- 1Department of Gastroenterology, Affiliated Hangzhou First People’s Hospital, Zhejiang University School of Medicine, Hangzhou, China
- 2School of Life Science and Technology, China Pharmaceutical University, Nanjing, China
- 3Department of Disease Control and Prevention, Center for Disease Control and Prevention of Eastern Theater Command, Nanjing, China
- 4The First School of Clinical Medicine, Nanjing Medical University, Nanjing, China
- 5Beijing Institute of Radiation Medicine, Beijing, China
The biliary microbiota is related to the pathogenesis of human bile duct stones. However, the extent to which a history of invasive endoscopic sphincterotomy (EST) affects the biliary bacterial community remains largely unknown. We collected bile samples from the common bile duct of 100 choledocholithiasis patients. We performed 16S rRNA sequencing to investigate and compare the biliary microbial community. The patients without antibiotic treatment (AT) were grouped into three clusters based on their biliary microbial compositions. The patients with a history of EST were significantly enriched in one cluster mainly consisting of gastrointestinal bacteria compared with the other two clusters consisting of oral and environmental bacteria. The β-diversities of patients with and without EST were also significantly different, whereas the α-diversities were comparable. The only significantly enriched bacterial genus associated with a history of EST was Pyramidobacter, while eight other genera were significantly decreased. For patients with AT, seven of these genera maintained their association with EST, including Pyramidobacter. However, after AT, the difference in β-diversities was diminished. EST induced a marked shift in the biliary microbial composition. A cluster of biliary bacteria was associated with a history of EST, and Pyramidobacter was specific to EST.
Introduction
Gallstones are a common disease of the human bile duct system. Approximately 15% of the population in the United States has cholelithiasis (Everhart et al., 1999), with choledocholithiasis patients accounting for approximately 10 to 20% of cholelithiasis patients (Williams et al., 2008). Common bile duct stone (CBDS) or choledocholithiasis can be divided into primary and secondary choledocholithiasis. Primary stones usually form in the bile duct and are prevalent in Asian populations, while secondary stones are common in Western countries. It is generally believed that primary choledocholithiasis could be caused by bacterial infection and cholestasis (Williams et al., 2008).
Researchers have endeavored to elucidate the biliary microbiota and its potential function in gallstone pathogenesis (Swidsinski and Lee, 2001; Stewart et al., 2002; Begley et al., 2005; Stewart et al., 2006; Boursi et al., 2016; Kose et al., 2018; Wang et al., 2018). Conventional techniques such as bacterial culture, polymerase chain reaction (PCR) targeting specific bacteria, and transmission electron microscopy have long been used to identify biliary bacteria including Escherichia, Pseudomonas, Klebsiella, Enterobacter, Enterococcus, Haemophilus, Veillonella, Citrobacter, and Acinetobacter (Brook, 1989; Swidsinski and Lee, 2001; Stewart et al., 2002; Yu et al., 2012). By using next-generation sequencing (NGS) technology, researchers can further reveal the composition and functions of microbiota and provide new insights into biliary bacteria. Wu et al. first applied 16S rRNA sequencing to bile and gallstone samples of patients with cholesterol gallstones (Wu et al., 2013). We previously performed unbiased metagenomic sequencing of bile samples from 15 patients with choledocholithiasis and identified 13 novel biliary bacteria (Shen et al., 2015; Ye et al., 2016). Recently, Kose et al. investigated the microbiota of pigmented and cholesterol gallstones from two patients (Kose et al., 2018).
Despite substantial advancements in the understanding of the biliary microbiota in recent years, the effect of invasive operations of the biliary duct, such as endoscopic sphincterotomy (EST), on the microbial community remains largely unclear. Although endoscopic retrograde cholangiopancreatography (ERCP) with EST is generally considered safe and effective, its invasive nature has been reported to be related to multiple short-term and long-term complications, including pancreatitis, cholangitis, recurrent CBDS, and post-procedural sphincter of Oddi inflammatory changes (Freeman et al., 1996; Tanaka et al., 1998; Konstantakis et al., 2017; Li et al., 2018; Nzenza et al., 2018; Zhou et al., 2019). It is generally believed that the function of the sphincter of Oddi cannot be well preserved after EST (Yasuda et al., 2001). The sphincter of Oddi plays an important role in protecting the biliary system from enteric bacterial invasion (Schneider et al., 2014). EST could lead to the dysfunction or damage of the sphincter of Oddi, which may further cause frequent duodenal-biliary reflux (DBR), i.e., regurgitation of the duodenal contents and gut bacteria into the bile duct, subsequent biliary tract infection and further choledocholithiasis recurrence (Sugiyama et al., 2004; Mu et al., 2015; Zhang et al., 2015).
Recently, Liang and colleagues performed 16S rDNA sequencing of bile samples from 18 cholangiolithiasis patients with sphincter of Oddi laxity (SOL) and found that they had more severe bacterial infections than patients without SOL (Liang et al., 2016). However, the infections did not differ between the patients with primary SOL or surgery-induced SOL. Comparatively, the biliary microbiota of patients with an intact sphincter undergoes a marked change in microenvironment after EST surgery, and thus, the alteration in the biliary microbiota in patients with a history of EST may be distinct from that in primary SOL patients. In this study, we focused on the influence of a history of EST on the biliary bacterial community and found EST led to a marked alteration in the biliary microbial composition which might be related with gallstone recurrence.
