
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Cell. Infect. Microbiol. , 09 March 2021
Sec. Clinical Microbiology
Volume 11 - 2021 | https://doi.org/10.3389/fcimb.2021.639065
This article is part of the Research Topic Drug Development against Protozoan Parasites View all 10 articles
 Arif Nurkanto1,2
Arif Nurkanto1,2 Ghulam Jeelani2
Ghulam Jeelani2 Herbert J. Santos2
Herbert J. Santos2 Yulia Rahmawati2
Yulia Rahmawati2 Mihoko Mori3,4
Mihoko Mori3,4 Yumi Nakamura5
Yumi Nakamura5 Kana Goto5
Kana Goto5 Yoko Saikawa5
Yoko Saikawa5 Takeshi Annoura6
Takeshi Annoura6 Yuzuru Tozawa7
Yuzuru Tozawa7 Takaya Sakura8
Takaya Sakura8 Daniel Ken Inaoka8
Daniel Ken Inaoka8 Kazuro Shiomi3
Kazuro Shiomi3 Tomoyoshi Nozaki2*
Tomoyoshi Nozaki2*Coenzyme A (CoA) is a well-known cofactor that plays an essential role in many metabolic reactions in all organisms. In Plasmodium falciparum, the most deadly among Plasmodium species that cause malaria, CoA and its biosynthetic pathway have been proven to be indispensable. The first and rate-limiting reaction in the CoA biosynthetic pathway is catalyzed by two putative pantothenate kinases (PfPanK1 and 2) in this parasite. Here we produced, purified, and biochemically characterized recombinant PfPanK1 for the first time. PfPanK1 showed activity using pantetheine besides pantothenate, as the primary substrate, indicating that CoA biosynthesis in the blood stage of P. falciparum can bypass pantothenate. We further developed a robust and reliable screening system to identify inhibitors using recombinant PfPanK1 and identified four PfPanK inhibitors from natural compounds.
Malaria is an infectious disease in humans caused by Plasmodium species and transmitted through the bites of female Anopheles mosquitoes. Among five Plasmodium species, P. falciparum, causes the highest mortality worldwide, and is responsible for 99.7% cases in Africa, 50% in South-Eastern Asia, 71% in the Eastern Mediterranean, and 65% in the Western Pacific. It is estimated that this parasite infects more than 200 million people and is responsible for 445,000 deaths in 2017 (WHO Report, 2017), with the number being slightly decreased to 405,000 in 2018 (WHO, 2019). Although many preventive and therapeutic efforts combating malaria helped in reducing its mortality in the last decade, the emergence and spread of antimalarial resistance does not seem to cease. Currently, artemisinin combination therapies (ACTs) are recommended as the standard first-line treatment for uncomplicated falciparum malaria (Visser et al., 2014) to repress malarial drug resistance. However, ACTs resistance were also reported in both Asian (Dondorp et al., 2009) and African countries (Lu et al., 2017).
A number of antimalarial drug candidates recently entered the preclinical and clinical trial stages. Some of them have been proven to have novel targets, while the precise mechanism of action of other candidates remains unknown (Wells et al., 2015; Tse et al., 2019). Current anti-malarial chemotherapy is based on standard therapies in which an artemisinin derivative and its partner drug with a preferably synergistic mechanism are used in combination. However, new antimalarials that are safe, effective with a single oral administration, have a novel mechanism of action with no cross-resistance to existing antimalarials, and active against multiple life stages including both asexual erythrocytic and liver stages in humans and sexual stages in the mosquito, with anti-relapse (P. vivax) and transmission-blocking activities (Fidock, 2010; Grimberg and Mehlotra, 2011; Burrows et al., 2017; Tse et al., 2019; Favuzza et al., 2020) are still needed.
Coenzyme A (CoA) is an essential cofactor in many metabolic processes, involving more than 9% of approximately 3,500 cellular activities (www.brenda-enzymes.info/). CoA biosynthesis has been attracting attention as a promising drug target (Fletcher et al., 2016). In P. falciparum, CoA is generated by four enzymatic reactions initiated by the conversion of pantothenate (Vitamin B5) to 4-phosphopantothenate, catalyzed by pantothenate kinase (PfPanK) (Saliba and Kirk, 2001). Other steps are mediated by phosphopantothenoylcysteine synthetase (PfPPCS)-phosphopantothenoylcysteine decarboxylase (PfPPCDC) bifunctional enzyme, phosphopantetheine adenylyltransferase (PfPPAT), and dephospho-CoA kinase (PfDPCK), with the last enzyme catalyzing the rate limiting step (Spry et al., 2008). Most prokaryotes and some eukaryotes including mammals and plants are reported to produce pantothenate de novo (Begley et al., 2001; Chakauya et al., 2008; Spry et al., 2008). In Plasmodium, it has been shown that pantothenate cannot be synthesized de novo, thus it is only obtained from the host (Spry et al., 2008; Spry and Saliba, 2009; Spry et al., 2010). The blood stage parasites of P. falciparum are absolutely dependent on pantothenate from human plasma or the culture medium (Divo et al., 1985; Mamoun et al., 2010; Augagneur et al., 2013; Bobenchik et al., 2013). In the whole CoA biosynthetic pathway, PfPanK has been best studied in P. falciparum (Saliba et al., 1998; Spry et al., 2008). All inhibitors against PfPanK that were so far identified are structurally related to pantothenate, including pantothenamide metabolites (Schalkwijk et al., 2019), pantothenol (PanOH), and CJ-15,801 (Saliba et al., 2005; Spry et al., 2005). Although these potential pantothenate analogs were presumed to target the CoA pathway, they apparently showed no direct inhibition against PfPanK and likely inhibit other targets such as PfPPCS and CoA-utilizing enzymes (Tjhin et al., 2018). Thus, no PfPanK specific inhibitors have been discovered until now.
Although PanK activity was previously measured in lysates from the P. falciparum blood stage parasites (Saliba et al., 2005; Spry et al., 2005; Spry et al., 2018; Tjhin et al., 2018), no previous report has described successful expression of bacterial recombinant PfPanK (Hart et al., 2016). In this study, we have developed the overexpression and purification of active PfPanK using an Escherichia coli expression system, and biochemically characterized PfPanK. We have also developed a robust enzymatic screening system using recombinant PfPanK and screened the natural compounds from microbes and plants against PfPanK to successfully identify inhibitors with structures unrelated to pantothenate.
Escherichia coli BL21Star™ (DE3) strain was purchased from Invitrogen (Carlsbad, CA, USA). All chemicals of analytical grade were purchased from Wako (Tokyo, Japan) or Sigma-Aldrich (Tokyo, Japan) unless otherwise stated. Magnesium-free ATP was purchased from DiscoverX (Fremont, CA, USA). Ni2+-NTA agarose was purchased from Novagen (Darmstadt, Germany). Structurally elucidated natural products with known efficacy. Gnetin A, gnetin C, and gnemonoside D were gift from Dr. Azuma Watanabe, AM, Fujisawa, Japan. Non-binding, 96-well microplates were purchased from Greiner Bio-One (Frickenhausen, Germany). Plasmodium falciparum sensitive strain 3D7 used in asexual blood stage phenotypic assay. AlbuMAX II was purchased from Gibco (Life Technologies, Carlsbad, CA, USA), hypoxanthine (Sigma), sodium l-lactate and nitro-tetrazolium blue chloride (NBT) (Fujifilm, Wako), APAD (Oriental Yeast, Japan).
PanK protein sequences (38 orthologues) from representative bacterial and eukaryotic taxa were retrieved from the non-redundant protein sequences (nr) database of the National Center for Biotechnology Information (NCBI, http://www.ncbi.nlm.nih.gov/). As a query, we used the PfPanK1 (PF3D7_1420600/XP_001348373) and only the sequences with an E-value lower than 1 x 10-10 were selected. Sequences were aligned using the Muscle program (Edgar, 2004) in SeaView package version 4.6.1 (Gouy et al., 2010). To select hit amino acid sequences for the model, the data matrices for phylogeny were subjected to the IQTREE program (Nguyen et al., 2015). The maximum likelihood (ML) analysis implemented in the RAxML program version 7.2.6 (Stamatakis, 2006) was used to infer ML tree. Bootstrap values higher than 50 (in percentages) are indicated on the corresponding internal branches of the ML tree constructed using FigTree program Version 1.4.2 (http://tree.bio.ed.ac.uk/software/figtree/).
