- 1Emergency and Critical Care Center, Beijing Friendship Hospital, Capital Medical University, Beijing, China
- 2Beijing Institute of Tropical Medicine, Beijing, China
- 3Warwick Medical School, Warwick University, Coventry, United Kingdom
- 4Department of Pediatrics and National School of Tropical Medicine, Baylor College of Medicine, Houston, TX, United States
Leishmaniasis is still a serious neglected tropical disease that may cause death in infected individuals. At present, the clinical diagnosis and treatment monitoring still rely on parasitological culture and microscopy that needs experienced technicians. The low sensitivity and inconvenience of microscopic examination could cause misdiagnosis and relapse of leishmaniasis. There is an urgent need for developing a sensitive and easily operated diagnostic method for the diagnosis and disease management of leishmaniasis. Thus, a quantitative real-time PCR (qPCR) based on the conversed regions of kinetoplast minicircle DNA (mkDNA) of Leishmania spp. was developed to detect different species of Leishmania. The designed mkDNA-based qPCR was able to detect as low as one copy of Leishmania mkDNA or DNA from single parasite. It also detected Pan-Leishmania protozoa including Leishmania donovani, Leishmania infantum and Leishmania major without cross-reaction with other pathogen DNAs available in our lab. This method was clinically applied to quantitatively detect skin lesion samples from 20 cutaneous leishmaniasis (CL) and bone marrow and/or PBMC samples from 30 current and cured visceral leishmaniasis (VL) patients, and blood samples from 11 patients with other infections and 5 normal donors as well. Total 20 skin lesion samples from current CL patients and 20 bone marrow and/or PBMC samples from current VL patients were all detected as positive with qPCR without cross-reaction with samples from patients with malaria, brucellosis and dengue or normal donors. Two VL patients with parasite converted to microscopically negative after treatment were detected positive with qPCR. The patients with bigger skin lesion in CL and higher level of immunoglobulin or splenomegaly in VL, had the higher parasite load detected by qPCR. The parasite load was significantly reduced after treatment. In conclusion, the mkDNA-based qPCR assay that we developed in this study can be used not only for diagnosis of both cutaneous and visceral leishmaniasis with high sensitivity and specificity, but also for evaluating the severity and treatment efficacy of this disease, presenting a rapid and accurate tool for clinical surveillance, treatment monitoring and the end point determination of leishmaniasis.
Introduction
Leishmaniasis is a serious neglected tropical disease caused by the protozoan parasite Leishmania. spp. transmitted by the bite of infected female phlebotomine sandflies (Burza et al., 2018). It has been estimated that there are 700,000–1 million cases of human infection with ~26,000–65,000 death reported annually (Burza et al., 2018). The reported cases are only small fraction of the real infection and many of them are neglected or misdiagnosed as other diseases, in addition to more than 350 million people living under high infection risk (Ghorbani and Farhoudi, 2018). The infection of Leishmania spp. causes a spectrum of diseases, including cutaneous leishmaniasis (CL), visceral leishmaniasis (VL), and mucocutaneous leishmaniasis (MCL) (Torres-Guerrero et al., 2017), with CL and VL representing the most common forms. Globally, it has been estimated that ~90% of CL cases are distributed in 10 worst affected countries including Afghanistan, Algeria, Colombia, Brazil, Iran, Syria, Ethiopia, North Sudan, Costa Rica, and Peru (Bailey et al., 2017). The Leishmania parasites causing CL can be divided into Old World species Leishmania major, Leishmania tropica, Leishmania aethiopica, and the New World species Leishmania amazonensis, Leishmania mexicana, Leishmania braziliensis, and Leishmania guyanensis (de Vries et al., 2015). The symptoms of CL usually present as nodule, patch/plaque and ulcerative skin lesions. Generally, CL is not life-threatening, however the parasite can spread through the lymphatic tissues, leading to mucocutaneous leishmaniasis or so-called “diffuse CL” that could result in extensive midfacial destruction and stigma (de Vries et al., 2015). Visceral leishmaniasis (VL), also known as kala-azar, is the most severe form of leishmaniasis. VL is mostly endemic in seven countries including India, Brazil, Ethiopia, Kenya, Somalia, Sudan, and South Sudan (Bi et al., 2018). The main etiological agents of VL are Leishmania donovani and Leishmania infantum (Bi et al., 2018). The main clinical presentations of VL include irregular fever at onset, followed by splenomegaly pancytopenia, hepatomegaly, hypergammaglobulinaemia and a permanent loss of weight. Without effective treatment the disease exacerbates by 2 years, typically with fatal consequences. In China, visceral leishmaniasis caused by L. donovani used to be seriously endemic in the central China. It has been eliminated in most endemic regions in 1960s since national control programs were launched in 1950s (Guan, 2009; Yang et al., 2014). However, some sporadic cases are still reported in the western part of China (Coordinating Office of the National Survey on the Important Human Parasitic Diseases, 2005) with two types of visceral leishmaniasis caused by L donovani transmitted by peridomestic Phlebotomus longiductus or by L. infantum transmitted by Phlebotomus chinensis (Wang et al., 2012). In recent years, leishmaniasis become re-emerging as more imported cases of both visceral and cutaneous leishmaniasis were reported in the major cities along east coast of China due to the increasing travel and business activities with African and other developing countries, which has brought more attention and concerns to physicians regarding the clinical diagnosis and treatment of this neglected tropical disease (Wang et al., 2017, 2019).