Materials and Methods
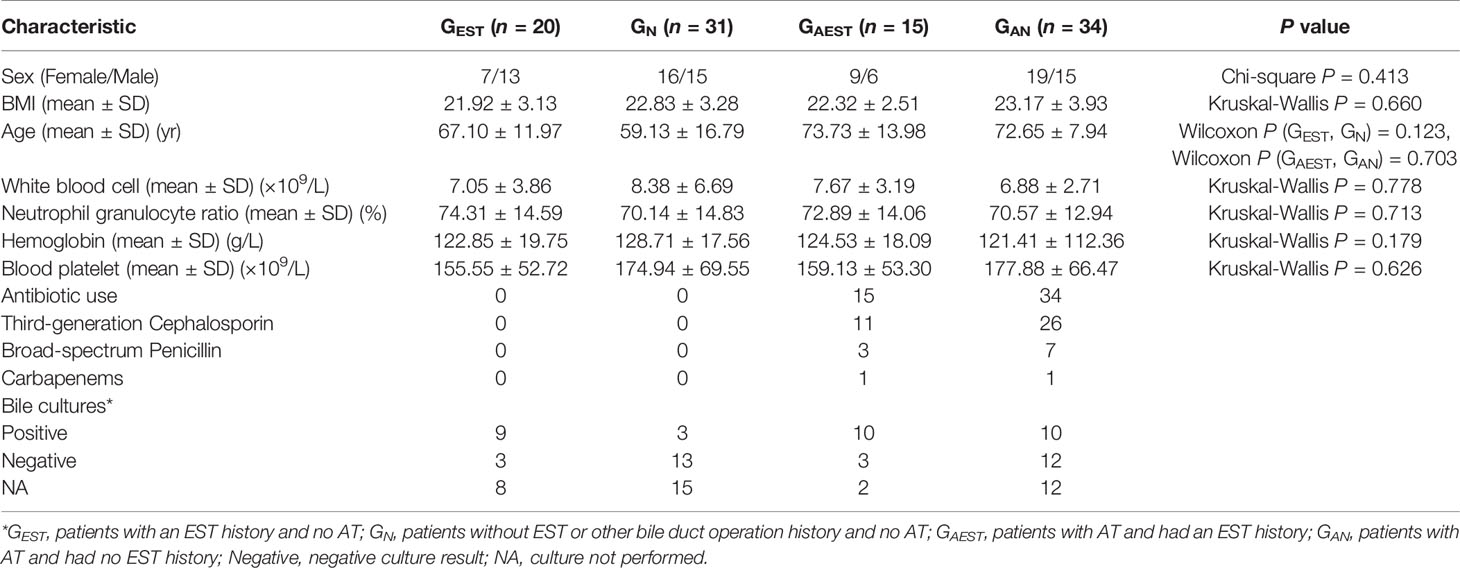
Study Design and Sample Collection
From October 2016 to January 2017, 100 patients who had been diagnosed with primary choledocholithiasis by computed tomography and B-mode ultrasonography at Affiliated Hangzhou First People’s Hospital, Zhejiang University School of Medicine were included in this study. These patients had no occurrences of gallbladder gallstones, hepatolithiasis, pancreatitis, or hepatobiliary tumors, otherwise they would be excluded from the enrollment. Patients with a history of cholecystectomy or diagnosed with secondary choledocholithiasis were also excluded from our study. Clinical metadata, including age, gender, BMI, antibiotic use, blood test results, and culture results of bile samples were also collected (Table 1). These patients were divided into four groups according to EST history and antibiotic treatment (AT) before the collection of bile samples (Table 1). (1) GEST, patients with an EST history and no AT, n = 20; (2) GN, patients without EST or other bile duct operation history and no AT, n = 31; (3) GAEST, patients with AT and had an EST history, n = 15; and (4) GAN, patients with AT and had no EST history, n = 34. For patients with antibiotic use, antibiotic treatment was ongoing at the timepoint of ERCP. All patients provided written informed consent upon enrollment. The study conformed to the ethical guidelines of the 1975 Declaration of Helsinki and was approved by the Institutional Review Board of Affiliated Hangzhou First People’s Hospital, Zhejiang University School of Medicine.
The samples were collected sterilely as previously described (Shen et al., 2015; Ye et al., 2016). In brief, strictly sterile side-viewing endoscopes (TJF240/JF-260V; Olympus Optical, Tokyo, Japan) were used to collect bile samples (2–5 ml) from each patient. Sterile sphincterotome catheters (V-SYSTEM; KD-V411M-0725; Olympus Optical), which passed through the work channel of the endoscopes, were used to aspirate bile samples from the common bile duct into sterile sputum cups. All samples were stored at −80°C until further use.
DNA Extraction
Total DNA was extracted from 200 μl of each bile sample by using the Invitrogen Purelink Genomic DNA Mini Kit (Life Technologies, Carlsbad, CA, USA) according to the manufacturer’s instructions. The DNA concentration and quality were tested by NanoDrop 2000 Spectrophotometer (Thermo Scientific, Waltham, MA, USA).
16S rRNA Amplicon Sequencing
The V3-V4 region of the 16S rRNA gene was amplified by using the following primers: forward primer: 5’- TCGTCGGCAGCGTCAGATGTGTATAAGAGACAGCCTACGGGNGGCWGCAG -3’ and reverse primer: 5’- GTCTCGTGGGCTCGGAGATGTGTATAAGAGACAGGACTACHVGGGTATCTAATCC -3’. The libraries were constructed according to the protocol (https://support.illumina.com/downloads/16s_metagenomic_sequencing_library_preparation.html) for MiSeq 16S Metagenomic Sequencing Library Preparation (Illumina, San Diego, CA, USA) with the Phanta Max Master Mix Kit (Vazyme, Jiangsu, China). As previously described (Ye et al., 2016), we used three tubes of sterile water as negative controls during the library construction process. No DNA products were detected in the negative controls when evaluated by the E-gel electrophoresis system (Life Technologies). The libraries were then sequenced on a MiSeq platform (Illumina) to generate 2 × 300-bp paired-end reads. Each sample was sequenced for one time.
Bioinformatic Analyses of 16S Sequencing Data
Quality control of the raw sequencing reads in “fastq” format was conducted by using cutadapt (v1.11). First, reads without V3-V4 primers were excluded. The pair-end reads were then merged into single sequences by PANDAseq (v2.9) (Masella et al., 2012) with an overlap of no less than 10 bp. Low-quality reads (with an average quality <Q20, having ≥1 N bases, or with a length <300 bp or >480 bp) were also filtered. Vsearch (v2.4.1) (Rognes et al., 2016) was employed to remove chimeras and cluster the remaining reads into operational taxonomic units (OTUs) at the 97% similarity level. Ribosomal Database Project classifier (v2.11) (Cole et al., 2009) then assigned a taxonomic rank (from phylum to genus) to each OTU. The analyses of the α- and β-diversities were performed by using QIIME (v1.8.0) (Caporaso et al., 2010). As previously described (Ye et al., 2016), taxonomic abundances and diversity indices were calculated by randomly resampling the reads of each sample to a uniform number (19,300 in this study) 1,000 times to remove potential bias introduced by varying sequencing depth of these samples. The β-diversities were compared by using the script “compare_categories.py” in QIIME. The predicted profiles of Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway and orthology terms were generated by Phylogenetic Investigation of Communities by Reconstruction of Unobserved States (PICRUSt) (Cox et al., 2014). A heatmap plot was drawn by using the pheatmap package in R with a base-10 logarithm transformation of each relative abundance (RA).
