
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Cell. Infect. Microbiol. , 06 October 2020
Sec. Fungal Pathogenesis
Volume 10 - 2020 | https://doi.org/10.3389/fcimb.2020.552394
 Nnamdi Ifechukwude Chidi1,2*
Nnamdi Ifechukwude Chidi1,2* Adedotun Adeyinka Adekunle1,2
Adedotun Adeyinka Adekunle1,2 Temitope Oluwaseun Samuel1,2
Temitope Oluwaseun Samuel1,2 Emmanuel Ifechukwude Eziashi1,2,3
Emmanuel Ifechukwude Eziashi1,2,3Fusarium wilt is caused by Fusarium oxysporum f. sp. elaeidis, and constitutes a severe threat to the oil palm industry in Africa. This study is aimed at surveying, identifying the secreted effector genes responsible for virulence during pathogenesis, and investigating the level of genetic diversity and cluster resolutions of alleles accountable for virulence in pathogenic strains of F. oxysporum f.sp. elaeidis from African countries. Fifty-eight fungal strains were isolated from acute and chronic Fusarium wilt diseased oil palms in Nigeria, Ghana and Cameroon. Morphological and sequencing analysis of the Internal Transcribed Spacer (ITS) region grouped all strains into nine dominant strains with a majority (41.37%) belonging to F. oxysporum, followed by F. solani (20.68%), F. equiseti (20.68%), F. verticilliodes (5.17%), F. proliferatum (3.44%), F. chlamydosporum (3.44%), F. nelsonii (1.72%), Fomes fomentarius, and Penicillium simplicissimum (1.72%). Disease incidence and severity showed varying levels of virulence with some Fusarium strains causing severe symptoms while others exhibited slight symptoms. ISSR evaluation disclosed a considerable level of genetic diversity among pathogenic F. oxysporum f.sp. elaeidis strains. Molecular characterization using defense gene primers revealed that the oil palm genotypes screened did not amplify defense genes. During pathogenesis, Fusarium strains produced GMC oxidoreductases, hypothetical proteins, FOIG 16629, FOXG 14258, and Pyranose dehydrogenase 3-like proteins using virulent effector gene primers. Polymerase Chain Reaction analysis using specific gene primers revealed that PRK02106, beta and BetA effector genes were secreted explicitly by F. oxysporum f.sp. elaeidis (4) and F. oxysporum f.sp. elaeidis (CRT) strains while screening tolerant oil palm genotypes. During screening susceptible oil palm genotypes, F. oxysporum f.sp. elaeidis (4) and F. oxysporum f.sp. elaeidis (CRT) strains produced FGGY_L-XK1, PRK10939, FGGY_N1, XylB1, XylB2, FGGY_L-XK2, XylB3, FGGY_N2, and XylB4 effector genes. Identifying these effector genes will provide the platform to study the basis of pathogenesis which will help breeders to modify breeding techniques for the improvement of oil palm genotypes in order to reduce oil palm loss in plantations and enhance food security.
Oil palm, which is also known as Elaeis guineensis Jacq., is a plant that is enduring and can last for an indefinite time. It is of African origin and grown by people for food and other uses, especially on a large farming scale. Oil palm is the ultimate oil-yielding plant in the world. Nigeria is the fourth significant palm oil producer in the world and first in Africa. More than 30% of vegetable oil is extracted from this plant, thereby making it the leader in world fat supply (United States Department of Agriculture, 2015). In as much as humans utilize oil palm produce for food, the extracted oil can also serve as a body insulator and an energy source to populations that live in developing countries (Goh et al., 2016).
The plant is in danger to so many diseases. The most destructive of these diseases in Africa is the Fusarium wilt disease whose causal agent is Fusarium oxysporum f.sp. elaeidis (Paterson et al., 2013; Noumouha et al., 2014). Fusarium wilt disease causes damage to oil palm seedlings in the nursery as well as to juvenile and mature palms in plantations. In the nursery seedlings, the symptoms commence with stunted growth, followed by yellowing of leaves, dryness and death. F. oxysporum strains can live in the soil and can be transmitted by both wind and rain (Leslie and Summerell, 2006). Africa's oil palm production is hugely declined due to the effects of Fusarium wilt disease (Ntsomboh et al., 2015). The damaging implications of this disease are as high as 70%, and the plantations where the condition had existed shows preference to the sudden emergence of the disease (Rival, 2017).
Plants are well-organized and are competent in protecting themselves against agents of disease through the commencement of very highly advanced and complicated defense systems (Bari and Jones, 2009). These fortifying or protective reactions encompass the release of defense-related proteins (Bowles, 1990). The effort of Fusarium strains in overcoming resistance is very insignificant if oil palm resistance genes are polygenic, but significant if it is based only on a few genes (Diener and Ausubel, 2005; Cooper and Rusli, 2014). The constitution of the primary cell wall makes it impenetrable to the entry of disease-causing agents. Traditionally, the enzymatic collapse of the cell wall has been linked with plant pathogenesis (Di-Pietro et al., 2003). The responsibility of pathogenesis-related (PR) proteins involved in stress response and metabolic regulations have been identified in some plants during the beginning of pathogen infection.
The diversity of F. oxysporum in the environment is enormous because of the extensive isolations from its natural range. This diversity can lead to variations found in the gene sequences during pathogen and host interactions (Di Pietro and Roncero, 1998). Since it is possible to confirm the pathogenesis of a disease-causing agent by the presence of a precise gene or a group of genes secreted as virulence effectors, it will be essential in this study to identify the virulent effector genes produced at the point of infection during pathogenesis using the approach of identification of the secreted effector proteins from pathogenic F. oxysporum f.sp. elaeidis strains. Furthermore, we will look at the genetic diversity, population structure among the pathogenic strains and the cluster resolutions of the virulent alleles.
The disease survey was conducted from 2012 to 2015 in three randomly selected African countries, namely Nigeria, Ghana, and Cameroon, in zones where the disease is widespread and severe. In Nigeria, samples were randomly collected from the Nigerian Institute for Oil Palm Research (NIFOR) central station, Oil Palm Producing Company in Ajagbodudu, and NIFOR substation in ABAK. In Ghana, samples were randomly collected from oil palm plantations at the Oil Palm Research Institute (OPRI), Kusi, Benso Oil Palm Plantation, and Ghana Oil Palm Development Company (GOPDC). Cameroon, depicted with plantations of an area of 16.86 hectares, random samples were collected from the Institute of Agricultural Research for Development (IRAD), Ekona (South West Region), and Cameroon Development Company (CDC) of the same South-West region. The disease incidence and severity were computed as described by Ntsomboh et al. (2015). Visual ratings of the symptoms were used to assess incidence and severity. The incidence of the disease was calculated as follows: Disease incidence = Nm /Nt x100; where Nm = Number of seedlings infected in the trial plot; Nt = total Number of seedlings (healthy + sick) in the trial plot. It was assessed by using the key; H = Healthy; LS = Leaf symptom only; L&R = Leaf and Root symptoms, and R= Root symptom only. Disease severity = summation of (a × b) /n ×100; where ∑(a × b) = sum of products of the number of the infected seedlings (a) corresponding to the degree of infection (b); n = Number of infected seedlings. The severity was determined using a scale of 0–5 (Table 1). The Geographic Information System (GIS) maps of the sample location points were designed.
The asexual resources investigated were 200 oil palm trees displaying chronic and acute symptoms of Fusarium wilt disease as described by Oritsejafor (1982); Tengoua (1994); Ntsomboh et al. (2012, 2015); Chidi et al. (2018). The external symptoms of the chronic and acute state of the disease include stunted growth, yellowing of the leaves, withering and rupture of the fronds. The death of the palms precedes these symptoms within 6 months (acute type) or a few years (chronic type). The internal symptoms include blackening, necrosis of the cortex and clogging of the vascular system.
F. oxysporum f.sp. elaeidis strains and secondary pathogens were isolated from the infected strands of plant tissues, petioles and soil samples of adult oil palms displaying chronic and acute symptoms of Fusarium wilt disease as described by Ntsomboh et al. (2015). The strands of plant tissues and petioles of infected oil palms were chopped into reduced portions with the aid of a sterile knife to expose the infected inner parts as described by Tengoua (1993). They were aseptically taken out and plated on previously prepared mycelium medium (MM) and Komada medium that has been cooled and supplemented with streptomycin antibiotic as described by Komada (1975); Tengoua (1993); Ntsomboh et al. (2015). Root samples were plated out, but not after the sliced portions had been superficially sanitized with 1% bleach combination. For the soil samples, the serial dilution method of Oritsejafor (1982) was employed. Incubation was conducted at room temperature of 26–29°C. The developing fungi colonies were aseptically subcultured into gelled prepared potato dextrose agar (PDA, Difco Laboratories, Detroit, USA) plates until pure cultures were gotten.
The induced fungal colonies were viewed under the light microscope. Then the images of the organisms were snapped using a Motic Camera, with model number 230 (1.3 Megapixel). The Fusarium strains and secondary pathogens were identified by comparing their morphology and color with fungi descriptions in Booth (1975); Leslie and Summerell (2006).
Fusarium strains and other associated fungi were grown overnight in a Fusarium liquid medium and transferred to Eppendorf tubes. The tubes were spun at 14,000 rpm for 2 min. The supernatants were discarded, and then 600 μl of 2X CTAB buffer was added to the pellets, which were later incubated at 65°C for 30 min. The samples were taken out and left to cool down to room temperature, after which chloroform was added. The samples were stirred together by gentle inversion of the tubes several times. After that, the samples were spun at 14,000 rpm for 15 min and then transferred into new Eppendorf tubes. An equivalent volume of cold Isopropanol was added to precipitate the DNA. The samples were put in the freezer for 1 h and later spun at 14,000 rpm for 10 min. The supernatants were discarded, and pellets were washed with 70% ethanol, which was later air-dried for 30 min on the bench. The pellets were re-suspended in 100 μl of sterile distilled water.
