
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Cell. Infect. Microbiol. , 29 January 2021
Sec. Parasite and Host
Volume 10 - 2020 | https://doi.org/10.3389/fcimb.2020.618430
This article is part of the Research Topic Molecular Basis of Stage Conversion in Apicomplexan Parasites View all 11 articles
Plasmodium sporozoites are transmitted to mammals by anopheline mosquitoes and first infect the liver, where they transform into replicative exoerythrocytic forms, which subsequently release thousands of merozoites that invade erythrocytes and initiate the malaria disease. In some species, sporozoites can transform into dormant hypnozoites in the liver, which cause malaria relapses upon reactivation. Transmission from the insect vector to a mammalian host is a critical step of the parasite life cycle, and requires tightly regulated gene expression. Sporozoites are formed inside oocysts in the mosquito midgut and become fully infectious after colonization of the insect salivary glands, where they remain quiescent until transmission. Parasite maturation into infectious sporozoites is associated with reprogramming of the sporozoite transcriptome and proteome, which depends on multiple layers of transcriptional and post-transcriptional regulatory mechanisms. An emerging scheme is that gene expression in Plasmodium sporozoites is controlled by alternating waves of transcription activity and translational repression, which shape the parasite RNA and protein repertoires for successful transition from the mosquito vector to the mammalian host.
Malaria is caused by protozoan parasites of the Plasmodium genus, and remains a major global health problem in endemic countries. P. falciparum is the deadliest species infecting humans, causing more than 200 million cases and 400,000 deaths every year, mainly in sub-Saharan Africa (World Health Organization, 2019). The absence of an available efficacious vaccine and the threat of parasite resistance to anti-malarial drugs emphasize the need for novel anti-malarial intervention strategies. To this end, a better understanding of the biology of the parasite is needed.
Plasmodium spp. have a complex life cycle that alternates between a mosquito vector and a vertebrate host (Figure 1). They are transmitted to mammals by the bite of infected female Anopheles mosquitoes, which deposit Plasmodium sporozoites in the host dermis (Sinnis and Zavala, 2012). The motile sporozoites actively migrate to the liver, where they invade hepatocytes for an initial and obligatory replication phase. Inside infected hepatocytes, Plasmodium resides in a specialized membrane-bound compartment, the parasitophorous vacuole (PV), where sporozoites transform into replicative exoerythrocytic forms (EEFs) (Frischknecht and Matuschewski, 2017). EEFs undergo a dramatic parasite multiplication, lasting 2 to 15 days depending on the parasite species and leading to the release of thousands of merozoites, which invade erythrocytes and initiate the pathogenic blood stage cycle. In some species such as P. vivax, a fraction of the parasites inside hepatocytes do not replicate immediately after invasion but, instead, transform into dormant stages called hypnozoites (Adams and Mueller, 2017). Activation of hypnozoites weeks or months after the infectious mosquito bite results in malaria relapses. During blood stage replication, a fraction of the parasites differentiates into sexual stage gametocytes, which upon ingestion by a blood-feeding anopheline mosquito can initiate sporogony in the insect.
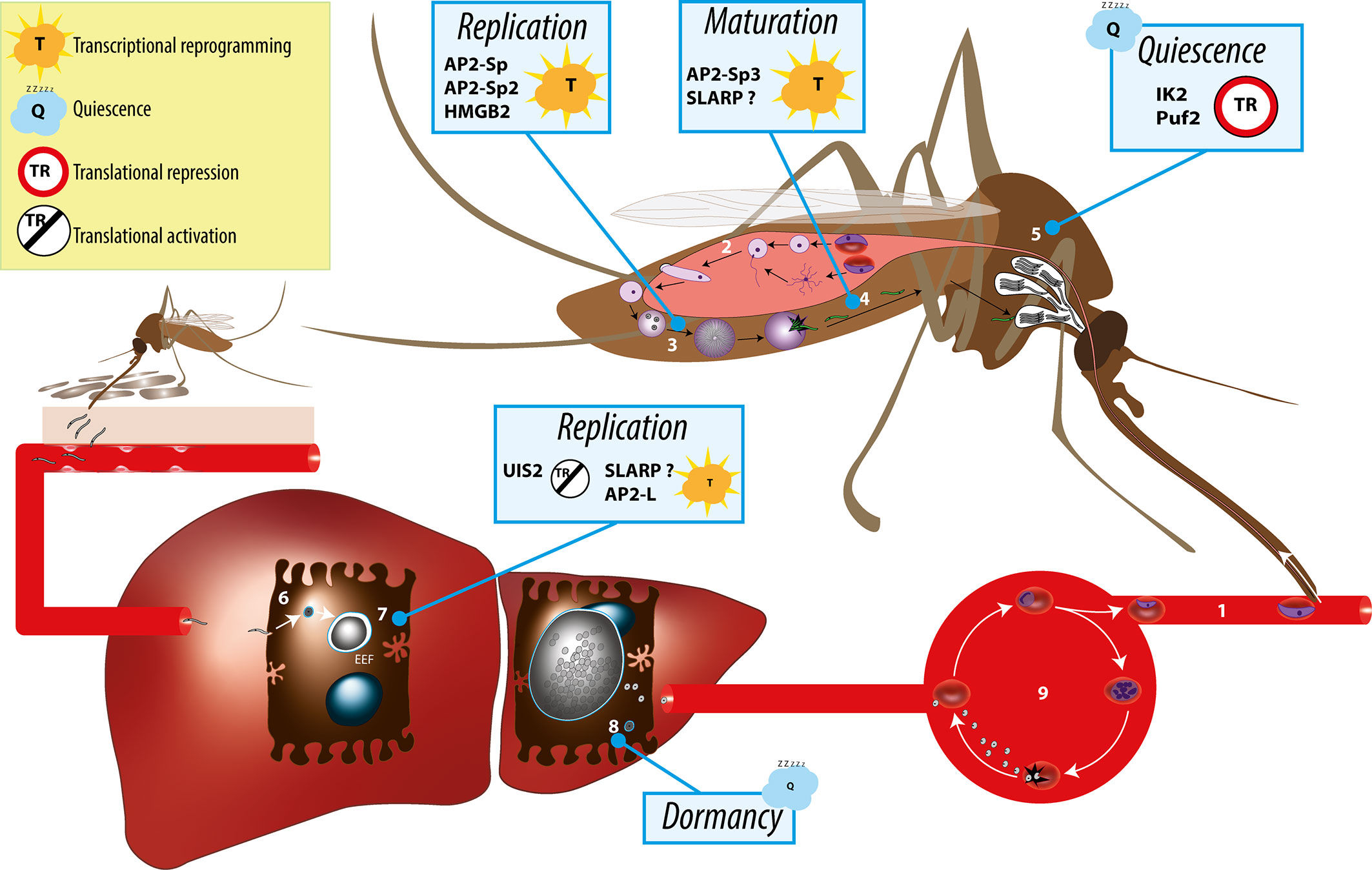
Figure 1 Overview of the Plasmodium life cycle and the gene regulation mechanisms controlling parasite transmission from the mosquito vector to the mammalian host. During blood feeding on an infected host, female anopheline mosquito ingest male and female gametocytes (1), which transform into male and female gametes in the insect midgut lumen, followed by fertilization (2) and formation of motile ookinetes that cross the midgut epithelium and transform into extracellular oocysts. Oocysts undergo intense parasite replication (3) until releasing thousands of immature sporozoites that traffic to the salivary glands and mature into infectious sporozoites (4). In the salivary glands, sporozoites persist in a poised state for several days or weeks (5), awaiting transmission, which occurs when the mosquito feeds for blood on a new host. Infectious sporozoites are injected in the host skin and migrate to the liver, where they invade hepatocytes (6) and transform into replicative exoerythrocytic forms (EEFs) (7). In species such as P. vivax, some of the parasites do not replicate immediately after invasion but, instead, transform into dormant stages called hypnozoites (8). Parasite replication in the liver culminates with the release of thousands of merozoites into the circulation, which invade erythrocytes and initiate the asexual blood stage cycle (9). Individual factors with functional evidence for a role in transcriptional or post-transcriptional gene regulation during sporozoite development, maturation, or establishment of infection in the liver are indicated.
Sporozoites develop within oocysts underneath the basal lamina of the mosquito midgut. Upon egress, they are released into the hemolymph and invade the insect salivary glands, where they persist in a poised state for several days or weeks, awaiting transmission, which occurs when the mosquito feeds for blood on a new host. The mosquito-mammal transition is a major bottleneck in the life of the parasite, as only a minor proportion (<20%) of sporozoites successfully invade the salivary glands (Hillyer et al., 2007), from which only a few dozen are inoculated during a mosquito bite (Frischknecht et al., 2004). Sporozoites released from oocysts are weakly infectious for the mammalian host, and gradually acquire infectivity while trafficking in the hemolymph, to become highly infectious after colonization of the insect salivary glands (Touray et al., 1992; Sato et al., 2014).
Comprehensive transcriptome and proteome surveys have shown that phenotypic maturation from immature oocyst-derived sporozoites in the mosquito midgut to mature infectious sporozoites in the insect salivary glands is associated with global changes in the parasite transcriptomes and proteomes (Matuschewski et al., 2002; Lasonder et al., 2008; Mikolajczak et al., 2008; Lindner et al., 2019). Studies based on RNAseq and LC-MS/MS performed with human-infecting species P. falciparum and rodent-infecting parasites P. yoelii revealed that around 3,500–4,200 genes (representing 60-70% of the genome) have detectable RNAs in sporozoites, while 1,500–2,000 proteins can be detected by mass spectrometry (about one third of the parasite proteins encoded by the genome) (Lindner et al., 2019). Comparison of the midgut and salivary gland sporozoite populations identified up-regulated in oocyst sporozoites (UOS) and up-regulated in infectious sporozoites (UIS) mRNAs and proteins (Matuschewski et al., 2002; Lasonder et al., 2008; Mikolajczak et al., 2008; Lindner et al., 2019). While these studies showed a correlation between RNA and protein abundance for many essential and conserved gene products, they also revealed extensive temporal dissociation between transcript and protein steady state abundance, with two overlapping and independent programs of translational repression that temporally regulate protein expression during sporozoite maturation and following transmission to the mammalian host (Lindner et al., 2019).
Plasmodium alternates between active proliferation, in the mosquito midgut and in mammalian hepatocytes and erythrocytes, and differentiation into transmission stages, the gametocytes and sporozoites (Figure 1). During progression through this complex life cycle, the parasite switches transcriptional programs. Like in other eukaryotes, gene expression is regulated at both transcriptional and post-transcriptional levels in Plasmodium. While gene regulation has been mostly investigated in the parasite asexual and sexual blood stages, fewer studies have looked into the sporozoite stage. In this review, we summarize the current understanding of the gene regulation mechanisms underpinning Plasmodium maturation into highly infectious sporozoites capable of invading the mammalian host hepatocytes, establish parasite reservoirs and transmit the disease.