Statistical Analysis
Categorical variables were tested by Chi-square or Fisher’s exact test in SPSS 22.0. Continuous variables were expressed as the mean ± SD when needed. Kruskal-Wallis and Wilcoxon rank-sum tests were employed to detect differences among samples from different groups in R (v3.5.1). When necessary, P-values were corrected to control the false-discovery rate (FDR) by using the method described previously (Benjamini and Hochberg, 1995). Differences with P < 0.05 or FDR < 0.05 were considered statistically significant.
Results
Patient Characteristics and 16S rRNA Sequencing
We included 100 patients with common bile duct stones and assigned them to four groups by history of EST and AT before sample collection. The distribution of gender, BMI, age, and the results of routine blood tests did not significantly differ between the relevant groups (Table 1). For patients without antibiotic use, 12 ones had positive bacterial culture results.
The bile samples of these patients were collected during ERCP and were submitted for 16S rRNA sequencing. We obtained 19,371 to 27,905 (mean 25,554) high-quality merged reads of the samples after quality control, chimera removal, and OTU clustering of the raw reads (Table S1). A total of 19,270 OTUs were identified and assigned taxonomic ranks in these samples. Rarefaction curves measured by the Chao1 and Shannon indices revealed that the sequencing depth reached a plateau as the read number increased (Figure S1).
The Biliary Microbiota of Patients Without Antibiotic Treatment
For the patients without AT before ERCP, we denoted the 20 patients with an EST history as the patient group GEST and the 31 patients with an intact sphincter of Oddi as GN. In GEST and GN, the identified biliary bacteria were mainly from six phyla, Firmicutes, Proteobacteria, Bacteroidetes, Fusobacteria, Actinobacteria, and Synergistetes (Figure 1). Proteobacteria was the most prevalent and had a mean RA of 53.12% (SD 30.24%), followed by Firmicutes (23.65% ± 21.02%). At the genus level, a total of 213 genera were observed with an RA ≥ 0.1%. Thirty-four prevalent genera existed in at least 10 (20%) samples (Figure 1, Table S2), and four of them, Streptococcus, Escherichia, Fusobacterium, and Pseudomonas, had a prevalence ≥70%. The prevalent bacterial genera were also identified in previous studies (Shen et al., 2015; Ye et al., 2016).
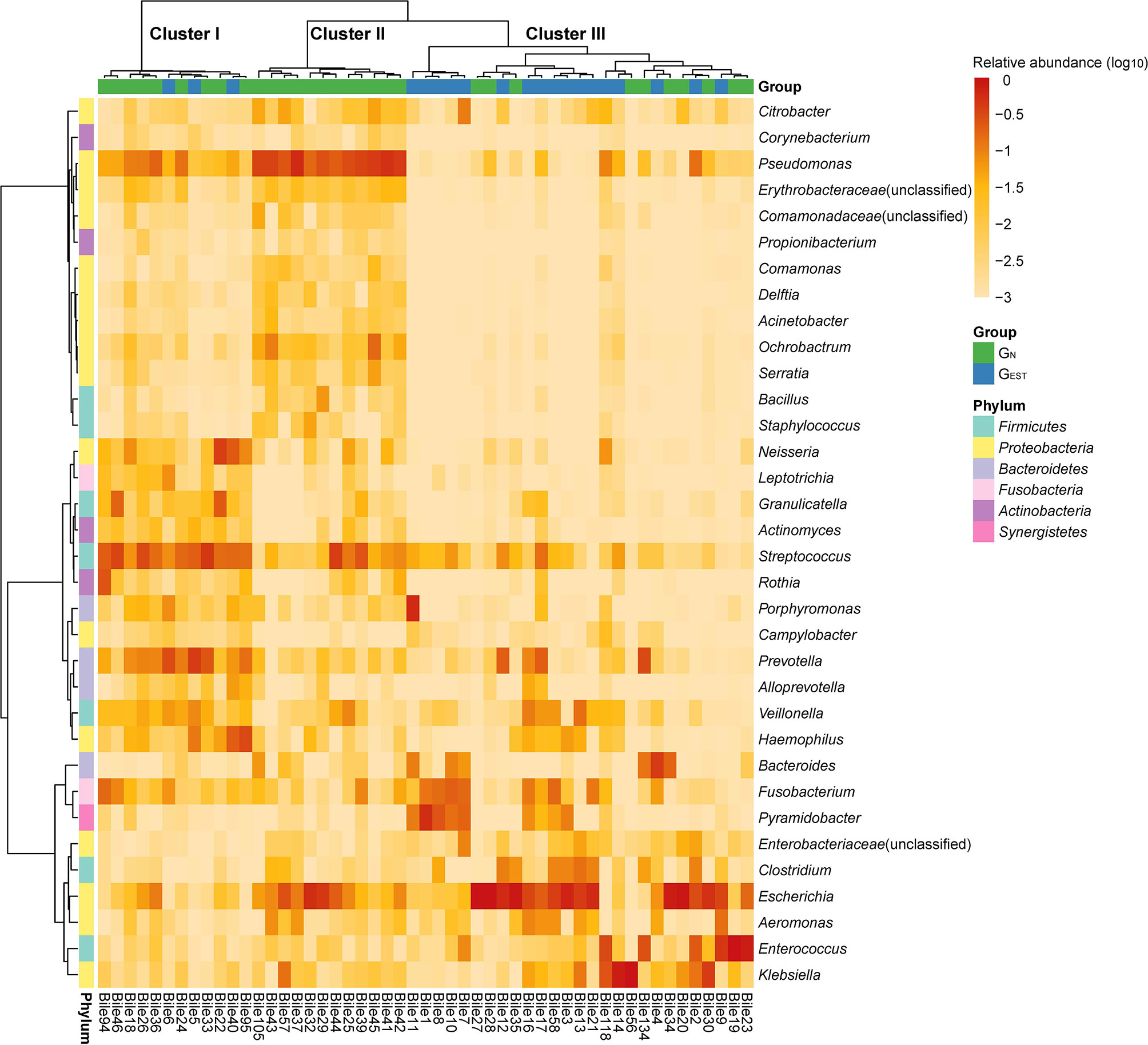
Figure 1 Distribution of highly prevalent genera in the GEST and GN groups. Only genera detected in at least 20% samples with a relative abundance ≥0.1% were displayed. The heatmap color scale quantifies the log10 relative abundance of each genus.