The genes encoding two putative PanKs, PfPanK1 and PfPanK2, were synthesized commercially after codon codon optimization for expression in E. coli or wheat-germ cell-free system, respectively (Eurofins Genomics, Tokyo, Japan). The E. coli codon-optimized PfPanK1 gene (NCBI accession number: MW331581) was inserted into the plasmid pCold™1 histidine-tag vector (Takara, Tokyo, Japan) to produce pCold™1-PfPanK1. The plasmid was introduced to BL21Star™ (DE3) chemically competent cells (ThermoFisher Scientific, Waltham, MA, USA) and the transformed E. coli strains were cultured at 37°C in Luria Bertani medium (Invitrogen) in the presence of 100 μg/ml ampicillin (Nacalai Tesque). The overnight culture was used to inoculate 1 L of freshly prepared medium. The culture was further incubated with 100 μg/ml ampicillin at 37°C with shaking at 180 rpm. Isopropyl β-D-thio galactopyranoside (IPTG) was added to the culture at the final concentration of 0.5 mM when A600 reached 0.8. Cultivation was continued for another 24 h at 15°C. E. coli cells were then harvested by centrifugation at 5,000 x g for 20 min at 4°C. The cell pellet collected was re-suspended in 40 mL of lysis buffer (50 mM Tris HCl, pH 8.0, 300 mM NaCl, and 10 mM imidazole) containing 0.1% Triton X-100 (v/v), 0.7 M trehalose, 100 μg/ml lysozyme, and 1 mM phenylmethylsulfonyl fluoride (PMSF), and incubated at room temperature for 30 min. The cell suspension was then passed through a French press (Ohtake, Tokyo) with high pressure at 800 kg/cm2 and centrifuged at 25,000 x g for 30 min at 4°C. The supernatant obtained was mixed with 2 ml of 50% Ni2+-NTA His-bind slurry (Qiagen, Germany) then incubated at 4°C with mild shaking for 1 h. The resin with bound recombinant enzyme was washed three times with 50 mM Tris-HCl, pH 8.0, 300 mM NaCl, containing 20 mM imidazole and 0.1% (v/v) Triton X-100. The bound enzyme was eluted with buffer containing stepwise gradient concentrations of imidazole (20–300 mM). The purity of the recombinant protein was confirmed with 12% SDS-PAGE analysis, followed by Coomassie Brilliant Blue (CBB) staining. A fraction containing the pure enzyme was dialyzed against a 300-fold volume buffer containing 50 mM Tris-HCl pH 8.0, 150 mM NaCl, containing 10% glycerol (v/v) supplemented with Complete Mini protease inhibitor cocktail (Roche, Mannheim, Germany) at 4°C for 18 h to remove imidazole. The enzyme was stored at -80°C with 20% glycerol in small aliquots until use. The wheat codon optimized PfPanK2 synthetic gene was cloned into pYT08 vector as previously described (Nozawa et al., 2007) to produce pY08-PfPanK2. Protein was expressed using the TnT® SP6 High-Yield Wheat Germ Protein Expression System (Promega) according to the manufacturer’s instructions. Since no tag was added to PfPanK2 in pYT08-PfPanK2, PanK activity for PfPanK2 was measured in the protein mixture of the wheat germ expression system.
PanK activities of PfPanK1 and PfPanK2 were estimated by measuring ADP production with a coupling assay using the ADP Hunter™ Plus Assay kit (DiscoverX, US) according to the manufacturer’s instructions. The assay mixture contained 15 mM HEPES, 20 mM NaCl, 10 mM MgCl2, 1 mM EGTA, 0.02% Tween 20, 0.1 mg/ml β-globulin, 2 mM pantothenante, 0.1 mM ATP, and 2.5 µg/ml of PfPanK recombinant enzyme in a 25 µl reaction mixture. Fluorescence intensities were measured to estimate the formation of resorufin at 37°C by excitation at 540 nm and emission at 590 nm. Kinetic data were estimated by curve fitting with the Michaelis–Menten equation using GraphPad Prism (GraphPad Software Inc., San Diego, USA). This experiment was performed in triplicate and kinetic values are presented as the means ± S.E. for three independent assays.
We screened 247 compounds from the Kitasato Natural Products Library (Mori et al., 2015; Nurkanto et al., 2018) against PfPanK recombinant enzyme. Compounds were dissolved in 50% DMSO at a final concentration of 1 mg/ml. Enzymatic reactions were carried out on a black 96-well microtiter plate with a 20 µl reaction mixture composed of 19 μL enzyme mix (50 μM pantothenate, 60 μM ATP, 50 ng of PfPanK recombinant enzyme in kinase buffer, described previously, and 1 μl of the individual compounds (final concentration 50 µg/ml) at 37°C for 2 h. ADP production was measured using the ADP Hunter™ Plus kinase assay kit, as described above. The inhibition constant was measured in triplicate. Compounds that showed > 50% inhibition at the primary screening were re-tested to confirm that they did not inhibit the enzyme in the coupled assay (pyruvate kinase, pyruvate oxidase, and peroxidase). Compounds that did not inhibit enzymes in the coupled assay were further subjected to the determination of the concentration showing 50% inhibition of PanK1 activity (IC50).
Selected hit compounds against PfPanK1 were also tested against P. falciparum cells. Before using, parasite culture was synchronized with 5% (w/v) d-sorbitol, as previously described (Lambros and Vanderberg, 1979). Ring stage 0.3% parasitemia (25 µl/well) were placed in a 384-well plate. Serial dilutions (50, 25, 12.5, 6.3, 3.1, 1.6, 0.8, and 0.4 μM) of each compound were used for calculating IC50. As the negative growth control, 50 µM of Mefloquine and 20 µM of Atovaquone were used. After 72 h of incubation, parasite growth was determined by diaphorase-coupled lactate dehydrogenase (LDH) assay, as previously described (Hartuti et al., 2018). Each well’s absorbance was measured at 650 nm using SpectraMax Paradigm®Multi-Mode microplate reader (Molecular Devices, San Jose, CA, USA). The IC50 values were analyzed and calculated with GraphPad PRISM 8.0 (San Diego, California USA).
Two genes that potentially encode PanK were identified in the genome database of P. falciparum 3D7 strain (http://PlasmoDB.org) [PF3D7_1420600 (PfPanK1) and PF3D7_1437400 (PfPanK2)]. The two genes contain open reading frames of 1,560 and 2,301 bp in length, which are presumed to encode 519- and 766-amino acid long proteins with the calculated molecular mass of 59.9 and 91.1 kDa, respectively. These two proteins show only 21% mutual identity. While PfPanK1 harbors all the signatures of eukaryotic PanKs for which enzymatic activity has been demonstrated, PfPanK2 lacks some critical amino acid residues implicated for PanK activity. PfPanK1 only shares 25%–28% positional amino acid identity to human PanK1-4 with the highest similarity to PanK3 (28% identity). PfPanK1 appears to be highly conserved among the Apicomplexa (Figure S1), whereas PfPanK2 may represent a divergent member as PfPanK2 is well separated from other PanKs by phylogenetic analysis and shares only a few motifs including DXXVXDXYGX and GLXXXXXASXFG (X is any amino acid) with PanKs from human and plants (Hart et al., 2016). These data, together with the lack of enzymatic activity (see below), indicate that PfPanK2 may be involved in a reaction other than that catalyzed by authentic PanK.
