- 1Changsha Hospital for Maternal and Child Health Care of Hunan Normal University, Changsha, China
- 2Institute of Food Safety and Nutrition, Jinan University, Guangzhou, China
- 3Institute of Biomedicine and Department of Cell Biology, Jinan University, Guangzhou, China
Clustered regularly interspaced short palindromic repeats (CRISPR) systems are a set of versatile gene-editing toolkit that perform diverse revolutionary functions in various fields of application such as agricultural practices, food industry, biotechnology, biomedicine, and clinical research. Specially, as a novel antiviral method of choice, CRISPR/Cas9 system has been extensively and effectively exploited to fight against human infectious viruses. Infectious diseases including human immunodeficiency virus (HIV), hepatitis B virus (HBV), human papillomavirus (HPV), and other viruses are still global threats with persistent potential to probably cause pandemics. To facilitate virus removals, the CRISPR/Cas9 system has already been customized to confer new antiviral capabilities into host animals either by modifying host genome or by directly targeting viral inherent factors in the form of DNA. Although several limitations and difficulties still need to be conquered, this technology holds great promises in the treatment of human viral infectious diseases. In this review, we will first present a brief biological feature of CRISPR/Cas9 systems, which includes a description of CRISPR/Cas9 structure and composition; thereafter, we will focus on the investigations and applications that employ CRISPR/Cas9 system to combat several human infectious viruses and discuss challenges and future perspectives of using this new platform in the preclinical and clinical settings as an antiviral strategy.
Introduction
Viruses are the deep cause for numerous acute and chronic diseases, some of which lead to severe situations, like the recent coronavirus disease 2019 (COVID-19) pandemic. However, some of them just produce minor diseases, like herpes simplex viruses. Currently, serious viral infectious illnesses such as human immunodeficiency virus (HIV), hepatitis B virus (HBV), and human papillomavirus (HPV) are potentially threating the human health and global stability (Morens and Fauci, 2013). They undoubtedly increase the socioeconomic burden on the public health systems throughout the world (Doerflinger et al., 2017). Compared to other health-relevant infectious viruses including herpes simplex virus, the three abovementioned viruses are more dangerous to human; once infected, it is difficult to cure, as the success rates with medical therapy are relatively lower. The therapy for fighting against viral infections is a challenging project, due to the overconsumption of cellular resources by viruses and the formation of latent viral reservoirs in the hosts (White et al., 2015). Moreover, many human viruses are capable of generating mutant strains to escape and even jump between different species resulting in pandemics (Parrish et al., 2008). Therefore, a series of antiviral strategies, such as synthesis drugs (Villa et al., 2017), herbal medicines (Castilla et al., 2010), animal-based medicines (Costa-Neto, 2005), antibody-based drugs (Kuprash et al., 2017), and genetically engineered drugs (Zündorf and Dingermann, 2000), have entered the preclinical and clinical fields one after another. Among the strategies, clustered regularly interspaced short palindromic repeats (CRISPR)/Cas genome editing technique, as a landmark discovery, has entered the field of biomedical research and gene therapy research, which holds great promise for tackling serious human infectious viruses.
Since 2013, the success of genome modifications via CRISPR/Cas9 apparatus in cultured human cells (Cho et al., 2013; Jinek et al., 2013; Mali et al., 2013) has opened up a new route for gene therapy in biomedical research. Gene editing is a combinational process of introducing site-specific DNA cleavages by nucleases and wielding the natural cellular pathways to repair the DNA breaks. Exogenous DNA double-strand breaks can be created in the genomes by means of various genome engineering platforms such as meganuclease (Maeder and Gersbach, 2016), zinc-finger nucleases (ZFNs), transcription activator-like effector nucleases (TALENs), and CRISPR/Cas nuclease systems (Ran et al., 2013; Hryhorowicz et al., 2017). Then, cell DNA repairs initiated by DNA lesions are completed through the homology-directed repair (HDR) pathway with repair templates or the nonhomologous end joining (NHEJ) pathway without repair templates (Mao et al., 2008). To date, CRISPR/Cas genome editing system has been developed as a robust instrument for targeted gene modifications in a broad range of animal species, gut microbiota (Citorik et al., 2014; Cresci et al., 2020), and their invading viruses (Ashfaq and Khalid, 2020), which cause changes in relationships between host and viruses. With the expansion of science interests in gene editing research, a new class of medicine based on CRISPR/Cas9 editing technology is entering clinical era for the treatment of viral infections (Hirakawa et al., 2020).
In this review, we will first outline a basic biological feature of CRISPR/Cas9 that focused on its structure and composition; then, we will prominently present the application investigations that employed CRISPR/Cas9 system to combat several human infectious viruses. Lastly, we will also discuss the known and potential limitations of CRISPR/Cas9 gene editing platform including off-target effects, delivery challenges, Cas9 cleavage activity, resistance to CRISPR/Cas9, viral escape problems, and ethical concerns.
Basic Biological Features of CRISPR/Cas9 Machinery
First described by Ishino et al. in 1987, CRISPR is a range of DNA repeat sequences with uncertain origin and unknown function in the Esherichia coli genome (Ishino et al., 1987). The CRISPR/Cas complexes existing in the prokaryotic organisms confer resistance to new incoming genetic elements such as plasmids, phages, and viruses (Jinek et al., 2012). According to the Cas protein contents and its amino acid sequences, the CRISPR/Cas systems have been initially artificially divided into three major types, namely, type I, type II, and type III (Wiedenheft et al., 2012; Chylinski et al., 2014). Until recently, three additional types of CRISPR/Cas system (types IV–VI) have been identified across bacterial genomes (Bayat et al., 2018), of which both type II and type V CRISPR/Cas systems are the similar apparatuses that consist of only single subunit RNA effector (Cas9 and Cas12, respectively) (Khadempar et al., 2019). The type II CRISPR/Cas system that is commonly known as CRISPR/Cas9 is a binary complex of endonuclease Cas9 and two small guide RNAs (gRNAs) [CRISPR RNA (crRNA) and transactivating CRISPR RNA (tracrRNA) (Gebre et al., 2018)] (Saayman et al., 2015), extensively used for RNA-programmable genome manipulating purpose in most cases. It just needs the optimization of Cas9 expression (Wright et al., 2015) and the matched design of gRNA to successfully function in its editing roles in cell genomes (Jiang et al., 2015). In such type of nuclease systems, the gRNA recognized by Cas9 proteins in a sequence independent manner (Nishimasu et al., 2014) directs Cas9 to recognize and cleave target specific DNA sequences with a short protospacer adjacent motif (PAM) of 17–20 nucleotides via the Watson–Crick base-pairing interactions (Garneau et al., 2010). The natural CRISPR/Cas9 systems possess a variety of structural variants and orthologues (Figure 1) (Ran et al., 2013; Lin et al., 2019), of which Streptococcus pyogenes Cas9 (SpCas9) and Staphylococcus aureus (SaCas9) are the two types that are widely used for research purposes. Furthermore, Cas9 nucleases from different bacterial species recognize different PAM sequences for seeking targets (Lino et al., 2018), with SpCas9 using “NGG” PAM as a binding target while SaCas9 employing “NNGRRT” PAM (Xie et al., 2018).
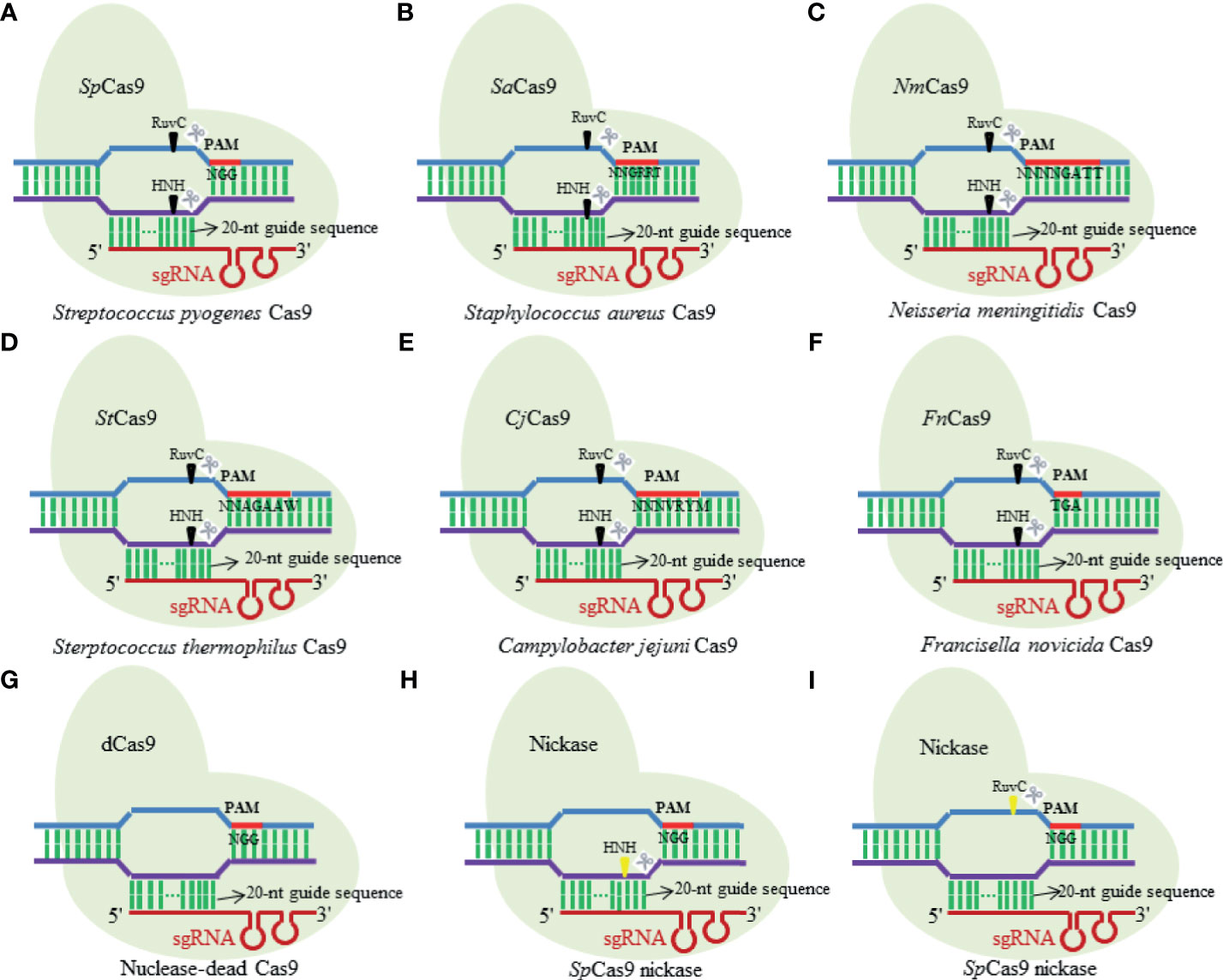
Figure 1 Diagram illustrating several structural variants and orthologues of natural Cas9 nuclease. (A) Streptococcus pyogenes Cas9; (B) Staphylococcus aureus Cas9; (C) Neisseria meningitidis Cas9; (D) Sterptococcus thermophilus Cas9; (E) Campylobacter jejuni Cas9; (F) Francisella novicida Cas9; (G) nuclease-dead Cas9; (H) SpCas9 nickase; (I) SpCas9 nickase.
In the last few years, the simple-designed CRISPR/Cas9 system has greatly expanded its application scopes in life science, and now, it is still in its rapidly developing stage. As an effective, highly specific and robust tool, CRISPR/Cas9 machinery holds great promise for targeting infectious viruses and removing their reservoirs to enhance human health.
Applications of CRISPR/Cas9 to Specifically Target Infectious Viruses
Theoretically, the CRISPR/Cas9 technology can be used not only to target any special nucleotide sequences in human genome but also to edit the double-stranded DNA (dsDNA) of viral invaders in in vivo and in vitro system (Figure 2) (Soppe and Lebbink, 2017). Moreover, the technologies of Cas9 equipped with multiple single guide RNAs (sgRNAs) have enabled the Cas9 endonucleases to target several different genomic loci in a single cell (Ota et al., 2014; Zhou et al., 2014). Besides, the Cas9 variants and orthologs also give the CRISPR/Cas system many more novel functions including targeted gene mutation, transcriptional activation and inhibition, epigenetic modification, imaging of DNA loci, and single base mutation (Cong et al., 2013; Barrangou and Doudna, 2016; Mohanraju et al., 2016; Hu et al., 2018; Miller et al., 2020). Furthermore, viral eradications from cells via CRISPR/Cas9 machinery could be theoretically applicable to any DNA or RNA virus with a DNA intermediate in its life cycle (Doudna and Charpentier, 2014; Khalili et al., 2015; Mohammadzadeh et al., 2020). Therefore, the CRISPR/Cas9 methodology with functional diversities holds huge potential promise for targeting different developmental phases of the viral life cycle and possess the ability to mediate an effective and sustained genetic therapy against human viruses. Herein, CRISPR/Cas9-based antiviral approach to manipulate major human infectious viruses including HIV, HBV, HPV, and other viruses will be discussed.
