- Institute of Microbiology of the Czech Academy of Sciences, Prague, Czechia
Pertussis, also known as whooping cough, is a resurging acute respiratory disease of humans primarily caused by the Gram-negative coccobacilli Bordetella pertussis, and less commonly by the human-adapted lineage of B. parapertussisHU. The ovine-adapted lineage of B. parapertussisOV infects only sheep, while B. bronchiseptica causes chronic and often asymptomatic respiratory infections in a broad range of mammals but rarely in humans. A largely overlapping set of virulence factors inflicts the pathogenicity of these bordetellae. Their genomes also harbor a pathogenicity island, named bsc locus, that encodes components of the type III secretion injectosome, and adjacent btr locus with the type III regulatory proteins. The Bsc injectosome of bordetellae translocates the cytotoxic BteA effector protein, also referred to as BopC, into the cells of the mammalian hosts. While the role of type III secretion activity in the persistent colonization of the lower respiratory tract by B. bronchiseptica is well recognized, the functionality of the type III secretion injectosome in B. pertussis was overlooked for many years due to the adaptation of laboratory-passaged B. pertussis strains. This review highlights the current knowledge of the type III secretion system in the so-called classical Bordetella species, comprising B. pertussis, B. parapertussis, and B. bronchiseptica, and discusses its functional divergence. Comparison with other well-studied bacterial injectosomes, regulation of the type III secretion on the transcriptional and post-transcriptional level, and activities of BteA effector protein and BopN protein, homologous to the type III secretion gatekeepers, are addressed.
Introduction
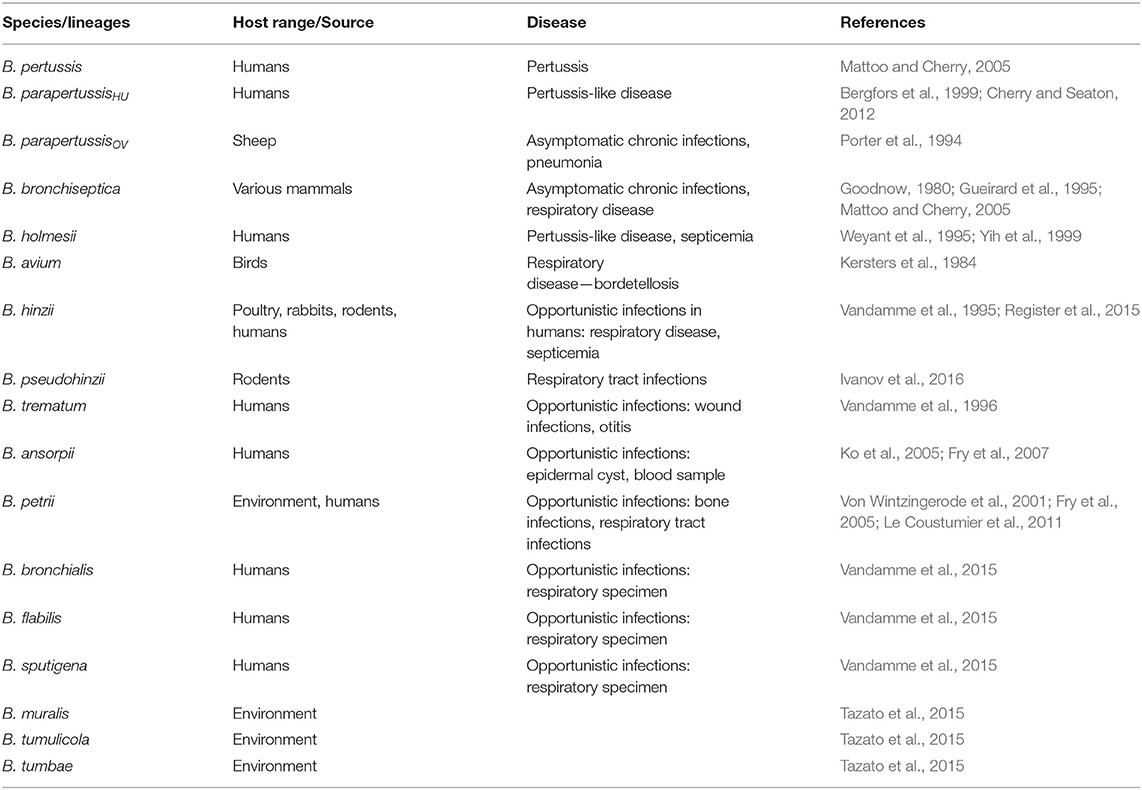
The genus Bordetella (Alcaligenaceae, phylum Betaproteobacteria) currently embraces 16 species of the Gram-negative coccobacilli. Its members include important respiratory pathogens of mammals and humans with limited genetic diversity, the so-called classical Bordetella species, namely B. pertussis, B. parapertussis, and B. bronchiseptica. The genus further comprises less extensively studied and phylogenetically distinct Bordetella species collectively referred to as non-classical bordetellae, comprising pathogens, opportunistic pathogens, and environmental isolates (Table 1). The phylogenetic analysis of the genus suggests that the animal-associated species likely evolved from their ancestors living in soil and/or water (Hamidou Soumana et al., 2017). The diversification and speciation in the genus were accompanied by the gain and loss of multiple genes, including genes for bacterial protein toxins, protein secretion systems, and other virulence factors (Linz et al., 2016, 2019). The presence of the genes encoding Bordetella protein toxins, consisting of adenylate cyclase toxin, pertussis toxin, and dermonecrotic toxin distinguishes classical Bordetella species from the non-classical bordetellae (Linz et al., 2016). However, pertussis toxin is solely produced by B. pertussis due to mutations in the ptx promoter (Arico and Rappuoli, 1987; Parkhill et al., 2003), and dermonecrotic toxin appears to be also imported into B. avium that causes respiratory disease of birds called bordetellosis (Linz et al., 2016). For detailed characterization of acquisition and loss of virulence-associated factors during the evolution of the genus Bordetella the reader is referred to a recent work of Linz B. and colleagues (Linz et al., 2016). This review aims to explore and discuss the type III secretion system (T3SS) in classical Bordetella species, its regulation and mechanism of action, and its role in Bordetella infections that is yet to be explored for B. pertussis. Remarkably, bsc-btr loci encoding Bordetella T3SS and its regulatory proteins are also present in B. ansorpii, but absent from genomes of all other non-classical bordetellae. A comparison of the genetic organization of bsc-btr loci of B. ansorpii and classical Bordetella species will also be provided.
The human-adapted B. pertussis is the primary causative agent of pertussis, also known as whooping cough, a contagious, prolonged respiratory illness that used to be the major cause of infant mortality in the pre-vaccine era (Mattoo and Cherry, 2005). Pertussis remains one of the least controlled vaccine-preventable infectious diseases. In the recent years, an increase in pertussis incidence and/or pertussis outbreaks have been experienced in a number of most developed countries with high vaccine coverage, including the Czech Republic, U.S., U.K., Netherlands, and Australia (Fabianova et al., 2010; Spokes et al., 2010; Burns et al., 2014; Sealey et al., 2016). Unrecognized or mildly symptomatic B. pertussis infections in adolescents and adults are common and represent a threat to unvaccinated infants to whom the disease can be fatal (Cherry, 2019). The key contributing factors of increased pertussis incidence are still under debate. Besides greater awareness, improved diagnostics and genetic changes in circulating B. pertussis strains, the major cause of pertussis resurgence appears to be the switch from whole-cell pertussis (wP) to the less reactogenic but less effective acellular pertussis (aP) vaccines. These confer significantly shorter-lasting protection and are not efficient in preventing the colonization of the vaccinated individuals by B. pertussis and pathogen spread in the population (reviewed in Mooi et al., 2014; Cherry, 2019; Kapil and Merkel, 2019).
Other Bordetella species, the human-adapted lineage of B. parapertussisHU and the non-classical species B. holmesii can also cause pertussis-like disease in humans, although generally accompanied by milder symptoms and shorter illness duration (Bergfors et al., 1999; Yih et al., 1999; Cherry and Seaton, 2012; Rodgers et al., 2013). The ovine-adapted lineage of B. parapertussisOV colonizes only sheep with no or little transmission to humans (Van Der Zee et al., 1996). In contrast, B. bronchiseptica infects a variety of mammals and causes diverse pathologies that range from typical chronic and often asymptomatic respiratory infections up to more acute diseases, such as the kennel cough in dogs, bronchitis in cats, bronchopneumonia and atrophic rhinitis in piglets, and snuffles in rabbits (Goodnow, 1980; Mattoo and Cherry, 2005). B. bronchiseptica infections in humans are rare and occur mostly in immunocompromised patients, children, and in elderly that are in contact with animals (Goodnow, 1980; Gueirard et al., 1995; Mattoo and Cherry, 2005). Nevertheless, clustering of B. bronchiseptica strains into two distinct B. bronchiseptica subpopulations, complex I, primarily of animal origin (68%), and complex IV that is primarily isolated from humans (80%) was reported based on multilocus sequence typing (MLST), distribution of insertion sequence elements (ISEs) and whole-genome sequence comparisons (Diavatopoulos et al., 2005; Park et al., 2012).
The classical Bordetella species are phylogenetically closely related, despite a different range of their mammalian hosts and diverse pathologies they cause. Hence, it was proposed to classify them as subspecies, rather than species (Musser et al., 1986). The B. pertussis, B. parapertussisHU, and B. parapertussisOV likely evolved very recently and independently from different lineages of B. bronchiseptica-like ancestors (Van Der Zee et al., 1996, 1997; Parkhill et al., 2003). Specifically, B. parapertussisHU and B. parapertussisOV appear to have evolved from a B. bronchiseptica complex I-like ancestor, whereas B. pertussis may have shared a common ancestor with complex IV strains of human-associated lineages of B. bronchiseptica (Diavatopoulos et al., 2005; Park et al., 2012). The speciation of B. pertussis and B. parapertussis has been accompanied by a large-scale gene loss and inactivation. This resulted in a respective reduction of their genome size by 22 and 9%, compared to B. bronchiseptica species (cf. B. pertussis Tohama I ~ 4.1 Mbp, B. parapertussisHU 12822 ~ 4.8 Mbp, B. parapertussisOV Bpp5 ~ 4.9 Mbp, B. bronchiseptica RB50 ~ 5.3 Mbp). These genomic rearrangements were apparently mediated by the acquisition and expansion of insertion sequence elements (ISEs) and recombination between their copies (Parkhill et al., 2003; Preston et al., 2004; Weigand et al., 2019). Interestingly, host adaptation and speciation of classical Bordetella species likely was a consequence of loss of function, rather than a gain of function. As a result, the differences in the virulence of the classical Bordetella species are related to the loss of regulatory or control functions and/or sequence polymorphism (Parkhill et al., 2003; Cummings et al., 2004).
A Brief Overview of the Virulence Factors of Classical Bordetellae
The three classical Bordetella species produce a largely overlapping array of virulence factors that are involved in the colonization of the host respiratory tract, immune evasion, and transmission to new hosts. These comprise (i) adhesins, such as filamentous hemagglutinin and fimbriae, (ii) a large number of autotransporters involved in the adhesion and/or resistance to complement, including the adhesion molecule pertactin and complement evasion factor Vag8, and (iii) protein toxins consisting of adenylate cyclase toxin and dermonecrotic toxin (Mattoo et al., 2001; Parkhill et al., 2003; Hovingh et al., 2017). The adenylate cyclase toxin is a potent immunomodulatory toxin that subverts host innate and adaptive immune defenses by its adenyl cyclase activity (reviewed in Fedele et al., 2017) while the dermonecrotic toxin is associated with induction of turbinate atrophy in pigs and appears to have neurotoxic activity (Brockmeier et al., 2002; Teruya et al., 2020). By contrast, pertussis toxin (PTX) that catalyzes the ADP-ribosylation of the alpha subunit of heterotrimeric G proteins of the Gi/o class is produced exclusively by B. pertussis species. The PTX production is responsible for systemic symptoms of pertussis disease, such as leukocytosis that is associated with the mortality in infants (Pierce et al., 2000; Carbonetti, 2010). The classical Bordetella species also differ in the levels of expression of the type III secretion system (T3SS) in vitro. Unlike B. bronchiseptica and ovine B. parapertussisOV, the human-adapted B. pertussis and B. parapertussisHU species have the expression of their T3SS blocked at a post-transcriptional level when grown in Stainer-Scholte medium (Mattoo et al., 2004). Therefore, the function of the T3SS in human-adapted Bordetella species was overlooked for many years. However, isolates of B. pertussis express the T3SS effector protein BteA/BopC (Hegerle et al., 2013). In addition, they produce a functional T3SS upon passage on an infected animal or eukaryotic cells (Fennelly et al., 2008; Gaillard et al., 2011; Bibova et al., 2015), or when grown in media with limiting glutamate and/or iron concentrations (Brickman et al., 2011; Hanawa et al., 2016).
Bordetella Injectosome
The T3SS injectosome is a sophisticated protein-export apparatus that enables the delivery of bacterial effector proteins directly from bacterial cytosol into the cytosol of the host cells through a conduit spanning the two bacterial membranes and the plasma membrane of the target cell. It consists of an extracellular needle-like appendage with a central channel of ~2 nm in diameter, which protrudes from the bacterial surface and is linked to a cell wall-embedded secretion system machinery steered by the associated cytoplasmic components, as depicted in Figure 1 (reviewed in Galan et al., 2014; Notti and Stebbins, 2016).