PfPanK1 was expressed using E. coli expression system with a standard protocol (Fig S2A), but mostly in an insoluble form. After optimization of expression vectors, extraction buffers, detergents, and stabilizing additives, recombinant PfPanK1 became partially soluble with a supplementation of 0.7 M trehalose in the extraction buffer (Fig S2B). The homogeneity of purified recombinant PfPanK1 of an estimated size of 62.5 kDa (59.9 kDa plus a 2.6 kDa histidine tag at the amino terminus) was confirmed and its purity was estimated to be >95%, as evaluated with SDS-PAGE gel followed by Coomassie Brilliant Blue staining (Figure S3A, Table S1) and immunoblot analysis using anti-histidine tag antibody (Figure S3B). The specific activity of PfPanK1 was estimated to be 9.6 µmole/min/mg (Table S1) when assayed under the standard conditions. PfPanK1 was catalytically active in a broad pH range with maximum activity obtained at pH 8.4 and 37°C (Figure S4A). On the other hand, production of recombinant PfPanK2 using E. coli expression system was unsuccessful. Instead, PfPanK2 was successfully expressed using wheat germ expression system (Figure S5A). Since PfPanK2 was not fused with the histidine tag, we attempted to measure activity in the crude lysate but no activity was detected (Figure S5B). Although there is a possibility that the produced PfPanK2 was improperly folded or truncated, which likely resulted in loss or reduction of activity, our data are consistent with the premise that PfPanK1, but not PfPanK2, is a functional enzyme, as previously suggested (Tjhin et al., 2018). Therefore, our downstream research was conducted only with PfPanK1.
PfPanK1 can catalyze phosphorylation of both pantothenate and pantetheine using ATP as a phosphoryl donor. The kinetic parameters such as Km, Vmax, and kcat values for PfPanK1 using pantothenate, pantetheine, and ATP as substrates were determined (Tables 1 and 2). PfPanK1 exhibited hyperbolic saturation kinetics when assayed over the substrate range of 4-128 μM for pantothenate with a saturated concentration (120 µM) of ATP (Figure S6A) and with 1-200 μM ATP and the saturated concentration (100 µM) of pantothenate (Figure S6B). Similar profiles were also obtained when pantetheine and ATP were used (Figures S6C, D, respectively). The apparent Km values for pantothenate and ATP were 44.5 ± 5.5 and 59.2 ± 15.9 μM, respectively (Table 1). Similarly, the Km values for pantetheine and ATP were 45.7 ± 6.9 and 43.4 ± 3.3 μM, respectively.
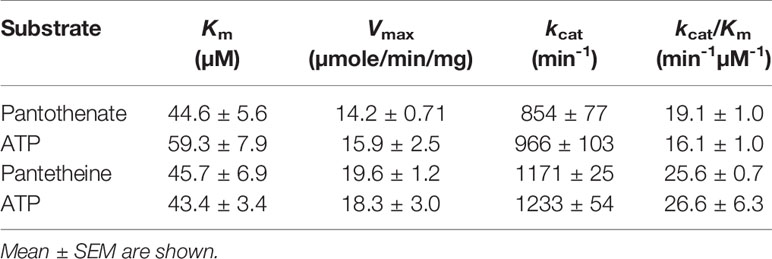
Table 1 Kinetic parameters of P. falciparum pantothenate kinase 1 with pantothenante, pantetheine, and ATP.
PfPanK1 utilizes various nucleoside triphosphates such as ATP, CTP, GTP, UTP, TTP, and dATP as a phosphate donor (Table 2). PfPanK1 showed a slightly higher activity with GTP compared to ATP. PfPanK1 showed an absolute requirement for a free divalent metal cofactor, with Mg2+ as the preferred cation for reactions using either pantothenate or pantetheine (Table 3). Ferrous cation supported a comparative activity with Mg2+ for phosphorylation of pantothenate, but approximately 60% of activity for phosphorylation of pantetheine. Other cations showed lower activity (Table 3). No significant difference was observed in the preference on nucleoside phosphates and metals between the reactions where pantothenate or pantetheine was used as a substrate.
It was reported that PanK from other organisms were subjected to regulation by allosteric inhibition with CoA, acetyl CoA, and malonyl CoA (Vallari et al., 1987; Calder et al., 1999; Takagi et al., 2010; Nurkanto et al., 2018). We examined if CoA and acetyl CoA inhibit PfPanK1 activity. PfPanK1 was inhibited by CoA in the presence of pantothenate or pantetheine as a substrate (Figure 1A). Acetyl CoA also inhibited PfPanK1 only when pantothenate was used as a substrate; however, relatively higher concentrations were needed (IC50 > 1 mM). However, acetyl CoA did not affect PfPanK1 activity when pantetheine was used (Figure 1B). In contrast, panthenol, a pantothenate analog, did not inhibit PfPanK1 activity regardless of the substrates, instead high concentrations of panthenol (e.g., 0.5 mM) slightly (up to 40%) increased enzyme activity (Figure 1C).
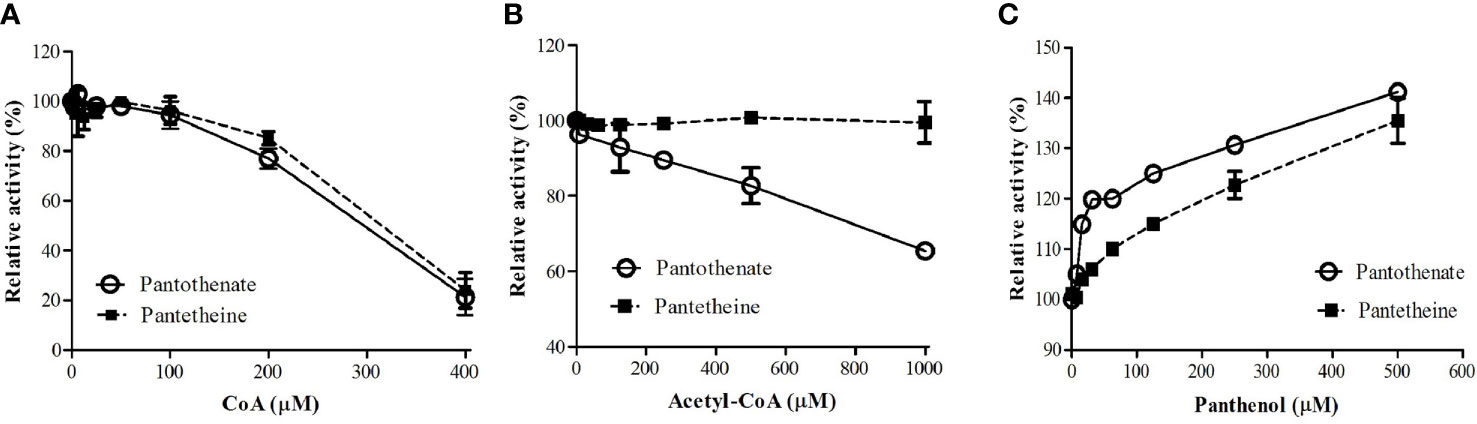
Figure 1 Effect of CoA (A), acetyl CoA (B), and panthenol (C) on P. falciparum pantothenate kinase activities. Relative activities of PfPanK1 in the presence of various concentrations of inhibitors to those without inhibitors are shown. The assay was performed in 15 mM HEPES, pH 8.4, 10 mM MgCl2, 20 mM NaCl, 1 mM EGTA, 0.02% Tween-20, 0.1 mg/ml γ−globulins, 0.2 mM pantothenate, and 100 μM ATP with various concentrations of CoA, acetyl CoA, or panthenol at 37°C. The assay was carried out three times independently, and the results are shown as means ± SEM of triplicates.
We developed an enzyme-based assay using recombinant PfPanK1, to identify its inhibitors by screening chemical and extract libraries. Our assay was proven to be highly sensitive and reproducible with Z’-factor (Zhang et al., 1999) being 0.85 and the signal-to-background ratio of 4.5 (Figure 2A). We screened 247 structurally elucidated natural products of Kitasato Natural Compound Library and plant origin. Twenty-five compounds showed > 40% inhibition against PfPanK1 at 50 µM final concentration (Figure 2B, Table S2). After eliminating the compounds that inhibited the coupling enzyme and those that failed to show inhibition in the reconfirmation assay, four compounds showed dose-dependent inhibition against PfPanK1 (Figure 3). These four PfPanK1 inhibitors, gnetin C, diacetylkinamycin C, gnemonoside D, and simaomicin α showed IC50 value of 20.3 ± 2.2, 36.2 ± 4.7, 57.5 ± 3.6, and 57.6 ± 4.8 µM, respectively (Figure 3, Table 4).