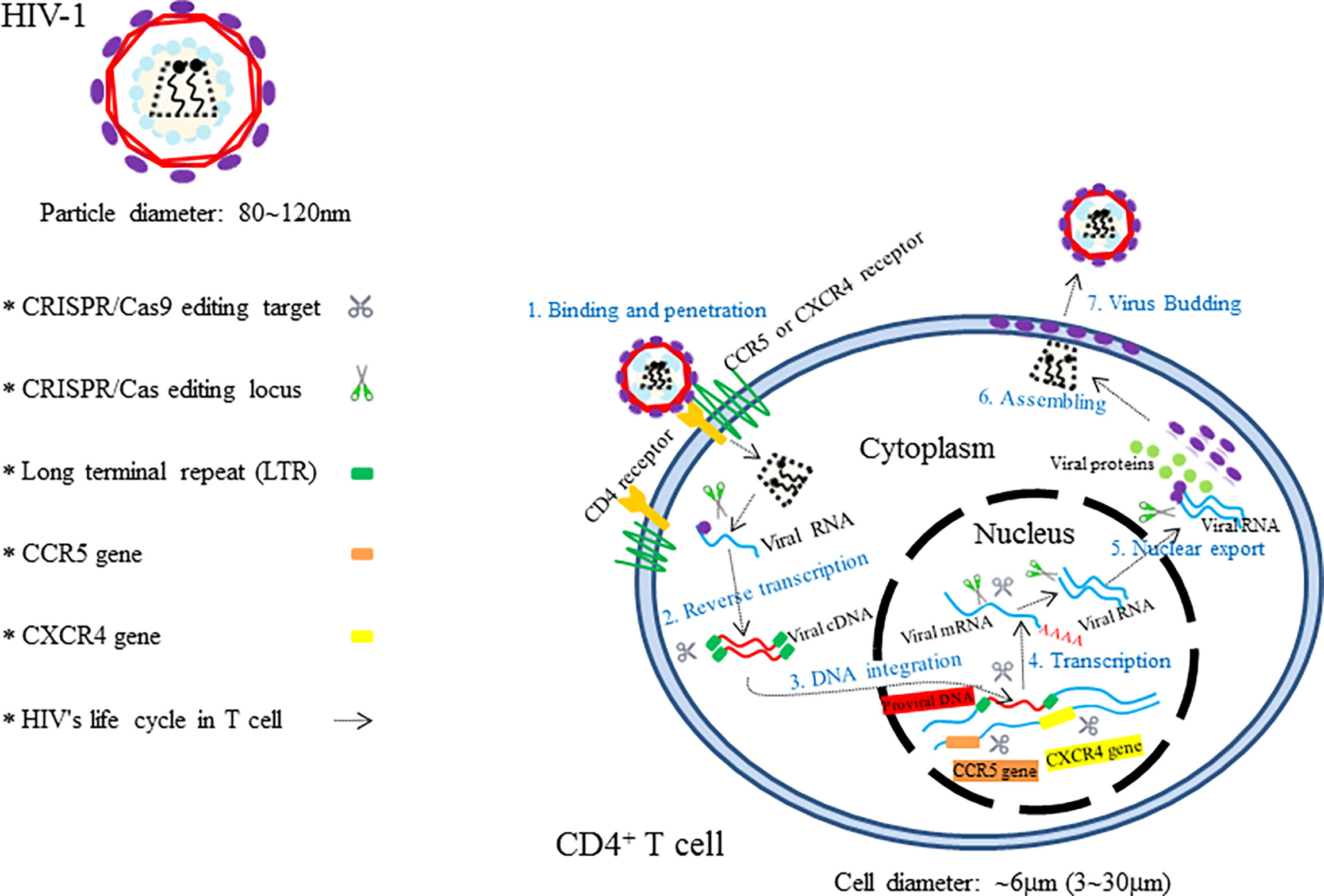
Figure 2 The illustration showing HIV’s invasion paths in cells and the therapeutic targets of CRISPR/Cas9.
Human Immunodeficiency Virus
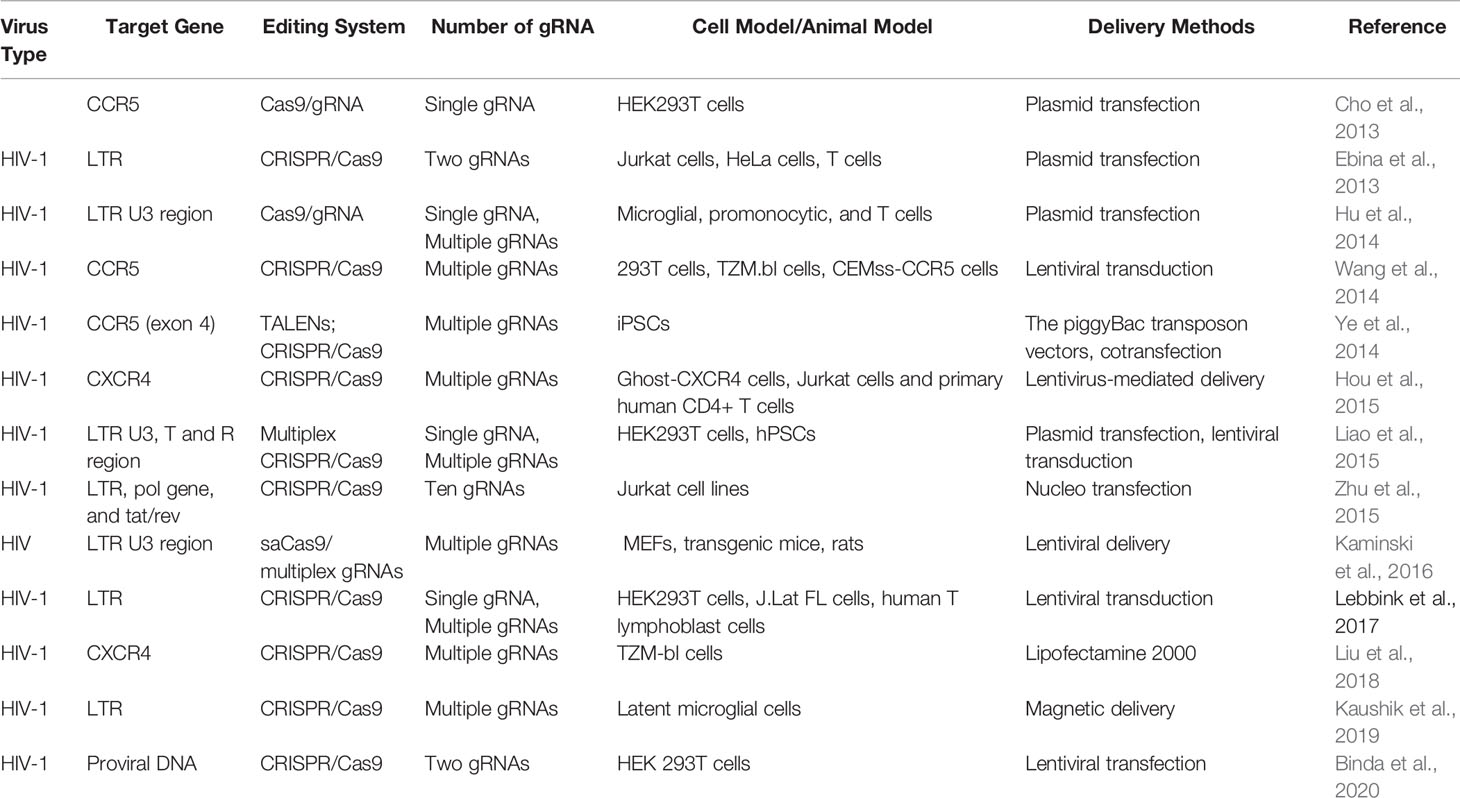
Acquired immunodeficiency syndrome (AIDS) caused by HIV infection is a kind of viral infectious disease. HIV, which mainly consists of HIV-1 and HIV-2, is an important global epidemic that requires advanced clinical remedies. According to the new report of United Nations Programme on HIV/AIDS (UNAIDS), more than 36.7 million people are infected with HIV in the whole world, and the new infection number is over 5,000 per day (Dash et al., 2019). Compared to HIV-2, HIV-1 is characterized by having higher transmissibility and pathogenicity in the human host (Campbell-Yesufu and Gandhi, 2011). Active HIV-1 reproduction in vivo causes severe CD4+ T-cell depletion, which ultimately results in the formation of the so-called chronic disease of AIDS (Alimonti et al., 2003; Doitsh et al., 2014). Great success has been achieved with the use of antiretroviral therapy (ART) and high active antiretroviral therapy (HAART) for the control of the deadly AIDS and even for lifesaving (Deng et al., 2018; Lu et al., 2018). However, these types of HIV therapeutics, designed to suppress various steps of the viral life cycle (Arribas and Eron, 2013), are still unable to cure the disease owing to the existence of permanent integration of HIV-1 into the host genome. In view of these facts, researchers have focused on the treatment of AIDS via CRISPR/Cas9-based gene editing systems, aiming to unlock many new possibilities for HIV-1 prevention and cure (Dampier et al., 2014). Since the first two CRISPR/Cas9-based applications in the prevention of HIV-1 have been reported by Cho and Ebina, respectively, in 2013 (Cho et al., 2013; Ebina et al., 2013), numerous studies that employ CRISPR/Cas9 technology as a method for the treatment of HIV-1/AIDS have been developed rapidly (Khalili et al., 2015). Up to now, targeting host genes and targeting viral genomes are two essential approaches for combating HIV-1 infection (Xiao et al., 2019). The attractive editing targets of CRISPR/Cas9 therapy mainly include C–C chemokine receptor 5 (CCR5) gene, C–C–C chemokine receptor 4 (CXCR4) gene, proviral DNA-encoding viral proteins, and the HIV 5′ and 3′ long terminal repeat (LTR) (Figure 2) (Manjunath et al., 2013; Bialek et al., 2016; Liu et al., 2017). Although Cas9/multiplexed-sgRNA technology has emerged, the use of CRISPR/Cas9 molecular scissor to precisely and jointly target two coreceptor genes, CCR5 and CXCR4, has not yet been seen in relevant reports. Here, Table 1 lists the research studies of HIV-1 infection via CRISPR/Cas9 techniques for editing the aforementioned gene sites (Cho et al., 2013; Ebina et al., 2013; Hu et al., 2014; Hou et al., 2015; Wang et al., 2014; Ye et al., 2014; Liao et al., 2015; Zhu et al., 2015; Kaminski et al., 2016; Soppe and Lebbink, 2017; Liu et al., 2018; Kaushik et al., 2019; Binda et al., 2020).
At present, purging of the latent viral reservoirs is the biggest hurdle for the effective management of HIV infection. As observed in HIV patients who are receiving ART therapy, latent viral reservoirs that mostly attach within resting memory CD4+ T cells, are able to stay for as long as 60 years (Siliciano et al., 2003).
It has been about 30 years since the discovery of HIV, but there is still no effective anti-HIV vaccine available (Haynes, 2015). The “Berlin patient” has been generally recognized as the only one case cured for HIV-1 for a decade (Hutter et al., 2009; Gupta et al., 2019), and now the “London patient” would probably be the second (Gupta et al., 2020). Stem cell transplantation (SCT) is scientifically not a standard treatment method for HIV/AIDS. Viewed from these two case reports, SCT used in the two patients is originally intended for treating cancer rather than HIV-1/AIDS. Fortunately, the accidental cures indeed brings hope for the future use of personalized gene therapy for AIDS.
Hepatitis B Virus
The population figure of chronic HBV carriers in the world (350–400 million people (Seo and Yano, 2014) suggests that hepatitis B is still an important health problem (Trépo et al., 2014). Hepatitis B virus (HBV), of the family Hepadnaviridae (Locarnini et al., 2013), is a hepatotropic DNA virus that replicates by reverse transcription in host hepatocytes at the stage of RNA intermediates and can lead to relatively high frequent occurrences of liver cirrhosis and liver cancer in chronic HBV infectors (Lee, 1997; El-Serag, 2012). Taxonomically, eight genotypes (A–H) of the HBV genome have been identified, in which over 8% of the nucleotides differences are present between any two (Sunbul, 2014). Given that the chances of HBV-infected persons acquiring sustained viral response (SVR) or cure are small, novel and more effective regimens against HBV need to be develop (Nassal, 2015). The rapid growth of the CRISPR/Cas9 technology provides opportunities for new approaches in the prevention and treatment of HBV infectious diseases. As we know, the persistence of covalently closed cyclic DNA (cccDNA) of HBV is the major obstacle hindering the eradication of chronic hepatitis B (CHB) under current antiviral treatments such as nucleoside analogues (NAs) and interferon-alpha (IFN-α) (Emery and Feld, 2017). So far, gene therapies have become the promising potential treatment for HBV infections especially in targeting of cccDNA effectively and hold high promise for entering clinical applications after overcoming some technical barriers (Maepa et al., 2015; Bloom et al., 2018). Suppression of HBV infection in preclinical applications through gene editing platform ZFNs or TALENs have been reported by two research groups independently (Weber et al., 2014; Dreyer et al., 2016). In 2014, the ground-breaking work to use CRISPR/Cas9 system in countering HBV infection in vitro and in vivo was first investigated by Lin et al. (2014). Thereafter, several studies have utilized designed Cas9/sgRNA (or Cas9/multiplex gRNA) combinations to edit only one locus (which is usually in the conserved region of HBV genome) for inhibiting the viral replication and production successfully (Dong et al., 2015; Karimova et al., 2015; Liu et al., 2015; Seeger and Sohn, 2016; Zhu et al., 2016; Li et al., 2017; Scott et al., 2017; Schiwon et al., 2018; Kostyusheva. et al., 2019b). In order to enhance the silence effects for targeted genes, multiple research teams have worked on the applications of CRISPR/Cas9 for the simultaneous targeting and cleavage of several functional loci [e.g., surface antigen region, X gene, reverse transcriptase (RT) gene, and episomal cccDNA] in HBV genomes via cell cultures or mouse models (Figure 3) (Seeger and Sohn, 2014; Kennedy et al., 2015; Ramanan et al., 2015; Wang et al., 2015; Zhen et al., 2015; Sakuma et al., 2016). In addition to the CRISPR/Cas9 system itself, several other studies associated with the combination of CRISPR/Cas9 and other methods (e.g., different molecules or inhibitory systems) have also been developed for the purpose of eradicating HBV genomes (Wang et al., 2017; Zheng et al., 2017; Kostyusheva et al., 2019a).
