- 1CAS Key Laboratory of Special Pathogens and Biosafety, Center for Biosafety Mega-Science, Wuhan Institute of Virology, Chinese Academy of Sciences, Wuhan, China
- 2International College, University of Chinese Academy of Sciences, Beijing, China
The rapid spread and emergence of multidrug-resistant Acinetobacter baumannii and other pathogenic Gram-negative bacteria spurred scientists and clinicians to look for alternative therapeutic agents to conventional antibiotics. In the present study, an A. baumannii bacteriophage p54 was isolated and characterized. Morphological and genome analysis revealed that bacteriophage p54 belongs to Myoviridae family with a genome size of 165,813 bps. A novel endolysin, namely LysAB54, showing low similarity with other well-known related endolysins, was cloned, expressed, and characterized from the bacteriophage p54. LysAB54 showed significant bactericidal activity against multidrug-resistant A. baumannii and other Gram-negative bacteria, including Pseudomonas aeruginosa, Klebsiella pneumoniae, and Escherichia coli, in the absence of outer membrane permeabilizers. Based on all those observations, LysAB54 could represent a potential agent for the treatment of multidrug-resistant Gram-negative superbugs.
Introduction
Bacterial infections deleteriously impact human health and are a leading cause of mortality worldwide. The emergence of antibiotic-resistant bacterial strains further exaggerates the present situation and alarming for humankind in various aspects of daily life. The occurrence of multi-drug resistant (MDR) Gram-negative bacterial pathogens, which mainly includes antibiotic-resistant strains of Acinetobacter baumannii, Pseudomonas aeruginosa, Klebsiella pneumoniae, and Escherichia coli, may lead to serious complications and nosocomial infections (Bonomo and Szabo, 2006; Spellberg et al., 2013; Heron, 2017; Collaborators, 2018). A. baumannii is an opportunistic Gram-negative pathogen that can initiate several serious nosocomial infections, which majorly includes wound infections, urinary tract infections, ventilator related pneumonia, and secondary meningitis. The high resistance of A. baumannii toward conventional antibiotics makes their management very cumbersome (Valencia et al., 2009; Pendleton et al., 2013; Potron et al., 2015). The high degree of antibiotic resistance conferred by A. baumannii and other pathogenic bacteria spurred scientists and clinicians to look for alternative therapeutic agents to antibiotics (Magiorakos et al., 2012).
Among various available alternative agents, bacteriophage encoded peptidoglycan hydrolases, popularly known as endolysins or lysins, represent a promising alternative for the treatment of drug-resistant bacteria. Endolysins are enzymatic proteins that are produced inside the bacterial host at the later stage of bacteriophage replication and can lyse bacterial hosts when applied externally (Fischetti, 2008; Nelson et al., 2012; Schmelcher et al., 2012; Yang et al., 2014; Gondil et al., 2020). Endolysin renders numerous advantages over other alternative antibacterial agents, including high activity against drug-resistant pathogens, low chances of resistance, and high specificity towards target bacterium (Loessner et al., 2002; Schuch et al., 2002; Fischetti, 2008; Briers et al., 2009). Several endolysin have been reported to harbor broad-spectrum activity against Gram-positive bacteria in vitro and in vivo, such as PlyC (Nelson et al., 2006), ClyR (Yang et al., 2015a), Cpl-1 (Loeffler et al., 2003), ClyJ (Yang et al., 2020), and ClyV (Huang et al., 2020).
On the Contrary, due to the presence of protective bacterial outer membrane, most endolysins fail to exhibit prominent activity against Gram-negative bacteria (Fischetti, 2005; Rodríguez-Rubio et al., 2016). However, treatment bacteria with outer membrane permeabilizers (OMPs), such as citric acid, trichloroethane (CHCl3), Triton X-100, and EDTA, can improve the antibacterial activity of endolysins and conquer the hindrance presented by the outer membrane of Gram-negative bacteria (Helander and Mattila-Sandholm, 2000; Briers et al., 2007; Briers et al., 2011; Briers et al., 2014; Yang et al., 2015b; Guo et al., 2017). In this context, several endolysins have showed improved or synergistic bactericidal activity against MDR A. baumannii and P. aeruginosa in the presence of OMPs, including LysSS (Kim et al., 2020b), Abtn-4 (Yuan et al., 2020), LysPA26 (Guo et al., 2017), PlyA (Yang et al., 2015b), ABgp46 (Oliveira et al., 2016), and OBPgp279 (Walmagh et al., 2012). However, lysins with OMP-independent antibacterial activity against Gram-negative microbes still un-adequately addressed since few of them are demonstrated effective in animal infection models.
In the present study, we report the comprehensive characterization of a novel A. baumannii bacteriophage p54 and its endolysin, namely, LysAB54. Our results demonstrated that LysAB54 has a high and rapid antibacterial activity against a range of antibiotic-resistant Gram-negative bacterial strains in the absence of OMPs.
Materials and Methods
Collection of Water Sample for Bacteriophage Isolation
The water sample was collected in sterile 50 ml screw tubes from the Tongji Hospital, Wuhan, Hubei Province, China. The water samples were kept in an ice bucket box to preserve the specimen. After removing the impurities by sterile syringe filter, samples were kept in 4°C for further use.
Isolation of Bacteriophage p54
The stored water sample was mixed 1:1 with phage buffer (50 mM Tris-HCl, 150 mM NaCl, 10 mM MgCl2, 2 mM CaCl2, pH 7.5). The A. baumannii strain WHG3083 was cultured in lysogeny broth (LB) at 37°C to logarithmic phase and then transferred to the conical flask. Conical flask solution containing water sample, phage buffer, and bacteria were incubated aerobically in shaking incubator on 160 rpm at 37°C for 2–3 days. After incubation, solution was centrifuged (1,000 rpm for 10 min, 4°C) and filtered through a sterile 0.22 µm syringe filter (Guangzhou Jet bio-filtration Co., Ltd). Then, 500 μl of the filtrate was mixed with 500 μl of A. baumannii in a sterile 1.5 ml microcentrifuge tube and incubated aerobically in a shaking incubator on 160 rpm at 37°C for 15–20 min. The mixture was mixed with 4 ml of 0.7% soft agar (approximately 50°C) in a sterile 15 ml tube (Guangzhou Jet bio-filtration Co., Ltd) and poured onto the LB agar plates and incubated overnight at 37°C.
Purification of the Bacteriophage p54
A single plaque was picked and resuspended in the phage buffer in a sterile 1.5 microcentrifuge tube. The microcentrifuge tube containing plaque was vortexed for approximately 2–3 min. The vortexed solution was further filtered through a sterile 0.22 µm syringe filter. 10 μl of the filtered solution (10-fold serially diluted) was mixed with the 1 ml of A. baumannii cell suspension and incubated on 160 rpm at 37°C for 15–30 min. Afterwards, bacterium-phage mixture was mixed with 4 ml of soft agar and poured onto LB agar plates and incubated overnight at 37°C. This cycle was repeated for five times to obtain purified bacteriophage p54.
