- 1Department of Dermatology, The First Affiliated Hospital of Jinan University, Guangzhou, China
- 2Institute of Mycology, Jinan University, Guangzhou, China
Macrophages provide the first-line defense against invasive fungal infections and, therefore, escape from macrophage becomes the basis for the establishment of Candida albicans invasive infection. Here, we found that deletion of ATP2 (atp2Δ/Δ) in C. albicans resulted in a dramatic decrease from 69.2% (WT) to 1.2% in the escape rate in vitro. The effect of ATP2 on macrophage clearance stands out among the genes currently known to affect clearance. In the normal mice, the atp2Δ/Δ cells were undetectable in major organs 72 h after systemic infection, while WT cells persisted in vivo. However, in the macrophage-depleted mice, atp2Δ/Δ could persist for 72 h at an amount comparable to that at 24 h. Regarding the mechanism, WT cells sustained growth and switched to hyphal form, which was more conducive to escape from macrophages, in media that mimic the glucose-deficient environment in macrophages. In contrast, atp2Δ/Δ cells can remained viable but were unable to complete morphogenesis in these media, resulting in them being trapped within macrophages in the yeast form. Meanwhile, atp2Δ/Δ cells were killed by oxidative stress in alternative carbon sources by 2- to 3-fold more than WT cells. Taken together, ATP2 deletion prevents C. albicans from escaping macrophage clearance, and therefore ATP2 has a functional basis as a drug target that interferes with macrophage clearance.
Introduction
Candida albicans is the most prevalent lethal fungal pathogen and can cause invasive infections in immunodeficiency patients, with variable rates from 5% to 70% (Bassetti et al., 2018; Pappas et al., 2018). The current antifungals target a limited number of cellular processes, and therefore, the development of different therapeutic approaches is urgently needed (Bassetti et al., 2018).
Macrophages provide the first-line defense against invasive fungal infections, and thus, C. albicans escape from macrophage becoming the basis for establishing systemic infection (Qian et al., 1994; Lionakis et al., 2013; Ngo et al., 2014; Weiss and Schaible, 2015). During the clearance, macrophages restrict the growth of and destroy C. albicans mainly by nutrient deprivation, a low pH, and oxidative stress in the phagosome (Austermeier et al., 2020; Williams and Lorenz, 2020). Furthermore, macrophages secrete cytokines to recruit more phagocytes to participate in pathogen clearance (Ngo et al., 2014). In contrast, similar to other successful intercellular pathogens (Mycobacterium tuberculosis, Histoplasma capsulatum, etc.), C. albicans has evolved elegant strategies to evade macrophage killing (Weiss and Schaible, 2015; Shen and Rappleye, 2020). These strategies can be divided into the following two categories: strategies that support pathogen survival within macrophages, including rapid conversion to metabolize alternative carbon sources, adaptation and neutralization of acidic phagosomes, and resistance to oxidative stress; and strategies that facilitate macrophage destruction, such as morphogenesis (Uwamahoro et al., 2014; Westman et al., 2019; Shen and Rappleye, 2020; Williams and Lorenz, 2020).
Transcriptomics and proteomics studies have identified some genes that affect the escape of C. albicans from macrophages (Lorenz et al., 2004; Kitahara et al., 2015; O’Meara et al., 2018). However, knocking out these genes associated with specific pathways may increase the clearance rate, but it does not completely block the escape of C. albicans from macrophages (Danhof et al., 2016; Jain et al., 2018; Williams and Lorenz, 2020). For example, knocking out genes related to non-glucose carbon source utilization or hyphae formation in C. albicans can only increase the clearance by only 20-40%, but blocking several pathways simultaneously has additive effects (Piekarska et al., 2006; Danhof and Lorenz, 2015; Jain et al., 2018; Williams and Lorenz, 2020). Therefore, searching for drug targets involved in the macrophage clearance process requires finding genes with a broader range of functions.
Oxidative phosphorylation is the central molecular node of the cellular metabolic network (Marcet-Houben et al., 2009). Here, we found that deletion of ATP2, which encodes the β subunit of F1Fo-ATP synthase, greatly reduced the ability of C. albicans to escape macrophages in vitro and in vivo. Regarding the mechanism, the ATP2 deletion impaired the adaptation of C. albicans to glucose-deficient environment within macrophages, and although they could remain viable for a short period of time, they were unable to form hyphae to escape from macrophages and were more susceptible to oxidative stress killing. Due to the high involvement of ATP2 in host-pathogen interactions, confirming that whether ATP2 can be used as a drug target deserves further investigation.
Materials and Methods
Strains, Cells and Culture Conditions
C. albicans strains used in this study were SC5314 (WT, ATCC) and ATP2 null mutant (atp2Δ/Δ) (Li et al., 2018). Strains were maintained on YPD (1% yeast extract, 2% peptone, 2% dextrose) agar at 30°C and stored in 50% glycerol at -80°C. For each assays, C. albicans cells were cultured in YPD liquid medium overnight at 30°C and 150 rpm, then collected, washed and normalized to appropriate concentrations.
Murine macrophage-like cell line RAW264.7 (ATCC) was used for all macrophage assays. Cells were maintained in Dulbecco’s modified Eagle medium (DMEM) supplemented with 10% heat-inactivated fetal bovine serum, 100 U/ml penicillin and 100 μg/ml streptomycin, at 37°C with 5% CO2.
Macrophage Killing Assay
The end-point dilution assay was performed to detect the killing effect of macrophages on C. albicans as described previously (She et al., 2013). RAW264.7 cells were seeded into 96-well plates at a density of 1×105 cells/well and adhered for 1h. Then 2×105 cells/well C. albicans were added and cocultured for 6, 12, 24 and 48 h. At each time point, the co-cultures were treated with 0.1% Triton X-100 for 2 min. A serial dilution was performed and plated onto YPD agar. Numbers of colony-forming units (CFU) were counted after 48 h incubation and the macrophage killing rate was determined by comparing cocultures to C. albicans control cultures without macrophages. Assays were performed in triplicate, and experiments were repeated three times.
Macrophage Cytotoxicity Assay
C. albicans-induced macrophage damage was assessed by measuring the release of lactate dehydrogenase (LDH) using a Non-Radioactive Cytotoxicity assay (Abcam) (She et al., 2013). RAW264.7 (1×105 cells) were co-incubated with C. albicans (1×105 cells) for 2, 4, 6, and 8 h. At each time point, the absorbance at 490 nm was recorded according to the manufacturer’s protocol. The release of LDH relative to maximum LDH release was then calculated and corrected for spontaneous release of LDH by the C. albicans or macrophages alone. The experiment was performed in triplicate.
Macrophage Phagocytosis Assay
Activated RAW264.7 cells were seeded in 96-well plates at 1×106 cells/ml (She et al., 2013). C. albicans cells were stained with 1.25 mM fluorescein isothiocyanate (FITC, Sigma) for 15 min, then diluted to 2 × 106 cells/ml in RPMI medium and cocultured with macrophages for 30, 90, 120 min. At each time point, 100 μl supernatant was removed and 100 μl trypan blue was added to quench the fluorescence of unengulfed strains. The fluorescence signal intensity (Ex/Em=495/525) of each well were detected with a microplate reader (Thermo Fisher Scientific). Assays were performed in triplicate, and experiments were repeated three times.
Fungal Burden in Clodronate Liposome Treated Mice
10-week-old female BALB/c mice (20 g) were injected intraperitoneally with 200 μl of clodronate- or PBS-liposome (http://clodronateliposomes.org) both 24 h before and 24 h after intravenous C. albicans infections (Wirnsberger et al., 2016). Mice were infected with 2×105 CFU of C. albicans cells intravenously. At 24 and 72 h after infection, organs were taken out and weighed, then grinded and measured fungal burden (CFU/g) by the end point dilution assay. There were three mice in each group, and experiment was repeated twice.
microPET/CT Scanning and Radioimmuno-γ Counting
10-week-old female BALB/c mice (20 g) were infected with 1× 106 CFU of C. albicans cells intravenously. Mice were fasted for 12 h before the imaging time point and only water was provided. Each mouse was injected 1% pentobarbital (15 ul per g mouse) intraperitoneally and 5 μCi/g of [18F]FDG intravenously 45 min before imaging. PET and CT images were obtained by the Inveon micro-PET/CT (Siemens) small-animal PET imagers with a static acquisition time of 15 min (Davis et al., 2009). After PET/CT scan, the ex vivo biodistribution of organs was detected by γ-counter (Perkin-Elmer) (Rolle et al., 2016). There were three mice in each group, and experiments were repeated three times.
RNA Extraction and Real‐time Quantitative PCR
Total RNA from co-cultured macrophages were extract using Trizol (Invitrogen), and total RNA from C. albicans were extract using the E.Z.N.A. Yeast RNA kit (Omega Bio-Tek) (Li et al., 2017). Approximately 0.8 μg of RNA was used to synthesize cDNA (Qiagen, Venlo, Netherlands). RT-qPCR was done in triplicate as previously described using Bio-Rad iQ5. Primers used are shown in Table S1. The expression levels of C. albicans genes were normalized to 18S rRNA levels, while the macrophage genes were normalized to GAPDH levels. The 2–ΔΔCT (where CT is the threshold cycle) method was used to determine the fold change in gene transcription. Each sample was performed in triplicate and experiments were repeated three times.
Growth Curve and Cell Viability Assay
C. albicans cells were collected and washed with PBS, then inoculated in 100 ml of macrophage-mimicking media (YNB liquid medium supplemented with 2% glucose, CAA, GLcNAc, oleic acid, or lactate) with an initial OD600 of 0.02. Shake cultures were grown at 30 °C and OD600 of each strain was measured every 2 h (Li et al., 2018). Cell viability was detected by the end-point dilution assay as describe previously. C. albicans cells were washed and resuspended in media mentioned above to a density of 1×106 cells/ml. Cultures were grown at 30 °C and 100 μl of liquid was taken out to perform end-point dilution assay every 2 h. All experiments were performed in triplicate and repeated three times.
Hyphal Morphogenesis of Phagocytosed C. albicans Cells
RAW264.7 cells (1×106 cells) were seeded to glass coverslips and incubated overnight (Vylkova and Lorenz, 2014). 500 nM MitoTracker Deep Red FM (Molecular Probes) was added and incubated for 30 min. C. albicans cells were stained with FITC as mentioned before, and cocultured with macrophages for 3 h. Images were taken using a confocal laser scanning microscope (Carl Zeiss LSM880), under the set of Ex644/Em655 (macrophages) and Ex488/Em525 (C. albicans). The percentage of hyphae cells were counted manually (the number of germinated cells versus the number of total cells). At least 50 cells per strain were counted. Experiments were repeated three times.