In current clinical practice, definitive diagnosis of leishmaniasis relies on a comprehensive analysis of the clinical manifestation, travel history and epidemiology, along with time/labor consuming laboratory tests (Aronson et al., 2016). Misdiagnosis of leishmania to other infection diseases is common due to the unspecific clinical manifestations especially for VL. The parasitological examination for the presence of the amastigote stage of the protozoa in the tissues of patients is considered as gold standard for the definitive diagnosis of leishmaniasis. However, it requires experienced and trained parasitologists or technicians, and the sensitivity of the parasitological examination of biopsy specimens by microscopy ranges only between 60 and 95% for cases of VLs and between 78.3 and 90.4% for cases of CL (Aronson et al., 2016; de Morais et al., 2016; Galluzzi et al., 2018). Furthermore, parasite can be identified by in vitro culture only in 39% VL samples and in 64.3% CL samples (Bahrami et al., 2018). Previous studies have shown that Leishmania antigens can be detected in urine, however both the sensitivity (28–82%) and specificity (49–53%) of this method were unacceptably low (Galluzzi et al., 2018; Abeijon et al., 2019; Mondal et al., 2019). An alternative immunological based assay is to detect antibody anti Leishmania rK39 antigen with sensitivity of 67–100% (Pagliano et al., 2016; van Griensven and Diro, 2019), however, this immunological assay is unable to differentiate between current and historical infection (Saliba et al., 2019). Regular PCR is sensitive, but cannot be used to monitor the parasite load during medical treatment, and so is not appropriate for ongoing patient/disease management, or evaluating the treatment efficacy (Mesa et al., 2020). Therefore, it is urgently needed to develop a diagnostic method with high sensitivity and quantitative measurement of infected parasite for clinical physicians.
In recent years molecular diagnostic tools for various infections have been developed for clinical application. These include PCR-based assays, which are recognized as rapid, sensitive method with ability to reliably discriminate between virus, bacteria, fungi, and parasites. We present here a new diagnostic method for leishmaniasis which uses quantitative real-time PCR (qPCR) technology to specifically detect Leishmania kinetoplast minicircle DNA (mkDNA). The mkDNA exhibits species-specific sequence divergence and therefore can be used as a marker for developing DNA probe-based diagnostic test for leishmaniasis and identification of Leishmania species that causes the disease (Spithill and Grumont, 1984; Rogers and Wirth, 1987). The mkDNA-based qPCR we developed in this study showed higher sensitivity than conventional parasite examination and high specificity for leishmaniasis. Except for its diagnostic purpose, it also showed the advantage as a tool to determine the disease severity and to evaluate the treatment efficacy and prognosis of the disease.
Methods
Ethics
This project was approved by the Ethics Committee of Beijing Friendship Hospital (Beijing, China) with approval number of 2020-P2-005-01. Informed consents were obtained from all involved patients.
Patients and Samples
The whole blood, bone marrow samples were collected from 23 clinically diagnosed current VL patients and 7 cured VL patients (with no parasite identified) at Beijing Friendship Hospital, Capital Medical University from July 2015 to Jan. 2020. The current VL patients presented with symptoms such as persistent fever, splenomegaly and/or hepatomegaly, positive in rK39 rapid diagnostic tests (RDT) and Leishmania amastigotes found in their blood and/or bone marrow samples under microscope. Skin lesion samples were obtained from 20 diagnosed CL patients. All patients' skin lesion appeared as ulcer and nodule/plaques features in which Leishmania amastigotes were identified. All tissue samples including skin lesion autopsy from CL patients, PBMC and bone marrow from VL patients were stored at liquid nitrogen till use.
In addition, 11 human blood samples were taken from patients with other infections including five with malaria, five with brucellosis, and one with dengue fever as non-leishmaniasis controls. Blood from five healthy volunteers were also included as normal control.
Primer and Probe Design
Total 25 mkDNA sequences from different species of Leishmania parasite were collected from GenBank and aligned using BIOEDIT software (v7.0.1, Ibis Biosciences, Carlsbad, CA, USA). Primers and probes were designed based on the conserved region of sequence shared by four common pathogenic species of L. donovani, L. infantum, L. major, and L. tropica using Primer Express 3.0.
DNA Extraction
DNA was extracted from cell and tissue samples using a TIANGEN DNA extraction kit (TIANGEN, DP705, Beijing, CHN) according to manufacturer's instructions. Briefly, about 20 mg lesion tissue or 200 μl of bone marrow or PBMC were digested with 20 μl proteinase K in 300 μl buffer GHA for 3 h at 56°C on a plate shaker (1,500 rpm), 300 μl lysis buffer GHL and 300 μl isopropyl alcohol were added and vortexed. The treated tissue samples were then mixed with magnetic beads in the kit to bind the DNA. The DNA-bound beads were subsequently washed and dried for 15 min at room temperature. Finally, 100 μl elution buffer was added and incubated at 56°C for 10 min to release the DNA from the magnetic beads. The beads were removed by centrifugation and the DNA containing supernatants were stored at −20°C till use.
Positive Control Plasmid Construction
The mkDNA target sequence fragment was PCR amplified using the primers designed as above and purified with DNA purification kit (TIANGEN, DP214, Beijing, CHN). The amplified mkDNA fragment was ligated into plasmid pUC19 (TAKARA, 3219, Tokyo, Japan) using EcoRI and HindIII sites. The correct insert of target DNA in recombinant plasmid DNA was confirmed by PCR amplification and DNA sequencing.
Quantitative Real-Time PCR Assay
All tests were conducted using the Applied Biosystems 7500 Fast Real-Time PCR System (ABI) in 10 μl reaction volume. The reaction mixtures contained the following; 5 μl of Promega GoTaq® Probe qPCR Master Mix (Promega, A6101, Madison, WI, USA), 200 nM Forward primer KD3, 200 nM Reverse primer KD4, 100 nM hydrolysis probes (5′FAM/3′TAMRA), CXR reference dye (30 nM), and 1 μl of template DNA (5–50 ng). The following cycle conditions were used; 95°C for 2 min followed by 40 cycles of 95°C for 15 s, 60°C for 30 s. A water only negative control (NTC-H2O) and a positive control using the mkDNA/pUC19 plasmid DNA were included in all experiments.
Sensitivity and Specificity Analysis
Analytical Sensitivity
The limit of detection (LOD) for the plasmid template was defined as the lowest number of detectable copies based on repeated tests (n = 3), using 1,000, 100, 10, 5, and 1 plasmid copies/reaction and the working assay precision (intra-assay SD <0.5 and inter-assay CV <5%). For testing the LOD for the parasite, cultured L. infantum promastigotes were enumerated by counting under a microscope. The Leishmania parasite suspension was then diluted in blood from healthy volunteer as 1, 5, 10, 50, 100 parasites in 200 μl blood and the total DNA was extracted from each dilution. The functional sensitivity was defined from the experimentally derived assay precision measurement (intra-assay SD <0.5 and inter-assay CV <5%). The sensitivity of qPCR was compared with the sensitivity of clinical diagnosis based on the typical clinical manifestation and parasite determination under microscope.
