
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Cell. Infect. Microbiol. , 28 January 2021
Sec. Microbiome in Health and Disease
Volume 10 - 2020 | https://doi.org/10.3389/fcimb.2020.619075
 Elisavet Stavropoulou1,2*
Elisavet Stavropoulou1,2* Konstantia Kantartzi3
Konstantia Kantartzi3 Christina Tsigalou4
Christina Tsigalou4 Theocharis Konstantinidis4
Theocharis Konstantinidis4 Chrissoula Voidarou5
Chrissoula Voidarou5 Theodoros Konstantinidis6
Theodoros Konstantinidis6 Eugenia Bezirtzoglou6
Eugenia Bezirtzoglou6Albeit the lungs were thought to be sterile, recent scientific data reported a microbial microbiota in the lungs of healthy individuals. Apparently, new developments in technological approachesincluding genome sequencing methodologies contributed in the identification of the microbiota and shed light on the role of the gut and lung microbiomes in the development of respiratory diseases. Moreover, knowledge of the human microbiome in health may act as a tool for evaluating characteristic shifts in the case of disease. This review paper discusses the development of respiratory disease linked to the intestinal dysbiosis which influences the lung immunity and microbiome. The gastrointestinal–lung dialogue provides interesting aspects in the pathogenesis of the respiratory diseases. Lastly, we were further interested on the role of this interconnection in the progression and physiopathology of newly emergedCOVID-19.
The function and structure of the microbial communities are determined by complex microbial interconnections and network patterns of unique microbiomes. The microbiome is a community of microorganisms colonizing a particular micro-environment in or out of the human body. Additionally, they participate actively in the metabolism and establish potent positive or negative interactions and relationships such as synergism, commensalism, parasitism, antagonism, and other which could explain to some extent the genetic diversity of microbial populations. Yet, following mutations or selective pressure functional microbial genes alter their functionality in the environment (Cordero and Polz, 2014), and via adaptive genes can increase colonizing ability (Hughes et al., 2008). Meanwhile, it was shown that phylogenetically close taxa were more frequently found in the same microenvironment (Chaffron et al., 2010).
Without any doubt, unraveling and exploring the involved microbial patterns and getting a better knowledge of the microbiota profile should clarify the role of the microbiome in health and disease and should lead to the development of more effective or even alternative therapeutic strategies. Outbreaks of various diseases seem to be more common, aggressive, and dangerous during the last years due to the changing environment and the climate warming. New pathogenic bacteria appeared and more than 60% of them originated from animals. Moreover, changes in land and agriculture practices and deforestation alter the environment. Mosquitoes, ticks, and other vector-borne insects are in a rise in the ‘previously’ mild climate European countries due to the shifting climate patterns which lead species to change their habitats. “Forgotten diseases” such as yaws and anthrax showed an emergence. For example, the Nipah virus causing severe encephalitis crossed the species border and passed from animal farms to humans in Malaysia in 1999 (Ang et al., 2018). On the other hand, calamitous rainfalls following climate shifts skyrocket rodent population and an outbreak of pulmonary syndrome caused by Hantavirus emerged in Panama in 1999 (Bayard et al., 2004). It is believed that more than 3,200 coronaviruses species are found in animal reservoirs such as bats and birds awaiting the favorable time to cross the species level and then pass to the humans (Shuman, 2010). Coronaviruses present a large range of disease states extending from simple flu-like illness and gastrointestinal disease to a severe acute respiratory syndrome (SARS). The novel coronavirus has spread rapidly to multiple countries and has been declared a pandemic by the World Health Organization showing the ‘terrifying awake of the nature’ over human civilization and politics (WHO). The most common symptoms of COVID-19 disease comprise mild fever, dry cough, fatigue, anosmia, sore throat, and diarrhea (Ferreira-Santos et al., 2020). However, there are an important number of disease carriers that silently spread the disease. Lung infection observed following SARS-CoV-2 virus invasion could finally lead to death, and the main complications include pneumonia, acute respiratory failure, acute respiratory distress syndrome, acute kidney damage complicated with acute liver damage and septic shock (Huang et al., 2020).
As stated, the COVID-19 infection caused specifically a broad spectrum of severe respiratory diseases and uncertainly the lung microbiome may play an important role to the development of the disease (Yuki et al., 2020). Data used for the present review were identified by a Medline database of systematic reviews and peer-review articles published in English since 1997. However, most articles concerning the main developed concept were recent publications. Articles that were consulted are included in the list of references if they presented an original approach. The main keywords used were: Gut, Lung, Microbiome, Lung microbiome, Gut–Lung axis, Lung in Health and Disease. Internet sites of paramount interest based mostly on the above keywords were identified such as WHO, NIH, Wiley on line library, Science Direct. Finally, important references mentioned in the selected articles were also explored and taken into account.
The subject of this review is divided by subheadings in order to focus in the different questions and thus to provide a concise and precise description of the current knowledge.
In order to improve understanding of the microbial flora involved in human health and disease, scientists proceeded to the characterization of the microbial communities of healthy individuals, across different body sites on the human body: nasal passages, oral cavity, skin, gastrointestinal tract, and urogenital tract. New technologies and specifically the 16S rRNA sequencing was applied to identify the complexity of microbial communities in the human body (NIH Human Microbiome Project - Home). The metagenomics Whole Genome Shotgun (WGS) sequencing brought to light the complexity of the human pathways and functions in relation to the human microbiome.
As known the most studied microbiome is the gut microbiome. Little was known on the lung microbiome which was considered as sterile in multiple studies and textbooks going back often without citations (Cox et al., 2019). The human newborn is deprived of bacteria before birth. The establishment of normal microflora is a continuous and complex pattern which initiates at delivery and goes on through consecutive stages under the influence of external and inner factors (Adlerberth, 2009; Bezirtzoglou and Stavropoulou, 2011; Arumugam et al., 2011). During vaginal delivery, bacteria of maternal origin colonize the newborn oral cavity (Lif Holgerson et al., 2011). Certainly, oral bacteria enrich and shape the lung microbiome (Segal et al., 2016). It was observed that in healthy people, bacterial DNA of several oral bacterial species, such as Prevotella and Veillonella was found in the lower respiratory tract owing to the topographical continuity (Charlson et al., 2011) as the oropharynx and the tracheobronchial tree are adjacent and continuous to the oral cavity (Mammen et al., 2000). Moreover, the exposure pattern during the neonatal period influence the microbial colonization and species variation (Bezirtzoglou, 1997; Rutayisire et al., 2016). The dominant genus found in lung are Prevotella, Veillonella, Streptococcus (Dickson et al., 2016a), Pseudomonas, Fusobacteria and rarely Haemophilus and Neisseria (Beck et al., 2012). These genera are easily colonizing the oxygen rich, damp ciliated mucosa of the larynx and the tracheobronchial tree in continuity to the oral purlieu. Yet, due to the air passage, mucus is continuously enriched with microorganisms. However, the respiratory mucus embedded with lipid-rich surfactants showed bacteriostatic effects against several bacteria and also it seems that there is an eternal fight between bacteria and alveolar macrophages in the lung (Wu et al., 2003).
In terms of phylum, varied phylogenetically microbial populations were found: Proteobacteria, Firmicutes, Fusobacteria, Actinobacteria and Bacteroidetes (Charlson et al., 2011; Moffatt and Cookson, 2017). Concerning the mycobiome in health individuals, Eremotheriumsinecaudum, Vanderwaltozymapolyspora, and Systenostrema alba of the Saccharomycetaceae family as well as several terrestrial microsporidia were isolated (Nguyen et al.).
Thus assuming, it is generally accepted actually that there is a lung microbiome in balance with different factors such as microbial penetration and expulsion from the airways and even growth of colonizing bacteria adapted to the local conditions and environment (Dickson et al., 2014).
The lungs are exposed to multiple hazards on a daily base. Virtually, the lung cells play a role in assessing whether they will respond or not. Originally, the upper respiratory tract role is dedicated to preventing entry of noxious particles. When sizeable particles deposit in the nasopharynx and tonsillar regions, they are cleared mechanically by coughing or sneezing. Besides this, the remaining non-eliminated particles are expelled smoothly via rhythmic movements of microscopic cilia to the upper airways (Beutler, 2004).
Antimicrobial peptides (AMPs) LL37 are expressed in the epithelial cells of the upper respiratory tract. AMPs are known to possess an antimicrobial activity against microorganisms either by direct binding to the bacterial surface or binding following microbial opsonization to be recognized by innate immune cells (Wang, 2014). The alveoli are tiny air sacs distributing oxygen within the body. They are located at the terminal branches of the lungs where most of the gaseous exchange occurs. Immune cells are poorly represented in the alveoli, and they are mainly consisting of alveolar macrophages (AMs) which provide the primary phagocytic activity against microorganisms without triggering inflammatory responses (Martin, 2000; Martin and Frevert, 2005).
Except for the upper airspaces part, the lung immune system is presented by the intraepithelial lymphoid tissue of lungs (ILTL). The immune response of ILTL in all stages is orchestrated by dendritic cells (DCs). Furthermore, the triad of DCs, lung microbiome and AM are actively participating in the bacterial recognition. T cells (predominantly cytotoxic), B lymphocytes, neutrophils, and rarely mast cells are observed in ILTL (Freeman and Curtis, 2017).
As known, epithelial cells are also involved in lung immunity by secretion of chemokines, cytokines, and antimicrobial compounds (Lacy, 2015). During dysbiosis, AM phagocytes adhere to the opsonins in order to facilitate phagocytosis as opsonized encapsulated bacteria should be ingested with more difficulty (Gordon and Rice, 1988; Arango Duque and Descoteaux, 2014; Murray and Stow, 2014). Additionally, immature DCs migrate into tiny airways, where after activation by AM, promote a pro-inflammatory microenvironment (Kawamura et al., 2005; van Rijt et al., 2011; Shaykhiev and Crystal, 2013).
Neutrophils and macrophages are involved in the innate immunity as first line of non-specific defense against pathogens. Their role is to engulf and destroy the pathogenic bacteria. Neutrophils’ recruitment and activation are observed in response to inflammation status. However, macrophages exhibit decreased phagocytosis in the large lung airways during the progression of chronic obstructive pulmonary disease (COPD) (Bu et al., 2020). Dropping of the phagocytic capacity of macrophages impair the lung function and produce a chronic inflammation status (Bu et al., 2020).
At the same time, in the small airways macrophages demonstrate high pro-inflammatory potential (Shapiro, 1999; Bu et al., 2020) (Figure 1). Moreover, Natural killer T (NKT) cells, eosinophils, and mast cells are incriminated to have a complementary function in the inflammatory process of small and mediate alveolar tissue that leads to chronic airway inflammation (O’byrne and Postma, 1999). Likewise, activated neutrophils are able to release Neutrophil Extracellular Traps (NETs) which promote a proteolytic microenvironment through enhanced expression of IL-17 resulting in the fibrotic repair of small airways (Bosch and Ramos-Casals, 2014; Chrysanthopoulou et al., 2014).