Bacteriophage p54 Genome Extraction and Sequencing Analysis
The bacteriophage p54 genome was extracted and sequenced following the protocol and procedures as previously described (Kering et al., 2020). The sequenced genome was compared with the available whole genome sequences of A. baumannii bacteriophages by the Neighbor-Joining method (Saitou and Nei, 1987). The bootstrap consensus tree inferred from 1,000 replicates is taken to represent the evolutionary history of the taxa analyzed (Felsenstein, 1985). Branches corresponding to partitions reproduced in less than 50% bootstrap replicates are collapsed. The percentage of replicate trees in which the associated taxa clustered together in the bootstrap test (1,000 replicates) are shown next to the branches (Felsenstein, 1985). The evolutionary distances were computed using the p-distance method and are in the units of the number of base differences per site. This analysis involved 10 nucleotide sequences and the codon positions included were 1st+2nd+3rd+Noncoding. All ambiguous positions were removed for each sequence pair. There was a total of 16,5813 positions in the final dataset. Evolutionary analyses were conducted in MEGA X software (Kumar et al., 2018).
Transmission Electron Microscopy (TEM) of the Bacteriophage p54
The bacteriophage p54 was purified and concentrated using cesium chloride density gradient ultracentrifugation on 35,000 rpm for 2 h at 4°C. The concentrated bacteriophage was stained with freshly prepared phosphotungstic acid solution (2%) for 3 min and spot dried on the copper grid for 2 h. The samples were examined on H-7000FA transmission electron microscope (Hitachi, Japan).
Bacterial Culture Conditions
All bacterial strains were cultured in LB broth aerobically at 37°C. The stocks of bacterial strains containing 20% glycerol was store in -80°C refrigerator for long time storage. E. coli BL21(DE3) (Takara) cells were used for cloning and protein expression.
Cloning of LysAB54 in the Expression Vector
The open-reading frame (ORF) encoding the putative endolysin LysAB54 was amplified by Polymerase Chain Reaction (PCR) with the primers (5′ to 3′): 54-Forward: TATACCATGGACGTTAAACCATTTTTTG (NcoI) and 54-Reverse: TATACTCGAGTTGTTCAAAATACGCTTTCTC (XhoI). The purified (Omega Bio-tek®) PCR product was digested with restriction endonuclease enzymes NcoI and XhoI (Thermo Scientific). Purified PCR product was ligated into pET28a (+) and transformed to the competent E. coli BL21(DE3) cells. The inserted gene was further confirmed by Sanger sequencing (Sangon Biotech, Shanghai).
Protein Expression and Purification
E. coli BL21 (DE3) containing the pET28a-LysAB54 was grown in 500 ml LB with 100 mg/ml of kanamycin for approximately 2–3 h until the OD600 reached to 0.5–0.6. Protein expression was induced with 1 mM Isopropyl-beta-D-thiogalactopyranoside (IPTG; Thermo Scientific) at 16°C for 16 h. The bacterial culture was harvested by centrifugation at 10,000 rpm at 4°C for 10 min. Bacterial cells were washed once by PBS (pH7.4) and centrifuged at 10,000 rpm at 4°C for 10 min. Cells were disrupted through sonication in ice cold environment. The insoluble cell fragments were removed through centrifugation at 10,000 rpm at 4°C for 10 min and the supernatant was collected by filter through 0.22 µm syringe filter. His-tagged LysAB54 was purified using nickel nitrilotriacetic acid affinity column chromatography (GE Healthcare, US) with gradient of imidazole solutions. After purification, proteins were dialyzed overnight at 4°C against Tris buffer (pH 7.4). The purity of protein was assessed using SDS-PAGE.
Antibacterial Activity Assay of Endolysin LysAB54
Antibacterial activity of endolysin LysAB54 was determined as defined previously (Yang et al., 2015b) with minor modifications. To determine the antibacterial activity of LysAB54, the bacteria were cultured in LB broth at 37°C for both the logarithmic phase (OD600 = 0.6–0.8) and stationary phase (OD600 = 1.4–1.6). Bacterial culture was centrifuged at 8,000 rpm at 4°C for 5 min and bacterial cells were washed and re-suspended in Tris buffer (pH 7.4). LysAB54 was added into bacterial suspension and incubated aerobically at 37°C for 60 min. Groups treated with the same amount of Tris buffer instead of LysAB54 were used as negative controls. The mixture was then serially diluted than plated on the LB agar petri dishes, incubated overnight aerobically at 37°C.
Killing Range of LysAB54
To evaluate the bactericidal activity of endolysin LysAB54 against clinical Gram-negative isolates, logarithmic phase bacteria were treated with 100 μg/ml of LysAB54 at 37°C for 1 h. The reduction in bacterial count in term of LogCFU/mL was defined as the antibacterial activity of LysAB54 against tested isolate.
The Activity of LysAB54 in Complex Medium
To test the antibacterial activity of endolysin LysAB54 against A. baumannii in complex medium, logarithmic phase bacteria were re-suspended in LB broth and human-serum (Sigma Aldrich, Shanghai, China), and treated with 100 μg/ml of LysAB54 at 37°C for 1 h. The mixture was serially diluted and plated on LB agar Petri dishes for viable cell count.
Statistical Analysis
The data were analyzed using GraphPad prism Version 5 software on Microsoft Windows 2019 operating system. All the experiments were performed in triplicates on different time points. The data was expressed in term of Mean ± Standard Deviation (SD) for statistical considerations.
Results
Characterization of Bacteriophage p54
Bacteriophage p54 was isolated and purified from the hospital water sample. Double-layer agar assay showed clear plaques on A. baumannii lawn postulating the lytic potential of isolated bacteriophage p54 (Figure 1A). Morphological analysis showed that the bacteriophage p54 contains an icosahedral head and a short tail, with a particle size of 24.4×55.8 nm (Figure 1B). Based on these observations, bacteriophage p54 can be classified to the family of Myoviridae according to the latest classification by the International Committee on the Taxonomy of Viruses (ICTV).
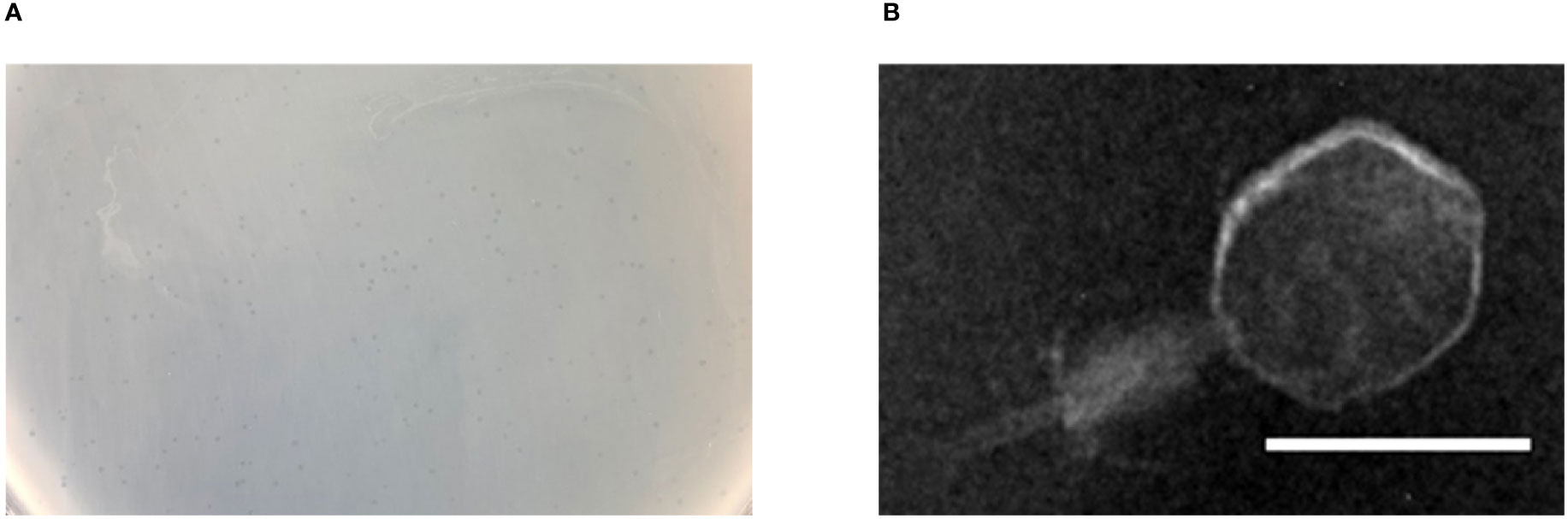
Figure 1 Characteristics of bacteriophage p54. (A) Double layer agar assay of the isolated bacteriophage p54 with host bacterium. (B) Morphological analysis of the bacteriophage p54 by transmission electron microscopy (TEM). Scale bar: 25 µm.
