- Division of Infectious Diseases, Department of Medicine, University of California, San Diego, La Jolla, CA, United States
N6-methyladenosine (m6A) is the most prevalent and internal modification of eukaryotic mRNA. Multiple m6A methylation sites have been identified in the viral RNA genome and transcripts of DNA viruses in recent years. m6A modification is involved in all the phases of RNA metabolism, including RNA stability, splicing, nuclear exporting, RNA folding, translational modulation, and RNA degradation. Three protein groups, methyltransferases (m6A-writers), demethylases (m6A-erasers), and m6A-binding proteins (m6A-readers) regulate this dynamic reversible process. Here, we have reviewed the role of m6A modification dictating viral replication, morphogenesis, life cycle, and its contribution to disease progression. A better understanding of the m6A methylation process during viral pathogenesis is required to reveal novel approaches to combat the virus-associated diseases.
Introduction
Methylation at N6 position of adenosine (m6A) is the most well-characterized and one of the most abundant internal modifications of cellular mRNAs, viral transcripts, long noncoding RNAs (lncRNAs), and microRNAs (miRNAs) (Fu et al., 2014; Meyer and Jaffrey, 2014). The term epitranscriptomic was coined to indicate this dynamic, reversible, co-transcriptional process of RNA synthesis. Pioneering studies in 1970s established m6A as the major form of internal methylation on mammalian mRNA, which occurs in the sequence-context of (G/A)(m6A)C (Desrosiers et al., 1974; Perry and Kelley, 1974; Schibler et al., 1977; Wei and Moss, 1977). With the recent advent of highly sensitive detection methods of antibody-mediated immunoprecipitation combined with high throughput sequencing methods leading to the identification of m6A sites in the transcriptome, functional significance of m6A has been intensely investigated (Dominissini et al., 2012; Meyer et al., 2012). These studies significantly intensified for viral genomic RNAs and transcripts of DNA viruses form the focus of this review.
The key principles revealed by the m6A-mapping studies have shown that m6A is a selective modification, based on its enrichment in certain mRNAs. There is a single m6A site in most of the m6A-modified mRNAs, where some mRNAs may contain 20 or more m6A sites. Transcriptome-wide m6A site mapping has provided greater details on its localization and prominence, revealing its frequency in thousands of transcripts. In different tissues and cell lines, most of the m6A sites appear to be constitutive and are distributed among the corresponding mRNAs in a very similar way (Dominissini et al., 2012; Meyer et al., 2012; Schwartz et al., 2014; Linder et al., 2015). In human and mice transcriptomes, m6A sites are distributed in a unique way preferentially around the stop codons and enriched at 3’ untranslated regions (3’ UTRs) (Dominissini et al., 2012; Meyer et al., 2012). The structure of a specific gene and abundance of m6A are correlated because the presence of long internal exon can be a strong inducer for m6A addition in the transcribed mRNA (Dominissini et al., 2012; Batista et al., 2014; Geula et al., 2015; Ke et al., 2015; Ke et al., 2017). Also, specific gene categories that regulate the development and cell fate specification may be associated with mRNAs containing an unreasonably high level of m6A (Dominissini et al., 2012; Meyer et al., 2012; Geula et al., 2015). After various cellular stresses or in different developmental states, changes in m6A levels have been detected in the 5’ UTRs of a variety of mRNAs (Dominissini et al., 2012; Meyer et al., 2012; Yoon et al., 2017). m6A modification can control the outcome of methylated RNAs at multiple steps, including cap-dependent translation, RNA splicing, mRNA stability, and miRNA biogenesis and m6A modified RNAs participate in many biological processes, like cellular reprogramming, stem cell differentiation, fertility, stress response, circadian cycle and cancer (Dominissini et al., 2012; Fustin et al., 2013; Li and Mason, 2014; Alarcon et al., 2015b; Chen et al., 2015b; Meyer et al., 2015; Zhou et al., 2015; Zhao and He, 2017). m6A modification not only exists in the RNAs of eukaryotic cells but also in genomic RNAs of viruses and of transcripts of DNA viruses. Deregulation of m6A modification is related to the diseases caused by pathogenic viruses. N6 methylation was discovered by Robert Perry in 1974 (Perry and Kelley, 1974). Early studies identified m6A residues in the mRNAs of Simian virus 40 (SV40), Influenza A virus (IAV), Adenovirus, Avian sarcoma virus, and Rous sarcoma virus (Hashimoto and Green, 1976; Krug et al., 1976; Beemon and Keith, 1977; Canaani et al., 1979). Biochemical analysis of different Influenza virus mRNAs showed a varied distribution of m6A in different hemagglutinin (HA) mRNAs (Narayan et al., 1987). But it was not possible to explore the function of m6A in RNA splicing and translation due to insufficient understanding of the experimental conditions and relevant knowledge of m6A for decades. The transcriptome-wide m6A mapping coupled with the development of the N6-methyladenosine-sequencing (m6A-seq) method, revealed this widespread RNA modification linked to various biological functions including differentiation, sex determination, metabolism, stress response, virus infections and cancer (Dominissini et al., 2012; Meyer et al., 2012; Gokhale et al., 2016; Kennedy et al., 2017). m6A-seq was followed by other newly developed techniques like PA- m6A-seq, miCLIP, m6A-LAIC-seq, microarray, and SELECT (Chen et al., 2015a; Li et al., 2015; Linder et al., 2015; Molinie et al., 2016; Xiao et al., 2018). Development of these tools led investigators to elucidate the role of m6A in viral epitranscriptomics. Other forms of epitranscriptomic modifications including, 5-methylcytidine and N4-acetylcytidine were also reported in viral genome but their functional significance remains elusive (Courtney et al., 2019; Tsai et al., 2020).
m6A modification in different pathogenic viruses is increasingly being studied in recent years to reveal its role in the regulation of viral life cycles. m6A modification can influence specific gene expression involved in viral life and can play a role in the inhibition or promotion of different pathogenic virus replication. In this review, we will summarize the emerging roles of m6A modifications described in the recent literature with a focus on viruses and discuss their functions and associated mechanisms related to the biological processes of different virus infections.
Cellular Machinery Associated With m6A Modification
Writers for m6A Methylation
Mammalian genome encodes various methyltransferases to establish m6A modification in distinct RNAs (Figure 1). Methyltransferase-like 3 (METTL3) and methyltransferase-like 14 (METTL14) form a stable core complex that co-transcriptionally installs m6A on mRNA (Bokar et al., 1997; Liu et al., 2014). In METTL3-METTL14 heterodimer, METTL3 is the enzymatic component and METTL14 acts as an allosteric activator to facilitate RNA binding (Sledz and Jinek, 2016; Wang P. et al., 2016; Wang X. et al., 2016). Wilms tumor 1-associated protein (WTAP), Vir like m6A methyltransferase associated (VIRMA), Zinc finger CCCH-type containing 13 (ZC3H13), and RNA binding motif protein 15/15B (RBM15/15B) are the additional subunits that contribute to the activity and specificity of m6A writers. WTAP binds to METTL3-METTL14 heterodimer and is necessary for the optimal substrate recruitment and localization of METTL3/14 complex. VIRMA is critical for the deposition of m6A specifically to the 3’ UTR, ZC3H13 facilitates nuclear localization of the writer complex and RBM15/15B binds to U-riched regions and can expedite methylation of certain mRNAs (Zhong et al., 2008; Ping et al., 2014; Patil et al., 2016; Wen et al., 2018; Yue et al., 2018). METTL3 forms a stable dimer with METTL14 while interacting with WTAP and was localized in the nuclear speckles of HeLa cells, although its cellular distribution can differ between cell lines and its redistribution can be induced by cellular stress. In both METTL3 and WTAP, functional nuclear localization signals (NLS) have been identified and mutations created in the key residues abolished preferential nuclear localization of ectopic METTL3 and WTAP. In multiple human cancer cell lines, a fraction of METTL3 protein has been observed in the cytoplasm at various proportions and the possible reasons could be due to varied amounts of protein abundance ratios between METTL3 and METTL14 with other adaptor subunits. Post-translational modifications (PTMs) could be the other reason which can change the interaction between METTL3 with its partner proteins, leading to cytoplasmic existence (Bokar et al., 1997; Ping et al., 2014; Alarcon et al., 2015b; Chen et al., 2015b; Lin et al., 2016; Barbieri et al., 2017; Knuckles et al., 2017; Xiang et al., 2017; Choe et al., 2018; Scholler et al., 2018).