Agarose gel electrophoresis was used to clarify the quality and integrity of the DNA by size fractionation on 1.0% agarose gels. Agarose gels were set up by dissolving and boiling 1.0 g agarose in 100 ml 0.5 X TBE buffer solutions. The gels were left to cool down to about 45°C. Then 10 μl of 5 mg/ml detergent free ethidium bromide was added, mixed before pouring into an electrophoresis compartment set with the combs inserted. After the gels had solidified, 3 μl of the DNA, plus 5 μl sterile distilled water, and 2 μl of 6X loading dye were mixed. These mixtures were later loaded in the wells created. Electrophoresis was done at 80 V for 2 h. The separated DNA bands were visualized and photographed under UV light source.
PCR analysis was run using two oligonucleotide primers for fungi ITS1 (5′-TCTGTAGGTGAACCTGCGG-3′) and ITS4 (5′-TCCTCCGCTTATTGATATGC-3′) to amplify the Internal Transcribed Spacer region. The PCR mix consists of 1 μl of 10X buffer, 0.4 μl of 50 mM MgCl2, 0.5 μl of 2.5 mMdNTPs, 0.5 μl 5 mM ITS1, 0.5 μl 5 mM ITS4, 0.05 μl of 5 units/μlTaq, with 2 μl of template DNA, and then 5.05 μl of distilled water to make-up 10 μl reaction mix. The PCR profile used had an initial denaturation temperature of 94°C for 3 min, followed by 30 cycles of 94°C for 60 s, 60°C for 60 s, 72°C for 120 s and the final extension temperature of 72°C for 5 min and 10°C hold.
The amplicons were further purified before sequencing using 2 M sodium acetate wash techniques. To about 10 μl of the PCR products, 1 μl 2 M NaAct pH 5.2 was supplemented, followed by 20 μl absolute ethanol, kept at −20°C for 1 hr. The amplicons were spun at 10,000 rpm for 10 min and then washed with 70% ethanol before air drying. Re-suspension was done in 5 μl sterile distilled water and keep at 4°C for sequencing.
The PCR mix used contained 0.5 μl of BigDye Terminator Mix, 1 μl of 5X sequencing buffer, 1 μl of ITS primer with 6.5 μl distilled water, and 1 μl of the PCR product, leading to a total of 10 μl. The PCR profile for sequencing was a Rapid profile. The initial Rapid thermal ramp was set at 96°C for 1 min, followed by 25 cycles of the Rapid thermal ramp at 96°C for 10 s; Rapid thermal ramp at 50°C for 5 s; and Rapid thermal ramp at 60°C for 4 min, then followed by a Rapid thermal ramp at 4°C hold.
The PCR sequence products were also purified before running the sequencing procedure using 2 M sodium acetate wash techniques. To 10 μl of the PCR product, 1 μl 2 M NaAct, with a pH of 5.2 was included, then 20 μl absolute ethanol, kept at −20°C for 1 hr was added. The entire mix was spun at 10,000 rpm for 10 min; washed with 70% ethanol, and then air-dried. The products were later re-suspended in 5 μl sterile distilled water and kept at 4°C for sequencing.
The Cocktail mix was a combination of 9 μl of Hi di Formamide, together with 1 μl of Purified sequence products, making a total of 10 μl. The samples were loaded on the ABI 3130 × l automated sequencing machine. The forward and reverse sequences were manually edited to avoid any ambiguities.
The BLAST searches were performed using the GenBank sequence database to confirm the identity of the sequence of the fungal strains (Geiser et al., 2004). The output from BLAST algorithms was used to query any unknown sequences against the database of all the fungal strains in the gene regions. MEGA X software (Tamura et al., 2013) was used to edit and align the nucleotide data, after which the alignments were improved manually.
Phylogenetic analyses were achieved using amplified PCR product generated from ITS 1 and ITS 4 primers. The BLAST algorithm performed the sequence similarity searches. MEGA X was used to generate the dendrogram as described by Nei and Kumar (2000); Tamura et al. (2013).
The seeds were sprouted by the dry heat treatment method. Harvested seeds from susceptible and tolerant oil palm trees were soaked in plastic bowls containing water for 1 day (Table 2). It was to ensure a moisture content level of 17–18%. The floated seeds were discarded. Seeds were later transferred to white transparent polythene bags, tightly sealed with a band. Polythene bags containing the seeds were weighed at intervals to continually check that the moisture content had not declined below 17%. Heat was then applied at 37–39°C for 50 days. After the expiration of 50 days, the seeds were soaked for 3 days to jack up the moisture content to 21–23%. After drying, seeds were subjected to mild fungicide treatment, which was succeeded by 8 h of air-drying. The seeds were finally put back in polythene bags and sealed to give it an ambient temperature of around 25–30°C. At the end of 1 month, seeds were inspected for germination and processed for field trials. The parents of the oil palm genotypes are represented in Table 3.
The germinating seeds obtained were planted with their plumules facing upwards at a depth of 2.54 cm in black polythene bags filled with topsoil (1 seed in 3 kg of soil). The poly bags containing the seeds were watered twice daily. After 2 months (two-leaf stage), initial heights measurements of the seedlings were taken using a meter rule and recorded.
Preparations of conidial suspensions were done by cutting 3 mm portions of the already grown F. oxysporum f.sp. elaeidis strains in Petri dishes. The cutoff portions of the cultures were inoculated into 250 ml conical flasks containing sterilized 50 ml of Armstrong medium. At each preparation, flasks were inoculated with F. oxysporum f.sp. elaeidis strain and incubated for 14 days at room temperature (26–29°C). After the incubation period had elapsed, contents in the flasks were poured out into a warren blender (Model MSE, London, serial no: 7702164) and macerated. The macerated mixtures were then filtered through a muslin cloth. The supernatant was decanted, and the suspension was filled with sterile water to the required spore concentration.
A drop of the inoculum was pipetted on a hemocytometer and covered with a coverslip. A phase-contrast illumination microscope with a 10x ocular and 4 mm objective for at least 20 squares was used to count the spores. The average number per square was determined. Three counts of diluted spore suspension were made, and their average was taken. The number per milliliter of the original spore suspension (undiluted) was calculated as follows:
(Average No. per square) × y × 4,000,000
Explanation of factors:
Area of square = 1/20 × 1/20 mm = 1/400 mm2
Volume over square =1/400 mm2 × 1/10 mm (depth)
= 1/4,000 mm3
= 1/4,000 mm3 × 1/1,000 ml/mm3
= 1/4,000,000 ml
Where dilution factor = y.
The conidia suspension of F. oxysporum f.sp. elaeidis strains were inoculated on 2 months old oil palm seedlings for pathogenesis, and to determine tolerance and susceptibility of the oil palm genotypes. The pathogenesis trial was carried out by separately inoculating the roots of oil palm genotypes with conidia suspensions. The roots were exposed by removing layers of soil covering the root system. Conidia suspensions of 3.2 × 106 spores/ml were dispensed into the root system. Soils were used to cover up the exposed roots. In the control experiments, sterile distilled water was used instead of spore suspension. After 1 month of inoculation, 5 g of NPK 15: 15: 20 fertilizer was applied to the inoculated seedlings. Six months post-inoculation, the seedlings were analyzed for disease symptoms.
Univariate Analysis of Variance with LSD plus Duncan Multiple Test was used to analyze the data for morphological parameters. The means were separated by the least significant difference at the 95% confidence interval for the difference. Bar charts were employed to compare the effect of the different F. oxysporum f.sp. elaeidis strains on the growth rate of the different oil palm genotypes.
The inocula of F. oxysporum f.sp. elaeidis strains were prepared as earlier described. The seeds of tolerant (6 and 7) and susceptible (1, 2, 3, 4, and 5) oil palm genotypes used for this study were sprouted using dry heat treatment method as described earlier. The roots of the oil palm seedlings were inoculated with the inoculum of the Fusarium oxysporum strains as described by Renard et al. (1972); Mcfadden et al. (2006). Samples for effector gene identification analysis were harvested when external symptoms of Fusarium wilt disease appeared within 6 months. All spots of infected seedlings on the bole showing discolouration were taken immediately for DNA extraction. The DNA extraction was carried out using a modified CTAB method.
Modifications were made to the PCR protocol involving specific primers used in this study. The PCR cocktail mix consists of 2.5 ul of 10x PCR buffer,1 ul of 25 mM MgCl2, 1 ul each of forward and reverse specific primers, 1 ul of DMSO, 2 ul of 2.5 mMDNTPs, 0.1 ul of 5 u/ul Taq DNA polymerase, and 3 ul of 10 ng/ul DNA. The whole reaction volume was made up to 24 ul using 13.4 ul Nuclease-free water. The PCR cycling parameter consists of a PCR profile as follows: Initial denaturation at 94°C for 5 min, nine cycles of denaturation at 94°C for 30 s, annealing at 65°C for 30 s and elongation at 72°C for 30 s succeeded by 35 cycle sequence of denaturation at 94°C for 30 s, annealing at 55°C for 30 s, and elongation time at 72°C for 30 s. followed by a final elongation step at 72°C for 7 min and hold temperature at 10°C. Amplified fragments were visualized on 1.5% agarose electrophoresis gels. The annealing temperatures varied for the defense-related gene and virulence effector genes primers used in Table 4. After the PCR procedure, sequencing was carried out as earlier described. The sequences generated were analyzed using the BlastX searches across multiple databases (Altschul et al., 1990). The identity of the sequences was matched with sequences homologous to the fungal sequence in the database. Thus, sequences that matched a known fungal sequence with an E-value equal to or lower than 10-10 corresponding to the genes were identified.