Genome Sequencing and ORF Annotation of Bacteriophage p54
Full genome sequencing analysis showed that bacteriophage p54 consists 165,813 bps with a GC content of 36.3%. A Blast-P analysis further revealed that bacteriophage p54 contains 248 predicted ORFs, with putative functions categorized into multiple functional modules, including phage structure, host lysis, phage DNA metabolism and modifications (Figure 2). Whole-genome based phylogenetic analysis showed that bacteriophage p54 possesses maximum similarity with A. baumannii phage AP22 (NCBI No: HE806280) among various phages analyzed (Figure S1), indicating the novelty of bacteriophage p54.
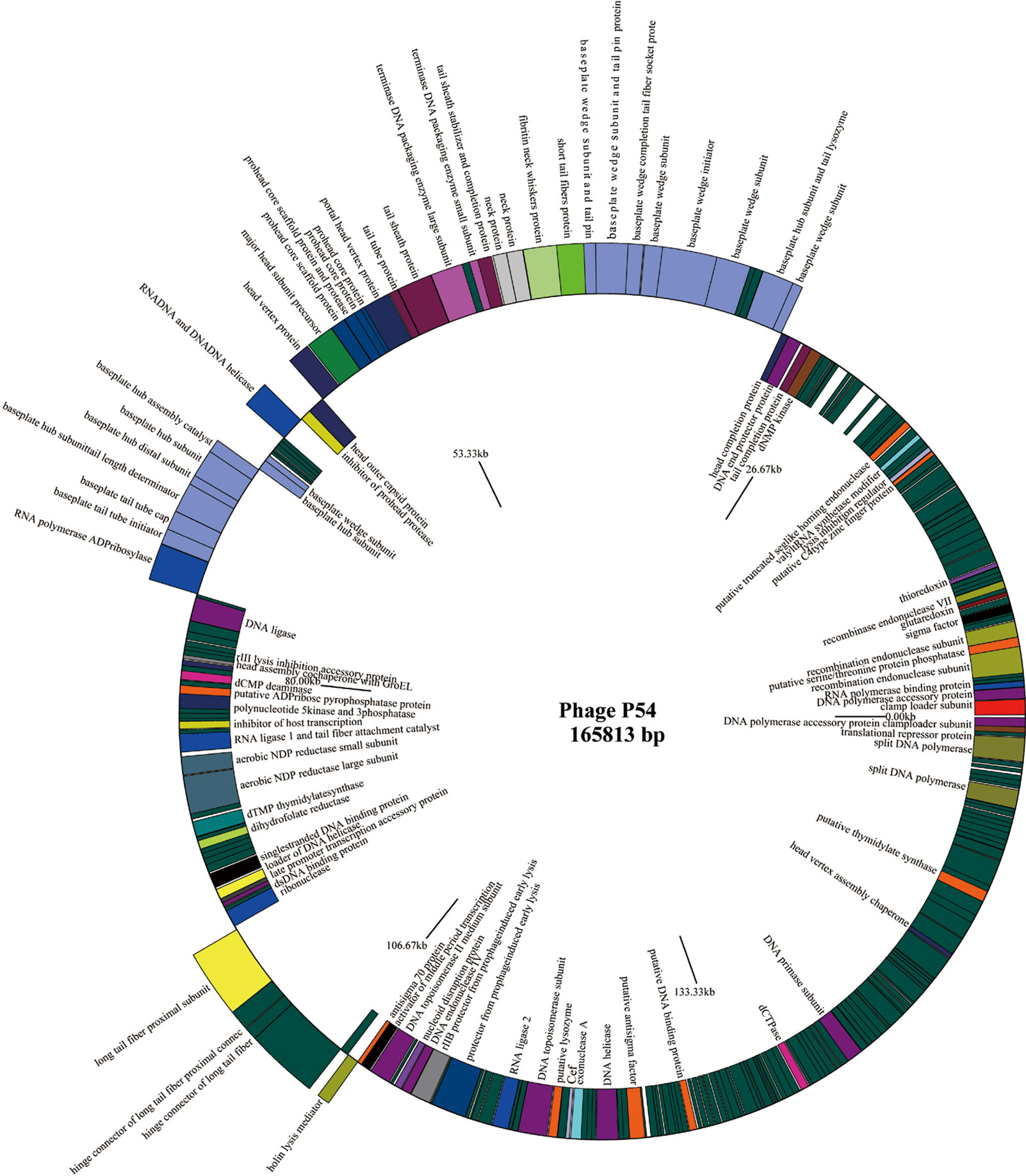
Figure 2 Genome map of the bacteriophage p54. Genes on the positive chain of the genome (Outer ring); Gene on the negative chain of the genome (Inner ring). The genome direction from 5′ to 3′ in the figure is drawn counterclockwise. Functions of putative open-reading frames (ORFs) are marked.
Bioinformatics Analysis of the Endolysin LysAB54
The putative endolysin LysAB54 of bacteriophage p54, i.e., ORF159 (120,967–121,530 bp), comprised 187 amino acids with a putative lysozyme activity. As shown in Figure 3A, a phylogenetic analysis of LysAB54 and other Gram-negative endolysins showed that LysAB54 has similarity with the other reported endolysin containing lysozyme domain, such as LysPA26 of A. baumannii phage (Guo et al., 2017). The similarity of LysAB54 with other closely related endolysins was further performed by the MultAlign server and revealed a lower level of similarity with known related endolysins, postulating the novelty of LysAB54 (Figure 3B). Phyre2 server-based structural prediction showed that LysAB54 contains multiple alpha-helix domains with the absence of predictable beta plated sheets (Figure 3C).