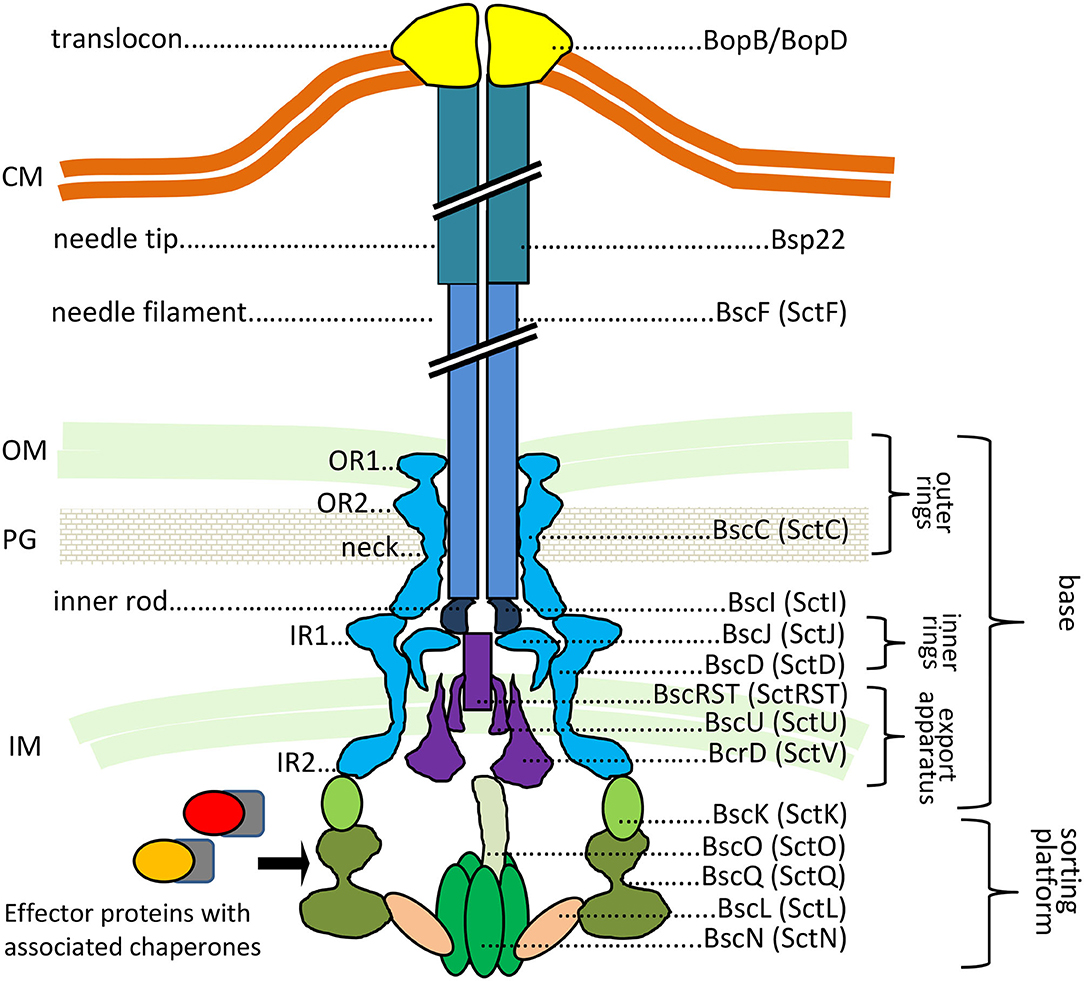
Figure 1. The predicted structure of Bordetella injectosome. The diagram is based on Galan et al. (2014), Hu et al. (2017), Park et al. (2018). Known or predicted functions and locations of Bordetella proteins are indicated. In Bordetella spp., the injectosome needle tip is formed by a helical assembly of the Bsp22 protein (Medhekar et al., 2009). See the text for further details. CM, cytoplasmic membrane; OM, outer membrane; PG, peptidoglycan; IM, inner membrane. The proteins are color-coded based on coding genes shown in Figure 3A.
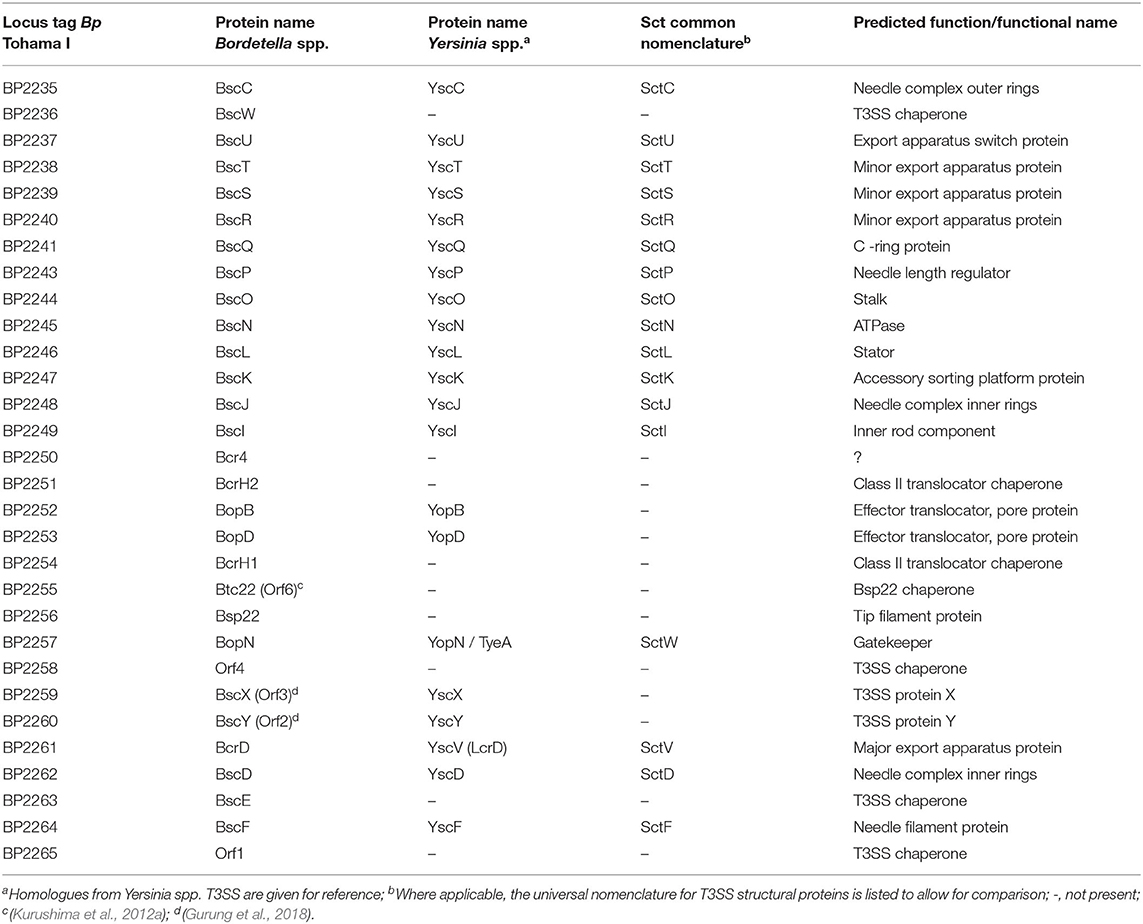
The genes for the components of the T3SS injectosome are generally clustered on a mobile genetic element, a pathogenicity island or a plasmid, and appear to have been acquired as an intact genetic block by horizontal gene transfer (Hueck, 1998; Hacker and Kaper, 2000). T3SS injectosomes are widespread among gram-negative bacteria. They are present in not only animal and plant pathogens but also in insect and amoeba pathogens, and are also essential to some symbionts (Troisfontaines and Cornelis, 2005). First genes in Bordetella spp. with a high degree of similarity to the genes coding for components of Yersinia injectosome were reported by Yuk et al. (1998). In the following years, the whole 22.5 kbp bsc (Bordetella secretion) locus encoding the Bordetella injectosome, and the adjacent 11 kbp btr (Bordetella type III regulation) locus, encoding injectosome-regulatory proteins, have been described (Kerr et al., 1999; Fauconnier et al., 2001; Mattoo et al., 2004; Ahuja et al., 2016). Genomic analysis of classical bordetellae showed that Bordetella injectosome is their most conserved secretion system (Park et al., 2012). The gene positions within the 33.5 kbp btr-bcs loci (btr BP2226-2234, bsc BP2235-BP2265) of B. pertussis Tohama I match the organization of the genes in bsc-btr loci (bsc BB1608-BB1637, btr BB1638-BB1646) of B. bronchiseptica RB50. In addition, the vast majority of nucleotide substitutions are silent or result in conservative amino acid substitutions, which implies the evolutionary pressure for the preservation of the T3SS in B. pertussis (Mattoo et al., 2004). However, compared to B. bronchiseptica RB50 genome, the B. pertussis Tohama I genomic region harboring the btr-bsc underwent inversion, likely due to an ISE-mediated rearrangement (see Figure 2). Interestingly as also depicted in Figure 2, although the bsc-btr loci (bsc BPP5_1370-BPP5_1399, btr BPP5_1400-BPP5_1408) of B. parapertussisOV Bpp5 are homologous and intact, the bsc-btr loci (bsc BPP2211-BPP2240, btr BPP2241-BPP2249) of the human-adapted B. parapertussisHU 12882 was suggested to contain pseudogenes for regulatory (BPP2241) and structural (BPP2215) proteins (Parkhill et al., 2003; Linz et al., 2016). The functionality of T3SS in human-adapted B. parapertussisHU thus remains to be clarified. Remarkably, the bsc-btr loci are also present in one non-classical Bordetella species, B. ansorpii that has so far been isolated only twice (Ko et al., 2005; Fry et al., 2007). The analysis of these loci in B. ansorpii NCTC1364 (bsc SAMEA1982600_01974-SAMEA1982600_02005, btr SAMEA1982600_02006-SAMEA1982600_02014) reveals a homologous organization, suggesting that B. ansorpii and classical bordetellae had a common ancestor. However, three different genes (tagged as SAMEA1982600_01983, SAMEA1982600_01984, SAMEA1982600_01985) are present at the position of the gene encoding the Bsp22 tip filament protein of classical bordetellae as highlighted in Figure 2.
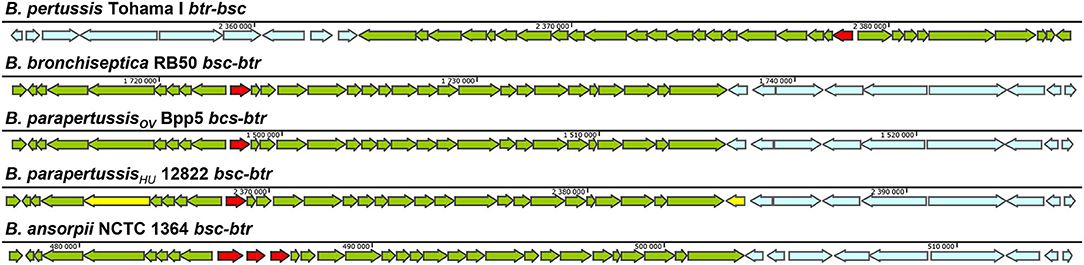
Figure 2. Genetic organization of the type III regulation btr and secretion bsc loci in different Bordetella species. The genetic organization of btr-bsc loci of B. pertussis Tohama I (NC_002929.2, btr BP2226—BP2234, bsc BP2235—BP2265) and bsc-btr loci of B. bronchiseptica RB50 (NC_002927.3, bsc BB1608-BB1637, btr BB1638-BB1646), B. parapertussisOV Bpp5 (NC_018828.1, bsc BPP5_1370-BPP5_1399, btr BPP5_1400-BPP5_1408), B. parapertussisHU 12822 (NC_002928.3, bsc BPP2211-BPP2240, btr BPP2241-BPP2249) and B. ansorpii NCTC13364 (FKBS01000014.1, contig: ERS088257SCcontig000014, bsc SAMEA1982600_01974-SAMEA1982600_02005, btr SAMEA1982600_02006-SAMEA1982600_02014) is shown. The genes of btr and bsc loci are depicted in blue and green, respectively. The putative pseudogenes BPP2215 and BPP2241 in bsc-btr loci of B. parapertussisHU 12822 are highlighted in yellow (Parkhill et al., 2003) whereas the Bsp22 tip filament protein gene in bsc loci of classical bordetellae and 3 orfs that replaced this gene in B. ansorpii are highlighted in red. Arrows indicate the direction of transcription.