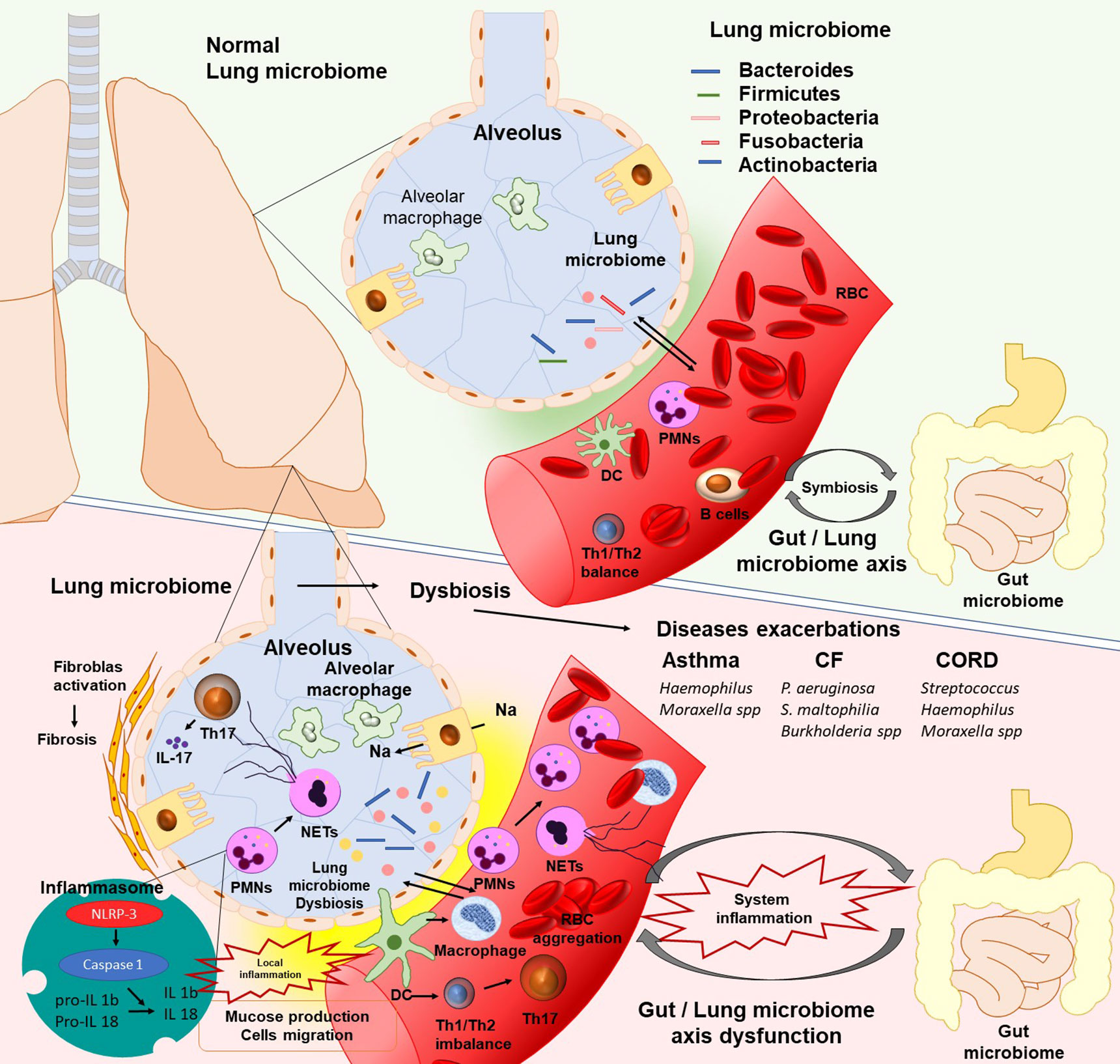
Figure 1 The existence of lung microbiota plays a critical role in lung homeostasis. In human’s lungs the bacterial load is increased from neonates to adulthood and contain Proteobacteria, Firmicutes, Fusobacteria, Bacteroidetes, and Actinobacteria bacterial phyla. Lung microbiota, in the same manner as gut microbiota, can promote the polarization of naïve T cells in the lungs from Th2 to Th1 to protect against asthma and allergy. Moreover, microbiota promote the differentiation of alveolar macrophages, inhibit the exaggerated inflammatory response, and promote protection against pathogens. The lung microbiome dysbiosis leads to dendritic cells activation and antigen presentation. Immune cells respond to microbial colonization through the pattern recognition receptors (PRRs). Activated cells migrate into the tissue, produce series of cytokines such as IL-1b and IL-8, and release Neutrophil Extracellular Traps (NETs), which contribute to local inflammatory response. Moreover, alterations in cytokines, especially IL 17(NETs or Th17 inducted), promote pathologic fibrotic remodeling (PMNs, polymorphonuclear leukocytes; DC, Dendritic cells; IL, Interleukins; CF, cystic fibrosis; CORD, Chronic obstructive pulmonary disease).
Epithelial cells seems to play a crucial role in the initiation of the immune response as they recognize viral antigens through cytoplasmic Pattern Recognition Receptors PRR stimulation, for instance Toll like receptors (TLRs) and intracellular sensors, including Retinoic acid-Inducible Gene-I-like receptors(RIG-I-like receptors) (Xiang and Fan, 2010; Whitsett and Alenghat, 2015). As a result, coupling induces a signaling avalanche effecting in the upregulation of type I and III interferons and initiating the inflammatory response and the differentiation of adaptive immunity involved cells (Xiang and Fan, 2010; Whitsett and Alenghat, 2015). Alveolar macrophages and DCs contribute to the secretion of the cytokines and chemokines (Kolli et al., 2014). Finally, PRR coupling and cytokine secretion induce DC maturation and adaptive immune response (Kranzer et al., 2004).
The adaptive immunity response is specific to the pathogen by clonal expansion of T and B lymphocytes and consists of a second line defense (Adams et al., 2020). This effect is highly specific and long maintained by memory T cells. Affinity maturation is expressed on TCRs found on T lymphocytes leading to a more specific antigen binding capacity (Zhu et al., 2015).
T lymphocytes carry the CD4 or CD8 receptors (Gao et al., 2020). The CD4+ T cells are known as T helper (Th) cells due to their capacity via cytokine production to stimulate immune cells (Zhu et al., 2015). Yet, the CD4 regulatory T cells (Tregs) are able to regulate immune feedback. Particularly, Tregs via the production of IL-10 suppress T cell differentiation (Ding et al., 2012). The CD8 T cell receptors are known as cytotoxic T cells due to their ability to produce molecules and cytokines (Altmann, 2018). This occurs in the lymph nodes of lung parenchyma, bronchioles, and trachea. CD4 T cells are divided in subsets following the secreted cytokines [T helper type 1 (Th1) cells and T helper type 2 (Th2) cells] which showed different functions (Gao et al., 2020). These cytokine production ends to a consistent local host protective response. However, thereafter most CD4+ T cells die (necrosis, apoptosis), and few CD+ cells are present as memory cells in the host (Maly and Schirmer, 2015).
Concerning the B cells’ specific response which is taking place in lymphoid tissues such as lymph nodes, mucosa-associated lymphoid tissue of broches and nose, it seems to promote IgA (Ladjemi et al., 2015). Affinity maturation is not usually expressed as B cells are short-lived (Zhu et al., 2015). The basic differentiation process of B cell is dependent on T cells and outcomes in the high-affinity antibody cells and memory B cells (Schuurman and Quesniaux, 1999; Zhu et al., 2015). Specifically, mature DCs fitting out viral antigens translocate to lymphoid tissues (Tang et al., 2010). These antigens are recognized, and differentiation proliferation processes are initiated (Tang et al., 2010). Specifically, antigen recognition is achieved through the CD4+ Th cells presented by the B cell via major histocompatibility complex class II and alternate Immunoglobulins isotype classes (IgM to IgG and IgA) (Couture et al., 2019). At this stage of the process, several B cells participate in the Germinal Center (GC) followed by hyper-mutation and affinity maturation (Anderson et al., 2009; Zhu et al., 2015). In this vein, affinity maturation is stimulated by CD4+ follicular helper T cells (TFH cells) (Zhu et al., 2015). TFH cells possess a pool of markers, such as chemokine receptor CXCR5, ICOS, CD28, PD-1, CD40L, and SAP as well as their driven specifying transcription factor Bcl-6 (Suh, 2015). TFH cells are found in GCs and DCs, and this permits their strait relation to B cells (Chen et al., 2012; Couture et al., 2019). TFH cells’ signals induce ulterior selective differentiation of B cells to low-affinity suboptimal clones (Chen et al., 2012; Suh, 2015). As stated TFH cells express multiple markers, shift immunoglobulins isotype classes to IgA thus stimulating Bcl-6 expression in B cells, and resulting in the memory generation (Suh, 2015; Zhu et al., 2015). Those TFH cells are responsible for a durable protective antibody response and in this way should be of paramount importance in the vaccines design (Suh, 2015; Zhu et al., 2015). Therefore, the cells TFHs role are not fully investigated. Recent research investigates novel ways as the microRNA cluster miR-17–92, for TFHs’ function, differentiation and migration to B cells (Masopust and Schenkel, 2013; Chiu and Openshaw, 2015). Moreover, it seems that microRNA cluster miR-17–92 inactivation impedes their functional role and the production of high affinity antibodies (Sakai et al., 2017). Without any doubt, investigation of T cell-specific expression of the microRNA cluster miR-17–92 and its importance on TFHs functional role should be promising in the development of effective vaccines (Baumjohann et al., 2013; Skinner et al., 2014).
The gut–lung axis (GLA) consists of a bidirectional communication between the two organs (Enaud et al., 2020). Modulation of the intestinal microbiota may cause lung disease. Gut microbes are recognized by the cells of the immune host by inducing the production of cytokines. After lung inflammation, the bloodstream is invaded by metabolites, immune signals, bacteria, and bacterial products that interact between the lungs and the intestine. In fact, the gut–lung axis concept implicates not only host–microbe interactions but also microbe–microbe interactions (Enaud et al., 2020). From an aspect, the gut–lung intercommunication maintains the host homeostasis in healthy individuals (Enaud et al., 2020).
However, most studies investigated the potential of the bacterial microbiota (bacteriobiota); it is of note that there is a fungal microbiota (mycobiota) that remains less studied (Nguyen et al.) as well as a viral component microbiota (virobiota). Viruses seem to be involved frequently in the development of multiple respiratory diseases (Mitchell and Glanville, 2018), and this is the case of the recent pandemic COVID-19 which affects mainly the respiratory system. Recent studies report that there is another communication between these three kingdoms (Enaud et al., 2020). Pseudomonas aeruginosa produces volatile sulfur compounds enhancing growth of Aspergillus fumigatus (Briard et al., 2019). There is a reciprocal communication between Streptococcus and Candida increasing biofilm formation or enhancing Candida pathogenicity (Diaz et al., 2012).
While the prevalent bacterial phyla are identical in lung and gut microbiomes, at the species level they are different. It is stated that a balanced intestinal microbiota is crucial for our health as gut microbiota possess an enormous metabolic profile that determines host health (McAleer and Kolls, 2018). Recent studies disclose the association of gut microbiota dysbiosis with multiple disease states (McAleer and Kolls, 2018). The GLA axis consists of a continuous interconnection between gut and lung that allows passage of gut microbial metabolites, endotoxins, cytokines, and hormones through the bloodstream to the lung. However, when lung inflammation occurs, changes in the gut microbiota are observed (Dumas et al., 2018) proving the bidirectional status. Without any doubt, understanding the impact of the gut microbiota on remote organs and systems is of accrued interest. However, it is not clarified yet if it is involved in the etiology or development of the lung disease (Zhang et al., 2020).
As known, antibiotics given in early life cause alterations of the gut communities and predispose to the development of allergy (Russell et al., 2012). The approach of germ-free animals is of high interest as they are bacteriologically sterile. Germ-free mice are extremely susceptible to acute lung infections caused by Pseudomonas aeruginosa, Streptococcus pneumoniae, and Klebsiella pneumoniae (Brown et al., 2017). In this vein, there is evidence that the gut microbiota amplifies the respiratory system defense potential via the granulocyte-macrophage colony-stimulating factor (GM-CSF) signaling (Brown et al., 2017) achieving pathogenic bacteria leveling and elimination through extracellular signal-regulated kinase signaling by alveolar macrophages (Brown et al., 2017). Finally, the intestinal microbiota via interleukin-17A induces increased pulmonary GM-CSF production in response to infection (Brown et al., 2017).