Studies of gene regulation in the malaria parasite have been greatly facilitated by the availability of genome sequence data, as illustrated by the first identification of a specific transcription factor (Myb1) in P. falciparum (Boschet et al., 2004; Gissot et al., 2005), rapidly followed by the discovery of the plant-like Apetala-2 (AP2) domain family of DNA-binding proteins (Balaji et al., 2005). However, the divergence of protein sequences compared to other eukaryotes, with nearly 30% of the parasite proteins still lacking an annotated function, has hindered the identification of key regulators. Genome-wide mutagenesis studies revealed that 60% of genes without a functional annotation in P. berghei show phenotypic defects during the erythrocytic stages, and 36% during the mosquito or liver stages (Bushell et al., 2017; Stanway et al., 2019), while in P. falciparum around 1,000 genes of unknown function were predicted to be essential in blood stages (Zhang et al., 2018). Prediction of domain architecture has been used to improve domain recognition methods in Plasmodium genomes, expanding new predicted domains up to 30% in P. falciparum (platform Plasmobase) (Bernardes et al., 2017; Briquet et al., 2018). While many transcription or translation associated proteins have been predicted in Plasmodium through bioinformatics (Roos et al., 2001; Bischoff and Vaquero, 2010; Bennink and Pradel, 2019), only a few of them have been biologically investigated, and it is plausible that other yet unidentified proteins play key roles in the gene regulation networks that concur to the progression of Plasmodium through its developmental stages. In addition, the genome extreme AT richness in some species such as P. falciparum, with more than 80% AT especially in intergenic regions, is a major obstacle for the identification of cis-regulatory elements.
Gene regulation has been mostly investigated in the context of asexual erythrocytic cycle and sexual conversion of Plasmodium parasites. In contrast, studies of the mosquito stages, including the sporozoite, are more scarse. Sporozoites must be isolated from anopheline mosquitoes, through hand dissection of the salivary glands of infected females. As a result, only limited numbers of sporozoites can be obtained (typically 104 to 105 per mosquito), resulting in limited sample amounts for downstream analysis of mRNA and/or proteins, with the additional issue of contamination with mosquito cellular content. Despite these limitations, highly sensitive next generation sequencing and mass spectrometry approaches, combined with methods to purify salivary gland sporozoites (Kennedy et al., 2012), have facilitated the deciphering of transcriptomes, epigenomes and proteomes of Plasmodium sporozoites (Lasonder et al., 2008; Lindner et al., 2013b; Gómez-Díaz et al., 2017; Swearingen et al., 2017; Zanghì et al., 2018; Lindner et al., 2019; Muller et al., 2019; Hamada et al., 2020).
Nevertheless, steady state mRNA levels result from a balance between mRNA synthesis and mRNA decay, which remain difficult to address in Plasmodium sporozoites. Importantly, detection of transcripts and proteins depends on their concentration and on the sensitivity of the analysis. A given messenger might be present in absence of its cognate protein because of translational repression or because the protein is expressed at low level even though it might be functionally competent. Reciprocally, mRNAs present below the detection limit may still be translated into functionally active proteins. Limited sample size is a major hindrance for proteomic detection of transcription factors, which are typically present at low concentrations in eukaryotic cells, allowing to switch on and off rapidly gene expression. Finally, while most studies have looked at bulk salivary gland sporozoites, single cell RNAseq approaches recently revealed some degree of transcriptome heterogeneity in sporozoite populations within the salivary glands (Howick et al., 2019), which could reflect continuing parasite maturation after invasion of the glands and until transmission. Liver stages, especially early exoerythrocytic forms, are typically present in extremely small numbers in experimental culture systems, and pose additional challenges as they cannot be easily isolated from the infected cell. For this reason, the mechanisms underlying stage conversion between quiescent sporozoites to proliferative liver stages remain poorly understood.
The steady state transcriptome of sporozoites is remarkably stable during residence of the parasite in the mosquito salivary glands (Gomes-Santos et al., 2011). This suggests that mature sporozoites are transcriptionally quiescent, a notion further supported by the observation that sporozoites retain infectivity after exposure to the transcription inhibitor Actinomycin D (Silvie et al., 2014). However, exposure of P. falciparum and P. vivax sporozoites to conditions that mimic the host environment result in profound transcriptome reprogramming, with up-regulation of genes associated with liver stage development (Siau et al., 2008; Roth et al., 2018), illustrating the plasticity of sporozoite transcriptional control.
Plasmodium shares with other eukaryotic organisms common mechanisms for transcriptional gene regulation, including remodeling of the chromatin, post-translational modifications of histones, recruitment of the basal transcription machinery and the concerted action of transcription factors (TFs) that recognize cis-regulatory elements in gene promoter regions (Bischoff and Vaquero, 2010; Toenhake and Bártfai, 2019). Many transcription associated proteins (TAPs) have been annotated in Plasmodium (Roos et al., 2001; Bischoff and Vaquero, 2010), although few have been biologically investigated in the parasite, especially in sporozoites. They can be classified in three main groups according to their putative function in transcription: general transcription factors (GTF), chromatin-related transcription factors (CTF) and specific transcription factors (STF) (Bischoff and Vaquero, 2010). The three groups are represented in the proteome of sporozoites, albeit at variable levels (Figure 2A). Table S1 lists annotated TAPs and protein detection by mass spectrometry in sporozoites from human and rodent Plasmodium species. The lower representation for some of the TAP subcategories could be explained by the absence of expression in sporozoites or by their low abundance in the minute biological material, below the detection limit of mass spectrometry. In eukaryotes, gene expression is largely governed at the level of transcription by stochastic molecular interactions between DNA and TAPs, linked to the fact that most of the proteins exist in cells in very small quantities, which allows switching on and off rapidly gene transcription (Kupiec, 1997). The constant association and dissociation events of DNA with interacting proteins, including histones, contributes to instability, while post-translational modifications of transcriptional regulators can stabilize stochastic gene expression (Kupiec, 1997; Nicolas et al., 2017).
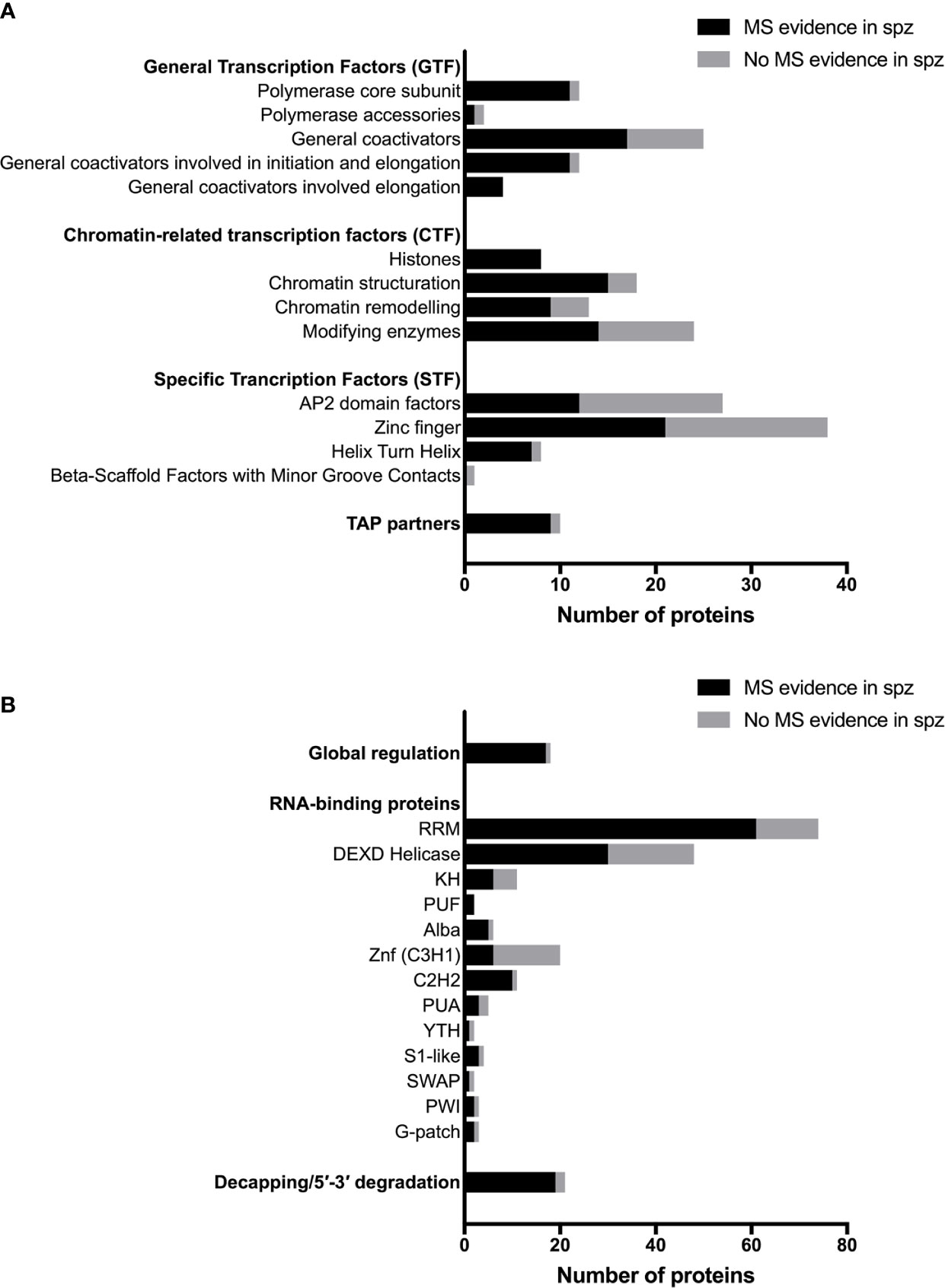
Figure 2 Transcriptional, post-transcriptional and translational regulators in Plasmodium sporozoites. (A) Number of transcriptional-associated proteins (TAPs), based on the classification of Bischoff and Vaquero (2010). Black bars represent proteins with mass spectrometry evidence in Plasmodium spp. sporozoites. Gray bars represent proteins not detected in sporozoites by mass spectrometry. For details on protein identity and mass spectrometry results, see Table S1. (B) Number of post-transcriptional and translational regulators, based on the classification of Reddy et al. (2015) and Bennink and Pradel (2019). Black bars represent proteins with mass spectrometry evidence in Plasmodium spp. sporozoites. Grey bars represent proteins not detected in sporozoites by mass spectrometry. For details on protein identity and mass spectrometry results, see Table S2.