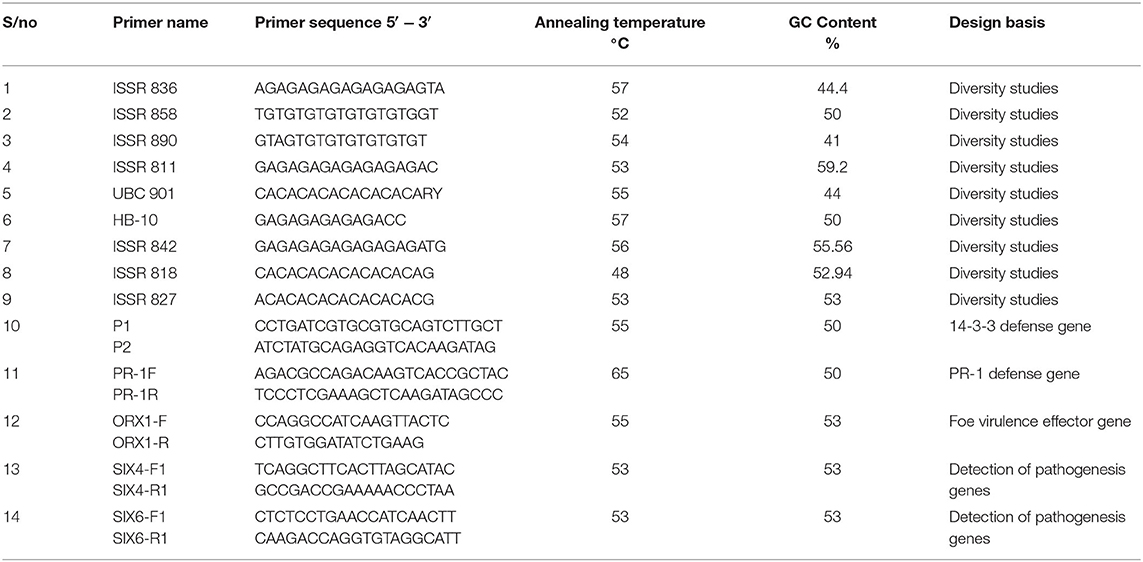
Table 4. ISSR primers used for genetic diversity study, defense-related gene, and putative virulence effector genes.
A total of seventeen F. oxysporum f.sp. elaeidis strains were used for this investigation. The Fusarium strains were grown overnight in Fusarium liquid medium containing a mixture of Glucose (30 g); Calcium nitrate quatrehydrate (Ca (NO3)2. 4H2O (8.4 g); Sodium nitrate (2 g); Potassium dihydrogen phosphate (KH2PO4) (1.09 g); Potassium chloride (KCL) (0.22 g); Iron III chloride (FeCl3) (0.2 μg); Magnesium sulfate (MgSO4) (0.75 g); Ferrous sulfate (FeSO4) (trace); Zinc sulfate (ZnSO4) (trace); Marmite (trace); Distilled water (1,000 ml) and Agar (1 g). The total genomic DNA extraction was carried out using ZYMOGEN KIT (Zymo Research Corporation, USA), according to its manufacturer's instructions.
Polymerase chain reaction (PCR) amplification was accomplished by mixing 1.50 μl of 50 mM MgCl2 (BIOLINE Massachusetts, USA), 2.00 μl of 2.50 mM dNTPs (BIOLINE, Massachusetts, USA), 0.20 μl 500 U Taq DNA polymerase (BIOLINE, Massachusetts, USA), 1.0 μl of 10 μM each of ISSR primer pair, 15.05 μl of 500 ml diethylpyrocarbonate (DEPC)-treated water (INVITROGEN, Carlsbad, CA, USA) and 2.0 μL 100 ng DNA, 2.5 μl of 10 × Taq Buffer (BIOLINE, Massachusetts, USA) to make up a volume of 24.25 μL. The list of ISSR markers, their sequences, and annealing temperatures are presented in Table 4. The PCR cycling profile employed for the reaction entailed an initial step at 94 °C for 5 min, succeeded by 35 cycles of 94°C for 30 s, 72°C for 1 min, and a 10 min last extension at 72°C. For the PCR reaction, eight (8) μl of the PCR products were dispensed in a 1.5% agarose gel comprising 0.5 mg/ml ethidium bromide. The bands were photographed on Transilluminator UV light (Fotodyne Incorporated, Analyst Express, USA).
The data matrix of ISSR profiles obtained from fragments of each amplicon was scored as 1 (presence of alleles) and 0 (absence of alleles). The data generated from the scoring of the ISSR amplicons were employed for phylogenetic reconstruction using Unweighted Pair Group Mean with Arithmetic (UPGMA) and dissimilarity index (Ojuederie et al., 2013). The analysis was carried out using NTSYSpc software version 2.02. Furthermore, genetic diversity, allele frequency, and the polymorphic information content (PIC) were analyzed using PowerMarker Version 3.25. Genetic diversity and population structure analyses of F. oxysporum f.sp. elaeidis strains were analyzed using POPGENE software version 1.32. Also, total gene diversity (Ht), gene diversity within the population (Hs), the level of gene flow (Nm), and the coefficient of gene differentiation (Gst) were calculated with POPGENE software version 1.32 (Yeh et al., 1999).
During the survey period, oil palm trees infected with Fusarium wilt disease displaying different states of chronic and acute symptoms were visited and felled (Figures 1A–D). The asexual resources investigated are represented in Figures 1E–G. The Geographical Positioning System (GPS) and the Geographical Information System (GIS) maps of the sample location points designed are presented in Supplementary Table 1, Figures 2A,B.
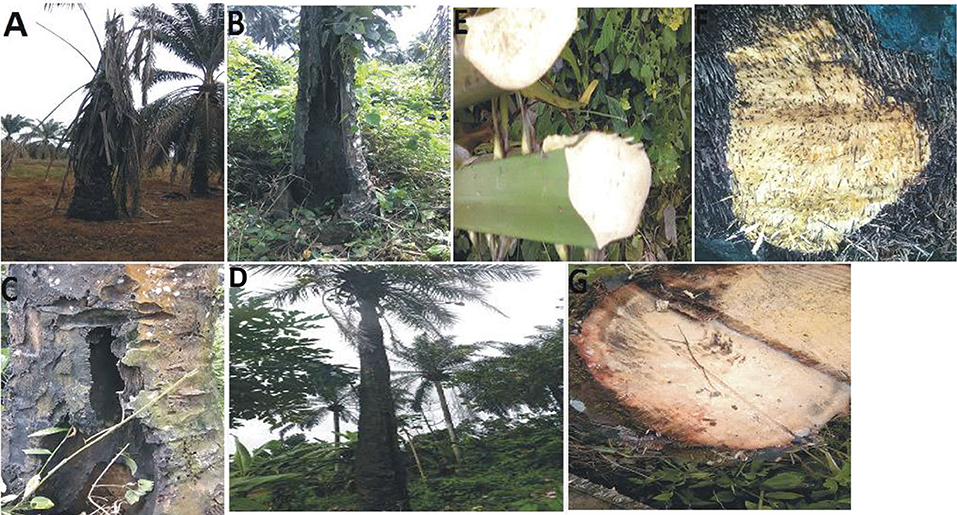
Figure 1. (A) Chronic symptoms of Fusarium wilt disease with wilting leaves. (B) Chronic symptoms of Fusarium wilt disease revealing extreme cracking of the trunk. (C) Chronic symptoms of Fusarium wilt disease with perforations in the vascular bundles. (D) Chronic symptoms of Fusarium wilt disease with a thin terminating trunk. (E) Infected petiole of an acute infected oil palm. (F) The cut transverse section of the vascular bundle of an acute Fusarium wilt infected oil palm. (G) The inner tissues of the vascular bundles of an acute Fusarium wilt infected oil palm.
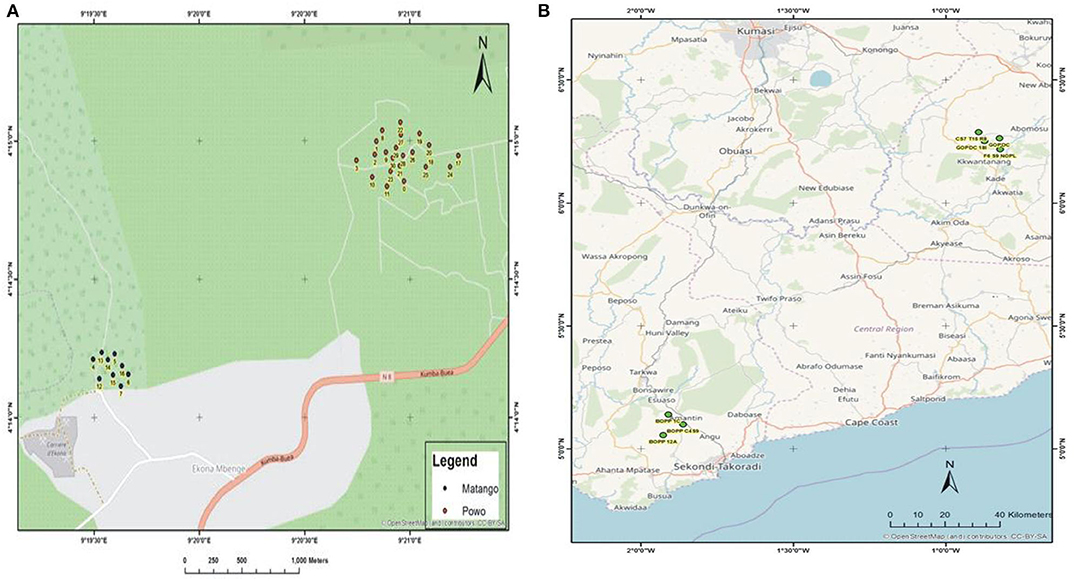
Figure 2. (A) The GIS map site of sample collection in Cameroon. (B) The GIS map site of sample collection in Ghana.