Nucleotide-binding oligomerization domain-like receptors (NOD-like receptors; NLRs) are known to be involved in the microbiota signals to regulate neutrophil functions (Clarke et al., 2010), inflammatory responses (Hergott et al., 2016) and hematopoiesis (Iwamura et al., 2017). The role of the gut microbiota and the receptor Nod2 in enhancing the mucosal adjuvant activity of Cholera Toxin (CT) was stated (Kim et al., 2016) in germ-free animals. Staphylococcus sciuri has proven to possess high Nod2-stimulatory activity. Thus, Staphylococcus sciuri mono-associated to germ-free mice produces a powerful Cholera Toxin (CT) activity (Kim et al., 2016).
As stated, NLR pattern recognition receptors can regulate inflammatory response (Hergott et al., 2016; Brown et al., 2017). Notably, the authors found the Nod2-activating recipients of the gut microbiota to adjust the antibacterial immunity of the lung and immune homeostasis (Brown et al., 2017). M. tuberculosis is the agent of tuberculosis (TB). Recent studies in patients with tuberculosis showed a gut microbiota with predominance of butyrate and propionate-producing bacteria such as Faecalibacterium, Phascolarctobacterium, Eubacterium and Roseburia (Hu et al., 2019a), while a decrease of short-chain fatty acid (SCFA)-producing bacteria was effective (Saitou et al., 2018).Moreover, in the intestine of tuberculosis patients a decrease in amino-acids and vitamins biosynthesis is observed (Saitou et al., 2018).
Prevotella and Lachnospira were detected in low levels in tuberculosis patients (Luo et al., 2017). However, patients undertaken tuberculosis treatment showed important shifts in their gut microbiota as Bacteroides fragilis and Bacteroides OTU230 numbers were found in increased levels, while the phylum Firmicutes and genus Clostridiales were considerably reduced (Hu et al., 2019b). Lastly, Helicobacter pylori was found to protect lung from M. tuberculosis infection (Schuijt et al., 2016; Tarashi et al., 2018).
The gut microbiota has also an impact upon Streptococcus pneumoniae lung infection (Henriques-Normark and Tuomanen, 2013). Recently, the interconnection between lung and Staphylocosccus aureus lung infection was investigated in mast cell-deficient mice. Gut dysbiosis and higher bacterial lung burden were found. Moreover, mast cells’ presence into the lung rebuilts the host protection against S. aureus and gut dysbiosis (Liu et al., 2019).
Respiratory viral infections seem to be influenced by the microbiota balance. Influenzae virus (IV) and respiratory syncytial virus (RSV) in animals showed a decrease in the phylum Firmicutes while Bacteroidetes were increased (Groves et al., 2018). Pneumocystis pneumonia infection in mice without CD4+ T cell modifies significantly the gut microbiota community (Samuelson et al., 2016).
Gut associated bacteria may enter the lungs across the bowel wall via a process called translocation (Mielcarek et al., 2011; Mukherjee and Hanidziar, 2018). The process seems to be enhanced by gut and alveolar permeability (Yacobi et al., 2010). Gut permeability is known to occur during acute intestinal conditions and sepsis (Vaishnavi, 2013). Permeability of the alveoli is observed in Acute Respiratory Distress Syndrome (ARDS) as a result of direct or indirect epithelial injury (Cardinal-Fernández et al., 2017). Critically ill patients’ present increased intestinal and alveolar permeability due eventually to the increased gut–lung translocation (Assimakopoulos et al., 2018). Authors reported (Xu et al., 2019) an interconnection between dysbiosis in critically ill patients and the risk of ARDS development. This interconnection of the gut and lung has been shown in both animal and human studies as discussed (Barcik et al., 2020). Gut-associated lymphoid tissue (GALT) and Inducible bronchus-associated lymphoid tissue (iBALT) seem to have similar morphology and function which is the immune response regulation (Barcik et al., 2020). Their main functions comprise the production and secretion of IgA at the mucosal tissues and cell T helper (Th) and cytotoxic (Tc) responses (Cesta, 2006).
The concept of this interconnection is stated on the fact that shifts in the intestinal microbiome influence lung disease burden and vice versa. Studies state the significance of the immune system maturation in early life (Sokolowska et al., 2018). It is also stated that childhood exposure to microorganisms, the so called hygiene hypothesis protects against allergy and disease (Stiemsma et al., 2015). In humans, histamine produced by intestinal bacteria showed important immunoregulatory functions (Smolinska et al., 2014). It is of note that the local immune reaction produced in GALT and iBALT lymphoid tissues can induce systemic immune responses, withal the mucosal immunity might act as a whole for producing immune response (Cesta, 2006). Epithelial cells are involved in lung immunity by secretion of chemokines, cytokines, and antimicrobial compounds (Whitsett and Alenghat, 2015) leading to modification of the immune response at distal sites (Budden et al., 2017). Yet, intestinal immune cells migrate via the mesenteric lymph system and blood to the lung where they have enhanced functions (Stubbington et al., 2017). The authors observed increase of the gut bacterial population following stimulation of the mouse lungs with lipopolysaccharide (LPS) (Anand and Mande, 2018). Short Chain Fatty acids (SCFAs) produced by the intestinal microbiome following dietary fiber fermentation by bacteria support immunity function as they effect as signaling molecules on resident antigen-presenting cells to attenuate the inflammatory response in the lungs (Anand and Mande, 2018; Cait, 2018).
They can be found in other sites circulating in the blood and thus they showed inhibitory effects on pro-inflammatory responses in the lungs (Stubbington et al., 2017). Moreover, liver appears to impede the innate immune response produced by SCFA ligation to G protein receptors or even by repressing the mevalonate pathway through HMGCoA reductase (Young and Hopkins, 2017). High amounts of SCFAs such as butyrate and propionate in children intestinal microbiota are less likely to develop lung pathologies (Roduit and on behalf of the PASTURE/EFRAIM study group, 2019). Likewise, administration of SCFAs in animals enhance the transcription factor FOXP3 by inhibiting deacetylation of histone, thus bracing T regulatory cells (Tregs) and production of IL-10 (Arpaia et al., 2013). Similarly, studies mentioned that intestinal metabolites such as oxylipins and biogenic amines have pro-inflammatory and anti-inflammatory potential (Ávila-Román et al., 2016).
Segmented filamentous bacteria (SFBs) in the gut microbiome of animals and humans are participating in the immune regulation (Yin et al., 2013; Hedblom et al., 2018). Specifically, SFBs regulate CD4+ T-cell polarization into the Th17 pathway involved in lung fungal infections response (McAleer et al., 2016; Bradley et al., 2017). Recent studies highlight the role of the gut innate lymphoid cells to be associated to the lungs tissue repair following inflammatory signals upon IL-25 (Liu et al., 2018).
Administration of the probiotic Bifidobacterium lactis HN019 (Gill et al., 2001) is correlated to the elevated number of mononuclear leukocytes as well as to an increased phagocytic and lytic activity (Gill et al., 2001). Reversely, in a global study (COPDMAP study) scientists reported that respiratory infections impact upon the intestinal microbiome mediated by Th17 cells (Wang and on behalf of COPDMAP study, 2017). Recently, it is shown that this bidirectional dialogue is not only of concern to the bacterial potential but it is related to fungal one as well (Li X. et al., 2020). The role of resident intestinal macrophages in airway inflammation and fungal dysbiosis is highlighted (Leonardi et al., 2018).
Considering the above, there is an evidence of bidirectional interconnection. Without any doubt gut microbiota holds a key role to regulate host homeostasis and promote resistance to respiratory infections (Brown et al., 2017). However, it is mainly thought to be associated with the PRR basal activity, specific ligands of the microbiota and intestinal dysbiosis (Chu and Mazmanian, 2013); there is a long way for clarifying the involved mechanisms and pathways.
Each one of us has our own unique microbiota as multiple factors such as diet, environment, ethnicity, hormonal status, hygienic habits which are crucial determinants to its composition (Bezirtzoglou, 1997). When microbial populations are disturbed, a negative impact called dysbiosis is produced. Similarly, higher abundance and species variation are observed in chronic disease states of the respiratory tract. Moreover, there is a shift in microbial populations. It is reported that several fungi associated with the intestinal dysbiosis could enhance the severity of asthma as intestine and lung communicate and work in tandem (Turturice et al., 2017). The study was carried out based on cytokine markers of inflammation (IFN-γ, IL-17F, TNF-α and MIP-1β and G-CSF) which were increased. Furthermore, the patient microbiome was diversified, and species such as Actinomyces ondontolyticus, Actinomyces oral taxon 180, Neisseria meningitidis, and Streptococcus pneumoniae were dominant (Mortaz et al., 2013). It must not also be neglected that antibiotic overuse or misuse could disrupt the microbiota balance (Bezirtzoglou et al., 2008).
Proteobacteria including genus of Haemophilus, Neisseria, Pseudomonas, Rickettsia and Moraxella species were pronounced in asthma and associated usually with the uncontrolled asthma. Surprisingly, Firmicutes with the genus Lactobacillus were isolated in several asthmatic patients (Park et al., 2014) as well as the genus Clostridium in children with airway allergies (Chiu et al., 2019). Serum and sputum inflammatory cytokines may be used as markers between bronchial asthma and chronic obstructive pulmonary disease (COPD) (Bai et al., 2019). IL-4, IL-5, IL-9, and IL-13 were shown in increasing levels in COPD, patients while in asthmatic recipients, the levels of TNF-α, IL-1β and IL-6 were found considerably increased (Bai et al., 2019). Asthma seems also to be related with particular phenotypes (Kuruvilla et al., 2019). In elderly asthmatic patients serum IL-33 and IL-31 levels were found lower, and this may contribute to less Th2 phenotype and asthma severity (Ulambayar et al., 2018). Bronchoalveolar lavage (BAL) fluids in asthmatic recipients showed elevated levels of IL-1RA, IL-1α, IL-1β, IL-2Rα, IL-5, IL-6, IL-7, IL-8, G-CSF, GROα (CXCL1), MIP-1β (CCL4), MIG (CXCL9), RANTES (CCL5) and TRAIL, eosinophils and neutrophils (Hosoki et al., 2015). Overall, neutrophils and IL-8 in BAL fluids seem to be the unique inflammatory markers to distinguish between controlled and uncontrolled asthma and disease severity (Hosoki et al., 2015).
Over-presented in asthmatic individuals was Malassezia genus (Nguyen et al.; Delhaes et al., 2012; van Woerden et al., 2013). Aspergillus penicillium and Cladosporium were also abundant in asthmatic cases (van Woerden et al., 2013). Several environmental Basidomycota species such as Psathyrella candolleana, Grifolasordulenta, and Termitomycesclypeatus were revealed from asthmatic individuals (Nguyen et al.).
We stated the importance of the newborn colonization (Bezirtzoglou and Stavropoulou, 2011). It is emphasized that early gut colonization by beneficial organisms such as Lactobacilli and Bifidobacteria is protecting us against different types of diseases (Ismail et al., 2016; Salminen et al., 2016). Early and elevated colonization by the bacterial genus of Clostridium, Bacteroides fragilis, Streptococcus and fungal populations of Saccharomyces and Pichia kudriavzenii predispose to asthmatic conditions (Nguyen et al.), as well as low amounts of the beneficial Bifidobacterium (Nguyen et al.; Ismail et al., 2016).