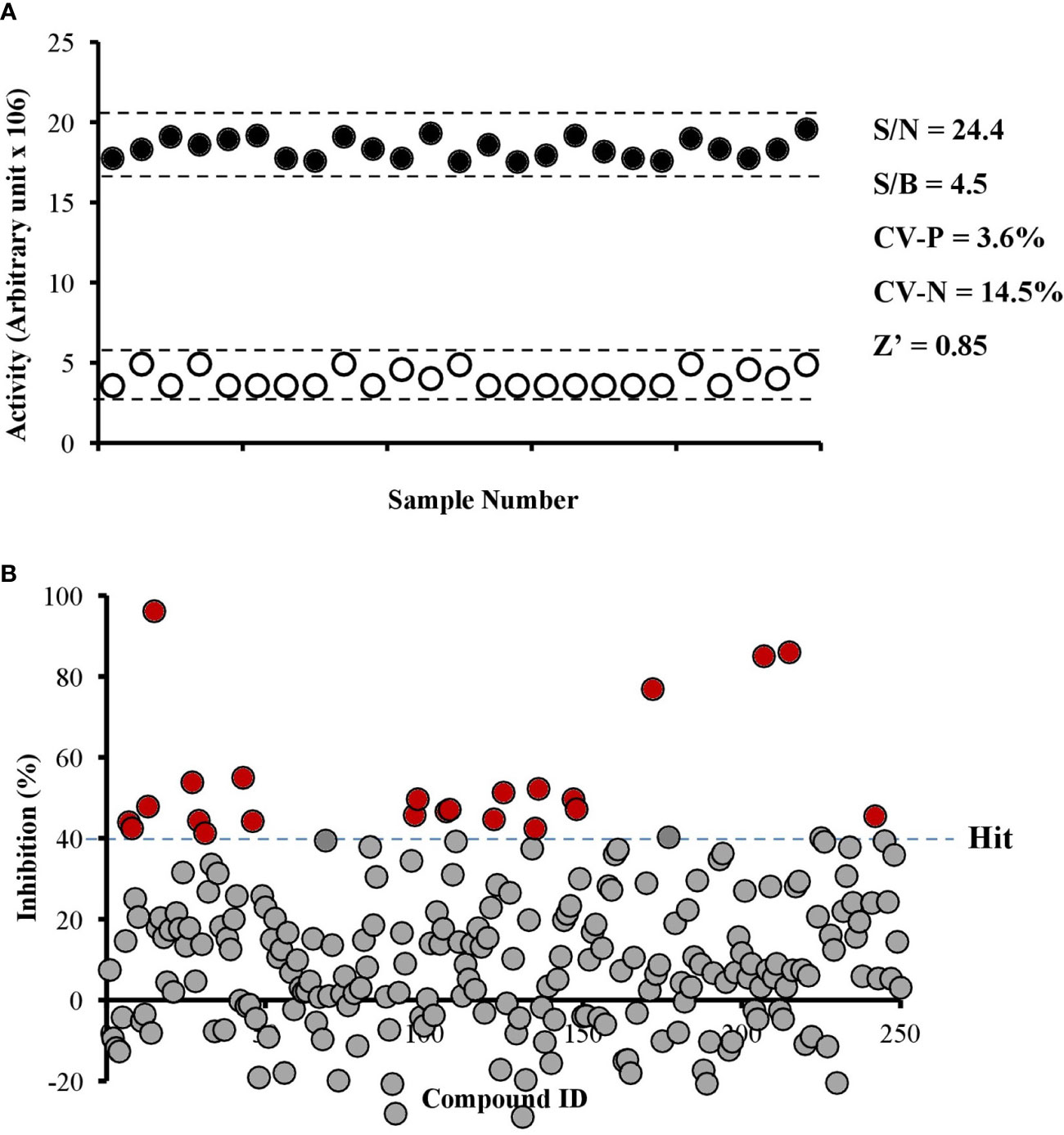
Figure 2 Robustness of PfPanK1 enzymatic assay used in the study (A) and an overview of PfPanK1 inhibition by 247 natural compounds (B). (A) Determination of Z-factor (Z’), signal-to-noise (S/N), signal-to-background ratio (S/B) and coefficient of variation in positive and negative (CV-P and CV-N, respectively) values of PfPanK1 assay on the 96-well plate format. Solid black and open white circles represent positive and negative (without PfPanK1) reaction, respectively enzyme. (B) Scatter plot of the percentage inhibition values of all compounds against PfPanK1 in the primary screening assay. Red solid circles indicate compounds showing >40% inhibition in both the primary and reconfirmation assays.
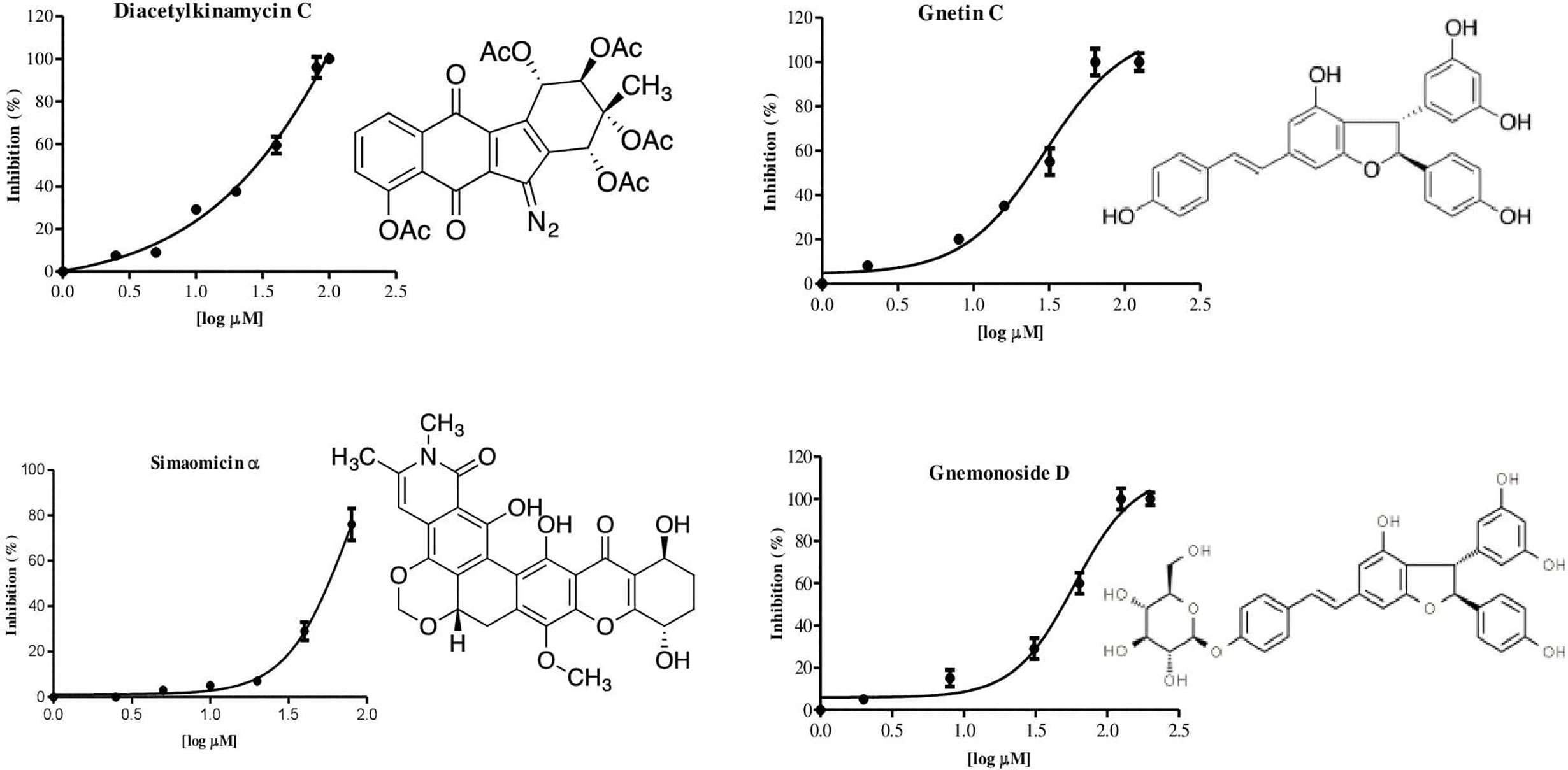
Figure 3 Chemical structure and dose-dependent inhibition for IC50 determination of four identified PfPanK1 inhibitors.