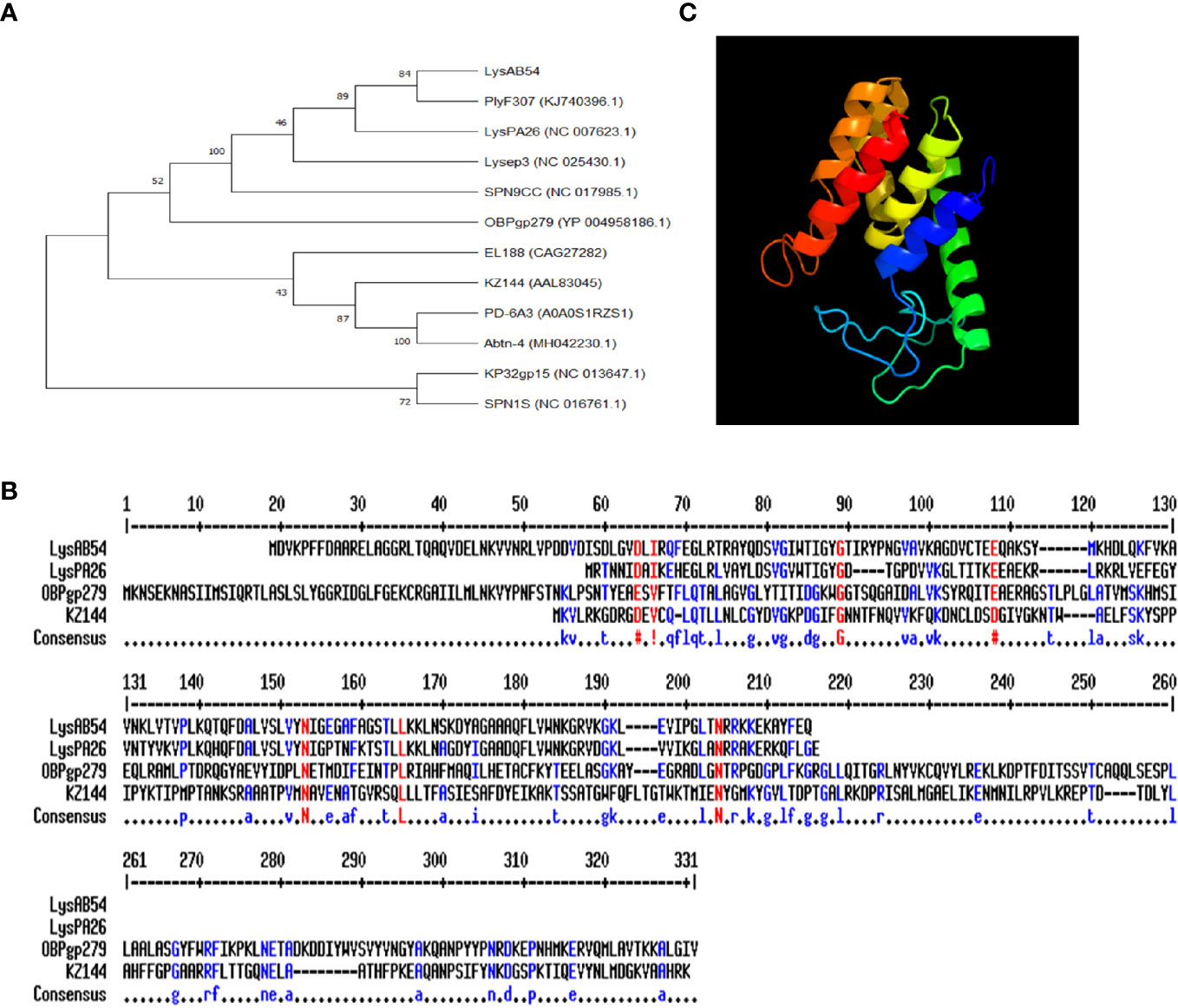
Figure 3 Phylogenetic analysis of LysAB54. (A) Phylogenetic relationship of various endolysins. The neighbor-joining trees were based on the ClustalW alignment of DNA sequences by MEGAX software. (B) Multi-alignment analysis of LysAB54 with other closely related Gram-negative endolysins. (C) Predicted three-dimensional structure of LysAB54. Three‐dimensional structure model by Phyre2 web server. NORMAL mode. Confidence in the model: 100.0%. 148 residues (79% of sequence) have been modeled with 100.0% confidence by the single highest scoring template. Model dimensions (Å): X: 35.277, Y: 41.465, Z: 46.142.
LysAB54 Shows High Bactericidal Activity Against A. baumannii
SDS-PAGE analysis showed a single band of LysAB54 after purification (Figure 4A), which was consistent with its predicted molecular mass of 24 kDa. The time-killing assay showed that the LysAB54 (100 μg/ml) could kill logarithmic A. baumannii with 0.6 logs of reduction in the first 1 min of incubation, while up to 4 logs of decrement in bacterial count was achieved after incubation for 10 min. These results showed that the LysAB54 exhibits robust as well as rapid bactericidal activity (Figure 4B).
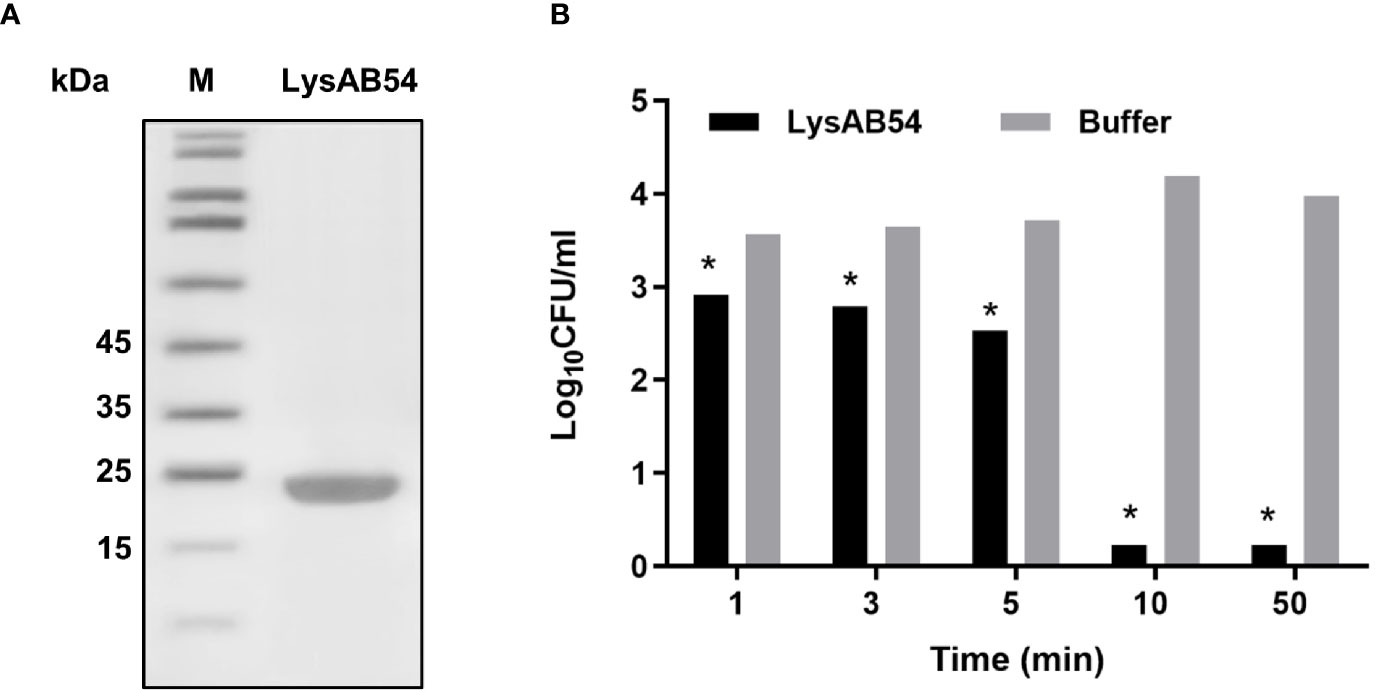
Figure 4 Characteristics of LysAB54. (A) Purified LysAB54 on SDS-PAGE gel. Lane 1 showing LysAB54 and lane 2 showing protein marker. (B) Time-killing activity of LysAB54 against logarithmic phase of A. baumannii WHG 40090 in Tris buffer. Data are shown as means ± standard deviations. *p < 0.05.