Hyphae Formation Assay
The hyphae formation assay was performed as described previously (Li et al., 2017). C. albicans cells were washed and resuspended in YNB liquid medium supplemented with 2% glucose, CAA, GLcNAc, oleic acid, or lactate to a density of 1×105 cells/ml. Cells were incubated in 24 well plates at 37°C for 3 h and images were taken by an inverted microscope (Olympus IX81).
H2O2 Killing Assay
The end-point dilution assay was performed to detect the killing effect of H2O2 on C. albicans (Wu et al., 2018). C. albicans cells were washed and resuspended in YNB liquid medium supplemented with 2% glucose, CAA, GLcNAc, oleic acid, and lactate to a concentration of 5×104 cells/ml. Then, 6 mM H2O2 was added except for the growth control well and incubated at 37°C for 6 h. A serial dilution was performed and plated onto YPD agar. CFU were counted after 48 h incubation and the mortality rate was determined by comparison with fungi recovered without exposure to H2O2.
Protein Extraction and LC-MS/MS Analysis
C. albicans protein was extracted by a liquid nitrogen grinding method as described previously (Kitahara et al., 2015). C. albicans cells were diluted into 1 L cultures at a starting OD = 0.5, cultured for 8 h at 30°C, and harvested at 4 °C. The samples were ground into cell powder, then lysis buffer (8 M urea, 1% Triton-100, 10 mM dithiothreitol, and 1% protease inhibitor) was added, followed by sonication three times on ice using a high-intensity ultrasonic processor (Scientz).
The proteins were reduced with 5 mM dithiothreitol for 30 min at 56°C and alkylated with 11 mM iodoacetamide for 15 min. The sample was diluted with urea, and trypsin was added at the mass ratio of trypsin: protein = 1:50 for the first overnight digestion and 1:100 for a second 4 h digestion. The tryptic peptides were fractionated by high pH reverse-phase HPLC using a Betasil™ C18 column (5 μm particles, 250 mm length, Thermo Scientific). The protein digests were then labeled with a Tandem Mass Tag kit (Thermo Scientific) (Kitahara et al., 2015). The peptides were desalted using a Strata X C18 SPE column (Phenomenex) and vacuum-dried. Then the peptides were separated into 60 fractions using a gradient of 8%–32% acetonitrile (pH 9.0) over 60 min. Finally, the peptides were combined into 10 fractions and dried by vacuum centrifugation.
The detailed protocol for LC‐MS/MS was described previously (Herrero-de-Dios et al., 2018; Truong et al., 2019). The tryptic peptides were dissolved in 0.1% formic acid (solvent A) and loaded on an analytical column (75 µm × 150 mm). The constant flow rate was set at 500 nL/min with the step gradients of mobile B (0.1% formic acid in 90% acetonitrile): 9%–26% for 38 min, 26%–35% for 14 min, 35%–80% for 4 min, and held at 80% for 4 min. The peptides were subjected to NSI source followed by tandem MS analysis in Q ExactiveTM Plus (Thermo) coupled online to the UPLC. The mass range was 400–1500 m/z, and tandem mass spectra were recorded in high sensitivity mode. In each cycle, a maximum of 20 precursors were selected for fragmentation, with the dynamic exclusion for 15 s. Protein identification and quantification were performed via the Maxquant search engine (v.1.5.2.8). The data were searched against a protein sequence database downloaded from UniProtKB for C. albicans SC5314 (total 6040 entries). The quantitative method was set to TMT-10plex, and the FDR for protein identification and PSM identification was adjusted to < 1% (Truong et al., 2019). The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE (Perez-Riverol et al., 2019) partner repository with the dataset identifier PXD024039.
Ethics Statement
All BABL/c mice were obtained from Guangdong Medical Laboratory Animal Center, Foshan, Guangdong, China. The animal experiments were performed in accordance with the recommendations in the Guide for the Care and Use of Laboratory Animals of the National Academy of Science. The protocol was carried out with permission from the Laboratory Animal Ethics Committee of Jinan University (NO. 2019670).
Results
Deletion of ATP2 in C. albicans Decreases Its Escape From Macrophages In Vitro
Escape from macrophage clearance is the basis by which C. albicans establishes systemic infection (Lionakis et al., 2013; Ngo et al., 2014; Weiss and Schaible, 2015). To investigate the impact of ATP2 on this pivotal procedure, we examined the proportion of C. albicans (WT and atp2Δ/Δ) killed by macrophages (RAW264.7) in a coculture system via the end-point dilution assay. When C. albicans and macrophages were cocultured at a ratio of 2:1 (multiplicity of infection [MOI] of 2), the atp2Δ/Δ and WT cells were phagocytosed at comparable rates (P > 0.05) (Figure 1A), but more atp2Δ/Δ cells were killed by macrophages and less macrophages were damaged at each time point than WT cells (P < 0.05) (Figures 1B, C). The mortality rate of atp2Δ/Δ was 98.2% at 24 h, but only 32.5% in the WT at this time point (P < 0.05) (Figure 1B). Then we compared the CFUs of WT and atp2Δ/Δ in medium without macrophages and found that atp2Δ/Δ grew only 31.1% less than WT at 24 h (Figure S1). This suggests that the deletion of ATP2 in C. albicans resulted in a significant reduction in the ability to escape macrophage clearance.
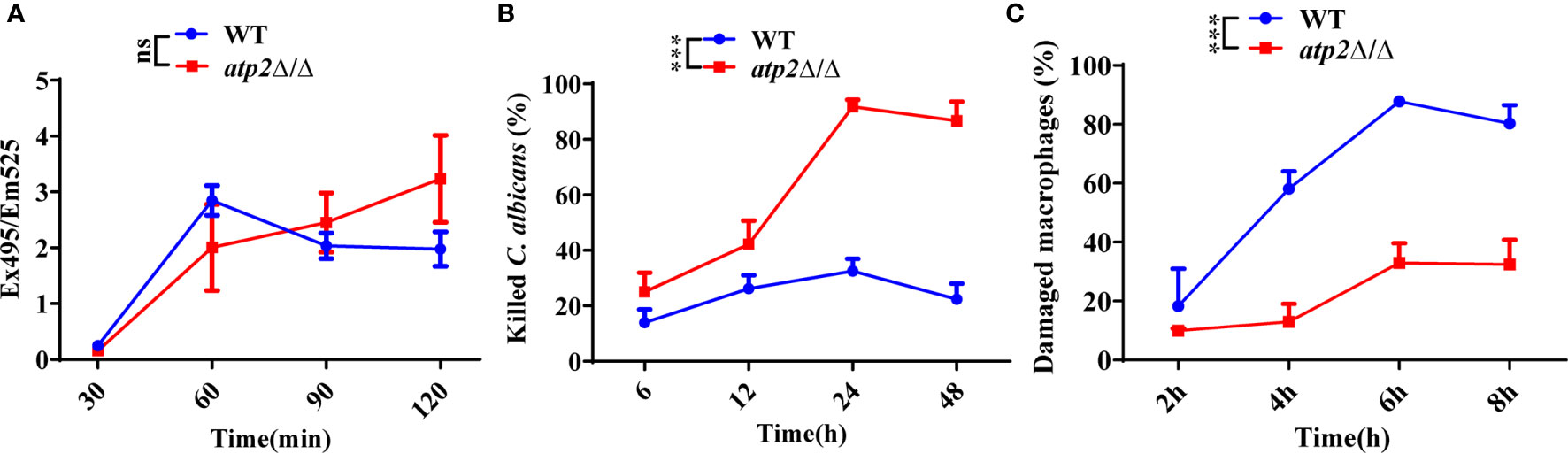
Figure 1 ATP2 modulates the C. albicans-macrophage interaction in vitro. (A) The phagocytosis of FITC-labeled WT and atp2Δ/Δ cells by macrophages was evaluated by measuring FITC fluorescence (Ex495/Em525) in macrophages. (B) Percentage of C. albicans cells killed by macrophages over time. WT and atp2Δ/Δ cells were cocultured with RAW264.7 cells at a ratio of 2:1 (MOI of 2), and the killing rate was determined by comparison with C albicans recovered without RAW264.7 cells at each time point. (C) Percentage of C. albicans-induced macrophage damage was assessed by measuring the release of LDH at each time point. In (A–C), assays were performed in triplicate. Data are shown as mean ± s.d. ***P < 0.001; ns, not significant; by two-way ANOVA.
Deletion of ATP2 in C. albicans Decreases Its Escape From Macrophages In Vivo
To further study the effect of ATP2 on the escape of C. albicans from macrophage clearance in vivo, we constructed macrophage normal/depleted mice with PBS/clodronate liposome, and detected the fungal burden at different time points after systemic infection with WT or atp2Δ/Δ. The results showed that the WT cells were not cleared in the normal mice (PBS liposome), and that the fungal burden in the kidney at 72 h was higher than that at 24 h post-infection (P < 0.05); however, the fungal burden in the liver, spleen and brain did not significantly differ from that at 24 h (P > 0.05). In contrast, the atp2Δ/Δ mutant cells were almost completely cleared 72 h after infection, with no detectable pathogens in the liver, kidney and spleen. Cells remained in the brain were far less than WT (Figures 2A–D) (P < 0.05).