Fifty-six (56) Fusarium strains and two (2) secondary pathogens were morphologically identified and further confirmed by sequencing the ITS region of the extracted genomic DNA of the fungal strains. The sequences generated from each of the strains were used to confirm the identity of the fungal strains. The results of the BLAST queries carried out on the GenBank sequence database confirmed the identity of the fungal strains as having 95–100% homology with highly similar sequences in the GeneBank (Supplementary Table 2) (Supplementary Figure 1). The sequences of 49 fungal strains showed 100% homology with the sequence of the accessions in the GenBank (Supplementary Figure 1). Twenty-three Fusarium strains showed homology with 100% similarity to F. oxysporum. Fifteen of these F. oxysporum strains are from Cameroon; three from Ghana, and five from Nigeria; although one Fusarium strain from Cameroon showed 95% homology to F. oxysporum. The sequenced data generated showed that out of the 56 Fusarium strains identified, F. oxysporum was the most prevalent strain in the regions that were sampled with a percentage of 41.37%. Other strains were identified as F. solani (20.68%), F. proliferatum (3.44%), F. equiseti (20.68%), F. verticilliodes (5.17%), F. chlamydosporum (3.44%), F. nelsonii (1.72%), and other fungi strains, Fomes fomentarius (1.72%), and Penicillium simplicissimum (1.72%). Four strains of F. oxysporum (1) from Cameroon; F. oxysporum (4) from Cameroon; F. oxysporum (CRT) from Ghana, and F. oxysporum (13) from Nigeria were selected for further studies based on their degree of virulence as compared to the other F. oxysporum strains screened.
The phylogenetic investigation of the fungal pathogens from the African countries indicates that the confidence probability (multiplied by 100) means of the interior branch length is > 0, as estimated using a bootstrap test (1,000 replicates shown next to the branches). The investigation of the ITS region positioned the majority of the fungal pathogens from the acute and chronic samples in the Fusarium genus (Figure 3). The investigation put all fungal strains into eight main clusters. Clade A clustered F. oxysporum strains into the F. oxysporum strain complex (FOSC) representing Cameroon and Ghana. The cluster contains a virulent F. oxysporum strain, CRT, from Ghana, and a low virulent strain, F. oxysporum (1) from Cameroon. The F. equiseti strains (MAT 15M, PW7B, MAT 16M, PW6M, PW2MA, PW8MP, 583, 44, PW11B, and 29) formed a distinct clade B assisted by 96% bootstrap values. Clade C consists of the F. solani strain complex (FSSC). Interestingly, the most virulent strain, F. oxysporum (4) and another strain, F. oxysporum (PW49A) are contained in clade D with some members of the Gibberella Fujikuroi species complex. Clade E contained the F. equiseti strains mainly from Cameroon. The members of the Fusarium oxysporum strain complex from Nigeria are grouped in clade F. It includes a low virulent strain, F. oxysporum (13). F. chlamydosporum (NG12, NG13) and Fusarium nelsonii (NG14) are grouped in clade G. Clade H includes members of the Fusarium solani strain complex from Nigeria. Penicillium simplicissimum (MAT4A) and Fomes fomentarius (PW'3A2) from Cameroon are in the out-group.

Figure 3. The phylogenetic tree of Fusarium strains and secondary pathogens using Neighbor- Joining method.
The mean disease incidence scores on the oil palm genotypes were different for all the genotypes (Figure 4) (Supplementary Table 3). The mean incidence scores on the oil palm genotypes indicate they were not significantly different from each other (p < 0.05) when inoculated with F. oxysporum f.sp. elaeidis (4) only. The control seedlings had no recorded incidence because they were not inoculated. In contrast, genotypes 6 and 7 had no disease incidence when inoculated with F. oxysporum f.sp. elaeidis strain (1). The effects of F. oxysporum f.sp. elaeidis strains (13) and (CRT) were not significantly different from each other on the genotypes. In contrast, the effect of F. oxysporum f.sp. elaeidis (4) differed on all other genotypes (Table 5).
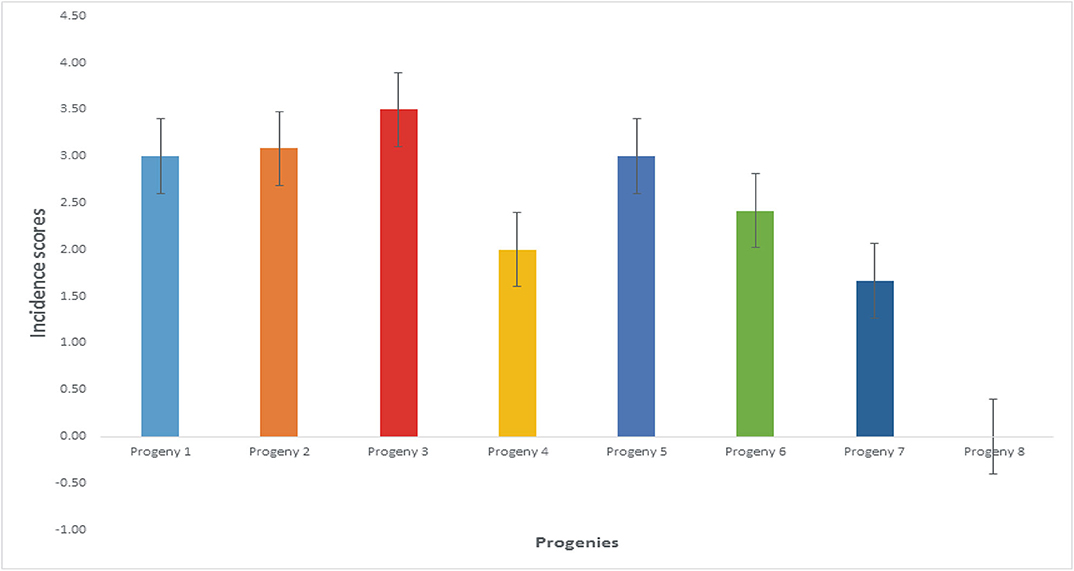
Figure 4. Mean incidence scores on oil palm genotypes as a result of the effects of Fusarium strains.
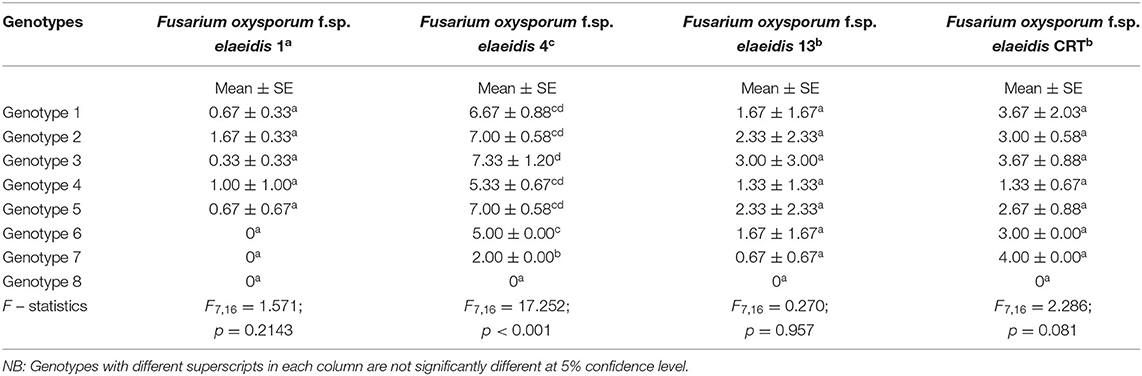
Table 5. Mean ± SE of Score Incidence of Fusarium oxysporum f.sp. elaeidis on oil palm genotypes in the leaf and root symptom category.
Fifty to sixty days after inoculation, external symptoms of Fusarium wilt disease began to appear. The severity of the disease caused by high-level aggressiveness of F. oxysporum f.sp. elaeidis strains is seen in the ability to colonize the roots and shoot of the oil palm seedlings (Figures 5A–G). The diseased seedlings exhibited suppressed growth and loss of vitality with chlorosis and necrosis (Figure 5B). The color of bole changed from being creamy to dark colored, which was seen in the internal tissue, a vivid sign of Fusarium wilt disease and symptom (Figures 5D,F). The control seedlings were differentiated with a standard height and creamy colored bole (Figures 5A–E) compared with the diseased seedlings exhibiting stunted growth and brownish discolouration (Figure 5G).

Figure 5. Phenotypic evaluation of oil palm genotypes. (A,C,E) Control of oil palm seedlings with normal height and creamy bole color. (B,D,F) Arrowheads indicate infected oil palm seedlings, external and internal symptoms of Fusarium wilt disease appearing sixty days after inoculations. (G) The control of oil palm seedlings differentiated itself with normal height and creamy bole color when put side by side with the diseased seedlings which exhibited stunted growth and brownish discolouration.