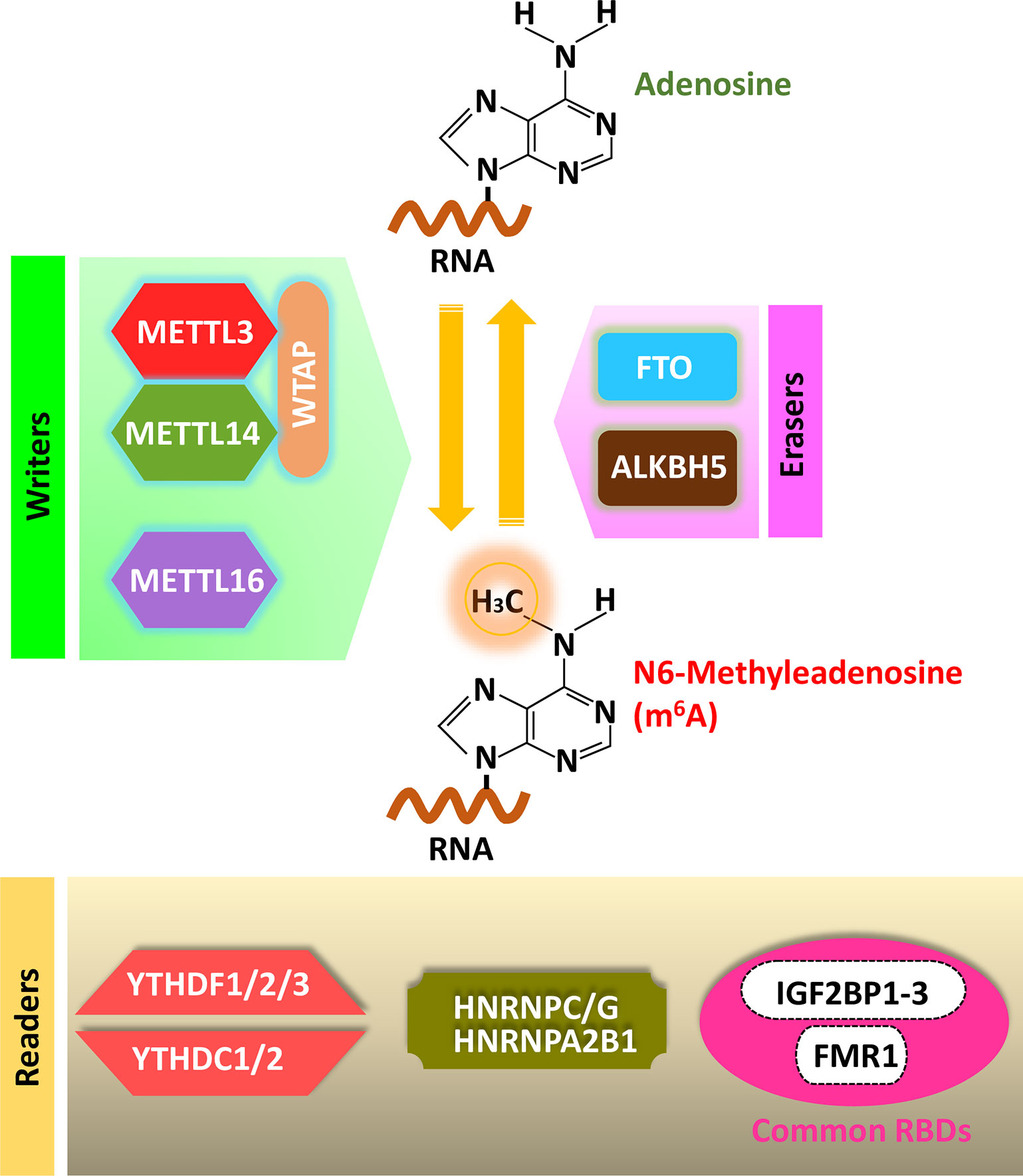
Figure 1 Cellular m6A machinery: Writers, Erasers and Readers. Writer complex is composed of core subunits METTL3 and METTL14 with some additional adaptor proteins. METTL16 is also known as writer. Erasers: FTO and ALKBH5 are the known m6A erasers. Readers: YTH-domain containing proteins (YTHDF1-3, YTHDC1-2) directly recognize m6A- containing RNAs. A local structure disrupted by the presence of m6A could favor RNA-binding events of several heterogeneous nuclear ribonucleoproteins including HNRNPC/G and HNRNPA2B1. RNA binding proteins including IGF2BP1-3 and FMR1 prefer m6A-modified RNAs.
m6A methylation is transcript-specific and the writer complex is recruited at desired chromatin loci through transcription factors and/or epigenetic marks (Shi et al., 2019). In response to stimuli and stress, a subset of m6A sites appears dynamic (Dominissini et al., 2012; Meyer et al., 2012). METTL3 was localized in the heat-shock genes in chromatin after heat shock and when the stress was finished m6A formation cleared from those heat-shock RNAs. METTL3/14 heterodimeric complex was localized in UV-induced damaged sites upon DNA UV damage, co-occurring with enhanced m6A intensity. A specialized adaptor protein might target the m6A writer in a distinct set of genes in the chromatin, which can lead to transcript-specificity of m6A methylation (Barbieri et al., 2017; Knuckles et al., 2017; Xiang et al., 2017).
Another m6A writer, methyltransferase-like 16 (METTL16), which forms a homodimer, installs m6A in a different sequence and structure context than METTL3/14 complex. METTL16 has a preference for UAC(m6A)GAGAA sequence in the bulge region of a stem-loop structured RNA and it has been confirmed by in vitro methylation selection assays, truncation/mutation tests, and a crystal structure of METTL16 protein with its RNA substrates. METTL16 has two authenticated substrates: U6 small nuclear RNA (snRNA) and a hairpin (hp1) in the 3’ UTR of human methionine adenosyltransferase 2A mRNA (MAT2A) which encodes for S-adenosyl methionine (SAM) synthetase (Liu et al., 2014; Pendleton et al., 2017; Doxtader et al., 2018; Mendel et al., 2018). ZCCHC4 is the recently identified m6A writer, mediates the methylation of A4220 on 28S rRNA within an AAC motif, and interacts with a subset of mRNAs (Ma et al., 2019).
Erasers for m6A Methylation
m6A demethylases fat mass and obesity-associated protein (FTO) and AlkB homolog 5 (AlkBH5) can reverse m6A methylation via active demethylation are considered as erasers (Figure 1). The first identified RNA demethylase, FTO can remove the methyl group of m6A in mRNA and N6,2’-O-dimethyladenosine (m6Am) in a portion of mRNA, both in vitro and inside the cells (Adams and Cory, 1975; Wei et al., 1975; Jia et al., 2011; Fu, 2012; Fu et al., 2013; Mauer et al., 2017). m6Am is a terminal modification at mRNA cap that exists in higher eukaryotes (Sun et al., 2019). Initially, FTO was described as a nuclear protein which contains a nuclear localization signal (NLS) in the N terminus and partially co-localize with nuclear speckles. But FTO was shown to localize both in the nucleus and cytoplasm in certain cell lines and its cellular distribution differs among other mammalian cell lines. A large portion of FTO protein localizes in the cytoplasmic part of certain acute myeloid leukemia (AML) cell lines, where it can demethylate cap-m6Am and up to 40% of all mRNA m6A. Also, 5% to 10% of mRNA m6A are demethylated by FTO in HeLa and HEK cell lines (Sanchez-Pulido and Andrade-Navarro, 2007; Jia et al., 2011; Gulati et al., 2014; Aas et al., 2017; Li et al., 2017; Su et al., 2018; Wei et al., 2018).
AlkBH5 is the second known RNA demethylase of m6A modification. AlkBH5 can regulate the nuclear export of target RNAs and this function is affected by the demethylation activity. In certain transcripts demethylation of 3’ UTR is mediated by AlkBH5. AlkBH5 has been reported to facilitate hypoxia-induced HIF-dependent breast cancer stem cell phenotype, regulation of glioblastoma proliferation, and tumorigenesis through the AlkBH5-FOXM1 pathway, and modulation of splicing and stability of long 3’ UTR mRNAs in male germ cells. AlkBH5 and FTO expression patterns are different and they participate in different biological pathways and interact with different protein partners (Zheng et al., 2013; Zhang et al., 2016; Zhang et al., 2017; Tang et al., 2018).
Readers for m6A Methylation
YT521-B homology (YTH) domain-containing proteins (YTHDF1-3 and YTHDC1–2) can bind m6A containing motifs and are known as m6A readers (Figure 1). YTHDF2 and YTHDF3 proteins were first recovered from an in vitro m6A-RNA pull-down experiment (Dominissini et al., 2012). Cytoplasmic YTHDF2 protein can recruit the CCR4-NOT (C-C motif chemokine receptor 4 - negative on TATA-less) deadenylase complex and promotes the degradation of target transcripts (Wang et al., 2014; Du et al., 2016). But, the other two cytoplasmic m6A readers YTHDF1 and YTHDF3 can recruit translation initiation factors and support the translation of target transcripts (Wang et al., 2015; Shi et al., 2017). YTHDC1 protein, an m6A reader localized in the nucleus performs several roles by enabling the nuclear export of mRNA, accelerating the decay of certain transcripts, and regulating mRNA splicing by recruiting certain splicing factors (Xiao et al., 2016; Roundtree et al., 2017b; Shima et al., 2017). Another nuclear m6A reader protein YTHDC2 regulates spermatogenesis and mediates both mRNA stability and translation (Hsu et al., 2017).
Another group of m6A readers can preferentially bind m6A-containing RNAs by utilizing common RNA binding domains (RBDs) such as K homology (KH) domains, RNA recognition motif (RRM) domains, and arginine/glycine-rich (RRG) domains. RNA protein interaction can be modulated by the presence of m6A, named “m6A-switch”, which can remodel the local RNA structure (Liu et al., 2015). Several heterogeneous nuclear ribonucleoproteins (HNRNPs) can regulate alternative splicing or processing of target transcripts, which includes HNRNPC, HNRNPG, and HNRNPA2B1 (Alarcon et al., 2015a; Liu et al., 2015; Liu N. et al., 2017; Wu et al., 2018). Fragile X mental retardation 1 (FMR1) protein binds m6A-containing RNA and contains three KH domains and one RGG domain. FMR1 interacts with m6A readers YTHDF1 and YTHDF2 and can modulate both RNA translation and stability (Edupuganti et al., 2017; Zhang et al., 2018). Although, presence of YTHDF2 protein makes the m6A containing mRNA unstable but another class of m6A reader, insulin-like growth factor 2 mRNA-binding proteins (IGF2BP1–3), stabilized the target mRNA in an m6A-dependent manner. Recently, another class of m6A reader has been identified, named proline-rich coiled-coil 2A (Prrc2a), which showed preferred binding to methylated probe and stabilized a critical m6A-modified transcript required for myelination. Still, it is unclear how m6A readers selectively bind m6A sites or certain m6A-modified transcripts. Probably reader proteins are localized to the different regions of mRNA via interaction with other RBPs that recognize distinct features of RNA (Huang et al., 2018; Wu et al., 2019).