While the regulation of gene expression via the binding of specific TFs on their cognate regulatory elements is a central mechanism that controls complex processes of development and differentiation, transcription is for a large part influenced by chromatin organization, in Plasmodium as in other eukaryotes (Toenhake and Bártfai, 2019). Chromatin remodeling relies on a variety of readers, writers and erasers, involved in post-translational modifications of histones and driving the highly dynamic structure of nucleosomes underlying gene reprogramming (Tyagi et al., 2016). Architectural remodeling provides plasticity by bringing DNA loops and bound TFs in close proximity to scaffold functional multi-protein complexes. Transcription and genome activity can also be modulated by incorporation of long non-coding RNAs (lncRNA). This well-organized environment, which depends on the number and affinity of regulatory elements, defines transcriptional ecosystems of connected genes that allow the fine tuning of transcription programs for a steady-state level of gene expression (Silveira and Bilodeau, 2018). Disturbance of these ecosystems, including through the action of environmental factors, can lead to reprogramming of gene expression.
Epigenetic regulation is a key control mechanism of Plasmodium sexual conversion. In P. falciparum, the heterochromatin protein 1 (HP1) plays a central role in epigenetic silencing of the master regulator of gametocytogenesis AP2-G (Brancucci et al., 2014). Derepression of ap2-g gene expression depends on gametocyte development 1 (GDV1), itself controlled by an antisense RNA (Filarsky et al., 2018). Furthermore, histone post-translational modifications (hPTMs) act as primary epigenetic regulators of monoallelic expression of variant gene families and phenotypic plasticity in P. falciparum erythrocytic stages (Lopez-Rubio et al., 2007; Lopez-Rubio et al., 2009). Recent studies in P. falciparum mosquito stages revealed that similar chromatin regulation mechanisms also operate in sporozoites. Analysis by ChIP-Seq of the genome-wide distribution of HP1 and the repressive histone H3 lysine 9 trimethylation (H3K9me3) mark showed that P. falciparum (NF54 strain) sporozoites expand heterochromatin at subtelomeric regions to silence blood-stage-specific genes, which correlates with the transcription status of clonally variant gene members (Zanghì et al., 2018). One single var gene remained euchromatic, associated with expression of the cognate PfEMP1 protein at the sporozoite surface. Similar results were obtained with P. falciparum sporozoites from field isolates, where transcription of a single (but distinct) var gene was observed, correlating with the presence of low levels of the repressive H3K9me3 mark in its promoter and with the expression of an antisense lncRNA (Gómez-Díaz et al., 2017). Consistent with observations in P. falciparum, the H3K9me3 heterochromatin marks primarily clustered in telomeric and subtelomeric regions in P. vivax sporozoites, while marks associated with active transcription, H3 lysine 4 trimethylation (H3K4me3) and H3 lysine 9 acetylation (H3K9ac), were distributed broadly across the chromosomes outside of the telomeric regions, with no overlap with regions under H3K9me3 suppression (Muller et al., 2019).
Sporozoites express several chromatin modifying enzymes, as shown by proteomic analysis, including chromatin writers (SET7 histone methyltransferases, GCN5 histone acetyl transferase) and erasers (HDAC1, Sir2A, Sir2B) (Figure 2A and Table S1). However, their biological function has not been investigated in sporozoites. One study showed that P. falciparum SET7 is distributed throughout the cytoplasm in salivary gland sporozoites, and localizes to distinct cytoplasmic foci adjacent to the nucleus in erythrocytic and liver stage parasites (Chen et al., 2016). Sporozoite proteomes also include chromatin readers, which participate in chromatin structuring and play an important role in transcriptional programming in eukaryotic cells. In P. falciparum, the bromodomain protein BDP1 has been reported to bind acetylated histone H3 and plays a role in regulating expression of genes encoding invasion factors in P. falciparum blood stages (Josling et al., 2015). BDP1 has been detected in sporozoites by mass spectrometry, as well as the SNF2 helicase and CHD1 chromodomain helicase (Table S1). Among TAP partners, the pleckstrin homology domain finger protein PHD1 is expressed in P. falciparum and P. berghei sporozoites, and could play a role in nodes of connectivity involving epigenetic complexes with bromodomain readers (Hoeijmakers et al., 2019). Again, none of these factors has been biologically investigated so far in sporozoites.
The final step leading to the recruitment of RNA polymerase II and the onset of transcription is chromatin remodeling by increasing nucleosome sliding and accessibility of the chromatin (Agresti and Bianchi, 2003). High-mobility group B (HMGB) proteins are typical members of this category, with architectural properties to recognize distorted DNA and induce stabilization of DNA bending. HMGB proteins appear to be associated with active chromatin (Németh and Längst, 2004; Briquet et al., 2006). HMGB and other chromatin remodeling factors are well represented in the sporozoite proteome (Figure 2A and Table S1). Interestingly, HMGB2 appears to be a critical regulator of parasite transmission to mosquitoes, since disruption of hmgb2 gene in P. yoelii caused a major reduction of oocyst numbers, associated with the down-regulation of a subset of gametocyte transcripts (Gissot et al., 2008). However, hmgb2(-) oocysts were still capable of producing viable sporozoites that were infectious to mice. Although mainly nuclear, HMGB2 was also detected in the cytoplasm of P. falciparum gametocytes (Briquet et al., 2006), consistent with the dual life of the protein both as a nuclear protein and as a secreted alarmin (Briquet et al., 2015).
Apicomplexan AP2 (ApiAP2) DNA-binding domain proteins form the largest known family of specific TFs in Plasmodium, with 27 members in P. falciparum (Balaji et al., 2005; Bischoff and Vaquero, 2010). This family includes sequence-specific TFs with homology to the plant apetala 2/ethylene response TF, and are characterized by the presence of one to three DNA-binding AP2 domains (Balaji et al., 2005; Painter et al., 2011). Plasmodium AP2 TFs are considered as the main specific TFs responsible for parasite progression through distinct developmental stages (Painter et al., 2011). Twelve AP2 factors have been detected by mass spectrometry in sporozoites from at least one species (Figure 2A and Table S1), including six that have been detected in all tested species. Reverse genetics studies in P. berghei and P. yoelii have identified several AP2 factors that play an important role for parasite development in mosquitoes. Four are essential for the formation of infectious ookinetes (AP2-O, AP2-O2, AP2-O3, and AP2-O4), and three are required for sporogony (AP2-Sp, AP2-Sp2, AP2-Sp3) (Yuda et al., 2009; Yuda et al., 2010; Modrzynska et al., 2017; Zhang et al., 2017). Among these, AP2-Sp (Apetala 2 in sporozoites, annotated as AP2-Exp in P. falciparum) was one of the first AP2 TFs to be functionally characterized in vivo in P. berghei with AP2-O. AP2-Sp is expressed from the late oocyst to the salivary gland sporozoite, where it regulates specific gene expression (Yuda et al., 2010). The single AP2 domain of P. falciparum and P. berghei AP2-Sp binds to the TGCATGCA motif, found in the promoter region of many sporozoite-specific genes (Campbell et al., 2010; Iwanaga et al., 2010; Westenberger et al., 2010), which acts as a cis-acting element specific for the sporozoite stage (Yuda et al., 2010; Klug et al., 2018). The AP2-Sp domain was shown to dimerize upon DNA binding (Lindner et al., 2010). P. berghei mutants lacking AP2-Sp form oocysts but fail to produce sporozoites (Yuda et al., 2010). Genes under control of AP2-Sp encode proteins associated with a wide range of functions, including sporozoite formation in the mosquito (CSP) (Ménard et al., 1997), sporozoite motility and invasion of the mosquito salivary glands (TRAP, MAEBL) (Sultan et al., 1997; Kariu et al., 2002), sporozoite cell traversal (SPECT, PLP1) (Ishino et al., 2004; Ishino et al., 2005a), invasion of mammalian host cells (P36, P52) (Ishino et al., 2005b) or liver stage development (UIS3, UIS4) (Mueller et al., 2005a; Mueller et al., 2005b). This strongly suggests that additional factors and possibly complex gene regulatory networks participate in the fine-tuning of gene expression during sporozoite development and maturation. Systematic genetic screens in P. berghei and P. yoelii identified two additional AP2 factors, AP2-Sp2, and AP2-Sp3, as essential for sporozoite production or maturation in oocysts, respectively (Modrzynska et al., 2017; Zhang et al., 2017). Another AP2 TF of the rodent malaria parasite P. berghei expressed in liver stages and designated AP2-L was reported to be necessary for parasite development in hepatocytes (Iwanaga et al., 2012). AP2-L disruption does not affect the ability of sporozoites to migrate through cells and to invade hepatocytes but induce an important delay in patency in mice. The AP2-L phenotype is associated with a decreased level of several transcripts specific to the early liver stages (UIS2, UIS3, UIS4, EXP1, and LISP1) but there was no evidence for a binding DNA motif shared within these co-regulated genes (Iwanaga et al., 2012). Finally, among P. berghei essential AP2 factors (Modrzynska et al., 2017), some are highly expressed in the sporozoite stage, including AP2-I, which regulates invasion-related genes in P. falciparum merozoites (Santos et al., 2017). In fact, among AP2 factors identified in the proteome of sporozoites, six were also identified in the first P. falciparum blood stage nuclear proteome, including AP2-I and AP2-Sp (Oehring et al., 2012), showing some overlap in regulatory components between distinct developmental stages of the parasite. AP2-I and other essential AP2 factors expressed in sporozoites possibly participate in the regulation of sporozoite development and/or maturation. As these AP2 factors are refractory to conventional knockout approaches, conditional genome editing will be required for functional characterization in mosquito stages.
Proteomic studies identified other putative specific TFs in sporozoites, i.e., zinc finger and helix turn helix (HTH) proteins (Figure 2A and Table S1), but their biological role remains unknown. This group includes HMGB3, the only HMGB factor which exhibits one Myb-binding domain (Plasmobase, Vaquero & Briquet personal communication). HMGB3 was detected in the proteome of all tested sporozoite species, as well as the HTH factor ADA2 (Alteration/Deficiency in Activation 2). The first specific TF identified in Plasmodium, Myb1 (Boschet et al., 2004; Gissot et al., 2005), is expressed in oocysts and in liver stages, at least at the mRNA level, but not in sporozoites (Howick et al., 2019). A Myb-like TF was recently identified as a master regulator of Toxoplasma differentiation, representing a counterpoint of the ApiAP2 factors that dominate the common view of Plasmodium gene regulation (Waldman et al., 2020).
In summary, Plasmodium sporozoites express many factors potentially involved in transcription regulation, very few of which have been functionally characterized. In addition, unconventional factors could also participate in gene regulation at the transcriptional level, as illustrated by SLARP, which is discussed in a specific section of this review, raising the possibility that non-canonical regulators remain to be discovered in the malaria parasite.