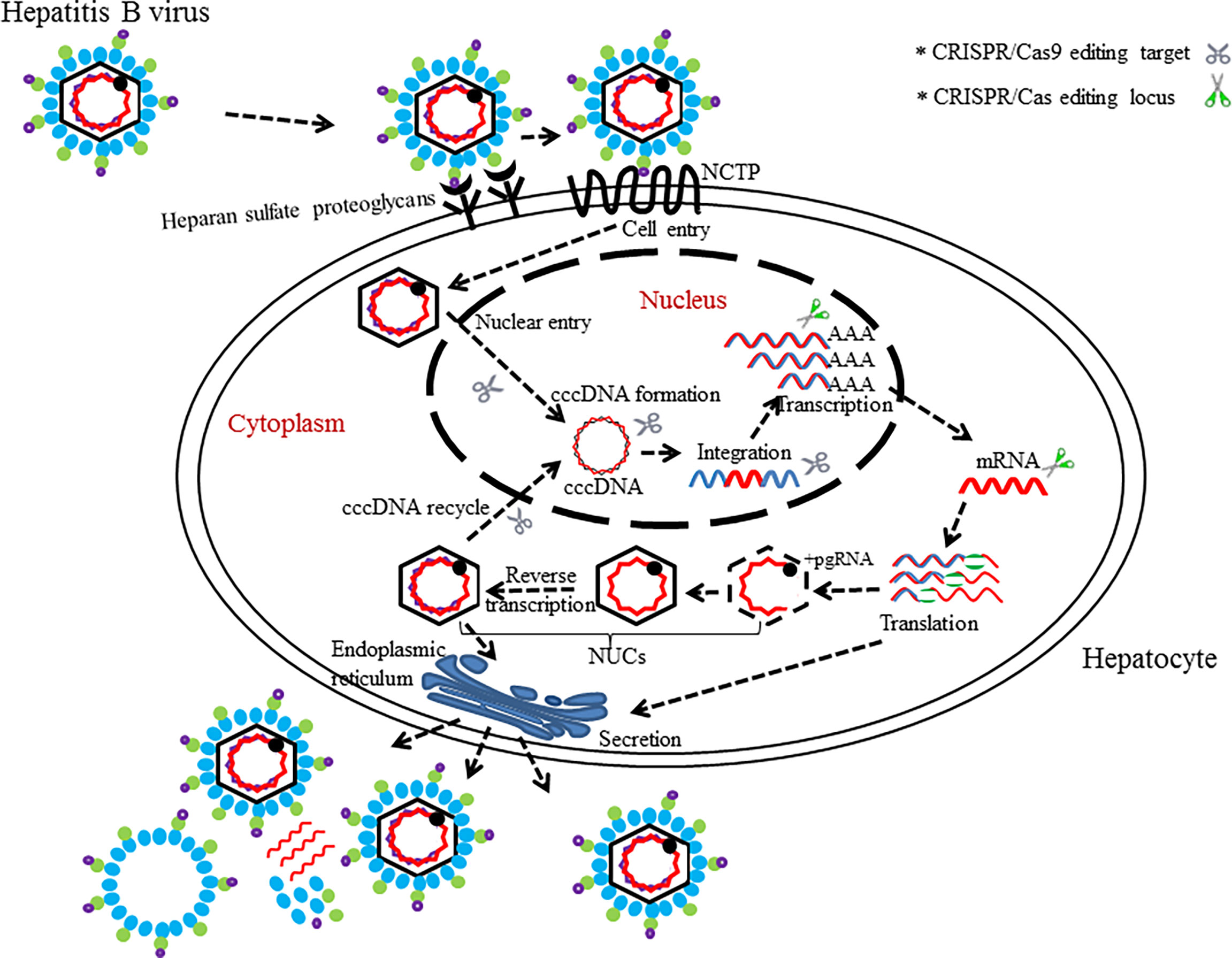
Figure 3 The life cycle of HBV with editing targets. HBV binds to surface receptors and enters the hepatocytes. Virus particle complete its process of growth and proliferation in host’s hepatocytes. CRISPR/Cas (or CRISPR/Cas9)-mediated disruption of the HBV life cycle can target several loci, which is necessary for the HBV life cycle.
Interestingly, a Cas9 variant called dead Cas9 (dCas9) has also been demonstrated to inhibit replication of HBV without dissection of HBV genome (Kurihara et al., 2017). It is noteworthy that one study was conducted recently to investigate a potent inhibitor of NHEJ named “NU7026,” which prevented the degradation of cccDNA mediated cleavages by CRISPR/Cas9 (Kostyushev et al., 2019). This study provides a verification methodology for the activity of CRISPR/Cas9 in destroying HBV genome.
Similarly to ZFNs and TALENs, there also exists a concern of viral escape mutants when there is therapeutic application of CRISPR/Cas9 systems in HBV-infected cells (Schinazi et al., 2018); despite of all these, nucleic acid editing tools could generate desired mutations on the target DNA (Pattanayak et al., 2013).
In summary, these artificial models (cell models or animal models) are only the simulations of persistent HBV infection in human hepatocytes and do not completely represent the actual HBV infection in vivo. These related studies do highlight the potentials of cccDNA disruption by endonuclease Cas9 protein in in vitro cells and in vivo mouse models. Nevertheless, additional studies are needed to ameliorate the CRISPR/Cas9 system so as to further destroy viral reproduction in vivo and to eradicate multiple HBV cccDNA copies residing in infected hepatocytes (Figure 3).
Human Papilloma Virus
HPVs are small double-stranded DNA viruses belonging to the Papovaviridae family, with approximately 150 identified types already described (Nguyen et al., 2014; McBride, 2017). The HPV genome is roughly 8 kbp in length, encodes 9 or 10 open reading frames (ORFs) and includes eight early viral regulatory proteins (E1−E8) and two late capsid proteins (L1 and L2) (Ebrahimi et al., 2019). Since HPVs present epithelia tissue tropism (Harden and Munger, 2017), sexual transmission (Ryndock and Meyers, 2014), and oncogenic property (Moens, 2018), their important status between human diseases and public health must be emphasized. Continued high-risk type HPV (e.g., HPV-16 and HPV-18) infection is highly associated with the development of cervical cancers in women (Gupta and Mania-Pramanik, 2019). HPV can also initiate other kinds of anogenital cancer, head and neck cancers, and genital warts in men and women (Chen et al., 2018). Currently, there is no clinical cure for HPV infection that can achieve a satisfactory effect due to the ability of the virus to reduce their activity in a host cell to circumvent a host immune surveillance, which makes it extremely difficult to remove a viral genome from an infected host cell in a latency state (Lee, 2019). Based on the existing literatures, HPV-driven tumor formations have been mostly attributed to the HPV E6 and E7 oncoproteins, whose corresponding genes are regarded as two prime therapeutic targets in gene therapy (Moody and Laimins, 2010; Hoppe-Seyler et al., 2018). Theoretically, HPV E6 and E7 genes serve the function of suppressing cellular tumor suppressors p53 and retinoblastoma protein (pRB), respectively (Kennedy and Cullen, 2017). Therefore, overexpression of E6 or E7 induced by HPVs can cause malignant transformation of human cells with high probability through the activation of cellular oncogenes (e.g., ras or fos) (McLaughlin-Drubin and Munger, 2009).
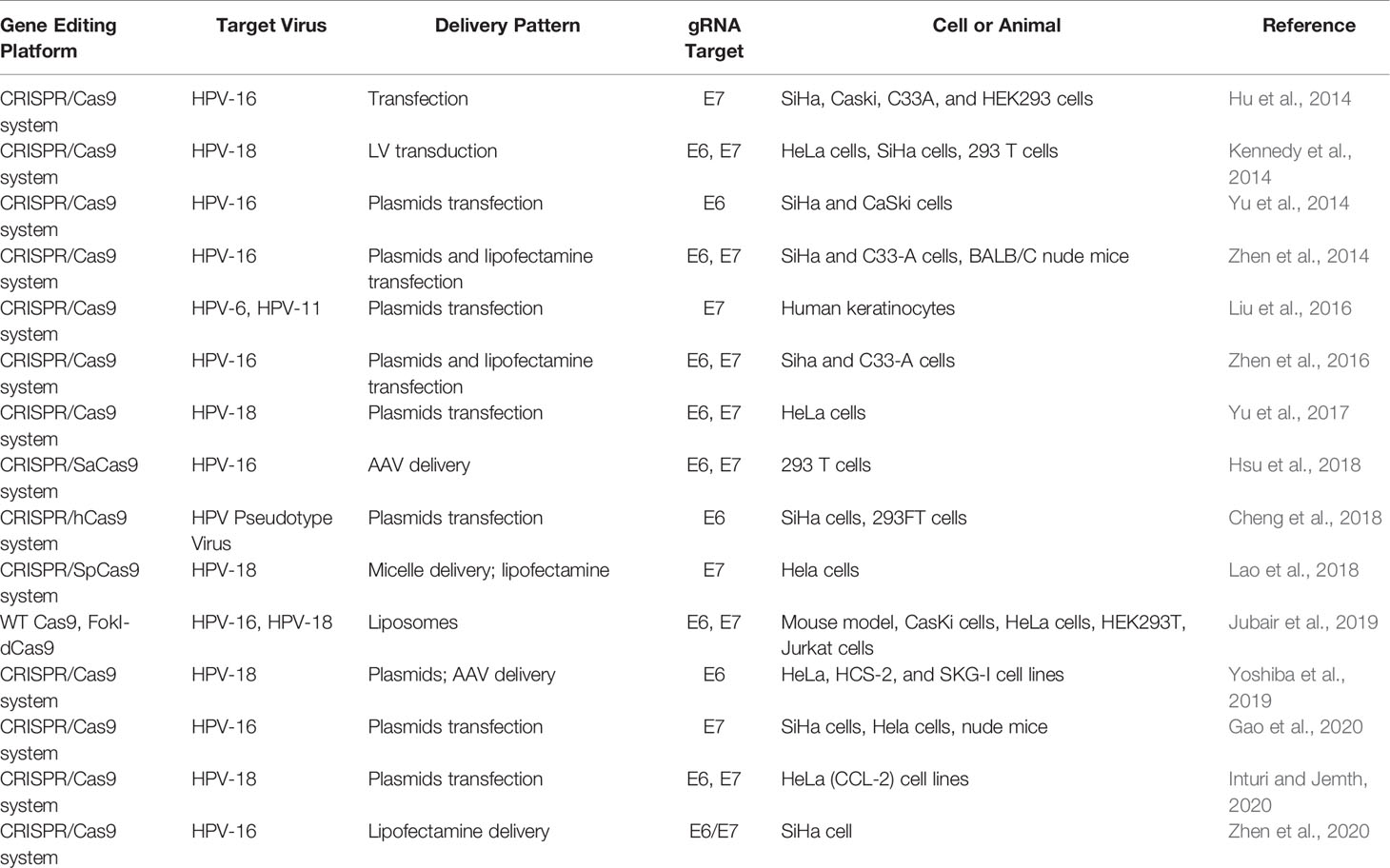
There is still an urgent need to develop novel effective therapies for HPV-associated carcinogenesis, although many progresses have been made in different treatments for HPV. Now, the technology of CRISPR/Cas9-based gene therapy for HPV infection has come into being in recent years. So far, several articles have reported anti-HPV applications of CRISPR/Cas9 system for the purpose of disruption of the HPV genome (Hu et al., 2014; Kennedy et al., 2014; Yu et al., 2014; Zhen et al., 2014; Liu et al., 2016; Yu et al., 2017; Cheng et al., 2018; Hsu et al., 2018; Lao et al., 2018; Jubair et al., 2019; Yoshiba et al., 2019; Gao et al., 2020; Inturi and Jemth, 2021; Zhen et al., 2016; Zhen et al., 2020) (Table 2). Based on the investigations, the CRISPR/Cas9 approach has much development potential to act as an effective therapy for HPV-associated diseases in clinical settings. Herein, several editing targets of CRISPR/Cas (CRISPR/Cas9) in HPV life cycle are shown in Figure 4. Both for HBV and HPV, CRISPR technology is a novel method for the treatment of such viral diseases because it can fill in the technical gaps in drug therapy when a vaccine has already existed. However, CRISPR-associated technologies still need to be developed to improve the therapeutic effects.
