
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Cell. Infect. Microbiol. , 29 April 2021
Sec. Molecular Bacterial Pathogenesis
Volume 11 - 2021 | https://doi.org/10.3389/fcimb.2021.645248
 Lore Van Damme1
Lore Van Damme1 Natasja Cox1
Natasja Cox1 Chana Callens1
Chana Callens1 Michelle Dargatz2
Michelle Dargatz2 Monika Flügel2
Monika Flügel2 Sarah Hark2
Sarah Hark2 Frank Thiemann2
Frank Thiemann2 Stefan Pelzer2
Stefan Pelzer2 Freddy Haesebrouck1
Freddy Haesebrouck1 Richard Ducatelle1
Richard Ducatelle1 Filip Van Immerseel1*
Filip Van Immerseel1* Evy Goossens1
Evy Goossens1Extracellular matrix (ECM) degrading enzymes produced by Clostridium perfringens may play an important role during the initial phases of avian necrotic enteritis by facilitating toxin entry in the intestinal mucosa and destruction of the tissue. C. perfringens is known to produce several ECM-degrading proteases, such as kappa toxin, an extracellular collagenase that is encoded by the colA gene. In this study, the colA gene sequence of a collection of 48 C. perfringens strains, including pathogenic (i.e. toxinotype G) and commensal (i.e. toxinotype A) chicken derived strains and strains originating from other host species, was analyzed. Although the colA gene showed a high level of conservation (>96% nucleotide sequence identity), several gene variants carrying different nonsense mutations in the colA gene were identified, leading to the definition of four truncated collagenase variant types (I-IV). Collagenase variant types I, III and IV have a (nearly) complete collagenase unit but lack parts of the C-terminal recruitment domains, whereas collagenase variant types II misses the N-terminal part of collagenase unit. Gene fragments encoding a truncated collagenase were mainly linked with necrotic enteritis associated C. perfringens type G strains with collagenase variant types I and II being the most prevalent types. Gelatin zymography revealed that both recombinant full-length and variant type I collagenase have active auto-cleavage products. Moreover, both recombinant fragments were capable of degrading type I as well as type IV collagen, although variant type I collagenase showed a higher relative activity against collagen type IV as compared to full-length collagenase. Consequently, these smaller truncated collagenases might be able to break down collagen type IV in the epithelial basement membrane of the intestinal villi and so contribute to the initiation of the pathological process leading to necrotic enteritis.
Collagens are major glycoproteins of the extracellular matrix and play an important role in maintaining the biological and structural integrity of various tissues and organs in both humans and animals (Rozario and DeSimone, 2010). Being ubiquitously present, collagens are attractive targets for various microorganisms to induce tissue destruction (Bober et al., 2010; Singh et al., 2012). Indeed, many different bacterial pathogens, including members of the Clostridium genus, have been shown to produce collagenolytic enzymes (Harrington, 1996; Duarte et al., 2016). By digesting the collagen-rich extracellular matrix (ECM), pathogens can facilitate their own colonization and spread through the host tissues (Matsushita and Okabe, 2001; Duarte et al., 2016). Additionally, digestion of ECM compounds contributes to the availability of amino acids for the pathogen’s survival and growth and can also favor toxin diffusion and tissue damage (Kalisz, 1988; Harrington, 1996; Singh et al., 2012; Tomlin and Piccinini, 2018).
Clostridium (C.) perfringens is one of the collagenase-producing Clostridia and is known to cause a variety of histotoxic and enterotoxic diseases in both humans and animals (Niilo, 1980; Uzal et al., 2010). Although toxicity of C. perfringens is mostly dependent on the production of specific toxins, extracellular proteolytic enzymes are thought to assist in the development of some diseases. C. perfringens has been shown to produce multiple gelatinolytic enzymes with molecular masses ranging from ~80 to approximately ~120 kDa (Kameyama and Akama, 1971). The 80 kDa collagenase, which was formerly designated as kappa toxin, is lethal for mice upon intravenous injection and has both hemorrhagic and dermonecrotic activities upon subcutaneous injection in guinea pigs (Kameyama and Akama, 1971). Since the mid-90s, the focus of scientific research has shifted from the 80 to the 120 kDa collagenase and the term kappa toxin was revised, now denoting the ~120 kDa enzyme (Obana et al., 2010; Obana et al., 2013). This enzyme is encoded by the chromosomal colA gene and is synthesized as a precursor enzyme (pre-pro-collagenase; 126 kDa) with an N-terminal stretch of 86 amino acids containing a putative signal sequence and a pro-region. Within this pro-region, a collagenase target sequence is present, suggesting that self-processing is involved in maturation of the collagenase enzyme (Kameyama and Akama, 1971). Removal of the signal peptide and pro-sequence generates a mature extracellular collagenase (116 kDa) composed of a collagen binding module, a polycystic kidney disease-like (PKD-like) domain and a collagenase module, which contains a zinc-binding sequence (HEXXH) at the center of the active site (Matsushita et al., 1994). The C. perfringens collagenases have been studied mostly with respect to their role in the pathogenesis of gangrene. Little research, however, has been conducted on the role of the clostridial collagenases in intestinal disease.
Necrotic Enteritis (NE) is an enteric disease of broiler chickens which is caused by avian-specific, NetB toxin producing strains of C. perfringens (i.e. toxinotype G strains) (Long et al., 1974; Rood et al., 2018). The secreted pore-forming NetB toxin is a key virulence factor in the development of necrotic ulcers which are typical for NE. Yet, where and how NetB initiates this process is not fully understood (Keyburn et al., 2008; Rood et al., 2016). A recent study focusing on the gene expression during colonization of the chicken intestinal tract by C. perfringens showed that NetB expression is not upregulated during the early stages of disease. It is currently believed that the toxin targets deeper layers of the intestinal mucosa rather than superficial structures (Parreira et al., 2016). As a consequence, initial breakdown or permeabilization of the intestinal mucosa might be needed in order for NetB to exert its action. This permeabilization might be caused by a combination of factors such as agents causing epithelial damage [e.g. Eimeria spp. (Al-Sheikhly and Al-Saieg, 1980) or mycotoxins (Ren et al., 2019)], either or not in combination with collagenolytic enzymes which could affect the connective tissue and basement membrane in the intestinal mucosal layer, and thus initiate lesions (Olkowski et al., 2006; Olkowski et al., 2008). C. perfringens strains isolated from field cases of NE secrete several potent collagenolytic enzymes and the expression of the colA gene is upregulated during the early stages of NE (Olkowski et al., 2008; Parreira et al., 2016), suggesting a potential role in the initial stages of the pathogenesis of NE.
The aim of the present study was to elucidate whether kappa toxin (~120 kDa collagenase encoded by colA) from avian pathogenic (NetB positive) C. perfringens strains (i.e. toxinotype G) differs from that of commensal (NetB negative) chicken-associated C. perfringens strains (i.e. toxinotype A) both in its sequence and function.
The colA gene sequences of 25 C. perfringens strains originating from chickens and 23 strains from non-chicken origin were used in this study (Table 1). This collection contains strains which were derived from a range of international locations and host disease statuses. From these strains, a selection of 21 chicken-associated C. perfringens strains were cultured, either for DNA extraction or for functionality assays (Table 1). C. perfringens strains were routinely grown on Columbia agar (Oxoid) plates supplemented with 5% defibrinated sheep blood or in Brain Heart Infusion broth (BHI, Oxoid, Basingstoke, UK) under anaerobic conditions at 37°C.
Escherichia (E.) coli TOP10 cells were used as the cloning host for recombinant expression of collagenase and were routinely grown on LB medium (Fisher Scientific, Merelbeke, Belgium) supplemented with 100 μg/ml of ampicillin. Terrific Broth (24% Tryptone, 42% Yeast extract, 4% glycerol, 0.72 M Na2HPO4, 0.16 M NaH2PO4) was used for recombinant protein production.
Growth curves were performed for a subset of 9 chicken-associated C. perfringens strains (Table 1). Overnight grown cultures were diluted 1/10,000 in fresh BHI broth and incubated anaerobically at 37°C. Bacterial growth was determined by measuring the optical density at 600 nm (Genesys 10S UV‐vis spectrophotometer, Thermo Scientific, Waltham, MA, USA) at 0, 1, 2, 3, 4, 5, 6 and 24 h after inoculation. At each timepoint, 1 mL aliquots from each culture were collected to obtain cell-free supernatants by centrifugation of the samples, followed by filtration of the supernatants through a 0.2 µm filter and stored at -20°C, until further use in zymography (see below).