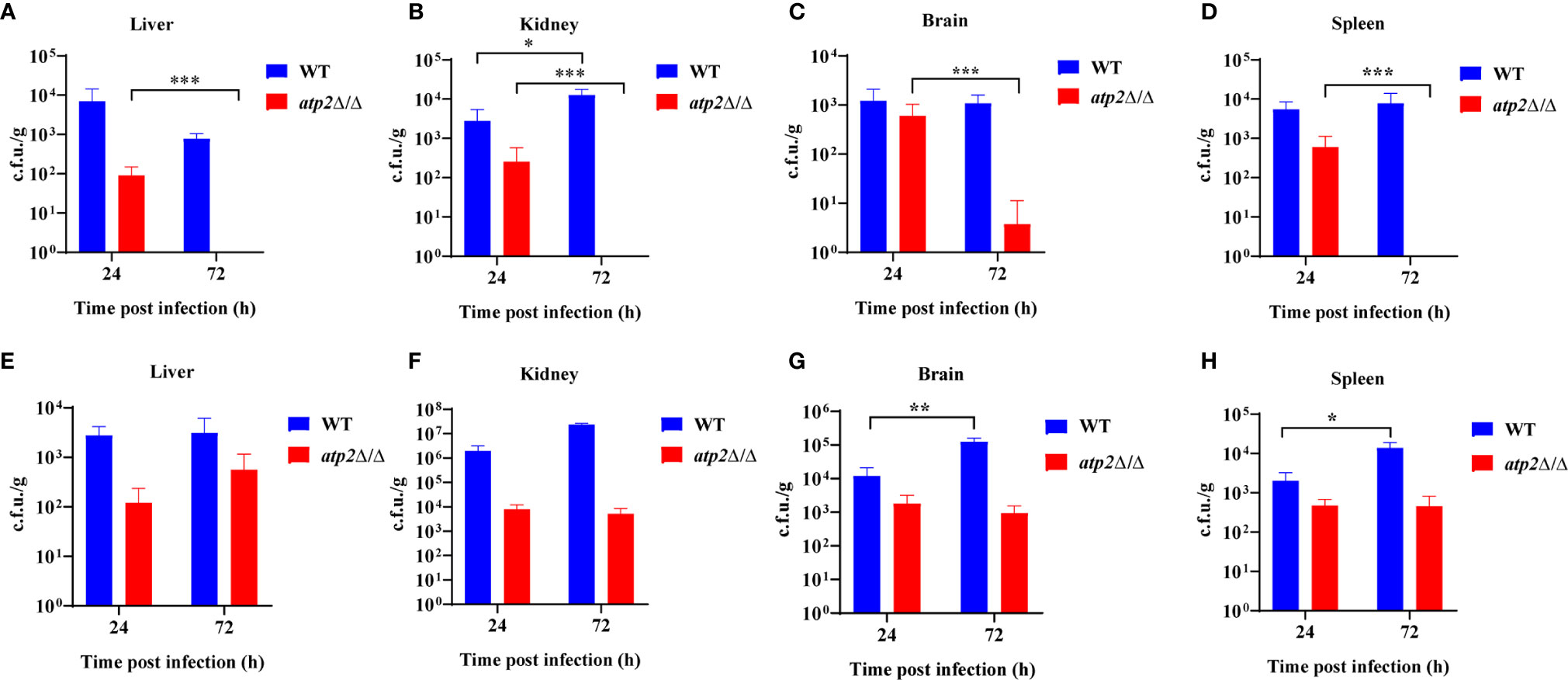
Figure 2 ATP2 is required for C. albicans to resist macrophage clearance in vivo. (A–D) Fungal burden in the liver (A), kidney (B), spleen (C), and brain (D) of mice injected with PBS liposomes 24 h before and 24 h after intravenous infection with C. albicans (2×105 CFU per mouse). (E–H) Fungal burden in the liver (E), kidney (F), spleen (G), and brain (H) of mice injected with clodronate liposomes 24 h before and 24 h after intravenous infection with C. albicans (2×105 CFU per mouse). Each group includes 3 mice. In (A–H), data are shown as mean ± s.d. *P < 0.05, **P < 0.01, ***P < 0.001; by Student’s t-test.
However, in the macrophage-depleted mice (clodronate liposome), the atp2Δ/Δ cells stably persisted in the liver, kidney, spleen, and brain at 24 h and 72 h after infection instead of being cleared. The number of WT cells was increased in the spleen (Figure 2H) and brain (Figure 2G) but unchanged in the liver (Figure 2E) and kidney (Figure 2F) 72 h post-infection compared with those at 24 h. These results suggest that C. albicans cells lacking ATP2 were unable to resist macrophage clearance in vivo.
Clearance of ATP2 Mutant Cells Does Not Result in Increased Recruitment of Phagocytes In Situ
The clearance of pathogens typically requires macrophages to recruit large numbers of phagocytes, including neutrophils and NK cells, to enhance the killing efficiency (Ngo et al., 2014; Netea et al., 2015). Is the efficient clearance of atp2Δ/Δ associated with increased recruitment of phagocytes? To answer this question, we injected mice with [18F]FDG, which accumulates in activated macrophages and neutrophils, and detected the [18F]FDG signal distribution by microPET/CT and γ counter in real time, dynamically and overall (Davis et al., 2009; Rolle et al., 2016). Surprisingly, the atp2Δ/Δ-infected mice never showed an increase in the [18F]FDG signal intensity throughout the body, and there was no significant difference between the atp2Δ/Δ and NS groups (Figures 3A–D) (P > 0.05). However, the [18F]FDG intensity in the WT-infected mice was significantly increased, especially in the target organ, i.e., kidney, where the signal intensity reached 7-fold of that observed in the atp2Δ/Δ and NS groups at 72 h (Figures 3E) (P < 0.05). The brain, heart, liver, and colon signals in the WT group were also stronger than those in the atp2Δ/Δ and NS groups (Figures 3C, F) (P < 0.05).
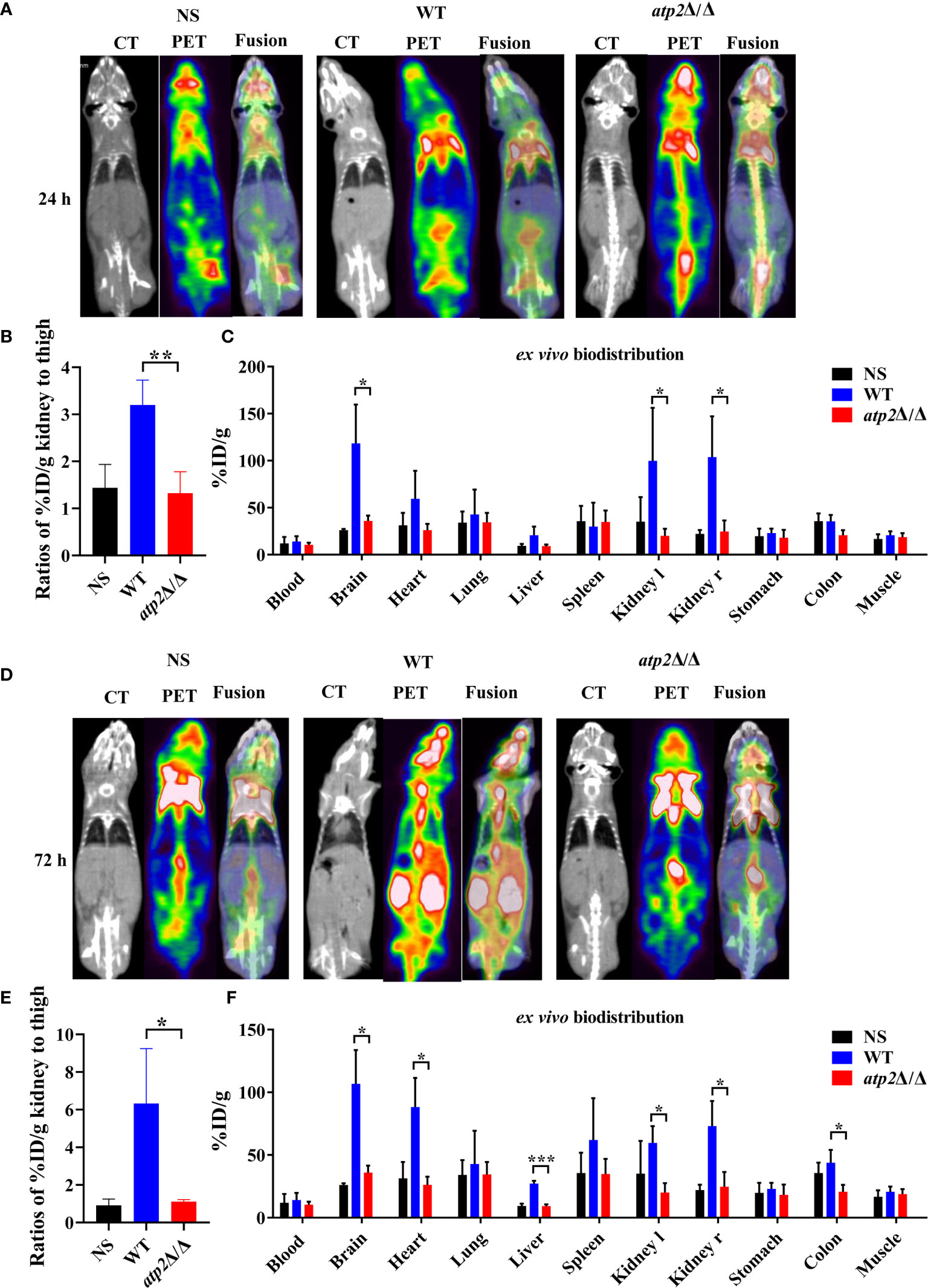
Figure 3 ATP2 affects the activation of phagocytes by C. albicans in situ. (A, B) Representative CT, PET, and fusion images (A) and quantitative statistics of kidney signals from PET images (B) of mice (n = 3) 24 h after systemic infection with NS, WT or atp2Δ/Δ (2 × 105 CFU per mouse). (C) Ex vivo biodistribution in different tissues detected by γ-counter immediately after micro-PET-CT examination of (A). (D, E) Representative CT, PET, and fusion images (D) and quantitative statistics of kidney signals from PET images (E) of mice 72 h after systemic infection. (F) Ex vivo biodistribution in different tissues detected by γ-counter immediately after microPET/CT examination of (E). In (A, D), one representative experiment of three independent experiments is shown. In (B, C, E, F), data are shown as mean ± s.d. *P < 0.05, **P < 0.01, ***P < 0.001; by Student’s t-test.
In alignment with the PET results, we did not observe significant inflammatory foci in the kidney sections from the atp2Δ/Δ-infected mice, whereas multiple foci of inflammatory cells containing mostly polymorphonuclear leukocytes (PMNs) were observed in the WT-infected mice (Figure 4). The inflammatory scores obtained from renal immune cell infiltration and tissue destruction were significantly lower than those in the WT group (Figure 4) (P < 0.05). These results suggest that the clearance of the atp2Δ/Δ mutant cells did not induce abnormal inflammatory cells recruitment in situ.
