
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
BRIEF RESEARCH REPORT article
Front. Cell. Infect. Microbiol. , 10 June 2021
Sec. Clinical Microbiology
Volume 11 - 2021 | https://doi.org/10.3389/fcimb.2021.630552
Background: Multiple RT-qPCR kits are available in the market for SARS-CoV-2 diagnosis, some of them with Emergency Use Authorization (EUA) by FDA or their country of origin agency, but many of them lack of proper clinical evaluation.
Objective: We evaluated the clinical performance of two Korean SARS-CoV-2 RT-PCR kits available in South America, AccuPower SARS-CoV-2 Multiplex RT-PCR kit (Bioneer, South Korea) and Allplex 2019-nCoV Assay (Seegene, South Korea), for RT-qPCR SARS-CoV-2 diagnosis using the CDC protocol as a gold standard.
Results: We found strong differences among both kits clinical performance and analytical sensitivity; while the Allplex 2019-nCoV Assay has sensitivity of 96.5% and an estimated limit of detection of 4,000 copies/ml, the AccuPower SARS-CoV-2 Multiplex RT-PCR kit has a sensitivity of 75.5% and limit of detection estimated to be bigger than 20,000 copies/ml.
Conclusions: AccuPower SARS-CoV-2 Multiplex RT-PCR kit and Allplex 2019-nCoV Assay are both made in South Korea but EUA by Korean CDC was only granted to the later. Our results support that Korean CDC EUA should be considered as a quality control proxy for Korean SARS-CoV-2 RT-PCR kits prior to importation by developing countries to guarantee high sensitivity diagnosis.
The COVID-19 pandemic has affected public health systems worldwide, challenging patient care quality and surveillance and control success. Moreover, to guarantee the quality of SARS-CoV-2–related diagnosis tools was compromised under an emergency use authorization (EUA) scenario. Multiple in vitro RT-qPCR diagnosis kits are available on the market for the detection of SARS-CoV-2, with EUA from the U.S. Food & Drug Administration (FDA) or other international agencies like Chinese CDC or Korean CDC, or without EUA and limited validations studies made by manufacturers. The CDC-designed FDA EUA 2019-nCoV CDC kit (IDT, USA) is based on N1 and N2 gene targets to detect SARS-CoV-2 and RNase P as an RNA extraction quality control, it is considered a gold standard worldwide for SARS-CoV-2 RT-PCR diagnosis (CDC, 2020; Nalla et al., 2020; Rhoads et al., 2020; Xiaoyan et al., 2020). Our lab has recently published a highly sensitive adaptation of the CDC protocol on a triplex assay format that is time and cost effective (Freire-Paspuel and Garcia-Bereguiain, 2021a).
Among the commercial kits available in South America for SARS-CoV-2 diagnosis, several are made in South Korea. For instance, Allplex 2019-nCoV Assay (Seegene, South Korea) is a multiplex SARS-CoV-2 RT-qPCR kit for E, N and RdRp genes that holds both FDA EUA and Korean CDC EUA. On the other hand, AccuPower SARS-CoV-2 Multiplex RT-PCR kit (Bioneer, South Korea) is also a multiplex SARS-CoV-2 RT-qPCR assay for E, N, and RdRp genes, which are not included neither in the FDA EUA list nor in the Korean CDC EUA list (FDA, 2020; Hong et al., 2020). Moreover, we have previously reported that other South Korean SARS-CoV-2 RT-PCR and RT-LAMP kits lacking Korean CDC EUA did not accomplish the clinical performance and analytical sensitivity described at the manufacturer’s manual (Freire-Paspuel et al., 2020a; Freire-Paspuel and Garcia-Bereguiain, 2020; Freire-Paspuel and Garcia-Bereguiain, 2021b).
We herein present a comparison of the analytical and clinical performance of AccuPower SARS-CoV-2 Multiplex RT-PCR kit (Bioneer, South Korea) and Allplex 2019-nCoV Assay (Seegene, South Korea) for SARS-CoV-2 RT-qPCR diagnosis from nasopharyngeal samples.
89 clinical specimens (nasopharyngeal swabs collected on 0.5 ml TE pH 8 buffer) were included in this study, coming from individuals selected for SARS-CoV-2 surveillance at the laboratory of “Universidad de Las Américas” in Quito (Ecuador). Also, negative controls (TE pH 8 buffer) were included as control for carryover contamination, one for each set of RNA extractions.