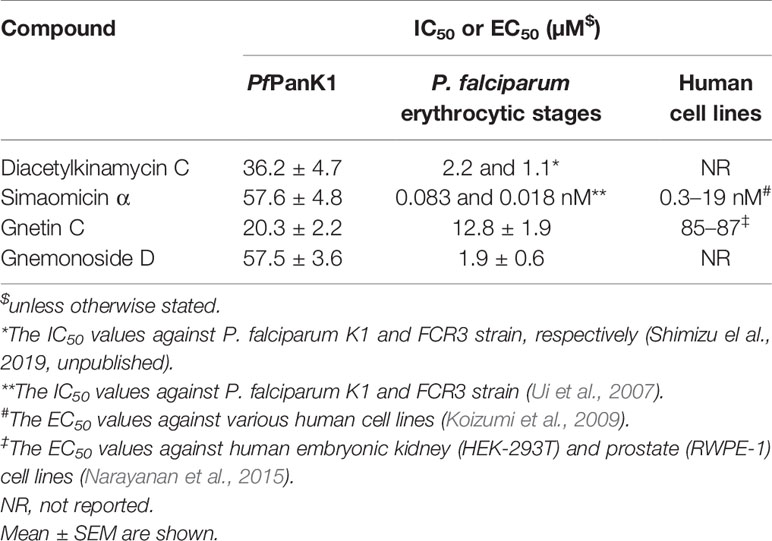
Table 4 Inhibitory activity (IC50 values) of four identified PfPanK1 inhibitors against PfPanK, P. falciparum erythrocytic culture, and human cell lines.
Two PfPanK1 inhibitors were further tested for their inhibitory activity to the P. falciparum erythrocytic stage parasites. Gnetin C and Gnemonoside D showed the IC50 values of 12.8 ± 1.9 and 1.9 ± 0.6 µM, respectively. It was previously shown that diacetylkinamycin C showed the IC50 values of 1.1–2.2 µM against P. falciparum drug-resistant K1 and drug-sensitive FCR3 strain (Shimizu et al., 2019; unpublished). Simaomicin α, been reported has very potent anti-plasmodial activity with the IC50 value was 0.083 and 0.018 nM against K1 and FCR3 strains (Ui et al., 2007). Therefore, four compounds that inhibit PfPanK1 also to inhibited P. falciparum erythrocytic stage parasites. However, while Gnetin C showed comparable IC50 values against PfPanK and the erythrocytic stage parasites, diacetylkinamycin C, Simaomicin α, and Gnemonoside D showed >10 fold lower IC50 values against P. falciparum cells compared to those against the enzyme. Thus, the efficacy of diacetylkinamycin C, Simaomicin α, and Gnemonoside D toward malaria parasites can be due to off-target effects. Gnetin C showed relatively low toxicity toward human cell lines (Narayanan et al., 2015) and an acceptable selectivity index (SI) (6.6-6.8).
PfPanK inhibitors were successfully identified using an assay system using recombinant PfPanK in our study. In the previous studies (Saliba et al., 2005; Spry et al., 2005; Pett et al., 2015), the crude enzyme from P. falciparum lysates were used for enzymological studies and identification of inhibitors. The development of a high throughput screening system in our study has provided. It will allow PfPanK inhibitors to be easily identified with no potential interference by parasite and host-derived factors.
Plasmodium falciparum has two putative PanK encoded by PfPanK1 and PfPanK2 genes. Both are transcribed in the blood stages, in which the steady state mRNA level of PfPanK1 is 12-fold higher than that of PfPanK2. It was previously suggested that only PfPanK1 is involved in the pantothenate-related metabolism in the blood stages of P. falciparum (Tjhin et al., 2018), because selection of resistant lines against a pantothenate analog led to mutations only in PfPanK1, but not PfPanK2, gene (Tjhin et al., 2018). Their study also suggests that PfPanK1 is most likely essential for the intraerythrocytic proliferation of P. falciparum. Our observation that recombinant PfPanK2 produced using the wheat germ cell-free expression system did not show PanK activity seems to agree to the previous reports (Hart et al., 2016; Tjhin et al., 2018). PfPanK2 apparently lacks the ATP binding motif (Hart et al., 2016), which is needed in all kinases that contain the highly conserved P-loop or Walker A sequence motif (GXXXXGKT/S) (Obmolova et al., 2001). Thus, PfPanK2 may serve as a scaffold for a protein complex containing PfPanK1 or other components of the CoA biosynthesis, as suggested (Park et al., 2015; Hart et al., 2016). In any event, the lack of PanK activity of recombinant PfPanK2 prompted us to conduct further studies on PfPanK1.
PfPanK1 can phosphorylate both pantothenate and pantetheine to generate 4-phosphopantothenate or 4-phosphopantethine using ATP or GTP as a phosphate donor. It was previously shown that in P. falciparum blood stage parasites, either pantothenate or pantetheine must be incorporated from the host cytoplasm because the biosynthetic pathways for pantothenate and pantetheine are lacking (Saliba et al., 1998). Incorporation of pantetheine allows a bypass of pantothenate to 4’-phosphopantetheine, by directly yielding 4’-phosphopantetheine from pantetheine by PfPanK, as demonstrated by the growth rescue of both sexual and asexual stages by pantethine supplementation in the culture (Fletcher et al., 2016). 4’-Phosphopantetheine subsequently generates CoA by the last two enzymes, phosphopantetheine adenylyltransferase (PfPPAT) and dephospho-CoA kinase (PfDPCK). Using recombinant PfPanK1 we demonstrated that PfPanK1 phosphorylates both pantothenate and pantetheine, and thus can bypass a few initial steps of CoA biosynthesis in this parasite.
We have shown that PfPanK1 is regulated by allosteric inhibition by CoA and acetyl CoA in a manner similar to other organisms (Calder et al., 1999; Brand and Strauss, 2005). Our previous report on Entamoeba histolytica PanK showed that CoA inhibits amebic PanK in a competitive manner with ATP and uncompetitive or non-competitive mode with pantothenate, while acetyl CoA seemed to inhibit it in a mixed fashion with both substrates (Nurkanto et al., 2018). It was shown that panthenol and a pantothenate analog hampered in vitro growth of P. falciparum erythrocytic stages by inhibition of pantothenate phosphorylation (Saliba et al., 2005; Spry et al., 2005; Fletcher and Avery, 2014). We have shown that panthenol unexpectedly increases PfPanK1 activity in vitro. Therefore, 4’-phosphopanthenol, which is produced by phosphorylation of panthenol may inhibit the downstream enzymes including phosphopantothenoylcysteine synthetase (PfPPCS) as previously proposed (Tjhin et al., 2018).
Many potential inhibitors that interfere with the CoA biosynthetic pathway in Plasmodium have been reported (Fletcher and Avery, 2014; Fletcher et al., 2016; Weidner et al., 2017). Although pantothenate analogs such as pantothenol (PanOH), CJ-15,801 (Saliba and Kirk, 2005; Saliba et al., 2005; Spry et al., 2005), N5-trz-C1-Pan, and N-PE-αMe-PanAm (Macuamule et al., 2015; Howieson et al., 2016) were expected to target PanK, they appeared to inhibit other enzymes than PanK. In the present study, we identified four best PanK inhibitors with the IC50 values being in a range of 20–50 µM from natural compounds by a robust enzyme-based assay using recombinant PfPanK1. None of these inhibitors are pantothenate analogs and they represent new scaffolds as PanK inhibitors although the IC50 values are >20 µM. Therefore, the mode of inhibition from these four compounds is assumed to be either noncompetitive or uncompetitive, not competitive. However, the exact mechanisms of inhibition by these compounds need to be clarified in the future. It is equally plausible that these inhibitors directly block the phosphorylation of pantothenate and pantethine, which, in both cases, results in the depletion of 4’-phosphopantothenate for the downstream reaction mediated by PfPPCS.