The injectosome of Bordetella spp. encoded in the aforementioned bsc locus does not fit comfortably into any of the seven phylogenetic families of the non-flagellar T3SS being classified by loci organization and amino acid sequences of the encoded proteins, and consisting of families Ysc, Inv-Mxi-Spa, Ssa-Esc, Hrc-Hrp 1, Hrc-Hrp 2, the Rhizobiales and the Chlamydiales (Troisfontaines and Cornelis, 2005). The Bsc system of Bordetella spp. could nevertheless form a subgroup within the Ysc family of injectosomes, which comprises the Ysc system of Yersinia spp., Asc system of Aeromonas spp., Lsc system of Photorhabdus luminescens, Psc system of Pseudomonas aeruginosa, and the Vsc system of Vibrio parahaemolyticus (Pallen et al., 2005; Troisfontaines and Cornelis, 2005). As depicted in Figures 3A,B, several homologous genes in the Bordetella bsc locus (e.g., bscI to bscL, as well as the bscN to bscU genes of Bordetella spp.) exhibit the same relative positions as the genes of the plasmid-encoded ysc-yop cluster of Y. enterocolitica W22703. However, the relative positions of other homologous genes differ, with bscN immediately following bscL in Bordetella, while yscN is not adjacent to yscL in Y. enterocolitica (Fauconnier et al., 2001). However, the bsc-encoded proteins still show strong sequence similarity to the well-described homologous proteins found in Yersinia spp. as listed in Table 2 (Yuk et al., 1998; Kerr et al., 1999; Fauconnier et al., 2001). The precise composition and function of the Bordetella injectosome still need to be verified experimentally. Nevertheless, based on studies of the homologous injectosomes of Yersinia and Salmonella (reviewed in Dewoody et al., 2013; Galan et al., 2014; Galan and Waksman, 2018; Wagner et al., 2018), its structural organization can be predicted with a high degree of confidence to resemble the arrangement depicted in Figure 1.
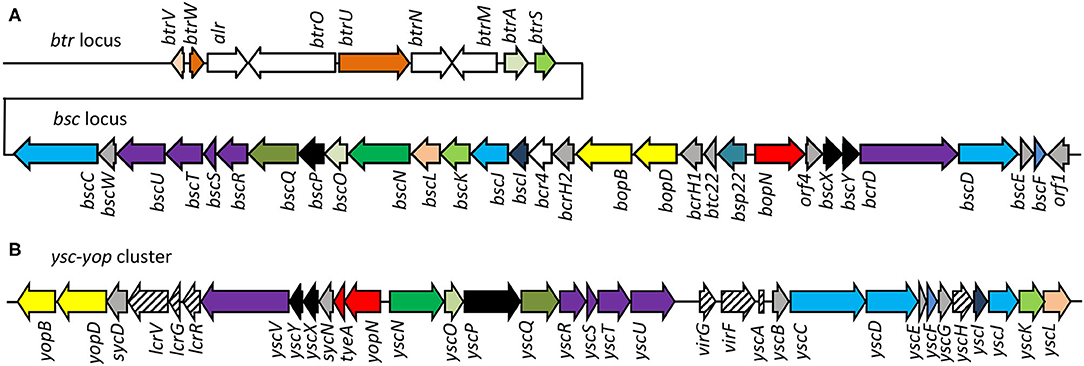
Figure 3. Comparison of genetic organization of B. pertussis btr-bsc loci and Yersinia Yop-Ysc cluster. (A) The type III regulation btr and secretion bsc loci of the B. pertussis Tohama I genome, accession number NC_002929.2, btr genes BP2226 – BP2234, bsc genes BP2235—BP2265. (B) The Yop-Ysc cluster of Yersinia enterocolitica plasmid pYVe227, accession number AF102990.1, between yopB and ycL. Arrows indicate the direction of transcription. Homologies of the encoded injectosome proteins are indicated by the same color code, chaperones are colored in gray.
By analogy, the base or the so-called basal body of Bordetella injectosome, embedded in the bacterial envelope, would consist of two membrane-spanning ring structures. The outer membrane ring would be built by oligomerization of BscC and the two concentric inner membrane rings would be formed by oligomerization of BscD on the outside and BscJ on the inside. The basal body would connect with the inner membrane export apparatus, formed by the BscRSTU and BcrD (SctV homolog) components. The latter would interact with the cytosolic sorting platform and additional regulatory proteins to allow for hierarchy in protein secretion, substrate unfolding, and export. By analogy to Salmonella, the sorting platform would be composed of three scaffolding proteins BscK, BscQ, BscL, and an ATPase BscN, linked to the export apparatus through another component, BscO (Hu et al., 2017). The needle filament composed of polymerized BscF would then attach to the basal body through the inner rod made from the BscI protein, and extend into the extracellular milieu forming a rigid hollow conduit for the secretion of proteins (Figure 1). The Bordetella needle filament on its distal end appears to be capped by another hollow helical assembly composed of the filament tip protein Bsp22 (Medhekar et al., 2009). The Bsp22 protein undergoes spontaneous polymerization and requires its chaperone Btc22 (formerly Orf6) for stabilization in the bacterial cytoplasm (Kurushima et al., 2012a; Villarino Romero et al., 2013). Bsp22 also binds directly to BopD, a component of the Bordetella translocon pore and it is essential for effector protein delivery into the target cells (Medhekar et al., 2009). It is thus assumed that the Bsp22 polymer forms a long flexible connecting channel that links the needle filament to the translocon pore inserted within the target cell membrane, as depicted in Figure 1. Indeed, similar needle extensions were described to be formed by the filament tip protein EspA of enteropathogenic Escherichia coli (EPEC) (Daniell et al., 2001; Sekiya et al., 2001; Wang et al., 2006). Intriguingly, the EspA filament appears to be eliminated upon attachment of EPEC to target cells (Knutton et al., 1998) and the function of the Bsp22 and EspA-formed needle extensions is currently unknown. It has been speculated that EspA filaments may cross the barrier of the mucous layer and help in adhesion to epithelial cells and/or in biofilm formation (Daniell et al., 2001; Cleary et al., 2004; Moreira et al., 2006). The Bordetella proteins BopD and BopB hetero-oligomerize with unknown stochiometry within the host plasma membrane and form the translocon pore that functions as the conduit for translocation of T3SS effector proteins into host cell cytosol (Kuwae et al., 2003; Nogawa et al., 2004), as depicted in Figure 1. Both BopD and BopB proteins are required for the pore-forming hemolytic activity of the injectosome on red blood cells but are not needed for in vitro secretion of other T3SS substrates (Kuwae et al., 2003; Nogawa et al., 2004).
Additional regulatory and structural components, presumably involved in the timing of protein secretion by the injectosome, are encoded within the Bordetella bsc locus. These include proteins BscX (formerly Orf3, homologous to T3SS protein X) and BscY (formerly Orf2, homologous to T3SS protein Y), BscP, and BopN (Table 2). The BscX and BscY proteins would by analogy with Yersinia YscX and YscY orchestrate the secretion of early substrates through their interaction with BcrD (Diepold et al., 2012). The YscX- and YscY-like proteins are unique to the Ysc family of injectosomes and are not encoded within other injectosome families (Gurung et al., 2018). BscP would by analogy with SctP protein control the length of BscF needle filament by a poorly understood mechanism (reviewed in Diepold and Wagner, 2014). Finally, the BopN protein would activate effector protein secretion upon contact with the host cell, in the so-called “second substrate switching event,” as deduced from its homology to the gatekeeper protein, SctW (reviewed in Portaliou et al., 2016). The BopN function in Bordetella injectosome, however, remains unclear and will be discussed in the section on effector proteins. The bsc locus further encodes the chaperones for the respective components of the injectosome (Table 2), e.g., Bsp22 chaperone Btc22 (formerly Orf6), the putative chaperone BcrH2 that co-immunoprecipitates with the BopB-BopD complex from bacterial cytosol, and one additional protein, called Bcr4, with unclear activity (Nogawa et al., 2004; Kurushima et al., 2012a; Nishimura et al., 2018).
Type III Secretion Regulation in Bordetella
The bsc-encoded genes of the Bordetella injectosome are induced during infection, and are responsive to blood or serum exposure, and increased CO2 concentrations (Gaillard et al., 2011; Hester et al., 2012; Bibova et al., 2015; Gestal et al., 2018; Van Beek et al., 2018; Wong et al., 2019). Other stimuli that can activate T3SS expression and secretion in bordetellae are the stringent response induced by iron limitation and/or starvation for carbon source, as depicted in Figure 4 (Brickman et al., 2011; Kurushima et al., 2012b; Hanawa et al., 2016). However, upon the internalization of bordetellae into macrophages the transcription of injectosome genes is down-regulated (Rivera et al., 2019; Petrackova et al., 2020). The bsc-encoded genes are under the control of Bordetella master virulence regulatory system BvgAS. This two-component system is composed of the membrane-bound sensor kinase BvgS and of its phosphorylation substrate, the DNA-binding response regulator protein BvgA, which coordinates expression of hundreds of genes (Hot et al., 2003; Cummings et al., 2006; Nicholson, 2007; Moon et al., 2017). The system appears to function as a “rheostat” controlling a spectrum of phenotypic modes in response to environmental clues. Nevertheless, the exact nature of the signals and the mechanism by which these signals are perceived and integrated into the BvgAS regulon remains unknown (reviewed in Mattoo et al., 2001; Chen and Stibitz, 2019).
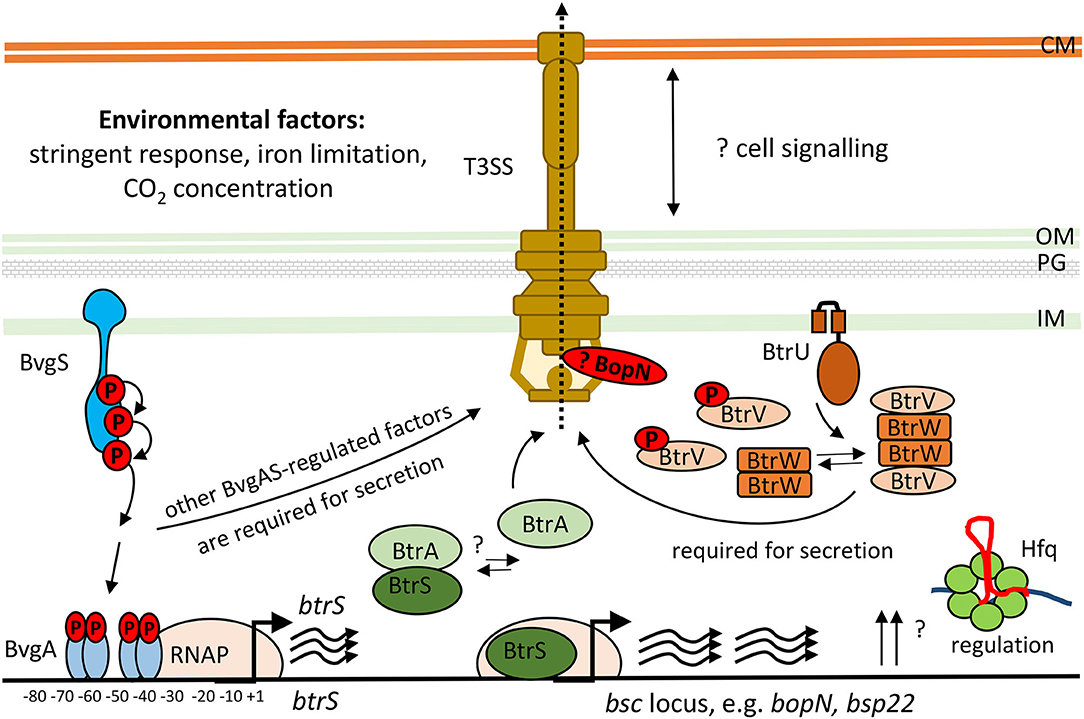
Figure 4. Scheme of transcriptional and post-transcriptional regulation of T3SS in Bordetella species. The two-component system, BvgAS, composed of the sensor kinase BvgS and the response regulator BvgA activates expression of the extracytoplasmic function (ECF) sigma factor BtrS and regulates additional factors necessary for T3SS function (Mattoo et al., 2004; Moon et al., 2017). The activity of BtrS leads to transcription of several bsc-encoded genes while being counteracted by the T3SS-exported anti-sigma factor, BtrA (Mattoo et al., 2004; Kurushima et al., 2012c; Ahuja et al., 2016). See text for further details. Besides, the partner-switcher proteins BtrU, BtrW, and BtrV control secretion through the injectosome via a series of serine phosphorylation and dephosphorylation events by a yet unknown mechanism (Mattoo et al., 2004; Kozak et al., 2005). BopN protein homologous to the gatekeeper StcW might also regulate the secretion post-transcriptionally. Also RNA chaperone Hfq is required for the expression of T3SS genes in B. pertussis (Bibova et al., 2015; Dienstbier et al., 2019). CM, cytoplasmic membrane; OM, outer membrane; PG, peptidoglycan; IM, inner membrane.
