meyer C. (2012). Short Communication: Streptococcus Species Isolated From Mastitis Milk Samples in Germany and Their Resistance to Antimicrobial Agents. J. Dairy Sci. 95, 6957–6962. doi: 10.3168/jds.2012-5852
Morach M., Stephan R., Schmitt S., Ewers C., Zschöck M., Reyes-Velez J., et al. (2018). Population Structure and Virulence Gene Profiles of Streptococcus Agalactiae Collected From Different Hosts Worldwide. Eur. J. Clin. Microbiol. Infect. Dis. 37, 527–536. doi: 10.1007/s10096-017-3146-x
National Mastitis Council (2017). Laboratory Handbook on Bovine Mastitis. 3rd ed (New Prague, MN, USA: National Mastitis Council).
Nocard N., Mollereau R. (1887). Sur Une Mammite Contagieuse Des Vaches Laitieres. Ann. Inst. Pasteur. 1, 109–126.
OIE - World Organization for Animal Health (2016) Annual Report on the Use of Antimicrobial Agents in Animals (First Report). Available at: https://www.oie.int/fileadmin/Home/eng/Our_scientific_expertise/docs/pdf/AMR/Survey_on_monitoring_antimicrobial_agents_Dec2016.pdf.
OIE - World Organization for Animal Health (2018) Annual Report on Antimicrobial Agents Intended for Use in Animals (Third Report). Available at: https://rr-africa.oie.int/wp-content/uploads/2019/09/annual_report_amr_3.pdf.
Otaguiri E. S., Morguette A. E. B., Tavares E. R., Dos Santos P. M. C., Morey A. T., Cardoso J. D., et al. (2013). Commensal Streptococcus Agalactiae Isolated From Patients Seen At University Hospital of Londrina, Paraná, Brazil: Capsular Types, Genotyping, Antimicrobial Susceptibility and Virulence Determinants. BMC Microbiol. 13, 297. doi: 10.1186/1471-2180-13-297
Pang M., Sun L., He T., Bao H., Zhang L., Zhou Y., et al. (2017). Molecular and Virulence Characterization of Highly Prevalent Streptococcus Agalactiae Circulated in Bovine Dairy Herds. Vet. Res. 48, 1–12. doi: 10.1186/s13567-017-0461-2
Pinto T. C. A., Costa N. S., Corrêa A. B., de A., de Oliveira I. C. M., de Mattos M. C., et al. (2014). Conjugative Transfer of Resistance Determinants Among Human and Bovine Streptococcus Agalactiae. Braz. J. Microbiol. 45, 785–789. doi: 10.1590/S1517-83822014000300004
Pol M., Ruegg P. L. (2007). Relationship Between Antimicrobial Drug Usage and Antimicrobial Susceptibility of Gram-Positive Mastitis Pathogens. J. Dairy Sci. 90, 262–273. doi: 10.3168/jds.S0022-0302(07)72627-9
Poyart C., Jardy L., Quesne G., Berche P., Trieu-Cuot P. (2003). Genetic Basis of Antibiotic Resistance in Streptococcus Agalactiae Strains Isolated in a French Hospital. Antimicrob. Agents Chemother. 47, 794–797. doi: 10.1128/AAC.47.2.794-797.2003
Radtke A., Bruheim T., Afset J. E., Bergh K. (2012). Multiple-Locus Variant-Repeat Assay (MLVA) is a Useful Tool for Molecular Epidemiologic Analysis of Streptococcus Agalactiae Strains Causing Bovine Mastitis. Vet. Microbiol. 157, 398–404. doi: 10.1016/j.vetmic.2011.12.034
Rajala-Schultz P. J., Smith K. L., Hogan J. S., Love B. C. (2004). Antimicrobial Susceptibility of Mastitis Pathogens From First Lactation and Older Cows. Vet. Microbiol. 102, 33–42. doi: 10.1016/j.vetmic.2004.04.010
Ramírez N. F., Keefe G., Dohoo I., Sánchez J., Arroyave O., Cerón J., et al. (2014). Herd- and Cow-Level Risk Factors Associated With Subclinical Mastitis in Dairy Farms From the High Plains of the Northern Antioquia, Colombia. J. Dairy Sci. 97, 4141–4150. doi: 10.3168/jds.2013-6815
Randis T. M., Gelber S. E., Hooven T. A., Abellar R. G., Akabas L. H., Lewis E. L., et al. (2014). Group B Streptococcus β-Hemolysin/Cytolysin Breaches Maternal-Fetal Barriers to Cause Preterm Birth and Intrauterine Fetal Demise In Vivo. J. Infect. Dis. 210, 265–273. doi: 10.1093/infdis/jiu067
Rato M. G., Bexiga R., Florindo C., Cavaco L. M., Vilela C. L., Santos-Sanches I. (2013). Antimicrobial Resistance and Molecular Epidemiology of Streptococci From Bovine Mastitis. Vet. Microbiol. 161, 286–294. doi: 10.1016/j.vetmic.2012.07.043
Reiß A., Braun J. S., Jäger K., Freyer D., Laube G., Bührer C., et al. (2011). Bacterial Pore-Forming Cytolysins Induce Neuronal Damage in a Rat Model of Neonatal Meningitis. J. Infect. Dis. 203, 393–400. doi: 10.1093/infdis/jiq047
Reyes J., Chaffer M., Sanchez J., Torres G., Macias D., Jaramillo M., et al. (2015). Evaluation of the Efficacy of Intramuscular Versus Intramammary Treatment of Subclinical Streptococcus Agalactiae Mastitis in Dairy Cows in Colombia. J. Dairy Sci. 98, 5294–5303. doi: 10.3168/jds.2014-9199