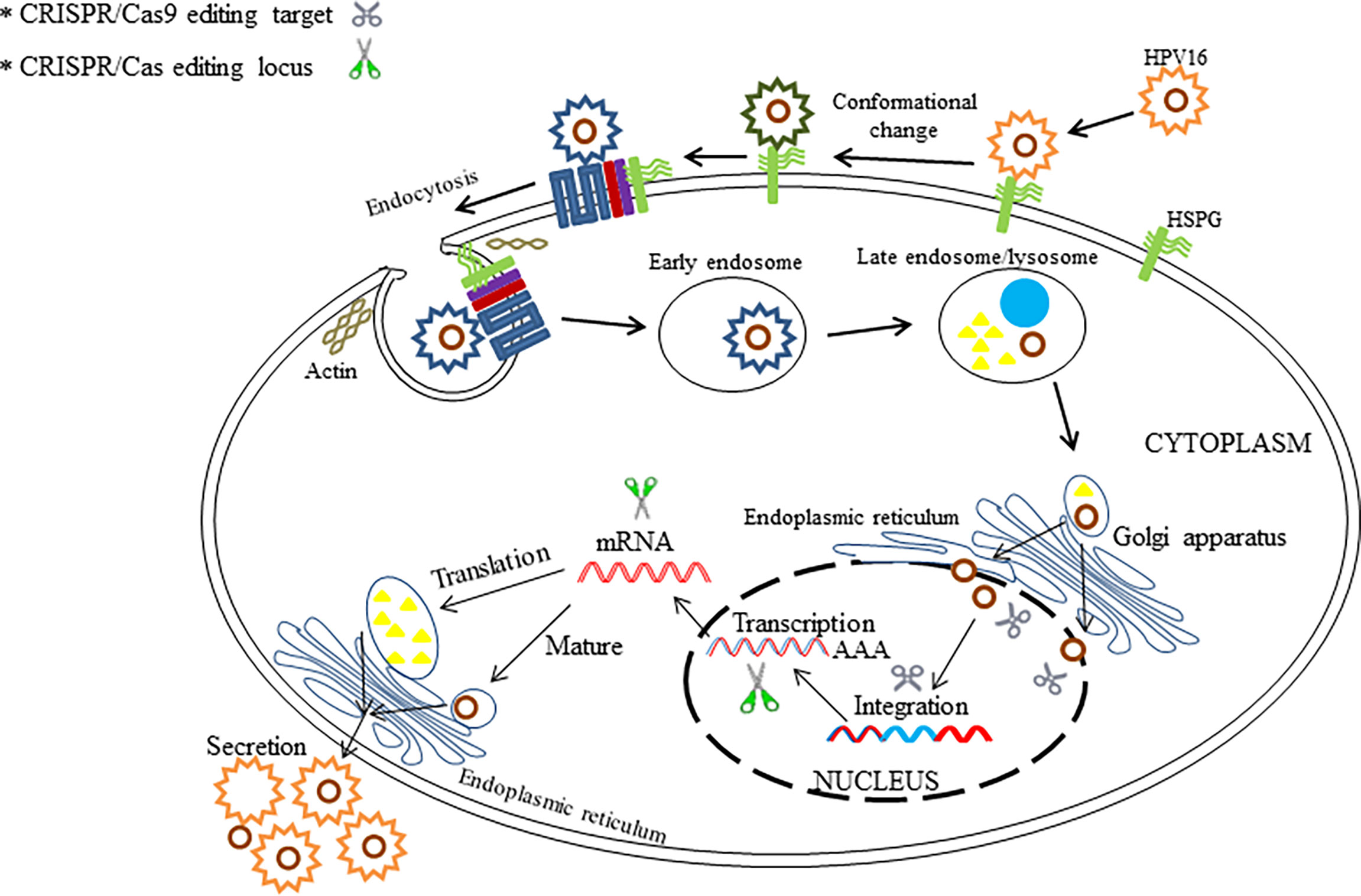
Figure 4 The illustration shows the potential editing and therapeutic targets in HPV life cycle by the use of CRISPR/Cas and CRISPR/Cas9.
Obstacles of CRISPR/Cas9 in the Treatment of Human Infectious Viruses
CRISPR/Cas9 system holds considerable potential for therapeutic applications of human infectious viruses in vivo, but certain questions have to be addressed before its use in clinical aspects. Generally, the off-target effect is the major concern associated with the use of this system. Other crucial challenges including delivery methods and strategies, Cas9 cleavage activity, resistance to Cas9/sgRNA system, viral escape problem, and ethical concerns still lie ahead (Figure 5).
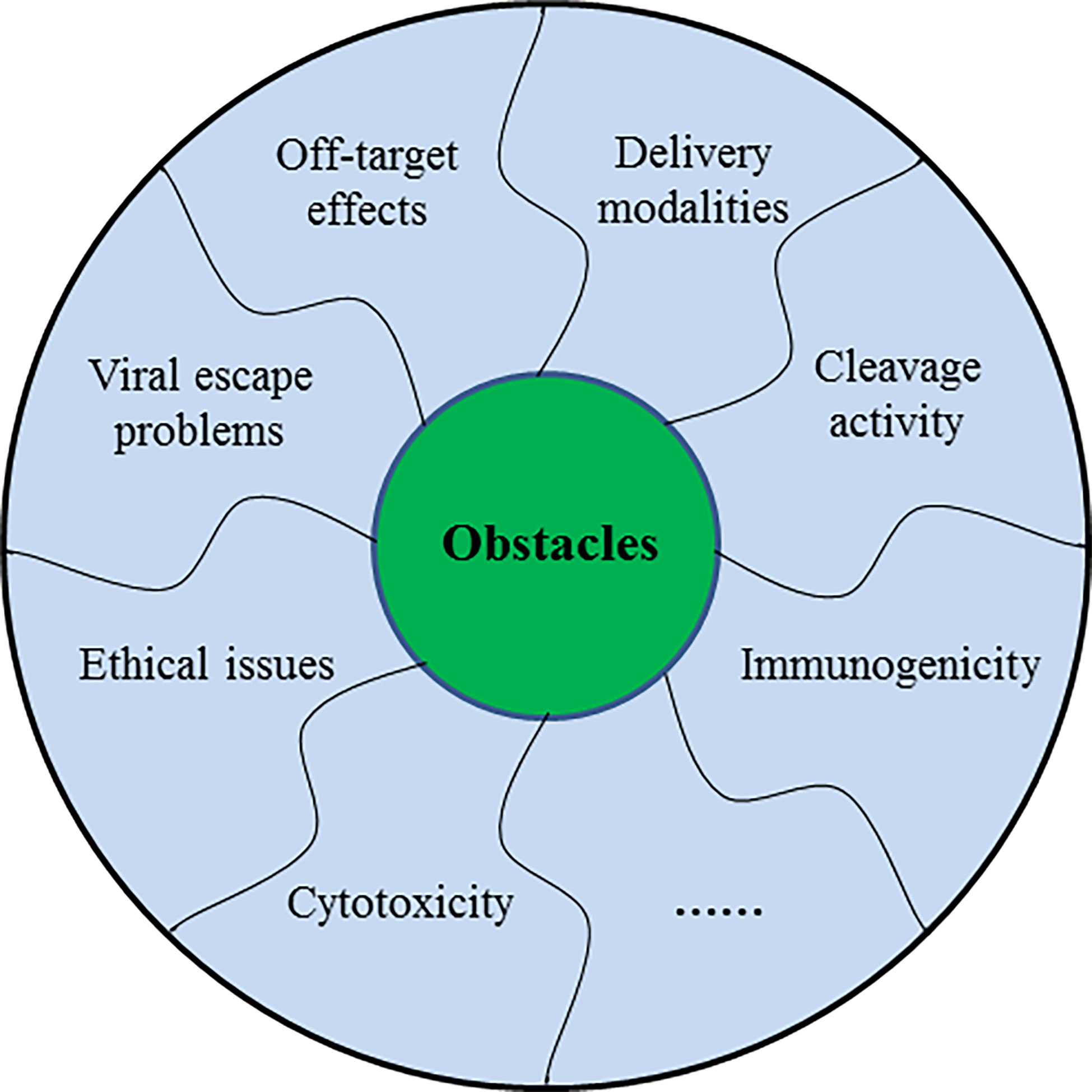
Figure 5 The schematic diagram showing several challenges of CRISPR/Cas9 in the treatment of human infectious viruses.
CRISPR/Cas9 Off-Target Effects
The CRISPR/Cas9 off-target concerns pose challenges for research advancements and therapeutic utilizations. Therefore, researchers have developed methods such as advanced versions or Cas9 nickases to minimize off-target activities (Bellizzi et al., 2019) and cytotoxicity (Wang et al., 2016). The two parts of CRISPR/Cas9 system, the optimizations of Cas9 proteins or the upgradations of gRNA, have equally contributed to the reductions in CRISPR/Cas9 off-target effects. On the one hand, many investigations have focused on reducing the unwanted off-target activities of CRISPR/Cas9 system (Eid and Mahfouz, 2016) through the amelioration of Cas9 nucleases. Recently, two versions of Cas9 variants termed “eSpCas9” (Slaymaker et al., 2016) and “SpCas9-HF1” (Kleinstiver et al., 2016) significantly optimize the CRISPR/Cas9 genome-editing toolbox with their own higher specificity and exceptional precision. On the other hand, it is very important to design special and compatible gRNA for the CRISPR/Cas9 systems. Nowadays, multiple website platforms are available for the optimal design of CRISPR gRNA (e.g., https://www.genscript.com/gRNA-design-tool.html and https://www.atum.bio/eCommerce/cas9/input). Besides, there are also other approaches whereby incorporation of chemical modifications into its structure improve gRNA stability and activity in the cell. Additionally, most CRISPR/Cas spacers that exist in bacterium naturally correspond to foreign nucleic acids (Hille et al., 2018), which precisely confer the bacterial immunity, but when this system works in animal cells, artificially designed CRISPR/Cas9 systems usually neglect this “implied condition” that can result in off-target effects, cytotoxicity, and cellular resistances. For example, Kim et al. reported that cytotoxicity caused by the tailored CRISPR gRNAs (5′-ppp gRNAs) triggers RNA-mediated innate immune responses in human and murine cells (Kim et al., 2018).
Delivery Modalities of CRISPR/Cas9
Delivery mode perhaps remains the biggest bottleneck to gene therapy. To enhance gene editing efficiency, in addition to the improvements of CRISPR/Cas9 reagents itself, another key factor is the delivery methods of this gene editing system. More importantly, delivery methods that maximize efficacy and minimize immune responses still need to be developed (Schinazi et al., 2018). For CRISPR/Cas system, Cas9 nucleases can be delivered to gain access to the genome of the target cells in the form of DNA, mRNA, or protein, while the gRNA could be transferred in the form of DNA or RNA (Yin et al., 2017). Therefore, Cas9 protein and gRNA can be transported either together or separately, which provides much more options for choosing the vehicles. For example, a single adeno-associated virus (AAV) vector (AAV’s packaging capacity is ∼4.8 kb) usually cannot accommodate SpCas9 gene (4.1 kb) and its gRNA sequence. The solution to this problem is to use a small SaCas9 (3.2 kb) to take the place of SpCas9 or use two vehicles to separately transport SpCas9 gene and gRNA sequence.
The delivery patterns for CRISPR/Cas9 system are generally categorized as viral vectors or non-viral vectors, both of which have their own unique advantages and limitations (Nelson and Gersbach, 2016). Viral-based vehicles commonly include adenovirus, lentivirus, AAV, and retrovirus (Tong et al., 2019). Other viral carriers less in use for delivery still include herpes simplex virus and poxvirus (Jin et al., 2014). Non-viral delivery modes can be basically divided into two groups: physical methods [e.g., electroporation, microinjection, sonoporation, and hydrodynamic delivery (Mellott et al., 2013; Ibraheem et al., 2014; Fajrial et al., 2020)] and chemical approaches [e.g., lipid particles (Yin et al., 2014), polymer nanoparticles (Kirtane and Panyam, 2013), gold nanoparticles (Ding et al., 2014), and cell-penetrating peptides (CPPs) (Farkhani et al., 2014)]. Taken together, the proper choice of delivery tool is essential for the safety of gene therapy using CRISPR/Cas9 platform.
Cas9 Cleavage Activity
To date, a growing series of wild-type and engineering Cas9 homologues and other CRISPR/Cas systems are expanding the gene-edited toolkits. However, Cas9 proteins with different species have distinct characteristics such as activity; scientists need to select the appropriate nucleases according to the research requirements and study protocols. For example, the SpCas9 enzyme is most commonly used for genome editing and genetic manipulation in eukaryotic cells while using CRISPR/Cas partly because of its high activity and comparative broad PAM compatibilities. Kim et al. listed 13 types of SpCas9 variants [wild-type SpCas9, eSpCas9 (1.1), SpCas9-HF1, HypaCas9, evoCas9, xCas9, Sniper-Cas9, and SpCas9-NG and the VQR, VRER, VRQR, VRQR-HF1, and QQR1 variants] for choice and made a comparison based on the activities, specificities, and PAM compatibilities (Choi et al., 2019). Eventually, the experimental results recorded on the overall activity could be ranked as SpCas9 ≥ Sniper-Cas9 > eSpCas9 (1.1) > SpCas9-HF1 > HypaCas9 ≈ xCas9 >> evoCas9 (Kim et al., 2020). Another example is that, when applying AAV as delivery vectors, small-volume genome-editing proteins with equal cleavage activity such as S. aureus Cas9 (SaCas9) or Campylobacter jejuni Cas9 (CjCas9), and other newly identified CRISPR/Cas enzymes may circumvent the limitation of packaging capacity (Doudna, 2020).
