- Office of Applied Research and Safety Assessment, Center for Food Safety and Applied Nutrition, U.S. Food and Drug Administration, Laurel, MD, United States
Ingestion of Shiga toxin-producing Escherichia coli (STEC) can result in a range of illness severity from asymptomatic to hemorrhagic colitis and death; thus risk assessment of STEC strains for human pathogenicity is important in the area of food safety. Illness severity depends in part on the combination of virulence genes carried in the genome, which can vary between strains even of identical serotype. To better understand how core genes are regulated differently among strains and to identify possible novel STEC virulence gene candidates that could be added to the risk assessment repertoire, we used comparative transcriptomics to investigate global gene expression differences between two STEC strains associated with severe illness and a commensal E. coli strain during in vitro intestinal epithelial cell (IEC) infections. Additionally, we compared a wide array of concomitant cytokine levels produced by the IECs. The cytokine expression levels were examined for a pattern representing STEC pathogenicity; however, while one STEC strain appeared to elicit a proinflammatory response, infection by the other strain produced a pattern comparable to the commensal E. coli. This result may be explained by the significant differences in gene content and expression observed between the STEC strains. RNA-Seq analysis revealed considerable disparity in expression of genes in the arginine and tryptophan biosynthesis/import pathways between the STEC strains and the commensal E. coli strain, highlighting the important role some amino acids play in STEC colonization and survival. Contrasting differential expression patterns were observed for genes involved in respiration among the three strains suggesting that metabolic diversity is a strategy utilized to compete with resident microflora for successful colonization. Similar temporal expression results for known and putative virulence genes were observed in the STEC strains, revealing strategies used for survival prior to and after initial adherence to IECs. Additionally, three genes encoding hypothetical proteins located in mobile genetic elements were, after interrogation of a large set of E. coli genomes, determined to likely represent novel STEC virulence factors.
Introduction
Shiga toxin-producing Escherichia coli (STEC) is a genomically diverse E. coli pathotype that produces Shiga toxin (Stx) and has been isolated from humans, animals, food, and many environmental sources (Garcia et al., 2010; Cooley et al., 2013). As a zoonotic foodborne pathogen, STEC primarily asymptomatically colonizes cattle; however, it has also been isolated from other domestic livestock and a variety of wild animals and birds (La Ragione et al., 2009; Cooley et al., 2013; Espinosa et al., 2018). Depending on their individual gene repertoire, STEC strains have the potential to be human pathogens associated with gastrointestinal illness, ranging from mild diarrhea to hemorrhagic colitis. In some cases, infection with STEC causes hemolytic uremic syndrome (HUS), a serious sequela that can progress to end-stage renal disease (ESRD) and lead to death (Kaper et al., 2004; Majowicz et al., 2014). Globally, based on data compiled between January 1990 and April 2012, there were an estimated 2.8 million illnesses associated with STEC, resulting in 3,890 cases of HUS, 270 cases of ESRD, and 230 deaths (Majowicz et al., 2014). Historically, STEC O157:H7 has been associated with outbreaks and severe illness (Kaper et al., 2004) and in 2018 and 2019 has been linked to multiple large outbreaks involving contaminated romaine lettuce (Centers for Disease Control and Prevention, 2020b). However, the proportion of non-O157 STEC illnesses has been increasing, and in fact, 58% of illnesses due to STEC infection were attributable to non-O157 serogroups in the US between 2008 and 2018 (Centers for Disease Control and Prevention, 2020a). Although at least 100 non-O157 serotypes have been known to cause human illness; those of serogroups O26, O45, O103, O111, O121, and O145 account for the greatest number of clinical cases (FAO/WHO STEC Expert Group, 2019; National Advisory Committee On Microbiological Criteria For Foods, 2019).
To cause illness, STEC must be ingested and survive passage through the gastrointestinal tract where it adheres to and colonizes the colon, followed by production of Stx. Prediction of the risk level as a human pathogen is important in the food industry since not all STEC strains cause illness in humans. STEC virulence genes are carried on mobile genetic elements, thus can be easily gained or lost. Therefore, while serotype is a useful indicator in epidemiological investigations, serotype alone is not considered a good predictor of clinical outcome. The best predictor of human pathogenicity is the set of virulence genes a particular STEC strain possesses, while also taking into consideration past illnesses linked with specific serotypes (Newell and La Ragione, 2018; FAO/WHO STEC Expert Group, 2019; National Advisory Committee On Microbiological Criteria For Foods, 2019). One virulence factor associated with severe clinical outcome is the type III secretion system (TTSS) encoded in the Locus of Enterocyte Effacement pathogenicity island (LEE PAI) that imparts, among other functions, the ability to adhere tightly to colonic intestinal epithelial cells (IECs) (Kaper et al., 2004). In risk assessments, the gene encoding the adhesin intimin, eae, is used as a marker for the presence of the LEE PAI. Enterohemolysin, encoded by ehxA, is also commonly considered when determining pathogenic potential, as it has been observed in many STEC strains causing severe disease (FAO/WHO STEC Expert Group, 2019; National Advisory Committee On Microbiological Criteria For Foods, 2019). However, there is not a perfect correlation between presence of ehxA and clinical outcome, which underscores the necessity of interrogating the genome for multiple factors that have been associated with virulence when determining risk, and this has become easier with the widespread use of whole genome sequencing (WGS) (Newell and La Ragione, 2018; Gonzalez-Escalona and Kase, 2019). Hybrid STEC pathotype strains have been increasingly reported (Leonard et al., 2016), the most important of which is the hybrid STEC/Enteroaggregative E. coli (EAEC) O104:H4 strain that caused a massive outbreak in 2011 with a high percentage of HUS (Rasko et al., 2011). EAEC carry aggR, encoding a transcriptional regulator of virulence genes including genes involved in adherence. The Stx subtype and presence/absence of eae, ehxA, and aggR are the principal virulence factors currently used in STEC risk assessments, although other adherence factors and toxins are taken into consideration. Progress on identification of novel STEC virulence genes has been slow; however, identification of novel STEC virulence factors is important since it would aid in refining suitable sets of genes to use in genome interrogations for risk determinations (Newell and La Ragione, 2018; FAO/WHO STEC Expert Group, 2019; National Advisory Committee On Microbiological Criteria For Foods, 2019). While WGS has been of great benefit to public health in the area of food safety (Allard et al., 2016), identification of additional STEC virulence factors by genome sequence comparisons alone remains elusive. For example, comparative genomics studies have revealed only previously identified virulence genes and were unable to differentiate STEC genomes by illness severity (Haugum et al., 2014; Baba et al., 2019).
In addition to the combination of virulence factors carried by an STEC strain, the dose ingested, and individual host responses can play a role in pathogenicity. Several reports have demonstrated that the interaction between the intestinal epithelium and different STEC factors, including lipopolysaccharides (Khan et al., 2006), flagellin (Berin et al., 2002; Rogers et al., 2003; Miyamoto et al., 2006), Stxs (Yamasaki et al., 1999; Thorpe et al., 2001), the TTSS factor EspT (Raymond et al., 2011), hemorrhagic coli pilus (HCP) (Ledesma et al., 2010), and long polar fimbriae (LPF) (Farfan et al., 2013; Vergara et al., 2014) can result in the induction of cytokines/chemokines that attract macrophages and polymorphonuclear leukocytes (PMNs) such as neutrophils in vitro and in vivo. Patients with STEC O157:H7 infections have been observed to possess high levels of PMNs in their feces (Slutsker et al., 1997), which is considered a possible risk marker of HUS development (Buteau et al., 2000). While most studies have focused on the induction of the neutrophil chemoattractant Interleukin-8 (IL-8), the induction of other cytokines and chemokines by intestinal epithelial cells (IECs) in response to STEC and its virulence factors has also been reported (Thorpe et al., 2001; Khan et al., 2006; Vergara et al., 2014; Stalb et al., 2018). Furthermore, depending on the experimental design, the cytokine profiles induced in various IEC models can vary greatly between different STEC strains and at times, but not always, from commensal E. coli (Hurley et al., 2001; Stalb et al., 2018). Comparison of induction levels of a wide variety of cytokines/chemokines by genetically diverse STECs known to have caused HUS and commensal E. coli may reveal patterns useful for predicting pathogenicity.
While STEC possesses virulence genes that are absent from commensal E. coli genomes, STEC and commensal strains share common core genes. Whether, and how, differences in global transcriptional responses of core genes contribute to human pathogenicity during survival and colonization in the gastrointestinal tract has not been elucidated. Previous reports have focused on either specific genes or global transcriptomic differences involving only one STEC O157:H7 strain adhered to epithelial cells (Jandu et al., 2009; Kim et al., 2009; Hu et al., 2013; Yang et al., 2015; Kumar and Sperandio, 2019). While these transcriptomic studies have been useful in determining regulation of select virulence genes and characterization of global responses in human infection, differences in transcriptional responses between STEC strains, and in contrast to commensal E. coli, upon adherence to IECs is unknown. Global transcriptomics studies involving sets of Enteropathogenic E. coli (EPEC) strains demonstrated phylogroup-specific responses to a variety of growth conditions but also individual differences between strains within the same phylogroup (Hazen et al., 2015; Hazen et al., 2017), thus highlighting the incomplete information gained from examining the transcriptional responses of only one strain in an E. coli pathotype.
In this study, we use an RNA-Seq approach to demonstrate differences and similarities among transcriptional responses of two non-O157 STEC strains associated with HUS that cluster with different phylogroups and a commensal E. coli strain in in vitro colonic IEC culture infections. In addition, we determine the concomitant cytokine/chemokine responses by the IECs, measuring an extensive panel of cytokine/chemokine levels in order to identify patterns that may be representative of pathogenic STEC. Through the RNA-Seq analysis combined with interrogation of a large set of E. coli genomes, we also identify hypothetical genes that represent potential novel STEC virulence factors.
Materials and Methods
Cell Culture
T84 cells (ATCC, Manassas, VA) were maintained at 37°C/5% CO2 in complete Dulbecco’s modified Eagle Medium/F12 (cDMEM/F12): DMEM/F12 (Invitrogen, Carlsbad, CA) supplemented with 10% heat-inactivated fetal bovine serum (FBS; Hyclone Laboratories, Logan, UT), 100 U/ml Penicillin/Streptomycin (Gibco, Gaithersburg, MD), and 2 mM L-glutamine (Gibco). T84 cells were passaged at no greater than 80% confluency. Caco-2 cells (ATCC) were maintained at 37°C/5% CO2 in complete DMEM (cDMEM): low glucose DMEM (Invitrogen) supplemented with 10% heat-inactivated FBS, 100 U/ml Penicillin/Streptomycin, and 2 mM L-glutamine.
Bacterial Isolates and Growth Conditions
Two STEC strains and one E. coli strain known to be commensal were used in this study. Draft whole genome sequences for STEC O26:H11 strain 97-3250 and STEC O145:H28 strain 4865/96 are publicly available with accession numbers JHEW01000000 and JHEY01000000, respectively. Annotation of the genomes was provided by NCBI. The genome sequence of the commensal E. coli O9:H4 strain HS is also publicly available (accession no. CP000802.1). For planktonic cultures, overnight cultures of the E. coli were grown in Luria broth at 37°C and used in subcultures at a ratio of 1:100 into 25 ml DMEM/F12 in a 250 ml flask. The flasks were shaken at 185 rpm and 37°C until the cultures were in late exponential growth phase. RNAprotect Bacteria Reagent (Qiagen, Germantown, MD) was used on aliquots of the planktonic cultures and pellets were stored at −80°C for RNA extraction. Biological triplicate cultures were grown for use in the RNA-Seq analysis. E. coli cultures were grown for use in in vitro infection experiments with polarized T84 IEC cultures using the same method as for planktonic cultures, except that to simulate passage through the small intestine, DMEM/F12 was supplemented with 0.4% w/v porcine bile extract (Sigma, St. Louis, MO). To obtain the correct density of E. coli cells for the infection experiments, aliquots of cultures were pelleted and resuspended in 250 µl of media as needed to yield a multiplicity of infection of 100:1 when added to the IECs. For STEC cultures to be used for both planktonic and adhered transcriptomics experiments using polarized Caco-2 IECs, the same procedure was used as for the T84 IEC experiments with the exception that low glucose DMEM was substituted for DMEM/F12.
T84 Cell Infection
T84 cells (1 × 106 cells/well), at passage >8, were seeded onto 6-well tissue culture plates containing collagen-coated (5 μg/cm2) transwell inserts (Corning, Lowell, MA). Media was changed every 2–3 days until transepithelial electrical resistance (TEER) measurements were at least 1,000 Ω (approximately 14 days) as measured by the EVOM2 Epithelial Voltohmmeter (World Precision Instruments, Sarasota, FL). E. coli cells were added in a volume of 250 μl into 1.75 ml of fresh cDMEM/F12 in the transwell insert. The plates were rocked back and forth and side to side to distribute bacteria evenly within the transwell insert. Each plate was centrifuged at 3,000 rpm for 1 min at RT and incubated at 37°C/5% CO2 for 3 h. Basolateral supernatants were collected and centrifuged at 14,000 rpm for 2 min at 4°C. Aliquots of basolateral supernatants were transferred to new microcentrifuge tubes and stored at −80°C for cytokine analysis. T84 cells and adherent bacteria remaining in the transwell inserts were rinsed two times with sterile 1× phosphate buffered saline (PBS) to remove residual non-adherent bacteria. To each transwell, 1 ml of RLT lysis buffer (RLT buffer containing β-mercaptoethanol) from the RNeasy mini kit (Qiagen) was added, and the adherent cells were gently scraped from the membrane using a sterile cell lifter (Corning). The contents from two transwells were combined for a total of 2 ml, vortexed 1 min, and centrifuged at 14,000 rpm for 2 min at 4°C to pellet adherent bacterial cells. Supernatants, which contained the contents of the lysed T84 cells, were transferred to new microcentrifuge tube and homogenized using 1 cc VanishPoint Syringes with 25 G × 5/8″ needles (Retractable Technologies, Inc, Little Elm, Texas). The remaining pellets of adherent bacterial cells were immediately resuspended in 300 µl PBS followed by the use of RNAprotect Bacteria Reagent to stabilize the RNA and stored at −80°C for RNA extraction. Biological triplicate infection experiments were performed starting with separate overnight cultures for each of the three E. coli strains.
Caco-2 Cell Infection
Caco-2 cells (0.5 × 106 cells/well), at passage >70, were seeded onto 6-well tissue culture plates (Corning). Media was changed every 2–3 days until TEER measurements were at least 400 Ω (approximately 21 days) as measured by the EVOM2 Epithelial Voltohmmeter. Three-hour infection experiments were performed in triplicate using the STEC strains. The same procedure was followed as for the T84 cell infections with the exception that low glucose DMEM media was used.
Bacterial RNA Isolation and Sequencing
Total RNA was extracted from both planktonic and adhered E. coli cell pellets using the RNeasy Mini Kit (Qiagen). The RNA was treated twice with DNase I from the Turbo DNA-free Kit (Ambion, Austin, TX) to remove contaminating DNA, followed by a clean-up step using the RNeasy Mini Kit. Depletion of bacterial rRNA from total RNA samples was accomplished using the MICROBExpress Bacterial mRNA Enrichment Kit (Ambion), after which RNA quality was assessed using a Bioanalyzer (Agilent, Santa Clara, CA). Sequencing libraries were constructed from the mRNA-enriched RNA samples using the TruSeq RNA Library Prep Kit v2 (Illumina, San Diego, CA), utilizing the protocol modifications as designated by the manufacturer for bacterial RNA. The libraries were sequenced using 75 bp paired-end sequencing on an Illumina MiSeq platform generating an average of 17.8 million reads per sample.