Supernatant aliquots of 10 µL were mixed with 2 × loading buffer (0.5 M Tris–HCl pH 6.8, 20% glycerol, 4% SDS, a pinch of bromophenol blue) and separated on 8% SDS page containing 0.1% gelatin under non-reducing conditions. After separation, the gel was incubated with renaturing buffer (2.5% Triton X-100, 30 min, room temperature) to remove SDS from the gel. This allows the separated enzymes in the gel to renature and restores enzymatic activity. Subsequently, the gel was washed with developing buffer (150 mM NaCl, 5 mM CaCl2, 0.05% NaN3 and 50 mM Tris–HCl buffer pH 7.5) and incubated with fresh developing buffer under continuous shaking at 37°C for 18 h. Afterwards, the gel was stained for 1h with Coomassie brilliant blue G-250 (Sigma-Aldrich, Bornem, Belgium) and destained for 20 minutes with destaining solution [40% methanol (v/v), 10% acetic acid (v/v)]. Activity of gelatin-degrading enzymes is visible as clear colourless bands against a blue background. Gels were scanned using a GS-800 calibrated densitometer and the intensities of the gelatinolytic bands were determined using the Quantity One software (BioRad, Hercules, CA, USA). Values were normalized against the background and expressed as arbitrary units
EHE-NE18 colA sequence was obtained from NCBI and a BLASTn search was performed for C. perfringens (tax ID: 1502) genomes across all annotated strains. ColA gene sequences of 28 C. perfringens strains (including 5 chicken-associated and 23 non-chicken-associated C. perfringens strains) were acquired from the NCBI genome database (Table 1).
The colA gene of a set of 20 chicken-associated C. perfringens strains from which no whole genome sequencing data was publicly available, was amplified by PCR and sequenced by Sanger sequencing (Table 1). Genomic DNA was extracted using alkaline lysis as previously described (Scheirlinck et al., 2007). Amplification of the colA genes was performed in a total reaction volume of 50 μl containing 0.5 µL dNTP Mix (100 mM), 2 μL DNA template, 1 µL Velocity DNA Polymerase, 1× Hi-Fi Buffer (Bioline, London, UK) and 0.4 μM of primers (Figure 1). Thermal cycling conditions: initial denaturing step of 98°C for 2 min, followed by 35 amplification cycles of 98°C for 30 s, 50°C for 1 min, and 72°C for 3 minutes and a final extension step of 72°C for 10 min. Presence of a single PCR band was verified by electrophoresis on 1.5% agarose gels. Sanger sequencing was performed by Eurofins Genomics (Ebersberg, Germany). As a single Sanger sequencing run generates a read of up to 1000 basepairs and the colA nucleotide sequence is ±3315 bp long, multiple internal sequencing primers were designed to make sure the entire gene sequence was covered. Design of all primers was based on a multiple sequence alignment of the publicly available colA sequences. All primers used in this work are shown in Figure 1. Nucleotide sequences are available in the GenBank sequence database under accession numbers MW393527 to MW393546.
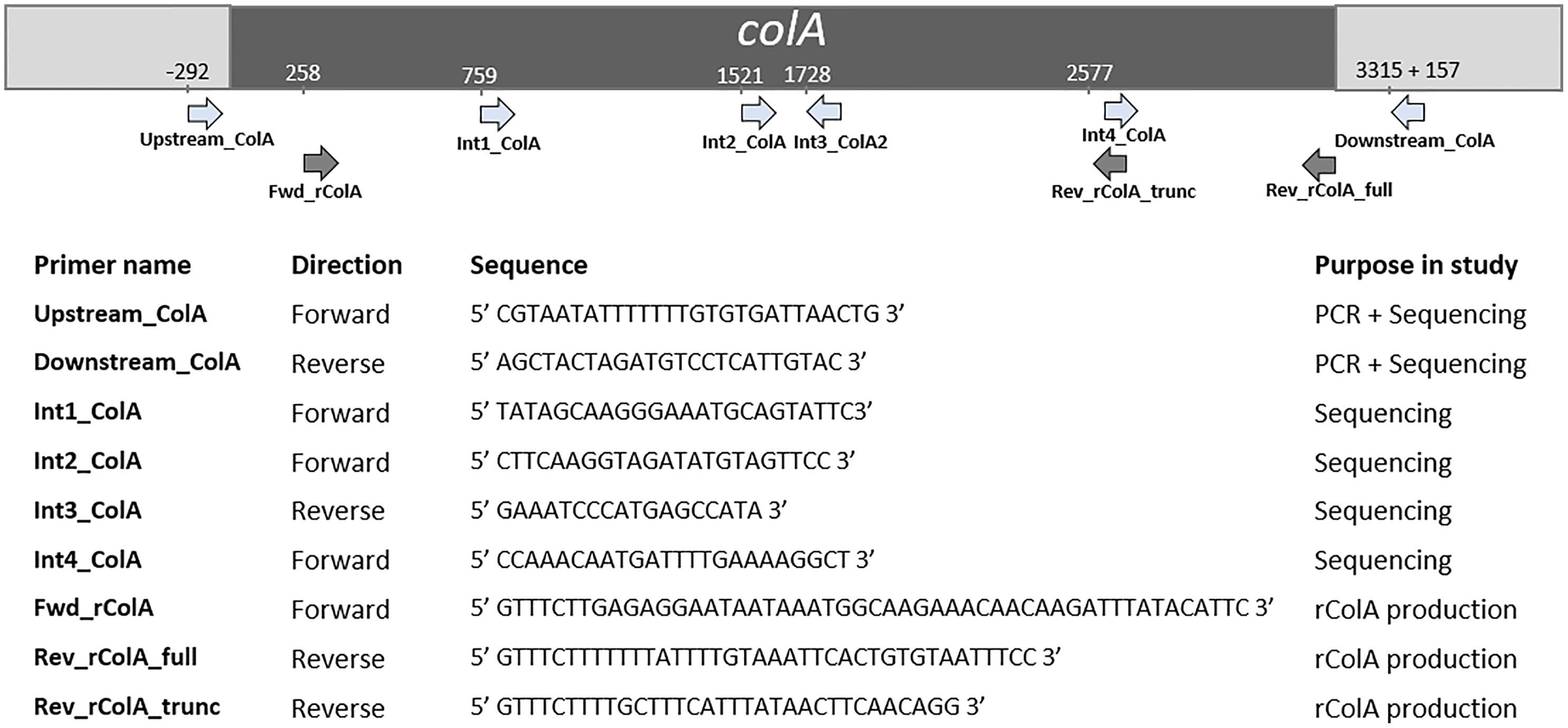
Figure 1 Primers used in this study and their relative position in colA. Positions are based on C. perfringens strain EHE-NE18 colA sequence.
The amino acid sequences encoded by the colA gene of 48 C. perfringens strains (Table 1) were deduced bioinformatically using the ExPASy Translate tool (https://web.expasy.org/translate). The domain architecture of the deduced protein fragments was retrieved by performing an NCBI Conserved Domain search and the molecular weights of the mature collagenase fragments (i.e. excluding the pre-pro domains) were calculated using the ExPASY pI/MW tool (https://web.expasy.org/compute_pi). Nucleotide and deduced amino acid sequence data were aligned by the ClustalW alignment algorithm using Mega version X and the alignment was visualized by BOXSHADE (Kumar et al., 2018). Dendrograms of full-length sequences of the colA genes (including both fragments when nonsense mutations leading to premature stop codons are present) were constructed by multiple alignment-based similarity coefficients and UPGMA (unweighted pair group method with arithmetic mean) cluster analysis using BioNumerics version 7.6.3 (Applied Maths, Sint-Martens-Latem, Belgium).
Full-length and truncated colA gene products were expressed in E. coli TOP10 cells using the pBAD TOPO® TA Expression Kit (Invitrogen, Paisley, UK). The desired fragments were amplified from the DNA of C. perfringens strains CP56 and CP24 (for respectively the full-length and truncated colA gene) by PCR using a DNA polymerase with proofreading activity (Velocity DNA Polymerase, Bioline, Brussels, Belgium). The forward primer (FWD_rColA, Figure 1) contained an in-frame stop codon and translation re-initiation sequence to remove the N-terminal leader sequence and allow native protein expression. The reverse primers (REV_rColA_full or REV_rColA_trunc, Figure 1) excluded the native colA gene stop codon and included the C-terminal V5 epitope and poly-histidine region for affinity purification. Amplifications were performed in a total reaction volume of 50 μl containing 0.5 µL dNTP Mix (100 mM), 2 μL DNA template, 1 µL Velocity DNA Polymerase, 1× Hi-Fi Buffer (Bioline) and 0.4 μM of primers. PCR conditions used for the amplification were an initial denaturation at 95°C for 4 min followed by 35 cycles of denaturation at 95°C for 1.5 min, annealing at 53.8°C for 30 s and extension at 72°C for 2 min, and a final extension step at 72°C for 10 min. The resulting PCR product was incubated with Taq polymerase for 10 min at 72°C (5 U; Promega, Madison, WI, USA) to add 3’ A-overhangs, cloned into the pBAD-TOPO expression vector (Invitrogen, Merelbeke, Belgium), and transformed into One Shot TOP10® E. coli (Invitrogen) according to the manufacturer’s instructions. The correct orientation of inserts was verified by Sanger sequencing.