Gnetin C is a naturally-occurring stilbenoid, a dimer of resveratrol and its structurally related compounds, gnemonosides, which are originally isolated from the Gnetum gnemon seeds, an edible plant native to Southeast Asia (Aiso-Sanada et al., 2018). It was previously reported that gnetin-C was reported to possess anti-cancer properties (Espinoza et al., 2017), and it also increased the number of circulating natural killer (NK) cells in the immunomodulatory system (Nakagami et al., 2019). Diacetylkinamycin C belongs to the polyketide family of metabolites mainly produced by Streptomyces, and acts as an antitumor agent (Hasinoff et al., 2006; Ballard and Melander, 2008) and anti-gram-positive bacteria. Simaomicin α was originally produced by actinomycetes, Actinomadura madura subspecies simaoensis, and also reported to possess very potent antibiotic activity (Lee et al., 1989).
All selected PfPanK1 inhibitors also inhibited blood stage P. falciparum cell in vitro. From four compounds, simaomicin α has the IC50 value against P. falciparum cell much lower than PfPanK1 enzyme, more than 10,000-fold. It probably indicated that inhibition of P. falciparum cell is not or not only due to the inhibition of PfPanK1. From previous report, this compound showed antimalarial activity in vitro and is known to be a cell-cycle effector in P. falciparum (Ishiyama et al., 2008), whereas the exact mechanism is unrevealed. in human cancer cell, target of simaomicin α has been described to suppress the retinoblastoma protein phosphorylation and promotes apoptosis (Koizumi et al., 2009). However, no molecular target has been identified from this compounds (Wang et al., 2013). Taken together, all four PfPanK1 inhibitors identified in this study were confirmed have anti P. falciparum phenotypic and also have a broad range of activities.
We established a high-throughput screening system against P. falciparum PanK, which catalyzes a rate-limiting step of the CoA biosynthesis, using the bacterial recombinant PfPanK1. We also enzymologically characterized PfPanK1. Finally, we identified four PfPanK natural inhibitors with various scaffolds. The screening system is readily available for large chemically defined compound and microbial/plant extract libraries to discover new inhibitors. Further studies are needed to optimize the inhibitors to improve PfPanK inhibitory activity.
The data presented in the study are deposited in the National Center for Biotechnology Information repository (https://www.ncbi.nlm.nih.gov/genbank/), accession number MW331581.
AN, the main contributor, designed and performed the experiments, analyzed the data, and wrote the manuscript. GJ designed the experiment and analyzed the data. HS performed the experiments and analyzed the data. YR, TS, and DI performed the experiments. MM and TN designed the experiments and analyzed the data. YN, KG, YS, and KS provided the chemical library and natural compounds. YT designed and performed the experiments and analyzed the data. TN conceived the project, acquired funding, wrote the manuscript, and supervised the study. All authors contributed to the article and approved the submitted version.
This research is funded by Grants-in-Aid for Scientific Research (B) (KAKENHI JP18H02650 to TN) and JSPS-LIPI Joint Research Program (JPJSBP120208201 to TN) from the Japan Society for the Promotion of Science, Grant for research on emerging and re-emerging infectious diseases from Japan Agency for Medical Research and Development (AMED, JP19fk0108046 and JP20fk0108138 to TN), and Grant for Science and Technology Research Partnership for Sustainable Development (SATREPS) from AMED and Japan International Cooperation Agency (JICA) (JP19jm0110009 and JP20jm0110022 to TN).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
We thank all members of the Nozaki laboratory at the University of Tokyo, Kumiko Nakada-Tsukui from the National Institute of Infectious Diseases Japan, the members of the Tozawa’s laboratory at Saitama University, Masato Iwatsuki and Yuki Shimizu from Research Center for Tropical Diseases, Kitasato University, Japan.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcimb.2021.639065/full#supplementary-material
Supplementary Figure 1 | Phylogenetic tree of PanK from P. falciparum and other species. Optimal ML tree was inferred by RAxML program with LG + Γ4 model is shown. 145 unambiguously aligned positions from 42 sequences were used for the analysis. Branch lengths are proportional to estimated numbers of substitutions. Bootstrap proportion (BP) values (shown in percentage) of higher than 50 are shown on the internal branches.
Supplementary Figure 2 | Expression and solubility optimization of PfPanK1 using E. coli expression system (A) Expression of PfPanK1 in E. coli was induced with IPTG for 18 h. PfPanK was equally well expressed in two independent bacterial clones (1 and 2). (B) PfPanK1 solubilized in various concentrations of trehalose, examined on SDS-PAGE, followed by immunoblot analysis using anti-His-tag antibody. A red arrow head indicates expressed PfPanK1 recombinant protein.
Supplementary Figure 3 | Purification of recombinant PfPanK1 in E. coli expression system. (A) Protein samples at each step of purification were subjected to 12% SDS-PAGE under reducing conditions, and the gel was stained with Coomassie Brilliant Blue. (B) Immunoblot analysis of recombinant PfPanK1 at each purification step using anti-His-tag antibody.
Supplementary Figure 4 | Specific activity of recombinant PfPanK1 at various pHs. Assays were performed as described in Materials and methods in 15 mM HEPES, pH 8.3, 20 mM NaCl, 10 mM MgCl2, 1 mM EGTA, 0.02% Tween-20, 0.1 mg/mL β−globulins, and 50 ng of PfPanK1 recombinant enzyme at 37°C. Data are shown in mean ± SEM of three replicates.
Supplementary Figure 5 | Expression (A) and activity of recombinant PfPanK2 (B). (A) Expression of PfPanK2 in wheat germ cell-free protein expression system. Red arrowheads indicate PfPanK2. PfPanK2-A and PfPanK2-B represent lysates from two independent bacteria clones. GFP, INH2 and INH3 are positive controls (black and grey arrowheads). (B) PanK activities detected in lysates containing PfPanK2. Assays were performed as described in Materials and methods, in a reaction of 100 µM ATP, 100 µM pantothenate in buffer contains 15 mM HEPES, pH 8.3, 20 mM NaCl, 10 mM MgCl2, 1 mM EGTA, 0.02% Tween-20, PfPanK2 lysate, and 0.1 mg/mL β−globulins at 37°C. The data are shown in mean ± SEM of three replicates in arbitrary unit.
Supplementary Figure 6 | Kinetic analysis of recombinant PfPanK1. (A) The specific activity of PfPanK1 at various pantothenate concentrations in the presence of 120 µM of ATP. (B) The specific activity of PfPanK at various ATP concentrations in the presence of 100 µM of pantothenate. (C) The specific activity of PfPanK at various pantetheine concentrations in the presence 120 µM ATP. (D) The specific activity of PfPanK2 at various ATP concentrations in the presence of 100 µM pantetheine. Assays were performed with 15 mM HEPES, pH 8.4, 20 mM NaCl, 10 mM MgCl2, 1 mM EGTA, 0.02% Tween-20, 50 µg/mL PfPanK, and 0.1 mg/mL β−globulins at 37°C. Data are shown in mean ± SEM of three replicates.
Supplementary Table 1 | Purification of recombinant PfPanK1. Enzyme activity was measured as described in Materials and methods.
Supplementary Table 2 | Inhibitory activities (> 40%) of PfPanK1 from natural products.