RNA-Seq Analysis
RNA-Seq analysis was performed using CLC Genomics Workbench version 12.0 (https://digitalinsights.qiagen.com) with default parameters. The sequence reads generated for each of the biological triplicate planktonic and adhered bacterial RNA samples were trimmed for quality, and adapter sequences were removed. The trimmed reads were mapped to the respective E. coli genomes imported as GenBank files, thus using the open reading frame annotations provided by NCBI. Differential expressions were calculated for adhered bacterial cells compared to planktonic bacterial cells for each E. coli strain, along with statistical analysis using the triplicate samples for each condition. CLC Genomics Workbench uses the normalization process to correct for sample library size used in edgeR (Robinson et al., 2010), and the False Discovery Rate (FDR) corrected p values were determined using the method of Benjamini and Hochberg. Differentially expressed genes were defined as those with fold adhered/planktonic changes of either >2 or <−2 and FDR (corrected p value) <0.05. The differential expression results were exported, and gene homolog results were used to compare differential expression for adhered versus planktonic cultures among the three E. coli strains.
E. coli Genome Comparisons
Shared genes were identified in the three E. coli genomes by pairwise megablast comparisons using a custom Python program. Genes were considered homologs if ≥90% of the gene length had ≥90% nucleotide identity and were reciprocal best matches. Amino acid sequences were used to assign genes to biochemical pathways in the KEGG database. To determine which E. coli genomes possess selected hypothetical genes found in the two STEC genomes, a megablast search of a database containing 25,527 publicly available E. coli genomes as well as Escherichia cryptic lineage 1 and 6 (the two cryptic lineages that cluster most closely with E. coli) was performed. Parameters used to determine gene presence were at least 90% similarity over at least 67% of the query length. E. coli genomes representing the entire E. coli genomic landscape were used, thus included genomes both with and without known molecular markers representing the E. coli pathotypes. The genomes were classified as belonging to one or more of six pathotypes, namely, Attaching and Effacing E. coli (AEEC), EAEC, Enteroinvasive E. coli (EIEC), Enterotoxigenic E. coli (ETEC), Extraintestinal pathogenic E. coli (ExPEC), and STEC. The pathotypes were defined as carrying one or more of the following molecular markers: AEEC (eae), EAEC (aggR, aafA), EIEC (ipaH, icsP, mkaD, ospB), ETEC (LT, ST), ExPEC (papD, afaD, cnf, hlyA), STEC (stx1, stx2). Genomes not falling into one or more of these pathotypes were classified as “other”.
Human Cytokine Analysis
Cell-free basolateral T84 supernatants were analyzed in duplicate using the Bio-Plex Pro™ Human Chemokine Panel, 40-Plex Kit (#171AK99MR2; Bio-Rad, Hercules, CA), and the Bio-Plex 200 Multiplex Reader (Bio-Rad), according to the manufacturer’s instructions. Concentration in range values were used for analysis to identify differences in cytokine expression among the uninfected and infected T84 cells. Samples from three independent experiments were analyzed in duplicate and graphed using GraphPad Prism 6 (GraphPad Software, La Jolla, CA). For samples where every value was out of range, indicating very low levels of cytokine production, a value of 0.1 was substituted for statistical analysis. The data are expressed as the means of the concentrations (pg/ml) ± standard errors of the mean (SEM). Statistical significance was assessed at a p value of <0.05 by one-way ANOVA with Tukey’s multiple comparisons test using Graphpad Prism 6.
Results
Gene Homologs in the E. coli Strains
Two non-O157 STEC strains and one commensal E. coli strain were selected for this study. Both STEC strains, STEC O26:H11 strain 97-3250, and STEC O145:H28 strain 4865/96, are LEE-positive strains with a known association with HUS. However, the two STEC strains belong to different E. coli phylogroups. STEC 97-3250 clusters with phylogroup B1, while STEC O145:H28 clusters with phylogroup E. The commensal E. coli strain HS belonging to phylogroup A was used for comparison. To enable transcriptional response comparisons among the strains, gene homologs were identified in the genomes of the three strains using megablast matching of nucleotide sequences of annotated open reading frames in the genomes (Figure 1A). The number of genes, after removal of rRNA genes, is 5,938, 5,023, and 4,604 for strains 97-3250, 4865/96, and HS, respectively (Table 1). Of these, 3,419 genes are shared between all three strains and 608 genes are shared between the two STEC strains, exclusive of HS. The genome of each strain contains over 800 unique genes.
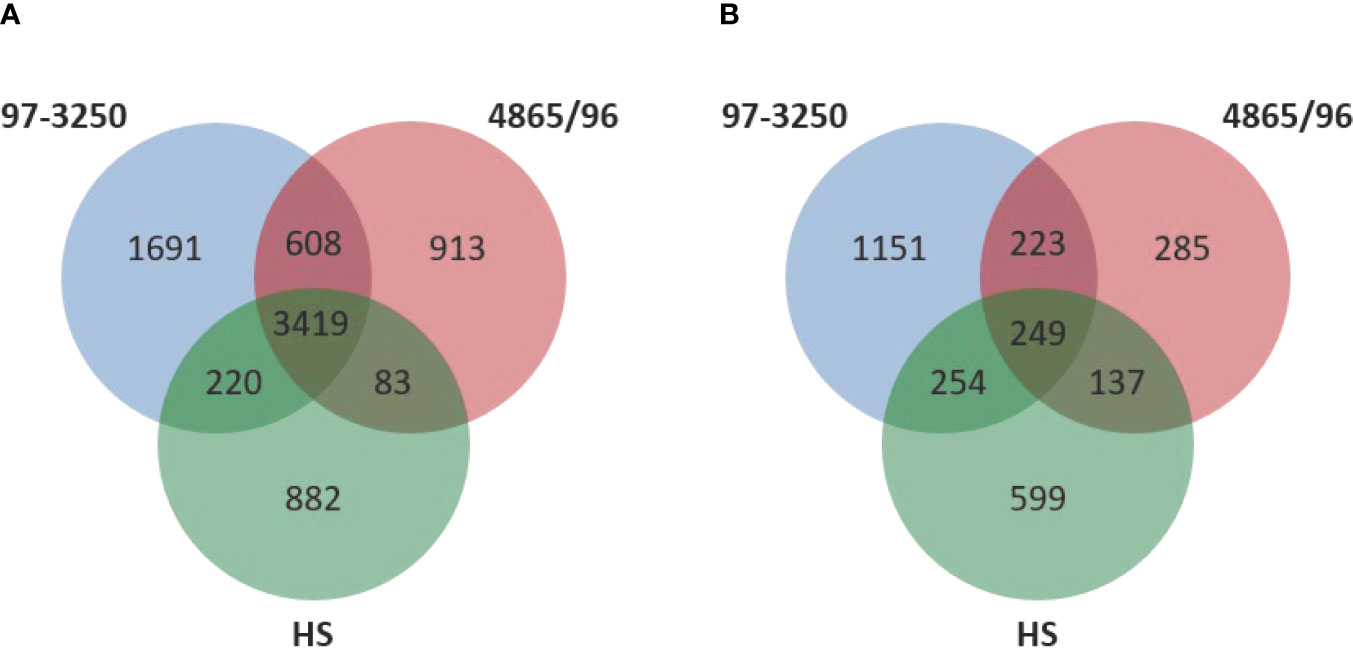
Figure 1 Comparison of genomic content and differentially expressed genes in the STEC strains and commensal E. coli HS. (A) Venn diagram comparing the number of shared and unique genes among the two STEC and HS strains. DNA homology of open reading frames among the strains was used to determine gene homologs. The rRNA genes are not included. (B) Venn diagram comparing the number of shared and unique differentially expressed genes for adhered compared to planktonic conditions among the STEC and HS strains.
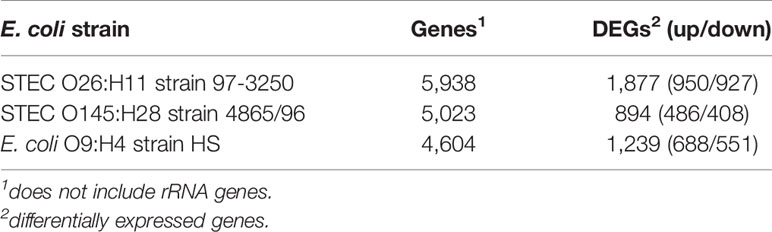
Table 1 Number of genes in the E. coli genomes and genes differentially expressed in adhered compared to planktonic conditions.
Global Transcriptional Responses
The STEC and commensal E. coli strains were used in in vitro infection experiments with polarized T84 colonic IECs. Genes within the LEE PAI are expressed during planktonic growth in virulence-promoting conditions as used in this study and found to be most highly expressed in late exponential growth phase but decreasing after adherence to IECs (Knutton et al., 1997; Hazen et al., 2015; Yang et al., 2015). We reasoned that some other virulence genes are also likely to exhibit greater expression differences between these two conditions. Thus, to maximize the possibility of discovering novel virulence genes in the STEC strains, we utilized RNA-Seq to compare E. coli transcriptomic results between late exponential planktonic growth and 3 h after infection with IECs. The global transcriptomes were determined for each of the three strains for both conditions, and significantly differentially expressed genes (DEGs) in adhered E. coli compared to planktonic E. coli were identified. Gene homolog matches among the three strains were used to determine shared DEGs (Figure 1B). RNA-Seq analysis identified 472 DEGs shared between the STEC strains, and of those, 76 do not have a homolog in HS. The HS genome does carry a homolog for the remaining 396 DEGs, but only 249 of those genes are also differentially expressed in HS. The number of DEGs for each strain were 1,877 (31.6%), 894 (17.8%), and 1,239 (26.9%) for strains 97-3250, 4865/96, and HS, respectively (Table 1). For all three strains, the number of upregulated genes was higher than the downregulated genes, but the difference was small, particularly in 97-3250 where it was only 23 genes. Variability in expression patterns for adhered compared to planktonic E. coli was observed among the strains. Of the 249 DEGs shared among all three strains, not all exhibited a fold change in the same direction and/or with similar magnitude. For some genes, disparity was observed between the transcriptional responses of the two STEC strains, and in others, the two STECs displayed a similar response that was different than that observed for HS. To determine the main biological functions of the DEGs, the genes were mapped to terms in the Gene Ontology (GO) database. For genes that could be classified, the percentage of DEGs was determined for each of the three main categories: biological process (Figure 2A), molecular function (Figure 2B), and cellular component (Figure 2C). In general, the percentages for the classifications in the three strains were similar, with metabolic process, cellular process, and localization having the highest percentages in the biological process category and binding, catalytic activity, and transporter activity in the molecular function category. Interestingly, for two of the subcategories, namely, localization within biological process and transporter activity within molecular function, there was a greater percentage of upregulated compared to downregulated genes for 4865/96, while the reverse was observed for HS. The greatest percentage of DEGs in the cellular component category was classified as membrane and intrinsic component of the membrane, and there was a higher percentage of upregulated compared to downregulated DEGs.
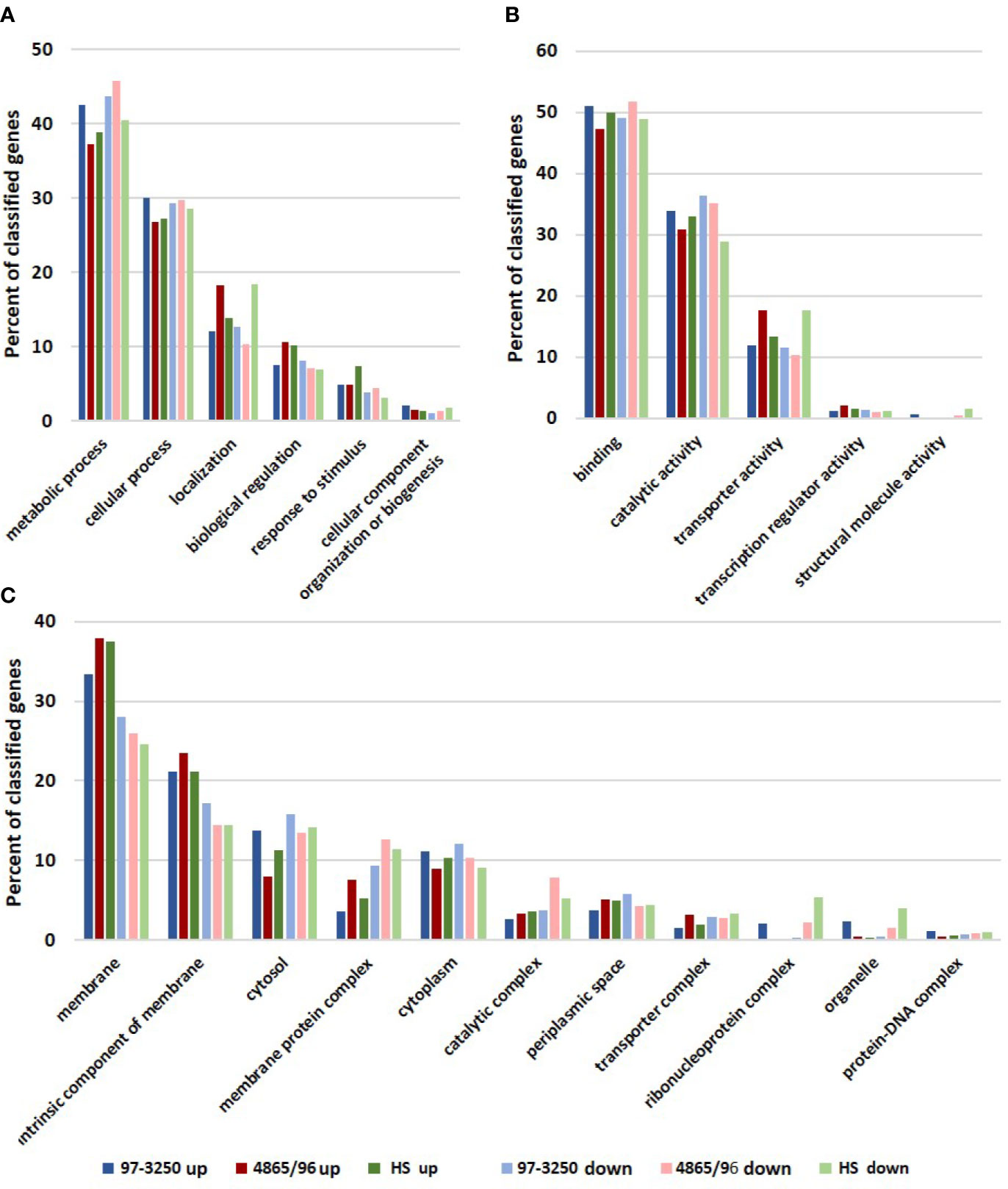
Figure 2 Biological functions of differentially expressed genes. Genes differentially expressed by each of the STEC or commensal E. coli adhered to T84 IECs compared to planktonic culture were classified using the Gene Ontology (GO) database. The number of upregulated and downregulated genes classified in each category were used to determine percentages in subcategories within the three main categories in the GO database: (A) biological process, (B) molecular function, and (C) cellular component.