Recombinant E. coli carrying the pBAD-collagenase vectors were grown in Terrific Broth supplemented with 100 µg/mL ampicillin at 37°C until an OD600 of 0.5 was reached. Expression of the recombinant full-length and truncated rColA was induced overnight with 0.2% L-arabinose (w/v). Bacteria were harvested by centrifugation and lysed enzymatically using BugBuster (Invitrogen, Merelbeke, Belgium). Recombinant proteins were purified on a Ni-sepharose column (His Gravitrap, GE Healthcare Bio-Sciences AB, Uppsala, Sweden) according to the manufacturer’s instructions. Subsequently, proteins were dialyzed against PBS, purity was analysed using SDS-PAGE, and protein concentration was measured using BCA protein assay (Fisher Scientific). Activity of the recombinant proteins was analysed by gelatin zymography (0.5 µg protein load) as described above.
The Molecular Probes EnzChek® Gelatinase/Collagenase Assay Kit was used to evaluate the activity of the recombinant collagenase fragments using DQ™ Gelatin (proxy for total collagenolytic activity), DQ™ Collagen I (activity against fibrillar collagen) and DQ™ Collagen IV (activity against non-fibrillar collagen). Different recombinant enzyme concentrations (5 µg/mL, 1 µg/mL or 0.1 µg/mL) were used to avoid effects of enzyme-substrate interactions. In a 96-well format, 50 µL of each recombinant sample was mixed with 130 μL of reaction buffer (0.5 M Tris–HCl, 1.5 M NaCl, 50 mM CaCl2 and 2 mM sodium azide at pH 7.6) and 20 µL of either fluorescein labelled substrate (DQ Collagen I (25 μg/mL), DQ Collagen IV (25 μg/mL) or DQ Gelatin (12.5 μg/mL)). Samples without enzyme activity served as control for auto-fluorescence. Reactions were incubated for 60 min at room temperature in the absence of light, and fluorescence was measured every 5 min (excitation 485 nm, emission 527 nm; Fluoroskan Ascent Fluorometer, Thermo Fisher Scientific). Activity of the recombinant collagenase fragments against each substrate was calculated through the slope of the linear phase (first 30 minutes) of the fluorescence resulting from the cleavage of the respective fluorescein labelled substrate over time
Subsequently, the activity of each recombinant fragment against collagen type I or IV was normalized against its total collagenolytic activity and the ratio
was calculated. One-sample t-tests were performed to evaluate whether the ratios differed significantly from 1, i.e. the ratio value that corresponds with an equal activity of both collagenase variant I and full-length collagenase. All statistical analyses were performed using GraphPad Prism software (version 5.03, San Diego, CA, USA).
To determine whether pathogenic C. perfringens type G strains harbor different collagenolytic properties as compared to commensal C. perfringens type A strains, the in vitro collagenase production of two type G strains with proven pathogenic activity (CP56 and EHE-NE18) was compared to two commensal C. perfringens type A isolates (JIR4857; proven to be non-pathogenic and CP24; isolated from a healthy chicken) (Gholamiandekhordi et al., 2006; Gharib-Naseri et al., 2019; Van Damme et al., 2020). Both C. perfringens type A strains showed 1 major band of activity at ± 116 kDa and 3 minor bands with molecular masses between 75 and 110 kDa. Remarkably, a different pattern was seen for the C. perfringens type G strains. For both of these strains a major band of molecular size 80-90 kDa and a fainter band of ±75 kDa was observed (Figure 2). The 116 kDa collagenase band, which is the predicted height of the colA gene product was absent in both tested pathogenic type G strains.
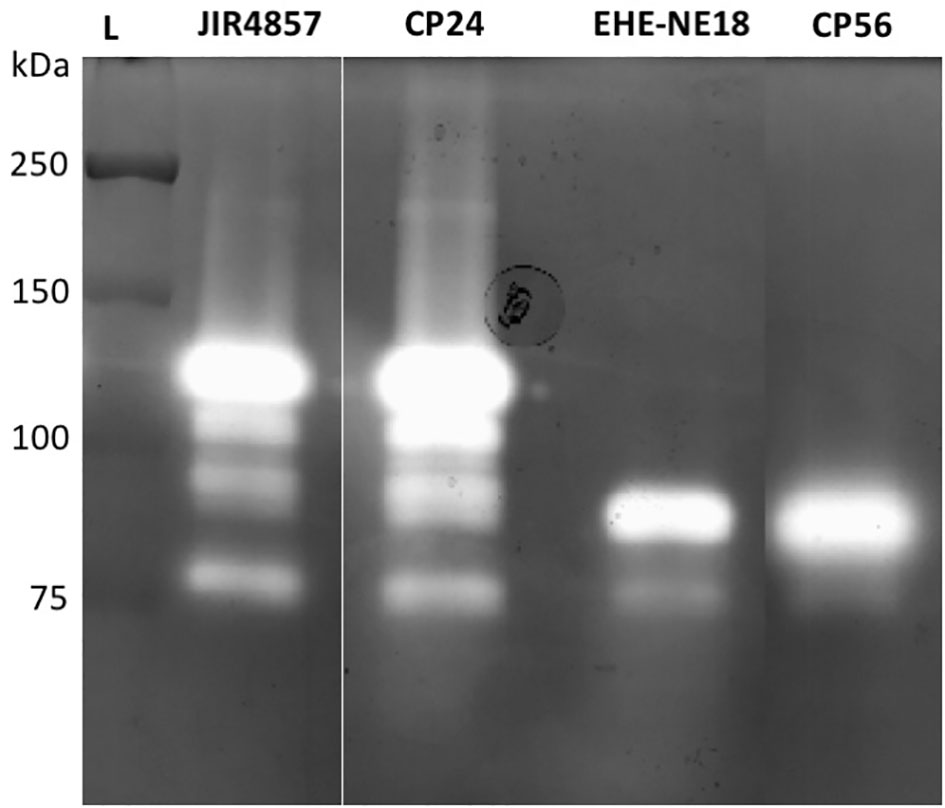
Figure 2 Gelatinolytic activities expressed by different C. perfringens strains. Supernatant samples of 2 C. perfringens type A strains (JIR4857, CP24) and 2 C. perfringens type G strains (EHE-NE18, CP56) were analyzed for proteolytic activity by gelatin zymography. Lane L, Protein Ladder (Precision Plus Protein All Blue Pre-stained Protein Standards, Biorad).
To explain the appearance of these different gelatinolytic profiles, the sequences of the colA gene of a collection of 48 C. perfringens strains were compared. Overall, the colA sequences were highly similar at nucleic acid level (> 96.6% sequence identity). Variations due to deletions, insertions and single-nucleotide polymorphisms were observed in 259 out of the 3316 nucleotide positions. Comparative analysis of the deduced amino acid sequences demonstrated that there were multiple variants containing nonsense mutations leading to the expression of truncated collagenase fragments (Supplementary Figure S1). Genetic variants were classified in different collagenase variant types (I to IV) based on the location of the nonsense mutation and the predicted protein domain structure of the corresponding protein fragments (Figure 3 and Table 1).