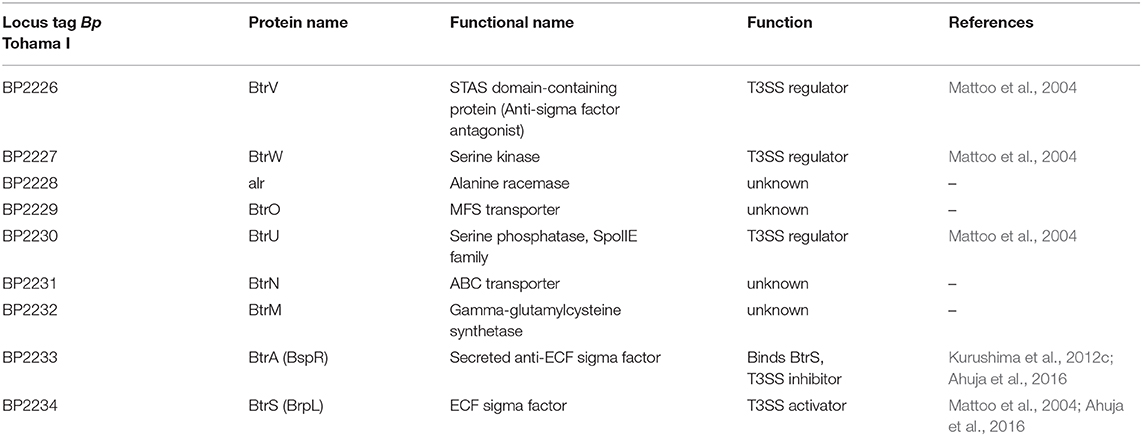
The BvgA-mediated activation of injectosome genes occurs at least for part of them indirectly through an extracytoplasmic function (ECF) sigma factor BtrS, annotated as BrpL, which is encoded in the btr locus (Figure 3A, Table 3) (Mattoo et al., 2004; Moon et al., 2017). As shown in Figure 4, in the BvgS-active mode, the membrane-bound phosphorelay sensor kinase BvgS phosphorylates the transcriptional activator BvgA, which in turn directly activates btrS transcription by binding as head-to-head BvgA dimers at positions centered at −41.5 and −63.5 upstream of the transcriptional start site (Moon et al., 2017). The BtrS can then activate transcription of bsp22 and bopN genes encoded in the bsc locus (Mattoo et al., 2004). However, the ectopic expression of btrS in the Bvg− locked mutant of B. bronchiseptica RB50 strain (ΔbvgS) does not allow for T3SS secretion. Additional unknown Bvg+ factors are thus required for the secretion process (Mattoo et al., 2004). Interestingly, two differently regulated gene clusters in the bsc locus of B. bronchiseptica RB50 strain were identified. The first cluster required the presence of BtrS for transcription (bscN to bsp22, and bopN to orf2) whereas genes in the other cluster (bscC to bscO, and bcrD to bscF) showed very little dependence, if any, on the presence of BtrS (Ahuja et al., 2016).
Besides of the ECF sigma factor BtrS, which controls also other regulatory networks (Gestal et al., 2019), additional genes encoded in the btr locus are linked to the injectosome function. These comprise the anti-sigma factor BtrA and the partner-switcher proteins BtrU, BtrW, and BtrV, respectively (Figure 3A, Table 3). As depicted in Figure 4, the secreted anti-sigma factor BtrA, also called BspR (Bordetella secreted protein regulator) was suggested to function as a secreted BtrS antagonist and establish a positive feedback loop that couples the injectosome secretion with the expression of T3SS genes in both B. pertussis and B. bronchiseptica species (Ahuja et al., 2016). The deletion of BtrA, indeed, enhances T3SS-dependent secretion and tissue culture phenotypes of both bacterial species (Kurushima et al., 2012c; Ahuja et al., 2016), and differential control over BtrA intracellular levels was suggested to contribute to the distinct T3SS activities of B. pertussis and B. bronchiseptica (Ahuja et al., 2016). However, the exact mechanism of BtrA-mediated inhibition of T3SS expression in Bordetella species remains to fully clarified. The work of Ahuja and colleagues used yeast two-hybrid system to show that BtrA interacts with BtrS, however, Kurushima and colleagues failed to detect this interaction using GST-pull down assay (Kurushima et al., 2012c; Ahuja et al., 2016).
Although studied so far only in B. bronchiseptica, the cascade that regulates T3SS secretion in bordetellae likely involves the partner-switcher regulatory proteins BtrU, BtrW, and BtrV (Figure 4, Table 3). These proteins exhibit homologies to partner-switching complexes of other bacteria that consist of a phosphatase, homologous to BtrU, a protein kinase/anti-sigma factor, homologous to BtrW, and an antagonist protein/anti-anti-sigma factor, homologous to BtrV. Nevertheless, the precise mechanism of regulation in Bordetella seems to differ from other bacteria, as all of the BtrU, BtrW, and BtrV proteins are required for Bordetella T3SS secretion and none of them acts as a negative regulator. BtrV seems to exert post-transcriptional control required for translation and/or protein stability, whereas BtrU and BtrW are assumed to specifically govern the secretion process (Mattoo et al., 2004; Kozak et al., 2005). Another level of complexity of regulation of T3SS production comes from the post-transcriptional regulator Hfq, a small hexameric RNA-binding protein (reviewed in Chao and Vogel, 2010). Indeed, the Hfq chaperone that anneals small RNA (sRNA) molecules to mRNA targets was found to be required for the expression of some of the T3SS genes (Figure 4) (Bibova et al., 2015; Dienstbier et al., 2019).
Effector Proteins of Bordetella Injectosome
The function of the T3SS injectosome consists in the transport of bacterial effector proteins into the host cell, where these modulate host cell functions by diverse molecular activities for the benefit of the bacteria. Only two effector proteins were so far reported to be present in classical bordetellae, namely the effector protein BteA, also called BopC, and the BopN protein, a homolog of a T3SS regulator (Panina et al., 2005; Kuwae et al., 2006; Nagamatsu et al., 2009).
The BteA effector protein was originally identified by Panina EM and colleagues in 2005 using a computational screen for chaperone-effector loci in B. bronchiseptica (Panina et al., 2005). Although the chaperone-effector pair, designated btcA-bteA, is located 2.5 Mbp away from the bsc locus, bteA expression is coordinated with the expression of injectosome genes and is activated by the BvgAS system and ECF sigma factor BtrS (Panina et al., 2005; Ahuja et al., 2016). As expected, BteA secretion depends on the T3SS ATPase BscN, and also on the BtrU, BtrW, BtrV partner-switcher proteins (Panina et al., 2005). Upon translocation into the host cells, BteA of B. bronchiseptica (Bb BteA) induces potent cytotoxicity. Compared to the wild type strains, inducing caspase 1-independent necrotic cell death, bteA-deficient strains of B. bronchiseptica exhibit negligible cytotoxicity levels similar to the type III secretion-deficient ΔbscN strains (Stockbauer et al., 2003; Panina et al., 2005; Kuwae et al., 2006; Ahuja et al., 2012). The Bb BteA effector protein alone is capable of inducing potent cytotoxicity in tissue culture and also yeast cells since even trace amounts of Bb BteA (undetectable by fluorescence microscopy or Western blot) are cytotoxic (Panina, 2007, dissertation thesis; French et al., 2009).
The 69 kDa BteA effector protein exhibits a modular architecture and is composed of an N-terminal multifunctional lipid raft targeting domain (LRT) of ~ 130 amino acid residues, and a cytotoxic C-terminal domain of ~528 amino acid residues, as depicted in Figure 5A. The LRT domain is rich in highly hydrophobic (~20% are Ile, Leu, Val) and positively charged (>10% are Arg, Lys) amino acid residues, resembling other known membrane localization domains (MLD) but targeting a specific portion of the plasma membrane. Therefore, the membrane localization domain of BteA was called the lipid raft targeting (LRT) domain (reviewed in Geissler, 2012). Within bacteria, the LRT domain binds the cognate chaperone BtcA that guides BteA for injectosome secretion upon recognition of the N-terminal secretion signal of the LRT (Panina et al., 2005; Kuwae et al., 2006). It is assumed that BtcA-BteA complex has a stoichiometry of 2:1 (chaperon:effector) like the chaperon-effector pair, InvB-SipA (Lilic et al., 2006; Guttman et al., 2013). Upon T3SS-mediated translocation into target cell cytosol, the LRT appears to mediate BteA localization into the cytosolic leaflet of lipid rafts of cell plasma membrane via phosphatidylinositol 4,5-bisphosphate (PIP2) binding (French et al., 2009; Yahalom et al., 2019). The crystal structure shows that LRT is an elongated four-helix bundle packed against two shorter perpendicular helices, the second of which caps the domain in a tip motif. The continuous positively charged surface of the second bundle helix was proposed to mediate a direct electrostatic interaction with the negatively charged PIP2 head while being supported by the structural tip helix (Yahalom et al., 2019). Interestingly, homologous domains responsible for lipid raft targeting, but surrounded by other domains, are also present in several known and predicted T3SS effectors and MARTX (multifunctional autoprocessing repeats-in-toxin) toxins, including Plu4750 and Plu3217 from Photorhabdus luminescens. The membrane-localization LRT domain thus seems to have been reshuffled during evolution (Panina et al., 2005; French et al., 2009). The C-terminal domain of BteA of ~ 528 amino acid residues (Figure 5A) is solely responsible for the BteA-mediated cytotoxicity (French et al., 2009; Kuwae et al., 2016). Whereas no reliable predictions of the mechanism of BteA-mediated cytotoxicity can be obtained by sequence homology searches, the deletion mutagenesis suggests that the cytotoxic domain of BteA harbors two separate cytotoxic activity-related regions that span over the amino acid residues 200–312 and 400–658, respectively (Kuwae et al., 2016). The last 14 amino acid residues of BteA are also critical for full cytotoxic activity of BteA (French et al., 2009). However, the mechanism underlying the cytotoxic action of BteA as well as its cellular targets remain unknown. The targets of BteA action were proposed to be associated with the cholesterol-rich domains of the host cell membrane since depletion of membrane cholesterol protected cells from the T3SS-dependent cytotoxic action of BteA (French et al., 2009). Nevertheless, the observed protection could have also been due to diminished translocation of BteA into cholesterol-depleted cells (Hayward et al., 2005). Besides, the previously reported T3SS-mediated dephosphorylation of tyrosine residues of proteins in infected mammalian cells appears to be a rather indirect consequence of the cytotoxic action of BteA (Yuk et al., 1998; Kuwae et al., 2006).
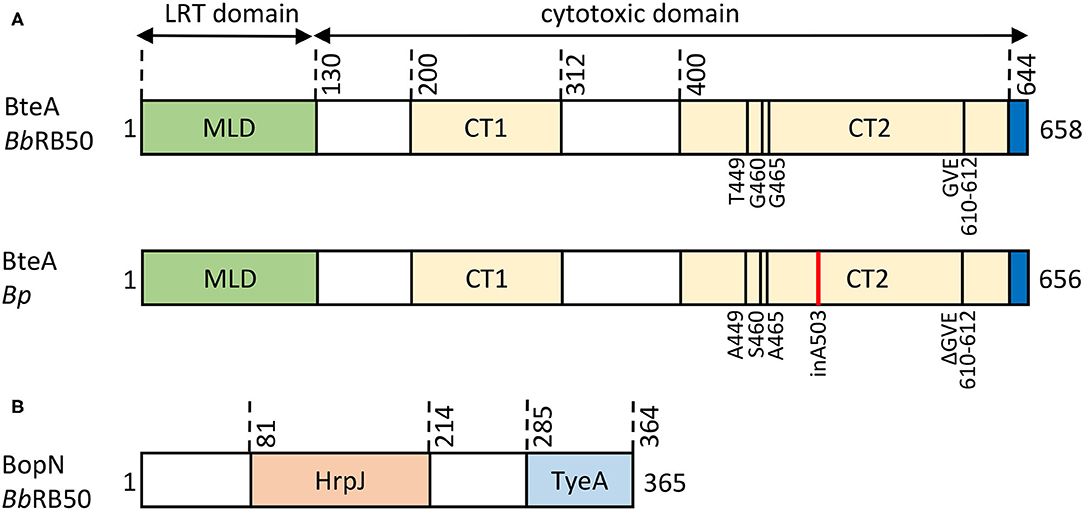
Figure 5. Structural organization of Bordetella T3SS effector proteins. (A) Schematic representation of the BteA effector protein of B. bronchiseptica RB50 (BbRB50 BteA, 658 aa) and B. pertussis Tohama I (Bp BteA, 656 aa). BteA is composed of two primary domains: a lipid-raft targeting domain (LRT) and a cytotoxic domain. The N-terminal LRT domain of ~ 130 residues is a multifunctional four helical bundle membrane localization domain (MLB) that harbors the secretion signal, comprises BtcA chaperone binding sites, and binds phosphatidylinositol 4,5-bisphosphate (PIP2) (French et al., 2009; Guttman et al., 2013; Yahalom et al., 2019). The C-terminal cytotoxic domain of ~ 528 aa contains 2 segments associated with cytotoxicity (BbRB50 BteA, CT1, aa 200-312, CT2, aa 400-658; Bp BteA, CT1, aa 200-312, CT2, aa 400-656) (Kuwae et al., 2016). In addition, the last 14 aa were shown to be critical for BteA cytotoxicity (French et al., 2009). Differences in the primary structure of BbRB50 BteA and Bp BteA are shown, insertion of A503 within Bp BteA is highlighted in red. (B) Schematic representation of the 365 residue-long BopN protein of B. bronchiseptica RB50 (BbRB50 BopN). The HrpJ (aa 81-214) and TyeA (aa 285-364) domains are indicated.