Specificity
The genomic DNA samples previously extracted from Plasmodium falciparum, Toxoplasma gondii, Rickettsia tsutsugamushi, Mycobacterium leprae, and Brucella melitensis, including DNAs extracted from 11 patients with other infections (mentioned above) were used to test the specificity of our developed Leishmania PCR/qPCR assay.
Clinical Sensitivity, Specificity, and Monitoring Performance of qPCR
Total 66 clinical samples including 20 skin lesion tissues from CL patients, 30 PBMC/bone marrow samples from VL patients, 11 blood samples from patients with other infections, and five normal donors, were analyzed using the designed qPCR assay for evaluating its sensitivity and specificity for leishmaniasis.
Statistical Analysis
The qPCR data analysis was performed using the ABI 7500 software 2.3. Statistical analysis was carried out using the SPSS software version 20.0 and visualized on Graphpad Prism version 5.0. The Mann-Whitney U-test was used to statistically compare between groups. P < 0.05 was considered as statistical significance.
Results
Design of a Quantitative Real-Time PCR Assay for Pan-Leishmania Detection
Alignments of mkDNA sequences from major Leishmania species reveals that mkDNAs of L. donovani, L. infantum, L. major, and L. tropica share sequence identity of 89.67%. Three regions of mkDNA sequence are conserved in all four species of Leishmania with consensus sequence of CSB-I (GGGCGT), CSB-II (CCCCGTTC), and CSB-III (GGGGTTGGTGTA). Primers were designed based on these consensus regions as forward KD3: 5′-TCCGGGTAGGGGCGTTCTG-3′ and reverse KD4: 5′-TTTACACCAACCCCCAGTTTCC-3′. A fragment DNA sequence within the amplified region was chosen for synthesizing probe: 5′-FAM-TTTGAACGGGATTTCTGCACCCAT-TAMRA-3′ for qPCR detection (Figure 1).
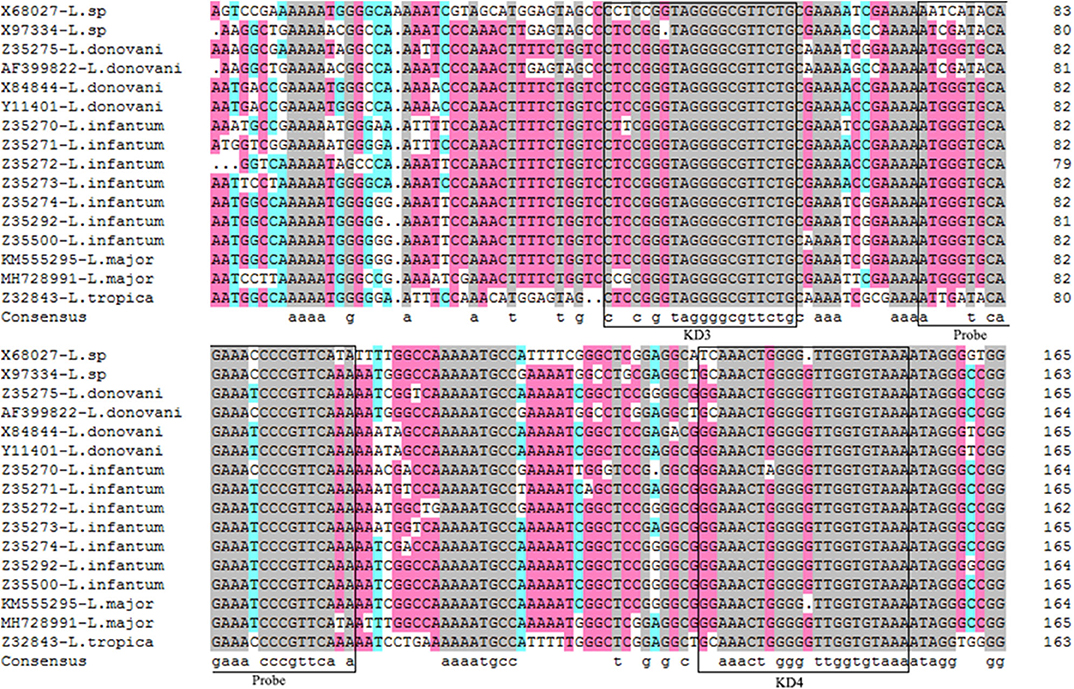
Figure 1. Alignments of mkDNA sequences from major Leishmania species. Sequences were aligned using CLUSTALW and prepared for display using BOXSHADE. Identical amino acids are shaded with different color. The consensus sequences were squared for making forward primer KD3 and reverse primer KD4, and probe. The gray mark represents the sequence homology 100%, the pink mark represents the sequence homology ≥75%, the blue mark represents the sequence homology ≥50%.
A PCR assay using this pair of primers was performed to detect DNA samples extracted from cultured L. donovani promastigote or from samples of patients infected with L. infantum or L. major. The results showed that the PCR based on the Leishmania mkDNA conserved sequence was able to amplify a 114 bp fragment from both visceral (L. donovani, L. infantum) and cutaneous Leishmania spp (L. major). However, these primers didn't recognize any DNA from samples of P. falciparum, T. gondii, R. tsutsugamushi, M. leprae, and B. melitensis (Figure 2).