Disease severity by Fusarium strains showed that the severity on the different genotypes differed from each other as a result of their variance in virulence. F. oxysporum f.sp. elaeidis strain (4) had the highest disease severity of 86% on genotype 1, and severity of 77.7, 85.9, 74.8, 84, 62, and 50% on genotypes 2, 3, 4, 5, 6, and 7, respectively. F. oxysporum f.sp. elaeidis (1 and 13) had low disease severity on the oil palm genotypes as compared to F. oxysporum f.sp. elaeidis (CRT) which caused high disease severity close to F. oxysporum f.sp. elaeidis (4). Supplementary Table 4.
The adequate permissions for this particular plate 2 have been obtained from the copyright holders.
The principal component analysis (PCA) of the generated amplicons resulted in four clusters (Figure 6). Each group was a representative of unique alleles of the strains of F. oxysporum f.sp. elaeidis. It shows the positions of unique alleles of F. oxysporum f.sp. elaeidis on the factorial axis which determined the positions of the Fusarium strains. Based on the positions, it separated the virulent Fusarium strains from other Fusarium strains on the factorial axis. It shows that Fusarium strain (4) and Fusarium strain (CRT) which were the most severe in the morphological screening are positioned up the principal component scale. In contrast, Fusarium strain (13) and Fusarium strain (1) which were less severe are positioned on a different factorial axis of the principal component scale.
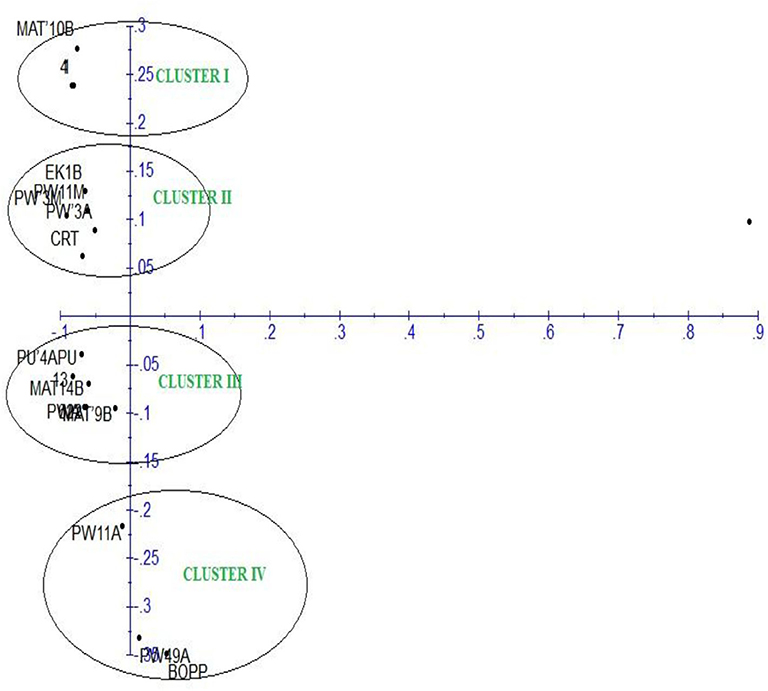
Figure 6. Principal component analysis of F. oxysporum f.sp. elaeidis strains based on Inter-simple sequence repeat markers.
The dendrogram of 17 strains of F. oxysporum f.sp. elaeidis was constructed using Unweighted Pair Group Mean Arithmetic (UPGMA) and dissimilarity index. It grouped the Fusarium strains into four major clusters, similar to the PCA (Figure 7). Cluster I grouped Fusarium strains MAT14B (Matango, Cameroon), CRT (Ghana), PU'4APU (Powo, Cameroon), PWA (Powo, Cameroon), 13(Nigeria) and 622(Ghana) at a bootstrap of 29%. Strain PU'4APU from Powo in Cameroon was the most genetically diverse in the group, followed by MAT14B and CRT. Cluster II contained PW3A (Powo, Cameroon) and EK1B (Ekona, Cameroon) both from Cameroon at a bootstrap of 31%. Cluster III was divided into subclusters I (SCI) and II (SCII) at a bootstrap of 30%. The SCI clustered four Fusarium strains at a bootstrap of 24%. The strains are 4(Cameroon) 1(Cameroon), MAT10B (Matango, Cameroon) and PW3M (Powo, Cameroon), all isolated from Cameroon. Fusarium strain PW3M (Powo, Cameroon) was the most genetically isolated strain in this subcluster. At 23%, the SGII clustered PW11M (Powo, Cameroon) and MAT9B (Matango, Cameroon) from Cameroon. Cluster IV had PW11A (Powo, Cameroon), PW49A (Powo, Cameroon) and BOPP (Ghana) isolated from Cameroon and Ghana. Fusarium strain BOPP was the most genetically dissimilar in this subcluster.
The genetic diversity between PW11A (Powo, Cameroon) and PW49A (Powo, Cameroon) strains were identified to be the highest when compared with other Fusarium strains. Their effective number of alleles (Ne), Nei's genetic diversity (H), and Shannon's information index (I) values of 1.9621, 0.4904, and 0.6835 are represented in Table 6. In contrast, MAT10B strain was found to have the lowest genetic diversity with Ne, H, and I values of 1.1484, 0.1292, and 0.2522 when compared with other Fusarium strains. The genetic diversity values of these strains were ranked in a descending order as MAT10B < (1, 4) < (PWA, PW11M) < PW3M < EK1B < CRT <13 < (PWA, PU'4APU, MAT9B) < (622, BOPP) < MAT 14B < (PW49A, PW11A) from low to high. The range values of Ne, H and I obtained were 1.1484–1.9621, 0.1292–0.4904, and 0.2522–0.6835, respectively. The overall mean values and standard deviations of Ne, H and I detected in the strains using ISSR were 1.6868 ± 0.2802, 0.3889 ± 0.1177, and 0.5705 ± 0.1369, respectively.
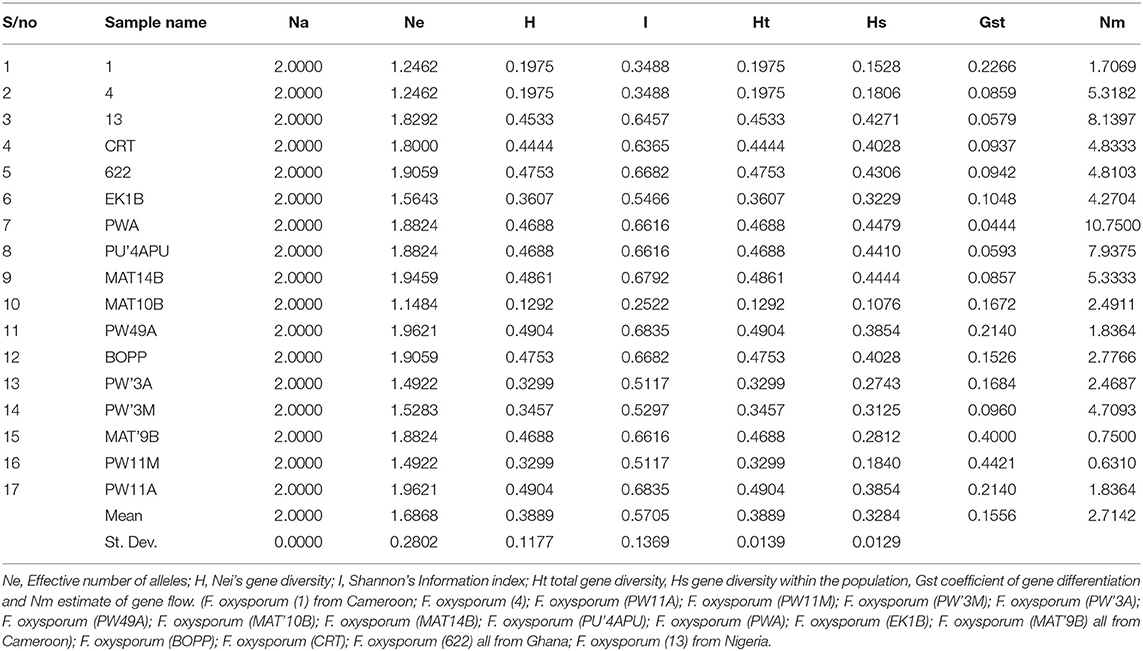
Table 6. Genetic diversity and genetic differentiation parameters generated from strains of F. oxysporum f.sp. elaeidis using ISSR markers.
The genetic variation among the Fusarium strains assessed shows that the mean values of total gene diversity (Ht), gene diversity within the population (Hs), coefficient of gene differentiation (Gst) and level of gene flow (Nm) are 0.3889, 0.3284, 0.1556, and 2.7142, respectively (Table 6). The allelic score count and frequencies were obtained from F. oxysporum f.sp. elaeidis strains using Inter-simple sequence repeat (ISSR) markers; Allele frequency, number of alleles, genetic diversity and polymorphic information content of ISSR markers (Supplementary Table 5).