Plasmodium sporozoites can persist and remain infectious within the salivary glands of the mosquito for several days until they are eventually transmitted to a mammalian host. Remarkably, the parasite mRNA repertoire remains stable during protracted sporozoite residency in the salivary glands (Gomes-Santos et al., 2011). Two control mechanisms are known to operate in sporozoites at the post-transcriptional level: global translation inhibition, at the level of the translation initiation complex, and mRNA-specific post-transcriptional silencing, which typically involves cis-regulatory RNA elements and mRNA-binding proteins. Other mechanisms could also be involved, such as mRNA decay mediated by ribonuclease complexes which plays a role in antigenic variation in P. falciparum (Zhang et al., 2014), but have not been characterized so far in sporozoites.
Translational regulation plays an important role in quiescent Plasmodium transmission stages, i.e., sporozoites and gametocytes, allowing rapid adjustment of gene expression and protein synthesis in response to the new environment that the parasite encounters when transiting from the mosquito vector to the mammalian host, and reciprocally (Bennink and Pradel, 2019). The eukaryote translation machinery is conserved in Plasmodium, where bioinformatics predict the presence of initiation, elongation and termination proteins (Cui et al., 2015; Bennink and Pradel, 2019). Many of the factors involved in translation regulation are expressed in sporozoites, as shown by mass spectrometry (Figure 2B and Table S2). Malaria parasites also express distinct rRNA genes at different developmental stages (Waters et al., 1989), with transcription of S-type rRNA genes taking place as the parasite begins to proliferate in the mosquito (Thompson et al., 1999).
The transient developmental arrest of sporozoites inside mosquito salivary glands implies efficient control mechanisms to prevent premature transformation before transmission. Sporozoite latency in the mosquito is controlled, at least in part, by global inhibition of protein synthesis, mediated through the phosphorylation of the translation initiation factor eIF2α by the sporozoite protein kinase IK2 (Zhang et al., 2010), also termed UIS1 (Matuschewski et al., 2002). P. berghei lacking IK2 display a partial loss of infectivity associated with premature transformation of sporozoites in the mosquito salivary glands (Zhang et al., 2010). In situ hybridization with oligo-d(T) or labelling with antibodies to the polyA-binding protein showed that sporozoites harbor cytoplasmic RNA granules (Zhang et al., 2010). Ik2(-) sporozoites lack such granules and show a general increase in protein synthesis (Zhang et al., 2010). Since global translation inhibition via eIF2α phosphorylation is a conserved stress response in eukaryotes, the premature transformation of sporozoites in the absence of IK2 possibly reflects an increased susceptibility of the mutant parasites to stress encountered during residence in the mosquito salivary glands.
Phosphorylation of eIF2α is regulated by the essential phosphatase UIS2, which is expressed in sporozoites and blood stages (Zhang et al., 2016). UIS2 conditional knockout P. berghei mutants have a defect in liver stage development, associated with increased levels of phosphorylation of eIF2α. UIS2 may promote reactivation of translation once the sporozoite entered the mammalian host, to allow sporozoite transformation and liver stage development. Intriguingly, UIS2 was recently identified as a PVM protein in P. falciparum asexual erythrocytic stages, indicating that UIS2 may have alternative roles (Khosh-Naucke et al., 2018).
Comparison of transcriptome and proteome datasets from P. yoelii and P. falciparum sporozoites revealed that translational repression affects a large subset of genes, including 70%–80% of UIS (Lindner et al., 2019). Transcriptomic and proteomic data from P. vivax sporozoites indicate that translational repression occurs in this species as well (Swearingen et al., 2017; Muller et al., 2019). Post-transcriptional silencing plays a crucial role in female gametocytes, where a defined population of mRNAs encoding ookinete proteins such as P25 and P28 are not translated but instead stably stored in quiescent messenger ribonucleoprotein particles (mRNPs) (Mair et al., 2006; Mair et al., 2010). Transcript-specific translational repression also operates in salivary gland sporozoites, where translation of one of the most abundant mRNAs, encoding UIS4, is inhibited (Silvie et al., 2014; Silva et al., 2016; Lindner et al., 2019). Inside hepatocytes, Plasmodium develops within a parasitophorous vacuole (PV) and remodels the membrane of its vacuole by inserting early transcribed membrane proteins such as UIS4 (Mueller et al., 2005a). uis4 transcripts accumulate in cytoplasmic granules and are translationally repressed in sporozoites, a process that depends on cis-regulatory elements contained in the open reading frame of uis4 (Silvie et al., 2014). UIS4 protein synthesis is activated only after host cell invasion, allowing PVM remodeling. Storage of translationally silent RNA probably permits swift stage conversion yet avoids premature expression of liver stage-specific proteins.
Activation of the translational repression machinery is linked with sporozoite maturation, at least in the case of UIS4 (Silvie et al., 2014), but the effectors remain to be characterized. One possible candidate is the RNA-binding protein Puf2 (Pumilio and fem-3 binding factor homology 2). Puf are evolutionary conserved proteins that typically bind to the 3´ UTR of target mRNAs and repress their translation or induce their degradation (Wickens et al., 2002; Quenault et al., 2011). Silva et al. reported that Puf2 binds to UIS4 mRNA in RNA-immunoprecipitation experiments, and observed an up-regulation of UIS4 protein levels in puf2(-) sporozoites (Silva et al., 2016). However, Puf2 is probably not the only effector of UIS4 repression, and overexpression of Puf2 in liver stages has no impact on parasite development in the liver (Lindner et al., 2013a), indicating that Puf2 is not sufficient to repress UIS4 protein expression. Puf2 localizes to cytoplasmic granules in P. berghei and P. yoelii sporozoites (Gomes-Santos et al., 2011; Lindner et al., 2013a; Silva et al., 2016), and could stabilize sporozoite mRNAs directly or indirectly by regulating other factors controlling RNA metabolism. These storage granules disappear after sporozoite invasion of hepatocytes, possibly releasing specifically bound mRNAs for selective and immediate translation in early EEFs (Lindner et al., 2013a). RNAseq documented genome wide transcriptional alterations in Puf2(-) as compared to WT sporozoites, indicating that Puf2, beyond specific translational repression, contributes to mRNA homeostasis in sporozoites (Lindner et al., 2013a). Puf2(-) sporozoites can invade hepatocytes and differentiate into EEFs, indicating that Puf2 is not required for liver stage development (Müller et al., 2011; Lindner et al., 2013a). However, Puf2 is essential for maintaining sporozoite infectivity during prolonged parasite residence in the mosquito salivary glands (Gomes-Santos et al., 2011; Müller et al., 2011; Lindner et al., 2013a), a function that relies on the protein RNA-binding domain (Lindner et al., 2013a). Over time, Puf2(-) sporozoites transform prematurely into round forms resembling EEFs and loose infectivity, a phenotype that is reminiscent of the behavior of ik2(-) mutants (Zhang et al., 2010). In fact, IK2 mRNAs are down-regulated in puf2(-) sporozoites, suggesting that their phenotype could be due, to a large extent, to depletion of IK2 (Gomes-Santos et al., 2011; Müller et al., 2011; Lindner et al., 2013a).
Sporozoites express many other putative RNA-binding proteins in addition to Puf proteins (Figure 2B and Table S2) (Reddy et al., 2015), some of which likely contribute to global or specific control of RNA homeostasis. As an example, ALBA proteins are DNA/RNA binding proteins that have been detected in DOZI/CITH mRNPs in gametocytes (Mair et al., 2010). In P. yoelii, ALBA4 has a dual function, first in suppressing the activation of male gametocytes, then allowing semi-synchronous development of sporozoites. ALBA4 localizes in cytoplasmic granules and deletion of alba4 gene modifies the steady state level of many genes in sporozoites, showing that ALBA4, like Puf2, contributes to mRNA homeostasis (Muñoz et al., 2017).
Once transmitted to the mammalian host, Plasmodium sporozoites invade hepatocytes and rapidly transform into round forms, which then grow and undergo multiple nuclear divisions. The Sporozoite and Liver Stage Asparagine-Rich Protein (SLARP, also called SAP1), which is conserved among Plasmodium species and specific for the genus, has been identified as a master regulator of liver stage development (Aly et al., 2008; Silvie et al., 2008). Gene deletion of slarp in P. berghei and P. yoelii does not affect parasite blood stage growth, transmission to mosquitoes nor production of salivary gland sporozoites. However, slarp(-) sporozoites are non-infective to mice, due to an early developmental arrest in the liver. slarp(-) sporozoites invade hepatocytes and transform into small uninuclear EEFs, but fail to replicate and are rapidly eliminated (Aly et al., 2008; Silvie et al., 2008; Manzoni et al., 2014).
SLARP plays an essential role in shaping the steady state mRNA repertoire of sporozoites in the mosquito (Aly et al., 2008; Silvie et al., 2008; Aly et al., 2011). Transcriptome analysis with oligonucleotide microarrays revealed 38 down-regulated and 14 up-regulated genes in P. yoelii slarp-deficient sporozoites compared to WT parasites (Aly et al., 2011). Several uis genes, including uis4, were enriched among down-regulated genes in P. berghei and P. yoelii slarp(-) sporozoites (Aly et al., 2008; Silvie et al., 2008). The mechanism of SLARP-mediated regulation of gene expression is still unclear. SLARP is a large protein containing long stretches of low complexity, with an overrepresentation of asparagine residues, but lacks any known functional domain such as DNA- or RNA-binding domains. The C-terminal portion of SLARP is highly conserved across Plasmodium species and contains two putative nuclear localization signals (Silvie et al., 2008). The cellular localization of SLARP remains controversial, as different studies documented a cytoplasmic distribution in P. yoelii sporozoites (Aly et al., 2008; Aly et al., 2011) and a nuclear localization in P. berghei sporozoites (Silvie et al., 2008). Based on 3’ and 5’ RACE PCR experiments in P. yoelii sporozoites, Aly et al. concluded that SLARP depletion causes specific mRNA degradation, suggesting a role at the post-transcriptional level (Aly et al., 2011). However, experiments with reporter centromeric plasmids established that SLARP controls gene expression by acting on the promoter region of target genes (Briquet & Silvie personal communication). Combined with the nuclear localization of the protein in P. berghei sporozoites and EEFs (Silvie et al., 2008), we propose that SLARP functions as a master transcriptional regulator in sporozoites and liver stages (Figure 1). The absence of nucleic acid binding domain identified in SLARP sequence suggests an indirect role, perhaps in complex with AP2 domain transcription factors or chromatin-associated regulators.