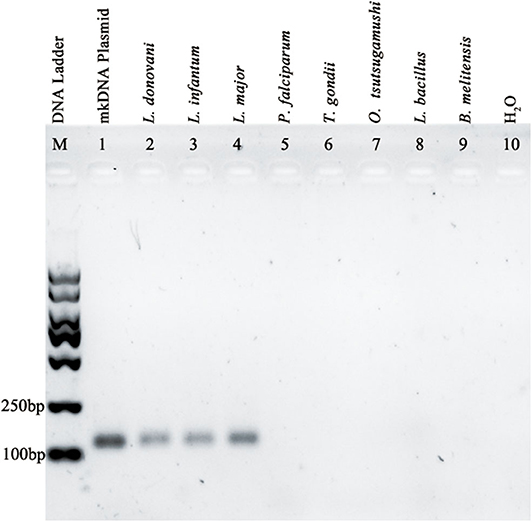
Figure 2. Amplification of Leishmania specific mkDNA in L. donovani, L. infantum, and L. major using mkDNA-based PCR method without cross-reaction with P. falciparum, T. gondii, R. tsutsugamushi, M. leprae, and B. melitensis.
Establishment of qPCR Assay for Testing Leishmania mkDNA
Using the primers and probe described above, we established a qPCR assay which was able to detect as little as single copy of Leishmania mkDNA plasmid or DNA from single Leishmania parasite with CV <5.0% (Table 1). Our results showed that the standard curve generated using serial copy number of plasmid DNA was linear over an 8-log range with a correlation coefficient (R2) of 0.998 (Figures 3A,B).
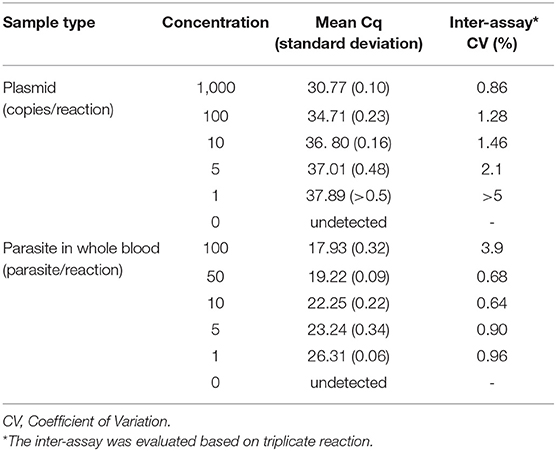
Table 1. Assessment of analytical sensitivity of qPCR assay to detect Leishmania mkDNA and parasite in blood.
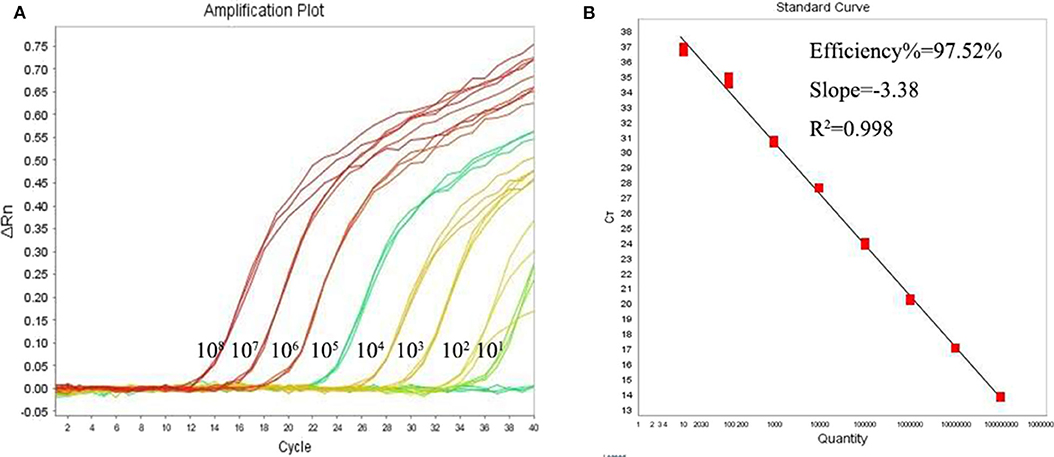
Figure 3. Quantitative correlation between mkDNA copy number and threshold cycle of the Leishmania qPCR assay. (A) Leishmania mkDNA plasmid was diluted in serial from 101 to 108 copies/reaction and subjected to qPCR. ΔRn = Rn (normalized reporter)—baseline. (B) Linear regression of Cq vs. lg copy number of mkDNA plasmid. Ct, Cycle threshold.
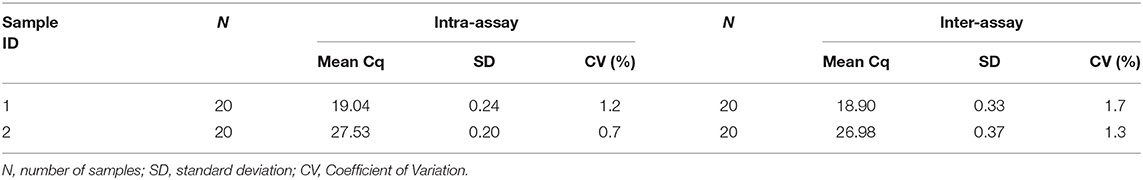
The intra assay and inter assay CV of Ct values for the 20 replicates was <2%, indicating a high precision in the assay (Table 2). Taken together, our results show that this qPCR assay is established with high sensitivity and precision. The qPCR was further used to test clinical specimens.
Sensitivity and Specificity of qPCR for Detecting Leishmania Parasite in Patients With Clinically Diagnosed Leishmaniasis
To test the sensitivity and specificity of established mkDNA-based qPCR to diagnose leishmaniasis, total DNAs were extracted from specimens from 50 patients with clinically diagnosed leishmaniasis including 20 CL and 30 VL. Among the 20 patients of CL, all are current patients with skin lesion and parasite identified under microscope. Among the 30 patients with VL, parasites have been identified in 21 patients either in blood or in bone marrow through direct microscopy examination or parasite culturing in vitro, nine patients were previously diagnosed as leishmaniasis by parasite examination and currently determined as cured without parasite identified in microscopic examination. After being tested by qPCR, the skin lesion samples from 20 CL patients were all positive. Among the 21-parasite confirmed VL patients, 20 were positive with qPCR, one with parasite identified only in the bone marrow (not in blood) was negative with qPCR for detecting blood sample. For other nine clinically cured VL patients (without parasite detected in blood and bone marrow), two of them were detected as positive in blood samples by qPCR (Table 3). The results indicate that the developed qPCR assay is sensitive (sensitivity 95.6%, 22/23) and the clinical diagnosis should combine the results of parasite detection in blood or bone marrow with qPCR test. Even if parasite examination converts to negative, it is still possible the trace parasite or its disrupted DNA could be detected using qPCR. The total sensitivity and diagnostic accuracy for all leishmaniosis can reach up to 97.6% (42/43) and 98.5% (65/66) (Table 3). To evaluate the specificity of qPCR for detecting Leishmania, DNAs were extracted from blood samples of 11 patients with infection of other pathogens including five with P. falciparum (malaria), five with B. melitensis (brucellosis) and one with Dengue virus, and five healthy normal people. All samples were detected as negative with the qPCR assay, indicating mkDNA-based qPCR has no cross reaction with other pathogens with 100% specificity for detecting Leishmania infections (Table 3).
