
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Cell. Infect. Microbiol. , 19 April 2021
Sec. Clinical Microbiology
Volume 11 - 2021 | https://doi.org/10.3389/fcimb.2021.634382
This article is part of the Research Topic Progress in Pathogen Identification Based on Mass Spectrometry View all 17 articles
 Mareike Bernhard1
Mareike Bernhard1 Navaporn Worasilchai2†
Navaporn Worasilchai2† Mourine Kangogo3
Mourine Kangogo3 Christine Bii4
Christine Bii4 Wioleta J. Trzaska5
Wioleta J. Trzaska5 Michael Weig1
Michael Weig1 Uwe Groß1
Uwe Groß1 Ariya Chindamporn2
Ariya Chindamporn2 Oliver Bader1*
Oliver Bader1*Yeasts of the Cryptococcus neoformans/gattii species complexes are human pathogens mostly in immune compromised individuals, and can cause infections from dermal lesions to fungal meningitis. Differences in virulence and antifungal drug susceptibility of species in these complexes indicate the value of full differentiation to species level in diagnostic procedures. MALDI-TOF MS has been reported to sufficiently discriminate these species. Here, we sought to re-evaluate sample pre-processing procedures and create a set of publicly available references for use with the MALDI Biotyper system. Peak content using four different pre-processing protocols was assessed, and database entries for 13 reference strains created. These were evaluated against a collection of 153 clinical isolates, typed by conventional means. The use of decapsulating protocols or mechanical disruption did not sufficiently increase the information content to justify the extra hands-on-time. Using the set of 13 reference entries created with the standard formic acid extraction, we were able to correctly classify 143/153 (93.5%) of our test isolates. The majority of the remaining ten isolates still gave correct top matches; only two isolates did not give reproducible identifications. This indicates that the log score cut-off can be lowered also in this context. Ease to identify cryptococcal isolates to the species level is improved by the workflow evaluated here. The database references are freely available from https://github.com/oliverbader/BioTyper-libraries for incorporation into local diagnostic systems.
The group of basidiomycetous yeast of the Cryptococcus neoformans/gattii complexes hosts a variety of human pathogenic species, causing infections from skin lesions to fatal meningitis [reviewed in (Kronstad et al., 2011)]. This mainly contributes to morbidity and mortality in patients with underlying immune deficiencies (e.g. HIV), but can also affect immunocompetent hosts. Species of the C. neoformans/gattii complexes are readily found in the environment, living, for example, on eucalyptus tree bark, and bird droppings.
The most prominent diagnostic feature of these species are the large capsules of most isolates [reviewed in (O’Meara and Alspaugh, 2012)], which can easily be visualized by, e.g., displacement of India ink stain. India ink does not penetrate the capsule and thus creates a halo around the cells visible in microscopy. The polysaccharides shed from the cell also give rise to efficient and specific serologic tests of cryptococcal infections through serum detection of galactomannan.
Species in this complex have traditionally been divided into four serotypes based on antigenicity of the capsule, forming three varieties: C. neoformans var. grubii (serotype A), var. gattii (serotypes B and C), and var. neoformans (serotype D). They are also able to form inter-species hybrids leading to, e.g., an AD serotype (Boekhout et al., 2001). Several genetic methods are available to stratify the different serotypes into further molecular types (Meyer et al., 2003) and characterize hybrid strains. Recently, it has been proposed to formally raise the non-hybrid molecular types to species level (Kwon-Chung et al., 2002; Hagen et al., 2015) and a fifth C. gattii lineage has recently been described (Farrer et al., 2019) from environmental and animal specimen.
In clinical samples from Europe most frequently serotype A is found, mainly from immunocompromised patients, e.g. those suffering from AIDS (Kronstad et al., 2011). Highly virulent isolates usually stem from the C. gattii complex, which also readily infect immuno-competent hosts. Differences in mean antifungal susceptibility between closely related molecular types have been reported (Trilles et al., 2012; Cogliati et al., 2018; Lee et al., 2019) and in vitro differences in cytokine responses (Herkert et al., 2018). Some molecular types, mainly VGII and VGIII, are more prone to be involved in outbreak scenarios (Kidd et al., 2004; Carriconde et al., 2011; Springer et al., 2014). A major difference between C. neoformans and C. gattii groups is the lack of growth inside macrophages among C. gattii isolates, with the notable exception of such outbreak lineages (Voelz et al., 2014).