Antibacterial Activity of LysAB54 Against Logarithmic Phase Gram-Negative Bacteria
The antibacterial activity of LysAB54 in Tris buffer against each ten different clinical isolates of A. baumannii, P. aeruginosa, E. coli, and K. pneumoniae was investigated. As shown in Figure 5, LysAB54 showed high bactericidal activity against all isolates of A. baumannii, E. coli, and K. pneumoniae tested. Moreover, 8 out of 10 P. aeruginosa strains were found to be highly susceptible to LysAB54 (Figure 5B). The difference in antibacterial activity of log reduction against these clinical strains may be due to the difference in the molecular architecture of the bacterial outer membrane. Notably, reduction of 4 logs (from 4.2 to 0), 2.17 logs (from 5.77 to 3.60), 2 logs (from 4.22 to 2.16), and 2.33 log (from 3.86 to 1.53) were observed in A. baumannii WHG 40090, P. aeruginosa WHG50023, E. coli WHG11023, and K. pneumoniae WHG11004, respectively. These results suggested that LysAB54 has a wide range of antibacterial activity against multi-drug resistant Gram-negative microbes.
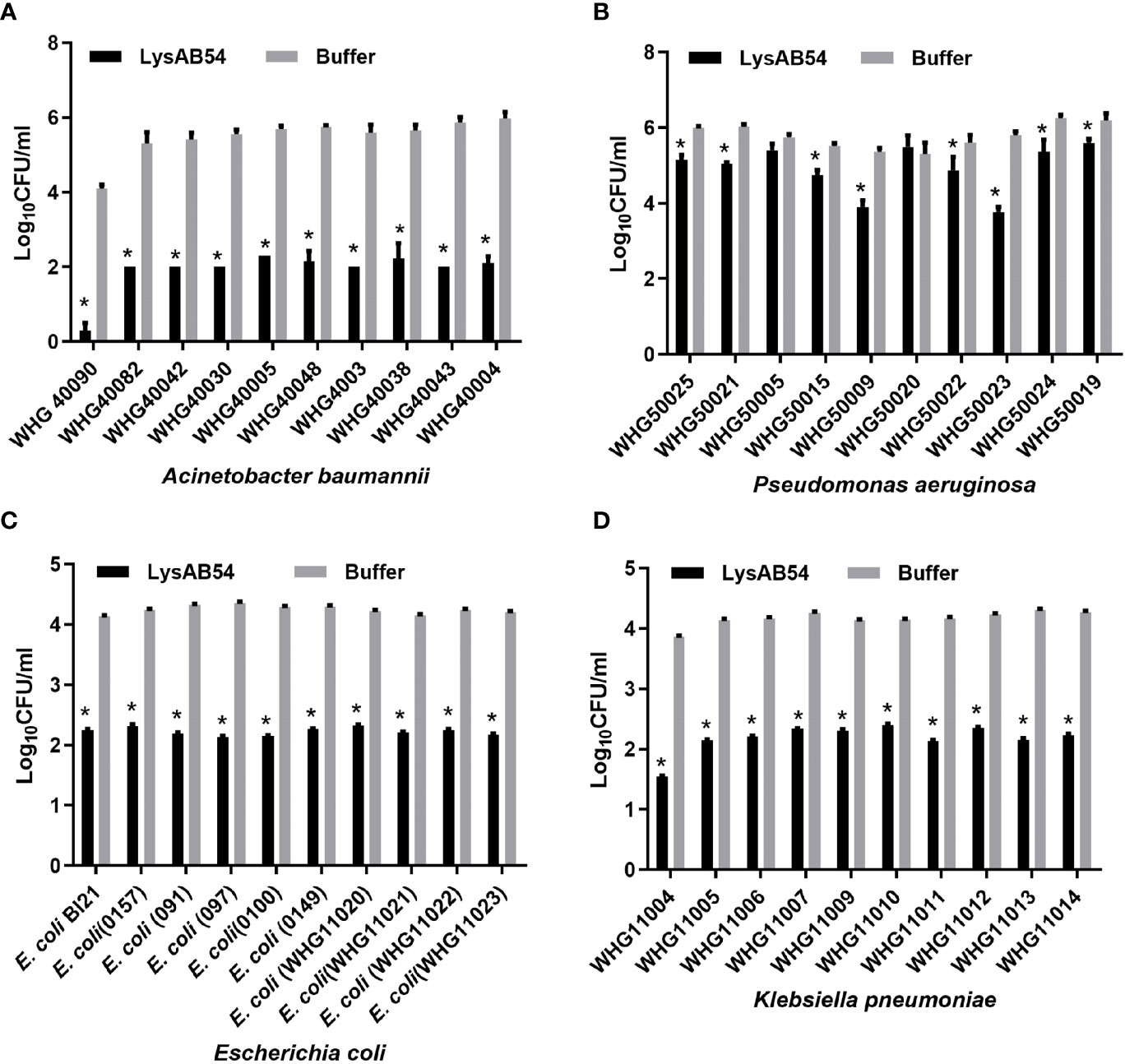
Figure 5 Antibacterial activities of LysAB54 against logarithmic phase of Gram-negative microbes in Tris buffer. Logarithmic phase strains of A baumannii (A), P. aeruginosa (B), E. coli (C), and K pneumoniae (D) were treated with 100 μg/ml of LysAB54 for 1 h at 37°C. The viable cell number was calculated on the LB agar plates. Data are shown as means ± standard deviations. *p < 0.05.
Antibacterial Activity of LysAB54 Under Various Conditions
Previous reports showed that multiple Gram-negative endolysins have good activity against the logarithmic phase bacteria but less or rare active against the stationary bacteria (Yang et al., 2015b; Guo et al., 2017). Therefore, we first evaluated the bactericidal activity of LysAB54 with bacteria under different growth phases. Results showed that LysAB54 showed similar antibacterial activity against both logarithmic as well as stationary phase of A. baumannii (Figure 6). These results indicates that the bacterial phase have minor effect on the antibacterial activity of LysAB54.