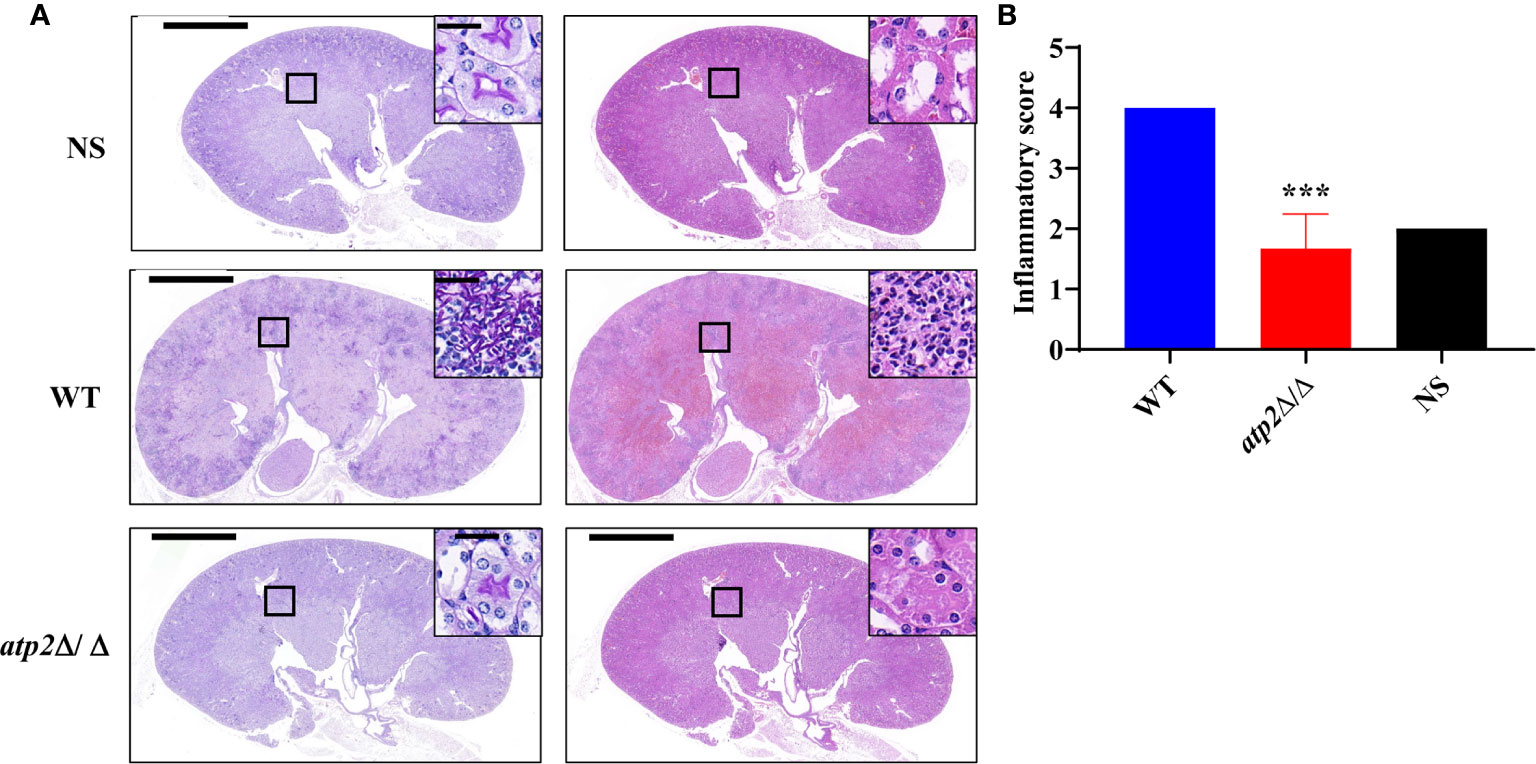
Figure 4 C. albicans cells lacking ATP2 do not cause abnormal phagocytes infiltration in the kidney. (A) Representative images of Periodic acid-Schiff-stained (PAS) and Hematoxylin-Eosin-stained (HE) kidney sections of mice (n = 3) 72 h after systemic infection with NS, WT or atp2Δ/Δ (2 × 105 CFU per mouse). (B) Inflammatory score based on renal immune cell infiltration and tissue destruction. In (A), one representative experiment of three mice is shown. In (B) data are shown as mean ± s.d. ***P < 0.001; by Student’s t-test. In (A), insets show higher-magnification images of boxed areas; scale bars, 1,000 µm, 50 µm (insets).
ATP2 Mutant Cells Can Remain Viable in Macrophage-Mimicking Environment
Once engulfed, C. albicans cells rapidly adapt the glucose-deficient environment, shifting to gluconeogenic growth and escaping from macrophages within 6-8 h (Lorenz et al., 2004; Wartenberg et al., 2014). To observe the effect of ATP2 on the adaptation of C. albicans to the environment, we first examined the growth and viability of WT and atp2Δ/Δ in the media that simulated glucose-deficient environment within macrophages. The results showed that the OD600 of WT group in macrophage-mimicking media (YNB medium with 2% CAA, GlcNAc, lactate, or oleic acid) increased, while the OD600 of atp2Δ/Δ group remained at the initial level (Figures 5A–D). More importantly, consistent with the growth results detected by OD600, the viable cells of WT were increased in these macrophage-mimicking media over time, reaching 3-5 times the initial value at 8 h. However, the number of viable atp2Δ/Δ in these media decreased by 20-40% at 8 h (Figures 5E–H). These results suggest that the atp2Δ/Δ cells stop proliferating, but can partially remain viable for a short period of time in the glucose-deficient environment.
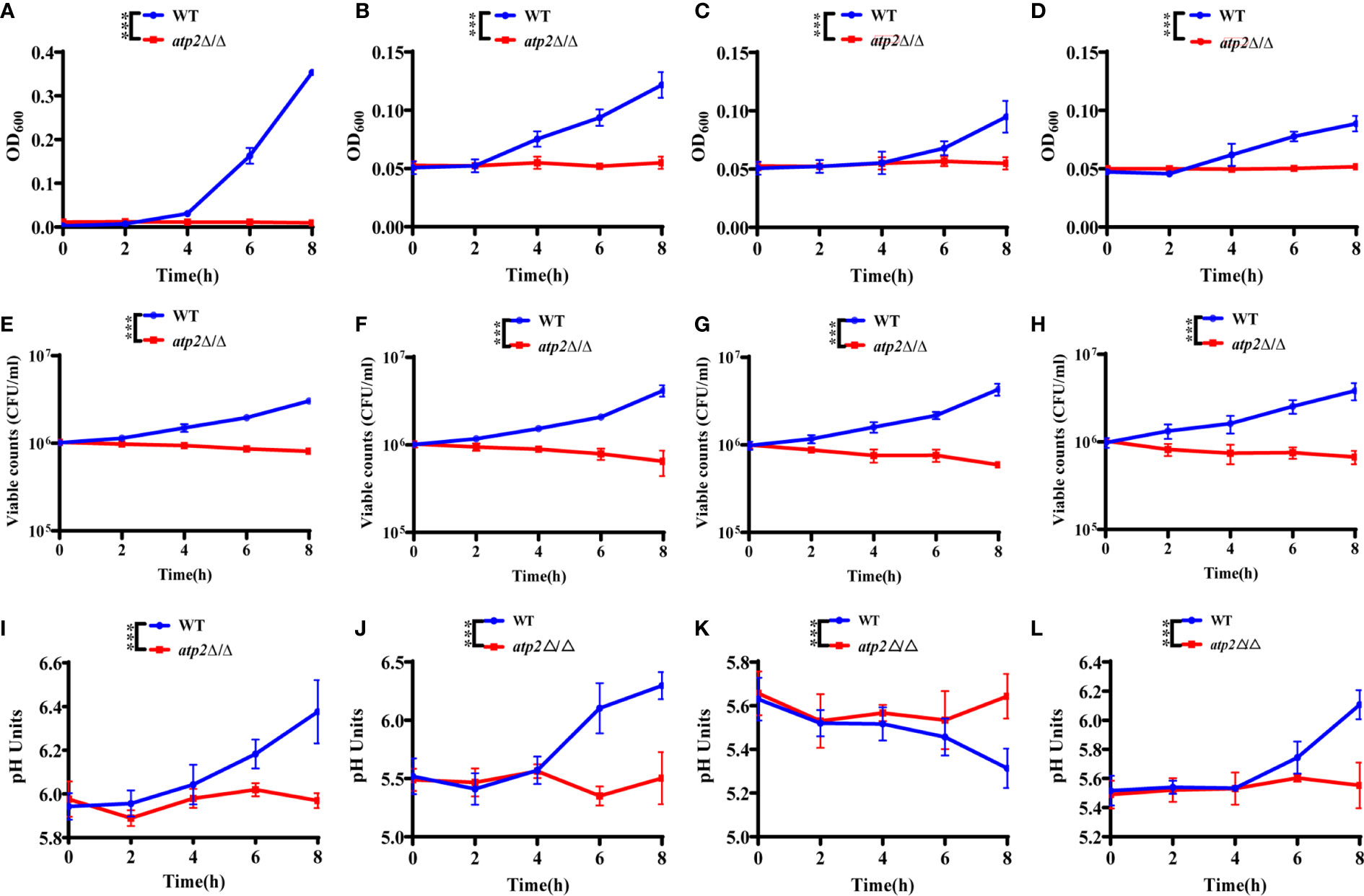
Figure 5 C. albicans cells lacking ATP2 are unable to utilize alternative carbon source. The growth (A–D), viability (E–H), and pH (I–L) of WT and atp2Δ/Δ cells in YNB medium plus 2% CAA, GLcNAc, oleic acid, and lactate over 8 h. In (A–L), the assays were performed in triplicate.***P < 0.001; by two-way ANOVA.
C. albicans can also adapt to or even alkalize the acidic environment within macrophages, creating more favorable conditions for its survival and escape (Vylkova and Lorenz, 2014). We examined the effect of WT and atp2Δ/Δ on the pH value of the environment in macrophage-mimicking media. The results showed that the pH values of the CAA (Figure 5I), GlcNAc (Figure 5J) and lactate (Figure 5L) medium increased simultaneously with the OD600 when the WT cells were cultured. The pH of the oleic acid medium gradually decreased when the oleic acid decomposed by WT cells (Figure 5K). In contrast, the pH values of the media with atp2Δ/Δ cells were not statistically different at each time point, the atp2Δ/Δ cells were unable to change the environment pH in all macrophage-mimicking media (Figures 5I–K). The above results suggest that deletion of ATP2 impaired the adaptation of C. albicans to the glucose-deficient environment within macrophages.
ATP2 Mutant Cells Are Unable to Undergo Morphogenesis in Macrophages
Morphogenesis is an important contributing factor for the escape from macrophages (Peroumal et al., 2019; Rogiers et al., 2019). To investigate the hyphae formation of these viable atp2Δ/Δ cells, we observed the status of FITC-stained C. albicans (green) cocultured with MitoTraker-preloaded macrophages (red) when cocultured for 3 h. The results showed that WT cells completed morphogenesis with a germination rate of 45.4%, whereas the atp2Δ/Δ mutant cells were trapped inside macrophages in the yeast phase (Figures 6A, B). Then, we extracted RNA from phagocytic C. albicans to conduct RT-qPCR and found that the expression levels of the genes involved in hyphal formation (ECE1, HGC1, HWP1 and ALS3) were significantly lower than those in the WT group (P < 0.05) (Figure 6C).