Together this underlines the benefit of methods easily discriminating between the major molecular types, not only in clinical contexts, but also for epidemiological studies which so far rely on laborious genetic typing [e.g. our own work (Tangwattanachuleeporn et al., 2013; Kangogo et al., 2015; Worasilchai et al., 2017) or others (Fang et al., 2020; Jin et al., 2020)]. MALDI-TOF MS has been established over the past years as a widely used clinical species identification tool and has been shown to be able to discriminate between the seven known molecular types within the C. neoformans/gattii complexes (McTaggart et al., 2011; Firacative et al., 2012; Posteraro et al., 2012; Hagen et al., 2015). For C. gatti and C. deuterogatii differential mass peaks have been described (Jin et al., 2020).
However, this has not been implemented in diagnostic systems, which remain at the point where only C. neoformans var. neoformans/grubii vs. C. gattii complexes can be identified. In part, this may be due to the observation that false species designations above the significance threshold can be observed (Posteraro et al., 2012), and reflect the complexity introduced by hybrid formation between the different linages.
In this study, we created a publicly available MALDI Biotyper database reference (“main spectrum projections”, MSPs) set from 13 type strains of seven recognized non-hybrid subtypes in the Cryptococcus neoformans/gattii complexes. Their performance using different preprocessing protocols is evaluated on a set of characterized isolates.
For long-term storage, Cryptococcus isolates were kept at -70°C in cryobank stocks (Mast Diagnostica, Reinfeld, Germany). After thawing, strains were propagated on Sabouraud’s (SAB) agar slants supplemented with 0.5% peptone (casein), 0.5% peptone (meat), and 2% glucose. Before sample preparation, strains were cultivated on SAB agar overnight at 30°C.
For the purpose of text clarity, only the species nomenclature according to Hagen et al. (Hagen et al., 2015) is adopted from here. As references, thirteen strains of the CBS collection (Westerdijk Fungal Biodiversity Institute) were used: three C. neoformans (CBS 8710 (molecular type VNI), CBS 10084 (VNII), CBS 10085(VNI)), two C. deneoformans (CBS 6900 and CBS 10079 (VNIV)), two C. gattii (CBS 6289,and CBS 10078, VGI), two C. deuterogattii (CBS 10082, and CBS 10514, VGII) two C. bacillisporus (CBS 6955 and CBS 10081, VGIII), one C. tetragattii (CBS 10101, VGIV), and C. decagattii (CBS 11687, VGIV).
A test set of 153 isolates was assembled from previously characterized collections. This included all Thai strains from Worasilchai et al. (2017), augmented with rare species isolates from Kenya (Kangogo et al., 2015) and the Birmingham laboratory collection, which include strain from various studies [e.g. (Voelz et al., 2014)]. All isolates were typed either previously (Kangogo et al., 2015; Worasilchai et al., 2017) or specifically for the purpose of this study using the URA5-RFLP method. The final set contained n=96 C. neoformans, n=6 C. deneoformans, n=5 C. gattii, n=18 C. bacillisporus, n=20 C. deuterogattii, and n=8 C. tetragattii isolates. A negative control group was assembled from mass spectra randomly chosen from those obtained during bacterial (n=86) of fungal (n=403) routine diagnostics.
Restriction fragment length polymorphisms were performed as described previously (Kangogo et al., 2015; Worasilchai et al., 2017). Briefly, genomic DNA was extracted from cells using phenol/chloroform and the URA5 gene was amplified using URA5 forward (5-ATGTCCTCCCAAGCCCTCGACTCCG-3) and SJ01 reverse (5-TTAAGACCTCTGAACACCGTACTC-3) primers (Meyer et al., 2003). The amplicons obtained were either simultaneously digested with HhaI (20 U/μl) and Sau96I (10 U/μl) or StuI (10 U/µl) alone for 8 hours (all from New England Biolabs). The digestion products were purified using a PCR clean-up kit (NucleoSpin, Macherey-Nagel, Düren, Germany) and visualized on a 3% agarose gel.