Heat shock stress and viral infection can cause cytoplasmic YTHDF proteins redistribution to the nucleus. It has been shown that within hours of heat shock stress, YTHDF2 gets highly upregulated at both transcript and protein levels and then the majority of the cytoplasmic YTHDF2 translocates to the nucleus (Zhou et al., 2015).
Role of m6A During Virus Infections
m6A modification in different pathogenic viruses has been increasingly being explored in recent years where numerous reports have revealed m6A modification to regulate the viral life cycle. m6A was identified in Influenza A virus (IAV) (Krug et al., 1976), Rous Sarcoma Virus (RSV) (Kane and Beemon, 1985), B77 Avian Sarcoma Virus (Stoltzfus and Dimock, 1976; Dimock and Stoltzfus, 1977) and Feline leukemia Virus (Thomason et al., 1976) decades ago. But the specific functions and mechanisms of m6A in RNA viruses remained to be characterized for a long time until recent advancement in new technologies of transcriptome-wide mapping and sequencing. In the following sections, we will summarize the recent studies addressing the role of functional m6A modifications in viral transcripts and genomic RNA of human viruses in the context of replicative viral life cycles and associated disease pathogenesis.
RNA Viruses
Human Immunodeficiency Virus I
At least four major studies reported the involvement of m6A during human immunodeficiency virus I (HIV-1) infection (Lichinchi et al., 2016a; Tirumuru et al., 2016; Kennedy et al., 2017; Lu et al., 2018). Lichinchi et al. first reported the presence of m6A methylation on HIV-1 RNA and characterized the function, topology, and molecular features of m6A modification in viral RNA (Lichinchi et al., 2016a). At least 14 methylation peaks were identified in coding and noncoding regions, splicing junctions, and splicing regulatory sequences. It was shown that m6A modification levels increased in both host and viral mRNAs upon infection and this modification promoted HIV-1 replication. m6A modification on HIV-1 RNA positively affected the interaction between HIV-1 Rev protein and Rev response element (RRE) RNA. This phenomenon promoted the formation of Rev-RRE complex and enhanced the nuclear export of viral RNA and increased viral replication (Lichinchi et al., 2016a). Kennedy et al. identified m6A residues in the 3’UTR of HIV-1 RNA and found that m6A modification facilitated enhanced mRNA expression and virus replication (Kennedy et al., 2017). Tirumuru et al. identified m6A peaks in the 5’ and 3’UTR of HIV-1 RNA and noted that m6A modification increased HIV-1 gag protein expression and YTHDF1–3 proteins inhibited HIV-1 infection by blocking viral reverse transcription and supported viral RNA degradation (Tirumuru et al., 2016). YTHDF proteins and HIV-1 Gag protein were shown to form a complex with viral RNA (Lu et al., 2018). However, there were discrepancies between these studies due to different experimental conditions and use of different mapping techniques. The differences between these studies requires further research. Overall, m6A of HIV RNAs impacts profoundly the RNA functions and various aspects of HIV life cycle.
Flaviviruses (HCV, ZIKV, DENV, WNV, and YFV)
m6A modification sites have been identified in approximately 19 regions of the Hepatitis C virus (HCV) RNA genome (Gokhale et al., 2016). m6A methyltransferases (METTL3/14) decreased the extracellular HCV RNA and production of virus particles. YTHDF proteins negatively regulated HCV virus particle production and re-localized the HCV genome to lipid droplet to inhibit viral particle assembly via interacting with the m6A-modified envelope region of the HCV genome. Mutation of m6A sites on the HCV E1 region dramatically increased virus production suggesting their role in virion production (Gokhale et al., 2016). During HCV infection, cellular m6A machinery was found to be involved with innate immune response. Modified nucleotides in HCV RNA affected the RIG-I signaling pathway (Durbin et al., 2016). HCV infection affected the m6A abundancy of host RNAs, including phosphatase and tensin homolog (PTEN) mRNA. HCV infection induced m6A modification of PTEN mRNA thus destabilized its RNA stability, leading to disrupt IFN synthesis. PTEN has been known to induce nuclear import of p-IRF3, thus affecting innate immunity to virus infection (Kim et al., 2020a). Kim et al. showed that m6A modified nucleotide 8766 near the HCV PAMP region reduced retinoic acid-induced gene I (RIG-I) recognition via the sequestration of RNA by YTHDF2 protein (Kim et al., 2020b). These results demonstrate that HCV evades immune response, among other mechanisms, by regulating host m6A machinery.
The RNA genomes of Zika virus (ZIKV), Dengue virus (DENV), West Nile virus (WNV), and Yellow Fever virus (YFV), were also m6A-mapped. Interestingly, the identified m6A peaks were localized to similar regions among NS3 and NS5B of viral genomes, suggesting that m6A modifications of the Flaviviridae family viruses may have a conserved role in post-transcriptional regulation (Gokhale et al., 2016). YTHDF2 protein can affect the stability of ZIKV RNA to regulate viral replication (Lichinchi et al., 2016b). The mechanism of m6A modification regulating ZIKV replication remained to be fully characterized.
Influenza A Virus
Although m6A modification on the Influenza A Virus (IAV) HA genome was identified in late 1970s but its role in infection remained undefined (Krug et al., 1976; Narayan et al., 1987). m6A residues were mapped in multiple locations on both the IAV mRNA and vRNA strands. Addition of m6A sites in IAV transcripts enhanced the viral gene expression and replication (Courtney et al., 2017). Ectopically expressed YTHDF2 protein increased IAV replication and infectious particle production. However, YTHDF1 and YTHDF3 proteins had no impact on the viral life cycle. Thus, m6A plays a substantial role in regulating viral gene expression during IAV infection.
Vesicular Stomatitis Virus
m6A modification of Vesicular Stomatitis Virus (VSV) RNA remains to be fully characterized. In normal situations, nuclear protein DDX46 is associated with pre-mRNA splicing and prespliceosome assembly but its role switched to a negative regulator of immune response upon viral infection. DDX46 protein can bind MAVS, Traf3, and Traf6 transcripts, which encode for signaling molecules involved in antiviral response. After VSV infection, DDX46 recruits the m6A demethylase AlkBH5 to remove m6A from these antiviral transcripts. Demethylation retains these antiviral transcripts inside the nucleus and reduces their translation which consequently disrupts antiviral immune response. Knockdown of AlkBH5 remarkably increased IFN production triggered by VSV infection, thus m6A modification might play a negative role in VSV life cycle (Zheng et al., 2017).
Enterovirus 71
Enterovirus 71 (EV71) transcripts contain several m6A peaks mostly localized in the coding regions and the expression and localization of m6A methyltransferases, demethylases, and YTH proteins are affected upon virus infection (Hao et al., 2019). Perturbation of the expression of METTL3, FTO, and YTH proteins can alter EV71 replication and mutation in the m6A site of viral RNA significantly reduced viral replication. Host METTL3 protein interacts with the viral polymerase 3D to enhance its sumoylation and ubiquitination, which facilitates the stability of 3D and increased viral replication. Thus, m6A modifications in EV71 transcripts played a positive role in viral replication.
Pneumoviruses
Human metapneumovirus (HMPV) causes acute respiratory infection in children, immunocompromised patients, and older individuals (Shafagati and Williams, 2018). m6A sites have been identified in HMPV genome, antigenome, and viral mRNAs. Five m6A peaks have been identified throughout the P and G regions of genomic RNA. m6A modification promoted HMPV replication and gene expression, whereas its abrogation resulted in the attenuation of HMPV. m6A-deficient HMPV RNA induced significantly higher Type I IFN in a RIG-I-dependent manner, suggesting that m6A modified RNAs are less sensitive to RIG-I (Lu et al., 2020). Thus, m6A modification served as a molecular marker for innate sensing by cells to discriminate self from non-self RNA. m6A modifications have been identified in Human respiratory syncytial virus (RSV) RNAs, which upregulated viral replication and pathogenesis (Xue et al., 2019).
DNA Viruses
Hepatitis B Virus
Hepatitis B virus (HBV) infection is one of the leading causes of chronic hepatitis that is associated with the elevated risk of severe liver disease including cirrhosis, fibrosis, and primary hepatocellular carcinoma (Ganem and Prince, 2004). HBV contains DNA genome but amplifies by reverse transcription of an intermediate pre-genomic RNA (pgRNA) (Seeger and Mason, 2015; Hu et al., 2019). Role of m6A has been defined in HBV transcripts and liver tissues collected from chronic HBV patients. A single m6A consensus DRACH motif (GGACA) was identified which resides within the epsilon stem-loop region (A1907) of all the HBV transcripts. This motif is repeated twice in the 5’ and 3’ ends of the pgRNA due to terminal redundancy but resides only once in the 3’ UTR of the subgenomic transcripts. m6A site located in the 3’ UTR of HBV transcripts reduced their stability, affecting corresponding viral protein expression. m6A reader YTHDF proteins were found to bind HBV RNAs and silencing of these m6A readers increased HBV protein expression by enhancing the stability of HBV RNA and similar results were observed when the m6A site was mutated within the 3’ epsilon loop of all HBV transcripts. m6A site at the 5’ epsilon stem-loop of viral pgRNA positively regulated the reverse transcription activity. Thus, m6A modification exerts a unique dual regulatory role during HBV life cycle, wherein it negatively regulates the stability of viral transcripts but positively affects the reverse transcription of the core-encapsidated pgRNA (Imam et al., 2018).