Selected Biochemical Pathways and Genes Exhibiting Different Expression Patterns Between the STEC Strains and the Commensal E. coli Strain
The RNA-Seq results were examined for genes that were regulated similarly in the STEC strains but either in the opposite direction than observed for HS or to a dissimilar extent. In particular, two biochemical pathways, the arginine biosynthesis/import and tryptophan biosynthesis/import pathways, demonstrated considerable disparity in differential expression patterns between the two STECs and the commensal HS (Figure 3, Table 2). Nearly all of the genes involved in arginine biosynthesis were more highly transcribed in adhered E. coli cells compared to planktonic cultures in the STEC strains compared to HS. In addition, the art genes, involved in importing arginine from the environment, and arcD, the gene encoding the arginine:ornithine antiporter that concurrently exports ornithine while importing arginine were more highly transcribed in the adhered compared to planktonic STEC cells in contrast to HS. However, argR, encoding the arginine repressor, was not differentially expressed. The polyamine putrescine can be synthesized from ornithine or agmatine (Figure 3) as well as imported from the environment. The transcription of genes involved in the conversion of arginine or ornithine to putrescine is unchanged in 4865/96 and HS, and speE, encoding the enzyme required for synthesis of spermidine from putrescine, is downregulated. However, the genes involved in the synthesis of putrescine and spermidine are upregulated in 97-3250. Importantly, although the transcription of genes used for the synthesis of polyamines is unchanged in HS, import of polyamines utilizing both the importers encoded by yeeF and the pot genes is downregulated, while it is upregulated in 97-3250 and either unchanged or downregulated to a lesser extent in 4865/96 than in HS. The tryptophan biosynthesis pathway was dramatically downregulated in adhered HS compared to planktonic culture, while for both STEC strains, transcription of the genes in the pathway was either increased, unchanged, or in the case of trpA, decreased but to a much lesser extent than observed for HS (Table 2). Transcription of mtr, encoding a tryptophan importer, was also significantly decreased in adhered HS, but increased in both STECs. Additionally, transport of other biomolecules was regulated differently in the STECs compared to HS. Dipeptide transport using the product of the dpp genes, as well as expression of ompF, was downregulated to a greater extent in HS in comparison to the STECs. The mgtA gene, encoding a transport protein mediating import of magnesium, was more highly expressed in adhered STEC compared to planktonic culture, while expression was unchanged in HS. Pronounced disparity in transcriptional responses between the STECs and HS were also observed for the pstSCAB-phoU operon and the two-component regulatory system encoded by phoBR that control the phosphate regulon. Aspartate kinase catalyzes the first reaction in the aspartate pathway in which aspartic acid is metabolized to produce the amino acids lysine, threonine, methionine, and isoleucine. The gene encoding aspartate kinase, thrA, was downregulated in the adhered STEC strains, while not differentially expressed in HS.
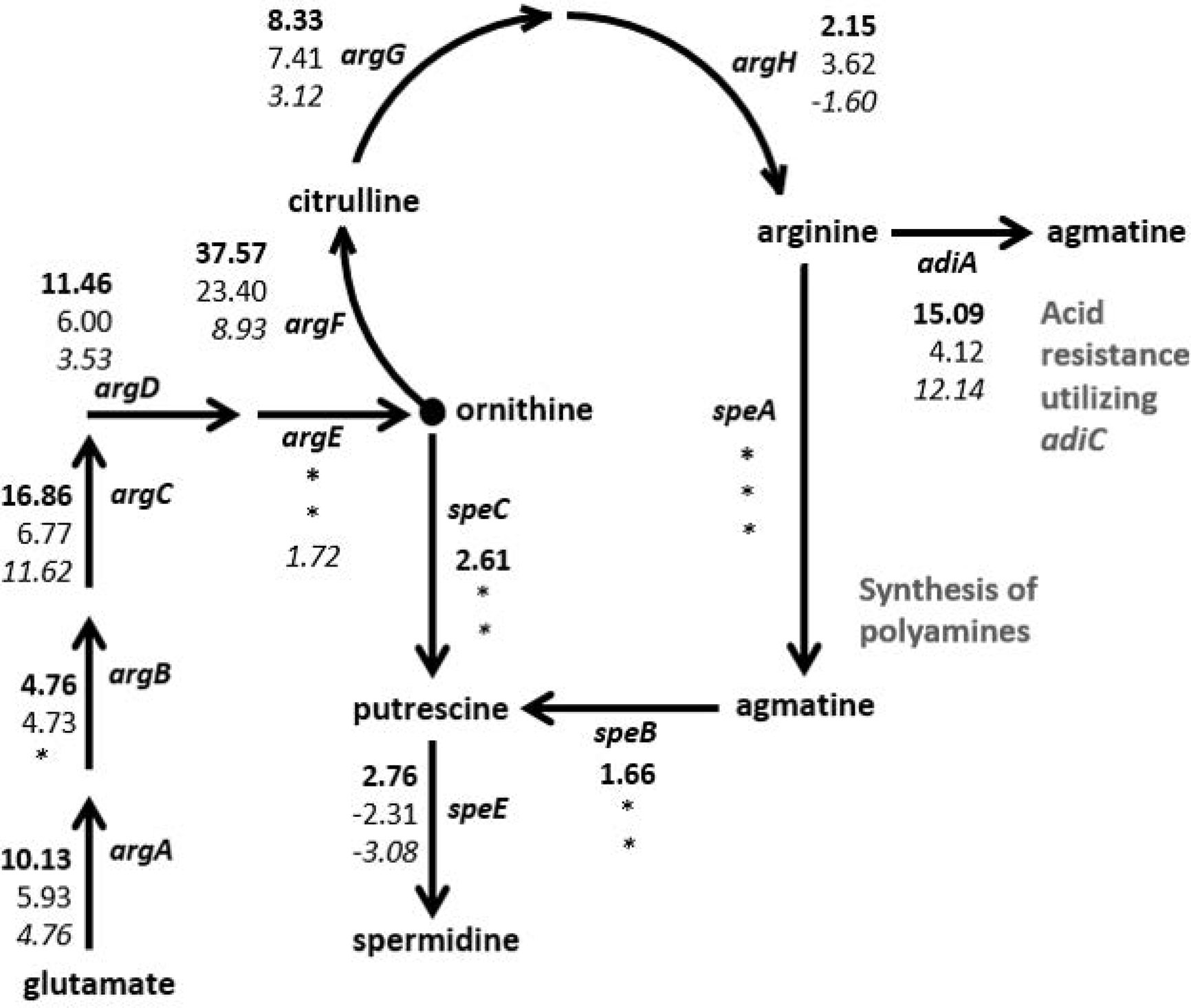
Figure 3 Differential expression of genes in the arginine and polyamine biosynthesis pathway in E. coli. Fold changes between the adhered and planktonic states for genes in the biosynthesis of arginine and polyamines were determined and are indicated next to the respective gene in the diagram. Numerical values for which the FDR adjusted p value < 0.05 are included, otherwise an asterisk is included instead. Fold change values are listed in descending order for strains 97-3250 (bold font), 4865/96 (standard font), and HS (italic font).
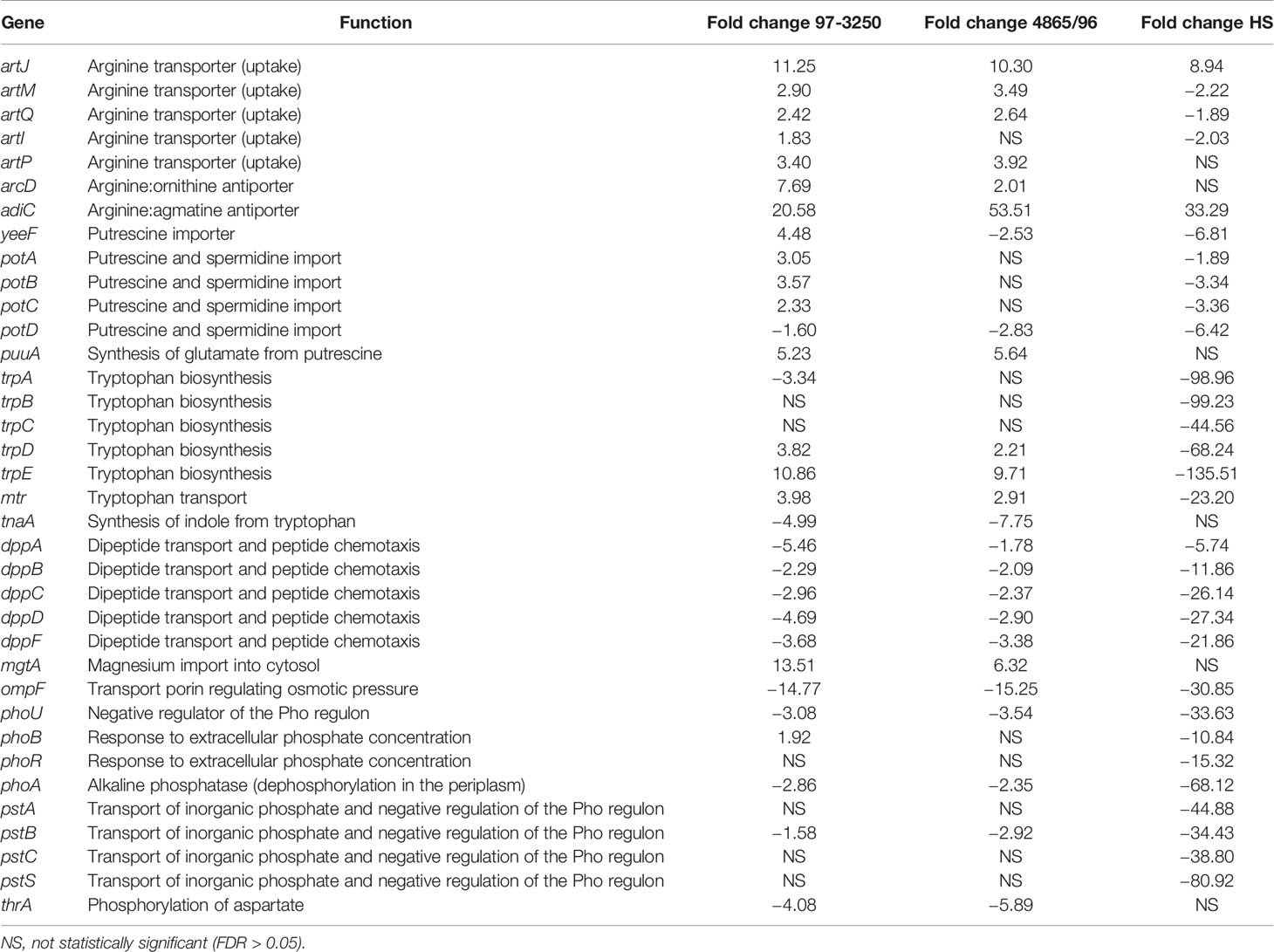
Table 2 Selected biochemical pathways or genes exhibiting similar differential expression patterns for adhered compared to planktonic conditions in the STEC strains and in contrast to the commensal HS strain.
Contrasting Expression Patterns Were Observed for Genes Involved in Respiration
Although all three E. coli strains displayed marked differences in the use of respiratory pathways when converting from a planktonic to an adhered lifestyle, there was variability in gene expression changes associated with aerobic and anerobic respiration among them (Table 3). Contrasting transcriptional responses were observed among the three strains for genes encoding the transcription factors fnr and arcA that facilitate metabolic shifts between aerobic and anaerobic conditions (Lin and Iuchi, 1991; Jiang et al., 2015). While there were genes or sets of genes encoding subunits of larger proteins that were regulated similarly in the STECs, but different than HS, overall there was no distinguishing transcriptional pattern differentiating the STEC strains from HS. For example, all of the nuo genes that encode the NADH ubiquinone oxidoreductase respiratory complex I involved in aerobic respiration were downregulated to a greater extent in HS than observed for the STEC strains. However, transcription of cytochrome oxidase, encoded by cyoABCD, was decreased to a greater extent in 4865/96 and HS than in 97-3250. Genes involved in anaerobic respiration utilizing, for example, glycerol, lactate, and formate are regulated to different magnitudes among the strains under the two conditions tested. At 3 h post infection, expression of the genes encoding formate hydrogenlyase, which is involved in anaerobic oxidation of formate, was considerably higher in 4865/96 and HS, but not differentially expressed in 97-3250. Expression of the glpABC operon, encoding the glycerol 3-phophate dehydrogenase utilized in anaerobic respiration was downregulated in both STECs, while not in HS. In contrast, glpD, involved in aerobic respiration, was upregulated in 97-3250 and HS, but not 4865/96. Additionally, the DEGs, narG and napA, involved in anaerobic nitrate respiration were transcribed with considerable differences between the STECs and HS. In general, the fold change values exhibited by 4865/96 and HS were greater in magnitude, suggesting a greater shift in metabolic pathways used for respiration between planktonic growth and adhered cells than for 97-3250.
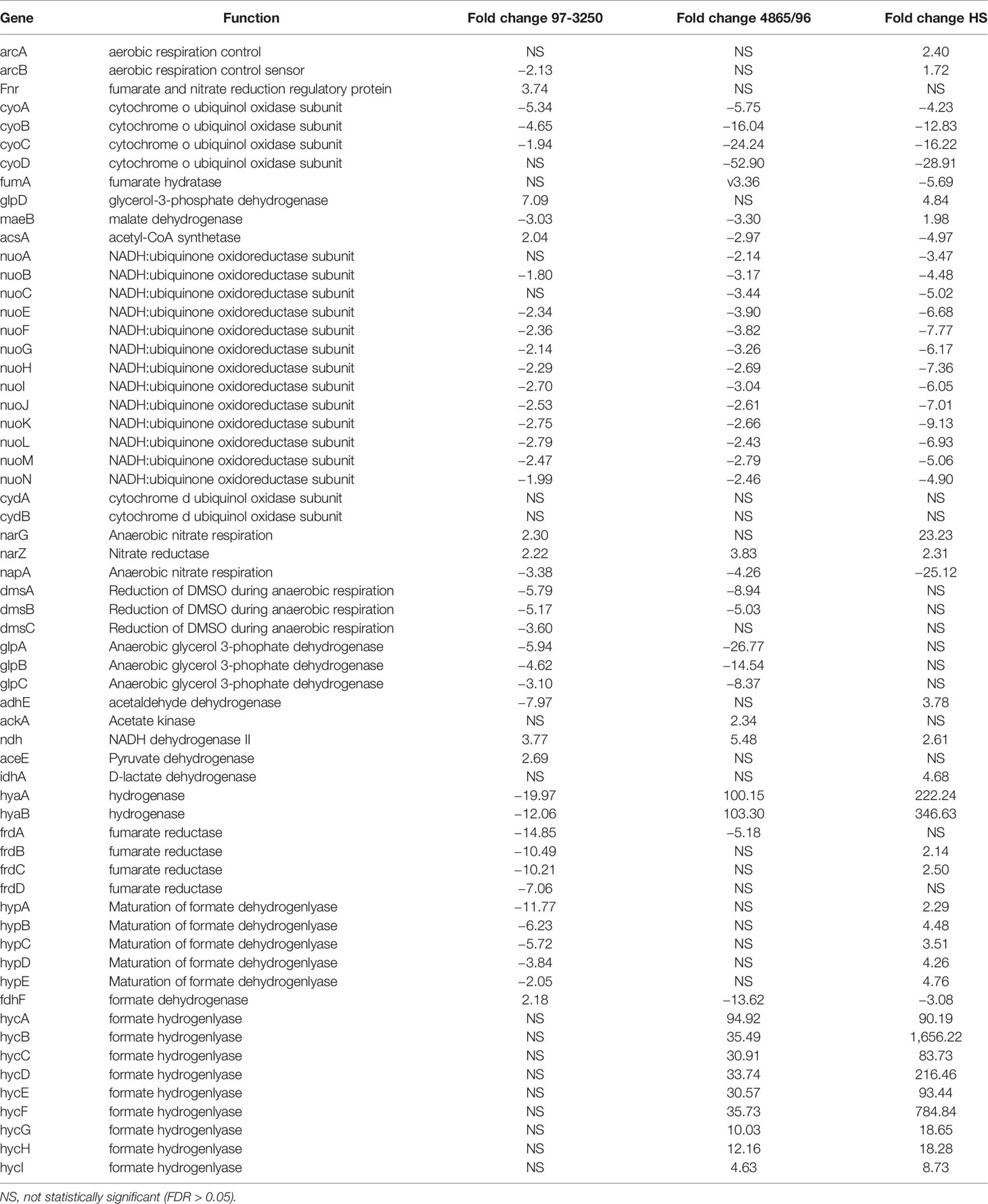
Table 3 Differential expression of E. coli genes involved in respiration for adhered compared to planktonic conditions.