The BteA effector proteins of classical Bordetella species were claimed to be functionally interchangeable (French et al., 2009). However, our recent study by Bayram and colleagues demonstrated that compared to its BteA homolog from the B. bronchiseptica, the BteA effector of B. pertussis (Bp BteA) exhibits a significantly reduced specific cytotoxic activity toward cultured cells (Bayram et al., 2020). This activity difference could be unambiguously attributed to the insertion of a single alanine residue at position 503 of the Bp BteA protein (Figure 5A). Indeed, the specific cytotoxic activity of the Bp BteA protein was strongly increased upon deletion of the A503 residue, and the activity of the Bb BteA protein was strongly reduced by insertion of an A503 residue, respectively (Bayram et al., 2020). This explains why low cytotoxicity was observed in cells infected by B. pertussis translocating BteA through a functional T3SS injectosome (Han et al., 2011; Ahuja et al., 2016). Remarkably, the analysis of amino acid sequences of BteA of the classical bordetellae revealed that the A503 residue is conserved across all B. pertussis lineages but is absent in the BteA of all distinct subpopulations and lineages of B. bronchiseptica and B. parapertussis species. This suggests that the acquisition of the A503 residue in the Bp BteA protein occurred early in the B. pertussis speciation to human hosts (Bayram et al., 2020). Interestingly, B. ansorpii does not appear to encode BteA effector protein homolog nor Bordetella master virulence regulatory system BvgAS.
The BopN protein was originally identified as a Bordetella T3SS-secreted protein that exhibits homology to the gatekeeper proteins, StcW (Table 2) (Yuk et al., 2000). Remarkably, in the Ysc family of injectosomes, the StcW protein is encoded as two polypeptides, e.g., YopN and its C-terminus-binding chaperone TyeA. There is a single protein corresponding to a chimeric product of YopN-TyeA in the other T3SS systems, including classical Bordetella species. The 39 kDa BopN protein (Figure 5B) thus highlights the divergence of Bordetella Bsc system away from the Ysc family of injectosomes (Pallen et al., 2005). The function of the StcW is to regulate translocator secretion and/or prevent a premature secretion of the effector proteins presumably through binding to the export apparatus in a protein complex that is released upon activating signal (sensing of the contact to a host cell) (Schubot et al., 2005; Portaliou et al., 2017; Yu et al., 2018). The SctW protein is then either secreted as is the case of Yersinia YopN and Shigella MxiC, or is degraded like the Salmonella SPI-II SsaL protein (Cheng et al., 2001; Botteaux et al., 2009; Yu et al., 2010). The fate of SctW protein if injected into the target host cell is not clear. It is assumed that the SctW does not exert any effector function, except for the Chlamydia CopN protein that was shown to induce G2/M cell cycle arrest by inhibiting tubulin polymerization (Huang et al., 2008; Archuleta et al., 2011; Nawrotek et al., 2014).
The function of the Bordetella BopN protein remains controversial (Figure 4). The protein was reported to be required for manifestation of the full BteA-mediated cytotoxicity in B. bronchiseptica infections of rat L2 pulmonary epithelial cells but was not required for cytotoxicity in mouse DC2.4 dendritic cells (Nagamatsu et al., 2009; Abe et al., 2017). Besides, the deletion of BopN did not affect the secretion of BteA into the culture supernatants of B. bronchiseptica grown in vitro (Abe et al., 2017). This would indicate that effector secretion in B. bronchiseptica does not require host cell contact and/or gatekeeper function, or that the used culture medium artificially mimics the contact to a cell, as previously observed for other T3SS-expressing bacterial species, e.g., enteropathogenic Escherichia coli (EPEC), Vibrio parahaemolyticus or Shigella flexnerii (Deng et al., 2005; Botteaux et al., 2009; Tandhavanant et al., 2018). Remarkably, the BopN protein was reported to be translocated into cells where it was suggested to localize to the cell nucleus (Nagamatsu et al., 2009; Abe et al., 2017). Its activity was also proposed to down-regulate MAPK signaling, block nuclear translocation of the NF-kBp65 subunit while promoting translocation of the NF-kBp50 subunit, and enhancing IL-10 production (Nagamatsu et al., 2009). However, the molecular basis of these processes remain unknown.
Type III Secretion in Infections With Bordetellae
The role of T3SS-mediated delivery of effector proteins is thoroughly characterized in animal infections with B. bronchiseptica. The elimination of T3SS function due to the deletion of the T3SS ATPse BscN results in defects of B. bronchiseptica persistence in the lower respiratory tract of rats, mice, and pigs (Figure 6) (Yuk et al., 1998, 2000; Nicholson et al., 2014). The functional T3SS of B. bronchiseptica was also reported to lower titres of anti-Bordetella serum antibodies and inhibit the generation of IFN-γ-producing splenocytes while enhancing the production of immunosuppressive IL-10 (Figure 6) (Yuk et al., 2000; Skinner et al., 2005; Pilione and Harvill, 2006; Nicholson et al., 2014). Since natural clearance of B. bronchiseptica depends on antibodies and production of IFN-γ while IL-10 promotes bacterial colonization, these results suggest that T3SS activity could favor bacterial persistence by altering the balance between IL-10 and IFN-γ, and hindering the antibody production (Kirimanjeswara et al., 2003; Skinner et al., 2005; Pilione and Harvill, 2006). Nevertheless, it appears that T3SS primarily targets innate immunity functions since ΔbscN strain was found to be hypervirulent in SCID-beige mice that are devoid of functional B cells and T cells (SCID) and NK cells (beige mutation) (Yuk et al., 2000). The T3SS may synergize with adenylate cyclase toxin to modulate macrophage and dendritic cell phenotypes and thereby subvert adaptive immune responses (Figure 6) (Skinner et al., 2004, 2005; Reissinger et al., 2005; Siciliano et al., 2006). Alternatively, or in combination, T3SS action may consist in inhibition of NF-kB activation in epithelial cells of the respiratory epithelia (Figure 6) (Yuk et al., 2000; Legarda et al., 2005; Ryan et al., 2018). Indeed, infection with wild type B. bronchiseptica, but not with the ΔbscN mutant, suppressed the activation of NF-κB and induction of β-defensin in primary bovine tracheal epithelial cells and/or during mice infection (Legarda et al., 2005; Ryan et al., 2018). Interestingly, though, T3SS-dependent phenotypes of B. bronchiseptica may vary between different phylogenetic lineages and/or isolates (Figure 6) (Buboltz et al., 2009; Ahuja et al., 2012). For example, enhanced expression of T3SS genes was reported to be partially responsible for the increased virulence of complex I B. bronchiseptica 1289 strain isolated from diseased host, as compared to the virulence of the RB50 strain isolated from an asymptomatic host (Cotter and Miller, 1994; Buboltz et al., 2009). Along the same line, in vitro T3SS-mediated hyper-cytotoxicity of a subset of complex IV B. bronchiseptica strains correlated with the increased ability of these strains to cause lethal pulmonary infections in mice (Ahuja et al., 2012).
Figure 6. Summary of the most important activities of B. bronchiseptica T3SS in animal models in vivo and on cells in vitro. See the text for further details and references.
The potential role of T3SS in B. pertussis infections was overlooked for many years due to adaptional block of injectosome expression in B. pertussis until the seminal studies of Fennelly et al. (2008); Gaillard et al. (2011). The activity of B. pertussis T3SS was then suggested to dampen the inflammatory responses in infected mice by inhibiting proinflammatory cytokine production and enhancing anti-inflammatory IL-10 production in the lungs early after infection, which correlated with lower antigen-specific IFN-γ, IL-17 and IgG responses later in the infection (Fennelly et al., 2008). These data, however, warrant further investigation. Surprisingly, compared with wild type bacteria, the T3SS-deficient B. pertussis mutant was significantly impaired in the capacity to colonize the lungs already 3 h after respiratory challenge (Fennelly et al., 2008).
The functionality of B. pertussis T3SS injectosome is intriguing, given the acute nature of pertussis disease, which is relatively short-lived. This contrasts with the rather chronic infections caused by B. bronchiseptica, which have been related to the action of the T3SS (Yuk et al., 1998, 2000; Nicholson et al., 2014) and/or BteA effector protein (Panina, 2007, dissertation thesis). The adaptation of T3SS of B. pertussis may have occurred in signaling cascades activating T3SS and/or BteA effector protein expression during in vivo infection. Alternatively, or in combination, a functional divergence could originate in the level of effector specific activities as recently described by Bayram et al. (2020). The Bp BteA has only residual cytotoxic activity as compared to Bb BteA, and the deletion of a differing A503 residue from the wild type Bp BteA (Figure 5A) increased T3SS-mediated B. pertussis cytotoxicity. Besides, the mutant B. pertussis bteAΔA503 exhibited a higher virulence in the mouse model of intranasal infection, while at a sublethal challenge dose it accounted for a reduced pathology in B. pertussis-infected mouse lungs. These data show that a more active BteAΔA503 was able to importantly intervene in the interactions of B. pertussis with the host defense and its action shaped the course and outcome of the infection (Bayram et al., 2020).
Concluding Remarks and Outstanding Questions
Since 1906, when the founding member of the Bordetella genus, B. pertussis, was isolated by Bordet and Gengou from Bordet's son suffering from pertussis, our comprehension of the bordetellae species has come a long way. Nevertheless, many unknowns remain and dozens of outstanding questions can be formulated in the very specific area of research on the type III secretion in bordetellae. Perhaps the most important question is the role of T3SS activity in the pathophysiology of human pertussis. Hopefully, the availability of a non-human primate in which the human pathology of pertussis and transmission of the pathogen can be reproduced in olive baboons will yield an answer (Pinto and Merkel, 2017; Zimmerman et al., 2018). It remains to be determined if the variation in the T3SS contributed to B. pertussis evolution and pertussis pathogenesis. A better insight into the regulation of the T3SS expression and injectosome functionality is also needed. It remains unclear what are the host signals that Bordetella spp. respond to and whether these signals and/or signaling cascades are different for B. pertussis and B. bronchiseptica. We still do not understand by which mechanism do B. pertussis bacteria turn off the T3SS activity upon passage on laboratory media (Gaillard et al., 2011) and why despite the potent cytotoxicity in cultured cells the in vivo B. bronchiseptica-colonized respiratory epithelia shows no damage (Cotter and Miller, 1994; Panina et al., 2005). Future research should also focus on the understanding of the Bordetella injectosome that differs by the presence of the Bsp22 helical assembly and YscX-like and YscY-like proteins from the well-studied injectosomes of Salmonella enterica and Shigella spp. Last not least, we need to get a better comprehension of the molecular details behind the actions of Bordetella effector protein/s, their function and interplay with other virulence factors of classical Bordetella species.
Author Contributions
The author confirms being the sole contributor of this work and has approved it for publication.
Funding
The work of the author is supported by Lumina Queruntur Fellowship LQ200202001 of the Czech Academy of Sciences, grant 18-16772Y of the Czech Science Foundation (www.gacr.cz) and project LM2018133 (Czech National Node to the European Infrastructure for Translational Medicine) of the Ministry of Education, Youth and Sports of the Czech Republic.
Conflict of Interest
The author declares that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The author wants to thank Peter Šebo (Insitute of Microbiology, Prague, Czech Republic) for critical reading of the manuscript and helpful advice.