For regular harvest and formic acid-extraction [preprocessing protocol (A) (Bader, 2017)], cells were taken from agar plates by scraping approximately a 1µl loop full of cells and re-suspending them in 300 µl water. 700 µl absolute ethanol was added to a final concentration of 70% (v/v) and vortexes. Cells were spun down at 8500xg for 5 min, the supernatant completely discarded and the cells lysed first with 50 µl 70% (v/v) formic acid, and 50 µl pure acetonitrile. Modifications to this protocol tested were for preprocessing protocol (B) that cells were collected in 300 µl 5% (v/v) DMSOad, for preprocessing protocol (C) that DMSO was included in the 70% ethanol washing step to a final volume of 5% (v/v), and for preprocessing protocol (D) that cells were collected in 300 µl water already including an equivalent of ~100 µl glass beads (0.5 mm diameter, Roth, Karlsruhe, Germany). Here, cells were mechanically disrupted in a FP120 fast prep machine (Bio101, Thermo Savant) at setting 4, for 30 sec during the formic acid step.
MSP references for the MALDI Biotyper were generated according to the manufacturer’s guidelines (Kostrzewa and Maier, 2017), using preprocessing protocol A. Spectra from 24 individual spots were gathered on a freshly calibrated (BTS reference standard) Autoflex III system (Bruker Daltonics, Bremen, Germany) using the automated acquisition mode of the Biotyper 3.1. Spectra were processed using the inbuilt MSP generation method, using the standard parameters.
The literature reports that both, removal of cryptococcal capsule can (Thomaz et al., 2016) or does not (Hagen et al., 2015) positively influence spectrum quality. Since the capsule material is soluble in DMSO, we devised pre-processing protocols that would deplete the capsule prior to the regular formic acid/acetonitrile extraction protocol. Both pre-processing protocols, B (Figures 1A, B) and C (not shown), efficiently removed capsules in all strains. However, subsequent measurement of mass spectra did not reveal any additional mass signals, or major differences in spectrum quality (Figure 1C).
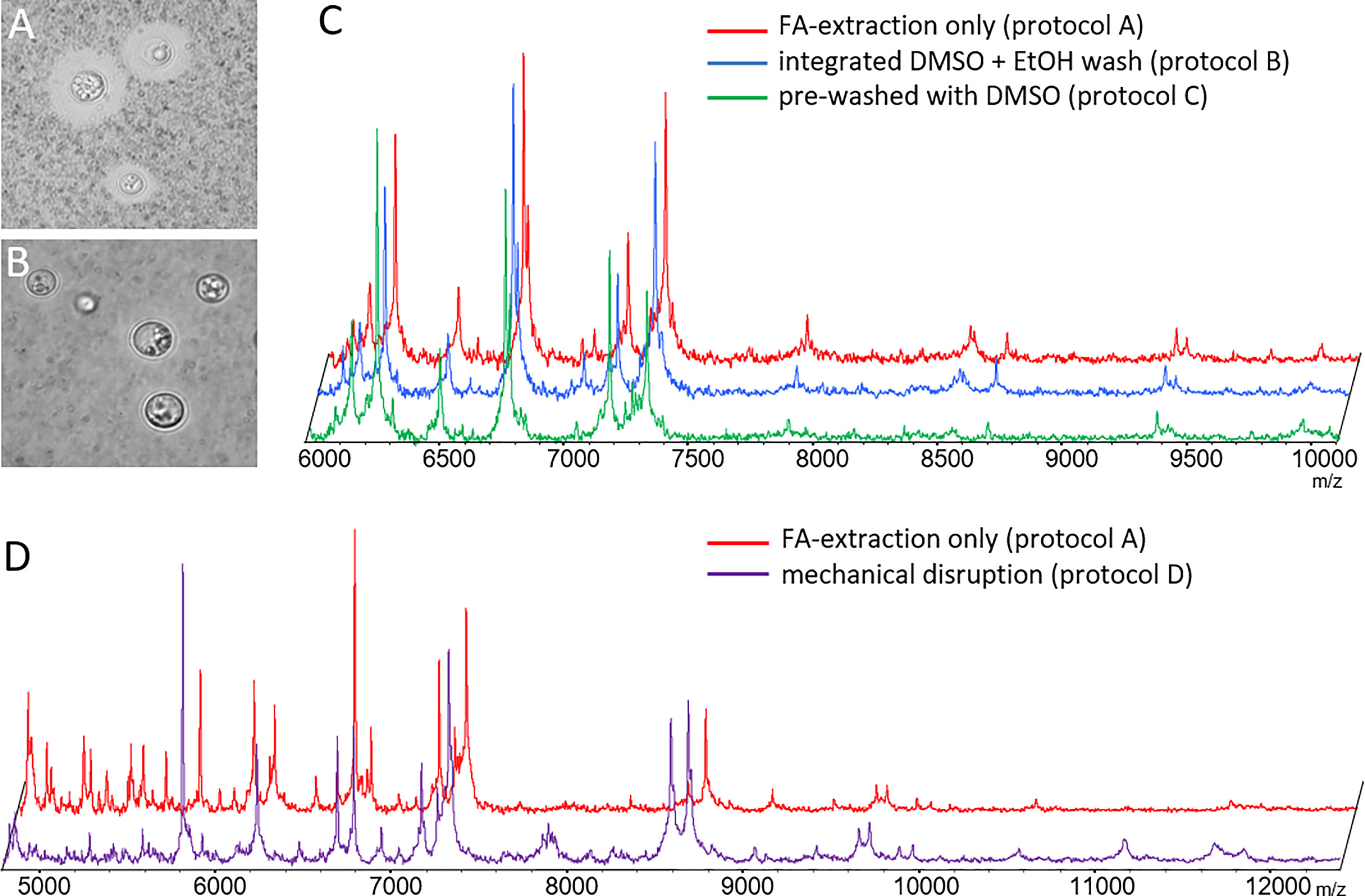
Figure 1 Optimization of sample pre-processing procedures. Phase contrast microscopy of ink-stained cryptococcal cells (A) without and (B) with DMSO treatment depleting capsular material (protocol B). (C) Spectra obtained from de-capsulated cells by protocols B and C were not different from those generated by formic acid extraction alone. (D) Spectra obtained from mechanically disrupted cells (protocol D) had similar masses, but differed in relative signal intensity for some, as compared to those obtained by formic acid extraction. Example results shown here for CBS 10485 are valid for all isolates. Signal intensities on y-axes in panels (C, D) mainly rely on number of spectra gathered in sum buffer. Spectra have been manually re-scaled on the y-axis for better visual comparison between different experimental conditions, and scaling has been omitted to reflect this fact.