Transcriptional Changes in STEC Virulence Genes Commonly Used for Risk Assessments
The subset of genes shared between the STEC strains, but absent from the HS genome, includes known E. coli virulence factors. We examined selected virulence factors commonly used to assess human pathogenicity for differential expression in STEC upon adherence for 3 h to T84 IECs. STEC strain 97-3250 carries two Stx alleles, stx1a and stx2a, while 4865/96 carries only stx2a. The transcript containing the A subunit of stx1a in 97-3250 was not differentially expressed, while expression of the B subunit was downregulated 2.64-fold. For stx2a, neither subunit was differentially expressed in either of the STECs, which is not unexpected. The LEE PAI contains genes involved in intimate adherence to colonic epithelial cells as well as genes encoding effector proteins. All the genes located within the LEE PAI were downregulated in both STECs, with fold change values ranging from −10.91 (ler) to −75.25 (escI) for 97-3250 and −3.85 (ler) to −48.05 (eae) for 4865/96. In addition to virulence genes carried in the STEC chromosome, ehxA, catalase-peroxidase (katP), and an extracellular serine protease autotransporter (espP) are carried on a large virulence plasmid harbored in many pathogenic STEC strains. The ehxA gene was downregulated in both STEC strains, with fold changes of −5.32 and −2.16 in 97-3250 and 4865/96, respectively. The katP gene on the plasmid harbored in 97-3250 was not differentially expressed and 4865/96 does not carry katP. The espP gene was not differentially expressed in STEC 4865/96, while it was downregulated 3.91-fold in 97-3250.
DEGs With No E. coli HS Homolog Potentially Involved in STEC Pathogenesis
In addition to the genes within the LEE PAI, the STEC strains shared 48 DEGs with no HS homolog. However, for six of the genes, the transcript was regulated in the opposite direction between the STEC strains, leaving 42 DEGs unique to the STEC strains with expression in both strains either increased or decreased upon 3 h adherence. To further corroborate that these 42 DEGs represent genes potentially contributing to pathogenesis in humans, we performed infection experiments utilizing polarized Caco-2 IECs, thereby reducing specific STEC-IEC and STEC-media type interaction effects. As for the transcriptomic experiments utilizing T84 IECs, DEGs were identified from a comparison of 3 h infections utilizing polarized Caco-2 IECs and STEC grown in planktonic culture. The resulting adhered versus planktonic transcriptional responses were compared to the T84 IEC results. Of the 42 DEGs, 20 genes were observed to be differentially expressed in both STECs and in the same direction as was observed in the T84 IEC experiments, with nine upregulated genes and 11 downregulated genes (Table 4). Protein sequences of the translated nucleotide sequences were used to verify or identify the genes utilizing BLASTp searches of the non-redundant protein sequence database at NCBI. Known virulence genes are included in the subset of genes, for example, ehxA and perC. Also included were the tellurite resistance genes terZ, terA, and terB, reported to also be involved in colicin resistance (Whelan et al., 1995) and increased resistance to oxidative stress caused by hydrogen peroxide (Valkova et al., 2007). There are two diacylglycerol kinase genes in both STECs, but only one copy in HS. The dgkA genes in the STEC strains with closest sequence homology to dgkA in HS were not differentially expressed; however, transcription of the second copy is reduced in the STECs (Table 4). Additionally, lpxR, a gene encoding a CAAX protease (Jandu et al., 2009), the P4 integrase gene, necessary for phage excision, and a gene with locus tag DA88_06695 encoding a protein with sequence homology to alpha/beta hydrolases but with unknown function, were expressed to a lesser extent in adhered compared to planktonic STEC cells. In contrast, other genes were upregulated after 3 h of infection. The immunoglobulin-binding regulator encoded by ibrB and a gene encoding colanic acid biosynthesis pyruvyl transferase, wcaK, demonstrated increased expression. A gene encoding a diguanylate cyclase was also upregulated, as well as yhiM, encoding an inner membrane protein with roles in acid resistance and in regulating growth of E. coli in conditions of low osmolarity and high temperature (Anderson et al., 2017). In addition, the gene identified as yahM, encoding an uncharacterized protein, was more highly transcribed. The IncFII plasmid replication initiator gene, repA, was also upregulated after 3 h of infection, however, mobC, involved in plasmid mobilization, was downregulated.
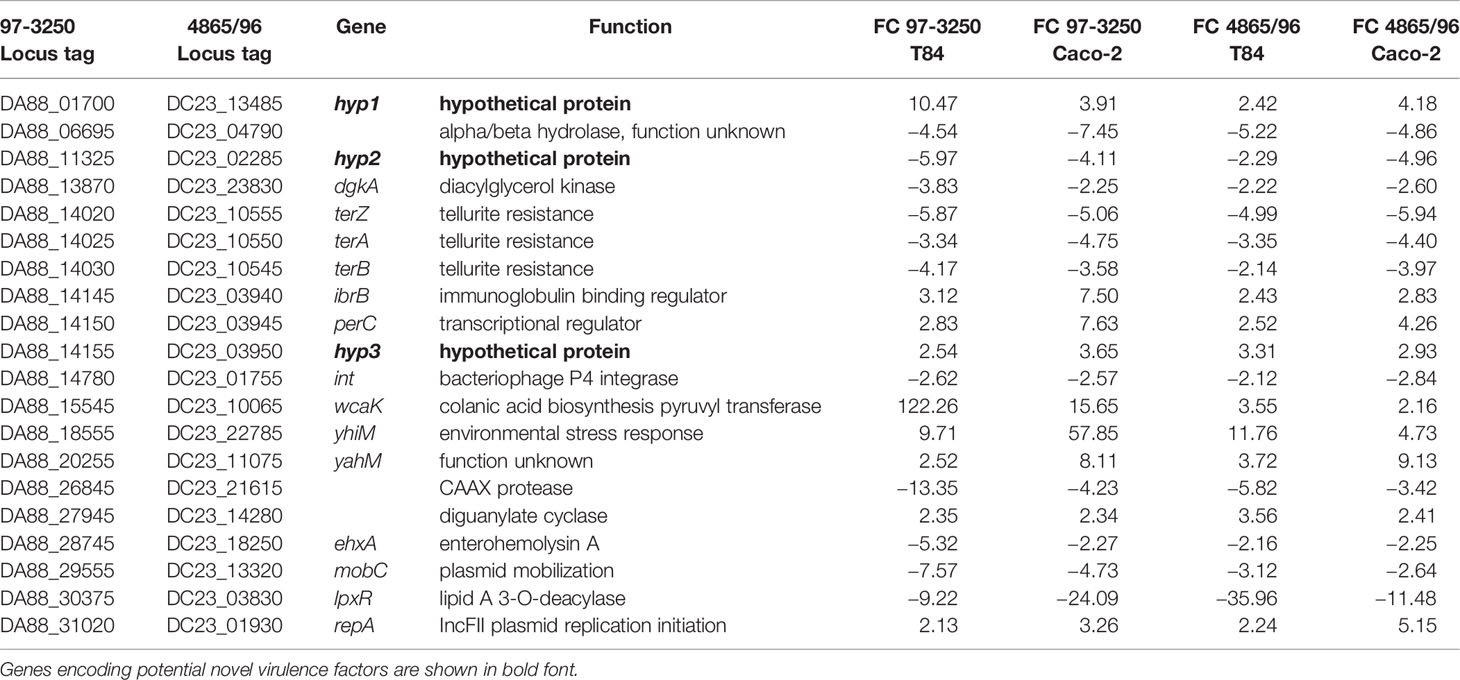
Table 4 Genes with no homolog in E. coli HS exhibiting differential expression in both STEC strains when adhered to both T84 and Caco-2 colonic epithelial cells compared to planktonic culture.
DEGs Representing Novel E. coli Virulence Genes
Along with genes of known or putative function that are differentially expressed in the same direction in both STEC strains upon adherence to both T84 and Caco-2 IECs, three of the DEGs represent hypothetical proteins (Table 4). These genes, referred to as hyp1, hyp2, and hyp3, are potentially novel virulence genes involved in pathogenesis in humans. However, the experiments undertaken in the present study include only two STEC strains and one commensal E. coli strain, thus, to substantiate the possibility that these genes may represent novel virulence genes, a total of 25,527 genomes were analyzed representing all known E. coli phylogroups as well as Escherichia cryptic lineages 1 and 6. To compare the presence of the three hypothetical genes in the different E. coli pathotypes with the presence of known virulence genes, all 20 genes in Table 4 were included in the analysis. Of the 25,527 genomes, hyp1, hyp2, and hyp3 were present in 3714, 3667, and 5475 E. coli genomes, respectively (Figure 4A). For the genomes possessing hyp1 and hyp2, over 88% were classified as AEEC and/or STEC, while less than 1% of the genomes were classified as “other”. Note that a subset of STEC carry the LEE PAI, thus are also classified as AEEC, resulting in an overlap between the pathotypes as defined. The hyp3 gene was present in a greater number of genomes and 58% were classified as STEC, while 14% of the genomes contain no pathotype-specific molecular markers. These results are consistent with those obtained for known virulence genes in the query set, however they do not demonstrate how widespread the three hyp genes are within the STEC pathotype. Thus, to further investigate, the percent of genomes of a given pathotype that are positive for each of the 20 genes was determined (Figure 4B). The results reveal that over 61% of STEC genomes carry at least one of the genes. In addition to STEC and AEEC, hyp2 is carried by 10.6% of ExPEC genomes. The hyp3 gene has a higher occurrence in a wider variety of pathotypes, notably in 82.6% of EAEC genomes. The hyp1 and hyp2 genes are almost exclusive to E. coli genomes that are classified as human pathogens, while hyp3 was discovered in 7.0% of genomes carrying no molecular markers defining the genome as a known E. coli pathotype. The genomic location of the three hypothetical genes was examined using the closed STEC O26:H11 strain 11368 genome (accession #AP010953.1) as a reference strain. Analysis revealed two of the genes to be carried on prophages. The hyp1 gene has homolog ECO26_2650 and is within prophage ECO26_P14 inserted at ryeB, and the hyp2 gene has homolog ECO26_1103 and is within prophage ECO26_P03 inserted at yccA. The other gene, hyp3, with homolog ECO26_1340, is located in the tellurite adherence island (TAI) ECO26_IE02 inserted at serX. The sequences of the contigs containing the three hyp genes in the draft genomes of 97-3250 and 4865/96 are consistent with the hyp genes having locations within these mobile genetic elements.
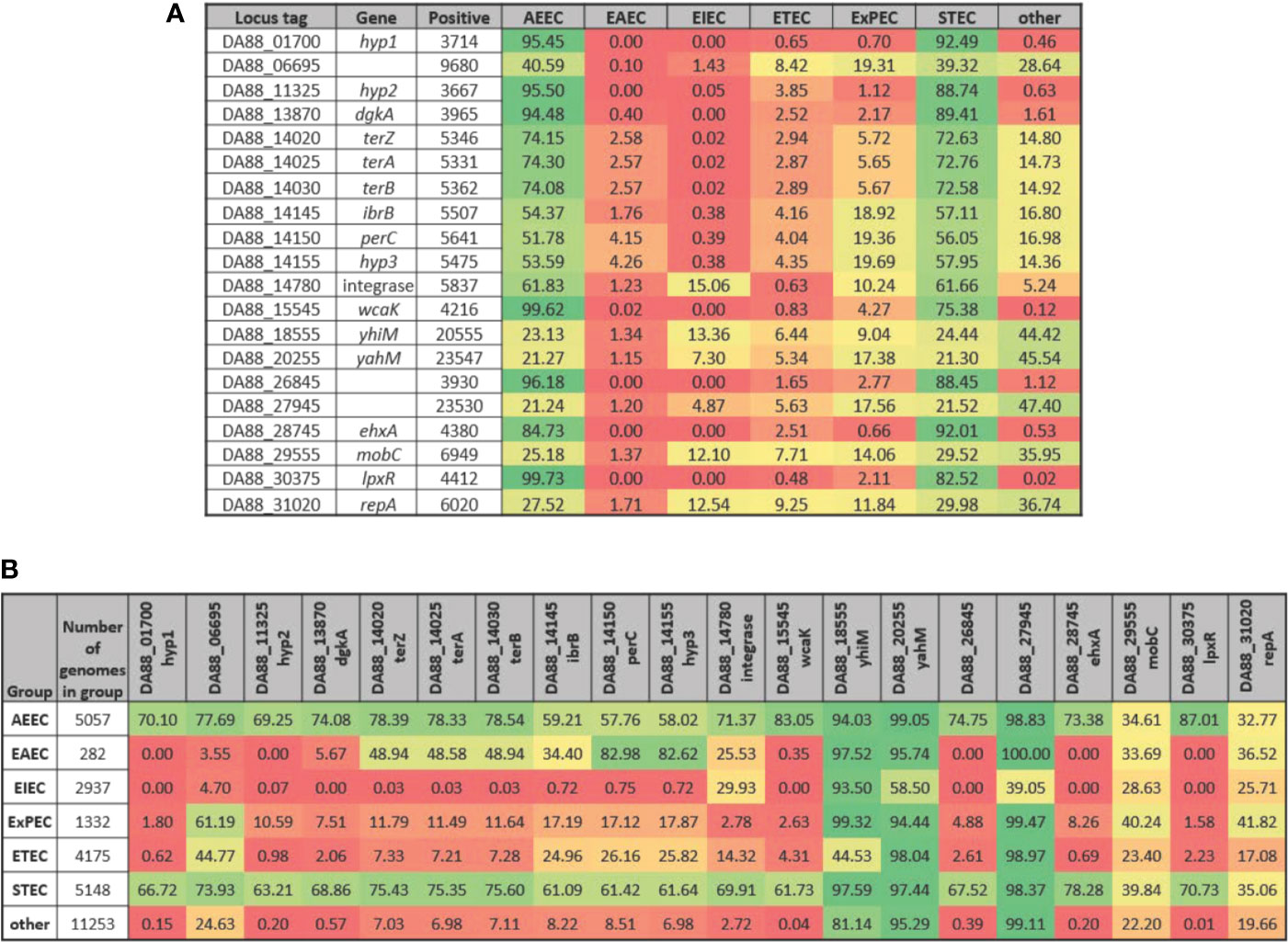
Figure 4 Presence of selected genes in E. coli genomes by pathotype. The STEC strains 97-3250 and 4865/96 were used in T84 and Caco-2 IEC infections and genes differentially expressed in infections for both IEC lines were determined. For each of the genes, the nucleotide sequences from 97-3250 were used in megablast queries performed on a database of 25,527 E. coli genomes. Percentages of (A) E. coli pathotypes comprising the positive matches and (B) genomes within a pathotype in which the gene is present were determined.