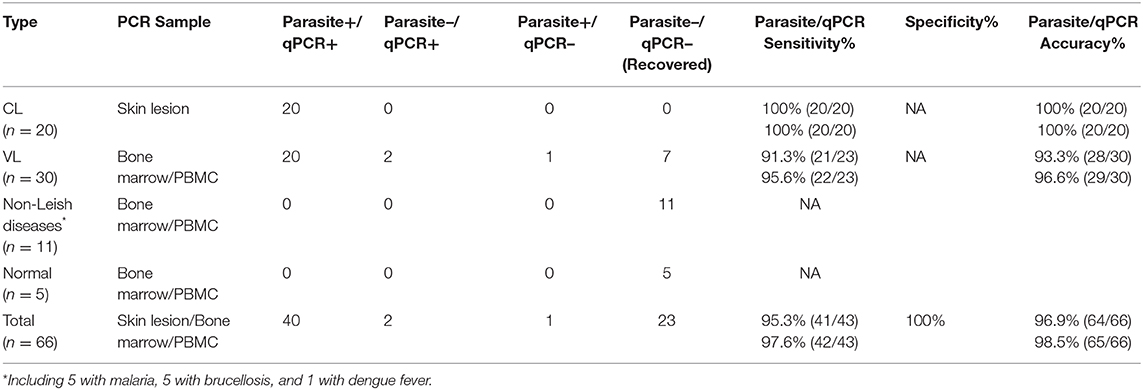
Table 3. Sensitivity and specificity of parasite detection and qPCR to diagnose cutaneous leishmaniasis (CL) and visceral leishmaniasis (VL).
Correlation Between Parasite Load Detected by qPCR With Clinical Severity of Cutaneous and Visceral Leishmaniasis
The established Pan-Leishmania qPCR assay was able to evaluate the severity of cutaneous or visceral leishmaniosis. Shown in Figure 4A, the parasite load detected by qPCR was correlated with the size of skin lesion in patients with CL. The bigger skin lesion, the higher parasite load was detected by qPCR (R2 = 0.794). The similar results were found in VL. The patients with higher level of serological immunoglobulin or bigger splenomegaly tested by ultrasonography, the common clinical signs of visceral leishmaniosis, the higher parasite burden was measured by qPCR (R2 = 0.823 and 0.871, respectively) (Figures 4B,C), indicating the mkDNA-based qPCR can be used not only for diagnosing leishmaniasis, but also for evaluating the severity of the infection.
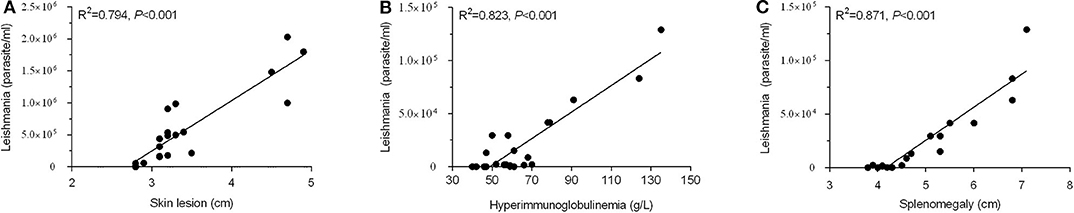
Figure 4. Correlation of parasite load detected by qPCR with severity of leishmaniasis. (A) Parasite load correlated with skin lesion diameter of CL. (B) Parasite load correlated with levels of serological immunoglobulin in VL. (C) Parasite load correlated with splenomegaly tested by ultrasonography in VL.
Assessment of the Treatment Efficacy and Prognosis of Cutaneous and Visceral Leishmaniasis by qPCR
After being treated with sodium stibogluconate for 1–30 weeks (6–211 days), the parasite load was dramatically reduced in both bone marrow and in blood (PBMC) of patients compared with the load before treatment (P < 0.05, Figures 5A,B), accompanied by significantly mitigated clinical manifestations. Besides, for testing samples from VL patients, although PBMC samples were easier to acquire in clinical practice than bone marrow, there was a higher detecting rate and parasite load in bone marrow sample than blood PBMC samples by qPCR (P < 0.05) (Figure 5B). The results indicate that the mkDNA-based qPCR is able to monitor and assess the treatment efficacy of visceral leishmaniosis. For CL, the skin sample was not collected for qPCR detection because the skin lesion was totally healed and cured after treatment.
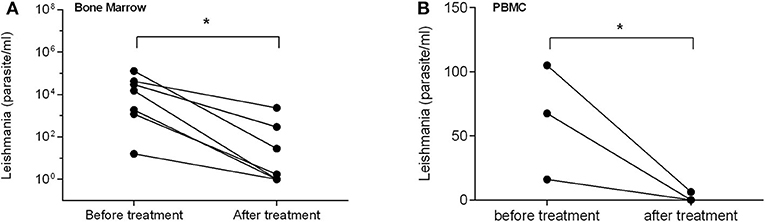
Figure 5. qPCR assessment of treatment efficacy of visceral leishmaniasis in samples of bone marrow (A) and PBMC (B). Each point shows qPCR measured parasite number from individual patient before and after treatment. The bar presents median parasite number. *P < 0.05.