Rubio-López V., Valdezate S., Álvarez D., Villalán P., Medina M. J., Salcedo C., et al. (2012). Molecular Epidemiology, Antimicrobial Susceptibilities and Resistance Mechanisms of Streptococcus Pyogenes Isolates Resistant to Erythromycin and Tetracycline in Spain, (1994-2006). BMC Microbiol. 12, 215. doi: 10.1186/1471-2180-12-215
Schwarz S., Silley P., Simjee S., Woodford N., van duijkeren E., Johnson A. P., et al. (2010). Editorial: Assessing the Antimicrobial Susceptibility of Bacteria Obtained From Animals. J. Antimicrob. Chemother. 65, 601–604. doi: 10.1093/jac/dkq037
Skoff T. H., Farley M. M., Petit S., Craig A. S., Schaffner W., Gershman K., et al. (2009). Increasing Burden of Invasive Group B Streptococcal Disease in Nonpregnant Adults 1990-2007. Clin. Infect. Dis. 49, 85–92. doi: 10.1086/599369
Slotved H. C., Kong F., Lambertsen L., Sauer S., Gilbert G. L. (2007). Serotype IX, a Proposed New Streptococcus Agalactiae Serotype. J. Clin. Microbiol. 45, 2929–2936. doi: 10.1128/JCM.00117-07
Smith T. C., Roehl S. A., Pillai P., Li S., Marrs C. F., Foxman B. (2007). Distribution of Novel and Previously Investigated Virulence Genes in Colonizing and Invasive Isolates of Streptococcus Agalactiae. Epidemiol. Infect. 135, 1046–1054. doi: 10.1017/S0950268806007515
Sweeney M. T., Lubbers B. V., Schwarz S., Watts J. L. (2018). Applying Definitions for Multidrug Resistance, Extensive Drug Resistance and Pandrug Resistance to Clinically Significant Livestock and Companion Animal Bacterial Pathogens. J. Antimicrob. Chemother. 73, 1460–1463. doi: 10.1093/jac/dky043
Tazi A., Disson O., Bellais S., Bouaboud A., Dmytruk N., Dramsi S., et al. (2010). The Surface Protein HvgA Mediates Group B Streptococcus Hypervirulence and Meningeal Tropism in Neonates. J. Exp. Med. 207, 2313–2322. doi: 10.1084/jem.20092594
Thomas V., De Jong A., Moyaert H., Simjee S., El Garch F., Morrissey I., et al. (2015). Antimicrobial Susceptibility Monitoring of Mastitis Pathogens Isolated From Acute Cases of Clinical Mastitis in Dairy Cows Across Europe: VetPath Results. Int. J. Antimicrob. Agents 46, 13–20. doi: 10.1016/j.ijantimicag.2015.03.013
Tian X. Y., Zheng N., Han R. W., Ho H., Wang J., Wang Y. T., et al. (2019). Antimicrobial Resistance and Virulence Genes of Streptococcus Isolated From Dairy Cows With Mastitis in China. Microb. Pathog. 131, 33–39. doi: 10.1016/j.micpath.2019.03.035
Tomazi T., de Souza Filho A. F., Heinemann M. B., dos Santos M. V. (2018). Molecular Characterization and Antimicrobial Susceptibility Pattern of Streptococcus Agalactiae Isolated From Clinical Mastitis in Dairy Cattle. PloS One 13, 1–18. doi: 10.1371/journal.pone.0199561
van Duin D., Paterson D. L. (2016). Multidrug-Resistant Bacteria in the Community: Trends and Lessons Learned. Infect. Dis. Clin. North Am. 30, 377–390. doi: 10.1016/j.idc.2016.02.004
Yang Y., Liu Y., Ding Y., Yi L., Ma Z., Fan H., et al. (2013). Molecular Characterization of Streptococcus Agalactiae Isolated From Bovine Mastitis in Eastern China. PloS One 8, 1–8. doi: 10.1371/journal.pone.0067755
Zadoks R. N., González R. N., Boor K. J., Schukken Y. H. (2004). Mastitis-Causing Streptococci are Important Contributors to Bacterial Counts in Raw Bulk Tank Milk. J. Food Prot. 67, 2644–2650. doi: 10.4315/0362-028X-67.12.2644
Zhao Z., Kong F., Martinez G., Zeng X., Gottschalk M., Gilbert G. L. (2006). Molecular Serotype Identification of Streptococcus Agalactiae of Bovine Origin by Multiplex PCR-based Reverse Line Blot (mPCR/RLB) Hybridization Assay. FEMS Microbiol. Lett. 263, 236–239. doi: 10.1111/j.1574-6968.2006.00428.x
Keywords: Streptococcus agalactiae, virulence, dairy cattle mastitis, multidrug resistance, serotypes
Citation: Hernandez L, Bottini E, Cadona J, Cacciato C, Monteavaro C, Bustamante A and Sanso AM (2021) Multidrug Resistance and Molecular Characterization of Streptococcus agalactiae Isolates From Dairy Cattle With Mastitis. Front. Cell. Infect. Microbiol. 11:647324. doi: 10.3389/fcimb.2021.647324
Received: 29 December 2020; Accepted: 09 April 2021;
Published: 30 April 2021.
Edited by:
Rodnei Dennis Rossoni, Sao Paulo State University, BrazilReviewed by:
Geraldo Costa, Universidade Federal de Lavras, BrazilCopyright © 2021 Hernandez, Bottini, Cadona, Cacciato, Monteavaro, Bustamante and Sanso. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Andrea Mariel Sanso, bXNhbnNvQHZldC51bmljZW4uZWR1LmFy
†These authors have contributed equally to this work and share second authorship
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.