Resistance to CRISPR/Cas9 (Cas9 Immunogenicity)
Curbing off-target activity has contributed immensely to the area of CRISPR/Cas gene therapeutics (Dolgin, 2020). Once the CRISPR/Cas9 system has been delivered into the target cell and being activated, there are limited means to lower or shut off its activity (Pawluk et al., 2016), which could raise new practical challenges and safety concerns to researchers. For example, excessive or prolonged Cas9 activity can exacerbate off-target effects. At present, some wild-typed Cas9-specific “anti-CRISPRs (Acr)” provide biotechnological tools that can be used to adjust the activities of CRISPR/Cas9 for gene engineering (Rauch et al., 2017). Recently, more than 50 anti-CRISPR protein families have been characterized, which provide various kinds of applications in genome engineering such as in posttranslational switches for control of Cas9 or dCas9 activity (Wiegand et al., 2020).
The Problem of Viral Escape
The problem of viral escape has been a serious source of concern in the field of virus research. Many viruses possess the capability to escape or inhibit the effect of pharmaceuticals (e.g., interferon) in their evolution. Specifically in CRISPR/Cas9 applications, viruses can escape from these suppressions through the acquisitions of specific mutations at the target site that prevent gRNA binding without hindering viral replications (Binda et al., 2020). Using the HIV-1 as example, as far as we know, the RNA interference (RNAi) technology for the treatment of HIV-1 has already reached the clinical stage (Bobbin et al., 2015; Swamy et al., 2016). As observed with RNAi techniques previously, CRISPR/Cas9-based therapy of HIV-1 could also generate the self-replicated viral mutants (White et al., 2016). Recent studies reported that PAM sequence mutations have been shown to allow phage to escape CRISPR/Cas system (Bikard and Barrangou, 2017; Strich and Chertow, 2019). However, viral escape is not insurmountable if an appropriate gene editing treatment measures are taken.
Ethical Issues
CRISPR/Cas9 technology is still in infancy stage, and many technical problems remain to be solved. However, the utilizations of CRISPR/Cas9 systems toward clinical applications will be confronted with some ethical questions. First, the misuse of this novel technology could likely create certain ethical controversies. Second, the safety induced by unwanted gene editing of CRISPR/Cas9 should be carefully improved and evaluated while clinically applying this system. However, in UK, scientists have gained license to edit human embryos with the use of CRISPR/Cas9 technology (Callaway, 2016), which potentially shows the technical strengths of this system in prospective clinical applications. Nevertheless, CRISPR/Cas9-associated clinics in the future must be strictly supervised with newly established regulations (Shinwari et al., 2017) so as to boost CRISPR gene editing technology to really serve humans.
Given the rapid progression of gene editing technologies, CRISPR/Cas9 is revolutionizing our ability to manipulate human genes and providing immense potentials and challenges for clinical trials. Hence, CRISPR/Cas9-based genomic methodologies will undoubtedly improve human life.
Howerver, CRISPR babies are currently not ready yet. As we know, significant progress has been made recently in CRISPR technology and has promoted the rapid development of biomedicine, agriculture, and animal husbandry. However, the off-target effect cannot be completely eradicated, the accuracy is not high enough, and the risks to growth are also unclear. At present, there is a worldwide consensus that in vitro gene editing research on embryonic development stage or germ line cells is allowed; CRISPR babies are expressly forbidden.
Conclusion
Virus–host interaction is a fluctuant and persistent process within the infectious life cycle. The existence of several tricky viruses has led to the continuous upgrade of anti-virus approaches. CRISPR/Cas-based genetic targeting technology represents an alternative solution for treatment applications of virus-related diseases in the future. To date, CRISPR/Cas9 technology has already demonstrated many potential applications to human illnesses including genetic disorders, tumors, and infectious viruses. In addition to the aforementioned viruses, this technology is becoming increasingly powerful and have already been extensively applied into study on preventing and combating additional human viruses including Epstein–Barr virus (EBV), hepatitis C viruses (HCV), Kaposi sarcoma virus (KSHV), JC virus (JCV), and Herpes simplex virus (HSV). Moreover, CRISPR/Cas9 technology has been utilized not only in the treatment of viral infections but also in the investigations of cellular mechanisms of viral carcinogenesis.
In summary, continued efforts on developing CRISPR/Cas systems will expand the toolbox, which enables us to acquire a greater understanding of complex biological processes associated with hosts and viruses. However, the future use of CRISPR/Cas9 for gene therapies need substantial improvements and perfections before clinical applications.
Author Contributions
HL, GL and XP wrote this manuscript. AD and LS help modify the manuscript. JH and TW direct the writing and support this project. All authors contributed to the article and approved the submitted version.
Funding
This work was collectively supported by grants from the National Key Research and Development Plan (2016YFD0500600), Guangdong Provincial Science and Technology Plan project (2017B020207004), Fundamental Research Funds for the Central Universities (21618309), Innovative province construction project--special topic on fighting against novel coronavirus pneumonia epidemic (2020SK3044), Youth fund project of Hunan natural science foundation (2019JJ50681), Changsha biological resources sample bank establishment project (20200365) and Fund project of Hunan Provincial Health Commission (20200365).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Alimonti, J. B., Ball, T. B., Fowke, K. R. (2003). Mechanisms of CD4+ T Lymphocyte Cell Death in Human Immunodeficiency Virus Infection and AIDS. J. Gen. Virol. 84, 1649–1661. doi: 10.1099/vir.0.19110-0
Arribas, J. R., Eron, J. (2013). Advances in Antiretroviral Therapy. Curr. Opin. HIV AIDS 8, 341–349. doi: 10.1097/COH.0b013e328361fabd
Ashfaq, U. A., Khalid, H. (2020). CRISPR/CAS9-Mediated Antiviral Activity: A Tool to Combat Viral Infection. Crit. Rev. Eukaryot. Gene Expression 30, 45–56. doi: 10.1615/CritRevEukaryotGeneExpr.2020028207
Barrangou, R., Doudna, J. A. (2016). Applications of CRISPR Technologies in Research and Beyond. Nat. Biotechnol. 34, 933941. doi: 10.1038/nbt.3659
Bayat, H., Modarressi, M. H., Rahimpour, A. (2018). The Conspicuity of CRISPR-Cpf1 System as a Significant Breakthrough in Genome Editing. Curr. Microbiol. 75, 107–115. doi: 10.1007/s00284-017-1406-8
Bellizzi, A., Ahye, N., Jalagadugula, G., Wollebo, H. S. (2019). A Broad Application of CRISPR Cas9 in Infectious Diseases of Central Nervous System. J. Neuroimmune. Pharmacol. 14, 578–594. doi: 10.1007/s11481-019-09878-7
Bialek, J. K., Dunay, G. A., Voges, M., Schafer, C., Spohn, M., Stucka, R., et al. (2016). Targeted HIV-1 Latency Reversal Using CRISPR/Cas9-Derived Transcriptional Activator Systems. PloS One 11, e0158294. doi: 10.1371/journal.pone.0158294
Bikard, D., Barrangou, R. (2017). Using CRISPR-Cas Systems as Antimicrobials. Curr. Opin. Microbiol. 37, 155–160. doi: 10.1016/j.mib.2017.08.005
Binda, C. S., Klaver, B., Berkhout, B., Das, A. T. (2020). CRISPR/Cas9 Dual-gRNA Attack Causes Mutation, Excision and Inversion of the HIV-1 Proviral DNA. Viruses 12 (3), 330. doi: 10.3390/v12030330
Bloom, K., Maepa, M. B., Ely, A., Arbuthnot, P. (2018). Gene Therapy for Chronic HBV-Can We Eliminate cccDNA? Genes (Basel) 9, 207. doi: 10.3390/genes9040207
Bobbin, M. L., Burnett, J. C., Rossi, J. J. (2015). RNA Interference Approaches for Treatment of HIV-1 Infection. Genome Med. 7, 50. doi: 10.1186/s13073-015-0174-y
Callaway, E. (2016). UK Scientists Gain Licence to Edit Genes in Human Embryos. Nature 530, 18. doi: 10.1038/nature.2016.19270
Campbell-Yesufu, O. T., Gandhi, R. T. (2011). Update on Human Immunodeficiency Virus (HIV)-2 Infection. Clin. Infect. Dis. 52, 780–787. doi: 10.1093/cid/ciq248
Castilla, V., Ramirez, J., Coto, C. E. (2010). Plant and Animal Steroids a New Hope to Search for Antiviral Agents. Curr. Med. Chem. 17, 1858–1873. doi: 10.2174/092986710791163975
Cheng, Y. X., Chen, G. T., Yang, X., Wang, Y. Q., Hong, L. (2018). Effects of HPV Pseudotype Virus in Cutting E6 Gene Selectively in SiHa Cells. Curr. Med. Sci. 38, 212–221. doi: 10.1007/s11596-018-1868-3
Chen, S., Yu, X., Guo, D. (2018). CRISPR-Cas Targeting of Host Genes as an Antiviral Strategy. Viruses 10, 40. doi: 10.3390/V10010040
Choi, G., Zhou, P., Yuen, C. T. L., Chan, B. K. C., Xu, F., Bao, S., et al. (2019). Combinatorial Mutagenesis En Masse Optimizes the Genome Editing Activities of Spcas9. Nat. Methods 16, 722730. doi: 10.1038/s41592-019-0473-0
Cho, S. W., Kim, S., Kim, J. M., Kim, J. S. (2013). Targeted Genome Engineering in Human Cells With the Cas9 RNA-Guided Endonuclease. Nat. Biotechnol. 31, 230–232. doi: 10.1038/nbt.2507
Chylinski, K., Makarova, K. S., Charpentier, E., Koonin, E. V. (2014). Classification and Evolution of Type II CRISPR-Cas Systems. Nucleic Acids Res. 42, 6091–6105. doi: 10.1093/nar/gku241
Citorik, R. J., Mimee, M., Lu, T. K. (2014). Sequence-Specific Antimicrobials Using Efficiently Delivered RNA-Guided Nucleases. Nat. Biotechnol. 32, 11411145. doi: 10.1038/nbt.3011
Cong, L., Ran, F. A., Cox, D., Lin, S., Barretto, R., Habib, N., et al. (2013). Multiplex Genome Engineering Using CRISPR/Cas Systems. Sci. (New. York. N.Y.) 339, 819–823. doi: 10.1126/science.1231143
Costa-Neto, E. M. (2005). Animal-Based Medicines: Biological Prospection and the Sustainable Use of Zootherapeutic Resources. Anais. Da. Acad. Bras. Cienc. 77, 33–43. doi: 10.1590/S0001-37652005000100004
Cresci, G. A. M., Lampe, J. W., Gibson, G. (2020). Targeted Approaches for In Situ Gut Microbiome Manipulation. JPEN. J. Parenter. Enteral. Nutr. 44, 581–588. doi: 10.1002/jpen.1779