IFN treatment of virus-infected cells leads to a reduction of replication mostly by the degradation of viral RNAs by an IFN-stimulated gene 20 (ISG20) exonuclease (Liu Y. et al., 2017). IFN-α induced ISG20 targets the m6A site in the HBV RNAs (Imam et al., 2020). ISG20 formed a complex with m6A reader YTHDF2, in which recruited ISG20 degraded m6A containing HBV transcripts. Mutation in m6A sites abrogates ISG20 mediated RNA decay. This work identified an unknown role of m6A modification of HBV RNA in IFN-α-induced viral RNA degradation and proposes a new role of YTHDF2 protein as a cofactor required for IFN-α mediated HBV RNA degradation (Imam et al., 2020). 5’ epsilon stem-loop structure of HBV pgRNA is recognized by RIG-I for IFN synthesis (Sato et al., 2015). Mutation of m6A site at 5’ epsilon stem-loop structure of HBV pgRNA increases RIG-I sensing, while the presence of m6A reduces RIG-I binding affinity (Kim et al., 2020b). Thus, m6A modification of HBV RNA may function as a molecular signature for distinguishing self from non-self RNA via the RNA sensor RIG-I. HBV dramatically enhances the m6A modification of host PTEN RNA among others, which leads to its degradation with a corresponding decrease in PTEN protein levels (Kim et al., 2020a). PTEN, being a tumor suppressor, its low expression may contribute to hepatocarcinogenesis associate with HBV infection.
Kaposi’s Sarcoma-Associated Herpesvirus (KSHV)
KSHV completes its life cycle in two separate phases; latent infection and lytic replication (Zhao et al., 2015; Liu L. et al., 2017). Most of the viral genome is suppressed through DNA methylation, repressive histone modifications, and other regulatory mechanisms during the latent condition of KSHV infection (Pantry and Medveczky, 2009; Gunther and Grundhoff, 2010; Lu et al., 2010; Toth et al., 2010; Purushothaman et al., 2015). Viruses can reactivate from latency to lytic cycle when the host cell microenvironment change and the inhibitory epigenetic marks are replaced by active ones to allow the transcription of viral lytic genes (Ye, 2017; Uppal et al., 2018). m6A modification was identified in KSHV transcripts and m6A modified mRNA levels significantly increased upon stimulation for lytic replication (Ye et al., 2017). METTL3 and YTHDF2 proteins promoted KSHV virion production (Hesser et al., 2018). Although opposing results showed m6A pathway to inhibit viral gene expression (Tan et al., 2018). By facilitating the degradation of viral transcripts, YTHDF2 protein inhibited KSHV lytic replication. Thus, m6A modifications play a critical role in KSHV life cycle.
Simian Virus 40 (SV40)
SV40 is a member of the Polyomavirus family and its gene expression is regulated in an early phase (encoding the regulatory proteins) and a late phase (encoding the structural proteins). Although m6A modification was reported in SV40 early in 1979, but the specific location of these m6A sites was only recently identified (Canaani et al., 1979). Two m6A sites were identified on viral early transcripts and eleven m6A sites on the late mRNAs. Mutating the m6A sites on late transcripts inhibited SV40 replication, without affecting the mRNA splicing. Similarly, mutational inactivation of the YTHDF2 or METTL3 gene showed a decrease in viral replication (Tsai et al., 2018). Overall m6A positively regulates SV40 life cycle.
Epstein - Barr Virus (EBV)
As a member of the Herpesvirus family, EBV was first isolated and identified from a Burkitt’s lymphoma patient in 1964 (Epstein et al., 1964). m6A sites have been identified on EBV latent and lytic transcripts, where the latent gene expression was enhanced and lytic gene expression was repressed (Lang et al., 2019). m6A methyltransferase METTL14 was dramatically increased during EBV latency while it was reduced during the lytic cycle. Also, METTL14 facilitated the cellular proliferation and colony formation of EBV transformed cells in vitro and increased the oncogenesis of EBV mediated tumorigenicity in vivo. Viral-encoded latent oncoprotein EBNA3C expression was upregulated by METTL14 mediated m6A modification, which is critical for the viral-mediated transformation of cells, and its expression led to a feedback loop by which METTL14 transcription was also induced, along with its protein stability. Latent antigens of EBV are the major contributors to EBV-associated malignancies and their reduced expression attenuates EBV-mediated tumorigenesis. Therefore, targeting METTL14 might be a plausible therapeutic strategy for treating EBV-associated cancer. Overall, m6A modification promoted growth and proliferation of EBV infected cells.
Human Cytomegalovirus (HCMV)
HCMV usually does not initiate any complications in the healthy individual; however, in immunocompromised patients, fetuses, and neonates, HCMV infection can cause an array of damaging clinical outcomes (Crough and Khanna, 2009). m6A machinery is required for HCMV propagation. In the absence of METTL3 followed by viral infection resulted in the modular and highly specific hundreds of IFN-stimulated genes (ISGs) production. Drug-induced blocking of IFN signaling restored viral proliferation in METTL3- and YTHDF2-depleted cells. Even, when the cells were stimulated with an ultraviolet-inactivated virus, the same modular ISG induction was also seen, which means this effect was not driven by viral mechanisms. Central cytokine (IFN-β) that drives type I IFN response is encoded by the gene IFNB. The mRNA of IFNB was modified by m6A and following depletion of METTL3 and YTHDF2, it was highly stabilized. Mice lacking the m6A reader YTHDF3, exhibited increased Ifna and Ifnb induction upon infection with virus. Here, m6A modification served as a negative regulator of IFN response by controlling the fast turnover of IFN mRNAs and consequently accelerates viral proliferation (Winkler et al., 2019).
Discussion and Future Directions
In this review, we have discussed the recent advances in identifying the role of m6A methylation in viral life cycles, including both the RNA and DNA viruses. m6A modifications were either supportive (proviral) or obstructive (antiviral) during viral infections, suggesting a complex pattern of epitranscriptomic regulation of viral gene expression. m6A modification affected viral life cycle in a wide variety of ways ultimately affecting disease pathogenesis. There were mechanistic inconsistencies in some of these studies reflecting different conclusions. These disparities may have originated from the use of different cell lines and experimental procedures of analyses. RNA methylation adds a new layer of regulation of RNA functions. m6A modification is a dynamic process that occurs co-transcriptionally as nascent mRNAs are being transcribed. Studies have shown that METTL3/14 methyltransferases are associated with open chromatin, the sites of transcription initiation (Barbieri et al., 2017; Knuckles et al., 2017; Xiang et al., 2017; Bertero et al., 2018; Huang et al., 2019). METTL enzymes have been localized both in the cytoplasm and nucleus. In cancer cells, the cytoplasmic METTLs have been found while in primary cells, it is mostly localized in the nucleus. For RNA genomes of RNA viruses such as HCV, ZIKA virus and others, RNA methylation likely occurs in the cytoplasm associated with replication complexes formed on organellar membranes. What other functions are associated with cytoplasmic methyltransferases in eukaryotic cells remains to be characterized. Various cellular signals may trigger the function of m6A in different ways in different cell lines. Subcellular localization of m6A writer, eraser and reader proteins and their associated binding partners may affect the biological events in different ways. Distinct reader proteins target different transcripts in different cells or tissues. Even the location of m6A site in certain transcript may have variable consequences in regulatory functions.
N6 methylation of viral RNAs provides potential antiviral therapeutic opportunities. In this context, 3-deazaadenosine (DAA) (a methylase inhibitor) and meclofenamic acid (MA) (an FTO inhibitor) have been tested. DAA can decreases the formation of S-adenosylmethionine (SAM) without affecting mRNA capping and reduces mRNA cap methylation and all types of SAM-dependent RNA methylations (Fustin et al., 2013). A wide variety of viruses have been suppressed by DAA (Bader et al., 1978; Fischer et al., 1990; Wyde et al., 1990; Mayers et al., 1995; Bray et al., 2000; Gordon et al., 2003; Courtney et al., 2017; Kennedy et al., 2017). Since, DAA inhibits all types of RNA modifications, it is difficult to conclude whether antiviral effect comes from inhibition of mRNA cap methylation or internal m6A methylation. As such currently, there is no drug that can specifically inhibit m6A. FTO inhibitor, MA has been reported to enhance the expression of KSHV lytic genes, by competing with m6A-containing substrate binding and inhibition of FTO driven demethylation (Ye et al., 2017). Therefore, the development of novel small molecule inhibitors or drugs specifically targeted for m6A machinery (writers, erasers, or readers) would be a highly significant venue for antiviral therapies.
Since m6A deficient HMPV can induce higher IFN production, live attenuated vaccine candidates for HMPV and other pneumoviruses were designed (Lu et al., 2020). Similar approaches can be applied to other viruses for the rational design of vaccine candidates. Apart from m6A modification, mammalian epitranscriptome contains 3 other internal modifications, Pseudouridine (ψ), N1-methyladenosine (m1A), and 5-methylcytosine (m5C) (Roundtree et al., 2017a). Future research can be expanded on these less explored RNA modifications to gain adequate information on their role in viral RNAs regulation and cellular antiviral responses.
Author Contributions
AS and HI conceptualized and wrote the manuscript. G-WK contributed to writing. All authors contributed to the article and approved the submitted version.