References
Abe, A., Nishimura, R., and Kuwae, A. (2017). Bordetella effector BopN is translocated into host cells via its N-terminal residues. Microbiol. Immunol. 61, 206–214. doi: 10.1111/1348-0421.12489
Ahuja, U., Liu, M., Tomida, S., Park, J., Souda, P., Whitelegge, J., et al. (2012). Phenotypic and genomic analysis of hypervirulent human-associated Bordetella bronchiseptica. BMC Microbiol. 12:167. doi: 10.1186/1471-2180-12-167
Ahuja, U., Shokeen, B., Cheng, N., Cho, Y., Blum, C., Coppola, G., et al. (2016). Differential regulation of type III secretion and virulence genes in Bordetella pertussis and Bordetella bronchiseptica by a secreted anti-sigma factor. Proc. Natl. Acad. Sci. U.S.A. 113, 2341–2348. doi: 10.1073/pnas.1600320113
Archuleta, T. L., Du, Y., English, C. A., Lory, S., Lesser, C., Ohi, M. D., et al. (2011). The Chlamydia effector chlamydial outer protein N (CopN) sequesters tubulin and prevents microtubule assembly. J. Biol. Chem. 286, 33992–33998. doi: 10.1074/jbc.M111.258426
Arico, B., and Rappuoli, R. (1987). Bordetella parapertussis and Bordetella bronchiseptica contain transcriptionally silent pertussis toxin genes. J. Bacteriol. 169, 2847–2853. doi: 10.1128/JB.169.6.2847-2853.1987
Bayram, J., Malcova, I., Sinkovec, L., Holubova, J., Streparola, G., Jurnecka, D., et al. (2020). Cytotoxicity of the effector protein BteA was attenuated in Bordetella pertussis by insertion of an alanine residue. PLoS Pathog. doi: 10.1371/journal.ppat.1008512
Bergfors, E., Trollfors, B., Taranger, J., Lagergard, T., Sundh, V., and Zackrisson, G. (1999). Parapertussis and pertussis: differences and similarities in incidence, clinical course, and antibody responses. Int. J. Infect. Dis. 3, 140–146. doi: 10.1016/S1201-9712(99)90035-8
Bibova, I., Hot, D., Keidel, K., Amman, F., Slupek, S., Cerny, O., et al. (2015). Transcriptional profiling of Bordetella pertussis reveals requirement of RNA chaperone Hfq for Type III secretion system functionality. RNA Biol. 12, 175–185. doi: 10.1080/15476286.2015.1017237
Botteaux, A., Sory, M. P., Biskri, L., Parsot, C., and Allaoui, A. (2009). MxiC is secreted by and controls the substrate specificity of the Shigella flexneri type III secretion apparatus. Mol. Microbiol. 71, 449–460. doi: 10.1111/j.1365-2958.2008.06537.x
Brickman, T. J., Cummings, C. A., Liew, S. Y., Relman, D. A., and Armstrong, S. K. (2011). Transcriptional profiling of the iron starvation response in Bordetella pertussis provides new insights into siderophore utilization and virulence gene expression. J. Bacteriol. 193, 4798–4812. doi: 10.1128/JB.05136-11
Brockmeier, S. L., Register, K. B., Magyar, T., Lax, A. J., Pullinger, G. D., and Kunkle, R. A. (2002). Role of the dermonecrotic toxin of Bordetella bronchiseptica in the pathogenesis of respiratory disease in swine. Infect. Immun. 70, 481–490. doi: 10.1128/IAI.70.2.481-490.2002
Buboltz, A. M., Nicholson, T. L., Weyrich, L. S., and Harvill, E. T. (2009). Role of the type III secretion system in a hypervirulent lineage of Bordetella bronchiseptica. Infect. Immun. 77, 3969–3977. doi: 10.1128/IAI.01362-08
Burns, D. L., Meade, B. D., and Messionnier, N. E. (2014). Pertussis resurgence: perspectives from the Working Group Meeting on pertussis on the causes, possible paths forward, and gaps in our knowledge. J. Infect. Dis. 209 (Suppl 1), S32–S35. doi: 10.1093/infdis/jit491
Carbonetti, N. H. (2010). Pertussis toxin and adenylate cyclase toxin: key virulence factors of Bordetella pertussis and cell biology tools. Future Microbiol. 5, 455–469. doi: 10.2217/fmb.09.133
Chao, Y., and Vogel, J. (2010). The role of Hfq in bacterial pathogens. Curr. Opin. Microbiol. 13, 24–33. doi: 10.1016/j.mib.2010.01.001
Chen, Q., and Stibitz, S. (2019). The BvgASR virulence regulon of Bordetella pertussis. Curr. Opin. Microbiol. 47, 74–81. doi: 10.1016/j.mib.2019.01.002
Cheng, L. W., Kay, O., and Schneewind, O. (2001). Regulated secretion of YopN by the type III machinery of Yersinia enterocolitica. J. Bacteriol. 183, 5293–5301. doi: 10.1128/JB.183.18.5293-5301.2001
Cherry, J. D. (2019). The 112-year odyssey of pertussis and pertussis vaccines-mistakes made and implications for the future. J. Pediatric Infect. Dis. Soc. 8, 334–341. doi: 10.1093/jpids/piz005
Cherry, J. D., and Seaton, B. L. (2012). Patterns of Bordetella parapertussis respiratory illnesses: 2008-2010. Clin. Infect. Dis 54, 534–537. doi: 10.1093/cid/cir860
Cleary, J., Lai, L. C., Shaw, R. K., Straatman-Iwanowska, A., Donnenberg, M. S., Frankel, G., et al. (2004). Enteropathogenic Escherichia coli (EPEC) adhesion to intestinal epithelial cells: role of bundle-forming pili (BFP), EspA filaments and intimin. Microbiology (Reading,. Engl) 150, 527–538. doi: 10.1099/mic.0.26740-0
Cotter, P. A., and Miller, J. F. (1994). BvgAS-mediated signal transduction: analysis of phase-locked regulatory mutants of Bordetella bronchiseptica in a rabbit model. Infect. Immun. 62, 3381–3390. doi: 10.1128/IAI.62.8.3381-3390.1994
Cummings, C. A., Bootsma, H. J., Relman, D. A., and Miller, J. F. (2006). Species- and strain-specific control of a complex, flexible regulon by Bordetella BvgAS. J. Bacteriol. 188, 1775–1785. doi: 10.1128/JB.188.5.1775-1785.2006
Cummings, C. A., Brinig, M. M., Lepp, P. W., Van De Pas, S., and Relman, D. A. (2004). Bordetella species are distinguished by patterns of substantial gene loss and host adaptation. J. Bacteriol. 186, 1484–1492. doi: 10.1128/JB.186.5.1484-1492.2004
Daniell, S. J., Takahashi, N., Wilson, R., Friedberg, D., Rosenshine, I., Booy, F. P., et al. (2001). The filamentous type III secretion translocon of enteropathogenic Escherichia coli. Cell. Microbiol. 3, 865–871. doi: 10.1046/j.1462-5822.2001.00168.x
Deng, W., Li, Y., Hardwidge, P. R., Frey, E. A., Pfuetzner, R. A., Lee, S., et al. (2005). Regulation of type III secretion hierarchy of translocators and effectors in attaching and effacing bacterial pathogens. Infect. Immun. 73, 2135–2146. doi: 10.1128/IAI.73.4.2135-2146.2005
Dewoody, R. S., Merritt, P. M., and Marketon, M. M. (2013). Regulation of the Yersinia type III secretion system: traffic control. Front. Cell. Infect. Microbiol. 3:4. doi: 10.3389/fcimb.2013.00004
Diavatopoulos, D. A., Cummings, C. A., Schouls, L. M., Brinig, M. M., Relman, D. A., and Mooi, F. R. (2005). Bordetella pertussis, the causative agent of whooping cough, evolved from a distinct, human-associated lineage of B. bronchiseptica. PLoS Pathog. 1:e45. doi: 10.1371/journal.ppat.0010045
Dienstbier, A., Amman, F., Stipl, D., Petrackova, D., and Vecerek, B. (2019). Comparative integrated omics analysis of the Hfq regulon in Bordetella pertussis. Int. J. Mol. Sci. 20:3073. doi: 10.3390/ijms20123073
Diepold, A., and Wagner, S. (2014). Assembly of the bacterial type III secretion machinery. FEMS Microbiol. Rev. 38, 802–822. doi: 10.1111/1574-6976.12061
Diepold, A., Wiesand, U., Amstutz, M., and Cornelis, G. R. (2012). Assembly of the Yersinia injectisome: the missing pieces. Mol. Microbiol. 85, 878–892. doi: 10.1111/j.1365-2958.2012.08146.x
Fabianova, K., Benes, C., and Kriz, B. (2010). A steady rise in incidence of pertussis since nineties in the Czech Republic. Epidemiol. Mikrobiol. Imunol. 59, 25–33.
Fauconnier, A., Veithen, A., Gueirard, P., Antoine, R., Wacheul, L., Locht, C., et al. (2001). Characterization of the type III secretion locus of Bordetella pertussis. Int. J. Med. Microbiol. 290, 693–705. doi: 10.1016/S1438-4221(01)80009-6
Fedele, G., Schiavoni, I., Adkins, I., Klimova, N., and Sebo, P. (2017). Invasion of dendritic cells, macrophages and neutrophils by the bordetella adenylate cyclase toxin: a subversive move to fool host immunity. Toxins (Basel) 9. doi: 10.3390/toxins9100293
Fennelly, N. K., Sisti, F., Higgins, S. C., Ross, P. J., Van Der Heide, H., Mooi, F. R., et al. (2008). Bordetella pertussis expresses a functional type III secretion system that subverts protective innate and adaptive immune responses. Infect. Immun. 76, 1257–1266. doi: 10.1128/IAI.00836-07
French, C. T., Panina, E. M., Yeh, S. H., Griffith, N., Arambula, D. G., and Miller, J. F. (2009). The Bordetella type III secretion system effector BteA contains a conserved N-terminal motif that guides bacterial virulence factors to lipid rafts. Cell. Microbiol. 11, 1735–1749. doi: 10.1111/j.1462-5822.2009.01361.x
Fry, N. K., Duncan, J., Malnick, H., and Cockcroft, P. M. (2007). The first UK isolate of 'Bordetella ansorpii' from an immunocompromised patient. J. Med. Microbiol. 56, 993–995. doi: 10.1099/jmm.0.47078-0
Fry, N. K., Duncan, J., Malnick, H., Warner, M., Smith, A. J., Jackson, M. S., et al. (2005). Bordetella petrii clinical isolate. Emerging Infect. Dis. 11, 1131–1133. doi: 10.3201/eid1107.050046
Gaillard, M. E., Bottero, D., Castuma, C. E., Basile, L. A., and Hozbor, D. (2011). Laboratory adaptation of Bordetella pertussis is associated with the loss of type three secretion system functionality. Infect. Immun. 79, 3677–3682. doi: 10.1128/IAI.00136-11
Galan, J. E., Lara-Tejero, M., Marlovits, T. C., and Wagner, S. (2014). Bacterial type III secretion systems: specialized nanomachines for protein delivery into target cells. Annu. Rev. Microbiol. 68, 415–438. doi: 10.1146/annurev-micro-092412-155725
Galan, J. E., and Waksman, G. (2018). Protein-injection machines in bacteria. Cell 172, 1306–1318. doi: 10.1016/j.cell.2018.01.034
Geissler, B. (2012). Bacterial toxin effector-membrane targeting: outside in, then back again. Front. Cell. Infect. Microbiol. 2:75. doi: 10.3389/fcimb.2012.00075
Gestal, M. C., Howard, L. K., Dewan, K., Johnson, H. M., Barbier, M., Bryant, C., et al. (2019). Enhancement of immune response against Bordetella spp. by disrupting immunomodulation. Sci. Rep. 9:20261. doi: 10.1038/s41598-019-56652-z
Gestal, M. C., Rivera, I., Howard, L. K., Dewan, K. K., Soumana, I. H., Dedloff, M., et al. (2018). Blood or serum exposure induce global transcriptional changes, altered antigenic profile, and increased cytotoxicity by classical bordetellae. Front. Microbiol 9:1969. doi: 10.3389/fmicb.2018.01969
Goodnow, R. A. (1980). Biology of Bordetella bronchiseptica. Microbiol. Rev. 44, 722–738. doi: 10.1128/MMBR.44.4.722-738.1980
Gueirard, P., Weber, C., Le Coustumier, A., and Guiso, N. (1995). Human Bordetella bronchiseptica infection related to contact with infected animals: persistence of bacteria in host. J. Clin. Microbiol. 33, 2002–2006. doi: 10.1128/JCM.33.8.2002-2006.1995
Gurung, J. M., Amer, A. A., Francis, M. K., Costa, T. R. D., Chen, S., Zavialov, A. V., et al. (2018). Heterologous complementation studies with the YscX and YscY protein families reveals a specificity for Yersinia pseudotuberculosis type III secretion. Front. Cell. Infect. Microbiol. 8:80. doi: 10.3389/fcimb.2018.00080
Guttman, C., Davidov, G., Yahalom, A., Shaked, H., Kolusheva, S., Bitton, R., et al. (2013). BtcA, A class IA type III chaperone, interacts with the BteA N-terminal domain through a globular/non-globular mechanism. PLoS ONE 8:e81557. doi: 10.1371/journal.pone.0081557
Hacker, J., and Kaper, J. B. (2000). Pathogenicity islands and the evolution of microbes. Annu. Rev. Microbiol. 54, 641–679. doi: 10.1146/annurev.micro.54.1.641