Dampier, W., Nonnemacher, M. R., Sullivan, N. T., Jacobson, J. M., Wigdahl, B. (2014). HIV Excision Utilizing CRISPR/Cas9 Technology: Attacking the Proviral Quasispecies in Reservoirs to Achieve a Cure. MOJ. Immunol. 1, 00022. doi: 10.15406/moji.2014.01.00022
Dash, P. K., Kaminski, R., Bella, R., Su, H., Mathews, S., Ahooyi, T. M., et al. (2019). Sequential LASER ART and CRISPR Treatments Eliminate HIV-1 in a Subset of Infected Humanized Mice. Nat. Commun. 10, 2753. doi: 10.1038/s41467-019-10366-y
Deng, Q., Chen, Z., Shi, L., Lin, H. (2018). Developmental Progress of CRISPR/Cas9 and its Therapeutic Applications for HIV-1 Infection. Rev. Med. Virol. 28, e1998. doi: 10.1002/rmv.1998
Ding, Y., Jiang, Z., Saha, K., Kim, C. S., Kim, S. T., Landis, R. F., et al. (2014). Gold Nanoparticles for Nucleic Acid Delivery. Mol. Ther. 22, 1075–1083. doi: 10.1038/mt.2014.30
Doerflinger, M., Forsyth, W., Ebert, G., Pellegrini, M., Herold, M. J. (2017). CRISPR/Cas9-The Ultimate Weapon to Battle Infectious Diseases? Cell Microbiol. 19, 1–31. doi: 10.1111/cmi.12693
Doitsh, G., Galloway, N. L., Geng, X., Yang, Z., Monroe, K. M., Zepeda, O., et al. (2014). Cell Death by Pyroptosis Drives CD4 T-Cell Depletion in HIV-1 Infection. Nature 505, 509–514. doi: 10.1038/nature12940
Dolgin, E. (2020). The Kill-Switch for CRISPR That Could Make Gene-Editing Safer. Nature 577, 308–310. doi: 10.1038/d41586-020-00053-0
Dong, C., Qu, L., Wang, H., Wei, L., Dong, Y., Xiong, S. (2015). Targeting Hepatitis B Virus cccDNA by CRISPR/Cas9 Nuclease Efficiently Inhibits Viral Replication. Antiviral Res. 118, 110–117. doi: 10.1016/j.antiviral.2015.03.015
Doudna, J. A. (2020). The Promise and Challenge of Therapeutic Genome Editing. Nature 578, 229236. doi: 10.1038/s41586-020-1978-5
Doudna, J. A., Charpentier, E. (2014). Genome Editing. The New Frontier of Genome Engineering With CRISPR-Cas9. Science 346, 1258096. doi: 10.1126/science.1258096
Dreyer, T., Nicholson, S., Ely, A., Arbuthnot, P., Bloom, K. (2016). Improved Antiviral Efficacy Using TALEN-Mediated Homology Directed Recombination to Introduce Artificial Primary miRNAs Into DNA of Hepatitis B Virus. Biochem. Biophys. Res. Commun. 478, 1563–1568. doi: 10.1016/j.bbrc.2016.08.152
Ebina, H., Misawa, N., Kanemura, Y., Koyanagi, Y. (2013). Harnessing the CRISPR/Cas9 System to Disrupt Latent HIV-1 Provirus. Sci. Rep. 3, 2510. doi: 10.1038/srep02510
Ebrahimi, S., Teimoori, A., Khanbabaei, H., Tabasi, M. (2019). Harnessing CRISPR/Cas 9 System for Manipulation of DNA Virus Genome. Rev. Med. Virol. 29, e2009. doi: 10.1002/rmv.2009
Eid, A., Mahfouz, M. M. (2016). Genome Editing: The Road of CRISPR/Cas9 From Bench to Clinic. Exp. Mol. Med. 48, e265. doi: 10.1038/emm.2016.111
El-Serag, H. B. (2012). Epidemiology of Viral Hepatitis and Hepatocellular Carcinoma. Gastroenterology 142, 1264–1273. doi: 10.1053/j.gastro.2011.12.061
Emery, J. S., Feld, J. J. (2017). Treatment of Hepatitis B Virus With Combination Therapy Now and in the Future. Best Pract. Res. Clin. Gastroenterol. 31, 347–355. doi: 10.1016/j.bpg.2017.04.007
Fajrial, A. K., He, Q. Q., Wirusanti, N. I., Slansky, J. E., Ding, X. (2020). A Review of Emerging Physical Transfection Methods for CRISPR/Cas9-Mediated Gene Editing. Theranostics 10, 5532–5549. doi: 10.7150/thno.43465
Farkhani, S. M., Valizadeh, A., Karami, H., Mohammadi, S., Sohrabi, N., Badrzadeh, F. (2014). Cell Penetrating Peptides: Efficient Vectors for Delivery of Nanoparticles, Nanocarriers, Therapeutic and Diagnostic Molecules. Peptides 57, 78–94. doi: 10.1016/j.peptides.2014.04.015
Gao, X., Jin, Z., Tan, X., Zhang, C., Zou, C., Zhang, W., et al. (2020). Hyperbranched Poly(Beta-Amino Ester) Based Polyplex Nanopaticles for Delivery of CRISPR/Cas9 System and Treatment of HPV Infection Associated Cervical Cancer. J. Controlled Release. 321, 654–668. doi: 10.1016/j.jconrel.2020.02.045
Garneau, J. E., Dupuis, M. E., Villion, M., Romero, D. A., Barrangou, R., Boyaval, P., et al. (2010). The CRISPR/Cas Bacterial Immune System Cleaves Bacteriophage and Plasmid DNA. Nature 468, 6771. doi: 10.1038/nature09523
Gebre, M., Nomburg, J. L., Gewurz, B. E. (2018). CRISPR-Cas9 Genetic Analysis of Virus-Host Interactions. Viruses 10, 1–22. doi: 10.3390/v10020055
Gupta, R. K., Abdul-Jawad, S., McCoy, L. E., Mok, H. P., Peppa, D., Salgado, M., et al. (2019). HIV-1 Remission Following CCR5Delta32/Delta32 Haematopoietic Stem-Cell Transplantation. Nature 568, 244–248. doi: 10.1038/s41586-019-1027-4
Gupta, S. M., Mania-Pramanik, J. (2019). Molecular Mechanisms in Progression of HPV-Associated Cervical Carcinogenesis. J. BioMed. Sci. 26, 28. doi: 10.1186/s12929-019-0520-2
Gupta, R. K., Peppa, D., Hill, A. L., Gálvez, C., Salgado, M., Pace, M., et al. (2020). Evidence for HIV-1 Cure After CCR5Δ32/Δ32 Allogeneic Haemopoietic Stem-Cell Transplantation 30 Months Post Analytical Treatment Interruption: A Case Report. Lancet HIV 7, e340–e347. doi: 10.1016/s2352-3018(20)30069-2
Harden, M. E., Munger, K. (2017). Human Papillomavirus Molecular Biology. Mutat. Res. Rev. Mutat. Res. 772, 312. doi: 10.1016/j.mrrev.2016.07.002
Haynes, B. F. (2015). New Approaches to HIV Vaccine Development. Curr. Opin. Immunol. 35, 39–47. doi: 10.1016/j.coi.2015.05.007
Hille, F., Richter, H., Wong, S. P., Bratovic, M., Ressel, S., Charpentier, E. (2018). The Biology of CRISPR-Cas: Backward and Forward. Cell 172, 1239–1259. doi: 10.1016/j.cell.2017.11.032
Hirakawa, M. P., Krishnakumar, R., Timlin, J. A., Carney, J. P., Butler, K. S. (2020). Gene Editing and CRISPR in the Clinic: Current and Future Perspectives. Biosci. Rep. 40, 1–37. doi: 10.1042/BSR20200127
Hoppe-Seyler, K., Bossler, F., Braun, J. A., Herrmann, A. L., Hoppe-Seyler, F. (2018). The HPV E6/E7 Oncogenes: Key Factors for Viral Carcinogenesis and Therapeutic Targets. Trends Microbiol. 26, 158–168. doi: 10.1016/j.tim.2017.07.007
Hou, P., Chen, S., Wang, S., Yu, X., Chen, Y., Jiang, M., et al. (2015). Genome Editing of CXCR4 by CRISPR/cas9 Confers Cells Resistant to HIV-1 Infection. Sci. Rep. 5 (1), 15577–15577. doi: 10.1038/srep15577
Hryhorowicz, M., Lipiński, D., Zeyland, J., Słomski, R. (2017). CRISPR/Cas9 Immune System as a Tool for Genome Engineering. Archivum. Immunol. Et. Therap. Experiment. 65, 233–240. doi: 10.1007/S00005-016-0427-5
Hsu, D. S., Kornepati, A. V., Glover, W., Kennedy, E. M., Cullen, B. R. (2018). Targeting HPV16 DNA Using CRISPR/Cas Inhibits Anal Cancer Growth. Future Virol. 13, 475–482. doi: 10.2217/fvl-2018-0010
Hu, J. H., Miller, S. M., Geurts, M. H., Tang, W., Chen, L., Sun, N., et al. (2018). Evolved Cas9 Variants With Broad PAM Compatibility and High DNA Specificity. Nature 556, 57–63. doi: 10.1038/nature26155
Hütter, G., Nowak, D., Mossner, M., Ganepola, S., Mussig, A., Allers, K., et al. (2009). Long-Term Control of HIV by CCR5 Delta32/Delta32 Stem-Cell Transplantation. N. Engl. J. Med. 360, 692–698. doi: 10.1056/NEJMoa0802905
Hu, Z., Yu, L., Zhu, D., Ding, W., Wang, X., Zhang, C., et al. (2014). Disruption of HPV16-E7 by CRISPR/Cas System Induces Apoptosis and Growth Inhibition in HPV16 Positive Human Cervical Cancer Cells. BioMed. Res. Int. 2014, 612823–612823. doi: 10.1155/2014/612823
Ibraheem, D., Elaissari, A., Fessi, H. (2014). Gene Therapy and DNA Delivery Systems. Int. J. Pharm. 459, 7083. doi: 10.1016/j.ijpharm.2013.11.041
Inturi, R., Jemth, P. (2021). CRISPR/Cas9-Based Inactivation of Human Papillomavirus Oncogenes E6 and E7 Induces Senescence in Cervical Cancer Cells. Virology 562, 92–102. doi: 10.1016/j.virol.2021.07.005
Ishino, Y., Shinagawa, H., Makino, K., Amemura, M., Nakata, A. (1987). Nucleotide Sequence of the IAP Gene, Responsible for Alkaline Phosphatase Isozyme Conversion in Escherichia Coli, and Identification of the Gene Product. J. Bacteriol. 169, 5429–5433. doi: 10.1128/jb.169.12.5429-5433.1987
Jiang, F., Zhou, K., Ma, L., Gressel, S., Doudna, J. A. (2015). STRUCTURAL BIOLOGY. A Cas9-Guide RNA Complex Preorganized for Target DNA Recognition. Sci. (New. York. N.Y.) 348, 1477–1481. doi: 10.1126/science.aab1452
Jinek, M., Chylinski, K., Fonfara, I., Hauer, M., Doudna, J. A., Charpentier, E. (2012). A Programmable Dual-RNA-Guided DNA Endonuclease in Adaptive Bacterial Immunity. Sci. (New. York. N.Y.) 337, 816–821. doi: 10.1126/science.1225829
Jinek, M., East, A., Cheng, A., Lin, S., Ma, E., Doudna, J. (2013). RNA-Programmed Genome Editing in Human Cells. Elife 2, e00471. doi: 10.7554/eLife.00471
Jin, L., Zeng, X., Liu, M., Deng, Y., He, N. (2014). Current Progress in Gene Delivery Technology Based on Chemical Methods and Nano-Carriers. Theranostics 4, 240–255. doi: 10.7150/thno.6914
Jubair, L., Fallaha, S., McMillan, N. A. J. (2019). Systemic Delivery of CRISPR/Cas9 Targeting HPV Oncogenes Is Effective at Eliminating Established Tumors. Mol. Ther. 27, 2091–2099. doi: 10.1016/j.ymthe.2019.08.012
Kaminski, R., Chen, Y., Fischer, T., Tedaldi, E., Napoli, A., Zhang, Y., et al. (2016). Elimination of HIV-1 Genomes From Human T-Lymphoid Cells by CRISPR/Cas9 Gene Editing. Sci. Rep. 6 (1), 22555–22555. doi: 10.1038/srep22555
Karimova, M., Beschorner, N., Dammermann, W., Chemnitz, J., Indenbirken, D., Bockmann, J. H., et al. (2015). CRISPR/Cas9 Nickase-Mediated Disruption of Hepatitis B Virus Open Reading Frame S and X. Sci. Rep. 5, 13734. doi: 10.1038/srep13734
Kaushik, A., Yndart, A., Atluri, V., Tiwari, S., Tomitaka, A., Gupta, P., et al. (2019). Magnetically Guided non-Invasive CRISPR-Cas9/gRNA Delivery Across Blood-Brain Barrier to Eradicate Latent HIV-1 Infection. Sci. Rep. 9 (1), 3928. doi: 10.1038/s41598-019-40222-4
Kennedy, E. M., Bassit, L. C., Mueller, H., Kornepati, A. V. R., Bogerd, H. P., Nie, T., et al. (2015). Suppression of Hepatitis B Virus DNA Accumulation in Chronically Infected Cells Using a Bacterial CRISPR/Cas RNA-Guided DNA Endonuclease. Virology 476, 196–205. doi: 10.1016/j.virol.2014.12.001
Kennedy, E. M., Cullen, B. R. (2017). Gene Editing: A New Tool for Viral Disease. Annu. Rev. Med. 68, 401411. doi: 10.1146/annurev-med-051215-031129
Kennedy, E. M., Kornepati, A. V., Goldstein, M., Bogerd, H. P., Poling, B. C., Whisnant, A. W., et al. (2014). Inactivation of the Human Papillomavirus E6 or E7 Gene in Cervical Carcinoma Cells by Using a Bacterial CRISPR/Cas RNA-Guided Endonuclease. J. Virol. 88, 11965–11972. doi: 10.1128/JVI.01879-14