All the samples were processed with the same RNA extraction kit “AccuPrep Viral RNA extraction kit” (Bioneer, South Korea), and the same RNA extraction was used for the three RT-PCR protocols. A maximum of a cycle of −80°C frozen/thawed was applied for some samples that could not be processed within the same day that the CDC protocol with the other commercial kits. We have also tested in our lab that RNase P values do not vary after cycles of frozen/thawed as no statistically significant differences were found for RNase P Ct values when an “inconclusive” (amplification of only N1 or N2 gene targets) sample has to be repeated. So, we can certainly assure that no RNA degradation happens for the experimental conditions described in our study.
All the samples included in the study were tested following an adapted version of the CDC protocol: (a) using Accupower Viral RNA extraction kit (Bioneer, South Korea) as an alternate RNA extraction method; (b) using CFX96 BioRad instrument (Freire-Paspuel et al., 2020a; Freire-Paspuel and Garcia-Bereguiain, 2020; Freire-Paspuel et al., 2020; Freire-Paspuel et al., 2020b; Freire-Paspuel et al., 2020; Freire-Paspuel and Garcia-Bereguiain, 2021b; Freire-Paspuel et al., 2021; Ortiz-Prado et al., 2021); (c) using a N1/N2/RNaseP multiplex assay (Freire-Paspuel and Garcia-Bereguiain, 2021a). The criteria for positivity was a Ct ≤ 40 for N1 and N2 targets simultaneously (CDC, 2020; Xiaoyan et al., 2020).
Same RNA extractions from all the samples included on the study were tested using AccuPower SARS-CoV-2 Multiplex RT-PCR kit, following the manufacturer’s manual (Ref: SCVM-2112 Lot: 200231G). Accordingly, a sample is considered positive if both E and RrRp-N targets amplify, or just only RdRp-N, with Ct ≤ 35 and Ct ≤ 34, respectively. If only E gene target amplify, the sample is considered “presumptive positive”. Although a master mix including internal control was prepared for each batch of tested samples and the internal control amplified in most of the samples, we only considered a CDC protocol SARS-CoV-2 positive sample as negative sample for AccuPower kit if the internal control amplified. So, for sensitivity calculation we excluded “invalid samples” (See Supplementary Table 1): SARS-CoV-2 positive samples according to the CDC protocol where neither E/RdRp-N targets viral targets nor the internal control amplified. So far, internal control amplification is not related to the quality of the sample as the RNaseP target on the CDC protocol, but to make our evaluation as fair as possible we excluded those samples defined as invalid.
Same RNA extractions from all the samples included on the study were tested using Allplex 2019-nCoV Assay, following the manufacturer’s manual (Ref: RP10243X Lot: RP4520GB6). Accordingly, a sample is considered positive if at least N or RdRp targets amplify with Ct ≤ 40. If only E gene target amplify with Ct ≤ 40, the sample is considered “presumptive positive”. Also, if the internal positive control has a Ct > 40 in the absence of E, RdRp or N amplification in a SARS-CoV-2 positive sample for to the CDC protocol, the sample is considered invalid.
Limit of detection (LoD) was performed using the commercially available 2019-nCoV N-positive control (IDT, USA); provided at 200,000 genome equivalents/ml, it was used for calibration curves to obtain the viral loads of the samples. Viral loads can be expressed as copies/µl of RNA extraction or copies/ml of sample; the conversion factor is 200, as 0.2 ml of sample is used for RNA extraction and 40 µl is used as final elution volume of RNA extraction.
89 samples were tested for SARS-CoV-2 following both Allplex 2019-nCoV Assay and CDC RT-PCR protocol, as described in Materials and Methods. For the CDC protocol, 57 samples tested positive and 32 samples tested negative (Table 1 and Supplementary Table 1). 30 out of 32 samples tested negative for the CDC protocol were also SARS-CoV-2 negative for Allplex 2019-nCoV Assay, so the specificity obtained in our study was 93.75%. The two “false positive” samples had Ct values > 36 (Samples 58 and 59 at Supplementary Table 1).