Gut bacterial burden is under the control of multiple factors, such as nutrition, environment, hormonal status, ageing (Turnbaugh et al., 2007), and are referred as gut microbiome. The gut microbiome is composed of trillions of bacteria and plays an important role in health and disease (Turnbaugh et al., 2007). The notion that the lung is sterile is abandoned (Huffnagle, 2015) as new molecular techniques gave evidence of bacterial populations in the lung (Huffnagle, 2015). However, bacterial concentration in the lung is shown humble compared to the gut communities (Hilty et al., 2010). Nevertheless, this cannot be generalized to the fungi kingdom as commensal fungi showed a protective effect on both local and systemic immunity due to their fungal wall mannans (Abt and Artis, 2013). As known sepsis is a potentially life-threatening state as a response to an infection due to chemicals produced into the blood against an infection. There is evidence in animals and humans that intestinal microbiota provide bacteria to the lung as abundance of Bacteroides observed in the lung following sepsis (Dickson et al., 2016b).
Shifts in microbial profile and decrease in the lung microbiota diversity is observed in chronic obstructive pulmonary disease (COPD) (Huang et al., 2010). The 16S rRNA PhyloChip analysis makes known that more than 1,200 bacterial taxa belonging to 140 distinct families were detected in the airways (Huang et al., 2010). The phylum Proteobacteria including families of Pseudomonadaceae, Enterobacteriaceae, and Helicobacteraceae was predominant (Wang et al., 2016). In intubated patients. Haemophilus influenzae and Pseudomonas aeruginosa were also found in COPD exacerbations (Huang et al., 2010). Moreover, rise in intestinal permeability and release of adrenal hormone metabolites were linked to short or long term mortality in COPD patients (Zurfluh et al., 2018).
A decrease in the lung microbiota diversity is observed in cystic fibrosis patients (Cox et al., 2010). Diversification was more pronounced in younger individuals than older ones with cystic fibrosis. Bacterial populations involved are members of Pseudomonadaceae, Xanthomonadaceae, Moraxellaceae and Enterobacteriaceae (Cox et al., 2010). The genus Streptococcus, Prevotella, Rothia, Veillonella, Acintomyces, Neisseria, Haemophilus, Gemellaare were isolated in cystic fibrosis during exacerbations in pediatric patients (Worlitzsch et al., 2009). However, Pseudomonas aeruginosa and Staphylococcus aureus are incriminated in most cystic fibrosis cases (Worlitzsch et al., 2009). Streptococcus, Prevotella, Rothia, Veillonella, Acinetomyces, Pseudomonas are more frequently involved in cystic fibrosis adult cases (Delhaes et al., 2012). Over-presented in cystic fibrosis were Aspergillus penicillium, Aspergillus fumigatus, Malassezia, Candida albicans and Candida parapsilosis (van Woerden et al., 2013). Cladosporium cladosporiodes has been isolated in immunocompromised patients and Malassezia pachydermatitis was also found in immunocompromised hosts and in atopic dermatitis (Nguyen et al.). Aspergillus fumigatus was associated with corticoid treatment (Fraczek et al., 2019). The authors stated a shift in bacterial and fungal population dominated by Proteobacteria phylum which includes many Gram-negative bacteria (Dickson et al., 2016b).
Our specific interest is aroused by the novel beta-coronavirus SARS-CoV-2 affecting seriously the respiratory system and causing an acute respiratory disease known as COVID-19. SARS-CoV-2 has recently taken over our attention due to the COVID-19 pandemic because of its contagiousness as well as unexpected mortality rates. SARS-CoV-2 belongs to the Coronaviridae which is a family of enveloped, positive-sense, single-stranded RNA (+ssRNA) viruses. SARS-CoV-2 contains four basic structural glycoproteins: spike (S), membrane (M), envelope (E), and nucleocapsid (N) (de Wit et al., 2016).
As little is known about the infection process of this novel virus, we turned our particular attention to this pathogen. As of November 2020, the number of COVID-19 confirmed cases exceeded 64 millions of cases with more than 1,500,000 deaths. The large majority of diseased seems to develop a mild disease or even being asymptomatic as they mount a suitable immune response. Nevertheless, several patients develop severe clinical images, and this is linked to the insufficient response of their immune system and underlying pathologies. In fact, the mechanisms involved in the progression of the disease remains yet obscure.
As stated previously, SARS-CoV-2 contains four structural glycoproteins. Among them the envelop glycoprotein E, the nucleocapsid protein N, and the membrane glycoprotein M are responsible for viral assemblage and binding to the host cells (Li, 2012).
The human angiotensin converting enzyme 2 (ACE2) was identified as the receptor for SARS-CoV-2 virus which permits access to the endocytic uptake (Petersen et al., 2020). This enzyme is expressed on the human lungs’ epithelium and catalyzes the conversion of angiotensin II (Letko et al., 2020; Ou et al., 2020). ACE 2 can be found in a wide variety of human tissues, such as kidney, intestine, heart, thyroid, adipose tissue and testis where viremia could happened in case of contamination (Li M. et al., 2020). Neurological symptoms are also reported due to CNS affection (Abboud et al., 2020).
The role of the spike glycoprotein S acts in connection with angiotensin. Specifically, this glycoprotein is cleaved (Anand et al., 2020) by a cellular derived protease into two glucoproteins S1 and S2. The glycoprotein S1 binds to the angiotensin, while S2 is activated by the host TMPRSS2 (Transmembrane Serine Protease) resulting in a membrane fusion which permits the virion to enter the host cells via receptor-mediated endocytosis (Petersen et al., 2020). In contrast to the majority of coronaviruses, SARS-CoV-2 expresses a furin-like protease. It is believed that this furin-like protease may contribute to the widened cell tropism and enhanced transmissibility of the virus (Shapiro et al., 1997). Moreover, it was shown that a Two-Pore Channel (TPC2) is crucial for the entry of SARS-CoV-2 into host cells (Ou et al., 2020). The Two-Pore Channels (TPCs) are entry channels into the endo-lysosomal system. In fact, the intracellular messenger nicotinic acid adenine dinucleotide phosphate (NAADP) mobilizes calcium from acidic organelles through two-pore channels (Calcraft et al., 2009). Moreover, the authors demonstrated that S-protein via the Transmembrane Serine Protease 2 (TMPRSS2) facilitates SARS-CoV-2 entry into host cells (Baughn et al., 2020; Hoffmann et al., 2020).
Following attachment to the ACE 2 receptor, there is membrane and viral fusion with the aid of glucoprotein S as stated previously (Petersen et al., 2020) which permits SARS-CoV-2 virus entry into the cells (Masters, 2006). After the virus enters the host cells, viral material is replicated by the aid of RNA polymerase, and another viral RNA released enzymes (Ratia et al., 2006). As known viral genome encodes the replicases PP1a and PP1ab which are cleaved by the 3CLPro (3C-Like Protease)and the PLPro (Parpain-Like Protease) into 16 nonstructural proteins as RNA dependent RNA polymerases (RdRp) shaping the replication complex (Ratia et al., 2006). During genome translation coronavirus replication induces ribosome frameshifting (Masters, 2006) before reassembly, encapsulation and exocytosis of the mature virions out of the host cell able to infect new host cells.
SARS-CoV-2 as an antigen exposed to the host Antigen Presenting Cells (APCs) produces releasing of inflammatory mediator cytokines such as interleukin-1 (IL-1), interleukin-6 (IL-6), CXCL-10 and tumor necrosis factor alpha (TNF-alpha). These cytokines are producing an excessive pro-inflammatory response called cytokine storm to the host which damages lung epithelium (Sanghai and Tranmer, 2020). Moreover, pathogens induce a pro-inflammatory response in epithelial cells by activating the transcription Nuclear Factor-κB (NF-kB) which regulates innate and adaptive immune functions, inflammation, and cell proliferation (Liu et al., 2017). It is of note that SARS-CoV-2 is a cytopathic virus which damages directly the alveolar epithelium in the lungs and induces epithelial cell death. This damage may occur to multiple organs causing a multi-organ failure of the host (Xu D. et al., 2020).
As it is known the expression of Pattern Recognition Receptors (PRRs) is enhanced in the lung cells during inflammation. As a response, macrophages, monocytes, and neutrophils increase levels of PAMPs (Pathogen-Associated Molecular patterns) and DAMPs (Danger-Associated Molecular Patterns) (Mogensen, 2009). PAMPs are nucleic acids or glycoproteins recognized by PRRs and expressing cytokines and other co-stimulatory components against the pathogenic virus which activate as well then antigen presenting cells and the specific adaptive immunity (Mogensen, 2009). DAMPs are found intracellularly and participates in the activation of the inflammasome as well as the conversion of proIL-1 to activeIL-1 (Law et al., 2009). Notably, the TLRs (Toll Like Receptors) membrane glycoproteins are the most known PRRs (Law et al., 2009).
On the other side, traditional biomarkers of acute infection such as C-Reactive Protein, ferritin, neutrophils, and leucocytes must be considered. Recent evidence stated that neutrophils and complement are involved in a maladaptive immune response as complement interacts with the platelet/neutrophil extracellular traps (NETs)/thrombin axis which leads to enhanced inflammation, ombotic microangiopathies and high mortality (Skendros et al., 2020). Similarly, the unbalance of the immune system due to the excessive cytokines releasing in response to SARS-CoV-2 invasion leads to an abnormal hypercoagulation with thrombotic events (Jose and Manuel, 2020).
Researchers argued the importance of the gut microbiota in relation to the development of immunity against SARS-CoV-2 and the recovery impact (Xu K. et al., 2020). Intestinal dysbiosis, with a decrease in the beneficial probiotic microbiota, is observed in subjects with COVID-19 (Xu K. et al., 2020).
As stated previously, the gut microbiome impact upon Influenzae virus (IV) and Respiratory Syncytial Virus (RSV) (Groves et al., 2018). Shifts in gut microbiome of COVID-19 patients were effectively (Zuo et al., 2020) characterized by depletion of the beneficial microbiota and accrued levels of opportunistic pathogens. Putrefactive bacteria such as several Clostridium species (C. ramosum, C.hathewayi) and Coprobacillus were present in abundance and correlated to the disease severity (Zuo et al., 2020). In contrast, the beneficial commensals Faecalibacterium prausnitzii, Eubacterium ventriosum Roseburia, Lachnospiraceae, Alistipes onderdonkii and Bacteroides ovatus were found in low levels in COVID-19 patients (Zuo et al., 2020). Patients fecal specimens showed an inverse correlation between SARS-CoV-2 levels and the presence of Bacteroides sp which are involved in downregulation expression of the angiotensin-converting enzyme 2(ACE2) (Zuo et al., 2020). ACE-2 receptors are the entry point into cells for SARS-CoV-2. It is of note that they are expressed in few distinct anatomical seats comprising the gut and the lungs.
The authors stated the accrued levels of Streptococcus in patients’ fecal microbiota to be correlated with infection risk by opportunistic pathogenic bacteria (Khatiwada and Subedi, 2020), such as Rothia, Veillonella, and Actinomyces (Khatiwada and Subedi, 2020).Moreover, opportunistic fungal pathogens such as Candida albicans, Candida auris, and Aspergillus flavus are detected in patients’ microbiome (Khatiwada and Subedi, 2020). The authors stated the value of the gut microbiota in COVID-19 patients as a dynamic diagnostic biomarker and therapeutic tool (Gu et al., 2020).
Other studies of bronchoalveolar lavage fluid, lung post-mortem biopsies and gut microbiota from COVID-19 patients showed important shifts in the gut microbiota balance as a mixed bacterial/fungal infection occurred (Fan et al., 2020).