Funding
National Institutes of Health NIH AI139234.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
Aas A., Isakson P., Bindesboll C., Alemu E. A., Klungland A., Simonsen A. (2017). Nucleocytoplasmic Shuttling of FTO Does Not Affect Starvation-Induced Autophagy. PloS One 12 (3), e0168182. doi: 10.1371/journal.pone.0168182
Adams J. M., Cory S. (1975). Modified nucleosides and bizarre 5’-termini in mouse myeloma mRNA. Nature 255 (5503), 28–33. doi: 10.1038/255028a0
Alarcon C. R., Goodarzi H., Lee H., Liu X., Tavazoie S., Tavazoie S. F. (2015a). HNRNPA2B1 Is a Mediator of m(6)A-Dependent Nuclear RNA Processing Events. Cell 162 (6), 1299–1308. doi: 10.1016/j.cell.2015.08.011
Alarcon C. R., Lee H., Goodarzi H., Halberg N., Tavazoie S. F. (2015b). N6-methyladenosine marks primary microRNAs for processing. Nature 519 (7544), 482–485. doi: 10.1038/nature14281
Bader J. P., Brown N. R., Chiang P. K., Cantoni G. L. (1978). 3-Deazaadenosine, an inhibitor of adenosylhomocysteine hydrolase, inhibits reproduction of Rous sarcoma virus and transformation of chick embryo cells. Virology 89 (2), 494–505. doi: 10.1016/0042-6822(78)90191-5
Barbieri I., Tzelepis K., Pandolfini L., Shi J., Millan-Zambrano G., Robson S. C., et al. (2017). Promoter-bound METTL3 maintains myeloid leukaemia by m(6)A-dependent translation control. Nature 552 (7683), 126–131. doi: 10.1038/nature24678
Batista P. J., Molinie B., Wang J., Qu K., Zhang J., Li L., et al. (2014). m(6)A RNA modification controls cell fate transition in mammalian embryonic stem cells. Cell Stem Cell 15 (6), 707–719. doi: 10.1016/j.stem.2014.09.019
Beemon K., Keith J. (1977). Localization of N6-methyladenosine in the Rous sarcoma virus genome. J. Mol. Biol. 113 (1), 165–179. doi: 10.1016/0022-2836(77)90047-x
Bertero A., Brown S., Madrigal P., Osnato A., Ortmann D., Yiangou L., et al. (2018). The SMAD2/3 interactome reveals that TGFbeta controls m(6)A mRNA methylation in pluripotency. Nature 555 (7695), 256–259. doi: 10.1038/nature25784
Bokar J. A., Shambaugh M. E., Polayes D., Matera A. G., Rottman F. M. (1997). Purification and cDNA cloning of the AdoMet-binding subunit of the human mRNA (N6-adenosine)-methyltransferase. RNA 3 (11), 1233–1247.
Bray M., Driscoll J., Huggins J. W. (2000). Treatment of lethal Ebola virus infection in mice with a single dose of an S-adenosyl-L-homocysteine hydrolase inhibitor. Antiviral Res. 45 (2), 135–147. doi: 10.1016/s0166-3542(00)00066-8
Canaani D., Kahana C., Lavi S., Groner Y. (1979). Identification and mapping of N6-methyladenosine containing sequences in simian virus 40 RNA. Nucleic Acids Res. 6 (8), 2879–2899. doi: 10.1093/nar/6.8.2879
Chen K., Lu Z., Wang X., Fu Y., Luo G. Z., Liu N., et al. (2015a). High-resolution N(6) -methyladenosine (m(6) A) map using photo-crosslinking-assisted m(6) A sequencing. Angew. Chem. Int. Ed. Engl. 54 (5), 1587–1590. doi: 10.1002/anie.201410647
Chen T., Hao Y. J., Zhang Y., Li M. M., Wang M., Han W., et al. (2015b). m(6)A RNA methylation is regulated by microRNAs and promotes reprogramming to pluripotency. Cell Stem Cell 16 (3), 289–301. doi: 10.1016/j.stem.2015.01.016
Choe J., Lin S., Zhang W., Liu Q., Wang L., Ramirez-Moya J., et al. (2018). mRNA circularization by METTL3-eIF3h enhances translation and promotes oncogenesis. Nature 561 (7724), 556–560. doi: 10.1038/s41586-018-0538-8
Courtney D. G., Kennedy E. M., Dumm R. E., Bogerd H. P., Tsai K., Heaton N. S., et al. (2017). Epitranscriptomic Enhancement of Influenza A Virus Gene Expression and Replication. Cell Host Microbe 22 (3), 377–386 e375. doi: 10.1016/j.chom.2017.08.004
Courtney D. G., Tsai K., Bogerd H. P., Kennedy E. M., Law B. A., Emery A., et al. (2019). Epitranscriptomic Addition of m(5)C to HIV-1 Transcripts Regulates Viral Gene Expression. Cell Host Microbe 26 (2), 217–227 e216. doi: 10.1016/j.chom.2019.07.005
Crough T., Khanna R. (2009). Immunobiology of human cytomegalovirus: from bench to bedside. Clin. Microbiol. Rev. 22 (1), 76–98, Table of Contents. doi: 10.1128/CMR.00034-08
Desrosiers R., Friderici K., Rottman F. (1974). Identification of methylated nucleosides in messenger RNA from Novikoff hepatoma cells. Proc. Natl. Acad. Sci. U. S. A. 71 (10), 3971–3975. doi: 10.1073/pnas.71.10.3971
Dimock K., Stoltzfus C. M. (1977). Sequence specificity of internal methylation in B77 avian sarcoma virus RNA subunits. Biochemistry 16 (3), 471–478. doi: 10.1021/bi00622a021
Dominissini D., Moshitch-Moshkovitz S., Schwartz S., Salmon-Divon M., Ungar L., Osenberg S., et al. (2012). Topology of the human and mouse m6A RNA methylomes revealed by m6A-seq. Nature 485 (7397), 201–206. doi: 10.1038/nature11112
Doxtader K. A., Wang P., Scarborough A. M., Seo D., Conrad N. K., Nam Y. (2018). Structural Basis for Regulation of METTL16, an S-Adenosylmethionine Homeostasis Factor. Mol. Cell 711001-1011 (6), e1004. doi: 10.1016/j.molcel.2018.07.025
Du H., Zhao Y., He J., Zhang Y., Xi H., Liu M., et al. (2016). YTHDF2 destabilizes m(6)A-containing RNA through direct recruitment of the CCR4-NOT deadenylase complex. Nat. Commun. 7, 12626. doi: 10.1038/ncomms12626
Durbin A. F., Wang C., Marcotrigiano J., Gehrke L. (2016). RNAs Containing Modified Nucleotides Fail To Trigger RIG-I Conformational Changes for Innate Immune Signaling. mBio 7 (5). doi: 10.1128/mBio.00833-16
Edupuganti R. R., Geiger S., Lindeboom R. G. H., Shi H., Hsu P. J., Lu Z., et al. (2017). N(6)-methyladenosine (m(6)A) recruits and repels proteins to regulate mRNA homeostasis. Nat. Struct. Mol. Biol. 24 (10), 870–878. doi: 10.1038/nsmb.3462
Epstein M. A., Achong B. G., Barr Y. M. (1964). Virus Particles in Cultured Lymphoblasts from Burkitt’s Lymphoma. Lancet 1 (7335), 702–703. doi: 10.1016/s0140-6736(64)91524-7
Fischer A. A., Muller K., Scholtissek C. (1990). Specific inhibition of the synthesis of influenza virus late proteins and stimulation of early, M2, and NS2 protein synthesis by 3-deazaadenosine. Virology 177 (2), 523–531. doi: 10.1016/0042-6822(90)90517-u
Fu Y., Jia G., Pang X., Wang R. N., Wang X., Li C. J., et al. (2013). FTO-mediated formation of N6-hydroxymethyladenosine and N6-formyladenosine in mammalian RNA. Nat. Commun. 4, 1798. doi: 10.1038/ncomms2822
Fu Y., Dominissini D., Rechavi G., He C. (2014). Gene expression regulation mediated through reversible m(6)A RNA methylation. Nat. Rev. Genet. 15 (5), 293–306. doi: 10.1038/nrg3724
Fu Y. (2012). Dynamic regulation of rna modifications by AlkB family dioxygenases (University of Chicago, Division of the Physical Sciences, Departmentof Chemistry).