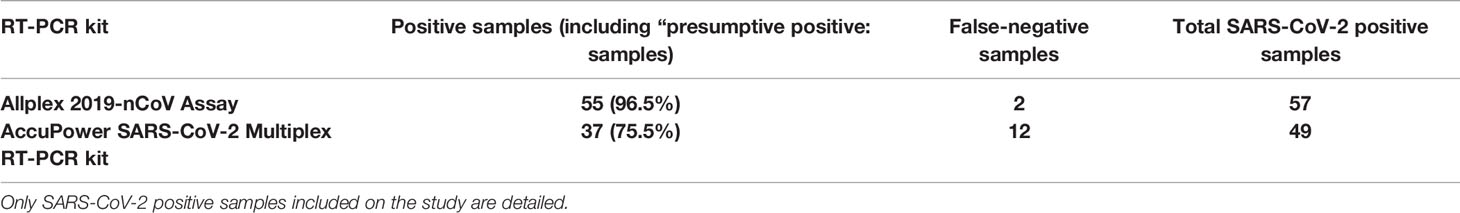
Table 1 Clinical performance of AccuPower SARS-CoV-2 Multiplex RT-PCR kit and Allplex 2019-nCoV Assay using the CDC protocol as a gold standard (% values: sensitivity).
For the 57 SARS-CoV-2–positive samples for the CDC RT-PCR protocol, 55 samples tested also positive for Allplex 2019-nCoV Assay, resulting a sensitivity of 96.5% (Table 1 and Supplementary Table 1). The viral loads for the two SARS-CoV-2 positive samples that tested negative for Allplex 2019-nCoV Assay were as low as 12.8 and 5.15 copies/µl (< 3000 copies/ml).
As the limit of detection (LoD) is defined as the lowest viral load in which all replicates are detected (100% sensitivity), our data indicate that the LoD for Allplex 2019-nCoV assay should be around 20 copies/µl of RNA extraction (4,000 viral RNA copies/ml of sample), as no failure to detect SARS-CoV-2–positive samples above this threshold was found (Supplementary Table 1).
89 samples were tested for SARS-CoV-2 following both using AccuPower SARS-CoV-2 Multiplex RT-PCR kit and the CDC RT-PCR protocol, as described on the methods. For the CDC protocol, 57 samples tested positive and 32 samples tested negative (Table 1 and Supplementary Table 1). None of the negative samples for the CDC RT-PCR protocol were positive for AccuPower SARS-CoV-2 Multiplex RT-PCR kit, so the specificity obtained in our study was 100%.
From the 57 samples that tested positive for the CDC RT-PCR protocol, eight of them were invalid for the AccuPower SARS-CoV-2 Multiplex RT-PCR kit, as neither the viral gene targets nor the internal positive control amplified (see Supplementary Table 1). For the remaining 49 SARS-CoV-2 positive samples, 35 samples were true positive and two were “presumptive positive” (only amplification of E gene target) for AccuPower SARS-CoV-2 Multiplex RT-PCR kit. Considering only true-positive samples, the sensitivity was 71.4% (35/49), but up to 75.5% (37/49) if we also included “presumptive positive” samples for AccuPower SARS-CoV-2 Multiplex RT-PCR kit (Table 1 and Supplementary Table 1).
As the limit of detection (LoD) is defined as the lowest viral load in which all replicates are detected (100% sensitivity), our data indicate that the LoD for AccuPower SARS-CoV-2 Multiplex RT-PCR kit should be bigger than 90 copies/µl of RNA extraction (18,000 viral RNA copies/ml of sample), as even two SARS-CoV-2 positives samples with viral loads of 203 and 90.4 copies/µl failed to be positive for AccuPower SARS-CoV-2 Multiplex RT-PCR kit. The sensitivity obtained for that viral load threshold was 94.3% (33/35) (see Supplementary Table 1).
Although the main limitations of our study is the sample size (89 specimens) and that a single lot of each of the kits was used for the evaluation, our results support that Allplex 2019-nCoV Assay had a great performance in terms on sensitivity compared to the CDC RT-PCR protocol, with values up to 96.5%. However, we found a relatively low specificity of 93.75%; as it has been previously reported that no cross reactivity with other respiratory viruses happened for Allplex 2019-nCoV Assay (Farfour et al., 2020), further studies are needed to set a most accurate specificity value as we only included 32 negative samples in ours. As we have described on the results, we could estimate de LoD of Allplex 2019-nCoV Assay on approximately 4,000 viral RNA copies/ml of sample which is similar to the LoD of 4,167 copies/ml detailed at manufacturer’s manual. This LoD is acceptable for a reliable SARS-CoV-2 diagnosis considering the viral load frequency population distributions (Kleiboeker et al., 2020; Lavezzo et al., 2020).