The PCR amplification of the ORX1 effector which amplifies coding regions of secreted effector proteins from plant pathogenic fungi were used to screen the DNA from F. oxysporum f.sp. elaeidis (1); F. oxysporum f.sp. elaeidis (4); F. oxysporum f.sp. elaeidis (CRT); F. oxysporum f.sp. elaeidis (13), F. oxysporum f.sp. elaeidis (MAT 10B, EK1B, PW11M, PW3M, PW3A, PU4APU, MAT14B, MAT9B, 622, PWA, PW11A, PW49A, and BOPP in order to detect the presence or absence of candidate pathogenesis genes associated with virulence toward Nigerian oil palm genotypes. The result shows that some strains of F. oxysporum f.sp. elaeidis possess putative virulent effector genes. As a result, the ORX1 effector genes were amplified in seven strains of F. oxysporum f.sp. elaeidis (4, CRT, MAT10B, EK1B, PWIIM, PW3M, and PW3A, while absent in the other F. oxysporum f.sp. elaeidis strains (Figure 8). The amplified sequences gotten were BLAST, and GMC oxidoreductases, hypothetical proteins FOIG 16629, FOXG 14258, FOVG 19709, FOVG 19549, and Pyranose dehydrogenase-3-like effector genes homologous to F. oxysporum in the data bank were identified (Supplementary Figure 2). The sequences generated from the PCR amplification product of the oil palm and F. oxysporum f.sp. elaeidis interaction were compared with the sequences of fungal EST's homologous to F. oxysporum which led to the identification of putative virulent effector genes. PRK02106, betA, and BetA effector genes were identified from susceptible oil palm genotypes. In contrast, FGGY_L-XK 1, PRK10939, FGGY_N 1, XylB 1, XylB 2, FGGY_L-XK 2, XylB 3, FGGY_N 2 and XylB 4 effector genes were identified from tolerant oil palm genotypes when screened with virulent strains of F. oxysporum f.sp. elaeidis (4 and CRT) (Table 7) (Supplementary Figure 3).
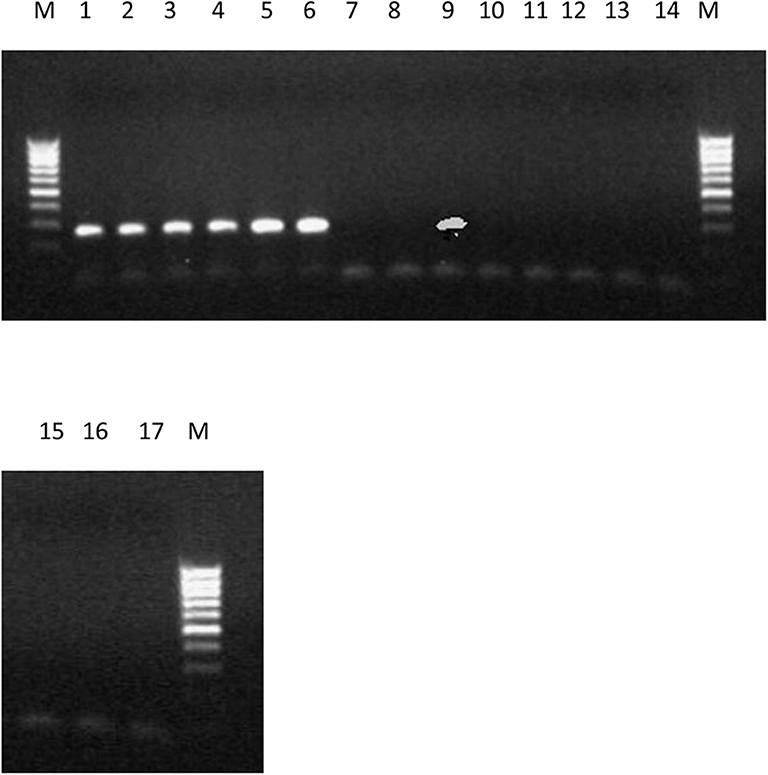
Figure 8. PCR products produced using primer set ORX-F1 and ORX-R1 and DNA from 17 strains of F. oxysporum f.sp. elaeidis from different African countries. The presence or absence of ORX1 effector genes indicates virulence. Lane 1-6(4), CRT, EK1B, PWIIM, PW3M, and PW3A. Lane 7-8(1, 13); lane 9 (MAT10B); lane 10-17(PU4APU, MAT14B, MAT9B, 622, PWA, PW11A, PW49A, and BOPP).
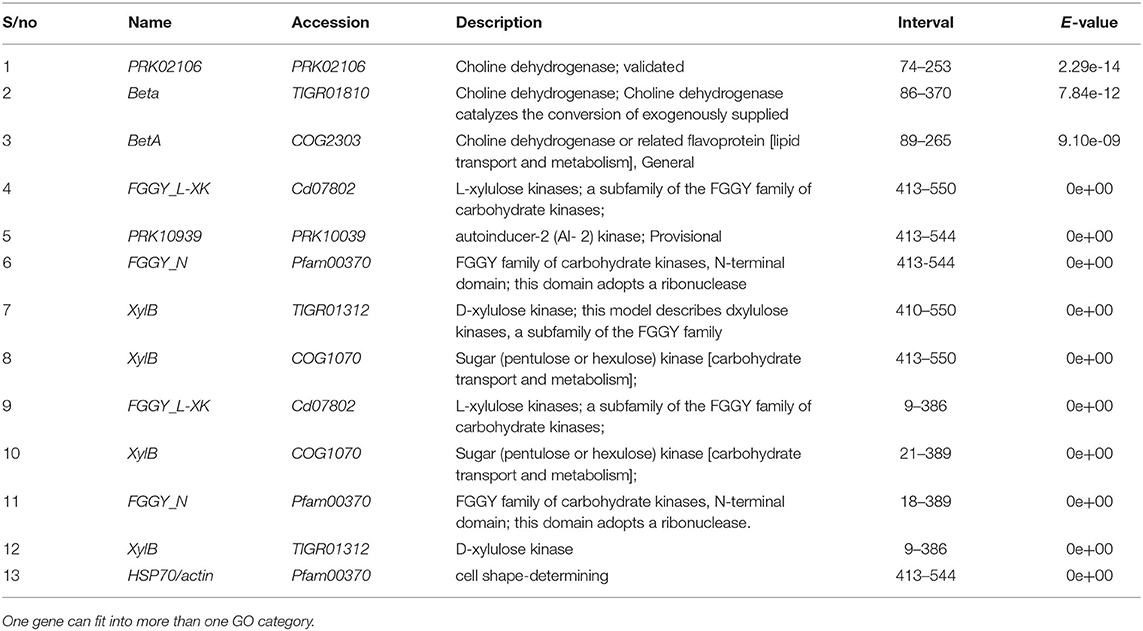
Table 7. Virulence effector genes identified from Fusarium oxysporum f.sp. elaeidis 4 and CRT post inoculation of tolerant and susceptible oil palm genotypes.
The amplification of the ORX1 effector genes in F. oxysporum f.sp. elaeidis (4, and CRT) confirms the virulence of these strains in the pathogenesis trial. It tallies with the results obtained from the disease incidence in Table 5. Since the ORX1 effector gene was not amplified in F. oxysporum f.sp. elaeidis (1, 13, PU4APU, MAT14B, MAT9B, 622, PWA, PW11A, PW49A, and BOPP strains, it implies that they are not pathogenic. This may be responsible for the low disease incidence caused by F. oxysporum f.sp. elaeidis (1, and 13) during pathogenesis. Additional PCR amplification was carried out on the oil palm genotypes using defense gene primers. The results indicate that there was no amplification of the P1 and P2 effector genes, as well as the PR-1 genes (Supplementary Figure 4). This is consistent with the results from the pathogenesis trail during screening oil palm genotypes using F. oxysporum f.sp. elaeidis (4, and CRT).
This study is the first within its limits to report the phylogenetic relationship among Fusarium strains infecting oil palms in some parts of Africa, and identifying the virulent effector genes possessed by African F. oxysporum f.sp. elaeidis strains during pathogenesis in screening Nigerian oil palm genotypes. Despite the strains being identified morphologically, some of the strains posed difficulty in proper identification. For instance, F. oxysporum f.sp. elaeidis (4) strain from Cameroon, which was identified morphologically as F. oxysporum had 95% homology to F. proliferatum when subjected to molecular analysis. Similarly, some fungi strains like Fomes fomentarius and Penicillium simplicissimum, which could not be identified using cultural techniques were identified by comparing their BLAST sequences with the hit in the database. The result of this study is similar to El-Rabbat et al. (2018). Abd Murad et al. (2016) and Zhenyue et al. (2014) likewise found limitations in the use of morphological characteristics in characterizing pathogens in the Gibberella fujikuroi species complex.
The planting of alternative crops, such as maize found in some of the locations where samples were collected could be the cause of the isolation of other Fusarium strains, like F. verticilliodes and F. proliferatum which are pathogenic to these alternative crops. F. proliferatum and F. verticilliodes had been isolated from the diseased roots of banana and other crops (Hsuan et al., 2011; Zakaria and Rahman, 2011).
The variations in the disease incidence on the leaves and roots of the oil palm genotypes during pathogenesis showed that the seedlings responded differently to the strains of F. oxysporum f.sp. elaeidis. Ntsomboh et al. (2015) also found disparities in the disease incidence on the seeds while carrying out pathogenesis. In this study, the disease incidence observed in some tolerant and susceptible oil palm genotypes remained until the termination of the experiment. This result is in contrast with some of the tolerant seedlings recovering from the leaf symptoms (Ntsomboh et al., 2015). The disease severity caused by pathogenic strains of F. oxysporum f.sp. elaeidis in this study could be attributed to the possession of virulent effector proteins secreted in the xylem of the oil palm genotypes. Tagoe (1995) disagrees in stating that the aggressiveness of strains on oil palm is linked to the oil palm genotypes coming from the same parental cross. The color change found in the leaves of the oil palm genotypes indicates the severity of Fusarium wilt disease. This is similar to previous results where disease severity was observed in the leaves of oil palm genotypes treated with a pathogen (Tengoua et al., 2014; Ntsomboh et al., 2015).