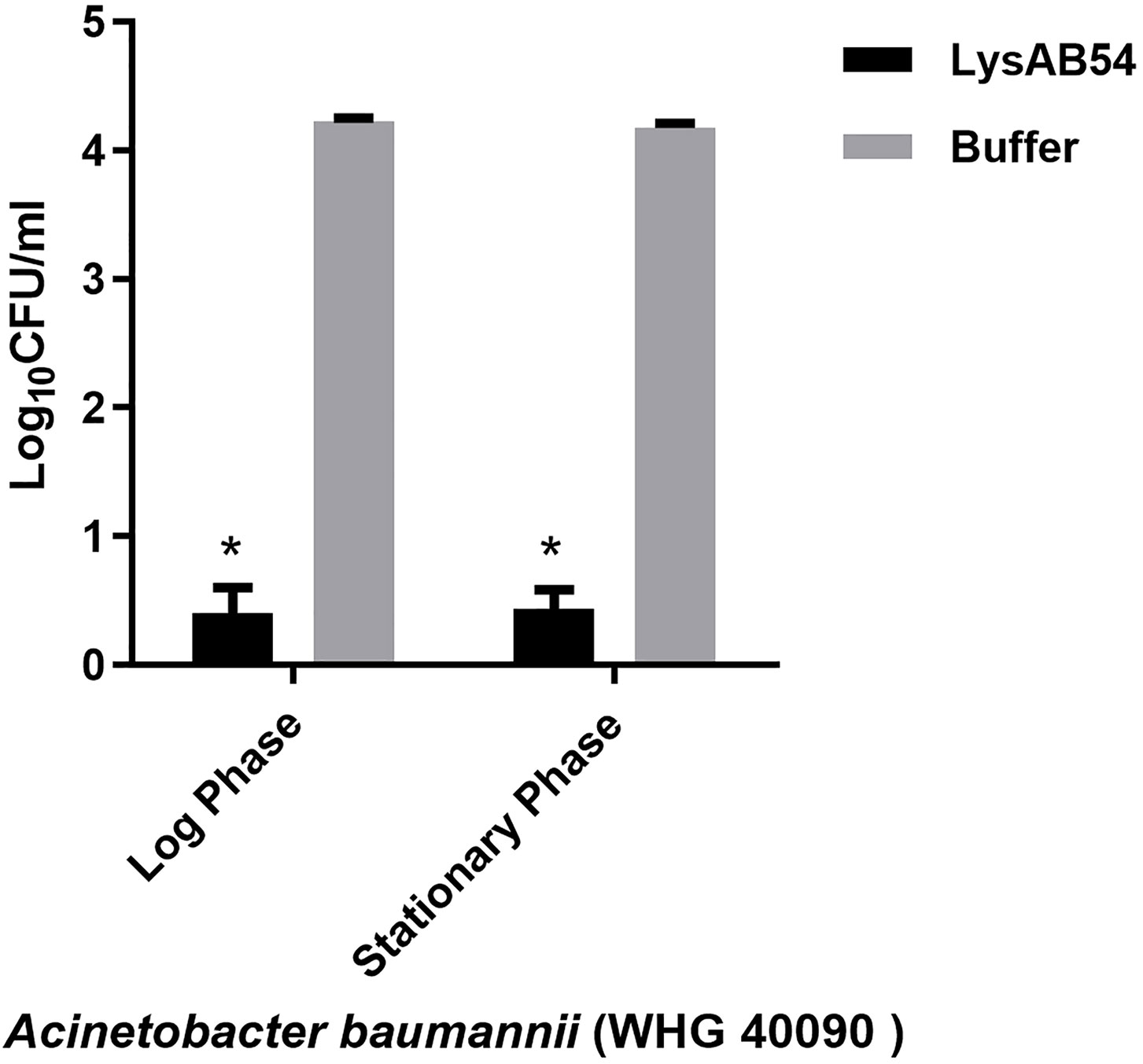
Figure 6 Antibacterial activities of LysAB54 against the logarithmic and stationary phase of A. baumannii. A. baumannii WHG 40090 cells under different growth phases were treated with 100 μg/ml of LysAB54 for 1 h at 37°C. The viable cell number was calculated on the LB agar plates. Data are shown as means ± standard deviations. *p < 0.05.
Another challenge for application of Gram-negative endolysin is the reduced or abolished killing activity in a complex medium like serum. Most of the reported endolysins lose their activity in serum. In this context, we checked out the antibacterial activity of LysAB54 against the logarithmic phase of A. baumannii in complex medium, including LB broth and human serum. As reported previously, insignificant bactericidal activity of LysAB54 was observed in the complex mediums (Figure S2). The possible reason for loss of bactericidal activity is the ion exchange in these complex media which can potentially neutralize the interaction between endolysin and the negatively charged outer membrane.
Discussion
The increasing infections caused by drug-resistant Gram-negative bacteria is a public health concern worldwide (Chopra et al., 2008; Scallan et al., 2011; Gondil et al., 2019). The Gram-negative bacterium A. baumannii which is responsible for multiple infections is an alarming threat to human health. Due to its intrinsic resistance and the overuse of antibiotics, some isolates of A. baumannii are now resistant to almost all known antibiotics (Manchanda et al., 2010). The increasing trend of antibiotic resistance in pathogenic bacteria necessitate the search for alternative antimicrobial agents to treat and cure such resistant pathogens mediated infections (Magiorakos et al., 2012). Currently, a very limited solutions are available in the treatment of MDR A. baumannii in clinical settings (Fischetti, 2010). Endolysin is a class of novel antimicrobial agents to treat and cure antibiotic-resistant bacteria due to its rapid bactericidal activity and low chances of antibiotic resistance (Nelson et al., 2012). Currently, several endolysins are being evaluated in clinical trials to explore their potential in treating Gram-positive bacterial infections (Kim et al., 2018). On the contrary, endolysins for Gram-negative bacterial infections are still under development. Although few studies have reported positive results in treating infections caused by Gram-negative bacteria in vitro and in vivo, including A. baumannii and P. aeruginosa (Ghose and Euler, 2020).
In this study, we characterized a novel bacteriophage endolysin LysAB54 encoded by a novel A. baumannii bacteriophage p54, which showed a broad range of antibacterial activity against Gram-negative bacteria irrespective of their growth phases, suggesting that the naturally occurring endolysin could be a promising alternative antimicrobial agent against number of MDR Gram-positive pathogens.
One excellent merit of the LysAB54 is its robust bactericidal activity against both logarithmic and stationary phase of Gram-negative bacteria in the absence of OMPs. Encouragingly, a reduction of over 4 logs in viable bacterial number was observed after treatment of A. baumannii cells with 100 μg/ml LysAB54 for 10 min.
To our knowledge, it is unconventional for an endolysin to render such rapid antibacterial activity against various antibiotic-resistant Gram-negative bacteria. Several natural known endolysins also exhibited antibacterial activity against Gram-negative bacteria in the absence of OMPs, such as OBPgp279 against P. aeruginosa, PlyF307 and LysAB21 against A. baumannii (Lai et al., 2011; Walmagh et al., 2012; Lood et al., 2015). It is demonstrated with increasing evidence that several endolysins are active against multiple Gram-negative bacteria, and few of them are reported active against Gram-positive pathogens in vitro, such as Abtn-4 (Yuan et al., 2020), LysSAP26 (Kim et al., 2020a), and LysSS (Kim et al., 2020b). Such lysins with broad host-range are commonly contain a lysozyme or glycoside hydrolase catalytic activity, which cleaves the β-1,4-glycosidic linkage between N-acetylmuramic acid (NAM) and N-acetylglucosamine (NAG) in peptidoglycans that is generally shared by various bacteria (Ghose and Euler, 2020). In this context, LysAB54 was predicted to contain a lysozyme activity, which is possibly the reason for its broad antibacterial activity against several other antibiotic-resistant Gram-negative bacteria, which includes P. aeruginosa, K. pneumoniae, and E. coli.
It is well known that environmental factors such as salts and proteins have a significant effect on the activity of Gram-negative endolysins. Our results showed that LysAB54 completely loses its antibacterial activity in the complex medium like LB broth and serum, which narrows down its application to topical infections, such as burn wound, or suture infections. One possible reason for the loss of LysAB54 activity in these media may be due to ions exchanging of endolysin in these environments, which could possibly neutralize the antibacterial activity of endolysins (Yang et al., 2015b).