Although AccuPower SARS-CoV-2 Multiplex RT-PCR kit has a great specificity (100%), our data showed that this kit had a low clinical performance in terms on sensitivity compared to the CDC RT-PCR protocol, with values of 75.5% and 71.4% depending if we consider “presumptive positive” (E gene amplification only) samples as true SARS-CoV-2 positive or not. Moreover, as we have described on the results, we could estimate de LoD of AccuPower SARS-CoV-2 Multiplex RT-PCR kit to be above on 18,000 viral RNA copies/ml of sample, as two SARS-CoV-2 positive samples above this threshold failed to test positive, resulting a sensitivity of 94.3% for that viral load threshold, below the 100% expected for the LoD. AccuPower SARS-CoV-2 Multiplex RT-PCR kit manufacturer’s manual reported a LoD of six copies/µl, equivalent to 1,200 copies/ml on our experimental conditions, but according to our data the LoD would be higher.
In Table 2, analytical parameters and other characteristics for Allplex 2019-nCoV Assay and AccuPower SARS-CoV-2 Multiplex RT-PCR kits are summarized. As we have detailed on the introduction, only Allplex 2019-nCoV Assay has both FDA EUA and Korean CDC EUA. Considering the better analytical sensitivity for Allplex 2019-nCoV Assay compared to AccuPower SARS-CoV-2 Multiplex RT-PCR kit, our study suggests that a good public policy for developing countries like Ecuador would be to use Korean CDC EUA as a quality control proxy to allow the importation of South Korean SARS-CoV-2 RT-PCR kits. Actually, we have already published other clinical evaluations for South Korean RT-PCR and RT-LAMP kits that endorse this same idea, as neither nCoV-QS (MiCo BioMed, South Korea) and AccuPower SARS-CoV-2 Real Time RT-PCR kit (Bioneer, South Korea) RT-PCR kits nor Isopollo COVID-19 detection kit (M Monitor, South Korea) RT-LAMP kit have Korean CDC EUA; and the three of them had a low clinical performance and a LoD much higher of what is indicated by the manufacturers. So far, we expressed our concern toward local regulatory agencies that allow these companies to export SARS-CoV-2 diagnosis kits to Ecuador and other South American countries despite their products lack of clinical use authorization in South Korea
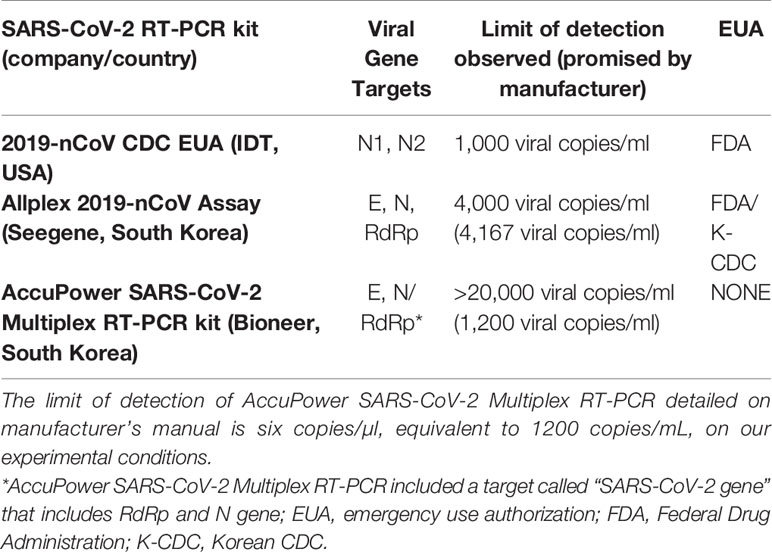
Table 2 Comparison of CDC-RT-PCR protocol, AccuPower SARS-CoV-2 Multiplex RT-PCR and Allplex 2019-nCoV Assay kits.
Considering the worldwide high demand of reagents for SARS-CoV RT-qPCR diagnosis, supplies shortage is a fact, EUA are necessary and regulations need to be more flexible. However, under this scenario, the role of Academia to carry out independent clinical performance and analytical sensitivity evaluations is crucial to guarantee the quality of the supplies in the market for every country in the world, particularly for developing countries usually lacking of reliable regulatory agencies. It is a matter of global justice and human rights.