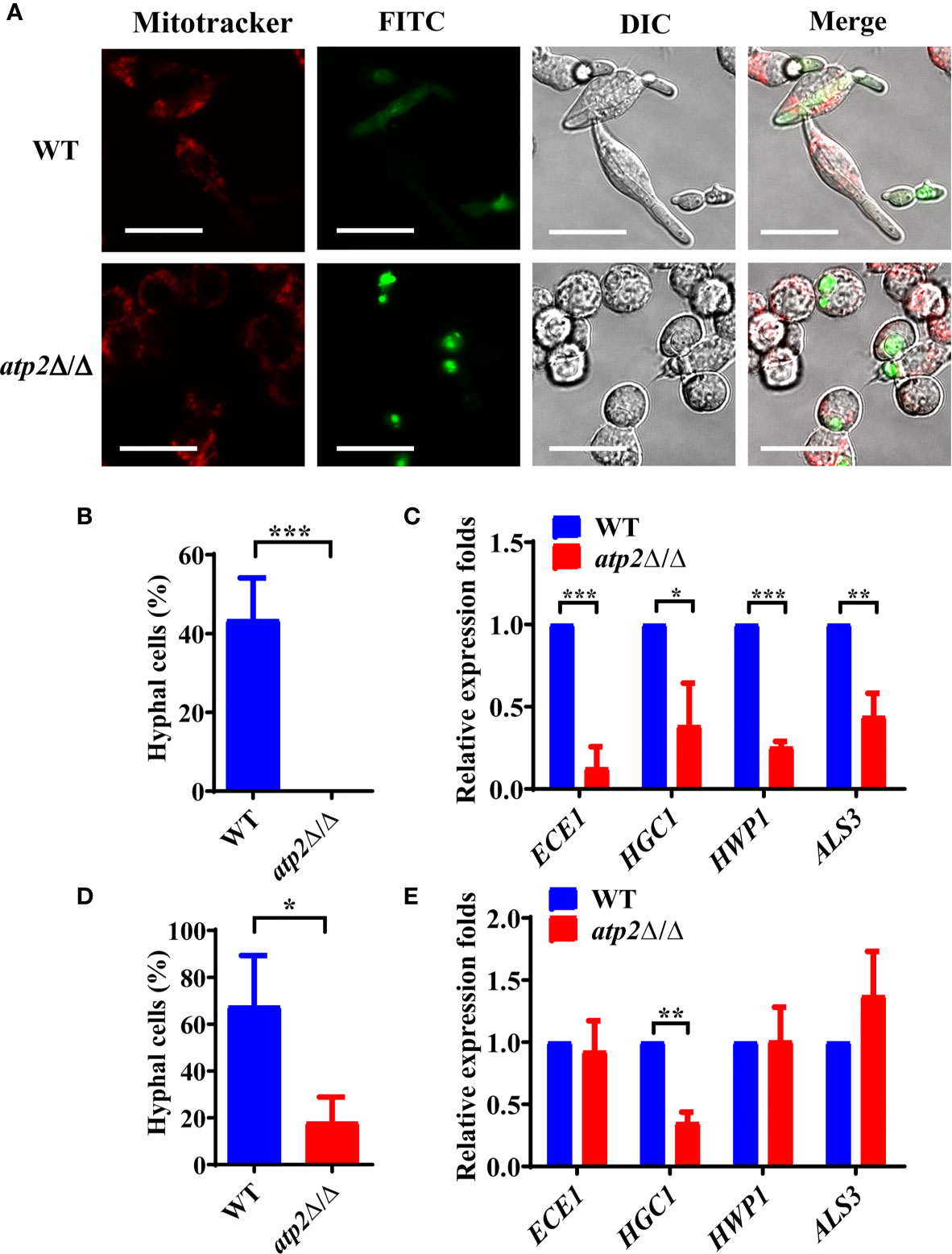
Figure 6 Hyphae formation of C. albicans inside and outside of the macrophages. (A) Representative images of FITC-stained WT and atp2Δ/Δ cells (green) co-cultured with MitoTracker Deep Red-preloaded RAW264.7 (red) at a 1:1 ratio for 3 h. Images were obtained by confocal laser scanning microscopy at channels Ex644/Em655, Ex488/Em525 and DIC, ×630. (B, D) The percentage of hyphal cells inside (B) and outside (D) macrophages was calculated from 50 cells each group. (C, E) The relative expression level of hyphal-related genes in WT and atp2Δ/Δ inside (C) and outside (E) macrophages after cocultured for 5 h. In (A), one representative experiment of three independent experiments is shown; scale bars, 20 µm. In (B–E) data are shown as mean ± s.d. *P < 0.05, **P < 0.01, ***P < 0.001; by Student’s t-test.
Interestingly, we found that the unengulfed atp2Δ/Δ mutant could undergo morphogenesis in the medium with a germination rate of 18.1% (Figure 6D). Under this condition, only HGC1 was expressed at a significantly lower level than that in the WT, while the expression levels of ECE1, HWP1 and ALS3 did not significantly differ (Figure 6E). The above results suggest that deletion of ATP2 prevents C. albicans from morphogenesis inside macrophages, but not outside of them.
Morphogenesis of ATP2 Mutant Cells Are Affected by the Presence or Absence of Glucose
Why atp2Δ/Δ cells were unable to form hyphae in macrophages? We performed hyphae formation assay in classic hyphae induction media (YPD+10% FBS, Spider, Lee’s), DMEM, and macrophage-mimicking media. The results showed that more than 90% of the WT cells formed elongated hyphae in all the media mentioned above, while the atp2Δ/Δ cells only formed short hyphae in media containing glucose (DMEM, YPD+10% FBS, and Lee’s) with germination rates around 10% (Figures 7A–D), and could not reach a level close to that of WT even with extended incubation time (Figure S2). In Spider medium (no glucose inside) and macrophage-mimicking media, the atp2Δ/Δ cells didn’t form hyphae (Figures 7A–D). Then we supplemented macrophage-mimicking media with 0.2% glucose, which was close to glucose level in blood, and found that hyphae formation of atp2Δ/Δ was partially restored (Figures 7E, F). The above results suggest that the hyphae formation of atp2Δ/Δ is reduced, but whether it forms hyphae also related to the presence or absence of glucose.
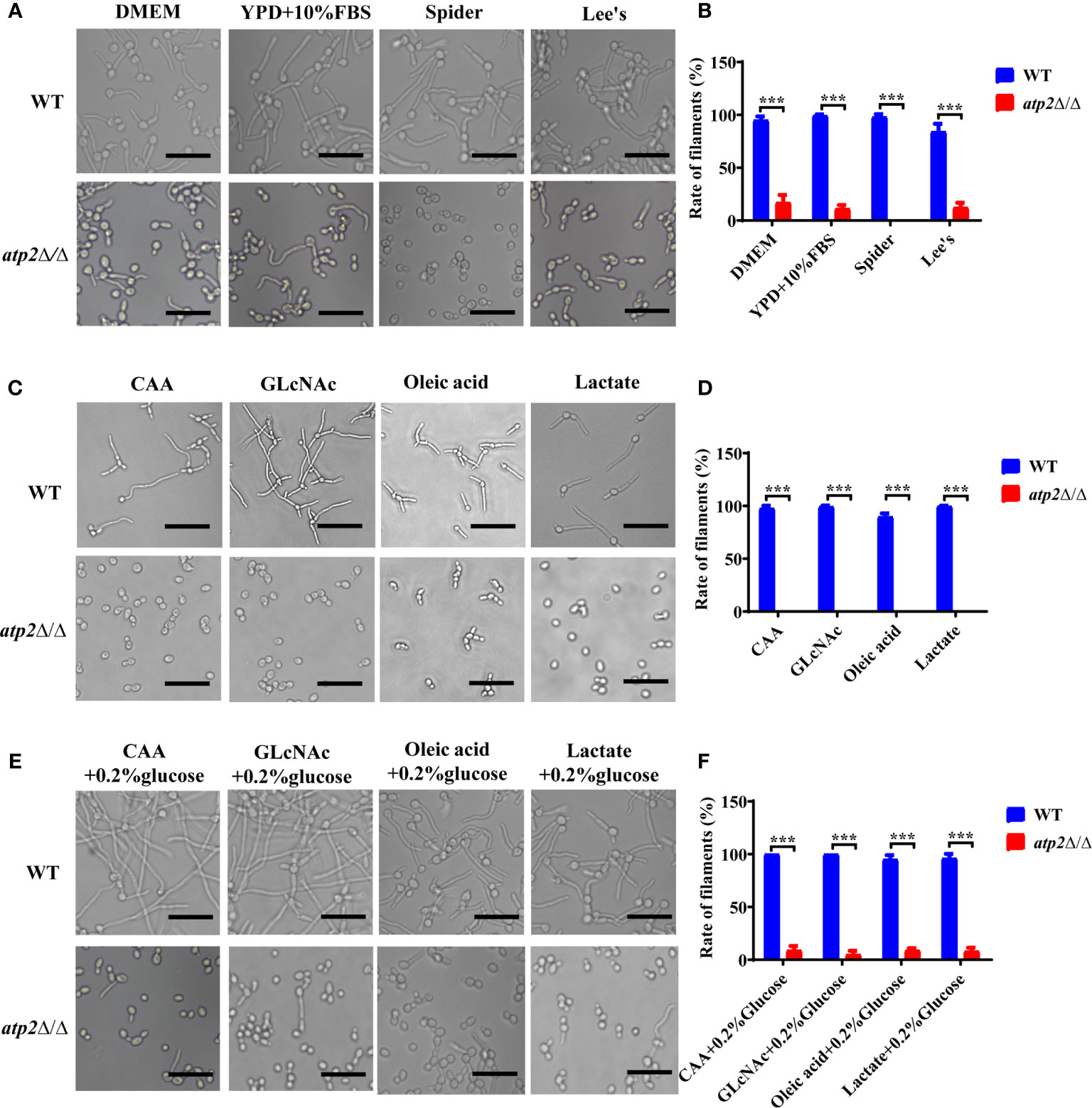
Figure 7 Hyphae formation of C albicans in various environments. (A, B) Representative images and hyphal cell percentages of WT and atp2Δ/Δ cultured in DMEM, YPD+10% FBS, Spider, and Lee’s medium at 37°C for 3 h. (C, D) Representative images and hyphal cell percentages of WT and atp2Δ/Δ cultured in CAA, GLcNAc, oleic acid, and lactate medium at 37°C for 3 h. (E, F) Representative images and hyphal cell percentages of WT and atp2Δ/Δ cultured in CAA, GLcNAc, oleic acid, and lactate medium supplemented with 0.2% glucose at 37°C for 3 h. In (B, D, F) the percentage of hyphal cells in each medium was calculated from at least 100 cells. The assays were performed in triplicate. ***P < 0.001; by Student’s t-test.
ATP2 Mutant Cells Are More Susceptible to Oxidative Stress
Oxidative stress within the phagosome creates a toxic environment that induces oxidative stress and programmed cell death in C. albicans (Dantas Ada et al., 2015). We further investigated the resistance of atp2Δ/Δ to oxidative stress (6 mM H2O2) in different environments. The results showed that 39.3% and 64.7% of the WT and atp2Δ/Δ cells were killed in the glucose medium at 6 h, respectively (P < 0.05) (Figure 8A). However, in the macrophage-mimicking media mentioned above, almost all atp2Δ/Δ cells were killed under the same H2O2 concentration, while the WT cells exhibited only slightly increased sensitivity to H2O2 in oleic acid (Figure 8A).