In conclusion, our study demonstrates the morphological and genomic characteristics of a newly isolated A. baumannii bacteriophage p54 and the bactericidal activity of its endolysin, LysAB54, against multiple drug-resistant Gram-negative bacteria under various growth phases and conditions. The OMP-independent activity of LysAB54 makes it a potential candidate for the treatment of infections caused by multiple drug resistant Gram-negative pathogens. However, molecular dissection and validation of LysAB54 in biofilms and animal models still need to be established to reveal the enigmatic aspects of the present endolysin.
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding authors.
Author Contributions
HY and HW designed the study. FK, VG, CL, MJ, and JL performed the experiments. FK, VG, JY, and HY performed data analysis. HY and HW contributed with reagents and/or funds for research. FK and VG wrote the draft manuscript. HY revised the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This work was financially supported by the Youth Innovation Promotion Association CAS (to HY) and the National Natural Science Foundation of China (no. 31770192, no. 32070187 to HY).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Mrs. Pei Zhang from the Core Facility and Technical Support, Wuhan Institute of Virology for her assistance in microscopy analysis.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcimb.2021.637313/full#supplementary-material
References
Bonomo R. A., Szabo D. (2006). Mechanisms of multidrug resistance in Acinetobacter species and. Pseudomonas Aeruginosa Clin. Infect. Dis. 43 Suppl 2, S49–S56. doi: 10.1086/504477
Briers Y., Volckaert G., Cornelissen A., Lagaert S., Michiels C. W., Hertveldt K., et al. (2007). Muralytic activity and modular structure of the endolysins of Pseudomonas aeruginosa bacteriophages phiKZ and EL. Mol. Microbiol. 65, 1334–1344. doi: 10.1111/j.1365-2958.2007.05870.x
Briers Y., Schmelcher M., Loessner M. J., Hendrix J., Engelborghs Y., Volckaert G., et al. (2009). The high-affinity peptidoglycan binding domain of Pseudomonas phage endolysin KZ144. Biochem. Biophys. Res. Commun. 383, 187–191. doi: 10.1016/j.bbrc.2009.03.161
Briers Y., Walmagh M., Lavigne R. (2011). Use of bacteriophage endolysin EL188 and outer membrane permeabilizers against Pseudomonas aeruginosa. J. Appl. Microbiol. 110, 778–785. doi: 10.1111/j.1365-2672.2010.04931.x
Briers Y., Walmagh M., Van Puyenbroeck V., Cornelissen A., Cenens W., Aertsen A., et al. (2014). Engineered endolysin-based “Artilysins”. Combat Multidrug-Resist. Gram-negative Pathog. mBio 5, e01379–e01314. doi: 10.1128/mBio.01379-14
Chopra I., Schofield C., Everett M., O’neill A., Miller K., Wilcox M., et al. (2008). Treatment of health-care-associated infections caused by Gram-negative bacteria: a consensus statement. Lancet Infect. Dis. 8, 133–139. doi: 10.1016/S1473-3099(08)70018-5
Collaborators G.B.D.L.R.I. (2018). Estimates of the global, regional, and national morbidity, mortality, and aetiologies of lower respiratory infections in 195 countrie-2016: a systematic analysis for the Global Burden of Disease Study 2016. Lancet Infect. Dis. 18, 1191–1210. doi: 10.1016/S1473-3099(18)30310-4
Felsenstein J. (1985). Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39, 783–791. doi: 10.1111/j.1558-5646.1985.tb00420.x
Fischetti V. A. (2005). Bacteriophage lytic enzymes: novel anti-infectives. Trends Microbiol. 13, 491–496. doi: 10.1016/j.tim.2005.08.007
Fischetti V. A. (2008). Bacteriophage lysins as effective antibacterials. Curr. Opin. Microbiol. 11, 393–400. doi: 10.1016/j.mib.2008.09.012
Fischetti V. A. (2010). Bacteriophage endolysins: a novel anti-infective to control Gram-positive pathogens. Int. J. Med. Microbiol. 300, 357–362. doi: 10.1016/j.ijmm.2010.04.002
Ghose C., Euler C. W. (2020). Gram-Negative Bacterial Lysins. Antibiotics (Basel) 9(2), 74. doi: 10.3390/antibiotics9020074
Gondil V. S., Kalaiyarasan T., Bharti V. K., Chhibber S. (2019). Antibiofilm potential of Seabuckthorn silver nanoparticles (SBT@AgNPs) against Pseudomonas aeruginosa. 3 Biotech. 9, 402. doi: 10.1007/s13205-019-1947-6
Gondil V. S., Harjai K., Chhibber S. (2020). Endolysins as emerging alternative therapeutic agents to counter drug-resistant infections. Int. J. Antimicrob. Agents 55, 105844. doi: 10.1016/j.ijantimicag.2019.11.001
Guo M., Feng C., Ren J., Zhuang X., Zhang Y., Zhu Y., et al. (2017). A novel antimicrobial endolysin, LysPA26, against Pseudomonas aeruginosa. Front. Microbiol. 8, 293. doi: 10.3389/fmicb.2017.00293
Helander I. M., Mattila-Sandholm T. (2000). Fluorometric assessment of gram-negative bacterial permeabilization. J. Appl. Microbiol. 88, 213–219. doi: 10.1046/j.1365-2672.2000.00971.x
Huang L., Luo D., Gondil V. S., Gong Y., Jia M., Yan D., et al. (2020). Construction and characterization of a chimeric lysin ClyV with improved bactericidal activity against Streptococcus agalactiae in vitro and in vivo. Appl. Microbiol. Biotechnol. 104, 1609–1619. doi: 10.1007/s00253-019-10325-z
Kering K. K., Zhang X., Nyaruaba R., Yu J., Wei H. (2020). Application of adaptive evolution to improve the stability of bacteriophages during storage. Viruses 12(4), 423. doi: 10.3390/v12040423
Kim N. H., Park W. B., Cho J. E., Choi Y. J., Choi S. J., Jun S. Y., et al. (2018). Effects of phage endolysin SAL200 combined with antibiotics on Staphylococcus aureus infection. Antimicrob. Agents Chemother. 62(10), e00731–18. doi: 10.1128/AAC.00731-18
Kim S., Jin J. S., Choi Y. J., Kim J. (2020a). LysSAP26, a new recombinant phage endolysin with a broad spectrum antibacterial activity. Viruses 12(11), 1340. doi: 10.3390/v12111340
Kim S., Lee D. W., Jin J. S., Kim J. (2020b). Antimicrobial activity of LysSS, a novel phage endolysin, against Acinetobacter baumannii and Pseudomonas aeruginosa. J. Glob. Antimicrob. Resist. 22, 32–39. doi: 10.1016/j.jgar.2020.01.005
Kumar S., Stecher G., Li M., Knyaz C., Tamura K. (2018). MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol. Biol. Evol. 35, 1547–1549. doi: 10.1093/molbev/msy096