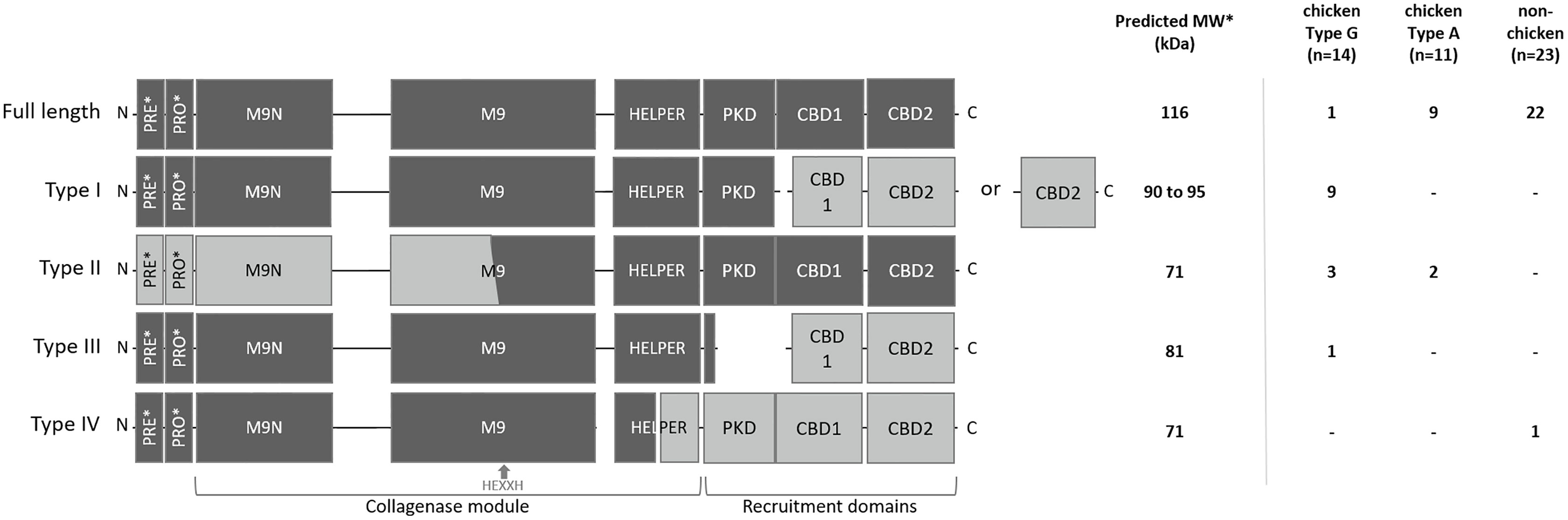
Figure 3 Schematic representation and distribution of collagenase variants. Schematic representation of the domain architecture of full-length and truncated collagenase types and their distribution among chicken (type G and type A strains) and non-chicken derived C. perfringens strains (respectively n=14, n=11 and n=23). PRE = signal sequence, PRO = pro-domain, M9N = activator domain, M9 = peptidase domain, HELPER = peptidase helper domain, PKD = polycystic kidney disease-like domain and CBD = collagen binding domains. Collagenase variant types are encoded by gene variants carrying a premature stop mutation in the colA gene which results in the occurrence of 2 protein-coding regions. Predicted active fragment containing the HEXXH motif is represented in dark grey, while the other fragment which is expected to be inactive is represented in light grey. *PRE & PRO domains are removed upon maturation. Molecular weight of the predicted mature active fragment is given.
The full-length collagenase is synthesized as a multimodular pre-pro-protein (predicted MW ~ 126 kDa) which is subsequently processed to its mature extracellular form (~ 116 kDa). This mature form consists of a collagenase module (containing an M9N activator domain, an M9 peptidase domain with an HEXXH zinc-binding motif and a helper domain), a polycystic kidney disease-like (PKD-like) domain and 2 collagen-binding domains (CBD1 and CBD2) (Kameyama and Akama, 1971). Full-length collagenase sequences were present in 95.65% of the non-chicken derived C. perfringens strains and 81.82% of the chicken-derived C. perfringens type A strains. By contrast, only 7.14% of the chicken-derived C. perfringens type G strains encode the full-length protein (Figure 3).
All truncated collagenase variant types are encoded by gene variants carrying a premature stop mutation in the colA gene which results in the occurrence of 2 coding sequences. For each of these variant types, the first coding sequence corresponds to the truncated protein at the N-terminal part of the full-length protein, which encodes a signal-sequence, while the second coding sequence corresponds to the truncated protein at the C-terminal part of the full-length protein, which lacks any secretion signal (Figure 3). As a consequence, only the fragment encoded by the first coding sequence will be secreted. For collagenase variant type I, this results in an active secreted fragment that contains a complete collagenase module and PKD domain yet lacks the 2 collagen binding domains. Depending on the location of the premature stop codon, two subtypes (e.g. type IA and IB) can be described which both encode a similar active collagenase protein (type I variant). The main difference between these subtypes is found in the second gene fragment, which encodes either both CBD domains (variant type IA) or only a single CBD domain (variant type IB). Both type I variants are exclusively present in C. perfringens type G strains (respectively present in 35.7% and 28.6% of the type G strains) (Figure 3).
Collagenase type II variants were present in 18.2% of the chicken-derived C. perfringens type A strains and 21.4% of the type G strains and were absent in non-chicken derived C. perfringens strains. Unlike all other collagenase variant types, the 2 coding sequences of collagenase type II variants are located on overlapping open reading frames. As a result, the M9 peptidase-domain is encoded in part by the first and in part by the second coding sequence. Only the second fragment is predicted to be active as it contains the conserved HEXXH motif which is characteristic for the catalytic center of a metalloproteinase. Besides a partial, HEXXH-containing M9 peptidase domain, this fragment also contains the collagenase helper domain, the PKD-like domain and 2 CBDs, yet it lacks secretion signal sequence, indicating that the protein cannot be secreted by the bacteria (Figure 3).
The third collagenase variant type (= type III) is uniquely present in one C. perfringens type G strain. In this type, the longest sequence encodes a fragment containing the pre-pro-region, the collagenase module and a small part of the PKD-like domain, while the other coding sequence only encodes the domains which are responsible for collagen binding.
In contrast to the collagenase variant types I, II and III, which were exclusively found in chicken-derived C. perfringens strains, the fourth collagenase variant type (type IV) was unique for one non-chicken derived C. perfringens strain. The colA gene of the collagenase type IV variant encodes a first fragment containing the pre-pro-region and a part of the collagenase module (i.e. half of the peptidase helper domain is missing) and a second fragment consisting of the other half of the helper domain, the PKD-like domain and both CBDs. The former fragment is expected to be active upon maturation (Figure 3).
A UPGMA phylogenetic tree was constructed based on the colA gene sequences (i.e. full gene sequence including both fragments when nonsense mutations leading to premature stop codons are present) of a collection of 48 C. perfringens strains (Figure 4). In general, C. perfringens strains encoding the same collagenase variant type clustered together showing very similar (e.g. type IB) or even identical (e.g. type IA or type II) nucleotide sequences. In contrast, C. perfringens strains encoding the full-length collagenase were found more scattered throughout the phylogeny and branch at various depths in the phylogeny, reflecting a higher diversity between these strains.
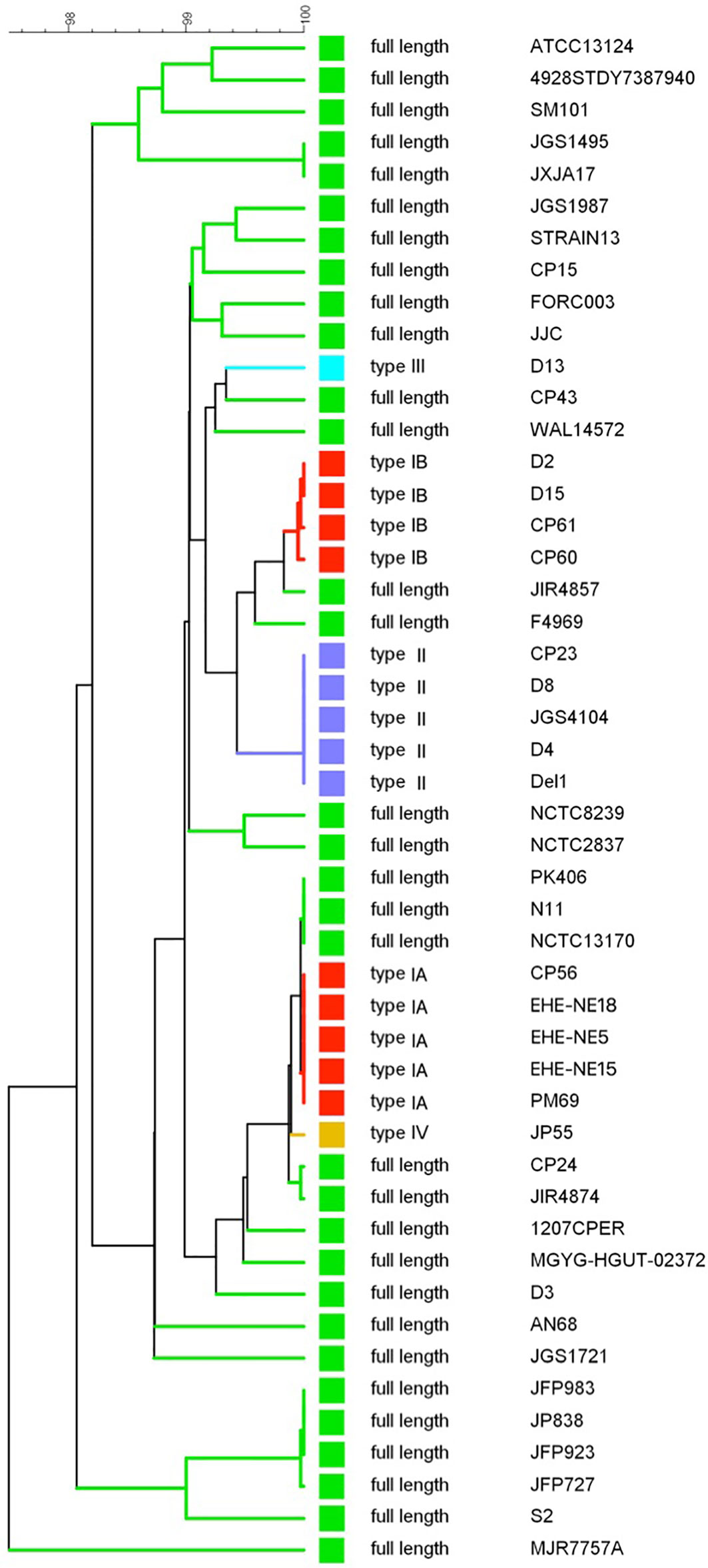
Figure 4 colA gene based dendrogram reflecting the similarity for 48 C. perfringens strains. A UPGMA phylogenetic tree was constructed based on the colA gene sequences (i.e. full gene sequences - from original start to original stop codon - including both fragments when nonsense mutations leading to premature stop codons are present). Sequence similarity percentages (top bar) are shown. Colored branches depict collagenase variant types (see Figure 3, Green, full-length; Red, collagenase type I variant; Blue, collagenase type II variant; Light blue, collagenase type III variant; Orange, collagenase type IV variant). Branch labels include the C. perfringens strains from which the colA gene sequence was derived.