Cytokine and Chemokine Expression in Polarized IECs
To determine whether the polarized IEC line T84 is a good model to screen for pathogenicity of individual non-O157 STEC strains, basolateral supernatants from uninfected T84 cells and T84 cells infected with HS, 97-3250, or 4865/96 were analyzed for cytokine biomarker expression using a multiplex assay of 40 cytokines (Table S1). Cytokine data sets having fewer than two data points or where the highest concentrations were less than 10 pg/ml, were excluded from further analyses. The remaining cytokine profiles (24 data sets) were compared for differences between uninfected and infected T84 cells as well as differences between IECs infected with HS and the STEC strains. In almost half of the cytokine profiles analyzed, infection with HS, 97-3250, or 4865/96 resulted in significantly lower levels of several cytokines (CCL27, CCL11, CXCL6, IL-10, IL-16, CCL7, CXCL9, CCL23, CCL17, & CCL19) compared to uninfected cells, and these cytokine levels were no greater than 20 pg/mL in infected cells (data not shown), except for CXCL1 and CXCL12 (Figure 5). For cytokine profiles CCL1, CCL20, and CCL25, the levels from uninfected T84 cells and those infected with 97-3250 were not significantly different (Figure 6). Similarly, the cytokine levels produced by T84 cells infected with HS and 4865/96 did not differ significantly. However, the cytokine levels from both uninfected and 97-3250-infected T84 cells were significantly different from both HS- and 4865/96-infected T84 cells (Figure 6). Only two cytokine profiles, CCL21 and CXCL5, demonstrated a pattern where cytokine levels produced by uninfected T84 cells were significantly higher than those infected with HS or 4865/96, but significantly lower than T84 cells infected with 97-3250 (Figures 5, 6). Conversely, for only one cytokine, MIF, a pattern was observed where cytokine levels in uninfected T84 cells were significantly lower than those infected with HS and 4865/96, but significantly higher than those infected with 97-3250 (Figure 5). In the remaining six cytokine profiles (CX3CL1, IL-1β, CXCL8, CXCL11, CCL15, and CXCL16), the cytokine levels from T84 cells infected with 97-3250 were significantly higher than levels from uninfected T84 cells and those infected with HS or 4865/96 (Figures 5, 6). Furthermore, the cytokine levels from the uninfected T84 cells and T84 cells infected with HS or 4865/96 were not significantly different from each other and were expressed at relatively low levels compared to cells infected with 97-3250 (Figures 5, 6).
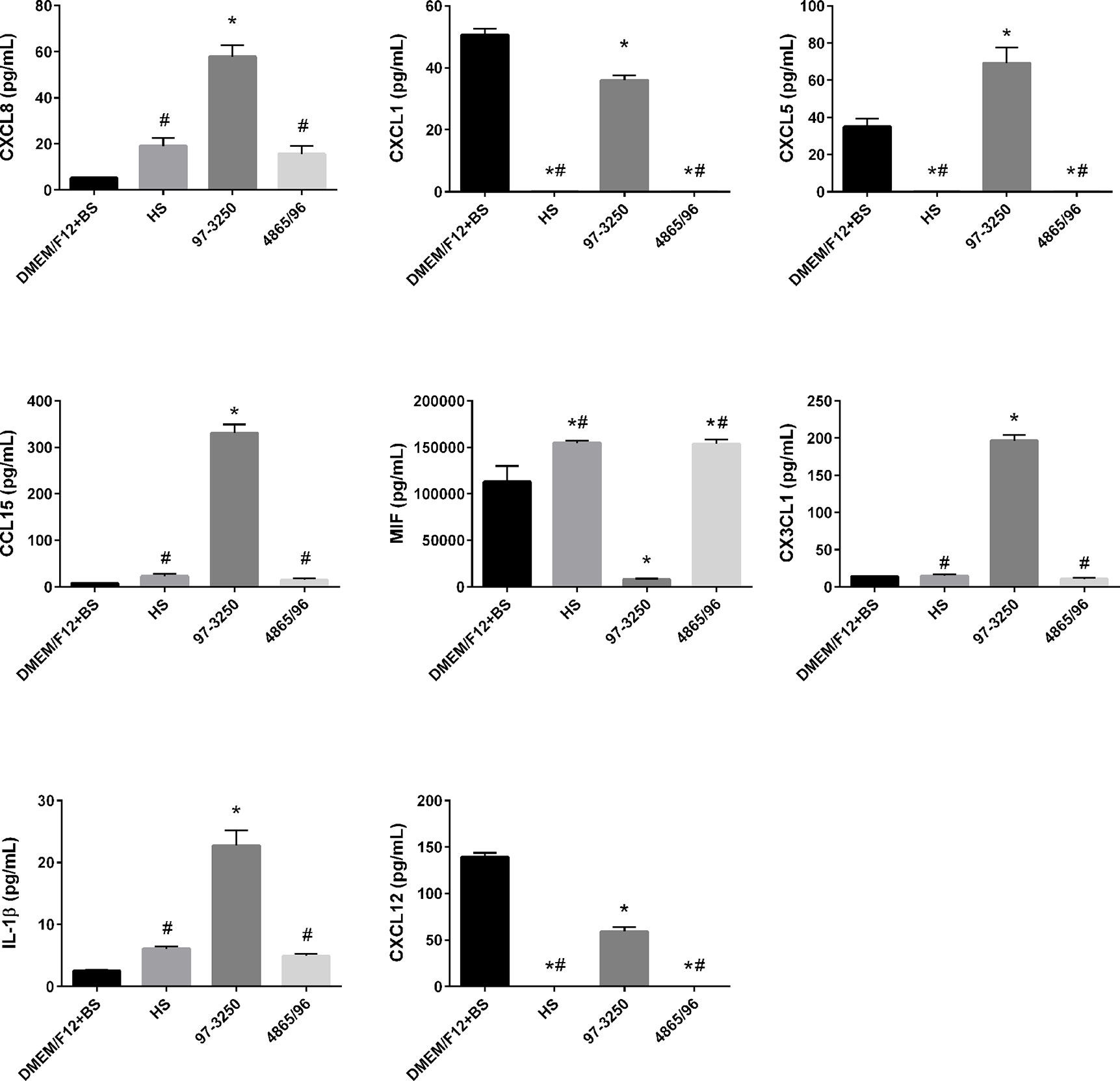
Figure 5 Secretion of PMN- and Monocyte-specific cytokines by polarized T84 cells. Polarized T84 cells were uninfected (DMEM/F12 + BS) or apically infected with commensal E. coli strain HS, or STEC strains 97-3250 or 4865/96 at a MOI of 100. Basolateral supernatants were collected after 3 h and analyzed using a human cytokine multiplex assay. Data are presented as a mean value of each cytokine ± standard error of the mean (n = 3). An asterisk (*) denotes significant differences from uninfected controls (p value < 0.01). A number sign (#) denotes significant differences between 97-3250 and either HS or 4865/96 infections (p value < 0.0001).
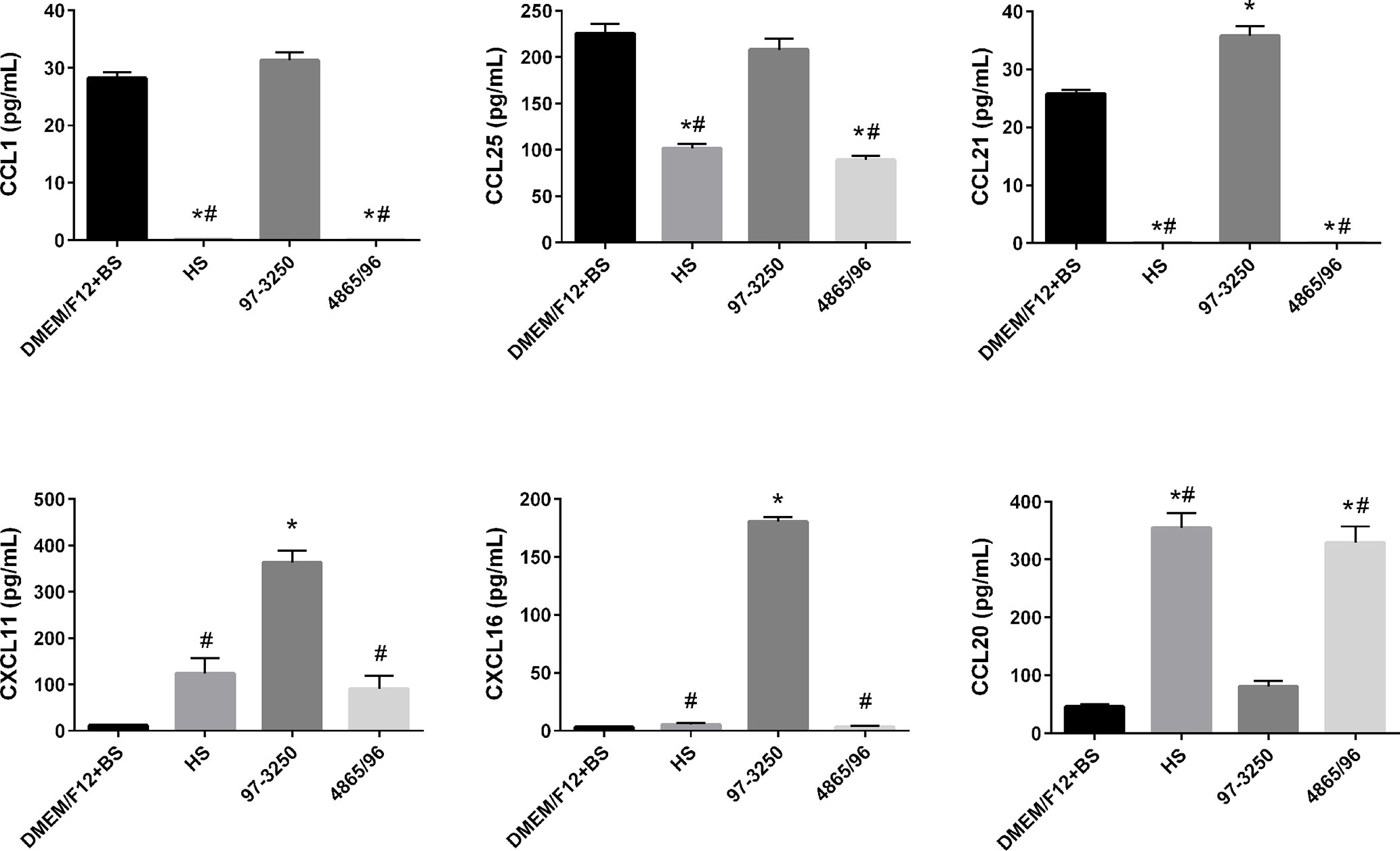
Figure 6 Secretion of T cell- and DC-specific cytokines by polarized T84 cells. Polarized T84 cells were uninfected (DMEM/F12 + BS) or apically infected with commensal E. coli strain HS, or STEC strains 97-3250 or 4865/96 at a MOI of 100. Basolateral supernatants were collected after 3 h and analyzed using a human cytokine multiplex assay. Data are presented as a mean value of each cytokine ± standard error of the mean (n = 3). An asterisk (*) denotes significant differences from uninfected controls (p value < 0.01). A number sign (#) denotes significant differences between 97-3250 and either HS or 4865/96 infections (p value < 0.0001).
Discussion
Expression Comparisons Between the STECs and Commensal E. coli HS
Interestingly, despite a larger gene repertoire, transcriptomic analysis demonstrated that the pathogenic STEC strain 4865/96 had a significantly lower percentage of DEGs than the commensal E. coli HS in transitioning between planktonic and adhered conditions, whereas STEC 97-3250 had a greater percentage (Table 1, Figure 1A). While we observed considerable differences in global transcriptional responses among the three strains as evidenced by the number of genes that were differentially expressed in only one or two strains (Figure 1B), the DEGs were classified similarly overall by GO analysis into categories that would be expected when transitioning from a planktonic to adhered lifestyle (Figures 2A–C). The Pho regulon includes a large network of genes associated with many metabolic processes and adaptive responses. In addition, numerous studies have demonstrated that the Pho regulon is linked to a more complex network of genes influencing bacterial pathogenesis including the LEE genes and other virulence attributes (Cheng et al., 2009; Crepin et al., 2011; Chekabab et al., 2014; Santos-Beneit, 2015). The disparate transcriptional responses of phoBR and pstSCAB between the STECs and HS (Table 2) may indicate that the Pho regulon plays a critical role in STEC survival and colonization in the human gut.
Metabolic pathways are a category where our RNA-Seq results demonstrated significant diversity among the three E. coli strains (Table 3). Metabolic diversity and/or shifts in metabolism during infection and successful colonization among STEC strains and among other E. coli pathotypes have been observed in various in vitro and in vivo studies (Alteri et al., 2009; Kim et al., 2009; Maltby et al., 2013; Pieper et al., 2013; Hazen et al., 2015; Schinner et al., 2015; Chen et al., 2020). Additional studies demonstrate that some Stx2a phages impact carbon source utilization by E. coli (Berger et al., 2019), and that PerC in EPEC also affects transcription of genes involved in metabolism (Mellies et al., 2017). The STEC strains utilized in our study carry multiple phages and perC homologs, cluster with different phylogroups, and have unique gene content, which taken together create the possibility of transcriptional responses of various genes contributing in ways that result in individual carbon usage patterns. Greater E. coli pathogenicity may not be associated with a particular pattern of metabolism, rather, metabolic diversity compared to resident commensal E. coli and other microflora would be advantageous and may allow an STEC strain with a unique carbon and nitrogen utilization pattern to gain a foothold and colonize the human colon. Our results are consistent with the idea that the two STEC strains used in our study employ differing metabolic transcriptional strategies for host infection.
Amino acids can serve as sources of carbon and nitrogen along with their role in protein synthesis. Arginine, in particular, plays additional vital roles in bacterial cell survival, such as its use in imparting acid resistance (Kanjee and Houry, 2013; Charlier and Bervoets, 2019) and as a substrate for the synthesis of the polyamines putrescine and spermidine (Igarashi and Kashiwagi, 2018; Charlier and Bervoets, 2019). With the increased transcription of arginine biosynthesis genes along with uptake of extracellular arginine demonstrated in our RNA-Seq results (Table 2, Figure 3), the STECs possess a greater quantity of arginine in the adhered state versus planktonic conditions than HS. Arginine can be used by E. coli for the synthesis of polyamines, and polyamines have been shown to increase general protein synthesis and in particular to stimulate the translation of a specific set of proteins involved in increasing cell viability and biofilm formation during stationary phase in E. coli (Igarashi and Kashiwagi, 2018). Extracellular arginine can be utilized by IECs to either synthesize polyamines, which are vital for IEC proliferation, or to generate nitric oxide (NO), a host defense against pathogens. While some pathogens modulate the quantity of NO produced by host cells either by expressing an arginase gene encoding an enzyme used in the pathway for polyamine biosynthesis from the substrate arginine, or by upregulating host cell arginase (Das et al., 2010), previous studies have demonstrated that the extracellular intestinal pathogen Giardia decreases NO production by IECs by actively importing arginine, thus reducing the arginine available to the IECs (Eckmann et al., 2000; Stadelmann et al., 2012; Stadelmann et al., 2013). This competition for arginine between the pathogen and IECs also reduces the arginine available for polyamine biosynthesis by the IECs, leading to reduced IEC turnover which may aid in pathogen colonization (Stadelmann et al., 2012). Our results demonstrate that in transitioning from planktonic growth to IEC adherence, the STEC strains scavenge both arginine and polyamines to a greater extent than HS, which downregulated their import (Table 2). Limiting the generation of NO by IECs serves yet another purpose in addition to cell survival for STEC. Inhibition of stx2 gene expression and phage synthesis by NO has been previously demonstrated (Vareille et al., 2007), thus attenuation of NO production by decreasing arginine availability to IECs affects not only the ability of STEC to colonize and survive, but also affects virulence.