Hamidou Soumana, I., Linz, B., and Harvill, E. T. (2017). Environmental origin of the genus bordetella. Front. Microbiol 8:28. doi: 10.3389/fmicb.2017.00028
Han, H. J., Kuwae, A., Abe, A., Arakawa, Y., and Kamachi, K. (2011). Differential expression of type III effector BteA protein due to IS481 insertion in Bordetella pertussis. PLoS ONE 6:e17797. doi: 10.1371/journal.pone.0017797
Hanawa, T., Kamachi, K., Yonezawa, H., Fukutomi, T., Kawakami, H., and Kamiya, S. (2016). Glutamate limitation, BvgAS activation, and (p)ppGpp regulate the expression of the bordetella pertussis Type 3 secretion system. J. Bacteriol. 198, 343–351. doi: 10.1128/JB.00596-15
Hayward, R. D., Cain, R. J., Mcghie, E. J., Phillips, N., Garner, M. J., and Koronakis, V. (2005). Cholesterol binding by the bacterial type III translocon is essential for virulence effector delivery into mammalian cells. Mol. Microbiol. 56, 590–603. doi: 10.1111/j.1365-2958.2005.04568.x
Hegerle, N., Rayat, L., Dore, G., Zidane, N., Bedouelle, H., and Guiso, N. (2013). In-vitro and in-vivo analysis of the production of the Bordetella type three secretion system effector A in Bordetella pertussis, Bordetella parapertussis and Bordetella bronchiseptica. Microbes Infect. 15, 399–408. doi: 10.1016/j.micinf.2013.02.006
Hester, S. E., Lui, M., Nicholson, T., Nowacki, D., and Harvill, E. T. (2012). Identification of a CO2 responsive regulon in Bordetella. PLoS ONE 7:e47635. doi: 10.1371/journal.pone.0047635
Hot, D., Antoine, R., Renauld-Mongenie, G., Caro, V., Hennuy, B., Levillain, E., et al. (2003). Differential modulation of Bordetella pertussis virulence genes as evidenced by DNA microarray analysis. Mol. Genet. Genomics 269, 475–486. doi: 10.1007/s00438-003-0851-1
Hovingh, E. S., Van Den Broek, B., Kuipers, B., Pinelli, E., Rooijakkers, S. H. M., and Jongerius, I. (2017). Acquisition of C1 inhibitor by Bordetella pertussis virulence associated gene 8 results in C2 and C4 consumption away from the bacterial surface. PLoS Pathog. 13:e1006531. doi: 10.1371/journal.ppat.1006531
Hu, B., Lara-Tejero, M., Kong, Q., Galan, J. E., and Liu, J. (2017). In situ molecular architecture of the salmonella Type III secretion machine. Cell 168, 1065–1074. doi: 10.1016/j.cell.2017.02.022
Huang, J., Lesser, C. F., and Lory, S. (2008). The essential role of the CopN protein in Chlamydia pneumoniae intracellular growth. Nature 456, 112–115. doi: 10.1038/nature07355
Hueck, C. J. (1998). Type III protein secretion systems in bacterial pathogens of animals and plants. Microbiol. Mol. Biol. Rev. 62, 379–433. doi: 10.1128/MMBR.62.2.379-433.1998
Ivanov, Y. V., Linz, B., Register, K. B., Newman, J. D., Taylor, D. L., Boschert, K. R., et al. (2016). Identification and taxonomic characterization of Bordetella pseudohinzii sp. nov. isolated from laboratory-raised mice. Int. J. Syst. Evol. Microbiol. 66, 5452–5459. doi: 10.1099/ijsem.0.001540
Kapil, P., and Merkel, T. J. (2019). Pertussis vaccines and protective immunity. Curr. Opin. Immunol. 59, 72–78. doi: 10.1016/j.coi.2019.03.006
Kerr, J. R., Rigg, G. P., Matthews, R. C., and Burnie, J. P. (1999). The Bpel locus encodes type III secretion machinery in Bordetella pertussis. Microb. Pathog 27, 349–367. doi: 10.1006/mpat.1999.0307
Kersters, K., Hinz, K.-H., Hertle, A., Segers, P., Lievens, A., Siegmann, O., et al. (1984). Bordetella avium sp. nov., isolated from the respiratory tracts of turkeys and other birds. Intern. J. System. Evol. Microbiol. 34, 56–70. doi: 10.1099/00207713-34-1-56
Kirimanjeswara, G. S., Mann, P. B., and Harvill, E. T. (2003). Role of antibodies in immunity to Bordetella infections. Infect. Immun. 71, 1719–1724. doi: 10.1128/IAI.71.4.1719-1724.2003
Knutton, S., Rosenshine, I., Pallen, M. J., Nisan, I., Neves, B. C., Bain, C., et al. (1998). A novel EspA-associated surface organelle of enteropathogenic Escherichia coli involved in protein translocation into epithelial cells. EMBO J. 17, 2166–2176. doi: 10.1093/emboj/17.8.2166
Ko, K. S., Peck, K. R., Oh, W. S., Lee, N. Y., Lee, J. H., and Song, J. H. (2005). New species of Bordetella, Bordetella ansorpii sp. nov., isolated from the purulent exudate of an epidermal cyst. J. Clin. Microbiol. 43, 2516–2519. doi: 10.1128/JCM.43.5.2516-2519.2005
Kozak, N. A., Mattoo, S., Foreman-Wykert, A. K., Whitelegge, J. P., and Miller, J. F. (2005). Interactions between partner switcher orthologs BtrW and BtrV regulate type III secretion in Bordetella. J. Bacteriol. 187, 5665–5676. doi: 10.1128/JB.187.16.5665-5676.2005
Kurushima, J., Kuwae, A., and Abe, A. (2012a). Btc22 chaperone is required for secretion and stability of the type III secreted protein Bsp22 in Bordetella bronchiseptica. FEMS Microbiol. Lett. 331, 144–151. doi: 10.1111/j.1574-6968.2012.02561.x
Kurushima, J., Kuwae, A., and Abe, A. (2012b). Iron starvation regulates the type III secretion system in Bordetella bronchiseptica. Microbiol. Immunol. 56, 356–362. doi: 10.1111/j.1348-0421.2012.00442.x
Kurushima, J., Kuwae, A., and Abe, A. (2012c). The type III secreted protein BspR regulates the virulence genes in Bordetella bronchiseptica. PLoS ONE 7:e38925. doi: 10.1371/journal.pone.0038925
Kuwae, A., Matsuzawa, T., Ishikawa, N., Abe, H., Nonaka, T., Fukuda, H., et al. (2006). BopC is a novel type III effector secreted by Bordetella bronchiseptica and has a critical role in type III-dependent necrotic cell death. J. Biol. Chem. 281, 6589–6600. doi: 10.1074/jbc.M512711200
Kuwae, A., Momose, F., Nagamatsu, K., Suyama, Y., and Abe, A. (2016). BteA secreted from the Bordetella bronchiseptica type III secetion system induces necrosis through an actin cytoskeleton signaling pathway and inhibits phagocytosis by macrophages. PLoS ONE 11:e0148387. doi: 10.1371/journal.pone.0148387
Kuwae, A., Ohishi, M., Watanabe, M., Nagai, M., and Abe, A. (2003). BopB is a type III secreted protein in Bordetella bronchiseptica and is required for cytotoxicity against cultured mammalian cells. Cell. Microbiol. 5, 973–983. doi: 10.1046/j.1462-5822.2003.00341.x
Le Coustumier, A., Njamkepo, E., Cattoir, V., Guillot, S., and Guiso, N. (2011). Bordetella petrii infection with long-lasting persistence in human. Emerging Infect. Dis. 17, 612–618. doi: 10.3201/eid1704.101480
Legarda, D., Klein-Patel, M. E., Yim, S., Yuk, M. H., and Diamond, G. (2005). Suppression of NF-kappaB-mediated beta-defensin gene expression in the mammalian airway by the Bordetella type III secretion system. Cell. Microbiol. 7, 489–497. doi: 10.1111/j.1462-5822.2004.00473.x
Lilic, M., Vujanac, M., and Stebbins, C. E. (2006). A common structural motif in the binding of virulence factors to bacterial secretion chaperones. Mol. Cell 21, 653–664. doi: 10.1016/j.molcel.2006.01.026
Linz, B., Ivanov, Y. V., Preston, A., Brinkac, L., Parkhill, J., Kim, M., et al. (2016). Acquisition and loss of virulence-associated factors during genome evolution and speciation in three clades of Bordetella species. BMC Genomics 17:767. doi: 10.1186/s12864-016-3112-5
Linz, B., Ma, L., Rivera, I., and Harvill, E. T. (2019). Genotypic and phenotypic adaptation of pathogens: lesson from the genus Bordetella. Curr. Opin. Infect. Dis. 32, 223–230. doi: 10.1097/QCO.0000000000000549
Mattoo, S., and Cherry, J. D. (2005). Molecular pathogenesis, epidemiology, and clinical manifestations of respiratory infections due to Bordetella pertussis and other Bordetella subspecies. Clin. Microbiol. Rev. 18, 326–382. doi: 10.1128/CMR.18.2.326-382.2005
Mattoo, S., Foreman-Wykert, A. K., Cotter, P. A., and Miller, J. F. (2001). Mechanisms of Bordetella pathogenesis. Front. Biosci. 6:741. doi: 10.2741/Mattoo
Mattoo, S., Yuk, M. H., Huang, L. L., and Miller, J. F. (2004). Regulation of type III secretion in Bordetella. Mol. Microbiol. 52, 1201–1214. doi: 10.1111/j.1365-2958.2004.04053.x
Medhekar, B., Shrivastava, R., Mattoo, S., Gingery, M., and Miller, J. F. (2009). Bordetella Bsp22 forms a filamentous type III secretion system tip complex and is immunoprotective in vitro and in vivo. Mol. Microbiol. 71, 492–504. doi: 10.1111/j.1365-2958.2008.06543.x
Mooi, F. R., Van Der Maas, N. A., and De Melker, H. E. (2014). Pertussis resurgence: waning immunity and pathogen adaptation - two sides of the same coin. Epidemiol. Infect. 142, 685–694. doi: 10.1017/S0950268813000071
Moon, K., Bonocora, R. P., Kim, D. D., Chen, Q., Wade, J. T., Stibitz, S., et al. (2017). The BvgAS regulon of Bordetella pertussis. MBio 8, 1–15. doi: 10.1128/mBio.01526-17
Moreira, C. G., Palmer, K., Whiteley, M., Sircili, M. P., Trabulsi, L. R., Castro, A. F., et al. (2006). Bundle-forming pili and EspA are involved in biofilm formation by enteropathogenic Escherichia coli. J. Bacteriol. 188, 3952–3961. doi: 10.1128/JB.00177-06
Musser, J. M., Hewlett, E. L., Peppler, M. S., and Selander, R. K. (1986). Genetic diversity and relationships in populations of Bordetella spp. J. Bacteriol. 166, 230–237. doi: 10.1128/JB.166.1.230-237.1986
Nagamatsu, K., Kuwae, A., Konaka, T., Nagai, S., Yoshida, S., Eguchi, M., et al. (2009). Bordetella evades the host immune system by inducing IL-10 through a type III effector, BopN. J. Exp. Med. 206, 3073–3088. doi: 10.1084/jem.20090494
Nawrotek, A., Guimaraes, B. G., Velours, C., Subtil, A., Knossow, M., and Gigant, B. (2014). Biochemical and structural insights into microtubule perturbation by CopN from Chlamydia pneumoniae. J. Biol. Chem. 289, 25199–25210. doi: 10.1074/jbc.M114.568436
Nicholson, T. L. (2007). Construction and validation of a first-generation Bordetella bronchiseptica long-oligonucleotide microarray by transcriptional profiling the Bvg regulon. BMC Genomics 8:220. doi: 10.1186/1471-2164-8-220
Nicholson, T. L., Brockmeier, S. L., Loving, C. L., Register, K. B., Kehrli, M. E. Jr., et al. (2014). The Bordetella bronchiseptica type III secretion system is required for persistence and disease severity but not transmission in swine. Infect. Immun. 82, 1092–1103. doi: 10.1128/IAI.01115-13
Nishimura, R., Abe, A., Sakuma, Y., and Kuwae, A. (2018). Bordetella bronchiseptica Bcr4 antagonizes the negative regulatory function of BspR via its role in type III secretion. Microbiol. Immunol. 62, 743–754. doi: 10.1111/1348-0421.12659
Nogawa, H., Kuwae, A., Matsuzawa, T., and Abe, A. (2004). The type III secreted protein BopD in Bordetella bronchiseptica is complexed with BopB for pore formation on the host plasma membrane. J. Bacteriol. 186, 3806–3813. doi: 10.1128/JB.186.12.3806-3813.2004
Notti, R. Q., and Stebbins, C. E. (2016). The structure and function of type III secretion systems. Microbiol. Spectr 4, 1–18. doi: 10.1128/9781555819286.ch9
Pallen, M. J., Beatson, S. A., and Bailey, C. M. (2005). Bioinformatics, genomics and evolution of non-flagellar type-III secretion systems: a Darwinian perspective. FEMS Microbiol. Rev 29, 201–229. doi: 10.1016/j.femsre.2005.01.001
Panina, E. M. (2007). Identification and Characterization of Type III Secretion Effector Proteins in Gram-negative Bacteria. [dissertation thesis]. Los Angeles, CA: University of California Los Angeles.