The results also show substantial heterogeneity in the bacterial distribution among non-treated patients, which was consistent with our previous findings (Shen et al., 2015). In the genera with an RA > 1% in at least one patient, we observed 29 genera in less than five patients (Table S3), 19 of which were patient-specific. Notably, among these genera, Edwardsiella, Akkermansia, Lactobacillus, Morganella, Helicobacter, and Clostridium XlVa had an RA higher than 10% in specific patients. Moreover, to the best of our knowledge, three of the rare genera, Tepidimonas, Aquabacterium, and Megamonas, were first discovered in the biliary samples of choledocholithiasis patients. The presence of the newly identified genera was reliable, as all had an RA ≥ 1.16% in at least one patient.
The results of bacterial cultivation were consistent with the results of NGS, but bacterial cultivation was much less sensitive than NGS in identifying bacteria. A total of 11 different bacterial species were cultured from 32 samples in the four patient groups (Table S4), and these samples all had corresponding OTUs at the genus level. In these samples validated by cultivation, 62.16% of the corresponding OTUs had an RA ≥ 10%, and 81.08% had an RA ≥ 1%. The majority of the cultured species belonged to the prevalent genera Escherichia, Klebsiella, Enterococcus, Citrobacter, Proteus, and Pseudomonas. Notably, species of two rare genera, Morganella morganii and Edwardsiella tarda, were also successfully cultivated.
Clusters of Biliary Microbial Compositions and Their Association With Endoscopic Sphincterotomy
Based on the biliary bacterial compositions, we performed an unsupervised clustering analysis of these samples (Figure 1). We identified three clusters that were characterized by the bacteria normally colonized in the oral cavity/respiratory tract (cluster I), environment/skin (cluster II), and gastrointestinal tract (cluster III) (Table S5). Cluster I and cluster III were consistent with our previous pilot study of the biliary microbiota (Shen et al., 2015), which failed to identify cluster II due to a limited sample size (n = 15). The abundant and enriched genera in each cluster were investigated. In cluster I, the top five abundant and significantly enriched bacterial genera (two-side Wilcoxon rank-sum test, FDR < 0.05) were Streptococcus (20.73 ± 11.44%), Prevotella (13.45 ± 12.44%), Neisseria (7.21 ± 11.62%), Haemophilus (6.83 ± 10.15%), and Granulicatella (4.62 ± 7.41%). Cluster II was highly abundant in Pseudomonas (35.37 ± 12.40%), followed by Ochrobactrum (3.86 ± 4.99%) and Serratia (1.09 ± 1.38%). These genera were significantly enriched (FDR < 0.05) in cluster II and have been reported to colonize natural water or soil. Propionibacterium, which is often isolated from human skin, was also enriched in cluster II (0.21 ± 0.15%; FDR < 0.05).
Compared with the most abundant genera in clusters I and II, the most abundant genera in cluster III were typical gastrointestinal inhabitants, such as Escherichia (33.78 ± 34.15%), Enterococcus (11.82 ± 24.74%), Klebsiella (10.80 ± 24.30%), Fusobacterium (4.56 ± 6.73%), and Bacteroides (3.70 ± 8.89%). However, although these genera had a high RA, they were not significantly enriched. In cluster III, Porphyromonas (2.34 ± 11.36%) and an unclassified Enterobacteriaceae (1.25 ± 2.22%) were significantly enriched (FDR < 0.05).
Intriguingly, samples from GN dominated cluster I (9 of 12) and cluster II (12 of 12), while cluster III was mainly enriched with samples from GEST (17 of 27). Thus, the distribution of GN and GEST were significantly biased among the three clusters (two-side Fisher’s exact test of a 2 × 3 contingency table, P = 0.00019). We combined clusters I and II and compared the pooled clusters with cluster III, which indicated that the association with EST history was also significant (two-side Chi-square test of a 2 × 2 contingency table, P = 0.00042). These results indicate that the history of EST was associated with the cluster III microbiota, which was characterized by abundant gastrointestinal bacteria.
Comparison Between Biliary Microbiota With and Without Endoscopic Sphincterotomy History
A comparative study between GEST and GN showed that biliary samples from the two groups had a comparable α-diversity but a significantly different β-diversity, namely, the biliary microbiota of patients with a history of EST did not have an elevated taxonomic diversity but was more likely to undergo a shift in bacterial composition than the biliary microbiota of patients without a history of EST. We measured the α-diversity of GN and GEST by using species richness, phylogenetic diversity, abundance, and evenness with three indices (Chao1, PD_whole_tree, and Shannon index). We found no statistically significant differences (Wilcoxon rank-sum test, P > 0.05, Figure S2). In contrast, analysis of similarities (ANOSIM) revealed that GN and GEST differed significantly in microbiota composition by using β-diversity indices of Bray-Curtis dissimilarity (P = 0.001, Figure 2), unweighted UniFrac distance (P = 0.001) and weighted UniFrac distance (P = 0.03). Adonis and PERMANOVA analyses exhibited similar results (Table S6). A principal coordinates analysis (PCoA) based on Bray-Curtis dissimilarity indicated that the first and second principal components (PC1 and PC2) contributed 24.87 and 14.32% of the total variance between GN and GEST (Figure 2).