Khadempar, S., Familghadakchi, S., Motlagh, R. A., Farahani, N., Dashtiahangar, M., Rezaei, H., et al. (2019). CRISPR-Cas9 in Genome Editing: Its Function and Medical Applications. J. Cell Physiol. 234, 57515761. doi: 10.1002/jcp.27476
Khalili, K., Kaminski, R., Gordon, J., Cosentino, L., Hu, W. (2015). Genome Editing Strategies: Potential Tools for Eradicating HIV-1/AIDS. J. Neurovirol. 21, 310–321. doi: 10.1007/s13365-014-0308-9
Kim, N., Kim, H. K., Lee, S., Seo, J. H., Choi, J. W., Park, J., et al. (2020). Prediction of the Sequence-Specific Cleavage Activity of Cas9 Variants. Nat. Biotechnol 38, 1328–1336. doi: 10.1038/s41587-020-0537-9
Kim, S., Koo, T., Jee, H. G., Cho, H. Y., Lee, G., Lim, D. G., et al. (2018). CRISPR RNAs Trigger Innate Immune Responses in Human Cells. Genome Res. 28, 367–373. doi: 10.1101/gr.231936.117
Kirtane, A. R., Panyam, J. (2013). Polymer Nanoparticles: Weighing Up Gene Delivery. Nat. Nanotechnol. 8, 805–806. doi: 10.1038/nnano.2013.234
Kleinstiver, B. P., Pattanayak, V., Prew, M. S., Tsai, S. Q., Nguyen, N. T., Zheng, Z., et al. (2016). High-Fidelity CRISPR-Cas9 Nucleases With No Detectable Genome-Wide Off-Target Effects. Nature 529, 490–495. doi: 10.1038/nature16526
Kostyusheva, A. P., Brezgin, S. A., Zarifyan, D. N., Chistyakov, D. S., Gegechkory, V. I., Bayurova, E. O., et al. (2019b). Hepatitis B Virus and Site-Specific Nucleases: Effects of Genetic Modifications in CRISPR/Cas9 on Antiviral Activity. Russian J. Infect. Immun. 9, 279–287. doi: 10.15789/2220-7619-2019-2-279-287
Kostyusheva, A. P., Kostyushev, D. S., Brezgin, S. A., Zarifyan, D. N., Volchkova, E. V., Chulanov, V. P. (2019a). Small Molecular Inhibitors of DNA Double Strand Break Repair Pathways Increase the ANTI-HBV Activity of CRISPR/Cas9. Mol. Biol. 53, 274–285. doi: 10.1134/s0026893319010072
Kostyushev, D., Kostyusheva, A., Brezgin, S., Zarifyan, D., Utkina, A., Goptar, I., et al. (2019). Suppressing the NHEJ Pathway by DNA-PKcs Inhibitor NU7026 Prevents Degradation of HBV cccDNA Cleaved by CRISPR/Cas9. Sci. Rep. 9, 1847. doi: 10.1038/s41598-019-38526-6
Kuprash, D. V., Garib, F. Y., Nedospasov, S. A. (2017). Antibody-Based Drugs and Other Recombinant Proteins for Diagnostics and Therapy of Viral Infections, Autoimmune Diseases and Cancer. Molekuliarnaia. Biol. 51, 883–885. doi: 10.7868/S0026898417060015
Kurihara, T., Fukuhara, T., Ono, C., Yamamoto, S., Uemura, K., Okamoto, T., et al. (2017). Suppression of HBV Replication by the Expression of Nickase- and Nuclease Dead-Cas9. Sci. Rep. 7, 6122. doi: 10.1038/s41598-017-05905-w
Lao, Y. H., Li, M., Gao, M. A., Shao, D., Chi, C. W., Huang, D., et al. (2018). HPV Oncogene Manipulation Using Nonvirally Delivered CRISPR/Cas9 or Natronobacterium Gregoryi Argonaute. Adv. Sci. (Weinh). 5, 1700540. doi: 10.1002/advs.201700540
Lee, W. M. (1997). Hepatitis B Virus Infection. New Engl. J. Med. 337, 1733–1745. doi: 10.1056/NEJM199712113372406
Lee, C. (2019). CRISPR/Cas9-Based Antiviral Strategy: Current Status and the Potential Challenge. Molecules 24, 1349. doi: 10.3390/molecules24071349
Liao, H. K., Gu, Y., Diaz, A., Marlett, J., Takahashi, Y., Li, M., et al. (2015). Use of the CRISPR/Cas9 System as an Intracellular Defense Against HIV-1 Infection in Human Cells. Nat. Commun. 6 (1), 6413. doi: 10.1038/ncomms7413
Lin, H., Deng, Q., Li, L., Shi, L. (2019). Application and Development of CRISPR/Cas9 Technology in Pig Research, Gene Editing-Technologies and Applications. IntechOpen. doi: 10.5772/intechopen.85540
Lino, C. A., Harper, J. C., Carney, J. P., Timlin, J. A. (2018). Delivering CRISPR: A Review of the Challenges and Approaches. Drug Deliv. 25, 1234–1257. doi: 10.1080/10717544.2018.1474964
Lin, S. R., Yang, H. C., Kuo, Y. T., Liu, C. J., Yang, T. Y., Sung, K. C., et al. (2014). The CRISPR/Cas9 System Facilitates Clearance of the Intrahepatic HBV Templates In Vivo. Mol. Ther. Nucleic Acids 3, e186. doi: 10.1038/mtna.2014.38
Li, H., Sheng, C., Wang, S., Yang, L., Liang, Y., Huang, Y., et al. (2017). Removal of Integrated Hepatitis B Virus DNA Using CRISPR-Cas9. Front. Cell Infect. Microbiol. 7, 91. doi: 10.3389/fcimb.2017.00091
Liu, Y. C., Cai, Z. M., Zhang, X. J. (2016). Reprogrammed CRISPR-Cas9 Targeting the Conserved Regions of HPV6/11 E7 Genes Inhibits Proliferation and Induces Apoptosis in E7-Transformed Keratinocytes. Asian J. Androl. 18, 475–479. doi: 10.4103/1008-682X.157399
Liu, Z., Chen, S., Jin, X., Wang, Q., Yang, K., Li, C., et al. (2017). Genome Editing of the HIV Co-Receptors CCR5 and CXCR4 by CRISPR-Cas9 Protects CD4(+) T Cells From HIV-1 Infection. Cell Biosci. 7, 47. doi: 10.1186/s13578-017-0174-2
Liu, X., Hao, R., Chen, S., Guo, D., Chen, Y. (2015). Inhibition of Hepatitis B Virus by the CRISPR/Cas9 System via Targeting the Conserved Regions of the Viral Genome. J. Gen. Virol. 96, 2252–2261. doi: 10.1099/vir.0.000159
Liu, S., Wang, Q., Yu, X., Li, Y., Guo, Y., Liu, Z., et al. (2018). HIV-1 Inhibition in Cells With CXCR4 Mutant Genome Created by CRISPR-Cas9 and Piggybac Recombinant Technologies. Sci. Rep. 8 (1), 8573–8573. doi: 10.1038/S41598-018-26894-4
Locarnini, S., Littlejohn, M., Aziz, M. N., Yuen, L. (2013). Possible Origins and Evolution of the Hepatitis B Virus (HBV). Semin. Cancer Biol. 23, 561–575. doi: 10.1016/j.semcancer.2013.08.006
Lu, D. Y., Wu, H. Y., Yarla, N. S., Xu, B., Ding, J., Lu, T. R. (2018). HAART in HIV/AIDS Treatments: Future Trends. Infect. Disord. Drug Targets 18, 15–22. doi: 10.2174/1871526517666170505122800
Maeder, M. L., Gersbach, C. A. (2016). Genome-Editing Technologies for Gene and Cell Therapy. Mol. Ther. 24, 430446. doi: 10.1038/MT.2016.10
Maepa, M. B., Roelofse, I., Ely, A., Arbuthnot, P. (2015). Progress and Prospects of Anti-HBV Gene Therapy Development. Int. J. Mol. Sci. 16, 17589–17610. doi: 10.3390/ijms160817589
Mali, P., Yang, L., Esvelt, K. M., Aach, J., Guell, M., DiCarlo, J. E., et al. (2013). RNA-Guided Human Genome Engineering via Cas9. Science 339, 823–826. doi: 10.1126/science.1232033
Manjunath, N., Yi, G., Dang, Y., Shankar, P. (2013). Newer Gene Editing Technologies Toward HIV Gene Therapy. Viruses 5, 2748–2766. doi: 10.3390/v5112748
Mao, Z., Bozzella, M., Seluanov, A., Gorbunova, V. (2008). DNA Repair by Nonhomologous End Joining and Homologous Recombination During Cell Cycle in Human Cells. Cell Cycle 7, 2902–2906. doi: 10.4161/cc.7.18.6679
McBride, A. A. (2017). Oncogenic Human Papillomaviruses. Philos. Trans. R. Soc. Lond. B. Biol. Sci. 372 (1732). doi: 10.1098/rstb.2016.0273
McLaughlin-Drubin, M. E., Munger, K. (2009). Oncogenic Activities of Human Papillomaviruses. Virus Res. 143, 195208. doi: 10.1016/j.virusres.2009.06.008
Mellott, A. J., Forrest, M. L., Detamore, M. S. (2013). Physical non-Viral Gene Delivery Methods for Tissue Engineering. Ann. BioMed. Eng. 41, 446–468. doi: 10.1007/s10439-012-0678-1
Miller, S. M., Wang, T., Randolph, P. B., Arbab, M., Shen, M. W., Huang, T. P., et al. (2020). Continuous Evolution of SpCas9 Variants Compatible With Non-G PAMs. Nat. Biotechnol. 38, 471–481. doi: 10.1038/s41587-020-0412-8
Moens, U. (2018). Human Polyomaviruses and Papillomaviruses. Int. J. Mol. Sci. 19, 2360. doi: 10.3390/ijms19082360
Mohammadzadeh, I., Qujeq, D., Yousefi, T., Ferns, G. A., Maniati, M., Vaghari-Tabari, M. (2020). CRISPR/Cas9 Gene Editing: A New Therapeutic Approach in the Treatment of Infection and Autoimmunity. IUBMB Life 72, 1603–1621. doi: 10.1002/iub.2296
Mohanraju, P., Makarova, K. S., Zetsche, B., Zhang, F., Koonin, E. V., van der Oost, J. (2016). Diverse Evolutionary Roots and Mechanistic Variations of the CRISPR-Cas Systems. Science 353, aad5147. doi: 10.1126/science.aad5147
Moody, C. A., Laimins, L. A. (2010). Human Papillomavirus Oncoproteins: Pathways to Transformation. Nat. Rev. Cancer 10, 550–560. doi: 10.1038/nrc2886
Morens, D. M., Fauci, A. S. (2013). Emerging Infectious Diseases: Threats to Human Health and Global Stability. PloS Pathog. 9, e1003467. doi: 10.1371/journal.ppat.1003467
Nassal, M. (2015). HBV cccDNA: Viral Persistence Reservoir and Key Obstacle for a Cure of Chronic Hepatitis B. Gut 64, 1972–1984. doi: 10.1136/gutjnl-2015-309809
Nelson, C. E., Gersbach, C. A. (2016). Engineering Delivery Vehicles for Genome Editing. Annu. Rev. Chem. Biomol. Eng. 7, 637–662. doi: 10.1146/annurev-chembioeng-080615-034711
Nguyen, H. P., Ramirez-Fort, M. K., Rady, P. L. (2014). The Biology of Human Papillomaviruses. Curr. Probl. Dermatol. 45, 1932. doi: 10.1159/000355959
Nishimasu, H., Ran, F. A., Hsu, P. D., Konermann, S., Shehata, S. I., Dohmae, N., et al. (2014). Crystal Structure of Cas9 in Complex With Guide RNA and Target DNA. Cell 156, 935–949. doi: 10.1016/j.cell.2014.02.001
Ota, S., Hisano, Y., Ikawa, Y., Kawahara, A. (2014). Multiple Genome Modifications by the CRISPR/Cas9 System in Zebrafish. Genes Cells 19, 555–564. doi: 10.1111/gtc.12154
Parrish, C. R., Holmes, E. C., Morens, D. M., Park, E. C., Burke, D. S., Calisher, C. H., et al. (2008). Cross-Species Virus Transmission and the Emergence of New Epidemic Diseases. Microbiol. Mol. Biol. Rev. 72, 457–470. doi: 10.1128/MMBR.00004-08
Pattanayak, V., Lin, S., Guilinger, J. P., Ma, E., Doudna, J. A., Liu, D. R. (2013). High-Throughput Profiling of Off-Target DNA Cleavage Reveals RNA-Programmed Cas9 Nuclease Specificity. Nat. Biotechnol. 31, 839–843. doi: 10.1038/nbt.2673
Pawluk, A., Amrani, N., Zhang, Y., Garcia, B., Hidalgo-Reyes, Y., Lee, J., et al. (2016). Naturally Occurring Off-Switches for CRISPR-Cas9. Cell 167, 1829–1838.e1829. doi: 10.1016/j.cell.2016.11.017
Ramanan, V., Shlomai, A., Cox, D. B., Schwartz, R. E., Michailidis, E., Bhatta, A., et al. (2015). CRISPR/Cas9 Cleavage of Viral DNA Efficiently Suppresses Hepatitis B Virus. Sci. Rep. 5, 10833. doi: 10.1038/srep10833
Ran, F. A., Hsu, P. D., Wright, J., Agarwala, V., Scott, D. A., Zhang, F. (2013). Genome Engineering Using the CRISPR-Cas9 System. Nat. Protoc. 8, 2281–2308. doi: 10.1038/nprot.2013.143
Rauch, B. J., Silvis, M. R., Hultquist, J. F., Waters, C. S., McGregor, M. J., Krogan, N. J., et al. (2017). Inhibition of CRISPR-Cas9 With Bacteriophage Proteins. Cell 168, 150–158. doi: 10.1016/j.cell.2016.12.009
Ryndock, E. J., Meyers, C. (2014). A Risk for non-Sexual Transmission of Human Papillomavirus? Expert Rev. Anti Infect. Ther. 12, 1165–1170. doi: 10.1586/14787210.2014.959497