Panina, E. M., Mattoo, S., Griffith, N., Kozak, N. A., Yuk, M. H., and Miller, J. F. (2005). A genome-wide screen identifies a Bordetella type III secretion effector and candidate effectors in other species. Mol. Microbiol. 58, 267–279. doi: 10.1111/j.1365-2958.2005.04823.x
Park, J., Zhang, Y., Buboltz, A. M., Zhang, X., Schuster, S. C., Ahuja, U., et al. (2012). Comparative genomics of the classical Bordetella subspecies: the evolution and exchange of virulence-associated diversity amongst closely related pathogens. BMC Genomics 13:545. doi: 10.1186/1471-2164-13-545
Park, D., Lara-Tejero, M., Waxham, M. N., Li, W., Hu, B., Galan, J. E., et al. (2018). Visualization of the type III secretion mediated Salmonella-host cell interface using cryo-electron tomography. Elife 7, 1–15. doi: 10.7554/eLife.39514.018
Parkhill, J., Sebaihia, M., Preston, A., Murphy, L. D., Thomson, N., Harris, D. E., et al. (2003). Comparative analysis of the genome sequences of Bordetella pertussis, Bordetella parapertussis and Bordetella bronchiseptica. Nat. Genet. 35, 32–40. doi: 10.1038/ng1227
Petrackova, D., Farman, M. R., Amman, F., Linhartova, I., Dienstbier, A., Kumar, D., et al. (2020). Transcriptional profiling of human macrophages during infection with Bordetella pertussis. RNA Biol. 17, 731–742. doi: 10.1080/15476286.2020.1727694
Pierce, C., Klein, N., and Peters, M. (2000). Is leukocytosis a predictor of mortality in severe pertussis infection? Intensive Care Med. 26, 1512–1514. doi: 10.1007/s001340000587
Pilione, M. R., and Harvill, E. T. (2006). The Bordetella bronchiseptica type III secretion system inhibits gamma interferon production that is required for efficient antibody-mediated bacterial clearance. Infect. Immun. 74, 1043–1049. doi: 10.1128/IAI.74.2.1043-1049.2006
Pinto, M. V., and Merkel, T. J. (2017). Pertussis disease and transmission and host responses: insights from the baboon model of pertussis. J. Infect. (74 Suppl 1), S114–S119. doi: 10.1016/S0163-4453(17)30201-3
Portaliou, A. G., Tsolis, K. C., Loos, M. S., Balabanidou, V., Rayo, J., Tsirigotaki, A., et al. (2017). Hierarchical protein targeting and secretion is controlled by an affinity switch in the type III secretion system of enteropathogenic Escherichia coli. EMBO J. 36, 3517–3531. doi: 10.15252/embj.201797515
Portaliou, A. G., Tsolis, K. C., Loos, M. S., Zorzini, V., and Economou, A. (2016). Type III secretion: building and operating a remarkable nanomachine. Trends Biochem. Sci. 41, 175–189. doi: 10.1016/j.tibs.2015.09.005
Porter, J. F., Connor, K., and Donachie, W. (1994). Isolation and characterization of Bordetella parapertussis-like bacteria from ovine lungs. Microbiology 140 (Pt 2), 255–261. doi: 10.1099/13500872-140-2-255
Preston, A., Parkhill, J., and Maskell, D. J. (2004). The bordetellae: lessons from genomics. Nat. Rev. Microbiol 2, 379–390. doi: 10.1038/nrmicro886
Register, K. B., Ivanov, Y. V., Harvill, E. T., Brinkac, L., Kim, M., and Losada, L. (2015). Draft genome sequences of six bordetella hinzii isolates acquired from avian and mammalian hosts. Genome Announc. 3, 1–2. doi: 10.1128/genomeA.00081-15
Reissinger, A., Skinner, J. A., and Yuk, M. H. (2005). Downregulation of mitogen-activated protein kinases by the Bordetella bronchiseptica Type III secretion system leads to attenuated nonclassical macrophage activation. Infect. Immun. 73, 308–316. doi: 10.1128/IAI.73.1.308-316.2005
Rivera, I., Linz, B., Dewan, K. K., Ma, L., Rice, C. A., Kyle, D. E., et al. (2019). Conservation of ancient genetic pathways for intracellular persistence among animal pathogenic bordetellae. Front. Microbiol. 10:2839. doi: 10.3389/fmicb.2019.02839
Rodgers, L., Martin, S. W., Cohn, A., Budd, J., Marcon, M., Terranella, A., et al. (2013). Epidemiologic and laboratory features of a large outbreak of pertussis-like illnesses associated with cocirculating Bordetella holmesii and Bordetella pertussis–Ohio, 2010-2011. Clin. Infect. Dis. 56, 322–331. doi: 10.1093/cid/cis888
Ryan, L. K., Wu, J., Schwartz, K., Yim, S., and Diamond, G. (2018). beta-defensins coordinate in vivo to inhibit bacterial infections of the trachea. Vaccines (Basel) 6:57. doi: 10.3390/vaccines6030057
Schubot, F. D., Jackson, M. W., Penrose, K. J., Cherry, S., Tropea, J. E., Plano, G. V., et al. (2005). Three-dimensional structure of a macromolecular assembly that regulates type III secretion in Yersinia pestis. J. Mol. Biol. 346, 1147–1161. doi: 10.1016/j.jmb.2004.12.036
Sealey, K. L., Belcher, T., and Preston, A. (2016). Bordetella pertussis epidemiology and evolution in the light of pertussis resurgence. Infect. Genet. Evol. 40, 136–143. doi: 10.1016/j.meegid.2016.02.032
Sekiya, K., Ohishi, M., Ogino, T., Tamano, K., Sasakawa, C., and Abe, A. (2001). Supermolecular structure of the enteropathogenic Escherichia coli type III secretion system and its direct interaction with the EspA-sheath-like structure. Proc. Natl. Acad. Sci. U.S.A. 98, 11638–11643. doi: 10.1073/pnas.191378598
Siciliano, N. A., Skinner, J. A., and Yuk, M. H. (2006). Bordetella bronchiseptica modulates macrophage phenotype leading to the inhibition of CD4+ T cell proliferation and the initiation of a Th17 immune response. J. Immunol. 177, 7131–7138. doi: 10.4049/jimmunol.177.10.7131
Skinner, J. A., Pilione, M. R., Shen, H., Harvill, E. T., and Yuk, M. H. (2005). Bordetella type III secretion modulates dendritic cell migration resulting in immunosuppression and bacterial persistence. J. Immunol. 175, 4647–4652. doi: 10.4049/jimmunol.175.7.4647
Skinner, J. A., Reissinger, A., Shen, H., and Yuk, M. H. (2004). Bordetella type III secretion and adenylate cyclase toxin synergize to drive dendritic cells into a semimature state. J. Immunol. 173, 1934–1940. doi: 10.4049/jimmunol.173.3.1934
Spokes, P. J., Quinn, H. E., and Mcanulty, J. M. (2010). Review of the 2008-2009 pertussis epidemic in NSW: notifications and hospitalisations. N. S. W. Public Health Bull. 21, 167–173. doi: 10.1071/NB10031
Stockbauer, K. E., Foreman-Wykert, A. K., and Miller, J. F. (2003). Bordetella type III secretion induces caspase 1-independent necrosis. Cell. Microbiol. 5, 123–132. doi: 10.1046/j.1462-5822.2003.00260.x
Tandhavanant, S., Matsuda, S., Hiyoshi, H., Iida, T., and Kodama, T. (2018). Vibrio parahaemolyticus Senses Intracellular K(+) To Translocate Type III secretion system 2 effectors Effectively. MBio 9, 1–14. doi: 10.1128/mBio.01366-18
Tazato, N., Handa, Y., Nishijima, M., Kigawa, R., Sano, C., and Sugiyama, J. (2015). Novel environmental species isolated from the plaster wall surface of mural paintings in the Takamatsuzuka tumulus: Bordetella muralis sp. nov., Bordetella tumulicola sp. nov. and Bordetella tumbae sp. nov. Int. J. Syst. Evol. Microbiol. 65, 4830–4838. doi: 10.1099/ijsem.0.000655
Teruya, S., Hiramatsu, Y., Nakamura, K., Fukui-Miyazaki, A., Tsukamoto, K., Shinoda, N., et al. (2020). Bordetella dermonecrotic toxin is a neurotropic virulence factor that uses CaV3.1 as the cell surface receptor. MBio 11, 1–15. doi: 10.1128/mBio.03146-19
Troisfontaines, P., and Cornelis, G. R. (2005). Type III secretion: more systems than you think. Physiology (Bethesda) 20, 326–339. doi: 10.1152/physiol.00011.2005
Van Beek, L. F., De Gouw, D., Eleveld, M. J., Bootsma, H. J., De Jonge, M. I., Mooi, F. R., et al. (2018). Adaptation of Bordetella pertussis to the respiratory tract. J. Infect. Dis. 217, 1987–1996. doi: 10.1093/infdis/jiy125
Van Der Zee, A., Groenendijk, H., Peeters, M., and Mooi, F. R. (1996). The differentiation of Bordetella parapertussis and Bordetella bronchiseptica from humans and animals as determined by DNA polymorphism mediated by two different insertion sequence elements suggests their phylogenetic relationship. Int. J. Syst. Bacteriol. 46, 640–647. doi: 10.1099/00207713-46-3-640
Van Der Zee, A., Mooi, F., Van Embden, J., and Musser, J. (1997). Molecular evolution and host adaptation of Bordetella spp.: phylogenetic analysis using multilocus enzyme electrophoresis and typing with three insertion sequences. J. Bacteriol. 179, 6609–6617. doi: 10.1128/JB.179.21.6609-6617.1997
Vandamme, P., Heyndrickx, M., Vancanneyt, M., Hoste, B., De Vos, P., Falsen, E., et al. (1996). Bordetella trematum sp. nov., isolated from wounds and ear infections in humans, and reassessment of Alcaligenes denitrificans Ruger and Tan 1983. Int. J. Syst. Bacteriol. 46, 849–858. doi: 10.1099/00207713-46-4-849
Vandamme, P., Hommez, J., Vancanneyt, M., Monsieurs, M., Hoste, B., Cookson, B., et al. (1995). Bordetella hinzii sp. nov., isolated from poultry and humans. Int. J. Syst. Bacteriol. 45, 37–45. doi: 10.1099/00207713-45-1-37
Vandamme, P. A., Peeters, C., Cnockaert, M., Inganas, E., Falsen, E., Moore, E. R. B., et al. (2015). Bordetella bronchialis sp. nov., Bordetella flabilis sp. nov. and Bordetella sputigena sp. nov., isolated from human respiratory specimens, and reclassification of Achromobacter sediminum Zhang et al. 2014 as Verticia sediminum gen. nov., comb. nov. Int. J. Syst. Evol. Microbiol. 65, 3674–3682. doi: 10.1099/ijsem.0.000473
Villarino Romero, R., Bibova, I., Cerny, O., Vecerek, B., Wald, T., Benada, O., et al. (2013). The Bordetella pertussis type III secretion system tip complex protein Bsp22 is not a protective antigen and fails to elicit serum antibody responses during infection of humans and mice. Infect. Immun. 81, 2761–2767. doi: 10.1128/IAI.00353-13
Von Wintzingerode, F., Schattke, A., Siddiqui, R. A., Rosick, U., Gobel, U. B., and Gross, R. (2001). Bordetella petrii sp. nov., isolated from an anaerobic bioreactor, and emended description of the genus Bordetella. Int. J. Syst. Evol. Microbiol. 51, 1257–1265. doi: 10.1099/00207713-51-4-1257
Wagner, S., Grin, I., Malmsheimer, S., Singh, N., Torres-Vargas, C. E., and Westerhausen, S. (2018). Bacterial type III secretion systems: a complex device for the delivery of bacterial effector proteins into eukaryotic host cells. FEMS Microbiol. Lett. 365, 1–13. doi: 10.1093/femsle/fny201
Wang, Y. A., Yu, X., Yip, C., Strynadka, N. C., and Egelman, E. H. (2006). Structural polymorphism in bacterial EspA filaments revealed by cryo-EM and an improved approach to helical reconstruction. Structure 14, 1189–1196. doi: 10.1016/j.str.2006.05.018
Weigand, M. R., Peng, Y., Batra, D., Burroughs, M., Davis, J. K., Knipe, K., et al. (2019). Conserved patterns of symmetric inversion in the genome evolution of bordetella respiratory pathogens. mSystems 4, 1–15. doi: 10.1128/mSystems.00702-19
Weyant, R. S., Hollis, D. G., Weaver, R. E., Amin, M. F., Steigerwalt, A. G., O'connor, S. P., et al. (1995). Bordetella holmesii sp. nov., a new gram-negative species associated with septicemia. J. Clin. Microbiol. 33, 1–7. doi: 10.1128/JCM.33.1.1-7.1995
Wong, T. Y., Hall, J. M., Nowak, E. S., Boehm, D. T., Gonyar, L. A., Hewlett, E. L., et al. (2019). Analysis of the in vivo transcriptome of bordetella pertussis during infection of mice. mSphere 4, 1–14. doi: 10.1128/mSphereDirect.00154-19
Yahalom, A., Davidov, G., Kolusheva, S., Shaked, H., Barber-Zucker, S., Zarivach, R., et al. (2019). Structure and membrane-targeting of a Bordetella pertussis effector N-terminal domain. Biochim Biophys Acta Biomembr 1861:183054. doi: 10.1016/j.bbamem.2019.183054
Yih, W. K., Silva, E. A., Ida, J., Harrington, N., Lett, S. M., and George, H. (1999). Bordetella holmesii-like organisms isolated from Massachusetts patients with pertussis-like symptoms. Emerging Infect. Dis. 5, 441–443. doi: 10.3201/eid0503.990317
Yu, X. J., Mcgourty, K., Liu, M., Unsworth, K. E., and Holden, D. W. (2010). pH sensing by intracellular Salmonella induces effector translocation. Science 328, 1040–1043. doi: 10.1126/science.1189000
Yu, X. J., Grabe, G. J., Liu, M., Mota, L. J., and Holden, D. W. (2018). SsaV interacts with SsaL to control the translocon-to-effector switch in the Salmonella SPI-2 type three secretion system. MBio 9, 1–16. doi: 10.1128/mBio.01149-18
Yuk, M. H., Harvill, E. T., Cotter, P. A., and Miller, J. F. (2000). Modulation of host immune responses, induction of apoptosis and inhibition of NF-kappaB activation by the Bordetella type III secretion system. Mol. Microbiol. 35, 991–1004. doi: 10.1046/j.1365-2958.2000.01785.x
Yuk, M. H., Harvill, E. T., and Miller, J. F. (1998). The BvgAS virulence control system regulates type III secretion in Bordetella bronchiseptica. Mol. Microbiol. 28, 945–959. doi: 10.1046/j.1365-2958.1998.00850.x
Keywords: pertussis, Bordetella, type III secretion system, effector protein, BteA/BopC, BopN
Citation: Kamanova J (2020) Bordetella Type III Secretion Injectosome and Effector Proteins. Front. Cell. Infect. Microbiol. 10:466. doi: 10.3389/fcimb.2020.00466
Received: 14 June 2020; Accepted: 29 July 2020;
Published: 04 September 2020.
Edited by:
Teresa Thurston, Imperial College London, United KingdomReviewed by:
Ondrej Cerny, Imperial College London, United KingdomAkio Abe, Kitasato University, Japan
Copyright © 2020 Kamanova. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jana Kamanova, a2FtYW5vdmFAYmlvbWVkLmNhcy5jeg==
 Jana Kamanova
Jana Kamanova