Aiso-Sanada H., Ishiguri F., Irawati D., Wahyudi I., Yokota S. (2018). Reaction wood anatomy and lignin distribution in Gnetum gnemon branches. J. Wood Sci. 64, 872–879. doi: 10.1007/s10086-018-1772-2
Augagneur Y., Jaubert L., Schiavoni M., Pachikara N., Garg A., Usmani-Brown S., et al. (2013). Identification and functional analysis of the primary pantothenate transporter, PfPAT, of the human malaria parasite Plasmodium falciparum. J. Biol. Chem. 288 (28), 20558–20567. doi: 10.1074/jbc.M113.482992
Ballard T. E., Melander C. (2008). Kinamycin-mediated DNA cleavage under biomimetic conditions. Tetrahedron Lett. 49 (19), 3157–3161. doi: 10.1016/j.tetlet.2008.03.019
Begley T. P., Kinsland C., Strauss E. (2001). The biosynthesis of coenzyme A in bacteria. Vitam. Horm. 61, 157–171. doi: 10.1016/s0083-6729(01)61005-7
Bobenchik A. M., Witola W. H., Augagneur Y., Lochlainn L. N., Garg A., Pachikara N., et al. (2013). Plasmodium falciparum phosphoethanolamine methyltransferase is essential for malaria transmission. Proc. Natl. Acad. Sci. U. S. A. 110 (45), 18262–18267. doi: 10.1073/pnas.1313965110
Brand L. A., Strauss E. (2005). Characterization of a new pantothenate kinase isoform from Helicobacter pylori. J. Biol. Chem. 280, 20185–20188. doi: 10.1074/jbc.C500044200
Burrows J. N., Duparc S., Gutteridge W. E., Hooft Van Huijsduijnen R., Kaszubska W., Macintyre F., et al. (2017). New developments in anti-malarial target candidate and product profiles. Malar. J 16 (26), 1–29. doi: 10.1186/s12936-016-1675-x
Calder R. B., Williams R. S. B., Ramaswamy G., Rock C. O., Campbell E., Unkles S. E., et al. (1999). Cloning and characterization of a eukaryotic pantothenate kinase gene (PanK) from Aspergillus nidulans. J. Biol. Chem. 274, 2014–2020. doi: 10.1074/jbc.274.4.2014
Chakauya E., Coxon K. M., Wei M., MacDonald M. V., Barsby T., Abell C., et al. (2008). Towards engineering increased pantothenate (vitamin B5) levels in plants. Plant Mol. Biol. 68, 493–503. doi: 10.1007/s11103-008-9386-5
Divo A. A., Geary T. G., Davis N. L., Jensen J. B. (1985). Nutritional Requirements of Plasmodium falciparum in Culture. I. Exogenously Supplied Dialyzable Components Necessary for Continuous Growth. J. Protozool. 32 (1), 59–64. doi: 10.1111/j.1550-7408.1985.tb03013.x
Dondorp A. M., Nosten F., Yi P., Das D., Phyo A. P., Tarning J., et al. (2009). Artemisinin resistance in Plasmodium falciparum malaria. N. Engl. J. Med 361, 455–467. doi: 10.1056/NEJMoa0808859
Edgar R. C. (2004). MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32, 1792–1797. doi: 10.1093/nar/gkh340
Espinoza J. L., Elbadry M. I., Taniwaki M., Harada K., Trung L. Q., Nakagawa N., et al. (2017). The simultaneous inhibition of the mTOR and MAPK pathways with Gnetin-C induces apoptosis in acute myeloid leukemia. Cancer Lett. 400, 127–136. doi: 10.1016/j.canlet.2017.04.027
Favuzza P., de Lera Ruiz M., Thompson J. K., Triglia T., Ngo A., Steel R. W. J., et al. (2020). Dual Plasmepsin-Targeting Antimalarial Agents Disrupt Multiple Stages of the Malaria Parasite Life Cycle. Cell Host Microbe 27 (4), 642–658. doi: 10.1016/j.chom.2020.02.005
Fidock D. A. (2010). Drug discovery: Priming the antimalarial pipeline. Nature 465 (7296), 297–298. doi: 10.1038/465297a
Fletcher S., Avery V. M. (2014). A novel approach for the discovery of chemically diverse anti-malarial compounds targeting the Plasmodium falciparum Coenzyme A synthesis pathway. Malar. J. 13, 343. doi: 10.1186/1475-2875-13-343
Fletcher S., Lucantoni L., Sykes M. L., Jones A. J., Holleran J. P., Saliba K. J., et al. (2016). Biological characterization of chemically diverse compounds targeting the Plasmodium falciparum coenzyme A synthesis pathway. Parasit. Vectors 9, 589. doi: 10.1186/s13071-016-1860-3
Gouy M., Guindon S., Gascuel O. (2010). SeaView Version 4: A Multiplatform Graphical User Interface for Sequence Alignment and Phylogenetic Tree Building. Mol. Biol. Evol. 27, 221–224. doi: 10.1093/molbev/msp259
Grimberg B. T., Mehlotra R. K. (2011). Expanding the antimalarial drug arsenal-now, but how? Pharmaceuticals 4 (5), 681–712. doi: 10.3390/ph4050681
Hart R. J., Cornillot E., Abraham A., Molina E., Nation C. S., Ben Mamoun C., et al. (2016). Genetic characterization of plasmodium putative pantothenate kinase genes reveals their essential role in malaria parasite transmission to the mosquito. Sci. Rep. 6, 33518. doi: 10.1038/srep33518
Hartuti E. D., Inaoka D. K., Komatsuya K., Miyazaki Y., Miller R. J., Xinying W., et al. (2018). Biochemical studies of membrane bound Plasmodium falciparum mitochondrial L-malate:quinone oxidoreductase, a potential drug target. Biochim. Biophys. Acta - Bioenerg. 1859 (3), 191–200. doi: 10.1016/j.bbabio.2017.12.004
Hasinoff B. B., Wu X., Yalowich J. C., Goodfellow V., Laufer R. S., Adedayo O., et al. (2006). Kinamycins A and C, bacterial metabolites that contain an unusual diazo group, as potential new anticancer agents: Antiproliferative and cell cycle effects. Anticancer Drugs 17 (7), 825–837. doi: 10.1097/01.cad.0000224442.78211.27
Howieson V. M., Tran E., Hoegl A., Fam H. L., Fu J., Sivonen K., et al. (2016). Triazole substitution of a labile amide bond stabilizes pantothenamides and improves their antiplasmodial potency. Antimicrob. Agents Chemother. 60 (12), 7146–7152. doi: 10.1128/AAC.01436-16
Ishiyama A., Otoguro K., Namatame M., Nishihara A., Furusawa T., Takahashi Y., et al. (2008). Simaomicin α: Effects on the cell cycle of synchronized, cultured Plasmodium falciparum. J. Antibiot. (Tokyo). doi: 10.1038/ja.2008.38
Koizumi Y., Tomoda H., Kumagai A., Zhou X. P., Koyota S., Sugiyama T. (2009). Simaomicin α, a polycyclic xanthone, induces G1 arrest with suppression of retinoblastoma protein phosphorylation. Cancer Sci. 61 (4), 254–257. doi: 10.1111/j.1349-7006.2008.01033.x
Lambros C., Vanderberg J. P. (1979). Synchronization of Plasmodium falciparum Erythrocytic Stages in Culture. J. Parasitol. 65 (3), 418–420. doi: 10.2307/3280287
Lee T. M., Carter G. T., Borders D. B. (1989). Structure determination of simaomicins α and β, extremely potent, novel anticoccidal agents produced by Actinomadura. J. Chem. Soc Chem. Commun. 22, 1771–1772. doi: 10.1039/C39890001771
Lu F., Culleton R., Zhang M., Ramaprasad A., Von Seidlein L., Zhou H., et al. (2017). Emergence of indigenous artemisinin-resistant Plasmodium falciparum in Africa. N. Engl. J. Med. 376 (10), 991–993. doi: 10.1056/NEJMc1612765
Macuamule C. J., Tjhin E. T., Jana C. E., Barnard L., Koekemoer L., De Villiers M., et al. (2015). A pantetheinase-resistant pantothenamide with potent, on-target, and selective antiplasmodial activity. Antimicrob. Agents Chemother. 59(6), 3666–3668. doi: 10.1128/AAC.04970-14
Mamoun C.B., Prigge S. T., Vial H. (2010). Targeting the lipid metabolic pathways for the treatment of malaria. Drug Dev. Res. 71 (1), 44–55. doi: 10.1002/ddr.20347
Mori M., Jeelani G., Masuda Y., Sakai K., Tsukui K., Waluyo D., et al. (2015). Identification of natural inhibitors of Entamoeba histolytica cysteine synthase from microbial secondary metabolites. Front. Microbiol. 6, 962. doi: 10.3389/fmicb.2015.00962