Tryptophan is another important amino acid involved in metabolic cross talk between host and pathogen (Ren et al., 2018). Depletion of tryptophan available to IECs has been demonstrated to lessen the host immune response (Kong et al., 2018; Ren et al., 2018). When transitioning from a planktonic to adhered lifestyle, the gene encoding the tryptophan importer Mtr was upregulated in the STEC strains while considerably downregulated in HS (Table 2), suggesting the STECs actively compete with IECs for tryptophan in the adhered state to a much greater extent than HS. Tryptophanase, encoded by tnaA and decreased in transcription in the STECs (Table 2), catabolizes tryptophan to indole, ammonia, and pyruvate which allows E. coli to utilize tryptophan as a source of carbon and nitrogen in addition to using it in protein synthesis (Yanofsky et al., 1991). Indole facilitates inter- and intracellular signaling among microbiota in the human intestine, and indole concentration affects expression of genes associated with colonization and virulence in pathogenic E. coli including the LEE genes and stx2a (Lee and Lee, 2010; Bommarius et al., 2013; Platenkamp and Mellies, 2018; Kumar and Sperandio, 2019). The pronounced disparity between the STECs and HS in the regulation of genes associated with tryptophan biosynthesis, import, and catabolism when transitioning to an adhered lifestyle will result in differing effects on the interplay between the human host and E. coli cells.
The DppBCDF permease, in conjunction with the DppA periplasmic substrate-binding protein, actively transports dipeptides across the inner cell membrane. In vivo and in vitro experiments have demonstrated the importance of dppA in UPEC infection of the bladder and kidneys (Alteri et al., 2009), and our results suggest the dppBCDF-dppA genes are also important in STEC infection of IECs. Interestingly, DppBCDF-DppA has been identified to function as an inner membrane heme transporter in E. coli (Letoffe et al., 2006), and an outer membrane heme receptor, ChuA, was identified in an STEC O157:H7 strain (Torres and Payne, 1997). The chuA gene is found more frequently in STEC isolates of serotypes typically associated with severe disease including O145 strains, and heme uptake is thought to be advantageous for colonization and contribute to pathogenicity in STEC (Bosilevac and Koohmaraie, 2011). Of the three E. coli strains included in this study, 4865/96 is the only strain carrying chuA and transcription is upregulated 10.39-fold after 3 h of infection. Despite lacking chuA, many O26 strains have been observed to utilize exogenous heme, suggesting an alternate gene for transport of heme through the outer membrane (Kresse et al., 2007). The hma gene was identified in UPEC as encoding an alternate heme receptor (Hagan and Mobley, 2009), but none of the E. coli strains utilized in our study carry a hma homolog. However, this does not preclude the presence of an as yet unidentified heme receptor. The dpp genes are downregulated to a lesser extent in the STECs compared to HS after IEC infection (Table 2), and this response may be due to a greater need for dipeptide transport in the STECs upon adherence compared to HS, but heme transport offers an alternate or additional explanation.
Expression of STEC Virulence Genes
The RNA-Seq results in this study are in agreement with many previous studies that demonstrate the importance of the LEE PAI and ehxA to STEC pathogenicity. The presence/absence patterns observed from examining 25,527 E. coli genomes for the three hypothetical DEGs identified as possible novel virulence genes are comparable to those of other known virulence genes, substantiating their identification as factors involved in STEC pathogenicity (Figure 4). Furthermore, their location in mobile genetic elements is consistent with identification as virulence genes. Collectively, our results indicate that these three genes represent novel E. coli virulence factors. Given that hyp2 is downregulated in adhered compared to planktonic STEC cells, the product of the hyp2 gene is likely important for survival and initial adherence in the human gastrointestinal tract, while the products of hyp1 and hyp3 would be predicted to be important for maintaining colonization after initial adherence.
The transcriptomics results in this study demonstrate not only the transcriptional regulation of commonly cited virulence factors but also highlight the fact that STEC utilizes other defense mechanisms to initially establish infection in the human host. The product of the gene encoded in 97-3250 and 4865/96 by locus tags DA88_26845 and DC23_21615, respectively, has been previously identified as a CAAX protease and potential virulence factor involved in subversion of IFNγ-Jak1,2-STAT-1 signaling (Jandu et al., 2009). Our results corroborate the finding that this protease is a virulence factor. The lipid A portion of lipopolysaccharide (LPS) is recognized by the host innate immune system resulting in production of pro-inflammatory cytokines. In an attempt to evade detection by the host, Helicobacter pylori, Yersinia enterocolitica, Vibrio cholerae, Salmonella, and many STEC strains remodel the lipid A structure, thereby attenuating the host inflammatory response (Reynolds et al., 2006; Cullen et al., 2011; Ogawa et al., 2018). Modification of lipid A is accomplished utilizing the 3'-O-deacylase encoded by lpxR. Expression of lpxR is regulated by Ler and Pch and is part of the virulence regulon including the LEE genes (Ogawa et al., 2018), thus the downregulation of both the LEE PAI and lpxR demonstrated in our work is internally consistent. Diacylglycerol kinase, encoded by dgkA, is a small enzyme spanning the inner cell membrane that plays a role in lipid recycling during membrane-derived oligosaccharide biosynthesis and in the recycling of diacylglycerol produced during LPS biosynthesis (Van Horn and Sanders, 2012). The second copy of dgkA, carried by the STECs but having less sequence homology to the dgkA gene found in HS, was downregulated in the STEC strains. An interrogation of 25,527 E. coli genomes demonstrated that this copy of dgkA is exclusive to pathogenic E. coli, predominantly STEC or AEEC (Figure 4B). Whether the second dgkA gene has the same physiological function as the dgkA found in all E. coli genomes is unknown. Finally, both the P4 integrase gene and mobC were expressed more highly in planktonic STEC cells than after adherence to IECs for 3 h, suggesting a higher rate of transfer of some mobile genetic elements prior to establishment of infection.
In contrast to virulence factors more highly expressed before adherence to IECs, transcription of other virulence factors was increased after 3 h of infection (Table 4). These included the transcriptional regulators perC and ibrB. The role of perC in virulence and niche adaptation has been well described (Mellies et al., 2017). In comparison to strains lacking ibrB, under inducing conditions the presence of ibrB in the genome activates enhanced expression of Eib on the bacterial cell surface (Sandt et al., 2002). The gene encoding IbrB is not restricted genomically to STEC but, similar to the terZAB and perC genes, is found in subsets of other E. coli pathotypes as well as approximately 8% of the E. coli genomes in our analysis carrying no pathotype-specific markers (Figure 4B). Transcription of the gene wcaK in the colanic acid biosynthesis pathway was also upregulated in the STECs after adherence to IECs (Table 4). Colanic acid is a secreted exopolysaccharide establishing a capsule around the bacterial cell that is protective from a variety of environmental stresses and plays a role in biofilm formation (Mao et al., 2001; Kocharunchitt et al., 2014; Miajlovic et al., 2014). Although the colanic acid capsule is produced and confers protection in the environment outside the human host, studies have demonstrated that it is protective against the bactericidal effects of serum in extraintestinal E. coli pathogens (Miajlovic et al., 2014; Ma et al., 2018). Interestingly, although even commensal E. coli can produce colanic acid, the sequence of wcaK carried in the STECs used in our work is fairly restricted to STEC and other AEEC genomes (Figure 4B). Whether this conveys any physiological relevance is unknown. Finally, there were genes found in most E. coli irrespective of pathotype, but not carried by HS, that were determined to be important for colonization, namely yhiM, yahM, and a diguanylate cyclase gene (Table 4, Figure 4B). E. coli genomes carry a variety of diguanylate cyclase genes. Diguanylate cyclase is involved in the synthesis of cyclic-di-GMP, a second messenger signaling molecule that regulates various physiological functions involved in the transition from a planktonic to sessile state (Hengge, 2009; Whiteley and Lee, 2015). Consistent with our work, several diguanylate cyclase genes were identified as DEGs in transcriptomic experiments comparing planktonic ETEC and ETEC adhered to Caco-2 IECs (Kansal et al., 2013). Additionally, reduced intracellular levels of cyclic-di-GMP have been demonstrated to reduce adherence of STEC O157:H7 to HT-29 epithelial cells and cattle colon explants (Hu et al., 2013).
Cytokine/Chemokine Responses from IECs
We observed the differential induction of a range of cytokines/chemokines following infection of polarized T84 IECs with pathogenic STEC or commensal E. coli. The PMN chemoattractants CXCL8, CXCL1, CXCL5, and CCL15 were differentially expressed in T84 cells infected with 97-3250 compared to HS or 4865/96 (Figure 5). While the role of epithelial-derived CXCL8/IL-8 in mucosal defense and inflammatory responses related to STECs may not be fully understood, its role in PMN recruitment during acute inflammation is considered an important early component of host defense against bacterial pathogens. Numerous studies have reported the involvement of CXCL8/IL-8 in STEC pathogenesis including intestinal barrier disruption through the induction of PMN transmigration into the intestinal lumen, leading to fecal leukocytes in infected patients, Stx access to the bloodstream, and PMN transport of Stxs to target cells and tissues (Elliott et al., 1994; Slutsker et al., 1997; Hurley et al., 2001; Karve et al., 2017). Several STEC factors such as Stxs, flagellin, LPF, and HCP can induce CXCL8/IL-8, CXCL1/GRO-α, and CXCL5/ENA-78 in a variety of intestinal epithelial cell lines (Yamasaki et al., 1999; Thorpe et al., 2001; Berin et al., 2002; Rogers et al., 2003; Miyamoto et al., 2006; Ledesma et al., 2010; Farfan et al., 2013). Our results demonstrate that 97-3250 may promote greater PMN infiltration during IEC infection than 4865/96 or HS.
In correlation with our PMN chemoattractant results, expression patterns of the monocyte/macrophage cytokines/chemokines macrophage migration inhibitory factor (MIF), CX3CL1/Fractalkine, IL-1β, and CXCL12/SDF-1 differed significantly between T84 cells infected with 97-3250 versus 4865/96 and HS (Figure 5). MIF is constitutively produced by untreated human IECs in vitro and in vivo, biologically active, and inhibits the migration of macrophages when there is no infection (Maaser et al., 2002). In our model, the reduced MIF levels induced by 97-3250 and increased MIF levels induced by 4865/96 and HS suggest that 97-3250 promotes while 4865/96 and HS reduce macrophage recruitment by IECs. IL-1β has been reported to modulate various aspects of immune function including the increased expression of adhesion molecules that can promote monocyte/macrophage and neutrophil infiltration to infection sites as well as the induction of CX3CL1/Fractalkine by IECs (Huang et al., 1996; Muehlhoefer et al., 2000; Dinarello, 2009). Recruited or circulating macrophages that encounter Stxs may produce additional cytokines, including IL-1β, leading to increased inflammation and disease (Ramegowda and Tesh, 1996; Harrison et al., 2004; Lee et al., 2013; Lee et al., 2016). Our IL-1β and CX3CL1/Fractalkine results (Figure 5) demonstrate that 97-3250 may promote a greater inflammatory response than 4865/96 through macrophage recruitment. In the intestine, CXCL12/SDF-1 may play a role in immune surveillance through the promotion of monocyte extravasation (Bleul et al., 1996). Although reduced compared to uninfected T84 cells, CXCL12/SDF-1 is still induced during 97-3250 infection and may allow for normal immune surveillance, while its absence during 4865/96 and HS infections may limit surveillance functions (Figure 5). Collectively, our monocyte/macrophage chemoattractant expression profiles suggest that 97-3250 infections may promote monocyte/macrophage infiltration to the intestine, and thus inflammation, while HS and 4865/96 may downregulate or fail to promote these responses.
The role of T cells and dendritic cells (DCs) in STEC pathogenesis in humans is not well understood or studied. Interestingly, we observed significantly higher levels of the T cell chemoattractants CX3CL1, CCL25, CCL1, CCL21, CXCL11, and CXCL16 but lower levels of the DC chemoattractant CCL20/MIP-3α in T84 cells infected with 97-3250 compared to cells infected with HS or 4865/96 (Figure 6). The role of these T cell chemoattractants in STEC pathogenesis is unknown; however, CCL25 (Wurbel et al., 2011; Trivedi et al., 2016; Hernandez-Ruiz and Zlotnik, 2017), CCL1 (Regan et al., 2013), CCL21 (Luther et al., 2002), CXCL11 (Dwinell et al., 2001), and CXCL16 (Diegelmann et al., 2010) have established roles in the maintenance of normal intestinal functions or during intestinal inflammation. The differential expression of these T cell chemoattractants observed during infection with HS or 4865/96 compared to 97-3250 suggests that 4865/96 may be recognized as a nonpathogenic E. coli strain, possibly as a way to evade detection. CCL20/MIP-3α can be expressed in inflamed epithelial crypts and at varying levels in IECs depending on the STEC serotype and their TTSS and flagellin (Dieu et al., 1998; Gobert et al., 2008). The differing CCL20/MIP-3α levels induced by 97-3250 and 4865/96 in our study are supported by these previous reports and further suggest that these two STEC serotypes are able to elicit opposing inflammatory responses on their path to causing disease.
Summary
In vitro infection models do not fully represent the overall in vivo condition, which includes a complex interplay between host cells, resident microbiota, and the invading STEC. Nevertheless, the cytokine and transcriptomics results presented in this study provide further insight into similarities and differences elicited by pathogenic STEC, a commensal E. coli strain, and IECs at the onset of infection. Although our infection model was not able to provide a single distinct cytokine profile indicative of STEC infection resulting in severe clinical outcome, it is a useful model for understanding individual cellular activities in response to STEC and highlights the variable strain-specific responses to pathogenic STEC that arise irrespective of host differences. Overall, our cytokine profiles suggest that 4865/96 either inhibits the proinflammatory response of polarized T84 IECs to resemble nonpathogenic E. coli commensals like HS, or lacks certain factors, which may be present in 97-3250, that elicit a proinflammatory response. While the transcriptomics results demonstrate biochemical pathways, regulons, and shared virulence genes that are regulated similarly in the STECs, STEC genomes contain a considerable mobilome that is often diverse and this is demonstrated by the number of unique genes found in the STECs used in this study (Figure 1A). Moreover, not all shared genes are transcriptionally regulated in an analogous way. These differences in STEC gene content and/or regulation support both the disparate IEC cytokine responses and DEGs associated with metabolism observed in this study and suggest there are multiple pathways involving different sets of genes that may lead to successful infection. However, through our focus on genes shared between the STECs, we have identified candidate genes likely representing novel STEC virulence factors that, if proven by further phenotypic study, may be added to the repertoire of genes considered for risk assessments.
Data Availability Statement
Sequence reads were deposited in the NCBI Sequence Read Archive (SRA) under the BioProject number PRJNA641161.
Author Contributions
LH and SL conceived of and designed the project and conducted experiments. SL performed the RNA-Seq analysis, LH analyzed the cytokine data, and DL and MM performed other analyses. LH and SL wrote the manuscript. All authors contributed to the article and approved the submitted version.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors would like to acknowledge Dennis W. Gaines for technical assistance during infection experiments as well as helpful discussions with Scott Jackson and Dr. Christopher Elkins in the beginning stages of this project. SL was supported by a fellowship appointment administered by Oak Ridge Institute for Science and Education during part of this work.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcimb.2020.575630/full#supplementary-material
Supplementary Table 1 | Panel of cytokines/chemokines included in the assay. *Data sets with fewer than 2 data points or where the highest concentrations were less than 10 pg/ml were excluded.