Saayman, S., Ali, S. A., Morris, K. V., Weinberg, M. S. (2015). The Therapeutic Application of CRISPR/Cas9 Technologies for HIV. Expert Opin. Biol. Ther. 15, 819–830. doi: 10.1517/14712598.2015.1036736
Sakuma, T., Masaki, K., Abe-Chayama, H., Mochida, K., Yamamoto, T., Chayama, K. (2016). Highly Multiplexed CRISPR-Cas9-Nuclease and Cas9-Nickase Vectors for Inactivation of Hepatitis B Virus. Genes Cells 21, 1253–1262. doi: 10.1111/gtc.12437
Schinazi, R. F., Ehteshami, M., Bassit, L., Asselah, T. (2018). Towards HBV Curative Therapies. Liver. Int. 38 (Suppl 1), 102–114. doi: 10.1111/liv.13656
Schiwon, M., Ehrke-Schulz, E., Oswald, A., Bergmann, T., Michler, T., Protzer, U., et al. (2018). One-Vector System for Multiplexed CRISPR/Cas9 Against Hepatitis B Virus cccDNA Utilizing High-Capacity Adenoviral Vectors. Mol. Ther. Nucleic Acids 12, 242–253. doi: 10.1016/j.omtn.2018.05.006
Scott, T., Moyo, B., Nicholson, S., Maepa, M. B., Watashi, K., Ely, A., et al. (2017). ssAAVs Containing Cassettes Encoding SaCas9 and Guides Targeting Hepatitis B Virus Inactivate Replication of the Virus in Cultured Cells. Sci. Rep. 7, 7401. doi: 10.1038/s41598-017-07642-6
Seeger, C., Sohn, J. A. (2014). Targeting Hepatitis B Virus With CRISPR/Cas9. Mol. Ther. Nucleic Acids 3, e216. doi: 10.1038/mtna.2014.68
Seeger, C., Sohn, J. A. (2016). Complete Spectrum of CRISPR/Cas9-Induced Mutations on HBV cccDNA. Mol. Ther. 24, 1258–1266. doi: 10.1038/mt.2016.94
Seo, Y., Yano, Y. (2014). Short- and Long-Term Outcome of Interferon Therapy for Chronic Hepatitis B Infection. World J. Gastroenterol. 20, 13284–13292. doi: 10.3748/wjg.v20.i37.13284
Shinwari, Z. K., Tanveer, F., Khalil, A. T. (2017). Ethical Issues Regarding CRISPR Mediated Genome Editing. Curr. Issues Mol. Biol. 26, 103–110. doi: 10.21775/CIMB.026.103
Siliciano, J. D., Kajdas, J., Finzi, D., Quinn, T. C., Chadwick, K., Margolick, J. B., et al. (2003). Long-Term Follow-Up Studies Confirm the Stability of the Latent Reservoir for HIV-1 in Resting CD4+ T Cells. Nat. Med. 9, 727–728. doi: 10.1038/nm880
Slaymaker, I. M., Gao, L., Zetsche, B., Scott, D. A., Yan, W. X., Zhang, F. (2016). Rationally Engineered Cas9 Nucleases With Improved Specificity. Science 351, 84–88. doi: 10.1126/science.aad5227
Soppe, J. A., Lebbink, R. J. (2017). Antiviral Goes Viral: Harnessing CRISPR/Cas9 to Combat Viruses in Humans. Trends Microbiol. 25, 833–850. doi: 10.1016/j.tim.2017.04.005
Strich, J. R., Chertow, D. S. (2019). CRISPR-Cas Biology and Its Application to Infectious Diseases. J. Clin. Microbiol. 57, e01307–18. doi: 10.1128/JCM.01307-18
Sunbul, M. (2014). Hepatitis B Virus Genotypes: Global Distribution and Clinical Importance. World J. Gastroenterol. 20, 5427–5434. doi: 10.3748/wjg.v20.i18.5427
Swamy, M. N., Wu, H., Shankar, P. (2016). Recent Advances in RNAi-Based Strategies for Therapy and Prevention of HIV-1/AIDS. Adv. Drug Deliv. Rev. 103, 174–86. doi: 10.1016/j.addr.2016.03.005
Tong, S., Moyo, B., Lee, C. M., Leong, K., Bao, G. (2019). Engineered Materials for In Vivo Delivery of Genome-Editing Machinery. Nat. Rev. Mater. 4, 726–737. doi: 10.1038/s41578-019-0145-9
Trépo, C., Chan, H. L. Y., Lok, A. (2014). Hepatitis B Virus Infection. Lancet 384, 2053–2063. doi: 10.1016/s0140-6736(14)60220-8
Villa, T. G., Feijoo-Siota, L., Rama, J. L. R., Ageitos, J. M. (2017). Antivirals Against Animal Viruses. Biochem. Pharmacol. 133, 97–116. doi: 10.1016/j.bcp.2016.09.029
Wang, J., Chen, R., Zhang, R., Ding, S., Zhang, T., Yuan, Q., et al. (2017). The gRNA-miRNA-gRNA Ternary Cassette Combining CRISPR/Cas9 With RNAi Approach Strongly Inhibits Hepatitis B Virus Replication. Theranostics 7, 3090–3105. doi: 10.7150/thno.18114
Wang, Y., Wei, D., Zhu, X., Pan, J., Zhang, P., Huo, L., et al. (2016). A ‘Suicide’ CRISPR-Cas9 System to Promote Gene Deletion and Restoration by Electroporation in Cryptococcus Neoformans. Sci. Rep. 6, 31145. doi: 10.1038/srep31145
Wang, J., Xu, Z. W., Liu, S., Zhang, R. Y., Ding, S. L., Xie, X. M., et al. (2015). Dual gRNAs Guided CRISPR/Cas9 System Inhibits Hepatitis B Virus Replication. World J. Gastroenterol. 21, 9554–9565. doi: 10.3748/wjg.v21.i32.9554
Wang, W., Ye, C., Liu, J., Zhang, D., Kimata, J. T., Zhou, P. (2014). CCR5 Gene Disruption via Lentiviral Vectors Expressing Cas9 and Single Guided RNA Renders Cells Resistant to HIV-1 Infection. PloS One 9, 126. doi: 10.1371/journal.pone.0115987
Weber, N. D., Stone, D., Sedlak, R. H., De Silva Feelixge, H. S., Roychoudhury, P., Schiffer, J. T., et al. (2014). AAV-Mediated Delivery of Zinc Finger Nucleases Targeting Hepatitis B Virus Inhibits Active Replication. PloS One 9, e97579. doi: 10.1371/journal.pone.0097579
White, M. K., Hu, W., Khalili, K. (2015). The CRISPR/Cas9 Genome Editing Methodology as a Weapon Against Human Viruses. Discov. Med. 19, 255262.
White, M. K., Hu, W., Khalili, K. (2016). Gene Editing Approaches Against Viral Infections and Strategy to Prevent Occurrence of Viral Escape. PloS Pathog. 12, e1005953. doi: 10.1371/journal.ppat.1005953
Wiedenheft, B., Sternberg, S. H., Doudna, J. A. (2012). RNA-Guided Genetic Silencing Systems in Bacteria and Archaea. Nature 482, 331–338. doi: 10.1038/nature10886
Wiegand, T., Karambelkar, S., Bondy-Denomy, J., Wiedenheft, B. (2020). Structures and Strategies of Anti-CRISPR-Mediated Immune Suppression. Annu. Rev. Microbiol. 74, 21–37. doi: 10.1146/annurev-micro-020518-120107
Wright, A. V., Sternberg, S. H., Taylor, D. W., Staahl, B. T., Bardales, J. A., Kornfeld, J. E., et al. (2015). Rational Design of a Split-Cas9 Enzyme Complex. Proc. Natl. Acad. Sci. U.S.A. 112, 2984–2989. doi: 10.1073/pnas.1501698112
Xiao, Q., Guo, D., Chen, S. (2019). Application of CRISPR/Cas9-Based Gene Editing in HIV-1/AIDS Therapy. Front. Cell Infect. Microbiol. 9, 69. doi: 10.3389/fcimb.2019.00069
Xie, H., Tang, L., He, X., Liu, X., Zhou, C., Liu, J., et al. (2018). SaCas9 Requires 5’-NNGRRT-3’ PAM for Sufficient Cleavage and Possesses Higher Cleavage Activity Than SpCas9 or FnCpf1 in Human Cells. Biotechnol. J. 13, e1700561. doi: 10.1002/biot.201700561
Ye, L., Wang, J., Beyer, A. I., Teque, F., Cradick, T. J., Qi, Z., et al. (2014). Seamless Modification of Wild-Type Induced Pluripotent Stem Cells to the Natural CCR5Δ32 Mutation Confers Resistance to HIV Infection. Proc. Natl. Acad. Sci. U.S.A. 111 (26), 9591–9596. doi: 10.1073/pnas.1407473111
Yin, H., Kanasty, R. L., Eltoukhy, A. A., Vegas, A. J., Dorkin, J. R., Anderson, D. G. (2014). Non-Viral Vectors for Gene-Based Therapy. Nat. Rev. Genet. 15, 541–555. doi: 10.1038/nrg3763
Yin, H., Kauffman, K. J., Anderson, D. G. (2017). Delivery Technologies for Genome Editing. Nat. Rev. Drug Discov. 16, 387–399. doi: 10.1038/nrd.2016.280
Yoshiba, T., Saga, Y., Urabe, M., Uchibori, R., Matsubara, S., Fujiwara, H., et al. (2019). CRISPR/Cas9-Mediated Cervical Cancer Treatment Targeting Human Papillomavirus E6. Oncol. Lett. 17, 2197–2206. doi: 10.3892/ol.2018.9815
Yu, L., Hu, Z., Gao, C., Feng, B., Wang, H. (2017). Deletion of HPV18 E6 and E7 Genes Using Dual sgRNA-Directed CRISPR/Cas9 Inhibits Growth of Cervical Cancer Cells. Int. J. Clin. Exp. Med. 10, 9206–9213.
Yu, L., Wang, X., Zhu, D., Ding, W., Wang, L., Zhang, C., et al. (2014). Disruption of Human Papillomavirus 16 E6 Gene by Clustered Regularly Interspaced Short Palindromic Repeat/Cas System in Human Cervical Cancer Cells. Onco. Targets Ther. 8, 37–44. doi: 10.2147/OTT.S64092
Zheng, Q., Bai, L., Zheng, S., Liu, M., Zhang, J., Wang, T., et al. (2017). Efficient Inhibition of Duck Hepatitis B Virus DNA by the CRISPR/Cas9 System. Mol. Med. Rep. 16, 7199–7204. doi: 10.3892/mmr.2017.7518
Zhen, S., Hua, L., Liu, Y. H., Gao, L. C., Fu, J., Wan, D. Y., et al. (2015). Harnessing the Clustered Regularly Interspaced Short Palindromic Repeat (CRISPR)/CRISPR-Associated Cas9 System to Disrupt the Hepatitis B Virus. Gene Ther. 22, 404–412. doi: 10.1038/gt.2015.2
Zhen, S., Hua, L., Takahashi, Y., Narita, S., Liu, Y. H., Li, Y. (2014). In Vitro and In Vivo Growth Suppression of Human Papillomavirus 16-Positive Cervical Cancer Cells by CRISPR/Cas9. Biochem. Biophys. Res. Commun. 450, 1422–1426. doi: 10.1016/j.bbrc.2014.07.014
Zhen, S., Lu, J., Liu, Y. H., Chen, W., Li, X. (2020). Synergistic Antitumor Effect on Cervical Cancer by Rational Combination of PD1 Blockade and CRISPR-Cas9-Mediated HPV Knockout. Cancer Gene Ther. 27, 168–178. doi: 10.1038/s41417-019-0131-9
Zhen, S., Lu, J. J., Wang, L. J., Sun, X. M., Zhang, J. Q., Li, X., et al. (2016). In Vitro and In Vivo Synergistic Therapeutic Effect of Cisplatin With Human Papillomavirus16 E6/E7 CRISPR/Cas9 on Cervical Cancer Cell Line. Transl. Oncol. 9, 498–504. doi: 10.1016/j.tranon.2016.10.002
Zhou, J., Shen, B., Zhang, W., Wang, J., Yang, J., Chen, L., et al. (2014). One-Step Generation of Different Immunodeficient Mice With Multiple Gene Modifications by CRISPR/Cas9 Mediated Genome Engineering. Int. J. Biochem. Cell Biol. 46, 49–55. doi: 10.1016/j.biocel.2013.10.010
Zhu, W., Lei, R., Duff, Y. L., Li, J., Guo, F., Wainberg, M. A., et al. (2015). The CRISPR/Cas9 System Inactivates Latent HIV-1 Proviral DNA. Retrovirology 12 (1), 22–22. doi: 10.1186/s12977-015-0150-z
Zhu, W., Xie, K., Xu, Y., Wang, L., Chen, K., Zhang, L., et al. (2016). CRISPR/Cas9 Produces Anti-Hepatitis B Virus Effect in Hepatoma Cells and Transgenic Mouse. Virus Res. 217, 125–132. doi: 10.1016/j.virusres.2016.04.003
Keywords: CRISPR/Cas9, delivery mode, HIV, HBV, HPV, infectious viruses, off-target effects
Citation: Lin H, Li G, Peng X, Deng A, Ye L, Shi L, Wang T and He J (2021) The Use of CRISPR/Cas9 as a Tool to Study Human Infectious Viruses. Front. Cell. Infect. Microbiol. 11:590989. doi: 10.3389/fcimb.2021.590989
Received: 03 August 2020; Accepted: 04 August 2021;
Published: 27 August 2021.
Edited by:
Gilles Darcis, University Hospital Center of Liège, BelgiumReviewed by:
Damien Ferhadian, National Institute of Allergy and Infectious Diseases, National Institutes of Health (NIH), United StatesDaniela Toro-Ascuy, Autonomous University of Chile, Chile
Copyright © 2021 Lin, Li, Peng, Deng, Ye, Shi, Wang and He. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Tuanmei Wang, MTM5NjMwODg2M0BxcS5jb20=; Jun He, MTkyNTI3MDJAcXEuY29t
†These authors have contributed equally to this work and share first authorship
 Huafeng Lin
Huafeng Lin Gang Li3†
Gang Li3†