Discussion
With globalization and rapid economic growth, people in developing or developed countries increase their economic activities and travel which significantly contribute to the increased world-wide transmission of some infections that are previously restricted in some specific endemic areas (Wang et al., 2019). Leishmaniasis is such a case which was prevalent in some endemic regions, but now identified in many other regions/countries with no history of the disease (Sakkas et al., 2016). The situation is further complicated by the fact that visceral and cutaneous leishmaniasis exhibit various non-specific clinical manifestations, making it hard to reliably diagnose and track the treatment. The definitive diagnosis of leishmaniasis usually replies on the identification of protozoa in the skin lesion (SL) or in blood/bone marrow (VL) that need experienced technicians or parasitologists. All these situations require a rapid, accurate and reliable diagnostic tool for leishmaniasis, especially in areas in which the disease is not endemic.
In Leishmania protozoa (genome size approximately 29–33 Mb), the kinetoplast organelle minicircle DNA (mkDNA) is conserved among the different Leishmania species (Cantacessi et al., 2015). This makes the mkDNA an attractive diagnostic biomarker for molecular detection of Leishmania parasite. Compared with other conserved sequences such as 18S rDNA, ITS or heat shock protein 70 KDa, the PCR based on mkDNA sequence yielded higher sensitivity and specificity than other targets, even for testing diverse clinical sample types from suspected leishmaniasis patients (Weirather et al., 2011; Real et al., 2013; Zampieri et al., 2016). In this study, we designed a new qPCR based on the mkDNA sequence regions conserved in all Leishmania species. The qPCR detected all three major pathogenic L. donovani, L. infantum, and L. major endemic or imported in China without cross-reaction with other pathogens such as P. falciparum, T. gondii, O. tsutsugamushi, L. bacillus, and B. melitensis. The pan detection of all Leishmania species indicates its application to detect Pan-Leishmania infections. The qPCR assay also has high sensitivity with ability to detect as low as one copy of mkDNA or one parasite in clinical sample.
The high sensitivity and specificity of developed Pan-Leishmania qPCR suggests this method could be used to better diagnose CL infected by L. major and VL infected mostly by L. donovani and L. infantum in China. Indeed, 20 skin lesion samples from clinically confirmed CL patients and 20 bone marrow and/or PBMC samples from clinically confirmed VL patients were tested by this new qPCR assay as all positive, with 100% sensitivity which is higher than previously reported for qPCR detection (69.23–98.53%)(Khosravi et al., 2012; El-Beshbishy et al., 2013) or microscopy smears (90–98%) (Mesa et al., 2020). We also confirmed the specificity of the assay not to cross-react with 11 clinical samples from patients with other infections with malaria, brucellosis, dengue fever and blood samples from five healthy volunteers. For nine samples from clinically cured VL patients infected with L. infantum (without parasite identified by microscopy or parasite culture), two of them were positively detected by qPCR in their blood samples, indicating the qPCR is a more sensitive and reliable assay than microscopy or parasite culture to diagnose leishmaniasis.
More importantly, this qPCR assay can be used not only to diagnose current infection of CL and VL patients, but also to evaluate the severity and treatment efficacy of leishmaniasis. The parasite load detected by qPCR was correlated with the severity of infection in terms of the size of lesion (CL) or the levels of serological immunoglobulin or splenomegaly tested by ultrasonography in visceral leishmaniasis. The patients with bigger skin lesion in CL and higher level of immunoglobulin or larger splenomegaly in VL, have the higher parasite load detected by qPCR. It is also noticed that the parasite load in PBMC/bone marrow of VL patients detected by this qPCR was significantly reduced after being treated with stibogluconate, accompanied with significantly mitigated clinical manifestations. The results indicate that the mkDNA-based qPCR is able to monitor and assess the treatment efficacy of visceral leishmaniosis. For CL, the skin sample was not collected for qPCR detection because the skin lesion was totally healed and cured after treatment.
The sensitive detection of Leishmania parasite is the key for avoiding relapse post-treatment (Martinez-Orellana et al., 2017). Previous studies have shown that a load of over 5–10 parasites/ml in the peripheral blood possibly caused clinical relapse except for other factors such as specific Leishmania strain, the immune status of the patient and prevalence of the disease in the region (Sudarshan et al., 2011; Sudarshan and Sundar, 2014). Thus, it is important to accurately measure the clinical therapeutic endpoint when patients are receiving therapy. Due to the high sensitivity and specificity, qPCR approach was strongly recommended by a consensus of experts to test patient sample after treatment to confirm the complete cure (Aronson et al., 2016; La Hoz and Morris, 2019; Sereno et al., 2019). Our results clearly revealed that this Leishmania-specific qPCR is a suitable tool not only for accurate diagnosis but also for evaluating the severity and prognosis of leishmaniasis. The qPCR assay is also useful for assess the prevalence of asymptomatic carriers of Leishmania in a specific population for better control of the infection in the region. Further work is needed to determine the threshold between asymptomatic and symptomatic carriage, as well as residual parasite infection level that might lead to leishmaniasis outbreak and relapse.
As a debilitating and potentially fatal neglected tropical disease, leishmaniasis needs to be well-managed and controlled in the endemic areas or non-endemic regions with potential imported cases. Since the Pan-Leishmania qPCR assay we developed in this study owns high sensitivity and specificity, with advantage of quick and easy operation without necessity of experienced technicians, it is a good tool for the control of leishmaniasis.
Data Availability Statement
All datasets generated for this study are included in the article/supplementary material.
Ethics Statement
The studies involving human participants were reviewed and approved by Ethics Committee of Beijing Friendship Hospital (Beijing, China). The patients/participants provided their written informed consent to participate in this study.
Author Contributions
YW, XT, MH, SL, and ZW performed experiments. YW, LW, BZ, and NS analyzed data and statistics. YW and LW prepared the figures and tables. YW, NW, BZ, LW, and GY wrote the manuscript. All authors reviewed the manuscript.