References
Allard M. W., Strain E., Melka D., Bunning K., Musser S. M., Brown E. W., et al. (2016). Practical value of food pathogen traceability through building a whole-genome sequencing network and database. J. Clin. Microbiol. 54 (8), 1975–1983. doi: 10.1128/JCM.00081-16
Alteri C. J., Smith S. N., Mobley H. L. (2009). Fitness of Escherichia coli during urinary tract infection requires gluconeogenesis and the TCA cycle. PloS Pathog. 5 (5), e1000448. doi: 10.1371/journal.ppat.1000448
Anderson M. A., Mann M. D., Evans M. A., Sparks-Thissen R. L. (2017). The inner membrane protein YhiM is necessary for Escherichia coli growth at high temperatures and low osmolarity. Arch. Microbiol. 199 (1), 171–175. doi: 10.1007/s00203-016-1288-3
Baba H., Kanamori H., Kudo H., Kuroki Y., Higashi S., Oka K., et al. (2019). Genomic analysis of Shiga toxin-producing Escherichia coli from patients and asymptomatic food handlers in Japan. PloS One 14 (11), e0225340. doi: 10.1371/journal.pone.0225340
Berger P., Kouzel I. U., Berger M., Haarmann N., Dobrindt U., Koudelka G. B., et al. (2019). Carriage of Shiga toxin phage profoundly affects Escherichia coli gene expression and carbon source utilization. BMC Genomics 20 (1), 504. doi: 10.1186/s12864-019-5892-x
Berin M. C., Darfeuille-Michaud A., Egan L. J., Miyamoto Y., Kagnoff M. F. (2002). Role of EHEC O157:H7 virulence factors in the activation of intestinal epithelial cell NF-kappaB and MAP kinase pathways and the upregulated expression of interleukin 8. Cell Microbiol. 4 (10), 635–648. doi: 10.1046/j.1462-5822.2002.00218.x
Bleul C. C., Fuhlbrigge R. C., Casasnovas J. M., Aiuti A., Springer T. A. (1996). A highly efficacious lymphocyte chemoattractant, stromal cell-derived factor 1 (SDF-1). J. Exp. Med. 184 (3), 1101–1109. doi: 10.1084/jem.184.3.1101
Bommarius B., Anyanful A., Izrayelit Y., Bhatt S., Cartwright E., Wang W., et al. (2013). A family of indoles regulate virulence and Shiga toxin production in pathogenic E. coli. PloS One 8 (1), e54456. doi: 10.1371/journal.pone.0054456
Bosilevac J. M., Koohmaraie M. (2011). Prevalence and characterization of non-O157 Shiga toxin-producing Escherichia coli isolates from commercial ground beef in the United States. Appl. Environ. Microbiol. 77 (6), 2103–2112. doi: 10.1128/AEM.02833-10
Buteau C., Proulx F., Chaibou M., Raymond D., Clermont M. J., Mariscalco M. M., et al. (2000). Leukocytosis in children with Escherichia coli O157:H7 enteritis developing the hemolytic-uremic syndrome. Pediatr. Infect. Dis. J. 19 (7), 642–647. doi: 10.1097/00006454-200007000-00012
Centers for Disease Control and Prevention (2020a). Pathogen surveillance. Available at: https://wwwn.cdc.gov/foodnetfast (Accessed April 28 2020).
Centers for Disease Control and Prevention (2020b). Reports of selected E. coli outbreak investigations. Available at: https://www.cdc.gov/ecoli/outbreaks.html (Accessed April 28 2020).
Charlier D., Bervoets I. (2019). Regulation of arginine biosynthesis, catabolism and transport in Escherichia coli. Amino Acids 51 (8), 1103–1127. doi: 10.1007/s00726-019-02757-8
Chekabab S. M., Jubelin G., Dozois C. M., Harel J. (2014). PhoB activates Escherichia coli O157:H7 virulence factors in response to inorganic phosphate limitation. PloS One 9 (4), e94285. doi: 10.1371/journal.pone.0094285
Chen L., Zhao X., Wu J., Liu Q., Pang X., Yang H. (2020). Metabolic characterisation of eight Escherichia coli strains including “Big Six” and acidic responses of selected strains revealed by NMR spectroscopy. Food Microbiol. 88, 103399. doi: 10.1016/j.fm.2019.103399
Cheng C., Tennant S. M., Azzopardi K. I., Bennett-Wood V., Hartland E. L., Robins-Browne R. M., et al. (2009). Contribution of the pst-phoU operon to cell adherence by atypical enteropathogenic Escherichia coli and virulence of Citrobacter rodentium. Infect. Immun. 77 (5), 1936–1944. doi: 10.1128/IAI.01246-08
Cooley M. B., Jay-Russell M., Atwill E. R., Carychao D., Nguyen K., Quinones B., et al. (2013). Development of a robust method for isolation of Shiga toxin-positive Escherichia coli (STEC) from fecal, plant, soil and water samples from a leafy greens production region in California. PloS One 8 (6), e65716. doi: 10.1371/journal.pone.0065716
Crepin S., Chekabab S. M., Le Bihan G., Bertrand N., Dozois C. M., Harel J. (2011). The Pho regulon and the pathogenesis of Escherichia coli. Vet. Microbiol. 153 (1–2), 82–88. doi: 10.1016/j.vetmic.2011.05.043
Cullen T. W., Giles D. K., Wolf L. N., Ecobichon C., Boneca I. G., Trent M. S. (2011). Helicobacter pylori versus the host: Remodeling of the bacterial outer membrane is required for survival in the gastric mucosa. PloS Pathog. 7 (12), e1002454. doi: 10.1371/journal.ppat.1002454
Das P., Lahiri A., Lahiri A., Chakravortty D. (2010). Modulation of the arginase pathway in the context of microbial pathogenesis: a metabolic enzyme moonlighting as an immune modulator. PloS Pathog. 6 (6), e1000899. doi: 10.1371/journal.ppat.1000899
Diegelmann J., Seiderer J., Niess J. H., Haller D., Goke B., Reinecker H. C., et al. (2010). Expression and regulation of the chemokine CXCL16 in Crohn’s disease and models of intestinal inflammation. Inflammation Bowel Dis. 16 (11), 1871–1881. doi: 10.1002/ibd.21306
Dieu M. C., Vanbervliet B., Vicari A., Bridon J. M., Oldham E., Ait-Yahia S., et al. (1998). Selective recruitment of immature and mature dendritic cells by distinct chemokines expressed in different anatomic sites. J. Exp. Med. 188 (2), 373–386. doi: 10.1084/jem.188.2.373
Dinarello C. A. (2009). Immunological and inflammatory functions of the interleukin-1 family. Annu. Rev. Immunol. 27, 519–550. doi: 10.1146/annurev.immunol.021908.132612
Dwinell M. B., Lugering N., Eckmann L., Kagnoff M. F. (2001). Regulated production of interferon-inducible T-cell chemoattractants by human intestinal epithelial cells. Gastroenterology 120 (1), 49–59. doi: 10.1053/gast.2001.20914
Eckmann L., Laurent F., Langford T. D., Hetsko M. L., Smith J. R., Kagnoff M. F., et al. (2000). Nitric oxide production by human intestinal epithelial cells and competition for arginine as potential determinants of host defense against the lumen-dwelling pathogen Giardia lamblia. J. Immunol. 164 (3), 1478–1487. doi: 10.4049/jimmunol.164.3.1478
Elliott E., Li Z., Bell C., Stiel D., Buret A., Wallace J., et al. (1994). Modulation of host response to Escherichia coli O157:H7 infection by anti-CD18 antibody in rabbits. Gastroenterology 106 (6), 1554–1561. doi: 10.1016/0016-5085(94)90410-3
Espinosa L., Gray A., Duffy G., Fanning S., McMahon B. J. (2018). A scoping review on the prevalence of Shiga-toxigenic Escherichia coli in wild animal species. Zoonoses Public Health 65 (8), 911–920. doi: 10.1111/zph.12508
FAO/WHO STEC Expert Group (2019). Hazard identification and characterization: Criteria for categorizing Shiga toxin-producing Escherichia coli on a risk basis†. J. Food Prot. 82 (1), 7–21. doi: 10.4315/0362-028X.JFP-18-291
Farfan M. J., Cantero L., Vergara A., Vidal R., Torres A. G. (2013). The long polar fimbriae of STEC O157:H7 induce expression of pro-inflammatory markers by intestinal epithelial cells. Vet. Immunol. Immunopathol. 152 (1–2), 126–131. doi: 10.1016/j.vetimm.2012.09.017
Garcia A., Fox J. G., Besser T. E. (2010). Zoonotic enterohemorrhagic Escherichia coli: A One Health perspective. ILAR J. 51 (3), 221–232. doi: 10.1093/ilar.51.3.221
Gobert A. P., Coste A., Guzman C. A., Vareille M., Hindre T., de Sablet T., et al. (2008). Modulation of chemokine gene expression by Shiga-toxin producing Escherichia coli belonging to various origins and serotypes. Microbes Infect. 10 (2), 159–165. doi: 10.1016/j.micinf.2007.10.018
Gonzalez-Escalona N., Kase J. A. (2019). Virulence gene profiles and phylogeny of Shiga toxin-positive Escherichia coli strains isolated from FDA regulated foods during 2010-2017. PloS One 14 (4), e0214620. doi: 10.1371/journal.pone.0214620
Hagan E. C., Mobley H. L. (2009). Haem acquisition is facilitated by a novel receptor Hma and required by uropathogenic Escherichia coli for kidney infection. Mol. Microbiol. 71 (1), 79–91. doi: 10.1111/j.1365-2958.2008.06509.x
Harrison L. M., van Haaften W. C., Tesh V. L. (2004). Regulation of proinflammatory cytokine expression by Shiga toxin 1 and/or lipopolysaccharides in the human monocytic cell line THP-1. Infect. Immun. 72 (5), 2618–2627. doi: 10.1128/iai.72.5.2618-2627.2004
Haugum K., Johansen J., Gabrielsen C., Brandal L. T., Bergh K., Ussery D. W., et al. (2014). Comparative genomics to delineate pathogenic potential in non-O157 Shiga toxin-producing Escherichia coli (STEC) from patients with and without haemolytic uremic syndrome (HUS) in Norway. PloS One 9 (10), e111788. doi: 10.1371/journal.pone.0111788
Hazen T. H., Daugherty S. C., Shetty A., Mahurkar A. A., White O., Kaper J. B., et al. (2015). RNA-Seq analysis of isolate- and growth phase-specific differences in the global transcriptomes of enteropathogenic Escherichia coli prototype isolates. Front. Microbiol. 6, 569. doi: 10.3389/fmicb.2015.00569
Hazen T. H., Daugherty S. C., Shetty A. C., Nataro J. P., Rasko D. A. (2017). Transcriptional variation of diverse enteropathogenic Escherichia coli isolates under virulence-inducing conditions. mSystems 2 (4), 1–19. doi: 10.1128/mSystems.00024-17
Hengge R. (2009). Principles of c-di-GMP signalling in bacteria. Nat. Rev. Microbiol. 7 (4), 263–273. doi: 10.1038/nrmicro2109
Hernandez-Ruiz M., Zlotnik A. (2017). Mucosal chemokines. J. Interferon. Cytokine Res. 37 (2), 62–70. doi: 10.1089/jir.2016.0076
Hu J., Wang B., Fang X., Means W. J., McCormick R. J., Gomelsky M., et al. (2013). c-di-GMP signaling regulates E. coli O157:H7 adhesion to colonic epithelium. Vet. Microbiol. 164 (3-4), 344–351. doi: 10.1016/j.vetmic.2013.02.023
Huang G. T., Eckmann L., Savidge T. C., Kagnoff M. F. (1996). Infection of human intestinal epithelial cells with invasive bacteria upregulates apical intercellular adhesion molecule-1 (ICAM)-1) expression and neutrophil adhesion. J. Clin. Invest. 98 (2), 572–583. doi: 10.1172/JCI118825
Hurley B. P., Thorpe C. M., Acheson D. W. (2001). Shiga toxin translocation across intestinal epithelial cells is enhanced by neutrophil transmigration. Infect. Immun. 69 (10), 6148–6155. doi: 10.1128/IAI.69.10.6148-6155.2001
Igarashi K., Kashiwagi K. (2018). Effects of polyamines on protein synthesis and growth of Escherichia coli. J. Biol. Chem. 293 (48), 18702–18709. doi: 10.1074/jbc.TM118.003465
Jandu N., Ho N. K., Donato K. A., Karmali M. A., Mascarenhas M., Duffy S. P., et al. (2009). Enterohemorrhagic Escherichia coli O157:H7 gene expression profiling in response to growth in the presence of host epithelia. PloS One 4 (3), e4889. doi: 10.1371/journal.pone.0004889
Jiang F., An C., Bao Y., Zhao X., Jernigan R. L., Lithio A., et al. (2015). ArcA controls metabolism, chemotaxis, and motility contributing to the pathogenicity of avian pathogenic Escherichia coli. Infect. Immun. 83 (9), 3545–3554. doi: 10.1128/IAI.00312-15
Kanjee U., Houry W. A. (2013). Mechanisms of acid resistance in Escherichia coli. Annu. Rev. Microbiol. 67, 65–81. doi: 10.1146/annurev-micro-092412-155708
Kansal R., Rasko D. A., Sahl J. W., Munson G. P., Roy K., Luo Q., et al. (2013). Transcriptional modulation of enterotoxigenic Escherichia coli virulence genes in response to epithelial cell interactions. Infect. Immun. 81 (1), 259–270. doi: 10.1128/IAI.00919-12
Kaper J. B., Nataro J. P., Mobley H. L. (2004). Pathogenic Escherichia coli. Nat. Rev. Microbiol. 2 (2), 123–140. doi: 10.1038/nrmicro818
Karve S. S., Pradhan S., Ward D. V., Weiss A. A. (2017). Intestinal organoids model human responses to infection by commensal and Shiga toxin producing Escherichia coli. PloS One 12 (6), e0178966. doi: 10.1371/journal.pone.0178966
Khan M. A., Ma C., Knodler L. A., Valdez Y., Rosenberger C. M., Deng W., et al. (2006). Toll-like receptor 4 contributes to colitis development but not to host defense during Citrobacter rodentium infection in mice. Infect. Immun. 74 (5), 2522–2536. doi: 10.1128/IAI.74.5.2522-2536.2006
Kim Y., Oh S., Park S., Kim S. H. (2009). Interactive transcriptome analysis of enterohemorrhagic Escherichia coli (EHEC) O157:H7 and intestinal epithelial HT-29 cells after bacterial attachment. Int. J. Food Microbiol. 131 (2–3), 224–232. doi: 10.1016/j.ijfoodmicro.2009.03.002
Knutton S., Adu-Bobie J., Bain C., Phillips A. D., Dougan G., Frankel G. (1997). Down regulation of intimin expression during attaching and effacing enteropathogenic Escherichia coli adhesion. Infect. Immun. 65 (5), 1644–1652. doi: 10.1128/IAI.65.5.1644-1652.1997
Kocharunchitt C., King T., Gobius K., Bowman J. P., Ross T. (2014). Global genome response of Escherichia coli O157:H7 Sakai during dynamic changes in growth kinetics induced by an abrupt downshift in water activity. PloS One 9 (3), e90422. doi: 10.1371/journal.pone.0090422
Kong S., Zhang Y. H., Zhang W. (2018). Regulation of intestinal epithelial cells properties and functions by amino acids. BioMed. Res. Int. 2018, 2819154. doi: 10.1155/2018/2819154
Kresse A. U., Rienacker I., Valle A. M., Steinruck H., Claus H., Payne S. M., et al. (2007). Enterohaemorrhagic Escherichia coli O157 and non-O157 serovars differ in their mechanisms for iron supply. Int. J. Med. Microbiol. 297 (1), 9–15. doi: 10.1016/j.ijmm.2006.11.002
Kumar A., Sperandio V. (2019). Indole signaling at the host-microbiota-pathogen interface. mBio 10 (3), 1–15. doi: 10.1128/mBio.01031-19