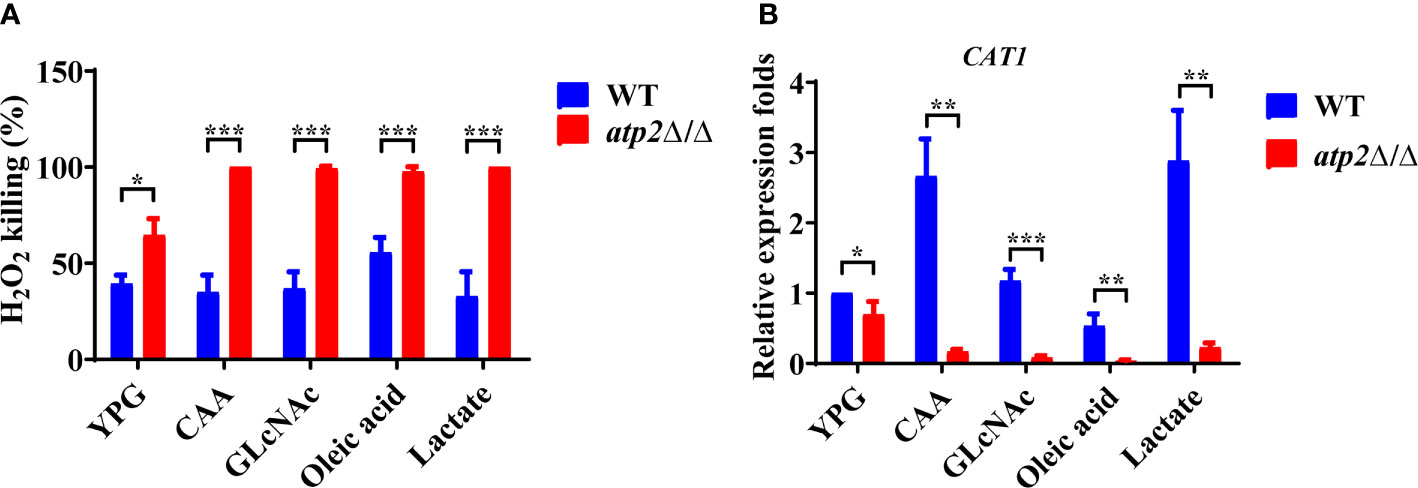
Figure 8 C albicans cells lacking ATP2 are more vulnerable to oxidative stress in macrophage-mimicking media. (A) The percentage of C albicans cells killed by 6 mM H2O2 in YNB medium plus 2% glucose, CAA, GLcNAc, oleic acid, and lactate at 6 h. The killing rate was determined by comparison with C albicans recovered without H2O2. (B) Relative expression levels of CAT1 in different media, and the expression level of WT in YNB + 2% glucose was set as 1. In (A, B), data are shown as mean ± s.d. *P < 0.05, **P < 0.01, ***P < 0.001; by Student’s t-test.
We further tested the transcription levels of the CAT1 gene (which protects C. albicans from oxidative stress) in the atp2Δ/Δ and WT cells under different conditions (Dantas Ada et al., 2015). Consistent with the phenotype, the expression level of CAT1 in the atp2Δ/Δ cells was decreased by 31.6% in the glucose medium compared with that in the WT (Figure 8B). However, the CAT1 levels in the atp2Δ/Δ cells were greatly downregulated in the alternative carbon source but were upregulated (CAA and lactate), unchanged (GlcNAc), or slightly downregulated (oleic acid) in the WT cells (P < 0.05) (Figure 8B). It can be seen that atp2Δ/Δ cells are more susceptible to oxidative stress, especially in the glucose-deficient environments.
Deletion of ATP2 in C. albicans Affects Proteins Involved in Its Escape From Macrophages
To further investigate the effect of ATP2 on multiple abilities associated with C. albicans escape from macrophages, we performed a proteomics study and found that deletion of ATP2 resulted in 112 proteins up-regulated and 268 proteins down-regulated (P < 0.05 and fold change > 1.5). Here, we focused on proteins involved in hyphae formation, stress responses, alternative carbon source utilization, etc.
When referred to the WT strain, several proteins required for hyphae formation were down-regulated, for example Ihd1p, Opt3p, and Ole1p were 0.21-, 0.54-, and 0.35-fold down-regulated in the atp2Δ/Δ mutant (Figure 9). In addition, putative glutathione peroxidase (Gpx2p) involved in Cap1p-dependent oxidative stress response was 0.28-fold down-regulated in atp2Δ/Δ mutant (Figure 9). However, Sod1p, which play a protective role against oxidative stress, was upregulated 3.3-fold in atp2Δ/Δ mutant (Figure 9). Proteins related to cell wall remodeling, GPI-anchored cell wall proteins (Exg2p, Rbt5p, and Crh11p) were up-regulated in atp2Δ/Δ mutant. Proteins associated with DNA damage repair were generally down-regulated.
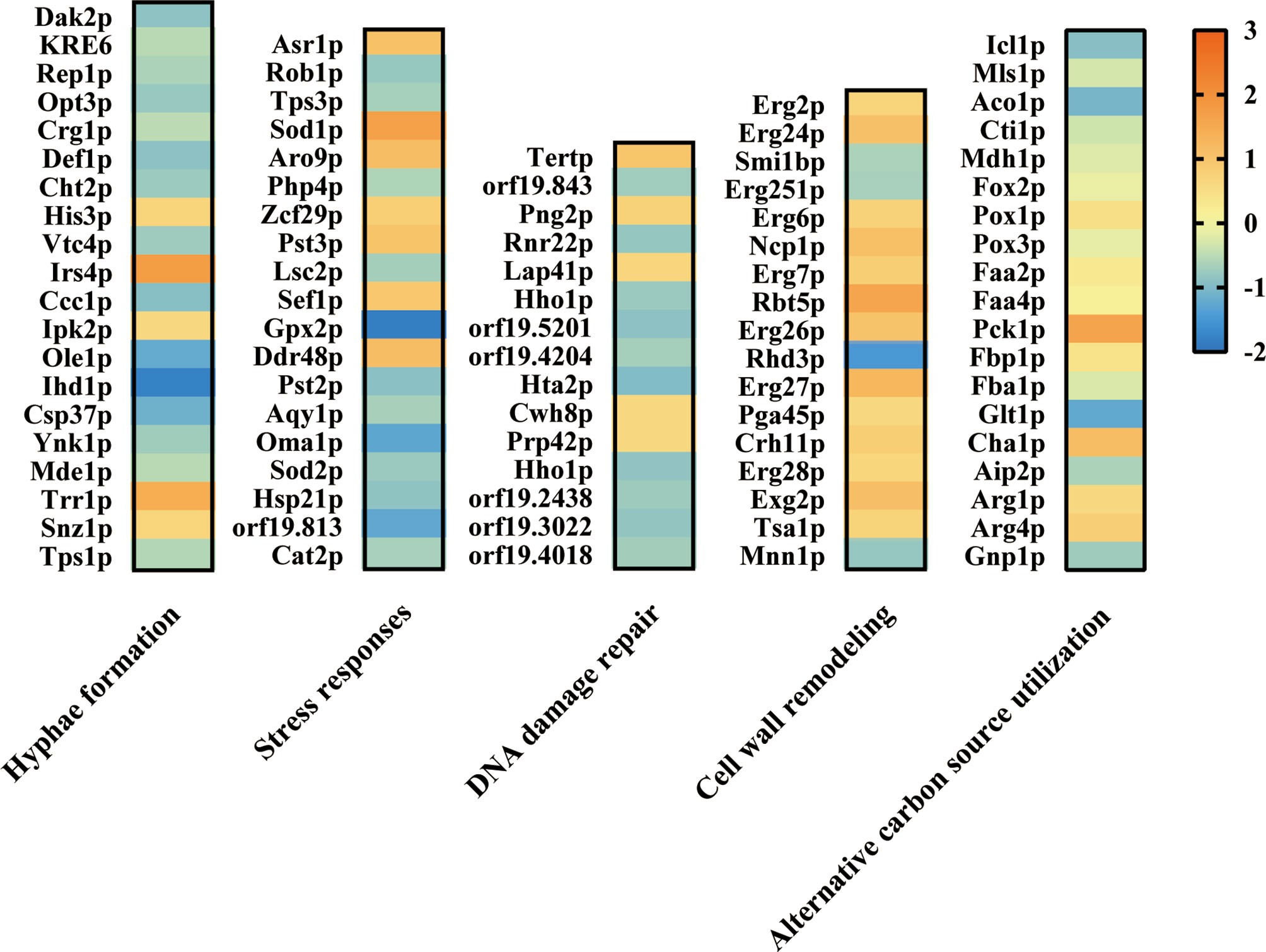
Figure 9 Effect of ATP2 on proteins associated with C. albicans escape from macrophage. Summary of differentially expressed proteins related to hyphae formation, stress responses, DNA damage repair, cell wall remodeling, and alternative carbon source utilization in proteomic results. Compared to WT, the differentially expressed proteins in atp2Δ/Δ mutant were identified at p < 0.05 and fold change > 1.5. The log2(FC) values were presented as a heat map.
As for proteins involved in alternative carbon source utilization, all enzymes (Icl1p, Mls1p, Aco1p, Cti1p, and Mdh1p) involved in the GC pathway were significantly repressed in the atp2Δ/Δ cells, two of the eight (Pck1p and Fbp1p) gluconeogenesis enzymes were upregulated, and there was no significant change in the β-oxidation pathway (Figure 9). The proteomic results were generally consistent with the phenotype, proteins related to multiple functions related to C. albicans escape from macrophages were repressed to some extent.
Discussion
The ability of C. albicans to persist in the human host and cause disease requires the capacity to evade and circumvent host defense mechanisms, especially macrophage-mediated clearance (O’Meara et al., 2018; Austermeier et al., 2020). Therefore, the C. albicans-macrophage interaction process offers promising targets for much-needed novel therapeutics to treat fungal infections and has not been exploited as a therapeutic target to date.
We found that the ATP2 deletion increased the macrophage clearance of C. albicans from 30.8% to 98.8%, almost completely preventing the escape of C. albicans. However, not only ATP2, there are other genes affecting the escape of C. albicans from macrophages. When calculated the variations in clearance rate using their parental strains as standard, the deletion of PHO4 (78%), TRK1 (70%), ALI1 (65.5%), or CYR1 (64.1%) caused changes in macrophage clearance rates of similar magnitude compared to ATP2 (67.4%) (Table S2) (Rocha et al., 2001; Ikeh et al., 2016; Llopis-Torregrosa et al., 2019; Williams and Lorenz, 2020). When directly comparing the percentage of killed mutant cells, the deletion of ALI1, CYR1, RTT109, COX4, or RAS1 increased the clearance of C. albicans to more than 90% (98.2% of the atp2Δ/Δ) (Table S2) (Marcil et al., 2002; Lopes da Rosa et al., 2010; Williams and Lorenz, 2020). Although different macrophage types, co-culture times, MOIs can lead to differences in the data from one study to another, it can provide us with some reference information when all mutants are not available for simultaneous experiments. Through this analysis, we found that ATP2, as well as two other genes related to mitochondrial function (ALI1 and CYR1) were at the forefront of the genes currently known to affect clearance.