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding author.
All samples have been submitted for routine patient care and diagnostics. Ethics approval was not sought because the study involves laboratory validation of test methods and the secondary use of anonymous pathological specimens that falls under the category ‘exempted’ by Comité de Etica para Investigación en Seres Humanos from “Universidad de Las Américas”.
BF-P and MG-B performed the experiments, analyzed the data, and wrote the manuscript. All authors contributed to the article and approved the submitted version.
This study was funded by Universidad de Las Americas (Quito, Ecuador).
The authors declare that their affiliation institution “Universidad de Las Américas” is involved in the commercialization of “ECUGEN SARS-CoV-2 RT-qPCR kit”. However, that kit was not used in the present study.
We thank Dr Tannya Lozada from “Dirección General de Investigación de la Universidad de Las Américas”, and also the authorities from Universidad de Las Américas, for logistic support to make SARS-CoV-2 diagnosis possible at UDLA laboratories.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcimb.2021.630552/full#supplementary-material
CDC. Interim Guidelines for Collecting, Handling, and Testing Clinical Specimens From Persons for Coronavirus Disease 2019 (Covid-19) (2020). Available at: https://www.cdc.gov/coronavirus/2019-ncov/lab/guidelines-clinical-specimens.html (Accessed last access 09/05/20).
FDA. (2020). Available at: https://www.fda.gov/medical-devices/emergency-situations-medical-devices/emergency-use-authorizations.
Farfour, E., Lesprit, P., Visseaux, B., Pascreau, T., Jolly, E., Houhou, N. (2020). The Allplex 2019-nCoV (Seegene) Assay: Which Performances are for SARS-CoV-2 Infection Diagnosis? Eur. J. Clin. Microbiol. Infect. Dis. 39, 1997–2000. doi: 10.1007/s10096-020-03930-8
Freire-Paspuel, B., Garcia-Bereguiain, M. A. (2020). Poor Sensitivity of AccuPower SARS-Cov-2 Real Time RT-PCR Kit (Bioneer, South Korea). Virol. J. 17 (1), 178. doi: 10.1186/s12985-020-01445-4
Freire-Paspuel, B., Garcia-Bereguiain, M. A. (2021). Analytical Sensitivity and Clinical Performance of a Triplex RT-qPCR Assay Using CDC N1, N2 and RP Targets for SARS-CoV-2 Diagnosis. Int. J. Infect. Dis. S1201-9712 (20), 32250–32255. doi: 10.1016/j.ijid.2020.10.047
Freire-Paspuel, B., Garcia-Bereguiain, M. A. (2021). Low Clinical Performance of “Isopollo COVID19 Detection Kit” (Monitor, South Korea) for RT-LAMP Sars-CoV-2 Diagnosis: A Call for Action Against Low Quality Products for Developing Countries”. Int. J. Infect. Dis. 104, 303–305. doi: 10.1016/j.ijid.2020.12.088
Freire-Paspuel, B., Vega-Mariño, P., Velez, A., Castillo, P., Cruz, M., Garcia-Bereguiain, M. (2020). Evaluation of nCoV-QS (Mico BioMed) for RT-qPCR Detection of SARS-CoV-2 From Nasopharyngeal Samples Using Cdc FDA EUA Qpcr Kit as a Gold Standard: An Example of the Need of Validation Studies. J. Clin. Virol. 128:104454. doi: 10.1016/j.jcv.2020.104454
Freire-Paspuel, B., Vega-Mariño, P., Velez, A., Castillo, P., Cruz, M., Garcia-Bereguiain, M. A. (2020). Sample Pooling of RNA Extracts to Speed Up SARS-CoV-2 Diagnosis Using CDC Fda EUA Rt-qPCR Kit. Virus Res. 290, 198173. doi: 10.1016/j.virusres.2020.198173
Freire-Paspuel, B., Vega-Mariño, P., Velez, A., Castillo, P., Gomez-Santos, E. E., Cruz, M., et al. (2020). Cotton-Tipped Plastic Swabs for SARS-CoV-2 RT-Qpcr Diagnosis to Prevent Supply Shortages. Front. Cell Infect. Microbiol. Jun 2310, 356. doi: 10.3389/fcimb.2020.00356