Next, we tested if mechanical disruption of the cells yielded more informative spectra using mechanical disruption (preprocessing protocol D). Indeed, mass spectra recorded from mechanically disrupted cells resulted in more evenly distributed peak intensities across the major mass signals. However, no additional mass signals of high intensity were found (Figure 1D).
In our hands removal of the capsule did not result in spectra with higher information content, at any time. Mechanical disruption did reveal some additional masses, but in favor of the lower hands-on-time the original pre-processing protocol A was subsequently used for MSP creation and testing.
Next, we created MSPs for 13 reference strains encompassing seven molecular types of the C. neoformans/gattii complexes (Meyer et al., 2003; Hagen et al., 2015), using the standard extraction procedure (pre-processing protocol A). Cluster analysis of the MSPs generated suggested sufficient distance to clearly distinguish between C. neoformans complex molecular types VNIV (C. deneoformans) and VNI/II, and possibly also between VNI and VNII themselves, but less so among molecular types within the C. gattii complex (Figure 2).
Mass spectra for all test isolates were obtained using preprocessing protocol A. Were MALDI-TOF results using the new MSP set deviated from previous data, URA5-RFLP typing was repeated as the gold standard (Figure 3A). All but two deviations could be resolved (see below). To discriminate between C. tetragattii and potential C. decagattii strains, we sequenced the URA5-amplicon obtained from CBS 11687 (C. decagattii, deposited at Genebank under the accession number MH605184) and compared it to the respective sequence of CBS 10101 (C. tetragattii, gene bank accession AY973155). Restriction with StuI was found, and experimentally confirmed, to discriminate the two species (Figure 3B). However, there were no further C. decagattii isolates among our strains. C. decagattii remains a rare species, and only a single isolate of this molecular type (CBS 11687) was available for this study, which was already included in the reference set. Therefore, the final test collection encompassed only six of the seven species used for generation of references.