The ATP2 mutant also displayed a reduced ability to escape macrophage clearance in vivo. We constructed macrophage-depleted mice by clodronate liposome, a simple, and stable method that has been widely used to study macrophage-pathogen interactions (Wirnsberger et al., 2016; Moreno, 2018). The persist of atp2Δ/Δ in macrophage-depleted mice suggested that it could survive, if not grow, in organs. Our results of growth in tissue homogenates supported this conclusion (Table S3). Thus, under the premise that atp2Δ/Δ could survive and even grow in vivo, the results of atp2Δ/Δ were undetectable in various organs of macrophage-normal mice suggesting that the mutant strain had a reduced ability to escape macrophage clearance in vivo.
Most C. albicans wild type cells can escape from the macrophage in 6-8 h (Wartenberg et al., 2014). In the early phase (1 h) of C. albicans-macrophages interaction, C. albicans switched to a slow gluconeogenic growth mode in the glucose-deficient environment, and in the late phase (6-8 h), along with hyphae formation and escape, C. albicans resumed rapid glycolytic growth (Lorenz et al., 2004; Tucey et al., 2018; Laurian et al., 2020). Why did ATP2 deletion lead to a substantial reduction of C. albicans escaping macrophage clearance? According to our results, atp2Δ/Δ cells had good glycolytic growth but were unable to undergo gluconeogenic growth. However, even if the mutant failed to proliferate, 60-80% of atp2Δ/Δ cells could remained viable for 8 h in glucose-deficient environments. Our previous work reported that viability of atp2Δ/Δ decreased substantially in glucose-deficient media after 24 h, but it could survive and proliferate in glucose-sufficient media (Li et al., 2018). Therefore, the ability to escape macrophages and resume glycolytic growth within 8 h is critical for atp2Δ/Δ to survive C. albicans-macrophages interaction.
Hyphal morphogenesis is a key factor promoting C. albicans escape from macrophages in a physical or inflammation dependent manner (Peroumal et al., 2019; Rogiers et al., 2019). We observed that atp2Δ/Δ cells were all trapped within macrophages in the yeast form. However, when we studied the morphogenesis ability of atp2Δ/Δ we found that the hyphae formation of the mutant was reduced but not absent in media containing glucose, and the expression level of related genes (ECE1, HWP1 and ALS3) was not even statistically different from WT. Therefore, the inability of atp2Δ/Δ cells to form hyphae within macrophages was likely due to their fitness defect rather than specific hyphae formation defect.
Unlike specific hyphae formation or carbon source utilization genes, ATP2 has a broader effect on the function involved in C. albicans-macrophages interaction. Specific hyphae formation defective mutant (cph1Δ/efg1Δ) was unable to escape from macrophages after 24 h, but no other functional defects allowed these cells to survive and still replicate in the yeast form (Wartenberg et al., 2014). Moreover, the sustained interaction of cph1Δ/efg1Δ with macrophages may lead to new variants capable of escape from macrophage (Wartenberg et al., 2014). Due to the broad effect of ATP2 on the ability of C. albicans associated with escape from macrophage, C. albicans cannot bypass or tolerate its inhibitory effects, rendering it a better target than specific functionally related genes.
Since F1Fo-ATP synthase is evolutionarily conserved in bacteria, fungi, and mammals, and using the β subunit (encoded by ATP2) or other subunits of F1Fo-ATP synthase as drug target may carry a risk of toxicity (Jonckheere et al., 2012). Bedaquiline is an FDA-approved anti-tuberculosis drug that targets the c-ring of F1Fo-ATP synthase, which is 20,000 times more sensitive to Mycobacterium tuberculosis than to mammal cells (Andries et al., 2005; Fiorillo et al., 2016). Although the β subunit has not yet been used as an anti-infection drug target, some small molecules and monoclonal antibodies targeting the β subunit have entered phase I or II clinical trials in antitumor studies, such as angiostatin, Hai178, and Aurovertin B, all of which have shown selective inhibition of tumor cells with low toxicity to normal cells (Moser et al., 2001; Huang et al., 2008; Chen et al., 2016).
In summary, deletion of ATP2 prevents C. albicans from escaping macrophage clearance in vitro and in vivo. ATP2 has the functional basis as a drug target of C. albicans- macrophages interaction and deserves further investigation in the future.
Data Availability Statement
The proteomics data presented in the study were deposited in the ProteomeXchange Consortium, accession number was PXD024039.
Ethics Statement
The animal study was reviewed and approved by Laboratory Animal Ethics Committee of Jinan University.
Author Contributions
YiZ, CT, and ZZ. contributed equally to the article. YiZ and SL developed the concept and designed the research plan. YiZ, CT, ZZ, YaZ, and LW performed experiments. YiZ, ZZ, and CT performed statistical analysis. YiZ, ZZ, and CT wrote the paper. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by the National Natural Science Foundation of China (81971913/81471995/81903675) and China Postdoctoral Science Foundation (2019M653291).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcimb.2021.643121/full#supplementary-material
References
Andries K., Verhasselt P., Guillemont J., Göhlmann H. W., Neefs J. M., Winkler H., et al. (2005). A diarylquinoline drug active on the ATP synthase of Mycobacterium tuberculosis. Science 307 (5707), 223–227. doi: 10.1126/science.1106753
Austermeier S., Kasper L., Westman J., Gresnigt M. S. (2020). I want to break free - macrophage strategies to recognize and kill Candida albicans, and fungal counter-strategies to escape. Curr. Opin. Microbiol. 58, 15–23. doi: 10.1016/j.mib.2020.05.007
Bassetti M., Righi E., Montravers P., Cornely O. A. (2018). What has changed in the treatment of invasive candidiasis? A look at the past 10 years and ahead. J. Antimicrob. Chemother. 73 (suppl_1), i14–i25. doi: 10.1093/jac/dkx445
Chen C., Liang H., Liao X., Pan J., Chen J., Zhao S., et al. (2016). A humanized chimeric antibody Hai178 targeted to the β subunit of F1F0 ATP synthase. Tumour Biol 37, 15903–15912. doi: 10.1007/s13277-016-5423-1
Danhof H. A., Lorenz M. C. (2015). The Candida albicans ATO Gene Family Promotes Neutralization of the Macrophage Phagolysosome. Infect. Immun. 83 (11), 4416–4426. doi: 10.1128/iai.00984-15
Danhof H. A., Vylkova S., Vesely E. M., Ford A. E., Gonzalez-Garay M., Lorenz M. C. (2016). Robust Extracellular pH Modulation by Candida albicans during Growth in Carboxylic Acids. mBio 7 (6), e01646–16. doi: 10.1128/mBio.01646-16
Dantas Ada S., Day A., Ikeh M., Kos I., Achan B., Quinn J. (2015). Oxidative stress responses in the human fungal pathogen, Candida albicans. Biomolecules 5 (1), 142–165. doi: 10.3390/biom5010142
Davis S. L., Nuermberger E. L., Um P. K., Vidal C., Jedynak B., Pomper M. G., et al. (2009). Noninvasive pulmonary [18F]-2-fluoro-deoxy-D-glucose positron emission tomography correlates with bactericidal activity of tuberculosis drug treatment. Antimicrob. Agents Chemother. 53 (11), 4879–4884. doi: 10.1128/AAC.00789-09
Fiorillo M., Lamb R., Tanowitz H. B., Cappello A. R., Martinez-Outschoorn U. E., Sotgia F., et al. (2016). Bedaquiline, an FDA-approved antibiotic, inhibits mitochondrial function and potently blocks the proliferative expansion of stem-like cancer cells (CSCs). Aging (Albany NY) 8 (8), 1593–1607. doi: 10.18632/aging.100983
Herrero-de-Dios C., Day A. M., Tillmann A. T., Kastora S. L., Stead D., Salgado P. S., et al. (2018). Redox Regulation, Rather than Stress-Induced Phosphorylation, of a Hog1 Mitogen-Activated Protein Kinase Modulates Its Nitrosative-Stress-Specific Outputs. mBio 9 (2), e02229–17. doi: 10.1128/mBio.02229-17
Huang T. C., Chang H. Y., Hsu C. H., Kuo W. H., Chang K. J., Juan H. F. (2008). Targeting therapy for breast carcinoma by ATP synthase inhibitor aurovertin B. J. Proteome Res. 7 (4), 1433–1444. doi: 10.1021/pr700742h
Ikeh M. A., Kastora S. L., Day A. M., Herrero-de-Dios C. M., Tarrant E., Waldron K. J., et al. (2016). Pho4 mediates phosphate acquisition in Candida albicans and is vital for stress resistance and metal homeostasis. Mol. Biol. Cell 27 (17), 2784–2801. doi: 10.1091/mbc.E16-05-0266
Jain P., Sethi S. C., Pratyusha V. A., Garai P., Naqvi N., Singh S., et al. (2018). Ras signaling activates glycosylphosphatidylinositol (GPI) anchor biosynthesis via the GPI-N-acetylglucosaminyltransferase (GPI-GnT) in Candida albicans. J. Biol. Chem. 293 (31), 12222–12238. doi: 10.1074/jbc.RA117.001225
Jonckheere A. I., Smeitink J. A., Rodenburg R. J. (2012). Mitochondrial ATP synthase: architecture, function and pathology. J. Inherit Metab. Dis. 35 (2), 211–225. doi: 10.1007/s10545-011-9382-9
Kitahara N., Morisaka H., Aoki W., Takeda Y., Shibasaki S., Kuroda K., et al. (2015). Description of the interaction between Candida albicans and macrophages by mixed and quantitative proteome analysis without isolation. AMB Express 5 (1):127. doi: 10.1186/s13568-015-0127-2