Funding
This study was supported by grants from the National Natural Science Foundation of China (No. 81702018) and Research Foundation of Friendship Hospital, Capital Medical University (No. yyqdkt2019-42). The funders of the study had no role in study design, data collection, data analysis, data interpretation, writing, or decision to publish of the manuscript.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
Abeijon, C., Alves, F., Monnerat, S., Wasunna, M., Mbui, J., Viana, A., et al. (2019). Development of a multiplexed assay for detection of Leishmania donovani and Leishmania infantum protein biomarkers in urine samples of patients with visceral leishmaniasis. J. Clin. Microbiol. 57:e02076-18. doi: 10.1128/JCM.02076-18
Aronson, N., Herwaldt, B. L., Libman, M., Pearson, R., Lopez-Velez, R., Weina, P., et al. (2016). Diagnosis and treatment of leishmaniasis: clinical practice guidelines by the infectious diseases society of America (IDSA) and the American society of tropical medicine and hygiene (ASTMH). Clin. Infect. Dis. 63, 202–264. doi: 10.1093/cid/ciw670
Bahrami, F., Harandi, A. M., and Rafati, S. (2018). Biomarkers of cutaneous leishmaniasis. Front. Cell. Infect. Microbiol. 8:222. doi: 10.3389/fcimb.2018.00222
Bailey, F., Mondragon-Shem, K., Hotez, P., Ruiz-Postigo, J., Al-Salem, W., Acosta-Serrano, A., et al. (2017). A new perspective on cutaneous leishmaniasis-Implications for global prevalence and burden of disease estimates. PLoS Negl. Trop. Dis. 11:e5739. doi: 10.1371/journal.pntd.0005739
Bi, K., Chen, Y., Zhao, S., Kuang, Y., and John, W. C. H. (2018). Current visceral leishmaniasis research: a research review to inspire future study. Biomed Res. Int. 2018:9872095. doi: 10.1155/2018/9872095
Burza, S., Croft, S. L., and Boelaert, M. (2018). Leishmaniasis. Lancet 392, 951–970. doi: 10.1016/S0140-6736(18)31204-2
Cantacessi, C., Dantas-Torres, F., Nolan, M. J., and Otranto, D. (2015). The past, present, and future of Leishmania genomics and transcriptomics. Trends Parasitol. 31, 100–108. doi: 10.1016/j.pt.2014.12.012
Coordinating Office of the National Survey on the Important Human Parasitic Diseases (2005). A national survey on current status of the important parasitic diseases in human population. Chin. J. Parasitol. Parasit. Dis. 23, 332–340.
de Morais, R. C., Da, C. O. C. N., de Albuquerque, S. C., Mendonca, T. S. L. A., Pessoa-E-Silva, R., Alves, D. C. H. L., et al. (2016). Real-time PCR for Leishmania species identification: evaluation and comparison with classical techniques. Exp. Parasitol. 165, 43–50. doi: 10.1016/j.exppara.2016.03.005
de Vries, H. J., Reedijk, S. H., and Schallig, H. D. (2015). Cutaneous leishmaniasis: recent developments in diagnosis and management. Am. J. Clin. Dermatol. 16, 99–109. doi: 10.1007/s40257-015-0114-z
El-Beshbishy, H. A., Al-Ali, K. H., and El-Badry, A. A. (2013). Molecular characterization of cutaneous leishmaniasis in Al-Madinah Al-Munawarah province, western Saudi Arabia. Int. J. Infect. Dis. 17, 334–338. doi: 10.1016/j.ijid.2012.11.015
Galluzzi, L., Ceccarelli, M., Diotallevi, A., Menotta, M., and Magnani, M. (2018). Real-time PCR applications for diagnosis of leishmaniasis. Parasit. Vectors 11:273. doi: 10.1186/s13071-018-2859-8
Ghorbani, M., and Farhoudi, R. (2018). Leishmaniasis in humans: drug or vaccine therapy? Drug Des. Devel. Ther. 12, 25–40. doi: 10.2147/DDDT.S146521
Guan, L. R. (2009). Present situation of visceral leishmaniasis and prospect for its control in China. Chin. J. Parasitol. Parasit. Dis. 27, 394–397.
Khosravi, S., Hejazi, S. H., Hashemzadeh, M., Eslami, G., and Darani, H. Y. (2012). Molecular diagnosis of Old World leishmaniasis: real-time PCR based on tryparedoxin peroxidase gene for the detection and identification of Leishmania spp. J. Vector Borne Dis. 49, 15–18.
La Hoz, R. M., and Morris, M. I. (2019). Tissue and blood protozoa including toxoplasmosis, Chagas disease, leishmaniasis, Babesia, Acanthamoeba, Balamuthia, and Naegleria in solid organ transplant recipients- guidelines from the American Society of Transplantation Infectious Diseases Community of Practice. Clin. Transplant. 33:e13546. doi: 10.1111/ctr.13546
Martinez-Orellana, P., Mari-Martorell, D., Montserrat-Sangra, S., Ordeix, L., Baneth, G., and Solano-Gallego, L. (2017). Leishmania infantum-specific IFN-gamma production in stimulated blood from dogs with clinical leishmaniosis at diagnosis and during treatment. Vet. Parasitol. 248, 39–47. doi: 10.1016/j.vetpar.2017.10.018
Mesa, L. E., Manrique, R., Muskus, C., and Robledo, S. M. (2020). Test accuracy of polymerase chain reaction methods against conventional diagnostic techniques for Cutaneous Leishmaniasis (CL) in patients with clinical or epidemiological suspicion of CL: systematic review and meta-analysis. PLoS Negl. Trop. Dis. 14:e7981. doi: 10.1371/journal.pntd.0007981
Mondal, D., Ghosh, P., Chowdhury, R., Halleux, C., Ruiz-Postigo, J. A., Alim, A., et al. (2019). Relationship of serum antileishmanial antibody with development of visceral leishmaniasis, post-kala-azar dermal leishmaniasis and visceral leishmaniasis relapse. Front. Microbiol. 10:2268. doi: 10.3389/fmicb.2019.02268
Pagliano, P., Ascione, T., Di Flumeri, G., Boccia, G., and De Caro, F. (2016). Visceral leishmaniasis in immunocompromised: diagnostic and therapeutic approach and evaluation of the recently released IDSA guidelines. Infez. Med. 24, 265–271.