The clustering of some F. oxysporum f.sp. elaeidis strains by principal component analysis (PCA) were based on their virulence and the representation of a unique allele. This study is in contrast with the clustering of F. oxysporum f.sp. lentis from Lentis plant not linked to the aggressiveness of the strains (Belabid et al., 2004). Furthermore, Sibounnavong (2012); Debbi et al. (2018) disagree with this study. The variations in the genetic diversity found in the strains could be attributed to ecotypic adaptations leading to mutations. The high genetic diversity shown in some of the Fusarium strains could be connected to the genetic recombination through a sexual cross between well-matched mating strains in the field.
In contrast, the low genetic diversity value found in F. oxysporum f.sp. elaeidis (MAT10B) could be linked to the less active asexual reproduction in that particular location where it was isolated. The coefficient of gene differentiation found in most of the Fusarium strains was high. This high values may be as a result of the host-pathogen relations, virulence ability and long-term resistance to unsuitable environmental conditions. This is similar to the high genetic diversity among M. nivale strains isolated from turfgrass (Mahuku et al., 1998).
The molecular relationship in disease are frequently not understood. An area of possible utilization involves pathogen protein effectors. When pathogens infect, they secrete a lot of virulent effectors to aid the colonization of the host. In this study, close to 193 F. oxysporum EST representing unique effector genes were identified as revealed in the NCBI data bank. These effector proteins represent close to 1.5% of the predicted genes in the genome. Most of these effector genes lack designated functions. This is similar to the inability to assign functions to a more significant part of the genes detected in F. oxysporum vasinfectum (Mcfadden et al., 2006). The PCR amplification of the ORX1 effector genes in some of the Fusarium strains shows that there are presences of pathogenesis genes linked to virulence toward the oil palm genotypes. Houterman et al. (2007) also indicated that the ORX1 genes were used as an indicator in detecting the presence or absence of pathogenicity genes. ORX1 (in planta-secreted oxidoreductase) are conserved in F. oxysporum f.sp. lycopersici strain initiating tomato wilt disease and found on the pathogenesis chromosome of F. oxysporum f.sp. lycopersici (Ma et al., 2010). Houterman et al. (2007) and Rep et al. (2004) also reported that the SIX genes encode for Avr3 protein that confers its virulence to susceptible tomato crops.
The SIX effector proteins were not amplified in all the F. oxysporum f.sp. elaeidis strains screened because the SIX proteins are limited within Fusarium oxysporum clade, and could be used to identify their host specificity. This is similar to F. oxysporum lycopersici which encompass genes encoding for SIX proteins (Lievens et al., 2009). Rep et al. (2004) found the majority of the SIX genes conserved in all F. oxysporum f.sp. lycopersici strains and not in other Fusarium strains. Chakrabarti et al. (2011) reported the SIX gene homolog in F. oxysporum f.sp. vasinfectum found in Australia. In this study, the oil palm genotypes screened did not amplify any of P1, P2, and PR-1 defense genes because of the virulent effector genes produced by the virulent strains of F. oxysporum f.sp. elaeidis during pathogenesis. This is similar to plant pathogenic microorganisms using minute secreted proteins called effectors which can suppress the immune responses from the host plants (Dowd et al., 2004; Houterman et al., 2009).
The Pyranose dehydrogenase 3-like protein detected in this study encodes the PDH genes, which enables the Fusarium strains to degrade the cellulose of the oil palm genotypes. In other studies, it was also found in the Basidiomycetes and wood-degrading fungi (Trametes sp. and Phanerochaete sp.). Jahr et al. (2000) likewise identified a protein which induces bacterial wilt symptoms in tomato crop when inoculated with phytopathogenic bacterium Clavibacter michiganensis. The presence of FGGY_L-XK 1, PRK10939, FGGY_N 1, XylB 1, XylB 2, FGGY_L-XK 2, XylB 3, FGGY_N 2, and XylB 4 detected in susceptible oil palm genotypes during the screening must have led to the increased browning found in the bole of the susceptible oil palm genotypes since the majority of these proteins are responsible for metabolic processes involved in oxidative reactions. Mcfadden et al. (2006) reported that the genes identified from F. oxysporum f.sp. vasinfectum encode for homolog proteins involved in metabolic activities. In the tolerant genotypes, the betA effector gene identified is also essential in the oxidative reaction found in both gram-positive and gram-negative bacteria, Escherichia coli, Staphylococcus xylosus and Sinorhizobium meliloti. The discrimination in the effector genes secreted by the pathogenic strains of F. oxysporum f.sp. elaeidis in the xylem of the susceptible and tolerant oil palm genotypes depends on the level of the compartmentalization of the cell wall.
Many methods can lead to the identification of secreted virulent effector proteins. The potential of using effector proteins has provided a platform to study the basis of pathogenesis. The comparison with genomes of different sequences could shed more light on any exceptional features linked to the pathogenesis of F oxysporum f.sp. elaeidis. However, detailed analysis involving expression of genes will be needed to reveal genes that are unique to F. oxysporum f.sp. elaeidis strains.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
NC and AA envisaged and designed the research. NC carried out the study. NC, AA, TS, and EE executed data evaluation. AA, TS, and EE reviewed the manuscript. All authors have read and approved the completed manuscript.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The authors are grateful to the Nigerian Institute for Oil Palm Research (NIFOR), the University of Lagos and the International Institute of Tropical Agriculture (IITA), and Ibadan for providing the facilities used in this study. The Oil palm Research Institute (OPRI), Kusi, Benso Oil Palm Plantation, Ghana Oil Palm Development Company (GOPDC), Institute of Agricultural Research for Development (IRAD), Ekona, Cameroon, and Cameroon Development Company (CDC) are also acknowledged. Mr. Afrim from the Oil palm Research Institute (OPRI), Kusi is appreciated. Mr. Jonah and Mr. Julius Obaseki from the Plant Pathology Division, NIFOR are exceptionally recognized for helping out in the sample collection. Dr. Tenguoa and Mr. Epoh Toussaint from the Institute of Agricultural Research for Development (IRAD), Ekona, Cameroon are also recognized.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcimb.2020.552394/full#supplementary-material
Abd Murad, N. B., Kusai, N. A., and Zainudin, I. M. (2016). Identification and diversity of Fusarium species isolated from tomato fruits. J. Plant Prot. Res. 56, 231–236. doi: 10.1515/jppr-2016-0032
Altschul, S. F., Gish, W., Miller, W., Eugene, W. M., and David, J. L. (1990). Basic local alignment tool. J. Mol. Biol. 215, 403–410. doi: 10.1016/S0022-2836(05)80360-2
Bari, R., and Jones, J. D. (2009). Role of plant hormones in plant defence responses. Plant Mol. Biol. 69, 473–488. doi: 10.1007/s11103-008-9435-0
Belabid, L., Baum, M., Fortes, Z., Bouznad, Z., and Eujayl, I. (2004). Pathogenic and genetic characterization of algerian isolates of Fusarium oxysporum f. sp. lentis by RAPD and AFLP analysis. Afr. J. Biotechnol. 3, 25–31. doi: 10.5897/AJB2004.000-2005
Booth, C. (1975). The present status of Fusarium taxonomy. Annu. Rev. Phytopathol. 13, 83–93. doi: 10.1146/annurev.py.13.090175.000503
Bowles, D. J. (1990). Defense-related proteins in higher plants. Annu. Rev. Biochem. 59, 873–907. doi: 10.1146/annurev.bi.59.070190.004301
Chakrabarti, A., Rep, M., Wang, B., Ashton, A., Dodds, P., and Ellis, J. (2011). Variation in potential effector genes distinguishing Australian and non-Australian isolates of the cotton wilt pathogen Fusarium oxysporum f. sp. vasinfectum. Plant Pathol. 60, 232–243. doi: 10.1111/j.1365-3059.2010.02363.x
Chidi, N. I., Adekunle, A. A., Eziashi, E. I., and Samuel, T. O. (2018). Evaluation of oil palm Elaeis guineensis Jacq. progenies for Fusarium wilt tolerance using African countries Fusarium oxysporium f.sp. elaeidis. J. Agric. Crop. Res. 6, 79–84.
Cooper, R. M., and Rusli, M. H. (2014). Threat from Fusarium wilt disease of oil palm to south-east Asia and suggested control measures. J. Oil Palm Res. 26, 109–119.
Debbi, A., Boureghda, H., Monte, E., and Hermosa, R. (2018). Distribution and genetic variability of Fusarium oxysporum associated with tomato diseases in algeria and a biocontrol strategy with indigenous Trichoderma spp. Microbiol 9, 82–93. doi: 10.3389/fmicb.2018.00282
Di Pietro, A., and Roncero, M. I. G. (1998). Cloning, expression, and role in pathogenicity of pg1 encoding the major extracellular endopolygalacturonase of the vascular wilt pathogen Fusarium oxysporum. Mol. Plant Microbe Interact. 11, 91–98. doi: 10.1094/MPMI.1998.11.2.91
Diener, A. C., and Ausubel, F. M. (2005). Resistance to Fusarium oxysporum 1, a dominant Arabidopsis disease-resistance gene, is not race specific. Genetics 171, 305–321. doi: 10.1534/genetics.105.042218
Di-Pietro, A., Madrid, M. P., Caracuel, Z., Delgado–Jarana, J., and Roncero, M. I. G. (2003). Fusarium oxysporum: exploring the molecular arsenal of a vascular wilt fungus. Mol. Plant Pathol. 4, 315–325. doi: 10.1046/j.1364-3703.2003.00180.x
Dowd, C., Wilson, L. W., and McFadden, H. (2004). Gene expression profile changes in cotton root and hypocotyl tissues in response to infection with Fusarium oxysporum f. sp. vasinfectum. Mol. Plant Microbe Interact. 17, 654–667. doi: 10.1094/MPMI.2004.17.6.654
El-Rabbat, S., El-Maghraby, O., El-Debaiky, S., and Haider, A. (2018). Isolation and molecular identification of fusarium species from some cereal grains and their products in Egypt. Int. J. Innov. Sci. Eng. Technol. 5, 2348–7968.