P. vivax, unlike P. falciparum, is characterized by relapsing infections, caused by the reactivation of dormant hypnozoites in the liver. Relapses account for a major proportion of P. vivax malaria cases, and constitute a major obstacle for malaria eradication (White et al., 2014). This phenotype is shared with a few other primate malaria parasites, including the sister species P. cynomolgi, which infects Asian macaques (Krotoski et al., 1982a; Krotoski et al., 1982b). The mechanisms underlying hypnozoite formation, persistence and reactivation remain unknown. In contrast with P. falciparum, in vitro culturing of P. vivax blood stages is not yet feasible (Gunalan et al., 2020), limiting the access to sporozoites since mosquitoes must be infected by feeding on blood collected from individuals carrying circulating gametocytes. In this context, P. cynomolgi provides a relevant model to investigate the hypnozoite biology, and recent progress in genetic manipulation of P. cynomolgi has opened new perspectives for the study of hypnozoite formation and reactivation (Voorberg-van der Wel et al., 2013; Voorberg-van der Wel et al., 2020).
It has been hypothesized that, in relapsing Plasmodium species, cell fate could be predetermined in sporozoites, which may exist in two forms in the salivary glands of infected mosquitoes, tachysporozoites that develop directly into replicating EEFs, and bradysporozoites, which transform into dormant hypnozoites (Lysenko et al., 1977; White et al., 2017). However, transcriptome and proteome analysis of P. vivax sporozoites revealed overall similar steady state mRNA and protein compositions as observed in non-relapsing P. falciparum and P. yoelii, with stage-specific expression of the same genes needed for hepatocyte invasion and liver stage development (Westenberger et al., 2010; Swearingen et al., 2017; Muller et al., 2019). Nevertheless, orthologs with significantly different expression levels between species were observed, as well as highly expressed species-specific P. vivax genes with no known ortholog (Westenberger et al., 2010; Swearingen et al., 2017). While the similar transcriptome and proteome profiles of P. vivax as compared to non-relapsing parasites does not favor the hypothesis of predetermined sporozoite fate, single cell approaches are needed to determine whether sporozoites form two distinct populations in relapsing species. Also, other mechanisms could differentially operate in sporozoite subpopulations, including chromatin regulation, which could contribute to stochastic generation of phenotypic diversity in sporozoites, as observed in P. falciparum blood stages with clonally variant gene expression (Cortés et al., 2012). Again, this hypothesis is difficult to explore without epigenome studies performed at the single cell level. Another possible model is that formation of hypnozoites takes place in the environment of infected hepatocytes, following sporozoite invasion. A phosphatidylinositol 4-kinase (PI4K) inhibitor reduced the number of hypnozoites in a prophylactic regimen but not when administered as a radical cure scheme (Zeeman et al., 2016), supporting the idea that the dormant phenotype is established post-invasion. Cell fate determination may be influenced by environmental factors in the host cell and/or stochastic regulatory events.
A still open question is whether hypnozoites represent a distinct stage or merely correspond to arrested early EEFs. To solve this question, comparison of hypnozoites and early non-arrested EEFs is still needed, yet would require a hypnozoite-specific marker to distinguish and sort dormant and non-dormant parasites. Unfortunately, there is to date no specific marker of hypnozoite identified. Candidate dormancy-related genes have been identified based on in silico studies (Carlton et al., 2008; Tachibana et al., 2012; de Souza Ribeiro et al., 2018), but none has been validated yet. The recent identification of LISP2 as an early marker of liver stage development should facilitate the differential analysis of hypnozoite versus developing early EEFs (Gupta et al., 2019). Indeed, transgenic P. cynomolgi expressing mCherry under control of lisp2 promoter allowed for the first time live imaging of hypnozoite reactivation events (Voorberg-van der Wel et al., 2020).
To circumvent the lack of hypnozoite-specific marker, several recent studies have unraveled the transcriptome of P. vivax or P. cynomolgi cultured hypnozoites in comparison with hepatic schizonts, either after pharmacological enrichment (Gural et al., 2018) or after isolation of hypnozoites and schizonts by laser capture microdissection (Cubi et al., 2017) or sorting of fluorescent P. cynomologi by flow cytometry (Voorberg-van der Wel et al., 2013; Voorberg-van der Wel et al., 2017; Bertschi et al., 2018). These studies revealed that hypnozoites express a lower number of genes compared to schizonts, representing a limited number of pathways. Not surprisingly, the level of gene expression was higher in schizonts than in hypnozoites. When comparing transcriptomes across distinct stages, hypnozoites appear to be a transition point between sporozoites and replicating schizonts, supporting the idea that hypnozoites correspond to arrested early EEFs rather than a distinct stage (Muller et al., 2019). These transcriptional studies failed to identify a specific transcriptional marker for hypnozoites, but revealed that despite a general metabolic shutdown hypnozoites continue to express several specific pathways likely involved in the maintenance of dormancy, including ATP homeostasis and chromatin maintenance (Voorberg-van der Wel et al., 2017; Bertschi et al., 2018).
P. cynomolgi hypnozoites display the acetylated H4K8 mark of open chromatin, indicating that they are metabolically active with persistent transcriptional activity (Voorberg-van der Wel et al., 2017). One AP2 domain protein (coined AP2-Q) was proposed as a candidate marker of quiescence, together with two other AP2 factors (AP2-O2 and AP2-G2) (Cubi et al., 2017). However, up-regulation of this putative AP2-Q was not confirmed in other studies (Voorberg-van der Wel et al., 2017; Gural et al., 2018). AP2-L, which plays a role during liver stage development (Iwanaga et al., 2012), was equally expressed in hypnozoites and schizonts in both P. vivax and P. cynomolgi. Another AP2 domain protein (PVP01_0916300) was found to be differentially expressed in P. vivax hypnozoites (Gural et al., 2018), and its ortholog was also found to be expressed in P. cynomolgi hypnozoites (Voorberg-van der Wel et al., 2017; Bertschi et al., 2018). Although not specific for relapsing malaria parasites, this particular AP2 factor could be involved in the maintenance of dormant liver stage parasites, together with other AP2 factors identified in hypnozoites (Voorberg-van der Wel et al., 2017).
Cubi et al. also noted an up-regulation of the eIF2 kinase IK2 in P. cynomolgi hypnozoites, suggesting a possible role of translational repression in liver stage dormancy (Cubi et al., 2017). Analysis of the phosphoproteome of P. vivax sporozoites revealed enrichment in DNA and RNA-binding proteins among phosphorylated proteins, raising the possibility that post-translational control may also participate in regulation of dormancy (Swearingen et al., 2017).
Dormancy could be regulated at least in part by chromatin modifications, since inhibitors of histone methyltransferases stimulate the reactivation of P. cynomolgi hypnozoites in simian hepatocyte cultures (Dembélé et al., 2014). In agreement with this idea, hypnozoites were found to express genes involved in histone acetylation and methylation as well as chaperone-mediated modulation of nucleosome-histone interactions, in addition to members of the alba gene family, which have DNA and RNA binding activities (Bertschi et al., 2018). Finally, non-AP2 transcriptional regulators could also participate in parasite dormancy, including Myb proteins and SLARP. A Myb-like transcription factor was recently identified as a master regulator of bradyzoite differentiation in Toxoplasma gondii (Waldman et al., 2020). Plasmodium genome encodes Myb-like proteins, including Myb1 (Gissot et al., 2005), for which transcripts have been found in P. berghei liver stages in single cell RNAseq experiments (Howick et al., 2019). Intriguingly, SLARP was detected in P. falciparum, P. berghei and P. yoelii sporozoites (Lindner et al., 2013b; Lindner et al., 2019; Hamada et al., 2020), but not in the P. vivax sporozoite proteome (Swearingen et al., 2017). Rodent parasites with targeted gene deletion of slarp show a complete developmental arrest in early liver stages, prior to replication, resulting in small hypnozoite-like forms in cell cultures (Aly et al., 2008; Silvie et al., 2008; Manzoni et al., 2014). SLARP is conserved across Plasmodium species, yet its contribution during liver stage dormancy is still unknown.
Transmission of Plasmodium from the insect vector to a mammalian host is a critical step of the parasite life cycle, and requires coordinated gene expression for development and maturation into infectious sporozoites that can establish infection in the host liver. Although the knowledge of the gene regulatory mechanisms operating in sporozoites is still fragmented, master transcriptional regulators of sporozoite development and maturation have been identified, which probably participate in more complex regulatory circuits that still need to be characterized. In addition, translational repression plays a crucial role in mosquito stages, ensuring that quiescent sporozoites awaiting transmission in the mosquito salivary glands are already prepared for swift transition to the next replicative stage in the liver of the mammalian host. Many questions remain to be addressed, including the role of chromatin regulators, non-coding RNAs and specific TFs in sporozoite development and maturation. Also, the mechanisms underlying sporozoite transformation into dormant liver stages and hypnozoite reactivation in the medically important P. vivax still need to be elucidated. Deciphering gene regulation in Plasmodium sporozoites not only will help us to better understand development of this unicellular parasite but will also provide insights into many parasite-specific phenomena such as antigenic variation, alternative invasion pathways and dormancy, which may ultimately lead to the development of novel control strategies targeting host-parasite interactions. More importantly, the panel of the parasite regulation machinery distinct from those of its mammalian hosts makes it a promising target for novel antimalarial chemotherapies.
SB, OS, and CV drafted the manuscript. CM and SB computed the proteome data. CM drew Figure 1. All authors contributed to the article and approved the submitted version.