Laurian R., Jacot-des-Combes C., Bastian F., Dementhon K., Cotton P. (2020). Carbon metabolism snapshot by ddPCR during the early step of Candida albicans phagocytosis by macrophages. Pathog. Dis. 78 (1), ftaa014. doi: 10.1093/femspd/ftaa014
Li S. X., Song Y. J., Zhang Y. S., Wu H. T., Guo H., Zhu K. J., et al. (2017). Mitochondrial Complex V α Subunit Is Critical for Candida albicans Pathogenicity through Modulating Multiple Virulence Properties. Front. Microbiol. 8:285. doi: 10.3389/fmicb.2017.00285
Li S.-X., Wu H.-T., Liu Y.-T., Jiang Y.-Y., Zhang Y.-S., Liu W.-D., et al. (2018). The F1Fo-ATP Synthase β Subunit Is Required for Candida albicans Pathogenicity Due to Its Role in Carbon Flexibility. Front. Microbiol. 9:1025. doi: 10.3389/fmicb.2018.01025
Lionakis M. S., Swamydas M., Fischer B. G., Plantinga T. S., Johnson M. D., Jaeger M., et al. (2013). CX3CR1-dependent renal macrophage survival promotes Candida control and host survival. J. Clin. Invest. 123 (12), 5035–5051. doi: 10.1172/JCI71307
Llopis-Torregrosa V., Vaz C., Monteoliva L., Ryman K., Engstrom Y., Gacser A., et al. (2019). Trk1-mediated potassium uptake contributes to cell-surface properties and virulence of Candida glabrata. Sci. Rep. 9 (1), 7529. doi: 10.1038/s41598-019-43912-1
Lopes da Rosa J., Boyartchuk V. L., Zhu L. J., Kaufman P. D. (2010). Histone acetyltransferase Rtt109 is required for Candida albicans pathogenesis. Proc. Natl. Acad. Sci. U. S. A. 107 (4), 1594–1599. doi: 10.1073/pnas.0912427107
Lorenz M. C., Bender J. A., Fink G. R. (2004). Transcriptional response of Candida albicans upon internalization by macrophages. Eukaryot Cell 3 (5), 1076–1087. doi: 10.1128/ec.3.5.1076-1087.2004
Marcet-Houben M., Marceddu G., Gabaldón T. (2009). Phylogenomics of the oxidative phosphorylation in fungi reveals extensive gene duplication followed by functional divergence. BMC Evol. Biol. 9, 295–295. doi: 10.1186/1471-2148-9-295
Marcil A., Harcus D., Thomas D. Y., Whiteway M. (2002). Candida albicans killing by RAW 264.7 mouse macrophage cells: effects of Candida genotype, infection ratios, and gamma interferon treatment. Infect. Immun. 70 (11), 6319–6329. doi: 10.1128/iai.70.11.6319-6329.2002
Martínez-Esparza M., Martínez-Vicente E., González-Párraga P., Ros J. M., García-Peñarrubia P., Argüelles J. C. (2009). Role of trehalose-6P phosphatase (TPS2) in stress tolerance and resistance to macrophage killing in Candida albicans. Int. J. Med. Microbiol. 299 (6), 453–464. doi: 10.1016/j.ijmm.2008.12.001
Moreno S. G. (2018). Depleting Macrophages In Vivo with Clodronate-Liposomes. Methods Mol. Biol. 1784, 259–262. doi: 10.1007/978-1-4939-7837-3_23
Moser T. L., Kenan D. J., Ashley T. A., Roy J. A., Goodman M. D., Misra U. K., et al. (2001). Endothelial cell surface F1-F0 ATP synthase is active in ATP synthesis and is inhibited by angiostatin. Proc. Natl. Acad. Sci. U. S. A. 98 (12), 6656–6661. doi: 10.1073/pnas.131067798
Netea M. G., Joosten L. A. B., van der Meer J. W. M., Kullberg B.-J., van de Veerdonk F. L. (2015). Immune defence against Candida fungal infections. Nat. Rev. Immunol. 15 (10), 630–642. doi: 10.1038/nri3897
Ngo L. Y., Kasahara S., Kumasaka D. K., Knoblaugh S. E., Jhingran A., Hohl T. M. (2014). Inflammatory monocytes mediate early and organ-specific innate defense during systemic candidiasis. J. Infect. Dis. 209 (1), 109–119. doi: 10.1093/infdis/jit413
O’Meara T. R., Duah K., Guo C. X., Maxson M. E., Gaudet R. G., Koselny K., et al. (2018). High-Throughput Screening Identifies Genes Required for Candida albicans Induction of Macrophage Pyroptosis. mBio 9 (4), e01581–18. doi: 10.1128/mBio.01581-18
Pappas P. G., Lionakis M. S., Arendrup M. C., Ostrosky-Zeichner L., Kullberg B. J. (2018). Invasive candidiasis. Nat. Rev. Dis. Primers 4, 18026. doi: 10.1038/nrdp.2018.26
Perez-Riverol Y., Csordas A., Bai J., Bernal-Llinares M., Hewapathirana S., Kundu D. J., et al. (2019). The PRIDE database and related tools and resources in 2019: improving support for quantification data. Nucleic Acids Res. 47 (D1), D442–d450. doi: 10.1093/nar/gky1106
Peroumal D., Manohar K., Patel S. K., Kumari P., Sahu S. R., Acharya N. (2019). Virulence and pathogenicity of a Candida albicans mutant with reduced filamentation. Cell Microbiol. 21 (12), e13103. doi: 10.1111/cmi.13103
Piekarska K., Mol E., van den Berg M., Hardy G., van den Burg J., van Roermund C., et al. (2006). Peroxisomal fatty acid beta-oxidation is not essential for virulence of Candida albicans. Eukaryot Cell 5 (11), 1847–1856. doi: 10.1128/ec.00093-06
Prigneau O., Porta A., Poudrier J. A., Colonna-Romano S., Noël T., Maresca B. (2003). Genes involved in beta-oxidation, energy metabolism and glyoxylate cycle are induced by Candida albicans during macrophage infection. Yeast 20 (8), 723–730. doi: 10.1002/yea.998
Qian Q., Jutila M. A., Van Rooijen N., Cutler J. E. (1994). Elimination of mouse splenic macrophages correlates with increased susceptibility to experimental disseminated candidiasis. J. Immunol. 152 (10), 5000–5008.
Rocha C. R., Schröppel K., Harcus D., Marcil A., Dignard D., Taylor B. N., et al. (2001). Signaling through adenylyl cyclase is essential for hyphal growth and virulence in the pathogenic fungus Candida albicans. Mol. Biol. Cell 12 (11), 3631–3643. doi: 10.1091/mbc.12.11.3631
Rogiers O., Frising U. C., Kucharíková S., Jabra-Rizk M. A., van Loo G., Van Dijck P., et al. (2019). Candidalysin Crucially Contributes to Nlrp3 Inflammasome Activation by Candida albicans Hyphae. mBio 10 (1), e02221–18. doi: 10.1128/mBio.02221-18
Rolle A.-M., Hasenberg M., Thornton C. R., Solouk-Saran D., Männ L., Weski J., et al. (2016). ImmunoPET/MR imaging allows specific detection of Aspergillus fumigatus lung infection in vivo. Proc. Natl. Acad. Sci. 113 (8), E1026–E1033. doi: 10.1073/pnas.1518836113
She X., Zhang L., Chen H., Calderone R., Li D. (2013). Cell surface changes in the Candida albicans mitochondrial mutant goa1Δ are associated with reduced recognition by innate immune cells. Cell Microbiol. 15 (9), 1572–1584. doi: 10.1111/cmi.12135
Shen Q., Rappleye C. A. (2020). Living Within the Macrophage: Dimorphic Fungal Pathogen Intracellular Metabolism. Front. Cell Infect. Microbiol. 10:592259. doi: 10.3389/fcimb.2020.592259
Truong T., Zeng G., Lim T. K., Cao T., Pang L. M., Lee Y. M., et al. (2019). Proteomics Analysis of Candida albicans dnm1 Haploid Mutant Unraveled the Association between Mitochondrial Fission and Antifungal Susceptibility. Proteomics 20 (1), e1900240. doi: 10.1002/pmic.201900240
Tucey T. M., Verma J., Harrison P. F., Snelgrove S. L., Lo T. L., Scherer A. K., et al. (2018). Glucose Homeostasis Is Important for Immune Cell Viability during Candida Challenge and Host Survival of Systemic Fungal Infection. Cell Metab. 27 (5), 988–1006.e1007. doi: 10.1016/j.cmet.2018.03.019
Uwamahoro N., Verma-Gaur J., Shen H. H., Qu Y., Lewis R., Lu J., et al. (2014). The pathogen Candida albicans hijacks pyroptosis for escape from macrophages. mBio 5 (2), e00003–e00014. doi: 10.1128/mBio.00003-14
Vylkova S., Lorenz M. C. (2014). Modulation of Phagosomal pH by Candida albicans Promotes Hyphal Morphogenesis and Requires Stp2p, a Regulator of Amino Acid Transport. PLoS Pathog. 10 (3), e1003995. doi: 10.1371/journal.ppat.1003995
Wartenberg A., Linde J., Martin R., Schreiner M., Horn F., Jacobsen I. D., et al. (2014). Microevolution of Candida albicans in macrophages restores filamentation in a nonfilamentous mutant. PLoS Genet. 10 (12), e1004824–e1004824. doi: 10.1371/journal.pgen.1004824
Weiss G., Schaible U. E. (2015). Macrophage defense mechanisms against intracellular bacteria. Immunol. Rev. 264 (1), 182–203. doi: 10.1111/imr.12266
Westman J., Hube B., Fairn G. D. (2019). Integrity under stress: Host membrane remodelling and damage by fungal pathogens. Cell. Microbiol. 21 (4), e13016. doi: 10.1111/cmi.13016
Williams R. B., Lorenz M. C. (2020). Multiple Alternative Carbon Pathways Combine To Promote Candida albicans Stress Resistance, Immune Interactions, and Virulence. mBio 11 (1), e03070–19. doi: 10.1128/mBio.03070-19
Wirnsberger G., Zwolanek F., Asaoka T., Kozieradzki I., Tortola L., Wimmer R. A., et al. (2016). Inhibition of CBLB protects from lethal Candida albicans sepsis. Nat. Med. 22 (8), 915–923. doi: 10.1038/nm.4134
Keywords: Candida albicans, ATP2 gene, macrophage, host-pathogen interaction, alternative carbon source, glyoxylate cycle
Citation: Zhang Y, Tang C, Zhang Z, Li S, Zhao Y, Weng L and Zhang H (2021) Deletion of the ATP2 Gene in Candida albicans Blocks Its Escape From Macrophage Clearance. Front. Cell. Infect. Microbiol. 11:643121. doi: 10.3389/fcimb.2021.643121
Received: 17 December 2020; Accepted: 30 March 2021;
Published: 16 April 2021.
Edited by:
Miguel Cacho Teixeira, University of Lisbon, PortugalReviewed by:
Sascha Brunke, Leibniz Institute for Natural Product Research and Infection Biology, GermanySandra Paiva, University of Minho, Portugal
Copyright © 2021 Zhang, Tang, Zhang, Li, Zhao, Weng and Zhang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Hong Zhang, dHpoYW5naEBqbnUuZWR1LmNu
†These authors have contributed equally to this work and share first authorship
 Yishan Zhang
Yishan Zhang Chuanyan Tang
Chuanyan Tang Zhanpeng Zhang1,2†
Zhanpeng Zhang1,2† Hong Zhang
Hong Zhang