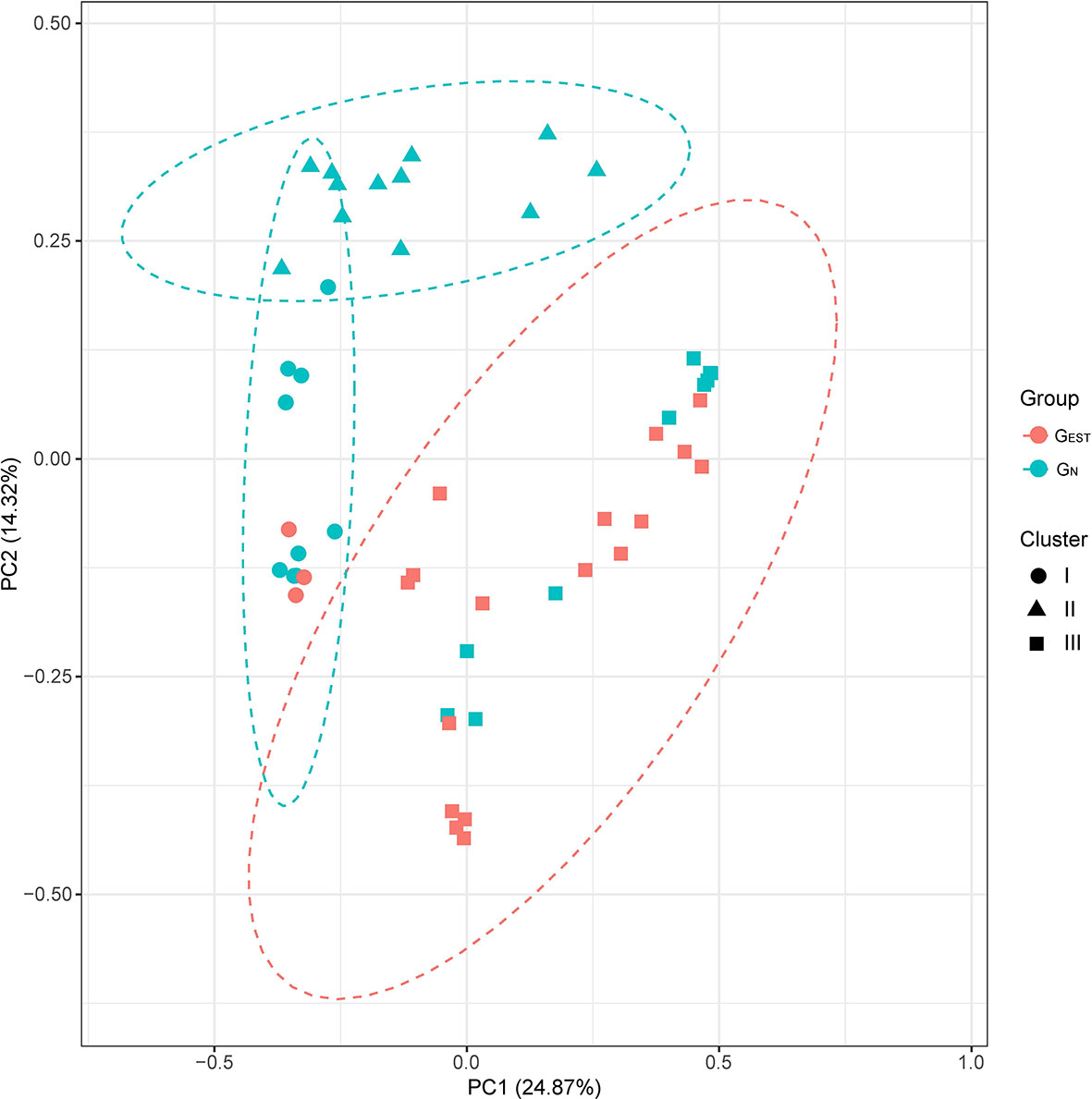
Figure 2 Principal coordinate analysis of the GEST and GN groups. The β-diversities were compared based on Bray-Curtis dissimilarity. The results of unsupervised clustering analysis are also presented.
Among the prevalent genera that existed in ≥20% patients, we found nine genera that had significantly different RAs between GN and GEST (Wilcoxon test, FDR < 0.05, Figure 3, Table S2). One genus, Pyramidobacter, was significantly enriched in GEST (7.72 ± 13.64%, FDR = 0.036), whereas the remaining eight genera had a reduced RA in GEST. Pyramidobacter was detected in 11 samples (55%) from GEST, mostly with a high RA (9 samples >1%). In contrast, in 24 of 31 GN samples, we failed to identify Pyramidobacter, and the rest had a low RA (0.1–1%). This result indicates that abundant Pyramidobacter in biliary samples was specific for patients with an EST history. Biliary Pyramidobacter was first discovered in a gallstone patient with an EST history (Shen et al., 2015). Representative species from Pyramidobacter were reported to be isolated from the oral cavity and small intestine (Downes et al., 2009; Marchandin et al., 2010; Rôças and Siqueira, 2010). We previously showed the coexistence of Pyramidobacter bile and gastric and duodenal fluid in six patients with an EST history, suggesting the possibility of retrograde infection of Pyramidobacter (Ye et al., 2016). Among the bacteria that exhibited a decline in RA in GEST (Figure 3), consistent with the clustering analysis, six out of eight originated from the environment or human skin.
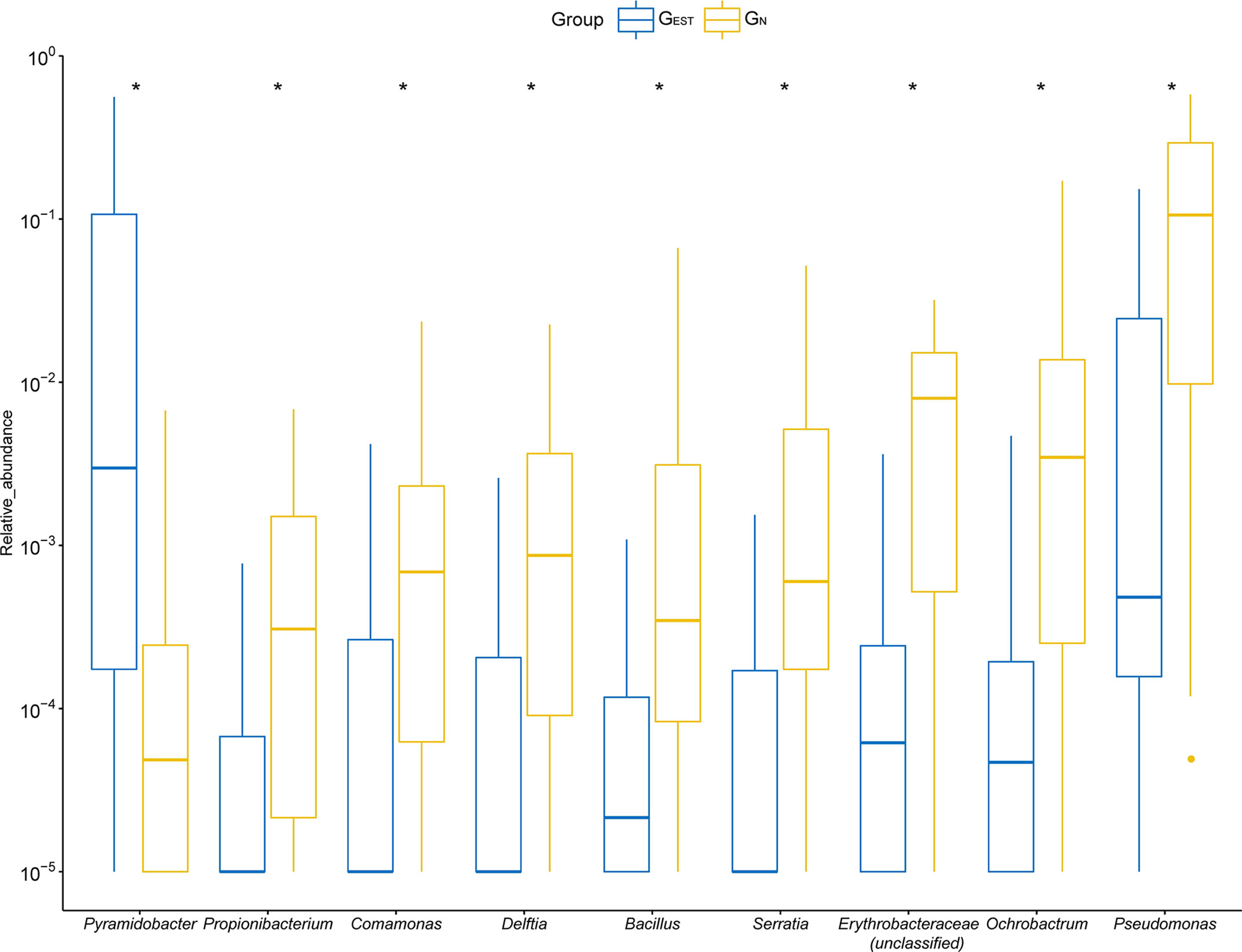
Figure 3 The differential genera detected between samples from the GEST and GN groups. For optimal visualization, a transformation of log10 (relative abundance + 0.00001) was employed. Boxes represent the interquartile range (IQR) between the first and third quartiles (25th and 75th percentiles, respectively). Lines inside denote the median, and whiskers denote the most extreme values within 1.5 times IQR from the first and third quartiles. Outlier values are represented as points. *FDR < 0.05.