La Ragione R. M., Best A., Woodward M. J., Wales A. D. (2009). Escherichia coli O157:H7 colonization in small domestic ruminants. FEMS Microbiol. Rev. 33 (2), 394–410. doi: 10.1111/j.1574-6976.2008.00138.x
Ledesma M. A., Ochoa S. A., Cruz A., Rocha-Ramirez L. M., Mas-Oliva J., Eslava C. A., et al. (2010). The hemorrhagic coli pilus (HCP) of Escherichia coli O157:H7 is an inducer of proinflammatory cytokine secretion in intestinal epithelial cells. PloS One 5 (8), e12127. doi: 10.1371/journal.pone.0012127
Lee J. H., Lee J. (2010). Indole as an intercellular signal in microbial communities. FEMS Microbiol. Rev. 34 (4), 426–444. doi: 10.1111/j.1574-6976.2009.00204.x
Lee M. S., Kim M. H., Tesh V. L. (2013). Shiga toxins expressed by human pathogenic bacteria induce immune responses in host cells. J. Microbiol. 51 (6), 724–730. doi: 10.1007/s12275-013-3429-6
Lee M. S., Kwon H., Lee E. Y., Kim D. J., Park J. H., Tesh V. L., et al. (2016). Shiga toxins activate the NLRP3 inflammasome pathway to promote both production of the proinflammatory cytokine interleukin-1beta and apoptotic cell death. Infect. Immun. 84 (1), 172–186. doi: 10.1128/IAI.01095-15
Leonard S. R., Mammel M. K., Rasko D. A., Lacher D. W. (2016). Hybrid Shiga toxin-producing and enterotoxigenic Escherichia sp. cryptic lineage 1 strain 7v harbors a hybrid plasmid. Appl. Environ. Microbiol. 82 (14), 4309–4319. doi: 10.1128/AEM.01129-16
Letoffe S., Delepelaire P., Wandersman C. (2006). The housekeeping dipeptide permease is the Escherichia coli heme transporter and functions with two optional peptide binding proteins. Proc. Natl. Acad. Sci. U.S.A. 103 (34), 12891–12896. doi: 10.1073/pnas.0605440103
Lin E. C., Iuchi S. (1991). Regulation of gene expression in fermentative and respiratory systems in Escherichia coli and related bacteria. Annu. Rev. Genet. 25, 361–387. doi: 10.1146/annurev.ge.25.120191.002045
Luther S. A., Bidgol A., Hargreaves D. C., Schmidt A., Xu Y., Paniyadi J., et al. (2002). Differing activities of homeostatic chemokines CCL19, CCL21, and CXCL12 in lymphocyte and dendritic cell recruitment and lymphoid neogenesis. J. Immunol. 169 (1), 424–433. doi: 10.4049/jimmunol.169.1.424
Ma J., An C., Jiang F., Yao H., Logue C., Nolan L. K., et al. (2018). Extraintestinal pathogenic Escherichia coli increase extracytoplasmic polysaccharide biosynthesis for serum resistance in response to bloodstream signals. Mol. Microbiol. 110 (5), 689–706. doi: 10.1111/mmi.13987
Maaser C., Eckmann L., Paesold G., Kim H. S., Kagnoff M. F. (2002). Ubiquitous production of macrophage migration inhibitory factor by human gastric and intestinal epithelium. Gastroenterology 122 (3), 667–680. doi: 10.1053/gast.2002.31891
Majowicz S. E., Scallan E., Jones-Bitton A., Sargeant J. M., Stapleton J., Angulo F. J., et al. (2014). Global incidence of human Shiga toxin-producing Escherichia coli infections and deaths: a systematic review and knowledge synthesis. Foodborne Pathog. Dis. 11 (6), 447–455. doi: 10.1089/fpd.2013.1704
Maltby R., Leatham-Jensen M. P., Gibson T., Cohen P. S., Conway T. (2013). Nutritional basis for colonization resistance by human commensal Escherichia coli strains HS and Nissle 1917 against E. coli O157:H7 in the mouse intestine. PloS One 8 (1), e53957. doi: 10.1371/journal.pone.0053957
Mao Y., Doyle M. P., Chen J. (2001). Insertion mutagenesis of wca reduces acid and heat tolerance of enterohemorrhagic Escherichia coli O157:H7. J. Bacteriol. 183 (12), 3811–3815. doi: 10.1128/JB.183.12.3811-3815.2001
Mellies J. L., Platenkamp A., Osborn J., Ben-Avi L. (2017). PerC manipulates metabolism and surface antigens in enteropathogenic Escherichia coli. Front. Cell Infect. Microbiol. 7, 32. doi: 10.3389/fcimb.2017.00032
Miajlovic H., Cooke N. M., Moran G. P., Rogers T. R., Smith S. G. (2014). Response of extraintestinal pathogenic Escherichia coli to human serum reveals a protective role for Rcs-regulated exopolysaccharide colanic acid. Infect. Immun. 82 (1), 298–305. doi: 10.1128/IAI.00800-13
Miyamoto Y., Iimura M., Kaper J. B., Torres A. G., Kagnoff M. F. (2006). Role of Shiga toxin versus H7 flagellin in enterohaemorrhagic Escherichia coli signalling of human colon epithelium in vivo. Cell Microbiol. 8 (5), 869–879. doi: 10.1111/j.1462-5822.2005.00673.x
Muehlhoefer A., Saubermann L. J., Gu X., Luedtke-Heckenkamp K., Xavier R., Blumberg R. S., et al. (2000). Fractalkine is an epithelial and endothelial cell-derived chemoattractant for intraepithelial lymphocytes in the small intestinal mucosa. J. Immunol. 164 (6), 3368–3376. doi: 10.4049/jimmunol.164.6.3368
National Advisory Committee On Microbiological Criteria For Foods (2019). Response to questions posed by the Food and Drug Administration regarding virulence factors and attributes that define foodborne Shiga toxin-producing Escherichia coli (STEC) as severe human pathogens†. J. Food Prot. 82 (5), 724–767. doi: 10.4315/0362-028X.JFP-18-479
Newell D. G., La Ragione R. M. (2018). Enterohaemorrhagic and other Shiga toxin-producing Escherichia coli (STEC): Where are we now regarding diagnostics and control strategies? Transbound Emerg. Dis. 65 Suppl 1, 49–71. doi: 10.1111/tbed.12789
Ogawa R., Yen H., Kawasaki K., Tobe T. (2018). Activation of lpxR gene through enterohaemorrhagic Escherichia coli virulence regulators mediates lipid A modification to attenuate innate immune response. Cell Microbiol. 20 (1), 1–13. doi: 10.1111/cmi.12806
Pieper R., Zhang Q., Clark D. J., Parmar P. P., Alami H., Suh M. J., et al. (2013). Proteomic view of interactions of Shiga toxin-producing Escherichia coli with the intestinal environment in gnotobiotic piglets. PloS One 8 (6), e66462. doi: 10.1371/journal.pone.0066462
Platenkamp A., Mellies J. L. (2018). Environment controls LEE regulation in enteropathogenic Escherichia coli. Front. Microbiol. 9, 1694. doi: 10.3389/fmicb.2018.01694
Ramegowda B., Tesh V. L. (1996). Differentiation-associated toxin receptor modulation, cytokine production, and sensitivity to Shiga-like toxins in human monocytes and monocytic cell lines. Infect. Immun. 64 (4), 1173–1180. doi: 10.1128/IAI.64.4.1173-1180.1996
Rasko D. A., Webster D. R., Sahl J. W., Bashir A., Boisen N., Scheutz F., et al. (2011). Origins of the E. coli strain causing an outbreak of hemolytic-uremic syndrome in Germany. N. Engl. J. Med. 365 (8), 709–717. doi: 10.1056/NEJMoa1106920
Raymond B., Crepin V. F., Collins J. W., Frankel G. (2011). The WxxxE effector EspT triggers expression of immune mediators in an Erk/JNK and NF-kappaB-dependent manner. Cell Microbiol. 13 (12), 1881–1893. doi: 10.1111/j.1462-5822.2011.01666.x
Regan T., Nally K., Carmody R., Houston A., Shanahan F., Macsharry J., et al. (2013). Identification of TLR10 as a key mediator of the inflammatory response to Listeria monocytogenes in intestinal epithelial cells and macrophages. J. Immunol. 191 (12), 6084–6092. doi: 10.4049/jimmunol.1203245
Ren W., Rajendran R., Zhao Y., Tan B., Wu G., Bazer F. W., et al. (2018). Amino acids as mediators of metabolic cross talk between host and pathogen. Front. Immunol. 9:319:319. doi: 10.3389/fimmu.2018.00319
Reynolds C. M., Ribeiro A. A., McGrath S. C., Cotter R. J., Raetz C. R., Trent M. S. (2006). An outer membrane enzyme encoded by Salmonella typhimurium lpxR that removes the 3’-acyloxyacyl moiety of lipid A. J. Biol. Chem. 281 (31), 21974–21987. doi: 10.1074/jbc.M603527200
Robinson M. D., McCarthy D. J., Smyth G. K. (2010). edgeR: A bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26 (1), 139–140. doi: 10.1093/bioinformatics/btp616
Rogers T. J., Paton A. W., McColl S. R., Paton J. C. (2003). Enhanced CXC chemokine responses of human colonic epithelial cells to locus of enterocyte effacement-negative Shiga-toxigenic Escherichia coli. Infect. Immun. 71 (10), 5623–5632. doi: 10.1128/iai.71.10.5623-5632.2003
Sandt C. H., Hopper J. E., Hill C. W. (2002). Activation of prophage eib genes for immunoglobulin-binding proteins by genes from the IbrAB genetic island of Escherichia coli ECOR-9. J. Bacteriol. 184 (13), 3640–3648. doi: 10.1128/jb.184.13.3640-3648.2002
Santos-Beneit F. (2015). The Pho regulon: A huge regulatory network in bacteria. Front. Microbiol. 6:402:402. doi: 10.3389/fmicb.2015.00402
Schinner S. A., Mokszycki M. E., Adediran J., Leatham-Jensen M., Conway T., Cohen P. S. (2015). Escherichia coli EDL933 requires gluconeogenic nutrients to successfully colonize the intestines of streptomycin-treated mice precolonized with E. coli Nissle 1917. Infect. Immun. 83 (5), 1983–1991. doi: 10.1128/IAI.02943-14
Slutsker L., Ries A. A., Greene K. D., Wells J. G., Hutwagner L., Griffin P. M. (1997). Escherichia coli O157:H7 diarrhea in the United States: Clinical and epidemiologic features. Ann. Intern. Med. 126 (7), 505–513. doi: 10.7326/0003-4819-126-7-199704010-00002
Stadelmann B., Merino M. C., Persson L., Svard S. G. (2012). Arginine consumption by the intestinal parasite Giardia intestinalis reduces proliferation of intestinal epithelial cells. PloS One 7 (9), e45325. doi: 10.1371/journal.pone.0045325
Stadelmann B., Hanevik K., Andersson M. K., Bruserud O., Svard S. G. (2013). The role of arginine and arginine-metabolizing enzymes during Giardia - host cell interactions in vitro. BMC Microbiol. 13, 256. doi: 10.1186/1471-2180-13-256
Stalb S., Barth S. A., Sobotta K., Liebler-Tenorio E., Geue L., Menge C. (2018). Pro-inflammatory capacity of Escherichia coli O104:H4 outbreak strain during colonization of intestinal epithelial cells from human and cattle. Int. J. Med. Microbiol. 308 (7), 899–911. doi: 10.1016/j.ijmm.2018.06.003
Thorpe C. M., Smith W. E., Hurley B. P., Acheson D. W. (2001). Shiga toxins induce, superinduce, and stabilize a variety of C-X-C chemokine mRNAs in intestinal epithelial cells, resulting in increased chemokine expression. Infect. Immun. 69 (10), 6140–6147. doi: 10.1128/IAI.69.10.6140-6147.2001
Torres A. G., Payne S. M. (1997). Haem iron-transport system in enterohaemorrhagic Escherichia coli O157:H7. Mol. Microbiol. 23 (4), 825–833. doi: 10.1046/j.1365-2958.1997.2641628.x
Trivedi P. J., Bruns T., Ward S., Mai M., Schmidt C., Hirschfield G. M., et al. (2016). Intestinal CCL25 expression is increased in colitis and correlates with inflammatory activity. J. Autoimmun. 68, 98–104. doi: 10.1016/j.jaut.2016.01.001
Valkova D., Valkovicova L., Vavrova S., Kovacova E., Mravec J., Turna J. (2007). The contribution of tellurite resistance genes to the fitness of Escherichia coli urophathogenic strains. Cent. Eur. J. Biol. 2 (2), 182–191. doi: 10.2478/s11535-007-0019-9
Van Horn W. D., Sanders C. R. (2012). Prokaryotic diacylglycerol kinase and undecaprenol kinase. Annu. Rev. Biophys. 41, 81–101. doi: 10.1146/annurev-biophys-050511-102330
Vareille M., de Sablet T., Hindre T., Martin C., Gobert A. P. (2007). Nitric oxide inhibits Shiga-toxin synthesis by enterohemorrhagic Escherichia coli. Proc. Natl. Acad. Sci. U.S.A. 104 (24), 10199–10204. doi: 10.1073/pnas.0702589104
Vergara A. F., Vidal R. M., Torres A. G., Farfan M. J. (2014). Long polar fimbriae participates in the induction of neutrophils transepithelial migration across intestinal cells infected with enterohemorrhagic E. coli O157:H7. Front. Cell Infect. Microbiol. 4:185:185. doi: 10.3389/fcimb.2014.00185
Whelan K. F., Colleran E., Taylor D. E. (1995). Phage inhibition, colicin resistance, and tellurite resistance are encoded by a single cluster of genes on the IncHI2 plasmid R478. J. Bacteriol. 177 (17), 5016–5027. doi: 10.1128/jb.177.17.5016-5027.1995
Whiteley C. G., Lee D. J. (2015). Bacterial diguanylate cyclases: structure, function and mechanism in exopolysaccharide biofilm development. Biotechnol. Adv. 33 (1), 124–141. doi: 10.1016/j.biotechadv.2014.11.010
Wurbel M. A., McIntire M. G., Dwyer P., Fiebiger E. (2011). CCL25/CCR9 interactions regulate large intestinal inflammation in a murine model of acute colitis. PloS One 6 (1), e16442. doi: 10.1371/journal.pone.0016442
Yamasaki C., Natori Y., Zeng X. T., Ohmura M., Yamasaki S., Takeda Y., et al. (1999). Induction of cytokines in a human colon epithelial cell line by Shiga toxin 1 (Stx1) and Stx2 but not by non-toxic mutant Stx1 which lacks N-glycosidase activity. FEBS Lett. 442 (2-3), 231–234. doi: 10.1016/s0014-5793(98)01667-6
Yang B., Feng L., Wang F., Wang L. (2015). Enterohemorrhagic Escherichia coli senses low biotin status in the large intestine for colonization and infection. Nat. Commun. 6, 6592. doi: 10.1038/ncomms7592
Keywords: STEC, transcriptomics, cytokines, virulence factors, pathogenesis
Citation: Harrison LM, Lacher DW, Mammel MK and Leonard SR (2020) Comparative Transcriptomics of Shiga Toxin-Producing and Commensal Escherichia coli and Cytokine Responses in Colonic Epithelial Cell Culture Infections. Front. Cell. Infect. Microbiol. 10:575630. doi: 10.3389/fcimb.2020.575630
Received: 23 June 2020; Accepted: 07 October 2020;
Published: 26 October 2020.
Edited by:
Jang Won Yoon, Kangwon National University, South KoreaReviewed by:
Fernando Navarro-Garcia, National Polytechnic Institute of Mexico (CINVESTAV), MexicoJorge Blanco, University of Santiago de Compostela, Spain
Copyright © 2020 Harrison, Lacher, Mammel and Leonard. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Susan R. Leonard, c3VzYW4ubGVvbmFyZEBmZGEuaGhzLmdvdg==
 Lisa M. Harrison
Lisa M. Harrison David W. Lacher
David W. Lacher Mark K. Mammel
Mark K. Mammel Susan R. Leonard
Susan R. Leonard