Increased lung loading with gut bacteria seems to be a predictive marker to the acute respiratory distress syndrome (ARDS) malefic consequences as dysbiosis is taking place dysregulating the immune response and leading to inflammation (Yang et al., 2020). The composition of the gut microbiome may be used as a predictive tool of the disease development and infection severity (Chu and Mazmanian, 2013). Based on the above, scientists developed a proteomic risk score’ (PRS) acting as a predictive tool using machine learning algorithms. Putrefactive bacteria such as Ruminococcus and Clostridium spp were found under dysbiosis in high numbers while beneficial probiotic bacteria such as Lactobacillus and Bifidobacterium were absent (Gou et al., 2020).
Further research focusing on the intestinal–respiratory microbiome interconnection is likely to reveal important aspects into the dynamics of the microbiomes. Getting an in-depth understanding on the role of intestinal dysbiosis may elucidate the pathogenesis of different diseases. Notably, it is crucial to bring more light in the interconnecting reciprocal dialogue between the lung and the gut as its microbial kingdom keeps a significant impact in this interconnection which needs further investigation. Without any doubt, new technologies including high-throughput and genome sequencing methodologies will enrich our knowledge about the role of the gut and lung microbiomes in the development of the respiratory diseases. Getting knowledge of the involved physio-pathological mechanisms will help the medical community to find solutions for the treatment of COVID-19 (Stavropoulou and Bezirtzoglou, 2020).
Medicines inactivating the enzymes involved in these mechanisms should be promising as potential antiviral treatments (Sze et al., 2014). Moreover, considering the role and the importance of the microbiota dynamics and homeostasis, the probiotic approach in the treatment of the respiratory disease seems to keep an important impact as prophylactic or therapeutic agents, especially when the failure of antibiotics occurs.
At this end, we conclude that more basic and clinical studies should be done in order to clarify the role of the physio-pathological mechanisms and human organs and systems dialogues in health and disease.
Conceptualization, original draft preparation, and editing, ES. Formal analysis, TK and KK. Investigation, ES. Resources, CV. Writing—original draft preparation and editing, CT. Editing, ThK. Supervision, original draft preparation, and editing, EB. All authors contributed to the article and approved the submitted version.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Abboud H., Abboud F. Z., Kharbouch H., Arkha Y., El Abbadi N., El Ouahabi A. (2020). COVID-19 and SARS-Cov-2 Infection: Pathophysiology and Clinical Effects on the Nervous System. World Neurosurg. 140, 49–53. doi: 10.1016/j.wneu.2020.05.193
Abt M. C., Artis D. (2013). The dynamic influence of commensal bacteria on the immune response to pathogens. Curr. Opin. Microbiol. 16, 4–9. doi: 10.1016/j.mib.2012.12.002
Adams N. M., Grassmann S., Sun J. C. (2020). Clonal expansion of innate and adaptive lymphocytes. Nat. Rev. Immunol. 20 (11), 694–707. doi: 10.1038/s41577-020-0307-4
Adlerberth I., Wold A. E. (2009). Establishment of the gut microbiota in Western infants. Acta Paediatr. 98, 229–238. doi: 10.1111/j.1651-2227.2008.01060.x
Altmann D. (2018). T-cell immunology of the lung: maintaining the balance between host defence and immune pathology. Immunology 156, 1–2. doi: 10.1111/imm.13029
Anand S., Mande S. S. (2018). Diet, Microbiota and Gut-Lung Connection. Front. Microbiol. 9, 2147. doi: 10.3389/fmicb.2018.02147
Anand P., Puranik A., Aravamudan M., Venkatakrishnan A., Soundararajan V. (2020). SARS-CoV-2 strategically mimics proteolytic activation of human ENaC. eLife 9, 1–7. doi: 10.7554/eLife.58603
Anderson S. M., Khalil A., Uduman M., Hershberg U., Louzoun Y., Haberman A. M., et al. (2009). Taking advantage: high-affinity B cells in the germinal center have lower death rates, but similar rates of division, compared to low-affinity cells. J. Immunol. (Baltimore Md. 1950) 183 (11), 7314–7325. doi: 10.4049/jimmunol.0902452
Ang B. S. P., Lim T. C. C., Wang L. (2018). Nipah Virus Infection. J. Clin. Microbiol. 56, 1–10. doi: 10.1128/JCM.01875-17
Arango Duque G., Descoteaux A. (2014). Macrophage cytokines: involvement in immunity and infectious diseases. Front. Immunol. 5, 491. doi: 10.3389/fimmu.2014.00491
Arpaia N., Campbell C., Fan X., Dikiy S., van der Veeken J., deRoos P., et al. (2013). Metabolites produced by commensal bacteria promote peripheral regulatory T-cell generation. Nature 504 (7480), 451–455. doi: 10.1038/nature12726
Arumugam M., Raes J., Pelletier E., Le Paslier D., Yamada T., Mende D. R., et al. (2011). Enterotypes of the human gut microbiome. Nature 473, 174–180. doi: 10.1038/nature09944
Assimakopoulos S. F., Triantos C., Thomopoulos K., Fligou F., Maroulis I., Marangos M., et al. (2018). Gut-origin sepsis in the critically ill patient: pathophysiology and treatment. Infection 46 (6), 751–760. doi: 10.1007/s15010-018-1178-5
Ávila-Román J., Talero E., Rodríguez-Luna A., García-Mauriño S., Motilva V. (2016). Anti- inflammatory effects of an oxylipin-containing lyophilised biomass from a microalga in a murine recurrent colitis model. Br. J. Nutr. 116 (12), 2044–2052. doi: 10.1017/S0007114516004189
Bai Y., Zhou Q., Fang Q Song L., Chen K. (2019). Inflammatory cytokines and T-lymphocyte subsets in serum and sputum in patients with bronchial asthma and chronic obstructive pulmonary disease. Med. Sci. Monit. 25, 2206–2210. doi: 10.12659/MSM.913703
Barcik W., Boutin R. C. T., Sokolowska M., Finlay B. B. (2020). The role of lung and gut microbiota in the pathology of asthma. Immunity 52 (2), P241–P255. doi: 10.1016/j.immuni.2020.01.007
Baughn L. B., Sharma N., Elhaik E., Sekulic A., Bryce A. H., Fonseca R. (2020). Targeting TMPRSS2 in SARS-CoV-2 Infection. Mayo Clin. Proc. 95, 1989–1999. doi: 10.1016/j.mayocp.2020.06.018
Baumjohann D., Kageyama R., Clingan J. M., Morar M. M., Patel S., de Kouchkovsky D., et al. (2013). The microRNA cluster miR-17∼92 promotes TFH cell differentiation and represses subset-inappropriate gene expression. Nat. Immunol. 14 (8), 840–848. doi: 10.1038/ni.2642
Bayard V., Kitsutani P. T., Barria E. O., Ruedas L. A., Tinnin D. S., Muñoz C., et al. (2004). Outbreak of Hantavirus Pulmonary Syndrome, Los Santos, Panama, 1999–2000. Emerg. Infect. Dis. 10, 1635–1642. doi: 10.3201/eid1009.040143
Beck J. M., Young V. B., Huffnagle G. B. (2012). The microbiome of the lung. Transl. Res. 160 (4), 258–266. doi: 10.1016/j.trsl.2012.02.005
Beutler B. (2004). Innate immunity: an overview. Mol. Immunol. 40 (12), 845–859. doi: 10.1016/j.molimm.2003.10.005
Bezirtzoglou E., Stavropoulou E. (2011). Immunology and probiotic impact of the newborn and young children intestinal microflora. Anaerobe 17, 369–374. doi: 10.1016/j.anaerobe.2011.03.010
Bezirtzoglou E., Alexopoulos A., Voidarou C. (2008). Apparent antibiotic misuse in environmental ecosystems and food. Microb. Ecol. Health Dis. 20, 197–198. doi: 10.1080/08910600802408103
Bezirtzoglou E. (1997). The Intestinal Microflora During the First Weeks of Life. Anaerobe 3, 173–177. doi: 10.1006/anae.1997.0102
Bosch X., Ramos-Casals M. (2014). “Neutrophil extracellular traps - an overview,” in The Autoimmune Diseases, 5th Edition.
Bradley C. P., Teng F., Felix K. M., Sano T., Naskar D., Block K. E., et al. (2017). Segmented filamentous bacteria provoke lung autoimmunity by inducing gut-lung axis Th17 cells expressing dual TCRs. Cell Host Microbe 22 (5), P697–P704.e4. doi: 10.1016/j.chom.2017.10.007
Briard B., Mislin G. L. A., Latgé J.-P., Beauvais A. (2019). Interactions between Aspergillus fumigatus and pulmonary bacteria: current state of the field, new data, and future perspective. J. Fungi (Basel) 5, 1–20. doi: 10.3390/jof5020048
Brown R. L., Sequeira R. P., Clarke T. B. (2017). The microbiota protects against respiratory infection via GM-CSF signaling. Nat. Commun. 8, 1512, 537. doi: 10.1038/s41467-017-01803-x
Bu T., Wang L. F., Yin Y. Q. (2020). How do innate immune cells contribute to airway remodeling in COPD progression? Int. J. Chron. Obstruct. Pulmon. Dis. 15, 107–116. doi: 10.2147/COPD.S235054
Budden K. F., Gellatly S. L., Wood D. L., Cooper M. A., Morrison M., Hugenholtz P., et al. (2017). Emerging pathogenic links between microbiota and the gut-lung axis. Nat. Rev. Microbiol. 15, 55–63. doi: 10.1038/nrmicro.2016.142
Cait A. (2018). The effect of the microbiome and short-chain fatty acid metabolites on early life immune development with long term consequences for atopy and asthma. Doctorate thesis. USA: University of British Columbia.
Calcraft P. J., Arredouani A., Ruas M., Pan Z., Cheng X., Hao X., et al. (2009). NAADP mobilizes calcium from acidic organelles through two-pore channels. Nature 459, 596–600. doi: 10.1038/nature08030
Cardinal-Fernández P., Lorente J. A., Ballén-Barragán, Matute-Bello G. (2017). Acute respiratory distress syndrome and diffuse alveolar damage. New insights on a complex relationship. Ann. Am. Thorac. Soc 14 (6), 844–850. doi: 10.1513/AnnalsATS.201609-728PS
Cesta M. F. (2006). Normal structure, function, and histology of mucosa-associated lymphoid tissue. Toxicol. Pathol. 34 (5), 599–608. doi: 10.1080/01926230600865531
Chaffron S., Rehrauer H., Pernthaler J. (2010). von Mering CA .Global network of coexisting microbes from environmental and whole-genome sequence data. Genome Res. 20, 947–959. doi: 10.1101/gr.104521.109
Charlson E. S., Bittinger K., Haas A. R., Fitzgerald A. S., Frank I., Yadav A., et al. (2011). Topographical continuity of bacterial populations in the healthy human respiratory tract. Am. J. Respir. Crit. Care Med. 184, 957–963. doi: 10.1164/rccm.201104-0655OC
Chen M., Guo Z., Ju W., Ryffel B., He X., Zheng S. G. (2012). The development and function of follicular helper T cells in immune responses. Cell Mol. Immunol. 9 (5), 375–379. doi: 10.1038/cmi.2012.18
Chiu C., Openshaw P. J. (2015). Antiviral B cell and T cell immunity in the lungs. Nat. Immunol. 16 (1), 18–26. doi: 10.1038/ni.3056
Chiu C.-Y., Chan Y.-L., Tsai M.-H., Wang C.-J., Chiang M.-H., Chiu C.-C. (2019). Gut microbial dysbiosis is associated with allergen-specific IgE responses in young children with airwayallergies. World Allergy Organ J. 12, 1–9. doi: 10.1016/j.waojou.2019.100021
Chrysanthopoulou A., Mitroulis I., Apostolidou E., Arelaki S., Mikroulis D., Konstantinidis T., et al. (2014). Neutrophil extracellular traps promote differentiation and function of fibroblasts. J. Pathol. 233, 294–307. doi: 10.1002/path.4359
Chu H., Mazmanian S. K. (2013). Innate immune recognition of the microbiota promotes host-microbial symbiosis. Nat. Immunol. 14, 668–675. doi: 10.1038/ni.2635
Clarke T. B., Davis K. M., Lysenko E. S., Zhou A. Y., Yu Y., Weiser J. N. (2010). Recognition of peptidoglycan from the microbiota by Nod1 enhances systemic innate immunity. Nat. Med. 16, 228–231. doi: 10.1038/nm.2087
Cordero O. X., Polz M. F. (2014). Explaining microbial genomic diversity in light of evolutionary ecology. Nat. Rev. Microbiol. 12, 263–273. doi: 10.1038/nrmicro3218
Couture A., Garnier A., Docagne F., Boyer O., Vivien D., Le-Mauff B., et al. (2019). HLA-Class II Artificial Antigen Presenting Cells in CD4+ T Cell-Based Immunotherapy. Front. Immunol. 10, 108. doi: 10.3389/fimmu.2019.0108
Cox M. J., Allgaier M., Taylor B., Baek M. S., Huang Y. J., Daly R. A., et al. (2010). Airway microbiota and pathogen abundance in age-stratified cystic fibrosis patients. PloS One 5, e11044. doi: 10.1371/journal.pone.0011044
Cox M. J., Ege M. J., Von Mutius E. (2019). The Lung Microbiome (Sheffield: European Respiratory Society).