Real, F., Vidal, R. O., Carazzolle, M. F., Mondego, J. M., Costa, G. G., Herai, R. H., et al. (2013). The genome sequence of Leishmania (Leishmania) amazonensis: functional annotation and extended analysis of gene models. DNA Res. 20, 567–581. doi: 10.1093/dnares/dst031
Rogers, W. O., and Wirth, D. F. (1987). Kinetoplast DNA minicircles: regions of extensive sequence divergence. Proc. Natl. Acad. Sci. U.S.A. 84, 565–569. doi: 10.1073/pnas.84.2.565
Sakkas, H., Gartzonika, C., and Levidiotou, S. (2016). Laboratory diagnosis of human visceral leishmaniasis. J. Vector Borne Dis. 53, 8–16.
Saliba, J. W., Lopes, K. F., Silva-Pereira, R. A., Teixeira, L. S., and Oliveira, E. (2019). Leishmania infantum exo-antigens: application toward serological diagnosis of visceral leishmaniasis. Parasitol. Res. 118, 2317–2323. doi: 10.1007/s00436-019-06352-4
Sereno, D., Harrat, Z., and Eddaikra, N. (2019). Meta-analysis and discussion on challenges to translate Leishmania drug resistance phenotyping into the clinic. Acta Trop. 191, 204–211. doi: 10.1016/j.actatropica.2019.01.009
Spithill, T. W., and Grumont, R. J. (1984). Identification of species, strains and clones of Leishmania by characterization of kinetoplast DNA minicircles. Mol. Biochem. Parasitol. 12, 217–236. doi: 10.1016/0166-6851(84)90137-3
Sudarshan, M., and Sundar, S. (2014). Parasite load estimation by qPCR differentiates between asymptomatic and symptomatic infection in Indian visceral leishmaniasis. Diagn. Microbiol. Infect. Dis. 80, 40–42. doi: 10.1016/j.diagmicrobio.2014.01.031
Sudarshan, M., Weirather, J. L., Wilson, M. E., and Sundar, S. (2011). Study of parasite kinetics with antileishmanial drugs using real-time quantitative PCR in Indian visceral leishmaniasis. J. Antimicrob. Chemother. 66, 1751–1755. doi: 10.1093/jac/dkr185
Torres-Guerrero, E., Quintanilla-Cedillo, M. R., Ruiz-Esmenjaud, J., and Arenas, R. (2017). Leishmaniasis: a review. F1000Res. 6:750. doi: 10.12688/f1000research.11120.1
van Griensven, J., and Diro, E. (2019). Visceral Leishmaniasis: recent advances in diagnostics and treatment regimens. Infect. Dis. Clin. North Am. 33, 79–99. doi: 10.1016/j.idc.2018.10.005
Wang, J. Y., Cui, G., Chen, H. T., Zhou, X. N., Gao, C. H., and Yang, Y. T. (2012). Current epidemiological profile and features of visceral leishmaniasis in people's republic of China. Parasit. Vectors. 5:31. doi: 10.1186/1756-3305-5-31
Wang, L., Zou, Y., Zhu, X., Bottazzi, M. E., Hotez, P., and Zhan, B. (2019). China's shifting neglected parasitic infections in an era of economic reform, urbanization, disease control, and the Belt and Road Initiative. PLoS Negl. Trop. Dis. 13:e6946. doi: 10.1371/journal.pntd.0006946
Wang, Y. L., Wang, X., Ren, R. Q., Zhou, L., Tu, W. W., Ni, D. X., et al. (2017). Epidemiology of imported infectious diseases in China, 2013-2016. Chin. J. Epidemol. 38, 1499–1503. doi: 10.3760/cma.j.issn.0254-6450.2017.11.012
Weirather, J. L., Jeronimo, S. M., Gautam, S., Sundar, S., Kang, M., Kurtz, M. A., et al. (2011). Serial quantitative PCR assay for detection, species discrimination, and quantification of Leishmania spp. in human samples. J. Clin. Microbiol. 49, 3892–3904. doi: 10.1128/JCM.r00764-11
Yang, G. J., Liu, L., Zhu, H. R., Griffiths, S. M., Tanner, M., Bergquist, R., et al. (2014). China's sustained drive to eliminate neglected tropical diseases. Lancet Infect. Dis. 14, 881–892. doi: 10.1016/S1473-3099(14)70727-3
Zampieri, R. A., Laranjeira-Silva, M. F., Muxel, S. M., Stocco, D. L. A. C., Shaw, J. J., and Floeter-Winter, L. M. (2016). High resolution melting analysis targeting hsp70 as a fast and efficient method for the discrimination of Leishmania species. PLoS Negl. Trop. Dis. 10:e4485. doi: 10.1371/journal.pntd.0004485
Keywords: human leishmaniasis, quantitative PCR, diagnosis, treatment efficacy, L. donovani, L. infantum, L. major
Citation: Wu Y, Tian X, Song N, Huang M, Wu Z, Li S, Waterfield NR, Zhan B, Wang L and Yang G (2020) Application of Quantitative PCR in the Diagnosis and Evaluating Treatment Efficacy of Leishmaniasis. Front. Cell. Infect. Microbiol. 10:581639. doi: 10.3389/fcimb.2020.581639
Received: 09 July 2020; Accepted: 11 September 2020;
Published: 07 October 2020.
Edited by:
Epke Le Rutte, Erasmus Medical Center, NetherlandsReviewed by:
Monica Varela, Leiden University, NetherlandsHirdesh Kumar, National Institutes of Health (NIH), United States
Copyright © 2020 Wu, Tian, Song, Huang, Wu, Li, Waterfield, Zhan, Wang and Yang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Lei Wang, d2x0b25namlAeWFob28uY29t; Guowei Yang, eWFuZ2d1b3dlaUBob3RtYWlsLmNvbQ==
†These authors have contributed equally to this work
 Yun Wu1,2†
Yun Wu1,2† Nan Song
Nan Song Nicholas R. Waterfield
Nicholas R. Waterfield Guowei Yang
Guowei Yang