Fustin J. M., Doi M., Yamaguchi Y., Hida H., Nishimura S., Yoshida M., et al. (2013). RNA-methylation-dependent RNA processing controls the speed of the circadian clock. Cell 155 (4), 793–806. doi: 10.1016/j.cell.2013.10.026
Ganem D., Prince A. M. (2004). Hepatitis B virus infection–natural history and clinical consequences. N. Engl. J. Med. 350 (11), 1118–1129. doi: 10.1056/NEJMra031087
Geula S., Moshitch-Moshkovitz S., Dominissini D., Mansour A. A., Kol N., Salmon-Divon M., et al. (2015). Stem cells. m6A mRNA methylation facilitates resolution of naive pluripotency toward differentiation. Science 347 (6225), 1002–1006. doi: 10.1126/science.1261417
Gokhale N. S., McIntyre A. B. R., McFadden M. J., Roder A. E., Kennedy E. M., Gandara J. A., et al. (2016). N6-Methyladenosine in Flaviviridae Viral RNA Genomes Regulates Infection. Cell Host Microbe 20 (5), 654–665. doi: 10.1016/j.chom.2016.09.015
Gordon R. K., Ginalski K., Rudnicki W. R., Rychlewski L., Pankaskie M. C., Bujnicki J. M., et al. (2003). Anti-HIV-1 activity of 3-deaza-adenosine analogs. Inhibition of S-adenosylhomocysteine hydrolase and nucleotide congeners. Eur. J. Biochem. 270 (17), 3507–3517. doi: 10.1046/j.1432-1033.2003.03726.x
Gulati P., Avezov E., Ma M., Antrobus R., Lehner P., O’Rahilly S., et al. (2014). Fat mass and obesity-related (FTO) shuttles between the nucleus and cytoplasm. Biosci. Rep. 34 (5), 621–628. doi: 10.1042/BSR20140111
Gunther T., Grundhoff A. (2010). The epigenetic landscape of latent Kaposi sarcoma-associated herpesvirus genomes. PloS Pathog. 6 (6), e1000935. doi: 10.1371/journal.ppat.1000935
Hao H., Hao S., Chen H., Chen Z., Zhang Y., Wang J., et al. (2019). N6-methyladenosine modification and METTL3 modulate enterovirus 71 replication. Nucleic Acids Res. 47 (1), 362–374. doi: 10.1093/nar/gky1007
Hashimoto S. I., Green M. (1976). Multiple methylated cap sequences in adenovirus type 2 early mRNA. J. Virol. 20 (2), 425–435. doi: 10.1128/JVI.20.2.425-435.1976
Hesser C. R., Karijolich J., Dominissini D., He C., Glaunsinger B. A. (2018). N6-methyladenosine modification and the YTHDF2 reader protein play cell type specific roles in lytic viral gene expression during Kaposi’s sarcoma-associated herpesvirus infection. PloS Pathog. 14 (4), e1006995. doi: 10.1371/journal.ppat.1006995
Hsu P. J., Zhu Y., Ma H., Guo Y., Shi X., Liu Y., et al. (2017). Ythdc2 is an N(6)-methyladenosine binding protein that regulates mammalian spermatogenesis. Cell Res. 27 (9), 1115–1127. doi: 10.1038/cr.2017.99
Hu J., Protzer U., Siddiqui A. (2019). Revisiting Hepatitis B Virus: Challenges of Curative Therapies. J. Virol. 93 (20). doi: 10.1128/JVI.01032-19
Huang H., Weng H., Sun W., Qin X., Shi H., Wu H., et al. (2018). Recognition of RNA N(6)-methyladenosine by IGF2BP proteins enhances mRNA stability and translation. Nat. Cell Biol. 20 (3), 285–295. doi: 10.1038/s41556-018-0045-z
Huang H., Weng H., Zhou K., Wu T., Zhao B. S., Sun M., et al. (2019). Histone H3 trimethylation at lysine 36 guides m(6)A RNA modification co-transcriptionally. Nature 567 (7748), 414–419. doi: 10.1038/s41586-019-1016-7
Imam H., Khan M., Gokhale N. S., McIntyre A. B. R., Kim G. W., Jang J. Y., et al. (2018). N6-methyladenosine modification of hepatitis B virus RNA differentially regulates the viral life cycle. Proc. Natl. Acad. Sci. U.S.A. 115 (35), 8829–8834. doi: 10.1073/pnas.1808319115
Imam H., Kim G. W., Mir S. A., Khan M., Siddiqui A. (2020). Interferon-stimulated gene 20 (ISG20) selectively degrades N6-methyladenosine modified Hepatitis B Virus transcripts. PloS Pathog. 16 (2), e1008338. doi: 10.1371/journal.ppat.1008338
Jia G., Fu Y., Zhao X., Dai Q., Zheng G., Yang Y., et al. (2011). N6-methyladenosine in nuclear RNA is a major substrate of the obesity-associated FTO. Nat. Chem. Biol. 7 (12), 885–887. doi: 10.1038/nchembio.687
Kane S. E., Beemon K. (1985). Precise localization of m6A in Rous sarcoma virus RNA reveals clustering of methylation sites: implications for RNA processing. Mol. Cell Biol. 5 (9), 2298–2306. doi: 10.1128/mcb.5.9.2298
Ke S., Alemu E. A., Mertens C., Gantman E. C., Fak J. J., Mele A., et al. (2015). A majority of m6A residues are in the last exons, allowing the potential for 3’ UTR regulation. Genes Dev. 29 (19), 2037–2053. doi: 10.1101/gad.269415.115
Ke S., Pandya-Jones A., Saito Y., Fak J. J., Vagbo C. B., Geula S., et al. (2017). m(6)A mRNA modifications are deposited in nascent pre-mRNA and are not required for splicing but do specify cytoplasmic turnover. Genes Dev. 31 (10), 990–1006. doi: 10.1101/gad.301036.117
Kennedy E. M., Bogerd H. P., Kornepati A. V. R., Kang D., Ghoshal D., Marshall J. B., et al. (2017). Posttranscriptional m(6)A Editing of HIV-1 mRNAs Enhances Viral Gene Expression. Cell Host Microbe 22 (6), 830. doi: 10.1016/j.chom.2017.11.010
Kim G. W., Imam H., Khan M., Mir S. A., Kim S. J., Yoon S. K., et al. (2020a). HBV-Induced Increased N6 Methyladenosine Modification of PTEN RNA Affects Innate Immunity and Contributes to HCC. Hepatology. doi: 10.1002/hep.31313
Kim G. W., Imam H., Khan M., Siddiqui A. (2020b). N (6)-Methyladenosine modification of hepatitis B and. J. Biol. Chem. 295 (37), 13123–13133. doi: 10.1074/jbc.RA120.014260
Knuckles P., Carl S. H., Musheev M., Niehrs C., Wenger A., Buhler M. (2017). RNA fate determination through cotranscriptional adenosine methylation and microprocessor binding. Nat. Struct. Mol. Biol. 24 (7), 561–569. doi: 10.1038/nsmb.3419
Krug R. M., Morgan M. A., Shatkin A. J. (1976). Influenza viral mRNA contains internal N6-methyladenosine and 5’-terminal 7-methylguanosine in cap structures. J. Virol. 20 (1), 45–53. doi: 10.1128/JVI.20.1.45-53.1976
Lang F., Singh R. K., Pei Y., Zhang S., Sun K., Robertson E. S. (2019). EBV epitranscriptome reprogramming by METTL14 is critical for viral-associated tumorigenesis. PloS Pathog. 15 (6), e1007796. doi: 10.1371/journal.ppat.1007796
Li S., Mason C. E. (2014). The pivotal regulatory landscape of RNA modifications. Annu. Rev. Genomics Hum. Genet. 15, 127–150. doi: 10.1146/annurev-genom-090413-025405
Li Y., Wang Y., Zhang Z., Zamudio A. V., Zhao J. C. (2015). Genome-wide detection of high abundance N6-methyladenosine sites by microarray. RNA 21 (8), 1511–1518. doi: 10.1261/rna.051474.115
Li Z., Weng H., Su R., Weng X., Zuo Z., Li C., et al. (2017). FTO Plays an Oncogenic Role in Acute Myeloid Leukemia as a N(6)-Methyladenosine RNA Demethylase. Cancer Cell 31 (1), 127–141. doi: 10.1016/j.ccell.2016.11.017
Lichinchi G., Gao S., Saletore Y., Gonzalez G. M., Bansal V., Wang Y., et al. (2016a). Dynamics of the human and viral m(6)A RNA methylomes during HIV-1 infection of T cells. Nat. Microbiol. 1, 16011. doi: 10.1038/nmicrobiol.2016.11
Lichinchi G., Zhao B. S., Wu Y., Lu Z., Qin Y., He C., et al. (2016b). Dynamics of Human and Viral RNA Methylation during Zika Virus Infection. Cell Host Microbe 20 (5), 666–673. doi: 10.1016/j.chom.2016.10.002
Lin S., Choe J., Du P., Triboulet R., Gregory R. I. (2016). The m(6)A Methyltransferase METTL3 Promotes Translation in Human Cancer Cells. Mol. Cell 62 (3), 335–345. doi: 10.1016/j.molcel.2016.03.021
Linder B., Grozhik A. V., Olarerin-George A. O., Meydan C., Mason C. E., Jaffrey S. R. (2015). Single-nucleotide-resolution mapping of m6A and m6Am throughout the transcriptome. Nat. Methods 12 (8), 767–772. doi: 10.1038/nmeth.3453
Liu J., Yue Y., Han D., Wang X., Fu Y., Zhang L., et al. (2014). A METTL3-METTL14 complex mediates mammalian nuclear RNA N6-adenosine methylation. Nat. Chem. Biol. 10 (2), 93–95. doi: 10.1038/nchembio.1432
Liu N., Dai Q., Zheng G., He C., Parisien M., Pan T. (2015). N(6)-methyladenosine-dependent RNA structural switches regulate RNA-protein interactions. Nature 518 (7540), 560–564. doi: 10.1038/nature14234
Liu L., Zhou Q., Xie Y., Zuo L., Zhu F., Lu J. (2017). Extracellular vesicles: novel vehicles in herpesvirus infection. Virol. Sin. 32 (5), 349–356. doi: 10.1007/s12250-017-4073-9
Liu N., Zhou K. I., Parisien M., Dai Q., Diatchenko L., Pan T. (2017). N6-methyladenosine alters RNA structure to regulate binding of a low-complexity protein. Nucleic Acids Res. 45 (10), 6051–6063. doi: 10.1093/nar/gkx141