de Wit E., van Doremalen N., Falzarano D., Munster V. J. (2016). SARS and MERS: recent insights into emerging coronaviruses. Nat. Rev. Microbiol. 14, 523–534. doi: 10.1038/nrmicro.2016.81
Delhaes L., Monchy S., Fréalle E., Hubans C., Salleron J., Leroy S., et al. (2012). The airway microbiota in cystic fibrosis: a complex fungal and bacterial community–implications for therapeutic management. PloS One 7, e36313. doi: 10.1371/journal.pone.0036313
Diaz P. I., Xie Z., Sobue T., Thompson A., Biyikoglu B., Ricker A., et al. (2012). Synergistic interaction between Candida albicans and commensal oral streptococci in a novel in vitro mucosal model. Infect. Immun. 80 (2), 620–632. doi: 10.1128/IAI.05896-11
Dickson R. P., Martinez F. J., Huffnagle G. B. (2014). The role of the microbiome in exacerbations of chronic lung diseases. Lancet 384, 691–702. doi: 10.1016/S0140-6736(14)61136-3
Dickson R. P., Erb-Downward J. R., Martinez F. J., Huffnagle G. B. (2016a). The microbiome and the respiratory tract. Annu. Rev. Physiol. 78, 481–504. doi: 10.1146/annurev-physiol-021115-105238
Dickson R. P., Singer B. H., Newstead M. W., Falkowski N. R., Erb-Downward J. R., Standiford T. J., et al. (2016b). Enrichment of the Lung Microbiome with Gut Bacteria in Sepsis and the Acute Respiratory Distress Syndrome. Nat. Microbiol. 1, 16113. doi: 10.1038/nmicrobiol.2016.113
Ding Y., Xu J., Bromberg J. S. (2012). Regulatory T cell migration during an immune response. Trends Immunol. 33 (4), 174–180. doi: 10.1016/j.it.2012.01.002
Dumas A., Bernard L., Poquet Y., Lugo-Villarino G., Neyrolles O. (2018). The role of the lung microbiota and the gut–lung axis in respiratory infectious diseases. Cell Microbiol. 20, e12966. doi: 10.1111/cmi.12966
Enaud R., Prevel R., Ciarlo E., Beaufils F., Wieërs G., Guery B., et al. (2020). Inter-kingdom crosstalks. Front. Cell Infect. Microbiol. 10 (9), 1–9. doi: 10.3389/fcimb.2020.00009
Fan J., Li X., Gao Y., Zhou J., Wang S., Huang B., et al. (2020). The lung tissue microbiota features of 20 deceased patients with COVID-19. J. Infect. 81 (3), e64–e67. doi: 10.1016/j.jinf.2020.06.047
Ferreira-Santos D., Maranhão P., Monteiro-Soares M. (2020). COVIDcids Identifying common baseline clinical features of COVID-19: a scoping review. BMJ Open 10, e041079. doi: 10.1136/bmjopen-2020-041079
Fraczek M. G., Zhao C., Dineen L., Lebedinec R., Bowyer P., Bromley M., et al. (2019). Fast and reliable PCR amplification from Aspergillus fumigatus spore suspension without traditional DNA extraction. Curr. Protoc. Microbiol. e89, 1–10. doi: 10.1002/cpmc.89
Freeman C. M., Curtis J. L. (2017). Lung dendritic cells: shaping immune responses throughout chronic obstructive pulmonary disease progression. Am. J. Respir. Cell Mol. Biol. 56, 152–159. doi: 10.1165/rcmb.2016-0272TR
Gao S., Chen J., Xie J., Wang J. (2020). The effects of BAFF on T lymphocytes in chronic obstructive pulmonary disease. Respir. Res. 21, 66. doi: 10.1186/s12931-020-01333-z
Gill H. S., Rutherfurd K. J., Cross M. L. (2001). Dietary probiotic supplementation enhances natural killer cell activity in the elderly: an investigation of age-related immunological changes. J. Clin. Immunol. 21 (4), 264–271. doi: 10.1023/a:1010979225018
Gordon D. L., Rice J. L. (1988). Opsonin-dependent and independent surface phagocytosis of S. aureus proceeds independently of complement and complement receptors. Immunology 64, 709–714. doi: 10.1111/j.1574-6968.1989.tb02433.x
Gou W., Fu Y., Yue L., Chen G., Cai, Shuai M., et al. (2020). Gut microbiota may underlie the predisposition of healthy individuals to COVID-19. medRxiv. doi: 10.1101/2020.04.22.2007609
Groves H. T., Cuthbertson L., James P., Moffatt M. F., Cox M. J., Tregoning J. S. (2018). Respiratory disease following viral lung infection alters the murine gut microbiota. Front. Immunol. 9, 182. doi: 10.3389/fimmu.2018.00182
Gu S., Chen Y., Wu Z., Chen Y., Gao H., Lv L., et al. (2020). Alterations of the gut microbiota in patients with coronavirus disease 2019 or H1N1 influenza. Clin. Infect. Dis. 71 (10), 2669–2678. doi: 10.1093/cid/ciaa709
Hedblom G. A., Reiland H. A., Sylte M. J., Johnson T. J., Baumler D. J. (2018). Segmented filamentous bacteria – Metabolism meets immunity. Front. Microbiol. 9, 1991. doi: 10.3389/fmicb.2018.01991
Henriques-Normark B., Tuomanen E. I. (2013). The Pneumococcus: Epidemiology, microbiology, and pathogenesis. CSH Perspect. Med., 1–31. doi: 10.1101/cshperspect.a010215
Hergott C. B., Roche A. M., Tamashiro E., Clarke T. B., Bailey A. G., Laughlin A., et al. (2016). Peptidoglycan from the gut microbiota governs the lifespan of circulating phagocytes at homeostasis. Blood 127, 2460–2471, 544. doi: 10.1182/blood-2015-10-675173
Hilty M., Burke C., Pedro H., Cardenas P., Bush A., Bossley C., et al. (2010). Disordered microbial communities in asthmatic airways. PLoS ONE. 5 (1), e8578. doi: 10.1371/journal.pone.0008578
Hoffmann M., Kleine-Weber H., Schroeder S., Krüger N., Herrler T., Erichsen S., et al. (2020). SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell 181 (2), P271–P280.e8. doi: 10.1016/j.cell.2020.02.052
Hosoki K., Ying S., Corrigan C., Qi H., Kurosky A., Jennings K., et al. (2015). Analysis of a panel of 48 cytokines in BAL fluids specifically identifies IL-8 levels as the only cytokine that distinguishes controlled asthma from uncontrolled asthma, and correlates inversely with FEV1. PloS One 10, 1–16. doi: 10.1371/journal.pone.0126035
Hu Y., Feng Y., Wu J., Liu F., Zhang Z., Hao Y., et al. (2019a). The gut microbiome signatures discriminate healthy from pulmonary tuberculosis patients. Front. Cell Infect. Microbiol. 9, 90. doi: 10.3389/fcimb.2019.00090
Hu Y., Yang Q., Liu B., Dong J., Sun L., Zhu Y., et al. (2019b). Gut microbiota associated with pulmonary tuberculosis and dysbiosis aused by anti-tuberculosis drugs. J. Infect. 78 (4), 317–322. doi: 10.1016/j.jinf.2018.08.006
Huang Y. J., Kim E., Cox M., Brodie E. L., Brown R., Wiener-Kronish J. P., et al. (2010). A Persistent and Diverse Airway Microbiota Present during Chronic Obstructive Pulmonary Disease Exacerbations. OMICS 14, 9–59. doi: 10.1089/omi.2009.0100
Huang C., Wang Y., Li X., Ren L., Zhao J., Hu Y., et al. (2020). Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet 395 (10223), P497–P506. doi: 10.1016/S0140-6736(20)30183-5
Huffnagle D. R. (2015). The Lung Microbiome: New principles for respiratory bacteriology in health and disease. PloS Pathog. doi: 10.1371/journal.ppat.1004923
Hughes A. R., Inouye B. D., Johnson M. T. J., Underwood N., Vellend M. (2008). Ecological consequences of genetic diversity. Ecol. Lett. 11, 609–623. doi: 10.1111/j.1461-0248.2008.01179.x
Ismail I. H., Boyle R. J., Licciardi P. V., Oppedisano F., Lahtinen S., Robins-Browne R. M., et al. (2016). Early gut colonization by Bifidobacterium breve and B. catenulatum differentially modulates eczema risk in children at high risk of developing allergic disease. Pediatr. Allergy Immunol. 27 (8), 838–846. doi: 10.1111/pai.12646
Iwamura C., Bouladoux N., Belkaid Y., Sher A., Jankovic D. (2017). Sensing of the microbiota by NOD1 in mesenchymal stromal cells regulates murine hematopoiesis. Blood 129, 171–176. doi: 10.1182/blood-2016-06-723742 547.