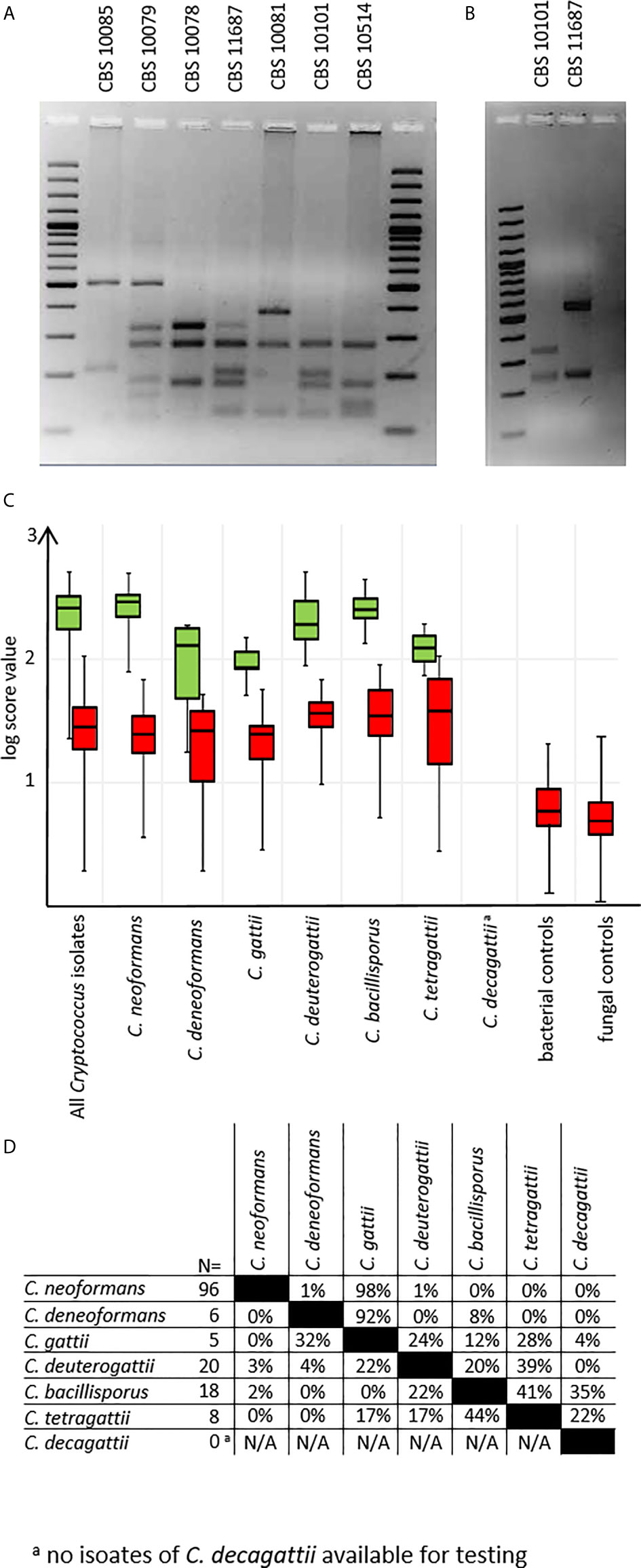
Figure 3 URA5-RFLPs. (A) URA5-RFLP patterns of control isolates. (B) CBS10101 (C. tetragattii) and CBS 11687 (C. decagattii) can be differentiated by digesting the URA5-amplicon with StuI. (C) log-score values stratified by species/URA5-RFLP and matching type. Green: scores of correct matches, red: highest scoring incorrect matches. (D) Pairwise percentages of highest scoring incorrect matches. a C. decagattii was not present among testing isolates, as the only strain available for this study was already used to create the MSP reference.