All collagenase variant types were predicted to be catalytically active as they all contain an intact HEFTH (HEXXH) motif (Supplementary Figure S1). In order to verify this, a zymographic analysis was performed on supernatant samples of 9 C. perfringens strains encoding for the most common collagenase types (i.e. full-length collagenase (n=4), variant type I (n=2 for subtype IA and n=1 for subtype IB) and variant type II strains (n=2) (Table 1 and Figure 5).
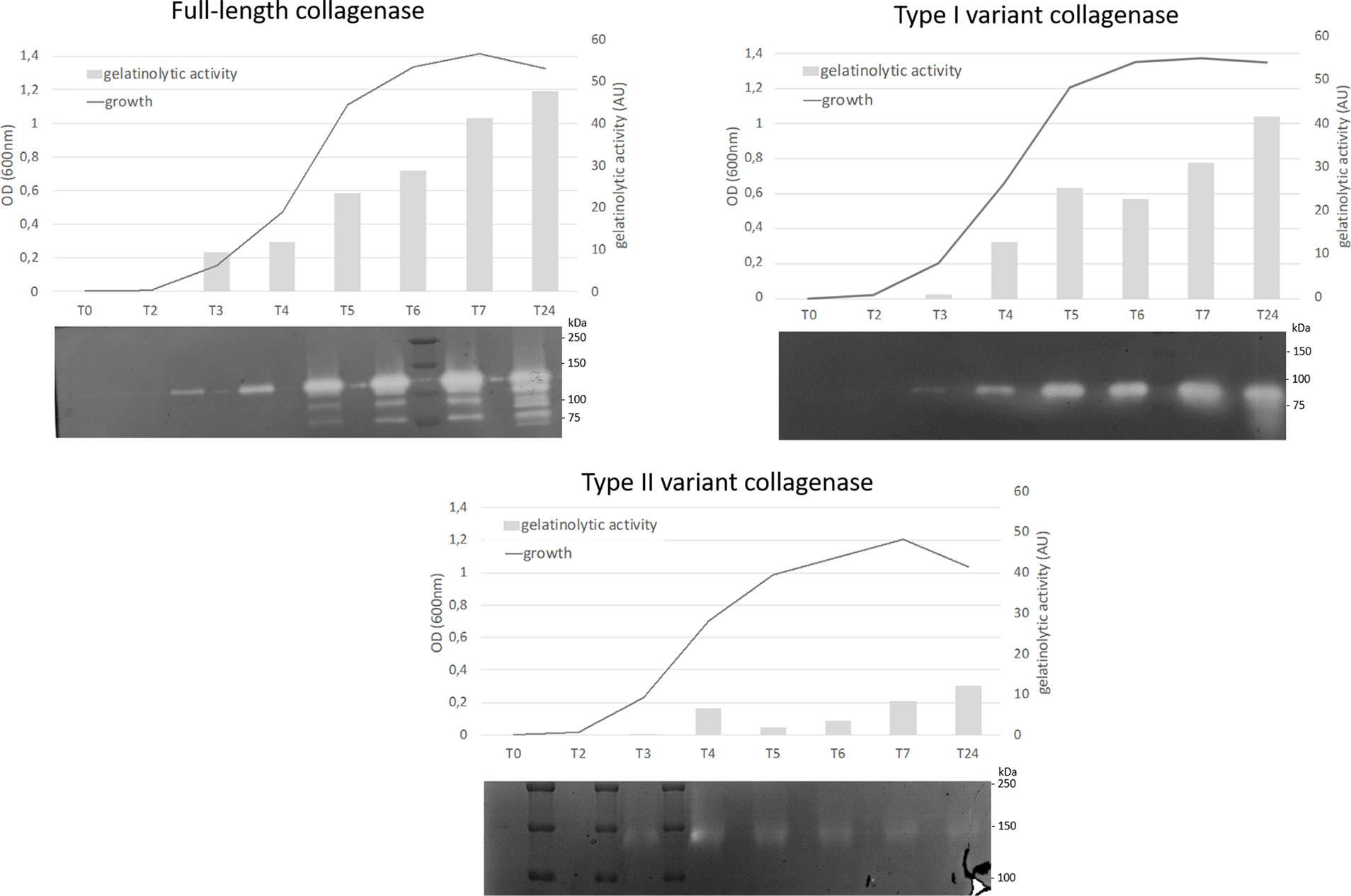
Figure 5 Analysis of proteolytic activity at different stages of growth of C. perfringens strains encoding for 3 different collagenase types. Growth was analyzed by measuring optical density at 600 nm at 8 timepoints (0, 2, 3, 4, 5, 6, 7 and 24 h post inoculation). At each timepoint supernatant samples were collected and analyzed by zymography. The observed bands were measured by Quantity One to determine the relative gelatinolytic activity. AU = arbitrary units. One representative image per collagenase variant type is shown (full-length collagenase: JIR4857, type I: CP56 and type II: D8).
All strains encoding the full-length collagenase showed proteolytic activity at the predicted molecular weight of 116 kDa in the early log phase. Later in growth, several catalytically active fragments with molecular weights ranging from 116 to approximately 80 kDa were observed. Likewise, strains encoding the collagenase type I variants, showed a single band of activity at the expected height (~ 90 kDa or ~ 95 kDa) early in growth, while on later time points one or more additional lower molecular weight band(s) (<75 kDa) became visible. In one strain, an additional band of higher molecular weight (90-110 kDa) was observed (strain S2, data not shown).
Type II variant collagenases lack a signaling peptide needed for protein secretion (described above), resulting in either no gelatinolytic activity observed in the supernatants (strain CP23, data not shown) or one very faint band of ± 140 kDa, which corresponds to the molecular weight of the dimeric form of the mature variant II collagenase fragment (monomer ~ 71 kDa) and might be released in the supernatants as the result of release of intracellular proteins from disrupted dead bacteria.
Gelatin zymography confirmed that both the full-length and type I recombinant collagenase fragments have gelatinolytic activity (Figure 6). Both proteins showed activity at the predicted height together with an active fragment at a lower molecular weight. For the full-length recombinant collagenase, both observed bands (~120 kDa and ~90-100 kDa) accounted for approximately half of the gelatinolytic activity (respectively ± 49% and ± 51%). For the type I recombinant collagenase, the predicted fragment (~94 kDa), only accounted for 26% of the activity, while the breakdown product (~75 kDa) accounted for 74% of the gelatinolytic activity, even though both fragments accounted for similar portions (respectively 27 and 33%) of the total protein detected on SDS-page.