Jose R. J., Manuel A. (2020). COVID-19 cytokine storm: the interplay between inflammation and coagulation. Lancet Respir. Med. 8, e46–e47. doi: 10.1016/S2213-2600(20)30216-2
Kawamura K., Iyonaga K., Ichiyasu H., Nagano J., Suga M., Sasaki Y. (2005). Differentiation, maturation, and survival of dendritic cells by osteopontin regulation. Clin. Diagn. Lab. Immunol. 121, 206–212. doi: 10.1128/cdli.12.1.206-212.2005
Khatiwada S., Subedi A. (2020). Lung microbiome and coronavirus disease 2019 796 (COVID-19): Possible link and implications. Hum. Microb. J. 17, 10007379. doi: 10.1016/j.humic.2020.100073
Kim D., Kim Y.-G., Seo S.-U., Kim D.-J., Kamada N., Prescott D., et al. (2016). Nod2-mediated recognition of the microbiota is critical for mucosal adjuvant activity of cholera toxin. Nat. Med. 22, 551,524–30. doi: 10.1038/nm.4075
Kolli D., Gupta M. R., Sbrana E., Velayutham T. S., Chao H., Casola A., et al. (2014). Alveolar macrophages contribute to the pathogenesis of human metapneumovirus infection while protecting against respiratory syncytial virus infection. Am. J. Respir. Cell Mol. Biol. 51 (4), 502–515. doi: 10.1165/rcmb.2013-0414oc
Kranzer K., Eckhardt A., Aigner M., Knoll G., Deml L., Speth C., et al. (2004). Induction of maturation and cytokine release of human dendritic cells by Helicobacter pylori. Infect. Immun. 72 (8), 4416–4423. doi: 10.1128/IAI.72.8.4416-4423.2004
Kuruvilla M. E., Lee F. E.-H., Lee G. B. (2019). Understanding asthma phenotypes, endotypes, and mechanisms of disease. Clin. Rev. Allergy Immunol. 56, 219–233. doi: 10.1007/s12016-018-8712-1
Lacy P. (2015). Editorial: Secretion of cytokines and chemokines by innate immune cells. Front. Immunol. 6, 190. doi: 10.3389/fimmu.2015.00190
Ladjemi M. Z., Lecocq M., Weynand B., Bowen H., Gould H. J., Van Snick J., et al. (2015). Increased IgA production by B-cells in COPD via lung epithelial interleukin-6 and TACI pathways. Eur. Respir. J. 45, 980–993. doi: 10.1183/09031936.00063914
Law H. K., Cheung C. Y., Sia S. F., Chan Y. O., Peiris J. M., Lau Y. L. (2009). Toll-like receptors, chemokine receptors and death receptor ligands responses in SARS coronavirus infected human monocyte derived dendritic cells. BMC Immunol. 10, 1–12. doi: 10.1186/1471-2172-10-35
Leonardi I., Li X., Iliev I. D. (2018). Macrophage interactions with fungi and bacteria in inflammatory bowel disease. Curr. Opin. Gastrenterol. 34 (6), 392–397. doi: 10.1097/MOG.0000000000000479
Letko M., Marzi A., Munster V. (2020). Functional assessment of cell entry and receptor usage for SARS-CoV-2 and other lineage B betacoronaviruses. Nat. Microbiol. 5, 562–569. doi: 10.1038/s41564-020-0688-y
Li X. J., You X. Y., Wang C. Y., Li X. L., Sheng Y. Y., Zhuang P. W., et al. (2020). Bidirectional Brain-gut-microbiota Axis in increased intestinal permeability induced by central nervous system injury. CNS Neurosci. Ther. 26 (8), 783–790. doi: 10.1111/cns.13401
Li M.-Y., Li L., Zhang Y., Wang X.-S. (2020). Expression of the SARS-CoV-2 cell receptor gene ACE2 in a wide variety of human tissues. Infect. Dis. Poverty. 9 (45), 737. doi: 10.1186/s40249-020-00662-x
Li F. (2012). Evidence for a common evolutionary origin of coronavirus spike protein receptor-binding subunits. J. Virol. 86, 2856–2858. doi: 10.1128/JVI.06882-11
Lif Holgerson P., Harnevik L., Hernell O., Tanner A. C. R., Johansson I. (2011). Mode of birth delivery affects oral microbiota in infants. J. Dent. Res. 90, 1183–1188. doi: 10.1177/0022034511418973
Liu T., Zhang L., Joo D., Sun S -C. (2017). NF-κB signaling in inflammation. Signal Transduct. Target Ther. 2, 1–9. doi: 10.1038/sigtrans.2017.23
Liu Y., Shao Z., Shangguan G., Bie Q., Zhang B. (2018). Biological properties and the role of IL-25 in disease pathogenesis. J. Immunol. Res. 2018, 6519465. doi: 10.1155/2018/6519465
Liu C., Yang L., Yu H., Ouyang W., Yin W., Xu F. (2019).Mast cells participate in regulation of lung-gut axis during Staphylococcus aureus pneumonia. In: Cell Proliferation. Available at: https://onlinelibrary.wiley.com/doi/full/10.1111/cpr.12565 (Accessed Oct 10, 2020).
Luo M., Liu Y., Wu P., Luo D.-X., Sun Q., Zheng H., et al. (2017). Alternation of gut microbiota in patients with pulmonary tuberculosis. Front. Physiol. 8, 822. doi: 10.3389/fphys.2017.00822
Maly K., Schirmer M. (2015). The story of CD4+CD28– T cells revisited: solved or still ongoing? J. Immunol. Res. 348746, 1–11. doi: 10.1155/2015/348746
Mammen M. J., Scannapieco F. A., Sethi S. (2000). Oral-lung microbiome interactions in lung diseases. Periodontol. (2020) 83, 234–241. doi: 10.1111/prd.12301
Martin T. R., Frevert C. W. (2005). Innate immunity in the lungs. Proc. Am. Thorac. Soc. 2, 403–411. doi: 10.1513/pats.200508-090JS
Martin T. R. (2000). Recognition of bacterial endotoxin in the lungs. Am. J. Respir. Cell Mol. Biol. 23, 128–132. doi: 10.1165/ajrcmb.23.2.f189
Masopust D., Schenkel J. (2013). The integration of T cell migration, differentiation and function. Nat. Rev. Immunol. 13, 309. doi: 10.1038/nri3442
Masters P. S. (2006). The molecular biology of coronaviruses. Adv. Virus Res. 66 (760), 193–192. doi: 10.1016/S0065-3527(06)66005-3
McAleer J. P., Kolls J. K. (2018). Contributions of the intestinal microbiome in lung immunity. Eur. J. Immunol. 48, 39–39. doi: 10.1002/eji.201646721
McAleer J. P., Nguyen N. L., Chen K., Kumar P., Ricks D. M., Binnie M., et al. (2016). Pulmonary Th17 Antifungal Immunity Is Regulated by the Gut Microbiome. J. Immunol. 197 (1), 97–107. doi: 10.4049/jimmunol.1502566
Mielcarek C., Romond P. C., Romond M. B., Bezirtzoglou E. (2011). Modulation of bacterial translocation in mice mediated through lactose and human milk oligosaccharides. Anaerobe 17 (6), 361–366. doi: 10.1016/j.anaerobe.2011.09.002
Mitchell A. B., Glanville A. R. (2018). Introduction to techniques and methodologies for characterizing the human respiratory virome. Methods Mol. Biol. 1838, 111–123. doi: 10.1007/978-1-4939-8682-8_9
Moffatt M. F., Cookson W. O. (2017). The lung microbiome in health and disease. Clin. Med. (Lond) 17, 525–529. doi: 10.7861/clinmedicine.17-6-525
Mogensen T. H. (2009). Pathogen recognition and inflammatory signaling in innate immune defenses. Clin. Microbiol. Rev. 22, 240–273. doi: 10.1128/CMR.00046-08
Mortaz E., Adcock I. M., Folkerts G., Barnes P. J., Paul Vos A., Garssen J. (2013). Probiotics in the management of lung diseases. Mediators Inflamm. 2013, 751068. doi: 10.1155/2013/751068
Mukherjee S., Hanidziar D. (2018). More of the Gut in the Lung: How two microbiomes meet in ARDS. Yale J. Biol. Med. 91 (2), 143–149.
Murray R. Z., Stow J. L. (2014). Cytokine secretion in macrophages: SNAREs, Rabs, and membrane trafficking. Front. Immunol. 5, 538. doi: 10.3389/fimmu.2014.00538
Nguyen L. D. N., Viscogliosi E., Delhaes L. (2015). The lung mycobiome: an emerging field of the human respiratory microbiome. Front. Microbiol. 6, 89. doi: 10.3389/fmicb.2015.00089
NIH Human Microbiome Project - Home. Available at: https://www.hmpdacc.org/ (Accessed on Sep 6, 2020).
O’byrne P. M., Postma D. S. (1999). The many faces of airway inflammation. Am. J. Respir. Crit. Care Med. 159, S1–S63. doi: 10.1164/ajrccm.159.supplement_2.mfa-1
Ou X., Liu Y., Lei X., Li P., Mi D., Ren L., et al. (2020). Characterization of spike glycoprotein of SARS-CoV-2 on virus entry and its immune cross-reactivity with SARS-CoV. Nat. Commun. 11, 1620. doi: 10.1038/s41467-020-15562-9
Park H., Shin J. W., Park S.-G., Kim W. (2014). Microbial Communities in the Upper Respiratory Tract of Patients with Asthma and Chronic Obstructive Pulmonary Disease. PloS One 9, 1–11. doi: 10.1371/journal.pone.0109710
Petersen O. H., Gerasimenko O. V., Gerasimenko J. V. (2020). Endocytic uptake of SARS-CoV-2: the critical roles of pH, Ca2+, and NAADP. Function 1, 727. doi: 10.1093/function/zqaa003
Ratia K., Saikatendu K. S., Santarsiero B. D., Barretto N., Baker S. C., Stevens R. C., et al. (2006). Severe acute respiratory syndrome coronavirus papain-like protease: structure of a viral deubiquitinating enzyme. Proc. Natl. Acad. Sci. U S A 103, 5717–5722. doi: 10.1073/pnas.0510851103
Roduit C., on behalf of the PASTURE/EFRAIM study group. (2019). High levels of butyrate and propionate in early life are associated with protection against atopy. Eur. J. Allergy Clin. Immunol. 1–11. doi: 10.1111/all.13660
Russell S. L., Gold M. J., Hartmann M., Willing B. P., Thorson L., Wlodarska M., et al. (2012). Early life antibiotic-driven changes in microbiota enhance susceptibility to allergic asthma. EMBO Rep. 13, 440–447. doi: 10.1038/embor.2012.32
Rutayisire E., Huang K., Liu Y., Tao F. (2016). The mode of delivery affects the diversity and colonization pattern of the gut microbiota during the first year of infants’ life: a systematic review. BMC Gastroenterol. 16, 86. doi: 10.1186/s12876-016-0498-0
Saitou M., Nemoto D., Utano K., Suzuki T., Lefor A. K., Togashi K., et al. (2018). Identification of intestinal abnormalities in patients with active pulmonary tuberculosis using small bowel capsule endoscopy. Endosc. Int. Open 06 (09), E1103–E1108. doi: 10.1055/a-0655-2086
Sakai A., Saitow F., Maruyama M., Miyake N., Miyake K., Shimada T., et al. (2017). MicroRNA cluster miR-17-92 regulates multiple functionally related voltage-gated potassium channels in chronic neuropathic pain. Nat. Commun. 8, 16079. doi: 10.1038/ncomms16079
Salminen S., Endo A., Isolauri E., Scalabrin D. (2016). Early gut colonization with Lactobacilli and Staphylococcus in infants. J. Pediatr. Gastroenterol. Nutr. 62 (1), 80–86. doi: 10.1097/MPG.0000000000000925
Samuelson D. R., Charles T. P., de la Rua N. M., Taylor C. M., Blanchard E. E., Luo M., et al. (2016). Analysis of the intestinal microbial community and inferred functional capacities during the host response to Pneumocystis pneumonia. Exp. Lung Res. 42, 425–439. doi: 10.1080/01902148.2016.1258442
Sanghai N., Tranmer G. K. (2020). Taming the cytokine storm: repurposing montelukast for the attenuation and prophylaxis of severe COVID-19 symptoms. Drug Discov. Today. 25, 2076–2079. doi: 10.1016/j.drudis.2020.09.013
Schuijt T. J., Lankelma J. M., Scicluna B. P., de Sousa e Melo F., Roelofs J. J., de Boer J. D., et al. (2016). The gut microbiota plays a protective role in the host defence against pneumococcal pneumonia. Gut 65 (4), 575–583. doi: 10.1136/gutjnl-2015-309728
Schuurman H.-J., Quesniaux V. F. J. (1999). “Development and maturation of T and B cells,” in Principles of Immunopharmacology, Chapter: A2. Eds. Nijkamp F. P., Parnham M. J. (Basel: Birkhäuser Verlag).