We further compared the predicted profiles of KEGG pathways and KEGG Orthology (KO) terms of biliary microbial communities. A total of 61 pathways were differentially expressed between the GN and GEST samples (Table S7). Notably, the glutathione metabolic pathway, which could be employed by some bacteria to address the induced oxidative stresses by bile acids, was depleted in recurrent patients, Among the 51 differential KO terms (Table S8), there were 23 ones associated with bacterial metabolism. Glycogen phosphorylase (K00688) with increased representation in recurrent patients was involved in biofilm formation of E. coli, indicating a potential role in gallstone formation.
The Biliary Microbiota After Antibiotic Treatment
Some patients received AT (denoted as AT patients) 1–5 days before ERCP due to the presence of acute suppurative cholangitis. We focused on the nine significantly distributed bacterial genera identified in GN and GEST. We found that for these AT patients, seven of the genera maintained an association with EST history but with a reduced significance (Table S2). For instance, Pyramidobacter was still the most significantly enriched genera among AT patients with a history of EST compared with those patients without (Wilcoxon rank-sum test, P = 0.00011, FDR = 0.062). Two genera, Fusobacterium and Citrobacter, had a markedly reduced RA in AT patients (both P < 0.05) such that the association with a history of EST was not significant or even reversed. After AT, the ANOSIM based on Bray-Curtis dissimilarity (β-diversity) could not separate the samples with or without EST history (P = 0.409). In summary, for patients treated with antibiotics before bile collection, the EST-associated pattern disappeared for overall biliary microbiota but was preserved for specific bacterial genera.
Discussion
By employing 16S rRNA sequencing technology, we found that the biliary microbial community of primary choledocholithiasis patients was associated with a history of an EST operation. We grouped the discovered biliary microbiota into three clusters, and the biliary microbiota of patients with a history of EST were significantly enriched in the cluster characterized by gastrointestinal bacteria. Although most genera that differed in samples from patients with a history of EST showed a reduced abundance, Pyramidobacter was the only significantly increased genus (FDR < 0.05) in the EST group and exhibited high specificity. In this study, we excluded patients with secondary choledocholithiasis, thus the impact of EST on the biliary microbiota of this sort of patients needs further investigation.
EST could be a vital factor that determines the shift in biliary bacteria. It could cause damage to the sphincter of Oddi and thus alter the microenvironment of the bile duct. Sphincter of Oddi damage after EST would lead to DBR. With the regurgitation of the duodenal contents, gut bacteria may enter the biliary duct and affect the ecological dynamics to a prolithogenic state, which may ultimately lead to gallstone recurrence. In a prospective case-control study, Zhang et al. found that DBR rates in patients with no, single, or multiple CBDS recurrences after EST were 15.6, 60.9, and 88.9%, respectively (Zhang et al., 2015). Therefore, DBR is correlated with CBDS recurrence in patients who had previously undergone ERCP. In our study, the frequency of duodenal-biliary reflux hasn’t been evaluated, which will be investigated in the further study.
Higher abundances of Pyramidobacter were observed in recurrent patients than in patients without EST history in our study. The archetype species P. piscolens encodes phospholipase proteins (GenBank: WP_009165038.1 and EFB90440.1), which might be involved in gallstone formation. With regard to bile resistance, the genome contains genes related to riboflavin metabolism, i.e., riboflavin synthase (WP_009165801.1), the riboflavin biosynthesis protein RibD (K11752, WP_050768911.1, and EFB89795.1), and the riboflavin biosynthesis protein RibF (K11753, WP_083798262.1, and EFB90920.1).
EST could also affect the functions of biliary bacteria in response to environmental modifications. Some gut bacteria might enter the biliary tract after EST operation, which would cause retrograde infection of the bile duct. These bacteria could use the glutathione metabolic pathway, riboflavin metabolic pathway, and cysteine/methionine metabolic pathway to address the induced oxidative stresses. The glutathione metabolic pathway was depleted, while two KEGG Orthology (KO) terms related to riboflavin metabolism were elevated in recurrent patients, suggesting the compensation of oxidative stress responses of the biliary bacteria in the absence of one particular pathway. There were 23 metabolism-related KO terms. Eleven genes were involved in the biosynthesis of secondary metabolites (ko01110), and eight KO terms were related to the biosynthesis of antibiotics (ko01130). Five members from microbial metabolism in diverse environments (ko01120) also had significant differences between the two groups, which suggested that the alteration in the biliary microenvironment might impact the bacterial responses to stresses.