OS is supported by the Laboratoire d’Excellence ParaFrap (ANR-11-LABX-0024), the Agence Nationale de la Recherche (ANR-16-CE15-0004 and ANR-16-CE15-0010), and the Fondation pour la Recherche Médicale (EQU201903007823).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcimb.2020.618430/full#supplementary-material
Adams J. H., Mueller I. (2017). The biology of plasmodium vivax. Cold Spring Harb. Perspect. Med. 7, a025585. doi: 10.1101/cshperspect.a025585
Agresti A., Bianchi M. E. (2003). HMGB proteins and gene expression. Curr. Opin. Genet. Dev. 13, 170–178. doi: 10.1016/S0959-437X(03)00023-6
Aly A. S. II, Mikolajczak S. A., Rivera H. S., Camargo N., Jacobs-Lorena V., Labaied M., et al. (2008). Targeted deletion of SAP1 abolishes the expression of infectivity factors necessary for successful malaria parasite liver infection. Mol. Microbiol. 69, 152–163. doi: 10.1111/j.1365-2958.2008.06271.x
Aly A. S. II, Lindner S. E., MacKellar D. C., Peng X., Kappe S. H. II (2011). SAP1 is a critical post-transcriptional regulator of infectivity in malaria parasite sporozoite stages. Mol. Microbiol. 79, 929–939. doi: 10.1111/j.1365-2958.2010.07497.x
Balaji S., Madan Babu M., Iyer L. M., Aravind L. (2005). Discovery of the principal specific transcription factors of Apicomplexa and their implication for the evolution of the AP2-integrase DNA binding domains. Nucleic Acids Res. 33, 3994–4006. doi: 10.1093/nar/gki709
Bennink S., Pradel G. (2019). The molecular machinery of translational control in malaria parasites. Mol. Microbiol. 112, 1658–1673. doi: 10.1111/mmi.14388
Bernardes J., Vaquero C., Carbone A. (2017). Plasmobase: a comparative database of predicted domain architectures for Plasmodium genomes. Malar. J. 16, 1–10. doi: 10.1186/s12936-017-1887-8
Bertschi N. L., Voorberg-Van der Wel A., Zeeman A. M., Schuierer S., Nigsch F., Carbone W., et al. (2018). Transcriptomic analysis reveals reduced transcriptional activity in the malaria parasite Plasmodium cynomolgi during progression into dormancy. Elife 7, e41081. doi: 10.7554/eLife.41081
Bischoff E., Vaquero C. (2010). In silico and biological survey of transcription-associated proteins implicated in the transcriptional machinery during the erythrocytic development of Plasmodium falciparum. BMC Genomics 11, 34. doi: 10.1186/1471-2164-11-34
Boschet C., Gissot M., Briquet S., Hamid Z., Claudel-Renard C., Vaquero C. (2004). Characterization of PfMyb1 transcription factor during erythrocytic development of 3D7 and F12 Plasmodium falciparum clones. Mol. Biochem. Parasitol. 138, 159–163. doi: 10.1016/j.molbiopara.2004.07.011
Brancucci N. M. B., Bertschi N. L., Zhu L., Niederwieser I., Chin W. H., Wampfler R., et al. (2014). Heterochromatin protein 1 secures survival and transmission of malaria parasites. Cell Host Microbe 16, 165–176. doi: 10.1016/j.chom.2014.07.004
Briquet S., Boschet C., Gissot M., Sevilla E., Franetich J., Hamid Z., et al. (2006). High-Mobility-Group Box Nuclear Factors of Plasmodium falciparum High-Mobility-Group Box Nuclear Factors of Plasmodium falciparum † Eukaryot Cell 5, 672–82. doi: 10.1128/EC.5.4.672
Briquet S., Lawson-Hogban N., Boisson B., Soares M. P., Péronet R., Smith L., et al. (2015). Disruption of parasite hmgb2 gene attenuates plasmodium berghei ANKA pathogenicity. Infect. Immun. 83, 2771–2784. doi: 10.1128/IAI.03129-14
Briquet S., Ourimi A., Pionneau C., Bernardes J., Carbone A., Chardonnet S., et al. (2018). Identification of Plasmodium falciparum nuclear proteins by mass spectrometry and proposed protein annotation. PloS One 13, e0205596. doi: 10.1371/journal.pone.0205596
Bushell E., Gomes A. R., Sanderson T., Anar B., Girling G., Herd C., et al. (2017). Functional Profiling of a Plasmodium Genome Reveals an Abundance of Essential Genes. Cell 170, 260–272.e8. doi: 10.1016/j.cell.2017.06.030
Campbell T. L., de Silva E. K., Olszewski K. L., Elemento O., Llinás M. (2010). Identification and Genome-Wide Prediction of DNA Binding Specificities for the ApiAP2 family of regulators from the malaria parasite. PloS Pathog. 6, e1001165. doi: 10.1371/journal.ppat.1001165
Carlton J. M., Adams J. H., Silva J. C., Bidwell S. L., Lorenzi H., Caler E., et al. (2008). Comparative genomics of the neglected human malaria parasite Plasmodium vivax. Nature 455, 757–763. doi: 10.1038/nature07327
Chen P. B., Ding S., Zanghì G., Soulard V., DiMaggio P. A., Fuchter M. J., et al. (2016). Plasmodium falciparum PfSET7: Enzymatic characterization and cellular localization of a novel protein methyltransferase in sporozoite, liver and erythrocytic stage parasites. Sci. Rep. 6, 21802. doi: 10.1038/srep21802
Cortés A., Crowley V. M., Vaquero A., Voss T. S. (2012). A View on the Role of Epigenetics in the Biology of Malaria Parasites. PloS Pathog. 8, e1002943. doi: 10.1371/journal.ppat.1002943
Cubi R., Vembar S. S., Biton A., Franetich J. F., Bordessoulles M., Sossau D., et al. (2017). Laser capture microdissection enables transcriptomic analysis of dividing and quiescent liver stages of Plasmodium relapsing species. Cell. Microbiol. 19, e12735. doi: 10.1111/cmi.12735
Cui L., Lindner S., Miao J. (2015). Translational regulation during stage transitions in malaria parasites. Ann. N. Y. Acad. Sci. 1342, 1–9. doi: 10.1111/nyas.12573
de Souza Ribeiro R., de Melo Resende D., Ruiz J. C., Ferreira Alves de Brito C. (2018). In silico analysis of putative dormancy genes in Plasmodium vivax. Acta Trop. 186, 24–34. doi: 10.1016/j.actatropica.2018.06.026
Dembélé L., Franetich J., Lorthiois A., Gego A., Zeeman A., Kocken C. H. M., et al. (2014). Persistence and activation of malaria hypnozoites in long-term primary hepatocyte cultures. Nat. Med. 20, 307–312. doi: 10.1038/nm.3461
Filarsky M., Fraschka S. A., Niederwieser I., Brancucci N. M. B., Carrington E., Carrió E., et al. (2018). GDV1 induces sexual commitment of malaria parasites by antagonizing HP1-dependent gene silencing. Science 359, 1259–1263. doi: 10.1126/science.aan6042
Frischknecht F., Matuschewski K. (2017). Plasmodium sporozoite biology. Cold Spring Harb. Perspect. Med. 7:a025478. doi: 10.1101/cshperspect.a025478
Frischknecht F., Baldacci P., Martin B., Zimmer C., Thiberge S., Olivo-Marin J. C., et al. (2004). Imaging movement of malaria parasites during transmission by Anopheles mosquitoes. Cell. Microbiol. 6, 687–694. doi: 10.1111/j.1462-5822.2004.00395.x
Gissot M., Briquet S., Refour P., Boschet C., Vaquero C. (2005). PfMyb1, a Plasmodium falciparum transcription factor, is required for intra-erythrocytic growth and controls key genes for cell cycle regulation. J. Mol. Biol. 346, 29–42. doi: 10.1016/j.jmb.2004.11.045
Gissot M., Ting L. M., Daly T. M., Bergman L. W., Sinnis P., Kim K. (2008). High mobility group protein HMGB2 is a critical regulator of Plasmodium oocyst development. J. Biol. Chem. 283, 17030–17038. doi: 10.1074/jbc.M801637200
Gomes-Santos C. S. S., Braks J., Prudêncio M., Carret C., Gomes A. R., Pain A., et al. (2011). Transition of Plasmodium sporozoites into liver stage-like forms is regulated by the RNA binding protein Pumilio. PloS Pathog. 7, e1002046. doi: 10.1371/journal.ppat.1002046
Gómez-Díaz E., Yerbanga R. S., Lefèvre T., Cohuet A., Rowley M. J., Ouedraogo J. B., et al. (2017). Epigenetic regulation of Plasmodium falciparum clonally variant gene expression during development in Anopheles gambiae. Sci. Rep. 7, 40655. doi: 10.1038/srep40655
Gunalan K., Rowley E. H., Miller L. H. (2020). A Way Forward for Culturing Plasmodium vivax. Trends Parasitol. 36, 512–519. doi: 10.1016/j.pt.2020.04.002
Gupta D. K., Dembele L., Voorberg-Van Der Wel A., Roma G., Yip A., Chuenchob V., et al. (2019). The plasmodium liver-specific protein 2 (LISP2) is an early marker of liver stage development. Elife 8, e43362. doi: 10.7554/eLife.43362
Gural N., Mancio-Silva L., Miller A. B., Galstian A., Butty V. L., Levine S. S., et al. (2018). In Vitro Culture, Drug Sensitivity, and Transcriptome of Plasmodium Vivax Hypnozoites. Cell Host Microbe 23, 395–406.e4. doi: 10.1016/j.chom.2018.01.002
Hamada S., Pionneau C., Parizot C., Silvie O., Chardonnet S., Marinach C. (2020). In-depth proteomic analysis of Plasmodium berghei sporozoites using trapped ion mobility spectrometry with parallel accumulation-serial fragmentation. BioRXiv. doi: 10.1101/2020.11.26.400192
Hillyer J. F., Barreau C., Vernick K. D. (2007). Efficiency of salivary gland invasion by malaria sporozoites is controlled by rapid sporozoite destruction in the mosquito haemocoel. Int. J. Parasitol. 37, 673–681. doi: 10.1016/j.ijpara.2006.12.007
Hoeijmakers W. A. M., Miao J., Schmidt S., Toenhake C. G., Shrestha S., Venhuizen J., et al. (2019). Epigenetic reader complexes of the human malaria parasite, Plasmodium falciparum. Nucleic Acids Res. 47, 11574–11588. doi: 10.1093/nar/gkz1044
Howick V. M., Russell A. J. C., Andrews T., Heaton H., Reid A. J., Natarajan K., et al. (2019). The malaria cell atlas: Single parasite transcriptomes across the complete Plasmodium life cycle. Science 365, eaaw2619. doi: 10.1126/science.aaw2619
Ishino T., Yano K., Chinzei Y., Yuda M. (2004). Cell-passage activity is required for the malarial parasite to cross the liver sinusoidal cell layer. PloS Biol. 2, 77–84. doi: 10.1371/journal.pbio.0020004