Segal L. N., Clemente J. C., Tsay J.-C. J., Koralov S. B., Keller B. C., Wu B. G., et al. (2016). Enrichment of the lung microbiome with oral taxa is associated with lung inflammation of a Th17 phenotype. Nat. Microbiol. 1, 16031. doi: 10.1038/nmicrobiol.2016.31
Shapiro J., Sciaky N., Lee J., Bosshart H., Angeletti R. H., Bonifacino J. S. (1997). Localization of endogenous furin in cultured cell lines. J. Histochem. Cytochem. 45, 3–12. doi: 10.1177/002215549704500102 747.
Shapiro S. D. (1999). The macrophage in chronic obstructive pulmonary disease. Am. J. Respir. Crit. Care Med. 160, S29–S32. doi: 10.1164/ajrccm.160.supplement_1.9
Shaykhiev R., Crystal R. G. (2013). Innate immunity and chronic obstructive pulmonary disease - A mini review. Gerontology 59, 481–489. doi: 10.1159/000354173
Skendros P., Mitsios A., Chrysanthopoulou A., Mastellos D. C., Metallidis S., Rafailidis P., et al. (2020). Complement and tissue factor-enriched neutrophil extracellular traps are key drivers in COVID-19 immunothrombosis. J. Clin. Invest. 130 (11), 6151–6157. doi: 10.1172/JCI141374
Skinner J. P., Keown A. A., Chong M. M. (2014). The miR-17 ∼ 92a cluster of microRNAs is required for the fitness of Foxp3+ regulatory T cells. PloS One 9 (2), e88997. doi: 10.1371/journal.pone.0088997
Smolinska S., Jutel M., Crameri R., O’Mahony L. (2014). Histamine and gut mucosal immune regulation. Allergy 69 (3), 273–281. doi: 10.1111/all.12330
Sokolowska M., Frei R., Lunjani N., Akdis C. A., O’Maho L. (2018). Microbiome and asthma. Asthma Res Pract 4, 1. doi: 10.1186/s40733-017-0037-y
Stavropoulou E., Bezirtzoglou E. (2020). Probiotics as a weapon in the fight against COVID-19. Front. Nutr. 7, 614986. doi: 10.3389/fnut.2020.614986
Stiemsma L., Reynolds L., Turvey S., Finlay B. (2015). The hygiene hypothesis: current perspectives and future therapies. Immunotargets Ther. 4, 143–157. doi: 10.2147/ITT.S61528
Stubbington M. J. T., Rozenblatt-Rosen O., Regev A., Teichmann S. A. (2017). Single-cell transcriptomics to explore the immune system in health and disease. Science 358 (6359), 58–63. doi: 10.1126/science.aan6828
Suh W. K. (2015). Life of T follicular helper cells. Mol. Cells 38 (3), 195–201. doi: 10.14348/molcells.2015.2331
Sze M. A., Tsuruta M., Yang S. W., Oh Y., Man S. F., Hogg J. C., et al. (2014). Changes in the bacterial microbiota in gut, blood, and lungs following acute LPS instillation into mice lungs. PloS One 9 (10), e111228. doi: 10.1371/journal.pone.0111228
Tang F., Du Q., Liu Y. J. (2010). Plasmacytoid dendritic cells in antiviral immunity and autoimmunity. Sci. China Life Sci. 53 (2), 172–182. doi: 10.1007/s11427-010-0045-0
Tarashi S., Ahmadi Badi S., Moshiri A., Nasehi M., Fateh A., Vaziri F., et al. (2018). The human microbiota in pulmonary tuberculosis: Not so innocent bystanders. Tuberculosis 113, 215–221. doi: 10.1016/j.tube.2018.10.010
Turnbaugh P. J., Ley R. E., Hamady M., Fraser-Liggett C. M., Knight R., Gordon J. I. (2007). The human microbiome project. Nature 449, 804–810. doi: 10.1038/nature06244
Turturice B. A., McGee H. S., Oliver B., Baraket M., Nguyen B. T., Ascoli C., et al. (2017). Atopic asthmatic immune phenotypes associated with airway microbiota and airway obstruction. PloS One 12, 1–18. doi: 10.1371/journal.pone.0184566
Ulambayar B., Lee S.-H., Yang E.-M., Ye Y.-M., Park H.-S. (2018). Association between epithelial cytokines and clinical phenotypes of elderly asthma. Allergy Asthma Immunol. Res. 11, 79–89. doi: 10.4168/aair.2019.11.1.79
Vaishnavi C. (2013). Translocation of gut flora and its role in sepsis. Indian J. Med. Microbiol. 31 (4), 334–342. doi: 10.4103/0255-0857.118870
van Rijt L. S., Vos N., Willart M., Muskens F., Tak P. P., van der Horst C., et al. (2011). Persistent activation of dendritic cells after resolution of allergic airway inflammation breaks tolerance to inhaled allergens in mice. Am. J. Respir. Crit. Care Med. 184, 303–311. doi: 10.1164/rccm.201101-0019OC
van Woerden H. C., Gregory C., Brown R., Marchesi J. R., Hoogendoorn B., Matthews I. P. (2013). Differences in fungi present in induced sputum samples from asthma patients and non-atopic controls: a community based case control study. BMC Infect. Dis. 13, 69. doi: 10.1186/1471-2334-13-69
Wang Z., on behalf of COPDMAP study. (2017). Sputum microbiome temporal variability and dysbiosis in chronic obstructive pulmonary disease exacerbations: An analysis of the COPDMAP study. Thorax 73 (4), 331–338. doi: 10.1136/thoraxjnl-2017-210741
Wang Z., Bafadhel M., Haldar K., Spivak A., Mayhew D., Miller B. E., et al. (2016). Lung microbiome dynamics in COPD exacerbations. Eur. Respir. J. 47, 1082–1092. doi: 10.1183/13993003.01406-2015
Wang G. (2014). Human antimicrobial peptides and proteins. Pharmaceuticals (Basel) 7, 545–594. doi: 10.3390/ph7050545
Whitsett J. A., Alenghat T. (2015). Respiratory epithelial cells orchestrate pulmonary innate immunity. Nat. Immunol. 16 (1), 27–5. doi: 10.1038/ni.3045
WHO The WHO Just Declared Coronavirus COVID-19 a Pandemic | Time Available online. Available at: https://time.com/5791661/who-coronavirus-pandemic-declaration/ (Accessed on Sep 5, 2020).
Worlitzsch D., Rintelen C., Böhm K., Wollschläger B., Merkel N., Borneff-Lipp M., et al. (2009). Antibiotic-resistant obligate anaerobes during exacerbations of cystic fibrosis patients. Clin. Microb. Infect. 15, 454–460. doi: 10.1111/j.1469-0691.2008.02659.x
Wu H., Kuzmenko A., Wan S., Schaffer L., Weiss A., Fisher J. H., et al. (2003). Surfactant proteins A and D inhibit the growth of Gram-negative bacteria by increasing membrane permeability. J. Clin. Invest. 111, 1589–1602. doi: 10.1172/JCI16889
Xiang M., Fan J. (2010). Pattern recognition receptor-dependent mechanisms of acute lung injury. Mol. Med. (Cambridge Mass.) 16 (1-2), 69–82. doi: 10.2119/molmed.2009.00097
Xu D., Zhou F., Sun W Chen L., Lan L., Li H., Xiao F., et al. (2020). Relationship between serum SARS-CoV-2 nucleic acid(RNAemia) and organ damage in COVID-19 patients: A Cohort Study. Clin. Infect. Dis. ciaa1085. doi: 10.1093/cid/ciaa1085
Xu R., Tan C., Zhu J., Zeng X., Gao X., Wu Q., et al. (2019). Dysbiosis of the intestinal microbiota in neurocritically ill patients and the risk for death. Crit. Care (London England) 23 (1), 195. doi: 10.1186/s13054-019-2488-4
Xu K., Cai H., Shen Y., Ni Q., Chen Y., Hu S., et al. (2020). [Management of corona virus disease-19 (COVID-19): the Zhejiang experience]. Zhejiang Da Xue Xue Bao Yi Xue Ban 49 (1), 147–157. doi: 10.3785/j.issn.1008-9292.2020.02.02
Yacobi N. R., Malmstadt N., Fazlollahi F., DeMaio L., Marchelletta R., Hamm-Alvarez S. F., et al. (2010). Mechanisms of alveolar epithelial translocation of a defined population of nanoparticles. Am. J. Respir. Cell Mol. Biol. 42 (5), 604–614. doi: 10.1165/rcmb.2009-0138OC
Yang D., Xing Y., Song X., Qian Y. (2020). The impact of lung microbiota dysbiosis on inflammation. Immunology 159 (2), 156–166. doi: 10.1111/imm.13139
Yin Y., Wang Y., Zhu L., Lis W., Liao N., Jiang M., et al. (2013). Comparative analysis of the distribution of segmented filamentous bacteria in humans, mice and chickens. ISME J. 7, 615–621. doi: 10.1038/ismej.2012.128
Young R. P., Hopkins R. J. (2017). The mevalonate pathway and innate immune hyper-responsiveness in the pathogenesis of COPD and lung cancer: potential for chemoprevention. Curr. Mol. Pharmacol. 10 (1), 46–59. doi: 10.2174/1874467209666160112130016
Yuki K., Fujiogi M., Koutsogiannaki S. (2020). COVID-19 pathophysiology: A review. Clin. Immunol. 215, 108427. doi: 10.1016/j.clim.2020.108427
Zhang D., Li S., Wang N., Tan H.-Y., Zhang Z., Feng Y. (2020). The Cross-Talk between gut microbiota and lungs in common lung diseases. Front. Microbiol. 11, 301. doi: 10.3389/fmicb.2020.00301
Zhu W., Germain C., Liu Z., Sebastian Y., Devi P., Knockaert S., et al. (2015). A high density of tertiary lymphoid structure B cells in lung tumors is associated with increased CD4+ T cell receptor repertoire clonality. Oncoimmunology A 4 (12), e1051922. doi: 10.1080/2162402X.2015.1051922
Zuo T., Zhang F., Lui G. C. Y., Yeoh Y. K., Li A. Y. L., Zhan H., et al. (2020). Alterations in gut microbiota of patients with COVID-19 during time of hospitalization. Gastroenterology 159, 944–955.e8. doi: 10.1053/j.gastro.2020.05.048
Keywords: lung microbiome, lung-gut axis, lung in health and disease, lung, microbiota, COVID-19, SARS-CoV-2, lung immunity
Citation: Stavropoulou E, Kantartzi K, Tsigalou C, Konstantinidis T, Voidarou C, Konstantinidis T and Bezirtzoglou E (2021) Unraveling the Interconnection Patterns Across Lung Microbiome, Respiratory Diseases, and COVID-19. Front. Cell. Infect. Microbiol. 10:619075. doi: 10.3389/fcimb.2020.619075
Received: 19 October 2020; Accepted: 11 December 2020;
Published: 28 January 2021.
Edited by:
Francesca Romana Ponziani, Hepatology Fondazione Policlinico Universitario Agostino Gemelli IRCCS, ItalyReviewed by:
Saroj Khatiwada, University of New South Wales, AustraliaCopyright © 2021 Stavropoulou, Kantartzi, Tsigalou, Konstantinidis, Voidarou, Konstantinidis and Bezirtzoglou. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Elisavet Stavropoulou, ZWxpc2FiZXRoLnN0YXZyb3BvdWxvdUBnbWFpbC5jb20=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.