Since the introduction of ERCP and EST nearly 50 years ago, in addition to their therapeutic effects, clinicians have performed many studies on their short-term and long-term complications, especially on the pathogenesis of long-term complications, such as cholangiocarcinoma and recurrent common bile duct stones. Many studies have observed that bacteria might play a key role in the development of these complications, most of which are due to the destruction of the function of sphincter of Oddi by EST. Nonetheless, little is known about the alterations in the bacterial flora in the human bile duct after the function of the papillary sphincter is damaged, and the potential underlying pathogenic role of bacteria in the bile duct remains largely unknown. This study has observed a shift in the biliary microbiota of gallstone patients induced by EST, which could provide evidence for the undesirable consequences due to this safe, effective, and yet invasive clinical procedure. This study also highlights the importance of protecting the normal functions of the sphincter of Oddi in future treatments.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found at: https://www.ncbi.nlm.nih.gov/, PRJNA543184.
Ethics Statement
The studies involving human participants were reviewed and approved by the Institutional Review Board of Affiliated Hangzhou First People’s Hospital, Zhejiang University School of Medicine. The patients/participants provided their written informed consent to participate in this study.
Author Contributions
MN and XZ conceived and designed the study. LF, JY, and QL collected the samples and patient information. JZ, DX, and HJ conducted the experiments. HS, FY, and MN generated the sequencing data. FY, MN, and HS analyzed and interpreted the data. FY and MN wrote the manuscript with contributions from all other authors. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by the Natural Science Foundation of Zhejiang Province (Grant No. LY17H030003), the National Natural Science Foundation of China (Grant Nos. 31701158, 31870079, and 81602325), the Zhejiang Medical and Health Science and Technology Plan (Grant Nos. WKJ-ZJ-2136 and 2019RC068), and the Hangzhou Medical and Health Science and Technology Plan (Grant Nos. 2016ZD01, OO20190610, and A20200174). MN was supported by the Beijing Nova Program (Grant No. Z181100006218114). The funders had no role in the study design, data collection and interpretation, or the decision to submit the work for publication.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcimb.2020.594778/full#supplementary-material
Supplementary Figure 1 | Rarefaction curves measured by the Chao1 (A) and Shannon (B) indices.
Supplementary Figure 2 | The comparison of α-diversities of GN and GEST samples. Chao1, PD_whole_tree and Shannon indices were employed.
References
Begley M., Gahan C. G., Hill C. (2005). The interaction between bacteria and bile. FEMS Microbiol. Rev. 29 (4), 625–651. doi: 10.1016/j.femsre.2004.09.003
Benjamini Y., Hochberg Y. (1995). Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Soc. Ser. B (Methodol.) 57 (1), 289–300. doi: 10.1111/j.2517-6161.1995.tb02031.x
Boursi B., Finkelman B., Giantonio B. J., Haynes K., Rustgi A. K., Rhim A. D., et al. (2016). A Clinical Prediction Model to Assess Risk for Pancreatic Cancer Among Patients With New-onset Diabetes. Gastroenterology 152 (4), 840–850. doi: 10.1053/j.gastro.2016.11.046
Brook I. (1989). Aerobic and anaerobic microbiology of biliary tract disease. J. Clin. Microbiol. 27 (10), 2373–2375. doi: 10.1128/JCM.27.10.2373-2375.1989
Caporaso J. G., Kuczynski J., Stombaugh J., Bittinger K., Bushman F. D., Costello E. K., et al. (2010). QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 7 (5), 335–336. doi: 10.1038/nmeth.f.303
Cole J. R., Wang Q., Cardenas E., Fish J., Chai B., Farris R. J., et al. (2009). The Ribosomal Database Project: improved alignments and new tools for rRNA analysis. Nucleic Acids Res. 37 (Database issue), D141–D145. doi: 10.1093/nar/gkn879
Cox L. M., Yamanishi S., Sohn J., Alekseyenko A. V., Leung J. M., Cho I., et al. (2014). Altering the intestinal microbiota during a critical developmental window has lasting metabolic consequences. Cell 158 (4), 705–721. doi: 10.1016/j.cell.2014.05.052
Downes J., Vartoukian S. R., Dewhirst F. E., Izard J., Chen T., Yu W.-H., et al. (2009). Pyramidobacter piscolens gen. nov., sp. nov., a member of the phylum ‘Synergistetes’ isolated from the human oral cavity. Int. J. Syst. Evol. Microbiol. 59 (5), 972–980. doi: 10.1099/ijs.0.000364-0
Everhart J. E., Khare M., Hill M., Maurer K. R. (1999). Prevalence and ethnic differences in gallbladder disease in the United States. Gastroenterology 117 (3), 632–639. doi: 10.1016/s0016-5085(99)70456-7
Freeman M. L., Nelson D. B., Sherman S., Haber G. B., Herman M. E., Dorsher P. J., et al. (1996). Complications of endoscopic biliary sphincterotomy. N. Engl. J. Med. 335 (13), 909–918. doi: 10.1056/NEJM199609263351301
Konstantakis C., Triantos C., Theopistos V., Theocharis G., Maroulis I., Diamantopoulou G., et al. (2017). Recurrence of choledocholithiasis following endoscopic bile duct clearance: Long term results and factors associated with recurrent bile duct stones. World J. Gastrointest. Endosc. 9 (1), 26–33. doi: 10.4253/wjge.v9.i1.26
Kose S. H., Grice K., Orsi W. D., Ballal M., Coolen M. J. L. (2018). Metagenomics of pigmented and cholesterol gallstones: the putative role of bacteria. Sci. Rep. 8 (1), 11218. doi: 10.1038/s41598-018-29571-8
Li S., Su B., Chen P., Hao J. (2018). Risk factors for recurrence of common bile duct stones after endoscopic biliary sphincterotomy. J. Int. Med. Res. 46 (7), 2595–2605. doi: 10.1177/0300060518765605