From the test collection, we were able to correctly identify 143/153 (93.5%) of the isolates on species-level using duplicate spots, with the top log score ≥ 2.000 (Figure 3C), as recommended by the manufacturer. Of the remaining ten isolates, eight still gave correct species matches at scores between 1.700 and 1.999, considered only genus-level by the manufacturer. Among the negative control set, there were no results higher than a log score of 1.300, indicating no false positives are to be expected under routine diagnostic conditions (Figure 3C). Inconsistent identifications were only observed for two C. tetragattii isolates where repetitively top matches of different spots of the same preparation were C. tetragattii, C. gattii, or C. deuterogattii, all at values above 1.999.
Because of this, and the close relations found during cluster analysis (Figure 2), we also inspected the log score difference from the correct to the highest scoring false match for each spot (Figure 3D) for those tests where a second species matched above the significance threshold. Only 3% of all tested spots (14 out of 428) matched a second MSP with a log score >1.999. As expected from the cluster analysis, these “best false” second matches were found only among species in the C. gattii complex. This was the case for three C. bacillisporus isolates giving a second best match with C. decagattii, with a log score difference between 0.1 to 0.4. In addition to the two inconsistent C. tetragattii isolates discussed above, one additional C. tetragattii isolate also gave a second best match with C. decagattii. The score values for both matches were near 2.000. The close relationships of the different species will likely also have implications on properly identifying hybrid isolates.
Cryptococcal typing and species identification is complicated by the ongoing discovery of new species (Farrer et al., 2019), and the formation of inter-species hybrids (Hagen et al., 2015). Nevertheless, our data confirms that proper routine identification of clinically relevant non-hybrid C. neoformans/gattii complex molecular types using MALDI-TOF is possible with the current algorithms and standard workflows. In our hands, the only exception was distinguishing the rarer types C. tetragattii and C. decagattii, which was not sufficiently possible. This may be due to the fact, that only low numbers of isolates of these linages were available for testing.
The MSP sets generated in this study are freely available from https://github.com/oliverbader/BioTyper-libraries for use with the molecular type- (Meyer et al., 2003) or the species nomenclatures (Hagen et al., 2015).
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article.
Performed experiments: MB, NW, MK, OB. Contributed typed strains: NW, MK, CB, WT, AC. Wrote the manuscript: MB, AC, OB. Prepared the revision: MB, OB. Supervised the study: MW, UG, AC, OB. All authors contributed to the article and approved the submitted version.
This study received funding from Thai-German mobility scheme “CryptoType” to AC and OB (grant number 01DP13001). Article publishing fees were covered by the Open-Access-publications funds of the Universitätsmedizin Göttingen.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The authors would like to thank Agnieszka Goretzki for expert technical assistance. This study was mainly funded by the Thai-German mobility scheme “CryptoType” to AC and OB.
Bader, O. (2017). Fungal Species Identification by MALDI-ToF Mass Spectrometry. Methods Mol. Biol. 1508, 323–337. doi: 10.1007/978-1-4939-6515-1_19