Nakagami Y., Suzuki S., Espinoza J. L., Quang L. V., Enomoto M., Takasugi S., et al. (2019). Immunomodulatory and metabolic changes after Gnetin-C supplementation in humans. Nutrients. doi: 10.3390/nu11061403
Narayanan N. K., Kunimasa K., Yamori Y., Mori M., Mori H., Nakamura K., et al. (2015). Antitumor activity of melinjo (Gnetum gnemon L.) seed extract in human and murine tumor models in vitro and in acolon-26 tumor-bearing mouse model in vivo. Cancer Med. 11 (6), 1403. doi: 10.1002/cam4.520
Nguyen L. T., Schmidt H. A., Von Haeseler A., Minh B. Q. (2015). IQ-TREE: A fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 32, 268–274. doi: 10.1093/molbev/msu300
Nozawa A., Nanamiya H., Miyata T., Linka N., Endo Y., Weber A. P. M., et al. (2007). A cell-free translation and proteoliposome reconstitution system for functional analysis of plant solute transporters. Plant Cell Physiol. 48 (12), 1815–1820. doi: 10.1093/pcp/pcm150
Nurkanto A., Jeelani G., Yamamoto T., Naito Y., Hishiki T., Mori M., et al. (2018). Characterization and validation of Entamoeba histolytica pantothenate kinase as a novel anti-amebic drug target. Int. J. Parasitol. Drugs Drug Resist. 8(1), 125–136. doi: 10.1016/j.ijpddr.2018.02.004
Obmolova G., Teplyakov A., Bonander N., Eisenstein E., Howard A. J., Gilliland G. L. (2001). Crystal structure of dephospho-coenzyme A kinase from Haemophilus influenzae. J. Struct. Biol. 136 (2), 119–125. doi: 10.1006/jsbi.2001.4428
Park C. H., Ryu H. G., Kim S. H., Lee D., Song H., Kim K. T. (2015). Presumed pseudokinase VRK3 functions as a BAF kinase. Biochim. Biophys. Acta - Mol. Cell Res 1853 (7), 1738–1748. doi: 10.1016/j.bbamcr.2015.04.007
Pett H. E., Jansen P. A., Hermkens P. H., Botman P. N., Beuckens-Schortinghuis C. A., Blaauw R. H., et al. (2015). Novel pantothenate derivatives for anti-malarial chemotherapy. Malar. J. 14, 169. doi: 10.1186/s12936-015-0673-8
Saliba K. J., Kirk K. (2001). H+-coupled Pantothenate Transport in the Intracellular Malaria Parasite. J. Biol. Chem. 276 (21), 18115–18121. doi: 10.1074/jbc.M010942200
Saliba K. J., Kirk K. (2005). CJ-15,801, a fungal natural product, inhibits the intraerythrocytic stage of Plasmodium falciparum in vitro via an effect on pantothenic acid utilisation. Mol. Biochem. Parasitol. 141 (1), 129–131. doi: 10.1016/j.molbiopara.2005.02.003
Saliba K. J., Horner H., Kirk K. (1998). Transport and metabolism of the essential vitamin pantothenic acid in human erythrocytes infected with the malaria parasite Plasmodium falciparum. J. Biol. Chem. 273, 10190–10195. doi: 10.1074/jbc.273.17.10190
Saliba K. J., Ferru I., Kirk K. (2005). Provitamin B5 (Pantothenol) inhibits growth of the intraerythrocytic malaria parasite. Antimicrob. Agents Chemother. 49 (2), 632–637. doi: 10.1128/AAC.49.2.632-637.2005
Schalkwijk J., Allman E. L., Jansen P. A. M., De Vries L. E., Verhoef J. M. J., Jackowski S., et al. (2019). Antimalarial pantothenamide metabolites target acetyl-coenzyme A biosynthesis in Plasmodium falciparum. Sci. Transl. Med. 11 (510), eaas9917. doi: 10.1126/scitranslmed.aas9917
Spry C., Saliba K. J. (2009). The human malaria parasite Plasmodium falciparum is not dependent on host coenzyme A biosynthesis. J. Biol. Chem. 284 (37), 24904–24913. doi: 10.1074/jbc.M109.025312
Spry C., Chai C. L. L., Kirk K., Saliba K. J. (2005). A class of pantothenic acid analogs inhibits Plasmodium falciparum pantothenate kinase and represses the proliferation of malaria parasites. Antimicrob. Agents Chemother. 49 (11), 4649–4657. doi: 10.1128/AAC.49.11.4649-4657.2005
Spry C., Kirk K., Saliba K. J. (2008). Coenzyme A biosynthesis: An antimicrobial drug target. FEMS Microbiol. Rev. 32 (1), 56–106. doi: 10.1111/j.1574-6976.2007.00093.x
Spry C., van Schalkwyk D. A., Strauss E., Saliba K. J. (2010). Pantothenate Utilization by Plasmodium as a Target for Antimalarial Chemotherapy. Infect. Disord. - Drug Targets 10 (3), 200–216. doi: 10.2174/187152610791163390
Spry C., Sewell A. L., Hering Y., Villa M. V. J., Weber J., Hobson S. J., et al. (2018). Structure-activity analysis of CJ-15,801 analogues that interact with Plasmodium falciparum pantothenate kinase and inhibit parasite proliferation. Eur. J. Med. Chem. 143, 1139–1147. doi: 10.1016/j.ejmech.2017.08.050
Stamatakis A. (2006). RAxML-VI-HPC: Maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics 22, 2688–2690. doi: 10.1093/bioinformatics/btl446
Takagi M., Tamaki H., Miyamoto Y., Leonardi R., Hanada S., Jackowski S., et al. (2010). Pantothenate kinase from the thermoacidophilic archaeon Picrophilus torridus. J. Bacteriol. 192, 233–241. doi: 10.1128/JB.01021-09
Tjhin E. T., Spry C., Sewell A. L., Hoegl A., Barnard L., Sexton A. E., et al. (2018). Mutations in the pantothenate kinase of Plasmodium falciparum confer diverse sensitivity profiles to antiplasmodial pantothenate analogues. PLoS Pathog. 14 (4), e1006918. doi: 10.1371/journal.ppat.1006918
Tse E. G., Korsik M., Todd M. H. (2019). The past, present and future of anti-malarial medicines. Malar. J. 18 (1), 93. doi: 10.1186/s12936-019-2724-z
Ui H., Ishiyama A., Sekiguchi H., Namatame M., Nishihara A., Takahashi Y., et al. (2007). Selective and potent in vitro antimalarial activities found in four microbial metabolites. J. Antibiot. (Tokyo) 60 (3), 220–222. doi: 10.1038/ja.2007.27
Vallari D. S., Jackowski S., Rock C. O. (1987). Regulation of pantothenate kinase by coenzyme A and its thioesters. J. Biol. Chem. 262, 2468–2471. doi: 10.1016/S0021-9258(18)61527-3
Visser B. J., Van Vugt M., Grobusch M. P. (2014). Malaria: An update on current chemotherapy. Expert Opin. Pharmacother. 15 (15), 2219–2254. doi: 10.1517/14656566.2014.944499
Wang Y., Wang C., Butler J. R., Ready J. M. (2013). Dehydrogenative coupling to enable the enantioselective total synthesis of (-)-simaomicin α. Angew. Chem. - Int. Ed. 52 (41), 10796–10799. doi: 10.1002/anie.201304812
Weidner T., Lucantoni L., Nasereddin A., Preu L., Jones P. G., Dzikowski R., et al. (2017). Antiplasmodial dihetarylthioethers target the coenzyme A synthesis pathway in Plasmodium falciparum erythrocytic stages. Malar. J. 16 (1), 192. doi: 10.1186/s12936-017-1839-3
Wells T. N. C., van Huijsduijnen R. H., Van Voorhis W. C. (2015). Malaria medicines: a glass half full? Nat. Rev. Drug Discov. 14, 424–442. doi: 10.1038/nrd4573
Keywords: coenzyme A, PfPanK1, PfPanK2, Plasmodium falciparum, inhibitors
Citation: Nurkanto A, Jeelani G, Santos HJ, Rahmawati Y, Mori M, Nakamura Y, Goto K, Saikawa Y, Annoura T, Tozawa Y, Sakura T, Inaoka DK, Shiomi K and Nozaki T (2021) Characterization of Plasmodium falciparum Pantothenate Kinase and Identification of Its Inhibitors From Natural Products. Front. Cell. Infect. Microbiol. 11:639065. doi: 10.3389/fcimb.2021.639065
Received: 08 December 2020; Accepted: 29 January 2021;
Published: 09 March 2021.
Edited by:
Yang Zhang, University of Pennsylvania, United StatesReviewed by:
Jie Chen, University of Pennsylvania, United StatesCopyright © 2021 Nurkanto, Jeelani, Santos, Rahmawati, Mori, Nakamura, Goto, Saikawa, Annoura, Tozawa, Sakura, Inaoka, Shiomi and Nozaki. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Tomoyoshi Nozaki, bm96YWtpQG0udS10b2t5by5hYy5qcA==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.