Liang T., Su W., Zhang Q., Li G., Gao S., Lou J., et al. (2016). Roles of Sphincter of Oddi Laxity in Bile Duct Microenvironment in Patients with Cholangiolithiasis: From the Perspective of the Microbiome and Metabolome. J. Am. Coll. Surg. 222 (3), 269–280 e210. doi: 10.1016/j.jamcollsurg.2015.12.009
Marchandin H., Damay A., Roudiere L., Teyssier C., Zorgniotti I., Dechaud H., et al. (2010). Phylogeny, diversity and host specialization in the phylum Synergistetes with emphasis on strains and clones of human origin. Res. Microbiol. 161 (2), 91–100. doi: 10.1016/j.resmic.2009.12.008
Masella A. P., Bartram A. K., Truszkowski J. M., Brown D. G., Neufeld J. D. (2012). PANDAseq: paired-end assembler for illumina sequences. BMC Bioinform. 13, 31. doi: 10.1186/1471-2105-13-31
Mu H., Gao J., Kong Q., Jiang K., Wang C., Wang A., et al. (2015). Prognostic Factors and Postoperative Recurrence of Calculus Following Small-Incision Sphincterotomy with Papillary Balloon Dilation for the Treatment of Intractable Choledocholithiasis: A 72-Month Follow-Up Study. Dig. Dis. Sci. 60 (7), 2144–2149. doi: 10.1007/s10620-015-3559-2
Nzenza T. C., Al-Habbal Y., Guerra G. R., Manolas S., Yong T., McQuillan T. (2018). Recurrent common bile duct stones as a late complication of endoscopic sphincterotomy. BMC Gastroenterol. 18 (1), 39. doi: 10.1186/s12876-018-0765-3
Rôças I. N., Siqueira J. F. (2010). Identification of bacteria enduring endodontic treatment procedures by a combined Reverse Transcriptase–Polymerase Chain reaction and Reverse-Capture Checkerboard approach. J. Endod. 36 (1), 45–52. doi: 10.1016/j.joen.2009.10.022
Rognes T., Flouri T., Nichols B., Quince C., Mahe F. (2016). VSEARCH: a versatile open source tool for metagenomics. PeerJ 4, e2584. doi: 10.7717/peerj.2584
Schneider J., De Waha P., Hapfelmeier A., Feihl S., Römmler F., Schlag C., et al. (2014). Risk factors for increased antimicrobial resistance: a retrospective analysis of 309 acute cholangitis episodes. J. Antimicrob. Chemother. 69 (2), 519–525. doi: 10.1093/jac/dkt373
Shen H., Ye F., Xie L., Yang J., Li Z., Xu P., et al. (2015). Metagenomic sequencing of bile from gallstone patients to identify different microbial community patterns and novel biliary bacteria. Sci. Rep. 5, 17450. doi: 10.1038/srep17450
Stewart L., Oesterle A. L., Erdan I., Griffiss J. M., Way L. W. (2002). Pathogenesis of pigment gallstones in Western societies: the central role of bacteria. J. Gastrointest. Surg. 6 (6), 891–904. doi: 10.1016/s1091-255x(02)00035-5
Stewart L., Grifiss J. M., Jarvis G. A., Way L. W. (2006). Biliary bacterial factors determine the path of gallstone formation. Am. J. Surg. 192 (5), 598–603. doi: 10.1016/j.amjsurg.2006.08.001
Sugiyama M., Suzuki Y., Abe N., Masaki T., Mori T., Atomi Y. (2004). Endoscopic retreatment of recurrent choledocholithiasis after sphincterotomy. Gut 53 (12), 1856–1859. doi: 10.1136/gut.2004.041020
Swidsinski A., Lee S. P. (2001). The role of bacteria in gallstone pathogenesis. Front. Biosci. 6 (1), 93–103. doi: 10.2741/swidsinski
Tanaka M., Takahata S., Konomi H., Matsunaga H., Yokohata K., Takeda T., et al. (1998). Long-term consequence of endoscopic sphincterotomy for bile duct stones. Gastrointest. Endosc. 48 (5), 465–469. doi: 10.1016/s0016-5107(98)70086-0
Wang Y., Qi M., Qin C., Hong J. (2018). Role of the biliary microbiome in gallstone disease. Expert Rev. Gastroenterol. Hepatol. 12 (12), 1193–1205. doi: 10.1080/17474124.2018.1533812
Williams E. J., Green J., Beckingham I., Parks R., Martin D., Lombard M. (2008). Guidelines on the management of common bile duct stones (CBDS). Gut 57 (7), 1004–1021. doi: 10.1136/gut.2007.121657
Wu T., Zhang Z., Liu B., Hou D., Liang Y., Zhang J., et al. (2013). Gut microbiota dysbiosis and bacterial community assembly associated with cholesterol gallstones in large-scale study. BMC Genomics 14 (1), 669. doi: 10.1186/1471-2164-14-669
Yasuda I., Tomita E., Enya M., Kato T., Moriwaki H. (2001). Can endoscopic papillary balloon dilation really preserve sphincter of Oddi function? Gut 49 (5), 686–691. doi: 10.1136/gut.49.5.686
Ye F., Shen H., Li Z., Meng F., Li L., Yang J., et al. (2016). Influence of the Biliary System on Biliary Bacteria Revealed by Bacterial Communities of the Human Biliary and Upper Digestive Tracts. PloS One 11 (3), e0150519. doi: 10.1371/journal.pone.0150519
Yu H., Guo Z., Xing W., Guo X., Liu F., Li B. (2012). Bile culture and susceptibility testing of malignant biliary obstruction via PTBD. Cardiovasc. Intervent. Radiol. 35 (5), 1136–1144. doi: 10.1007/s00270-011-0263-2
Zhang R., Luo H., Pan Y., Zhao L., Dong J., Liu Z., et al. (2015). Rate of duodenal-biliary reflux increases in patients with recurrent common bile duct stones: evidence from barium meal examination. Gastrointest. Endosc. 82 (4), 660–665. doi: 10.1016/j.gie.2015.03.1908
Keywords: choledocholithiasis, bile, biliary microbiota, endoscopic sphincterotomy, amplicon sequencing
Citation: Shen H, Zhu J, Ye F, Xu D, Fang L, Yang J, Lv H, Lou Q, Jin H, Ni M and Zhang X (2021) Biliary Microbial Structure of Gallstone Patients With a History of Endoscopic Sphincterotomy Surgery. Front. Cell. Infect. Microbiol. 10:594778. doi: 10.3389/fcimb.2020.594778
Received: 14 August 2020; Accepted: 09 December 2020;
Published: 27 January 2021.
Edited by:
Gabriel Sandblom, Karolinska Institutet (KI), SwedenReviewed by:
Naminatsu Takahara, The University of Tokyo Hospital, JapanJunbo Hong, First Affiliated Hospital of Nanchang University, China
Karl Svennersten, Stockholm County Council, Sweden
Copyright © 2021 Shen, Zhu, Ye, Xu, Fang, Yang, Lv, Lou, Jin, Ni and Zhang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ming Ni, bmltaW5nQGJtaS5hYy5jbg==; Xiaofeng Zhang, ODM3ODM3QHpqdS5lZHUuY24=
†These authors have contributed equally to this work
 Hongzhang Shen
Hongzhang Shen Juanjuan Zhu2†
Juanjuan Zhu2† Fuqiang Ye
Fuqiang Ye Ming Ni
Ming Ni