Boekhout, T., Theelen, B., Diaz, M., Fell, J. W., Hop, W. C., Abeln, E. C., et al. (2001). Hybrid genotypes in the pathogenic yeast Cryptococcus neoformans. Microbiology 147, 891–907. doi: 10.1099/00221287-147-4-891
Carriconde, F., Gilgado, F., Arthur, I., Ellis, D., Malik, R., Van De Wiele, N., et al. (2011). Clonality and alpha-a recombination in the Australian Cryptococcus gattii VGII population–an emerging outbreak in Australia. PloS One 6, e16936. doi: 10.1371/journal.pone.0016936
Cogliati, M., Prigitano, A., Esposto, M. C., Romano, L., Grancini, A., Zani, A., et al. (2018). Epidemiological trends of cryptococcosis in Italy: Molecular typing and susceptibility pattern of Cryptococcus neoformans isolates collected during a 20-year period. Med. Mycol. 56, 963–971. doi: 10.1093/mmy/myx152
Fang, L. F., Zhang, P. P., Wang, J., Yang, Q., Qu, T. T. (2020). Clinical and microbiological characteristics of cryptococcosis at an university hospital in China from 2013 to 2017. Braz. J. Infect. Dis. 24, 7–12. doi: 10.1016/j.bjid.2019.11.004
Farrer, R. A., Chang, M., Davis, M. J., Van Dorp, L., Yang, D. H., Shea, T., et al. (2019). A New Lineage of Cryptococcus gattii (VGV) Discovered in the Central Zambezian Miombo Woodlands. mBio 10 (6), e02306-19. doi: 10.1128/mBio.02306-19
Firacative, C., Trilles, L., Meyer, W. (2012). MALDI-TOF MS enables the rapid identification of the major molecular types within the Cryptococcus neoformans/C. gattii species complex. PloS One 7, e37566. doi: 10.1371/journal.pone.0037566
Hagen, F., Khayhan, K., Theelen, B., Kolecka, A., Polacheck, I., Sionov, E., et al. (2015). Recognition of seven species in the Cryptococcus gattii/Cryptococcus neoformans species complex. Fungal Genet. Biol. 78, 16–48. doi: 10.1016/j.fgb.2015.02.009
Herkert, P. F., Dos Santos, J. C., Hagen, F., Ribeiro-Dias, F., Queiroz-Telles, F., Netea, M. G., et al. (2018). Differential In Vitro Cytokine Induction by the Species of Cryptococcus gattii Complex. Infect. Immun. 86 (4), e00958-17. doi: 10.1128/IAI.00958-17
Jin, L., Cao, J. R., Xue, X. Y., Wu, H., Wang, L. F., Guo, L., et al. (2020). Clinical and microbiological characteristics of Cryptococcus gattii isolated from 7 hospitals in China. BMC Microbiol. 20, 73. doi: 10.1186/s12866-020-01752-4
Kangogo, M., Bader, O., Boga, H., Wanyoike, W., Folba, C., Worasilchai, N., et al. (2015). Molecular types of Cryptococcus gattii/Cryptococcus neoformans species complex from clinical and environmental sources in Nairobi, Kenya. Mycoses 58, 665–670. doi: 10.1111/myc.12411
Kidd, S. E., Hagen, F., Tscharke, R. L., Huynh, M., Bartlett, K. H., Fyfe, M., et al. (2004). A rare genotype of Cryptococcus gattii caused the cryptococcosis outbreak on Vancouver Island (British Columbia, Canada). Proc. Natl. Acad. Sci. U.S.A. 101, 17258–17263. doi: 10.1073/pnas.0402981101
Kostrzewa, M., Maier, T. (2017). “Criteria for Development of MALDI-TOF Mass Spectral Database,” in MALDI-TOF and Tandem MS for Clinical Microbiology, one ed. Eds. Shah, H. N., Garbia, S. E. (Hoboken, New Jersey: John Wiley & Sons Ltd), 39–54.
Kronstad, J. W., Attarian, R., Cadieux, B., Choi, J., D’souza, C. A., Griffiths, E. J., et al. (2011). Expanding fungal pathogenesis: Cryptococcus breaks out of the opportunistic box. Nat. Rev. Microbiol. 9, 193–203. doi: 10.1038/nrmicro2522
Kwon-Chung, K. L., Boekhout, T., Fell, J. W., Diaz, M. (2002). Proposal to conserve the name Cryptococcus gattii against C. hondurianus and C. bacillisporus (Basidiomycota, Hymenomycetes, Tremenomycetidae). Taxon 51, 804–806. doi: 10.2307/1555045
Lee, G. A., Arthur, I., Merritt, A., Leung, M. (2019). Molecular types of Cryptococcus neoformans and Cryptococcus gattii in Western Australia and correlation with antifungal susceptibility. Med. Mycol. 57 (8), 1004–1010. doi: 10.1093/mmy/myy161