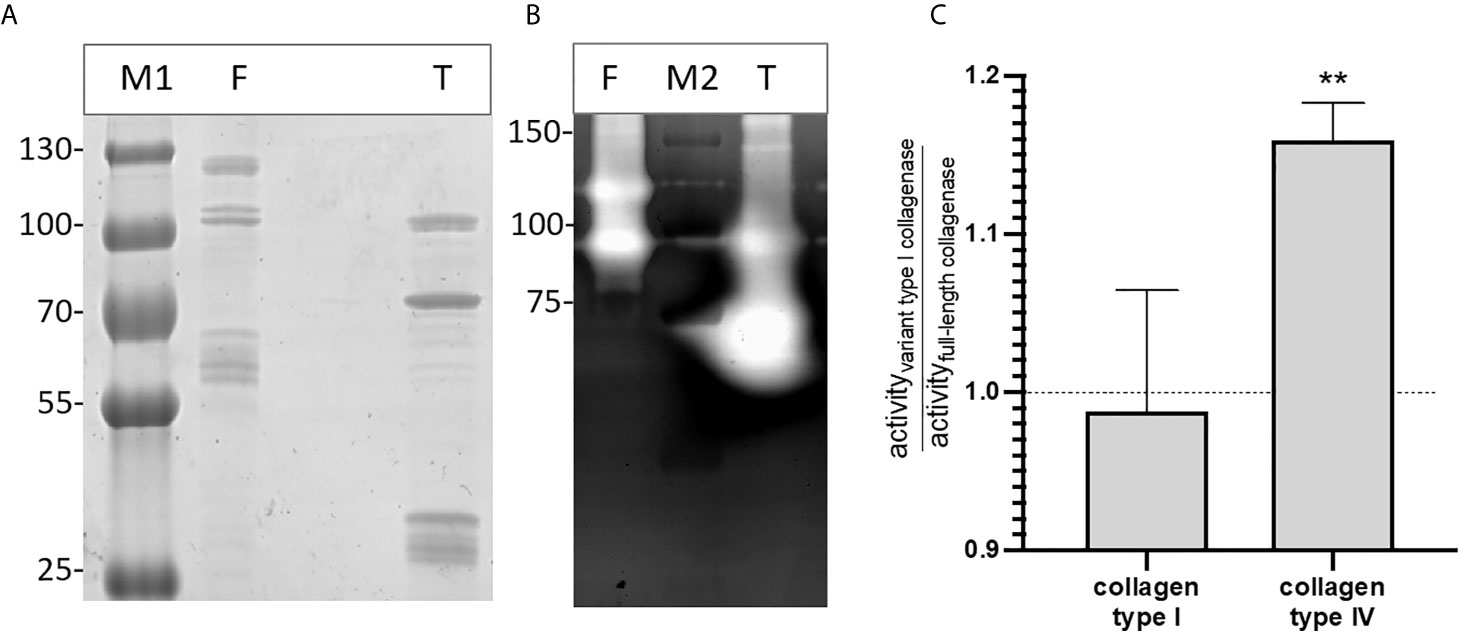
Figure 6 Recombinant full-length and variant type I collagenase have active auto-cleavage products. (A) SDS page and (B) Gelatin zymography of the recombinantly expressed collagenase fragments. F = full-length collagenase, expected fragment 120 kDa; T = Type I variant collagenase, expected fragment 94 kDa; M1= Pageruler plus pre-stained, M2 = Precision Plus protein All blue Pre-stained Protein standard. (C) Substrate-degrading activity of both the full-length collagenase and variant type I collagenase towards collagen type I or collagen type IV was measured using the EnzChek collagenase assay. The bar shows the activity of the variant type I collagenase relative to the full-length collagenase towards each of the tested substrates. Data represents mean ± standard deviations (n=3). One-sample t-tests were performed to evaluate whether the ratios differed significantly from 1, i.e. the ratio value that corresponds with an equal activity of both collagenase variant I and full-length collagenase. **p ≤ 0,01.
The relative enzyme activity of recombinant full-length and truncated variant type I collagenase against interstitial (type I) and non-fibrillar (type IV) collagen was determined. Both recombinant fragments were capable of degrading type I and type IV collagen. Moreover, the variant type I collagenase showed a higher relative activity against collagen type IV as compared to full-length collagenase (t=11.69, df=2, p=0,0072), whereas no statistically significant difference in activity was seen against collagen type I (t=0.2750, df=2, p=0,8091) (Figure 6C).
Bacterial collagenases are enzymes involved in the degradation of the extracellular matrix due to their ability to digest native collagen. These enzymes might play a role in C. perfringens-induced NE by facilitating toxin entry in the intestinal mucosa and by destruction of the tissue structure (Kalisz, 1988; Harrington, 1996; Singh et al., 2012; Tomlin and Piccinini, 2018). In the past, C. perfringens has been shown to produce various gelatinolytic enzymes with molecular masses ranging from approximately 120 to approximately 80 kDa. Yet, only one gene (colA) encoding the 120-kDa collagenase was identified and characterized (Kameyama and Akama, 1971).
In this study, several gene variants carrying different nonsense mutations in the colA gene were identified which were mainly linked to C. perfringens type G strains, that are pathogenic for broiler chickens. Indeed, although the colA gene showed a high level of conservation (> 96% nucleotide sequence identity), truncating variants were identified encoding four different collagenase variant types (type I to IV). These truncated collagenase variants were detected in 13 out of the 14 investigated C. perfringens type G strains, which originated from four different countries, on three different continents (USA, Europe and Australia). This indicates that the observed association of truncated collagenase versions with Type G strains is widespread and not due to single mutation event. The most prevalent collagenase variant types were type I (9/48) and II (5/48), whereas both collagenase type III and IV were present in only 1 of the analyzed C. perfringens strains. All collagenase variant types were predicted to be active as they all contain an intact HEXXH motif which is characteristic for the catalytic center of a metalloproteinase. However, little to no gelatinolytic activity was observed in the supernatant of strains encoding type II variant collagenases. This might be due to the fact that this collagenase type lacks a signaling peptide needed for protein secretion, or because it is inefficiently transcribed. The active fragments of collagenase variant types I, III and IV have a (nearly) complete collagenase module but lack parts of the recruitment domains (i.e. type I and III miss the CBD domains, type IV misses both PKD and CBD domains). Studies using the ColG collagenase from Clostridium histolyticum showed that these recruitment domains are needed to degrade dense collagen networks, such as fibrils, but are not required for the degradation of triple helical collagen (Eckhard and Brandstetter, 2011; Eckhard et al., 2011). However, in this study we showed that recombinant variant type I collagenase - which misses the CBD domains - was able to degrade both interstitial (collagen type I) and non-fibrillar (collagen type IV) collagens. It also had a higher relative activity against collagen type IV as compared to recombinant full-length collagenase. Type IV collagen is the major structural scaffold of basement membranes (Sand et al., 2019). Consequently, these truncated collagenases might be able to break down collagen type IV in the epithelial basement membrane of the intestinal villi. As such, they can contribute to the pathogenesis of NE, as the process leading to intestinal mucosal necrosis has been shown to start at the level of the basement membrane and the lateral domain of the enterocytes (Olkowski et al., 2006; Olkowski et al., 2008). Moreover, the loss of the collagen binding domains in these collagenase variants might allow them to more freely and transiently interact with collagens in the extracellular matrix, potentially leading to a higher non-specific activity resulting in an undefined breakdown of the ECM surrounding the cells, and so contributing to the pathogenesis. Even more, epithelial damage because of predisposing factors such as Eimeria might cause easy accessibility to structures underlying the epithelial cells, so that these collagenases can cause further damage to the initial lesion.
Based on the results of our genetic analysis, we showed that genes encoding for a truncated collagenase are more frequently found in pathogenic C. perfringens type G strains as compared to commensal chicken-derived C. perfringens type A strains or strains originating from other host species, which indicates that having a smaller collagenase might provide an advantage for the bacteria. Indeed, using recombinant collagenase fragments, we showed that both the full-length and variant type I collagenase undergo (auto)proteolysis which results in smaller, but still active collagenase fragments. Additionally, we showed that the cleavage product of the type I collagenase accounts for more of its gelatinolytic activity than the original fragment, despite both fragments being present in approximately equal amounts. It is plausible that these smaller collagenases (e.g. truncated variants) might more efficiently migrate in the mucus layer and/or host tissue. This is an issue for future work to explore.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Study design: LV, RD, FV, EG, MD, MF, SH, FT, and SP. Bioinformatic analysis: EG and LV. In vitro experiments: LV, NC, and CC. Preparation of the manuscript: LV, FH, RD, FV, and EG. All authors offered a critical review of the manuscript. All authors contributed to the article and approved the submitted version.
EG is supported by the Research Foundation Flanders (FWO) under grant number [12W8919N]. The authors declare that this study received funding from Evonik Operations GmbH, Division Nutrition & Care. The funder had the following involvement in the study: Development of the study design. The funder was not involved in the collection, analysis and interpretation of the data or the writing of the article.