Liu Y., Nie H., Mao R., Mitra B., Cai D., Yan R., et al. (2017). Interferon-inducible ribonuclease ISG20 inhibits hepatitis B virus replication through directly binding to the epsilon stem-loop structure of viral RNA. PloS Pathog. 13 (4), e1006296. doi: 10.1371/journal.ppat.1006296
Lu F., Stedman W., Yousef M., Renne R., Lieberman P. M. (2010). Epigenetic regulation of Kaposi’s sarcoma-associated herpesvirus latency by virus-encoded microRNAs that target Rta and the cellular Rbl2-DNMT pathway. J. Virol. 84 (6), 2697–2706. doi: 10.1128/JVI.01997-09
Lu W., Tirumuru N., St Gelais C., Koneru P. C., Liu C., Kvaratskhelia M., et al. (2018). N(6)-Methyladenosine-binding proteins suppress HIV-1 infectivity and viral production. J. Biol. Chem. 293 (34), 12992–13005. doi: 10.1074/jbc.RA118.004215
Lu M., Zhang Z., Xue M., Zhao B. S., Harder O., Li A., et al. (2020). N(6)-methyladenosine modification enables viral RNA to escape recognition by RNA sensor RIG-I. Nat. Microbiol. 5 (4), 584–598. doi: 10.1038/s41564-019-0653-9
Ma H., Wang X., Cai J., Dai Q., Natchiar S. K., Lv R., et al. (2019). N(6-)Methyladenosine methyltransferase ZCCHC4 mediates ribosomal RNA methylation. Nat. Chem. Biol. 15 (1), 88–94. doi: 10.1038/s41589-018-0184-3
Mauer J., Luo X., Blanjoie A., Jiao X., Grozhik A. V., Patil D. P., et al. (2017). Reversible methylation of m(6)Am in the 5’ cap controls mRNA stability. Nature 541 (7637), 371–375. doi: 10.1038/nature21022
Mayers D. L., Mikovits J. A., Joshi B., Hewlett I. K., Estrada J. S., Wolfe A. D., et al. (1995). Anti-human immunodeficiency virus 1 (HIV-1) activities of 3-deazaadenosine analogs: increased potency against 3’-azido-3’-deoxythymidine-resistant HIV-1 strains. Proc. Natl. Acad. Sci. U. S. A. 92 (1), 215–219. doi: 10.1073/pnas.92.1.215
Mendel M., Chen K. M., Homolka D., Gos P., Pandey R. R., McCarthy A. A., et al. (2018). Methylation of Structured RNA by the m(6)A Writer METTL16 Is Essential for Mouse Embryonic Development. Mol. Cell 71986-1000 (6), e1011. doi: 10.1016/j.molcel.2018.08.004
Meyer K. D., Jaffrey S. R. (2014). The dynamic epitranscriptome: N6-methyladenosine and gene expression control. Nat. Rev. Mol. Cell Biol. 15 (5), 313–326. doi: 10.1038/nrm3785
Meyer K. D., Saletore Y., Zumbo P., Elemento O., Mason C. E., Jaffrey S. R. (2012). Comprehensive analysis of mRNA methylation reveals enrichment in 3’ UTRs and near stop codons. Cell 149 (7), 1635–1646. doi: 10.1016/j.cell.2012.05.003
Meyer K. D., Patil D. P., Zhou J., Zinoviev A., Skabkin M. A., Elemento O., et al. (2015). 5’ UTR m(6)A Promotes Cap-Independent Translation. Cell 163 (4), 999–1010. doi: 10.1016/j.cell.2015.10.012
Molinie B., Wang J., Lim K. S., Hillebrand R., Lu Z. X., Van Wittenberghe N., et al. (2016). m(6)A-LAIC-seq reveals the census and complexity of the m(6)A epitranscriptome. Nat. Methods 13 (8), 692–698. doi: 10.1038/nmeth.3898
Narayan P., Ayers D. F., Rottman F. M., Maroney P. A., Nilsen T. W. (1987). Unequal distribution of N6-methyladenosine in influenza virus mRNAs. Mol. Cell Biol. 7 (4), 1572–1575. doi: 10.1128/mcb.7.4.1572
Pantry S. N., Medveczky P. G. (2009). Epigenetic regulation of Kaposi’s sarcoma-associated herpesvirus replication. Semin. Cancer Biol. 19 (3), 153–157. doi: 10.1016/j.semcancer.2009.02.010
Patil D. P., Chen C. K., Pickering B. F., Chow A., Jackson C., Guttman M., et al. (2016). m(6)A RNA methylation promotes XIST-mediated transcriptional repression. Nature 537 (7620), 369–373. doi: 10.1038/nature19342
Pendleton K. E., Chen B., Liu K., Hunter O. V., Xie Y., Tu B. P., et al. (2017). The U6 snRNA m(6)A Methyltransferase METTL16 Regulates SAM Synthetase Intron Retention. Cell 169824-835 (5), e814. doi: 10.1016/j.cell.2017.05.003
Perry R. P., Kelley D. E. (1974). Existence of methylated messenger RNA in mouse L cells. Cell 1 (1), 37–42. doi: 10.1016/0092-8674(74)90153-6
Ping X. L., Sun B. F., Wang L., Xiao W., Yang X., Wang W. J., et al. (2014). Mammalian WTAP is a regulatory subunit of the RNA N6-methyladenosine methyltransferase. Cell Res. 24 (2), 177–189. doi: 10.1038/cr.2014.3
Purushothaman P., Uppal T., Verma S. C. (2015). Molecular biology of KSHV lytic reactivation. Viruses 7 (1), 116–153. doi: 10.3390/v7010116
Roundtree I. A., Evans M. E., Pan T., He C. (2017a). Dynamic RNA Modifications in Gene Expression Regulation. Cell 169 (7), 1187–1200. doi: 10.1016/j.cell.2017.05.045
Roundtree I. A., Luo G. Z., Zhang Z., Wang X., Zhou T., Cui Y., et al. (2017b). YTHDC1 mediates nuclear export of N(6)-methyladenosine methylated mRNAs. Elife 6. doi: 10.7554/eLife.31311
Sanchez-Pulido L., Andrade-Navarro M. A. (2007). The FTO (fat mass and obesity associated) gene codes for a novel member of the non-heme dioxygenase superfamily. BMC Biochem. 8, 23. doi: 10.1186/1471-2091-8-23
Sato S., Li K., Kameyama T., Hayashi T., Ishida Y., Murakami S., et al. (2015). The RNA sensor RIG-I dually functions as an innate sensor and direct antiviral factor for hepatitis B virus. Immunity 42 (1), 123–132. doi: 10.1016/j.immuni.2014.12.016
Schibler U., Kelley D. E., Perry R. P. (1977). Comparison of methylated sequences in messenger RNA and heterogeneous nuclear RNA from mouse L cells. J. Mol. Biol. 115 (4), 695–714. doi: 10.1016/0022-2836(77)90110-3
Scholler E., Weichmann F., Treiber T., Ringle S., Treiber N., Flatley A., et al. (2018). Interactions, localization, and phosphorylation of the m(6)A generating METTL3-METTL14-WTAP complex. RNA 24 (4), 499–512. doi: 10.1261/rna.064063.117
Schwartz S., Mumbach M. R., Jovanovic M., Wang T., Maciag K., Bushkin G. G., et al. (2014). Perturbation of m6A writers reveals two distinct classes of mRNA methylation at internal and 5’ sites. Cell Rep. 8 (1), 284–296. doi: 10.1016/j.celrep.2014.05.048
Seeger C., Mason W. S. (2015). Molecular biology of hepatitis B virus infection. Virology 479-480, 672–686. doi: 10.1016/j.virol.2015.02.031
Shafagati N., Williams J. (2018). Human metapneumovirus - what we know now. F1000Res 7, 135. doi: 10.12688/f1000research.12625.1
Shi H., Wang X., Lu Z., Zhao B. S., Ma H., Hsu P. J., et al. (2017). YTHDF3 facilitates translation and decay of N(6)-methyladenosine-modified RNA. Cell Res. 27 (3), 315–328. doi: 10.1038/cr.2017.15
Shi H., Wei J., He C. (2019). Where, When, and How: Context-Dependent Functions of RNA Methylation Writers, Readers, and Erasers. Mol. Cell 74 (4), 640–650. doi: 10.1016/j.molcel.2019.04.025
Shima H., Matsumoto M., Ishigami Y., Ebina M., Muto A., Sato Y., et al. (2017). S-Adenosylmethionine Synthesis Is Regulated by Selective N(6)-Adenosine Methylation and mRNA Degradation Involving METTL16 and YTHDC1. Cell Rep. 21 (12), 3354–3363. doi: 10.1016/j.celrep.2017.11.092
Sledz P., Jinek M. (2016). Structural insights into the molecular mechanism of the m(6)A writer complex. Elife 5. doi: 10.7554/eLife.18434
Stoltzfus C. M., Dimock K. (1976). Evidence of methylation of B77 avian sarcoma virus genome RNA subunits. J. Virol. 18 (2), 586–595. doi: 10.1128/JVI.18.2.586-595.1976
Su R., Dong L., Li C., Nachtergaele S., Wunderlich M., Qing Y., et al. (2018). R-2HG Exhibits Anti-tumor Activity by Targeting FTO/m(6)A/MYC/CEBPA Signaling. Cell 172 (1-2), 90–105.e123. doi: 10.1016/j.cell.2017.11.031
Sun H., Zhang M., Li K., Bai D., Yi C. (2019). Cap-specific, terminal N(6)-methylation by a mammalian m(6)Am methyltransferase. Cell Res. 29 (1), 80–82. doi: 10.1038/s41422-018-0117-4
Tan B., Liu H., Zhang S., da Silva S. R., Zhang L., Meng J., et al. (2018). Viral and cellular N(6)-methyladenosine and N(6),2’-O-dimethyladenosine epitranscriptomes in the KSHV life cycle. Nat. Microbiol. 3 (1), 108–120. doi: 10.1038/s41564-017-0056-8
Tang C., Klukovich R., Peng H., Wang Z., Yu T., Zhang Y., et al. (2018). ALKBH5-dependent m6A demethylation controls splicing and stability of long 3’-UTR mRNAs in male germ cells. Proc. Natl. Acad. Sci. U. S. A. 115 (2), E325–E333. doi: 10.1073/pnas.1717794115
Thomason A. R., Brian D. A., Velicer L. F., Rottman F. M. (1976). Methylation of high-molecular-weight subunit RNA of feline leukemia virus. J. Virol. 20 (1), 123–132. doi: 10.1128/JVI.20.1.123-132.1976