Freire-Paspuel, B., Vega-Mariño, P., Velez, A., Castillo, P., Masaquiza, C., Cedeño-Vega, R., et al. (2020). “One Health” Inspired SARS-CoV-2 Surveillance: The Galapagos Islands Experience. One Health 11, 100185. doi: 10.1016/j.onehlt.2020.100185
Freire-Paspuel, B., Vega-Mariño, P., Velez, A., Cruz, M., Perez, F., Garcia-Bereguiain, M. A. (2021). Analytical and Clinical Comparison of Viasure (Certest Biotec) and 2019-nCOV CDC (Idt) RT-qPCR Kits for SARS-CoV2 Diagnosis. Virology. 553, 154–156. doi: 10.1016/j.virol.2020.10.010
Hong, K. H., Lee, S. W., Kim, T. S., Huh, H. J., Lee, J., Kim, S. Y., et al. (2020). On Behalf of Korean Society for Laboratory Medicine, COVID-19 Task Force and the Center for Laboratory Control of Infectious Diseases, the Korea Centers for Disease Control and Prevention. Guidelines for Laboratory Diagnosis of Coronavirus Disease 2019 (Covid-19) in Korea. Ann. Lab. Med. 40 (5), 351–360. doi: 10.3343/alm.2020.40.5.351
Kleiboeker, S., Cowden, S., Grantham, J., Nutt, J., Tyler, A., Berg, A., et al. (2020). SARS-Cov-2 Viral Load Assessment in Respiratory Samples. J. Clin. Virol. 19, 104439. doi: 10.1016/j.jcv.2020.104439
Lavezzo, E., Franchin, E., Ciavarella, C., Cuomo-Dannenburg, G., Barzon, L., Del Vecchio, C., et al. (2020). Suppression of a SARS-CoV-2 Outbreak in the Italian Municipality of Vo’. Nature 2020 Volume 584 (7821), 425–429. doi: 10.1038/s41586-020-2488-1
Nalla, A. K., Castob, A. M., Huang, M.-L. W., Perchetti, G. A., Sampoleo, R., Shrestha, L., et al. (2020). Comparative Performance of SARS-CoV-2 Detection Assays Using Seven Different Primer/Probe Sets and One Assay Kit. J. Clin. Microbiol. 58 (6), e00557–e00520. doi: 10.1128/JCM.00557-20
Ortiz-Prado, E., Henriquez-Trujillo, A. R., Rivera-Olivero, I. A., Lozada, T., Garcia-Bereguiain, M. A., the UDLA-COVID-19 team (2021). High Prevalence of SARS-CoV-2 Infection Among Food Delivery Riders. A Case Study From Quito, Ecuador. Sci. Total Environ. 770, 145225. doi: 10.1016/j.scitotenv.2021.145225
Rhoads, D. D., Cherian, S. S., Roman, K., Stempak, L. M., Schmotzer, C. L., Sadri, N. (2020). Comparison of Abbott Id Now, Diasorin Simplexa, and CDC Fda EUA Methods for the Detection of SARS-CoV-2 From Nasopharyngeal and Nasal Swabs From Individuals Diagnosed With COVID-19. J. Clin. Microbiol. 58 (8), e00760–e00720. doi: 10.1128/JCM.00760-20
Keywords: SARS-CoV-2, RT-PCR, Bioneer, Seegene, developing countries
Citation: Freire-Paspuel B and Garcia-Bereguiain MA (2021) Analytical and Clinical Evaluation of “AccuPower SARS-CoV-2 Multiplex RT-PCR kit (Bioneer, South Korea)” and “Allplex 2019-nCoV Assay (Seegene, South Korea)” for SARS-CoV-2 RT-PCR Diagnosis: Korean CDC EUA as a Quality Control Proxy for Developing Countries. Front. Cell. Infect. Microbiol. 11:630552. doi: 10.3389/fcimb.2021.630552
Received: 17 November 2020; Accepted: 10 February 2021;
Published: 10 June 2021.
Edited by:
Jorge Eugenio Vidal, University of Mississippi Medical Center, United StatesReviewed by:
Srinivasan Velusamy, Centers for Disease Control and Prevention (CDC), United StatesCopyright © 2021 Freire-Paspuel and Garcia-Bereguiain. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Miguel Angel Garcia-Bereguiain, bWFnYmVyZWd1aWFpbkBnbWFpbC5jb20=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.