Ishino T., Chinzei Y., Yuda M. (2005a). A Plasmodium sporozoite protein with a membrane attack complex domain is required for breaching the liver sinusoidal cell layer prior to hepatocyte infection. Cell. Microbiol. 7, 199–208. doi: 10.1111/j.1462-5822.2004.00447.x
Ishino T., Chinzei Y., Yuda M. (2005b). Two proteins with 6-cys motifs are required for malarial parasites to commit to infection of the hepatocyte. Mol. Microbiol. 58, 1264–1275. doi: 10.1111/j.1365-2958.2005.04801.x
Iwanaga S., Khan S. M., Kaneko I., Christodoulou Z., Newbold C., Yuda M., et al. (2010). Functional Identification of the Plasmodium Centromere and Generation of a Plasmodium Artificial Chromosome. Cell Host Microbe 7, 245–255. doi: 10.1016/j.chom.2010.02.010
Iwanaga S., Kaneko I., Kato T., Yuda M. (2012). Identification of an AP2-family Protein That Is Critical for Malaria Liver Stage Development. PloS One 7, e47557. doi: 10.1371/journal.pone.0047557
Josling G. A., Petter M., Oehring S. C., Gupta A. P., Dietz O., Wilson D. W., et al. (2015). A Plasmodium Falciparum Bromodomain Protein Regulates Invasion Gene Expression. Cell Host Microbe 17, 741–751. doi: 10.1016/j.chom.2015.05.009
Kariu T., Yuda M., Yano K., Chinzei Y. (2002). MAEBL is essential for malarial sporozoite infection of the mosquito salivary gland. J. Exp. Med. 195, 1317–1323. doi: 10.1084/jem.20011876
Kennedy M., Fishbaugher M. E., Vaughan A. M., Patrapuvich R., Boonhok R., Yimamnuaychok N., et al. (2012). A rapid and scalable density gradient purification method for Plasmodium sporozoites. Malar. J. 11:421. doi: 10.1186/1475-2875-11-421
Khosh-Naucke M., Becker J., Mesén-Ramírez P., Kiani P., Birnbaum J., Fröhlke U., et al. (2018). Identification of novel parasitophorous vacuole proteins in P. falciparum parasites using BioID. Int. J. Med. Microbiol. 308, 13–24. doi: 10.1016/j.ijmm.2017.07.007
Klug D., Kehrer J., Frischknecht F., Singer M. (2018). A synthetic promoter for multi-stage expression to probe complementary functions of Plasmodium adhesins. J. Cell Sci. 131, jcs210971. doi: 10.1242/jcs.210971
Krotoski W. A., Collins W. E., Bray R. S., Garnham P. C., Cogswell F. B., Gwadz R. W., et al. (1982a). Demonstration of hypnozoites in sporozoite-transmitted Plasmodium vivax infection. Am. J. Trop. Med. Hyg. 31, 1291–1293. doi: 10.4269/ajtmh.1982.31.1291
Krotoski W. A., Garnham P. C. C., Bray R. S., Killick-Kendrick R., Draper C. C., Targett G. A., et al. (1982b). Observations on early and late post-sporozoite tissue stages in primate malaria. I. Discovery of a new latent form of Plasmodium cynomolgi (the hypnozoite), and failure to detect hepatic forms within the first 24 hours after infection. Am. J. Trop. Med. Hyg. 31, 24–35. doi: 10.4269/ajtmh.1982.31.24
Kupiec J. J. (1997). A Darwinian theory for the origin of cellular differentiation. Mol. Gen. Genet. 255, 201–208. doi: 10.1007/s004380050490
Lasonder E., Janse C. J., Van Gemert G. J., Mair G. R., Vermunt A. M. W., Douradinha B. G., et al. (2008). Proteomic profiling of Plasmodium sporozoite maturation identifies new proteins essential for parasite development and infectivity. PloS Pathog. 4, e1000195. doi: 10.1371/journal.ppat.1000195
Lindner S. E., De Silva E. K., Keck J. L., Llinás M. (2010). Structural Determinants of DNA Binding by a P. falciparum ApiAP2 Transcriptional Regulator. J. Mol. Biol. 395, 558–567. doi: 10.1016/j.jmb.2009.11.004
Lindner S. E., Mikolajczak S. A., Vaughan A. M., Moon W., Joyce B. R., Sullivan W. J. Jr., et al (2013a). Perturbations of Plasmodium Puf2 expression and RNA-seq of Puf2-deficient sporozoites reveal a critical role in maintaining RNA homeostasis and parasite transmissibility. Cell Microbiol. 15, 1266–1283. doi: 10.1111/cmi.12116
Lindner S. E., Swearingen K. E., Harupa A., Vaughan A. M., Sinnis P., Moritz R. L., et al. (2013b). Total and putative surface proteomics of malaria parasite salivary gland sporozoites. Mol. Cell Proteomics 12, 1127–1143. doi: 10.1074/mcp.M112.024505
Lindner S. E., Swearingen K. E., Shears M. J., Walker M. P., Vrana E. N., Hart K. J., et al. (2019). Transcriptomics and proteomics reveal two waves of translational repression during the maturation of malaria parasite sporozoites. Nat. Commun. 10, 4964. doi: 10.1038/s41467-019-12936-6
Lopez-Rubio J. J., Gontijo A. M., Nunes M. C., Issar N., Hernandez Rivas R., Scherf A. (2007). 5′ flanking region of var genes nucleate histone modification patterns linked to phenotypic inheritance of virulence traits in malaria parasites. Mol. Microbiol. 66, 1296–1305. doi: 10.1111/j.1365-2958.2007.06009.x
Lopez-Rubio J. J., Mancio-Silva L., Scherf A. (2009). Genome-wide Analysis of Heterochromatin Associates Clonally Variant Gene Regulation with Perinuclear Repressive Centers in Malaria Parasites. Cell Host Microbe 5, 179–190. doi: 10.1016/j.chom.2008.12.012
Lysenko A. J., Beljaev A. E., Rybalka V. M. (1977). Population studies of Plasmodium vivax. I. The theory of polymorphism of sporozoites and epidemiological phenomena of tertian malaria. Bull. World Health Organ. 55, 541–549.
Mair G. R., Braks J. A., Garver L. S., Wiegant J. C., Hall N., Dirks R. W., et al. (2006). Regulation of sexual development of Plasmodium by translational repression. Science 313, 667–669. doi: 10.1126/science.1125129
Mair G. R., Lasonder E., Garver L. S., Franke-Fayard B. M., Carret C. K., Wiegant J. C., et al. (2010). Universal features of post-transcriptional gene regulation are critical for Plasmodium zygote development. PloS Pathog. 6, e1000767. doi: 10.1371/journal.ppat.1000767
Manzoni G., Briquet S., Risco-Castillo V., Gaultier C., Topcu S., Ivanescu M. L., et al. (2014). A rapid and robust selection procedure for generating drug-selectable marker-free recombinant malaria parasites. Sci. Rep. 4, 4760. doi: 10.1038/srep04760
Matuschewski K., Ross J., Brown S. M., Kaiser K., Nussenzweig V., Kappe S. H. II (2002). Infectivity-associated changes in the transcriptional repertoire of the malaria parasite sporozoite stage. J. Biol. Chem. 277, 41948–41953. doi: 10.1074/jbc.M207315200
Ménard R., Sultan A. A., Cortes C., Altszuler R., Van Dijk M. R., Janse C. J., et al. (1997). Circumsporozoite protein is required for development of malaria sporozoites in mosquitoes. Nature 385, 336–339. doi: 10.1038/385336a0
Mikolajczak S. A., Silva-rivera H., Peng X., Tarun A. S., Camargo N., Jacobs-lorena V., et al. (2008). Distinct Malaria Parasite Sporozoites Reveal Transcriptional Changes That Cause Differential Tissue Infection Competence in the Mosquito Vector and Mammalian Host Mol Cell Biol. 28, 6196–207. doi: 10.1128/MCB.00553-08
Modrzynska K., Pfander C., Chappell L., Yu L., Suarez C., Dundas K., et al. (2017). A Knockout Screen of ApiAP2 Genes Reveals Networks of Interacting Transcriptional Regulators Controlling the Plasmodium Life Cycle. Cell Host Microbe 21, 11–22. doi: 10.1016/j.chom.2016.12.003
Mueller A. K., Camargo N., Kaiser K., Andorfer C., Frevert U., Matuschewski K., et al. (2005a). Plasmodium liver stage developmental arrest by depletion of a protein at the parasite-host interface. Proc. Natl. Acad. Sci. U. S. A. 102, 3022–3027. doi: 10.1073/pnas.0408442102
Mueller A. K., Labaled M., Kappe S. H. II, Matuschewski K. (2005b). Genetically modified Plasmodium parasites as a protective experimental malaria vaccine. Nature 433, 164–167. doi: 10.1038/nature03188
Müller K., Matuschewski K., Silvie O. (2011). The Puf-family RNA-binding protein Puf2 controls sporozoite conversion to liver stages in the malaria parasite. PloS One 6, e19860. doi: 10.1371/journal.pone.0019860
Muller I., Jex A. R., Kappe S. H. II, Mikolajczak S. A., Sattabongkot J., Patrapuvich R., et al. (2019). Transcriptome and histone epigenome of Plasmodium vivax salivary-gland sporozoites point to tight regulatory control and mechanisms for liver-stage differentiation in relapsing malaria. Int. J. Parasitol. 49, 501–513. doi: 10.1016/j.ijpara.2019.02.007
Muñoz E. E., Hart K. J., Walker M. P., Kennedy M. F., Shipley M. M., Lindner S. E. (2017). ALBA4 modulates its stage-specific interactions and specific mRNA fates during Plasmodium yoelii growth and transmission. Mol. Microbiol. 106, 266–284. doi: 10.1111/mmi.13762
Németh A., Längst G. (2004). Chromatin higher order structure: Opening up chromatin for transcription. Briefings Funct. Genomics Proteomics 2, 334–343. doi: 10.1093/bfgp/2.4.334
Nicolas D., Phillips N. E., Naef F. (2017). What shapes eukaryotic transcriptional bursting? Mol. Biosyst. 13, 1280–1290. doi: 10.1039/c7mb00154a
Oehring S. C., Woodcroft B. J., Moes S., Wetzel J., Dietz O., Pulfer A., et al. (2012). Organellar proteomics reveals hundreds of novel nuclear proteins in the malaria parasite Plasmodium falciparum. Genome Biol. 13, R108. doi: 10.1186/gb-2012-13-11-r108
Painter H. J., Campbell T. L., Llinás M. (2011). The Apicomplexan AP2 family: integral factors regulating Plasmodium development. Mol. Biochem. Parasitol. 176, 1–7. doi: 10.1016/j.molbiopara.2010.11.014
Quenault T., Lithgow T., Traven A. (2011). PUF proteins: repression, activation and mRNA localization. Trends Cell Biol. 21, 104–112. doi: 10.1016/j.tcb.2010.09.013
Reddy B. N., Shrestha S., Hart K. J., Liang X., Kemirembe K., Cui L., et al. (2015). A bioinformatic survey of RNA-binding proteins in Plasmodium. BMC Genomics 16, 890. doi: 10.1186/s12864-015-2092-1