McTaggart, L. R., Lei, E., Richardson, S. E., Hoang, L., Fothergill, A., Zhang, S. X. (2011). Rapid identification of Cryptococcus neoformans and Cryptococcus gattii by matrix-assisted laser desorption ionization-time of flight mass spectrometry. J. Clin. Microbiol. 49, 3050–3053. doi: 10.1128/JCM.00651-11
Meyer, W., Castaneda, A., Jackson, S., Huynh, M., Castaneda, E., Iberoamerican Cryptococcal Study Group (2003). Molecular typing of IberoAmerican Cryptococcus neoformans isolates. Emerg. Infect. Dis. 9, 189–195. doi: 10.3201/eid0902.020246
O’Meara, T. R., Alspaugh, J. A. (2012). The Cryptococcus neoformans capsule: a sword and a shield. Clin. Microbiol. Rev. 25, 387–408. doi: 10.1128/CMR.00001-12
Posteraro, B., Vella, A., Cogliati, M., De Carolis, E., Florio, A. R., Posteraro, P., et al. (2012). Matrix-Assisted Laser Desorption Ionization-Time of Flight Mass Spectrometry-Based Method for Discrimination between Molecular Types of Cryptococcus neoformans and Cryptococcus gattii. J. Clin. Microbiol. 50, 2472–2476. doi: 10.1128/JCM.00737-12
Springer, D. J., Billmyre, R. B., Filler, E. E., Voelz, K., Pursall, R., Mieczkowski, P. A., et al. (2014). Cryptococcus gattii VGIII isolates causing infections in HIV/AIDS patients in Southern California: identification of the local environmental source as arboreal. PloS Pathog. 10, e1004285. doi: 10.1371/journal.ppat.1004285
Tangwattanachuleeporn, M., Somparn, P., Poolpol, K., Gross, U., Weig, M., Bader, O. (2013). Prevalence and Antifungal Susceptibility of Cryptococcus neoformans Isolated from Pigeon Excreta in Chon Buri Province, Eastern Thailand. Med. Mycol. J. 54, 303–307. doi: 10.3314/mmj.54.303
Thomaz, D. Y., Grenfell, R. C., Vidal, M. S., Giudice, M. C., Del Negro, G. M., Juliano, L., et al. (2016). Does the Capsule Interfere with Performance of Matrix-Assisted Laser Desorption Ionization-Time of Flight Mass Spectrometry for Identification of Cryptococcus neoformans and Cryptococcus gattii? J. Clin. Microbiol. 54, 474–477. doi: 10.1128/JCM.02635-15
Trilles, L., Meyer, W., Wanke, B., Guarro, J., Lazera, M. (2012). Correlation of antifungal susceptibility and molecular type within the Cryptococcus neoformans/C. gattii species complex. Med. Mycol. 50, 328–332. doi: 10.3109/13693786.2011.602126
Voelz, K., Johnston, S. A., Smith, L. M., Hall, R. A., Idnurm, A., May, R. C. (2014). ‘Division of labour’ in response to host oxidative burst drives a fatal Cryptococcus gattii outbreak. Nat. Commun. 5, 5194. doi: 10.1038/ncomms6194
Keywords: MALDI-TOF MS, identification, capsule, Cryptococcus neoformans complex, Cryptococcus gattii complex
Citation: Bernhard M, Worasilchai N, Kangogo M, Bii C, Trzaska WJ, Weig M, Groß U, Chindamporn A and Bader O (2021) CryptoType – Public Datasets for MALDI-TOF-MS Based Differentiation of Cryptococcus neoformans/gattii Complexes. Front. Cell. Infect. Microbiol. 11:634382. doi: 10.3389/fcimb.2021.634382
Received: 27 November 2020; Accepted: 30 March 2021;
Published: 19 April 2021.
Edited by:
Di Xiao, National Institute for Communicable Disease Control and Prevention (China CDC), ChinaReviewed by:
Bryan Schmitt, Indiana University Bloomington, United StatesCopyright © 2021 Bernhard, Worasilchai, Kangogo, Bii, Trzaska, Weig, Groß, Chindamporn and Bader. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Oliver Bader, b2xpdmVyLmJhZGVyQG1lZC51bmktZ29ldHRpbmdlbi5kZQ==
†Present address: Navaporn Worasilchai, Department of Transfusion Medicine and Clinical Microbiology, Faculty of Allied Health Sciences, Chulalongkorn University, Bangkok, Thailand
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.