Tirumuru N., Zhao B. S., Lu W., Lu Z., He C., Wu L. (2016). N(6)-methyladenosine of HIV-1 RNA regulates viral infection and HIV-1 Gag protein expression. Elife 5. doi: 10.7554/eLife.15528
Toth Z., Maglinte D. T., Lee S. H., Lee H. R., Wong L. Y., Brulois K. F., et al. (2010). Epigenetic analysis of KSHV latent and lytic genomes. PloS Pathog. 6 (7), e1001013. doi: 10.1371/journal.ppat.1001013
Tsai K., Courtney D. G., Cullen B. R. (2018). Addition of m6A to SV40 late mRNAs enhances viral structural gene expression and replication. PloS Pathog. 14 (2), e1006919. doi: 10.1371/journal.ppat.1006919
Tsai K., Jaguva Vasudevan A. A., Martinez Campos C., Emery A., Swanstrom R., Cullen B. R. (2020). Acetylation of Cytidine Residues Boosts HIV-1 Gene Expression by Increasing Viral RNA Stability. Cell Host Microbe 28306-312 (2), e306. doi: 10.1016/j.chom.2020.05.011
Uppal T., Sarkar R., Dhelaria R., Verma S. C. (2018). Role of Pattern Recognition Receptors in KSHV Infection. Cancers (Basel) 10 (3). doi: 10.3390/cancers10030085
Wang X., Lu Z., Gomez A., Hon G. C., Yue Y., Han D., et al. (2014). N6-methyladenosine-dependent regulation of messenger RNA stability. Nature 505 (7481), 117–120. doi: 10.1038/nature12730
Wang X., Zhao B. S., Roundtree I. A., Lu Z., Han D., Ma H., et al. (2015). N(6)-methyladenosine Modulates Messenger RNA Translation Efficiency. Cell 161 (6), 1388–1399. doi: 10.1016/j.cell.2015.05.014
Wang P., Doxtader K. A., Nam Y. (2016a). Structural Basis for Cooperative Function of Mettl3 and Mettl14 Methyltransferases. Mol. Cell 63 (2), 306–317. doi: 10.1016/j.molcel.2016.05.041
Wang X., Feng J., Xue Y., Guan Z., Zhang D., Liu Z., et al. (2016b). Structural basis of N(6)-adenosine methylation by the METTL3-METTL14 complex. Nature 534 (7608), 575–578. doi: 10.1038/nature18298
Wei C. M., Moss B. (1977). Nucleotide sequences at the N6-methyladenosine sites of HeLa cell messenger ribonucleic acid. Biochemistry 16 (8), 1672–1676. doi: 10.1021/bi00627a023
Wei C. M., Gershowitz A., Moss B. (1975). Methylated nucleotides block 5’ terminus of HeLa cell messenger RNA. Cell 4 (4), 379–386. doi: 10.1016/0092-8674(75)90158-0
Wei J., Liu F., Lu Z., Fei Q., Ai Y., He P. C., et al. (2018). Differential m(6)A, m(6)Am, and m(1)A Demethylation Mediated by FTO in the Cell Nucleus and Cytoplasm. Mol. Cell 71973-985 (6), e975. doi: 10.1016/j.molcel.2018.08.011
Wen J., Lv R., Ma H., Shen H., He C., Wang J., et al. (2018). Zc3h13 Regulates Nuclear RNA m(6)A Methylation and Mouse Embryonic Stem Cell Self-Renewal. Mol. Cell 691028-1038 (6), e1026. doi: 10.1016/j.molcel.2018.02.015
Winkler R., Gillis E., Lasman L., Safra M., Geula S., Soyris C., et al. (2019). m(6)A modification controls the innate immune response to infection by targeting type I interferons. Nat. Immunol. 20 (2), 173–182. doi: 10.1038/s41590-018-0275-z
Wu B., Su S., Patil D. P., Liu H., Gan J., Jaffrey S. R., et al. (2018). Molecular basis for the specific and multivariant recognitions of RNA substrates by human hnRNP A2/B1. Nat. Commun. 9 (1), 420. doi: 10.1038/s41467-017-02770-z
Wu R., Li A., Sun B., Sun J. G., Zhang J., Zhang T., et al. (2019). A novel m(6)A reader Prrc2a controls oligodendroglial specification and myelination. Cell Res. 29 (1), 23–41. doi: 10.1038/s41422-018-0113-8
Wyde P. R., Ambrose M. W., Meyer H. L., Zolinski C. L., Gilbert B. E. (1990). Evaluation of the toxicity and antiviral activity of carbocyclic 3-deazaadenosine against respiratory syncytial and parainfluenza type 3 viruses in tissue culture and in cotton rats. Antiviral Res. 14 (4-5), 215–225. doi: 10.1016/0166-3542(90)90003-p
Xiang Y., Laurent B., Hsu C. H., Nachtergaele S., Lu Z., Sheng W., et al. (2017). RNA m(6)A methylation regulates the ultraviolet-induced DNA damage response. Nature 543 (7646), 573–576. doi: 10.1038/nature21671
Xiao W., Adhikari S., Dahal U., Chen Y. S., Hao Y. J., Sun B. F., et al. (2016). Nuclear m(6)A Reader YTHDC1 Regulates mRNA Splicing. Mol. Cell 61 (4), 507–519. doi: 10.1016/j.molcel.2016.01.012
Xiao Y., Wang Y., Tang Q., Wei L., Zhang X., Jia G. (2018). An Elongation- and Ligation-Based qPCR Amplification Method for the Radiolabeling-Free Detection of Locus-Specific N(6) -Methyladenosine Modification. Angew. Chem. Int. Ed. Engl. 57 (49), 15995–16000. doi: 10.1002/anie.201807942
Xue M., Zhao B. S., Zhang Z., Lu M., Harder O., Chen P., et al. (2019). Viral N(6)-methyladenosine upregulates replication and pathogenesis of human respiratory syncytial virus. Nat. Commun. 10 (1), 4595. doi: 10.1038/s41467-019-12504-y
Ye F., Chen E. R., Nilsen T. W. (2017). Kaposi’s Sarcoma-Associated Herpesvirus Utilizes and Manipulates RNA N(6)-Adenosine Methylation To Promote Lytic Replication. J. Virol. 91 (16). doi: 10.1128/JVI.00466-17
Ye F. (2017). RNA N(6)-adenosine methylation (m(6)A) steers epitranscriptomic control of herpesvirus replication. Inflammation Cell Signal 4 (3). doi: 10.14800/ics.1604
Yoon K. J., Ringeling F. R., Vissers C., Jacob F., Pokrass M., Jimenez-Cyrus D., et al. (2017). Temporal Control of Mammalian Cortical Neurogenesis by m(6)A Methylation. Cell 171877-889 (4), e817. doi: 10.1016/j.cell.2017.09.003
Yue Y., Liu J., Cui X., Cao J., Luo G., Zhang Z., et al. (2018). VIRMA mediates preferential m(6)A mRNA methylation in 3’UTR and near stop codon and associates with alternative polyadenylation. Cell Discov. 4:10. doi: 10.1038/s41421-018-0019-0
Zhang C., Samanta D., Lu H., Bullen J. W., Zhang H., Chen I., et al. (2016). Hypoxia induces the breast cancer stem cell phenotype by HIF-dependent and ALKBH5-mediated m(6)A-demethylation of NANOG mRNA. Proc. Natl. Acad. Sci. U. S. A. 113 (14), E2047–E2056. doi: 10.1073/pnas.1602883113
Zhang S., Zhao B. S., Zhou A., Lin K., Zheng S., Lu Z., et al. (2017). m(6)A Demethylase ALKBH5 Maintains Tumorigenicity of Glioblastoma Stem-like Cells by Sustaining FOXM1 Expression and Cell Proliferation Program. Cancer Cell 31591-606 (4), e596. doi: 10.1016/j.ccell.2017.02.013
Zhang F., Kang Y., Wang M., Li Y., Xu T., Yang W., et al. (2018). Fragile X mental retardation protein modulates the stability of its m6A-marked messenger RNA targets. Hum. Mol. Genet. 27 (22), 3936–3950. doi: 10.1093/hmg/ddy292
Zhao B. S., He C. (2017). “Gamete On” for m(6)A: YTHDF2 Exerts Essential Functions in Female Fertility. Mol. Cell 67 (6), 903–905. doi: 10.1016/j.molcel.2017.09.004
Zhao Q., Liang D., Sun R., Jia B., Xia T., Xiao H., et al. (2015). Kaposi’s sarcoma-associated herpesvirus-encoded replication and transcription activator impairs innate immunity via ubiquitin-mediated degradation of myeloid differentiation factor 88. J. Virol. 89 (1), 415–427. doi: 10.1128/JVI.02591-14
Zheng G., Dahl J. A., Niu Y., Fedorcsak P., Huang C. M., Li C. J., et al. (2013). ALKBH5 is a mammalian RNA demethylase that impacts RNA metabolism and mouse fertility. Mol. Cell 49 (1), 18–29. doi: 10.1016/j.molcel.2012.10.015
Zheng Q., Hou J., Zhou Y., Li Z., Cao X. (2017). The RNA helicase DDX46 inhibits innate immunity by entrapping m(6)A-demethylated antiviral transcripts in the nucleus. Nat. Immunol. 18 (10), 1094–1103. doi: 10.1038/ni.3830
Zhong S., Li H., Bodi Z., Button J., Vespa L., Herzog M., et al. (2008). MTA is an Arabidopsis messenger RNA adenosine methylase and interacts with a homolog of a sex-specific splicing factor. Plant Cell 20 (5), 1278–1288. doi: 10.1105/tpc.108.058883
Keywords: viral epitranscriptomics, m6A-writer, m6A-eraser, m6A-binding protein, m6A modification
Citation: Imam H, Kim G-W and Siddiqui A (2020) Epitranscriptomic(N6-methyladenosine) Modification of Viral RNA and Virus-Host Interactions. Front. Cell. Infect. Microbiol. 10:584283. doi: 10.3389/fcimb.2020.584283
Received: 16 July 2020; Accepted: 27 October 2020;
Published: 24 November 2020.
Edited by:
Haitao Guo, University of Pittsburgh, United StatesReviewed by:
Koichi Watashi, National Institute of Infectious Diseases (NIID), JapanJianrong Li, The Ohio State University, United States
Copyright © 2020 Imam, Kim and Siddiqui. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Aleem Siddiqui, YXNpZGRpcXVpQGhlYWx0aC51Y3NkLmVkdQ==
 Hasan Imam
Hasan Imam Geon-Woo Kim
Geon-Woo Kim Aleem Siddiqui
Aleem Siddiqui