Geiser, D. M., Jiménez-Gasco, M., Kang, S., Makalowska, I., Veeraraghavan, N., Ward, T. J., et al. (2004). FUSARIUM-ID v. 1.0: A DNA sequence database for identifying Fusarium. Eur. J. Plant Pathol. 110, 473–479. doi: 10.1023/B:EJPP.0000032386.75915.a0
Goh, C. S., Wicke, B., Faaij, A., Bird, D. N., Schwaiger, H., and Junginger, M. (2016). Linking carbon stock change from land-use change to consumption of agricultural products: alternative perspectives. J. Environ. Manage. 182, 542–556. doi: 10.1016/j.jenvman.2016.08.004
Houterman, P. M., Ma, L., Ooijen, G. V., Vroomen, M. J. D., Cornelissen, B. J. C., and Takken, F. L. W. (2009). The effector protein Avr2 of the xylem-colonizing fungus Fusarium oxysporum activates the tomato resistance protein I-2 intracellularly. Plant J. 25, 970–978. doi: 10.1111/j.1365-313X.2009.03838.x
Houterman, P. M., Speijer, D., Dekker, H. L., de Koster, C. G., Cornelissen, B. J. C., and Rep, M. (2007). The mixed xylem sap proteome of Fusarium oxysporum-infected tomato plants. Mol. Plant Pathol. 8, 215–221. doi: 10.1111/j.1364-3703.2007.00384.x
Hsuan, H. M., Salleh, B., and Zakaria, L. (2011). Molecular identification of Fusarium species in Gibberella fujikuroi species complex from rice, sugarcane and maize from peninsular Malaysia. Int. J. Mol. Sci. 12, 6722–6732. doi: 10.3390/ijms12106722
Jahr, H., Dreier, J., Meletzus, D., Bahro, R., and Eichenlaub, R. (2000). The endo-beta-1, 4-glucanase CelA of Clavibacter michiganensis sub sp michiganensis is a pathogenicity determinant required for induction of bacterial wilt of tomato. Mol. Plant Microbe Interact. 13, 703–714. doi: 10.1094/MPMI.2000.13.7.703
Komada, H. (1975). Development of a selective medium for quantitative isolation of Fusarium oxysporum from natural soil. Rev. Plant Prot. Res. 8, 114–124.
Leslie, J. F., and Summerell, B. A. (2006). The Fusarium Laboratory Manual, 1st ed. Asia State Avenue: Blackwell Publishing.
Lievens, B., Houterman, P. M., and Rep, M. (2009). Effector gene screening allows unambiguous identification of Fusarium oxysporum f. sp. lycopersici races and discrimination from other formae speciales. FEMS Microbiol. Lett. 300, 201–215. doi: 10.1111/j.1574-6968.2009.01783.x
Ma, L. J., van der Does, H. C., Borkovich, K. A., Coleman, J. J., Daboussi, M. J., Di Pietro, A., et al. (2010). Comparative genomics reveals mobile pathogenicity chromosomes in Fusarium. Nature 464, 367–373. doi: 10.1038/nature08850
Mahuku, G. S., Hsiang, T., and Yang, L. (1998). Genetic diversity of Microdochium nivale isolates from turfgrass. Mycol. Res. 102, 559–567. doi: 10.1017/S0953756297005340
Mcfadden, H. G., Wilson, I. W., Chapple, R. M., and Dowd, C. (2006). Fusarium wilt (Fusarium oxysporum f. sp. vasinfectum) genes expressed during infection of cotton (Gossypium hirsutum). Mol. Plant pathol. 7, 87–101. doi: 10.1111/j.1364-3703.2006.00327.x
Nei, M., and Kumar, S. (2000). Molecular Evolution and Phylogenetics. New York, NY: Oxford University Press.
Noumouha, E. N., Ghislain, N. E. N., Désiré, A., Benjamin, A., Jean-Noël, K., Emmanuel, I. A., et al. (2014). Assessment of nigerian wild oil palm (Elaeis guineensis Jacq.) populations in crosses with delitesters. J. Plant Breed. Genet. 2, 77–86. doi: 10.15580/GJPBCS.2016.1.111015157
Ntsomboh, N. G., Madi, G., Nyaka, N. A., Nsimi, M. A., Epoh, N. T., Kato, S. N., et al. (2015). Vascular wilt disease tolerance status of some oil palm (Elaeis guineensis Jacq.) Progenies in relation to local strains of Fusarium oxysporum f. sp. elaeidis in cameroon. Int. J. Curr. Res. Biosci. Plant Biol. 2, 111–122.
Ntsomboh, N. G., Ngando Ebongue, G. F., Koona, P., Bell, J. M., Youmbi, E., Ngalle Hermine, B., et al. (2012). Control approaches against vascular wilt disease of Elaeis guineensis jacq. caused by Fusarium oxysporum f.sp. elaeidis. J. Biol. Life Sci. 3, 160–173. doi: 10.5296/jbls.v3i1.992
Ojuederie, O. B., Igwe, D. O., Okuofu, S. I., and Faloy, B. (2013). Assessment of genetic diversity in some Moringa oleifera Lam. Landraces from Western Nigeria using RAPD markers. African J. Plant Sci. Biotechnol. 7, 15–20.
Oritsejafor, J. J. (1982). Factors Affecting the Survival of Fusarium oxysporum f.sp. elaeidis in the Soil. (Ph.D. Thesis), University of Ibadan.
Paterson, R. R. M., Sariah, M., and Lima, N. (2013). How will climate change affect oil palm fungal diseases? Crop Protect. 46, 113–120. doi: 10.1016/j.cropro.2012.12.023
Renard, J. L., Gascon, J. P., and Bachy, A. (1972). Recherches sur la fusariose du palmier à huile (Bilingue fr. –angl.). Oléagineux 27, 581–591.
Rep, M., Does, H. C. V. D., Meijer, M., Wijk, R. V., Houterman, P. M., Dekker, H. L., et al. (2004). A small, cysteine-rich protein secreted by Fusarium oxysporum during colonization of xylem vessels is required for I-3-mediated resistance in tomato. Mol. Microbiol. 53, 1373–1383. doi: 10.1111/j.1365-2958.2004.04177.x
Rival, A. (2017). Breeding the oil palm (Elaeis guineensis Jacq.) for climate change. Oilseeds Fats Crops Lipids 24:107. doi: 10.1051/ocl/2017001
Sibounnavong, P. (2012). Screening of Emericella nidulans for biological control of tomato Fusarium wilt in Lao PDR. J. Agri. Technol. 8, 241–260.
Tagoe, S. M. A. (1995). Reaction of Some Oil Palm (Elaeis guineensis Jacq.) Tenera Progenies to Fusarium oxysporum f. sp. elaeidis Causal Agent of Vascular Wilt Disease of Oil Palm. (Master of Philosophy thesis), Legon: University of Ghana.
Tamura, K., Stecher, G., Peterson, D., Filipski, A., and Kumar, S. (2013). MEGA6: molecular evolutionary genetics analysis, version 6.0. J. Mol. Biol. Evol. 30, 2725–2729. doi: 10.1093/molbev/mst197
Tengoua, F. F. (1994). Project STD3 Fusariose. Rapport Bilan de la Prospection de la Maladie Dans les Palmeraies du Cameroun. Ekona: Ekona Regional Research Centre.
Tengoua, F. F., Hanafi, M. M., Idris, A. S., Jugah, K., Azwa, J. N. M., Hasmah, M., et al. (2014). Effects of micronutrients-enriched fertilizers on basal stem rot disease incidence and severity on oil palm (Elaeis guineensis Jacq.) seedlings. Am. J. Appl. Sci. 11, 1841–1859. doi: 10.3844/ajassp.2014.1841.1859
Yeh, F. C., Boyle, R., Yang, R. C., Ye, Z., Mao, J. X., Yeh, D. P. O. P. G. E. N. E., et al. (1999). Available online at: http://www.ualberta.ca/~fyeh/popgene.html (accessed May 16, 2018).
Zakaria, L., and Rahman, N. H. A. (2011). Endophytic Fusarium spp. from wild banana (Musa acuminata) roots. African J. Microbiol. Res. 5, 3600–3602. doi: 10.5897/AJMR11.298
Keywords: Fusarium oxysporum f.sp. elaeidis, Fusarium wilt, pathogenesis, oil palm, virulence, incidence, severity, genetic diversity
Citation: Chidi NI, Adekunle AA, Samuel TO and Eziashi EI (2020) Molecular Identification of Secreted Effector Genes Involved in African Fusarium oxysporum f.sp. elaeidis Strains Pathogenesis During Screening Nigerian Susceptible and Tolerant Oil Palm (Elaeis guineensis Jacq.) Genotypes. Front. Cell. Infect. Microbiol. 10:552394. doi: 10.3389/fcimb.2020.552394
Received: 16 April 2020; Accepted: 20 August 2020;
Published: 06 October 2020.
Edited by:
Brian Wickes, The University of Texas Health Science Center at San Antonio, United StatesReviewed by:
Lukasz Stepien, Institute of Plant Genetics (PAN), PolandCopyright © 2020 Chidi, Adekunle, Samuel and Eziashi. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Nnamdi Ifechukwude Chidi, c3dpc3Nzd2lzc3lAeWFob28uY28udWs=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.