Roos D. S., Kissinger J. C., Milgram A. J., Fraunholz M. J., Roos D. S., Brunk B. P., et al. (2001). PlasmoDB: An integrative database of the Plasmodium falciparum genome. Tools for accessing and analyzing finished and unfinished sequence data. Nucleic Acids Res. 29, 66–69. doi: 10.1093/nar/29.1.66
Roth A., Adapa S. R., Zhang M., Liao X., Saxena V., Goffe R., et al. (2018). Unraveling the Plasmodium vivax sporozoite transcriptional journey from mosquito vector to human host. Sci. Rep. 8, 12183. doi: 10.1038/s41598-018-30713-1
Santos J. M., Josling G., Ross P., Joshi P., Orchard L., Campbell T., et al. (2017). Red Blood Cell Invasion by the Malaria Parasite Is Coordinated by the PfAP2-I Transcription Factor. Cell Host Microbe 21, 731–741.e10. doi: 10.1016/j.chom.2017.05.006
Sato Y., Montagna G. N., Matuschewski K. (2014). Plasmodium berghei sporozoites acquire virulence and immunogenicity during mosquito hemocoel transit. Infect. Immun. 82, 1164–1172. doi: 10.1128/IAI.00758-13
Siau A., Silvie O., Franetich J.-F., Yalaoui S., Marinach C., Hannoun L., et al. (2008). Temperature shift and host cell contact up-regulate sporozoite expression of Plasmodium falciparum genes involved in hepatocyte infection. PloS Pathog. 4, e1000121. doi: 10.1371/journal.ppat.1000121
Silva P. A., Guerreiro A., Santos J. M., Braks J. A. M., Janse C. J., Mair G. R. (2016). Translational control of UIS4 protein of the host-parasite interface is mediated by the RNA binding protein Puf2 in Plasmodium berghei sporozoites. PloS One 11, e0147940. doi: 10.1371/journal.pone.0147940
Silveira M. A. D., Bilodeau S. (2018). Defining the Transcriptional Ecosystem. Mol. Cell 72, 920–924. doi: 10.1016/j.molcel.2018.11.022
Silvie O., Goetz K., Matuschewski K. (2008). A sporozoite asparagine-rich protein controls initiation of Plasmodium liver stage development. PloS Pathog. 4, e1000086. doi: 10.1371/journal.ppat.1000086
Silvie O., Briquet S., Müller K., Manzoni G., Matuschewski K. (2014). Post-transcriptional silencing of UIS4 in Plasmodium berghei sporozoites is important for host switch. Mol. Microbiol. 91, 1200–1213. doi: 10.1111/mmi.12528
Sinnis P., Zavala F. (2012). The skin: where malaria infection and the host immune response begin. Semin. Immunopathol. 34, 787–792. doi: 10.1007/s00281-012-0345-5
Stanway R. R., Bushell E., Chiappino-Pepe A., Roques M., Sanderson T., Franke-Fayard B., et al. (2019). Genome-Scale Identification of Essential Metabolic Processes for Targeting the Plasmodium Liver Stage. Cell 179, 1112–1128.e26. doi: 10.1016/j.cell.2019.10.030
Sultan A. A., Thathy V., Frevert U., Robson K. J. H., Crisanti A., Nussenzweig V., et al. (1997). TRAP is necessary for gliding motility and infectivity of plasmodium sporozoites. Cell 90, 511–522. doi: 10.1016/S0092-8674(00)80511-5
Swearingen K. E., Lindner S. E., Flannery E. L., Vaughan A. M., Morrison R. D., Patrapuvich R., et al. (2017). Proteogenomic analysis of the total and surface-exposed proteomes of Plasmodium vivax salivary gland sporozoites. PloS Negl. Trop. Dis. 11, e0005791. doi: 10.1371/journal.pntd.0005791
Tachibana S. II, Sullivan S. A., Kawai S., Nakamura S., Kim H. R., Goto N., et al. (2012). Plasmodium cynomolgi genome sequences provide insight into Plasmodium vivax and the monkey malaria clade. Nat. Genet. 44, 1051–1055. doi: 10.1038/ng.2375
Thompson J., Van Spaendonk R. M. L., Choudhuri R., Sinden R. E., Janse C. J., Waters A. P. (1999). Heterogeneous ribosome populations are present in Plasmodium berghei during development in its vector. Mol. Microbiol. 31, 253–260. doi: 10.1046/j.1365-2958.1999.01167.x
Toenhake C. G., Bártfai R. (2019). What functional genomics has taught us about transcriptional regulation in malaria parasites. Brief. Funct. Genomics 18, 290–301. doi: 10.1093/bfgp/elz004
Touray M. G., Warburg A., Laughinghouse A., Krettli A. U., Miller L. H. (1992). Developmentally Regulated Infectivity of Malaria Sporozoites for Mosquito Salivary Glands and the Vertebrate Host. J. Exp. Med. 175, 1607–1612. doi: 10.1084/jem.175.6.1607
Tyagi M., Imam N., Verma K., Patel A. K. (2016). Chromatin remodelers: We are the drivers! Nucleus 7, 388–404. doi: 10.1080/19491034.2016.1211217
Voorberg-van der Wel A., Zeeman A.-M., van Amsterdam S. M., van den Berg A., Klooster E. J., Iwanaga S., et al. (2013). Transgenic fluorescent Plasmodium cynomolgi liver stages enable live imaging and purification of Malaria hypnozoite-forms. PloS One 8, e54888. doi: 10.1371/journal.pone.0054888
Voorberg-van der Wel A., Roma G., Gupta D. K., Schuierer S., Nigsch F., Carbone W., et al. (2017). A comparative transcriptomic analysis of replicating and dormant liver stages of the relapsing malaria parasite Plasmodium cynomolgi. Elife 6, e29605. doi: 10.7554/elife.29605
Voorberg-van der Wel A. M., Zeeman A. M., Nieuwenhuis I. G., van der Werff N. M., Klooster E. J., Klop O., et al. (2020). A dual fluorescent Plasmodium cynomolgi reporter line reveals in vitro malaria hypnozoite reactivation. Commun. Biol. 3, 7. doi: 10.1038/s42003-019-0737-3
Waldman B. S., Schwarz D., Wadsworth M. H., Saeij J. P., Shalek A. K., Lourido S. (2020). Identification of a Master Regulator of Differentiation in Toxoplasma. Cell 180, 359–372.e16. doi: 10.1016/j.cell.2019.12.013
Waters A. P., Syin C., McCutchan T. F. (1989). Developmental regulation of stage-specific ribosome populations in Plasmodium. Nature 342, 438–440. doi: 10.1038/342438a0
Westenberger S. J., McClean C. M., Chattopadhyay R., Dharia N. V., Carlton J. M., Barnwell J. W., et al. (2010). A systems-based analysis of Plasmodium vivax lifecycle transcription from human to mosquito. PloS Negl. Trop. Dis. 4, e653. doi: 10.1371/journal.pntd.0000653
White M. T., Karl S., Battle K. E., Hay S. II, Mueller I., Ghani A. C. (2014). Modelling the contribution of the hypnozoite reservoir to Plasmodium vivax transmission. Elife 3, 1–19. doi: 10.7554/eLife.04692
White M., Amino R., Mueller I. (2017). Theoretical Implications of a Pre-Erythrocytic Plasmodium vivax Vaccine for Preventing Relapses. Trends Parasitol. 33, 260–263. doi: 10.1016/j.pt.2016.12.011
Wickens M., Bernstein D. S., Kimble J., Parker R. (2002). A PUF family portrait: 3′UTR regulation as a way of life. Trends Genet. 18, 150–157. doi: 10.1016/S0168-9525(01)02616-6
World Health Organization (2019). World Malaria Report 2019. 1–232. Available at: https://www.who.int/publications/i/item/9789241565721
Yuda M., Iwanaga S., Shigenobu S., Mair G. R., Janse C. J., Waters A. P., et al. (2009). Identification of a transcription factor in the mosquito-invasive stage of malaria parasites. Mol. Microbiol. 71, 1402–1414. doi: 10.1111/j.1365-2958.2009.06609.x
Yuda M., Iwanaga S., Shigenobu S., Kato T., Kaneko I. (2010). Transcription factor AP2-Sp and its target genes in malarial sporozoites. Mol. Microbiol. 75, 854–863. doi: 10.1111/j.1365-2958.2009.07005.x
Zanghì G., Vembar S. S., Baumgarten S., Ding S., Guizetti J., Bryant J. M., et al. (2018). A Specific PfEMP1 Is Expressed in P. falciparum Sporozoites and Plays a Role in Hepatocyte Infection. Cell Rep. 22, 2951–2963. doi: 10.1016/j.celrep.2018.02.075
Zeeman A. M., Lakshminarayana S. B., Van Der Werff N., Klooster E. J., Voorberg-Van Der Wel A., Kondreddi R. R., et al. (2016). PI4 kinase is a prophylactic but not radical curative target in Plasmodium vivax-type malaria parasites. Antimicrob. Agents Chemother. 60, 2858–2863. doi: 10.1128/AAC.03080-15
Zhang M., Fennell C., Ranford-Cartwright L., Sakthivel R., Gueirard P., Meister S., et al. (2010). The Plasmodium eukaryotic initiation factor-2alpha kinase IK2 controls the latency of sporozoites in the mosquito salivary glands. J. Exp. Med. 207, 1465–1474. doi: 10.1084/jem.20091975
Zhang Q., Siegel T. N., Martins R. M., Wang F., Cao J., Gao Q., et al. (2014). Exonuclease-mediated degradation of nascent RNA silences genes linked to severe malaria. Nature 513, 431–435. doi: 10.1038/nature13468
Zhang M., Mishra S., Sakthivel R., Fontoura B. M. A., Nussenzweig V. (2016). UIS2: A Unique Phosphatase Required for the Development of Plasmodium Liver Stages. PloS Pathog. 12, e1005370. doi: 10.1371/journal.ppat.1005370
Zhang C., Li Z., Cui H., Jiang Y., Yang Z., Wang X., et al. (2017). Systematic CRISPR-Cas9-mediated modifications of plasmodium yoelii ApiAP2 genes reveal functional insights into parasite development. MBio 8, e01986-17. doi: 10.1128/mBio.01986-17
Keywords: Plasmodium, malaria, sporozoite, gene regulation, quiescence, transcription, translational repression
Citation: Briquet S, Marinach C, Silvie O and Vaquero C (2021) Preparing for Transmission: Gene Regulation in Plasmodium Sporozoites. Front. Cell. Infect. Microbiol. 10:618430. doi: 10.3389/fcimb.2020.618430
Received: 16 October 2020; Accepted: 16 December 2020;
Published: 29 January 2021.
Edited by:
Katarzyna Kinga Modrzynska, University of Glasgow, United KingdomReviewed by:
Gunnar Mair, Iowa State University, United StatesCopyright © 2021 Briquet, Marinach, Silvie and Vaquero. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Olivier Silvie, b2xpdmllci5zaWx2aWVAaW5zZXJtLmZy; Catherine Vaquero, Y2F0aGVyaW5lLnZhcXVlcm9Ac29yYm9ubmUtdW5pdmVyc2l0ZS5mcg==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.