Authors MD, MF, SH, FT, and SP were employed by the company Evonik Operations GmbH.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
We thank Prof. Dr. K. Gevaert for critically reading the manuscript and suggesting substantial improvements.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcimb.2021.645248/full#supplementary-material
NE, necrotic enteritis; C. perfringens, Clostridium perfringens; ECM, extracellular matrix.
Al-Sheikhly, F., Al-Saieg, A. (1980). Role of Coccidia in the Occurrence of Necrotic Enteritis of Chickens. Avian Dis. 24, 324–333. doi: 10.2307/1589700
Biosample database at NCBI (2019a) Strain 4928STDY7387940. Available at: https://www.ncbi.nlm.nih.gov/biosample/?term=4928STDY7387940 (Accessed 1 Sep 2020).
Biosample database at NCBI (2019b) Strain Mgyg-Hgut-02372. Available at: https://www.ncbi.nlm.nih.gov/biosample/?term=MGYG-HGUT-02372 (Accessed 2 Sep 2020).
Biosample database at NCBI (2019c)Strain NCTC13170. Available at: https://www.ncbi.nlm.nih.gov/biosample/?term=4928STDY7387940 (Accessed 1 Sep 2020).
Biosample database at NCBI (2020)Strain JXJA17. Available at: https://www.ncbi.nlm.nih.gov/biosample/14228639 (Accessed 2 Sep 2020).
Bober, M., Enochsson, C., Collin, M., Mörgelin, M. (2010). Collagen VI is a Subepithelial Adhesive Target for Human Respiratory Tract Pathogens. J. Innate. Immun. 2, 160–166. doi: 10.1159/000232587
Cooper, K. K., Songer, J. G. (2010). Virulence of Clostridium Perfringens in an Experimental Model of Poultry Necrotic Enteritis. Vet. Microbiol. 142, 323–328. doi: 10.1016/j.vetmic.2009.09.065
Cornillot, E., Saint-Joanis, B., Daube, G., Katayama, S., Granum, P. E., Canard, B., et al. (1995). The Enterotoxin Gene (Cpe) of Clostridium Perfringens Can Be Chromosomal or Plasmid-Borne. Mol. Microbiol. 15, 639–647. doi: 10.1111/j.1365-2958.1995.tb02373.x
Duarte, A. S., Correia, A., Esteves, A. C. (2016). Bacterial Collagenases—a Review. Crit. Rev. Microbiol. 42, 106–126. doi: 10.3109/1040841X.2014.904270
Eckhard, U., Brandstetter, H. (2011). Polycystic Kidney Disease-Like Domains of Clostridial Collagenases and Their Role in Collagen Recruitment. Biol. Chem. 392, 1039–1045. doi: 10.1515/BC.2011.099
Eckhard, U., Schönauer, E., Nüss, D., Brandstetter, H. (2011). Structure of Collagenase G Reveals a Chew-and-Digest Mechanism of Bacterial Collagenolysis. Nat. Struct. Mol. Biol. 18, 1109–1114. doi: 10.1038/nsmb.2127
Fisher, D. J., Fernandez-Miyakawa, M. E., Sayeed, S., Poon, R., Adams, V., Rood, J. I., et al. (2006). Dissecting the Contributions of Clostridium Perfringens Type C Toxins to Lethality in the Mouse Intravenous Injection Model. Infect. Immun. 74, 5200–5210. doi: 10.1128/IAI.00534-06
Gharib-Naseri, K., Kheravii, S. K., Keerqin, C., Morgan, N., Swick, R. A., Choct, M., et al. (2019). Two Different Clostridium Perfringens Strains Produce Different Levels of Necrotic Enteritis in Broiler Chickens. Poult. Sci. 98, 6422–6432. doi: 10.3382/ps/pez480
Gholamiandekhordi, A. R., Ducatelle, R., Heyndrickx, M., Haesebrouck, F., Van Immerseel, F. (2006). Molecular and Phenotypical Characterization of Clostridium Perfringens Isolates From Poultry Flocks With Different Disease Status.Vet. Microbiol. 113, 143–152. doi: 10.1016/j.vetmic.2005.10.023
Harrington, D. J. (1996). Bacterial Collagenases and Collagen-Degrading Enzymes and Their Potential Role in Human Disease. Infect. Immun. 64, 1885–1891. doi: 10.1128/IAI.64.6.1885-1891.1996
Hassan, K. A., Elbourne, L. D. H., Tetu, S. G., Melville, S. B., Rood, J. I., Paulsen, I. T. (2015). Genomic Analyses of Clostridium Perfringens Isolates From Five Toxinotypes. Res. Microbiol. 166, 255–263. doi: 10.1016/j.resmic.2014.10.003
Hauduroy, P., Ehringer, G., Urbain, A., Guillot, G., Magrou, J. (1937). Dictionnaire Des Bactéries Pathogènes Pour L’homme, Les Animaux Et Les Plantes (Paris: Masson et cie).
Hobbs, B. C., Smith, M. E., Oakley, C. L., Warrack, G. H., Cruickshank, J. C. (1953). Clostridium Welchii Food Poisoning. J. Hyg. 51, 75–101. doi: 10.1017/S0022172400015515
Kalisz, H. M. (1988). Microbial Proteinases. Adv. Biochem. Eng. Biotechnol. 36, 1–65. doi: 10.1007/BFb0047944
Kameyama, S., Akama, K. (1971). Purification and Some Properties of Kappa Toxin of Clostridium Perfringens. Jpn. J. Med. Sci. Biol. 24, 9–23. doi: 10.7883/yoken1952.24.9
Keyburn, A. L., Boyce, J. D., Vaz, P., Bannam, T. L., Ford, M. E., Parker, D., et al. (2008). Netb, a New Toxin That is Associated With Avian Necrotic Enteritis Caused by Clostridium Perfringens. PloS Pathog. 4 (2), e26. doi: 10.1371/journal.ppat.0040026
Kiu, R., Caim, S., Alexander, S., Pachori, P., Hall, L. J. (2017). Probing Genomic Aspects of the Multi-Host Pathogen Clostridium Perfringens Reveals Significant Pangenome Diversity, and a Diverse Array of Virulence Factors. Front. Microbiol. 8, 2485. doi: 10.3389/fmicb.2017.02485
Kumar, S., Stecher, G., Li, M., Knyaz, C., Tamura, K. (2018). MEGA X: Molecular Evolutionary Genetics Analysis Across Computing Platforms. Mol. Biol. Evol. 35, 1547–1549. doi: 10.1093/molbev/msy096
Lepp, D., Roxas, B., Parreira, V. R., Marri, P. R., Rosey, E. L., Gong, J., et al. (2010). Identification of Novel Pathogenicity Loci in Clostridium Perfringens Strains That Cause Avian Necrotic Enteritis. PloS. One 5 (5), e10795. doi: 10.1371/journal.pone.0010795
Li, J., Freedman, J. C., Evans, D. R., McClane, B. A. (2017). Cody Promotes Sporulation and Enterotoxin Production by Clostridium Perfringens Type a Strain SM101. Infect. Immun. 85 (3), e00855–16. doi: 10.1128/IAI.00855-16
Li, C., Lillehoj, H. S., Gadde, U. D., Ritter, D., Oh, S. (2017). Characterization of Clostridium Perfringens Strains Isolated From Healthy and Necrotic Enteritis-Afflicted Broiler Chickens. Avian Dis. 61, 178–185. doi: 10.1637/11507-093016-Reg.1
Li, C. Z., Yan, X., Lillehoj, H. S. (2020). Comparison Genomics of Two Closely Related Isolates of Clostridium Perfringens Full Genome Sequences Reveals Regions of Genome Plasticity With Prevention Potential. J. Immunol. 198, 226.9. doi: 10.1186/s13099-017-0217-6
Long, J. R., Pettit, J. R., Barnum, D. A. (1974). Necrotic Enteritis in Broiler Chickens. II. Pathology and Proposed Pathogenesis. Can. J. Comp. Med. Rev. Can. Med. Comp. 38, 467–474.