Lai M. J., Lin N. T., Hu A., Soo P. C., Chen L. K., Chen L. H., et al. (2011). Antibacterial activity of Acinetobacter baumannii phage ϕAB2 endolysin (LysAB2) against both gram-positive and gram-negative bacteria. Appl. Microbiol. Biotechnol. 90, 529–539. doi: 10.1007/s00253-011-3104-y
Loeffler J. M., Djurkovic S., Fischetti V. A. (2003). Phage lytic enzyme Cpl-1 as a novel antimicrobial for pneumococcal bacteremia. Infect. Immun. 71, 6199–6204. doi: 10.1128/IAI.71.11.6199-6204.2003
Loessner M. J., Kramer K., Ebel F., Scherer S. (2002). C-terminal domains of Listeria monocytogenes bacteriophage murein hydrolases determine specific recognition and high-affinity binding to bacterial cell wall carbohydrates. Mol. Microbiol. 44, 335–349. doi: 10.1046/j.1365-2958.2002.02889.x
Lood R., Winer B. Y., Pelzek A. J., Diez-Martinez R., Thandar M., Euler C. W., et al. (2015). Novel phage lysin capable of killing the multidrug-resistant gram-negative bacterium Acinetobacter baumannii in a mouse bacteremia model. Antimicrob. Agents Chemother. 59, 1983–1991. doi: 10.1128/AAC.04641-14
Magiorakos A. P., Srinivasan A., Carey R. B., Carmeli Y., Falagas M. E., Giske C. G., et al. (2012). Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: an international expert proposal for interim standard definitions for acquired resistance. Clin. Microbiol. Infect. 18, 268–281. doi: 10.1111/j.1469-0691.2011.03570.x
Manchanda V., Sanchaita S., Singh N. (2010). Multidrug resistant acinetobacter. J. Glob. Infect. Dis. 2, 291–304. doi: 10.4103/0974-777X.68538
Nelson D., Schuch R., Chahales P., Zhu S., Fischetti V. A. (2006). PlyC: a multimeric bacteriophage lysin. Proc. Natl. Acad. Sci. U. S. A. 103, 10765–10770. doi: 10.1073/pnas.0604521103
Nelson D. C., Schmelcher M., Rodriguez-Rubio L., Klumpp J., Pritchard D. G., Dong S., et al. (2012). Endolysins as antimicrobials. Adv. Virus Res. 83, 299–365. doi: 10.1016/B978-0-12-394438-2.00007-4
Oliveira H., Vilas Boas D., Mesnage S., Kluskens L. D., Lavigne R., Sillankorva S., et al. (2016). Structural and enzymatic characterization of ABgp46, a novel phage endolysin with broad anti-gram-negative bacterial activity. Front. Microbiol. 7, 208. doi: 10.3389/fmicb.2016.00208
Pendleton J. N., Gorman S. P., Gilmore B. F. (2013). Clinical relevance of the ESKAPE pathogens. Expert Rev. Anti Infect. Ther. 11, 297–308. doi: 10.1586/eri.13.12
Potron A., Poirel L., Nordmann P. (2015). Emerging broad-spectrum resistance in Pseudomonas aeruginosa and Acinetobacter baumannii: Mechanisms and epidemiology. Int. J. Antimicrob. Agents 45, 568–585. doi: 10.1016/j.ijantimicag.2015.03.001
Rodríguez-Rubio L., Chang W. L., Gutiérrez D., Lavigne R., Martínez B., Rodríguez A., et al. (2016). ‘Artilysation’ of endolysin λSa2lys strongly improves its enzymatic and antibacterial activity against streptococci. Sci. Rep. 6, 35382. doi: 10.1038/srep35382
Saitou N., Nei M. (1987). The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4, 406–425. doi: 10.1093/oxfordjournals.molbev.a040454
Scallan E., Griffin P. M., Angulo F. J., Tauxe R. V., Hoekstra R. M. (2011). Foodborne illness acquired in the United States–unspecified agents. Emerg. Infect. Dis. 17, 16–22. doi: 10.3201/eid1701.P21101
Schmelcher M., Donovan D. M., Loessner M. J. (2012). Bacteriophage endolysins as novel antimicrobials. Future Microbiol. 7, 1147–1171. doi: 10.2217/fmb.12.97
Schuch R., Nelson D., Fischetti V. A. (2002). A bacteriolytic agent that detects and kills Bacillus anthracis. Nature 418, 884–889. doi: 10.1038/nature01026
Spellberg B., Bartlett J. G., Gilbert D. N. (2013). The future of antibiotics and resistance. N. Engl. J. Med. 368, 299–302. doi: 10.1056/NEJMp1215093
Valencia R., Arroyo L. A., Conde M., Aldana J. M., Torres M. J., Fernández-Cuenca F., et al. (2009). Nosocomial outbreak of infection with pan-drug-resistant Acinetobacter baumannii in a tertiary care university hospital. Infect. Control. Hosp. Epidemiol. 30, 257–263. doi: 10.1086/595977
Walmagh M., Briers Y., Dos Santos S. B., Azeredo J., Lavigne R. (2012). Characterization of modular bacteriophage endolysins from Myoviridae phages OBP, 201φ2-1 and PVP-SE1. PLoS One 7, e36991. doi: 10.1371/journal.pone.0036991
Yang H., Yu J., Wei H. (2014). Engineered bacteriophage lysins as novel anti-infectives. Front. Microbiol. 5, 542. doi: 10.3389/fmicb.2014.00542
Yang H., Linden S. B., Wang J., Yu J., Nelson D. C., Wei H. (2015a). A chimeolysin with extended-spectrum streptococcal host range found by an induced lysis-based rapid screening method. Sci. Rep. 5, 17257. doi: 10.1038/srep17257
Yang H., Wang M., Yu J., Wei H. (2015b). Antibacterial activity of a novel peptide-modified lysin against Acinetobacter baumannii and Pseudomonas aeruginosa. Front. Microbiol. 6, 1471. doi: 10.3389/fmicb.2015.01471
Yang H., Luo D., Etobayeva I., Li X., Gong Y., Wang S., et al. (2020). Linker editing of pneumococcal lysin ClyJ conveys improved bactericidal activity. Antimicrob. Agents Chemother. 64(2), e01610–19. doi: 10.1128/AAC.01610-19
Keywords: bacteriophage, endolysin, Acinetobacter baumannii, Gram-negative “superbugs”, antimicrobial resistance
Citation: Khan FM, Gondil VS, Li C, Jiang M, Li J, Yu J, Wei H and Yang H (2021) A Novel Acinetobacter baumannii Bacteriophage Endolysin LysAB54 With High Antibacterial Activity Against Multiple Gram-Negative Microbes. Front. Cell. Infect. Microbiol. 11:637313. doi: 10.3389/fcimb.2021.637313
Received: 03 December 2020; Accepted: 21 January 2021;
Published: 02 March 2021.
Edited by:
Jingmin Gu, Jilin University, ChinaReviewed by:
Jinquan Li, Huazhong Agricultural University, ChinaShuai Le, Army Medical University, China
Copyright © 2021 Khan, Gondil, Li, Jiang, Li, Yu, Wei and Yang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Hongping Wei, aHB3ZWlAd2guaW92LmNu; Hang Yang, eWFuZ2hAd2guaW92LmNu
 Fazal Mehmood Khan
Fazal Mehmood Khan Vijay Singh Gondil
Vijay Singh Gondil Changchang Li1,2
Changchang Li1,2 Mengwei Jiang
Mengwei Jiang Junping Yu
Junping Yu Hongping Wei
Hongping Wei Hang Yang
Hang Yang