Matsushita, O., Okabe, A. (2001). Clostridial Hydrolytic Enzymes Degrading Extracellular Components. Toxicon. Off. J. Int. Soc Toxinol. 39, 1769–1780. doi: 10.1016/S0041-0101(01)00163-5
Matsushita, O., Yoshihara, K., Katayama, S., Minami, J., Okabe, A. (1994). Purification and Characterization of Clostridium Perfringens 120-Kilodalton Collagenase and Nucleotide Sequence of the Corresponding Gene. J.Bacteriol. 176, 149–156. doi: 10.1128/JB.176.1.149-156.1994
Medvecky, M., Cejkova, D., Polansky, O., Karasova, D., Kubasova, T., Cizek, A., et al. (2018). Whole Genome Sequencing and Function Prediction of 133 Gut Anaerobes Isolated From Chicken Caecum in Pure Cultures. BMC Genomics 19 (1), 561. doi: 10.1186/s12864-018-4959-4
Mehdizadeh Gohari, I., Kropinski, A. M., Weese, S. J., Parreira, V. R., Whitehead, A. E., Boerlin, P., et al. (2016). Plasmid Characterization and Chromosome Analysis of Two Netf+ Clostridium Perfringens Isolates Associated With Foal and Canine Necrotizing Enteritis. PloS One 11 (2), e0148344. doi: 10.1371/journal.pone.0148344
Mehdizadeh Gohari, I., Kropinski, A. M., Weese, S. J., Whitehead, A. E., Parreira, V. R., Boerlin, P., et al. (2017). Netf-Producing Clostridium Perfringens: Clonality and Plasmid Pathogenicity Loci Analysis. Infect. Genet. Evol. J. Mol. Epidemiol. Evol. Genet. Infect. Dis. 49, 32–38. doi: 10.1016/j.meegid.2016.12.028
Möllby, R., Holme, T., Nord, C. E., Smyth, C. J., Wadström, T. (1976). Production of Phospholipase C (Alpha-Toxin), Haemolysins and Lethal Toxins by Clostridium Perfringens Types a to D. J. Gen. Microbiol. 96, 137–144. doi: 10.1099/00221287-96-1-137
Niilo, L. (1980). Clostridium Perfringens in Animal Disease: A Review of Current Knowledge. Can. Vet. J. 21, 141–148.
Obana, N., Nomura, N., Nakamura, K. (2013). Structural Requirement in Clostridium Perfringens Collagenase Mrna 5′ Leader Sequence for Translational Induction Through Small RNA-Mrna Base Pairing. J. Bacteriol. 195, 2937–2946. doi: 10.1128/JB.00148-13
Obana, N., Shirahama, Y., Abe, K., Nakamura, K. (2010). Stabilization of Clostridium Perfringens Collagenase Mrna by VR-RNA-Dependent Cleavage in 5’ Leader Sequence. Mol. Microbiol. 77, 1416–1428. doi: 10.1111/j.1365-2958.2010.07258.x
Olkowski, A. A., Wojnarowicz, C., Chirino-Trejo, M., Drew, M. D. (2006). Responses of Broiler Chickens Orally Challenged With Clostridium Perfringens Isolated From Field Cases of Necrotic Enteritis. Res. Vet. Sci. 81, 99–108. doi: 10.1016/j.rvsc.2005.10.006
Olkowski, A. A., Wojnarowicz, C., Chirino-Trejo, M., Laarveld, B., Sawicki, G. (2008). Sub-Clinical Necrotic Enteritis in Broiler Chickens: Novel Etiological Consideration Based on Ultra-Structural and Molecular Changes in the Intestinal Tissue. Res. Vet. Sci. 85, 543–553. doi: 10.1016/j.rvsc.2008.02.007
Parreira, V. R., Russell, K., Athanasiadou, S., Prescott, J. F. (2016). Comparative Transcriptome Analysis by Rnaseq of Necrotic Enteritis Clostridium Perfringens During in Vivo Colonization and in Vitro Conditions. BMC Microbiol. 16 (1), 186. doi: 10.1186/s12866-016-0792-6
Ren, Z., Guo, C., Yu, S., Zhu, L., Wang, Y., Hu, H., et al. (2019). Int. J. Mol. Sci. 20 (11), 2777. doi: 10.3390/ijms20112777
Ribeiro, F. J., Przybylski, D., Yin, S., Sharpe, T., Gnerre, S., Abouelleil, A., et al. (2012). Finished Bacterial Genomes From Shotgun Sequence Data. Genome Res. 22, 2270–2277. doi: 10.1101/gr.141515.112
Roach, D. J., Burton, J. N., Lee, C., Stackhouse, B., Butler-Wu, S. M., Cookson, B. T., et al. (2015). A Year of Infection in the Intensive Care Unit: Prospective Whole Genome Sequencing of Bacterial Clinical Isolates Reveals Cryptic Transmissions and Novel Microbiota. PloS Genet. 11 (7), e1005413. doi: 10.1371/journal.pgen.1005413
Rood, J. I., Adams, V., Lacey, J., Lyras, D., McClane, B. A., Melville, S. B., et al. (2018). Expansion of the Clostridium Perfringens Toxin-Based Typing Scheme. Anaerobe. 53, 5–10. doi: 10.1016/j.anaerobe.2018.04.011
Rood, J. I., Keyburn, A. L., Moore, R. J. (2016). Netb and Necrotic Enteritis: The Hole Movable Story. Avian Pathol. J. WVPA 45, 295–301. doi: 10.1080/03079457.2016.1158781
Rozario, T., DeSimone, D. W. (2010). The Extracellular Matrix in Development and Morphogenesis: A Dynamic View. Dev. Biol. 341, 126–140. doi: 10.1016/j.ydbio.2009.10.026
Sand, J. M. B., Genovese, F., Gudmann, N. S., Karsdal, M. A. (2019). “Chapter 4 - Type IV Collagen,” in Biochemistry of Collagens, Laminins and Elastin, 2nd ed. Ed. M.A. Karsdal, M. A. (Cambridge, Massachusetts, USA: Cambridge, Academic Press), 37–49.
Scheirlinck, I., Van der Meulen, R., Van Schoor, A., Vancanneyt, M., De Vuyst, L., Vandamme, P., et al. (2007). Influence of Geographical Origin and Flour Type on Diversity of Lactic Acid Bacteria in Traditional Belgian Sourdoughs. Appl. Environ. Microbiol. 73, 6262–6269. doi: 10.1128/AEM.00894-07
Sheedy, S. A., Ingham, A. B., Rood, J. I., Moore, R. J. (2004). Highly Conserved Alpha-Toxin Sequences of Avian Isolates of Clostridium Perfringens. J. Clin. Microbiol. 42, 1345–1347. doi: 10.1128/JCM.42.3.1345-1347.2003
Shimizu, T., Ohshima, S., Ohtani, K., Shimizu, T., Hayashi, H. (2001). Genomic Map of Clostridium Perfringens Strain 13. Microbiol. Immunol. 45, 179–189. doi: 10.1111/j.1348-0421.2001.tb01278.x
Singh, B., Fleury, C., Jalalvand, F., Riesbeck, K. (2012). Human Pathogens Utilize Host Extracellular Matrix Proteins Laminin and Collagen for Adhesion and Invasion of the Host. FEMS. Microbiol. Rev. 36, 1122–1180. doi: 10.1111/j.1574-6976.2012.00340.x
Tomlin, H., Piccinini, A. M. (2018). A Complex Interplay Between the Extracellular Matrix and the Innate Immune Response to Microbial Pathogens. Immunology. 155, 186–201. doi: 10.1111/imm.12972
Uzal, F. A., Vidal, J. E., McClane, B. A., Gurjar, A. A. (2010). Clostridium Perfringens Toxins Involved in Mammalian Veterinary Diseases. Open Toxinol. J. 2, 24–42. doi: 10.2174/1875414701003020024
Van Damme, L., Cox, N., Callens, C., Haesebrouck, F., Dargatz, M., Ducatelle, R., et al. (2020). C. Perfringens Challenge Reduces Matrix Metalloproteinase Activity in the Jejunal Mucosa of Eimeria-Infected Broiler Chickens. Vet. Res. 51 (1), 100. doi: 10.1186/s13567-020-00825-6
Keywords: Clostridium perfringens, kappa toxin, collagenase, nonsense mutation, necrotic enteritis disease
Citation: Van Damme L, Cox N, Callens C, Dargatz M, Flügel M, Hark S, Thiemann F, Pelzer S, Haesebrouck F, Ducatelle R, Van Immerseel F and Goossens E (2021) Protein Truncating Variants of colA in Clostridium perfringens Type G Strains. Front. Cell. Infect. Microbiol. 11:645248. doi: 10.3389/fcimb.2021.645248
Received: 22 December 2020; Accepted: 09 April 2021;
Published: 29 April 2021.
Edited by:
Haider H. Dar, University of Pittsburgh, United StatesReviewed by:
Horst Posthaus, University of Bern, SwitzerlandCopyright © 2021 Van Damme, Cox, Callens, Dargatz, Flügel, Hark, Thiemann, Pelzer, Haesebrouck, Ducatelle, Van Immerseel and Goossens. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Filip Van Immerseel, RmlsaXAuVmFuSW1tZXJzZWVsQFVHZW50LmJl
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.